-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaProbing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding Domain
Transcription factors have two functional constraints on their evolution:
(1) their binding sites must have enough information to be distinguishable from all other sequences in the genome, and (2) they must bind these sites with an affinity that appropriately modulates the rate of transcription. Since both are determined by the biophysical properties of the DNA–binding domain, selection on one will ultimately affect the other. We were interested in understanding how plastic the informational and regulatory properties of a transcription factor are and how transcription factors evolve to balance these constraints. To study this, we developed an in vivo selection system in Escherichia coli to identify variants of the helix-turn-helix transcription factor MarA that bind different sets of binding sites with varying degrees of degeneracy. Unlike previous in vitro methods used to identify novel DNA binders and to probe the plasticity of the binding domain, our selections were done within the context of the initiation complex, selecting for both specific binding within the genome and for a physiologically significant strength of interaction to maintain function of the factor. Using MITOMI, quantitative PCR, and a binding site fitness assay, we characterized the binding, function, and fitness of some of these variants. We observed that a large range of binding preferences, information contents, and activities could be accessed with a few mutations, suggesting that transcriptional regulatory networks are highly adaptable and expandable.
Published in the journal: . PLoS Genet 8(3): e32767. doi:10.1371/journal.pgen.1002614
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002614Summary
Transcription factors have two functional constraints on their evolution:
(1) their binding sites must have enough information to be distinguishable from all other sequences in the genome, and (2) they must bind these sites with an affinity that appropriately modulates the rate of transcription. Since both are determined by the biophysical properties of the DNA–binding domain, selection on one will ultimately affect the other. We were interested in understanding how plastic the informational and regulatory properties of a transcription factor are and how transcription factors evolve to balance these constraints. To study this, we developed an in vivo selection system in Escherichia coli to identify variants of the helix-turn-helix transcription factor MarA that bind different sets of binding sites with varying degrees of degeneracy. Unlike previous in vitro methods used to identify novel DNA binders and to probe the plasticity of the binding domain, our selections were done within the context of the initiation complex, selecting for both specific binding within the genome and for a physiologically significant strength of interaction to maintain function of the factor. Using MITOMI, quantitative PCR, and a binding site fitness assay, we characterized the binding, function, and fitness of some of these variants. We observed that a large range of binding preferences, information contents, and activities could be accessed with a few mutations, suggesting that transcriptional regulatory networks are highly adaptable and expandable.Introduction
The precise regulation of gene expression depends upon the specific binding of transcription factors to their cognate binding sites. For this process to be accurate, the sites for each factor need to be separable from all other sequences in the genome [1], [2]. Many groups have studied specific protein-DNA interactions, and while nucleotide preferences are starting to be understood at the biophysical level for some DNA binding domains [3]–[5], no universal DNA-recognition code has been discovered [6]. What has emerged is a consistent picture of binding site degeneracy. That is, for most factors there is a single consensus binding site that is bound with the highest affinity and an increasing number of lower affinity sites that vary from the consensus. At some point the degeneration is so great that all remaining sites show the same non-specific binding energy [7]–[9]. Using information theory, the amount of conservation within a set of binding sites (information content), as well as the amount of information needed to specifically locate N sites in a genome of length L, can be quantified [1], [10]. In bacteria, it has been shown that these values are identical for many factors, suggesting that the size of a factor's regulon constrains how specific it needs to be [1], [11], [12]. This relationship does not hold as well for individual transcription factors in eukaryotes though [13], [14], where gene regulation is often under the control of cooperatively acting factors [15].
Once bound to their target sequence, transcription factors can modulate the rate of expression over a range of activities. Differences in expression levels have been suggested and shown to vary with binding site strength [16]–[19]. Given this relationship, the range and continuity of binding affinities for a factor partially define the range and continuity of potential outputs for that factor [19], [20]. These outputs in turn can significantly affect the phenotype and fitness of the cell and are selected to maximize cellular gain while minimizing cost [19], [21], [22]. Therefore, there is not only a selective advantage for transcription factors to specifically recognize and bind their target sites, but to bind them with an affinity that produces the maximally fit transcriptional output. Since both specific binding preferences and transcriptional activity are dependent on the distribution of binding energies for a factor, selection on one will ultimately affect the other.
We are interested in understanding how plastic the informational and regulatory properties of a transcription factor are, and how transcription factors evolve to balance these functions. To address this, we developed an in vivo selection system in E. coli to select for functional variants of the transcription factor MarA with altered binding preferences, whose binding properties and activity could be further characterized. By functional, we mean that a variant could modulate the level of transcriptional output within a physiological range. This is in contrast to in vitro selection assays, like phage display, that generally select for high affinity binding to a single target sequence, and disregard the impact of these mutations on transcriptional activity.
To do these selections, we wanted to use a monomeric, transcriptional activator whose binding sites have been characterized and structure had been solved. MarA fit these criteria. It is a monomeric, helix-turn-helix transcription factor in the AraC family [23] that can both activate and repress transcription in E. coli [24]–[26]. It regulates the expression of approximately 20 genes involved in exporting low levels of drugs and organic solvents from the cell [24], [27]. The structure of the MarA-DNA complex suggests that specific recognition occurs through two alpha-helices that bind the major groove [28], [29]. Additionally, MarA has two homologues in E. coli, Rob and SoxS, that have similar binding preferences [30], suggesting that the MarA binding domain can be selected to recognize additional sites.
Results
MarA binding domain and sites
We generated a sequence logo from the 16 E. coli MarA binding sites summarized in Martin et al. [24] to visualize the natural binding preference of the protein and the relative contribution of each contacting residue to binding specificity (Figure 1). Sequence conservation follows a sine wave as seen for other transcription factors [31], [32]. MarA specifically contacts the DNA through helices 3 and 6. Bases contacted by helix 3 (red helix on structure, DNA positions to ) have a greater information content than do those contacted by helix 6 (blue helix on structure that intersects the sine wave, DNA positions to ), suggesting that helix 3 is more important for specific DNA recognition. This is consistent with alanine-scanning mutagenesis data for MarA [33].
Fig. 1. MarA logo and structure. 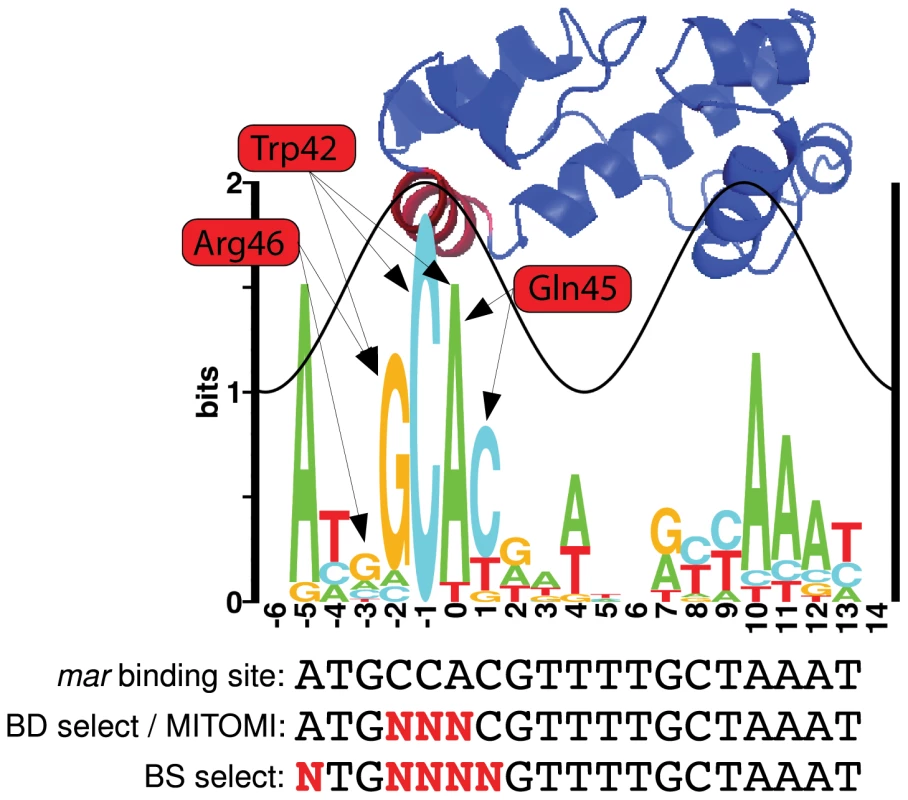
The height of each letter in the sequence logo is proportional to the frequency of that base at that position. The height of the stack at each position is the information content [39]. The sine wave on the logo has the same helical twist as B-form DNA (10.6 bp) [32] and its position was assigned based on the MarA-DNA cocrystal structure [28]. The structure of E. coli MarA is positioned above the logo to show which bases each helix specifically binds [28]. Three residues in helix 3 (red helix on the structure) specifically contact DNA bases. Arrows show which bases these residues specify. We randomized these three residues and selected for variants that had altered affinity. Binding domain selections (BD select), MITOMI experiments, and in vivo binding site selections (BS select) were performed with variants of the mar MarA binding site. A red ‘N’ specifies bases that were varied for each experiment. For binding site and binding domain selections, these variants were cloned into the selection plasmid in the MarA binding site (Figure 2B). The information content for this logo is 12.60.9 bits. Three residues in helix 3 (Trp42, Gln45, and Arg46) specifically contact DNA bases according to the MarA-DNA structure [28] (Figure 1). Interestingly, the structure does not predict a specific contact at position , but the sequence logo indicates a strong preference for ‘A’ at this position. The ‘C’ at position is completely conserved and only contacted by the tryptophan at residue 42, suggesting this is a highly specific amino acid.
Selection of MarA binding domain variants
To identify variants of MarA that have altered binding preferences, we randomized the three specifically contacting residues in helix 3 and selected for mutants that could bind a target DNA sequence and initiate transcription of the tetracycline resistance gene (tet) on the selection plasmid shown in Figure 2. Both the promoter of the tet gene and helix 3 of the MarA protein were flanked by restriction sites that allowed promoter and binding domain variants to be cloned into the plasmid (Figure 2). Functional MarA protein-binding site pairs within this system activated tet and allowed for cell survival in tetracycline. As we increased the concentration of drug, we selected for higher affinity interactions [19]. Additional parameters can affect the rate of transcriptional initiation, most notably the position of the binding site relative to the polymerase [24]. Since we vary the binding site within a fixed promoter context, our selection should just be on the strength of the DNA-protein interaction. We performed our selection in the E. coli strain N8453 (mar, sox-8::cat, rob::kan, see Materials and Methods) to prevent activation by wild type MarA, or by the MarA E. coli homologues Rob and SoxS. Expression of MarA on the plasmid was controlled by an L-arabinose inducible promoter [34].
