-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
An enormous number of alternative pre–mRNA splicing patterns in multicellular organisms are coordinately defined by a limited number of regulatory proteins and cis elements. Mutually exclusive alternative splicing should be strictly regulated and is a challenging model for elucidating regulation mechanisms. Here we provide models of the regulation of two sets of mutually exclusive exons, 4a–4c and 7a–7b, of the Caenorhabditis elegans uncoordinated (unc)-32 gene, encoding the a subunit of V0 complex of vacuolar-type H+-ATPases. We visualize selection patterns of exon 4 and exon 7 in vivo by utilizing a trio and a pair of symmetric fluorescence splicing reporter minigenes, respectively, to demonstrate that they are regulated in tissue-specific manners. Genetic analyses reveal that RBFOX family RNA–binding proteins ASD-1 and FOX-1 and a UGCAUG stretch in intron 7b are involved in the neuron-specific selection of exon 7a. Through further forward genetic screening, we identify UNC-75, a neuron-specific CELF family RNA–binding protein of unknown function, as an essential regulator for the exon 7a selection. Electrophoretic mobility shift assays specify a short fragment in intron 7a as the recognition site for UNC-75 and demonstrate that UNC-75 specifically binds via its three RNA recognition motifs to the element including a UUGUUGUGUUGU stretch. The UUGUUGUGUUGU stretch in the reporter minigenes is actually required for the selection of exon 7a in the nervous system. We compare the amounts of partially spliced RNAs in the wild-type and unc-75 mutant backgrounds and raise a model for the mutually exclusive selection of unc-32 exon 7 by the RBFOX family and UNC-75. The neuron-specific selection of unc-32 exon 4b is also regulated by UNC-75 and the unc-75 mutation suppresses the Unc phenotype of the exon-4b-specific allele of unc-32 mutants. Taken together, UNC-75 is the neuron-specific splicing factor and regulates both sets of the mutually exclusive exons of the unc-32 gene.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003337
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003337Summary
An enormous number of alternative pre–mRNA splicing patterns in multicellular organisms are coordinately defined by a limited number of regulatory proteins and cis elements. Mutually exclusive alternative splicing should be strictly regulated and is a challenging model for elucidating regulation mechanisms. Here we provide models of the regulation of two sets of mutually exclusive exons, 4a–4c and 7a–7b, of the Caenorhabditis elegans uncoordinated (unc)-32 gene, encoding the a subunit of V0 complex of vacuolar-type H+-ATPases. We visualize selection patterns of exon 4 and exon 7 in vivo by utilizing a trio and a pair of symmetric fluorescence splicing reporter minigenes, respectively, to demonstrate that they are regulated in tissue-specific manners. Genetic analyses reveal that RBFOX family RNA–binding proteins ASD-1 and FOX-1 and a UGCAUG stretch in intron 7b are involved in the neuron-specific selection of exon 7a. Through further forward genetic screening, we identify UNC-75, a neuron-specific CELF family RNA–binding protein of unknown function, as an essential regulator for the exon 7a selection. Electrophoretic mobility shift assays specify a short fragment in intron 7a as the recognition site for UNC-75 and demonstrate that UNC-75 specifically binds via its three RNA recognition motifs to the element including a UUGUUGUGUUGU stretch. The UUGUUGUGUUGU stretch in the reporter minigenes is actually required for the selection of exon 7a in the nervous system. We compare the amounts of partially spliced RNAs in the wild-type and unc-75 mutant backgrounds and raise a model for the mutually exclusive selection of unc-32 exon 7 by the RBFOX family and UNC-75. The neuron-specific selection of unc-32 exon 4b is also regulated by UNC-75 and the unc-75 mutation suppresses the Unc phenotype of the exon-4b-specific allele of unc-32 mutants. Taken together, UNC-75 is the neuron-specific splicing factor and regulates both sets of the mutually exclusive exons of the unc-32 gene.
Introduction
Alternative splicing of pre-mRNAs is a major source of proteomic complexity in metazoans. More than 90% of human multi-exon genes undergo alternative pre-mRNA processing and many alternative splicing events are controlled in tissue - and cell-type dependent manners [1]. Mis-splicing of pre-mRNAs underlie many inherited diseases [2]. A variety of auxiliary trans-acting factors and cis-acting elements regulating alternative splicing have been identified [3], [4], [5], [6]. Recent genome-wide studies of protein-RNA interactions for trans-acting splicing factors led to creation of RNA splicing maps [7]. Combinations of hundreds of RNA features were used to assemble ‘splicing codes’ to predict splicing patterns in four major tissues to a significant extent [8]. However, much of our knowledge of splicing regulation relies on experiments utilizing cultured cells, and therefore complex mechanisms of the tissue-specific regulation of pre-mRNA splicing by coordination of multiple trans-factors and cis-elements in living organisms remain less understood.
Mutually exclusive splicing should consist of multiple steps of strictly regulated splicing events and offers good models for elucidating regulation mechanisms for alternative pre-mRNA splicing [9], [10]. Among them, fibroblast growth factor receptor (FGFR) genes have been well studied because tissue-specific and mutually exclusive selection of exons encoding a part of the extracellular domain determines the ligand specificity of the receptors [11], [12], [13], [14]. The most extraordinary examples of the mutually exclusive exons are in the Drosophila Dscam gene [9], [10], which has four clusters of mutually exclusive exons. Selection of only one exon out of 48 candidate exons at a time for the exon 6 cluster is considered to be regulated by a complex system of competing RNA structures and a globally-acting cluster-specific splicing repressor [15], [16]. However, the molecular mechanisms governing the selection patterns for the entire Dscam mRNA remain poorly understood [10].
A nematode Caenorhabditis elegans is intron-rich like vertebrates and is an excellent model organism for studying the regulation mechanisms of pre-mRNA processing in vivo [17]. Up to 25% of its protein-coding genes are estimated to undergo alternative pre-mRNA processing and hundreds of the events are developmentally regulated [18]. We developed a fluorescence alternative splicing reporter system and visualized spatio-temporal selection patterns of mutually exclusive exons in living worms [19], [20], [21]. Through genetic and biochemical analyses, we successfully identified evolutionarily-conserved and broadly-expressed RBFOX (named after RNA binding protein, fox-1 homolog (C. elegans)) family splicing regulators ASD-1 and FOX-1 and a muscle-specific RNA-binding protein SUP-12 as the co-regulators of the muscle-specific selection of exon 5B of the egl-15 gene encoding the sole homolog of the FGFRs in C. elegans [19], [22].
The unc-32 gene of C. elegans, analyzed in this study, encodes the a subunit of V0 complex of vacuolar-type H+-ATPases considered to be proton pumps that acidify intracellular organelles [23], [24]. The unique property of the unc-32 gene as a model for studying alternative splicing regulation is that it has two sets of mutually exclusive exons (Figure 1A). Only one exon at a time is selected from three exons 4a, 4b and 4c; only one exon is selected at a time from two exons 7a and 7b. Of the six possible combinations of exons 4 and 7, the three isoforms UNC-32A (4a/7b), UNC-32B (4b/7a) and UNC-32C (4c/7b) were predominantly detected [25] and appear to be developmentally regulated [18], raising questions about the exact selection patterns and the regulation mechanisms in vivo. In the present study, we demonstrate that unc-32 exon 4 and exon 7 are selected in tissue-specific manners and that a neuron-specific RNA-binding protein UNC-75 regulates the neuron-specific selection of exons 4b and 7a.
Fig. 1. The mutually exclusive exons of the unc-32 gene are regulated in tissue-specific manners. 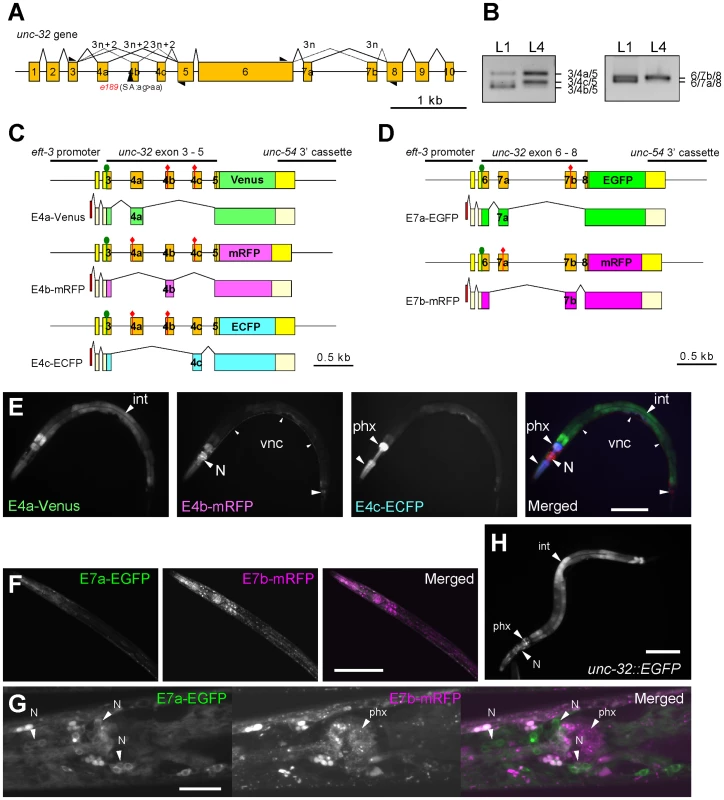
(A) Schematic structure of the unc-32 gene. Numbered boxes indicate exons. The position of the 3′-splice site mutation in the unc-32 (e189) allele is indicated. Black triangles indicate positions and directions of the exonic primers used in the RT-PCR analyses shown in (B). (B) RT-PCR analyses of unc-32 exon 4 (left) and exon 7 (right) at the L1 and L4 larval stages. (C, D) Schematic illustration of the symmetric trio of the exon 4 reporter minigenes (C) and the symmetric pair of the exon 7 reporter minigenes (D) and the mRNA isoforms derived from them. The cDNA cassettes and the predicted ORFs for Venus/EGFP, mRFP and ECFP are colored in green, magenta and cyan, respectively. Green circles and red diamonds indicate the artificially-introduced initiation and termination codons, respectively. (E) Fluorescence images of an L4 larva of the exon 4 reporter worm ybIs1891 [eft-3::unc-32E4a-Venus eft-3::unc-32E4b-mRFP eft-3::unc-32E4c-ECFP]. E4a-Venus, E4b-mRFP and E4c-ECFP images in black-white and a merged image with pseudo colors. (F, G) Confocal images of the exon 7 reporter worms ybEx1440 [eft-3::unc-32E7-EGFP eft-3::unc-32E7b-mRFP]. (H) A fluorescence image of the unc-32 transcriptional fusion reporter worm ybEx1896 [unc-32::EGFP] containing 1.7-kb of the unc-32 promoter. int, intestine; N, neurons in head ganglia; phx, pharynx. Scale bars in (E), (F) and (H), 100 µm; in (G), 20 µm. Results
Fluorescence splicing reporters for unc-32 mutually exclusive exons exhibited tissue-specific patterns
We first confirmed mutually exclusive selection of endogenous unc-32 exon 4 and exon 7 by RT-PCR (Figure 1B). Consistent with the previous report based on microarray profiling and high-throughput sequencing of mRNAs from synchronized worms [18], the splicing patterns of both exon 4 and exon 7 appeared to be developmentally regulated; the relative amounts of the exon 4b isoform and the exon 7a isoform dramatically decreased at the L4 stage (Figure 1B).
