-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMultifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
Methylation of histone H3 at lysine 4 (H3K4me) is a well-documented mark associated with euchromatin. In this study, we investigate the contributions of the histone methyltransferase Set1 (KMT2) and its associated Set1C/COMPASS complex in the fission yeast Schizosaccharomyces pombe to histone H3 lysine 4 methylation (H3K4me), transcriptional repression, and genome organization. We show that Set1 exhibits multiple modes of transcriptional repression at different types of repetitive elements, requiring distinct domains of Set1 and other Set1C subunits. Despite high conservation of subunits between the S. pombe and S. cerevisiae Set1C complexes, there are considerable differences in contributions to H3K4me by several individual subunits. Furthermore, unlike a recent report in S. cerevisiae, the abundance of Set1 proteins in S. pombe is generally not coupled to either the status of H3K4 methylation or H2B ubiquitination, further highlighting critical differences in Set1 regulation between the two yeast species. We describe a role for the Set1C complex in the nuclear organization of dispersed retrotransposons into Tf bodies. Set1C maintains Tf body integrity by employing H3K4me to antagonize the activities of the H3K4 acetyltransferase Mst1. Collectively, our findings dramatically expand the regulatory landscape controlled by the Set1C complex, an important and highly conserved chromatin-modifying complex with diverse roles in genome control and development.
Published in the journal: . PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004740
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004740Summary
Methylation of histone H3 at lysine 4 (H3K4me) is a well-documented mark associated with euchromatin. In this study, we investigate the contributions of the histone methyltransferase Set1 (KMT2) and its associated Set1C/COMPASS complex in the fission yeast Schizosaccharomyces pombe to histone H3 lysine 4 methylation (H3K4me), transcriptional repression, and genome organization. We show that Set1 exhibits multiple modes of transcriptional repression at different types of repetitive elements, requiring distinct domains of Set1 and other Set1C subunits. Despite high conservation of subunits between the S. pombe and S. cerevisiae Set1C complexes, there are considerable differences in contributions to H3K4me by several individual subunits. Furthermore, unlike a recent report in S. cerevisiae, the abundance of Set1 proteins in S. pombe is generally not coupled to either the status of H3K4 methylation or H2B ubiquitination, further highlighting critical differences in Set1 regulation between the two yeast species. We describe a role for the Set1C complex in the nuclear organization of dispersed retrotransposons into Tf bodies. Set1C maintains Tf body integrity by employing H3K4me to antagonize the activities of the H3K4 acetyltransferase Mst1. Collectively, our findings dramatically expand the regulatory landscape controlled by the Set1C complex, an important and highly conserved chromatin-modifying complex with diverse roles in genome control and development.
Introduction
In eukaryotic cells, DNA-based processes operate within the context of a chromatin template [1], [2]. Chromatin-modifying complexes targeting select residues of histones for posttranslational modifications exert various levels of genome control including chromatin assembly, transcription, DNA repair, replication and recombination [1], [3]. Furthermore, modified histone marks contribute to shaping the genome landscape into distinct chromatin domains. Most notable is the methylation of histone H3 at lysine 4 (H3K4me), which distinguishes euchromatin from heterochromatin, which is marked by H3 lysine 9 methylation (H3K9me) [4], [5]. H3K4me can exist as mono - (H3Kme1), di - (H3K4me2), or tri - (H3K4me3) methylation, and is catalyzed by a number of SET-containing histone methyltransferases that are parts of Set1C/COMPASS and MLL complexes [6], [7]. The roles of individual Set1C/COMPASS subunits have been revealed through studies primarily in the budding yeast Saccharomyces cerevisiae, with loss of individual subunits of Set1C having different effects on the stability of the complex and the states of H3K4me [8], [9], [10], [11]. Interestingly, there is a positive codependency between the levels of H3K4me and those of Set1 proteins [12]. Consistent with the prevalent enrichment of H3K4me2 and H3K4me3 throughout the gene-rich euchromatin [13], [14] Set1C has been shown to localize to active RNA Polymerase II (Pol II) genes [15], [16]. However, accumulating evidence implicates a variety of genetic elements under the repressive control of Set1 and H3K4me. In budding yeast the silencing of Ty1 retrotransposons [17], long noncoding RNAs [18], and antisense regulatory noncoding RNAs [19] requires Set1 and H3K4me. In addition, transcriptional profiling analysis of Set1C/COMPASS mutants supports repressive roles for H3K4me3 at ribosomal genes during multiple stresses [20] and for H3K4me2 and H3K4me3 through promotion of 3'end antisense transcription [21].
In the fission yeast Schizosaccharomyces pombe, Set1 (KMT2) is the sole histone methyltransferase responsible for H3K4me [22], [23]. Biochemical purification identified Set1 as the core subunit of the Set1C complex whose components have orthologs in budding yeast and humans [24]. However, the contributions of Set1 and individual Set1C subunits to H3K4me and their roles in transcriptional repression are not well-characterized in S. pombe. Previous genome-wide mapping shows that the S. pombe genome is dominated by a euchromatin landscape marked with H3K4me2 [14]. Heterochromatin domains distinguished by H3K9me are restricted to prominent genome landmarks including pericentromeres, subtelomeres, ribosomal DNA arrays, and the silent mating-type locus (mat) [14]. These domains contain repetitive elements that help direct RNAi-mediated heterochromatin assembly [25]. The S. pombe genome also contains repetitive elements in the forms of long terminal repeat (LTR) Tf2 retrotransposons and their LTR remnants interspersed across euchromatin and not normally targeted for heterochromatic silencing [14], [26]. Instead, repression of Tf2 retrotransposons, which are enriched for H3K4me2 [14], requires Set1 [27].
In this study, we investigate the contributions of various protein domains of Set1 and its associated Set1C subunits to H3K4me and their roles in the regulation of repetitive elements associated with euchromatin and heterochromatin. We find that S. pombe Set1 possesses multiple modes of regulation that are dependent and independent of H3K4me and Set1C. Set1-mediated repression of Tf2s and pericentromeric repeats is maintained in mutant cells deficient in Set1C subunits or Set1 domain mutants with defects in H3K4me activity. In contrast, intact H3K4me by the Set1C complex is required to maintain repression at the mat and subtelomeric regions. We show that the contributions of several individual Set1C subunits to the levels of H3K4me and Set1 proteins are notably different from those of S. cerevisiae orthologs. Whereas a recent study identifies a feedback mechanism between H3K4me and Set1 protein levels in S. cerevisiae, we find that the stability of Set1 proteins is not coupled to the levels of H3K4me or HULC complex-mediated H2B ubiquitination. Finally, we describe a surprising role for the Set1C complex in the nuclear organization of Tf2 elements into Tf bodies. Set1C employs H3K4me to limit the levels of H3K4 acetylation at Tf2s by antagonizing the function of the histone H3K4 acetyltransferase Mst1. Our study considerably expands the regulatory repertoire of an important histone modifier and highlights the multifaceted function by a highly conserved chromatin-modifying complex with diverse roles in genome control.
Results
Set1 domains and methyltransferase activity play distinct roles in transcriptional silencing of Tf2 retrotransposons and heterochromatic repeats
The protein architecture of fission yeast Set1 is highly conserved [22], [24], containing two putative RNA-recognition motifs (RRMs) termed RRM1 and RRM2 near the N-terminus [28], [29], an nSET domain responsible for interaction with certain COMPASS subunits [11], [30], a catalytic SET domain and a short post-SET (pSET) domain near its C-terminus [28] (Figure 1A). Previous studies from budding yeast have shown that H3K4me is affected by the loss of various domains of Set1 [11], [28], [29], [31]. We examined the status of H3K4me in S. pombe mutant strains lacking individual domains of set1. Loss of RRM1 abolished H3K4me3, substantially diminished H3K4me2 [22], and slightly decreased H3K4me1 compared to wildtype (Figures 1B and S1). An RRM2 deletion resulted in no appreciable decrease in H3K4me levels. Cells expressing Set1 with a deleted nSET, SET or pSET domain displayed a complete loss of H3K4me. S. cerevisiae cells expressing Set1 with a C-terminal TAP epitope showed reduced H3K4me levels [32] while an affinity-purified S. pombe equivalent Set1-TAP protein retained in vitro H3K4me activity [24]. We found that H3K4me was completely abolished in cells containing a FLAG (3×) epitope attached to the C-terminus of Set1 (set1FH3K4me-), likely a result of the epitope interfering with the interaction of the SET or pSET domain with the H3K4 substrate [33], [34].
