-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
Diphthamide is a highly modified histidine residue in eukaryal translation elongation factor 2 (eEF2) that is the target for irreversible ADP ribosylation by diphtheria toxin (DT). In Saccharomyces cerevisiae, the initial steps of diphthamide biosynthesis are well characterized and require the DPH1-DPH5 genes. However, the last pathway step—amidation of the intermediate diphthine to diphthamide—is ill-defined. Here we mine the genetic interaction landscapes of DPH1-DPH5 to identify a candidate gene for the elusive amidase (YLR143w/DPH6) and confirm involvement of a second gene (YBR246w/DPH7) in the amidation step. Like dph1-dph5, dph6 and dph7 mutants maintain eEF2 forms that evade inhibition by DT and sordarin, a diphthamide-dependent antifungal. Moreover, mass spectrometry shows that dph6 and dph7 mutants specifically accumulate diphthine-modified eEF2, demonstrating failure to complete the final amidation step. Consistent with an expected requirement for ATP in diphthine amidation, Dph6 contains an essential adenine nucleotide hydrolase domain and binds to eEF2. Dph6 is therefore a candidate for the elusive amidase, while Dph7 apparently couples diphthine synthase (Dph5) to diphthine amidation. The latter conclusion is based on our observation that dph7 mutants show drastically upregulated interaction between Dph5 and eEF2, indicating that their association is kept in check by Dph7. Physiologically, completion of diphthamide synthesis is required for optimal translational accuracy and cell growth, as indicated by shared traits among the dph mutants including increased ribosomal −1 frameshifting and altered responses to translation inhibitors. Through identification of Dph6 and Dph7 as components required for the amidation step of the diphthamide pathway, our work paves the way for a detailed mechanistic understanding of diphthamide formation.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003334
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003334Summary
Diphthamide is a highly modified histidine residue in eukaryal translation elongation factor 2 (eEF2) that is the target for irreversible ADP ribosylation by diphtheria toxin (DT). In Saccharomyces cerevisiae, the initial steps of diphthamide biosynthesis are well characterized and require the DPH1-DPH5 genes. However, the last pathway step—amidation of the intermediate diphthine to diphthamide—is ill-defined. Here we mine the genetic interaction landscapes of DPH1-DPH5 to identify a candidate gene for the elusive amidase (YLR143w/DPH6) and confirm involvement of a second gene (YBR246w/DPH7) in the amidation step. Like dph1-dph5, dph6 and dph7 mutants maintain eEF2 forms that evade inhibition by DT and sordarin, a diphthamide-dependent antifungal. Moreover, mass spectrometry shows that dph6 and dph7 mutants specifically accumulate diphthine-modified eEF2, demonstrating failure to complete the final amidation step. Consistent with an expected requirement for ATP in diphthine amidation, Dph6 contains an essential adenine nucleotide hydrolase domain and binds to eEF2. Dph6 is therefore a candidate for the elusive amidase, while Dph7 apparently couples diphthine synthase (Dph5) to diphthine amidation. The latter conclusion is based on our observation that dph7 mutants show drastically upregulated interaction between Dph5 and eEF2, indicating that their association is kept in check by Dph7. Physiologically, completion of diphthamide synthesis is required for optimal translational accuracy and cell growth, as indicated by shared traits among the dph mutants including increased ribosomal −1 frameshifting and altered responses to translation inhibitors. Through identification of Dph6 and Dph7 as components required for the amidation step of the diphthamide pathway, our work paves the way for a detailed mechanistic understanding of diphthamide formation.
Introduction
Regulation of biological processes by posttranslational modification can involve the function, distribution and interaction capabilities of the modified protein [1]–[3]. Though most modification pathways such as phosphorylation and ubiquitin conjugation target many different proteins, some exceptional ones uniquely target just a single polypeptide [4]. One prominent example is diphthamide formation on eukaryal translation elongation factor 2 (eEF2) [5]. Strikingly, this modification is pathobiologically important because it is hijacked for eEF2 inhibition by sordarin fungicides and by diphtheria toxin (DT) produced by pathovarieties of Corynebacterium diphtheriae that cause the severe human disease syndrome diphtheria [6]–[8]. Both agents efficiently block protein synthesis by inactivating the essential function of the modified translation factor, via stalling the diphthamide modified form of eEF2 on ribosomes and irreversible ADP ribosylation of eEF2, respectively [9]–[12]. Diphthamide itself is a highly modified histidine residue on eEF2 – 2-[3-carboxyamido-3-(trimethylamino)-propyl]-histidine – which is conserved from yeast (H699) to man (H715) (Figure 1) [5], [8], [13]. Intriguingly, it is absent from the bacterial eEF2 analog, EF-G, thus conferring immunity on the DT producer.
Fig. 1. The biosynthetic pathway for modification of eEF2 by diphthamide. 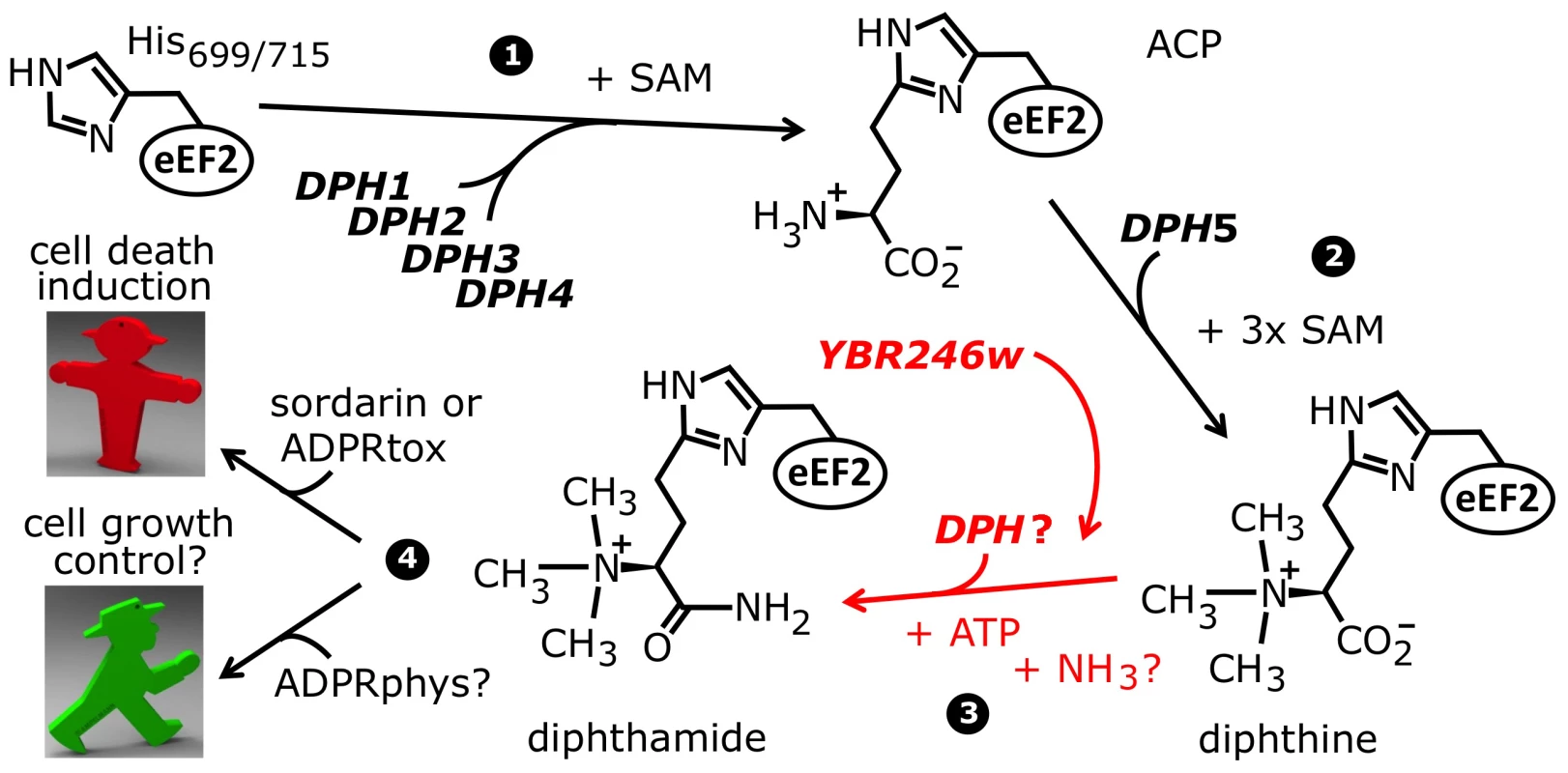
For roles played by the bona fide diphthamide genes DPH1–DPH5 in steps 1 and 2 of the pathway, see main text. The ill-defined step 3, conversion of diphthine to diphthamide by amidation, is highlighted (red label). It likely involves ATP and ammonium cofactors in a reaction catalyzed by unidentified DPH gene product(s). Step 4 indicates diphthamide can be hijacked for eEF2 inactivation and cell death induction by antifungals, i.e. sordarin and bacterial ADP ribosylase toxins (ADPRtox); alternatively, it has been reported to undergo cell growth related physiological ADP ribosylation (ADPRphys?) by elusive cellular modifier(s). ACP, 2-[3-amino-carboxyl-propyl]-histidine; SAM: S-adenosylmethionine. Among the archaea and eukarya, diphthamide formation involves a conserved biosynthetic pathway, which has been extensively dissected in Saccharomyces cerevisiae via isolation of mutant strains that confer resistance to growth inhibition by DT and sordarin. This has led to the identification of the diphthamide synthesis genes DPH1-DPH5 [7], [12], [14]–[16] (Figure 1). The first step in diphthamide synthesis involves transfer of a 3-amino-3-carboxypropyl (ACP) radical from S-adenosyl-methionine (SAM) to the histidine imidazole ring, generating the ACP modified intermediate of eEF2 [17]–[19]. ACP radical transfer requires the proteins Dph1-Dph4 [16], where Dph1 and Dph2 are paralogous iron-sulfur cluster containing partner proteins that copurify and interact with Dph3, potentially as part of a multimeric complex [6], [20]–[22]. Dph3 and Dph4 are thought to chaperone Dph1-Dph2 by maintaining their iron-sulfur clusters in redox states required for proper ACP radical generation. In line with this, Dph3 and Dph4 have electron carrier activities [23], [24], while Dph3 (also known as Kti11 [25]) additionally partners with Elongator subunit Elp3 [6], [20], an iron-sulfur cluster and radical SAM enzyme with roles in protein and tRNA modifications [26]–[28].