Next we visualized the selection patterns of unc-32 exon 4 and exon 7 in vivo by applying our fluorescence alternative splicing reporter system [20]. A trio of symmetric reporter minigenes for exon 4 was constructed by cloning the genomic fragment spanning from exon 3 through exon 5 upstream of one of three fluorescent protein cDNA cassettes and by introducing artificial termination codons into two of the three mutually exclusive exons in each construct (Figure 1C). From these minigenes, we expect expression of Venus-fusion protein (E4a-Venus), monomeric red fluorescent protein (mRFP)-fusion protein (E4b-mRFP) and enhanced cyan fluorescent protein (ECFP)-fusion protein (E4c-ECFP) only when exon 4a alone, 4b alone and 4c alone are selected, respectively (Figure 1C). In the same way, a pair of symmetric exon 7 reporter minigenes was constructed by cloning the genomic fragment spanning from exon 6 through exon 8 upstream of either of two fluorescent protein cDNA cassettes and by introducing an artificial termination codon into one of the two mutually exclusive exons in each construct (Figure 1D). From these minigenes, we expect expression of enhanced green fluorescent protein (EGFP)-fusion protein (E7a-EGFP) and mRFP-fusion protein (E7b-mRFP) when exon 7a and exon 7b are selected, respectively (Figure 1D).
We utilized a ubiquitous promoter to drive expression of the minigenes and generated transgenic reporter worms (Figure 1E–1G). Expression of the three fluorescent proteins in the exon 4 reporter worms varied among tissues; intestine, the nervous system and pharynx predominantly or exclusively expressed E4a-Venus, E4b-mRFP and E4c-ECFP, respectively (Figure 1E). Expression of the two fluorescent proteins in the exon 7 reporter worms also showed tissue-specificity. Most tissues predominantly expressed E7b-mRFP and therefore the worms appear almost Red (Figure 1F). Confocal microscopy revealed that neurons in head ganglia predominantly expressed E7a-EGFP (Figure 1G). The expression patterns of the exon 4 and exon 7 reporters were consistent throughout development. We suspected that lack of the developmental change in the reporter expression was due to ectopic expression of the reporters in tissues that do not express the endogenous unc-32 gene. A transcriptional fusion reporter, however, revealed that the unc-32 promoter drives expression in intestine, neurons and pharynx (Figure 1H), the major tissues where the exon 4 and exon 7 reporters were expressed. We therefore concluded that the mutually exclusive exons of the unc-32 exon 4 and exon 7 reporter minigenes are selected in tissue-specific and not developmentally regulated manners.
The RBFOX family and UGCAUG stretch in intron 7b are required for exon 7a selection from the unc-32 exon 7 reporter in the nervous system
To focus on the neuron-specific selection of exon 7a, we utilized the rgef-1 (also known as F25B3.3) promoter to drive pan-neuronal expression of the exon 7 reporter. As expected, transgenic worms with an integrated reporter allele ybIs1622 [rgef-1::unc-32E7a-EGFP rgef-1::unc-32E7b-mRFP] predominantly expressed E7a-EGFP in the nervous system and appeared Green with a dual-bandpass filter (Figure 2A). We therefore used the rgef-1 promoter for further analyses described below.
Fig. 2. The UGCAUG stretch and the RBFOX family proteins are involved in the neuron-specific selection of exon 7a from the unc-32 exon 7 reporter. 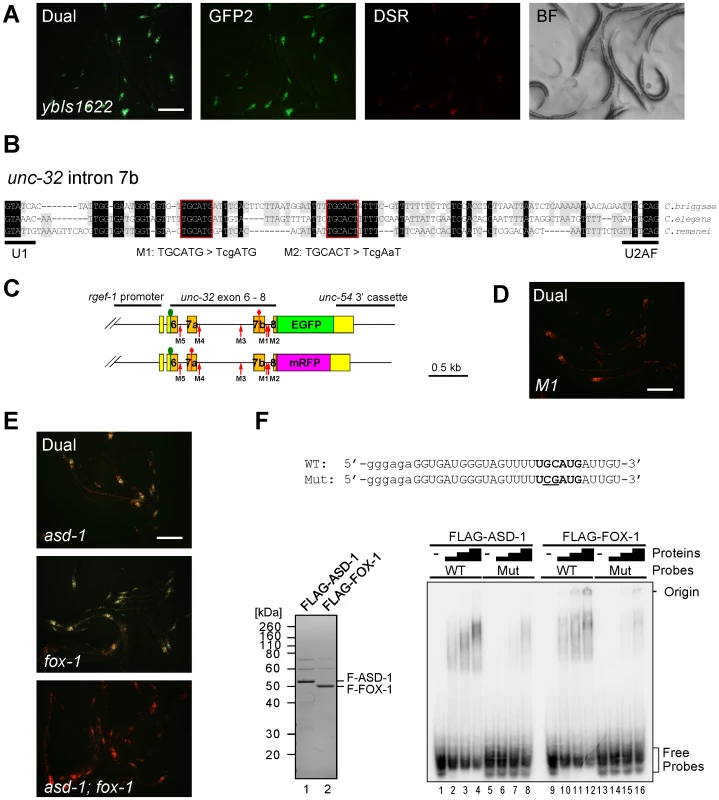
(A) Fluorescence images of the exon 7 reporter worms ybIs1622 [rgef-1::unc-32E7a-EGFP rgef-1::unc-32E7b-mRFP] with dual-bandpass (Dual), green (GFP2) and red (DSR) filters and a bright field image (BF). Note that individual neurons differentially express E7a-EGFP and E7b-mRFP in a cell-type-specific pattern. (B) Nucleotide sequence alignment of intron 7b from C. briggsae, C. elegans and C. remanei. Residues shaded in black and gray are conserved among three and two species, respectively. Conserved stretches are boxed in red and the sequences of the M1 and M2 mutant pairs of the reporter minigenes are indicated. (C) Schematic illustrations of the mutated pairs of the exon 7 reporter minigenes. Red arrows indicate the positions of the modification. (D) A fluorescence image of the M1 mutant reporter worms with a dual-bandpass filter. (E) Fluorescence images of the ybIs1622 worms in the asd-1 (yb978) (top), fox-1 (e2643) (middle) and asd-1; fox-1 (bottom) backgrounds with a dual-bandpass filter. Scale bars, 200 µm in (A), (D) and (E). (F) Top, radiolabelled wild-type (WT) and mutant (Mut) intron 7b probes. The substituted bases are underlined. Lowercase indicates the sequence derived from the T7 promoter. Bottom left, Neutral PAGE and CBB staining of the recombinant FLAG-tagged ASD-1 and FOX-1 proteins. Bottom right, EMSA using the WT and Mut probes without (−) or with 4-fold dilution series of FLAG-ASD-1 and -FOX-1. As cis-elements regulating alternative splicing are often evolutionarily conserved in the genus Caenorhabditis [19], [21], [22], [26], we first focused on the five stretches in flanking introns of exons 7a and 7b conserved among C. elegans, C. briggsae and C. remanei (Figure 2B, Figure S1A). We constructed five pairs of mutagenized exon 7 reporter minigenes M1 to M5 (Figure 2C, Figure S1A) and found that disruption of the UGCAUG stretch in intron 7b (M1) changed the color of the exon 7 reporter from Green to Orange (Figure 2D), while disruption of the other stretches had no apparent effect (Figure S1B). We therefore concluded that the UGCAUG stretch in intron 7b is required for the neuron-specific selection of exon 7a.
