-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGenetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
Most previous genetic epidemiology studies within the field of osteoporosis have focused on the genetics of the complex trait areal bone mineral density (aBMD), not being able to differentiate genetic determinants of cortical volumetric BMD (vBMD), trabecular vBMD, and bone microstructural traits. The objective of this study was to separately identify genetic determinants of these bone traits as analysed by peripheral quantitative computed tomography (pQCT). Separate GWA meta-analyses for cortical and trabecular vBMDs were performed. The cortical vBMD GWA meta-analysis (n = 5,878) followed by replication (n = 1,052) identified genetic variants in four separate loci reaching genome-wide significance (RANKL, rs1021188, p = 3.6×10−14; LOC285735, rs271170, p = 2.7×10−12; OPG, rs7839059, p = 1.2×10−10; and ESR1/C6orf97, rs6909279, p = 1.1×10−9). The trabecular vBMD GWA meta-analysis (n = 2,500) followed by replication (n = 1,022) identified one locus reaching genome-wide significance (FMN2/GREM2, rs9287237, p = 1.9×10−9). High-resolution pQCT analyses, giving information about bone microstructure, were available in a subset of the GOOD cohort (n = 729). rs1021188 was significantly associated with cortical porosity while rs9287237 was significantly associated with trabecular bone fraction. The genetic variant in the FMN2/GREM2 locus was associated with fracture risk in the MrOS Sweden cohort (HR per extra T allele 0.75, 95% confidence interval 0.60–0.93) and GREM2 expression in human osteoblasts. In conclusion, five genetic loci associated with trabecular or cortical vBMD were identified. Two of these (FMN2/GREM2 and LOC285735) are novel bone-related loci, while the other three have previously been reported to be associated with aBMD. The genetic variants associated with cortical and trabecular bone parameters differed, underscoring the complexity of the genetics of bone parameters. We propose that a genetic variant in the RANKL locus influences cortical vBMD, at least partly, via effects on cortical porosity, and that a genetic variant in the FMN2/GREM2 locus influences GREM2 expression in osteoblasts and thereby trabecular number and thickness as well as fracture risk.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003247
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003247Summary
Most previous genetic epidemiology studies within the field of osteoporosis have focused on the genetics of the complex trait areal bone mineral density (aBMD), not being able to differentiate genetic determinants of cortical volumetric BMD (vBMD), trabecular vBMD, and bone microstructural traits. The objective of this study was to separately identify genetic determinants of these bone traits as analysed by peripheral quantitative computed tomography (pQCT). Separate GWA meta-analyses for cortical and trabecular vBMDs were performed. The cortical vBMD GWA meta-analysis (n = 5,878) followed by replication (n = 1,052) identified genetic variants in four separate loci reaching genome-wide significance (RANKL, rs1021188, p = 3.6×10−14; LOC285735, rs271170, p = 2.7×10−12; OPG, rs7839059, p = 1.2×10−10; and ESR1/C6orf97, rs6909279, p = 1.1×10−9). The trabecular vBMD GWA meta-analysis (n = 2,500) followed by replication (n = 1,022) identified one locus reaching genome-wide significance (FMN2/GREM2, rs9287237, p = 1.9×10−9). High-resolution pQCT analyses, giving information about bone microstructure, were available in a subset of the GOOD cohort (n = 729). rs1021188 was significantly associated with cortical porosity while rs9287237 was significantly associated with trabecular bone fraction. The genetic variant in the FMN2/GREM2 locus was associated with fracture risk in the MrOS Sweden cohort (HR per extra T allele 0.75, 95% confidence interval 0.60–0.93) and GREM2 expression in human osteoblasts. In conclusion, five genetic loci associated with trabecular or cortical vBMD were identified. Two of these (FMN2/GREM2 and LOC285735) are novel bone-related loci, while the other three have previously been reported to be associated with aBMD. The genetic variants associated with cortical and trabecular bone parameters differed, underscoring the complexity of the genetics of bone parameters. We propose that a genetic variant in the RANKL locus influences cortical vBMD, at least partly, via effects on cortical porosity, and that a genetic variant in the FMN2/GREM2 locus influences GREM2 expression in osteoblasts and thereby trabecular number and thickness as well as fracture risk.
Introduction
Meta-analyses of genome-wide association studies (GWAS) have identified a large number of loci associated with areal bone mineral density (aBMD) [1]–[6]. aBMD is a complex trait, obtained from a 2-dimensional projectional scan of the given bone with dual x-ray absorptiometry (DXA). Skeletal sites which are measured in this way, such as the lumbar spine and hip, comprise a mixture of cortical bone (compact bone comprising the outer shell), and trabecular bone (a network of thin interconnecting plates within the marrow cavity of vertebrae and the end of long bones). The lumbar spine has a relatively high proportion of trabecular bone, whereas the hip has a higher proportion of cortical bone. DXA-measured aBMD depends not only on bone cross-sectional size but also on apparent volumetric bone mineral density which is largely determined by trabecular microstructure and cortical thickness [7]. Although aBMD is the gold standard for diagnosing osteoporosis, it fails to provide a detailed skeletal phenotype necessary to discern traits such as trabecular volumetric BMD (vBMD), cortical vBMD and bone microstructural parameters. Previous studies using DXA have demonstrated that age is a major predictor of fracture risk independent of aBMD. Although this aBMD independent effect of age has been attributed to poor bone “quality”, the structural basis for this remains unclear [8]. Age-related changes in bone include microstructural deterioration, such as trabecular perforation, thinning, and loss of connectivity, as well as increased cortical porosity [8], [9]. Quantitative computed tomography (QCT) analysis has the capacity to reveal unique information about these bone traits.
Standard peripheral QCT (pQCT) with a resolution of 500 µm has the advantage of being able to separately analyse trabecular and cortical vBMDs. The correlation between trabecular and cortical vBMDs is low (rs 0.11 in the young adult men of the GOOD cohort; [10]), supporting the notion that the determinants of these two bone parameters differ. Cortical vBMD but not trabecular vBMD reflects material density while trabecular vBMD mainly is influenced by trabecular number and thickness. In addition, the correlations of these vBMD parameters with femoral neck aBMD are low (cortical vBMD, rs 0.04) or moderate (trabecular vBMD rs 0.65), suggesting that cortical and trabecular vBMDs are at least partly influenced by genetic determinants not possible to identify by a GWAS of aBMD [10]. The heritability for trabecular vBMD has been reported to be as high as 59% while the heritability for cortical vBMD was slightly lower (40%) [11]. GWAS have revealed differences in genetic associations with lumbar and hip aBMD, providing some evidence that cortical and trabecular bone have distinct genetic influences [2].
We have in a previous smaller-scale GWAS meta-analysis (n = 1,934) identified a genetic variant in the RANKL locus to be significantly associated with cortical vBMD [10]. The genetic determinants of trabecular vBMD have not yet been evaluated using GWAS.
High resolution pQCT (HRpQCT) not only allows the separation of the trabecular and cortical bone compartments but also the assessment of bone microstructure. HRpQCT has an isotrophic voxel size of 82 µm and shows excellent correlation with ex vivo μCT imaging (resolution 20 µm or better) [8], [12], [13]. Importantly, HRpQCT analysis recently demonstrated that younger and older subjects with the same aBMD differed in cortical porosity, a key parameter not captured by DXA [8]. The genetic determinants of trabecular and cortical bone microstructure parameters as analysed by HRpQCT are unknown.
The objective of the present study was to identify genetic determinants of vBMDs and bone microstructure parameters separately for the cortical and trabecular bone compartments as analyzed by pQCT and HRpQCT. As our assembled discovery cohort was larger for the pQCT measurements (cortical vBMD n = 5,878, trabecular vBMD n = 2,500) than for the HRpQCT measurements (n = 729), we aimed to first identify genome-wide significant genetic variants for cortical and trabecular vBMDs separately and then to evaluate the impact of the identified variants on trabecular and cortical bone microstructure parameters in the HRpQCT cohort.
Results
Genome-wide association (GWA) meta-analyses of cortical and trabecular vBMDs
Table 1 displays the anthropometrics and bone traits for the four cohorts (ALSPAC discovery, GOOD baseline discovery, YFS discovery, and MrOS Sweden replication) evaluated. The association between cortical vBMD and trabecular vBMD was rather modest (Spearman's rank correlation coefficient [rho] GOOD baseline r = 0.11 [10]; GOOD five year follow-up r = −0.01). Separate GWA meta-analyses for cortical and trabecular vBMD were performed including all three discovery cohorts for cortical vBMD while trabecular vBMD was available in the YFS and GOOD cohorts.
Tab. 1. Characteristics of the cohorts included in the discovery GWA meta-analyses and replications. 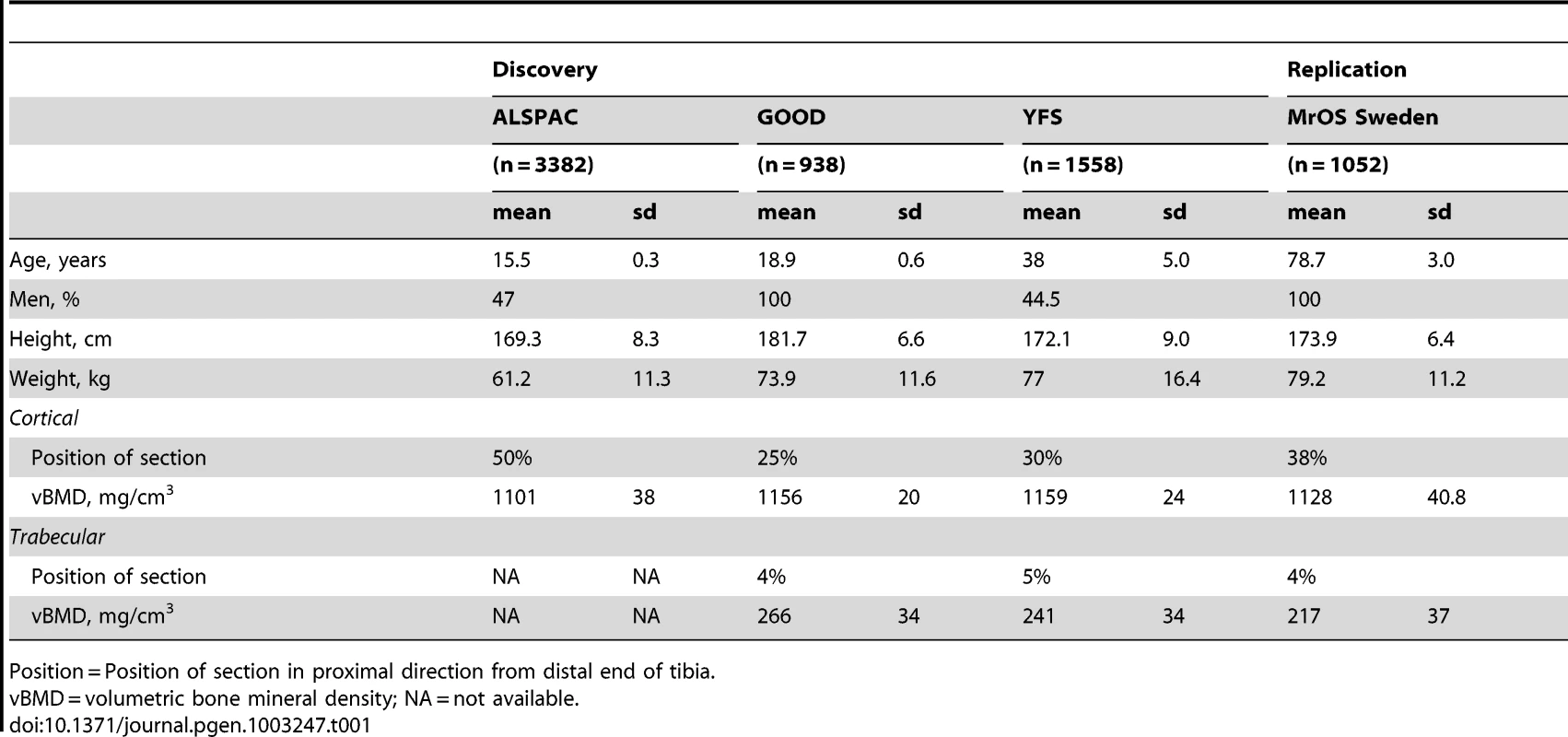
Position = Position of section in proximal direction from distal end of tibia. GWA meta-analysis of cortical vBMD
Inverse variance weighted fixed-effect model meta-analysis of study-specific results was performed. In the cortical vBMD GWA meta-analysis of the ALSPAC, GOOD and YFS cohorts there was little systematic inflation of test statistics (Overall λ = 1.012 (1.023 for ALSPAC; 1.013 for GOOD; 1.023 for YFS)), but a marked deviation from the null distribution amongst the lowest observed p-values (Figure 1A). We identified genetic variants in four separate loci reaching genome-wide significance (Figure 1B). The greatest evidence for association between genetic variation and cortical vBMD was seen for rs1021188 (0.15 SD decrease per C allele; p = 1.4×10−12) on chromosome 13, slightly upstream of the RANKL gene (TNFSF11; Table 2, Figure 2A, Table S1). The second strongest genetic signal for cortical vBMD (rs271170; 0.11 SD decrease per T allele, p = 2.9×10−11) is a novel bone-related locus, located on chromosome 6, upstream of LOC285735 (Table 2, Figure 2B, Table S1). The third strongest signal (rs7839059, 0.10 SD decrease per A allele, p = 4.1×10−9) was located on chromosome 8, upstream of OPG (TNFRSF11B; Table 2, Figure 2C, Table S1). The fourth genome-wide signal (rs6909279, 0.09 SD decrease per allele G, p = 1.0×10−8) was located on chromosome 6, in C6orf97 upstream and close to estrogen receptor-α (ESR1; Table 2, Figure 2D, Table S1).
