-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
The gene encoding the GroEL chaperonin is duplicated in nearly 30% of bacterial genomes; and although duplicated groEL genes have been comprehensively determined to have distinct physiological functions in different species, the mechanisms involved have not been characterized to date. Myxococcus xanthus DK1622 has two copies of the groEL gene, each of which can be deleted without affecting cell viability; however, the deletion of either gene does result in distinct defects in the cellular heat-shock response, predation, and development. In this study, we show that, from the expression levels of different groELs, the distinct functions of groEL1 and groEL2 in predation and development are probably the result of the substrate selectivity of the paralogous GroEL chaperonins, whereas the lethal effect of heat shock due to the deletion of groEL1 is caused by a decrease in the total groEL expression level. Following a bioinformatics analysis of the composition characteristics of GroELs from different bacteria, we performed region-swapping assays in M. xanthus, demonstrating that the differences in the apical and the C-terminal equatorial regions determine the substrate specificity of the two GroELs. Site-directed mutagenesis experiments indicated that the GGM repeat sequence at the C-terminus of GroEL1 plays an important role in functional divergence. Divergent functions of duplicated GroELs, which have similar patterns of variation in different bacterial species, have thus evolved mainly via alteration of the apical and the C-terminal equatorial regions. We identified the specific substrates of strain DK1622's GroEL1 and GroEL2 using immunoprecipitation and mass spectrometry techniques. Although 68 proteins bound to both GroEL1 and GroEL2, 83 and 46 proteins bound exclusively to GroEL1 or GroEL2, respectively. The GroEL-specific substrates exhibited distinct molecular sizes and secondary structures, providing an encouraging indication for GroEL evolution for functional divergence.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003306
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003306Summary
The gene encoding the GroEL chaperonin is duplicated in nearly 30% of bacterial genomes; and although duplicated groEL genes have been comprehensively determined to have distinct physiological functions in different species, the mechanisms involved have not been characterized to date. Myxococcus xanthus DK1622 has two copies of the groEL gene, each of which can be deleted without affecting cell viability; however, the deletion of either gene does result in distinct defects in the cellular heat-shock response, predation, and development. In this study, we show that, from the expression levels of different groELs, the distinct functions of groEL1 and groEL2 in predation and development are probably the result of the substrate selectivity of the paralogous GroEL chaperonins, whereas the lethal effect of heat shock due to the deletion of groEL1 is caused by a decrease in the total groEL expression level. Following a bioinformatics analysis of the composition characteristics of GroELs from different bacteria, we performed region-swapping assays in M. xanthus, demonstrating that the differences in the apical and the C-terminal equatorial regions determine the substrate specificity of the two GroELs. Site-directed mutagenesis experiments indicated that the GGM repeat sequence at the C-terminus of GroEL1 plays an important role in functional divergence. Divergent functions of duplicated GroELs, which have similar patterns of variation in different bacterial species, have thus evolved mainly via alteration of the apical and the C-terminal equatorial regions. We identified the specific substrates of strain DK1622's GroEL1 and GroEL2 using immunoprecipitation and mass spectrometry techniques. Although 68 proteins bound to both GroEL1 and GroEL2, 83 and 46 proteins bound exclusively to GroEL1 or GroEL2, respectively. The GroEL-specific substrates exhibited distinct molecular sizes and secondary structures, providing an encouraging indication for GroEL evolution for functional divergence.
Introduction
Chaperonins are essential cellular components that are responsible for protein folding, assembly and transport [1]–[6]. Chaperonins are also a major group of heat shock proteins that are over-expressed at high temperatures and have fundamental roles in growth and survival at non-permissive temperatures [6]–[8]. GroEL is a type I chaperonin, and in Escherichia coli, the GroEL chaperonin is required in vivo for the proper folding, at all temperatures, of approximately 300 newly translated polypeptides (accounting for approximately 10% of the total) that participate in various physiological processes [9]. Because of its importance in many cellular processes, the groEL gene is ubiquitously distributed in bacteria. Most bacterial species, such as E. coli, possess a single groEL gene, whereas other species (nearly 30% of bacteria with sequenced genomes) have evolved multiple groEL copies [1]. The paralogous GroEL proteins are highly similar in sequence and, most likely, in structure. However, some differences exist between duplicated groEL genes, and these duplicated GroEL proteins have evolved to play divergent roles in many different cellular processes in different bacterial species [10]–[15]. Although the mechanisms of functional divergence are important for our understanding of the complexity of evolution, these mechanisms have not been characterized to date.
Myxobacteria are δ-proteobacteria with unique and complex multicellular behaviors, such as movement in swarms on solid surfaces, cooperative feeding on macromolecules or other microbial cells and the development of multicellular fruiting bodies containing numerous myxospores against adversity conditions [16], [17]. Myxococcus xanthus DK1622 is a model myxobacterium with a large genome (9.14 Mbp) that includes many duplicated crucial genes [18]. It has been suggested that such duplication is responsible for the complex social behavior of these cells, although this hypothesis has not been experimentally validated. There are two copies of the groEL gene in the genome of M. xanthus DK1622. Previous studies indicate that either of the two paralogous groEL genes can be deleted in strain DK1622 without affecting cellular viability, although the deletion does result in distinct defects in the cellular heat-shock response, predation and development [15]. In this study, we investigated the effects of the substrate selectivity of the GroEL proteins and the expression levels of the duplicated groEL genes on the functional divergence of heat-shock responses, development and predation. We performed a comparative proteomics analysis of the substrate specificity of the two GroELs, and the relationships between the structural differences and substrate specificity were investigated using bioinformatics, molecular swapping and site-directed mutagenesis.
Results
Effects of groEL expression levels on functional divergence
Otani et al. found that, although GroEL1 and GroEL2 are among the major proteins induced by heat shock, the density of GroEL2 spots in two-dimensional electrophoretic gels is much lower than that of GroEL1 [19]. It was also noted that the expression levels of groEL1 and groEL2 were not equal in the wild-type strain DK1622 in CTT growth medium and TPM development medium and that the two groEL genes played distinct roles in heat-shock responses, development and predation [15]. To assess the changes in groEL1 or groEL2 expression in groEL-deletion mutants and whether these changes contribute to functional divergence, we inserted groEL1 or groEL2, each with its own promoter, into the genome of groEL1 - or groEL2-deletion mutants at the attB integration site using pSWU30, producing four groEL-complemented strains (Table S1). The groEL expression levels and cell viability were compared between these mutants and the wild-type strain DK1622 following heat shock at 42°C for 30 min. Quantitative PCR assays indicated that the expression of the groEL genes was regulated in a complex manner in different mutants upon heat shock (Figure 1). In the wild-type strain DK1622, the groEL2 expression level was only one-quarter of that of groEL1 after heat shock. The deletion of groEL1 (strain YL0301) led to an increase in the groEL2 expression level (approximately twofold). The expression of groEL1 inserted in YL0301 (strain YL0901) was approximately half that in DK1622 under the heat shock conditions, but the presence of exogenous groEL1 had no obvious effect on the expression level of groEL2 (P>0.05). Thus, the total expression of all the groEL genes in YL0901 was similar to that in DK1622. In YL0902, which contained an additional groEL2 gene, the total expression of groEL2 also doubled, reaching a level equal to four times that of groEL2 in DK1622. In YL0302, the deletion of the groEL2 gene led to reduced expression of the groEL1 gene under the heat shock conditions (approximately 60% of that in DK1622). Transformation of the YL0302 mutant (strain YL0906) with another groEL1 gene increased the total groEL expression level to that of DK1622 (P>0.05). The total expression level of groEL1 and groEL2 in the groEL2-complemented YL0302 mutant (strain YL0907) also reached the level in DK1622 (P>0.05).
