-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDown-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
Interhomolog recombination plays a critical role in promoting proper meiotic chromosome segregation but a mechanistic understanding of this process is far from complete. In vegetative cells, Rad51 is a highly conserved recombinase that exhibits a preference for repairing double strand breaks (DSBs) using sister chromatids, in contrast to the conserved, meiosis-specific recombinase, Dmc1, which preferentially repairs programmed DSBs using homologs. Despite the different preferences for repair templates, both Rad51 and Dmc1 are required for interhomolog recombination during meiosis. This paradox has recently been explained by the finding that Rad51 protein, but not its strand exchange activity, promotes Dmc1 function in budding yeast. Rad51 activity is inhibited in dmc1Δ mutants, where the failure to repair meiotic DSBs triggers the meiotic recombination checkpoint, resulting in prophase arrest. The question remains whether inhibition of Rad51 activity is important during wild-type meiosis, or whether inactivation of Rad51 occurs only as a result of the absence of DMC1 or checkpoint activation. This work shows that strains in which mechanisms that down-regulate Rad51 activity are removed exhibit reduced numbers of interhomolog crossovers and noncrossovers. A hypomorphic mutant, dmc1-T159A, makes less stable presynaptic filaments but is still able to mediate strand exchange and interact with accessory factors. Combining dmc1-T159A with up-regulated Rad51 activity reduces interhomolog recombination and spore viability, while increasing intersister joint molecule formation. These results support the idea that down-regulation of Rad51 activity is important during meiosis to prevent Rad51 from competing with Dmc1 for repair of meiotic DSBs.
Published in the journal: . PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1004005
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004005Summary
Interhomolog recombination plays a critical role in promoting proper meiotic chromosome segregation but a mechanistic understanding of this process is far from complete. In vegetative cells, Rad51 is a highly conserved recombinase that exhibits a preference for repairing double strand breaks (DSBs) using sister chromatids, in contrast to the conserved, meiosis-specific recombinase, Dmc1, which preferentially repairs programmed DSBs using homologs. Despite the different preferences for repair templates, both Rad51 and Dmc1 are required for interhomolog recombination during meiosis. This paradox has recently been explained by the finding that Rad51 protein, but not its strand exchange activity, promotes Dmc1 function in budding yeast. Rad51 activity is inhibited in dmc1Δ mutants, where the failure to repair meiotic DSBs triggers the meiotic recombination checkpoint, resulting in prophase arrest. The question remains whether inhibition of Rad51 activity is important during wild-type meiosis, or whether inactivation of Rad51 occurs only as a result of the absence of DMC1 or checkpoint activation. This work shows that strains in which mechanisms that down-regulate Rad51 activity are removed exhibit reduced numbers of interhomolog crossovers and noncrossovers. A hypomorphic mutant, dmc1-T159A, makes less stable presynaptic filaments but is still able to mediate strand exchange and interact with accessory factors. Combining dmc1-T159A with up-regulated Rad51 activity reduces interhomolog recombination and spore viability, while increasing intersister joint molecule formation. These results support the idea that down-regulation of Rad51 activity is important during meiosis to prevent Rad51 from competing with Dmc1 for repair of meiotic DSBs.
Introduction
Meiotic recombination is a highly conserved process that is critical for the accurate segregation of homologous chromosomes to opposite poles at the first meiotic division. Crossovers between non-sister chromatids, in combination with sister chromatid cohesion, physically connect homologous chromosomes, thereby allowing them to align properly at Metaphase I [1]. In the absence of crossovers, homologs segregate randomly to generate chromosomally imbalanced gametes, resulting in infertility or birth defects [2].
Meiotic recombination is initiated by the introduction of programmed double-strand breaks (DSBs) catalyzed by a meiosis-specific, evolutionarily conserved, topoisomerase-like protein, Spo11 [3]. Resection of the 5′ ends of the breaks results in 3′ single stranded tails. In many organisms, including yeast and mammals, these single stranded ends are bound by two conserved RecA-like recombinases, Rad51 and Dmc1, to form nucleoprotein filaments that mediate strand invasion of homologous DNA duplexes [4], [5]. Rad51 is present in both vegetative and meiotic cells, while Dmc1 is meiosis-specific. Subsequent processing of the resulting recombination intermediates results in the formation of either crossover or non-crossover chromosomes [6].
Rad54 and Rdh54/Tid1 are key accessory factors that physically interact with Rad51 and Dmc1 [7]–[10]. These paralogs are members of the Swi/Snf chromatin remodeling family of proteins and affect multiple steps of recombination, including stabilization of the Rad51 filament, the promotion of strand invasion, and the removal of recombinases after strand invasion to allow extension of the 3′ ends by DNA synthesis [11]. Rdh54 is also important in meiosis for removing Dmc1 from double stranded DNA that has not experienced DSBs [12]. There can be crosstalk between Rad54/Rdh54 and Rad51/Dmc1 (meaning, for example, that Rad54 may function with both Rad51 and Dmc1), as rad54Δ rdh54Δ double mutants have more severe meiotic phenotypes than either rad54Δ or rdh54Δ alone [13], [14]. However, recent biochemical studies using recombinant Dmc1, Rad51, Rad54 and Rdh54 proteins have demonstrated that Dmc1-Rdh54 and Rad51–Rad54 work as functionally distinct pairs for strand invasion in vitro [15].
In each meiosis, there are ∼160 DSBs generated by Spo11 [16]. The majority of these breaks are repaired by interhomolog interactions either as crossovers (COs) or noncrossovers (NCOs), while the remainder are presumably repaired using sister chromatids as templates [17], [18]. In yeast, physical analyses of joint molecule recombination intermediates have indicated a bias of approximately 4∶1 for interhomolog versus intersister recombination events [19]. This bias for interhomolog recombination is unique to meiosis: in vegetative cells, Rad51 acts to repair DNA damage preferentially using sister chromatids [20], [21].
Axial elements are generated when sister chromatids condense through the formation of loops tethered at their bases by meiosis-specific proteins [22]. In yeast, these proteins include Hop1, Red1 and Mek1 [23]–[25]. Diploids containing deletions of these genes exhibit reduced interhomolog recombination and increased levels of intersister recombination, demonstrating the importance of axial element components in promoting interhomolog recombination [26]–[31]. While Hop1 and Red1 are structural chromosomal components of axial elements, Mek1 is a serine/threonine protein kinase [32].
Prior to DSB formation, sequences within the loops of DNA created during axial element formation are recruited to the axes to form “tethered loop axis complexes” [33], [34]. Breaks are then created on the axes where Red1, Hop1 and Mek1 are present. This is important because DSBs result in the localized activation of Mek1 kinase activity via checkpoint kinase Mec1 phosphorylation of Hop1 [32], [35], [36]. It has been proposed that one end of each break remains tethered to the sister chromatid while the other end forms a “tentacle” that searches for the homologous chromosome. Mek1 facilitates interhomolog interactions in part by antagonizing sister chromatid cohesion mediated by meiosis-specific cohesin complexes [27].
Although both rad51Δ and dmc1Δ reduce interhomolog recombination, additional phenotypes of rad51Δ and dmc1Δ mutants are different, suggesting they play discrete roles in meiotic recombination [19], [37]–[39], [40]. RAD51 is required to efficiently recruit Dmc1 to DSBs [37], [41]. In budding yeast, strains in which Dmc1 is the only recombinase (i.e. rad51Δ mutants) do not show interhomolog bias, indicating that the presence of the Rad51 protein is necessary to promote Dmc1 strand invasion of homologs [19], [38], [42]. This idea was confirmed by studies using a mutant of RAD51 that is specifically defective in strand exchange. This mutant, rad51-II3A, exhibits a wild-type ratio of interhomolog∶intersister joint molecules, indicating that the presence of inactive Rad51 protein is sufficient for Dmc1 to mediate the bulk of meiotic recombination [43].
DSBs in dmc1Δ mutants are not repaired and become hyperresected in the SK1 strain background where the phenotype is most severe [44]. These breaks trigger the meiotic recombination checkpoint and cells arrest prior to the Meiosis I division [45]. Rad51 foci persist in dmc1Δ diploids, indicating the recombinase is present at DSBs but is not able to repair them [37], [40]. Inactivation of an analog-sensitive version of Mek1, called Mek1-as, after DSBs have formed in dmc1Δ cells allows Rad51-dependent repair of the breaks using sister chromatids [32], [46]. Therefore Mek1 phosphorylation of protein substrates is essential to inhibit Rad51 activity in the absence of DMC1.
A key step in downregulating Rad51 activity in dmc1Δ diploids is the inhibition of Rad51–Rad54 complex formation. There are two independent, meiosis-specific mechanisms by which formation of this complex can be suppressed. First, a meiosis-specific protein called Hed1 binds to Rad51 and excludes Rad54 [47], [48]. Ectopic expression of HED1 in vegetative cells makes cells sensitive to DNA-damaging agents, indicating that the presence of Hed1 is sufficient to impair DNA repair by Rad51 [48]. Second, Rad54 is an in vivo target of Mek1. Phosphorylation of threonine 132 on Rad54 reduces the affinity of Rad54 for Rad51 in vitro, as the negative charge conferred by phosphorylation makes Rad51–Rad54 complex formation more difficult [49]. Consistent with this idea, substitution of aspartic acid, a negatively charged, phosphomimetic amino acid, for Rad54 threonine 132 increases sensitivity to DNA damaging agents in vegetative cells, while substitution with either alanine or lysine does not. Conditions that allow formation Rad51–Rad54 complexes in meiosis, such as over-expression of RAD51 or RAD54, deletion of HED1 or mutation of RAD54-T132 to alanine, result in partial suppression of the dmc1Δ interhomolog recombination and spore viability defects [48]–[51]. Rad51-mediated interhomolog recombination in these strains requires Mek1, indicating there are additional Mek1 substrates that direct filaments containing Rad51–Rad54 to homologs [49].
