-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaVariation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
Segregation of chromosomes during the first meiotic division relies on crossovers established during prophase. Although crossovers are strictly regulated so that at least one occurs per chromosome, individual variation in crossover levels is not uncommon. In an analysis of different inbred strains of male mice, we identified among-strain variation in the number of foci for the crossover-associated protein MLH1. We report studies of strains with “low” (CAST/EiJ), “medium” (C3H/HeJ), and “high” (C57BL/6J) genome-wide MLH1 values to define factors responsible for this variation. We utilized immunofluorescence to analyze the number and distribution of proteins that function at different stages in the recombination pathway: RAD51 and DMC1, strand invasion proteins acting shortly after double-strand break (DSB) formation, MSH4, part of the complex stabilizing double Holliday junctions, and the Bloom helicase BLM, thought to have anti-crossover activity. For each protein, we identified strain-specific differences that mirrored the results for MLH1; i.e., CAST/EiJ mice had the lowest values, C3H/HeJ mice intermediate values, and C57BL/6J mice the highest values. This indicates that differences in the numbers of DSBs (as identified by RAD51 and DMC1) are translated into differences in the number of crossovers, suggesting that variation in crossover levels is established by the time of DSB formation. However, DSBs per se are unlikely to be the primary determinant, since allelic variation for the DSB-inducing locus Spo11 resulted in differences in the numbers of DSBs but not the number of MLH1 foci. Instead, chromatin conformation appears to be a more important contributor, since analysis of synaptonemal complex length and DNA loop size also identified consistent strain-specific differences; i.e., crossover frequency increased with synaptonemal complex length and was inversely related to chromatin loop size. This indicates a relationship between recombination and chromatin compaction that may develop as DSBs form or earlier during establishment of the meiotic axis.
Published in the journal: . PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1004125
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004125Summary
Segregation of chromosomes during the first meiotic division relies on crossovers established during prophase. Although crossovers are strictly regulated so that at least one occurs per chromosome, individual variation in crossover levels is not uncommon. In an analysis of different inbred strains of male mice, we identified among-strain variation in the number of foci for the crossover-associated protein MLH1. We report studies of strains with “low” (CAST/EiJ), “medium” (C3H/HeJ), and “high” (C57BL/6J) genome-wide MLH1 values to define factors responsible for this variation. We utilized immunofluorescence to analyze the number and distribution of proteins that function at different stages in the recombination pathway: RAD51 and DMC1, strand invasion proteins acting shortly after double-strand break (DSB) formation, MSH4, part of the complex stabilizing double Holliday junctions, and the Bloom helicase BLM, thought to have anti-crossover activity. For each protein, we identified strain-specific differences that mirrored the results for MLH1; i.e., CAST/EiJ mice had the lowest values, C3H/HeJ mice intermediate values, and C57BL/6J mice the highest values. This indicates that differences in the numbers of DSBs (as identified by RAD51 and DMC1) are translated into differences in the number of crossovers, suggesting that variation in crossover levels is established by the time of DSB formation. However, DSBs per se are unlikely to be the primary determinant, since allelic variation for the DSB-inducing locus Spo11 resulted in differences in the numbers of DSBs but not the number of MLH1 foci. Instead, chromatin conformation appears to be a more important contributor, since analysis of synaptonemal complex length and DNA loop size also identified consistent strain-specific differences; i.e., crossover frequency increased with synaptonemal complex length and was inversely related to chromatin loop size. This indicates a relationship between recombination and chromatin compaction that may develop as DSBs form or earlier during establishment of the meiotic axis.
Introduction
Recombination is a defining event of meiosis, resulting in the physical exchange of DNA between homologous chromosomes. It is generally thought that this is essential for proper alignment and subsequent segregation of homologs during meiosis I and, indeed, evidence from yeast [1], [2], Caenorhabditis elegans [3], Drosophila melanogaster [4], and mammals [5] indicates that alterations in the number or positioning of recombination events increase the likelihood of meiotic nondisjunction. For this reason, it might be expected that recombination levels are strictly regulated but, surprisingly, substantial inter-individual variation in recombination is observed in most mammalian species. For example, linkage studies have demonstrated extensive variation in recombination rates and/or recombination hotspot usage in human males and females (e.g., [6], [7], [8]), and cytological studies of recombination indicate 15–25% individual differences in genome-wide recombination levels in rhesus and human males [9], [10], and similar levels of variation in different inbred strains of mice [11], [12]. Further, the frequency and location of exchanges vary between the sexes; e.g., in humans the female genetic map is approximately 1.6 fold longer than that of males, with interstitial exchanges being more common in females than males (e.g., [13], [14], [15]). These differences beg an obvious question: what is responsible for the variation in recombination levels observed among individuals or inbred strains?
Arguably, there are at least three different time-points in the recombination process at which variation in recombination levels could arise. First, variation could be induced at the beginning of the recombination pathway, when double-strand breaks (DSBs) are formed. For example, some individuals might have a greater number of DSBs than others and, assuming that similar proportions of DSBs are converted into crossovers, the end result would be variation in the number of crossovers. On the surface, recent studies in mice would seem to eliminate this possibility: as in other model organisms [16], crossover homeostasis operates in male mice to ensure the presence of a minimum number of crossovers over a wide range of DSBs, suggesting that variation in DSBs does not translate into variation in crossovers [17]. However, these analyses were intended to assess variation in recombination among mice with different numbers of functional alleles for the DSB-inducing locus Spo11, and do not preclude the possibility that genetic background differences may influence the overall number of crossovers. Second, variation could arise at some stage in the processing of recombination intermediates. For example, genotypic differences at RNF212, encoding a protein with homology to crossover promoting proteins in S. cerevisiae and C. elegans, is the best characterized determinant of individual variation in genome-wide recombination rates in humans (e.g., [18]). Recently, analyses of mice deficient for RNF212 indicate that it acts downstream of the initiation of DSBs, stabilizing joint DNA molecules and promoting the resolution of DSBs as crossovers [19]. Taken together, these observations provide evidence that events occurring after the formation DSBs can, indeed, affect the eventual number of recombination events, although it is not clear that this accounts for all of the among-individual or among-strain differences that have been reported in mammals. Third, differences might arise at the end of the recombination pathway, owing to individual variation in proteins such as MLH1 and MLH3 that are important in the resolution of double Holliday junctions into crossovers (e.g., [20]).