The UGCAUG stretches are known to be specifically recognized by the RBFOX family splicing regulators in metazoans including C. elegans [27]. We have previously reported that the RBFOX family proteins in C. elegans, ASD-1 and FOX-1, redundantly repress egl-15 exon 5B by specifically binding to the UGCAUG stretch in the upstream intron [19]. The asd-1; fox-1 double mutant is defective in expression of a muscle-specific fibroblast growth factor receptor (FGFR) isoform EGL-15(5A) and shows the egg-laying-defective (Egl-d) phenotype [19]. To test whether ASD-1 and FOX-1 also regulate the neuron-specific selection of unc-32 exon 7a, we crossed the reporter allele ybIs1622 with the asd-1 and fox-1 mutants. As expected, the reporter worms turned the color from Green to Yellow in the single mutant backgrounds (Figure 2E, top and middle) and to Orange in the double (Figure 2E, bottom), confirming that ASD-1 and FOX-1 are redundantly involved in the neuron-specific selection of exon 7a from the exon 7 reporter.
To confirm direct and specific binding of ASD-1 and FOX-1 to the UGCAUG stretch in intron 7b in vitro, we performed an electrophoretic mobility shift assay (EMSA) using the radiolabelled RNA probes with an intact (WT) and a mutagenized (M1) sequence as in the reporters (Figure 2F, top). Recombinant full-length ASD-1 and FOX-1 proteins (Figure 2F, bottom left) efficiently shifted the mobility of the WT probe (Figure 2F, bottom right, lanes 1–4, 9–12) and less efficiently of the M1 probe (lanes 5–8, 13–16) in a dose-dependent manner, demonstrating direct and specific binding of ASD-1 and FOX-1 to the UGCAUG stretch. These results led to the conclusion that ASD-1 and FOX-1 regulate the selection of exon 7a from the unc-32 exon 7 reporter via the UGCAUG stretch in intron 7b in the nervous system.
UNC-75 is required for exon 7a selection from the unc-32 exon 7 reporter in the nervous system
To identify other regulator(s) that confer the neuron-specificity to the exon 7 reporter, we mutagenized the ybIs1622 strain to screen for mutants exhibiting altered colors. We successfully isolated many homozygous viable strains with Yellow, Orange or Red phenotype (Figure 3A, Figure S2). In some other strains, most neurons turned red while some remained green (Red/Green) (Figure 3A, Figure S2). The color phenotypes were completely penetrated within the strains. Notably, all the Red and Red/Green strains also showed an uncoordinated (Unc) phenotype while the Orange or Yellow strains did not.
Fig. 3. UNC-75 is required for the selection exon 7a from the unc-32 exon 7 reporter in the nervous system. 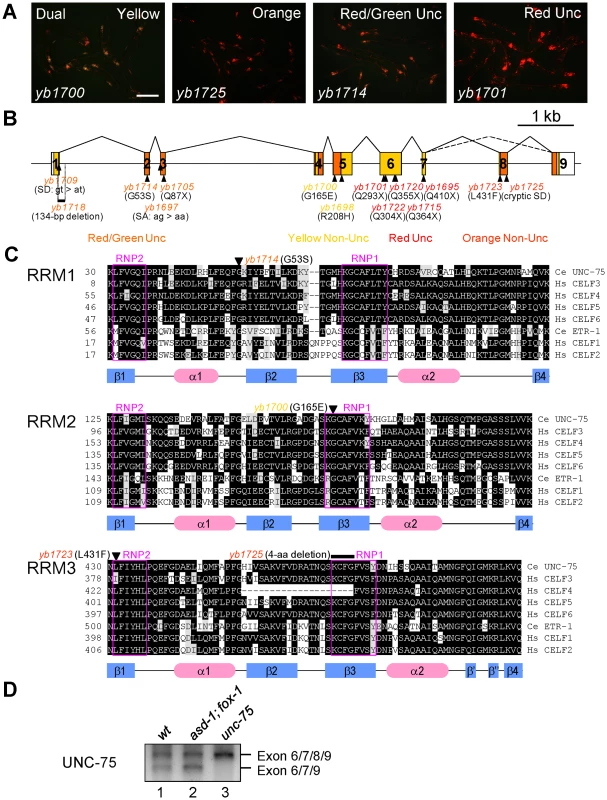
(A) Fluorescence images of the ybIs1622 worms with the unc-75 mutant alleles yb1700, yb1725, yb1714 and yb1701 with a dual-bandpass filter. Scale bar, 200 µm. (B) Schematic structure of the unc-75 gene and the positions and consequence of the mutations. The ORF is colored in yellow and the regions corresponding to the three RRMs are in orange. The allele names are in the colors representing the color phenotypes. SA, splice acceptor; SD, splice donor. (C) Amino acid sequence alignments of the three RRMs from the CELF3–6 subfamily members C. elegans UNC-75, human CELF3, CELF4, CELF5 and CELF6 and the CELF1–2 subfamily members C. elegans ETR-1, human CELF1 and CELF2. Conserved residues are shaded in black. Residues with similar properties to the consensus are shaded in gray. The secondary structure elements determined for human CELF1 [51], [52] are depicted with the blue rectangles (β-sheets) and the red ellipses (α-helices) below the alignments. The highly conserved RNP1 and RNP2 motifs in each RRM are boxed in magenta. The positions of the missense mutations and the short deletion are indicated with the allele names. Note that yb1725 generated a cryptic splice donor site almost exclusively used in the mutant, which resulted in the in-frame deletion of the 4-amino acid stretch indicated. (D) RT-PCR analysis of the UNC-75 mRNAs from the synchronized L1 larvae of N2 (wt), asd-1 (yb978); fox-1 (e2643) and unc-75 (yb1701). By single-nucleotide polymorphism (SNP)-based mapping and sequencing candidate genes, we identified mutations in the unc-75 gene in the color mutants. The unc-75 gene was originally identified as the gene responsible for the Unc phenotype caused by defects in synaptic transmission [28]. The exon 7 reporter allele ybIs1622 crossed with an existing null allele unc-75 (e950), which lacks exon 1 through exon 5 and exhibits the Unc phenotype [28], showed the RedUnc phenotype (data not shown), confirming that the color phenotype is caused by loss of function of the unc-75 gene.
UNC-75 belongs to the CUG-BP and ETR-3-like factor (CELF) family of RNA-binding proteins, which have two N-terminal RNA recognition motifs (RRMs) followed by a so-called divergent domain and the third RRM at the C-terminus. The CELF family can be divided into two subfamilies CELF1–2 and CELF3–6 according to sequence similarities [29] and UNC-75 is the sole member of the CELF3–6 subfamily in C. elegans [29]. Although UNC-75 has been shown to be expressed exclusively in the nervous system and localized to subnuclear speckles [28], it is still unknown what process UNC-75 is involved in.
The mutations identified in the unc-75 gene are summarized in Figure 3B. All of the five alleles with the RedUnc phenotype have nonsense mutations in exon 6 or exon 7 (Figure 3B). Figure 3C shows amino acid sequence alignments of the three RRMs from the CELF family members in C. elegans and human. A missense mutation (yb1714) in the conserved glycine residue in the α1β2 loop of RRM1 and four other mutations (yb1697, yb1705, yb1709 and yb1718) in the region between exon 1 and exon 3 were associated with the Red/GreenUnc phenotype (Figure 3B and 3C, top). A missense mutation (yb1700) in the conserved glycine residue in the RNP1 motif of RRM2 (Figure 3B and 3C, middle) and a missense mutation (yb1698) in the conserved arginine residue in the divergent domain (Figure S3) were associated with the Yellow phenotype. A missense mutation (yb1723) in the RNP2 motif and a 4-aa deletion (yb1725) in the RNP1 motif in RRM3 were associated with the Orange phenotype (Figure 3B and 3C, bottom). These results suggested that all the three RRMs and the divergent domain are required for UNC-75 to properly regulate the selection of exon 7a in the nervous system.
During the course of cDNA cloning, we found another UNC-75 mRNA isoform lacking exon 8 corresponding to the anterior half of RRM3 (Figure 3B and 3D, lane 1). Although the skipping of exon 8 does not cause a frame-shift or nonsense-mediated mRNA decay (NMD), the deletion of the half of RRM3 would more significantly affect the function of UNC-75 than the yb1723 and yb1725 mutations (Figure 3C, bottom). As many splicing factors are known to regulate their own expression at the pre-mRNA splicing level, we analyzed the effect of the nonsense mutation in the unc-75 gene on its own mRNAs. The splicing patterns of the UNC-75 mRNAs were not affected in the asd-1; fox-1 mutant (Figure 3D, lane 2), while the Δexon 8 isoform was undetected in the unc-75 (yb1701) mutant (lane 3), consistent with the idea that UNC-75 negatively regulates its own expression by repressing exon 8.
The C-termini of ASD-1, FOX-1, and UNC-75 function as the sole nuclear localization signals
We noticed that the C-termini of the CELF family proteins as well as the RBFOX family proteins are evolutionarily conserved and match the consensus of the hydrophobic PY nuclear localization signal (PY-NLS) [30] (Figure 4A). To test this idea, we analyzed the effect of substitution or deletion of the C-terminal motifs upon subcellular localization of the proteins (Figure 4B–4G). The substitution of the three residues in the PY element of UNC-75 (Figure 4A) disrupted the nuclear localization of UNC-75 (Figure 4B, 4C), confirming that the C-terminal motif of UNC-75 functions as the PY-NLS. In the same way, the deletion of the 7 and 16 residues from the C-termini of ASD-1 and FOX-1, respectively (Figure 4A), disrupted the nuclear localization of the proteins (Figure 4D–4G), indicating that the C-terminal portions of ASD-1 and FOX-1 are the sole NLSs.
