-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAt Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
The ends of chromosomes are protected by specialized structures called telomeres, which prevent their recognition as DNA breaks and enable recruitment of telomerase, the reverse transcriptase that maintains telomere length by replacing terminal TG-repeat sequences lost during successive rounds of DNA replication. Chromosomal DNA is replicated from initiation sites called origins, which are activated in a reproducible temporal sequence. Replication origins close to telomeres are subject to specialized temporal control that contributes to telomere stabilization: origins close to normal-length telomeres initiate replication late, while those close to shortened telomeres initiate early. Here we uncover the control mechanism that links telomere length with replication timing. Rif1, one of the components of telomeric chromatin, directs late replication of normal telomeres by delaying the activation of nearby origins. Our experiments show that a kinase called Tel1, which is recruited to shortened telomeres, neutralizes the origin-delaying activity of Rif1. We also find that Tel1 phosphorylates Rif1 at short telomeres, although our investigation shows this phosphorylation is not the sole mechanism through which Tel1 prevents Rif1-mediated replication delay. Since correct telomere replication timing control is important for telomerase-mediated length maintenance, this discovery represents an important step towards understanding the molecular mechanisms that ensure proper long-term stabilization of chromosome ends, as well as the controls over the DNA replication temporal program.
Published in the journal: . PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004691
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004691Summary
The ends of chromosomes are protected by specialized structures called telomeres, which prevent their recognition as DNA breaks and enable recruitment of telomerase, the reverse transcriptase that maintains telomere length by replacing terminal TG-repeat sequences lost during successive rounds of DNA replication. Chromosomal DNA is replicated from initiation sites called origins, which are activated in a reproducible temporal sequence. Replication origins close to telomeres are subject to specialized temporal control that contributes to telomere stabilization: origins close to normal-length telomeres initiate replication late, while those close to shortened telomeres initiate early. Here we uncover the control mechanism that links telomere length with replication timing. Rif1, one of the components of telomeric chromatin, directs late replication of normal telomeres by delaying the activation of nearby origins. Our experiments show that a kinase called Tel1, which is recruited to shortened telomeres, neutralizes the origin-delaying activity of Rif1. We also find that Tel1 phosphorylates Rif1 at short telomeres, although our investigation shows this phosphorylation is not the sole mechanism through which Tel1 prevents Rif1-mediated replication delay. Since correct telomere replication timing control is important for telomerase-mediated length maintenance, this discovery represents an important step towards understanding the molecular mechanisms that ensure proper long-term stabilization of chromosome ends, as well as the controls over the DNA replication temporal program.
Introduction
Chromosomal DNA replication occurs according to a regulated program, with some replication origins initiating early and others late in S phase [1], [2]. S. cerevisiae telomeres provide a good model for understanding molecular controls over the temporal regulation of DNA replication. The replication time of S. cerevisiae telomeric regions is regulated by telomere length; chromosome regions close to telomeres with a normal length terminal TG1–3 tract generally replicate late, but those close to telomeres with a shortened TG1–3 tract replicate early [3], [4]. This control is mediated through altered initiation time of replication origins. Telomeres may be replicated either by replication forks from an origin within the subtelomeric repeat sequences (X or Y′ ARS elements), or by a fork arriving from a nearby telomere-proximal origin (such as ARS522, close to chromosome V-right; previously known as ARS501) [5]–[7]. Normal length telomeres can direct the late activation of such origins, while telomeric and telomere-proximal origins activate earlier if next to a shortened telomere—as demonstrated by experiments using recombination-based excision of TG1–3 repeats or the mutation yku70Δ that causes shortened telomeres [3], [4], [8]. Telomere repeat length can affect origins up to 40 kb from the chromosome end [4]. Earlier replication is proposed to favor telomerase recruitment and TG1–3 repeat lengthening [9]–[11]. However, how cells detect and respond to telomere length in order to control the replication time of telomeres remains unclear.
The end-replication problem causes shortening of terminal TG1–3 tracts in successive cell cycles, and a network of controls detects critically short telomeres and ensures they are preferentially elongated by telomerase enzyme [12]. The mechanisms that detect TG1–3 tract length to control replication timing are likely to overlap with mechanisms that ensure preferential elongation of short telomeres. Indeed, the Rif1 protein is already implicated in both pathways. S. cerevisiae Rif1 binds to the TG1–3 repeat recognition factor Rap1 and with Rif2 regulates telomerase recruitment in response to telomere length [13], [14]. Rif1 and Rif2 appear to ‘count’ the telomeric repeats and repress telomerase recruitment if the TG1–3 tract does not require extension. Cells lacking either Rif1 or Rif2 have abnormally long telomeres due to uncontrolled lengthening by telomerase [13]. The molecular mechanism by which Rif1 represses telomerase recruitment is still under investigation. Long and short telomeres bind similar amounts of Rif1 [15]; one proposal is that molecular modifications occurring selectively at short telomeres may relieve the repressive effect of Rif1 on telomerase recruitment [16]. For Rif2, number of molecules may determine the repressive effect on telomerase, since more Rif2 molecules are present at long than short telomeres [17].
As well as acting in the pathway that recognizes short telomeres for lengthening, Rif1 is involved in controlling telomere replication time in response to length [4]. Specifically, in cells lacking Rif1 the link between telomere length and replication time is broken, since the telomeres of a rif1Δ mutant replicate early despite being abnormally long. Recently, Rif1 has been implicated as a regulator of replication timing more generally, having a repressive effect on genome-wide DNA replication mediated through recruitment of Protein Phosphatase 1 [18]–[23].
Tel1, a PIK (phosphatidylinositol 3-kinase)-related checkpoint kinase, is involved in short telomere recognition. Tel1 binds to short telomeres and contributes to their preferential recruitment of telomerase [15], [24]–[26]. Tel1 is recruited by interacting with the C-terminus of Xrs2, a subunit of the MRX (Mre11-Rad50-Xrs2) nuclease complex, which is also enriched at short telomeres. The kinase activity of Tel1 is required for its role in telomere maintenance [27]. Potential targets for Tel1 phosphorylation at telomeres include Xrs2, Mre11 [28] and the telomeric single-stranded binding protein Cdc13 [29], but it is unclear whether phosphorylation of these targets is important for telomere lengthening [30]; discussed in [16]. There is some overlap in function between Tel1 and Mec1, the other yeast PIK checkpoint kinase, but while telomeres in tel1Δ mutant cells are extremely short, lack of Mec1 causes only a mild telomere length defect [31], [32]. In general, Tel1 seems to play the primary role in regulating telomere function while Mec1 is the major checkpoint kinase.
Since Tel1 is preferentially recruited to short telomeres, we investigated whether Tel1 is also involved in the pathway that detects short telomeres to specify early replication. We show here that Tel1 is required to drive the early replication of short telomeres, and that it acts upstream of Rif1 in the pathway that controls telomere replication timing. We tested whether Tel1 phosphorylates Rif1, and identified two SQ (i.e. Tel1 consensus) sites that are preferentially phosphorylated in a short telomere mutant. Phosphorylation of one of the sites, Serine-1308, is completely dependent on the presence of Tel1. Mutation of these sites did not prevent the early replication of short telomeres, suggesting that Rif1 phosphorylation is not the sole mechanism through which Tel1 drives early replication. Our results are consistent with a model in which Tel1 that is recruited to short telomeres counteracts the repressive effect of Rif1 on replication initiation at nearby origins, to promote early origin activation and advance the replication time of short telomeres.
Results
Tel1 is required for the early replication of shortened telomeres
To investigate the mechanism linking telomere length with replication timing, we examined the role of Tel1, since this kinase is implicated in telomere length detection. tel1Δ cells have very short telomeres as shown in Fig. 1A, due to defective telomerase recruitment. If telomere replication time is still correctly coupled to TG1–3 tract length in this mutant, we would expect the short telomeres of a tel1Δ strain to replicate early—like the telomeres of a yku70Δ mutant, which replicate earlier than normal because they are short [4].
