-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaKeeping mtDNA in Shape between Generations
Since the unexpected discovery that mitochondria contain their own distinct DNA molecules, studies of the mitochondrial DNA (mtDNA) have yielded many surprises. In animals, transmission of the mtDNA genome is explicitly non-Mendelian, with a very high number of genome copies being inherited from the mother after a drastic bottleneck. Recent work has begun to uncover the molecular details of this unusual mode of transmission. Many surprising variations in animal mitochondrial biology are known; however, a series of recent studies have identified a core of evolutionarily conserved mechanisms relating to mtDNA inheritance, e.g., mtDNA bottlenecks during germ cell development, selection against specific mtDNA mutation types during maternal transmission, and targeted destruction of sperm mitochondria. In this review, we outline recent literature on the transmission of mtDNA in animals and highlight the implications for human health and ageing.
Published in the journal: . PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004670
Category: Review
doi: https://doi.org/10.1371/journal.pgen.1004670Summary
Since the unexpected discovery that mitochondria contain their own distinct DNA molecules, studies of the mitochondrial DNA (mtDNA) have yielded many surprises. In animals, transmission of the mtDNA genome is explicitly non-Mendelian, with a very high number of genome copies being inherited from the mother after a drastic bottleneck. Recent work has begun to uncover the molecular details of this unusual mode of transmission. Many surprising variations in animal mitochondrial biology are known; however, a series of recent studies have identified a core of evolutionarily conserved mechanisms relating to mtDNA inheritance, e.g., mtDNA bottlenecks during germ cell development, selection against specific mtDNA mutation types during maternal transmission, and targeted destruction of sperm mitochondria. In this review, we outline recent literature on the transmission of mtDNA in animals and highlight the implications for human health and ageing.
Introduction
Evidence continues to accumulate that a fusion event between an α-proteobacteria and an archaebacteria is the defining event in the evolution of the eukaryotic cell. Though it has been speculated that other endosymbiotic events may have been involved in the organellar structure of the eukaryotic cell, genomic evidence to date supports the presence of only α-proteobacterial and archaebacterial genomes at the beginning of this fusion [1]. Indeed, all eukaryotes sampled to date contain an organelle of mitochondrial origin and have genes of the mitochondrial ancestor within their nuclear genomes [2]. As with most endosymbiotic associations, the mitochondrial ancestral genome quickly began to lose genes that were no longer needed within their new host cell environment. Though we often shorthand the reassignment of mitochondrial genes as “being transferred to the nucleus”, this is only literally true for a minority of genes. An early study in yeast showed that only about 10% of the nuclear genes encoding mitochondrial proteins are clearly derived from the ancestral α-proteobacterial genome [3]. In mice and humans, only ∼35% of the gene products targeted to the mitochondria have good matches to the proteome of Rickettsia [4], whereas the remaining mitochondrial-targeted proteins mostly are derived from nuclear genes of nonmitochondrial origin that have displaced or complemented functions lost as the mitochondrial genome contracted. Other mitochondrial proteins are clearly the result of unusual horizontal gene transfer events, such as the replacement of the genes involved in mtDNA replication and transcription by T-odd bacteriophage enzymes [5].
Throughout eukaryotic life, very different evolutionary states of the mitochondria have arisen, and nearly all generalities to describe the mitochondria have been broken in various taxa [6]. All aerobic mitochondria appear to have retained a small mtDNA genome [7]–[9], encoding at least the cox1, cox3, and cytb genes as well as remnants of the rRNA genes [10], [11]. Anaerobic mitochondria differ in their requirements of an independent mtDNA molecule. There are anaerobic mitochondria and hydrogen-producing mitochondria, which have altered function of their respiratory chain, but still maintain mtDNA. However, the loss of mtDNA in hydrogenosomes or mitosomes is also well documented [2]. The path to an mtDNA-free organelle appears to have occurred independently in multiple lineages, and preliminary reports suggest that even a recently described anaerobic animal has an organelle, which visually resembles a hydrogenosome [12], [13], although detailed biochemical and genetic characterization of this organelle still remains to be done.
We will constrain this review to a discussion of mitochondrial genetics in animals, with special focus on principles for inheritance and purifying selection of mtDNA in the mammalian maternal germline. Most animal mitochondrial genomes conform to a specific genome composition, but even within animals, a surprising level of variation has recently been observed [14]. The genes encoded by the mtDNA are typically packed tightly together, with minimal noncoding DNA. One conspicuously large noncoding region goes by various names, including the control region, D-loop region, or large noncoding region. This region contains regulatory elements for transcription and replication of mtDNA [15]. The mammalian mtDNA encodes 13 protein components of the oxidative phosphorylation system (subunits of complexes I, III, IV, and V), highly reduced small and large rRNAs, and a minimal array of 22 tRNAs to decode the simplified animal mitochondrial genetic codes [16]. Occasionally, the observation of incomplete complements of tRNAs in some animal groups [17] have led to the hypothesis of mitochondrial tRNA import [18]. Furthermore, recent RNA sequencing has shown high levels of nuclear encoded tRNAs in RNA preparations from purified mitochondria [19]. However, the observation of cryptically encoded tRNAs in diverse animals groups [20]–[23] questions the true absence of a complete set of mtDNA-encoded tRNAs. Additionally, groups within the Porifera and Cnideria have lost all of the tRNA genes except one or two that are essential for maintaining their mitochondrial genetic codes [24], [25]. Presumably, after the origin of RNA import into the mitochondria in these animals, the mitochondrially-encoded tRNAs, which share their codons with imported nuclear-derived tRNAs, would have become functionally redundant and spared the evolutionary pressure to conserve their sequence. The elevated mutational load would therefore have led to loss of functional mtDNA genes encoding these redundant tRNAs. Our experience is that it is extremely challenging, if not impossible, to obtain mtDNA preparations free of nuclear DNA contamination, and it is therefore unlikely that mitochondrial RNA pools completely free of cytosolic contamination can be isolated [26]. Validation of the import hypothesis in animals will require the identification of the molecular pathway dedicated to the import of cytosolic tRNAs, as found in other eukaryotic groups that utilize cytosolic tRNAs for mitochondrial translation [27].
