-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEvolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
Seminal fluid proteins transferred from males to females during copulation are required for full fertility and can exert dramatic effects on female physiology and behavior. In Drosophila melanogaster, the seminal protein sex peptide (SP) affects mated females by increasing egg production and decreasing receptivity to courtship. These behavioral changes persist for several days because SP binds to sperm that are stored in the female. SP is then gradually released, allowing it to interact with its female-expressed receptor. The binding of SP to sperm requires five additional seminal proteins, which act together in a network. Hundreds of uncharacterized male and female proteins have been identified in this species, but individually screening each protein for network function would present a logistical challenge. To prioritize the screening of these proteins for involvement in the SP network, we used a comparative genomic method to identify candidate proteins whose evolutionary rates across the Drosophila phylogeny co-vary with those of the SP network proteins. Subsequent functional testing of 18 co-varying candidates by RNA interference identified three male seminal proteins and three female reproductive tract proteins that are each required for the long-term persistence of SP responses in females. Molecular genetic analysis showed the three new male proteins are required for the transfer of other network proteins to females and for SP to become bound to sperm that are stored in mated females. The three female proteins, in contrast, act downstream of SP binding and sperm storage. These findings expand the number of seminal proteins required for SP's actions in the female and show that multiple female proteins are necessary for the SP response. Furthermore, our functional analyses demonstrate that evolutionary rate covariation is a valuable predictive tool for identifying candidate members of interacting protein networks.
Published in the journal: . PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1004108
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004108Summary
Seminal fluid proteins transferred from males to females during copulation are required for full fertility and can exert dramatic effects on female physiology and behavior. In Drosophila melanogaster, the seminal protein sex peptide (SP) affects mated females by increasing egg production and decreasing receptivity to courtship. These behavioral changes persist for several days because SP binds to sperm that are stored in the female. SP is then gradually released, allowing it to interact with its female-expressed receptor. The binding of SP to sperm requires five additional seminal proteins, which act together in a network. Hundreds of uncharacterized male and female proteins have been identified in this species, but individually screening each protein for network function would present a logistical challenge. To prioritize the screening of these proteins for involvement in the SP network, we used a comparative genomic method to identify candidate proteins whose evolutionary rates across the Drosophila phylogeny co-vary with those of the SP network proteins. Subsequent functional testing of 18 co-varying candidates by RNA interference identified three male seminal proteins and three female reproductive tract proteins that are each required for the long-term persistence of SP responses in females. Molecular genetic analysis showed the three new male proteins are required for the transfer of other network proteins to females and for SP to become bound to sperm that are stored in mated females. The three female proteins, in contrast, act downstream of SP binding and sperm storage. These findings expand the number of seminal proteins required for SP's actions in the female and show that multiple female proteins are necessary for the SP response. Furthermore, our functional analyses demonstrate that evolutionary rate covariation is a valuable predictive tool for identifying candidate members of interacting protein networks.
Introduction
Sexual reproduction is a fundamental biological process by which many eukaryotic organisms transmit their genetic material to the next generation. While the end result of a successful mating is the fusion of the gametes, other molecular interactions must occur to allow this fusion. In internally fertilizing animals, males transfer to females not only sperm, but also a suite of seminal fluid proteins (Sfps) that are essential for reproductive success. Across diverse taxa, Sfps are required for: the mobilization of sperm and their storage within the female; increasing the reproductive capacity of the female; affecting the outcome of sperm competition between multiple males; and, facilitating the union of the gametes [reviewed in 1]. In insects, Sfps also alter female behaviors and physiology [2]. Effects of Sfps can be caused by interactions between specific Sfps, between Sfps and proteins on the sperm, and between Sfps and proteins native to the female reproductive tract. Thus, characterizing the functions and interactions of Sfps is important for understanding how the sexes together ensure the successful production of progeny.