Much of the work looking at Rad51 regulation has occurred in the dmc1Δ strains, as it simplifies the analysis to have only one recombinase present. However, the fact that hed1Δ and/or RAD54-T132A have little to no effect on spore viability in the presence of wild-type DMC1 raises the question of whether down-regulation of Rad51 activity is part of normal meiosis [48], [49]. Several proposals have been made that suggest inhibition of Rad51 activity is specific to dmc1Δ: (1) Hed1 only inactivates Rad51 when DMC1 is absent, (2) Mek1 regulation of Rad54 occurs only under checkpoint induced conditions and (3) the accumulation of single stranded DNA from the failure of DSB repair in dmc1Δ mutants causes hyperactivation of Mek1, resulting in a global inhibition of recombination [27], [48], [52]. This work addresses the question of whether down-regulation of Rad51 is important in wild-type meiosis by examining recombination in situations where Rad51 is activated but where Dmc1 is still functional and meiotic checkpoint arrest does not occur. We demonstrate that under certain conditions, Rad51, if not down-regulated, is capable of competing with Dmc1 during meiosis for DSB repair in yeast. We propose that down-regulation of Rad51 activity is therefore important to promote Dmc1-dependent interhomolog recombination.
Results
Mutation of a Conserved Amino Acid Creates a Hypomorphic dmc1 Mutant
The use of dmc1Δ to study Rad51 inhibition has raised the possibility that it is the absence of DMC1 or a pathological situation created by checkpoint arrest that is responsible for the down-regulation of Rad51 activity. One way to circumvent this problem is to create a mutant of DMC1 that is still able to function in interhomolog recombination and therefore does not arrest, but is less efficient than wild type. If Rad51 and Dmc1 can compete for repair of DSBs, then combining up-regulated Rad51 with a hypomorphic version of DMC1 may result in reduced interhomolog recombination and spore viability.
A hypomorphic DMC1 allele was generated by mutation of threonine 159 to alanine. This threonine is highly conserved in both the Dmc1 and Rad51 protein families [53] (A. Neiman, personal communication). T159 is located two amino acids away from a conserved glutamate that is required for hydrolysis of ATP. In addition, T159 is equivalent to T164 in human Dmc1, which is located immediately adjacent to an asparagine (N163) that is important for monomer-monomer contacts [53]. We therefore hypothesized that the T159A mutation might make oligomerization of Dmc1 less efficient. Dmc1 interacts with itself in the two-hybrid system [7]. The T159A mutation was introduced into DMC1 fused to either the Gal4 DNA binding domain (GBD) or the Gal4 activation domain (GAD), the plasmids were transformed into a strain containing a GAL1-lacZ reporter construct and protein-protein interactions were assessed by β-galactosidase assays. Consistent with published results, interaction between the two wild-type alleles of DMC1 resulted in >100 units of β-galactosidase activity, while the control containing GBD-DMC1 and GAD alone, exhibited background levels of activity [7] (Table 1). Plasmid combinations containing DMC1/dmc1-T159A exhibited only ∼30% the levels of wild-type β-galacotosidase activity, while no interaction was detected in cells containing both GBD-dmc1-T159A and GAD-dmc1-T159A. These results support the idea that the substitution of threonine 159 for alanine affects Dmc1 oligomerization.
Tab. 1. Two-hybrid interactions between DMC1 and dmc1-T159A. 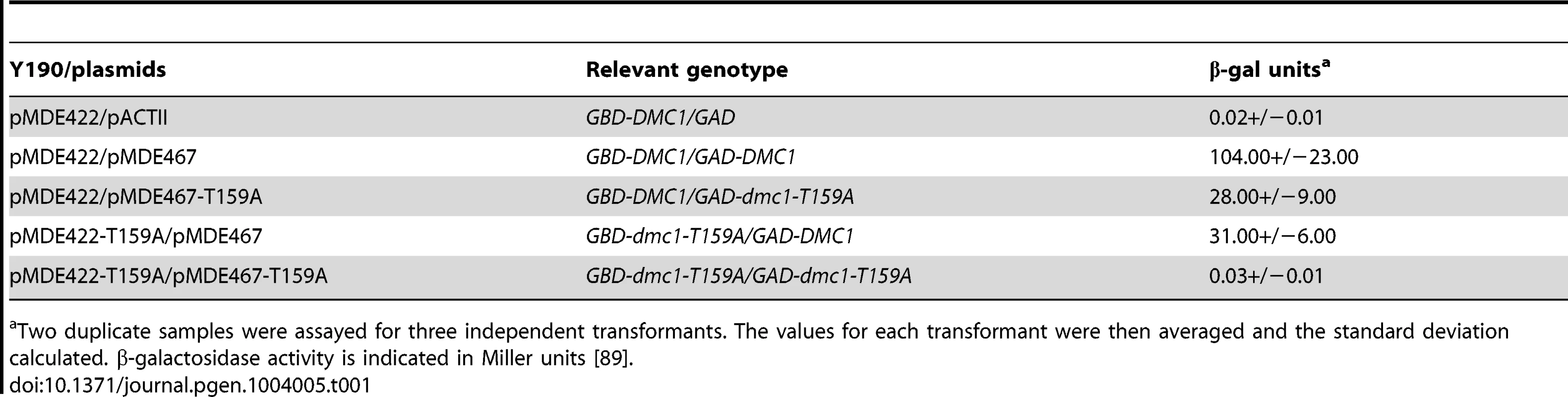
a Two duplicate samples were assayed for three independent transformants. The values for each transformant were then averaged and the standard deviation calculated. β-galactosidase activity is indicated in Miller units [89]. To determine whether any biochemical properties are affected by the T159A mutation, His6-tagged Dmc1 and Dmc1-T159A were purified from bacteria using a new procedure developed in the Sung laboratory [54]. Both wild-type and mutant proteins gave equivalent yields of good purity, indicating that the T159A mutation does not destabilize the protein (Figure 1A). To look at filament stability, an RPA competition assay was used [55]. This assay involves preloading Dmc1 onto single stranded (ss) DNA that is attached to magnetic beads through a biotin-streptavidin connection, adding RPA, a single-stranded DNA binding protein complex, and monitoring the amount of Dmc1 that is retained on the beads. If Dmc1 is stably bound to the ssDNA, then it will not be competed off by RPA. The experiment was performed under three different Ca2+ conditions. High Ca2+ (1 mM) has previously been shown to stabilize Dmc1 filaments [56]. The bulk of both the Dmc1 and Dmc1-T159A proteins remained associated with the ssDNA/beads under high Ca2+ conditions (Figure 1B). When the Ca2+ concentration was lowered to either 10 or 25 µM, RPA was able to compete off the Dmc1-T159A protein more readily than wild type, indicating that filaments made from the mutant protein are less stable (Figure 1B). To test whether this filament instability affects Dmc1-T159A activity, strand exchange reactions were performed. In the strand exchange reaction, one strand of a piece of duplex DNA is radioactively labeled and the duplex DNA is then incubated with an unlabeled single strand of DNA along with the recombinase (Figure 1C, panel i). Exchange of the unlabeled strand for the radioactive strand in the duplex is then detected by slower migration on polyacrylamide gels. Consistent with a lack of stable filament formation, only background levels of product were observed for Dmc1-T159A in strand exchange reactions at 10 µM Ca2+ (Figure 1C, panel ii). This defect is due to unstable filaments and not to a defect in catalytic function, as a wild-type level of strand exchange activity for Dmc1-T159A is restored at 1 mM Ca2+, where the filaments are stabilized.
