-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaExpression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
We discovered a novel interaction between phage P22 and its host Salmonella Typhimurium LT2 that is characterized by a phage mediated and targeted derepression of the host dgo operon. Upon further investigation, this interaction was found to be instigated by an ORFan gene (designated pid for phage P22 encoded instigator of dgo expression) located on a previously unannotated moron locus in the late region of the P22 genome, and encoding an 86 amino acid protein of 9.3 kDa. Surprisingly, the Pid/dgo interaction was not observed during strict lytic or lysogenic proliferation of P22, and expression of pid was instead found to arise in cells that upon infection stably maintained an unintegrated phage chromosome that segregated asymmetrically upon subsequent cell divisions. Interestingly, among the emerging siblings, the feature of pid expression remained tightly linked to the cell inheriting this phage carrier state and became quenched in the other. As such, this study is the first to reveal molecular and genetic markers authenticating pseudolysogenic development, thereby exposing a novel mechanism, timing, and populational distribution in the realm of phage–host interactions.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003269
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003269Summary
We discovered a novel interaction between phage P22 and its host Salmonella Typhimurium LT2 that is characterized by a phage mediated and targeted derepression of the host dgo operon. Upon further investigation, this interaction was found to be instigated by an ORFan gene (designated pid for phage P22 encoded instigator of dgo expression) located on a previously unannotated moron locus in the late region of the P22 genome, and encoding an 86 amino acid protein of 9.3 kDa. Surprisingly, the Pid/dgo interaction was not observed during strict lytic or lysogenic proliferation of P22, and expression of pid was instead found to arise in cells that upon infection stably maintained an unintegrated phage chromosome that segregated asymmetrically upon subsequent cell divisions. Interestingly, among the emerging siblings, the feature of pid expression remained tightly linked to the cell inheriting this phage carrier state and became quenched in the other. As such, this study is the first to reveal molecular and genetic markers authenticating pseudolysogenic development, thereby exposing a novel mechanism, timing, and populational distribution in the realm of phage–host interactions.
Introduction
Due to billions of years of co-evolution and their overpowering abundance in the biosphere, viruses of bacteria (i.e. bacteriophages or phages) have a profound impact on the conduct and ecology of their hosts [1], [2]. Lytic proliferation of phages for example can affect host mutation rates [3], structure microbial consortia [4], and contribute significantly to the global biogeochemical carbon flux [5]. Lysogenic proliferation as stable prophages, on the other hand, increases the genetic repertoire and genome plasticity of the host, thereby often extending its adaptive potential in terms of virulence and ecological fitness [6].
While the basic molecular events and genetic circuitry behind lytic and lysogenic development have traditionally received a lot of attention and are reasonably well understood for a number of model phages [7]–[9], the increasing wealth of novel phage genes with no known homologs and function nevertheless suggests an unforeseen intricacy in phage – host interactions [1], [10]. Furthermore, in many ecological niches phage – host associations often appear to defy the classical bifurcation into strict lytic or lysogenic development, as a large number of reports indicate a lysogeny-independent but stable co-existence between phages and their hosts. These phenomena are often vaguely referred to as pseudolysogeny, and hypothesize the existence of stable “phage carrier” cells in which the incoming phage has temporarily refrained from lytic or lysogenic development [11]. This suspended state is believed to play an important role in the long term survival strategy of viruses, as it might (i) prevent poor replication or even degradation of the phage chromosome in a host that is too starved to support further steps in lytic or lysogenic development, and/or (ii) provide a transient intracellular refuge for the phage chromosome in environments characterized by low host densities and short capsid half-lives [12], [13]. Despite its ecological importance [11], [14], however, no formal molecular evidence currently exists for the presence of such a state, let alone its possible impact on the physiology of the cell.
In this study, we extend on the intricacy of phage – host interactions and provide both genetic and direct cell biological evidence for the existence of a dedicated pseudolysogenic state in the Salmonella Typhimurium – phage P22 model system.
Results
MudK mutagenesis of Salmonella Typhimurium LT2 reveals a clone that responds to infection by phage P22
During routine screening of a MudK based lacZ promoter-trap library in Salmonella Typhimurium LT2 on LB X-Gal agar plates, our attention was drawn to a colony displaying an inhomogeneous distribution of LacZ activity (i.e. blue coloration; Figure 1A) that was neither symmetrical, nor sectorial. Moreover, after streaking out on new LB X-Gal plates, this particular clone segregated both into plain white colonies and colonies with an irregular blue coloration similar to that of the parent colony (Figure 1B). Interestingly, however, when the latter colonies were replica-plated on green indicator agar, the blue patches on LB X-Gal agar overlapped perfectly with the dark green sites of cell lysis that were revealed by the green indicator agar (compare Figure 1B and 1C). As we reasonably assumed this cell lysis to stem from infection by residual P22 HT105/1 int201 transducing phage that was initially used to deliver the MudK element during construction of the library, we hypothesized LacZ activity of the isolated clone to be triggered by exposure to phage P22.
Fig. 1. Initial observation of an LT2 clone (designated LT2K7) carrying a lacZ translational fusion that responds to P22 infection. 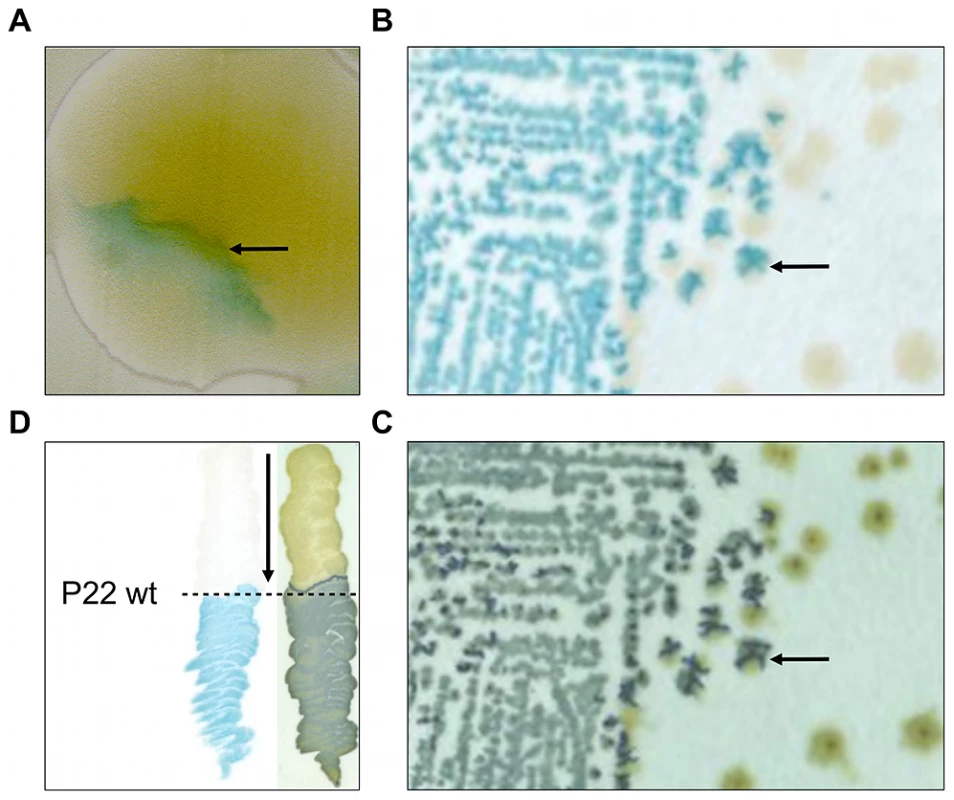
In clockwise orientation: (A) Photograph of a P22 HT105/1 int201 infected colony of LT2K7, showing inhomogeneous distribution of LacZ activity as visualized by the blue color that results from hydrolysis of X-Gal on LB X-Gal agar (cfr. arrow). (B) Typical streak out on LB X-Gal agar of a colony shown in panel A. (C) An exact replica on green indicator agar of the colonies shown in panel B. The arrow in B and C indicates the position of the same colony, indicating LacZ activity (shown in panel B) to overlap with phage infection (shown in panel C). (D) Streaking of LT2K7 (following the direction of the arrow) across a suspension of wild-type P22 (P22 wt; indicated by a dashed line) on LB X-Gal agar to reveal LacZ activity (left panel), and green indicator agar to reveal phage infection (right panel). In order to further examine this phenotype, the MudK insertion of the corresponding clone was transduced into a fresh LT2 strain and a phage-free transductant (designated LT2K7) was streaked across wild-type P22 (P22 wt) on LB X-Gal agar (Figure 1D). As a result, we found LT2K7 to turn from white to blue upon encountering P22, suggesting that phage P22 is causally involved in triggering lacZ expression in LT2K7.