Fig. 1. Set1 represses different classes of repetitive elements via distinct functional domains. 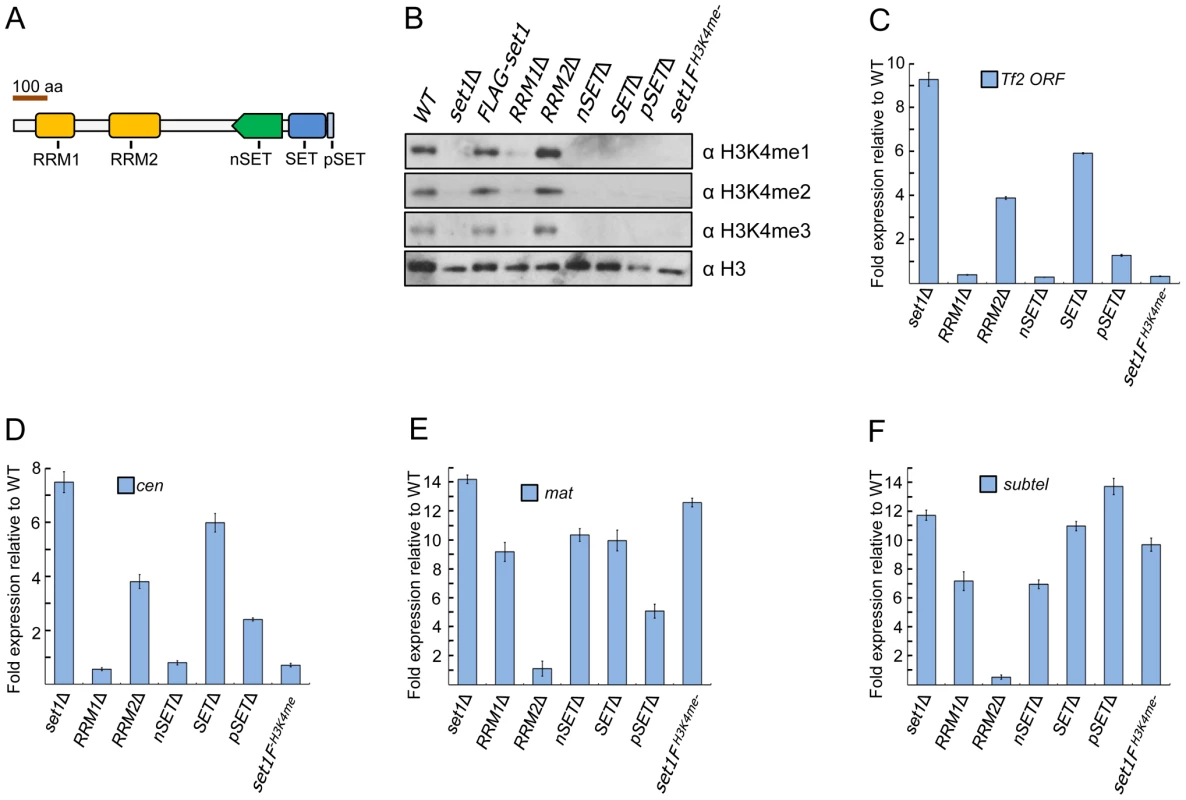
(A) Schematic of S. pombe Set1 protein domain architecture. (B) H3K4 methylation (H3K4me) in set1 domain mutants. Mono (H3K4me1), di (H3K4me2), and tri (H3K4me3) methylation of H3K4 was analyzed from histone extracts of indicated set1 mutant strains by western blotting. Full-length FLAG-set1 and the indicated deleted domain mutants of set1 contain an N-terminal FLAG (3×) epitope. set1FH3K4me- denotes an H3K4me null mutant due to the presence of a FLAG (3×) epitope at the C-terminus of set1. (C) Tf2 repression requires intact RRM2 and SET domains. (D–F) Set1 represses heterochromatic loci dependent and independent of H3K4me. Expression of Tf2 ORF and heterochromatic loci was analyzed by qRT-PCR. Fold changes relative to wildtype were normalized by act1 expression. (s.d., error bars; n = 3). Pericentromeric repeat dg (cen), silent mating type cenH (mat), subtelomeric prl70 (subtel). Deletion of a protein domain could affect the stability of Set1 proteins [11], [12]. To examine this possibility, we constructed strains expressing either full-length or domain-deleted Set1 that contains a fused FLAG epitope at the N-terminus of Set1. Unlike certain S. cerevisiae domain mutants in which Set1 protein level was undetectable [12], our western blot analysis readily detected Set1 expression of all domain mutants (Figure S2). However, there were noticeably reduced levels of Set1 proteins lacking either the RRM1 or nSET domain, suggesting that H3K4me defects observed in RRM1 and nSET mutants could partly be due to reduced amount of Set1 proteins in these mutants. Slight decreases in Set1 protein levels were seen in mutants deficient in RRM2, SET, and pSET domains (Figure S2).
We next performed reverse transcription followed by realtime PCR (qRT-PCR) analysis to examine the effect of various set1 mutations on Tf2 expression. Cells deficient in H3K4me (set1FH3K4me-) or lacking either the RRM1, nSET or pSET domain exhibited little change in transcript levels of Tf2s (Figure 1C). However, Tf2 expression was substantially increased in cells lacking either the RRM2 or SET domain. These results support a catalytic mode of Set1-mediated repression independent of H3K4me that requires intact RRM2 and SET domains. Loss of set1 has been shown to compromise centromeric and telomeric silencing of a reporter gene [23]. We performed qRT-PCR in set1 mutant strains to assess the status of transcription at known heterochromatic regions. Intriguingly, similar to Tf2s, derepression of pericentromeric repeats was observed only in set1Δ and in cells lacking either the RRM2 or SET domain (Figure 1D), suggesting a common mode of Set1-mediated repression for retrotransposons and pericentromeric repeats. In contrast, derepression at the mat locus and subtelomeres was observed in all mutants with defects in H3K4me including set1FH3K4me- (Figures 1E and 1F).
The S. pombe genome encodes three functional copies of histone H3 at distinct loci [35], [36]. We previously observed that repression of Tf2s was maintained in H3K4 mutants in which lysine 4 on all three copies of histone H3 was substituted for either alanine (H3K4A) or arginine (H3K4R) [27]. A previous study reported upregulation of pericentromeric repeats in a H3K4R mutant containing only one functional copy of histone H3 [37]. However, we saw little change in repression at the three major heterochromatin domains in our H3K4 mutants (Figure S3), suggesting a role for histone gene dosage acting together with modifications at certain histone residues (i.e., H3K4) to maintain heterochromatic silencing. Collectively, these results suggest that Set1 utilizes distinct modes to repress different classes of repetitive elements via H3K4me -dependent and -independent pathways.
Loss of the RRM1 or nSET domain impairs Set1 localization at a house-keeping gene and Tf2s, but is dispensable at pericentromeric repeats
We have previously shown that Set1 localizes at Tf2s [27]. Whether Set1 localizes at heterochromatic repeats is not known. We utilized chromatin immunoprecipitation (ChIP) to monitor Set1 enrichment at known euchromatin and heterochromatin targets in set1 mutant strains. In strains deficient in either the RRM1 or nSET domain, there was reduced Set1 enrichment at the housekeeping actin gene act1 and Tf2s (Figures 2A and 2B). Loss of the RRM2, SET domain or H3K4me function did not appear to hamper Set1 localization at these elements. Surprisingly, we detected Set1 localization at pericentromeric repeats (Figure 2C). Unlike the two examined euchromatic targets (i.e., act1 and Tf2s), Set1 localization at pericentromeric repeats was generally not affected in strains deficient in any one of the domains. We did not detect Set1 enrichment at the mat locus or subtelomeric repeats either in wildtype or domain mutants (Figures 2D and 2E). These results suggest that the absence of H3K4me does not adversely affect Set1 localization at certain euchromatic and heterochromatic targets and that H3K4me-dependent repression might not require stable interaction of Set1 with its target loci.
