-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEncodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
Land plants have evolved increasingly complex regulatory modes of their flowering time (or heading date in crops). Rice (Oryza sativa L.) is a short-day plant that flowers more rapidly in short-day but delays under long-day conditions. Previous studies have shown that the CO-FT module initially identified in long-day plants (Arabidopsis) is evolutionary conserved in short-day plants (Hd1-Hd3a in rice). However, in rice, there is a unique Ehd1-dependent flowering pathway that is Hd1-independent. Here, we report isolation and characterization of a positive regulator of Ehd1, Early heading date 4 (Ehd4). ehd4 mutants showed a never flowering phenotype under natural long-day conditions. Map-based cloning revealed that Ehd4 encodes a novel CCCH-type zinc finger protein, which is localized to the nucleus and is able to bind to nucleic acids in vitro and transactivate transcription in yeast, suggesting that it likely functions as a transcriptional regulator. Ehd4 expression is most active in young leaves with a diurnal expression pattern similar to that of Ehd1 under both short-day and long-day conditions. We show that Ehd4 up-regulates the expression of the “florigen” genes Hd3a and RFT1 through Ehd1, but it acts independently of other known Ehd1 regulators. Strikingly, Ehd4 is highly conserved in the Oryza genus including wild and cultivated rice, but has no homologs in other species, suggesting that Ehd4 is originated along with the diversification of the Oryza genus from the grass family during evolution. We conclude that Ehd4 is a novel Oryza-genus-specific regulator of Ehd1, and it plays an essential role in photoperiodic control of flowering time in rice.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003281
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003281Summary
Land plants have evolved increasingly complex regulatory modes of their flowering time (or heading date in crops). Rice (Oryza sativa L.) is a short-day plant that flowers more rapidly in short-day but delays under long-day conditions. Previous studies have shown that the CO-FT module initially identified in long-day plants (Arabidopsis) is evolutionary conserved in short-day plants (Hd1-Hd3a in rice). However, in rice, there is a unique Ehd1-dependent flowering pathway that is Hd1-independent. Here, we report isolation and characterization of a positive regulator of Ehd1, Early heading date 4 (Ehd4). ehd4 mutants showed a never flowering phenotype under natural long-day conditions. Map-based cloning revealed that Ehd4 encodes a novel CCCH-type zinc finger protein, which is localized to the nucleus and is able to bind to nucleic acids in vitro and transactivate transcription in yeast, suggesting that it likely functions as a transcriptional regulator. Ehd4 expression is most active in young leaves with a diurnal expression pattern similar to that of Ehd1 under both short-day and long-day conditions. We show that Ehd4 up-regulates the expression of the “florigen” genes Hd3a and RFT1 through Ehd1, but it acts independently of other known Ehd1 regulators. Strikingly, Ehd4 is highly conserved in the Oryza genus including wild and cultivated rice, but has no homologs in other species, suggesting that Ehd4 is originated along with the diversification of the Oryza genus from the grass family during evolution. We conclude that Ehd4 is a novel Oryza-genus-specific regulator of Ehd1, and it plays an essential role in photoperiodic control of flowering time in rice.
Introduction
Flowering is a profound transition from vegetative to reproductive development in plants, and is largely determined by genetic pathways that integrate endogenous and environmental signals [1]. Plants control flowering by perceiving their surroundings, such as day-length (photoperiod) and temperature that is synchronized with seasonal changes, in order to maximize their reproductive fitness [2]. Flowering time or heading date in crops is also a critical agronomic trait that determines the cropping season and regional adaptability of plants. Thus, control of flowering time has been extensively studied by plant breeders and scientists for more than 100 years [3].
Photoperiod control of flowering refers to the ability of plants to measure day-length and use it as an indicator to initiate flowering. Extensive studies in a model long-day plant (LDP), Arabidopsis thaliana, have revealed that light regulation of the GIGANTEA (GI)-CONSTANT (CO)-FLOWERING LOCUS T (FT) pathway is essential for integrating cellular signals from light signaling transduction and the circadian clock to promote flowering under long-day conditions (LDs) [4]–[6]. Phytochrome A (phyA), phytochrome B (phyB) and cryptochrome 2 (cry2) regulate FT expression by post-translationally regulating CO protein [7], [8]. In addition, blue light promotes CO expression by stabilizing the FLAVIN-binding KELCH DOMAIN F BOX PROTEIN1 (FKF1)-GI protein complex [9], [10]. CO, a zinc finger transcription factor, promotes FT expression under LDs by directly binding to its promoter [11], [12]. FT, a small mobile protein functioning as the ‘florigen’, is synthesized in the phloem of leaves, and is then transported to the apical meristem where it initiates flowering by inducing the expression of the floral meristem identity genes, such as AP1 [13]–[15].
Rice (Oryza sativa L.) is an important source of calories for mankind and a model short-day plant (SDP) that flowers more rapidly in short-day conditions (SDs) but delays under LDs with a critical day-length response [16], [17]. Previous studies have revealed that rice flowering is regulated both by a “SD-activation pathway” and a “LD-suppression pathway”. OsGIGANTEA (OsGI), Heading date 1 (Hd1) and Heading date 3a (Hd3a) have been identified as the counterpart of GI, CO and FT, respectively [18]–[20]. Hd1 executes dual function that promotes flowering by regulating Hd3a (the major SD ‘florigen’) expression under SDs, but suppresses it through unknown mechanisms under LDs [19], [21], [22]. However, the OsGI-Hd1-Hd3a pathway only plays a limited role in flowering time control in rice because there is a high degree of polymorphism in Hd1 and non-functional alleles of Hd1 are associated with only moderate phenotypic changes [23].
Rice has a unique, Hd1-independent flowering pathway that is mediated by Early heading date 1 (Ehd1). Ehd1 encodes a B-type response regulator that is highly conserved in cultivated rice, but has no homolog in Arabidopsis [23], [24]. It has been shown that Ehd1 positively regulates the expression of Hd3a and RICE FLOWERING LOCUS T 1 (RFT1), the closest paralog of Hd3a that works as a LD ‘florigen’ 22,24,25. Circumstantial evidence suggests that Ehd1 is a critical convergence point of regulation by multiple signaling pathways. Among them, OsphyB inhibits flowering under both SDs and LDs by suppressing Hd3a expression through posttranslational modification of HD1 protein function and transcriptional suppression of Ehd1 expression [25]–[27]. The OsphyB-mediated suppression of Ehd1 is regulated by OsCOL4, which encodes a protein containing two B-box zinc finger domains and one CCT domain and it also acts as a constitutive suppressor of flowering in rice under both SD and LD conditions [26], [27]. In addition, both Ghd7 (Grain number, plant height and heading date 7), encoding a CCT domain protein [28], and DTH8 (Days to heading 8), encoding a putative HAP3 subunit of the CCAAT-box-binding transcription factor, down-regulate Ehd1 expression and delay flowering under LDs [29]. On the other hand, Ehd1 expression is promoted by a number of positive regulators. Among them, OsMADS51 encodes a type I MADS-box protein and induces Ehd1 expression under SDs [30], whereas a rice homolog of Arabidopsis SOC1 (Suppressor of Overexpression of Constant1), OsMADS50, was identified as a promoter of Ehd1 expression under LDs [31]. Recently, it was shown that Ehd1 expression could be independently up-regulated by Early heading date 2/Rice Indeterminate 1/Oryza sativa Indeterminate 1 (referred to as Ehd2 hereafter) and Early heading date 3 (Ehd3) under both SDs and LDs [32]–[34]. The former encodes a Cys2/His2-type zinc finger protein with high homology to maize Indeterminate1 [35], while the latter encodes a putative plant homeodomain (PHD) finger-containing protein. Notably, loss of function of OsMADS50, Ehd2 and Ehd3 showed a never-flowering phenotype under LDs [25], [32], [34], [36]. Thus, it appears that OsMAD50, Ehd2, Ehd3 and Ehd1 may constitute a “LD-activation pathway” in rice. Although these studies have revealed much insight into the photoperiodic flowering of rice, the underlying molecular mechanisms are still not well understood.
Here we report the identification of Early heading date 4 (Ehd4) using a mutagenesis approach and its positional cloning. Ehd4 encodes a novel CCCH (C-X7-C-X5-C-X3-H)-type zinc finger protein and it acts as a critical regulator promoting flowering under both SDs and LDs, particularly under LDs. Mutation in Ehd4 causes a never-flowering phenotype under natural long-day conditions (NLDs). EHD4 protein is localized to the nucleus and it has nucleic acid-binding and transcriptional activation properties, consistent with a plausible function as a transcription factor. We show that Ehd4 promotes flowering by up-regulating the expression of Hd3a and RFT1 through stimulation of Ehd1 expression. Interestingly, Ehd4 is highly conserved in the Oryza genus and it has no homologs in other plant species. Thus, our findings identified a novel, highly conserved rice-specific regulator of flowering time.