Formation of diphthine, the second pathway intermediate (Figure 1), requires trimethylation of the amino group in ACP and is catalyzed in yeast by diphthine synthase Dph5, using SAM as methyl donor [29]–[31]. Intriguingly, reconstitution of archaeal Dph5 activity has shown that the trimethylamino group formed in diphthine is prone to elimination in vitro [32]. Finally, the carboxyl group of diphthine is amidated by an elusive ATP dependent diphthamide synthetase (Figure 1). Once fully modified, diphthamide can be efficiently targeted by NAD+-dependent ADP ribosylase toxins including DT, Pseudomonas exotoxin A [33] and Vibrio cholix toxin [34]. However, the intermediate diphthine is also a very weak substrate for inhibitory ADP ribosylation [29], [31]. Together with data showing that growth inhibition by sordarin also depends on DPH1-DPH5 [6], [7], translation factor eEF2 constitutes an ‘Achilles heel’ for yeast, study of which has provided important insight into the pathobiological relevance of posttranslational protein modification [35].
Physiologically, the function of the diphthamide modification is enigmatic. Yeast mutants unable to synthesize diphthamide confer elevated frequency of ribosomal frameshifting [6], [36] but are viable and grow normally [14], although substitution of the modified histidine in eEF2 by other amino acids confers growth defects in some instances [37]. However, loss of diphthamide synthesis leads to delayed development and is embryonic lethal in homozygous DPH3 knockout mice [38]–[40]. Together with the association of mammalian DPH1 with tumorigenesis [16], [38] as well as neuronal and embryonic development, this indicates that diphthamide modification plays an important biological role. Whether or not this implies structural or regulatory roles for diphthamide modified eEF2 remains to be seen, but the latter notion is intriguing given the possibility of endogenous cellular ADP ribosylases that target eEF2 [4].
Interestingly, no DT resistant yeast mutants have been identified to date that affect the final amidation step in the pathway, probably because diphthine is targetable, albeit inefficiently, by ADP ribosylation [29], [31]. Thus amidase-deficient mutants may display DT sensitivity in vivo and thereby escape identification in screens for DT resistant yeast mutants.
Indication that additional proteins are involved in diphthamide biosynthesis has come from recent work on WDR85 and its potential yeast ortholog YBR246w [41], [42], while our preliminary investigation of the yeast DPH1 genetic interaction network [13] implicated both YBR246w and YLR143w as novel proteins potentially involved in the diphthamide pathway. Here we further exploit yeast genome-wide genetic interaction and chemical genomics databases [43], [44] to demonstrate that YLR143w (DPH6) and YBR246W (DPH7) cluster tightly with all known members of the diphthamide gene network. We find that dph6 and dph7 mutants phenocopy sordarin and DT traits typical of the bona fide dph1-dph5 mutants, which are defective in the first two steps of diphthamide synthesis. Importantly, we show that DPH6 and DPH7 deletions block the final amidation step of the diphthamide pathway, cause diphthine modified forms of eEF2 to accumulate and consequently abolish ADP ribose acceptor activity upon DT treatment. Thus conversion of diphthine to diphthamide depends on Dph6 and Dph7.
Results
Yeast Gene Interaction Databases Predict Diphthamide Functions for YLR143w (DPH6) and YBR246w (DPH7)
To identify factors involved in the terminal amidation step of the diphthamide modification pathway (Figure 1), we took advantage of synthetic genetic array (SGA) screens, which previously enabled systematic mapping of genetic interactions among yeast deletion collections using high-density arrays of double mutants [45], [46]. SGA analysis provides the set of genetic interactions for a given gene – the genetic interaction profile – and thereby the phenotypic signatures indicative of functions of both known genes and unassigned ORFs [47]. For example, genes with similar interaction profiles are often functionally related in shared biochemical pathways and/or protein complexes [48], [49]. Consistent with this, SGA analysis revealed that the diphthamide gene network members have highly correlated interaction profiles and tightly cluster in the global genetic interaction landscape from yeast [45].
Since our preliminary examination of the yeast genetic interaction landscape placed two uncharacterized yeast ORFs, YLR143w and YBR246w, within the diphthamide gene network [13], we next examined this network in more detail by mining the SGA DRYGIN database for quantitative S. cerevisiae genetic interactions [44], [50]. We compared DPH1, DPH2, DPH4, DPH5, YLR143w and YBR246w gene interactions with every array ORF represented in the SGA network and deposited at DRYGIN, ranking the similarity between all possible pairwise profiles according to their Pearson correlation coefficient (PCC; see Table S1 for full details). As expected, the other known DPH genes scored significantly highly among the correlation profiles generated for each specific DPH query gene, consistently being ranked among the top ten genetic interactors (Figure 2A). Strikingly, YLR143w and YBR246w were among the top interactors of DPH1, DPH2, DPH4 and DPH5, while the most correlated interactors for YLR143w and YBR246w included each other and several bone fide DPH genes (Figure 2A). Such highly correlated interaction patterns suggest that YLR143w and YBR246w are both functionally interrelated and qualify as candidate ORFs of the pathway for eEF2 modification by diphthamide. In line with this notion, the two eEF2 encoding gene copies, EFT1 and EFT2, also ranked among the top ten interactors of DPH1, DPH2 and DPH5 (Figure 2A).
Fig. 2. Genome-wide gene interaction databases identify additional diphthamide related candidate genes: YLR143w/DPH6 and YBR246w/DPH7. 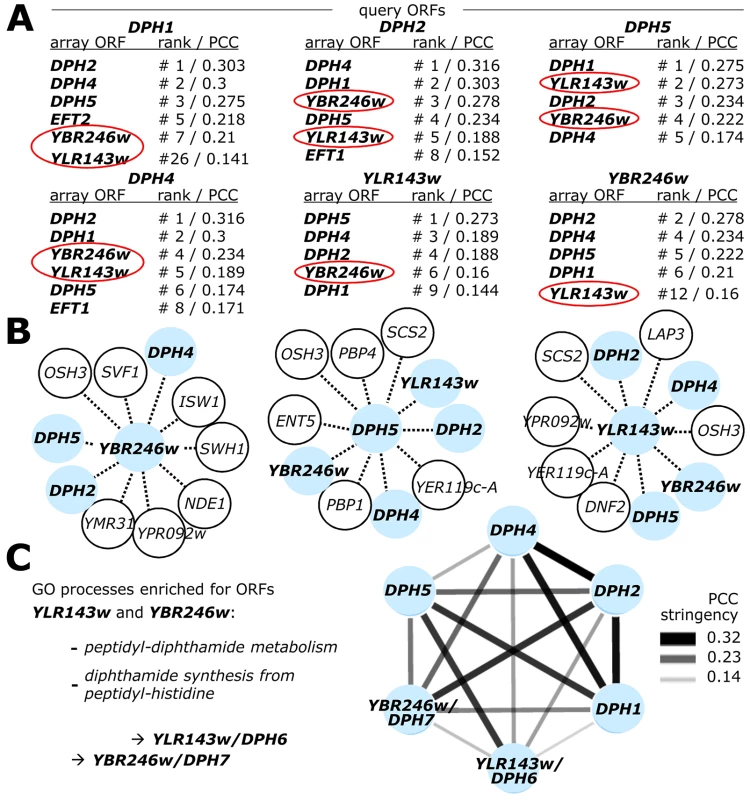
(A) SGA database (DRYGIN). Genetic interaction profiles among DPH1, DPH2, DPH4, DPH5, YBR246w and YLR143w query gene deletion strains and 3885 or 4457 array ORF mutants were extracted from data for a total of ∼1700 query strains deposited at DRYGIN (for full details, see excel spread sheet in Table S1). Ranking of top interactors for each query ORF was according to PCC (Pearson correlation coefficient) determination. For simplicity, array ORFs DPH1, DPH2, DPH4, DPH5, EFT1, EFT2 (shown in bold) as well as potentially diphthamide related candidate loci YLR143w and YBR246w (red circles) are listed that score repeatedly as significantly high interactors of the query ORFs. (B) Yeast Fitness database (FitDB). Genes whose deletions phenocluster with the six query ORFs above were extracted from FitDB, which is based on genome-scale co-fitness defect analysis of homozygous yeast deletion mutants in response to greater than 1144 different conditions. For simplicity, the top ten interactors for three of the six query genes (DPH5, YLR143w and YBR246w: pale blue central nodes) above are depicted. (C) Representation of the tightly clustered and expanded DPH1-DPH7 gene network where nodes (pale blue) correspond to individual DPH gene family members and edges connect gene pairs by PCC>0.14. Enhanced gene interaction strength is proportional to PCC stringency. Enriched GO process likelihoods in the diphthamide modification pathway are listed as P-values for the identified candidates DPH6/YLR143w and DPH7/YBR246w. For independent validation of these correlations, we searched the FitDB yeast fitness database [51], which contains genome-scale phenotypic profiles for diploid yeast deletion collections in response to more than 1100 different growth conditions [43], [52]. Here, scoring gene interaction profiles by homozygous co-sensitivity revealed that among the top loci to phenocluster with YBR246w are DPH2, DPH4 and DPH5, while top interactors of YLR143w include DPH4, DPH5, YBR246w and DPH2 (Figure 2B). A similar pattern of interaction is shown by DPH5 (Figure 2B), DPH2 and DPH4 (data not shown). Based on correlated interaction profiles, FitDB ascribes GO terms enriched for processes concerning peptidyl-diphthamide biosynthesis from peptidyl-histidine to YLR143w and YBR246w with p-values of 2×10−3 and 9×10−4 respectively (Figure 2C). Collectively, the FitDB and DRYGIN profiles thus provide robust phenotypic signatures suggesting novel roles in the diphthamide pathway for YBR246w and YLR143w, which are tightly clustered within the DPH gene network (Figure 2C). This notion is consistent with a recent report that YBR246w and its mammalian homolog, WDR85, have a diphthamide related function [41], [42]. Since YLR143w is as yet unassigned in the Saccharomyces genome database (SGD), based on the evidence below that YLR143w and YBR246w are indeed diphthamide synthesis genes we have named them DPH6 (YLR143w) and DPH7 (YBR246w).
