-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
CtIP plays an important role in homologous recombination (HR)–mediated DNA double-stranded break (DSB) repair and interacts with Nbs1 and BRCA1, which are linked to Nijmegen breakage syndrome (NBS) and familial breast cancer, respectively. We identified new CDK phosphorylation sites on CtIP and found that phosphorylation of these newly identified CDK sites induces association of CtIP with the N-terminus FHA and BRCT domains of Nbs1. We further showed that these CDK-dependent phosphorylation events are a prerequisite for ATM to phosphorylate CtIP upon DNA damage, which is important for end resection to activate HR by promoting recruitment of BLM and Exo1 to DSBs. Most notably, this CDK-dependent CtIP and Nbs1 interaction facilitates ATM to phosphorylate CtIP in a substrate-specific manner. These studies reveal one important mechanism to regulate cell-cycle-dependent activation of HR upon DNA damage by coupling CDK - and ATM-mediated phosphorylation of CtIP through modulating the interaction of CtIP with Nbs1, which significantly helps to understand how DSB repair is regulated in mammalian cells to maintain genome stability.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003277
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003277Summary
CtIP plays an important role in homologous recombination (HR)–mediated DNA double-stranded break (DSB) repair and interacts with Nbs1 and BRCA1, which are linked to Nijmegen breakage syndrome (NBS) and familial breast cancer, respectively. We identified new CDK phosphorylation sites on CtIP and found that phosphorylation of these newly identified CDK sites induces association of CtIP with the N-terminus FHA and BRCT domains of Nbs1. We further showed that these CDK-dependent phosphorylation events are a prerequisite for ATM to phosphorylate CtIP upon DNA damage, which is important for end resection to activate HR by promoting recruitment of BLM and Exo1 to DSBs. Most notably, this CDK-dependent CtIP and Nbs1 interaction facilitates ATM to phosphorylate CtIP in a substrate-specific manner. These studies reveal one important mechanism to regulate cell-cycle-dependent activation of HR upon DNA damage by coupling CDK - and ATM-mediated phosphorylation of CtIP through modulating the interaction of CtIP with Nbs1, which significantly helps to understand how DSB repair is regulated in mammalian cells to maintain genome stability.
Introduction
DNA double-strand breaks (DSBs) are often generated during normal cellular processes such as the development of immune systems and meiotic recombination, and they can also be induced by DNA damaging agents [1]–[4]. DSBs must be repaired properly to maintain cell viability and to prevent genome instability. A number of inherited human diseases associated with genome instability and cancer are found to be caused by mutations in the DNA DSB repair pathways. For instance, Mre11, Nbs1, ATM and BRCA1, which play critical roles in DNA damage checkpoint and DSB repair are linked ataxia-telangiectasia-like disorder (ATLD), Nijmegen breakage syndrome (NBS), ataxia-telangiectasia and familial breast cancer, respectively [5]–[9]. However, the exact mechanisms of how these proteins regulate DSB repair in mammalian cells are not fully understood.
DSBs can be repaired by Ku-dependent non-homologous end joining (NHEJ) and homologous recombination (HR)-mediated DSB repair [10], [11]. While NHEJ is active in all phases of the cell cycle, HR is activated only when cells enter S-phase, which is mainly attributed to the essential function of cyclin-dependent kinases (CDKs) to promote end resection at DSB ends [12]. NHEJ can be error-prone, but HR is highly accurate in preserving all genetic information by using the identical sister chromatids as templates to repair DSBs.
The Mre11/Rad50/Nbs1 (MRN) complex binds to DSB ends and plays important roles in facilitating ATM activation and promoting end resection to initiate HR-mediated DSB repair [13]–[15]. CtIP (CtBP-interacting protein), which is associated with MRN and BRCA1, is also a critical player in the regulation of HR [16]–[20]. In mammalian cells, CtIP binds to MRN through Nbs1 [16], [18], [21]. Such interaction was also observed in fission yeast, where the FHA/BRCT domains of Nbs1 interact with the phospho-motifs at the CK2 sites on Ctp1, and this interaction is important for recruiting Ctp1 to DSB ends by Nbs1 [22], [23]. In budding yeast, the CtIP homologue Sae2 exhibits endonuclease activities to cleave the ssDNA (single-strand DNA)/dsDNA (double-strand DNA) junctions and the sites close to DNA hairpin loops [24]. Although the nuclease activity of CtIP itself was not identified, CtIP was found to promote the nuclease activity of MRN to process DSB ends in mammalian cells [16].
Various studies suggest that CtIP and its homologues are important for regulating end resection during the cell cycle [16], [17], [25]–[27]. CtIP is directly phosphorylated by CDKs, and phosphorylation of CtIP at the CDK site S327 induces its interaction with BRCA1, which is important for HR-mediated DSB repair [18], [20], [26]. Furthermore, phosphorylation of a conserved CDK site S267 on Sae2 is essential for HR in budding yeast [27], and its corresponding site T847 on CtIP is also required for end resection in mammalian cells [17], although the mechanisms underlying this regulation are not clear.
In this study, we identified a new cluster of CDK sites situated in the middle of CtIP and demonstrated that phosphorylation of these CDK sites is a prerequisite for ATM to phosphorylate CtIP upon DNA damage. We also showed that ATM-mediated phosphorylation of CtIP, mainly through the newly identified conserved site T859, is essential for promoting end resection and HR, suggesting a mechanism of how CDK-mediated phosphorylation activates damage-induced HR. We further showed that phosphorylation of these identified CDK sites on CtIP induces associations of CtIP with the FHA/BRCT domains of Nbs1. Interestingly, this CDK-dependent interaction of CtIP with the FHA/BRCT domains of Nbs1 facilitates ATM to phosphorylate CtIP, thereby coupling CDK-phosphorylation with ATM-mediated phosphorylation of CtIP to promote end resection and HR.
Results
CtIP is phosphorylated by ATM in response to DNA damage
DNA damage induces CtIP phosphorylation, as revealed by phosphatase-sensitive mobility shift of CtIP on SDS-PAGE after ionization radiation (IR) or camptothecin (CPT) treatment (Figure 1A and data not shown). By using ATM inhibitor (ATMi) KU-55933 or expressing ATM shRNAs, we found that IR - and CPT-induced CtIP phosphorylation is dependent on ATM (Figure 1B and data not shown). In vitro kinase assays showed that purified CtIP can be phosphorylated by ATM (Figure S1A). Mutating all eight putative ATM kinase sites (SQ/TQ) on CtIP (CtIP-8A-ATM) completely abolished damage-induced CtIP phosphorylation shift (Figure 1C), further supporting that damage-induced phosphorylation is mediated by ATM.
Fig. 1. ATM is important for CtIP hyper-phosphorylation after DNA damage. 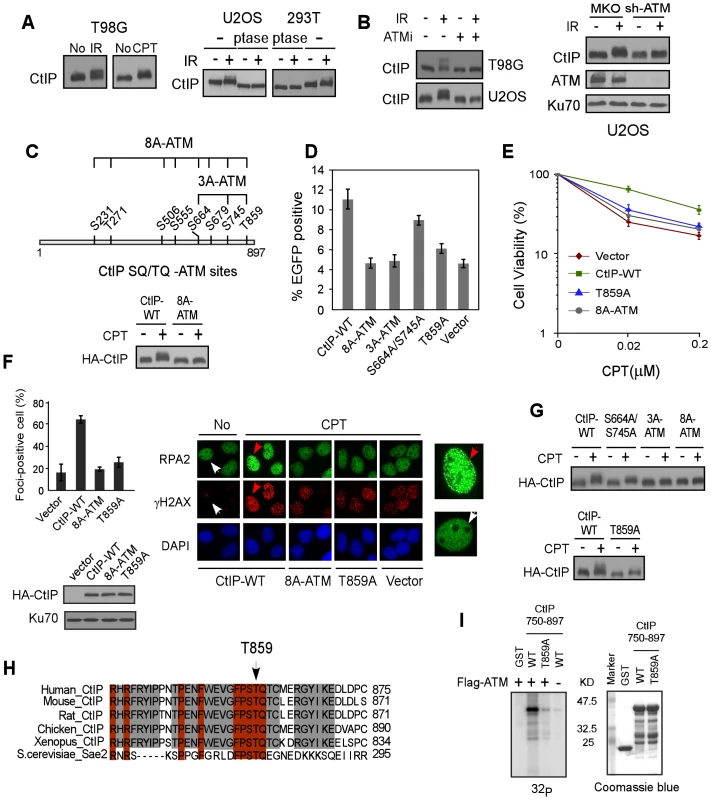
A. Indicated cell lines were treated with IR (10 Gy) or CPT (2 µM, 2 h) or No and lysed. Western blotting of CtIP was performed using cell lysates incubated with lambda phosphatase (ptase) or without ptase (-). B. U2OS and/or T98G cells were treated with ATM inhibitor (ATMi) KU-55933 (20 µM, 1 h) or infected with retroviruses encoding shRNAs for ATM or vector control MKO, treated with IR (10 Gy) and lysed 1 h later. Western blotting was performed with indicated antibodies. C. Schematic drawing of eight putative ATM consensus sites (SQ/TQ motifs) on CtIP. U2OS cells stably expressing HA-CtIP wild-type (WT) or HA-CtIP 8A-ATM mutant (all SQ/TQ sites mutated to AQ) with endogenous CtIP silenced by shRNAs and siRNA, were treated with CPT (2 µM, 2 h) followed by anti-HA Western blotting. D–F. EGFP-based HR assay, clonogenic survival and RPA foci formation assays were performed with U2OS cells stably expressing indicated CtIP variants, with endogenous CtIP silenced. Data shown represents the mean of three independent experiments; error bars, s.d. In panel F, indicated cells were treated with DMSO (No) or CPT (1 µM, 2 h) and fixed for immunostaining. The red and white arrows indicate representative RPA2 and γH2AX foci-positive and foci-negative cells, respectively, with RPA2-immunostained cells enlarged in right-side panels. The percentage of RPA2 foci-positive cells among γH2AX-positive cells for each sample is shown. G. Western blot of U2OS cells expressing indicated CtIP variants with endogenous CtIP silenced and treated with CPT (2 µM, 2 h). H. The C-terminus protein sequence of human CtIP was aligned with CtIP homologues from other indicated species, with newly identified conserved T859 site shown. I. ATM in vitro kinase assay was performed with purified GST-tagged CtIP fragments (residues 750–897), containing either WT or the T859A mutation. Coomassie Blue staining indicates protein loading. A newly identified conserved TQ site T859 on CtIP is phosphorylated by ATM and is important for end resection and HR
By using EGFP-based HR assays (Figure S1B), we demonstrated that the CtIP-8A-ATM mutant is significantly impaired in HR (Figure 1D). We further showed that the CtIP-S664A/S745A/T859A (CtIP-3A-ATM) mutant carrying mutations at three C-terminal SQ/TQ sites exhibits a strong defect in HR, but not the mutants CtIP-S231A/T271A and CtIP-S506A/S555A/S679A (Figure S1C and Figure 1D). While mutating the previously described S664 and S745 sites caused limited HR defect (Figure 1D, [28]), a single site mutation at T859 strongly reduced HR. Consistently, the CtIP-T859A mutant exhibited strong sensitivity to CPT, and reduced CPT-induced RPA foci formation (Figure 1E and 1F). These data suggest that ATM-mediated phosphorylation on CtIP mainly at T859 is important for promoting end resection and HR-mediated DSB repair.
