-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAffects Plant Architecture by Regulating Local Auxin Biosynthesis
Plant architecture is one of the key factors that affect plant survival and productivity. Plant body structure is established through the iterative initiation and outgrowth of lateral organs, which are derived from the shoot apical meristem and root apical meristem, after embryogenesis. Here we report that ADP1, a putative MATE (multidrug and toxic compound extrusion) transporter, plays an essential role in regulating lateral organ outgrowth, and thus in maintaining normal architecture of Arabidopsis. Elevated expression levels of ADP1 resulted in accelerated plant growth rate, and increased the numbers of axillary branches and flowers. Our molecular and genetic evidence demonstrated that the phenotypes of plants over-expressing ADP1 were caused by reduction of local auxin levels in the meristematic regions. We further discovered that this reduction was probably due to decreased levels of auxin biosynthesis in the local meristematic regions based on the measured reduction in IAA levels and the gene expression data. Simultaneous inactivation of ADP1 and its three closest homologs led to growth retardation, relative reduction of lateral organ number and slightly elevated auxin level. Our results indicated that ADP1-mediated regulation of the local auxin level in meristematic regions is an essential determinant for plant architecture maintenance by restraining the outgrowth of lateral organs.
Published in the journal: . PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1003954
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003954Summary
Plant architecture is one of the key factors that affect plant survival and productivity. Plant body structure is established through the iterative initiation and outgrowth of lateral organs, which are derived from the shoot apical meristem and root apical meristem, after embryogenesis. Here we report that ADP1, a putative MATE (multidrug and toxic compound extrusion) transporter, plays an essential role in regulating lateral organ outgrowth, and thus in maintaining normal architecture of Arabidopsis. Elevated expression levels of ADP1 resulted in accelerated plant growth rate, and increased the numbers of axillary branches and flowers. Our molecular and genetic evidence demonstrated that the phenotypes of plants over-expressing ADP1 were caused by reduction of local auxin levels in the meristematic regions. We further discovered that this reduction was probably due to decreased levels of auxin biosynthesis in the local meristematic regions based on the measured reduction in IAA levels and the gene expression data. Simultaneous inactivation of ADP1 and its three closest homologs led to growth retardation, relative reduction of lateral organ number and slightly elevated auxin level. Our results indicated that ADP1-mediated regulation of the local auxin level in meristematic regions is an essential determinant for plant architecture maintenance by restraining the outgrowth of lateral organs.
Introduction
Higher plants have a diverse range of body structures. Phyllotaxis of lateral organs, branching pattern, as well as size, shape and position of lateral organs all contribute to the overall architecture of a plant. Plant architecture is the most obvious morphology of mature plants and has long served as an important criterion for systematic and taxonomic classification of plant species [1], [2]. Plant architecture is largely determined by genetic programs and, to some extent, by environmental cues, such as light, humidity, temperature, nutrition, and plant density. Detailed studies have been focused on genetic factors that are crucial for maintenance of shoot apical meristem (SAM), initiation and outgrowth of axillary meristem (AM), proper growth rate for lateral organ development, and correct timing for reproduction and senescence [2]. Research on plant architecture has important agronomic implications because it has a direct impact on the suitability and productivity of a plant. One of the most successful modifications of plant architecture is the Green Revolution, which is based on the selection of wheat cultivars with shorter and sturdier stems, resulting in plants with enhanced yield via improved resistance to wind and rain [3]. Over the past several years, branching patterns have been intensively investigated in rice, since the formation of tillers and panicle branches will greatly affect the efficiency of light absorption, which will in turn influence the adaptation of plants to the environment [4]–[6]. Understanding the genetic and molecular mechanisms of the regulation of plant architecture would help us to modify agronomically useful traits and thus facilitate the breeding of high-yield crops.
The success of the Green Revolution mainly results from selection of plants with altered biosynthesis and/or signaling of plant hormones, among which auxin is a determinant for plant architecture. Auxin is a critical factor controlling a wide variety of developmental processes, including embryogenesis, maintenance of apical dominance, and formation of lateral organs [7], [8]. Active auxin, mainly indole-3-acetic acid (IAA), is reported to be synthesized de novo by tryptophan (Trp)-dependent and/or independent pathways in the shoot apex, young leaves, and root apex [7], [9]–[14]. After synthesis, auxin is transported by the polar transport machinery [7], so that an appropriate distribution of auxin is established to maintain normal plant architecture. Disruption of auxin gradient, either by changing auxin biosynthesis, transport, or signaling, will lead to alteration of organ growth patterns and changes of plant architecture. For example, over-expression of the auxin biosynthesis genes YUCCA1 (YUC1) and YUCCA6 (YUC6) led to auxin over-production, resulting in increased apical dominance [15], [16], whereas the quadruple knock-out mutant yuc1,2,4,6 showed abnormal flower development and loss of apical dominance, i.e., increased branching [17], [18]. The double mutant pgp1-1 pgp19-1 (mdr1-1), which exhibited a 70%–80% reduction in polar auxin transport, displayed pleiotropic phenotypes such as curly leaves, dwarfism and decreased fertility [19], [20]. Moreover, the auxin response mutant axr1-12 and the tir1 afb1 afb2 afb3 quadruple receptor mutant produced highly branched inflorescences at maturity. The quadruple mutant occasionally had no roots or produced only a single cotyledon, leading to lethality at early stages [21]–[23]. Thus, maintenance of proper auxin response and/or homeostasis is critical for normal plant architecture.
In this paper, we identified a dominant Arabidopsis mutant with an abnormal architecture, which we named adp1-D (altered development program 1 - Dominant). The architecture of adp1-D was greatly altered at maturity, with increased number of axillary branches, flowers, and lateral roots. The growth rate of the mutant was accelerated throughout its life cycle. We discovered that the mutant phenotypes were caused by over-expression of ADP1 gene. ADP1 encodes a protein with sequence similarity to the multidrug and toxic compound extrusion (MATE) transporter family, which is found in prokaryotes and eukaryotes. MATE transporters are reported to be involved in a variety of important biological processes, since they function in the exclusion of toxic organic cation and disease resistance and exhibit multi-substrate specificity [24], [25].
Here we provide molecular and genetic evidence to demonstrate that the phenotypes of adp1-D were caused by reduction of the local auxin levels in the meristematic regions. The reduction was probably due to decreased levels of auxin biosynthesis in the local meristematic regions. When expression levels of ADP1 and its three closest homologs were down-regulated in Arabidopsis, the resulting quadruple mutant exhibited growth retardation and a slight reduction of lateral organ number. Our results indicated that ADP1 and its homologous genes play important roles in maintaining normal plant architecture, possibly by regulating local auxin biosynthesis.
Results
The adp1-D Mutant Displayed Pleiotropic Phenotypes
We screened an Arabidopsis activation tagging mutant collection for mutants with altered plant architectures. The activation tagging mutant collection was generated using the activation tagging vector pSKI015 as described previously [26]. A dominant mutant with abnormal plant architecture, later designated adp1-D, was identified. The mutant displayed pleiotropic phenotypes, including accelerated growth rate of rosette leaves (Figure 1A, 1B and 1C), early flowering (Figure 1B and 1D), increased number of lateral roots (Figure S1A and Figure S1B). At maturity, the mutant had significantly more axillary branches (Figure 1D), including first-order rosette branches (RI, Figure 1E and 1F), higher-order rosette and cauline branches (RII and CII, Figure S1C). The lengths of first-order branches (RI and CI) were almost the same at different node positions (Figure S1D), suggesting the loss of apical dominance in the mutant. Occasionally, the axillary inflorescences were found in the axil of the cotyledons in the mutant (Figure 1H), which is unique because wild-type plants do not produce axillary inflorescences on the axil of the cotyledons (Figure 1G).
Fig. 1. The adp1-D mutant displayed pleiotropic phenotypes. 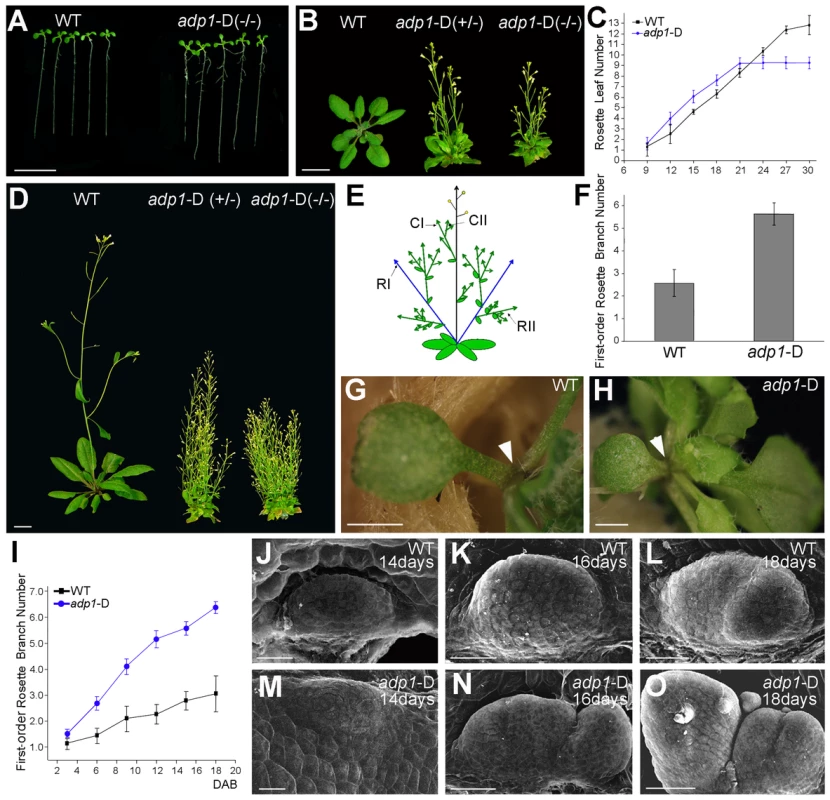
(A) Twelve-day-old seedlings and (B) 28-day-old plants grown under long-day conditions. The symbol +/− represents heterozygous mutants, while −/− represents homozygous mutants. (C) Emergence rate of rosette leaves in wild-type and adp1-D plants. At least 20 plants were measured for each genotype. (D) Six-week-old plants grown under long-day conditions. (E) Schematic diagram of Arabidopsis branching pattern. CI, first order cauline branches; CII, higher order cauline branches; RI, first order rosette branches; RII, higher order rosette branches. (F) First-order rosette branch number of two-month-old wild-type and adp1-D plants. At least 40 plants of each genotype were measured. (G) and (H) Detection of cotyledon petiole from 20-day-old plants of (G) the wild type and (H) the adp1-D mutant. Although no axillary bud was produced in the cotyledon axil in wild-type plants, about 30% of the adp1-D mutants produced an inflorescence in the cotyledon axil (indicated by arrowheads). (I) Generation rate of rosette branches in wild-type and adp1-D plants. At least 20 plants were measured for each genotype. Axillary buds longer than 2 mm were considered as axillary branches. (J) to (O) Scanning electron micrographs of developing axillary buds in the axils of the first pair of rosette leaves of wild-type (J–L), and adp1-D (M–O) plants. Plants were fixed after 14–18 days growth under long-day conditions. Development of the axillary buds can be divided into three stages. In stage 1, the axillary shoot bulged outwards, forming a semicircular zone. In stage 2, the outgrowth increased in size, forming an axillary leaf primordia. In stage 3, the axillary buds start to grow outwards. Under the growth conditions in this study, only one primordium was formed in the leaf petiole axil in wild-type plants, and most of the primordia stagnate in stage 2, whereas the axillary buds in adp1-D mutants developed faster (comparing K with N) and were at more advanced stages (comparing L with O). For (A) (B) and (D), bar = 1 cm; for (G) and (H), bar = 1 mm; for (J) to (O), bar = 40 µm. adp1-D mutant was almost sterile, producing very few seeds. Sterile phenotypes have been previously reported to be associated with induction of axillary branch outgrowth [27], [28]. In order to clarify whether the bushy phenotype of this mutant was a secondary effect of the sterile phenotype or not, we analyzed the kinetics of the initiation rate of the first-order rosette branches (RI). Our result showed that the difference between the mutant and wild type appeared as early as 3 days after reproductive transition, and the difference became larger with time (Figure 1I), until sterility appeared. However, exogenous application of GR24 [29], [30], a strigolactone-like inhibitor of shoot branching, revealed that the first-order rosette branches (RI) could be completely inhibited by GR24 application (Figure S2B to S2E), whereas higher-order cauline branches (CII) were not affected (Figure S2B to S2F), suggesting that the higher order branches were probably the secondary effect of sterility. Therefore in this study, we only focused on the first-order rosette branches.