DPH6 and DPH7 Deletions Cause Phenotypes Typical of Bona Fide Diphthamide Mutants
To verify the predicted roles for DPH6 and DPH7 in the diphthamide pathway, we next examined strains deleted for these ORFs for phenotypes specifically linked to defects in diphthamide formation on eEF2, namely sordarin resistance and response to DT [6], [7]. Sordarin traps eEF2 on the 80S ribosome [53], blocking mRNA translation elongation and yeast cell growth [54] in a fashion that depends on diphthamide synthesis [6], [7]. As a result, diphthamide mutants dph1-dph5 efficiently protect against sordarin inhibition [6], [7]. Like dph1-dph5, dph6 and dph7 mutants showed robust resistance towards sordarin at 10 µg/ml, a concentration inhibitory to the wild-type (Figure 3A). This resistance was comparable to that shown by eEF2 substitution mutants eft2H699I and eft2H699N (Figure 3A), which lack the His699 acceptor residue for diphthamide modification [37]. Thus DPH6 and DPH7 are novel sordarin effectors, a feature they share with the diphthamide synthesis genes DPH1-DPH5 [6], [7].
Fig. 3. DPH6 and DPH7 deletion strains copy traits typically related to the bona fide diphthamide mutants dph1-dph5. 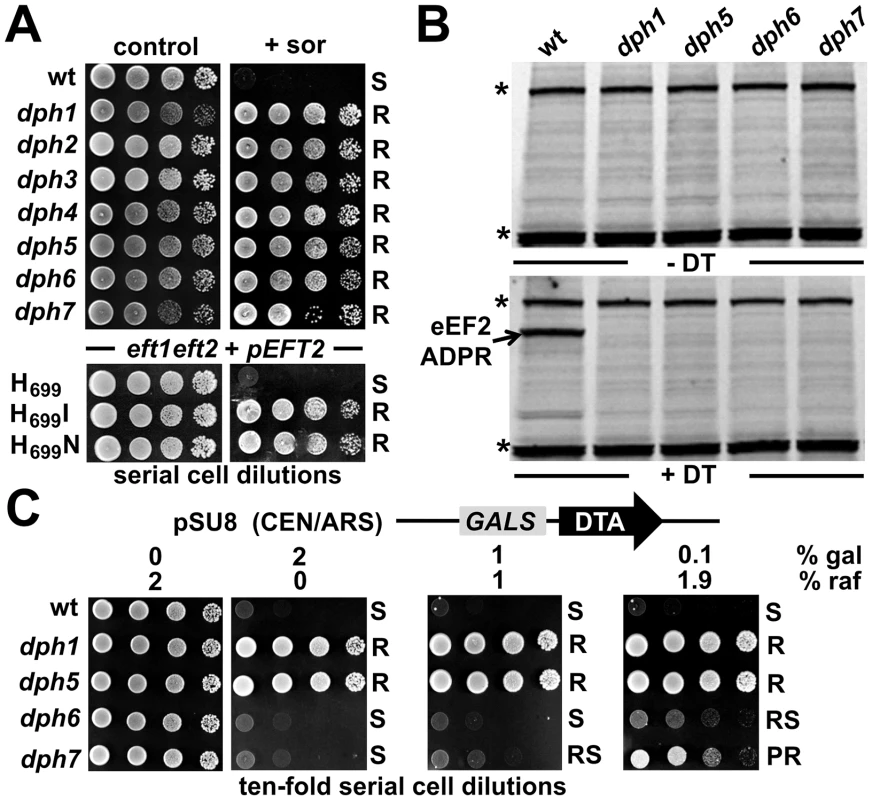
(A) Sordarin resistance. Ten-fold serial cell dilutions of the indicated yeast strains, BY4741 wild-type (wt) background and its dph1-dph7 gene deletion derivatives (upper panels) as well an MKK-derived eft1 eft2 double deletion background maintaining plasmid pEFT2 wild-type or H699 substitution (H699 N and H699I) alleles of EFT2 (lower panels), were grown on YPD plates in the absence (control) or presence (+sor) of 10 µg ml−1 sordarin. Growth was assayed for 3 d at 30°C. Sordarin resistant (R) and sensitive (S) responses are indicated. (B) Lack of in vitro ADP ribose acceptor activity of eEF2. Cell extracts obtained from dph1, dph5, dph6 and dph7 mutant and wild-type (wt) strains were incubated with (+DT) or without (−DT) 20 nM diphtheria toxin in the presence of biotin-NAD (10 µM) at 37°C for 1 hour. The transfer of biotin-ADP-ribose to eEF2 was detected by Western blotting using a streptavidin-conjugate. Two unknown non-specific bands (indicated by *) served as internal controls for even sample loading. (C) DT phenotype. As indicated, yeast dph mutants and wild-type control (wt) were tested for sensitivity to intracellular expression of DTA, the cytotoxic ADP ribosylase fragment of DT. This in vivo assay involved galactose-inducible expression from vector pSU8 (see Materials and Methods). Serial cell dilutions were replica spotted onto raffinose (2% raf) and galactose-inducing conditions using concentrations (2, 0.2 and 0.1% gal) suited to achieve gradual down-regulation of DTA toxicity. Growth was for 3 days at 30°C. DTA sensitive (S) resistant (R), partially resistant (PR) and reduced sensitive (RS) phenotypes are indicated. Diphthamide modification plays a key effector role for inhibitory ADP ribosylation of eEF2 by DT, hence dph1-dph5 mutants in both yeast and mammalian cells confer resistance towards DT [14], [16]. We therefore compared DT-dependent ADP ribosylation of eEF2 in vitro between wild-type cells and dph1, dph5, dph6 and dph7 mutants. While the translation factor from wild-type cells was efficiently modified by the toxic ADP ribosylase (Figure 3B), eEF2 extracted from dph1, dph5, dph6 and dph7 mutants could not be ADP ribosylated by exogenously added DT under the conditions used (Figure 3B). Such lack of ADP ribose acceptor activity in vitro strongly suggests defects in the diphthamide pathway and that DPH6 and DPH7 encode novel functions required for diphthamide formation. To further address this experimentally, we assayed the response of dph6 and dph7 mutants to intracellular expression of the ADP ribosylase domain of DT (DTA) using GALS, a truncated variant of the GAL1 promoter [55]. When DTA expression was induced by 0.1% galactose, dph6 and dph7 mutants were indeed found to show some protection against DTA in contrast to wild-type cells (Figure 3C), consistent with defects in diphthamide formation. However, at a higher level of expression on 2% galactose, they showed wild-type like sensitivity to DTA whereas dph1 and dph5 mutants remained fully resistant (Figure 3C). This suggests that eEF2 forms from dph6 or dph7 mutants, although not substrates in vitro (Figure 3B), can nonetheless be ADP ribosylated in vivo if DTA is expressed at a sufficiently high level [30]. While our work was in progress, eEF2 from a ybr246w/dph7 mutant was shown to be a very weak substrate for ADP ribosylation when treated with 10 mM DT [42], a 500-fold increase in concentration over that used in our in vitro ADP ribosylation assays (Figure 3B). Thus eEF2 from the dph6 or dph7 mutants is resistant to sordarin and shows a vastly reduced ability to be ADP-ribosylated by DT, strongly suggesting that the diphthamide pathway is defective. Since the intermediate diphthine can serve as a sub-optimal substrate for ADP ribosylation using excess levels of DT or upon overexpressing its toxic ADP ribosylase domain from inside cells [29], [31], the properties of eEF2 from dph6 and dph7 mutants are consistent with a defect in the final step of the pathway that converts diphthine to diphthamide. Our analysis is therefore entirely consistent with the above database predictions and indicates DPH6 and DPH7 constitute novel candidate loci for diphthamide biosynthesis.
Mass Spectrometry Reveals Diphthine Accumulation in dph6 and dph7 Mutants Due to a Block in the Terminal Amidation Step of the Diphthamide Pathway
Given the above evidence, we next examined whether eEF2 prepared from cells deleted for either DPH6 or DPH7 carried any modification on His699, the eEF2 residue that is modified to generate diphthamide. eEF2 preparations made from wild-type and gene deletion strains expressing His6-tagged eEF2 were digested with trypsin and examined by mass spectrometry. The His6-tagged form was chosen as the source of eEF2 since expression rescued the inviability of an eft1 eft2 double mutant lacking eEF2 function, and it is thus considered to be biologically active [56]. Strains lacking either DPH1, in which the first step of diphthamide biosynthesis is blocked, or lacking DPH5 (encoding diphthine synthase), were used respectively as controls for complete lack of modification and presence of ACP, the first intermediate in the diphthamide pathway [14], [16], [30], [32]. All strains expressed similar levels of His6-tagged eEF2 (data not shown).
The modified histidine in eEF2 (His699) is located in the tryptic peptide 686-VNILDVTLHADAIHR-700 and, as expected, unmodified versions of this peptide were readily detected in eEF2 prepared from the dph1 mutant (Figure S1C). Unmodified peptide was also found in eEF2 prepared from dph5, dph6 and dph7 deletion strains as well as from wild-type cells (Figures S1 and S2). Thus even in wild-type cells not all of the eEF2 is modified by diphthamide. In addition to the unmodified peptide, we readily detected diphthamide-modified peptide in eEF2 prepared from the wild-type strain (Figure 4A), but failed to detect this in any of the mutants. Instead, ACP-modified peptide was found in eEF2 prepared from the dph5 mutant (Figure 4B), as expected given its known role in generating diphthine [32] from the ACP intermediate in the pathway.
