-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe miR-35-41 Family of MicroRNAs Regulates RNAi Sensitivity in
RNA interference (RNAi) utilizes small interfering RNAs (siRNAs) to direct silencing of specific genes through transcriptional and post-transcriptional mechanisms. The siRNA guides can originate from exogenous (exo–RNAi) or natural endogenous (endo–RNAi) sources of double-stranded RNA (dsRNA). In Caenorhabditis elegans, inactivation of genes that function in the endo–RNAi pathway can result in enhanced silencing of genes targeted by siRNAs from exogenous sources, indicating cross-regulation between the pathways. Here we show that members of another small RNA pathway, the mir-35-41 cluster of microRNAs (miRNAs) can regulate RNAi. In worms lacking miR-35-41, there is reduced expression of lin-35/Rb, the C. elegans homolog of the tumor suppressor Retinoblastoma gene, previously shown to regulate RNAi responsiveness. Genome-wide microarray analyses show that targets of endo–siRNAs are up-regulated in mir-35-41 mutants, a phenotype also displayed by lin-35/Rb mutants. Furthermore, overexpression of lin-35/Rb specifically rescues the RNAi hypersensitivity of mir-35-41 mutants. Although the mir-35-41 miRNAs appear to be exclusively expressed in germline and embryos, their effect on RNAi sensitivity is transmitted to multiple tissues and stages of development. Additionally, we demonstrate that maternal contribution of miR-35-41 or lin-35/Rb is sufficient to reduce RNAi effectiveness in progeny worms. Our results reveal that miRNAs can broadly regulate other small RNA pathways and, thus, have far reaching effects on gene expression beyond directly targeting specific mRNAs.
Published in the journal: . PLoS Genet 8(3): e32767. doi:10.1371/journal.pgen.1002536
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002536Summary
RNA interference (RNAi) utilizes small interfering RNAs (siRNAs) to direct silencing of specific genes through transcriptional and post-transcriptional mechanisms. The siRNA guides can originate from exogenous (exo–RNAi) or natural endogenous (endo–RNAi) sources of double-stranded RNA (dsRNA). In Caenorhabditis elegans, inactivation of genes that function in the endo–RNAi pathway can result in enhanced silencing of genes targeted by siRNAs from exogenous sources, indicating cross-regulation between the pathways. Here we show that members of another small RNA pathway, the mir-35-41 cluster of microRNAs (miRNAs) can regulate RNAi. In worms lacking miR-35-41, there is reduced expression of lin-35/Rb, the C. elegans homolog of the tumor suppressor Retinoblastoma gene, previously shown to regulate RNAi responsiveness. Genome-wide microarray analyses show that targets of endo–siRNAs are up-regulated in mir-35-41 mutants, a phenotype also displayed by lin-35/Rb mutants. Furthermore, overexpression of lin-35/Rb specifically rescues the RNAi hypersensitivity of mir-35-41 mutants. Although the mir-35-41 miRNAs appear to be exclusively expressed in germline and embryos, their effect on RNAi sensitivity is transmitted to multiple tissues and stages of development. Additionally, we demonstrate that maternal contribution of miR-35-41 or lin-35/Rb is sufficient to reduce RNAi effectiveness in progeny worms. Our results reveal that miRNAs can broadly regulate other small RNA pathways and, thus, have far reaching effects on gene expression beyond directly targeting specific mRNAs.
Introduction
The ability of double stranded RNA (dsRNA) to induce silencing of specific genes was discovered in the nematode Caenorhabditis elegans and dubbed RNA interference (RNAi) [1]. RNAi was subsequently identified in many organisms, including mammals, providing a powerful experimental tool for inactivating specific genes [2], [3]. In worms, RNAi can be initiated by long dsRNAs from exogenous sources, such as injected or ingested bacterially produced dsRNA [1], [4]. Cellular factors process the dsRNAs into small interfering RNAs (exo–siRNAs) of ∼21 nucleotides long, which target complementary mRNAs for degradation [5]. Exo–siRNAs form complexes with Argonaute proteins that mediate target mRNA cleavage or degradation through other mechanisms [6]. Recently, a nuclear RNAi pathway was discovered in worms where the Argonaute NRDE-3 silences target genes through a co-transcriptional mechanism of silencing [7], [8].
Comparable to exo–RNAi, endogenous dsRNAs can enter processing pathways that produce endo–siRNAs that silence target genes through base-pairing interactions [6], [9]. Endo–siRNAs have been identified by small RNA cloning in C. elegans, Drosophila and mouse and are predicted to target thousands of endogenous genes, particularly mRNAs present in germline and embryos [10], [11], [12], [13], [14], [15]. In C. elegans, both the exo - and endo–siRNA pathways require the endoribonuclease enzyme Dicer (DCR-1) to process the initiating dsRNAs into ∼21 nt siRNAs, but distinct Argonaute proteins usually bind the different types of siRNAs to mediate gene silencing [16], [17]. For example the Argonaute RDE-1 is required for RNAi initiated by exogenous but not endogenous dsRNAs [17].
In nematodes and plants, RNAi-mediated silencing can be amplified through the production of secondary siRNAs by RNA dependent RNA polymerases (RdRPs). In C. elegans, loss of the RdRP RRF-3 results in greatly reduced levels of endo–siRNAs and an enhanced response to exogenous dsRNA 18,19. Likewise, the exonuclease ERI-1 is required for the accumulation of some endo–siRNAs and worms with mutations in eri-1 display hypersensitivity to exo–siRNAs [18], [20]. Another class of mutants with enhanced RNAi is represented by lin-35/Rb, which encodes the worm homolog of the Retinoblastoma tumor suppressor, and includes other genes in the lin-35/Rb pathway such as lin-15, dpl-1 (mammalian DP), and hpl-2 (mammalian HP1) [21], [22], [23]. The molecular mechanism by which lin-35/Rb negatively regulates the RNAi pathway remains to be fully understood. Worms with mutations in lin-35/Rb have up-regulated levels of mRNAs corresponding to cloned endo–siRNAs, suggesting decreased levels or function of endo–siRNAs in these mutants [24]. Reduced endo–RNAi activity may free limiting factors for the exo–RNAi pathway. Furthermore, some of the up-regulated genes in lin-35/Rb mutants encode Argonaute proteins that might also contribute to enhanced exo–RNAi [24]. Additionally, lin-35/Rb mutants have increased expression of germline-specific genes in somatic tissues [23]. This expression pattern is predicted to enhance RNAi sensitivity by providing factors normally utilized in the germline for silencing pathways [23].
MicroRNAs (miRNAs) represent another class of small RNAs that derive from endogenously expressed transcripts [25]. Typically, miRNAs are transcribed as long primary transcripts containing hairpin structures that are released by Drosha cleavage. The resulting precursor undergoes processing by Dicer to produce the mature ∼22 nt miRNA, which forms a complex with Argonaute proteins. While mammalian miRNAs seem to distribute evenly among the four Argonaute proteins, worm and fly miRNAs mostly associate with specific Argonautes [6]. For example, Argonaute Like Genes 1 and 2 (ALG-1/2) bind worm miRNAs, but not exo - or endo–siRNAs [26]. In contrast to exo and endo–siRNAs, animal miRNAs pair with imperfect sequence complementarity to their target mRNAs, causing target degradation and translational repression [27]. The common factor for most small RNA pathways is Dicer. Loss of Dicer activity in worm or mammalian cells results in defective RNAi and severely reduced levels of most miRNAs and endo–siRNAs [16], [18], [28], [29], [30]. However, mutations in other genes required for endo - or exo–RNAi seem to have little effect on the miRNA pathway in C. elegans [16], [18].
In an effort to understand the function of specific miRNAs, a large collection of deletion mutants was generated in C. elegans [31]. While most individual mutants displayed no obvious phenotypes, simultaneous deletion of multiple family members (miRNAs with identical sequences at positions 2–7) often resulted in developmental defects and lethality [32], [33]. For example, the miR-35-41 miRNAs are clustered within ∼800 nt of a common transcript and share a high degree of sequence homology [34]. Deletion of this miRNA cluster results in defective germ and intestinal cell proliferation and temperature sensitive embryonic lethality [32], [35]. While the genes regulated by miR-35-41 that are responsible for these phenotypes are yet to be determined, several direct targets of these miRNAs were recently validated in embryonic extracts [36]. Although the miR-35-41 miRNAs have only been found in worms and planaria [34], [37], poorly conserved miRNA clusters have also been observed to be highly expressed and function in early development in other species, including Drosophila, mouse and humans [38], [39], [40].
Here we show that the miR-35-41 miRNAs regulate other small RNA pathways in C. elegans. Deletion of the miRNA cluster results in worms with enhanced RNAi sensitivity in multiple developmental stages and tissues. This effect is likely related to decreased endo–RNAi activity in mir-35-41 mutants, as they also exhibit up-regulation of many endo–siRNA targets. We found that the miR-35-41 miRNAs negatively regulate the exo–RNAi pathway through lin-35/Rb and that this function can be supplied maternally to the progeny. These results point to a new level of cross-regulation between small RNA pathways, whereby the expression of specific miRNAs can impact the efficiency of RNAi mediated by exo – and endo–siRNAs.