Fig. 4. The C-termini of UNC-75, ASD-1, and FOX-1 are the evolutionarily conserved nuclear localization signals. 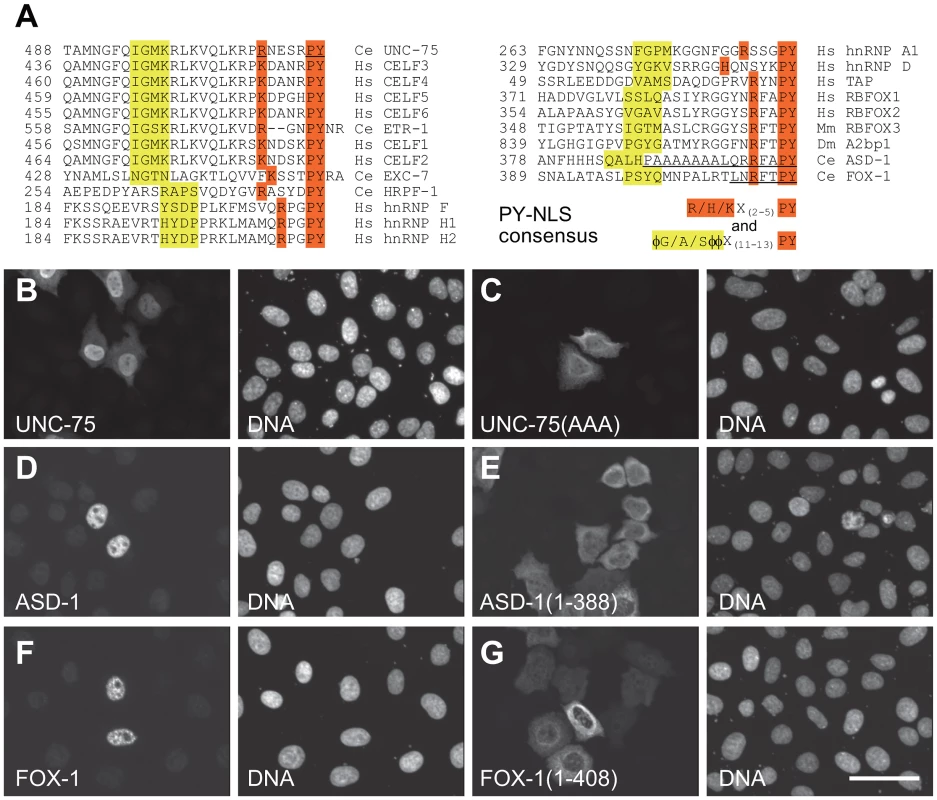
(A) Amino acid sequence alignment of the C-terminal ends of the CELF family proteins and the putative PY-NLSs from the related proteins C. elegans EXC-7, HRPF-1, human hnRNP F, hnRNP H1 and hnRNP H2 (left), human hnRNP A1, hnRNP D and TAP and the C-terminal ends of the RBFOX family proteins human RBFOX1, RBFOX2, mouse RBFOX3, Drosophila A2bp1, C. elegans ASD-1 and FOX-1 (right). The consensus sequences of the PY-NLS are indicated. The amino acid residues that match the consensus are shaded in yellow or orange. φ, hydrophobic residues. The underlines in the UNC-75 sequence indicate residues substituted with alanine in UNC-75(AAA) used in (C). The underlines in the ASD-1 and FOX-1 sequences indicate residues deleted in the truncated constructs used in (E) and (G), respectively. (B, C) Fluorescence images of the HeLa cells transfected with full-length UNC-75 (B) and UNC-75(AAA) (C) stained with anti-UNC-75 (left panels) and Hoechst 33258 (right panels). (D–G) Confocal images of the HeLa cells transfected with HA-tagged full-length ASD-1 (D), ASD-1(1–388) (E), full-length FOX-1 (F) and FOX-1(1–408) (G) stained with anti-ASD-1 (D and E, left panels) or anti-FOX-1 (F and G, left panels) and TO-PRO3 (right panels). Scale bar, 50 µm. UNC-75 directly and specifically binds to a short fragment in unc-32 intron 7a in vitro
To determine the element(s) in the exon 7 cluster region that UNC-75 directly and specifically recognizes in vitro, we performed EMSAs with the radiolabelled RNA probes schematically illustrated in Figure 5A (top panel). Recombinant full-length UNC-75 protein shifted the mobility of Probe 2 (Figure 5B, lanes 3,4) and Probe 2-1 (lanes 9–12) and not of the other probes (Figure 5B). As more than half of Probe 2-1 overlapped with Probe 1 or Probe 2-2, we prepared a shorter probe 2-1-1 (Figure 5A) containing most of the sequence unique to Probe 2-1. UNC-75 shifted the mobility of Probe 2-2-1 (Figure 5C, lanes 1–4, 25–28), demonstrating that UNC-75 directly and specifically binds to the 2-1-1 fragment in this region.
Fig. 5. UNC-75 directly and specifically binds to a short fragment in unc-32 intron 7a in vitro. 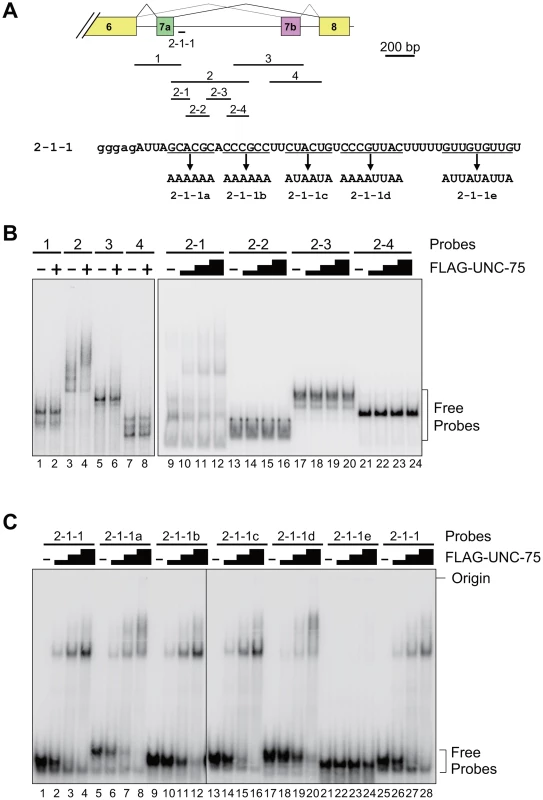
(A) Top, schematic illustration of the radiolabelled RNA probes 1 to 4, 2-1 to -4 and 2-1-1 used in the EMSAs. Bottom, sequences of Probe 2-1-1 and its mutants 2-1-1a to -1e. The modified stretches are underlined and the mutant sequences are indicated. Lowercase indicates the sequence derived from the T7 promoter. (B) Left, EMSA using the probes 1 to 4 without (−) or with (+) FLAG-tagged recombinant UNC-75 (FLAG-UNC-75). Right, EMSA using the probes 2-1 to 2-4 without (−) or with 2-fold dilution series of FLAG-UNC-75. (C) EMSAs using Probe 2-1-1 and its mutants shown in (A) without (−) or with 4-fold dilution series of FLAG-UNC-75. To further specify the element(s) necessary for the UNC-75-binding, we prepared the five mutant probes 2-1-1a to -1e, in each of which G and C residues in a short stretch were replaced with A (Figure 5A, bottom panel). UNC-75 shifted the mobility of the probes 2-1-1a to -1d (lanes 5–20) similarly to Probe 2-1-1, while the mobility of Probe 2-1-1e was unaffected by UNC-75 (lane 21–24), indicating that the UUGUUGUGUUGU stretch disrupted in Probe 2-1-1e is essential for UNC-75 to specifically recognize the 2-1-1 fragment.
RRM3 of UNC-75 mediates the specific binding to the UUGUUGUGUUGU stretch in unc-32 intron 7a
To test whether all the three RRMs of UNC-75 are involved in the recognition of the 2-2-1 fragment, we performed EMSAs using three mutant recombinant proteins UNC-75 (G53S), UNC-75 (G165E) and UNC-75 (L431F) (Figure 6A, left), each of which had a single missense mutation in one of the three RRMs as found in the mutant alleles. UNC-75 (G53S) and UNC-75 (G165E) less efficiently shifted the mobility of Probe 2-1 and Probe 2-1-1 than wild-type UNC-75 (Figure S4, lanes 1–10; Figure 6A, right, lanes 1–13). UNC-75 (L431F) failed to shift the mobility of these probes (Figure S4, lanes 11–13; Figure 6A, right, lanes 14–17). These results indicated that the missense mutations affected the RNA-binding properties of UNC-75 in vitro and that all the three RRMs of UNC-75 are required for the specific recognition of unc-32 intron 7a.