MudK of LT2K7 maps to the dgoT gene
The MudK insertion site of LT2K7 was mapped to the dgoRKAT operon. DNA sequence analysis revealed that the MudK insertion resulted in a translational fusion of the lacZ reporter gene to dgoT (Figure 2A). The dgoR gene located at the beginning of the operon is predicted to function as an autorepressor [15] and indeed LT2K7 ΔdgoR constitutively expressed the dgoT::MudK fusion (Figure 2B), regardless of infection by P22 wt. Furthermore, increasing the level of DgoR by providing the corresponding gene on a multicopy plasmid (pFPV-dgoR) was able to abolish induction of dgoT::MudK by P22 wt, but had no obvious effect on phage infection per se (Figure 2C). These data suggest that infection by P22 interferes with autorepression of the dgo operon in LT2.
Fig. 2. The dgoRKAT operon and its derepression in LT2. 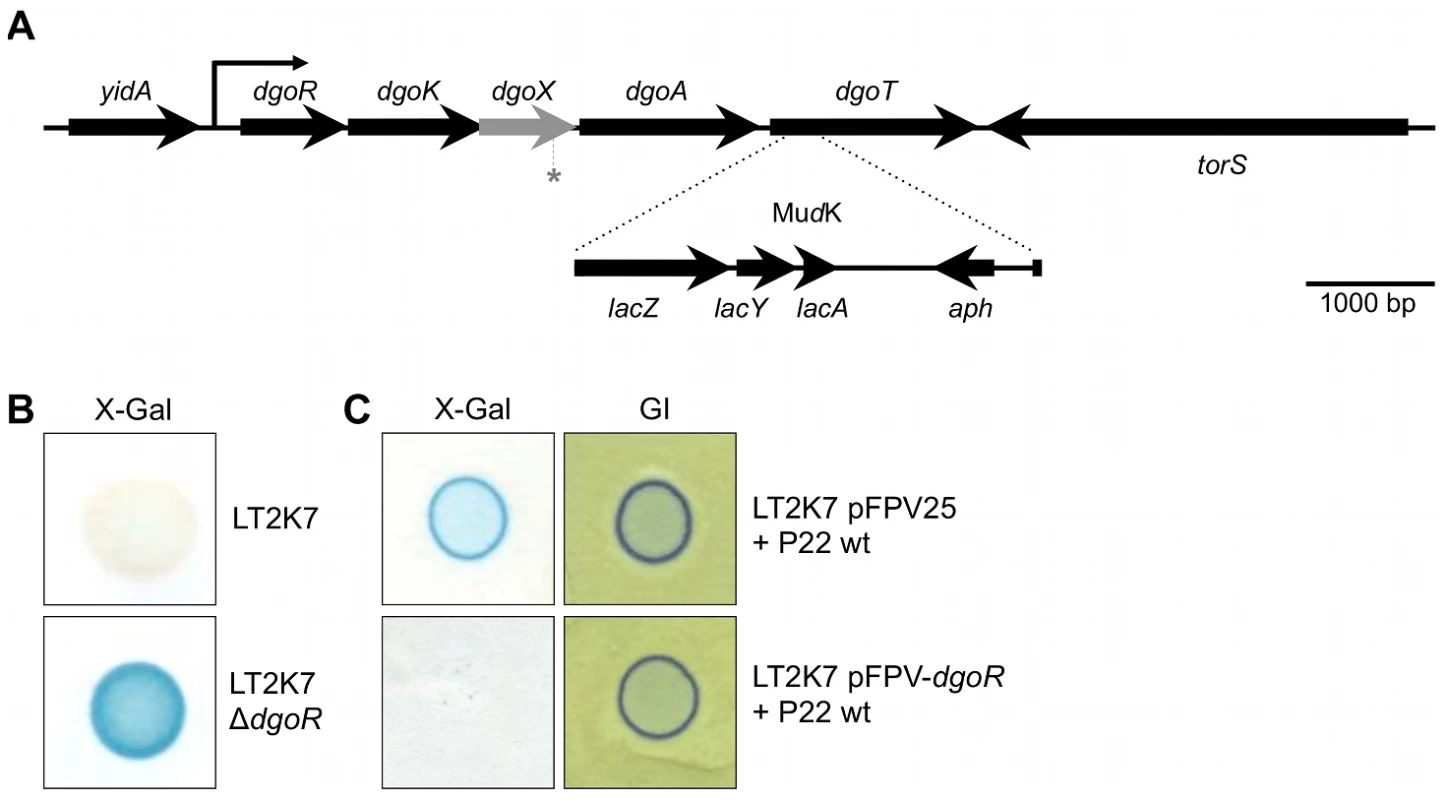
(A) Scheme showing the position of the MudK element (not drawn to scale) generating a translational lacZ reporter fusion to the dgoT gene in strain LT2K7 (i.e. dgoT::MudK). Please note that the grey arrow corresponds to an open reading frame compromised by a −1 frame shift at the position marked with an asterisk (*). (B) Deletion of the dgoR gene in LT2K7 yields constitutive expression of the dgoT::MudK fusion, as shown by the difference in LacZ activity (i.e. blue color) between a spot of LT2K7 and its ΔdgoR derivative grown on LB X-Gal agar. (C) Overexpression of dgoR interferes with activation of the dgoT::MudK fusion by P22 infection, as a plaque of P22 wt grown on a lawn of LT2K7 pFPV-dgoR fails to display LacZ activity (i.e. blue color) on LB X-Gal agar (left panel). A plaque of P22 wt grown on LT2K7 pFPV25 (i.e. empty vector) was included as a control. A similar experiment performed on green indicator agar (GI; right panel) confirms the actual infection of both strains by P22 wt. A P22 moron locus is responsible for specifically triggering dgo expression in LT2
Subsequently, we noticed that derepression of dgoT::MudK upon phage infection was a feature supported by P22, but not by another S. enterica specific temperate phage such as ES18 (Figure S1). This raised the possibility that induction of the dgo operon stemmed from a genetic circuit in P22, rather than from a generic host response to phage infection. To examine this, a plasmid library of random P22 genomic fragments was screened for loci able to render the LT2K7 indicator strain blue on LB X-Gal. As such, a 521 bp P22 fragment could eventually be obtained that triggered dgoT::MudK upon conditional expression (using the arabinose inducible PBAD promoter) in LT2K7. More specifically, this fragment was found to correspond to a small and unannotated locus situated between orf25 and orf80 in the late region of the P22 genome [16], [17] (Figure 3A, boxed region), and was subsequently designated as pid (for phage P22 encoded instigator of dgo expression).
Fig. 3. Genomic context of the P22 pid locus. 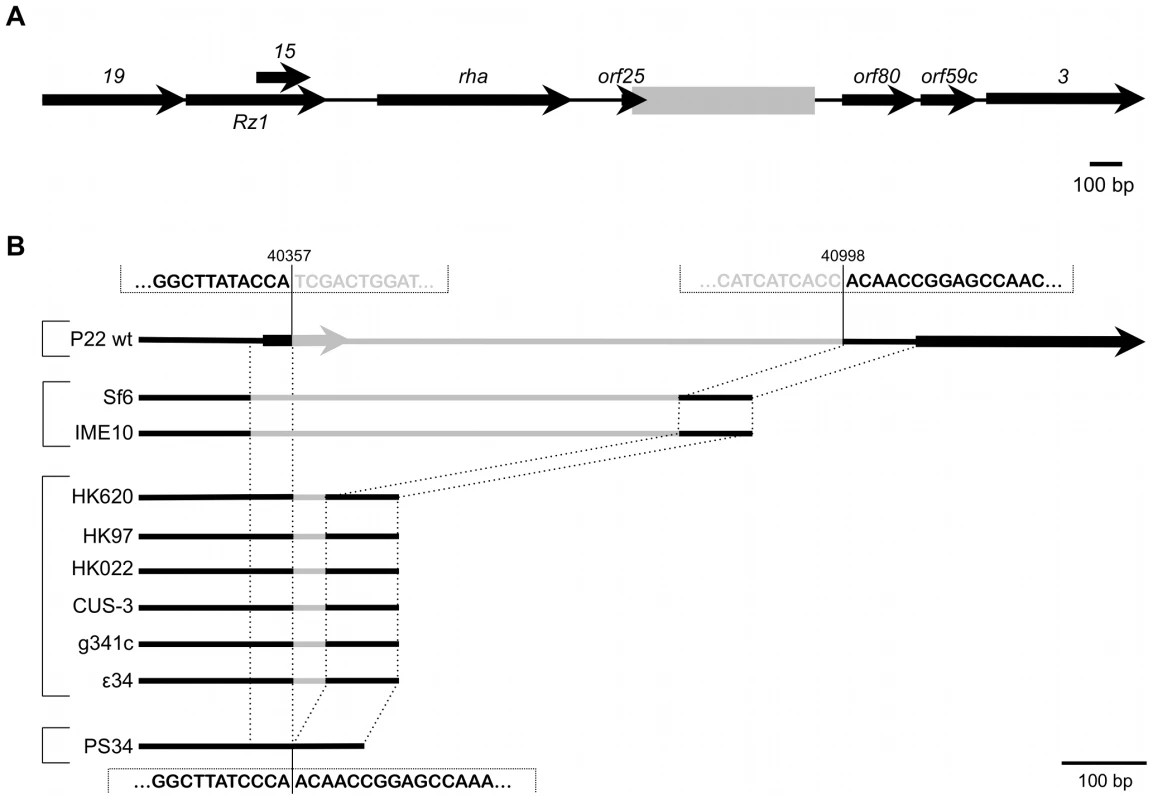
(A) Position of the region containing the pid locus (boxed in gray), situated between orf25 and orf80 in the late region of the P22 wt genome. Expression of pid proceeds in the direction opposite to that of its surrounding genes. (B) Comparison of this specific locus, situated between nucleotide 40357 and 40998 of the P22 genome (NCBI Reference Sequence: NC_002371.2), throughout related phages. Black lines indicate regions of homology while grey lines are only homologous within phages grouped by brackets. Interestingly, close inspection of the pid region revealed it to be a genuine moron locus [6], as it is integrated at a site where related phages have either no (cfr. PS34 in Figure 3B) or another insert (Figure 3B). In addition, the pid locus is further characterized (i) by the fact that it is divergently transcribed relative to its surrounding genes, indicating that its regulatory control might deviate from that of the late region, and (ii) by a 3′ Rho-independent transcriptional termination site.
