-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCoordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
Because cohesion prevents sister-chromatid separation and spindle elongation, cohesion dissolution may trigger these two events simultaneously. However, the relatively normal spindle elongation kinetics in yeast cohesin mutants indicates an additional mechanism for the temporal control of spindle elongation. Here we show evidence indicating that S-phase CDK (cyclin dependent kinase) negatively regulates spindle elongation. In contrast, mitotic CDK promotes spindle elongation by activating Cdc14 phosphatase, which reverses the protein phosphorylation imposed by S-phase CDK. Our data suggest that S-phase CDK negatively regulates spindle elongation partly through its phosphorylation of a spindle pole body (SPB) protein Spc110. We also show that hyperactive S-phase CDK compromises the microtubule localization of Stu2, a processive microtubule polymerase essential for spindle elongation. Strikingly, we found that hyperactive mitotic CDK induces uncoupled spindle elongation and sister-chromatid separation in securin mutants (pds1Δ), and we speculate that asynchronous chromosome segregation in pds1Δ cells contributes to this phenotype. Therefore, the tight temporal control of spindle elongation and cohesin cleavage assure orchestrated chromosome separation and spindle elongation.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003319
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003319Summary
Because cohesion prevents sister-chromatid separation and spindle elongation, cohesion dissolution may trigger these two events simultaneously. However, the relatively normal spindle elongation kinetics in yeast cohesin mutants indicates an additional mechanism for the temporal control of spindle elongation. Here we show evidence indicating that S-phase CDK (cyclin dependent kinase) negatively regulates spindle elongation. In contrast, mitotic CDK promotes spindle elongation by activating Cdc14 phosphatase, which reverses the protein phosphorylation imposed by S-phase CDK. Our data suggest that S-phase CDK negatively regulates spindle elongation partly through its phosphorylation of a spindle pole body (SPB) protein Spc110. We also show that hyperactive S-phase CDK compromises the microtubule localization of Stu2, a processive microtubule polymerase essential for spindle elongation. Strikingly, we found that hyperactive mitotic CDK induces uncoupled spindle elongation and sister-chromatid separation in securin mutants (pds1Δ), and we speculate that asynchronous chromosome segregation in pds1Δ cells contributes to this phenotype. Therefore, the tight temporal control of spindle elongation and cohesin cleavage assure orchestrated chromosome separation and spindle elongation.
Introduction
In eukaryotic cells, spindle elongation and sister-chromatid separation are two critical mitotic events, and the coordination of these two events is essential for the fidelity of chromosome segregation. During mitosis, the cleavage of sister chromatid cohesion allows the sister chromatids to move toward the respective spindle pole, then the spindle elongates further to pull sister chromatids into two daughter cells [1]. Because sister-chromatid cohesion prevents sister chromatid separation and spindle elongation, these two events could be coupled by the cleavage of cohesin. Before anaphase entry, cohesin cleavage is prohibited by the presence of securin (Pds1), which binds to and inhibits separase (Esp1) that cleaves cohesin Scc1/Mcd1 [2], [3]. The degradation of Pds1 before anaphase entry alleviates the inhibition of Esp1, resulting in the robust cohesin cleavage and simultaneous sister-chromatid separation [2], [4], [5]. However, yeast cells lacking either Pds1 or cohesin do not show premature spindle elongation, indicating that a cohesion-independent mechanism controls the timing of spindle elongation [6].
CDKs are the key driving force for cell cycle progression. In budding yeast, S-phase cyclins Clb5 and Clb6 appear during S-phase, which is consistent with their function in DNA synthesis. Compared to mitotic CDK Clb2-Cdk1, Clb5-Cdk1 shows stronger kinase activity toward a subset of CDK substrates [7]. In addition to proteins involved in DNA synthesis, other Clb5-specific substrates, such as Sli15, Fin1, Ase1, and Spc110, associate with the SPB or microtubules, indicating that S-phase CDK may also regulate spindle dynamics [8]–[10].
The tightly regulated activity of CDK and phosphatase enables unique temporal phosphorylation kinetics of each CDK substrate during the cell cycle. The periodical expression of cyclins controls the activity and substrate specificity of the CDK, while a conserved protein phosphatase Cdc14 reverses the phosphorylation of these CDK substrates [11]. In budding yeast, the regulation of Cdc14 activity is achieved through its subcellular localization. Before anaphase entry, Cdc14 is sequestered within the nucleolus by binding to a nucleolar protein Net1 [12], [13]. The dephosphorylation of Net1 by PP2ACdc55 ensures a strong Net1-Cdc14 interaction, while during early anaphase the phosphorylation of Net1 by mitotic CDK, Clb2-Cdk1, triggers the release of Cdc14 from the nucleolus [14]–[17]. The degradation of Pds1 frees separase Esp1, which not only cleaves cohesin rings but also down-regulates PP2ACdc55 with the assistance of other FEAR components [15]. The increased ratio of Clb2-Cdk1/PP2ACdc55 leads to Net1 phosphorylation and the subsequent Cdc14 release. Because S-phase cyclin Clb5 is also degraded before anaphase entry [18], the combination of increased Cdc14 and decreased Clb5-Cdk1 activity during early anaphase results in the dephosphorylation of Clb5-specific substrates [19], [20]. Therefore, mitotic CDK may promote mitotic progression by reversing the phosphorylation imposed by S-phase CDK.
Recent work in the Amon lab has shown that the loss of function of both mitotic cyclins Clb1 and Clb2 in clb1Δ clb2-IV mutant cells prevents spindle elongation, indicating the essential role of mitotic CDK in this process. Moreover, neither the loss of sister-chromatid cohesion nor deletion of securin Pds1 is able to rescue the spindle elongation defect in clb1Δ clb2-IV mutant cells, further supporting the direct role of mitotic cyclins in spindle elongation, but the CDK substrates involved in this process remain unknown [21]. If mitotic CDK promote spindle elongation, cells overexpressing these cyclins are expected to show premature spindle elongation, but these cells exhibit relatively normal spindle elongation, although defect in mitotic exit was noticed [22], [23]. The presence of the CDK inhibitory kinase Swe1 may prevent hyper-activation of mitotic CDK when CLB2 is overexpressed, as Swe1 specifically inhibits mitotic CDK [24]–[27].
Here we show evidence indicating that overexpression of mitotic cyclin CLB2 induces premature spindle elongation in swe1Δ mutant cells. We further found that FEAR mutants suppress this premature spindle elongation, suggesting that Clb2 induces spindle elongation through FEAR that facilitates Cdc14 release during early anaphase. In contrast to mitotic cyclins, we found that S phase cyclin Clb5 plays a negative role in spindle elongation. Therefore, mitotic CDK likely activates the FEAR pathway to alleviate the inhibitory effect of S-phase CDK on spindle elongation. Our data further suggest that the phosphorylation of a SPB protein Spc110 by S-phase CDK contributes at least partially to the inhibition of spindle elongation. Moreover, high levels of S-phase CDK prevent microtubule localization of Stu2, a microtubule-plus-end tracking protein essential for spindle elongation. Strikingly, overexpression of CLB2 in securin mutants pds1Δ leads to uncoupled sister-chromatid separation and spindle elongation. Given the established role of securin Pds1 in the synchronous chromosome segregation [5], our data support the possibility that this securin-dependent synchrony and the temporal control of spindle elongation by the balance of mitotic versus S-phase CDK ensure the sequential order of sister-chromatid separation and spindle elongation, which is critical for faithful chromosome segregation.
Results
Overexpression of mitotic cyclin Clb2 results in premature spindle elongation
Clb2 is the major mitotic cyclin in budding yeast, but its overexpression from a galactose-inducible promoter does not cause obvious premature mitosis in wild-type (WT) cells. Because Swe1 kinase phosphorylates and inhibits mitotic CDK, it is possible that the presence of Swe1 prevents the hyper-activation of mitotic CDK after CLB2 overexpression. Therefore, we overexpressed CLB2 from a galactose-inducible promoter in swe1Δ mutant cells and examined the cell growth. The Western blotting result confirmed the high level expression of Clb2 protein after galactose induction (Figure S1A). Compared to the control cells, swe1Δ mutants with a PGALCLB2 plasmid showed obvious growth defect on galactose plates. Overexpression of Clb1, which is closely related to Clb2, also caused sick growth phenotype in swe1Δ cells, but overexpression of Clb3, Clb4, or S phase cyclin Clb5, Clb6 was not toxic (Figure 1A and S1B). Therefore overexpression of mitotic cyclins Clb1 and Clb2 is toxic to swe1Δ cells. To confirm that mitotic CDK is hyperactive in swe1Δ cells after CLB2 overexpression, we compared the phosphorylation kinetics of Pol12 in synchronized WT and swe1Δ cells overexpressing CLB2, as Pol12 is a known substrate of mitotic CDK required for DNA replication [24]. Our results showed that CLB2 overexpression induces premature Pol12 phosphorylation in both WT and swe1Δ cells based on the band-shift, and the phosphorylation became more significant in swe1Δ cells as indicated by the increased slow migrating band (Figure S1C). Therefore, the absence of Swe1 indeed causes hyper-activation of mitotic CDK after CLB2 overexpression.
