-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaComprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
More than 800 published genetic association studies have implicated dozens of potential risk loci in Parkinson's disease (PD). To facilitate the interpretation of these findings, we have created a dedicated online resource, PDGene, that comprehensively collects and meta-analyzes all published studies in the field. A systematic literature screen of ∼27,000 articles yielded 828 eligible articles from which relevant data were extracted. In addition, individual-level data from three publicly available genome-wide association studies (GWAS) were obtained and subjected to genotype imputation and analysis. Overall, we performed meta-analyses on more than seven million polymorphisms originating either from GWAS datasets and/or from smaller scale PD association studies. Meta-analyses on 147 SNPs were supplemented by unpublished GWAS data from up to 16,452 PD cases and 48,810 controls. Eleven loci showed genome-wide significant (P<5×10−8) association with disease risk: BST1, CCDC62/HIP1R, DGKQ/GAK, GBA, LRRK2, MAPT, MCCC1/LAMP3, PARK16, SNCA, STK39, and SYT11/RAB25. In addition, we identified novel evidence for genome-wide significant association with a polymorphism in ITGA8 (rs7077361, OR 0.88, P = 1.3×10−8). All meta-analysis results are freely available on a dedicated online database (www.pdgene.org), which is cross-linked with a customized track on the UCSC Genome Browser. Our study provides an exhaustive and up-to-date summary of the status of PD genetics research that can be readily scaled to include the results of future large-scale genetics projects, including next-generation sequencing studies.
Published in the journal: . PLoS Genet 8(3): e32767. doi:10.1371/journal.pgen.1002548
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002548Summary
More than 800 published genetic association studies have implicated dozens of potential risk loci in Parkinson's disease (PD). To facilitate the interpretation of these findings, we have created a dedicated online resource, PDGene, that comprehensively collects and meta-analyzes all published studies in the field. A systematic literature screen of ∼27,000 articles yielded 828 eligible articles from which relevant data were extracted. In addition, individual-level data from three publicly available genome-wide association studies (GWAS) were obtained and subjected to genotype imputation and analysis. Overall, we performed meta-analyses on more than seven million polymorphisms originating either from GWAS datasets and/or from smaller scale PD association studies. Meta-analyses on 147 SNPs were supplemented by unpublished GWAS data from up to 16,452 PD cases and 48,810 controls. Eleven loci showed genome-wide significant (P<5×10−8) association with disease risk: BST1, CCDC62/HIP1R, DGKQ/GAK, GBA, LRRK2, MAPT, MCCC1/LAMP3, PARK16, SNCA, STK39, and SYT11/RAB25. In addition, we identified novel evidence for genome-wide significant association with a polymorphism in ITGA8 (rs7077361, OR 0.88, P = 1.3×10−8). All meta-analysis results are freely available on a dedicated online database (www.pdgene.org), which is cross-linked with a customized track on the UCSC Genome Browser. Our study provides an exhaustive and up-to-date summary of the status of PD genetics research that can be readily scaled to include the results of future large-scale genetics projects, including next-generation sequencing studies.
Introduction
Parkinson's disease (PD) is the second most common neurodegenerative disease with a prevalence of ∼1% over 60 years of age [1]. Approximately 5–10% of the patients show an autosomal dominant or recessive mode of inheritance, and several causative genes have been identified, e.g. SNCA, LRRK2, PARK2, and PINK1 (for review see ref. [2]). Recently, two other novel autosomal dominant PD genes, VPS35 and EIF4G1 [3]–[5], have been identified, the former via application of next-generation sequencing techniques. It can be anticipated that causal mutations in additional genes will emerge within the next years. However, the vast majority of patients suffer from non-Mendelian forms of PD, which are likely caused by the combined effects of genetic and environmental factors. In order to decipher the genetic architecture underlying PD susceptibility, more than 800 genetic association studies have been performed over the past 20 years. While early candidate gene studies and subsequent meta-analyses provided conclusive evidence showing that polymorphisms in SNCA [6] (encoding alpha-synuclein), LRRK2 [7] (leucine-rich repeat kinase 2), MAPT [8] (microtubule-associated protein tau), and GBA [9] (acid beta-glucosidase) significantly impact PD susceptibility, most association studies in the field provided inconclusive or even conflicting results.
During the last few years, genome-wide association studies (GWAS) [10]–[19] have postulated additional PD loci. While the early GWAS and a GWAS-meta-analysis [20] were of limited sample sizes and yielded mostly inconsistent results, more recent studies have identified a number of loci that were independently confirmed in follow-up studies (e.g. GAK, BST1, and PARK16, see Table 1 for all proposed GWAS findings across GWAS publications). Very recently, a GWAS meta-analysis [21] implicated several other new putative PD loci which currently await further validation. Despite this progress, approximately 40% or more of the population-attributable risk probably remains unexplained by today's most promising PD loci [21]. To this end, genetic association studies remain one of the mainstays of PD genetics research. However, GWAS and other large-scale association studies typically only highlight the most promising results and often do not provide data on variants showing suggestive evidence for association, or previously implied variants that could not be confirmed in the GWAS setting. As a result, the cumulative genetic evidence in favor of or against association with certain variants in the PD field is becoming increasingly difficult to follow, evaluate and interpret. To address this problem, we have comprehensively collected, catalogued and systematically meta-analyzed the data from all genetic association studies published in the field of non-Mendelian PD, including GWAS, and made all results publicly available on a regularly updated online database, “PDGene” (http://www.pdgene.org).
