-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPhosphorylation of the RNA–Binding Protein HOW by MAPK/ERK Enhances Its Dimerization and Activity
Drosophila melanogaster Held Out Wings (HOW) is a conserved RNA–binding protein (RBP) belonging to the STAR family, whose closest mammalian ortholog Quaking (QKI) has been implicated in embryonic development and nervous system myelination. The HOW RBP modulates a variety of developmental processes by controlling mRNA levels and the splicing profile of multiple key regulatory genes; however, mechanisms regulating its activity in tissues have yet to be elucidated. Here, we link receptor tyrosine kinase (RTK) signaling to the regulation of QKI subfamily of STAR proteins, by showing that HOW undergoes phosphorylation by MAPK/ERK. Importantly, we show that this modification facilitates HOW dimerization and potentiates its ability to bind RNA and regulate its levels. Employing an antibody that specifically recognizes phosphorylated HOW, we show that HOW is phosphorylated in embryonic muscles and heart cardioblasts in vivo, thus documenting for the first time Serine/Threonine (Ser/Thr) phosphorylation of a STAR protein in the context of an intact organism. We also identify the sallimus/D-titin (sls) gene as a novel muscle target of HOW–mediated negative regulation and further show that this regulation is phosphorylation-dependent, underscoring the physiological relevance of this modification. Importantly, we demonstrate that HOW Thr phosphorylation is reduced following muscle-specific knock down of Drosophila MAPK rolled and that, correspondingly, Sls is elevated in these muscles, similarly to the HOW RNAi effect. Taken together, our results provide a coherent mechanism of differential HOW activation; MAPK/ERK-dependent phosphorylation of HOW promotes the formation of HOW dimers and thus enhances its activity in controlling mRNA levels of key muscle-specific genes. Hence, our findings bridge between MAPK/ERK signaling and RNA regulation in developing muscles.
Published in the journal: . PLoS Genet 8(3): e32767. doi:10.1371/journal.pgen.1002632
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002632Summary
Drosophila melanogaster Held Out Wings (HOW) is a conserved RNA–binding protein (RBP) belonging to the STAR family, whose closest mammalian ortholog Quaking (QKI) has been implicated in embryonic development and nervous system myelination. The HOW RBP modulates a variety of developmental processes by controlling mRNA levels and the splicing profile of multiple key regulatory genes; however, mechanisms regulating its activity in tissues have yet to be elucidated. Here, we link receptor tyrosine kinase (RTK) signaling to the regulation of QKI subfamily of STAR proteins, by showing that HOW undergoes phosphorylation by MAPK/ERK. Importantly, we show that this modification facilitates HOW dimerization and potentiates its ability to bind RNA and regulate its levels. Employing an antibody that specifically recognizes phosphorylated HOW, we show that HOW is phosphorylated in embryonic muscles and heart cardioblasts in vivo, thus documenting for the first time Serine/Threonine (Ser/Thr) phosphorylation of a STAR protein in the context of an intact organism. We also identify the sallimus/D-titin (sls) gene as a novel muscle target of HOW–mediated negative regulation and further show that this regulation is phosphorylation-dependent, underscoring the physiological relevance of this modification. Importantly, we demonstrate that HOW Thr phosphorylation is reduced following muscle-specific knock down of Drosophila MAPK rolled and that, correspondingly, Sls is elevated in these muscles, similarly to the HOW RNAi effect. Taken together, our results provide a coherent mechanism of differential HOW activation; MAPK/ERK-dependent phosphorylation of HOW promotes the formation of HOW dimers and thus enhances its activity in controlling mRNA levels of key muscle-specific genes. Hence, our findings bridge between MAPK/ERK signaling and RNA regulation in developing muscles.
Introduction
Regulation of gene expression at the level of RNA is often mediated through the activities of RNA-binding proteins (RBPs), which control different aspects of RNA metabolism of target genes [1], [2]. Drosophila melanogaster Held Out Wings (HOW) is an RBP that belongs to a family of evolutionarily conserved “Signal Transduction and Activation of RNA” (STAR) proteins [3]. STAR family members control a wide range of tissue differentiation processes. For example, in mammals, Sam68 controls spermatogenesis [4], and Quaking (QKI) regulates myelination by Schwann cells and oligodendrocytes [5], [6], [7] as well as muscle fiber maturation in Zebrafish [8]. In C. elegans, the STAR protein GLD-1 promotes germ cell differentiation [9], while ASD-2 is required for alternative splicing [10].
HOW, the Drosophila protein orthologous to mammalian QKI, is highly expressed in muscles, tendons [11], [12], [13], [14] and glial cells [15], [16], where it plays an essential role during development by controlling the mRNA levels of an array of target genes [17]. HOW performs various activities on its target RNAs: it facilitates the alternative splicing of stripe A, a transcription factor essential for tendon cell maturation [18], and mediates specific splicing of the septate junction constituent, nrxIV, thereby controlling glial cell maturation [15]. It also functions by reducing mRNA levels of various targets. For example, during gastrulation, HOW-dependent downregulation of cdc25/string, a cell cycle promoting phosphatase, is essential to inhibit cell division in invaginating mesodermal cells [19].
Structurally, HOW contains a single maxi-KH RNA binding motif that is flanked by two additional conserved domains, QUA1 and QUA2 [14]. While the QUA2 motif, located C terminally to the KH domain, takes part in RNA-binding and contributes to the specificity of RNA recognition [20], [21], [22], the QUA1 motif, located N terminally to the KH domain, was shown to mediate protein dimerization in GLD-1, Sam68 and QKI [23], [24], [25]. Notably, despite the fact that this domain is not essential for RNA-binding, its deletion in GLD-1 nevertheless reduces the affinity of the protein to the RNA binding motif TGE (Tra-2 and GLI response element), by about ten-fold [26], suggesting that dimerization of STAR proteins might enhance their affinity to RNA. To date, it is not clear what regulates the degree of STAR protein dimer formation.
Consistent with their expression in a wide range of tissues during development, the activity of STAR proteins is highly regulated at distinct post-translational levels, including phosphorylation by various kinases (reviewed in [27]). These modifications likely impinge on the activity, subcellular distribution, or the formation of protein complexes of specific STAR proteins. Phosphorylation of STAR proteins could couple regulation of RNA metabolism with distinct signaling cascades operating in a spatial and temporal restricted manner.
While Tyrosine phosphorylation of the C terminal regions of both Sam68 [28] and QKI [29] has been established, Serine/Threonine (Ser/Thr) phosphorylation has yet to be demonstrated with respect to the QKI-subfamily of proteins. A more evolutionarily distant STAR protein,Sam68, was shown to be Ser/Thr phosphorylated, both by Cdc2 [30] and by ERK1/2 in various culture lines, promoting differential activities. Specifically, Sam68 phosphorylation by ERK1/2 in a lymphoma cell line enhances its ability to regulate alternative splicing [31], while in mouse spermatocytes and in HEK293 cells it induces cytoplasmic accumulation correlated with its association with polyribosomes [4], [32]. We therefore postulated that ERK-dependent phosphorylation of STAR proteins might affect their activity in a tissue-specific manner.
In the present manuscript, we show that the Drosophila STAR protein HOW is phosphorylated on conserved Thr residues. Importantly, we demonstrate that in embryos in vivo, a particular HOW isoform is phosphorylated on Thr, in a tissue-specific manner. Moreover, we identify the major Z-disc gene product sallimus (kettin/D-titin, sls [33], [34]) as a specific target for HOW in muscle cells and show that sls regulation is dependent on MAPK phosphorylation of HOW. Mechanistically, we demonstrate that HOW phosphorylation is essential for its efficient homodimerization and RNA binding capability. Taken together, our results reveal a molecular mechanism linking muscle-specific MAPK-dependent phosphorylation of HOW to its ability to homodimerize, bind its targets and regulate them, and thereby contribute to muscle sarcomerization.
Results
HOW contains conserved consensus sites for MAPK/ERK phosphorylation
Examination of HOW revealed two putative MAPK/ERK consensus sites, comprised of Thr followed by Proline (Pro) (TP) at residues 59 and 64 (Figure 1A). These could also serve as putative sites for other Ser/Thr kinases such as Cyclin Dependent Kinases (CDKs). Importantly, T64 is conserved throughout HOW(L) sequences in all annotated Drosophila species, while T59 is found only in closely related species (Figure 1B).
Fig. 1. HOW possesses conserved MAPK/ERK consensus phospho-acceptor sites and docking domains. 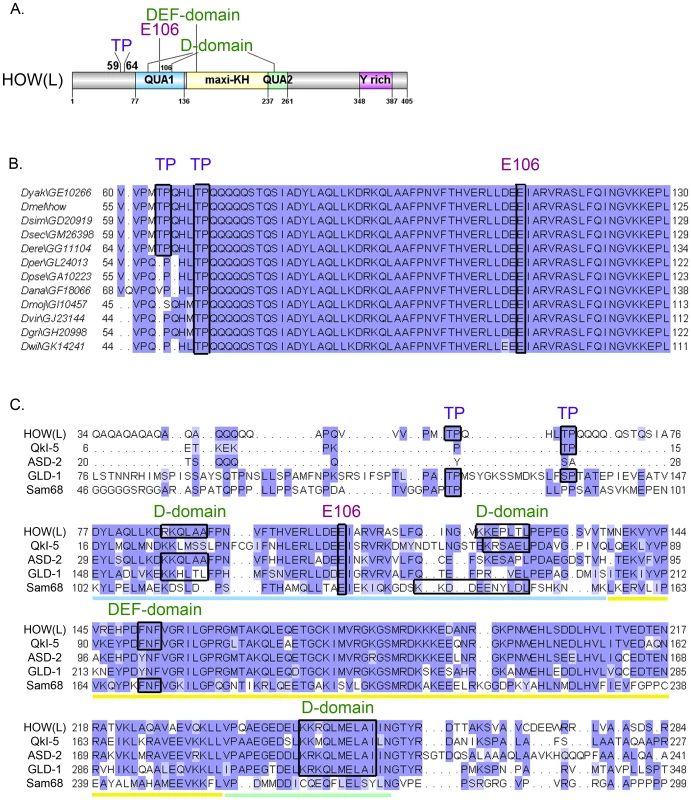
A. A scheme of HOW(L) protein domains. The locations of the TP sites at positions 59 and 64, the Glutamic acid at position 106 required for homodimerization, and the four potential MAPK/ERK docking sites are indicated. The STAR/GSG domain is indicated by colored squares: The QUA1 domain (light blue), the maxi-KH domain (yellow), and the QUA2 domain (green). Also indicated is the Tyrosine rich area in the C-terminus (pink). B. Alignment of HOW(L) amino acid sequences from 12 Drosophila species. While T64 is conserved throughout all the species, T59 is only conserved in five of them (conserved residues are demarcated by black frames). E106 is also fully conserved, as are the putative docking sites (not shown). C. Alignment of the HOW(L) protein with the mammalian proteins QKI-5 and Sam68, and the nematode homologs, GLD-1 and ASD-2. The Qua1 domain is underlined in light blue, the maxi-KH domain in yellow, and the Qua2 in green. Note the conserved TP sites, E106 and various docking sites, demarcated by frames. Both the DEF domain and the second D-domain (at a.a. 121) are conserved in the mammalian proteins QKI and Sam68, whereas the first (at a.a. 85) and third (at a.a. 245) D-domains are conserved in C. elegans proteins (the latter is also conserved in QKI). In addition to these conserved phosphorylation sites, HOW also contains putative MAPK docking sites. These “D” [35] or “DEF” [36] domains frequently mediate high-affinity interactions between MAPK and its substrates, allowing efficient phosphorylation of the substrate [37]. HOW harbors three such potential D-domain motifs, 85RKQLAA, 121KKEPLTL, and 245KKRQLMELAI, as well as a single DEF domain motif, 151FNF. All four motifs are fully conserved throughout Drosophila species, and partially conserved in other STAR proteins (Figure 1C). The presence of these domains, together with the occurrence of potential phosphorylation sites, led us to test the possibility that HOW is phosphorylated by MAPK/ERK.
