-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGenome-Wide Association and Functional Follow-Up Reveals New Loci for Kidney Function
Chronic kidney disease (CKD) is an important public health problem with a genetic component. We performed genome-wide association studies in up to 130,600 European ancestry participants overall, and stratified for key CKD risk factors. We uncovered 6 new loci in association with estimated glomerular filtration rate (eGFR), the primary clinical measure of CKD, in or near MPPED2, DDX1, SLC47A1, CDK12, CASP9, and INO80. Morpholino knockdown of mpped2 and casp9 in zebrafish embryos revealed podocyte and tubular abnormalities with altered dextran clearance, suggesting a role for these genes in renal function. By providing new insights into genes that regulate renal function, these results could further our understanding of the pathogenesis of CKD.
Published in the journal: . PLoS Genet 8(3): e32767. doi:10.1371/journal.pgen.1002584
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002584Summary
Chronic kidney disease (CKD) is an important public health problem with a genetic component. We performed genome-wide association studies in up to 130,600 European ancestry participants overall, and stratified for key CKD risk factors. We uncovered 6 new loci in association with estimated glomerular filtration rate (eGFR), the primary clinical measure of CKD, in or near MPPED2, DDX1, SLC47A1, CDK12, CASP9, and INO80. Morpholino knockdown of mpped2 and casp9 in zebrafish embryos revealed podocyte and tubular abnormalities with altered dextran clearance, suggesting a role for these genes in renal function. By providing new insights into genes that regulate renal function, these results could further our understanding of the pathogenesis of CKD.
Introduction
Chronic kidney disease (CKD) affects nearly 10% of the global population [1], [2], and its prevalence continues to increase [3]. Reduced estimated glomerular filtration rate (eGFR), the primary measure used to define CKD (eGFR<60 ml/min/1.73 m2) [4], is associated with an increased risk of cardiovascular morbidity and mortality [5], acute kidney injury [6], and end stage renal disease (ESRD) [6], [7].
Using genome-wide association studies (GWAS) in predominantly population-based cohorts, we and others have previously identified more than 20 genetic loci associated with eGFR and CKD [8]–[11]. Although most of these genetic effects seem largely robust across strata of diabetes or hypertension status [9], evidence suggests that some of the loci such as the UMOD locus may have heterogeneous effects across these strata [11]. We thus hypothesized that GWAS in study populations stratified by four key CKD risk factors - age, sex, diabetes or hypertension status - may permit the identification of novel eGFR and CKD loci. We carried this out by extending our previous work [9] to a larger discovery sample of 74,354 individuals with independent replication in additional 56,246 individuals, resulting in a total of 130,600 individuals of European ancestry. To assess for potential heterogeneity, we performed separate genome-wide association analyses across strata of CKD risk factors, as well as in a more extreme CKD phenotype.
Results
Meta-analyses of GWAS on the 22 autosomes were performed for: 1) eGFR based on serum creatinine (eGFRcrea) and CKD (6,271 cases) in the overall sample, 2) eGFRcrea and CKD stratified by the four risk factors, and 3) CKD45, a more severe CKD phenotype defined as eGFRcrea <45 ml/min/1.73 m2 in the overall sample (2,181 cases). For the stratified analyses, in addition to identifying loci that were significant within each stratum, we performed a genome-wide comparison of the effect estimates between strata of the four risk factors. A complete overview of the analysis workflow is given in Figure S1. All studies participating in the stage 1 discovery and stage 2 replication phases are listed in Tables S1 and S2. The characteristics of all stage 1 discovery samples by study are reported in Table S3, and information on study design and genotyping are reported in Table S4. Results of the eGFRcrea analyses are summarized in the Manhattan and quantile-quantile plots reported in Figures S2 and S3. A total of 21 SNPs from the discovery stage were carried forward for replication in an independent set of 56,246 individuals (Tables S5 and S6). These SNPs were selected for replication for the following (Figure S1): 5 reached genome-wide significance in either eGFRcrea overall or stratified analyses, 1 based on a test of direction-consistency of SNP-eGFR associations across the discovery cohorts for eGFRcrea overall, 4 demonstrated a P value≤10−6 and high between-study homogeneity (I2<25%) in the CKD45 analysis (Table S7), and 11 demonstrated between-strata P value≤5×10−5 along with a P value≤5×10−5 for association with eGFRcrea in at least one of the two strata (Table S8).
While none of the loci identified for CKD45 or the test for between-strata difference analyses replicated, all 6 loci identified from the eGFRcrea overall analysis, stratified analyses, and the direction test did (Table 1). These 6 loci were identified and replicated in the overall analysis (rs3925584, located upstream of the MPPED2 gene; rs6431731 near the DDX1 gene), in the diabetes-free sub-group (rs2453580 in an intron of the SLC47A1 gene), in the younger age stratum (rs11078903 in an intron of the CDK12 gene; rs12124078 located near the CASP9 gene), and the direction test (rs2928148, located in the INO80 gene, see Methods for details). In the combined meta-analysis of all 45 studies used in the discovery and replication stages, all six SNPs met the genome-wide significance threshold of 5×10−8, with individual P values ranging from 4.3×10−8 to 8.4×10−18 (Table 1). The imputation quality of these SNPs is reported in Table S9, and Figure S4 shows the regional association plots for each of the 6 loci. We also confirmed all previously identified renal function loci in the current data (Table S10). Brief descriptions of the genes included within the 6 new loci uncovered can be found in Table S11. Forest plots for the associations between the index SNP at each of the 6 novel loci and eGFR across all discovery studies and all strata are presented in Figures S5 and S6. Most of the 6 new loci had similar associations across strata of CKD risk factors except for the CDK12 locus, which revealed stronger association in the younger (≤65 years of age) as compared to the older age group (>65 years of age).