The P22 pid locus encodes a small ORFan protein that triggers expression of the LT2 dgo operon
During our efforts to discriminate whether the pid locus encoded a small regulatory RNA or a small protein, we discovered the appearance of a distinct low molecular weight protein band on SDS-PAGE upon triggering transcription of the locus from a plasmid (pFPV-PBAD-pid) (Figure 4A). Moreover, sequencing of this protein indeed revealed peptide signatures encoded by one of the possible reading frames of the moron locus (Figure 4B). While the stop codon of this open reading frame could be inferred, the start codon was predicted by the presence of an upstream canonical Shine-Dalgarno sequence (AAGGAG) [18] (Figure 4C). Importantly, introduction of a −1 frame shift in the start codon (Figure 4C) simultaneously abolished both expression of the characteristic protein band and induction of dgoT::MudK in LT2K7 (Figure 4D), establishing this 86 amino acid and 9.23 kDa protein (termed Pid; Figure 4B) (and not a small RNA species putatively originating from the same locus) as the actual trigger of the interaction. It should be noted that subsequent deletion of the pid open reading frame in P22 correspondingly abolished induction of the dgo operon upon infection (Figure 4E), but had no noticeable impact on the ability to develop lytically or lysogenically.
Fig. 4. Identification of the P22 Pid protein. 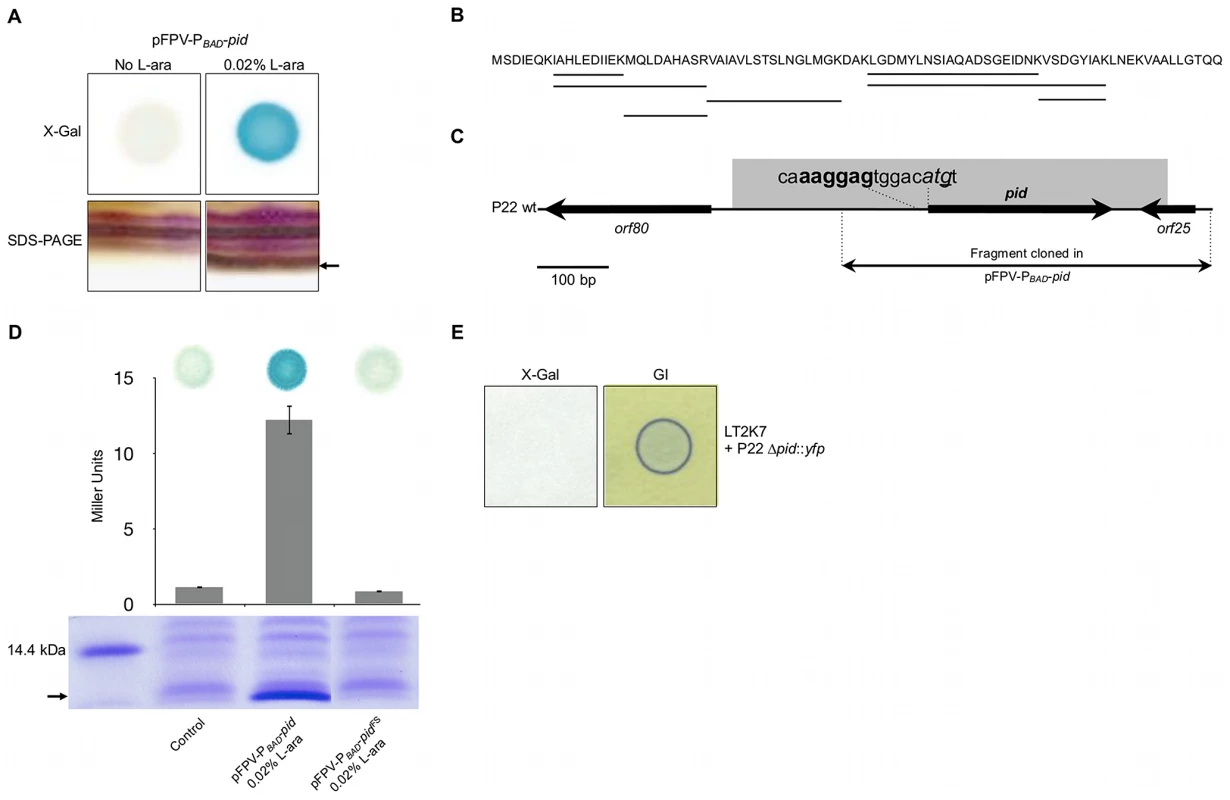
(A) Conditional expression with 0.02% L-arabinose (L-ara) of the pid locus from pFPV-PBAD-pid yields (i) LacZ activity (i.e. blue color) caused by induction of the dgoT::MudK fusion in a spot of LT2K7 pFPV-PBAD-pid grown on LB X-Gal agar (upper right panel) as opposed to a non-induced control (upper left panel), and (ii) the appearance of a small protein (its position is marked by the arrow) on a silver stained SDS-PAGE of total protein extract of LT2 pFPV-PBAD-pid (lower right panel) as opposed to a non-induced control (lower left panel). (B) Protein sequence of Pid, in which peptide signatures identified by mass-spectrometry are underlined. (C) Location of the pid gene on the P22 genome, with indication of its predicted start codon (shown in italic) and Shine-Dalgarno sequence (shown in bold). The nucleotide that was deleted to introduce a −1 frame-shift (yielding pFPV-PBAD-pidFS) is underlined. (D) Plasmids pFPV-PBAD-pid and pFPV-PBAD-pidFS were compared with respect to their ability to yield (i) Pid protein in LT2, as determined by SDS-PAGE of total protein extracts (lower panel; position of Pid is marked by the arrow), and (ii) LacZ activity caused by induction of the dgoT::MudK fusion in LT2K7, as determined qualitatively by blue coloration on LB X-Gal agar (upper panel) and quantitatively by Miller Units (middel panel; means ± standard error of three independent experiments are shown). For comparison, an uninduced pFPV-PBAD-pid plasmid was used as a control. (E) Deletion of the pid gene in P22 (i.e. P22 Δpid::yfp) abolishes induction of the dgoT::MudK fusion upon infection of LT2K7, as indicated by absence of LacZ activity (i.e. blue color) in the corresponding plaque on LB X-Gal agar (left panel), but does not interfere with phage infection, as indicated by green indicator agar (GI; right panel). The Pid/dgo interaction is not supported during strict lytic or lysogenic propagation of P22
Since upon infection the propagation of P22 wt can either proceed lytically or lysogenically, we wondered which of these two distinct developmental routes would actually mount the Pid/dgo interaction (Figure 5A) in the cell. Surprisingly, however, dgoT::MudK expression was completely absent both when LT2K7 was subjected to obligate lytic infection with P22 c2 (Figure 5C) or when the reporter strain carried P22 wt as a prophage (Figure 5D). The latter finding is in fact consistent with our initial observation of the Pid/dgo interaction being fully supported by the P22 HT105/1 int-201 transducing phage (Figure 1A, 1B and Figure 5B) despite its inability to integrate in the host chromosome as a prophage.