HOW is phosphorylated by MAPK/ERK in vitro
To start investigating phosphorylation of HOW by MAPK/ERK, we generated a HOW construct in which the putative phosphoacceptor sites T59 and T64 were mutated to Alanine (HOWTTAA), rendering it non-phosphorylatable.
Next, we tested whether MAPK/ERK could phosphorylate HOW in vitro. Briefly, HOW(L)WT and HOW(L)TTAA were transcribed and translated in vitro in the presence of S35-Methionine, incubated with recombinant activated ERK2, and run on an SDS gel. Incubation of in vitro translated Yan, an established MAPK substrate protein [38] with active Erk2 resulted in a protein mobility shift on SDS Page as compared to the unphosphorylated protein (Figure 2A, lanes 1–4). Importantly, HOW(L)WT but not HOW(L)TTAA also displayed a MAPK-dependent mobility shift (lane 5,7), indicating that MAPK phosphorylates HOW and that the phosphorylation event occurs on the predicted residues (Figure 2A, arrow). However, under these conditions, HOW underwent only partial phosphorylation, as a relatively small fraction of HOW was shifted (see below, Figure 3).
Fig. 2. HOW(L) is phosphorylated in vitro and in S2R+ cells by MAPK/ERK on one or more TP sites. 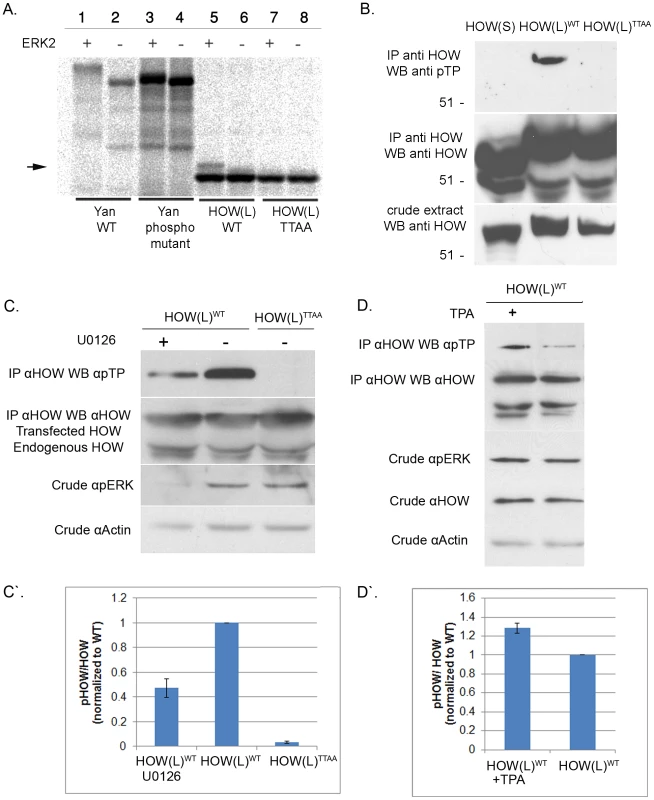
A. ERK2 phosphorylates HOW in vitro. HOW(L)WT and HOW(L)TTAA (phospho mutant) constructs were in vitro translated (using S35 labeling) and incubated with activated ERK2. The reaction was resolved on an SDS gel. The left part of the panel (lanes 1–4) shows phosphorylation of the Yan (Aop) protein as a control. HOW(L)WT protein, but not the HOW(L)TTAA mutant (lanes 7–8) exhibits a molecular weight shift of a fraction of the protein when incubated with the kinase (lane 5) relative to HOW without the kinase (lane 6). B. HOW(L), but not HOW(S), is phosphorylated in S2R+ cells. HOW proteins were immunoprecipitated using an anti-HOW antibody and reacted with anti-pTP (top panel) and anti-HOW (middle panel) antibodies. HOW proteins in the crude extract are shown in the bottom panel. From the left: HOW(S)WT, HOW(L)WT and HOW(L)TTAA. C. Treatment with the MAPKK/MEK inhibitor U0126 decreases HOW phosphorylation. HOW proteins were immunoprecipitated with an anti-HOW antibody from cells transfected with how(l)WT treated (or not) with U0126, or with how(l)TTAA and reacted with anti-pTP antibody (top panel) or with anti-HOW antibody (2nd from the top). The crude extract was reacted with anti-pERK antibody (2nd from the bottom), to confirm the extent of U0126 inhibition, and anti-Actin (bottom panel) as a loading control. C′. Quantification of the reduction of phosphorylation following treatment with U0126. For each sample, the ratio between the pTP band measurement and the total HOW band was calculated, and normalized to the HOW(L)WT (non-treated sample) ratio. Results shown are the average of three experiments; error bars indicate SEM. Following U0126 treatment, HOW phosphorylation was reduced to 0.47±0.07 relative to non-treated cells (P = 0.0021, unpaired t-test, n = 3). D. Same as in C, except cells were treated with TPA/PMA. A representative experiment is presented. D′. Quantification of three TPA treatment experiments, in which HOW phosphorylation was increased by 1.28±0.06 (P = 0.0076, unpaired t-test, n = 3). Fig. 3. HOW(L) phosphorylation correlates with its homo-dimerization. 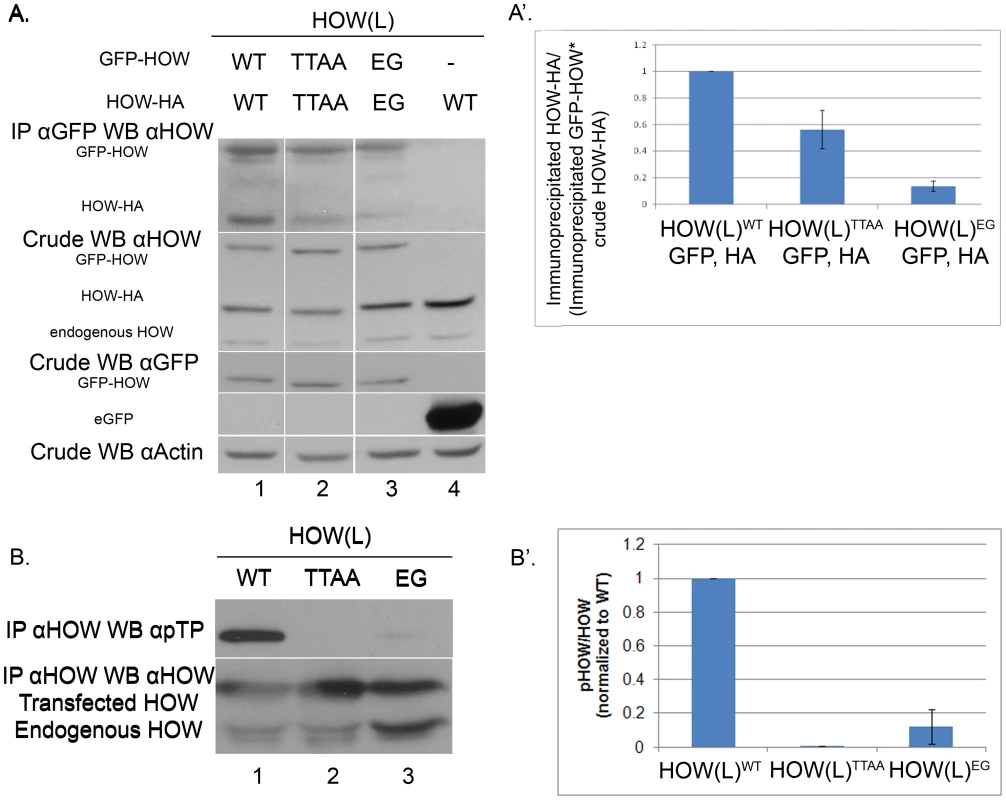
A. Phospho-mutant HOW(L)TTAA oligomerizes to a lesser extent than HOW(L)WT. S2R+ cells were co-transfected with expression vectors for the following: gfp- how(l)WT and how(l)WT-HA (1) gfp- how(l)TTAA with how(l)TTAA-HA (2); gfp-how(l)EG with how(l)EG-HA (3); or gfp with how(l)WT-HA (4) as a control. IP was carried out using anti-GFP antibody and Western blotting was performed with anti-HOW. Crude lysate was reacted with anti-HOW (2nd panel from the top), anti-GFP (to visualize GFP-HOW (lanes 1–3) and the GFP control (lane 4)), as well as with anti-Actin. A′. Quantification of lanes 1–3 from three experiments as described in A. For each sample, the ratio between the Co-immunoprecipitated HA protein and its total crude level was normalized to the amount of the GFP protein that pulled it down. HOWTTAA (2) exhibited partial dimerization (0.56±0.14, P = 0.039 unpaired t-test, n = 3). The HOWEG (3) is almost completely non-dimerized (0.13±0.04, P<0.0001, unpaired t-test, n = 3). B. HOW(L) protein that is unable to homo-dimerize undergoes significantly less phosphorylation. S2R+ cells were transfected with either how(l)WT, how(l)TTAA or the how(l)EG mutant construct. Proteins were immunoprecipitated using anti-HOW antibody. The amount of lysate used for the IP of the HOW(L)EG protein was doubled, in order to obtain a comparable amount of immunoprecipitated HOW protein (since it does not dimerize, less protein precipitates). B′. Quantification of two of several experiments in which the total levels of the different proteins were comparable. For each sample, a ratio between the pTP band measurement and the total HOW band was calculated, and normalized to the HOW(L)WT ratio. Error bars indicate SEM (HOW(L)EG 0.12±0.10). HOW is phosphorylated in Drosophila S2R+ cells on MAPK/ERK consensus sites
We next tested whether HOW is phosphorylated on its MAPK/ERK consensus sites using Drosophila S2R+ cells. HOW(L)WT and HOW(L)TTAA were expressed in these cells, precipitated with an anti-HOW antibody, and subjected to Western blot analysis using an anti-phospho-Thr-Pro (anti-pTP) antibody (Figure 2B). HOW(L)WT protein reacted with the anti-pTP antibody, whereas the HOW(L)TTAA form did not. This indicated that in unstimulated cells HOW is phosphorylated at least on one TP site.