To investigate the origin of the abnormal branches, we compared the early stage of axillary bud outgrowth in the first pair of leaves using scanning electron microscopy (SEM). In the wild type, although the axillary buds initiated from the epidermal cells in the semicircular zone, and then bulged outwards to form the meristems, in most cases, the axillary buds ceased development at this stage (Figure 1J to 1L). However, most of the axillary meristems of the mutant appeared to be larger than those of the wild type, and they continued to develop into inflorescences (Figure 1M to 1O). In many cases, the mutant had increased number of axillary meristems (Figure 1O). Since the developmental program of the mutant had been changed from the beginning to the end of the entire life cycle, and since the plant architecture had been greatly altered, we named the mutant as adp1-D (altered development program 1 - Dominant).
The adp1-D Mutant Is a Gain-of-Function Mutant
To investigate the cause for the pleiotropic phenotypes in adp1-D, we performed thermal asymmetric interlaced PCR (TAIL-PCR), and identified a single T-DNA insertion in the intergenic region between At4g29130 and At4g29140 (Figure 2A). To examine whether the T-DNA insertion co-segregated with adp1-D phenotypes, we genotyped the T3 plants produced by heterozygous T2 mutants. Among 420 T3 plants, 102 were wild type without the T-DNA insertion, 106 were homozygous, and 212 were heterozygous with the T-DNA insertion (Figure 2B). All of the plants that were homozygous and heterozygous with the T-DNA insertion showed accelerated growth and increased number of lateral organs, whereas all of the plants without the T-DNA insertion appeared normal, suggesting that the pleiotropic phenotypes in adp1-D were caused by this single T-DNA insertion. To determine which gene was altered in its expression level, we examined the expression levels of all the genes within 10 kb upstream and downstream of the insertion site by quantitative RT-PCR. Only one gene, At4g29140, was over-expressed (by about 25-fold), while the expression of all the other genes remained mostly unchanged (Figure 2C), suggesting that over-expression of At4g29140 could be responsible for the adp1-D phenotypes. To confirm this, we over-expressed At4g29140 in wild-type Arabidopsis under its own promoter with four copies of the 35S enhancer (Figure 2D). The transgenic plants recapitulated all the phenotypes in adp1-D. About 10% of the transgenic plants showed more severe phenotypes of smaller plant size, highly compact leaves, and many more branches (Figure 2E and 2F). The expression level of At4g29140 correlated with the severity of the phenotypes (Figure 2E to 2G). These data indicate that the pleiotropic phenotypes of adp1-D were indeed caused by over-expression of At4g29140.
Fig. 2. Characterization of the ADP1 gene. 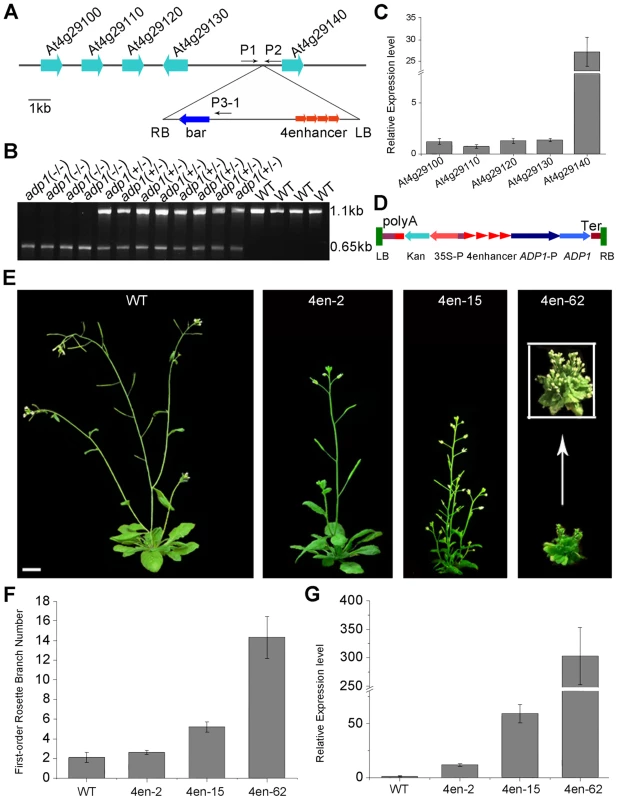
(A) Schematic diagram of the genomic region flanking the T-DNA insertion site in adp1-D. The arrow direction represents the transcriptional orientation of the gene. The four red arrowheads represent the four 35S enhancers from pSKI015. LB, T-DNA left border; bar, Basta resistance gene; 4Enhancers, CaMV 35S enhancer tetrad; RB, T-DNA right border. (B) Linkage analysis of the T-DNA insertion and the bushy phenotypes. The primers P1 and P2 amplified an 1123-bp fragment from the wild type, and P1 and LBb1 amplified a 650-bp fragment from the homozygous adp1-D mutant. (C) Expression of genes flanking the insertion site in the wild type and the homozygous adp1-D mutant measured by quantitative RT-PCR with a Tubulin gene as an internal control. The expression levels of each gene in the wild type were set as 1.0. Error bars represent the SD of three biological replicates. (D) Schematic diagram of the construct for At4g29140 over-expression in plants driven by four 35S enhancers. LB, T-DNA left border; polyA, CaMV 35S poly(A); KanR, kanamycin resistance gene NPT II; 35S-P, CaMV 35S promoter; 4Enhancer, CaMV 35S enhancer tetrad; ADP1-P, promoter of ADP1; ADP1, open reading frame of ADP1; Ter, nopaline synthase terminator; RB, T-DNA right border. (E) Transgenic plants over-expressing At4g29140 driven by four 35S enhancers showed the accelerated growth rate and bushy phenotypes. Bar = 1 cm. (F) First-order rosette branch number in transgenic plants. (G) Quantitative analysis of the ADP1 expression level in transgenic plants. The expression level of the transgenic plants was in accordance with the severity of the phenotypes. Error bars represent three biological replicates. ADP1 has no intron and encodes a protein of 532 amino acid residues (Supplemental Figure 3A), sharing sequence similarity with the Arabidopsis MATE proteins. These proteins are characterized by 11 to 13 transmembrane helixes and two typical MATE domains [31]. The Arabidopsis genome contains 58 putative MATE transporters, which are grouped into five groups [32]. ADP1 belongs to a clade, which has eight members sharing high sequence identity in the conserved MATE domain (Figure S3B and S3C).
ADP1 Is Expressed in the Meristematic Regions
To characterize the expression pattern of ADP1, a 2 kb promoter fragment upstream of ADP1 start codon was fused with β-glucuronidase (GUS) reporter gene and transformed into wild-type Arabidopsis. GUS staining analysis of the homogenous transgenic lines showed that the promoter activity of ADP1 was mainly detected in tissues where cells were actively dividing, such as leaf primordia and young leaves (Figure 3A), the junction between lateral root and the primary root (Figure 3B), root cap (Figure 3C), hydathodes (Figure 3D), the junction between secondary inflorescence and the main inflorescence (Figure 3D), young stamen and young siliques (Figure 3E).