Tab. 1. Overview of genome-wide association studies (GWAS) published in PD until March 31, 2011. 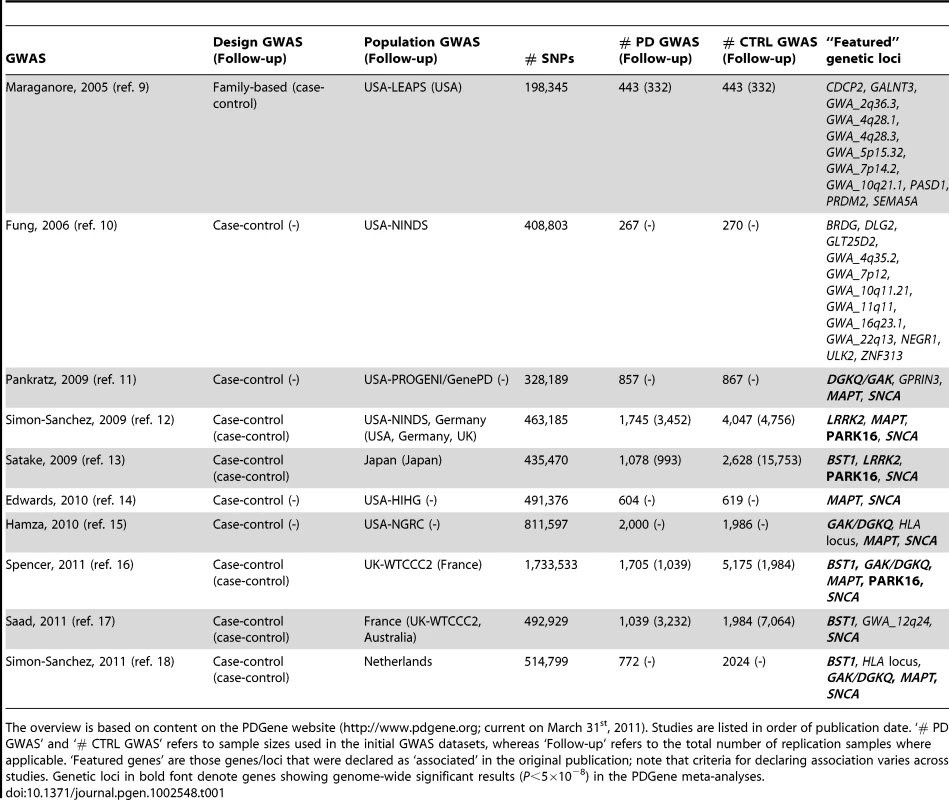
The overview is based on content on the PDGene website (http://www.pdgene.org; current on March 31st, 2011). Studies are listed in order of publication date. ‘# PD GWAS’ and ‘# CTRL GWAS’ refers to sample sizes used in the initial GWAS datasets, whereas ‘Follow-up’ refers to the total number of replication samples where applicable. ‘Featured genes’ are those genes/loci that were declared as ‘associated’ in the original publication; note that criteria for declaring association varies across studies. Genetic loci in bold font denote genes showing genome-wide significant results (P<5×10−8) in the PDGene meta-analyses. Results
Database content
The results of this research synopsis are based on a freeze of the PDGene database content on March 31st 2011 (available upon request from the authors). At that time, PDGene included details on 828 individual studies across more than 50 different countries and six continents reporting on 3,382 polymorphisms in 890 genetic loci. Data for more than 2,000 SNPs were supplemented by results derived from up to three publicly available GWAS datasets [10], [12], [13] following extensive quality control and imputation. Ultimately, this procedure yielded a total of 867 polymorphisms across ∼300 genetic loci that met our criteria for meta-analysis (see Methods). Additional independent GWAS data for 147 SNPs yielding P values of ≤0.1 in these initial meta-analyses were provided by researchers of all remaining currently published Caucasian GWAS datasets [13], [15]–[19], [22]. Following the identification of genome-wide significant association with an intronic SNP (rs7077361) in ITGA8 after addition of these data, we obtained additional data from the same GWAS datasets on ∼1,400 SNPs in the chromosomal region encompassing ITGA8 (chr10 : 15346353–15801533, hg18). Finally, independent replication data in Caucasian and Asian populations from the GEO-PD consortium [23] generated for ten recently described PD loci [21] were made available for inclusion. As a result, we were able to substantially increase the sample size (up to 16,452 PD cases and 48,810 controls) for a large number of some of the most promising PD loci. For instance, we were able to add data from up to 48,861 previously not analyzed combined cases and controls to meta-analyses of some of the recently proposed PD loci [21] (median sample size 14,896, see Table 2 and Table S1 for details). In addition to these focused analyses, PDGene displays meta-analysis results for more than seven million additional SNPs originating from up to three publicly available GWAS datasets [10], [12], [13]. The results are available online (e.g. as summarized in http://www.pdgene.org/largescalemeta.asp), where they are cross-linked to a customized and fully browsable track on the UCSC Genome Browser.
Tab. 2. Genome-wide significant summary meta-analysis results of the PDGene database in populations of Caucasian and Asian decent. 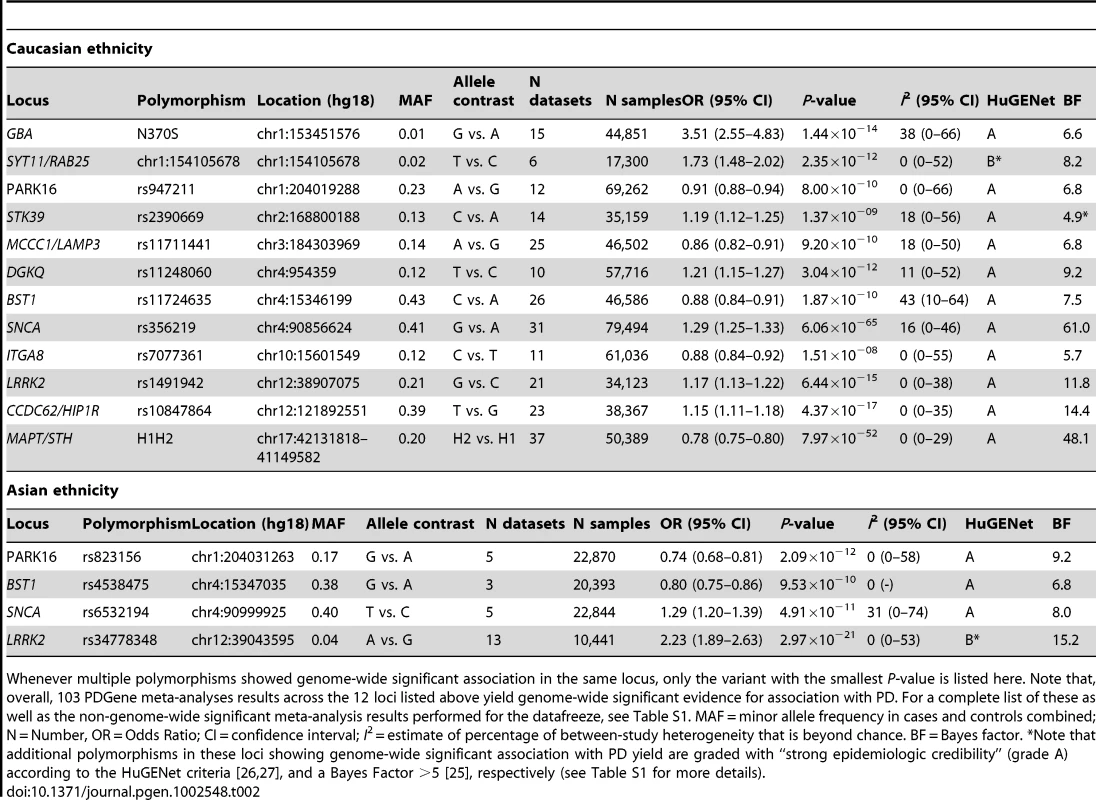
Whenever multiple polymorphisms showed genome-wide significant association in the same locus, only the variant with the smallest P-value is listed here. Note that, overall, 103 PDGene meta-analyses results across the 12 loci listed above yield genome-wide significant evidence for association with PD. For a complete list of these as well as the non-genome-wide significant meta-analysis results performed for the datafreeze, see Table S1. MAF = minor allele frequency in cases and controls combined; N = Number, OR = Odds Ratio; CI = confidence interval; I2 = estimate of percentage of between-study heterogeneity that is beyond chance. BF = Bayes factor. *Note that additional polymorphisms in these loci showing genome-wide significant association with PD yield are graded with “strong epidemiologic credibility” (grade A) according to the HuGENet criteria [26], [27], and a Bayes Factor >5 [25], respectively (see Table S1 for more details). PDGene meta-analysis results
The PDGene meta-analyses of the 867 core polymorphisms were based on a median of 7,680 subjects (interquartile range 4,612–16,726). Additional meta-analyses were performed after stratification for Caucasian and Asian ancestry (for details on sample size and included ethnicities for individual meta-analyses see Table S1). In addition, we also performed random-effects meta-analyses across all three publicly available GWAS datasets [10], [12], [13] following genotype imputation using data from the International HapMap Consortium and 1000 Genomes Project. Ultimately this yielded 7,123,920 SNPs that could be meta-analyzed across at least two GWAS datasets (see Figure S1 for a quantile-to-quantile plot of the GWAS-only meta-analyses). All 867 core meta-analysis results are available online on PDGene as forest plots, summarizing the relative contributions of each dataset to the most current summary effect estimate, and in the form of cumulative plots, illustrating how summary ORs evolve over time. All meta-analysis results are plotted in Figure 1 (green dots) alongside the GWAS-only meta-analysis results (black and grey dots).
