-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
Expansions of trinucleotide CAG/CTG repeats in somatic tissues are thought to contribute to ongoing disease progression through an affected individual's life with Huntington's disease or myotonic dystrophy. Broad ranges of repeat instability arise between individuals with expanded repeats, suggesting the existence of modifiers of repeat instability. Mice with expanded CAG/CTG repeats show variable levels of instability depending upon mouse strain. However, to date the genetic modifiers underlying these differences have not been identified. We show that in liver and striatum the R6/1 Huntington's disease (HD) (CAG)∼100 transgene, when present in a congenic C57BL/6J (B6) background, incurred expansion-biased repeat mutations, whereas the repeat was stable in a congenic BALB/cByJ (CBy) background. Reciprocal congenic mice revealed the Msh3 gene as the determinant for the differences in repeat instability. Expansion bias was observed in congenic mice homozygous for the B6 Msh3 gene on a CBy background, while the CAG tract was stabilized in congenics homozygous for the CBy Msh3 gene on a B6 background. The CAG stabilization was as dramatic as genetic deficiency of Msh2. The B6 and CBy Msh3 genes had identical promoters but differed in coding regions and showed strikingly different protein levels. B6 MSH3 variant protein is highly expressed and associated with CAG expansions, while the CBy MSH3 variant protein is expressed at barely detectable levels, associating with CAG stability. The DHFR protein, which is divergently transcribed from a promoter shared by the Msh3 gene, did not show varied levels between mouse strains. Thus, naturally occurring MSH3 protein polymorphisms are modifiers of CAG repeat instability, likely through variable MSH3 protein stability. Since evidence supports that somatic CAG instability is a modifier and predictor of disease, our data are consistent with the hypothesis that variable levels of CAG instability associated with polymorphisms of DNA repair genes may have prognostic implications for various repeat-associated diseases.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003280
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003280Summary
Expansions of trinucleotide CAG/CTG repeats in somatic tissues are thought to contribute to ongoing disease progression through an affected individual's life with Huntington's disease or myotonic dystrophy. Broad ranges of repeat instability arise between individuals with expanded repeats, suggesting the existence of modifiers of repeat instability. Mice with expanded CAG/CTG repeats show variable levels of instability depending upon mouse strain. However, to date the genetic modifiers underlying these differences have not been identified. We show that in liver and striatum the R6/1 Huntington's disease (HD) (CAG)∼100 transgene, when present in a congenic C57BL/6J (B6) background, incurred expansion-biased repeat mutations, whereas the repeat was stable in a congenic BALB/cByJ (CBy) background. Reciprocal congenic mice revealed the Msh3 gene as the determinant for the differences in repeat instability. Expansion bias was observed in congenic mice homozygous for the B6 Msh3 gene on a CBy background, while the CAG tract was stabilized in congenics homozygous for the CBy Msh3 gene on a B6 background. The CAG stabilization was as dramatic as genetic deficiency of Msh2. The B6 and CBy Msh3 genes had identical promoters but differed in coding regions and showed strikingly different protein levels. B6 MSH3 variant protein is highly expressed and associated with CAG expansions, while the CBy MSH3 variant protein is expressed at barely detectable levels, associating with CAG stability. The DHFR protein, which is divergently transcribed from a promoter shared by the Msh3 gene, did not show varied levels between mouse strains. Thus, naturally occurring MSH3 protein polymorphisms are modifiers of CAG repeat instability, likely through variable MSH3 protein stability. Since evidence supports that somatic CAG instability is a modifier and predictor of disease, our data are consistent with the hypothesis that variable levels of CAG instability associated with polymorphisms of DNA repair genes may have prognostic implications for various repeat-associated diseases.
Introduction
At least 14 neurodegenerative and neuromuscular diseases are caused by expansions of CAG/CTG repeats including Huntington's disease (HD) and myotonic dystrophy type 1. An inverse correlation between the length of CAG repeat tracts and age-of-onset is observed in HD families [1], [2]. The expanded CAG repeat is unstable in several organs, undergoing progressive length increases over time, coincident with disease progression [3]–[7]. Within the brain, somatic CAG expansions are region-specific with the greatest instability observed in striatum and cortex, which show the most severe neuropathology in HD patients [3]–[7]. The potential contribution of somatic repeat instability to HD/DM1 disease age-of-onset, severity and progression [1], [7], [8], make it imperative to understand the process of instability as it is a therapeutic target [9].
Several transgenic mouse models have contributed to our understanding of the mechanisms of CAG/CTG instability [10]–[18]. Both cis-elements and trans-factors that modify CAG/CTG repeat instability have been identified. Cis-elements include flanking sequence context such as CTCF binding sites, CpG-methylation, DNA sequence, G+C-content, DNA replication direction and progression, and transcription levels and direction [9], [11], [17]–[23]. Trans-factors that have been linked to CAG/CTG instability include DNA replication, repair and recombination proteins. Of those tested in mice, Fen1, Rad52, Rad54, Xpc, appear to have no effect [24]–[27], while Ogg1, Neil1, Csb, Lig1, Xpa, Msh6, or Pms2 show partial effects [16], [28]–[33]. However, the mismatch repair (MMR) genes Msh2 and Msh3 have been shown to be absolutely required for repeat expansions [13], [16], [26], [27], [34]–[36].
In addition to being the strongest modifiers of repeat instability identified to date, [9], [37] the roles of the MMR proteins MSH2 and MSH3 have recently been extended to CAG/CTG instability in human HD and DM1 stem cells [38]. MMR is a pathway dedicated to protecting against mutations arising from mispaired nucleotides and insertion/deletion loops [39]. There are two heterodimeric protein complexes that recognize unpaired DNAs: MutSα consists of MSH2-MSH6, and MutSβ is formed by MSH2–MSH3. MutSα is predominantly required to repair base-base mismatches, and MutSβ, with some functional redundancy with MutSα, is predominantly involved in the repair of insertion/deletion loops (1-12 nucleotides) [40]–[44]. MutSβ, more so than MutSα, is required to repair short CAG/CTG slip-outs [42]. Recent evidence revealed that the levels of MSH2, MSH3, and MSH6 protein varied widely between 14 different murine tissue types, and MSH3 protein levels were greater than MSH6 levels in most tissues analyzed [45]. MMR typically functions to protect against mutations; however, in the case of long CAG/CTG repeat alleles, MSH2 and MSH3 are required for additional repeat expansion mutations [37]. Msh2 deficiency stabilized CAG/CTG repeat tracts from inherent expansions in somatic tissues of R6/1 mice transgenic for exon 1 of the HD gene [13], [46], HdhQ111 knock-in mice [27], [36], and several DM1 mouse models [16], [26], [47]. The MSH3 protein, like MSH2, is required for the expansion-biased CAG/CTG repeat instability in somatic tissues [16], [27], [35]. The absence of Msh3 blocks CAG/CTG expansions in tissues from HD mice [16], [27], [35]. The absence of one Msh3 allele (Msh3/null mice) is sufficient to decrease CAG expansion frequencies in HD and DM1 mice, suggesting that MSH3 may be a limiting factor in the process leading to the formation of expansions, and that CAG instability could tightly depend on MSH3 protein levels [16], [27], [35]. An absence of MSH6 increased CAG/CTG expansions [16], [27], [35], probably due to the competition between MSH3 and MSH6 for binding to MSH2 to form functional complexes [16], [48].
Several models have been proposed through which MutSβ can drive CAG/CTG expansions. In vivo mouse models suggest that MutSβ is required to drive CAG expansions [13], [27], [35] and to protect against repeat contractions [16], [26], [27], [34]–[36], [49]. The role of MutSβ in expansions extends beyond its ability to bind slipped-DNAs [50] as an ATPase-functional MutSβ complex is necessary for CAG expansions [47] and downstream mismatch repair proteins, like PMS2 are partially required for instability [28]. The MutSβ complex may act on CAG repeats during errors at replication forks or during transcription, as both processes can enhance instability in a MMR-dependant manner [51], [52]. Instability in non-proliferating tissues may arise when attempted repair events by MutSβ on clustered short CAG/CTG slip-outs is arrested [42]. Arrested repair along these clusters may allow for strand displacement, slippage, further out-of-register mispairing, and repair synthesis resulting in expansions on un-repaired clustered slip-outs [42]. Reiterations of such events, using the aberrant repair products as substrates, could lead to continuous expansions. Perturbed levels of MutSβ decreased repair of short CTG slip-outs, allowing them to be integrated as expansions [42]. The sensitivity of short TNR slip-out repair to MutSβ concentration is similar to other reports of repair protein levels affecting repeat instability [53]–[55].
In this study we used the R6/1 HD transgenic mouse model [10], [56]. The R6/1 HD transgenic mice were generated by using a construct with ∼1000 bp of the human Huntingtin gene HTT promoter, the entire HTT exon-1, including ∼116 CAG repeats, and 262 bp of HTT intron-1 [56]. The R6/1 transgene has been reported to be integrated as a head to tail dimer on chromosome 3 [57]. However, in our colony the transgene appears to harbour only a single CAG repeat tract length as assessed by SP-PCR (see below). The transgene expresses the expanded CAG transcript and is translated to produce a HTT exon 1 fragment with an expanded polyglutamine tract. Males show limited CAG instability upon transmission and females are infertile, as reported [10], [56]. R6/1 mice have been used extensively to assess both HD pathogenesis and CAG instability, where the latter results have found tissue-specific instability dependent upon Msh2 and Msh3 [13], partially-dependent upon Ogg1 and Neil1 [31]. R6/1 mice have also been found to be protected from instability by Csb, and unaffected by Fen1 [25], [32]. Furthermore, CAG instability in R6/1 mice has been shown to be sensitive to transcription progression [58] and tissue-specific stoichiometric levels of base excision repair proteins [54].
Several studies have reported the existence of other modifiers of CAG/CTG repeat instability, as different mouse strains harbouring the same HD or DM1 CAG/CTG transgene have variable levels of repeat instability [16], [59], [60]. Similarly, extreme repeat changes in some Huntington's families suggests the existence of family-specific instability modifiers that may be heritable [61]. However, none of these studies have proposed a candidate factor as a source for strain-specific variations in CAG/CTG instability patterns. Here we have identified the source of variable CAG repeat instability between two inbred mouse strains, C57BL/6J (B6) and BALB/cByJ (CBy), congenic for an HTT exon 1 transgene (R6/1). Using both congenic and reciprocal congenic mice, we identified coding variations in the Msh3 gene as sources of the variable levels of somatic CAG instability in the different strains of R6/1 transgenic mice. The B6 MSH3 protein variant is highly expressed and associated with expansion biased mutations, while the CBy MSH3 protein variant is expressed at low levels and is associated with CAG tract stability.