Fig. 6. RRM3 of UNC-75 mediates specific binding to the UUGUUGUGUUGU stretch in unc-32 intron 7a. 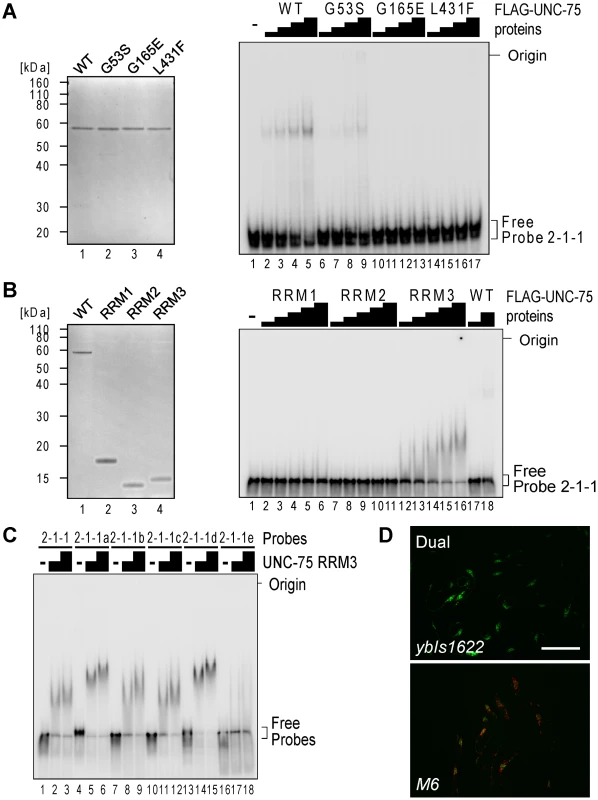
(A) Left, neutral PAGE and CBB staining of the recombinant FLAG-tagged wild-type (WT) UNC-75 protein and the mutant proteins UNC-75(G53S), UNC-75(G165E) and UNC-75(L431F). Right, EMSA using Probe 2-1-1 without or with 2-fold dilution series of the wild-type and mutant UNC-75 proteins. (B) Left, CBB staining of the recombinant FLAG-tagged full-length UNC-75 protein (WT; lane 1) and UNC-75(1–114) (RRM1; lane 2), UNC-75(121–213) (RRM2; lane 3) and UNC-75(417–514) (RRM3; lane 4) proteins. Right, EMSA using Probe 2-1-1 and 2-fold dilution series of the three UNC-75 RRM proteins and the full-length protein. (C) EMSA using Probe 2-2-1 (lanes 1–3) and five mutant probes (lanes 4–18) without (−) or with 2-fold dilution series of UNC-75 (417–514) protein. (D) Fluorescence images of the wild-type (ybIs1622; top) and M6 mutant (bottom) of the exon 7 reporter worms with a dual-bandpass filter. Scale bar, 200 µm. To specify which of the three RRMs of UNC-75 mediates the specific recognition of the elements in Probe 2-1-1, we prepared recombinant proteins for each of the three RRMs and performed an EMSA (Figure 6B). The RRM3 protein (Figure 6B, right, lanes 12–16) as well as full-length UNC-75 (lanes 17,18) shifted the mobility of Probe 2-1-1, while the RRM1 or RRM2 protein did not (lanes 1-11), indicating that only RRM3 can bind to Probe 2-1-1 by itself. So we used only RRM3 protein for a further EMSA with the mutant 2-1-1 probes. The RRM3 protein shifted the mobility of Probe 2-2-1 (Figure 6C, lanes 1–3) and the mutant probes 2-2-1a to -1d (lanes 4–15) and not of Probe 2-2-1e (lanes 16–18), indicating that RRM3 specifically recognizes the UUGUUGUGUUGU stretch.
To test the requirement of the UUGUUGUGUUGU stretch for the splicing regulation in vivo, we constructed another mutant pair of the exon 7 reporter minigenes M6 that has the same substitutions as in Probe 2-2-1e and generated transgenic worms. The disruption of the UUGUUGUGUUGU stretch turned the color into Orange (Figure 6D), confirming that the stretch is essential for the selection of exon 7a in the nervous system in vivo.
The RBFOX family and UNC-75 differentially regulate the exon 7a selection
Next we analyzed the effects of the RBFOX family and UNC-75 on the endogenous unc-32 gene. In the wild-type L1 larvae, the exon 7a and exon 7b mRNA isoforms were almost equally detected (Figure 7A, left, lane 1). The relative amount of the exon 7a isoform was reduced in the asd-1; fox-1 double mutant (lane 2) and unc-75 mutant (lane 3) backgrounds. A double inclusion isoform or a double skipping isoform was not detected in either of the mutants. These results are consistent with their color phenotypes and the splicing patterns of the exon 7 reporter expressed in the nervous system (Figure 7A, right) and confirm that the RBFOX family and UNC-75 regulate the mutually exclusive splicing of exons 7a and 7b of the endogenous unc-32 gene.
Fig. 7. The RBFOX family and UNC-75 differentially regulate the intron excision from the unc-32 pre-mRNA. 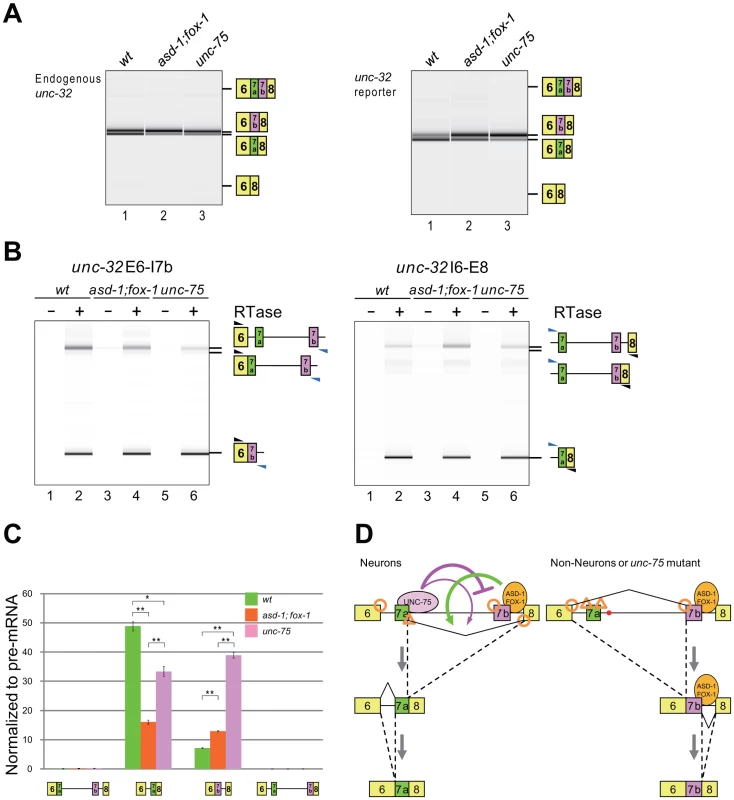
(A) RT-PCR analysis of the mature mRNAs from the endogenous unc-32 gene (left) and the exon 7 reporter transgene ybIs1622 (right) in the wild-type (lane 1), asd-1 (yb978); fox-1 (e2643) double (lane 2) and unc-75 (yb1725) (lane 3) backgrounds. Note that the isoform with double inclusion or double skipping of exons 7a and 7b was not detected. (B) RT-PCR analyses of the partially spliced RNAs from ybIs1622 in the wild-type, asd-1; fox-1 and unc-75 backgrounds. The schematic structures of the RT-PCR products are indicated on the right. Black and blue arrowheads indicate the positions and directions of the exonic and the intronic primers, respectively. RTase, reverse transcriptase. (C) Averages of the relative amounts of the partially spliced RNAs to the pre-mRNA. Error bars indicate S.E.M. (n = 3). *p<0.005 and **p<0.001, Student's t-test. (D) Schematic models of the mutually exclusive selection of unc-32 exons 7a and 7b. See Discussion for detail. For mutually exclusive alternative splicing, upstream and downstream flanking introns should be sequentially excised (Figure S5A). To obtain insight into the orders of intron removal for the production of the exon 7a and 7b mRNA isoforms, we analyzed the relative amounts of the four partially spliced RNA species to the unspliced RNA from the exon 7 reporter expressed in the nervous system by RT-PCR using two pairs of an intronic primer and a reporter-specific exonic primer. With one primer set, the partially spliced RNA in which exon 6 was spliced to exon 7b (E6/E7b–E8) was detected but the other partially spliced RNA in which intron 6 was removed (E6/E7a-E7b-E8) was almost undetectable in the wild-type, asd-1; fox-1 double mutant and unc-75 mutant worms (Figure 7B, left). With the other primer set, the partially spliced RNA in which exon 7a was spliced to exon 8 (E6–E7a/E8) was detected but the other partially spliced RNA in which intron 7b was removed (E6-E7a-E7b/E8) was almost undetectable in these worms (Figure 7B, right). The relative amounts of the four partially spliced RNAs to the unspliced RNA are summarized in Figure 7C. Of the two partially spliced RNAs that are the putative intermediates for the exon 7a isoform, E6–E7a/E8 was predominantly detected and its relative amount was decreased in the mutants. Of the two partially spliced RNAs that are the putative intermediates for the exon 7b isoform, E6/E7b–E8 was predominantly detected and its relative amount was increased in the mutants. Although these partially spliced RNAs may not necessarily be the processing intermediates but instead dead-end products, the changes in the relative amounts of the partially spliced RNAs are in good correlation with the changes in the amounts of the mature mRNA isoforms in the mutants. These results suggest that E6–E7a/E8 and E6/E7b–E8 are the major processing intermediates for the exon 7a and exon 7b isoforms, respectively. Notably, the mutations in the RBFOX family genes and unc-75 differentially affected the relative amounts of these partially spliced RNAs, suggesting their differential roles in the alternative splicing regulation of unc-32 exon 7.