To discriminate among these possibilities in a mammalian system, we took advantage of strain-specific differences in genome-wide rates of meiotic recombination in the male mouse [11]. Specifically, we used immunofluorescence to examine the localization patterns of early-, mid-, and late-acting meiotic recombination proteins, asking whether the patterns were the same or different among inbred strains known to have “low”, “mid”, or “high” genome-wide rates of recombination. For each strain, we analyzed the number of foci for RAD51 and DMC1 (strand invasion proteins acting shortly after double-strand break formation; [21]), MSH4 (part of the complex stabilizing double Holliday junctions; [22]), and BLM (thought to have anti-crossover activity; e.g., see [23]), and compared these observations with results of analyses of the CO-associated protein MLH1 [20].
Our results demonstrate that inter-strain differences in crossovers (MLH1 foci) are preceded by proportionally similar differences in early-acting recombination proteins, indicating that the variation is established at, or before, the time of DSB formation. Subsequent analyses of males heterozygous for the gene encoding the DSB-inducing protein SPO11 [24], allowed us to eliminate DSBs per se as the source of the variation, since Spo11 heterozygotes exhibited a decrease in DSBs, but not in MLH1 foci.
In analyses of chromatin loop size and synaptonemal complex (SC) length, we detected striking differences among the three inbred strains, but not between Spo11 heterozygotes and their wildtype littermates. Taken together with the observations on recombination proteins, our results suggest that strain-specific differences in chromatin architecture, presumably established prior to the initiation of recombination, are important determinants of variation in crossover frequency.
Results
Strain-specific variation in MLH1 distribution
In previous studies of recombination in male mice [11], we identified strain-specific differences in the number of foci per cell of the DNA mismatch repair protein MLH1, known to mark the vast majority of sites of crossing-over [9], [26], [27]. We decided to exploit these differences to investigate the basis of the variation. Accordingly, we examined three inbred strains –C57BL/6J (“B6”), CAST/Ei (“CAST”) and C3H/HeJ (“C3H”) – assaying a minimum of 15 pachytene stage cells per mouse, and at least five mice per strain, scoring the number of autosomal MLH1 foci per cell (Figure 1A).
Fig. 1. Inter-strain variation in mean MLH1 values. 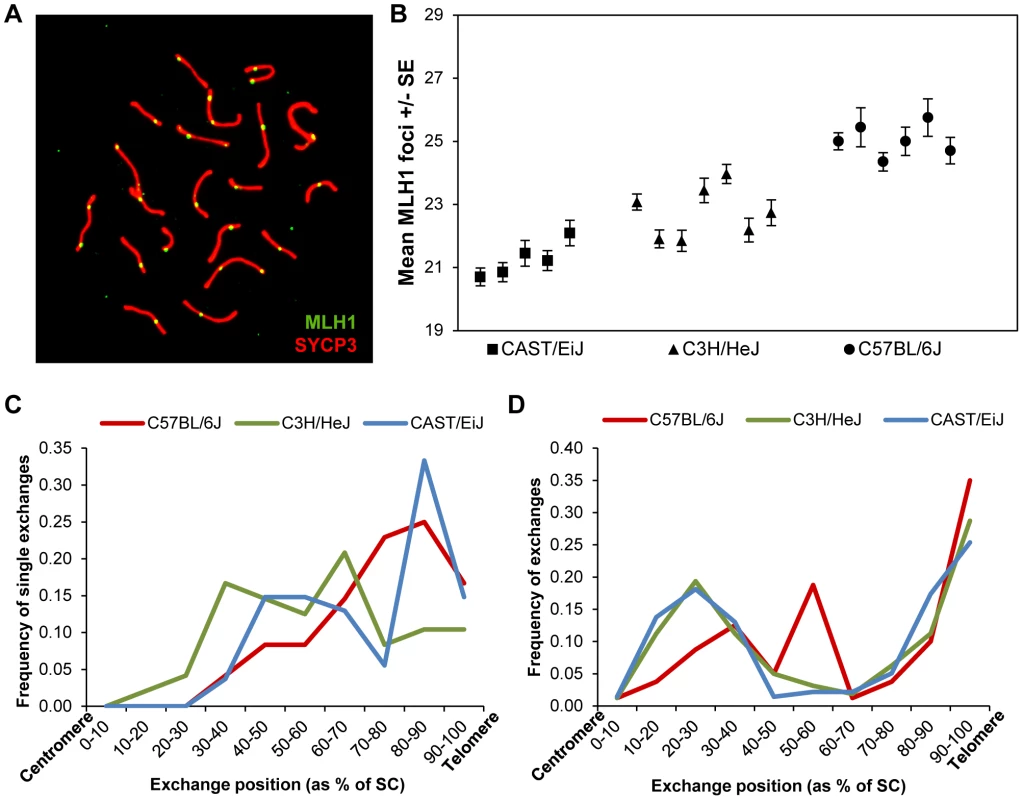
(A) Pachytene cell from B6 male immunostained with antibodies to MLH1 (green) and SYCP3 (red). The number of MLH1 foci per cell were counted and used as a surrogate for meiotic recombination events. (B) The mean number of MLH1 foci per spermatocyte for 5 CAST (n of cells = 105), 7 C3H (n of cells = 209) and six B6 (n of cells = 110) males varied significantly among the three inbred strains. (C, D) For each strain, slides were hybridized with FISH probes for chromosomes 19 and 1 and the positions of MLH1 foci were calculated as a percent of SC length (centromere = 0%; telomere = 100%). (C) On chromosomes 19 with a single MLH1 focus, the focus was typically medially or distally placed; data are based on analysis of 50 chromosomes/strain. (D) On chromosomes 1 with two MLH1 foci, typically one was proximally located and the other distally placed, consistent with positive interference. Data based on analysis of 50 cells/strain. Two of the inbred strains, CAST and B6, had previously been found to have “low” and “high” genome-wide MLH1 values, respectively [11]. Our re-analysis produced virtually identical results: the mean number +/ − S.D. of autosomal MLH1 foci per cell was 21.3+/−1.6 for CAST (n = 105 cells) and 25.0+/−2.2 for B6 (n = 102 cells) (Figure 1B; Table S1). Subsequently, we analyzed C3H males, and observed that this strain had mean MLH1 values that were intermediate to the other two strains: i.e., 22.7+/−1.9 (n = 209 cells) (Figure 1B; Table S1). Because the number of MLH1 foci per cell is not normally distributed (i.e., typically each bivalent has at least one focus, thus constraining the autosomal foci per cell to 19 or more), inter-strain differences have to be interpreted with caution. Nevertheless, ANOVA analyses are typically robust in the face of modest departures from normality, and the magnitude of the differences we observed (F = 105.1; p<0.0001), make it likely that the variation is real. Thus, these observations confirm our previous conclusion of variation in genome-wide MLH1 values – and presumably MLH1-driven crossovers – among males of different mouse strains.