Fig. 5. The Pid/dgo interaction is not supported during strict lytic or lysogenic development of P22. 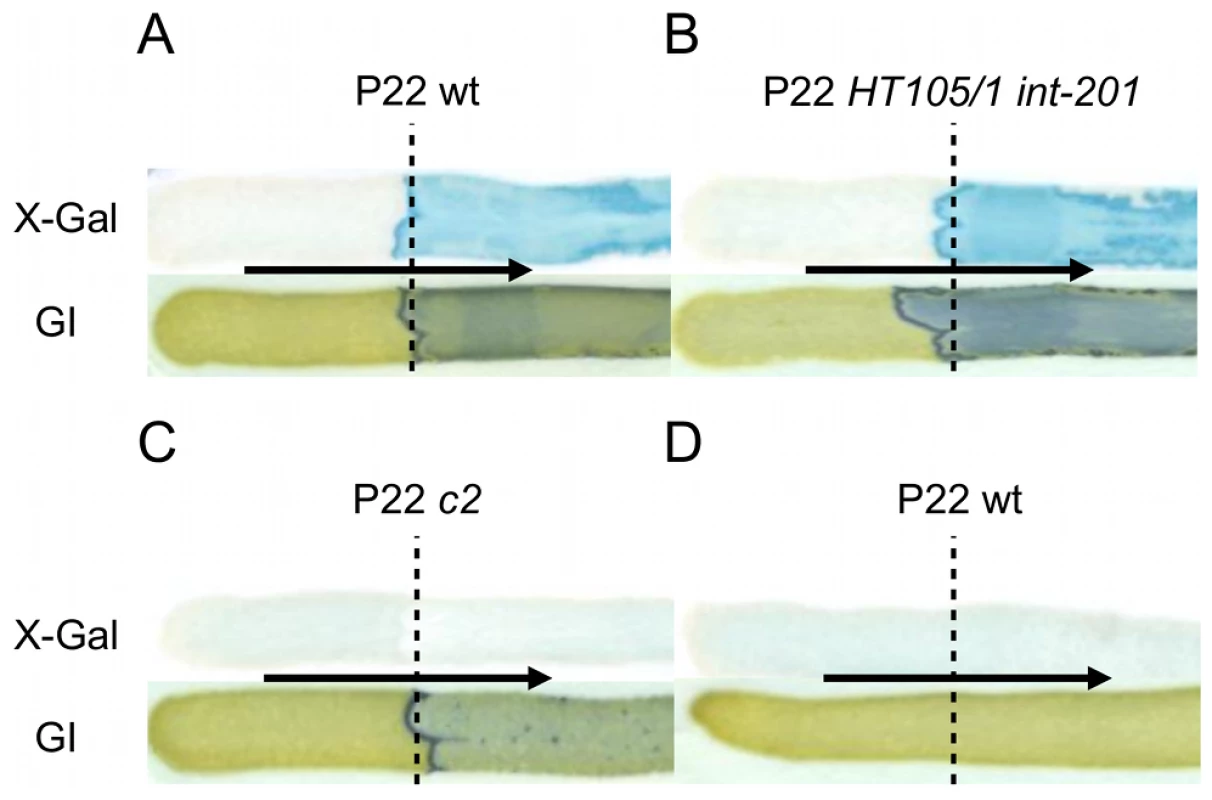
LT2K7 (A, B, C) and a P22 wt lysogen of LT2K7 (D) was streaked (in the direction of the arrow) across a suspension (indicated by a dashed line) of P22 wt (A, D), P22 HT105/1 int-201 (B) and P22 c2 (C) on either LB X-Gal agar (upper panels) in order to visualize LacZ activity caused by induction of the dgoT::MudK fusion, or green indicator agar (lower panels) in order to visualize phage infection. To further corroborate this finding, we extended the P22 pid open reading frame with a strep-tag encoding sequence (leading to P22 pid-strep) to facilitate Pid detection by western blot, and checked whether the observed absence of dgoT::MudK expression also correlated with attenuated levels of Pid. In agreement with the results above (Figure 5), Pid production was abundant in LT2 infected with P22 pid-strep (Figure 6A), while it was severely attenuated in LT2 infected with the obligate lytic P22 c2 pid-strep derivative (Figure 6B) and completely absent in LT2 carrying P22 pid-strep as a prophage (Figure 6C).
Fig. 6. Expression of Pid protein during P22 infection. 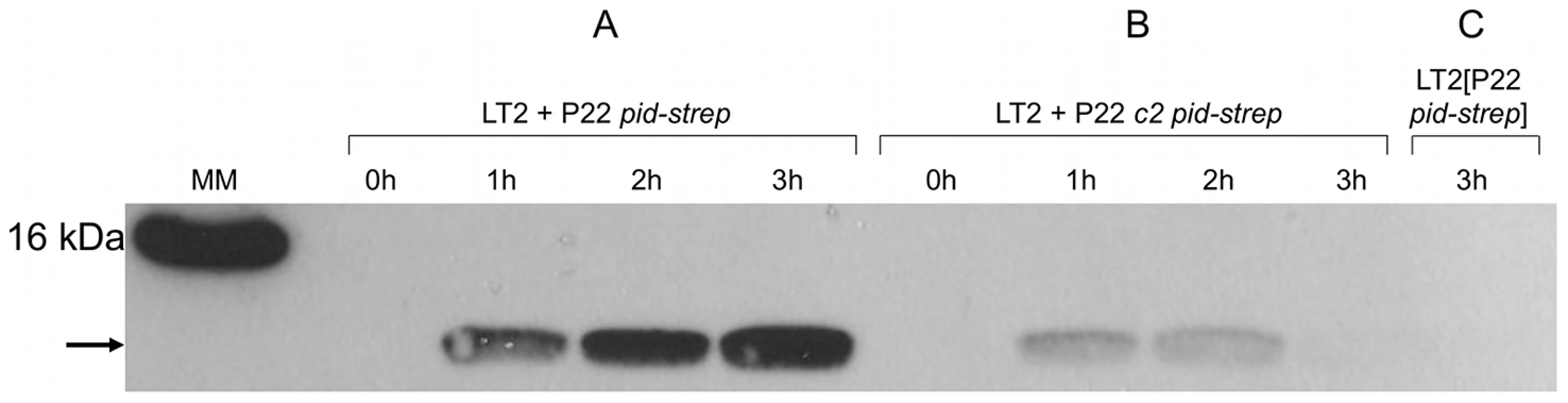
Western blot analysis of strep-tagged Pid produced during the indicated time after infecting exponential phase cultures of LT2 at MOI = 0.1 with (A) P22 pid-strep or (B) P22 c2 pid-strep, or (C) in early exponential phase cultures of LT2 carrying P22 pid-strep as a prophage. The position of Pid is indicated by a black arrow, while the molecular marker (MM) shows the position of the 16 kDa reference. To determine whether or not compromised Pid production stemmed from attenuated pid transcription, the pid open reading frame of P22 was replaced with the yfp fluorescent reporter gene, and the resulting phage (i.e. P22 Δpid::yfp, carrying yfp under the control of the native pid promoter) was used to interact with LT2. In agreement with our previous findings (Figure 5 and Figure 6), cells infected with an obligate lytic derivative of P22 Δpid::yfp (i.e. P22 c2 Δpid::yfp) only displayed very faint fluorescence in the few minutes before cell lysis (Figure 7C), while cells carrying P22 Δpid::yfp as a prophage displayed no detectable fluorescence (Figure 7D). On the contrary, cells infected with P22 Δpid::yfp (Figure 7A) or its int derivative (i.e. P22 Δint Δpid::yfp) (Figure 7B) clearly showed a plethora of cells exhibiting YFP expression to different extents.