Post-mating changes in physiology and behavior induced by Sfps have been extensively characterized in Drosophila melanogaster [2], [3]. In response to the receipt of Sfps, females produce, ovulate and lay eggs [4]–[6]; store sperm in specialized storage organs [7]–[10]; show altered immune responses [11], [12]; undergo changes in sleeping, feeding and excretion behavior [13]–[16]; and, become refractory to male courtship [17], [18]. Several of these behavioral changes – egg production, sperm storage and release, and refractoriness to remating – persist in females for several days after mating and have thus been termed the long-term response [19]–[21]. The proximate cause of these changes is a short (36 amino acid) seminal protein called sex peptide (SP) [17], [18]. While most Sfps are no longer detectable in females several hours after mating [22], SP persists in females for days by binding to stored sperm [19]. Gradually, the C-terminal portion of the peptide is proteolytically cleaved to release it from sperm into the female reproductive tract [19]. This C-terminal portion of SP can then signal through its receptor, sex peptide receptor (SPR), which prolongs at least some behavioral changes in the female [23]–[26]. Indeed, SP cleavage is required for the protein to affect female behavior for more than one day [19] and for sperm to be released efficiently from storage [27].
We have previously used RNA interference (RNAi) or gene knockout lines to test 32 Sfps for function in the SP-mediated long-term response [4], [7], [10], [20], [28], [29]. These studies identified five proteins that are required for SP to function over the long term in mated females: two C-type lectins, CG1652 and CG1656; a serine protease homolog, CG9997; a cysteine-rich secretory protein, CG17575; and, a serine protease, seminase (CG10586). These proteins act in a network in which each member is required for SP to become bound to sperm [21], [28]. Loss of any network protein causes an early resumption of female receptivity to remating and a decrease in long-term fecundity. Such loss also impairs the release of sperm from the seminal receptacle in the days following mating [27]. Specific members of the network act interdependently on one another. For example, males that do not produce CG9997 are unable to transfer CG1652 and CG1656 to the female, while CG1652 and CG1656 are required to slow the rate at which CG9997 is processed in the female. Thus, while SP-SPR signaling is the proximate cause of the female post-mating response, several additional Sfps are required for this signaling to persist over the long term. We refer to this set of seven proteins as the SP network.
While genomic and proteomic analyses in D. melanogaster have identified hundreds of proteins from sperm [30], [31], seminal fluid [32]–[35], and the female sperm storage organs [36]–[40], we know of few examples of how these proteins interact to cause the dramatic post-mating phenotypes observed in females [21], [26], [28]. Biochemical approaches to identify interacting proteins are challenging due to the small amount of protein per fly, and exhaustive genetic screening of each known reproductive protein would be laborious. Here, we demonstrate a successful effort to prioritize male and female proteins for functional testing by examining covariation in their rates of evolution among species.
Evolutionary Rate Covariation (ERC) is a new metric that bioinformatically infers functional relationships between proteins based solely on their evolutionary rates across an array of species [41]. ERC operates from the hypothesis that functionally related proteins will experience correlated rate changes, because forces governing protein evolutionary rate are expected to influence entire pathways simultaneously. Evolutionary rate depends on several factors including a protein's expression level, its essentiality, and its interactions with other proteins [42]–[49]. Pathway-wide fluctuation in each of these factors has been associated with correlated rate changes (i.e., ERC) between functionally related proteins [41], [50]–[53].
In practice, an ERC value is calculated by computing the correlation between the rates of change of two proteins across all branches of a phylogeny. ERC values range from 1 to −1 for a perfect positive or negative correlation, respectively, with the genome-wide ERC distribution between all protein pairs centered at zero [41]. Functionally related pairs of proteins have been observed to have more positive ERC values in taxa as diverse as eubacteria, fungi, invertebrates and mammals [41], [50], [51], [54]–[58]. This finding holds for proteins that share physical or genetic interactions and proteins that are found in common complexes or metabolic pathways [41], [59]. Generally, a high ERC value is best interpreted as a potential functional link, which could have resulted from a common evolutionary force acting on both proteins. Accordingly, we can infer that proteins with correlated rates may be functionally related.