Results
CAG repeat instability in C57BL/6J and BALB/cByJ mice
To assess CAG repeat instability in mice with different genetic backgrounds, we backcrossed B6CBA-Tg(HDexon1)61Gpb(R6/1) transgenic mice [10] to B6 and CBy inbred mice to obtain B6.Cg-Tg(HDexon1)61Gpb (B6.Cg-R6/1) and CBy.Cg-Tg(HDexon1)61Gpb (CBy.Cg-R6/1) congenic lines, respectively. These congenic lines were typed at each generation for the presence of the R6/1 transgene, thus after 10 backcross generations, it was predicted that 99.8% of the genome was homozygous for the inbred line (B6 or CBy), while the remaining 0.2% of the genome remained heterozygous. The B6.Cg-R6/1 and CBy.Cg-R6/1 congenic mice contained (CAG)98 and (CAG)94, respectively - so these mice and their progeny should be well matched for HD transgene effects with the same flanking cis-elements. Genome-wide SNP analysis confirmed that the HTT transgene had integrated into chromosome 3 [57] and showed minimal contamination of adjacent regions in the congenic strains (Figure S1A, Table S1). We analysed CAG instability by SP-PCR in liver, striatum, tail and heart from 20 week-old mice. B6.Cg-R6/1 mice showed a high level of somatic instability biased toward expansions in liver and striatum (Figure 1A), while the repeat was relatively stable in heart and tail, as previously described [10], [13]. Surprisingly, the CAG repeats were very stable in all of these four tissues from age-matched CBy.Cg-R6/1 mice, including in liver and striatum (Figure 1A). The stabilizing effect of the CBy background was as striking as the genetic deficiency of the MMR protein MSH2, as previously described [13]. Thus, the level of somatic CAG expansions can be dramatically different between B6.Cg-R6/1 and CBy.Cg-R6/1 mice, revealing that CAG expansions are affected by genetic background.
Fig. 1. Representative CAG repeat distributions, and Msh3 variations in B6 and CBy mice. 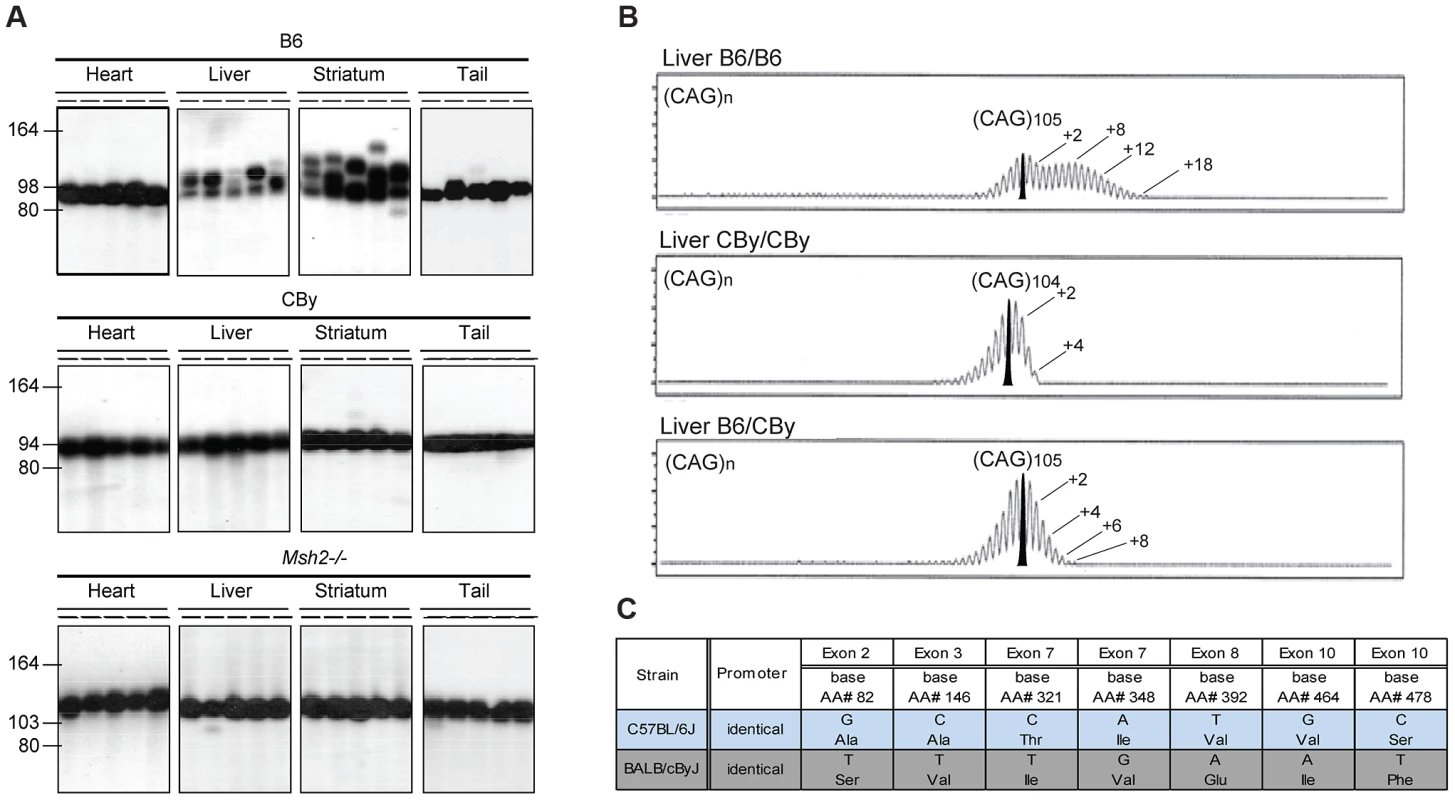
A) The autoradiographs show representative SP-PCR analyses of DNA, extracted from heart, liver, striatum and tail. At weaning the B6.Cg-R6/1 (B6) and CBy.Cg-R6/1 (CBy) congenic mice contained in tail DNA (CAG)98 and (CAG)94, respectively. For comparison the profiles of the Msh2−/− mouse is shown. About 5–10 DNA amplifiable molecules were amplified in each reaction with primers MS-1F and MS-1R. Animals were 20-weeks old. B) Congenic CBy.Cg-R6/1 mice were crossed to B6 and the resulting F1 progeny were crossed to produce F2 mice with all possible genotypes at the Msh3 locus. Repeat instability was assayed by amplifying 10 ng genomic DNA using fluorescently labelled primers and resolving the fragments by capillary gel electrophoresis (Figure 1B). Using this high-resolution approach repeat length distributions present with the typical ‘hedgehog’ pattern (e.g. [10], [13], [15], [16]. This pattern reflects both somatic mosaicism within the sample and PCR artefacts generated by Taq polymerase slippage [62], [63]. The PCR artefacts are predominantly repeat contractions, hence these are not considered here. The pattern of CAG repeat instability depended on genotype at the MSH3 locus. B6 homozygosity resulted in the greatest instability, CBy homozygosity resulted in lack of expansion, while heterozygosity resulted in an intermediate instability, indicative of a gene dosage effect of the Msh3 locus. Numbers indicate the CAG repeat size corresponding to major peaks. In addition, on the B6 tracing, a second number indicates the highest CAG repeat number detected. C) Msh3 polymorphisms in Msh3 gene from C57BL/6 (B6) and BALB/cBy (CBy) mice. Promoters were identical. SNPs were identified or confirmed to those in dbSNP by sequencing the Msh3 gene. CAG repeat instability difference is likely a single-gene/locus effect
Towards identifying modifiers of CAG instability, we performed a F2 intercross between CBy.Cg-R6/1 and B6, and tested offspring for differential CAG instability patterns in the liver as this tissue displayed considerably different patterns of CAG instability between the R6/1 congenic lines (Figure 1A). Repeat instability was assayed blind (to remove bias) by high-resolution capillary gel electrophoresis where repeat length distributions present a typical ‘hedgehog’ pattern (e.g. [10], [13], [15], [16]) (Figure 1B). In R6/1 mice the overall level of somatic instability is generally relatively low and the inherited or progenitor allele is usually defined as the modal allele within the distribution of peaks (see bold-filled peak in Figure 1B) and is conserved between tissues from the same mouse. As Taq polymerase slippage during PCR generates repeat contractions [62], [63], we concentrated on defining an instability phenotype based on the expanded alleles. Using this approach, three distinct patterns of CAG instability in liver DNA of F2 mice were observed (Figure 1B). Firstly, as in the parental B6.Cg.R6/1 mice, some F2 mice presented with high levels of instability with a broad bimodal distribution profile with a second peak at ∼+7–9 repeats and a long tail extending out to greater than +15 repeats. Secondly, as in the parental CBy.Cg.R6/1 mice, some F2 mice presented with only very low levels of CAG mosaicism with a unimodal negatively skewed distribution with a tail of expanded alleles that extended only to +3 or +4 repeats and ended very abruptly. Thirdly, we detected an intermediate instability phenotype in which the distributions were unimodal, but more normally distributed, without the pronounced negative skew, and in which the tail of expansions extended out to +7 to +8 repeats (Figure 1B). These distinct patterns of CAG instability between F2 offspring suggested that they may contain varying dosages of a specific modifier gene(s) of CAG instability. Of 81 mice assessed, 20 had highly unstable, 24 stable and 37 intermediate levels of CAG repeat instability. This phenotypic distribution fits with the 1∶2∶1 segregation ratio expected for a single modifier gene with a semi-dominant allele (Chi-Square analysis (X2(2, N = 81) = 1.0, p = 0.61)).
Since Msh3 is one of the strongest known drivers of CAG expansions [9], [37], and it also shows a gene dosage effect [16], [27], [35], we considered the possibility that the Msh3 gene variants between the CBy and B6 mouse strains may account for the variations in CAG instability patterns between the congenic strains. Towards this end, we genotyped the locus containing the Msh3 gene using microsatellite markers flanking the gene (D13Mit159 and D13Mit147) in the offspring of the F2 intercross. All mice showing high levels of CAG expansions were homozygous for B6 alleles at the Msh3 locus, while those with the stable CAG tract were homozygous for CBy alleles at the Msh3 locus and those with the intermediate CAG instability were heterozygous at the Msh3 locus. These data firmly link variation in Msh3 or a nearby gene on mouse chromosome 13 with the differential repeat instability phenotypes (LOD score = 48.8, (θ = 0), see Materials and Methods).
Msh3 polymorphisms between mouse strains
In an effort to identify Msh3 gene polymorphisms, we sequenced the exons and promoter of the Msh3 gene of the CBy and B6 strains. We identified 7 polymorphisms that resulted in non-synonymous amino acid changes, within exons 2, 3, 7, 8 and 10 (Figure 1C), between B6 and CBy. There was no sequence variation of the Msh3 promoter between the CBy and B6 mice. The polymorphic, coding Msh3 variants between the CBy and B6 mice may therefore be responsible for the variable CAG instabilities between the mouse lines. It is highly unlikely that the original non-synonymous polymorphisms were unlinked and became linked during the course of the construction of the inbred lines, as we sequenced the Msh3 gene of the strains from colonies originating independent from those used in our breedings.