Fig. 1. Genome-wide meta-analysis of cortical vBMD. 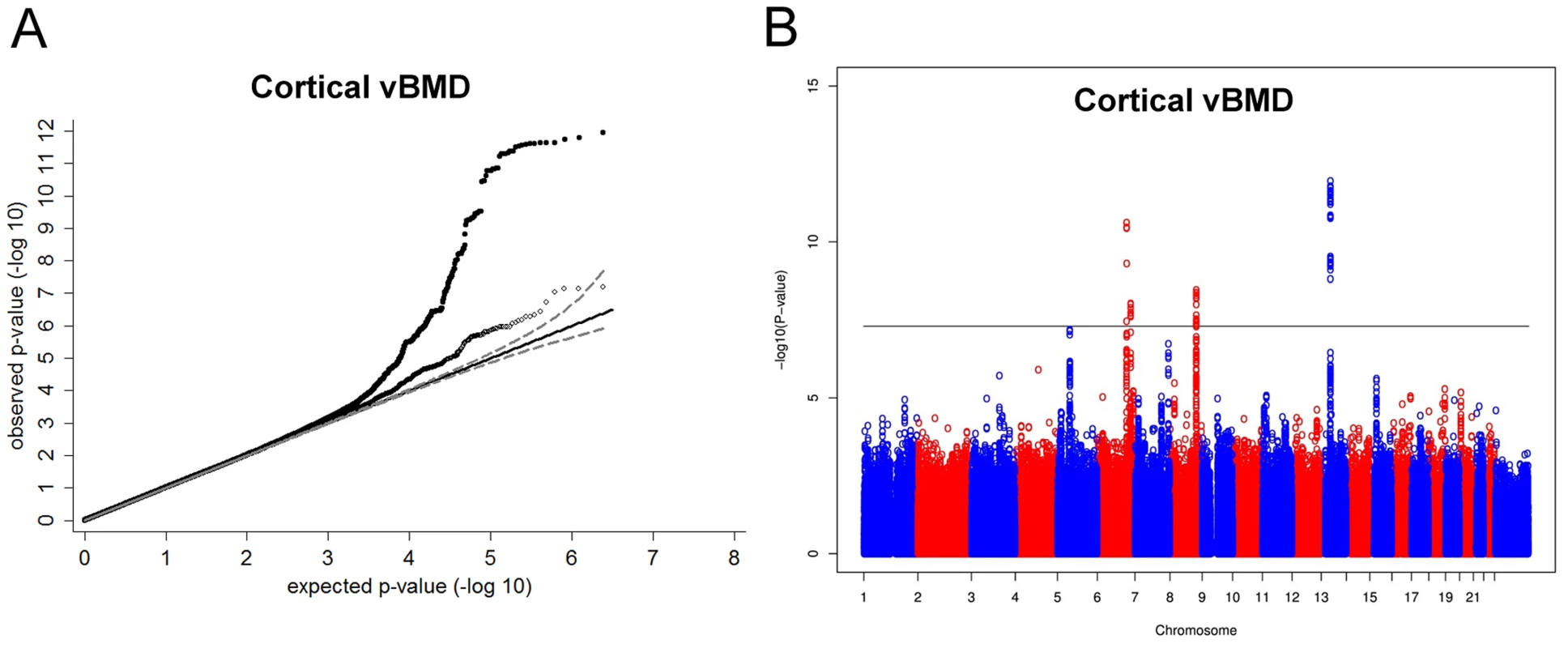
(A) QQ plots of the genome-wide meta-analysis of cortical vBMD without (filled circles) or with (open diamonds) removal of the genome-wide significant loci (All SNPs excluded ±1 Mb around the hits). (B) Manhattan plot of the genome-wide meta-analysis of cortical vBMD. Fig. 2. Regional association plots for the 5 independent signals from the discovery genome-wide meta-analysis of cortical vBMD. 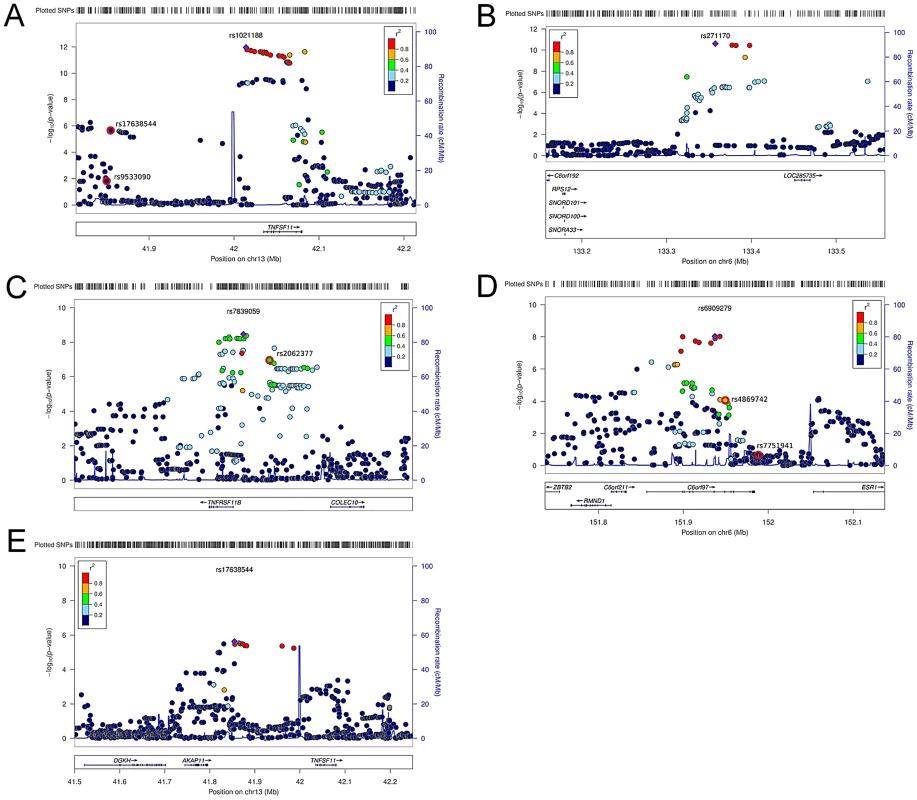
(A) rs1021188, (B) rs271170, (C) rs7839059, (D) rs6909279, (E) rs17638544. Circles show the GWA meta-analysis p-values, with different colors indicating varying linkage disequilibrium with the indicated SNP (diamond). SNPs in the same region identified in a recent large-scale GWA meta-analysis of aBMD are indicated by a red outer circle [2]. LocusZoom: http://csg.sph.umich.edu/locuszoom/. Tab. 2. Top cortical and trabecular vBMD signals from pQCT GWA meta-analyses followed by replication. 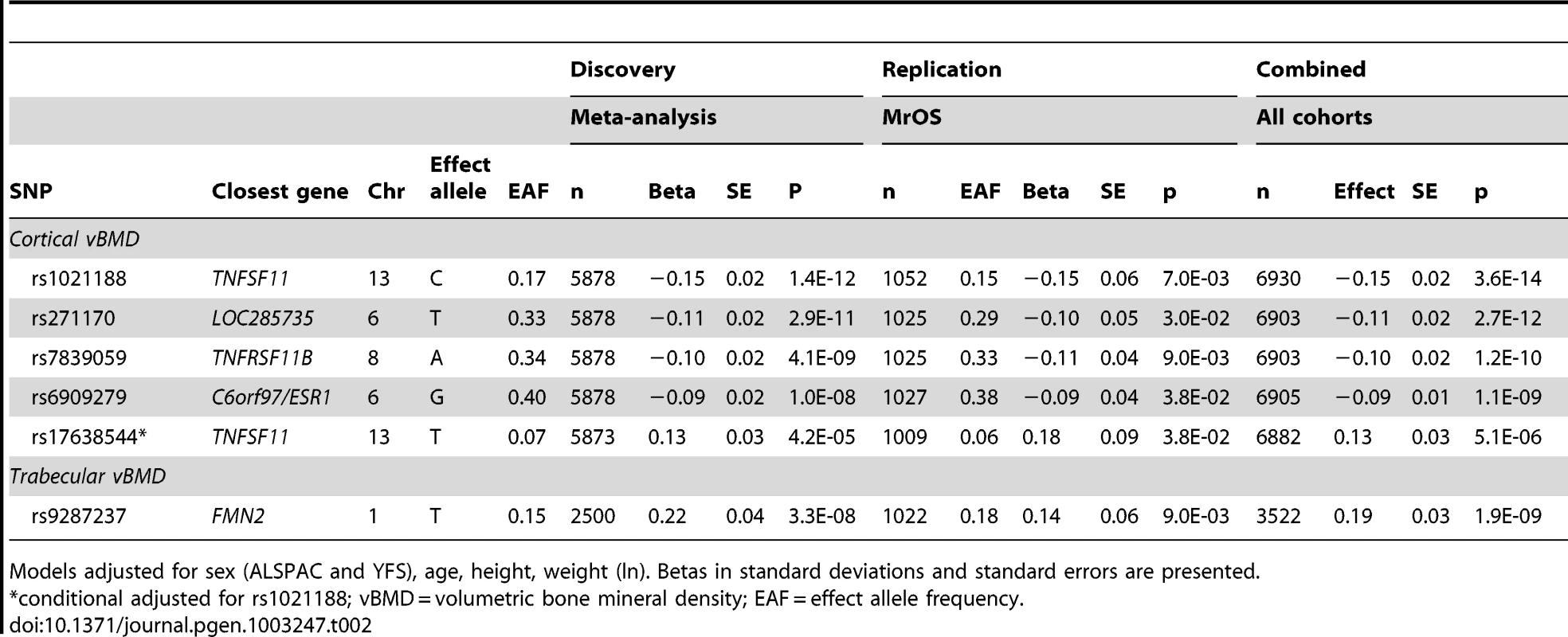
Models adjusted for sex (ALSPAC and YFS), age, height, weight (ln). Betas in standard deviations and standard errors are presented. We selected our top four regions and carried out analyses conditional on the most associated SNPs in each region. When conditioning on the most significant SNP in the RANKL region (rs1021188) an additional suggestive signal (rs17638544 close to AKAP1 and upstream of RANKL, p = 4.2×10−5) appeared, but did not achieve genome-wide significance (Table 2, Figure 2E, Table S1). Using similar conditional analysis, no additional SNPs with an independent signal appeared in the other three evaluated regions (p<5×10−5).