Fig. 4. MS/MS spectra of diphthamide-, ACP-, and diphthine-modified EF2 peptide 686-VNILDVTLHADAIHR-700 from wild-type and mutant yeast strains. 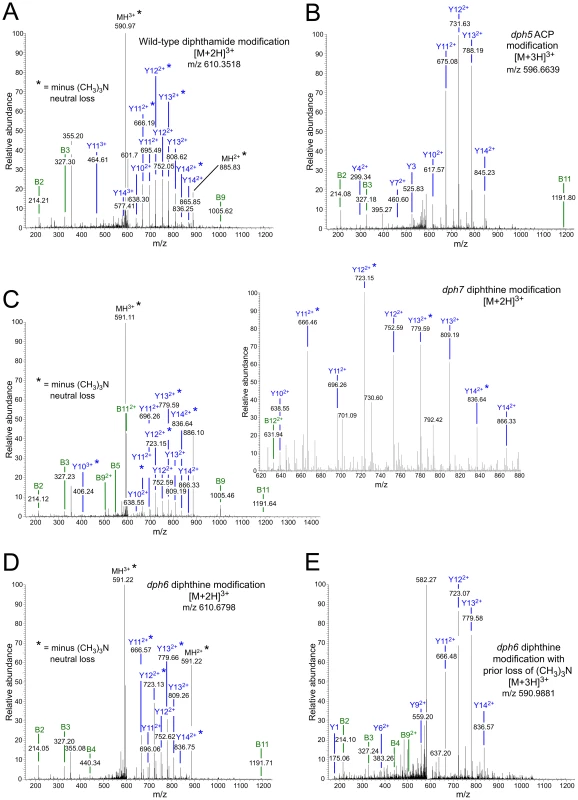
Spectra are shown for (A) diphthamide-modified peptide from the wild-type yeast strain; (B) ACP-modified peptide from the dph5Δ mutant; (C) diphthine-modified peptide in the dph7Δ strain; (D) diphthine-modified peptide in the dph6Δ strain; (E) diphthine-modified peptide in the dph6Δ strain with loss of the trimethylamino group before analysis in the mass spectrometer indicated by the parent ion m/z. In each case the parent ion m/z and charge state is indicated. In (A), (C) and (D), * indicates neutral loss of trimethylamino during MS/MS. The inset in (C) shows greater detail for the more crowded part of the MS/MS spectrum. Figure S2A indicates how the B and Y ions are derived from the peptide sequence. In contrast, eEF2 from the dph7 mutant generated spectra consistent with the presence of diphthine on His699, in which the m/z values for both the parent ions and the daughter ions in the MS/MS spectra were higher in a manner consistent with the 0.984 Da extra mass associated with presence of a carboxyl group in diphthine rather than the amide group in diphthamide (Figure 4C). Thus each of the doubly-charged daughter ions in Figure 4C is larger by an m/z of ∼0.5 than the corresponding ion in the wild-type spectrum (Figure 4A). Furthermore, the quite different elution times of the diphthine-modified and diphthamide-modified peptide that are evident from the extracted ion chromatograms (Figure S3) are consistent with differently modified forms of eEF2. As noted in previous studies [32], [33], [36], some of the ions in our MS/MS spectra had undergone neutral loss of the trimethylamino group during MS/MS, as indicated by loss of 59.110 mass units.
Two types of spectra corresponding to the peptide with modified His699 were seen when eEF2 from the dph6 mutant was analyzed. In some spectra (Figure 4D), the parent ion m/z and MS/MS data indicated the presence of diphthine as in the dph7 mutant, with some daughter ions again showing neutral loss of the trimethylamino group during MS/MS as noted above. However, we also detected peptide forms in which elimination of the trimethylamino group had occurred prior to analysis, as indicated by the lower parent ion m/z (Figure 4E) and an MS/MS spectrum in which all assignable peaks corresponded to ions lacking the trimethylamino group. Such trimethylamino elimination prior to mass spectrometry was observed previously when diphthine-modified Pyrococcus horikoshii EF2 was generated in an in vitro reaction [32], indicating that this modification might be unstable. However, we failed to detect any pre-mass spectrometry loss of the trimethylamino group when eEF2 from the dph7 mutant was analyzed. Thus while eEF2 from both mutants carries diphthine, the modification appears to be more labile in the dph6 mutant and may be protected from trimethylamino elimination by the absence of Dph7.
Figure S3 shows extracted ion chromatograms for ions with m/z values corresponding to the His699 containing peptide modified with diphthamide, diphthine or with ACP, indicating that the ACP modified peptide was only present in the dph5 mutant, the diphthine modified peptide was only present in dph6 and dph7 mutants, and diphthamide-modified peptide was only seen in wild-type cells. Our mass spectrometry analysis therefore shows that in yeast strains lacking either DPH6 or DPH7, modification of His699 progresses only as far as diphthine. Thus both loci indeed qualify as novel diphthamide synthesis genes with likely roles in conversion of diphthine to diphthamide.
Protein–Protein Interactions Between Dph6, Dph7, Dph5, and EF2
Although Dph6 and Dph7 appear to function within the same step of the diphthamide synthesis pathway, using co-immune precipitation they were not found to interact either with one another or with Dph2 and Dph5, players involved in the two earlier pathway steps (Figure S4; Figure S5 and data not shown). However, in support of our evidence that Dph6 is a diphthamide biosynthetic factor, we observed by co-immune precipitation that Dph6-HA bound to a fraction of (His)6-tagged eEF2 (Figure 5A). Intriguingly, this interaction was independent of Dph7 (Figure 5A), suggesting Dph7 may not mediate interaction between Dph6 and the translation factor. Dph7 is also unlikely to play an indirect role through regulation of DPH6 gene expression because Dph6 protein levels were unaltered in the DPH7 deletion strain (Figure 5A).
Fig. 5. Co-immune precipitations reveal eEF2 interactions with Dph6 and Dph5. 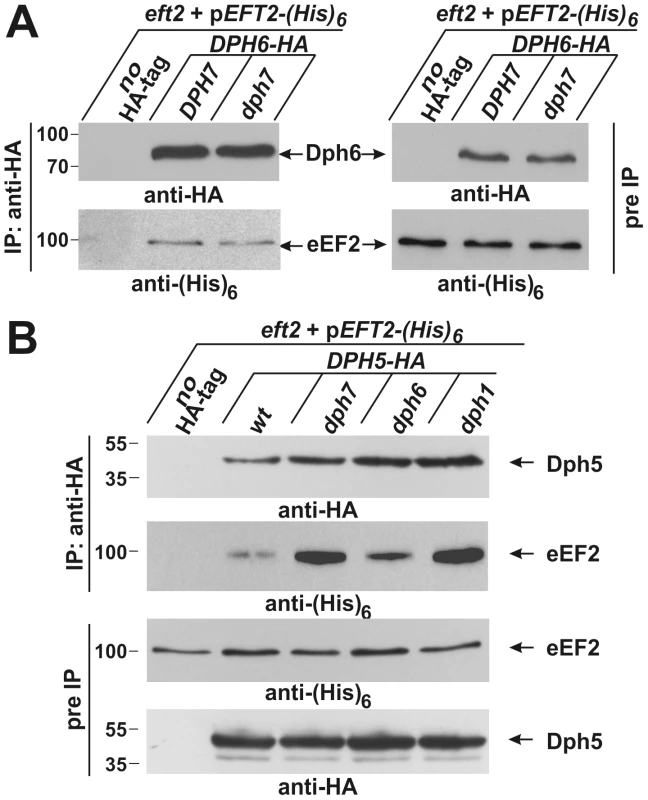
(A) eEF2 interacts with Dph6 in a fashion that is independent of Dph7. (B) eEF2 interaction with Dph5 is dramatically enhanced by elimination of Dph7 or Dph1. Yeast strains co-expressing (His)6-tagged eEF2 with Dph6-HA (A) or Dph5-HA (B) in the background of wild-type (A: DPH7 and B: wt) and dph mutant strains (A: dph7; B: dph1, dph6 and dph7) were subjected to immune precipitations (IP) using the anti-HA antibody. Strains expressing (His)6-tagged eEF2 on their own served as IP controls (A and B: no HA-tag). Subsequently, the precipitates were probed with anti-HA (A: top left panel; B: first panel) and anti-(His)6 antibodies (A: bottom left panel) to check for the content of Dph6-HA (A) and Dph5-HA (B), respectively (all indicated by arrows). The content of HA-tagged Dph6 (A) and Dph5 (B) as well as (His)6-marked eEF2 (A and B) in the protein extracts prior to IP (pre-IP) was examined on individual Western blots using anti-HA (A: top right panel; B: fourth panel) and anti-(His)6 antibodies (A: bottom right panel; B: third panel), respectively. While absence of Dph7 hardly affected the Dph6•eEF2 interaction (A), Dph5•eEF2 interaction was strongly enhanced by inactivating DPH7 or DPH1 (B). Inactivation of WDR85, the mammalian homolog of Dph7, was recently shown to dramatically enhance association of diphthine synthase Dph5 with eEF2 [41]. We therefore examined whether Dph7 impacts on the interaction between Dph5 and eEF2 in budding yeast. We found that a much higher level of affinity tagged eEF2 could be co-immune precipitated with HA-tagged Dph5 from extracts of the dph7 mutant in comparison to wild-type extracts (Figure 5B). A smaller increase was also seen with the dph6 mutant (Figure 5B). This strongly suggests a conserved role for Dph7/WDR85 as a regulator of the Dph5•eEF2 interaction. Remarkably, we also found similarly enhanced binding of Dph5 to eEF2 in the dph1 mutant, which has a defect in the first step of the diphthamide pathway and therefore lacks the ACP modification that is the immediate substrate of diphthine synthase (Figure 5B). Strikingly, DPH5 overproduction from a galactose-inducible promoter was found to be highly detrimental to cells deleted for DPH7 and to all mutants blocked at the first step of the pathway, but had little effect on the dph6 mutant and no effect on wild-type or dph5 cells (Figure 6A). Intriguingly, this cytotoxicity goes hand in hand with the enhanced Dph5•eEF2 interaction profiles we observed in dph1, dph6 and dph7 cells under conditions of wild-type DPH5 copy number and normal Dph5 expression levels (Figure 5B). Taken together, our results suggest that binding of Dph5 to incompletely modified eEF2 may be inhibitory to the function of the translation factor. Our data also indicate that both unmodified eEF2, and diphthine-modified eEF2 in the absence of Dph7, show strongly enhanced binding to Dph5. Furthermore, since we failed to detect association between Dph5 and Dph6 despite demonstrating interaction of each with eEF2, it is likely that Dph5 and Dph6 do not bind concurrently to eEF2 and that their binding may therefore be mutually exclusive.