Fig. 1. Tel1 is required for early replication of short telomeres. 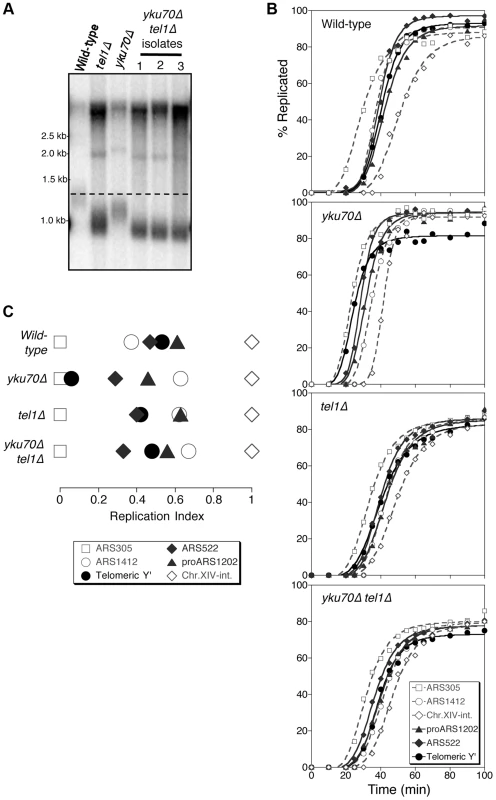
(A) Telomere length analysis in wild-type (YKU70 TEL1), tel1Δ, yku70Δ, and yku70Δ tel1Δ strains. Terminal chromosome fragments were detected by probing a Southern blot of XhoI-digested genomic DNA for TG1–3 sequence. Smear represents average length of Y′ telomeres. (B) Replication kinetics of various genomic sequences in wild-type and short telomere mutants yku70Δ, tel1Δ and yku70Δ tel1Δ. Telomere-proximal sequences shown are Y′ (solid line with filled circles), ARS522 (solid line with filled diamonds), and proARS1202 (solid line with filled triangles). Non-telomeric marker sequences (dashed lines) are early origins ARS305 (open squares), late origin ARS1412 (open circles), and Chr XIV-internal sequences (open diamonds). Strains were released from α-factor block at 30°C. (C) Replication indices (RI) values from experiments in B, where replication times are normalized to early origin ARS305 (RI = 0) and Chr.XIV-int (RI = 1). Strains are BB14-3a (wild-type), ASY5 (tel1Δ), AW99 (yku70Δ) and ASY13 (yku70Δ tel1Δ; corresponding to second isolate in part A); all are in A364a background as listed in Table S1. Replication time can be measured using the dense isotope transfer method, in which cells blocked in G1 phase with α-factor are transferred from isotopically dense to light medium. Upon release into S phase the transition of specific sequences from heavy∶heavy to heavy∶light DNA fractions on cesium gradient centrifugation is then monitored. Replication kinetics of particular sequences are plotted (Fig. 1B), and replication time assigned as the time at which half the final level of replication has occurred. Since kinetics of α-factor release show some variability between experiments, the replication program can be usefully summarized using ‘replication indices’ (Fig. 1C), with the various replication times normalized to early and late-replicating marker sequences (ARS305 and Chr XIV-int respectively) [33]. Replication times plotted relative to ARS305 are shown in Fig. S1.
In wild-type cells, the subtelomeric Y′ repeat sequences (indicative of average telomeric replication) replicate late in S phase (Fig. 1B; top panel, solid line with filled circles, Fig. 1C&S1, filled circle), 3.4 min later than the internal late replication origin ARS1412 [33], [34]. In the yku70Δ mutant that has shortened TG1–3 repeat sequences, the Y′ sequences replicate much earlier, at a similar time to early origin ARS305 (Fig. 1B,C&S1) [4], [8]. Examining replication kinetics in a tel1Δ mutant strain revealed that Y′ sequences replicated late, close to their normal replication time (Fig. 1B; third panel from top & Fig. 1C). Telomere-proximal sequences (ARS522 and proARS1202) show a similar trend (solid curve with filled diamonds and solid curve with filled triangles, respectively; Fig. 1B), so that overall the replication program of the tel1Δ mutant resembles that of wild-type cells (Fig. 1C). Since the tel1Δ mutant has very short telomeres (even shorter than those of yku70Δ; Fig. 1A) this result suggests that in the absence of Tel1 kinase, the replication time of telomeric regions is uncoupled from telomere length.
Telomeres of a yku70Δ mutant are short due to defects in telomere capping and extension. yku70Δ cells can detect telomere length status—since yku70Δ telomeres replicate early and Tel1 is correctly recruited to the short telomeres of a yku70Δ strain [35]. Early telomere replication in a yku70Δ mutant appears to result from telomere shortness, since restoring telomeres to wild-type length in a yku70Δ background leads to recovery of normal, late telomere replication [4]. The yku70Δ mutant therefore provides a convenient tool to investigate the controls linking telomere length to replication control. Note that the strength of the effect on telomere length of the yku70Δ mutation differs in A364a (used for timing replication in this study) and BY4741 yeast strain backgrounds (Fig. S2). Strain dependence of the effect of yku70Δ on telomere length was previously observed (compare [36], [37] with [38]–[40]). The reason for the strain dependence is not known, but the effects of the yku70Δ mutation on replication timing appear similar regardless of whether the effect on telomere length is weak or strong [8], [41], [42].
To understand whether Tel1 is required to transmit the signal for the early replication of short telomeres, we examined the replication program of a yku70Δ tel1Δ double mutant. This mutant has extremely short telomeres similar to a tel1Δ mutant (Fig. 1A), but subtelomeric (Y′) and telomere-proximal (ARS522 and proARS1202) sequences replicated later than in yku70Δ single mutant, with replication timing similar to that in wild-type or tel1Δ cells (Fig. 1B,C&S1). While precise replication times and order show some variability between experiments [33], repetition of these experiments confirmed the general trends (Fig. S3). Overall, these results suggest that in the absence of Tel1, yku70Δ telomeres are no longer sensed as short and hence not replicated early, implying that Tel1 is involved in specifying early replication of short telomeres.
Tel1 stimulates the early initiation of a replication origin next to an induced short telomere
We cannot exclude the possibility that effects shown above result from mutant phenotypes unrelated to telomere length. For example, the effect on replication timing of telomere uncapping in yku70Δ has not been tested. We therefore examined whether Tel1 promotes early telomere replication using an alternative mode of telomere shortening. We utilized a yeast strain in which a short telomere can be created by induction of HO endonuclease in cells blocked in G1 phase, as illustrated in Fig. 2A and similar to the construct previously described [43]. In this system, an HO cut site close to the left end of chromosome VII is flanked by short (80 bp) and long (250 bp) TG1–3 tracts on its centromere - and telomere-proximal sides respectively. Cutting with HO endonuclease in G1-blocked cells creates a single shortened telomere which, following release into S phase, stimulates earlier initiation at the neighboring, normally late-replicating origin ARS700.5. ARS700.5 is located 18 kb from unmodified telomere VII-left and lies 5.3 kb from the HO cut site in this construct [Cooley & Bianchi, personal communication]. In a small-scale experiment we found that HO cutting levels exceeded 67% 5.5 hr after galactose addition, confirming that short telomere induction occurred in the majority of cells (Fig. S4A).