Fig. 1. Overexpression of mitotic cyclin CLB2 results in premature spindle elongation in swe1Δ mutants. 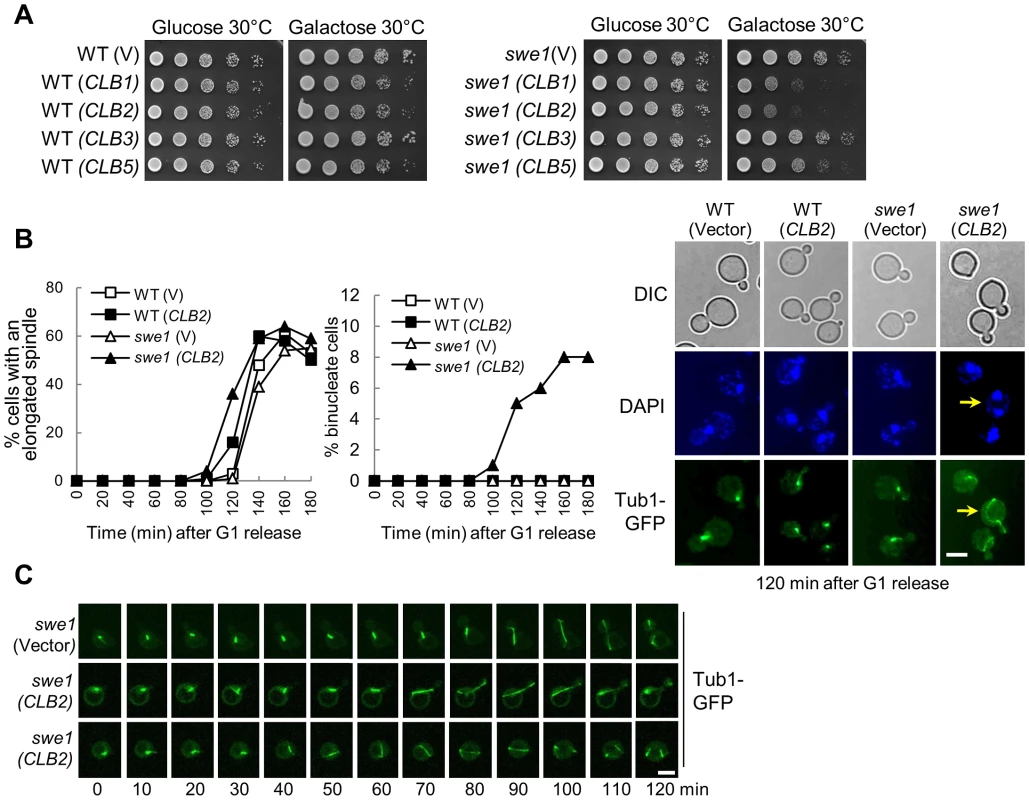
A. Overexpression of CLB2 is toxic to swe1Δ mutants. WT and swe1Δ mutant cells with a control vector or PGALCLB1, PGALCLB2, PGALCLB3, and PGALCLB5 plasmids were grown to saturation in glucose medium, 10-fold diluted, and spotted onto glucose or galactose plates. The plates were scanned after incubation at 30°C for 3 days. B. Overexpression of CLB2 in swe1Δ mutant cells leads to premature spindle elongation. G1-arrested PDS1-Myc TUB1-GFP and swe1Δ PDS1-Myc TUB1-GFP cells with a vector or a PGALCLB2 plasmid were released into 30°C galactose medium to induce CLB2 overexpression. Cells were collected over time and fixed to examine GFP signal and for DAPI staining. Spindles longer than 3 µm were counted as elongated. The percentage of cells with an elongated spindle is shown in the left panel (n>100). The percentage of binucleate cells is shown in the middle panel and the spindle morphology in cells at 120 min time point is shown in the right panel. The arrow indicates a binucleate cell with premature spindle elongation. Scale bar, 5 µm. The budding index, FACS analysis and Pds1 protein level are shown in Figure S2. C. Live-cell imaging shows the premature spindle elongation in swe1Δ mutants overexpressing CLB2. swe1Δ TUB1-GFP cells with a vector or a PGALCLB2 plasmid were arrested in G1-phase in raffinose medium. After released into galactose medium for 1.5 hr, the cells were transferred to an agarose pad on a microscope slide to perform live-cell imaging at 25°C. Scale bar, 5 µm. To understand the cause of this sick growth phenotype, G1-arrested WT and swe1Δ cells with a control vector or a PGALCLB2 plasmid were released into galactose medium to induce CLB2 overexpression and we compared the cell cycle progression in these cells. Both WT and swe1Δ cells showed almost identical budding index and DNA synthesis kinetics either with or without CLB2 overexpression (Figure S2A and S2B). These cells also exhibited similar cell cycle regulated fluctuation of Pds1 protein levels (Figure S2C). However, we noticed premature spindle elongation in some small-budded and unbudded swe1Δ cells overexpressing CLB2, but this phenotype is much less significant in WT cells (Figure 1B). After G1 release into galactose medium for 120 min, 14% of WT cells with a PGALCLB2 plasmid had elongated spindles (>3 µM), while 36% of swe1Δ cells showed elongated spindles. Interestingly, about 8% swe1Δ cells became binucleate after G1 release for 160 min, i.e. two nuclei were observed in a single cell body (the arrow in Figure 1B). Among them, half were small-budded, while the others were unbudded.
Previous data indicate that overexpression of a single copy of a Clb2 destruction box mutant prevents bud formation and results in binucleate cells [28]. Indeed, we found that some cells overexpressing CLB2 remained unbudded after G1 release (Figure S2A). To further define the role of the inhibition of budding and premature spindle elongation in the formation of binucleate cells, we performed live-cell imaging to examine the dynamics of spindle elongation. G1-arrested swe1Δ cells with a control vector or a PGALCLB2 plasmid were released into galactose medium. The spindle elongation in swe1Δ (PGALCLB2) cells initiated ∼20 min earlier compared to the cells with a control vector. Interestingly, we observed premature spindle elongation in both small-budded and unbudded swe1Δ cells (Figure 1C), indicating that both budding inhibition and premature spindle elongation may contribute to the formation of binucleate cells. Because we also observed the binucleate phenotype in both unbudded and small budded cells after Clb2 overproduction (Figure 1B), we reason that the inhibition of budding is not essential for the formation of binucleate cells.
If a cell elongates spindle when cohesion is still present, some sister chromatids may remain together after spindle elongation. However, all the swe1Δ cells overexpressing CLB2 with an elongated spindle showed separated sister chromatids (Figure S3A), indicating that spindle elongation did not occur prior to cohesion dissolution. Alternatively, hyperactive mitotic CDK may promote cohesin cleavage. Thus, we examined Scc1 proteins in WT and swe1Δ cells with and without CLB2 overexpression, but all these cells exhibited similar Scc1 cleavage kinetics based on the appearance of the short Scc1 fragments (Figure S3B), arguing against the possibility that Clb2 induces spindle elongation through cohesin cleavage. We speculate that both hyperactive mitotic CDK and cohesion dissolution are essential for spindle elongation. If that is the case, overexpression of CLB2 may cause more dramatic premature spindle elongation in cohesin mutant cells. We first found that scc1-73 mutant cells with PGALCLB2 plasmids grew more slowly on galactose plates at 25°C compared to control cells (Figure S4A). Moreover, after G1 release, CLB2 overexpression caused premature spindle elongation in scc1-73 cells (Figure S4B), and this phenotype became more pronounced in swe1Δ scc1-73 cells (Figure S4C). Therefore, hyperactive mitotic CDK cause more dramatic premature spindle elongation in cells with compromised sister chromatid cohesion.
Mitotic CDK promotes spindle elongation through the FEAR pathway
One of the substrates of mitotic CDK is the nucleolar Cdc14-binding protein Net1, whose phosphorylation triggers the dissociation of Cdc14 from Net1 and the release of Cdc14 from the nucleolus. It is possible that hyperactive mitotic CDK stimulates spindle elongation by activating FEAR. Because the replacement of 6 CDK phosphorylation sites in Net1 with alanine generates net1-6Cdk mutant, which prevents FEAR activation [14], we first compared the growth of swe1Δ and swe1Δ net1-6Cdk cells after CLB2 overexpression. The swe1Δ net1-6Cdk cells grew much better than the single mutant cells after CLB2 overexpression. Another FEAR mutant spo12Δ showed an even stronger suppression of the sick growth phenotype of swe1Δ (Figure 2A). The nuclear morphology was also compared in swe1Δ, swe1Δ spo12Δ and swe1Δ net1-6Cdk mutant cells overexpressing CLB2, both spo12Δ and net1-6Cdk mutants suppressed the formation of binucleate cells (Figure 2B), suggesting that the activation of FEAR pathway contributes to the growth defect in swe1Δ cells overexpressing CLB2. To directly determine if the toxicity of CLB2 overexpression to swe1Δ cells is due to hyperactive Cdc14, we examined the growth of swe1Δ cdc14-1 cells overexpressing CLB2. We found that cdc14-1 swe1Δ cells with PGALCLB2 plasmids grew better than swe1Δ cells on galactose plates at 25°C (Figure 2A). In addition, the cdc14-1 mutant partially suppressed binucleate phenotype of swe1Δ cells (Figure 2C). We reason that the incomplete suppression is due to the presence of partial functional Cdc14 in cdc14-1 mutant. Therefore, CLB2 overexpression likely induces premature spindle elongation by activating Cdc14.