The variation in overall MLH1 frequency was reflected by highly significant strain-specific differences in the proportion of chromosomes with zero, one, two, or three MLH1 foci (χ2 = 292.0, p<0.00001; Table S1). In large part, the difference was attributable to differing ratios of chromosomes with one vs. two MLH1 foci in the three strains; i.e., 6.8∶1, 3.9∶1 and 2.2∶1 for CAST, C3H and B6, respectively (Table S1). However, intriguingly, the strain with the highest MLH1 average value (B6), also had the highest proportion of bivalents lacking MLH1 foci; indeed, this value was significantly increased over that for CAST (χ2 = 12.1, p<0.001) and for C3H (χ2 = 13.9, p<0.001) males.
Subsequently, we analyzed the placement of MLH1 foci among the three strains, asking whether variation in MLH1 levels might be linked to differences in the location of the foci. Initially, we simply pooled results from all homologs and found no obvious differences among the strains; i.e., consistent with previous results, distally located MLH1 foci predominated among all three strains [11]. However, because the strain-specific differences in the proportion of homologs with one, two and three MLH1 foci complicate interpretations of these data, we conducted a second set of studies in which we analyzed individual chromosomes. Specifically, for each strain, we analyzed the placement of MLH1 foci on chromosomes 19 and 1 on SCs that exhibited one and two foci, respectively (Figure 1C, D). No obvious differences were observed among the strains.
Localization patterns of proteins involved in repair of DSBs vary among strains
The MLH1 data demonstrate inter-strain differences in recombination, but provide no information on when the variation originates. To address this question, we examined the abundance and distribution of signals for proteins involved at different stages of DSB repair, as follows:
Single end invasion is the earliest stage of recombination that can be consistently assayed using immunofluorescence. The ubiquitous RAD51 protein forms a complex around the resected ends of DSBs and facilitates invasion of the intact homologous chromosome [28]. We assayed localization patterns of RAD51 foci on zygotene stage chromosomes (Figure 2A) when RAD51 activity is high [29]; at this stage most RAD51 foci localize to synapsed regions, although in a few instances they (and also DMC1 foci, see below) are associated with unsynapsed axial elements. From Figure 2A, it is clear that there was considerable among-animal variation, particularly in the C3H and B6 strains. Nevertheless, considering the pooled data, the mean numbers of RAD51 foci per cell were highly significantly different among the three strains (F = 113.7; p<0.0001), with the variation mirroring that observed for MLH1; i.e., CAST had the lowest mean value (163.0+/−18.6), C3H an intermediate value (179.9+/−28.0), and B6 the highest value (222.1+/−33.8) (Figure 2A; Table S2). Further, considering those animals for which we had both RAD51 and MLH1 values (see Tables S1 and S2), the ratios of RAD51:MLH1 foci were similar among the three strains, with values of 7.7∶1 for CAST, 7.3∶1 for C3H and 8.8∶1 for B6.
Fig. 2. Inter-strain variation in early- and mid-stage meiotic recombination proteins. 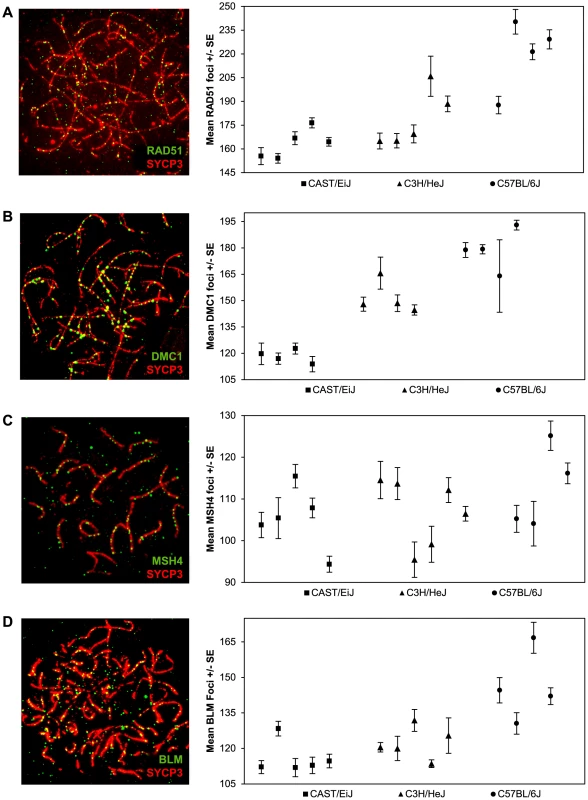
The number and location of foci for proteins acting upstream of MLH1 in the recombination pathway [i.e., (A) RAD51 (B) DMC1 (C) MSH4] or anti-recombination pathway [i.e., (D) BLM] were determined for each of the three strains and inter-strain values compared. For each of the four proteins, mean numbers of foci per cell varied significantly among the three strains. Data for each protein were based on 4–6 animals per strain, and a minimum of 60 cells per strain (see Supplemental Tables 2–5 for data). Other than RAD51, relatively few animals were scored for both MLH1 and another recombination protein (i.e., either DMC1, MSH4 or BLM) in each of the three strains; thus, we were not able to directly compare the ratios of MLH1:DMC1, MLH1:MSH4 or MLH1:BLM among the three strains. Nevertheless, the pooled data were consistent with the results from MLH1 and RAD51. For example, similar strain-specific differences were observed for zygotene stage cells scored for the meiosis specific strand invasion protein DMC1, which attaches to DSB sites shortly after RAD51 [29], [30]. Specifically, the mean values rose from 119.5+/−16.4 for CAST to 149.3+/−18.1 for C3H and 181.8+/−21.1 for B6 (F = 214.3; p<0.0001) (Figure 2B; Table S3). Consistent with previous reports on DMC1 localization in mice [29], all three strains exhibited fewer DMC1 than RAD51 foci. While the reason for this variation in numbers of DMC1 and RAD51 foci is not entirely clear, it presumably reflects the different roles the two proteins play in the early stages of the recombination pathway [28], [31], [32] or possibly, differences in the time that the proteins remain complexed to the recombination nodules.