Fig. 2. Tel1 stimulates activation of an origin neighboring an induced short telomere upon release into hydroxyurea. 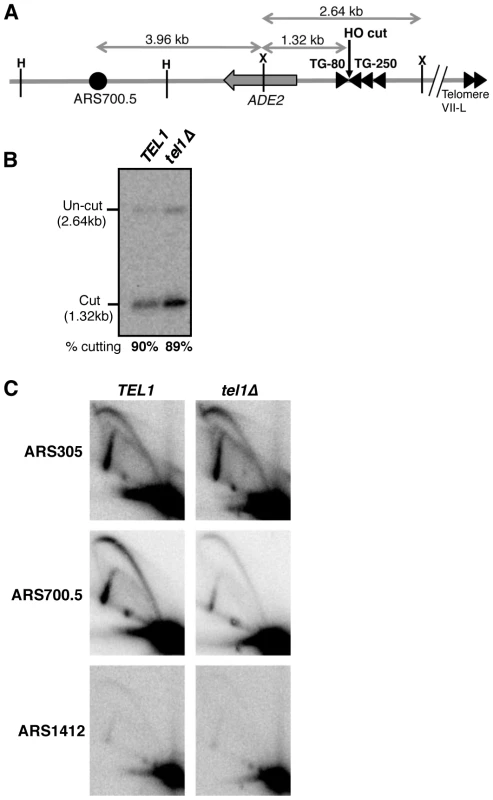
(A) Cartoon showing HO endonuclease-inducible short telomere construct on the left arm of Chr. VII, with positions of HindIII (H) and XmnI (X) restriction sites. Triangles represent TG repeat sequences and the filled circle, ARS700.5. Not to scale. (B) HO endonuclease cutting efficiency in the hydroxyurea-arrested cultures used for 2D gel analysis in C. Cells were arrested with α-factor then galactose added to induce HO cutting, followed by release into hydroxyurea. (C) 2D gel analysis of replication intermediates present at early origin ARS305 (upper panels), ARS700.5 (middle panels), and late origin ARS1412 (lower panels), in TEL1 (left) and tel1Δ (right) strains. The same blot of HindIII-digested DNA was probed sequentially for the three origins. Strains used are YAB1410, SMKY10 (TEL1) and SMKY13 (tel1Δ). When S. cerevisiae cells attempt S phase in the presence of the replication inhibitor hydroxyurea (HU), early origins are activated but late origin initiation is repressed by the Rad53-mediated S phase checkpoint [44]–[46]. Two-dimensional gel analysis of origin activation levels [47] after release into hydroxyurea therefore provides a proxy for differences in origin initiation time.
The short telomere was induced by HO cutting and cells were then released into HU-containing medium (during which cutting levels reached 90%; Fig. 2B). At the control early-initiating replication origin ARS305, 2-dimensional gel analysis revealed strong bubble arcs in both TEL1 and tel1Δ strains (Fig. 2C, upper panels). In contrast only low levels of replication intermediates were observed at the control late origin ARS1412 (Fig. 2C, lower panels), due to checkpoint-mediated late origin repression. At ARS700.5 close to the induced short telomere, a strong bubble arc was observed in the TEL1 strain, consistent with stimulation of early ARS700.5 initiation as expected. Bubble arc intensity was however substantially reduced in the tel1Δ mutant (Fig. 2C, middle panels), revealing that Tel1 is needed to drive early, checkpoint-resistant initiation of ARS700.5 following nearby short telomere induction. Quantitation of the bubble arc signal (as shown in Fig. S4B&C) revealed 4.8-fold-reduction in bubble arc intensity at ARS700.5 in the tel1Δ strain. In a construct with ARS700.5 placed proximal to a long telomere repeat a bubble arc was almost undetectable (Fig. S5), confirming that early activation of this origin depends on the nearby induced short telomere.
Our 2-dimensional gel analysis therefore confirmed that after nearby short telomere induction, the absence of Tel1 changes the character of ARS700.5 from that of an early-initiating origin to that of a late replication origin. The results were therefore consistent with the replication timing analyses in Figs. 1 & S3, showing that Tel1 is required to specify early replication of chromosomal regions in proximity to a short telomere.
We also attempted to use isotope labeling-based replication timing analysis to examine ARS700.5 replication following short telomere induction, but inefficient and variable HO cutting after growth in the minimal medium required for this technique prevented satisfactory analysis of replication timing.
Tel1 acts upstream of Rif1 in controlling telomere replication timing
Rif1 is implicated in the control of replication timing in response to telomere length, since in a rif1Δ mutant the link between telomere length and replication time is uncoupled. Specifically, in a rif1Δ mutant the TG1–3 tracts are over-extended (Fig. 3A), but cells fail to detect the length of their telomeres and replicate them inappropriately early (Fig. 3B,C & Fig. S6A) [4]. Consistently, ARS700.5 initiates prior to the S phase checkpoint in a rif1Δ mutant with an induced short telomere (Fig. S6B). Early replication of the long rif1Δ telomeres presents an interesting reversal of the effect in tel1Δ, where cells fail to detect the shortness of their telomeres and replicate them inappropriately late (Fig. 1). The opposite nature of these phenotypes implies that Tel1 and Rif1 have opposing actions in the pathway that controls telomere replication timing, with Rif1 enforcing the late replication of long or normal length telomeres, while Tel1 signals early replication of telomeres that are shortened. Loss of Rif1 impacts replication timing of many genomic regions [48] with subtelomeric regions most strongly affected [4], probably because telomeres are the main genomic Rif1 binding locations [18].
Fig. 3. Rif1 acts downstream of Tel1 in regulating telomere replication time. 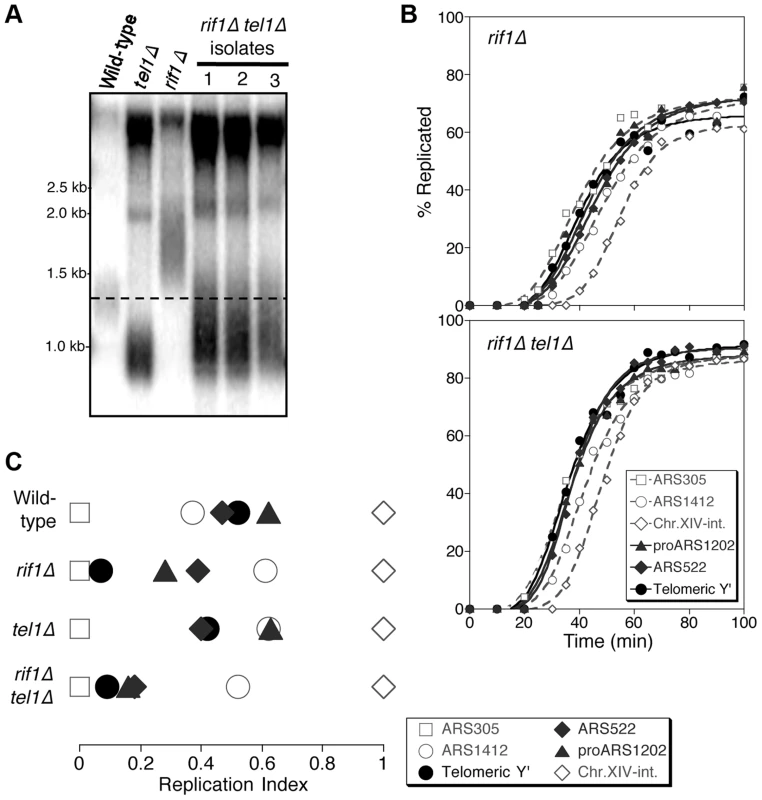
(A) Telomere length analysis in wild-type, tel1Δ, rif1Δ and rif1Δ tel1Δ strains. Southern blot analysis carried out as in Fig. 1A. (B) Replication kinetics of various genomic sequences in rif1Δ and rif1Δ tel1Δ strains. Plots and symbols as in Fig. 1B. (C) Replication indices from experiments in B, along with values from wild-type and tel1Δ experiments from Fig. 1. Strains are HYLS44 (rif1Δ) and ASY14 (rif1Δ tel1Δ; corresponding to first isolate in part A). To test the relationship of Tel1 and Rif1 in the telomere replication timing control, we examined a rif1Δ tel1Δ double mutant. Deleting RIF1 somewhat relieves the short telomere phenotype of tel1Δ (Fig. 3A), presumably reflecting an effect of Rif1 on the backup mechanisms that recognize critically short telomeres in the absence of Tel1 [24]. We tested whether the rif1Δ tel1Δ strain replicates its telomeres early (as in rif1Δ) or late (as in tel1Δ). We found that in rif1Δ tel1Δ cells, both Y′ and telomere-proximal sequences replicate very early, similar to their replication time in a rif1Δ single mutant (Fig. 3B & C; replication times shown in Fig. S6). The rif1Δ mutation is therefore epistatic to tel1Δ in control of telomere replication—consistent with the idea that Tel1 counteracts Rif1-mediated delay to telomere replication timing.
