-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaConditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
The RAD9-RAD1-HUS1 (9-1-1) complex is a heterotrimeric PCNA-like clamp that responds to DNA damage in somatic cells by promoting DNA repair as well as ATR-dependent DNA damage checkpoint signaling. In yeast, worms, and flies, the 9-1-1 complex is also required for meiotic checkpoint function and efficient completion of meiotic recombination; however, since Rad9, Rad1, and Hus1 are essential genes in mammals, little is known about their functions in mammalian germ cells. In this study, we assessed the meiotic functions of 9-1-1 by analyzing mice with germ cell-specific deletion of Hus1 as well as by examining the localization of RAD9 and RAD1 on meiotic chromosomes during prophase I. Hus1 loss in testicular germ cells resulted in meiotic defects, germ cell depletion, and severely compromised fertility. Hus1-deficient primary spermatocytes exhibited persistent autosomal γH2AX and RAD51 staining indicative of unrepaired meiotic DSBs, synapsis defects, an extended XY body domain often encompassing partial or whole autosomes, and an increase in structural chromosome abnormalities such as end-to-end X chromosome-autosome fusions and ruptures in the synaptonemal complex. Most of these aberrations persisted in diplotene-stage spermatocytes. Consistent with a role for the 9-1-1 complex in meiotic DSB repair, RAD9 localized to punctate, RAD51-containing foci on meiotic chromosomes in a Hus1-dependent manner. Interestingly, RAD1 had a broader distribution that only partially overlapped with RAD9, and localization of both RAD1 and the ATR activator TOPBP1 to the XY body and to unsynapsed autosomes was intact in Hus1 conditional knockouts. We conclude that mammalian HUS1 acts as a component of the canonical 9-1-1 complex during meiotic prophase I to promote DSB repair and further propose that RAD1 and TOPBP1 respond to unsynapsed chromatin through an alternative mechanism that does not require RAD9 or HUS1.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003320
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003320Summary
The RAD9-RAD1-HUS1 (9-1-1) complex is a heterotrimeric PCNA-like clamp that responds to DNA damage in somatic cells by promoting DNA repair as well as ATR-dependent DNA damage checkpoint signaling. In yeast, worms, and flies, the 9-1-1 complex is also required for meiotic checkpoint function and efficient completion of meiotic recombination; however, since Rad9, Rad1, and Hus1 are essential genes in mammals, little is known about their functions in mammalian germ cells. In this study, we assessed the meiotic functions of 9-1-1 by analyzing mice with germ cell-specific deletion of Hus1 as well as by examining the localization of RAD9 and RAD1 on meiotic chromosomes during prophase I. Hus1 loss in testicular germ cells resulted in meiotic defects, germ cell depletion, and severely compromised fertility. Hus1-deficient primary spermatocytes exhibited persistent autosomal γH2AX and RAD51 staining indicative of unrepaired meiotic DSBs, synapsis defects, an extended XY body domain often encompassing partial or whole autosomes, and an increase in structural chromosome abnormalities such as end-to-end X chromosome-autosome fusions and ruptures in the synaptonemal complex. Most of these aberrations persisted in diplotene-stage spermatocytes. Consistent with a role for the 9-1-1 complex in meiotic DSB repair, RAD9 localized to punctate, RAD51-containing foci on meiotic chromosomes in a Hus1-dependent manner. Interestingly, RAD1 had a broader distribution that only partially overlapped with RAD9, and localization of both RAD1 and the ATR activator TOPBP1 to the XY body and to unsynapsed autosomes was intact in Hus1 conditional knockouts. We conclude that mammalian HUS1 acts as a component of the canonical 9-1-1 complex during meiotic prophase I to promote DSB repair and further propose that RAD1 and TOPBP1 respond to unsynapsed chromatin through an alternative mechanism that does not require RAD9 or HUS1.
Introduction
The requirement for effective genome maintenance is particularly notable in germ cells, which must transmit high quality DNA to future generations. Therefore, germ cells must employ DNA damage response mechanisms that are at least as stringent as those present in somatic cells. Meiosis, which includes the intentional generation and subsequent repair of DNA double-strand breaks (DSBs), involves a variety of DNA repair mechanisms as well as cell cycle checkpoints that monitor chromosomal integrity; however, much remains unknown about how these mechanisms operate in mammalian cells. In this study, we investigated how an essential DNA repair and DNA damage checkpoint complex, the RAD9-RAD1-HUS1 (9-1-1) complex, functions to maintain genome integrity in the germline.
The 9-1-1 complex is a heterotrimeric ring that shares extensive structural similarity with PCNA, the sliding clamp that functions in DNA replication and repair [1]. In mammalian somatic cells, the best characterized role of 9-1-1 is in activation of ATR pathway DNA damage checkpoint signaling following replication-associated DNA damage [2]. In response to stalled replication forks or lesions involving single-stranded DNA, the 9-1-1 complex is loaded onto 5′ recessed ends by the RAD17-RFC clamp loader complex. The ATR kinase is independently loaded onto RPA-coated single-stranded DNA through the interaction of ATR-interacting protein (ATRIP) with both ATR and RPA [3]. The phosphorylated RAD9 C-terminus physically recruits the ATR activator TOPBP1, which then stimulates the kinase activity of ATR [reviewed in 4]–[6]. Once active, ATR phosphorylates downstream effectors such as CHK1 that promote cell cycle arrest, DNA repair, or apoptosis.
In addition to its known checkpoint signaling function, the 9-1-1 clamp physically interacts with a variety of DNA repair proteins, indicating that, like PCNA, 9-1-1 may also function as a scaffold for recruiting DNA repair proteins to damage sites. Indeed, 9-1-1 has been shown to interact with RAD51 in human cells [7], and 9-1-1 complexes from yeast, mouse, and human physically interact with, and in some cases, stimulate the activity of, translesion polymerases [8] as well as base excision repair factors, including DNA Polymerase β [9], [10], FEN1 [11], [12], APE1 [10], DNA ligase I [13], and the NEIL1, TDG, OGG1, and MutY glycosylases [14]–[18]. Other 9-1-1 physical interactors are as varied as HDAC1 histone deacetylase [19], WRN helicase [20], mismatch repair factors MSH2, MSH3, MSH6, and MLH1 [21], [22], and the ATR co-activator RHINO [23]. That 9-1-1 interacts with and stimulates numerous DNA repair factors points to a direct role of 9-1-1 in facilitating DNA repair apart from its ATR-dependent checkpoint signaling activities, though in vivo evidence for this proposed function for the mammalian 9-1-1 complex remains limited.
During meiosis, homologous chromosomes pair, synapse, and undergo recombination. The ATR signaling pathway has been proposed to monitor both meiotic chromosomal synapsis and recombination as part of the pachytene DNA damage checkpoint that is believed to prevent cells with unrepaired breaks or synapsis defects from progressing beyond meiotic prophase I. ATR and its activator TOPBP1 localize to unsynapsed autosome cores in zygotene spermatocytes and to the sex body domain during pachytene [24]–[26], where the mostly unsynapsed X and Y chromosomes in males are packaged into a unique chromatin territory [27], [28]. Phosphorylation of histone H2AX to γH2AX, which marks the entire sex body domain, is thought to be ATR-dependent, whereas H2AX phosphorylation at DSB sites on autosomes during leptotene and early zygotene stages appears to depend primarily on another checkpoint kinase, ATM [26], [29]. In the sex body, ATR promotes transcriptional silencing of the X and Y chromosomes in a process termed meiotic sex chromosome inactivation (MSCI) [30], which is required for male fertility [31]. ATR and TOPBP1 also promote meiotic silencing of unsynapsed chromatin (MSUC), a process fundamentally similar to MSCI that functions at sites of autosomal asynapsis during pachytene [24], [25], [30], [32]. MSUC and MSCI require BRCA1 loading at asynapsed chromatin, and both BRCA1 and the HORMA domain-containing protein HORMAD2 must be present at asynapsed axes in order for ATR to be recruited [26], [33]. While TOPBP1 localizes to the sex body and sites of failed autosomal synapsis and is a known ATR activator, it is unclear whether it is strictly required for MSCI and MSUC, and also whether its recruitment is dependent upon the 9-1-1 complex, as during somatic responses to replication stress. The canonical model for mammalian ATR activation predicts a requirement for 9-1-1 in meiotic ATR activation, perhaps after HORMAD2 or BRCA1 loading, although this has remained untested until this study.
Studies in several organisms indicate that the 9-1-1 complex plays critical roles during meiosis, not only for activation of the pachytene meiotic checkpoint in response to DNA damage or synapsis defects [34]–[36], but also in DSB repair. Yeast rad17 (mRad1) mutants exhibit persistent RAD51 foci [37], in addition to increased rates of ectopic recombination and altered crossover frequencies during meiosis [38], and DDC1 (mRAD9) colocalizes with RAD51 [35]. In Drosophila, meiotic DSBs are not repaired efficiently in the absence of Hus1 [39]. Additionally, Spo11-deficient mouse mutants, which lack meiotic DSBs, have reduced Hus1 expression, suggesting that 9-1-1 functions depend upon DSB formation [40]. The same is true for expression of Mre11 and Brca2, which are known to be involved in DSB repair, further supporting the idea that 9-1-1 may function at meiotic DSBs.
The existing evidence from lower eukaryotes, together with reported physical interactions with mammalian DNA repair and meiotic proteins, suggests a possible critical role for the 9-1-1 complex during mammalian meiosis, either through activation of ATR-dependent checkpoint functions or direct roles in DNA repair at damage sites. Although mouse RAD1 has been determined to localize to both synapsed and unsynapsed meiotic chromosomes [41], little is known about the requirements for 9-1-1 complex components in mammalian germ cells because mutation of Rad9, Rad1, or Hus1 results in embryonic lethality [42]–[45]. We addressed this by producing mice with germ cell-specific Hus1 deletion. Two conditional knockout models were generated, utilizing Stra8-Cre expressed in spermatogonia and Spo11-Cre expressed in spermatocytes, and in both cases, Hus1 inactivation in testicular germ cells resulted in persistent meiotic DNA damage, chromosomal defects, and germ cell depletion. Meiotic silencing, on the other hand, appeared unaffected, indicating that HUS1 is dispensable for at least some meiotic ATR functions, including meiotic sex chromosome inactivation. Intriguingly, Hus1 inactivation resulted in complete loss of RAD9 but not RAD1 foci from meiotic chromosomes, suggesting that RAD1 has additional meiotic functions, likely related to the monitoring of unsynapsed chromatin based on its localization pattern. Together, these data indicate that efficient DSB repair, proper maintenance of the sex body domain, and preservation of chromosome integrity during meiosis require HUS1, and additionally highlight novel roles for 9-1-1 components outside of the conventional heterotrimeric complex.
Results
Generation of germ cell-specific Hus1 conditional knockout mice
Several lines of evidence from non-vertebrate organisms suggest that the mammalian 9-1-1 complex, the components of which are highly expressed in mouse and human testis, is likely to be critical for normal germ cell development and completion of meiosis. In order to assess the meiotic functions of the 9-1-1 complex, we generated mice in which the Hus1 gene was conditionally inactivated specifically in testicular germ cells. We combined a conditional floxed Hus1 allele (Figure S1A; [46]) with two Cre-expressing lines, Stra8-Cre and Spo11-Cre. Stra8-Cre mice begin to express the CRE recombinase in spermatogonia [47], which undergo several rounds of mitotic division prior to meiosis, whereas CRE expression in Spo11-Cre mice begins in spermatocytes that have initiated meiosis (Figures S1 and S2; Text S1). We focused our analysis on mice made with Stra8-Cre and used the Spo11-Cre line to confirm our findings and to assess whether defects we observed originated during pre-meiotic or meiotic processes. Hus1 deletion in the testis was confirmed by Southern blot detection of Hus1Δ2,3, the null allele produced by CRE-mediated recombination (Figures S1C, S1D and S2D). As shown in Figure 1A and Figure S3B, Western blotting of whole testis lysates further confirmed that Hus1 gene deletion using Stra8-Cre resulted in a drastic reduction in HUS1 protein level in the testis. Despite high genomic Hus1 deletion efficiency, the reduction in HUS1 protein levels was more subtle in Spo11-Cre Hus1 conditional knockout (CKO) mice, consistent with expectation that CRE expression from the Spo11 promoter would result in HUS1 loss in a more restricted subset of cells and with delayed kinetics relative to that in mice with Stra8-Cre (Figure S3C). Hus1 loss also led to a significant reduction in total RAD9 and RAD1 protein levels in testes from Stra8-Cre Hus1 CKO animals (Figure S3A, S3B), indicating that the entire 9-1-1 complex was destabilized in the absence of HUS1.