Results
Characterization of the late flowering mutant ehd4
In an effort to isolate genes that are essential for promoting flowering time in rice, we generated a large T-DNA population in a day-length neutral, early flowering variety Kita-ake (O. sativa ssp. japonica). Kita-ake (Kit) has been widely used in rice transformation experiments because of its short life cycle. Kit flowers about two months after germination under both SDs (10 h light/14 h dark) and LDs (14.5 h light/9.5 h dark) conditions in the controlled growth chamber, as well as under natural long-day field conditions (NLDs) in Beijing (39°54′N, 116°23′E), North China (Figure 1A and 1B). To understand the day-length neutral nature of Kita-ake, we cloned ten genes reported to have significant effect on flowering time in rice, including seven genes that promote flowering (Ehd 1 to 3, OsMADS50, OsMADS51, Hd3a and RFT1) and three genes that suppress flowering under LDs (Hd6, Hd1 and Ghd7), and compared them with the corresponding genes in Nipponbare (Nip), a japonica variety that is sensitive to day-length. Those flowering-promoting genes are identical in Kit and Nip varieties, except OsMADS51 that contains one amino acid variation (Figure S1). In contrast, Kit has an immature stop in Ghd7 and a 36-bp insertion and two amino acid changes in Hd1 (Figure S1). Although there is no difference in Hd6 sequences between Kit and Nip, both of them have an early stop compared with the allele of the indica variety Kasalash (Figure S1), which delays flowering in Nip background under LDs [37], [38]. Therefore, complete or partial loss of function of those three genes in Kit could at least partially explain its insensitivity to day-length.
Fig. 1. Characterization of Ehd4. 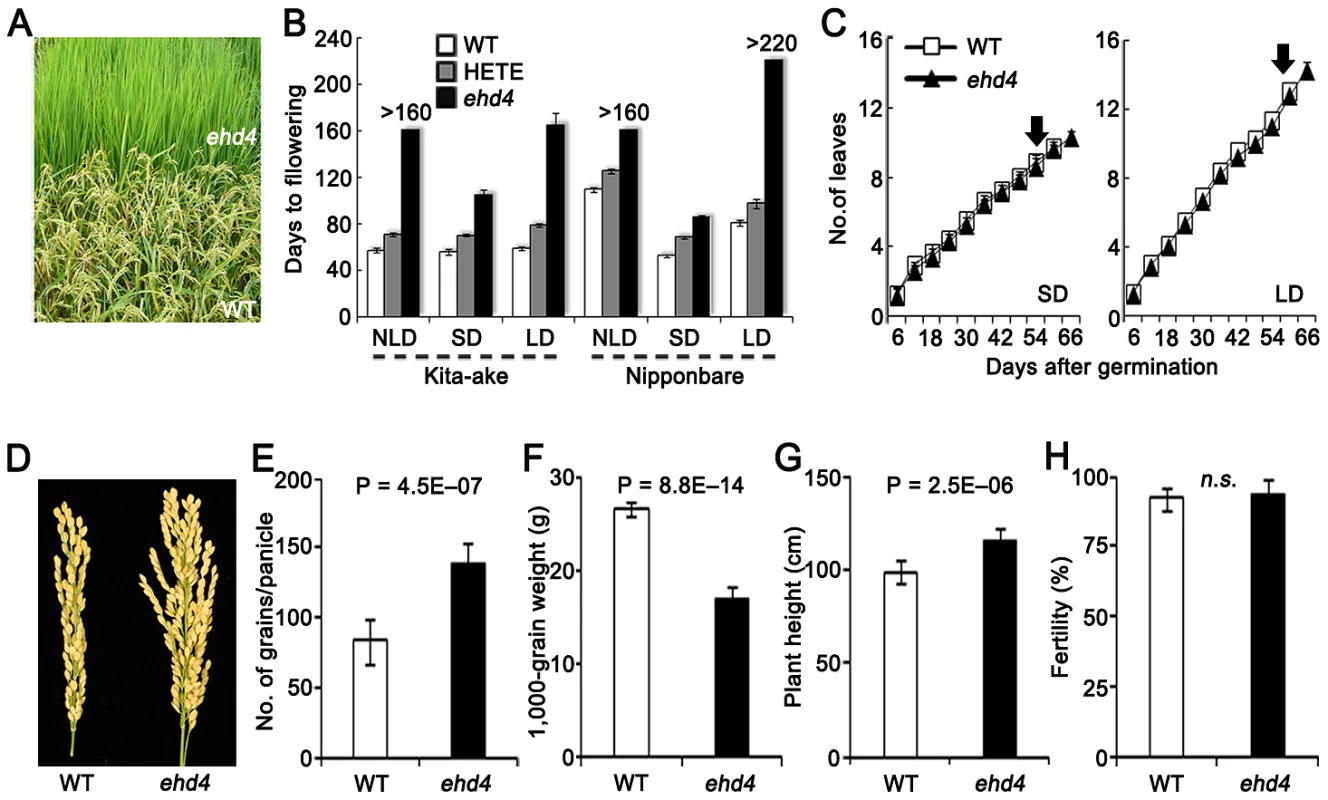
(A) Never-flowering phenotype of ehd4 mutants in field (Top). WT, Kita-ake wild-type plants (Bottom). (B) Flowering time of ehd4, heterozygote (HETE) and WT plants under different day length conditions in Kita-ake (day-length neutral) and Nipponbare (day-length sensitive) backgrounds (n = 12). ND, natural-day; SD, short-day; LD, long-day. (C) ehd4 plants had the same leaf emergence rate as WT (Kita-ake) under both SDs and LDs (n = 8). Arrow indicates the flowering time of WT plants. (D) Panicle morphology of WT and ehd4 plants. (E) to (H) Comparisons of grain number per panicle (E), 1000-grain weight (F), plant height (G) and fertility (H) between WT and ehd4 plants. Values are means±s.d. (standard deviations) (n = 15). **Significant at 1% level; n.s., not significant. We screened our T-DNA population in the NLDs and identified a mutant that failed to flower during the 160 days of growing season (from late April to early October, 2006), whereas the wild-type (WT) Kit flowered 55 days after germination (Figure 1A and 1B). We were able to produce seeds by moving this mutant plant to a controlled SDs. Plants from an F2 population derived from a cross of the mutant and WT segregated in field conditions into three categories based on their flowering time (days after germination): 56.9±1.8, 70.8±1.8 and never flowering mutants in a ratio of 1∶2∶1 (χ2[1∶2∶1] = 0.415<χ20.05,2 = 5.99, n = 200). This result indicates that the mutation is semidominant and is controlled by a single gene. We named this locus Ehd4 (Early heading date 4). Compared with WT, ehd4 delayed flowering time by 49 d and 106 d under SDs and LDs, respectively (Figure 1B). Consistent with field observations, flowering time of the heterozygotes was also delayed under both SDs and LDs (Figure 1B). Notably, ehd4 had a similar leaf emergence rate to WT under both SDs and LDs (Figure 1C), indicating that the late flowering phenotype is not caused by retardation in growth rate. The mature ehd4 plants were taller, producing more but smaller seeds. The fertility of ehd4 plants was similar to that of WT (Figure 1D–1H).
To test whether the delayed flowering phenotype is genetic background-dependent, we introduced the ehd4 locus into Nip by backcrossing five times, followed by selfing. The ehd4-NIP plants (BC5F3) delayed flowering by 23 d under SDs compared to the WT NIP, but did not flower in NLDs or LDs (Figure 1B). Flowering time of the heterozygotes was also significantly delayed (Figure 1B). Thus, ehd4 has a profound effect on flowering time, especially under LDs, in both a day-length neutral and a day-length sensitive genetic backgrounds.