Fig. 2. Mitotic CDK promotes spindle elongation through FEAR pathway. 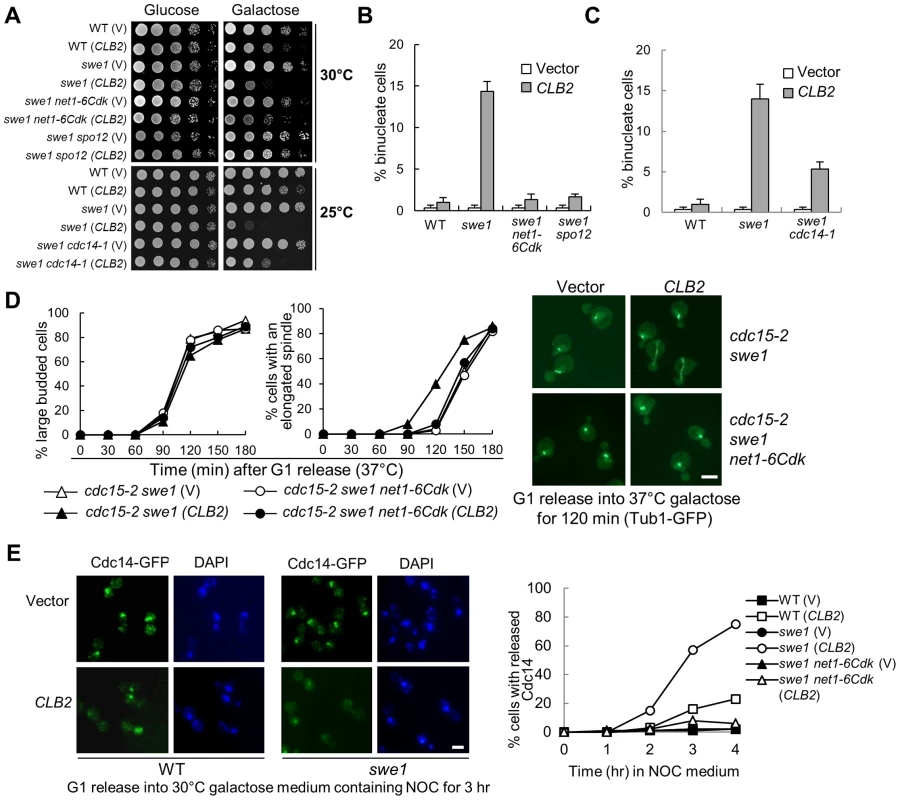
A. FEAR and cdc14-1 mutants suppress the growth defects in swe1Δ cells overexpressing CLB2. A control vector or a PGALCLB2 plasmid was transformed into WT, swe1Δ, swe1Δ net1-6Cdk, and swe1Δ spo12Δ cells. The growth of the transformants on 30°C glucose and galactose plates was examined as described in Figure 1A. The growth of WT, swe1Δ, swe1Δ cdc14-1 with a vector control of PGALCLB2 plasmid on glucose and galactose plates at 25°C is also shown. B. FEAR mutants suppress the binucleate phenotype in swe1Δ cells overexpressing CLB2. Cells with the indicated genotypes were grown to mid-log phase in raffinose medium and then shifted to galactose medium. The cells were collected after 4 hr incubation at 30°C and fixed for DAPI staining. The percentage of binucleate cells is shown. The experiments were repeated for 3 times and at least 100 cells were counted for each sample. C. cdc14-1 mutants suppress the binucleate phenotype in swe1Δ cells overexpressing CLB2. WT, swe1Δ and cdc14-1 swe1Δ cells with a control vector or a PGALCLB2 plasmid were grown to mid-log phase in raffinose medium and then switched to galactose medium. After 4 hr incubation at 37°C, the cells were fixed for DAPI staining. The percentage of binucleate cells from three independent experiments is shown (n>100). D. FEAR mutants suppress the premature spindle elongation in swe1Δ cells overexpressing CLB2. cdc15-2 swe1Δ and cdc15-2 swe1Δ net1-6Cdk cells with a control vector or a PGALCLB2 plasmid were arrested in G1 phase in raffinose medium at 25°C and then released into galactose medium at 37°C. The budding index and the percentage of cells with an elongated spindle are shown in the left panels (n>100). The spindle morphology after 120 min release from G1 is shown in the right panel. The scale bar is 5 µm. E. Overexpression of CLB2 in swe1Δ mutant results in premature Cdc14 release. G1-arrested CDC14-5GFP, swe1Δ CDC14-5GFP and swe1Δ net1-6Cdk CDC14-5GFP cells with a vector or a PGALCLB2 plasmid were released into 30°C galactose medium containing 20 µg/ml nocodazole (NOC). Cells were collected over time and fixed to examine Cdc14 localization. The Cdc14 localization in cells at 3 hr time point is shown in the left panel. The percentage of cells with released Cdc14 was counted over time (right panel, n>100). Scale bar, 5 µm. Unlike the FEAR pathway, the mitotic exit network (MEN) induces Cdc14 release in late anaphase [22], [29]. To further test if Clb2-Cdk1 promotes spindle elongation by activating FEAR or MEN, we examined Clb2-induced premature spindle elongation in cdc15-2 swe1Δ cells. The abolishment of MEN function in cdc15-2 mutant did not block Clb2-induced premature spindle elongation. However, the introduction of either net1-6Cdk or spo12Δ mutation in swe1Δ cdc15-2 cells abolished premature spindle elongation completely (Figure 2D and Figure S5), indicating that Clb2-Cdk1 induces this phenotype through FEAR but not MEN. To directly determine if CLB2 overexpression in swe1Δ cells causes premature FEAR activation, the localization of Cdc14 was examined after G1-arrested swe1Δ cells were released into galactose medium containing microtubule poison nocodazole that arrests cells in metaphase. Strikingly, 75% of swe1Δ cells with a PGALCLB2 plasmid showed released Cdc14 after incubation in galactose medium for 4 hrs, while only 23% of WT cells showed this phenotype. In contrast, swe1Δ cells with a control vector did not show any Cdc14 release. net1-6Cdk mutant suppressed the premature Cdc14 release in swe1Δ cells (Figure 2E). All these data indicate that excess mitotic CDK likely trigger premature spindle elongation by activating Cdc14 through the FEAR pathway. However, we cannot exclude the possibility that Clb2-Cdk1 also phosphorylates other substrates to promote spindle elongation.
Excess S-phase CDK delays spindle elongation
Overexpression of CLB5 slows cell growth, suggesting that hyperactive Clb5-Cdk1 may have a negative effect on the cell cycle (Figure 1A). To test if hyperactive S-phase CDK inhibits spindle elongation, we examined the spindle structure in WT cells overexpressing CLB5. After incubation in galactose medium for 4 hrs, more yeast cells with a PGALCLB5 plasmid arrested with a large bud and a very short spindle structure (Figure 3A). Because these spindles are very short, one explanation is that the failure of SPB separation contributes to the spindle elongation defect. Therefore, we examined the spindle elongation kinetics in cells overexpressing CLB5 after release from hydroxyurea (HU) arrest. HU blocks DNA synthesis and HU-arrested cells have a short spindle with separated SPBs [30]. After HU wash off, CLB5 overexpression also caused a clear spindle elongation delay as indicated by the accumulation of cells with a bar-like short spindle structure (Figure 3B). This result suggests that the short spindle observed in cells with hyperactive S-phase CDK is not due to SPB separation defect. As we have shown that CLB5 overexpression blocks nuclear division in FEAR mutants, such as slk19Δ and spo12Δ [19], we further examined the spindle elongation kinetics in spo12Δ mutants with and without CLB5 overexpression after HU release. Obviously, spindle elongation was largely blocked by CLB5 overexpression in spo12Δ mutant cells (Figure 3B).
Fig. 3. S-phase CDK negatively regulates spindle elongation. 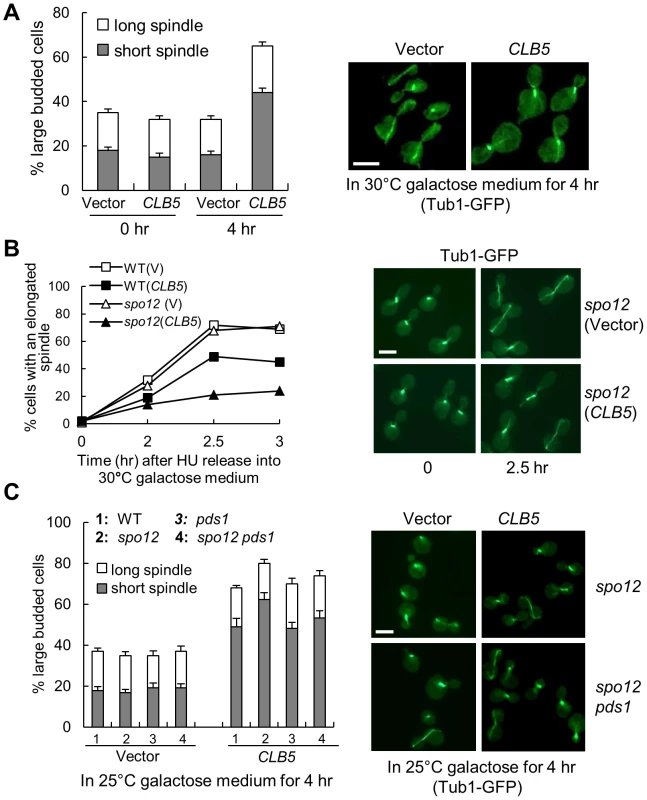
A. Overexpression of S-phase cyclin CLB5 leads to accumulation of cells with a short spindle. TUB1-GFP cells with a control vector or a PGALCLB5 plasmid were grown in raffinose medium to log phase and then shifted into 30°C galactose medium for 4 hrs. The budding index and the percentage of cells with a short (<3 µm) or long spindle are shown in the left panel. The spindle morphology in some representative cells (4 hr in galactose) is shown in the right panel. The experiment was repeated 3 times. The scale bar is 5 µm. B. Overexpression of CLB5 blocks spindle elongation in cells released from HU arrest. The G1-arrested cells with the indicated genotypes were released into 200 mM HU medium and incubated at 30°C for 2 hr. After HU was washed off, the cells were released into 30°C galactose medium and collected over time to examine spindle morphology. The percentage of cells with elongated spindle (>3 µm) is shown in the left panel (n>100). The spindle morphology in some representative cells at time 0 and 2.5 hr is shown in the right panel. Scale bar, 5 µm. C. Deletion of PDS1 gene does not suppress the spindle elongation defects in cells with high levels of Clb5. TUB1-GFP cells with the indicated genotypes were grown in raffinose medium and then shifted to 25°C galactose medium for 4 hr. The experiment was repeated 3 times and the percentage of large budded cells with a short or long spindle is shown in the left panel. The spindle morphology in some representative cells is shown in the right panel. Scale bar, 5 µm. The spindle elongation defect could be due to the activation of the DNA damage or the spindle checkpoint that prevents anaphase onset. Because the activation of these checkpoints depends on the stabilization of securin Pds1 [31], [32], deletion of PDS1 will abolish these checkpoints. We therefore compared the spindle elongation kinetics in WT, pds1Δ, spo12Δ, and spo12Δ pds1Δ mutant cells when CLB5 is overexpressed. Like WT and spo12Δ mutants, pds1Δ and spo12Δ pds1Δ mutants also exhibited accumulation of large-budded cells with a short spindle structure after CLB5 overexpression (Figure 3C), indicating that hyperactive S-phase CDK prevents spindle elongation in a checkpoint-independent manner. Another possibility is that high levels of S-phase CDK block the cleavage of sister chromatid cohesin to prevent spindle elongation. Strikingly, cells overexpressing CLB5 showed almost identical kinetics for cohesin cleavage compared to control cells (Figure S6A). Moreover, overexpression of CLB5 also caused delayed spindle elongation in cohesin mutants incubated at the non-permissive temperature (Figure S6B). Therefore, we conclude that S-phase CDK negatively regulates spindle elongation, and this function is unlikely due to the inability of cohesion resolution.