Fig. 7. Evidence for pid transcription during P22 infection at the single cell level. 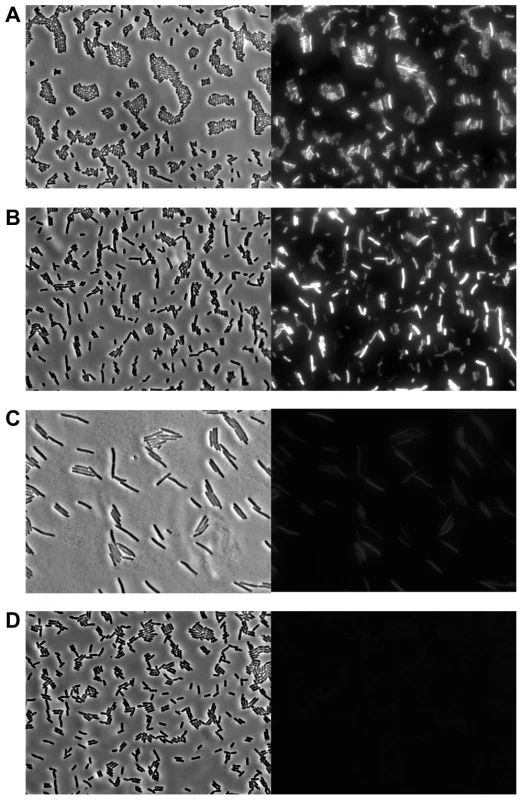
Phase contrast (left panels) and corresponding YFP epifluorescence (right panels) micrographs of exponential phase cultures of LT2 either (A) 3 h after infection with P22 Δpid::yfp, (B) 3 h after infection with P22 Δint Δpid::yfp, (C) 2 h after infection with P22 c2 Δpid::yfp (just before general cell lysis occurred), or (D) carrying P22 Δpid::yfp as a prophage. Infections with P22 (A, B and C) were performed at MOI = 0.1. Interestingly, the finding that expression of pid and subsequent derepression of the dgo operon are not supported during lytic or lysogenic propagation of P22 strongly suggests that the Pid/dgo interaction might be dedicated to a different state of P22 development.
pid expression is tightly linked with cells in the phage carrier state
Spurred by the above observations, time-lapse fluorescence microscopy was used to more closely examine the timing and dynamics of pid expression during infection of LT2 with P22 Δpid::yfp at single cell resolution. While this approach demonstrated that the pid locus indeed became expressed in lineages emerging from non-lytic infection with the reporter phage, it also revealed that this expression was a feature that subsequently segregated asymmetrically between siblings (Figure 8). Surprisingly, in fact, only one individual within the growing lineage consistently displayed the ability to express pid, thereby revealing an unprecedented timing and populational distribution of this phage – host interaction. It should also be noted that disruption of the int gene in P22 Δpid::yfp did not affect the timing nor the asymmetric distribution of pid expression (Figure S2), corroborating that the actual chromosomal integration event leading to the establishment of a prophage was not required for this phenomenon.
Fig. 8. Expression of P22 pid segregates asymmetrically between siblings. 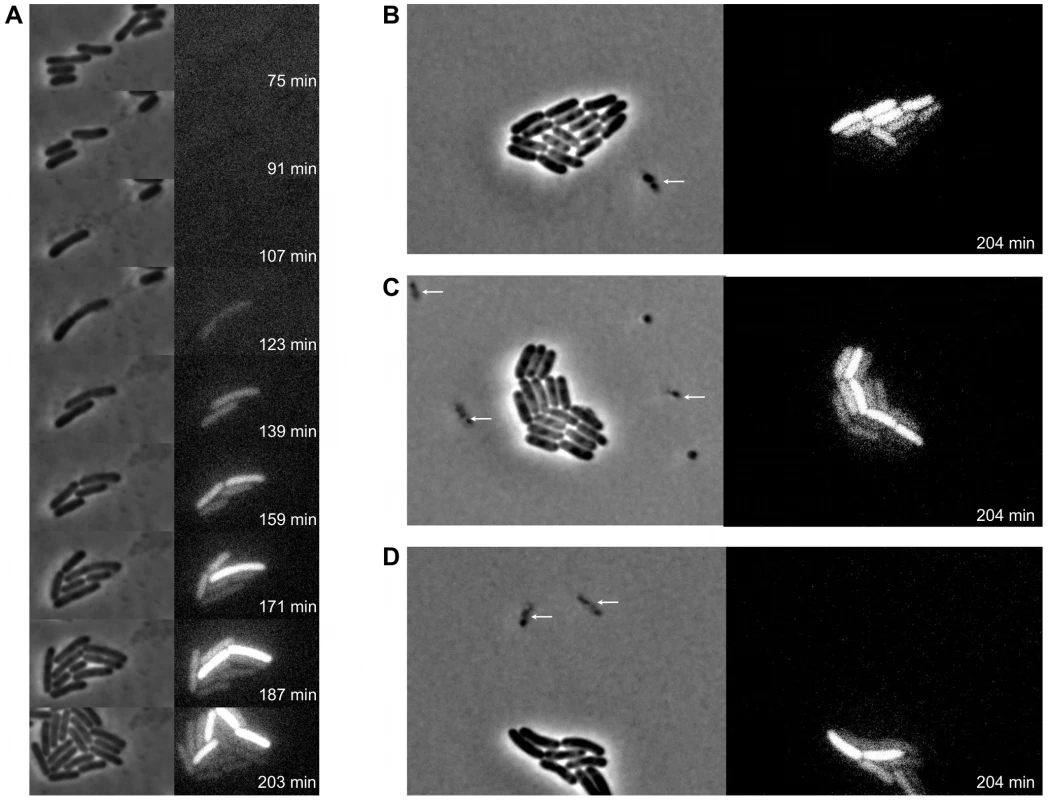
Exponential phase cultures of LT2 were infected with P22 Δpid::yfp (MOI = 0.1) and chased after 30 minutes with the virulent P22 H5 mutant (MOI = 20) to lyse cells not destined for non-lytic development of P22 Δpid::yfp. (A) Time-lapse fluorescence microscopy image sequence of an LT2 cell destined for non-lytic development of P22 Δpid::yfp. (B–D) Images from clonal microcolonies similar to the one emerging from a single cell in panel A, and also displaying asymmetrical segregation of yfp expression. Please note that the surrounding cells (indicated by arrows) are lysing due to lytic infection with either P22 Δpid::yfp or P22 H5. Phase contrast (left panels) and corresponding YFP epifluorescence (right panels) images are shown, and the time after infection with P22 Δpid::yfp is indicated on the frames. In order to more closely examine the possible role of the P22 chromosome in this peculiar asymmetric segregation phenotype, P22 Δpid::yfp was equipped with a parS site (resulting in P22 Δpid::yfp parS), allowing its whereabouts during infection to become fluorescently tractable in an LT2 strain expressing the ParB protein fused to mCherry (i.e. LT2 pCW-mCherry-parB). Interestingly, soon after infection of LT2 pCW-mCherry-parB with P22 Δpid::yfp parS, a single and coherent mCherry cloud appeared in cells destined for non-lytic infection (Figure 9A and 9B), indicative for the presence of one (or possibly more) P22 chromosome(s). Furthermore, upon subsequent cell divisions, this cloud became asymmetrically segregated between siblings, with pid expression remaining tightly linked to the cell inheriting and carrying the unintegrated P22 chromosome(s) (Figure 9). The gradual dilution of YFP molecules in siblings not inheriting this phage carrier state is consistent with the heterogeneity in YFP fluorescence in liquid cultures of LT2 infected with P22 Δpid::yfp observed earlier (cfr. Figure 7).
Fig. 9. Pid expression is linked to phage carrier cells. 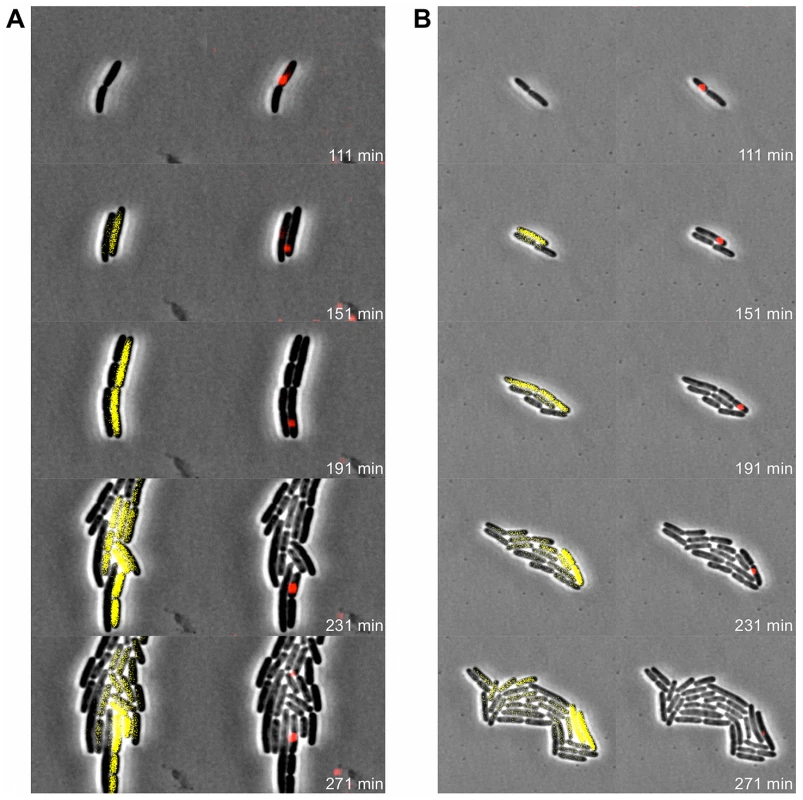
Exponential phase cultures of LT2 pCW-mCherry-parB were infected with P22 Δpid::yfp parS (MOI = 0.1) and chased after 30 minutes with the virulent P22 H5 mutant (MOI = 20) to lyse cells not destined for non-lytic development of P22 Δpid::yfp parS. Two independent time-lapse fluorescence microscopy image sequences (A and B) of an LT2 pCW-mCherry-parB cell destined for non-lytic development of P22 Δpid::yfp parS are shown. The corresponding phase contrast images are either superimposed with YFP epifluorescence images visualizing pid expression (left panels) or mCherry epifluorescence images visualizing the presence of the P22 chromosome(s) (right panels), and the time after infection is indicated on the frames. Please note that only the carrier cell produces new YFP, while non-carrier segregants passively acquire preformed YFP by cytoplasmic diffusion, and further dilute it upon subsequent cell divisions. Discussion
Given the penetration and importance of bacteriophages in global ecology, understanding their possible associations with a host is of tremendous importance. In this report, the S. Typhimurium – phage P22 model system yielded both molecular and genetic evidence authenticating the existence of a dedicated phage carrier state in which an unintegrated phage chromosome is stably maintained in the cell and asymmetrically inherited by only one of the siblings upon further divisions. This behavior differs fundamentally from cells undergoing lytic or lysogenic phage development, which are forced either to lyse after the production of new virions or to symmetrically segregate the prophage chromosome (integrated in the host chromosome or existing as a stable episome) among siblings [19], [20], respectively.
