-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
Accumulation of DNA damage has been implicated in aging. Many premature aging syndromes are due to defective DNA repair systems. The endonuclease XPG is involved in repair of helix-distorting DNA lesions, and XPG defects cause the cancer-prone condition xeroderma pigmentosum (XP) alone or combined with the severe neurodevelopmental progeroid disorder Cockayne syndrome (CS). Here, we present a novel (conditional) Xpg−/ − mouse model which -in a C57BL6/FVB F1 hybrid background - displays many progressive progeroid features, including early cessation of growth, cachexia, kyphosis, osteoporosis, neurodegeneration, liver aging, retinal degeneration, and reduced lifespan. In a constitutive mutant with a complex phenotype it is difficult to dissect cause and consequence. We have therefore generated liver - and forebrain-specific Xpg mutants and demonstrate that they exhibit progressive anisokaryosis and neurodegeneration, respectively, indicating that a cell-intrinsic repair defect in neurons can account for neuronal degeneration. These findings strengthen the link between DNA damage and the complex process of aging.
Published in the journal: . PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004686
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004686Summary
Accumulation of DNA damage has been implicated in aging. Many premature aging syndromes are due to defective DNA repair systems. The endonuclease XPG is involved in repair of helix-distorting DNA lesions, and XPG defects cause the cancer-prone condition xeroderma pigmentosum (XP) alone or combined with the severe neurodevelopmental progeroid disorder Cockayne syndrome (CS). Here, we present a novel (conditional) Xpg−/ − mouse model which -in a C57BL6/FVB F1 hybrid background - displays many progressive progeroid features, including early cessation of growth, cachexia, kyphosis, osteoporosis, neurodegeneration, liver aging, retinal degeneration, and reduced lifespan. In a constitutive mutant with a complex phenotype it is difficult to dissect cause and consequence. We have therefore generated liver - and forebrain-specific Xpg mutants and demonstrate that they exhibit progressive anisokaryosis and neurodegeneration, respectively, indicating that a cell-intrinsic repair defect in neurons can account for neuronal degeneration. These findings strengthen the link between DNA damage and the complex process of aging.
Introduction
If DNA damage, either inflicted from exogenous or endogenous sources, cannot be repaired, this has detrimental consequences for an organism ranging from transcription blocks, permanent cell cycle arrest and mutations, to cell death. In the end, this unrepaired DNA damage contributes to the onset and progression of the aging process, as well as to cancer [1]–[3]. Cells are equipped with a set of elaborate DNA repair mechanisms integrated into a complex DNA damage response machinery that jointly attempt to fix the unrepaired DNA [4]. One such DNA repair mechanism is the Nucleotide Excision Repair (NER) pathway that removes a wide category of helix-distorting lesions, such as those induced by UV and bulky chemical adducts, in a tightly coordinated process involving over 30 proteins [5]–[7]. NER can be divided into two subpathways based on the mode of damage recognition. The Global Genome (GG-)NER subpathway specifically involves the XPC and XPE protein complexes, and probes the entire genome for lesions that disrupt base-pairing [5], [7]–[9]. Transcription-Coupled (TC-)NER, on the other hand, detects helix-distorting lesions that stall transcription in the transcribed strand of expressed genes, and hence enables resumption of transcription. TC-NER is independent of XPC and XPE and specifically involves proteins such as CSA, CSB and UVSSA [8], [10], [11]. After lesion recognition, the subsequent ‘cut-and-patch’ core repair reaction encompasses local opening of the DNA helix and lesion verification, performed by the TFIIH complex together with XPA. Both correctly position the structure-specific endonucleases ERCC1/XPF and XPG for strand-specific excision of the lesion as part of a 22–30 bp oligonucleotide [5], [7], [12]. Finally, the gap is filled by repair synthesis and closed by ligation [5], [7], [12].
Multiple NER proteins have been attributed additional roles, both in DNA repair pathways other than NER, and in transcription regulation. For instance, TFIIH is an essential component of the general transcription machinery [13], [14], but also other NER factors, including XPG and CSB, have been implicated in transcription regulation [15]–[18]. The 5′ endonuclease ERCC1/XPF participates in the repair of interstrand crosslinks [19], [20] and subpathways of DNA double-strand break repair [21]. XPC, CSB, and XPG have been individually implicated in promoting base excision repair (BER) of oxidative DNA damage [22]–[30]. TFIIH and XPG are, together with CSB, thought to be involved in the early steps of Transcription-Coupled Repair (TCR), and XPG interacts directly with both CSB and RNA Polymerase II [31]. Although still controversial, there are accumulating reports that TCR not only directs NER to blocked transcription but may also recruit BER for preferential repair of oxidative DNA damage in transcribed strands [32]–[34]. Such a mechanism might be related to the roles of both CSB and XPG in promoting BER more globally. If correct, it could explain the much greater consequences for the organism of TCR defects compared to defects in NER alone (see below) [35].
A number of rare, autosomal recessive disorders resulting from mutations in NER genes underscore the importance of genome maintenance for the prevention of cancer as well as aging [3]. NER-associated diseases are characterized by sun (UV) hypersensitivity and include xeroderma pigmentosum (XP), UV-sensitivity syndrome (UVSS), Cockayne syndrome (CS), cerebro-oculo-facio-skeletal (COFS) syndrome, XPF-ERCC1 (XFE) progeroid syndrome, trichothiodystrophy (TTD) and disorders that combine the symptoms of these syndromes, including XP/CS [35]–[39]. XP originates from defects in GG-NER or total NER activity and is characterized by an over 2000-fold increased risk of cancer in sun-exposed skin and, to a much lesser extent, in internal organs [36]. XP patients may also develop progressive neurological symptoms and neuronal degeneration depending on the severity of the total NER deficiency [36], [39], [40]. UVSS is characterized by skin UV hypersensitivity without actually developing skin cancer. UVSS results from the selective loss of TC-NER function as a consequence of mutations in the proteins involved in detection of UV-induced transcription-blocking DNA lesions, i.e. UVSSA, CSA, and CSB [11], [35], [41]–[44]. Mutations in CSA and CSB, however, generally cause CS, a heterogeneous multisystem disorder that, in addition to UV-sensitivity, is characterized by severe growth failure and cachexia, accelerated aging features, short lifespan, and progressive sensori-neuronal abnormalities [38], [45]. The severe symptoms of CS cannot be explained by the loss of TC-NER function as they do not occur in fully NER-deficient XP patients and TC-NER deficient UVSS patients. Therefore, CS symptoms have been linked to additional, yet incompletely, defined functions of CSA and CSB in DNA repair, transcription regulation, other processes, or a combination of deficiencies [3], [46], [47]. The same applies for mutations in the down-stream NER factors XPB, XPD, XPF, ERCC1 and XPG that cause combined XP/CS, or severe developmental/degenerative multisystem disorders such as COFS and XFE that share multiple features with severe CS forms [35], [48]–[51]. Thus CS symptoms can result from mutations in multiple proteins that operate together in NER, but the symptoms caused by these mutations cannot be explained by NER deficiency alone, raising questions about the identities of these non-NER activities underlying CS symptoms and the extent to which different symptoms reflect deficits of different cellular processes [35], [46].
Mutations in the structure-specific NER 3′-endonuclease XPG are rare, with less than 20 patients and 25 mutant alleles described so far [52]–[55], and cause a spectrum of disease phenotypes varying from XP to XP/CS and COFS [53]. Point mutations that selectively eliminate XPG nuclease activity cause XP, while C-terminal truncations, destabilizing point mutations, and mutations that abolish the interaction between XPG and the basal transcription factor TFIIH cause XP/CS and COFS [52]–[58]. These data support the notion that a deficient function of XPG outside NER is responsible for the severe CS symptoms [15], [52], [53], [55]–[57].
For most NER disorders, mouse mutants have been generated that mimic the genetic defect found in patients, and to various extents reproduce XP and CS-like features as well as the progeroid hallmarks found in the corresponding human syndrome [59]–[62]. Accordingly, Xpg-null (Xpg−/−) mice were found to develop a severe phenotype characterized by growth deficiency and very short lifespan, resembling severe XP/CS [63]. In contrast, Xpg mutant mice carrying amino acid substitutions that selectively abolish the nuclease function of XPG (XpgE791A and XpgD811A) show severe UV-sensitivity but normal lifespan, hence, reproducing the XP phenotype [64], [65]. In addition, a mutant XPG construct containing a C-terminal truncation lacking the last 360 amino acids that was made to mimic the genotype of some XP-G/CS patients, developed a growth deficiency and short-living phenotype resembling that of Xpg−/− mice, albeit somewhat milder [65]. Yet another C-terminal truncation mutant lacking the last 180 amino acids showed a normal lifespan, but produced a CS-like growth-deficient short-living phenotype after crossing with Xpa−/− mice that are already fully NER-deficient [65], [66]. Significantly, the same conversion of a normal lifespan into a short-living mouse model is observed after crossing CSA - or CSB-deficient CS mice with total NER - (Xpa−/−) or GG-NER (Xpc−/−) deficient mouse models [67]–, but not by crossing NER-deficient XpgD811A with Xpa−/− mice [66]. Together these data indicate a deleterious synergistic interaction between NER deficiency and loss of non-NER activities that underlie CS. Furthermore, they show that the C-terminus of XPG could play a role in the CS symptoms, and that the XPG-deficient Xpg−/− mice may reproduce the phenotype of Xpa−/−Csb−/−, Xpc−/−Csb−/− or Xpa−/−Csa−/− double mutant mice [62], [66]–[69].
In previous analyses we clearly observed progeroid characteristics in many NER mutant mouse models including Xpa/Csb, Xpb, Xpd, and Ercc1 mutants [51],[68],[70]–[72], yet the occurrence of progeroid features in Xpg−/− mice has hitherto been poorly established, mostly due to their very short lifespan. Since we are particularly interested in the effect of Xpg deletion on organ-specific aging, we generated a conditional Xpg mutant. As genetic background can have a significant effect on phenotype development, we first re-examined the pathological characteristics of Xpg−/− mice in a C57BL6/FVB F1 hybrid background, as was previously described for Ercc1 mutant mice [51], [72]. In this hybrid background Xpg−/− mice lived longer and presented progeroid features including cachexia and osteoporosis with pronounced degenerative phenotypes in both liver and brain. We next studied the effect of liver - and forebrain-specific inactivation of Xpg, showing that the observed phenotypes are indeed due to lack of XPG protein. Together our data show that, consistent with previous data in ERCC1 - and CSB/XPA-deficient mice, Xpg−/− mice develop a multisystem progeroid degenerative phenotype.
Results
Generation of NER-deficient Xpg−/− mice
To generate a Cre-inducible Xpg knockout allele we flanked the third exon of Xpg with LoxP sites (Figure 1A). Deletion of this exon causes a frame shift and a premature translational stop immediately after exon 2 at amino acid residue 89 (instead of the full-length 1170). After transfection to 129 ES cells and selection of properly targeted clones (Figure 1B and Materials and Methods), two independent transfected clones were used to generate germ-line transmitting chimeras (Figure 1C). Heterozygous males, carrying the conditional Xpg allele, were crossed to females ubiquitously expressing Flp for excision of the Neomycin cassette and to yield mice that are heterozygous for the floxed Xpg (Xpgf) allele. Xpgf/+ mice were backcrossed and maintained in FVB/N background. To generate Xpg mice carrying a knockout allele (Xpg−, Figure 1A), Xpgf/+ mice were crossed to Cag-Cre mice, which ubiquitously express Cre recombinase from germline [73], yielding heterozygous Xpg+/− animals (Figure 1C). Xpg+/− animals were>10 times backcrossed into C57BL6 or FVB/N genetic backgrounds. Unless otherwise stated, experiments were performed with Xpg−/− mice in the C57BL6/FVB F1 hybrid background obtained from intercrossing C57BL6 Xpg+/−×FVB/N Xpg+/− animals to minimalize background specific effects (see below).
Fig. 1. Generation of Xpg−/− mice. 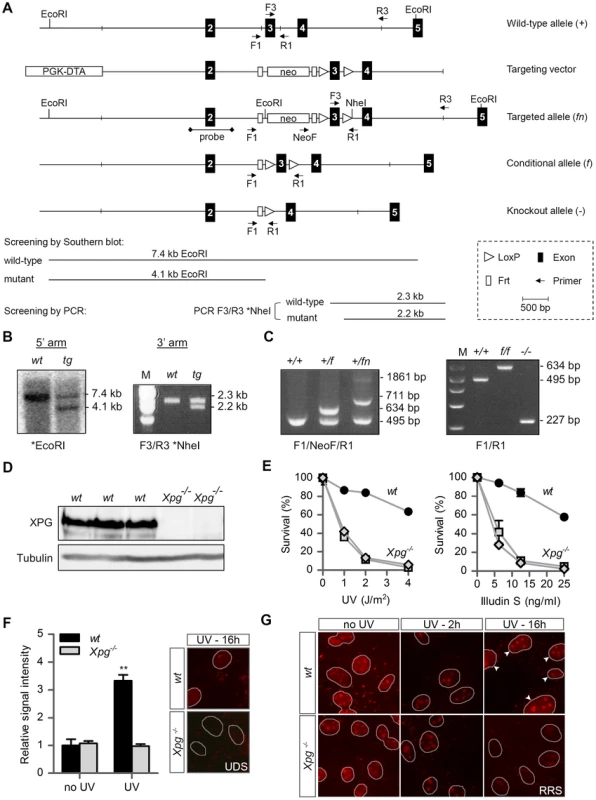
(A) Genomic organization and disruption strategy for Xpg depicting the wild type allele (+), the targeting construct, the targeted allele (fn), the conditional allele after Flp-mediated recombination of Frt sites (f) and the targeted Xpg allele following subsequent Cre-mediated recombination of LoxP sites (−). Exons 2–5 are indicated by black boxes. PCR primers are shown as arrows. (B) Southern blot and PCR analysis of an ES clone showing the correct insertion of the targeting construct. ES cell genomic DNA was digested with EcoRI for Southern blot analysis and hybridized with a 0.9 kb DpnI probe. The wild type (wt) allele yields a 7.4-kb fragment whereas the targeted (tg) allele yields a 4.1-kb fragment. The NheI-digested PCR product shows the 2.3-kb and 2.2-kb bands corresponding with the wt and tg allele, respectively (see also panel A). (C) PCR detection of mouse genotypes using the primers F1, NeoF and R1 as indicated as in A. (D) Immunoblot analysis of extracts from Xpg−/− and wt MDFs using a rabbit polyclonal antibody raised against a peptide conserved between human and mouse XPG. Tubulin is used as loading control. (E) Primary Xpg−/− and wt MDFs, cultured at low (3%) O2 levels were irradiated with the indicated doses of UV-C (left) or treated with the indicated doses of Illudin S for 1 h (right). After 48 h recovery, survival was assessed by cell count. (F) UV-induced UDS in primary Xpg−/− and wt MDFs reveals a severe GG-NER defect in Xpg−/− cells. MDFs were irradiated with 16 J/m2 of UV-C. UDS levels are expressed relative to the non-irradiated wt cells. (G) UV-induced RRS in primary Xpg−/− and wt MDFs reveals a severe TC-NER defect in Xpg−/− cells. MDFs were irradiated with 16 J/m2 of UV-C. 16 h after UV irradiation the wt cells show recovery of RNA synthesis, while Xpg−/− MDFs only show residual activity in nucleoli (rRNA transcription). Arrowheads indicate nuclei. Error bars indicate standard error of the mean. **p<0.01. The presence of a premature stop codon in the Xpg− allele was confirmed by sequencing Xpg cDNA from liver of Xpg−/− mice (Figure S1A). Accordingly, Western immunoblot analysis with an antibody raised against the central spacer region (R-domain) of XPG shows the absence of XPG protein product in mouse dermal fibroblasts (MDFs) isolated from Xpg−/− mice (Figure 1D). Next, we tested DNA repair deficiency of Xpg−/− MDFs. In accordance with complete NER deficiency, Xpg−/− MDFs showed an almost 10-fold hypersensitivity to UV, similar to fully NER-defective MDFs derived from Xpa−/− mice (Figure 1E) [74]. In addition, Xpg−/− MDFs were hypersensitive to treatment with Illudin S (Figure 1E), consistent with the loss of TC-NER function [75], and were deficient in UV-induced unscheduled DNA synthesis in line with loss of GG-NER activity (Figure 1F). Also, recovery of RNA synthesis after UV exposure was almost completely abolished in Xpg−/− MDFs, further demonstrating loss of TC-NER activity (Figure 1G). Xpg−/− MDFs showed no increased sensitivity to potassium bromate (KBrO3) which causes oxidative DNA lesions, and a minimal increased sensitivity to the cross-linking agent cisplatin (Figure S1B).