We have also examined the potential phosphorylation of a shorter HOW isoform, HOW(S), which differs from HOW(L) only at the C terminus [13], . Interestingly, although HOW(S)WT possesses all of the sites predicted to be phosphorylated by, and to bind MAPK, it did not react with the anti-pTP antibody when tested in a similar manner (Figure 2B). This difference may stem from the distinct subcellular localization of the different HOW isoforms; while HOW(L) is nuclear and is thus probably more accessible to active MAPK/ERK, HOW(S) is mostly expressed in the cytoplasm. We therefore limited our analysis to the phosphorylation of HOW(L).
MAPK/ERK is phosphorylating HOW in S2R+ cells
We further confirmed the identity of the kinase phosphorylating HOW in S2R+ cells by employing the specific MAPKK/MEK inhibitor, U0126 [40] (Figure 2C). Treatment of the cells with U0126 led to a significant decrease in pERK levels (compare the left and middle lanes in Figure 2C). Notably, in the treated cells a marked reduction was observed in the phosphorylation of immunoprecipitated HOW detected by anti-pTP antibody (top panel) in comparison to non-treated cells (phosphorylation was reduced to 0.47±0.07 fold of control, n = 3, Figure 2C′). This result indicates that HOW phosphorylation is dependent on MAPKK/MEK. Interestingly, the phosphorylation of HOW on the TP sites was not completely eliminated, as expected from the effective reduction in pERK levels, suggesting that another kinase might also be involved in the phosphorylation of at least one of the TP sites.
In a complementary approach, we stimulated MAPK/ERK signaling using Phorbol 12-Myristate 13-Acetate (TPA/PMA). This compound is an activator of protein kinase C [41], which in turn leads to activation of MAPK/ERK [42]. Treatment with TPA increased HOW phosphorylation, as indicated by stronger reaction with the pTP antibody (Figure 2D, quantification of 3 experiments is shown in Figure 2D′).
Taken together, these experiments demonstrate that HOW is phosphorylated by MAPK/ERK in Drosophila S2R+ cells.
MAPK/ERK-dependent phosphorylation stabilizes HOW dimers
What could be the reason for the partial phosphorylation of in vitro translated HOW by activated ERK (Figure 2A)? We considered the possibility that the incomplete phosphorylation is due to a limited degree of HOW dimerization that occurred under our experimental conditions.
Accordingly, we examined whether MAPK/ERK phosphorylation was related to the degree of HOW dimerization by performing dimerization experiments with HOWWT and HOWTTAA. Both HOW variants were fused to GFP and expressed in S2R+ cells together with either HA-tagged HOWWT or HOWTTAA, respectively. We immunoprecipitated the GFP-HOW(L) protein using an anti-GFP antibody, and tested, using anti-HOW antibody, the co-precipitation of HOW(L)-HA (Figure 3A). We differentiated between GFP-HOW, HOW-HA and the endogenous HOW by virtue of their different molecular weights. Indeed, HOWWT-HA readily co-immunoprecipitated with GFPWT-HOW (Figure 3A, lane1), indicating the two proteins oligomerized, presumably as dimers. A negative control protein (eGFP) did not precipitate HOW(L)-HA, demonstrating the specificity of the interaction (Figure 3A, lane 4). Interestingly, the non-phosphorylatable HOWTTAA co-immunoprecipitated less efficiently (lane 2). Quantification of the experiments, while normalizing the co-precipitated HOW-HA levels to its levels in the crude extract as well as to the immunoprecipitated HOW-GFP, showed that co-precipitation of the phospho-mutant HOW was reduced by about 40% compared to that of HOWWT (Figure 3A′). In addition, we attempted to examine the effect of enhanced phosphorylation on HOW dimerization, using a putative phospho-mimicking mutant variant, in which the two Thr residues were replaced by Aspartic acid (HOW(L)TTDD). However, this mutation did not appear to mimic phosphorylated HOW, in this as well as in other assays (data not shown). As a control for the dimerization assay, we mutated HOW on a Glutamic acid residue, changing it to Glycine (E106G, HOWEG) and similarly tagged it with HA or GFP. This residue was previously shown to be essential for QKI [23] and GLD-1 dimerization [24]. As expected, the amount of co-precipitated HOWEG-HA was reduced to about 10% of wild-type levels (Figure 3A lane 3 and Figure 3A′), confirming the reliability of our dimerization assay. Strikingly, testing the phosphorylation state of HOW dimerization mutant HOWEG using anti-pTP antibody revealed that this HOW mutant essentially does not undergo phosphorylation by MAPK/ERK (Figure 3B lane 3, 3B′).
Collectively, the above experiments strongly suggest that MAPK-dependent phosphorylation occurs only on HOW dimers, although we cannot exclude a possible change of conformation in the monomeric structure of HOWEG [24]. Our results also further support the idea that phosphorylation stabilizes HOW dimerization since the extent of dimer formation of HOWTTAA is significantly reduced.
Phosphorylation of HOW strengthens its binding to RNA
To address whether the phosphorylation of HOW influences its ability to bind RNA, we performed an RNA binding assay. In this experiment, wild-type HA-tagged HOW(L) was affinity purified from S2R+ cells that were either grown in normal medium (in which HOW is phosphorylated, see Figure 2C) or in medium containing the MEK inhibitor U0126 (in which HOW phosphorylation is reduced, shown in Figure 2C). The RNA-binding activity of purified HOW-HA was then tested using biotin-labeled RNA oligomers that either contained or did not contain the HOW response element (HRE) [43]. A significant reduction of about 70% in the binding of HOW to the RNA was detected following treatment with the MEK inhibitor U0126 (Figure 4A, left lane). In addition, HOW(L)EG, the mutant form that does not dimerize, exhibited an extremely low RNA binding activity, supporting a role for the dimerization of HOW in RNA binding (Figure 4A, right lane). Notably, the non-phosphorylatable HOW(L)TTAA only showed slightly reduced binding (Figure 4A, second lane from the right, about 20% reduction), possibly due to its ability to dimerize with endogenous, phosphorylated HOW protein (contrary to the U0126-treated cells, where all HOW proteins are less phosphorylated). This experiment demonstrates that ERK-dependent phosphorylation of HOW enhances not only its homodimerization but also its RNA binding activity, suggesting that both functions are linked.
Fig. 4. HOW phosphorylation strengthens its affinity to RNA, while not affecting its subcellular localization. 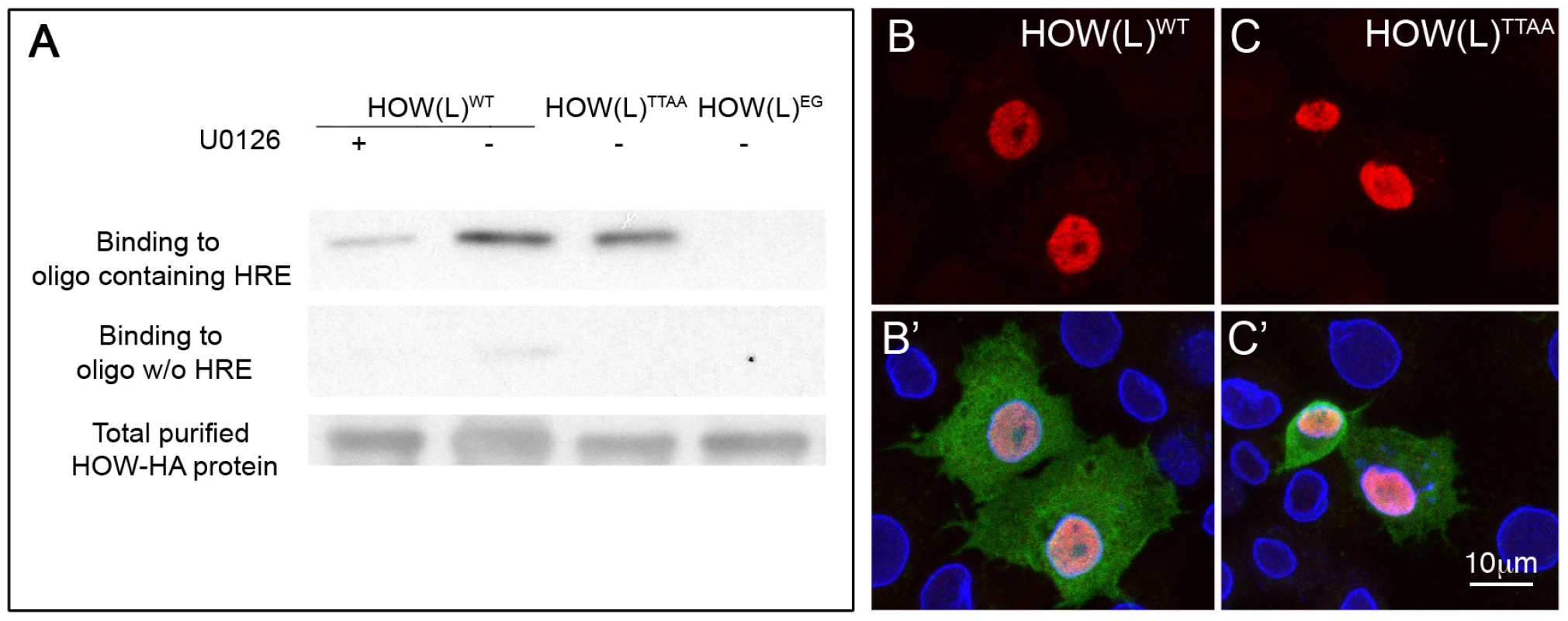
A. In vitro RNA-protein binding assay. HA tagged HOW(L)WT, HOW(L)TTAA or HOW(L)EG constructs were purified from S2R+ cells by immunoprecipitation of the HA tag, followed by elution with free HA peptide. HOW(L)WT transfected cells were either treated with U0126 or not treated. Equal volumes of purified proteins were incubated with biotin labeled 12 nt oligomers that either bind HOW (top panel) or do not bind HOW (middle panel). The complexes were incubated with avidin beads, eluted by boiling in sample buffer and reacted with anti HOW in a Western blot. The amounts of HOW protein used for each reaction were comparable (bottom panel). B–C′. Localization of HOW(L) was not altered due to the mutations in the Thr residues. Cells expressing either HOW(L)WT (B,B′) or HOW(L)TTAA (C,C′) were stained for HOW (red, B–C). Merge images (B′–C′) also present GFP (green, marks transfected cells) and Lamin (blue). Thr phosphorylation does not alter the subcellular localization of HOW
Phosphorylation alters the subcellular localization of Sam68 [4], [32], raising the possibility that, via this mode of regulation, MAPK impinges on HOW's ability to bind and regulate its RNA targets. To address this issue, we transfected S2R+ cells with HOWWT or HOWTTAA tagged with HA, and stained with an anti-HA antibody. We did not detect any alteration in the subcellular localization of HOW(L)TTAA relative to HOW(L)WT (Figure 4B–4C′). Thus, HOW Thr phosphorylation does not alter its subcellular localization.