Fig. 1. Manhattan plot of all meta-analysis results performed in PDGene. 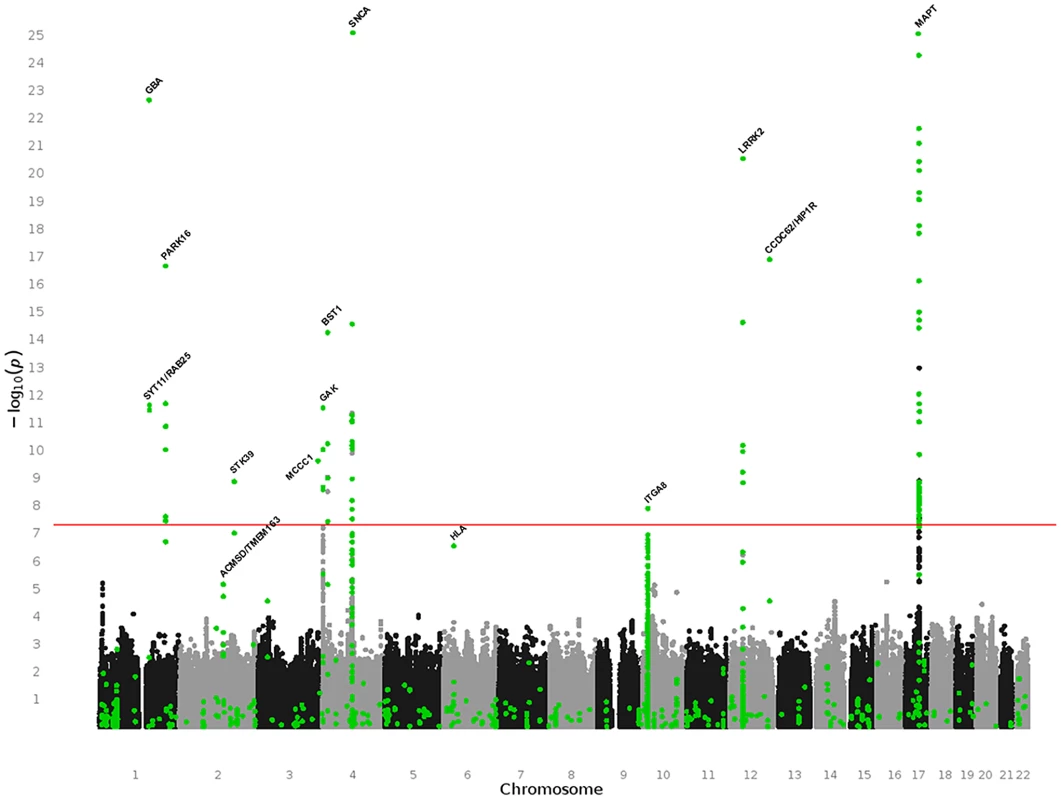
This summary combines association results from 7,123,986 random-effects meta-analyses based on the March 31st 2011 datafreeze of the PDGene database. Results are plotted as −log10 P-values (y-axis) against physical chromosomal location (x-axis). Black and grey dots indicate results originating exclusively from the three fully publicly available GWAS datasets [10], [12], [13] (see Methods), while green dots are based on a combination of smaller scale studies, supplemented by GWAS datasets (where applicable). Gene annotations are provided for genes highlighted in the main text. One-hundred-three meta-analyses across 12 genetic loci (BST1, CCDC62/HIP1R, DGKQ/GAK, GBA, ITGA8, LRRK2, MAPT, MCCC1/LAMP3, PARK16, SNCA, STK39, SYT11/RAB25) yielded summary ORs suggesting a genome-wide significant (P≤5×10−8) increase or decrease in PD risk in all ethnicities and/or after stratification for ethnic ancestry (Table 2, Table S1, and Figure S2 [forest plots]). None of these loci contained more than one SNP independently associated at genome-wide significance (as judged by pair-wise linkage disequilibrium assessments using ‘SNAP’ and r2-values of 0.2 as cut off http://www.broadinstitute.org/mpg/snap/). The majority of polymorphisms tested in the genome-wide significant loci do not show evidence for publication bias (Table S1). Finally, all genome-wide significant signals were robust against potential undetected sample overlap using a recently proposed procedure [24] (see Table S2 for more details). Combined sample sizes for all 12 loci were substantially larger here as compared to any previously published meta-analysis (Table S1), providing unequivocal evidence for an involvement of these loci in PD susceptibility. While power to detect genome-wide significance was excellent for most of these loci (>80% based on an OR of 1.15, and a minor allele frequency down to 0.05 using the Genetic Power Calculator, http://pngu.mgh.harvard.edu/~purcell/gpc/), power was less for a large number of other meta-analyses due to smaller sample sizes and allele frequencies (see Table S1 for details). Thus, no simple statistic can summarize the overall power of our study.
The above list includes an intronic polymorphism in ITGA8 located on chromosome 10p13 for which we identified novel evidence for genome-wide association with PD risk (OR 0.88, P = 1.3×10−8, I2 = 0, see Table 2, and Figure 2). This SNP had previously been proposed to be associated with PD risk at sub-genome-wide significance by Simon-Sanchez et al [13]. After obtaining and meta-analyzing GWAS data from ∼1,400 additional SNPs in this region derived from all Caucasians GWAS datasets [10], [12], [13], [15]–[19], [21], [22], rs7077361 remained the most significantly associated SNP in this region (Figure S3).
