-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGenome-Wide Mapping of Yeast RNA Polymerase II Termination
Transcription termination is an important regulatory event for both non-coding and coding transcripts. Using high-throughput sequencing, we have mapped RNA Polymerase II's position in the genome after depletion of termination factors from the nucleus. We found that depletion of Ysh1 and Sen1 cause build up of polymerase directly downstream of coding and non-coding genes, respectively. Depletion of Nrd1 causes an increase in polymerase that is distributed up to 1,000 bases downstream of non-coding genes. The depletion of Nrd1 helped us to identify more than 250 unique termination regions for non-coding RNAs. Within this set of newly identified non-coding termination regions, we are further able to classify them based on sequence motif similarities, suggesting a functional role for different terminator motifs. The role of these factors in transcriptional termination of coding and/or non-coding transcripts can be inferred from the effect of polymerase's position downstream of given termination sites. This method of depletion and sequencing can be used to further elucidate other factors whose importance to transcription has yet to be determined.
Published in the journal: . PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004632
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004632Summary
Transcription termination is an important regulatory event for both non-coding and coding transcripts. Using high-throughput sequencing, we have mapped RNA Polymerase II's position in the genome after depletion of termination factors from the nucleus. We found that depletion of Ysh1 and Sen1 cause build up of polymerase directly downstream of coding and non-coding genes, respectively. Depletion of Nrd1 causes an increase in polymerase that is distributed up to 1,000 bases downstream of non-coding genes. The depletion of Nrd1 helped us to identify more than 250 unique termination regions for non-coding RNAs. Within this set of newly identified non-coding termination regions, we are further able to classify them based on sequence motif similarities, suggesting a functional role for different terminator motifs. The role of these factors in transcriptional termination of coding and/or non-coding transcripts can be inferred from the effect of polymerase's position downstream of given termination sites. This method of depletion and sequencing can be used to further elucidate other factors whose importance to transcription has yet to be determined.
Introduction
Termination of RNA Polymerase II (Pol II) transcription plays an essential role in the transcription cycle and has been the subject of several recent reviews [1], [2]. Disruption of the elongation complex at terminators recycles Pol II maintaining a pool of free enzyme able to compete for unoccupied promoters. Correct termination also prevents Pol II from interfering with expression of downstream genes either by colliding with oncoming Pol II elongation complexes or by dislodging transcription factors from the downstream DNA template [3]–[5]. Termination can also serve a regulatory purpose. Several yeast genes are regulated by premature termination [6]–[8] and genes involved in yeast nucleotide metabolism are regulated by the choice of alternative transcription start sites, one of which leads to premature termination [7], [9]. More recent studies have shown that correct termination is also necessary for efficient re-initiation at the same gene through the formation of a loop between the 3′ and 5′ ends [10], [11].
In addition to mRNAs, Pol II transcribes a diverse set of non-coding RNAs including snoRNA, some snRNAs and several classes of ncRNA with unknown functions. In yeast, this pervasive transcription [12]–[14] falls into several classes. Cryptic unstable transcripts (CUTs) are turned over rapidly by the nuclear exosome [15]–[20]. In contrast, another class of yeast ncRNA, termed stable uncharacterized transcripts (SUTs) are observed in the presence of an active nuclear exosome [21].
Pol II terminates coding and non-coding transcripts by different mechanisms [1], [2], [22]–[24]. Coding transcripts and possibly some SUTs are processed at the 3′-end by the cleavage and polyadenylation (pA) machinery. This reaction is coupled to termination occurring downstream of the processing site [1], [2]. In contrast, ncRNAs are terminated and processed by an alternative pathway that, in yeast, requires the RNA-binding proteins Nrd1 and Nab3 and the RNA helicase Sen1 [25]–[29].
Yeast Pol II terminators contain short RNA sequences that bind proteins within large complexes associated with the elongating Pol II. Loosely conserved pA signal sequences downstream of protein-coding genes bind to components of the CF1 complex leading to assembly of the cleavage and polyadenylation machinery [1]. Termination is coupled to cleavage in a manner that has not yet been completely resolved. Non-pA termination components Nrd1 and Nab3 recognize RNA sequence elements downstream of snoRNAs and CUTs [8], [28], [30]–[34] and this leads to the association of a complex that contains the DNA/RNA helicase Sen1 and the nuclear exosome [35]. The mechanism of termination of these ncRNA transcripts has also not yet been determined.
Several possible mechanisms for Pol II termination have been proposed. The “torpedo” model postulates that cleavage at the pA site exposes an uncapped 5′ end on the nascent transcript that acts as a substrate for the 5′→3′ RNA exonuclease Rat1 in yeast or Xrn2 in metazoans [36]–[38]. The exonuclease degrades the nascent transcript and upon reaching the Pol II elongation complex facilitates termination by an unknown mechanism. Another model postulates that an allosteric change in Pol II occurs upon assembly of the pA complex [39]. A member of this complex, Pcf11 has been shown to dismantle an elongation complex in vitro [40], [41] and it is possible that Nrd1 plays a similar role. The DNA/RNA helicase Sen1 interacts with the Pol II CTD [42] and has been proposed to act like the bacterial termination factor rho and track along the nascent transcript and pry off the elongating Pol II [1], [25], [26], [43], [44]. Mutation of Sen1 has been shown to lead to readthrough of both coding and non-coding transcripts in vivo [45] and in vitro can arrest transcription [46].
Part of the uncertainty in delineating termination mechanisms is identifying which factors operate at which terminators in vivo. In this study we have used PAR-CLIP to map Pol II on the yeast genome in living cells [8], [47]–[49] after depletion from the nucleus of components of the different termination pathways [50]. By comparing the Pol II maps with and without nuclear depletion we are able to map the location of termination to narrow regions of the genome downstream of coding and non-coding genes. This approach avoids the complexity of using transcripts to map terminators as the effect of RNA turnover is eliminated. We show here that depletion of Ysh1, the protein that cleaves nascent pre-mRNA transcripts at the pA site, enhances accumulation of Pol II at the 3′-ends of protein-coding genes. In contrast, both Nrd1 and Sen1 depletion lead to readthrough transcription of ncRNAs and our data has allowed us to map the position of non-pA termination facilitated by these factors.
Results
Mapping in vivo Pol II termination
To map in vivo termination we have used the anchor-away (AA) system [50] to deplete termination factors from the nucleus. In this approach rapamycin (rap) induces a complex between a protein tagged with a FKBP12-rapamycin binding (FRB) domain and an anchor protein tagged with an FKBP12 domain. In our case the FKBP12 domain is on the ribosomal protein RPL13A leading to rap-dependent depletion of FRB-tagged protein from the nucleus. Previous work has shown that a Pol II subunit RPB1-FRB strain shows greater than 90% depletion of Pol II at the PMA1 gene within 40 min [51]. We show here that in NRD1-FRBGFP or YSH1-FRBGFP strains that also contain, RPB3-TAGRFP, Nrd1 and Ysh1 are depleted from the nucleus with similar kinetics (Figure 1A–B). We were unable to observe Sen1-FRBGFP because it is present only in ∼100–2,000 copies per cell. When over-expressed 10–100-fold from a Gal promoter Sen1-FRBGFP is visible and is depleted from the nucleus with similar kinetics (Figure 1C). Given the limitations of background signals in live-cell imaging we cannot rule out the possibility that some small amount of Nrd1, Ysh1 or Sen1 remain in the nucleus. Growth curves show that all three strains grow normally for several divisions after administration of rap and we carried out PAR-CLIP analysis well before any changes in growth were observed (Figure S1A, shaded box). When cells are plated on media containing rap we observe no growth for YSH1-FRB and NRD1-FRB, while SEN1-FRB grows very slowly (Figure 1D).