Mitochondrial Transmission
The prokaryotic origins of the mitochondria also influence their genetics. In animals, most cells contain many copies of mtDNA, and the transmission can be better imagined as thousands of independent genomes being segregated by stochastic mechanisms and relaxed, uncoupled replication, which is in stark contrast to the structured meiosis and mitosis of eukaryotic nuclear genomes [28]. Despite the high copy number of mtDNA, it was noted very early that mtDNA of vertebrates can switch from one type to another in a very short period of time, even in a single generation [29]–[31]. To explain this very rapid shift, the concept of the mitochondrial bottleneck was developed (Figure 1). In the development of the germline, a large population of mtDNAs are subsampled to a relatively small number. This subselected population may differ markedly from the source population, allowing rare alleles to, by chance, suddenly come to dominate the mtDNA pool. With a sufficiently small sample, as observed in Holstein cows, single-generation shifts of mtDNA genotype are possible [30], [31]. Such a bottleneck phenomenon has since been detected widely in vertebrates [32]–[34] and even in Drosophila [35]–[37], though the size of this subsampling bottleneck appears to be an order of magnitude larger in flies than in vertebrates.
Fig. 1. Schematic representation of the mitochondrial bottleneck. 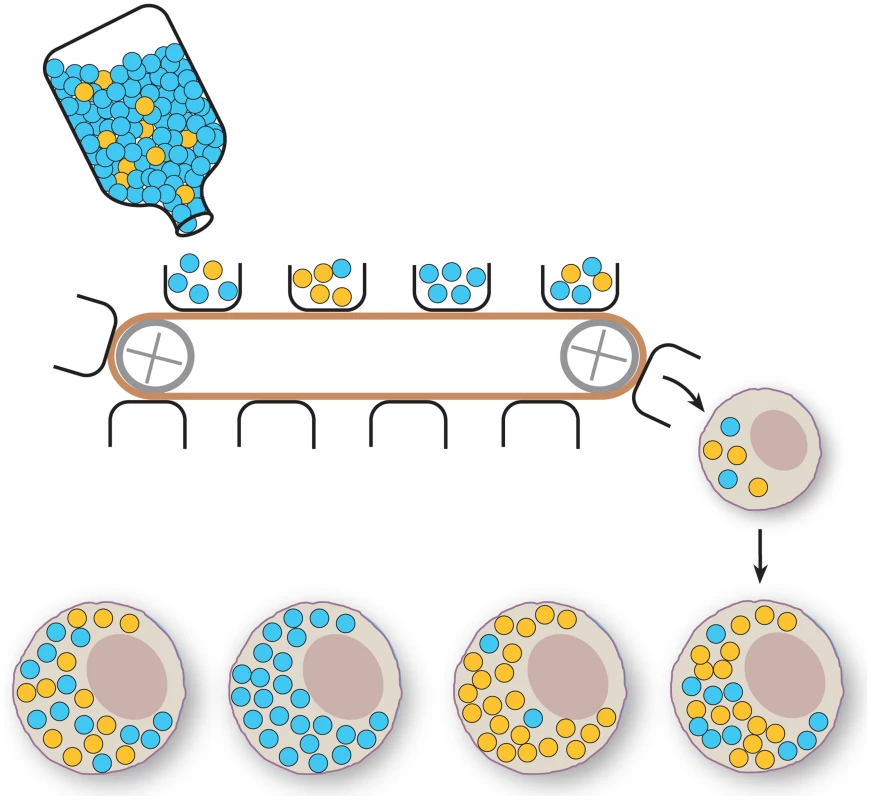
A large number of mtDNA molecules are present in the maternal mtDNA pool (bottle). The figure depicts two genotypes (blue and yellow circles). The generation of an oocyte involves subsampling of just a few mtDNA molecules from the maternal mtDNA pool (buckets on conveyor belt), which will be transferred to the developing oocyte and extensively replicated. The effect of this poorly understood bottleneck mechanism is that the proportion of the two genotypes can vary widely between oocytes. Usually, animal mtDNAs are inherited solely through the female oocyte. Upon fertilization, molecular mechanisms that identify and eliminate the sperm mitochondria have been described [38]. A similar phenomenon has been recently described in Drosophila [39] and Caenorhabditis elegans [40], [41], implying that it may be an ancestral feature to most animals. This targeted destruction of paternal mtDNA has clearly been lost in some species such as the Mytilis mussels, in which double uniparental inheritance of mtDNA occurs [42]. Like most molecular mechanisms, these systems can be circumvented. Interspecific crosses of mice lead to transmission of low levels of paternal mtDNA in early embryos [43]. However, the paternally transmitted mtDNA in these interspecific crosses is overrun by the maternal mtDNA during development to create offspring that only contain maternally transmitted mtDNA [43], [44]. There are some reports of recombination between the divergent maternal and paternal mtDNAs [45], [46], and recombined molecules appear to be present in the double uniparental mtDNA inheritance mussels as well, but at much lower rates than those resulting from homologous recombination during nuclear meiosis [47]. However, recombination appears to be conspicuously absent in many vertebrate groups. Mice that segregate neutral alleles appear to lack recombination in PCR-free assays [48], even when maintained in a constant heteroplasmic state for more than 50 generations [49]. However, mice heteroplasmic for one deleterious mtDNA type and a second mtDNA type from a distantly related species appear to have small patches of sequence consistent with recombination events [48]. It should be mentioned that the presence of rare recombinant mtDNA molecules does not necessarily necessitate the existence of a molecular machinery for homologous recombination. Instead, such recombinant molecules may be the product of copy choice recombination whereby the mitochondrial DNA polymerase starts replication on one mtDNA template and shifts to another template to complete replication. It remains possible that in deleterious backgrounds, these types of rare events may lead to a fitness advantage for the organism or organelle, leading to the fixation of these rare events, as seen in the interspecies hybrids [45], [46].