We also analyzed the partially spliced RNAs from the endogenous unc-32 gene with endogenous RNA-specific pairs of primers. The result revealed consistent but weaker effects of the mutations in the RBFOX family genes and unc-75 on the partially spliced RNAs (Figure S5B–S5C). Considering that the endogenous unc-32 gene is expressed not only in the nervous system but also in pharynx and intestine that select exon 7b, this result is consistent with the idea that the RBFOX family and UNC-75 regulate the selection exon 7a from the endogenous unc-32 gene in the same way as from the reporter in the nervous system. Taking the relative strength of the splice sites in this region (Figure S5D) into account, Figure 7D summarizes the schematic models for the mutually exclusive selection of unc-32 exon 7, which will be discussed later (see Discussion).
UNC-75 regulates the neuron-specific selection of unc-32 exon 4b
As unc-32 exon 4b is also selected in a neuron-specific manner (Figure 1F), we tested whether the RBFOX family and UNC-75 are also involved in the regulation of the exon 4 cluster. Consistent with the absence of a (U)GCAUG stretch in the exon 4 cluster region, the asd-1; fox-1 double mutation did not affect the splicing patterns of exon 4 of the endogenous unc-32 gene (Figure 8A, lanes 1, 2). On the other hand, the unc-75 mutation caused marked reduction of the exon 4b isoform (lane 3). Furthermore, the neuron-specific expression of E4b-mRFP from the exon 4 reporter ybIs1891 was also abolished in the unc-75 mutant (Figure 8B, compare with Figure 1F). These results indicated that UNC-75 is required for the selection of exon 4b in the nervous system. We performed an EMSA to localize the UNC-75-binding site(s) with four overlapping probes in the exon 4 cluster region, but none of the probes were shifted as effectively as Probe 2 in Figure 5B by full-length UNC-75 (data not shown). We speculate that other cooperative factors may be required for the specific recognition of the exon 4 cluster region by UNC-75.
Fig. 8. UNC-75 regulates the neuron-specific selection of unc-32 exon 4b. 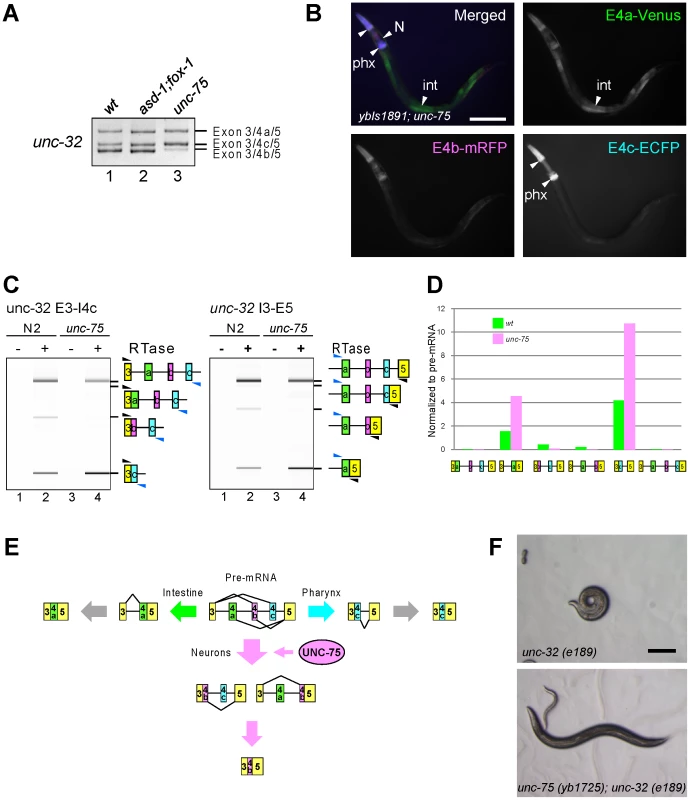
(A) RT-PCR analysis of endogenous unc-32 exon 4 in the synchronized L1 worms of N2 (wt; lane 1), asd-1 (yb978); fox-1 (e2643) (lane 2) and unc-75 (yb1701) (lane 3). (B) Fluorescence images of an L4 worm of the ybIs1891 reporter allele in the unc-75 (yb1701) background as in Figure 1F. Scale bar, 100 µm. (C) RT-PCR analyses of the partially spliced RNAs from the endogenous unc-32 gene in N2 and unc-75 (yb1701) as in Figure 7B. (D) Relative amounts of the six partially spliced RNAs normalized to the pre-mRNA analyzed in (C). (E) Schematic illustration of the neuron-specific selection of exon 4b by UNC-75. See Discussion for detail. (F) Microphotoimages of the unc-32 (e189) (left) and the unc-75 (yb1725); unc-32 (e189) (right) worm. Scale bar, 200 µm. Note that the unc-32 worm exhibits the coiler Unc phenotype. We next analyzed the amounts of the six theoretical partially spliced RNAs or putative processing intermediates (Figure S6) from the endogenous unc-32 gene in the wild type and unc-75 mutant. Both of the two putative processing intermediate RNAs for the exon 4b isoform were detected in the wild type (Figure 8C, left and right panels, lanes 1, 2) but almost undetectable in the unc-75 mutant (lanes 3, 4) consistently with the amount of the mature exon 4b isoform. Only one (E3–E4a/E5) of the two partially spliced RNAs that are the putative intermediate RNAs for the exon 4a isoform was detected and its relative amount was increased in the unc-75 mutant (Figure 8C–8D). Only one (E3/E4c–E5) of the two partially spliced RNAs that are the putative intermediate RNAs for the exon 4c isoform was detected and its relative amount was increased in the unc-75 mutant (Figure 8C–8D). These results propose a model schematically illustrated in Figure 8E; UNC-75 represses splicing of exon 3 to exon 4c and exon 4a to exon 5 and promotes splicing of exon 4b to exons 3 and 5.
The exon 4b-specific mutation in the unc-32 (e189) allele causes the uncoordinated (Unc) phenotype (Figure 1A) [25] and our results demonstrated that exon 4b is specifically selected in the nervous system in an UNC-75-dependent manner. So we speculated that the mutations in unc-75 should bypass the requirement of exon 4b in the nervous system. Consistent with this idea, the OrangeNon-Unc allele unc-75 (yb1725) suppressed the Unc phenotype of the unc-32 (e189) mutant (Figure 8F). As neuron-specific ectopic expression of any of the three major isoforms can rescue unc-32 (e189) (Figure S7), we reasoned that unc-75 (yb1725) suppressed unc-32 (e189) via the ectopic expression of the exon 4a or exon 4c isoform in the nervous system. Thus, UNC-75 is the critical splicing factor for the nervous system to specifically select unc-32 exon 4b in vivo.
Discussion
Regulation of the mutually exclusive alternative splicing of the unc-32 gene
In this study, we demonstrated that the two sets of the mutually exclusive exons of the unc-32 gene are independently regulated in tissue-specific manners by utilizing the fluorescence alternative splicing reporters. Our study revealed that intestine, neurons and pharynx express the UNC-32A (4a/7b), UNC-32B (4b/7a) and UNC-32C (4c/7b) isoforms, respectively. The expression patterns are consistent with the previous report that these three are the major isoforms and that the translational fusion reporter consisting of the unc-32 promoter through exon 4b is expressed in the nervous system [25]. The neuron-specific isoforms become relatively less abundant in elder stages in the RT-PCR experiments (Figure 1B) probably due to decrease in the relative population and/or mass of the nervous system. Our study thus demonstrated the importance of carefully analyzing alternative splicing patterns at a single cell resolution in vivo.
Figure 7D illustrates the proposed models of the neuron-specific selection of exon 7a. In the non-neuronal tissues, exon 7a is skipped presumably due to its weak splice sites and exon 6 is readily spliced to exon 7b (right panel). In neurons, UNC-75 specifically binds to its cis-elements in intron 7a to repress exon 7b and the RBFOX family and UNC-75 activate splicing between exon 7a and exon 8 (left panel). The models may explain why the mutations in unc-75 exerted more sever effects on the selection of exon 7a in the nervous system than the disruption of the RBFOX family genes; in the absence of UNC-75, exon 7b would be readily spliced to exon 6, where the target exon of the RBFOX family is no longer left (right panel).
Figure 8E illustrates the proposed model of the mutually exclusive selection of unc-32 exon 4. In neurons, UNC-75 activates splicing both between exon 3 and exon 4b and between exon 4b and exon 5 so that exon 4b alone is selected. In intestine and pharynx, splicing between exon 4a and exon 5 and between exon 3 and exon 4c, respectively, occurs first to determine the fate of the pre-mRNA presumably depending on other tissue-specific factor(s). The proposed order of intron excision for each isoform in this model explains the fidelity of the mutually exclusive selection from the three exons of the unc-32 exon 4 cluster.
The number of the mutually exclusive exons in a cluster is at most two in mammals. The fidelity of the mutually exclusive splicing relies on steric hindrance due to close proximity of the mutually exclusive exons [31], incompatibility between U2-type and U12-type splice sites [32], splicing regulators that repress one exon and activate the other [12], [33] and/or mRNA surveillance system [34]. We have previously raised regulation models for two genes with mutually exclusive exons in C. elegans. In the case of egl-15, the RBFOX family and SUP-12 cooperatively repress the splice acceptor of the upstream exon [22]. In the case of let-2, ASD-2 activates the splice donor of the downstream exon [21]. In the present study, we demonstrate novel types of regulation; for unc-32 exons 7a and 7b, UNC-75 and the RBFOX family switch the first splicing from E6/E7b to E7a/E8; for unc-32 exons 4a, 4b and 4c, UNC-75 activates both the splice acceptor and the donor of exon 4b. It has been recently suggested that the mutually exclusive exons in the slo-1 gene are regulated in intragenic coordination with downstream alternative splicing events although the splicing patters are not analyzed at a single cell resolution [35]. Thus, the order of intron excision and the modes of regulation for the mutually exclusive exons vary from case to case even in the simple model organism.