Somatic CAG instability in Msh3 locus reciprocal congenic mice
In order to test the potential role of MSH3 protein variants on CAG instability, we created Msh3-locus reciprocal congenic mice carrying the B6 Msh3 variant on a CBy genetic background (CBy.B6-msh3B6/B6), and a CBy Msh3 variant in the B6 genetic background (B6.CBy-msh3CBy/CBy). Each line was backcrossed to the recipient strain 10 times as in the creation of the R6/1 congenic lines. Next they were inter-crossed as appropriate with the R6/1 congenic lines to create mice CBy homozygous at the Msh3 locus on a B6 genetic background and hemizygous for the R6/1 transgene (B6.CBy-msh3CBy/CBy, R6/1) and mice B6 homozygous at the Msh3 locus on a CBy genetic background and hemizygous for the R6/1 transgene (CBy.B6-msh3B6/B6, R6/1). With these mice we could better isolate the effect of each Msh3 variant on both mouse backgrounds on CAG stability. Genome-wide SNP genotyping revealed minimal donor haplotype contamination in the reciprocal congenic strains B6.CBy-msh3CBy/CBy and CBy.B6-msh3B6/B6 and their corresponding R6/1 congenic strains B6.Cg-R6/1 and CBy.Cg-R6/1 (Figure S1B, Table S1). Outside of the genomic region flanking chromosome 3 integration site of the R6/1 transgene [57], there appears to be no contamination of donor DNA in the CBy background line, and only minor areas of residual heterozygosity in the B6 background lines on chromosomes 6, 15 and 17. The contaminating regions linked to the Msh3 gene in the reciprocal congenics contain a limited number of genes (Figure S7), none of which have an obvious or documented role in CAG repeat instability. The regions linked to the Msh3 gene in the CBy.B6-Msh3 R6/1 reciprocal congenic mice span 43 Mbp and include 314 genes, of which 233 are protein-coding (Figure S7). In the B6.CBy-Msh3 R6/1 strain, the linked genes cover a region of approximately 22 Mbp, which lies within the 43 Mbp region of the CBy.B6-Msh3 R6/1 strain. A total of 151 genes are found within this region with 104-protein coding transcripts (Figure S7). Therefore, differences in CAG instability between and within the strains were interpreted to be a consequence of the introgressed Msh3 allele variants. At 16–20 weeks of age, a high level of CAG expansion was present in the liver from mice containing the B6 Msh3 gene for both B6 and CBy backgrounds. This instability was evident as a broad bimodal distribution profile whereas the liver DNA from mice with the CBy Msh3 gene showed a low level of instability with a unimodal distribution (Figure 2A). A similar pattern of CAG instability in the striatum further indicated greater levels of CAG instability in mice with the B6 Msh3 gene than those with the CBy Msh3 gene (Figure 2B). The striking differences in the levels of instability between mice harbouring B6 Msh3 compared to CBy Msh3, regardless of background, supports the concept that the B6 Msh3 gene variant drives CAG expansions to a greater degree than does the CBy Msh3 gene variant.
Fig. 2. Representative CAG repeat distributions from reciprocal Msh3 congenic lines of mice. 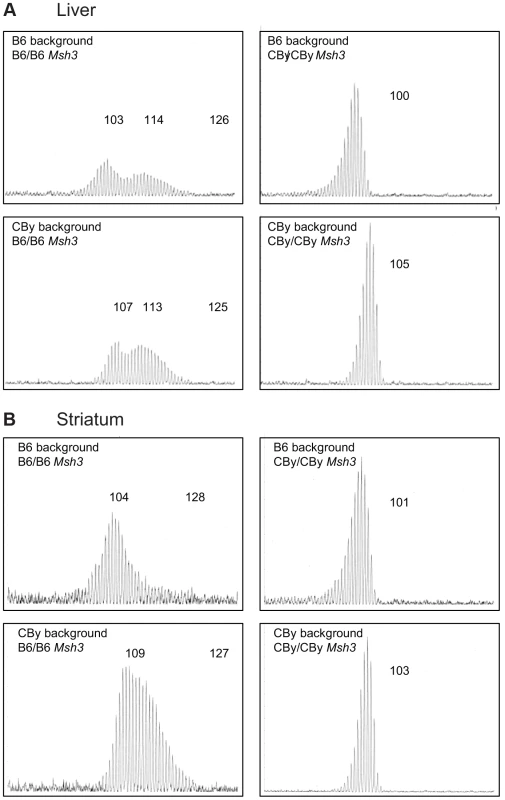
Typical GeneScan traces for sizing of the CAG repeat as outlined in Figure 1B. Liver (A) and Striatum (B) from 16–20 week old R6/1 transgenic mice showing the effect of homozygosity at the Msh3 locus on the pattern of expansion in the reciprocal congenic mice. Regardless of genetic background, CBy homozygosity at the congenic locus results in loss of somatic expansion, while B6 homozygosity is permissive of somatic expansion. We also assessed CAG repeat instability in testes and sperm from 12-week old and 24-week old mice (Figure S2). The CAG repeats were relatively stable in the germline of both mouse lines, regardless of age, consistent with the relatively low levels of transmitted mutations observed in our colony and consistent with previous reports of R6/1 mice [10], [13]. A few changes of a single repeat unit were observed in the testes of 24-week old B6 and a similar range was observed in the SP-PCR analysis of sperm DNA. These small changes were not obviously observed in the germline of CBy mice (Figure S2). However, the R6/1 transgenic mice from which the CBy.Cg-R6.1 line was derived initially had ∼115 CAG repeats which decreased to ∼95 repeats over the course of ∼12 years of transmissions (not shown). This observation is consistent with a tendency for CAG contractions to occur in the presence of reduced levels of MMR proteins [16], [26], [27], [34]–[36], [49]. Typically, the R6/1 line gives rise to occasional expansions of 1–2 repeat units/transmission and rarer large contractions [64].
MSH3, but not MSH2 or MSH6, protein levels are Msh3 gene variant-dependent
To test the possibility that Msh3 polymorphisms may affect the expression of MMR proteins, which subsequently lead to variable levels of CAG instability between mouse strains, we assessed MMR protein levels in mouse tissues by Western blotting [38], [42], [45]. In liver, the levels of MSH2 and MSH6 were similar between all mouse strains (Figure 3A). However, the level of MSH3 protein varied widely between mice, with high expression in mice carrying the B6 Msh3 gene, and undetectable levels in mice carrying the CBy Msh3 gene (Figure 3A). An intermediate level of MSH3 was reproducibly observed in mice heterozygous for the B6 and CBy Msh3 genes, on both B6 and CBy genetic backgrounds, thus indicating a gene dosage effect between Msh3 variant alleles. This pattern did not vary with age (Figure 3A; compare 4 weeks with 16 weeks). The same MSH3 expression patterns were observed using a MSH3-specific antibody alone (Figure 3B, right panel). The striatum displayed the same strain-specific MSH3 expression pattern, where mice homozygous for the B6 Msh3 gene showed the highest levels of MSH3 protein, while mice homozygous for the CBy Msh3 gene expressed the lowest level, and mice heterozygous for the Msh3 allele displayed intermediate MSH3 protein expression (Figure 3B, right panel). It is notable that MSH3 levels varied in a manner that depended on the Msh3 variant and was independent of mouse strain background. The spleen, thymus, cortex and cerebellum also showed a similar Msh3 gene variant-specific pattern of MSH3 protein expression (Figure S3). Towards ensuring that the apparent expression variations were not due to differential ability of the antibody to recognize its epitope, we analyzed MSH3 protein expression using an independent monoclonal MSH3 antibody (5A5, which recognizes an epitope within exon 4 compared to 2F11 which recognizes an epitope in exon 1, neither of which have amino acid differences between B6 and CBy mice), as described by [65]. We observed the same expression patterns, suggesting that the MSH3 levels observed in tissues are independent of the binding site of the antibody on MSH3 (Figure S4). Thus, regardless of genetic background, the level of MSH3 protein expression depended upon whether the mouse carried the B6 Msh3 variant (high) or the CBy Msh3 variant (low).
Fig. 3. Western blot analysis of MMR and DHFR protein levels. 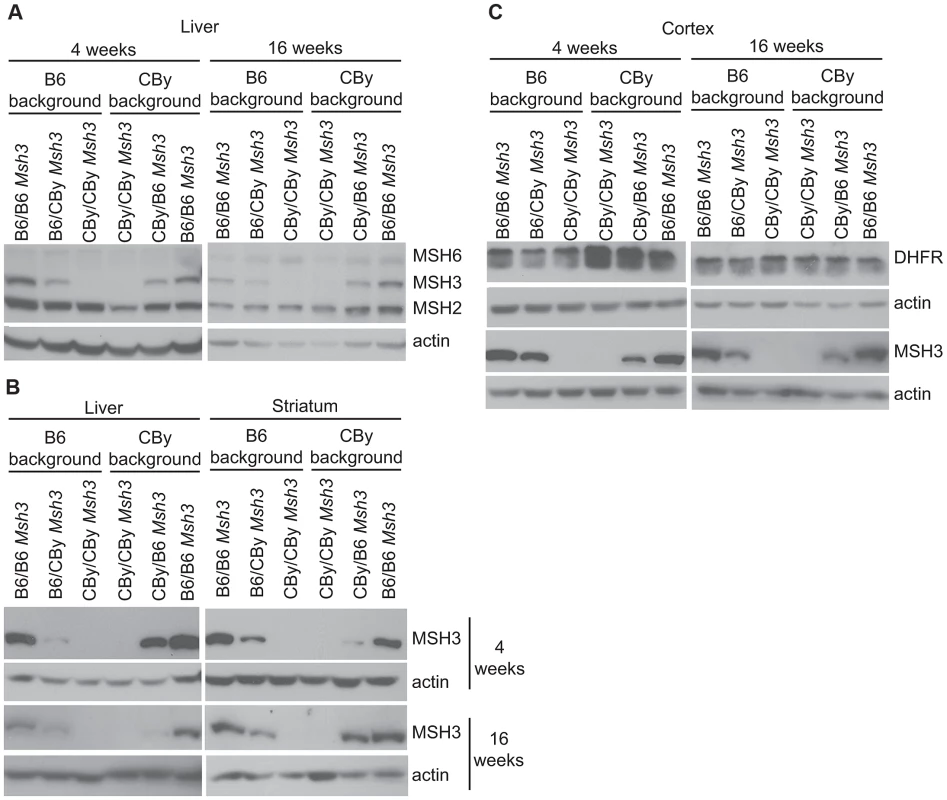
MMR expression in liver and striatum from 4 and 16 week-old mouse. Actin was used as a loading control. MSH2: 104 kD, MSH6: 160 kD, MSH3: 127 kD (Ab = 2F11) and actin: 42 kD. DHFR expression in cortex from 4 and 16 week-old mice DHFR: 21 kD. A) Simultaneous Western blot using MSH2-, MSH3-, MSH6- and actin-specific antibodies in liver. For antibody dilutions see Materials and methods. B) Western blot using only anti-MSH3 (Ab = 2F11) and actin antibodies in liver and striatum. C) Western blot analysis of DHFR in cortex from 4 and 16 week-old mice. DHFR expression in Msh3 locus reciprocal congenic mice
The Msh3 and dihydrofolate reductase (Dhfr) genes are arranged in a head-to-head orientation and share a common promoter that divergently drives transcription [66]–[68]. The levels of both transcripts are produced at similar levels in various mouse tissues [67], [68]. We analyzed DHFR expression (Figure 3C) from R6/1 congenic and Msh3 locus reciprocal congenic mice carrying either homozygous B6 Msh3 variants, or CBy Msh3 variants, or B6/CBy variants. DHFR protein levels did not vary between mouse strains, unlike the MSH3 protein (Figure 3C). These results suggest that the variation of MSH3 protein levels between the B6 - and CBy-Msh3 gene variants are not regulated by promoter, which is identical between variants (Figure 1C), but by the coding variations of the Msh3 gene.