Fig. 1. Conditional Hus1 inactivation in the mouse testis results in reduced testis size and sperm count. 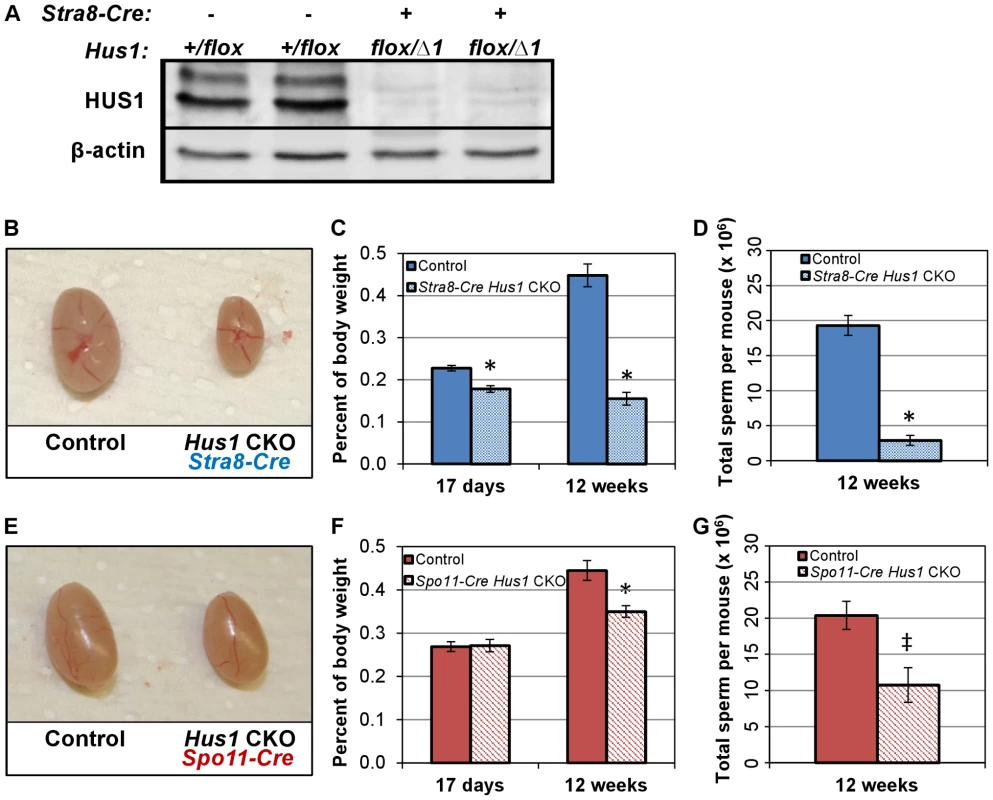
A. Western blot analysis of HUS1 protein in adult (12-week old) control and Hus1 CKO testes. B, E. Photographs of testes from 12-week old Hus1 CKO and control mice from Stra8-Cre and Spo11-Cre crosses, respectively. In these representative images, the Stra8-Cre control is Cre-positive Hus1+/flox, whereas the Spo11-Cre control is Cre-negative Hus1flox/Δ1. C, F. Testis weights of 17-day and 12-week old adult Hus1 CKO and control mice from both Stra8-Cre and Spo11-Cre crosses, shown as the mean testis weight relative to body weight ± SEM. D, G. Epididymal sperm counts from Stra8-Cre and Spo11-Cre Hus1 CKO 12-week old males, shown as the mean ± SEM. Statistically significant differences between Hus1 CKO and the respective control as determined by Student's t-test are indicated (* at p<0.001; ‡ at p<0.01). Conditional Hus1 inactivation results in reduced testis size, reduced epididymal sperm count, and severely compromised fertility
In both the Stra8-Cre and Spo11-Cre Hus1 CKO models, Hus1 loss resulted in reduced adult testis size (Figure 1B, 1E). As expected, the phenotype was more severe in Stra8-Cre Hus1 CKOs with earlier Hus1 loss than Spo11-Cre Hus1 CKOs. Testis weights were significantly reduced in Stra8-Cre Hus1 CKO males as early as postnatal day 17 (Figure 1C), raising the possibility of roles for Hus1 during early meiotic and/or pre-meiotic stages, including during spermatogonial DNA replication given the role of 9-1-1 during S-phase in somatic cells [2], [48]–[50]. Spo11-Cre Hus1 CKO males, on the other hand, exhibited normal testis sizes at 17 and 28 days but significantly reduced testis weight in adults (Figure 1F; data not shown), consistent with meiotic defects beginning at the spermatocyte stage when the Spo11 promoter is active. Due at least in part to greater fecundity in the FVB strain background, experimental mice were produced more readily using Stra8-Cre (FVB) compared to Spo11-Cre (129/B6) mice, and we therefore focused our further analysis of meiotic chromosome stability on the Stra8-Cre Hus1 CKO model. As described below, our data indicate that the majority of meiotic phenotypes associated with Hus1 inactivation are similar in both models.
The reduction in testis size in Stra8-Cre and Spo11-Cre Hus1 conditional knockout animals was accompanied by significant reductions in the production of spermatozoa. The number of sperm in the caudal epididymis of 12-week old Stra8-Cre or Spo11-Cre conditional Hus1 knockout males was reduced by approximately 10-fold and 2-fold, respectively (Figure 1D, 1G). To test sperm function in these mice, we mated Stra8-Cre Hus1 conditional knockout males to wild-type female mice. While control matings produced 18 pregnancies and 154 viable pups, 49 matings with 5 different Stra8-Cre Hus1 CKO males produced only 2 pregnancies and no viable pups (Table 1). Overall, we conclude that HUS1 is required for normal spermatogenesis and fertility.
Tab. 1. <i>Hus1</i> inactivation in the mouse testis results in severely reduced fertility. 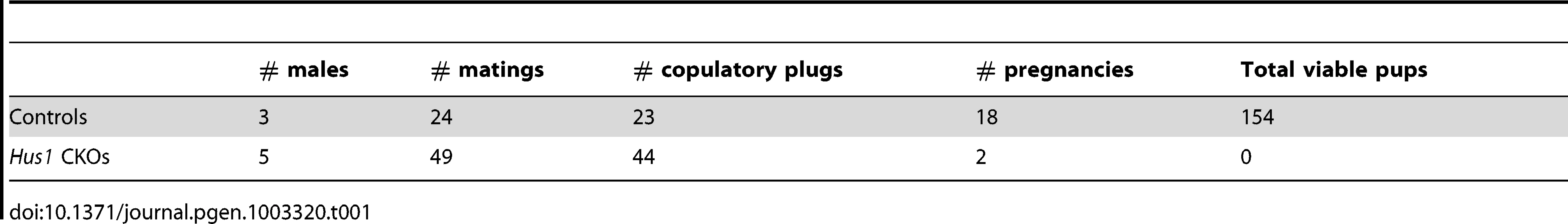
Hus1 loss results in germ cell depletion
To determine the underlying cause of the fertility defects in Hus1-deficient mice, we analyzed testes from Stra8-Cre Hus1 CKO and control animals. As shown in Figure 2A, Stra8-Cre Hus1 CKO adult testes exhibited a marked decrease in tubule size and cellularity, with many tubules lacking spermatogenic cells within the lumen, and an abundance of both pyknotic nuclei as well as multinucleate spermatid giant cells. We additionally performed immunohistochemical staining with GCNA1 antibody to detect germ cells in testis sections. Stra8-Cre Hus1 CKO testes exhibited significant loss of germ cells, including spermatogonia (Figure 2B). To further assess germ cell loss, we utilized TUNEL staining to detect fragmented DNA in Stra8-Cre Hus1 CKO nuclei. Hus1 mutant testes exhibited a significant increase in TUNEL-positive nuclei at both 17 days (Figure S4) and 12 weeks (Figure 2C–2E). Interestingly, a significant proportion of TUNEL-positive nuclei appeared to have progressed beyond pachytene stage to the first meiotic division (arrows, Figure 2E, right panel). 21% of all TUNEL-positive tubules contained apoptotic metaphase cells, although the majority of TUNEL-positive tubules (75%) contained apoptotic cells of a morphology consistent with zygotene/pachytene spermatocytes (Figure 2E, arrows, center panel). Spo11-Cre Hus1 CKO animals, in which Hus1 inactivation occurred later in germ cell development, also exhibited a decrease in testis cellularity and an increase in both multinucleate spermatid giant cells and TUNEL-positive cells (Figure S5). Altogether, these data indicate a requirement for Hus1 and the 9-1-1 complex for germ cell maintenance and development.
