-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaFeeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
Animals dramatically modify their chemosensory behaviors to attractive and noxious chemical stimuli when starved. This could allow them to alter and optimize their food-search strategies to increase their survival and reproduction. Changes in the gene expression of chemoreceptors specialized in detecting environmental stimuli is observed in fish, insects and nematodes, and may be a general mechanism underlying the changes in chemosensory behaviors observed in starved animals. To elucidate this mechanism, we have developed an in vivo reporter assay in C. elegans for monitoring the expression of a candidate chemoreceptor gene in a single sensory neuron type, called ADL, as a function of feeding state. Using this reporter assay, we show that sensory inputs into ADL and neural outputs from ADL, as well as inputs from the RMG interneuron, which is electrically connected to ADL, are required to fine-tune expression of chemoreceptor genes in ADL. Sensory and circuit-mediated regulation of chemoreceptor gene expression is dependent on multiple pathways, including the neuropeptide receptor, NPR-1, and the DAF-2 insulin-like receptor. Our results reveal mechanisms underlying chemoreceptor gene expression, and provide insight into how expression changes in chemoreceptor genes may contribute to changes in chemosensory behavior as a function of feeding state.
Published in the journal: . PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004707
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004707Summary
Animals dramatically modify their chemosensory behaviors to attractive and noxious chemical stimuli when starved. This could allow them to alter and optimize their food-search strategies to increase their survival and reproduction. Changes in the gene expression of chemoreceptors specialized in detecting environmental stimuli is observed in fish, insects and nematodes, and may be a general mechanism underlying the changes in chemosensory behaviors observed in starved animals. To elucidate this mechanism, we have developed an in vivo reporter assay in C. elegans for monitoring the expression of a candidate chemoreceptor gene in a single sensory neuron type, called ADL, as a function of feeding state. Using this reporter assay, we show that sensory inputs into ADL and neural outputs from ADL, as well as inputs from the RMG interneuron, which is electrically connected to ADL, are required to fine-tune expression of chemoreceptor genes in ADL. Sensory and circuit-mediated regulation of chemoreceptor gene expression is dependent on multiple pathways, including the neuropeptide receptor, NPR-1, and the DAF-2 insulin-like receptor. Our results reveal mechanisms underlying chemoreceptor gene expression, and provide insight into how expression changes in chemoreceptor genes may contribute to changes in chemosensory behavior as a function of feeding state.
Introduction
An animals' feeding state (i.e. fed versus starved) and food availability dramatically alters the responsiveness of chemosensory neurons and behavioral output to suit the needs of the animal to, for instance, locate food, find mates and avoid predators under different environmental conditions. Although these state-dependent changes in chemosensory behaviors have long been thought to arise from plasticity in central processes, there is now growing evidence that feeding state gates responses in peripheral chemosensory neurons themselves, thereby directly modulating changes in chemosensory behaviors [1]. One particular mechanism by which feeding state could alter the response properties of chemosensory neurons, is by dynamically changing the transcript levels of chemoreceptor genes. For example, in mosquitoes, olfactory neuron responsiveness to host-specific odors after a blood-feeding is highly correlated with small changes in transcript levels of the corresponding olfactory receptor [2]–[5], which may allow them to alter their sensitivity to these odors, thereby altering its host-seeking behavior. Thus, dynamic changes in the expression levels of chemoreceptor genes may provide a simple mechanism by which chemosensory neurons may alter their responses to specific chemical stimuli under different feeding state conditions.
The Caenorhabditis elegans sensory system provides an ideal system to explore the mechanisms by which chemosensory gene expression and behavior is altered by changes in its environment. As in other animals, C. elegans is able to rapidly and reversibly modify its chemosensory behavior to specific chemicals according to its feeding state. For example, starved animals increase their adaptation towards particular attractive odors and they discriminate more classes of food-related odors than fed animals [6]. Starved animals also change their response to certain volatile repulsive odors, and this modulation occurs through serotonergic and dopaminergic signaling [7], [8]. The neuropeptide receptor, NPR-1, and insulin-like receptor, DAF-2, in C. elegans are also involved in a multitude of food-dependent behaviors. For example, pre-exposure of C. elegans to salt in the absence of food switches the normally attractive salt response to avoidance, and this modulation is dependent on insulin signaling from the AIA interneurons via DAF-2 acting in ASE sensory neurons [9], [10]. Recent elegant work has shown that the neuropeptide receptor NPR-1 plays a key role in the RMG interneuron to regulate avoidance responses of sensory neurons to pheromones, a response that appears to be mediated by food [11], [12]. Thus, insulin and neuropeptide signaling from interneurons play an important role in translating multiple aspects of food and feeding state information to peripheral chemosensory neurons to fine-tune their responses.
As described above, dynamic changes in the expression levels of chemoreceptor genes could, at least in part, contribute to modifications in chemosensory behaviors, but it is unknown whether or how neuromodulators, such as neuropeptides, insulin and monoamines, alter expression levels of chemoreceptor genes as a function of feeding state. In C. elegans, individual and distinct subsets of chemoreceptor genes are regulated by several mechanisms, including developmental changes, sensory activity, and levels of pheromones [13],[14]. In addition, our previous work showed that KIN-29 regulates a subset of chemoreceptors in chemosensory neurons [15] by phosphorylating the HDA-4 class II histone deacetylase (HDAC), and inhibiting the gene repressive functions of the MEF-2 transcription factor [16]. KIN-29 is a member of the salt-inducible kinase (SIK) family, which plays a major role in the regulation of lipolysis and gluconeogenic gene expression in response to feeding and fasting [17]–[20]. For example, during feeding, Drosophila SIK3 is activated by insulin to regulate fat stores through phosphorylation of HDAC4 [21], a function that appears to be conserved in C. elegans [16]. Upon starvation, Drosophila SIK3 is inactivated resulting in HDAC4 dephosphorylation and subsequent activation of FOXO-mediated gene expression [21]. Thus, SIKs are critical regulators of feeding and fasting states, suggesting that a role for KIN-29 in feeding-state dependent regulation of gene expression may be conserved.
Here, we identified a KIN-29-dependent chemoreceptor, srh-234, whose expression levels in the ADL sensory neuron type of C. elegans is downregulated upon starvation. We find that this starvation-modulation is likely a consequence of both sensory inputs associated with a decrease in food presence and an internal state of starvation due to a decrease in food ingestion. We show that in addition to KIN-29, expression levels of srh-234 are regulated by multiple pathways, including signaling mediated by the DAF-2 insulin-like receptor, OCR-2 TRPV channel, and NPR-1 neuropeptide receptor. We show that intact cilia and dendrites of ADL, as well as neural output from ADL are required for srh-234 expression. Cell - and tissue-specific rescue experiments show that DAF-2 and OCR-2 act in ADL neurons, whereas NPR-1 acts in RMG interneurons to regulate srh-234 expression and this regulation is dependent on unc-7/9 gap-junctions. While MEF-2 and DAF-16 FOXO transcription factors act downstream of KIN-29 and DAF-2, respectively, in regulating srh-234 expression, OCR-2 and NPR-1 pathways act independently from MEF-2 and DAF-16. Taken together, our results suggest that integration of sensory and circuit inputs via multiple signaling pathways allows animals to precisely modulate chemoreceptors genes according to their feeding status, providing insights into the gene expression mechanisms that contribute to chemosensory plasticity in C. elegans.
Results
Expression of srh-234 in ADL neurons is downregulated in starved animals
Since SIKs regulate feeding state-dependent gene expression, we investigated whether feeding and starvation regulates the expression of kin-29-dependent chemoreceptor genes. Expression of gfp driven under the regulatory sequences of candidate chemoreceptor genes, str-1, sra-6 and srh-234 is strongly downregulated in AWB, ASH and ADL neurons, respectively [15], [16]. We found that gfp expression driven under only 165 bp of the regulatory sequence of srh-234 (srh-234p::gfp) is strongly expressed in ADL in fed animals (Figure 1A, upper panel), but when animals were starved for long-periods of time (>6 hours), srh-234p::gfp was significantly downregulated (Figure 1A, lower panel). Similar results were found for another independent integrated transgenic array of srh-234p::gfp (oyIs57) (Table S2). The expression of str-1::gfp and sra-6::gfp was unaffected in starved animals. We further confirmed the starvation-induced change in expression by examining the endogenous levels of srh-234 with help of qRT-PCRs, and found that the transcript levels of srh-234 were similarly downregulated but not abolished in starved animals (Figure 1B). The effect of starvation on srh-234 expression is reversible as L1 larvae or adult animals starved for 12 hours and then re-fed with E. coli food restore expression to near wild-type levels within 6 hours (Figure S1A). Moreover, when starved L1 larvae were grown on a nutrient-rich minimal media for 24 hours that is axenic, i.e. in the absence of any bacterial food, allowing developmental growth albeit delayed {Szewczyk, 2003 #4559], no increase in srh-234 expression was observed (Figure S1B). As bacterial food can alter the production of pheromones in C. elegans [22], which in turn can regulate chemoreceptor gene expression [13], we examined srh-234 expression in the absence and presence of pheromones. However, no effect on srh-234 expression was found in daf-22(m130) mutants, which is required for pheromone biosynthesis [23], and in the presence of crude pheromone extracts (Table S2), suggesting that pheromones do not alter srh-234 expression. Thus, starvation reduces the expression of the kin-29-dependent chemoreceptor, srh-234, in ADL, and this modulation is reversible upon re-feeding with food.