HOW is phosphorylated on T64 in the embryo in vivo
To follow the pattern of HOW phosphorylation in vivo, we generated a polyclonal antibody designed to specifically recognize HOW only when it is phosphorylated on the more conserved Thr residue, T64 (we refer to the antibody as anti-pHOW (pT64) (see Materials and Methods)). We first confirmed the specificity of the antibody by transfecting S2R+ cells with how(l)WT, how(l)TTAA, and by treating a sample of the HOW(L)WT expressing cells with the MAPKK/MEK inhibitor U0126. We performed immunoprecipitation (IP) with an anti-HOW antibody and used anti-pHOW (pT64) antibodies in Western blot analysis (Figure 5A). The antibody reacted with the immunoprecipitated HOW(L)WT (lane 2) but not with HOW(L)TTAA (lane 4) nor with non-transfected cells (lane 1). In the sample treated with U0126 (lane 3), the reactivity of the antibody was significantly reduced, suggesting the antibody indeed detects HOW phosphorylation, particularly on T64.
Fig. 5. Phosphorylated HOW is detected in the nuclei of somatic muscles and cardioblasts. 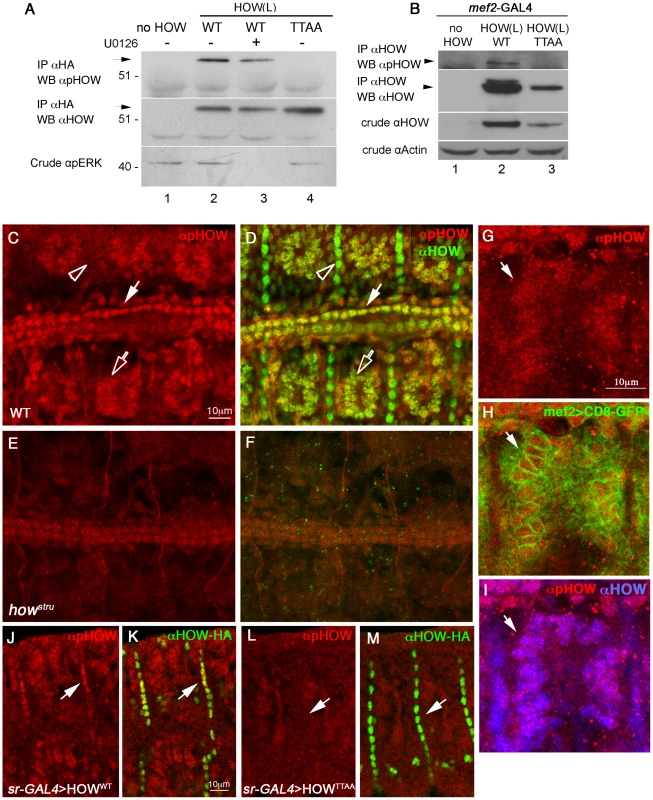
A. Anti-pHOW (pT64) antibody reacts with HOW, in a phosphorylation-dependent manner. HOW protein was immunoprecipitated with an anti-HOW antibody from S2R+ cells transfected with either HOW(L)WT, treated (or not) with U0126 (lanes 3, 2 respectively), with HOW(L)TTAA (lane 4), or no HOW (lane 1). The IP was reacted with anti-pHOW (pT64) antibody (top) or with anti-HOW antibody (middle). The crude extract was reacted with anti-pERK antibody (bottom), confirming U0126 inhibition. Note specific reactivity of the anti-pHOW antibody. B. Anti-pHOW (pT64) antibody reacts with HOW protein overexpressed in embryonic muscles and heart. Transgenic UAS-HOW(L)WT (lane 2) and UAS-HOW(L)TTAA (lane 3) flies or wild-type controls (lane 1) were crossed to the mef2-GAL4 driver line. Embryos were collected, and protein extracts were subjected to IP with an anti-HA antibody followed by Western with anti-pHOW (pT64) antibody (top), and anti-HOW antibody (2nd from top). Crude extracts were reacted with anti-HOW (2nd from bottom), and anti-Actin (bottom) as a loading control. C-F. Drosophila embryos stained with anti-pHOW (pT64) exhibit specific staining in somatic muscles and heart cardioblasts. Stage 16 WT (C,D) and how stru mutant (E,F) embryos were stained with anti-pHOW (pT64) (red, C–F) and anti-HOW (green in merge D,F). Embryos are oriented dorsal to the top and anterior to the left. (C,D) Note the staining in muscles (hollow arrow), heart cardioblasts (white arrow) and the lack of pHOW staining in the muscle attachment sites (arrowhead). (E,F) Weak staining in cardioblasts in how stru−/− mutants. G–I. Drosophila embryos expressing CD8-GFP, which localizes at the plasma membrane, but also often concentrates in the ER surrounding the nuclei, in muscles under mef2-GAL4, were stained with anti-pHOW (red, G) shown merged with GFP (green, H) and with HOW (blue, I). J–M. HOW(L) overexpressed in embryonic tendon cells reacts with the anti-pHOW (pT64) antibody. Embryos expressing UAS-HOW(L)WT-HA (J,K) or UAS-HOW(L)TTAA–HA (L,M) in the tendon cells under sr–GAL4 were stained with anti-pHOW (pT64) (red, J–M) and anti-HA (green, K, M, merged with the anti-pHOW staining). Arrows mark tendon cells reactive with anti-pHOW upon expression of HOW(L)WT (J,K), or non-reactive upon expression of the phospho-mutant (L,M). We used the anti-pHOW (pT64) antibody to identify tissues in which HOW is phosphorylated (Figure 5C). To this end, we stained wild-type Drosophila embryos, employing how mutant (howstru−/−) embryos, in which zygotic HOW is not produced, as a control for antibody specificity. The embryos were also co-stained with a general anti-HOW antibody. Specific anti-pHOW staining was detected in the nuclei of somatic muscles (Figure 5C, open arrow) as well as in the heart cardioblasts (Figure 5C, white arrow). This staining co-localized with anti-HOW staining (Figure 5D, corresponding arrows) and was reduced to background levels in howstru mutants (Figure 5E, 5F). Importantly, phosphorylated HOW was confined to the nuclei of the somatic muscles that were marked with muscle-specific expression of CD8-GFP (which localizes to the cell membrane but often concentrates in the ER surrounding the nuclei), as shown in Figure 5G–5I. This result indicates that HOW is phosphorylated on T64 in muscle nuclei.
Intriguingly, the anti-pHOW (pT64) antibody did not label tendon cells, which exhibited a strong anti-HOW staining (Figure 5C, 5D, open arrowheads), further validating the specificity of our antibody. The lack of pHOW staining is not simply due to low levels of MAPK in these cells, since previous analysis demonstrated high level of phospho-ERK in tendon cells at this stage [44]. We therefore hypothesized that the HOW(L) isoform found to be specifically phosphorylated by MAPK/ERK (Figure 2) is not expressed at high levels in tendon cells at this developmental stage and that the anti-HOW staining is mainly detecting HOW(S), which does not undergo phosphorylation in S2R+ cells (Figure 2B). To directly address this possibility, we generated transgenic flies expressing HA-tagged versions of HOW(L)WT or HOW(L)TTAA under UAS-GAL4 binding sequences. The expression of HOW(L)WT or HOW(L)TTAA was driven in embryonic tendon cells by the tendon-specific driver, sr-GAL4, and the embryos were stained for pHOW (Figure 5J–5M) Whereas over expression of HOW(L)WT in tendon cells led to positive nuclear staining with the anti-pHOW antibody (Figure 5J, 5K arrows), overexpression of HOW(L)TTAA did not result in such a staining (Figure 5L, 5M arrows). This experiment shows that the kinase required for HOW phosphorylation on T64 is activated in tendon cells, and that the lack of reactivity of the antibody in wild-type tendon cells at late embryonic stages is due to low levels of HOW(L) in this tissue. This is also consistent with the cytoplasmic HOW staining characteristic of the HOW(S) isoform present at stage 16 embryos [39].
Based on these results, we conclude that HOW(L) is phosphorylated on T64 in the nuclei of somatic muscles and heart cardioblasts. To further verify that HOW undergoes Thr phosphorylation in embryonic somatic muscles, we also expressed HA–tagged HOW(L)WT or HOW(L)TTAA in muscles using the mef2-GAL4 driver. The HOW variants were subsequently immunoprecipitated using anti-HA conjugated beads and reacted with anti-pHOW (pT64) antibodies on Western blots (Figure 5B, lane 2). Only HOW(L)WT but not HOW(L)TTAA (lane 3), was detectable by the anti-pHOW antibody. In addition, an anti-HOW antibody identified an additional upper band that might represent phosphorylated HOW (lane 2). To conclude, these findings strongly indicate that HOW(L) undergoes phosphorylation on T64 in embryonic somatic and cardiac muscles and that phosphorylated HOW in muscles is localized specifically to the nucleus.
HOW(L) downregulates Sallimus in muscles in a phosphorylation-dependent manner
To further characterize the physiological significance of MAPK/ERK-dependent phosphorylation of HOW, we focused on somatic muscles, a tissue in which a high degree of HOW phosphorylation was detected. Recently, HOW was shown to be essential for muscle sarcomerization [45]. Specifically, in larvae where how was knocked down (using how RNAi expressed specifically in muscles), the sarcomeric organization is aberrant. In these larvae, the Z discs appear discontinuous and spotty, a phenotype similar to that caused by the knock down of several genes encoding sarcomeric proteins [45]. To address the possibility that this phenotypic resemblance is a result of regulation of one or more of these proteins by HOW, we assessed the levels of different sarcomeric proteins in 3rd instar larvae, in which we reduced HOW levels using RNAi mediated knock-down in muscles. To enhance the effect of how down-regulation, the larvae used were also heterozygous for howstru.
Under these conditions, we observed an increase in the 500 kDa isoform of Sls, a giant protein that serves as a scaffold in the sarcomere, linking the Z-discs to the thick filaments [33] (Figure 6A). In contrast, the levels of MSP-300 (isoform 800 kDa), a Z-disc protein [46], [47], were decreased (Figure 6A). This suggests that in wild-type larval muscles, Sls is down-regulated by HOW, while MSP-300 is elevated. Although these results do not distinguish between direct and indirect effects, nonetheless, we used the levels of these two proteins in muscles as readout of the activity of phosphorylated (HOWWT) versus non-phosphorylated HOW (HOWTTAA) in this developmental process.