Genetic background affects perinatal viability and lifespan of Xpg−/− mice
In view of a significant effect of genetic background on embryonic lethality and lifespan in ERCC1-deficient mice [51], [72], we examined whether a similar genetic background effect occurred in Xpg−/− mice, by comparing birth frequencies and lifespan of Xpg−/− mice in a C57BL6, FVB/N or a C57BL6/FVB F1 hybrid background. In C57BL6 background birth frequencies were below Mendelian expectations (∼8%, Table 1), whereas in the FVB/N and C57BL6/FVB F1 hybrid background birth frequencies were Mendelian and near-Mendelian, respectively (Table 1). Also the lifespan of Xpg−/− animals was strongly dependent on genetic background, with C57BL6 Xpg−/− mice showing a lifespan of 3 weeks, and Xpg−/− animals in FVB/N and C57BL6/FVB F1 hybrid background living for 15–18 weeks (Figure 2A).
Fig. 2. Progeroid characteristics of Xpg−/− mice. 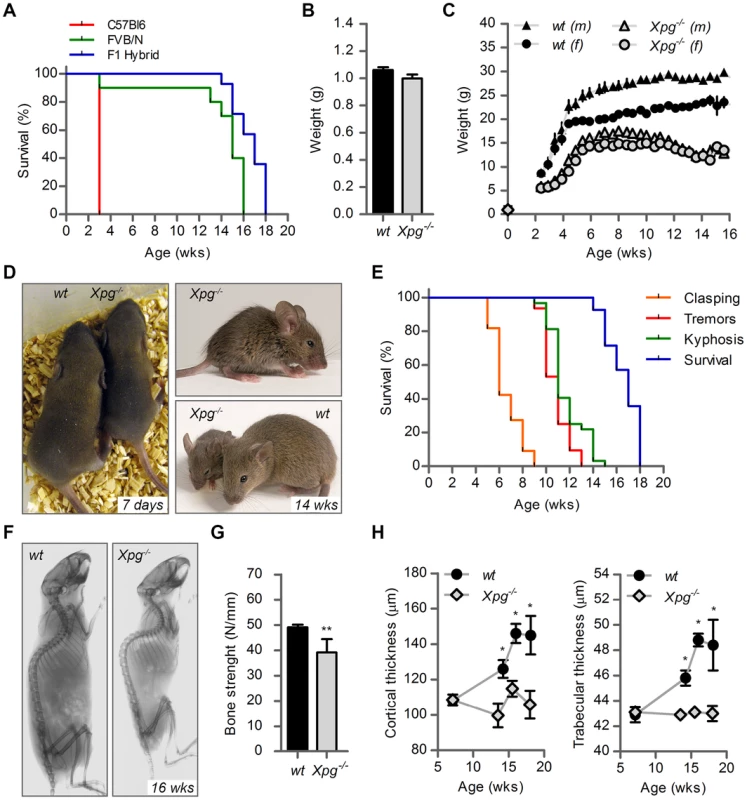
(A) Survival of Xpg−/− mice in a C57Bl6 (red), FVB/N (green) or C57Bl6/FVB F1 hybrid (blue) background; n = 5 (C57Bl6), n = 10 (FVB/N), n = 14 (C57Bl6/FVB F1 hybrid). (B) Average body weight of embryonic 17.5-day old F1 hybrid Xpg−/− and wild type (wt) littermates; n≥12 animals/group. (C) Average body weight of F1 hybrid wt males (black triangles), wt females (black circles), Xpg−/− males (grey triangles), and Xpg−/− females (grey circles); n≥4 animals/group. (D) Left: Photograph of a 7-day old F1 hybrid Xpg−/− and wt littermate, showing no apparent differences except a slightly smaller size. Top right: Photograph of a 14-week old Xpg−/− mouse. Bottom right: Side by side comparison of the same 14-week old Xpg−/− and wt littermate showing a pronounced growth deficiency of the Xpg−/− mouse. (E) Onset of hind limb clasping (orange), tremor (red) and kyphosis (green) with age and survival of F1 hybrid Xpg−/− mice; n = 33 (clasping, tremor and kyphosis), n = 14 (survival). (F) CT-scan of a 16-week old F1 hybrid wt (left) and Xpg−/− (right) mouse showing prominent curvature of the spine (kyphosis) in the Xpg−/− mouse. (G) Bone strength of F1 hybrid Xpg−/− and wt mice analyzed by a 3-point-bending assay of the femur at an average age of 15 weeks; n≥6 animals/group. (H) Cortical (left) and trabecular (right) thickness of the femora of F1 hybrid Xpg−/− and wt mice at different ages; n = 4 animals/group. Error bars indicate standard error of the mean. *p<0.05, **p<0.01. Tab. 1. Xpg−/− mice are born below Mendelian ratio in a C57BL6 background. 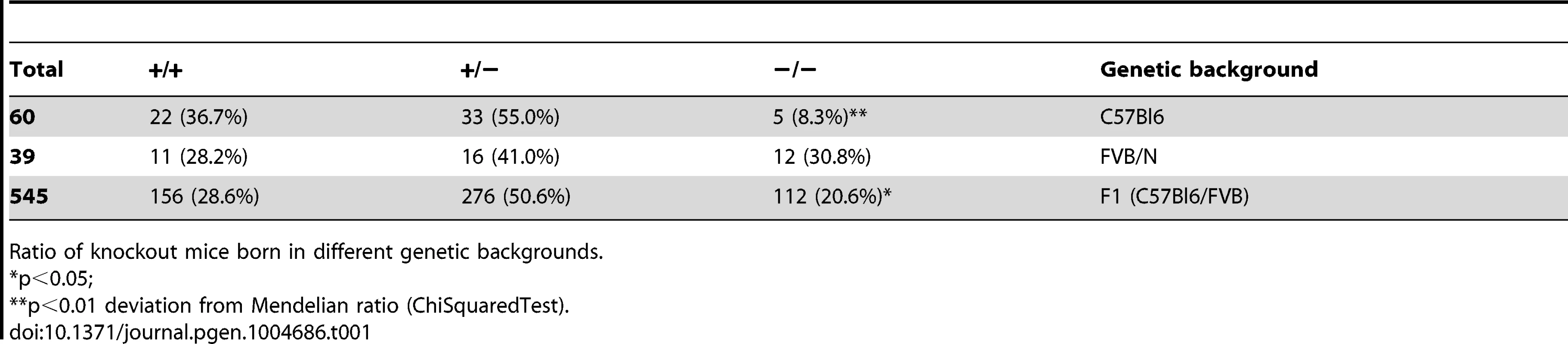
Ratio of knockout mice born in different genetic backgrounds. Cachexia, short lifespan and osteoporosis in Xpg−/− mice
Further analysis of C57BL6/FVB F1 Xpg−/− mice showed that they had the same size and weight as wild type and heterozygote littermates at late embryonic stage (E17.5; Figure 2B and C). However, after birth, Xpg−/− mice showed reduced growth and weight gain compared to controls, and stopped growing at 6–8 weeks when their body weight was about 65–70% of that of wild type littermates (Figure 2C). From 10–11 weeks, body weights declined and the Xpg−/− mice became progressively cachectic (Figure 2C and D). At 14 weeks all Xpg−/− mice were severely runted (Figure 2D), and the mice died a few weeks thereafter between 15–18 weeks of age (Figure 2A and E). The growth deficiency was paralleled by the development of kyphosis (Figure 2D and F). In addition, Xpg−/− mice progressively developed neurological symptoms, including clasping of the hind-limbs when lifted by their tails (Figure S2A), and at a later time point fine tremors (Figure 2E). Accelerating rotarod and grip strength tests in 14-week old Xpg−/− mice revealed severe motor deficits and muscle weakness at this age (Figure S2B and C).
Computed tomography (CT) confirmed severe kyphosis in Xpg−/− mice at 16 weeks of age (Figure 2F). To further examine skeletal abnormalities and the occurrence of osteoporosis as observed in other NER-deficient mouse models [68], [76]–[79], we measured several bone parameters using femoral bones. Analysis of bone strength revealed decreased strength of the Xpg−/− femoral bone at 14–16 weeks (Figure 2G). Micro-CT analysis showed that the thickness of the trabeculae and cortex of the femoral bones was significantly smaller in Xpg−/− compared to wild type mice at 14–17 weeks, but not yet at 7 weeks (Figure 2H). Overall, these data indicate a progressive increase of age-related features such as osteoporosis.
Mild progeroid features and a ‘survival-like’ stress response in livers of Xpg−/− mice
Weight loss and reduced size of Xpg−/− mice was associated with reduced weight of internal organs (Figure S3A) and with a strong reduction in the amount of subcutaneous fat (Figure S3B). The Xpg−/− mice previously reported by Harada et al. [63] showed developmental abnormalities of the gastro-intestinal tract. These gastro-intestinal abnormalities were proposed to be a major contributor of the post-natal growth failure and short lifespan (<3 weeks) of their animals [63]. However, in contrast to their data, the gastro-intestinal tract of our Xpg−/− mice had a normal size and macroscopic appearance, and showed a normal histological appearance in HE-stained sections (Figure 3A). Furthermore, staining for the proliferative cell marker Ki-67 indicated that the number of proliferative cells in the intestinal epithelium was similar between Xpg−/− and wild type mice (Figure 3A). In accord with normal function of the gastro-intestinal tract we found that food intake per gram body weight was similar between wild type and Xpg−/− animals (Figure S3C).
Fig. 3. Intestine and liver phenotype of Xpg−/− mice. 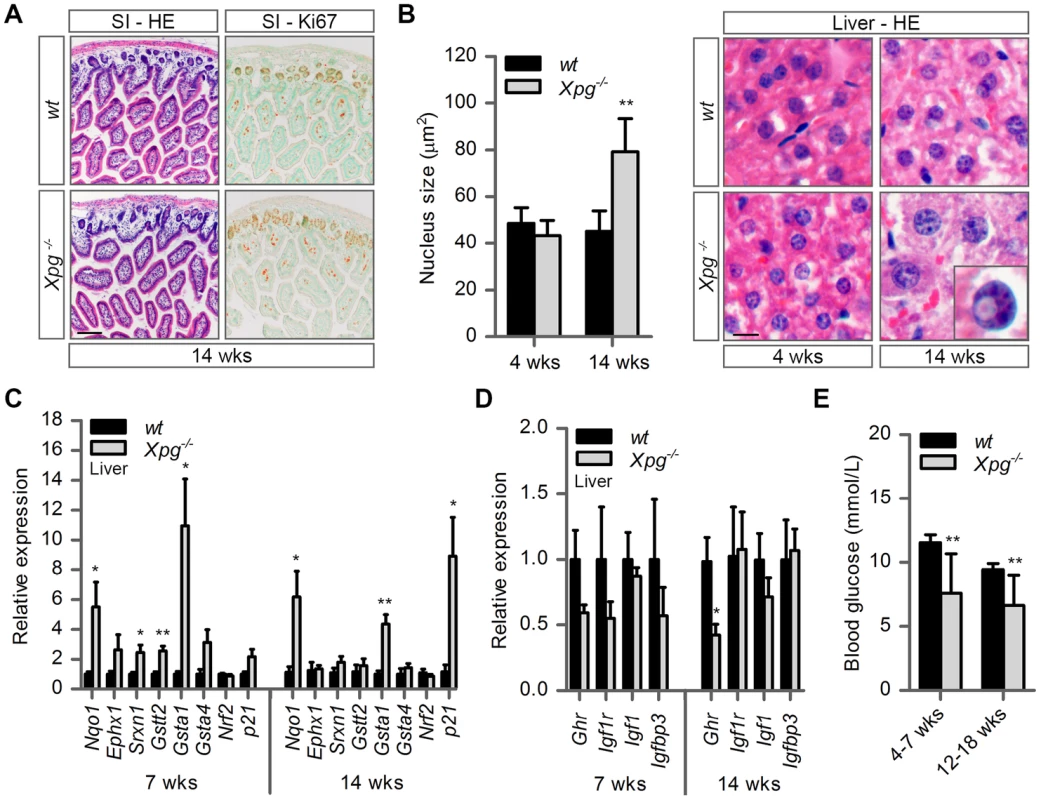
(A) Representative images of HE and Ki67 stained small intestine (SI) of 14-week old Xpg−/− and wild type (wt) mice showing no gross morphological differences. (B) Average nucleus size of hepatocytes in the liver of 4- and 14-week old Xpg−/− and wt mice; n≥3 animals/group. Bottom right: magnification of a nuclear inclusion found sporadically in liver sections of 14-week old Xpg−/− mice. (C) Relative mRNA expression levels of several antioxidant genes and the DNA damage response gene p21 in liver tissue of 7- and 14-week old Xpg−/− and wt mice. All values are corrected for TubG2, Hprt, and Rps9 (Table S1) expression as internal standard and normalized to the 7-week old wt expression levels; n = 4 animals/group. (D) Relative expression levels of the somatotrophic genes Ghr, Igf1r, Igf1, and Igfbp3 in liver tissue of 7- and 14-week old Xpg−/− and wt mice. All values are corrected for TubG2, Hprt, and Rps9 expression and normalized to the 7-week old wt expression levels; n = 4 animals/group. (E) Average basal blood glucose levels in groups of 4–7 and 12–18 week old Xpg−/− and wt mice; n≥15 animals/group. Scale bars: 50 µm (A), 10 µm (B). Error bars indicate standard error of the mean. *p<0.05, **p<0.01. The liver is a central organ in many aspects of metabolic control, including regulation of circulating glucose levels and detoxification, and it plays a key role in regulation of IGF1-somatotrophic axis signaling. Previous studies have shown that ERCC1/XPF-deficient mice develop multiple liver abnormalities, in particular anisokaryosis resulting from polyploidy, and intranuclear inclusions [72], [80]–[83]. Analysis of HE-stained liver sections of our Xpg−/− mice revealed mild anisokaryosis, and increased mean nuclear size at 14 weeks, but not at 4 weeks of age (Figure 3B). In addition, sporadically, hepatocytes had intranuclear inclusions. These liver nuclear changes are a well characterized phenomenon in the aging liver, and indicate that Xpg−/− mice show features of accelerated aging in the liver similar to Ercc1Δ/− mice [72], [82].