The phage carrier (or pseudolysogenic) state is believed to have a tremendous impact on phage ecology, as the ability to postpone the commitment to lytic or lysogenic development might improve phage survival in inhospitable environments [11]–[14]. Specifically with regard to the biology of phage P22, our findings at the single cell level are in remarkable agreement with very early observations made by Zinder, who anticipated that upon infection P22 could be maintained in a pseudolysogenic form during several generations before integrating itself as a prophage [21]. Despite the long-standing assumption of its alleged existence and its ecological importance, however, the phage carrier state has so far hardly been documented from a molecular or genetic point of view. In fact, although it has been proposed that the phage remains idle or inert while being in this state [12], [13], our results on the contrary provide the first evidence that a dedicated phage – host interaction (as exemplified by Pid/dgo) can be mounted in phage carrier cells. Clearly, the existence of dedicated genetic programs that are executed solely in phage carrier cells substantiates their biological significance and allows them to differentiate from uninfected cells or cells destined for lytic or lysogenic development.
On itself, the induction of the LT2 dgo operon by the P22 Pid ORFan protein is also peculiar, since only a very limited number of phage – host interactions have so far been discovered in which the phage deliberately and specifically interferes with host gene expression. Indeed, in currently recognized interactions, phage encoded functions either (i) hijack cellular machinery and generally shut down host gene expression to support phage reproduction during lytic proliferation [22], or (ii) contribute virulence factors that support the pathogenicity of the host during lysogenic development [6], [23]. A notable exception was only recently described for λ lysogens of E. coli, in which the λ CI repressor was shown to compromise cellular gluconeogenesis by physically obstructing the host pckA promoter [24].
Interestingly, the dgo operon encodes proteins involved in the uptake and metabolism of D-galactonate, which is considered to be an important source of carbon and energy during intracellular survival and proliferation of Salmonella spp. [25]. Moreover, a dgoT knock-out was correspondingly found to attenuate the virulence of S. enterica serovar Choleraesuis in pigs [26]. It remains to be established, however, how exactly the Pid/dgo interaction is mounted within the carrier state, and whether it would endow carrier cells with increased virulence or rather constitutes a way for the phage to decide on how long to maintain this state.
In summary, our results authenticate the existence of the phage carrier state as a distinct developmental route in phage biology that differs from strict lytic or lysogenic propagation. The phenotypic consequences of the interactions taking place in phage carrier cells are likely to provide the missing link in the proper and accurate interpretation of phage – host dynamics occurring throughout microbial ecosystems.
Materials and Methods
Strains and growth conditions
Bacterial strains, phages and plasmids used throughout this study are listed in Table 1. For culturing bacteria, Lysogeny Broth (LB; [27]) medium was used either as a broth or as agar plates after the addition of 15% (for spreading plates) or 7% (for soft-agar plates) agar. Cultures were grown in LB broth for 15–20 h at 37°C under well-aerated conditions (200 rpm on a rotary shaker) to reach stationary phase. Exponential phase cultures were in turn prepared by diluting stationary phase cultures 1/100 or 1/1000 in fresh pre-warmed broth, and allowing further incubation at 37°C. When appropriate, the following chemicals (Applichem, Darmstadt, Germany) were added to the growth medium at the indicated final concentrations: ampicillin (100 µg/ml; Ap100), chloramphenicol (30 µg/ml; Cm30), kanamycin (50 µg/ml; Km50), tetracycline (20 µg/ml; Tc20), glucose (0.02%), L-arabinose (0.02%), and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal; 40 µg/ml).
Tab. 1. Strains, phages, and plasmids used in this study. 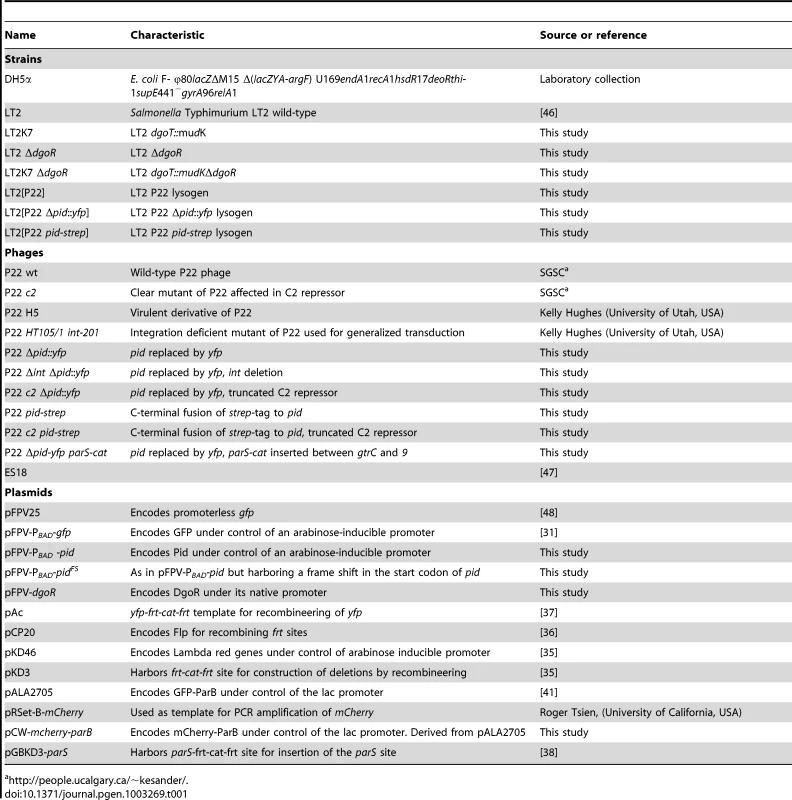
http://people.ucalgary.ca/~kesander/. Phages were propagated on S. Typhimurium LT2 as plaques in LB soft-agar or as lysates in LB broth as described previously [28]. Phage stocks were filter sterilized with 0.2 µm filters (Fisher Scientific, Aalst, Belgium) and chloroform was added to maintain sterility. Generalized transduction was performed with phage P22 HT105/1 int-201 as described previously [28], [29]. This mutant is unable to integrate into the host chromosome as a prophage due to the lack of integrase (Int) activity. To discriminate phage infected from uninfected colonies, plates containing green indicator (GI; [28]) agar were used to indicate cell lysis. The latter medium contains glucose as a carbon source, and a pH indicator dye that turns dark green at sites where phage infection causes cell lysis and the concomitant release of organic acids.
Please note that for clear visualization of spotted or (cross-)streaked bacterial and/or phage populations, agar plates were printed to Whatman filter papers (GE Healthcare, Diegem, Belgium) before photographing.
β-galactosidase assay
Expression of β-galactosidase (LacZ) was inferred from the hydrolysis of either 5-bromo-4-chloro-3-indolyl-β-galactopyranoside (X-Gal) or o-nitrophenyl-β-D-galactoside (ONPG). X-Gal was typically added to agar plates (40 µg/ml), where its hydrolysis by β-galactosidase yielded an insoluble blue precipitate. For quantitative measurements of lacZ expression, Miller units were determined as described previously [30] using the CHCl3-sodium dodecyl sulfate permeabilization procedure.