Fig. 2. Forest plot of the meta-analysis of rs7077361 in ITGA8. 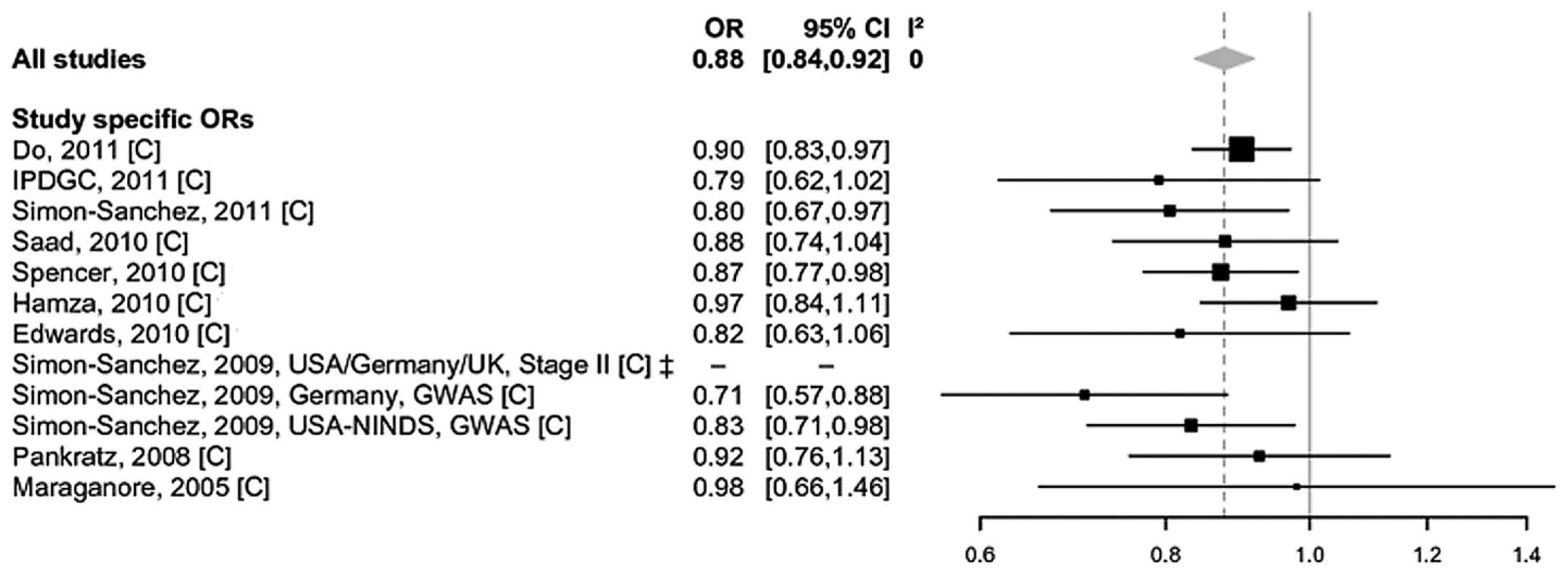
Study-specific allelic odds ratios (ORs, black squares) and 95% confidence intervals (CIs, lines) were calculated for each included dataset. The summary OR and CI was calculated using the DerSimonian Laird random-effects model (grey diamond) [31]. C = Caucasian ancestry. In addition to using random-effects models, we also performed exploratory fixed-effect meta-analyses on all eligible polymorphisms. These analyses did not reveal genome-wide significant effect sizes for any additional locus, except ACMSD/TMEM163 (most significant SNP rs6723108, OR 0.91, P = 1.3×10−9, I2 = 46% [95% CI 0–73%], Figure S4, panel 1) and HLA (most significant SNP chr6 : 32609909, OR 0.78, P = 8.8×10−15, I2 = 84% [95% CI 70–91%], Figure S4, panel 2), both of which were reported to be associated with PD risk at genome-wide significance in previous work [16], [21]. In both instances, the lack of genome-wide significance in the random-effects models (Table S1) was due to relatively pronounced heterogeneity of effect estimates across studies. However, the heterogeneity across the 11 datasets in the ACMSD/TMEM163 meta-analysis is almost entirely due to variance of effect size estimates in the same direction (see Figure S4, panel 1), making it likely that ACMSD/TMEM163 represents a genuine PD risk locus. For the SNP tested in the HLA locus (chr6 : 32609909, Figure S4, panel 2), heterogeneity is more pronounced and more complex owing to ORs on either side of 1. This could be due to a number of reasons, e.g. subtle and uncorrected population substructure and/or different LD patterns between the analyzed SNP and the actual functional variant(s) [16]. Thus, although the evidence is currently not as conclusive as for ACMSD/TMEM163 it still appears quite possible that there is one or more PD association signals in the HLA region. Regardless of these considerations, additional data are needed to more firmly assess the role of both loci in contributing to PD susceptibility.
Ethnicity-specific meta-analysis results
SNCA, LRRK2, BST1, and PARK16 show evidence for genome-wide significance in meta-analyses restricted to Caucasian and Asian populations (Table 2). Furthermore, data obtained from the GEO-PD consortium [23] suggest that the effect estimates for some of the recently discovered PD loci (i.e. CCDC62/HIP1R, MCC1, and STK39) [21] may be comparable in Caucasian and Asian populations (Table S1), although additional datasets are needed to establish genome-wide significance in populations of Asian-descent for these loci. Conversely, only insufficient data are currently available to assess the effect sizes of GAK and SYT11/RAB25 on PD risk in Asians: GAK rs6599388 violated Hardy-Weinberg equilibrium in Asian datasets from the GEO-PD consortium and was thus excluded from further analyses on that ethnic group [23]. SYT11/RAB25 chr1 : 154105678 was excluded from all analyses due to technical reasons in the study by the GEO-PD consortium [23]. Moreover, none of the reported SYT11/RAB25 and GAK SNPs from the recent GWAS meta-analysis [21] were captured directly or by proxy (with an r2≥0.8) in the Japanese GWAS dataset [14], [23]. Finally, Asian-descent populations cannot be appropriately assessed for PD association with the MAPT-H1/H2 haplotype, rs10928513 in ACMSD, and rs7077361 in ITGA8 owing to monomorphicity at these sites [14], [23].
Evaluating the credibility of significant associations
To estimate the epidemiologic credibility of associations with polymorphisms showing sub-genome-wide significant association with PD (P>5×10−8), we applied two “credibility” measures for each such result. First, we calculated Bayes factors (BF, expressed here as log10-values, “logBF”) assuming an average non-null odds ratio of 1.15, as approximation of a typical “complex disease effect size”, and a spike and smear prior distribution of effects [25]. Our second assessment was based on the Human Genome Epidemiology Network's (HuGENet) interim criteria for the assessment of cumulative epidemiologic evidence in genetic association studies [26], [27]. The results of these analyses are summarized in Table S1.
There was strong epidemiologic support in both assessments for all loci showing genome-wide significant association. This included several additional polymorphisms in these same loci that only showed sub-genome-wide significant association. However, there was no additional sub-genome-wide significantly associated locus that received unequivocally strong support from both credibility assessments (Table S1). In this list, the strongest support was assigned to SNP chr6 : 32588205 in the HLA locus receiving the best possible grade in the HuGENet criteria (grade A), but more moderate support in the Bayesian analyses (logBF = 4.4). However, the relevance of this assessment needs to be evaluated as the underlying analysis was only based on four GWAS datasets.
