-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
Crown gall tumors develop after integration of the T-DNA of virulent Agrobacterium tumefaciens strains into the plant genome. Expression of the T-DNA–encoded oncogenes triggers proliferation and differentiation of transformed plant cells. Crown gall development is known to be accompanied by global changes in transcription, metabolite levels, and physiological processes. High levels of abscisic acid (ABA) in crown galls regulate expression of drought stress responsive genes and mediate drought stress acclimation, which is essential for wild-type-like tumor growth. An impact of epigenetic processes such as DNA methylation on crown gall development has been suggested; however, it has not yet been investigated comprehensively. In this study, the methylation pattern of Arabidopsis thaliana crown galls was analyzed on a genome-wide scale as well as at the single gene level. Bisulfite sequencing analysis revealed that the oncogenes Ipt, IaaH, and IaaM were unmethylated in crown galls. Nevertheless, the oncogenes were susceptible to siRNA–mediated methylation, which inhibited their expression and subsequently crown gall growth. Genome arrays, hybridized with methylated DNA obtained by immunoprecipitation, revealed a globally hypermethylated crown gall genome, while promoters were rather hypomethylated. Mutants with reduced non-CG methylation developed larger tumors than the wild-type controls, indicating that hypermethylation inhibits plant tumor growth. The differential methylation pattern of crown galls and the stem tissue from which they originate correlated with transcriptional changes. Genes known to be transcriptionally inhibited by ABA and methylated in crown galls became promoter methylated upon treatment of A. thaliana with ABA. This suggests that the high ABA levels in crown galls may mediate DNA methylation and regulate expression of genes involved in drought stress protection. In summary, our studies provide evidence that epigenetic processes regulate gene expression, physiological processes, and the development of crown gall tumors.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003267
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003267Summary
Crown gall tumors develop after integration of the T-DNA of virulent Agrobacterium tumefaciens strains into the plant genome. Expression of the T-DNA–encoded oncogenes triggers proliferation and differentiation of transformed plant cells. Crown gall development is known to be accompanied by global changes in transcription, metabolite levels, and physiological processes. High levels of abscisic acid (ABA) in crown galls regulate expression of drought stress responsive genes and mediate drought stress acclimation, which is essential for wild-type-like tumor growth. An impact of epigenetic processes such as DNA methylation on crown gall development has been suggested; however, it has not yet been investigated comprehensively. In this study, the methylation pattern of Arabidopsis thaliana crown galls was analyzed on a genome-wide scale as well as at the single gene level. Bisulfite sequencing analysis revealed that the oncogenes Ipt, IaaH, and IaaM were unmethylated in crown galls. Nevertheless, the oncogenes were susceptible to siRNA–mediated methylation, which inhibited their expression and subsequently crown gall growth. Genome arrays, hybridized with methylated DNA obtained by immunoprecipitation, revealed a globally hypermethylated crown gall genome, while promoters were rather hypomethylated. Mutants with reduced non-CG methylation developed larger tumors than the wild-type controls, indicating that hypermethylation inhibits plant tumor growth. The differential methylation pattern of crown galls and the stem tissue from which they originate correlated with transcriptional changes. Genes known to be transcriptionally inhibited by ABA and methylated in crown galls became promoter methylated upon treatment of A. thaliana with ABA. This suggests that the high ABA levels in crown galls may mediate DNA methylation and regulate expression of genes involved in drought stress protection. In summary, our studies provide evidence that epigenetic processes regulate gene expression, physiological processes, and the development of crown gall tumors.
Introduction
The bacterial pathogen A. tumefaciens genetically engineers the host plant by transferring its T-DNA, a piece of DNA from the tumor-inducing (Ti) plasmid, into the plant genome. Expression of the T-DNA-encoded oncogenes IaaH, IaaM, and Ipt results in increased synthesis of both auxin and cytokinin [1], [2]. High concentrations of these phytohormones not only facilitate proliferation of transformed plant cells, but also differentiation of specialized cell types within the resulting crown gall tumor [3]. In addition to auxin and cytokinin, elevated levels of the phytohormones salicylic acid, ethylene and abscisic acid (ABA) have been observed in crown galls on A. thaliana stems [4]–[6]. In particular, ABA was shown to be important for drought stress acclimation to ensure wildtype-like crown gall growth [7]. Moreover, approximately 20% of protein coding genes are differentially transcribed in these plant tumors compared to tumor-free stem tissue [8]. The massive changes in gene expression, together with the cooperative action of phytohormones, fulfill distinct roles in differentiation, pathogen defense, metabolic changes, and physiological adaptations in crown galls [3], [7], [8]. In recent years, there has been increased interest in the role of epigenetic events in regulating biotic and abiotic stress responses in plants [9]. Environmental stresses have been shown to influence epigenetic processes, inducing the release of transcriptional silencing of transgenes and several endogenous A. thaliana gene loci.
Changes in the DNA methylation pattern have also been reported in cases where, similar to A. thaliana crown galls, foreign DNA is integrated into the mammalian genome prior to tumor formation. For example, mammalian tumors induced by adenovirus type 12 display extensive genome-wide hypermethylation [10]. These widespread differences in the methylation pattern during mammalian tumor formation indicate that they may be a common feature of neoplastic growth, possibly also during plant tumor development. Such an epigenetic contribution to crown gall formation was already suggested by Braun 50 years ago [11]. To date, only the integrated T-DNA has been examined with respect to DNA methylation. The T-DNA of different crown gall lines was shown to be frequently methylated. At least one T-DNA copy in each tumor genome remained unmethylated [12], [13], which allowed expression of oncogenes and thereby crown gall proliferation. T-DNA methylation can be induced by siRNAs which are produced by dicer activities on long dsRNA. Synthesis of the latter RNAs results from bidirectional or read-through transcription of rearranged or integrated T-DNAs. While siRNAs corresponding to T-DNA oncogenes accumulate in A. tumefaciens-infected plant tissue, synthesis of siRNA is specifically inhibited in developing tumors resulting in a potent antisilencing state [14].
In the A. thaliana genome, the highest levels of methylation are found in transposon-rich heterochromatic regions. This methylation pattern is in agreement with a primary function for methylation in transposon silencing. However, DNA methylation of protein coding genes also frequently occurs. Methylation is depleted at promoters and gene ends, indicating that it interferes with important regulatory functions in these gene segments [15]. Endoreduplication is also known to cause methylation changes as a result of increased ploidy levels in A. thaliana [16]. Furthermore, endoreduplication is a phenomenon known to occur in specialized cell types of animals and in different tissues of many plant species [17]. It extensively occurs in A. thaliana, especially if the levels of auxin and cytokinin are increased, such as in crown galls [18], [19].
Methylation in plants differs from that in mammals in its sequence context. In plants, cytosines are methylated in three different sequence contexts (CG, CHG and CHH, where H = A, C, T), whereas methylation at CG dinucleotides predominates in mammals [20]. DNA methylation in A. thaliana is established by DRM1 and DRM2 (DOMAINS-REARRANGED-METHYLASE) methyltransferases in all sequence contexts [21]. Methylation of specific genomic regions can be targeted by DRM proteins through interaction with ARGONAUTE4 (AGO4). Small RNAs of 21–24 nt are incorporated in AGO4 and guide DRM activity to the corresponding genomic sequences [22]. Both DRM methyltransferases and AGO4 are also essential for transgene silencing which is inducible by hairpin constructs complementary to a transgene promoter [23]. CG methylation is maintained during genome replication by the activity of MET1 (METHYLTRANSFERASE1; [24]), while the plant-specific DNA methyltransferase CMT3 (CHROMOMETHYLASE) primarily methylates cytosines in the CHG context [25]. Furthermore, subsets of genomic DNA methylation patterns are influenced by the activity of the demethylating enzymes ROS1 (REPRESSOR OF SILENCING), DME (DEMETER), DML2 (DEMETER-LIKE) and DML3 [26].
Our study provides the results of a genome-wide methylation analysis of stem-derived A. thaliana crown galls in comparison with mock-inoculated stem tissue. This analysis indicates that the crown gall tumor genome is globally hypermethylated, while promoter regions are hypomethylated. These changes in the DNA methylation pattern seem to exert an inhibitory influence on growth of crown gall tumors, since A. thaliana mutants with reduced DNA methylation, like drm1/drm2/cmt3 (ddc) and ago4, developed significantly larger crown galls. The global differences in DNA methylation between the crown gall and the tumor-free stem genome were in agreement with the transcriptomic changes of protein coding genes. For example, genes involved in ABA-dependent drought stress protection were promoter methylated and transcriptionally silenced in crown gall tumors. A. thaliana seedlings treated with the stress phytohormone demonstrated ABA-dependent methylation of the promoters of these genes. Taken together, our studies provide evidence for a role of epigenetic processes in controlling gene expression, development and physiology in crown gall tumors.
Results
Methylation of the T-DNA–encoded oncogenes
Earlier studies have shown that the T-DNA-encoded oncogenes of the virulent A. tumefaciens strain C58 are always actively transcribed in crown gall tumors of A. thaliana stems [8]. However, transgenes are known to be frequently methylated. Therefore, we analyzed the cytosine methylation pattern of a 5,429 bp T-DNA segment of the pTiC58 plasmid. This segment consists of the coding sequences (CDS) from the oncogenes IaaH, IaaM and Ipt as well as the two intergenic regions between them (IGR1 and IGR2, Figure 1A). Methylated cytosines in this region were determined by bisulfite sequencing of genomic DNA preparations of stem-derived A. thaliana crown galls. This analysis revealed that only 0.94% of all cytosines were methylated in the three coding sequences (CDS) of the oncogenes, whereas the two IGRs were completely devoid of methylated cytosines (Figure 1B).