Tab. 1. Novel loci associated with eGFRcrea. 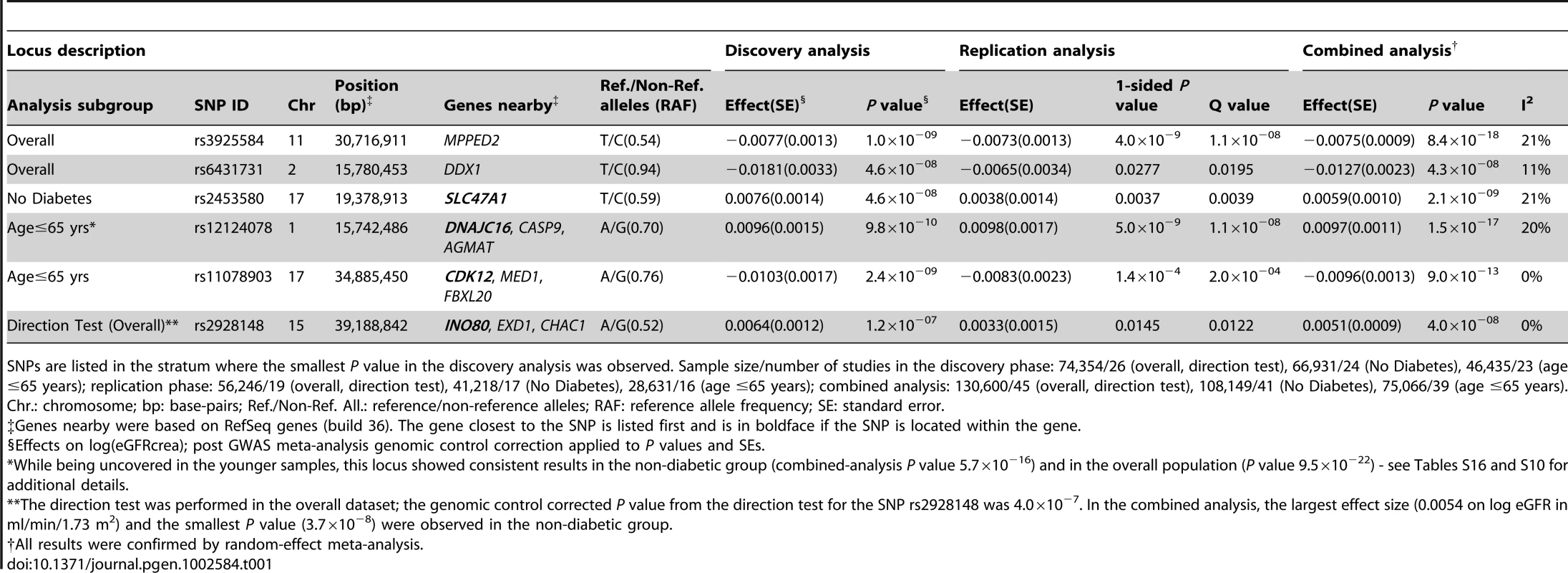
SNPs are listed in the stratum where the smallest P value in the discovery analysis was observed. Sample size/number of studies in the discovery phase: 74,354/26 (overall, direction test), 66,931/24 (No Diabetes), 46,435/23 (age ≤65 years); replication phase: 56,246/19 (overall, direction test), 41,218/17 (No Diabetes), 28,631/16 (age ≤65 years); combined analysis: 130,600/45 (overall, direction test), 108,149/41 (No Diabetes), 75,066/39 (age ≤65 years). We further examined our findings in 8,110 African ancestry participants from the CARe consortium [12] (Table 2). Not surprisingly, given linkage disequilibrium (LD) differences between Europeans and African Americans, none of the 6 lead SNPs uncovered in CKDGen achieved significance in the African American samples. Next, we interrogated the 250 kb flanking regions from the lead SNP at each locus, and showed that 4 of the 6 regions (MPPED2, DDX1, SLC47A1, and CDK12) harbored SNPs that achieved statistical significance after correcting for multiple comparisons based on the genetic structure of each region (see Methods for details). Figure 1 presents the regional association plots for MPPED2, and Figure S7 presents the plots of the remaining loci in the African American sample. Imputation scores for the lead SNPs can be found in Table S12. We observed that rs12278026, upstream of MPPED2, was associated with eGFRcrea in African Americans (P value = 5×10−5, threshold for statistical significance: P value = 0.001). While rs12278026 is monomorphic in the CEU population in HapMap, rs3925584 and rs12278026 have a D′ of 1 (r2 = 0.005) in the YRI population, suggesting that these SNPs may have arisen from the same ancestral haplotype.
Fig. 1. Genetic association and LD distribution of the MPPED2 gene locus in European and African ancestry populations. 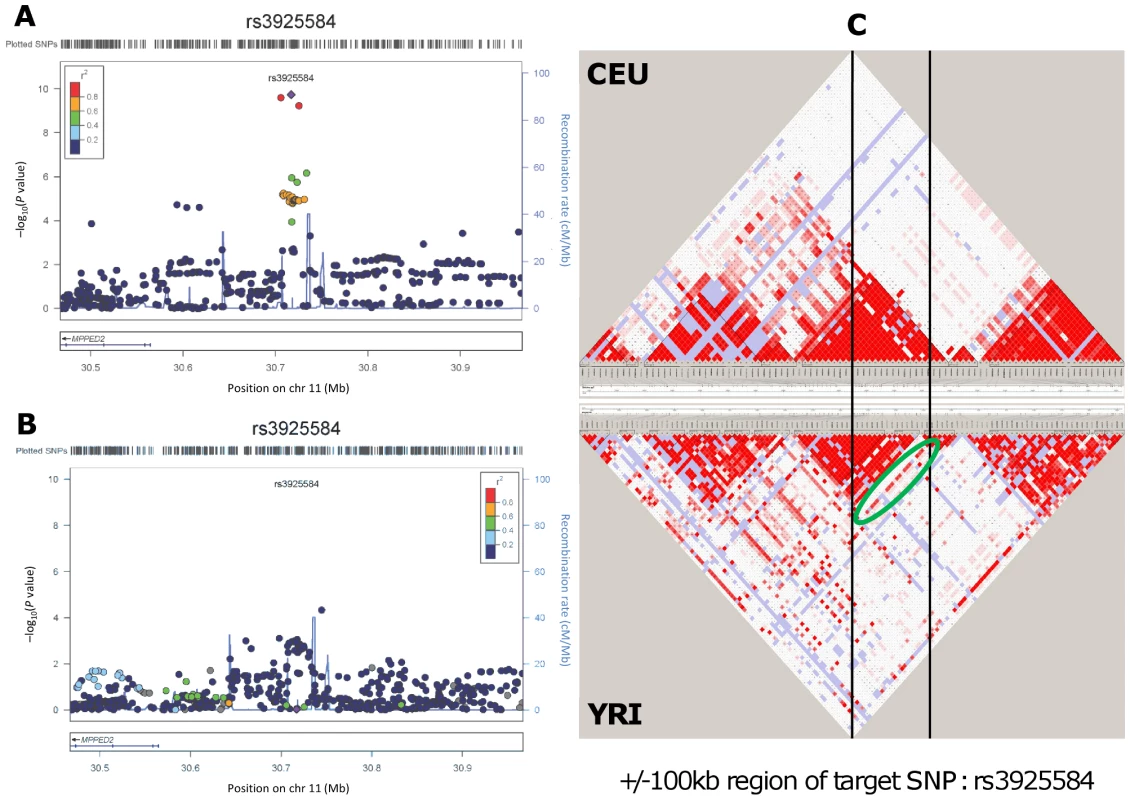
Regional association plots in the CKDGen European ancestry discovery analysis (N = 74,354) (A) and in the CARe African ancestry discovery analysis (N = 8,110) (B). LD structure: comparison between the HapMap release II – CEU and YRI samples in the region included within +/−100 kb from the target SNP rs3925584 identified in the CKDGen GWAS. The green circle highlights a stream of high LD connecting the two blocks, indicating the presence of common haplotypes (C). Tab. 2. Interrogation of the six novel loci uncovered in the European ancestry (EA) individuals (CKDGen consortium) in individuals of African ancestry (AA) from the CARe consortium for the trait eGFRcrea. 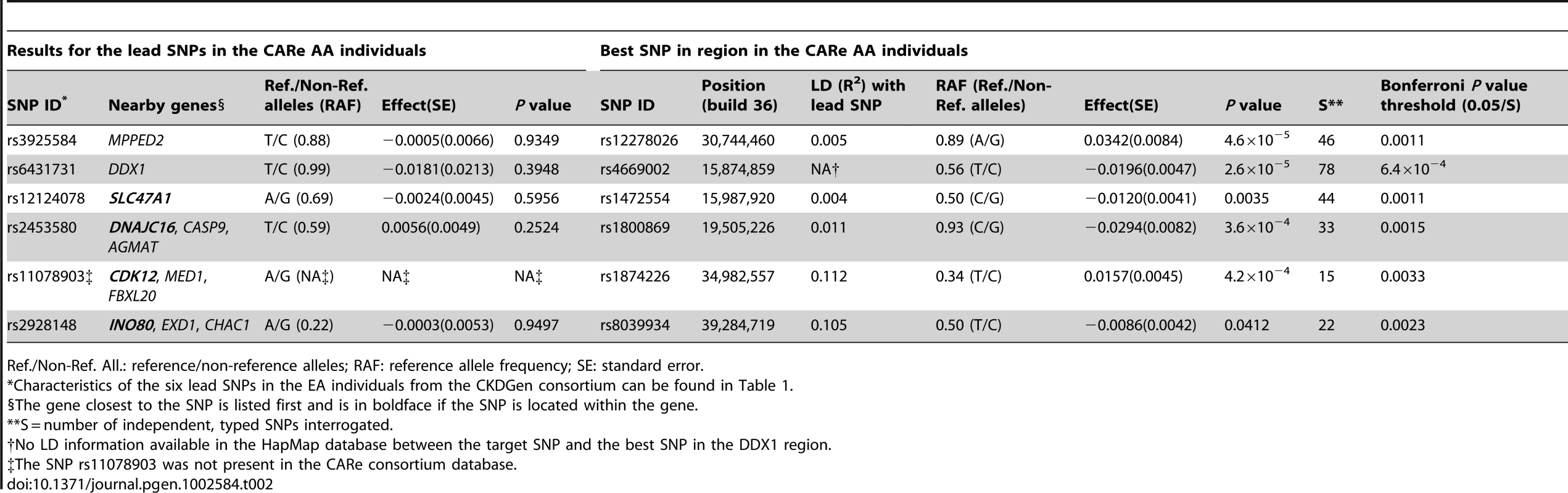
Ref./Non-Ref. All.: reference/non-reference alleles; RAF: reference allele frequency; SE: standard error. We also performed eQTL analyses of our 6 newly identified loci using known databases and a newly created renal eSNP database (see Methods) and found that rs12124078 was associated with cis expression of the nearby CASP9 gene in myocytes, which encodes caspase-9, the third apoptotic activation factor involved in the activation of cell apoptosis, necrosis and inflammation (P value for the monocyte eSNP of interest = 3.7×10−13). In the kidney, caspase-9 may play an important role in the medulla response to hyperosmotic stress [13] and in cadmium-induced toxicity [14]. The other 5 SNPs were not associated with any investigated eQTL. Additional eQTL analyses of 81 kidney biopsies (Table S13) did not reveal further evidence of association with eQTLs (Table S14).