Molecular cloning of Ehd4
Flowering time control in rice is regulated by the interaction of multiple QTLs and the environments. Generally, flowering time of F2 population derived from japonica×indica displays a normal distribution pattern. Therefore, we crossed ehd4 with 93-11, an indica variety with an available genome sequence [39], and generated a BC1F2 population for mapping the ehd4 locus by backcrossing the F1 with 93-11. The ehd4 locus was initially mapped to the short arm of chromosome 3 (Figure 2A). Using 871 extremely late flowering plants from approximately 25,000 BC1F2 plants grown in Hainan Island (18°48′N, 110°02′E, average day length 11 hours), South China, during the winter of 2008, we further delimited Ehd4 to a 103 kb region, between the markers EJ-4 and EJ-5 (Figure 2B). This region contains 16 annotated ORFs (http://rapdb.dna.affrc.go.jp) (Figure 2C). Sequencing of the genomic DNA of all these genes revealed that there is a single nucleotide substitution (G to A) in the first exon of LOC_Os03g02160, which is predicted to encode a CCCH-Type zinc finger protein. The nucleotide change creates a premature stop codon at the very beginning of the predicted coding region (Figure 2D). Genomic sequence of this gene is identical between Kit and Nip (Figure S1). Quantitative real-time PCR (qRT-PCR) assay showed comparable expression of LOC_Os03g02160 in wild type, heterozygote and ehd4 mutant plants (Figure S2). Transgenic plants carrying the full-length cDNA of LOC_Os03g02160, driven by the maize Ubiquitin-1 promoter, fully complemented the ehd4 phenotype under both SDs and LDs. Further, cDNA driven by its native promoter (2.7 kb upstream from ATG) also partially rescued the ehd4 phenotype. The phenotypes of these transgenic lines (days to flowering) appeared to correlate with the expression level of Ehd4 (Figure 2E and Figure S2). Thus, we concluded that the LOC_Os03g02160 locus corresponds to Ehd4.
Fig. 2. Map-based cloning of Ehd4. 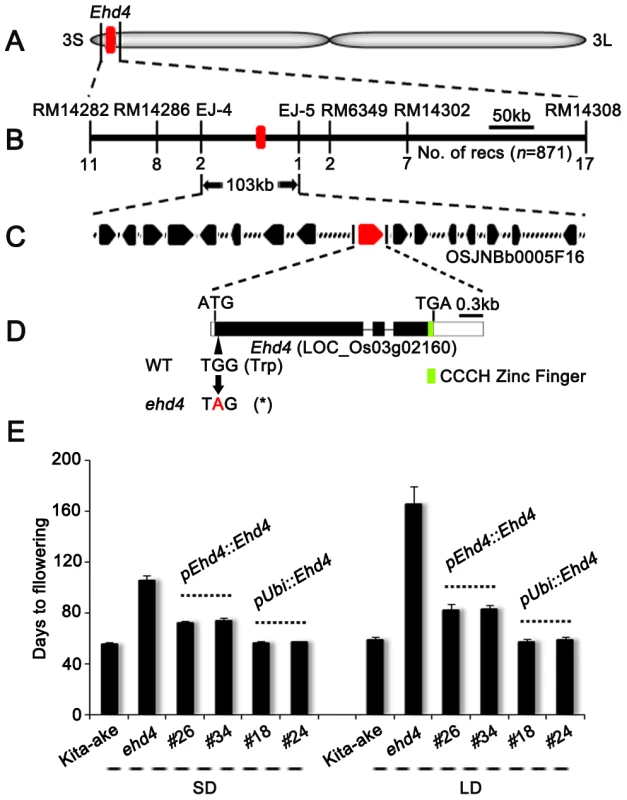
(A) Location of the Ehd4 locus on rice chromosome 3. (B) High-resolution linkage map of Ehd4. (C) Candidate genes on BAC OSJNBb0005F16. (D) Structure of the Ehd4 gene. Lines, black and white boxes represent introns, exons and untranslated regions, respectively. The base change from G to A creates an early stop codon (Asterisk). (E) Complementation of ehd4. Ehd4 was driven by either the native promoter (pEhd4::Ehd4) or the maize Ubiquitin-1 promoter (pUbi::Ehd4). T2 plants of two pEhd4::Ehd4 lines (#26 and #34) and two pUbi::Ehd4 lines (#18 and #24) were measured (n = 10). All plants were grown under both SD and LD conditions. Expression of Ehd4 is constitutive and diurnal
We examined the expression levels of Ehd4 in various tissues and at different stages of leaf development (Figure 3A) by using qRT-PCR. Ehd4 transcripts were detected in all tissues examined, but the highest expression was found in emerging young leaves and the lowest level in fully expanded leaves (Figure 3B). Histochemical staining of transgenic plants carrying the GUS reporter gene driven by the Ehd4 promoter indicated that GUS was expressed in all tissues examined and was most abundant in the vascular tissue and apical meristem (Figure 3C–3I). The expression of Ehd4 showed a diurnal expression pattern in leaves. It accumulates after dusk, reaching a peak at dawn, and damping rapidly thereafter under both SDs and LDs (Figure 3J). Moreover, Ehd4 was expressed constantly during the vegetative growth from the second week to the 10th week after germination (Figure 3K).
Fig. 3. Expression pattern of Ehd4. 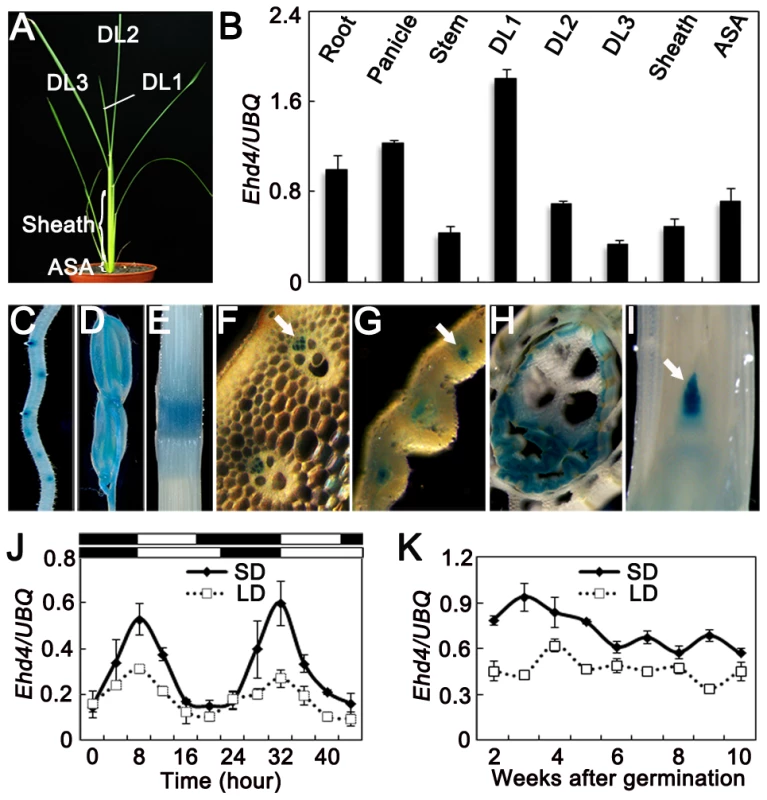
(A) 30-d old wild-type plants (Kita-ake) grown under SDs were used for quantitative RT-PCR. DL1, newly emerging leaf; DL2, expending leaf; DL3, fully expended leaf; ASA, around the shoot apex. (B) Ehd4 transcript levels in various organs (means±s.d, n = 3). (C) to (I) GUS staining of various organs in pEHD4::GUS transgenic plants. (C) Root; (D) Floret; (E) Stem; (F) to (H) Transverse sections of stem, immature leaf and sheath, respectively; (I) Longitudinal section of the shoot apical meristem (SAM). Arrow indicates phloem in (F) and (G) and SAM in (I). (J) and (K) Rhythmic and developmental expression of Ehd4. The rice Ubiquitin-1 (UBQ) gene was used as the internal control. Values are shown as mean±s.d of three independent experiments and two biological replicates. The open and filled bars at the bottom represent the light and dark periods, respectively. s.d: standard deviations. EHD4 may act as a transcriptional regulator
In higher plants, CCCH-type zinc finger proteins have been shown to regulate gene expression by binding to DNA or RNA molecules in the nucleus [40]–[42]. We fused Ehd4 with GFP and transiently expressed the EHD4-GFP fusion protein in rice leaf protoplasts. EHD4-GFP was exclusively co-localized with the OsMADS3-mCherry fusion protein (a nuclear marker) in the nucleus (Figure 4A–4C), indicating that EHD4 functions in the nucleus. We further fused EHD4 and its various deletions with the GAL4 DNA binding domain and investigated if EHD4 has transcriptional activation activity in yeast. Full-length wild type EHD4 and an EHD4 variant with only the CCCH motif removed were able to activate the reporter gene expression (Figure 4D). Further deletion of the C terminal region resulted in a dramatic reduction of the activation activity, whereas deletion of both the N-terminal and CCCH motif only had mild effects (Figure 4D). These observations suggest that the activation domain is located in the middle region close to the C-terminal of EHD4. In addition, a nucleic acid binding assay demonstrated that the C-terminal region, but not the N-terminal region, can bind to both double - and single-stranded calf thymus DNA and ribohomopolymers in vitro, and that removal of the CCCH motif from the C-terminal abolished the binding activity (Figure 4E). These results strongly support the notion that EHD4 likely functions as a transcriptional activator and that the CCCH motif is essential for its nucleic acid binding activity.