Fig. 2. Hus1 loss results in germ cell depletion. 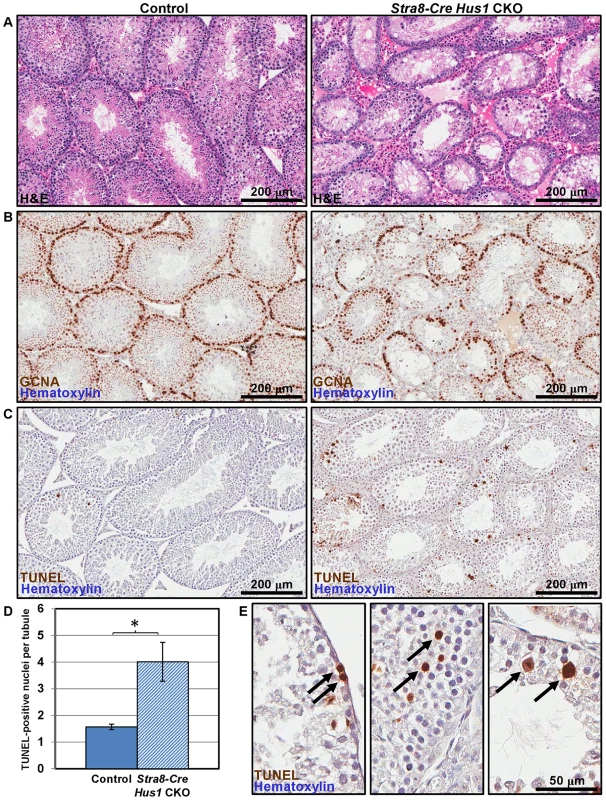
A. 100× images of H&E-stained histological sections from 12-week old control (left; Cre+ Hus1+/flox) and Stra8-Cre Hus1 CKO (right) testes. B. GCNA1 staining of germ cells in control and Stra8-Cre Hus1 CKO testes indicating germ cell loss. C. TUNEL staining of control and Stra8-Cre Hus1 CKO adult testes indicating germ cell apoptosis. D. Quantification of TUNEL staining shown in C, shown as the mean ± SEM. Asterisk indicates statistically significant difference between Hus1 CKO and control animals (p<0.05, Student's t-test). E. Higher magnification (400×) images of TUNEL staining in Stra8-Cre Hus1 CKO adult testes. Arrows highlight TUNEL-positive cells that appeared to be pre-meiotic (left), mid-prophase I (middle), or at meiotic metaphase (right). Hus1 loss results in abnormal γH2AX localization and increased CHK1 phosphorylation
Because the 9-1-1 complex protects chromosomal integrity in somatic cells, we hypothesized that germ cell loss in Hus1 CKOs might be due to DNA damage accumulation. We prepared meiotic chromosome spreads and assessed the localization of γH2AX, the phosphorylated form of the histone variant H2AX, by indirect immunofluorescence. γH2AX marks DSBs as well as unsynapsed chromatin, and is phosphorylated by the ATM and ATR kinases [26], [29]. We found that γH2AX staining during leptotene and zygotene, when meiotic DSBs are generated and processed, was similar in Hus1 CKOs and controls. Furthermore, γH2AX localized normally to the sex body domain in pachytene Hus1 CKO spermatocytes (Figure 3), suggesting that ATR-dependent H2AX phosphorylation is independent of HUS1 as is also true during responses to replication stress [51]. However, in Hus1 mutant spermatocytes, the sex body domain marked by γH2AX was enlarged and extended, and often encompassed parts of or entire autosomes (Figure 3B, 3C, 3F). 56% of mutant pachytene spermatocytes showed some type of sex body abnormality (compared to 13% of control nuclei), with 39% of Stra8-Cre Hus1 CKO nuclei having sex body extensions or protrusions and 33% with autosome inclusion (compared to 4% and 9%, respectively, in control spermatocytes; Table 2). Similar phenotypes were also observed in Spo11-Cre Hus1 CKO spermatocytes (Table 2). Autosomes included in the sex body domain often appeared synapsed (based on chromosome number and SYCP3 staining intensity), though asynapsed chromosomes and parts of chromosomes were also included in the sex body, as shown in Figure 3F. Additionally, a substantial number of meiotic nuclei harbored apparent end-to-end fusions between the X chromosome and an autosome (Figure 3E). In such cases, the X and Y chromosomes were synapsed at the pseudoautosomal region (PAR) and were contained within the sex body domain containing γH2AX, while the autosome remained outside of the sex body domain and was devoid of γH2AX. The frequency of X-autosome fusions was significantly increased in Stra8-Cre Hus1 CKO pachytene spermatocytes, from 2% in controls to 12% in CKO mice (p<0.001; Table 2).
Fig. 3. Hus1 inactivation results in abnormal accumulation of γH2AX on autosomes, an extended sex body domain, inclusion of autosomes within the sex body, and X-autosome fusions. 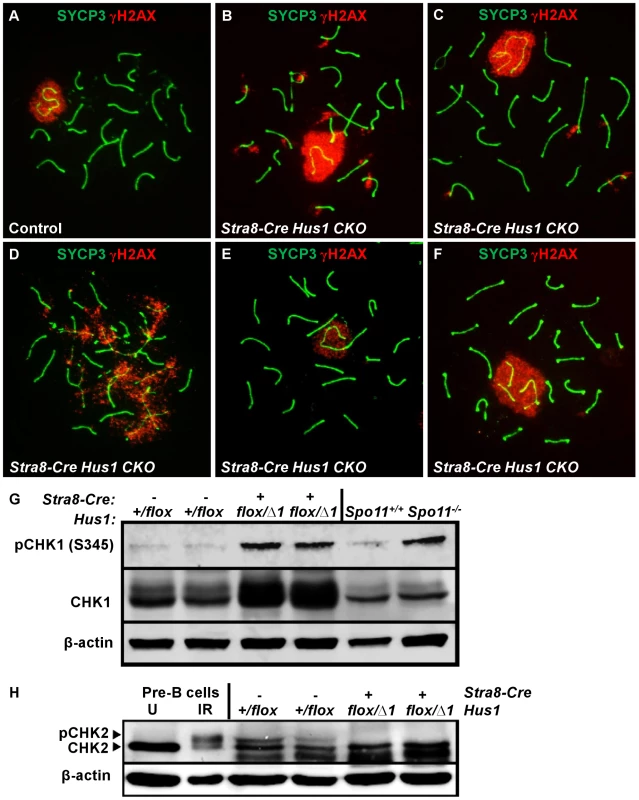
A–F. Immunofluorescence staining for γH2AX and SYCP3. γH2AX staining persisted in a perpendicular pattern on synapsed autosomes in pachytene (B) and diplotene stage (C), and in clouds surrounding unsynapsed chromosomal regions (D). The sex body domain marked by γH2AX was extended in Stra8-Cre Hus1 CKOs (B,F), and often contained whole or partial autosomes (B,F). Some Stra8-Cre Hus1 CKO nuclei exhibited apparent X chromosome-autosome end-to-end fusions (E). G,H. Western blot analysis of CHK1 (G) and CHK2 (H) in total testis extracts from mice of the indicated genotypes. In the CHK2 immunoblot, pre-B cells that were untreated (U) or treated with 5Gy ionizing radiation (IR) are included as controls. β-actin is shown as a loading control. In Hus1 CKO samples, total CHK1 levels were increased on average 2.9-fold over controls, and phosphorylated CHK1 (S345) levels were increased 4.1-fold. CHK2 phosphorylation was not detected in the absence of Hus1. Tab. 2. Patterns of γH2AX localization in Stra8-Cre and Spo11-Cre Hus1 CKO spermatocytes. 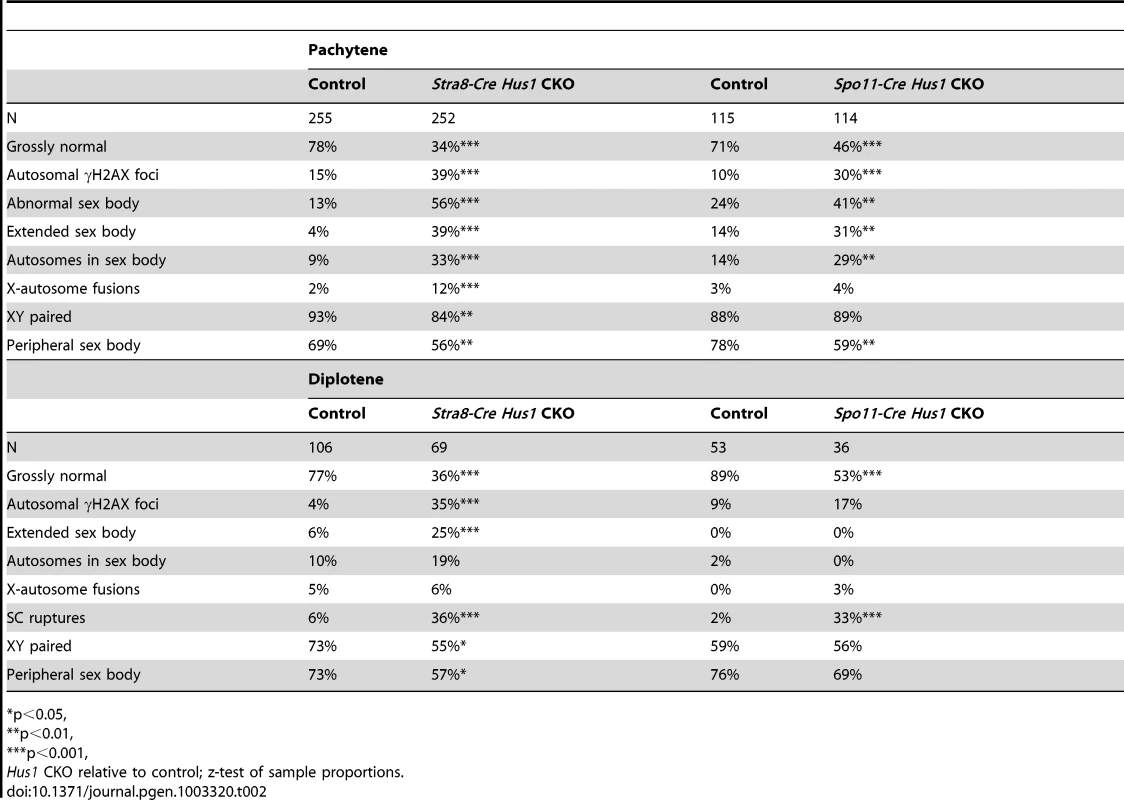
p<0.05, Extended sex body domains marked by γH2AX persisted in diplotene-stage Stra8-Cre Hus1 CKOs (25% of Hus1 CKO nuclei versus 6% of controls; Table 2), indicating that these cells did not arrest at the pachytene/diplotene transition despite chromatin abnormalities. Additionally, the sex body domain in both pachytene and diplotene spermatocytes was located at the nuclear periphery less often in both Stra8-Cre and Spo11-Cre Hus1 CKOs than in controls (Table 2), indicating a perturbation in the normal compartmentalization or localization of the sex chromatin. Although we cannot entirely rule out the possibility that the 9-1-1 complex directly promotes sex body maintenance through stimulation of ATR activity or other roles, we favor the idea that defects in maintenance of sex body integrity following Hus1 loss could instead be related to aberrant responses to meiotic DSBs. In cases where autosomes were asynapsed, clouds of γH2AX surrounded the unsynapsed regions (Figure 3D), supporting the idea that ATR-mediated responses to unsynapsed chromatin remain intact despite Hus1 loss. Consistent with the idea that expanded sex body domains marked by γH2AX reflect upregulation of ATR kinase activity, we also observed that Hus1 loss was associated with increased levels of both total and phosphorylated CHK1 (Figure 3G and Figure S3), the latter being an established indicator of ATR activity in somatic cells [52]. Increased phosphorylated CHK1 was also observed in extracts prepared from Spo11-deficient testes (Figure 3G), which have severe defects in chromosome synapsis.
A significant percentage of pachytene Stra8-Cre Hus1 CKO nuclei exhibited γH2AX staining on fully synapsed autosomes (39% versus 15%, p<0.001; Table 2), generally in a burst-like pattern perpendicular to the synaptonemal complex, indicative of unrepaired DSBs (Figure 3B). This eruption-like pattern of γH2AX staining on Hus1 CKO cores is consistent with previously described γH2AX L-foci [53], which are proposed to be sites of delayed or unregulated DSB repair events, and have been observed in several other meiotic mutants with break repair defects [54]–[56]. A similar pattern of γH2AX staining was observed in Spo11-Cre- and Stra8-Cre-driven Hus1 CKO mice (Table 2), suggesting that many of the persistent breaks originate during meiosis and are not due exclusively to aberrant pre-meiotic replication in the absence of Hus1. The autosomal γH2AX staining in Hus1 CKOs persisted into diplotene (Figure 3C), with 35% of Hus1 mutant nuclei containing bursts of autosomal γH2AX at this stage compared to 4% of control nuclei (Table 2). Persistence of these γH2AX foci into diplotene indicates that homologous chromosomes began to desynapse despite incomplete DSB repair, and that these aberrant spermatocytes did not arrest in the pachytene stage in the absence of Hus1. Consistent with the possibility of impaired DSB-induced DNA damage checkpoint signaling following Hus1 loss, we observed that the phosphorylated form of the DSB-responsive checkpoint kinase CHK2, which was evident in extracts from control testes, was absent in those from adult Hus1 CKO mice (Figure 3H and Figure S3B).