Of the 6 novel loci identified, 2 (MPPED2 and DDX1) were in regions containing only a single gene, and 1 (CASP9) had its expression associated with the locus lead SNP. Thus, to determine the potential involvement of these three genes during zebrafish kidney development, we independently assessed the expression of 4 well-characterized renal markers following morpholino knockdown: pax2a (global kidney) [15], nephrin (podocyte) [16], slc20a1a (proximal tubule) [17], and slc12a3 (distal tubule) [17]. While we observed no abnormalities in ddx1 morphants (Figure S8), mpped2 and casp9 knockdown resulted in expanded pax2a expression in the glomerular region in 90% and 75% of morphant embryos, respectively, compared to 0% in controls (P value<0.0001 for both genes; Figure 2A versus 2F and 2K; 2B versus 2G and 2L; and 2P). Significant differences were also observed in expression of the podocyte marker nephrin (Figure 2C versus 2H and 2M; 80% and 74% abnormalities for mpped2 and casp9, respectively, versus 0% in controls, P value<0.0001 for both genes). For mpped2, no differences were observed in expression of the proximal or distal tubular markers slc20a1a and slc12a3 (P value = 1.0; Figure 2D versus 2I and 2E versus 2J). Casp9 morphants and controls showed no differences in proximal tubular marker expression (Figure 2D versus 2N), but abnormalities were observed in distal tubular marker expression in casp9 knockdown embryos (30% versus 0%; Figure 2E versus 2O; P value = 0.0064).
Fig. 2. Mpped2 and casp9 knockdowns result in defective kidney development. 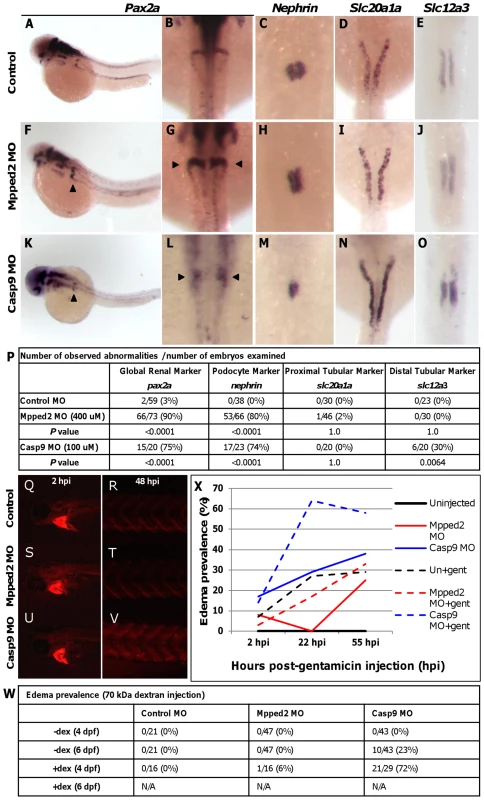
(A–E) Whole mount in situ hybridization in control embryos demonstrates normal expression of the global kidney marker pax2a (A: lateral view; B: dorsal view), the glomerular marker nephrin (C), and the tubular markers slc20a1a (proximal tubule, D), and slc12a3 (distal tubule, E) at 48 hours post fertilization (hpf). (F–J) Mpped2 morpholino (MO) knockdown embryos develop glomerular gene expression defects (F–H, arrowheads), but tubular marker expression is normal (I, J). (K–O) Casp9 MO knockdown embryos demonstrate reduced glomerular gene expression (K–M, arrowheads) and shortened distal tubules (O). (P) Quantification of observed abnormalities per number of embryos reveal significant differences in expression of pax2a and nephrin in response to knockdown of both mpped2 and casp9 (Fisher's exact test). (Q–V) Embryos were injected with control, mpped2, or casp9 MO at the one-cell stage and subsequently injected with 70,000 MW fluorescent rhodamine dextran at 80 hpf. Dextran fluorescence was monitored over the next 48 hours. All dextran-injected embryos show equal loading into the cardiac sinus venosus at 2 hours post-injection (2 hpi/82 hpf; Q, S, U). Compared to control MO-injected embryos (R) and mpped2 knockdown embryos (T), knockdown of casp9 resulted in reduced dextran clearance at 48 hpi as shown by increased trunk fluorescence (V). (W) Casp9 knockdown results in increased susceptibility to edema formation both spontaneously (−dex) (P value = 0.0234, Fisher's exact test) and after dextran challenge (+dex) (P value<0.0001). Embryos injected with both MO and dextran did not survive to 6 dpf (N/A). (X) Edema develops earlier and with higher frequency in casp9 morphants following injection of the nephrotoxin gentamicin. Casp9 morphants displayed diminished clearance of 70,000 MW fluorescent dextran 48 hours after injection into the sinus venosus compared to controls, revealing significant functional consequences of casp9 knockdown (Figure 2Q–2V). No clearance abnormalities were observed in mpped2 morphants. The occurrence of abdominal edema is a non-specific finding that is frequently observed in zebrafish embryos with kidney defects. We examined the occurrence of edema in mpped2 and casp9 knockdown embryos at 4 and 6 days post fertilization (dpf), both in the absence and presence of dextran, and observed a significant increase in edema prevalence in casp9 with (P value<0.0001) and without (P value = 0.0234) dextran challenge but not in mpped2 morphants (Figure 2W).
In order to further demonstrate differences in kidney function in response to knockdown of mpped2 and casp9, we injected the nephrotoxin gentamicin which predictably causes edema in a subset of embryos. Casp9 morphants were more susceptible to developing edema compared to both controls and mpped2 morphants (Figure 2X). In addition, edema developed earlier and was more severe, encompassing a greater area of the entire embryo (Figure S9). Together, these findings suggest that casp9 and mpped2 knockdowns result in altered kidney gene expression and function. Specifically, abnormal expression of pax2a and nephrin in casp9 morphants in addition to dextran retention and edema formation suggest loss of casp9 impacts glomerular development and function.