Fig. 1. Biochemical characterization of recombinant Dmc1-T159A protein. 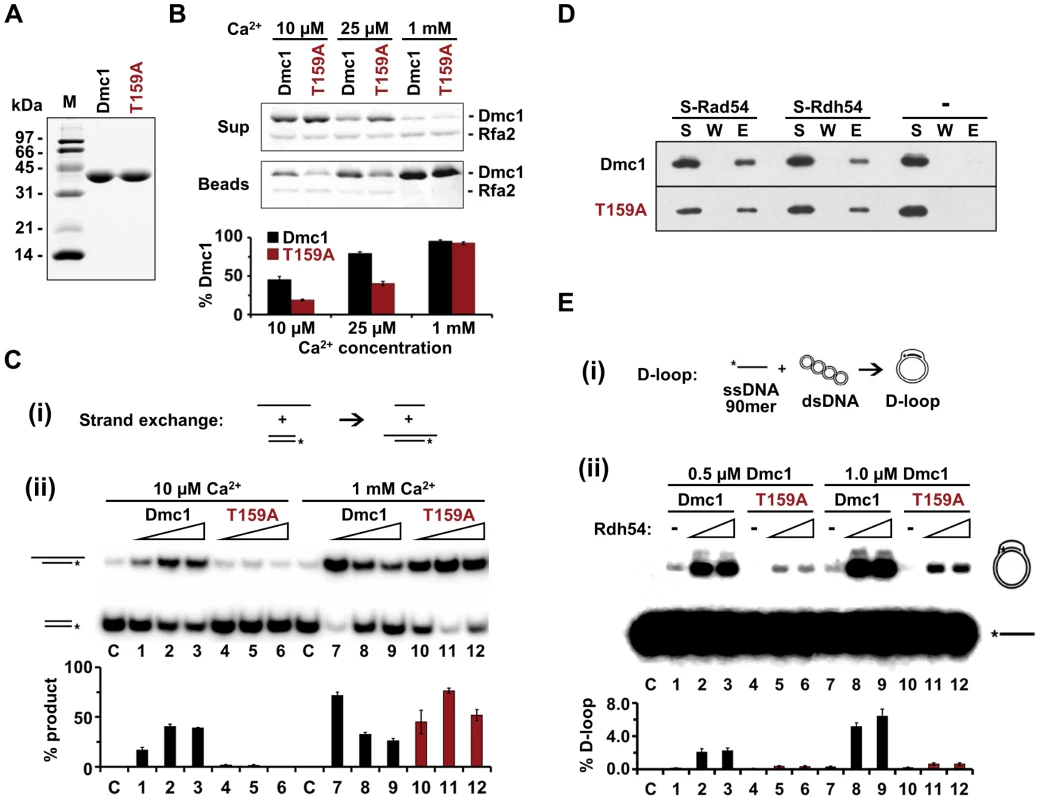
A. For Dmc1 and Dmc1-T159A, 2.5 µg of each protein were analyzed by SDS-polyacrylimide gel electrophoresis (PAGE) and Coomassie staining. B. The stability of Dmc1 filaments on bead-immobilized ssDNA was assessed by exposing the filaments to RPA and measuring the amount of Dmc1 retained on the beads (n = 3, +/− standard error). Proteins were monitored by SDS-PAGE and Coomassie staining. The experiments were performed with different Ca2+ concentrations as indicated. Rfa2, the 30 kDa subunit of RPA is indicated. The histogram shows the percent of Dmc1 protein that remained associated with the beads after challenge by RPA as determined by band desitometry. C. (i) Schematic of the strand exchange recombination assay. (ii) Strand exchange activity of Dmc1 and Dmc1-T159A was monitored using 2, 4, or 8 µM protein in the presence of either 10 µM or 1 mM Ca2+. The histogram indicates the percent of radioactively labeled oligonucleotide that was incorporated into the slower migrating product by strand exchange (n = 3, +/− standard error). At 1 mM Ca2+, the inhibition of strand exchange seen at elevated concentrations of wild-type Dmc1 is likely due to coating of the dsDNA substrate by the recombinase, thereby blocking access of the ssDNA filament. D. Dmc1 and Dmc1-T159A interactions with Rad54 and Rdh54 were assayed by pull-down experiments. S-tagged Rad54 or Rdh54 (2 µg each) was incubated with 1.2 µg Dmc1 or Dmc1-T159A. The S-tagged protein was captured on S-protein agarose resin, which was washed and the protein eluted with SDS. S = supernatant after collecting the beads, W = supernatant obtained from washing the beads, E = eluate from beads. The “-” indicates that no tagged protein was added to the reaction. E. (i) Schematic of the D-loop recombination assay. (ii) Homologous DNA pairing activity of Dmc1 and Dmc1-T159A was assessed by a D-loop formation assay (n = 3, +/− standard error). 0.5 or 1.0 µM of Dmc1 or Dmc1-T159A was combined with 0, 150, or 250 nM of Rdh54. The histogram shows the percent of radioactive ssDNA that is incorporated into the slower migrating D-loop product via homologous pairing. Dmc1 exhibits physical interactions with Rad54 and Rdh54 both in vitro and in vivo [7], [15], [57]. Pull-down experiments show that Dmc1-T159A interacts as well as wild-type Dmc1 with both Rad54 and Rdh54 (Figure 1D). Homologous pairing activity can be assayed by examining D-loop formation, where a radioactively labeled single strand of DNA is annealed to a complementary sequence within a circular plasmid, thereby displacing the sequence of like polarity (Figure 1E, panel i). The ability of Dmc1 to form D-loops is stimulated by Rdh54 [15]. For Rad51, RDH54 mutants specifically defective in interactions with the recombinase are unable to stimulate Rad51's homologous pairing activity [58]. Therefore, the observation that Rdh54 stimulates D-loop formation mediated by Dmc1-T159A indicates that Rdh54 is associated with the Dmc1-T159A presynaptic filament in a functionally competent manner (Figure 1E, panel ii). A reduced amount of D-loop product formed by Dmc1-T159A compared to Dmc1, both with and without Rdh54, is observed because D-loop formation is a more stringent measure of homologous pairing than the strand exchange assay in Figure 1C. We conclude that Dmc1-T159A is able to mediate homologous pairing but is less efficacious than wild type, presumably due to less efficient filament formation.
To examine whether the less stable filaments observed for Dmc1-T159A in vitro result in phenotypes in vivo, strains homozygous for dmc1-T159A were compared to wild type in two different strain backgrounds (SK1 and S288c/YJM789). dmc1-T159A rescues the sporulation defect of dmc1Δ in both backgrounds (Figure 2A and D). Furthermore, the spores produced by dmc1-T159A diploids have near wild-type levels of viability (Figure 2B and E). Meiotic progression is delayed ∼ two hours in the dmc1-T159A mutant (Figure 3A and S1A). DSBs arise with similar timing as wild type but accumulate to higher levels and persist longer, consistent with the idea that Dmc1-T159A is less efficient at DNA repair compared to Dmc1 (Figure 3B and C).
Fig. 2. Characterization of various meiotic phenotypes in diploids containing dmc1-T159A. 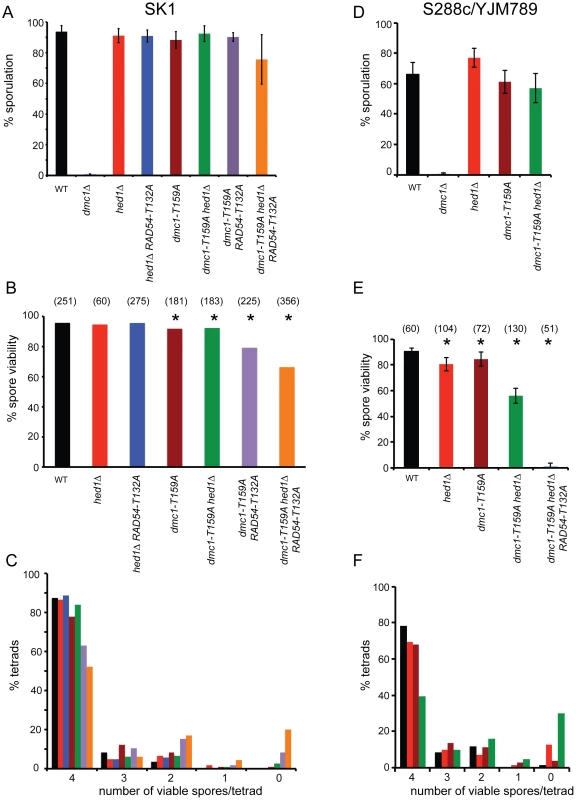
A. Sporulation in SK1 diploids: Wild-type (NH716), dmc1Δ (NH792), hed1Δ (NH1065), hed1Δ RAD54-T132A (NH1065::pHN104(S/N)2, dmc1-T159A (NH792::pNH301-T159A2), dmc1-T159A hed1Δ (NH942::pNH301-T159A2), dmc1-T159A RAD54-T132A (NH2231) and dmc1-T159A hed1Δ RAD54-T132A (NH2184) cells were transferred to Spo medium on plates for two days at 30°C and the percent sporulation was determined by phase contrast microscopy. 200 cells from at least four independent colonies were examined. Error bars represent the standard error. B. Spore viability in SK1 strains was assayed by tetrad dissection from at least four independent colonies. Numbers in parentheses indicate the number of tetrads dissected. * indicates that the spore viability is statistically significantly different from wild type by χ2 analysis. The p values are dmc1-T159A (<0.001); dmc1-T159A hed1Δ (<0.006); dmc1-T159A RAD54-T132A (<0.0001); dmc1-T159A hed1Δ RAD54-T132A (<0.0001) C. The distribution of viable spores in tetrads from the asci dissected for Panel B. D. Sporulation in S288c/YJM789 diploids: Wild-type (NH1053), dmc1Δ (NH2030), hed1Δ (NH2038), dmc1-T159A (NH2142), dmc1-T159A hed1Δ (NH2145), and dmc1-T159A hed1Δ RAD54-T132A (NH2146) cells were assayed for sporulation after four days on Spo plates at 30°C. E. Spore viability was assayed by dissection of at least 3 independent colonies. * indicates that the spore viability is statistically significantly different from wild type by χ2 analysis. The p values are hed1Δ (<0.001); dmc1-T159A (<0.02); dmc1-T159A hed1Δ (<0.0001); dmc1-T159A hed1Δ RAD54-T132A (<0.0001). F. The distribution of viable spores in tetrads from the asci dissected for Panel E. Fig. 3. Meiotic progression and crossover formation in various dmc1-T159A SK1 strains. 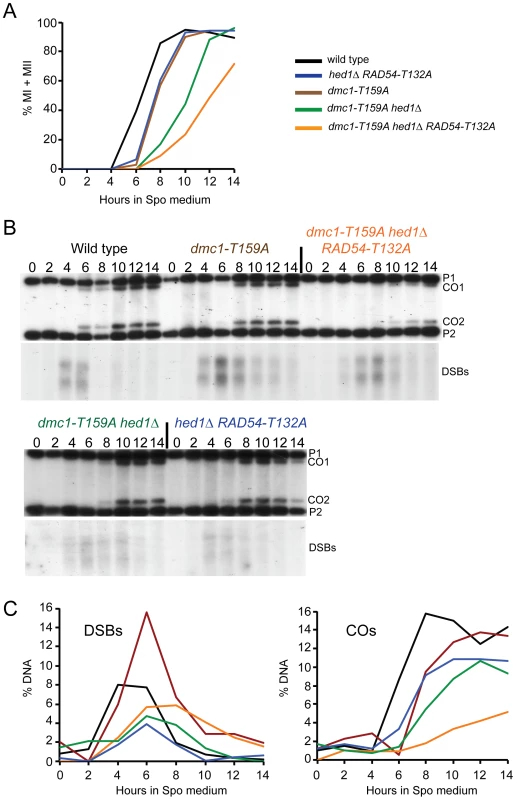
Wild-type, hed1Δ RAD54-T132A, dmc1-T159A, dmc1-T159A hed1Δ and dmc1-T159A hed1Δ RAD54-T132A diploids were transferred to Spo medium at 30°C at 0 hr and samples were taken at two hour intervals. Color coding is the same as in Figure 2. A. Meiotic progression was measured by staining the nuclei with DAPI and counting the fraction of bi-nucleate (MI) and tetranucleate (MII) cells. B. Crossovers and DSBs at the HIS4/LEU2 hotspot. The DNA was digested with XhoI and probed as described in [64]. P1 and P2 represent the parental fragments and CO1 and CO2 represent the two products of reciprocal recombination. Numbers above each lane indicate the hours after transfer to Spo medium. C. Quantitation of the crossover and DSBs bands shown in Panel B. A replicate of this experiment is shown in Figure S1. To test whether dmc1-T159A is defective in recombination, interhomolog crossovers were monitored by physical analyses using the HIS4/LEU2 hotspot. The HIS4/LEU2 hotspot is comprised of a DSB site flanked by restriction site polymorphisms that allow the detection of recombinant products using Southern blots [59]. In the wild-type diploid, crossovers were first detected at 6 hours (Figure 3B and C). Consistent with the meiotic progression data, crossovers in the dmc1-T159A diploid were reproducibly delayed by about two hours. Crossover levels are similar or only slightly reduced compared to wild type (Figure 3B and C; S1B and C).