Fig. 6. HOW regulates the levels of Sls RNA and protein in a phosphorylation-dependent manner. 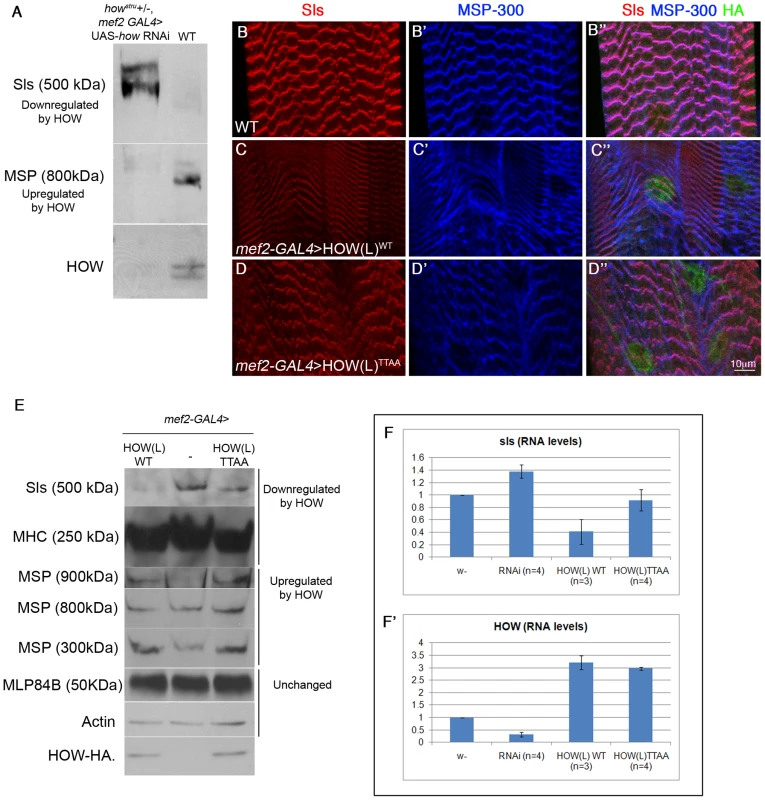
A. Sls and MSP-300 are directly or indirectly regulated by HOW. Extracts from larvae in which HOW was down-regulated by expressing how RNAi (aimed against all HOW isoforms) under mef2-GAL4, on the background of howstru heterozygotes, were reacted with anti-Sls, MSP-300, and HOW antibodies and compared to larvae heterozygotes for mef2-GAL4 in Western blot analysis. Down-regulation of HOW (lower panel) led to upregulation of Sls (upper panel) and down-regulation of MSP-300 (middle panel). B–D. Overexpression of HOW leads to reduction of Sls protein levels, in a phosphorylation-dependent manner. Body wall muscles from 3rd instar larvae expressing mef2-GAL4 alone (B,B′,B″), UAS-HOW(L)WT-HA, or UAS-HOW(L)TTAA-HA driven by the mef2-GAL4 driver (C,C′,C″ and D,D′,D″, respectively) were stained with anti-Sls (B,C,D - red), anti-MSP-300 (B′,C′,D′- blue) and anti-HA (B″,C″,D″ - green, shown within merge). Quantification of at least three larvae from each genotype showed that HOW(L)WT over-expression reduced Sls intensity to 0.71±0.05 of the intensity of the mef2 -GAL4 heterozygotes, while the HOW(L)TTAA protein only reduced it to 0.83±0.03 of control (P = 0.049, ANOVA test). E. Overexpression of HOW(L) in larval muscles alters the levels of multiple muscle proteins, of which only Sls is affected in a HOW phosphorylation-dependent manner. Protein extracts from 2nd instar larvae expressing mef2-GAL4 alone (middle), or either HOW(L)WT (left) or HOW(L)TTAA (right) driven by mef2-GAL4 were analyzed by Western blotting with the following antibodies (from top to bottom): anti-Sls, anti-MHC, anti-MSP300 (reacts with three different isoforms), anti-MLP84B, anti-Actin, and anti-HOW. Note the comparable levels of HOW(L)WT and HOW(L)TTAA expression, and the relatively mild reduction in Sls when HOW(L)TTAA is overexpressed. F–F′. HOW regulates Sls at the RNA level. RNA was extracted from single 3rd instar larvae, and real-time PCR was performed with sls primers (F) and how primers (F′), both normalized to rp49 as a control. From left to right: mef2-GAL4 heterozygotes (w−), howstru+/−, mef2>how RNAi (RNAi), and mef2-GAL4 driving either HOW(L)WT or HOW(L)TTAA. Error bars indicate SEM. While reduction of HOW levels elevated sls (1.38±0.11, P = 0.039), overexpression of HOW(L)WT reduced it more efficiently (0.54±0.20, P = 0.08) than HOW(L)TTAA (0.92±0.18, P = 0.78) (Student's t-test, n = 3 or 4). Accordingly, we expressed comparable amounts of either HOWWT or HOWTTAA in muscles (using the mef2-GAL4 driver) and followed the expression of Sls and MSP-300 by fluorescent labeling (Figure 6B–6D) or by Western blot analysis (Figure 6E) in 3rd instar larvae. As expected, over-expression of HOW(L)WT resulted in reduced levels of Sls (Figure 6C). This was confirmed by measuring the immunofluorescence intensity of Sls in Z-discs (see legend to Figure 6). Importantly, overexpression of HOW(L)TTAA exhibited a significantly milder effect on Sls levels (Figure 6D). In contrast, over-expression of either HOWWT or HOWTTAA resulted in a similar aberrant distribution of MSP-300 (Figure 6C′, 6D′).
We also quantified the changes in protein levels by employing Western blot analysis of extracts from the body walls of several larvae, expressing driver alone (mef2-GAL4), or together with HOWWT or HOWTTAA (Figure 6E). Consistent with the immunofluorescent staining, the anti-Sls antibody reacted with a major band of 500 kDa that was downregulated following HOW(L)WT overexpression. A much milder effect was observed following HOW(L)TTAA overexpression (Figure 6E, upper band). Myosin Heavy Chain (MHC) [48], [49] was reduced following expression of both HOWWT and HOWTTAA (Figure 6E second band from top). Of the three distinct bands representing MSP-300 (900 kDa, 800 kDa, 300 kDa), the 900 kDa and the 300 kDa bands were elevated, whereas that of 800 kDa was unaffected following HOWWT overexpression. No significant differences were found between the effect of HOWWT and HOWTTAA on all three bands. The levels of two additional muscle proteins, MLP84B [46] and Actin remained unchanged in these genetic backgrounds.
To conclude, the down-regulation of the newly-identified HOW target, Sls, is dependent on the phosphorylation state of HOW, whereas MSP-300 and MHC are regulated by HOW in a phosphorylation-independent manner. This discrepancy may be due to differential activities of HOW (e.g. regulation at the level of RNA degradation versus alternative splicing).
Given that HOW is an RBP, we next sought to determine whether HOW controls sls RNA levels, and if so, whether phosphorylation is important for this type of regulation. To this end, we purified total RNA from the body walls of single 3rd instar larvae and performed real-time PCR to quantify the levels of the sls and how transcripts (rp49 RNA served as control) using the SYBR green method. We compared larvae from a wild-type background (w−), to larvae heterozygous for howstru which expressed how RNAi in muscles, and to larvae over-expressing either HOW(L)WT or HOW(L)TTAA in muscles. In larvae expressing lower levels of how (Figure 6F′, second left bar, RNAi), sls mRNA was upregulated (Figure 6F). Importantly, HOW(L)WT was more efficient in the down-regulation of sls RNA relative to HOW(L)TTAA (Figure 6F right bars). Although this result was not statistically significant, the trend seen at the level of RNA is consistent with the results obtained in the protein analysis. Thus, our data demonstrate that HOW(L) has a role in the down-regulation of sls mRNA and protein levels in muscles, and that this process is dependent on HOW phosphorylation.
Downregulation of MAPK signaling in larval muscles results in diminished phosphorylation of HOW and elevation of Sallimus protein levels
We next examined whether HOW phosphorylation in muscles, and its resulting elevated activity in the regulation of Sls levels (Figure 6), are indeed dependent on MAPK signaling. To test this idea, we expressed RNAi for the Drosophila MAPK gene rolled in larval muscles under mef2-GAL4 regulation, immunoprecipitated HOW from protein extracts of these larvae and examined HOW phosphorylation using Western blot analysis with an anti-pTP antibody (Figure 7A). Importantly, in the larvae where rolled was down regulated (note the reduction in the levels of pERK in Figure 7B), we observed diminished phosphorylation of HOW, compared to wild-type larvae (Figure 7A). The phosphorylation of HOW in wild-type larvae was present in a band corresponding to the HOW(L) protein. Thus, phosphorylation of HOW(L) in larval muscles in vivo is dependent on MAPK signaling, as in S2R+ cells and in an in vitro kinase assay (Figure 2).
Fig. 7. Drosophila MAPK rolled is directing HOW phosphorylation in muscles and regulation of Sallimus levels. 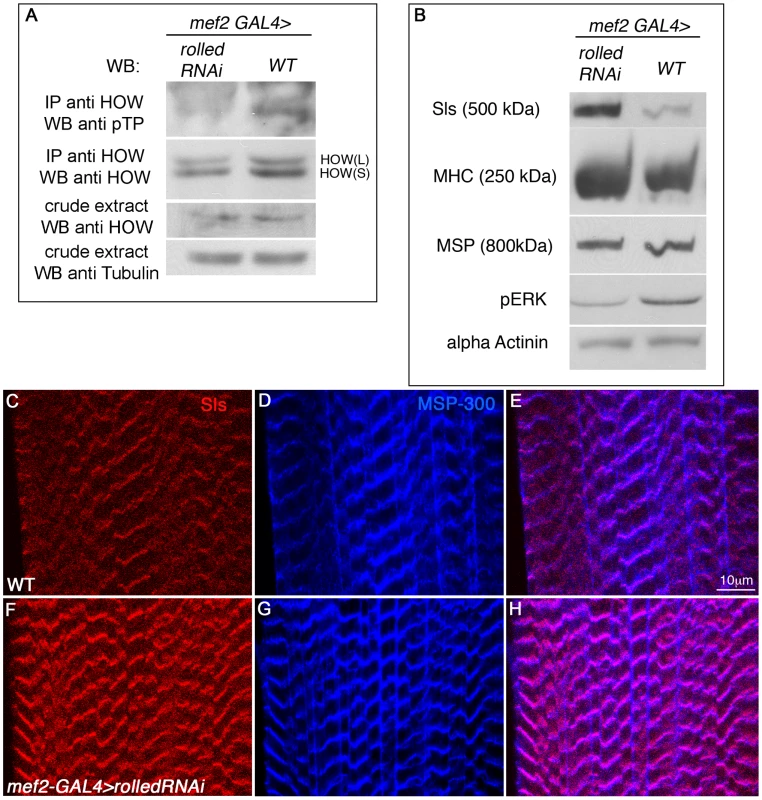
A. HOW was immunoprecipitated from lysates of 3rd instar larvae either expressing rolled RNAi in muscles under mef2-GAL4 (left) or of wild-type mef2-GAL4 heterozygotes (w−, right) and reacted with a pTP antibody to observe its phosphorylation level and with anti-HOW to compare total HOW levels. The crude extract was reacted with anti-HOW and anti-Tubulin as a loading control, showing that the total quantity of HOW is not significantly altered by MAPK/Rolled levels. Note the reduction in HOW phosphorylation following expression of rolled RNAi. B. Reduction in MAPK/Rolled levels elevates Sls protein levels. Same genotypes as in A were analyzed by Western with the following antibodies (from top to bottom): anti-Sls, anti-MHC, anti-MSP300, anti-pERK (to verify the reduction in activated MAPK in these larvae) and anti-α-Actinin as a loading control. C–H. Wild-type (C–E) and mef2-GAL4>rolled RNAi (F–H) larvae were stained with anti-Sls antibody (red, C,F) and anti MSP-300 (blue, D,G). Merge is shown in E,H. Note the significant elevation in Sls levels following down regulation of MAPK/Rolled levels. Since rolled down regulation resulted in decreased HOW(L) phosphorylation, we hypothesized that HOW(L) activity will be lowered in these larvae, resulting in elevated levels of Sls. Indeed, using a Western blot analysis (Figure 7B) we find that in larvae expressing rolled RNAi under mef2-GAL4, Sls levels are significantly elevated. The increase in Sls levels is specific, as only a mild increase is observed in MHC levels, while MSP and α-Actinin levels are unaffected (Figure 7B). The marked elevation of Sls levels following reduction of rolled in the larval muscles is also readily detectable in individual muscles (Figure 7C, 7F), while MSP-300 levels remain constant (Figure 7D, 7G, merge in Figure 7E, 7H). Thus, we conclude that in wild-type larval muscles, MAPK signaling is required for HOW(L) phosphorylation and reduction of Sls protein levels.