MSH3 expression in different mouse strains
The higher levels of MSH3 in the B6 variant may be due to stabilizing amino acid sequences or alternatively, the lower levels of MSH3 in the CBy variant may be due to destabilizing amino acid sequences. Since the levels of MSH2 were consistent between the congenic and reciprocal congenics we presume that the contribution of MSH2 variants upon MSH3 levels is less than that of MSH3. Towards identifying Msh3 gene polymorphisms that may affect MSH3 protein levels, we sequenced the Msh3 gene from 12 other inbred mouse lines for promoter and exon 2, 3, 7, 8 and 10 variations (A, AKR, C3H, CBA, FVB, DBA/2, 129P2, 129S1, 129S2, 129S6, 129T2, & 129X1). These mouse lines contained variant amino acids similar to either CBy or B6 (Figure 4A, Table S2). We next assessed the MMR protein levels in various strains that harboured the B6 and CBy Msh3 gene coding polymorphisms (Figure 4B). MSH3 expression varied between strains. MSH3 was barely detectable in CBy and was the highest in B6 and C3H/HEJ (Figure 4B). These MSH3 levels are similar to the lower and higher levels observed in our reciprocal congenic mice with the CBy - and B6-Msh3 alleles, respectively. MSH3 is highest in B6 and C3H/HEJ mice, which share alleles in exon 3, exon 7 and exon 10 suggesting that these may contribute positively to MSH3 levels. MSH3 levels were intermediate in DBA/2J, CBA/J and 129/S1 (Figure 4B), and these all share the B6 variants at exon 10, which provides additional support for a stabilizing association of exon 10. This is further supported by the higher MSH3 expression in DBA/2 than CBy since DBA/2 differs from CBy by two polymorphisms in exon 10 (Figure 4A). Our results indicate that polymorphisms within exon 3, exon 7 and exon 10 may modulate the level of MSH3 protein in mouse tissue.
Fig. 4. MSH3 coding polymorphisms and protein expression in different mouse strains. 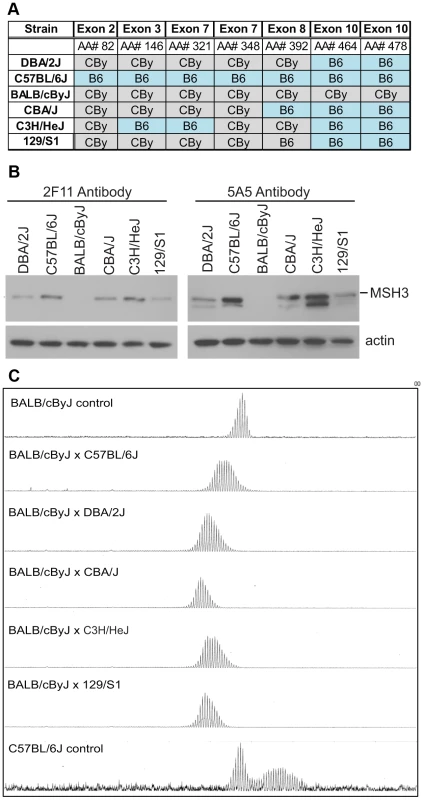
A) Msh3 polymorphisms in Msh3 gene from C57BL/6 (B6) and BALB/cBy (CBy) mice. Promoters were identical. SNPs were identified or confirmed to those in dbSNP by sequencing the Msh3 gene. In DBA/2J, exon 8, AA#392 was correctly identified to be T/Valine. For a given amino acid the same codon was used for the variants. The complete set of MSH3 protein polymorphisms in 14 mouse strains is in Table S2. B) MSH3 expression in spleen extract from different background using two different MSH3 antibodies [65]. The faster migrating band for 5A5 was a non-specific cross-reacting product, as described for 5A5 but not 2F11 [65]. All other figures in this study used 2F11. C) Typical GeneScan traces for sizing of the CAG repeat as outlined in Figure 1B. Representative CAG repeat distributions from liver of F1 progeny between CBy and other inbred strains of mice. The top, bottom and second panel show the controls CBy (stable), B6 (unstable), and CBy X B6 (intermediate) CAG profiles, respectively. Note: Western blot data comes from inbred mice. The higher levels of MSH3 in C3H and B6 are halved in the cross to CBy. In further support for the CAG repeat-stabilizing effect of the CBy Msh3 variant, we crossed the CBy.Cg-R6/1 mice to the above noted 12 strains of mice including B6, which contained different Msh3 gene variants (Figure 4A, Table S2). All F1 mice regained an intermediate level of CAG instability in their liver and/or striatum, which is consistent with this set of Msh3 variants being the source of altered CAG/CTG instability (Figure 4C). Notably, in 3 independent crosses of CBy.Cg-R6/1 to C3H/HeJ, which showed the highest expression of MSH3, all F1 mouse livers showed the same pattern of CAG instability with a broad distribution of expanded alleles extending to as many as +12 repeats. This dosage effect is consistent with a dominant effect of MSH3 levels upon CAG instability. Further support for a MSH3 dosage effect is the near complete absence of MSH3 protein in either tail or heart of either CBy or B6 mice with the exception of tail tissue of 4 week-old mice. This expression profile correlates with the relatively stable repeat tract observed in these tissues, regardless of mouse strain (Figure S5, see also Figure 1). These findings are consistent with a direct association of tissue-specific MSH3 levels with levels of tissue-specific CAG stability.
T321I MSH3 variant is highly conserved and may destabilize MSH3 protein
In order to uncover potential amino acid changes, which could be contributing to loss of MSH3 protein expression in the CBy variants, we have examined both sequence and structural features of MSH3 homologs. Sequence alignment has revealed that most of the B6-CBy variants are well conserved, but occur where amino acid changes are not predicted to have physiochemical consequences, or occur within poorly conserved regions, suggesting those regions minimally contribute to structure/function of the protein (Figure 5A). One exception is the T321I variant, which is conserved in 16/17 of the mammalian homologs and yeast. Further, in this one exception (in both giant panda and yeast), the Threonine is replaced by physiochemically-similar Serine, so that a hydroxyl group at this position is observed to be highly conserved (Figure 5A and Figure S6). Importantly, the T321 variant occurs within a Type I β-Turn (Figure 5B), where Isoleucine is extremely unfavoured (Figure 5B) [69]. Despite the large evolutionary distance, a Type I β-Turn also occurs in E. coli MutS (Figure 5B) [70], suggesting the importance of this region to overall function. β-Turns are thought to be crucial to the protein folding process [71], [72], where they may direct nucleation of secondary structure elements towards hydrophobic collapse [73]. The change of Threonine to disfavoured Isoleucine at the ‘i+2’ site within the turn of MSH3 may disrupt the β-Turn, representing a significant barrier to protein folding, potentially leading to proteolysis. The full effect of the T321I change upon MSH3 protein stability may require some of the other amino acid changes, which will require experimental assessment.
Fig. 5. Structural and sequence analysis of MSH3. 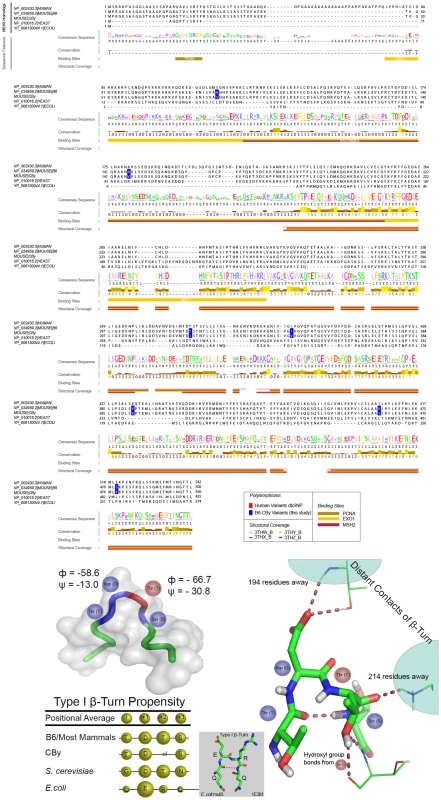
A: Multiple sequence alignment of MSH3. Jalview created visualization [120] using the first 500 amino acids of the mouse B6 MSH3 (NP_034959.2). Conservation values and consensus sequence are based on alignment of S. cerevisiae Msh3p, E. coli MutS and 17 mammalian MSH3 homologs; values range from 0–9, where 0 is lowest and 9 is the highest. Protein interacting domains indicated pertain to those regions of the human MSH3 protein. This panel only shows an abbreviated set of the species of MSH3 sequence, the full set analysed is shown in Figure S5. B: MSH3 variant within β-turn. The T321I variant occurs within a Type I β-turn, as determined by specific backbone turn angles [117], [118] from the human MSH3 structure (3THW_B). Top left: hMSH3 tube diagram of Cα atoms of β-turn (blue), i+2 (T) residue (red) and additional three residues on N- and C-terminal ends (green). Bottom left table shows the β-turn propensity is relatively strong throughout MutS/MSH3 homologs, while the CBy variant (Isoleucine at i+2 position) is extremely disfavored (table bottom left) [69]. Right: Ball and stick diagram of contact sites of Asp (D) and Thr (T) residues in β-turn with residues 194 and 214 respectively. Line diagram of Thr (T) hydroxyl group contact with neighbouring Threonine residue at position 365. The absence of the Threonine hydroxyl group may be important to stabilizing the β-turn itself, and/or may change the conformation of the turn, potentially disrupting distant contacts important for proper protein folding. MSH3 visualizations created using PyMol (PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC). Discussion
Trinucleotide repeat instability is governed by cis-elements and trans-acting modifiers. Repeat length, sequence of repeat, purity of the repeat, genomic context and DNA metabolizing proteins can contribute to patterns of repeat instability in mouse models of trinucleotide repeat diseases such as DM1, HD and SCA7 [19]. Since the R6/1 transgene is common to each of the mouse lines described herein, the variable levels of CAG instability between strains are unlikely to be the result of a cis-element, and most likely result from the different Msh3 gene variants. Further support for a trans-factor as the source of the variable CAG stability is that B6, FVB, and 129 mouse strains did not influence HTT mRNA levels for either knock-in or YAC HD mice [59] thereby arguing against a role for transcription as an in cis source for the inter-strain variations of instability. To date there has been no report of a naturally occurring mouse strain-specific factor that modifies repeat instability.