The analysis of a single hotspot suggests that the Dmc1-T159A recombinase, although less efficient, is still able to produce nearly as many crossovers as wild-type. This idea was confirmed using a more global approach, whole genome sequencing, to examine all of the recombination events in individual tetrads. The dmc1-T159A mutation was introduced into two haploid strains (S288c and YJM789) that differ by 0.6% in their nucleotide sequences [17], [18]. All four spores from six dmc1-T159A tetrads were sequenced and then a software program called ReCombine was used to determine the numbers of COs and NCOs throughout the genome [60]. Because only four-spore viable tetrads were used for sequencing, a slight selection bias for meioses with higher numbers of COs theoretically may exist. However an actual bias has only previously been demonstrated when the sporulation frequency was particularly poor (<0.4% 4-spore asci) ([17], see zip1 mutant), which is not the case here. (All sequences from this paper can be accessed at http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA217886). In addition, the ReCombine data files are available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.8gh60). The dmc1-T159A diploid exhibits 93% and 77% the wild-type levels of COs and NCOs, respectively (Table 2). The decrease in COs is not statistically significant, while the decrease in NCOs is. Crossover homeostasis occurs when the CO number is maintained at the expense of NCOs [61]. While the specific decrease in NCOs is suggestive that crossover homeostasis is occurring, one needs to look at the coefficient of variation (CV) and the correlation coefficient (CC) to determine whether CO homeostasis is intact. We are precluded from getting an accurate assessment of CV and CC because the sample size is too small.
Tab. 2. Various phenotypes obtained from sequencing tetrads from various strains derived from the S288c/YJM789 background. 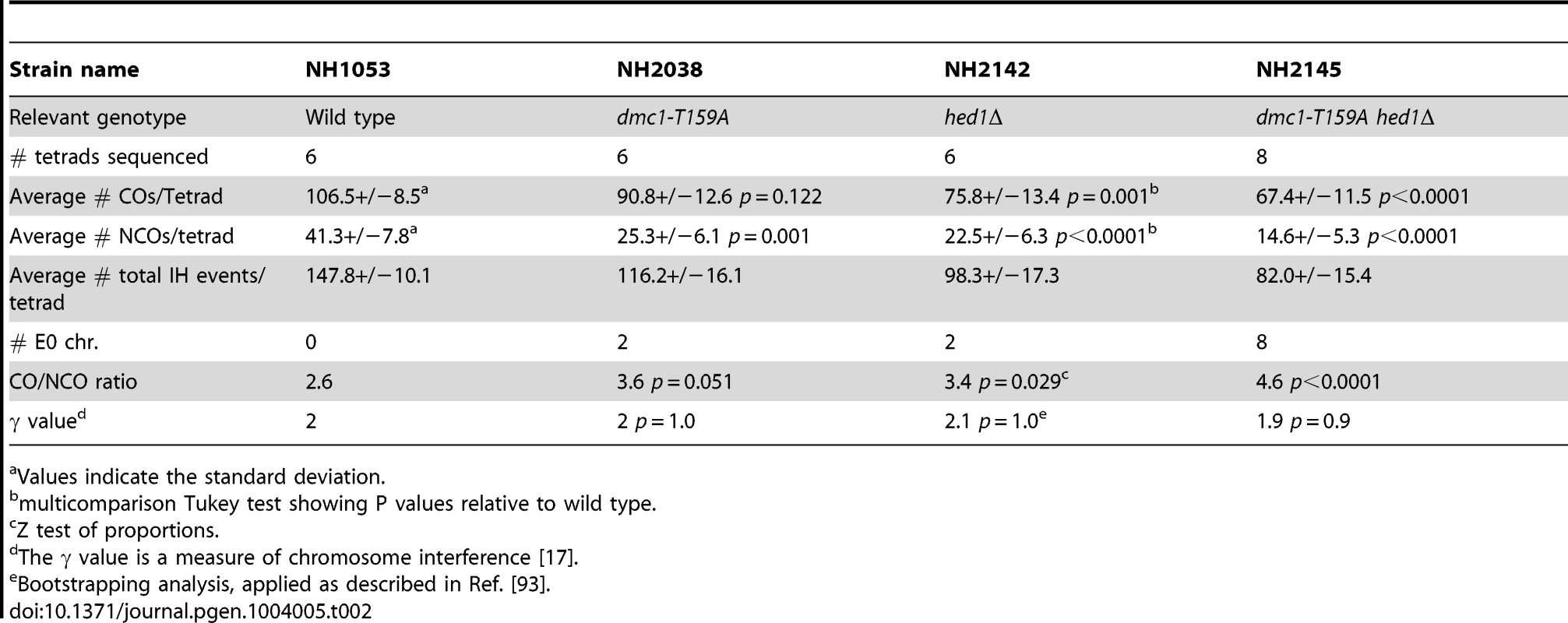
a Values indicate the standard deviation. Crossovers are distributed throughout the genome by a process called interference, in which a crossover in one region lowers the probability of a crossover in an adjacent region [62]. Genome-wide data can be used to calculate interference by measuring inter-CO distances to calculate a value called γ. A lack of interference results in a value of γ = 1, while γ values >1 indicate positive interference [17]. The γ values for the wild-type and dmc1-T159A diploids are basically identical, indicating that the crossovers generated using the hypomorphic recombinase are distributed properly (Table 2). The dmc1-T159A mutant therefore provides a good tool for testing whether up-regulated Rad51 can compete with weakened Dmc1 in vivo.
Elimination of Meiosis-Specific Constraints on Rad51–Rad54 Complex Formation Decreases Interhomolog Recombination in the Presence of dmc1-T159A
Combining hed1Δ and RAD54-T132A together, thereby removing the meiosis-specific constraints on Rad51–Rad54 interaction, has no deleterious effect on spore viability, suggesting that COs are not affected or only modestly affected (Figure 2B) [49]. Time course analysis confirms this prediction, with COs formed at the HIS4/LEU2 hotspot at levels only 30% reduced from wild type (Figure 3B and C). There is, however, a delay in meiotic progression and CO formation in hed1Δ RAD54-T132A diploids, suggesting that allowing Rad51 and Rad54 to interact does affect meiotic recombination even in the presence of DMC1 (Figure 3A). One explanation for the failure to see a strong CO defect in the hed1Δ RAD54-T132A diploid is that Dmc1 is a more efficient recombinase than Rad51 for interhomolog recombination. If true, then hed1Δ and/or RAD54-T132A may exhibit mutant phenotypes when combined with the less efficient dmc1-T159A. In SK1 strains, spore viability is similar for the dmc1-T159A and dmc1-T159A hed1Δ diploids, while a stronger decrease in spore viability was observed for dmc1-T159A RAD54-T132A (Figure 2B). Deletion of HED1 in the dmc1-T159A diploid results in a modest decrease in COs, similar to what has previously been observed for hed1Δ alone [48] (Figure 3B and C). hed1Δ removes only one impediment to Rad51–Rad54 interaction, as Rad54 T132 phosphorylation by Mek1 is still occurring. Combining both hed1Δ and RAD54-T132A with dmc1-T159A results in a further decrease in spore viability (Figure 2B). The difference in spore viability between the triple mutant and either dmc1-T159A hed1Δ or dmc1-T159A RAD54-T132A is significant (χ2 analysis; p<0.0001), confirming the proposed functional redundancy between HED1 and Rad54 T132 phosphorylation in the downregulation of Rad51 activity. Furthermore, the number of tetrads with four viable spores is decreased in the triple mutant with a corresponding increase in tetrads with either two or zero viable spores, a pattern indicative of Meiosis I chromosome non-disjunction (Figure 2C) [63]. This result suggests that up-regulating Rad51 activity does not interfere with repair (which would lead to broken chromosomes and a different spore viability pattern), but instead alters repair in such a way that interhomolog crossovers are decreased. In fact, COs are substantially reduced in the triple mutant compared to the other three strains (Figure 3B and C; S1B and C). Although the dmc1-T159A hed1Δ RAD54-T132A diploid sporulates well, addition of RAD54-T132A results in a further delay in meiotic progression than is observed for either dmc1-T159A or dmc1-T159A hed1Δ (Figure 3A and S1A).
In contrast to the SK1 background, the dmc1-T159A hed1Δ combination in the S288c/YJM789 background exhibits a synergistic decrease in spore viability compared to either dmc1-T159A or hed1Δ alone (Figure 2E). As with the SK1 diploid, the distribution of viable spores in the dmc1-T159A hed1Δ tetrads indicates that spore lethality is likely due to Meiosis I nondisjunction (Figure 2F). dmc1-T159A hed1Δ RAD54-T132A in the sequencing background reduced spore viability even further, making it difficult to obtain four viable spores for sequencing (Figure 2E). Therefore, we were unable to analyze the triple mutant using the sequencing assay. Sequence analysis of the genomes from the spores of eight dmc1-T159A hed1Δ tetrads shows statistically significant reductions in both COs and NCOs to 68% and 48% of the wild-type levels, respectively (Table 2). The γ value for dmc1-T159A hed1Δ is identical to wild type, however, indicating that interference is normal. Chromosomes without any crossovers are defined as E0 chromosomes. In 26 wild-type tetrads analyzed by Chen et al. (2008) and the six additional tetrads included here, no E0 chromosomes were observed. In contrast, eight E0 chromosomes were observed in the eight tetrads sequenced for dmc1-T159A hed1Δ (Table 2).