Results
Enhanced RNAi in mir-35-41 mutants
In C. elegans the mir-35-41 miRNAs are required for embryonic viability and proliferation of certain cell types, but the targets relevant for these phenotypes have not yet been established [31], [32], [35]. In the mir-35-41(gk262) and mir-35-41(nDf50) strains all seven members of the miRNA cluster are deleted, resulting in temperature sensitive embryonic lethality (Figure S1) [31], [32]. While using RNAi to identify factors that genetically interact with mir-35-41(gk262), we discovered that this strain exhibits enhanced sensitivity to RNAi. In agreement with previous studies [1], [4], wild type worms fed bacteria expressing double stranded RNA targeting the muscle gene unc-22 resulted in a high penetrance of worms with the twitching phenotype, while only 2% resembled the null unc-22 loss-of-function phenotype of paralysis (Figure 1A and Table 1) [41]. In contrast, the majority (83%) of mir-35-41(gk262) worms were paralyzed by unc-22(RNAi) (Figure 1A and Table 1). This same treatment produced paralysis in about 20% of the established RNAi hypersensitive strain, rrf-3(pk1426) (Table 1), consistent with previous studies [19], [20]. We confirmed that the enhanced RNAi phenotype of mir-35-41(gk262) was due to loss of the miRNA gene instead of the overlapping anti-sense Y62F5A.9 protein-coding gene. A transgene encoding only the mir-35-41 locus rescued the hypersensitivity of this strain to unc-22(RNAi) (Figure 1A) and supported miRNA expression in mir-35-41(gk262) worms (Figure 1B).
Fig. 1. Deletion of the mir-35-41 miRNA cluster results in RNAi hypersensitivity. 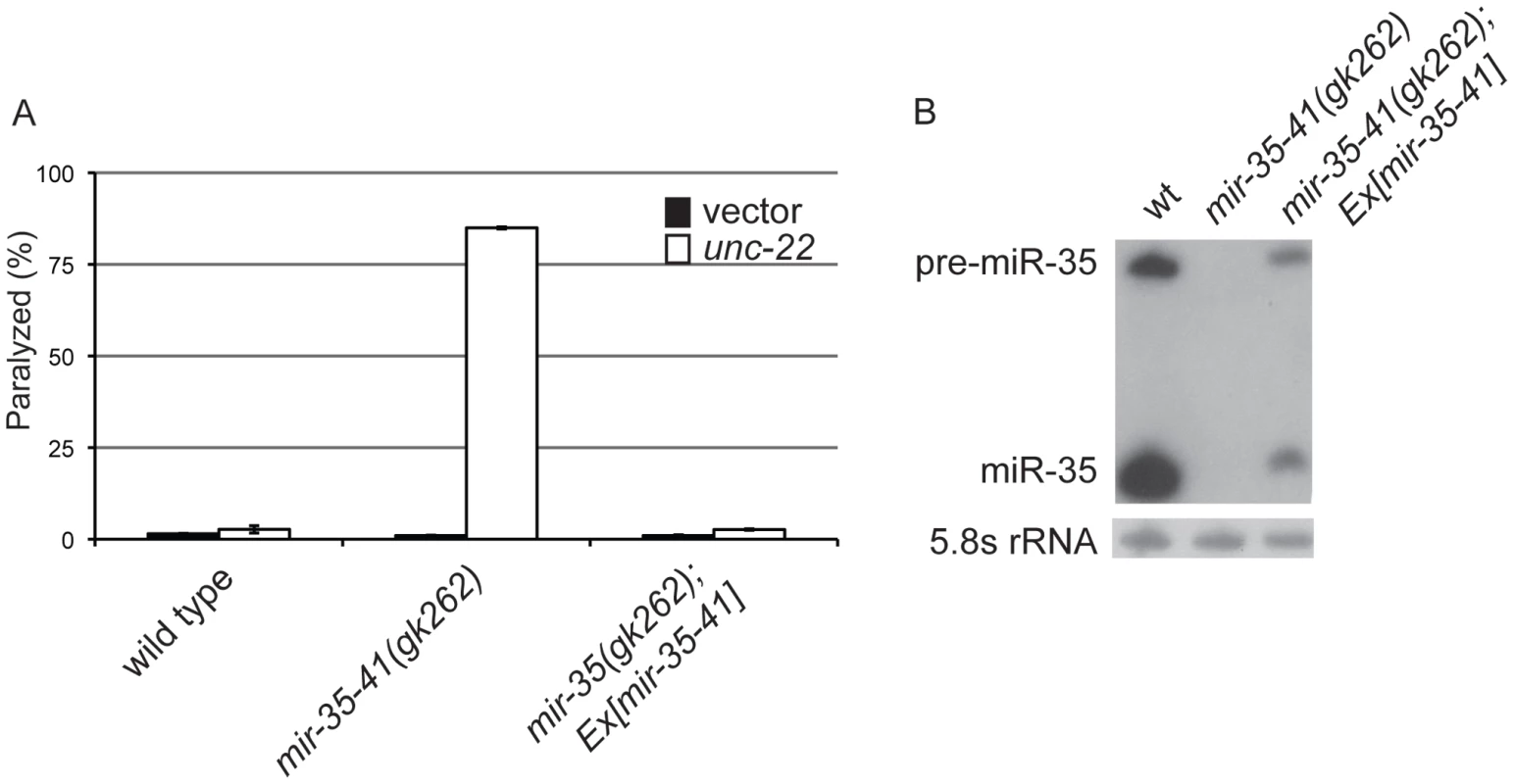
(A) L4 stage wild type, mir-35-41(gk262) and mir-35-41(gk262); apEx160 [mir-35-41] strains were grown on bacteria expressing unc-22 dsRNA (open columns) or the empty RNAi vector L4440 (black columns). The percentages of paralyzed worms after 28 h of exposure to RNAi conditions are graphed as the mean and standard deviation from three independent experiments. (B) PAGE Northern blot analysis of RNA from wild type, mir-35-41(gk262) and mir-35-41(gk262); apEx160 [mir-35-41] embryos shows restored expression of precursor and mature mir-35 miRNA in the transgenic strain. 5.8S rRNA levels are shown as loading controls. Tab. 1. The mir-35-41(gk262) mutant worms show enhanced RNAi in multiple tissues and stages of development. 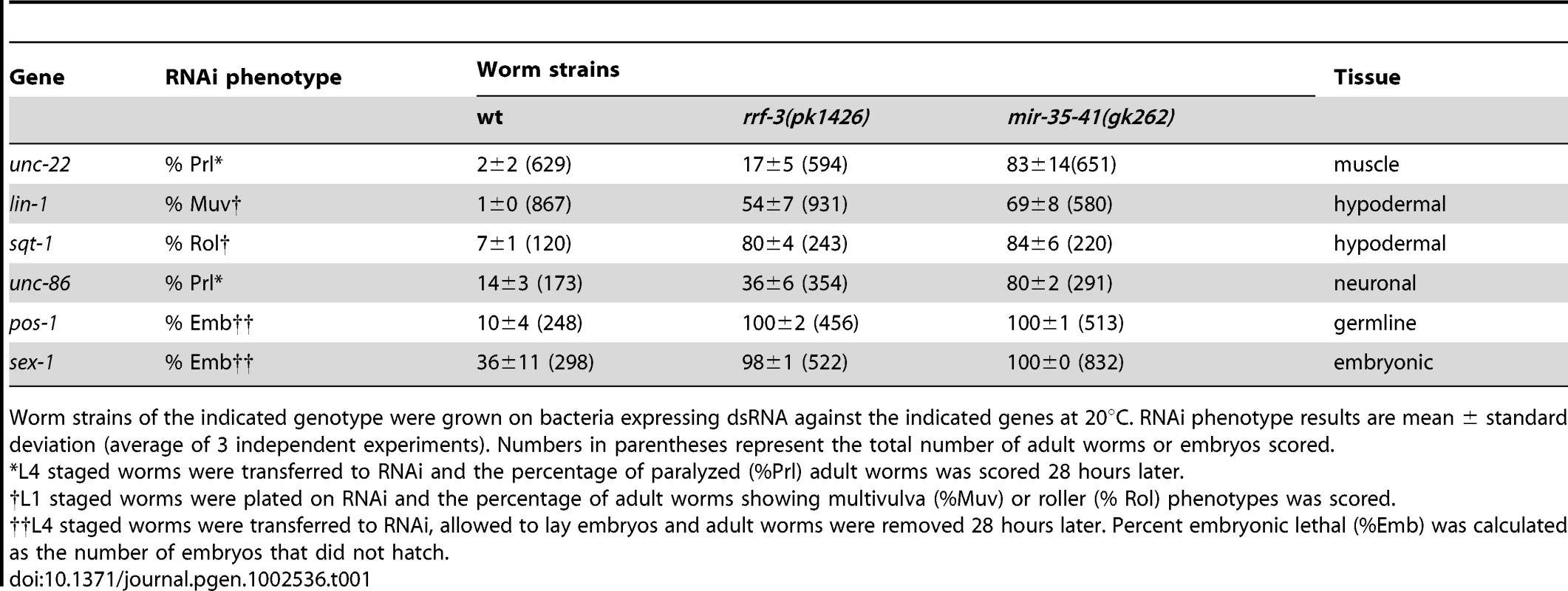
Worm strains of the indicated genotype were grown on bacteria expressing dsRNA against the indicated genes at 20°C. RNAi phenotype results are mean ± standard deviation (average of 3 independent experiments). Numbers in parentheses represent the total number of adult worms or embryos scored. Loss of mir-35-41 results in enhanced RNAi in multiple tissues and stages of development. We observed that RNAi knockdown of lin-1 (hypodermal), sqt-1 (hypodermal), unc-86 (neuronal), pos-1 (germline) and sex-1 (embryonic) resulted in enhanced phenotypes in mir-35-41(gk262) relative to wild type worms (Table 1). Furthermore, the effects were similar or stronger in mir-35-41(gk262) compared to the RNAi hypersensitive rrf-3 mutants. These results indicate that expression of the miR-35-41 miRNAs negatively impacts the efficiency of exo–RNAi in C. elegans.