ERC and related methods have primarily been used to study proteins that are already known to interact functionally or physically; the use of such methods for functional prediction is only now starting to emerge [60]. We tested the utility of applying ERC prospectively by examining proteins required for Drosophila SP function. Because proper function of the SP network is essential for fertility, we reasoned that members of this network could have experienced shared evolutionary selective pressures over time and might thus show patterns of ERC across the phylogeny of sequenced Drosophila species [61]. To test this hypothesis, we created an ERC dataset specific to Drosophila. This analysis revealed significant levels of ERC between known members of the SP network. We then screened for new members of this network by searching for elevated ERC between known network proteins and sets of uncharacterized Sfps and female reproductive proteins. RNAi tests of 18 top candidates revealed three female and three male proteins required for network function. Through molecular genetic analysis, we placed five of these proteins into specific positions in the SP network, and we observed that the steps in the network in which these new proteins act are largely consistent with their evolutionary correlations. Our results demonstrate that signatures of ERC can be used prospectively to predict members of a protein network, suggesting that this method may be broadly applicable for identifying novel protein interactions.
Results
Proteins in the SP network show correlated evolutionary rate variation
We first calculated Evolutionary Rate Covariation (ERC) values for all pairs of orthologous proteins (reproductive and otherwise) from 12 Drosophila species. Briefly, we assembled orthologous protein sequences for each gene from each species for which they were available, resulting in 11,100 multiple alignments. For each pair of alignments, we calculated the correlation coefficient between their branch-specific evolutionary rates (see Methods and Figure S1). The resulting ERC values ranged from −1 to 1 and reflect the degree to which evolutionary rates correlate for any particular pair of proteins. Typically, ERC values between functionally related protein pairs are elevated compared to unrelated pairs [55]. We observed this same pattern for the seven previously known members of the Drosophila SP network. ERC values calculated for all possible pairs of these seven proteins had a mean of 0.3115, compared to the proteome-wide mean of 0.0019 (Figure S1A). The highly significant elevation between SP network proteins (permutation p = 0.000154) suggests that ERC could be used to predict additional SP network proteins. However, since proteins that are expressed at similar levels or in similar patterns can also show correlated evolution [43], we also tested whether reproductive proteins as a class had elevated ERC values. To do so, we examined a set of 664 proteins found in seminal fluid, sperm, or female sperm storage organs (see Methods and Figure S1A; we refer to these proteins below as “reproductive” but note that some are also expressed in non-reproductive tissues and could thus have other functions). The mean ERC value between all reproductive proteins was 0.0326, a highly significant elevation for sets of this size (permutation p<0.0001). This elevation could be driven by direct functional relationships and/or more indirect relationships such as expression patterns [41].
To control for this elevation in ERC across all reproductive proteins when evaluating correlations between individual pairs of proteins, we factored out the broad relationship between them. To do so, we recalculated ERC using only the 664 reproductive proteins to estimate the background rate of evolution, instead of all 11,100 proteins (see Methods and Figure S1B). After this adjustment, the mean pairwise ERC between all proteins in the reproductive set fell to 0.0047. By contrast, the mean correlation between the seven known SP network proteins remained significantly elevated (mean = 0.2806; permutation p = 0.001002). These results suggest that while shared patterns of expression or function can cause a significant increase in ERC, a much stronger signal is shared by the specific set of proteins that act together in the SP network.
Several of the strongest pairwise correlations between known members of the SP network were found between proteins with recognized genetic interactions. For example, males that do not produce network protein CG9997 are unable to transfer CG1652 and CG1656 to females during mating [21]. These pairs of proteins show ERC values in the top 5 percent of all pairwise correlations (CG9997-CG1652: r = 0.62, empirical p = 0.03; CG9997-CG1656: r = 0.62, empirical p = 0.03; Figure 1). In other instances, we did not observe strong correlations between proteins that might be expected to coevolve, such as SP and SPR. However, this particular lack of correlation may be explained by the fact that SPR has additional, non-reproductive ligands besides SP [62], [63], which may constrain its evolution. Nonetheless, the overall signature of correlated evolution throughout the SP network, the high proportion of positive pairwise correlations in the group (i.e., 16 of the 21 pairwise correlations in Table 1 are positive), and the significant correlations between specific group members suggest that members of the SP network show significant levels of evolutionary rate covariation.