Mutating S664 and S745 or T859 alone reduced, but mutating all three S664, S745 and T859 sites (CtIP-3A-ATM) almost completely abolished damage-induced phosphorylation as revealed by damage-induced phosphorylation shift on SDS-PAGE (Figure 1G), suggesting that the ATM sites S664, S745 and T859 are indeed phosphorylated in vivo. Alignment of CtIP from different species revealed that the newly identified TQ site T859 is conserved (Figure 1H). In vitro kinase assay showed that ATM phosphorylates the wild-type CtIP-750-897 fragment but not the fragment carrying T859A mutation, suggesting that ATM can directly phosphorylate this site (Figure 1I). Collectively, our studies suggest that ATM-mediated phosphorylation of CtIP is important for end resection during HR, and the conserved ATM site T859 identified in this study is critical for this function.
CDK-mediated phosphorylation of CtIP is required for ATM to phosphorylate CtIP upon DNA damage
Treating cell lysates with phosphatase or mutating 12 putative CDK sites (SP/TP sites) increased CtIP mobility on SDS-PAGE to a similar level (Figure 2A and Figure 2B, middle panel), suggesting that CtIP is mainly phosphorylated by CDKs during the cell cycle. However, the CtIP mutants S327A or T847A/S889A only slightly reduced the phosphorylation-dependent shift (Figure 2B, middle and bottom panels), suggesting that CDK sites other than previously identified S327 and T847 [17], [20] are also phosphorylated in vivo. Indeed, mutating a middle cluster of seven CDK sites (CtIP-7A-CDK: S233A, T245A, S276A, T315A, S347A, S549A and S568A, excluding S327) almost completely abolished the CtIP shift (Figure 2B, bottom panel).
Fig. 2. CtIP is phosphorylated by CDKs at multiple sites. 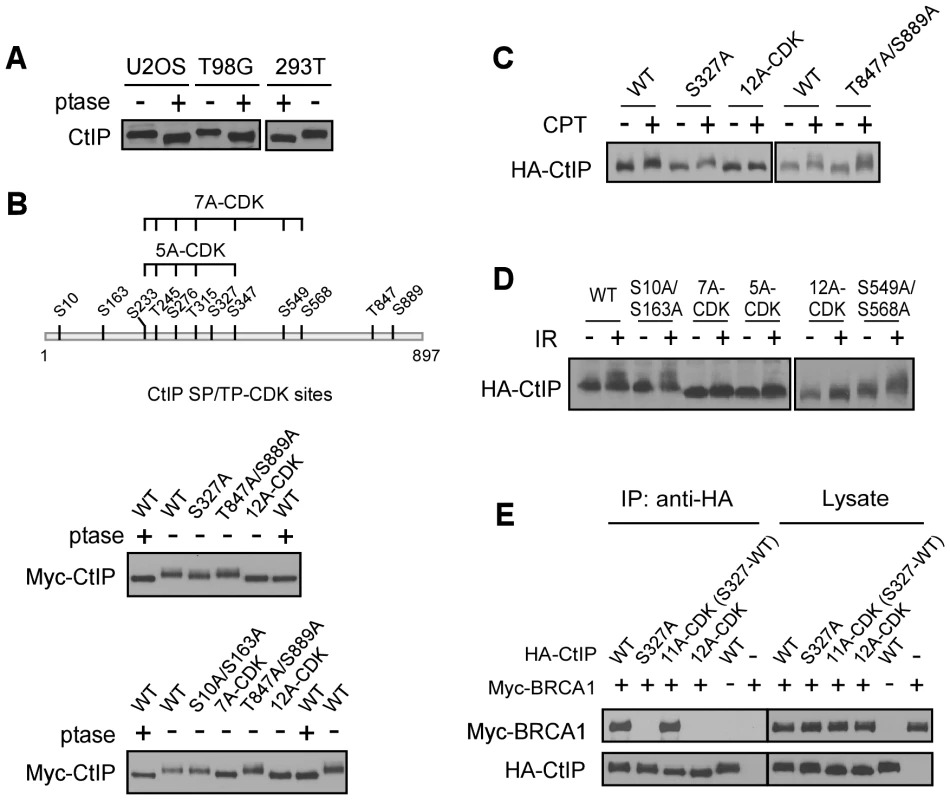
A. Lysates from indicated cell lines were collected and treated with (+) or without (−) lambda phosphatase (ptase), followed by immunoblotting for endogenous CtIP. B. Schematic drawing of twelve putative CDK consensus sites (SP/TP motifs) on CtIP, with the middle cluster of sites 7A-CDK and 5A-CDK indicated. Myc-CtIP WT or indicated mutants were expressed in 293T cells, and cell lysates were treated with or without lambda phosphatase, followed by anti-Myc immunoblotting. C. and D. U2OS cells stably expressing HA-CtIP WT or indicated mutants with endogenous CtIP silenced, were treated with or without CPT (2 µM, 2 h) or IR (10 Gy, recovered for 1 h), followed by immunoblotting for anti-HA. E. Myc-BRCA1 and/or HA-CtIP WT or indicated mutants were expressed in 293T cells and co-immunoprecipitation was performed. To confirm that the CDK sites on CtIP are indeed phosphorylated in vivo, we performed mass spectrometry analysis of endogenous CtIP purified from HeLa cells. Peptides containing phospho-Ser and phospho-Thr at multiple CDK putative SP/TP sites including S10, S163, S233, T245, S276, T315, S327, S347, S549, S568 and S889 were recovered, confirming that these putative CDK sites are indeed phosphorylated in vivo (Table S1). However, the previously identified CDK site T847 was not revealed in this analysis [17], which may be attributed to instability of the phosphorylation at this site.
Strikingly, upon DNA damage including CPT and IR, the CtIP-12A-CDK mutant failed to be further phosphorylated by ATM as revealed by mobility shift on SDS-PAGE (Figure 2C and 2D), suggesting that CDK-mediated CtIP phosphorylation is a prerequisite for CtIP to be phosphorylated after DNA damage. Further analysis showed that the CDK-site mutants S10A/S163A, S327A, S549A/S568A and T847A/S889A exhibited normal damage-induced CtIP phosphorylation, but CtIP-5A-CDK (S233A, T245A, S276A, T315A and S347A) and CtIP-7A-CDK mutants were strongly impaired in this damage-induced phosphorylation (Figure 2C and 2D). These studies suggest that phosphorylation of the 5 CDK sites situated in the middle of CtIP (5mCDK sites: 5 middle CDK sites S233, T245, S276, T315 and S347) is required for damage-induced phosphorylation of CtIP by ATM. By synchronizing T98G cells through serum starvation, we showed that IR-induced CtIP hyper-phosphorylation does not occur in G1 cells and only occurs when cells enter S-phase (Figure S2). These data further support that CDK activity is important for damage-induced CtIP phosphorylation by ATM. Furthermore, we showed that while S327 is critical for BRCA1 binding [20], mutating the other 11 CDK sites on CtIP [all CDK sites except S327 : 11A-CDK(S327-WT)] does not influence BRCA1 association (Figure 2E), suggesting that the interaction of CtIP with BRCA1 is not required for regulating damage-induced CtIP phosphorylation.
CDK-mediated CtIP phosphorylation is important for end resection and HR
Consistent with that CDK-mediated phosphorylation of the middle cluster of SP/TP sites is required for ATM to phosphorylate CtIP upon DNA damage, the CDK mutants CtIP-5A-CDK and CtIP-7A-CDK were found to be defective in HR to a similar level as the CtIP-T847A/S889A mutant containing the previously mapped CDK site T847 [17], while the mutants CtIP-S10A/S163A and S549A/S568A did not exhibit such defects (Figure 3A). In addition, the CDK mutants CtIP-5A-CDK and CtIP-12A-CDK were sensitive to CPT (Figure 3B). As revealed by damage-induced RPA foci formation, end resection was defective in the CtIP-5A-CDK mutant to a level comparable to the CtIP-12A-CDK mutant with all CDK sites mutated (Figure 3C), and consequently IR-induced ATR activation as judged by Chk1 phosphorylation was also compromised (Figure 3D). These studies suggest that the 5mCDKs are important for end resection and HR-mediated DSB repair.