To determine if the enhanced RNAi phenotype of mir-35-41(gk262) was dependent on core exo–RNAi factors, we crossed this strain to mutants in the RNAi pathway. Upon unc-22(RNAi) treatment, the single mutants defective in RNAi (rde-1, rde-4 and rrf-1) showed no phenotypic response, as expected (Table 2) [42], [43], [44]. Likewise, addition of mutations in rde-1, rde-4 or rrf-1 rendered mir-35-41(gk262) mutants completely RNAi defective (Table 2). These data show that mir-35-41(gk262) mutants require rde-1, rde-4 and rrf-1 to exhibit an RNAi response. While this might be expected, there is precedence for an RNAi hypersensitive strain (lin-15B(n744)) being independent of rrf-1 activity [23].
Tab. 2. The RNAi sensitivity of mir-35-41(gk262) mutant worms is dependent on core RNAi factors. 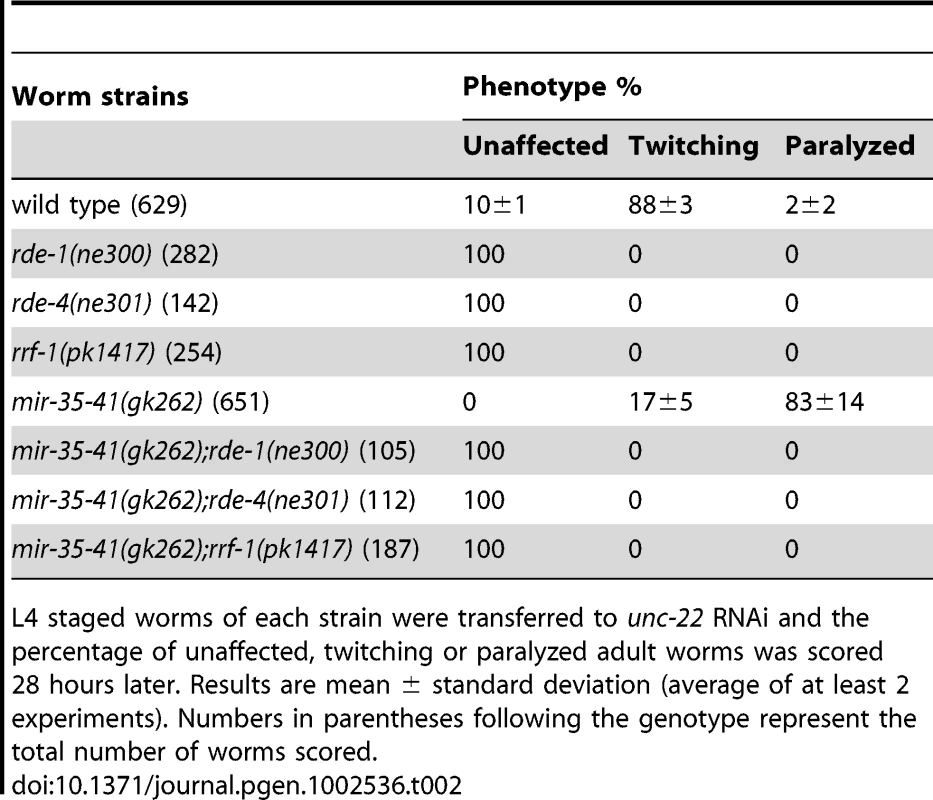
L4 staged worms of each strain were transferred to unc-22 RNAi and the percentage of unaffected, twitching or paralyzed adult worms was scored 28 hours later. Results are mean ± standard deviation (average of at least 2 experiments). Numbers in parentheses following the genotype represent the total number of worms scored. The response of mir-35-41(gk262) to unc-22(RNAi) is comparable to that of the exceptionally RNAi hypersensitive strain lin-35(n745), which carries a putative null mutation [21], [22], [23], [45]. About 20–30% of the population of the enhanced RNAi strains rrf-3(pk1426), eri-1(mg366) and ergo-1(tm1860) exhibited paralysis in response to unc-22(RNAi) (Table 3). In comparison, over 80% of the mir-35-41(gk262) and lin-35(n745) strains were paralyzed by this RNAi treatment (Table 3). The rrf-3, eri-1 and ergo-1 genes are part of the endogenous RNAi pathway in C. elegans and loss of these genes results in decreased levels of endo–siRNAs [16], [17], [18], [46]. Diminished activity of the endo–RNAi pathway is one explanation for enhanced RNAi sensitivity to dsRNA from exogenous sources. The Argonaute protein NRDE-3 serves as a sensor for endo–siRNAs, as nuclear localization of this protein is dependent on the presence of siRNAs [8]. In contrast to wild type worms that express endo–siRNAs, GFP::NRDE-3 was mostly cytoplasmic in the seam cells of eri-1(mg366) worms, which are defective in endo–siRNA accumulation (Figure 2A) [8], [16], [18]. As in wild type, localization of GFP::NRDE-3 was nuclear in the mir-35-41(gk262) and lin-35(n745) mutant strains (Figure 2A). Additionally, the hypersensitivity of mir-35-41 mutants continued in the absence of nrde-3 activity (Figure 2B). These results show that in mir-35-41 mutants sufficient endo–siRNAs are present to target NRDE-3 to the nucleus and the enhanced RNAi phenotype of these mutants does not require NRDE-3 activity.
Fig. 2. The RNAi hypersensitivity of mir-35-41 mutants is independent of nrde-3 activity. 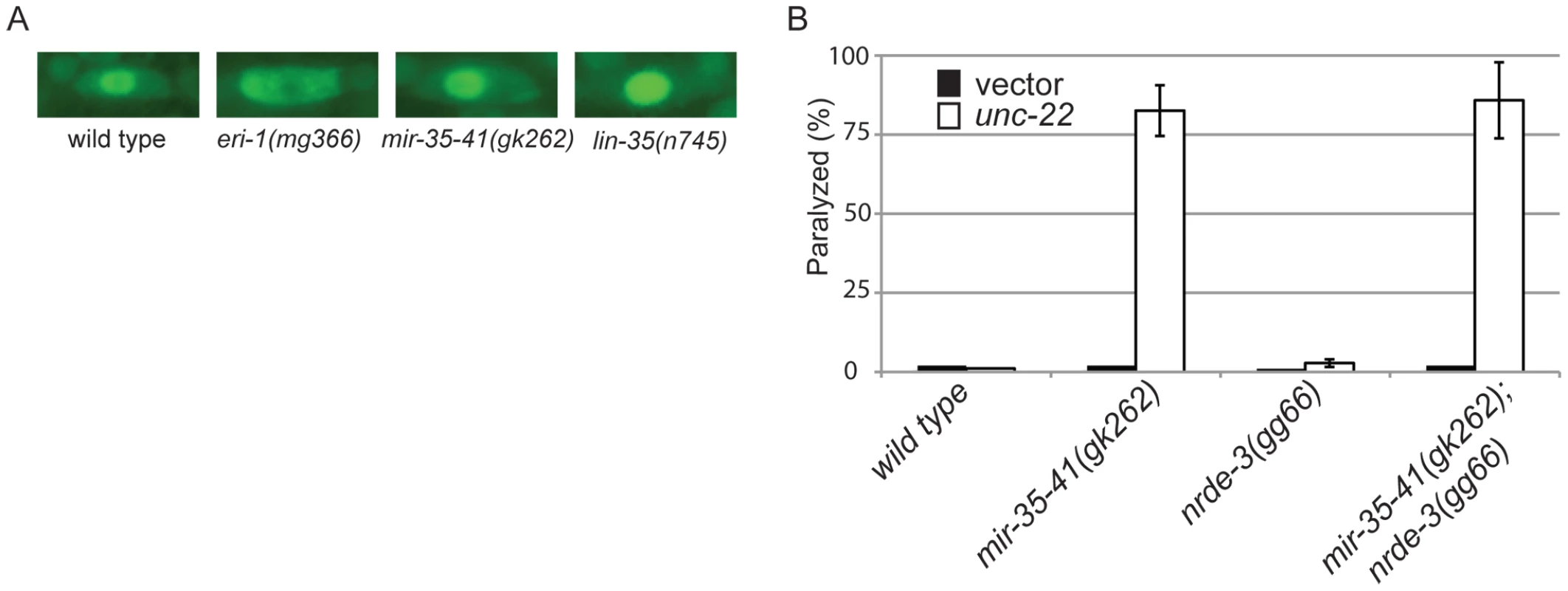
(A) Fluorescent microscopy showing subcellular localization of GFP::NRDE-3 in seam cells of the indicated genotypes. Pictures are representative of 50 worms analyzed for each strain. (B) Histogram representing the percentages of paralyzed worms for the indicated strains fed unc-22 dsRNA for 28 hours from the L4 to adult stage. The means and standard deviations from three independent experiments are graphed. Tab. 3. The RNAi hypersensitivity of mir-35-41(gk262) is comparable to that of lin-35(n745). 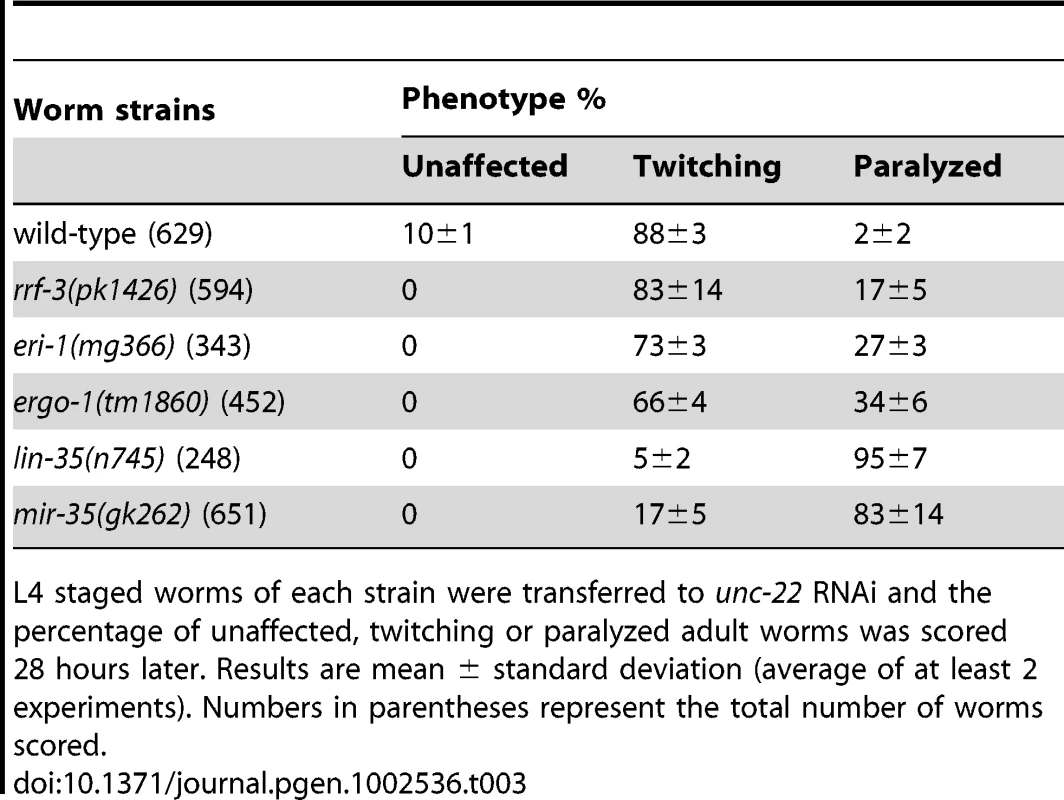
L4 staged worms of each strain were transferred to unc-22 RNAi and the percentage of unaffected, twitching or paralyzed adult worms was scored 28 hours later. Results are mean ± standard deviation (average of at least 2 experiments). Numbers in parentheses represent the total number of worms scored. Up-regulation of endogenous siRNA targets in mir-35-41 mutants