Liver cells of ERCC1-deficient and other progeroid NER-deficient mouse mutants display changes in gene expression that encompass a downregulation of catabolic and oxidative metabolism and an upregulation of antioxidant and stress defense pathways, suggestive of a compensatory survival response to cope with increased DNA damage [51], [68], [84], [85]. To determine whether Xpg−/− liver cells also display a ‘survival-like’ stress response, we determined expression levels of selected antioxidant and somatotrophic genes by real-time PCR. Indeed, mRNA levels from a subset of antioxidant effector genes, including Nqo1, Srxn1, Gstt2 and Gsta1, were significantly increased in liver homogenates of young (7-week old) Xpg−/− animals compared to controls. At 14 weeks of age, we observed a similar significant increased expression of Nqo1 and Gsta1 while mRNA from the other antioxidant effector genes tested showed unaltered expression (Figure 3C). Expression levels of Nrf2, which is a potent inducer of the antioxidant response element (ARE), were unaltered, in line with the fact that NRF2-activation is largely achieved by post-translational mechanisms [86]. As increased expression of antioxidant genes could be an indication of increased genotoxic stress, we also checked the expression of the p53-responsive kinase inhibitor p21, a master regulator of cell survival and death [87], which is generally increased after DNA damage and was previously shown to be elevated in livers of Ercc1 mutant mice [88]. Expression levels of p21 doubled at the age of 7 weeks and were massively increased at the age of 14 weeks, indicative of genotoxic stress caused by the absence of XPG. To determine changes in somatotrophic gene expression we examined mRNA levels of Ghr, Igf1r, Igf1 and Igfbp3. We found a two-fold suppression of Ghr and Igf1r mRNA expression at 7 weeks, and a significant downregulation of Ghr mRNA levels at 14 weeks of age (Figure 3D). Together the data indicate that the Xpg−/− liver in part reproduces gene expression changes observed in other short-living NER-deficient mice, which we refer to as a survival-like stress response. Finally, consistent with other NER-deficient progeroid mice [51], [85], we found significantly reduced steady-state blood glucose levels in Xpg−/− mice (Figure 3E).
Age-related accumulation of neurodegenerative changes in Xpg−/− central nervous system
The occurrence of neurological abnormalities and impaired motor behavior in Xpg−/− mice (Figure 2E and S2), as well as the abundant neurodegenerative features in ERCC1-deficient and combined XP/CS mouse models [89]–[91], prompted us to investigate the central nervous systems of Xpg−/− animals for neurodegenerative changes. Macroscopically, the brains and spinal cords of Xpg−/− mice showed a normal appearance, albeit somewhat smaller. In addition, the gross histological organization analyzed in thionin-stained sections appeared normal in all brain regions. As a first step to examine the occurrence of neurodegenerative changes, we examined the brains of 4 - and 14-week old Xpg−/− mice immunohistologically for glial acidic filament protein (GFAP) expression, which outlines reactive astrocytosis in response to neuronal injury. A mild increase in GFAP immunostaining occurred in patches in multiple nervous system areas at 4 weeks (Figure 4A and S4A). Instead, at 14 weeks, Xpg−/− mice showed a prominent ubiquitous increase in GFAP staining throughout the entire central nervous system including spinal cord, indicative of widespread astrocytosis (Figure 4A and S4A). Double-labelling of GFAP and the microglia cell marker Iba-1 showed that the increased GFAP staining was paralleled by microglia activation, characterized by increased Iba-1 immunoreactivity and the transformation of resting microglia cells into activated cells with thicker processes and larger cell bodies (Figure S4B and C).
Fig. 4. Increased cell death, degeneration and stress responses in post-mitotic tissues of Xpg−/− mice. 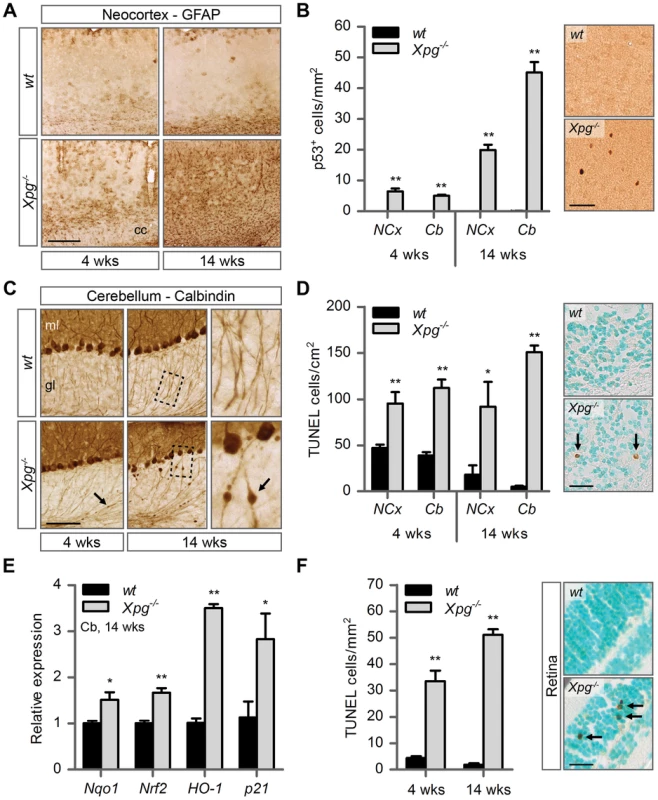
(A) Representative images of GFAP immunostained sagittal neocortex sections of 4- and 14-week old Xpg−/− and wild type (wt) mice showing progressive astrocytosis in Xpg−/− mice. cc: corpus callosum. (B) Quantification of p53-positive cells per mm2 in neocortex (NCx) and cerebellum (Cb) sections of 4- and 14-week old Xpg−/− and wt mice; n = 3 (14 weeks) and the average of five sections of a 4-week old Xpg−/− and wt animal. (C) Representative images of calbindin immunostained sagittal cerebellum sections of 4- and 14-week old Xpg−/− and wt mice. Right panel: Magnification of the areas marked with dotted black boxes. Arrows indicate cerebellar torpedoes. ml: molecular layer, gl: granular layer. (D) Quantification of TUNEL-positive cells per cm2 in neocortex and cerebellum sections of 4- and 14-week old Xpg−/− and wt mice; n≥3 animals/group. Arrows indicate positive cells. (E) Relative mRNA expression levels of the antioxidant genes Nqo1, Nrf2, and HO-1 and the DNA damage response gene p21 in 14-week old Xpg−/− and wt cerebellum tissue. All values are corrected for TubG2 expression and normalized to wt expression levels; n = 4 animals/group. (F) Quantification of TUNEL-positive cells per mm2 in retinal sections of 4- and 14-week old Xpg−/− and wt mice; n = 6 animals/group. Arrows indicate positive cells. Scale bars: 250 µm (A), 50 µm (B), 100 µm (C), 25 µm (D, F). Error bars indicate standard error of the mean. *p<0.05, **p<0.01. Next, to determine whether Xpg−/− central nervous system cells experience genotoxic stress, we studied the expression of the transcription factor p53, which is activated by multiple types of DNA damage and is expressed in neurons and macroglia of many NER-deficient mouse models including mice defective in Ercc1, Csa or Csb [89]–[91]. Immunohistochemistry revealed p53-positive cells in all central nervous system regions. Analysis of the p53 density in neocortex and cerebellum indicated an increase in number of p53-positive cells in brains of 14-week old compared to 4-week old Xpg−/− mice (Figure 4B). Similar to our findings in other NER mutant mice [89]–[91], double labelling of p53 with neuronal (NeuN) and astrocytic (GFAP, S100β) markers, indicated that these p53-positive cells include neurons, astrocytes (GFAP+ or S100β+; Figure S4D), and oligodendrocytes. Although not systematically investigated, we also noted that, as in other NER-deficient mice, in neocortex and cerebellar cortex the majority of p53-positive cells were neurons, while in spinal cord a large proportion of p53-positive cells were astrocytes (Figure S4E).
To obtain evidence for the occurrence of neuronal death, we analyzed calbindin staining in cerebellar cortex where it outlines Purkinje cells and enables easy detection of the degeneration of these cells [90]–[92]. Calbindin staining revealed degeneration and loss of Purkinje cells in 14-week old Xpg−/− mice (Figure 4C). Also, calbindin staining revealed sporadic Purkinje cells with abnormal dendritic morphologies and, more frequently, Purkinje cells with swellings in their proximal axon (Figure 4C). Axonal swellings (also designated torpedoes or axonal spheroids) are a common feature in neurodegenerative disorders and aging [93], that is also well documented for Purkinje cell axons of CS and XP/CS patients [94]. The presence of axonal pathology indicates that many surviving Purkinje cells in 14-week old Xpg−/− mice display compromised health. Notably, few small axonal swellings occurred in Purkinje cells axons in 4-week old Xpg−/− mice.
To further examine the extent to which neurons in Xpg−/− mice show compromised health, we examined the morphology of the Golgi apparatus in motor neurons. In a previous study in Ercc1 mutant mice we noted that motor neurons displayed a variety of morphological abnormalities of the Golgi apparatus, and we proposed that these abnormalities reflect a heterogeneity of cellular deficits resulting from stochastic DNA damage [90]. Immunostaining for the cis-Golgi marker GM130 showed that motor neurons in Xpg−/− mice developed the same heterogeneity in morphological abnormalities of the Golgi apparatus as observed in Ercc1Δ/− mice. Double labelling of GM130 and p53 indicated that only a small subset of neurons with abnormal Golgi apparatus is p53 positive. This variability in p53 expression further illustrates the heterogeneity of degenerative events that may occur in Xpg−/− neurons (Figure S4F).
TUNEL staining to determine the amount of apoptotic cells showed a significant increase in both the cerebrum and the cerebellum at 4 as well as 14 weeks of age (Figure 4D). Finally, real-time PCR in Xpg−/− cerebellum revealed an upregulation of the p53-responsive kinase inhibitor p21 consistent with the activation of genotoxic stress pathways (Figure 4E), and increased expression of several oxidative stress response genes (Figure 4E).
In addition to the brain and spinal cord, we also investigated the retina, as retinal degeneration is a frequent symptom of CS and XP/CS patients [38], that is also reproduced in CSA - and CSB-deficient mice [23]. TUNEL staining revealed cell death in both the inner and outer nuclear layers of the retina of 4 - and 14-week old Xpg−/− mice (Figure 4F). Hence, Xpg−/− mice display loss of photoreceptor cells as well as degeneration of the retinal circuitry. Together these data indicate the occurrence of widespread progressive degenerative changes in Xpg−/− nervous system, strongly resembling the phenotype of ERCC1-deficient mice.
Liver-specific inactivation of XPG results in progeroid features in the liver without causing early death or reduced body weight
Transgenic expression of ERCC1 in the liver has been shown to alleviate growth deficiency and to extend lifespan of ERCC1-deficient mice [83], suggesting that liver abnormalities are an important determinant of the reduced lifespan of these mice. To determine the importance of liver pathology in the runting and the reduced lifespan of our Xpg−/− mice, we generated mice with liver-specific inactivation of the Xpg gene by crossing Xpgf/+ mice carrying the floxed Xpg allele with heterozygous Xpg+/− mice that also express the albumin-Cre recombinase transgene that drives Cre expression specifically in hepatocytes [95] to yield Xpgf/−/Alb-Cre+ mice, hereafter designated Alb-Xpg mice. This Alb-Xpg mouse also has the advantage that it allows to study the effect of liver XPG-deficiency in the absence of abnormalities in other tissues.
A cohort of Alb-Xpg mice was allowed to reach the age of one year. All Alb-Xpg mice displayed normal growth and weight gain, and none of them died prematurely (Figure 5A). Livers of Alb-Xpg mice had an increased size compared to wild type, while brain, kidney and spleen displayed unaltered size and weight (Figure S5A). Albumin and glucose blood levels were the same as in control mice (Figure S5B and C). Histological analysis revealed anisokaryosis with karyomegaly in the liver of Alb-Xpg mice analyzed at 26 and 52 weeks (Figure 5B). The observed karyomegaly was more prominent than that observed in 14-week old Xpg−/− mice, and cells with intranuclear inclusions were more frequent. In addition, we identified p53-positive cells, increased cell death and increased cell proliferation in Alb-Xpg liver consistent with a progeroid degenerative phenotype (Figure 5C–E). Furthermore, real-time PCR showed that livers of Alb-Xpg mice displayed a massive induction of the DNA damage response gene p21 as well as increased expression of several antioxidant effector genes (Figure 5F). We also observed a trend of reduced expression of Ghr and Igf1r (Figure 5G), hence, reproducing gene expression changes determined in livers of Xpg−/− mice. Activation of the Nrf2 antioxidant response genes, reduction of the IGF1 axis, and increased proliferation shown by Ki67-staining are all consistent with liver regeneration after tissue damage [96]. As an additional control we showed that the expression of these genes is unaltered in livers from Emx1-Xpg mice that are XPG-deficient in the dorsal forebrain (see below; Figure S5D and E).