Fig. 6. dph mutants show sensitivity to elevated diphthine synthase levels and confer reduced translational accuracy. 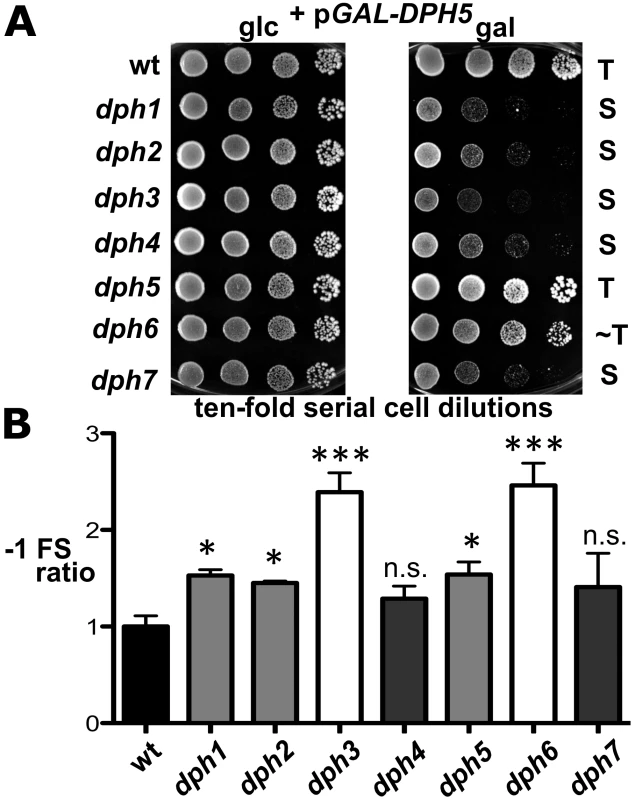
(A) DPH5 overexpression in dph1-dph4 and dph7 mutants causes cytotoxicity and a severe cell growth defect. Cells of yeast strains with the indicated genetic backgrounds and maintaining plasmid pGAL-DPH5 for galactose inducible overexpression of diphthine synthase Dph5 were serially diluted and replica spotted onto glucose (2% glc) and galactose (2% gal) media to assay their response to DPH5 overexpression. Growth was for 3 days at 30°C. Unaltered (T), slightly weakened tolerance (∼T) and sensitive (S) responses are indicated. Note that dph1-dph4 and dph7 mutants are extremely sensitive to DPH5 overexpression. (B) Ribosomal frameshift analysis reveals erroneous translation in dph1-dph7 mutants. Strains with the indicated genetic backgrounds were transformed with control (pJD240.0) or lacZ −1 frameshift (pJD240.−1) plasmids [59] to monitor lacZ expression through β-galactosidase (β-Gal) production using O-nitrophenol-D- galactopyranoside assays and to score translation efficiency (pJD240.0) and fidelity (pJD240.−1). Ribosomal −1 frameshifts are expressed relative to the level of overall translation efficiency with statistical significance determined by one-way ANOVA followed by Dunnett's multiple comparison. With the exception of dph4 and dph7, post-hoc comparison found that all other mutant backgrounds showed a significant increase in ribosomal −1 frameshifting relative to wild-type (wt) yeast cells (* = P<0.05; *** = P<0.001; ns. = not significant). Physiological Implications of the Diphthamide Modification on eEF2
Although the precise biological function of diphthamide is unclear, its location at the tip of the eEF2 anticodon mimicry domain IV predicts a potentially important role in translation. Consistent with this, structure-function studies have shown that domain IV is sufficiently proximal for interaction with tRNA in the decoding P-site of the ribosome [57] and alterations of invariant tip residues, including H699 substitutions that cannot be diphthamide modified, confer biologically significant traits including thermosensitive growth defects [37], [58]. Nonetheless, when compared to their wild-type parental strain, we found no significant changes in the growth performance of dph1-dph7 mutants in either liquid or on solid media and at standard cultivation temperatures of 30°C (Figure S6). Even increasing the cultivation temperatures to 39°C had no discernable effect on dph cell growth except for the dph3/kti11 mutant (Figure S6), which is known to be thermosensitive due to additional functions unrelated to diphthamide [6].
However, intrigued by previous reports that diphthamide defects can induce ribosomal frame-shifts [6], [36], we next studied whether DPH6 and DPH7 deletions affect the accuracy of eEF2 in the translation process (Figure 6B). Using lacZ-based reporters to monitor programmed +1 and −1 frameshift signals derived from Ty elements [36], [59], dph1-dph7 mutants failed to induce significant ribosomal +1 frameshifts (data not shown). However, dph1, dph2, dph3, dph5 and dph6 mutants significantly enhanced lacZ expression dependent on a −1 frameshift, with dph6 and dph3 cells scoring as the top −1 frameshifters followed by lower but statistically significant effects in dph1, dph2 and dph5 mutants (Figure 6B). This confirms increased −1 frameshifting in dph2 and dph5 mutants seen previously [36] and demonstrates an even larger defect in dph3 and dph6 strains. Ribosomal −1 frameshift induction by dph7 and dph4, though slightly increased in relation to wild-type controls, was considered statistically insignificant (Figure 6B). The −1 frameshifting phenotype shared between dph6 and bona fide dph mutants is consistent with a role for diphthamide in promoting translational accuracy of eEF2. In line with a role for diphthamide in the fine tuning of translation elongation, growth assays performed under thermal and/or chemical stress conditions showed that certain dph mutants including DPH6 and DPH7 deletion strains displayed altered responses to translation elongation indicator drugs such as hygromycin, anisomycin or paromomycin (Figure S7). In conclusion, our data indicate that diphthamide mutant strains such as dph6 increase ribosomal errors typical of −1 translational frameshifts and that the diphthamide modification function of Dph6, which is required for completion of diphthamide synthesis, is likely to assist eEF2 in reading frame maintenance during translation.
Discussion
We have presented genetic, phenotypic, mass spectrometric and biochemical analyses that clearly identify Dph6 as a novel protein required for the final step of diphthamide biosynthesis and that confirm a similar role for Dph7 as reported recently [41], [42]. Thus in yeast strains lacking either DPH6 or DPH7, modification of His699 on eEF2 progresses only as far as diphthine and these gene products are required for amidation of diphthine to generate diphthamide. Our findings are consistent both with a recent bioinformatics analysis that predicted a role for Dph6 in the diphthine to diphthamide conversion [60] and with the identification of Dph6 as yeast diphthamide synthetase reported by Su et al. [61] while we were revising our manuscript.
Dph6
Dph6 contains three conserved domains consistent with it functioning as an enzyme (Figure S8). The amino-terminal 225 residues constitute an Alpha_ANH_like_IV domain (cd1994 in the NCBI Conserved Domain Database [62], also known as DUF71), a member of the adenine nucleotide alpha hydrolase superfamily that is predicted to bind ATP. Many DUF71 proteins from archaea to mammals contain the highly conserved motif –E215GG(D/E)XE220 – (Dph6 numbering), which has been proposed to be involved in substrate binding and catalysis and which is replaced by –ENGE(F/Y)H – in a group of related DUF71 proteins implicated in biotin synthesis [60]. Based on this we generated a dph6 allele encoding two substitutions in this region (G216N, E220A) and tested its functionality by monitoring complementation of sordarin resistance in a yeast dph6 knockout strain. Figure 7 clearly shows that this small change completely inactivates the function of Dph6, demonstrating that the Alpha_ANH_like_IV domain is critical for the conversion of diphthine to diphthamide. The C-terminal portion of Dph6 contains two domains related to the YjgF-YER057c-UK114 protein family (eu_AANH_C1: cd06155 and eu_AANH_C2: cd06166) that may promote homotrimerisation and formation of an inter-subunit cleft that has been proposed to bind small molecule ligands [63]–[65]. Several key residues in human UK114 required for homotrimerisation and ligand binding [66] are present in Dph6 (Figure S8) including arg-107, which in E. coli TdcF forms a bidentate salt bridge with the carboxylic acid group of bound ligands [63]. Deletion of residues 335–415 encompassing much of the YjgF-YER057c-UK114 region abolished the function of Dph6 as monitored by sordarin resistance (Figure 7), while truncation of Dph6 at the first of the two conserved domains by insertion of a myc tag also eliminated Dph6 function (Figure 7) despite detectable expression of the truncated polypeptide (data not shown), indicating that the YjgF-YER057c-UK114 domains are also important for Dph6 function and that the Alpha_ANH_like_IV domain is nonfunctional on its own. Since Salmonella enterica YjgF has an enamine/imine deaminase activity that is conserved in human UK114 [67] it is possible that the YjgF-YER057c-UK114 domains in Dph6 are used to generate ammonia for diphthamide formation.
Fig. 7. Both the Alpha_ANH_like_IV and YjgF-YER057c-UK114 domains in Dph6 are essential for its functionality. 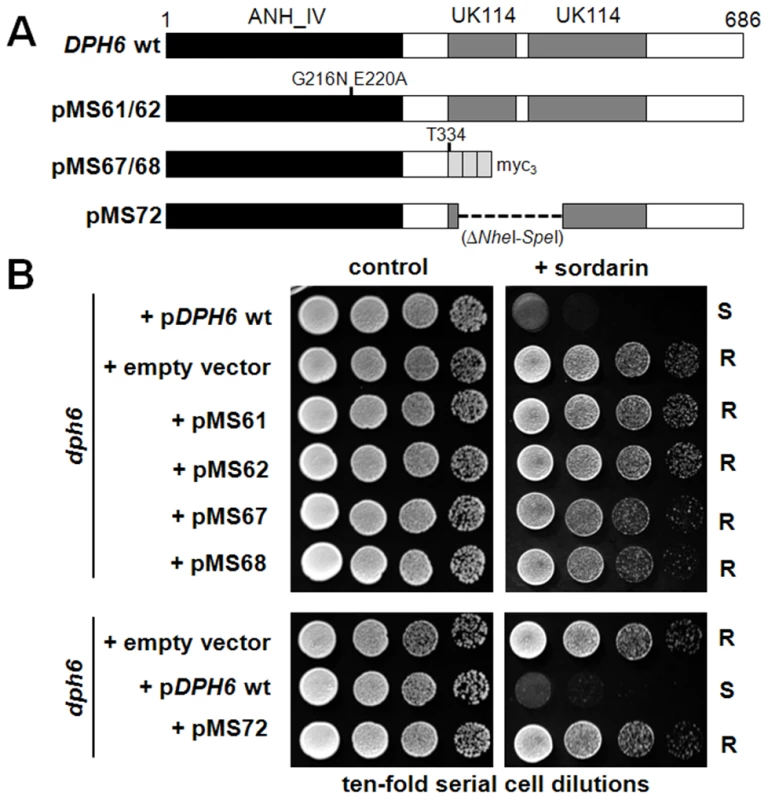
(A) Diagram showing the DPH6 wild-type and mutant constructs tested in (B), indicating the Alpha_ANH_like_IV (ANH) and YjgF-YER057c-UK114 (UK114) domains and the position of point mutations, an in-frame deletion (- - - - -) and triple myc epitope tag (myc3) as appropriate. (B) Ten-fold serial cell dilutions of a dph6 deletion strain carrying the constructs shown in (A) or the corresponding empty vector (top panel, pSU6 [wt DPH6]; lower panel, pSU7 [wt DPH6]: Table S3) were spotted onto SCD-Leu plates with or without 10 µg/ml sordarin and grown at 30°C for 3 days. Taken together, these properties suggest a direct, ATP-dependent role for Dph6 in diphthine amidation proceeding via an adenylated intermediate and with ammonia acting as the source of the amide group. Such a direct role has now been demonstrated by Su et al., who have used an in vitro assay system to show that Dph6 has diphthamide synthetase activity [61]. Although proteins showing Dph6-like domain organization are readily identified in fungi, plants, amphibians and insects (Figure S8), they are largely absent from archaeal and mammalian proteomes. However, mammals and archaea have separate proteins showing strong similarity to either the adenine nucleotide alpha hydrolase domain or to the YjgF-YER057c-UK114 related regions (Figure S8 and data not shown), suggesting Dph6 functionality may be split between different polypeptides in these cases. It is therefore surprising that expression of the human DPH6 ortholog in a yeast dph6 mutant can restore diphthamide biosynthesis [61] despite lacking the YjgF-YER057c-UK114 domains that are essential in the yeast protein (Figure 7; [61]). Thus while the core function of the enzyme must therefore reside in the Alpha_ANH_like_IV domain, it will be interesting to determine the role of the YjgF-YER057c-UK114 domains in Dph6 from lower eukaryotes.