Fig. 3. ATM-mediated phosphorylation of CtIP is important for end resection in a CDK-dependent manner. 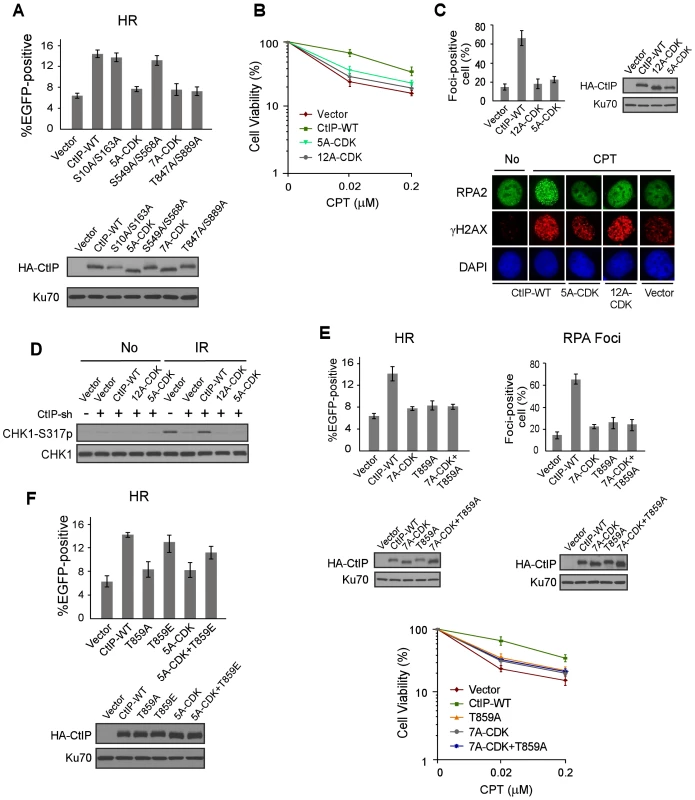
A. EGFP-HR assay was performed with U2OS cells stably expressing HA-CtIP WT or indicated mutants, with endogenous CtIP silenced. Data represent the mean ± s.d. of three independent experiments. Western blot shows expression of HA-CtIP variants. B. Clonogenic survival assay after treatment with indicated doses of CPT (1 h) was performed with U2OS cells stably expressing indicated CtIP variants with endogenous CtIP silenced. Data represent the mean ± s.d. of three independent experiments. C. RPA foci formation was examined in U2OS cells with indicated CtIP variants, before or after CPT treatment. The percentage of RPA foci-positive cells was determined as in Figure 1F. D. U2OS cells stably expressing indicated CtIP variants with endogenous CtIP silenced were treated with IR (10 Gy, recovered for 1 h), lysed, and immunoblotting was performed. E. EGFP-HR repair assay, clonogenic survival and RPA foci formation assays were performed in U2OS cells expressing HA-CtIP-WT or indicated mutants with endogenous CtIP silenced. F. EGFP-HR assay was performed with U2OS cells stably expressing HA-CtIP WT or indicated mutants, with endogenous CtIP silenced. Data represent the mean ± s.d. of three independent experiments. Western blot shows expression of HA-CtIP variants. To show that CDK-mediated phosphorylation of CtIP is in the same pathway as ATM-mediated CtIP phosphorylation to regulate end resection and HR, we combined the mutations of CtIP-7A-CDK with the ATM site mutation T859A, and the resultant mutant CtIP-7A-CDK/T859A exhibited similar defects in HR, end resection and CPT sensitivity as the separate CDK and ATM mutants, CtIP-7A-CDK and CtIP-T859A (Figure 3E). These data support the conclusion that HR defects observed in the CtIP-7A-CDK mutant is due to its impaired function to promote ATM-mediated CtIP phosphorylation upon DNA damage. To determine whether CtIP phosphorylation by ATM can bypass the requirement of CDK-mediated phosphorylation of CtIP, we introduced the phospho-mimic mutation T859E to the CtIP-5A-CDK mutant. The HR defects observed in the CtIP-5A-CDK were largely suppressed in the CtIP-5A-CDK/T859E mutant (Figure 3F). The interaction of Nbs1 with CtIP phospho-mimic mutants CtIP-8E-ATM (mutating all 8 SQ/TQ sites to EQ) and CtIP-T859E remains at the similar levels as wild-type CtIP (Figure S4C). These data further support that CDK-mediated phosphorylation of CtIP is important for CtIP to be phosphorylated by ATM to mediate the DNA damage response.
Nbs1 promotes ATM-mediated phosphorylation of CtIP through its FHA/BRCT domains in a CDK phosphorylation-dependent manner
Our studies suggest that CDK-mediated phosphorylation of CtIP directly or indirectly modulates damage-induced phosphorylation of CtIP by ATM. Consistent with the previous findings that CtIP is dispensable for ATM activation [16], [29], no defects of ATM activation were detected in the CtIP-7A-CDK-mutant as revealed by damage-induced ATM auto-phosphorylation and Chk2 phosphorylation (Figure 4A). As with many other ATM substrates, damage-induced CtIP phophorylation depends on Nbs1 and Mre11 (Figure 4B), likely due to the role of MRN in promoting ATM activation [30]. Interestingly, however, when we used an Nbs1 FHA/BRCT mutant, Nbs1-RRHK (R28A/R43A/H45A/K160A) carrying mutations at the critical sites for binding to the phospho-motifs in the Nbs1 FHA/BRCT domains [31], [32], we found that damage-induced CtIP phosphorylation was impaired, but ATM auto-phosphorylation and ATM-mediated phosphorylation of Chk2 was normal (Figure 4D). This suggests that the FHA/BRCT domains of Nbs1 are not required for ATM activation per se but are specifically required for ATM to phosphorylate CtIP in response to DNA damage.
Fig. 4. The FHA/BRCT domains of Nbs1 are important for ATM-mediated phosphorylation of CtIP. 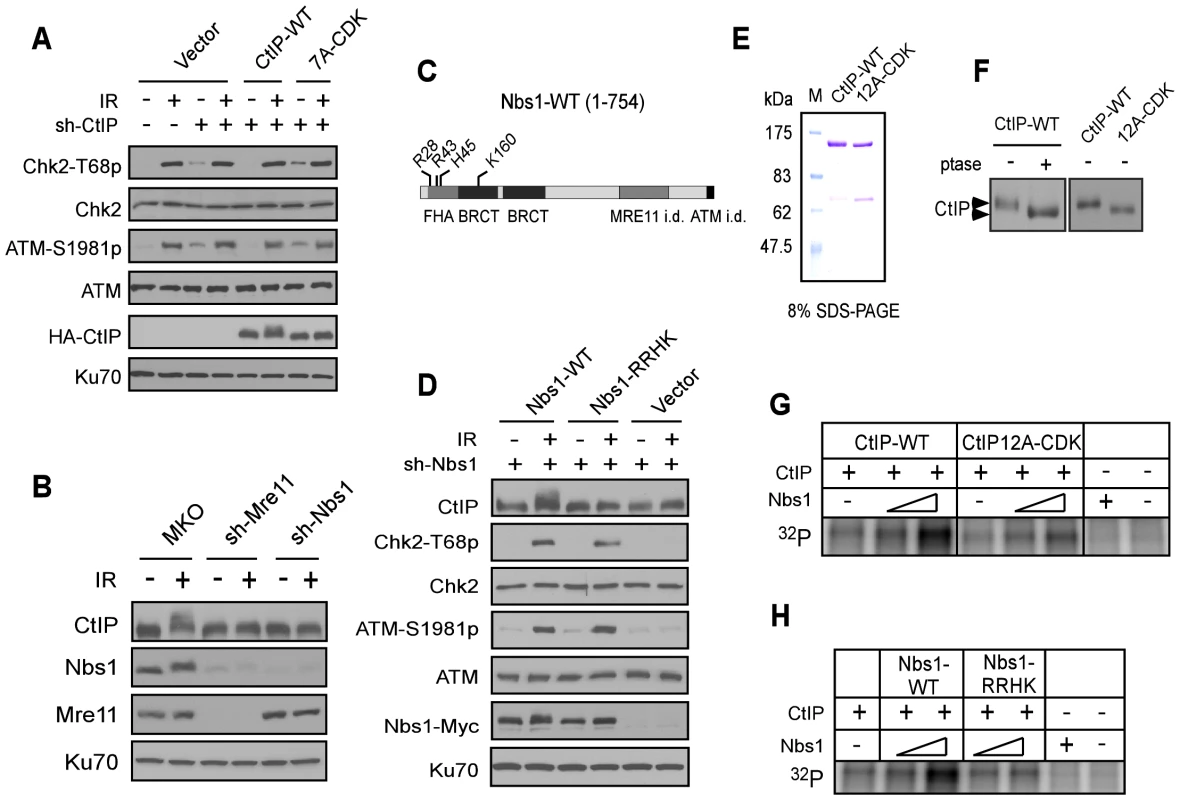
A. The CtIP-7A-CDK mutant does not show defects in ATM and checkpoint activation. U2OS cells stably expressing HA-CtIP WT or 7A-CDK mutant, with endogenous CtIP silenced, were treated with or without IR (10 Gy) and allowed to recover for 1 h. Cells were then collected and lysed for Western blot analysis with indicated antibodies. B. U2OS cells stably expressing control vector MKO, sh-Mre11 or sh-Nbs1 were treated with IR (10 Gy, recovered for 1 h), lysed, and immunoblotting was performed. C. Schematic drawing of human Nbs1 protein domains [FHA, BRCT and Mre11- and ATM interacting domains (i.d.)], with indicated phospho-binding sites on the FHA/BRCT domains [31], [32]. D. U2OS cells stably expressing C-terminus Myc-tagged Nbs1 alleles, Nbs1-WT and Nbs1-RRHK (R28A, R43A, H45A and K160M) or empty vector with endogenous Nbs1 silenced by shRNAs were treated with or without IR (10 Gy, recovered for 1 h), lysed, and immunoblotting was performed. E. Purified GST-CtIP WT and GST-CtIP-12A-CDK from Sf9 insect cells were analyzed by SDS-PAGE with Coomassie blue staining. F. CtIP expressed in insect cells is phosphorylated by CDKs. Purified GST-CtIP WT and 12A-CDK mutant protein from insect cells were treated without or with lambda phosphatase (ptase), and anti-CtIP Western blot analysis was performed. G and H. Baculovirus-expressed and purified CtIP-WT and CtIP-12A-CDK mutant proteins (100 ng) were used as substrates for in vitro ATM kinase assays with or without addition of purified Nbs1-WT or Nbs1-RRHK mutant proteins (100 ng and 500 ng). The radiolabeled CtIP was visualized following SDS-PAGE. To further understand the role of Nbs1 to facilitate ATM-mediated phosphorylation of CtIP, we performed in vitro ATM kinase assays using purified GST-CtIP-His (Figure 4E). Human CtIP, expressed in insect cells but not in bacteria, was phosphorylated by CDKs as revealed by reduced mobility on SDS-PAGE upon phosphatase treatment or after mutating 12 putative CDK sites on CtIP (Figure 4F). We also performed mass spectrometry analysis and found that the same CDK sites are phosphorylated on CtIP when expressed in insect cells as in mammalian cells (Table S2). Although ATM can phosphorylate CtIP in vitro (Figure S1A), addition of Nbs1, purified from insect cells (Figure S3A), significantly promoted such ATM-mediated phosphorylation of CtIP when less amounts of CtIP and ATM were used (Figure 4G). Interestingly, this Nbs1-promoted CtIP phosphorylation by ATM was observed only when CtIP-WT, but not CtIP-12A-CDK was used. Furthermore, the Nbs1 FHA/BRCT mutant Nbs1-RRHK also failed to promote ATM-mediated phosphorylation of CtIP in vitro (Figure 4H). These studies suggest that Nbs1 promotes ATM phosphorylation of CtIP in a manner dependent on CDK-phosphorylation of CtIP and the FHA/BRCT domains of Nbs1.