We investigated a later stage of recombination by assaying MSH4, a member of the MSH4-MSH5 complex that is thought to stabilize recombination intermediates (e.g., [19], [22], [33]. Since MSH4 localizes only to synapsed chromosome regions (Figure 2C), we counted foci in cells at the zygotene/pachytene boundary. We observed substantial variation in the values among the different individual mice but, similar to the results for RAD51 and DMC1, we saw an increase in the mean number of MSH4 foci from CAST (105.3+/−12.8) to C3H (109.0+/−14.8) and B6 (112.7+/−17.5) males (Figure 2C; Table S4). These differences were more modest than those identified for RAD51 and DMC1 and because there was considerable overlap in individual values from strain to strain, they need to be interpreted with caution. Nevertheless, the mean values for the “low” (CAST) and “high” (B6) strains were still highly significantly different from one another (t = 2.9; p<0.01); for CAST vs C3H and C3H vs B6 the values were non-significantly different (t = 1.5, p>0.13 and t = 1.5, p>0.14, respectively)
Finally, we analyzed zygotene cells for the helicase BLM, a regulator of recombination intermediates suggested to have anti-crossover activity ([23]; Figure 2D; Table S5). Strain specific differences in average numbers of BLM foci were similar to those observed for the positive regulators of crossovers; i.e., the mean value was lowest for CAST (114.4+/−15.9), intermediate for C3H (120.0+/−12.4), and highest for B6 (146.4+/−26.3) (F = 62.3; p<0.0001).
In summary, our observations on strain-specific variation for five different recombination pathway proteins (RAD51, DMC1, MSH4, BLM and MLH1) are consistent with one another; i.e., in each instance the mean number of foci per cell was lowest for CAST males, intermediate for C3H males, and highest for B6 males. Unfortunately, we were not able to collect data on each protein from each animal, limiting our ability to directly compare values among the different strains and, accordingly, to address other obvious questions, such as: which protein is the best predictor of MLH1 values, are the ratios of foci for different proteins the same among all three strains, and does the ratio of recombinogenic∶anti-recombinogenic proteins (e.g., RAD51:BLM) vary among strains? Nevertheless, taken together, the data provide strong evidence that, at least in males of these strains, a similar proportion of DSBs are translated into MLH1-associated crossovers.
Variation in chromatin configuration is correlated with MLH1 levels
In subsequent studies, we were interested in determining whether the strain differences were accompanied by variation in the configuration of the meiotic axis and/or DNA loops. Accordingly, for each strain we assayed total autosomal SC lengths in pachytene stage cells. We observed highly significant differences in mean SC length among the strains; i.e., for CAST 156.7+/−2.0 µm, for C3H 161.5+/−1.8 µm, and for B6 170.7+/−1.9 µm (F = 13.8; p<0.0001). These results are similar to the strain-specific observations for MLH1, suggesting that the strain differences in crossover levels are linked to variation in length of the meiotic axis (Figure 3A).
Fig. 3. Inter-strain variation in SC length and DNA loop size. 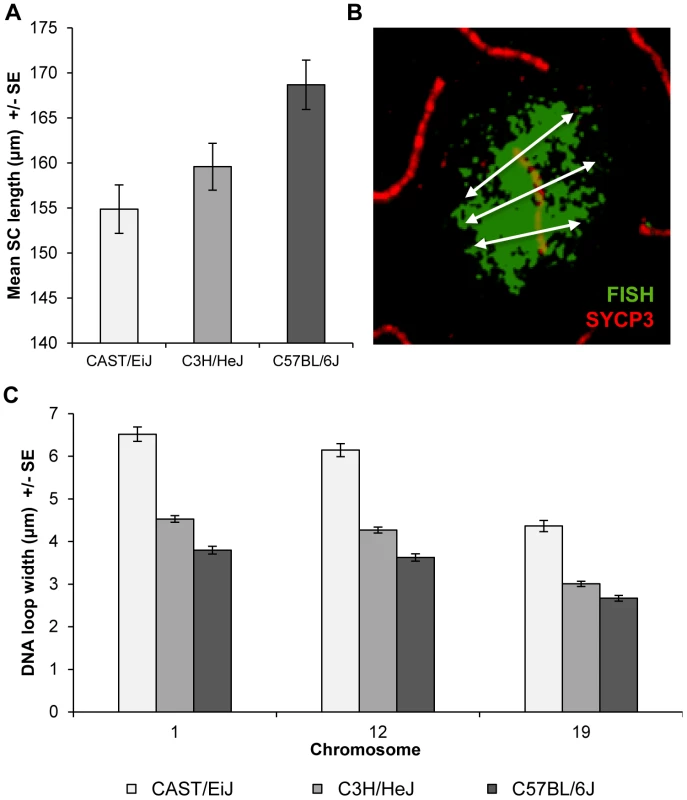
(A) Mean autosomal SC lengths varied in direct relationship to MLH1 values; i.e., the strain with the lowest mean MLH1 values (CAST) had the shortest SCs, while the strain with the highest MLH1 values (B6) had the longest SCs. Data are based on analyses of mid-pachytene spermatocytes from 2 CAST (n = 8 cells), 3 C3H (n = 11 cells) and 2 B6 (n = 21 cells) males. (B) DNA loop sizes for individual chromosomes were calculated as the average of the width of the FISH signal at three points along the SC: at the centromere, the midpoint, and the telomere; image shows example for chromosome 12. (C) Chromosomes 1, 12, and 19 were examined and for each, we observed an inverse relationship between strain-specific mean MLH1 values and mean DNA loop sizes. As a surrogate for DNA loop size, we examined the width of the signal from whole chromosome paint probes on representative large, medium-sized and small chromosomes; i.e., chromosomes, 1, 12 and 19, respectively (see Figure 3B for an example of a chromosome 12 paint probe). DNA loop sizes differed significantly among the strains for each of the three chromosomes: i.e., for chromosome 1, CAST = 6.5+/−1.3 µm, C3H = 4.5+/−0.6 µm and B6 = 3.8+/−0.7 µm (F = 183.5; p<0.0001); for chromosome 12, CAST = 6.2+/−1.2 µm, C3H = 4.3+/−0.6 µm and B6 = 3.6+/−0.7 µm (F = 176.5; p<0.0001); and for chromosome 19, CAST = 4.4+/−1.0 µm, C3H = 3.0+/−0.5 µm and B6 = 2.7+/−0.5 µm (F = 116.5; p<0.0001) (Figure 3C). Together with the observations on SC length, these analyses indicate that increasing MLH1 values are associated with smaller DNA loops and longer SCs.
DSB number per se does not regulate MLH1 levels or chromatin morphology
The correlation between the number of foci for “early”, “mid”, and “late” recombination proteins and strain-specific recombination levels raises the possibility that the number of DSBs per se regulates recombination. We tested this by comparing meiotic profiles of males on the same genetic background but with different rates of DSB formation. Specifically, we compared wildtype B6 males with siblings heterozygous for a null allele of Spo11, the type II topoisomerase-like protein responsible for programmed DSB formation in meiocytes [24]. Notably, the results from the wildtype Spo11 males were somewhat different than those of the B6 male described above, presumably due to variation from maintenance of the stocks at different facilities.