Fig. 3. ADP1 expression pattern. 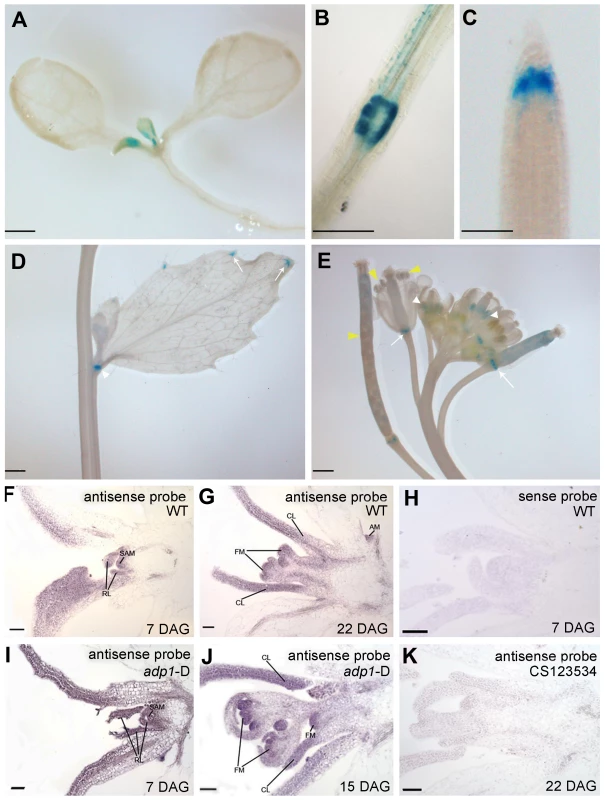
(A) to (E) ADP1 expression pattern by Promoter:GUS staining. ADP1 was expressed mainly in actively dividing tissues, such as leaf primordium and young leaf (A), the junction between lateral root and main root (B), root cap (C), the junction between cauline branch and the main inflorescence (D), the young stamen primordium and siliques (E, white and yellow arrow heads, respectively) and the junction between siliques and petiole (E, white arrows). (F) to (K) Wax-embedded sections of vegetative shoots and reproductive shoots from wild-type (F and G) and adp1-D (I and J) hybridized with the antisense probe. ADP1 was expressed mainly in meristematic regions, such as the shoot apical meristem (SAM), axillary meristem (AM) in cauline leaf petiole and flower meristem (FM). RL, rosette leaf; CL, cauline leaf. (F) and (I), 8-day-old seedlings; (G), 22-day-old wild-type seedlings; (J), 15-day-old adp1-D seedlings. (H) Section from an 8-day-old wild-type seedling hybridized with the sense probe. (K) Section from a 22-day-old CS123534 seedling hybridized with the antisense probe. Bar = 20 µm. Since the adp1-D phenotypes were apparently associated with shoot apical meristem (SAM) activity, we performed in situ hybridization for both wild-type and adp1-D seedlings. The transcript signals of ADP1 were analyzed in shoot apical tissues, using a 300-bp ADP1 cDNA fragment as the antisense probe. ADP1 transcripts were detected primarily in the meristematic regions (e.g., SAM), young leaves and flowers (Figure 3F and 3G). Comparison of adp1-D and the wild type showed that the overall distribution of mRNA was similar, but the signal in the adp1-D mutant was much stronger (Figure 3I and 3J compared with Figure 3F and 3G). No hybridization signals were detected for the control sense probe in wild type (Figure 3H), or the antisense probe in the ADP1 loss-of-function mutant CS123534 (Figure 3K). Taken together these expression results correlated well with adp1-D phenotypes and indicated that ADP1 transcript levels were up-regulated in adp1-D only within the regions where ADP1 transcript was originally detected.
ADP1 Is Localized in Endo-Membrane Structures
Antisera were raised against ADP1 and were used to immunolocalize ADP1 protein in the root apical meristem region. ADP1 was localized to small intracellular structures in the root cap and the junction between lateral root and primary root (Figure 4A to 4D). To further characterize these structures, transgenic lines were generated to express ADP1 in fusion with GFP or RFP on its N-terminus, under the CaMV 35S promoter. Approximately 30% of the transgenic lines with these tags recapitulated the mutant phenotypes to varying extents, suggesting that the fusion proteins were functional (Figure S4A to S4C). We found that, in the transgenic lines with recapitulated mutant phenotypes, the fluorescent signals of the fusion proteins were localized to punctuated particles with different sizes and shapes, which were distributed ubiquitously in the cells (Figure 4E), suggesting that ADP1 fusion proteins are retained in the endo-membrane organelles. To investigate the nature of these small particles, we crossed the endo-membrane marker lines with these transgenic lines, and found that ADP1 could co-localize with the endosome marker RabF2a-GFP [33] (Figure 4E to 4G) but not with TGN or ER markers (Figure S4D to S4J). Next, we used the fluorescent dye FM4-64 [34] to trace the endocytic dynamics of the fusion protein. After a half-hour treatment with the sterol dye FM4-64, the green signals (GFP tagged fusion protein) were partially co-localized with FM4-64 particles (Figure 4H to 4J). Next we treated the roots for one hour with brefeldin A (BFA), a fungal toxin that targets a subclass of ARF GEFs and is often used as an inhibitor of vesicle transport [35]. As a result, we found that some of the ADP1-GFP granules aggregated into larger bodies which co-localized with FM4-64, while the rest remained scattering in the cytoplasm (Figure 4K to 4M). Taken together, these results indicate that ADP1 resides in endo-membrane vesicles.
Fig. 4. Subcellular localization of ADP1. 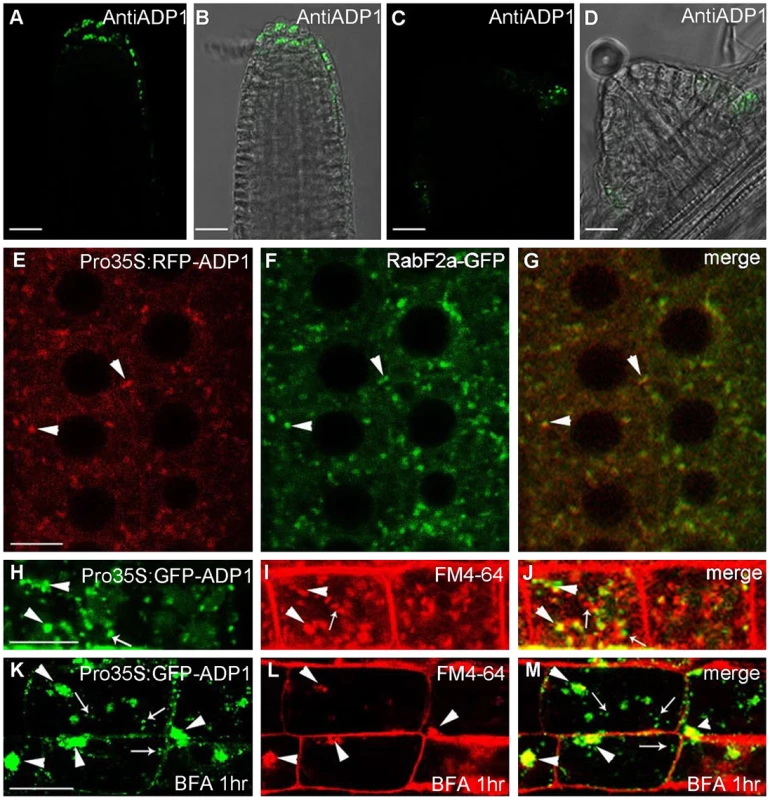
(A) to (D) Subcellular localization of ADP1 in root cap (A and B) and the junction between lateral root and the main root (C and D). (E) to (G) RFP-ADP1 was co-localized with RabF2a-GFP. Typical particles were indicated by arrowheads. (F) The merged image of the two fluorescence signals. (H) to (J) GFP-ADP1 was partially co-localized with FM4-64 staining particles. The overlapped granules are indicated by arrowheads, and the non-overlapped granules are indicated by arrows. (K) to (M) GFP-ADP1 was partially resistant to BFA treatment. The overlapped particles with BFA bodies of FM4-64 are indicated by arrowheads, and the resistant ADP1 granules are indicated by arrows. For (D) to (L), bar = 15 µm. Auxin Signals Were Decreased in Meristematic Regions in adp1-D
The phenotypes of adp1-D (i.e., accelerated growth rate, highly branched shoots and increased number of lateral organs) resemble those mutants of auxin synthesis, transport, and response. To test whether auxin pathways are defective in adp1-D, we first examined hypocotyl length in a temperature shift experiment. The rationale was that decreased level of auxin, either by alteration of auxin biosynthesis, transport and/or signaling, would prevent hypocotyls from elongating under high temperature condition [36]. As shown in Figure 5A and 5B, after shifting the plants from 22°C to 29°C, the hypocotyl length of the wild-type seedlings increased by three to four folds, while that of adp1-D remained unchanged, suggesting that auxin synthesis, transport, or signaling was affected in the mutant. We then crossed adp1-D to DR5:GUS auxin-responsive reporter lines [37] and observed the GUS signal in different tissues of the F3 homozygous plants. In the wild type, DR5:GUS signals were detected mainly in the actively growing regions, such as the SAM, leaf tips, petiole bases, emerging axillary buds and flower primordia (Figure 5C to 5G). However, in adp1-D, DR5:GUS signals were substantially decreased in almost all the meristematic tissues (Figure 5H to 5L).
Fig. 5. Evidence of reduced auxin levels in adp1-D mutants. 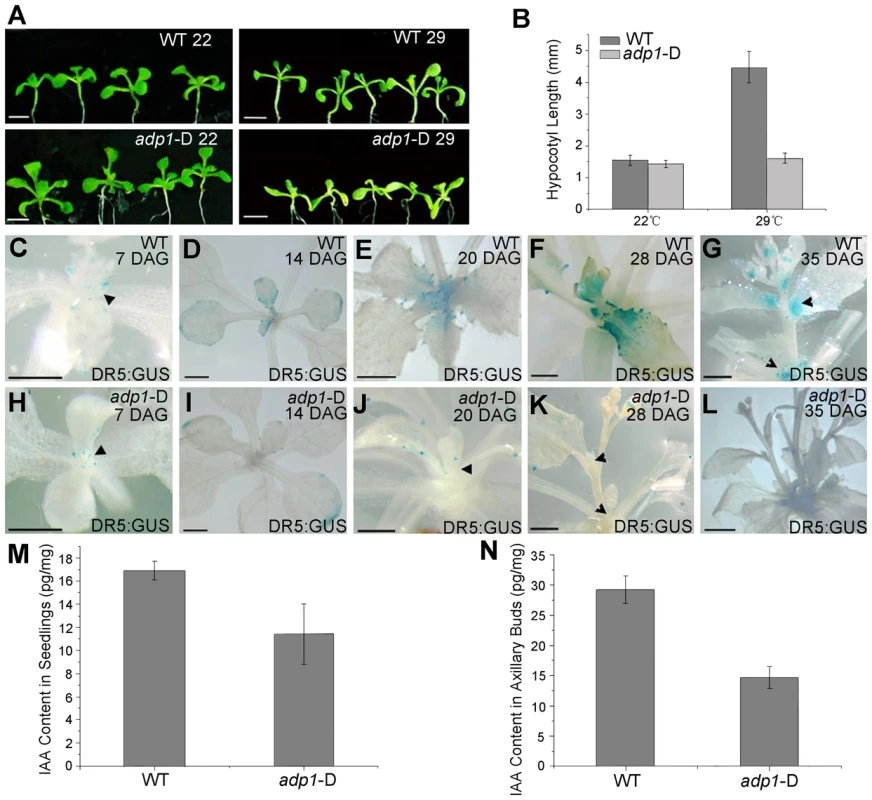
(A) Morphology of wild type (WT) and adp1-D seedlings after growth for 9 days at 22°C (left) and 29°C (right) under long-day conditions. The white vertical line indicates the average hypocotyl length of wild-type and adp1-D seedlings. Bar = 5 mm. (B) Hypocotyl length of wild-type and adp1-D seedlings grown at 22°C and 29°C. At least 30 seedlings of each genotype were measured. The error bars represent the SD. (C) to (L) Detection of DR5:GUS signal in wild-type (C–G) and adp1-D (H–L) plants at different developmental stages. DR5:GUS stained mainly in the shoot apical meristem, young leaves and axillary buds in the wild-type plants (indicated by black arrowheads), whereas the signal in corresponding tissues of adp1-D was reduced dramatically. Bar = 0.5 mm. (M) and (N) Free IAA content in young seedlings (M) and axillary buds (N). 200 mg of corresponding tissues were dissected for the measurements and each experiment had three biological replications. The decreased DR5:GUS signals indicated either decreased cellular auxin levels or altered auxin signaling. Root growth and the expression levels of two auxin-responsive genes (IAA1 and IAA5) were examined in adp1-D after auxin treatment; an adp1-D axr1-12 double mutant was also generated to test the genetic interactions of ADP1 with auxin signaling mechanisms [21], [22], [38]. The results of these experiments indicated that ADP1 does not interact directly with auxin signaling mechanisms (Figure S5).