The absence of S-phase cyclins leads to premature spindle elongation
If S-phase CDK plays a negative role in spindle elongation, we expect that the loss of S-phase cyclins will cause premature spindle elongation. However, both clb5Δ and clb5Δ clb6Δ mutants showed normal spindle elongation kinetics. It is possible that other redundant mechanism, such as the presence of sister-chromatid cohesion, prevents premature spindle elongation in these mutant cells. To test this possibility, we need to examine the spindle elongation kinetics in clb5Δ clb6Δ mutants in the absence of cohesion. Because the spindle is unstable in cohesin mutant cells and deletion of the spindle checkpoint, such as MAD2, suppresses the spindle instability [33], we determined the spindle elongation kinetics in scc1-73 mad2Δ and clb5Δ clb6Δ scc1-73 mad2Δ mutants. Consistent with previous data, scc1-73 mad2Δ cells elongated spindle with kinetics similar to WT cells, but clb5Δ clb6Δ scc1-73 mad2Δ mutant cells showed premature spindle elongation (Figure S7A). Therefore, the absence of S-phase cyclins leads to premature spindle elongation in the absence of cohesion.
Our data indicate that S-phase and mitotic CDK may play opposing roles in spindle elongation. If that is true, we expect CLB2 overexpression to cause premature spindle elongation in the absence of S-phase cyclins. Therefore, we examined spindle elongation in clb5Δ clb6Δ mutant cells with a PGALCLB2 plasmid and control cells. As expected, clb5Δ clb6Δ cells showed premature spindle elongation when CLB2 is overexpressed and these cells grew slowly on galactose medium (Figure S7B). Together, these data support the conclusion that S-phase CDK plays a negative role in spindle elongation, while mitotic CDK plays a positive role in this process.
The phosphorylation of Spc110 by S-phase CDK inhibits spindle elongation
Functional FEAR is required for Clb2-induced premature spindle elongation, and the FEAR promotes Cdc14 release to dephosphorylate Clb5 substrates, such as Ase1 and Fin1 [7], [34], but we found that ase1Δ or fin1Δ mutant did not suppress Clb2-induced premature spindle elongation. A previous study suggests that Clb5-Cdk1-induced phosphorylation of Spc110, one of the SPB proteins, also modulates spindle dynamics [9]. Interestingly, a phospho-mimetic spc11018D91D mutant, in which the CDK phosphorylation sites at Thr18 and Ser91 were mutated to aspartic acid, showed dramatic suppression of the binucleate phenotype in swe1Δ cells overexpressing CLB2 (Figure 4A), indicating that the dephosphorylation of Spc110 might be required for Clb2-induced premature spindle elongation. We further compared the spindle elongation kinetics in WT and spc11018D91D mutant cells and found that the mutant cells did exhibit delayed spindle elongation, although the delay was not pronounced (Figure S8). Nevertheless, the spc11018D91D mutant failed to rescue the sick growth phenotype of swe1Δ cells with PGALCLB2 on galactose medium, suggesting that phosphorylation of other Clb5 substrates can prevent spindle elongation as well. Alternatively, other unidentified defects induced by Clb2 overexpression may also lead to the sick growth.
Fig. 4. The dephosphorylation of Spc110 promotes spindle elongation. 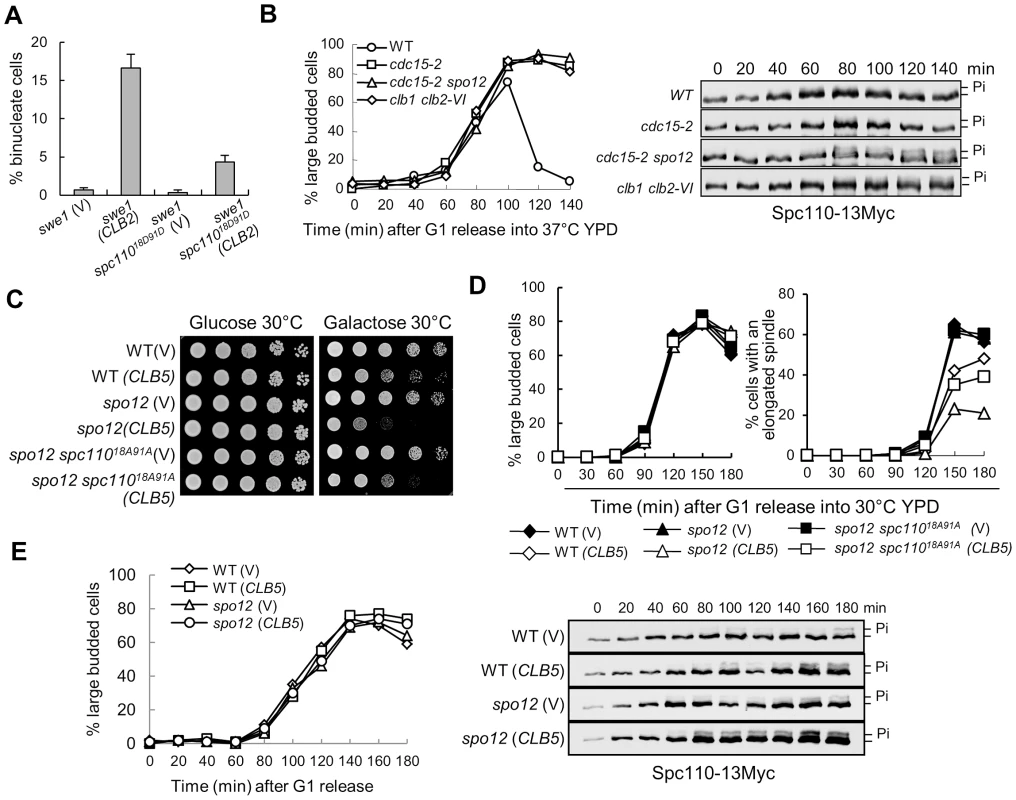
A. spc11018D91D mutants suppress the binucleate phenotype in swe1Δ cells after CLB2 overexpression. swe1Δ and swe1Δ spc11018D91D cells with a control vector or a PGALCLB2 plasmid were grown to mid-log phase in raffinose medium and then switched to galactose medium. After 4 hr incubation at 30°C, the cells were fixed for DAPI staining. The percentage of binucleate cells is shown after 3 repeats (n>100). B. The dephosphorylation of Spc110 is FEAR dependent. WT, cdc15-2, cdc15-2 spo12Δ, and clb1Δ clb2-VI cells with Spc110-13myc were arrested in G1 phase at 25°C and then released into cell cycle at 37°C. The protein samples were prepared every 20 min and Western blotting was performed to detect Spc110 phosphorylation. The budding index is shown in the left panel; the Spc110 protein levels and band-shift are shown in the right panel. C. spc11018A91A mutants partially suppress the growth defects of spo12Δ mutant cells overexpressing CLB5. Saturated cells with the indicated genotypes were 10-fold diluted and the growth on glucose and galactose plates was examined after 3 day incubation at 30°C. D. spc11018A91A mutants partially suppress the spindle elongation defects in spo12Δ mutants overexpressing CLB5. Cells with the indicated phenotypes were arrested in G1 phase in raffinose medium and then released into 30°C galactose medium. The percentages of large budded cells and cells with an elongated spindle are shown (n>100). E. Overexpression of CLB5 in spo12Δ mutant cells leads to more dramatic Spc110 phosphorylation. SPC110-13Myc and spo12Δ SPC110-13Myc cells with a vector or a PGALCLB5 plasmid were arrested in G1 phase and released into 30°C galactose medium. The protein samples were prepared every 20 min and Western blotting was performed to detect Spc110 phosphorylation. The budding index is shown in the left panel; the Spc110 protein levels and band-shift are shown in the right panel. The phosphorylation of Spc110 by Clb5-Cdk1 produces a band-shift on protein gels detectable by Western blotting [9]. We have demonstrated that the dephosphorylation of some Clb5-Cdk1 substrates depends on functional FEAR [19]. To test if the FEAR pathway is also required for the dephosphorylation of Spc110, we compared the band-shift of Spc110 protein in WT and mutant cells lacking functional MEN or FEAR. Significant Spc110 phosphorylation was not observed in synchronous WT and MEN mutant cells cdc15-2, but we noticed more obvious Spc110 phosphorylation in cdc15-2 spo12Δ mutant wherein the function of both MEN and FEAR is abolished, indicating that functional FEAR may be required for Spc110 dephosphorylation. Because mitotic CDK activates the FEAR by phosphorylating Net1, we also examined Spc110 phosphorylation in clb1Δ clb2-VI temperature sensitive mutants. These mutant cells exhibited more Spc110 phosphorylation, which supports the notion that mitotic CDK promotes Spc110 dephosphorylation (Figure 4B).