The FHA/BRCT domains of Nbs1 interact with CtIP through the middle cluster of CDK sites, which promotes ATM-mediated phosphorylation of CtIP
FHA and BRCT domains often mediate the binding of phospho-proteins through specific interactions of phosphorylated motifs [33]–[35]. To understand how Nbs1 facilitates CtIP phosphorylation by ATM through its FHA/BRCT domains in a CDK phosphorylation-dependent manner, we characterized the interactions of Nbs1 with CtIP. As shown in Figure 5A, full-length Nbs1 binds to CtIP-WT and CtIP-12A-CDK at similar levels. Further analysis revealed that both the N-terminus of Nbs1 [Nbs1-(1–335), containing the FHA/BRCT domains] and C-terminus of Nbs1 [Nbs1-(336-end)] bind to CtIP (Figure 5B), suggesting that CtIP and Nbs1 interact at more than one site.
Fig. 5. CtIP interacts with the FHA/BRCT domains of Nbs1 in a CDK phosphorylation-dependent manner. 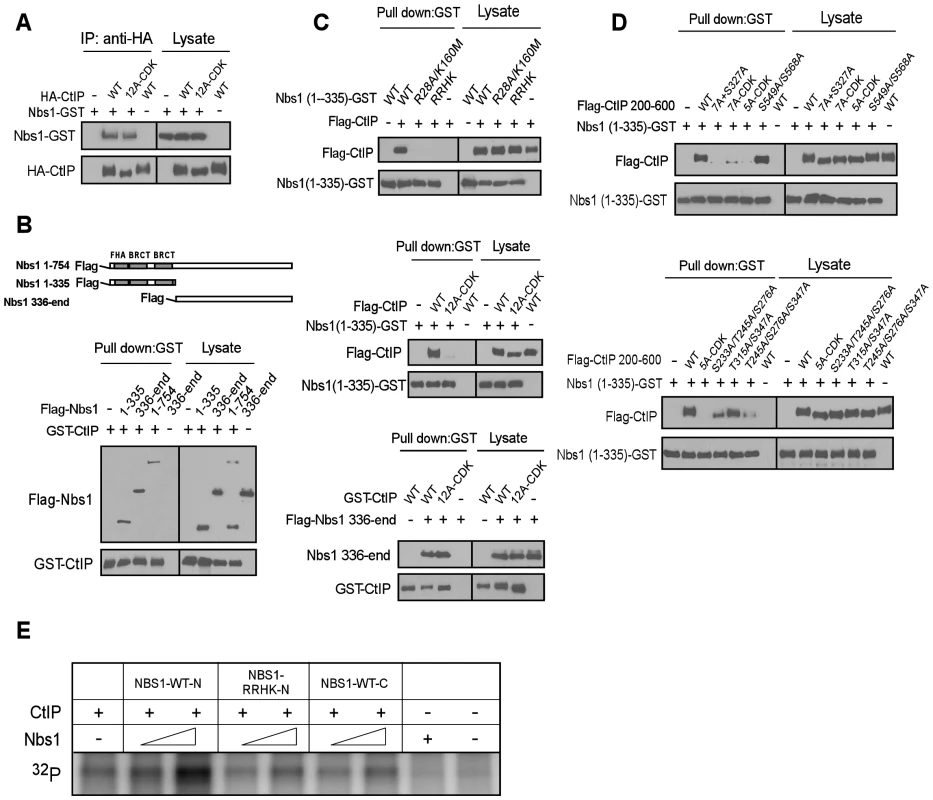
A to D. Characterizing the interaction of CtIP and Nbs1. Indicated Nbs1 and CtIP constructs were co-expressed in Sf21 insect cells by baculovirus infection, followed by anti-HA IP or GST pull-down assays and immunoblotting. A. Full-length CtIP-12A-CDK mutant associates with Nbs1 containing C-terminal GST-tag. B. Both the N-terminus and C-terminus of Nbs1 bind to CtIP. C. Top: The phospho-binding sites in the Nbs1 FHA/BRCT domains are important for mediating the interaction with CtIP. Middle: The CtIP-12A-CDK mutant fails to bind to the Nbs1 FHA/BRCT domains. Bottom: Both CtIP-WT and CtIP-12A-CDK mutant interact with the C-terminus of Nbs1. D. The CtIP 5mCDK sites are important for interaction with Nbs1 FHA/BRCT domains. Top: The CtIP 200–600 fragment containing the 5A-CDK mutations fails to bind to Nbs1 FHA/BRCT domains. Bottom: The 5mCDK sites redundantly mediate the interaction of CtIP with Nbs1 FHA-BRCT domains. E. Purified CtIP (100 ng) was used for in vitro ATM kinase assays in the presence or absence of Nbs1-WT-N (residues 1–335), Nbs1-RRHK-N mutant (residues 1–335) or Nbs1-WT-C (residues 336-end) (100 ng and 500 ng). Strikingly, the Nbs1-(1–335)-GST fragment containing the FHA/BRCT domains with GST tagged at the C-terminus, but not the corresponding FHA/BRCT mutants Nbs1-(1–335)-R28A/K160M-GST and Nbs1-(1–335)-RRHK-GST, bind CtIP efficiently (Figure 5C, top). Furthermore, Nbs1-(1–335)-GST binds to Flag-tagged CtIP-WT but not the CtIP-CDK-12A mutant (Figure 5C, middle), while the C-terminus Nbs1-(336-end) fragment binds to CtIP-WT and CtIP-12A-CDK at approximately equal levels (Figure 5C, bottom). By using purified proteins, we further demonstrated that the interaction of Nbs1-(1–335) with CtIP is direct, and such interaction can be disrupted by the RRHK mutations in the Nbs1 FHA domain and the CDK-site mutations (12A-CDK) in CtIP (Figure S3B).
We also found that Nbs1-(1–335), but not Nbs1-(1–335)-RRHK interacts with the middle region of CtIP [CtIP-(200–600)], and mutating all CDK sites (7A+S327A) in this CtIP-(200–600) fragment abolished the interactions (Figure S4A and Figure 5D top). On the other hand, the C-terminus Nbs1-(336-end) fragment binds to the C-terminus of CtIP [CtIP-(462-end)] (Figure S4B). These studies suggest that Nbs1 interacts with CtIP through two distinct means: one is through the FHA/BRCT domains of Nbs1 to bind to the middle cluster of CDK phospho-motifs on CtIP, and the other is a constitutive interaction through the C-termini of Nbs1 and CtIP.
To further analyze CDK-phosphorylation-mediated interaction of CtIP with the FHA/BRCT domains of Nbs1, we mutated different CDK sites in the CtIP-(200–600) fragment. While the CtIP - (200–600)-S549A/S568A mutant still binds to the Nbs1 FHA/BRCT domains [Nbs1-(1–335)-GST], the CtIP-(200–600) fragment containing the 7A-CDK or 5A-CDK mutation fails to bind (Figure 5D, top), suggesting that the 5mCDK sites are important for mediating the interactions with the Nbs1 FHA/BRCT domains. We then separated the mutations in the 5A-CDK mutant, and found that the CtIP-(200–600) fragments containing mutations of S233A/S245A/S276A, T315A/S347A or T245A/S276A/S347A all reduce, but do not abolish the binding to Nbs1-(1–335) (Figure 5D, bottom). It has been suggested that the FHA domain mediates phosphorylated threonine binding, while the BRCT domains often bind to phosphorylated serines [36], [37]. We thus examined the interaction of Nbs1-(1–335)-R28A-GST carrying mutations in the FHA domain with the CtIP-(200–600)-S233A/S276A/S347A mutant, and found that the interaction of the BRCT domains with CtIP is abolished by non-phosphorylation mutations at these three serine residues (Figure S5A, left). The interaction of Nbs1-(1–335)-K160M-GST carrying mutations in the second BRCT domain with CtIP-(200–600)-T245A/T315A was also reduced (Figure S5A, right). These data suggest that S233, S276 and S347 are important for binding to the BRCT domains, and T245 and T315 contributes to the binding to the Nbs1 FHA domain. Consistent with that both FHA and BRCT domains of Nbs1 are involved in the interactions with the CDK sites on CtIP, we also showed that introducing mutations to both FHA and BRCT domains of Nbs1 in the Nbs1-(1–335) fragment (Nbs1-RRHK and Nbs1-R28A/K160M) abolished its interactions with CtIP-(200–600), while mutating either FHA domain (Nbs1-R28A) or BRCT domain (Nbs1-K160M) only reduced the interaction (Figure S5B).
We demonstrated that full-length Nbs1 promotes ATM to phosphorylate CtIP in a manner dependent on the FHA/BRCT domains of Nbs1 and CDK-phosphorylation of CtIP (Figure 4G and 4H). To examine whether this activity depends on the Nbs1 and CtIP interaction, purified Nbs1-(1–335) or Nbs1-(336-end) was added into the in vitro ATM kinase assay (Figure S3A). Interestingly, only Nbs1-(1–335) but not Nbs1-(336-end) promoted ATM to phosphorylate CtIP, and Nbs1-(1–335) carrying the FHA/BRCT mutations did not show this Nbs1-dependent stimulation of ATM phosphorylation of CtIP (Figure 5E). These data suggest that the specific interactions of the FHA/BRCT domains of Nbs1 with 5mCDK sites on CtIP modulate CtIP phosphorylation by ATM.