Fig. 2. MarA binding domain selection system. 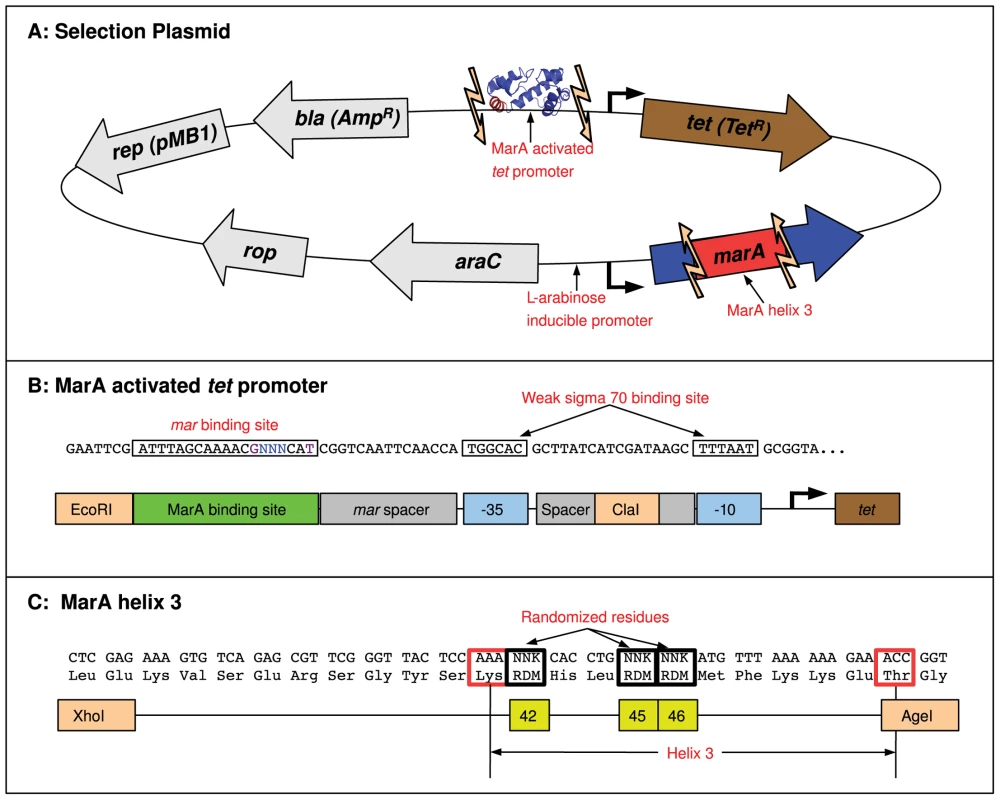
(A) A schematic representation of the MarA selection plasmid. Expression of the marA gene is controlled by an AraC repressed, L-arabinose inducible promoter [34]. Unique restriction sites (light orange arrows) flank the promoter region of the tetracycline resistance gene tet and helix 3 of the marA gene. Promoter and binding domain variants can be cloned into this plasmid and functional binding domain-binding site pairs can be identified by selection in tetracycline+L-arabinose. (B) The sequence of the MarA-activated tet promoter (top) and a cartoon marking each component (bottom). This construct is based on the promoter used in [19]. Bases that were varied in binding domain selection experiments are designated by a blue ‘N’. Additional bases that were randomized in the binding site selection are shown in purple. Orange boxes mark the restriction sites used to clone in these constructs. The mar binding site has the opposite orientation as in Figure 1. (C) The sequence of the MarA binding domain variants (top) and a cartoon marking components within this region. The three residues that were randomized are marked with yellow boxes. The boundaries of helix 3 are marked with red boxes. We needed to identify a promoter that was only functional when activated to have tet expression and cell survival dependent upon MarA binding. To identify one, we randomized the of the tet promoter construct (Figure 2B) and selected for a promoter sequence that allowed cell growth on tetracycline plates with L-arabinose (induced expression of MarA) but not on plates without it (see Materials and Methods). The 6.5 bit binding site that we identified is marked in Figure 2B. The strength of this site was predicted using the model presented in [18], and is an average site compared to all sites in the genome. In a single construct, we cloned 3 in-frame and 2 out-of-frame stop codons into helix 3 of the MarA binding domain and tested if the resulting truncated protein could express tet with this promoter. At 15 g/ml tetracycline and 0.1% L-arabinose, we observed significant growth with wild type MarA, and no growth with the truncated mutant (data not shown), suggesting that in this condition activation of tet and cell survival is dependent upon binding by MarA.
The MarA regulon in E. coli includes the arcAB operon, which when over-expressed shows increased tolerance to many antibiotics including tetracylcine [35], [36]. To ensure that we are selecting for variants that directly activate tet, we performed a selection against the anti-consensus MarA binding site (the worst possible binding site according to Figure 1: CGTTTGACCCGCCAGGGCG). We could not identify any protein variants that allowed for survival in 20 or 30 g/ml tetracycline, suggesting that differential regulation of the MarA regulon is not sufficient for cell viability. This does not exclude the possibility that the over-expression of the arcAB operon may reduce the selective pressure on tet production. Selection in this system is somewhat similar to selection in a natural system, where the fitness of a binder is dependent upon the relative contribution of multiply expressed genes. We have in essence added tet to the MarA regulon. Because of the high concentration of tetracycline used for selection, the fitness gain for expressing tet is probably much greater than for any other gene that it regulates.
MarA binding domain mutants were selected against three variants of the 15.3 bit mar binding site (Figure 1) that is found upstream of the mar operon in E. coli [24]. The three target sequences we selected against are named ‘GCA’, ‘GAA’ and ‘GAC’ according to the bases present at positions , and (Figure 1 and Figure 2B). We varied these bases because they are the most highly conserved ones contacted by helix 3. Binding domain libraries were made as described in Materials and Methods. We transformed the N8453 cells with each library and selected for growth on plates at 20 and 30 g/ml of tetracycline +0.1% L-arabinose. Individual colonies were sequenced.
Sequences of viable MarA binding domain variants are shown in Table 1 and sequence logos generated from these variants are shown in Figure 3. Each binding domain is referenced by residues 42, 45 and 46. For example, wild type MarA is noted as WQR. Of the 18 sequenced binding domains selected against the MarA consensus ‘GCA’ binding site at 20 g/ml tetracycline, we identified 13 different variants, including that of the wild type protein, that could initiate tet transcription to a sufficiently high level for cell survival. Only 5 different variants were observed at 30 g/ml tetracycline, and no new variants were observed at this higher concentration as expected. Three of the 13 binding domains were represented by multiple codon sets further supporting that these variants are functional. Interestingly, only the ‘TCK’ variant selected against ‘GCA’ lacks an arginine at position 46, but it retains a positively charged lysine residue at that position.
Fig. 3. Sequence logos of natural and selected variants of the MarA binding domain. 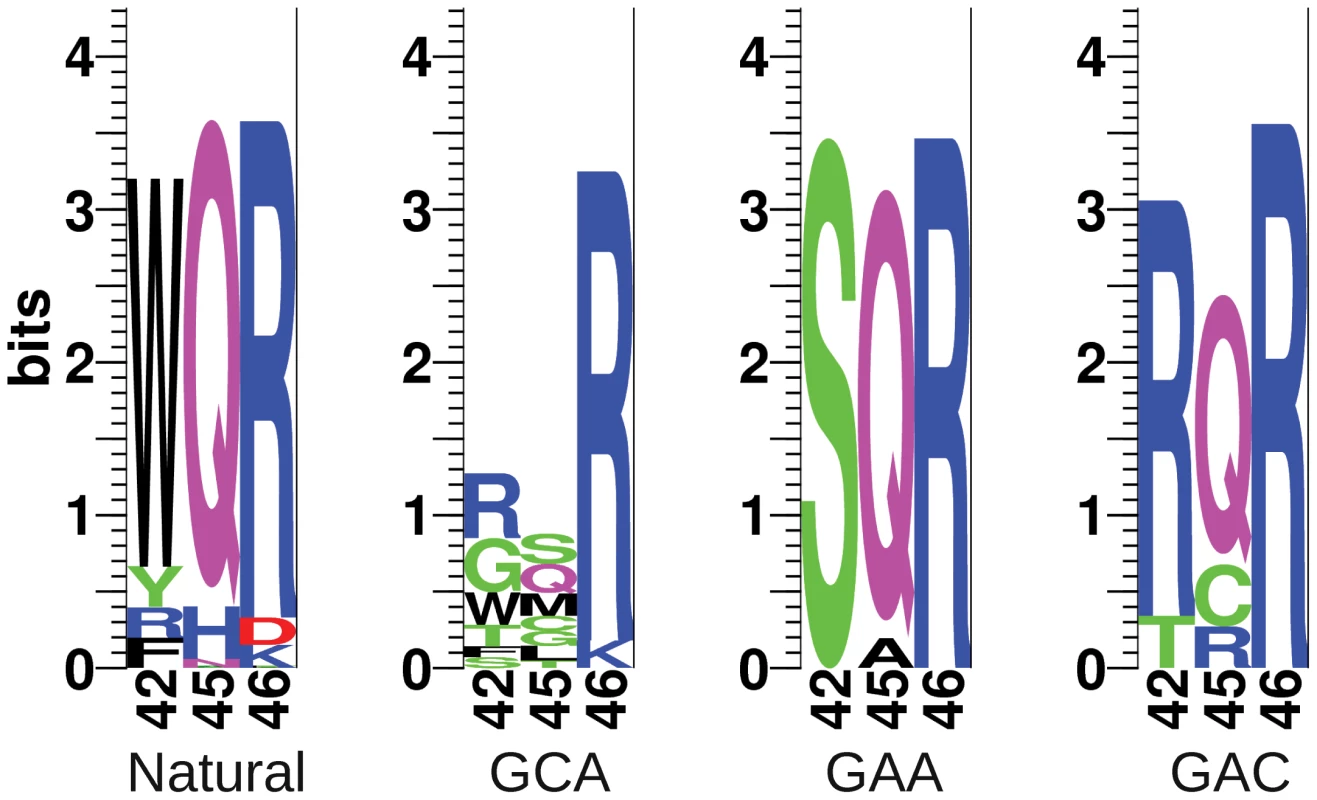
The sequence logos show the degree of variability at residues 42, 45 and 46 in functional binders selected against different binding sites. ‘Natural’ is the natural variability in positions 42, 45 and 46 for MarA homologues. ‘GCA’, ‘GAA’ and ‘GAC’ designate which sequence the binding domain was selected against. These logos are made from the sequences in Table 1. Tab. 1. Functional MarA binding domain variants selected against different binding sites. 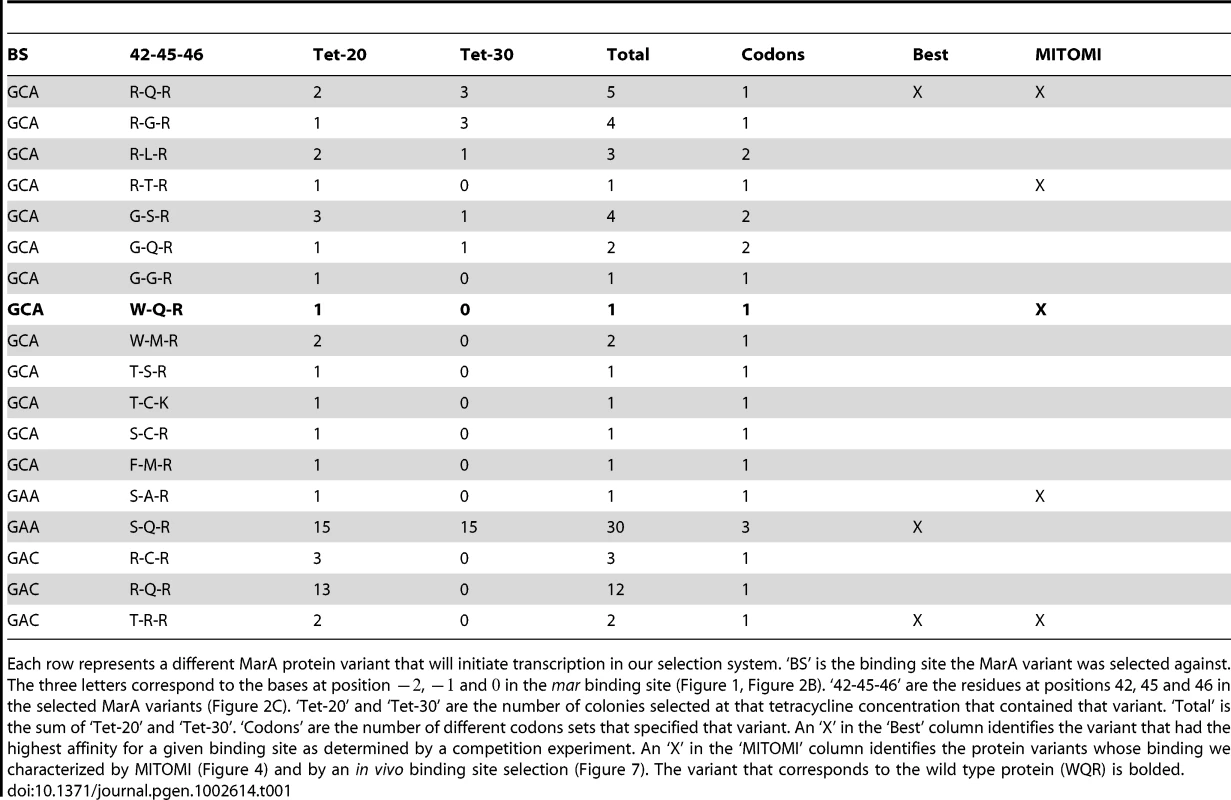
Each row represents a different MarA protein variant that will initiate transcription in our selection system. ‘BS’ is the binding site the MarA variant was selected against. The three letters correspond to the bases at position , and in the mar binding site (Figure 1, Figure 2B). ‘42-45-46’ are the residues at positions 42, 45 and 46 in the selected MarA variants (Figure 2C). ‘Tet-20’ and ‘Tet-30’ are the number of colonies selected at that tetracycline concentration that contained that variant. ‘Total’ is the sum of ‘Tet-20’ and ‘Tet-30’. ‘Codons’ are the number of different codons sets that specified that variant. An ‘X’ in the ‘Best’ column identifies the variant that had the highest affinity for a given binding site as determined by a competition experiment. An ‘X’ in the ‘MITOMI’ column identifies the protein variants whose binding we characterized by MITOMI (Figure 4) and by an in vivo binding site selection (Figure 7). The variant that corresponds to the wild type protein (WQR) is bolded. Selection against the ‘GAA’ and ‘GAC’ binding sites showed much less variability in the number of identified functional MarA variants. We only identified two mutants that could activate the ‘GAA’ binding site and three that could activate ‘GAC’. No colonies were observed when we selected against ‘GAC’ at the higher tetracycline concentration of 30 g/ml.