Rif1 is phosphorylated at Tel1 consensus sites in a mutant with short telomeres
Since Tel1 is actively recruited to shortened telomeres, we hypothesized that Tel1 may act to prevent or ‘switch off’ the delaying effect of Rif1 on nearby replication origins. The Rif1 protein sequence contains multiple S/TQ motifs, corresponding to the consensus sequence for Tel1-mediated phosphorylation [27], [29], so Rif1 is a potential target for Tel1 kinase activity. We therefore tested by mass spectrometry whether Tel1 phosphorylates Rif1. Using Myc-tagged Rif1 that retains almost complete protein functionality (as assayed by telomere length, Fig. 4A), we devised an immunoprecipitation procedure to pull down the majority of cellular Rif1 (Fig. 4B). Initial high-resolution mass spectrometry identified multiple phosphorylated peptides in Rif1 from both YKU70 and yku70Δ strains, including two phosphorylation sites corresponding to Tel1 consensus sequences, one at Serine-1308 (within the sequence…KVDSQDIQ…) and the other at Serine-1351 (…MNSSQQE…) (Fig. 4C). Rif1 S-1308 phosphorylation is not previously described; while S-1351 was identified as phosphorylated in response to DNA damage by MMS [49].
Fig. 4. Rif1 is phosphorylated at Tel1 consensus sites in the short telomere mutant yku70Δ. 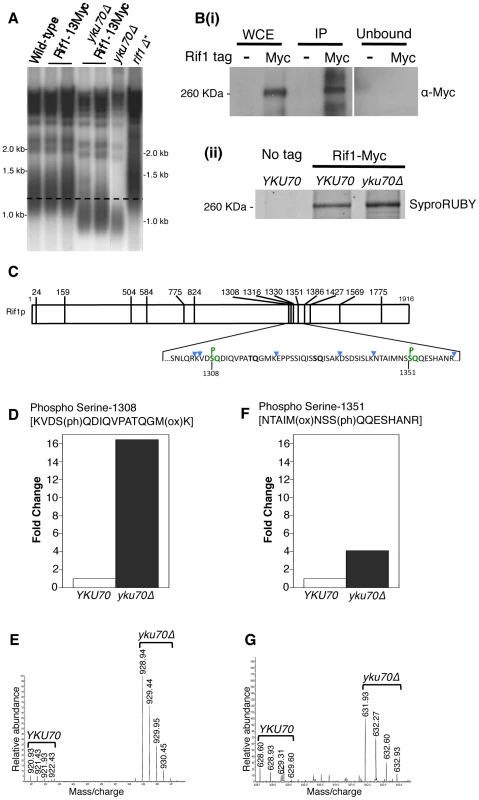
(A) Telomere length gel confirms Myc-tagged Rif1 protein is functional. (B) Upper panel (i): Western blot analyzing Rif1-Myc protein in Whole Cell Extract (WCE), immunoprecipitated sample (IP) and supernatant (Unbound). All lanes show equivalent cell loading. Lower panel (ii): SyproRUBY-stained gel showing Rif1-Myc isolated from YKU70 and yku70Δ strains, and mock IP from untagged control sample. Rif1 was quantified based on SyproRUBY gel bands and equivalent quantities mixed for SILAC mass spectrometry analysis. 260 kD marker position is indicated. The predicted size of Rif1-13Myc is 232 kD; Rif1-13Myc migration is slightly retarded relative to its predicted mass. (C) Cartoon of Rif1p sequence, illustrating the position of the 14 S/TQ sites. In enlarged sequence below S/TQ sequences are bold, and colored green are the two sites identified as phosphorylated in an initial mass spectrometry run (carried out using Rif1-Myc from YKU70 and yku70Δ strains). Blue arrowheads indicate trypsin digestion sites. (D) Plot of SILAC ratio for phosphorylated peptide containing S-1308 in yku70Δ relative to YKU70 (16.4× increased). In this and similar plots relative values are normalized during processing to the median H/L ratio of all Rif1 peptides. (E) MS spectrum showing raw results for the same S-1308 phosphorylated peptide [KVDS(ph)QDIQVPATQGM(ox)K], with light (R0K0) peptide from YKU70 on left and heavy (R10K8) peptide from yku70Δ on right. (F) & (G) show equivalent SILAC analysis for the S-1351 phosphorylated peptide NTAIM(ox)NSS(ph)QQESHANR (4.1× increased in Δyku70 relative to YKU70). Strains (BY4741 strain background) are SHY201 (untagged wild-type), ASY25 (YKU70 RIF1-13Myc), ASY30 (yku70Δ RIF1-13Myc), Y00870 (untagged yku70Δ) and HYLS44 (rif1Δ*; asterisk indicating A364a strain background). An initial, non-SILAC, mass spectrometry analysis depicted in C used W303 Rad5+ strains YSM20 (YKU70 RIF1-13Myc) and ASY17 (yku70Δ RIF1-13Myc). Identification of these phosphorylated SQ sites suggests that Rif1 may indeed be a target for Tel1 kinase, perhaps specifically at shortened telomeres which recruit Tel1. We used the comparative proteomic method of SILAC to compare phosphorylation levels in wild-type cells with the short telomere yku70Δ mutant. Phosphorylation at both sites were increased in the yku70Δ mutant, by about 16-fold at S-1308, and about 4-fold at S-1351 (Fig. 4D–G; Dataset S1). The corresponding unphosphorylated peptides were not increased in the yku70Δ strain (Fig. S7; Dataset S1). These results show that phosphorylation of these Rif1 SQ motifs is increased in the shortened telomere context of yku70Δ.
Tel1 is required for phosphorylation of Rif1 Serine-1308
To address whether Tel1 kinase mediates phosphorylation of Rif1 at S-1308 and S-1351, we used a similar SILAC strategy to test whether the phosphorylation levels are decreased when Tel1 is unavailable. This experiment was carried out in the yku70Δ background where peptides containing phosphorylated S-1308 and S-1351 residues are reliably detected. Peptides from heavy-labeled yku70Δ tel1Δ cells were compared with those from light-labeled yku70Δ cells. The S-1308 phosphorylated peptide was abundant in the yku70Δ mutant, but was 10-fold reduced in the yku70Δ tel1Δ strain (Fig. 5A; Dataset S2). A longer peptide covering the same phosphorylated S-1308 residue was also greatly reduced in yku70Δ tel1Δ (Fig. S8A; Dataset S2), while its unphosphorylated equivalent showed no significant change (Fig. S8C; Dataset S2). In contrast, levels of the S-1351 phosphorylated peptide were largely unchanged in yku70Δ tel1Δ when compared to yku70Δ (Fig. 5B). Based on this analysis, we propose that Tel1 directly phosphorylates S-1308. However, we cannot exclude the possibility that Tel1 activates a different SQ-directed kinase that phosphorylates Rif1 S-1308 at short telomeres. If Tel1 contributes to phosphorylation at S-1351, its role can be substituted by a different kinase (probably Mec1 since [49] showed S-1351 phosphorylation requires one or other of Mec1 and Tel1). Alternatively, Mec1 may be solely responsible for S-1351 phosphorylation.
