-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaOligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
The sustained production of functional motile sperm is critical for male fertility. In recent years, a dramatic increase of cases of male infertility were reported, with the most common cause represented by the production of morphologically abnormal spermatozoa with low motility. Several genetic and environmental factors have been proven to impact on sperm development. In particular, preliminary studies on samples from fertile and sterile individuals suggested that the deregulation of a class of small noncoding RNAs, called microRNAs, might be detrimental for sperm formation. To this end, we investigated the expression of Dicer, a core microRNA pathway component, in male germ cells and observed a peak of expression during meiosis. We performed a microRNA-expression screening and identified 5 members of the miR-34 family (miR-34bc and miR-449abc) as highly expressed from late meiosis to the sperm stage. Deletion of miR-34bc and miR-449 leads to sterility due to the production of abnormal spermatozoa with reduced motility. Thus our work proves for the first time the importance of a microRNA family in sperm formation and male fertility.
Published in the journal: . PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004597
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004597Summary
The sustained production of functional motile sperm is critical for male fertility. In recent years, a dramatic increase of cases of male infertility were reported, with the most common cause represented by the production of morphologically abnormal spermatozoa with low motility. Several genetic and environmental factors have been proven to impact on sperm development. In particular, preliminary studies on samples from fertile and sterile individuals suggested that the deregulation of a class of small noncoding RNAs, called microRNAs, might be detrimental for sperm formation. To this end, we investigated the expression of Dicer, a core microRNA pathway component, in male germ cells and observed a peak of expression during meiosis. We performed a microRNA-expression screening and identified 5 members of the miR-34 family (miR-34bc and miR-449abc) as highly expressed from late meiosis to the sperm stage. Deletion of miR-34bc and miR-449 leads to sterility due to the production of abnormal spermatozoa with reduced motility. Thus our work proves for the first time the importance of a microRNA family in sperm formation and male fertility.
Introduction
Spermatogenesis is a complex developmental program that supports the generation of spermatozoa and fertility throughout the adult male life. Spermatogenesis can be divided into three principal phases, a mitotic phase, meiosis and spermiogenesis [1]. The mitotic stages of spermatogenesis encompass the spermatogonial stem cell (SSCs) as well as differentiating spermatogonia. SSCs underpin testicular homeostasis whereas the differentiating spermatogonia act as transit amplifying cells generating a large pool of cells that will undergo several terminal differentiation processes [2]. From one round of DNA replication followed by two subsequent sets of chromosomal divisions, meiosis generates round spermatids with haploid recombined genomes [3]. These round spermatids then undergo the morphogenic process of spermiogenesis that transforms these round shaped cells through an intermediate known as elongating spermatids into spermatozoa [1]. Interestingly, the meiotic stages of lepto/zygotene as well as the terminal stages of spermiogenesis are mostly transcriptionally inert suggesting the majority of the regulation of gene expression must occur at the post-transcriptional level [4], [5]. After chromosomal pairing is completed at the end of zygotene, transcription resumes in early pachytene cells [4], [5]. The full complement and importance of mechanisms that underlie the regulation of gene expression during these periods of transcriptional quiescence/reemergence remains undefined.
MiRNAs are genome encoded small 21–23 nt non-coding RNAs that negatively post-transcriptionally regulate gene expression, either through the degradation of target mRNAs or inhibition of translation [6]. MiRNAs encoding transcripts are sequentially processed by the action of two type III ribonucleases, Drosha and Dicer [7]–[10]. Drosha forms the catalytic core of the nuclear microprocessor complex that cleaves primary miRNA transcripts to yield the precursor-miRNA (pre-miR), a 60–70 nt stem loop structure [8]. Upon genesis the pre-miR is exported to the cytoplasm where it is processed by Dicer within the RNA induced silencing complex (RISC), which cleaves the terminal loop to generate an intermediate 21–22 nucleotide miRNA duplex [9]–[11]. Subsequently one strand of this duplex, the nascent miRNA, gets incorporated into an Argonaute (Ago) protein that is a key component of RISC and the execution of miRNA function [12], [13]. The miRNA defines the target specificity of RISC through base pairing with complementary RNAs [13], [14]. The majority of miRNAs display imperfect complementarity with their target transcripts and the specificity is primarily defined by the ‘seed’ (bases 2–8) sequence of the miRNA [14], [15]. MiRNA are also classified on the basis of their seed sequences, miRNA loci that share the same seed sequences are grouped into families [16]. Upon RISC binding miRNAs initially inhibit translation of the transcript followed by destabilization through the recruitment of the CCR4-NOT deadenlyation complex [17], [18]. MiRNAs exert modest changes on target gene expression ranging from 1.5–3 fold on the transcript level [19]–[21]. An interesting facet of this ubiquitous post-transcriptional gene-silencing pathway is that a given miRNA can have multiple target transcripts and thus fine-tune or buffer gene expression of numerous genes within a cell [19]–[21]. While miRNAs have been found to regulate a plethora of developmental and physiological processes, none to date have been found that regulate mammalian spermatogenesis.
Results and Discussion
The importance of post-transcriptional regulation of gene expression in spermatogenesis prompted us to examine the contribution of the miRNA pathway to this process. The RNase III Dicer catalyzes the last step of canonical miRNA biogenesis and thus its expression levels within testicular germ cell populations would be indicative of where this pathway or the biogenesis of miRNAs for current or later use would be important. Since antibodies against mouse Dicer that function for tissue immunofluorescence are lacking, we therefore generated a knock-in allele in mice that carry an N-terminal Flag-HA2 tagged Dicer (DcrFH) (Fig. 1A–C). The Flag-HA2 tag did not adversely impact on the function of Dicer as mice homozygous for the DcrFH allele are viable and fertile. Visualization in adult testis sections of Flag-HA2-Dicer with anti-HA antibodies revealed abundant expression of Dicer in the mitotic spermatogonia and the early meiotic stages of pre-leptotene and leptotene. Thereafter Dicer was up regulated in zygotene reaching a maximum expression in early pachytene spermatocytes (Fig. 1D). From mid-pachytene onwards Dicer was downregulated but still detected in the later stages of spermiogenesis (Fig. 1D). The expression pattern of Dicer would suggest a critical function for the miRNA pathway in meiosis as well as during haploid germ cell development. While non-canonical miRNA biogenesis pathways do exist, only a single miRNA (miR-451) has been shown to be Dicer independent [22]–[24]. In addition to miRNAs, the other Dicer products, the endogenous siRNAs, have thus far only been found in oocytes and ESCs [25]–[27]. While the failure to detect siRNAs in the male germ cells cannot formally exclude their presence therein, the loss of Dicer can more than likely be used to explore the function of the miRNA pathway in post-mitotic spermatogenesis. The importance of Dicer in early germ cell development was shown through its conditional ablation during early embryogenesis in primordial germ cells (PGCs) using the TNAP-Cre [28]. This loss of Dicer results in proliferative defects in PGCs with either absent or retarded spermatogenesis in adult seminiferous tubules [28]. To understand whether Dicer is required during meiosis, we combined the Dicer LoxP (DcrFl) allele with the Stra8Cre transgene that deletes in differentiating spermatogonia to generate meiotic Dicer conditional knockouts (DicerC-KO) [29]–[31]. Fertility was lost in some of these animals; genotyping of pups sired by fertile DicerC-KO mice revealed the presence of the undeleted DcrFl allele, indicating the incomplete deletion in these animals. Histological examination of DicerC-KO testis sections revealed the presence of highly abnormal seminiferous tubules with a high apoptotic index (Fig. 1E–F). Thus the impairment of Dicer function has major impact on post-mitotic male germ cell development.
