-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaNotch Controls Cell Adhesion in the Drosophila Eye
Sporadic evidence suggests Notch is involved in cell adhesion. However, the underlying mechanism is unknown. Here I have investigated an epithelial remodeling process in the Drosophila eye in which two primary pigment cells (PPCs) with a characteristic ‘kidney’ shape enwrap and eventually isolate a group of cone cells from inter-ommatidial cells (IOCs). This paper shows that in the developing Drosophila eye the ligand Delta was transcribed in cone cells and Notch was activated in the adjacent PPC precursors. In the absence of Notch, emerging PPCs failed to enwrap cone cells, and hibris (hbs) and sns, two genes coding for adhesion molecules of the Nephrin group that mediate preferential adhesion, were not transcribed in PPC precursors. Conversely, activation of Notch in single IOCs led to ectopic expression of hbs and sns. By contrast, in a single IOC that normally transcribes rst, a gene coding for an adhesion molecule of the Neph1 group that binds Hbs and Sns, activation of Notch led to a loss of rst transcription. In addition, in a Notch mutant where two emerging PPCs failed to enwrap cone cells, expression of hbs in PPC precursors restored the ability of these cells to surround cone cells. Further, expression of hbs or rst in a single rst - or hbs-expressing cell, respectively, led to removal of the counterpart from the membrane within the same cell through cis-interaction and forced expression of Rst in all hbs-expressing PPCs strongly disrupted the remodeling process. Finally, a loss of both hbs and sns in single PPC precursors led to constriction of the apical surface that compromised the ‘kidney’ shape of PPCs. Taken together, these results indicate that cone cells utilize Notch signaling to instruct neighboring PPC precursors to surround them and Notch controls the remodeling process by differentially regulating four adhesion genes.
Published in the journal: . PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1004087
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004087Summary
Sporadic evidence suggests Notch is involved in cell adhesion. However, the underlying mechanism is unknown. Here I have investigated an epithelial remodeling process in the Drosophila eye in which two primary pigment cells (PPCs) with a characteristic ‘kidney’ shape enwrap and eventually isolate a group of cone cells from inter-ommatidial cells (IOCs). This paper shows that in the developing Drosophila eye the ligand Delta was transcribed in cone cells and Notch was activated in the adjacent PPC precursors. In the absence of Notch, emerging PPCs failed to enwrap cone cells, and hibris (hbs) and sns, two genes coding for adhesion molecules of the Nephrin group that mediate preferential adhesion, were not transcribed in PPC precursors. Conversely, activation of Notch in single IOCs led to ectopic expression of hbs and sns. By contrast, in a single IOC that normally transcribes rst, a gene coding for an adhesion molecule of the Neph1 group that binds Hbs and Sns, activation of Notch led to a loss of rst transcription. In addition, in a Notch mutant where two emerging PPCs failed to enwrap cone cells, expression of hbs in PPC precursors restored the ability of these cells to surround cone cells. Further, expression of hbs or rst in a single rst - or hbs-expressing cell, respectively, led to removal of the counterpart from the membrane within the same cell through cis-interaction and forced expression of Rst in all hbs-expressing PPCs strongly disrupted the remodeling process. Finally, a loss of both hbs and sns in single PPC precursors led to constriction of the apical surface that compromised the ‘kidney’ shape of PPCs. Taken together, these results indicate that cone cells utilize Notch signaling to instruct neighboring PPC precursors to surround them and Notch controls the remodeling process by differentially regulating four adhesion genes.
Introduction
Pattern formation in developing tissues requires cell signaling. A small number of signaling pathways are repeatedly utilized for cell fate decisions in developing tissues (reviewed in [1]). In addition, cell signaling is also known to play a role in controlling cell sorting. For example, in the Drosophila wing, Hh signaling regulates cell segregation between anterior and posterior compartments (reviewed in [2]), while Notch signaling is required for establishing a boundary that separates dorsal and ventral cells (reviewed in [3]). In the Drosophila eye, Notch is required for a variety of developmental steps including rearranging pigment cells into hexagonal arrays [4]. All these observations raise the question of how Notch is involved in tissue remodeling. The observation that Notch is expressed in an epithelial sheet in the Drosophila embryo and continuously required for embryonic development after cell fate decision has led to speculation that Notch is involved in cell adhesion [5]. The behavior of primary pigment cells in the pupal eye also supports this view [4]. However, how Notch is involved in cell adhesion remains unclear.
Evidence accumulated to date supports the notion that cell adhesion plays a direct role in tissue remodeling. As first noted by J. Holtfreter and later formulated in “Differential Adhesion Hypothesis” (DAH) by M. Steinberg: sorting behaviors of cells are driven by interfacial free energy arising from differential adhesion among cells [6], [7], [8], [9]. In vivo observations support the DAH model. For example, in the Drosophila egg chamber, differential expression of E-cadherin determines localization of oocytes [10], [11]. In the eye epithelium, homophilic interactions mediated by E - and N-cadherin direct a group of four cone cells to arrange in a pattern that minimizes surface free energy [12]. In the chick spinal cord, MN-cadherin is involved in sorting out motor neurons [13]. All these examples show that cadherins are directly responsible for cell sorting in a variety of tissues through homophilic interactions. On the other hand, more complex patterns involve more intricate mechanisms. For example, in the Drosophila pupal eye organizing pigment cells into hexagonal arrays requires two groups of heterophilic-interacting adhesion molecules: Hibris (Hbs) and Sticks-and-Stones (Sns) from the Nephrin group; Roughest (Rst) and Kin of Irre (Kirre) from the Neph1 group [14]. Nephrin and Neph1 are adhesion molecules of the IRM family within the immunoglobulin (Ig) superfamily and both proteins are essential for maintaining specialized junctions during kidney development in mammals [15]. Despite mounting evidence linking cell adhesion to cellular patterns, how cell-cell adhesion is regulated in developing tissues to generate a variety of cellular patterns remains unclear.
This work describes a mechanism underlying an epithelial remodeling process in the Drosophila eye in which two primary pigment cells (PPCs) enwrap and isolate a group of cone cells from inter-ommatidial cells (IOCs). This paper shows that Notch signaling controls transcription of two groups of adhesion genes in the Drosophila eye. Notch activates adhesion genes of the Nephrin group but suppresses those of the Neph1 group. Differential distribution of two groups of adhesion molecules is further facilitated by removal of one group of adhesion molecules by another group through cis-interactions, leading to complementary distribution of four adhesion molecules within two populations of cells. This work uncovers a link between cell signaling and tissue remodeling.
Results
1. Notch is required for organization of ommatidial cells
The Drosophila eye derives from an invaginated epithelium at the embryonic stage [16]. Photoreceptor neurons and lens-secreting cone cells are specified at late larval and early pupal stages. At 18 h after puparium formation (APF), cone cells are surrounded by 4–5 inter-ommatidial cells (IOCs), which have relaxed apical profiles (Fig. 1A–A′). Shortly, two cells adjacent to cone cells start to expand apical contacts with cone cells in most ommatidia. At 20 h APF, these two cells completely enwrap cone cells with a ‘kidney’ shape and they become two primary pigment cells (PPCs) (Fig. 1B–B′). As a result, cone cells are fully isolated from the rest of IOCs within the epithelial plane. Further rearrangement of IOCs gives rise to a one-cell wide hexagonal lattice of IOCs that fully separates ommatidia. Separation of ommatidia by IOCs will eventually serve to optically insulate the ommatidial array across the eye (Fig. 1C–C′).