RAD51 foci persist in Hus1-deficient spermatocytes, primarily at sites lacking MLH1
Since increased numbers of γH2AX foci were present on Hus1 CKO meiotic chromosomes, we further investigated whether the RAD51 strand exchange protein accumulated normally on Hus1-deficient chromosomes. During the early stages of meiotic DSB repair in zygotene and early pachytene spermatocytes, RAD51 properly localized to punctate structures along meiotic chromosome cores in the absence of Hus1 (Figure 4A). By late pachytene, when most RAD51 foci are normally lost from autosomal and XY chromosome cores, all Hus1 CKO spermatocytes exhibited persistent localization of RAD51 to the autosomes as well as the X chromosome, indicative of a DSB repair defect (Figure 4C). These foci persisted into diplotene as well (Figure 4D), demonstrating that nuclei with unrepaired DSBs were also able to progress beyond the pachytene stage. RAD51 focus numbers were quantified at each of these stages (Figure 4F–4I), and the number of foci in Hus1 CKO nuclei was significantly increased in late pachytene and diplotene. While the number of persistent RAD51 foci per nucleus was modest, the total number of nuclei with at least one RAD51 focus was high in Hus1 CKOs, at 100% of Hus1 CKO nuclei versus 52% of controls in late pachytene, and 73% of Hus1 CKOs versus 17% of controls in diplotene (Figure 4). Further, the reduction in the frequency of cells with RAD51 foci from 100% to 73% of Hus1 CKO nuclei from late pachytene to diplotene could indicate that Hus1 mutants can repair DSBs, albeit inefficiently in some cases, such that repair is significantly delayed. We conclude that HUS1 is critical for efficient completion of DNA repair at a subset of meiotic DSBs.
Fig. 4. RAD51 foci persist in late pachytene and diplotene spermatocytes in the absence of Hus1. 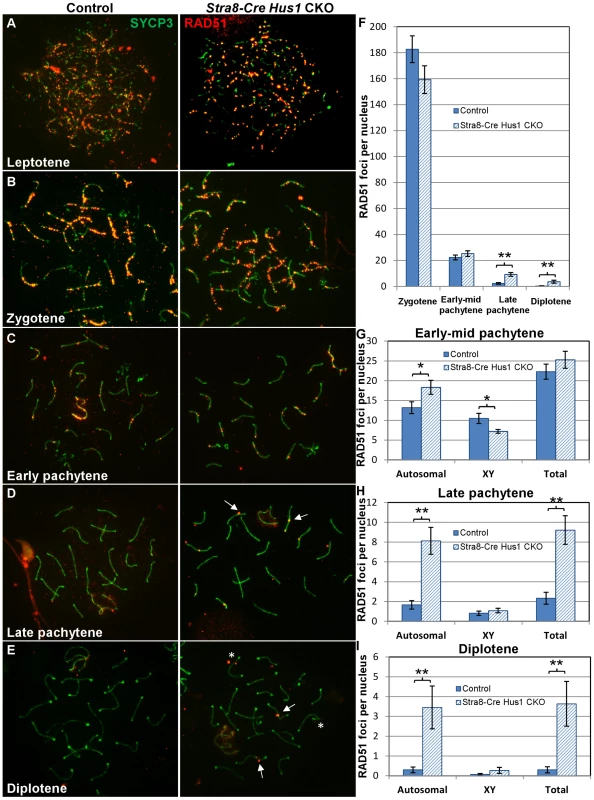
A–E. Immunofluorescence staining for RAD51/DMC1 from zygotene through diplotene stage of meiosis in control (Cre-negative Hus1flox/Δ1) and Stra8-Cre Hus1 CKO males. Arrows indicate persistent foci, and asterisks indicate symmetrical foci located on either side of chromosomal cores. F–I. Quantification of RAD51 foci at the indicated stages. ** indicates p<0.01; * indicates p<0.05. To determine whether Hus1 loss alters later stages in DSB repair such as crossover formation, we assessed MLH1 localization in pachytene spermatocytes. The mismatch repair complex MLH1-MLH3 is required for the formation of Class I crossover events, which account for approximately 90% of all meiotic crossovers in mice [57], [58]. Mouse and human RAD9 proteins have been shown to interact with MLH1 [21], although it is unclear whether this interaction is relevant for meiotic crossover recombination or whether it is limited primarily to post-replicative DNA mismatch repair. As shown in Figure S6A, the total number of MLH1 foci per nucleus was not significantly different in Hus1 CKO cells relative to controls, with one to two MLH1 foci per homolog pair. Hus1 CKOs exhibited a mean of 25.4±0.3 MLH1 foci per nucleus, compared to 25.9±0.3 in controls (Figure S6B), suggesting that Hus1 loss does not significantly impair crossover formation. However, a general impairment of meiotic DSB repair in the absence of functional 9-1-1 complex might manifest as only a modest reduction in the number of MLH1 foci per nucleus, as 10% of crossovers are not dependent on MLH1-MLH3 and the majority of meiotic DSBs are repaired as noncrossovers [59]. To determine whether the persistent RAD51 foci observed in Hus1 CKOs were primarily associated with noncrossover or crossover events, we assessed the relative localization of RAD51 and MLH1. As shown in Figure S6C, the majority of persistent RAD51 foci in late pachytene were exclusive of MLH1, though an occasional single RAD51 focus localized at or adjacent to an MLH1 focus. These data indicate that while some meiotic DSBs are not repaired efficiently in the absence of Hus1, these unrepaired breaks consist primarily of MLH1-independent repair events (most likely noncrossovers) but can include MLH1-dependent crossovers.
Since a significant proportion of TUNEL-positive Hus1 CKO germ cells appeared to be in meiotic metaphase, it remained possible that Hus1 CKOs might be deficient in crossover formation, despite the normal complement of MLH1 foci we observed. Thus, we prepared meiotic diakinesis chromosome spreads from control and Hus1 CKO testes to assess chromosome integrity at the end of prophase I, when the SC has disintegrated and chiasmata are first evident. As shown in Figure S6D–S6E, the majority of nuclei had 20 bivalents, indicating that each homolog pair received at least the one obligate crossover necessary for proper chromosome segregation. Taken together, these results indicate that while HUS1 is critical for repair of a subset of meiotic DSBs, it appears to be dispensable for crossover formation.
Hus1 inactivation results in meiotic chromosome defects
To investigate additional causes for germ cell loss in Hus1 CKOs, we assessed the localization of various meiotic markers on chromosome spreads from control and Stra8-Cre Hus1 CKO males. Meiotic chromosome synapsis appeared normal in most Hus1 mutant nuclei, with colocalization of the synaptonemal complex proteins SYCP1 and SYCP3 in pachytene spermatocytes and the presence of 19 pairs of fully synapsed homologs in addition to the X and Y chromosomes paired at the pseudoautosomal region. However, as shown in Figure 5B, a significant number of Stra8-Cre Hus1 CKO nuclei exhibited synapsis defects, usually involving the X chromosome (14% of Hus1 CKOs versus 3% of controls; N = 161 and 185, respectively; p<0.001). Notably, Hus1 CKOs also frequently displayed ruptures in the synaptonemal complex during diplotene (36% of Stra8-Cre Hus1 CKOs and 33% of Spo11-Cre Hus1 CKOs compared to 2–6% of controls; Table 2). In these cases, there were clear discontinuities in synaptonemal complex protein staining and the ends of the broken SC were spatially separated (Figure 5C, arrows), indicating either a defect in SC integrity or breakage of chromosomal DNA in the absence of HUS1.
Fig. 5. Conditional Hus1 knockout meiotic chromosomes display synapsis defects and ruptures in the synaptonemal complex. 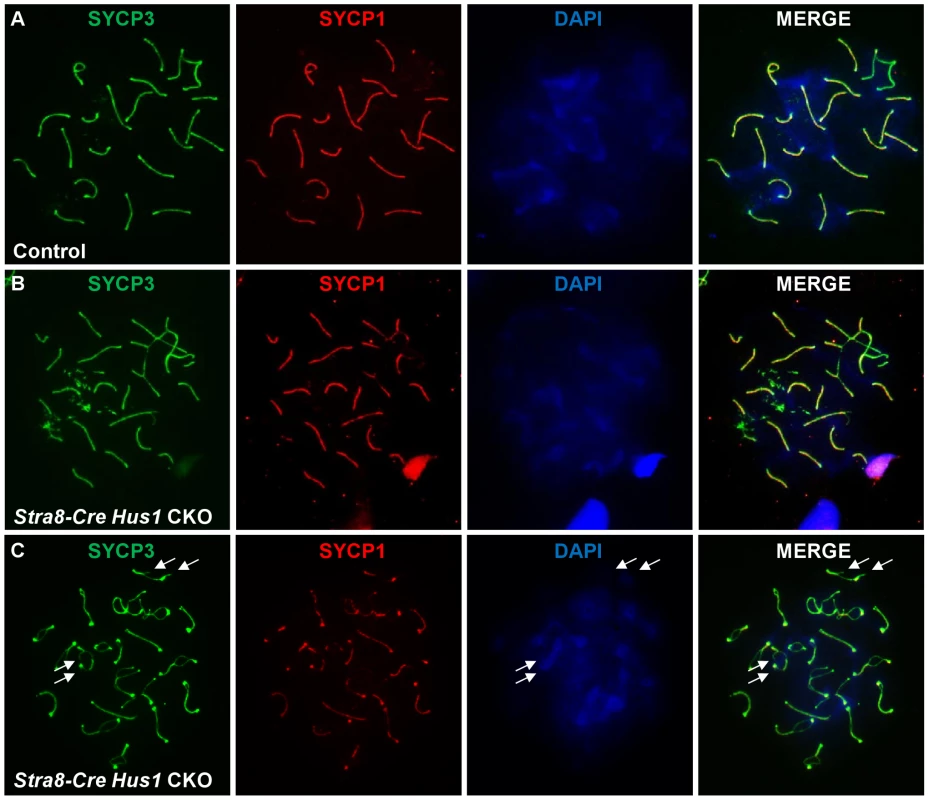
Meiotic chromosome spreads from control (Cre-negative Hus1flox/Δ1) and Stra8-Cre Hus1 CKO mice were stained for SYCP1 and SYCP3, as well as with DAPI. A. Normal synapsis of pachytene chromosomes in control males, as indicated by SYCP1 and SYCP3 immunofluorescence. B. Chromosomal asynapsis in pachytene-stage Stra8-Cre Hus1 CKO nuclei, with unsynapsed chromosomal regions devoid of SYCP1. C. Diplotene Stra8-Cre Hus1 CKO chromosomes with ruptures in the SC lateral elements, as indicated by arrows. TOPBP1 localization and RNA Pol II exclusion from the sex body remain intact in the absence of Hus1
During meiosis, TOPBP1 normally colocalizes with ATR at sites of unsynapsed chromatin, including unsynapsed autosomes in early spermatocytes as well as the XY chromatin throughout prophase I [24], [60]. In mammalian somatic cells, the best characterized function of 9-1-1 is to recruit TOPBP1 to sites of DNA damage, where it physically interacts with and stimulates the kinase activity of ATR [61]–[63]. Therefore, we next tested whether Hus1 loss affected the meiotic localization of TOPBP1. In contrast to the 9-1-1-dependent mechanism elucidated in somatic cells, TOPBP1 localization to the asynapsed sex chromatin was unperturbed in Hus1 CKO mice (Figure 6). Additionally, TOPBP1 localized to autosomes that resided within the sex body domain (Figure 6B, asterisk), and to perpendicular foci on some autosomes in Stra8-Cre Hus1 CKOs (Figure 6C) similar to the γH2AX eruptions described above, indicating that HUS1 loss does not preclude assembly of TOPBP1 on chromatin and results in TOPBP1 localization to abnormal autosomes as well as to unsynapsed sex chromatin.