The reduced auxin levels in adp1-D were further confirmed by direct quantitation of free IAA levels. Free IAA levels were measured in seedling shoot apices and axillary buds after bolting with quantification methods described previously [39]. The results showed that the free IAA content was indeed reduced in active dividing tissues in adp1-D (Figure 5M and 5N). Furthermore, sensitive mass spectrometry-based method of auxin metabolome profiling was conducted [40] to show that levels of several precursors [indole-3-acetonitrile (IAN), indole-3-acetamide (IAM), indole-3-pyruvic acid (IPyA) and indole-3-acetaldehyde (IAAld)] of auxin biosynthesis in adp1-D were decreased (Table S1). The same trend was also found in adp1-D, in terms of the free IAA level. These results are consistent with the hypothesis that enhanced outgrowth of axillary meristem might be caused by reduction of local auxin levels in adp1-D.
Although the decreased DR5:GUS signals might be caused by down-regulated auxin biosynthesis, the bushy phenotype of adp1-D is somewhat reminiscent of that of the pgp1 pgp19 (abcb1 abcb19) mutants with impaired polar auxin transport. Furthermore, in abcb19 mutant, that is defective in rootward polar auxin transport, levels of IAA at the shoot apex were decreased while levels of oxIAA and oxIAA-Glc were highly increased, as a result of long-term IAA pooling in this region. Accordingly, the polar auxin transport capacity in inflorescences and seedlings of adp1-D and wild type [41] were examined, revealing no differences for 3H-IAA (Figure S6). Due to the fact that this type of transport assay might not reveal differences in the transport capacity in the shoot apical meristem region, microscale transport assays [42] were used to determine if auxin transport capacity out of the meristem/cotyledonary node is affected in adp1-D. No difference between wild type and the mutant was detected by this method (data not shown), indicating that auxin transport is not impaired in adp1-D.
Analyses of auxin biosynthetic mutants suggested a connection of the observed phenotypes to the YUCCA family which belongs to flavin monooxygenase enzyme proteins functioning in auxin biosynthesis [10], [13]–[15], [17]. YUCCA6 has been shown to be associated with an endomembrane compartment [16]. The yuc1 yuc4 double mutants and yuc1,2,4,6 quadruple mutants, which are defective in auxin biosynthesis [17], [18], also exhibited reduced DR5:GUS signal in the SAM and in leaf petioles where axillary meristems were initiated (Figure S7A to S7H). These higher order yuc mutants also exhibited enhanced shoot branching (Figure S7I). Next, adp1-D was crossed with ProYUCCA1:GUS transgenic marker lines. The GUS signal was decreased in almost all the meristematic regions in adp1-D, compared to that in the wild type (Figure 6A to 6H). This result indicated that ADP1 over-expression affected auxin biosynthesis through the down-regulation of YUCCA expression, at least by YUCCA1. Furthermore, we examined the expression levels of all the YUCCA gene family members by qRT-PCR in the axillary buds after bolting, where ADP1 was highly expressed. Results showed that the expression levels of all the YUCCA genes were decreased by three to ten folds in the axillary buds of adp1-D (Figure 6I to 6S). These results confirmed that the auxin biosynthesis pathway might be down-regulated in adp1-D.
Fig. 6. Auxin Biosynthesis was reduced in adp1-D mutants. 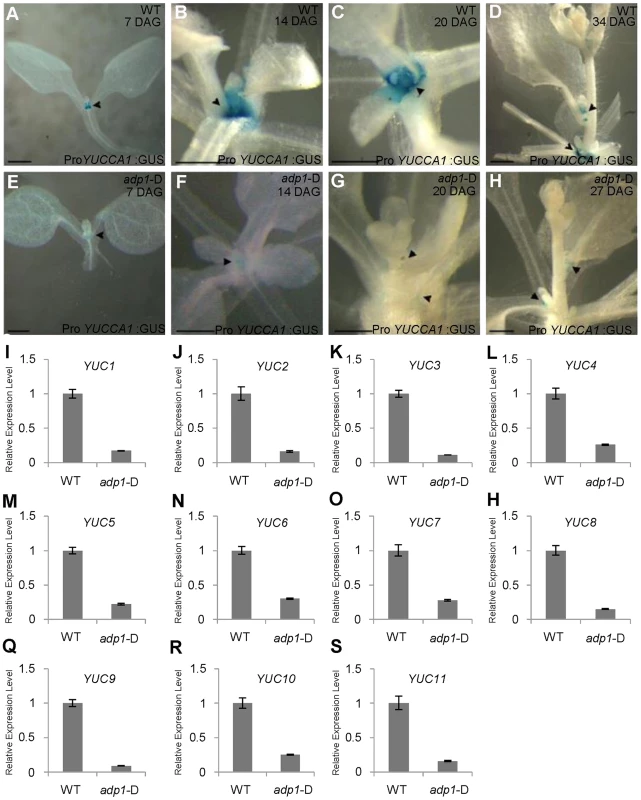
(A) to (H) Detection of ProYUCCA1:GUS signal in wild-type (A–D) and adp1-D (E–H) plants at different developmental stages. The ProYUCCA1:GUS staining pattern was similar to that of DR5:GUS, i.e., mostly in young meristematic regions, as indicated by black arrowheads. The signal in corresponding tissues of adp1-D (indicated by black arrowheads) was reduced markedly. Bar = 0.5 mm. (I) to (S) Relative expression levels of all the YUCCA genes in wild-type and adp1-D. Axillary buds of adult plant (3 days after bolting) were collected as samples, and Elongation factor-1α was used as internal control. The expression levels of each gene in the wild type were set as 1.0. Error bars represent three biological replicates. Transgenic Restoration of Auxin Biosynthesis in adp1-D Restores Wild-Type Growth
Crosses of adp1-D with a Pro35S:YUCCA1 transgenic line that overproduces auxin [15] largely restored wild type growth (i.e., both the rosette branch number and leaf initiation rate) in double homozygous F2 plants (Figure 7A to 7C). This result indicated that global increases of auxin levels through YUCCA1 overexpression could rescue the pleiotropic phenotypes of the adp1-D mutant. In an effort to restrict the over-production of auxin to the expression domains of ADP1, the bacterial indoleacetic acid-tryptophan monooxygenase (iaaM) gene was expressed under the control of the ADP1 promoter in wild type. iaaM catalyzes the conversion of Trp into indole-3-acetamide (the IAA biosynthetic precursor) [42], [43] and has been used to recapitulate Pro35S:YUCCA1 phenotypes in Arabidopsis [15]. Domain-specific iaaM overexpression resulted in more epinastic leaves and aerial rosettes as well as reduced first-order rosette branch number (Figure S8A to S8I). qRT-PCR analysis showed that the severity of the phenotypes was correlated with iaaM expression levels (Figure S8H).
Fig. 7. Correlation of auxin level with first-order rosette branch number. 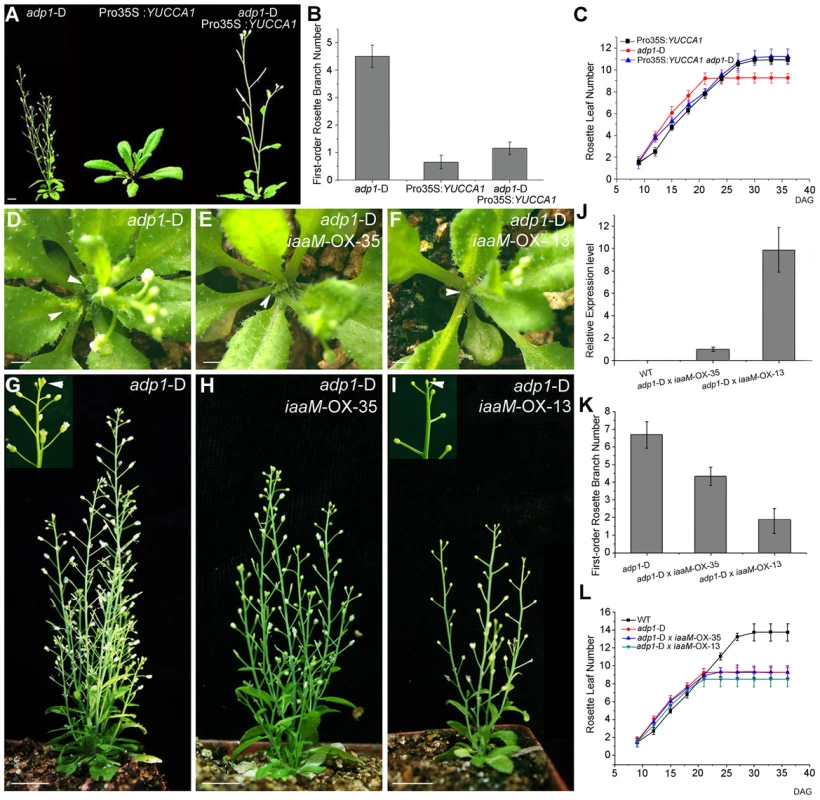
(A) Morphology of 35-day-old plants of adp1-D, Pro35S:YUCCA1 and double mutants of adp1-D Pro35S:YUCCA1. Bar = 1 cm. (B) Quantitative analysis of first-order rosette branch from 2-month-old plants of adp1-D, Pro35S:YUCCA1 and double mutants of adp1-D Pro35S:YUCCA1. Thirty plants of each genotype were measured. The error bars represent the SD. (C) Quantitative analysis of emergence rate of rosette leaves in adp1-D, Pro35S:YUCCA1 and double mutants of adp1-D Pro35S:YUCCA1. Thirty plants of each genotype were measured. The error bars represent the SD. (D) to (I) Morphology of crossed lines between adp1-D and ProADP1:iaaM transgenic lines at 25 days after germination (D to F) and two months after germination (G to I). The axillary buds developed much slower in the crossed lines compared with adp1-D, indicated by arrowheads from (D) to (F). At maturity, the crossed lines with stronger iaaM expression level even showed terminated flowers, as indicated by the inserted frame in (I). (J) Expression quantity of iaaM in the wild type and the crossed lines shown in (D) to (I) analyzed by real-time-quantitative PCR (qPCR). (K) First-order rosette branch number of the wild type and the crossed lines shown in (D) to (I). (L) Emergence rate of rosette leaves in the wild type and the crossed lines shown in (D) to (I). However, flower number and fertility were also reduced in ProADP1:iaaM lines, and, in some lines with severe phenotypes, the inflorescences were pin-formed or produced 3–4 undifferentiated flower meristems before termination (Figure S8B to S8D). These results suggest that overproduction of auxin in the ADP1 expression domain can disrupt the sequential formation of auxin gradients which are required for normal floral development and phyllotactic growth [44]–[46]. Furthermore, these results indicate that auxin synthesis in the ADP1 expression domains does not fully compensate for ADP1 transporter function in the endo-membrane system.