Fig. 1. Intergenic regions (IGRs) of the T-DNA–encoded oncogenes become methylated upon an siRNA trigger. 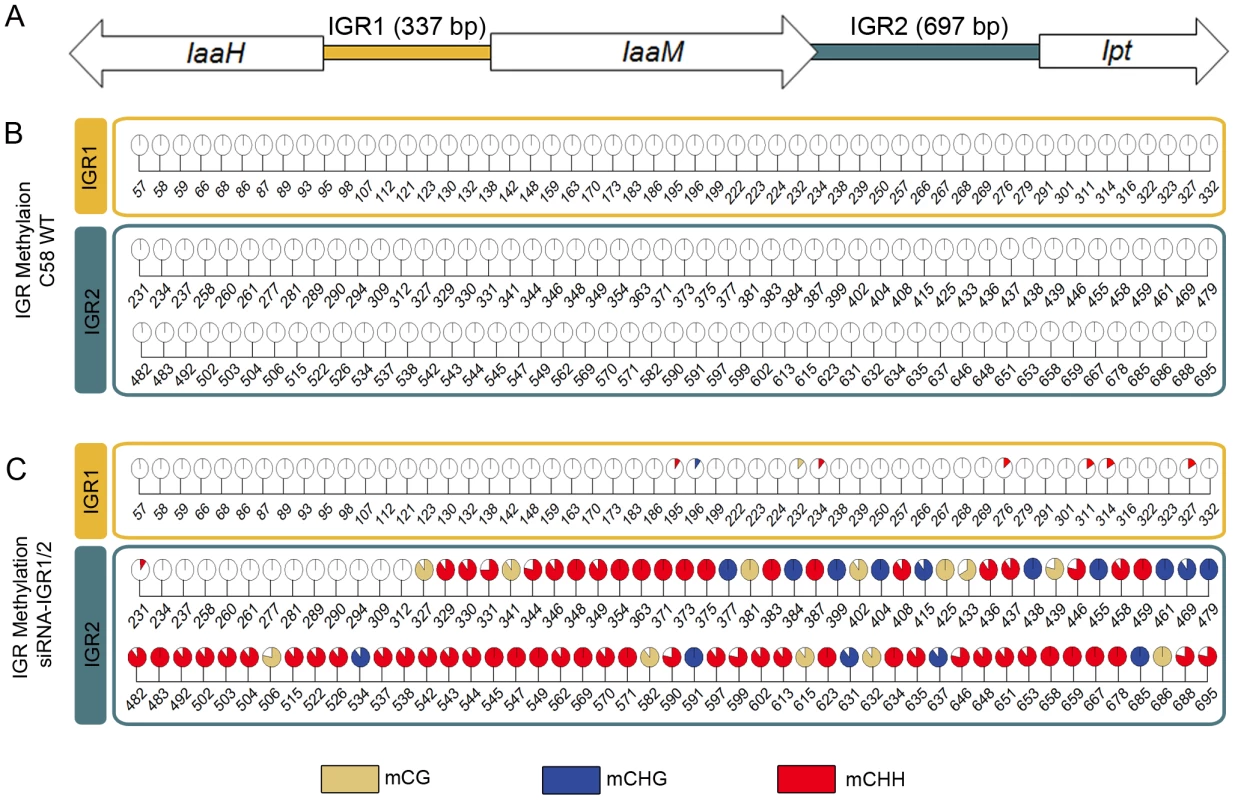
(A) IGRs upstream of the oncogenes IaaH, IaaM (IGR1) and Ipt (IGR2) from the T-DNA of pTiC58 were analyzed by applying bisulte sequencing (model not drawn to scale). Coding sequences for IaaH, IaaM and Ipt are depicted as arrows, colored bars illustrate the two IGRs. (B) Detailed map of all methylated cytosines within IGR1 and IGR2 in the crown gall tumor induced by the wildtype A. tumefaciens strain C58 (C58 WT) and (C) of plant material inoculated with the same strain C58, but in addition harboring a binary vector with a hairpin construct directed against both IGRs (siRNA-IGR1/2). Percentages of methylation at each position are visualized by pie charts filled with different colors for the three methylation motifs (mCG brown, mCHG blue, mCHH red). Numbers below pie charts indicate nucleotide positions from the start of the analyzed region. Cytosines outside of the displayed regions were unmethylated. Percentages were calculated from bisulfite sequencing results of multiple independent clones. The extremely low degree of T-DNA methylation in crown gall cells suggests that this is a prerequisite to maintain the expression levels of oncogenes required for tumor formation. This hypothesis was tested by the induction of oncogene promoter methylation, making use of the endogenous siRNA-directed plant methylation pathway. Plasmids containing a hairpin construct directed against the IGRs upstream of the oncogene CDSs, each fused to the CaMV35S promoter (Figure S1), were transferred into the crown gall genome by using the virulent A. tumefaciens strain C58. Development of crown gall tumors was strongly impaired (Figure 2A) when A. thaliana was inoculated with strain C58 that contained hairpin sequences directed against both IGRs (siRNA-IGR1/2, Figure S1A). In contrast, no growth inhibition occurred on plants inoculated with strain C58 if only one IGR was addressed by a hairpin construct (Figure S1B). Bisulfite sequencing analysis of both IGRs revealed that the IGR1/2 hairpin induced methylation of cytosines in all three sequence contexts upstream of the Ipt and IaaH CDS (Figure 1C). However, IGR1 was only marginally methylated in contrast to IGR2.
Fig. 2. Tumor development and oncogene transcription are inhibited by RNAi–mediated DNA methylation. 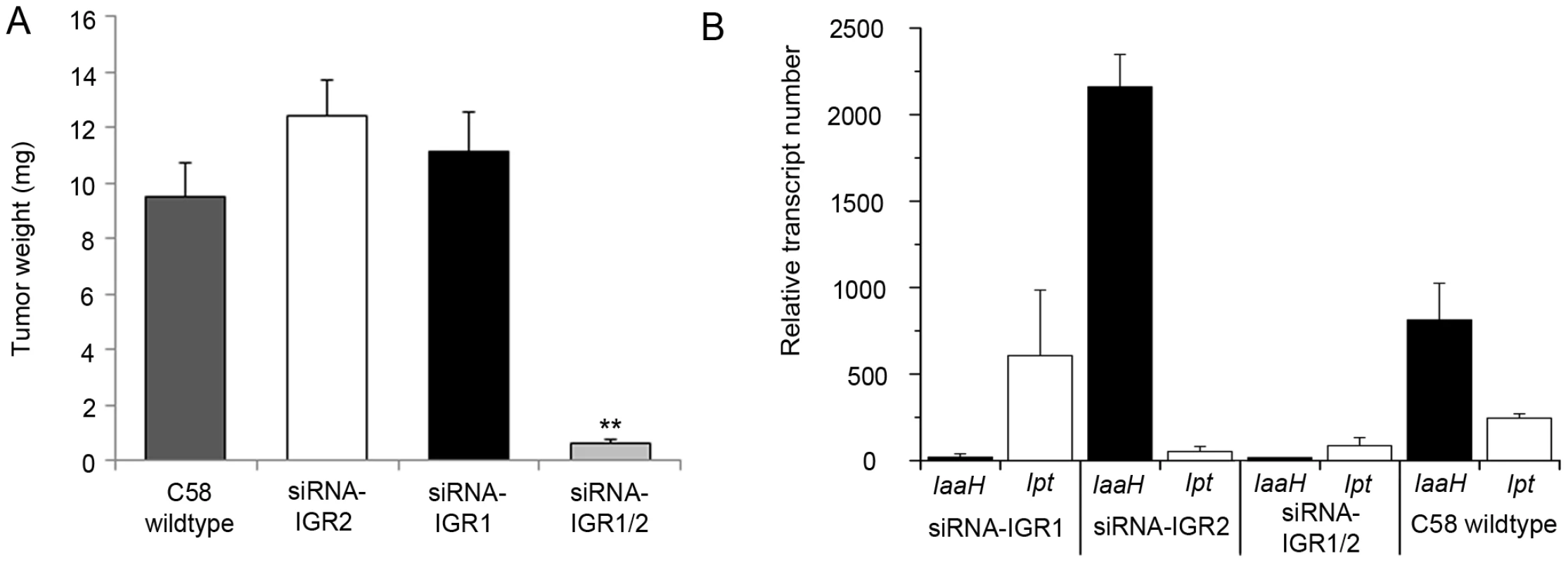
(A) Crown gall tumor weights were determined four weeks after injection of recombinant virulent A. tumefaciens strains of C58 into A. thaliana stems (ecotype WS-2). The recombinant strain C58 harbored either an empty pHellsgate12 vector (C58) or a hairpin construct directed against IGR1 (siRNA-IGR1) or IGR2 (siRNA-IGR2) or against both IGRs (siRNA-IGR1/2). Bars represent mean values (± SEM) of at least 38 inoculated plants per A. tumefaciens genotype. Statistical analysis was performed using student's t-test: p-value<0.01 (**). (B) Relative transcript numbers of the oncogenes of strain C58 in A. thaliana stem tissue six days after inoculation with the A. tumefaciens genotypes used for the crown gall tumor growth assay in (A). Relative transcript numbers were quantified by real time qRT-PCR and normalized to 10.000 molecules of ACTIN2/8. Bars represent mean values (± SD) of three independent experiments. Induction of IGR methylation by hairpin constructs suggests that expression of the oncogenes may be hindered. Quantification of IaaH and Ipt transcripts by qRT-PCR in A. thaliana stems six days after inoculation demonstrated that the transcription of oncogenes was indeed inhibited whenever strain C58 contained a hairpin construct directed against the corresponding IGR (Figure 2B). Transcriptional silencing of IaaH was achieved despite a low cytosine methylation level, which suggests that other factors such as siRNA-mediated histone modification may also play role in silencing of this gene [27], [28]. Wildtype A. tumefaciens lacking siRNA induced expression of the two oncogene transcripts as expected. Overall, the results demonstrate that the T-DNA sequence is susceptible to DNA methylation and crown gall development can be efficiently prevented by transcriptional silencing of oncogenes.
The crown gall tumor genome is globally hypermethylated
In order to analyze whether A. tumefaciens causes genome-wide DNA methylation changes in the plant genome, we determined the methylation pattern of A. tumefaciens-induced A. thaliana crown gall tumors. Tumor growth was induced on A. thaliana stems, and tumor-free stem material was used as reference tissue. Genomic DNA from each of three independent tumor and stem samples, was randomly fragmented. Thereafter it was subjected to methylcytosine immunoprecipitation (mCIP), resulting in an enrichment of methylated DNA fragments. Each mCIP sample (3× tumor and 3× stem) was separately hybridized to an Affymetrix A. thaliana Tiling 1.0R array in parallel with three non-enriched input controls of each tissue type. This tiling array consists of oligonucleotide probes that represent the A. thaliana genome with an average resolution of 35 bp. Hybridization signals allowed the detection of genomic regions consisting of at least five genomically adjacent probes that displayed signal intensities above the local background. These regions were assumed to be enriched by the mCIP procedure and therefore considered to contain methylated cytosines.