Fig. 5. Aging features observed in the liver of liver-specific Xpg knockout mice. 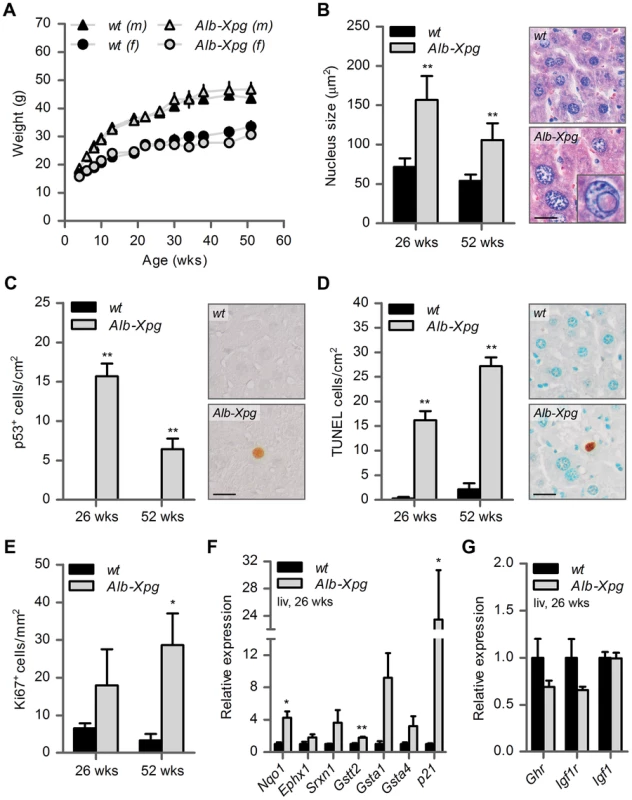
(A) Average body weight of C57Bl6/FVB F1 hybrid wild type (wt) males (black triangles), wt females (black circles), liver specific XPG-deficient (Alb-Xpg) males (gray triangles) and Alb-Xpg females (grey circles); n = 4 males/group, n = 2 females/group. (B) Average nucleus size of hepatocytes in the liver of 26- and 52-week old Alb-Xpg and wt mice; n = 4 animals/group. Bottom right: magnification of a nuclear inclusion found regularly in liver sections of 26- and 52-week old Alb-Xpg mice. (C) Quantification of p53-positive cells per cm2 in the liver of 26- and 52-week old Alb-Xpg and wt mice; n = 3 animals/group. (D) Quantification of TUNEL-positive cells per cm2 in the liver of 26- and 52-week old Alb-Xpg and wt mice; n = 3 animals/group. (E) Quantification of Ki67-positive cells per mm2 in the liver of 26- and 52-week old Alb-Xpg and wt mice; n = 3 animals/group. (F) Relative mRNA expression levels of several antioxidant genes and the DNA damage response gene p21 in liver tissue of 26-week old Alb-Xpg and wt mice. All values are corrected for TubG2, Hprt, and Rps9 expression and normalized to wt expression levels; n = 3 animals/group. (G) Relative expression levels of the somatotrophic genes Ghr, Igf1r and Igf1 in liver tissue of 26-week old Alb-Xpg and wt mice. All values are corrected for TubG2, Hprt, and Rps9 expression as internal standard and normalized to wt expression levels; n = 3 animals/group. Scale bars: 25 µm (B, C, D). Error bars indicate standard error of the mean. *p<0.05, **p<0.01. Together the data from the Alb-Xpg mice show that progeroid and gene expression changes in the liver triggered by the absence of XPG are not sufficient to explain the runted short-living phenotype of Xpg−/− mice.
Dorsal forebrain specific inactivation of XPG results in a mixed macroglia/neuronopathy in cortex and hippocampus
Our neuropathological analyses of Xpg−/− mice uncovered severe neurodegenerative changes at 14 weeks of age, compatible with neurological and motor deficits in these mice. Importantly, the presence of p53 in glia and abundant astrocytosis and microgliosis in the white matter indicate that abnormalities in Xpg−/− mice are not limited to neurons, but also involve glia cells. This is consistent with our findings in CS mouse models [62], [91], and with the neuropathological changes found in CS and XP/CS patients that is dominated by white matter pathology, in addition to neuronal, glial and vascular pathology, and, in severe cases, developmental abnormalities [39], [94], [97]–[99]. In previous studies, using a Cre-lox approach with CamKIIα-Cre and L7-Cre transgenic mice that drive Cre expression in post-mitotic forebrain neurons and Purkinje cells, respectively, we showed that neuron-specific deficiency of ERCC1 or combined deficiency of XPA and CSB was sufficient to trigger stochastic degeneration of these neuronal populations [89], [91], [92]. These studies showed that neurodegenerative changes in ubiquitous ERCC1 - and XPA/CSB-deficient mice are not a consequence of developmental abnormalities, vascular problems, or degenerative changes in other organs. Furthermore, these neuron-specific mice enabled us to follow the degenerative process beyond the normal lifespan of the short-living ubiquitous ERCC1 - and XPA/CSB-deficient mice [89], [91], [92]. In the present study we therefore used a similar approach, but with an Emx1-Cre transgenic line that drives Cre expression in progenitor cells of the dorsal telencephalon, to achieve inactivation of the Xpg gene not only in excitatory neurons of the neocortex and hippocampus, but also of astrocytes and oligodendrocytes in these brain areas [100]. Analysis of a cohort of Emx1-Xpg mice that were allowed to age for one year revealed no early death and showed that body weights were indistinguishable from that of wild type littermates until the age of 30 weeks. At older age the mean weight gain of Emx1-Xpg mice was significantly smaller than in control littermates (Figure 6A). This difference in weight was associated with a proportional reduced weight of internal organs (Figure S6A). Basal blood glucose concentrations were the same as in controls, indicating that reduced weight of old Emx1-Xpg mice is not a consequence of reduced energetic intake (Figure S6B).
Fig. 6. Age-related increase of neuronal stress in forebrain-specific Xpg knockout mice. 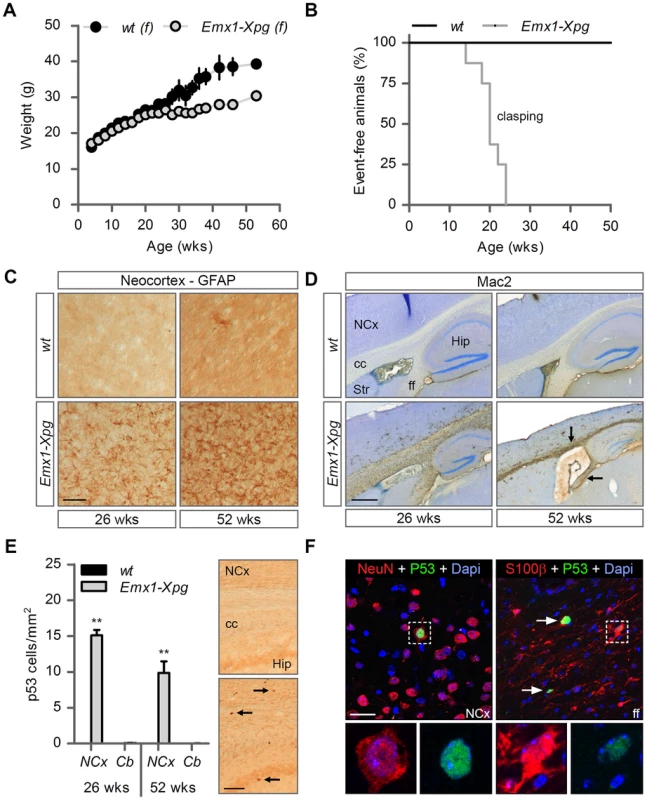
(A) Average body weight of C57Bl6/FVB F1 hybrid wild type (wt) females (black circles) and forebrain-specific XPG-deficient (Emx1-Xpg) females (gray circles); n≥4 animals/group. (B) Onset of clasping of the hind limbs in Emx1-Xpg mice; n = 7 animals/group. (C) Representative images of GFAP immunostained sagittal neocortex sections of 26- and 52-week old Emx1-Xpg and wt mice showing progressive astrocytosis in Emx1-Xpg mice. (D) Representative images of Mac2 immunostained sagittal brain sections of 26- and 52-week old Emx1-Xpg and wt mice showing Mac2-positive microgliosis and a progressive decrease in size of the cerebral cortex and hippocampus of Emx1-Xpg mice. Arrows indicate microgliosis in corpus callosum and fimbria fornix. A thionin counterstaining was used. (E) Quantification of p53-positive cells in neocortex and cerebellum of 26- and 52-week old Emx1-Xpg and wt mice. Values are the average of four sections per genotype. Arrows indicate p53 positive cells. (F) Representative confocal images showing double labeled p53-NeuN cells in the neocortex (left) and p53-S100ß in the fimbria fornix (right) of 26-week old Emx1-Xpg mice. Arrows indicate p53 positive cells. NCx: neocortex, cc: corpus callosum, Str: striatum, ff: fimbria fornix, Hip: hippocampus. Scale bars: 50 µm (C), 500 µm (D), 200 µm (E) and 20 µm (F). Error bars indicate standard error of the mean. **p<0.01. To obtain a crude impression of the development of neurological symptoms, we examined the time of onset of clasping of the hind limbs when animals were lifted by their tails. This abnormality developed in Emx1-Xpg between 14–24 weeks of age and was not observed in control littermates (Figure 6B). The Emx1-Xpg mice did not develop tremors and deficits in accelerating rotarod performance (Figure S6C). Both the absence of these motor deficits and the delayed onset of clasping in comparison to the Xpg−/− mice can be explained by the selective inactivation of Xpg in neocortex and hippocampus, avoiding the bulk of circuitries controlling motor behavior in mice [101].
Macroscopic inspection of brains of Emx1-Xpg mice at 26 - and 52-week already revealed that the neocortex was considerably smaller. Histological analysis confirmed that the neocortex was thinner, and showed that also the hippocampus was dramatically smaller, while other brain regions were unaltered (Figure S6D). GFAP immunohistochemistry showed a very strong increase in GFAP staining indicative of astrocytosis in both cortex and hippocampus (Figure 6C), and no changes in other brain regions. As in the Xpg−/− nervous system, astrocytosis was paralleled by microgliosis, identified by immunohistology with an Iba-1 antibody. Staining for Mac2 (also known as galectin-3), to outline phagocytosing microglia cells [102], revealed very high levels of Mac2-positive cells in the corpus callosum and the fimbria fornix of Emx1-Xpg mice (Figure 6D and S6D,E), and a moderate amount of phagocytosing microglia in the neocortex and hippocampus (Figure 6D). Remarkably, the presence of Mac2 could not be explained by axonal degeneration of cortical and hippocampal neurons solely, as we did not observe this phenomenon in our CamKIIα-Ercc1 mice [89]. Furthermore, the capsula interna of Emx1-Xpg mice, which contains the descending corticofugal axons and shows severe axonal degeneration, did not show this dramatic increase of Mac2 staining (Figure 6D and S6D). Hence, the presence of high levels of Mac2 labelling may reflect the same phenomenon that we observed in our CS mice, i.e. the presence of phagocytosing microglia in the absence of axonal degeneration [91]. Accordingly, we also found an upregulation of Hsp25 expression in astrocytes in the corpus callosum and fimbria fornix (Figure S6F), a phenomenon that we also observed in the white matter of CS mouse models [91]. Furthermore, analysis of p53 expression in Emx1-Xpg showed that multiple p53-positive cells populated the fimbria-fornix and the corpus callosum (Figure 6E). Double-labelling of p53 with NeuN or the glia marker GFAP and S100β showed that some p53-positve cells were neurons (NeuN), but a large proportion were astrocytes (GFAP+, S100β+), or oligodendrocytes as identified on the basis of nuclear morphology (Figure 6F). Together the data indicate that Emx1-Xpg mice develop a combined neuro - and gliopathy of cortex and hippocampus.
Discussion
XPG is a 3′-endonuclease that is essential for the excision step of the GG-NER and TC-NER subpathways of NER and that has additional, yet incompletely defined, roles in other repair processes and possibly in transcription [15], [16], [31], [53], [103]–[105]. Consistent with multiple diverse functions, mutations in XPG cause a spectrum of disease phenotypes varying from the UV-sensitivity disorder XP to the multisystem developmental/degenerative disorders XP/CS and COFS [52]–[55], [57]. Several Xpg mouse models have been generated that partially recapitulate the spectrum of genetic defects and disorders found in patients [63]–[66]. To better define the effect of the complete absence of XPG, in the present study we have generated mice with a Cre-inducible Xpg knockout allele and examined the phenotypes of total (designated Xpg−/−), liver-specific (Alb-Xpg), and dorsal forebrain-specific (Emx1-Xpg) XPG-deficient mice. Consistent with previous data [63] we found that Xpg−/− mice are born with normal size and weight, but progressively fail to grow and to gain weight after birth, combined with a short lifespan. In addition, we found that lifespan was influenced by genetic background, in that Xpg−/− mice displayed a much shorter lifespan in C57BL6 background (3 weeks), compared to FVB or C57BL6/FVB F1 hybrid background (16–18 weeks). An effect of genetic background is well known for many mouse models of disease and was also noted in our Ercc1-mutant mice [51], [72]. Differences in genetic background likely explain the differences in lifespan of our C57BL6/FVB F1 hybrid Xpg−/− mice and the Xpg−/− mice of Harada and coworkers [63], which all died at 3–4 weeks of age similar to our C57BL6 Xpg−/− mice. However, what mechanism defines the short lifespan of our C57BL6 Xpg−/− mice remains to be determined. Importantly, the effect of genetic background in mice suggests that genetic context may also impact the disease phenotype in CS and XP/CS patients. Interestingly, preliminary analysis of neuropathological changes of our C57BL6 Xpg−/− mice at 3 weeks of age revealed subtle degenerative changes (Figure S7) that are comparable in severity to the neurodegenerative changes of C57BL6/FVB Xpg−/− mice of 4 weeks of age, and considerably milder than those in 14-week old C57BL6/FVB F1 hybrid Xpg−/− mice (compare Figure S7 with Figure 4). These data suggest time-dependence of the neurodegenerative phenotype and indicate that the much shorter lifespan of C57BL6 Xpg−/− mice cannot be explained by enhanced neurodegeneration. An alternative possibility suggested by the data of Harada and coworkers [63] is that C57BL6 Xpg−/− mice display an exaggerated developmental phenotype with more prominent developmental deficits, such as an improperly developed gastro-intestinal tract, that would reduce their ability to cope with extra-uterine life. Recent evidence from XPA/CSA-deficient mice bred in C57BL6 background suggest that providing mutant pups with soft food before weaning is sufficient to increase survival from 3–4 weeks to 12–18 weeks [106]. So far, we found no beneficial effect of soft food in our C57BL6 Xpg−/− mice, but this remains to be further examined. Nevertheless, together the data suggest that the ability of short-living NER mutant mice to survive beyond weaning might also depend on the ability to cope with the transition from mother milk to other food.