Fig. 1. Notch is required for development of primary pigment cells (PPCs). 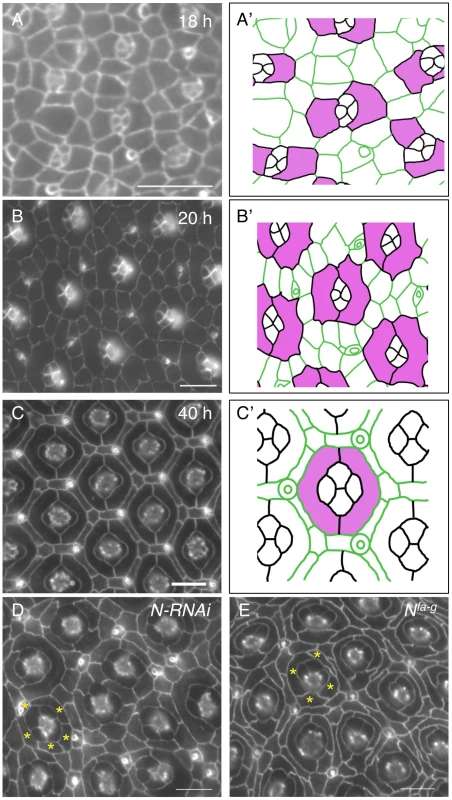
Eyes were stained using an anti-E-cadherin antibody in this figure. A–C) Wild type eyes at 18 h (A), 20 h (B) and 40 h (C) are shown. The tracings of eyes are shown in A′–C′, where PPC or PPCs precursors are highlighted in pink. D) Knockdown of Notch using a Notch RNAi transgene led to a failure of PPCs to develop. Typically, 3–5 cells (asterisks) were found adjacent to a cone cell cluster at 40 h APF. E) In the Nfa-g mutant, PPCs failed to develop. Frequently, 3–5 cells (asterisks) were contacting cone cells at 40 h APF. Scale bars, 10 µm. When Notch was depleted in all IOCs using RNAi, PPC precursors failed to enwrap cone cells. As a result, at 40 h APF, the cone cell cluster was found typically in direct contact with 4∼5 IOCs in an ommatidium (Fig. 1D), indicating Notch is required for the assembly of ommatidia (cone cells and PPCs). This phenotype is very reminiscent of the one seen in Nfa-g (Fig. 1E). Nfa-g is a loss-of-function Notch allele in which the activity of Notch is retained throughout larval stages but lost within the pupal stage [17]. As a result, cone cells in the eye are not affected by the mutation. Previous studies indicate that in Nfa-g mutants two PPC precursors initially touch each other at both ends but they fail to establish contacts [4], [18], suggesting weakened adhesion between PPCs and/or adhesion between PPCs and cone cells. However, it has remained unclear how Notch is involved in cell-cell adhesion.
2. Notch is activated in PPCs
The receptor Notch is broadly expressed in all cells in the early pupal eye [19], [20], [21]. In contrast, expression of the ligand Delta (Dl) is often cell type - specific and the protein is predominantly found within endocytic vesicles [21]. Consistent with the previous study [21], Dl was detected in cone cells at 18 h APF (Fig. 2A). Using a Dl-specific reporter, Dl transcript was detected in cone cells (Fig. 2B). Especially, anterior cone cells had the highest level of Dl expression at this early stage (Fig. 2B). By 24 h APF, although expression in the posterior cone cells was slightly increased, Dl expression in the anterior cone cells still remained the highest within the cone cell cluster (Fig. 2C). To identify the cell types that receive active Notch signaling, a Notch activity reporter GBE-Su(H)m8-lacZ [22] was used. Consistent with expression of the Dl reporter, the Notch activity was detected in a significantly higher level in two cells adjacent to anterior-posterior cone cells than in other cells at 18 h APF (Fig. 2D). These two cells were presumably the two PPC precursors. In particular, the highest Notch reporter activity was detected in the PPC precursor adjacent to the anterior cone cell within each ommatidium (Fig. 2D). By 24 h APF, GBE-Su(H)m8-lacZ expression was found in both PPCs and the difference in the level of lacZ expression between these cells became less obvious than earlier stages (Fig. 2E). Therefore, Dl transcription within the anterior and posterior cone cells is correlated with a high level of the Notch activity in the two PPC precursors.
Fig. 2. Notch is activated in PPC precursors. 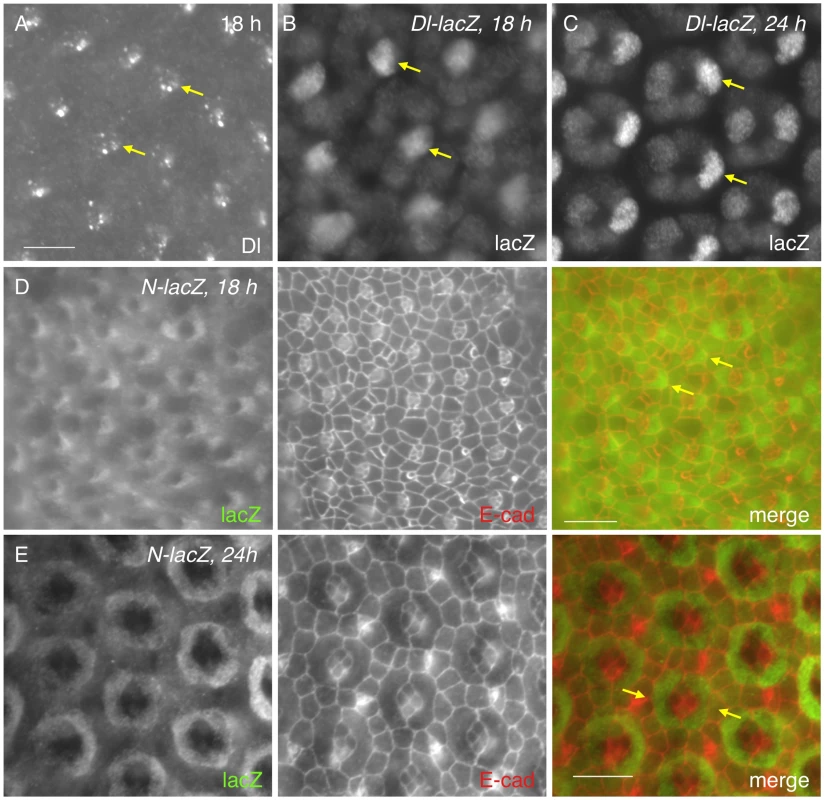
A) The Delta (Dl) protein was detected in cone cells (arrows) at 18 h APF as assessed with an anti-Dl antibody. B–C) Expression of a Dl reporter at 18 h (B) and 24 h APF (C) confirms its cone cell specificity. LacZ was detected at the highest level in anterior cone cells (arrows). D–E) Expression of GBE-Su(H)m8-lacZ, a reporter for Notch activity at 18 h (D) and 24 h APF (E). LacZ staining is shown on the left panel and E-cad channel in the middle. Merged views are shown on the right. Notch activity was high in cells adjacent to the anterior cone cells at 18 h APF (arrows, D). The difference of Notch activity between PPC precursors became less obvious at 24 h APF than earlier stages (arrows, E). Scale bars, 10 µm. 3. Notch signaling activates transcription of hbs and sns
Previously it has been shown that hbs and sns, two genes from the Nephrin group, are transcribed in PPCs [14], [23]. The pattern of the Notch activity is very reminiscent of hbs and sns expression. When an intracellular domain of Notch (NICD, an activated form of Notch) was expressed in a single PPC in the eye using a FLP-out technique [24], the Rst protein level was increased 128% at the border between the target PPC and neighboring IOCs compared with wild type borders (Fig. 3A and Table 1), a phenotype very similar to over-expression of hbs in a PPC [14]. To test whether hbs transcription was activated upon activation of Notch, NICD was expressed in a single IOC. Upon activation of Notch, an ectopic activity of the hbs reporter P[w+]36.1 was observed in the target IOC (Fig. 3B). These results indicate Notch is sufficient to activate hbs transcription.