Fig. 6. TOPBP1 localization to and RNA Pol II exclusion from the sex body domain remain unperturbed following Hus1 inactivation. 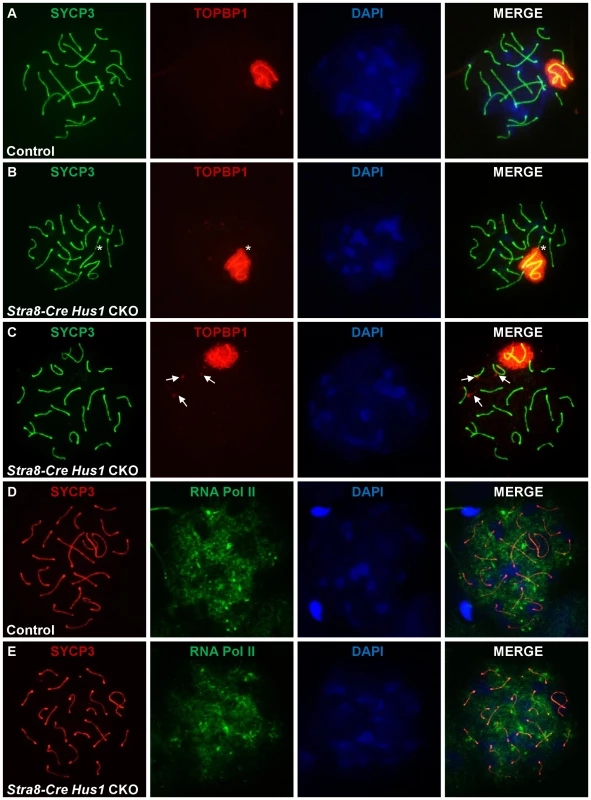
A–E. Immunofluorescence staining for TOPBP1 (A–C) and RNA Polymerase II (D–E) in control (Cre-negative Hus1flox/Δ1) and Stra8-Cre Hus1 CKO pachytene spermatocytes. The asterisk in B indicates an autosome partially included within the sex body domain and coated with TOPBP1. Arrows in C indicate abnormal TOPBP1 staining in a perpendicular pattern on autosomes in pachytene Hus1 CKOs. Because the downstream consequence of ATR action at asynapsed meiotic chromatin is meiotic silencing [30], [32], we also assessed the localization of RNA Polymerase II, which indicates sites of active transcription. Consistent with the apparently normal TOPBP1 localization following Hus1 deletion in germ cells, RNA Pol II immunofluorescence staining revealed no difference in transcriptional silencing of the XY chromatin, as indicated by a similar lack of RNA Pol II signal in the sex body domain of cells from both Hus1 CKO and control mice (Figure 6D, 6E). Together, these results indicate that despite the absence of the HUS1 component of the 9-1-1 complex in Hus1 CKO spermatocytes, TOPBP1 - and ATR-dependent responses to unsynapsed chromatin remained intact. These findings raise the possibility that ATR activation in response to at least some signals during mammalian meiosis, such as unsynapsed chromatin, may occur through a distinct mechanism independent of the canonical 9-1-1 complex.
RAD9 localizes to DSB sites on meiotic chromosomes during prophase I
Since Hus1 disruption resulted in chromosomal defects without affecting TOPBP1 localization, we next analyzed RAD9 localization to meiotic chromosomes to gain insights into the normal functions of 9-1-1. As shown in Figure 7A–7C, RAD9 localized to meiotic chromosome cores during prophase I, with many foci on both unsynapsed and synapsed chromosomes during zygotene stage, fewer autosomal foci and prominent X chromosome foci in early pachytene, and even fewer foci by mid - to late-pachytene stage when most staining was confined to bright foci along the X chromosome core. By diplotene stage, RAD9 foci were not detectable on meiotic chromosomes. Thus, the RAD9 subunit of 9-1-1 is appropriately positioned for a potential role in genome maintenance during meiosis.
Fig. 7. RAD9 localizes to meiotic chromosomes during early prophase I and colocalizes with a subset of RAD51 foci. 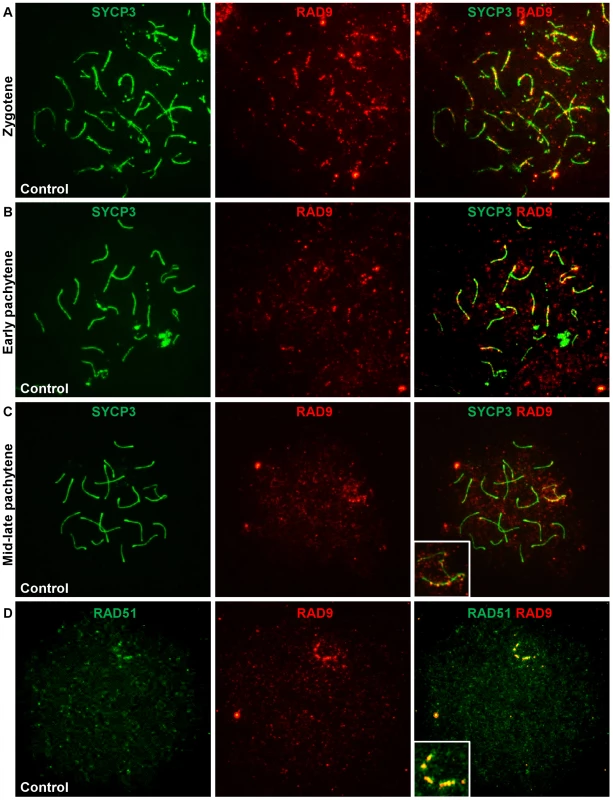
A–C. Meiotic chromosome spreads from control animals were stained for RAD9 and SYCP3. In wild-type adult males, RAD9 localized along the synaptonemal complex of synapsed and unsynapsed chromosomes during zygotene (A) and pachytene (B, C). RAD9 localized to autosomes and the sex chromosomes in early pachytene (B), was confined primarily to the X chromosome by mid-late pachytene (C), and was absent by diplotene. D. Meiotic chromosome spreads from control animals were stained for RAD9 and RAD51. RAD9 colocalized with a subset of RAD51 foci, particularly along the X chromosome in pachytene-like nuclei. The pattern and timing of the RAD9 signal on meiotic chromosomes was reminiscent of that of the DSB repair factor RAD51, particularly with respect to its presence at late-persisting breaks along the X chromosome core. To determine whether RAD9 colocalized with RAD51, we simultaneously stained meiotic chromosomes for RAD9 and RAD51. In pachytene-like nuclei, the majority of RAD9 foci colocalized with RAD51 foci, most notably along what is presumably the X chromosome axial element (Figure 7D). Thus, RAD9 appears to localize to DSBs undergoing homologous recombinational repair. To further assess RAD9 function in DSB repair, we also analyzed RAD9 localization in Spo11−/− mutants [64], which lack meiotic DSBs, and Dmc1−/− mutants [65], which harbor persistent unrepaired DSBs. As shown in Figure S7, RAD9 staining was much reduced in the absence of meiotic DSBs and elevated in the presence of increased DSBs. Thus, RAD9, and likely its partner HUS1 (see below), function together at meiotic DSBs at a stage overlapping with RAD51/DMC1.
The distribution of RAD9 and RAD1 on meiotic prophase I chromosomes is only partially overlapping
The RAD1 subunit of 9-1-1 was previously determined to localize to meiotic chromosomes, where RAD1 foci outnumber RAD51 foci and nearly continuously coat pachytene chromosome cores, especially along the unsynapsed X and Y [41]. To reconcile this staining with our observations of fewer, more punctate RAD9 foci, we assessed the colocalization of RAD1 and RAD9 subunits in control nuclei. These experiments revealed that RAD1 had a broader distribution than RAD9, with RAD1 foci present along the entire unsynapsed regions of the X and Y while RAD9 was detected primarily in bright foci along the X. Overall, only 23% of RAD1 foci colocalized with RAD9 (16% of autosomal and 46% of XY RAD1 foci colocalized with RAD9). A larger percentage (64%) of RAD9 foci colocalized with RAD1, particularly on the sex chromosomes (52% of autosomal and 86% of XY RAD9 foci colocalized with RAD1). We next assessed both RAD1 and RAD9 localization on abnormal chromosomes in Slx4mut/mut and Msh4−/− mutant spermatocytes, which exhibit asynapsis as well as inclusion of autosomes within the sex body domain [55], [66]. Consistent with the co-staining results, RAD1 localized along synapsed autosomal cores and to a greater extent along asynapsed autosomal and XY cores (Figure 8B, 8D). RAD9 on the other hand localized in a more punctate pattern along chromosome cores in the sex body domain and on asynapsed autosomes (Figure 8C, 8E). Together, these results indicate that RAD1 and RAD9 partially colocalize along abnormal chromosomes and the sex chromosome cores, and that a subset of RAD1-containing sites lack RAD9.
Fig. 8. RAD9 and RAD1 localize in overlapping yet distinct patterns on meiotic chromosomes. 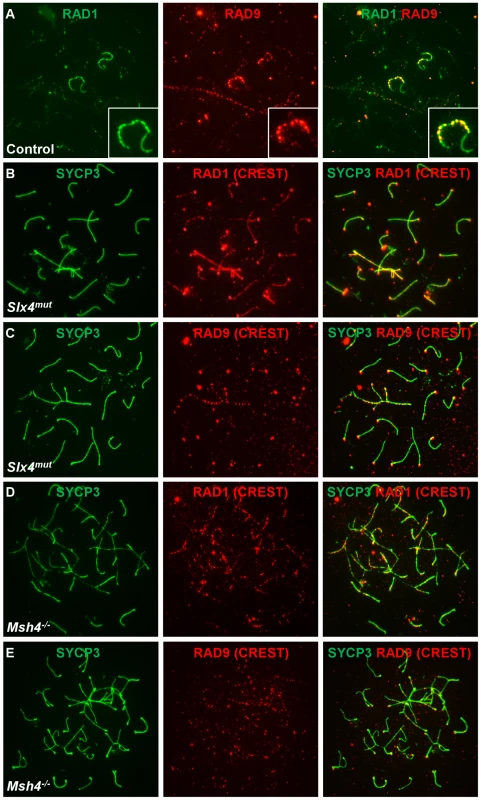
A. Meiotic chromosome spreads from control animals were stained for RAD9 and RAD1. RAD9 colocalized with a subset of RAD1 foci in control spermatocytes. A region containing one X-Y pair is shown at higher magnification. B–E. Meiotic chromosome spreads from Slx4mut/mut (B, C) or Msh4−/− (D, E) mice were stained for SYCP3 and RAD1 (B, D) or RAD9 (C, E). RAD1 localized more continuously along sex chromosome cores and autosomes and to asynaptic sites in Slx4 and Msh4 mutants, whereas RAD9 formed fewer, more punctate foci along asynapsed chromosome cores. Chromosome spreads in panels B–E were additionally stained using human CREST serum, which marks centromeric regions. CREST signal is detected in the red channel (middle column) and in the merged images. Hus1 loss differentially affects the localization of RAD9 and RAD1 on meiotic chromosomes
Prompted by the differences in RAD1 and RAD9 localization described above, we next assessed localization of these 9-1-1 subunits in Hus1 CKO spermatocytes. Since the chromatin association and nuclear localization of the 9-1-1 complex in somatic cells requires all three subunits [48], we expected RAD9 and RAD1 to be depleted from chromosomes in the absence of HUS1. However, the more extensive RAD1 staining observed in normal and aberrant spermatocytes suggested a possible separation of functions for the individual subunits. Remarkably, bright RAD1 staining of XY cores as well as punctate staining of autosomes persisted upon Hus1 deletion (Figure 9A). By contrast, RAD9 foci were undetectable in nuclei from Hus1 CKO mice (Figure 9B). Whereas RAD9 staining was readily detected in all control nuclei, approximately 99% of Stra8-Cre Hus1 CKO pachytene nuclei lacked RAD9 foci. The continued presence of RAD1 on meiotic chromosomes following Hus1 loss despite the significant reduction in total RAD1 levels (Figure S3) may indicate that a fraction of RAD1 protein participates in chromatin-associated complexes that remain stable independently of HUS1, while the remaining pool of RAD1 is unstable in the absence of HUS1.