Dph7
Dph7 has four well-defined WD40 repeats (Figure S9) and its predicted structure consists exclusively of β-sheet elements [41], [68]. Although its human homolog WDR85 has been implicated in the first step of diphthamide biosynthesis [41], our work and that of Su et al. [42] show that the pathway can proceed as far as diphthine in the absence of DPH7 and that the block is therefore in conversion of diphthine to diphthamide. Furthermore, this block cannot be bypassed simply by introducing DPH6 on a multicopy plasmid to increase the level of diphthamide synthetase (data not shown). How then might Dph7 contribute to diphthine amidation? Its domain structure suggests it could act as an adaptor molecule for diphthine amidation [42], but this notion is at odds with our failure to detect interaction between Dph7 and Dph6 (see above). Our intriguing finding that eEF2 binds much more Dph5 in the absence of Dph7 suggests an alternative role, namely that Dph7 is needed to displace Dph5 from diphthine-modified eEF2 to allow the amidation reaction to occur. Similar findings in mammalian cells upon inactivation of WDR85 support this notion [41]. Together with our data showing that viability of dph1-dph4 and dph7 cells is extremely sensitive to excess Dph5 in comparison to wild-type or dph6 cells, it appears that binding of Dph5 to eEF2 is inhibitory to the function of the translation factor and negatively interferes with cell growth unless eEF2 carries the completed diphthamide modification. Perhaps in addition to catalyzing methylation of ACP-modified eEF2, Dph5 binds to newly-synthesised eEF2 to exclude it from functioning in translation until the diphthine amidation step takes place (Figure 8). Consistent with this proposal is our observation that the level of Dph5 associated with eEF2 in the dph1 mutant, in which modification of His699 cannot be initiated, is drastically increased and virtually indistinguishable from the enhanced Dph5-eEF2 interaction seen when Dph7 is absent. Dph7 may be needed to displace Dph5 once diphthine has been generated so that Dph6 can carry out the diphthine to diphthamide conversion (Figure 8), a notion consistent with the sensitivity of the dph7 mutant to DPH5 overexpression. In contrast, the dph6 mutant may tolerate Dph5 overexpression because Dph7 is present to displace it.
Fig. 8. Model for the diphthamide pathway incorporating the proposed novel roles of Dph5, Dph6, and Dph7. 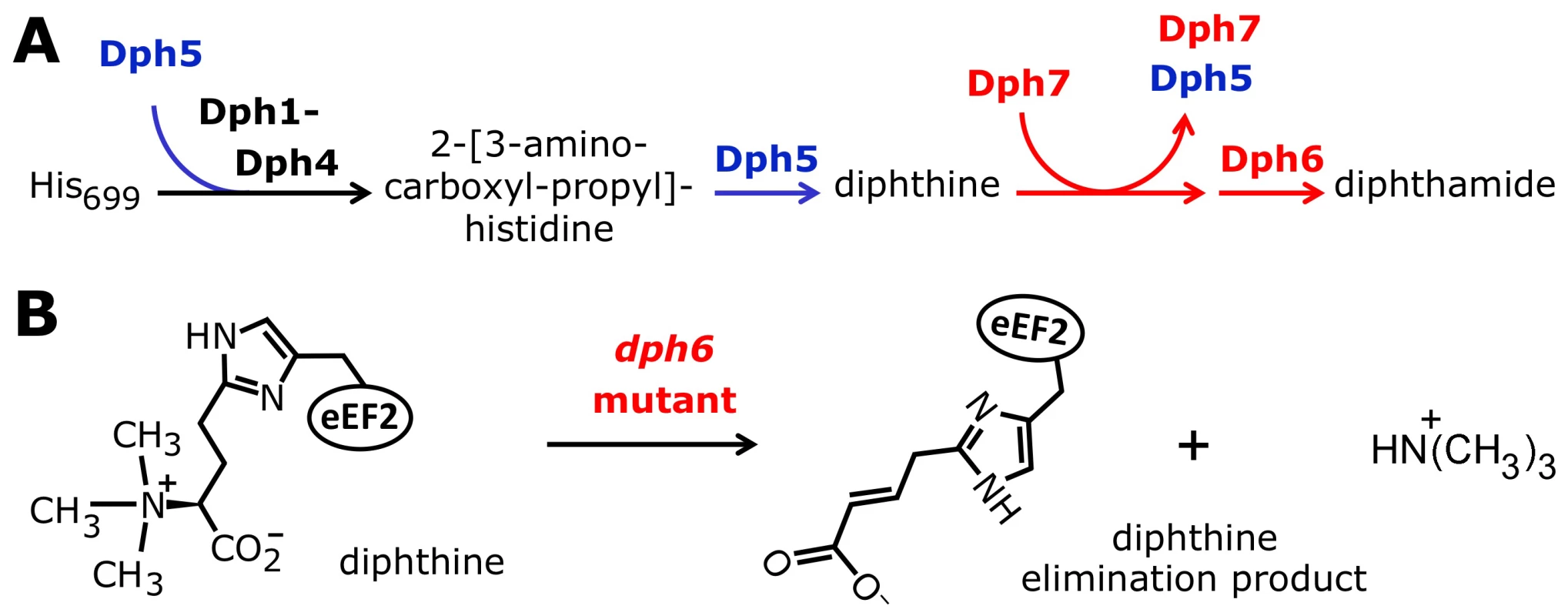
(A) Diphthamide pathway showing interaction of Dph5 with unmodified eEF2 and the proposed role of Dph7 in displacement of Dph5 prior to diphthine amidation. (B) Elimination of the trimethylamino group in the absence of the proposed amidase Dph6 suggesting lability of diphthine in its absence. Two other seemingly unrelated functions have been previously proposed for DPH7. Firstly, it emerged from a genetic screen as a potential negative regulator of RNA polymerase I (Rrt2), although no other DPH genes were similarly identified [69]. Secondly, DPH7 has been implicated in retromer mediated endosomal recycling and named ERE1 [68]. The connection between endosomal recycling and diphthamide biosynthesis is currently unclear and it remains to be determined whether Dph7 is multifunctional or if these other roles are linked to its eEF2 modification function.
Diphthamide on eEF2 is the target for bacterial ADP-ribosylase toxins and also affects toxicity of sordarin and ricin, a ribosome inhibiting protein toxin from plants [70]. Although this emphasizes its pathological relevance, the physiological significance of diphthamide remains enigmatic and elusive. Nonetheless, the evolutionary conservation of the diphthamide pathway among eukaryotes and the embryonic lethality of mice that cannot synthesize diphthamide [38] strongly suggest that it is important in translation related processes. In support of this notion, evidence presented here and by others shows that diphthamide mutants cause increased translational frameshifting, a defect also observed in mammalian cells [6], [36], [71]. Diphthamide modification may have particular importance in multicellular organisms or when cells are stressed [4]. Mutation of mammalian diphthamide synthesis genes affects cell proliferation and development: inactivation of DPH3/KTI11 is associated with tRNA modification defects and neurodegeneration and mutations in DPH1/OVCA1 revealed a tumor suppressor role for this diphthamide synthesis gene in ovarian cancer [27], [38]–[40], [72]. Regardless of its physiological functions, our data indicate that the diphthamide pathway is more complex than originally anticipated and comprises, in addition to Dph1-Dph5, two further components, Dph6 and Dph7, which operate in the terminal amidation step (Figure 8). While it is now clear that Dph6 is diphthamide synthetase [61], in the future it will be important to understand why the archaeal and mammalian orthologs can dispense with the otherwise conserved YjgF-YER057c-UK114 domains and to define the precise role of Dph7. It will also be crucial to explore the potential role of diphthine synthase (Dph5) as a potential regulator of the entire pathway and the reasons for apparent lability of diphthine in the dph6 mutant that is suggested by our data (Figure 8).
Materials and Methods
Strains, Media, Growth Conditions, and Growth Assays
Yeast strains used in this study are listed in Table S2 and plasmids in Table S3. Cultures were grown in complete (YPD) or minimal (SD) media [73] at 30°C unless otherwise stated. For phenotypic assays, YPD was supplemented with 10 µg/ml sordarin sodium salt from Sordaria araneosa (Sigma-Aldrich). Yeast transformations with plasmid DNAs were performed following the lithium acetate protocol [74]. Diphtheria toxin (DT) growth assays in vivo involved expression of the toxin's cytotoxic ADP ribosylase fragment (DTA) from vector pSU8 (p415-GALS-DTA), essentially as previously described for dph1-dph5 mutants [6]. pSU8 was made by cloning the BamHI fragment encoding DTA from pLMY101 [30] into plasmid p415-GALS, a single-copy E. coli-yeast shuttle vector with a truncated GAL promoter [55], which allows for conditional DTA induction on galactose-containing media. [55]. The translational frameshift reporter assay essentially involved previously published protocols together with the described lacZ reporter plasmids pJD204.0 (wild-type control), pJD204.−1 (−1 frame) and pJD204.+1 (+1 frame) [36], [59]; the pJD204 plasmid series was kindly provided by T. Kinzy (UMDNJ, USA). The relative values for +1 and −1 frameshifting were statistically analyzed using one-way ANOVA followed by Dunnett's multiple comparison post test and was performed with Graphpad Prism 5.0 software essentially as previously described [75].