Discussion
The PDGene database represents a comprehensive, regularly updated and freely available online research synopsis of genetic association studies in PD. Detailed summaries of the most compelling findings are provided within an easy-to-use, dedicated online framework, displaying forest plots, cumulative meta-analyses, and an up-to-date ranking of “Top Results”. To allow comparison of PDGene results with association findings from other complex diseases and to facilitate their interpretation with respect to functional genetics data, all meta-analysis results have been ported as a customized track onto the UCSC Genome Browser. This will also allow for a integration and visualization [28] of association results from large-scale resequencing data (e.g. from whole-exome or whole-genome studies) into PDGene once these become available.
To the best of our knowledge, our study represents the most comprehensive research synopsis in the field of PD genetics. In addition, it represents the first disease-specific genetic database that allows a systematic and exhaustive inclusion of GWAS data, and may serve as a model for similar databases in other complex genetic diseases. Owing to our multi-pronged data retrieval and analysis protocol we were able to perform meta-analyses on the vast majority of PD risk-gene candidates, including those “featured” as top association results in all published GWAS. In particular, this includes the five novel loci recently featured in the recent GWAS meta-analysis [21]. Through collaboration with other PD genetics laboratories we obtained independent summary data for these and 142 additional SNPs, substantially extending the hitherto available evidence. Taken together, our analyses provide unequivocal evidence that BST1, CCDC62/HIP1R, DGKQ/GAK, GBA, ITGA8, LRRK2, MAPT, MCCC1/LAMP3, PARK16, SNCA, STK39, SYT11/RAB25 represent genuine PD risk loci, while the role of several other loci (e.g. ACMSD/TMEM163, and the HLA locus) remains to be determined. The unpublished data aggregated here from various PD genetics groups for selected candidate genes represents the first step towards a systematic meta-analysis across the full GWAS datasets from the same populations. Once completed, the results of this “mega” meta-analysis will be posted on the PDGene database, allowing users to browse the complete results via the customized genome browser track already in place.
Of particular interest are loci with unusually large effect sizes. While most loci in PDGene have only small effects on PD risk (with ORs ranging from 1.10 to 1.35, which are typical for complex diseases), for some loci much larger ORs were estimated (i.e. GBA [OR 3.51 in Caucasians], LRRK2 [OR 2.23 in Asians], and SYT11/RAB25 [OR 1.73 in Caucasians], see Table 2). The risk-allele frequencies at these polymorphisms are typically rather small (i.e. below 0.05), resulting in low population attributable risks for these loci (for the above mentioned loci individually less than 2%).
Interestingly, the meta-analysis results of GBA N370S as well as the LRRK2 rs34778348 are solely based on candidate-gene approaches since these SNPs are not on any of the current GWAS arrays or imputation reference panels. Thus, even in the “GWAS era” smaller-scale, non-GWAS but “focused” genetic studies, will likely continue to play an important role. This is also true when it comes to providing independent replication of proposed disease associations and/or when validating imputation-derived results by direct genotyping in sufficiently sized datasets. PDGene systematically concatenates all these different types of data into one database framework, vastly facilitating an assessment of the overall evidence for any given SNP or locus.
The strength of our approach is further exemplified by the identification of genome-wide significant association between disease risk and a SNP in ITGA8, which was not featured as a relevant PD gene in any previous study. ITGA8 (encoding integrin alpha 8, a type-I transmembrane protein) is functionally interesting as it is expressed in brain [29], mediates cell-cell interactions and regulates neurite outgrowth of sensory and motor neurons [30]. Additional studies are needed to further assess the potential role of this gene in PD pathogenesis. Furthermore, PDGene shows that two additional loci, not highlighted by the recent GWAS meta-analysis [21], yield genome-wide signficiant results in the PDGene meta-analyses, i.e. PARK16, originally implicated as a PD susceptibility locus in an Asian GWAS [14] but not highlighted in the recent GWAS meta-analysis on Caucasian samples [21] and GBA, a gene that was found soley by candidate-gene approaches. Another strength of our study is that it combines genetic data from currently more than 50 different countries allowing a systematic assessment of genetic associations across populations of different ethnic descent. For instance, these analyses suggest that variants in BST1, LRRK2, the PARK16 locus, and SNCA show genome-wide significant association with PD risk in both Caucasian and Asian-descent samples. Furthermore, the recently described Caucasian GWAS loci CCDC62/HIP1R, MCC1, and STK39 [21] also show similar effect size estimates in populations of Asian-descent [23]. PD association data originating from other ethnic groups are still relatively scarce. However, they could easily be added to the already existing data on the respective polymorphisms available on PDGene.
In summary, we have created a continuously updated online resource for genetic association studies in the field of PD. Synthesizing essentially all available data in the field led to the identification of ITGA8 as a novel potential PD risk locus. Our quantitative approach to data integration across a multitude of different study designs can be readily scaled to include large-scale resequencing efforts that will emerge over the coming years, making the complex field of PD genetics accessible to a broad range of investigators.
Methods
Note that the following section only provides a brief summary of the methods applied to our study. A much more detailed description can be found in Text S1.
Literature searches
Inclusion criteria
For inclusion in PDGene, a study has to meet three criteria: 1) It must evaluate the association between a bi-allelic genetic polymorphism (minor allele frequency ≥0.01 in the healthy control population of at least one study) and Parkinson's disease (PD) risk in datasets comprised of both affected (defined as clinically and/or neuropathologically diagnosed “Parkinson's disease”) and unaffected individuals; 2) it must be published in a peer-reviewed journal; 3) it must be published in English. For this manuscript, we also included data on ten SNPs generated in the GEO-PD Consortium datasets [14], [23] and obtained data for the newly identified SNP rs7077361 in ITGA8 from the Japanese GWAS dataset [14].
Exclusion criteria
In brief, genetic association data of the following studies were excluded from the meta-analyses (see Text S1 for details): family-based studies without available subject-level data (however, unrelated case-control data enriched for familial cases were not excluded), studies investigating only disease controls, multi-allelic polymorphisms, and studies of polymorphisms in mitochondrial DNA. We also excluded genetic data of apparently “poor” quality if discrepancies could not be resolved after contacting the study authors (e.g. inadequate genotyping/sequencing protocols or discrepancies in terms of allele names or frequencies when compared with public databases; more details can be found in Text S1).
Search strategies
Our literature searches until March 31st, 2011, yielded 27,210 articles, which were screened for eligibility using the title, abstract, or full-papers, as necessary. Additional screening of bibliographies in reviews, published meta-analyses, and original genetic association studies were also performed. Overall, full text versions of 1,534 articles were obtained. Following the inclusion and exclusion criteria outlined above, 828 articles were included in PDGene until March 31st 2011 (also see Figure 3).