To gain insight into the role of the mir-35-41 miRNAs in regulating the exo–RNAi pathway, we performed microarray analysis of gene expression in WT versus mir-35-41(gk262) embryos. We chose statistically significant changes in gene expression of at least 1.5-fold differences between the triplicate sample averages (p value≤0.005 (two-sample t-test)) (Table S1). Of the 550 up-regulated genes in mir-35-41(gk262), only a small fraction (4%) were predicted miR-35 targets based on a list of 606 genes from four different algorithms (Targetscan, PicTar, RNA22 and MiRWIP). Furthermore, obvious genes that might explain the heightened RNAi sensitivity of mir-35-41(gk262) were not mis-regulated at the mRNA level. Comparing the microarray results to a list of 6,469 endo–siRNA targets obtained from four independent studies revealed significant enrichment of endo–siRNA targets in the up-regulated genes (37%, p value 1.3e-05 two tailed exact Fisher's significance test) (Table S1) [11], [13], [18], [47]. In contrast, endo–siRNA targets were under-represented in the list of genes down-regulated in mir-35-41(gk262) (13% p value 2.2e-16 exact Fisher's significance test) (Table S1). Differential expression of the established endo–siRNA target E01G4.5 was confirmed by RT-qPCR (Figure 3). Similar to the previously reported effect of eri-1 on this gene, both the spliced and unspliced forms of E01G4.5 were up-regulated in mir-35-41 mutants (Figure 3) [8].
Fig. 3. Mis-regulation of the E01G4.5 endo–siRNA target in mir-35-41 mutants. 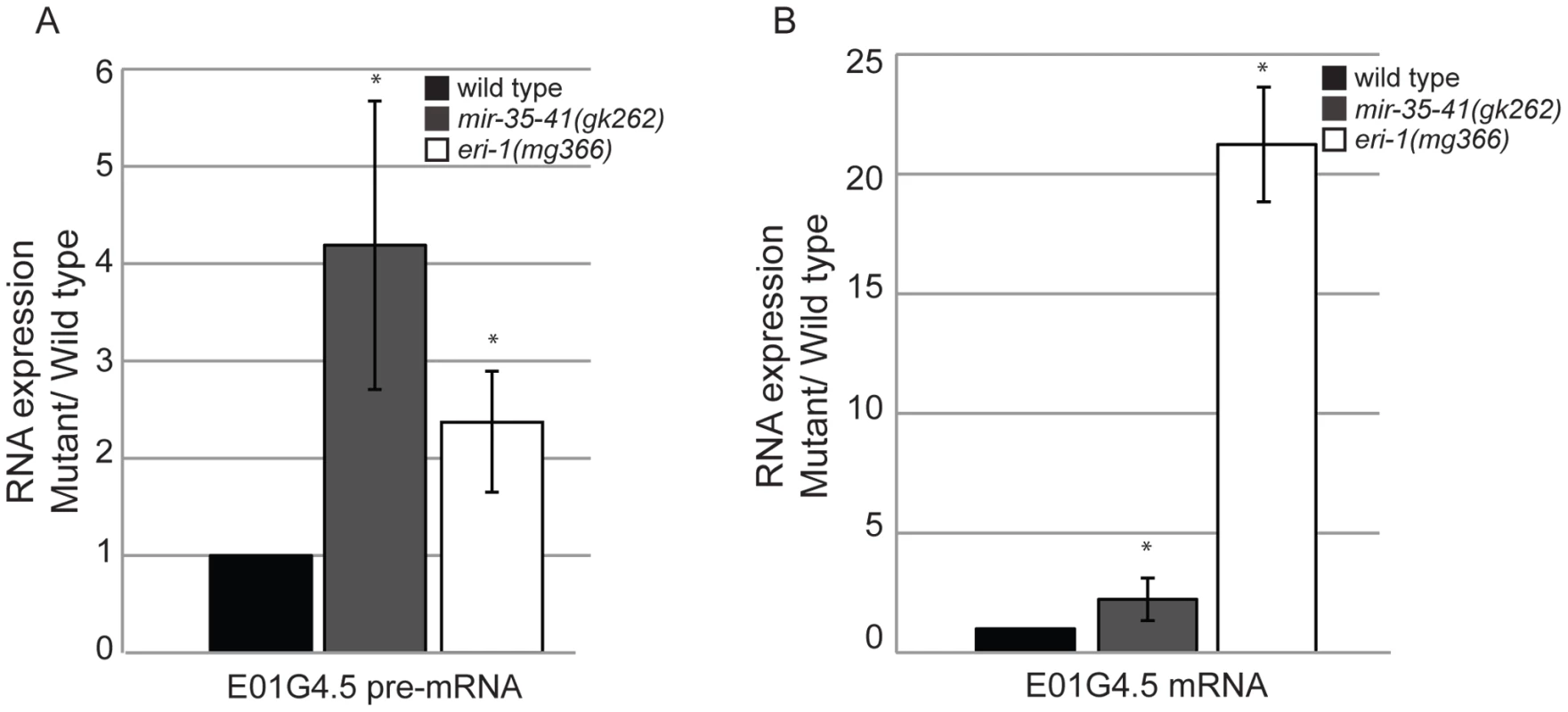
RT-qPCR of E01G4.5 pre-mRNA (A) and mature mRNA (B) normalized to 18S rRNA and compared to wild type levels (mean ± s.e.m., n = 3, *, P<0.05). Decreased LIN-35/Rb contributes to the RNAi hypersensitivity of mir-35-41 mutants
The mir-35-41 and lin-35/Rb mutant strains both show strongly enhanced RNAi and up-regulation of endo–siRNA targets. Also both strains resemble WT for NRDE-3 localization, which is dependent on siRNAs for nuclear residence (Table 3, Table S1, Figure 2) [21], . These similarities led us to investigate if the mir-35-41 and lin-35/Rb genes might regulate each other. Although lin-35/Rb mRNA levels were not significantly different, protein levels of this gene were reduced to about 20% in mir-35-41(gk262) relative to wild type worm embryos (Figure 4A and 4B), suggesting that mir-35-41 positively regulates the accumulation of LIN-35 protein. In contrast, mir-35 miRNA levels were unaltered in lin-35(n745) mutant embryos (Figure 4C). The decreased levels of LIN-35 protein could be responsible for the RNAi hypersensitivity of mir-35-41 mutants. To test this possibility, we crossed an extrachromosomal array expressing lin-35 into mir-35-41(gk262). Worms carrying the array, Ex[lin-35(+); sur-5::GFP], are distinguished by GFP expression [48]. In the mir-35-41(gk262) worms with extra copies of lin-35 provided by the array (GFP+), the hypersensitivity to unc-22(RNAi) was rescued with the majority of worms showing the twitching instead of paralysis phenotype (Figure 4D). The non-GFP (GFP−) siblings were comparable to the original mir-35-41(gk262) strain for sensitivity to unc-22(RNAi) (Figure 4D). Other GFP transgenes that lack lin-35 did not affect the RNAi hypersensitivity of mir-35 mutants, consistent with the idea that lin-35 is required for the rescue (Figure 4D). The extra copies of lin-35 also reduced the RNAi hypersensitivity of lin-35(n745) genetic mutants (Figure 4D). Rescue of the RNAi phenotype by extra copies of lin-35 was stronger in mir-35-41(gk262) than in lin-35(n745), possibly due to the reduction versus complete absence of LIN-35 protein in these mutants, respectively. To test if extra copies of lin-35 specifically rescues the RNAi hypersensitivity of mir-35-41(gk262), we introduced the transgene into other enhanced RNAi mutants. The response to unc-22(RNAi) was unaffected by the addition of the lin-35(+) transgene to eri-1 or rrf-3 mutants, supporting the conclusion that the RNAi hypersensitivity of the mir-35-41 mutants can be attributed to reduced lin-35/Rb. A further prediction of this model is that a mir-35-41;lin-35 double mutant would exhibit the same RNAi response as the lin-35 single mutant. However, the double mutant proved to be inviable and could not be tested for RNAi sensitivity.