Fig. 3. Notch signaling activates hbs and sns expression. 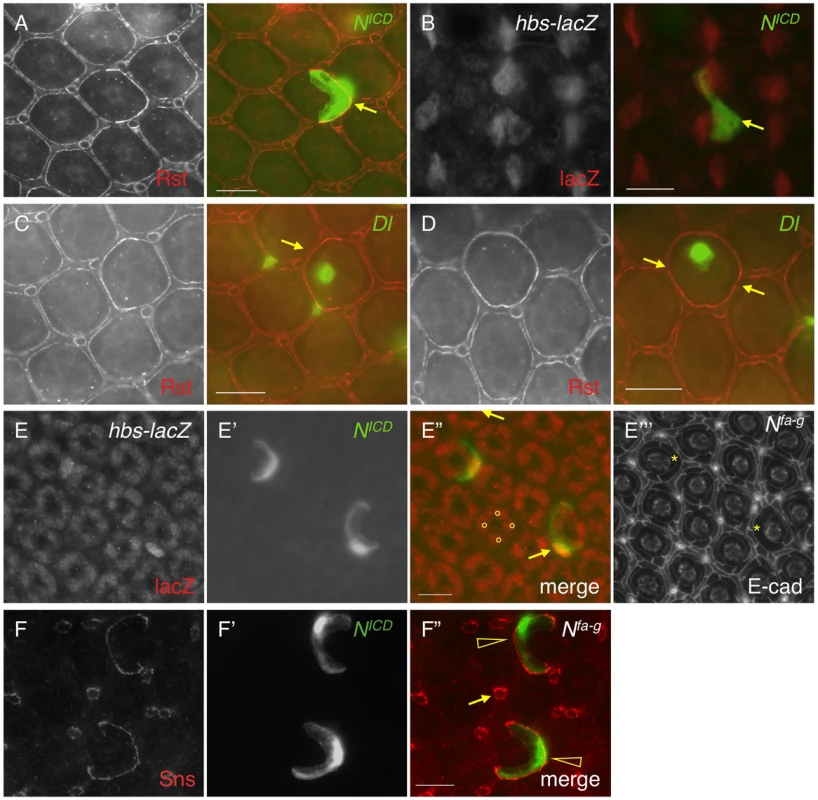
A–D) NICD or Dl (green) was over-expressed in single cells marked by GFP. The eyes were stained with an anti-Rst (left, A, C and D) or anti-lacZ antibody (left, B). Merged views are shown on the right. A) Over-expression of NICD increased Rst on the membrane. Arrows point to a PPC-IOC border with elevated Rst. B) Ectopic NICD induced ectopic hbs transcription as assessed using a hbs reporter (hbs-lacZ). Upon expression of NICD in a single IOC, ectopic lacZ was observed (arrow). C) When Dl was over-expressed in a posterior cone cell, ectopic Rst was detected at the border between the posterior PPC and its neighboring IOCs (arrow). D) When Dl was over-expressed in a polar cone cell, ectopic Rst was found at all borders surrounding the two PPCs (arrows). E–E″′) Notch is required for hbs transcription. In the Nfa-g mutant, hbs transcription (E) as assessed by the hbs reporter activity, was only detected in cone cells (bullets, E″) but lost in PPCs. In the Notch mutant, when Notch was activated in single IOCs by expressing NICD (E′), hbs transcription (arrows) as well as the characteristic ‘kidney’ shape of PPCs (asterisks) was restored. The lacZ and NICD channels are shown in E and E′, respectively, and the merged view in E″. The E-cadherin channel is shown in E″′. F–F″) Notch is required for sns expression. In the Nfa-g mutant, the Sns protein (F) was lost in PPCs. Sns expression in bristle groups (arrows) was not affected. In this mutant, when Notch was activated in single IOCs by expressing NICD (F′), the Sns protein (open arrowheads) was restored. The Sns channel is shown in F and the NICD channel in F′. The merged view is shown in F″. Scale bars, 10 µm. Tab. 1. Quantification of changes in the level of Rst or Hbs upon genetic manipulations. 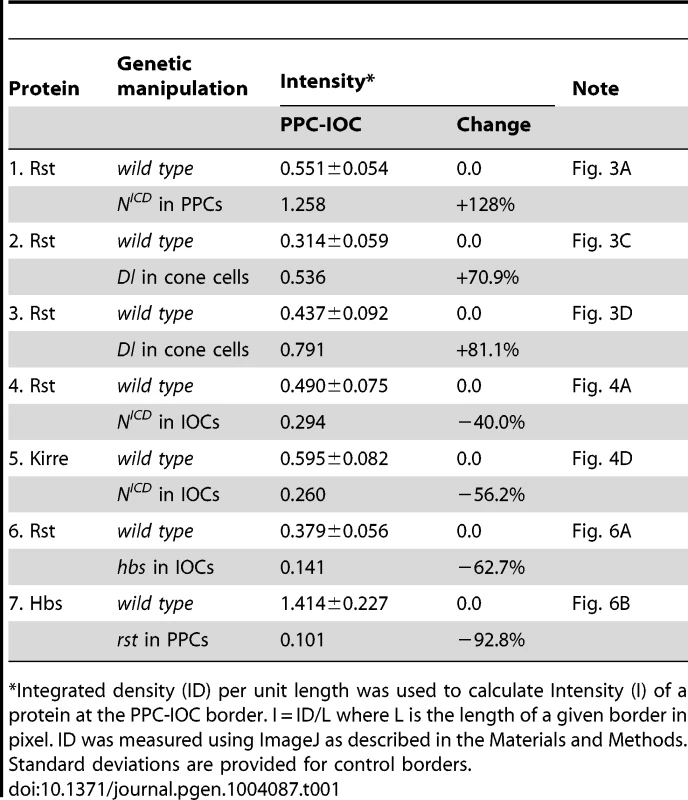
Integrated density (ID) per unit length was used to calculate Intensity (I) of a protein at the PPC-IOC border. I = ID/L where L is the length of a given border in pixel. ID was measured using ImageJ as described in the Materials and Methods. Standard deviations are provided for control borders. Consistently, when the Notch ligand Delta (Dl) was over-expressed in a single cone cell (either anterior or posterior), the Rst level was increased about 71% at the border between the adjacent PPC and its neighboring IOCs (Fig. 3C and Table 1). When Dl was over-expressed in a single polar or equatorial cone cell, the Rst level was elevated about 81% at the two PPC-IOC borders encircling two PPCs (Fig. 3D). These results indicate that the ligand Dl in cone cells is sufficient to activate hbs transcription in neighboring PPCs.
To test the necessity of Notch in control of hbs transcription, Nfa-g mutant was used along with the hbs reporter P[w+]36.1. In the wild type eye, the hbs reporter was detected in emerging PPCs as well as in cone cells [14]. In Nfa-g mutants, the hbs reporter activity was retained in cone cells but lost in PPC precursors at 40 h APF (Fig. 3E). When GFP alone was expressed in single IOCs in the Nfa-g mutant, 12% of clones (n = 86, 4 eyes) exhibited a kidney-shape seen in wild type PPCs. In contrast, when NICD (activated Notch) was expressed in single IOCs in the same mutant, 100% of clones (n = 92, 4 eyes) exhibited kidney shape (Fig. 3E″′). In addition, these cells also expressed the hbs reporter (Fig. 3E–E″). These results indicate that Notch signaling is required for activation of hbs transcription. A similar effect was also observed with sns when Notch was activated in single IOCs in the Nfa-g mutant (Fig. 3F–F″). Taken together, these data indicate that Notch is both sufficient and necessary to activate transcription of both hbs and sns, the adhesion genes of the Nephrin group.