Fig. 1. Starvation downregulates the expression of srh-234 in ADL neurons. 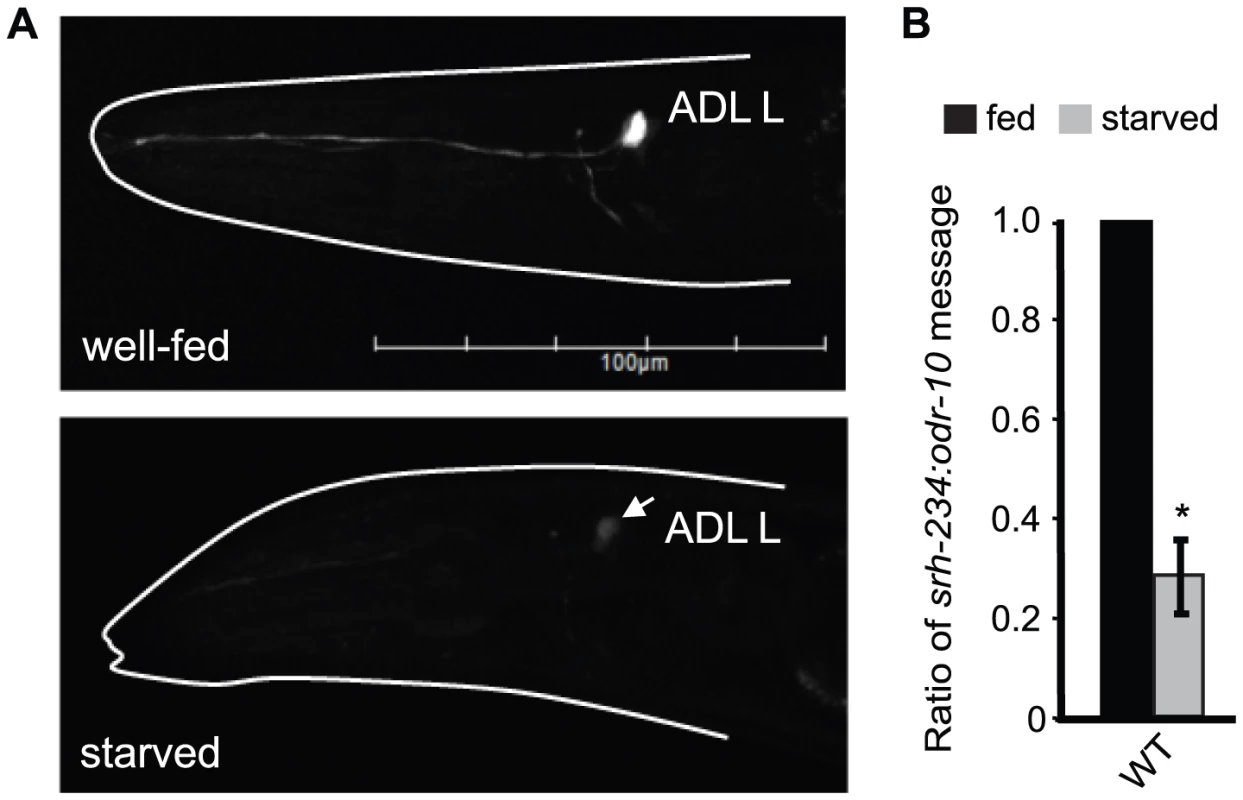
A) Expression of srh-234p::gfp in an ADL neuron of wild-type animals when well-fed in the presence of E. coli OP50 food (upper panel) and starved (lower panel) in the absence of food for 12 hours. The head of the animal is indicated with a white line. Arrow points to the ADL cell body. Images were acquired at the same exposure time. Lateral view: anterior is on the left. Scale, 100 µm. B) Levels of endogenous srh-234 messages are downregulated in starved animals compared to fed animals. Shown is the ratio of endogenous srh-234 message to endogenous odr-10 [65] message as quantified by qRT–PCR in fed and starved animals. In hermaphrodite animals, expression of odr-10 is unaffected by starvation. The mean of the ratios from two independent experiments is shown. * indicates values that are different from fed wild-type animals at P<0.01 using a two-sample t-test. Error bars denote the SEM. We next sought to identify additional chemoreceptor genes in ADL regulated by fed and starved conditions. Of the 11 chemoreceptor genes tested, only gfp driven by the cis-regulatory region of the chemoreceptor srh-34 in ADL was altered in fed and starved animals with an opposite phenotype than observed for srh-234; srh-34 is expressed in starved animals but not in fed animals (Table S1). Thus, the feeding status (fed versus starved) of C. elegans alters the expression levels of at least two chemoreceptor genes in ADL. We focused our subsequent studies on using the srh-234p::gfp reporter assay to further explore the mechanisms underlying starvation modulation of chemoreceptor gene expression in ADL.
External food presence and an internal state of starvation modulate srh-234 expression
The reduced srh-234 expression levels we observed in starved wild-type animals could arise as a consequence of an internal state of starvation triggered by a decrease in the ingestion of food, or alternatively, by an external sensory response as a result of a decrease in the perception of food. To distinguish between these possibilities, we exposed fed and starved wild-type adult animals carrying the srh-234p::gfp reporter to E. coli food treated with the antibiotic aztreonam, which results in bacteria that grow in long chains that C. elegans cannot eat due its large size, but still can smell and touch comparable to regular non-treated bacteria [24]. We found that fed animals placed on aztreonam-treated E. coli OP50 (inedible food) for 24 and 48 hours significantly reduces srh-234 expression mimicking the effects of starvation when compared to fed animals placed on non-treated E. coli OP50 (edible food) (Figure 2A). Consistent with these findings, loss-of-function (lf) mutations in eat-2 that result in animals with a pharyngeal pumping defect compromising their food ingestion, also reduce srh-234 expression on edible food (Figure 2B). Thus, the reduced srh-234 expression is likely due to an internal state response triggered by a decreased food ingestion. However, when we placed starved adult animals (in the absence of food for 6–12 hours) on aztreonam-treated E. coli OP50 (inedible food), we found that the srh-234 expression phenotype was not significantly different from their non-treated E. coli OP50 (edible food) counterpart as if they sense food presence correctly even when they cannot eat this food (Figure 2A). It is possible that starved animals in our experiments can ingest some of the inedible food but at a reduced amount. However, we found that animals placed on aztreonam-treated E. coli food have a starved appearance similar to starved animals placed on plates without any food. Moreover, we verified that aztreonam-treated E. coli cannot be eaten properly as we find that L1 larvae exposed to treated food used in our experiments do not sustain growth (98% of L1 animals placed on aztreonam-treated bacteria for 24 hours were arrested, as compared to 0% of L1 larvae placed on edible food). Thus, the perception of inedible food can override the effects of starvation on reducing srh-234 expression levels. In summary, our results suggest that the starvation-induced downregulation of srh-234 expression is likely a consequence of both sensory inputs associated with a decreased food presence, and an internal state of starvation triggered by a decrease in food ingestion.
Fig. 2. External food presence and internal state signals alter srh-234 expression levels. 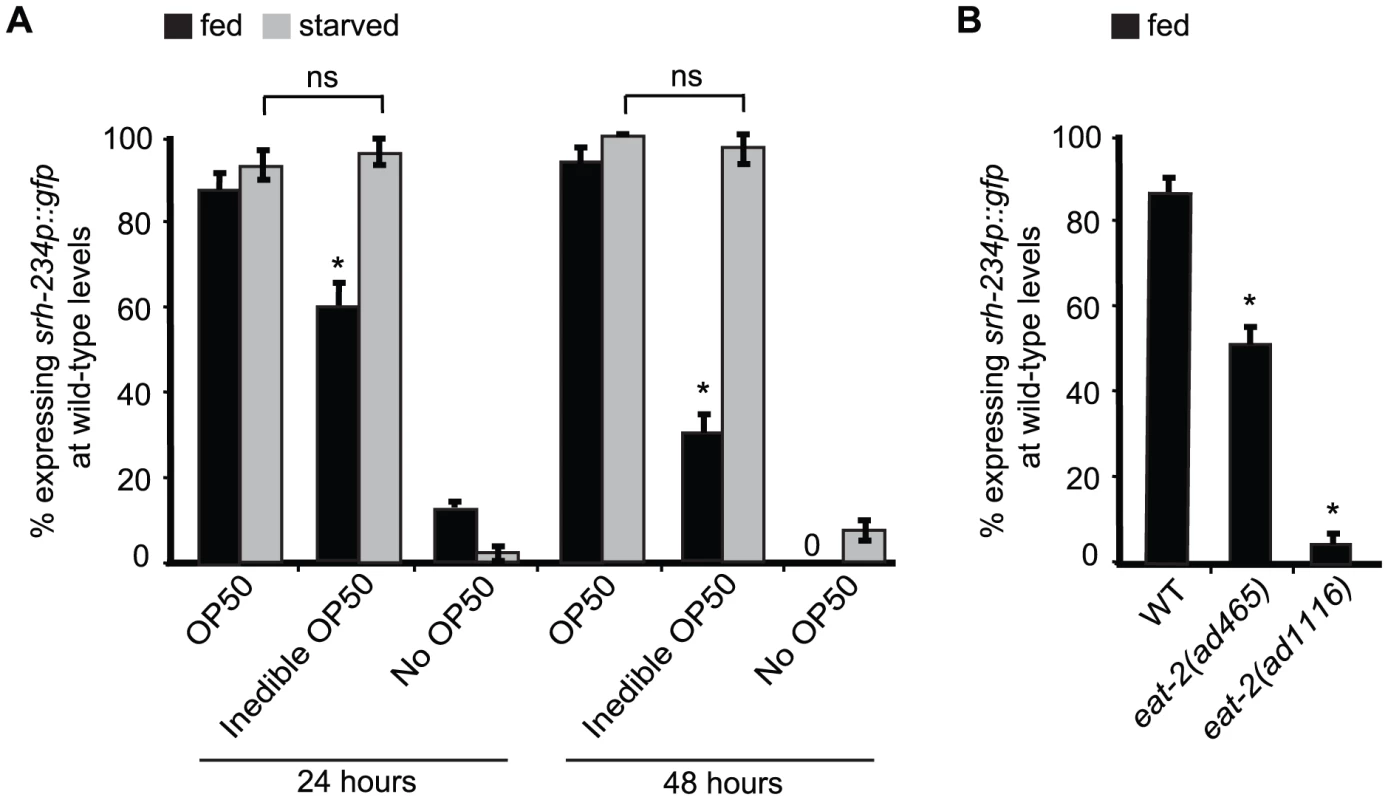
A) Percentage of animals expressing srh-234p::gfp at wild-type levels when fed with E. coli OP50 food (OP50), aztreonam-treated E. coli OP50 food (inedible OP50), or no food (no OP50) for 24 and 48 hours. Young adult animals grown on edible E. coli OP50 food were divided into two groups: a fed group maintained in the presence of food, and a starved group maintained in the absence of food for 6–12 hours. We confirmed that srh-234 expression levels upon starvation were reduced. Subsequently, adults were picked onto new NGM plates seeded with either edible E. coli OP50, no E. coli OP50, or inedible E. coli OP50 food (see Material and Methods). B) Percentage of eat-2(lf) mutants defective in food intake expressing srh-234p::gfp at wild-type levels. In all experiments, wild-type expression of srh-234p::gfp was defined as expression levels that allowed visualization of both the cell bodies and processes of at least one ADL neuron (see Material and Methods). Animals (n>150) were examined at 150× magnification for each condition or genotype. * indicates values that is different from that of wild-type animals at P<0.001, and n.s. indicates the values that are not significantly different between the different food conditions compared by brackets using a χ2 test of independence. Error bars denote the SEP. DAF-2/DAF-16 acts cell-autonomously in ADL to regulate srh-234 expression
As internal state signals in C. elegans are conveyed through an insulin signaling pathway with DAF-2 being the main insulin-like receptor [25], we explored whether insulin signaling plays a role in the regulation of srh-234. Consistent with low daf-2 insulin signaling being associated with a starved state, we found that daf-2(e1307) mutants reduce srh-234 expression in ADL in fed conditions, similar to starved wild-type animals (Figure 3A). Since daf-2 activates insulin signaling by repressing the daf-16 FOXO transcription factor, and loss of daf-16 function results in active insulin signaling [26], we next examined whether daf-16(mu86) suppressed the daf-2 - and starvation-induced downregulation of srh-234 expression. Indeed, we found that both daf-16(mu86) mutants and daf-2(e1307); daf-16(mu86) double mutants showed a significant increase in srh-234 expression during starvation compared to starved wild-type animals (Figure 3A), suggesting that starved animals reduce srh-234 expression by lowering DAF-2 signaling and activating DAF-16.