For the Spo11 animals, consistent with previous reports [34], [35], zygotene spermatocytes from Spo11+/− males displayed significantly fewer DSBs (estimated by the number of RAD51 foci) than spermatocytes from wildtype littermates (mean values = 152.2+/−20.6 and 200.5+/−21.5, respectively; t = 5.8, p<0.0001) (Figure 4A;Table S6). However, this did not translate into a difference in MLH1 values, with mean values of 23.9+/−1.9 and 23.8+/−1.6 for heterozygotes and wildtype littermates, respectively (Figure 4B; Table S7).
Fig. 4. Spo11+/+ and Spo11+/− animals. 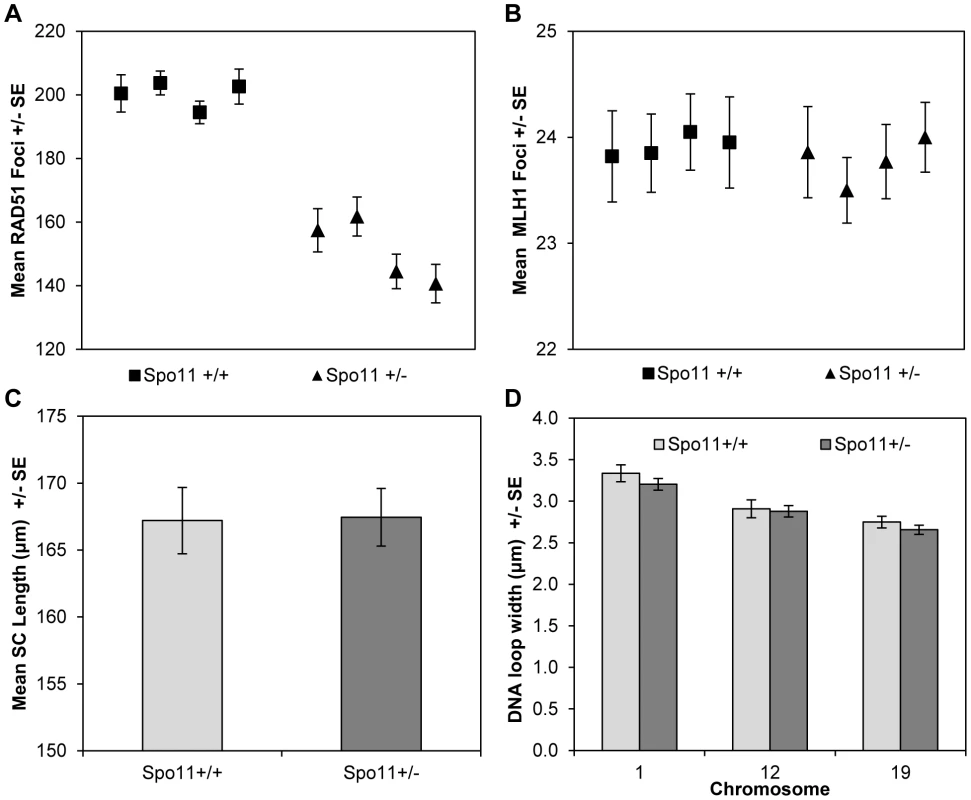
(A) Mice heterozygous for a null allele of Spo11 exhibited a significant decrease in RAD51 foci (a marker of DSBs) by comparison to wildtype littermates. However, (B) the mean number of MLH1 foci (a marker of COs), (C) mean SC lengths and (D) mean DNA loop sizes were not different between the two genotypes. Although it altered RAD51 values, Spo11 heterozygosity had no obvious effect on chromatin morphology. Specifically, total SC lengths per cell were virtually identical for Spo11+/+ and Spo11+/− littermates (167.1+/−3.3 µm and 169.7+/−4.8 µm, respectively; t = 0.46 p>0.65) (Figure 4C). Further, there were no consistent differences in DNA loop size on individual chromosomes between Spo11+/+ and Spo11+/− males [i.e, 3.3+/−0.5 µm and 3.2+/−0.5 µm, respectively, for chromosome 1 (t = 1.01, p = 0.31); 2.9+/−0.5 µm and 2.9+/−0.5 µm, respectively, for chromosome 12 (t = 0.27, p = 0.78), and 2.8+/−0.3 µm and 2.7+/−0.4 µm, respectively, for chromosome 19 (t = 1.00, p = 0.32)] (Figure 4D). Taken together with the results from the RAD51 and MLH1 assays, this indicates that the variation in crossover level among strains is not simply due to variation in the number of DSBs but more likely reflects differences in chromatin morphology.
Discussion
The purpose of this study was to investigate the basis for individual variation in recombination rates. Over the past 10–15 years a number of tools have become available to investigate the biology of meiotic recombination in mammals; e.g., knockout mice have been used to identify and characterize the functions of numerous meiotic genes (e.g.,[20], [36], [37], [38], [39]); genotypic analysis of individual and pooled sperm samples has led to the identification of small, discrete regions of high recombination activity [40], [41], [42], [43]; linkage and linkage disequilibrium studies have revealed the presence of thousands of recombination hotspots in mammalian genomes [7], [44], [45]; and genome-wide analyses of genetic polymorphisms have led to the identification of genes involved in modulating hotspot activity [46], [47], [48] or genome-wide recombination levels [13], [18].
Nevertheless, our understanding of the origin of among-individual variation in recombination levels in mammals remains rudimentary. In recent studies, genotypic differences at three loci have been linked to variation in the recombination phenotype. Specifically, allelic variation in the gene encoding the meiosis-specific histone methytransferase PRDM9 has been associated with hotspot activity in both mice and humans [46], [47], [48]; allelic variation in RNF212, a homolog of the C. elegans synapsis/cross-over associated gene zhp-3, has been shown to affect genome-wide recombination levels in human males and females [13], [14], [18]; and the presence of an inversion at 17q21.31 affects recombination rates in human females [13], [14], [49]. However, several lines of evidence indicate that these individual loci may be relatively unimportant, or at least not the only, determinants of variation in CO levels. First, in studies of PRDM9 in humans, allelic variation has been shown to influence hotspot usage but appears to have relatively little effect on genome-wide recombination levels [8], [46], [47], [50]. Second, the magnitude of the effects attributable to the other two loci (RNF212 and inv17q21.31) is insufficient to account for the level of variation in genome-wide rates identified in humans or in the present study [13], [18]. Finally, in recent analyses of inbred strains of mice with differing levels of genome-wide recombination conducted by us [51] and others [52], putative QTL-containing chromosome regions did not include either Prdm9 or Rnf212. Thus, the available evidence suggests that other, as yet unidentified, loci, are responsible for generating most of the variation in recombination rates among individuals or inbred strains.