Fig. 3. Flowchart of literature search, data extraction, and analysis strategies applied for PDGene. 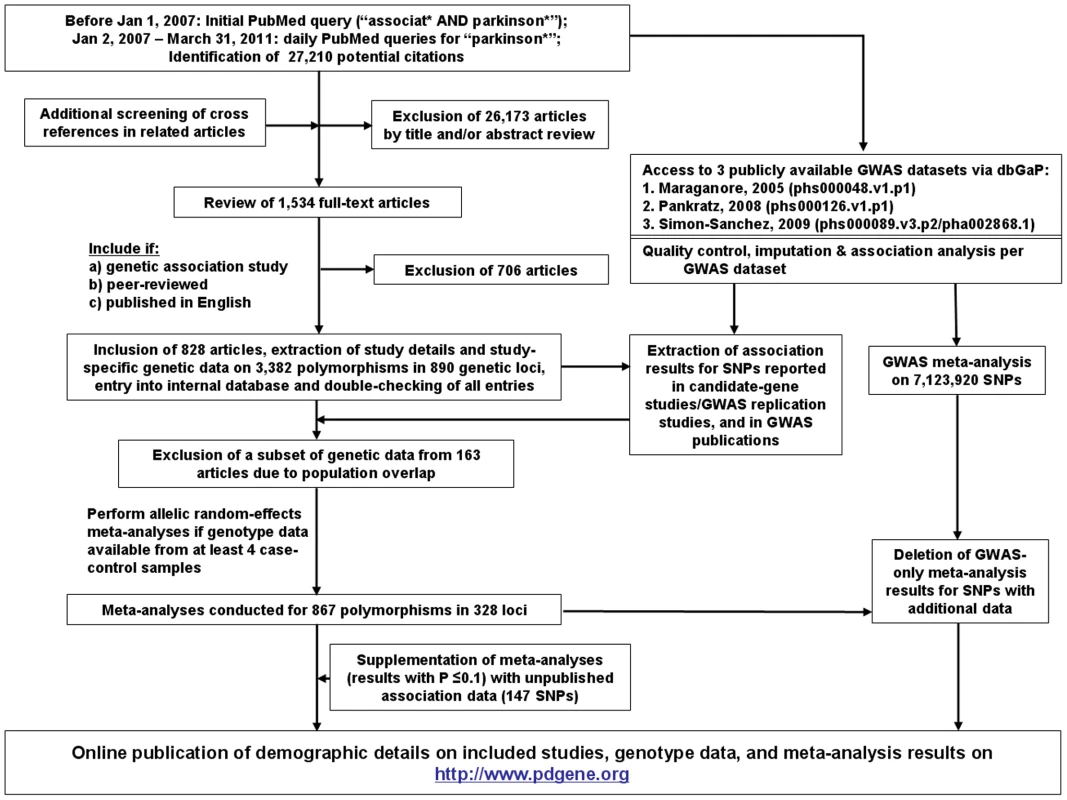
Statistical analyses
Meta-analyses
Random-effects allelic meta-analyses [31] were performed if a minimum of four independent datasets existed per polymorphism. Summary odds ratios [ORs] and 95% confidence intervals [CIs] were calculated irrespective of ethnic descent as well as for distinct ethnic groups (i.e. Caucasians, and Asians) if sufficient data were available. In addition, we performed a number of sensitivity analyses (excluding the initial studies and datasets in which HWE was violated in control individuals), systematically assessed between-study heterogeneity (via I2), and assessed the credibility of each at least nominally significant meta-analysis result by calculating Bayes factors (BF; here expressed as log10(BF)="logBF”) [25] and by determining a grading score developed by the Human Genome Epidemiology Network (HuGENet) [26], [27].
Assessment of small-study bias/publication bias
This is of particular importance in meta-analyses of published association data and was carefully addressed here: First, we added publicly available GWAS data [10], [12], [13] to the vast majority of SNPs. Since these data are typically unbiased, this should decrease the potential for small-study bias/publication bias. Secondly, for 147 SNPs of the core PDGene meta-analyses that showed statistically suggestive results (P≤0.1), we obtained additional data from all currently published, but not publicly available GWAS datasets, further decreasing a potential impact of small-study bias/publication bias. Thirdly, we directly assessed the evidence for small study bias by applying a recently proposed regression test [32] on all nominally significant (P<0.05) meta-analysis results. The results of these analyses are fully displayed in Table S1.
GWAS-only meta-analyses
We obtained individual-level genotype data for all publicly available PD GWAS datasets from NCBI's “dbGAP” database (a total of three [10], [12], [13] at the time of the datafreeze, March 31st, 2011). Genotype data were cleaned using standard procedures, followed by imputation of untested genotypes (using reference panels from HapMap and the 1000 Genomes Project), and association analyses incorporating imputation uncertainty (case-control datasets only), age, sex, and population stratification. Overall, this procedure led to a total of 7,723,931 unique SNPs, 7,123,920 of which were present in at least two, and 711,271 in at least three datasets. Meta-analyses (either combining test-statistics and standard errors using random-effects models, or by combining P-values weighted by sample size, see Text S1 for more details) were performed on the 7,123,920 SNPs present in at least two of the GWAS datasets.
Online database
After completion of all data-management and analysis steps, all study-specific variables, genotype data (except for GWAS), and meta-analysis plots are posted on a dedicated, publicly available, online adaptation of the PDGene database using the same software and code as our databases for Alzheimer's disease [33] and schizophrenia [34]. The online database is hosted by the “Alzheimer Research Forum” and can be accessed via its own designated URL (http://www.pdgene.org).
Database code
The database software can easily be ported to other genetically complex diseases and will be made available on a collaborative basis to interested researchers upon request.