Fig. 1. Expression and function of Dicer in adult spermatogenesis. 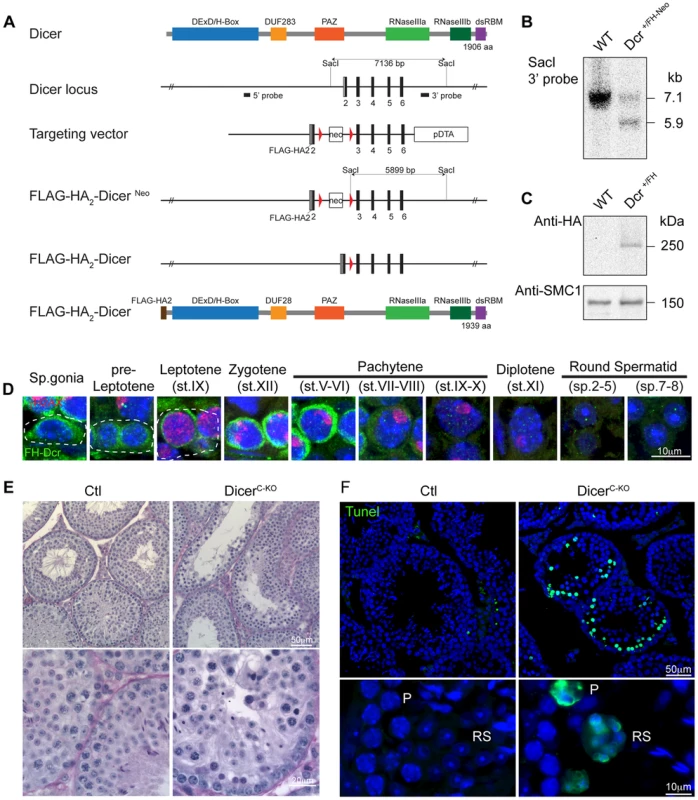
(A) Domain structure of the Dicer protein is shown. The organization of the 5′ portion of Dicer locus is depicted. The targeting vector used for introduction of FlagHA2 into the Dicer locus and the schematic map of the targeted Dicer gene before and after Cre mediated-recombination are shown. Triangles represent loxP sites as indicated. Rectangles indicate the position of Neomycin (Neo) and Diptheria toxin A (DTA) selection marker genes. The SacI restriction sites are indicated as well as the respective Southern fragments detected by the 3′probe. A schematic diagram of the resulting FlagHA2-Dicer protein is shown. (B) Southern blot of tail derived SacI-digested DNA from wild-type and Dcr+/FH-Neo mice is shown with the 3′ probe indicated in A. (C) Western blot using anti-HA and anti-SMC1 antibodies on extracts from adult wild type and Dcr+/FH testis is shown. (D) Immunofluorescence using anti-HA and anti-γH2AX antibodies on Dcr+/FH testis germ cells from adult testis sections is shown. Scale bar = 10 µm. (E) Hematoxylin and eosin stained testis section from adult DcrCtl and DcrC-KO mice with representative tubules shown. Scale bars = 50 µm and 20 µm in the upper and lower panel, respectively. (F) Increased apoptosis in DcrC-KO testis. A TUNEL assay counterstained with DAPI is shown on testis sections from adult DcrCtl and DcrC-KO mice. The apoptotic cells stain in green. Scale bars = 50 µm and 10 µm in the upper and lower panel, respectively. Abbreviations: P, pachytene and RS, round spermatid. Representative images are shown from at least 3 mice analyzed in panels D–F. The expression and function of Dicer during adult spermatogenesis indicates a critical role for not just the collective miRNA pathway but also potentially for individual miRNAs. We therefore decided to perform a miRNA expression screen to identify such individual miRNA loci. To this end in vitro cultured SSC cell lines representative of the mitotic phase of spermatogenesis as well as ex vivo isolated meiotic spermatocytes and post-meiotic round spermatids were selected for miRNA profiling. Interestingly the post-mitotic spermatocyte and round spermatid populations show very similar miRNA expression profiles, that was very distinct from the mitotic SSCs with the bidirectional regulation of many miRNA loci across these developmental stages observed (Fig. 2A). MiR-34b/c stood out from this analysis due to the binary nature of their expression, essentially being absent in SSCs to representing one of the most abundantly expressed miRNAs in post mitotic germ cells (Fig. 2A–B). The miR-34b/c miRNAs are part of a miR-34 family encompassing six miRNAs (miR-34a, b, c and 449a, b, c) encoded by three distinct loci (miR-34a, miR-34b/c and miR-449) (Fig. 2B). The miR-34a locus showed ubiquitous and low levels of expression across spermatogenesis and in general ubiquitous tissue expression (Fig. 2A–C). In contrast, miR-449a displayed the binary expression as miR-34b/c during spermatogenesis but had an overall lower expression (Fig. 2A–B). Both the miR-34b/c and miR-449 showed highly restricted expression profiles across an assortment of mouse tissues (Fig. 2C) [32]. Next we wanted to determine the precise onset of miR-34b/c and miR-449a during spermatogenesis and we decided to take advantage of the first wave of spermatogenesis, as it proceeds in a near synchronous manner with the appearance of successive spermatogenic populations across juvenile mouse development (Fig. S1A). Northern blotting of testicular RNA revealed the robust expression of miR-34b/c and miR-449a at postnatal day 14, a time when the appearance of pachytene spermatocytes is observed. miRNA in situ coupled with immunostaining of γH2AX as a meiotic marker revealed the onset of miR-34c expression in early pachytene spermatocytes within the first wave (Fig. 2E). The same onset of expression in the adult was observed with sustained miR-34c expression detected throughout meiosis and spermiogenesis (Fig. 2F). Our analysis identifies miR-34b/c and miR-449 loci as specifically and abundantly expressed in post-mitotic germ cells.