The delay in meiotic progression observed for dmc1-T159A indicates that the checkpoint may still be activated by the inefficient recombinase, even though a pathological arrest is not occurring. To test whether hed1Δ alone affects recombination, the genomes from six hed1Δ tetrads were sequenced. In this strain, DMC1 is wild-type so the meiotic recombination checkpoint should not be triggered.
A small, but statistically significant reduction in spore viability was observed in the hed1Δ sequencing diploid, indicating this genetic background is more sensitive to the absence of HED1 than SK1 or BR strains (Figure 2E). The average numbers of COs and NCOs were reduced, exhibiting 78% and 57% of the wild-type levels, respectively (Table 2). These reductions are statistically significant. This reduction is likely due to a change in repair and not because fewer breaks are being initiated, as hed1Δ does not affect DSB formation [48]. Interference is functioning, as indicated by the γ value. Two E0 chromosomes were observed for the six hed1Δ tetrads. This number is less than the 8 E0 chromosomes exhibited by dmc1-T159A hed1Δ. While this difference is intriguing, the sample size is currently too small to definitively say whether this difference is meaningful or not.
Up-Regulation of Rad51 Activity in the dmc1-T159A Background Increases Intersister Recombination
The dmc1-T159A hed1Δ RAD54-T132A SK1 diploid exhibits decreased COs and spore viability with increased Meiosis I nondisjunction. One explanation for these results is that allowing Rad51–Rad54 complex formation in the presence of a less efficient Dmc1 results in more intersister repair. This idea was directly tested by examining formation of interhomolog and intersister joint molecules by two-dimensional gel analysis using the HIS4/LEU2 hotspot. To prevent joint molecules from coming apart by branch migration, the DNA was crosslinked with psoralen prior to extraction [64]. After digestion with XhoI, the DNA was separated in one dimension by mass and in the second dimension by mass and shape. Probing a Southern blot of the DNA then reveals three spots that represent one interhomolog joint molecule (JM) intermediate flanked by the two intersister JM intermediates. Because DSB repair occurs at different rates in the different mutant diploids, this analysis was carried out in strains deleted for the middle meiotic gene transcription factor, NDT80. ndt80Δ diploids arrest in pachytene with unresolved double Holliday junctions, thereby allowing JMs to accumulate [65]–[68]. Cells were arrested after incubation in Spo medium for nine hours and the samples processed. In the ndt80Δ diploid, an ∼4∶1 ratio of interhomolog∶ intersister joint molecules was observed, consistent with the literature [19] (Figure 4). The IH∶IS ratio was reduced approximately two-fold by the combination of hed1Δ and RAD54-T132A. Therefore removing the meiosis-specific barriers to Rad51–Rad54 interaction does increase intersister recombination, although not to the extent that an effect on spore viability is observed. A wild-type ratio was observed for the dmc1-T159A strain, demonstrating that this mutant affects the rate of recombination without affecting interhomolog bias (Figure 4). The two-fold reduction in interhomolog bias in the dmc1-T159A hed1Δ diploid is similar to that observed for hed1Δ RAD54-T132A but is accompanied by a mild spore viability defect (Figure 2B; Figure 4). These data support the interpretation that the reduction of interhomolog events observed by genomic sequencing in the absence of hed1Δ is due to more DSBs being repaired using sister chromatids as templates. Finally the interhomolog∶intersister JM ratio was reduced even further (eight fold) in the dmc1-T159A hed1Δ RAD54-T132A diploid, consistent with the more severe phenotypes observed for this strain (Figure 4).
Fig. 4. Meiotic joint molecule analysis in various SK1 dmc1-T159A ndt80Δ strains. 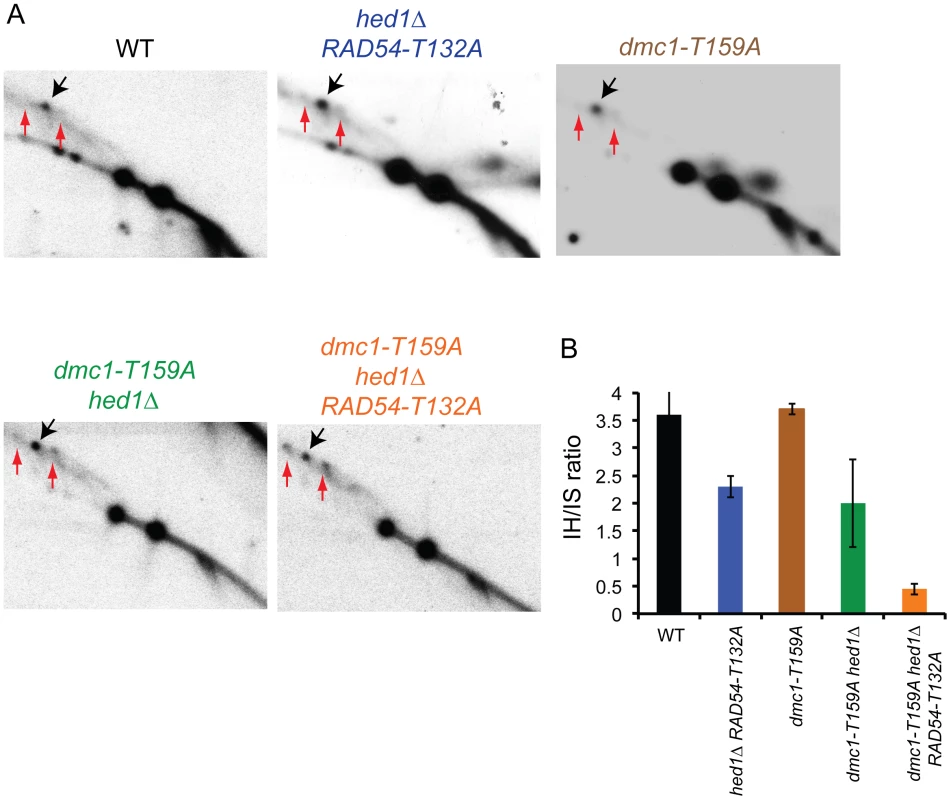
ndt80Δ (NH2188), hed1Δ ndt80Δ RAD54-T132A (NH2223::pHN104(S/N)2, dmc1-T159A ndt80Δ (NH2235), dmc1-T159A hed1Δ ndt80Δ (NH2190) and dmc1-T159A hed1Δ ndt80Δ RAD54-T132A (NH2193) diploids were transferred to Spo medium for nine hours to arrest the cells in pachytene and the DNA was crosslinked with psoralen, extracted and digested with XhoI. Color coding is the same as in Figure 2. A. Southern blots of two-dimensional gels probed to detect interhomolog JMs (indicated by black arrows) and intersister JMs (indicated by red arrows) as described in [64]. B. Quantitation of the ratio of interhomolog:intersister joint molecules in the gels shown in A averaged with a second replicate. Error bars indicate the standard error. Discussion
Given that Rad51 strand exchange activity is not required for interhomolog recombination, the question arises as to whether having Rad51 active when interhomolog recombination is occurring is deleterious to the cell due to Rad51's preference for repairing DSBs using sister chromatids [14], [20], [21], [43]. In yeast, Rad51 is prevented from repairing DSBs in dmc1Δ mutants both by Hed1 and Mek1 phosphorylation of Rad54 [46], [69] [47]–[49]. However the lack of obvious phenotypes observed for hed1Δ RAD54-T132A diploids has raised questions about whether down-regulation of Rad51 activity occurs normally during meiosis or whether it only occurs after triggering of the meiotic recombination checkpoint. One argument supporting the idea that Rad51 strand exchange activity is inhibited normally during meiosis is that phosphorylation of Rad54 T132 by Mek1 occurs during wild-type meiosis [70]. Furthermore cytological studies using wild-type cells show that Hed1 focus formation on chromosome is dependent upon RAD51 (but not DMC1) and that 98% of Rad51 foci co-localize with Hed1 [48]. While these studies demonstrate that meiotic impediments to Rad51–Rad54 complex formation are present during wild-type meiosis, they do not address whether this downregulation is functionally important. To answer this question, whole genome sequencing of tetrads from a hed1Δ diploid was performed and revealed a decrease in both interhomolog COs and NCOs, even though DMC1 is wild type. Furthermore, joint molecule experiments showed that intersister recombination is increased when down-regulation of Rad51 activity is abolished, particularly when combined with dmc1-T159A. These results indicate that regulation of Rad51–Rad54 complex formation by both Hed1 and Rad54 T132 phosphorylation normally occurs during meiosis.
In the sequencing strain background, hed1Δ decreases spore viability slightly on its own and synergistically in combination with dmc1-T159A, a phenotype that was not observed in SK1. The decreased spore viability of the double mutant is likely due, in part, to a greater reduction in crossovers compared to hed1Δ. In addition, the frequency of chromosomes without crossovers appears to be increased, although interference is wild-type. This suggests that, while the dmc1-T159A hed1Δ mutant is proficient in the distribution of crossovers along individual chromosomes, it may be defective in crossover assurance, which is the regulation that ensures that every chromosome sustains at least one crossover. If true, this could also account for the decreased spore viability relative to hed1Δ. Further work is necessary, however, to increase the sample size of E0 chromosomes before any definitive conclusions concerning this point can be made.
Multiple Mechanisms Promote Interhomolog Bias during Meiosis
Crossovers between homologs are critical for proper alignment and segregation of homologous pairs of sister chromatids to opposite poles at the first meiotic division. This importance is underlined by the multiple, non-overlapping meiosis-specific mechanisms that cells use to change the bias for repair from sisters in vegetative cells to homologs in meiotic cells. First, in vegetative cells, intersister DSB repair is mediated by Rad51 and promoted by the recruitment of cohesin molecules to the DSBs [71]–[73]. In contrast, meiotic cells utilize cohesin complexes containing a meiosis-specific kleisin subunit, Rec8, and cohesin function at DSBs is antagonized by Mek1, thereby promoting interhomolog recombination [27], [74]. Second, the meiosis-specific axial element structures into which sister chromatids are packaged create additional constraints that promote interhomolog strand invasion in a variety of organisms. Functional orthologs of Hop1 in nematodes, plants and mammals all promote interhomolog bias, suggesting that at least some of the fundamental mechanisms for promoting crossovers between homologs during meiosis are conserved [75]–[79]. Third, many organisms utilize the meiosis-specific Dmc1 recombinase, which in budding yeast has been shown to be sufficient for the bulk of meiotic recombination [43], [80]. Dmc1 may be better at performing interhomolog recombination than Rad51 due to intrinsic properties of the protein. For example, the D-loops formed by Dmc1 are more resistant to dissociation than those formed by Rad51 [57]. In addition Dmc1 utilizes different accessory factors such as Rdh54 that promote interhomolog recombination [15]. This work demonstrates yet another mechanism for promoting interhomolog bias: the down-regulation of Rad51 activity by the prevention of Rad51–Rad54 interaction.