4. Notch signaling suppresses transcription of rst and kirre
When Notch was activated in a single IOC by expressing NICD, as expected, the Rst level was increased at IOC-IOC borders (Fig. 4A–A″′). Unexpectedly, the Rst level was reduced 40% at IOC-PPC borders (Fig. 4A″′ and Table 1). To test whether a reduction of the Rst protein seen at the PPC-IOC border is due to a reduction in rst transcription, a rst reporter (rstF6-lacZ) was used to monitor the rst activity. Upon activation of Notch in a single IOC, rstF6-lacZ was lost in the target cell (Fig. 4B–B″′), indicating Notch is sufficient to suppress rst transcription in IOCs. Since Notch is normally activated in PPC precursors, this result suggests that in the wild-type eye Notch suppresses rst in developing PPCs. Consistently, in Nfa-g mutants, rstF6-lacZ was expanded to all pigment cells surrounding cone cells (Fig. 4C–C″), indicating that Notch is necessary for suppressing rst in emerging PPCs. A similar effect was also seen with kirre when Notch activities were altered (Fig. 4D–D″ and Table 1). Taken together, these results indicate that Notch is both sufficient and necessary to suppress rst and kirre transcription in developing PPCs.
Fig. 4. Notch signaling suppresses rst and kirre expression. 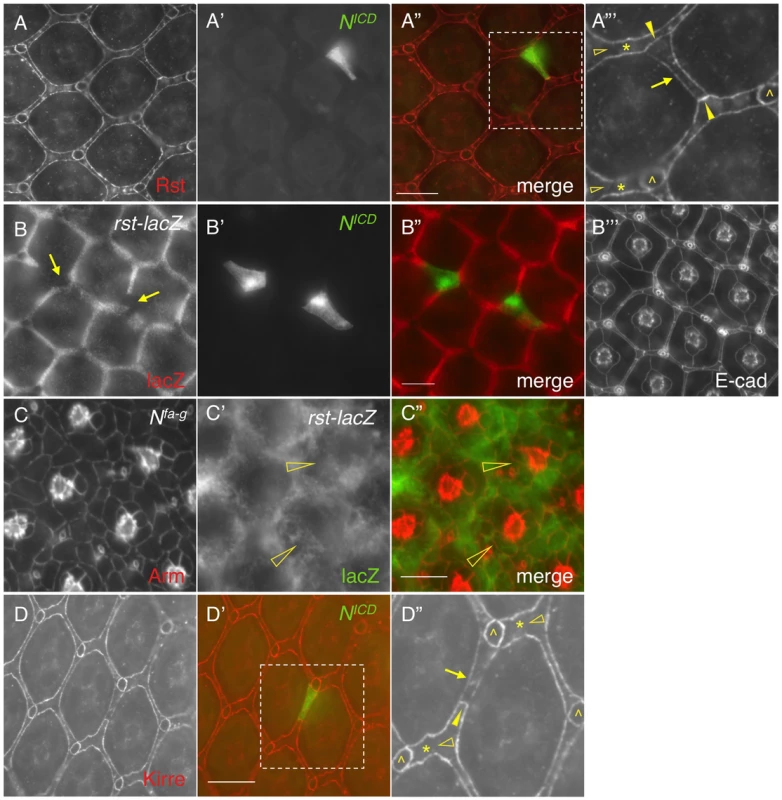
A–A″′) NICD (A′) was over-expressed in a single IOC and the Rst protein (A) was assessed using an anti-Rst antibody. The merged view is shown in A″. The enlarged view of a boxed region in A″ is shown in A″′. For clarity, only Rst channel is shown in A″′. Rst was increased at IOC-IOC borders (arrowheads) but reduced at the PPC-IOC border (arrow). Rst was undetectable at wild type IOC-IOC borders (open arrowheads). Single IOCs (asterisks) and bristle groups (carets) are indicated. B–B″′) NICD (B′) was over-expressed in a single IOC and the rst transcript (B) was assessed using a rst reporter (rstF6-lacZ). Merged view is shown in B″ and cell shape was visualized using an anti-DE-cadherin antibody (B″′). Arrows point to IOCs that lost lacZ staining. C–C″) Notch is required to suppress rst transcription. In the Nfa-g mutant, the rst transcript was detected in cells adjacent to cone cells (open arrowheads). Cell morphology was visualized using an anti-Armadillo (Arm) antibody (C). rst transcription was assessed using the rst reporter rstF6-lacZ (C′). The merged view is shown in C″. D–D″) NICD (green, D′) was over-expressed in a single IOC and the eye was stained with an anti-Kirre antibody (D). The merged view is shown in D′. The enlarged view of a boxed region in D′ is shown in D″. For clarity, only Kirre channel is shown in D″. The Kirre protein was increased at an IOC-IOC border (arrowhead) but reduced at the PPC-IOC border (arrow). Kirre was undetectable at wild type IOC-IOC borders (open arrowheads). Single IOCs (asterisks) and bristle groups (carets) are indicated. Scale bars, 10 µm. 5. Distribution dynamics of adhesion molecules
It has been shown previously that genes coding for adhesion molecules of the IRM family are expressed in complementary cell types during cell rearrangement (e.g., 24 h APF): hbs and sns in PPCs; rst and kirre in IOCs [14], [23]. The patterns of hbs and rst transcription at 18 h are similar to those at later stages (e.g., 27 h APF) based on the hbs and rst reporters (Fig. 5A–C). However, immune-staining using specific antibodies revealed striking differences in the distribution patterns of the Hbs and Rst proteins in the eye between 18 h and 40 h APF. Especially, both Hbs and Rst were present ubiquitously at a high level at all borders among epithelial cells at 18 h APF (Fig. 5D–D″′). This is in drastic contrast to later stages (e.g., 27–40 h APF) when both Hbs and Rst were diminished at IOC-IOC and PPC-cone borders (see below). At 20 h APF when two PPCs fully enwrapped the four cone cells, both Hbs and Rst remained at a high level at PPC-PPC and PPC-cone borders but slightly reduced at IOC-IOC borders (Fig. 5E–E″′). At 27 h APF, similar to earlier stages, both Hbs and Rst were enriched at PPC-IOC borders. In contrast, these proteins were reduced at PPC-PPC and PPC-cone borders and diminished at IOC-IOC borders (Fig. 5F–F″′). At 40 h APF, both Hbs and Rst proteins were again enriched at IOC-PPC borders but diminished at PPC-PPC and PPC-cone borders (Fig. 5G–G″′). They were undetectable at IOC-IOC borders (Fig. 5G–G″′). A similar dynamics in protein distribution was also observed with Sns and Kirre (data not shown). These results indicate that four adhesion molecules are initially present in all epithelial cells at 18–20 h APF in the eye and removed from one group of cells at later stages. Therefore, distribution of Hbs, Sns, Rst and Kirre proteins undergoes a transition from ubiquitous to complementary distribution during epithelial remodeling.