The RANKL, OPG and ESR1 regions have earlier been reported to be associated with aBMD in large scale GWA meta-analyses [1]–[6]. To evaluate if the identified SNPs associated with cortical vBMD in these regions are independent from the previously reported aBMD related SNPs, conditional analyses were performed (RANKL region, rs1021188 and rs17638544 were conditioned on the known aBMD hit rs9533090; OPG region, rs7839059 was conditioned on the known aBMD hit rs2062377; ESR1 region, rs6909279 was conditioned on known aBMD hits rs7751941 and rs4869742; [2]). The two cortical vBMD RANKL signals (rs1021188 and rs17638544) were distinct from the previously reported aBMD signal (rs9533090; [2]) in this region, supported by the fact that (i) rs9533090 was not significantly associated with cortical vBMD (Figure 2A), (ii) adjustment for rs9533090 did not influence the associations for rs1021188 or rs17638544 with cortical vBMD and the two cortical vBMD signals displayed a low r2 (<0.04) with rs9533090 (Table S2). It is difficult to determine if the identified cortical vBMD signal in the OPG region is separate from the previous reported aBMD signal in this region (rs2062377; [2]) as this previous aBMD signal also was significantly associated with cortical vBMD (Figure 2C), the r2 between the two SNPs was 0.39, and adjustment for rs2062377 slightly but not completely attenuated the association for rs7839059 with cortical vBMD (Table S2).
The identified cortical vBMD SNP in the ESR1 region (rs6909279) is independent from one of the previous reported aBMD signals (rs7751941) while the other reported independent aBMD SNP in this region (rs4869742 [2]) displayed a relatively high r2 with rs6909279 (r2 = 0.60) (Figure 2D). However, adjustment for rs4869742 only slightly attenuated the association for rs6909279 with cortical vBMD (Table S2).
GWA meta-analysis of trabecular volumetric BMD
In the trabecular vBMD GWA meta-analysis there was little systematic inflation of test statistics (Overall λ = 1.005 (1.020 for GOOD; 1.018 for YFS)), but a significant deviation from the null distribution amongst the lowest observed p-values (Figure 3A). We identified one novel bone-related genetic variant reaching genome-wide significance (Figure 3B). The greatest evidence for association between genetic variation and trabecular vBMD was seen for rs9287237 (0.22 SD increase per T allele; p = 3.3×10−8) on chromosome 1, in the formin 2 gene (FMN2 gene; Table 2, Figure 4, Table S1). When conditioning on the most significant SNP in the FMN2 region, no additional SNPs with an independent signal appeared (p<5×10−5).
Fig. 3. Genome-wide meta-analysis of trabecular vBMD. 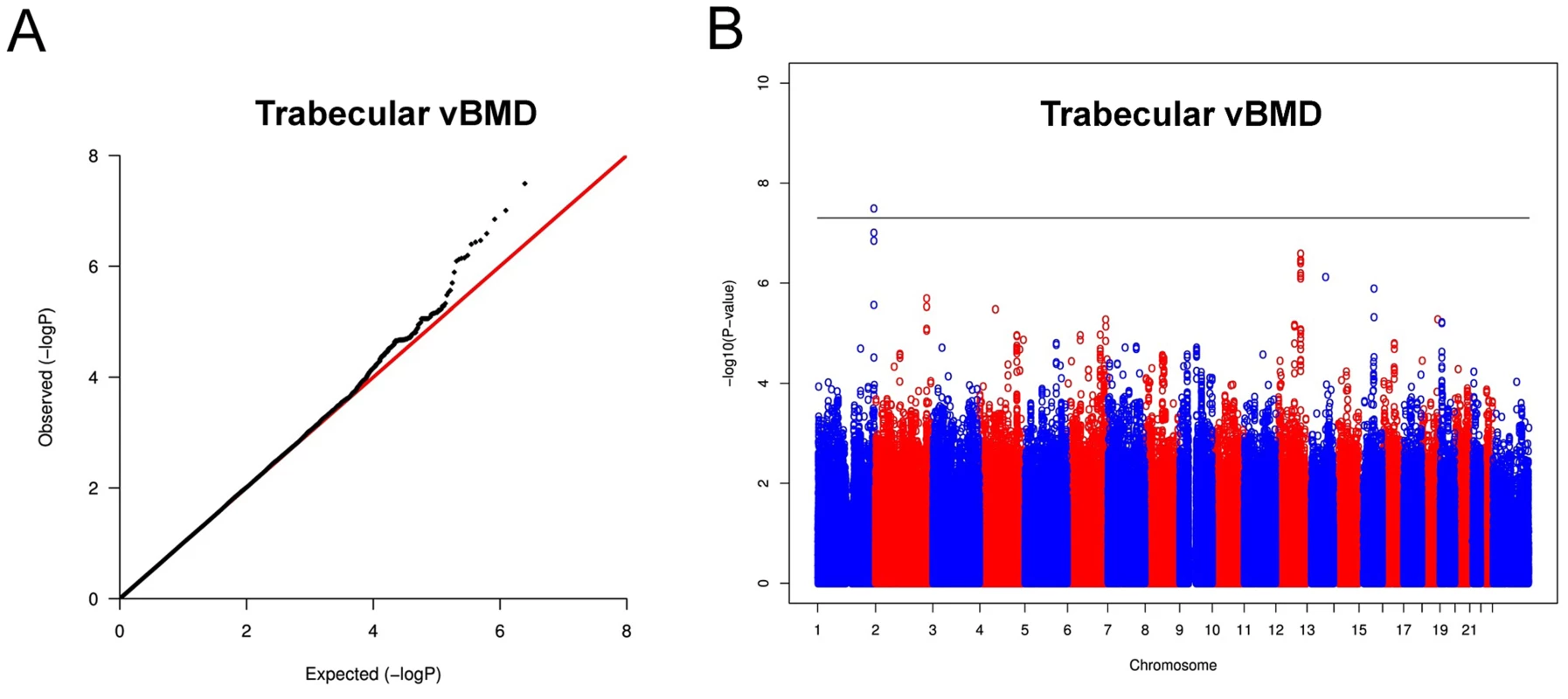
(A) QQ plot of the genome-wide meta-analysis of trabecular vBMD. (B) Manhattan plot of the genome-wide meta-analysis of trabecular vBMD. Fig. 4. Regional association plot for rs9287237 of the discovery genome-wide meta-analysis of trabecular vBMD. 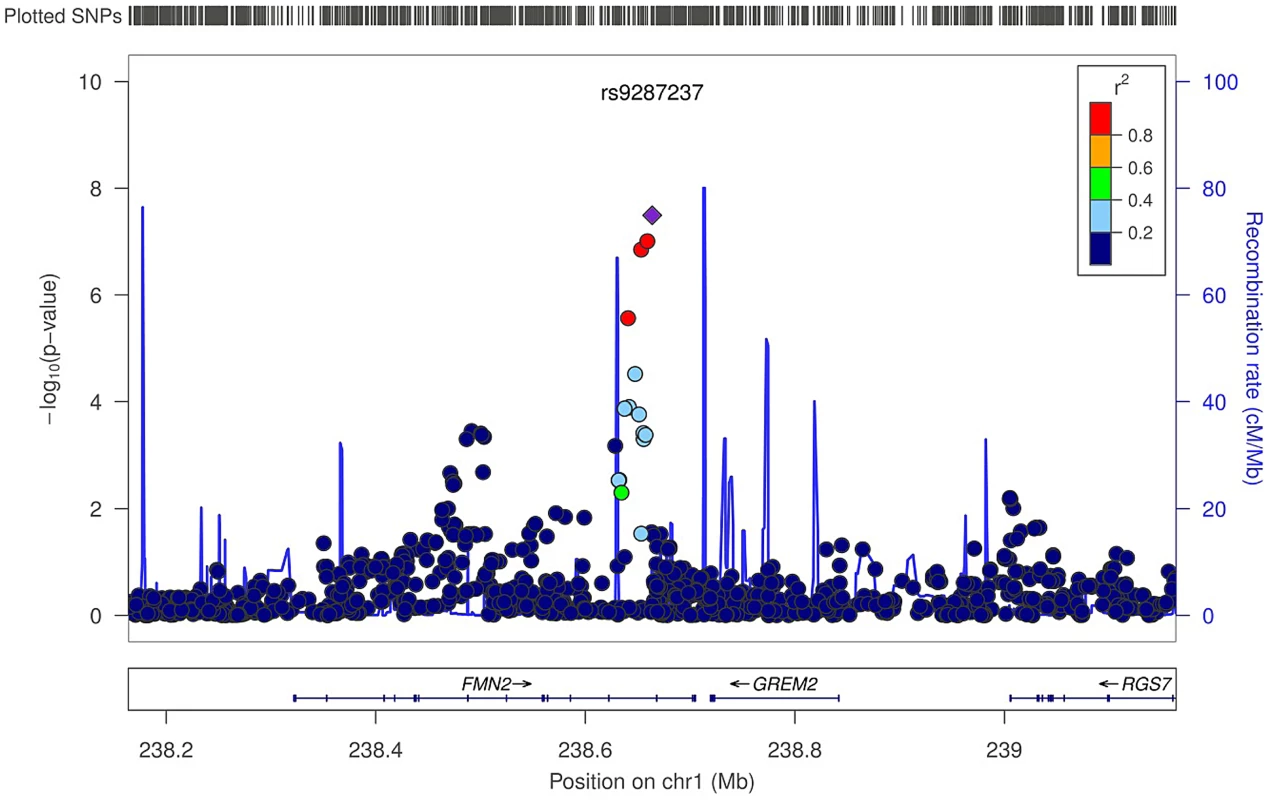
Circles show the GWA meta-analysis p-values, with different colors indicating varying linkage disequilibrium with rs9287237 (diamond). LocusZoom: http://csg.sph.umich.edu/locuszoom/. The rs9287237 SNP significantly associated with trabecular vBMD was not associated with cortical vBMD, and three of the four genome-wide significant cortical vBMD SNPs were not significantly associated with trabecular vBMD (Table 3). However, the cortical vBMD SNP rs7839059 upstream of OPG displayed a modest association with trabecular vBMD (0.06 SD decrease per A allele; p = 4.7×10−2) in the same direction as seen for the genome-wide significant association with cortical vBMD (Table 3).