Alternative splicing regulation by the CELF3–6 subfamily protein UNC-75
In this study, we identified the first endogenous alternative splicing events regulated by the CELF3–6 subfamily. A recent splicing-sensitive microarray analysis of the unc-75 mutant suggested only one affected gene, lec-3 [36], but the selection patterns of the putative target exons in each tissue in vivo are not known yet and the function of UNC-75 in the splicing regulation of the lec-3 gene are to be experimentally defined. In vertebrates, the CELF1–2 subfamily proteins CELF1 (also known as CUG-BP1) and CELF2 (also known as ETR-3 and CUG-BP2) are broadly expressed, highest in heart, skeletal muscle and brain, and their biological functions and biochemical properties are well characterized [29], [37]. On the other hand, CELF3 to CELF6 are predominantly expressed in the nervous system [38], [39], [40], [41], [42] and have been shown to regulate alternative splicing in heterologous minigene systems [33], [38], [43], [44], [45], [46]. However, the in vivo functions and biochemical properties of the CELF3–6 subfamily are less characterized [29] presumably due to their functional redundancy.
We identified the short fragment specifically recognized by UNC-75 in unc-32 intron 7a and provided the genetic and biochemical evidence that all the three RRMs are required for the recognition and regulation of the unc-32 pre-mRNA (Figure 3C, Figure 6A). Among them, RRM3 recognizes the UUGUUGUGUUGU stretch in the target element by itself (Figure 6C). On the other hand, the stretches that RRM1 and RRM2 recognize could not be determined, although our data shown in Figure 5 and Figure 6 do not preclude the possibility that RRM1 and/or RRM2 also recognize the UUGUUGUGUUGU stretch. These results suggest that recognition of target RNAs by RRM1 and RRM2 is context-dependent or cooperative, which may explain why it is difficult to determine the precise binding sites or consensus sequences for RRM1 and RRM2. The CELF1–2 subfamily has been shown to bind to a variety of UG-rich and related sequences via the three RRMs in a context-dependent manner [47], [48], [49], [50], [51], [52]. Considering the amino acid sequence similarities between the two subfamilies (Figure 3C), it is reasonable that UNC-75 also recognizes the UG-rich sequences. Collection of the unc-75 mutant alleles revealed that the conserved stretch in the N-terminal portion of the divergent domain is also involved in the recognition and/or splicing regulation of unc-32 (Figure S3). This is consistent with the previous reports that the N-terminal portion of the divergent domain of CELF4 is involved in the RNA recognition and/or splicing regulation in minigene contexts [44], [46].
The RedUnc mutant alleles have nonsense mutations in unc-75 exon 6 or 7 (Figure 3A, 3B), while some other mutants show the Red/Green phenotype (Figure 3A, Figure S2), suggesting cell-type-dependent remaining activity of UNC-75 in such mutants. Paradoxically, most of the Red/Green alleles have nonsense mutations or splice site mutations in exon 1, 2 or 3 (Figure 3B), indicative of fatal effects on the UNC-75 expression. The remaining activity of UNC-75 in certain neurons might derive from the use of alternative promoters in the upstream region or in intron 3 to bypass exons 1–3, although we have not experimentally identified such mRNA isoforms from the unc-75 gene.
PY-NLS in the RNA–binding proteins
We demonstrated that the C-termini of all the CELF family and the RBFOX family proteins match the consensus of the PY-NLS and that the C-termini are indeed required for the proper nuclear localization of UNC-75, ASD-1 and FOX-1 (Figure 4). As RRM3 of the CELF family resides at the C-terminus, the PY-NLS is overlapping with RRM3 and is highly conserved. It has been reported that deletion of a C-terminal KRP stretch affected the nuclear localization of UNC-75 in neurons [28], consistent with our finding. In contrast to the PY-NLSs in the RBFOX and CELF families, the PY-NLS was originally identified in the internal portion of hnRNP A1 and other RNA-binding proteins including hnRNP D, TAP, HuR, hnRNP F and hnRNP M [30]. Most of the PY-NLSs predicted in many other proteins are structurally divergent and reside in the internal portion [30]. Evolutionary conservation of the sequences and positions of the PY-NLSs in the RBFOX and CELF families may suggest importance of their positions for the functions of these proteins.
Cooperative regulation of the tissue-specific alternative splicing by the RBFOX family and other splicing regulators
In this and previous studies, we demonstrated that the broadly-expressed RBFOX family proteins ASD-1 and FOX-1 regulate the neuron - and muscle-specific alternative splicing events in a target-specific manner in combination with the neuron-specific RNA-binding protein UNC-75 and the muscle-specific RNA-binding protein SUP-12 [22], respectively. Similarly, an RBFOX family protein RBFOX2 is expressed in a variety of cell types in mammals, yet it can regulate the epithelium-specific alternative splicing of the FGFR2 gene in coordination with epithelium-specific splicing factors ESRP1 and ESRP2 [11], [12]. The RBFOX family splicing regulators have only one RNA-binding domain that can specifically recognize the (U)GCAUG stretch in the target pre-mRNAs [27], [53]. Therefore, the presence of the (U)GCAUG stretch in the pre-mRNAs is not the sole determinant of the tissue-specificity but can be considered to offer an opportunity for the combinatorial and context-dependent regulation of alternative splicing. Considering their broad expression, the RBFOX family may regulate alternative splicing with a variety of tissue-specificity in cooperation with other tissue-specific factors in both mammals and C. elegans.
Materials and Methods
Reporter minigene construction
To construct the unc-32 exon 7 reporter cassettes, the unc-32 genomic fragment was cloned upstream of either mRFP1 [54] or EGFP (Clontech) cDNA in the Entry vectors by using In-Fusion system (BD Biosciences) and the artificial termination codons were introduced with QuickChange (Stratagene). The reporter minigenes for unc-32 exon 4 and the unc-32 transcriptional fusion were constructed as described previously [20]. The sequences of the primers used in the plasmid construction are available in Table S1.
Worm culture and microscopy
The worms were cultured following standard methods. Generation of transgenic worms, mutant screening and mapping were performed as described previously [20]. The images of the fluorescence reporter and mutant worms were captured using fluorescence stereoscopes (MZ16FA and M205FA, Leica) equipped with color, cooled CCD cameras (DP71, Olympus and DFC310FX, Leica) or a confocal microscope (FV500, Olympus) and processed with Metamorph (Molecular Devices) or Photoshop (Adobe).
RT–PCR
The RT-PCRs were performed essentially as described previously for amplifying the mature mRNAs [20] and the partially spliced RNAs [55]. The RT-PCR products were analyzed by standard agarose gel electrophoresis or by using BioAnalyzer (Agilent) and the sequences of the RT-PCR products were confirmed by direct sequencing or cloning and sequencing. The sequences of the primers used in the RT-PCR assays are available in Table S2.
Amino acid sequence alignment
The amino acid sequences of the proteins used in the alignments were retrieved from the protein sequence databases derived from GenBank and RefSeq. The accession numbers are as follows: human CELF1, NP_006551; CELF2, NP_001020247; CELF3, AAK07474; CELF4, NP_064565; CELF5, NP_068757; CELF6, NP_443072; hnRNP A1, AAH02355; hnRNP D, BAA09525; TAP, AAD20016; hnRNP F, NP_004957; hnRNP H1, NP_005511; hnRNP H2, NP_062543; RBFOX1, Q9NWB1; RBFOX2, NP_001026865; mouse RBFOX3, NP_001034256; Drosophila A2bp1, AAQ22527; C. elegans UNC-75, AAQ19851; ETR-1, NP_493673; EXC-7, CAA85327; HRPF-1, AAK21490; ASD-1, NP_497841; FOX-1, NP_508446. The amino acid sequences were aligned by Clustal W using Lasergene (DNASTAR).
Antibodies and immunocytochemistry
The rabbit polyclonal anti-UNC-75 antiserum (9493R2R) was generated by using denatured His-tagged full-length UNC-75 protein as described previously [55]. The rabbit polyclonal anti-ASD-1 (RbD8211) and -FOX-1 (RbD8209) antisera were generated with the mixtures of synthetic peptides TVEKLNDFDYKVAL+C and C+RGVPQPGRIPTSTA for anti-ASD-1 and C+GKVKDDPNSDYDLQ and C+LPSYQMNPALRTLN for anti-FOX-1 by Operon Biotechnologies (Tokyo, Japan). The expression vectors for untagged UNC-75 and HA-tagged ASD-1 and FOX-1 were constructed by using Destination vectors pDEST-cDNA3 and pDEST-ME18S-3HA (H.K.), respectively. HeLa cells were transfected with the expression vectors by utilizing GeneJuice (Novagen). For UNC-75, the cells were stained with anti-UNC-75 (9493R2R), Alexa488-conjugated goat anti-rabbit IgG (Molecular Probes) and DAPI (Vector Laboratories) and fluorescence images were captured with a compound microscope (Eclipse E600, Nikon) and a CCD camera (DP71, Olympus). For ASD-1 and FOX-1, the cells were stained with anti-ASD-1 (RbD8211) or -FOX-1 (RbD8209), Cy3-conjugated goat anti-rabbit IgG (Jackson) and TO-PRO3 (Molecular Probes) and the confocal images were acquired with FV1000 (Olympus).