Fig. 5. Distribution of Hbs and Rst is dynamic in the eye epithelium. 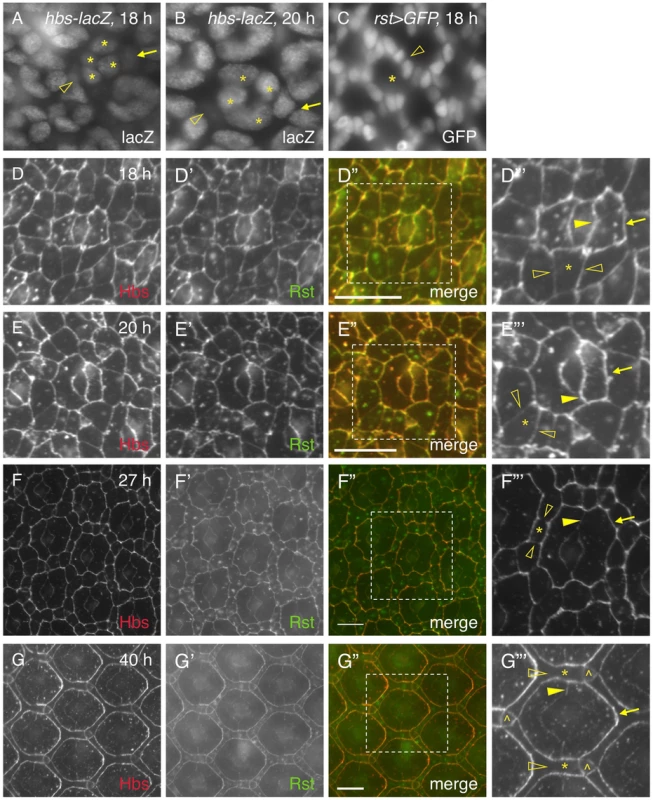
The activity of a hbs reporter was assessed using an anti-lacZ antibody (A–B). Cone cells are marked (asterisks). A) The activity of the hbs reporter was detected in emerging PPCs at 18 h APF. The nucleus of a PPC precursor (arrow) was rising half way to the level of cone cell nuclei while the nucleus of the second PPC precursor was lagging behind (open arrowhead). B) The activity of the hbs reporter was detected in PPC precursors at 20 h APF. The nucleus of a PPC precursor (arrow) had arisen to the level of those of cone cells while the nucleus of the second PPC precursor was still below the plane (open arrowhead). C) rst was transcribed in IOCs (open arrowhead) at 18 h APF. rst-Gal4 was used to drive expression of nuclear GFP(rst>GFP). The location for an ommatidium is indicated (asterisk). D–G″′) Distribution of Hbs and Rst is dynamic during 18–40 h APF. The Hbs channels (red) are shown in D–G and the Rst channels (green) in D′–G′. The merged views are shown on D″–G″. The enlarged views of boxed regions in D″–G″ are shown in D″′–G″′. For clarity, only Hbs channels are shown in D″′–G″′. Single IOCs (asterisks) and bristle groups (carets) are indicated. D) At 18 h APF, both Hbs and Rst were found at all borders surrounding PPC precursors including PPC-IOC borders (arrows) and PPC-cone borders (arrowheads). These proteins were also present at IOC-IOC borders (open arrowheads). E) At 20 h APF, PPCs fully enwrapped cone cells. Both Hbs and Rst were enriched at PPC-IOC (arrows), PPC-cone and PPC-PPC borders (arrowheads) while these proteins were slightly reduced at IOC-IOC borders (open arrowheads). F) At 27 h APF, both Hbs and Rst were enriched at PPC-IOC borders (arrows) while these proteins were reduced at PPC-PPC and PPC-cone borders (arrowheads). Hbs and Rst were diminished at IOC-IOC borders (open arrowheads). G) At 40 h APF, both Hbs and Rst proteins were enriched at IOC-PPC borders (arrows) while they were diminished at PPC-PPC and PPC-cone borders (arrowhead). Hbs and Rst were undetectable at IOC-IOC borders (open arrowheads). Scale bars, 10 µm. 6. cis-interactions destabilize the adhesion complex
Hbs and Sns from the Nephrin group and Rst and Kirre from the Neph1 group co-localize at the border between PPCs and IOCs, and heterophilic interactions between these two groups of proteins in trans (interactions between proteins from two adjacent cells or trans-interactions) stabilize the adhesion complex on the membrane [14], [23]. The observation that both Hbs and Rst were found in all IOCs at the beginning of cell rearrangement (Fig. 5D–E″′) raises the question of how these IRM adhesion molecules interact with each other when placed in the same cell (cis-interaction). To assess the effect of cis-interaction, Hbs was mis-expressed in the cells that normally express the counterparts Rst and Kirre. Upon expression of Hbs in a single IOC, the level of Rst was reduced about 63% at the PPC-IOC border and the number of vesicles was increased significantly in the target IOC (Fig. 6A–A″ and Table 1). Nevertheless, transcription of rst as assessed by the rst reporter rstF6-lacZ was not altered in the clone (data not shown). Similarly, when Rst was mis-expressed in a single PPC that normally transcribes hbs and sns, the Hbs level on the membrane was reduced 93% and the number of vesicles increased markedly in the target PPC (Fig. 6B–B″ and Table 1). Similarly, the activity of the hbs reporter P[w+]36.1 was unchanged in the clone (data not shown). These results suggest that, while heterophilic interactions between two groups of IRM adhesion molecules in trans stabilize both proteins on the membrane, interactions among these proteins in cis destabilize proteins on the membrane and promote turnover of these proteins.
Fig. 6. cis-interactions promote protein turnover. 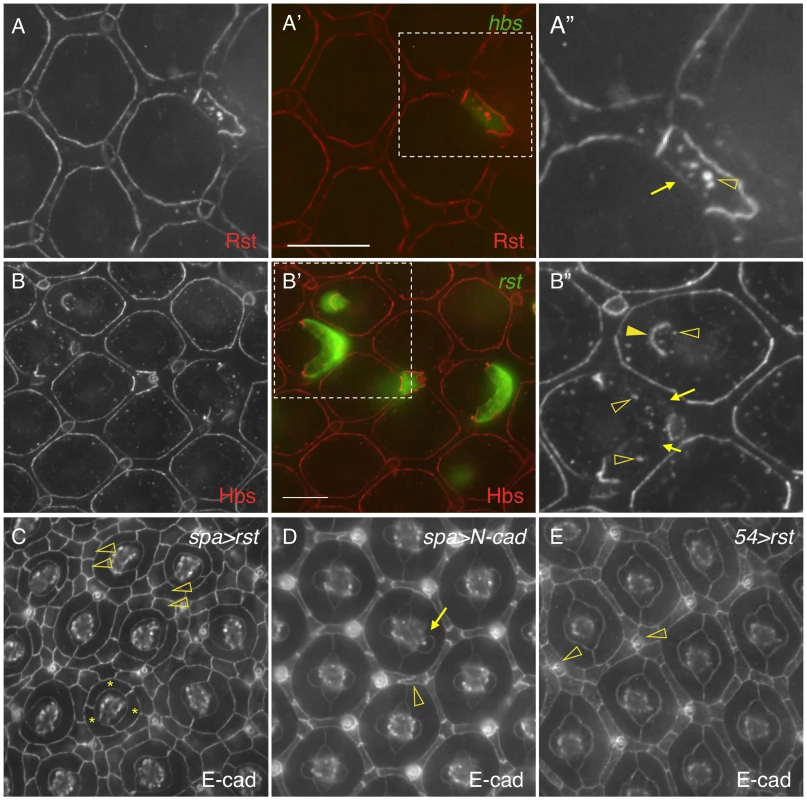
A–A″) Hbs promotes turnover of Rst in the same cell. hbs was mis-expressed in a single IOC (A′) and the eye was stained with an anti-Rst antibody (A). Levels of Rst were reduced at the border between the target IOC and its neighboring PPC (arrow) and increased in vesicles (open arrowhead). Merged view is shown in A′. The enlarged view of a boxed region in A′ with the Rst channel is shown in A″. B–B″) Rst promotes turnover of Hbs in the same cell. rst was mis-expressed in single cells (B′) and the eye stained with an anti-Hbs antibody (B). The Hbs level was reduced at PPC-IOC borders (arrows). A higher level of the Hbs protein was observed in vesicles (open arrowheads). When Rst was expressed in a cone cell, the Hbs level was elevated at the cone-PPC border (arrows). The target cone cell also had a higher level of Hbs in vesicles (open arrowheads). Merged view is shown in B′. The enlarged view of a boxed region in B′ with the Hbs channel is shown in B″. C) Interference of Hbs by mis-expressing Rst in PPCs (spa>rst) led to severe disruption of the hexagonal pattern of the eye. Three cells surrounding a cone cell cluster are highlighted (asterisks). IOCs failed to sort into a single file (open arrowheads). D) Over-expression of N-cadherin in cone cells (spa>N-cadherin) had a mild effect on tissue remodeling. An abnormal cone cell was highlighted (arrow) along with a defective IOC (open arrowhead). E) Over-expression of Rst in IOCs (54>rst) had a mild effect on tissue remodeling. Several IOCs formed a cluster around a bristle group (open arrowheads). Scale bars, 10 µm. To assess the effect of cis-interactions on pattern formation, rst was mis-expressed in all PPCs using spa-Gal4. Spa-Gal4 is known to drive expression of transgenes in cone cells and PPCs [25]. Upon expression of rst in cone cells and PPCs (spa>rst), the hexagonal pattern of the eye was severely disrupted. While spatial organization of cone cells was mildly affected, various numbers of PPCs (typically ranging from 1 to 3) were found adjacent to cone cells. More strikingly, IOCs failed to sort into single file. As a result, 2–3 rows of IOCs scattered in between ommatidia across the eye and the eye was extremely rough (Fig. 6C). To exclude the possibility that the effect of over-expression was simply due to enhanced adhesion among cone cells and/or PPCs, N-cadherin was over-expressed in these cells using the same spa-Gal4. N-cadherin is known to mediate adhesion among cone cells through homophilic interactions [12]. In contrast to Rst, over-expression of N-cadherin (spa>N-cadherin) only led to mild defects in IOCs and cone cells with largely intact PPCs (Fig. 6D). To exclude the possibility that the severe defects seen in spa>rst are simply due to detrimental effects of the protein on the cells when expressed at a high level, Rst was over-expressed in IOCs using Gal4-54 (54>rst). In the 54>rst eye, IOCs were occasionally found in cluster with a bristle group. Nevertheless, the hexagonal pattern was only mildly affected (Fig. 6E). Although we cannot exclude the possibility that different protein levels also contribute to the different phenotypes seen in these experiments, the results presented here strongly suggest that interference of IRM adhesion molecules by cis-interactions has a strong impact on the establishment of the hexagonal pattern.