Fig. 1. Nuclear depletion of Pol II termination factors. 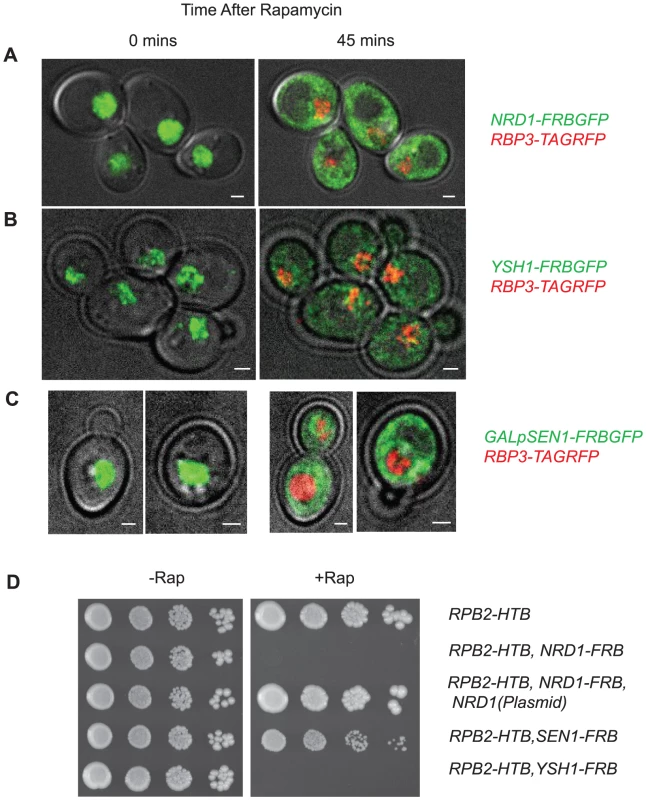
Live yeast cell imaging of: A. NRD1-FRBGFP, RPB3-TAGRFP; B. YSH1-FRBGFP, RPB3-TAGRFP; C. GALpSEN1-FRBGFP, RPB3-TAGRFP strains. Time after rap treatment is indicated above the pictures. D. Growth of indicated FRB strains on CSM plates with and without rap. Growing yeast cultures were cross-linked as previously described [8], [49] with modifications described in Methods. Briefly, 4-thiouracil (4tU) was added to a growing culture for 15 minutes to allow equilibration of uracil pools before the addition of rap. After thirty minutes of rap treatment cultures were irradiated with 365 nm UV for 15 min (Figure S1B). Incubation of yeast cells in 4tU does not significantly affect growth during the course of the experiment as cells continue to grow beyond the time frame of the cross-linking for at least one more doubling (0.2 OD600 to 0.7 OD600 as measured by BioTek scanner) indicating that 4tU has a less drastic effect on yeast growth than it does in mammalian cells treated for a longer time [52].
To map the position of elongating Pol II we isolated Pol II-bound RNA using a dual 6×His-biotin tagged Pol II Rpb2 subunit (Rpb2-HTB) as previously described [8]. Duplicate libraries were derived from the RPB2-HTB NRD1-FRB strain grown in the presence or absence of rap. We also created libraries of Rpb2-bound RNA from SEN1-FRB and the parental RPB2-HTB strains grown in the presence of rap and a single library from YSH1-FRB. Replicate libraries were multiplexed and sequenced by Illumina Hi-seq. Each library yielded between 15–39 million unique reads (Table S1). Biological replicates were strongly correlated (ρ>.99) and thus were added together for analysis (Figure S2). The WT dataset correlates very well to NET-seq [53] and GRO-seq [54] datasets (Figure S3). Thus, PAR-CLIP data accurately represents the position of Pol II elongation complexes on the yeast genome.
Ysh1-dependent termination
To map pA-dependent termination we carried out PAR-CLIP on Rpb2 after depletion of Ysh1, the factor that has been shown to be required for cleavage of the nascent mRNA at the GAL7 and CYC1 pA sites [55]. We reasoned that failure to cleave the nascent transcript would prevent Rat1 degradation thus disabling the “torpedo” mechanism. While we cannot rule out the possibility that other factors are depleted along with Ysh1 we do observe a failure to cleave at the pA site for several genes (Figure S4). The most dramatic effect of Ysh1 depletion is seen in a buildup of reads just downstream of the major pA site of highly expressed protein-coding genes (Figure 2A and 2C). The peak of reads is present in most of the highly expressed protein-coding genes as seen in figure 2B. No similar effect is seen downstream of snoRNAs or CUTS. One surprising aspect of Ysh1 depletion is that readthrough Pol II does not extend further downstream but seems to pause or terminate within 200 bp of the pA site (Figure 2A). This result indicates that Pol II that reads through termination signals fails to elongate but is unable to efficiently terminate.
Fig. 2. Ysh1 depletion causes readthrough at pA sites. 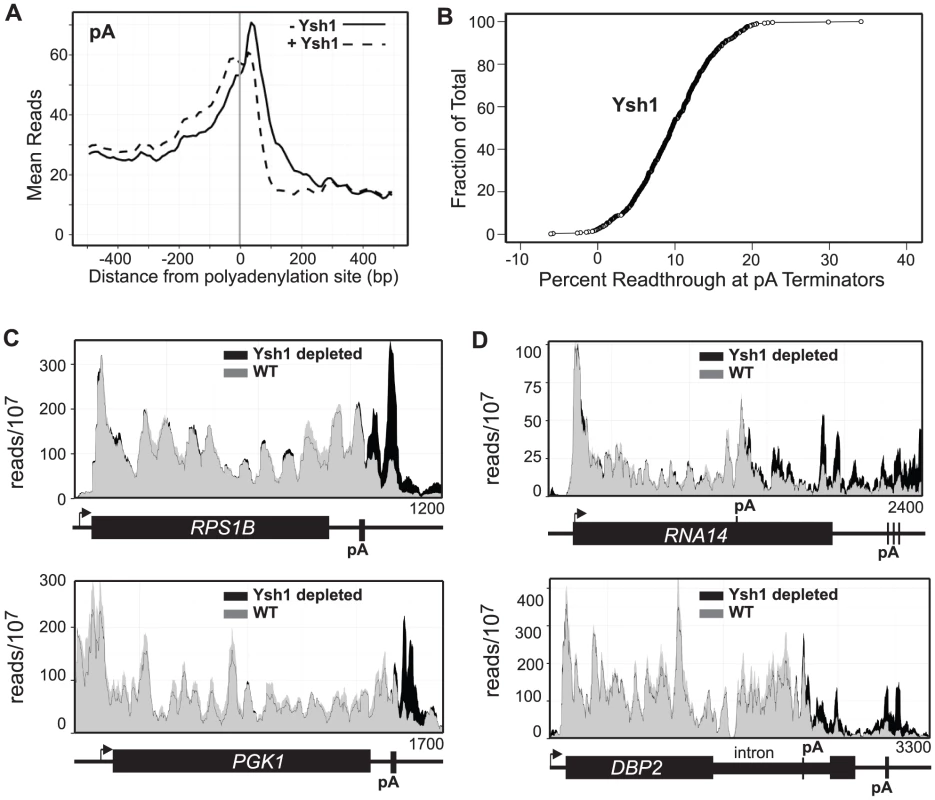
A. Mean reads every 10 bp for the 500 most frequently used pA sites with (dotted) and without (solid) Ysh1. B. Plot showing percent readthrough of each of 500 pA terminators as a fraction of the total 500 pA terminators. C and D. Histograms representing normalized reads with Ysh1 (grey) and without Ysh1 (black) at the given genomic locations. The TSS with the direction of transcription is indicated by an arrow. Genes and pA sites are represented below each graph and the length of the genome depicted is given in the lower right hand corner. We do, however, observe several instances of Pol II extending 1 kb or more downstream of apparent terminators in response to Ysh1 depletion. The RNA14 and DBP2 genes show an increase of Pol II toward the 3′ end of the coding region indicating the presence of a Ysh1-dependent terminator upstream of the main pA site (Figure 2D). Rna14p is part of the CF1 complex required for recognition of pA sites [1] and previous studies have shown that in addition to the mature 2.2 kb mRNA several smaller mRNAs are present under normal conditions [56], [57]. The terminator we map in RNA14 is located in the same region as the 3′ end of the shorter (1 kb) mRNA (Figure 2D).
Dbp2p is a DEAD-box RNA helicase that plays a role in assembly of mRNP complexes and in RNA quality control [58], [59]. The DBP2 gene is unique in yeast in having a long (1 kb) intron localized toward the 3′ end of the coding region and previous work has shown that the gene is autoregulated through sequences in the intron [60]. In Figure 2D we show that depletion of Ysh1 leads to an increase in Pol II cross-linking downstream of the intron suggesting the presence of an upstream terminator. Consistent with this view polyadenylation sites have been mapped to this location in the Dbp2 intron [61]–[63].
Nrd1-dependent termination
Previous work has shown that Nrd1p is required for proper termination of ncRNAs like snoRNAs and CUTs as well as premature termination of coding transcripts of genes regulated by attenuation [6], [7], [9], [19], [28], [35], [64]. Here we show that depletion of Nrd1 in the presence of rap leads to readthrough transcription of a large number of these ncRNAs. Similar results have recently been obtained by a different protocol [65].