Based on phenotypes and relative gene expression levels, a stronger and a weaker ProADP1:iaaM transgenic lines were selected and crossed with adp1-D. Domain-specific expression of iaaM rescued the bushy phenotype in a manner corresponding to the expression level of iaaM in the parental lines (Figure 7D to 7K), but did not restore wild-type rosette leaf emergence rates (Figure 7L). These results suggest that increasing the auxin content in ADP1 expression domains is sufficient to restore apical dominance, but is insufficient to overcome developmental defects resulting from reduced auxin production in adp1-D that produce accelerated first-order rosette branches.
ADP1 Accumulation Rescued the Pin-Formed Phenotypes of pin1 and pinoid
The experiments described above indicated that ADP1 functions primarily in regulating auxin biosynthesis in the shoot apex and does not impact long-distance auxin transport capacity or auxin transport out of the SAM/cotyledonary node region. The pinformed1 (pin1) mutant exhibits pin-formed inflorescences due to loss of the local auxin gradients that are mediated by the PIN1 auxin transporter at the shoot apex [47], [48]. Developmental and cell biology studies indicate that polar orientation of PIN1 in the SAM region follows and “canalizes” auxin gradients, which was generated by localized auxin biosynthesis and was associated with initiating floral primordia [49], [50]. Modeling analysis with PIN1-fluorescent fusion proteins and fluorescent auxin reporter indicates that these local gradients are formed successively to maintain phyllotactic growth [51], [52]. Since the PINOID (PID) kinase regulates PIN1 trafficking, polar localization of PIN1 is perturbed in pid, making pid form partial pin-formed inflorescences similar to pin1 [53]–[55].
We hypothesize that the domain-specific decreases in auxin biosynthesis observed in adp1-D would weaken these auxin gradients, and thus would enhance the severity of the pin-formed phenotypes. Unexpectedly, the adp1-D pid double mutant displayed an overall appearance similar to adp1-D (Figure 8A, 8B and 8D), except that the mutant produced many flowers with fused petals (Figure 8F and 8H). Statistical analysis showed that the number of the flowers produced on the main inflorescence was at least three-fold more in the adp1-D pid double mutant than that in the pid mutant (Figure 8J), and the first-order rosette branch number was also increased by about two fold in the double mutant (Figure 8L). ADP1 overexpression also largely rescued the pin1 shoot phenotype, with greatly increased flower generation frequency in the inflorescence (Figure 8E, 8I and 8K). However, the first-order rosette branch number of the double mutants was not much increased (Figure 8M), probably due to the fact that pin1 itself already has increased first-order rosette branch number, resulting from 30% decrease in auxin transport out of the shoot apex [20]. Taken together, these data indicate that increased ADP1 activity at the shoot apex is sufficient to overcome a loss of PIN1-mediated canalization required for phyllotactic growth.
Fig. 8. Overexpression of ADP1 partially recovered pin1 and pid phenotypes. 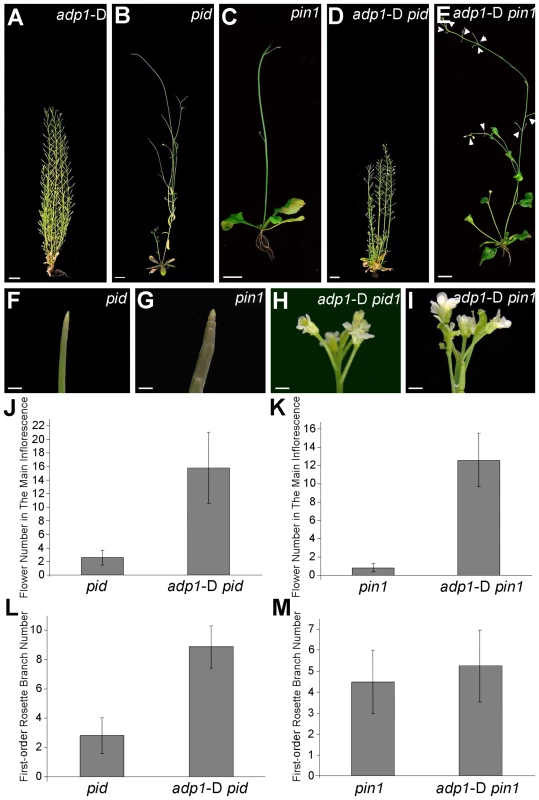
(A) to (E) Phenotypes of six-week-old adp1-D (A), pid (B), pin1 (C) and double homozygous mutants adp1-D pid (D) and adp1-D pin1 (E). Bar = 1 cm. (F) to (I) Close-up view of shoot apical tissue of pid (F), pin1 (G), double homozygous mutants adp1-D pid (H) and adp1-D pin1 (I). Hardly any flowers were formed in pid and pin1, but in the adp1-D pid and adp1-D pin1 double mutants, much more flowers were produced on the inflorescence stem. Bar = 1 mm. (J) Flower number on the inflorescence stem of pid and pid adp1-D. At least 20 plants of each genotype were measured. The error bars represent the SD. (K) Flower number on the inflorescence stem of pin1 and pin1 adp1-D. At least 20 plants of each genotype were measured. The error bars represent the SD. (L) First-order rosette branch number of pid and pid adp1-D. At least 20 plants of each genotype were measured. The error bars represent the SD. (M) First-order rosette branch number of pin1 and pin1 adp1-D. At least 20 plants of each genotype were measured. The error bars represent the SD. Higher-Order Loss-of-function Mutants Exhibit Retarded Growth and Slightly Reduced Number of Lateral Organs
Seven homologous proteins were clustered with ADP1 in the same clade in the phylogenetic tree (Figure S3B and S3C), suggesting the possibility of functional redundancy. This is supported by an over-expression experiment in which each of these seven genes was driven under the CaMV 35S promoter to over-express each gene of interest in Arabidopsis plants. All of the transgenic plants recapitulated the adp1-D phenotypes in their T1 generation to different extents (Figure S9A to S9G).
To further elucidate the function of ADP1 and its homologous genes, higher-order loss-of-function mutants were generated between the T-DNA insertion lines of ADP1 and its closest homologs, i.e., At5g19700, At2g38510 and At5g52050 (Figure S3B). The double mutants of each combination had no obvious phenotypes. However, some combinations of the triple mutants and the quadruple mutants exhibited developmental defects. In contrast to the gain-of-function mutant adp1-D, the quadruple mutants showed retarded growth from early developmental stages to maturation (Figure 9C to 9I and 9K to 9M). First-order rosette branch number was also slightly reduced in quadruple mutants compared to wild type (Figure 9J), and first-order rosette branches were generated much more slowly than the wild type (Figure 9N).