We previously showed that overexpression of S-phase cyclin CLB5 is toxic to FEAR mutants and delays nuclear separation in the mutant cells [19]. If Clb5-induced phosphorylation of Spc110 contributes to this phenotype, a nonphosphorylatable spc110 mutant will suppress this phenotype. We first found that a nonphosphorylatable spc11018A91A mutant partially restored the growth of FEAR mutants (spo12Δ) with a PGALCLB5 plasmid on galactose medium (Figure 4C). The spindle elongation dynamics was also examined in spo12Δ and spo12Δ spc11018A91A mutants overexpressing CLB5. Clb5 overproduction in spo12Δ mutants significantly delayed spindle elongation. After G1 release for 150 min, 45% of WT cells exhibited elongated spindles, while only 23% of spo12Δ mutant cells did. However, 35% of spo12Δ spc11018A91A mutant cells displayed elongated spindle at 150 min (Figure 4D), indicating that active S-phase CDK prevents spindle elongation at least partially through Spc110 phosphorylation. Consistently, more significant Spc110 phosphorylation was observed in spo12Δ cells overexpressing CLB5 (Figure 4E). In these cells, the kinetics of DNA synthesis is indistinguishable with or without CLB5 overexpression based on the FACS analysis. Collectively, these data reveal the possibility of a negative role of Clb5-dependent Spc110 phosphorylation in spindle elongation.
Overexpression of CLB5 decreases the microtubule localization of Stu2
Our data suggest that the phosphorylation of SPB component Spc110 plays a role in the timing control of spindle elongation, and this phosphorylation is regulated by the balance of S-phase and mitotic CDKs. As a SPB component, however, it is likely that the phosphorylation of Spc110 regulates the spindle elongation via other microtubule-associated protein(s). Stu2 is the yeast homologue of the XMAP215 protein that binds to the microtubule plus-end [35], [36]. This protein is a processive microtubule polymerase essential for spindle elongation [37], [38]. One possibility is that the CDK activity controls the timing of spindle elongation by regulating the activity of Stu2. Interestingly, the temperature sensitive mutant stu2-10 dramatically suppressed the toxicity of CLB2 overexpression to swe1Δ mutant cells when incubated at 25°C (Figure 5A), indicating that intact Stu2 function is required for CLB2-induced premature spindle elongation. In contrast, stu2-10 mutant cells were more sensitive to CLB5 overexpression than WT cells (Figure 5B), indicating that Clb5 may negatively regulates Stu2 function. As we have showed that the phosphorylation of Spc110 by Clb5-Cdk1 plays a negative role in spindle elongation (Figure 4C and 4D), we further compared the growth and spindle elongation in stu2-10 and stu2-10 spc11018A91A at 35°C. The results showed that nonphosphorylatable spc110 mutant partially suppressed the temperature sensitivity and the spindle elongation defect of stu2-10 mutant cells (Figure 5C), suggesting that S-phase CDK-dependent Spc110 phosphorylation may down-regulate Stu2 function.
Fig. 5. Overexpression of CLB5 impairs the localization of Stu2 on spindle and cytoplasmic microtubules. 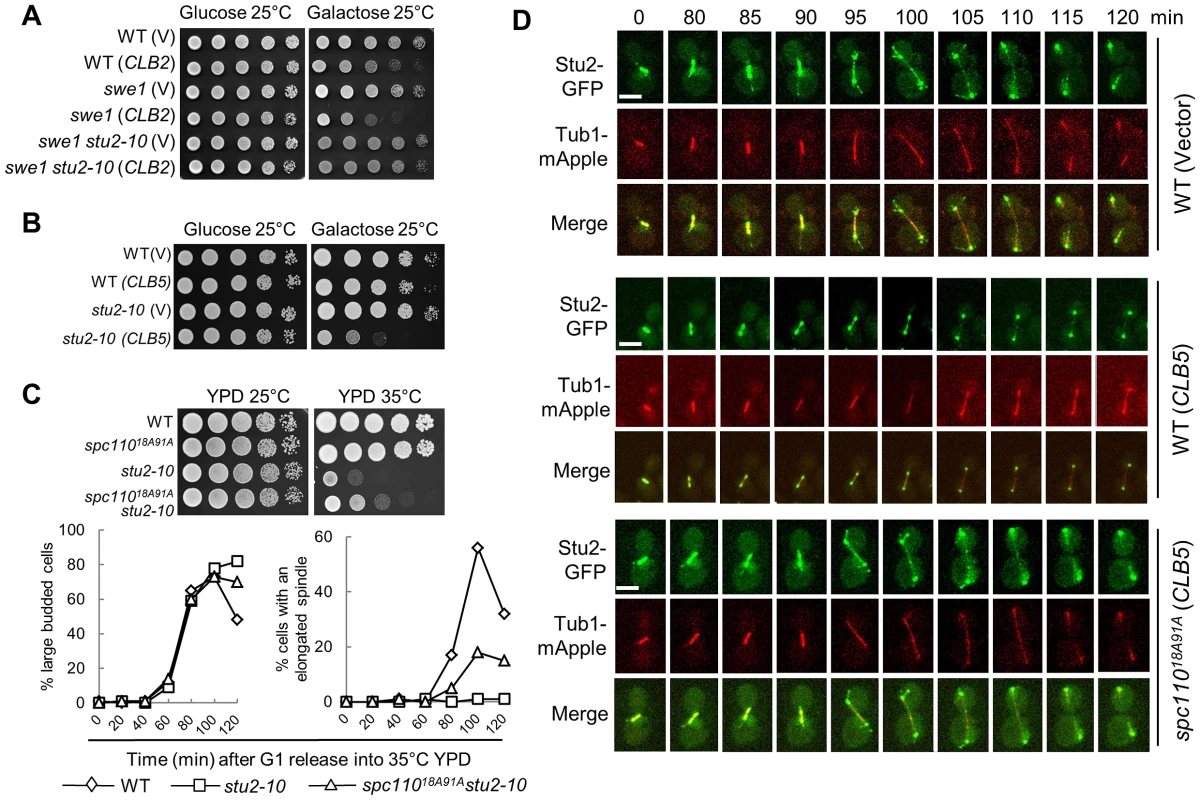
A. stu2-10 temperature-sensitive mutants suppress the growth defect of swe1Δ cells overexpressing CLB2. The cell cultures with indicated genotypes were 10-fold diluted and then spotted onto glucose and galactose plates. The growth was examined after 4 day incubation at 25°C. B. Overexpression of CLB5 is toxic to stu2-10 mutants. The growth of WT and stu2-10 mutant cells with a control vector or PGALCLB5 plasmids at 25°C were examined as described in A. C. spc11018A91A mutants partially suppress the temperature sensitivity and spindle elongation defects of stu2-10 mutants. The growth of cells with the indicated genotypes at 25°C and 35°C is shown in the top panel. The cells with indicated genotypes were arrested at G1 phase at 25°C and then released into YPD medium at 35°C. The budding index and the percentage of cells with an elongated spindle (>3 µm) are shown in the bottom panel (n>100). D. Overexpression of CLB5 impairs the localization of Stu2 on spindle and cytoplasmic microtubules. WT and spc11018A91A cells with STU2-GFP TUB1-mApple harboring a control vector or a PGALCLB5 plasmid were arrested with 200 mM HU in raffinose medium for 2.5 hrs. After the cells were released into galactose medium for 1 hr, they were subjected to live-cell microscopy at 25°C. The Stu2 localization and spindle morphology are shown (a reprehensive from more than 10 cells). The scale bar is 5 µm. To further test if the spindle localization of Stu2 is impaired in cells overexpressing CLB5, live-cell imaging was performed in cells with and without CLB5 overexpression. Consistent with previous reports, we found that Stu2 localized on both spindle poles and spindle at metaphase and early anaphase [36]. At some cell cycle stages, the localization of Stu2 on the cytoplasmic microtubules was also clearly observed in the control cells (Figure 5D). Although the SPB-localization of Stu2 remained similar, we noticed that overexpression of CLB5 significantly decreased the Stu2 localization on spindle and cytoplasmic microtubules. However, spc11018A91A mutant restored the microtubule-localization of Stu2 in cells overexpressing CLB5 (Figure 5D), which is consistent with the result that spc11018A91A partially suppressed the temperature sensitivity of stu2-10. Therefore, we speculate that the phosphorylation of Spc110 by S-phase CDK prevents the localization of Stu2 on spindle and cytoplasmic microtubules, presumably at the plus-ends. In contrast, Spc110 dephosphorylation likely facilitates the microtubule localization of Stu2, which promotes spindle elongation.