Fig. 3. daf-2 and daf-16a are required in ADL, but not in the intestine, to regulate srh-234 expression. 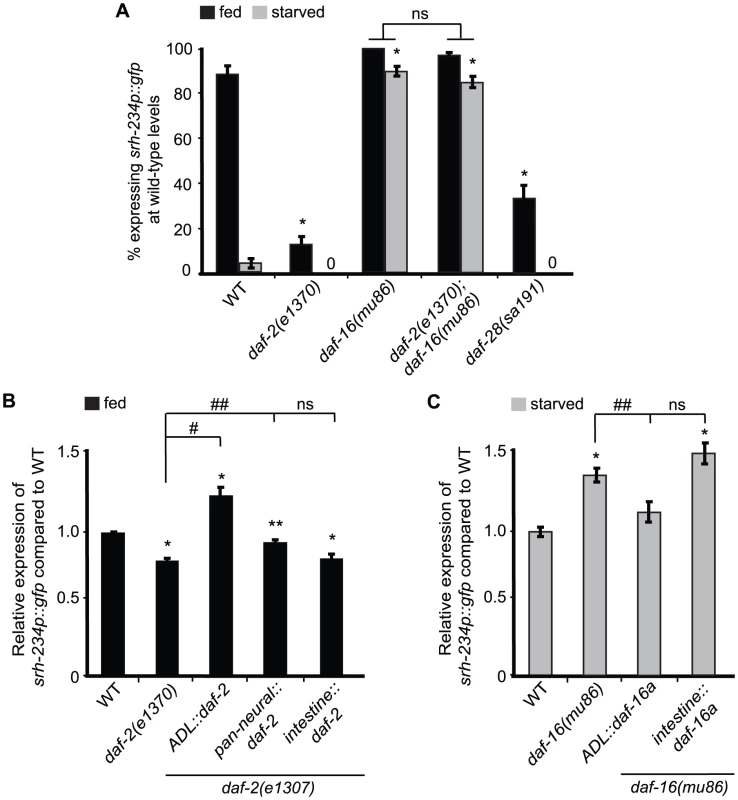
A) Percentage of mutant animals with defects in insulin signaling expressing srh-234p::gfp at wild-type levels. daf-2(e1307) is a temperature sensitive allele. In all experiments, animals were raised at 15°C (permissive temperature) and shifted to the 25°C (restrictive temperature) as L4 larvae. Animals (n>150) were examined at 150× magnification for each genotype. B, C) Relative expression of srh-234p::gfp in daf-2 or daf-16 mutants carrying compared to wild-type when fed or starved. For strains carrying ADL::daf-2 cDNA, pan-neural::daf-2 cDNA, intestine::daf-2 cDNA, ADL::daf-16a cDNA and intestine::daf-16a cDNA extrachromosomal arrays (see Material and Methods), data shown is for at least two independent transgenic lines. Animals (n = 18–25) were examined at 400× magnification for each genotype. * indicates values that are different from that of wild-type animals at P<0.001, # and ## indicates the values that are different between the genotypes compared by brackets at P<0.001, and P<0.05, respectively, using either a two-sample t-test or a χ2 test of independence. Error bars denote the SEM or SEP. To further refine the site of action of the DAF-2/DAF-16 insulin signaling pathway in regulating srh-234 expression, we introduced cDNAs of daf-2 and daf-16 under different cell - and tissue-specific promoters in daf-2 and daf-16 mutants, respectively, and measured their effects on srh-234 expression in either fed or starved conditions. We found that expression of daf-2 in ADL neurons fully restored the reduced srh-234 expression phenotype of daf-2(e1307) mutants during feeding to near wild-type levels, whereas daf-2 expression in the nervous system had a partial effect (Figure 3B). Expression of daf-2 in the intestine did not result in a restoration of the reduced srh-234 expression phenotype of daf-2(e1307) mutants (Figure 3B). These results suggest that DAF-2 signaling acts in ADL to regulate srh-234 expression. There are three functionally characterized DAF-16 isoforms, DAF-16a, DAF-16b and DAF-16df, and all of these isoforms show neuronal expression in developing larvae [27]–[29]. We found that expression of daf-16a cDNA specifically in ADL restored the increased srh-234 expression of daf-16(mu86) mutants during starvation back to wild-type levels, but similar to daf-2, expression of daf-16a cDNA in the intestine had no effect (Figure 3C). Together, these results suggest that both DAF-2 and DAF-16 act cell-autonomously in ADL to regulate srh-234 expression levels.
There are over 40 insulin-like peptides (ILPs) expressed in C. elegans. The daf-28 ILP, is a known agonist for DAF-2 and is expressed at high levels only when food is present [30],[31]. Interestingly, a semi-dominant mutation, sa191, in daf-28, thought to block other agonistic ILPs through stereo-hindrance [32], partially reduces srh-234 expression in ADL during feeding (Figure 3A). These results suggest that DAF-28 or other ILPs regulate srh-234 expression in ADL, likely through the DAF-2/DAF-16 insulin pathway.
NPR-1 acts in RMG to regulate srh-234 expression
The neuropeptide receptor, NPR-1, in C. elegans regulates a range of food-related behaviors. For example, mutants lacking npr-1 move rapidly, avoid high oxygen concentrations and aggregate in groups in a food-dependent manner [33]–[35]. We therefore examined whether loss of NPR-1 activity alters the expression levels of srh-234 in ADL. Indeed, we found a strong reduction in srh-234 expression in ADL in lf mutants of npr-1 (alleles ad609, ky13 and ok1447), and in a reduction-of-function npr-1 allele, g320, in fed conditions, with the g320 allele having the weakest effect (Figure 4A; Table S2). lf mutations in flp-18 and flp-21 encoding NPR-1 ligands as well as double mutants inactivating both ligands did not alter srh-234 expression (Table S2), suggesting that neuropeptides other than FLP-18/FLP-21 may act on NPR-1 to regulate srh-234 expression. Expression of npr-1 under control of its own promoter fully restored the reduced srh-234 expression phenotype of npr-1(ad609) mutants back to wild-type levels (Figure 4A). However, ADL-specific expression of npr-1 using the sre-1 promoter did not restore the reduced srh-234 expression phenotype of npr-1(ad609) mutants (Figure 4A), suggesting that npr-1 activity is required in neurons other than ADL to regulate srh-234 expression.
Fig. 4. Reducing npr-1 activity in RMG promotes srh-234 expression levels. 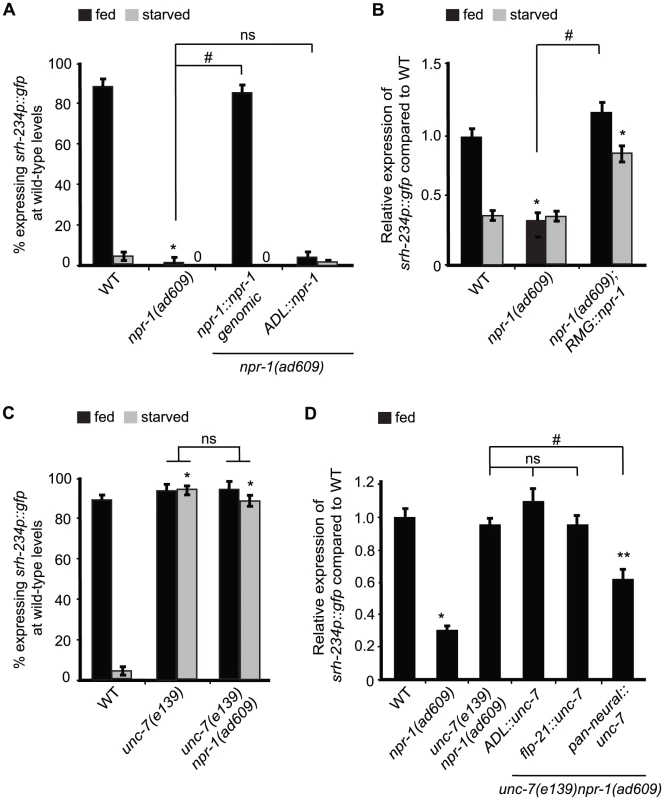
A) Percentage of npr-1 mutant animals expressing srh-234p::gfp at wild-type levels. For strains carrying the npr-1::npr-1 genomic and ADL::npr-1 extrachromosomal arrays (see Material and Methods), data shown is the average of at least two independent transgenic lines. Animals (n>150) were examined at 150× magnification for each genotype. B) Relative expression of srh-234p::gfp in npr-1 mutants compared to wild-type. For strains carrying RMG::npr-1 extrachromosomal arrays (see Material and Methods), data shown is for at least two independent transgenic lines. Animals (n = 20–23) were examined at 400× magnification for each genotype. C) Percentage of animals of the indicated genotypes expressing srh-234p::gfp at wild-type levels. Animals (n>150) were examined at 150× magnification for each genotype. D) Relative expression of srh-234p::gfp in unc-7 npr-1 double mutants compared to wild-type. For strains carrying ADL::unc-7L cDNA, flp-21::unc-7L cDNA and pan-neural::unc-7L cDNA extrachromosomal arrays (see Material and Methods), data shown is for at least two independent transgenic lines. Animals (n = 15–22) were examined at 400× magnification for each genotype. In all experiments, * indicates values that is different from that of wild-type animals at P<0.001, and # indicates the values that are different between the genotypes compared by brackets at P<0.001 using either a χ2 test of independence or using a two-sample t-test. n.s. indicates the values between brackets that are not significantly different. Error bars denote the SEM or SEP. We next sought to identify the cells where npr-1 activity is required for regulation of srh-234 expression in ADL. npr-1 is expressed in at least 20 cells [36]; 4 of these form chemical synapses with ADL, and 3 form gap-junctions with ADL, including RMG interneurons [37]. Previous studies have shown that RMG is the major site for NPR-1 in regulating aggregative behavior and pheromone responses [11], [12]. Based on these findings, we asked whether similarly RMG is the site of action for NPR-1 to modulate srh-234 expression in ADL. Cell-specific expression of npr-1 in RMG using a previously described Cre-Lox system (referred further as RMG::npr-1) [11] in npr-1(ad609) mutants completely restored the reduced srh-234 expression phenotype of npr-1 mutants to wild-type levels during feeding, and increased srh-234 expression when starved (Figure 4B). This increase is likely due to overexpression effects of the RMG::npr-1 transgene, which could overwhelm inhibitory regulation of srh-234 by starvation. Thus, npr-1 is necessary and sufficient in RMG to promote srh-234 expression.
We next asked how NPR-1 in RMG interneurons affects srh-234 expression in ADL? Reasoning by analogy to the recently proposed RMG hub-and-spoke circuit [11] we examined whether loss of gap-junction function alters srh-234 expression in ADL. The innexins, unc-7 and unc-9, are widely expressed in neurons and muscles and form electrical synapses in the locomotory system [38], and unc-9 is involved in the modulation of ADL-mediated pheromone responses [12]. We found that unc-7(e139) and unc-9(e101) fully suppressed the reduced srh-234 expression phenotype of npr-1(ad609) mutants in fed conditions (Figure 4C), but no suppression was observed in a daf-2(e1307) or kin-29(oy38) mutant background (Table S2). These results suggest that unc-7/9 gap junctions are necessary for npr-1-mediated regulation of srh-234 expression; however, other signaling pathways such as ocr-2 and daf-2 may act in parallel on the srh-234 promoter. We also observed that srh-234 expression in starved conditions is significantly upregulated in unc-7(e139) and unc-9(e101) mutants, as well as in unc-7 npr-1 and unc-9 npr-1 double mutants when compared to starved wild-type animals (Figure 4C; Table S2), suggesting that loss of unc-7/9 gap-junctions can suppress the effects of starvation on reducing srh-234 expression.
We next investigated whether UNC-7/9 are directly involved in the RMG gap-junction circuit to regulate srh-234 expression. We therefore expressed the cDNA of the unc-7 specifically in ADL neurons (ADL::unc-7), in flp-21-expressing cells that include RMG interneurons (flp-21::unc-7), and in all neurons (pan-neural::unc-7) in unc-7(e139) npr-1(ad609) double mutants, and examined their effect on srh-234 expression. Surprisingly, however, we did not observe a restoration of the reduced srh-234 expression phenotype of unc-7 npr-1 double mutants back to npr-1 levels for either transgene in fed conditions (Figure 4D). Moreover, ADL-specific knock down of unc-7 or unc-9 by RNAi did not suppress the reduced srh-234 expression phenotype of npr-1(ad609) mutants in fed conditions (Figure S3). Only pan-neural expression of unc-7 cDNA showed a partial suppression of the reduced srh-234 expression phenotype of unc-7 npr-1 double mutants (Figure 4D). Thus, UNC-7/9 may have indirect effects on the regulation of srh-234 expression mediated by NPR-1, although they could have subtle roles within the RMG gap-junction circuit.