Fig. 2. Set1 localization at euchromatic and heterochromatic targets is largely unaffected by certain domain deletions or H3K4me status. 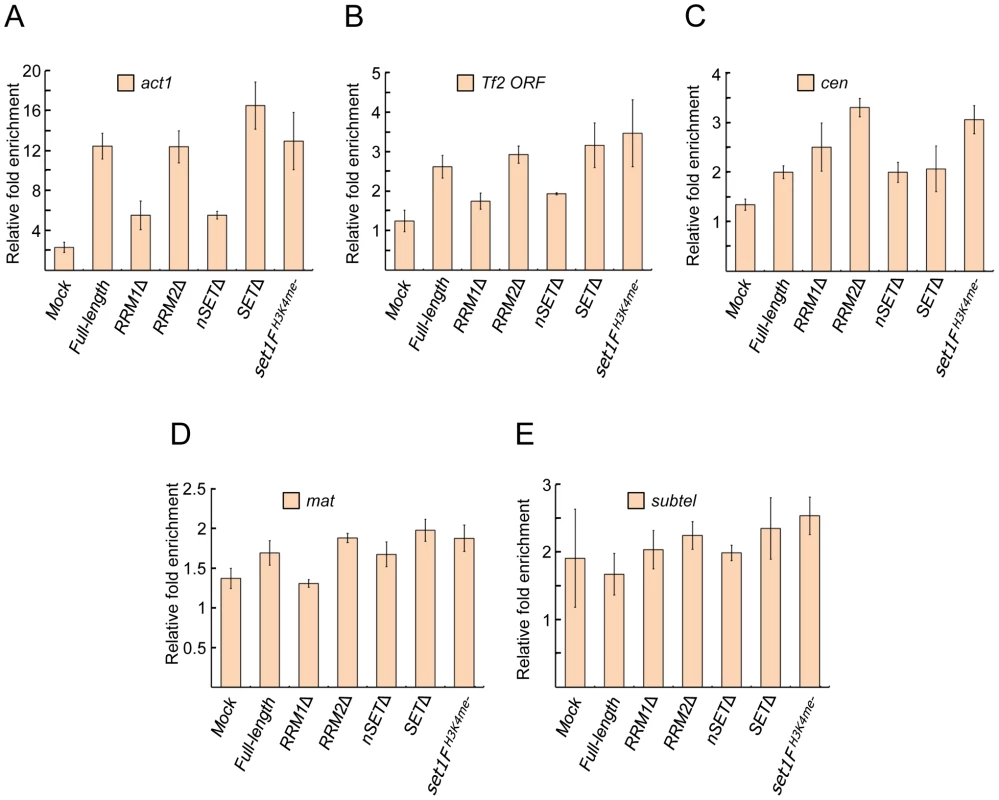
Enrichment of Set1 was determined by chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qPCR) using primers targeting (A) the highly expressed actin gene (act1) promoter, (B) the 5′ end of Tf2 open reading frames, (C) the pericentromeric dg repeat of chromosome II (cen), (D) the silent mating type cenH (mat), and (E) the chromosome I subtelomeric prl70 (subtel). Mock denotes untagged wildtype Set1 strains. Percent enrichment of target amplification compared to input (whole cell extract) was calculated using the 2−ΔΔCt method following normalization by primers targeting mitochondrial DNA (see Materials and Methods). (s.d., error bars; n = 2 biological×2 qPCR replicates). S. pombe Set1C/COMPASS subunits affect H3K4 methylation distinctly from their S. cerevisiae orthologs
The contribution of individual Set1C subunits to H3K4me has been well characterized in S. cerevisiae [8], [9], [10], [38], [39], [40], [41]. However, aside from H3K4me2 [24], the roles of Set1C subunits in S. pombe in H3K4 methylation are not well explored. We therefore assessed the status of H3K4me in cells deficient for individual Set1C components. Cells deficient for set1, swd1 or swd3 exhibit a complete loss of H3K4me (Figures 3A and S3), similar to results observed in S. cerevisiae mutant orthologs [41]. spp1 (spf1) is essential for all three forms of H3K4me in S. pombe. In contrast, the loss of S. cerevisiae SPP1 only diminishes [9], [31] or abolishes H3K4me3 [8]. Loss of swd2, which is lethal in S. cerevisiae [42], abolished H3K4me3 and diminished the levels of H3K4me2 and H3K4me1 in S. pombe. Cells lacking ash2 exhibited relatively intact levels of H3K4me2 and H3K4me1. Although H3K4me3 was detectable within individual cells in the ash2 mutant (Figure S4), we were unable to detect H3K4me3 at bulk histone levels (Figure 3A). In the sdc1 mutant, only H3K4me3 level was slightly diminished, while H3K4me2 and H3K4me1 levels were largely unaffected. These results differ from budding yeast findings, in which loss of ASH2 or SDC1 reduced all three states of H3K4me [9], [10] or completely abolished H3K4me2 [40]. All three forms of H3K4me appeared to be intact in cells deficient for shg1. Collectively, our results show that several components of S. pombe Set1C make different contributions to H3K4me compared to their orthologs in S. cerevisiae.
Fig. 3. Set1 represses Tf2s and heterochromatic loci dependent and independent of Set1C subunits. 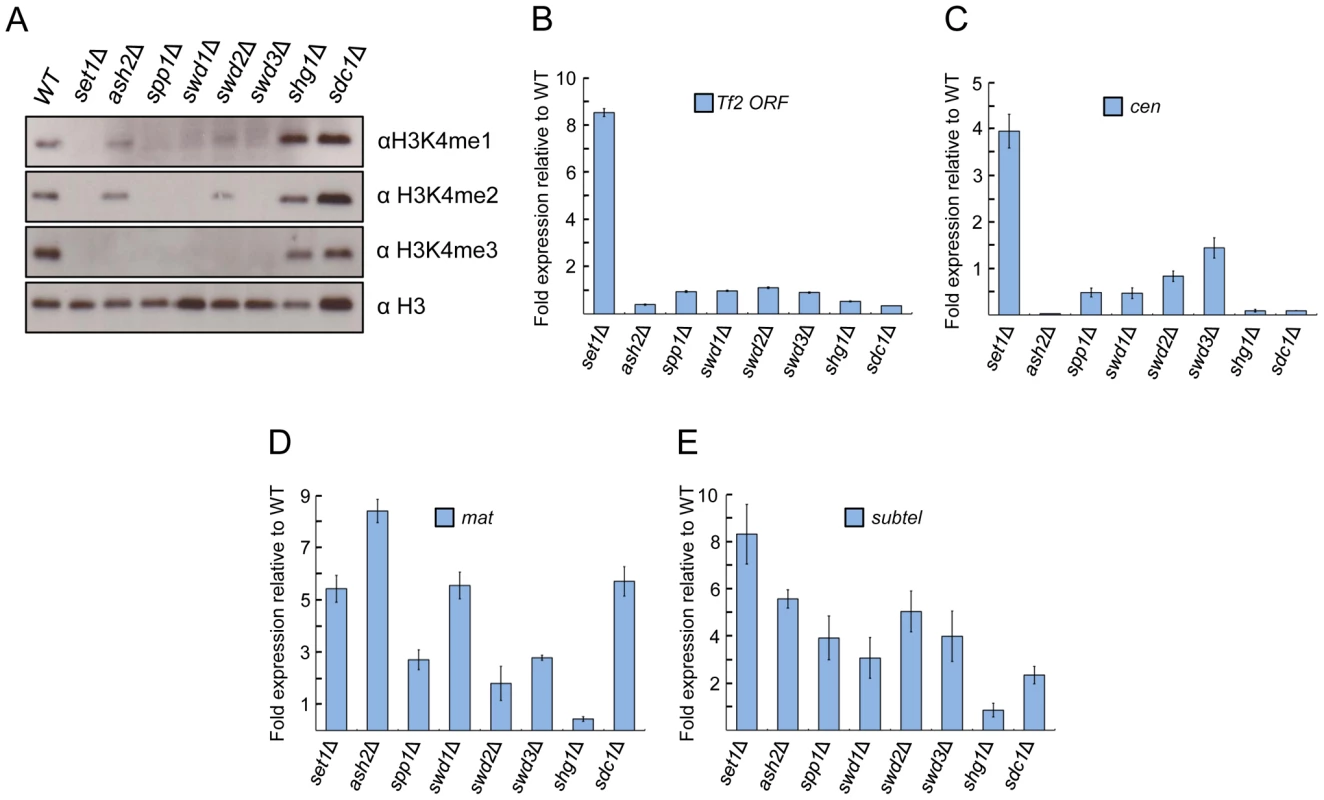
(A) Set1C components contribute differently to H3K4me. H3K4me1, H3K4me2, and H3K4me3 were analyzed from histone extracts of indicated Set1C mutant strains by western blotting. (B) Set1-mediated repression of Tf2s is little affected in other Set1C mutant strains. (C–E) Set1 represses heterochromatic loci dependent and independent of Set1C. qRT-PCR was performed as in Figure 1 (s.d., error bars; n = 3). Pericentromeric repeat dg (cen), silent mating type cenH (mat), subtelomeric prl70 (subtel). Set1C subunits exhibit distinct effects on transcriptional repression of Tf2s and heterochromatic repeats
To gain insights into potential mechanisms underlying the repressive function of Set1, we investigated the role of individual Set1C subunits in the repression of Tf2s and heterochromatic repeats. We found that loss of repression at Tf2s and pericentromeric repeats was observed only in the set1Δ mutant (Figures 3B and 3C). However, at the mating-type and subtelomeric regions, cells deficient for any of the Set1C components except shg1 displayed a noticeable derepression (Figures 3D and 3E). These findings indicate that depending on genomic context, Set1 can dispense or act together with Set1C components to repress different classes of repetitive elements.