Fig. 5. Phosphorylation of Rif1 Serine-1308 depends on Tel1. 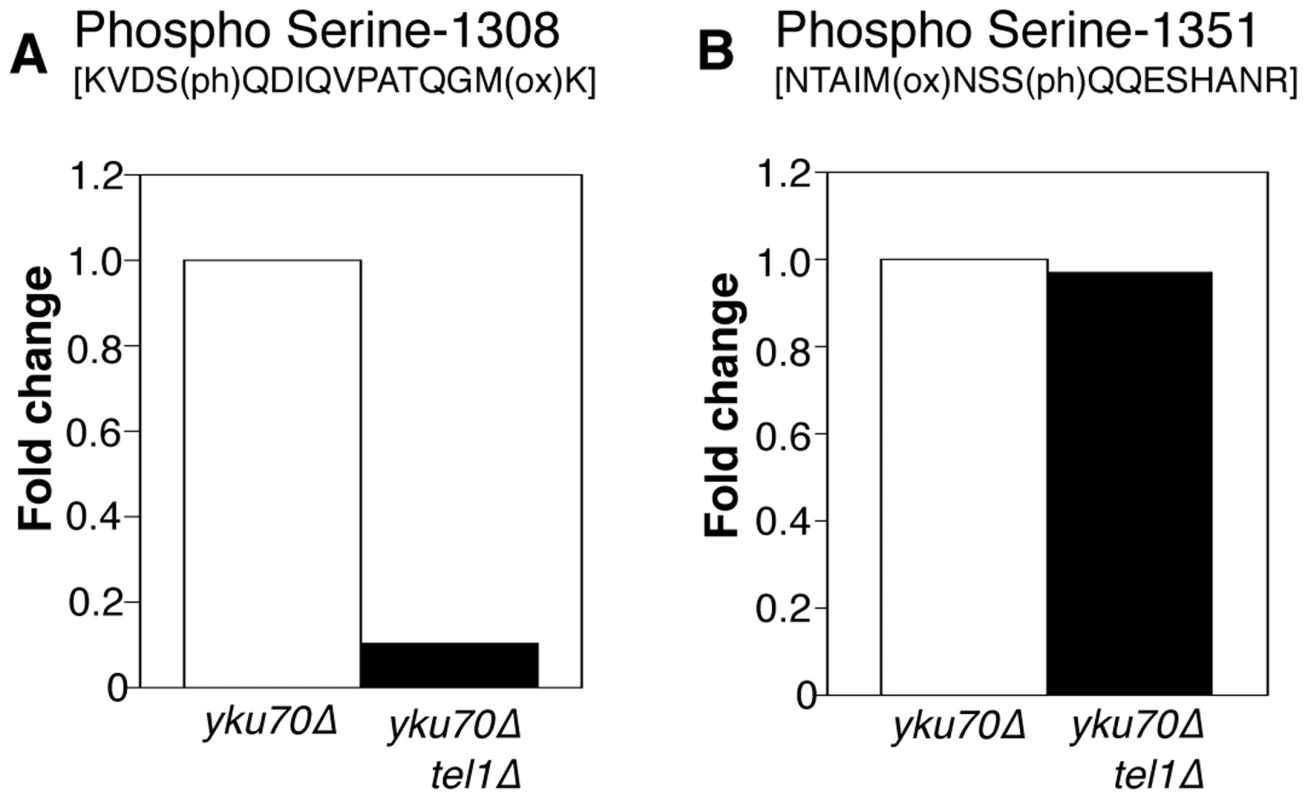
(A) Plots shows relative levels of the S-1308 phosphorylated peptide [KVDS(ph)QDIQVPATQGM(ox)K] in yku70Δ (Light-labeled R0K0) and yku70Δ tel1Δ (Heavy-labeled R10K8) strains. H/L ratio is 0.10. (B) Equivalent plot for S-1351 phosphorylated peptide NTAIM(ox)NSSQQESHANR. H/L ratio is 0.97048. Strains used are ASY30 (yku70Δ RIF1-13Myc), and ASY46 (yku70Δ tel1Δ RIF1-13Myc). Fig. S9 provides a summary of the phosphorylation sites identified on Rif1 in these proteomic analyses. This study identified a cluster of phosphorylated DDK and CDK consensus sites close to the Rif1 N-terminus, which in a separate investigation were shown to regulate Protein Phosphatase 1 recruitment by Rif1 [21].
Mutation of the Rif1 phosphorylated S/TQ cluster does not prevent the effect of telomere length on replication timing
Our results suggest a model in which Tel1-mediated phosphorylation of Rif1 antagonizes the delaying effect of Rif1 on telomeric and telomere-proximal replication origins at short telomeres. We constructed a Rif1 mutant where the relevant serine residues are replaced by alanine, to test whether this non-phosphorylatable construct constitutively delays replication, preventing early replication of short telomeres. We replaced the serine or threonine residue with alanine at all seven of the potential Tel1 phosphorylation sites (SQ and TQ motifs) between 1308 and 1569 in the Rif1 amino acid sequence, to construct a rif1-7S→A allele. We mutated the entire cluster of S/TQ motifs since it could contain phosphorylation sites not detected proteomically and because preventing phosphorylation of one of these residues might re-direct kinase activity to a nearby consensus site. Telomere length was hardly affected by this rif1-7S→A allele, or by an phospho-mimetic equivalent rif1-7S→E glutamate substitution allele, in either YKU70 or yku70Δ backgrounds (Fig. 6A)—confirming that these substitutions do not ablate Rif1 protein function. We examined the replication program of the rif1-7S→A mutant in the short telomere (yku70Δ) background. We found that telomeres still replicate early (Fig. 6B), with Y′ elements replicating at a similar time to the early marker sequence ARS305 (Fig. 6C & S10), equivalent to the yku70Δ mutant. The non-phosphorylatable RIF1 allele therefore does not prevent the early replication of short telomeres, implying that phosphorylation of the Rif1 S/TQ cluster is not essential for Tel1 to drive early replication of short telomeres. The rif1-7S→A mutation similarly caused minimal change to the replication timing program in a YKU70 background (Fig. S11). We also tested the replication program of the rif1-7S→E allele designed to mimic a phosphorylated form of Rif1. In this rif1-7S→E mutant telomeres still replicate at approximately the same time as late origin ARS1412 (Fig. S12).
Fig. 6. Non-phosphorylatable rif1-7S→A does not delay early replication of short yku70Δ telomeres. 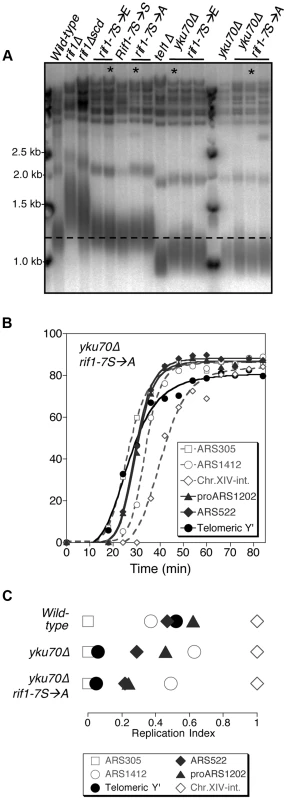
(A) Telomere length analysis of Rif1 phospho-site mutants. Genomic DNA was extracted from the indicated strains and telomere length analyses performed as described. Smear indicates average length of Y′ telomeres. rif1Δscd represents an internal deletion within the RIF1 C-terminal region made as a strain construction intermediate (see Supplementary Materials & Methods). RIF1-7S→S represents a RIF1 reconstruction, where wild-type sequence was re-inserted into rif1Δscd to control for telomere length recovery. (B) Replication program of yku70Δ rif1-7S→A, released from an α-factor block at 30°C. Sequences analyzed are as in Fig. 1. (C) Replication indices from yku70Δ rif1-7S→A experiment shown in B, along with values from wild-type and yku70Δ experiments from Fig. 1B&C. Strains in part A are BB14-3a (wild-type), HYLS44 (rif1Δ), ASY5 (tel1Δ), AW99 (yku70Δ), ASY51 (rif1Δscd); ASY81 (RIF1-7S→S). For rif1-7S→A asterisk indicates ASY69, used for replication timing in Fig. S11; for rif1-7S→E asterisk indicates ASY73, used for replication timing in Fig. S12; for yku70Δ rif1-7S→A asterisk indicates ASY76, used for replication timing in Fig. 6 B&C and S10; for yku70Δ rif1-7S→E asterisk indicates ASY78. The phenotypic analyses of the yku70Δ rif1-7S→A and rif1-7S→E mutant therefore suggest that Tel1-mediated phosphorylation of the Rif1 S/TQ cluster is not necessary or sufficient to drive early replication. They do not however exclude the possibility that Tel1-mediated Rif1 S/TQ cluster phosphorylation could contribute to early replication of short telomeres. Indeed a very slight advancement (3–4 min; Fig. S12 B&C) in telomere replication time in the rif1-7S→E allele may be consistent with this idea. One possibility is that phosphorylation of Rif1 S-1308, S-1351 and nearby S/TQ sites is integrated with other, redundant mechanisms to ensure that shortened telomeres replicate early.