7. Adhesion restores spatial pattern of cell clusters
This work demonstrates that Notch controls transcription of IRM adhesion genes. On the other hand, Notch is also known to control transcription of multiple other genes during eye development. It is not clear whether loss of adhesion molecules is responsible for the PPC defects seen in Notch mutants. To address this issue, a rescue experiment was performed using Nfa-g as a background mutant and UAS-hbs as a rescue construct. When Hbs was expressed in a single cell adjacent to cone cells, 83% of the target cells (n = 257, 13 eyes) elongated and the interface between the target cell and IOCs expanded in a manner similar to a wild type PPC (Fig. 7A). Further, when Hbs was expressed in two cells adjacent to cone cells, in nearly all cases examined so far, Hbs positive cells fully enwrapped the cone cell group from the anterior and posterior sides resembling two wild type PPCs (Fig. 7B). These results indicate that adhesion is sufficient to restore spatial relationship of cell clusters in Notch mutants.
Fig. 7. Spatial organization of primary pigment cells requires Hbs and Sns. 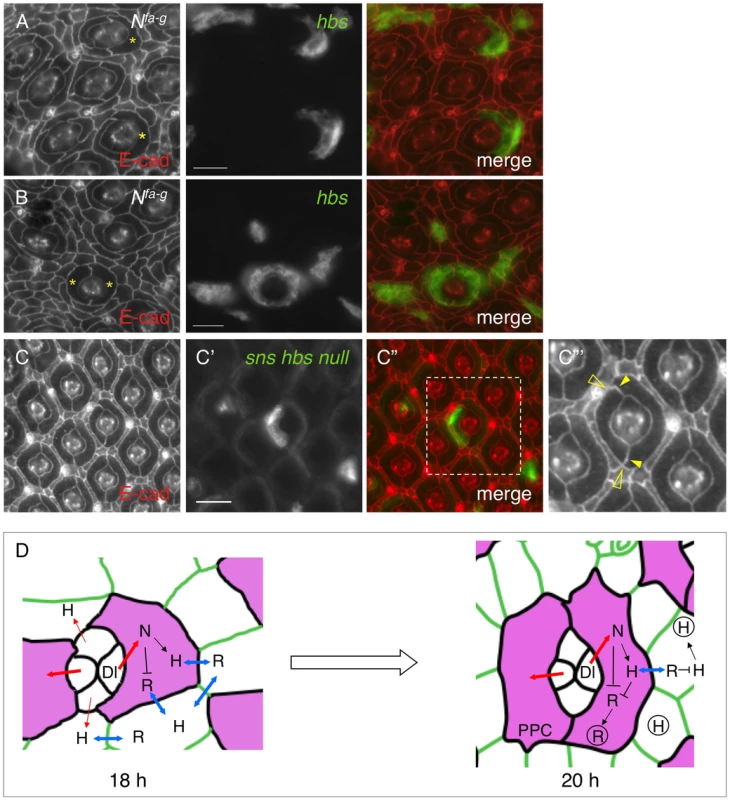
Eyes were stained with an anti-E-cadherin antibody (red, left). Target (mutant or over-expression) cells are marked by GFP (green, middle). The merged views are shown on the right. A–B) Expression of Hbs in a single cell restored the ‘kidney’ shape of PPCs. In the Nfa-g eye, hbs was expressed in single IOCs. When hbs was expressed in single cells adjacent to cone cells, the cell (asterisks) spread around cone cells (A). When hbs was introduced into two cells adjacent to cone cells, these cells (asterisks) fully enwrapped cone cells (B). C–C″′) Hbs and Sns are required for organization of PPCs. In a single PPC mutant for both hbs and sns (green, C′), the cell reduced the apical surface area and PPC-IOC border. The enlarged view of a boxed region in C″ with the E-cadherin channel is shown in C″′. Open arrowheads mark the shortened PPC-IOC border while arrowheads highlight the curved PPC-PPC borders. D) A model for control of PPC recruitment by cell signaling and cell adhesion. At 18 h APF, all IOCs that contact cone cells have access to Dl and express Hbs. However, IOCs adjacent to anterior-posterior cone cells receive a high level of Notch signaling (thick red lines) than other IOCs (thin red lines). These cells express a higher level of Hbs than other IOCs. Hbs boosts the ability of these cells to enwrap cone cells and gain more access to Dl. Therefore, Notch signaling and Hbs create a positive feedback loop so that initially a small difference in Notch signaling is amplified. As a result, two cells adjacent to anterior and posterior cone cells outcompete other IOCs and enwrap cone cells as PPC precursors. At 20 h APF, PPC precursors gain full access to Dl and constantly produce Hbs while shutting down Rst production. The remaining Rst in PPCs is removed by cis-interactions. In the meantime, other IOCs that are now denied access to Dl constantly supply Rst. The remaining Hbs in IOCs are cleared out by cis-interactions, leading to complementary distribution of two groups of adhesion molecules. For simplicity, only Hbs and Rst are shown. H = Hbs; R = Rst; N = Notch; Dl = Delta. Double-headed arrows represent trans-interactions between Hbs and Rst; Hbs and Rst in circle represent proteins in vesicles. Scale bars, 10 µm. To test the necessity of cell-cell adhesion for formation of the spatial pattern of PPCs, hbs and sns double mutant was generated using snsZF1.4 and hbs459 mutant alleles (see Materials and Methods). Large clonal patches generated using this double mutant together with ey-FLP led to extremely rough eyes in adults (data not shown). Single PPCs mutant for both sns and hbs had a shorten PPC-IOC border and reduced apical surface (Fig. 7C–C″′). In addition, PPC-PPC border became curved. As a result, the apical profile of the target PPC became more rounded. These results indicate that sns and hbs are required for the normal ‘kidney’ shape of PPCs.
Discussion
In developing tissues, one way to isolate a small group of cells from other groups is to induce a few neighboring cells to surround them. This paper shows that Notch provides an instructive signal in inducing neighboring cells to spread around and eventually surround centrally localized cone cells in the Drosophila eye. This work demonstrates that Notch functions in this process by differentially regulating four adhesion genes.