Set1 stability is differentially affected in the absence of Set1C subunits
In budding yeast, reduced levels of Set1 proteins have been observed in mutants deficient in SWD1, SWD2, SWD3, or SPP1 [10], [11], [38], [43], [44]. We examined the levels of Set1 proteins in S. pombe strains lacking each of the other Set1C components. The levels of Set1 were minimally affected by the loss of ash2, sdc1 and shg1, somewhat diminished in swd1Δ and swd3Δ, and most noticeably reduced in swd2Δ and spp1Δ mutants (Figure 4A). The loss of individual Set1C subunits on Set1 protein stability is likely to occur at the posttranslational level, as Set1 transcript levels were relatively unaffected in Set1C mutants (Figure S5).
Fig. 4. Stability of Set1 proteins is uncoupled from the status of H3K4me and H2Bub. 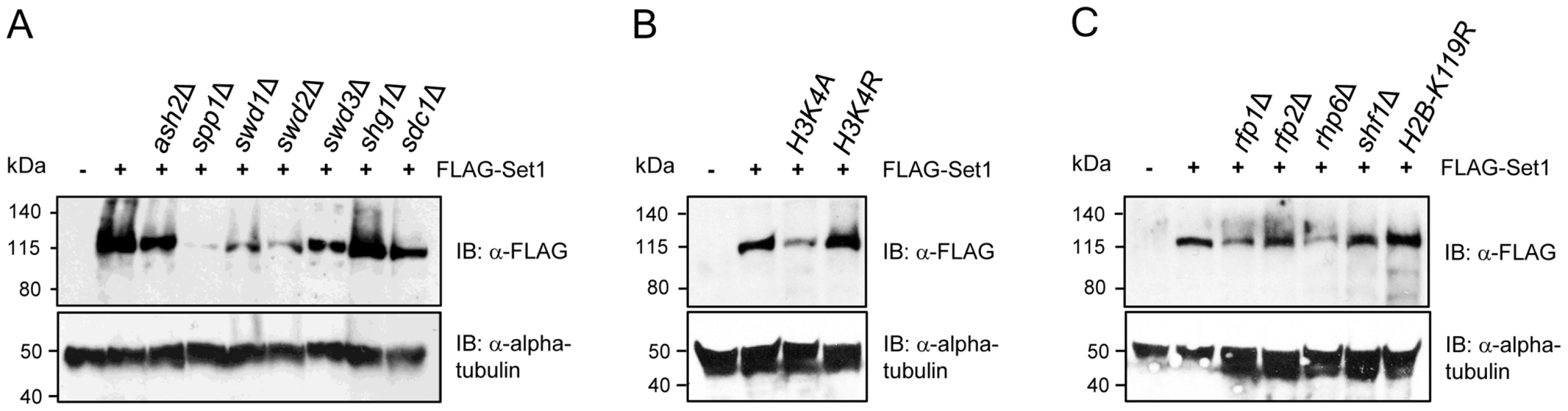
Set1 proteins containing an N-terminal FLAG epitope were analyzed by immunoblotting (IB) in cells deficient in (A) individual components of Set1C, (B) histone H3K4 mutants, or (C) HULC/H2Bub mutants. Alpha tubulin (loading control) was detected by anti-tubulin antibody (tat-1). Set1 protein abundance is not coupled to H3K4 methylation or H2B ubiquitination
Soares et al. recently reported that there is feedback control linking the stability of Set1 proteins to the status of H3K4me in S. cerevisiae [12]. Our analysis showed that despite the complete loss of H3K4me in set1F H3K4me- or set1 mutants lacking either the SET or pSET domain, the levels of these Set1 mutant proteins remain comparable to that of wildtype (Figure S2). However, except for ash2Δ, reduced Set1 protein levels in Set1C mutants (i.e., spp1Δ, swd1Δ, swd2Δ, swd3Δ) generally correlated with a loss of H3K4me, in particular, H3K4me2/3 (see Figure 3A). To disentangle the effects of Set1C component deficiency or Set1 mutations on the stability of Set1 proteins from their contributions to Set1 activity toward H3K4me, we analyzed Set1 proteins in H3K4 mutant cells. We found that Set1 proteins are readily detectable in either H3K4A or H3K4R strains though with apparently reduced levels in H3K4A mutant (Figure 4B). Mono-ubiquitination of histone H2B (H2Bub) has been shown to contribute to H3K4 methylation [45], [46], [47], [48], [49]. In S. pombe H2Bub is mediated by a histone H2B-conjugating complex termed HULC, consisting of a rad6 ortholog Rhp6, two RING finger proteins Rfp1 and Rfp2 similar to budding yeast Bre1, and a serine-rich protein Shf1 [50], [51]. Deficiency of HULC components resulted in drastic reduction in H3K4me levels [50], [51]. We analyzed the loss of H2Bub or HULC subunits on the abundance of Set1 proteins. We could detect Set1 proteins in all HULC/H2Bub mutants. Similar to H3K4A, Set1 levels were reduced in rhp6Δ (Figure 4C). Our results suggest that in contrast to what has been observed in S. cerevisiae, the regulation of Set1 protein abundance in S. pombe is largely independent of the status of H3K4 methylation and H2B ubiquitination.
Set1 regulates the nuclear organization of Tf2s distinct from its transcriptional repressor function
We have previously identified a novel role for Set1 in the nuclear organization of Tf2s into Tf bodies [27]. To dissect potential mechanisms underlying Set1-mediated clustering of Tf2s, we performed fluorescence in situ hybridization (FISH) analysis to monitor the status of Tf bodies in various set1 mutants. In contrast to wildtype cells with intact Tf bodies, set1 mutants lacking the RRM1, nSET, SET or pSET domain exhibited defects in Tf bodies to the same extent as set1Δ (Figure 5A). Because H3K4me is severely compromised in these set1 mutants (see Figure 1B and. S1), H3K4me might be required to maintain the integrity of Tf bodies. Indeed, Tf2 elements were observed to decluster in the set1FH3K4me- cells which lack H3K4me altogether. In the set1 mutant containing the RRM2 deletion, which resulted in intermediately increased levels of Tf2 expression (see Figure 1C), the integrity of Tf bodies was modestly affected. These results suggest that Set1 relies on disparate domains and possibly different catalytic activities (see discussion below) to exert control over different aspects of Tf2 regulation.
Fig. 5. Set1C and H3K4me are required for the integrity of Tf bodies. 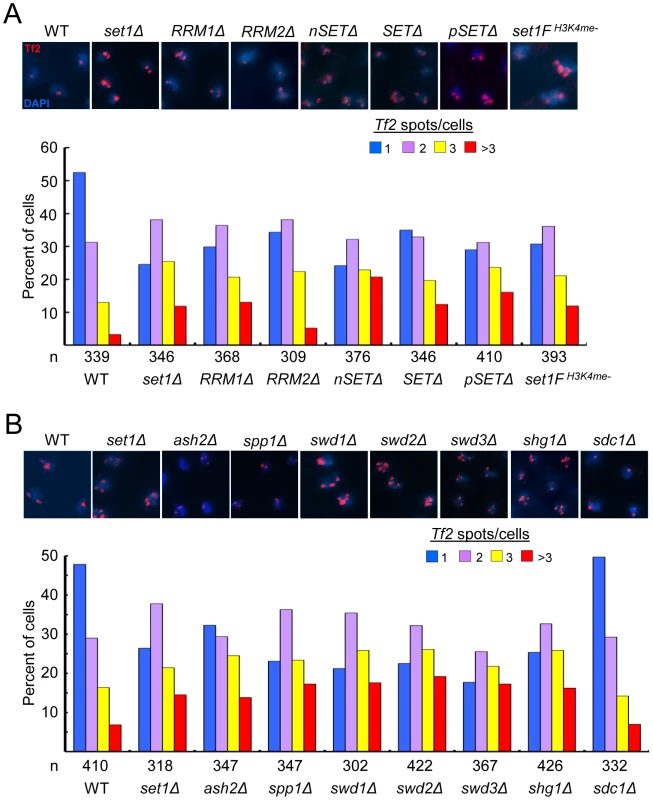
(A, B) Declustering of Tf2s in (A) set1 mutant with defects in H3K4me and (B) Set1C mutants. Fluorescence in situ hybridization (FISH) analysis was performed using a FISH probe corresponding to the ∼3.6 kb coding region of Tf2 elements. Representative FISH images from indicated strains (top panels). Quantitative FISH analysis of observed Tf2 foci/cell in indicated strains (bar graph; bottom panels). Number of cells analyzed per strain (n). Except for sdc1Δ, Tf2 declustering in all mutant strains compared to WT was significant (p<0.005, chi-square test). The Set1C complex maintains Tf body organization by antagonizing the H3K4 histone acetyltransferase Mst1
The dependence on H3K4 methylation to maintain the integrity of Tf bodies suggests a role for Set1C subunits in the organization of Tf2s within the nucleus. We utilized Tf2 FISH analysis to monitor the status of Tf bodies in strains with null mutations for each of the Set1C components. Similar to what was observed in set1 mutants with impaired H3K4me, Set1C mutants with gross defects in H3K4me displayed declustering of Tf2s compared to wildtype (Figure 5B). Only in shg1 mutant cells did we observe a disruption of Tf body integrity that did not correspond with the loss of H3K4me (Figure 3A), suggesting a distinct role for Shg1 in nuclear organization.