Tab. 3. Association of cortical and trabecular vBMD with top cortical and trabecular vBMD SNPs. 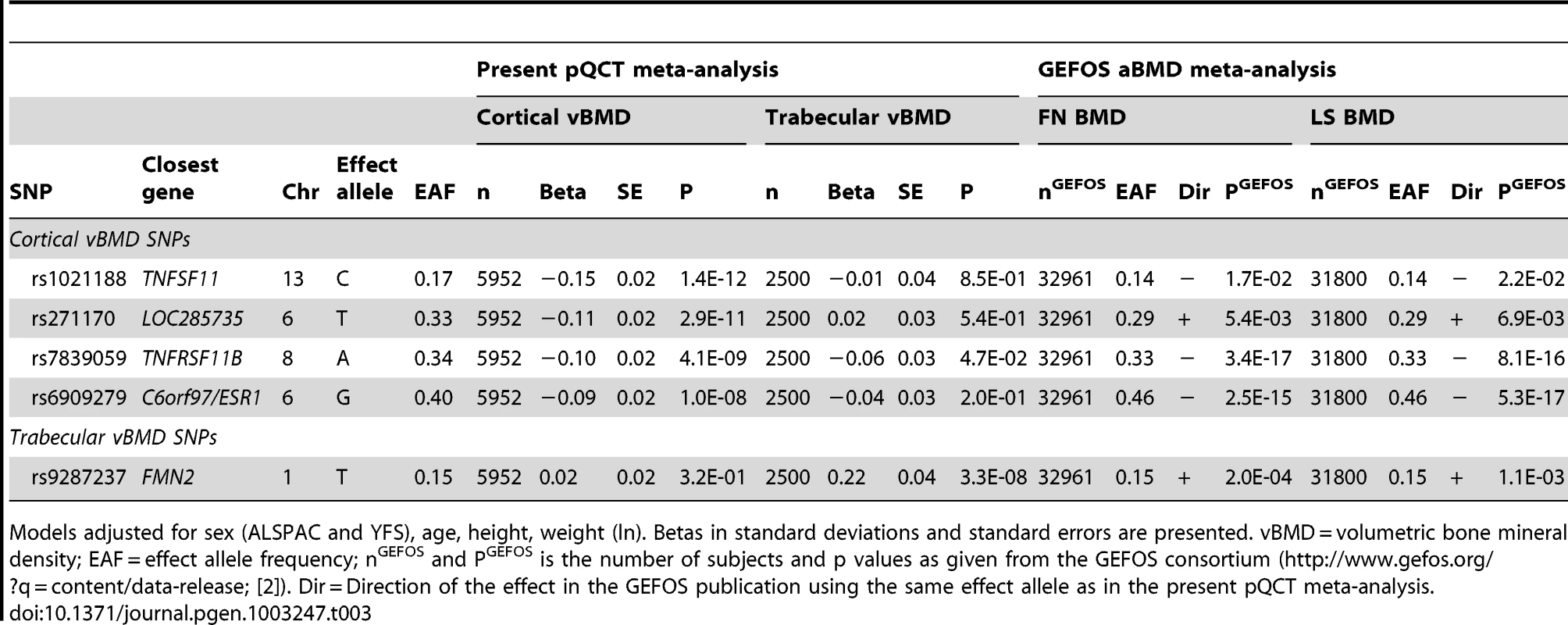
Models adjusted for sex (ALSPAC and YFS), age, height, weight (ln). Betas in standard deviations and standard errors are presented. vBMD = volumetric bone mineral density; EAF = effect allele frequency; nGEFOS and PGEFOS is the number of subjects and p values as given from the GEFOS consortium (http://www.gefos.org/?q=content/data-release; [2]). Dir = Direction of the effect in the GEFOS publication using the same effect allele as in the present pQCT meta-analysis. Replication study of top SNPs for cortical and trabecular vBMD
Cortical vBMD
Five SNPs (four genome-wide significant [rs1021188, rs271170, rs7839059 and rs6909279] and one identified in the conditional analyses rs17638544) were selected for replication in the MrOS Sweden cohort. In this replication analysis all five SNPs were significantly associated with cortical vBMD in the same direction and with similar effect sizes as found in the discovery GWA meta-analysis (Table 2; Combined all cohorts, rs1021188 0.15 SD decrease per C allele, p = 3.6×10−14; rs271170 0.11 SD decrease per T allele, p = 2.7×10−12; rs7839059 0.10 SD decrease per A allele, p = 1.2×10−10; rs6909279 0.09 SD decrease per G allele, p = 1.1×10−9; rs17638544 0.13 SD increase per T allele, p = 5.1×10−6). The four GWA significant cortical vBMD SNPs rs1021188, rs271170, rs7839059 and rs6909279 explained 0.6%, 0.4%, 0.6% and 0.3%, respectively of the variation in cortical vBMD in the elderly men of the MrOS Sweden replication cohort. When including all four SNPs together, 1.5% of the variation in cortical vBMD was explained in the MrOS Sweden replication cohort. There was evidence for heterogeneity in the meta-analyses of the associations with rs1021188 (het p = 0.028) and rs17638544 (het p = 0.011). Random effects meta-analysis results for rs1021188 and rs17638544 were 0.18 SD per allele (SE = 0.05, 95% CI 0.08 to 0.28, p = 0.0003) and 0.14 SD per allele (SE = 0.07, 95% CI 0.00 to 0.28, p = 0.056), respectively.
Trabecular vBMD
One SNP (rs9287237) was replicated in the MrOS Sweden cohort and it was significantly associated with trabecular vBMD also in the replication cohort (Table 2; Combined all cohorts, 0.19 SD increase per T allele, p = 1.9×10−9). rs9287237 explained 0.7% of the variation in trabecular vBMD in the MrOS Sweden replication cohort.
Sex-specific analyses
Sex-specific analyses demonstrated that all five cortical vBMD SNPs were significantly associated with cortical vBMD in both men and women with effects in the same direction. However, the magnitude of the effect sizes differed significantly according to sex for three of the identified hits (Figure 5A; rs1021188 in RANKL region, men 0.21 SD and women 0.06 SD decrease per C allele, p = 4.3×10−5; rs6909279 in ESR1 region, men 0.12 SD and women 0.05 SD decrease per G allele, p = 3.0×10−3; rs7839059 in OPG region, men 0.12 SD and women 0.07 SD decrease per A allele, p = 3.0×10−2). The sex difference may explain the heterogeneity observed in the unstratified analyses for rs1021188 (both sexes have a het p>0.05). Significant heterogeneity is still observed in the males for rs17638544 (het p = 0.016).
Fig. 5. The genome-wide meta-analyses according to sex. 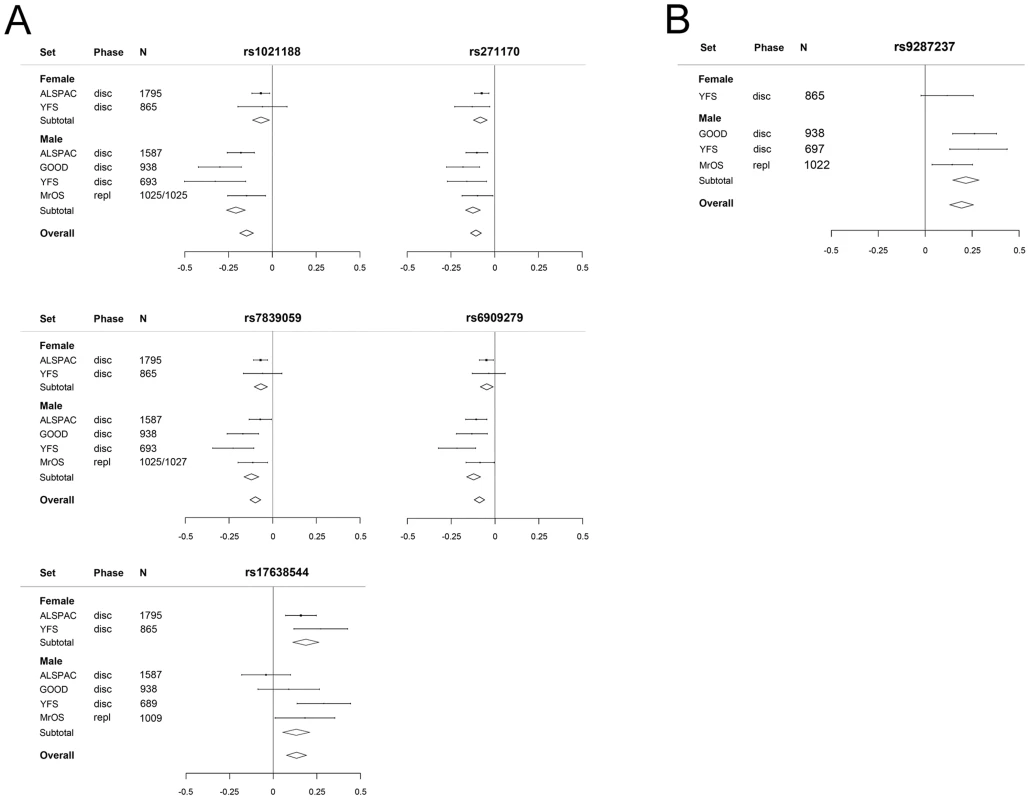
(A) Cortical vBMD (rs1021188 effect allele = C, rs271170 effect allele = T, rs7839059 effect allele = A, rs6909279 effect allele = G, rs17638544 effect allele = T). (B) Trabecular vBMD for rs9287237 (effect allele = T). Mean and standard error vBMD z-scores are shown for each cohort, stratified by sex. Diamonds show the combined z-scores estimates per genotype (the width of the diamond represents the combined standard error). Disc = discovery cohort, repl = replication cohort. The genetic variant associated with trabecular vBMD (rs9287237) was significantly associated with trabecular vBMD in men (Figure 5B, 0.22 SD increase per T allele) and a similar non-significant tendency in the same direction was observed in females (0.12 SD increase per T allele). It should be emphasized that for trabecular vBMD, we had a low statistical power to detect sex differences in effect sizes as the number of females included was rather low (n = 865).
Genetic association versus HRpQCT measured bone microstructure
At the five year follow-up of the GOOD cohort both pQCT analysis, giving information about cortical and trabecular vBMD, and HRpQCT analyses, giving information about trabecular bone microstructure and cortical porosity, were available in the tibia for 729 subjects with genotype data available (Table 4). To determine the impact of the identified genome-wide significant cortical and trabecular vBMD signals for bone microstructure parameters, their associations with HRpQCT parameters were evaluated in the GOOD cohort. Trabecular vBMD as analysed by pQCT was strongly (r = 0.94) associated with trabecular bone fraction (BV/TV) as analysed by HRpQCT. The pQCT-derived cortical vBMD was moderately inversely correlated to cortical porosity as analysed by HRpQCT (r = −0.21).
Tab. 4. Characteristics of the GOOD five year follow-up cohort. 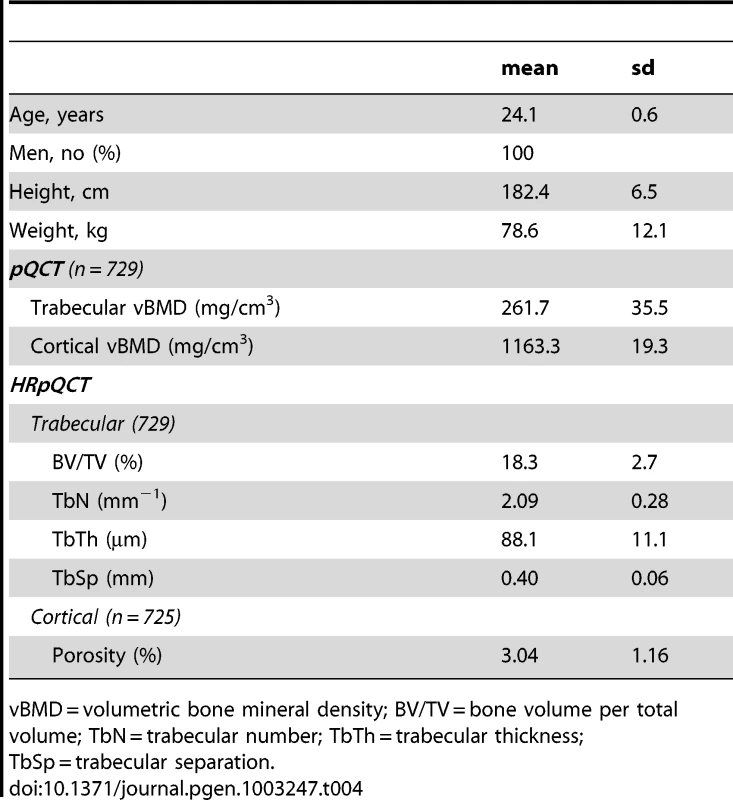
vBMD = volumetric bone mineral density; BV/TV = bone volume per total volume; TbN = trabecular number; TbTh = trabecular thickness; TbSp = trabecular separation. Cortical vBMD SNPs
The four genome-wide significant cortical vBMD SNPs were all associated with (p<0.05) cortical but not trabecular vBMD at the five year follow-up visit of the GOOD cohort and their effect sizes for cortical vBMD were of similar magnitude and direction as seen for the GOOD cohort at the baseline visit (Tables S1 and S3, Figure 6). Interestingly, rs1021188, being the SNP explaining most of the cortical vBMD variation, was also associated with cortical porosity (0.15 SD increase per C allele, p = 3.0×10−2) but, as expected, in the inverse direction compared with the association with cortical vBMD (Figure 6 and Table S3).