Discussion
In investigating controls over telomere replication timing, we discovered that Tel1 specifies the early replication of short telomeres, as assessed either using a short telomere mutant (yku70Δ; Fig. 1) or by analyzing origin activation close to an induced short telomere (Fig. 2). Rif1 specifies late replication of normal telomeres, and epistatic analysis indicated that Tel1 counteracts the delaying effect of Rif1 on telomere replication time. Phosphoproteomic analysis of endogenous S. cerevisiae Rif1 revealed at least two SQ motifs to be phosphorylated. Phosphorylation at these sites is increased in a short telomere mutant, with phosphorylation at Serine-1308 completely dependent on the presence of Tel1. However, corresponding Rif1 alanine substitution mutants did not prevent early replication of telomeres in a yku70Δ background, indicating that phosphorylation of Rif1 by Tel1 at S-1308, S-1351, or nearby consensus sites within the Rif1 S/TQ cluster domain, cannot be the sole mechanism by which Tel1 drives early replication at short telomeres. While Rif1 phosphorylation could potentially contribute, Tel1 must mediate early replication of short telomeres through additional, possibly redundant, pathways.
S. pombe, S. cerevisiae and human Rif1 proteins all negatively regulate DNA replication genome-wide [18]–[21], and very recently it was shown that Rif1 recruits Protein Phosphatase 1 to control DNA replication [21]–[23]. The stimulatory effect of removing S. cerevisiae Rif1 on the overall replication program is reflected by a shortened S phase (Fig. 3B & S6A: S phase duration is 21.5 min in wild-type but 15.3 min in rif1Δ). Within the generally shortened S phase of the rif1Δ mutant telomeres are more dramatically affected, with telomere-associated sequences shifting their replication time from the latter half to the early part of S phase (Fig. 3C). Proximity of Rif1 binding sites has been suggested to determine the susceptibility of replication origin initiation to Rif1-mediated repression [18], and the delaying effect of Rif1 on replication is probably focused at chromosome ends by the preferential association of Rif1 with telomeres, as illustrated in Fig. 7A, explaining why telomere regions show the largest shift in replication timing when Rif1 is removed (Fig. 3C). It is possible that non-telomeric Rif1 also contributes to the late replication of telomere regions.
Fig. 7. Model of replication timing control by Tel1 and Rif1. 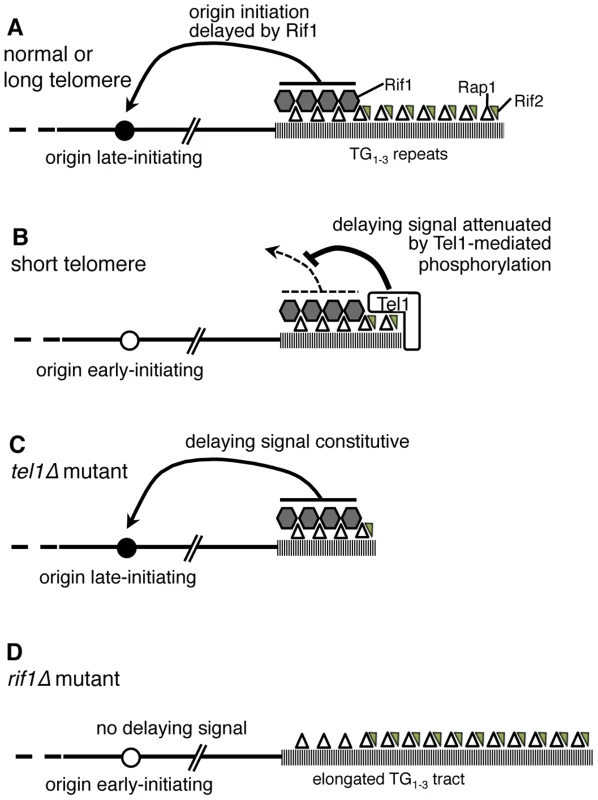
(A) In wild-type cells, terminal TG1–3 tract is bound by Rap1 (open triangles) which recruits Rif1 (grey hexagons) and Rif2 (small grey triangles). If the telomere is normal in length, Rif1 signals to nearby origins (such as telomere-proximal Y′ or ARS522 origins) specifying their late replication time (filled circle). (B) If telomeres are short (as in a yku70Δ mutant) Tel1 kinase is recruited and neutralizes the Rif1 delaying signal, so that nearby origins initiate early (white circle). (C) In tel1Δ mutant cells, the delaying effect of Rif1 cannot be neutralized so that nearby origins initiate late despite the short telomeres. (D) A rif1Δ mutant lacks the delaying signal, with the result that nearby origins initiate replication early despite their extended TG1–3 repeat length. Removing both Tel1 and Rif1 leads to a phenotype that is essentially equivalent to a rif1Δ single mutant—that is, in the absence of Rif1, it becomes largely irrelevant for telomere replication timing whether Tel1 is present (Fig. 3C). For this reason, our results support a model where Tel1 affects replication timing by counteracting the delaying action of Rif1 on telomere replication, as illustrated in Fig. 7A & B. If non-telomeric Rif1 contributes to late replication of subtelomeric regions, its effect is presumably also neutralized by Tel1.
We envisage two modes through which Tel1 could counteract the delaying effect of Rif1 on origin initiation. First, phosphorylation of Rif1 by Tel1 at SQ sites might ‘switch off’ the Rif1 repressive effect. We identified Rif1 as a target of Tel1 phosphorylation at shortened telomeres, but mutating the sites identified, along with neighboring potential phosphorylation sites, did not dramatically impact telomere replication timing. This observation argues that Rif1 phosphorylation cannot be solely responsible for Tel1-driven early telomere replication, while leaving open the possibility that Rif1 phosphorylation acts redundantly with other control mechanisms. It is possible that Rif1 contains additional functionally critical Tel1 phosphorylation sites not identified by our proteomic analysis. It is also conceivable that phosphorylation of Rif1 by Tel1 at non-consensus (i.e. non-S/TQ) sites might contribute to replication timing control. A previous study [28] showed that a Dun1 substrate lacking any SQ consensus was still phosphorylated by Tel1 kinase, and noted that ATM (the mammalian homolog of Tel1) phosphorylates non-canonical sites in the tumor suppressor BRCA1 [50]. Intriguingly, in the yku70Δ mutant we observed a 2 to 4-fold increase in phosphorylation levels of five serine or threonine residues that are not followed by glutamine (Rif1 S-1338, S-1355, S-1362, T-1367, and S-1694; Fig. S9 & Dataset S1).