Fig. 4. Decreased LIN-35/Rb contributes to the RNAi hypersensitivity of mir-35-41(gk262) worms. 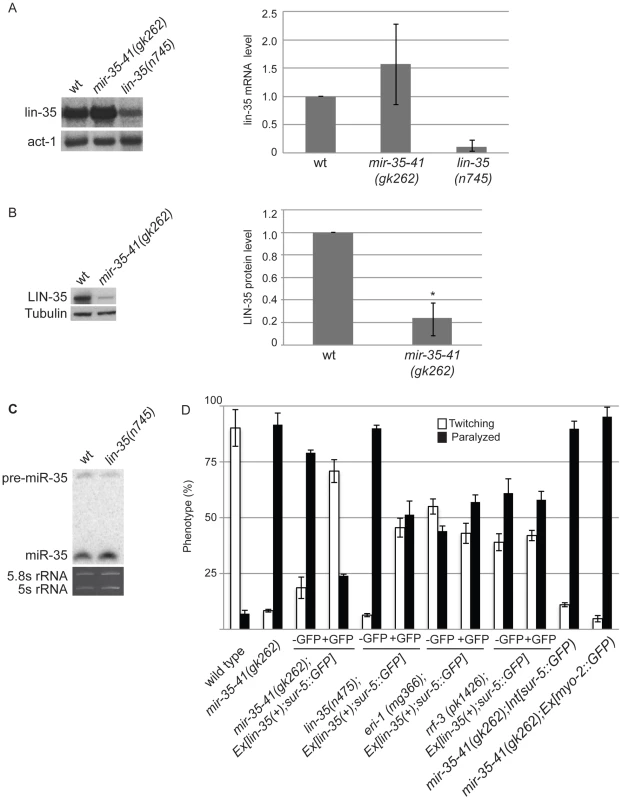
(A) Northern blot analyses of lin-35/Rb mRNA levels in WT, mir-35-41(gk262) and lin-35(n745) embryos. After normalization to actin mRNA the average and standard deviation from 3 independent experiments was calculated with wild type levels set to one. (B) LIN-35 protein is decreased in mir-35-41(gk262) embryos compared to wild type, as shown by western blotting. After normalization to tubulin the average and standard deviation from 4 independent experiments was calculated with wild type levels set to one (Student's t-test *p<4×10−5). (C) PAGE Northern blot analysis of RNA from wild type and lin-35(n475) embryos shows similar levels of pre- and mature miR-35 expression. The rRNAs are shown as loading controls. Results are representative of 3 independent experiments. (D) The indicated worm strains were grown on bacteria expressing unc-22 dsRNA. Ex[lin-35; sur-5::GFP] is an extrachromosomal array that expresses lin-35 in GFP positive (+) worms. Int[sur-5::GFP] is an integrated array that expresses GFP in all worms and Ex[myo-2::GFP] is an extrachromosomal array that expresses GFP in worms that inherit the array. Phenotype was scored as percent of twitching or paralyzed worms after 28 h of exposure to RNAi from the L4 to adult stage. Error bars represent the standard error of the mean (s.e.m) for at least two independent experiments. The lin-35/Rb gene is a member of the synthetic multivulva B (synMuv B) family, which includes transcriptional repressor and chromatin modifying genes [45]. Worms with combined mutations in synMuv B and synMuv A genes produce multiple vulva structures because of cell lineage defects [49]. In addition to lin-35, some of the other synMuv B genes, such as lin-53, lin-9 and dpl-1, also negatively regulate the exo–RNAi pathway and cause enhanced RNAi when mutated [22], [23]. We tested if the decreased levels of lin-35/Rb in mir-35-41(gk262) are sufficient to produce the multiple vulva (Muv) phenotype when combined with the synMuv A mutants lin-15a(n767) or lin-8(n111). Double mutants consisting of mir-35-41(gk262) and either of the synMuv A genes did not display the Muv phenotype (n = 230 adult worms/strain). In comparison, 100% of the lin-35(n745); lin-15a(n767) and the lin-35(n745); lin-8(n111) double mutants were Muv (n = 240 adult worms/strain). Taken together, these results suggest that RNAi efficiency is more sensitive than the vulva formation pathway to reduction in lin-35/Rb levels or that tissues dependent on lin-35/Rb for vulva formation produce sufficient protein in the absence of the miR-35-41 miRNAs.
Maternal rescue of RNAi hypersensitivity in mir-35-41 and lin-35/Rb mutants
Previous work has shown that the miR-35-41 cluster of miRNAs is predominantly expressed in embryos with comparatively little expression in larval and adult somatic tissues [34]. In fact, miRNAs from this cluster make up about 75% of all mature miRNAs present in early embryos [50]. Since the miR-35-41 miRNAs are present in one-cell stage embryos before zygotic transcription by RNA Polymerase II has initiated [50], [51], the miRNAs or their precursors are likely supplied by maternal germ cells. Thus, we tested if maternal contribution of mir-35-41 activity would be sufficient to regulate RNAi sensitivity. Since our results suggest that the RNAi hypersensitivity of mir-35-41 mutants is through lin-35/Rb, we also analyzed maternal rescue of lin-35(n745). Crossing of mir-35-41(gk262) or lin-35(n745) hermaphrodites to wild type males rescued the unc-22(RNAi) paralyzed phenotype to ∼20% in the F1 progeny compared to ∼80% in the parental strains (Figure 5). These heterozygous F1's were allowed to self-fertilize and the resulting F2 progeny were scored for the paralysis phenotype and then genotyped. Regardless of the zygotic genotype, the worms that had come from mothers with one wild type allele for mir-35-41 or lin-35 were less sensitive to unc-22(RNAi) than the original mutant strains (Figure 5). Thus, maternal contribution of mir-35-41 or lin-35/Rb is sufficient to regulate RNAi sensitivity.