Fig. 1. Quantitative PCR analysis of the groEL1 and groEL2 expression levels and survival rates after heat shock at 42°C for 30 min. 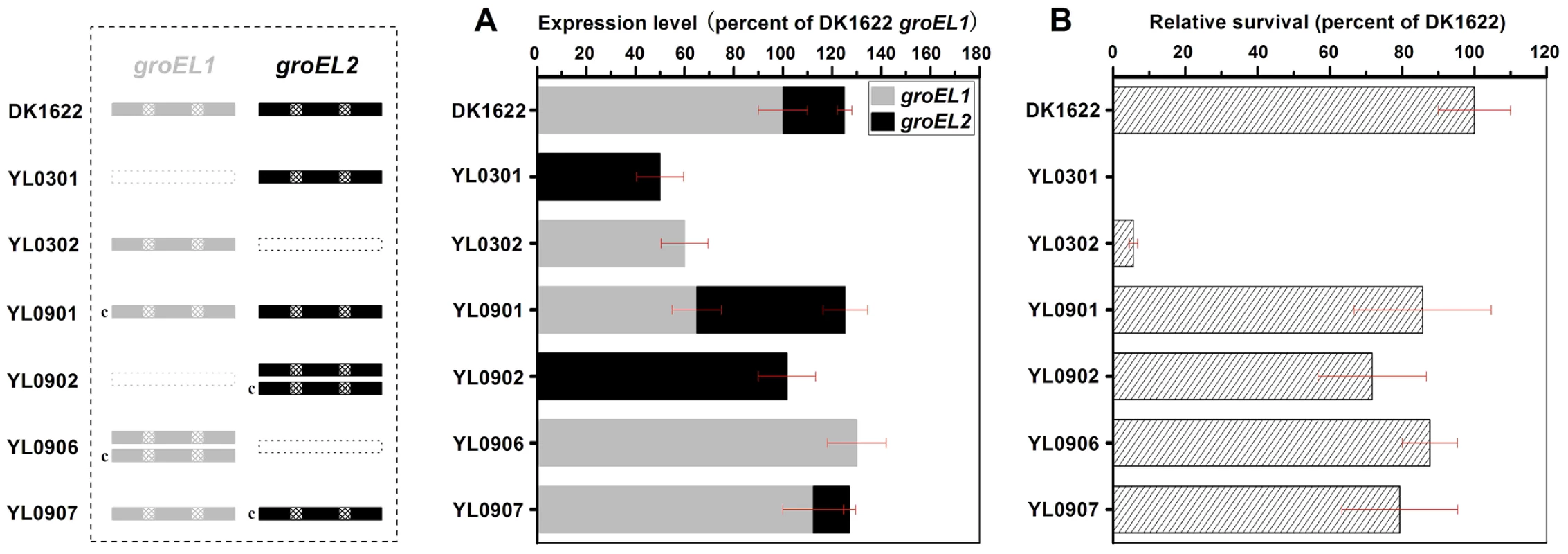
(A) Gene expression levels. (B) Survival rates after heat shock. The left panel is a schematic diagram of the groELs gene present in the different strains. The values for each groEL gene are shown as levels relative to the expression of groEL1 in DK1622, which was defined as 100%. The survival rates of the different strains were calculated as a percentage of the survival rate of DK1622, which was 1.01×10−2 after the heat shock treatment and was defined as 100%. The error bars show the standard deviation of three replicates. DK1622, wild-type strain; YL0301, groEL1-deletion mutant; YL0302, groEL2-deletion mutant; YL0901, YL0301 complemented with GroEL1; YL0902, YL0301 complemented with GroEL2; YL0906, YL0302 complemented with GroEL1; YL0907, YL0302 complemented with GroEL2. “c” denotes “complement.” Interestingly, although YL0902 had two groEL2 genes and no groEL1, the survival rates of both YL0901 and YL0902 were similarly increased after heat shock at 42°C for 30 min, paralleling the increase in the total groEL expression level in these two mutants (Figure 1). The survival rates of the YL0906 and YL0907 mutants after the heat shock treatment also corresponded to an increase in the total groEL expression level. These results suggest that the lethal nature of the heat shock in the groEL1 deletion mutant (YL0301) and the increased sensitivity of the groEL2 deletion mutant (YL0302) to high temperatures [15] result from a significant decrease in the total expression of GroEL, leading to an insufficient level of GroEL proteins to facilitate the refolding of denatured proteins. This is consistent with a model where there is a threshold level of GroEL beneath which cells cannot survive. When the total expression of groEL is higher than the threshold, there is a positive correlation between the survival rate and groEL expression in M. xanthus DK1622 cells after heat shock (r = 0.98, P<0.01) (Figure S1), a result which supports the hypothesis that, after duplication, both groEL1 and groEL2 retain fundamental functions by balancing their expression dosage [20], [21].
Because the deletion of groEL1 and the deletion of groEL2 result in deficiencies in development and predation, respectively [15], we performed a development assay on DK1622, YL0301, YL0901 and YL0902 and predation assays on DK1622, YL0302, YL0906 and YL0907. Figure 2A shows the expression levels of groEL1 and groEL2 in different strains after 12, 36 and 60 h of incubation on TPM development medium. When groEL1 was inserted into the genome of the groEL1-deletion mutant (strain YL0901), the developmental defect was mostly reversed, with sporulation reaching 70%–80% of that of DK1622. However, although YL0902 (containing two copies of groEL2) had a total groEL expression level similar to that of YL0901 at different developmental stages, YL0902 displayed a development defect similar to that of YL0301, and the sporulation ability of YL0902 was only approximately 20% of that of the wild-type strain DK1622 (Figure 2B and 2C).
Fig. 2. The expression levels of groEL genes, the development and sporulation analysis of different strains. 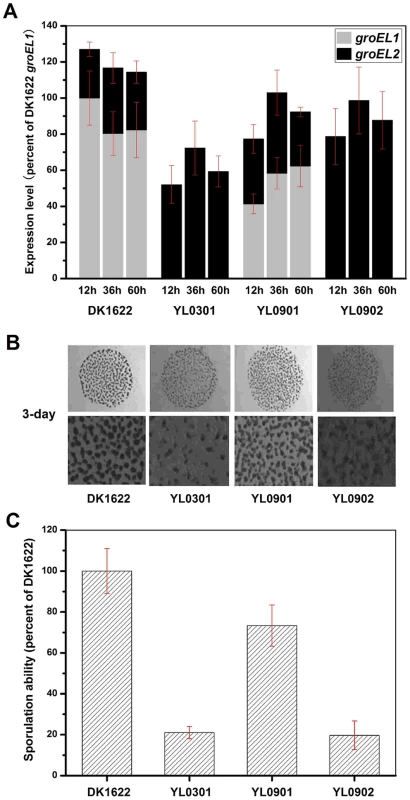
(A) The expression levels of groEL genes at three developmental time points. (B) The development of fruiting bodies. (C) The sporulation of different strains on TPM plates after three days. The expression levels for each groEL gene in the different strains are shown as levels relative to the expression of DK1622 groEL1 after 12 hours, which was defined as 100%. The sporulation ability of the different strains was calculated as a percentage of the sporulation by DK1622, which was defined as 100%. DK1622, wild-type strain; YL0301, groEL1-deletion mutant; YL0901, YL0301 complemented with GroEL1; YL0902, YL0301 complemented with GroEL2; YL0903, YL0301 complemented with GroEL2-equatorial-NGroEL1; YL0904, YL0301 complemented with GroEL2-apicalGroEL1; YL0905, YL0301 complemented with GroEL2-equatorial-CGroEL1. The insertion of groEL1 into YL0302 (strain YL0906) did not improve the predation feeding ability of cells on an E. coli mat, and the swarming time of YL0906 to the edge of the E. coli colony was 60–65 h, which is similar to that of YL0302 (P>0.05). When groEL2 was inserted into YL0302 (strain YL0907), the swarming time to the E. coli colony edge decreased to 40 h (Figure 3). Because the presence of living E. coli cells in the E. coli predation experiments might affect the qPCR assay, we instead performed the analysis using a liquid feeding assay with casein as the only nutrient [15]. The total expression level of groEL1 and groEL2 was also similar in the YL0906 and YL0907 mutants, suggesting that the changes in expression are not the major contributors to functional divergence (Figure S2).