Electophoretic mobility shift assay (EMSA)
The expression vectors for FLAG-tagged ASD-1, FOX-1 and UNC-75 proteins were constructed using the primers listed in Table S3 and the recombinant proteins were prepared as previously described [55]. The 32P-labelled RNA probes were prepared as described previously [55] using the template oligo DNAs listed in Table S4 and the PCR products amplified with the primes in Table S5. The EMSAs were performed as described previously [55].
Supporting Information
Zdroje
1. WangET, SandbergR, LuoS, KhrebtukovaI, ZhangL, et al. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456 : 470–476.
2. HammondSM, WoodMJ (2011) Genetic therapies for RNA mis-splicing diseases. Trends Genet 27 : 196–205.
3. BlackDL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72 : 291–336.
4. NilsenTW, GraveleyBR (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463 : 457–463.
5. ChenM, ManleyJL (2009) Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10 : 741–754.
6. KalsotraA, CooperTA (2011) Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet 12 : 715–729.
7. WittenJT, UleJ (2011) Understanding splicing regulation through RNA splicing maps. Trends Genet 27 : 89–97.
8. BarashY, CalarcoJA, GaoW, PanQ, WangX, et al. (2010) Deciphering the splicing code. Nature 465 : 53–59.
9. SmithCW (2005) Alternative splicing–when two's a crowd. Cell 123 : 1–3.
10. HemaniY, SollerM (2012) Mechanisms of Drosophila Dscam mutually exclusive splicing regulation. Biochem Soc Trans 40 : 804–809.
11. TakeuchiA, HosokawaM, NojimaT, HagiwaraM (2010) Splicing reporter mice revealed the evolutionally conserved switching mechanism of tissue-specific alternative exon selection. PLoS ONE 5: e10946 doi:10.1371/journal.pone.0010946
12. WarzechaCC, SatoTK, NabetB, HogeneschJB, CarstensRP (2009) ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 33 : 591–601.
13. BaraniakAP, ChenJR, Garcia-BlancoMA (2006) Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol 26 : 1209–1222.
14. CarstensRP, WagnerEJ, Garcia-BlancoMA (2000) An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol Cell Biol 20 : 7388–7400.
15. OlsonS, BlanchetteM, ParkJ, SavvaY, YeoGW, et al. (2007) A regulator of Dscam mutually exclusive splicing fidelity. Nat Struct Mol Biol 14 : 1134–1140.
16. GraveleyBR (2005) Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell 123 : 65–73.
17. Zahler AM (2012) Pre-mRNA splicing and its regulation in Caenorhabditis elegans. WormBook. 2012/04/03 ed. pp. 1–21.
18. RamaniAK, CalarcoJA, PanQ, MavandadiS, WangY, et al. (2011) Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res 21 : 342–348.
19. KuroyanagiH, KobayashiT, MitaniS, HagiwaraM (2006) Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat Methods 3 : 909–915.
20. KuroyanagiH, OhnoG, SakaneH, MaruokaH, HagiwaraM (2010) Visualization and genetic analysis of alternative splicing regulation in vivo using fluorescence reporters in transgenic Caenorhabditis elegans. Nat Protoc 5 : 1495–1517.
21. OhnoG, HagiwaraM, KuroyanagiH (2008) STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes Dev 22 : 360–374.
22. KuroyanagiH, OhnoG, MitaniS, HagiwaraM (2007) The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol Cell Biol 27 : 8612–8621.
23. SyntichakiP, SamaraC, TavernarakisN (2005) The vacuolar H+ -ATPase mediates intracellular acidification required for neurodegeneration in C. elegans. Curr Biol 15 : 1249–1254.
24. OkaT, ToyomuraT, HonjoK, WadaY, FutaiM (2001) Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. Cell-specific expression during development. J Biol Chem 276 : 33079–33085.
25. PujolN, BonnerotC, EwbankJJ, KoharaY, Thierry-MiegD (2001) The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J Biol Chem 276 : 11913–11921.
26. KabatJL, Barberan-SolerS, McKennaP, ClawsonH, FarrerT, et al. (2006) Intronic alternative splicing regulators identified by comparative genomics in nematodes. PLoS Comput Biol 2: e86 doi:10.1371/journal.pcbi.0020086
27. KuroyanagiH (2009) Fox-1 family of RNA-binding proteins. Cell Mol Life Sci 66 : 3895–3907.
28. LoriaPM, DukeA, RandJB, HobertO (2003) Two neuronal, nuclear-localized RNA binding proteins involved in synaptic transmission. Curr Biol 13 : 1317–1323.
29. DasguptaT, LaddAN (2012) The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip Rev RNA 3 : 104–121.
30. LeeBJ, CansizogluAE, SuelKE, LouisTH, ZhangZ, et al. (2006) Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126 : 543–558.
31. SmithCW, Nadal-GinardB (1989) Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell 56 : 749–758.
32. LetunicI, CopleyRR, BorkP (2002) Common exon duplication in animals and its role in alternative splicing. Hum Mol Genet 11 : 1561–1567.
33. GromakN, MatlinAJ, CooperTA, SmithCW (2003) Antagonistic regulation of alpha-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA 9 : 443–456.
34. JonesRB, WangF, LuoY, YuC, JinC, et al. (2001) The nonsense-mediated decay pathway and mutually exclusive expression of alternatively spliced FGFR2IIIb and -IIIc mRNAs. J Biol Chem 276 : 4158–4167.
35. GlauserDA, JohnsonBE, AldrichRW, GoodmanMB (2011) Intragenic alternative splicing coordination is essential for Caenorhabditis elegans slo-1 gene function. Proc Natl Acad Sci U S A 108 : 20790–20795.
36. Barberan-SolerS, MedinaP, EstellaJ, WilliamsJ, ZahlerAM (2011) Co-regulation of alternative splicing by diverse splicing factors in Caenorhabditis elegans. Nucleic Acids Res 39 : 666–674.
37. GalloJM, SpickettC (2010) The role of CELF proteins in neurological disorders. RNA Biol 7 : 474–479.
38. LaddAN, NguyenNH, MalhotraK, CooperTA (2004) CELF6, a member of the CELF family of RNA-binding proteins, regulates muscle-specific splicing enhancer-dependent alternative splicing. J Biol Chem 279 : 17756–17764.
39. LaddAN, CharletN, CooperTA (2001) The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol 21 : 1285–1296.
40. YangY, MahaffeyCL, BerubeN, MaddatuTP, CoxGA, et al. (2007) Complex seizure disorder caused by Brunol4 deficiency in mice. PLoS Genet 3: e124 doi:10.1371/journal.pgen.0030124
41. BrimacombeKR, LaddAN (2007) Cloning and embryonic expression patterns of the chicken CELF family. Dev Dyn 236 : 2216–2224.
42. WuJ, LiC, ZhaoS, MaoB (2010) Differential expression of the Brunol/CELF family genes during Xenopus laevis early development. Int J Dev Biol 54 : 209–214.
43. BarronVA, ZhuH, HinmanMN, LaddAN, LouH (2010) The neurofibromatosis type I pre-mRNA is a novel target of CELF protein-mediated splicing regulation. Nucleic Acids Res 38 : 253–264.
44. SinghG, CharletBN, HanJ, CooperTA (2004) ETR-3 and CELF4 protein domains required for RNA binding and splicing activity in vivo. Nucleic Acids Res 32 : 1232–1241.
45. KinoY, WashizuC, OmaY, OnishiH, NezuY, et al. (2009) MBNL and CELF proteins regulate alternative splicing of the skeletal muscle chloride channel CLCN1. Nucleic Acids Res 37 : 6477–6490.
46. HanJ, CooperTA (2005) Identification of CELF splicing activation and repression domains in vivo. Nucleic Acids Res 33 : 2769–2780.
47. FaustinoNA, CooperTA (2005) Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol 25 : 879–887.
48. MoriD, SasagawaN, KinoY, IshiuraS (2008) Quantitative analysis of CUG-BP1 binding to RNA repeats. J Biochem 143 : 377–383.
49. TakahashiN, SasagawaN, SuzukiK, IshiuraS (2000) The CUG-binding protein binds specifically to UG dinucleotide repeats in a yeast three-hybrid system. Biochem Biophys Res Commun 277 : 518–523.
50. MarquisJ, PaillardL, AudicY, CossonB, DanosO, et al. (2006) CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem J 400 : 291–301.
51. TsudaK, KuwasakoK, TakahashiM, SomeyaT, InoueM, et al. (2009) Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res 37 : 5151–5166.
52. TeplovaM, SongJ, GawHY, TeplovA, PatelDJ (2010) Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure 18 : 1364–1377.
53. AuweterSD, FasanR, ReymondL, UnderwoodJG, BlackDL, et al. (2006) Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J 25 : 163–173.
54. CampbellRE, TourO, PalmerAE, SteinbachPA, BairdGS, et al. (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci U S A 99 : 7877–7882.
55. OhnoG, OnoK, TogoM, WatanabeY, OnoS, et al. (2012) Muscle-Specific Splicing Factors ASD-2 and SUP-12 Cooperatively Switch Alternative Pre-mRNA Processing Patterns of the ADF/Cofilin Gene in Caenorhabditis elegans. PLoS Genet 8: e1002991 doi:10.1371/journal.pgen.1002991
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