To address this problem in a somewhat different way, we utilized inbred strains of mice with varying levels of genome-wide recombination to identify the temporal window during which the variation is generated. Surprisingly, our results provide strong evidence that the variation is attributable to processes acting at, or upstream of, DSB formation and repair. Specifically, our observations on foci number for each of three recombination-promoting proteins (RAD51, DMC1, and MSH4) and for the anti-recombination protein BLM were similar in finding a direct correlation between foci numbers and levels of recombination. In each of the three strains, the number of DSBs (measured as RAD51 foci) exceeded the number of crossovers (MLH1 foci) by approximately ten-fold. This is consistent with observations from previous studies of male mice (e.g, [53]) and indicates that as in other organisms, the vast majority of DSBs are repaired as non-crossovers. Further, it suggests that, at least for the inbred strains we examined, events occurring downstream of DSBs are relatively unimportant in mediating genetic background-dependent variation in recombination rates. This does not mean that these processes are irrelevant – e.g., RNF212, which is known to affect recombination levels in humans, has recently been show to stabilize proteins important in processing recombination intermediates in male mice [19] – only that there are other, more important determinants.
While our observations indicate that the variation in recombination levels is established by the time of DSB formation, our analyses of Spo11 deficient animals suggest that this is not attributable to DSBs per se. That is, despite having significantly different numbers of RAD51 foci, wildtype and heterozygous animals had virtually identical mean MLH1 values. These results are similar to those recently reported by Cole and colleagues [17], in which they demonstrated a positive correlation between the number of functional Spo11 genes and the number of DMC1 and RAD51 foci, but no accompanying change in the number of MLH1 foci. Thus, as in other model organisms (e.g., [16], [54]), mammalian meiosis appears to have homeostatic controls that operate to maintain the number of crossovers in the face of variation in the number of DSBs.
Taken together, our analyses of different inbred strains of mice and of mice varying at a single locus but on an otherwise isogenic background demonstrate that DSB number itself is not the driver of variation in CO levels. What other processes might be responsible? One obvious possibility is the way in which the chromatin is packaged, an idea consistent with the observations of the present report. For example, the best predictor of the number of MLH1 foci across the five different categories of mice that we examined (i.e., CAST, C3H, B6, Spo11+/+ and Spo11+/−) was SC length, since in each instance there was an average of approximately 7 µm SC per MLH1 focus. Similarly, the relationship between DNA loop sizes on individual chromosomes and the number of MLH1 foci was comparable in animals of different genetic background or Spo11 genotype. An obvious caveat to this interpretation is the fact that the observations on SC length and DNA loop size were based on pachytene cells while, presumably, the determination of the relative level of COs occurs much earlier. However, in unrelated studies of human males and females, we found that sex-specific differences in chromatin morphology in pachytene stage meiocytes were mirrored in leptotene preparations [55]. Thus, we think it unlikely that the observations of the present report simply reflect pachytene cells. Accordingly, we suggest that modification of chromatin morphology – but not DSB numbers – is a primary determinant of CO levels. This is consistent with recent analyses of Petukhova and Camerini-Otero and colleagues [56], [57], as they found that trimethylation of H3K4 at potential PRDM9 recombination hotspots is not dependent on SPO11; i.e., the establishment of hotspot-associated marks occurs regardless of the presence of DSBs.
What genetic loci might be at work to influence chromatin packaging and, ultimately, variation in CO levels? In two recent QTL analyses of recombination in different inbred strains of [51], [52], several QTLs were shared between the studies; i.e., overlapping regions were identified on chromosomes 4, 15 and 17, and nearly overlapping regions on chromosome 3 and the X chromosome. Notably, in both studies the highest lod scores were observed on the X chromosome, consistent with previous studies linking X-linked loci with variation in genome-wide recombination rates [12], [58]. Because inactivation of the sex chromosomes (MSCI) appears to be essential for normal male meiosis [59], it is generally thought that few, if any, X-linked loci are necessary for spermatogenesis. The results of the analyses of Murdoch et al [51] and Dumont and Payseur [52], as well as studies identifying spermatogenic functions for X-linked genes (e.g., Tex11; [60]), suggests that this is not an absolute rule, and that high resolution analyses of the X chromosome may uncover important recombination-associated loci.
Materials and Methods
Ethics statement
All animal experiments and procedures conducted at Washington State University were performed using protocols approved by the Institutional Animal Care and Use Committee (WSU IACUC number 03737-014).
Mice
Breeding stocks of three inbred strains, C57BL/6J, C3H/HeJ, and CAST/EiJ, were obtained from The Jackson Laboratory and maintained by brother-sister mating. Spo11+/+ and Spo11+/− animals (on a C57BL/6J background) were kindly provided by Drs. Maria Jasin and Scott Keeney, Memorial Sloan Kettering Cancer Center. All animals were housed in polysulfone cages on ventilated racks or static cages. Experiments were approved by the Washington State University (WSU) Institutional Animal Care and Use Committee. WSU is fully accredited by the American Association for Accreditation of Laboratory Animal Care and all investigations were conducted in accordance with the Guide for the Care and Use of Laboratory Animals.
Tissue collection
Adult male mice between the ages of 6 and 40 weeks were killed by cervical dislocation and the testes removed. Seminiferous tubules were removed from the testes and surface spread preparations of spermatocytes for immunofluorescence studies prepared as described previously [61].
Immunostaining
Slides were immunostained using similar methodology to that of Anderson et al. [26]. Antibodies were diluted in sterile filtered 1×ADB consisting of 10 ml normal donkey serum (Jackson ImmunoResearch), 3 g BSA (Sigma-Aldrich), 50 µl Triton X-100 (Alfa Aesar), and 990 ml PBS. Incubations were performed in a dark humid chamber at 37°C.
Slides were incubated overnight in a dark humid chamber at 37°C with one of the following primary antibodies: RAD51 (rabbit anti-human, Santa Cruz Biotechnology) diluted 1∶75, DMC1 (rabbit anti-human, Santa Cruz Biotechnology) diluted 1∶200, BLM (rabbit anti-human, Santa Cruz Biotechnology) diluted 1∶100, MSH4 (rabbit anti-human, provided by Dr. Chengtao Her) diluted 1∶75, or MLH1 (rabbit anti-human, Calbiochem) diluted 1∶75. All slides were incubated with a primary antibody for SYCP3 (goat anti-mouse, Santa Cruz Biotechnology) diluted 1∶100 for 2 hours. Fluorescein-labeled donkey anti-rabbit (Jackson ImmunoResearch) secondary antibody was diluted 1∶75 and incubated overnight, followed by an incubation for 45 minutes with rhodamine-labeled donkey anti-goat secondary antibodies (Jackson ImmunoResearch), diluted 1∶200. Slides were counterstained and fixed using Prolong Gold antifade reagent with DAPI (Invitrogen) and sealed with rubber cement.