We were interested in how the variability in the selected mutants compared to the natural variability at these residues. We blasted the E. coli MarA sequence against all bacterial genomes using BlastP with non-redundant protein sequences and default search parameters [37]. The top 250 hits were aligned by ClustalX [38] and sequence logos were generated using the Delila programs [39] (Figure 3, Natural). Both the natural and the experimentally selected binding domain variants show a strong preference for arginine at position 46. Interestingly, tryptophan is highly conserved at position 42 in the natural binding domains, whereas it was only observed in two selected variants (Table 1). In a similar selection for specifically contacting residues in the engrailed homeodomain by phage display, experimentally and naturally selected variability correlated well [40]. Engrailed binds a more specific set of sequences than does MarA. Therefore, natural selection on binding by engrailed is probably directed to maintain high affinity to a single or small set of sites as was experimentally selected. Conversely, MarA has probably been selected to maintain affinity to a more degenerate set of sequences, which may explain the discordance between the naturally and experimentally selected binding domains.
To identify the highest affinity MarA mutant for each of the three DNA binding sites, the protein binding domains in each library were competed against each other in liquid culture containing 30 g/ml tetracycline+L-arabinose for 24 hours. The competed cultures were mini-prepped, retransformed and individual variants were sequenced (Materials and Methods). We expected the mutant that produced the highest tet output to be represented at the highest frequency in the competed population as seen in a similar experiment [19]. We sequenced 8 individuals from each library and observed only one protein variant for each target binding site: RQR for ‘GCA’, SQR for ‘GAA’ and TRR for ‘GAC’ (Table 1, marked with ‘X’ in Best column). Interestingly, wild type MarA (WQR) was not identified as the most fit variant for its naturally evolved consensus binding site ‘GCA’.
High-throughput measurement of DNA binding preferences for MarA mutants
We determined the relative affinity of wild type MarA and four selected MarA variants to 64 different binding sites using MITOMI (Figure 4). MITOMI (Mechanically Induced Trapping of Molecular Interactions) measures the relative thermodynamic association constant of a single transcription factor for a large number of DNA sequences using a microfluidics based approach. The relative amount of fluorescently-labeled protein associated with fluorescently-labeled DNA is quantified by microscopy for each binding site to determine interaction strengths [8].
Fig. 4. Binding affinities for 5 MarA variants to 64 binding sites. 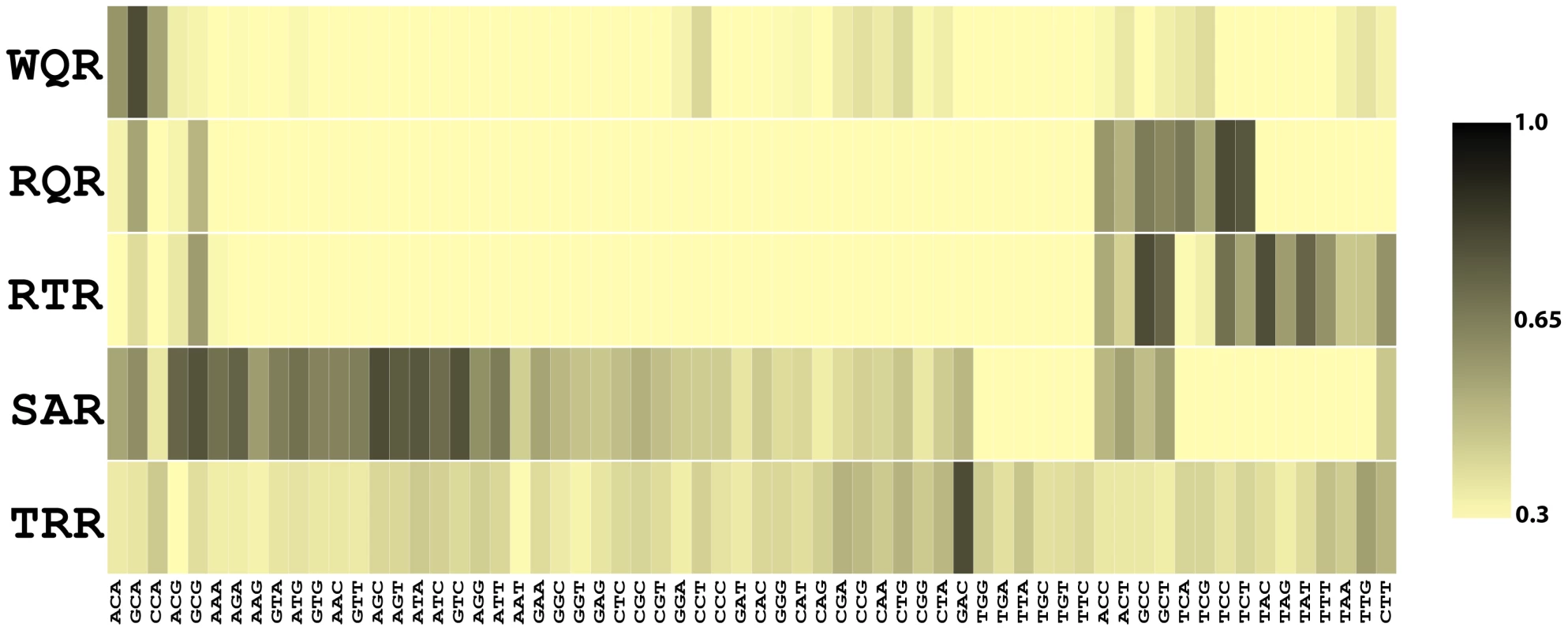
The heat map shows the relative binding affinities of wild type MarA (WQR) and 4 selected variants to 64 variations of the mar binding site (Figure 1). Each MarA variant (y-axis) is named according to its residues at positions 42, 45 and 46. Each DNA sequence (x-axis) was substituted for ‘NNN’ in the mar binding site (Figure 1). Data for all variants were normalized so the highest affinity site was set to 1 (black). All other sites are colored relative to that site according to the color scale. All sites below 0.3 were colored the same as 0.3. The 64 sequences we measured binding to covered all combinations of bases at positions , and in the mar binding site (Figure 1). The 5 transcription factor variants chosen were wild type MarA (WQR), the most fit binder for the wild type consensus binding site (RQR), a double mutant that binds to the wild type consensus (RTR), a double mutant that activates the ‘GAA’ site (SAR), and the most fit mutant for the binding site ‘GAC’ (TRR). We did not obtain reliable binding data for SQR, the most fit mutant for ‘GAA’, and therefore did not include it in this study. For each of these five transcription factor variants, we set the binding affinity of the strongest site to 1 and scaled the strength of all other sites relative to that (Figure S1). To identify sequences that are similarly bound for each mutant, we clustered the DNA binding sites according to their relative affinities using Cluster [41] (Figure 4). Additionally, we we generated energy-based position weight matrices and logos [42] (Figure 5), and calculated the degree of similarity between all matrices as Kullback-Leibler Divergences (KLD) using the program MatCompare [43] (see Materials and Methods). A KLD generally indicates that two matrices are significantly similar, and a KLD of 0 indicates that they are identical. All measured binding affinities, position weight matrices, and pair-wise KLD values are reported in Table S1.