In HD and DM1 mice, engineered null alleles of Msh2 and Msh3 were identified as the strongest modifiers of trinucleotide repeat instability suggesting an important role of MutSβ in trinucleotide repeat instability [9], [37]. MMR deficiencies stabilized CAG/CTG repeat tracts from spontaneous expansions in two different kinds of HD mice [13], [27], [36], [46] and three different DM1 mouse models [16], [26], [28], [34], [35], [74]. These results indicate that the effects of MMR proteins on CAG/CTG instability are independent of cis-elements and sequence context. We observed two distinct patterns of somatic CAG instability in two different M.musculus backgrounds, CBy and B6 and sequenced the Msh3 gene in those strains and found seven polymorphisms in exons 2, 3, 7, 8 and 10, which differ between the strains. Thus, the differences in CAG/CTG instability between the two strains may be modulated by these Msh3 polymorphisms. By generating reciprocal congenic mice for the Msh3 gene, we demonstrated that CAG/CTG repeat instability appears to be modulated by Msh3 variants, where expansion levels are the highest in liver and striatum of mice homozygous for the B6 Msh3 gene. Mice homozygous for the CBy Msh3 gene show an absence of CAG instability. We also showed that MSH3 protein expression depends upon the Msh3 gene variant, independent of genetic background outside the Msh3 locus: The B6 MSH3 protein variant was expressed at high levels, whereas the CBy MSH3 variant was expressed at nearly undetectable levels. The protein expression patterns of MSH3 correlated positively with the level of somatic CAG expansions. The loss of one B6 Msh3 allele in mice heterozygous for both variants was sufficient to decrease CTG/CAG instability; consistent with results which shows that MSH3 protein levels are a limiting factor in CAG/CTG repeat expansions in DM1 and HD mouse models, where MSH3/null mice have less expansions than MSH3/MSH3 mice, but more than null/null mice [16], [27], [35]. Interestingly, the loss of one Msh3 allele (Msh3/null) was more dramatic than the loss of one Msh2 allele [13], [27], [35], suggesting that CAG instability may be exquisitely sensitive to MSH3 levels. In a repair assay, the levels of human MSH3 protein altered the ability to repair slipped-DNAs formed by CAG/CTG repeats [42]. The distinct levels of MSH3 protein between B6 and CBy strains are unlikely to be due to varying transcription levels, as we detected similar levels of the DHFR protein between strains, whose transcript is driven from the same divergently transcribed promoter as the Msh3 gene [66]–[68]. Furthermore, considerable evidence indicates that the levels of MMR transcripts is not always reflective of MMR protein levels [48]. The stability of MSH3 and MSH6 proteins is dependent on the ability of these proteins to form heterodimeric complexes [48]; in mice the genetic absence of Msh2 led to undetectable levels of MSH3 protein [45]. However, the levels of MSH2 protein did not vary between the B6 and CBy strain (Figure 3), and MSH3 protein levels (low or high) persisted in the reciprocal congenic mice; arguing against variations of MSH3 levels by either strain-specific MSH2 expression levels or MSH2 variants.
Polymorphisms in the MSH3 coding region may alter the stability of the MSH3 protein directly or by altering its interaction with MSH2 [47], [48]. In particular, although our homology modeling results did not offer insight into which variants resided in regions critical to overall protein structure nor did the polymorphism reside in known protein-binding domains (Figure 5A and Figure S6), the highly conserved T321I variant occurs within a Type I β-Turn which could be critical for protein folding [71], [72]. Changes in β-Turn sequences modulate protein stability [71], [72], [75], [76], where unfavourable sequence changes can dramatically decrease protein folding rate [77], [78] and in some cases completely ablate protein expression [79]. In addition to potential changes brought by the T321I variant, the CBy strain gains a potential serine phosphosite at amino acid 79 as experimentally determined in the homologous human MSH3 protein (site 116 in hMSH3) [80], which could impact overall protein conformation, its protein-binding capacity and stability [81]. While the actual contribution of any of the MSH3 amino acid variants, alone or coincident with the others, will require experimental support, together our findings support an effect upon protein stability.
Here we have shown that naturally occurring genetic variation in an MMR gene, like engineered genetic deficiencies of MMR genes, can lead to changes in the direction and pattern of CAG/CTG repeat instability. A loss of Msh2 and Msh3 have led to both a loss of expansions and increased CAG/CTG contractions; suggesting that MMR proteins may both drive expansions as well as protect against contractions [16], [26], [27], [34]–[36], [49]. The pattern of CAG instability is also affected by MMR genes – possibly reflected by changes in the number of repeat units involved in a mutagenic event. It is possible that there are two different mechanisms involved in large expansions; the accumulation of many short (single-repeat) length changes per mutagenic event; or salutatory large (many-repeat) jumps per mutagenic event. In vivo evidence suggesting the existence of two distinct mechanisms was the observation of bimodal distribution of repeat length in certain tissues of HD and DM1 mice [13], [27], which is also evident in some patient tissues and may be due to cell lineage-specific instabilities [4], [82]–[84]. This bimodal distribution of repeat lengths was only observed herein with homozygosity for the B6 Msh3 gene (Figure 1 and Figure 2). Recent modeling studies of HD mice suggest the involvement of distinct short and large mutagenic events [25]. Similarly, it was reported that two distinct modes of repeat instability occur at dinucleotide repeats in MMR-defective (hMSH2, hMLH1, hPMS2) deficient tumours of humans but not mice, those with changes of < or = 6 repeats and those with changes of >8 repeats [85]–[87]. In cultured cells of patients suffering a CAG/CTG disease and certain tissues the mutation events appear to be short increments, of 1 to 3 CTG/CAG units per mutation event [88], similar to that occurring at other simple repeats such as (CA)n and (A)n [85], [89], [90]. Interestingly, the bimodal distribution of CAG expansions was lost in mice harbouring a CBy Msh3 gene, which might suggest that MSH3 is involved in larger repeat expansions. However, the requirement of hMutSβ for the repair of short slip-outs of a single repeat unit, but not of longer slip-outs (>3 units), strongly supports the concept that the MSH3-sensitive expansions we observe in the mice are in fact the accumulation of many single-repeat expansion events [42].
An effect of mouse genetic backgrounds on the dynamics of CAG/CTG expansions was suggested in HD and DM1 mouse models, but a candidate for the source for the variation was not suggested [16], [59]. van den Broek et al., (2002) showed the greatest CTG instability when present on the C3H background, while Lloret et al., (2006) observed the highest CAG instability on a B6 background and low levels of instability in the 129Sv background [16], [59]. These observations with independent transgenic mice showing the highest repeat instability in mice with the B6 Msh3 gene (C3H and B6) and lower instability in mice with the CBy Msh3 gene (129) are consistent with our findings that the B6 MSH3 variant is a major driver of CAG expansions, and is also consistent with the high levels of MSH3 protein in B6 and C3H mice. It is unclear why these Msh3 polymorphisms, which appear to affect MSH3 protein levels, exist in the various mouse lineages. We propose that differences in the MSH3 protein between mouse strains may provide a molecular explanation for some of the strain-specific differences observed in somatic CAG instability seen by other labs [16], [59].
DNA polymorphisms in other DNA metabolizing proteins might affect CAG/CTG instability patterns, and such family-specific instability modifiers have been suggested to exist in HD families [61]. Many trans-factors have been considered for their role in CAG/CTG instability, and few have been assessed for the possible contribution of their polymorphic variants. Neither human FEN1 mutants nor its polymorphic variants were linked to CAG instability in HD patients [91]. OGG1 has been reported to play a partial role in CAG instability in R6/1 mice [33]. Huntington's subjects having the Cys326-OGG1 allele were reported to have increased HD CAG tract lengths and significantly earlier disease onset than HD individuals with the Ser326 variant [92]. However, this association was not observed in a study using a larger sample size [93]. Our recent observation that the human mismatch repair protein MLH1 is required to repair short CTG slip-outs and arrested on clustered slip-outs, might suggest that MLH1 is involved in CAG/CTG expansions, and MLH1 variants may have differential effects [94]. Polymorphisms in human MSH2 have been identified in patients with hereditary non-polyposis colorectal cancer that are thought to inactivate the function of the MSH2–MSH3 complex but not the MSH2–MSH6 complex; leading to altered frameshift mutations in yeast [95], [96]. Polymorphic variants of hMSH3 have been significantly linked to cancer and radiation sensitivity [97]–[100]. However, in no case has there been any demonstration of altered genetic variation with a particular hMSH3 variant, nor any direct link of an hMSH3 variant with a mutagenic outcome.
Might polymorphisms of Msh3 affect the instability of other repeat sequences? Mismatch repair proteins act in distinct manners upon the instability/stability of different repeats. A loss of Msh3 can lead to varying levels of changes of single repeat units (predominantly losses) at mono - di-, tri - and tetranucleotide repeat tracts [101] and references therein). The role of mismatch repair proteins in the instability of expanded repeat sequences including the FRDA disease-associated GAA tracts, the murine Ms6-hm (also known as Pc-1) (CAGGG)n and Hm-2 (GGCA)n repeats can vary widely from their effects upon CAG/CTG repeats [102]–[106]. Together these findings support the contention that the role of MMR can vary dramatically across different repeat sequences. Thus, the effect MMR gene polymorphisms on different repeat sequences will need to be determined for each sequence. However, since the Msh3 variants appeared to have similar effects upon CAG/CTG instability in various transgenic contexts, the effect of MMR gene polymorphisms may be similar for each of the 14 different CAG/CTG disease loci including HD, DM1, SCA7, and others.
Our data provide the first evidence that Msh3 polymorphic variants associate with levels of CAG/CTG trinucleotide instability in HD mice. This discovery may lead to the identification of human polymorphic variants that could explain the extreme variability of CAG/CTG instability observed in HD and DM1 patients. Since somatic repeat expansions through an individual's life may contribute to disease severity and progression, factors that affect this could have clinical relevance [1], [7], [8], [9]. Unknown genetic factors modify the onset and severity of disease in HD families and HD mice [60], [107]–[109]. Un-explored variations in the levels of somatic CAG instability between HD families or individuals may be the source for these clinical variations. Polymorphic variants in DNA repair genes that lead to enhanced somatic CAG/CTG expansions could ultimately lead to increased disease progression and severity. Similarly, variants that lead to reduced somatic expansions could be less deleterious. Identification of such variants in individuals affected with any one of the 14 CAG/CTG diseases may have prognostic implications. Furthermore, attempts to modulate MMR to modulate CAG/CTG-repeat associated diseases [9] would be wise to consider any particular variants of MMR proteins that may differentially affect instability levels.
Materials and Methods
Mouse breeding
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the Wadsworth Center (Public Health Service Animal Welfare Assurance Number A3183-01).
Creation of R6/1 congenic lines of mice (CBy.Cg-R6/1 and B6.CBy-R6/1): male B6CBA-Tg(HDexon1)61Gpb mice originally purchased from Jackson Labs (Bar Harbor, ME), were bred to recipient strain CBy and B6 females to create F1 progeny. Male R6/1 transgenic F1 progeny were then backcrossed as appropriate to CBy or B6 females. Male R6/1 progeny from this cross (N2 generation) were selected for backcross to the appropriate recipient strain until 10 backcrosses had occurred (N10 generation). All R6/1 mice were genotyped from tail tip biopsy taken at weaning, using primers 1594 (CCGCTCAGGTTCTGCTTTTA) and 1596 (TGGAAGGACTTGAGGGACTC). PCR conditions were initial denaturation for 2 min at 95°C, followed by 30 cycles (94°C - 30 sec., 54°C - 45 sec., 72°C – 1 min), followed by 10 min at 72°C. Qiagen Taq polymerase (cat #201225) was used as recommended by manufacturer.