Fig. 2. miR-34b/c and miR-449 are selectively expressed in post-mitotic spermatogenesis. 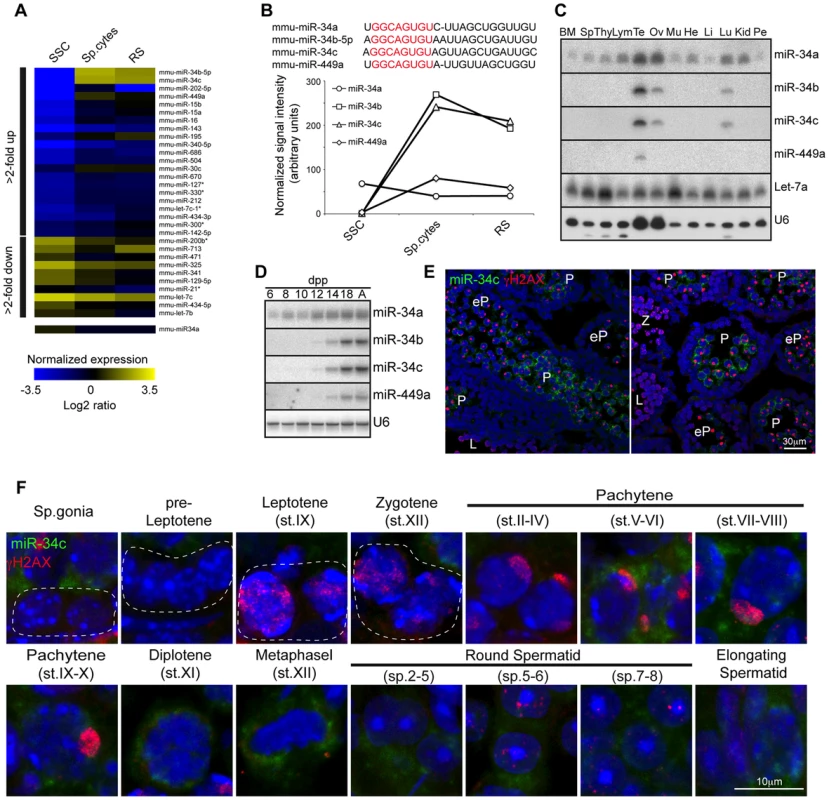
(A) Heat diagram summarizing the expression of miRNAs in mitotic in vitro cultured spermatogonial stem cell (SSC) lines, ex vivo isolated meiotic spermatocytes (sp.cytes) and spermiogenic round spermatids (RS). The average expression of biological replicates is shown. (B) The mature sequence of the murine miR-34 family miRNAs is shown with the seed sequence highlighted in red. The expression of the miR-34 family members is summarized from the array data. (C) The expression of miR-34 family was determined by Northern blotting of RNA derived from a broad panel of tissues. U6 snRNA and Let-7a was used as loading controls. Abbreviations: BM, bone marrow; Sp, spleen; Thy, Thymus; Lym, lymph node; Te, testis; Ov, ovary; Mu, muscle, He, heart; Li, liver; Lu, lung; Kid. Kidney and Pe, peritoneal cavity cells. (D) Expression of miR-34 family members was determined in the first wave of spermatogenesis by Northern blotting of total RNA from testis at the indicated day post partum (dpp), Ad indicates adult. U6 snRNA was used as a loading control. Representative data is shown from two independent experiments in panel C and D. (E) The onset of miR-34c expression in early pachytene spermatocytes during the first wave of spermatogenesis. The spatial expression of miR-34c (Green) is shown by in situ hybridization on sections of 14 dpp mouse testis, the section were counterstained with anti-γH2AX (Red) antibodies to precisely identify the meiotic stage. Scale bar = 30 µm. Abbreviations: L, leptotene; Z, zygotene; eP, early pachytene and P, pachytene. (F) The expression of miR-34c (Green) by in situ hybridization in the indicated adult spermatogenic populations is shown. Anti-γH2AX (Red) was used as in (E). Scale bar = 10 µm. Representative images from one of three independent experiments are shown for panel E and F. The miR-34 family genes are proven important regulators of cell fate and physiology. MiR-34a and miR-34b/c loci are direct p53 target genes with the ability to repress induced reprogramming [33]–[35]. The miR34a locus also regulates cardiac function upon aging, however none of the individual miR-34 family gene disruptions affects fertility in mice (Fig. S2) [32], [34]–[36]. With the similarity of expression of miR-34b/c and miR-449 loci and their potential to be functionally redundant with respect to spermatogenesis, we generated miR-34bc−/−;449−/− mice (Fig. 3A) that were born in Mendelian ratios. Both male and female miR-34bc−/−;449−/− mice were infertile when mated with wild type mice (Fig. 3B). Histological analysis of epididymis revealed a dramatic reduction of spermatozoa: quantitatively this reflected in a precipitous 60-fold drop in sperm counts in the miR-34bc−/−;449−/− mice which had an appreciable sperm count (Fig. 3C–D). Other miR-34bc−/−;449−/− mice had so few sperm that the count approached zero (Fig. 3D). Moreover not only was a drop in quantity of mature sperm observed but also in the quality. MiR-34bc−/−;449−/− sperm was of aberrant morphology with separation of spermatozoa heads and tails observed (Fig. 3E). Accordingly the motility of miR-34bc−/−;449−/− sperm was severely affected. Thus miR-34bc−/−;449−/− mice presented infertility due to low sperm count as well as spermatozoa that were immotile and of aberrant morphology. This phenotype is classified as oligoasthenoteratozoospermia and is the major cause of male infertility in humans [37].