Xpg−/− mice as a progeria model
The main goal of the present study was to determine the extent to which XPG deficiency in mice results in multisystem progeroid degenerative changes as observed in CS and XP/CS patients, as well as in other NER-deficient mouse models including Xpa/Csb, Xpd, and Ercc1 mutants [48], [51], [68], [69], [72], [77], [107]. Our data show that, although seemingly normal at late embryonic stage, and showing only mild degenerative features at 4 weeks of age, C57BL6/FVB Xpg−/− at 14–16 weeks showed abundant degenerative changes in multiple tissues, indicative of accelerated aging. These degenerative/accelerated aging features included kyphosis, cachexia, osteoporosis, liver aging, and abundant nervous system degenerative changes. In addition, the data from our liver - and dorsal forebrain-specific Xpg mice showed that a much more severe tissue-specific degenerative phenotype could be obtained in these mice because of survival beyond the maximal lifespan of Xpg−/− mice. Hence, the data indicate that Xpg−/− mice indeed develop a multisystem premature aging phenotype reminiscent of the phenotypes of some of our other NER-deficient mouse models, in particular the Ercc1Δ/− mice, which show a slightly longer lifespan and comparable pathological changes in liver and nervous system (see below). Moreover, the Xpg−/− mouse phenotype shares many features with XP-G/CS patients [17], [94], [108] including a cachectic ‘frail’ appearance with loss of subcutaneous fat and signs of osteoporosis (Table 2).
Tab. 2. Comparison of the Xpg−/− phenotype to that of XP/CS patients. 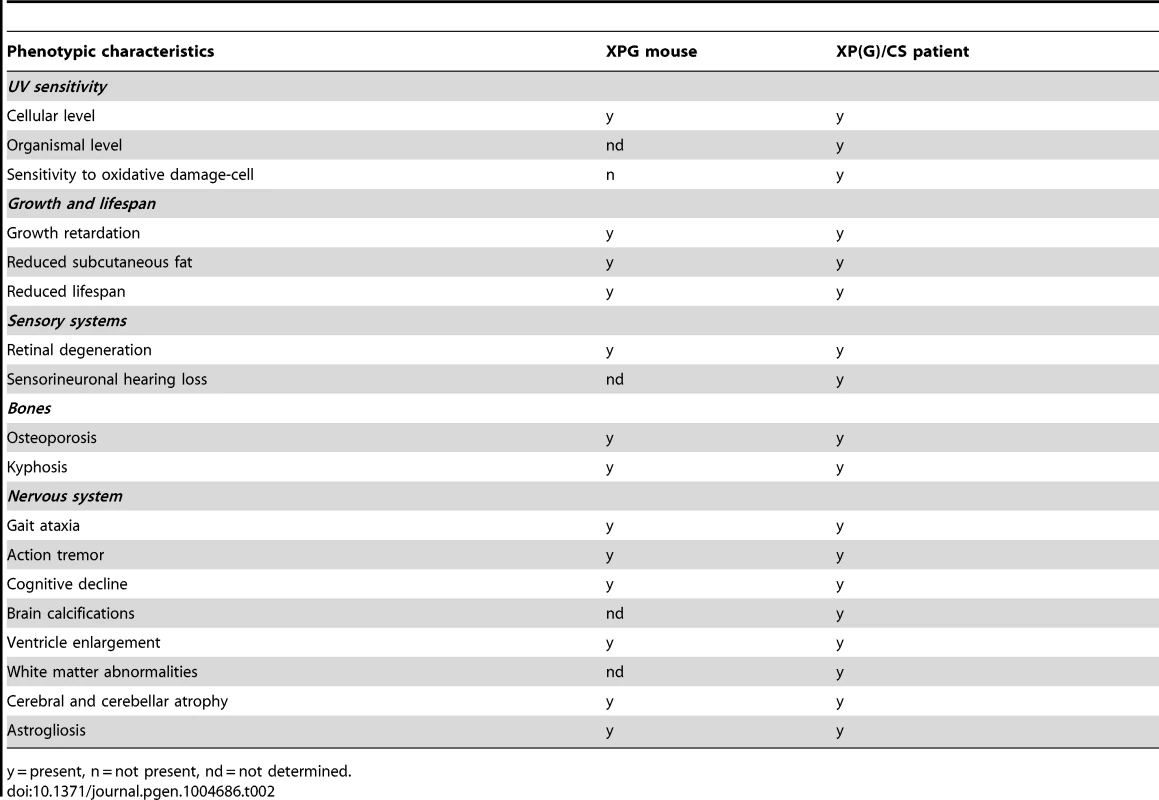
y = present, n = not present, nd = not determined. Compensatory survival response in Xpg−/− liver
Previously we have shown that transcription-blocking lesions trigger a “survival response” involving somatotroph attenuation and increased resistance to oxidative stress, which resembles the response triggered by dietary restriction and is associated with delaying many aspects of aging and increasing lifespan [51], [68], [84]. This mechanism has been observed in response to direct but persistent DNA damage as well as during the course of aging, both in natural aging as well as in specific progeroid mouse models, and is activated in long-lived mutant mice and centenarians [51], [68], [109]. A central hallmark of this survival response is the downregulation of genes involved in the GH/IGF axis, combined with upregulation of genes involved in the antioxidant response. Using real-time PCR for selected genes of these pathways in the present study, we show that Xpg−/− mice display survival-response-like changes in gene expression in liver cells, including a decrease in Ghr mRNA levels and a robust increase in expression of the antioxidant defense effector genes Nqo1, Srxn1, Gstt2, and Gsta1. Similar changes were observed in the liver of liver-specific XPG-deficient mice. Consistent with previous observations concerning the survival response, we found significantly reduced circulating glucose levels in the Xpg−/− mouse. However, we found no change in glucose in the liver-specific mouse, indicating that reduced glucose levels are not necessarily a consequence of gene expression changes in the liver. We did not observe significantly lower levels of Igf1 and Igf1r, as previously found for several other progeroid DNA repair mutants [51], [84], although we did find a trend of reduced expression.
Increased expression of antioxidant genes was also observed in the central nervous system (i.e. cerebellum) of Xpg−/− mice, but the identity and the degree of changed expression is somewhat different. For instance, Nrf2 shows a relatively increased expression in cerebellum but not in liver, while Nqo1 shows a very large relative increase in liver, and a modest relative increase in cerebellum. These differences may be explained by altered stress responses in different cell types [110]. In addition, in the nervous system, changes in gene expression may result from the death and disappearance of neurons and reactive proliferation of glial cells and as a consequence, reduced and increased expression of neuronal and glial genes, respectively.
Progeroid features in the livers of Xpg−/− animals are not sufficient to cause reduced growth and lifespan
The liver of Xpg−/− mice showed several characteristics also found in aging liver, i.e. increased nuclear size and the presence of nuclear inclusions. These changes were much more prominent and associated with additional degenerative changes in liver-specific XPG-deficient mice. However, despite prominent liver pathology, the liver-specific Xpg−/− mice at 52 weeks did not show altered weight, nor altered glucose and albumin levels suggestive of metabolic problems. Although at this point we cannot exclude that our liver-specific mutants will develop health problems in their second year of life and might develop a shorter than normal lifespan, it is safe to conclude that liver problems on their own cannot be the main culprit of the reduced lifespan and the small cachectic appearance of Xpg−/− mice. In contrast to our data, findings in Ercc1 mutant mice show that transgenic overexpression of ERCC1 in the liver alleviates growth deficiency and extends lifespan [83], indicating that liver abnormalities are an important determinant of the reduced lifespan of these mice. A possible explanation is that liver pathology is more prominent in Ercc1 mutant mice, putatively as a consequence of additional defects in interstrand crosslink and double-strand break repair [48]. An alternative explanation for the differences in results could be that the impact of liver pathology on survival depends on the severity of pathology in other organs. This hypothesis is testable by studying survival of liver-specific ERCC1-deficient mice, and vice versa by generating liver-corrected Xpg−/− mice using a transgenic ‘rescue’ strategy as reported by Selfridge et al. [83]. A possible conclusion from these studies could be that Ercc1 and Xpg mutant mice die prematurely as a consequence of a synergistic deleterious effect of multi-organ failure. In view of the severe neurological and neurodegenerative deficits in 14–16 week old Xpg−/− mice, yet an alternative scenario is that Xpg−/− mice die as a consequence of nervous system abnormalities (see below). This scenario is realistic for a subset of human NER-deficient patients, in particular those developing XP with neurological abnormalities [36], [40]. Interestingly, our dorsal forebrain-specific XPG-deficient mice also showed reduced weight-gain after 30 weeks of age. At this age these mice displayed severe neuronal degeneration in neocortex and hippocampus, as a result of which the mice would be expected to have severe cognitive deficits, whereas basal motor functions remain unaltered. Weight loss was not associated with altered glucose levels or gene expression changes in the liver (Figures S5 and S6). We noted the same reduced weight-gain in our forebrain neuron-specific XPA/CSB-deficient mouse model [91]. The data suggest that severe disruption of cortical or hippocampal circuitries may result in the weight loss, but the precise mechanism remains to be defined.
XPG deficiency causes age-dependent degeneration of neurons and macroglia
Our data show that Xpg−/− mice, within their relatively short lifespan (16–18 weeks), develop neurological abnormalities of increasing severity in association with degenerative changes throughout the nervous system. These nervous system degenerative changes seemingly are more severe than those previously reported for another line of Xpg−/− mice [111]. The differences in nervous system neurodegenerative changes can be explained by the very short lifespan (3 weeks) of previously reported Xpg−/− mice, since our C57BL6 Xpg−/− mice, which show the same short lifespan, also displayed a low level of nervous system pathology at the end of life (see above and Figure S7). Together with our demonstration that dorsal forebrain-specific XPG-deficient mice, allowed to age for one year, display very severe neurodegenerative changes, our data indicate that age is a key determinant in the development of neurodegenerative changes in Xpg−/− mice. Our data also indicate that XPG deficiency does not result in obvious neurodevelopmental abnormalities, although subtle developmental abnormalities cannot be excluded at this point.
The neurodegenerative changes in our Xpg−/− mice strongly resemble those of incomplete ERCC1-deficient Ercc1Δ/− mice that have a slightly longer lifespan (24–30 weeks). Our studies with Ercc1Δ/− mice and neuron-specific Ercc1 knock-out animals indicate that ERCC1-deficient neurons in time stochastically accumulate structural and functional deficits to eventually die and disappear [89], [90], [92], [112]. A stochastic accumulation of deficits also appears to occur in Xpg−/− neurons. Thus, the widespread distribution of astrocytosis and microgliosis, as well as p53 and TUNEL staining, indicates that degenerative changes in the Xpg−/− nervous system affect all neuronal populations. The increased size and frequency of axonal spheroids in Purkinje cells, and a diversity of Golgi apparatus morphological abnormalities in motor neurons, on the other hand, illustrate that Xpg−/− neurons may asynchronously accumulate a variety of degenerative features over time. This widespread and asynchronous accumulation of cellular damage in Xpg−/− neurons is consistent with a model in which neurons are afflicted by stochastic DNA lesions that deregulate gene expression, as we have proposed for Ercc1Δ/− mice [89]. A prime role for genotoxic stress in causing the degenerative phenotype in the Xpg−/− nervous system is further suggested by the presence of stochastically distributed p53-positive cells and increased expression of the DNA damage responsive p21 gene.
Together the degenerative changes in the Xpg−/− nervous system favor a pathogenic model involving deficient DNA repair in the same way as proposed for ERCC1-deficient mice [48], [89]. The central role of DNA damage in the Xpg−/− nervous system raises questions about the identity of the DNA lesions involved. Importantly, the absence of a significant neurodegenerative phenotype in entirely NER-deficient Xpa−/− mice has led to the conclusion that DNA lesions that accumulate in the nervous system in the absence of NER are not sufficient to trigger neuronal degeneration within the normal lifespan of mice [74], [91], [113]. In the case of ERCC1-deficient mice, it therefore has been proposed that the degenerative nervous system changes result from combined deficiencies in NER and other DNA repair pathways, i.e. interstrand crosslink repair and double-strand break repair [48], [89]. It is possible that a similar mechanism may contribute to the severe neurodegenerative phenotype caused by XPG deficiency, as XPG has been reported to play a role in repair of crosslinks induced by mitomycin C [114]. An alternative possibility is that XPG deficiency reproduces the severe degenerative phenotype resulting from crossing NER-deficient Xpa−/− with the Csb−/− or Csa−/− CS mouse models [67]–[69], [91], [106]. In this scenario XPG deficiency combines the deleterious synergistic interaction between NER deficiency and loss of the yet incompletely understood non-NER activities that underlie CS. CS proteins operate together at the interface of DNA repair and transcription regulation, and several mechanisms have been put forward to explain CS symptoms [46].