Fig. 3. The predation of an E. coli mat by different strains. 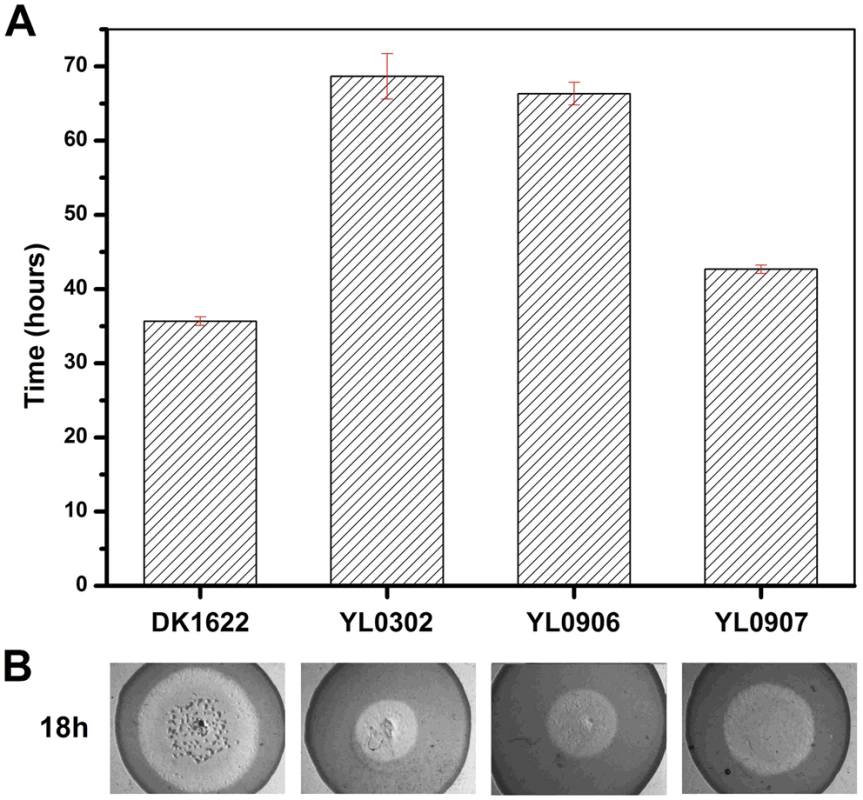
(A) The time for the different strains to reach the edge of the E. coli mat. (B) Zones of predation after 18 h and 36 h for the knockout and complemented mutants. DK1622, wild-type strain; YL0302, groEL2-deletion mutant; YL0906, YL0302 complemented with GroEL1; YL0907, YL0302 complemented with GroEL2; YL0908, YL0302 complemented with GroEL1-equatorial-NGroEL2; YL0909, YL0302 complemented with GroEL1-apicalGroEL2; YL0910, YL0302 complemented with GroEL1-equatorial-CGroEL2. Evolutionary analysis of duplicated GroELs
The above results indicate that although the distinct heat-shock responses of the groEL1 and groEL2 mutants were determined by the total groEL expression level, the divergent functions of groEL1 and groEL2 in development and predation are the result of the substrate specificity of the corresponding GroEL chaperonins. To explore the evolutionary relationships of groEL, we compared the M. xanthus GroEL sequences with those of ten genome-sequenced bacteria, including three Myxobacteria, three Actinobacteria, three Cyanobacteria, and E. coli (Figure 4 and Table S2). With the exception of E. coli, these species possess duplicated groEL genes. The maximum likelihood tree showed that the GroELs from Myxobacteria, Actinobacteria and Cyanobacteria clustered separately (Figure 4A), suggesting that the groEL gene duplication originated from three independent evolutionary events in these three taxa. We further calculated the Ka/Ks values of these orthologous groEL genes (Figure 4B and Table S3). The average Ka/Ks values for Actinobacteria groEL2 and Cyanobacteria groEL1 were less than 0.1, suggesting that they are highly evolutionarily conserved. It has been reported that groEL2 in the three Actinobacteria species [12], [22]–[24] and groEL1 in the three Cyanobacteria species [25]–[27] are housekeeping genes, which is consistent with their Ka/Ks values. In M. xanthus, the Ka/Ks value for groEL1 was significantly lower than that for groEL2 (P<0.05) but significantly higher than that for the housekeeping groEL genes in Actinobacteria or Cyanobacteria (P<0.01). These results suggest that both of groEL1 and groEL2 in M. xanthus are suffered weak selection pressure, consistent to the finding that the deletion of either gene alone does not affect cell viability [15].