Fig. 9. RAD1 and RAD9 localization to meiotic chromosomes is differentially affected by Hus1 loss. 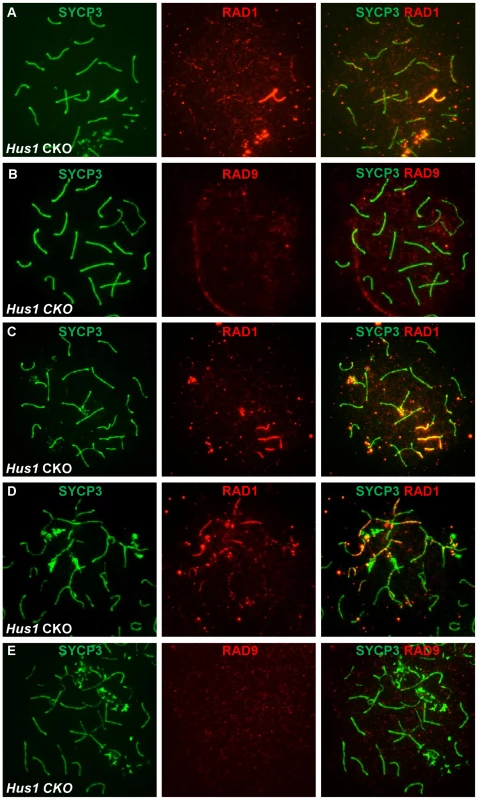
Meiotic chromosome spreads from Stra8-Cre Hus1 CKO mice were stained for SYCP3 and RAD1 (A, C, D) or RAD9 (B, E). A,C,D. RAD1 continued to localize to sex chromosomes and aberrant meiotic chromosomes in the absence of Hus1. Most notably, RAD1 localization to asynapsed autosomes and chromosomes with the sex body domain persisted in cells from Hus1 CKO mice. By contrast, RAD9 localization to both normal and aberrant chromosome structures was abolished following Hus1 loss (B,E). The occasional occurrence of asynapsed homologs and the presence of autosomes within the sex body domain of pachytene Hus1 CKO cells additionally allowed for evaluation of the dependency on Hus1 for RAD9 and RAD1 recruitment to abnormal structures during meiosis. Similar to our observation that the localization of TOPBP1 was unperturbed in Hus1 CKOs, RAD1 clearly localized to autosomes within the vicinity of the sex body domain (Figure 9C) and to asynapsed autosomes (Figure 9D) in the absence of Hus1. The localization of RAD9 on the other hand appeared strictly Hus1-dependent, as RAD9 was not detected on asynapsed chromosomes or sex chromosome cores in Hus1 CKOs (Figure 9E), although it did localize to such structures in Hus1-expressing cells (Figure 7 and Figure 8). Together with the results described above, these data indicate that RAD9 and RAD1 have overlapping Hus1-dependent functions that intersect with RAD51, and that RAD1 has additional meiotic functions that may involve TOPBP1 but are independent of its traditional binding partners, HUS1 and RAD9.
Discussion
In order to examine the role of the 9-1-1 complex during mammalian meiosis, we analyzed mice with germ cell-specific Hus1 deletion and evaluated the localization of the RAD9 and RAD1 subunits to normal and aberrant meiotic chromosomes. Most notably, we report that the RAD9 and RAD1 subunits of the 9-1-1 complex exhibit overlapping yet distinct localization patterns along mammalian meiotic chromosomes and are differentially affected by loss of the HUS1 subunit. RAD1 forms more foci along chromosome cores than RAD9 (this study) or RAD51 [41], and we show that it preferentially localizes to the axial elements of asynapsed chromosomes or autosomes included within the sex body domain in a manner that is independent of Hus1. Previous electron microscopy analysis indicated that while RAD1 localizes to meiotic chromosomes cores, it does not colocalize with DMC1 [41], which is consistent with our finding of RAD1 at asynaptic sites. We show here that only a subset of RAD1 foci colocalize with RAD9, raising the possibility that RAD1 is present both at DSBs, as part of the canonical 9-1-1 complex, and at other chromosomal sites.
We propose a model in which meiotic genome maintenance involves several distinct checkpoint clamp complexes, some of which function in DSB repair, and others of which carry out separate functions, possibly including ATR activation through TOPBP1 in response to asynapsis. Of particular note are the previous findings that the mouse and human genomes contain paralogs of RAD9 and HUS1, termed RAD9B and HUS1B, respectively, that are highly expressed in the testis [67], [68]. Given that RAD1 localized to some chromosomal sites that lacked RAD9 and did so following genetic ablation of Hus1, it is tempting to speculate that RAD9B and HUS1B associate with RAD1 to form an alternative 9-1-1-like heterotrimer, although it remains possible that RAD1 localizes to chromosomes on its own or in conjunction with other cofactors. In this regard, it is worth noting that a substantial portion of RAD1 protein in cultured human cells exists in monomeric form, independent of the heterotrimeric 9-1-1 complex [69]. Overall, our results are consistent with formation of the canonical 9-1-1 complex composed of RAD9, RAD1, and HUS1 at meiotic DSBs, as well as with RAD1 assembling on chromosomes independently of HUS1 and RAD9 in patterns overlapping with TOPBP1 and ATR.
Despite a well-established role for 9-1-1 in ATR activation in somatic cells, we observed unperturbed localization of the 9-1-1-interacting, ATR-activating TOPBP1 protein and similarly no change in RNA Polymerase II exclusion from the XY body domain in Hus1 CKO spermatocytes. These findings indicate that ATR-dependent responses to unsynapsed chromatin, including meiotic silencing via MSUC and MSCI, do not require HUS1, which is in distinct contrast to the canonical model of mammalian ATR activation in which 9-1-1-mediated TOPBP1 recruitment to damaged DNA allows TOPBP1 to contact and activate ATR. Since RAD1 localization to asynapsed meiotic cores was also unperturbed in the absence of Hus1, it is possible that a RAD1-containing complex functions along with TOPBP1 to activate ATR at such sites, in a manner that may be at least in part distinct from established mechanisms for 9-1-1 loading and function at DNA damage sites. The meiosis-specific HORMAD1 and HORMAD2 proteins are required for ATR recruitment to sites of unsynapsed chromatin [33], [70], and together with BRCA1, which is also required for ATR recruitment [26], these proteins could provide an alternative mechanism for ATR activation that is independent of 9-1-1 and typical substrates for 9-1-1 loading. We favor the idea that alternative complexes involving RAD1 and possibly RAD9B and HUS1B might engage in a 9-1-1-like interaction with TOPBP1. It remains to be determined whether the recruitment of such a complex to asynaptic sites is facilitated by particular DNA structures or binding partners that would not attract conventional 9-1-1 complexes.
Conditional Hus1 deletion in germ cells resulted in numerous chromosomal defects, including asynapsis, unrepaired meiotic DSBs, X-autosome end-to-end fusions, autosomes incorporated into the sex body domain (which presumably are inappropriately transcriptionally silenced), and SC ruptures. We propose that some of these defects (asynapsed chromosomes, X-autosome fusions, and autosomes incorporated into the sex body domain) lead to loss of cells during the pachytene stage in Hus1 CKOs, consistent with an absence of cells with such abnormalities in diplotene (Table 2), whereas other defects (unrepaired DSBs, extended sex body domains, and SC ruptures; Table 2) either may not be actively monitored or may normally require HUS1 for checkpoint-dependent clearance and therefore persist beyond pachytene into diplotene in Stra8-Cre Hus1 CKOs. The latter set of phenotypes was less prominent in Spo11-Cre Hus1 CKO mice, with fewer defective cells persisting into diplotene, suggesting that HUS1-dependent checkpoint monitoring may be an earlier function that is less affected by mid-prophase I loss of Hus1. Notably, SC ruptures were still prominent in diplotene cells of both Stra8-Cre and Spo11-Cre Hus1 CKOs, affecting one-third of the diplotene cell population. Thus, HUS1 is required for some aspect of chromosome integrity that affects the SC, the failure of which may contribute to the high level of metaphase germ cell loss. This SC rupture phenotype (Figure 5C; Table 2) is particularly striking and may be a key driver of the fertility defect observed following Hus1 loss, as no significant change was observed in the number of MLH1 foci in Hus1 mutant pachytene spermatocytes or in the number of bivalents seen at diakinesis (Figure S6). The molecular cause of the SC rupture defect remains unresolved, although it could be related to incomplete repair of meiotic DSBs.
It is still unclear why Hus1 CKO cells have a small number of persistent DSBs, and why many of these are associated with death at metaphase I. Perhaps the 4–8 remaining unrepaired breaks are not sufficient to trigger the pachytene checkpoint (or cannot, due to a requirement for HUS1), but lead to a defect that is recognized at metaphase by the spindle checkpoint. It is possible that the persistent breaks are not simple D-loop intermediates containing RAD51 and γH2AX, but are complex multi-chromatid intermediates, such as those seen in Blm/Sgs1 helicase mutants [71], [72]. The symmetrical RAD51 foci seen on either side of the chromosome cores in 9% of diplotene nuclei from Hus1 CKOs (see Figure 4, asterisks) would support this, pointing to RAD51/DMC1 presence on more than one homolog during homolog separation. Clearly, the extent of the DNA repair defect in Hus1 CKO mice is less severe than that observed in other meiotic mutants. For instance, Trip13 mutant spermatocytes have been reported to exhibit between 99 and 138 RAD51 foci during pachytene compared to 11–18 foci in controls [54], [73], whereas we observed an average of 25 RAD51 foci in early to mid-pachytene Hus1 CKOs (compared to 22 foci in controls) and 9 RAD51 foci in late pachytene (compared to 2 foci/nucleus in controls). These results are consistent with the idea that HUS1 has a relatively late and restricted role in DSB repair, perhaps related to the completion of a subset of late recombination events.
The persistent γH2AX and RAD51 foci on Hus1-deficient meiotic chromosomes as well as the colocalization of RAD9 with RAD51 on normal chromosomes suggest that the 9-1-1 complex is critical for the efficient repair of a subset of meiotic DSBs. Several lines of evidence in somatic cells also indicate that 9-1-1 functions directly in homologous recombination (HR) repair of DSBs. In human cells, 9-1-1 is reported to physically interact with RAD51, and Rad9 knockdown results in reduced HR [7]. In a separate system using human cells with conditional Rad9 repression, RAD9 enhances survival and DNA repair in response to ionizing radiation [74], and similarly, mouse Rad9−/− ES cells are sensitive to γ-irradiation [43]. Reducing Hus1 expression in mouse cells via siRNA also decreases the efficiency of HR repair [75]. In contrast to the situation in yeast where 9-1-1 is proposed to be important for the loading or assembly of RAD51 complexes onto meiotic chromosomes [37], we propose that mammalian 9-1-1 may be important for later steps of meiotic recombination, since we did not observe delayed loading of RAD51, and RAD51 foci persisted in the absence of Hus1. Among the known 9-1-1 binding partners are DNA ligase I [13] and DNA Polymerase β [9], [10], the latter of which is known to play critical roles during mammalian meiosis [76], [77], raising the possibility that 9-1-1 may recruit Pol β and DNA ligase to recombination intermediates to complete repair. Alternatively, interactions between the 9-1-1 complex and translesion synthesis polymerases [8], which have been implicated in HR in somatic cells [78]–[80], could promote the extension of the 3′ ends subsequent to RAD51-mediated strand exchange. The requirement for 9-1-1 could be accentuated due to unique features of genome maintenance in meiotic cells. For instance, non-homologous end joining, a major mechanism for DSB repair in somatic cells, is suppressed during meiosis [29], [81]. In addition, the unique structure of SPO11-induced meiotic DSBs may create a greater demand for 9-1-1 complex-mediated repair functions than a typical mitotic DSB. The repair of most meiotic DSBs occurred normally in the absence of HUS1, suggesting that the 9-1-1 complex may be necessary to deal with only a subset of breaks, perhaps ones that prove difficult to repair because of the sequence context, chromatin structure, or other factors.