To map the position of termination we used an approach similar to that used to calculate a travelling ratio [66] or Escape Index [38], [65]. Reads in 500 bp windows upstream and downstream of a fixed point were tallied and a ratio of reads in the downstream window to total reads in both windows were calculated for that point. This ratio was determined for each point in the genome on both strands for both control and rap treated cells. Subtracting the fraction of readthrough of the control data from the rap-treated data resulted in a Readthrough Index that allowed us to rank order the regions of the genome showing the highest level of Nrd1-dependent readthrough (Table S2).
Once these regions were identified, we created plots of the difference between the reads in the control and reads with rap (Figure 3A). Nrd1 depletion leads to both a reduction of Pol II upstream and an increase of Poll II downstream of apparent termination sites. This is consistent with previous reports that Pol II pauses prior to termination [67]–[70]. In the absence of Nrd1, this apparent pause is eliminated and Pol II is able to transcribe through the termination site. Fitting this difference plot with a spline function allowed us to determine the point at which the difference plot crosses the X-axis. We have called this the termination site but this represents the center point of a narrow (about 50 nt) region over which Pol II is released from the template.
Fig. 3. Mapping Nrd1-dependent terminators. 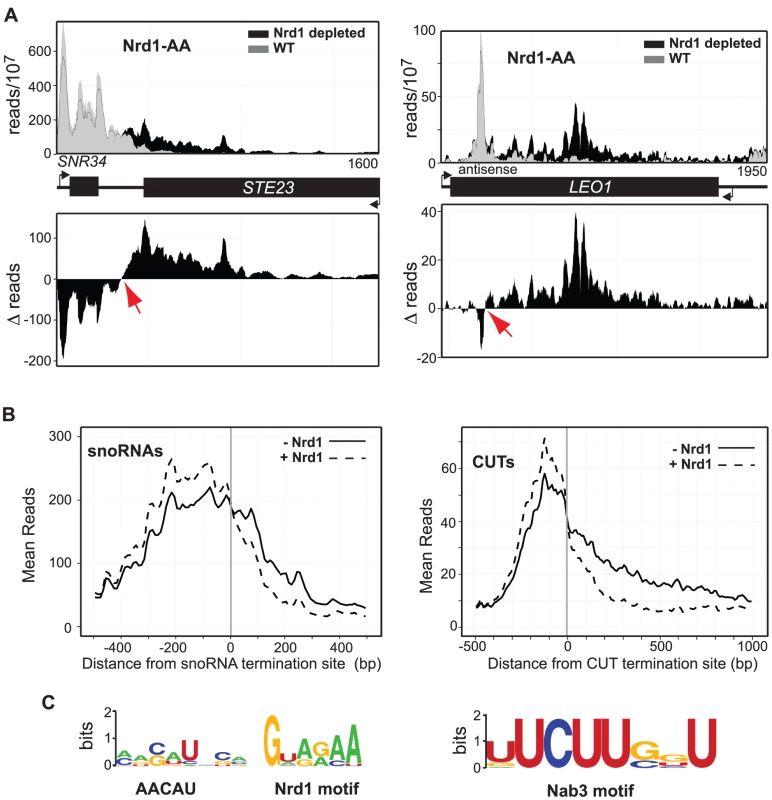
A. Histograms representing of normalized reads for NRD1-FRB with Nrd1 (grey) and without Nrd1 (black) at the given genomic locations. The difference between WT and Nrd1-depleted at every nucleotide is represented as a histogram below each graph. B. Mean reads every 10 bp for 49 snoRNA termination sites showing the highest level of readthrough and 144 CUT termination sites with Nrd1 (dotted) and without Nrd1 (solid). C. Most abundant Nrd1 motif and the most abundant Nab3 motif in the 150 nt 5′ of the snoRNA and CUT terminators, respectively. MEME input parameters were any number of motifs of length 4–15 nt. Results are presented using WebLogo [92]. In Figure 3A we show the difference plots for snoRNA snR34 and the antisense CUT to the LEO1 gene demonstrating how the termination sites were identified. We have carried out this analysis for 49 snoRNAs and for 144 CUTs showing the highest level of readthrough transcription (Tables S2 and S3). Comparing these termination sites to the sites determined by Pol II ChIP [65] reveals that 70% of the Schulz et al. sites are more than 100 nt downstream of our termination sites and 25% are more than 200 nt downstream (Figure S5). We do not know the reason for this systematic discrepancy but the PAR-CLIP analysis has greater resolution than Pol II ChIP and unlike ChIP gives strand-specificity. The metagene analysis of the ncRNAs for which we have mapped termination sites is shown in Figure 3B demonstrating a more significant increase in readthrough transcription for Nrd1-depletion when compared to the increase in readthrough transcription due to Ysh1 depletion (Figure 2A). SnoRNA and CUT Nrd1-dependent readthrough extends over 500 bp and 1 kb, respectively.
Examining DNA sequences upstream of the termination site for these sets of snoRNAs and CUTs revealed an interesting difference in the occurrence of Nrd1 and Nab3 binding motifs. The top hit in the MEME analysis [71] of the top 27 snoRNA upstream regions (Figure 3C) include the GUA[A/G] sequence previously identified as a Nrd1 binding site [8], [31]–[34], [72]. This analysis also identifies a loosely conserved sequence AACUA centered about seven nucleotides upstream of the GUA[A/G] sequence that has not previously been reported. The second most significant motif upstream of the snoRNA termination site was the UCUU sequence that binds Nab3, consistent with the presence of a Nrd1-Nab3 heterodimer [29], [31]. In sequences upstream of CUT terminators the most significant hit was UCUUG which contains the previously identified Nab3-binding sequence but with a downstream G as has been observed previously [8], [33], [34]. Among the 144 CUT terminators the Nrd1 binding motif was not present above background. Together, these observations suggest a significant difference in the manner of recognition of snoRNA and CUT terminators.
U-rich sequences in the Nrd1-dependent termination region
In Figure 4 we show NET-seq [53] and PAR-CLIP data demonstrating that Pol II levels decline in the SNR13-TRS31 intergenic region. NET-seq reads for several peaks, indicated by bars, are reduced several-fold immediately downstream from the 3′-end of the SNR13 gene and more than 6-fold further than 100 nt downstream. PAR-CLIP reads from cells grown in the absence of rap decline at the same region reaching a minimum at about 150 nt downstream of the SNR13 3′-end. A similar correspondence between NET-seq and PAR-CLIP data is seen for other snoRNA genes (Figure S6). Taken together, these observations indicate that Pol II normally terminates in a Nrd1-dependent fashion in a narrow region located downstream of the snoRNA gene. The sequence from this region for SNR13, located from 50–150 nt downstream, is shown below the figure and a series of runs of U residues is indicated in red. This is not unexpected as intergenic regions in S. cerevisiae are AT-rich. Sequences of termination regions of several antisense CUTs are shown below the SNR13 sequence. In these cases we also observe runs of U residues. This is unexpected as these termination regions fall within coding regions (on the opposite strand) which in general are not AT-rich. We propose that these U-rich regions constitute part of the non-poly(A) terminator.