Molecular Basis of the mtDNA Bottleneck
In mice, the mitochondrial life cycle starts with ∼2×106 copies of mtDNA in a single oocyte (Figure 2). After fertilization, designation of the primordial germ cells (PGCs) is delayed until after implantation of the embryo [50], [51]. During this stage, there is no gain in the amount of mtDNA in the embryo as a whole, effectively diluting the per-cell levels of mtDNA with each cell division [51]–[53]. Finally, mtDNA replication is re-initiated sometime between embryonic day (E)6 and E7.5, and the mtDNA copy number per cell begins to increase throughout embryogenesis [34], [51], [52]. There is further increase of mtDNA copy number during oocyte development in the postnatal period to reach the ∼2×106 copies of mtDNA that are present in mature oocytes and passed on to the next generation [52], [54]. Though the oocyte development in Drosophila displays marked differences, a similar period without mtDNA replication has been observed, which could be expected to lead to an mtDNA bottleneck, albeit of a far less drastic nature than in mammals [36], [37].
Fig. 2. The life cycle of mtDNA within the female germline of mice. 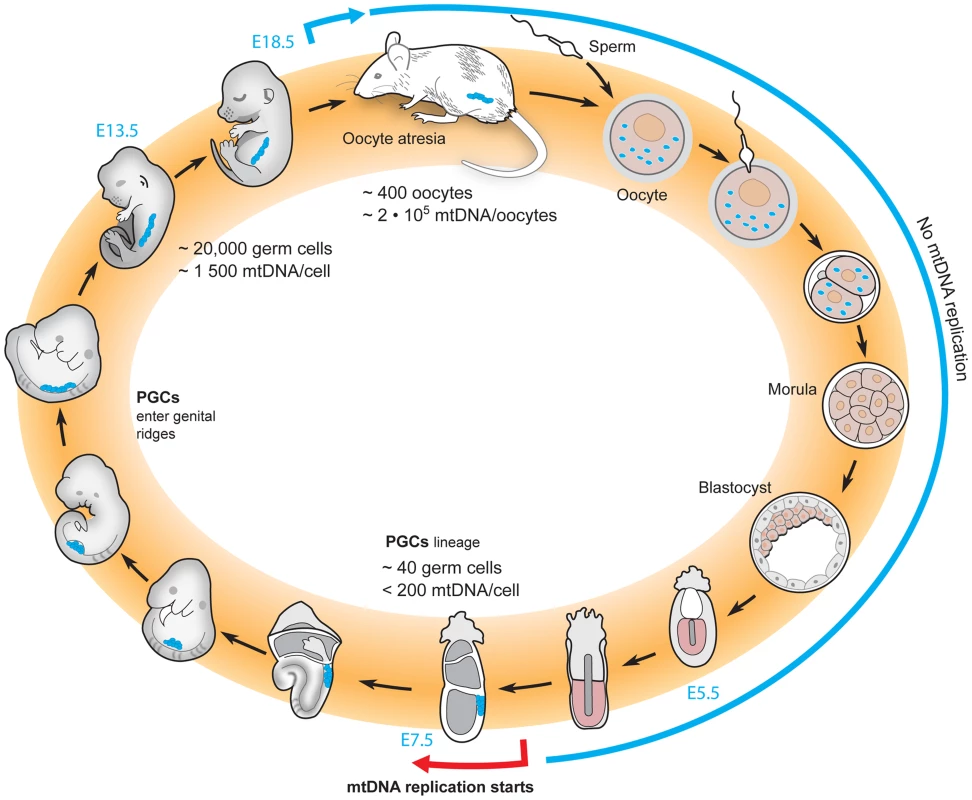
The total amount of mtDNA remains unchanged from the oocyte to the blastocyst stage, resulting in a dilution of the number of mtDNA molecules per cell with each cell division. Re-initiation of mtDNA replication does not occur until the postimplantation stage. At approximately E7.5, a physical bottleneck with a very low number of mtDNA molecules per cell has been reported, and thereafter replication reinitiates to increase the mtDNA content of the embryo. During the late stages of embryonic life and early postnatal life, oocytes are lost due to atresia. There is an ongoing debate regarding when the mtDNA bottleneck occurs in the life cycle of mammalian mtDNA. Three different groups have used qRT-PCR based methods to assess the mtDNA copy number in the developing mouse germline, but have failed to reach a consensus [34], [53]–[57]. Two groups have concluded that a low number of mtDNAs is present at the early stages of the primordial germ cells [34], [54], and this low number approximates the bottleneck size estimated by measuring the variance of two neutral alleles segregating in mice [32]. One group argues that this event explains the mtDNA bottleneck [34], [57]. However, another group argues that the number of mtDNAs in early primordial germ cells is an order of magnitude higher than this low estimate and have reported that the bottleneck instead occurs via a subselection of mtDNAs that are preferentially replicated to fill the developing oocyte [53], [55]. Yet another group claims that the bottleneck occurs after birth, by a subpopulation of mtDNAs that supply the final replicative burst that populates the mature oocytes [54], [56]. Obviously, clarifying this point continues to be of great interest to the field. So far, only correlative data have addressed this important question and experiments will be needed to determine how changes in mtDNA copy number in the maternal germline influence the size of the bottleneck.