Fig. 4. Evolutionary analysis of GroELs. 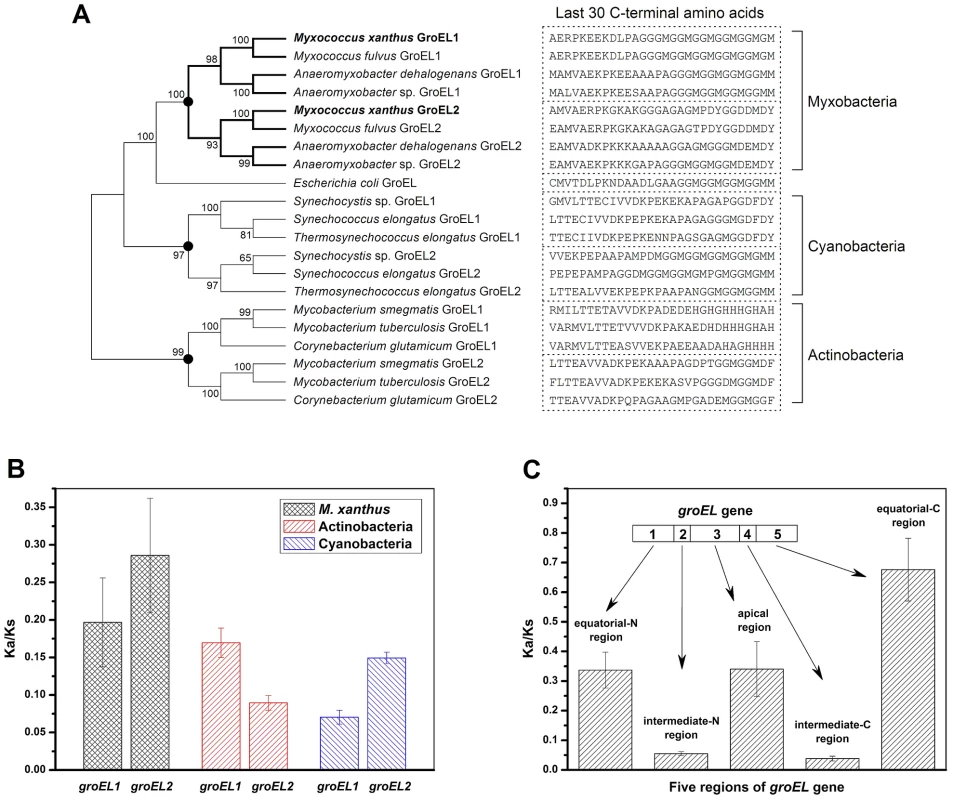
(A) Maximum likelihood tree of the M. xanthus GroELs and the GroELs from ten other bacteria. The sequence of the last 30 C-terminal amino acids of each GroEL is shown. The black dots in the tree represent potential duplication events. (B) Mean Ka/Ks values of the different bacteria, as calculated using orthologous groEL genes. (C) Mean Ka/Ks values of five regions of the duplicated groEL genes from four myxobacterial species, as calculated using paralogous groEL genes. Based on their structural characteristics and sequence conservation, the GroEL protein sequences have been divided into five regions, i.e., an N-terminal equatorial region, an N-intermediate region, an apical region, a C-intermediate region and a C-terminal equatorial region (Figure S3) [28]. The two intermediate regions have the highest level of conservation between M. xanthus DK1622 GroEL1 and GroEL2, i.e., 97.7% and 97.2% identities for the N - and C-intermediate regions, respectively. The identities for the other three regions are 81% for the N-terminal equatorial region, 75.4% for the apical region, and 62.6% for the C-terminal equatorial region. Further Ka/Ks analysis showed similar sequence characteristics for the five GroEL1 and GroEL2 regions in the four Myxobacterial species referred to above (Figure 4C). For example, the Ka/Ks values of the N - and C-intermediate regions were very low (<0.05), suggesting that these two regions are highly conserved; in contrast, the other three regions had higher Ka/Ks values (>0.3), suggesting these regions are most likely involved in the functional divergence of GroEL1 and GroEL2. A sequence alignment showed that the high Ka/Ks values of the C-terminal equatorial regions were largely due to the variability of the C-terminal tail sequences. For example, the C-terminal tail of GroEL1 in M. xanthus was composed of six repeated GGM motifs, similar to that of E. coli GroEL, whereas the C-terminal tail of GroEL2 is greatly different. It was also noted that there are substantial differences in the C-terminal sequences between the duplicated GroELs (Figure 4A) [1].
Region swapping and site-directed mutagenesis assays
To clarify the relationships between the structural and functional divergence, we designed a series of single region-swaps between the groEL1 and groEL2 genes to determine the roles of the regions and their contributions to functional divergence. The swapped regions included the N-terminal equatorial, apical, and C-terminal equatorial regions between GroEL1 and GroEL2; the two highly similar intermediate regions were not included. The groEL2 hybrids containing the N-terminal equatorial, apical or C-terminal equatorial region of groEL1 were inserted into the genome of the groEL1-deletion mutant YL0301 using pSWU30, producing the region-swapped strains YL0903, YL0904 and YL0905, respectively. Similarly, the groEL1 hybrids with a swapped N-terminal equatorial, apical or C-terminal equatorial region of groEL2 were inserted into the genome of the groEL2-deletion mutant YL0302 to produce the mutant strains YL0908, YL0909 and YL0910, respectively (Figure 5A). Because the groEL2 mutant displays defective cellular predation and the groEL1 mutant displays deficient development and sporulation [15], region swapping was performed in YL0301 using the groEL1 chimeras and YL0302 using the groEL2 chimeras. Detailed descriptions of these mutants are listed in Tables S1 and S4 and Figure S4.
Fig. 5. Social behavior feature of region-swapping mutants, GGM deletion, and shorten mutants. 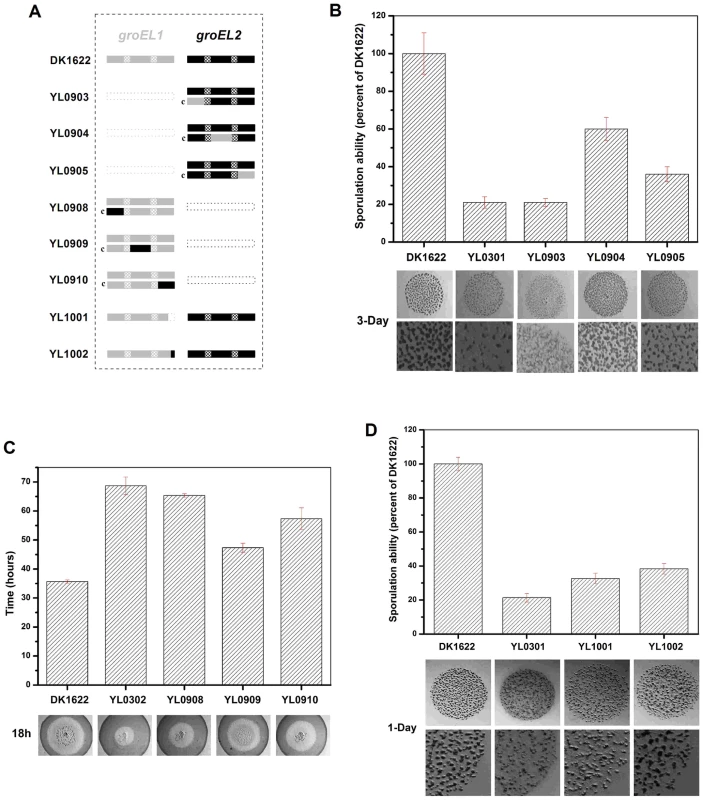
(A) is a schematic diagram of the groELs present in the different strains. (B) Sporulation ability and development of region-swapping mutants. (C) Predation speed of region-swapping mutants. (D) Sporulation ability and development of GGM deletion and shorten mutants. The sporulation ability of each strain was calculated as a percentage of the sporulation ability of DK1622, which was defined as 100%. The development and predation phenotypes of the region-swapped mutants were assayed using the intact groEL1 - and groEL2-complemented mutants as controls. The results showed that the developmental defect of the groEL1-deletion mutant was not reversed by GroEL2-equatorial-NGroEL1 (YL0903). The sporulation ability of YL0903 was approximately 20% of that of DK1622, which was the same as that of YL0301. However, the fruiting bodies of YL0904 (YL0301 complemented with GroEL2-apicalGroEL1) were more similar to the fruiting bodies of the wild-type strain DK1622 than to the fruiting bodies of YL0301, and the sporulation ability also increased to 55%–65% of that of DK1622. The sporulation of the strain complemented with GroEL2-equatorial-CGroEL1 (YL0905) was 30%–40% of that of DK1622 (Figure 5B). In the predation experiments, the single swapped region in YL0908 did not noticeably improve the predation defects, which were similar to those of YL0906 (P>0.05). However, the YL0909 strain significantly recovered its predation ability, which was similar to that of YL0907. These two mutants spread to the edge of the E. coli colonies within 40–45 h. The predation defect was also improved to some extent in YL0910, which required 55–60 h to reach the edge of the E. coli mat (Figure 5C). Accordingly, the apical region and the C-terminal equatorial region determine the substrate preference, thus causing the functional divergence of the duplicated chaperonins with respect to development and predation; conversely, the N-terminal equatorial region has almost no effect.