Fig. 4. SNR13 termination region. 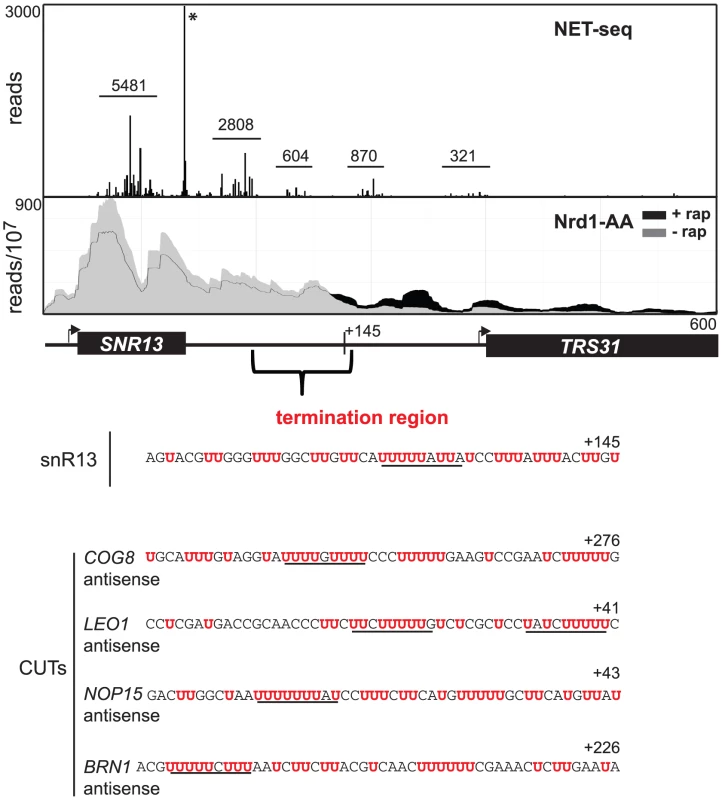
NET-Seq data from Churchman and Weissman [53] are shown above PAR-CLIP data from the SNR13-TRS31 locus. The number of NET-seq reads from peaks of Pol II are shown above the lines. The asterisk indicates reads derived from mature snR13 RNA that contaminates the NET-seq library. The calculated SNR13 termination region has been expanded to show U-rich sequences (red) surrounding the termination point. The sequences of several antisense CUT termination regions are shown below the SNR13 sequence. Nrd1 and RNA turnover
We estimate that SNR13 readthrough transcripts are present at less than one copy per cell in wild-type cells and this is consistent with the low level of readthrough transcripts seen in the absence of rap in the Northern blot shown in Figure 5A. Despite this low level of steady-state readthrough transcripts we observe significant cross-linking to Rpb2 in the region downstream of SNR13 (Figure 5B) even in the presence of Nrd1. Presumably the RNA synthesized by these polymerases is rapidly degraded by the nuclear exosome. Figure 5A shows that snR13 readthrough transcripts detected by Northern blot or RT-PCR are increased more than 10-fold by 15 min and more than 100-fold by 30 min of Nrd1 depletion. However, depletion of Nrd1 results in only a 2–3-fold increase in PAR-CLIP signal for Rpb2 (Figure 5B) indicating that depletion of Nrd1 not only allows readthrough of ncRNA transcripts but also results in their stabilization by uncoupling degradation by the nuclear exosome.
Fig. 5. Nrd1 increases the stability of the readthrough transcripts. 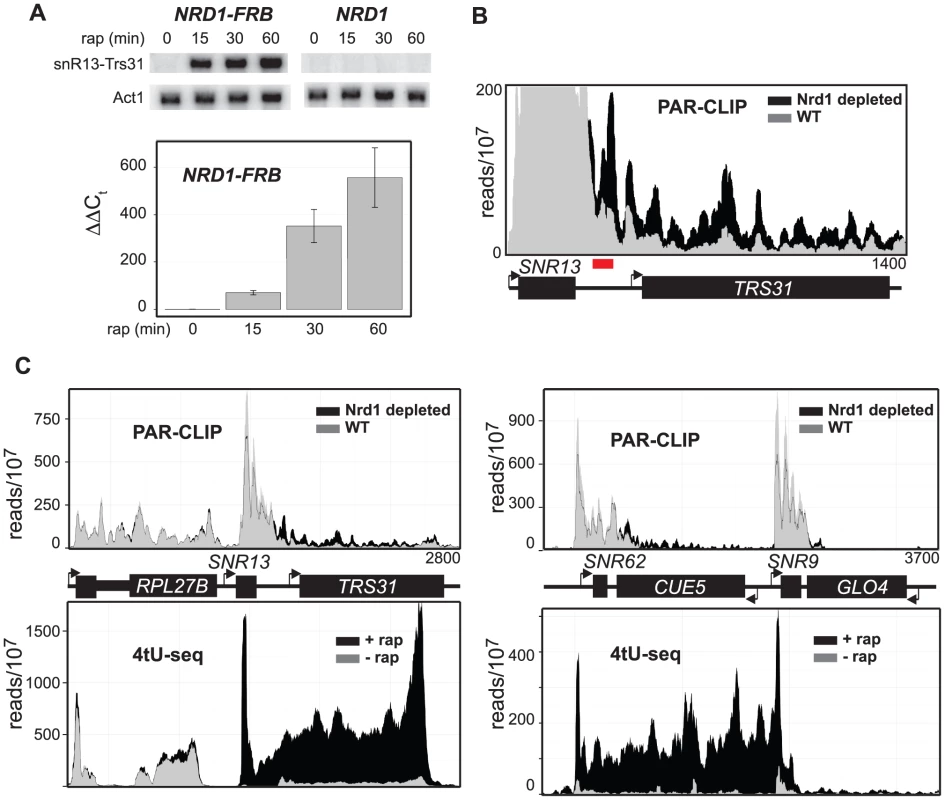
A. Northern blot and qPCR at different times after addition of rap using the amplicon highlighted in red. B. Histograms representing normalized reads with Nrd1 (grey) and without Nrd1 (black) at the given genomic locations. The Y-axis has been changed to emphasize the differences between treatment with (black) and without rap (grey) for the SNR13-TRS31 locus. C. Similar to A but Schultz et. al. [65] 4tU-seq data is also represented in as a histogram below each graph for the same region of the genome with Nrd1 (grey) and without Nrd1 (black). Our analysis of steady-state RNA levels is consistent with recently published 4tU-seq data [65]. Figure 5C shows the comparison of our data and that of Schulz et al for SNR13 and the upstream RPL27B gene and for the SNR62 and SNR9 genes. 4tU-seq data show a dramatic increase in SNR13 and SNR62 readthrough transcription compared to PAR-CLIP data. The most likely explanation for this difference is that 4tU labeling is not completely restricted to nascent transcripts. Labeling cells for six minutes with 4tU allows for multiple rounds of transcription especially for heavily transcribed genes like snoRNAs and ribosomal protein genes. The earliest synthesized transcripts are subject to processing as is clearly evident in the paucity of reads derived from the intron of RPL27B in the 4tU-seq compared to the PAR-CLIP (Figure 5C) and a number of other intron-containing genes (Figure S7). Depletion of Nrd1 does not affect all snoRNAs as is seen in Figure 5C where neither PAR-CLIP nor 4tU-seq detect appreciable readthrough transcription at SNR9.
Sen1-dependent termination
A role for Sen1p in termination of both coding and ncRNA transcripts has been proposed [26], [28], [45]. Our current data show that depletion of Sen1 results in no change in the localization of Pol II at the 3′-ends of most protein-coding genes (Figure 6A) including those previously shown [45] to be dependent on Sen1 (Figure S8). Thus, despite our previous observation that Sen1 cross-links to these transcripts [8], there is little evidence for a role for Sen1 in pA termination. Sen1 depletion does result in readthrough transcription at both snoRNAs and CUTS (Figure 6B–C) which is consistent with previous observations [15], [19], [28]. The pattern of Pol II readthrough in response to Sen1 depletion differs, however, from that of Nrd1. Rather than an extended region of readthrough downstream of the terminator, Sen1 depletion often results in an increase just downstream by a few hundred nucleotides. This type of pattern is seen in Figure 6D for SNR47 and the CUT antisense to the COG8 gene and in Figure S9 for several protein-coding genes regulated by Nrd1-dependent attenuation. Figure 6E compares the percent readthrough downstream of all 266 non-pA terminators in Sen1 depleted and Nrd1 depleted cells. The percent readthrough after Nrd1 depletion is twice that for Sen1 depletion and these values are not dominated by a subset of terminators. This observation suggests that Sen1 may play a different function than Nrd1 at these genes and that loss of Sen1 results in less processive or more pause prone readthrough transcription.