Fig. 9. Growth retardance in the quadruple mutants. 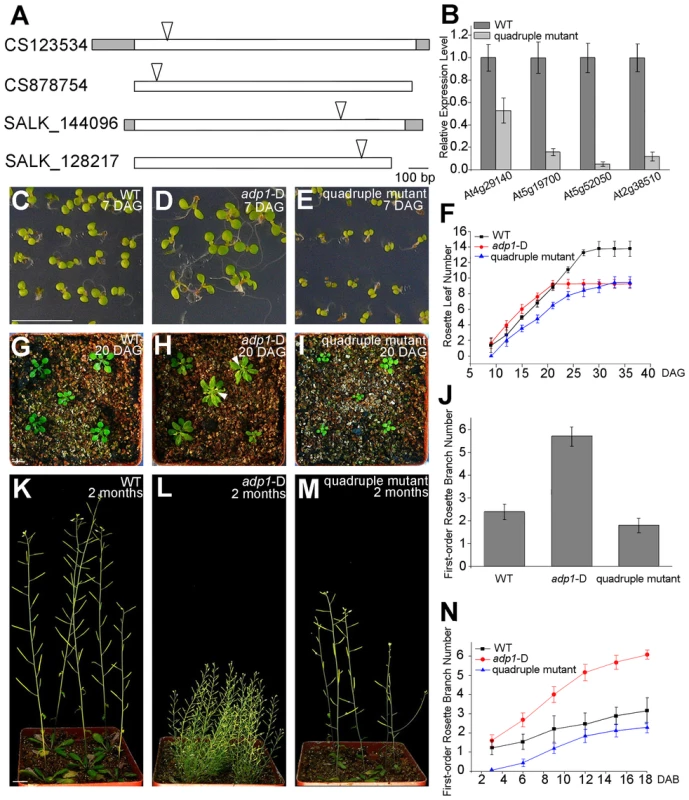
(A) T-DNA insertion sites in the CS123534, CS878754, SALK_144096 and SALK_128217 genes. Grey box, untranslated regions; white box, exon; arrowheads, T-DNA insertion sites. (B) Expression of At4g29140, At5g19700, At5g52050 and At2g38510 in wild-type seedlings and quadruple mutants as detected by real-time-qPCR. (C) to (E), (G) to (I) and (K) to (M) Morphology of wild-type seedlings, adp1-D and quadruple mutants at 7 days (C to E), 18 days (G to I) and 2 months (K to M) after germination, respectively. Bar = 1 cm. (F) Growth rate as measured by the record of rosette leaf emerging rate in wild-type plants, adp1-D and quadruple mutants. Forty plants of each genotype were measured. The error bars represent the SD. (J) First-order rosette branch number in mature plants of the wild type, adp1-D and quadruple mutants. Forty plants of each genotype were measured. The error bars represent the SD. (N) The generation rate of first-order rosette branch in the wild type, adp1-D and quadruple mutants. Forty plants of each genotype were measured. The error bars represent the SD. To clarify the origin of the aberrant growth pattern, we first examined the shoot apical regions of the wild type, adp1-D and the quadruple mutants by SEM. The results showed that the difference of the phenotypes between wild type, the quadruple mutants and adp1-D started as early as three days after germination (DAG). While adp1-D increased the size of the shoot apical region and produced more leaf primordia, the quadruple mutants showed much reduction of shoot apical size and retarded leaf initiation at 3 DAG (Figure 10A to 10C). This difference persisted from 3 DAG to 5 DAG (Figure 10D to 10F), suggesting that the developmental defects in the quadruple mutants and adp1-D reflect ADP1 function rather than a concomitant developmental effect. Free IAA content in young seedlings and axillary buds of the quadruple mutant was measured by quantification methods described previously [39], showing that the free IAA levels of the quadruple mutants was not significantly different from that of wild type (Figure 10G and 10H). Same results (Table S1) were obtained by using the sensitive mass spectrometry-based method of auxin metabolome profiling [40], which might be due to additional redundancy of untested MATE transporters. However, we were able to detect slightly increased levels of several precursors (IAN, IAM and IPyA) of auxin biosynthesis (Table S1) by the mass spectrometry-based method. These results were consist with the phenotypes of the epinastic cotyledon and slightly increased hypocotyl length observed in the quadruple mutants (Figure S10), since the same phenotypes were also observed in Pro35S:YUCCA1 plants which was believed to have increased auxin levels [15].
Fig. 10. Morphology of Shoot apical meristems and detection of auxin level in adp1-D and quadruple mutants. 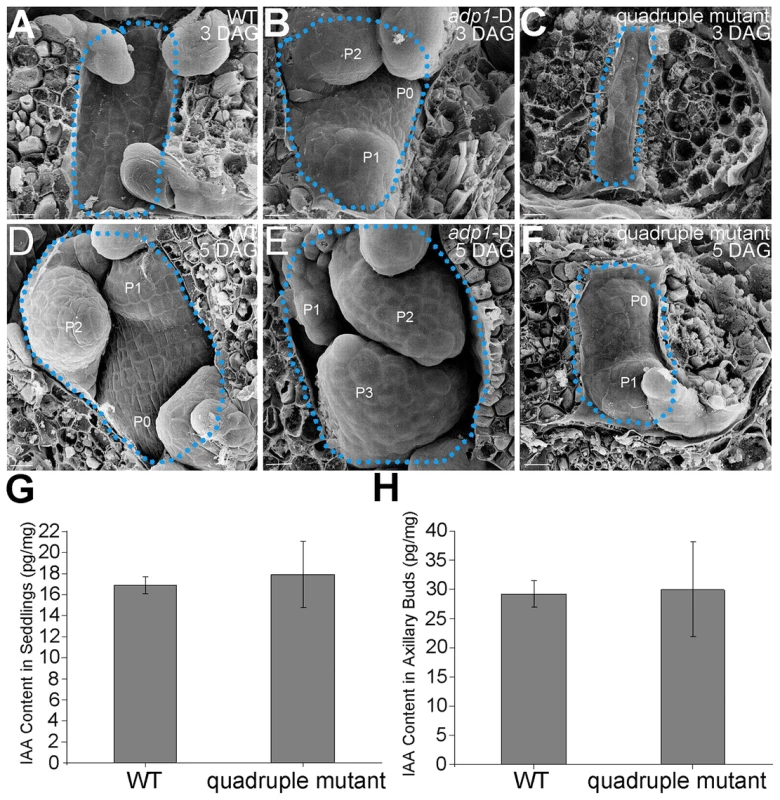
(A) to (C) Scanning electronic micrographs of 3-day-old seedlings of (A) the wild type, (B) adp1-D, and (C) quadruple mutant. The area of apical tissues was enclosed by dashed lines. P0 to P3 indicate the leaf meristem by emergence order. Bar = 10 µm. (D) to (F) Scanning electronic micrographs of 5-day-old seedlings of (D) the wild type, (E) adp1-D and (F) quadruple mutant. Bar = 10 µm. (G) Measurement of IAA content in shoot apical tissues of seedlings from wild type and quadruple mutants. 200 mg of each genotype was dissected for the measurements and bars represent three biological replications. (H) Measurement of IAA content in axillary buds of wild type and quadruple mutants. 200 mg of each genotype was dissected for the measurements and bars represent three biological replications. Discussion
Branching pattern is one of the main factors contributing to plant architecture. In Arabidopsis, the number of the first-order and higher-order branches determines the light harvesting efficiency. While the first-order rosette branch number is controlled by strigolactones, the mechanism that regulates higher-order branches is largely unknown. Because many bushy mutants also exhibit reduced fertility, it is proposed that the increased number of higher order branches may be the consequence of sterility. The evidence presented here demonstrates that, in the bushy mutant adp1-D, increased first-order rosette branch number is a directly result of ADP1 overproduction.
Reduced auxin levels, transport, and signaling have long been associated with overproduction of rosette branches. Higher order yucca (auxin biosynthesis), abcb/pgp (auxin transport), and tir1/afb (auxin perception) mutants develop more rosette branches [17], [23], [56]. In the present study, adp1-D mutants produced an increased number of first-order rosette branches (Figure 1D and 1F) and ProADP1:iaaM plants showed a reduced number of first-order rosette branches (Figure S8I). Furthermore, lower levels of free IAA in adp1-D and the rescue of the adp1-D rosette branch phenotype by crosses adp1-D with ProADP1:iaaM transformants confirmed that reduction of local auxin levels in the active dividing meristems caused the bushy phenotype of adp1-D.
The growth retardation observed in the quadruple mutants was opposite to adp1-D phenotypes, suggesting that the genes in the same clade are functionally redundant in the regulation of lateral organ outgrowth in Arabidopsis. Although axillary shoot branching was not obviously changed in the quadruple mutants, the generation rate of the first-order rosette branches was much more slower in the mutants, compared to wild type, which is also opposite to that in adp1-D. In terms of free auxin levels, no significant difference was found in quadruple mutants, compared with wild-type (Figure 10G, 10H, and Table S1). This is probably due to gene redundancy, since the other four homologous genes of ADP1 are still functioning in the quadruple mutants, which may be able to maintain a proper auxin homeostasis in the whole plants. However, levels of the several precursors (IAM, IAN, and IPyA) of auxin biosynthesis were indeed increased in the quadruple mutants (Table S1). Taken together with the excess-auxin phenotypes observed in quadruple mutants (slightly increased hypocotyls and epinastic cotyledons), the auxin biosynthesis in quadruple mutants might be slightly increased. The fact that both gain-of-function and loss-of-function mutants change the plant growth pattern indicates that temporally and spatially appropriate expression of these MATE genes are essential for maintaining plant architecture.
The partial rescue of pid and pin1 pin-formed phenotypes by crosses with adp1-D is more difficult to rationalize. It is quite possible that over-expression of ADP1 actually increases auxin levels within the ADP1 expression domain. This increase is probably sufficient to overcome a loss of PIN canalization in some cell types, but leads to increased auxin catabolism in the majority of cells in the ADP1 expression domain. Similar situation was found in the abcb19/pgp19 auxin transport mutant in which decreased auxin levels and increased oxIAA-Hex levels were observed and attributed to an effect of auxin pooling near the SAM [41]. Alternatively, the decrease of IAA levels in adp1-D could alleviate repression of primordia induction by auxin, or, more likely, generate micro-gradients that stimulate development of new floral primordia.
ADP1 is a member of the large MATE family of transporters that have been implicated in mobilization of ions, toxins and secondary metabolites. As with many other transporter families, MATEs have expanded in plants and function in many aspects: the sequestration of a diverse range of secondary metabolites in vacuoles or their excretion out of the cells, and defense against herbivores and microbial pathogens [57]–[61]. One of the best characterized MATE transporters is TRANSPARENT TESTA12 (TT12), which has been shown to transport proanthocyanidin across the tonoplast [57], [61]. However, a number of MATE proteins appear to regulate the transport of organic acids. FRD3/AtDTX43 controls responses of iron deficiency in plant and is thought to mediate citrate secretion into the xylem and the rhizosphere [60]. The EDS5/AtDTX47 MATE protein functions in salicylic acid signaling for disease resistance [59]. ALF5/AtDTX19 was reportedly involved in the regulation of lateral root formation [58], suggesting a potential role in auxin signaling.
The results presented here indicate that ADP1 is localized to a post-Golgi endomembrane compartment and acts upstream of, or co-ordinately with, YUCCAs in auxin biosynthesis. YUCCA6 was localized to a similar endo-membrane compartment [16], suggesting that ADP1 may function in mobilization of IAA precursors to YUCCAs (for conversion to IAA), or, less likely, movement of IAA out of the endosomal compartments. The function of ADP1 may be homeostatic and involve reversible activity, because a prokaryotic MATE, NorM, which has been crystallized from Vibrio cholera, may exhibit conformational change on substrate binding [32]. Alternatively, ADP1 may simply prevent adsorption of hydrophobic indolic compounds into endosomal membranes or export IAA out of the endo-membrane vesicles. ADP1 may also be involved in auxin cellular homeostasis, which is maintained by PIN5 [62] and PILS auxin transporter [63]. Since so far no auxin exporter in ER has been reported except pollen specific PIN8 [64], it would be interesting to investigate in the future whether ADP1 could balance auxin homeostasis in ER. The lack of successful ADP1 protein expression in multiple heterologous expression systems (e.g., different vectors in different E. coli strains, S. cerevisiae, Pichea pastoris, and SF9 insect cells) has prevented more detailed biochemical characterization.