In addition, we deleted the repeated GGM region (GGMGGMGGMGGMGGMGM) from GroEL1 in M. xanthus DK1622, producing the YL1001 mutant (Figure 5A). Compared with the wild-type DK1622, the mutant was markedly defective in development, and the sporulation ability of YL1001 was only 32.6% of that of DK1622 (Figure 5D). Furthermore, we swapped three of the six GGM repeats with YGGDDMDY in DK1622, the corresponding sequence in GroEL2, producing the YL1002 mutant. Similar to YL1001, YL1002 was also defective in development and exhibited a decreased sporulation ability (38.1% of that of DK1622) (Figure 5D). These results demonstrate that the GGM repeated region is necessary for GroEL1 to perform its normal functions in development.
GroEL1 and GroEL2 substrates and their characteristics
To identify the proteins that interact with GroEL1 and GroEL2 in M. xanthus DK1622, immunoprecipitation assays were performed using the groEL1 - and groEL2-deletion mutants (strains YL0301 and YL0302), and the bound proteins were subjected to mass spectral identification. Most of the non-specific substrates identified using two negative controls (see Methods) were ribosomal proteins (Table S5). This result was consistent with the results for E. coli [9]. After removing the non-specifically bound proteins, 151 and 114 proteins were found to be bound by GroEL1 and GroEL2, respectively. Of the bound proteins, 68 were bound to both GroEL1 and GroEL2 (GroEL1/2), whereas 83 and 46 proteins bound exclusively to GroEL1 and GroEL2, respectively (Table S5). Of the functionally annotated GroEL1/2 substrates (58/68, 85.3%), many had functions or predicted functions related to fundamental physiological cellular processes; examples of such substrates are succinyl coenzyme A synthetase and isocitrate dehydrogenase, two key enzymes of the citric acid cycle [29], [30]. This result is consistent with the fact that either the groEL1 or groEL2 gene could be deleted without affecting cellular growth but that the double deletion of groEL1 and groEL2 resulted in inviable cells [15]. However, except for PilA, no proteins involved in M. xanthus social behavior were found to bind to both GroEL1 and GroEL2. In contrast, of those annotated proteins that were exclusively bound by GroEL1 or GroEL2 (accounting for 75.9% and 76.1% of bound proteins, respectively), a considerable number are involved in the social behaviors of M. xanthus DK1622 (Table S5). For example, the frz signal transduction system is well known to play important roles in development process of M. xanthus DK1622 [31]. The frizzy aggregation protein FrzCD [31] is in the substrate list of GroEL1. Besides, Flp pilus assembly protein CpaB [18], sensor histidine kinase/response regulator CheA4 [32] and Type IV pilus secretin PilQ [33] were found to be specific substrates of GroEL1, whereas type IV pilus assembly ATPase PilB [34], gliding motility protein MglA [35], type IV pilus biogenesis protein PilM [36] and several proteins related to the biosynthesis of secondary metabolites were found to be exclusively bound by GroEL2. These results are consistent with the hypothesis that GroEL is an essential component and that the duplicated groEL genes evolved to participate in various complex physiological processes in Myxococcus cells.
The structural characteristics of the substrate proteins were further analyzed by comparing their secondary structures with the known protein domain classification database CATH [37]. After excluding the proteins that had low E-values (>0.001), we obtained 36 reliable secondary structures for the 68 identified GroEL1/2 substrates, 38 for the 83 GroEL1-specific substrates and 34 for the 46 GroEL2-specific substrates. It is known that proteins with β-sheets exposed to the hydrophobic surface and packed with the hydrophobic surfaces of α-helices (called the αβ domain) have high-affinity interactions with the apical region of GroEL and are normally present as substrates of GroELs [38]. As expected, most GroEL1/2 substrates (34 of 36) contain at least one αβ domain. It is interesting that, although 31 of the 34 (91.18%) GroEL2-specific substrates possess at least one αβ domain, only 27 of the 38 (71.05%) GroEL1-specific substrates contain an αβ domain (Figure 6A, Table S5).
Fig. 6. Substrate features of GroEL1 and GroEL2. 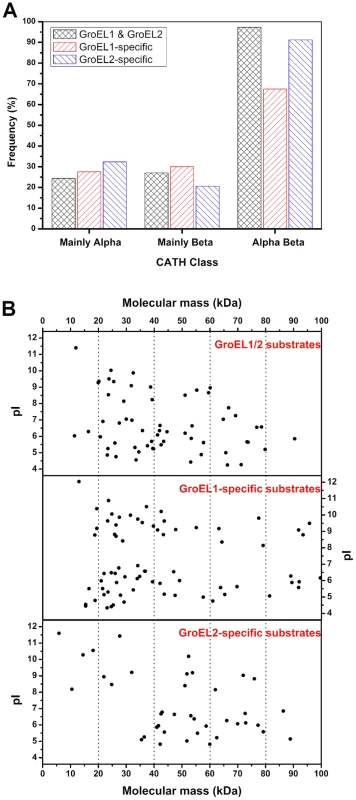
(A) αβ-domain, (B) molecular weight distribution and pI distributions. The details of the substrate proteins of GroEL1 and GroEL2 are shown in Table S5. Another interesting difference is the difference in the molecular sizes of the GroEL1 and GroEL2 substrates. According to the current model of GroEL [28], [39]–[45], substrate selection is heavily dependent upon the adaptation of a substrate molecule to the cavity volume of the GroEL chaperonin, which may change in response to GroEL sequence changes. Previous studies have shown that GroEL strongly prefers to act on proteins with a molecular weight ranging from 20 kDa to 60 kDa [46]. The average molecular weight of GroEL1-specific substrates was significantly smaller than that of GroEL2-specific substrates (Figure 6B; P<0.05). For example, while 51.8% (43 of 83) of the GroEL1-specific substrate proteins were less than 40 kDa, the molecular weights of only 21.7% (10 of 46) of the GroEL2-specific proteins were less than 40 kDa. A third important characteristic is the pI value of the substrate; there was no significant difference between the GroEL1 and GroEL2 substrates with respect to pI (Figure 6B; P>0.05).