Gene Deletion and Epitope Tagging
Details of all primers used in numerous PCR dependent genomic manipulation experiments can be found in Table S4. Gene deletions were performed using in vivo PCR-based one step-gene disruption protocols in combination with marker plasmids YDpKl-L, YDpKl-U or YDpSp-H [76] and knockout primers (Table S4) including those previously described [6], [25], [77]. Gene deletions were confirmed via diagnostic PCR on genomic DNA preparations using target ORF-specific primer pairs (Table S4) as well as sordarin response assays. C-terminal tagging of DPH1, DPH2, DPH5, DPH6/YLR143w and DPH7/YBR246 was performed according to previously published in vivo PCR-based epitope tagging protocols [78] using appropriate S3/S2 primer pairs (Table S4). Tagged genes were confirmed by Western blot detection with anti-HA or anti-c-Myc antibodies (Santa Cruz Biotechnology A-14 and F7, respectively). Detection of HA - or c-Myc-tagged Dph1, Dph2, Dph5, Dph6 and Dph7 as well as Dph3 and Elp2 in co-immune precipitation (Co-IP) assays were performed as previously described [6], [77], [79].
DPH6 Constructs
pSU6 was generated by insertion into YCplac111 [80] of a genomic PCR fragment including DPH6 together with 829 bp of upstream and 59 bp of downstream sequence flanked by EcoRI and BamHI sites incorporated using PCR primers (Table S4). The insert was verified by sequencing and shown to complement a dph6 knockout strain. pSU7 was made by cloning the DPH6 insert from pSU6 into YEplac181 [80]. To generate a G216N E220A dph6 mutant, pSU6 was digested with AgeI and BsmBI and the small DPH6 fragment replaced by an identical synthetic fragment (Integrated DNA Technologies) carrying the G216N E220A mutations, generating independent clones pMS61 and pMS62. The replaced region was verified by DNA sequencing. pMS67 and pMS68 were generated from pSU6 by replacing the BsmBI-SalI fragment carrying the C-terminal region of DPH6 and downstream sequence with a synthetic BsmBI-SalI fragment in which codons 335–685 were replaced by sequence encoding the linker and triple myc tag from pYM23 [81]. To generate pMS72, the smaller NheI-SpeI fragment of pSU7 was excised and the large fragment ligated to generate an in-frame fusion that removed DPH6 codons 347–471, checking the resulting fusion by DNA sequencing.
In Vitro ADP Ribosylation Assay
Yeast cell extracts were prepared as described previously [15]. ADP ribosylation reactions were performed at 37°C for 1 hour in a volume of 40 µl ADP ribosylation buffer (20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 50 mM DTT) containing 50 µg of yeast extract, 50 ng fully-nicked DT and 10 µM 6-biotin-17-NAD (Trevigen). Samples were then mixed with SDS sample buffer, boiled for 5 min and run on 4–25% SDS-PAGE gradient gels (Invitrogen). The proteins were transferred to nitrocellulose membranes and Western blotting was performed using streptavidin-IR conjugate (Rockland Immunochemicals, Gilbertsville, PA) and scanned on an Odyssey Infrared Imager (LICOR Biosciences, Lincoln, NE).
Expression and Purification of Affinity-Tagged eEF2-(His)6
BY4741 wild-type yeast cells as well as dph1, dph5, ylr142w/dph6 and ybr246w/dph7 mutants thereof carrying an eft2 null-allele were transformed with plasmid pTKB612 (a kind gift from A. R. Merrill, University of Guelph, Ontario, Canada), which expresses a (His)6-tagged form of translation elongation factor eEF2 (Table S3) that is fully functional and able to complement an eft1 eft2 double mutant [56]. In order to express and purify (His)6-tagged eEF2 for MS/MS analysis, 750 ml of yeast culture were grown in YPD to an OD600 2.0 and harvested by centrifugation. The pellet was resuspended in 3 ml B60 buffer (50 mM HEPES-KOH pH 7.3, 60 mM KOAc, 5 mM Mg(OAc)2, 0.1% Triton X100, 10% (v/v) glycerol, 1 mM NaF, 20 mM glycerophosphate, complete protease inhibitor [Roche]) without DTT and cells were lysed in a bead beater. The lysate was centrifuged twice at 13,500 rpm for 30 min. and the protein concentration measured with a NanoDrop spectrophotometer. Five mg total protein was applied to 2 mg anti-(His)6-tag Dynabeads (Invitrogen, #101-03D) and purified according to manufacturer's instructions. The identity of purified eEF2 fraction was confirmed by SDS-PAGE and Western blot analysis using an anti-(His)6 antibody (Abcam, #ab18184).
Analysis of Diphthamide Pathway Modifications on eEF2 by Mass Spectrometry
Crude yeast eEF2 preparations from wild-type and dph mutants strains were separated by SDS-PAGE using 4–12% Bis-Tris precast gels (Invitrogen, Carlsbad, USA) and the area of the gel containing eEF2 was excised after staining with Instant Blue Coomassie (Expedeon, Cambridge, UK). In-gel digests were performed using trypsin, subsequent to reduction and alkylation with dithiothreitol and iodoacetamide, with the resulting peptides cleaned over C18 columns. Peptides were then analyzed via HPLC-MS/MS using a Dionex U300 HPLC (Dionex California) with a 15 cm PepMap C18 column coupled to a Thermo Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). The peptides were eluted from the C18 column at 300 nL/min over 120 min using a linear 5–90% (v/v) acetonitrile gradient. The Orbitrap Velos was operated in positive ion mode, with an ion source voltage of 1.2 kV and capillary temperature 200°C, using a lock mass of 445.120024. The initial survey scan was performed at 60000 resolution, FTMS scanning from 335–1800 Da. The top 15 most intense ions were selected for MS/MS sequencing, using collision-induced dissociation (CID; MS/MS charge state 1+ rejected, >2+ accepted). Protein identification was performed using MaxQuant 1.2.2.5 [82] against a proteome database generated from the Saccharomyces Genome database [83]. Manual annotation of the modified peptide spectra corresponding to the modified EF2 peptide and generation of extracted ion chromatograms were done using the Thermo Xcalibur software for spectra visualization.
Supporting Information
Zdroje
1. AhrneE, MullerM, LisacekF (2010) Unrestricted identification of modified proteins using MS/MS. Proteomics 10 : 671–686.
2. SeetBT, DikicI, ZhouMM, PawsonT (2006) Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol 7 : 473–483.
3. WalshCT, Garneau-TsodikovaS, GattoGJJ (2005) Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl 44 : 7342–7372.
4. GreganovaE, AltmannM, ButikoferP (2011) Unique modifications of translation elongation factors. FEBS J 278 : 2613–2624.
5. Van NessBG, HowardJB, BodleyJW (1980) ADP-ribosylation of elongation factor 2 by diphtheria toxin. NMR spectra and proposed structures of ribosyl-diphthamide and its hydrolysis products. J Biol Chem 255 : 10710–10716.
6. BärC, ZabelR, LiuS, StarkMJ, SchaffrathR (2008) A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes: Kti11/Dph3. Mol Microbiol 69 : 1221–1233.
7. BotetJ, Rodriguez-MateosM, BallestaJP, RevueltaJL, RemachaM (2008) A chemical genomic screen in Saccharomyces cerevisiae reveals a role for diphthamidation of translation elongation factor 2 in inhibition of protein synthesis by sordarin. Antimicrob Agents Chemother 52 : 1623–1629.
8. Van NessBG, HowardJB, BodleyJW (1980) ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J Biol Chem 255 : 10717–107120.
9. DominguezJM, Gomez-LorenzoMG, MartinJJ (1999) Sordarin inhibits fungal protein synthesis by blocking translocation differently to fusidic acid. J Biol Chem 274 : 22423–22427.
10. JørgensenR, OrtizPA, Carr-SchmidA, NissenP, KinzyTG, et al. (2003) Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat Struct Biol 10 : 379–385.
11. OppenheimerNJ, BodleyJW (1981) Diphtheria toxin. Site and configuration of ADP-ribosylation of diphthamide in elongation factor 2. J Biol Chem 256 : 8579–8581.
12. PappenheimerAMJr (1977) Diphtheria toxin. Annu Rev Biochem 46 : 69–94.
13. Uthman S, Liu S, Giorgini F, Stark MJR, Costanzo M, et al.. (2012) Diphtheria disease and genes involved in formation of diphthamide, key effector of the diphtheria toxin. In: Priti R, editor. Insight and Control of Infectious Disease in Global Scenario. Rijeka: InTech.
14. ChenJY, BodleyJW, LivingstonDM (1985) Diphtheria toxin-resistant mutants of Saccharomyces cerevisiae. Mol Cell Biol 5 : 3357–3360.
15. LiuS, LepplaSH (2003) Retroviral insertional mutagenesis identifies a small protein required for synthesis of diphthamide, the target of bacterial ADP-ribosylating toxins. Mol Cell 12 : 603–613.
16. LiuS, MilneGT, KuremskyJG, FinkGR, LepplaSH (2004) Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol Cell Biol 24 : 9487–9497.
17. DunlopPC, BodleyJW (1983) Biosynthetic labeling of diphthamide in Saccharomyces cerevisiae. J Biol Chem 258 : 4754–4758.
18. LinH (2011) S-adenosylmethionine-dependent alkylation reactions: when are radical reactions used? Bioorg Chem 39 : 161–170.
19. MoehringJM, MoehringTJ, DanleyDE (1980) Posttranslational modification of elongation factor 2 in diphtheria-toxin-resistant mutants of CHO-K1 cells. Proc Natl Acad Sci USA 77 : 1010–1014.
20. FichtnerL, JablonowskiD, SchierhornA, KitamotoHK, StarkMJ, et al. (2003) Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol Microbiol 49 : 1297–1307.
21. ZhangY, ZhuX, TorelliAT, LeeM, DzikovskiB, et al. (2010) Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature 465 : 891–896.
22. ZhuX, DzikovskiB, SuX, TorelliAT, ZhangY, et al. (2011) Mechanistic understanding of Pyrococcus horikoshii Dph2, a [4Fe-4S] enzyme required for diphthamide biosynthesis. Mol Biosyst 7 : 74–81.
23. ProudfootM, SandersSA, SingerA, ZhangR, BrownG, et al. (2008) Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain. J Mol Biol 375 : 301–315.
24. ThakurA, ChitoorB, GoswamiAV, PareekG, AtreyaHS, et al. (2012) Structure and mechanistic insights into novel iron-mediated moonlighting functions of human J-protein cochaperone, Dph4. J Biol Chem 287 : 13194–13205.
25. FichtnerL, SchaffrathR (2002) KTI11 and KTI13, Saccharomyces cerevisiae genes controlling sensitivity to G1 arrest induced by Kluyveromyces lactis zymocin. Mol Microbiol 44 : 865–875.
26. GreenwoodC, SelthLA, Dirac-SvejstrupAB, SvejstrupJQ (2009) An iron-sulfur cluster domain in Elp3 important for the structural integrity of elongator. J Biol Chem 284 : 141–149.