Fig. 6. Sen1 depletion causes readthrough at non-pA terminators. 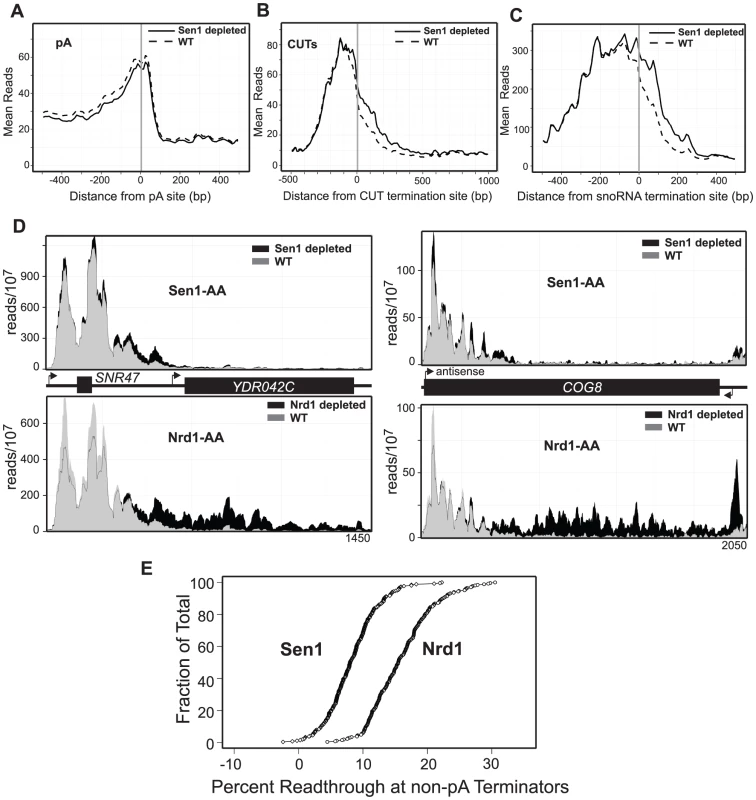
A.–C. Mean reads every 10 bp for the top 500 polyA,144 calculated CUT termination sites, and 49 snoRNA termination sites, with Sen1 (dotted) and without Sen1 (solid). D. Histograms representing normalized reads with Sen1 (grey) and without Sen1 (black) at the given genomic locations. The same region is also represented in as a histogram below each graph with Nrd1 (grey) and without Nrd1 (black). E. Percent readthrough of non-pA terminators (ordered from highest to lowest) represented as a fraction of the total number of calculated non-pA terminators. SEN1-FRB is represented as diamonds and NRD1-FRB is represented as circles. Discussion
In this paper we describe the development and implementation of a technique for mapping the position of in vivo Pol II termination. Depletion of components of the yeast Pol II termination machinery followed by Pol II cross-linking to nascent RNA has allowed the identification of both pA and non-pA termination regions. The site of pA termination has been mapped to within a few hundred bases of the pA site as has previously been observed for the Cyc1 terminator [73]. We have identified non-pA terminators using an algorithm that identifies points in the genome where Pol II readthrough increases in response to Nrd1 depletion.
While many of the components of both the pA and non-pA termination complexes have been identified, the mechanism of termination is still unclear. Two general models have been proposed [1], [2], [22], [74]. The allosteric model proposes a conformational change in Pol II that slows elongation [39], [75], [76] while the torpedo model proposes that cleavage at the pA site creates a substrate for a 5′→3′ exonuclease (Rat1 in yeast and Xrn2 in metazoa) that degrades the nascent transcript like a “fuse” and upon reaching elongating Pol II somehow facilitate termination [36], [37], [77]. Neither model adequately explains all experimental observations leading to a hybrid model that includes both allosteric and topedo mechanisms [78].
pA termination
Our data on Ysh1 depletion supports a two-step pause and release model for termination that invokes both allosteric and torpedo mechanisms [79]. The buildup of Pol II just downstream of the uncleaved pA site suggests that Pol II has paused but is deficient in the subsequent release step. This is consistent with a role for Ysh1 in providing the substrate for the Rat1 exonuclease that facilitates removal of Pol II from the template [80]. Ysh1 apparently is not required for the allosteric change that results in pausing downstream of the pA site as seen in the lack of readthrough at most pA sites. In contrast, mutation of the pA site [81] or mutations in RNA14, RNA15, PCF11, CLP1 and GLC7 result in readthrough transcription more than 500 nt downstream [73], [82], [83] arguing for a role for these factors in the allosteric change that renders Pol II immobilized.
Depletion of Ysh1 has also revealed several genes that are apparently regulated by premature termination through the pA pathway. RNA14 and DBP2 contain pA sites upstream from the 3′ pA site. Loss of Ysh1 leads to processive elongation ending at the downstream pA site indicating that Ysh1 is required for termination at these upstream sites. Why the putative allosteric step does not function efficiently at these upstream sites is not clear. Perhaps the phosphorylation pattern of the CTD at these sites precludes the assembly of factors like Pcf11. Alternatively, chromatin structure modification induced by the Pol II that elongates through these terminators may suppress normal termination. Finally, it is possible that use of the downstream terminator by Pol II that ignores the upstream pA site creates a gene loop that favors termination at the downstream site.
Non-pA termination
Nrd1 and Nab3 bind specific RNA sequences and act as sensors to detect non-pA terminator sequences in the nascent transcript. While a number of studies have characterized the short motifs recognized by Nrd1 and Nab3 the relative orientation and abundance of these sequences varies widely among non-pA terminators [8], [31]–[34], [49], [65]. In this study we have localized termination downstream of 49 snoRNAs and 144 CUT transcripts. This has allowed a search for sequences that may define these two sets of terminators. We find that the most significant motif in this set of snoRNA terminators contains the GUA[A/G] motif previously identified for Nrd1 in the context of a longer sequence that indicates the possible involvement of another, unidentified RNA-binding protein. The second most significant motif is the Nab3 binding sequence UCUUG. In the case of the CUT terminators the only significant motif is the Nab3 binding sequence. Thus, it appears that while both Nrd1 and Nab3 binding contribute to recognition of snoRNA terminators Nab3 predominates for CUTs. This does not preclude Nrd1 from playing a critical role in termination at these CUTs. We have previously shown that Nrd1 is necessary for termination of the CUT antisense to FMP40 presumably by acting as an adaptor to couple the Nrd1-Nab3-Sen1 complex to the elongating Pol II through interaction between the Nrd1 CID and the CTD [15].
No other significant motifs were observed upstream of these terminators but we did observe that the sequences surrounding the termination site are U-rich and contain multiple runs of U residues. This is not unexpected for snoRNA downstream sequences as intergenic regions are AT-rich in S. cerevisiae. The U-rich sequences surrounding the termination sites (Figure 4) of antisense CUTs are more significant as these sequences occur in the context of a coding region on the opposite strand. U-rich sequences have been shown to form unstable rU:dA base pairs [84] and in the hybrid binding site of Pol II transcribing the antisense CUT such an unstable hybrid sequence may increase the probability of termination [85], [86].
In addition to its role in Pol II termination, our data highlight the role Nrd1 plays in the turnover of ncRNAs. Nrd1 depletion by anchor away leads to accumulation of readthrough transcripts in 4tU-seq to a much higher degree than observed by our Rpb2-HTB PAR-CLIP. We show here by Northern and RT-PCR that after 40 min of Nrd1 depletion the level of snR13 readthrough transcripts increases over 100-fold. However, after 40 minutes of depletion the amount of readthrough as observed by PAR-CLIP is less than 10-fold. This difference is likely due to an important role for Nrd1 in coupling termination to turnover of the completed transcript. Nrd1 is found in a complex with components of the TRAMP and exosome complexes [35] and previous studies have indicated that nrd1 mutants are deficient in RNA turnover [15], [19], [35].
Pulse labeling RNA with 4tU was recently used to analyze newly synthesized transcripts in a Nrd1 anchor away mutant and this data identified over 1500 Nrd1-dependent transcripts [65], far more than the number of terminators we describe here. Part of the reason for this discrepancy is that the six-minute pulse used in the 4tU-seq protocol is substantially longer than the time needed to synthesize short RNAs. Thus, 4tU-seq over-estimates the effect on termination and may ascribe termination functions to cases where the role in turnover is independent of termination.
Depletion of the DNA/RNA helicase Sen1 has allowed us to place this factor squarely in the Nrd1-Nab3 non-pA termination pathway. We observe readthrough transcription downstream of both snoRNAs and CUTs but not downstream of pA sites. Pol II does not progress far downstream after Sen1 depletion indicating that this factor may act after the allosteric change has occurred. This is consistent with Sen1 acting in place of Rat1 to provide the activity that dislodges the paused Pol II [46] and supports previous work showing the difference in the requirement for these factors in pA and non-pA termination [24].
The results presented here are summarized in a model shown in Figure 7. We have been able to demonstrate a difference in the global effect of depleting different components of the yeast termination machinery. These results have been interpreted in the context of a two state model for termination in which terminator sequences in the nascent RNA trigger assembly of protein complexes that signal to the elongating Pol II to decrease its elongation rate and processivity. These anti-processivity factors include Pcf11 for pA terminators and Nrd1 for non-pA terminators. Loss of either of these factors leads to runaway processive elongation. In contrast, depletion of Ysh1 or Sen1 does not block the transition to non-processive elongation but does result in a defect in removal of Pol II from the template. For Sen1 this likely occurs through loss of a rho-like function [44], [46] while Ysh1 depletion leads to a lack of cleavage at the pA site thereby limiting access of Rat1 and preventing the torpedo mechanism of Pol II release [80].
Fig. 7. Schematic representation of Pol II termination after removal of non-pA and pA termination factors. 