Fig. 6. The associations of the SNPs explaining most of the cortical vBMD (rs1021188) and trabecular vBMD variations (rs9287237), respectively, with bone parameters in the GOOD cohort at the follow-up visit (n = 729). 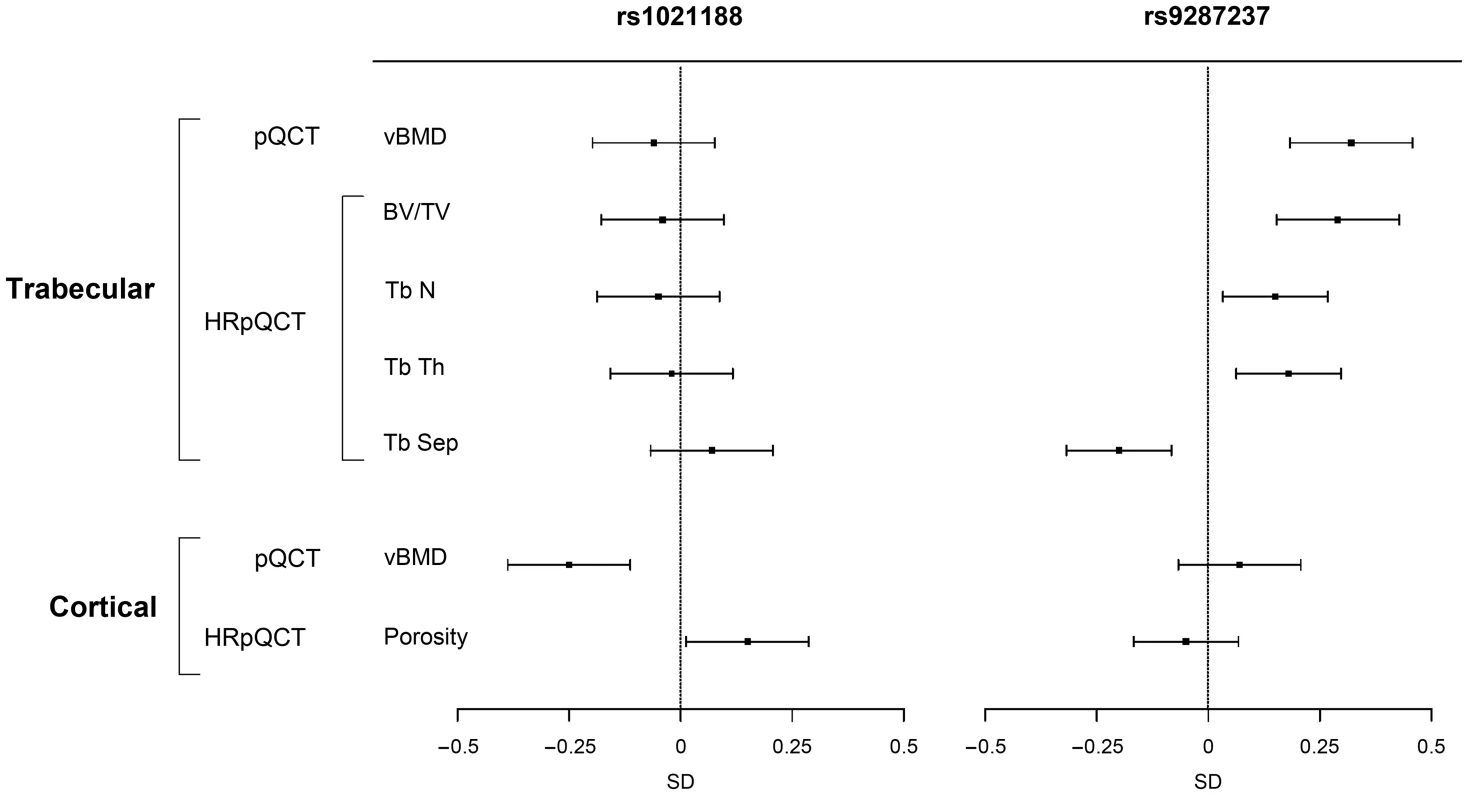
Mean and standard error z-scores are shown for trabecular and cortical vBMDs as analyzed by pQCT, and for trabecular bone volume per total volume (BV/TV), trabecular number (TbN), trabecular thickness (TbTh), trabecular separation (TbSp) and cortical porosity as analyzed by HRpQCT. Trabecular vBMD SNP
The genome-wide significant trabecular vBMD SNP rs9287237 was significantly associated with trabecular but not cortical vBMD at the five year follow-up visit of the GOOD cohort and the effect size (0.32 SD increase per T allele, p = 2.6×10−6) for trabecular vBMD was of similar magnitude and direction as seen for the GOOD cohort at the baseline visit (Tables S1 and S3, Figure 6). This SNP was also significantly associated with trabecular bone fraction (BV/TV) as analyzed by HRpQCT (0.29 SD increase per T allele, p = 1.8×10−5) while it was not significantly associated with cortical porosity (Figure 6). Detailed analysis of trabecular bone microstructure revealed that rs9287237 was not only associated with trabecular bone fraction but also with trabecular number (0.15 SD increase per T allele, p = 1.6×10−2), trabecular thickness (0.18 SD increase per T allele, p = 5.0×10−3) and trabecular spacing (0.20 SD decrease per T allele, p = 1.2×10−3; Figure 6).
Estimation of the genetic correlation between cortical and trabecular vBMD
Although there appeared to be no overlap in the identity of the genome-wide significant SNPs between cortical and trabecular vBMD, it is still possible that there are genetic variants lower down the distribution of tests statistics which do not meet the stringent criteria for genome-wide significance, but nevertheless affect both traits pleiotropically. In order to investigate this possibility we ran a bivariate REML analysis using the GCTA software package in the GOOD cohort, having both cortical and trabecular vBMDs measurements available [14]. GCTA estimated the genetic correlation between trabecular and cortical BMD as rG = 0.0 (SE = 0.39) suggesting an absence of common genetic variants affecting both traits and consistent with our results from the genome-wide association analysis. However, we note that there are relatively few individuals in this analysis and consequently the standard errors on this estimate are very wide. In order to be more definitive with respect to the possible existence of pleiotropy one would need to perform the analysis in a much larger sample of individuals to yield precise estimates of the genetic correlation between the two traits.
Comparison of the impact of identified genome-wide significant SNPs for vBMD and previously described aBMD SNPs
All five genome-wide significant vBMD SNPs were nominally significantly associated (p<0.05) with both femoral neck and lumbar spine aBMD as provided in the public data release from the discovery phase (n≅32,000) of the recent aBMD analyses from the GEFOS consortium (Table 3; http://www.gefos.org/?q=content/data-release) [2]. The direction of the effect was the same when comparing vBMDs and aBMD for four of the SNPs while it was opposite to the one described for aBMD for the cortical vBMD SNP rs271170.
When evaluating the 64 genome-wide significant aBMD SNPs recently identified by the GEFOS consortium [2] it was found that 15 of these were also significantly associated (p<0.05) with cortical vBMD and 15 were significantly associated with trabecular vBMD. Four of these SNPs were associated with both cortical and trabecular vBMDs (Table S4).
eQTL analysis in human osteoblasts
In an attempt to assess the underlying functional mechanism of our identified loci we examined their potential role in regulating gene expression using expression quantitative trait locus (eQTL) data from resting (i.e. untreated) and induced (i.e. dexamethasone, BMP-2 and PGE2 treated) primary human osteoblasts [15], [16]. Expression of genes in close proximity to the five genome-wide significant SNPs (defined as located within the gene ±250 kb) was tested for association (Table S5). We found that the trabecular vBMD-associated SNP (rs9287237) was the strongest SNP significantly associated (P = 2.3×10−4) with expression of the nearby GREM2 gene. No significant effects on gene expression were noted at the additional four loci (Bonferroni adjusted P>0.05 corresponding to 0.05/88 = 5.7×10−4; Table S5).
Association with fractures in MrOS Sweden
Overall, 388 men had at least one validated incident fracture after an average follow-up of 5.4 years in the MrOS Sweden cohort (Table S6). The trabecular vBMD SNP rs9287237, but none of the four cortical vBMD SNPs, was significantly associated with risk of all fractures (HR per extra T allele 0.75, 95% confidence interval (CI) 0.60–0.93) and hip fractures (HR per extra T allele 0.59, 95% CI 0.36–0.98; Table S7).
In two centers of MrOS Sweden, Gothenburg and Malmö, overall, 1445 men had a spine X-ray at baseline, which identified 225 men with prevalent vertebral fractures (15.6%; Table S6). The trabecular vBMD SNP rs9287237, but none of the four cortical vBMD SNPS, was significantly associated with prevalent X-ray verified vertebral fractures (OR per extra T allele 0.68, 95% CI 0.50–0.94; Table S7).
Discussion
Osteoporosis is a common highly heritable skeletal disease characterized by reduced aBMD and deteriorated bone microstructure, resulting in an increased risk of fragility fracture [17]. Most previous genetic epidemiology studies have focused on the genetics of the complex trait aBMD, not being able to separate genetic determinants of trabecular vBMD, cortical vBMD and bone microstructure. However, all these structural traits are determinants of aBMD [7]. We, herein, provide evidence that the genetic determinants of cortical and trabecular vBMDs differ. Separate GWAS meta-analyses of cortical and trabecular vBMDs demonstrated that genetic variants in the RANKL, LOC285735, OPG and ESR1 loci were associated with cortical vBMD while a genetic variant in the FMN2/GREM2 locus was associated with trabecular vBMD. Follow-up analyses in a HRpQCT cohort providing bone microstructural traits revealed that a genetic variant in the RANKL locus is associated with not only cortical vBMD but also with cortical porosity. The FMN2/GREM2 locus, on the other hand, is robustly associated with not only trabecular vBMD but also trabecular number and thickness as well as GREM2 expression in human osteoblasts and fracture risk.
Using the same pQCT cohorts as evaluated in the present study, we have recently identified a missense variant in the WNT16 gene to be associated with cortical bone thickness [18]. Subsequent functional studies revealed that Wnt16 inactivated mice had reduced cortical bone thickness, providing strong evidence that WNT16 is an important regulator of cortical bone thickness. In the present discovery GWAS-meta-analysis, we identified genome-wide significant cortical vBMD signals in four loci and a trabecular vBMD signal in one locus and all these signals were significantly replicated in the MrOS Sweden cohort. One of the identified cortical vBMD loci (LOC285735) and the identified trabecular vBMD locus (FMN2) are novel bone-related loci while the remaining three identified cortical vBMD loci have been previously reported to be associated with aBMD. [2], [6], [19]. Although the number of subjects included in the present discovery GWAS meta-analyses of vBMD (cortical vBMD n = 5878; trabecular vBMD n = 2500) was substantially lower than the number of subjects included in a recent large-scale aBMD GWA meta-analysis (discovery cohort n = 32,961; [2]), the identification of novel bone-related loci in the trabecular and cortical vBMD analyses is not surprising as the correlations between aBMD on the one side and cortical and trabecular vBMDs on the other side are low/modest. In addition, the genetic variants associated with cortical and trabecular bone parameters, respectively, differed, and were also distinct from the WNT16 locus recently identified to be associated with cortical bone thickness [18], underscoring the complexity of the genetics of bone parameters.
Conditional analysis revealed a secondary signal in the RANKL locus and both these cortical vBMD signals were independent of the previously reported aBMD signal (rs9533090; [2]) in this region, demonstrating that separate signals within the same region can have an impact on different bone traits ( = allelic heterogeneity). RANKL exerts its biological effects on bone by stimulating osteoclast differentiation following interactions with its receptor, RANK; how distinct genetic pathways might influence this functionality in different ways, so as to influence distinct phenotypic traits, is currently unclear. Alternatively, one of these signals may be in LD with a marker at a different gene responsible for mediating the genetic effect in question, or else represent a variant which although trans to a structural gene, affects transcription at other sites [20]. The cortical vBMD SNPs rs7839059 (TNFRSF11B locus) was also nominally (p<0.05) significantly associated with trabecular vBMD, although with less pronounced effect size, suggesting that this SNP does not exclusively have an impact on cortical bone. The present report describing two independent RANKL signals and one OPG signal with an impact on cortical vBMD provides further evidence that the RANK/RANKL/OPG axis affects the skeleton at least in part by influencing volumetric apparent density of cortical bone. It is tempting to speculate that changes in cortical vBMD contribute to the recent observations that the RANKL inhibitor denosumab reduces fracture risk [10], [21], [22]. Consistent with this possibility, administration of denosumab has been found to increase femoral cortical vBMD in mice with a knock-in of humanized RANKL [23].