Notch controls cell adhesion
This work demonstrates that Notch is involved in cell-cell adhesion by regulating transcription of adhesion genes. Notch signaling is known to play a pleiotropic role in controlling cell fate during animal development [26]. The requirement of Notch during Drosophila embryonic development after cell fate decision has led to speculation that Notch is involved in cell adhesion [5]. This notion is supported by the behavior of PPCs in the pupal eye [4]. However, clear evidence linking Notch to cell adhesion has been lacking. This study shows that in the pupal eye Notch differentially controls transcription of four IRM adhesion genes. Notch activates transcription of hbs and sns but represses rst and kirre, leading to differential expression of IRM adhesion genes in two populations of cells: IOCs by default express rst and kirre; PPCs by activation of Notch signaling express hbs and sns. Heterophilic interactions between Hbs/Sns and Rst/Kirre proteins mediate preferential adhesion between IOCs and PPCs [14], [23]. Therefore, Notch signaling sets up differential expression of adhesion genes (Fig. 7D).
This work also illustrates how a single signaling pathway transforms an initially homogeneous population of cells into two morphologically distinct groups of cells. In the wild-type eye, PPCs are polarized since PPCs without exception enwrap cone cells from anterior/posterior rather than from polar/equatorial sides. Data presented in this work suggest that asymmetric distribution of Dl in cone cells sets up PPC polarity. At the beginning of cell rearrangement (∼18 h APF), all IOCs that contact cone cells have access to Dl and express Hbs. However, asymmetric expression of Dl in cone cells creates a bias. IOCs that contact anterior-posterior cone cells receive a high level of Notch signaling (thick red lines, Fig. 7D) and produce more Hbs, which in turn boosts the ability of these cells to enwrap cone cells and gain more access to Notch signaling. In contrast, other IOCs that initially receive a low level of Notch signaling (thin red lines, Fig. 7D) are at a disadvantage and quickly lose competition to PPC precursors in enwrapping cone cells. As a result, Notch and Hbs create a positive feedback loop through which an initial small difference in Notch signaling is amplified, giving rise to PPCs exclusively enwrapping cone cells from anterior and posterior sides (Fig. 7D).
cis-interactions promote protein turnover
This work provides evidence that interactions between adhesion molecules from the Nephrin group and those from the Neph1 group in cis promote protein turnover. IRM adhesion molecules are known to form heterophilic interactions. Proteins from the Nephrin group bind in trans to proteins from the Neph1 group and trans-interactions among IRM adhesion molecules stabilize proteins on the membrane [14], [23]. In contrast, cis-interactions among these proteins destabilize proteins on the membrane (this work). Results presented herein support a model that cis-interactions provide a mechanism for removing counterpart proteins from the same cells (Fig. 7D). After two PPC precursors completely surround the cone cell group (e.g., 20 h APF), these cells gain full access to Dl. In response to Notch signaling, PPC precursors constantly produce Hbs, which removes Rst from the same cells through cis-interaction. By the same mechanism, all other IOCs that are now denied access to Dl by default constantly produce Rst, which in turn clears Hbs from IOCs. Therefore, a combination of transcriptional regulation by Notch and post-translational mechanism by cis-interactions provides a mechanism for the transformation of initially ubiquitous distribution into complementary distribution of four adhesion molecules within two populations of cells (Fig. 7D).
cis-interactions observed in the Drosophila eye are very reminiscent of interactions between the Notch receptor and its ligand Dl. It has been shown that, in the Drosophila embryo and the eye imaginal disk, an increase of Dl in a Notch-expressing cell inhibits Notch signaling in a cell-autonomous fashion via cis-interaction [27], [28]. In the Notch-Dl case, the level of Dl within a Notch-expressing cell determines the intensity of Notch signaling that cells receive, which in turn determines cell fates [27], [28]. In the case of IRM adhesion molecules, the level of a protein from one group in a cell determines the amount of counterpart proteins from the other group on the membrane of the same cell, which alters cell-cell adhesion. More specifically, in the Drosophila eye cis-interactions remove remnant proteins and facilitate the differential distribution of IRM adhesion molecules without affecting cell fate. Despite different impact of cis-interactions on cell-cell interactions in both cases (N-Dl versus IRM adhesion molecules), they share one common feature: presence of one protein interferes with the function of the counterpart protein in the same cell. What structural elements are involved in cis-interactions between these proteins and how cis-interactions lead to a reduction of protein activity still remain questions for further investigation.
Although evidence presented in this work suggests a simple relationship among cell signaling, cell adhesion and cell shape, two observations highlight the complexity of PPC recruitment in the developing Drosophila eye. First, hbs can restore the ‘kidney’ shape of PPCs at a lower frequency than Notch (or NICD) in the Nfa-g mutant. This observation suggests that adhesion genes may not represent all the function of Notch in recruiting PPCs. Notch is known to have a wide range of target genes. In particular, several transcription factors are known targets of Notch for the determination of PPC cell fate [29]. Therefore, it is possible that additional effectors of Notch signaling are also involved in conferring on PPCs the ability to enwrap cone cells. Second, this work suggests a positive feedback loop that promotes selection of PPC precursors in the developing eye (Fig. 7D). On the other hand, it has been shown recently that Hbs promotes Notch signaling by interacting with presenilin [30]. A potential more direct impact of Hbs on Notch signaling raises the possibility that there may exist a second positive feedback loop between Notch and Hbs: Notch activates hbs transcription and Hbs in return enhances Notch signaling, whereby initially a small difference of Notch signaling among IOCs is amplified, leading to separation of Hbs/Sns-expressing cells from those expressing Rst/Kirre. Whether the second positive feedback loop plays a role in PPC recruitment remains to be tested.
This paper illustrates how a small number of cells utilize a single signaling pathway to instruct neighboring cells to surround them, whereby the centrally localized cells are isolated from other cells. Since isolation of a group of cells by another is commonly seen in developing tissues, a correlation between cell signaling and cell adhesion may be a more general mechanism for organizing cells during organ formation.
Materials and Methods
1. Drosophila genetics
The sns and hbs double mutant snsZF1.4 hbs459 was generated for this work by recombining snsZF1.4 and hbs459, a loss-of-function allele of sns and hbs, respectively, onto the second chromosome. Nfa-g, UAS-Notch RNAi, Dl-lacZ, y w hsFLP, UAS-nlsGFP and Act5C>y+>Gal4 UAS-GFP were provided by the Bloomington Stock Center. rst-Gal4 was obtained from National Institute of Genetics Fly Stock Center (Japan). Other flies used: rstF6-lacZ [31], spa-Gal4 [25], UAS-N-cadherin [12], snsZF1.4 and UAS-sns (gift of Susan Abmayr), UAS-NICD (gift of Cedric Wesley), UAS-NICD-lexA (gift of Toby Lieber), P[w+]36.1 and hbs459 (gift of Mary Baylies), UAS-hbs (gift of Helen Sink), GBE-Su(H)m8-lacZ (N-lacZ) [22], Gal-54 [23], UAS-rst (gift of Karl-F. Fischbach), UAS-kirre/duf (gift of Marc Ruiz-Gomez), UAS-Dl (gift of Marek Mlodzik) and hsFLP MKRS (gift of Matthew Freeman).