The lead SNP at the MPPED2 locus is located approximately 100 kb upstream of the gene metallophosphoesterase domain containing 2 (MPPED2), which is highly evolutionary conserved and encodes a protein with metallophosphoesterase activity [18]. It has been recognized for a role in brain development and tumorigenesis [19] but thus far not for kidney function.
To determine whether the association at our newly identified eGFRcrea loci was primarily due to creatinine metabolism or renal function, we compared the relative associations between eGFRcrea and eGFR estimated using cystatin C (eGFRcys) (Figure S10, File S1). The new loci showed similar effect sizes and consistent effect directions for eGFRcrea and eGFRcys, suggesting a relation to renal function rather than to creatinine metabolism. Placing the results of these 6 loci in context with our previously identified loci [8], [9] (23 known and 6 novel), 18 were associated with CKD at a 0.05 significance level (odds ratio, OR, from 1.05 to 1.26; P values from 3.7×10−16 to 0.01) and 11 with CKD45 (OR from 1.08 to 1.34; P values from 1.1×10−5 to 0.047; Figure S11 and Table S15).
When we examined these 29 renal function loci by age group, sex, diabetes and hypertension status (Tables S16, S17, S18, and S19), we observed consistent associations with eGFRcrea for most loci across all strata, with only two exceptions: UMOD had a stronger association in older individuals (P value for difference 8.4×10−13) and in those with hypertension (P value for difference 0.002), and CDK12 was stronger in younger subjects (P value for difference 0.0008). We tested the interaction between age and rs11078903 in one of our largest studies, the ARIC study. The interaction was significant (P value = 0.0047) and direction consistent with the observed between-strata difference.
Finally, we tested for associations between our 6 new loci and CKD related traits. The new loci were not associated with urinary albumin-to-creatinine ratio (UACR) or microalbuminuria [20] (Tables S20 and S21), with blood pressure from the ICBP Consortium [21] (Table S22) or with myocardial infarction from the CARDIoGRAM Consortium [22] (Table S23).
Discussion
We have extended prior knowledge of common genetic variants for kidney function [8]–[11], [23] by performing genome-wide association tests within strata of key CKD risk factors, including age, sex, diabetes, and hypertension, thus uncovering 6 loci not previously known to be associated with renal function in population-based studies (MPPED2, DDX1, CASP9, SLC47A1, CDK12, INO80). In contrast to our prior genome-wide analysis [8], [9], the majority of the new loci uncovered in the present analysis have little known prior associations with renal function. This highlights a continued benefit of the GWAS approach by using large sample sizes to infer new biology.
Despite our hypothesis that genetic effects are modified by CKD risk factors, most of the identified variants did not exhibit strong cross-strata differences. This highlights that many genetic associations with kidney function may be shared across risk factor strata. The association of several of these loci with kidney function in African Americans underscores the generalizability of identified renal loci across ethnicities. Zebrafish knockdown of mpped2 resulted in abnormal podocyte anatomy as assessed by expression of glomerular markers, and loss of casp9 led to altered podocyte and distal tubular marker expression, decreased dextran clearance, edema, and enhanced susceptibility to gentamicin-induced kidney damage. These findings demonstrate the potential importance of these genes with respect to renal function and illustrate that zebrafish are a useful in vivo model to explore the functional consequences of GWAS-identified genes.
Despite these strengths, there are some limitations of our study that warrant discussion. Although we used cystatin C to separate creatinine metabolism from true filtration loci, SNPs within the cystatin C gene cluster have been shown to be associated with cystatin C levels [8], which might result in some degree of misclassification in absolute levels. While we used standard definitions of diabetes and hypertension in the setting of population-based studies, these may differ from those definitions used in clinical practice. In addition, we were unable to differentiate the use of anti-hypertension medications from other clinical indications of these agents or type 1 from type 2 diabetes. The absence of association between our six newly discovered SNPs and the urinary albumin to creatinine ratio, blood pressure, and cardiovascular disease may have resulted from disparate genetic underpinnings of these traits, the overall small effect sizes, or the cross-sectional nature of our explorations; and we were unable to differentiate between these potential issues. Finally, power was modest to detect between-strata heterogeneity.
With increased sample size and stratified analyses, we have identified additional loci for kidney function that continue to have novel biological implications. Our primary findings suggest that there is substantial generalizability of SNPs associations across strata of important CKD risk factors, specifically with hypertension and diabetes.
Materials and Methods
Phenotype definition
Serum creatinine and cystatin C were measured as detailed in Tables S1 and S2. To account for between-laboratory variation, serum creatinine was calibrated to the US nationally representative National Health and Nutrition Examination Study (NHANES) standards in all discovery and replication studies as described previously [8], [24], [25]. GFR based on serum creatinine (eGFRcrea) was estimated using the four-variable MDRD Study equation [26]. GFR based on cystatin C (eGFRcys) was estimated as eGFRcys = 76.7×(serum cystatin C)−1.19 [27]. eGFRcrea and eGFRcys values<15 ml/min/1.73 m2 were set to 15, and those >200 were set to 200 ml/min/1.73 m2. CKD was defined as eGFRcrea <60 ml/min/1.73 m2 according to the National Kidney Foundation guidelines [28]. A more severe CKD phenotype, CKD45, was defined as eGFRcrea <45 ml/min/1.73 m2. Control individuals for both CKD and CKD45 analyses were defined as those with eGFRcrea >60 ml/min/1.73 m2.
Covariate definitions
In discovery and replication cohorts, diabetes was defined as fasting glucose ≥126 mg/dl, pharmacologic treatment for diabetes, or by self-report. Hypertension was defined as systolic blood pressure ≥140 mmHg or diastolic blood pressure ≥90 mmHg or pharmacologic treatment for hypertension.
Discovery analyses
Genotyping was conducted as specified in Table S4. After applying quality-control filters to exclude low-quality SNPs or samples, each study imputed up to ∼2.5 million HapMap-II SNPs, based on the CEU reference samples. Imputed genotypes were coded as the estimated number of copies of a specified allele (allelic dosage). Additional, study-specific details can be found in Table S1.
Primary association analysis
A schematic view of our complete analysis workflow is presented in Figure S1. Using data from 26 population-based studies of individuals of European ancestry, we performed GWA analyses of the following phenotypes: 1) loge(eGFRcrea), loge(eGFRcys), CKD, and CKD45 overall and 2) loge(eGFRcrea) and CKD stratified by diabetes status, hypertension status, age group (≤/>65 years), and sex. GWAS of loge(eGFRcrea) and loge(eGFRcys) were based on linear regression. GWAS of CKD and CKD45 were performed in studies with at least 25 cases (i.e. all 26 studies for CKD and 11 studies for CKD45) and were based on logistic regression. Additive genetic effects were assumed and models were adjusted for age and, where applicable, for sex, study site and principal components. Imputation uncertainty was accounted for by including allelic dosages in the model. Where necessary, relatedness was modeled with appropriate methods (see Table S1 for study-specific details). Before including in the meta-analysis, all GWA data files underwent to a careful quality control, performed using the GWAtoolbox package in R (www.eurac.edu/GWAtoolbox.html) [29].
Meta-analyses of study-specific SNP-association results, assuming fixed effects and using inverse-variance weighting, i.e.: the pooled effect is estimated as , where is the effect of the SNP on the outcome in the ith study, K is the number of studies, and is the weight given to the ith study. The meta-analyses were performed using METAL [30], with genomic control correction applied across all imputed SNPs [31] if the inflation factor λ>1 at both the individual study level and after the meta-analysis. SNPs with minor allele frequency (MAF)<1% were excluded. All SNPs with a meta-analysis P value≤5×10−8 for any trait or any stratum were deemed genome-wide significant [32].