Fig. 5. Energy logos for MarA variant binding sites. 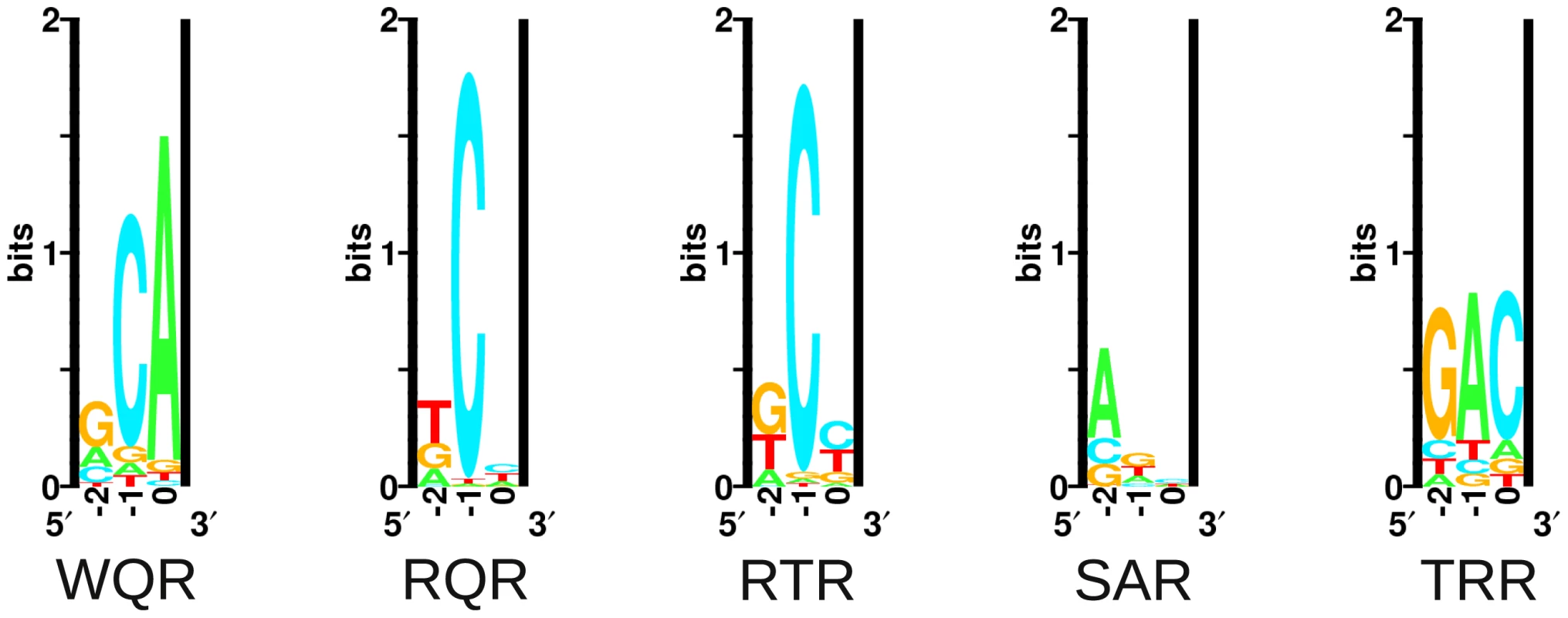
Energy logos were generated from the MITOMI data for the 5 variants in Figure 4, using the enoLogos Webserver [42] (see Materials and Methods). MITOMI data for wild type MarA are consistent with the MarA sequence logo (Figure 1). Three sequences are tightly bound, ‘GCA’‘ACA’‘CCA’, as seen in natural sites. A single mutation from a Trp at position 42 to an Arg has a dramatic effect on the binding preferences of the factor (Figure 5, KLD = 1.53). The RQR mutant still specifically recognizes ‘GCA’, but with a 1.6 fold reduced affinity relative to its most tightly bound site ‘TCC’. As with wild type MarA, RQR has a strong preference for ‘C’ at position , but overall RQR is a less specific binder; the information content () [1] for positions to is 3.03 and 2.27 bits for WQR and RQR respectively (Figure 5, Table 2). The 2.46 bit RTR logo is significantly similar to the RQR logo (KLD = 0.15), but shows a slight decrease in degeneracy at position , as well as a switch in preference for ‘G’ over ‘T’ at position . Interestingly, the RQR and RTR mutants maintained the same relative difference in affinity between the bound sequences ‘GCA’, ‘ACA’ and ‘CCA’ as wild type ( for both, data not shown), suggesting that the core binding preferences of wild type are somehow preserved in these variants although they are no longer the highest affinity sites.
Tab. 2. Specific binding energies and information contents. 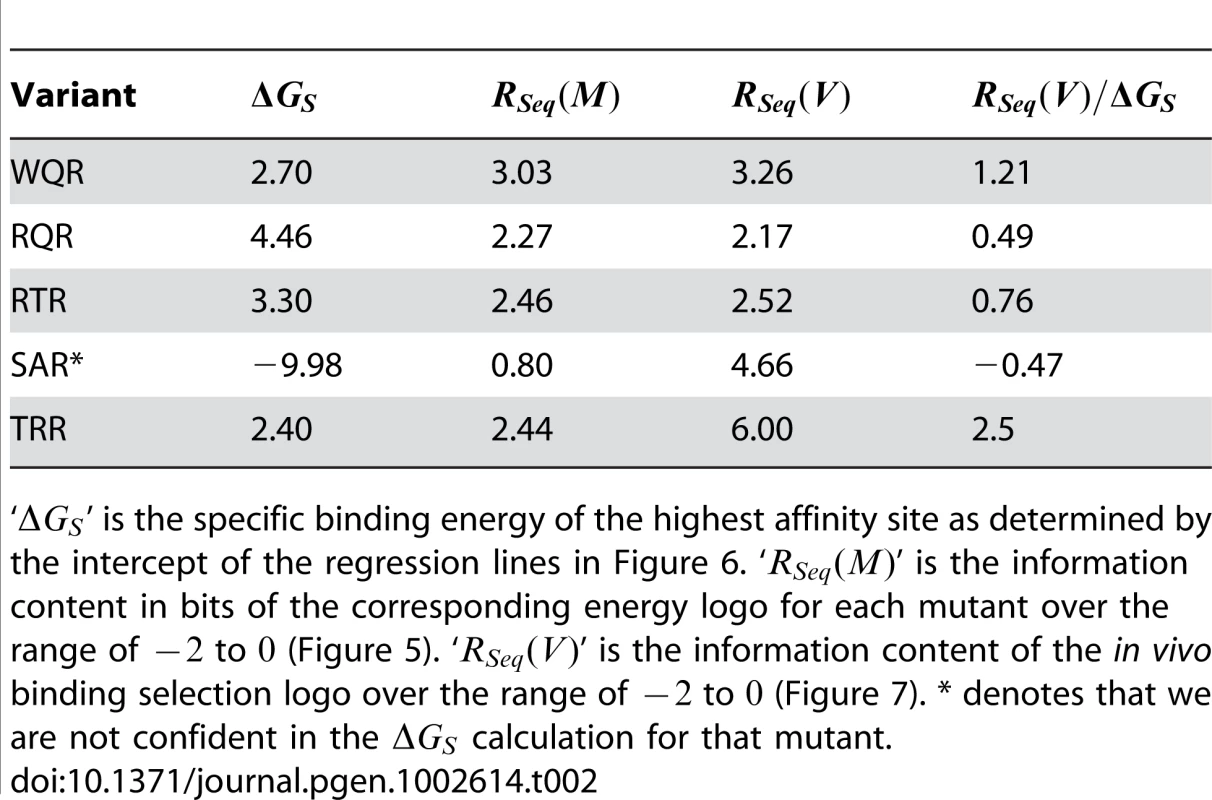
‘’ is the specific binding energy of the highest affinity site as determined by the intercept of the regression lines in Figure 6. ‘’ is the information content in bits of the corresponding energy logo for each mutant over the range of to (Figure 5). ‘’ is the information content of the in vivo binding selection logo over the range of to (Figure 7). * denotes that we are not confident in the calculation for that mutant. SAR is the least specific of the variants ( bits). It shows a preference for ‘A’ or ‘G’ at position , and almost no preference at positions and 0. It does not strongly bind ‘GAA’, the site it was selected against. Conversely, TRR appears to only bind its selected target site ‘GAC’ (Figure S1). While TRR is specific for this sequence, the relative difference in binding strength between ‘GAC’ and the non-specific background () is much less than observed for WQR, RQR and RTR (Figure S1). As the logos in Figure 5 are generated from the calculated differences in binding energy from the strongest bound site to all single base-pair mutants (see Materials and Methods), a low would result in a logo with a weak equiprobable conservation of all non-specifically bound bases at each position as observed for TRR.
Given the MITOMI data, we can test two assumptions that underlie most thermodynamic DNA binding models: (1) that the energetic contribution of each nucleotide at each position is independent of neighboring bases and (2) that this contribution is purely additive to the overall binding affinity [7], [44], [45]. Using Scan, an information theory based program that predicts binding affinities based on an independent and additive model, we calculated the predicted affinity for each protein mutant to all 64 sequences [44], and plotted this against the corresponding measured of binding (Figure 6, see Materials and Methods). Theoretically sites with an bits are predicted to be bound non-specifically, as [9], [44].
Fig. 6. An independent and additive thermodynamic binding model fits the MITOMI data with varying degrees of success. 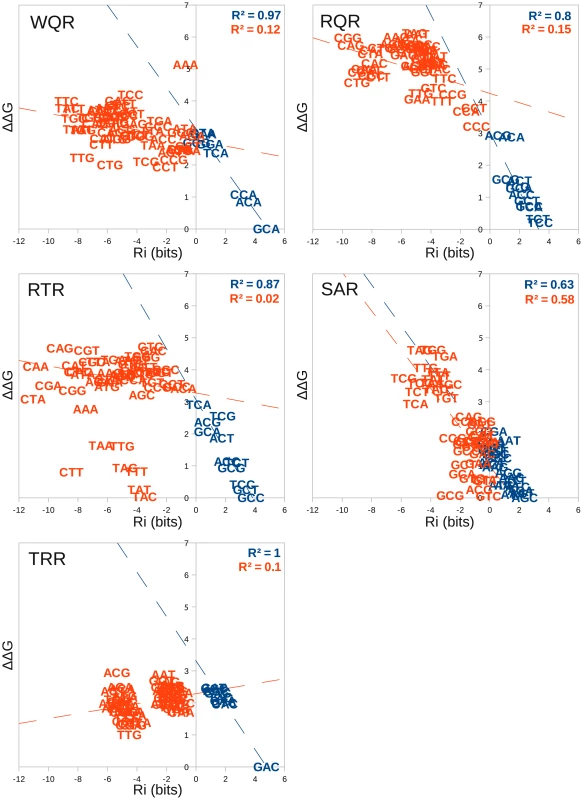
The relative binding affinity of each mutant to each binding site () was calculated using the models presented in Figure 5. This was plotted against the of binding for each sequence as determined by the difference in binding energy between that sequence and the highest affinity site (kJ/mol). A linear regression line was fit to sites with an bits (blue sequences) and sites bits (red sequences). The intercept of these lines were used to approximate the boundary between specifically and non-specifically bound sites and the corresponding values are reported in Table 2. values for each regression line are given in the upper right hand corner in the same color. For all mutants, except for SAR, predicted binding strength is highly correlated with actual binding for sites bits (blue sequences in Figure 6), and is poorly correlated for sites bits (red sequences in Figure 6). The experimental measurement of binding affinity for weakly bound sites has previously been shown to be less accurate than for strongly bound ones [9]. Because of this, we are not surprised by the weak correlation for the sites with an bits. If these sequences are truly bound non-specifically though, we would also expect the slope of the regression line to be 0. For WQR, RQR and RTR we observe a slightly negative slope (, and respectively), which suggests that to a small degree, binding energy does change as a function of sequence (bound specifically) for a fraction of these sites. This is evident for RQR, where sites bits lie close to the regression line for the positively bound sequences (Figure 6). We expect the specific/non-specific boundary to be closer to bits for this binding domain. Likewise, for TRR the non-specific boundary is probably at bits, but this deviation from 0 bits can be explained by the low , and subsequently biased model for TRR as previously mentioned.
To approximate the non-specific binding energy for each mutant, we determined the intercept of the positive and negative site regression lines (Table 2). SAR appears to be almost completely non-specific from the MITOMI data, and we are not confident in the identified boundary between specific and non-specific binding for this mutant.