Fig. 1. Proteins in the SP network show a significantly elevated signature of ERC. 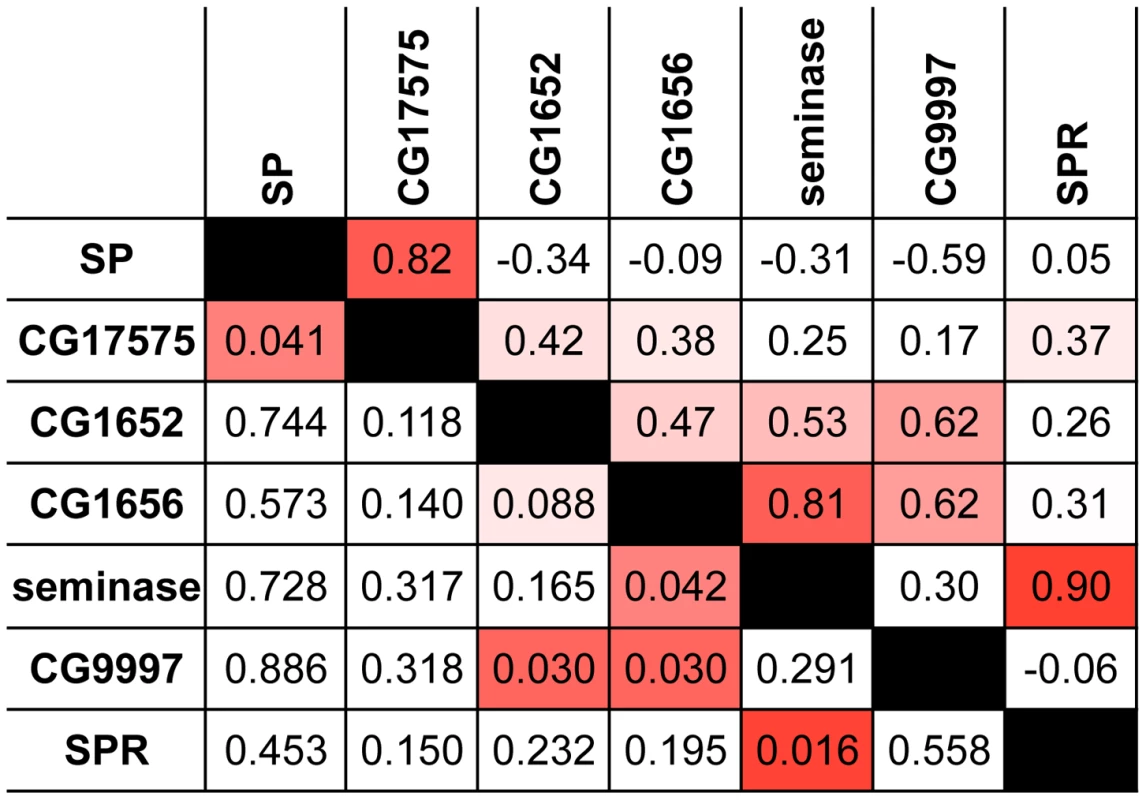
This pairwise matrix shows ERC values (above diagonal) and their corresponding empirical p-values (below diagonal) between the seven known members of the SP network. Red shading indicates correlations with empirical p<0.05; more intense shading indicates a stronger correlation. Tab. 1. Candidates identified by ERC and tested for effects on 4-day remating receptivity. 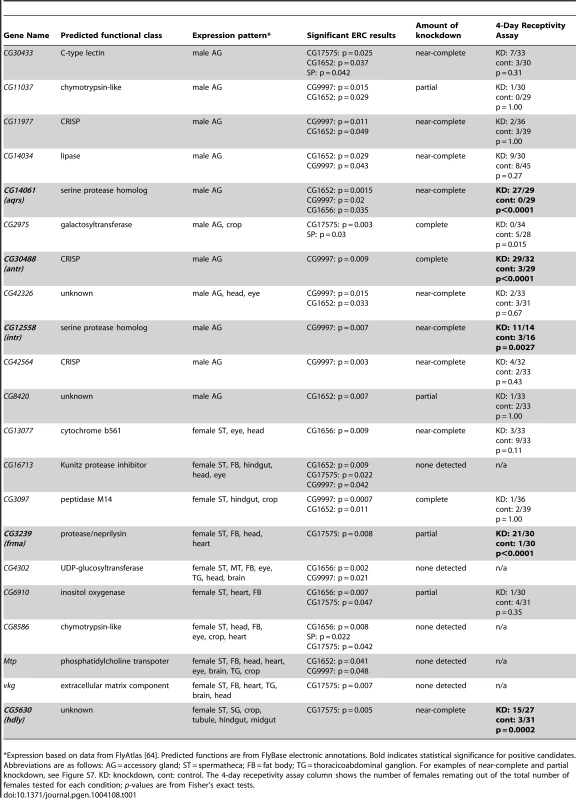
Expression based on data from FlyAtlas [64]. Predicted functions are from FlyBase electronic annotations. Bold indicates statistical significance for positive candidates. Abbreviations are as follows: AG = accessory gland; ST = spermatheca; FB = fat body; TG = thoracicoabdominal ganglion. For examples of near-complete and partial knockdown, see Figure S7. KD: knockdown, cont: control. The 4-day recepetivity assay column shows the number of females remating out of the total number of females tested for each condition; p-values are from Fisher's exact tests. ERC reveals new candidate SP network proteins
Since we detected positive evolutionary correlations between known SP network proteins, we applied the ERC method prospectively to identify new candidate network members. For this analysis, we calculated pairwise correlations using the reproductive protein data set described above, and we focused specifically on correlations between the known SP network proteins and the 434 proteins that comprised the sets of secreted Sfps and proteins present in the female reproductive tract. To identify candidates, we queried each of five network proteins (CG1652, CG1656, CG9997, CG17575 and SP) against the 434 Sfp and female proteins. SPR was not used as a query because it has additional ligands that do not appear to function in reproduction [62], [63]. Thus, SPR may need to maintain interactions with multiple proteins, which may dampen signals of correlated evolution with any single interacting partner. Seminase was excluded because unambiguous orthologs were found in only five species, which would cause low statistical power.