In total 15,431 distinct genomic regions were methylated in either tissue type. These regions cover 26,287 kb (22.06%) of the A. thaliana nuclear genome. In order to verify the reliability of the methylation profile analysis the distribution of methylation signals was examined for both the tumor and stem genome in four categories of annotated loci: Protein coding genes, transposable elements, pseudogenes and non-coding RNAs (ncRNA). For this purpose the proportion (%) of methylated genes out of the total number of genes showing methylation was calculated at 60 positions from 1 kb upstream to 1 kb downstream of genes and plotted along an abstracted model sequence for each gene category. The highest percentages of methylation at all positions were detected in pseudogenes, where they were almost evenly distributed along the entire sequence (Figure S2). In protein coding genes, methylation was especially enriched in the 3′-half of the transcribed region, whereas it decreased towards the transcription start (TSS) and transcription end sites (TES). The methylation pattern was similar for ncRNAs, except that the overall methylated proportion was higher and did not increase in the transcribed region. In accordance with its presumed function in transposon silencing [28], the transcribed regions of transposable elements were highly methylated. Overall, in the genomes of both tissue types the methylation patterns of the mentioned gene categories are well in agreement with those reported in earlier studies [15], [29].
In order to identify differences in DNA methylation between crown gall tumor and stem tissue, differentially methylated regions (DMRs) were determined in the four categories (Figure 3; for details see Materials and Methods). This analysis revealed that 2,876 annotated loci differed in methylation levels between crown galls and the tissue from which they originate. The majority of these loci overlapped with regions found to be hypermethylated in crown galls (1,822), whereas 1,100 hypomethylated regions were located in the proximity or inside of annotated loci. The sum of hyper - and hypomethylated regions is higher than that of all affected loci because one locus may contain several DMRs. With respect to the total number of DMRs between crown gall and stem tissue, the majority could be assigned to protein coding genes (71.3%) and transposable elements (25.3%, Figure 3A), which together account for nearly 97% of all DMRs. DMRs were also present in pseudogenes (1.8%) and ncRNAs (1.5%). However, when separately calculating the number of DMRs for each of the four categories of annotated loci, the highest proportion was detected in protein coding genes (7.7%), followed by pseudogenes (6.1%) and ncRNAs (3.4%), whereas only 2.4% of transposable elements were differentially methylated between the two tissue types (Figure 3B). Hypermethylation was more prominent than hypomethylation in protein coding genes (5.1% vs. 1.6%), pseudogenes (3.8% vs. 2.3%) and ncRNAs (2.3% vs. 1.1%), but not in transposable elements (1.2% vs. 1.2%). Taken together, the genome of the crown gall tumor is globally hypermethylated. The changes in the genome wide methylation pattern reflected the increased expression of genes involved in DNA methylation (MET1, DRM2, CMT3 and AGO4) as well as demethylation (ROS1/DML1) in crown galls (Table S1).
Fig. 3. Distribution of differentially methylated regions (DMRs) within four gene types. 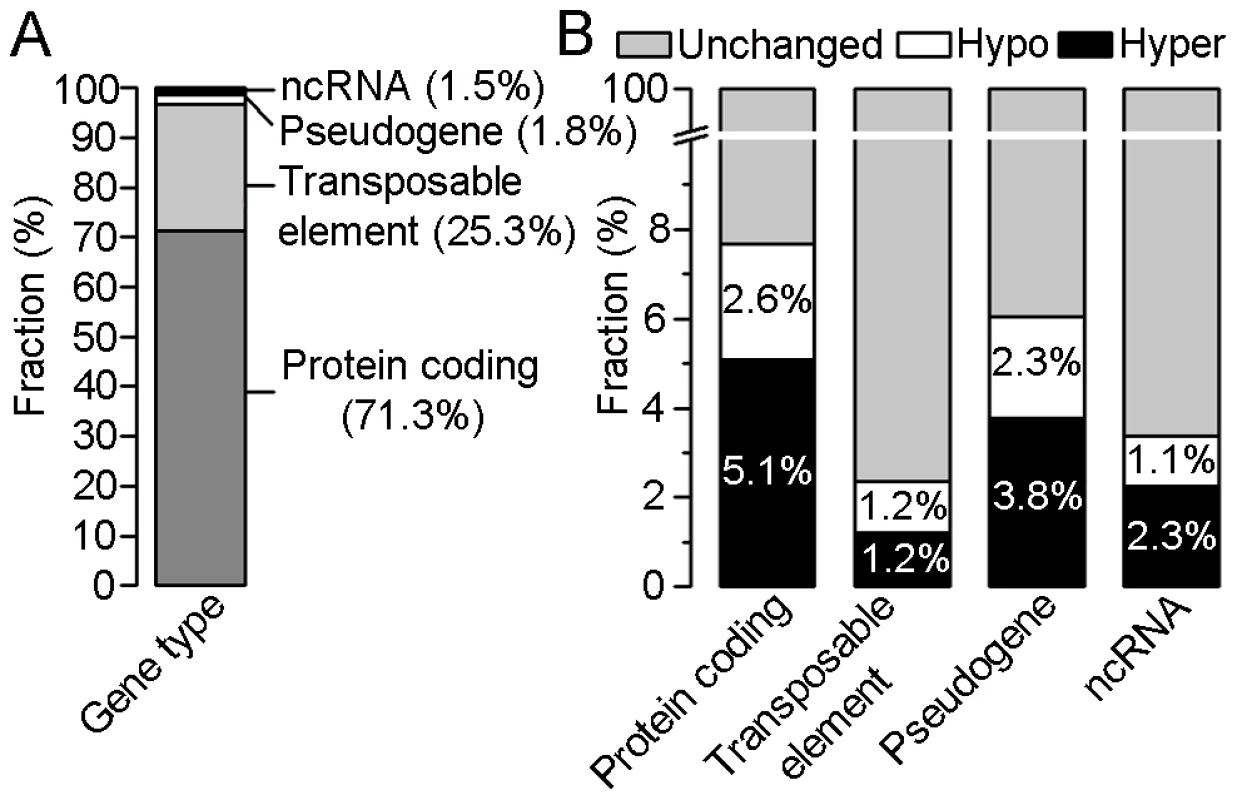
(A) Percentages of DMRs between crown gall tumors and mock-inoculated stems in four types of annotated loci (protein coding, transposable elements, pseudogene and non-coding (nc)RNA). (B) Percentages of unchanged, hyper- and hypomethylated regions within each of the four gene types. The calculation is based on 2876 DMRs out of which 2052 belong to the group of protein coding genes, 729 to transposable elements, 53 to pseudogenes and 42 to ncRNAs. In order to verify methylation differences by an independent method, one DMR was randomly chosen from each of the five A. thaliana chromosomes and analyzed by bisulfite sequencing. The directions of methylation changes at the tested loci (hypermethylation: At5g58370, At4g12460 and hypomethylation: At3g19250, At1g20850) were in agreement with the results achieved by tiling array analysis, except for the locus At2g16595 from chromosome 2 (Figure S3). This locus was enriched in immunoprecipitated crown gall DNA, which indicates an increase in methylation. However, bisulfite sequencing revealed that only CG and CHH methylation was increased at this locus, but this was accompanied by a decrease in CHG methylation.
Changes in the methylation pattern may be a result of altered ploidy levels which frequently occur in A. thaliana. Therefore, the (endo)ploidy levels of A. thaliana crown gall cells as well as tumor-free stem cells were determined by flow-cytometry (Figure S4). In both cases the first DNA peak (2C) was found at similar positions, excluding a ploidy change towards tetraploidy in the crown gall tumor cells. Neither the histogram nor the cycle value, defined as the mean number of endoreduplication cycles per nucleus [30], indicated an increased endopolyploidization rate in crown gall (0.784) versus tumor-free (0.899) tissues. Furthermore, we observed no peak shifts or changes in the peak width on the histograms derived from the crown gall tissue which might have indicated aneuploidy. These data demonstrate that hypermethylation in A. thaliana crown gall tumors is not due to an increased DNA content per nucleus. The significantly decreased 4C/2C ratio in crown galls (1.11) compared to stem tissue (2.32) indicates an increased number of 2C nuclei (Figure S4B) and thus an elevated rate of cell division in crown galls. Whereas in the non-tumor tissue more and more cells switch from the initial mitotic divisions to endoreduplication cycles, tumor cells tend to proliferate mitotically resulting in 2C cells at the end of each cycle.
Methylation changes occur mainly at non–CG motifs and affect crown gall tumor development
In contrast to the animal genome, a substantial amount of cytosine methylation occurs in non-CG contexts in plants. To identify the sequence motifs which were mostly affected by differential DNA methylation in A. thaliana crown gall tumors, all methylated genomic regions were grouped into three classes (hypomethylated, unchanged or hypermethylated). These classes indicate the methylation levels of crown gall DNA compared to control tissue. Pairwise Wilcoxon rank sum tests were conducted to determine whether the frequencies of the three sequence motifs (CG, CHG or CHH) differ between the three classes. Differences in methylation frequency were much less significant for the CG motif (P-value>0.01, Figure S5) than for CHG - and CHH motifs (P-value<0.01). This suggests that methylation changes in the crown gall tumor mainly occurred at CHG and CHH motifs and to a lower extent at CG nucleotides.