Fig. 3. Oligoasthenoteratozoospermia and infertility in miR-34bc−/−;449−/− mice. 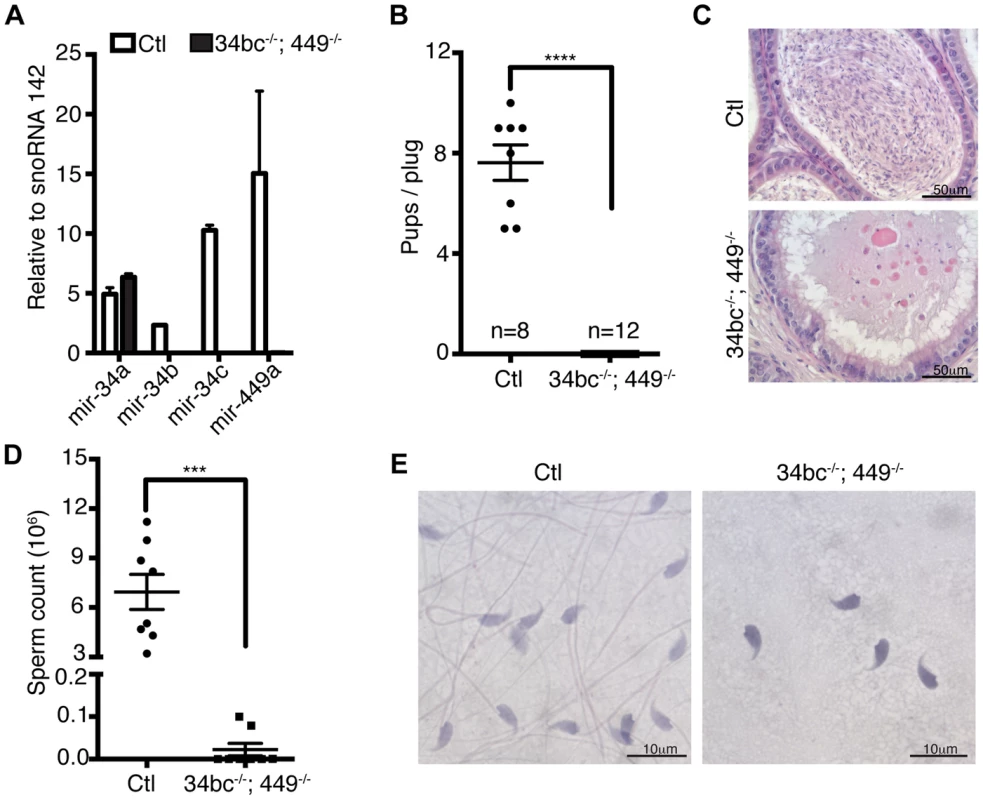
(A) qRT-PCR of miR-34a, miR-34b, miR-34c and miR-449a from control (Ctl) and miR-34bc−/−;449−/− adult testis. (B) miR-34bc−/−;449−/− male mice are infertile. The number of pups born per plug from wild type and miR-34bc−/−;449−/− mice is shown. The number of animals tested and the mean ±s.e.m. are indicated. (C) Hematoxylin and eosin stained epididymis section from control (Ctl) and miR-34bc−/−;449−/− adult mice is shown. A representative image of 5 mice analyzed is shown. Scale bar = 50 µm. (D) Reduced sperm count in miR-34bc−/−;449−/− mice. Mean sperm count ±s.e.m. from control and miR-34bc−/−;449−/− adult mice is shown (n = 8). (E) Sperm morphology from the indicated genotypes is shown. Scale bar = 10 µm. *** and **** indicates a p value (unpaired t test with Welch correction) of <0.001 and <0.0001 respectively. Having established that loss of both miR-34b/c and miR-449 loci results in oligoasthenoteratozoospermia, we next wanted to define the etiology of this disorder. To this end we studied the impact of miR-34bc/449 deficiency on spermatogenesis. Immediately obvious was the thinning of epithelium within the seminiferous tubules in miR-34bc−/−;449−/− mice (Fig. 4A). In approximately 5% of the tubules in the testis, cells at the zygotene stage were the most advanced spermatogenic cells that could be detected (Fig. 4B). In the majority of tubules, histological examination revealed the apparently normal appearance of germ cells until the pachytene stage of development, thereafter several spermatogenic defects were observed (Fig. 4A). Specifically a reduction in the number of germ cells after pachytene stage was evident. Moreover, in miR-34bc−/−;449−/− mice the development of the remaining round spermatids appeared to proceed normally until the final stages of spermiogenesis, when a dramatic decrease of elongating spermatids was observed (Fig. 4A). Accordingly, a high incidence of apoptosis was specifically detected in pachytene stages of meiosis as well as in elongating spermatids (Fig. 4C). The onset of the phenotype in miR-34bc−/−;449−/− mice perfectly coincided with the expression domain of miR34b/c observed in wild type adult spermatogenesis. To precisely quantitate the impact of miR-34bc/449 deficiency on the respective germ cell populations in the testis we utilized Hoechst staining of testicular cells analyzed by FACS that can effectively discriminate between lepto-zygotene, pachytene-diplotene, round spermatid and elongating spermatid populations (Fig. 4D) [38], [39]. This analysis revealed an overall decrease in the cellularity of miR-34bc−/−;449−/− testis but this decrease was unevenly distributed across developmental stages (Fig. 4E). Normal amounts for lepto-zygotene cells were observed in miR-34bc−/−;449−/− testis, thereafter a significant reduction in subsequent stages was evident (Fig. 4E). Thus in combination with the histological analysis we can conclude that the miR-34 family has multiple functions during spermatogenesis both in regulating meiosis as well as the later stages of spermiogenesis (Fig. 4F).