Fig. 5. Maternal rescue of RNAi hypersensitivity in mir-35-41 and lin-35/Rb mutants. 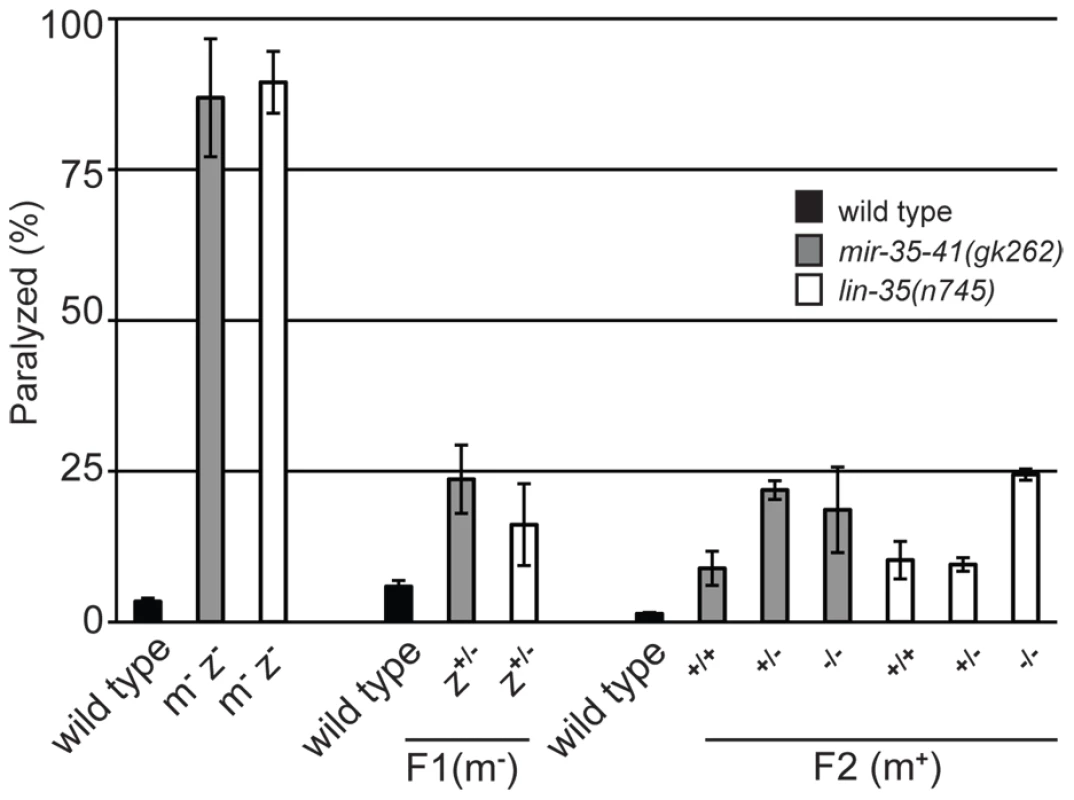
Histogram representing the percent of paralyzed worms for the indicated strains fed unc-22 dsRNA for 28 hours from the L4 to adult stage. Wild type and the mutants mir-35-41(gk262) and lin-35(n745) (maternal−, zygotic−: m−/z−) were crossed to wild type males containing a GFP expressing transgene to generate green heterozygous F1 progeny (m−/z+), which were transferred to unc-22 RNAi and scored for paralysis. To test for maternal rescue of RNAi sensitivity, heterozygous F1s were singled and allowed to self fertilize. The F2 progeny were transferred to unc-22 RNAi and each worm was scored for paralysis and then genotyped. Error bars represent the standard error of the mean (s.e.m) for two independent experiments. Discussion
We have shown that the mir-35-41 miRNA gene inhibits the exogenous RNAi pathway by positively regulating the expression of LIN-35/Rb protein. The effect of the miR-35-41 miRNAs on LIN-35/Rb levels is likely indirect as miRNAs typically repress gene expression and obvious binding sites for the miRNAs are not present in the lin-35 sequence. Regulation of LIN-35/Rb appears to be through a post-transcriptional mechanism since mRNA levels were unaffected in mir-35-41 mutants. We, and others, have been unable to identify direct targets of mir-35-41 that could explain the mis-regulation of LIN-35 levels, RNAi hypersensitivity, or embryonic lethal phenotypes of mir-35-41 mutant strains [32]. Positive regulation of the endo–siRNA pathway by mir-35-41 is likely indirect through targets of this miRNA family. Although few predicted targets of miR-35-41 were up-regulated upon loss of these miRNAs, this result is consistent with a recent study showing that embryonic targets of miR-35-41 undergo deadenylation while the rest of the mRNA remains stable in many cases [36]. Thus, microarrays lack the sensitivity to detect genes mis-regulated in mir-35-41 mutant embryos. The seven miRNAs in the miR-35-41 cluster share a common seed sequence and may regulate many genes that affect the viability and RNAi sensitivity phenotypes of mir-35-41 mutants, which could prevent individual targets from being discovered through genetic approaches.
The reason for enhanced exo–RNAi in lin-35 mutant strains is yet to be fully understood. Consistent with its role as a transcriptional repressor, loss of lin-35/Rb activity results in mis-expression of germline specific genes in somatic cells [23], [24], [52]. However, this effect alone cannot explain the enhanced RNAi, as lin-35 mutants exhibit hypersensitivity in both germline and somatic cells [22], [52]. Several genes for Argonaute proteins that function in the RNAi pathway are also up-regulated in lin-35/Rb mutants [24]. Additionally, targets of endo–siRNAs are overexpressed in the absence of lin-35/Rb, indicating that the endo–RNAi pathway is defective in lin-35/Rb mutants [24]. Thus, the increased expression of RNAi factors and reduced competition with the endo–RNAi pathway may underlie the improved efficiency of RNAi in lin-35 mutants. We found that sufficient endo–siRNAs are produced to target NRDE-3 to the nucleus in lin-35/Rb and mir-35-41 mutants, suggesting that these genes regulate the function, but not accumulation, of endo–siRNAs. Furthermore, both the spliced and unspliced forms of the endo–siRNA target E01G4.1 were up-regulated in mir-35-41 mutants. Since RNAi can silence targets at the transcriptional level in worms, mir-35-41 may also regulate RNAi effectiveness through this pathway [7], [8], [53].
RNAi initiated from exogenous dsRNA can propagate across generations in worms and other organisms [54], [55], [56], [57]. While the Argonaute RDE-1 and dsRNA binding protein RDE-4 are required to activate the RNAi response, they are dispensable for maintenance of RNAi [55], [57]. Instead, chromatin remodeling factors seem to mediate the inheritance of RNAi-induced phenotypes [53], [57]. Thus, the original RNA signal that directs post-transcriptional silencing of complementary targets may also stimulate chromatin remodeling events that repress gene expression across generations. Our results indicate that mir-35-41, through regulation of lin-35/Rb, decreases exo–RNAi potency and that this effect can be maternally contributed. The RNAi hypersensitivity of lin-35/Rb mutants can also be maternally rescued when the gene is expressed from the chromosomal locus but not from a transgene (Figure 4D and Figure 5). The inability of Ex[lin-35(+); sur-5::GFP] transgenes to provide maternal rescue has previously been observed and is likely due to the generally poor expression of transgenes in the germline [48], [58].
The mir-35-41 miRNAs are expressed in oocytes and present in early embryos prior to the onset of zygotic transcription [32], [50]. This expression pattern is consistent with our demonstration that maternal contribution is sufficient to regulate exo–RNAi sensitivity. However, this activity can also be provided by the wave of zygotic expression of mir-35-41 during embryogenesis [50], since mating of mir-35-41(gk262) to wild type worms rescues the RNAi hypersensitivity phenotype. Although the sequences of the mir-35-41 miRNAs are not conserved across species, many animals express high levels of poorly conserved miRNAs from clusters in embryonic stem cells and during early embryogenesis [38], [59], [60], [61]. One role for these miRNAs is to target maternal mRNAs for degradation [36], [38], [59], [62]. Our results show that miRNAs can also affect the activity of other small RNA pathways, demonstrating a broad function in regulating gene expression through both direct and indirect silencing mechanisms.
Materials and Methods
Nematode strains
C. elegans worm strains were maintained on NGM plates seeded with OP50 bacteria, under standard conditions [63]. Worms were synchronized by hypochlorite treatment of gravid hermaphrodites followed by overnight hatching of embryos at 20°C. Strains used in this study include the following: wild type (WT) Bristol N2 strain, NL2098 rrf-1(pk1417)I, MT10430 lin-35(n745)I, VC514 mir-35-41(gk262)II, NL2099 rrf-3(pk1426)II, MT111 lin-8(n111)II, WM49 rde-4(ne301)III, GR1373 eri-1(mg366)IV, WM158 ergo-1(tm1860)V, WM27 rde-1(ne219)V, MT1806 lin-15A(n767)X, YY158 nrde-3(gg66)X. Double mutants: PQ300 [mir-35-41(gk262);rde-1(ne219)], PQ301 [mir-35-41(gk262);rde-4(ne301)], PQ302 [mir-35-41(gk262); eri-1(mg366)], PQ303 [mir-35-41(gk262);rrf-1(pk1417)], PQ304 [mir-35-41(gk262); ergo-1(tm1860)], PQ421 [mir-35(gk262);lin-8(n111)], PQ422 [mir-35-41(gk262);lin-15A(n767)], PQ423 [lin-35(n745);lin-8(n111)], PQ424 [lin-35(n745); lin-15A(n767)], PQ459 [mir-35-41(gk262);nrde-3(gg66)]. Non-integrated transgenic strains: PQ20 [mir-35-41(gk262); apEX160 [mir-35-41]], MH1461 [lin-35(n745); kuEx119 [lin-35(+); sur-5::GFP]], PQ416 [mir-35-41(gk262); kuEx119 [lin-35(+), sur-5::GFP]], PQ439 [mir-35-41(gk262); Ex [myo-2::GFP)], PQ456 [eri-1(mg366); kuEx119 [lin-35(+), sur-5::GFP]], PQ458 [rrf-3 (pk1426); kuEx119 [lin-35(+); sur-5::GFP]]. Integrated transgenic strains: YY174 [ggIS1 [nrde-3p::3xflag::gfp::nrde-3]], YY178 eri-1(mg366); [ggIS1 [nrde-3p::3xflag::gfp::nrde-3]], PQ427 [mir-35-41 (gk262); yy1774 [ggIS1 [nrde-3p::3xflag::gfp::nrde-3]], PQ428 [mir-35-41(gk262);nrIs20 [sur-5::nls-gfp]], PD4251 ccIs4251I [myo-3::Ngfp-lacZ].
Worm viability was assayed by synchronizing worms and plating L1 hatchlings at 20°C. Worms were transferred to individual plates at the L4 stage and allowed to lay embryos for 24 h at 20°C or 25°C. Parents were then removed from the plates and embryos were counted. Viable worms were counted 40 hours later. Percent viable progeny represents the number of viable worms/total number of embryos laid.