Creation of Msh3 reciprocal congenic lines of mice (CBy.B6-msh3 and B6.CBy-msh3): similar to creation of the R6/1 congenic mice, B6 and CBy inbred mice were intercrossed to produce F1 progeny. F1 progeny were backcrossed to recipient B6 and CBy inbred lines until attainment of the N10 generation. Starting with N2 progeny, mice were genotyped with markers that flanked the Msh3 gene. D13Mit159 (Forward: CCCATTGTCCCTGTTCAGAT, Reverse: AAACCCACCATGAATTAAATGC, position: Chr13 : 92953376–92953513 bp) and D13Mit147 (Forward: CATCCAGGAAGGCAATAAGG, Reverse: CAAATGCACAGTGCCGAG, position: Chr13 : 98359080–98359187 bp), were used for genotyping the locus. The Msh3 gene is located at Chr13 : 93121206–93121391 bp. Animals heterozygous for both markers at each generation were selected as breeders. Qiagen Taq polymerase (cat #201225) was used as recommended by the manufacturer.
Creation of double congenic lines: CBy.B6-msh3B6/B6 females were crossed to CBy.Cg-R6/1 males, and progeny were genotyped at both D13Mit markers and for the R6/1 transgene. Females heterozygous at the Msh3 locus and males heterozygous at the Msh3 locus and carrying the R6/1 transgene were selected for mating. Female progeny of this mating typing either homozygous B6 or CBy at the Msh3 locus were mated with a male B6 or CBy Msh3 homozygotes who also carried the R6/1 transgene to establish CBy.B6-msh3B6/B6 R6/1 and to derive a control CBy.Cg-R6/1 (homozygous CBy at the Msh3 locus) lines. The same procedure was used to create B6.CBy-msh3CBy/CBy R6/1 and control B6.Cg-R6/1 lines.
Genome-wide SNP genotyping
The efficacy of our congenic and reciprocal congenic mice was assessed by SNP genotyping with a medium-density SNP array (Mouse MD Linkage panel #GT-18–131, Illumina, San Diego, CA) on DNA samples isolated from mouse tail clips using the GoldenGate Genotyping Assay according to the manufacturer's protocol. This allowed us to precisely map the recombination boundaries and test for contaminating regions. The microarray detects 1449 loci where 796 are informative between C57BL/6 and BALB/cBy, excluding those on the X chromosome. These loci span the entire mouse genome with approximately three SNPs per 5 Mb intervals. Briefly, 250 ng of DNA (5 uL at 50 ng/uL) was hybridized to locus-specific oligonucleotides, extended, ligated and amplified before hybridizing to universal 1,536-plex 12-sample BeadChip microarrays. The arrays were then scanned with default settings using the Illumina iScan. Analysis and intra-chip normalization of resulting image files was performed using Illumina's GenomeStudio Genotyping Module software v.2011 with default parameters. Genotype calls were generated by clustering project samples with a manual review of each SNP plot. The identified contaminating SNPs were visualized by ideogram using the Ideographica web-based software [110].
CAG repeat analysis
Genescan Analysis: purified gDNA from tissue and 10 ng was amplified using primers HDSizeF (6FAM-ATGAAGGCCTTCGAGTCCCTCAAGTCCTTC) and HDSizeR (CGGCGGTGGCGGCTGTTG). PCR conditions were initial denaturation for 3 minutes at 94°C, followed by 30 cycles (94°C (30 s.), 58°C (1 min.), 72°C (1 min), followed by 10 minutes at 72°C. Qiagen Taq polymerase (cat #201225) was used as recommended by manufacturer. Products were processed in the Applied Genomics Technologies Core at the Wadsworth Center on an ABI3730, and analysed using PeakScanner Software (Applied Biosystems).
Small pool-PCR: DNA from testis was extracted by phenol-chloroform and sperm was extracted as described [26],[34],[74]. SP-PCR was performed as described [111]. DNA samples were digested with HindIII and SP-PCR was performed with MS-1F (GCCCAGAGCCCCATTCATT) and MS-1R (GGCTACGGCGGGGATGGCGG) primers. The DNA was denatured by heating to 94°C (5 min.) and amplified through 30 cycles of 94°C (1 min.), 62°C (1 min.) and 72°C (1 min.) with a chase of 10 minutes at 72°C. The products of the PCR were resolved by electrophoresis on 40 cm long 1.5% agarose gels in 0.5× TBE at 180 V for 18 hours. The products were then transferred to nylon membrane by Southern blotting and detected by hybridization using a radiolabelled CAG repeat containing probe.
To assess degrees of instability we used the following criteria: The presence of instability was evidence by multiple PCR products with varying lengths of repeats. The degree of instability between different tissues was assessed based upon the size range and the relative amount of the expanded product was different from the major sized product in the stable tissues (presumed as the progenitor allele; most studies indicate the tail as representative of the progenitor allele). The degree of instability for the same tissue between different mouse lineages was assessed in a similar manner. The above were done on an age-matched basis. Relative between age-matched and tissue matched mice, an assessment of the size range and the intensity of the fragments as previously outlined [13], [17], [18], [29], [47], [111].
LOD score calculation
LOD score was calculated using numbers of mice from the F1 intercross = 81. The F1 parents were all by definition heterozygous, therefore the 81 mice derive from 2 * 81 = 162 informative meiosis. Assuming the phenotype is mediated by two linked semi-dominant alleles, then there are no recombinants observed (i.e., all the mice have the phenotype expected consistent with their genotype). Thus the number of recombinants = 0 and the number of non-recombinants = 162. The odds of getting this outcome assuming linkage and 0% recombination (θ = 0%) = 1. The odds of getting this outcome if the loci were not linked = 0.5162 = 1.7×10–49. The LOD score is then calculated as the log of the odds of observing this pattern assuming no linkage/assuming linkage = log (1/1.7×10–49) = 48.8 (θ = 0)
Protein sample preparation and Western blotting
Tissues were collected from 4 and 16-week-old-mice with different Msh3 genotypes. Protein extractions and Western blotting were performed as described in Tomé et al., (2013) [45]. Briefly, proteins were extracted by mechanical homogenisation in lysis buffer (0.125 M, Tris-HCl (pH 6.8), 4% SDS, 10% glycerol) containing protease inhibitor cocktail (Roche, complete Mini 7× cat. no. 04 693 124 001). Protein concentration was determined using the Pierce BCA protein assay kit (cat. no. 23225). Proteins (40 µg) were denatured for 5 minutes at 95°C in loading buffer and 10% β-mercaptoethanol added extemporaneously, resolved by electrophoresis on an 8% (MMR proteins and actin expression) and 12% (DHFR expression) SDS-PAGE. Membranes were blocked for one hour at room temperature in 5% (m/v) dried milk for MMR antibodies incubation and 10% (m/v) dried milk for DHFR incubation in TBST pH 7.5, then incubated overnight at 4°C in antibodies anti-MSH2 (Ab-2) mouse mAb (FE11) (Calbiochem, Ab-2; cat. no. NA27, 1∶200), mouse anti-MSH6 (BD Laboratories, cat. no. 610918, 1∶200), monoclonal mouse anti-MSH3 (2F11 and 5A5 clones, gifts from Glen Morris and Ian Holt, at 1∶750) [65], Rabbit anti-DHFR [112] (gift from Joseph R. Bertino, 1∶500) and mouse anti-Actin Ab-5 (BD laboratories, cat.no. 612656, 1∶500). The anti-MSH3 antibodies 2F11 and 5A5 recognize epitopes in exons 1 and 4, respectively [65]. The membranes were incubated for 1 h in mouse secondary antibody (sheep anti-mouse-HRP, cat. no. 515-035-062, 1∶5,000) at room temperature for MSH2, MSH3, MSH6 and for actin and Rabbit secondary antibody (Abcam, Rabbit polyclonal to goat IgG H and L, HRP, cat. no. ab6741) for DHFR. Antibody binding was visualized using ECL plus Western blotting detection system (Amersham, cat. no. RPN2132).
Spleen extracts were prepared from fresh spleens placed in 3 ml cold PBS with 2% FBS to rinse away excess erythrocytes. Spleens were passed through nylon mesh filter (BD Falcon cell strainer, 40 mm, REF352340) containing fresh cold PBS with 2% FBS and 2 mM EDTA on ice, then centrifuged at 1200 rpm at 4°C for 5 minutes, and processed as outlined [42].
Sequence alignment
Mammalian homologs of MSH3 were obtained using 5-iteration PSI-BLAST [113] with E-value set to 1e-05 against mouse MSH3 (NP_304959.2). Results were filtered to exclude MSH6 proteins and partial or low quality proteins, leaving 17 mammalian MSH3 homologs (including mouse). Mammalian sequences were chosen to provide consistent dataset where structural features of MSH3 are likely conserved. Saccharomyces cerevisiae 288c Msh3p, Escherichia coli str. K-12 substr. MG1655 MutS sequences and the 17 mammalian homologs were aligned using MAFFT with default settings [114]. Mammalian homologs were aligned using MAFFT [114].
Structure modeling
The hMHS3 structure 3THW_B from the Protein Data Bank (PDB) [115], offered the greatest coverage of the MSH3 sequence and has an extremely high percent identity to mouse MSH3 (87.1%), strongly suggesting similar structures for both mouse and human MSH3. Efficient side-chain packing of 3THW_B was achieved using SCWLR4 software [116] and the DSSP program [117] was used to assign secondary structure and phi/psi bond angles. β-Turn type was determined based on [118] and confirmed using PROMOTIF [69], [119]. 3THW_B structures were visualized using PyMol. Protein bonds were assigned with PyMol and distant contacts confirmed using an in-house Python script.
Supporting Information
Zdroje
1. LeeJM, RamosEM, LeeJH, GillisT, MysoreJS, et al. (2012) CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology 78 : 690–695.
2. RosenblattA, KumarBV, MoA, WelshCS, MargolisRL, et al. (2012) Age, CAG repeat length, and clinical progression in Huntington's disease. Mov Disord 27 : 272–276.
3. TeleniusH, KremerB, GoldbergYP, TheilmannJ, AndrewSE, et al. (1994) Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet 6 : 409–414.
4. De RooijKE, De Koning GansPA, RoosRA, Van OmmenGJ, Den DunnenJT (1995) Somatic expansion of the (CAG)n repeat in Huntington disease brains. Hum Genet 95 : 270–274.
5. KennedyL, EvansE, ChenCM, CravenL, DetloffPJ, et al. (2003) Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet 12 : 3359–3367.
6. ShelbournePF, Keller-McGandyC, BiWL, YoonSR, DubeauL, et al. (2007) Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum Mol Genet 16 : 1133–1142.
7. SwamiM, HendricksAE, GillisT, MassoodT, MysoreJ, et al. (2009) Somatic expansion of the Huntington's disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum Mol Genet 18 : 3039–3047.
8. MoralesF, CoutoJM, HighamCF, HoggG, CuencaP, et al. (2012) Somatic instability of the expanded CTG triplet repeat in myotonic dystrophy type 1 is a heritable quantitative trait and modifier of disease severity. Hum Mol Genet 21 : 3558–3567.
9. Lopez CastelA, ClearyJD, PearsonCE (2010) Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol 11 : 165–170.
10. MangiariniL, SathasivamK, MahalA, MottR, SellerM, et al. (1997) Instability of highly expanded CAG repeats in mice transgenic for the Huntington's disease mutation. Nat Genet 15 : 197–200.