Mutations in mtDNA
The most common types of mutations observed in the mtDNA are single nucleotide substitutions, single base insertions, single base deletions [58], and large deletions that result in smaller, circular mtDNA molecules [59], [60]. Other mutation types, such as multimers of sequences associated with replication elements [61], have been observed in the aging human brain [62], while deleted linear mtDNA molecules have only been seen in disease states or severely afflicted mouse models [63], [64]. The mtDNA copies within a cell may all be identical (homoplasmy) or contain two or more variants (heteroplasmy). It is common to refer to individual organisms as homoplasmic for an mtDNA type based on Sanger sequencing of amplified mtDNA pools. However, it is becoming clear that most individuals carry appreciable numbers of low-level mtDNA variants in their tissues [65]–[67]. One of us has recently reviewed the role of mtDNA mutations in ageing [68], and some recent additional insights will be further discussed here.
According to the classic mitochondrial theory of ageing, reactive oxygen species from the OXPHOS (Oxidative Phosphorylation) system will be mutagenic and lead to the accumulation of mtDNA mutations over time [69], [70]. However, recent studies have begun to cast doubt on this model and argue that normally occurring replication errors may be a more important source of mutations than unrepaired damage. Oxidative damage to DNA is predicted to create G>T transversions and C>T transitions. Unfortunately, only the G>T transversions are diagnostic when sequencing mtDNA because C>T transitions also appears to be a common replication error. It has been known for some time that there is a preponderance of transition mutations in human cell cultures, arguing that the observed mutational spectra are consistent with errors occurring during normal replication by mitochondrial DNA polymerase [71], [72]. The role of the oxidative adduct 8-oxo-deoxyguanosine in mtDNA mutagenesis was recently experimentally assessed in flies that were engineered to have a down-regulation of two base excision repair enzymes (OGG1/MUTYh) and mitochondrial superoxide dismutase [63]. This combined intervention is predicted to drastically increase 8-oxo-deoxyguanosine adducts and the formation of G>T transversion mutations, but, quite surprisingly, there was no increase in mtDNA mutations in these flies [73]. In fact, the observed mtDNA mutation pattern closely resembled the mitochondrial DNA polymerase error-based mutation pattern observed in wildtype flies [58]. Also, studies in ageing mice [26] and humans [62], [74] have failed to detect a transversion mutation bias as would be expected if elevated levels of oxidative damage would be the main source of mtDNA mutations.
A mutation will initially affect a single copy of mtDNA and each mutational event will therefore be exceptionally rare upon its generation, as most mammalian cells contain thousands of copies of mtDNA. However, the lack of coordination between the cell cycle and replication of mtDNA allows mitotic segregation of the mutated mtDNA copy, which thereby can undergo clonal expansion and over time overrun the mtDNA pool to cause physiological consequences. In fact, age-associated somatic mtDNA mutations, both point mutations and large deletions, are typically clonally expanded to create a mosaic pattern of respiratory chain deficient cells in ageing humans [75]–[77]. Characterization of point mutations in ageing humans have confirmed their apparently stochastic nature as sequencing shows no signature of purifying selection [78], [79]. A purely stochastic mutation profile is also observed in the coding regions of mtDNA isolated from the mtDNA mutator mouse, a substitution-prone model where the mitochondrial DNA polymerase has had its proofreading function disrupted [63], [80], [81]. Also, studies of patients with genetic disorders due to mtDNA mutations do not show a targeted reduction in the disease-causing deleterious alleles over time. These observations challenge the assertion that mitochondrial quality control mechanisms are able to target specific deleterious mtDNA molecules in somatic tissues.
However, there appears to be selection against somatic mtDNA mutations that affect sequences needed for mtDNA replication. Recent work has identified specific mutations, most commonly observed near the replication elements of the control region, which appear to undergo tissue-specific propagation in humans with age [82]. Similar mtDNA variants leading to replicative advantage have been observed in invertebrates [83], [84]. Negative selection against mutations in the origin of light strand replication, which controls initiation of lagging strand replication, has been observed in somatic tissues of mtDNA mutator mice [85]. Negative selection may also explain the decreased mtDNA mutation rate in the control region [63], [80].
If mtDNA mutations are present in the germline and pass through the oocyte, they rapidly expand to appreciable levels and end up in all tissues of the resulting organism [86]. Using the mtDNA mutator mice, we have recently shown that these germline–transmitted mtDNA mutations can severely affect the health of the resulting organism. By breeding successive generations of females that are heterozygous for the mtDNA mutator allele, we observed reduced litter sizes, impaired health, and shortened lifespan of the offspring. These effects in the offspring can be immediately reversed by the introduction of nonmutated mtDNA into the heterozygous mtDNA mutator females [86]. We have also been able to generate mice hemizygous for the mtDNA mutator allele that are functionally equivalent to classical mtDNA mutator mice, but lack maternally transmitted mutations from their mothers [86]. These mice are healthier than the standard mtDNA mutator mice and live, on average, ten weeks longer, thus showing that low levels of mtDNA mutations transmitted through the germline worsen the effects of somatic mutagenesis of mtDNA [86].