Fig. 4. EHD4 is a nuclear protein with intrinsic transcriptional activation and nucleic acid binding activities. 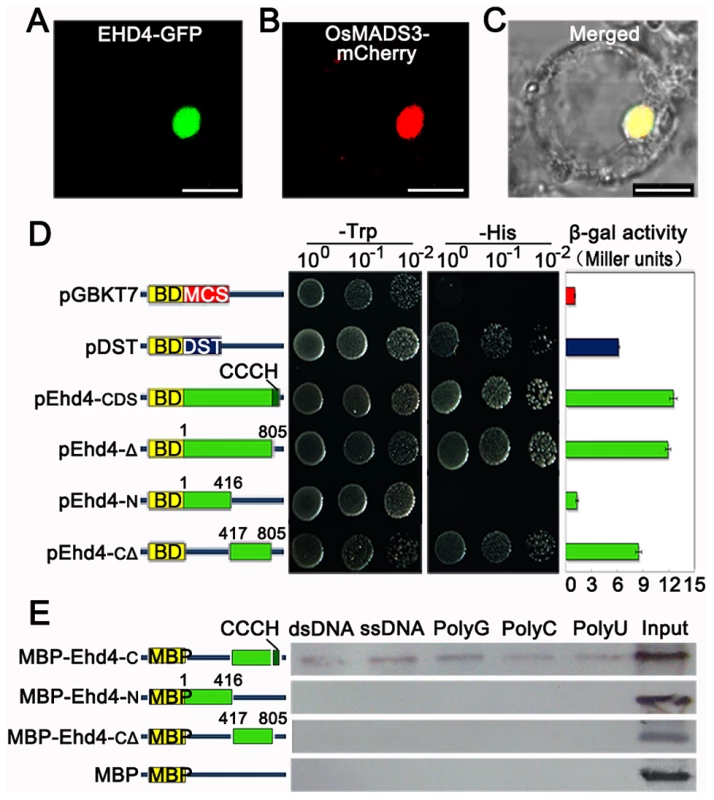
(A) Sub-cellular localization of EHD4-GFP fusion protein. (B) The nuclear marker MADS3-mCherry fusion protein. (C) Merged image of (A) and (B) under bright field. Scale bar = 10 µm in (A) to (C). (D) Transactivation assays of EHD4 and its deletion derivatives in the yeast GAL4 system. Full length EHD4 and several deletion derivatives of EHD4 (pEhd4-Δ, pEhd4-N and pEhd4-CΔ) were used in assays. The empty vector (BD-MCS) and BD-DST [56] were used as negative and positive control, respectively. Transformants were dropped onto SD/Trp- and SD/His- plates to allow growth of 48 hours before taking pictures. Values in β-galactosidase activity are means of three independent experiments. Bars stand for standard deviations. BD, DNA-binding domain of GAL4. (E) The CCCH motif is essential for binding to nucleic acids. C terminal, N terminal or C terminal without CCCH motif of EHD4 was expressed in E.coli and purified for binding assays. Deletion of the CCCH motif abolished the binding to ribohomopolymers and both double- and single-stranded calf thymus DNA. Ehd4 regulates expression of the “florigen” genes through Ehd1
Photoperiodic induction of the floral transition in rice requires transcriptional activation of Hd3a and RFT1, the two ‘florigen’ genes, in leaves [20]–[22]. The diurnal expression pattern of Ehd4 implies that it could be involved in photoperiodic control of flowering. To test this, we examined mRNA abundance of Hd3a and RFT1 in ehd4 and WT plants by qRT-PCR. The expression levels of Hd3a and RFT1 were undetectable in ehd4 under both SDs and LDs at all-time points examined during the 48 h period (Figure 5A–5D). Subsequently, expression of the downstream genes OsMADS1, OsMADS14 and OsMADS15 (three floral meristem identity genes; [22], [25]) was severely impaired in the ehd4 mutants (Figure S3).
Fig. 5. The rhythmic expression pattern of Hd3a, RFT1, and Ehd1, but not Hd1, was abolished in the ehd4 mutant plants under both SDs and LDs. 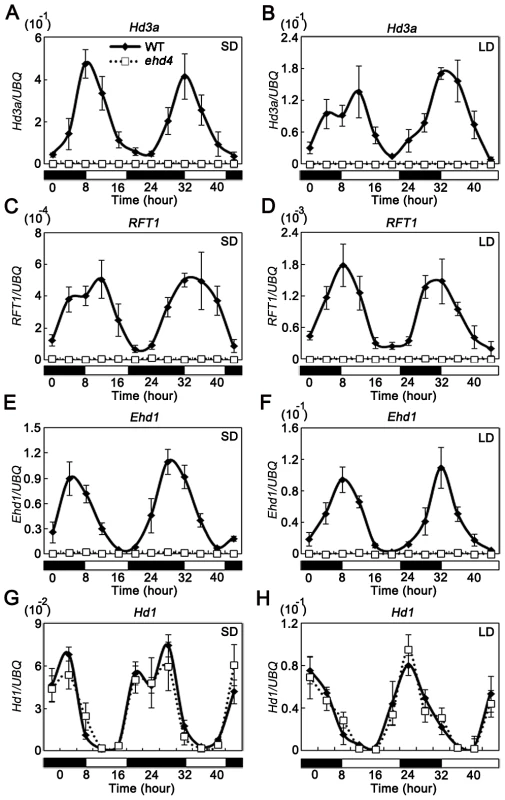
SDs (A, C, E and G); LDs (B, D, F and H). The open and filled bars at the bottom represent the light and dark periods, respectively. The rice Ubiquitin-1 (UBQ) gene was used as the internal control. Values are shown as mean±s.d. (standard deviations) of three independent experiments and two biological replicates. Hd1 and Ehd1 are known to regulate Hd3a and RFT1 [19], [24]. To investigate whether the activation of ‘florigen’ genes by Ehd4 is mediated by Hd1 and/or Ehd1, we compared their mRNA levels between ehd4 and WT plants. Strikingly, expression of Ehd1, but not Hd1, was abolished in ehd4 mutants, indicating that Ehd4 is essential for Ehd1 expression (Figure 5E–5H). Ehd4 has a diurnal expression pattern similar to that of Ehd1, typically peaking at dawn (Compare Figure 3J with Figure 5E and 5F). Next, we examined whether Ehd4 affects the expression of other known regulators of Ehd1. To our surprise, the transcription levels of five positive regulators (Ehd2, Ehd3, OsMADS50, OsGI and OsMADS51) and four negative regulators of Ehd1 (OsphyB, OsCOL4, DTH8 and Ghd7) were not significantly affected in ehd4 (Figure 6A and 6B). These observations suggest that Ehd4 functions upstream of Ehd1, but largely independent of other known regulators of Ehd1. Consistent with this, down regulation of Ehd1, Hd3a and RFT1 in ehd4 was also seen in the Nipponbare background and constantly seen at different stages during plant development (Figure 6B and Figure S4).