Sensory inputs into ADL and neural outputs from ADL regulate srh-234 expression levels
In addition to inputs from RMG facilitated by NPR-1, our experiments with aztreonam-treated E. coli food that can be sensed but not eaten suggest that srh-234 expression is also modulated by sensory inputs from food. The presence of food could be perceived directly by ADL, or through other sensory neurons that connect to ADL. To examine these possibilities, we first performed physical and genetic manipulations of ADL sensory dendrites, thereby eliminating the ability of ADL neurons to perceive any sensory inputs from the environment. We analyzed srh-234 expression in wild-type animals in which dendrites of the bilateral ADL pair are physically cut with a femtosecond laser (Figure 5A). This subcellular laser surgery exhibits precision with sub-micrometer resolution and has been successfully used in C. elegans to analyze the role of AFD sensory dendrites in temperature sensation [39]. We found that cutting either the ADLL or ADLR sensory dendrite showed a significant reduction in srh-234 expression over time (Figure 5B). This is in contrast to AWB neurons where expression of str-1p::gfp is maintained after severing sensory dendrites [40]. Thus, ADL dendrites are necessary to promote srh-234 expression in fed conditions.
Fig. 5. Expression levels of srh-234 are modulated by sensory inputs into ADL, neural outputs from ADL and OCR-2 activity. 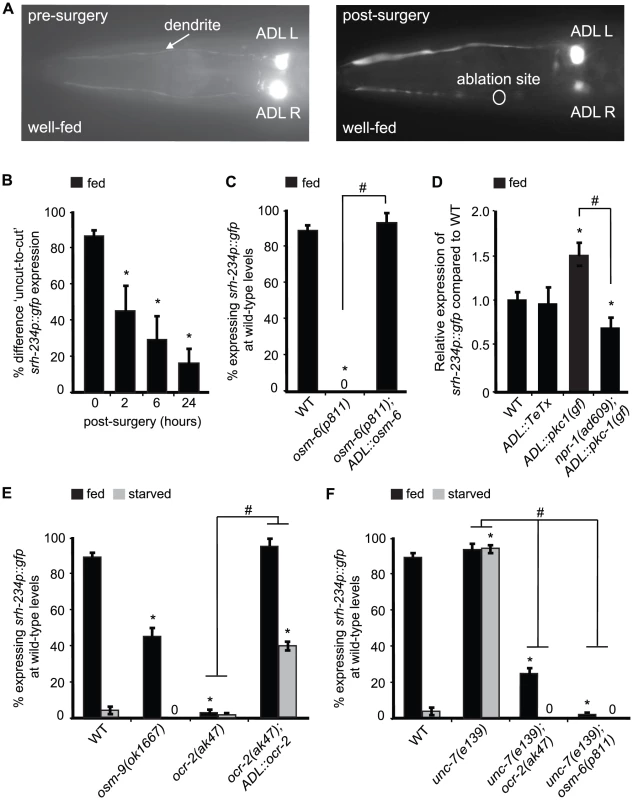
A) Pictures show examples of fed animals expressing srh-234p::gfp before (left panel) and after surgery (right panel; 24 hr) of the right ADL (ADLR) dendrite using a femtosecond laser (see Material and Methods). The ablation site is indicated as a circle. B) Difference in fed animals expressing srh-234p::gfp between severed and non-severed (‘cut-to-uncut’) ADL sensory dendrites. Animals (n = 8–17) were examined at 400× magnification for each genotype. C) Percentage of mutant animals with defects in cilia formation expressing srh-234p::gfp at wild-type levels. For strains carrying the ADL::osm-6 extrachromosomal array (see Material and Methods), data shown is the average of two independent transgenic lines. Animals (n>150) were examined at 150× magnification. D) Relative expression of srh-234p::gfp in the indicated genotypes compared to wild-type animals. For strains carrying ADL::pkc-1(gf) or ADL::TeTx extrachromosomal arrays, data shown is for two independent transgenic lines. Animals (n = 10–17) were examined at 400× magnification for each genotype. E, F) Percentage of animals of the indicated genotypes expressing srh-234p::gfp at wild-type levels. For strains carrying the ADL::ocr-2 extrachromosomal array (see Material and Methods), data shown is the average of two independent transgenic lines. Animals (n>150) were examined at 150× magnification. * indicates values that are different from that of wild-type animals at P<0.001, and # between the genotypes compared by brackets at P<0.001 using either a two-sample t-test, or the using a χ2 test of independence. Error bars denote the SEP or SEM. Although these dendritic cuts indicate a direct role for ADL sensory neurons in regulating srh-234 expression, it is possible that these subcellular cuts could damage ADL and thus compromise their physiology. To further confirm whether sensory inputs into ADL regulate srh-234 expression, we examined osm-5(p813) and osm-6(p811) mutants that lack functional sensory cilia [41], [42], and found that these cilia defective mutants strongly reduce srh-234 expression in ADL in fed conditions (Figure 5C). Functional reconstitution of ADL cilia in osm-6(p811) mutants by expressing osm-6 fused to mCherry under control of the sre-1 promoter was sufficient to restore wild-type srh-234 expression in fed conditions (Figure 5C). We confirmed that ADL formed proper cilia by visually inspecting their morphology with help of mCherry expression, and found a significant correlation between upregulation of srh-234 expression and wild-type cilia morphology of ADL (97% of ADL neurons with wild-type cilia expressed srh-234p::gfp expression at wild-type levels, n = 36). These results suggest that sensory inputs from food presence specifically through ADL cilia and dendrites are essential to promote srh-234 expression.
We next asked whether altering the neural output of ADL neurons also effects the expression levels of srh-234. We therefore generated transgenic animals that express the pkc-1(gf) and the tetanus toxin (TeTx) cDNA under control of the ADL-specific sre-1 promoter (ADL::pkc-1(gf) and ADL::TeTx). The pkc-1(gf) is a constitutively active protein kinase C that enhances synaptic output by promoting secretion of dense-core vesicles containing neuropeptides [11], [43]–[45], while TeTx prevents secretion of small neurotransmitter molecules by blocking synaptobrevin-mediated fusion of small, synaptic vesicles [46]. We found that expression of ADL::pkc-1(gf) strongly increased srh-234 expression in fed wild-type animals compared to non-transgenic siblings (Figure 5D), while blocking ADL synaptic output by expressing TeTx specifically in ADL (ADL::TeTx) did not significantly change levels of srh-234 expression (Figure 5D). Interestingly, the increased srh-234 expression phenotype of ADL::pkc-1(gf) is completely suppressed by npr-1(ad609) (Figure 5D), suggesting that npr-1 may act downstream of pkc-1(gf)-enhanced neuropeptide secretion from ADL to regulate srh-234 expression. Consistent with these findings, lf mutations in genes that disrupt the release and processing of neuropeptides globally (e.g. egl-3, unc-31 [47]–[49]), as well as an inhibitor of vesicle release (e.g. tom-1 [50]), also modulate srh-234 expression levels (Table S2). No changes in srh-234 expression are observed in mutants with defects in monoamine synthesis including serotonin, octopamine and dopamine, or upon exogenous exposure to these amines (Table S2). Together, these results suggest that perhaps release of neuropeptides from ADL may in turn modulate NPR-1 in RMG to regulate srh-234 expression.
OCR-2 acts in ADL to regulate srh-234 expression
We showed that intact ADL cilia are required to properly express srh-234 in fed conditions, suggesting that a cilia-localized mechanism for sensing food presence may be important for the regulation of srh-234. The TRPV channels, OCR-2 and OSM-9, localize to the cilia of a subset of sensory neurons, including ADL [51], [52], and OCR-2 may function in coupling the perception of food presence to starvation survival [53]. Based on these findings, we wondered whether OCR-2 and OSM-9 may transduce sensory inputs from food into ADL to modulate srh-234 expression. Indeed, we found that lf mutations in ocr-2 (alleles ak47 and yz5) strongly reduce srh-234 expression in ADL in fed animals, whereas osm-9(ok1667) did so more weakly (Figure 5E; Table S2). Thus, ocr-2 and to a lesser extent osm-9 promote srh-234 expression in fed conditions. It is possible that ocr-2(ak47) and osm-9(ok1667) mutants reduce srh-234 expression when fed as a result of a decrease in food ingestion instead of a decrease in food perception; however, consistent with previous findings [54], both ocr-2 and osm-9 mutants ingest food similar to wild-type animals as measured by pumping rates (231.0±4 and 225±5 pumps/min for ocr-2 and osm-9 mutants, respectively, as compared to 235±5 pumps/min for wild-type animals, n = 25). ADL-specific expression of ocr-2 under control of the sre-1 promoter (ADL::ocr-2) fully restored srh-234 expression in ocr-2(ak47) mutants during feeding, and is slightly upregulated when starved (Figure 5E); an increase likely caused by overexpression effects of the transgene. Thus, OCR-2 activity in ADL is necessary during feeding and sufficient when starved to regulate srh-234.
We further investigated the relationship between the OCR-2 and NPR-1 pathways in regulating srh-234 expression. As expected, double mutants for ocr-2(ak47); npr-1(ad609) showed a reduced srh-234 expression phenotype similar to that of npr-1(ad609) or ocr-2(ak47) mutants alone in both fed and starved conditions (Table S2). Interestingly, ocr-2(ak47) and osm-6(p811) fully suppressed the unc-7(e139) phenotype of upregulated srh-234 expression in starved conditions (Figure 5F), suggesting that sensory inputs and OCR-2 activity are essential for the gap junction-mediated reduction of srh-234 expression in response to starvation. Consistent with these findings, sensory inputs from inedible food that can be sensed but not eaten (Figure 2A), and ADL-specific overexpression of OCR-2 (Figure 5D), can override the effects of starvation on reducing srh-234 expression. Thus, starvation-modulation of srh-234 expression mediated by gap-junctions is likely dependent on sensory inputs.
Increased calcium signaling can bypass the requirement for OCR-2, NPR-1, DAF-2 and KIN-29 in regulating the expression of srh-234
As calcium signaling plays an important role in the regulation of the KIN-29-dependent str-1 chemoreceptor in AWB neurons [16], we explored the possibility that calcium signaling is also important for regulating srh-234. gf mutations in the voltage-gated calcium channel, egl-19, are predicted to prolong depolarization and result in sustained calcium influx [55]. We found that egl-19(gf) suppressed the starvation-induced downregulation in a wild-type background (Figure 6A), whereas lf mutations in egl-19 and unc-36, but not unc-2, encoding other voltage-gated calcium channels, partially reduce srh-234 expression during feeding (Figure S2). Thus, increased calcium signaling can override the effects of starvation on reducing srh-234 expression. We further show that egl-19(gf) can suppress the reduced srh-234 expression phenotype of npr-1(ad609), daf-2(e1307) and osm-9(ok1667) mutants (Figure 6A) as well as kin-29(oy38) mutants (Table S2). Expression of egl-19(gf) specifically in ADL neurons (ADL::egl-19(gf)) also suppressed the reduced srh-234 expression phenotype of ocr-2(ak47) and npr-1(ad609) mutants in fed conditions (Figure 6B). These results suggest that increased calcium signaling is sufficient in ADL to bypass the requirement of OCR-2, DAF-2 and KIN-29, and NPR-1 pathways in regulating srh-234 expression.