In the eGFRcrea analyses, after excluding loci that were previously reported [8], [9], we selected for replication all SNPs with P value<5×10−8 in any trait or stratum that were independent (defined by pairwise r2<0.2), in the primary association analysis. This yielded five SNPs in five independent loci. The same criterion was applied to the CKD analysis, where no SNPs passed the selection threshold. Given the smaller number of cases with severe CKD resulting in less statistical power, a different selection strategy was adopted for the CKD45 analysis: selected for replication were SNPs with discovery P value≤5×10−6, MAF≥5%, and homogeneous effect size across studies (I2≤25%). Four additional SNPs were thereby selected for replication from the CKD45 analysis.
Direction test to identify SNPs for replication
In addition to identifying SNPs for replication based on the genome-wide significance threshold from a fixed effect model meta-analysis, we performed a “direction test” to identify additional SNPs for which between-study heterogeneity in effect size might have obscured the overall association that was nevertheless highly consistent in the direction of allelic effects. Under the null hypothesis of no association, the a priori probability that a given effect allele of a SNP has either a positive or negative association with eGFRcrea is 0.5. Because the meta-analysis includes independent studies, the number of concordant effect directions follows a binomial distribution. Therefore, we tested whether the number of discovery cohorts with the same sign of association (i.e. direction of effect) was greater than expected by chance given the binomial distribution and a null expectation of equal numbers of associations with positive and negative sign. The test was only applied for eGFRcrea in the overall analysis. Multiple testing was controlled by applying the same P value threshold of 5×10−8 as in the overall GWAS. Given that no SNP met this criterion, we selected for replication one novel SNP with the lowest P value of 4.0×10−7.
Genome-wide between-strata difference test to identify SNPs for replication
Based on the results of the stratified GWAS of eGFRcrea and CKD, for each SNP we tested the hypothesis whether the effect of a SNP on eGFRcrea or CKD was the same between strata (null hypothesis), i.e. diabetes versus non-diabetes subjects, hypertensive versus normotensive, younger versus older, females versus males. We used a two-sample test defined as Z = (b1−b2)/(SE(b1)2+SE(b2)2)0.5, with b1 and b2 indicating the effect estimates in the two strata and SE(b1) and SE(b2) their standard errors [33]. For large samples, the test statistic follows a standard normal distribution. SNPs were selected for replication if they had a between-stratum difference P value≤5×10−5, an association P value≤5×10−5 in one of the two strata, and MAF≥10%. Independent loci were defined using the same criteria as described above. Eleven further SNPs, one per locus, were selected for replication from the between-strata difference test.
Replication analysis
Replication was performed for a total of 21 SNPs including 5 from the overall and stratified eGFRcrea analyses, 1 from the direction test on eGFRcrea, 4 from the overall CKD45 analysis, and 11 from the between-strata difference test. Replication studies used the same phenotype definition, and had available genotypes from imputed in silico genome-wide SNP data or de novo genotyping. The same association analyses including the identical stratifications were performed as in discovery studies. Details can be found in the Tables S2, S5 and S6. Study-specific replication results for the selected SNPs were combined using the same meta-analysis approach and software as in the discovery stage. One-sided P values were derived with regard to the effect direction found in the discovery stage. Based on the P value distribution of all SNPs submitted for replication (the 10 from eGFRcrea and CKD45 and the 11 from the between strata difference test), we estimated the False Discovery Rate as a q-value using the QVALUE [34] package in R. SNPs with q-value<0.05 were called significantly replicating, thus specifying a list of associations expected to include not more than 5% false positives.
Finally, study-specific results from both the discovery and replication stage were combined in a joint inverse-variance weighted fixed-effect meta-analysis and the two-sided P values were compared to the genome-wide significance threshold of 5×10−8 to test whether a SNP was genome-wide significant. Between-study heterogeneity of replicated SNPs was quantified by the I2 statistic [35].
Replication genotyping
For de novo genotyping in 10,446 samples from KORA F3, KORA F4, SAPHIR and SAPALDIA, the MassARRAY system at the Helmholtz Zentrum (München, Germany) was used, using Assay Design v3.1.2 and the iPLEX chemistry (Sequenom, San Diego, USA). Assay design failed for rs1322199 and genotyping was not performed. Ten percent of the spectra were checked by two independent, trained persons, and 100% concordance between investigators was obtained. SNPs with a P value<0.001 when testing for Hardy-Weinberg equilibrium (rs10490130, rs10068737, rs11078903), SNPs with call rate <90% (rs500456 in KORA F4 only) or monomorphic SNPs (rs2928148) were excluded from analyses without attempting further genotyping. The call rates of rs4149333 and rs752805 were near 0% on the MassARRAY system. These SNPs were thus genotyped on a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, USA). Mean call rate across all studies and SNPs ranged from 96.8% (KORA F4) to 99% (SAPHIR). Duplicate genotyping was performed in at least 14% of the subjects in each study with a concordance of 95–100% (median 100%). In the Ogliastra Genetic Park Replication Study (n = 3000) de novo genotyping was conducted on a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, USA), with a mean call rate of 99.4% and 100% concordance of SNPs genotyped in duplicate.
Between-strata analyses for candidate SNPs in replication samples
Twenty-nine SNPs, including the 6 novel loci reported in the current manuscript along with 23 previously confirmed to be associated with renal function [9], were tested for differential effects between the strata. The same Z statistics as described for discovery (above) was used and the Bonferroni-adjusted significance level was set to 0.10/29 = 0.003.
SNP-by-age interaction, for the one SNP showing significantly different effects between strata of age, was tested in the ARIC study by fitting a linear model on log(eGFRcrea) adjusted for sex, recruitment site, the first and the seventh genetic principal components (only these two were associated with the outcome at P value<0.05). Both the interaction term and the terms for the main effects of age and the SNP were included in the model.
Power to assess between-strata effect difference
To assess genome-wide between-strata differences, with alpha = 5×10−8 and power = 80%, the maximum detectable difference was 0.025 when comparing nonDM versus DM and 0.015 when comparing nonHTN versus HTN. Similarly, when testing for between-strata differences the 29 known and new loci (Bonferroni-corrected alpha = 0.003) in the combined sample (n = ∼125,000 in nonDM and n = ∼13,000 in DM) we had 80% power to detect differences as large as 0.035.
Look-up in African Americans (CARe)
For each of the 6 lead SNPs identified in our European ancestry samples, we extracted eGFR association statistics from a genome-wide study in the CARe African ancestry consortium [12]. We further investigated potential allelic heterogeneity across ethnicities by examining the 250 kb flanking region surrounding each lead SNP to determine whether other SNPs with stronger associations exist in each region. A SNP with the smallest association P value with MAF>0.03 was considered the top SNP in the African ancestry sample. We defined statistical significance of the identified lead SNP in African ancestry individuals based on a region-specific Bonferroni correction. The number of independent SNPs was determined based on the variance inflation factor (VIF) with a recursive calculation within a sliding window of 50 SNPs and pairwise r2 of 0.2. These analyses were performed using PLINK.