RNAi experiments
RNAi plates were prepared using 25 ug/mL carbenicillin and 6 mM IPTG (Isopropyl β-D-1-thiogalactopyranoside, Apex). RNAi clones were obtained from the Ahringer feeding RNAi library [64]. For most experiments synchronized L1 worms were cultured on OP50 until the L4 stage (40 hours) and then transferred to RNAi food. After 28 hours adults were scored for unc-22, lin-1 and sqt-1 phenotypes. For pos-1 and sex-1 RNAi, L1 worms were directly plated on RNAi food and allowed to grow to adults. Parents were then transferred to individual RNAi plates and allowed to lay about 50 embryos. Percent embryonic lethality was calculated as the number of embryos that did not hatch by 40 hours later. For paternal/zygotic rescue experiments, wild-type male worms containing a body muscle GFP marker (PD4251) were crossed to mir-35(gk262) (mir-35−/−) mutant hermaphrodites. Parents were removed from the mating plate after 24 hours. Young GFP positive worms represented heterozygous mir-35−/+ F1 cross progeny. F1s were allowed to grow to the L4 stage and 50 GFP positive worms were transferred to unc-22(RNAi). F1s were scored for phenotypes 28 hours later. For maternal rescue, GFP positive F1's were transferred to individual plates and allowed to lay self-fertilized F2 embryos. Fifty F2 worms at the L4 stage were transferred to unc-22(RNAi). Each worm was scored for phenotypes 28 hours later, lysed and genotyped.
RNA and protein analyses
Trizol reagent (Invitrogen) was used to extract total RNA from frozen embryo pellets of the wild type (N2) and mir-35(gk262) strains. RNA samples were reverse-transcribed and hybridized to arrays following the Affymetrix manufacturer's protocol. Samples from three independent replicates of each strain were hybridized to the Affymetrix GeneChip C. elegans Genome Arrays representing 22,500 transcripts. Microarray samples were processed by the UCSD GeneChip Microarray Core. The raw data was normalized and t-statics were computed using R and Bioconductor (www.bioconductor.org) with the “affy” package and Benjamini-Hochberg (BH) correction method for multiple comparisons [65]. RNA levels that changed at least 1.5-fold with a probability of p<0.005 after BH correction were considered significantly different in mir-35(gk262) mutants relative to wild-type. Quantitative real-time PCR of reverse transcribed RNA (RT-qPCR) was performed with DNase treated RNA and the primers listed in Table S2. Northern blots to detect mRNAs and miRNAs and Western blots to detect proteins were performed as previously described [66]. The antibody for LIN-35 was provided by the Horvitz lab [45].
Supporting Information
Zdroje
1. FireAXuSMontgomeryMKKostasSADriverSE 1998 Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 806 811
2. CeruttiHCasas-MollanoJA 2006 On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet 50 81 99
3. MeisterGTuschlT 2004 Mechanisms of gene silencing by double-stranded RNA. Nature 431 343 349
4. TimmonsLFireA 1998 Specific interference by ingested dsRNA. Nature 395 854
5. FischerSE 2010 Small RNA-mediated gene silencing pathways in C. elegans. Int J Biochem Cell Biol 42 1306 1315
6. CzechBHannonGJ 2011 Small RNA sorting: matchmaking for Argonautes. Nature reviews Genetics 12 19 31
7. GuangSBochnerAFBurkhartKBBurtonNPavelecDM 2010 Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465 1097 1101
8. GuangSBochnerAFPavelecDMBurkhartKBHardingS 2008 An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321 537 541
9. OkamuraKLaiEC 2008 Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 9 673 678
10. AmbrosVLeeRCLavanwayAWilliamsPTJewellD 2003 MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13 807 818
11. RubyJGJanCPlayerCAxtellMJLeeW 2006 Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127 1193 1207
12. AsikainenSHeikkinenLWongGStorvikM 2008 Functional characterization of endogenous siRNA target genes in Caenorhabditis elegans. BMC Genomics 9 270
13. PakJFireA 2007 Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315 241 244
14. TamOHAravinAASteinPGirardAMurchisonEP 2008 Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453 534 538
15. GhildiyalMSeitzHHorwichMDLiCDuT 2008 Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320 1077 1081
16. DuchaineTFWohlschlegelJAKennedySBeiYConteDJr 2006 Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124 343 354
17. YigitEBatistaPJBeiYPangKMChenCC 2006 Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127 747 757
18. LeeRCHammellCMAmbrosV 2006 Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12 589 597
19. SimmerFTijstermanMParrishSKoushikaSPNonetML 2002 Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol 12 1317 1319
20. KennedySWangDRuvkunG 2004 A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427 645 649
21. CeronJRualJFChandraADupuyDVidalM 2007 Large-scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity. BMC Dev Biol 7 30
22. LehnerBCalixtoACrombieCTischlerJFortunatoA 2006 Loss of LIN-35, the Caenorhabditis elegans ortholog of the tumor suppressor p105Rb, results in enhanced RNA interference. Genome Biol 7 R4
23. WangDKennedySConteDJrKimJKGabelHW 2005 Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436 593 597
24. GrishokAHoerschSSharpPA 2008 RNA interference and retinoblastoma-related genes are required for repression of endogenous siRNA targets in Caenorhabditis elegans. Proc Natl Acad Sci U S A 105 20386 20391
25. KrolJLoedigeIFilipowiczW 2010 The widespread regulation of microRNA biogenesis, function and decay. Nature reviews Genetics 11 597 610
26. CorreaRLSteinerFABerezikovEKettingRF 2010 MicroRNA-directed siRNA biogenesis in Caenorhabditis elegans. PLoS Genet 6 e1000903 doi:10.1371/journal.pgen.1000903
27. HuntzingerEIzaurraldeE 2011 Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature reviews Genetics 12 99 110
28. GrishokAPasquinelliAEConteDLiNParrishS 2001 Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106 23 34
29. KanellopoulouCMuljoSAKungALGanesanSDrapkinR 2005 Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19 489 501
30. KettingRFFischerSEBernsteinESijenTHannonGJ 2001 Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & development 15 2654 2659
31. MiskaEAAlvarez-SaavedraEAbbottALLauNCHellmanAB 2007 Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet 3 e215 doi:10.1371/journal.pgen.0030215
32. Alvarez-SaavedraEHorvitzHR 2010 Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol 20 367 373
33. ShawWRArmisenJLehrbachNJMiskaEA 2010 The conserved miR-51 microRNA family is redundantly required for embryonic development and pharynx attachment in Caenorhabditis elegans. Genetics 185 897 905
34. LauNCLimLPWeinsteinEGBartelDP 2001 An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294 858 862
35. LiuMLiuPZhangLCaiQGaoG 2011 mir-35 is involved in intestine cell G1/S transition and germ cell proliferation in C. elegans. Cell Res
36. WuEThiviergeCFlamandMMathonnetGVashishtAA 2010 Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Molecular cell 40 558 570
37. LuYCSmielewskaMPalakodetiDLovciMTAignerS 2009 Deep sequencing identifies new and regulated microRNAs in Schmidtea mediterranea. RNA 15 1483 1491
38. BushatiNStarkABrenneckeJCohenSM 2008 Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol 18 501 506
39. SeitzHRoyoHBortolinMLLinSPFerguson-SmithAC 2004 A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome research 14 1741 1748
40. WalserCBLipshitzHD 2011 Transcript clearance during the maternal-to-zygotic transition. Current opinion in genetics & development 21 431 443
41. MoermanDGBaillieDL 1979 Genetic Organization in CAENORHABDITIS ELEGANS: Fine-Structure Analysis of the unc-22 Gene. Genetics 91 95 103
42. SijenTFleenorJSimmerFThijssenKLParrishS 2001 On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107 465 476
43. TabaraHSarkissianMKellyWGFleenorJGrishokA 1999 The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99 123 132
44. TabaraHYigitESiomiHMelloCC 2002 The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109 861 871
45. LuXHorvitzHR 1998 lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95 981 991
46. VasaleJJGuWThiviergeCBatistaPJClaycombJM 2010 Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci U S A 107 3582 3587
47. GuWShirayamaMConteDJrVasaleJBatistaPJ 2009 Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36 231 244
48. FayDSKeenanSHanM 2002 fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev 16 503 517
49. FergusonELHorvitzHR 1989 The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123 109 121
50. StoeckiusMMaaskolaJColomboTRahnHPFriedlanderMR 2009 Large-scale sorting of C. elegans embryos reveals the dynamics of small RNA expression. Nat Methods 6 745 751
51. EdgarLGWolfNWoodWB 1994 Early transcription in Caenorhabditis elegans embryos. Development 120 443 451
52. CuiMKimEBHanM 2006 Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet 2 e74 doi:10.1371/journal.pgen.0020074
53. GrishokASinskeyJLSharpPA 2005 Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev 19 683 696
54. AlcazarRMLinRFireAZ 2008 Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics 180 1275 1288
55. GrishokATabaraHMelloCC 2000 Genetic requirements for inheritance of RNAi in C. elegans. Science 287 2494 2497
56. RassoulzadeganMGrandjeanVGounonPVincentSGillotI 2006 RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 441 469 474
57. VastenhouwNLBrunschwigKOkiharaKLMullerFTijstermanM 2006 Gene expression: long-term gene silencing by RNAi. Nature 442 882
58. FayDSLargeEHanMDarlandM 2003 lin-35/Rb and ubc-18, an E2 ubiquitin-conjugating enzyme, function redundantly to control pharyngeal morphogenesis in C. elegans. Development 130 3319 3330
59. GiraldezAJMishimaYRihelJGrocockRJVan DongenS 2006 Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312 75 79
60. HoubaviyHBMurrayMFSharpPA 2003 Embryonic stem cell-specific MicroRNAs. Dev Cell 5 351 358
61. SuhMRLeeYKimJYKimSKMoonSH 2004 Human embryonic stem cells express a unique set of microRNAs. Dev Biol 270 488 498
62. TangFKanedaMO'CarrollDHajkovaPBartonSC 2007 Maternal microRNAs are essential for mouse zygotic development. Genes Dev 21 644 648
63. BrennerS 1974 The genetics of Caenorhabditis elegans. Genetics 77 71 94
64. KamathRSFraserAGDongYPoulinGDurbinR 2003 Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231 237
65. YeoGWXuXLiangTYMuotriARCarsonCT 2007 Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol 3 e196 doi:10.1371/journal.pcbi.0030196
66. BaggaSBrachtJHunterSMassirerKHoltzJ 2005 Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122 553 563
Štítky
Genetika Reprodukční medicína
Článek Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1Článek Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / TranscriptionČlánek Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding DomainČlánek Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin ComplexesČlánek An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood ObesityČlánek Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAsČlánek Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 3
-
Všechny články tohoto čísla
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
- Genomic Analysis of the Hydrocarbon-Producing, Cellulolytic, Endophytic Fungus
- Networks of Neuronal Genes Affected by Common and Rare Variants in Autism Spectrum Disorders
- Akirin Links Twist-Regulated Transcription with the Brahma Chromatin Remodeling Complex during Embryogenesis
- Too Much Cleavage of Cyclin E Promotes Breast Tumorigenesis
- Imprinted Genes … and the Number Is?