Surprisingly, there appears to be a di-nucleotide binding preference for the RTR mutant (Figure 6). RTR binds ‘GC-C’‘GC-T’‘GC-G’‘GC-A’ and ‘TA-C’‘TA-T’‘TA-G’‘TA-A’ with almost equivalent energies between sites that have the same nucleotide at the third position ( = 0.99). A simple independent and additive model would predict that a single mutation of a ‘G’ to ‘T’ at position or a ‘C’ to ‘A’ at position would not affect the binding energy of the site. Indeed, ‘TC-C’‘TC-T’‘TC-G’‘TC-A’ and is highly correlated to the equivalent ‘GC-N’ and ‘TA-N’ sites ( = 0.84 and 0.91 respectively), but ‘GA-N’ sites are not correlated and all sites have a greater than the RTR non-specific binding threshold of 3.30 kJ/mol. This clearly violates a simple independence assumption.
In vivo binding site selection for MarA variants
To identify the in vivo binding preferences of the 5 MarA protein variants, we generated a library of selection plasmids for each mutant where positions , , , 0 and in the mar binding site were randomized (Figure 2). We transformed N8453 cells with these libraries and competed them against each other in 5 ml LB+50 g/ml tetracycline+ L-arabinose for 24 hours. The competed populations were mini-prepped and sequenced in a single sequencing reaction (Figure S2). Sequence logos were generated for all mutants as described in Materials and Methods (Figure 7). Higher affinity binding sites should be more fit and represented at a higher frequency in the competed population [19]. While the relative peak height for a given base at a given position within the chromatogram is correlated with the base frequency in the population, it can be biased by the identify of the neighboring bases. Therefore, this is a semi-quantitative representation of positional nucleotide frequency.
Fig. 7. In vivo binding site selection logos. 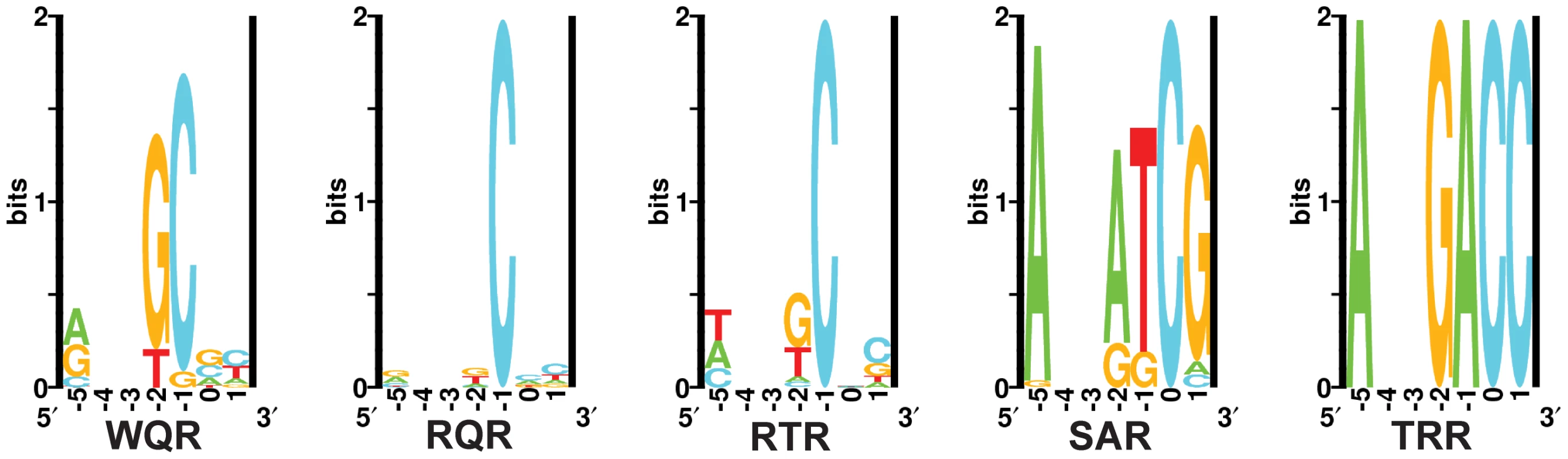
Sequence logos were generated from the chromatograms in Figure S2. Positions and were not randomized in the selection and therefore are left blank in the logos. In vivo binding preferences identified by this selection method are consistent with our MITOMI results. The wild type MarA protein (WQR) requires a ‘C’ at position and shows a strong preference for a ‘G’ at position . Unlike the MITOMI data, there is more variability at position in the selected sites, resulting in a large Kullback-Leibler Divergence between the corresponding WQR logos of 1.67, but a decrease in KLD between WQR and the RQR and RTR mutants (Table S1). The RQR in vivo selected sites have an increased variability at positions and relative to the MITOMI data, but overall the resulting logos are nearly identical (KLD = 0.15). Similar results are observed for RTR (KLD = 0.15), which only shows a slight decrease in degeneracy at position in the experimentally selected sites. Interestingly ‘A’ is not observed at position in the RTR in vivo sites, even though ‘TAA’ is tightly bound according to the MITOMI data.
The SAR mutant shows substantially less variability in the in vivo binding site selection as compared to the MITOMI data; the for positions to = 4.66 and 0.80 bits respectively. The concentration of tetracycline used for selection, imposes an energetic minimum that the factor must bind its site above to be viable [19]. This lack of variability in the SAR in vivo binding site selection suggests that unlike WQR, RQR and RTR, few SAR sites are above this threshold (i.e. weakly bound). SAR is the only mutant to show a strong preference for ‘G’ at position , while all other mutants preferred a cytosine there. Differences in the SAR binding preferences observed in vivo and in vitro may also be accounted for by the presence of a unfavorable ‘C’ at position in the MITOMI binding site library (Figure 1), which could significantly reduce the binding affinity of all sites. TRR binds to a single site, ‘GAC’, as expected.
Interestingly, we observed a wide range of degeneracy at position , which does not appear to be directly contacted by any of the varied residues. There is a preference for ‘A’ at this position for all mutants, and it is completely conserved for SAR and TRR. We expect that the amount of observed variation at is not dependent upon specific contacts at that base, but on the energetic contribution of the rest of the binding site. That is, weak binding at positions , and by residual differences requires a base with a higher affinity (‘A’) at position for the site to be sufficiently strong in this selection. This suggests that degeneracy at a single position in a site is not completely defined by the residue that contacts it, but by the energy of the other contacts in the site.
To quantify the extent of overlap in sites specifically bound by all mutants in vivo, we calculated the predicted binding strength () of each mutant to the 64 potential binding site variants at positions to , and directly compared these affinities (Figure 8). Since the RQR logo has the lowest information content, we compared all mutants to it. Sequences that fall in the upper right quadrants in Figure 8 are predicted to be specifically bound by the two mutants compared (positive for both). Sites in the lower left are predicted to not be bound by either. The remaining quadrants contain sites that are only bound by one mutant. As the RQR and RTR logos are significantly similar (KLD = 0.14), it is not surprising that their predicted affinities are highly correlated (). Only a few sequences specifically bound by RQR are not bound by RTR (lower right quadrant) and no unique sequences are bound by RTR (upper left quadrant) suggesting that RTR is merely binding a subset of the sites bound by RQR (Figure 8A). A similar result is observed for WQR, except that it binds a further reduced subset of the specifically bound RQR sites. There is no overlap in specifically bound sites by SAR and TRR with RQR, suggesting that these bind a completely orthogonal set of sequences (Figure 8B).
Fig. 8. Binding domain mutations can reduce binding targets or generate orthogonal regulators. 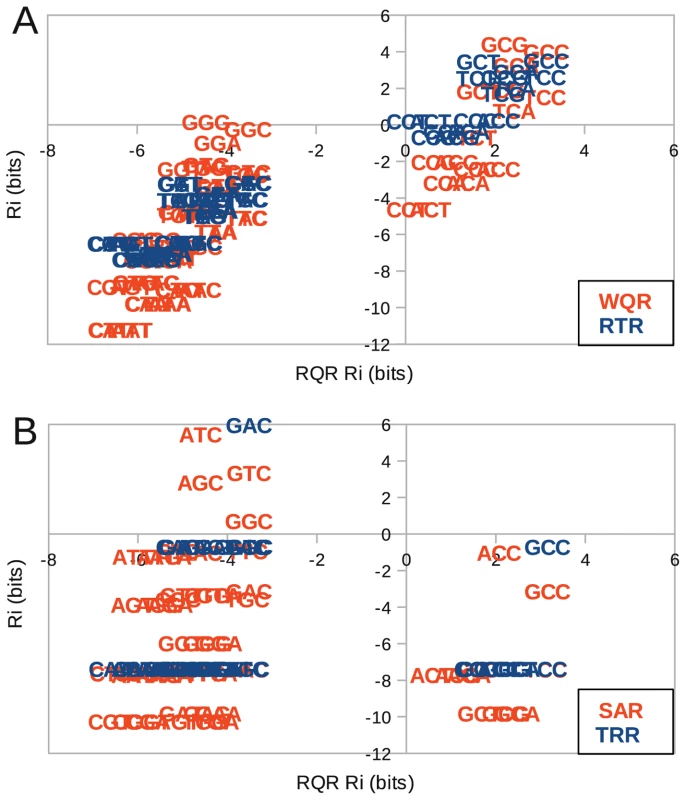
(A) Comparison of corresponding predicted binding strengths () between highly overlapping MarA variants WQR and RTR with RQR. (B) Similar comparison between orthogonal binders SAR and TRR with RQR. The of each mutant to each binding site was calculated using the logos presented in Figure 7 over the range −2 to 0. Transcriptional output
To better understand how mutations in the binding domain affect the transcriptional activity of MarA, we measured the expression of tet under the control of wild type MarA (WQR) with 11 different binding sites, and under the control of RQR with 15 different binding sites using quantitative PCR (Figure 9). We chose binding sites for each variant that covered a range of binding strengths based on the MITOMI data. For convenience, we normalized the output so that the relative expression of the ‘GCA’ binding site by WQR is 1.
Fig. 9. The RQR mutant accesses a much larger output space than wild-type MarA. 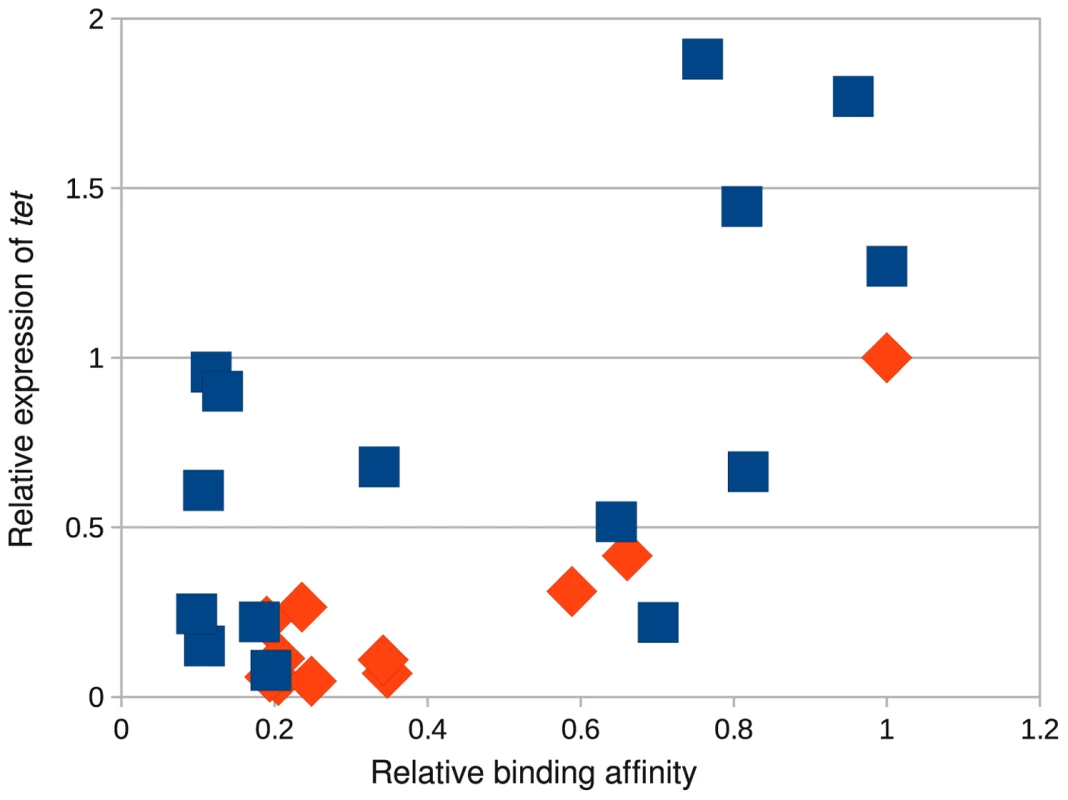
Relative affinity of a MarA variant for a given site as determined by MITOMI (Figure 4) vs. the quantity of tet gene expressed. Expression levels were monitored by Q-PCR. We show data for wild type WQR MarA (orange diamonds) and the RQR mutant (blue squares). The transcriptional output was normalized with wild type MarA bound to its consensus site of ‘GCA’ = 1. For the WQR binding sites, the expression data correlate well with binding site strength ( for all sites, for the 3 tightly bound sites). The non-specifically bound sites show minor variability in their measured output. The expression data for the RQR bound sites do correlate with binding affinity but not as well ( for all sites) and we observed much more variability in the non-specifically bound sites. The transcriptional output from the highest affinity RQR site is almost twice that of the strongest WQR site, suggesting that functionally this mutant can access a much larger dynamic range of outputs.