Analyses of related phenotypes
For each replicating SNP, we obtained association results for urinary albumin-to-creatinine ratio and microalbuminuria from our previous genome-wide association analysis [20], and for blood pressure and myocardial infarction from genome-wide association analysis from the ICBP [21] and CARDIoGRAM [22] consortia, respectively.
eSNP analysis
Significant renal SNPs were searched against a database of expression SNPs (eSNP) including the following tissues: fresh lymphocytes [36], fresh leukocytes [37], leukocyte samples in individuals with Celiac disease [38], lymphoblastoid cell lines (LCL) derived from asthmatic children [39], HapMap LCL from 3 populations [40], a separate study on HapMap CEU LCL [41], peripheral blood monocytes [42], [43], adipose [44], [45] and blood samples [44], 2 studies on brain cortex [42], [46], 3 large studies of brain regions including prefrontal cortex, visual cortex and cerebellum (Emilsson, personal communication), liver [45], [47], osteoblasts [48], skin [49] and additional fibroblast, T cell and LCL samples [50]. The collected eSNP results met criteria for statistical significance for association with gene transcript levels as described in the original papers.
A second expression analysis of 81 biopsies from normal kidney cortex samples was performed as described previously [51], [52]. Genotyping was performed using Affymetrix 6.0 Genome-wide chip and called with GTC Software (Affymetrix). For eQTL analyses, expression probes (Affymetrix U133set) were linked to SNP probes with >90% call-rate using RefSeq annotation (Affymetrix build a30). P values for eQTLs were calculated using linear multivariable regression in both cohorts and then combined using Fisher's combined probability test (see also [52]). Pairwise LD was calculated using SNAP [53] on the CEU HapMap release 22.
Zebrafish functional experiments
Zebrafish were maintained according to established IACUC protocols. Briefly, we injected zebrafish embryos with newly designed (mpped2, ddx1) or previously validated (casp9 [54]) morpholino antisense oligonucleotides (MO, GeneTools, Philomath OR) at the one-cell stage at various doses. We fixed embryos in 4% PFA at the appropriate stages for in situ hybridization (http://zfin.org/ZFIN/Methods/ThisseProtocol.html). Different anatomic regions of the kidney were visualized using a panel of 4 established markers: pax2a (global kidney marker) [15], nephrin (podocyte marker) [16], slc20a1a (proximal tubule) [17], and slc12a3 (distal tubule marker) [17]. Abnormalities in gene expression were independently scored by two investigators. We compared the number of abnormal morphant embryos to control embryos, injected with a standard control MO designed by GeneTools, with the Fisher's exact test, at the Bonferroni-corrected significance level of 0.0125, i.e.: 0.05/4 markers. We documented the development of gross edema at 4 and 6 days post-fertilization in live embryos.
We performed dextran clearance experiments following previously described protocols [55]. Briefly, 80 hours after MO injection, we anesthetized embryos in 4 mg/ml Tricaine in embryo water (1∶20 dilution), then positioned embryos on their back in a 1% agarose injection mold. We injected an equal volume of tetramethylrhodamine dextran (70,000 MW; Invitrogen) into the cardiac sinus venosus of each embryo. We then returned the embryos to fresh embryo water. Using fluorescence microscopy, we imaged the embryos at 2 hours post-injection (82 hpf) to demonstrate equal loading, then at 48 hours post-injection (128 hpf) to evaluate dextran clearance.
Embryos were injected with control, mpped2, or casp9 MOs at the one-cell stage. At 48 hpf, embryos were manually dechorionated, anesthetized in a 1∶20 dilution of 4 mg/ml Tricaine in embryo water, and oriented on a 1% agarose injection mold. As previously described [56], embryos were injected with equal volumes of 10 mg/ml gentamicin (Sigma) in the cardiac sinus venosus, returned to fresh embryo water, and subsequently scored for edema (prevalence, time of onset) over the next 3 days.
Supporting Information
Zdroje
1. Meguid El NahasABelloAK 2005 Chronic kidney disease: The global challenge. Lancet 365 9456 331 340
2. ImaiEMatsuoS 2008 Chronic kidney disease in asia. Lancet 371 9631 2147 2148
3. CoreshJSelvinEStevensLAManziJKusekJW 2007 Prevalence of chronic kidney disease in the united states. JAMA 298 17 2038 2047
4. LeveyASde JongPECoreshJEl NahasMAstorBC 2011 The definition, classification, and prognosis of chronic kidney disease: A KDIGO controversies conference report. Kidney Int 80 1 17 28
5. van der VeldeMMatsushitaKCoreshJAstorBCWoodwardM 2011 Lower estimated glomerular filtration rate and higher albuminuria are associated with all-cause and cardiovascular mortality. A collaborative meta-analysis of high-risk population cohorts. Kidney Int 79 12 1341 1352
6. GansevoortRTMatsushitaKvan der VeldeMAstorBCWoodwardM 2011 Lower estimated GFR and higher albuminuria are associated with adverse kidney outcomes. A collaborative meta-analysis of general and high-risk population cohorts. Kidney Int 80 1 93 104
7. AstorBCMatsushitaKGansevoortRTvan der VeldeMWoodwardM 2011 Lower estimated glomerular filtration rate and higher albuminuria are associated with mortality and end-stage renal disease. A collaborative meta-analysis of kidney disease population cohorts. Kidney Int 79 12 1331 1340
8. KottgenAGlazerNLDehghanAHwangSJKatzR 2009 Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet 41 6 712 717
9. KottgenAPattaroCBogerCAFuchsbergerCOldenM 2010 New loci associated with kidney function and chronic kidney disease. Nat Genet 42 5 376 384
10. ChambersJCZhangWLordGMvan der HarstPLawlorDA 2010 Genetic loci influencing kidney function and chronic kidney disease. Nat Genet 42 5 373 375
11. GudbjartssonDFHolmHIndridasonOSThorleifssonGEdvardssonV 2010 Association of variants at UMOD with chronic kidney disease and kidney stones-role of age and comorbid diseases. PLoS Genet 6 e1001039 doi:10.1371/journal.pgen.1001039
12. LiuCTGarnaasMKTinAKottgenAFranceschiniN 2011 Genetic association for renal traits among participants of african ancestry reveals new loci for renal function. PLoS Genet 7 e1002264 doi:10.1371/journal.pgen.1002264
13. AllanLAClarkePR 2009 Apoptosis and autophagy: Regulation of caspase-9 by phosphorylation. FEBS J 276 21 6063 6073
14. GobeGCraneD 2010 Mitochondria, reactive oxygen species and cadmium toxicity in the kidney. Toxicol Lett 198 1 49 55
15. DrummondIAMajumdarAHentschelHElgerMSolnica-KrezelL 1998 Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development 125 23 4655 4667
16. Kramer-ZuckerAGWiessnerSJensenAMDrummondIA 2005 Organization of the pronephric filtration apparatus in zebrafish requires nephrin, podocin and the FERM domain protein mosaic eyes. Dev Biol 285 2 316 329
17. WingertRASelleckRYuJSongHDChenZ 2007 The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet 3 e189 doi:10.1371/journal.pgen.0030189