The Dmc1-T159A protein is defective in efficiently forming filaments while maintaining the ability to mediate strand exchange and interaction with Rad54 and Rdh54. The delay in DSB repair, crossover formation and meiotic progression confirms that this mutation makes Dmc1 less efficient as a recombinase. dmc1-T159A cells produce nearly wild-type levels of crossovers which are distributed normally throughout the genome, as well as being wild-type for interhomolog bias, consistent with biochemical experiments indicating that the mutant affects filament formation. One possibility is that it may take longer for Dmc1-T159A containing filaments to form, but having done so, they then function normally in recombination. It is important however, that although meiotic progression is delayed by dmc1-T159A, the cells do not arrest, thereby avoiding the proposed hyperactivation of Mek1 [52]. The fact that decreased spore viability, increased Meiosis I non-disjunction, and reduced interhomolog recombination are strongly apparent only when up-regulation of Rad51 activity was combined with the less efficient dmc1-T159A mutant, supports the idea that Dmc1 is fundamentally a more effective interhomolog recombinase than Rad51 in meiosis. The amino acid mutated in Dmc1, T159, is conserved in Dmc1 and Rad51 proteins from all species that have been analyzed, suggesting that it may be possible to construct similar hypomorphic mutants for these recombinases in other organisms.
Recent work has shown that DSBs occur on chromosome axes where localized activation of Mek1 is presumed to occur [34]. In plants the localization of Rad51 and Dmc1 to DSBs is asymmetric, with Rad51 loaded onto one end and Dmc1 loaded onto the other end [41]. The asymmetric loading model is appealing because of the asymmetry inherent in the recombination reaction that generates COs. During CO formation, the two ends of the DSB play different roles. The first end must undergo strand invasion of a homologous duplex, while the second end undergoes annealing to the newly displaced strand such that a double Holliday junction can subsequently be formed [6]. One model is that local antagonism of sister chromatid cohesion allows the release of one end of the DSB, presumably the one bound by Dmc1, while the other end remains tethered to the sister chromatid [27]. After the Dmc1 filament stably invades the homolog, the second end must be released to allow its annealing to the displaced strand at the nascent interhomolog joint. Whether this release results from a signal arising from individual recombination events, or is a globally regulated event, is not known.
We propose that preventing Rad51 from binding to Rad54 is important for keeping the second end quiescent until the first end has engaged the homolog. When HED1 is removed and Rad54-T132 phosphorylation is prevented, Rad51 is now active. Although there is a two-fold increase in intersister recombination, the efficiency of Dmc1 in making stable connections with the homolog prevents a large reduction in crossovers. The inefficiency of Dmc1-T159A in mediating strand invasion (possibly due to a delay in filament formation) may provide the end containing Rad51 more time to mediate the stable strand invasion of the sister chromatid. This would then send the signal for the Dmc1-containing end to anneal to the displaced strand at the sister chromatid. The lack of coordination between the two ends could further slow DSB repair, explaining the meiotic delay of the dmc1-T159A hed1Δ RAD54-T132A diploid.
Although it has been suggested that Dmc1 and Rad51 are bound to different sides of DSBs in yeast, similar to the situation in plants [81], this has not been definitively established. In contradiction to this idea, recent experiments have shown that Rad51 can act as an accessory factor for Dmc1 in vitro, suggesting instead that the two recombinases may co-localize on the same ends of DSBs in yeast [43]. In this case, our model that activation of Rad51 allows one end to engage in intersister recombination still holds, although the mechanism that determines the asymmetry of the ends (strand invasion vs. annealing to the displaced strand) remains to be determined.
It should be noted that interhomolog recombination does not require that there be two recombinases. Organisms such as Drosophila and C. elegans do not contain DMC1 in their genomes and therefore meiotic recombination is mediated solely by Rad51 [80]. Furthermore even in organisms that utilize both recombinases, mutants exist which allow interhomolog recombination to occur by either Rad51 alone or Dmc1 alone [41], [48], [49]. However, the frequencies of interhomolog recombination in these latter situations are not wild-type. Therefore, the optimal situation in organisms with both Rad51 and Dmc1 is to have both recombinases present. This may be because of a requirement for Rad51 to load Dmc1 or to stimulate Dmc1's enzymatic activities [41], [43], [81]. But while the presence of Rad51 is important for recombination, it is also important that Rad51's capacity to perform strand invasion be turned off while interhomolog recombination is occurring to prevent competition with Dmc1 for repair of DSBs.
Materials and Methods
Strains
The genotypes of all the strains used in this work can be found in Table 3. Genes were deleted by polymerase chain reaction (PCR)-based methods using the kanMX6, natMX4 or hphMX4 markers that confer resistance to G418, nourseothricin and Hygromycin B, respectively [82]–[84]. All deletions were confirmed by colony PCR. For genomic sequencing of tetrads, isogenic diploids were generated in which one parent is a derivative of S288c, JCF4413, and the other is a clinical isolate, YJM789 [17]. YJM789 has a tendency to become aneuploid, therefore manipulations of this strain were limited as much as possible. To create a diploid homozygous for dmc1-T159A, DMC1 was first deleted from both haploids using kanMX6. The dmc1-T159A URA3 plasmid, pNH301-T159A, was digested with HindIII to target integration upstream of the DMC1 open reading frame, and transformed into JCF4413 dmc1. Transformants were grown in YPD and plated on 5-fluoro-orotic acid to select for popouts of the plasmid [85]. Strains that retained the dmc1-T159A allele were selected based on their sensitivity to G418. The presence of the dmc1-T159A allele was further confirmed by colony PCR. To introduce a second copy of dmc1-T159A into this haploid, pNH301-T159A was integrated as before upstream of dmc1-T159A. JCF4413 dmc1-T159A::URA3::dmc1-T159A was then mated to YJM789 dmc1 to make the diploid, NH2142. A similar procedure was followed using JCF4413 dmc1 hed1 and YJM789 dmc1 hed1 to make NH2145. Prior to sequencing, each spore colony was checked using allele-specific colony PCR to confirm that no aneuploidies had arisen during the growth of the strains [86].
Tab. 3. S. cerevisiae strainsa. 
a Superscript “2” indicates that the plasmid was integrated into both haploid parents and the diploid is therefore homozygous. The two-hybrid reporter strain,Y190, was generously provided by Michael Dresser (Oklahoma Medical Research Foundation). All other strains were derived from NHY1210 and NHY1215, SK1 strains that contain the HIS4/LEU2 hotspot (generously provided by Neil Hunter). The wild-type diploid created by mating NHY1210 and NHY1215 is called NH716 [42]. Strains containing dmc1-T159A were generated by first substituting DMC1 with kanMX6 and then transforming the strains with pNH301-T159A to integrate dmc1-T159A upstream of dmc1Δ::kanMX6. For some strains, the dmc1Δ::kanMX6 allele was recombined out as described above so that dmc1-T159A haploids could subsequently be transformed with pHN104(Sph1/NruI), which contains the RAD54-T132A allele [49].
Plasmids
The GBD-DMC1 and GAD-DMC1 plasmids (pMDE422 and pMDE467, respectively; provided by Michael Dresser) contain codons 3-334 of DMC1 fused to either the Gal4 DNA binding or activation domains [7]. The T159A mutation changes codon 159 from ACT to GCT and was introduced into various plasmids using the QuikChange kit (Stratagene). For purification of Dmc1 and Dmc1-T159A from E.coli, the T159A mutation was introduced into the DMC1 expression plasmid, pNRB150scDMC1 [87], to generate pNRB150scDMC1-T159A. To introduce the dmc1-T159A mutation into yeast cells, DMC1 was first cloned into a URA3 integrating plasmid by moving a 2.4 kb NotI/XhoI fragment from pRS316-DMC1 (generously provided by J. Engebrecht) into NotI/XhoI-digested pRS306 [88]. The resulting plasmid, pNH301, was used for site-directed mutagenesis to make pNH301-T159A. All mutated alleles were sequenced at the Stony Brook University DNA Sequencing Facility to confirm that no unexpected mutations were introduced during the mutagenesis. Digestion of pNH301-T159A with HindIII targets the plasmid to integrate approximately 600 bp upstream of the DMC1 gene. To introduce RAD54-T132A, pHN104(Sph1/NruI), a URA3 RAD54-T132A integrating plasmid, was digested with BsiWI to target integration upstream of RAD54 and transformed into the appropriate haploids.
Whole Genome DNA Sequencing
DNA was prepared from each spore colony as described in [86] and sequenced at either the Vincent J. Coates Genomics or University of California San Francisco Sequencing Facilities using the Illumina HiSeq 2000 platform with 50 nt single end reads. Analysis of the sequences was performed using the ReCombine suite of programs to determine the number of crossovers and non-crossovers and the interference values [60]. The sequencing data can be found at NCBI SRA Bioproject, Accession number PRJNA217886 (http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA217886). The ReCombine user manual and software package can be accessed at http://sourceforge.net/projects/recombine/. The ReCombine data files used to determine the number of COs and NCOs are available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.8gh60).