The significant changes in the DNA methylation pattern prompted us to test its impact on crown gall development. Several A. thaliana mutants with no obvious growth phenotype but with defects in either methylation or demethylation processes were inoculated with the virulent A. tumefaciens strain C58. The fresh weight of mature crown galls from both mutant and wildtype plants was compared after 28 days (Figure 4). The ddc triple mutant, in which CHG and CHH methylation are strongly impaired, displayed significantly enhanced crown gall growth. A similar difference in tumor growth was found between wildtype plants and the ago4 mutant, which is impaired in RNA-dependent methylation processes. Note that the differences in tumor weights between ago4 plants and the ddc mutant are likely to be based on their genetic background. Plants in the Ler background of ago4 are known to develop much smaller crown gall tumors than plants in the Col-0 background of ddc. The growth of crown gall tumors was not altered in the rdd mutant, which demonstrates that demethylation pathways are not essential for A. tumefaciens-induced tumor development. Enhanced growth of crown galls on mutants that are affected in non-CG methylation pathways suggests that hypomethylation at CHG and CHH motifs facilitates plant tumor proliferation. Together with the increased differences in non-CG motif frequency in DMRs, these results provide further evidence for a prominent role of non-CG methylation during crown gall development. Nevertheless, we cannot rule out an involvement of CG methylation, because homozygous met1-3 mutants are not suitable for tumor growth assays due to severe developmental abnormalities.
Fig. 4. Tumor growth is enhanced in DNA methylation mutants. 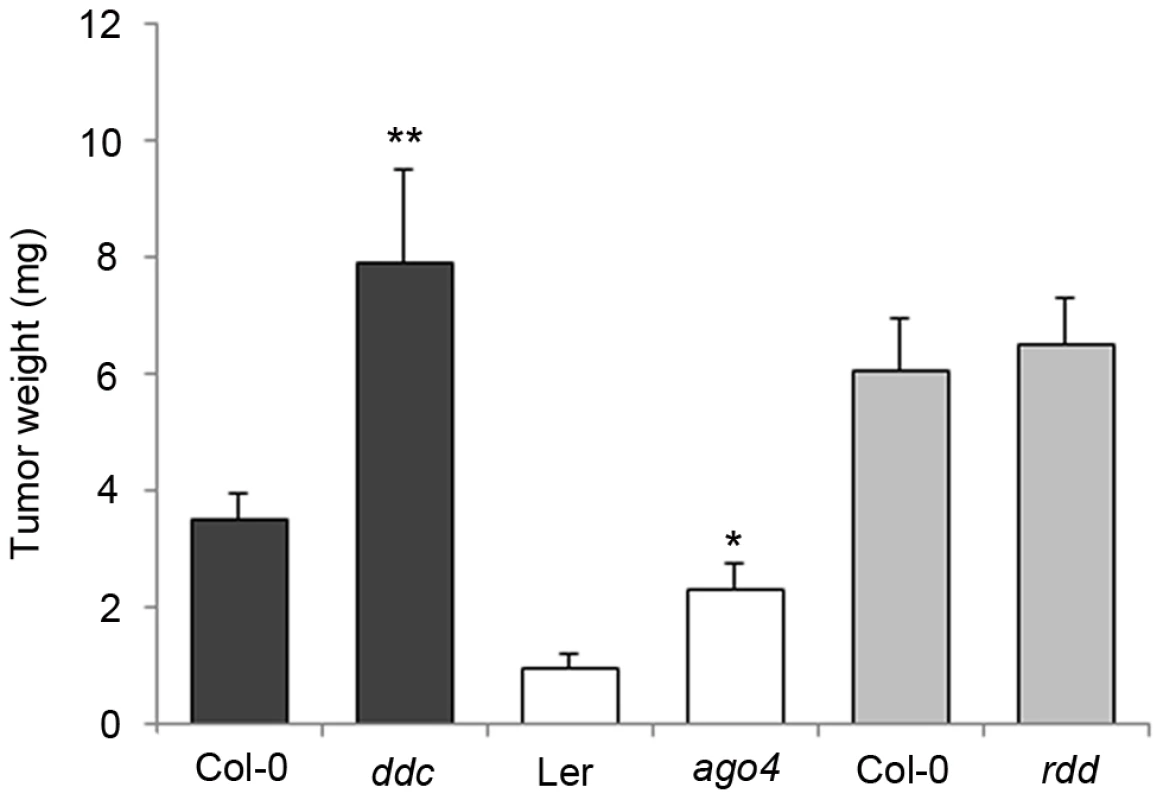
Tumor weights were determined four weeks after infection of methylation mutants (ddc and ago4) and the rdd demethylation mutants. Plants were inoculated with the virulent A. tumefaciens strain C58 at the base of the inflorenscence stem. Error bars represent mean values (± SEM) of at least 45 plants per A. thaliana genotype. Statistical analysis was performed using student's t-test: p-value<0.05 (*); p-value<0.01 (**). Distribution of DNA methylation changes and its impact on gene expression
DNA methylation especially at transcriptional start sites (TSS) and transcriptional end sites (TES) of protein encoding genes and in the transcribed region of transposable elements is known to affect both gene expression and transposon mobility in plants. In order to determine where the changes in DNA methylation preferentially occur, the percentages of hyper - and hypomethylated regions out of all DMRs were calculated for the tumor genome in comparison to the uninfected stem at 60 positions from 1 kb upstream to 1 kb downstream of genes. The distribution of DMRs was plotted along a model sequence for protein coding genes and transposable elements. In the crown gall genome, hypomethylated regions dominated in transcribed regions of transposable elements, while the distal 5′ - and 3′-flanking sequences were rather hypermethylated (Figure S6A). Protein coding genes were preferentially hypermethylated in the 3′-half of the transcribed region, whereas both the upstream sequence and the 5′-half of transcribed region were hypomethylated in crown galls compared to stems (Figure S6B). The proportion of hypomethylation in the upstream sequence and around the TSS was relatively high, which may be a mechanism to regulate gene expression in the plant tumor. A comparison of the methylome with transcriptome data from a previous study [8] supported this hypothesis. Methylation changes at the TSS or TES had an inverse effect on gene expression for the majority of genes (negative logFC product). In contrast, differential methylation within the gene body was preferentially associated with gene expression changes in the same direction (positive logFC product; Figure 5).
Fig. 5. Comparison of differential DNA methylation and differential gene expression. 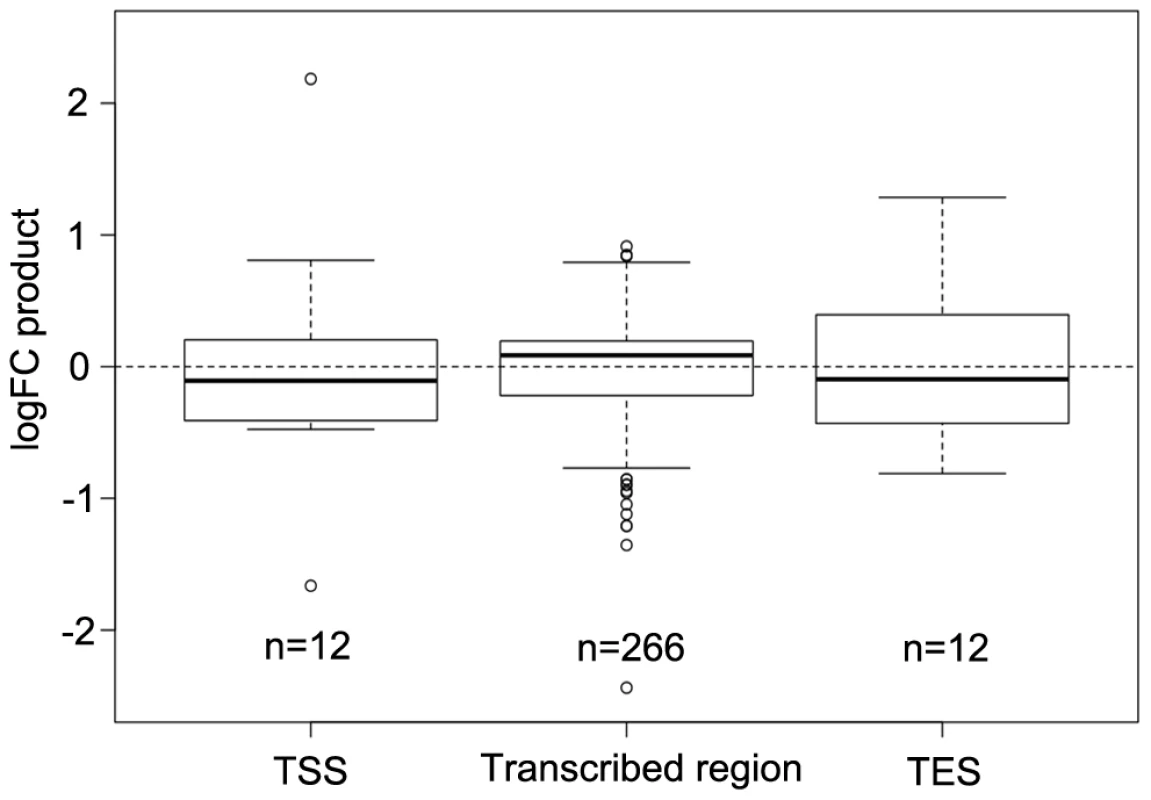