We found 111 proteins (55 Sfps, 56 female proteins) that showed a significant correlation (p<0.05) with at least one of the five network proteins. From this group, we selected 21 candidates for further testing, each of which showed a significant (p<0.05) level of ERC with multiple SP network proteins and/or a highly significant (p<0.01) level of ERC with at least one network protein (Table 1). We tested each candidate in Table 1 by using RNAi to knockdown expression of the gene in the appropriate sex; five of the 21 candidates showed no evidence of knockdown by RT-PCR and were excluded from further analysis. For the remaining 16 candidates, we screened for genes whose knockdown caused a significant increase in female remating receptivity four days after an initial mating.
Of the 16 candidates that were at least partially knocked down by RNAi, five showed highly significant effects on 4-day remating receptivity (Table 1). Knockdown of the remaining 11 candidates caused no significant increase in female receptivity. This latter result could be due in some cases to insufficient knockdown or to functional redundancy with other Sfps or female proteins. Alternatively, these proteins may not function in the SP network. Of the positive candidates, three genes (CG14061, CG30488 and CG12558) are expressed specifically in the male accessory glands [64]; at least two of them (CG14061 and CG30488) encode proteins that are transferred to females as Sfps at mating [33]. The other two positive candidates, CG3239 and CG5630, are each expressed in the female's spermathecae, as well as in other non-reproductive locations [64]. CG5630 is also expressed in the female's seminal receptacle [39].
ERC signatures, but not genomic location, predicts an additional SP network protein
One striking feature of several of the new candidate network genes was their genomic positioning next to previously known SP network genes (Table S2). This pattern was previously observed for the SP network lectins, CG1652 and CG1656, which are believed to have arisen from an ancient gene duplication event [33], [34]. We found that three additional pairs of network genes (CG9997 and CG14061, CG17575 and CG30488, and CG3239 and SPR) are also located in tandem with one another. For two of these pairs, the tandemly-located genes encode proteins in the same biochemical category (CG9997 and CG14061 each encode predicted serine protease homologs, and CG17575 and CG30488 each encode predicted CRISPs), but in contrast to the situation with the lectins CG1652 and CG1656, we do not find unambiguous evidence that either the protease or the CRISP cluster arose by tandem gene duplication. However, regardless of each cluster's origin, it is possible that such genomic clustering enables the co-regulation of genes that function in a common pathway [65].