There are multiple ADP1 homologues in rice, maize and sorghum, sharing 50% to 65% identity at the amino acid level. It is reasonable to speculate that over-expression of these ADP1 genes could also change plant architecture in these crops. Crop plant architecture determines planting density in the field which, to a large extent, affects the light harvest, disease resistance, use of nutrients, and lodging [2]. Understanding the molecular mechanisms in the regulation of plant architecture will therefore provide a basis for modification of the plant architecture of crops, ultimately facilitating crop production.
Methods
Plant Materials and Growth Conditions
Seeds of Arabidopsis thaliana, ecotype Columbia, were surface sterilized with 15%? NaClO, stratified for 3 days at 4°C before incubation on Murashige and Skoog (MS) medium containing 1% sucrose at 22±2°C under long-day conditions (16 h light/8 h dark) for 1 week. Seeds of adp1-D mutants were sown on MS medium containing DL-phosphinothricin and drug-resistant seedlings were transferred to soil and grown under the same conditions.
For the temperature transferal experiment, plants were germinated and grown on MS medium for the wild type, and on MS medium containing DL-phosphinothricin for the adp1-D mutant, for 3 days under long-day conditions and then transferred to the same MS medium in a test chamber at 20°C and 29°C for another 7 days before measurement.
For assay of root elongation as an auxin sensitivity test, seedlings were grown on vertically placed MS medium for the wild type, and on MS medium containing DL-phosphinothricin for the adp1-D mutant, under long-day conditions for 4 days before transferal to medium containing 0, 20 nM, 40 nM, 60 nM, 80 nM and 100 nM of synthetic auxin 2,4-D. Root length was measured after incubation for 6 days.
For GR24 treatment, adp1-D were germinated and grown on MS medium containing DL-phosphinothricin for 7 days under long-day conditions and then transferred to MS medium containing 5 µM GR24 or 5 µM acetone for 40 days before phenotype analysis.
Primers and PCR Conditions
Genomic DNA was extracted from homozygous and heterozygous adp1-D mutants and the flanking sequence of the T-DNA insertion was determined by thermal asymmetric interlaced PCR [65]. The specific degenerate primers in the T-DNA border and the random primers for three sequential PCRs were used as described previously [26]. Three primers (P1, P2 and P3-1) were designed for co-segregation analysis. P1 and P2 corresponded to the genomic sequence flanking the T-DNA insertion and P3-1 corresponded to the T-DNA vector sequence (Figure 2A). The primer sequences were as follows: P1 (5′-ATC CCA CTA AAG CAC TGT CA-3′); P2 (5′-TTT AAG CTA CTT ACC GTT GA-3′); and P3-1 (5′-TTG GTA ATT ACT CTTTCT TTT CCT CC-3′). For cloning of ADP1, the primer pair ADP1-F (5′-ATG TGT AACCCA TCA ACA ACA-3′) and ADP1-R (5′-TTA ATA AAG CAC CGT GAT GC-3′) were designed according to the cDNA sequence from the National Center for Biotechnology Information database (accession number NM_119058). Two primers, ADP1-RT-F (5′-CGA ACC GGA CTC TTC CTC GA-3′) and ADP1-RT-R (5′-GGT GAG CAC CGAAGG CTT GA-3′), were designed based on the coding region sequences to detect transgene transcripts in overexpression lines. The primers designed for amplification of the auxin-responsive genes IAA1 and IAA5 were IAA1-F (5′-GCG TCA GAA GCA ACAAGC G-3′); IAA1-R (5′-TCC TTT GTA GCC TTC TCT CTC GGA-3′); IAA5-F (5′-AGA TCT TGC TTC CGC TCT GCA A-3′) and IAA5-F (5′-CCC AAG GAA CATCTC CAG CAA GC-3′), respectively. The primers for detecting the transcription level of endogenous auxin transporters were as follows: PIN1-F (5′-TAC GGC GGC GGACTT CTA CC-3′); PIN1-R (5′-CGG CGA GGA AAC GGA GGT TC-3′); PIN2-F (5′-AATGGC CGT GAA CCC CTC CA-3′); PIN2-R (5′-TTG ACG TTC TCG GCG TCA CG-3′); PIN3-F (5′-CGG TAG CCT CGA GTG GAG CA-3′); PIN3-R (5′-CCG CCG GAC CGAAAT TGG AG-3′); PIN7-F (5′-TCT ACA CCG TCC TCA CGG CG-3′); PIN7-R(5′-AAG TTC GAA AGC CGG CCA CC-3′). The TUB2 (β-tubulin) gene was used as an internal control in real-time PCR (qRT-PCR); the primers for TUB2 used were those described previously (Qin et al., 2005). The qRT-PCR procedure comprised 40 cycles as described previously (Qin et al., 2003). PCR reactions were performed for 26–35 cycles (94°C for 30 s, 58°C for 30 s, and 72°C for 30 s to 1.5 min).
Overexpression Constructs and Arabidopsis Transformation
The ADP1 cDNA was amplified from wild-type Arabidopsis by RT-PCR and cloned into the EcoR V site of the pBluescript SK+ vector (designated pBADP1). The presence of ADP1 in the recombinant plasmids was confirmed by sequencing in both sense and antisense orientations. The ADP1 promoter was amplified from genomic DNA using the primers 5′-GCT CAC AGG AGC CTT ACT TAT-3′ and 5′-GAC GGT GAT GAT GATGAT GGT-3′ and cloned into the EcoR V site of pBluescript SK+ (designated pBADP1P).
The CaMV 35S enhancer tetrad was amplified from pSKI015 as described previously [66] and was designated pA4Ehancer. The iaaM gene was amplified from the plasmid pBJ36-iaaM with the primers 5′-ATG TCA GCT TCA CCT CTC CT-3′ and 5′-TAA TTT CTA GTG CGG TAG TTA - 3′ and then cloned into the EcoRV site of pBluescript SK+ (designated pBiaaM). For the construction of the plant expression vector, the pQG110 vector [66], pJIM19 vector and pBI101.3 vector were used. 4Enhancer-ADP1 was constructed by ligation of four DNA fragments: the HindIII-XbaI fragment from pQG110, the HindIII-EcoRI enhancer tetrad fragment from pA4Ehancer, the EcoRI-KpnI ADP1 promoter fragment from pBADP1P, and the KpnI-XbaI ADP1 fragment from pBADP1. The Pro35S:ADP1 construct was obtained by ligation of two DNA fragments: the KpnI-SacI fragment from pJIM19, and the KpnI-SacI ADP1 fragment from pBADP1P. Pro:ADP1-iaaM was constructed by ligation of three DNA fragments: the XbaI-SacI fragment from pBI101, the XbaI-KpnI ADP1 promoter fragment from pBADP1P, and the KpnI-SacI iaaM fragment from pBiaaM. Wild-type plants were used for Agrobacterium-mediated transformation by the floral dip method. The seeds of transgenic wild-type plants were screened on MS medium containing 50 mg/mL kanamycin. The resistant seedlings were transferred to soil.
In situ Hybridization
In situ hybridization was performed as described previously [67]. Antisense and sense probes were synthesized with digoxigenin-11-UTP (Roche Diagnostics) using T7 and T3 RNA polymerases, respectively. The primers used to amplify the DNA template for the probe synthesis were as follows: ADP1-INSITU-F (5′-ATG TGT AAC CCA TCA ACA ACA-3′) and ADP1-INSITU-R (5′-CGG TTA TGTTAG CAA AGG CAA T-3′).
Histochemical Assays
GUS staining was performed in the following steps. Samples were first fixed in 90% acetone on ice for 20 min, then washed thoroughly three times with staining buffer (0.1 M Na3PO4, pH 7.0, 10 mM EDTA, 0.5 mM ferricyanide, 0.5 mM K ferrocyanide, and 0.1% Triton X-100) on ice, vacuum-infiltrated briefly, and incubated in staining buffer containing 50–100 mg X-gluc per 100 mL for 3–12 h.
Tissue Sectioning and Microscopy
Tissue sectioning was performed as described previously [68]. Inflorescence stems were collected from the most basal 5 cm of stems of wild-type and adp1-D plants and fixed in FAA solution (50% ethanol, 5% acetic acid, and 3.7% formaldehyde). After dehydration with an ethanol gradient series, the samples were embedded in Historesin (Leica). Sectioning was performed using a Leica microtome and 7 µm sections were mounted on slides. The sections were stained with 0.25% (w/v) toluidine blue O (Sigma-Aldrich) and observed under an Olympus BX51 microscope as previously described [69]. Digital images were captured with a SPOT camera (Diagnostic Instruments) and processed using Adobe Photoshop.
Marker Gene Analysis
DR5:GUS, ProYUC1:GUS, ProPIN1:PIN1:GFP, ProPIN2:PIN2:GFP, ProPIN3:PIN3:GFP, ProPIN7:PIN7:GFP, ProPIN1:GUS and ProPIN1:PIN1:GUS marker lines were crossed to adp1-D and the homozygous lines in the T3 generation were used for analysis. In all analyses, the parental lines were used for comparison with those in the mutant background. Endo-membrane organelle localization and endocytotic dynamic analysis were performed as described previously [70].
Generation of adp1-D Double Mutants
The double mutants adp1-D axr1-12, adp1-D pin1 and adp1-D pid were generated by crossing heterozygous adp1-D with axr1-12, pin1 or pid. The double mutants were identified from the F2 progeny grown in soil by comparison with the parental phenotypes and through PCR-based molecular analyses.
Auxin Transport Assay
Auxin transport in the inflorescence stem was assayed as described previously [71]. Inflorescence stems of 6-week-old plants were cut into 2.5 cm segments, submerged with one end in a 1.5 ml microcentrifuge tube containing 30 µl MES buffer (5 mM MES, 1% [w/v] sucrose, pH5.5) with 100 nM 3H-IAA in 1.45 µM total IAA at room temperature in the dark for 24 h. Basipetal or acropetal auxin transport was measured in accordance with the orientation of the inflorescence segments. After incubation, the segments were removed and the terminal 5 mm of the non-submerged ends were excised and placed into a scintillation vial containing 2.5 ml scintillation fluid for 18 h before counting with a liquid scintillation counter. Microscale auxin transport assays in seedlings were conducted as described previously [70].
Measurement of IAA Content
For measurement of IAA content in seedlings, 7 days seedlings of adp1-D, wild type and quadruple mutants in long day conditions were sectioned with a sharp blade, and collected 200 mg tissues (including upper hypocotyls, shoot apical meristems and young leaves without cotyledons) for each genotype. For measurement of IAA content in axillary buds, 200 mg tissues of each genotype were collected three days after bolting, including 1 mm basal leave petiole and the attached newly produced axillary meristems. IAA content measurement was conducted as previously reported [39]. Mass spectrometry-based method of profiling the auxin metabolome [40] was conducted also, which requires much less amount of sample (approximately 20 mg). The samples were collected as the same way as for the above-mentioned method, except that only about 20 mg of each genotype was collected, and three biological replicates were conducted.