Germline Purifying Selection
The high mutation rate of mtDNA, relative to the nuclear rate [58], [71], [72], and the absent or infrequent recombination of mtDNA are predicted to strongly increase the mutational burden of mtDNA [87]. The mitochondrial threshold effect allows disease alleles to accumulate without causing any disease phenotypes, but once the relative level of mutated mtDNA exceeds a critical threshold, devastating disease will occur [88]. Mothers of patients with mtDNA disease are often healthy despite having substantial levels of the disease-causing allele. However, the mtDNA bottleneck phenomenon allows the levels of a disease allele to be drastically elevated in a single generation, thus leading to mitochondrial disease in the offspring [89].
The famous geneticist Hermann Muller predicted that asexually transmitted genomes, such as mtDNA, would accumulate deleterious mutations over time, a phenomenon referred to as Muller's ratchet [90], [91]. Work during recent years has revealed strong mechanisms for purifying selection of mtDNA in the maternal germline that counteract a mutational meltdown. In mice, deleterious alleles of mtDNA that lead to amino acid substitutions are strongly selected against in the female germline. In experimental work, we introduced a random set of mtDNA mutations from mtDNA mutator mice into lineages of mice with a wildtype nuclear genome. Sequencing of mtDNA showed evidence of strong purifying selection because the levels of mtDNA mutations affecting the amino acid, altering first or second codon positions of protein coding genes, were much lower than the levels of mutations affecting the normally synonymous third codon position or tRNA genes [92]. The speed at which this selection occurred was very fast and was observable two generations after the introduction of mtDNA mutations from the mtDNA mutator mouse. These alleles are thus selected against when they are still very rare in the mtDNA pool and therefore can have no gross phenotypic effects on the organism itself. Independent support for purifying selection against mutations causing deleterious amino acid substitutions has been reported in mice with a deletion in mt-nd6, which was selectively eliminated in favor of a compensatory insertion mutation that corrected the reading frame [93].
The situation with mutations in protein coding genes is in stark contrast to mutations in tRNA genes of mtDNA. We studied a single-base deletion in the tRNAMet gene, where mice could only be bred to harbor a maximum of 90% mutated mtDNA [84]. This mutation could accumulate to very high levels (∼100%) in the germline, but was selected against postfertilization in the embryonic period [94]. These experimental results showing quite different behaviors of mtDNA mutations affecting tRNA or protein coding genes likely provide an explanation for why tRNA mutations are frequent causes of mitochondrial disease in humans, whereas mutations in protein coding genes are rare. This discrepancy is especially striking when one takes into account that tRNA genes occupy 9% and protein coding genes 68% of the human mtDNA sequence.
Two recent studies in Drosophila have illustrated the deep conservation of mechanisms in the germline selecting against mtDNA mutations causing amino acid substitutions and have also added some interesting insights into how purifying selection can operate. These studies utilized a temperature sensitive mutation in the mt-co1 gene, which has no phenotype at permissive temperatures, but is lethal when the flies are raised at 29°C [36], [37]. This temperature sensitivity allowed the generation of lines with very high relative levels of the mutant allele at permissive temperatures. By switching the line to high temperature, the allele was transmitted less efficiently and there was a selective lowering of its relative levels over a few generations. However, the allele was maintained at low levels for a very large number of generations, consistent with the predicted less drastic mtDNA bottleneck effect in the fruit fly [35]–[37].
The temperature sensitive nature of the mutation also allowed determination of when the allele was selected against in the germline. The selective event occurred with germline development in the flies and coincided with the re-initiation of mtDNA replication [37]. High levels of the deleterious allele led to the inability of the mitochondria to re-initiate mtDNA replication in the germline. These results led to the hypothesis that a decrease in the mitochondrial membrane potential will decrease import of nuclear gene products necessary for mtDNA replication. Such a mechanism could hamper the transmission of mtDNA mutations causing amino acid substitutions during the rapid mitochondrial biogenesis stage of oogenesis.
Now that the evolutionary conservation of this germline selection has been established, a more mechanistic study of the molecular or cell biological basis of the purifying selection is in order. A comparative genomics approach will be very useful in unraveling the mechanistic details of this phenomenon. Using the powerful genetics of Drosophila, it will be feasible to screen for the molecular pathways that could disrupt (or perhaps enhance) the selection against various deleterious alleles. Suitable mutant or RNAi stocks are available in Drosophila, including inducible mutants, such as the temperature sensitive mutants [36], [37]. For instance, the selection in Drosophila has already been determined to occur independently of the PARKIN pathway, as flies with PARK RNAi knockdown, or a null mutant of the PARKIN gene still showed germline selection against mutated mtDNA [36]. Results obtained in flies should then allow us to test for conserved mechanisms in mammals, such as the mouse, or eventually in humans.
Concluding Remarks
The mtDNA is asexually transmitted in metazoans and should, in principle, be sensitive to the Muller ratchet phenomenon with irreversible mtDNA mutation accumulation between generations. However, studies in mammals have clarified that there are several mechanisms that can completely prevent or decrease the transmission of deleterious mtDNA mutations from one generation to the next: (I) The bottleneck phenomenon allows a mother with low levels of a pathogenic mtDNA mutation to produce offspring lacking mutated mtDNA, (II) there is a strong germline purifying selection against mtDNA mutations causing amino acid substitutions [82], (III) there is a strong purifying selection during embryogenesis against high levels of mtDNA mutations impairing tRNA function [84], and (IV) females with mtDNA mutations in the germline have a dose-dependent reduced capacity to reproduce [77]. Although recent progress has dramatically increased our knowledge of mechanisms that may reset or decrease the mtDNA mutation rate between generations, the involved molecular components are not at all understood. The challenge for the future will, therefore, be to understand how these mechanisms operate at a molecular level and how they have evolved.