Fig. 6. mef-2 mutations and increased calcium signaling suppress the starvation-induced downregulation of srh-234 expression. 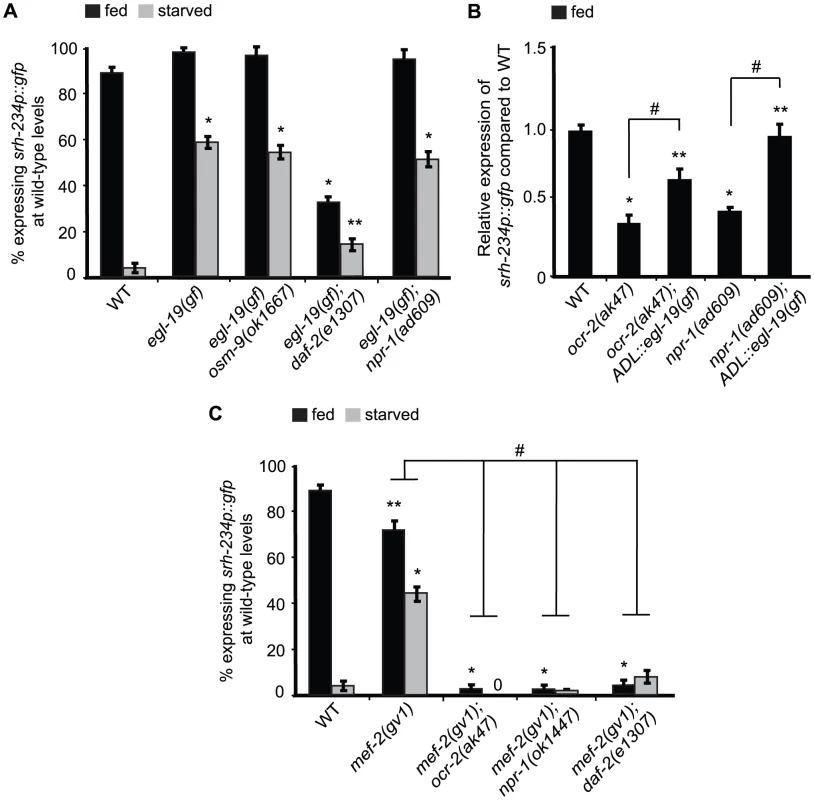
A, C) Percentage of animals of the indicated genotypes expressing srh-234p::gfp at wild-type levels. Animals (n>150) were examined at 150× magnification for each genotype. B) Relative expression of srh-234p::gfp in ocr-2 and npr-1 mutants compared to wild-type. For strains carrying ADL::egl-19(gf) extrachromosomal arrays (see Material and Methods), data shown is for two independent transgenic lines. Animals (n = 15–20) were examined at 400× magnification for each genotype. * and ** indicates values that are different from that of wild-type animals at P<0.001, and P<0.05, respectively, and # indicates the values that are different between the genotypes compared by brackets at P<0.05 using either a two-sample t-test or a χ2 test of independence. Error bars denote the SEP or SEM. To further test the hypothesis whether calcium levels in ADL may be modulated by feeding and starvation responses, we measured intracellular calcium dynamics specifically in ADL neurons in fed and starved wild-type animals in the presence of the C9-pheromone (asc-ΔC9; ascr#3) using the genetically encoded Ca2+ sensor GCaMP3. ADL is known to sense C9 as determined by calcium imaging and behavioral assays [12]. However, we found that C9-induced calcium transients were not significantly different in wild-type animals starved for 6 hours when compared to fed animals (Figure S3). Moreover, when we quantified the fluorescence intensity of the ADL::GCaMP3 reporter in the absence of any stimuli in wild-type animals starved for 6, 12 or 24 hours, we also observed no significant differences in ADL::GCaMP3 expression when compared to control fed animals (Figure S4). Thus, although we were unable to detect changes in calcium in fed and starved animals, our experiments with egl-19(gf) suggest that increased calcium signaling is correlated with increased srh-234 expression.
Mutations in mef-2 suppress the starvation-induced downregulation of srh-234 expression
We previously showed that lf mutations in mef-2 encoding the MEF2 transcription factor can suppress the reduced srh-234 expression phenotype of kin-29 mutants [16], suggesting that KIN-29 antagonizes the function of MEF-2 in regulating str-1. We show that mef-2(gv1) failed to suppress the reduced srh-234 expression phenotype of ocr-2(ak47), npr-1(ad609) and daf-2(e1307) mutants in fed and starved conditions (Figure 6C), suggesting that MEF-2 does not act genetically downstream of OCR-2, DAF-2 and NPR-1. In addition, we show that kin-29(oy38) mutants can suppress the increased srh-234 expression phenotype when overexpressing OCR-2 and NPR-1 (Figure 4B; Table S2), suggesting that these pathways likely act interdependently to regulate srh-234. Interestingly, mef-2(gv1) suppressed the starvation-induced downregulation of srh-234 expression, but had no major effect on srh-234 expression during feeding (Figure 6C), suggesting that the inhibitory regulation of srh-234 expression by starvation signals is dependent on MEF-2 function.
Since insulin signaling appears to be compromised in kin-29 mutants in the regulation of dauer formation and life-span [15], we next asked whether DAF-16 acts as a downstream effector of KIN-29 to regulate srh-234 expression. However, daf-16(mu86) did not suppress the srh-234 expression phenotype of kin-29(oy38) mutants as well as of ocr-2(ak47) and npr-1(ok1447) mutants (Table S2). Together, these results suggest that MEF-2 and DAF-16 may act as state-dependent transcriptional regulators of srh-234 expression downstream of KIN-29 and DAF-2, respectively, while OCR-2 and NPR-1 act via MEF-2 and DAF-16-independent pathways.
Discussion
In this study, we identified a chemoreceptor gene, srh-234, in the ADL sensory neuron type of C. elegans, whose expression levels is altered by feeding state conditions. Expression levels of srh-234 are regulated by sensory signals associated with food presence and internal starvation signals via integration of signals by multiple pathways. In ADL neurons, signaling mediated by the kin-29 salt-inducible kinase, the daf-2 insulin-like receptor, and the ocr-2 TRPV channel converge their regulation on srh-234 expression, while the npr-1 neuropeptide receptor acts in RMG interneurons to regulate srh-234 expression in ADL sensory neurons (Figure 7). This sensory - and circuit-mediated regulation of chemoreceptor genes may allow animals to precisely regulate and fine-tune their chemosensory responses as a function of feeding state.
Fig. 7. Model for sensory and circuit-mediated regulation of srh-234 expression. 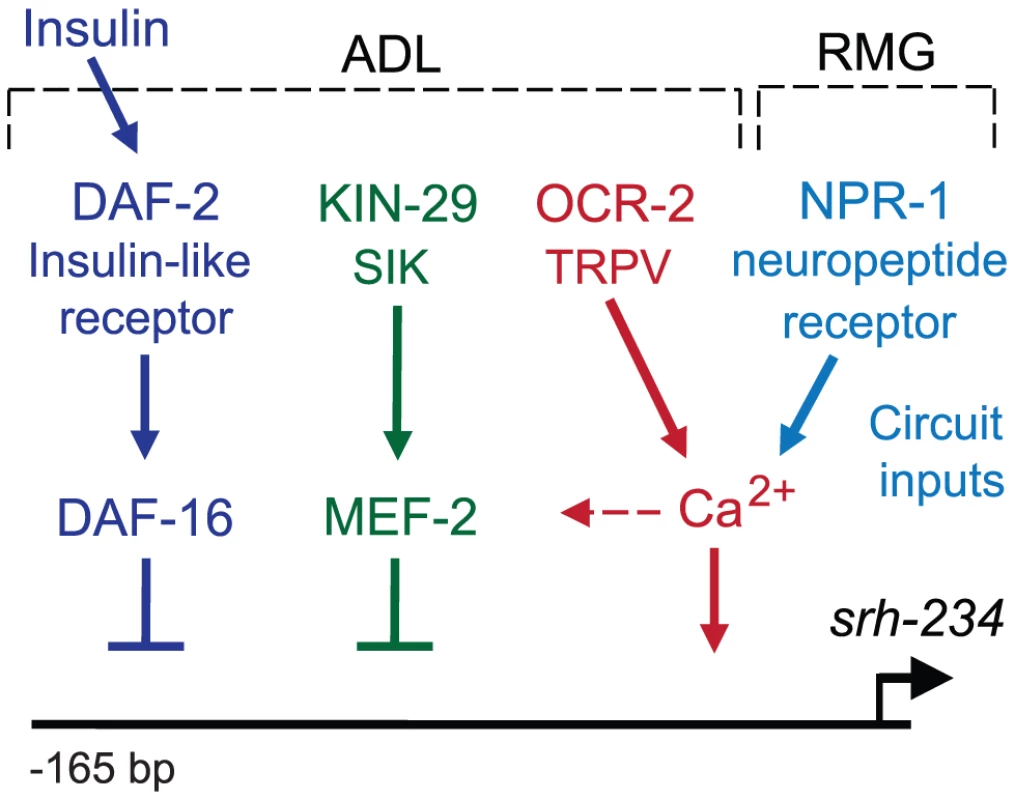
Expression levels of srh-234 are modulated by integration of sensory and internal feeding state signals via multiple pathways. During feeding, srh-234 expression is promoted by kin-29 salt-inducible kinase, daf-2 insulin-like receptor, and ocr-2 TRPV channel in ADL, and the npr-1 neuropeptide receptor in RMG interneurons. Negative transcriptional regulators of srh-234 expression, mef-2 and daf-16, act genetically downstream of kin-29 and daf-2, respectively, and likely act in parallel to ocr-2 and npr-1 pathways. The epistatic relationship between the different signaling pathways in ADL neurons remains to be fully defined. Unknown insulin-like peptides secreted by ADL or other neurons lead to activation of daf-2. Signaling mediated by kin-29, ocr-2, and npr-1, but less daf-2, converge on Ca2+ signaling, which likely affects activity-dependent transcription of chemoreceptor genes. After prolonged starvation, mef-2 and daf-16 may repress srh-234 expression, while yet unknown transcription factors may drive srh-234 expression during feeding. Sensory inputs from food into ADL, and an internal state of starvation modulates chemoreceptor gene expression
Expression of srh-234 is increased in fed animals and dramatically reduced in starved animals. When wild-type fed animals were presented with inedible E. coli food (bacterial food that can be sensed but not eaten after treatment with the aztreonam antibiotic), or when eat-2(lf) mutants were exposed to edible food, the expression of srh-234 decreased in ADL, suggesting that the signal needed to reduce srh-234 expression is probably an internal metabolic signal arising after food ingestion. However, when starved animals were presented with either edible or inedible E. coli food, expression of srh-234 is increased in ADL, suggesting that animals perceive the availability of food, even when they are unable to eat the inedible food. Thus, inedible food likely blocks signals associated with starvation to regulate srh-234 expression. Interestingly, this finding is similar to a previous study, showing that the repressive effects of starvation on C. elegans mating can be partially blocked by placing males on inedible food [24].
Sensory inputs are known to alter chemoreceptor gene expression in a subset of sensory neurons. We show that loss of all sensory inputs into ADL, as demonstrated by physically cutting the ADL dendrites, causes fed animals to reduce srh-234 expression. Moreover, restoring only ADL cilia in osm-6(lf) mutants that have severe truncations of all ciliated sensory neurons [41], [42], rescues the reduced srh-234 expression phenotype of osm-6(lf) mutants. These results suggest that sensory inputs through ADL sensory endings are essential to regulate srh-234 expression. Thus, we propose that depending on food availability and feeding state, srh-234 expression in ADL is regulated by two inputs, through external signals from food presence mediated directly by ADL and internal state signals resulting from food ingestion.