Overexpression of mitotic cyclin CLB2 in pds1Δ mutants results in uncoupled sister-chromatid separation and spindle elongation
Our data indicate that the balance of mitotic and S-phase CDKs regulates the timing of spindle elongation. In addition, the presence of sister chromatid cohesion prevents spindle elongation. Although cohesion mutant cells (scc1-73) exhibit premature spindle elongation when CLB2 is overexpressed, most of the cells have a successful mitosis because they are viable after CLB2 overexpression. We suspect that an additional mechanism also plays a role in the coordination of cohesion cleavage and spindle elongation.
Securin Pds1 binds to and inhibits separase Esp1, whose activity is required for the cleavage of cohesion Scc1/Mcd1 and the subsequent anaphase onset [2]. Thus, cell cycle-regulated Pds1 protein levels control the timing of cohesion cleavage and anaphase onset. A previous study showed that deletion of a nonessential gene CDH1 caused lethality in pds1Δ cells and expression of an extra copy of SWE1 suppressed this lethality [39]. Because CDH1 encodes an APC activator required for Clb2 degradation, it is possible that the high Clb2 levels contribute to the synthetic lethality of cdh1 pds1 mutants. Therefore, we examined the growth of pds1Δ cells overexpressing CLB2 and found that overexpression of CLB2 was very toxic to pds1Δ cells (Figure 6A). One possibility is that high levels of mitotic CDK cause chromosome biorientation defects [40], which require the intact spindle assembly checkpoint for survival. However, overexpression of CLB2 was not toxic to checkpoint mutant mad2Δ (Figure S9), excluding the possibility that the checkpoint defect in pds1Δ contributes to the lethality. After CLB2 expression for 4 hrs, 68% of pds1Δ cells lost viability (Figure 6B). A FEAR mutant spo12Δ failed to suppress the growth defect of pds1Δ (PGALCLB2) cells on galactose plates, but it partially suppressed the lethality of pds1Δ (PGALCLB2) cells after incubation in liquid galactose media (Figure 6B). Therefore, we conclude that high level of Clb2 proteins causes the lethality in pds1Δ mutant cells.
Fig. 6. Overexpression of CLB2 in pds1Δ mutants causes uncoupled sister-chromatid separation and spindle elongation. 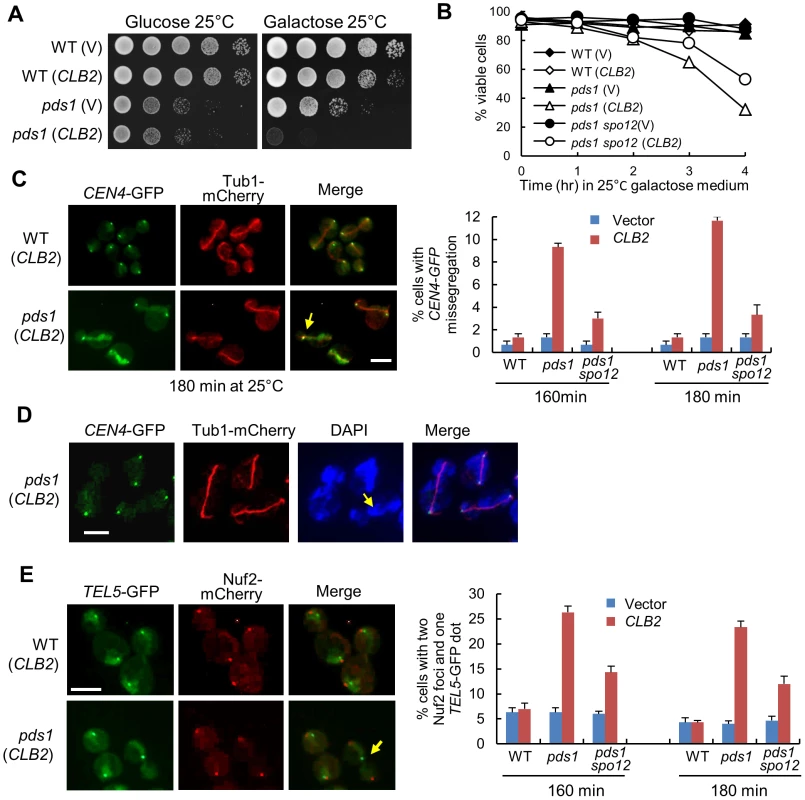
A. Saturated cells with the indicated genotypes were examined for the growth on glucose and galactose plates after 4 day incubation at 25°C. B. Cells with the indicated genotypes were grown to mid-log phase in raffinose medium and then shifted to galactose medium. Cells were collected over time and the viability was examined using platting efficiency (n>300). C. WT, pds1Δ, and pds1Δ spo12Δ cells in CEN4-GFP TUB1-mCherry background with a vector or a PGALCLB2 plasmid were arrested in G1-phase and then released into 25°C galactose medium to induce CLB2 overexpression. Cells were collected at 160 and 180 min and fixed to examine chromosome segregation. The spindle morphology and CEN4-GFP segregation in some representative cells at 180 min are shown in the left panel. The arrow indicates a cell with an elongated spindle and mis-segregated CEN4-GFP. The average percentage of cells with mis-segregated CEN4-GFP at 160 and 180 min from three experiments is shown in the right panel (n>100). Scale bar, 5 µm. D. WT and pds1Δ cells in CEN4-GFP TUB1-mCherry background with a vector or PGALCLB2 plasmid were grown to mid-log phase in raffinose medium and then shifted to galactose medium. The cells were collected after 4 hr incubation at 25°C and fixed for DAPI staining. The segregation of CEN4-GFP as well as the spindle and nuclear morphology in some representative cells is shown. The scale bar is 5 µm. E. WT, pds1Δ, and pds1Δ spo12Δ cells in TEL5-GFP NUF2-mCherry background were treated as described in C. The experiment was repeated for 3 times. The segregation of telomere of chromosome V (TEL5-GFP) in cells at 180 min is shown in the left panel. The arrow indicates a cell with unseparated TEL5. The average percentage of cells with two Nuf2 foci and one TEL5-GFP dot at 160 and 180 min is shown in the right panel after three repeats (n>100). Scale bar, 5 µm. The presence of Pds1 prevents the activation of separase Esp1 and the abrupt Pds1 degradation prior to anaphase onset allows a robust cohesion cleavage, resulting in synchronous dissolution of all chromosome pairs. The absence of Pds1 decreases this synchrony [5], thus, the loss of this synchrony may cause catastrophic mitosis when CLB2 is overexpressed. To test this possibility, we first examined the spindle elongation and sister centromere separation in pds1Δ cells overexpressing CLB2. After pds1Δ cells were released from G1-arrest into galactose medium for 3 hrs, about 12% cells exhibited an elongated spindle with a single CEN4-GFP dot, indicating the failure of chromosome IV separation after spindle elongation (Figure 6C). Given that this number is only for one of the 16 chromosomes, the defect in chromosome segregation should be very dramatic and quick viability loss supports this speculation. We found that the mis-segregation of CEN4-GFP was largely suppressed by spo12Δ, which is consistent with the notion that CLB2 promotes spindle elongation through FEAR (Figure 6C).
We further used DAPI staining to examine the chromosome segregation in pds1Δ cells overexpressing CLB2. Strikingly, most of the cells failed to show two clear DNA masses after spindle elongation. Instead, they exhibited lagged DNA along the elongated spindle (Figure 6D), indicating a remarkable chromosome segregation defect. The observation of cells with an elongated spindle and a single CEN4-GFP dot supports this speculation. We further examined the segregation of the telomere of chromosome V (TEL5-GFP) in pds1Δ cells overexpressing CLB2. After G1 release into galactose for 180 min, 23% of pds1Δ cells exhibited a single TEL5-GFP dot with two kinetochore clusters (Nuf2-mCherry) separated to two daughter cells, indicating the failure of telomere separation for chromosome V (Figure 6E). spo12Δ also showed partial suppression of TEL5-GFP mis-segregation (Figure 6E). Interestingly, the percentage of pds1Δ cells with a single TEL5-GFP dot after CLB2 overexpression is obviously higher than that with unseparated CEN4-GFP (23% vs. 11%). Our explanation is that both unseparated and partially separated chromosomes contribute to the telomere separation defect. The results support the possibility that overexpression of CLB2 in pds1Δ cells induces spindle elongation when cohesin still holds a few chromosomes or some parts of a chromosome. Nevertheless the small amount of cohesin is unable to restrain the premature spindle elongation induced by CLB2 overexpression, thereby resulting in the failure for the segregation of entire or part of a chromosome. Premature spindle elongation in pds1Δ mutant cells, therefore, induces uncoupled chromosome segregation and spindle elongation, which leads to significant chromosome mis-segregation.
Discussion
The key to a successful cell division is the coordination of various cell cycle events. For efficient chromosome segregation, spindle elongation should follow the dissolution of sister-chromatid cohesion in an orderly fashion. The molecular mechanism that ensures this sequential order remains unclear. The absence of premature spindle elongation in cells lacking cohesion indicates a cohesion-independent mechanism that controls the timing of spindle elongation. Here we show that S-phase CDK negatively regulates spindle elongation, while mitotic CDK actives the FEAR pathway to trigger Cdc14 release, which reverses S-phase CDK-dependent protein phosphorylation and simulates spindle elongation. Therefore, the balance of mitotic vs. S-phase CDK activity is critical for the timing control of spindle elongation. We also show that S-phase CDK prevents spindle elongation in part by phosphorylating a SPB component Spc110, while dephosphorylation of Spc110 by Cdc14 likely facilitates the localization of Stu2, a plus-end tracking protein, to spindle microtubules, which may directly promotes spindle elongation by enhance microtubule polymerization [37]. Furthermore, hyperactive mitotic CDK in pds1Δ cells, where the synchrony of chromosome segregation is compromised, leads to uncoupled sister-chromatid separation and spindle elongation, resulting in chromosome mis-segregation and cell death. A model illustrating and integrating this cell cycle regulatory network is shown in Figure 7.