ATM-mediated phosphorylation of CtIP is important for promoting recruitment of BLM and Exo1 to DSBs to initiate HR
In fission yeast, the interaction of Nbs1 FHA/BRCT domains with the CK2 sites on Ctp1 is required for Ctp1 to be recruited to DSBs [23]. In mammalian cells, CtIP recruitment to DSBs depends on Nbs1 and ATM [29]. To monitor whether CDK - and ATM-mediated phosphorylation regulates CtIP recruitment, we performed live-cell imaging to examine the recruitment of EGFP-tagged CtIP to DSBs upon laser microirradiation. With endogenous CtIP silenced by shRNAs, we showed that the initial recruitments of the CtIP-5A-CDK and CtIP-3A-ATM (S664A/S745A/T859A) mutants to chromosomal DSBs are comparable to CtIP-WT (Figure 6A). However at later time points, CtIP-WT started to disassociate from DSBs faster than the CtIP-5A-CDK and CtIP-3A-ATM mutants, likely due to compromised DSB repair in these mutants. We also monitored CtIP localization to DSB ends in the absence of γH2AX-dependent recruitment to DSB-flanking regions. As expected, inactivation of H2AX significantly reduced CtIP recruitment to laser-generated DSB-containing regions, but γH2AX-independent recruitment of CtIP-5A-CDK and CtIP-3A-ATM mutants to DSB ends was also similar to that of CtIP-WT at the initial stages after laser treatment (Figure 6A). Thus, CDK-phosphorylation of CtIP is not necessarily required for CtIP recruitment to DSBs in mammalian cells.
Fig. 6. ATM-mediated phosphorylation of CtIP promotes the recruitment of BLM and Exo1 to laser-induced DSBs. 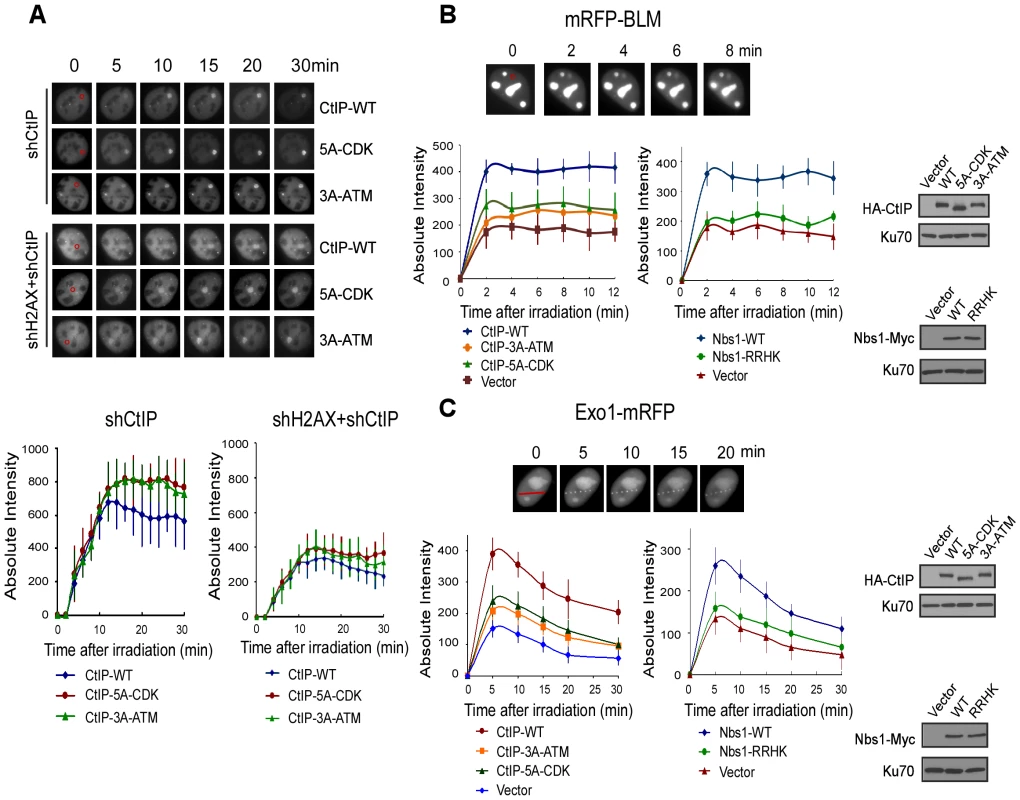
A. EGFP-CtIP variants and mRFP-PCNA were co-expressed in U2OS cells with endogenous CtIP or both CtIP and H2AX silenced by shRNAs. Live-cell imaging of EGFP-CtIP in S-phase cells, marked by PCNA S-phase-associated replication foci [68], was performed following laser-induced microirradiation. Representative cells show the recruitment of EGFP-CtIP WT or indicated mutants to laser-microirradiated disk regions, which are indicated by red circles in the 0 min cell images. Absolute intensity of EGFP-CtIP fluorescence signals at damage sites was determined; error bars, s.d. B and C. Recruitment of mRFP-BLM (B) and Exo1-mRFP (C) to DSBs was monitored in U2OS cells expressing HA-CtIP or Nbs1-Myc variants, with endogenous CtIP or Nbs1 silenced. Representative cells show the recruitment of mRFP-BLM or Exo1-mRFP to microirradiated regions (a disk region for mRFP-BLM marked by a red circle and dots on a line for Exo1-mRFP marked by a red line in 0 min cell images). Absolute intensities of mRFP-BLM or Exo1-mRFP fluorescence signals were determined; error bars, s.d. Western blots show expression of indicated proteins, with Ku70 as a loading control. BLM and Exo1 play important roles in end resection to promote HR, and the recruitments of BLM and Exo1 to DSBs are dependent on CtIP (Figure 6B, 6C: compare CtIP-WT and vector, [38]–[41]). By monitoring mRFP-tagged BLM and Exo1 localization to chromosomal DSBs in live cells, we found that the recruitment of BLM and Exo1 to DSBs was significantly reduced in the CtIP-5A-CDK and CtIP-3A-ATM mutant cell lines (Figure 6B left and 6C left). Furthermore, BLM and Exo1 recruitments were also compromised when the FHA/BRCT domains of Nbs1 are mutated (Figure 6B right and 6C right). These observations suggest that ATM-mediated phosphorylation of CtIP, which requires the interactions of the FHA/BRCT domains of Nbs1 with the 5mCDK sites, is important for promoting the recruitment of BLM and Exo1 to DSBs. Impaired recruitment of BLM and Exo1 to DSBs is thus an important factor causing the defects of the CtIP CDK - and ATM-phosphorylation mutants in end resection and HR.
The interactions of the Nbs1 FHA/BRCT domains with CtIP and with MDC1 are involved in different mechanisms to repair DSBs
It was described that the Nbs1 FHA/BRCT domains bind to MDC1 through multiple phosphorylated CK2 sites on MDC1 in mammalian cells [42]–[45]. The binding of Nbs1 to multiple CtIP CDK sites shows similar characteristics, where the 5mCDK sites redundantly contribute to the binding. We thus compared the role of Nbs1/MDC1 and Nbs1/CtIP complexes in DSB repair. First, the interaction of the Nbs1 FHA/BRCT domains with phosphorylated CK2 sites on MDC1 is important for Nbs1 to be recruited to DSB-flanking site in a γH2AX-dependent manner [42]–[46]. However, we found that ATM-dependent phosphorylation of CtIP depends on the Nbs1 FHA/BRCT domains, but not on MDC1 and H2AX (Figure 4D and Figure 7A), suggesting that the interactions of the FHA/BRCT domains of Nbs1 with CtIP at CDK sites likely occur directly at DSB ends independently of γH2AX and MDC1 to promote ATM-mediated phosphorylation of CtIP. Therefore, the Nbs1/CtIP and Nbs1/MDC1 complexes are likely formed at different chromosomal locations with respect to the DNA lesions. In addition, overexpression of MDC1 does not influence ATM-mediated phosphorylation of CtIP (Figure S6A), and thus does not affect the interaction of Nbs1 with CtIP, which further supports that the interactions of Nbs1 with CtIP and with MDC1 are independent events. Second, we found that although both Nbs1/MDC1 and Nbs1/CtIP complexes are needed for HR, they play different roles in microhomology-mediated end joining (MMEJ), which is a major pathway of Ku-independent alternative NHEJ (alt-NHEJ, [47], [48]). To monitor MMEJ, we used an EGFP-based repair substrate with 9-bp duplication around the I-Sce1 cleavage site (Figure S6B and [49]). While HR was significantly reduced when MDC1 was inactivated by shRNAs, MMEJ was not affected (Figure 7B). However, when we expressed the CDK mutant CtIP-5A-CDK or CtIP-3A-ATM with endogenous CtIP silenced by shRNAs, both HR and MMEJ were reduced (Figure 7C, Figure 1D and Figure 3A). Consistently, both MMEJ and HR were reduced in the Nbs1 FHA/BRCT domain mutant Nbs1-RRHK with endogenous Nbs1 inactivated by shRNAs (Figure 7D). These data suggest that the associations of the FHA/BRCT domains of Nbs1 with CtIP and with MDC1 are engaged in different mechanisms to regulate DSB repair. While the Nbs1/MDC1 complex is important only for HR, the Nbs1/CtIP complex is needed for both MMEJ and HR.