27. HuangB, JohanssonMJ, ByströmAS (2005) An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11 : 424–436.
28. ParaskevopoulouC, FairhurstSA, LoweDJ, BrickP, OnestiS (2006) The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol Microbiol 59 : 795–806.
29. ChenJY, BodleyJW (1988) Biosynthesis of diphthamide in Saccharomyces cerevisiae. Partial purification and characterization of a specific S-adenosylmethionine:elongation factor 2 methyltransferase. J Biol Chem 263 : 11692–11696.
30. MattheakisLC, ShenWH, CollierRJ (1992) DPH5, a methyltransferase gene required for diphthamide biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol 12 : 4026–4037.
31. MoehringTJ, DanleyDE, MoehringJM (1984) In vitro biosynthesis of diphthamide, studied with mutant Chinese hamster ovary cells resistant to diphtheria toxin. Mol Cell Biol 4 : 642–650.
32. ZhuX, KimJ, SuX, LinH (2010) Reconstitution of diphthine synthase activity in vitro. Biochemistry 49 : 9649–9657.
33. ZhangY, LiuS, LajoieG, MerrillAR (2008) The role of the diphthamide-containing loop within eukaryotic elongation factor 2 in ADP-ribosylation by Pseudomonas aeruginosa exotoxin A. Biochem J 413 : 163–174.
34. JørgensenR, PurdyAE, FieldhouseRJ, KimberMS, BartlettDH, et al. (2008) Cholix toxin, a novel ADP-ribosylating factor from Vibrio cholerae. J Biol Chem 283 : 10671–10678.
35. Uthman S, Kheir E, Bär C, Jablonowski D, Schaffrath R (2011) Growth inhibition strategies based on antimicrobial microbes/toxins. In: A M-V, editor. Science against Microbial Pathogens: Communicating Current Research and Technological Advances: Formatex Research Center, Spain. pp. 1321–1329.
36. OrtizPA, UlloqueR, KiharaGK, ZhengH, KinzyTG (2006) Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J Biol Chem 281 : 32639–32648.
37. KimataY, KohnoK (1994) Elongation factor 2 mutants deficient in diphthamide formation show temperature-sensitive cell growth. J Biol Chem 269 : 13497–13501.
38. ChenCM, BehringerRR (2004) Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev 18 : 320–332.
39. LiuS, WigginsJF, SreenathT, KulkarniAB, WardJM, et al. (2006) Dph3, a small protein required for diphthamide biosynthesis, is essential in mouse development. Mol Cell Biol 26 : 3835–3841.
40. WebbTR, CrossSH, McKieL, EdgarR, VizorL, et al. (2008) Diphthamide modification of eEF2 requires a J-domain protein and is essential for normal development. J Cell Sci 121 : 3140–3145.
41. CaretteJE, GuimaraesCP, VaradarajanM, ParkAS, WuethrichI, et al. (2009) Haploid genetic screens in human cells identify host factors used by pathogens. Science 326 : 1231–1235.
42. SuX, ChenW, LeeW, JiangH, ZhangS, et al. (2012) YBR246W is required for the third step of diphthamide biosynthesis. J Am Chem Soc 134 : 773–776.
43. HillenmeyerME, EricsonE, DavisRW, NislowC, KollerD, et al. (2010) Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. Genome Biol 11: R30.
44. KohJL, DingH, CostanzoM, BaryshnikovaA, ToufighiK, et al. (2010) DRYGIN: a database of quantitative genetic interaction networks in yeast. Nucleic Acids Res 38: D502–D507.
45. CostanzoM, BaryshnikovaA, BellayJ, KimY, SpearED, et al. (2010) The genetic landscape of a cell. Science 327 : 425–431.
46. TongAH, EvangelistaM, ParsonsAB, XuH, BaderGD, et al. (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294 : 2364–2368.
47. TongAH, LesageG, BaderGD, DingH, XuH, et al. (2004) Global mapping of the yeast genetic interaction network. Science 303 : 808–813.
48. CostanzoM, BaryshnikovaA, MyersCL, AndrewsB, BooneC (2011) Charting the genetic interaction map of a cell. Curr Opin Biotechnol 22 : 66–74.
49. DixonSJ, CostanzoM, BaryshnikovaA, AndrewsB, BooneC (2009) Systematic mapping of genetic interaction networks. Annu Rev Genet 43 : 601–625.
50. DRYGIN [http://drygin.ccbr.utoronto.ca/]
51. FitDB [http://drygin.ccbr.utoronto.ca/]
52. HillenmeyerME, FungE, WildenhainJ, PierceSE, HoonS, et al. (2008) The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320 : 362–365.
53. JusticeMC, HsuMJ, TseB, KuT, BalkovecJ, et al. (1998) Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J Biol Chem 273 : 3148–3151.
54. SøeR, MosleyRT, JusticeM, Nielsen-KahnJ, ShastryM, et al. (2007) Sordarin derivatives induce a novel conformation of the yeast ribosome translocation factor eEF2. J Biol Chem 282 : 657–666.
55. MumbergD, MullerR, FunkM (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22 : 5767–5768.
56. JørgensenR, Carr-SchmidA, OrtizPA, KinzyTG, AndersenGR (2002) Purification and crystallization of the yeast elongation factor eEF2. Acta Crystallogr D Biol Crystallogr 58 : 712–715.
57. SpahnCM, Gomez-LorenzoMG, GrassucciRA, JorgensenR, AndersenGR, et al. (2004) Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 23 : 1008–1019.
58. OrtizPA, KinzyTG (2005) Dominant-negative mutant phenotypes and the regulation of translation elongation factor 2 levels in yeast. Nucleic Acids Res 33 : 5740–5748.
59. HargerJW, MeskauskasA, NielsenJ, JusticeMC, DinmanJD (2001) Ty1 retrotransposition and programmed +1 ribosomal frameshifting require the integrity of the protein synthetic translocation step. Virology 286 : 216–224.
60. de Crecy-LagardV, ForouharF, Brochier-ArmanetC, TongL, HuntJF (2012) Comparative genomic analysis of the DUF71/COG2102 family predicts roles in diphthamide biosynthesis and B12 salvage. Biology Direct 7 : 32.
61. SuX, LinZ, ChenW, JiangH, ZhangS, et al. (2012) Chemogenomic approach identified yeast YLR143W as diphthamide synthetase. Proc Natl Acad Sci USA 109 : 19983–19987.
62. Marchler-BauerA, LuS, AndersonJB, ChitsazF, DerbyshireMK, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229.
63. BurmanJD, StevensonCE, SawersRG, LawsonDM (2007) The crystal structure of Escherichia coli TdcF, a member of the highly conserved YjgF/YER057c/UK114 family. BMC Struct Biol 7 : 30.
64. SinhaS, RappuP, LangeSC, MantsalaP, ZalkinH, et al. (1999) Crystal structure of Bacillus subtilis YabJ, a purine regulatory protein and member of the highly conserved YjgF family. Proc Natl Acad Sci USA 96 : 13074–13079.
65. VolzK (1999) A test case for structure-based functional assignment: the 1.2 A crystal structure of the yjgF gene product from Escherichia coli. Protein Sci 8 : 2428–2437.
66. MistinieneE, PozdniakovaiteN, PopendikyteV, NaktinisV (2005) Structure-based ligand binding sites of protein p14.5, a member of protein family YER057c/YIL051c/YjgF. Int J Biol Macromol 37 : 61–68.
67. LambrechtJA, FlynnJM, DownsDM (2012) Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5′-phosphate (PLP)-dependent enzyme reactions. J Biol Chem 287 : 3454–3461.
68. ShiY, StefanCJ, RueSM, TeisD, EmrSD (2011) Two novel WD40 domain-containing proteins, Ere1 and Ere2, function in the retromer-mediated endosomal recycling pathway. Mol Biol Cell 22 : 4093–4107.
69. HontzRD, NiedererRO, JohnsonJM, SmithJS (2009) Genetic identification of factors that modulate ribosomal DNA transcription in Saccharomyces cerevisiae. Genetics 182 : 105–119.
70. GuptaPK, LiuS, BataviaMP, LepplaSH (2008) The diphthamide modification on elongation factor-2 renders mammalian cells resistant to ricin. Cell Microbiol 10 : 1687–1694.
71. LiuS, BachranC, GuptaP, Miller-RandolphS, WangH, et al. (2012) Diphthamide modification on eukaryotic elongation factor 2 is needed to assure fidelity of mRNA translation and mouse development. Proc Natl Acad Sci USA 109 : 13817–13822.
72. NobukuniY, KohnoK, MiyagawaK (2005) Gene trap mutagenesis-based forward genetic approach reveals that the tumor suppressor OVCA1 is a component of the biosynthetic pathway of diphthamide on elongation factor 2. J Biol Chem 280 : 10572–10577.
73. ShermanF (1991) Getting started with yeast. Methods Enzymol 194 : 3–21.
74. GietzD, St JeanA, WoodsRA, SchiestlRH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20 : 1425.
75. RoyV, GhaniK, CarusoM (2010) A dominant-negative approach that prevents diphthamide formation confers resistance to Pseudomonas exotoxin A and diphtheria toxin. PLoS ONE 5: e15753 doi:10.1371/journal.pone.0015753.
76. JablonowskiD, FichtnerL, MartinVJ, KlassenR, MeinhardtF, et al. (2001) Saccharomyces cerevisiae cell wall chitin, the Kluyveromyces lactis zymocin receptor. Yeast 18 : 1285–1299.
77. FrohloffF, JablonowskiD, FichtnerL, SchaffrathR (2003) Subunit communications crucial for the functional integrity of the yeast RNA polymerase II elongator (gamma-toxin target (TOT)) complex. J Biol Chem 278 : 956–961.
78. KnopM, SiegersK, PereiraG, ZachariaeW, WinsorB, et al. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15 : 963–972.
79. ZachariaeW, ShinTH, GalovaM, ObermaierB, NasmythK (1996) Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science 274 : 1201–1204.
80. GietzRD, SuginoA (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74 : 527–534.
81. JankeC, MagieraMM, RathfelderN, TaxisC, ReberS, et al. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21 : 947–962.
82. CoxJ, MannM (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26 : 1367–1372.
83. CherryJM, HongEL, AmundsenC, BalakrishnanR, BinkleyG, et al. (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40: D700–D705.
84. FrohloffF, FichtnerL, JablonowskiD, BreunigKD, SchaffrathR (2001) Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J 20 : 1993–2003.
85. JablonowskiD, SchaffrathR (2007) Zymocin, a composite chitinase and tRNase killer toxin from yeast,. Biochem Soc Trans 35 : 1533–1537.
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