Discussion
It is becoming increasingly clear that differences in transcriptional regulation are an important driving force in species diversification and evolution [46], [47]. Fine scale differences in the expression level of an individual gene can be easily achieved by mutations in transcription factor binding sites contained within the associated cis-regulatory region [19]. Larger scale effects on the transcriptional network, and subsequently cellular phenotype, can be accessed through mutations in transcription factor binding domains which will impact the expression levels of all genes within their regulons [48]. As the systematic effects of transcription factor mutations are more difficult to characterize, few experimental studies have been done to probe their evolvability [5]. Since both the informational and regulatory properties of a transcription factor are determined by its binding site energy distribution [1], [20], we developed an in vivo selection assay to select for variants with altered binding preferences that still maintain a physiologically relevant transcriptional activity. Further in vivo and in vitro characterization of a subset of these mutants revealed that a large range of binding preferences, information contents and activities could be accessed with a few mutations suggesting that transcriptional regulatory networks may be easily adaptable.
One way in which regulatory networks are believed to evolve is through the duplication of an existing transcription factor gene that is subsequently selected to recognize a unique set of targets [49], [50]. It is unclear how readily this can happen. Maerkl and Quake observed that a relatively limited range of binding preferences could be accessed by single mutations in the basic helix-loop-helix protein MAX [5]. For MarA, we observed that we could get an orthogonal regulator with two mutations. The double mutant TRR is the most dramatic example. It is absolutely specific for ‘GAC’, which no other variant specifically bound (Figure S1, Figure 8B). Likewise SAR bound its own unique set of sites that do not overlap wild type (Figure 8B). Interestingly, both SAR and TRR have a lower for their highest affinity sites compared to mutants that bind the wild type consensus sequence. This suggests that a novel regulator may emerge or be engineered relatively easily, but may be initially limited in its range of potential activities.
Gene duplication may not be the only pathway by which orthogonal regulators can evolve. WQR, RQR and RTR appear to have largely overlapping binding sites, where RTR and RQR have an incrementally increasing number of specifically bound sites (Figure 8A). This suggests that a transcription factor could evolve to have an increased or decreased information content (become more or less specific), while still maintaining the majority of its binding targets. An orthogonal regulator could potentially evolve through an intermediate with broader specificity like RQR or RTR (Figure 10). A mutation of this type would impact the relative expression levels of the genes controlled by the transcription factor, as seen in Figure 9, and initially compromise the fitness of the cell [21], but would presumably have a significant advantage over a mutation that leads to the loss of potential targets. Further selection could re-specify the transcription factor after becoming promiscuous to regulate a new set of sequences. As this broadening of specificity can be done relatively easily (WQR can be converted to RQR by a single nucleotide mutation), this pathway may be highly tractable by evolution and useful for engineering regulatory networks. As previously mentioned, the information content of a transcription factor's binding sites is highly correlated to the amount of information needed to specifically locate its binding sites in the genome for bacterial systems [1]. This suggests that as the size of a bacterial factor's regulon increases or decreases, so does the selective pressure on binding site information. The decrease in information from WQR to RTR to RQR, also suggests that a transcription factor can easily evolve to expand or contract the size of its regulon.
Fig. 10. The respecification of an orthogonal regulator may occur through a despecified intermediate. 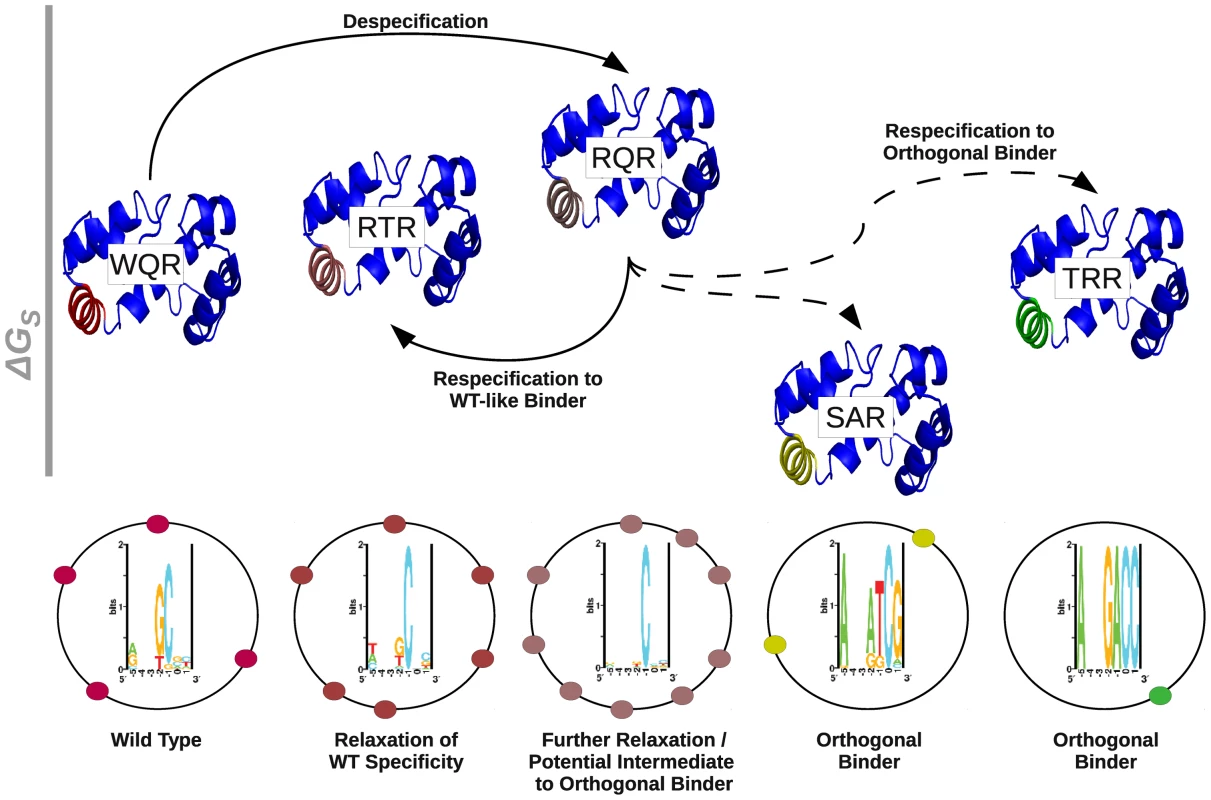
A schematic representation of the respecification of wild type MarA (WQR) to an orthogonal binder through a broadly specific intermediate. Each mutant is represented by the MarA protein structure [28]. Helix 3 in each structure is colored to highlight similarities in binding preferences between mutants. As WQR, RTR and RQR have largely overlapping binding sites, they have a similar coloration. The relative height of each protein structure is determined by the value reported in Table 2. As we are not confident in our estimate for the for SAR, we gave it a value of 0 kJ/mol. Solid lines between variants indicate single amino acid differences, dashed lines indicate double mutants. The sequence logo for each variant as determined by the in vivo binding site selection assay (Figure 7) are shown directly below that mutant. The black circles surrounding the logo represent the E. coli genome, and the colored circles represent hypothetical binding sites for each respective variant. The overlap in binding sites between WQR, RQR and RTR may not be surprising as all were selected to bind the wild type consensus sequence ‘GCA’. The dominant feature for these three mutants is a highly conserved ‘C’ at position (Figure 5, Figure 7). One possibility is that the for this base is increased from WQR to RTR to RQR, and a stronger individual contact here compensates for a greater number of energetically unfavorable mismatches at positions and , decreasing the information content (Table 2). Interestingly, expanding the number of specifically bound sites for RQR also expands the range of transcriptional outputs nearly two fold (Figure 9). If RQR has a much greater range of potential activities, and largely similar binding preferences to wild type MarA (WQR), why is it not observed in nature? WQR has a greater information content to ratio than both RQR and RTR (Table 2), suggesting that it encodes the fewest number of specifically bound sites for its range of binding energies (Table 2). It also appears to have a large energetic gap between its three highest affinity sites and the background, which the other variants lack (Figure 4). These properties of the wild type MarA binding site distribution, and not just overall affinity, may be evolutionarily advantageous and thus selected, as an increased for all sites would decrease the likelihood of the factor binding the wrong location [51], [52], and fewer recognized sites would decrease the probability of spurious sites emerging in the genome [53]. Directly assaying the global effects of these mutations by RNA profiling and chromatin immunoprecipitation would dramatically improve our understanding of their cellular implications.
Materials and Methods
MarA selection system and library construction
We modified the plasmid-based selection system described in [19] to select for and characterize MarA variants that have altered binding preferences (Figure 2). Griffith et al. generated an L-arabinose inducible MarA expression pBAD18 variant (pBAD18-hisMarA) [34]. We cloned the marA gene, the AraC regulated promoter and the araC gene from this plasmid into our pBR322-based selection system, allowing for us to control the expression of MarA by the addition of L-arabinose (Figure 2A). An XhoI site was introduced about 10 residues upstream of the start of helix 3 by modifying the ‘CTG’ codon encoding the leucine at residue 30 to the synonymous codon ‘CTC’ by QuickChange [54] (Figure 2). An AgeI site exists immediately downstream of helix 3. To make this a unique restriction site, we removed a second AgeI site present in a non-regulatory region upstream of the marA gene by QuickChange.
To generate variants of the MarA-activated tet promoter (Figure 2B), the selection plasmid was simultaneously digested with EcoRI and ClaI restriction enzymes for 2 hours at 37C (NEB). Inserted promoter variants and libraries were generated by DNA synthesis (Integrated DNA Technologies). We synthesized both strands of the DNA, and designed oligos to contain the appropriate overhang to be cloned into the EcoRI and ClaI sites. Digested plasmid and synthesized inserts were ligated overnight at 14C using T4 DNA ligase (NEB).