Fluorescence in-situ hybridization
StarFish whole chromosome paint probes (Cambio) were used according to the manufacturer's instructions to estimate DNA loops of specific chromosomes. Briefly, previously immunostained slides were dehydrated in a series of brief ethanol washes (75%, 90%, 100%), denatured in a 70% dionized formamide/2×SSC solution for 5 minutes at 65°C, quenched in 70% ethanol at −20°C, and dehydrated again with a series of ethanol washes. Paint probes for chromosomes 1, 12, and 19 were denatured for 10 minutes at 65°C and applied to the slides overnight in a dark humid chamber at 37°C. Following incubation, slides were soaked twice for 5 minutes in 50% dionized formamide/1×SSC solution at 45°C, washed twice in 1×SSC at 45°C for 5 minutes, and soaked for three cycles in 25% Detergent DT/4×SSC (Cambio) for 4 minutes at 45°C. Slides were allowed to drain and mounted with Reagent MD (Cambio) and sealed with rubber cement.
Microscopy and scoring
For immunofluorescence analysis, cells were identified using DAPI and the sub-stage of meiotic prophase determined on the basis of the morphology of the SC (visualized with SYCP3 antibody). For each cell and each protein analyzed, we scored the number of foci localizing to the axial element or SC at the appropriate sub-stage of meiotic prophase; i.e., for RAD51, DMC1, and BLM prior to complete synapsis during zygotene, MSH4 at the zygotene-pachytene transition, and MLH1 during pachytene prior to desynapsis of the XY bivalent. For analysis of MLH1 foci we restricted our scoring to autosomal chromosomes, since the appearance and disappearance of foci on the XY bivalent and on the autosomes are temporally uncoupled.
Position data for MLH1 foci was collected by measuring along the fully synapsed SC from the centromere to each MLH1 focus. The placement of MLH1 was calculated by dividing the centromere-MLH1 distances by the full length of each individual SC. Values are represented as a percentage of total SC length, with 0% being the centromeric end and 100% the telomeric end of the SC.
Genome-wide SC length was measured in fully synapsed pachytene cells by manually tracing the length of the SYCP3 signal for all SCs except the XY bivalent. The genome-wide SC length per cell was calculated as the sum of the SYCP3 signal lengths for the 19 autosomes.
DNA loop sizes were assayed on three representative chromosomes, 1, 12, and 19, identified by FISH paint probes, using an approach similar to that previously described by Novak et al [62] FISH images were then overlaid on immunofluorescence images of the SC and loop. Loop size was assayed by taking linear measurements of the width of the FISH signal perpendicular to the SC at the centromere, telomere, and mid-point of the SC. The three lengths for each chromosome were averaged and compared on a chromosome-specific basis among the strains.
All slides were imaged on a Zeiss Axio Imager epifluorescence microscope and analyzed by blinded observers using Zeiss Axiovision software (version 4.7).
Statistical analysis
Comparisons in the average numbers of foci between different strains or genotypes of mice were tested by analysis of variance or Student t-test analysis, pooling the results from multiple mice in a strain or genotype. For post-hoc comparisons of mean values between specific strains, we used the Bonferroni correction to account for the multiple tests. Mean values +/ − SD are provided in the text and tables and, for illustrative purposes, as mean values +/ − SE in the figures. Differences in the numbers of E0 bivalents were compared by chi-square tests. All statistical analyses were performed with JMP software, version 7.0.1 (SAS Institute Inc.).
Supporting Information
Zdroje
1. RockmillB, RoederGS (1994) The yeast med1 mutant undergoes both meiotic homolog nondisjunction and precocious separation of sister chromatids. Genetics 136 : 65–74.
2. MolnarM, ParisiS, KakiharaY, NojimaH, YamamotoA, et al. (2001) Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics 157 : 519–532.
3. ZetkaMC, RoseAM (1995) Mutant rec-1 eliminates the meiotic pattern of crossing over in Caenorhabditis elegans. Genetics 141 : 1339–1349.
4. HawleyRS, TheurkaufWE (1993) Requiem for distributive segregation: achiasmate segregation in Drosophila females. Trends Genet 9 : 310–317.
5. HassoldT, HuntP (2001) To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2 : 280–291.
6. CheungVG, BurdickJT, HirschmannD, MorleyM (2007) Polymorphic variation in human meiotic recombination. American journal of human genetics 80 : 526–530.
7. CoopG, WenX, OberC, PritchardJK, PrzeworskiM (2008) High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319 : 1395–1398.
8. KongA, ThorleifssonG, GudbjartssonDF, MassonG, SigurdssonA, et al. (2010) Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467 : 1099–1103.
9. LynnA, KoehlerKE, JudisL, ChanER, CherryJP, et al. (2002) Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296 : 2222–2225.
10. HassoldT, HansenT, HuntP, VandeVoortC (2009) Cytological studies of recombination in rhesus males. Cytogenetic and genome research 124 : 132–138.
11. KoehlerKE, CherryJP, LynnA, HuntPA, HassoldTJ (2002) Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics 162 : 297–306.
12. DumontBL, WhiteMA, SteffyB, WiltshireT, PayseurBA (2011) Extensive recombination rate variation in the house mouse species complex inferred from genetic linkage maps. Genome research 21 : 114–125.
13. ChowdhuryR, BoisPR, FeingoldE, ShermanSL, CheungVG (2009) Genetic analysis of variation in human meiotic recombination. PLoS genetics 5: e1000648.
14. Fledel-AlonA, LefflerEM, GuanY, StephensM, CoopG, et al. (2011) Variation in human recombination rates and its genetic determinants. PloS one 6: e20321.
15. MatiseTC, ChenF, ChenW, De La VegaFM, HansenM, et al. (2007) A second-generation combined linkage physical map of the human genome. Genome Res 17 : 1783–1786.
16. MartiniE, DiazRL, HunterN, KeeneyS (2006) Crossover homeostasis in yeast meiosis. Cell 126 : 285–295.
17. ColeF, KauppiL, LangeJ, RoigI, WangR, et al. (2012) Homeostatic control of recombination is implemented progressively in mouse meiosis. Nature cell biology 14 : 424–430.
18. KongA, ThorleifssonG, StefanssonH, MassonG, HelgasonA, et al. (2008) Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319 : 1398–1401.
19. ReynoldsA, QiaoH, YangY, ChenJK, JacksonN, et al. (2013) RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nature Genetics 45 : 269–278.
20. BakerSM, PlugAW, ProllaTA, BronnerCE, HarrisAC, et al. (1996) Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over [see comments]. Nat Genet 13 : 336–342.