The second strongest genetic signal for cortical vBMD was located on chromosome 6 (rs271170), 93.4 kb upstream of LOC285735. This is a novel bone-related signal and further targeted sequencing efforts and functional studies are required to characterize this signal. Several clinical and preclinical studies have clearly demonstrated that ESR1 is an important regulator of both female and male bone health [24]–[28] but the present study is first to provide genetic evidence that this receptor influences the volumetric apparent density of cortical bone. This finding is of importance as Khosla and co-workers recently proposed that the main physiological target for estrogen in bone is cortical and not trabecular bone [24].
A significant signal (rs9287237) for trabecular vBMD was identified on chromosome 1 located in the intron region of the FMN2 gene. The combined effect size of this signal was substantial with an increase of 0.19 SD per T allele. FMN2 is a gene that is expressed in oocytes and is required for progression through metaphase of meiosis 1 but it is not previously reported to influence the skeleton [29]. However, a genetic variant within FMN2 has been associated with coronary heart disease [30]. The rs9287237 SNP is located slightly (55.7 kb) downstream of GREM2 ( = PRDC), which is an extracellular antagonist of bone morphogenetic proteins (BMPs) and it inhibits osteoblastic differentiation [31], [32], making it an alternative plausible candidate gene underlying the rs9287237 association with trabecular vBMD. Importantly, eQTL analyses in human osteoblasts demonstrated that the trabecular vBMD-associated SNP (rs9287237) was significantly associated with expression of the nearby GREM2 gene, indicating that GREM2 is a strong candidate for mediating the trabecular vBMD association at rs9287237. However, further targeted sequencing efforts and functional studies are required to characterize this signal.
There are known sex differences in bone traits in mice [33]–[36]. Similarly, some genome-wide linkage analyses in humans have reported sex-specific results. In a whole–genome linkage analysis stratified by sex, sex-specific QTLs were found in the Framingham sample [37]. Furthermore, in a meta-analysis that included data from nine whole-genome linkage scans for aBMD, several sex-specific QTLs were observed [38]. To our knowledge there is only one reported genome-wide significant aBMD signal, located on the X-chromosome (Xp22.31), which displays significant sex heterogeneity [2]. This signal was only significant in men and the same signal was also shown to be associated with male serum testosterone levels [39]. Sex-specific analyses in the present study revealed that all identified cortical vBMD signals were significantly associated with cortical vBMD in both men and women with effects in the same direction. Nevertheless, the magnitude of the effect sizes differed significantly according to sex for three of the identified hits. Importantly, the effect sizes of the RANKL and ESR1 signals were more than three (0.21 SD vs. 0.06 SD) and two (0.12 vs. 0.05 SD) times larger, respectively, in men than in women. The smaller effect within females observed for rs1021188 in the RANKL region is mainly driven by ALSPAC, and there may be other reasons (such as younger age) why this study showed a smaller effect. However, the consistent results between ALSPAC and the YFS provide some evidence against the differences being driven primarily by age. The relative strong ESR1 signal in men supports experimental and clinical studies, demonstrating that estrogens are crucial for male bone health [24], [25], [27], [40].
We examined genetic effects across cohorts encompassing a relatively broad age range, including 15 year old participants from ALSPAC who were still attaining peak bone mass, to older men from MrOS Sweden starting to show age-related bone loss. Inclusion of an older cohort had the advantage of providing an opportunity to study relationships with fracture risk. However, this design may have reduced the power to detect genetic associations by introducing greater heterogeneity. In contrast to aBMD, vBMD has been reported to change relatively little from adolescence to mid-life suggesting that analyses combining cohorts of different ages might be more informative when based on this trait [41]. However, recent follow up studies based on the GOOD cohort revealed substantial changes in cortical vBMD in the late teens and early twenties, at least in males [42]. Hence the suggestion that certain genetic associations with cortical vBMD were weaker in ALSPAC compared with other cohorts may reflect attenuation of effect during the consolidation of cortical bone whilst attaining peak bone mass.
Age-related changes in bone include microstructural deterioration, such as trabecular perforation, thinning, and loss of connectivity, as well as increased cortical porosity [8], [9]. These bone microstructural parameters are believed to have an aBMD-independent influence on fracture risk and they can be analyzed by HRpQCT. The present study is the first to identify genetic loci associated with cortical and trabecular bone microstructural parameters as analyzed by HRpQCT. The SNP in the RANKL region with the strongest association with cortical vBMD was also significantly associated with cortical porosity but, as expected, in the reverse direction. This finding suggests that a genetic variant in the RANKL locus influences cortical vBMD, at least partly, via effects on cortical porosity. Importantly, this signal in the RANKL region was independent from the previously reported aBMD signal in the same region [2]. Analyses of trabecular bone microstructure demonstrated that the trabecular vBMD SNP rs9287237 in the FMN2/GREM2 locus was significantly associated with several trabecular but not cortical bone microstructure parameters. When evaluated in the five-year follow-up visit in the GOOD cohort, each T allele of this SNP resulted in a substantial increase in trabecular vBMD (0.32 SD), trabecular bone fraction (BV/TV 0.29 SD), trabecular number (0.15 SD), and trabecular thickness (0.18 SD). Thus, a genetic variant in the FMN2/GREM2 locus influences trabecular vBMD via substantial effects on both trabecular number and thickness. Although, the present study is the first to report on genetic variants associated with microstructural bone-parameters, the analyses were candidate-based as a follow-up of our initial cortical and trabecular vBMD GWA meta-analyses. In order to identify novel genetic loci for bone microstructural parameters in a hypothesis-free manner, well-powered HRpQCT cohorts with genome-wide genotype data available need to be established.
We believe that our study provides strong evidence that previous large-scale GWA meta-analyses of the complex bone trait aBMD did not have the capability to identify a number of important loci with an impact on aspects of micro-architecture which may have important effects on fracture risk but be poorly reflected by overall aBMD measurements. We, therefore, propose that future well-powered pQCT and HRpQCT GWA meta-analyses of these specific bone structural traits will add useful information and might result in the identification of novel osteoporosis drug targets and provide novel aBMD-independent genetic markers for the prediction of fracture risk.
The implication of our results suggesting that cortical and trabecular bone compartments are under distinct genetic control is consistent with the fact that patients with idiopathic osteoporosis may present with a predominantly trabecular or cortical bone phenotype [43]. Although the lumbar spine and hip both comprise a mixture of bone types, the former has a relatively high proportion of trabecular bone, whereas the hip has a higher proportion of cortical bone. Hence, patients presenting with a disproportionate decrease in lumbar spine aBMD, which are well recognized, presumably have greater reductions in trabecular compared to cortical BMD [44]. Further studies are required to determine whether genetic variation in the FMN2/GREM2 locus helps to explain this type of presentation.
The genetic variant in the FMN2/GREM2 locus was associated with fracture risk and prevalent X-ray verified vertebral fractures in the MrOS Sweden cohort. However, further large-scale studies are required to replicate the fracture findings of this SNP. Collectively our data demonstrate that each extra T allele of rs9287237 is associated with decreased expression of the BMP antagonist GREM2 in osteoblasts, increased trabecular vBMD and decreased fracture risk. As previous in vitro studies have demonstrated that GREM2 inhibits osteoblast differentiation, we propose that rs9287237 is involved in the regulation of GREM2 expression in osteoblasts, which in turn regulates osteoblast differentiation and thereby the amount of trabecular bone and fracture risk [31], [32].
In conclusion, we identified five genetic loci associated with trabecular or cortical vBMD. Two of these (FMN2/GREM2 and LOC285735) are novel bone-related loci while the other three have previously been reported to be associated with aBMD. The genetic variants reported to be associated with cortical and trabecular bone parameters differed and were also distinct from the WNT16 locus recently identified to be associated with cortical thickness [18], underscoring the complexity of the genetics of bone parameters. Finally, we propose that a genetic variant in the RANKL locus influences cortical vBMD, at least partly, via effects on cortical porosity, and that a genetic variant in the FMN2/GREM2 locus influences trabecular vBMD and fracture risk via substantial effects on both trabecular thickness and number.
Materials and Methods
Ethics statement
All study participants provided informed written consent. Approval by local institutional review boards was obtained in all studies.
GOOD cohort
Participants
The Gothenburg Osteoporosis and Obesity Determinants (GOOD) study was initiated to determine both environmental and genetic factors involved in the regulation of bone and fat mass [45], [46]. Young, Caucasian men were randomly identified in the greater Gothenburg area in Sweden using national population registers, contacted by telephone, and invited to participate. Enrolled subjects were between 18 and 20 years of age. There were no other exclusion criteria, and 49% of the study candidates agreed to participate (n = 1,068). Five years later, the study subjects in the original GOOD study were contacted by letter and telephone and invited to participate in the 5-yr follow-up study. Of the original 1,068 subjects, 833 men, 24.1±0.6 yr of age, were included in the follow-up study (78% of the original population) [42]. The GOOD study was approved by the local ethics committee at University of Gothenburg. Oral and written informed consent was obtained from all study participants. Weight was measured to the nearest 0.1 kg and height was measured using a wall-mounted stadiometer.
pQCT measurements
Cortical volumetric BMD (not including the bone marrow) was measured on a single tibial diaphyseal slice (at 25% of the bone length in the proximal direction of the distal end) using the Stratec XCT2000 (Germany) at both the baseline visit and the five-year follow-up visit in the GOOD cohort [42], [46]. A threshold routine was used for defining cortical bone, which specified a voxel with a density >710 mg/cm3 as cortical bone. Trabecular vBMD (mg/cm3) was measured using a scan through the metaphysis (at 4% of the bone length in the proximal direction of the distal end) of the tibia. Tibia length was measured from the medial malleolus to the medial condyle. The CVs were <1% for all pQCT measurements. Both pQCT measurements and genotype information were available for 938 study subjects.
HRpQCT measurements
A high-resolution 3D pQCT device (XtremeCT; Scanco Medical AG, Brüttisellen, Switzerland) was used to scan the ultradistal tibia of the nondominant leg at the five-year follow-up visit in the GOOD cohort [47]. Anatomically formed carbon fiber shells, especially designed for each type of limb (Scanco Medical), were used to immobilize the subject's leg during the scan. The measurements of the volume of interest in the ultradistal tibia, 1 cm in the proximal direction and the whole cross-section in transversal direction, were carried out according to a standardized protocol previously described [47]. Briefly, a reference line was manually placed at the center of the end plate of the distal tibia. The first tomographic slice started 22.5 mm proximal to the reference line. A total of 110 parallel slices, with a nominal isotropic resolution (voxel size) of 82 µm, were obtained, delivering a 3D representation of an approximately 9-mm section of the tibia in the proximal direction. The entire volume of interest was automatically separated into cortical and trabecular region. From this separation and by previously described methods to process the data, we obtained trabecular bone volume fraction (BV/TV, percent), trabecular number (TbN; millimeters−1), trabecular thickness (TbTh; micrometers) and trabecular separation (TbSp; micrometers). For the cortical porosity, we used the manufacturer's software, which uses an approach published recently by several groups [48], [49] and described in detail by Burghardt and colleagues [8], [50]. The coefficients of variation (CVs) for the used bone measurements were obtained by three repeated measurements according to the standardized protocol on two subjects. The CVs ranged from 0.04 to 1.6%. Both HRpQCT measurements and genotype information were available for 729 study subjects.
Discovery set genotyping
Genotyping was performed with Illumina HumanHap610 arrays at the Genetic Laboratory, Department of Internal Medicine, Erasmus Medical Center, Rotterdam, the Netherlands. Genotypes were called using the BeadStudio calling algorithm. Genotypes from 938 individuals passed the sample quality control criteria (exclusion criteria: sample call rate <97.5%, excess autosomal heterozygosity >0.33 (FDR <0.1%), duplicates and/or first degree relatives identified using IBS probabilities (>97%), ethnic outliers (3 SD away from the population mean) using multi-dimensional scaling analysis with four principal coordinates). Across 22 duplicate samples, genotype concordance exceeded 99.9%. We carried out imputation to HapMap (NCBI build 36, release 21 and 22 for X chromosome and autosomes, respectively) (after excluding SNPs with MAF <1%, SNP call rate <98% and HWE p value<1×10−6) using Mach 1.0, Markov Chain Haplotyping, giving a total of 2,608,508 SNPs.