Fig. 6. Quantitative RT–PCR analysis of representative flowering-related genes and Ehd4 in various flowering-time mutants or their NILs (near-isogenic lines) and corresponding WTs under SDs and LDs. 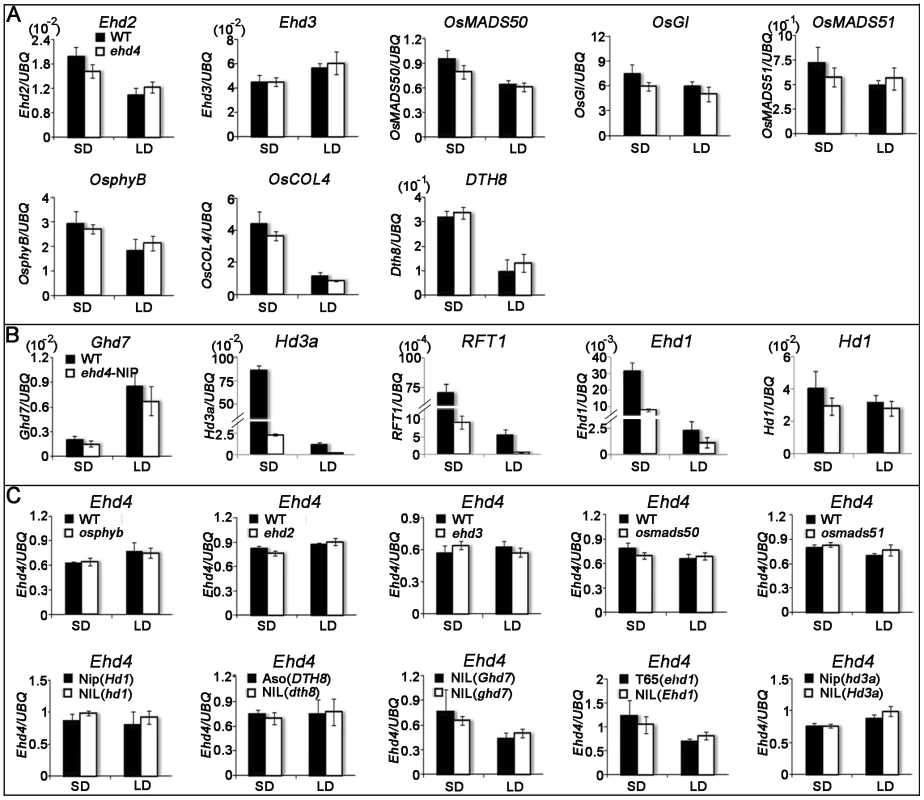
(A) Transcript level of Ehd2, Ehd3, OsMADS50, OsGI, OsMADS51, OsphyB, OsCOL4 and DTH8 in WT (Kita-ake) and ehd4 plants. (B) Transcript level of Ghd7, Hd3a, RFT1, Ehd1 and Hd1 in WT (Nipponbare) and ehd4-Nip plants. (C) Transcript level of Ehd4 in various flowering-time mutants or their NILs (near-isogenic lines) and corresponding WTs. Dongjin and the osphyb mutant [26]; Tohoku IL9 and the ehd2 mutant [34]; Tohoku IL9 and the ehd3 mutant [36]; Dongjin and the osmads50 mutant [31]; Dongjin and the osmads51 mutant [30]; Nipponbare and a NIL carrying a non-functional Hd1 allele [19]; Asominori and a NIL carrying a nonfunctional DTH8 allele [29]; A NIL carrying a functional Ghd7 allele and a NIL carrying a non-functional in the Shanyou 63 background [28]. Taichun 65 carrying a non-functional Ehd1 allele and a NIL carrying a functional Ehd1 allele [24]; Nipponbare carrying a partially functional Hd3a allele and a NIL carrying a functional Hd3a allele [20]; Penultimate leaves were harvested around reported peak expression level of each gene during the 24 hrs photoperiod - at dawn for OsphyB, OsCOL4, Ehd1, Ehd2, Hd3a, RFT1 and Ehd4, 3 h after dawn for Ghd7, 8 h after dawn for Ehd3, OsMADS50, OsMADS51 and DTH8 and immediately after dusk for OsGI and Hd1 from 28 d-old (SDs) and 35 d-old (LDs) plants. The rice Ubiquitin-1 (UBQ) gene was used as the internal control. Values are shown as mean±s.d (standard deviations) of three independent experiments and two biological replicates. To investigate whether Ehd4 expression is regulated by other flowering genes, we examined the expression of Ehd4 in osphyb, ehd2, ehd3, osmads50 and osmads51 mutants and near-isogenic lines (NILs) which carrying a deficient Hd1, Ghd7 or DTH8 alleles. Notably, we detected no significant differences of Ehd4 expression in these mutants or NILs, as compared to their corresponding WT plants (Figure 6C). In addition, no significant change of Ehd4 expression was seen in NILs deficient in Ehd1 or Hd3a either (Figure 6C). These results suggest that Ehd4 acts independent of other Ehd1 regulators we examined. Together, these observations suggest that Ehd4 regulates the expression of Hd3a and RFT1 through Ehd1. This notion was also supported by the observation that over-expression of Ehd1 fully rescued the late flowering phenotype of ehd4 under SDs (Figure 7).
Fig. 7. Complementation of ehd4 by over-expression of Ehd1. 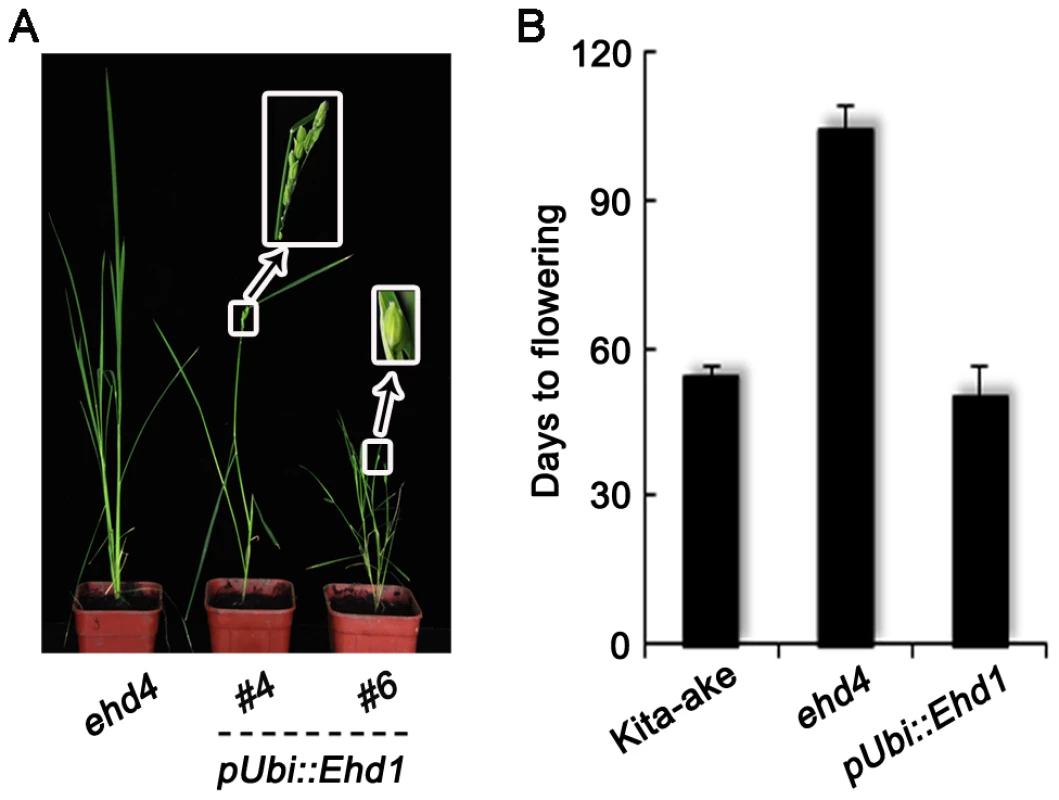
(A) Phenotypes of the mutant and transgenic plants at heading stage. (B) Ehd1, when driven by the maize Ubiquitin-1 promoter (pUbi::Ehd1), was able to rescue the flowering phenotype of ehd4 (T0 plants, n = 8). All plants were grown under SDs. Since EHD4 has a transcriptional activation and nucleic acid binding activity and it promotes Ehd1 expression, we next carried out a yeast one-hybrid assay and a transient transcription assay [43], [44] to test whether Ehd1 is a direct downstream target of EHD4. However, only OsLFL1 [45], but not EHD4, was able to interact with the Ehd1 promoter (Figure S5), indicating that Ehd1 is likely an indirect target of Ehd4. In addition, yeast three-hybrid assay [46] also failed to detect a direct binding of EHD4 to the Ehd1 mRNA (Figure S6). Moreover, neither EHD2 nor EHD3, directly binds to the Ehd1 promoter (Figure S5). Our yeast two-hybrid assay showed that there was no direct interaction among the EHD2, EHD3 and EHD4 proteins (Figure S7). Together, these results suggest that Ehd2, Ehd3 and Ehd4 likely act through distinct pathways to promote the expression of Ehd1.
Transcriptome analysis of ehd4 plants
To further reveal the molecular basis of the flowering phenotype of ehd4, we performed a transcriptome analysis of ehd4 and wild-type plants using RNA-seq to identify genes downstream of Ehd4. RNA samples were extracted from the penultimate leaves (collected at dawn) of 30 d-old ehd4 and WT plants (Kita-ake) grown under LDs. We obtained 2.5 M tags and found a total of 256 genes altered in expression with an estimated false-discovery rate of 0.1% and the absolute value of log2Ratio at 3.15 under the Bayesian model (Table S1; [47]). We found that the transcript numbers of Hd3a, RFT1, Ehd1, OsMADS1, OsMADS14 and OsMADS15 reduced dramatically in ehd4 plants (Table S1), consistent with the qRT-PCR results (Figure 5A–5F and Figure S3). Our qRT-PCR analysis with other four genes (with a log2 Ratio of −11.73, −10.33, −5.80 and −3.15, respectively) also further confirmed the reliability of the RNA-seq results (Figure S8). Notably, we found that among the genes down-regulated in end4, 25 of them are known or putative transcription factors, including MADS box, Zinc finger, MYB, SBP and B3 proteins (Table S1). These genes could be potential candidates involved in the Ehd4-Ehd1-Hd3a/RFT1 pathway to regulate photoperiodic flowering in rice.