Elongating Pol II (green) terminates pA transcripts (A) after an allosteric change (red) that reduces processivity. (B) Depletion of Ysh1 leads to minimally extended readthrough transcripts but does not block the allosteric change in Pol II. (C) Nrd1 and Nab3 binding recruit Sen1 for termination of non-pA transcripts. (D) Pol II elongation complex lacking Nrd1 does not recognize termination sequences in the nascent transcript and thus does not facilitate the allosteric transition in Pol II. This leads to processive readthrough. (E) Nrd1 and Nab3 recognize terminator sequences allowing the allosteric change in Pol II but depletion of Sen1 blocks removal of Pol II from the template. The data presented here demonstrate that PAR-CLIP analysis of Pol II in living yeast cells can define the site of Pol II termination regions and when coupled with depletion of termination factors offers an avenue for examining the roles of different factors in termination genome-wide. Such experiments will illuminate the variety of mechanisms the cell uses to prevent Pol II from transcribing beyond genetically determined 3′ boundaries.
Materials and Methods
Strain construction
Yeast strains for anchor away were constructed using the parental strain HHY168 [50] (Euroscarf #Y40343). RPB2 was C-terminally tagged with an HTB (6×HIS, TEV, and Biotin) tag as described previously [8], [87]. NRD1, YSH1, and SEN1 were C-terminally tagged with FRB and FRBGFP as described in Haruki et al. [50]. The SEN1 promotor was replaced with the GAL1 promotor using a cassette from pFA6a-kanMX6-PGAL1-HBH [87]. All strains used in these experiments are listed in Table S4.
Cell growth
Cells were grown in Complete Synthetic Media (CSM) supplemented with 2% glucose or galactose and 40 mg/l adenine. For RNA analysis, cells were seeded to an OD600 of 0.1 from a 5 ml or 50 ml overnight culture. The cells were then incubated at 30°C to an OD600 of 1. 1 µg/ml of rap (LC laboratories) was then added to the cultures and the cells were allowed to grow for the indicated times in the experiment. For growth curves, a BioTek Infinity 2 (BioTek) was used to incubate a 24 well plate containing CSM media with and without 1 µg/ml rap (added after 6 hrs of growth) with orbital shaking (Slow, 1 mm amplitude). Cells were diluted to an OD600 of 0.1 per well and an OD600 was measured every 10 min for 24 hours.
RNA purification and cDNA synthesis
Total RNA was extracted from yeast with hot acid phenol as previously described [8]. Strand specific northern blots were done as described previously [88] with the following modifications. Ultrahyb (Invitrogen) was used instead of the described hybridization buffer. Real-time PCR analysis was done on a CFX96 instrument (Biorad) in triplicate as described previously [49]. Primers used can be found in Table S5.
Live cell imaging of FRBGFP strains
The strains containing NRD1-FRBGFP, YSH1-FRBGFP, or GALpSEN1-FRBGFP, RPB3-TAGRFP were grown to an OD600 of 1 at 30°C. Live cell imaging was done on a Deltavision microscope (Applied Precision) as described previously [89] with the following modification: 1 µg/ml rapamyacin was added to the agarose/media pad. Galactose was substituted for glucose with the GALpSEN1-FRBGFP strain. Pictures were taken of GFP at 0 mins and 45 mins post rap exposure and 45 mins for RFP.
Anchor away and PAR-CLIP
Cells were grown overnight 30°C in 50 ml of YPD (Yeast Extract Peptone with 2% glucose) media. Four 500 ml flasks of sterile cross-linking media (CSM–Ura supplemented with 2% glucose, 40 mg/l adenine, 60 µM uracil, and 1 µM biotin were seeded to an OD600 of 0.1 and incubated at 30°C until an OD600 of 1. Crosslinking was carried out by the protocol described in Figure S1B. 4-thiouracil (4tU) was added to a final concentration of 4 mM and incubated at 30°C for 15 min. 1 µg/ml of rapamycin was then added to the cells and incubated at 30°C for 30 min. All four 500 ml cultures were then pooled into one 2 l beaker and irradiated from a distance of two cm with 365 nm UV (∼1 W/cm2) from an LC-L5 LED UV Lamp (Hamamatsu) for 15 min with gentle stirring. The top ∼2 mm of the culture appears luminescent indicating that the UV light does not penetrate further into the culture. Assuming an even mixture of the culture we calculate that the average cell is cross-linked for only ∼10 seconds within the 15 min crosslinking period. The cultures were then filtered through 0.45 micron nitrocellulose filters (Millipore) and cells were scraped into 5 ml Buffer 1 (300 mM NaCl, 0.5% NP-40, 50 mM NaPO4 pH 7.2, 10 mM immidazole, 6 M Guanidine HCl, Protease inhibitor Cocktail VII [RPI]) and frozen in liquid nitrogen.
HTB tagged Rpb2 purification was adapted from our previously published protocol [49]. Cells were lysed in liquid nitrogen using a SamplePrep 6870 freezer mill (Spex) with 15 cycles per second of cracking for 15 cycles of 1 min with 2 min of cooling between each cycle. Lysates were then incubated with 1 ml of Ni-NTA agarose (Qiagen), which was equilibrated in buffer 1 for 2 hours at room temperature. The Ni-NTA agarose was then added to an empty 10 mL plastic column (Biorad) and washed with 20 ml of Buffer 1 followed by 10 ml of Buffer 2 (20 mM NaPO4 pH 7.2, 300 mM NaCl, 0.5% NP40, 10 mM imidazole, 4 M Urea). The protein was eluted off of the Ni-NTA agarose with 5 ml Buffer 2+250 mM immidazole and into 5 ml Buffer 2+120 µl Protease inhibitor Cocktail VII+100 µl of hydrophilic streptavidin magnetic beads (NEB). The slurry was allowed to incubate for 4 hours at room temperature. The strepavadin magnetic beads were then resuspended in 500 ul of Buffer 3 (50 mM Tris pH 7.4, 200 mM NaCl, 4 M Urea), transferred to a 1.5 ml siliconized tube and washed with 3×1 ml of Buffer 3 followed by 3×1 ml of T1 Buffer (50 mM Tris pH 7.4, 150 mM NaCl, 2 mM EDTA). Beads were resuspended in 200 µl of T1 buffer. 0.15 U/µl of Rnase T1 (Fermentas) was added to the bead slurry and allowed to incubate at 25°C for 15 min. The beads were then washed 3×1 ml T1 wash buffer (50 mM Tris pH 7.4, 500 mM NaCl, 1% NP-40, 0.5% Na deoxycholate) followed by 3×1 ml washes in PNK Buffer (50 mM Tris pH 7.2, 50 mM NaCl, 10 mM MgCl2). The beads were resuspended in 200 µl of TSAP reaction solution (0.15 U/ul Thermosensitive Alkaline Phosphatase [TSAP, Promega], 1 U/µl SupeRase Inhibitor [Invitrogen], 1 mM DTT in PNK Buffer) and incubated at 37°C for 30 min. The beads were washed 1×1 ml Buffer 3, 2×1 ml T1 Wash Buffer, and 3×1 ml PNK Buffer.
cDNA library preparation
Streptavadin beads were resuspended in 200 ul of PNK reaction solution (1 U/µl T4 PNK [NEB], 32P γ-ATP, 5 mM DTT in PNK Buffer) and incubated at 37°C for 30 min. 1 mM of cold ATP was added to the reaction and incubated at 37°C for 10 min. The beads were washed 1×1 ml Buffer 3, 1×1 ml T1 wash, 3×1 ml T4 RNA Ligase Buffer (50 mM Tris pH 7.4, 10 mM MgCl2). The beads were resuspended in 44 µl of T4 RNA Ligase 2 reaction solution (25% PEG 8000, 5 µM 3′ adaptor [AppAGATCGGAAGAGCACACGTCTddC, IDT], 10 U/µl T4 RNA Ligase 2, truncated K227Q [NEB], 2 U/µl RNase Inhibitor [Invitrogen], 1 mM DTT in RNA Ligase Buffer) and incubated at 25°C for 4 hours. The beads were then washed with 1×1 ml Buffer 3 and 5×1 ml T4 RNA Ligase Buffer. The beads were resuspended in 50 ul of T4 RNA Ligase reaction solution (5 uM 5′ Adaptor [GUUCAGAGUUCUACAGUCCGACGAUC, IDT], 1.2 U/µl RNase Inhibitor, 1 mM ATP, 0.5 U/µl T4 RNA Ligase [NEB], 1 mM DTT in RNA Ligase Buffer) and incubated at 16°C overnight. The beads were then washed 5×1 ml Proteinase K Buffer (100 mM Tris pH 7.4, 150 mM NaCl, 12.5 mM EDTA). The beads were resuspended in 200 µl of Proteinase K Buffer +2% SDS and 12 µl of Proteinase K (NEB). The suspension was incubated for 30 min at 37°C at which point the supernatant was removed and saved and the beads were resuspended in 200 µl of Proteinase K Buffer +2% SDS and 12 µl of Proteinase K. The beads were then incubated 30 min at 37°C and the supernatants were pooled. The RNA was recovered by acid phenol/chloroform extraction followed by two serial ethanol precipitations. The resulting pellet was allowed to dry and resuspended in 15 µl of Nuclease-Free Water [Invitrogen]. The RNA was split into 5 µL aliquots and frozen at −80°C for storage.