Construction and screening of a P22 shotgun library
Particles of phage P22 were purified by passing a lysate through a 0.45 µm pore-size filter, after which particles were concentrated by centrifugation (4,000×g, 20 min) in the presence of polyethylene glycol (PEG) 8,000 (8%, w/v) and 1 M NaCl. Subsequently, further purification was attained by ultracentrifugation (140,000×g, 3 hours) using a layered CsCl step gradient of 1.33, 1.45, 1.50 and 1.70 g/ml. This resulted in a distinct blue band containing the concentrated P22 particles. This band was subsequently collected and dialysed against phage buffer (10 mM Tris-HCl pH 7, 10 mM MgSO4, 150 mM NaCl) three times using a Slide-A-Lyzer dialysis cassette (Pierce, Rockford, IL, USA). For DNA extraction, the purified and dialysed phage particles were incubated at 56°C for 1 h in the presence of 0.5% SDS (w/v), 20 mM EDTA and 2 µg/ml proteinase K. Subsequently, DNA was extracted and purified from this mixture by phenol/chloroform [27] and precipitated with Na-acetate/ethanol. Finally, the sample was treated with RNase A (0.1 mg/ml) (Fermentas, St. Leon-Rot, Germany) for 1 h at room temperature to remove any residual RNA. Next, the resulting purified P22 genomic DNA was partially digested with the blunt 4 bp-cutter BsuRI restriction enzyme (Fermentas) and separated by agarose gel electophoresis (1% agarose), after which fragments between 1–2 kb were isolated from the gel using the GeneJET Gel Extraction Kit (Fermentas).
Parallel to this, pFPV-PBAD–gfp (pAA100; [31]) was digested with XbaI and HindIII (Fermentas) to remove gfp, and treated with calf intestinal alkaline phosphatase (Fermentas) to prevent self-ligation. The genomic P22 DNA fragments and the cut pFPV-PBAD vector were subsequently ligated after blunting with T4 ligase and Klenow polymerase (Fermentas), and transformed by electroporation into LT2K7. After plating on LB Ap100, this random P22 shotgun library was replica-plated on LB Ap100 X-Gal with and without 0.02% arabinose to screen for plasmids able to trigger LacZ expression in LT2K7.
Whole-genome alignment
Phages containing homologous regions to the region surrounding pid were selected with nucleotide Blast [32]. Whole genome alignment was performed manually and was based on the Blast-search results. The resulting conclusions were later confirmed by a progressive Mauve alignment [33] on the full genomes using default settings.
Construction of bacterial and phage mutants
Strain LT2K7 stems from a random MudK library, generated as described previously [34], and harbors a translational lacZ fusion to the LT2 dgoT gene (i.e. dgoT::MudK). In strain LT2 ΔdgoR, the dgoR gene was deleted via recombineering [35], using an amplicon (Phusion DNA polymerase; Fermentas) prepared on pKD3 [35] with the primers dgoR_pkd3_Fw and dgoR_pkd3_Rev (Table 2). The cat cassette replacing dgoR was flipped out using pCP20-borne Flp to recombine the two frt-sites [36], resulting in a small frt-scar followed by a new ribosome binding site [35]. Strain LT2K7 ΔdgoR was subsequently constructed by transducing dgoT::MudK to LT2 ΔdgoR.
Tab. 2. Primers used throughout the study. 
When relevant, primer attachment sites are indicated in bold. Relevant restriction enzyme sites are shown in italic. Recombination regions in regular font and the strep-tag and two stop-codons introduced in c2 are underlined. For the construction of P22 Δpid::yfp, the yfp-frt-cat-frt cassette was PCR amplified (Phusion DNA polumerase; Fermentas) from plasmid pAc [37] with primers pid_YFP_Cm_Fw and pid_YFP_Cm_Rev (Table 2), and used to replace the pid gene in LT2 lysogenized with wild-type P22 via recombineering [35]. Subsequently, the cat cassette was flipped out using pCP20-borne Flp to recombine the two frt-sites [36], and the resulting P22 Δpid::yfp phage was isolated and purified from the corresponding lysogen.
For the construction of P22 Δint Δpid::yfp, the integrase gene (int) in LT2 lysogenized with P22 Δpid::yfp was deleted by recombineering, using a PCR amplicon prepared on pKD3 with primers P22_Int_Fw and P22_Int_Rev (Table 2) [35]. Please note that the frt-flanked cat cassette was not removed by site specific Flp recombination, since this would interfere with the frt-scar already present in the pid locus. The resulting P22 Δint Δpid::yfp phage could be released by amplifying rare excision events through growth on wild-type LT2 in order to allow detection and purification of plaques. Please note that these phages produced normal turbid plaques and were unable to from true lysogens on LT2.
For the construction of P22 Δpid::yfp parS, the parS-frt-cat-frt cassette was PCR amplified from pGBKD3-parS [38] with primers P22_parS_Fw and P22_parS_Rev (Table 2), and inserted between the gtrC and 9 genes in LT2 lysogenized with P22 Δpid::yfp via recombineering [35]. Please note that the frt-flanked cat cassette was not removed by site specific Flp recombination, since this would interfere with the frt-scar already present in the pid locus.
For the construction of P22 pid-strep, a strep-tag encoding sequence (strep) was added to the 3′ end of the pid open reading frame by recombineering a strep-frt-cat-frt amplicon prepared on pKD3 [35] with primers pid_Strep_Fw and pid_Strep_Rev (Table 2) in LT2 lysogenized with wild-type P22. Subsequently, the cat cassette was flipped out using pCP20-borne Flp to recombine the two frt-sites [36], and the resulting P22 pid-strep phage was isolated and purified from the corresponding lysogen. Please note that the C-terminal addition of the strep-tag to Pid was shown to have no effect on the ability of Pid to trigger the dgo-operon.
Finally, clear mutants P22 c2 Δpid::yfp and P22 c2 pid-strep were constructed by oligo-mediated mutagenesis [39] of the corresponding P22 Δpid::yfp and P22 pid-strep lysogens in LT2, using olignucleotide Oligo_C2_Stop (Table 2). This oligo introduced two flanking stop-codons after the first 11 amino acids of the P22 C2 repressor. After recombination, transformants were inoculated in LB with wild type LT2 and grown overnight at 37°C to amplify the corresponding clear mutants. Afterwards, P22 c2 Δpid::yfp and P22 c2 pid-strep were isolated by plaquing on LT2, and the c2 mutation was verified by sequencing.
Construction of plasmids
Plasmid pFPV-PBAD-pid was constructed by digesting pFPV-PBAD–gfp with XbaI and HindIII (Fermentas), and subsequently replacing gfp with pid. The latter amplicon (Phusion DNA polymerase; Fermentas) was obtained using primers pid_Fw and pid_Rev (Table 2), digested with XbaI and HindIII prior to ligation. Plasmid pFPV-dgoR expresses the LT2 dgoR gene under the control of its own promoter, and was constructed by ligating an XbaI digested PCR amplicon of the LT2 dgoR locus, obtained with primers dgoR_Fw and dgoR_Rev, into the XbaI site of pFPV25 [40]. Plasmid pCW-mCherry-parB was constructed by first making an amplicon of the pALA2705 vector [41] with primers pALA_Out_Left and pALA_Out_Right. These primers amplify the entire plasmid except its gfp gene, and added an EcoRI and a SacI restriction site at the end of the amplicon. Subsequently, the mCherry gene was amplified from pRSet-B-mCherry (kind gift from Roger Tsien, University of California, USA) with primers Mcherry_Fw and Mcherry_Rev (Table 2), and both amplicons were digested with EcoRI and SacI prior to being ligated to each other. The resulting plasmid, pCW-mCherry-parB, expresses an N-terminal fusion of mCherry to ParB under control of an IPTG inducible promoter. Please note, however, that leaky expression of the latter promoter in the absence of IPTG was already sufficient, as mentioned previously [42].
Site-directed mutagenesis
For site-directed mutagenesis, the “Phusion Site-Directed Mutagenesis Kit” protocol (Thermo Scientific, Epsom, United Kingdom) was followed. As such, plasmid pFPV-PBAD-pid was used as a template for amplification with primers pid_FS_Fw and pid_FS_Rev (Table 2) for constructing a frame shift mutation in the actual pid start codon. After phosphorylating the 5′ ends of the primers according to the manufacturer's instruction, the primer pair was used to PCR amplify pFPV-PBAD-pid (Phusion polymerase; Fermentas). The resulting linear fragment was purified from an agarose gel using the GeneJET Gel Extraction Kit (Fermentas), subsequently self ligated, and finally transformed by electroporation to E. coli DH5α. The resulting pFPV-PBAD-pidFS plasmid from transformants selected on LB Ap100 was further confirmed by sequencing, prior to transformation to LT2 and LT2K7.
Protein identification and Western blotting
Samples were lysed in standard lysis buffer containing 50 µl/ml Bugbuster (Novagen, Darmstadt, Germany). Total protein concentration was assessed by the BCA protein assay kit (Novagen) and SDS-PAGE was performed as described previously by Sambrook and Russel [27]). Finally, gels were stained with coomassie [27] and when necessary, silver staining was employed as previously described [43].