Logarithmic fold changes (logFCs) were determined for differentially methylated regions (DMR) as well as for differentially expressed genes. Methylation logFCs for DMRs and logFCs of differential transcript levels mapping to the transcriptional start site (TSS), the transcribed region and the transcriptional end site (TES) were multiplied and the products displayed as boxplots. Negative logFC products indicate that changes in methylation are associated with gene expression changes in the opposite direction. Positive logFC products imply changes of methylation and gene expression in the same direction. Due to the relationship between the methylation patterns and gene expression levels, a gene ontology analysis was performed to determine which pathways are mostly affected by differential methylation. According to the MapMan software [31], [32] a large number of DMRs that target protein coding genes were enriched in the functional category “development” (FDR≤0.002; Table 1). DMRs were also overrepresented (FDR<0.1) in the categories “cell” (particularly in the subcategory “cell division”), as well as “signaling”, “biotic stress” and “cytochrome P450”. Most of the genes in these categories are involved in processes that are associated with crown gall development; such as transcriptional regulation, cell cycle, chromosome condensation, redox and disease resistance. These differences in methylation correlated with the differences in transcription, as exemplified in Figure 6 for genes involved in embryo development (At2g22870, Figure 6A), microtubule-based movement (At1g63640, Figure 6B), cysteine rich receptor kinase signaling (At4g11480, Figure 6C), and pathogenesis-related protein signaling (At1g78780, Figure 6D). A complete list of all genes affected by differential methylation in the respective pathways is present in Table S2
Fig. 6. Differences in the degree of methylation in upstream regions correlate with differential transcription. 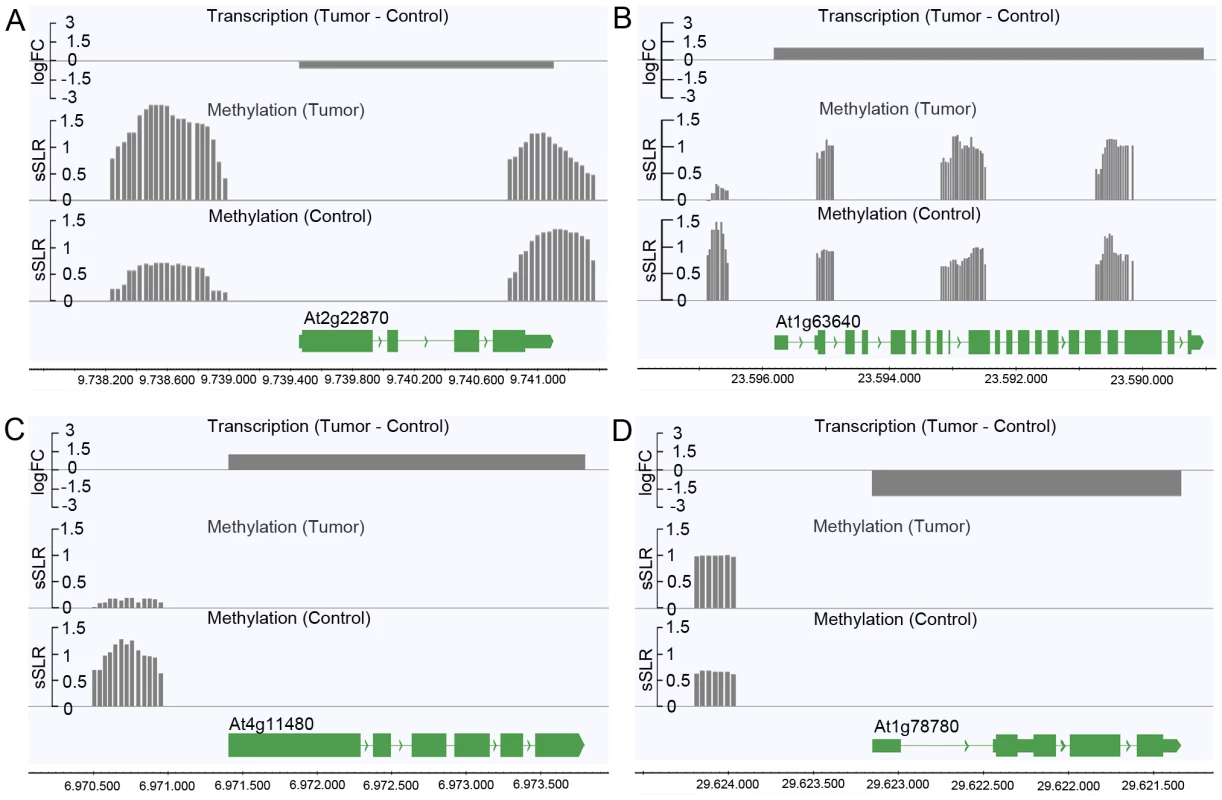
Differential transcription between crown galls and mock-inoculated stem tissue (control) and the DNA methylation status are depicted for selected genes listed in Table S2. The selected genes belong to the following functional categories of Table 1: (A) Development (At2g22870), (B) cell (At1g63640), (C) signaling (At4g11480), and (D) biotic stress (At1g78780). Transcriptional differences were calculated as logarithmic fold changes (logFC, grey bars in the top row). Regions of DNA methylation in tumor and control tissue (grey bars in the 2nd and 3rd row from the top) are shown as smoothed signal log ratios (sSLRs, mCIP versus input). The gene models (green) are displayed according to their corresponding genomic positions at the bottom of each subfigure. Tab. 1. Enrichment of protein coding genes with differentially methylated regions (DMRs) in functional categories. 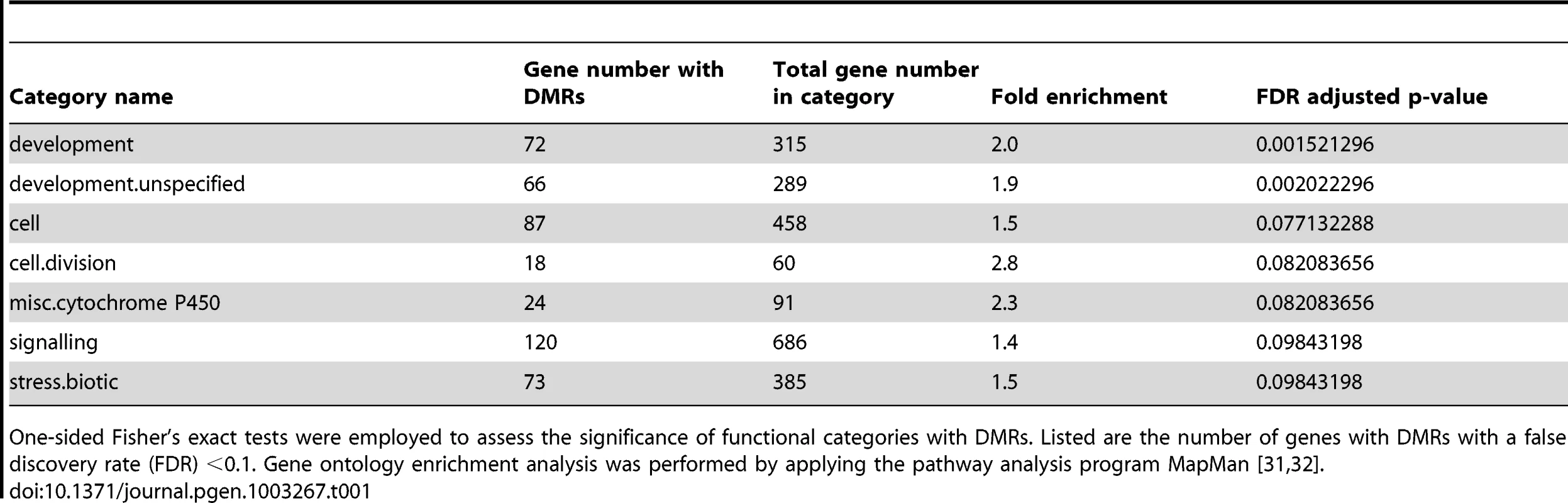
One-sided Fisher's exact tests were employed to assess the significance of functional categories with DMRs. Listed are the number of genes with DMRs with a false discovery rate (FDR) <0.1. Gene ontology enrichment analysis was performed by applying the pathway analysis program MapMan [31], [32]. Abscisic acid induces promoter methylation
The significant changes in DNA methylation in crown gall tumors and their role in tumor development piqued our interest regarding the control of physiological processes by DNA methylation. Previously we had shown that the lack of an intact epidermis causes induction of ABA-dependent protection against drought stress in crown galls. Drought stress acclimation is associated with altered transcript levels of many genes involved in ABA-dependent signaling. Acclimation to drought stress is also important for crown gall tumor growth, which has been shown to be impaired in ABA-deficient or -signaling mutants [7]. In order to assess whether ABA influences DNA methylation processes in crown galls, genes known to be strongly transcriptionally repressed by ABA according to the Genevestigator database [33], [34] were selected. In addition to transcriptional regulation by ABA, these genes were known to be significantly downregulated and highly methylated in their promoter sequence in the crown gall tumor. The genes are involved in chloroplast-specific processes, such as cyclic electron flow around photosystem I (NDF4), light-dependent transcription of the photosystem II subunit proteins D2 (SIG5) and alpha-/beta-hydrolase activity in the chloroplast (F12A4.4).
The influence of ABA on promoter methylation of the selected genes was studied in germination experiments with A. thaliana seeds, because these experimental conditions were used in the original study from the Genevestigator data set [34]. Methylation patterns of the promoters of the selected genes were analyzed by applying bisulfite sequencing. ABA treatment of .A. thaliana seeds provoked increased methylation levels in upstream regions (Figure S7) of NDF4 (logFC: 1), SIG5 (logFC: 0.52) and F12A4.4 (logFC: 0.96) and additionally caused severely reduced transcript levels (Figure 7A). This result reflects the situation observed in A .thaliana tumors, where elevated ABA levels and reduced transcription were accompanied by upstream hypermethylation of the tested genes (Figure 7B). Apparently, part of the difference in methylation patterns and transcription between crown galls and the tissue of origin can be ascribed to elevated ABA levels within the tumor.