Fig. 7. The working model for the timing control of spindle elongation by the balance of mitotic versus S-phase CDKs. 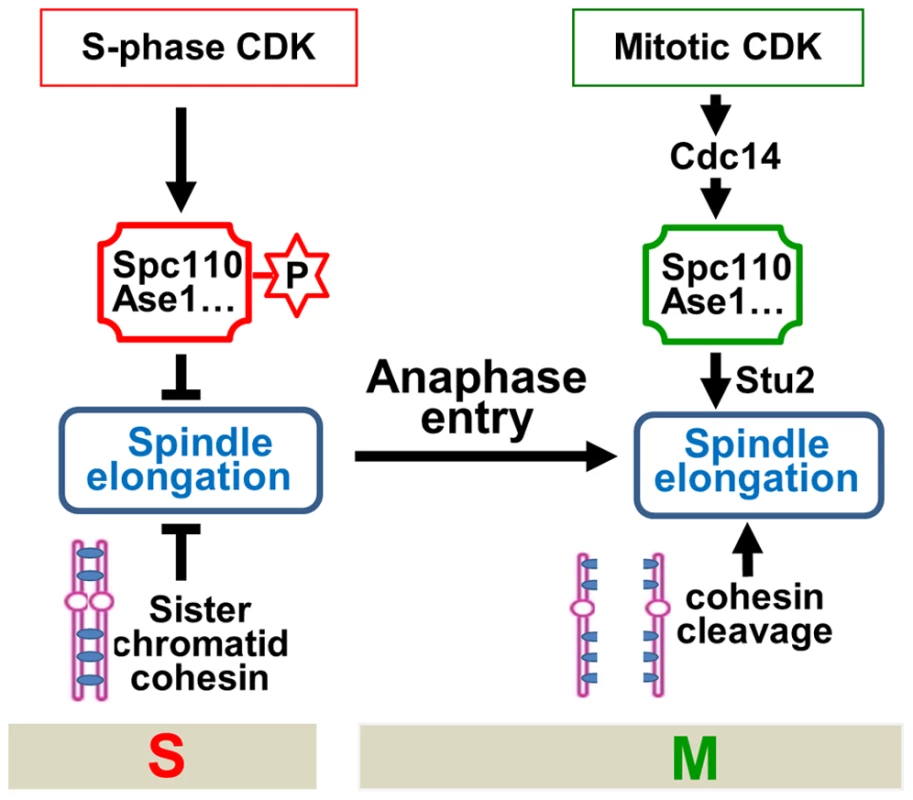
These findings reveal at least two mechanisms that prevent premature spindle elongation. Firstly, before anaphase onset, S-phase CDK phosphorylates several microtubule-associated proteins to prevent spindle elongation. Therefore, in addition to promoting DNA synthesis, S-phase CDK also negatively regulates mitosis to ensure the correct order of S and M phase spindle function [41]. Secondly, sister-chromatid cohesion restrains spindle elongation as well. To allow spindle elongation, both the loss of sister-chromatid cohesion and the reversion of protein phosphorylation imposed by S-phase CDK have to be achieved. Before anaphase onset, the destruction of Pds1 frees separase Esp1 that cleaves cohesin rings. Nevertheless, the reversion of S-phase CDK-dependent phosphorylation requires the destruction of S-phase cyclin Clb5 as well as the activation of phosphatase Cdc14. Clb5 is degraded before anaphase onset along with Pds1 through APCCdc20 [18]. The release of Esp1 from the inhibition by Pds1 not only triggers cohesin cleavage but also activates FEAR to release Cdc14 [15]. Therefore, this mechanism ensures that cohesin cleavage and spindle elongation are coordinated temporally (Figure 7).
The tightly regulation of the protein levels of securin Pds1 during cell cycle not only avoids the dissolution of sister-chromatid cohesion prior to anaphase entry [42], but also ensures that all chromosome pairs disjoin almost simultaneously [5]. Loss of this switch-like mechanism in pds1Δ mutant cells results in asynchronous cohesion dissolution. Therefore, in pds1Δ mutant cells, it is possible that only a few sister chromatids are linked by cohesin to restrain spindle elongation while cells have initiated spindle elongation. Surprisingly, most pds1Δ cells are able to segregate chromosomes successfully. The temporal control mechanism described above may prevent spindle elongation when cohesion is partially dissolved in pds1Δ mutants, but deregulation of this temporal control mechanism could lead to catastrophic mitosis. Indeed, we showed that overexpression of CLB2 in pds1Δ mutants results in the failure of the separation of some sister chromatids after spindle elongation. In addition to high levels of mitotic cyclins, constitutive activation of FEAR by deletion of CDC55 or the deletion of S-phase cyclins has also been shown to be lethal to pds1Δ mutants [39], [43], [44], and the induction of premature spindle elongation is likely the cause of the lethality. Because only some sister-chromatids are linked by cohesin rings when spindles elongate in these cells, the pulling force may break the kinetochore-microtubule interaction, resulting in chromosome mis-segregation. It is also possible that cohesin only exists in part of a chromosome, such as the arm or telomere regions while the spindle is elongating. Under this situation, sister centromeres segregate successfully, but the telomeres of this chromosome still stay together.
Previous work validates the role of mitotic CDK in spindle elongation, as cells lacking both Clb1 and Clb2 fail to elongate the spindle. Because overexpression of Cdc14 phosphatase cannot suppress the spindle elongation defect in clb1Δ clb2-IV mutant, it was speculated that mitotic CDK promotes spindle elongation in FEAR-independent manner [21]. Our observation that FEAR mutants completely suppress Clb2-induced premature spindle elongation and toxicity to swe1Δ cells strongly supports the conclusion that mitotic CDK promotes spindle elongation through the FEAR pathway. However, we cannot exclude the possibility that additional mitotic CDK targets might also be involved in spindle elongation.
An interesting question is whether cohesin cleavage and spindle elongation occur at the same time. We found that spindle elongation happens earlier in swe1Δ mutant cells overexpressing CLB2, but the cells with elongated spindle always show separated sister chromatids. Moreover, our result indicates that CLB2 overexpression does not leads to premature cohesin cleavage. These results suggest that spindle elongation may occur later than cohesin cleavage, but hyperactive CDK eliminates this lag. Because FEAR mutants block Clb2-induced premature spindle elongation in swe1Δ mutants, we speculate that the activation of FEAR occurs later than cohesin cleavage, which contributes to the time difference. Therefore, an important function of the FEAR pathway is likely to ensure the sequential order of cohesin cleavage and spindle elongation. Although the hyperactive FEAR in cdc55Δ mutant cells did not result in dramatic mitotic defect, cdc55Δ bub2Δ double mutant cells exhibit catastrophic mitosis, wherein both FEAR and MEN are hyperactive [16].
Several microtubule-associated proteins, such as Ase1, Fin1, and Ask1, are phosphorylated more efficiently by S-phase CDK and their phosphorylation regulates spindle dynamics [8], [34], [45]. Here we show that S-phase CDK-mediated phosphorylation of Spc110 also plays an important role in spindle elongation. We found that the phospho-mimetic spc11018D91D mutant partially abolishes Clb2-induced premature spindle elongation. In contrast, the nonphosphorylatable spc11018A91A mutant suppresses the Clb5-induced delay in spindle elongation. Although spc11018D91D mutant cells show a noticeable delay in spindle elongation, the delay is not significant. We reason that the phosphorylation of a group of proteins by Clb5-Cdk1 prevents spindle elongation, but the phospho-mimetic mutant for each single protein may not be sufficient to block spindle elongation completely. In contrast to Ase1 and Fin1, which directly bind to microtubules and regulate microtubule dynamics, Spc110 is a SPB component, so it is likely that Spc110 phosphorylation regulates spindle elongation in an indirect way. We show that overexpression of CLB5 clearly impairs the localization of Stu2 on spindle and cytoplasmic microtubules. The dephosphorylation of Spc110 and/or other Clb5 substrates likely promotes the localization of Stu2 to the plus-end of spindle microtubules, where Stu2 triggers the polymerization and spindle elongation. Further studies are needed to understand how S-phase CDK prevents microtubule localization of Stu2.
In summary, our data reveal the molecular mechanism that coordinates chromosome segregation and spindle elongation. Before anaphase, S-phase CDK and sister chromatid cohesion prevent spindle elongation. The establishment of chromosome bipolar attachment triggers the degradation of both securin Pds1 and S-phase cyclin Clb5. The disappearance of Pds1 activates Esp1 that cleaves cohesin and triggers Cdc14 release through FEAR. Consequently, the loss of sister chromatid cohesion and the reversion of S-phase CDK-dependent protein phosphorylation trigger spindle elongation. This mechanism becomes essential in cells with decreased synchrony of chromosome segregation, as induction of premature spindle elongation in these cells results in unseparated or partially separated chromosomes after spindle elongation. Like budding yeast Clb5, the S-phase cyclin (Cyclin A) in mammalian cells also exhibits substrate specificity [46]. Moreover, the proteins involved in this regulation, such as Ase1, Spc110, and Stu2, are well conserved in higher eukaryotic cells. For example, XMAP215, the Stu2 homologue in mammalian cells, is required for microtubule polymerization and spindle elongation [47]. Therefore, the mechanism of the temporal control of spindle elongation described in budding yeast could be conserved. We represent data showing that the temporal control of spindle elongation is critical for accurate chromosome segregation, suggesting that defects in this network may contribute to aneuploidy that is associated with cancer progression.