18. TyagiRShenoyARVisweswariahSS 2009 Characterization of an evolutionarily conserved metallophosphoesterase that is expressed in the fetal brain and associated with the WAGR syndrome. J Biol Chem 284 8 5217 5228
19. SchwartzFEisenmanRKnollJGesslerMBrunsG 1995 cDNA sequence, genomic organization, and evolutionary conservation of a novel gene from the WAGR region. Genomics 29 2 526 532
20. BogerCAChenMHTinAOldenMKottgenA 2011 CUBN is a gene locus for albuminuria. J Am Soc Nephrol 22 3 555 570
21. EhretGBMunroePBRiceKMBochudM The International Consortium for Blood Pressure Genome-Wide Association Studies 2011 Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478 7367 103 109
22. SchunkertHKonigIRKathiresanSReillyMPAssimesTL 2011 Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 43 4 333 338
23. PattaroCDe GrandiAVitartVHaywardCFrankeA 2010 A meta-analysis of genome-wide data from five european isolates reveals an association of COL22A1, SYT1, and GABRR2 with serum creatinine level. BMC Med Genet 11 41
24. FoxCSLarsonMGLeipEPCulletonBWilsonPW 2004 Predictors of new-onset kidney disease in a community-based population. JAMA 291 7 844 850
25. CoreshJAstorBCMcQuillanGKusekJGreeneT 2002 Calibration and random variation of the serum creatinine assay as critical elements of using equations to estimate glomerular filtration rate. Am J Kidney Dis 39 5 920 929
26. LeveyASBoschJPLewisJBGreeneTRogersN 1999 A more accurate method to estimate glomerular filtration rate from serum creatinine: A new prediction equation. modification of diet in renal disease study group. Ann Intern Med 130 6 461 470
27. StevensLACoreshJSchmidCHFeldmanHIFroissartM 2008 Estimating GFR using serum cystatin C alone and in combination with serum creatinine: A pooled analysis of 3,418 individuals with CKD. Am J Kidney Dis 51 3 395 406
28. National Kidney Foundation. 2002 K/DOQI clinical practice guidelines for chronic kidney disease: Evaluation, classification, and stratification. Am J Kidney Dis 39 2 Suppl 1 S1 266
29. FuchsbergerCTaliunDPramstallerPPPattaroC on behalf of the CKDGen consortium 2011 GWAtoolbox: An R package for fast quality control and handling of GWAS meta-analysis data. Bioinformatics 10.1093/bioinformatics/btr679
30. WillerCJLiYAbecasisGR 2010 METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26 17 2190 2191
31. DevlinBRoederK 1999 Genomic control for association studies. Biometrics 55 4 997 1004
32. Pe'erIYelenskyRAltshulerDDalyMJ 2008 Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol 32 4 381 385
33. CohenA 1983 Comparing regression coefficients across subsamples. Sociol Methods Res 12 77 94
34. StoreyJDTibshiraniR 2003 Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100 16 9440 9445
35. HigginsJPThompsonSGDeeksJJAltmanDG 2003 Measuring inconsistency in meta-analyses. BMJ 327 7414 557 560
36. GoringHHCurranJEJohnsonMPDyerTDCharlesworthJ 2007 Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet 39 10 1208 1216
37. IdaghdourYCzikaWShiannaKVLeeSHVisscherPM 2010 Geographical genomics of human leukocyte gene expression variation in southern morocco. Nat Genet 42 1 62 67
38. HeapGATrynkaGJansenRCBruinenbergMSwertzMA 2009 Complex nature of SNP genotype effects on gene expression in primary human leucocytes. BMC Med Genomics 2 1
39. DixonALLiangLMoffattMFChenWHeathS 2007 A genome-wide association study of global gene expression. Nat Genet 39 10 1202 1207
40. StrangerBENicaACForrestMSDimasABirdCP 2007 Population genomics of human gene expression. Nat Genet 39 10 1217 1224
41. KwanTBenovoyDDiasCGurdSProvencherC 2008 Genome-wide analysis of transcript isoform variation in humans. Nat Genet 40 2 225 231
42. HeinzenELGeDCroninKDMaiaJMShiannaKV 2008 Tissue-specific genetic control of splicing: Implications for the study of complex traits. PLoS Biol 6 e1 doi:10.1371/journal.pbio.1000001
43. ZellerTWildPSzymczakSRotivalMSchillertA 2010 Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS ONE 5 e10693 doi:10.1371/journal.pone.0010693
44. EmilssonVThorleifssonGZhangBLeonardsonASZinkF 2008 Genetics of gene expression and its effect on disease. Nature 452 7186 423 428
45. GreenawaltDMDobrinRChudinEHatoumIJSuverC 2011 A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res 21 7 1008 1016
46. WebsterJAGibbsJRClarkeJRayMZhangW 2009 Genetic control of human brain transcript expression in alzheimer disease. Am J Hum Genet 84 4 445 458
47. SchadtEEMolonyCChudinEHaoKYangX 2008 Mapping the genetic architecture of gene expression in human liver. PLoS Biol 6 e107 doi:10.1371/journal.pbio.0060107
48. GrundbergEKwanTGeBLamKCKokaV 2009 Population genomics in a disease targeted primary cell model. Genome Res 19 11 1942 1952
49. DingJGudjonssonJELiangLStuartPELiY 2010 Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet 87 6 779 789
50. DimasASDeutschSStrangerBEMontgomerySBBorelC 2009 Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325 5945 1246 1250
51. RodwellGESonuRZahnJMLundJWilhelmyJ 2004 A transcriptional profile of aging in the human kidney. PLoS Biol 2 e427 doi:10.1371/journal.pbio.0020427
52. WheelerHEMetterEJTanakaTAbsherDHigginsJ 2009 Sequential use of transcriptional profiling, expression quantitative trait mapping, and gene association implicates MMP20 in human kidney aging. PLoS Genet 5 e1000685 doi:10.1371/journal.pgen.1000685
53. JohnsonADHandsakerREPulitSLNizzariMMO'DonnellCJ 2008 SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24 24 2938 2939
54. SidiSSandaTKennedyRDHagenATJetteCA 2008 Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, bcl-2, and caspase-3. Cell 133 5 864 877
55. HentschelDMMengelMBoehmeLLiebschFAlbertinC 2007 Rapid screening of glomerular slit diaphragm integrity in larval zebrafish. Am J Physiol Renal Physiol 293 5 F1746 50
56. HentschelDMParkKMCilentiLZervosASDrummondI 2005 Acute renal failure in zebrafish: A novel system to study a complex disease. Am J Physiol Renal Physiol 288 5 F923 9
57. PruimRJWelchRPSannaSTeslovichTMChinesPS 2010 LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 26 18 2336 2337
Štítky
Genetika Reprodukční medicína
Článek Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1Článek Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / TranscriptionČlánek Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding DomainČlánek Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin ComplexesČlánek An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood ObesityČlánek Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAsČlánek Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 3
-
Všechny články tohoto čísla
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
- Genomic Analysis of the Hydrocarbon-Producing, Cellulolytic, Endophytic Fungus
- Networks of Neuronal Genes Affected by Common and Rare Variants in Autism Spectrum Disorders
- Akirin Links Twist-Regulated Transcription with the Brahma Chromatin Remodeling Complex during Embryogenesis
- Too Much Cleavage of Cyclin E Promotes Breast Tumorigenesis
- Imprinted Genes … and the Number Is?