YFS cohort
Participants
The Cardiovascular Risk in Young Finns Study (YFS) is an ongoing multi-centre follow-up of atherosclerosis risk factors in young Finns of Caucasian origin [51]. The first cross-sectional survey conducted in 1980 comprised a total of 3,596 subjects (83% of those invited) aged 3, 6, 9, 12, 15 and 18 years. The subjects were randomly selected from the national population register from five university cities in Finland (Helsinki, Turku, Tampere, Kuopio and Oulu) and the rural municipalities in their vicinity. In 2008, 1,884 subjects (1,058 women and 826 men) aged 31–46 years participated in pQCT measurements organized in five study centers (Turku, Helsinki, Tampere, Oulu and Kuopio) between February and December 2008. Trained technologists in each center performed the measurements. The same pQCT device was used in all study centers (Stratec XCT 2000R). Pregnant women were excluded from the pQCT measurements. Subjects gave written informed consent. The study protocol was approved by the local ethics committees of the participating universities and complied with national legislation.
pQCT measurements
Cortical volumetric BMD was measured on a single tibia diaphyseal slice (at 30% of the bone length in the proximal direction of the distal endplate ot the tibia) using the Stratec XCT2000 (Germany). A threshold routine was used for defining cortical bone, which specified a voxel with a density >710 mg/cm3 as cortical bone. Trabecular vBMD (mg/cm3) was measured using a scan through the metaphysis (at 5% of the bone length in the proximal direction of the distal endplate) of the tibia. Tibia length was measured from the medial malleolus to the medial condyle. The CVs were <2.6% for all pQCT measurements. Both pQCT measurements and genotype information were available for 1558 study subjects.
Discovery set genotyping
Genomic DNA was extracted from peripheral blood leukocytes using a commercially available kit and Qiagen BioRobot M48 Workstation according to the manufacturer's instructions (Qiagen, Hilden, Germany). Genotyping was done for 2556 samples using custom build Illumina Human 670 k BeadChip at the Welcome Trust Sanger Institute. Genotypes were called using the Illuminus clustering algorithm. 56 samples failed Sanger genotyping pipeline QC criteria (i.e., duplicated samples, heterozygosity, low call rate, or Sequenom fingerprint discrepancy). From the remaining 2500 samples one sample failed gender check, three were removed due to low genotyping call rate (<0.95) and 54 samples for possible relatedness (pi-hat>0.2). 11,766 SNPs were excluded based on HWE test (p< = 1×10−6), 7,746 SNPs failed a missingness test (call rate <0.95) and 34,596 SNPs failed frequency test (MAF<0.01). After quality control there were 2,442 samples and 546,677 genotyped SNPs available for further analysis. Genotype imputation was performed using MACH 1.0 and HapMap II CEU (NCBI build 36, release 21 and 22 for X chromosome and autosomes, respectively) samples as the reference set.
ALSPAC cohort
Participants
The Avon Longitudinal Study of Parents and their Children (ALSPAC) is a geographically based birth cohort study investigating factors influencing the health, growth, and development of mainly Caucasian children [52]. All pregnant women resident within a defined part of the former county of Avon in South West England with an expected date of delivery between April 1991 and December 1992 were eligible for recruitment, of whom 14,541 were enrolled (http://www.alspac.bris.ac.uk). Both mothers and children have been extensively followed from the 8th gestational week onwards using a combination of self-reported questionnaires, medical records and physical examinations. Ethical approval was obtained from the ALSPAC Law and Ethics committee and relevant local ethics committees, written informed consent was provided by all parents. Blood samples were taken and DNA extracted as previously described [53]. Height was measured using a Harpenden stadiometer (Holtain Limited, Wales) and weight using a Tanita Body Fat Analyser.
pQCT measurements
pQCT scans were performed on approximately 4,500 children when they attended the age 15 research clinic. Cortical vBMD was measured on a single slice at the mid tibia (50%) using the Stratec XCT2000L (Germany). A threshold routine was used for defining cortical bone, which specified a voxel with a density >650 mg/cm3 as cortical bone. Of the 4500 pQCT scans obtained in ALSPAC 89 were rejected as being of insufficient quality. The CVs for cortical volumetric BMD based on 139 ALSPAC subjects scanned a mean of 31 days apart, was 1.3%.
Discovery set genotyping
9,912 ALSPAC individuals were genotyped using the Illumina HumanHap550 quad genome-wide SNP genotyping platform by 23andMe, subcontracting the Wellcome Trust Sanger Institute, Cambridge, UK and the Laboratory Corporation of America, Burlington, NC, US. Markers with <1% minor allele frequency >5% missing genotypes or which failed an exact test of Hardy-Weinberg equilibrium (HWE) (p<5×10−7) were excluded from further analysis. We also excluded any individuals who did not cluster with the CEU individuals in multidimensional scaling analysis, who had >3% missing data, minimal or excessive heterozygosity (>34.5% or <32% for the Sanger data and >33% or <31% for the LabCorp data), evidence of cryptic relatedness (>10% IBD) and any individuals with incorrect gender assignments. After data cleaning we were left with 8365 unrelated individuals with genome-wide genotyping, and 500,541 SNPs. We carried out imputation using MACH 1.0.16, Markov Chain Haplotyping, using CEPH individuals from phase 2 of the HapMap project as a reference set (NCBI build 36, release 21 and 22 for X chromosome and autosomes, respectively). Of the 8,365 individuals with imputed genotype data, 3,382 also had pQCT data.
MrOS Sweden cohort (replication)
Participants
The Osteoporotic Fractures in Men (MrOS) study is a prospective multicenter study including older Caucasian men in Sweden (n = 3,014), Hong Kong (>2,000), and the United States (>6,000). In the present study, associations between candidate polymorphisms and skeletal parameters were investigated in the Swedish cohort, which consists of three sub-cohorts from three different Swedish cities (n = 1,005 in Malmö, n = 1,010 in Gothenburg, and n = 999 in Uppsala) [54]. Study subjects were randomly identified using national population registers, contacted and asked to participate. To be eligible for the study, the subjects had to be able to walk without assistance, provide self-reported data, and sign an informed consent; there were no other exclusion criteria. The study was approved by the ethics committees at the Universities of Gothenburg, Lund, and Uppsala. Informed consent was obtained from all study participants.
pQCT measurements
Cortical volumetric BMD (not including the bone marrow) was measured on a single tibial diaphyseal slice (at 38% of the bone length in the proximal direction of the distal end) using the Stratec XCT2000 (Germany) [10]. A threshold routine was used for defining cortical bone, which specified a voxel with a density >710 mg/cm3 as cortical bone. Trabecular vBMD (mg/cm3) was measured using a scan through the metaphysis (at 4% of the bone length in the proximal direction of the distal end) of the tibia. Tibia length was measured from the medial malleolus to the medial condyle. The CVs were <1% for all pQCT measurements. Adjustments for study centre were performed.
Replication set genotyping
Genotyping of SNPs identified in the GWAS meta-analysis was carried out at KBioscience using a competitive allele specific PCR (KASP) genotyping chemistry. The genotyping call rate was >97%.
eQTL analysis in human osteoblasts
SNPs associated with vBMD at the genome-wide significance level as reported here were tested for association with resting or induced gene expression of neighbouring gene transcripts, in primary human osteoblasts derived from 113 (51 female and 62 male donors, respectively) unrelated Swedish donors. Detailed cell culture and analysis methods have been described in detail [15], [16]. Briefly, expression profiling of untreated, dexamethasone, BMP-2 and PGE2-treated cells each with up to three biological replicates was performed using the Illumina HumRef-8 BeadChips according to the protocol supplied by the manufacturer. Genotyping for genotype-expression association was performed using Illumina HapMap 550 k Duo chip. Individuals with low genotyping rate and SNPs showing significant deviation from Hardy-Weinberg equilibrium (P<0.05) were excluded. Similarly low frequency (MAF<0.05) SNPs and SNPs with high rates of missing data were excluded. Genotypes from samples that passed quality control (N = 103) were imputed for all SNPs (n = 478,805) oriented to the positive strand from phased (autosomal) chromosomes of the HapMap CEU Phase II panel (release 22, build 36) using MACH 1.0) [55]. A cut-off of R2<0.3 were used to remove poorly imputed markers. Association of imputed genotypes using estimated genotype probabilities with nearby expression traits (defined as ±250 kb window flanking the gene) were performed using a linear regression model implemented in the MACH2QTL software with sex and age as covariates.
Assessment of incident fractures in MrOS Sweden
Participants were followed for 5.4 years on average after the baseline examination. The follow-up time was recorded from the date of the baseline visit to the date of the first fracture, the date of death, or end of the present follow-up interval. When a subject sustained a first fracture at different sites during the follow-up, the various fractures and the follow-up time for each respective first fracture type were included in the analyses. Complete follow-up was possible because central registers covering all Swedish citizens were used to identify the subjects and the time of death for all subjects who died during the study, and these analyses were performed after the time of fracture validation. At the time of fracture evaluation, the computerized X-ray archives in Malmö, Göteborg, and Uppsala were searched for new fractures occurring after the baseline visit, using the unique personal registration number, which all Swedish citizens have. All fractures reported by the study subject after the baseline visit were confirmed by physician review of radiology reports. Fractures reported by the study subject, but not possible to confirm by X-ray analyses report, were not included in this study. All validated fractures and hip fractures were evaluated. Information about the type of trauma associated with the incident fractures was not available. Fracture rates were expressed as the number of subjects with first fractures per 1000 person-years (Table S6).
Assessment of prevalent vertebral fractures in MrOS Sweden
In Gothenburg and Malmö, 1445 men had an X-ray of the lateral thoracic and lumbar spine at baseline. All vertebral fractures were evaluated by an expert radiologist. If the vertebral body had a reduced height of 3 mm or more compared with the vertebra above, it was classified as a vertebral fracture [56].
Statistical methods
The ALSPAC (n = 3382), YFS (n = 1558) and GOOD (n = 938) discovery cohorts contributed to the cortical vBMD genome-wide meta-analysis while the YFS and GOOD discovery cohorts contributed to the trabecular vBMD genome-wide meta-analysis. We analyzed only those imputed SNPs which had a minor allele frequency of >0.01 and an r2 imputation quality score of >0.3 in all 3 sets (n = 2,401,124). We carried out genome-wide association analyses for cortical and trabecular vBMDs using additive linear regression in Mach2QTL for ALSPAC, ProbABEL [57] for YFS and Mach2QTL on GRIMP [19] for the GOOD analyses. We included age, sex, height and weight(ln) as covariates. We carried out meta-analyses of the results from the three cohorts using the inverse variance method in METAL. Standardized betas and standard errors from each study were combined using a fixed effect model which weights the studies using the inverse variance and applying genomic control to individual studies and the combined results. Genome-wide significance was taken to be p<5×10−8. We also repeated the analyses in each of the three discovery cohorts, conditional on these top SNPs, to identify any additional independent associations in the regions. We selected one SNP for replication in the MrOS Sweden cohort from each independent region that had a p<5×10−8 as well as a secondary SNP from the RANKL region which appeared to influence cortical vBMD. Additive linear regression analyses were carried out for the associations between these SNPs and cortical and trabecular vBMDs in SPSS Statistics 17.0 for MrOS Sweden, using age, sex, height and weight(ln) as covariates. The results of all four cohorts were combined using a fixed effects inverse-variance meta-analysis in Stata (version 11.2). The SNPs showing evidence for heterogeneity (as assessed by a chi-squared test) were also meta-analysed using the DerSimonian & Laird random effects method. Correlations between bone traits in the GOOD cohort were tested and presented as Spearman's rank correlation coefficients (rho). The difference of the allelic association effects between males and females was tested using a two sample z-test.
Cox proportional hazards models were used to study the associations between SNPs and incident fractures. Prevalent vertebral fractures were analyzed using binary logistic regression models.
Estimation of the genetic correlation between cortical and trabecular vBMD
In order to estimate the genetic correlation between cortical and trabecular vBMD we ran a bivariate REML analysis using the GCTA software package in the GOOD cohort, having both cortical and trabecular vBMDs measurements available [14]. This method essentially decomposes the covariance between the two traits into a part explained by common genetic variation that is tagged by markers on the SNP chip and a residual part that is not. It is then possible to use these estimates to estimate a genetic correlation between the two.