The production of haploid gametes during meiosis clearly raises challenges for genome maintenance, many of which are distinct from those in somatic cells. Based on the findings reported here, we propose that mammalian 9-1-1 components have acquired specialized roles during meiosis, with the canonical RAD9-RAD1-HUS1 complex functioning at DSBs and an alternative RAD1-containing complex functioning at sites of asynapsis. The mouse models described here represent powerful systems to elucidate how the mammalian 9-1-1 complex promotes meiotic chromosome integrity, in some cases distinct from the well-established roles of 9-1-1 in TOPBP1 - and ATR-dependent checkpoint signaling, and highlight the intriguing possibility of alternative checkpoint clamps functioning in various capacities in the mammalian germline.
Materials and Methods
Ethics statement
All animals used in this study were handled in accordance with federal and institutional guidelines, under a protocol approved by the Cornell University Institutional Animal Care and Use Committee (IACUC).
Mouse strains and husbandry
Mice harboring two conditional Hus1 alleles (Hus1flox/flox) were crossed to Cre-positive mice harboring one null Hus1 allele (Stra8-Cre+ Hus1+/Δ1 or Spo11-Cre+ Hus1+/Δ1) to generate experimental germ cell-specific Hus1 conditional knockout mice (Cre+ Hus1flox/Δ1) as well as littermate control animals (Cre+ Hus1+/flox and Cre - Hus1flox/Δ1), as shown in Figure S1A. Conditional and null Hus1 alleles were described previously [46]. Stra8-Cre transgenic FVB mice were kindly provided by Bob Braun (The Jackson Laboratory; [47]). Spo11-Cre mice were generated as described in Figure S2 and in Text S1. Msh4−/− mice were kindly provided by Winfried Edelmann (Albert Einstein College of Medicine; [66]), and Dmc1−/− [65] and Spo11−/− [64] mutant mice were generously provided by John Schimenti (Cornell University). Slx4mut/mut mice were derived from the previously reported Btbd12tm1a(EUCOMM)Wtsi strain and carried an intact β-geo cassette and germline Slx4 exon 3 deletion [55]. For fertility testing, 8 - to 12-week old Stra8-Cre Hus1 CKO and control males were singly housed with wild-type 129 or FVB females. Copulatory plugs were monitored daily, and plugged females were removed to separate cages and monitored for pregnancy. Viable pups were counted on the first day of life.
Western blotting
Flash frozen testes from 17-day old, 20-day old, or adult animals (12–14 weeks, unless labeled otherwise) were homogenized in RIPA buffer supplemented with protease inhibitors and sodium orthovanadate using a Tissuelyzer, sonicated at 24–27W twice for 2 minutes each, then cleared by centrifugation. Antibodies included rabbit polyclonal anti-HUS1 HM199 (rabbit polyclonal antiserum generated against purified recombinant GST-tagged full-length mouse HUS1 protein), anti-phosphoCHK1 (Ser345; Cell Signaling #2341), anti-CHK1 (Santa Cruz), anti-CHK2 (clone 7, Millipore #05-649), anti-RAD9 HM456 (rabbit polyclonal antiserum generated against purified recombinant HIS-tagged full-length mouse RAD9 protein), affinity purified rabbit anti-RAD1 [41], and anti-β-actin (Sigma).
Histology and immunohistochemistry
Testes were fixed overnight at 4°C in Bouin's fixative (for H&E and GCNA1 staining) or at room temperature in 10% neutral-buffered formalin (for TUNEL staining), embedded in wax, and sectioned at 5 µm. Immunohistochemical staining for germ cell nuclear antigen [GCNA1; 82] was performed using anti-GCNA1 antibody provided by George Enders. TUNEL assays were performed using the Apoptag kit (Millipore) as per the manufacturer's instructions. TUNEL data were quantified by counting the number of TUNEL-positive cells per tubule in at least 50 tubules from the testes of at least 3 different mice of each genotype, and differences between controls and Hus1 CKOs were analyzed statistically via Student's t-test.
Epididymal sperm counts
Both caudal epididymides from each 12-week old mouse were minced with fine forceps in a petri dish with 37°C PBS, incubated, and fixed in 10% neutral-buffered formalin (1∶25 dilution). Sperm counts shown in Figure 1 are the mean of 6 to 9 mice per group ± SEM, analyzed statistically using a Student's t-test.
Meiotic chromosome spreading and immunofluorescence staining
Surface-spread nuclei were prepared from 12-week old male mice as described previously [58], with the exception of Dmc1−/− and control littermate samples which were prepared similarly from flash-frozen testes. Briefly, tubules were incubated in hypotonic extraction buffer on ice for 1 hour, minced in 100 mM sucrose, and spread on slides dipped in 1% PFA with 0.15% Triton X-100. Slides were incubated in a humid chamber for 2.5 hours, dried, and washed in PBS and water containing Photoflo (Kodak). Immunofluorescence staining was performed following blocking in 10% goat or donkey serum and 3% BSA, with primary antibodies incubated overnight at room temperature and secondary antibodies incubated at 37°C for one hour in the dark. Slides were mounted with coverslips using homemade anti-fade mounting medium (2.3% DABCO, 20 mM Tris pH 8.0, 8 µg DAPI in 90% glycerol).
Primary antibodies used for immunofluorescence staining included those recognizing: γH2AX (1∶5000; Upstate/Millipore), SYCP1 (1∶500), SYCP3 (1∶5000; [83]), RAD9 HM456 (1∶600; see above), RAD1 (1∶17; [41]), TOPBP1 (1∶500; [84]), RAD51 (1∶500; Oncogene Research Products/EMD Biosciences), RNA Polymerase II (1∶500; Millipore), and MLH1 (1∶50; BD Pharmingen). For co-staining of RAD9 and RAD1, we additionally used affinity purified sheep anti-RAD1 (sheep polyclonal antiserum generated against purified recombinant 6X HIS full-length human RAD1) generated in the Freire laboratory, and for co-staining of RAD9 with RAD51, we used mouse monoclonal anti-RAD51 (Abcam). For MLH1/SYCP3/RAD51 co-staining, anti-SYCP3 antibody was used at 1∶50,000 as described Lipkin et al. [85]. Human CREST serum was used to detect centromeres in some experiments as previously described [86]. Secondary antibodies were used at 1∶1000 dilution and included goat anti-mouse Alexafluor 488, goat anti-rabbit Alexafluor 555, donkey anti-sheep Alexafluor 488, and donkey anti-rabbit Alexafluor 555 (Invitrogen). Microscopy and imaging was performed as described previously [56].
For quantification of phenotypes shown in Table 2, a “grossly normal” nucleus was defined as one with normal synapsis and γH2AX staining confined to the sex body, and an “abnormal sex body” was defined as one with extended γH2AX signal, inclusion of whole or partial autosomes, and/or the presence of an apparent X chromosome-autosome end-to-end fusion. P-values for differences in γH2AX staining (Table 2), sex body abnormalities (Table 2), and synapsis defects (text) were calculated using a z-test of sample proportions, comparing Hus1 CKOs to control animals. Diakinesis chromosome spreads (Figure S6D) were generated as described previously [56] from two independent 4 - to 5-week old mice per genotype.
Supporting Information
Zdroje
1. EichingerCS, JentschS (2011) 9-1-1: PCNA's specialized cousin. Trends Biochem Sci 36 : 563–568.
2. WeissRS, LederP, VaziriC (2003) Critical role for mouse Hus1 in an S-phase DNA damage cell cycle checkpoint. Mol Cell Biol 23 : 791–803.
3. ZouL, CortezD, ElledgeSJ (2002) Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev 16 : 198–208.
4. Navadgi-PatilVM, BurgersPM (2009) A tale of two tails: Activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1. DNA Repair (Amst) 8 : 996–1003.
5. XuY-j, LeffakM (2010) ATRIP from TopBP1 to ATR—in vitro activation of a DNA damage checkpoint. Proc Natl Acad Sci USA 107 : 13561–13562.
6. Parrilla-CastellarER, ArlanderSJH, KarnitzL (2004) Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 3 : 1009–1014.
7. PanditaRK, SharmaGG, LaszloA, HopkinsKM, DaveyS, et al. (2006) Mammalian Rad9 plays a role in telomere stability, S - and G2-phase-specific cell survival, and homologous recombinational repair. Mol Cell Biol 26 : 1850–1864.
8. SabbionedaS, MinesingerBK, GiannattasioM, PlevaniP, Muzi-FalconiM, et al. (2005) The 9-1-1 checkpoint clamp physically interacts with Polzeta and is partially required for spontaneous Polzeta-dependent mutagenesis in Saccharomyces cerevisiae. J Biol Chem 38657–38665.
9. ToueilleM, El-AndaloussiN, FrouinI, FreireR, FunkD, et al. (2004) The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase β and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res 32 : 3316–3324.
10. GembkaA, ToueilleM, SmirnovaE, PoltzR, FerrariE, et al. (2007) The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase β in long patch base excision repair. Nucleic Acids Res 35 : 2596–2608.
11. WangW, BrandtP, RossiML, Lindsey-BoltzL, PodustV, et al. (2004) The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc Natl Acad Sci USA 101 : 16762–16767.
12. Friedrich-HeinekenE, ToueilleM, TännlerB, BürkiC, FerrariE, et al. (2005) The two DNA clamps Rad9/Rad1/Hus1 complex and Proliferating Cell Nuclear Antigen differentially regulate Flap Endonuclease 1 activity. J Mol Biol 353 : 980–989.
13. SongW, PascalJM, EllenbergerT, TomkinsonAE (2009) The DNA binding domain of human DNA ligase I interacts with both nicked DNA and the DNA sliding clamps, PCNA and hRad9-hRad1-hHus1. DNA Repair (Amst) 8 : 912–919.
14. GuanX, MadabushiA, ChangD-Y, FitzgeraldME, ShiG, et al. (2007) The human checkpoint sensor Rad9 Rad1 Hus1 interacts with and stimulates DNA repair enzyme TDG glycosylase. Nucleic Acids Res 35 : 6207–6218.
15. GuanX, BaiH, ShiG, TheriotCA, HazraTK, et al. (2007) The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates NEIL1 glycosylase. Nucleic Acids Res 35 : 2463–2472.
16. ChangD-Y, LuA-L (2005) Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J Biol Chem 280 : 408–417.
17. ShiG, ChangD-Y, ChengC-C, GuanX, VenclovasÄ, et al. (2006) Physical and functional interactions between MutY glycosylase homologue (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem J 400 : 53–62.
18. ParkMJ, ParkJ-H, HahmS-H, KoSI, LeeYR, et al. (2009) Repair activities of human 8-oxoguanine DNA glycosylase are stimulated by the interaction with human checkpoint sensor Rad9–Rad1–Hus1 complex. DNA Repair (Amst) 8 : 1190–1200.
19. CaiRL, Yan-NealeY, CuetoMA, XuH, CohenD (2000) HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J Biol Chem 275 : 27909–27916.