Discussion
Duplication is a major source of new genes and is equally important in Bacteria, Archaea and Eukarya [47], [48]. The duplication of the GroEL chaperonin gene has occurred in many different bacterial cells as part of the evolution of complexity [49]. M. xanthus DK1622 is well known for its complex multicellular behaviors [16], [17], and this strain possesses a large genome (9.14 Mb) in which there are many duplicated genes, including two copies of groEL [18]. In addition to participating in fundamental processes involved in cellular growth, the two duplicated groEL genes have been demonstrated to play distinct roles in heat-shock responses, development and predation in DK1622 [15]. The results described in this report show that the divergent functions of GroEL1 and GroEL2 in various physiological processes result from different mechanisms. The groEL expression level is the key reason for the difference in the heat-shock response after the deletion of groEL1 or groEL2, suggesting that the duplicated groEL genes in Myxococcus have similar functions in cell survival. These functions are most likely similar to their fundamental function in cell growth at normal temperatures. Either of the two groEL genes can be deleted without significantly affecting cell growth, but at least one groEL gene is required for cell survival [15]. In contrast, the functional divergence of the duplicated GroELs with respect to their roles in development and predation processes reflects in their substrate specificity, which has been suggested to evolutionarily relate to the unusual social behavior of Myxococcus. The co-substrates of GroEL1 and GroEL2 have consistently been shown to be essential cellular components, but the duplicated GroEL chaperonins have also evolved their own substrate preferences related to late-appearing cellular processes, such as social behaviors and PKS/NRPS biosynthesis. The evolutionary models for the functional divergence of duplicated genes are likely to include neofunctionalization, subfunctionalization, or a combination of thereof [20], [47], [50]. Thus, the functional divergence of the duplicated groELs in M. xanthus is likely a combination of neofunctionalization and subfunctionalization, i.e., the subneofunctionalization model [50].
Although extensive studies have demonstrated that the duplicated groEL genes play distinct roles in different cellular physiological processes [10]–[15], an understanding of the mechanisms involved in their functional divergence will provide insight into bacterial evolution. The GroEL proteins have been divided into five regions based on structural characteristics and sequence conservation [28]. Previous studies have shown that GroEL chimeras bearing equatorial or apical regions exchanged between M. tuberculosis and E. coli retained the normal chaperonin functions of GroEL [51]. Bioinformatics analyses indicate that the duplicated GroELs from different bacteria share similar characteristics: the N - and C-intermediate regions are highly conserved, suggesting that these regions have essential functions in maintaining the functional structure of GroELs, and the apical, N-terminal and C-terminal regions are much more flexible, suggesting their possible roles in functional divergence. The region-swapping experiments indicated that the functional divergence of the duplicated GroELs in M. xanthus was caused by the apical and C-terminal regions. The GGM repeat at the tail of the GroEL1 C-terminal region, which is similar to that of E. coli GroEL, is important for the distinct functions of GroEL1 in development and sporulation. These results are consistent with the positions of these two regions in the GroEL oligomeric complex, i.e., the apical region is at the opening through which substrates enter the central cavity, and the C-terminal equatorial region is at the bottom of the cavity [43]. However, it remains unclear whether region swapping has effects on in vivo chaperonin functions. To address the question, we assayed the survival rates of the mutants YL0903, YL0904 and YL0905 in response to heat shock and found that all the mutants rescued the lethality of heat-shock observed for YL0301. However, the survival rates of YL0903, YL0904, and YL0905 were low compared with the strains complemented with an intact groEL gene (Figure S5). This result suggests that the region-swapped GroELs function in M. xanthus cells but that these functions were affected, at least at non-permissive temperatures. It is still unclear whether the chimeras interact with intact GroELs and whether the in vivo functions of the chimeras result from mixed GroEL complexes. Furthermore, although the protein substrates of the M. xanthus GroELs were consistent with those of the single E. coli GroEL with regard to their secondary structural features [9], the substrate spectra varied significantly. This variation is most likely due to the low level of sequence similarity between the E. coli GroEL and the M. xanthus GroELs (E. coli GroEL is 67.3% and 65.2% similar to M. xanthus GroEL1 and GroEL2, respectively) and to the difference in the protein substrates between these two bacteria. Therefore, there are many questions related to the GroEL chaperonins and their functional divergence that need to be addressed.
Methods
Cultures, plasmids, and growth conditions
The strains and plasmids used in this study are listed in Table S1. For the growth assays, the M. xanthus strains were cultivated in the Casitone-based nutrient-rich CTT medium [52]. The E. coli strains were routinely grown on Luria-Bertani (LB) agar or in LB broth. E. coli was grown at 37°C, whereas the Myxococcus strains were incubated at 30°C. When required, 40 µg/ml of kanamycin (Km) and 10 µg/ml of tetracycline (Tet) (Sigma) were added to the medium.
Expression analysis of groEL genes during heat shock, predation, and development
The groEL expression levels during heat shock and liquid predation were analyzed using quantitative real-time PCR. M. xanthus DK1622 and other mutants were harvested after 18 h and exposed to 42°C for 1 h. The RNA was extracted immediately using a total RNA extraction kit following the manufacturer's instructions (Promega). Contaminating DNA was removed with a DNAfree kit (Ambion). The purified RNA was transcribed to yield cDNA, which was stored at −70°C. The quantitative real-time PCR was performed using a Bio-Rad sequence detection system with 250 nM primers, 10 µl of SYBR Green PCR Master Mix (Bio-Rad), 7 µl of RNase-free water, and 2 µl of cDNA template. The PCR was performed for 3 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 59°C, and 15 s at 72°C. The 16S rRNA was used as a normalization signal. Calibration curves (groEL1, groEL2, and 16S RNA) were generated using 10-fold dilutions of M. xanthus DK1622 genomic DNA. The following pairs of forward and reverse primer pairs were used: groEL1, 5′-CACCGAGACGGAGATGAAGG-3′ and 5′-TGAGGCAGCGGATGTAGGC-3′; groEL2, 5′-ATCCGCACGCAGATTGAC-3′ and 5′-GCfCTTCTTCTCCTTCATCTCC-3′; and 16S rRNA, 5′-CGCCGTAAACGATGAGAA-3′ and 5′-TTGCGTCGAATTAAACCAC-3′. The groEL expression levels during predation were analyzed using quantitative real-time PCR. The strains were cultured for 50 h in medium containing casein as a substrate instead of hydrolyzed proteins, and the RNA was extracted immediately. The method and the primers used were the same as those described above. The groEL expression level during different developmental stages was analyzed by measuring the β-galactosidase activity, as described by Li et al. [15], [53], with minor modifications. The cells were broken using a Mini-Beadbeater (BioSpec) at a speed of 2500 rpm. The β-galactosidase activity was determined using o-nitrophenyl-β-galactopyranoside (Sigma), and the samples were analyzed at 420 nm. The total protein concentration was determined using the bicinchoninic acid protein assay (Pierce). The specific activity was calculated as follows: specific activity = 213×A420/(sample volume×protein concentration×reaction time) [15], [54].
Development assays
M. xanthus cells were harvested at mid-logarithmic phase and suspended to a final density of 5×109 cells/ml in TPM buffer. Aliquots (10 µl) were spotted onto TPM agar, and the plates were cultivated at 30°C and observed every 24 h to monitor the formation of fruiting bodies. The sporulation rate was measured after 5 days as previously described. The assays were performed at least three times [15].
Predation assays
The predation assays were performed according to the method used in a previous study [15]. E. coli and M. xanthus cultures were harvested at mid-logarithmic phase and washed three times with 10 mM MOPS buffer (pH 7.6). The final cell densities of the cultures were 5×109 cells/ml for M. xanthus and 1×1011 cells/ml for E. coli. Then, 50 µl of E. coli was pipetted onto a plate to form a 1-cm-diameter colony, and 2 µl of M. xanthus was added to the center of the E. coli colony, with an inoculation diameter of 0.15 cm. The assay was repeated at least three times. The plates were incubated at 30°C for 6 days, during which time the size of the M. xanthus growth area was recorded every 12 h. The predation ability of M. xanthus was reported as the time required for M. xanthus to spread to the edge of the E. coli colonies.