Discussion
STAR proteins regulate tissue differentiation in a wide range of species including nematodes [50], flies [17], Zebrafish [8], mice, and humans [51]. They function by controlling diverse posttranscriptional events, often forming specific protein-RNA complexes mediated by 3′UTR sequences of their target mRNAs, or with alternatively spliced introns [2]. In this study we reveal a molecular mechanism regulating the activity of the STAR protein HOW. Our findings demonstrate that HOW is phosphorylated on Thr residues embedded within conserved MAPK consensus sequences, both in cultured cells as well as in muscle cells and heart cardioblasts (Figure 2, 5), and that this phosphorylation is executed by MAPK/ERK (Figure 2, 7). Significantly, our results provide novel molecular insights to the importance of this phosphorylation, demonstrating that phosphorylation stabilizes HOW dimer formation (Figure 3). Moreover, because phosphorylation presumably occurs only on dimers, and phospho-dimers are more stable, we propose that a feed-forward loop ensures that a large fraction of HOW dimers are formed following a short temporal burst of MAPK/ERK activation. Importantly, since STAR proteins are evolutionarily conserved, the novel mode of regulation that we have uncovered might have important implications for other members of the QKI sub-family.
Two of the four potential MAPK/ERK docking sites reside within the HOW QUA1 domain. The close proximity between these sites, the MAPK/ERK phosphorylation sites, and the E106 residue critical for dimerization ([23], [24], Figure 3), all of which are highly evolutionarily conserved (Figure 1), is consistent with local conformational changes in the QUA1 domain induced by phosphorylation, which further lead to dimer formation and stabilization. HOW dimerization might be essential for a number of its characteristics; first, it enables binding to several HREs present on a single target RNA, leading to an overall higher affinity of HOW to its target RNA. For example, the 3′UTR of the HOW target stripe contains three consecutive HREs and a half site that resides between the first and second HREs. We have previously shown that a higher affinity of HOW is observed when multiple HREs are clustered together [43]. HOW dimerization and binding to two sites may also contribute to the formation of a secondary RNA structure, thus facilitating RNA processing. Interestingly, HOW dimerization apparently enhances its RNA binding ability even to a single binding site (Figure 4A).
Additionally, HOW dimerization might also potentiate the recruitment of other proteins/enzymes to the vicinity of the targeted mRNA. This may occur either if one subunit associates with the target RNA, and the other with a specific enzyme that induces modification/degradation of the target RNA, or if the formation of HOW dimers enables recruitment of proteins that would not associate with its monomeric form. A similar mechanism has been proposed to take place when GLD-1 dimers associate with their RNA target(s) close to the polyadenylation site, possibly recruiting an E3 ubiquitin ligase complex to this site. Ubiquitination of the polyadenylation complex would subsequently lead to shortening of the poly-A tail [24]. A mechanism for HOW activity that includes protein interactions with binding partners has yet to be described, but such an interaction is highly likely, since HOW does not contain any recognizable catalytic domains that could independently lead to mRNA degradation.
We show that a HOW mutant unable to form dimers exhibits a greatly diminished RNA binding capacity (Figure 4A, HOWEG mutant). This is also in line with our findings that HOW binding to RNA is significantly reduced when it is hypophosphorylated, and that phosphorylated HOW has a higher tendency to form dimers. We conclude that dimeric HOW binds RNA with higher affinity than monomeric HOW. This conclusion is in line with the observations that GLD-1 lacking its QUA1 domain has a lower affinity to RNA [26], and possibly applies to other STAR proteins.
To our knowledge, this is the first example of a positive effect of phosphorylation of a STAR protein on its RNA binding capacity. Phosphorylation of Sam68 by MAPK/ERK was demonstrated to have a subtle negative influence on its RNA binding [32], [52], while Tyrosine phosphorylation was shown to more severely impair RNA binding of both QKI [29] and Sam68 [53]. As the Tyrosine residues in HOW are highly conserved with those of QKI, it is highly likely that they are also phosphorylated (Kirenberg and Volk, unpublished data), and that this phosphorylation may have an opposite, negative effect on the ability of HOW to bind its targets. Hence, it is interesting to note that changes in cellular signaling may have the capacity to fine-tune, both positively and negatively, the ability of an RNA binding protein to bind its targets, thus modulating the levels of a variety of mRNAs.
A requirement for HOW in developing muscles had been demonstrated previously [45], but its target RNAs in this tissue have not been characterized. In this study, we identify three different muscle proteins, Sls, MSP-300 and MHC, whose levels are altered by the expression of HOW in this developmental setting. We still do not know whether the mRNAs of these proteins are all directly bound by HOW, and at what level the regulation by HOW occurs, i.e. via control of specific alternative splicing or by regulation of overall mRNA levels. Since sls mRNA levels respond to both HOW overexpression and knock-down, and given that the sls transcript contains several potential binding sites for HOW (both at the 3′UTR as well as within several introns; not shown), it likely represents a true direct RNA target of HOW.
Even though Sls is a structural protein, its fast turnover in sarcomeres might be essential for maintenance of the sarcomeric architecture [54]. To fulfill this requirement, its protein and RNA half-life should be short. Thus, HOW might play an essential role in promoting destabilization of sls mRNA to promote fast exchange of newly formed Sls protein at the Z-disc. Indeed, in muscles where MAPK signaling was downregulated using rolled RNAi, a significant elevation in Sls levels is clearly evident (Figure 7). This result is in line with tight regulation of Sls levels occurring in wild-type larvae.
Although the precise RTK signaling pathway that regulates HOW phosphorylation in muscles is yet to be elucidated, the FGFR Heartless, which is expressed by muscle cells throughout their development [55], [56], is an attractive candidate. It is possible that continuous FGFR activation in muscles promotes HOW phosphorylation by MAPK, rendering it more active in controlling the levels of its target mRNAs in this tissue.
In summary, in this study we have unraveled and characterized a novel molecular mechanism at the basis of the activity of the STAR protein, HOW. By linking its activity to MAPK/ERK-dependent phosphorylation and regulation, we provide a mechanistic linkage between HOW phosphorylation, the degree of its dimerization and its biological activity/function. We propose that this mechanism may apply to other STAR proteins, in which the dimerization domain and phosphorylation sites are evolutionarily conserved.
Materials and Methods
Expression constructs for cell lines and flies
Mutant HOW(L) constructs (TTAA, EG) were created by site directed mutagenesis (Stratagene), following the manufacturer's protocols. pUAST-HOW(L) constructs tagged with HA or fused with GFP were generated in the pTWH and pTGW Drosophila Gateway vectors, respectively (T. Murphy, Carnegie Institution of Washington) using the Gateway cloning system (Invitrogen).
Fly strains
Fly stocks used in this study include w−, mef2-GAL4 (Bloomington stock center), mef2-GAL4, UAS-CD8 GFP (F. Schnorrer, Martinsried, Germany), stripe-GAL4 (G. Morata, Madrid, Spain), howstru [57], how RNAi line (ds-HOW) [43], rolled RNAi (TRiP HMS00173, Bloomington stock 34855). UAS-how(l)WT-3HA and TTAA-3HA were injected to flies by Genetic Services, Sudbury, MA, USA.
Antibodies
Primary antibodies used in this study include mouse anti-phospho-Thr-Pro (p-Thr-Pro-101) (Cell Signaling Technology), rabbit and rat anti-HOW [39], guinea pig anti-MSP-300 [47], rat anti-Sls (Kettin) (Klg16, MAC155, Abcam), rat anti α-Actinin (Abcam), rabbit anti-MHC (P. Fisher, Stony Brook, NY), mouse anti-Actin (Sigma), rabbit anti-Mlp84bB [58], mouse anti-GFP (Roche), mouse anti-HA (Roche), chick anti-HA (Aves Labs), mouse anti-Lamin ([59], gift from Y. Gruenbaum, Hebrew University), mouse anti-pERK (gift from B. Shilo, Weizmann Institute). The polyclonal anti-pHOW (pT64) antibody was raised in rats against the phospho-peptide PQHL(p)TPQQ (generated by Sigma), corresponding to amino acids 60–67 of HOW. Immunizations were performed by the antibody unit at the Weizmann Institute. The serum was cleaned on beads conjugated to a similar peptide, PQHLQPQQ, in an attempt to reduce background staining that was suspected to be due to another protein containing this sequence. Secondary antibodies used in this study include various Cy3, Cy2, Cy5 and HRP-conjugated antibodies (Jackson ImmunoResearch Laboratories, USA).
Tissue culture and transfection
S2R+ cells were grown and transfected essentially as previously described [43], except that cells were usually collected for analysis 36 hours after transfection. For inhibition of MAPK/ERK activity, 10 µM U0126 (Sigma) in DMSO (or only DMSO for controls) was administered with the serum-containing media, about 18 h after the transfection. Since U0126 led to a decrease in the efficiency of recovery from transfection, the control cells were transfected with smaller amounts of how DNA. Phorbol 12-Myristate 13-Acetate (TPA/PMA) (Sigma) treatment (10 µM in DMSO, or only DMSO for controls) was performed on starved cells, 20 h after transfection, for 15 min.
Cell lysis
Cells were scraped off the flasks and collected in PBS, washed twice and lysed in 1% NP40 lysis buffer (50 mM Tris pH 7.5, 1% NP40, 100 mM NaCl, 1% Protease inhibitor cocktail (P8340, Sigma), 0.5% Phosphatase Inhibitor Cocktail 1(P2850, Sigma), 20 mM β -glycerol phosphate).