11. GourdonG, RadvanyiF, LiaAS, DurosC, BlancheM, et al. (1997) Moderate intergenerational and somatic instability of a 55-CTG repeat in transgenic mice. Nat Genet 15 : 190–192.
12. MoncktonDG, CoolbaughMI, AshizawaKT, SicilianoMJ, CaskeyCT (1997) Hypermutable myotonic dystrophy CTG repeats in transgenic mice. Nat Genet 15 : 193–196.
13. ManleyK, ShirleyTL, FlahertyL, MesserA (1999) Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet 23 : 471–473.
14. ShelbournePF, KilleenN, HevnerRF, JohnstonHM, TecottL, et al. (1999) A Huntington's disease CAG expansion at the murine Hdh locus is unstable and associated with behavioural abnormalities in mice. Hum Mol Genet 8 : 763–774.
15. WheelerVC, AuerbachW, WhiteJK, SrinidhiJ, AuerbachA, et al. (1999) Length-dependent gametic CAG repeat instability in the Huntington's disease knock-in mouse. Hum Mol Genet 8 : 115–122.
16. van Den BroekWJ, NelenMR, WansinkDG, CoerwinkelMM, te RieleH, et al. (2002) Somatic expansion behaviour of the (CTG)(n) repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet 11 : 191–198.
17. LibbyRT, MoncktonDG, FuYH, MartinezRA, McAbneyJP, et al. (2003) Genomic context drives SCA7 CAG repeat instability, while expressed SCA7 cDNAs are intergenerationally and somatically stable in transgenic mice. Hum Mol Genet 12 : 41–50.
18. LibbyRT, HagermanKA, PinedaVV, LauR, ChoDH, et al. (2008) CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet 4: e1000257 doi:10.1371/journal.pgen.1000257.
19. PearsonCE, Nichol EdamuraK, ClearyJD (2005) Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 6 : 729–742.
20. WarbySC, MontpetitA, HaydenAR, CarrollJB, ButlandSL, et al. (2009) CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am J Hum Genet 84 : 351–366.
21. BrockGJ, AndersonNH, MoncktonDG (1999) Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: associations with flanking GC content and proximity to CpG islands. Hum Mol Genet 8 : 1061–1067.
22. NestorCE, MoncktonDG (2011) Correlation of inter-locus polyglutamine toxicity with CAG•CTG triplet repeat expandability and flanking genomic DNA GC content. PLoS ONE 6: e28260 doi:10.1371/journal.pone.0028260.
23. ClearyJD, PearsonCE (2003) The contribution of cis-elements to disease-associated repeat instability: Clinical and experimental evidence. Cytogenetics and Genome Research 100 : 25–55.
24. van den BroekWJ, NelenMR, van der HeijdenGW, WansinkDG, WieringaB (2006) Fen1 does not control somatic hypermutability of the (CTG)(n)•(CAG)(n) repeat in a knock-in mouse model for DM1. FEBS Lett 580 : 5208–5214.
25. MollersenL, RoweAD, LarsenE, RognesT, KlunglandA (2010) Continuous and periodic expansion of CAG repeats in Huntington's disease R6/1 mice. PLoS Genet 6: e1001242 doi:10.1371/journal.pgen.1001242.
26. SavouretC, BrissonE, EssersJ, KanaarR, PastinkA, et al. (2003) CTG repeat instability and size variation timing in DNA repair-deficient mice. Embo J 22 : 2264–2273.
27. DragilevaE, HendricksA, TeedA, GillisT, LopezET, et al. (2009) Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiol Dis 33 : 37–47.
28. Gomes-PereiraM, FortuneMT, IngramL, McAbneyJP, MoncktonDG (2004) Pms2 is a genetic enhancer of trinucleotide CAG.CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum Mol Genet 13 : 1815–1825.
29. TomeS, PanigrahiGB, Lopez CastelA, FoiryL, MeltonDW, et al. (2011) Maternal germline-specific effect of DNA ligase I on CTG/CAG instability. Hum Mol Genet 20 : 2131–2143.
30. HubertLJr, LinY, DionV, WilsonJH (2011) Xpa deficiency reduces CAG trinucleotide repeat instability in neuronal tissues in a mouse model of SCA1. Hum Mol Genet 20 : 4822–4830.
31. MollersenL, RoweAD, IlluzziJL, HildrestrandGA, GerholdKJ, et al. (2012) Neil1 is a genetic modifier of somatic and germline CAG trinucleotide repeat instability in R6/1 mice. Hum Mol Genet
32. KovtunIV, JohnsonKO, McMurrayCT (2011) Cockayne syndrome B protein antagonizes OGG1 in modulating CAG repeat length in vivo. Aging (Albany NY) 3 : 509–514.
33. KovtunIV, LiuY, BjorasM, KlunglandA, WilsonSH, et al. (2007) OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 447 : 447–452.
34. SavouretC, Garcia-CordierC, MegretJ, te RieleH, JunienC, et al. (2004) MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol Cell Biol 24 : 629–637.
35. FoiryL, DongL, SavouretC, HubertL, te RieleH, et al. (2006) Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum Genet 119 : 520–526.
36. WheelerVC, LebelLA, VrbanacV, TeedA, Te RieleH, et al. (2003) Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum Mol Genet 12 : 273–281.
37. SleanMM, PanigrahiGB, RanumLP, PearsonCE (2008) Mutagenic roles of DNA “repair” proteins in antibody diversity and disease-associated trinucleotide repeat instability. DNA Repair (Amst) 7 : 1135–1154.
38. SeriolaA, SpitsC, SimardJP, HilvenP, HaentjensP, et al. (2011) Huntington's and myotonic dystrophy hESCs: down-regulated trinucleotide repeat instability and mismatch repair machinery expression upon differentiation. Hum Mol Genet 20 : 176–185.
39. JiricnyJ (2006) The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol 7 : 335–346.
40. GenschelJ, LittmanSJ, DrummondJT, ModrichP (1998) Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem 273 : 19895–19901.
41. LittmanSJ, FangWH, ModrichP (1999) Repair of large insertion/deletion heterologies in human nuclear extracts is directed by a 5′ single-strand break and is independent of the mismatch repair system. J Biol Chem 274 : 7474–7481.
42. PanigrahiGB, SleanMM, SimardJP, GileadiO, PearsonCE (2010) Isolated short CTG/CAG DNA slip-outs are repaired efficiently by hMutSbeta, but clustered slip-outs are poorly repaired. Proc Natl Acad Sci U S A 107 : 12593–12598.
43. HarringtonJM, KolodnerRD (2007) Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base-base mispairs. Mol Cell Biol 27 : 6546–6554.
44. TianL, GuL, LiGM (2009) Distinct nucleotide binding/hydrolysis properties and molar ratio of MutSalpha and MutSbeta determine their differential mismatch binding activities. J Biol Chem 284 : 11557–11562.
45. TomeS, SimardJP, SleanMM, HoltI, MorrisGE, et al. (2013) Tissue-specific mismatch repair protein expression: MSH3 is higher than MSH6 in multiple mouse tissues. DNA Repair (Amst) 12 : 46–52.
46. ManleyK, PughJ, MesserA (1999) Instability of the CAG repeat in immortalized fibroblast cell cultures from Huntington's disease transgenic mice. Brain Res 835 : 74–79.
47. TomeS, HoltI, EdelmannW, MorrisGE, MunnichA, et al. (2009) MSH2 ATPase domain mutation affects CTG•CAG repeat instability in transgenic mice. PLoS Genet 5: e1000482 doi:10.1371/journal.pgen.100048.
48. ChangDK, RicciardielloL, GoelA, ChangCL, BolandCR (2000) Steady-state regulation of the human DNA mismatch repair system. J Biol Chem 275 : 29178.
49. KovtunIV, ThornhillAR, McMurrayCT (2004) Somatic deletion events occur during early embryonic development and modify the extent of CAG expansion in subsequent generations. Hum Mol Genet 13 : 3057–3068.
50. PearsonCE, EwelA, AcharyaS, FishelRA, SindenRR (1997) Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum Mol Genet 6 : 1117–1123.
51. ClearyJD, TomeS, Lopez CastelA, PanigrahiGB, FoiryL, et al. (2010) Tissue - and age-specific DNA replication patterns at the CTG/CAG-expanded human myotonic dystrophy type 1 locus. Nat Struct Mol Biol 17 : 1079–1087.
52. LinY, DionV, WilsonJH (2006) Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol 13 : 179–180.
53. Lopez CastelA, TomkinsonAE, PearsonCE (2009) CTG/CAG repeat instability is modulated by the levels of human DNA ligase i and its interaction with proliferating cell nuclear antigen: a distinction between replication and slipped-DNA repair. J Biol Chem 284 : 26631–26645.
54. GoulaAV, BerquistBR, WilsonDM3rd, WheelerVC, TrottierY, et al. (2009) Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum In Huntington's disease transgenic mice. PLoS Genet 5: e1000749 doi:10.1371/journal.pgen.1000749.
55. GoulaAV, PearsonCE, Della MariaJ, TrottierY, TomkinsonAE, et al. (2012) The nucleotide sequence, DNA damage location, and protein stoichiometry influence the base excision repair outcome at CAG/CTG repeats. Biochemistry 51 : 3919–3932.
56. MangiariniL, SathasivamK, SellerM, CozensB, HarperA, et al. (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87 : 493–506.
57. ChiangC, JacobsenJC, ErnstC, HanscomC, HeilbutA, et al. (2012) Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet 44 : 390–397, S391.
58. GoulaAV, StysA, ChanJP, TrottierY, FestensteinR, et al. (2012) Transcription elongation and tissue-specific somatic CAG instability. PLoS Genet 8: e1003051 doi:10.1371/journal.pgen.1003051.
59. LloretA, DragilevaE, TeedA, EspinolaJ, FossaleE, et al. (2006) Genetic background modifies nuclear mutant huntingtin accumulation and HD CAG repeat instability in Huntington's disease knock-in mice. Hum Mol Genet 15 : 2015–2024.
60. CowinRM, BuiN, GrahamD, GreenJR, GrueningerS, et al. (2011) Onset and progression of behavioral and molecular phenotypes in a novel congenic R6/2 line exhibiting intergenerational CAG repeat stability. PLoS ONE 6: e28409 doi:10.1371/journal.pone.0028409.
61. RamosEM, CerqueiraJ, LemosC, Pinto-BastoJ, AlonsoI, et al. (2012) Intergenerational instability in Huntington disease: extreme repeat changes among 134 transmissions. Mov Disord 27 : 583–585.
62. ZhangY, MoncktonDG, SicilianoMJ, ConnorTH, MeistrichML (2002) Age and insertion site dependence of repeat number instability of a human DM1 transgene in individual mouse sperm. Hum Mol Genet 11 : 791–798.
63. HaugeXY, LittM (1993) A study of the origin of ‘shadow bands’ seen when typing dinucleotide repeat polymorphisms by the PCR. Hum Mol Genet 2 : 411–415.
64. VatsavayaiSC, DalleracGM, MilnerwoodAJ, CummingsDM, RezaieP, et al. (2007) Progressive CAG expansion in the brain of a novel R6/1-89Q mouse model of Huntington's disease with delayed phenotypic onset. Brain Res Bull 72 : 98–102.