Two-Hybrid Assays
Y190 was co-transformed with the indicated plasmids and protein-protein interactions were monitored using liquid β-galactosidase assays. Transformants were inoculated into 5 ml SD-leu-trp and grown overnight at 30°C. Cells were diluted 1∶10 in SD-leu-trp and the OD600 was measured to determine cell density. Two 1.5 ml aliquots of culture from each transformant were pelleted in microfuge tubes and washed once in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4.H20, 10 mM KCl, 1 mM MgSO4.7H20, 4 mM 2-mercaptoethanol). The cells were then resuspended in 150 µl Z buffer, vortexed and lysed by the addition of 50 µl chloroform and 20 µl 0.1% SDS. After vortexing for 30 sec, the tubes were equilibrated at 30°C for five minutes. 700 µl of 1.2 mg/ml ortho-Nitrophenyl-β-galactoside (ONPG) made up in Z buffer was added to each tube which were then placed at 30°C. As soon as a yellow color appeared, the time was noted and the reactions were stopped by the addition of 500 µl 1M Na2CO3 and put on ice. Any reactions that had not turned yellow by two hours were stopped at that time. Cells were pelleted and the OD420 of the supernatants was determined. Miller units were calculated using the following formula: (1000×A420)/(A600×time (min)×vol (ml) [89]. Two replicates from each transformant were averaged and these numbers were then used to calculate the average and standard deviation from three transformants.
Protein Purification
Recombinant budding yeast Dmc1 proteins (WT and T159A) were purified according to a new procedure developed by the Sung laboratory [54], which results in Dmc1 preparations that have a higher specific recombinase activity than the published protocol [87]. Two independent preparations of Dmc1-T159A were analyzed to ensure consistency of results. The proteins were concentrated in an Amicon Ultra micro-concentrator (Millipore), snap-frozen in liquid nitrogen, and stored at −80°C. RPA was purified as described in [90]. S-Rad54 and S-Rdh54 were purified as described in [10] and [58], respectively.
Strand Exchange Assay
Oligonucleotide-based DNA pairing and strand exchange assay was conducted as described previously [91]. Briefly, Dmc1 (2, 4, and 8 µM) was incubated with 150-mer ssDNA oligonucleotide (6 µM nucleotides) in 10.5 µl of buffer A (50 mM Tris-HCl, pH 7.5, 1 mM DTT, 20 mM KCl, 2 mM ATP, 5 mM MgCl2) with the indicated amount of CaCl2 for 5 min at 37°C. 1 µl of 50 mM spermidine and 1 µl of 32P-labeled homologous 40-mer dsDNA (for 0.8 µM base pairs final concentration) were added to initiate the reaction. The reactions were incubated for 30 min at 37°C. The samples were deproteinized by the addition of 1% SDS and 1 mg/ml proteinase K, and subjected to electrophoresis in a 10% polyacrylamide gel run in TAE buffer (40 mM Tris acetate, pH 7.4, 0.5 mM EDTA). Products were quantitated using a phosphoimager.
RPA Challenge Assay to Test for Dmc1 Filament Stability
This assay was modified from one developed by [55] for Rhp51. Dmc1 protein (2.4 µM) was added to biotinylated dT 83-mer ssDNA (4.3 µM nucleotides) coupled to magnetic streptavidin beads (Roche) in 10 µl of buffer B (35 mM Tris-HCl, pH 7.5, 1 mM DTT, 20 mM KCl, 2 mM ATP, 5 mM MgCl2, 100 µg/ml BSA). The reactions were incubated at 37°C for 5 min to permit filament formation. Unbound Dmc1 protein was removed by magnetic separation, and 10 µl of buffer B with 0.4 µM of RPA was added to the beads. The reactions were mixed and incubated at 30°C for 5 min followed by magnetic separation of the supernatant and bead fractions. Both fractions were analyzed by SDS-PAGE, Coomassie Blue staining, and band densitometry. The indicated amount of CaCl2 was included in both the binding and RPA challenge buffers.
Affinity Pull-Down Assay
Dmc1 (1.2 µg) was incubated with or without S-tagged Rad54 or S-tagged Rdh54 (2 µg each) in 30 µl of buffer C (40 mM K2HPO4, pH 7.5, 0.5 mM EDTA, 10% glycerol, 150 mM KCl, 0.01% IGEPAL, 1 mM DTT) for 30 min at 4°C. The reactions were mixed with 10 µl of S-protein agarose beads (Novagen) and incubated for 30 min at 4°C with agitation. Beads were washed twice with 200 µl of buffer C and bound protein was eluted with 2% SDS. Supernatant (S), elution (E), and wash (W) fractions were analyzed by 10% SDS-PAGE followed by western blot with α-hDMC1 antibody (Santa Cruz Biotechnology, catalog # 22768). This experiment was performed twice with similar results.
D-loop Assay
The D-loop reaction was conducted as described [10], [87]. Briefly, Dmc1 (0.5 or 1.0 µM) was incubated with 32P-labeled 90-mer oligonucleotide substrate (1.5 µM nucleotides) at 37°C for 5 min. Next, Rdh54 (0, 150, or 250 nM) was added along with pBluescript SK replicative form I DNA (72 µM base pairs). The reaction (12.5 µl final volume) had a buffer composition of 50 mM Tris-HCl, pH 7.5, 1 mM DTT, 72 mM KCl, 1 mM MgCl2, 5 mM CaCl2, and 4 mM ATP with an ATP regenerating system (20 mM creatine phosphate, 30 µg/ml creatine kinase). After a 15 min incubation at 30°C, SDS (1%) and proteinase K (1 mg/ml) were added, followed by a 5 min incubation at 37°C. The deproteinized samples were subjected to electrophoresis in a 1% agarose gel and analyzed by phosphorimaging.
Timecourses and Physical Analyses of Recombination
Cells were pregrown in YPA and transferred to Spo medium (2% potassium acetate) and incubated at 30°C as described in [92]. At the appropriate time points, an aliquot of cells was fixed with formaldehyde and stained with 4′,6-diamidino-2-phenylindole (DAPI). Meiotic progression was monitored by fluorescent microscopy to determine the number of binucleate (MI) and tetranucleate cells (MII). At least 200 cells were counted for each colony. Physical analyses were performed as described in [64]. For the one-dimensional gel analysis shown in Figure 3B, DNA was isolated using the MasterPure Yeast DNA Purification kit (Epicentre, Cat. # MPY80200). For the CO analysis shown in Figure S1 as well as the two-dimensional gel experiments, cells were treated with psoralen and crosslinked with ultra-violet light. DNA was extracted and digested with XhoI and probed after fractionation on a one-dimensional gel to look at crossover formation. To look at joint molecules, ndt80Δ diploids were arrested nine hours after transfer to Spo medium and the psoralen crosslinked, XhoI-digested DNA was fractionated in two-dimensions prior to probing on Southern blots. The interhomolog JMs were normalized to the total DNA, as was the sum of the two intersister JMs, and these values were used to calculate the interhomolog: intersister JM ratios. Quantitation was performed using the MultiGauge software with a Fujifilm FLA-7000 phosphoimager. Timecourses analyzing COs were conducted three times for all strains except NH1065 (hed1Δ RAD54-T132A), which was only examined twice. All five strains shown in Figure 3 are from the same timecourse, while for Figure S1, the NH1065 strain was performed on a different day as the other four strains. DSBs were only examined once in the Figure 3 timecourse.
Supporting Information
Zdroje
1. PetronczkiM, SiomosMF, NasmythK (2003) Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112 : 423–440.
2. HassoldT, HuntP (2001) To err (meiotically) is human: the genesis of human aneuploidy. Nature reviews 2 : 280–291.
3. KeeneyS (2008) Spo11 and the formation of DNA double-strand breaks in meiosis. Genome dynamics and stability 2 : 81–123.
4. NealeMJ, KeeneyS (2006) Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442 : 153–158.
5. SheridanS, BishopDK (2006) Red-Hed regulation: recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev 20 : 1685–1691.
6. Hunter N (2007) Meiotic Recombination; Aguilera A, Rothstein R, editors. Heidelberg: Springer-Verlag. 381–462 p.
7. DresserME, EwingDJ, ConradMN, DominguezAM, BarsteadR, et al. (1997) DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147 : 533–544.
8. PetukhovaG, SungP, KleinH (2000) Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev 14 : 2206–2215.
9. PetukhovaG, Van KomenS, VerganoS, KleinH, SungP (1999) Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J Biol Chem 274 : 29453–29462.
10. RaschleM, Van KomenS, ChiP, EllenbergerT, SungP (2004) Multiple interactions with the Rad51 recombinase govern the homologous recombination function of Rad54. J Biol Chem 279 : 51973–51780.
11. CeballosSJ, HeyerWD (2011) Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochimica et biophysica acta 1809 : 509–523.
12. HolzenTM, ShahPP, OlivaresHA, BishopDK (2006) Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev 20 : 2593–2604.
13. KleinHL (1997) RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147 : 1533–1543.
14. ShinoharaM, Shita-YamaguchiE, BuersteddeJ-M, ShinagawaH, OgawaH, et al. (1997) Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homolog of RAD54, RDH54/TID1 in mitosis and meiosis. Genetics 147 : 1545–1556.
15. NimonkarAV, DombrowskiCC, SiinoJS, StasiakAZ, StasiakA, et al. (2012) Saccharomyces cerevisiae Dmc1 and Rad51 preferentially function with Tid1 and Rad54, respectively, to promote DNA strand invasion during genetic recombination. J Biol Chem 287 : 28727–28737.
16. PanJ, SasakiM, KniewelR, MurakamiH, BlitzblauHG, et al. (2011) A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144 : 719–731.
17. ChenSY, TsubouchiT, RockmillB, SandlerJS, RichardsDR, et al. (2008) Global analysis of the meiotic crossover landscape. Dev Cell 15 : 401–415.
18. ManceraE, BourgonR, BrozziA, HuberW, SteinmetzLM (2008) High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454 : 479–485.
19. SchwachaA, KlecknerN (1997) Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90 : 1123–1135.
20. BzymekM, ThayerNH, OhSD, KlecknerN, HunterN (2010) Double Holliday junctions are intermediates of DNA break repair. Nature 464 : 937–941.
21. KadykLC, HartwellLH (1992) Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132 : 387–402.
22. PageSL, HawleyRS (2004) The genetics and molecular biology of the synaptonemal complex. Annual review of cell and developmental biology 20 : 525–558.