The MYST family histone acetyltransferase Mst1 has been shown to acetylate lysine 4 of histone H3 (H3K4ac) [37]. Declustering of Tf2 elements in H3K4me mutants could be due to inappropriate Mst1 activity such as heightened H3K4ac at Tf2s. Consistent with this idea, mutation of mst1 in cells lacking H3K4me abrogated defects in the integrity of Tf bodies (Figure 6A) and diminished the elevated H3K4ac levels at Tf2s and the housekeeping gene act1 observed in the absence of H3K4me (Figure 6B and 6C). Together, these results reveal that Set1 controls Tf2 repression independent of the Set1C complex, but relies on Set1C-mediated H3K4me to maintain Tf body integrity by antagonizing the H3K4 acetyltransferase activity of Mst1.
Fig. 6. Set1C-mediated H3K4me contributes to the integrity of Tf bodies by antagonizing the H3K4 acetyltransferase Mst1. 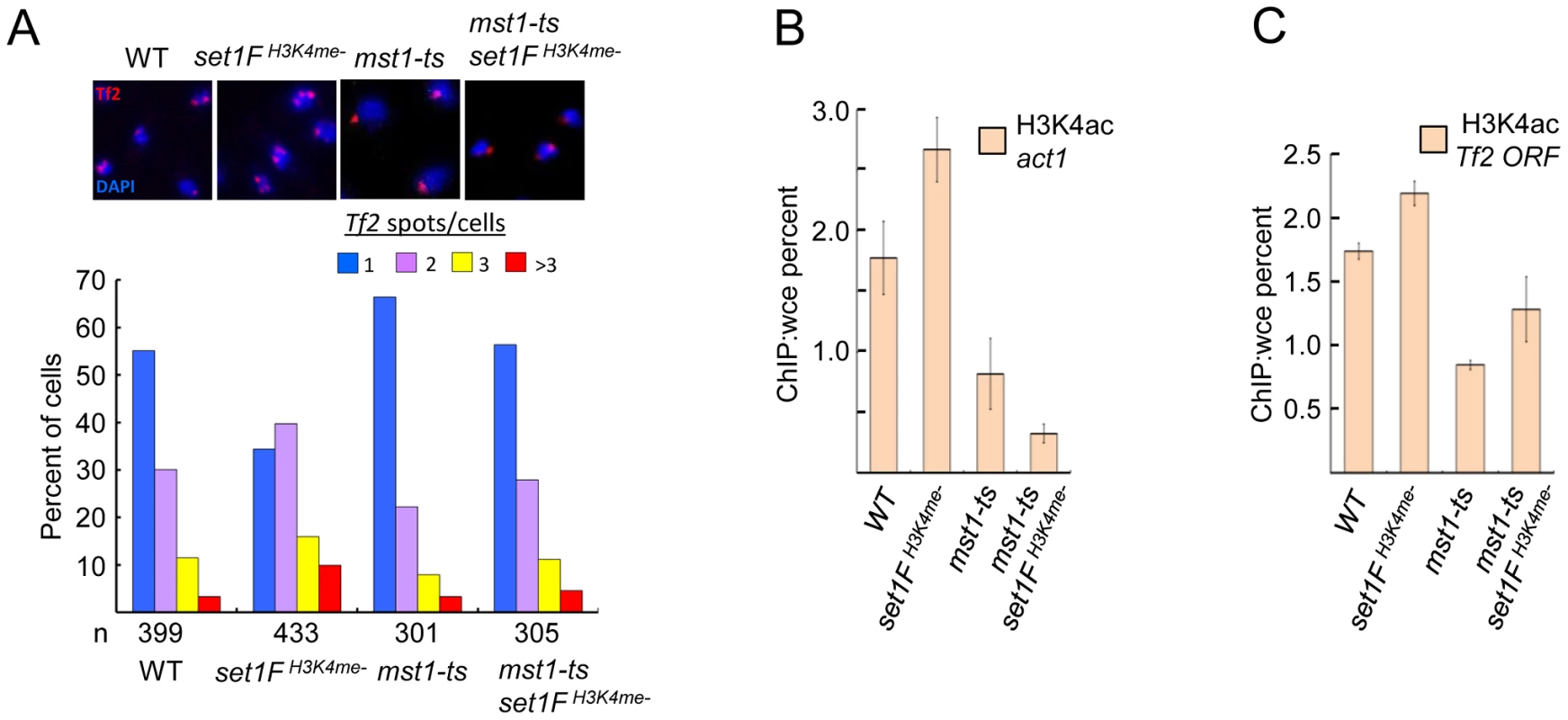
(A) An mst1 mutation (mst1-ts) alleviates Tf body defects seen in an H3K4me mutant strain. Representative FISH images from indicated strains (top panels). Quantitative FISH analysis of observed Tf2 foci/cell in indicated strains (bar graph; bottom panels). Tf2 declustering for set1FH3K4me- strain was significant (p<0.005, chi-square test). H3K4ac enrichment at (B) the housekeeping gene act1 and (C) Tf2 ORF in indicated mutant strains was analyzed by ChIP followed by qPCR. Discussion
The contributions of Set1 domains and Set1C subunits to H3K4 methylation in S. pombe
Unlike other chromatin-modifying enzymes such as the Clr4/Suv39h H3K9 methyltransferase not universally present in eukaryotes, the protein architecture of Set1 and its associated complex subunits are remarkably conserved across known eukaryotic lineages [7]. The multi-domain structure of Set1 suggests its ability to interact with multiple proteins and integrate opposing inputs. Works from budding yeast have yielded many insights into the roles of various Set1 domains in H3K4me and domain interactions with various components of the Set1C/COMPASS complex [10], [11], [28], [29], [30]. Our analysis of Set1C in S. pombe reveals additional insights. We found that the nSET, SET, and pSET domains are essential for all three states of H3K4me, similar to results previously noted in budding yeast [12], [28]. In the RRM1 mutant, we observed defects in H3K4me2 as previously reported [22], in addition to the complete loss of H3K4me3 and slightly reduced H3K4me1, though the degree to which these defects are due to reduced Set1 abundance in this mutant is unclear. Reduced levels of Set1 proteins in RRM1 and nSET mutants could also account for the relatively lower enrichment of these mutant proteins at euchromatic targets observed in ChIP experiments. In contrast, the loss of RRM2 did not appear to affect the protein levels of Set1 or hamper the ability of Set1 to methylate all three states of H3K4me. In fact, we detected slight increases in overall H3K4me levels in the RRM2 mutant (Figure 1B), suggesting it might have an inhibitory role against H3K4me, similar to the roles of the central autoinhibitory region noted in budding yeast Set1 [28].