Circuit inputs modulate chemoreceptor gene expression
Modulation of chemoreceptor gene expression is thought to be defined primarily by external environmental inputs perceived by sensory neurons and not by circuit inputs from other neurons. Our results described here reveal a novel role for circuit-mediated regulation of chemoreceptor genes in sensory neurons. First, we show that npr-1 in RMG interneurons, which forms gap-junctions with ADL [56], was both necessary and sufficient for srh-234 expression in ADL. Second, this npr-1-mediated regulation of srh-234 is dependent on the function of unc-7/9 gap-junctions. Third, we show that enhancing the secretion of dense-core vesicles that contain neuropeptides by expressing pkc-1(gf) in ADL neurons promotes expression of srh-234, and this regulation is dependent on npr-1. Fourth, we show that mutants with defects in neuropeptide processing and secretion alter the expression of srh-234. Lastly, insulin signals from other neurons may act on DAF-2 in ADL to regulate srh-234 as suggested by the srh-234 expression phenotype of daf-28(gf) mutants, and daf-2/daf-16 signaling acting cell-autonomously in ADL.
The specific mechanisms by which npr-1 in RMG interneurons alters the expression of srh-234 in ADL remains to be determined. Current understanding suggests that NPR-1-mediated regulation of aggregative behavior and pheromone responses requires RMG [11], [12], which forms the hub of a circuit that is connected to spoke sensory neurons including ADL via gap junctions [56]. We show that unc-7 and unc-9 gap-junctions are essential for npr-1-mediated regulation of srh-234. Surprisingly, however, cell-specific rescue and RNAi knock down experiments of unc-7/9 in either ADL or RMG neurons suggest that they may not be directly involved in forming gap-junctions between RMG and ADL neurons. One possibility is that unc-7/9 are required in other neurons besides RMG and ADL for regulating srh-234 expression, which is consistent with the partial rescue of the srh-234 expression phenotype of unc-7 npr-1 double mutants by driving unc-7 cDNA in the nervous system (Figure 4D). Alternatively, any of the other innexins in C. elegans may act as gap-junction components within the RMG circuit.
A role for insulin signaling in regulating chemoreceptor gene expression
Insulin signals reflect the feeding state of an animal, which has been suggested to directly affect chemosensory sensitivity to chemical cues. Our results suggest a new role for insulin-mediated regulation of chemoreceptor genes. Consistent with low daf-2 signaling being associated with a starved state of C. elegans, we show that srh-234 expression is strongly reduced in daf-2(lf) mutants in a daf-16-dependent fashion. Rescue experiments with various promoters suggest that both DAF-2 and DAF-16 act cell-autonomously in ADL to regulate srh-234 expression. A gf mutation in the daf-28 insulin-like peptide (ILP), which has been proposed to block other agonistic ILPs from binding to DAF-2 [32], reduces srh-234 expression in fed conditions. These results indicate that ILPs from other yet unknown neurons may target DAF-2 in ADL, which in turn regulates srh-234 expression. C. elegans contains 40 ILPs, and for some specific functions in particular sensory neurons have been reported. For example, in salt-chemotaxis learning, DAF-2 acts in ASE sensory neurons, and is targeted by the INS-1 ILP released from AIA interneurons [9]. Similarly, release of INS-1 from AIA regulates food-dependent AWC responses [57]. However, we show that ins-1 mutants did not change srh-234 expression in fed or starved conditions, suggesting there may be other ILPs involved in the regulation of srh-234 expression.
A role for OCR-2 and calcium in regulating chemoreceptor gene expression
The OCR-2 and OSM-9 TRPV channels in C. elegans have been implicated in regulating the expression of sensory neuronal genes. For example, loss of these TRPV channels reduces expression of the odr-10 chemoreceptor gene in AWA neurons [51], [52], while they reduce the expression of the key enzyme in serotonin synthesis, tph-1, in AFD neurons [54]. We reveal here the first description of a chemoreceptor regulated by ocr-2 in ADL neurons. TRPV channels in mammals are permeable to calcium [58], and in C. elegans, the reduced tph-1 expression phenotype of ocr-2(lf) mutants can be suppressed by activating downstream calcium-dependent pathways [54]. Similarly, we show that increased calcium signaling using egl-19(gf) can bypass the requirement of OCR-2 in regulating the expression of srh-234. Such activity-dependent transcriptional regulation of chemoreceptors may allow TRPV channels to regulate the sensitivity of sensory neurons to particular chemical stimuli as a function of feeding state. MEF2 plays a critical role in activity-dependent transcription in the nervous system [59], but we show that mef-2 does not suppress the srh-234 reduced expression phenotype of ocr-2 mutants. Thus, TRPV-mediated transcriptional regulation of srh-234 is independent of MEF-2 function, suggesting other possible downstream effectors of OCR-2 in regulating srh-234 expression. A possible target is the CREB-regulated transcriptional coactivator CRTC1, which recently was shown to regulate TRPV-mediated longevity [60], and is a known target for SIK phosphorylation [20].
A role for MEF-2 and DAF-16 as state-dependent regulators of chemoreceptor gene expression
The specific transcriptional mechanisms by which feeding and starvation regulate chemoreceptor gene expression remain to be deciphered; however, our genetic experiments implicate a key role for the MEF-2 and DAF-16 transcription factors acting downstream of the KIN-29 and DAF-2 pathways, respectively. We show that mef-2(lf) and daf-16(lf) suppress the starvation-induced downregulation of srh-234 expression, but the expression of srh-234 is not altered during feeding. On the basis of these genetic experiments, we suggest that under starved conditions, MEF-2 and DAF-16 are required to repress but not activate srh-234 expression, suggesting that other yet unknown transcription factors may be required to drive srh-234 expression in ADL. Our previous work showed that MEF-2 is able to directly bind a MEF-2 sequence motif upstream of the str-1 chemoreceptor [16]. Searching the upstream sequence of srh-234 revealed a similar MEF2 binding motif as well as an E-box sequence motif (Figure S5). E-box motifs are found in the cis-regulatory region of genes expressed in ADL neurons [61], which are known to bind transcription factors of the basic helix-loop-helix (bHLH). Interestingly, MEF2 has been shown to interact with bHLH factors at E-boxes to regulate myogenic gene expression [62]. Thus, a similar mechanism may operate in C. elegans, in which MEF-2 function is essential for the starvation-induced reduction in srh-234 expression levels by repressing a bHLH transcription factor that drives expression of srh-234 in ADL via the E-box. The molecular mechanism by which DAF-16 regulates srh-234 remains unclear as no canonical DAF-16 binding element (DBE) appears to be present in the 165 bp promoter sequence necessary for feeding-state regulation of srh-234 (pers. comm. M.G and A.M.V). In Drosophila, SIK3 activity can be activated by insulin, which antagonizes FOXO-activated gene expression [21]. However, we show that KIN-29 antagonizes the function of MEF-2 but not DAF-16 in regulating the expression of srh-234, suggesting that DAF-16 mainly acts downstream of insulin signaling.
Functional consequences of regulating chemoreceptor genes in ADL
C. elegans expresses multiple chemoreceptors in each chemosensory neuron. We and others have proposed that to selectively modify a response to a single chemical may rely on changing the expression of individual or subsets of chemoreceptor genes, rather than altering the response of the entire neuron, which would inadvertently result in changing the response to all stimuli sensed by that neuron. Selectively modulating distinct populations of chemoreceptors in this manner may allow C. elegans to modulate ‘long-term’ changes in chemosensory responses in changing environmental conditions. This expression strategy may be particularly important in ADL, which mediates avoidance responses to a wide variety of chemical signals in its environment, such as odors [63], pheromones [12], and heavy metals [64].
Our results show that the expression of least two candidate chemoreceptor genes in ADL, srh-234 and srh-34, are regulated by fed and starved conditions, but it is unknown what chemical or subset of chemicals are detected by these chemoreceptors. Only a few chemoreceptors with known chemical ligands have been found in C. elegans, such as the chemoreceptor gene, odr-10, expressed in the AWA olfactory neuron type for the attractive chemical diacetyl [65]. When the odr-10 gene was introduced into the repulsive AWB neuron, C. elegans changes its normally attractive response to diacetyl to trigger avoidance of diacetyl [66], suggesting that chemoreceptors expressed in a specific neuron type are linked to a common odor response that is determined by the identity of the neuron. Similarly, all chemoreceptors expressed in the ADL neuron type may be linked to a common avoidance response to chemicals detected by ADL, although in some cases a sensory neuron has the capacity to switch its behavioral preference towards odors [45]. The presence and absence of food is known to rapidly and reversibly alter responses of animals to repulsive chemical cues, and part of this response appears to be mediated by ADL [7]. It is therefore tempting to speculate that increased srh-234 expression in ADL, allows fed animals to be less tolerant of aversive stimuli detected by srh-234, whereas decreased srh-234 expression in ADL following starvation may allow starved animals to be more tolerant to these aversive stimuli. This dynamic modulation in chemoreceptor gene expression could allow starved C. elegans animals to sample different environments, perhaps increasing their chances of finding food under stress-full conditions.
Plasticity in chemoreceptor gene expression in other systems
Other invertebrates and vertebrates alter chemoreceptor gene expression in response to environmental and developmental signals. For example, expression of olfactory receptor genes in the zebrafish Danio rerio is induced in temporal waves, which may reflect a mechanism for odorant sensitivity during development [67]. Changes in chemoreceptor gene expression is also dependent on the sex of an animal. After mating, female flies of Drosophila rapidly modify their chemosensory behaviors to lower their attraction to males such that they can focus on reproduction, and these behavioral changes are accompanied by modulation of different chemoreceptor genes [68]. Interestingly, in mosquitoes, the regulated expression of a subset of olfactory receptor genes before and after a blood feeding has been correlated with transitions between host-seeking and leaving behavior [2]–[5]. Parasitic nematodes also actively seek out their host using chemical cues (reviewed in [69]); however, little is known about host-seeking behaviors in parasitic nematodes, and even less is known about whether expression of chemoreceptor genes are modulated by its feeding state. The function of certain chemosensory neuron types in parasitic nematodes have been examined in only a few cases. For example, similar to C. elegans, ablation of ASE and ASH neurons of the parasitic nematode S. stercoralis showed that these sensory neuron types mediate attraction and repulsion to soluble chemicals, respectively [70]. Given that C. elegans has high anatomical and functional similarity to certain parasitic nematodes, it would be important to determine whether similar mechanisms of feeding state-regulated chemoreceptor gene expression presented here operate in parasitic nematodes. Moreover, identification of additional regulators and mechanisms underlying chemoreceptor gene expression will lead to a better understanding of how animals modify their chemosensory behavior in response to changes in external and internal conditions.