- Genetic Architecture of Highly Complex Chemical Resistance Traits across Four Yeast Strains
- Exploring the Complexity of the HIV-1 Fitness Landscape
- MNS1 Is Essential for Spermiogenesis and Motile Ciliary Functions in Mice
- A Fundamental Regulatory Mechanism Operating through OmpR and DNA Topology Controls Expression of Pathogenicity Islands SPI-1 and SPI-2
- Evidence for Positive Selection on a Number of MicroRNA Regulatory Interactions during Recent Human Evolution
- Variation in Modifies Risk of Neonatal Intestinal Obstruction in Cystic Fibrosis
- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Critical Evaluation of Imprinted Gene Expression by RNA–Seq: A New Perspective
- A Meta-Analysis and Genome-Wide Association Study of Platelet Count and Mean Platelet Volume in African Americans
- Mouse Genetics Suggests Cell-Context Dependency for Myc-Regulated Metabolic Enzymes during Tumorigenesis
- Transcriptional Control in Cardiac Progenitors: Tbx1 Interacts with the BAF Chromatin Remodeling Complex and Regulates
- Synthetic Lethality of Cohesins with PARPs and Replication Fork Mediators
- APOBEC3G-Induced Hypermutation of Human Immunodeficiency Virus Type-1 Is Typically a Discrete “All or Nothing” Phenomenon
- Interpreting Meta-Analyses of Genome-Wide Association Studies
- Error-Prone ZW Pairing and No Evidence for Meiotic Sex Chromosome Inactivation in the Chicken Germ Line
- -Dependent Chemosensory Functions Contribute to Courtship Behavior in
- Diverse Forms of Splicing Are Part of an Evolving Autoregulatory Circuit
- Phenotypic Plasticity of the Drosophila Transcriptome
- Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1
- Precocious Metamorphosis in the Juvenile Hormone–Deficient Mutant of the Silkworm,
- Igf1r Signaling Is Indispensable for Preimplantation Development and Is Activated via a Novel Function of E-cadherin
- Accurate Prediction of Inducible Transcription Factor Binding Intensities In Vivo
- Mitochondrial Oxidative Stress Alters a Pathway in Strongly Resembling That of Bile Acid Biosynthesis and Secretion in Vertebrates
- Mammalian Neurogenesis Requires Treacle-Plk1 for Precise Control of Spindle Orientation, Mitotic Progression, and Maintenance of Neural Progenitor Cells
- Tcf7 Is an Important Regulator of the Switch of Self-Renewal and Differentiation in a Multipotential Hematopoietic Cell Line
- REST–Mediated Recruitment of Polycomb Repressor Complexes in Mammalian Cells
- Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / Transcription
- Age-Dependent Brain Gene Expression and Copy Number Anomalies in Autism Suggest Distinct Pathological Processes at Young Versus Mature Ages
- A Genome-Wide Association Study Identifies Variants Underlying the Shade Avoidance Response
- -by- Regulatory Divergence Causes the Asymmetric Lethal Effects of an Ancestral Hybrid Incompatibility Gene
- Genome-Wide Association and Functional Follow-Up Reveals New Loci for Kidney Function
- A Natural System of Chromosome Transfer in
- Cell Size and the Initiation of DNA Replication in Bacteria
- Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding Domain
- Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin Complexes
- Temporal Transcriptional Profiling of Somatic and Germ Cells Reveals Biased Lineage Priming of Sexual Fate in the Fetal Mouse Gonad
- Rapid Analysis of Genome Rearrangements by Multiplex Ligation–Dependent Probe Amplification
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- The Atypical Calpains: Evolutionary Analyses and Roles in Cellular Degeneration
- The Silkworm Coming of Age—Early
- Development of a Panel of Genome-Wide Ancestry Informative Markers to Study Admixture Throughout the Americas
- Balanced Codon Usage Optimizes Eukaryotic Translational Efficiency
- The Min System and Nucleoid Occlusion Are Not Required for Identifying the Division Site in but Ensure Its Efficient Utilization
- Neurobeachin, a Regulator of Synaptic Protein Targeting, Is Associated with Body Fat Mass and Feeding Behavior in Mice and Body-Mass Index in Humans
- Statistical Analysis of Readthrough Levels for Nonsense Mutations in Mammalian Cells Reveals a Major Determinant of Response to Gentamicin
- Gene Reactivation by 5-Aza-2′-Deoxycytidine–Induced Demethylation Requires SRCAP–Mediated H2A.Z Insertion to Establish Nucleosome Depleted Regions
- The miR-35-41 Family of MicroRNAs Regulates RNAi Sensitivity in
- Genetic Basis of Hidden Phenotypic Variation Revealed by Increased Translational Readthrough in Yeast
- An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood Obesity
- Modelling Human Regulatory Variation in Mouse: Finding the Function in Genome-Wide Association Studies and Whole-Genome Sequencing
- Novel Loci for Adiponectin Levels and Their Influence on Type 2 Diabetes and Metabolic Traits: A Multi-Ethnic Meta-Analysis of 45,891 Individuals
- Polycomb-Like 3 Promotes Polycomb Repressive Complex 2 Binding to CpG Islands and Embryonic Stem Cell Self-Renewal
- Insulin/IGF-1 and Hypoxia Signaling Act in Concert to Regulate Iron Homeostasis in
- EMF1 and PRC2 Cooperate to Repress Key Regulators of Arabidopsis Development
- Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAs
- Contrasted Patterns of Molecular Evolution in Dominant and Recessive Self-Incompatibility Haplotypes in
- A Machine Learning Approach for Identifying Novel Cell Type–Specific Transcriptional Regulators of Myogenesis
- Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
- Nos2 Inactivation Promotes the Development of Medulloblastoma in Mice by Deregulation of Gap43–Dependent Granule Cell Precursor Migration
- Intracranial Aneurysm Risk Locus 5q23.2 Is Associated with Elevated Systolic Blood Pressure
- Heritability and Genetic Correlations Explained by Common SNPs for Metabolic Syndrome Traits
- A Genome-Wide Association Study of Nephrolithiasis in the Japanese Population Identifies Novel Susceptible Loci at 5q35.3, 7p14.3, and 13q14.1
- DNA Damage in Nijmegen Breakage Syndrome Cells Leads to PARP Hyperactivation and Increased Oxidative Stress
- DNA Resection at Chromosome Breaks Promotes Genome Stability by Constraining Non-Allelic Homologous Recombination
- Genetic Analysis of Floral Symmetry in Van Gogh's Sunflowers Reveals Independent Recruitment of Genes in the Asteraceae
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Promoter Nucleosome Organization Shapes the Evolution of Gene Expression
- The Nucleoside Diphosphate Kinase Gene Acts as Quantitative Trait Locus Promoting Non-Mendelian Inheritance
- The Ciliogenic Transcription Factor RFX3 Regulates Early Midline Distribution of Guidepost Neurons Required for Corpus Callosum Development
- Phosphorylation of the RNA–Binding Protein HOW by MAPK/ERK Enhances Its Dimerization and Activity
- A Genome-Wide Scan of Ashkenazi Jewish Crohn's Disease Suggests Novel Susceptibility Loci
- Parkinson's Disease–Associated Kinase PINK1 Regulates Miro Protein Level and Axonal Transport of Mitochondria
- LMW-E/CDK2 Deregulates Acinar Morphogenesis, Induces Tumorigenesis, and Associates with the Activated b-Raf-ERK1/2-mTOR Pathway in Breast Cancer Patients
- Mapping the Hsp90 Genetic Interaction Network in Reveals Environmental Contingency and Rewired Circuitry
- Autoregulation of the Noncoding RNA Gene
- The Human Pancreatic Islet Transcriptome: Expression of Candidate Genes for Type 1 Diabetes and the Impact of Pro-Inflammatory Cytokines
- Spo0A∼P Imposes a Temporal Gate for the Bimodal Expression of Competence in
- Antagonistic Regulation of Apoptosis and Differentiation by the Cut Transcription Factor Represents a Tumor-Suppressing Mechanism in
- A Downstream CpG Island Controls Transcript Initiation and Elongation and the Methylation State of the Imprinted Macro ncRNA Promoter
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