The contributions of several S. pombe Set1C subunits to H3K4me diverge from those in S. cerevisiae. Among these are Ash2, Sdc1, Swd2, and Spp1 (Spf1). S. pombe spp1 is essential not only for H3K4me2 [24], but H3K4me3 and H3K4me1. This pattern is somewhat similar to that seen in S. cerevisiae spp1Δ mutant expressing a Set1 C-terminal fragment containing only the nSET, SET, and pSET domains [11], [30]. These data point to a more critical role for Spp1 in conferring the H3K4me activity within the Set1C complex in S. pombe compared to S. cerevisiae, perhaps by more effectively countering the inhibitory effect of the RRM2 domain on H3K4me and/or by influencing the conformation of the nSET domain within the Set1C complex to effect H3K4me [11], [30]. However, the substantially reduced amount of Set1 protein levels in S. pombe spp1 null cells could also contribute to the complete loss of all three forms of H3K4me. Differences in the role of the Swd2 subunit in previous reports in budding yeast and our study were also noted. S. cerevisiae mutant cells carrying temperature-sensitive alleles of SWD2 exhibit severe reductions in H3K4me2/3 [43], [52], and an S. cerevisiae swd2Δ mutant overexpressing a C-terminal fragment of Sen1 that suppresses the lethality of swd2Δ displays similar defects in H3K4me2/3 but no significant change to H3K4me1 [44]. In contrast, we found reduction for all three states of H3K4me in S. pombe swd2Δ cells, with H3K4me3 most affected, followed by H3K4me2 and H3K4me1. Similar H3K4me2/3 defects in both yeast species deficient in swd2 could be due to the requirement of swd2 to maintain sufficient levels of Set1 protein abundance [43], [44]. However, H3K4me1 defects seen in S. pombe swd2Δ might reflect a more dedicated role for Swd2 in Set1C function compared to its S. cerevisiae counterpart, which is also a subunit of the essential transcription termination factor APT [53], and proposed to be needed to overcome antagonism by Set1C [54]. Despite discrepancies in findings from several groups [9], [10], [11], [40], [44], S. cerevisiae ash2Δ and sdc1Δ mutants exhibit similar H3K4me defects, likely reflecting their function as a heterodimer within the Set1C complex [10], [11]. Our study suggests a more extensive role for S. pombe Ash2 than Sdc1 in maintaining various states of H3K4me, in particular H3K4me3 and H3K4me2. In addition, loss of either ash2 or sdc1 did not produce H3K4me defects as severe as those seen in the equivalent budding yeast mutants. These differences might reflect divergence in the functions of Ash2 and Sdc1 in S. pombe, likely due to their associations with the histone H3K4 demethylase Lid2 complex, which is not present in S. cerevisiae [24], [55].
Set1 stability and its relation to H3K4me
Set1 has been documented to be a highly unstable protein in budding yeast [44]. This instability of Set1 has been shown to be coupled to the levels of H3K4me [12], complicating efforts to untangle the specific roles of various Set1 domains and Set1C subunits to H3K4me from their direct contributions to the stability of Set1. Our study indicates that Set1 is inherently more stable in S. pombe, and is readily detectable in whole cell extracts from wildtype and mutant strains deficient for either the individual Set1 domains or Set1C complex components. It is worth noting that the RRM1 and nSET domains and certain Set1C subunits (Swd1, Swd2, Swd3, Spp1) appear to contribute to Set1 stability. However, the regulation of Set1 protein levels in S. pombe is largely independent of H3K4me abundance and H2Bub levels, further highlighting the considerable divergence in the regulation of Set1 in S. pombe versus that of S. cerevisiae.
Set1-mediated repression of Tf2s and heterochromatic loci
The role of Set1 as a transcriptional repressor has been widely documented in budding yeast [17], [18], [20], [32], [46], [56], [57], [58]. However, these studies ascribed Set1 repressor function solely to H3K4me2 and/or H3K4me3 [19], [20], [21], [58]. We have previously shown that Set1 mediates repression of Tf2 retrotransposons independent of H3K4me [27]. Our current study reveals an unanticipated complexity in the repressive function of Set1, in that the requirement of H3K4me in transcriptional silencing depends upon the genomic context (Figure 7). The complete loss of H3K4me does not appear to hamper the ability of Set1 to localize to and maintain repression at Tf2s and pericentromeric repeats, while at the mat locus and subtelomeric repeats Set1-mediated H3K4me contributes to repression. These findings were consistent with analyses using Set1C subunit deletion mutants. Loci that depend on H3K4me-mediated repression (mat and subtelomeric repeats) also require Set1C components needed for maintaining proper H3K4me (all Set1C subunits except Shg1). Repression of Tf2s and pericentromeric repeats, on the other hand, is maintained in all Set1C mutants (except set1Δ).
Fig. 7. Model for the roles of Set1C in genome control in S. pombe. 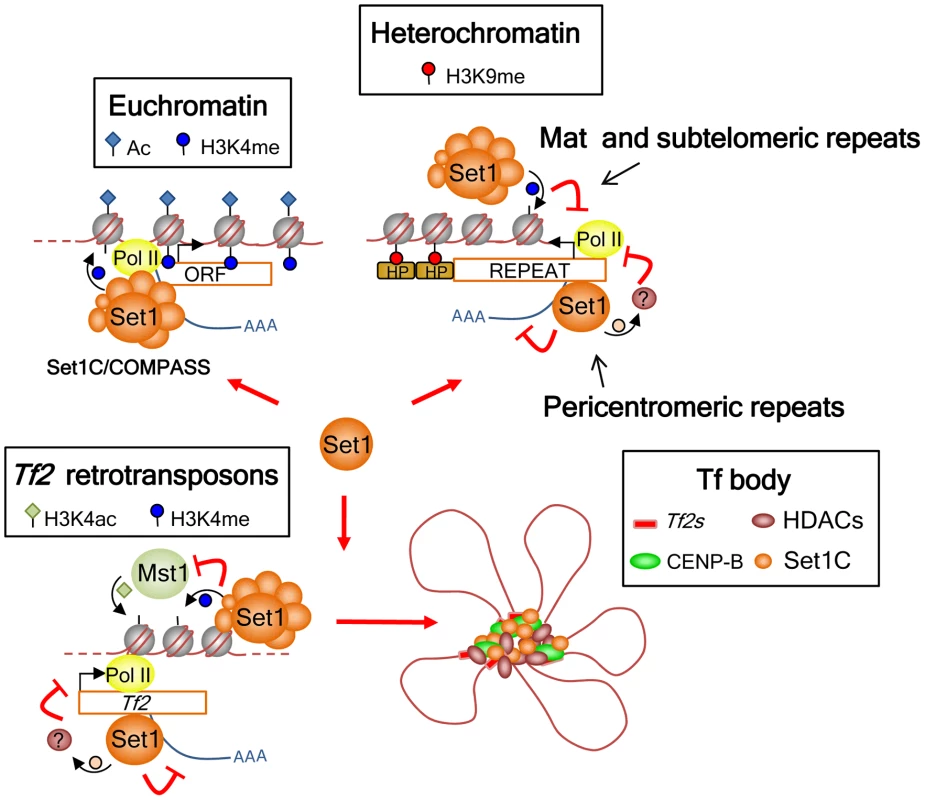
Set1 exerts its multifaceted genome control at euchromatin and heterochromatin dependent and independent of H3K4 methylation and the Set1C complex. At euchromatin, Set1 operates as part of the Set1C/COMPASS complex that is responsible for H3K4me distribution at active RNA polymerase II genes. At interspersed Tf2s and heterochromatic loci, Set1 has a repressive role mediated through two distinct pathways: H3K4me/Set1C-dependent repression at the silent mat locus and subtelomeres, and H3K4me/Set1C-independent repression at Tf2s and pericentromeric heterochromatin. Our findings also anticipate the presence of novel non-histone H3K4 substrates involved in the repression of Tf2s and pericentromeric repeats that could be partly mediated via Set1 association with Pol II nascent transcripts. In addition, Set1C and H3K4me have a distinct genome organization role at Tf2s by antagonizing the activity of the histone H3K4 acetyltransferase Mst1 to maintain the integrity of Tf bodies. Our study identifies a novel mode of Set1 function that does not depend on H3K4me and an intact Set1C complex. The requirement of the SET domain but not H3K4me activity suggests that Set1 mediates repression of Tf2s and pericentromeric repeats via methylation of a novel substrate(s). In budding yeast, Set1 has been shown to methylate Dam1, a component of the kinetochore DASH complex [59]. Although methylation of Dam1 is independent of H3K4me, its methylation requires other Set1C subunits [40]. S. pombe encodes a dam1 ortholog, though it appears not to contain conserved Set1 methylation sites [60]. Considering that Set1 represses Tf2s and pericentromeric repeats independent of H3K4me and other Set1C subunits, it is possible that repression of these repeats might involve Set1 binding to RNA via its RRM2 domain and methylation of targets associated with either transcription and/or RNA processing.
Heterochromatic repeats and Tf2s in certain genetic backgrounds are targeted for RNAi-mediated heterochromatic and exosome-mediated silencing [26], [61], [62]. It has been shown that histone deacetylases (HDACs) cooperate with RNAi to assemble heterochromatin at pericentromeres [63]. Even though loss of set1 does not appear to affect the levels of H3K9 methylation and siRNAs at pericentromeric heterochromatin (Figure S6A) [37], there were noticeable increased levels of H3K9 acetylation at that region (Figure S6B). Several HDAC mutants (i.e., sir2, clr3) are known to retain robust levels of siRNAs and H3K9me and yet exhibit increased levels of certain histone acetylation marks at pericentromeres [63], [64], [65]. Thus, it is likely that in the absence of set1, HDACs, RNAi and exosome act in redundant pathways to help maintain heterochromatin.