XPG can interact directly with both CSB and RNA Polymerase II [31], and may be implicated in the repair of transcription blocking oxidative lesions, for instance by recruiting base excision repair factors [32]–[34]. Accordingly, cells from CS patients including XP-G/CS cells have been found to display increased vulnerability to inducers of oxidative DNA lesions [55], [115], [116]. Importantly, Soltys et al. showed oxidative damage sensitivity of XP-G/CS, but not of XP-G patient cells [55]. Yet, in accordance with findings by Harada and coworkers [63], we found that cells from our Xpg−/− mice do not display increased vulnerability to inducers of oxidative DNA lesions such as KBrO3 (Figure S1B). This is despite clear indications of endogenous damage in, for example, the retina of these mice, as has been previously observed for Csb/Xpa mice that are sensitive to oxidative damage [23]. It is currently unclear whether this is a peculiarity of these particular mouse cells in culture, as we have reported that cells from CSB-deficient mice are sensitive to IR and paraquat [117], [118]. It has been argued that cultured cells may build up defense responses that mask the increased vulnerability of these cells in vivo [119].
To what extent do Xpg−/− mice reproduce the nervous system abnormalities of patients carrying XPG mutations? Roughly, the progressive widespread neurodegenerative changes of Xpg−/− mice are reminiscent of neuropathological changes of patients with ‘XP-type neurological degeneration’ [39], [40], [97], [120]. In well documented cases these patients, carrying XPA mutations resulting in complete NER deficiency, develop a wide array of neurological symptoms that show early juvenile onset, over time become more severe, and ultimately cause premature death in mid-adult life [39], [40], [97]. However, the limited documented cases indicate that XP-G patients either develop no neurological symptoms, or reproduce mild to severe neurological and neuropathological features of CS [39], [40], [57], [94], [120]. In CS and XP/CS patients, neuronal degeneration generally is less prominent. Instead, these patients, including documented XP-G/CS cases, show prominent white matter degeneration, vascular pathology, calcium depositions, and, in severe cases, developmental abnormalities [39], [94], [97]–[99]. We have recently noted that CSA - and CSB-deficient CS mouse models, in addition to mild neurodegenerative changes, develop subtle white matter abnormalities and glial pathology reminiscent of the glia and white matter degenerative changes of CS patients, albeit milder [62], [91]. The higher levels of neuronal degeneration in Xpg−/− mice hamper detection of primary glial and white matter pathology, due to secondary glial pathology caused by neuronal degeneration. However, the severe white matter pathology in the corpus callosum and fimbria-fornix in our dorsal forebrain specific XPG-deficient mice strongly indicates that XPG deficiency triggers CS-like white matter pathology in mice.
Concluding remarks
In this study, we show that Xpg−/− mice from young age onwards develop a multisystem degenerative phenotype and die before the age of 20 weeks. This phenotype strongly resembles the progeroid features of CS and XP/CS patients. In addition, the Xpg−/− mouse model shows a number of similarities to other NER-based mouse models of progeria such as Xpa/Csb and Ercc1 mutants [3], [62], pointing to the importance of NER in multiple tissues. In particular, a detailed analysis of commonalities and differences between Xpg−/− and Ercc1 mutant mice may aid in our understanding of the contribution of different types of DNA damage and DNA repair defects in the accelerated aging process, since both endonucleases have a joint role in the damage excision step of NER but have divergent additional non-NER roles. Together our findings further stress the relationship between compromised DNA repair and acceleration of specific aging features, as well as progressive neurodegeneration. Finally, the neurodegenerative phenotype indicates that Xpg−/− mice may serve as a model to test intervention strategies aimed at reducing the formation of detrimental DNA lesions in neurons.
Materials and Methods
Generation of a floxed Xpg allele
The Xpg targeting construct was generated using multiple elements. First, a cassette consisting of a Neomycin (NEO) resistance marker, flanked by Frt sites, and followed by a single LoxP site was cloned into a modified pBlueScript SK+ vector containing a PGK-DTA negative selection marker, making use of a klenow blunted ApaI (insert)/XbaI(vector) and a NotI restriction site. Second, Xpg homologous arms were PCR amplified from C57BL6 genomic DNA (originating from BAC clone RP24-343K18) and cloned into the same plasmid. The following primers (non-homologous regions indicated in italics; the LoxP sequence is underlined) were used for amplification of the 5′ and 3′ arm, respectively: LAF2 (5′-CGCACCCGGGTGTGATCCTGTGGTCCTGTAGT-3′) and LAR2 (5′-CCATCGATATCCTCAGAAAGGTATCTCTTAAGCA-3′), yielding a 3.2-kb XmaI-ClaI fragment; RAF1 (5′-CCCTGCTAGCGGGATGAGGAATCGTGACTAAGGAG-3′) and RAR1 (5′-CCGCAGCGGCCGCAAACAAGGGACCCAAATGTAGGCT-3′), yielding a 2.0-kb NheI-NotI fragment, where the restriction sites were introduced in the PCR primers. Last, the third exon of Xpg followed by a PCR-introduced LoxP site was amplified using the primers Ex3LoxF2 (5′-GGGAACCGGTTTGAGTGTCCTTGGTGACAGG-3′) and Ex3LoxR2 (5′-CCCTGCTAGCATAACTTCGTATAGCATACATTATACGAAGTT ATCC-3′), yielding a 350-bp AgeI-NheI fragment, which was inserted between the neomycin cassette and the 5′ homology arm.
Next, a total of 10 µg of NotI-linearized targeting vector was electroporated to Ola129 ES cells, and the targeted clones were selected through the use of the Neomycin selection marker (G418 200 µg/ml). Clones resistant for G418 were initially screened by PCR, using a forward primer in exon 3 (F3 5′-GAGACAGGCTCTGAAAACTGCTT-3′) and a reverse primer outside the 3′ homologous region (R3 5′-CACTGAACAAACAAGGGACCCAAA-3′). ES clones showing a 2.2-kb fragment in addition to the wild type 2.3-kb fragment after NheI digestion of the PCR product were further screened by Southern blot. ES genomic DNA was digested with EcoRI and hybridized with a 0.9-kb DpnI restriction fragment from BAC RP24-343K18, spanning the 2nd exon of Xpg. The probe hybridizes to a 7.4-kb fragment in wild type DNA and to an additional 4.1-kb fragment in targeted DNA.
ES cells from two independent targeted clones were micro-injected into C57BL6 blastocysts. Heterozygous mutant mice were generated by crossing the male chimeras with C57BL6 females and verified by coat color and PCR genotyping. The Neomycin (NEO) resistance gene was flanked by Frt sites to allow specific elimination of this dominant selectable marker by an Flp recombinase to avoid potential undesired influence of the Neo gene on Xpg transcription or mRNA processing. The NEO cassette was removed by crossing mice carrying the targeted allele with Cag-Flp recombinase FVB/N transgenic animals [121]. These mice carry the floxed allele, and are referred to as Xpgf throughout this paper. Thereafter, the F3 offspring was crossed with a Cag-Cre C57BL6 transgenic [73], resulting in Cre-mediated recombination and excision of the third exon. Xpg+/− animals were backcrossed to C57BL6 and FVB/N in parallel, at least ten times, and interbred to obtain C57BL6, FVB/N and C57BL6/FVB F1 hybrid Xpg−/− mice. To achieve liver specific Xpg gene inactivation, a transgenic line with Cre recombinase under the control of the albumin promoter (hereafter referred to as Alb-Cre) was used [95]. Female Alb-Cre+ mice were crossed with male Xpg+/− mice (both in a C57BL6 background). Female Xpg+/− Alb-Cre+ mice, obtained from these breedings, were crossed with male Xpgf/f FVB/N mice to yield hybrid Xpgf/− Alb-Cre+ mice. Xpgf/− Alb-Cre+ mice (in a C57BL6/FVB F1 hybrid background) are heterozygous for Xpg in their entire body, except for the hepatocytes in the liver, which are homozygous for Xpg after Cre excision of the floxed allele. All littermates, with and without Cre-recombinase expression were used as controls (referred to as wt).
FVB/N Xpgf/f animals were similarly bred to the female offspring from C57BL6 Xpg+/− and C57BL6 Emx1-Cre mice [100] to obtain forebrain specific XPG knock-out animals (referred to as Emx1-Xpg). All animals used in the studies described in this paper were of the same C57BL6/FVB F1 hybrid background (unless otherwise stated) and had ad libitum access to water and standard mouse food (CRM pellets, SDS BP Nutrition Ltd.; gross energy content 4.39 kcal/g dry mass, digestible energy 3.2 kcal/g or AIN93G synthetic pellets, Research Diet Services B.V.; gross energy content 4.9 kcal/g dry mass, digestible energy 3.97 kcal/g). Since the Xpg knockout animals were smaller, food was administered within the cages and water bottles with long nozzles were used from around two weeks of age. Experiments were performed in accordance with the Principles of Laboratory Animal Care (National Institutes of Health publication no. 86-23) and with the guidelines approved by the Erasmus University Animal Care Committee.
Genotyping
For PCR genotyping the following primers were used: F1 forward primer (5′-TCTGTTTAGGTGGTGCCCATTT-3′) annealing 5′ of the third exon of Xpg; NeoF forward primer (5′-GCTTCCTCGTGCTTTACGGTAT-3′) located in the Neomycin resistance marker; R1 reverse primer (5′-CGACAGCACTTCTTTCTCCTTAGT-3′) annealing 3′ of the third exon of Xpg. A 711-bp fragment was generated from the targeted allele using primers NeoF and R1, whereas a 495-bp and 227-bp fragment are amplified from the wild type and knockout allele, respectively, using the F1/R1 primerset. Cycling conditions were 95°C for 45 sec, 58°C for 45 sec, 72°C for 1 min (35 cycles), followed by an extension at 72°C for 5 min.
Sequencing
Total RNA was extracted from wild type and Xpg−/− liver using TRIzol reagent and reverse transcribed with SuperScript II Reverse Transcriptase (Life technologies), according to the manufacturer's instructions, to generate cDNA. A 0.4-kb PCR fragment ranging from the 2nd to 6th of Xpg was produced using the following primers: Ex2F (5′-GCTCATCTTCTCACATTATTCC-3′) and Ex6R (5′ - GGTAAACTCTTTCATGTCAGTC-3′) and analyzed by Sanger sequencing.
Phenotype scoring
The mice were weighed and visually inspected weekly, and were scored for the onset of various phenotypical parameters. Clasping was measured by suspending mice from their tails for 20 seconds. A clasping event was scored when retraction of both hind limbs towards the body was observed for at least 5 seconds. Whole body tremor was scored if mice were trembling for a combined total of at least 10 seconds, when put on a flat surface for 20 seconds. Mice showing an abnormal curvature of the spine were scored as having kyphosis.
Isolation and analysis of MDFs
Primary MDFs were isolated from the tail of 12–14 week old Xpg−/− animals and wild type littermates. Minced tail skin was immersed in F10/DMEM culture medium supplemented with 20% fetal calf serum, 50 µg/ml penicillin/streptomycin, and 1.6 mg/ml type II collagenase (Gibco, Life Technologies). After incubation at 37°C for 24 hours, MDFs were filtered through a 40 µm cell strainer, centrifuged for removal of the collagenase, and cultured at 37°C, 5% CO2, and 3% O2.
Immunoblot analysis of XPG protein in MDFs
Whole cell extracts were prepared from cultured MDFs using cells isolated from four wild type mice and four Xpg−/− littermates. Proteins from 30 µl of each extract were separated by electrophoresis on 7% SDS-PAGE gels and transferred overnight onto a nitrocellulose membrane. As previously described [31], XPG protein was detected with a rabbit polyclonal antibody designated R2 (97727) that was raised against a conserved peptide from the spacer region (R-domain) of XPG corresponding to residues 267–281 of the human protein, which are identical with the same residues in mouse XPG except for amino acid 267, which is E in the human protein but Q in mouse. Whole cell extract from human embryonic kidney 293 cells was used as a positive control for XPG protein. As a loading control, tubulin was detected using a commercial antibody.
Cell survival assays
MDFs were seeded in triplicates at equal densities and treated 24 h after seeding as indicated. After 48 h recovery cell survival was determined by cell count on Beckman Coulter, Z2 Coulter particle count and size analyzer.
Unscheduled DNA Synthesis (UDS)
UDS was performed using the Click-iT EdU imaging kit (Life technologies). MDFs were seeded on coverslips and 24 h later washed with PBS and irradiated with 16 J/m2 UV-C (Philips) or mock treated. Cells were directly labelled for 3 h in thymidine-free Ham's F10 medium supplemented with 10% dialyzed serum, 50 µg/ml penicillin/streptomycin, 20 µM Ethynyl-deoxyuridine and 1 µM Fluoro-desoxyuridine. After a PBS wash, the cells were chased with 10 µM non-labelled thymidine in normal medium for 15 minutes and fixed with 3.7% formaldehyde. Slides were washed 3× with PBS/3%BSA and permeabilized with PBS/0.5%Triton-X100 for 20 min. The Click-iT reaction, linking azide-conjugated Alexa dye to ethynyl groups was performed for 30 min in a dark, humid environment. After 3× PBS/3%BSA and 2× PBS wash, the slides were mounted with vectashield containing DAPI (Vector Laboratories) to stain nuclei.
Recovery of RNA Synthesis (RRS)
RRS was performed using the Click-iT EU imaging kit (Life Technologies). MDFs were seeded on coverslips and 8 h later washed with PBS and irradiated with 16 J/m2 UV-C (Philips) or mock treated. After 14 h recovery, the cells were labelled for 2 h in 0.1 mM EU-containing medium (Ham's F10 medium supplemented with 10% dialyzed serum, 50 µg/ml penicillin/streptomycin and 20 mM HEPES). After a PBS wash, the cells were fixed and processes as described for UDS.
Mechanical testing of bone strength
Force-deflection curves from the left femora of 14-week-old and 16-week-old mice were acquired in a three-point bending assay using a Chatillon TCD series mechanical test frame (Technex BV, The Netherlands), equipped with 3 mm hemi-cylindrical supports with a 8.5 mm total span. Width between the supports was adjusted according to the anatomical landmarks of the femur, i.e. lesser trochanter and condyles. The femora were aligned such that the femoral head was in the horizontal plane and the posterior aspect of the condyles was facing down. All samples were preconditioned for five cycles to 2 Newton (N) at a rate of 0.6 mm/min before testing to failure at a rate of 0.1 mm/min. The obtained force-deflection curves were analyzed for bone strength (N/mm), which was represented by the Δ force/deflection of the linear part of the curve.