Embryos and larvae were collected and crushed in RIPA buffer (1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 0.15M NaCl, 0.05M Tris pH 7.0, supplemented with the same inhibitors).
Western blotting and immunoprecipitation (IP)
For IP, equal amounts of protein lysate were incubated with protein A/G beads (SC-2003, Santa Cruz) coupled with either rabbit anti-HOW polyclonal antibody or mouse anti-GFP monoclonal antibody, or agarose HA conjugated beads (A2095, Sigma) for 1–2 h at 4°C. Beads were washed three times with the NP40 lysis buffer, and boiled in protein sample buffer to elute the proteins.
MAPK/ERK in vitro phosphorylation assay was performed as described [60]. High molecular weight proteins were analyzed by SDS–PAGE using 2.5% acrylamide gels strengthened with 1.5% agarose, essentially as in [61].
Fixation and immunostaining
Fixation and staining of embryos were done following standard procedures. Larval Flat Preparations were performed essentially as described [62], except the fixation was done in 4% PFA, the primary antibody staining was carried out for 2 h and the secondary for 1 h. Quantification of average intensity of larval muscles images were performed using MATLAB.
S2R+ cells were seeded on Ibidi u-Slide 8 well (0.2*106 cells per well), fixed with 3% PFA for 5 min, permeabilized using 3% PFA+0.1% TritonX-100, stained with primary antibody (1∶200) for 30 min, and secondary antibody (1∶400) for 30 min.
Visualization was carried out using a Zeiss LSM710 confocal system.
RT–PCR analysis of RNA extracts
Single larvae were flipped over and their interior was cleaned, leaving the carcass only. RNA was extracted by the Nucleospin Purification Kit (Macherey-Nagel) according to the manufacturer's instructions. Equal amounts were used as a template for cDNA preparation using the Verso cDNA kit (Thermo Scientific). Real time PCR was carried out using Fast SYBR Green Mastermix (Applied Biosystems) in a StepOne plus machine (Applied Biosystems). The following primers were used:
sls (TGCCCATGCCGAAGACA) and (TGTCTTGTTTGCTGTTACGTTTACAG),
rp49 (GACCATCCGCCCAGCATAC) and (CCATTTGTGCGACAGCTTAGC),
how (AACTTTGTCGGTCGCATTTT) and (CGTCCTCCTTCTTCTTGTCG).
RNA–protein binding
In vitro RNA–protein binding assay was performed essentially as described [43], only that the proteins were not in vitro translated, but were purified from S2R+ cells using immunoprecipitation with HA conjugated beads, followed by elution with an HA peptide (Sigma, I2149).
Software
Phosphorylation sites were predicted by the GPS2.1 program [63]. Protein scheme was generated by DOG2.0 software [64]. Student's t-tests were performed using GraphPad software.
Zdroje
1. GlisovicTBachorikJLYongJDreyfussG 2008 RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582 1977 1986
2. LaskoP 2003 Gene regulation at the RNA layer: RNA binding proteins in intercellular signaling networks. Sci STKE 2003 RE6
3. VernetCArtztK 1997 STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet 13 479 484
4. ParonettoMPMessinaVBianchiEBarchiMVogelG 2009 Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J Cell Biol 185 235 249
5. SidmanRLDickieMMAppelSH 1964 Mutant Mice (Quaking and Jimpy) with Deficient Myelination in the Central Nervous System. Science 144 309 311
6. SuzukiKZagorenJC 1977 Quaking mouse: an ultrastructural study of the peripheral nerves. J Neurocytol 6 71 84
7. EbersoleTAChenQJusticeMJArtztK 1996 The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat Genet 12 260 265
8. LobbardiRLambertGZhaoJGeislerRKimHR 2011 Fine-tuning of Hh signaling by the RNA-binding protein Quaking to control muscle development. Development 138 1783 1794
9. FrancisRBartonMKKimbleJSchedlT 1995 gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139 579 606
10. OhnoGHagiwaraMKuroyanagiH 2008 STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes Dev 22 360 374
11. BaehreckeEH 1997 who encodes a KH RNA binding protein that functions in muscle development. Development 124 1323 1332
12. FyrbergCBeckerJBarthmaierPMahaffeyJFyrbergE 1997 A Drosophila muscle-specific gene related to the mouse quaking locus. Gene 197 315 323
13. LoPCFraschM 1997 A novel KH-domain protein mediates cell adhesion processes in Drosophila. Dev Biol 190 241 256
14. ZaffranSAstierMGratecosDSemerivaM 1997 The held out wings (how) Drosophila gene encodes a putative RNA-binding protein involved in the control of muscular and cardiac activity. Development 124 2087 2098
15. EdenfeldGVolohonskyGKrukkertKNaffinELammelU 2006 The splicing factor crooked neck associates with the RNA-binding protein HOW to control glial cell maturation in Drosophila. Neuron 52 969 980
16. ReuvenyAElhananyHVolkT 2009 Enhanced sensitivity of midline glial cells to apoptosis is achieved by HOW(L)-dependent repression of Diap1. Mech Dev 126 30 41
17. VolkT 2010 Drosophila star proteins: what can be learned from flies? Adv Exp Med Biol 693 93 105
18. VolohonskyGEdenfeldGKlambtCVolkT 2007 Muscle-dependent maturation of tendon cells is induced by post-transcriptional regulation of stripeA. Development 134 347 356
19. Nabel-RosenHToledano-KatchalskiHVolohonskyGVolkT 2005 Cell divisions in the drosophila embryonic mesoderm are repressed via posttranscriptional regulation of string/cdc25 by HOW. Curr Biol 15 295 302
20. LiuZLuytenIBottomleyMJMessiasACHoungninou-MolangoS 2001 Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294 1098 1102
21. MaguireMLGuler-GaneGNietlispachDRaineARZornAM 2005 Solution structure and backbone dynamics of the KH-QUA2 region of the Xenopus STAR/GSG quaking protein. J Mol Biol 348 265 279
22. GarreySMCassDMWandlerAMScanlanMSBerglundJA 2008 Transposition of two amino acids changes a promiscuous RNA binding protein into a sequence-specific RNA binding protein. RNA 14 78 88
23. ChenTRichardS 1998 Structure-function analysis of Qk1: a lethal point mutation in mouse quaking prevents homodimerization. Mol Cell Biol 18 4863 4871
24. BeuckCSzymczynaBRKerkowDECarmelABColumbusL 2010 Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation. Structure 18 377 389
25. MeyerNHTripsianesKVincendeauMMadlTKatebF 2010 Structural basis for homodimerization of the Src-associated during mitosis, 68-kDa protein (Sam68) Qua1 domain. J Biol Chem 285 28893 28901
26. RyderSPFraterLAAbramovitzDLGoodwinEBWilliamsonJR 2004 RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat Struct Mol Biol 11 20 28
27. SetteC 2010 Post-translational regulation of star proteins and effects on their biological functions. Adv Exp Med Biol 693 54 66
28. WongGMullerOClarkRConroyLMoranMF 1992 Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell 69 551 558
29. ZhangYLuZKuLChenYWangH 2003 Tyrosine phosphorylation of QKI mediates developmental signals to regulate mRNA metabolism. Embo J 22 1801 1810
30. ResnickRJTaylorSJLinQShallowayD 1997 Phosphorylation of the Src substrate Sam68 by Cdc2 during mitosis. Oncogene 15 1247 1253
31. MatterNHerrlichPKonigH 2002 Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420 691 695
32. ParonettoMPZalfaFBottiFGeremiaRBagniC 2006 The nuclear RNA-binding protein Sam68 translocates to the cytoplasm and associates with the polysomes in mouse spermatocytes. Mol Biol Cell 17 14 24
33. BurkartCQiuFBrendelSBenesVHaagP 2007 Modular proteins from the Drosophila sallimus (sls) gene and their expression in muscles with different extensibility. J Mol Biol 367 953 969
34. LakeyALabeitSGautelMFergusonCBarlowDP 1993 Kettin, a large modular protein in the Z-disc of insect muscles. EMBO J 12 2863 2871
35. BardwellLThornerJ 1996 A conserved motif at the amino termini of MEKs might mediate high-affinity interaction with the cognate MAPKs. Trends Biochem Sci 21 373 374
36. JacobsDGlossipDXingHMuslinAJKornfeldK 1999 Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev 13 163 175
37. SharrocksADYangSHGalanisA 2000 Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem Sci 25 448 453
38. RebayIRubinGM 1995 Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell 81 857 866
39. Nabel-RosenHDorevitchNReuvenyAVolkT 1999 The balance between two isoforms of the Drosophila RNA-binding protein how controls tendon cell differentiation. Mol Cell 4 573 584
40. DunciaJVSantellaJB3rdHigleyCAPittsWJWityakJ 1998 MEK inhibitors: the chemistry and biological activity of U0126, its analogs, and cyclization products. Bioorg Med Chem Lett 8 2839 2844
41. CastagnaMTakaiYKaibuchiKSanoKKikkawaU 1982 Direct activation of calcium-activated, phospholipid-dependent protein kinase by tumor-promoting phorbol esters. J Biol Chem 257 7847 7851
42. SchonwasserDCMaraisRMMarshallCJParkerPJ 1998 Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel, and atypical protein kinase C isotypes. Mol Cell Biol 18 790 798
43. IsraeliDNirRVolkT 2007 Dissection of the target specificity of the RNA-binding protein HOW reveals dpp mRNA as a novel HOW target. Development 134 2107 2114
44. GabayLSegerRShiloBZ 1997 In situ activation pattern of Drosophila EGF receptor pathway during development. Science 277 1103 1106
45. SchnorrerFSchonbauerCLangerCCDietzlGNovatchkovaM 2010 Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 464 287 291
46. ClarkKABlandJMBeckerleMC 2007 The Drosophila muscle LIM protein, Mlp84B, cooperates with D-titin to maintain muscle structural integrity. J Cell Sci 120 2066 2077
47. VolkT 1992 A new member of the spectrin superfamily may participate in the formation of embryonic muscle attachments in Drosophila. Development 116 721 730
48. BernsteinSIMogamiKDonadyJJEmersonCPJr 1983 Drosophila muscle myosin heavy chain encoded by a single gene in a cluster of muscle mutations. Nature 302 393 397
49. RozekCEDavidsonN 1983 Drosophila has one myosin heavy-chain gene with three developmentally regulated transcripts. Cell 32 23 34
50. LeeMHSchedlT 2010 C. elegans star proteins, GLD-1 and ASD-2, regulate specific RNA targets to control development. Adv Exp Med Biol 693 106 122
51. ArtztKWuJI 2010 STAR trek: An introduction to STAR family proteins and review of quaking (QKI). Adv Exp Med Biol 693 1 24
52. TisserantAKonigH 2008 Signal-regulated Pre-mRNA occupancy by the general splicing factor U2AF. PLoS ONE 3 e1418 doi:10.1371/journal.pone.0001418
53. WangLLRichardSShawAS 1995 P62 association with RNA is regulated by tyrosine phosphorylation. J Biol Chem 270 2010 2013
54. HaasKFWoodruffE3rdBroadieK 2007 Proteasome function is required to maintain muscle cellular architecture. Biol Cell 99 615 626
55. MichelsonAMGisselbrechtSZhouYBaekKHBuffEM 1998 Dual functions of the heartless fibroblast growth factor receptor in development of the Drosophila embryonic mesoderm. Dev Genet 22 212 229
56. ShishidoEOnoNKojimaTSaigoK 1997 Requirements of DFR1/Heartless, a mesoderm-specific Drosophila FGF-receptor, for the formation of heart, visceral and somatic muscles, and ensheathing of longitudinal axon tracts in CNS. Development 124 2119 2128
57. ProutMDamaniaZSoongJFristromDFristromJW 1997 Autosomal mutations affecting adhesion between wing surfaces in Drosophila melanogaster. Genetics 146 275 285
58. StronachBESiegristSEBeckerleMC 1996 Two muscle-specific LIM proteins in Drosophila. J Cell Biol 134 1179 1195
59. MillerKGKarrTLKelloggDRMohrIJWalterM 1985 Studies on the cytoplasmic organization of early Drosophila embryos. Cold Spring Harb Symp Quant Biol 50 79 90
60. KimYCoppeyMGrossmanRAjuriaLJimenezG 2010 MAPK substrate competition integrates patterning signals in the Drosophila embryo. Curr Biol 20 446 451
61. TatsumiRHattoriA 1995 Detection of giant myofibrillar proteins connectin and nebulin by electrophoresis in 2% polyacrylamide slab gels strengthened with agarose. Anal Biochem 224 28 31
62. SubramanianAProkopAYamamotoMSugimuraKUemuraT 2003 Shortstop recruits EB1/APC1 and promotes microtubule assembly at the muscle-tendon junction. Curr Biol 13 1086 1095
63. XueYRenJGaoXJinCWenL 2008 GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics 7 1598 1608
64. RenJWenLGaoXJinCXueY 2009 DOG 1.0: illustrator of protein domain structures. Cell Res 19 271 273
Štítky
Genetika Reprodukční medicína
Článek Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1Článek Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / TranscriptionČlánek Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding DomainČlánek Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin ComplexesČlánek An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood ObesityČlánek Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAs
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 3
-
Všechny články tohoto čísla
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
- Genomic Analysis of the Hydrocarbon-Producing, Cellulolytic, Endophytic Fungus
- Networks of Neuronal Genes Affected by Common and Rare Variants in Autism Spectrum Disorders
- Akirin Links Twist-Regulated Transcription with the Brahma Chromatin Remodeling Complex during Embryogenesis
- Too Much Cleavage of Cyclin E Promotes Breast Tumorigenesis
- Imprinted Genes … and the Number Is?