Materials and Methods
Yeast strains and growth
The yeast strains used in this study are listed in Table S1. All strains are isogenic to Y300, a W303 derivative. Yeast cells were grown in YPD (Yeast extract, Peptone, Dextrose) or indicated synthetic medium. To arrest cells in G1 phase, 5 µg/ml α-factor was added into cell cultures. After 2 hr incubation, the G1-arrested cells were washed twice with water and then released into fresh medium to start the cell cycle. Nocodazole was used at 20 µg/ml in 1% DMSO. To induce cyclin overexpression, galactose was added to the medium to a final concentration of 2%.
Cytological techniques
Cells with GFP, mCherry or mApple-tagged proteins were fixed with 3.7% formaldehyde for 5 min at room temperature, and then washed twice with 1× PBS buffer and resuspended in PBS buffer for fluorescence microscopy (Carl Zeiss MicroImaging, Inc.). The spindle morphology was monitored by using TUB1-GFP, TUB1-mApple, or TUB1-mCherry strains and we count the spindles longer than 3 µm as elongated spindles. For DAPI staining, cells were fixed with 3.7% formaldehyde for 5 min at room temperature, and then resuspended in 100% MEOH at −20°C for 30 min. The cells were incubated in DAPI solution (final concentration of 2.5 µg/ml) for 1 min at room temperature. For each experiment, we repeated 3 times and at least 100 cells were counted for each sample.
Live-cell microscopy was carried out with a Nikon Eclipse Ti imaging system (Andor). We used a glass depression slide to prepare an agarose pad filled with synthetic complete medium with the addition of galactose. All live-cell images were acquired at 25°C with an ×100 objective lens. Twelve Z-sections were collected at each time point, and each optical section was set at 0.5 µm thick. The time-lapse interval was set at 5 min. Maximum projection from applicable time points were created using Andor IQ2 software.
FACS analysis
G1 phase-arrested cells in raffinose medium were released into galactose medium. Samples were taken at various time points and fixed in 70% EtOH overnight at 4°C. Cells were then incubated in Tris pH 7.8 buffer with 0.2 mg/ml RNase A at 37°C for 4 hrs and stained with 30 µg/ml propidium iodide at 4°C overnight. FACS analysis was performed using FACSCanto equipped with the FACSDiva software.
Protein techniques
Cell pellets from 1.5 ml of cell culture were resuspended in 200 µl 0.1 N NaOH and incubated at room temperature for 5 min. After centrifugation, the cells were resuspended in equal volume (30 µl) of ddH2O and SDS protein-loading buffer. The samples were then boiled for 5 min and resolved with 8% SDS-polyacrylamide gel. Proteins were detected with ECL (Perkin Elmer LAS, Inc.) after probing with anti-Myc or anti-HA antibodies (Covance Research Products, Inc.) and HRP-conjugated secondary antibody (Jackson ImmunoResearch, Inc.).
Supporting Information
Zdroje
1. StraightAF, MarshallWF, SedatJW, MurrayAW (1997) Mitosis in living budding yeast: anaphase A but no metaphase plate. Science 277 : 574–578.
2. UhlmannF, LottspeichF, NasmythK (1999) Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400 : 37–42.
3. UhlmannF, WernicD, PoupartMA, KooninEV, NasmythK (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103 : 375–386.
4. Cohen-FixO, KoshlandD (1999) Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes Dev 13 : 1950–1959.
5. HoltLJ, KrutchinskyAN, MorganDO (2008) Positive feedback sharpens the anaphase switch. Nature 454 : 353–357.
6. MichaelisC, CioskR, NasmythK (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91 : 35–45.
7. LoogM, MorganDO (2005) Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434 : 104–108.
8. WoodburyEL, MorganDO (2007) Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol 9 : 106–112.
9. HuismanSM, SmeetsMF, SegalM (2007) Phosphorylation of Spc110p by Cdc28p-Clb5p kinase contributes to correct spindle morphogenesis in S. cerevisiae. J Cell Sci 120 : 435–446.
10. PereiraG, SchiebelE (2003) Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302 : 2120–2124.
11. GrayCH, GoodVM, TonksNK, BarfordD (2003) The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. Embo J 22 : 3524–3535.
12. ShouW, SeolJH, ShevchenkoA, BaskervilleC, MoazedD, et al. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97 : 233–244.
13. VisintinR, HwangES, AmonA (1999) Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398 : 818–823.
14. AzzamR, ChenSL, ShouW, MahAS, AlexandruG, et al. (2004) Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science 305 : 516–519.
15. QueraltE, LehaneC, NovakB, UhlmannF (2006) Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell 125 : 719–732.
16. WangY, NgTY (2006) Phosphatase 2A Negatively Regulates Mitotic Exit in Saccharomyces cerevisiae. Mol Biol Cell 17 : 80–89.
17. YellmanCM, BurkeDJ (2006) The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell 17 : 658–666.
18. ShirayamaM, TothA, GalovaM, NasmythK (1999) APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 402 : 203–207.
19. JinF, LiuH, LiangF, RizkallahR, HurtMM, et al. (2008) Temporal control of the dephosphorylation of Cdk substrates by mitotic exit pathways in budding yeast. Proc Natl Acad Sci U S A 105 : 16177–16182.
20. BouchouxC, UhlmannF (2011) A Quantitative Model for Ordered Cdk Substrate Dephosphorylation during Mitotic Exit. Cell 147 : 803–814.
21. RahalR, AmonA (2008) Mitotic CDKs control the metaphase-anaphase transition and trigger spindle elongation. Genes Dev 22 : 1534–1548.
22. WangY, ShiroganeT, LiuD, HarperJW, ElledgeSJ (2003) Exit from exit: resetting the cell cycle through Amn1 inhibition of G protein signaling. Cell 112 : 697–709.
23. CrossFR, SchroederL, KruseM, ChenKC (2005) Quantitative characterization of a mitotic cyclin threshold regulating exit from mitosis. Mol Biol Cell 16 : 2129–2138.
24. LiuH, WangY (2006) The function and regulation of budding yeast Swe1 in response to interrupted DNA synthesis. Mol Biol Cell 17 : 2746–2756.
25. KeatonMA, BardesES, MarquitzAR, FreelCD, ZylaTR, et al. (2007) Differential susceptibility of yeast S and M phase CDK complexes to inhibitory tyrosine phosphorylation. Curr Biol 17 : 1181–1189.
26. HuF, AparicioOM (2005) Swe1 regulation and transcriptional control restrict the activity of mitotic cyclins toward replication proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 102 : 8910–8915.
27. LiangF, JinF, LiuH, WangY (2009) The molecular function of the yeast polo-like kinase Cdc5 in Cdc14 release during early anaphase. Mol Biol Cell 20 : 3671–3679.
28. SuranaU, AmonA, DowzerC, McGrewJ, ByersB, et al. (1993) Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. Embo J 12 : 1969–1978.
29. StegmeierF, VisintinR, AmonA (2002) Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108 : 207–220.
30. LiuH, LiangF, JinF, WangY (2008) The coordination of centromere replication, spindle formation, and kinetochore-microtubule interaction in budding yeast. PLoS Genet 4: e1000262 doi:10.1371/journal.pgen.1000262.
31. Cohen-FixO, PetersJM, KirschnerMW, KoshlandD (1996) Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev 10 : 3081–3093.
32. SanchezY, BachantJ, WangH, HuF, LiuD, et al. (1999) Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286 : 1166–1171.
33. SeverinF, HymanAA, PiattiS (2001) Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J Cell Biol 155 : 711–718.
34. KhmelinskiiA, RoostaluJ, RoqueH, AntonyC, SchiebelE (2009) Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev Cell 17 : 244–256.
35. WangPJ, HuffakerTC (1997) Stu2p: A microtubule-binding protein that is an essential component of the yeast spindle pole body. J Cell Biol 139 : 1271–1280.
36. WolyniakMJ, Blake-HodekK, KoscoK, HwangE, YouL, et al. (2006) The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol Biol Cell 17 : 2789–2798.
37. BrouhardGJ, StearJH, NoetzelTL, Al-BassamJ, KinoshitaK, et al. (2008) XMAP215 is a processive microtubule polymerase. Cell 132 : 79–88.
38. SeverinF, HabermannB, HuffakerT, HymanT (2001) Stu2 promotes mitotic spindle elongation in anaphase. J Cell Biol 153 : 435–442.
39. RossKE, Cohen-FixO (2003) The role of Cdh1p in maintaining genomic stability in budding yeast. Genetics 165 : 489–503.
40. TsukaharaT, TannoY, WatanabeY (2010) Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature
41. JinF, RichmondD, WangY (2009) The multilayer regulation of the metaphase-to-anaphase transition. Cell Cycle 8 : 700–704.
42. CioskR, ZachariaeW, MichaelisC, ShevchenkoA, MannM, et al. (1998) An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93 : 1067–1076.
43. CliftD, BizzariF, MarstonAL (2009) Shugoshin prevents cohesin cleavage by PP2A(Cdc55)-dependent inhibition of separase. Genes Dev 23 : 766–780.
44. HsuWS, EricksonSL, TsaiHJ, AndrewsCA, VasAC, et al. (2011) S-phase cyclin-dependent kinases promote sister chromatid cohesion in budding yeast. Mol Cell Biol 31 : 2470–2483.
45. HiguchiT, UhlmannF (2005) Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature 433 : 171–176.
46. CrossFR, JacobsonMD (2000) Conservation and function of a potential substrate-binding domain in the yeast Clb5 B-type cyclin. Mol Cell Biol 20 : 4782–4790.
47. BrennanIM, PetersU, KapoorTM, StraightAF (2007) Polo-like kinase controls vertebrate spindle elongation and cytokinesis. PLoS ONE 2: e409 doi:10.1371/journal.pone.0000409.
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