Zdroje
1. RochetteNC, Brochier-ArmanetC, GouyM (2014) Phylogenomic Test of the Hypotheses for the Evolutionary Origin of Eukaryotes. Mol Biol Evol 4 : 832–845.
2. MullerM, MentelM, van HellemondJJ, HenzeK, WoehleC, et al. (2012) Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev 76 : 444–495.
3. KarlbergO, CanbackB, KurlandCG, AnderssonSG (2000) The dual origin of the yeast mitochondrial proteome. Yeast 17 : 170–187.
4. PagliariniDJ, CalvoSE, ChangB, ShethSA, VafaiSB, et al. (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134 : 112–123.
5. MeisingerC, SickmannA, PfannerN (2008) The mitochondrial proteome: from inventory to function. Cell 134 : 22–24.
6. BurgerG, GrayMW, Franz LangB (2003) Mitochondrial genomes: anything goes. Trends Genet 19 : 709–716.
7. NassMM, NassS (1963) Intramitochondrial Fibers with DNA Characteristics. I. Fixation and Electron Staining Reactions. J Cell Biol 19 : 593–611.
8. NassS, NassMM (1964) Intramitochondrial Fibers with Deoxyribonucleic Acid Characteristics: Observations of Ehrlich Ascites Tumor Cells. J Natl Cancer Inst 33 : 777–798.
9. NassMM, NassS, AfzeliusBA (1965) The General Occurence of Mitochondrial DNA. Exp Cell Res 37 : 516–539.
10. HikosakaK, KitaK, TanabeK (2013) Diversity of mitochondrial genome structure in the phylum Apicomplexa. Mol Biochem Parasitol 188 : 26–33.
11. SlamovitsCH, SaldarriagaJF, LarocqueA, KeelingPJ (2007) The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J Mol Biol 372 : 356–368.
12. LevinLA (2010) Anaerobic metazoans: no longer an oxymoron. BMC Biol 8 : 31.
13. DanovaroR, Dell'AnnoA, PuscedduA, GambiC, HeinerI, et al. (2010) The first metazoa living in permanently anoxic conditions. BMC Biol 8 : 30.
14. BerntM, BrabandA, SchierwaterB, StadlerPF (2013) Genetic aspects of mitochondrial genome evolution. Mol Phylogenet Evol 69 : 328–338.
15. FalkenbergM, LarssonNG, GustafssonCM (2007) DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem 76 : 679–699.
16. BooreJL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27 : 1767–1780.
17. YangJS, YangWJ (2008) The complete mitochondrial genome sequence of the hydrothermal vent galatheid crab Shinkaia crosnieri (Crustacea: Decapoda: Anomura): a novel arrangement and incomplete tRNA suite. BMC Genomics 9 : 257.
18. DornerM, AltmannM, PääboS, MorlM (2001) Evidence for import of a lysyl-tRNA into marsupial mitochondria. Mol Biol Cell 12 : 2688–2698.
19. MercerTR, NephS, DingerME, CrawfordJ, SmithMA, et al. (2011) The human mitochondrial transcriptome. Cell 146 : 645–658.
20. BeckenbachAT, JoyJB (2009) Evolution of the Mitochondrial Genomes of Gall Midges (Diptera: Cecidomyiidae): Rearrangement and Severe Truncation of tRNA Genes. Genome Biol Evol 1 : 278–287.
21. MastaSE, BooreJL (2008) Parallel evolution of truncated transfer RNA genes in arachnid mitochondrial genomes. Mol Biol Evol 25 : 949–959.
22. MastaSE, BooreJL (2004) The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol Biol Evol 21 : 893–902.
23. SegoviaR, PettW, TrewickS, LavrovDV (2011) Extensive and evolutionarily persistent mitochondrial tRNA editing in Velvet Worms (phylum Onychophora). Mol Biol Evol 28 : 2873–2881.
24. ShaoZ, GrafS, ChagaOY, LavrovDV (2006) Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): A linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene 381 : 92–101.
25. WangX, LavrovDV (2008) Seventeen new complete mtDNA sequences reveal extensive mitochondrial genome evolution within the Demospongiae. PLoS ONE 3: e2723.
26. AmeurA, StewartJB, FreyerC, HagströmE, IngmanM, et al. (2011) Ultra-Deep Sequencing of Mouse Mitochondrial DNA: Mutational Patterns and Their Origins. PLoS Genet 7: e1002028.
27. SeidmanD, JohsnonD, GerbasiV, GoldenD, OrlandoR, et al. (2012) A Mitochondrial membrane complex that contains proteins necessary for tRNA import in Trypanosoma brucei. J Biol Chem 12 : 8892–8903.
28. ChinneryPF, SamuelsDC (1999) Relaxed replication of mtDNA: A model with implications for the expression of disease. Am J Hum Genet 64 : 1158–1165.
29. AshleyMV, LaipisPJ, HauswirthWW (1989) Rapid segregation of heteroplasmic bovine mitochondria. Nucleic Acids Res 17 : 7325–7331.
30. OlivoPD, Van de WalleMJ, LaipisPJ, HauswirthWW (1983) Nucleotide sequence evidence for rapid genotypic shifts in the bovine mitochondrial DNA D-loop. Nature 306 : 400–402.
31. HauswirthWW, LaipisPJ (1982) Mitochondrial DNA polymorphism in a maternal lineage of Holstein cows. Proc Natl Acad Sci U S A 79 : 4686–4690.
32. JenuthJP, PetersonAC, FuK, ShoubridgeEA (1996) Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet 14 : 146–151.
33. WolffJN, WhiteDJ, WoodhamsM, WhiteHE, GemmellNJ (2011) The strength and timing of the mitochondrial bottleneck in salmon suggests a conserved mechanism in vertebrates. PLoS ONE 6: e20522.