21. MassonJY, WestSC (2001) The Rad51 and Dmc1 recombinases: a non-identical twin relationship. Trends in biochemical sciences 26 : 131–136.
22. Santucci-DarmaninS, WalpitaD, LespinasseF, DesnuelleC, AshleyT, et al. (2000) MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 14 : 1539–1547.
23. JessopL, RockmillB, RoederGS, LichtenM (2006) Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS genetics 2: e155.
24. KeeneyS, GirouxCN, KlecknerN (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 : 375–384.
25. KeeneyS, BaudatF, AngelesM, ZhouZH, CopelandNG, et al. (1999) A mouse homolog of the Saccharomyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics 61 : 170–182.
26. AndersonLK, ReevesA, WebbLM, AshleyT (1999) Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151 : 1569–1579.
27. SvetlanovA, BaudatF, CohenPE, de MassyB (2008) Distinct functions of MLH3 at recombination hot spots in the mouse. Genetics 178 : 1937–1945.
28. SheridanSD, YuX, RothR, HeuserJE, SehornMG, et al. (2008) A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic acids research 36 : 4057–4066.
29. MoensPB, KolasNK, TarsounasM, MarconE, CohenPE, et al. (2002) The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. Journal of cell science 115 : 1611–1622.
30. TarsounasM, MoritaT, PearlmanRE, MoensPB (1999) RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J Cell Biol 147 : 207–220.
31. BarchiM, MahadevaiahS, Di GiacomoM, BaudatF, de RooijDG, et al. (2005) Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Molecular and cellular biology 25 : 7203–7215.
32. CallenderTL, HollingsworthNM (2010) Mek1 suppression of meiotic double-strand break repair is specific to sister chromatids, chromosome autonomous and independent of Rec8 cohesin complexes. Genetics 185 : 771–782.
33. BornerGV, KlecknerN, HunterN (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117 : 29–45.
34. BellaniMA, RomanienkoPJ, CairattiDA, Camerini-OteroRD (2005) SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm-/ - spermatocytes. Journal of cell science 118 : 3233–3245.
35. BarchiM, RoigI, Di GiacomoM, de RooijDG, KeeneyS, et al. (2008) ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS genetics 4: e1000076.
36. BakerSM, BronnerCE, ZhangL, PlugAW, RobatzekM, et al. (1995) Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell 82 : 309–319.
37. YuanL, LiuJG, HojaMR, WilbertzJ, NordqvistK, et al. (2002) Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science 296 : 1115–1118.
38. BaudatF, ManovaK, YuenJP, JasinM, KeeneyS (2000) Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking spo11. Mol Cell 6 : 989–998.
39. EdelmannW, CohenPE, KneitzB, WinandN, LiaM, et al. (1999) Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nature Genetics 21 : 123–127.
40. BadgeRM, YardleyJ, JeffreysAJ, ArmourJA (2000) Crossover breakpoint mapping identifies a subtelomeric hotspot for male meiotic recombination. Human Molecular Genetics 9 : 1239–1244.
41. LienS, SzydaJ, SchechingerB, RappoldG, ArnheimN (2000) Evidence for heterogeneity in recombination in the human pseudoautosomal region: high resolution analysis by sperm typing and radiation-hybrid mapping. American journal of human genetics 66 : 557–566.
42. JeffreysAJ, MurrayJ, NeumannR (1998) High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol Cell 2 : 267–273.
43. JeffreysAJ, MayCA (2004) Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nature Genetics 36 : 151–156.
44. McVeanGA, MyersSR, HuntS, DeloukasP, BentleyDR, et al. (2004) The fine-scale structure of recombination rate variation in the human genome. Science 304 : 581–584.
45. MyersS, BottoloL, FreemanC, McVeanG, DonnellyP (2005) A fine-scale map of recombination rates and hotspots across the human genome. Science 310 : 321–324.
46. BaudatF, BuardJ, GreyC, Fledel-AlonA, OberC, et al. (2010) PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327 : 836–840.
47. BergIL, NeumannR, LamKW, SarbajnaS, Odenthal-HesseL, et al. (2010) PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nature Genetics 42 : 859–863.
48. ParvanovED, PetkovPM, PaigenK (2010) Prdm9 controls activation of mammalian recombination hotspots. Science 327 : 835.
49. StefanssonH, HelgasonA, ThorleifssonG, SteinthorsdottirV, MassonG, et al. (2005) A common inversion under selection in Europeans. Nature Genetics 37 : 129–137.
50. HinchAG, TandonA, PattersonN, SongY, RohlandN, et al. (2011) The landscape of recombination in African Americans. Nature 476 : 170–175.
51. MurdochB, OwenN, ShirleyS, CrumbS, BromanKW, et al. (2010) Multiple loci contribute to genome-wide recombination levels in male mice. Mammalian genome : official journal of the International Mammalian Genome Society 21 : 550–555.
52. DumontBL, PayseurBA (2011) Genetic analysis of genome-scale recombination rate evolution in house mice. PLoS Genet 7: e1002116.
53. MoensPB, MarconE, ShoreJS, KochakpourN, SpyropoulosB (2007) Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. Journal of cell science 120 : 1017–1027.
54. RosuS, LibudaDE, VilleneuveAM (2011) Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science 334 : 1286–1289.
55. GruhnJ, RubioC, BromanK, HuntP, HassoldT (2013) Cytological studies of human meiosis: sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PloS one 8(12): e85075.
56. SmagulovaF, GregorettiIV, BrickK, KhilP, Camerini-OteroRD, et al. (2011) Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472 : 375–378.
57. BrickK, SmagulovaF, KhilP, Camerini-OteroRD, PetukhovaGV (2012) Genetic recombination is directed away from functional genomic elements in mice. Nature 485 : 642–645.
58. de La Casa-EsperonE, Loredo-OstiJC, Pardo-Manuel de VillenaF, BriscoeTL, MaletteJM, et al. (2002) X chromosome effect on maternal recombination and meiotic drive in the mouse. Genetics 161 : 1651–1659.
59. RoyoH, PolikiewiczG, MahadevaiahSK, ProsserH, MitchellM, et al. (2010) Evidence that meiotic sex chromosome inactivation is essential for male fertility. Current biology : CB 20 : 2117–2123.
60. AdelmanCA, PetriniJH (2008) ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS genetics 4: e1000042.
61. PetersAH, PlugAW, van VugtMJ, de BoerP (1997) A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 5 : 66–68.
62. NovakI, WangH, RevenkovaE, JessbergerR, ScherthanH, et al. (2008) Cohesin Smc1beta determines meiotic chromatin axis loop organization. The Journal of cell biology 180 : 83–90.
Štítky
Genetika Reprodukční medicína
Článek Unwrapping BacteriaČlánek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 1
-
Všechny články tohoto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