For protein identification, the corresponding protein band was excised and trypsin-digested according to the method described earlier [44]. Subsequently, the digested peptides were identified by LC–ESI MS/MS (Thermo Electron, San Jose, CA) and further analyzed using Mascot (Matrix Sciences, London, UK) against the NCBI database (http://www.ncbi.nlm.nih.gov/).
For western-blotting, equal amounts of proteins were separated with PAGE and transferred to a nitrocellulose membrane (Hybond-C Extra; GE Healthcare) by semi-dry electroblotting for 1 hour at 0.15 A using a Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad Laboratories) and transfer buffer (50 mM Tris; 40 mM glycine; 0.075% SDS; 20% Methanol). Strep-tagged Pid was subsequently detected by StrepMAB-Classic, an anti-strep monoclonal antibody conjugated with Horse radish peroxidase (IBA, Göttingen, Germany). Horse radish peroxidase activity was assessed with Pierce ECL Western Blotting Substrate (Thermo Scientific), and detected on photo-sensitive film (Hyperfilm ECL; GE Healthcare). The strep-tagged protein ladder (IBA) was used as a molecular ruler and positive control of the blotting process.
Fluorescence microscopy
Fluorescence microscopy and time-lapse fluorescence microscopy were performed with a temperature controlled (Okolab Ottaviano, Italy) Ti-Eclipse inverted microscope (Nikon, Champigny-sur-Marne, France) equiped with a TI-CT-E motorised condensor, a YFP filter (Ex 500/24, DM 520, Em 542/27), an mCherry filter (Ex 562/40, Dm 593, Em 641/75), and a CoolSnap HQ2 FireWire CCD-camera. For imaging, cells were placed between LB agar pads and a cover glass, essentially as described previously [45], and incubated at 37°C. Please note that for experiments involving LT2 pCW-mCherry-parB, cells were grown on agar pads of AB-minimal media supplemented with 0.02% D-glucose, 100 µg/ml Uracil and 100 µg/ml Thiamine, Ap100, and incubated at 30°C, as described previously [42]. Images were acquired using NIS-Elements (Nikon) and resulting pictures were further handled with open source software ImageJ (Downloaded from http://rsbweb.nih.gov/ij/).
Supporting Information
Zdroje
1. RohwerF (2003) Global phage diversity. Cell 113 : 141.
2. BreitbartM, RohwerF (2005) Here a virus, there a virus, everywhere the same virus? Trends Microbiol 13 : 278–284.
3. PalC, MaciáMD, OliverA, SchacharI, BucklingA (2007) Coevolution with viruses drives the evolution of bacterial mutation rates. Nature 450 : 1079–1081.
4. BreitbartM (2012) Marine Viruses: Truth or Dare. Annu Rev Marine Sci 4 : 425–448.
5. SuttleCA (2007) Marine viruses — major players in the global ecosystem. Nat Rev Microbiol 5 : 801–812.
6. BrüssowH, CanchayaC, HardtW-D (2004) Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68 : 560–602.
7. Ptashne M (2004) A genetic switch. Gann A, Inglis J, Dickerson M, Frey M, Schaefer S, editors. New York: Cold Spring Harbor Laboratory Press. 154p.
8. SternbergN, HoessR (1983) The molecular genetics of bacteriophage P1. Annu Rev Genet 17 : 123–154.
9. SusskindMM, BotsteinD (1978) Molecular genetics of bacteriophage P22. Microbiol Rev 42 : 385–413.
10. SuttleCA (2005) Viruses in the sea. Nature 437 : 356–361.
11. ŁosM, WegrzynG (2012) Pseudolysogeny. Adv Virus Res 82 : 339–349.
12. RippS, MillerRV (1997) The role of pseudolysogeny in bacteriophage-host interactions in a natural freshwater environment. Microbiology 143 : 2065–2070.
13. RippS, MillerRV (1998) Dynamics of the pseudolysogenic response in slowly growing cells of Pseudomonas aeruginosa. Microbiology 144 : 2225–2232.
14. ClokieMR, MillardAD, LetarovAV, HeaphyS (2011) Phages in nature. Bacteriophage 1 : 31–45.
15. RigaliS, DerouauxA, GiannottaF, DusartJ (2002) Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem 277 : 12507–12515.
16. Vander BylC, KropinskiAM (2000) Sequence of the genome of Salmonella bacteriophage P22. J Bacteriol 182 : 6472–6481.
17. PedullaML, FordME, KarthikeyanT, HoutzJM, HendrixRW, et al. (2003) Corrected sequence of the bacteriophage P22 genome. J Bacteriol 185 : 1475–1477.
18. HayesWS, BorodovskyM (1998) Deriving ribosomal binding site (RBS) statistical models from unannotated DNA sequences and the use of the RBS model for N-terminal prediction. Pac Symp Biocomput 279–290.
19. LiY, AustinS (2002) The P1 plasmid in action: time-lapse photomicroscopy reveals some unexpected aspects of plasmid partition. Plasmid 48 : 174–178.
20. LehnherrH, MaguinE, JafriS, YarmolinskyMB (1993) Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol 233 : 414–428.
21. ZinderN (1958) Lysogenization and superinfection immunity in Salmonella. Virology 5 : 291–326.
22. RoucourtB, LavigneR (2009) The role of interactions between phage and bacterial proteins within the infected cell: a diverse and puzzling interactome. Environmental Microbiology 11 : 2789–2805.
23. Figueroa-BossiN, BossiL (1999) Inducible prophages contribute to Salmonella virulence in mice. Mol Microbiol 33 : 167–176.
24. ChenY, GoldingI, SawaiS, GuoL, CoxEC (2005) Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biol 3: e229 doi:10.1371/journal.pbio.0030229.
25. ErikssonS, LucchiniS, ThompsonA, RhenM, HintonJCD (2003) Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47 : 103–118.
26. KuYW, McDonoughSP, PalaniappanRUM, ChangCF, ChangYF (2005) Novel attenuated Salmonella enterica serovar Choleraesuis strains as live vaccine candidates generated by signature-tagged mutagenesis. Infect Immun 73 : 8194–8203.
27. Sambrook J, Russel DW (2001) Molecular cloning (a laboratory manual). New York: Cold Spring Harbor Laboratory press. 2344p.
28. Davis R, Botstein D, Roth J (1980) Advanced bacterial genetics. New York: Cold Spring Harbor Laboratory Press. 254p.
29. SchmiegerH (1972) Phage P22-mutants with increased or decreased transduction abilities. Mol Genet Genomics 119 : 75–88.
30. Miller JH (1992) A short course in bacterial genetics. New York: Cold Spring Harbor Laboratory Press. 876p.
31. AertsenA, Tesfazgi MebrhatuM, MichielsCW (2008) Activation of the Salmonella Typhimurium Mrr protein. Biochem Biophys Res Commun 367 : 435–439.
32. AltschulSF, GishW, MillerW, MyersEW, LipmanDJ (1990) Basic local alignment search tool. J Mol Biol 215 : 403–410.
33. DarlingAE, MauB, PernaNT (2010) progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5: e11147 doi:10.1371/journal.pone.0011147.
34. HughesKT, RothJR (1988) Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics 119 : 9–12.
35. DatsenkoKA, WannerBL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97 : 6640–6645.
36. CherepanovPP, WackernagelW (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158 : 9–14.
37. LindnerAB, MaddenR, DemarezA, StewartEJ, TaddeiF (2008) Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proceedings of the National Academy of Sciences 105 : 3076–3081.
38. EspeliO, MercierR, BoccardF (2008) DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol 68 : 1418–1427.
39. SawitzkeJA, CostantinoN, LiX-T, ThomasonLC, BubunenkoM, et al. (2011) Probing cellular processes with oligo-mediated recombination and using the knowledge gained to optimize recombineering. J Mol Biol 407 : 45–59.
40. ValdiviaRH, FalkowS (1996) Bacterial genetics by flow cytometry: rapid isolation of Salmonella Typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol 22 : 367–378.
41. AustinS, LiY (2002) The P1 plasmid is segregated to daughter cells by a “capture and ejection” mechanism coordinated with Escherichia coli cell division. Mol Microbiol 46 : 63–74.
42. NielsenHJ, LiY, YoungrenB, HansenFG, AustinS (2006) Progressive segregation of the Escherichia coli chromosome. Mol Microbiol 61 : 383–393.
43. HeukeshovenJ, DernickR (1988) Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis 9 : 28–32.
44. ShevchenkoA, WilmM, VormO, MannM (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68 : 850–858.
45. StewartEJ, MaddenR, PaulG, TaddeiF (2005) Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol 3: e45 doi:10.1371/journal.pbio.0030045.
46. McClellandM, SandersonKE, SpiethJ, CliftonSW, LatreilleP, et al. (2001) Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413 : 852–856.
47. CasjensSR (2005) Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol 8 : 451–458.
48. ValdiviaRH, FalkowS (1997) Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277 : 2007–2011.
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