Fig. 7. ABA induces methylation and reduces transcript levels. 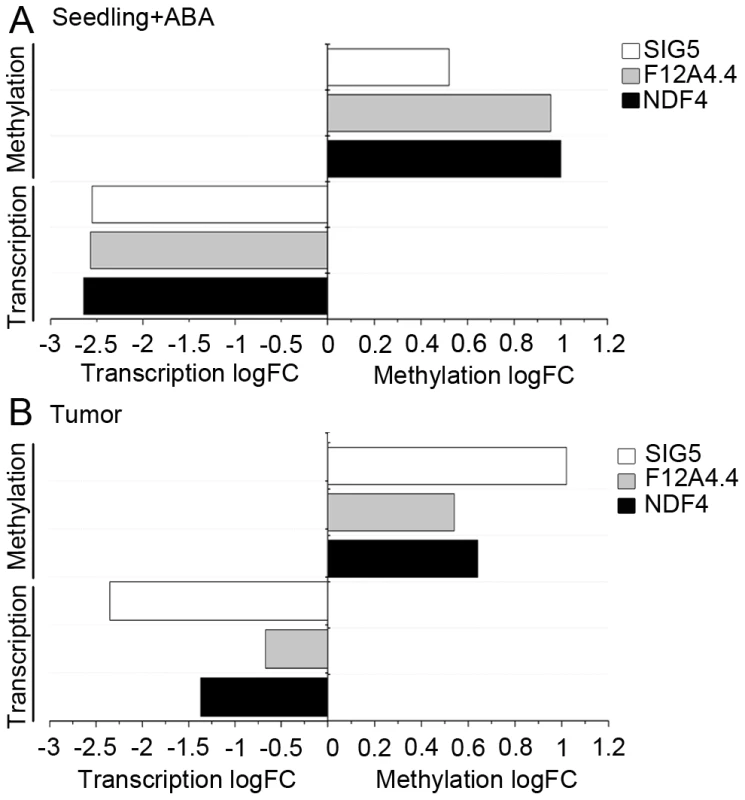
(A) The difference in the levels of transcription (Transcription logFC) and DNA methylation (Methylation logFC) were determined from samples treated with ABA versus samples not treated with ABA for the A. thaliana genes NDF4 (At3g16250), SIG5 (At5g24120) and F12A4.4 (At1g35420). Transcript levels were determined by qRT-PCR and methylation profiles of upstream regions by bisulfite sequencing. For these experiments A. thaliana seedlings were treated with ABA for two days according to the protocol of Nishimura et al. [34]. (B) Changes in methylation and transcription in the crown gall tumor compared to mock-iocculated stems according to methylome (Methylation logFC) and transcriptome data (Transcription logFC) of a previous study [8]. Each data set is based on at least three independent biological replicates and the logarithmic of fold changes (logFC) were calculated from the mean values. Discussion
In recent years, epigenetic processes such as DNA methylation have received increasing attention focused on their function in development, biotic and abiotic stress responses, as well as genome defense [35]–[37]. In our studies we have focused on the impact of DNA methylation on development and physiology of A. tumefaciens-induced crown gall tumors. A precondition for crown gall tumor formation is expression of the oncogenes IaaH, IaaM and Ipt, which are encoded on the agrobacterial T-DNA that is integrated into the plant genome. Therefore, it is not surprising that post-transcriptional gene silencing (PTGS) of these genes by means of RNAi caused resistance to crown gall tumorigenesis in previous experiments [38]–[40]. In these studies the Ipt and Iaa oncogenes were silenced using hairpin constructs directed against their coding sequences. Apart from such PTGS-dependent processes, transcriptional gene silencing can be induced by de novo methylation. Previously this epigenetic mechanism has been used to initiate methylation of the agrobacterial nopaline synthase promoter [23]. However, synthesis of siRNA directed against T-DNA-encoded genes like IaaM and agropine synthase is specifically inhibited in developing crown gall tumors [14]. This observation is in agreement with our finding that the sequence of the oncogene cluster IaaH, IaaM and Ipt was unmethylated in A. thaliana crown galls [23], despite the proposed ancient role of methylation in genome defense [37]. Thus, suppression of siRNA-mediated oncogene silencing as well as methylation-mediated silencing of oncogene promoters may guarantee unimpeded expression of T-DNA oncogenes which is indispensable for tumor growth. The view that promoter methylation induces transcriptional silencing was further supported by introducing siRNAs complementary to oncogene promoter regions. SiRNA-directed methylation of both promoter regions prevented crown gall development, whereas targeting of only one promoter region still allowed proliferation. The latter result indicates that expression of either oncogene is sufficient for tumor growth, which is in line with earlier studies [41], [42]. Transcriptional gene silencing by promoter methylation is thus an effective tool in transgene silencing, as it was previously demonstrated for the nopaline synthase promoter [43]. Induction of siRNA synthesis complementary to oncogene promoters probably outweighs the antisilencing state induced by A. tumefaciens. Consequently, it may provide a potent mechanism to suppress tumor development after A. tumefaciens infection.
Methylation analysis of A. thaliana crown gall tumors revealed that the genome was globally hypermethylated. Hypermethylation may be attributable to increased expression of DRM2, CMT3 and AGO4 in crown galls, all of which are involved in RNA-directed DNA methylation pathways. Transcription levels of demethylating enzymes probably do not impact the global methylation level since they affect only subsets of genomic loci [26]. In the crown gall genome mainly CHG and CHH motifs were altered, whereas CG methylation was less affected. This suggests that CG methylation does not play an important role in plant tumor development, although MET1 expression is also increased in crown galls. CG methylation is known to play a major role in generating meiotically stable epialleles due to spontaneous gain or loss of DNA methylation after propagation of A. thaliana over several generations. These transgenerational effects are probably a result of epigenetic reprogramming that occurs after fertilization [44], [45]. In contrast, reprogramming of the transformed plant cells begins with the expression of the T-DNA-encoded oncogenes, causing proliferation of the plant tumor. Epigenetic reprogramming during crown gall tumor formation is more likely to be induced by DRM and CMT3 methyltransferases which govern methylation of CHG and CHH sequence motifs [46]. Consistent with this hypothesis, A. thaliana mutants (ddc and ago4), severely defective in non-CG methylation, developed much larger tumors than the wildtype controls. In the ddc mutant, for example, CHG and CHH methylation are reduced from 22% in the wildtype to only 1% and 7% of the total methylcytosines, respectively [47]. This indicates that wildtype plants restrict tumor growth by changes in the methylation pattern of cellular genes which are altered in ddc and ago4 mutants. As the integrated oncogene sequences are already unmethylated in wildtype tumors, growth restriction is most likely a result of differential methylation of endogenous plant genes.
In A. thaliana crown galls, the majority of DMRs are located in protein coding genes and transposable elements. However, differential methylation within the group of transposable elements is much lower than that of protein coding genes. This is in agreement with a report demonstrating that methylation of transposon sequences is much more stable than that of protein coding genes [48]. The higher stability of transposable element methylation is not surprising considering that loss of methylation of transposable elements has been shown to result in activation of their movement with occasionally mutagenic consequences [49]–[51].
Methylation changes in A. thaliana are known to be caused by endoreduplication which results in increased ploidy levels [12]. This is unlikely to happen in A. thaliana crown galls as the genome is rather stable in terms of ploidy level alterations. In contrast to the observed global hypermethylation, promoter sequences of protein coding genes in A. thaliana tumors were found to be rather hypomethylated and showed an increased level of gene expression. Methylation changes in transcribed regions were associated with transcriptional changes in the same direction. These observations are in accordance with previous studies of DNA methylation in A. thaliana [15], [29]. The studies revealed that gene body methylation was preferentially found in highly expressed genes while methylation near the TSS was associated with low gene expression levels. It is widely accepted that DNA methylation at the TSS inhibits transcription by interfering with transcriptional initiation. More controversy surrounds the role of DNA methylation in transcribed regions that may be important in preventing spurious transcription from internal promoters [29] or exon definition [52]. Overall, the results of this study implicate epigenetic processes, among others, as one mechanism to control gene expression in crown galls.
Little is known about plant signals that may affect the DNA methylation patterns and thereby gene expression. A process associated with A. thaliana crown gall development is ABA-dependent drought stress acclimation which was shown to be important for wildtype-like crown gall growth [7]. A. thaliana mutants in ABA-signaling (abi1-1, abi2-1, abi4-1) and -synthesis (aba3-1) display severely reduced crown gall growth. Under drought stress conditions photosynthesis is very much reduced [53]. Accordingly, in crown galls, which undergo increased water loss due to the lack of an intact epidermal layer [54], genes involved in photosynthetic light reactions are significantly downregulated [8]. The idea that biotic and abiotic stresses give rise to an epigenetic modification by ABA signaling has been put forward earlier [36]. For example, the pea genome has been shown to be hypermethylated as a response to water deficit [55]. In addition, ABA has previously been suggested to play a role in DNA methylation in a study from Khraiwesh et al. [56], who found that expression of stress-related genes in Physcomitrella patens is regulated by ABA in a methylation-dependent manner. In our studies, ABA-mediated methylation of promoters from photosynthesis-related genes caused their transcriptional silencing. The observed methylation and gene expression patterns were similar to those found in A. thaliana crown galls accumulating high levels of ABA. Thus, an increase of ABA levels induces promoter methylation and reduces gene expression. These observations suggest that ABA signaling pathways are interconnected with methylation processes in A. thaliana crown galls as a response to environmental stress. Unraveling the molecular mechanism underlying ABA-dependent DNA methylation will be an important task for future studies.
The global methylation patterns of mammalian tumors that are induced by integration of viral DNA into chromosomes, such as from Adenovirus type 12 (Ad12), differ from those of most other mammalian tumors. In Ad12-induced tumors, DNA integration into the eukaryotic genome causes de novo methylation in cis and trans, resulting in a hypermethylated genome [10]. Accordingly, hypermethylation in crown galls may be a consequence of the integration of bacterial DNA into the plant genome and the resulting acquisition of constitutive pathways favoring crown gall formation. Alterations in DNA methylation are most likely attributable to the methylation pattern of the transformed plant cells since it has been shown that almost every cell in crown galls expresses the T-DNA-encoded oncogenes and therefore has the potential for proliferation [8].
Until now, infection of A. thaliana with Pseudomonas syringae is the only plant-pathogen interaction which has been extensively studied with respect to DNA methylation changes [57]. It is accompanied by alterations in the methylation pattern and gene expression, preferentially of defense genes. In contrast to P. syringae, infections with virulent A. tumefaciens strains induce cell proliferation and crown gall growth. Consequently, the most severe DNA methylation changes in crown gall tumors were detected in genes involved in development, cell division and signaling. Differential methylation of cell division-related genes in A. thaliana crown galls is also in line with the observed increased rate of cell division. For example, the gene encoding a kinesin motor protein (AT1G63640) may contribute to cytoskeleton organization during cell division and cell growth as it is both less promoter-methylated and increasingly expressed in the tumor. Methylation seems to be linked to abiotic stress responses. Both the protein kinase CRK32 (At4g11480), which is upregulated in response to abiotic stress [58], and ABA-dependent photosynthesis-related genes (NDF4, SIG5, F12A4.4) are involved in stress-dependent signaling pathways and are differentially methylated in crown galls. The latter genes are severely downregulated in tumors, which are characterized by a heterotrophic metabolism and strong inhibition of photosynthesis genes. Thus, physiological and developmental adaptations during crown gall tumor growth seem to be controlled by epigenetic processes. Furthermore, these results suggest that in accordance with the prevailing response of the host towards a pathogen (development or defense), distinct sets of genes are regulated by DNA methylation in crown gall tumors and in tissues infected with P. syringae.