The Reverse Transcription, PCR, and gel extraction of the cDNA Library were carried out as described previously [49] with the following modifications. For reverse transcription, the primer used was AGACGTGTGCTCTTCCGATCT (IDT). For PCR analysis, biological repeats were multiplexed with the following primers. The forward primer was AATGATACGGCGACCACCGAGATCTACACGTTGAGAGTTCTACAGTCCG*A (where a * denotes and phosphorothioate bond). The first reverse primer was CAAGCAGAAGACGGCATACGAGATATTGGCGTGACTGGAGTTCAGACGTGTGCTCTTCGGATC*T for the first biological repeat and the second reverse primer was CAAGCAGAAGACGGCATACGAGATTACAAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC*T for the second biological repeat (IDT).
Sequencing and bioinformatics
Sequencing and demultiplexing was done at UC Riverside on an Illumina HiSeq. Trimming of the resulting sequences as described previously [49]. Briefly, raw reads were trimmed of the 3′ adaptor using a wrapper for R-bioconductor [90] developed by Sarah Wheelan. Raw trimmed reads were condensed (defined as no more than one of the same exact sequence) to eliminate any PCR artifacts within the data. Bowtie 1.0.0 [91] was run with the following arguments (-y –best -v 2) and aligned to the SacCer3 (R64) genome. Reads mapping to tRNA genes were removed as these likely represent artifactual binding during the affinity purification steps. Reads were then converted to a wig format where each read was multiplied by a factor that was defined as the total number of reads aligned per 107 reads. PAR-CLIP datasets have been submitted to GEO with the accession number GSE56435.
Calculation of the top 500 polyA sites
To calculate the best polyA sites per gene we used the sites provided by Moqtaderi et al. [63]. We set a hard cutoff of at least 200 raw reads. If, however, there were multiple very strong polyA sites within 200 base pairs of each other, we calculated the read ratio between the two most significant polyA sites. If the read ratio was less than 0.4, we ignored them.
Global readthrough percentage
Global readthrough percentage was calculated for each point in the genome as the percentage readthrough (reads downstream 500 bp/reads downstream 500 bp + reads upstream 500 bp) in the treated sample minus control sample. All points with a percentage readthrough of greater than 10%, at least a total of 1000 reads in either sample, and greater reads in the treatment vs the control were extracted. The list was further refined by taking the difference of every point 2 kb around non-overlapping termination sites (approx 500 total), graphing the region, and then fitting a spline function over a 25 bp window to find the best point of inflection. These points of inflection were then used as a focal point for the meta gene analysis.
Supporting Information
Zdroje
1. KuehnerJN, PearsonEL, MooreC (2011) Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12 : 283–294.
2. MischoHE, ProudfootNJ (2013) Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochim Biophys Acta 1829 : 174–185.
3. CloutierSC, WangS, MaWK, PetellCJ, TranEJ (2013) Long noncoding RNAs promote transcriptional poising of inducible genes. PLoS Biol 11: e1001715.
4. CastelnuovoM, RahmanS, GuffantiE, InfantinoV, StutzF, et al. (2013) Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat Struct Mol Biol 20 : 851–858.
5. ShearwinKE, CallenBP, EganJB (2005) Transcriptional interference–a crash course. Trends Genet 21 : 339–345.
6. ArigoJT, CarrollKL, AmesJM, CordenJL (2006) Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell 21 : 641–651.
7. KuehnerJN, BrowDA (2008) Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol Cell 31 : 201–211.
8. CreamerTJ, DarbyMM, JamonnakN, SchaughencyP, HaoH, et al. (2011) Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet 7: e1002329.
9. ThiebautM, ColinJ, NeilH, JacquierA, SeraphinB, et al. (2008) Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol Cell 31 : 671–682.
10. Tan-WongSM, ZauggJB, CamblongJ, XuZ, ZhangDW, et al. (2012) Gene loops enhance transcriptional directionality. Science 338 : 671–675.
11. GrzechnikP, Tan-WongSM, ProudfootNJ (2014) Terminate and make a loop: regulation of transcriptional directionality. Trends Biochem Sci 39 : 319–27.
12. JacquierA (2009) The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 10 : 833–844.
13. JensenTH, JacquierA, LibriD (2013) Dealing with pervasive transcription. Mol Cell 52 : 473–484.
14. BerrettaJ, MorillonA (2009) Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep 10 : 973–982.
15. ArigoJT, EylerDE, CarrollKL, CordenJL (2006) Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell 23 : 841–851.
16. DavisCA, AresMJr (2006) Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 103 : 3262–3267.
17. GudipatiRK, XuZ, LebretonA, SeraphinB, SteinmetzLM, et al. (2012) Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell 48 : 409–421.
18. NeilH, MalabatC, d'Aubenton-CarafaY, XuZ, SteinmetzLM, et al. (2009) Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457 : 1038–1042.
19. ThiebautM, Kisseleva-RomanovaE, RougemailleM, BoulayJ, LibriD (2006) Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol Cell 23 : 853–864.
20. WyersF, RougemailleM, BadisG, RousselleJC, DufourME, et al. (2005) Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121 : 725–737.
21. XuZ, WeiW, GagneurJ, PerocchiF, Clauder-MunsterS, et al. (2009) Bidirectional promoters generate pervasive transcription in yeast. Nature 457 : 1033–1037.
22. BuratowskiS (2005) Connections between mRNA 3′ end processing and transcription termination. Curr Opin Cell Biol 17 : 257–261.
23. HsinJP, ManleyJL (2012) The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26 : 2119–2137.
24. KimM, VasiljevaL, RandoOJ, ZhelkovskyA, MooreC, et al. (2006) Distinct pathways for snoRNA and mRNA termination. Mol Cell 24 : 723–734.
25. UrsicD, ChinchillaK, FinkelJS, CulbertsonMR (2004) Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res 32 : 2441–2452.
26. SteinmetzEJ, BrowDA (1996) Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol 16 : 6993–7003.
27. SteinmetzEJ, BrowDA (1998) Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc Natl Acad Sci U S A 95 : 6699–6704.
28. SteinmetzEJ, ConradNK, BrowDA, CordenJL (2001) RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413 : 327–331.
29. ConradNK, WilsonSM, SteinmetzEJ, PatturajanM, BrowDA, et al. (2000) A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154 : 557–571.
30. SteinmetzEJ, NgSB, ClouteJP, BrowDA (2006) cis - and trans-Acting determinants of transcription termination by yeast RNA polymerase II. Mol Cell Biol 26 : 2688–2696.
31. CarrollKL, GhirlandoR, AmesJM, CordenJL (2007) Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA 13 : 361–373.
32. CarrollKL, PradhanDA, GranekJA, ClarkeND, CordenJL (2004) Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol 24 : 6241–6252.
33. PorruaO, HoborF, BoulayJ, KubicekK, D'Aubenton-CarafaY, et al. (2012) In vivo SELEX reveals novel sequence and structural determinants of Nrd1-Nab3-Sen1-dependent transcription termination. EMBO J 31 : 3935–3948.
34. WlotzkaW, KudlaG, GrannemanS, TollerveyD (2011) The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J 30 : 1790–1803.
35. VasiljevaL, BuratowskiS (2006) Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell 21 : 239–248.
36. KimM, KroganNJ, VasiljevaL, RandoOJ, NedeaE, et al. (2004) The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432 : 517–522.
37. WestS, GromakN, ProudfootNJ (2004) Human 5′→3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432 : 522–525.
38. BrannanK, BentleyDL (2012) Control of Transcriptional Elongation by RNA Polymerase II: A Retrospective. Genet Res Int 2012 : 170173.