Micro-computed tomographic (micro-CT) quantification of bone thickness
Xpg−/− and wild type mice were sacrificed by cervical dislocation at scheduled ages (7, 14, 16, 18 weeks), femora were excised and non-osseous tissue was removed. Left femora were placed in PBS and stored at −20°C until further use for mechanical testing and the right femora were fixated (4% formalin). Two days post-fixation, the right femora were scanned using the Skyscan 1076 in vivo X-Ray computed tomography (Skyscan, Kontich, Belgium) with a voxel size of 8.88 µm. Osseous tissue was distinguished from non-osseous tissue by segmenting the reconstructed grayscale images with an automated algorithm using local thresholds [122]. The region of interest (ROI), i.e. distal metaphysis of the femora, was selected by using 3D data analysis software. To compensate for bone length differences between the Xpg−/− and wild type mice, the length of each ROI was determined relative to the largest specimen femur of the cohort.
Cortex and trabeculae of the metaphysis were separated using in-house developed automated software. Thickness of the trabeculae and cortices were assessed using 3D analysis software as described [123] using the CT analyzer software package (Skyscan). A bone specimen with known bone morphometrics was included within each scan as a quantitative control.
Real-time PCR
TRIzol reagent (Life Technologies) was used to isolate total RNA from mouse tissue specimens. 4 µg RNA was reverse transcribed using SuperScript II (Life Technologies). Real-time PCR was performed on a Bio-Rad CFX96 thermocycler using SYBR Green (Sigma-Aldrich) and Platinum Taq polymerase (Life Technologies). Generation of specific PCR products was confirmed by melting-curve analysis and gel electrophoresis. For data analysis, the induction of target cDNA was calculated by the method described by [124]. p-values were determined using two-tailed t-tests. The used gene specific real-time PCR primers are listed in Supplementary Table S1.
Blood glucose and albumin levels
Mice were euthanized by CO2 asphyxiation and blood was immediately collected from the heart. Glucose levels were measured using a Freestyle mini blood glucose meter. Albumin levels were measured in blood plasma using a mouse albumin ELISA kit (Immunology Consultants Laboratory, Inc.).
Antibodies
Primary antibodies (supplier; dilutions) used in this study were as follows: rabbit anti-Calbindin (Swant; 1∶10,000); goat anti-ChAT (Millipore; 1∶500); rabbit anti-GFAP (DAKO; 1∶8,000); mouse anti-GM130 (BD Transduction; 1∶100); rabbit anti-Hsp25 (Enzo; 1∶8,000); rabbit anti-Iba-1 (Wako; 1∶5,000); rat anti-Ki67 (DAKO; 1∶200); rat anti-Mac2 (Cedarlane;1∶2,000); mouse anti-NeuN (Millipore; 1∶1,000); rabbit anti-p53 (Leica; 1∶1,000). For avidin – biotin–peroxidase immunocytochemistry biotinylated secondary antibodies from Vector Laboratories, diluted 1∶200 were used. FITC-, Cy3-, and Cy5-conjugated secondary antibodies raised in donkey (Jackson ImmunoResearch) diluted at 1∶200 were used for confocal immunofluorescence.
Histological procedures
TUNEL staining on liver, brain and retina: To quantify apoptotic cells, specimens were fixed overnight in 10% buffered formalin, paraffin-embedded, sectioned at 5 µm, and mounted on Superfrost Plus slides. Paraffin sections were employed for TdT-mediated dUTP Nick-End Labeling (TUNEL) assay using a commercial kit (Apoptag Plus Peroxidase in situ apoptosis detection kit, Millipore). Sections were deparaffinized and incubated as described by the manufacturer. HE staining on liver, skin and small intestine: Liver, skin and intestine specimens were processed using the same fixation and sectioning methods as described for the TUNEL staining. Paraffin sections were deparaffinized, rehydrated in decreasing concentrations of ethanol and stained with haematoxilin and eosin.
For immunohistochemistry, paraffin sections of liver and intestine specimens were deparaffinized, rehydrated in decreasing concentrations of ethanol, treated for 10 minutes with 3% H2O2 to quench endogenous peroxidase activity and heated to 100°C for 1 h in 10 mM sodium citrate buffer, pH 6, for antigen retrieval. The amount of damaged (p53) and proliferating (Ki67) cells were subsequently detected using the avidin–biotin–immunoperoxidase complex method (ABC, Vector Laboratories, USA) with diaminobenzidine (0.05%) as chromogen.
Gelatin sections of brain and spinal cord: Mice were anaesthetized with pentobarbital and perfused transcardially with 4% paraformaldehyde. Brain and spinal cord were dissected out, post-fixed for 1 h in 4% paraformaldehyde, cryoprotected, embedded in 12% gelatin, rapidly frozen, and sectioned at 40 µm using a freezing microtome or stored at −80°C until use. Frozen sections were processed free floating using the ABC method (ABC, Vector Laboratories, USA) or single-, double-, and triple-labelling immunofluorescence. Immunoperoxidase-stained sections were analyzed and photographed using an Olympus BX40 microscope. Immunofluorescence sections were analyzed using a Zeiss LSM700 confocal.
Rotarod performance
Average time spent on an accelerating rotarod (Ugo Basile). Xpg−/− mice and wild type controls were given four consecutive trials of maximally 5 minutes with inter-trial intervals of 1 hour. Emx1-Xpg mice were given two trials per day with a 1 hour inter-trial interval for four consecutive days.
Grip strength
Grip strength was determined by placing mice with forelimbs or all limbs on a grid attached to a force gauge, and steadily pulling the mice by their tail. Grip strength is defined as the maximum strength produced by the mouse before releasing the grid. For each value the test is performed in triplicate.
Statement of ethical approvement
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Erasmus MC (Permit Numbers: 139-03-08, 139-09-03, 139-12-18).
Supporting Information
Zdroje
1. HanahanD, WeinbergRA (2011) Hallmarks of cancer: the next generation. Cell 144 : 646–674.
2. Lopez-OtinC, BlascoMA, PartridgeL, SerranoM, KroemerG (2013) The hallmarks of aging. Cell 153 : 1194–1217.
3. HoeijmakersJH (2009) DNA damage, aging, and cancer. N Engl J Med 361 : 1475–1485.
4. HoeijmakersJH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411 : 366–374.
5. FagbemiAF, OrelliB, ScharerOD (2011) Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair (Amst) 10 : 722–729.
6. FriedbergEC, AguileraA, GellertM, HanawaltPC, HaysJB, et al. (2006) DNA repair: from molecular mechanism to human disease. DNA Repair (Amst) 5 : 986–996.
7. ScharerOD (2013) Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol 5: a012609.
8. HanawaltPC (2008) Emerging links between premature ageing and defective DNA repair. Mech Ageing Dev 129 : 503–505.
9. NaegeliH, SugasawaK (2011) The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair (Amst) 10 : 673–683.
10. FousteriM, MullendersLH (2008) Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res 18 : 73–84.
11. VermeulenW, FousteriM (2013) Mammalian transcription-coupled excision repair. Cold Spring Harb Perspect Biol 5: a012625.
12. StaresincicL, FagbemiAF, EnzlinJH, GourdinAM, WijgersN, et al. (2009) Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J 28 : 1111–1120.
13. EglyJM, CoinF (2011) A history of TFIIH: two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair (Amst) 10 : 714–721.
14. Giglia-MariG, CoinF, RanishJA, HoogstratenD, TheilA, et al. (2004) A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet 36 : 714–719.
15. ItoS, KuraokaI, ChymkowitchP, CompeE, TakedachiA, et al. (2007) XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell 26 : 231–243.
16. Le MayN, FradinD, IltisI, BougneresP, EglyJM (2012) XPG and XPF Endonucleases Trigger Chromatin Looping and DNA Demethylation for Accurate Expression of Activated Genes. Mol Cell 47 : 622–632.
17. ScharerOD (2008) XPG: its products and biological roles. Adv Exp Med Biol 637 : 83–92.
18. LakeRJ, FanHY (2013) Structure, function and regulation of CSB: a multi-talented gymnast. Mech Ageing Dev 134 : 202–211.
19. SuY, OrelliB, MadireddyA, NiedernhoferLJ, ScharerOD (2012) Multiple DNA binding domains mediate the function of the ERCC1-XPF protein in nucleotide excision repair. J Biol Chem 287 : 21846–21855.
20. Klein DouwelD, BoonenRA, LongDT, SzypowskaAA, RaschleM, et al. (2014) XPF-ERCC1 Acts in Unhooking DNA Interstrand Crosslinks in Cooperation with FANCD2 and FANCP/SLX4. Mol Cell 54 : 460–471.
21. AhmadA, RobinsonAR, DuensingA, van DrunenE, BeverlooHB, et al. (2008) ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol 28 : 5082–5092.
22. D'ErricoM, ParlantiE, TesonM, de JesusBM, DeganP, et al. (2006) New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J 25 : 4305–4315.
23. GorgelsTG, van der PluijmI, BrandtRM, GarinisGA, van SteegH, et al. (2007) Retinal degeneration and ionizing radiation hypersensitivity in a mouse model for Cockayne syndrome. Mol Cell Biol 27 : 1433–1441.
24. MelisJP, LuijtenM, MullendersLH, van SteegH (2011) The role of XPC: implications in cancer and oxidative DNA damage. Mutat Res 728 : 107–117.
25. TrappC, ReiteK, KlunglandA, EpeB (2007) Deficiency of the Cockayne syndrome B (CSB) gene aggravates the genomic instability caused by endogenous oxidative DNA base damage in mice. Oncogene 26 : 4044–4048.
26. MenoniH, HoeijmakersJH, VermeulenW (2012) Nucleotide excision repair-initiating proteins bind to oxidative DNA lesions in vivo. J Cell Biol 199 : 1037–1046.
27. StevnsnerT, MuftuogluM, AamannMD, BohrVA (2008) The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mech Ageing Dev 129 : 441–448.
28. KlunglandA, HossM, GunzD, ConstantinouA, ClarksonSG, et al. (1999) Base excision repair of oxidative DNA damage activated by XPG protein. Mol Cell 3 : 33–42.
29. BesshoT (1999) Nucleotide excision repair 3′ endonuclease XPG stimulates the activity of base excision repair enzyme thymine glycol DNA glycosylase. Nucleic Acids Res 27 : 979–983.
30. OyamaM, WakasugiM, HamaT, HashidumeH, IwakamiY, et al. (2004) Human NTH1 physically interacts with p53 and proliferating cell nuclear antigen. Biochem Biophys Res Commun 321 : 183–191.
31. SarkerAH, TsutakawaSE, KostekS, NgC, ShinDS, et al. (2005) Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol Cell 20 : 187–198.
32. BanerjeeD, MandalSM, DasA, HegdeML, DasS, et al. (2011) Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J Biol Chem 286 : 6006–6016.
33. ReisAM, MillsWK, RamachandranI, FriedbergEC, ThompsonD, et al. (2012) Targeted detection of in vivo endogenous DNA base damage reveals preferential base excision repair in the transcribed strand. Nucleic Acids Res 40 : 206–219.
34. GuoJ, HanawaltPC, SpivakG (2013) Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res 41 : 7700–7712.
35. MarteijnJA, LansH, VermeulenW, HoeijmakersJH (2014) Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 15 : 465–481.
36. DiGiovannaJJ, KraemerKH (2012) Shining a light on xeroderma pigmentosum. J Invest Dermatol 132 : 785–796.
37. StefaniniM, BottaE, LanzafameM, OrioliD (2010) Trichothiodystrophy: from basic mechanisms to clinical implications. DNA Repair (Amst) 9 : 2–10.
38. LaugelV, DallozC, DurandM, SauvanaudF, KristensenU, et al. (2010) Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat 31 : 113–126.
39. KraemerKH, PatronasNJ, SchiffmannR, BrooksBP, TamuraD, et al. (2007) Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience 145 : 1388–1396.
40. AnttinenAKL, NikoskelainenE, PortinR, KurkiT, ErkinjunttiM, et al. (2008) Neurological symptoms and natural course of xeroderma pigmentosum. Brain 131 : 1979–1989.
41. NakazawaY, SasakiK, MitsutakeN, MatsuseM, ShimadaM, et al. (2012) Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat Genet 44 : 586–592.
42. SchwertmanP, LagarouA, DekkersDH, RaamsA, van der HoekAC, et al. (2012) UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet 44 : 598–602.
43. ZhangX, HoribataK, SaijoM, IshigamiC, UkaiA, et al. (2012) Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet 44 : 593–597.
44. FeiJ, ChenJ (2012) KIAA1530 is recruited by cockayne syndrome complementation group protein A (CSA) to participate in transcription-coupled repair (TCR). J Biol Chem 287 : 35118–35126.
45. NataleV (2011) A comprehensive description of the severity groups in Cockayne syndrome. Am J Med Genet A 155A: 1081–1095.
46. BrooksPJ (2013) Blinded by the UV light: how the focus on transcription-coupled NER has distracted from understanding the mechanisms of Cockayne syndrome neurologic disease. DNA Repair (Amst) 12 : 656–671.
47. ChoI, TsaiPF, LakeRJ, BasheerA, FanHY (2013) ATP-dependent chromatin remodeling by Cockayne syndrome protein B and NAP1-like histone chaperones is required for efficient transcription-coupled DNA repair. PLoS Genet 9: e1003407.
48. GreggSQ, RobinsonAR, NiedernhoferLJ (2011) Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease. DNA Repair (Amst) 10 : 781–791.
49. JaspersNG, RaamsA, SilengoMC, WijgersN, NiedernhoferLJ, et al. (2007) First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am J Hum Genet 80 : 457–466.
50. KashiyamaK, NakazawaY, PilzDT, GuoC, ShimadaM, et al. (2013) Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am J Hum Genet 92 : 807–819.
51. NiedernhoferLJ, GarinisGA, RaamsA, LalaiAS, RobinsonAR, et al. (2006) A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 444 : 1038–1043.
52. LehmannJ, SchubertS, SchaferA, ApelA, LaspeP, et al. (2014) An unusual mutation in the XPG gene leads to an internal in-frame deletion and a XP/CS complex phenotype. Br J Dermatol E-pub ahead of print. doi:10.1111/bjd.13035
53. ScharerOD (2008) Hot topics in DNA repair: the molecular basis for different disease states caused by mutations in TFIIH and XPG. DNA Repair (Amst) 7 : 339–344.
54. SchaferA, SchubertS, GratchevA, SeebodeC, ApelA, et al. (2013) Characterization of three XPG-defective patients identifies three missense mutations that impair repair and transcription. J Invest Dermatol 133 : 1841–1849.
55. SoltysDT, RochaCR, LernerLK, de SouzaTA, MunfordV, et al. (2013) Novel XPG (ERCC5) mutations affect DNA repair and cell survival after ultraviolet but not oxidative stress. Hum Mutat 34 : 481–489.