To generate binding domain variants (Figure 2C), we used a similar method. Plasmid was digested with XhoI and AgeI simultaneously for 2 hours at 37C (NEB). The digested plasmid was gel purified and ligated to complementary synthesized inserts that had XhoI and AgeI overhangs. To randomize the residues 42, 45 and 46, we synthesized the oligos with an equal mixture of all four bases at the first two positions of the codon, and an equal mixture of ‘G’ and ‘T’ at the third position of the codon to generate a more equal distribution of amino acids at each position. The ligated promoter and binding domain libraries were transformed into DH10B cells, recovered for 1 hour in LB, and plated on 100 ml LB+30 g/ml ampicillin plates. Cells were suspended from the plates in 10 ml LB and mini-prepped using the QIAquick miniprep kit (Qiagen).
MarA binding domain and binding site selections
To prevent activation of the tet gene by the endogenous MarA, Rob or SoxS proteins, selections were performed in the E. coli strain N8453 (mar, sox-8::cat, rob::kan variant of GC4468) prepared by J.L. Rosner and R.G. Martin and obtained from R.E. Wolf. To identify a binding site that was only functional when activated, we transformed a library with a variant of the promoter construct shown in Figure 2B that contain the mar MarA binding site (Figure 1) and a randomized hexamer. These plasmids also contain the wild type MarA protein. The library was transformed in N8453 cells by electroporation, recovered for 1 hour in 500 l LB at 37C, shaken at 225 rpm and plated on 5 g/ml tetracycline LB plates + L-arabinose. Individual colonies were picked and streaked on on 10, 15 and 20 g/ml tetracycline LB plates+/ − L-arabinose. Colonies that only grew on L-arabinose containing plates were sequenced.
To identify binding domain variants that specifically bound different DNA sequences, libraries were transformed into N8453 cells by electroporation, recovered for 1 hour in 500 l LB at 37C, shaken at 225 rpm and plated on 100 ml LB plates containing 30 g/ml ampicillin + L-arabinose. Colonies that grew on the plates overnight were suspended in 10 ml LB containing 30 g/ml ampicillin + L-arabinose and grown at 37C, shaken at 225 rpm for 8 hours. 70 l of these cells were then plated on 25 ml LB agar plates containing 20 or 30 g/ml of tetracycline + L-arabinose. Individual colonies were picked, grown overnight, miniprepped by the QIAquick miniprep kit and sequenced.
To identify the binding domain for each site that could produce the most tet transcript, libraries were transformed by electroporation into N8453 cells and plated on 5 g/ml tetracycline LB plates and grown overnight. These colonies were suspended in 5 ml LB with 5 g/ml tetracycline + L-arabinose and allowed to grow in liquid culture overnight. The following morning fresh 5 ml 30 g/ml tetracycline + L-arabinose cultures were inoculated with 100 l of the overnight culture and competed for 24 h. The competed library was miniprepped by a QIAquick miniprep kit, transformed into DH10B cells and plated on 30 g/ml ampicillin plates. Individual colonies were picked, grown up overnight, miniprepped and sequenced as described above.
Binding site competitions for the 5 MarA selected variants were performed as described previously [19], except that the libraries were transformed into N84533 cells and all media contained L-arabinose. Libraries were competed in 50 g/ml tetracycline for 24 hours and sequenced on a 96 capillary 3730xl DNA Analyzer (Applied Biosystems). Nucleotide variation in the population of competed promoters was visualized using Finch TV (Geospiza Inc). To generate sequence logos from these data (Figure 7), we measured the peak height of each base at each position in a chromatogram (Figure S2), and divided this height by the summed heights of all peaks at the position to calculate a relative nucleotide frequency. A standard position weight matrix was generate from these frequencies, and represented as a sequence logo using the Delila programs [39].
MITOMI data acquisition and analysis
MITOMI (Mechanically Induced Trapping of Molecular Interactions) was performed according to Maerkl et al. [8]. The 64 variants of the mar binding site (Figure 1) were synthesized by Integrated DNA Technologies. In vitro transcription and translation was done using the RTS E. coli HY kit (Roche). Fluorescently labeled lysines were incorporated into the protein during in vitro translation by addition of tRNA-lys-bodipy-fl (Promega). Protein and DNA fluorescence was measured using Genepix (Molecular Devices).
The of binding for each variant to each binding site was calculated using , where is the ideal gas constant, is the temperature of the experiment (295K) and is the association constant as measured by MITOMI. The of binding was calculated for each binding site by subtracting the of binding for that site from the of binding from the highest affinity site for a protein variant.
To generate the energy logos, we calculated a matrix for each variant by determining the difference in binding energy between the strongest bound site for that factor (the consensus site) and all single base-pair mutants. For example, to calculate the relative weights of each base at position for wild type MarA, we subtracted the measured binding energies of ‘ACA’, ‘CCA’, ‘GCA’ and ‘TCA’ from ‘GCA’. We used the enoLogos webserver to convert these energies into a log-likelihood matrix [42] and generated logos using the Delila programs [39]. The matrices for all logos are given in Table S1.
Comparison and applications of binding models
To quantify the similarity in binding preferences between MarA variants, we used the program MatCompare to calculate the Kullback-Leiber Divergence (KLD) between the inferred sequence logos [43]. All pair-wise KLD values are reported in Table S1. The relative affinity () of a given binding model to all DNA sequences was calculated using the information theory based program Scan [44].
Q–PCR
A library of mar binding sites was cloned into plasmids containing either the wild type MarA protein, or the RQR mutant. The library was transformed into N8453 cells, plated on 30 g/ml ampicillin and grown overnight. Individual colonies were grown overnight in 5 ml LB+30 g/ml ampicillin. Glycerol was added to 200 l of cells to a final concentration of 20% and stored at C. The remaining culture was mini-prepped and sequenced to determine which binding site was present. 11 different binding sites covering a range of affinities as determined by MITOMI were chosen for wild type MarA and 15 were chosen for the RQR mutant. These were not the same sites for both factors.
Cultures were inoculated with the frozen samples and grown overnight in 5 ml LB cultures with 30 g/ml ampicillin and L-arabinose. A fresh 5 ml LB+30 g/ml ampicillin+L-arabinose culture was started at and grown to an . cells were added to RNAprotect Bacteria reagent (Qiagen), and RNA was purified using the RNeasy Mini kit with on-column DNase digestion (Qiagen). cDNA was made from 2 g of RNA using the Superscript III RT kit (Invitrogen). QPCR was performed with the SYBR green mix from NEB. QPCR primers specific to the tet and marA gene were both used. The relative expression of the tet gene was determined by the ratio of tet abundance over marA abundance for each sample.
Supporting Information
Zdroje
1. SchneiderTDStormoGDGoldLEhrenfeuchtA 1986 Information content of binding sites on nucleotide sequences. J Mol Biol 188 415 431
2. ItzkovitzSTlustyTAlonU 2006 Coding limits on the number of transcription factors. BMC genomics 7 239
3. PaboCOPeisachEGrantRA 2001 Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem 70 313 340
4. BenosPVLapedesASStormoGD 2002 Probabilistic code for DNA recognition by proteins of the EGR family. J Mol Biol 323 701 727
5. MaerklSQuakeS 2009 Experimental determination of the evolvability of a transcription factor. Proceedings of the National Academy of Sciences 106 18650
6. PaboCONekludovaL 2000 Geometric analysis and comparison of protein-DNA interfaces: why is there no simple code for recognition? J Mol Biol 301 597 624
7. von HippelPHBergOG 1986 On the specificity of DNA-protein interactions. Proc Natl Acad Sci USA 83 1608 1612
8. MaerklSJQuakeSR 2007 A systems approach to measuring the binding energy landscapes of transcription factors. Science 315 233 237
9. ShultzabergerRKRobertsLRLyakhovIGSidorovIAStephenAG 2007 Correlation between binding rate constants and individual information of E. coli Fis binding sites. Nucleic Acids Res 35 5275 5283
10. ShannonCE 1948 A Mathematical Theory of Communication. Bell System Tech J 27 379 423 623–656
11. SchneiderTD 2000 Evolution of biological information. Nucleic Acids Res 28 2794 2799
12. SenguptaADjordjevicMShraimanB 2002 Specificity and robustness in transcription control networks. Proceedings of the National Academy of Sciences of the United States of America 99 2072
13. WunderlichZMirnyL 2009 Different gene regulation strategies revealed by analysis of binding motifs. Trends in genetics 25 434 440
14. ShultzabergerRKChiangDYMosesAMEisenMB 2007 Determining physical constraints in transcriptional initiation complexes using DNA sequence analysis. PLoS ONE 2 e1199 doi:10.1371/journal.pone.0001199
15. LemonBTjianR 2000 Orchestrated response: a symphony of transcription factors for gene control. Genes & development 14 2551 2569
16. McClureWR 1985 Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem 54 171 204
17. BintuLBuchlerNGarciaHGerlandUHwaT 2005 Transcriptional regulation by the numbers: models. Current opinion in genetics & development 15 116 124
18. ShultzabergerRKChenZLewisKASchneiderTD 2007 Anatomy of Escherichia coli σ70 promoters. Nucleic Acids Res 35 771 788
19. ShultzabergerRMalashockDKirschJEisenM 2010 The Fitness Landscapes of cis-Acting Binding Sites in Different Promoter and Environmental Contexts. PLoS Genet 6 e1001042 doi:10.1371/journal.pgen.1001042
20. MustonenVKinneyJCallanCLassigM 2008 Energy-dependent fitness: A quantitative model for the evolution of yeast transcription factor binding sites. Proceedings of the National Academy of Sciences 105 12376
21. DekelEAlonU 2005 Optimality and evolutionary tuning of the expression level of a protein. Nature 436 588 592
22. GerlandUHwaT 2002 On the selection and evolution of regulatory DNA motifs. Journal of molecular evolution 55 386 400
23. MartinRGRosnerJL 2001 The AraC transcriptional activators. Curr Opin Microbiol 4 132 137
24. MartinRGGilletteWKRheeSRosnerJL 1999 Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol Microbiol 34 431 441
25. MartinRGRosnerJL 2002 Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol Microbiol 44 1611 1624
26. SchneidersTBarbosaTMMcMurryLMLevySB 2004 The Escherichia coli transcriptional regulator MarA directly represses transcription of purA and hdeA. J Biol Chem 279 9037 9042
27. AlekshunMLevyS 1999 The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends in Microbiology 7 410 413
28. RheeSMartinRGRosnerJLDaviesDR 1998 A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci U S A 95 10413 10418
29. DangiBPelupesseyPMartinRGRosnerJLLouisJM 2001 Structure and dynamics of MarA-DNA complexes: an NMR investigation. J Mol Biol 314 113 127
30. MartinRRosnerJ 2002 Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Molecular Microbiology 44 1611 1624
31. SchneiderTD 1996 Reading of DNA sequence logos: Prediction of major groove binding by information theory. Meth Enzym 274 445 455
32. SchneiderTD 2001 Strong minor groove base conservation in sequence logos implies DNA distortion or base flipping during replication and transcription initiation. Nucleic Acids Res 29 4881 4891
33. GilletteWKMartinRGRosnerJL 2000 Probing the Escherichia coli transcriptional activator MarA using alanine-scanning mutagenesis: residues important for DNA binding and activation. J Mol Biol 299 1245 1255
34. GriffithKLWolfREJr 2004 Genetic evidence for pre-recruitment as the mechanism of transcription activation by SoxS of Escherichia coli : the dominance of DNA binding mutations of SoxS. J Mol Biol 344 1 10
35. McmurryLOethingerMLevyS 1998 Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiology Letters 166 305 309
36. OkusuHMaDNikaidoH 1996 AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. Journal of Bacteriology 178 306 308
37. AltschulSGishWMillerWMyersELipmanD 1990 Basic local alignment search tool. J mol Biol 215 403 410
38. LarkinMBlackshieldsGBrownNChennaRMcGettiganP 2007 Clustal W and Clustal X version 2.0. Bioinformatics 23 2947
39. SchneiderTDStephensRM 1990 Sequence logos: A new way to display consensus sequences. Nucleic Acids Res 18 6097 6100
40. SimonMDSatoKWeissGAShokatKM 2004 A phage display selection of engrailed homeodomain mutants and the importance of residue Q50. Nucleic Acids Res 32 3623 3631
41. EisenMSpellmanPBrownPBotsteinD 1998 Cluster analysis and display of genome-wide expression patterns
42. WorkmanCYinYCorcoranDIdekerTStormoG 2005 enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic acids research 33 W389
43. SchonesDSumazinPZhangM 2005 Similarity of position frequency matrices for transcription factor binding sites. Bioinformatics 21 307 313
44. SchneiderTD 1997 Information content of individual genetic sequences. J Theor Biol 189 427 441
45. BenosPVBulykMLStormoGD 2002 Additivity in protein-DNA interactions: how good an approximation is it? Nucleic Acids Res 30 4442 4451
46. KingMWilsonA 1975 Evolution at two levels in humans and chimpanzees. Science 188 107 116
47. CarrollS 2005 Evolution at two levels: on genes and form. PLoS Biol 3 e245 doi:10.1371/journal.pbio.0030245
48. AlperHMoxleyJNevoigtEFinkGStephanopoulosG 2006 Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314 1565
49. TeichmannSBabuM 2004 Gene regulatory network growth by duplication. Nature Genetics 36 492 496
50. Madan BabuMTeichmannS 2003 Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Research 31 1234
51. SlutskyMMirnyL 2004 Kinetics of protein-DNA interaction: facilitated target location in sequence-dependent potential. Biophysical journal 87 4021 4035
52. GerlandUMorozJHwaT 2002 Physical constraints and functional characteristics of transcription factor–DNA interaction. Proceedings of the National Academy of Sciences of the United States of America 99 12015
53. BergJWillmannSLässigM 2004 Adaptive evolution of transcription factor binding sites. BMC Evolutionary Biology 4 42
54. ZhengLBaumannUReymondJ 2004 An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic acids research 32 e115
Štítky
Genetika Reprodukční medicína
Článek Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1Článek Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / TranscriptionČlánek Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin ComplexesČlánek An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood ObesityČlánek Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAsČlánek Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 3
-
Všechny články tohoto čísla
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
- Genomic Analysis of the Hydrocarbon-Producing, Cellulolytic, Endophytic Fungus
- Networks of Neuronal Genes Affected by Common and Rare Variants in Autism Spectrum Disorders
- Akirin Links Twist-Regulated Transcription with the Brahma Chromatin Remodeling Complex during Embryogenesis
- Too Much Cleavage of Cyclin E Promotes Breast Tumorigenesis
- Imprinted Genes … and the Number Is?