Second, Tel1 could prevent the Rif1-mediated replication delay by phosphorylating a different telomeric protein. A number of telomeric proteins have been identified as likely or possible targets of Tel1 phosphorylation, including Cdc13 [29], Xrs2, and Mre11 [28]. While they cannot be formally excluded, none of these proteins is directly implicated in controlling replication origin activation. It seems more likely that Tel1 counteracts the Rif1-mediated delay by phosphorylating an unidentified component of the molecular pathway through which Rif1 restrains origin activation. Such a mechanism could act redundantly with Tel1-mediated Rif1 phosphorylation to neutralize the Rif1 replication-delaying signal. Tel1 appears to have multiple targets at telomeres [28], [29], which may act in concert to produce biological function, so that ablating any particular phosphorylation event has rather mild effects.
A third possibility is that telomere replication timing control depends on multiple mechanisms some of which do not involve Rif1, although the strong effect of Rif1 loss on telomere replication (Fig. 3) does suggest it is the most central controller of telomere replication time. H2A-S129 phosphorylation depends on Tel1 in telomere-proximal regions [51], and a non-phosphorylatable (H2A-S129A) allele caused a slight delay to telomere replication in a yku70Δ background (unpublished observations); however, H2A-S129 phosphorylation is not elevated at shortened telomeres [51], inconsistent with H2A-S129 phosphorylation being a critical mediator of the early replication of short telomeres.
Phosphorylation of Rif1 may contribute to other telomeric functions. One possibility is that Tel1-mediated Rif1 phosphorylation counteracts repression of telomerase recruitment, favoring TG1–3 tract extension. Telomere length is not greatly altered by the rif1-7S→A or rif1-7S→E mutants (Fig. 6A)—although very slight telomere lengthening in some rif1-7S→E isolates hints that Rif1 phosphorylation might contribute to telomerase recruitment. As with replication timing, Rif1 phosphorylation may be one of a series of redundant mechanisms through which Tel1 regulates telomerase recruitment—another potential pathway being phosphorylation of Cdc13 [16]. A further role for Rif1 phosphorylation might involve regulation of anti-checkpoint function at telomeric DNA damage sites [43], [52].
To summarize, we have identified an important new function for Tel1—namely, driving the early replication of shortened telomeres. Our results suggest that Tel1 exerts this function by neutralizing the delaying effect of telomeric Rif1 on nearby replication origins. Tel1 also directs phosphorylation of Rif1, which may contribute to replication timing control along with other mechanisms that impact on origin initiation time. Since Rif1 and Tel1 are conserved and play similar roles in replication timing control and coordination of DNA repair in higher eukaryotes as in yeast, our discoveries are likely to illuminate general functions of these proteins.
Materials and Methods
Yeast strains
Yeast strains are listed in Supplemental Table S1. Gene knockouts and tagging used standard PCR-based insertion methods, confirmed by PCR analysis; see Supplemental material (Text S1) for details of specific strain constructions. Primer sequences are available on request.
Analysis of replication time
The replication time of specific sequences was measured using the dense isotope transfer procedure [53], [54] in cells released from α-factor at 30°C, probing for genomic EcoRI fragments as described previously [8].
Two-dimensional gel analysis of replication intermediates
Inducible HO cut strains were initially grown in YP medium containing 2% raffinose with 0.01% glucose (to allow adaptation to raffinose), and then grown for 24–48 hours in 2% raffinose at 30°C before blocking with 200 nM α-factor. Then galactose was added to obtain a final concentration of 4%, to induce HO endonuclease. After 5.5–6 hr, the cells were then released by the addition of pronase with simultaneous addition of 200 mM hydroxyurea, and harvested 2 h later. DNA was prepared using the NIB-n-grab method [55] digested with HindIII followed by 2-dimensional gel electrophoresis under standard conditions [47]. HO cutting efficiency was confirmed by Southern blot analysis of XmnI-digested DNA.
Immunoprecipitation of Rif1-Myc
Immunoprecipitation of Rif1 was carried out as described [56] with modifications as described in Supplementary Material (Text S1). Protein concentrations were estimated using the RCDC kit (Bio-rad).
Western blotting and SyproRUBY staining
Immunoprecipitated proteins were eluted in 1× SDS sample buffer (Invitrogen) with 5% 2-Mercaptoethanol. Cellular equivalent protein samples were separated by SDS PAGE (Novex 8–16% Tris-Glycine gels, Precast; Invitrogen) and wet blotted using 1× Towbin buffer with 10% Methanol onto PVDF membrane (Hybond-P, GE Healthcare). Rabbit anti-Myc (ab9106, Abcam) was used to detect epitope-tagged RIF1, with secondary antibody AP-conjugated anti-Rabbit IgG (S3731, Promega). Detection substrate was CDP-Star (Perkin Elmer) using Medical X-ray (Fuji) film. For quantification of amount of Rif1 protein, a similar gel was stained overnight using SyproRUBY total protein staining solution (Bio-rad) and quantified with a Fuji Phosphorimager (FLA3000) at 473 nm with O580 filter and FujiFILM ImageGauge (software V4.21).
SILAC sample preparation and mass spectrometry analysis
SILAC samples were prepared based on the procedure described [57]. To compare yku70Δ with wild-type (Fig. 4), yeast strain AYS30 was labeled with heavy L-ARGININE:HCL (U-13C6: U-15N4; CNLM-539-H; Cambridge Isotope Laboratory) and L-LYSINE:2HCL (U-13C6; U-15N2, CNLM-291-H; Cambridge Isotope Laboratory) [R10K8] and ASY25 was labeled with light alternatives [R0K0] for at least ten generations. To compare yku70Δ tel1Δ with yku70Δ (Fig. 5), ASY46 was labeled with heavy Lysine and Arginine [R10K8] and ASY30 was labeled with light alternatives [R0K0] for at least ten generations, and subjected to immunoprecipitation as described above. Immunoprecipitated Rif1 was quantified by SYPRORuby staining. Equal masses of Rif1 were then mixed and run on a Novex 8–16% Tris-Glycine gel, and the Rif1 band was excised for analysis by high-resolution mass spectrometry (FingerPrints Proteomics, University of Dundee) as described in Supplementary Information (Text S1).
Telomere length analysis
Genomic DNA was digested with XhoI, separated on a 1.5% agarose gel and transferred to neutral membrane (MP Biomedicals) by Southern blotting. Terminal restriction fragments were detected using a probe directed against the TG repeats.
Supporting Information
Zdroje
1. FergusonBM, FangmanWL (1992) A position effect on the time of replication origin activation in yeast. Cell 68 : 333–339.
2. YamazakiS, HayanoM, MasaiH (2013) Replication timing regulation of eukaryotic replicons: Rif1 as a global regulator of replication timing. Trends Genet 29 : 449–460.
3. BianchiA, ShoreD (2007) Early replication of short telomeres in budding yeast. Cell 128 : 1051–1062.
4. LianHY, RobertsonED, HiragaS, AlvinoGM, CollingwoodD, et al. (2011) The effect of Ku on telomere replication time is mediated by telomere length but is independent of histone tail acetylation. Mol Biol Cell 22 : 1753–1765.
5. StevensonJB, GottschlingDE (1999) Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev 13 : 146–151.
6. NieduszynskiCA, HiragaS, AkP, BenhamCJ, DonaldsonAD (2007) OriDB: a DNA replication origin database. Nucleic Acids Res 35: D40–46.
7. SiowCC, NieduszynskaSR, MullerCA, NieduszynskiCA (2012) OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res 40: D682–686.
8. CosgroveAJ, NieduszynskiCA, DonaldsonAD (2002) Ku complex controls the replication time of DNA in telomere regions. Genes Dev 16 : 2485–2490.
9. BianchiA, ShoreD (2007) Increased association of telomerase with short telomeres in yeast. Genes Dev 21 : 1726–1730.
10. GallardoF, LaterreurN, CusanelliE, OuenzarF, QueridoE, et al. (2011) Live cell imaging of telomerase RNA dynamics reveals cell cycle-dependent clustering of telomerase at elongating telomeres. Mol Cell 44 : 819–827.