2. Clonal analyses
Single cell clones for over-expressing a target gene were generated using a FLP-out technique as described previously [14]. To induce clones, pupae at 12 h APF were heat-shocked at 37°C in a water bath for 20 min. Clones were marked by GFP. The genotypes of clones are shown as follows:
-
UAS-NICD/Act5C>y+>Gal4 UAS-GFP; hsFLP MKRS/+ (Figs. 3A, 4A and 4D)
-
P[w+]36.1/Act5C>y+>Gal4 UAS-GFP; UAS - NICD/hsFLP MKRS (Fig. 3B)
-
UAS-Dl/Act5C>y+>Gal4 UAS-GFP; hsFLP MKRS/+ (Figs. 3C–D)
-
Nfa-g; P[w+]36.1/Act5C>y+>Gal4 UAS-GFP; UAS - NICD/hsFLP MKRS (Fig. 3E)
-
Nfa-g; UAS - NICD/Act5C>y+>Gal4 UAS-GFP; hsFLP MKRS/+ (Fig. 3F)
-
rstF6-lacZ/Act5C>y+>Gal4 UAS-GFP; UAS - NICD/hsFLP MKRS (Fig. 4B)
-
UAS-hbs/Act5C>y+>Gal4 UAS-GFP; hsFLP MKRS/+ (Figs. 6A)
-
UAS-rst/Act5C>y+>Gal4 UAS-GFP; hsFLP MKRS/+ (Figs. 6B)
-
Nfa-g; UAS-hbs/Act5C>y+>Gal4 UAS-GFP; hsFLP MKRS/+ (Fig. 7A–B)
Loss-of-function clones were generated using a MARCM technique [32]. Clones were induced by heat-shocking third instar larvae at 37°C for 1 h. Clones were marked by GFP. The genotype of clones: yw hsFLP; FRT42D snsZF1.4 hbs459/FRT42D Gal80; tub-Gal4 UAS-mCD8-GFP/+ (Fig. 7C).
3. Immunohistochemistry
Immunostaining of the pupal eye was carried out as described [14]. Rat anti-Kirre (1∶5000) and rabbit anti-Hbs AS14 (1∶2500) were used as previously described [23]. Other primary antibodies: mouse anti-Rst Mab24A5.1(1∶100) [33], rabbit anti-Sns (1∶300) [34] and rabbit anti-lacZ (1∶2000; 5 Prime→3 Prime). Rat anti-DE-cadherin (1∶20), mouse anti-Armadillo (1∶20) and mouse anti-Dl 9B (1∶20) were provided by Developmental Studies Hybridoma Bank at the University of Iowa. Secondary antibodies: Alexa 488 and Alexa 568 conjugated secondary antibodies (1∶5000; Molecular Probes); Cy5 conjugated secondary antibodies (1∶1000; Jackson ImmunoResearch Laboratories). All images were captured using an Axioplan2 epi-fluorescence microscope equipped with an Axiocam digital camera (Carl Zeiss, Inc.).
4. Quantification of membrane protein levels
Levels of membrane proteins were quantified using ImageJ [35]. Briefly, a long and narrow stripe that surrounded and closely followed the target border was carefully traced. The integrated density (ID1) of the selected region was recorded using ImageJ. The background integrated density (ID0) was recorded by moving the same selection box to a background region. The membrane protein level (I) reflected by integrated intensity per unit length was determined following I = 2*(ID1−ID0)/L, where L is the perimeter of the selected region in pixel. For each experiment, the average intensity from 6 neighboring wild type borders was calculated and used as a control.
Zdroje
1. BaroloS, PosakonyJW (2002) Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev 16 : 1167–1181.
2. DahmannC, BaslerK (1999) Compartment boundaries: at the edge of development. Trends Genet 15 : 320–326.
3. IrvineKD, VogtTF (1997) Dorsal-ventral signaling in limb development. Curr Opin Cell Biol 9 : 867–876.
4. CaganRL, ReadyDF (1989) Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev 3 : 1099–1112.
5. HoppePE, GreenspanRJ (1986) Local function of the Notch gene for embryonic ectodermal pathway choice in Drosophila. Cell 46 : 773–783.
6. HoltfreterJ (1939) Gewebeaffinität, ein Mittel der embryonal Formbildung. Arch Exptl Zellforsch Gewebezucht 23 : 169–209.
7. HoltfreterJ (1944) A study of the mechanics of gastrulation: Part II. J Exp Zool 95 : 171–212.
8. SteinbergMS (1963) Reconstruction of tissues by dissociated cells. Some morphogenetic tissue movements and the sorting out of embryonic cells may have a common explanation. Science 141 : 401–408.
9. SteinbergMS (1970) Does differential adhesion govern self-assembly processes in histogenesis? Equilibrium configurations and the emergence of a hierarchy among populations of embryonic cells. J Exp Zool 173 : 395–433.
10. GodtD, TepassU (1998) Drosophila oocyte localization is mediated by differential cadherin-based adhesion. Nature 395 : 387–391.
11. Gonzalez-ReyesA, St JohnstonD (1998) The Drosophila AP axis is polarised by the cadherin-mediated positioning of the oocyte. Development 125 : 3635–3644.
12. HayashiT, CarthewRW (2004) Surface mechanics mediate pattern formation in the developing retina. Nature 431 : 647–652.
13. PriceSR, De Marco GarciaNV, RanschtB, JessellTM (2002) Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell 109 : 205–216.
14. BaoS, CaganR (2005) Preferential adhesion mediated by Hibris and Roughest regulates morphogenesis and patterning in the Drosophila eye. Dev Cell 8 : 925–935.
15. FischbachKF, LinneweberGA, AndlauerTF, HertensteinA, BonengelB, et al. (2009) The irre cell recognition module (IRM) proteins. J Neurogenet 23 : 48–67.
16. Garcia-BellidoA, MerriamJR (1969) Cell lineage of the imaginal discs in Drosophila gynandromorphs. J Exp Zool 170 : 61–75.
17. ShellenbargerDL, MohlerJD (1975) Temperature-sensitive mutations of the notch locus in Drosophila melanogaster. Genetics 81 : 143–162.
18. LarsonDE, LibermanZ, CaganRL (2008) Cellular behavior in the developing Drosophila pupal retina. Mech Dev 125 : 223–232.
19. FehonRG, JohansenK, RebayI, Artavanis-TsakonasS (1991) Complex cellular and subcellular regulation of notch expression during embryonic and imaginal development of Drosophila: implications for notch function. J Cell Biol 113 : 657–669.
20. KoohPJ, FehonRG, MuskavitchMA (1993) Implications of dynamic patterns of Delta and Notch expression for cellular interactions during Drosophila development. Development 117 : 493–507.
21. ParksAL, TurnerFR, MuskavitchMA (1995) Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech Dev 50 : 201–216.
22. FurriolsM, BrayS (2001) A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr Biol 11 : 60–64.
23. BaoS, FischbachKF, CorbinV, CaganRL (2010) Preferential adhesion maintains separation of ommatidia in the Drosophila eye. Dev Biol 344 : 948–956.
24. StruhlG, FitzgeraldK, GreenwaldI (1993) Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell 74 : 331–345.
25. FuW, NollM (1997) The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev 11 : 2066–2078.
26. Artavanis-TsakonasS, RandMD, LakeRJ (1999) Notch signaling: cell fate control and signal integration in development. Science 284 : 770–776.
27. JacobsenTL, BrennanK, AriasAM, MuskavitchMA (1998) Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development 125 : 4531–4540.
28. MillerAC, LyonsEL, HermanTG (2009) cis-Inhibition of Notch by endogenous Delta biases the outcome of lateral inhibition. Curr Biol 19 : 1378–1383.
29. NagarajR, BanerjeeU (2007) Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development 134 : 825–831.
30. SinghJ, MlodzikM (2012) Hibris, a Drosophila nephrin homolog, is required for presenilin-mediated Notch and APP-like cleavages. Dev Cell 23 : 82–96.
31. ApitzH, KambacheldM, HohneM, RamosRG, StraubeA, et al. (2004) Identification of regulatory modules mediating specific expression of the roughest gene in Drosophila melanogaster. Dev Genes Evol 214 : 453–459.
32. LeeT, LuoL (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22 : 451–461.
33. SchneiderT, ReiterC, EuleE, BaderB, LichteB, et al. (1995) Restricted expression of the irreC-rst protein is required for normal axonal projections of columnar visual neurons. Neuron 15 : 259–271.
34. BourBA, ChakravartiM, WestJM, AbmayrSM (2000) Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev 14 : 1498–1511.
35. SchneiderCA, RasbandWS, EliceiriKW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9 : 671–675.
Štítky
Genetika Reprodukční medicína
Článek Unwrapping BacteriaČlánek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 1
-
Všechny články tohoto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