Ehd4 is unique and highly conserved in rice
Ehd4 is a single copy gene in the rice genome. It is predicted to code for a polypeptide of 832 amino acids long, which contains a CCCH (C-X7-C-X5-C-X3-H)-type zinc finger motif at the C-terminus (Figure S9). A blast search (http://www.ncbi.nlm.nih.gov/) found that EHD4 has no clear homologs in other plant or animal species. Thus it appears that Ehd4 represents a unique regulator of flowering time in rice.
To investigate the evolutionary history of the gene in rice, we analyzed the Ehd4 sequences from 86 rice accessions with wide geographic distribution and diverse genetic backgrounds, including 32 wild rice species (O. rufipogon and O. nivara) and 54 cultivated rice (Table S2; [48]). Ehd4 appears to be highly conserved across these accessions (share >99.2% or higher amino acid sequence identities) (Table S2). Sequence analysis identified 25 haplotypes among these accessions. Strikingly, 21 haplotypes were identified in 32 wild rice accessions (O. rufipogon and O. nivara) but only 8 haplotypes were identified in 54 cultivated rice accessions analyzed in this study. The dramatic reduction in genetic diversity at this locus suggests that Ehd4 might subject to bottleneck effect [49]. Notably, 4 haplotypes (Hap_2, 3, 6 and 7) are shared in cultivated and wild rice (Figure 8A, Table S2), and among them, Hap_2 and Hap_3 together account 25% of the 32 wild rice and 85% of the 54 cultivated rice (O. sativa) respectively (Figure 8A, 8B and Table S2). Among the cultivated rice, 20 (77%) indica and 6 (23%) japonica accessions belong to Hap_2, while 19 (95%) japonica and 1 (5%) indica accessions belong to Hap_3 (Figure 8A and Table S2). This result suggests that the Hap_2 and Hap_3 represent the two major haplotypes at the Ehd4 locus in cultivated rice and that they exist in wild rice before domestication. The distribution pattern of Hap_2 and Hap_3 in indica (mostly distributed in lower latitude and elevation zones) and japonica (mostly distributed in higher latitude and elevation zones) (Figure 8C) implies a likely correlation between the geographic distribution and the functional differences of Ehd4 haplotypes among these cultivated accessions analyzed.
Fig. 8. Natural variations in the Ehd4 coding region among rice germplasm core collection. 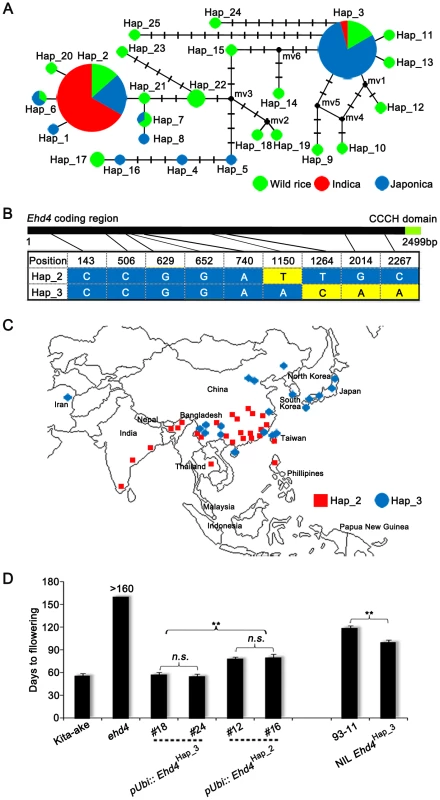
(A) Haplotype network of the Ehd4 alleles in 86 rice accessions. Haplotype frequencies are proportional to the area of the circles. The proportion of wild rice and two cultivated subgroups (indica and japonica) in each haplotype is represented by different colors. (B) The polymorphic nucleotides of Hap_2 and Hap_3 of Ehd4 gene in the core collection. The number on the top shows the position of nucleotide polymorphisms in the coding region starting from the ATG start codon. (C) Geographic distribution of the cultivated rice accessions belonging to Hap_2 and Hap_3. (D) Flowering time of transgenic plants carrying two major haplotypes of Ehd4 driven by the maize Ubiquitin-1 promoter in ehd4 (Kita-ake background) and NIL carrying Ehd4Hap3 compared with the 93-11 parental plants. T2 plants of two pUbi::Ehd4Hap3 (#18 and #24) and two pUbi::Ehd4Hap2 (#12 and #16) lines were measured (n = 15). All plants were grown in the natural long day field conditions. Values are means±s.d. (standard deviations) (n = 15). **Significant at 1% level; n.s., not significant. To test the possible functional differences of these two haplotypes, we introduced the full-length Ehd4 cDNA of indica variety 93-11 (Hap_2) and Kita-ake (a japonica landrace, Hap_3) driven by the maize Ubiquitin-1 promoter into the ehd4 mutant (Kita-ake background). Strikingly, over-expression of Hap_3, but not Hap_2, fully complemented the ehd4 phenotype under NLDs, although their expression levels are comparable (Figure 8D and Figure S10). This result suggests that Hap_3 of Ehd4 is functionally more potent in promoting flowering than Hap_2. As another test of this notion, we introduced the Hap_3 allele from Kita-ake into 93-11 by backcrossing five times, followed by selfing. Strikingly, the NIL Ehd4Hap_3 plants (BC5F3) flowered earlier by 19 d under NLDs compared to the parental 93-11 plants (Figure 8D). Together, these results suggest that the functional differences of Ehd4 haplotypes might play a role in geographic adaptation of cultivated rice.
Discussion
In this study, we have uncovered Ehd4, which codes for a CCCH-type zinc finger protein essential for promoting flowering under both SD and LD conditions in rice, irrespective of genetic backgrounds. We demonstrated that Ehd4 promotes flowering by positively regulating the expression of Hd3a and RFT1 through Ehd1 but independent of other known important Ehd1 regulators. We further showed that the late-flowering phenotype of ehd4 is more profound in Kita-ake (Kit) (a day-length neutral variety) than in Nipponbare (Nip) (a day-length sensitive variety) under SDs, but ehd4 plants flowered eventually in Kit (164 days after germination) but not in Nip under LDs (Figure 1B). It is known that Hd1 acts to promote flowering under SDs but delay flowering under LDs [19]. We found that Hd1 has a 36-bp insertion and two SNPs in Kit compared to Nip (Figure S1), implying that in Kit, the promoting role under SDs and repressing role under LDs of Hd1 may be impaired. This may at least partially explains why the late-flowering phenotype of ehd4-Nip is less severe under SDs but more severe under LDs. In addition, we found that Kit carries a truncated allele of Ghd7, a repressor of Ehd1 under LDs, whereas Nip carries a partially functional Ghd7 allele (Figure S1; [28]). Therefore, the never-flowering phenotype of ehd4 in Nip under LDs could be due to the repressive effect of Hd1 and Ghd7. Strikingly, even in the absence of the functional Hd1 and Ghd7 alleles in Kit, ehd4 alone delayed flowering time by three folds under LDs (Figure 1B), suggesting that Ehd4 plays a major role in promoting flowering in rice, particularly under LDs.
Previous studies revealed that rice Ehd1 is a critical convergence point of flowering time regulation by multiple signaling pathways and that Ehd1 acts independently of Hd1. Ehd1 encodes a B-type response regulator that is highly conserved in cultivated rice, but has no homolog in Arabidopsis [23], [24]. Up to date, 12 genes have been shown to regulate Ehd1 expression, including 5 positive regulators (Ehd2, Ehd3, OsGI, OsMADS50 and OsMADS51) and 7 negative regulators (SE5, OsphyB, Ghd7, DTH8, OsLFL1, OsCOL4 and OsMADS56). We demonstrated that Ehd4 promotes flowering by positively regulating the expression of Hd3a and RFT1 through Ehd1, but independently of these known Ehd1 regulators.