23. BailisJM, RoederGS (1998) Synaptonemal complex morphogenesis and sister-chromatid cohesion require Mek1-dependent phosphorylation of a meiotic chromosomal protein. Genes Dev 12 : 3551–3563.
24. HollingsworthNM, GoetschL, ByersB (1990) The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61 : 73–84.
25. SmithAV, RoederGS (1997) The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol 136 : 957–967.
26. HollingsworthNM, ByersB (1989) HOP1: a yeast meiotic pairing gene. Genetics 121 : 445–462.
27. KimKP, WeinerBM, ZhangL, JordanA, DekkerJ, et al. (2010) Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143 : 924–937.
28. Mao-DraayerY, GalbraithAM, PittmanDL, CoolM, MaloneRE (1996) Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics 144 : 71–86.
29. RockmillB, RoederGS (1990) Meiosis in asynaptic yeast. Genetics 126 : 563–574.
30. RockmillB, RoederGS (1991) A meiosis-specific protein kinase homologue required for chromosome synapsis and recombination. Genes Dev 5 : 2392–2404.
31. ThompsonDA, StahlFW (1999) Genetic control of recombination partner preference in yeast meiosis. Isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics 153 : 621–641.
32. NiuH, WanL, BaumgartnerB, SchaeferD, LoidlJ, et al. (2005) Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell 16 : 5804–5818.
33. BlatY, KlecknerN (1999) Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98 : 249–259.
34. PanizzaS, MendozaMA, BerlingerM, HuangL, NicolasA, et al. (2011) Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146 : 372–383.
35. CarballoJA, JohnsonAL, SedgwickSG, ChaRS (2008) Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132 : 758–770.
36. NiuH, LiX, JobE, ParkC, MoazedD, et al. (2007) Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol Cell Biol 27 : 5456–5467.
37. BishopDK (1994) RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79 : 1081–1092.
38. LaoJP, OhSD, ShinoharaM, ShinoharaA, HunterN (2008) Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol Cell 29 : 517–524.
39. LiW, ChenC, Markmann-MulischU, TimofejevaL, SchmelzerE, et al. (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Nat Acad Sci USA 101 : 10596–10601.
40. ShinoharaA, GasiorS, OgawaT, KlecknerN, BishopDK (1997) Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2 : 615–629.
41. KurzbauerMT, UanschouC, ChenD, SchlogelhoferP (2012) The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell 24 : 2058–2070.
42. CallenderTL, HollingsworthNM (2010) Mek1 suppression of meiotic double-strand break repair is specific to sister chromatids, chromosome autonomous and independent of Rec8 cohesin complexes. Genetics 185 : 771–782.
43. CloudV, ChanY-L, GrubbJ, BudkeB, BishopDK (2012) Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337 : 1222–1225.
44. BishopDK, ParkD, XuL, KlecknerN (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell 69 : 439–456.
45. LydallD, NikolskyY, BishopDK, WeinertT (1996) A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383 : 840–843.
46. WanL, de los SantosT, ZhangC, ShokatK, HollingsworthNM (2004) Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic DSB repair in budding yeast. Mol Biol Cell 15 : 11–23.
47. BusyginaV, SehornMG, ShiIY, TsubouchiH, RoederGS, et al. (2008) Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev 22 : 786–795.
48. TsubouchiH, RoederGS (2006) Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev 20 : 1766–1775.
49. NiuH, WanL, BusyginaV, KwonY, AllenJA, et al. (2009) Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol Cell 36 : 393–404.
50. BishopDK, NikolskiY, OshiroJ, ChonJ, ShinoharaM, et al. (1999) High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells 4 : 425–443.
51. TsubouchiH, RoederGS (2003) The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev Cell 5 : 915–925.
52. GoldfarbT, LichtenM (2010) Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biology 8: e1000520.
53. KinebuchiT, KagawaW, EnomotoR, TanakaK, MiyagawaK, et al. (2004) Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein Dmc1. Mol Cell 14 : 363–374.
54. BusyginaV, GainesW, XuY, KwonY, WilliamsG, et al. (2013) Functional attributes of the S. cerevisiae meiotic recombinase Dmc1. DNA Repair 12 : 707–712.
55. KurokawaY, MurayamaY, Haruta-TakahashiN, UrabeI, IwasakiH (2008) Reconstitution of DNA strand exchange mediated by Rhp51 recombinase and two mediators. PLoS biology 6: e88.
56. LeeMH, ChangYC, HongEL, GrubbJ, ChangCS, et al. (2005) Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J Biol Chem 280 : 40980–40984.
57. BugreevDV, PezzaRJ, MazinaOM, VoloshinON, Camerini-OteroRD, et al. (2011) The resistance of DMC1 D-loops to dissociation may account for the DMC1 requirement in meiosis. Nat Struct Mol Biol 18 : 56–60.
58. ChiP, KwonY, SeongC, EpshteinA, LamI, et al. (2006) Yeast recombination factor Rdh54 functionally interacts with the Rad51 recombinase and catalyzes Rad51 removal from DNA. J Biol Chem 281 : 26268–26279.
59. HunterN, KlecknerN (2001) The single-end invasion: an asymmetric intermediate at the double - strand break to double-holliday junction transition of meiotic recombination. Cell 106 : 59–70.
60. AndersonCM, ChenSY, DimonMT, OkeA, DeRisiJL, et al. (2011) ReCombine: a suite of programs for detection and analysis of meiotic recombination in whole-genome datasets. PloS One 6: e25509.
61. MartiniE, DiazRL, HunterN, KeeneyS (2006) Crossover homeostasis in yeast meiosis. Cell 126 : 285–295.
62. SturtevantAH (1913) The linear arrangment of six sex-linked factors in Drosophila, as shown by their mode of association. J Exptl Zool 14 : 43–59.
63. HollingsworthNM, PonteL, HalseyC (1995) MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev 9 : 1728–1739.
64. OhSD, JessopL, LaoJP, AllersT, LichtenM, et al. (2009) Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods Mol Biol 557 : 209–234.
65. AllersT, LichtenM (2001) Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106 : 47–57.
66. ChuS, HerskowitzI (1998) Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell 1 : 685–696.
67. SourirajanA, LichtenM (2008) Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev 22 : 2627–2632.
68. XuL, AjimuraM, PadmoreR, KleinC, KlecknerN (1995) NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol 15 : 6572–6581.
69. XuL, WeinerBM, KlecknerN (1997) Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev 11 : 106–118.
70. ChuangCN, ChengYH, WangTF (2012) Mek1 stabilizes Hop1-Thr318 phosphorylation to promote interhomolog recombination and checkpoint responses during yeast meiosis. Nucleic acids research 40 : 11416–11427.
71. StromL, LindroosHB, ShirahigeK, SjogrenC (2004) Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell 16 : 1003–1015.
72. UnalE, Arbel-EdenA, SattlerU, ShroffR, LichtenM, et al. (2004) DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell 16 : 991–1002.
73. WatrinE, PetersJM (2006) Cohesin and DNA damage repair. Exp Cell Res 312 : 2687–2693.
74. KleinF, MahrP, GalovaM, BuonomoSBC, MichaelisC, et al. (1999) A central role for cohesins in sister chromatid cohesion, formation of axial elements and recombination during meiosis. Cell 98 : 91–103.
75. CouteauF, NabeshimaK, VilleneuveA, ZetkaM (2004) A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr Biol 14 : 585–592.
76. DanielK, LangeJ, HachedK, FuJ, AnastassiadisK, et al. (2011) Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat Cell Biol 13 : 599–610.
77. FerdousM, HigginsJD, OsmanK, LambingC, RoitingerE, et al. (2012) Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet 8: e1002507.
78. LiXC, Bolcun-FilasE, SchimentiJC (2011) Genetic evidence that synaptonemal complex axial elements govern recombination pathway choice in mice. Genetics 189 : 71–82.
79. Sanchez-MoranE, SantosJL, JonesGH, FranklinFC (2007) ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev 21 : 2220–2233.
80. VilleneuveAM, HillersKJ (2001) Whence meiosis? Cell 106 : 647–650.
81. ShinoharaM, GasiorSL, BishopDK, ShinoharaA (2000) Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc Natl Acad Sci U S A 97 : 10814–10819.
82. GoldsteinAL, McCuskerJH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 : 1541–1553.
83. LongtineMS, McKenzieA3rd, DemariniDJ, ShahNG, WachA, et al. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 : 953–961.
84. Tong A, Boone C (2005) Synthetic Genetic Array (SGA) analysis in Saccharomyces cerevisiae. Yeast Protocols, Methods in Molecular Biology. Second ed. Totowa, NJ, USA: The Humana Press, Inc. pp. 171–192.
85. RothsteinR (1991) Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol 194 : 281–301.
86. ChenSY, FungJC (2011) Mapping of crossover sites using DNA microarrays. Meth Mol Biol 745 : 117–134.
87. HongEJ, ShinoharaA, BishopDK (2001) Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J Biol Chem 276 : 41906–41912.
88. SikorskiRS, HieterP (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 : 19–27.
89. Miller JH (1972) Experiments in Molecular Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory.
90. Van KomenS, MacrisM, SehornMG, SungP (2006) Purification and assays of Saccharomyces cerevisiae homologous recombination proteins. Methods Enzymol 408 : 445–463.
91. San FilippoJ, ChiP, SehornMG, EtchinJ, KrejciL, et al. (2006) Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem 281 : 11649–11657.
92. de los SantosT, HollingsworthNM (1999) Red1p: A MEK1-dependent phosphoprotein that physically interacts with Hop1p during meiosis in yeast. J Biol Chem 274 : 1783–1790.
93. Efron B, Tibshirani RJ (1993) An introduction to the Bootstrap. Boca Raton: Chapman and Hall/CRC.
94. WinzelerEA, RichardsDR, ConwayAR, GoldsteinAL, KalmanS, et al. (1998) Direct allelic variation scanning of the yeast genome. Science 281 : 1194–1197.
Štítky
Genetika Reprodukční medicína
Článek Unwrapping BacteriaČlánek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 1
-
Všechny články tohoto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