Materials and Methods
Strains
Nematodes were grown at 20°C under standard conditions [71] on nematode growth medium (NGM) with E. coli OP50 as the primary food source unless indicated otherwise. The wild-type strain used was C. elegans variety Bristol, strain N2. Mutant strains used in this study were obtained from the Caenorhabditis Genetics Center (CGC) unless indicated otherwise: PR811 osm-6(p811), PR813 osm-5(p813), DA609 npr-1(ad609), RB1330 npr-1(ok1447), CX4148 npr-1(ky13); CX4057 npr-1(g320); VC2106 flp-18(gk3063), RB982 flp-21(ok889), VC461 egl-3(gk328), CB169 unc-31(e169), CB55 unc-2(e55), CB251 unc-36(e251), CB1370 daf-2(e1370), JT191 daf-28(sa191), CF1038 daf-16(mu86), LC33 bas-1(tm351), CB1112 cat-2(e1112), RB1161 tbh-1(ok1196), RB993 tdc-1(ok914), GR1321 tph-1(mg280), VC1262 osm-9(ok1677), CX4544 ocr-2(ak47), JY243 ocr-2(yz5), DA465 eat-2(ad465), DA1116 eat-2(ad1116), KM134 mef-2(gv1), PY1476 kin-29(oy38), CB139 unc-7(e139), CB101 unc-9(e101), MT6129 egl-19(n2368)gf, MT1212 egl-19(n582), VC223 tom-1(ok285), and DR476 daf-22(m130). Transgenic strains used in this study were: VDL3 oyIs56[srh-234p::gfp, unc-122p::rfp], VDL143 oyIs57[srh-234p::gfp, unc-122p::rfp], and BOL171 npr-1(ad609); lin-15[ncs-1p::Cre flp-21p::loxPstoploxP::npr-1 SL2 gfp, lin-15(+)] (RMG::npr-1) [11], and sre-1::GCaMP3 [12]. Double mutant strains were constructed using standard genetic methods, and the presence of each mutation was confirmed via PCR or sequencing.
Real-time qRT PCR
Total RNA was isolated from a growth-synchronized population of adult animals and reverse transcribed using oligo(dT) primers. Real-time quantitative reverse transcription–PCR (qRT–PCR) was performed with a Corbett Research Rotor-Gene 3000 real-time cycler, Platinum Taq polymerase (Invitrogen), and primers specific for srh-234 and odr-10 coding sequences. Primer sequences for srh-234 are 5′-GGACAATTGAAATGCAACACA-3′ and 5′-GACGGGGACAATAAAGAGCA-3′. Primer sequences for odr-10 are 5′-GAGAATTGTGGATTACCCTAG-3′ and 5′-CTCAATATGCATTATAGGTCGTAATATG-3′.
Measurement and quantification of srh-234p::gfp
Animals carrying srh-234p::gfp reporters were cultured and grown at 20°C on standard nematode growth media (NGM) plates seeded with E. coli OP50 as the bacterial food source unless indicated otherwise. Young adult animals were washed with M9 buffer (to remove any bacteria in the gut) and transferred onto plates with E. coli OP50 food (Fed) or without E. coli OP50 food for 12–24 hours (Starved) unless indicated otherwise. Levels of srh-234p::gfp expression in animals were measured under a dissection microscope equipped with epifluorescence as described [16]. For quantification, gfp was scored as “bright” if levels of gfp fluorescence allowed visualization of one of the ADL cell bodies and dendritic process, and “dim” if gfp expression could not be detected or could be detected weakly at the same magnification. For more precise measurements, we mounted animals on a DM5500 compound microscope, and used Volocity analytical software to quantify gfp expression levels emanating from the srh-234p::gfp reporter. Statistical analyses of srh-234 expression were performed using either the two-sample t-test, or the χ2 test of independence to test for statistically significant differences between proportions in the categories “bright” and “dim” for different genotypes (d.f. 1). A proportion of 0% was set to a default of 1.
Analysis of srh-234p::gfp expression
Different food conditions: We exposed fed and starved animals carrying srh-234p::gfp reporters to aztreonam-treated E. coli OP50 (inedible food). For generating inedible food, E. coli OP50 was treated with the aztreonam antibiotic as previously described [24]. In brief, E. coli OP50 was grown in LB to log phase at 37°C with shaking. Cultures were mixed with the aztreonam antibiotic (Sigma) to a final concentration of 10 µg/ml and incubated for an additional 3 hours at 37°C with minimal shaking to prevent bacterial shearing. Bacteria was spread onto NGM plates containing 10 µg/ml aztreonam and immediately dried and used the same day, since the septum inhibitory effects of aztreonam are short lived. Expression levels of srh-234p::gfp were measured and quantified following exposure to the different food conditions as described above.
Sephadex-beads: We exposed starved wild-type animals carrying srh-234p::gfp reporters to 1 ml of 30 mg/ml Illustra Sephadex G-50 DNA Grade Fine (GE Healthcare UK Limited) suspended in water spread on NGM agar plates without bacterial food as described previously [72]. After at least 6 hours of exposure to Sephadex beads, animals were rapidly transferred into 35% sucrose solution to separate animals from the Sephadex beads. Animals floating on the surface of the sucrose solution were collected in 1× M9 buffer and immediately transferred to NGM agar plates without food for measurement and quantification of srh-234p::gfp expression levels. We confirmed that the sucrose floating procedure alone does not alter srh-234p::gfp expression levels.
Monogenic amines: We exposed fed or starved wild-type animals carrying srh-234p::gfp reporters to NGM plates with or without 3 mg/ml octopamine-hydrochloride (Sigma) in the presence of food, and 1 mg/ml serotonin creatinine sulphate (Sigma) in the absence of food. For octopamine treatment, an overnight culture of E. coli OP50 in LB was spun down and resuspended in 1/20 volume of water. About 50 µl of the concentrated OP50 was spread on the assay plates and was left until the surface of the plates became dry. After 6 hours of exposure to serotonin and octopamine, srh-234p::gfp expression levels were measured and quantified.
Pheromones: We exposed fed wild-type animals carrying srh-234p::gfp reporters to low levels of crude dauer pheromone in the presence of UV-killed E. coli food as described previously [13]. Dauer pheromone was prepared according to Golden and Riddle [22].
Expression constructs and generation of transgenic animals
Expression vectors were generated by amplifying either the wild-type genomic sequences of osm-6, npr-1, ocr-2, egl-19(gf), or cDNAs of daf-2 ([73], a kind gift from Shreekanth Chalasani), daf-16a (this study), unc-7, unc-9 (this study), pkc-1(gf) and tetanus toxin (TeTx) (kind gift from Cori Bargmann, [12]). This resulted in the generation of the constructs pMG1 sre-1p::osm-6 genomic::mCherry (ADL::osm-6), pMG2 sre-1p::npr-1 genomic::mCherry (ADL::npr-1), pMG3 sre-1p::ocr-2 genomic::mCherry (ADL::ocr-2), pMG4 sre-1::pkc-1(gf) cDNA (ADL::pkc-1(gf)), pMG5 sre-1p::tetanus toxin cDNA (ADL::TeTx), pVDL14 sre-1p::daf-2 cDNA SL2::mCherry (ADL::daf-2), pMG14 sre-1p::daf-16a cDNA SL2::mCherry (ADL::daf-16a), pMG24 sre-1p::unc-7 cDNA SL2::mCherry (ADL::unc-7), and pMG39 flp-21::unc-7 cDNA SL2::mCherry (flp-21::unc-7), pMG40 unc-31p::unc-7 cDNA SL2::mCherry (pan-neural::unc-7), and pMG37 sre-1p::egl-19(gf) genomic SL2::mCherry (ADL::egl-19(gf)). Expression constructs rgef-1p::daf-2 (pJH664) (pan-neural::daf-2), ges-1p::daf-2 (pJH668) (intestine:daf-2), and ges-1p::gfp::daf-16a (pJH2973) (intestine::daf-16a) are kind gifts from Mei Zhen [32]. For generating the npr-1p::npr-1 expression construct, we fused 2.5 Kb of the npr-1 regulatory sequence and the complete npr-1 wild-type genomic sequence to gfp as previously described [74]. Transgenic animals were generated using the unc-122p::dsRed (50–100 ng/µl) or the pRF4 rol-6(su1006) co-injection markers injected at 150 ng/µl. Expression constructs were injected at 20 ng/µl. For generating the cell-specific knock down constructs, we fused the sre-1 promoter to a 1 kb exon-rich genomic fragment of either unc-7 or unc-9 in the sense and anti-sense (sas) orientation as described [75]. Equal amounts of PCR products of either ADL::unc-7(sas) or ADL::unc-9(sas) were mixed and microinjected together with the pRF4 rol-6(su1006) co-injection markers injected at 90 ng/µl. All amplified products in the generated constructs were sequenced to confirm the absence of errors generated via the amplification procedure.
Subcellular laser ablation
Laser microsurgery was carried out as previously described [39]. A titanium::sapphire laser system (Mantis PulseSwitch Laser, Coherent Inc., Santa Clara, CA) generated a 1 kHz train of near infrared (λ = 800 nm) pulses that were ∼100 fs in duration and had a pulse energy of 5–15 nJ. The laser beam was focused to a diffraction-limited spot (using a 60× microscope objective) and used to disrupt sensory dendrites of ADL neurons expressing srh-234p::gfp. Following brief laser exposure, the targeted dendrite was inspected for a visual break to confirm that is was severed. Prior to laser surgery of either the ADLL or ADLR sensory dendrite, fed animals were aestheticized on 2% agar pads with 3 mM Sodium Azide, removed post-surgery for recovery and returned to NGM agar plates containing E. coli OP50 for measuring and quantifying srh-234p::gfp expression levels 2, 4, 6 and 24 hours following surgery in fed animals. Expression of srh-234 in severed (“cut”) neurons was compared to controls (“un-cut”) neurons in the same animals that were aestheticized for imaging but received no laser surgery.
Quantification of sre-1p::GCaMP3 in fed and starved animals
To measure fluorescence intensity of GCaMP3 expressed specifically in ADL neurons under either fed or starved conditions, three L4 staged sre-1p::GCaMP3 integrated transgenic animals [12] were grown on a E. coli OP50 seeded plate for 1.5 days to obtain about 100 eggs. The eggs were grown until they became young adults at 20°C. The young adult animals were washed in 1× M9 buffer and divided and placed on two NGM plates (one plate in the presence of E. coli OP50 food: “fed”, and one plate in the absence of E. coli OP50 food, “starved”). Images of GCaMP3 expression in the ADL neurons were captured under fixed exposure time (500 ms) with a Zeiss Axioplan microscopy using a 40× objective and Zeiss AxioCam HR camera at 0 hr, 6 hr, 12 hr or 24 hr in either fed or starved conditions (n = 40–60 for each). The fluorescence intensity of GCaMP3 in ADL was quantified using the Image J software (NIH).
Imaging of C9 ascaroside-induced Ca2+ responses in fed and starved animals
Ca2+ imaging experiments in the presence of C9 (asc-ΔC9; ascr#3) pheromone were carried out as previously described [12] with custom-made microfluidics chips [76]. Imaging was performed on a Zeiss Axioplan microscopy using a 40× objective and a Zeiss AxioCam HR camera. The images were analyzed using Image J software (NIH), and a custom-written MATLAB (The Mathworks) script.
Supporting Information
Zdroje
1. SenguptaP (2012) The belly rules the nose: feeding state-dependent modulation of peripheral chemosensory responses. Curr Opin Neurobiol 23 : 68–75.
2. FoxAN, PittsRJ, RobertsonHM, CarlsonJR, ZwiebelLJ (2001) Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc Natl Acad Sci U S A 98 : 14693–14697.
3. HallemEA, Nicole FoxA, ZwiebelLJ, CarlsonJR (2004) Olfaction: mosquito receptor for human-sweat odorant. Nature 427 : 212–213.
4. RinkerDC, PittsRJ, ZhouX, SuhE, RokasA, et al. (2013) Blood meal-induced changes to antennal transcriptome profiles reveal shifts in odor sensitivities in Anopheles gambiae. Proc Natl Acad Sci U S A 110 : 8260–8265.
5. QiuYT, van LoonJJ, TakkenW, MeijerinkJ, SmidHM (2006) Olfactory Coding in Antennal Neurons of the Malaria Mosquito, Anopheles gambiae. Chem Senses 31 : 845–863.
6. ColbertHA, BargmannCI (1997) Environmental signals modulate olfactory acuity, discrimination, and memory in Caenorhabditis elegans. Learn Mem 4 : 179–191.