It is also of interest to note that the majority of Ehd1 regulators uncovered thus far are nuclear proteins and many of them act as transcriptional regulators, including GHD7, DTH8, OsMAD50, OsMAD51, EHD2, EHD3, OsLFL1, OsMAD56 and OsCOL4 [26], [28]–[34], [36], [45], [50]. Map-based cloning revealed that Ehd4 encodes a novel CCCH-type zinc finger protein also localized to the nucleus. The CCCH-type zinc finger protein family is defined as a group of proteins containing 1–6 copies of the canonical C-X-C-X-C-X-H motif (C-X6–14-C-X4–5-C-X3-H, where X is any amino acid) [51]. This type of proteins has been found in organisms ranging from human to yeast and many of them have been shown to have either an RNA binding function involved in RNA processing or DNA binding activity [40]–[42]. There are at least 68 CCCH-type genes in Arabidopsis and 67 in rice, respectively [52]. However, only a few plant CCCH proteins have been functionally characterized. EHD4 is the first CCCH-type protein found to regulate photoperiodic flowering. We found that EHD4 is capable of binding to nucleic acids in vitro and transactivate transcription in yeast, suggesting that it likely functions as a transcription factor. Further, our transcriptome analysis revealed that a significant portion of Ehd4-regulated downstream genes are also transcription factors, including several previously identified flowering regulators, such as Ehd1, OsLFL1 OsMADS1, OsMADS14 and OsMADS15, and other putative transcription factors, including MADS box, Zinc finger, MYB, SBP and B3 proteins (Table S1). These findings together suggest that transcriptional regulation plays a critical role in photoperiodic regulation of flowering in rice. However, despite we have demonstrated that EHD4 has double-stranded DNA and ribohomopolymer binding activity and transactivation activity in yeast, we have not been able to identify the direct target genes of EHD4 in this study. It is also possible that EHD4 may bind to RNA molecules and degrades transcripts of unknown Ehd1 repressors. Further studies are required to elucidate the biochemical function of EHD4 and its functional relationship with other nuclear regulators of flowering time.
Rice is known as a short day plant. However, cultivated rice (O. sativa) is grown widely in Asia, with a northern limit of nearly 53°N in northern Asia (Northern provinces of China and Korea, where natural day length during rice cultivation is nearly 15 hours light; [53]), whereas O. rufipogon, a wild rice that is the most relative ancestor of O. sativa, is mainly distributed at tropical latitudes with a northern limit about 28°N [54]. The northward expansion of cultivated rice into higher latitudes must be accompanied by human selection of the flowering time trait during rice domestication and breeding, to secure a harvest before cold weather approaches. Strikingly, Ehd4 has no obvious homologs in other plant species including Arabidopsis, maize and sorghum, suggesting that Ehd4 originated along with the diversification of the Oryza genus from the grass family during evolution. Amino acid sequence comparison of EHD4 showed identities at 99.2% or higher among a core collection of rice germplasm with wide geographic distribution and diverse genetic backgrounds, including wild rice species ([48]; Table S2). Interestingly, we found two major haplotypes of Ehd4, Hap_2 (the major haplotype in indica) and Hap_3 (the major haplotype in japonica) and that Hap_3 is functional more potent in promoting flowering under NLDs. Since indica rice is known to distribute mostly in lower latitude and elevation zones (between the latitude 3°S-35°N), while japonica varieties are mostly distributed in higher latitude and elevation zones (between the latitude 15°N-53°N), our findings suggest that Ehd4 may have contributed to the northward expansion and regional adaptation of cultivated rice into higher latitudes.
Materials and Methods
Plant material and growth conditions
The ehd4 mutant was initially identified from a tissue culture-derived population of rice cv Kita-ake (japonica) under natural-day conditions in a paddy field in Beijing (39°54′N, 116°23′E), China (2006). To generate ehd4-nip plants, the mutant locus was introgressed into Nipponbare (japonica) background by crossing and backcrossing for five generations (BC5), where ehd4 mutant is the donor parent and Nipponbare is the recurrent parent, by using marker assisted selection (MAS).
Plants were grown in controlled-growth chambers (Conviron) under SDs (10 h light at 30°C/14 h dark at 25°C) or LDs (14.5 h light at 30°C/9.5 h dark at 25°C) with a relative humidity of ∼70%. The light intensity was ∼800 µmol m−2 s−1.
Map-based cloning
To map the ehd4 locus, the mutant was crossed with the indica cv 93-11 and then the F1 plants were backcrossed with 93-11 to produce a BC1F2 population. We used two DNA pools generated from 15 BC1F2 late-flowering and 15 normal plants, respectively, for rough mapping. For fine mapping, 871 never-flowering plants segregated in the BC1F2 population were used.
Vector construction and plant transformation
For the complementary test, the Ehd4 full-length cDNA driven by its native (2.7-kb) or the maize Ubiquitin-1 (Ubi) promoter were cloned into the binary vector pCAMBIA1390 by using In-Fusion Advantage PCR Cloning Kits (Clontech) to create pEhd4::Ehd4 and pUbi::Ehd4, respectively. The 2.7-kb long promoter was also cloned into pCAMBIA-1305.1 to create pEhd4::GUS. The resultant plasmids were transformed into the Agrobacterium tumefaciens strain EHA105 and then introduced into ehd4 (for complementary test) or Kita-ake WT plants (pEhd4::GUS). At least 15 transgenic events were produced for each construct.
Subcellular localization of EHD4
GFP was fused to the C-terminus of EHD4 under the control of the 35S CaMV promoter in the pA7 vector. The EHD4-GFP fusion and the nucleus marker OsMADS3-mCherry were transiently co-expressed in rice leaf protoplasts by PEG (polyethylene glycol) treatment [55]. Fluorescence was observed using a Leica TCS-SP4 confocal microscope.
Transactivation activity assay
Transactivation activity assay was performed using the Matchmaker GAL4 Two-Hybrid System 3 (Clontech). Plasmids containing GAL4 DNA binding domain fused with EHD4 deletions were transformed into the yeast strain AH109. The substrate chlorophenol red-β-D-galactopyranoside (CPRG; Roche Biochemicals) was used to measure the β-galactosidase activity according to the Yeast Protocols Handbook (Clontech).
In vitro nucleic acid binding assay
EHD4 deletions were cloned into the pMAL-c2x (NEB) and expressed in E. coli. 0.5 mg purified protein was incubated with 20 mL of poly rG, poly rC, or poly rU attached to agarose beads or double - or single-stranded calf thymus DNA attached to cellulose beads (Sigma) in 500 mL of RHPA binding buffer (10 mM Tris, pH 7.4, 2.5 mM MgCl2, 0.5% Triton X-100, NaCl at various concentrations) with 1 mg/mL heparin. After incubation at 4°C for 10 min, the beads were washed five times in the RHPA buffer and then boiled in the SDS loading buffer. Binding of fusion proteins to RNA or DNA was confirmed by protein gel blot using anti-MBP antibodies (NEB).
Yeast one-hybrid assay
Yeast one-hybrid assay was performed according to the method described in [43]. To generate GAD-EHD4, GAD-EHD2, GAD-EHD3 and GAD-OsLFL1, their full-length cDNAs were cloned into pJG4-5 vector. To generate the Ehd1p::LacZ reporter gene, a 3.2 kb fragment of Ehd1 promoter (including the 5′-UTR) was amplified from Nipponbare genomic DNA and inserted into the corresponding sites of the reporter plasmid pLacZi2μ. Plasmids were co-transformed into the yeast strain EGY48. Transformants were grown onto SD/Trp-/Ura plates for 48 hours and then transferred onto X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) plates for blue color development.
Yeast three-hybrid assay
Yeast three-hybrid assay was performed according to the method described in [46]. Full-length Ehd4 cDNA was cloned into pACTII vector to generate GAD-EHD4. A series DNA fragments (about 140 bp) that transcripting Ehd1 mRNA sequence were cloned into pMS2-1 vector. Plasmids were co-transformed into the yeast strain YBZ-1. Transformants were grown on plates containing selective media (SD/Ura-/Leu-/His-+0, 2 or 8 mM 3-aminotriazole) for 48 hours before assay.
Bioluminescence assays
Isolation of rice leaf protoplast and PEG-mediated transfection were performed as described [55]. The reporter construct pGreen-Ehd1p-LUC and effector plasmids (pEGAD-MycEHD4, EHD2, EHD3 or OsLFL1) were co-transformed into protoplasts. After transformation, the protoplasts were incubated in darkness for 12–16 h. Bioluminescence assay was performed according to the method described in [44].
Quantitative real-time RT-PCR
Total RNA was extracted using the RNeasy Plant Mini Kit (QIAGEN). For quantitative real-time RT-PCR, the first-strand cDNA was synthesized using the QuantiTect Reverse Transcription Kit (QIAGEN) and then PCR was performed using gene-specific primers and SYBR Premix ExTaq reagent (Takara) with an ABI Prism 7900 HT Sequence Detection System (Applied Biosystems) according to the manufacturer's instructions. PCR reactions were carried out in triplicate for each sample from two independent biological replicates and the rice Ubiquitin-1 gene was used as the internal control.