Heat-shock assays
M. xanthus cultures were harvested as described above. The cells were heat shocked for 30 min at 42°C, serially diluted and plated on CTT agar. After 6 days incubation, the CFUs were calculated [15].
Evolutionary analysis
The groEL gene sequences from ten genome-sequenced bacterial strains were retrieved from the NCBI database (Table S2), and the amino acid sequences were aligned using the protein sequence alignment program in CLUSTALW [55]. A maximum likelihood tree was constructed using MEGA5 [56]. The Ka/Ks values among orthologous groEL genes or among paralogous groEL genes were calculated using KaKs_Calculator 1.2 [57] with the NG, MLWL and MLPB models [58], [59].
Vector construction for the region swapping of groELs and mutant identification
The region-swapping assay was conducted according to a previously published method [51]. The regions responsible for the developmental defects of YL0301 and the predation defects of YL0302 were investigated by incorporating single groEL regions into YL0301 or YL0302. The complementation mutants were constructed with the site-specific integration plasmid pSWU30. The apical region of groEL1 was inserted into YL0301 to obtain YL0904 (YL0301::pSWU - groEL2-apicalgroEL1). Briefly (Figure S4), 0.5 kb of the upstream sequence and the N-terminal region (bp 1–597) of groEL2 and the C-terminal (597 to the end) of groEL1 were amplified by PCR. The two fragments were spliced by fusion PCR, digested with XbaI and BamHI, and ligated into pSWU30 digested with XbaI and BamHI. The plasmid was transferred to E. coli λ-pir cells, and the plasmid DNA was extracted from the Tet-resistant transformants using the eZNA Plasmid Mini Kit I (Omega Bio-Tek) according to the manufacturer's instructions. The correct plasmid was used as the template in the second round of fusion PCR. The plasmid containing the correct sequence was transferred by electroporation into YL0301, and individual Tet-resistant colonies were screened. The mutant phenotypes were observed to determine the effects of the apical region on development. The same method was used to replace other regions. The primers used are listed in Table S4.
Vector construction for the deletion of the GGM region and the truncation of groEL1
The GGM region deletion mutants were constructed using the positive-negative KG cassettes described by Ueki et al. Briefly, the upstream sequence (before the GGM sequence) and the downstream sequence (after the GGM sequence) were amplified by PCR. The two fragments were fused to the XbaI restriction site to construct homologous fragments with in-frame deletions. These homologous fragments were ligated into SmaI-digested pBJ113. The resulting plasmid containing the correct sequence was transferred by electroporation into DK1622. The second round of screening was then performed on CTT plates containing 1% galactose (Sigma). The deletion mutants that grew on galactose but were sensitive to kanamycin were identified and verified by PCR and sequencing. The GGM sequence-swapping mutants were constructed in a similar manner. The upstream sequence (739–1623 of groEL1+the DNA sequence corresponding to the last eight amino acids in groEL2) and the downstream sequence (the DNA sequence corresponding to the last eight amino acids in groEL2+815 bp downstream of groEL1) were PCR amplified. The two fragments were fused to the XbaI site to construct in-frame deletion fragments that were ligated into SmaI-digested pBJ113. The resulting plasmid was subjected to two rounds of screening to obtain the GGM-swapping mutant. The phenotypes of the mutants were observed to determine the effects of the GGM repeat region on development. The primers used are listed in Table S6.
Immunoprecipitation assay
groEL1-ko and groEL2-ko mutants were resuspended in Tris buffer (50 mM Tris, 150 mM NaCl, 5 mM EDTA, and 20 µl PMSF) after cultivation and washed three times. The cells were lysed using a high-pressure homogenizer. Aliquots of 50 µl of protein A/G beads were added to 10 ml of supernatant solution to remove the proteins that non-specifically bound to the beads. An anti-GroEL antibody (3 mg/ml) was added, and the solution was shaken at low speed at 4°C. Aliquots of 100 µl of protein A/G beads were mixed with 50 ml of solution and incubated for another 2 h. The beads bound by the GroEL substrates were washed with Tris buffer three times, and the beads and proteins were separated with lysis buffer. The substrates of GroELs were identified by high-pressure liquid chromatography-tandem mass spectrometry (LC-MS/MS) by Shanghai Zhongke Biotech Company. A negative control assay was performed with DK1622 using the same protocol in the absence of the antibody against GroEL to exclude non-specific binding between the beads and proteins [38]. To exclude non-specific binding between the antibody and proteins and to exclude non-specific binding between GroEL and proteins after cell lysis, another negative control was performed by adding 0.1% SDS to the lysis buffer to separate GroEL from its substrates [60]. The solution was diluted 50 fold after cell lysis and the addition of extra GroEL1 and GroEL2 protein. The antibody against GroEL was then added to identify non-specifically bound proteins [38], [60].
Supporting Information
Zdroje
1. LundPA (2009) Multiple chaperonins in bacteria—why so many? FEMS Microbiol Rev 33 : 785–800.
2. LundPA (2001) Microbial molecular chaperones. Adv Microb Physiol 44 : 93–140.
3. RansonNA, WhiteHE, SaibilHR (1998) Chaperonins. Biochem J 333 (Pt 2) 233–242.
4. RadfordSE (2006) GroEL: More than Just a folding cage. Cell 125 : 831–833.
5. LinZ, RyeHS (2006) GroEL-mediated protein folding: making the impossible, possible. Crit Rev Biochem Mol Biol 41 : 211–239.
6. Bernal ZLHaRA (2012) Chaperonin-mediated folding of viral proteins. Adv Exp Med Biol 726 : 307–324.
7. VanBogelenRA, ActonMA, NeidhardtFC (1987) Induction of the heat shock regulon does not produce thermotolerance in Escherichia coli. Genes Dev 1 : 525–531.
8. FayetO, ZiegelhofferT, GeorgopoulosC (1989) The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol 171 : 1379–1385.
9. KernerMJ, NaylorDJ, IshihamaY, MaierT, ChangHC, et al. (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122 : 209–220.
10. GeorgeR, KellySM, PriceNC, ErbseA, FisherM, et al. (2004) Three GroEL homologues from Rhizobium leguminosarum have distinct in vitro properties. Biochem Biophys Res Commun 324 : 822–828.
11. Rodriguez-QuinonesF, MaguireM, WallingtonEJ, GouldPS, YerkoV, et al. (2005) Two of the three groEL homologues in Rhizobium leguminosarum are dispensable for normal growth. Arch Microbiol 183 : 253–265.
12. OjhaA, AnandM, BhattA, KremerL, JacobsWRJr, et al. (2005) GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123 : 861–873.
13. BittnerAN, FoltzA, OkeV (2007) Only one of five groEL genes is required for viability and successful symbiosis in Sinorhizobium meliloti. J Bacteriol 189 : 1884–1889.
14. GouldPS, BurgarHR, LundPA (2007) Homologous cpn60 genes in Rhizobium leguminosarum are not functionally equivalent. Cell Stress Chaperones 12 : 123–131.
15. LiJ, WangY, ZhangCY, ZhangWY, JiangDM, et al. (2010) Myxococcus xanthus viability depends on groEL supplied by either of two genes, but the paralogs have different functions during heat shock, predation, and development. J Bacteriol 192 : 1875–1881.