Fig. 4. miR-34bc/449 are required for multiple stages of post-mitotic spermatogenesis. 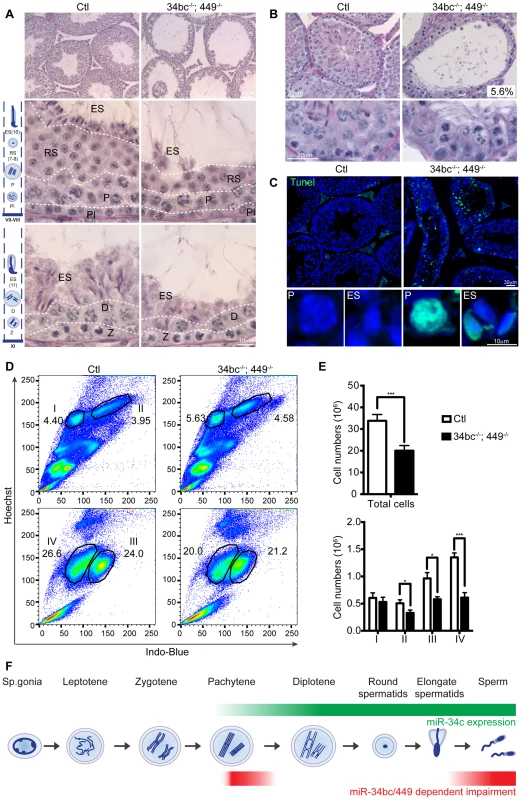
(A) PAS stained testis section from adult control and miR-34bc−/−;449−/− mice is shown. Overview of several tubules is shown in upper panels. Magnified and staged tubules are presented in the lower panels, the schematic diagram summarizes the spermatogenic content of tubules in wild type mice. Abbreviations: Pl, preleptotene; Z, zygotene; P, pachytene; D, diplotene;; RS, round spermatid and ES, elongating spermatid. Scale bars = 50 µm and 10 µm in the upper and lower panels, respectively. (B) MiR-34bc−/−;449−/− testis sections and percentages of tubules with meiotic arrest at zygotene stage is shown. Scale bars = 30 µm and 20 µm in the upper and lower panel, respectively. Representative images are shown from 6 mice analysed is shown in panel A and B. (C) Increased apoptosis in miR-34bc−/−;449−/− testis. A TUNEL assay counterstained with DAPI (Blue) is shown on testis sections from adult control and miR-34bc−/−;449−/− mice (upper panel). The apoptotic cells stain in green. Apoptotic miR-34bc−/−;449−/− pachytene (P) and elongating spermatid (ES) are shown in the lower panel along with non-apoptotic control cells. Scale bars = 30 µm and 10 µm in the upper and lower panel, respectively. Representative images are shown from 3 mice analysed is shown. (D) FACS plot of adult testis shown, gated populations in upper panel I (lepto-zygotene) and II (pachytene-diplotene), in lower panel III (round spermatids) and IV (elongating spermatids). Numbers indicated the overall percentage of the respective populations. (E) Comparative enumeration of spermatogenic populations of control (Ctl) and miR-34bc/499−/− mice. Total testicular cell numbers (upper) are shown. Numbers plotted for the developmentally defined subpopulations indicated in (D) by roman numerals (lower panel). 8 animals per genotype were analyzed by FACS. Mean ±s.e.m. values are shown in the graph. (F) Schematic diagram indicating the expression and impact of loss of miR-34bc/449 expression. * and *** indicates a p value (unpaired t test with Welch correction) of <0.05 and <0.001 respectively. We next wanted to explore the mechanisms by which miR-34bc/449 supports spermatogenesis. The binding of miRNA:RISC to target mRNAs results in transcript destabilization, this facet of miRNA silencing has been used to reliably identify miRNA targets from the analysis of cellular transcriptomes with gain or loss of a specific miRNA function [19]–[21]. miRNAs exert a relatively small impact in the order of 1.5–3 fold change of target mRNAs, therefore the isolation of pure populations of cells from wild type and mutant mice is critical for comparative transcriptomic analysis and the identification of miRNA target genes. MiR-34bc/449 deficiency impacts both meiosis and the latter stages of spermiogenesis, FACS can be used to sort both of these populations, however in the case of elongating spermatids cells are damaged during the process losing both their tails and cytoplasm. We therefore decided to profile pachytene spermatocytes from control and miR-34bc−/−;449−/− mice. This population also has the added advantage in that it is the cell type where the onset of miR-34bc/449 expression is first observed and thus likely the most promising stage to define the primary impact of miR-34bc/449-deficiency. The comparison of wild type and miR-34bc−/−;449−/− pachytene cells revealed relatively minor changes in the transcriptome (Fig. 5A), setting a threshold of 1.4 fold (Log2>0.5 fold) and with significance value greater than 0.05, we found 22 genes upregulated and 2 genes downregulated in the mutant (Fig. 5A). In miRNA loss of function experiments, the expectation is the loss of repression and concomitant increase in target dosage. Strikingly, 13 of the 22 upregulated genes contained 3′UTR miR-34 ‘seed’ matches and were predicted targets of the miR-34 family (Fig. 5 A–B). Deregulation of 9 from the 13 predicted targets could be confirmed by qRT-PCR (Fig. 5C). We next employed Sylamer to search for significant enrichment of 7 nucleotide motifs corresponding to all known miRNA seed motifs across the 3′UTRs of all genes arranged from most upregulated to most downregulated in the mutant (Fig. 5D). This unbiased approach revealed a highly significant enrichment (p = 2.44×10−9) for the complementary seed match of miR-34 family (CACTGCC) in the cohort of most unregulated genes (Fig. 5D). Most importantly, no other significant miRNA seed matches were identified in this analysis. From the 9 validated miR-34 target genes identified, the forkhead transcription factor FoxJ2 merits special interest as it contains two highly conserved miR-34 binding sites and has been shown that transgenic levels of FoxJ2 overexpression are incompatible with male fertility [40]. Together, these analysis show that the loss of miR-34bc/449 has an intrinsic impact on the meiotic transcriptome and identifies a small cohort of likely direct miR-34bc/449 target genes.