34. CreeLM, SamuelsDC, de Sousa LopesSC, RajasimhaHK, WonnapinijP, et al. (2008) A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet 40 : 249–254.
35. zPetitN, TourailleS, DebiseR, MorelF, RenouxM, et al. (1998) Developmental changes in heteroplasmy level and mitochondrial gene expression in a Drosophila subobscura mitochondrial deletion mutant. Curr Genet 33 : 330–339.
36. MaH, XuH, O'FarrellPH (2014) Transmission of mitochondrial mutations and action of purifying selection in Drosophila melanogaster. Nat Genet 46 : 393–397.
37. HillJH, ChenZ, XuH (2014) Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat Genet 46 : 389–392.
38. ShitaraH, KanedaH, SatoA, InoueK, OguraA, et al. (2000) Selective and continuous elimination of mitochondria microinjected into mouse eggs from spermatids, but not from liver cells, occurs throughout embryogenesis. Genetics 156 : 1277–1284.
39. DeLucaSZ, O'FarrellPH (2012) Barriers to male transmission of mitochondrial DNA in sperm development. Dev Cell 22 : 660–668.
40. SatoM, SatoK (2011) Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 334 : 1141–1144.
41. Al RawiS, Louvet-ValleeS, DjeddiA, SachseM, CulettoE, et al. (2011) Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 334 : 1144–1147.
42. StewartDT, SaavedraC, StanwoodRR, BallAO, ZourosE (1995) Male and female mitochondrial DNA lineages in the blue mussel (Mytilus edulis) species group. Mol Biol Evol 12 : 735–747.
43. GyllenstenU, WhartonD, JosefssonA, WilsonAC (1991) Paternal inheritance of mitochondrial DNA in mice. Nature 352 : 255–257.
44. GyllenstenU, WhartonD, WilsonAC (1985) Maternal inheritance of mitochondrial DNA during backcrossing of two species of mice. J Hered 76 : 321–324.
45. UjvariB, DowtonM, MadsenT (2007) Mitochondrial DNA recombination in a free-ranging Australian lizard. Biol Lett 3 : 189–192.
46. GuoX, LiuS, LiuY (2006) Evidence for recombination of mitochondrial DNA in triploid crucian carp. Genetics 172 : 1745–1749.
47. LadoukakisED, TheologidisI, RodakisGC, ZourosE (2011) Homologous recombination between highly diverged mitochondrial sequences: examples from maternally and paternally transmitted genomes. Mol Biol Evol 28 : 1847–1859.
48. SatoA, NakadaK, AkimotoM, IshikawaK, OnoT, et al. (2005) Rare creation of recombinant mtDNA haplotypes in mammalian tissues. Proc Natl Acad Sci U S A 102 : 6057–6062.
49. HagstromE, FreyerC, BattersbyBJ, StewartJB, LarssonNG (2014) No recombination of mtDNA after heteroplasmy for 50 generations in the mouse maternal germline. Nucleic Acids Res 42 : 1111–1116.
50. PesceM, ScholerHR (2001) Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells 19 : 271–278.
51. PikoL, TaylorKD (1987) Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev Biol 123 : 364–374.
52. WaiT, AoA, ZhangX, CyrD, DufortD, et al. (2010) The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod 83 : 52–62.
53. CaoL, ShitaraH, HoriiT, NagaoY, ImaiH, et al. (2007) The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat Genet 39 : 386–390.
54. WaiT, TeoliD, ShoubridgeEA (2008) The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet 40 : 1484–1488.
55. CaoL, ShitaraH, SugimotoM, HayashiJ, AbeK, et al. (2009) New evidence confirms that the mitochondrial bottleneck is generated without reduction of mitochondrial DNA content in early primordial germ cells of mice. PLoS Genet 5: e1000756.
56. WaiT, ShoubridgeEA (2010) Reply to “Reassessing evidence for a postnatal mitochondrial genetic bottleneck”. Nat Genet 42 : 472–473.
57. SamuelsDC, WonnapinijP, CreeLM, ChinneryPF (2010) Reassessing evidence for a postnatal mitochondrial genetic bottleneck. Nat Genet 42 : 471–472 author reply 472–473.
58. Haag-LiautardC, CoffeyN, HouleD, LynchM, CharlesworthB, et al. (2008) Direct Estimation of the Mitochondrial DNA Mutation Rate in Drosophila melanogaster. PLoS Biol 6: e204.
59. PikoL, HoughamAJ, BulpittKJ (1988) Studies of sequence heterogeneity of mitochondrial DNA from rat and mouse tissues: evidence for an increased frequency of deletions/additions with aging. Mech Ageing Dev 43 : 279–293.
60. KatayamaM, TanakaM, YamamotoH, OhbayashiT, NimuraY, et al. (1991) Deleted mitochondrial DNA in the skeletal muscle of aged individuals. Biochem Int 25 : 47–56.
61. WilliamsSL, HuangJ, EdwardsYJ, UlloaRH, DillonLM, et al. (2010) The mtDNA mutation spectrum of the progeroid Polg mutator mouse includes abundant control region multimers. Cell Metab 12 : 675–682.
62. WilliamsSL, MashDC, ZuchnerS, MoraesCT (2013) Somatic mtDNA mutation spectra in the aging human putamen. PLoS Genet 9: e1003990.
63. TrifunovicA, WredenbergA, FalkenbergM, SpelbrinkJN, RovioAT, et al. (2004) Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429 : 417–423.