- Genetic Architecture of Highly Complex Chemical Resistance Traits across Four Yeast Strains
- Exploring the Complexity of the HIV-1 Fitness Landscape
- MNS1 Is Essential for Spermiogenesis and Motile Ciliary Functions in Mice
- A Fundamental Regulatory Mechanism Operating through OmpR and DNA Topology Controls Expression of Pathogenicity Islands SPI-1 and SPI-2
- Evidence for Positive Selection on a Number of MicroRNA Regulatory Interactions during Recent Human Evolution
- Variation in Modifies Risk of Neonatal Intestinal Obstruction in Cystic Fibrosis
- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Critical Evaluation of Imprinted Gene Expression by RNA–Seq: A New Perspective
- A Meta-Analysis and Genome-Wide Association Study of Platelet Count and Mean Platelet Volume in African Americans
- Mouse Genetics Suggests Cell-Context Dependency for Myc-Regulated Metabolic Enzymes during Tumorigenesis
- Transcriptional Control in Cardiac Progenitors: Tbx1 Interacts with the BAF Chromatin Remodeling Complex and Regulates
- Synthetic Lethality of Cohesins with PARPs and Replication Fork Mediators
- APOBEC3G-Induced Hypermutation of Human Immunodeficiency Virus Type-1 Is Typically a Discrete “All or Nothing” Phenomenon
- Interpreting Meta-Analyses of Genome-Wide Association Studies
- Error-Prone ZW Pairing and No Evidence for Meiotic Sex Chromosome Inactivation in the Chicken Germ Line
- -Dependent Chemosensory Functions Contribute to Courtship Behavior in
- Diverse Forms of Splicing Are Part of an Evolving Autoregulatory Circuit
- Phenotypic Plasticity of the Drosophila Transcriptome
- Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1
- Precocious Metamorphosis in the Juvenile Hormone–Deficient Mutant of the Silkworm,
- Igf1r Signaling Is Indispensable for Preimplantation Development and Is Activated via a Novel Function of E-cadherin
- Accurate Prediction of Inducible Transcription Factor Binding Intensities In Vivo
- Mitochondrial Oxidative Stress Alters a Pathway in Strongly Resembling That of Bile Acid Biosynthesis and Secretion in Vertebrates
- Mammalian Neurogenesis Requires Treacle-Plk1 for Precise Control of Spindle Orientation, Mitotic Progression, and Maintenance of Neural Progenitor Cells
- Tcf7 Is an Important Regulator of the Switch of Self-Renewal and Differentiation in a Multipotential Hematopoietic Cell Line
- REST–Mediated Recruitment of Polycomb Repressor Complexes in Mammalian Cells
- Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / Transcription
- Age-Dependent Brain Gene Expression and Copy Number Anomalies in Autism Suggest Distinct Pathological Processes at Young Versus Mature Ages
- A Genome-Wide Association Study Identifies Variants Underlying the Shade Avoidance Response
- -by- Regulatory Divergence Causes the Asymmetric Lethal Effects of an Ancestral Hybrid Incompatibility Gene
- Genome-Wide Association and Functional Follow-Up Reveals New Loci for Kidney Function
- A Natural System of Chromosome Transfer in
- Cell Size and the Initiation of DNA Replication in Bacteria
- Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding Domain
- Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin Complexes
- Temporal Transcriptional Profiling of Somatic and Germ Cells Reveals Biased Lineage Priming of Sexual Fate in the Fetal Mouse Gonad
- Rapid Analysis of Genome Rearrangements by Multiplex Ligation–Dependent Probe Amplification
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- The Atypical Calpains: Evolutionary Analyses and Roles in Cellular Degeneration
- The Silkworm Coming of Age—Early
- Development of a Panel of Genome-Wide Ancestry Informative Markers to Study Admixture Throughout the Americas
- Balanced Codon Usage Optimizes Eukaryotic Translational Efficiency
- The Min System and Nucleoid Occlusion Are Not Required for Identifying the Division Site in but Ensure Its Efficient Utilization
- Neurobeachin, a Regulator of Synaptic Protein Targeting, Is Associated with Body Fat Mass and Feeding Behavior in Mice and Body-Mass Index in Humans
- Statistical Analysis of Readthrough Levels for Nonsense Mutations in Mammalian Cells Reveals a Major Determinant of Response to Gentamicin
- Gene Reactivation by 5-Aza-2′-Deoxycytidine–Induced Demethylation Requires SRCAP–Mediated H2A.Z Insertion to Establish Nucleosome Depleted Regions
- The miR-35-41 Family of MicroRNAs Regulates RNAi Sensitivity in
- Genetic Basis of Hidden Phenotypic Variation Revealed by Increased Translational Readthrough in Yeast
- An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood Obesity
- Modelling Human Regulatory Variation in Mouse: Finding the Function in Genome-Wide Association Studies and Whole-Genome Sequencing
- Novel Loci for Adiponectin Levels and Their Influence on Type 2 Diabetes and Metabolic Traits: A Multi-Ethnic Meta-Analysis of 45,891 Individuals
- Polycomb-Like 3 Promotes Polycomb Repressive Complex 2 Binding to CpG Islands and Embryonic Stem Cell Self-Renewal
- Insulin/IGF-1 and Hypoxia Signaling Act in Concert to Regulate Iron Homeostasis in
- EMF1 and PRC2 Cooperate to Repress Key Regulators of Arabidopsis Development
- Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAs
- Contrasted Patterns of Molecular Evolution in Dominant and Recessive Self-Incompatibility Haplotypes in
- A Machine Learning Approach for Identifying Novel Cell Type–Specific Transcriptional Regulators of Myogenesis
- Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
- Nos2 Inactivation Promotes the Development of Medulloblastoma in Mice by Deregulation of Gap43–Dependent Granule Cell Precursor Migration
- Intracranial Aneurysm Risk Locus 5q23.2 Is Associated with Elevated Systolic Blood Pressure
- Heritability and Genetic Correlations Explained by Common SNPs for Metabolic Syndrome Traits
- A Genome-Wide Association Study of Nephrolithiasis in the Japanese Population Identifies Novel Susceptible Loci at 5q35.3, 7p14.3, and 13q14.1
- DNA Damage in Nijmegen Breakage Syndrome Cells Leads to PARP Hyperactivation and Increased Oxidative Stress
- DNA Resection at Chromosome Breaks Promotes Genome Stability by Constraining Non-Allelic Homologous Recombination
- Genetic Analysis of Floral Symmetry in Van Gogh's Sunflowers Reveals Independent Recruitment of Genes in the Asteraceae
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Promoter Nucleosome Organization Shapes the Evolution of Gene Expression
- The Nucleoside Diphosphate Kinase Gene Acts as Quantitative Trait Locus Promoting Non-Mendelian Inheritance
- The Ciliogenic Transcription Factor RFX3 Regulates Early Midline Distribution of Guidepost Neurons Required for Corpus Callosum Development
- Phosphorylation of the RNA–Binding Protein HOW by MAPK/ERK Enhances Its Dimerization and Activity
- A Genome-Wide Scan of Ashkenazi Jewish Crohn's Disease Suggests Novel Susceptibility Loci
- Parkinson's Disease–Associated Kinase PINK1 Regulates Miro Protein Level and Axonal Transport of Mitochondria
- LMW-E/CDK2 Deregulates Acinar Morphogenesis, Induces Tumorigenesis, and Associates with the Activated b-Raf-ERK1/2-mTOR Pathway in Breast Cancer Patients
- Mapping the Hsp90 Genetic Interaction Network in Reveals Environmental Contingency and Rewired Circuitry
- Autoregulation of the Noncoding RNA Gene
- The Human Pancreatic Islet Transcriptome: Expression of Candidate Genes for Type 1 Diabetes and the Impact of Pro-Inflammatory Cytokines
- Spo0A∼P Imposes a Temporal Gate for the Bimodal Expression of Competence in
- Antagonistic Regulation of Apoptosis and Differentiation by the Cut Transcription Factor Represents a Tumor-Suppressing Mechanism in
- A Downstream CpG Island Controls Transcript Initiation and Elongation and the Methylation State of the Imprinted Macro ncRNA Promoter
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