39. LoganJ, Falck-PedersenE, DarnellJEJr, ShenkT (1987) A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc Natl Acad Sci U S A 84 : 8306–8310.
40. ZhangZ, GilmourDS (2006) Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol Cell 21 : 65–74.
41. ZhangZ, KlattA, HendersonAJ, GilmourDS (2007) Transcription termination factor Pcf11 limits the processivity of Pol II on an HIV provirus to repress gene expression. Genes Dev 21 : 1609–1614.
42. ChinchillaK, Rodriguez-MolinaJB, UrsicD, FinkelJS, AnsariAZ, et al. (2012) Interactions of Sen1, Nrd1, and Nab3 with multiple phosphorylated forms of the Rpb1 C-terminal domain in Saccharomyces cerevisiae. Eukaryot Cell 11 : 417–429.
43. KimHD, ChoeJ, SeoYS (1999) The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry 38 : 14697–14710.
44. BrowDA (2011) Sen-sing RNA terminators. Mol Cell 42 : 717–718.
45. SteinmetzEJ, WarrenCL, KuehnerJN, PanbehiB, AnsariAZ, et al. (2006) Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell 24 : 735–746.
46. PorruaO, LibriD (2013) A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat Struct Mol Biol 20 : 884–891.
47. HafnerM, LandthalerM, BurgerL, KhorshidM, HausserJ, et al. (2010) PAR-CliP–a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp
48. HafnerM, LandthalerM, BurgerL, KhorshidM, HausserJ, et al. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141 : 129–141.
49. JamonnakN, CreamerTJ, DarbyMM, SchaughencyP, WheelanSJ, et al. (2011) Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA 17 : 2011–2025.
50. HarukiH, NishikawaJ, LaemmliUK (2008) The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31 : 925–932.
51. FanX, MoqtaderiZ, JinY, ZhangY, LiuXS, et al. (2010) Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3′-end formation. Proc Natl Acad Sci U S A 107 : 17945–17950.
52. BurgerK, MuhlB, KellnerM, RohrmoserM, Gruber-EberA, et al. (2013) 4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol 10.
53. ChurchmanLS, WeissmanJS (2011) Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469 : 368–373.
54. PelechanoV, ChavezS, Perez-OrtinJE (2010) A complete set of nascent transcription rates for yeast genes. PLoS One 5: e15442.
55. ZhelkovskyA, TacahashiY, NasserT, HeX, SterzerU, et al. (2006) The role of the Brr5/Ysh1 C-terminal domain and its homolog Syc1 in mRNA 3′-end processing in Saccharomyces cerevisiae. RNA 12 : 435–445.
56. BrendoliseC, RouillardJM, DufourME, LacrouteF (2002) Expression analysis of RNA14, a gene involved in mRNA 3′ end maturation in yeast: characterization of the rna14-5 mutant strain. Mol Genet Genomics 267 : 515–525.
57. MandartE (1998) Effects of mutations in the Saccharomyces cerevisiae RNA14 gene on the abundance and polyadenylation of its transcripts. Mol Gen Genet 258 : 16–25.
58. CloutierSC, MaWK, NguyenLT, TranEJ (2012) The DEAD-box RNA helicase Dbp2 connects RNA quality control with repression of aberrant transcription. J Biol Chem 287 : 26155–26166.
59. MaWK, CloutierSC, TranEJ (2013) The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly. J Mol Biol 425 : 3824–3838.
60. BartaI, IggoR (1995) Autoregulation of expression of the yeast Dbp2p ‘DEAD-box’ protein is mediated by sequences in the conserved DBP2 intron. EMBO J 14 : 3800–3808.
61. OzsolakF, KapranovP, FoissacS, KimSW, FishilevichE, et al. (2010) Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143 : 1018–1029.
62. WilkeningS, PelechanoV, JarvelinAI, TekkedilMM, AndersS, et al. (2013) An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res 41: e65.
63. MoqtaderiZ, GeisbergJV, JinY, FanX, StruhlK (2013) Species-specific factors mediate extensive heterogeneity of mRNA 3′ ends in yeasts. Proc Natl Acad Sci U S A 110 : 11073–11078.
64. VasiljevaL, KimM, MutschlerH, BuratowskiS, MeinhartA (2008) The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol 15 : 795–804.
65. SchulzD, SchwalbB, KieselA, BaejenC, TorklerP, et al. (2013) Transcriptome Surveillance by Selective Termination of Noncoding RNA Synthesis. Cell
66. RahlPB, LinCY, SeilaAC, FlynnRA, McCuineS, et al. (2010) c-Myc regulates transcriptional pause release. Cell 141 : 432–445.
67. GromakN, WestS, ProudfootNJ (2006) Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol 26 : 3986–3996.
68. HymanLE, MooreCL (1993) Termination and pausing of RNA polymerase II downstream of yeast polyadenylation sites. Mol Cell Biol 13 : 5159–5167.
69. KazerouniniaA, NgoB, MartinsonHG (2010) Poly(A) signal-dependent degradation of unprocessed nascent transcripts accompanies poly(A) signal-dependent transcriptional pausing in vitro. RNA 16 : 197–210.
70. LarsonDR, ZenklusenD, WuB, ChaoJA, SingerRH (2011) Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332 : 475–478.
71. BaileyTL, BodenM, BuskeFA, FrithM, GrantCE, et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–208.
72. BacikovaV, PasulkaJ, KubicekK, SteflR (2014) Structure and semi-sequence-specific RNA binding of Nrd1. Nucleic Acids Res 42 : 8024–38.
73. BirseCE, Minvielle-SebastiaL, LeeBA, KellerW, ProudfootNJ (1998) Coupling termination of transcription to messenger RNA maturation in yeast. Science 280 : 298–301.
74. LuoW, BentleyD (2004) A ribonucleolytic rat torpedoes RNA polymerase II. Cell 119 : 911–914.
75. CalvoO, ManleyJL (2001) Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol Cell 7 : 1013–1023.
76. NagA, NarsinhK, KazerouniniaA, MartinsonHG (2006) The conserved AAUAAA hexamer of the poly(A) signal can act alone to trigger a stable decrease in RNA polymerase II transcription velocity. RNA 12 : 1534–1544.
77. ConnellyS, ManleyJL (1988) A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev 2 : 440–452.
78. LuoW, JohnsonAW, BentleyDL (2006) The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev 20 : 954–965.
79. NagA, NarsinhK, MartinsonHG (2007) The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat Struct Mol Biol 14 : 662–669.
80. PearsonEL, MooreCL (2013) Dismantling promoter-driven RNA polymerase II transcription complexes in vitro by the termination factor Rat1. J Biol Chem 288 : 19750–19759.
81. KimM, AhnSH, KroganNJ, GreenblattJF, BuratowskiS (2004) Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23 : 354–364.
82. SchreieckA, EasterAD, EtzoldS, WiederholdK, LidschreiberM, et al. (2014) RNA polymerase II termination involves C-terminal-domain tyrosine dephosphorylation by CPF subunit Glc7. Nat Struct Mol Biol 21 : 175–179.
83. Al HusiniN, KudlaP, AnsariA (2013) A role for CF1A 3′ end processing complex in promoter-associated transcription. PLoS Genet 9: e1003722.
84. HuangY, WengX, RussuIM (2010) Structural energetics of the adenine tract from an intrinsic transcription terminator. J Mol Biol 397 : 677–688.
85. MartinFH, TinocoIJr (1980) DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res 8 : 2295–2299.
86. KireevaML, KomissarovaN, WaughDS, KashlevM (2000) The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem 275 : 6530–6536.
87. TagwerkerC, ZhangH, WangX, LarsenLS, LathropRH, et al. (2006) HB tag modules for PCR-based gene tagging and tandem affinity purification in Saccharomyces cerevisiae. Yeast 23 : 623–632.
88. MarquardtS, HazelbakerDZ, BuratowskiS (2011) Distinct RNA degradation pathways and 3′ extensions of yeast non-coding RNA species. Transcription 2 : 145–154.
89. RinesDR, ThomannD, DornJF, GoodwinP, SorgerPK (2011) Live cell imaging of yeast. Cold Spring Harb Protoc 2011
90. GentlemanRC, CareyVJ, BatesDM, BolstadB, DettlingM, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80.
91. LangmeadB (2010) Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 11: Unit 11 17.
92. CrooksGE, HonG, ChandoniaJM, BrennerSE (2004) WebLogo: a sequence logo generator. Genome Res 14 : 1188–1190.
Štítky
Genetika Reprodukční medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 10
-
Všechny články tohoto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