Taken together, this study demonstrates that essential processes during crown gall development are regulated by methylation, which alters the gene expression pattern and controls tumor development. We propose that hypermethylation of the plant tumor genome is a mechanism which restricts tumor growth, for example by affecting genes which are necessary for development and physiological adaptions. Growth restriction allows long-term coexistence of a developing tumor with the host plant and guarantees its survival.
Materials and Methods
Plant material
A. thaliana plants were cultivated in growth chambers at 22°C during the light and 16°C during the dark period in 12 h intervals. Plants used in this study included the wildtype accessions WS-2, Col-0 and Ler as well as the mutants ddc (drm1–2 drm2–2 cmt3–11, [59]), rdd (ros1-3; dml2-1; dml3-1, [26]) and ago4-1 [27].
Tumor induction, ABA treatment, and DNA extraction
Tumors were induced by injecting the nopaline-utilizing A. tumefaciens strain C58noc (nopaline catabolism; no. 584; Max-Planck-Institute for Plant Breeding Research) into the base of young inflorescence stalks (2 to 5 cm). Tumor tissue was separated from the host inflorescence stalk 28 d after inoculation under a stereo-zoom microscope (Leica MZ6, Leica Microsystems GmbH) using a scalpel. Mock-injected segments of tumor-free inflorescence stalks of the same age were used as reference tissue.
ABA treatment was conducted according to the protocol of Nishimura et al. [34]. In brief, A. thaliana seeds (ecotype WS-2) were stratified at 4°C for 4 days and were allowed to germinate on 0.8% agar supplemented with full strength Murashige and Skoog salts and 2% sucrose in the presence or absence of 0.5 µM ABA for two days.
Genomic DNA of all plant material was isolated by applying the DNeasy Plant Mini Kit (Qiagen) as outlined in the manufacturer's protocol.
Immunoprecipitation of methylated DNA fragments
Genomic DNA (1.1 µg) was sonicated with a Bioruptor (Diagenode) until fragments of approximately 600 bp were obtained. The DNA fragments were heated for 10 min at 99°C and immediately cooled on ice for 10 min. One hundred nanograms of genomic DNA fragments were used as input samples for array hybridization. Immunoprecipitation was performed by incubating 1 µg of sonicated DNA with 10 µg of 5-mC monoclonal antibody (Diagenode) in 600 µl IP-Buffer (10 mM Na-Phosphate Buffer, 0.14 M NaCl, 0.05% Triton X-100, pH 7.0) at 4°C for 12 h. Thereafter 100 µl of Dynabeads Protein G were added (Life Technologies), incubated at 4°C for 3 h and washed twice with 600 µl IP-Buffer for 10 min. DNA elution was performed by vortexing the Dynabeads three times in 200 µl TE buffer with increasing SDS-concentrations (0.1%, 0.5% and 1.5%). The DNA was purified by phenol-chloroform extraction and ethanol precipitation.
Tiling array analysis
Genomic input and mCIP DNA samples were labeled using the GeneChip Mapping 10K Xba kit and hybridized to GeneChip A. thaliana Tiling 1.0R arrays (both from Affymetrix) according to the instructions of the manufacturer. Locations of genomic probes were mapped to the TAIR9 version of the A. thaliana nuclear genome sequence. Raw array data of input and mCIP pairs were loess normalized. Signal log ratios (SLRs) of mCIP versus input were calculated for crown gall and mock inoculated stem tissue samples and finally quantiles normalized. Probe SLRs from the three biological replicates of each of the two groups were then subjected to a 500 bp sliding window median smoothing in order to create a robust and smoothed SLR (sSLR) for each probe position in each group. An implementation of the CMARRT algorithm [60], [61] was used for detection of genomic regions with consistently increased sSLRs across at least five consecutive probes for each of the two groups. For all regions displaying signal enrichments, log fold changes (logFCs) of crown gall tumor versus non-tumorous samples were calculated based on median region sSLRs. Thereafter, the distribution of logFCs was determined for the regions found to be enriched in the crown gall tumor as well as the tumor-free group (Dnull). Tumor-enriched regions of sSLRs with logFCs greater than the 75% quantile of Dnull were classified as hypermethylated, whereas those of sSLRs enriched in the tumor-free group with logFCs less than the 25% quantile of Dnull were defined as hypomethylated. These hyper - and hypomethylated regions were classified as differentially methylated regions (DMRs). All other regions were considered unchanged. The analyses were performed in R (http://www.r-project.org) along with the packages IRanges, Ringo and Starr (http://www.bioconductor.org).
Bisulfite sequencing
Bisulfite conversion of methylated cytosine nucleotides was conducted using the Epitect Bisulfite Kit (Qiagen) according to the manufacturer's protocol. A different number of PCR products, depending on the length of the analyzed DNA fragment were generated from the bisulfite-treated DNA, inserted into the pGEM-T easy vector (Promega) and cloned in E. coli XL1-Blue MRF′ cells. For each DNA locus multiple independent clones were sequenced, ten clones each for verification of microarrays, ABA-dependent methylation samples and analysis of oncogene methylation in tumors induced by transgenic Agrobacteria, five clones each for oncogene analysis wildtype tumors. Analysis of oncogene methylation of wildtype tumors was performed by analysis of 15 separate fragments covering the sequences of IaaH, IaaM, and Ipt. Primers used for bisulfite sequencing are listed in Table S3.
Constructs for oncogene silencing
The full lengths of the two IGRs between the coding sequences of IaaH, IaaM and Ipt, comprising 337 bp for IGR1 and 697 bp for IGR2 (Figure 1A) were separately cloned into pHellsgate12 [62] in sense and antisense orientation (Figure S1). This vector expresses the two self-complementary sequences of Ipt IGR and/or IaaH IGR separated by a PDK intron under control of the 35S promoter in planta. For generation of the construct with both IGR sequences, the hairpin cassette including 35S promoter, IGR1 hairpin and OCS terminator was PCR-amplified with primers containing SpeI adapter sequences (Table S3). Subsequently, this fragment was inserted into the recombinant pHellsgate plasmid already containing the hairpin construct of IGR2 using SpeI restriction and ligation. All recombinant pHellsgate12 plasmids were transformed into the virulent A. tumefaciens strain C58.
Flow cytometric measurements
Nuclei of non-tumorous stem and crown gall tumor tissue were isolated one month after mock injection or injection of A. tumefaciens strain C58, stained with propidium iodide (PI) as described previously [63] and analyzed using a FACStarPLUS flow cytometer (BD Biosciences) equipped with an INNOVA 90-C argon laser (Coherent). PI fluorescence was excited with 500 mW at 514 nm and measured in the FL1 channel using a 630 nm band-pass filter. Usually 10.000 nuclei per sample were analyzed.
RNA extraction, reverse transcription (RT), and qRT–PCR
RNA extraction, reverse transcription and qRT-PCR were conducted as described previously [64].
Data deposition
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE37680).
Supporting Information
Zdroje
1. ThomashowLS, ReevesS, ThomashowMF (1984) Crown gall oncogenesis: evidence that a T-DNA gene from the Agrobacterium Ti plasmid pTiA6 encodes an enzyme that catalyzes synthesis of indoleacetic acid. Proc Natl Acad Sci USA 81 : 5071–5075.
2. AkiyoshiDE, KleeH, AmasinoRM, NesterEW, GordonMP (1984) T-DNA of Agrobacterium tumefaciens encodes an enzyme of cytokinin biosynthesis. Proc Natl Acad Sci USA 81 : 5994–5998.
3. UllrichCI, AloniR (2000) Vascularization is a general requirement for growth of plant and animal tumours. J Exp Bot 51 : 1951–1960.
4. AloniR, WolfA, FeigenbaumP, AvniA, KleeHJ (1998) The Never ripe Mutant Provides Evidence That Tumor-Induced Ethylene Controls the Morphogenesis of Agrobacterium tumefaciens-Induced Crown Galls on Tomato Stems. Plant Physiol 117 : 841–849 doi:10.1104/pp.117.3.841.
5. HansenH, GrossmannK (2000) Auxin-induced ethylene triggers abscisic acid biosynthesis and growth inhibition. Plant Physiol 124 : 1437–1448 doi:10.1104/pp.124.3.1437.
6. VeselovD, LanghansM, HartungW, AloniR, FeussnerI, et al. (2003) Development of Agrobacterium tumefaciens C58-induced plant tumors and impact on host shoots are controlled by a cascade of jasmonic acid, auxin, cytokinin, ethylene and abscisic acid. Planta 216 : 512–522 doi:10.1007/s00425-002-0883-5.
7. EfetovaM, ZeierJ, RiedererM, LeeC-W, StinglN, et al. (2007) A Central Role of Abscisic Acid in Drought Stress Protection of Agrobacterium-Induced Tumors on Arabidopsis. Plant Physiol 145 : 853–862 doi:10.1104/pp.107.104851.
8. DeekenR, EngelmannJC, EfetovaM, CzirjakT, MüllerT, et al. (2006) An integrated view of gene expression and solute profiles of Arabidopsis tumors: a genome-wide approach. Plant Cell 18 : 3617–3634 doi:10.1105/tpc.106.044743.
9. MirouzeM, PaszkowskiJ (2011) Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol 14 : 267–274 doi:10.1016/j.pbi.2011.03.004.
10. HellerH, KämmerC, WilgenbusP, DoerflerW (1995) Chromosomal Insertion of Foreign (adenovirus Type 12, Plasmid, or Bacteriophage Lambda) DNA Is Associated with Enhanced Methylation of Cellular DNA Segments. Proc Natl Acad Sci USA 92 : 5515–5519.
11. BraunAC (1962) Tumor Inception and Development in the Crown Gall Disease. Annu Rev Plant Physiol 13 : 533–558 doi:10.1146/annurev.pp.13.060162.002533.
12. GelvinS, KarcherSJ, DiRitaVJ (1983) Methylation of the T-DNA in Agrobacterium tumefaciens and in several crown gall tumors. Nucleic Acids Res 11 : 159–174.
13. HepburnAG, ClarkeLE, PearsonL, WhiteJ (1983) The role of cytosine methylation in the control of nopaline synthase gene expression in a plant tumor. J Mol Appl Genet 2 : 315–329.
14. DunoyerP, HimberC, VoinnetO (2006) Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat Genet 38 : 258–263 doi:10.1038/ng1722.