- Genetic Architecture of Highly Complex Chemical Resistance Traits across Four Yeast Strains
- Exploring the Complexity of the HIV-1 Fitness Landscape
- MNS1 Is Essential for Spermiogenesis and Motile Ciliary Functions in Mice
- A Fundamental Regulatory Mechanism Operating through OmpR and DNA Topology Controls Expression of Pathogenicity Islands SPI-1 and SPI-2
- Evidence for Positive Selection on a Number of MicroRNA Regulatory Interactions during Recent Human Evolution
- Variation in Modifies Risk of Neonatal Intestinal Obstruction in Cystic Fibrosis
- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Critical Evaluation of Imprinted Gene Expression by RNA–Seq: A New Perspective
- A Meta-Analysis and Genome-Wide Association Study of Platelet Count and Mean Platelet Volume in African Americans
- Mouse Genetics Suggests Cell-Context Dependency for Myc-Regulated Metabolic Enzymes during Tumorigenesis
- Transcriptional Control in Cardiac Progenitors: Tbx1 Interacts with the BAF Chromatin Remodeling Complex and Regulates
- Synthetic Lethality of Cohesins with PARPs and Replication Fork Mediators
- APOBEC3G-Induced Hypermutation of Human Immunodeficiency Virus Type-1 Is Typically a Discrete “All or Nothing” Phenomenon
- Interpreting Meta-Analyses of Genome-Wide Association Studies
- Error-Prone ZW Pairing and No Evidence for Meiotic Sex Chromosome Inactivation in the Chicken Germ Line
- -Dependent Chemosensory Functions Contribute to Courtship Behavior in
- Diverse Forms of Splicing Are Part of an Evolving Autoregulatory Circuit
- Phenotypic Plasticity of the Drosophila Transcriptome
- Physiological Notch Signaling Maintains Bone Homeostasis via RBPjk and Hey Upstream of NFATc1
- Precocious Metamorphosis in the Juvenile Hormone–Deficient Mutant of the Silkworm,
- Igf1r Signaling Is Indispensable for Preimplantation Development and Is Activated via a Novel Function of E-cadherin
- Accurate Prediction of Inducible Transcription Factor Binding Intensities In Vivo
- Mitochondrial Oxidative Stress Alters a Pathway in Strongly Resembling That of Bile Acid Biosynthesis and Secretion in Vertebrates
- Mammalian Neurogenesis Requires Treacle-Plk1 for Precise Control of Spindle Orientation, Mitotic Progression, and Maintenance of Neural Progenitor Cells
- Tcf7 Is an Important Regulator of the Switch of Self-Renewal and Differentiation in a Multipotential Hematopoietic Cell Line
- REST–Mediated Recruitment of Polycomb Repressor Complexes in Mammalian Cells
- Intronic -Regulatory Modules Mediate Tissue-Specific and Microbial Control of / Transcription
- Age-Dependent Brain Gene Expression and Copy Number Anomalies in Autism Suggest Distinct Pathological Processes at Young Versus Mature Ages
- A Genome-Wide Association Study Identifies Variants Underlying the Shade Avoidance Response
- -by- Regulatory Divergence Causes the Asymmetric Lethal Effects of an Ancestral Hybrid Incompatibility Gene
- Genome-Wide Association and Functional Follow-Up Reveals New Loci for Kidney Function
- A Natural System of Chromosome Transfer in
- Cell Size and the Initiation of DNA Replication in Bacteria
- Probing the Informational and Regulatory Plasticity of a Transcription Factor DNA–Binding Domain
- Repression of Germline RNAi Pathways in Somatic Cells by Retinoblastoma Pathway Chromatin Complexes
- Temporal Transcriptional Profiling of Somatic and Germ Cells Reveals Biased Lineage Priming of Sexual Fate in the Fetal Mouse Gonad
- Rapid Analysis of Genome Rearrangements by Multiplex Ligation–Dependent Probe Amplification
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- The Atypical Calpains: Evolutionary Analyses and Roles in Cellular Degeneration
- The Silkworm Coming of Age—Early
- Development of a Panel of Genome-Wide Ancestry Informative Markers to Study Admixture Throughout the Americas
- Balanced Codon Usage Optimizes Eukaryotic Translational Efficiency
- The Min System and Nucleoid Occlusion Are Not Required for Identifying the Division Site in but Ensure Its Efficient Utilization
- Neurobeachin, a Regulator of Synaptic Protein Targeting, Is Associated with Body Fat Mass and Feeding Behavior in Mice and Body-Mass Index in Humans
- Statistical Analysis of Readthrough Levels for Nonsense Mutations in Mammalian Cells Reveals a Major Determinant of Response to Gentamicin
- Gene Reactivation by 5-Aza-2′-Deoxycytidine–Induced Demethylation Requires SRCAP–Mediated H2A.Z Insertion to Establish Nucleosome Depleted Regions
- The miR-35-41 Family of MicroRNAs Regulates RNAi Sensitivity in
- Genetic Basis of Hidden Phenotypic Variation Revealed by Increased Translational Readthrough in Yeast
- An Alu Element–Associated Hypermethylation Variant of the Gene Is Associated with Childhood Obesity
- Modelling Human Regulatory Variation in Mouse: Finding the Function in Genome-Wide Association Studies and Whole-Genome Sequencing
- Novel Loci for Adiponectin Levels and Their Influence on Type 2 Diabetes and Metabolic Traits: A Multi-Ethnic Meta-Analysis of 45,891 Individuals
- Polycomb-Like 3 Promotes Polycomb Repressive Complex 2 Binding to CpG Islands and Embryonic Stem Cell Self-Renewal
- Insulin/IGF-1 and Hypoxia Signaling Act in Concert to Regulate Iron Homeostasis in
- EMF1 and PRC2 Cooperate to Repress Key Regulators of Arabidopsis Development
- Three Essential Ribonucleases—RNase Y, J1, and III—Control the Abundance of a Majority of mRNAs
- Contrasted Patterns of Molecular Evolution in Dominant and Recessive Self-Incompatibility Haplotypes in
- A Machine Learning Approach for Identifying Novel Cell Type–Specific Transcriptional Regulators of Myogenesis
- Genomic Tools for Evolution and Conservation in the Chimpanzee: Is a Genetically Distinct Population
- Nos2 Inactivation Promotes the Development of Medulloblastoma in Mice by Deregulation of Gap43–Dependent Granule Cell Precursor Migration
- Intracranial Aneurysm Risk Locus 5q23.2 Is Associated with Elevated Systolic Blood Pressure
- Heritability and Genetic Correlations Explained by Common SNPs for Metabolic Syndrome Traits
- A Genome-Wide Association Study of Nephrolithiasis in the Japanese Population Identifies Novel Susceptible Loci at 5q35.3, 7p14.3, and 13q14.1
- DNA Damage in Nijmegen Breakage Syndrome Cells Leads to PARP Hyperactivation and Increased Oxidative Stress
- DNA Resection at Chromosome Breaks Promotes Genome Stability by Constraining Non-Allelic Homologous Recombination
- Genetic Analysis of Floral Symmetry in Van Gogh's Sunflowers Reveals Independent Recruitment of Genes in the Asteraceae
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Promoter Nucleosome Organization Shapes the Evolution of Gene Expression
- The Nucleoside Diphosphate Kinase Gene Acts as Quantitative Trait Locus Promoting Non-Mendelian Inheritance
- The Ciliogenic Transcription Factor RFX3 Regulates Early Midline Distribution of Guidepost Neurons Required for Corpus Callosum Development
- Phosphorylation of the RNA–Binding Protein HOW by MAPK/ERK Enhances Its Dimerization and Activity
- A Genome-Wide Scan of Ashkenazi Jewish Crohn's Disease Suggests Novel Susceptibility Loci
- Parkinson's Disease–Associated Kinase PINK1 Regulates Miro Protein Level and Axonal Transport of Mitochondria
- LMW-E/CDK2 Deregulates Acinar Morphogenesis, Induces Tumorigenesis, and Associates with the Activated b-Raf-ERK1/2-mTOR Pathway in Breast Cancer Patients
- Mapping the Hsp90 Genetic Interaction Network in Reveals Environmental Contingency and Rewired Circuitry
- Autoregulation of the Noncoding RNA Gene
- The Human Pancreatic Islet Transcriptome: Expression of Candidate Genes for Type 1 Diabetes and the Impact of Pro-Inflammatory Cytokines
- Spo0A∼P Imposes a Temporal Gate for the Bimodal Expression of Competence in
- Antagonistic Regulation of Apoptosis and Differentiation by the Cut Transcription Factor Represents a Tumor-Suppressing Mechanism in
- A Downstream CpG Island Controls Transcript Initiation and Elongation and the Methylation State of the Imprinted Macro ncRNA Promoter
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PIF4–Mediated Activation of Expression Integrates Temperature into the Auxin Pathway in Regulating Hypocotyl Growth
- Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations
- A Splice Site Variant in the Bovine Gene Compromises Growth and Regulation of the Inflammatory Response
- Comprehensive Research Synopsis and Systematic Meta-Analyses in Parkinson's Disease Genetics: The PDGene Database
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