7. ChaoMY, KomatsuH, FukutoHS, DionneHM, HartAC (2004) Feeding status and serotonin rapidly and reversibly modulate a Caenorhabditis elegans chemosensory circuit. Proc Natl Acad Sci U S A 101 : 15512–15517.
8. EzcurraM, TanizawaY, SwobodaP, SchaferWR (2011) Food sensitizes C. elegans avoidance behaviours through acute dopamine signalling. EMBO J 30 : 1110–1122.
9. TomiokaM, AdachiT, SuzukiH, KunitomoH, SchaferWR, et al. (2006) The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron 51 : 613–625.
10. OdaS, TomiokaM, IinoY (2011) Neuronal plasticity regulated by the insulin-like signaling pathway underlies salt chemotaxis learning in Caenorhabditis elegans. J Neurophysiol 106 : 301–308.
11. MacoskoEZ, PokalaN, FeinbergEH, ChalasaniSH, ButcherRA, et al. (2009) A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 458 : 1171–1175.
12. JangH, KimK, NealSJ, MacoskoE, KimD, et al. (2012) Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron 75 : 585–592.
13. PeckolEL, TroemelER, BargmannCI (2001) Sensory experience and sensory activity regulate chemosensory receptor gene expression in C. elegans. Proc Natl Acad Sci USA
14. NolanKM, Sarafi-ReinachTR, HorneJG, SafferAM, SenguptaP (2002) The DAF-7 TGF-beta signaling pathway regulates chemosensory receptor gene expression in C. elegans. Genes Dev 16 : 3061–3073.
15. LanjuinA, SenguptaP (2002) Regulation of chemosensory receptor expression and sensory signaling by the KIN-29 Ser/Thr kinase. Neuron 33 : 369–381.
16. van der LindenAM, NolanKM, SenguptaP (2007) KIN-29 SIK regulates chemoreceptor gene expression via an MEF2 transcription factor and a class II HDAC. EMBO J 26 : 358–370.
17. DentinR, LiuY, KooSH, HedrickS, VargasT, et al. (2007) Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449 : 366–369.
18. KooSH, FlechnerL, QiL, ZhangX, ScreatonRA, et al. (2005) The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437 : 1109–1111.
19. WangB, GoodeJ, BestJ, MeltzerJ, SchilmanPE, et al. (2008) The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab 7 : 434–444.
20. ChoiS, KimW, ChungJ (2011) Drosophila salt-inducible kinase (SIK) regulates starvation resistance through cAMP-response element-binding protein (CREB)-regulated transcription coactivator (CRTC). J Biol Chem 286 : 2658–2664.
21. WangB, MoyaN, NiessenS, HooverH, MihaylovaMM, et al. (2011) A hormone-dependent module regulating energy balance. Cell 145 : 596–606.
22. GoldenJW, RiddleDL (1984) The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol 102 : 368–378.
23. ButcherRA, RagainsJR, LiW, RuvkunG, ClardyJ, et al. (2009) Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc Natl Acad Sci U S A 106 : 1875–1879.
24. GruningerTR, GualbertoDG, GarciaLR (2008) Sensory perception of food and insulin-like signals influence seizure susceptibility. PLoS Genet 4: e1000117.
25. KimuraKD, TissenbaumHA, LiuY, RuvkunG (1997) daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277 : 942–946.
26. LinK, DormanJB, RodanA, KenyonC (1997) daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278 : 1319–1322.
27. OggS, ParadisS, GottliebS, PattersonGI, LeeL, et al. (1997) The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389 : 994–999.
28. LibinaN, BermanJR, KenyonC (2003) Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115 : 489–502.
29. KwonES, NarasimhanSD, YenK, TissenbaumHA (2010) A new DAF-16 isoform regulates longevity. Nature 466 : 498–502.
30. LiW, KennedySG, RuvkunG (2003) daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev 17 : 844–858.
31. RitterAD, ShenY, Fuxman BassJ, JeyarajS, DeplanckeB, et al. (2013) Complex expression dynamics and robustness in C. elegans insulin networks. Genome Res 23 : 954–965.
32. HungWL, WangY, ChitturiJ, ZhenM (2014) A Caenorhabditis elegans developmental decision requires insulin signaling-mediated neuron-intestine communication. Development 141 : 1767–1779.
33. de BonoM, BargmannCI (1998) Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94 : 679–689.
34. RogersC, PerssonA, CheungB, de BonoM (2006) Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr Biol 16 : 649–659.
35. CheungBH, CohenM, RogersC, AlbayramO, de BonoM (2005) Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr Biol 15 : 905–917.
36. CoatesJC, de BonoM (2002) Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature 419 : 925–929.
37. WhiteJG, SouthgateE, ThomsonJN, BrennerS (1986) The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Transact R Soc Lond B 314 : 1–340.
38. StarichTA, XuJ, SkerrettIM, NicholsonBJ, ShawJE (2009) Interactions between innexins UNC-7 and UNC-9 mediate electrical synapse specificity in the Caenorhabditis elegans locomotory nervous system. Neural Dev 4 : 16.
39. ChungSH, ClarkDA, GabelCV, MazurE, SamuelAD (2006) The role of the AFD neuron in C. elegans thermotaxis analyzed using femtosecond laser ablation. BMC Neurosci 7 : 30.
40. WuZ, Ghosh-RoyA, YanikMF, ZhangJZ, JinY, et al. (2007) Caenorhabditis elegans neuronal regeneration is influenced by life stage, ephrin signaling, and synaptic branching. Proc Natl Acad Sci U S A 104 : 15132–15137.
41. ColletJ, SpikeCA, LundquistEA, ShawJE, HermanRK (1998) Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics 148 : 187–200.
42. PerkinsLA, HedgecockEM, ThomsonJN, CulottiJG (1986) Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol 117 : 456–487.
43. SieburthD, MadisonJM, KaplanJM (2007) PKC-1 regulates secretion of neuropeptides. Nat Neurosci 10 : 49–57.
44. OkochiY, KimuraKD, OhtaA, MoriI (2005) Diverse regulation of sensory signaling by C. elegans nPKC-epsilon/eta TTX-4. EMBO J 24 : 2127–2137.
45. TsunozakiM, ChalasaniSH, BargmannCI (2008) A behavioral switch: cGMP and PKC signaling in olfactory neurons reverses odor preference in C. elegans. Neuron 59 : 959–971.
46. SchiavoG, BenfenatiF, PoulainB, RossettoO, Polverino de LauretoP, et al. (1992) Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature 359 : 832–835.
47. KassJ, JacobTC, KimP, KaplanJM (2001) The EGL-3 proprotein convertase regulates mechanosensory responses of Caenorhabditis elegans. J Neurosci 21 : 9265–9272.
48. HussonSJ, ClynenE, BaggermanG, JanssenT, SchoofsL (2006) Defective processing of neuropeptide precursors in Caenorhabditis elegans lacking proprotein convertase 2 (KPC-2/EGL-3): mutant analysis by mass spectrometry. J Neurochem 98 : 1999–2012.
49. SpeeseS, PetrieM, SchuskeK, AilionM, AnnK, et al. (2007) UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in Caenorhabditis elegans. J Neurosci 27 : 6150–6162.
50. GrachevaEO, BurdinaAO, TouroutineD, Berthelot-GrosjeanM, ParekhH, et al. (2007) Tomosyn negatively regulates CAPS-dependent peptide release at Caenorhabditis elegans synapses. J Neurosci 27 : 10176–10184.
51. ColbertHA, SmithTL, BargmannCI (1997) OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci 17 : 8259–8269.
52. TobinD, MadsenD, Kahn-KirbyA, PeckolE, MoulderG, et al. (2002) Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron 35(2): 307–318.
53. LeeBH, AshrafiK (2008) A TRPV channel modulates C. elegans neurosecretion, larval starvation survival, and adult lifespan. PLoS Genet 4: e1000213.
54. ZhangS, SokolchikI, BlancoG, SzeJY (2004) Caenorhabditis elegans TRPV ion channel regulates 5HT biosynthesis in chemosensory neurons. Development 131 : 1629–1638.
55. LeeRY, LobelL, HengartnerM, HorvitzHR, AveryL (1997) Mutations in the alpha1 subunit of an L-type voltage-activated Ca2+ channel cause myotonia in Caenorhabditis elegans. Embo J 16 : 6066–6076.
56. WhiteJG, SouthgateE, ThomsonJN, BrennerS (1976) The structure of the ventral nerve cord of Caenorhabditis elegans. Phil Transact R Soc Lond B 275 : 327–348.
57. ChalasaniSH, KatoS, AlbrechtDR, NakagawaT, AbbottLF, et al. (2010) Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat Neurosci 13 : 615–621.
58. PedersenSF, OwsianikG, NiliusB (2005) TRP channels: an overview. Cell Calcium 38 : 233–252.
59. FlavellSW, CowanCW, KimTK, GreerPL, LinY, et al. (2006) Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311 : 1008–1012.
60. RieraCE, HuisingMO, FollettP, LeblancM, HalloranJ, et al. (2014) TRPV1 pain receptors regulate longevity and metabolism by neuropeptide signaling. Cell 157 : 1023–1036.
61. McCarrollSA, LiH, BargmannCI (2005) Identification of transcriptional regulatory elements in chemosensory receptor genes by probabilistic segmentation. Curr Biol 15 : 347–352.
62. MolkentinJD, BlackBL, MartinJF, OlsonEN (1995) Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83 : 1125–1136.
63. TroemelER, ChouJH, DwyerND, ColbertHA, BargmannCI (1995) Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 83 : 207–218.
64. SambongiY, NagaeT, LiuY, YoshimizuT, TakedaK, et al. (1999) Sensing of cadmium and copper ions by externally exposed ADL, ASE, and ASH neurons elicits avoidance response in Caenorhabditis elegans. Neuroreport 10 : 753–757.
65. SenguptaP, ChouJH, BargmannCI (1996) odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 84 : 899–909.
66. TroemelER, KimmelBE, BargmannCI (1997) Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91 : 161–169.
67. BarthAL, JusticeNJ, NgaiJ (1996) Asynchronous onset of odorant receptor expression in the developing zebrafish olfactory system. Neuron 16 : 23–34.
68. ZhouS, StoneEA, MackayTF, AnholtRR (2009) Plasticity of the chemoreceptor repertoire in Drosophila melanogaster. PLoS Genet 5: e1000681.
69. ChaissonKE, HallemEA (2012) Chemosensory behaviors of parasites. Trends Parasitol 28 : 427–436.
70. ForbesWM, AshtonFT, BostonR, ZhuX, SchadGA (2004) Chemoattraction and chemorepulsion of Strongyloides stercoralis infective larvae on a sodium chloride gradient is mediated by amphidial neuron pairs ASE and ASH, respectively. Vet Parasitol 120 : 189–198.
71. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
72. SuoS, CulottiJG, Van TolHH (2009) Dopamine counteracts octopamine signalling in a neural circuit mediating food response in C. elegans. EMBO J 28 : 2437–2448.
73. LeinwandSG, ChalasaniSH (2013) Neuropeptide signaling remodels chemosensory circuit composition in Caenorhabditis elegans. Nat Neurosci 16 : 1461–1467.
74. HobertO (2002) PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32 : 728–730.
75. EspositoG, Di SchiaviE, BergamascoC, BazzicalupoP (2007) Efficient and cell specific knock-down of gene function in targeted C. elegans neurons. Gene 395 : 170–176.
76. ChalasaniSH, ChronisN, TsunozakiM, GrayJM, RamotD, et al. (2007) Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature 450 : 63–70.
Štítky
Genetika Reprodukční medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 10
-
Všechny články tohoto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