65. HoltI, Thanh LamL, TomeS, WansinkDG, Te RieleH, et al. (2011) The mouse mismatch repair protein, MSH3, is a nucleoplasmic protein that aggregates into denser nuclear bodies under conditions of stress. J Cell Biochem 112 : 1612–1621.
66. CrouseGF, LeysEJ, McEwanRN, FrayneEG, KellemsRE (1985) Analysis of the mouse dhfr promoter region: existence of a divergently transcribed gene. Mol Cell Biol 5 : 1847–1858.
67. LintonJP, YenJY, SelbyE, ChenZ, ChinskyJM, et al. (1989) Dual bidirectional promoters at the mouse dhfr locus: cloning and characterization of two mRNA classes of the divergently transcribed Rep-1 gene. Mol Cell Biol 9 : 3058–3072.
68. WatanabeA, IkejimaM, SuzukiN, ShimadaT (1996) Genomic organization and expression of the human MSH3 gene. Genomics 31 : 311–318.
69. HutchinsonEG, ThorntonJM (1994) A revised set of potentials for beta-turn formation in proteins. Protein Sci 3 : 2207–2216.
70. LamersMH, PerrakisA, EnzlinJH, WinterwerpHH, de WindN, et al. (2000) The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature 407 : 711–717.
71. TakanoK, YamagataY, YutaniK (2000) Role of amino acid residues at turns in the conformational stability and folding of human lysozyme. Biochemistry 39 : 8655–8665.
72. MarcelinoAM, GieraschLM (2008) Roles of beta-turns in protein folding: from peptide models to protein engineering. Biopolymers 89 : 380–391.
73. FullerAA, DuD, LiuF, DavorenJE, BhabhaG, et al. (2009) Evaluating beta-turn mimics as beta-sheet folding nucleators. Proc Natl Acad Sci U S A 106 : 11067–11072.
74. SavouretC, JunienC, GourdonG (2004) Analysis of CTG Repeats Using DM1 Model Mice. Methods Mol Biol 277 : 185–198.
75. FuH, GrimsleyGR, RazviA, ScholtzJM, PaceCN (2009) Increasing protein stability by improving beta-turns. Proteins 77 : 491–498.
76. TrevinoSR, ScholtzJM, PaceCN (2007) Amino acid contribution to protein solubility: Asp, Glu, and Ser contribute more favorably than the other hydrophilic amino acids in RNase Sa. J Mol Biol 366 : 449–460.
77. ChenRP, HuangJJ, ChenHL, JanH, VelusamyM, et al. (2004) Measuring the refolding of beta-sheets with different turn sequences on a nanosecond time scale. Proc Natl Acad Sci U S A 101 : 7305–7310.
78. McCallisterEL, AlmE, BakerD (2000) Critical role of beta-hairpin formation in protein G folding. Nat Struct Biol 7 : 669–673.
79. YbeJA, HechtMH (1996) Sequence replacements in the central beta-turn of plastocyanin. Protein Sci 5 : 814–824.
80. OlsenJV, VermeulenM, SantamariaA, KumarC, MillerML, et al. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3: ra3.
81. JohnsonLN, LewisRJ (2001) Structural basis for control by phosphorylation. Chem Rev 101 : 2209–2242.
82. TakanoH, OnoderaO, TakahashiH, IgarashiS, YamadaM, et al. (1996) Somatic mosaicism of expanded CAG repeats in brains of patients with dentatorubral-pallidoluysian atrophy: cellular population-dependent dynamics of mitotic instability. Am J Hum Genet 58 : 1212–1222.
83. WatanabeY, Haugen-StranoA, UmarA, YamadaK, HemmiH, et al. (2000) Complementation of an hMSH2 defect in human colorectal carcinoma cells by human chromosome 2 transfer. Mol Carcinog 29 : 37–49.
84. HashidaH, GotoJ, SuzukiT, JeongS, MasudaN, et al. (2001) Single cell analysis of CAG repeat in brains of dentatorubral-pallidoluysian atrophy (DRPLA). J Neurol Sci 190 : 87–93.
85. OdaS, MaeharaY, IkedaY, OkiE, EgashiraA, et al. (2005) Two modes of microsatellite instability in human cancer: differential connection of defective DNA mismatch repair to dinucleotide repeat instability. Nucleic Acids Res 33 : 1628–1636.
86. GiuntiL, CeticaV, RicciU, GiglioS, SardiI, et al. (2009) Type A microsatellite instability in pediatric gliomas as an indicator of Turcot syndrome. Eur J Hum Genet 17 : 919–927.
87. ThibodeauSN, BrenG, SchaidD (1993) Microsatellite instability in cancer of the proximal colon. Science 260 : 816–819.
88. YangZ, LauR, MarcadierJL, ChitayatD, PearsonCE (2003) Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells. Am J Hum Genet 73 : 1092–1105.
89. WeberJL (1990) Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics 7 : 524–530.
90. BlakeC, TsaoJL, WuA, ShibataD (2001) Stepwise deletions of polyA sequences in mismatch repair-deficient colorectal cancers. Am J Pathol 158 : 1867–1870.
91. OttoCJ, AlmqvistE, HaydenMR, AndrewSE (2001) The “flap” endonuclease gene FEN1 is excluded as a candidate gene implicated in the CAG repeat expansion underlying Huntington disease. Clin Genet 59 : 122–127.
92. CoppedeF, MigheliF, CeravoloR, BregantE, RocchiA, et al. (2010) The hOGG1 Ser326Cys polymorphism and Huntington's disease. Toxicology 278 : 199–203.
93. Taherzadeh-FardE, SaftC, WieczorekS, EpplenJT, ArningL (2010) Age at onset in Huntington's disease: replication study on the associations of ADORA2A, HAP1 and OGG1. Neurogenetics 11 : 435–439.
94. PanigrahiGB, SleanMM, SimardJP, PearsonCE (2012) Human Mismatch Repair Protein hMutLalpha Is Required to Repair Short Slipped-DNAs of Trinucleotide Repeats. J Biol Chem 287 : 41844–41850.
95. MartinezSL, KolodnerRD (2010) Functional analysis of human mismatch repair gene mutations identifies weak alleles and polymorphisms capable of polygenic interactions. Proc Natl Acad Sci U S A 107 : 5070–5075.
96. KumarC, PiacenteSC, SibertJ, BukataAR, O'ConnorJ, et al. (2011) Multiple factors insulate Msh2–Msh6 mismatch repair activity from defects in Msh2 domain I. J Mol Biol 411 : 765–780.
97. MangoniM, BisanziS, CarozziF, SaniC, BitiG, et al. (2011) Association between genetic polymorphisms in the XRCC1, XRCC3, XPD, GSTM1, GSTT1, MSH2, MLH1, MSH3, and MGMT genes and radiosensitivity in breast cancer patients. Int J Radiat Oncol Biol Phys 81 : 52–58.
98. DongX, LiY, HessKR, AbbruzzeseJL, LiD (2011) DNA mismatch repair gene polymorphisms affect survival in pancreatic cancer. Oncologist 16 : 61–70.
99. CondeJ, SilvaSN, AzevedoAP, TeixeiraV, PinaJE, et al. (2009) Association of common variants in mismatch repair genes and breast cancer susceptibility: a multigene study. BMC Cancer 9 : 344.
100. MichielsS, DanoyP, DessenP, BeraA, BouletT, et al. (2007) Polymorphism discovery in 62 DNA repair genes and haplotype associations with risks for lung and head and neck cancers. Carcinogenesis 28 : 1731–1739.
101. HaugenAC, GoelA, YamadaK, MarraG, NguyenTP, et al. (2008) Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. Cancer Res 68 : 8465–8472.
102. BurrKL, van Duyn-GoedhartA, HickenbothamP, MongerK, van BuulPP, et al. (2007) The effects of MSH2 deficiency on spontaneous and radiation-induced mutation rates in the mouse germline. Mutat Res 617 : 147–151.
103. DuJ, CampauE, SoragniE, KuS, PuckettJW, et al. (2012) Role of mismatch repair enzymes in GAA.TTC triplet-repeat expansion in Friedreich ataxia induced pluripotent stem cells. J Biol Chem 287 : 29861–29872.
104. EzzatizadehV, PintoRM, SandiC, SandiM, Al-MahdawiS, et al. (2012) The mismatch repair system protects against intergenerational GAA repeat instability in a Friedreich ataxia mouse model. Neurobiol Dis 46 : 165–171.
105. KimHM, NarayananV, MieczkowskiPA, PetesTD, KrasilnikovaMM, et al. (2008) Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. Embo J 27 : 2896–2906.
106. KuS, SoragniE, CampauE, ThomasEA, AltunG, et al. (2010) Friedreich's ataxia induced pluripotent stem cells model intergenerational GAATTC triplet repeat instability. Cell Stem Cell 7 : 631–637.
107. LiJL, HaydenMR, AlmqvistEW, BrinkmanRR, DurrA, et al. (2003) A genome scan for modifiers of age at onset in Huntington disease: The HD MAPS study. Am J Hum Genet 73 : 682–687.
108. WexlerNS, LorimerJ, PorterJ, GomezF, MoskowitzC, et al. (2004) Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington's disease age of onset. Proc Natl Acad Sci U S A 101 : 3498–3503.
109. CowinRM, BuiN, GrahamD, GreenJR, Yuva-PaylorLA, et al. (2012) Genetic background modulates behavioral impairments in R6/2 mice and suggests a role for dominant genetic modifiers in Huntington's disease pathogenesis. Mamm Genome 23 : 367–377.
110. KinT, OnoY (2007) Idiographica: a general-purpose web application to build idiograms on-demand for human, mouse and rat. Bioinformatics 23 : 2945–2946.
111. Gomes-PereiraM, BidichandaniSI, MoncktonDG (2004) Analysis of unstable triplet repeats using small-pool polymerase chain reaction. Methods Mol Biol 277 : 61–76.
112. HsiehYC, SkacelNE, BansalN, ScottoKW, BanerjeeD, et al. (2009) Species-specific differences in translational regulation of dihydrofolate reductase. Mol Pharmacol 76 : 723–733.
113. AltschulSF, GishW, MillerW, MyersEW, LipmanDJ (1990) Basic local alignment search tool. J Mol Biol 215 : 403–410.
114. KatohK, MisawaK, KumaK, MiyataT (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30 : 3059–3066.
115. BermanHM, WestbrookJ, FengZ, IypeL, SchneiderB, et al. (2003) The nucleic acid database. Methods Biochem Anal 44 : 199–216.
116. KrivovGG, ShapovalovMV, DunbrackRLJr (2009) Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77 : 778–795.
117. KabschW, SanderC (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 : 2577–2637.
118. VenkatachalamCM (1968) Stereochemical criteria for polypeptides and proteins. VI. Non-bonded energy of polyglycine and poly-L-alanine in the crystalline beta-form. Biochim Biophys Acta 168 : 411–416.
119. HutchinsonEG, ThorntonJM (1996) PROMOTIF–a program to identify and analyze structural motifs in proteins. Protein Sci 5 : 212–220.
120. ClampM, CuffJ, SearleSM, BartonGJ (2004) The Jalview Java alignment editor. Bioinformatics 20 : 426–427.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