16. Dworkin M, and Kaiser D. (1993) Myxobacteria II. American Society for Microbiology, Wshington, DC.
17. ShimketsLJ (1990) Social and developmental biology of the myxobacteria. Microbiol Rev 54 : 473–501.
18. GoldmanBS, NiermanWC, KaiserD, SlaterSC, DurkinAS, et al. (2006) Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci U S A 103 : 15200–15205.
19. OtaniM, TabataJ, UekiT, SanoK, InouyeS (2001) Heat-shock-induced proteins from Myxococcus xanthus. J Bacteriol 183 : 6282–6287.
20. LynchM, ForceA (2000) The probability of duplicate gene preservation by subfunctionalization. Genetics 154 : 459–473.
21. QianW, LiaoBY, ChangAY, ZhangJ (2010) Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet 26 : 425–430.
22. HuY, HendersonB, LundPA, TormayP, AhmedMT, et al. (2008) A Mycobacterium tuberculosis mutant lacking the groEL homologue cpn60.1 is viable but fails to induce an inflammatory response in animal models of infection. Infect Immun 76 : 1535–1546.
23. BarreiroC, Gonzalez-LavadoE, BrandS, TauchA, MartinJF (2005) Heat shock proteome analysis of wild-type Corynebacterium glutamicum ATCC 13032 and a spontaneous mutant lacking GroEL1, a dispensable chaperone. J Bacteriol 187 : 884–889.
24. KimAI, GhoshP, AaronMA, BibbLA, JainS, et al. (2003) Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene. Mol Microbiol 50 : 463–473.
25. KovacsE, van der ViesSM, GlatzA, TorokZ, VarvasovszkiV, et al. (2001) The chaperonins of Synechocystis PCC 6803 differ in heat inducibility and chaperone activity. Biochem Biophys Res Commun 289 : 908–915.
26. SatoM, Nimura-MatsuneK, WatanabeS, ChibazakuraT, YoshikawaH (2007) Expression analysis of multiple dnaK genes in the cyanobacterium Synechococcus elongatus PCC 7942. J Bacteriol 189 : 3751–3758.
27. SatoS, IkeuchiM, NakamotoH (2008) Expression and function of a groEL paralog in the thermophilic cyanobacterium Thermosynechococcus elongatus under heat and cold stress. FEBS Lett 582 : 3389–3395.
28. BrocchieriL, KarlinS (2000) Conservation among HSP60 sequences in relation to structure, function, and evolution. Protein Sci 9 : 476–486.
29. BridgerWA, WolodkoWT, HenningW, UptonC, MajumdarR, et al. (1987) The subunits of succinyl-coenzyme A synthetase–function and assembly. Biochem Soc Symp 54 : 103–111.
30. DalzielK (1980) Isocitrate dehydrogenase and related oxidative decarboxylases. FEBS Lett 117Suppl: K45–55.
31. McBrideMJ, ZusmanDR (1993) FrzCD, a methyl-accepting taxis protein from Myxococcus xanthus, shows modulated methylation during fruiting body formation. J Bacteriol 175 : 4936–4940.
32. VlamakisHC, KirbyJR, ZusmanDR (2004) The Che4 pathway of Myxococcus xanthus regulates type IV pilus-mediated motility. Mol Microbiol 52 : 1799–1811.
33. WallD, KolenbranderPE, KaiserD (1999) The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J Bacteriol 181 : 24–33.
34. JakovljevicV, LeonardyS, HoppertM, Sogaard-AndersenL (2008) PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J Bacteriol 190 : 2411–2421.
35. ThomassonB, LinkJ, StassinopoulosAG, BurkeN, PlamannL, et al. (2002) MglA, a small GTPase, interacts with a tyrosine kinase to control type IV pili-mediated motility and development of Myxococcus xanthus. Mol Microbiol 46 : 1399–1413.
36. NudlemanE, WallD, KaiserD (2006) Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol Microbiol 60 : 16–29.
37. OrengoCA, PearlFM, BrayJE, ToddAE, MartinAC, et al. (1999) The CATH Database provides insights into protein structure/function relationships. Nucleic Acids Res 27 : 275–279.
38. HouryWA, FrishmanD, EckerskornC, LottspeichF, HartlFU (1999) Identification of in vivo substrates of the chaperonin GroEL. Nature 402 : 147–154.
39. TyagiNK, FentonWA, HorwichAL (2009) GroEL/GroES cycling: ATP binds to an open ring before substrate protein favoring protein binding and production of the native state. Proc Natl Acad Sci U S A 106 : 20264–20269.
40. SkjaervenL, MugaA, ReuterN, MartinezA (2012) A dynamic model of long-range conformational adaptations triggered by nucleotide binding in GroEL-GroES. Proteins 80 : 2333–2346.
41. TaguchiH (2005) Chaperonin GroEL meets the substrate protein as a “load” of the rings. J Biochem 137 : 543–549.
42. KrishnaKA, RaoGV, RaoKR (2007) Chaperonin GroEL: structure and reaction cycle. Curr Protein Pept Sci 8 : 418–425.
43. AzemA, KesselM, GoloubinoffP (1994) Characterization of a functional GroEL14(GroES7)2 chaperonin hetero-oligomer. Science 265 : 653–656.
44. BraigK, OtwinowskiZ, HegdeR, BoisvertDC, JoachimiakA, et al. (1994) The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature 371 : 578–586.
45. TangYC, ChangHC, RoebenA, WischnewskiD, WischnewskiN, et al. (2006) Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125 : 903–914.
46. HouryWA (2001) Mechanism of substrate recognition by the chaperonin GroEL. Biochem Cell Biol 79 : 569–577.
47. Ohno S (1970) Evolution by Gene Duplication. New York: Springer-Verlag.
48. ZhangJ (2003) Evolution by gene duplication: an update. Trends in Ecology & Evolution 18 : 292–298.
49. GoyalK, QamraR, MandeSC (2006) Multiple gene duplication and rapid evolution in the groEL gene: functional implications. J Mol Evol 63 : 781–787.
50. HeX, ZhangJ (2005) Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169 : 1157–1164.
51. KumarCM, KhareG, SrikanthCV, TyagiAK, SardesaiAA, et al. (2009) Facilitated oligomerization of mycobacterial GroEL: evidence for phosphorylation-mediated oligomerization. J Bacteriol 191 : 6525–6538.
52. HodgkinJ, KaiserD (1977) Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A 74 : 2938–2942.
53. BodeHB, RingMW, SchwarG, KroppenstedtRM, KaiserD, et al. (2006) 3-Hydroxy-3-methylglutaryl-coenzyme A (CoA) synthase is involved in biosynthesis of isovaleryl-CoA in the myxobacterium Myxococcus xanthus during fruiting body formation. J Bacteriol 188 : 6524–6528.
54. KroosL, KuspaA, KaiserD (1986) A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol 117 : 252–266.
55. ThompsonJD, HigginsDG, GibsonTJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 : 4673–4680.
56. TamuraK, PetersonD, PetersonN, StecherG, NeiM, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28 : 2731–2739.
57. ZhangZ, LiJ, ZhaoXQ, WangJ, WongGK, et al. (2006) KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4 : 259–263.
58. NeiM, GojoboriT (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3 : 418–426.
59. TzengYH, PanR, LiWH (2004) Comparison of three methods for estimating rates of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 21 : 2290–2298.
60. EwaltKL, HendrickJP, HouryWA, HartlFU (1997) In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell 90 : 491–500.
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