Fig. 7. The interactions of Nbs1 FHA/BRCT domains with CtIP or with MDC1 involve different mechanisms to regulate DSB repair. 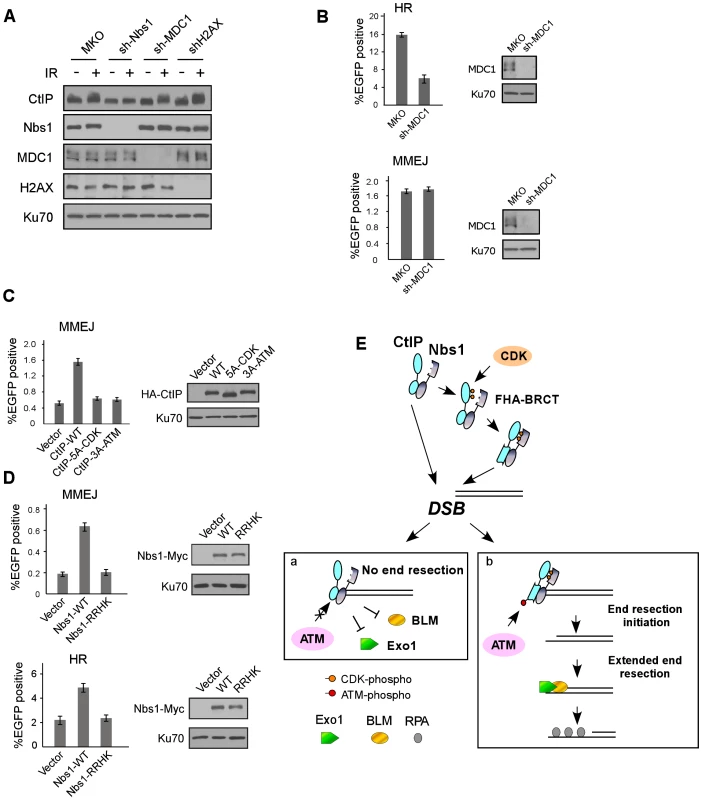
A. U2OS cells stably expressing control MKO or indicated shRNAs were treated with or without IR (10 Gy, recovered for 1 h), lysed, and immunoblotting was performed. B. EGFP-HR and EGFP-MMEJ assays were performed with U2OS cells stably expressing MKO or sh-MDC1. Western blotting was performed to show silencing of MDC1, with Ku70 as a loading control. C. EGFP-MMEJ assay was performed in U2OS cells expressing indicated CtIP variants, with endogenous CtIP silenced. Western blotting shows expression of HA-CtIP variants with Ku70 as a loading control. D. EGFP-MMEJ and EGFP-HR assays were performed in U2OS cells expressing Nbs1-WT-Myc or Nbs1-RRHK-Myc mutant with endogenous Nbs1 silenced. Western blotting shows expression of Nbs1-Myc variants with Ku70 as a loading control. E. A model to describe a role for CDK- and ATM-mediated phosphorylation of CtIP in the regulation of end resection and DSB repair (see Discussion). Discussion
Substantial evidence suggests that CDKs play a critical role in promoting end resection to activate HR, and CtIP is an important target of CDKs in this regulation [14], [17], [26], [50]. In addition to previously identified CDK sites S327 and T847 [17], [20], we identified novel CDK phosphorylation sites on CtIP and found that the FHA/BRCT domains of Nbs1 interact with these sites after CDK phosphorylation, which in turn promotes ATM-mediated phosphorylation of CtIP upon DNA damage to activate end resection and HR. Our further analyses showed that although the CDK sites S327, T847 and the newly identified 5mCDK sites are all involved in regulating CtIP function in end resection, the underlying mechanisms are different. While the CtIP-5A-CDK mutant is severely impaired in ATM-mediated phosphorylation of CtIP, mutating S327 or T847 does not show such defect. On the other hand, loss of CDK phosphorylation at S327 almost completely abolishes CtIP interaction with BRCA1, but the CtIP-5A-CDK and T847A mutants do not show defects in BRCA1 association. Therefore, CDK-mediated phosphorylation of CtIP regulates end resection through multiple levels of regulation.
We identified a novel ATM phosphorylation site T859 and showed that this site is conserved and located in the conserved RHR motif at the C-terminal Sae2-like domain [16], [25]. Along with two previously mapped ATM phosphorylation sites S664 and S745 [28], we found that T859 plays an important role in activating end resection and HR, suggesting that CtIP is a critical target for ATM to regulate end resection and HR. We further showed that phospho-mimic mutation at T859 (T859E) can bypass the requirement of CDK-mediated phosphorylation of CtIP for HR (Figure 3F), suggesting an important role of CDK-mediated phosphorylation of CtIP to promote CtIP phosphorylation by ATM at T859 to mediate DNA damage response. Damage-induced phosphorylation of Sae2 and Ctp1 was also observed in yeast and the phosphorylation is dependent on Tel1 and Mec1/Rad3 [51], [52]. Since ATM-mediated phosphorylation of CtIP does not require functional MDC1 and H2AX (Figure 7A), CtIP is likely phosphorylated by ATM at DSB ends independent of γH2AX-mediated recruitment of repair proteins to DSB-flanking chromatin.
It is intriguing that CDK-mediated phosphorylation promotes ATM to phosphorylate CtIP upon DNA damage. In this aspect, we found that in addition to a constitutive binding of Nbs1 with CtIP through the C-terminus of both proteins, phosphorylation of 5mCDK sites on CtIP induces a new interaction between these CDK sites with the FHA/BRCT domains of Nbs1. By using in vitro ATM kinase assays, we demonstrated that Nbs1 promotes ATM to phosphorylate CtIP in a manner dependent on its FHA/BRCT domains as well as the CDK sites on CtIP, suggesting that the interactions of the FHA/BRCT domains of Nbs1 with phosphorylated 5mCDK sites regulate ATM phosphorylation of CtIP. Since mutating the middle cluster of CDK sites on CtIP and the FHA/BRCT domains on Nbs1 does not affect ATM activation per se and phosphorylation of other ATM substrates such as Chk2 (Figure 4A and 4D), this CDK-dependent interaction of CtIP with Nbs1 is specifically required for facilitating ATM to phosphorylate CtIP. Based on these studies, we propose that upon DNA damage, CtIP is recruited to DSBs through constitutive interactions with MRN at the C-terminus of Nbs1 (Figure 7E). However, only those CtIP species that have been phosphorylated at the 5mCDK sites form additional associations with the FHA/BRCT domains of Nbs1, which may trigger conformational changes of CtIP and convert CtIP to a more favorable substrate for ATM by inducing exposure of the ATM phosphorylation sites such as S664, S745 and T859 at the C-terminus of CtIP. After being phosphorylated by ATM, CtIP is activated and functions to promote end resection and HR. Since CDK-mediated phosphorylation occurs only when cells enter S-phase, the requirement of CDK-dependent association of CtIP with Nbs1 to promote CtIP phosphorylation by ATM couples cell cycle regulation with DNA damage responses.
Based on the studies in yeast, it was proposed that DNA end resection is carried out via two steps: the initial end resection by the Mre11 complex and Sae2, and the extended end resection by Sgs1/Exo1 and Dna2 [53], [54]. In mammalian cells, it was described that BLM (human homologue of Sgs1) and Exo1 are important for promoting extended end resection to activate HR [39]–[41]. We observed that ATM-mediated phosphorylation of CtIP as well as CDK phosphorylation at 5mCDK sites on CtIP is required for the recruitments of BLM and Exo1 to DSBs. This study suggests that promoting BLM and Exo1 recruitment to DSBs is one important mechanism underlying ATM-mediated phosphorylation of CtIP to regulate end resection and HR. One possibility is that phosphorylation of CtIP by ATM promotes the initial end resection, which is required for BLM and Exo1 to be loaded onto DSBs to further promote end resection and activate HR (Figure 7E). In this aspect, we observed that ATM-mediated phosphorylation of CtIP is needed for both HR and MMEJ. In our designed MMEJ substrate, only limited end resection is needed to allow 9-bp microhomology to anneal for the end joining process, which implies that CtIP phosphorylation by ATM may be important for the initial limited end resection step. The exact mechanism of ATM activation of CtIP to promote initial end resection is still not clear. It is noteworthy that T859 is located in the conserved Sae2-like domain and ATM-mediated phosphorylation of T859 may modulate conformational change of this domain to activate CtIP function. Alternatively, ATM-mediated phosphorylation of CtIP may regulate the loading of BLM/Exo1 to DSBs through modulating protein-protein interactions. For instance, CtIP binds to Exo1 [38], and ATM-mediated phosphorylation of CtIP may modulate this interaction. It is also possible that ATM-mediated phosphorylation of CtIP induces CtIP to interact with other proteins, which in turn promote the recruitment of BLM and Exo1 to DSBs.
In mammalian cells, the FHA/BRCT domains of Nbs1 interact with phosphorylated CK2 sites on MDC1, which is important for Nbs1 recruitment to DSB-flanking regions [42]–[45]. In this study, we showed that the FHA/BRCT domains of human Nbs1 also bind to CtIP through the 5mCDK sites. Similar to MDC1 binding, the 5mCDK sites on CtIP redundantly mediate its interaction with the Nbs1 FHA/BRCT domains. Although sharing similar features, the bindings of the FHA/BRCT domains of Nbs1 with CtIP and with MDC1 appear to be independent and contribute to different modes of regulating DSB repair. While the Nbs1/MDC1 complex is recruited to the chromatin around DSBs by γH2AX, the Nbs1/CtIP complex likely binds directly to DSB ends and promotes ATM-mediated phosphorylation of CtIP in a manner independent of H2AX and MDC1. Furthermore, the CDK-dependent CtIP and Nbs1 interaction as well as ATM-mediated phosphorylation of CtIP are important for both HR and MMEJ, but MDC1 is only required for HR and not for MMEJ. One possibility for this difference is that the Nbs1/CtIP complex participates in both initial end resection required for MMEJ and subsequent extended end resection for HR, but the Nbs1/MDC1 complex only participates in the step to promote extended end resection.
Our studies identify new CDK sites on CtIP, and reveal a novel mechanism which couples CDK - and ATM-mediated phosphorylation of CtIP to promote end resection and HR. The involvement of multiple CDK phosphorylation events (S327, 5mCDK sites and T847) as well as ATM-mediated phosphorylation to regulate CtIP function in DSB repair supports the notion that CtIP is a key regulator for cell cycle-dependent selection of DSB repair pathways. The regulated interaction of CtIP and Nbs1 to modulate ATM-mediated phosphorylation of CtIP also provides new information as to how these proteins function coordinately with each other to promote appropriate DSB repair pathways during the cell cycle to avoid genome instability.
Materials and Methods
Cell culture and antibodies
Human U2OS, T98G, HeLa and 293T cells were cultured at 37°C in Dulbecco's modified eagle's medium with 10% fetal bovine serum (FBS) in the presence of antibiotics and 5% CO2. Insect cell line Sf21 was cultured in Grace's insect medium (Gibco) supplemented with 10% fetal bovine serum; Sf9 insect cell line was cultured in Sf-900II SFM medium (Gibco). T98G cells were synchronized via serum starvation by culturing cells in DMEM containing 0.1% FBS for 48 hours, followed by release into complete medium containing 10% FBS [55], [56]. Cell cycle profile analysis of synchronized T98G cells was performed as described [55], [56].
Monoclonal anti-CtIP antibody was generously provided by R. Bear [Columbia University, [57]. CtIP polyclonal antibodies were described previously [49]. The polyclonal antibodies against Mre11 (D27, 1∶500 dilution) and monoclonal anti-Nbs1 antibody (EE15, 1∶500 dilution) were described previously [18]. Other antibodies used include anti-Flag-M2 (Sigma, 1∶1000 dilution), anti-Myc-9E10 (Novus Biologicals, 1∶1000 dilution), anti-HA-11 (Covance, 1∶1000 dilution), anti-Ku70 (Santa Cruz Biotechnology, 1∶3000 dilution), anti-RPA2 (Oncogene, 1∶200 dilution), anti-H2AX-S319-p, anti-Chk1-S317-p (Cell Signaling Technology, 1∶200 and 1∶500 dilution), anti-Chk1 (R&D Systems, 1∶800 dilution), anti-Chk2-T68 (GeneTex, 1∶600 dilution), anti-Chk2 (Santa Cruz Biotechnology, 1∶800 dilution), anti-ATM (GeneTex, 1∶500 dilution), anti-ATM-S1981-p (GeneTex, 1∶500 dilution), anti-MDC1 (Sigma, 1∶500 dilution), anti-His (Invitrogen, 1∶500 dilution), anti-GAPDH (RDI, 1∶500 dilution).