The roles of Set1C in genome organization
We previously reported that Set1 has a novel role in genome organization by clustering interspersed Tf2 elements into Tf bodies [27]. Even though declustering of Tf2s was not observed in H3K4 mutant strains (H3K4A, H3K4R) [27], a role for H3K4me in Tf2 clustering could not be excluded due to the loss of both H3K4 acetylation and methylation in those H3K4 mutants. Our current study supports an active role for the Set1C complex in maintaining the integrity of Tf bodies by antagonizing the function of the H3K4 acetyltransferase Mst1. Set1 has been shown to limit the abundance of H3K4ac at gene promoters in S. cerevisiae [66]. Thus, H3K4me catalyzed by Set1C could compete with Mst1-mediated H3K4ac at Tf2s to maintain the integrity of Tf bodies. However, as loss of H3K4me also results in increased H3K4ac at the house keeping gene act1, Tf2 declustering in set1 mutants could reflect global changes in genome organization due to heightened levels of H3K4ac across multiple loci. HDACs recruited by CENP-B proteins to Tf2s have also been shown to contribute to Tf2 clustering [27], [67]. Cells may therefore exploit dynamic competition between Set1C, HATs, and HDACs to regulate the various states of H3K4, which could in turn facilitate rapid genome reorganization in response to acute environmental changes [68].
Materials and Methods
Strain construction
Null mutant and C-terminal FLAG (3×) strains were constructed using a Kanamycin cassette [69]. Double mutants were generated by standard genetic cross methods [70]. Full-length and domain mutants of set1 containing an N-terminal FLAG (3×) epitope were generated by a two-step site-directed mutagenesis (SDM). First, the set1 gene was replaced with a ura5 lys7 cassette [71]. Second, an SDM PCR fragment containing either full-length or domain deleted FLAG-Set1 was transformed into the above set1 null strain (set1Δ::ura5 lys7 ura5-14 lys7-2), and transformants were scored by growth on the uracil counter selective agent 5-Fluoroorotic acid (5-FOA) and sensitivity to lysine minus media [71]. Proper insertions were confirmed by PCR and DNA sequencing. Liquid cultures were grown at 30°C in standard rich media supplemented with 225 mg/L adenine (YEA).
Quantitative Reverse Transcription Real-time PCR (qRT-PCR)
RNA was isolated by a hot acid phenol method [72] and converted to cDNA with Superscript III reverse transcriptase and anchored oligo-dT primer (Life Technologies). cDNA was subjected to qPCR analysis using DyNAzyme™ II PCR Master Mix (Finnzymes) with SYBR green on the Applied Biosystems 7500 Fast Real-Time PCR System. Fold expression changes of mutant versus wildtype cells relative to act1 gene were determined using the 2−ΔΔCt method in Microsoft Excel.
Histone extraction and detection
Cells from 50 ml culture (OD∼0.5) were washed in 10 ml NIB buffer (15 mM PIPES pH 6.8, 0.25M sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 0.8% Triton X-100, 10 ng/µl TSA, 1 mM PMSF, Roche protease inhibitor mini tablet), lysed with acid-washed glass beads in a bead beater, and centrifuged at 11,000× g for 10 min [22]. Cell extract pellets were resuspended in 0.4M H2SO4, incubated on ice for 1 h with occasional mixing, and the supernatant was collected following centrifugation at 8,000× g for 5 min. Histone extract was concentrated by trichloroacetic acid (TCA) precipitation, washed in acetone and resuspended in 100 µl LDS buffer (Life Technologies) and quantitated using the BCA method (Pierce). 5 µg of histone extracts were resolved on 14–22% Tricine SDS PAGE and transferred to a nitrocellulose membrane using the iBlot system (Life Technologies). Histone H3 and modified residues were detected with antibodies against H3 (Abcam, ab1791), H3K4me1 (Abcam, ab8895), H3K4me2 (Fisher, 07030MI), or H3K4me3 (Fisher, 07-473MI).
Protein extractions and western blotting
S. pombe cells (OD 1–2) were lysed in HCS buffer (150 mM HEPES pH 7.2, 250 mM NaCl, 0.1% NP-40, 1 mM EDTA, 1 mM dithiothreitol, 1 mM PMSF) and protein inhibitor tablet (Roche) by acid-washed beads in a bead beater (three times 30 s with 2 min interval on ice). 50 µg of protein extracts were run on a PAGE gel (Express Plus 4–20% Bis-tris (MOPS), Genescript) and subjected to overnight western blot transfer at 4°C. Set1 was detected using anti-FLAG antibody (Genescript, A00187).
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed as previously described [27]. qPCR was performed using Phire Hot Start II DNA Polymerase (Thermo Scientific) supplemented with SYBR green (Life Technologies) on the Applied Biosystems 7500 Fast Real-Time PCR System. Enrichment of ChIP vs. input DNA was determined using the 2−ΔΔCt method in Microsoft Excel.
Immunofluorescence (IF) and Fluorescence In Situ Hybridization (FISH)
IF and FISH assays were performed as previously described [27], [68]. Briefly, S. pombe cells were grown in 10 ml YEA media until OD595 ∼0.5–1. 10 ml of 2.4 M sorbitol YEA solution was added to culture, and cells were immediately cross-linked with 2.9 ml of freshly made 30% paraformaldehyde/YEA solution for 30 min in a 18°C water bath shaker. Cross-linked reaction was quenched with 1.2 ml of 2.5 M glycine. Cells were transferred to a microcentrifuge tube, subjected to cell wall digestion in 0.5 mg/ml zymolyase solution (Associated of Cape Cod, 100T) at 37°C for 30 min, blocked with PEMBAL (100 mM PIPES pH 6.9, 1 mM EGTA, 1 mM MgSO4, 1% BSA, 0.1 M L-lysine) for 1 hr and subjected to either IF or FISH analysis. For IF analysis, cells were incubated overnight at room temperature with antibodies against either H3K4me1 (Abcam, ab8895), H3K4me2 (Fisher, 07030MI), or H3K4me3 (Fisher, 07-473MI). Cells were then incubated with anti-mouse Alexa Fluor 488 (Invitrogen) followed by DAPI staining to visualize H3K4me signal in the nucleus. For FISH analysis, PEMBAL-treated cells were treated with RNase A (0.1 mg/ml) at 37°C for 3 h. Hybridization was carried out at 40°C for 12–14 h with 100–150 ng of Tf2-orf probes in 100 µl hybridization buffer (50% formamide, 2× SSC, 5× Denhart's solution, 10% dextran sulfate). Cells were washed three times in 100 ml 2× SSC for 30 min each. Images were obtained using a Zeiss Axioplan 2 microscope. The chi-square test of homogeneity was used to determine whether declustering of Tf2 elements seen in mutant cells relative to wildtype was significant.
Supporting Information
Zdroje
1. CamposEI, ReinbergD (2009) Histones: annotating chromatin. Annu Rev Genet 43 : 559–599.
2. BadeauxAI, ShiY (2013) Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol 14 : 211–224.
3. KouzaridesT (2007) Chromatin modifications and their function. Cell 128 : 693–705.
4. NomaK, AllisCD, GrewalSI (2001) Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293 : 1150–1155.
5. LittMD, SimpsonM, GasznerM, AllisCD, FelsenfeldG (2001) Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293 : 2453–2455.
6. KuschT (2012) Histone H3 lysine 4 methylation revisited. Transcription 3 : 310–314.
7. ShilatifardA (2012) The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81 : 65–95.
8. MorillonA, KarabetsouN, NairA, MellorJ (2005) Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell 18 : 723–734.
9. SchneiderJ, WoodA, LeeJS, SchusterR, DuekerJ, et al. (2005) Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell 19 : 849–856.
10. DehePM, DichtlB, SchaftD, RoguevA, PamblancoM, et al. (2006) Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem 281 : 35404–35412.
11. KimJ, KimJA, McGintyRK, NguyenUT, MuirTW, et al. (2013) The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol Cell 49 : 1121–1133.
12. SoaresLM, Radman-LivajaM, LinSG, RandoOJ, BuratowskiS (2014) Feedback control of Set1 protein levels is important for proper H3K4 methylation patterns. Cell Rep 6 : 961–972.
13. PokholokDK, HarbisonCT, LevineS, ColeM, HannettNM, et al. (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122 : 517–527.
14. CamHP, SugiyamaT, ChenES, ChenX, FitzGeraldPC, et al. (2005) Comprehensive analysis of heterochromatin - and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37 : 809–819.