Generation of Quadruple Mutants
The quadruple mutants were generated first by crossing CS123534 and CS878754, and crossing SALK_144096 and SALK_128217 to get the F1 heterozygous generation. Next, the two F2 homozygous mutants from the F1 selfed generation were crossed to obtain the F3 heterozygous mutants. Finally, the F3 selfed generation was screened by PCR to obtain the homozygous quadruple mutants in the F4 generation.
Accession Data
Sequence data for ADP1, YUCCA1, PIN1, PIN2, PIN3, PIN7, AXR1 and PINOID can be found in the GenBank/EMBL data libraries under accession numbers NM_119058, NM_119406.2, NM_106017.3, NM_125091.3, NM_105762.2, NM_102156.1, NM_001035893 and NM_129019.
Supporting Information
Zdroje
1. ReinhardtD, KuhlemeierC (2002) Plant architecture. EMBO Rep 3 : 846–851.
2. WangY, LiJ (2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59 : 253–279.
3. PengJ, RichardsDE, HartleyNM, MurphyGP, DevosKM, et al. (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400 : 256–261.
4. LinH, WangR, QianQ, YanM, MengX, et al. (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21 : 1512–1525.
5. JiaoY, WangY, XueD, WangJ, YanM, et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42 : 541–544.
6. WangY, LiJ (2011) Branching in rice. Curr Opin Plant Biol 14 : 94–99.
7. McSteenP, LeyserO (2005) Shoot branching. Annu Rev Plant Biol 56 : 353–374.
8. VannesteS, FrimlJ (2009) Auxin: a trigger for change in plant development. Cell 136 : 1005–1016.
9. NormanlyJ (2010) Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol 2: a001594.
10. ZhaoY (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61 : 49–64.
11. MashiguchiK, TanakaK, SakaiT, SugawaraS, KawaideH, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci U S A 108 : 18512–18517.
12. StepanovaAN, AlonsoJM (2011) Bypassing transcription: a shortcut in cytokinin-auxin interactions. Dev Cell 21 : 608–610.
13. ZhaoY (2012) Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 5 : 334–338.
14. WonC, ShenX, MashiguchiK, ZhengZ, DaiX, et al. (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci U S A 108 : 18518–18523.
15. ZhaoY, ChristensenSK, FankhauserC, CashmanJR, CohenJD, et al. (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 : 306–309.
16. KimHB, LeeH, OhCJ, LeeNH, AnCS (2007) Expression of EuNOD-ARP1 encoding auxin-repressed protein homolog is upregulated by auxin and localized to the fixation zone in root nodules of Elaeagnus umbellata. Mol Cells 23 : 115–121.
17. ChengY, DaiX, ZhaoY (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 : 1790–1799.
18. ChengY, QinG, DaiX, ZhaoY (2007) NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc Natl Acad Sci U S A 104 : 18825–18829.
19. GeislerM, BlakesleeJJ, BouchardR, LeeOR, VincenzettiV, et al. (2005) Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J 44 : 179–194.
20. BlakesleeJJ, BandyopadhyayA, LeeOR, MravecJ, TitapiwatanakunB, et al. (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19 : 131–147.
21. StirnbergP, ChatfieldSP, LeyserHM (1999) AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol 121 : 839–847.
22. BookerJ, ChatfieldS, LeyserO (2003) Auxin acts in xylem-associated or medullary cells to mediate apical dominance. Plant Cell 15 : 495–507.
23. DharmasiriN, DharmasiriS, WeijersD, LechnerE, YamadaM, et al. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9 : 109–119.
24. OmoteH, HiasaM, MatsumotoT, OtsukaM, MoriyamaY (2006) The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci 27 : 587–593.
25. SunX, GilroyEM, ChiniA, NurmbergPL, HeinI, et al. (2011) ADS1 encodes a MATE-transporter that negatively regulates plant disease resistance. New Phytol 192 : 471–482.
26. QinG, KangDM, DongYY, ShenYP, ZhangL, et al. (2003) Obtaining and analysis of flanking sequences from T-DNA transformants of Arabidopsis. Plant Sci 165 : 941–949.
27. KurataN, MiyoshiK, NonomuraK, YamazakiY, ItoY (2005) Rice mutants and genes related to organ development, morphogenesis and physiological traits. Plant Cell Physiol 46 : 48–62.
28. GautamPK, RathoreRKS, SutarAR, MangleBB (2011) Characterization of different male sterile lines on morphological characters of India mustard [Brassica juncea (L.) Czern & Coss ] along with their maintainer. Intl Multidiscipl Res J 1 : 4–8.
29. Gomez-RoldanV, FermasS, BrewerPB, Puech-PagesV, DunEA, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455 : 189–194.
30. UmeharaM, HanadaA, YoshidaS, AkiyamaK, AriteT, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455 : 195–200.
31. YazakiK (2005) Transporters of secondary metabolites. Curr Opin Plant Biol 8 : 301–307.
32. HeX, SzewczykP, KaryakinA, EvinM, HongWX, et al. (2010) Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467 : 991–994.
33. VoigtB, TimmersAC, SamajJ, MullerJ, BaluskaF, et al. (2005) GFP-FABD2 fusion construct allows in vivo visualization of the dynamic actin cytoskeleton in all cells of Arabidopsis seedlings. Eur J Cell Biol 84 : 595–608.
34. PenalvaMA (2005) Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet Biol 42 : 963–975.
35. DonaldsonJG, FinazziD, KlausnerRD (1992) Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature 360 : 350–352.
36. JensenPJ, HangarterRP, EstelleM (1998) Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol 116 : 455–462.
37. UlmasovT, MurfettJ, HagenG, GuilfoyleTJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 : 1963–1971.
38. LeyserHM, LincolnCA, TimpteC, LammerD, TurnerJ, et al. (1993) Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364 : 161–164.
39. AndersenSU, BuechelS, ZhaoZ, LjungK, NovakO, et al. (2008) Requirement of B2-type Cyclin-Dependent Kinases for mersitem integrity in Arabidopsis thaliana. Plant Cell 20 : 88–100.
40. NovakO, HenykovaE, SairanenI, KowalczykM, PospisilT, et al. (2012) Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J 72 : 523–536.
41. OkadaK, UedaJ, KomakiMK, BellCJ, ShimuraY (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3 : 677–684.
42. RomanoCP, RobsonPR, SmithH, EstelleM, KleeH (1995) Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol Biol 27 : 1071–1083.
43. KleeHJ, HorschRB, HincheeMA, HeinMB, HoffmannNL (1987) The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Genes Dev 1 : 86–96.
44. HeislerMG, OhnoC, DasP, SieberP, ReddyGV, et al. (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15 : 1899–1911.
45. KuhlemeierC (2007) Phyllotaxis. Trends Plant Sci 12 : 143–150.
46. BraybrookSA, KuhlemeierC (2010) How a plant builds leaves. Plant Cell 22 : 1006–1018.
47. FrimlJ, YangX, MichniewiczM, WeijersD, QuintA, et al. (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 : 862–865.
48. PetrasekJ, FrimlJ (2009) Auxin transport routes in plant development. Development 136 : 2675–2688.
49. SauerM, BallaJ, LuschnigC, WisniewskaJ, ReinohlV, et al. (2006) Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev 20 : 2902–2911.
50. BenkovaE, IvanchenkoMG, FrimlJ, ShishkovaS, DubrovskyJG (2009) A morphogenetic trigger: is there an emerging concept in plant developmental biology? Trends Plant Sci 14 : 189–193.
51. BayerEM, SmithRS, MandelT, NakayamaN, SauerM, et al. (2009) Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev 23 : 373–384.
52. HeislerMG, HamantO, KrupinskiP, UyttewaalM, OhnoC, et al. (2010) Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol 8: e1000516.
53. MichniewiczM, ZagoMK, AbasL, WeijersD, SchweighoferA, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130 : 1044–1056.
54. MichniewiczM, BrewerPB, FrimlJI (2007) Polar auxin transport and asymmetric auxin distribution. Arabidopsis Book 5: e0108.
55. GaoX, NagawaS, WangG, YangZ (2008) Cell polarity signaling: focus on polar auxin transport. Mol Plant 1 : 899–909.
56. NohB, MurphyAS, SpaldingEP (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13 : 2441–2454.
57. DebeaujonI, PeetersAJ, Leon-KloosterzielKM, KoornneefM (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13 : 853–871.
58. DienerAC, GaxiolaRA, FinkGR (2001) Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins. Plant Cell 13 : 1625–1638.
59. NawrathC, HeckS, ParinthawongN, MetrauxJP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 : 275–286.
60. RogersEE, GuerinotML (2002) FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell 14 : 1787–1799.
61. MarinovaK, PourcelL, WederB, SchwarzM, BarronD, et al. (2007) The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19 : 2023–2038.
62. MravecJ, SkupaP, BaillyA, HoyerovaK, KreckP, et al. (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459 : 1136–1140.
63. BarbezE, KubesM, RolcikJ, BeziatC, et al. (2012) A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485 : 119–122.
64. DingZ, WangB, MorenoI, DuplakovaN, SimonS, et al. (2012) ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat Commun 3 : 941.
65. LiuYG, HangN (1998) Efficient amplification of insert end sequences from bacterial artificial chromosome clones by thermal asymmetric interlaced PCR. Plant Mol Biol Rep 16 : 175–181.
66. QinG, GuH, ZhaoY, MaZ, ShiG, et al. (2005) An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17 : 2693–2704.
67. LiuJ, ZhangY, QinG, TsugeT, SakaguchiN, et al. (2008) Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20 : 1538–1554.
68. GuoY, QinG, GuH, QuLJ (2009) Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell 21 : 3518–3534.
69. QinG, MaZ, ZhangL, XingS, HouX, et al. (2007) Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res 17 : 249–263.
70. ChristieJM, YangH, RichterGL, SullivanS, ThomsonCE, et al. (2011) phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol 9: e1001076.
71. DaiY, WangH, LiB, HuangJ, LiuX, et al. (2006) Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis. Plant Cell 18 : 308–320.
Štítky
Genetika Reprodukční medicína
Článek Unwrapping BacteriaČlánek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 1
-
Všechny články tohoto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