Plasmids, mutagenesis, and shRNA/RNAi
CtIP cDNA [20] was subcloned into pUC19 at BamHI/SalI and used to generate CtIP variants by site-directed mutagenesis (Stratagene). CtIP wild-type and indicated mutants were then subcloned into mammalian expression vectors pcDNA3 or pBabepuro containing HA, Myc or Flag epitopes. EGFP-tagged CtIP or indicated mutants were generated using EGFP-C1 expression vector (Clontech). GST-fused CtIP fragments were generated using pGEX4T-1 (GE Healthcare). Recombinant baculoviruses expressing GST-, HA - or Flag-tagged CtIP or Nbs1 were generated by using Bac-to-Bac Baculovirus expression systems (Invitrogen). Nbs1-RRHK mutant was made by site-directed mutagenesis (Stratagene), and Nbs1 truncation mutants were generated by PCR amplification of indicated fragments. Nbs1 wild-type and indicated mutants were subcloned into pBabepuro or pFastBAC-HTb (Invitrogen) vectors containing in-frame C-terminal Myc-, 10×His-, 6×His - or GST-tags. EGFP-Exo1 was a gift from Dr. Kum Kum Khanna [58] and used to make Exo1-mRFP-N1. mRFP-BLM was a gift from Dr. Marek Rusin [59]. HA-MDC1 was a gift from Dr. Jiri Lukas [42].
Silencing of endogenous CtIP by shRNAs and siRNAs was performed as described [49]. shRNA - and siRNA - resistant CtIP wild-type and indicated mutants were constructed by mutating four nucleotides at the shRNA/siRNA targeting sequences by site-directed mutagenesis (Stratagene). All other shRNAs were generated in pMKO vector using the following RNAi target sequences as designed by Dharmacon: for shH2AX GGGACGAAGCACUUGGUAA; for shMDC1 GUCUCCCAGAAGACAGUGAUU; for shNbs1 GAAGAAACGUGAACUCAAGUU; for shKu70 GAAGGAGGUUGCAGCAUUGUG and for shATM GGGCAUUACGGGUGUUGAA.
Immunoprecipitation, in vitro binding assay, immunostaining, and clonogenic survival assay
Whole cell lysis, co-immunoprecipitation and Western blotting were performed as described [55]. Cells were lysed in NETN [150 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl (pH 8.0), 0.5% NP-40] containing protease inhibitors. Immunoprecipitation was conducted by incubating primary antibodies with cell lysates at 4°C for 4 h, followed by the addition of protein A-agarose for one additional hour. For in vitro binding assay, GST-CtIP, Flag-Nbs1, Nbs1-GST, HA-CtIP or Flag-CtIP was expressed alone or in combination in insect cells, and binding assay was performed using NETN buffer.
For immunostaining, cells were cultured on sterile coverslips and treated with CPT as indicated. Cells were fixed with 4% paraformaldehyde, followed by immunostaining as described [18], [49]. After blocking with 5% goat serum in phosphate-buffered saline (PBS), fixed cells were incubated with indicated primary antibodies at 4°C overnight followed by incubation for 1 h at room temperature with either fluorescein isothiocyanate-conjugated anti-mouse and/or rhodamine-conjugated anti-rabbit secondary antibodies (Jackson ImmunoResearch Laboratories), followed by DAPI staining for nuclei. Fluorescence microscopy was performed using a Nikon Eclipse E800 upright fluorescence microscope (60×/1.4 oil Plan-APO objective, with FITC, Rhodamine and DAPI fluorochromes). Images were captured with a digital camera (AG Heinze, Co., Model RT-SE6) and analyzed using SPOT Advanced imaging software (Diagnostic Instruments, Inc).
For clonogenic survival assay, indicated cell lines were treated with camptothecin for 1 h, and allowed to culture in complete media for 10–14 days. Colonies were stained with a solution containing 0.5% crystal violet and 20% ethanol then counted, with the percentage of cell viability shown. Error bars are s.d. of three independent experiments [16].
Homologous recombination (HR) and microhomology-mediated end-joining (MMEJ) assay
HR and MMEJ assays for DSB repair were described previously [49]. U2OS cells carrying EGFP-HR (Figure S1B) or EGFP-MMEJ (Figure S6B), with or without expressing indicated CtIP or Nbs1 variants, and/or indicated shRNAs, were transfected with I-SceI-IRES-dsRedNLS and plated in non-selective medium. After 72 h, cells were trypsinized and collected for fluorescence-activated cell sorting (FACS) analysis of EGFP-positive events, using a BD Accuri C6 flow cytometer and accompanying analysis software (Becton-Dickinson, MA, USA).
Laser-induced micro-irradiation and live-cell imaging
U2OS cells expressing indicated EGFP - or mRFP-tagged proteins, with or without expressing indicated shRNAs, were cultured in sterile glass-bottom dishes (MatTek) in DMEM medium containing 10% FBS. Dishes were directly placed onto the microscope stage for image acquisition at room temperature and processed within a 1 h maximum time period for the experimental duration of any given dish. DSBs were generated in live-cell nuclei by laser-induced microirradiation using a picosecond short-pulsed green laser (a diode-pumped second harmonic 532 nm Nd∶YAG laser microbeam with 76 MHz repetition rate, 12 ps pulse duration). The average laser power used for DNA cutting was 16 milliwatts (post-objective), and the total energy delivered per focused laser spot was 480 mJ. Live-cell, time-lapse images were taken by the robotic laser microscopy system (RoboLase II) built on a Zeiss Axiovert 200 M inverted fluorescence microscope (63×,1.4 oil Plan-Apo objective, using FITC and Rhodamine fluorochromes) and digital camera (Hamamatsu ORCA-EA) [49], [60]. Images were captured using RoboLase II system software, and raw images (16-bit) were imported into ImageJ software (NIH of USA) for processing. Fluorescence intensities of the micro-irradiated area were determined by measuring the mean absolute intensity of the microirradiated areas, with the mean cellular background intensity subtracted [49], [61]. Each data point shown is the average of 10 independent measurements.
Protein purification and in vitro kinase assay
GST-fused CtIP C-terminal region (residues 750–897) was expressed in BL21 and affinity-purified with glutathione-Sepharose 4B (GE Healthcare). CtIP wild-type and indicated mutants were expressed in Sf9 insect cells and purified by Ni-NTA and GST tandem affinity purification. C-terminal His-tagged Nbs1 wild-type and indicated mutant were expressed in Sf9 insect cells and purified by Ni-NTA column followed by MonoQ column. ATM in vitro kinase assays were previously described [49]. Briefly, FLAG-tagged ATM was transiently transfected into 293T cells and immunoprecipitated using anti-FLAG M2-agarose (Sigma). Immunoprecipitates were extensively washed and then incubated with purified proteins in the presence of 10 µCi γ-32P-ATP in ATM kinase buffer [25 mM HEPES (pH 7.4), 50 mM NaCl, 10 mM MnCl2, 10 mM MgCl2, 1 mM DTT, 5 µM ATP]. The kinase reaction was conducted at 30°C for 30 min. Phosphorylated proteins were separated by SDS-PAGE and visualized using a PhophorImage scanner (GE, Typhoon Trio). To test the effects of Nbs1 WT and mutants on CtIP phosphorylation by ATM in vitro, indicated amounts of CtIP and Nbs1 proteins were pre-incubated for 15 min on ice and then combined with Flag-ATM immunoprecipitates and γ-32P-ATP to perform the ATM kinase reaction.
Mass spectrometry
Mass spectrometry was described before [62]. Five liters of HeLa cell suspension was lysed with NETN containing 1 µg/ml pepstatin A and aprotinin, and endogenous CtIP was affinity-purified for analysis using mammalian cells. For analysis using insect cells, recombinant human CtIP was expressed in SF9 insect cells, and purified by Ni-NTA and GST tandem affinity purification. Protein samples were precipitated by mixing cold sample solution (1 vol) with 1/3 vol of 100% (w/v) trichloroacetic acid (TCA) (6.1 N) to a final TCA concentration (conc.) of 25%, incubated on ice for 3 h, pelleted by cold centrifugation with acetone washes, and dried.
Proteins were dissolved in 60 µL of 100 mM Tris-HCl, pH 8.5 containing 8 M urea. The protein was reduced by adding 500 mM tris (2-carboxyethyl) phosphine to a final conc. of 5 mM, followed by carboxyamidomethylation of cysteines with 500 mM iodoacetamide (final conc. 10 mM). Protein was prepared for analysis of phosphorylation as described [62]. The protein sample was equally split for separate digestions by Subtilisin (Promega), Elastase, and Trypsin. The resulting peptides from the three digests were dissolved in 20% acetonitrile and 2% formic acid, combined, and subjected to TiO2 enrichment and LC-MS/MS analysis.
For TiO2 enrichment of phosphopeptide, a TiO2 column was made by pressure-slurry packing TiO2 (5-µ partisphere, Whatman, Clifton, NJ) into fused-silica capillary (250 µm i.d.). The column was washed with buffer A [water/acetonitrile/formic acid (95∶5∶0.1, v/v/v)] and buffer B [water/acetonitrile/formic acid (20∶80∶0.1, v/v/v)], then eluted using 250 mM ammonium bicarbonate onto an analytical column, which was then inserted into an Agilent 1200 quaternary HPLC pump for mass spectrometry analysis.
Data-dependent tandem mass spectrometry (MS/MS) analysis was performed with a LTQ-Orbitrap mass spectrometer (ThermoFisher). Full MS and tandem mass spectra were extracted and searched against an EBI-IPI human protein database (database released on June 28, 2007). A decoy database containing reversed sequences [63] was used to estimate peptide probabilities and identify false positives. Tandem mass spectra were matched to sequences using the ProLuCID algorithm [64].