11. DehePM, RogO, FerreiraMG, GreenwoodJ, CooperJP (2012) Taz1 enforces cell-cycle regulation of telomere synthesis. Mol Cell 46 : 797–808.
12. PfeifferV, LingnerJ (2013) Replication of telomeres and the regulation of telomerase. Cold Spring Harb Perspect Biol 5: a010405.
13. MarcandS, GilsonE, ShoreD (1997) A protein-counting mechanism for telomere length regulation in yeast. Science 275 : 986–990.
14. ShiT, BunkerRD, MattarocciS, RibeyreC, FatyM, et al. (2013) Rif1 and Rif2 shape telomere function and architecture through multivalent Rap1 interactions. Cell 153 : 1340–1353.
15. SabourinM, TuzonCT, ZakianVA (2007) Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell 27 : 550–561.
16. WellingerRJ, ZakianVA (2012) Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 191 : 1073–1105.
17. McGeeJS, PhillipsJA, ChanA, SabourinM, PaeschkeK, et al. (2010) Reduced Rif2 and lack of Mec1 target short telomeres for elongation rather than double-strand break repair. Nat Struct Mol Biol 17 : 1438–1445.
18. HayanoM, KanohY, MatsumotoS, Renard-GuilletC, ShirahigeK, et al. (2012) Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev 26 : 137–150.
19. CornacchiaD, DileepV, QuivyJP, FotiR, TiliF, et al. (2012) Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J 31 : 3678–3690.
20. YamazakiS, IshiiA, KanohY, OdaM, NishitoY, et al. (2012) Rif1 regulates the replication timing domains on the human genome. EMBO J 31 : 3667–3677.
21. HiragaS, AlvinoGM, ChangF, LianHY, SridharA, et al. (2014) Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev 28 : 372–383.
22. DaveA, CooleyC, GargM, BianchiA (2014) Protein Phosphatase 1 Recruitment by Rif1 Regulates DNA Replication Origin Firing by Counteracting DDK Activity. Cell Rep 7 : 53–61.
23. MattarocciS, ShyianM, LemmensL, DamayP, AltintasDM, et al. (2014) Rif1 Controls DNA Replication Timing in Yeast through the PP1 Phosphatase Glc7. Cell Rep 7 : 62–69.
24. ArnericM, LingnerJ (2007) Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep 8 : 1080–1085.
25. HiranoY, FukunagaK, SugimotoK (2009) Rif1 and rif2 inhibit localization of tel1 to DNA ends. Mol Cell 33 : 312–322.
26. GoudsouzianLK, TuzonCT, ZakianVA (2006) S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol Cell 24 : 603–610.
27. MalloryJC, PetesTD (2000) Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc Natl Acad Sci U S A 97 : 13749–13754.
28. MalloryJC, BashkirovVI, TrujilloKM, SolingerJA, DominskaM, et al. (2003) Amino acid changes in Xrs2p, Dun1p, and Rfa2p that remove the preferred targets of the ATM family of protein kinases do not affect DNA repair or telomere length in Saccharomyces cerevisiae. DNA Repair (Amst) 2 : 1041–1064.
29. TsengSF, LinJJ, TengSC (2006) The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res 34 : 6327–6336.
30. GaoH, ToroTB, PaschiniM, Braunstein-BallewB, CervantesRB, et al. (2010) Telomerase recruitment in Saccharomyces cerevisiae is not dependent on Tel1-mediated phosphorylation of Cdc13. Genetics 186 : 1147–1159.
31. RitchieKB, MalloryJC, PetesTD (1999) Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol 19 : 6065–6075.
32. RayA, RungeKW (1999) Varying the number of telomere-bound proteins does not alter telomere length in tel1Delta cells. Proc Natl Acad Sci U S A 96 : 15044–15049.
33. FriedmanKL, DillerJD, FergusonBM, NylandSV, BrewerBJ, et al. (1996) Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev 10 : 1595–1607.
34. FergusonBM, BrewerBJ, ReynoldsAE, FangmanWL (1991) A yeast origin of replication is activated late in S phase. Cell 65 : 507–515.
35. HectorRE, ShtofmanRL, RayA, ChenBR, NyunT, et al. (2007) Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell 27 : 851–858.
36. RayA, RungeKW (2001) Yeast telomerase appears to frequently copy the entire template in vivo. Nucleic Acids Res 29 : 2382–2394.
37. HangLE, LiuX, CheungI, YangY, ZhaoX (2011) SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat Struct Mol Biol 18 : 920–926.
38. BoultonSJ, JacksonSP (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J 17 : 1819–1828.
39. GravelS, LarriveeM, LabrecqueP, WellingerRJ (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science 280 : 741–744.
40. PorterSE, GreenwellPW, RitchieKB, PetesTD (1996) The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res 24 : 582–585.
41. HiragaS, RobertsonED, DonaldsonAD (2006) The Ctf18 RFC-like complex positions yeast telomeres but does not specify their replication time. EMBO J 25 : 1505–1514.
42. HiragaS, BotsiosS, DonaldsonAD (2008) Histone H3 lysine 56 acetylation by Rtt109 is crucial for chromosome positioning. Journal of Cell Biology 183 : 641–651.
43. RibeyreC, ShoreD (2012) Anticheckpoint pathways at telomeres in yeast. Nat Struct Mol Biol 19 : 307–313.
44. PaulovichAG, HartwellLH (1995) A Checkpoint Regulates the Rate of Progression through S-Phase in Saccharomyces cerevisiae in Response to DNA-Damage. Cell 82 : 841–847.
45. CrabbeL, ThomasA, PantescoV, De VosJ, PaseroP, et al. (2010) Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat Struct Mol Biol 17 : 1391–1397.
46. KubotaT, HiragaS, YamadaK, LamondAI, DonaldsonAD (2011) Quantitative proteomic analysis of chromatin reveals that Ctf18 acts in the DNA replication checkpoint. Mol Cell Proteomics 10: M110.005561.
47. FriedmanKL, BrewerBJ (1995) Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. DNA Replication 262 : 613–627.
48. PeaceJM, Ter-ZakarianA, AparicioOM (2014) Rif1 regulates initiation timing of late replication origins throughout the S. cerevisiae genome. PLoS One 9: e98501.
49. SmolkaMB, AlbuquerqueCP, ChenSH, ZhouH (2007) Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A 104 : 10364–10369.
50. CortezD, WangY, QinJ, ElledgeSJ (1999) Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286 : 1162–1166.
51. KimJA, KruhlakM, DotiwalaF, NussenzweigA, HaberJE (2007) Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. Journal of Cell Biology 178 : 209–218.
52. XueY, RushtonMD, MaringeleL (2011) A novel checkpoint and RPA inhibitory pathway regulated by Rif1. PLoS Genet 7: e1002417.
53. DonaldsonAD, RaghuramanMK, FriedmanKL, CrossFR, BrewerBJ, et al. (1998) CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell 2 : 173–182.
54. McCarrollRM, FangmanWL (1988) Time of replication of yeast centromeres and telomeres. Cell 54 : 505–513.
55. PayenC, Di RienziSC, OngGT, PogacharJL, SanchezJC, et al. (2014) The Dynamics of Diverse Segmental Amplifications in Populations of Saccharomyces cerevisiae Adapting to Strong Selection. G3 (Bethesda) 4 : 399–409.
56. MorohashiH, MaculinsT, LabibK (2009) The amino-terminal TPR domain of Dia2 tethers SCF(Dia2) to the replisome progression complex. Curr Biol 19 : 1943–1949.
57. KubotaT, SteadDA, HiragaS, ten HaveS, DonaldsonAD (2012) Quantitative proteomic analysis of yeast DNA replication proteins. Methods 57 : 196–202.
Štítky
Genetika Reprodukční medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 10
-
Všechny články tohoto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