Supporting Information
Zdroje
1. de LauLMLBretelerMMB 2006 Epidemiology of Parkinson's disease. Lancet Neurol 5 525 535 doi:10.1016/S1474-4422(06)70471-9
2. HardyJLewisPReveszTLeesAPaisan-RuizC 2009 The genetics of Parkinson's syndromes: a critical review. Curr Opin Genet Dev 19 254 265 doi:10.1016/j.gde.2009.03.008
3. Vilariño-GüellCWiderCRossOADachselJCKachergusJM 2011 VPS35 mutations in Parkinson disease. Am J Hum Genet 89 162 167 doi:10.1016/j.ajhg.2011.06.001
4. ZimprichABenet-PagèsAStruhalWGrafEEckSH 2011 A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet 89 168 175 doi:10.1016/j.ajhg.2011.06.008
5. Chartier-HarlinM-CDachselJCVilariño-GüellCLincolnSJLeprêtreF 2011 Translation initiator EIF4G1 mutations in familial Parkinson disease. Am J Hum Genet 89 398 406 doi:10.1016/j.ajhg.2011.08.009
6. MaraganoreDMde AndradeMElbazAFarrerMJIoannidisJP 2006 Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA 296 661 670 doi:10.1001/jama.296.6.661
7. ZabetianCPYamamotoMLopezANUjikeHMataIF 2009 LRRK2 mutations and risk variants in Japanese patients with Parkinson's disease. Mov Disord 24 1034 1041 doi:10.1002/mds.22514
8. GorisAWilliams-GrayCHClarkGRFoltynieTLewisSJG 2007 Tau and alpha-synuclein in susceptibility to, and dementia in, Parkinson's disease. Ann Neurol 62 145 153 doi:10.1002/ana.21192
9. SidranskyENallsMAAaslyJOAharon-PeretzJAnnesiG 2009 Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N Engl J Med 361 1651 1661 doi:10.1056/NEJMoa0901281
10. MaraganoreDMde AndradeMLesnickTGStrainKJFarrerMJ 2005 High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet 77 685 693 doi:10.1086/496902
11. FungH-CScholzSMatarinMSimón-SánchezJHernandezD 2006 Genome-wide genotyping in Parkinson's disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol 5 911 916 doi:10.1016/S1474-4422(06)70578-6
12. PankratzNWilkJBLatourelleJCDeStefanoALHalterC 2009 Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet 124 593 605 doi:10.1007/s00439-008-0582-9
13. Simón-SánchezJSchulteCBrasJMSharmaMGibbsJR 2009 Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet 41 1308 1312 doi:10.1038/ng.487
14. SatakeWNakabayashiYMizutaIHirotaYItoC 2009 Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet 41 1303 1307 doi:10.1038/ng.485
15. EdwardsTLScottWKAlmonteCBurtAPowellEH 2010 Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann Hum Genet 74 97 109 doi:10.1111/j.1469-1809.2009.00560.x
16. HamzaTHZabetianCPTenesaALaederachAMontimurroJ 2010 Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson's disease. Nat Genet 42 781 785 doi:10.1038/ng.642
17. SpencerCCAPlagnolVStrangeAGardnerMPaisan-RuizC 2011 Dissection of the genetics of Parkinson's disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet 20 345 353 doi:10.1093/hmg/ddq469
18. SaadMLesageSSaint-PierreACorvolJ-CZelenikaD 2011 Genome-wide association study confirms BST1 and suggests a locus on 12q24 as the risk loci for Parkinson's disease in the European population. Hum Mol Genet 20 615 627 doi:10.1093/hmg/ddq497
19. Simón-SánchezJvan HiltenJJvan de WarrenburgBPostBBerendseHW 2011 Genome-wide association study confirms extant PD risk loci among the Dutch. Eur J Hum Genet 19 655 661 doi:10.1038/ejhg.2010.254
20. EvangelouEMaraganoreDMIoannidisJPA 2007 Meta-analysis in genome-wide association datasets: strategies and application in Parkinson disease. PLoS ONE 2 e196 doi:10.1371/journal.pone.0000196
21. NallsMAPlagnolVHernandezDGSharmaMSheerinU-M 2011 Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet 377 641 649 doi:10.1016/S0140-6736(10)62345-8
22. DoCBTungJYDorfmanEKieferAKDrabantEM 2011 Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson's disease. PLoS Genet 7 e1002141 doi:10.1371/journal.pgen.1002141
23. SharmaMIoannidisJPAAaslyJOAnnesiGBriceA n.d. Large-scale replication and heterogeneity in Parkinson disease genetic loci. Neurology in press
24. LinD-YSullivanPF 2009 Meta-analysis of genome-wide association studies with overlapping subjects. Am J Hum Genet 85 862 872 doi:10.1016/j.ajhg.2009.11.001
25. IoannidisJPA 2008 Effect of formal statistical significance on the credibility of observational associations. Am J Epidemiol 168 374 383; discussion 384–390 doi:10.1093/aje/kwn156
26. IoannidisJPABoffettaPLittleJO'BrienTRUitterlindenAG 2008 Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol 37 120 132 doi:10.1093/ije/dym159
27. KhouryMJBertramLBoffettaPButterworthASChanockSJ 2009 Genome-wide association studies, field synopses, and the development of the knowledge base on genetic variation and human diseases. Am J Epidemiol 170 269 279 doi:10.1093/aje/kwp119
28. KentWJZweigASBarberGHinrichsASKarolchikD 2010 BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26 2204 2207 doi:10.1093/bioinformatics/btq351
29. MyersAJGibbsJRWebsterJARohrerKZhaoA 2007 A survey of genetic human cortical gene expression. Nat Genet 39 1494 1499 doi:10.1038/ng.2007.16
30. Varnum-FinneyBVenstromKMullerUKyptaRBackusC 1995 The integrin receptor alpha 8 beta 1 mediates interactions of embryonic chick motor and sensory neurons with tenascin-C. Neuron 14 1213 1222
31. DerSimonianRLairdN 1986 Meta-analysis in clinical trials. Control Clin Trials 7 177 188
32. HarbordRMEggerMSterneJAC 2006 A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat Med 25 3443 3457 doi:10.1002/sim.2380
33. BertramLMcQueenMBMullinKBlackerDTanziRE 2007 Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet 39 17 23 doi:10.1038/ng1934
34. AllenNCBagadeSMcQueenMBIoannidisJPAKavvouraFK 2008 Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet 40 827 834 doi:10.1038/ng.171
Štítky
Genetika Reprodukční medicína
Článek Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1Článek Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / TranscriptionČlánek Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding DomainČlánek Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin ComplexesČlánek An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood ObesityČlánek Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAsČlánek Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 3
-
Všechny články tohoto čísla
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
- Genomic Analysis of the Hydrocarbon-Producing, Cellulolytic, Endophytic Fungus
- Networks of Neuronal Genes Affected by Common and Rare Variants in Autism Spectrum Disorders
- Akirin Links Twist-Regulated Transcription with the Brahma Chromatin Remodeling Complex during Embryogenesis
- Too Much Cleavage of Cyclin E Promotes Breast Tumorigenesis
- Imprinted Genes … and the Number Is?