ProLuCID searches were performed on an Intel Xeon 80-processor cluster (search tolerance set to 3 Da). The Cysteine mass was modified by +57.02146 Da to account for carboxyamidomethylation, and Serine, Threonine and Tyrosine were modified by +79.9663 Da for phosphorylation. Since no enzymatic cleavage conditions were imposed, the search included all candidates within the mass tolerance window, regardless of tryptic status [65]. Validation of peptide/spectrum matches (PSM) was assessed in DTASelect [66] using two SEQUEST [67] defined parameters, the cross-correlation score (XCorr), and normalized difference in cross-correlation scores (DeltaCN). Search results were grouped by charge state (+1, +2, +3, greater than +3) and tryptic status (full, half, and non) into 12 distinct sub-groups, and distribution of XCorr, DeltaCN, and DeltaMass values for direct and decoy database PSMs were obtained and separated by discriminant analysis. Peptide match probabilities were calculated based on a non-parametric fit of direct and decoy score distributions (minimum threshold of 90%). After filtering for false positives based on the percentage of reverse decoy PSMs, we estimate that both the protein and peptide false discovery rates were reduced to 0.0%–0.5%.
Supporting Information
Zdroje
1. KeeneyS, NealeMJ (2006) Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans 34 : 523–525.
2. JungD, GiallourakisC, MostoslavskyR, AltFW (2006) Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol 24 : 541–570.
3. BassingCH, AltFW (2004) The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst) 3 : 781–796.
4. KhannaKK, JacksonSP (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27 : 247–254.
5. MatsuuraS, TauchiH, NakamuraA, KondoN, SakamotoS, et al. (1998) Positional cloning of the gene for Nijmegen breakage syndrome. Nat Genet 19 : 179–181.
6. VaronR, VissingaC, PlatzerM, CerosalettiKM, ChrzanowskaKH, et al. (1998) Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93 : 467–476.
7. StewartGS, MaserRS, StankovicT, BressanDA, KaplanMI, et al. (1999) The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99 : 577–587.
8. SavitskyK, Bar-ShiraA, GiladS, RotmanG, ZivY, et al. (1995) A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268 : 1749–1753.
9. MikiY, SwensenJ, Shattuck-EidensD, FutrealPA, HarshmanK, et al. (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266 : 66–71.
10. LieberMR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79 : 181–211.
11. MoynahanME, JasinM (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 11 : 196–207.
12. SymingtonLS, GautierJ (2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45 : 247–271.
13. D'AmoursD, JacksonSP (2002) The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol 3 : 317–327.
14. JazayeriA, FalckJ, LukasC, BartekJ, SmithGC, et al. (2006) ATM - and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8 : 37–45.
15. BuisJ, WuY, DengY, LeddonJ, WestfieldG, et al. (2008) Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135 : 85–96.
16. SartoriAA, LukasC, CoatesJ, MistrikM, FuS, et al. (2007) Human CtIP promotes DNA end resection. Nature 450 : 509–514.
17. HuertasP, JacksonSP (2009) Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284 : 9558–9565.
18. ChenL, NieveraCJ, LeeAY, WuX (2008) Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem 283 : 7713–7720.
19. YuX, WuLC, BowcockAM, AronheimA, BaerR (1998) The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem 273 : 25388–25392.
20. YuX, ChenJ (2004) DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol 24 : 9478–9486.
21. YuanJ, ChenJ (2009) N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J Biol Chem 284 : 31746–31752.
22. LloydJ, ChapmanJR, ClappertonJA, HaireLF, HartsuikerE, et al. (2009) A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell 139 : 100–111.
23. WilliamsRS, DodsonGE, LimboO, YamadaY, WilliamsJS, et al. (2009) Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 139 : 87–99.
24. LengsfeldBM, RattrayAJ, BhaskaraV, GhirlandoR, PaullTT (2007) Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28 : 638–651.
25. LimboO, ChahwanC, YamadaY, de BruinRA, WittenbergC, et al. (2007) Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28 : 134–146.
26. YunMH, HiomK (2009) CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459 : 460–463.
27. HuertasP, Cortes-LedesmaF, SartoriAA, AguileraA, JacksonSP (2008) CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455 : 689–692.
28. LiS, TingNS, ZhengL, ChenPL, ZivY, et al. (2000) Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature 406 : 210–215.
29. YouZ, ShiLZ, ZhuQ, WuP, ZhangYW, et al. (2009) CtIP links DNA double-strand break sensing to resection. Mol Cell 36 : 954–969.
30. LeeJH, PaullTT (2007) Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26 : 7741–7748.
31. HariFJ, SpycherC, JungmichelS, PavicL, StuckiM (2010) A divalent FHA/BRCT-binding mechanism couples the Mre11-Rad50-Nbs1 complex to damaged chromatin. EMBO Rep 11 : 387–392.
32. CerosalettiKM, ConcannonP (2003) Nibrin forkhead-associated domain and breast cancer C-terminal domain are both required for nuclear focus formation and phosphorylation. J Biol Chem 278 : 21944–21951.
33. DurocherD, SmerdonSJ, YaffeMB, JacksonSP (2000) The FHA domain in DNA repair and checkpoint signaling. Cold Spring Harb Symp Quant Biol 65 : 423–431.
34. HuangM, ElledgeSJ (2000) The FHA domain, a phosphoamino acid binding domain involved in the DNA damage response pathway. Cold Spring Harb Symp Quant Biol 65 : 413–421.
35. GloverJN, WilliamsRS, LeeMS (2004) Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem Sci 29 : 579–585.
36. DurocherD, HenckelJ, FershtAR, JacksonSP (1999) The FHA domain is a modular phosphopeptide recognition motif. Mol Cell 4 : 387–394.
37. MohammadDH, YaffeMB (2009) 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 8 : 1009–1017.
38. EidW, StegerM, El-ShemerlyM, FerrettiLP, Pena-DiazJ, et al. (2010) DNA end resection by CtIP and Exonuclease 1 prevents genomic instability. EMBO Rep 11 : 962–968.
39. GravelS, ChapmanJR, MagillC, JacksonSP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22 : 2767–2772.
40. NimonkarAV, GenschelJ, KinoshitaE, PolaczekP, CampbellJL, et al. (2011) BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25 : 350–362.
41. NimonkarAV, OzsoyAZ, GenschelJ, ModrichP, KowalczykowskiSC (2008) Human Exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci U S A 105 : 16906–16911.
42. MelanderF, Bekker-JensenS, FalckJ, BartekJ, MailandN, et al. (2008) Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of Nbs1 at the DNA damage-modified chromatin. J Cell Biol 181 : 213–226.
43. SpycherC, MillerES, TownsendK, PavicL, MorriceNA, et al. (2008) Constitutive phosphorylation of MDC1 physically links the Mre11-Rad50-Nbs1 complex to damaged chromatin. J Cell Biol 181 : 227–240.
44. ChapmanJR, JacksonSP (2008) Phospho-dependent interactions between Nbs1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep 9 : 795–801.
45. WuL, LuoK, LouZ, ChenJ (2008) MDC1 regulates intra-S-phase checkpoint by targeting Nbs1 to DNA double-strand breaks. Proc Natl Acad Sci U S A 105 : 11200–11205.
46. StuckiM, ClappertonJA, MohammadD, YaffeMB, SmerdonSJ, et al. (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123 : 1213–1226.
47. McVeyM, LeeSE (2008) MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet 24 : 529–538.
48. NussenzweigA, NussenzweigMC (2007) A backup DNA repair pathway moves to the forefront. Cell 131 : 223–225.
49. WangH, ShaoZ, ShiLZ, HwangPY, TruongLN, et al. (2012) CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded Breaks. J Biol Chem 287 : 21471–21480.
50. IraG, PellicioliA, BalijjaA, WangX, FioraniS, et al. (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431 : 1011–1017.
51. AkamatsuY, MurayamaY, YamadaT, NakazakiT, TsutsuiY, et al. (2008) Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol Cell Biol 28 : 3639–3651.
52. BaroniE, ViscardiV, Cartagena-LirolaH, LucchiniG, LongheseMP (2004) The functions of budding yeast Sae2 in the DNA damage response require Mec1 - and Tel1-dependent phosphorylation. Mol Cell Biol 24 : 4151–4165.
53. ZhuZ, ChungWH, ShimEY, LeeSE, IraG (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134 : 981–994.
54. MimitouEP, SymingtonLS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455 : 770–774.
55. OlsonE, NieveraCJ, LiuE, LeeAY, ChenL, et al. (2007) The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol Cell Biol 27 : 6053–6067.
56. LeeAY, ChibaT, TruongLN, ChengAN, DoJ, et al. (2012) Dbf4 is direct downstream target of ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) protein to regulate intra-S-phase checkpoint. J Biol Chem 287 : 2531–2543.
57. YuX, BaerR (2000) Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J Biol Chem 275 : 18541–18549.
58. BoldersonE, TomimatsuN, RichardDJ, BoucherD, KumarR, et al. (2010) Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res 38 : 1821–1831.
59. VaitiekunaiteR, ButkiewiczD, KrzesniakM, PrzybylekM, GrycA, et al. (2007) Expression and localization of Werner syndrome protein is modulated by SIRT1 and PML. Mech Ageing Dev 128 : 650–661.
60. BotvinickEL, BernsMW (2005) Internet-based robotic laser scissors and tweezers microscopy. Microsc Res Tech 68 : 65–74.
61. UematsuN, WeteringsE, YanoK, Morotomi-YanoK, JakobB, et al. (2007) Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol 177 : 219–229.
62. MacCossMJ, McDonaldWH, SarafA, SadygovR, ClarkJM, et al. (2002) Shotgun identification of protein modifications from protein complexes and lens tissue. Proc Natl Acad Sci U S A 99 : 7900–7905.
63. PengJ, EliasJE, ThoreenCC, LickliderLJ, GygiSP (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2 : 43–50.
64. XuT, VenableJD, ParkSK, CociorvaD, LuB, et al. (2006) ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Molecular & Cellular Proteomics 5: S174–S174.
65. LuB, XuT, ParkSK, YatesJR3rd (2009) Shotgun protein identification and quantification by mass spectrometry. Methods Mol Biol 564 : 261–288.
66. TabbDL, McDonaldWH, YatesJR3rd (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1 : 21–26.
67. EngJK, MccormackAL, YatesJR (1994) An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. Journal of the American Society for Mass Spectrometry 5 : 976–989.
68. SomanathanS, SuchynaTM, SiegelAJ, BerezneyR (2001) Targeting of PCNA to sites of DNA replication in the mammalian cell nucleus. J Cell Biochem 81 : 56–67.
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