15. NgHH, RobertF, YoungRA, StruhlK (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11 : 709–719.
16. KroganNJ, DoverJ, WoodA, SchneiderJ, HeidtJ, et al. (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11 : 721–729.
17. BerrettaJ, PinskayaM, MorillonA (2008) A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev 22 : 615–626.
18. CamblongJ, BeyrouthyN, GuffantiE, SchlaepferG, SteinmetzLM, et al. (2009) Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev 23 : 1534–1545.
19. van DijkEL, ChenCL, d'Aubenton-CarafaY, GourvennecS, KwapiszM, et al. (2011) XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475 : 114–117.
20. WeinerA, ChenHV, LiuCL, RahatA, KlienA, et al. (2012) Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol 10: e1001369.
21. MargaritisT, OrealV, BrabersN, MaestroniL, Vitaliano-PrunierA, et al. (2012) Two Distinct Repressive Mechanisms for Histone 3 Lysine 4 Methylation through Promoting 3′-End Antisense Transcription. PLoS Genet 8: e1002952.
22. NomaK, GrewalSI (2002) Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc Natl Acad Sci U S A 99 Suppl 4 : 16438–16445.
23. KanohJ, FrancesconiS, ColluraA, SchramkeV, IshikawaF, et al. (2003) The fission yeast spSet1p is a histone H3-K4 methyltransferase that functions in telomere maintenance and DNA repair in an ATM kinase Rad3-dependent pathway. J Mol Biol 326 : 1081–1094.
24. RoguevA, SchaftD, ShevchenkoA, AaslandR, StewartAF (2003) High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J Biol Chem 278 : 8487–8493.
25. GrewalSI (2010) RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 20 : 134–141.
26. YamanakaS, MehtaS, Reyes-TurcuFE, ZhuangF, FuchsRT, et al. (2013) RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature 493 : 557–560.
27. LorenzDR, MikheyevaIV, JohansenP, MeyerL, BergA, et al. (2012) CENP-B Cooperates with Set1 in Bidirectional Transcriptional Silencing and Genome Organization of Retrotransposons. Mol Cell Biol 32 : 4215–4225.
28. SchlichterA, CairnsBR (2005) Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. EMBO J 24 : 1222–1231.
29. TresauguesL, DehePM, GueroisR, Rodriguez-GilA, VarletI, et al. (2006) Structural characterization of Set1 RNA recognition motifs and their role in histone H3 lysine 4 methylation. J Mol Biol 359 : 1170–1181.
30. ThorntonJL, WestfieldGH, TakahashiYH, CookM, GaoX, et al. (2014) Context dependency of Set1/COMPASS-mediated histone H3 Lys4 trimethylation. Genes Dev 28 : 115–120.
31. TakahashiYH, LeeJS, SwansonSK, SarafA, FlorensL, et al. (2009) Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol Cell Biol 29 : 3478–3486.
32. KroganNJ, DoverJ, KhorramiS, GreenblattJF, SchneiderJ, et al. (2002) COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277 : 10753–10755.
33. SouthallSM, WongPS, OdhoZ, RoeSM, WilsonJR (2009) Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell 33 : 181–191.
34. TakahashiYH, WestfieldGH, OleskieAN, TrievelRC, ShilatifardA, et al. (2011) Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc Natl Acad Sci U S A 108 : 20526–20531.
35. MatsumotoS, YanagidaM (1985) Histone gene organization of fission yeast: a common upstream sequence. EMBO J 4 : 3531–3538.
36. MelloneBG, BallL, SukaN, GrunsteinMR, PartridgeJF, et al. (2003) Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr Biol 13 : 1748–1757.
37. XhemalceB, KouzaridesT (2010) A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev 24 : 647–652.
38. StewardMM, LeeJS, O'DonovanA, WyattM, BernsteinBE, et al. (2006) Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol 13 : 852–854.
39. MuellerJE, CanzeM, BrykM (2006) The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics 173 : 557–567.
40. LathamJA, ChosedRJ, WangS, DentSY (2011) Chromatin signaling to kinetochores: transregulation of Dam1 methylation by histone H2B ubiquitination. Cell 146 : 709–719.
41. MersmanDP, DuHN, FingermanIM, SouthPF, BriggsSD (2012) Charge-based interaction conserved within histone H3 lysine 4 (H3K4) methyltransferase complexes is needed for protein stability, histone methylation, and gene expression. J Biol Chem 287 : 2652–2665.
42. NagyPL, GriesenbeckJ, KornbergRD, ClearyML (2002) A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A 99 : 90–94.
43. DichtlB, AaslandR, KellerW (2004) Functions for S. cerevisiae Swd2p in 3′ end formation of specific mRNAs and snoRNAs and global histone 3 lysine 4 methylation. RNA 10 : 965–977.
44. NedeaE, NalbantD, XiaD, TheoharisNT, SuterB, et al. (2008) The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol Cell 29 : 577–587.
45. SunZW, AllisCD (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418 : 104–108.
46. BriggsSD, BrykM, StrahlBD, CheungWL, DavieJK, et al. (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 15 : 3286–3295.
47. DoverJ, SchneiderJ, Tawiah-BoatengMA, WoodA, DeanK, et al. (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277 : 28368–28371.
48. HwangWW, VenkatasubrahmanyamS, IanculescuAG, TongA, BooneC, et al. (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 11 : 261–266.
49. WoodA, KroganNJ, DoverJ, SchneiderJ, HeidtJ, et al. (2003) Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 11 : 267–274.
50. TannyJC, Erdjument-BromageH, TempstP, AllisCD (2007) Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev 21 : 835–847.
51. ZofallM, GrewalSI (2007) HULC, a histone H2B ubiquitinating complex, modulates heterochromatin independent of histone methylation in fission yeast. J Biol Chem 282 : 14065–14072.
52. ChengH, HeX, MooreC (2004) The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol Cell Biol 24 : 2932–2943.
53. RoguevA, SchaftD, ShevchenkoA, PijnappelWW, WilmM, et al. (2001) The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 20 : 7137–7148.
54. SoaresLM, BuratowskiS (2012) Yeast Swd2 Is Essential Because of Antagonism between Set1 Histone Methyltransferase Complex and APT (Associated with Pta1) Termination Factor. J Biol Chem 287 : 15219–15231.
55. ShevchenkoA, RoguevA, SchaftD, BuchananL, HabermannB, et al. (2008) Chromatin Central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol 9: R167.
56. NislowC, RayE, PillusL (1997) SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell 8 : 2421–2436.
57. BrykM, BriggsSD, StrahlBD, CurcioMJ, AllisCD, et al. (2002) Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr Biol 12 : 165–170.
58. KimT, BuratowskiS (2009) Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137 : 259–272.
59. ZhangK, LinW, LathamJA, RieflerGM, SchumacherJM, et al. (2005) The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell 122 : 723–734.
60. WoodV, HarrisMA, McDowallMD, RutherfordK, VaughanBW, et al. (2012) PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res 40: D695–699.
61. MoazedD (2011) Mechanisms for the inheritance of chromatin states. Cell 146 : 510–518.
62. Reyes-TurcuFE, GrewalSI (2012) Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev 22 : 156–163.
63. YamadaT, FischleW, SugiyamaT, AllisCD, GrewalSI (2005) The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell 20 : 173–185.
64. ShankaranarayanaGD, MotamediMR, MoazedD, GrewalSI (2003) Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol 13 : 1240–1246.
65. AlperBJ, JobG, YadavRK, ShankerS, LoweBR, et al. (2013) Sir2 is required for Clr4 to initiate centromeric heterochromatin assembly in fission yeast. EMBO J 32 : 2321–2335.
66. GuillemetteB, DrogarisP, LinHH, ArmstrongH, Hiragami-HamadaK, et al. (2011) H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet 7: e1001354.
67. TanakaA, TanizawaH, SriswasdiS, IwasakiO, ChatterjeeAG, et al. (2012) Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol Cell 48 : 532–546.
68. CamHP, NomaK, EbinaH, LevinHL, GrewalSI (2008) Host genome surveillance for retrotransposons by transposon-derived proteins. Nature 451 : 431–436.
69. BahlerJ, WuJQ, LongtineMS, ShahNG, McKenzieA3rd, et al. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14 : 943–951.
70. MorenoS, KlarA, NurseP (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194 : 795–823.
71. MudgeDK, HoffmanCA, LubinskiTJ, HoffmanCS (2012) Use of a ura5+-lys7+ cassette to construct unmarked gene knock-ins in Schizosaccharomyces pombe. Curr Genet 58 : 59–64.
72. LyneR, BurnsG, MataJ, PenkettCJ, RusticiG, et al. (2003) Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4 : 27.
Štítky
Genetika Reprodukční medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 10
-
Všechny články tohoto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