RNA–seq (next-generation sequencing of RNA)
We used the Illumina HiSeq 2000 Genome Analyzer to get tags with CATG site, in which the adapter sequences are 2*100 bp. With Illumina's digital gene expression assay, we obtained 11.7 million sequence tags per sample. After removing low quality reads and low quality bases of quality value, clean reads were mapped to the O. ssp. japonica reference sequences using SOAPaligner/soap2. Mismatches of not more than one base were allowed in the alignment and we generated 8.9 million perfect match tags (76.08%) for each sample. Initially, we determine 27020 genes of significant differences in expression between the groups of wild-type and mutants by a Student's t-test. With a dedicated Bayesian model, we found 256 transcripts of differential expression with an estimated false-discovery rate of 0.1% and the absolute value of log2Ratio is more than 3.15.
All primers used in this study are listed in Table S3.
Accession numbers
Data deposition: The Ehd4 sequence reported in this paper has been deposited in the GenBank database accession no. JQ828863 (cDNA).
Supporting Information
Zdroje
1. SimpsonGG, DeanC (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296 : 285–289.
2. IzawaT (2007) Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J Exp Bot 58 : 3091–3097.
3. SrikanthA, SchmidM (2011) Regulation of flowering time: all roads lead to Rome. Cellular and Molecular Life Sciences 68 : 2013–2037.
4. KobayashiY, WeigelD (2007) Move on up, it's time for change–mobile signals controlling photoperiod-dependent flowering. Genes Dev 21 : 2371–2384.
5. ImaizumiT, KaySA (2006) Photoperiodic control of flowering: not only by coincidence. Trends in plant science 11 : 550–558.
6. TurckF, FornaraF, CouplandG (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59 : 573–594.
7. YanovskyMJ, KaySA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 : 308–312.
8. ValverdeF, MouradovA, SoppeW, RavenscroftD, SamachA, et al. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science Signalling 303 : 1003.
9. SawaM, NusinowDA, KaySA, ImaizumiT (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science Signalling 318 : 261.
10. FornaraF, PanigrahiK, GissotL, SauerbrunnN, RühlM, et al. (2009) Arabidopsis DOF Transcription Factors Act Redundantly to Reduce CONSTANS Expression and Are Essential for a Photoperiodic Flowering Response. Developmental cell 17 : 75–86.
11. PutterillJ, RobsonF, LeeK, SimonR, CouplandG (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 : 847–857.
12. TiwariSB, ShenY, ChangHC, HouY, HarrisA, et al. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis‐element. New Phytologist 187 : 57–66.
13. AbeM, KobayashiY, YamamotoS, DaimonY, YamaguchiA, et al. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science Signalling 309 : 1052.
14. WiggePA, KimMC, JaegerKE, BuschW, SchmidM, et al. (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science Signalling 309 : 1056.
15. CorbesierL, VincentC, JangS, FornaraF, FanQ, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science Signalling 316 : 1030.
16. TAKIMOTOA, IKEDAK (1961) Effect of twilight on photoperiodic induction in some short day plants. Plant and Cell Physiology 2 : 213–229.
17. ItohH, NonoueY, YanoM, IzawaT (2010) A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nature genetics 42 : 635–638.
18. HayamaR, YokoiS, TamakiS, YanoM, ShimamotoK (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422 : 719–722.
19. YanoM, KatayoseY, AshikariM, YamanouchiU, MonnaL, et al. (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. The Plant Cell Online 12 : 2473–2483.
20. KojimaS, TakahashiY, KobayashiY, MonnaL, SasakiT, et al. (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant and Cell Physiology 43 : 1096–1105.
21. TamakiS, MatsuoS, WongHL, YokoiS, ShimamotoK (2007) Hd3a protein is a mobile flowering signal in rice. Science Signalling 316 : 1033.
22. KomiyaR, IkegamiA, TamakiS, YokoiS, ShimamotoK (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135 : 767–774.
23. TakahashiY, TeshimaKM, YokoiS, InnanH, ShimamotoK (2009) Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proceedings of the National Academy of Sciences 106 : 4555–4560.
24. DoiK, IzawaT, FuseT, YamanouchiU, KuboT, et al. (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18 : 926–936.
25. KomiyaR, YokoiS, ShimamotoK (2009) A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136 : 3443–3450.
26. LeeYS, JeongDH, LeeDY, YiJ, RyuCH, et al. (2010) OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. The Plant Journal 63 : 18–30.
27. IzawaT, OikawaT, SugiyamaN, TanisakaT, YanoM, et al. (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16 : 2006–2020.
28. XueW, XingY, WengX, ZhaoY, TangW, et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature genetics 40 : 761–767.
29. WeiX, XuJ, GuoH, JiangL, ChenS, et al. (2010) DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiology 153 : 1747–1758.
30. KimSL, LeeS, KimHJ, NamHG, AnG (2007) OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiology 145 : 1484–1494.
31. LeeS, KimJ, HanJJ, HanMJ, AnG (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS‐LIKE 20 (SOC1/AGL20) ortholog in rice. The Plant Journal 38 : 754–764.
32. WuC, YouC, LiC, LongT, ChenG, et al. (2008) RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proceedings of the National Academy of Sciences 105 : 12915–12920.
33. ParkSJ, KimSL, LeeS, JeBI, PiaoHL, et al. (2008) Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. The Plant Journal 56 : 1018–1029.
34. MatsubaraK, YamanouchiU, WangZX, MinobeY, IzawaT, et al. (2008) Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiology 148 : 1425–1435.
35. ColasantiJ, YuanZ, SundaresanV (1998) The indeterminate Gene Encodes a Zinc Finger Protein and Regulates a Leaf-Generated Signal Required for the Transition to Flowering in Maize. Cell 93 : 593–603.
36. MatsubaraK, YamanouchiU, NonoueY, SugimotoK, WangZX, et al. (2011) Ehd3, encoding a plant homeodomain finger‐containing protein, is a critical promoter of rice flowering. The Plant Journal 66 : 603–612.
37. TakahashiY, ShomuraA, SasakiT, YanoM (2001) Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proceedings of the National Academy of Sciences 98 : 7922–7927.
38. OgisoE, TakahashiY, SasakiT, YanoM, IzawaT (2010) The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiology 152 : 808–820.
39. YuJ, HuS, WangJ, WongGKS, LiS, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 : 79–92.
40. LiJ, JiaD, ChenX (2001) HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. The Plant Cell Online 13 : 2269–2281.
41. KimDH, YamaguchiS, LimS, OhE, ParkJ, et al. (2008) SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. The Plant Cell Online 20 : 1260–1277.
42. HurtJA, ObarRA, ZhaiB, FarnyNG, GygiSP, et al. (2009) A conserved CCCH-type zinc finger protein regulates mRNA nuclear adenylation and export. The Journal of Cell Biology 185 : 265–277.
43. LinR, DingL, CasolaC, RipollDR, FeschotteC, et al. (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science Signalling 318 : 1302.
44. LiuH, YuX, LiK, KlejnotJ, YangH, et al. (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science Signalling 322 : 1535.
45. PengLT, ShiZY, LiL, ShenGZ, ZhangJL (2007) Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochemical and biophysical research communications 360 : 251–256.
46. SeayD, HookB, EvansK, WickensM (2006) A three-hybrid screen identifies mRNAs controlled by a regulatory protein. Rna 12 : 1594–1600.
47. ChinL, HahnWC, GetzG, MeyersonM (2011) Making sense of cancer genomic data. Genes Dev 25 : 534–555.
48. GarrisAJ, TaiTH, CoburnJ, KresovichS, McCOUCHS (2005) Genetic structure and diversity in Oryza sativa L. Genetics. 169 : 1631–1638.
49. ZhuQ, ZhengX, LuoJ, GautBS, GeS (2007) Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice. Molecular Biology and Evolution 24 : 875–888.
50. RyuCH, LeeS, ChoLH, KimSL, LeeYS, et al. (2009) OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell and Environment 32 : 1412–1427.
51. BergJM, ShiY (1996) The galvanization of biology: a growing appreciation for the roles of zinc. Science 271 : 1081–1085.
52. WangD, GuoY, WuC, YangG, LiY, et al. (2008) Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9 : 44.
53. Vaughan D. Collection, conservation, and potential use of the wild relatives of rice in Asia and Australia; 1989. IRRI.
54. LondoJP, ChiangYC, HungKH, ChiangTY, SchaalBA (2006) Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci U S A 103 : 9578–9583.
55. BartR, ChernM, ParkCJ, BartleyL, RonaldPC (2006) A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2 : 13.
56. HuangXY, ChaoDY, GaoJP, ZhuMZ, ShiM, et al. (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23 : 1805–1817.
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