56. NouspikelT, LalleP, LeadonSA, CooperPK, ClarksonSG (1997) A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function. Proc Natl Acad Sci U S A 94 : 3116–3121.
57. EmmertS, SlorH, BuschDB, BatkoS, AlbertRB, et al. (2002) Relationship of neurologic degeneration to genotype in three xeroderma pigmentosum group G patients. J Invest Dermatol 118 : 972–982.
58. SchaferA, GratchevA, SeebodeC, HofmannL, SchubertS, et al. (2013) Functional and molecular genetic analyses of nine newly identified XPD-deficient patients reveal a novel mutation resulting in TTD as well as in XP/CS complex phenotypes. Exp Dermatol 22 : 486–489.
59. FriedbergEC, MeiraLB (2006) Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage Version 7. DNA Repair (Amst) 5 : 189–209.
60. WijnhovenSW, HoogervorstEM, de WaardH, van der HorstGT, van SteegH (2007) Tissue specific mutagenic and carcinogenic responses in NER defective mouse models. Mutat Res 614 : 77–94.
61. NiedernhoferLJ (2008) Nucleotide excision repair deficient mouse models and neurological disease. DNA Repair (Amst) 7 : 1180–1189.
62. JaarsmaD, van der PluijmI, van der HorstGT, HoeijmakersJH (2013) Cockayne syndrome pathogenesis: Lessons from mouse models. Mech Ageing Dev 134 : 180–195.
63. HaradaYN, ShiomiN, KoikeM, IkawaM, OkabeM, et al. (1999) Postnatal growth failure, short life span, and early onset of cellular senescence and subsequent immortalization in mice lacking the xeroderma pigmentosum group G gene. Mol Cell Biol 19 : 2366–2372.
64. TianM, JonesDA, SmithM, ShinkuraR, AltFW (2004) Deficiency in the Nuclease Activity of Xeroderma Pigmentosum G in Mice Leads to Hypersensitivity to UV Irradiation. Mol Cell Biol 24 : 2237–2242.
65. ShiomiN, KitoS, OyamaM, MatsunagaT, HaradaYN, et al. (2004) Identification of the XPG Region That Causes the Onset of Cockayne Syndrome by Using Xpg Mutant Mice Generated by the cDNA-Mediated Knock-In Method. Mol Cell Biol 24 : 3712–3719.
66. ShiomiN, MoriM, KitoS, HaradaYN, TanakaK, et al. (2005) Severe growth retardation and short life span of double-mutant mice lacking Xpa and exon 15 of Xpg. DNA Repair (Amst) 4 : 351–357.
67. LaposaRR, HuangEJ, CleaverJE (2007) Increased apoptosis, p53 up-regulation, and cerebellar neuronal degeneration in repair-deficient Cockayne syndrome mice. Proc Natl Acad Sci U S A 104 : 1389–1394.
68. van der PluijmI, GarinisGA, BrandtRM, GorgelsTG, WijnhovenSW, et al. (2007) Impaired genome maintenance suppresses the growth hormone–insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol 5: e2.
69. MuraiM, EnokidoY, InamuraN, YoshinoM, NakatsuY, et al. (2001) Early postnatal ataxia and abnormal cerebellar development in mice lacking Xeroderma pigmentosum Group A and Cockayne syndrome Group B DNA repair genes. Proc Natl Acad Sci U S A 98 : 13379–13384.
70. AndressooJO, WeedaG, de WitJ, MitchellJR, BeemsRB, et al. (2009) An Xpb mouse model for combined xeroderma pigmentosum and cockayne syndrome reveals progeroid features upon further attenuation of DNA repair. Mol Cell Biol 29 : 1276–1290.
71. AndressooJO, MitchellJR, de WitJ, HoogstratenD, VolkerM, et al. (2006) An Xpd mouse model for the combined xeroderma pigmentosum/Cockayne syndrome exhibiting both cancer predisposition and segmental progeria. Cancer Cell 10 : 121–132.
72. WeedaG, DonkerI, de WitJ, MorreauH, JanssensR, et al. (1997) Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol 7 : 427–439.
73. SakaiK, MiyazakiJ (1997) A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun 237 : 318–324.
74. NakaneH, TakeuchiS, YubaS, SaijoM, NakatsuY, et al. (1995) High incidence of ultraviolet-B-or chemical-carcinogen-induced skin tumours in mice lacking the xeroderma pigmentosum group A gene. Nature 377 : 165–168.
75. JaspersNG, RaamsA, KelnerMJ, NgJM, YamashitaYM, et al. (2002) Anti-tumour compounds illudin S and Irofulven induce DNA lesions ignored by global repair and exclusively processed by transcription - and replication-coupled repair pathways. DNA Repair (Amst) 1 : 1027–1038.
76. VoN, SeoHY, RobinsonA, SowaG, BentleyD, et al. (2010) Accelerated aging of intervertebral discs in a mouse model of progeria. J Orthop Res 28 : 1600–1607.
77. de BoerJ, AndressooJO, de WitJ, HuijmansJ, BeemsRB, et al. (2002) Premature aging in mice deficient in DNA repair and transcription. Science 296 : 1276–1279.
78. NicolaijeC, DiderichKE, BotterSM, PriemelM, WaarsingJH, et al. (2012) Age-related skeletal dynamics and decrease in bone strength in DNA repair deficient male trichothiodystrophy mice. PLoS ONE 7: e35246.
79. DiderichKE, NicolaijeC, PriemelM, WaarsingJH, DayJS, et al. (2012) Bone fragility and decline in stem cells in prematurely aging DNA repair deficient trichothiodystrophy mice. Age (Dordr) 34 : 845–861.
80. TianM, ShinkuraR, ShinkuraN, AltFW (2004) Growth Retardation, Early Death, and DNA Repair Defects in Mice Deficient for the Nucleotide Excision Repair Enzyme XPF. Mol Cell Biol 24 : 1200–1205.
81. McWhirJ, SelfridgeJ, HarrisonDJ, SquiresS, MeltonDW (1993) Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat Genet 5 : 217–224.
82. GreggSQ, GutierrezV, RobinsonAR, WoodellT, NakaoA, et al. (2012) A mouse model of accelerated liver aging caused by a defect in DNA repair. Hepatology 55 : 609–621.
83. SelfridgeJ, HsiaKT, RedheadNJ, MeltonDW (2001) Correction of liver dysfunction in DNA repair-deficient mice with an ERCC1 transgene. Nucleic Acids Res 29 : 4541–4550.
84. GarinisGA, UittenboogaardLM, StachelscheidH, FousteriM, van IjckenW, et al. (2009) Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat Cell Biol 11 : 604–615.
85. van de VenM, AndressooJO, HolcombVB, von LindernM, JongWM, et al. (2006) Adaptive stress response in segmental progeria resembles long-lived dwarfism and calorie restriction in mice. PLoS Genet 2: e192.
86. KenslerTW, WakabayashiN, BiswalS (2007) Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47 : 89–116.
87. el-DeiryWS (1998) Regulation of p53 downstream genes. Semin Cancer Biol 8 : 345–357.
88. ChipchaseMD, O'NeillM, MeltonDW (2003) Characterization of premature liver polyploidy in DNA repair (Ercc1)-deficient mice. Hepatology 38 : 958–966.
89. BorgesiusNZ, de WaardMC, van der PluijmI, OmraniA, ZondagGC, et al. (2011) Accelerated age-related cognitive decline and neurodegeneration, caused by deficient DNA repair. J Neurosci 31 : 12543–12553.
90. de WaardMC, van der PluijmI, Zuiderveen BorgesiusN, ComleyLH, HaasdijkED, et al. (2010) Age-related motor neuron degeneration in DNA repair-deficient Ercc1 mice. Acta Neuropathol 120 : 461–475.
91. JaarsmaD, van der PluijmI, de WaardMC, HaasdijkED, BrandtR, et al. (2011) Age-related neuronal degeneration: complementary roles of nucleotide excision repair and transcription-coupled repair in preventing neuropathology. PLoS Genet 7: e1002405.
92. de GraafEL, VermeijWP, de WaardMC, RijksenY, van der PluijmI, et al. (2013) Spatio-temporal analysis of molecular determinants of neuronal degeneration in the aging mouse cerebellum. Mol Cell Proteomics 12 : 1350–1362.
93. AdalbertR, ColemanMP (2013) Review: Axon pathology in age-related neurodegenerative disorders. Neuropathol Appl Neurobiol 39 : 90–108.
94. WeidenheimKM, DicksonDW, RapinI (2009) Neuropathology of Cockayne syndrome: Evidence for impaired development, premature aging, and neurodegeneration. Mech Ageing Dev 130 : 619–636.
95. PosticC, ShiotaM, NiswenderKD, JettonTL, ChenY, et al. (1999) Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem 274 : 305–315.
96. Beyer TAXW, TeupserD, auf dem KellerU, BugnonP, HildtE, et al. (2008) Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J 9 : 212–223.
97. ItohM, HayashiM, ShiodaK, MinagawaM, IsaF, et al. (1999) Neurodegeneration in hereditary nucleotide repair disorders. Brain Dev 21 : 326–333.
98. KoobM, LaugelV, DurandM, FothergillH, DallozC, et al. (2010) Neuroimaging in Cockayne syndrome. AJNR Am J Neuroradiol 31 : 1623–1630.
99. HayashiM, Miwa-SaitoN, TanumaN, KubotaM (2012) Brain vascular changes in Cockayne syndrome. Neuropathology 32 : 113–117.
100. IwasatoT, NomuraR, AndoR, IkedaT, TanakaM, et al. (2004) Dorsal telencephalon-specific expression of Cre recombinase in PAC transgenic mice. Genesis 38 : 130–138.
101. LemonRN, JohanssonRS, WestlingG (1995) Corticospinal control during reach, grasp, and precision lift in man. J Neurosci 15 : 6145–6156.
102. RotshenkerS (2009) The role of Galectin-3/MAC-2 in the activation of the innate-immune function of phagocytosis in microglia in injury and disease. J Mol Neurosci 39 : 99–103.
103. LeeSK, YuSL, PrakashL, PrakashS (2002) Requirement of yeast RAD2, a homolog of human XPG gene, for efficient RNA polymerase II transcription. implications for Cockayne syndrome. Cell 109 : 823–834.
104. ThorelF, ConstantinouA, Dunand-SauthierI, NouspikelT, LalleP, et al. (2004) Definition of a short region of XPG necessary for TFIIH interaction and stable recruitment to sites of UV damage. Mol Cell Biol 24 : 10670–10680.
105. TregoKS, ChernikovaSB, DavalosAR, PerryJJ, FingerLD, et al. (2011) The DNA repair endonuclease XPG interacts directly and functionally with the WRN helicase defective in Werner syndrome. Cell Cycle 10 : 1998–2007.
106. BraceLE, VoseSC, VargasDF, ZhaoS, WangXP, et al. (2013) Lifespan extension by dietary intervention in a mouse model of Cockayne syndrome uncouples early postnatal development from segmental progeria. Aging Cell 12 : 1144–1147.
107. AndressooJO, JansJ, de WitJ, CoinF, HoogstratenD, et al. (2006) Rescue of progeria in trichothiodystrophy by homozygous lethal Xpd alleles. PLoS Biol 4: e322.
108. VermeulenW, JaekenJ, JaspersNG, BootsmaD, HoeijmakersJH (1993) Xeroderma pigmentosum complementation group G associated with Cockayne syndrome. Am J Hum Genet 53 : 185–192.
109. SchumacherB, van der PluijmI, MoorhouseMJ, KosteasT, RobinsonAR, et al. (2008) Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet 4: e1000161.
110. VermeijWP, HoeijmakersJHJ, PothofJ (2014) Aging: Not All DNA Damage is Equal. Curr Opin Genet Dev In press.
111. SunXZ, HaradaYN, TakahashiS, ShiomiN, ShiomiT (2001) Purkinje cell degeneration in mice lacking the xeroderma pigmentosum group G gene. J Neurosci Res 64 : 348–354.
112. VeghMJ, de WaardMC, van der PluijmI, RidwanY, SassenMJ, et al. (2012) Synaptic proteome changes in a DNA repair deficient ercc1 mouse model of accelerated aging. J Proteome Res 11 : 1855–1867.
113. MelisJP, WijnhovenSW, BeemsRB, RoodbergenM, van den BergJ, et al. (2008) Mouse models for xeroderma pigmentosum group A and group C show divergent cancer phenotypes. Cancer Res 68 : 1347–1353.
114. LeeYJ, ParkSJ, CicconeSL, KimCR, LeeSH (2006) An in vivo analysis of MMC-induced DNA damage and its repair. Carcinogenesis 27 : 446–453.
115. D'ErricoM, ParlantiE, TesonM, DeganP, LemmaT, et al. (2007) The role of CSA in the response to oxidative DNA damage in human cells. Oncogene 26 : 4336–4343.
116. SpivakG, HanawaltPC (2006) Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair (Amst) 5 : 13–22.
117. de WaardH, de WitJ, GorgelsTG, van den AardwegG, AndressooJO, et al. (2003) Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair (Amst) 2 : 13–25.
118. de WaardH, de WitJ, AndressooJO, van OostromCT, RiisB, et al. (2004) Different effects of CSA and CSB deficiency on sensitivity to oxidative DNA damage. Mol Cell Biol 24 : 7941–7948.
119. HalliwellB (2003) Oxidative stress in cell culture: an under-appreciated problem? FEBS Letters 540 : 3–6.
120. TotonchyMB, TamuraD, PantellMS, ZalewskiC, BradfordPT, et al. (2013) Auditory analysis of xeroderma pigmentosum 1971–2012: hearing function, sun sensitivity and DNA repair predict neurological degeneration. Brain 136 : 194–208.
121. SchaftJ, Ashery-PadanR, van der HoevenF, GrussP, StewartAF (2001) Efficient FLP recombination in mouse ES cells and oocytes. Genesis 31 : 6–10.
122. WaarsingJH, DayJS, WeinansH (2004) An improved segmentation method for in vivo microCT imaging. J Bone Miner Res 19 : 1640–1650.
123. BotterSM, GlassonSS, HopkinsB, ClockaertsS, WeinansH, et al. (2009) ADAMTS5−/ − mice have less subchondral bone changes after induction of osteoarthritis through surgical instability: implications for a link between cartilage and subchondral bone changes. Osteoarthritis Cartilage 17 : 636–645.
124. PfafflMW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45.
Štítky
Genetika Reprodukční medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 10
-
Všechny články tohoto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