- Genetic Architecture of Highly Complex Chemical Resistance Traits across Four Yeast Strains
- Exploring the Complexity of the HIV-1 Fitness Landscape
- MNS1 Is Essential for Spermiogenesis and Motile Ciliary Functions in Mice
- A Fundamental Regulatory Mechanism Operating through OmpR and DNA Topology Controls Expression of Pathogenicity Islands SPI-1 and SPI-2
- Evidence for Positive Selection on a Number of MicroRNA Regulatory Interactions during Recent Human Evolution
- Variation in Modifies Risk of Neonatal Intestinal Obstruction in Cystic Fibrosis
- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Critical Evaluation of Imprinted Gene Expression by RNA–Seq: A New Perspective
- A Meta-Analysis and Genome-Wide Association Study of Platelet Count and Mean Platelet Volume in African Americans
- Mouse Genetics Suggests Cell-Context Dependency for Myc-Regulated Metabolic Enzymes during Tumorigenesis
- Transcriptional Control in Cardiac Progenitors: Tbx1 Interacts with the BAF Chromatin Remodeling Complex and Regulates
- Synthetic Lethality of Cohesins with PARPs and Replication Fork Mediators
- APOBEC3G-Induced Hypermutation of Human Immunodeficiency Virus Type-1 Is Typically a Discrete “All or Nothing” Phenomenon
- Interpreting Meta-Analyses of Genome-Wide Association Studies
- Error-Prone ZW Pairing and No Evidence for Meiotic Sex Chromosome Inactivation in the Chicken Germ Line
- -Dependent Chemosensory Functions Contribute to Courtship Behavior in
- Diverse Forms of Splicing Are Part of an Evolving Autoregulatory Circuit
- Phenotypic Plasticity of the Drosophila Transcriptome
- Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1
- Precocious Metamorphosis in the Juvenile Hormone–Deficient Mutant of the Silkworm,
- Igf1r Signaling Is Indispensable for Preimplantation Development and Is Activated via a Novel Function of E-cadherin
- Accurate Prediction of Inducible Transcription Factor Binding Intensities In Vivo
- Mitochondrial Oxidative Stress Alters a Pathway in Strongly Resembling That of Bile Acid Biosynthesis and Secretion in Vertebrates
- Mammalian Neurogenesis Requires Treacle-Plk1 for Precise Control of Spindle Orientation, Mitotic Progression, and Maintenance of Neural Progenitor Cells
- Tcf7 Is an Important Regulator of the Switch of Self-Renewal and Differentiation in a Multipotential Hematopoietic Cell Line
- REST–Mediated Recruitment of Polycomb Repressor Complexes in Mammalian Cells
- Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / Transcription
- Age-Dependent Brain Gene Expression and Copy Number Anomalies in Autism Suggest Distinct Pathological Processes at Young Versus Mature Ages
- A Genome-Wide Association Study Identifies Variants Underlying the Shade Avoidance Response
- -by- Regulatory Divergence Causes the Asymmetric Lethal Effects of an Ancestral Hybrid Incompatibility Gene
- Genome-Wide Association and Functional Follow-Up Reveals New Loci for Kidney Function
- A Natural System of Chromosome Transfer in
- Cell Size and the Initiation of DNA Replication in Bacteria
- Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding Domain
- Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin Complexes
- Temporal Transcriptional Profiling of Somatic and Germ Cells Reveals Biased Lineage Priming of Sexual Fate in the Fetal Mouse Gonad
- Rapid Analysis of Genome Rearrangements by Multiplex Ligation–Dependent Probe Amplification
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- The Atypical Calpains: Evolutionary Analyses and Roles in Cellular Degeneration
- The Silkworm Coming of Age—Early
- Development of a Panel of Genome-Wide Ancestry Informative Markers to Study Admixture Throughout the Americas
- Balanced Codon Usage Optimizes Eukaryotic Translational Efficiency
- The Min System and Nucleoid Occlusion Are Not Required for Identifying the Division Site in but Ensure Its Efficient Utilization
- Neurobeachin, a Regulator of Synaptic Protein Targeting, Is Associated with Body Fat Mass and Feeding Behavior in Mice and Body-Mass Index in Humans
- Statistical Analysis of Readthrough Levels for Nonsense Mutations in Mammalian Cells Reveals a Major Determinant of Response to Gentamicin
- Gene Reactivation by 5-Aza-2′-Deoxycytidine–Induced Demethylation Requires SRCAP–Mediated H2A.Z Insertion to Establish Nucleosome Depleted Regions
- The miR-35-41 Family of MicroRNAs Regulates RNAi Sensitivity in
- Genetic Basis of Hidden Phenotypic Variation Revealed by Increased Translational Readthrough in Yeast
- An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood Obesity
- Modelling Human Regulatory Variation in Mouse: Finding the Function in Genome-Wide Association Studies and Whole-Genome Sequencing
- Novel Loci for Adiponectin Levels and Their Influence on Type 2 Diabetes and Metabolic Traits: A Multi-Ethnic Meta-Analysis of 45,891 Individuals
- Polycomb-Like 3 Promotes Polycomb Repressive Complex 2 Binding to CpG Islands and Embryonic Stem Cell Self-Renewal
- Insulin/IGF-1 and Hypoxia Signaling Act in Concert to Regulate Iron Homeostasis in
- EMF1 and PRC2 Cooperate to Repress Key Regulators of Arabidopsis Development
- Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAs
- Contrasted Patterns of Molecular Evolution in Dominant and Recessive Self-Incompatibility Haplotypes in
- A Machine Learning Approach for Identifying Novel Cell Type–Specific Transcriptional Regulators of Myogenesis
- Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
- Nos2 Inactivation Promotes the Development of Medulloblastoma in Mice by Deregulation of Gap43–Dependent Granule Cell Precursor Migration
- Intracranial Aneurysm Risk Locus 5q23.2 Is Associated with Elevated Systolic Blood Pressure
- Heritability and Genetic Correlations Explained by Common SNPs for Metabolic Syndrome Traits
- A Genome-Wide Association Study of Nephrolithiasis in the Japanese Population Identifies Novel Susceptible Loci at 5q35.3, 7p14.3, and 13q14.1
- DNA Damage in Nijmegen Breakage Syndrome Cells Leads to PARP Hyperactivation and Increased Oxidative Stress
- DNA Resection at Chromosome Breaks Promotes Genome Stability by Constraining Non-Allelic Homologous Recombination
- Genetic Analysis of Floral Symmetry in Van Gogh's Sunflowers Reveals Independent Recruitment of Genes in the Asteraceae
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Promoter Nucleosome Organization Shapes the Evolution of Gene Expression
- The Nucleoside Diphosphate Kinase Gene Acts as Quantitative Trait Locus Promoting Non-Mendelian Inheritance
- The Ciliogenic Transcription Factor RFX3 Regulates Early Midline Distribution of Guidepost Neurons Required for Corpus Callosum Development
- Phosphorylation of the RNA–Binding Protein HOW by MAPK/ERK Enhances Its Dimerization and Activity
- A Genome-Wide Scan of Ashkenazi Jewish Crohn's Disease Suggests Novel Susceptibility Loci
- Parkinson's Disease–Associated Kinase PINK1 Regulates Miro Protein Level and Axonal Transport of Mitochondria
- LMW-E/CDK2 Deregulates Acinar Morphogenesis, Induces Tumorigenesis, and Associates with the Activated b-Raf-ERK1/2-mTOR Pathway in Breast Cancer Patients
- Mapping the Hsp90 Genetic Interaction Network in Reveals Environmental Contingency and Rewired Circuitry
- Autoregulation of the Noncoding RNA Gene
- The Human Pancreatic Islet Transcriptome: Expression of Candidate Genes for Type 1 Diabetes and the Impact of Pro-Inflammatory Cytokines
- Spo0A∼P Imposes a Temporal Gate for the Bimodal Expression of Competence in
- Antagonistic Regulation of Apoptosis and Differentiation by the Cut Transcription Factor Represents a Tumor-Suppressing Mechanism in
- A Downstream CpG Island Controls Transcript Initiation and Elongation and the Methylation State of the Imprinted Macro ncRNA Promoter
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