20. PichierriP, NicolaiS, CignoloL, BignamiM, FranchittoA (2011) The RAD9-RAD1-HUS1 (9.1.1) complex interacts with WRN and is crucial to regulate its response to replication fork stalling. Oncogene doi:10.1038/onc.2011.1468
21. HeW, ZhaoY, ZhangC, AnL, HuZ, et al. (2008) Rad9 plays an important role in DNA mismatch repair through physical interaction with MLH1. Nucleic Acids Res 36 : 6406–6417.
22. BaiH, MadabushiA, GuanX, LuAL (2010) Interaction between human mismatch repair recognition proteins and checkpoint sensor Rad9-Rad1-Hus1. DNA Repair (Amst) 9 : 478–487.
23. Cotta-RamusinoC, McDonaldER, HurovK, SowaME, HarperJW, et al. (2011) A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science 332 : 1313–1317.
24. PereraD, Perez-HidalgoL, MoensPB, ReiniK, LakinN, et al. (2004) TopBP1 and ATR colocalization at meiotic chromosomes: Role of TopBP1/Cut5 in the meiotic recombination checkpoint. Mol Biol Cell 15 : 1568–1579.
25. MoensPB, TarsounasM, MoritaT, HabuT, RottinghausST, et al. (1999) The association of ATR protein with mouse meiotic chromosome cores. Chromosoma 108 : 95–102.
26. TurnerJMA, AprelikovaO, XuX, WangR, KimS, et al. (2004) BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol 14 : 2135–2142.
27. HandelMA (2004) The XY body: a specialized meiotic chromatin domain. Exp Cell Res 296 : 57–63.
28. Hoyer-FenderS (2004) Molecular aspects of XY body formation. Cytogenet Genome Res 103 : 245–255.
29. BellaniMA, RomanienkoPJ, CairattiDA, Camerini-OteroRD (2005) SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm-/ - spermatocytes. J Cell Sci 118 : 3233–3245.
30. TurnerJMA (2007) Meiotic sex chromosome inactivation. Development 134 : 1823–1831.
31. RoyoH, PolikiewiczG, MahadevaiahSK, ProsserH, MitchellM, et al. (2010) Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol 20 : 2117–2123.
32. TurnerJMA, MahadevaiahSK, Fernandez-CapetilloO, NussenzweigA, XuX, et al. (2005) Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 37 : 41–47.
33. WojtaszL, CloutierJM, BaumannM, DanielK, VargaJ, et al. (2012) Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes Dev 26 : 958–973.
34. LydallD, NikolskyY, BishopDK, WeinertT (1996) A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383 : 840–843.
35. HongE-JE, RoederGS (2002) A role for Ddc1 in signaling meiotic double-strand breaks at the pachytene checkpoint. Genes Dev 16 : 363–376.
36. AbduU, KlovstadM, Butin-IsraeliV, BakhratA, SchüpbachT (2007) An essential role for Drosophila hus1 in somatic and meiotic DNA damage responses. J Cell Sci 120 : 1042–1049.
37. ShinoharaM, SakaiK, OgawaT, ShinoharaA (2003) The mitotic DNA damage checkpoint proteins Rad17 and Rad24 are required for repair of double-strand breaks during meiosis in yeast. Genetics 164 : 855–865.
38. GrushcowJM, HolzenTM, ParkKJ, WeinertT, LichtenM, et al. (1999) Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics 153 : 607–620.
39. PeretzG, ArieLG, BakhratA, AbduU (2009) The Drosophila hus1 gene is required for homologous recombination repair during meiosis. Mech Dev 126 : 677–686.
40. SmirnovaNA, RomanienkoPJ, KhilPP, Camerini-OteroRD (2006) Gene expression profiles of Spo11−/ − mouse testes with spermatocytes arrested in meiotic prophase I. Reproduction 132 : 67–77.
41. FreireR, MurguiaJR, TarsounasM, LowndesNF, MoensPB, et al. (1998) Human and mouse homologs of Schizosaccharomyces pombe rad1+ and Saccharomyces cerevisiae RAD17: linkage to checkpoint control and mammalian meiosis. Genes Dev 12 : 2560–2573.
42. BrownEJ, BaltimoreD (2000) ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 14 : 397–402.
43. HopkinsKM, AuerbachW, WangXY, HandeMP, HangH, et al. (2004) Deletion of mouse Rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol Cell Biol 24 : 7235–7248.
44. WeissRS, EnochT, LederP (2000) Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev 14 : 1886–1898.
45. HanL, HuZ, LiuY, WangX, HopkinsK, et al. (2010) Mouse Rad1 deletion enhances susceptibility for skin tumor development. Mol Cancer 9 : 67.
46. LevittPS, LiuH, ManningC, WeissRS (2005) Conditional inactivation of the mouse Hus1 cell cycle checkpoint gene. Genomics 86 : 212–224.
47. Sadate-NgatchouPI, PayneCJ, DearthAT, Robert EBraun (2008) Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46 : 738–742.
48. WangX, GuanJ, HuB, WeissRS, IliakisG, et al. (2004) Involvement of Hus1 in the chain elongation step of DNA replication after exposure to camptothecin or ionizing radiation. Nucleic Acids Res 32 : 767–775.
49. LevittPS, ZhuM, CassanoA, YazinskiSA, LiuH, et al. (2007) Genome maintenance defects in cultured cells and mice following partial inactivation of the essential cell cycle checkpoint gene Hus1. Mol Cell Biol 27 : 2189–2201.
50. ZhuM, WeissRS (2007) Increased common fragile site expression, cell proliferation defects, and apoptosis following conditional inactivation of mouse Hus1 in primary cultured cells. Mol Biol Cell 18 : 1044–1055.
51. WardIM, ChenJ (2001) Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 276 : 47759–47762.
52. ZhaoH, Piwnica-WormsH (2001) ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Molecular and Cellular Biology 21 : 4129–4139.
53. ChicheporticheA, Bernardino-SgherriJ, de MassyB, DutrillauxB (2007) Characterization of Spo11-dependent and independent phospho-H2AX foci during meiotic prophase I in the male mouse. J Cell Sci 120 : 1733–1742.
54. LiXC, SchimentiJC (2007) Mouse pachytene checkpoint 2 (Trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet 3: e130 doi:10.1371/journal.pgen.0030130.
55. HollowayJK, MohanS, BalmusG, SunX, ModzelewskiA, et al. (2011) Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genet 7: e1002094 doi:10.1371/journal.pgen.1002094.
56. HollowayJK, MorelliMA, BorstPL, CohenPE (2010) Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol 188 : 779–789.
57. HollowayJK, BoothJ, EdelmannW, McGowanCH, CohenPE (2008) MUS81 generates a subset of MLH1-MLH3–independent crossovers in mammalian meiosis. PLoS Genet 4: e1000186 doi:10.1371/journal.pgen.1000186.
58. KolasNK, SvetlanovA, LenziML, MacalusoFP, LipkinSM, et al. (2005) Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J Cell Biol 171 : 447–458.
59. BaudatF, de MassyB (2007) Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res 15 : 565–577.
60. ReiniK, UittoL, PereraD, MoensP, FreireR, et al. (2004) TopBP1 localises to centrosomes in mitosis and to chromosome cores in meiosis. Chromosoma 112 : 323–330.
61. Lindsey-BoltzLA, SancarA (2011) Tethering DNA damage checkpoint mediator proteins Topoisomerase IIβ-binding Protein 1 (TopBP1) and Claspin to DNA activates Ataxia-Telangiectasia Mutated and RAD3-related (ATR) phosphorylation of Checkpoint Kinase 1 (Chk1). J Biol Chem 286 : 19229–19236.
62. NamEA, CortezD (2011) ATR signalling: more than meeting at the fork. Biochem J 436 : 527–536.
63. DelacroixS, WagnerJM, KobayashiM, YamamotoK-i, KarnitzLM (2007) The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 21 : 1472–1477.
64. RomanienkoPJ, Camerini-OteroRD (2000) The mouse Spo11 gene is required for meiotic chromosome synapsis. Molecular Cell 6 : 975–987.
65. PittmanDL, CobbJ, SchimentiKJ, WilsonLA, CooperDM, et al. (1998) Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell 1 : 697–705.
66. KneitzB, CohenPE, AvdievichE, ZhuL, KaneMF, et al. (2000) MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes & Development 14 : 1085–1097.
67. HangH, ZhangY, DunbrackRLJr, WangC, LiebermanHB (2002) Identification and characterization of a paralog of human cell cycle checkpoint gene HUS1. Genomics 79 : 487–492.
68. DufaultVM, OestreichAJ, VromanBT, KarnitzLM (2003) Identification and characterization of RAD9B, a paralog of the RAD9 checkpoint gene. Genomics 82 : 644–651.
69. BurtelowMA, Roos-MattjusPMK, RauenM, BabendureJR, KarnitzLM (2001) Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage responsive checkpoint complex. Journal of Biological Chemistry 276 : 25903–25909.
70. DanielK, LangeJ, HachedK, FuJ, AnastassiadisK, et al. (2011) Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat Cell Biol 13 : 599–610.
71. OhSD, LaoJP, HwangPY-H, TaylorAF, SmithGR, et al. (2007) BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130 : 259–272.
72. De MuytA, JessopL, KolarE, SourirajanA, ChenJ, et al. (2012) BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Molecular Cell 46 : 43–53.
73. RoigI, DowdleJA, TothA, de RooijDG, JasinM, et al. (2010) Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet 6: e1001062 doi:10.1371/journal.pgen.1001062
74. BrandtPD, HeltCE, KengPC, BambaraRA (2006) The Rad9 protein enhances survival and promotes DNA repair following exposure to ionizing radiation. Biochem Biophys Res Comm 347 : 232–237.
75. WangX, HuB, WeissRS, WangY (2006) The effect of Hus1 on ionizing radiation sensitivity is associated with homologous recombination repair but is independent of nonhomologous end-joining. Oncogene 25 : 1980–1983.
76. KidaneD, JonasonAS, GortonTS, MihaylovI, PanJ, et al. (2010) DNA polymerase [beta] is critical for mouse meiotic synapsis. EMBO J 29 : 410–423.
77. PlugAW, ClairmontCA, SapiE, AshleyT, SweasyJB (1997) Evidence for a role for DNA polymerase β in mammalian meiosis. Proc Natl Acad Sci USA 94 : 1327–1331.
78. KawamotoT, ArakiK, SonodaE, YamashitaYM, HaradaK, et al. (2005) Dual roles for DNA polymerase η in homologous DNA recombination and translesion DNA synthesis. Mol Cell 20 : 793–799.
79. McIlwraithMJ, VaismanA, LiuY, FanningE, WoodgateR, et al. (2005) Human DNA polymerase η promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell 20 : 783–792.
80. SharmaS, HicksJK, ChuteCL, BrennanJR, AhnJ-Y, et al. (2012) REV1 and polymerase ζ facilitate homologous recombination repair. Nucleic Acids Res 40 : 682–691.
81. GoedeckeW, EijpeM, OffenbergHH, AalderenMv, HeytingC (1999) Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet 23 : 194–198.
82. EndersGC, MayJJ (1994) Developmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female mice. Dev Biol 163 : 331–340.
83. LenziML, SmithJ, SnowdenT, KimM, FishelR, et al. (2005) Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis I in human oocytes. Am J Hum Genet 76 : 112–127.
84. DanielsenJMR, LarsenDH, SchouKB, FreireR, FalckJ, et al. (2009) HCLK2 is required for activity of the DNA damage response kinase ATR. J Biol Chem 284 : 4140–4147.
85. LipkinSM, MoensPB, WangV, LenziM, ShanmugarajahD, et al. (2002) Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet 31 : 385–390.
86. KanR, SunX, KolasNK, AvdievichE, KneitzB, et al. (2008) Comparative analysis of meiotic progression in female mice bearing mutations in genes of the DNA mismatch repair pathway. Biology of Reproduction 78 : 462–471.
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