Supporting Information
Zdroje
1. DuncanEL, DanoyP, KempJP, LeoPJ, McCloskeyE, et al. (2011) Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet 7: e1001372 doi:10.1371/journal.pgen.1001372.
2. EstradaK, StyrkarsdottirU, EvangelouE, HsuYH, DuncanEL, et al. (2012) Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 44 : 491–501.
3. RivadeneiraF, StyrkarsdottirU, EstradaK, HalldorssonBV, HsuYH, et al. (2009) Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet 41 : 1199–1206.
4. StyrkarsdottirU, HalldorssonBV, GretarsdottirS, GudbjartssonDF, WaltersGB, et al. (2008) Multiple genetic loci for bone mineral density and fractures. N Engl J Med 358 : 2355–2365.
5. StyrkarsdottirU, HalldorssonBV, GretarsdottirS, GudbjartssonDF, WaltersGB, et al. (2009) New sequence variants associated with bone mineral density. Nat Genet 41 : 15–17.
6. RichardsJB, RivadeneiraF, InouyeM, PastinenTM, SoranzoN, et al. (2008) Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet 371 : 1505–1512.
7. SievanenH (2000) A physical model for dual-energy X-ray absorptiometry–derived bone mineral density. Invest Radiol 35 : 325–330.
8. NicksKM, AminS, AtkinsonEJ, RiggsBL, MeltonLJ3rd, et al. (2012) Relationship of age to bone microstructure independent of areal bone mineral density. J Bone Miner Res 27 : 637–644.
9. ZebazeRM, Ghasem-ZadehA, BohteA, Iuliano-BurnsS, MiramsM, et al. (2010) Intracortical remodelling and porosity in the distal radius and post-mortem femurs of women: a cross-sectional study. Lancet 375 : 1729–1736.
10. PaternosterL, LorentzonM, VandenputL, KarlssonMK, LjunggrenO, et al. (2010) Genome-wide association meta-analysis of cortical bone mineral density unravels allelic heterogeneity at the RANKL locus and potential pleiotropic effects on bone. PLoS Genet 6: e1001217 doi:10.1371/journal.pgen.1001217.
11. HavillLM, MahaneyMC, TLB, SpeckerBL (2007) Effects of genes, sex, age, and activity on BMC, bone size, and areal and volumetric BMD. J Bone Miner Res 22 : 737–746.
12. LaibA, RuegseggerP (1999) Calibration of trabecular bone structure measurements of in vivo three-dimensional peripheral quantitative computed tomography with 28-microm-resolution microcomputed tomography. Bone 24 : 35–39.
13. MacNeilJA, BoydSK (2007) Accuracy of high-resolution peripheral quantitative computed tomography for measurement of bone quality. Med Eng Phys 29 : 1096–1105.
14. YangJ, BenyaminB, McEvoyBP, GordonS, HendersAK, et al. (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42 : 565–569.
15. GrundbergE, AdoueV, KwanT, GeB, DuanQL, et al. (2011) Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS Genet 7: e1001279 doi:10.1371/journal.pgen.1001279.
16. GrundbergE, KwanT, GeB, LamKC, KokaV, et al. (2009) Population genomics in a disease targeted primary cell model. Genome Res 19 : 1942–1952.
17. Consensus Development Conference on Osteoporosis. Hong Kong, April 1–2, 1993. Am J Med 95 : 1S–78S.
18. ZhengHF, TobiasJH, DuncanE, EvansDM, ErikssonJ, et al. (2012) WNT16 Influences Bone Mineral Density, Cortical Bone Thickness, Bone Strength, and Osteoporotic Fracture Risk. PLoS Genet 8: e1002745 doi:10.1371/journal.pgen.1002745.
19. EstradaK, AbuseirisA, GrosveldFG, UitterlindenAG, KnochTA, et al. (2009) GRIMP: a web - and grid-based tool for high-speed analysis of large-scale genome-wide association using imputed data. Bioinformatics 25 : 2750–2752.
20. GrundbergE, SmallKS, HedmanAK, NicaAC, BuilA, et al. (2012) Mapping cis - and trans-regulatory effects across multiple tissues in twins. Nat Genet 44 : 1084–1089.
21. CummingsSR, San MartinJ, McClungMR, SirisES, EastellR, et al. (2009) Denosumab for prevention of fractures in postmenopausal women with osteoporosis. N Engl J Med 361 : 756–765.
22. SeemanE, DelmasPD, HanleyDA, SellmeyerD, CheungAM, et al. (2010) Microarchitectural deterioration of cortical and trabecular bone: differing effects of denosumab and alendronate. J Bone Miner Res 25 : 1886–1894.
23. KostenuikPJ, NguyenHQ, McCabeJ, WarmingtonKS, KuraharaC, et al. (2009) Denosumab, a fully human monoclonal antibody to RANKL, inhibits bone resorption and increases BMD in knock-in mice that express chimeric (murine/human) RANKL. J Bone Miner Res 24 : 182–195.
24. KhoslaS, MeltonLJ3rd, RiggsBL (2011) The unitary model for estrogen deficiency and the pathogenesis of osteoporosis: is a revision needed? J Bone Miner Res 26 : 441–451.
25. MellstromD, VandenputL, MallminH, HolmbergAH, LorentzonM, et al. (2008) Older men with low serum estradiol and high serum SHBG have an increased risk of fractures. J Bone Miner Res 23 : 1552–1560.
26. SmithEP, BoydJ, FrankGR, TakahashiH, CohenRM, et al. (1994) Estrogen resistance caused by a mutation in the estrogen-receptor gene in a man. N Engl J Med 331 : 1056–1061.
27. VandenputL, OhlssonC (2009) Estrogens as regulators of bone health in men. Nat Rev Endocrinol 5 : 437–443.
28. VanderschuerenD, VandenputL, BoonenS, LindbergMK, BouillonR, et al. (2004) Androgens and bone. Endocr Rev 25 : 389–425.
29. LeaderB, LimH, CarabatsosMJ, HarringtonA, EcsedyJ, et al. (2002) Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nat Cell Biol 4 : 921–928.
30. Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447 : 661–678.
31. IdenoH, TakanabeR, ShimadaA, ImaizumiK, ArakiR, et al. (2009) Protein related to DAN and cerberus (PRDC) inhibits osteoblastic differentiation and its suppression promotes osteogenesis in vitro. Exp Cell Res 315 : 474–484.
32. ImJ, KimH, KimS, JhoEH (2007) Wnt/beta-catenin signaling regulates expression of PRDC, an antagonist of the BMP-4 signaling pathway. Biochem Biophys Res Commun 354 : 296–301.
33. BeamerWG, ShultzKL, Ackert-BicknellCL, HortonLG, DelahuntyKM, et al. (2007) Genetic dissection of mouse distal chromosome 1 reveals three linked BMD QTLs with sex-dependent regulation of bone phenotypes. J Bone Miner Res 22 : 1187–1196.
34. EdderkaouiB, BaylinkDJ, BeamerWG, ShultzKL, WergedalJE, et al. (2007) Genetic regulation of femoral bone mineral density: complexity of sex effect in chromosome 1 revealed by congenic sublines of mice. Bone 41 : 340–345.
35. IshimoriN, StylianouIM, KorstanjeR, MarionMA, LiR, et al. (2008) Quantitative trait loci for BMD in an SM/J by NZB/BlNJ intercross population and identification of Trps1 as a probable candidate gene. J Bone Miner Res 23 : 1529–1537.
36. OrwollES, BelknapJK, KleinRF (2001) Gender specificity in the genetic determinants of peak bone mass. J Bone Miner Res 16 : 1962–1971.
37. KarasikD, CupplesLA, HannanMT, KielDP (2003) Age, gender, and body mass effects on quantitative trait loci for bone mineral density: the Framingham Study. Bone 33 : 308–316.
38. IoannidisJP, NgMY, ShamPC, ZintzarasE, LewisCM, et al. (2007) Meta-analysis of genome-wide scans provides evidence for sex - and site-specific regulation of bone mass. J Bone Miner Res 22 : 173–183.
39. OhlssonC, WallaschofskiH, LunettaKL, StolkL, PerryJR, et al. (2011) Genetic determinants of serum testosterone concentrations in men. PLoS Genet 7: e1002313 doi:10.1371/journal.pgen.1002313.
40. VidalO, LindbergMK, HollbergK, BaylinkDJ, AnderssonG, et al. (2000) Estrogen receptor specificity in the regulation of skeletal growth and maturation in male mice. Proc Natl Acad Sci U S A 97 : 5474–5479.
41. HenryYM, FatayerjiD, EastellR (2004) Attainment of peak bone mass at the lumbar spine, femoral neck and radius in men and women: relative contributions of bone size and volumetric bone mineral density. Osteoporos Int 15 : 263–273.
42. OhlssonC, DarelidA, NilssonM, MelinJ, MellstromD, et al. (2011) Cortical consolidation due to increased mineralization and endosteal contraction in young adult men: a five-year longitudinal study. J Clin Endocrinol Metab 96 : 2262–2269.
43. LarocheM (2012) Pattern of bone mineral density in idiopathic male osteoporosis. Rheumatol Int 32 : 3093–3096.
44. LeslieWD, LixLM, JohanssonH, OdenA, McCloskeyE, et al. (2011) Spine-hip discordance and fracture risk assessment: a physician-friendly FRAX enhancement. Osteoporos Int 22 : 839–847.
45. LorentzonM, MellstromD, OhlssonC (2005) Age of attainment of peak bone mass is site specific in Swedish men–The GOOD study. J Bone Miner Res 20 : 1223–1227.
46. LorentzonM, SwansonC, AnderssonN, MellstromD, OhlssonC (2005) Free testosterone is a positive, whereas free estradiol is a negative, predictor of cortical bone size in young Swedish men: the GOOD study. J Bone Miner Res 20 : 1334–1341.
47. NilssonM, OhlssonC, SundhD, MellstromD, LorentzonM (2010) Association of physical activity with trabecular microstructure and cortical bone at distal tibia and radius in young adult men. J Clin Endocrinol Metab 95 : 2917–2926.
48. BurghardtAJ, KazakiaGJ, RamachandranS, LinkTM, MajumdarS (2010) Age - and gender-related differences in the geometric properties and biomechanical significance of intracortical porosity in the distal radius and tibia. J Bone Miner Res 25 : 983–993.
49. NishiyamaKK, MacdonaldHM, BuieHR, HanleyDA, BoydSK (2010) Postmenopausal women with osteopenia have higher cortical porosity and thinner cortices at the distal radius and tibia than women with normal aBMD: an in vivo HR-pQCT study. J Bone Miner Res 25 : 882–890.
50. BurghardtAJ, BuieHR, LaibA, MajumdarS, BoydSK (2010) Reproducibility of direct quantitative measures of cortical bone microarchitecture of the distal radius and tibia by HR-pQCT. Bone 47 : 519–528.
51. RaitakariOT, JuonalaM, RonnemaaT, Keltikangas-JarvinenL, RasanenL, et al. (2008) Cohort profile: the cardiovascular risk in Young Finns Study. Int J Epidemiol 37 : 1220–1226.
52. BoydA, GoldingJ, MacleodJ, LawlorDA, FraserA, et al. (2012) Cohort Profile: The ‘Children of the 90s’–the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol
53. JonesRW, RingS, TyfieldL, HamvasR, SimmonsH, et al. (2000) A new human genetic resource: a DNA bank established as part of the Avon longitudinal study of pregnancy and childhood (ALSPAC). Eur J Hum Genet 8 : 653–660.
54. MellstromD, JohnellO, LjunggrenO, ErikssonAL, LorentzonM, et al. (2006) Free testosterone is an independent predictor of BMD and prevalent fractures in elderly men: MrOS Sweden. J Bone Miner Res 21 : 529–535.
55. LiYA, G.R. (2006) Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet S79 : 2290.
56. JutbergerH, LorentzonM, Barrett-ConnorE, JohanssonH, KanisJA, et al. (2010) Smoking predicts incident fractures in elderly men: Mr OS Sweden. J Bone Miner Res 25 : 1010–1016.
57. AulchenkoYS, StruchalinMV, van DuijnCM (2010) ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 11 : 134.
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