- Genetic Architecture of Highly Complex Chemical Resistance Traits across Four Yeast Strains
- Exploring the Complexity of the HIV-1 Fitness Landscape
- MNS1 Is Essential for Spermiogenesis and Motile Ciliary Functions in Mice
- A Fundamental Regulatory Mechanism Operating through OmpR and DNA Topology Controls Expression of Pathogenicity Islands SPI-1 and SPI-2
- Evidence for Positive Selection on a Number of MicroRNA Regulatory Interactions during Recent Human Evolution
- Variation in Modifies Risk of Neonatal Intestinal Obstruction in Cystic Fibrosis
- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Critical Evaluation of Imprinted Gene Expression by RNA–Seq: A New Perspective
- A Meta-Analysis and Genome-Wide Association Study of Platelet Count and Mean Platelet Volume in African Americans
- Mouse Genetics Suggests Cell-Context Dependency for Myc-Regulated Metabolic Enzymes during Tumorigenesis
- Transcriptional Control in Cardiac Progenitors: Tbx1 Interacts with the BAF Chromatin Remodeling Complex and Regulates
- Synthetic Lethality of Cohesins with PARPs and Replication Fork Mediators
- APOBEC3G-Induced Hypermutation of Human Immunodeficiency Virus Type-1 Is Typically a Discrete “All or Nothing” Phenomenon
- Interpreting Meta-Analyses of Genome-Wide Association Studies
- Error-Prone ZW Pairing and No Evidence for Meiotic Sex Chromosome Inactivation in the Chicken Germ Line
- -Dependent Chemosensory Functions Contribute to Courtship Behavior in
- Diverse Forms of Splicing Are Part of an Evolving Autoregulatory Circuit
- Phenotypic Plasticity of the Drosophila Transcriptome
- Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1
- Precocious Metamorphosis in the Juvenile Hormone–Deficient Mutant of the Silkworm,
- Igf1r Signaling Is Indispensable for Preimplantation Development and Is Activated via a Novel Function of E-cadherin
- Accurate Prediction of Inducible Transcription Factor Binding Intensities In Vivo
- Mitochondrial Oxidative Stress Alters a Pathway in Strongly Resembling That of Bile Acid Biosynthesis and Secretion in Vertebrates
- Mammalian Neurogenesis Requires Treacle-Plk1 for Precise Control of Spindle Orientation, Mitotic Progression, and Maintenance of Neural Progenitor Cells
- Tcf7 Is an Important Regulator of the Switch of Self-Renewal and Differentiation in a Multipotential Hematopoietic Cell Line
- REST–Mediated Recruitment of Polycomb Repressor Complexes in Mammalian Cells
- Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / Transcription
- Age-Dependent Brain Gene Expression and Copy Number Anomalies in Autism Suggest Distinct Pathological Processes at Young Versus Mature Ages
- A Genome-Wide Association Study Identifies Variants Underlying the Shade Avoidance Response
- -by- Regulatory Divergence Causes the Asymmetric Lethal Effects of an Ancestral Hybrid Incompatibility Gene
- Genome-Wide Association and Functional Follow-Up Reveals New Loci for Kidney Function
- A Natural System of Chromosome Transfer in
- Cell Size and the Initiation of DNA Replication in Bacteria
- Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding Domain
- Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin Complexes
- Temporal Transcriptional Profiling of Somatic and Germ Cells Reveals Biased Lineage Priming of Sexual Fate in the Fetal Mouse Gonad
- Rapid Analysis of Genome Rearrangements by Multiplex Ligation–Dependent Probe Amplification
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- The Atypical Calpains: Evolutionary Analyses and Roles in Cellular Degeneration
- The Silkworm Coming of Age—Early
- Development of a Panel of Genome-Wide Ancestry Informative Markers to Study Admixture Throughout the Americas
- Balanced Codon Usage Optimizes Eukaryotic Translational Efficiency
- The Min System and Nucleoid Occlusion Are Not Required for Identifying the Division Site in but Ensure Its Efficient Utilization
- Neurobeachin, a Regulator of Synaptic Protein Targeting, Is Associated with Body Fat Mass and Feeding Behavior in Mice and Body-Mass Index in Humans
- Statistical Analysis of Readthrough Levels for Nonsense Mutations in Mammalian Cells Reveals a Major Determinant of Response to Gentamicin
- Gene Reactivation by 5-Aza-2′-Deoxycytidine–Induced Demethylation Requires SRCAP–Mediated H2A.Z Insertion to Establish Nucleosome Depleted Regions
- The miR-35-41 Family of MicroRNAs Regulates RNAi Sensitivity in
- Genetic Basis of Hidden Phenotypic Variation Revealed by Increased Translational Readthrough in Yeast
- An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood Obesity
- Modelling Human Regulatory Variation in Mouse: Finding the Function in Genome-Wide Association Studies and Whole-Genome Sequencing
- Novel Loci for Adiponectin Levels and Their Influence on Type 2 Diabetes and Metabolic Traits: A Multi-Ethnic Meta-Analysis of 45,891 Individuals
- Polycomb-Like 3 Promotes Polycomb Repressive Complex 2 Binding to CpG Islands and Embryonic Stem Cell Self-Renewal
- Insulin/IGF-1 and Hypoxia Signaling Act in Concert to Regulate Iron Homeostasis in
- EMF1 and PRC2 Cooperate to Repress Key Regulators of Arabidopsis Development
- Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAs
- Contrasted Patterns of Molecular Evolution in Dominant and Recessive Self-Incompatibility Haplotypes in
- A Machine Learning Approach for Identifying Novel Cell Type–Specific Transcriptional Regulators of Myogenesis
- Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
- Nos2 Inactivation Promotes the Development of Medulloblastoma in Mice by Deregulation of Gap43–Dependent Granule Cell Precursor Migration
- Intracranial Aneurysm Risk Locus 5q23.2 Is Associated with Elevated Systolic Blood Pressure
- Heritability and Genetic Correlations Explained by Common SNPs for Metabolic Syndrome Traits
- A Genome-Wide Association Study of Nephrolithiasis in the Japanese Population Identifies Novel Susceptible Loci at 5q35.3, 7p14.3, and 13q14.1
- DNA Damage in Nijmegen Breakage Syndrome Cells Leads to PARP Hyperactivation and Increased Oxidative Stress
- DNA Resection at Chromosome Breaks Promotes Genome Stability by Constraining Non-Allelic Homologous Recombination
- Genetic Analysis of Floral Symmetry in Van Gogh's Sunflowers Reveals Independent Recruitment of Genes in the Asteraceae
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Promoter Nucleosome Organization Shapes the Evolution of Gene Expression
- The Nucleoside Diphosphate Kinase Gene Acts as Quantitative Trait Locus Promoting Non-Mendelian Inheritance
- The Ciliogenic Transcription Factor RFX3 Regulates Early Midline Distribution of Guidepost Neurons Required for Corpus Callosum Development
- Phosphorylation of the RNA–Binding Protein HOW by MAPK/ERK Enhances Its Dimerization and Activity
- A Genome-Wide Scan of Ashkenazi Jewish Crohn's Disease Suggests Novel Susceptibility Loci
- Parkinson's Disease–Associated Kinase PINK1 Regulates Miro Protein Level and Axonal Transport of Mitochondria
- LMW-E/CDK2 Deregulates Acinar Morphogenesis, Induces Tumorigenesis, and Associates with the Activated b-Raf-ERK1/2-mTOR Pathway in Breast Cancer Patients
- Mapping the Hsp90 Genetic Interaction Network in Reveals Environmental Contingency and Rewired Circuitry
- Autoregulation of the Noncoding RNA Gene
- The Human Pancreatic Islet Transcriptome: Expression of Candidate Genes for Type 1 Diabetes and the Impact of Pro-Inflammatory Cytokines
- Spo0A∼P Imposes a Temporal Gate for the Bimodal Expression of Competence in
- Antagonistic Regulation of Apoptosis and Differentiation by the Cut Transcription Factor Represents a Tumor-Suppressing Mechanism in
- A Downstream CpG Island Controls Transcript Initiation and Elongation and the Methylation State of the Imprinted Macro ncRNA Promoter
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