15. ZhangX, YazakiJ, SundaresanA, CokusS, ChanSW-L, et al. (2006) Genome-wide High-Resolution Mapping and Functional Analysis of DNA Methylation in Arabidopsis. Cell 126 : 1189–1201 doi:10.1016/j.cell.2006.08.003.
16. LeeHS, ChenZJ (2001) Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc Natl Acad Sci USA 98 : 6753–6758 doi:10.1073/pnas.121064698.
17. LeeHO, DavidsonJM, DuronioRJ (2009) Endoreplication: polyploidy with purpose. Genes Dev 23 : 2461–2477 doi:10.1101/gad.1829209.
18. GalbraithDW, HarkinsKR, KnappS (1991) Systemic Endopolyploidy in Arabidopsis thaliana. Plant Physiol 96 : 985–989 doi:10.1104/pp.96.3.985.
19. IshidaT, AdachiS, YoshimuraM, ShimizuK, UmedaM, et al. (2010) Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137 : 63–71 doi:10.1242/dev.035840.
20. HendersonIR, JacobsenSE (2007) Epigenetic inheritance in plants. Nature 447 : 418–424 doi:10.1038/nature05917.
21. CaoX, JacobsenS (2002) Role of the Arabidopsis DRM Methyltransferases in De Novo DNA Methylation and Gene Silencing. Curr Biol 12 : 1138–1144 doi:10.1016/S0960-9822(02)00925-9.
22. ZilbermanD, CaoX, JohansenLK, XieZ, CarringtonJC, et al. (2004) Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol 14 : 1214–1220 doi:15242620.
23. MetteMF, AufsatzW, van der WindenJ, MatzkeMA, MatzkeAJM (2000) Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J 19 : 5194–5201 doi:10.1093/emboj/19.19.5194.
24. SazeH, ScheidOM, PaszkowskiJ (2003) Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34 : 65–69 doi:10.1038/ng1138.
25. JacksonJP, LindrothAM, CaoX, JacobsenSE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 : 556–560 doi:10.1038/nature731.
26. PentermanJ, ZilbermanD, HuhJH, BallingerT, HenikoffS, et al. (2007) DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA 104 : 6752–6757 doi:10.1073/pnas.0701861104.
27. ZilbermanD, CaoX, JacobsenS (2003) ARGONAUTE4 Control of Locus-Specific siRNA Accumulation and DNA and Histone Methylation. Science 299 : 716–719 doi:10.1126/science.1079695.
28. BenderJ (2004) Chromatin-based silencing mechanisms. Current Opinion in Plant Biology 7 : 521–526 doi:10.1016/j.pbi.2004.07.003.
29. ZilbermanD, GehringM, TranRK, BallingerT, HenikoffS (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39 : 61–69 doi:10.1038/ng1929.
30. BarowM, MeisterA (2003) Endopolyploidy in seed plants is differently correlated to systematics, organ, life strategy and genome size. Plant Cell Environ 26 : 571–584 doi:10.1046/j.1365-3040.2003.00988.x.
31. ThimmO, BläsingO, GibonY, NagelA, MeyerS, et al. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 : 914–939 doi:10.1111/j.1365-313X.2004.02016.x.
32. UsadelB, NagelA, ThimmO, RedestigH, BlaesingOE, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138 : 1195–1204 doi:10.1104/pp.105.060459.
33. HruzT, LauleO, SzaboG, WessendorpF, BleulerS, et al. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008 : 420747 doi:10.1155/2008/420747.
34. NishimuraN, YoshidaT, KitahataN, AsamiT, ShinozakiK, et al. (2007) ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50 : 935–949 doi:10.1111/j.1365-313X.2007.03107.x.
35. XiaoW, CustardKD, BrownRC, LemmonBE, HaradaJJ, et al. (2006) DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 18 : 805–814 doi:10.1105/tpc.105.038836.
36. ChinnusamyV, GongZ, ZhuJ-K (2008) Abscisic Acid-mediated Epigenetic Processes in Plant Development and Stress Responses. J Integr Plant Biol 50 : 1187–1195 doi:10.1111/j.1744-7909.2008.00727.x.
37. MatzkeMA, MetteMF, MatzkeAJ (2000) Transgene silencing by the host genome defense: implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol Biol 43 : 401–415 doi:10.1023/A:1006484806925.
38. EscobarM, CiveroloE, SummerfeltK, DandekarA (2001) RNAi-mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proc Natl Acad Sci USA 98 : 13437–13442 doi:10.1073/pnas.241276898.
39. EscobarM, CiveroloE, PolitoV, PinneyK, DandekarA (2003) Characterization of oncogene-silenced transgenic plants: implications for Agrobacterium biology and post-transcriptional gene silencing. Mol Plant Pathol 4 : 57–65 doi:10.1046/j.1364-3703.2003.00148.x.
40. LeeH, HumannJL, PitrakJS, CuperusJT, ParksTD, et al. (2003) Translation Start Sequences Affect the Efficiency of Silencing of Agrobacterium tumefaciens T-DNA Oncogenes. Plant Physiol 133 : 966–977 doi:10.1104/pp.103.026534.
41. OomsG, HooykaasPJ, MoolenaarG, SchilperoortRA (1981) Grown gall plant tumors of abnormal morphology, induced by Agrobacterium tumefaciens carrying mutated octopine Ti plasmids; analysis of T-DNA functions. Gene 14 : 33–50.
42. ReamLW, GordonMP, NesterEW (1983) Multiple mutations in the T region of the Agrobacterium tumefaciens tumor-inducing plasmid. Proc Natl Acad Sci USA 80 : 1660–1664.
43. AufsatzW, MetteMF, van der WindenJ, MatzkeAJM, MatzkeM (2002) RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA 99 : 16499–16506 doi:10.1073/pnas.162371499.
44. SlotkinRK, VaughnM, TanurdžicM, BorgesF, BeckerJD, et al. (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136 : 461–472 doi:10.1016/j.cell.2008.12.038.
45. MosherRA, MelnykCW, KellyKA, DunnRM, StudholmeDJ, et al. (2009) Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460 : 283–286 doi:10.1038/nature08084.
46. CaoX, AufsatzW, ZilbermanD, MetteMF, HuangMS, et al. (2003) Role of the DRM and CMT3 Methyltransferases in RNA-Directed DNA Methylation. Curr Biol 13 : 2212–2217 doi:10.1016/j.cub.2003.11.052.
47. ListerR, O'MalleyRC, Tonti-FilippiniJ, GregoryBD, BerryCC, et al. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133 : 523–536 doi:10.1016/j.cell.2008.03.029.
48. VaughnMW, TanurdžićM, LippmanZ, JiangH, CarrasquilloR, et al. (2007) Epigenetic Natural Variation in Arabidopsis thaliana. PLoS Biol 5: e174 doi:10.1371/journal.pbio.0050174.
49. MiuraA, NakamuraM, InagakiS, KobayashiA, SazeH, et al. (2009) An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J 28 : 1078–1086 doi:10.1038/emboj.2009.59.
50. SingerT, YordanC, MartienssenRA (2001) Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev 15 : 591–602 doi:10.1101/gad.193701.
51. KatoM, MiuraA, BenderJ, JacobsenS, KakutaniT (2003) Role of CG and Non-CG Methylation in Immobilization of Transposons in Arabidopsis. Current Biology 13 : 421–426 doi:10.1016/S0960-9822(03)00106-4.
52. ChodavarapuRK, FengS, BernatavichuteYV, ChenP, StroudH, et al. (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466 : 388–392 doi:10.1038/nature09147.
53. FlexasJ, MedranoH (2002) Drought‐inhibition of Photosynthesis in C3 Plants: Stomatal and Non‐stomatal Limitations Revisited. Ann Bot 89 : 183–189 doi:10.1093/aob/mcf027.
54. SchurrU, SchuberthB, AloniR, PradelKS, SchmundtD, et al. (1996) Structural and functional evidence for xylem-mediated water transport and high transpiration in Agrobacterium tumefaciens-induced tumors of Ricinus communis. Bot Acta 109 : 405–411.
55. LabraM, GhianiA, CitterioS, SgorbatiS, SalaF, et al. (2002) Analysis of Cytosine Methylation Pattern in Response to Water Deficit in Pea Root Tips. Plant Biology 4 : 694–699 doi:10.1055/s-2002-37398.
56. KhraiweshB, ArifMA, SeumelGI, OssowskiS, WeigelD, et al. (2010) Transcriptional Control of Gene Expression by MicroRNAs. Cell 140 : 111–122 doi:10.1016/j.cell.2009.12.023.
57. DowenR, PelizzolaM, SchmitzRJ, ListerR, DowenJM, et al. (2012) Widespread Dynamic DNA Methylation in Response to Biotic Stress. Proc Natl Acad Sci USA 109: E2183–2191 doi:10.1073/pnas.1209329109.
58. WrzaczekM, BroschéM, SalojärviJ, KangasjärviS, IdänheimoN, et al. (2010) Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol 10 : 95 doi:10.1186/1471-2229-10-95.
59. ChanS, HendersonI, ZhangX, ShahG, ChienJ, et al. (2006) RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PLoS Genet 2: e83 doi:10.1371/journal.pgen.0020083.
60. KuanPF, ChunH, KeleşS (2008) CMARRT: a tool for the analysis of ChIP-chip data from tiling arrays by incorporating the correlation structure. Pac Symp Biocomput 515–526.
61. ZacherB, KuanPF, TreschA (2010) Starr: Simple Tiling ARRay analysis of Affymetrix ChIP-chip data. BMC Bioinformatics 11 : 194 doi:10.1186/1471-2105-11-194.
62. HelliwellC, WaterhouseP (2003) Constructs and methods for high-throughput gene silencing in plants. Methods 30 : 289–295 doi:10.1016/S1046-2023(03)00036-7.
63. SreenivasuluN, RadchukV, AlawadyA, BorisjukL, WeierD, et al. (2010) De-regulation of abscisic acid contents causes abnormal endosperm development in the barley mutant seg8. Plant J 64 : 589–603 doi:10.1111/j.1365-313X.2010.04350.x.
64. LeeC-W, EfetovaM, EngelmannJC, KramellR, WasternackC, et al. (2009) Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell 21 : 2948–2962 doi:10.1105/tpc.108.064576.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