Fig. 5. miR-34bc/449 regulates a small cohort of genes in spermatocytes. 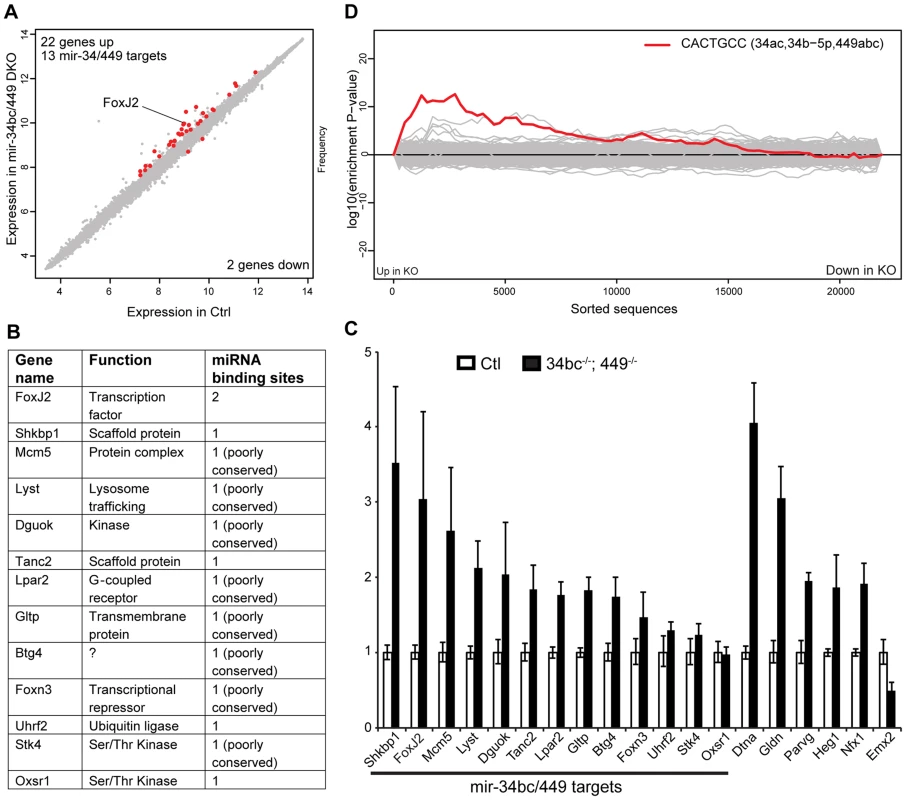
(A) Expression scatterplot showing relative average expression of affymetrix probes between control (x-axis) and miR-34bc−/−;449−/− (y-axis). Significantly deregulated (p = 0.05) genes with a log2 fold change of 0.5 (red) are shown. (B) The list of the 13 upregulated genes with predicted miR-34 seed binding sites is shown. Also indicated is the gene function as well as number of miR-34 binding sites. (C) qRT-PCR expression analysis of representative miR-34 family seed-containing deregulated genes identified. Normalized data are plotted as relative fold change in miR-34bc−/−;449−/− versus wild type pachytene spermatocytes. Standard error is shown and the asterisk indicates significantly upregulated expression (P<0.05). Other genes identified from the array that change in expression are also presented. The data in all panels are from biological quadruplicates of each genotype. (D) Sylamer enrichment landscape plot for all 876 7 nt words complementary to canonical mouse miRNA seed regions. The y-axis represents the sorted genelist of 21,560 genes from most up-regulated to most down-regulated in the miR-34bc−/−;449−/− pachytene spermatocytes. Each 7mer word was tested for significant enrichment across the 3′UTRs of genes in this list. The word corresponding to seed matching miR-34 family (Red) is enriched in the up-regulated genes. Here we explored the contribution of the miRNA pathway to the post-mitotic stages of spermatogenesis. The increase in Dicer expression reaching a maximum in pachytene spermatocytes is indicative of the importance of this pathway within meiosis, indeed the loss of Dicer results in spermatogenic defects. It is interesting to note that the resumption of transcription during meiosis occurs as cells enter pachytene, therefore the high levels of Dicer therein may be required to process a burst in pre-miRNAs for current or later use. Indeed, the complement of miRNAs expressed in spermatocytes and round spermatids is very similar, thus the elevated expression of Dicer observed may be required to generate large quantities of miRNA not only for meiosis but also thereafter. In addition, the challenges presented by the resumption of transcription in pachytene cells necessitate the miRNA pathway to fine tune gene expression. This is evident from the deregulated gene expression in miR-34bc−/−;449−/− spermatocytes and its phenotypic consequences. Our study identifies the miR-34b/c and miR-449 as the first miRNA loci required for mammalian spermatogenesis. The loss of miR-34bc/449 did not block the process of spermatogenesis per se but impairs several developmental transitions resulting in a low sperm count and sperm of aberrant morphology and motility. These are the phenotypic hallmarks of oligoasthenoteratozoospermia, the most common cause of reduced male fertility or infertility in humans [37]. Thus our study presents the loss of specific miRNAs as one definitive causal event in the genesis of oligoasthenoteratozoospermia. These observations bear importance for the etiology of this disorder as well as potential future basis for molecular diagnostic and therapeutic strategies.
Materials and Methods
Mouse strains
For the DcrFH allele, we introduced immediately after the starting ATG of Dicer located within exon 2 the sequence encoding Flag-HA-HA (FlagHa2) epitope tags. To generate this allele, a targeting construct was generated that contains the 5′ 5.1 kb and 3′ 3.4 kb homology arms, an loxP flanked neo cassette placed in intron 2 and FlagHA2 sequence inserted into exon 2 as described above. Southern blotting of the individual ES cell clones-derived genomic SacI digested DNA with an external 3′ probe was used to identify homologous recombinants. A 7.1 kb DNA fragment corresponds to the wild-type Dcr locus, integration of the neo cassette site 5′ of exon 3 introduces an additional SacI site, thus decreasing the size of the SacI DNA fragment recognized to 5.9 kb. The integration of the FlagHA2 tag was confirmed by sequencing of targeted clones. Cre-mediated removal of the loxP flanked neo cassette resulted in the generation of the FlagHa2-Dicer (DcrFH) allele.
The miR-34b and miR-34c miRNAs are derived from a single non-coding transcriptional unit. For the miR-34bc loss of function allele, the targeting strategy allows for Cre-mediated deletion of the hairpins that encode both miR-34b and miR-34c. To generate this allele, a targeting construct was generated that contains the 5′ 3.65 kb and 3′ 4.6 kb homology arms, an frt flanked neo cassette with a loxP site 5′ of the miR-34b/c encoding sequences and a second 3′ loxP site. Southern blotting of the individual ES cell clones-derived genomic HindIII-digested DNA with an external 5′ probe was used to identify homologous recombinants. A 9 kb DNA fragment corresponds to the wild-type miR-34b/c locus, integration of the loxP site 3′ of introduces an additional HindIII site, thus decreasing the size of the HindIII DNA fragment recognized to 5 kb. Flp-mediated and subsequent Cre recombination results in results in the generation miR-34bc flox (miR-34bcFl) and miR-34bc null (miR-34bc−) alleles, respectively.