64. KornblumC, NichollsTJ, HaackTB, ScholerS, PeevaV, et al. (2013) Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet 45 : 214–219.
65. PayneBA, WilsonIJ, Yu-Wai-ManP, CoxheadJ, DeehanD, et al. (2013) Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet 22 : 384–390.
66. LiM, SchonbergA, SchaeferM, SchroederR, NasidzeI, et al. (2010) Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet 87 : 237–249.
67. HeY, WuJ, DressmanDC, Iacobuzio-DonahueC, MarkowitzSD, et al. (2010) Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 464 : 610–614.
68. LarssonNG (2010) Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem 79 : 683–706.
69. HarmanD (1972) The biologic clock: the mitochondria? J Am Geriatr Soc 20 : 145–147.
70. HarmanD (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol 11 : 298–300.
71. ZhengW, KhrapkoK, CollerHA, ThillyWG, CopelandWC (2006) Origins of human mitochondrial point mutations as DNA polymerase gamma-mediated errors. Mutat Res 599 : 11–20.
72. KhrapkoK, CollerHA, AndrePC, LiXC, HanekampJS, et al. (1997) Mitochondrial mutational spectra in human cells and tissues. Proc Natl Acad Sci U S A 94 : 13798–13803.
73. ItsaraLS, KennedySR, FoxEJ, YuS, HewittJJ, et al. (2014) Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PLoS Genet 10: e1003974.
74. KennedySR, SalkJJ, SchmittMW, LoebLA (2013) Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet 9: e1003794.
75. FayetG, JanssonM, SternbergD, MoslemiAR, BlondyP, et al. (2002) Ageing muscle: clonal expansions of mitochondrial DNA point mutations and deletions cause focal impairment of mitochondrial function. Neuromuscul Disord 12 : 484–493.
76. CottrellDA, IncePG, WardellTM, TurnbullDM, JohnsonMA (2001) Accelerated ageing changes in the choroid plexus of a case with multiple mitochondrial DNA deletions. Neuropathol Appl Neurobiol 27 : 206–214.
77. TaylorRW, BarronMJ, BorthwickGM, GospelA, ChinneryPF, et al. (2003) Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest 112 : 1351–1360.
78. GreavesLC, ElsonJL, NooteboomM, GradyJP, TaylorGA, et al. (2012) Comparison of mitochondrial mutation spectra in ageing human colonic epithelium and disease: absence of evidence for purifying selection in somatic mitochondrial DNA point mutations. PLoS Genet 8: e1003082.
79. PereiraL, SoaresP, MaximoV, SamuelsDC (2012) Somatic mitochondrial DNA mutations in cancer escape purifying selection and high pathogenicity mutations lead to the oncocytic phenotype: pathogenicity analysis of reported somatic mtDNA mutations in tumors. BMC Cancer 12 : 53.
80. TrifunovicA, HanssonA, WredenbergA, RovioAT, DufourE, et al. (2005) Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc Natl Acad Sci U S A 102 : 17993–17998.
81. KujothGC, HionaA, PughTD, SomeyaS, PanzerK, et al. (2005) Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309 : 481–484.
82. SamuelsDC, LiC, LiB, SongZ, TorstensonE, et al. (2013) Recurrent tissue-specific mtDNA mutations are common in humans. PLoS Genet 9: e1003929.
83. RandDM (2011) Population genetics of the cytoplasm and the units of selection on mitochondrial DNA in Drosophila melanogaster. Genetica 139 : 685–697.
84. LiauWS, Gonzalez-SerricchioAS, DeshommesC, ChinK, LaMunyonCW (2007) A persistent mitochondrial deletion reduces fitness and sperm performance in heteroplasmic populations of C. elegans. BMC Genet 8 : 8.
85. WanrooijS, Miralles FusteJ, StewartJB, WanrooijPH, SamuelssonT, et al. (2012) In vivo mutagenesis reveals that OriL is essential for mitochondrial DNA replication. EMBO Rep 13 : 1130–1137.
86. RossJM, StewartJB, HagstromE, BreneS, MourierA, et al. (2013) Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature 501 : 412–415.
87. LynchM (1996) Mutation accumulation in transfer RNAs: molecular evidence for Muller's ratchet in mitochondrial genomes. Mol Biol Evol 13 : 209–220.
88. TaylorRW, TurnbullDM (2005) Mitochondrial DNA mutations in human disease. Nat Rev Genet 6 : 389–402.
89. LarssonNG, TuliniusMH, HolmeE, OldforsA, AndersenO, et al. (1992) Segregation and manifestations of the mtDNA tRNA(Lys) A→G(8344) mutation of myoclonus epilepsy and ragged-red fibers (MERRF) syndrome. Am J Hum Genet 51 : 1201–1212.
90. MullerHJ (1964) The Relation of Recombination to Mutational Advance. Mutat Res 1 : 2–9.
91. FelsensteinJ (1974) The evolutionary advantage of recombination. Genetics 78 : 737–756.
92. StewartJB, FreyerC, ElsonJL, WredenbergA, CansuZ, et al. (2008) Strong Purifying Selection in Transmission of Mammalian Mitochondrial DNA. PLoS Biol 6: e10.
93. FanW, WaymireKG, NarulaN, LiP, RocherC, et al. (2008) A Mouse Model of Mitochondrial Disease Reveals Germline Selection Against Severe mtDNA Mutations. Science 319 : 958–962.
94. FreyerC, CreeLM, MourierA, StewartJB, KoolmeisterC, et al. (2012) Variation in germline mtDNA heteroplasmy is determined prenatally but modified during subsequent transmission. Nat Genet 44 : 1282–1285.
Štítky
Genetika Reprodukční medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 10
-
Všechny články tohoto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