The miR-449a, miR-449b and miR-449c miRNAs are encoded in 1.6 kb of sequence within an intron of 20 Kb of the coding Cdc20B gene. To generate mice lacking all miR-449 miRNAs, we replaced the hairpins that encode all miR-449s with loxP flanked neo cassette. A targeting construct was generated that contains the 5′ 4.9 kb and 3′ 4.7 kb homology arms, an loxP flanked neo cassette that replaces the sequences the encoding miR-449a, b and c. Southern blotting of the individual ES cell clones-derived genomic BamHI-digested DNA with an external 3′ probe was used to identify homologous recombinants. A 8 kb DNA fragment corresponds to the wild-type miR-449 locus, integration of loxP flanked neo cassette of introduces an additional BamHI site, thus decreasing the size of the BamHI DNA fragment recognized to 6.6 kb in the miR-449 targeted allele. Cre-mediated recombination removes the neo cassette resulting in a single scarring loxP site leaving the remainder of the intron Cdc20B intact. This strategy is designed to remove the miR-449 without affecting the Cdc20B gene.
The DcrFH, miR-34bc and miR-449 targeting constructs were electroporated into A9 ES cells (ESCs) and manipulated to generate mice fully derived from ESCs [41]. The miR-34b/c-targeted mice were then crossed to the FLP expressing transgenic mice (FLPeR) [42] to remove the frt flanked neor cassette, resulting in the generation of the miR-34bcFl allele. Mice heterozygous for the miR-34bcFl, targeted Dcr and miR-449 targeted alleles were crossed to Deleter Cre [43] to generate the miR-34b−, DcrFH and miR-449− alleles, respectively. Adult mice between two and four months of age on a mixed C57Bl/6 and 129 genetic background were analyzed in this study. The Stra8-Cre [29], [30] allele was also used in this study. For the miR-34bc−/−;449−/− experiments, miR-34bc+/−;449+/− or miR-34bc+/− or miR-449+/− were used as control mice. These control animals were normally littermates but if this was not possible age matched control mice were used. All of the mice were bred and maintained in EMBL Mouse Biology Unit, Monterotondo in accordance with current Italian legislation (Art. 9, 27. Jan 1992, n°116) under license from the Italian health ministry.
Antibodies
A mouse monoclonal antibody against the HA epitope (Covance HA.11 Clone 16B12) was used for WB and IF (1∶1000 and 1∶100 respectively) experiments. A rabbit polyclonal anti-γH2AX (ICH, ICH-00059) (1∶250) was used for immunofluorescence and in combination with RNA in situ hybridization. A rabbit polyclonal anti-Smc1a (Bethyl A300-55A) (1∶10000) was used in this study.
Immunofluorescence
Adult testes were collected and fixed in 4% paraformaldehyde overnight and embedded in paraffin. 6 µm sections were cut for HA and γH2AX immunostaining. Sections were subjected to antigen retrieval using steam vapor for 30 minutes in antigen unmasking solution (Vector Lab) and then permeabilized for 10 minutes at room temperature in 0.1% triton-X. Sections were blocked 30 minutes at room temperature in 10% normal donkey serum, 2% BSA and 0.1M glycine (Sigma). Primary antibody incubation was done overnight at 4°C in the blocking buffer. Appropriate Alexa secondary antibodies (Invitrogen) (1∶1000) were used. Hoechst 33342 (5 µg/ml) (Sigma) was used to stain DNA. Leica TCS SP5 confocal microscope was used to acquire all images. Photoshop was used for cropping and other modifications that were equally performed on control or experimental samples.
Histology and detection of apoptotic cells
Testes were fixed in Bouin's fixative overnight at 4°C temperature, paraffin embedded and sectioned at 8-µm thickness. Sections were then stained with hematoxylin and eosin or by periodic acid Schiff and hematoxylin by using routine methods. Detection of apoptotic cells was performed on paraformaldehyde-fixed paraffin embedded testis sections using the in situ cell death detection kit (Roche). Sections were then stained with DAPI (5 µg/ml) (Sigma) for the identification of different germ cell populations.
Germ cell isolation
Germ cell populations are isolated and analyzed by FACS precisely as described [38], [39], [44].
miRNA and mRNA expression analysis
RNA was isolated using Trizol (Invitrogen) according to manufacturer's instructions. 500 ng of total RNA was labeled and hybridized to miRCURY LNA microRNA arrays V.11 (Exiqon) for miRNA profiling. Northern blotting of miRNAs was performed as described (Rasmussen, 2010). For miRNA in situ, testes were harvested from 4% paraformaldehyde perfused animals of various ages (6 dpp, 8 dpp, 10 dpp, 12 dpp, 14 dpp, 18 dpp, and 3 months). Tissues were further fixed by immersion in 4% paraformaldehyde overnight, cryoprotected in 30% sucrose/PBS, frozen, and sectioned at 7 µm onto Superfrost Plus slides. In situ hybridization was performed using LNA-probes with 3′-DIG label (Exiqon) for mir-34c. Hybridization with scramble LNA-probes were used as negative controls. Briefly, sections were digested with proteinase K for 5 min, acetylated, and hybridized with the probes in 50% formammide, 5× SSC, 5× Denhardt's solution, 500 µg/ml salmon sperm DNA, and 250 µg/ml tRNA overnight at 52.5°C. After post-hybridization washes with 50% formammide, 2× SSC at 52.5°C, and with 2× SSC at ambient temperature, sections were then blocked and incubated overnight with anti-digoxigenin-POD (Roche; at 1∶500). Signal detection was done using TSA-Plus Fluorescein system. The slides were subsequently incubated with rabbit anti-γH2AX (1∶250) and goat anti-rabbit Alexa 546 as secondary antibody. For miRNA qRT PCR, 10 ng of total RNA were reverse-transcribed using the TaqMan MicroRNA Reverse Transcription kit (4366569, Invitrogen) following manufacturer's instructions. qRT-PCRs were performed using the TaqMan Universal Master Mix II, no UNG (4440040; Invitrogen). For the reverse transcription and the qRT-PCR specific TaqMan miRNA assays (Applied Biosystem) for mir-34a, mir-34b, mir-34c, mir-449a and control snoRNA142 were used.
For mRNA microarray analysis, total RNA was isolated from FACS-sorted pachytene cells from control and mir-34bc−/−;449−/− male mice. The RNA was hybridized to Mouse Gene 2.0 ST Arrays from Affymetrix. Data were analyzed with R/bioconductor using the limma package [45], [46]. The data was normalized and corrected from background using the Robust Multi-Array Average expression measure [47] function (rma) from the Affymetrix package. MiRNA binding motifs enrichment was analyzed using Sylamer [48].
Accession numbers
All array data are deposited in ArrayExpress under the accession number E-MTAB-2668 and E-MTAB-2676.
Supporting Information
Zdroje
1. EddyEM (2002) Male germ cell gene expression. Recent Prog Horm Res 57 : 103–128.
2. de RooijDG, RussellLD (2000) All you wanted to know about spermatogonia but were afraid to ask. J Androl 21 : 776–798.
3. HandelMA, SchimentiJC (2010) Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11 : 124–136.
4. MonesiV (1964) Ribonucleic Acid Synthesis during Mitosis and Meiosis in the Mouse Testis. J Cell Biol 22 : 521–532.
5. ParonettoMP, MessinaV, BarchiM, GeremiaR, RichardS, et al. (2011) Sam68 marks the transcriptionally active stages of spermatogenesis and modulates alternative splicing in male germ cells. Nucleic Acids Res 39 : 4961–4974.
6. O'CarrollD, SchaeferA (2013) General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology 38 : 39–54.
7. LeeY, AhnC, HanJ, ChoiH, KimJ, et al. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425 : 415–419.
8. LandthalerM, YalcinA, TuschlT (2004) The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14 : 2162–2167.
9. BernsteinE, CaudyAA, HammondSM, HannonGJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409 : 363–366.
10. HutvagnerG, McLachlanJ, PasquinelliAE, BalintE, TuschlT, et al. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293 : 834–838.
11. YiR, QinY, MacaraIG, CullenBR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17 : 3011–3016.
12. MeisterG, LandthalerM, PetersL, ChenPY, UrlaubH, et al. (2005) Identification of novel argonaute-associated proteins. Curr Biol 15 : 2149–2155.
13. SchwarzDS, HutvagnerG, HaleyB, ZamorePD (2002) Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol Cell 10 : 537–548.
14. GrimsonA, FarhKK, JohnstonWK, Garrett-EngeleP, LimLP, et al. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27 : 91–105.
15. DoenchJG, SharpPA (2004) Specificity of microRNA target selection in translational repression. Genes Dev 18 : 504–511.
16. AmbrosV, BartelB, BartelDP, BurgeCB, CarringtonJC, et al. (2003) A uniform system for microRNA annotation. RNA 9 : 277–279.
17. Behm-AnsmantI, RehwinkelJ, DoerksT, StarkA, BorkP, et al. (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20 : 1885–1898.
18. OrbanTI, IzaurraldeE (2005) Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11 : 459–469.
19. FarhKK, GrimsonA, JanC, LewisBP, JohnstonWK, et al. (2005) The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310 : 1817–1821.
20. BaekD, VillenJ, ShinC, CamargoFD, GygiSP, et al. (2008) The impact of microRNAs on protein output. Nature 455 : 64–71.
21. LimLP, LauNC, Garrett-EngeleP, GrimsonA, SchelterJM, et al. (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433 : 769–773.
22. YangJS, MaurinT, RobineN, RasmussenKD, JeffreyKL, et al. (2010) Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A 107 : 15163–15168.
23. CheloufiS, Dos SantosCO, ChongMM, HannonGJ (2010) A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465 : 584–589.
24. CifuentesD, XueH, TaylorDW, PatnodeH, MishimaY, et al. (2010) A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328 : 1694–1698.
25. BabiarzJE, RubyJG, WangY, BartelDP, BlellochR (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22 : 2773–2785.
26. WatanabeT, TotokiY, ToyodaA, KanedaM, Kuramochi-MiyagawaS, et al. (2008) Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453 : 539–543.
27. TamOH, AravinAA, SteinP, GirardA, MurchisonEP, et al. (2008) Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453 : 534–538.
28. HayashiK, Chuva de Sousa LopesSM, KanedaM, TangF, HajkovaP, et al. (2008) MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One 3: e1738.
29. Sadate-NgatchouPI, PayneCJ, DearthAT, BraunRE (2008) Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46 : 738–742.
30. HobbsRM, FagooneeS, PapaA, WebsterK, AltrudaF, et al. (2012) Functional antagonism between Sall4 and Plzf defines germline progenitors. Cell Stem Cell 10 : 284–298.
31. YiR, O'CarrollD, PasolliHA, ZhangZ, DietrichFS, et al. (2006) Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet 38 : 356–362.
32. BaoJ, LiD, WangL, WuJ, HuY, et al. (2012) MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J Biol Chem 287 : 21686–21698.
33. HeL, HeX, LimLP, de StanchinaE, XuanZ, et al. (2007) A microRNA component of the p53 tumour suppressor network. Nature 447 : 1130–1134.
34. ChoiYJ, LinCP, HoJJ, HeX, OkadaN, et al. (2011) miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol 13 : 1353–1360.
35. ConcepcionCP, HanYC, MuP, BonettiC, YaoE, et al. (2012) Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet 8: e1002797.
36. BoonRA, IekushiK, LechnerS, SeegerT, FischerA, et al. (2013) MicroRNA-34a regulates cardiac ageing and function. Nature 495 : 107–110.
37. HirshA (2003) Male subfertility. BMJ 327 : 669–672.
38. BastosH, LassalleB, ChicheporticheA, RiouL, TestartJ, et al. (2005) Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A 65 : 40–49.
39. Di GiacomoM, ComazzettoS, SainiH, De FazioS, CarrieriC, et al. (2013) Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell 50 : 601–608.
40. Martin-de-LaraF, Sanchez-AparicioP, Arias de la FuenteC, Rey-CamposJ (2008) Biological effects of FoxJ2 over-expression. Transgenic Res 17 : 1131–1141.
41. De FazioS, BartonicekN, Di GiacomoM, Abreu-GoodgerC, SankarA, et al. (2011) The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480 : 259–263.
42. FarleyFW, SorianoP, SteffenLS, DymeckiSM (2000) Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28 : 106–110.
43. SchwenkF, BaronU, RajewskyK (1995) A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 23 : 5080–5081.
44. MahadevaiahSK, TurnerJM, BaudatF, RogakouEP, de BoerP, et al. (2001) Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27 : 271–276.
45. SmythGK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3.
46. SmythGK, MichaudJ, ScottHS (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21 : 2067–2075.
47. IrizarryRA, HobbsB, CollinF, Beazer-BarclayYD, AntonellisKJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 : 249–264.
48. van DongenS, Abreu-GoodgerC, EnrightAJ (2008) Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods 5 : 1023–1025.
Štítky
Genetika Reprodukční medicína
Článek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 10
-
Všechny články tohoto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