In the CG17575/CG30488 cluster, we found a third annotated gene that encodes a seminal fluid protein of the same predicted functional class as the other cluster members: CG30486, which encodes a predicted CRISP. Similarly, we observed a known Sfp gene encoding a predicted serine protease homolog, CG34295, immediately upstream of CG12558. While neither CG30486 nor CG34295 was identified by our ERC analysis, we hypothesized that their shared locations with known or candidate SP network members could indicate their involvement in the SP network. However, when each of these additional genes was knocked down individually, we observed no effect on female remating receptivity 4 days after mating (Table 2). Thus, either these neighboring genes are uninvolved in the SP network, or they function in the network in a completely redundant role. Alternatively, their degrees of knockdown may have been insufficient to produce a phenotype.
Tab. 2. Tests of neighboring genes and additional ERC candidates for 4-day receptivity phenotypes. 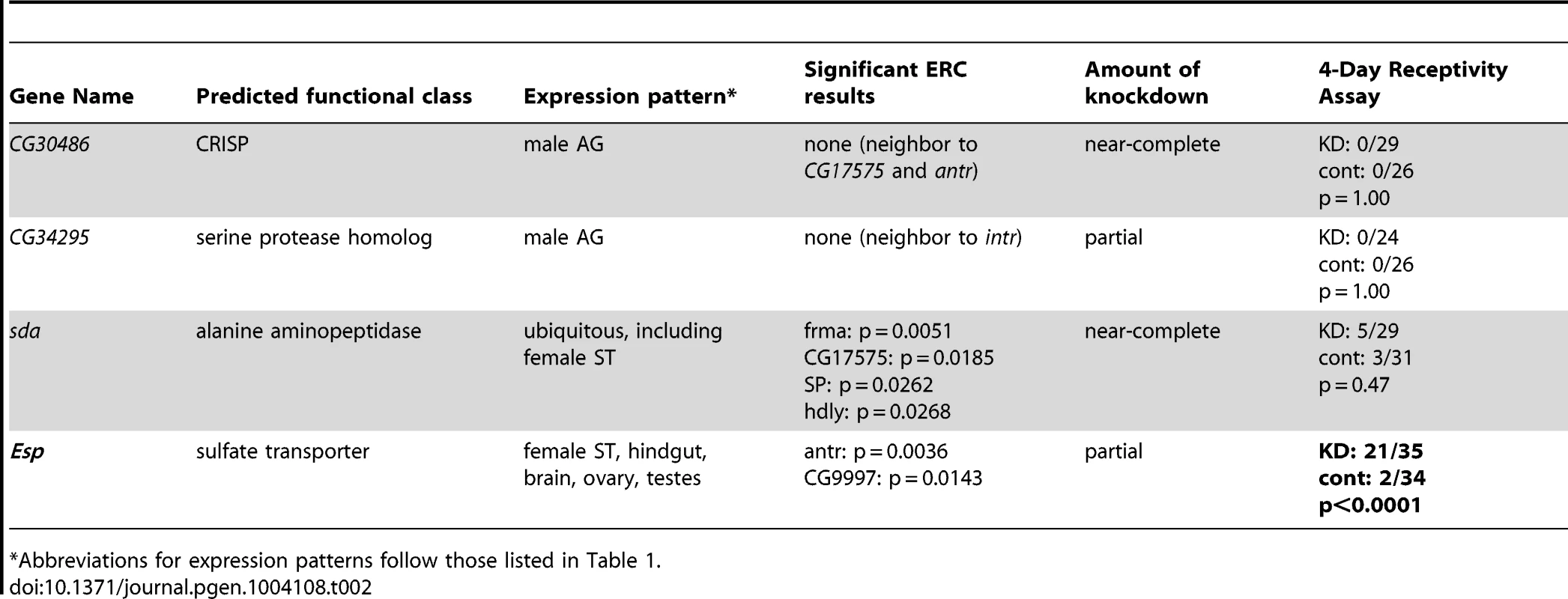
Abbreviations for expression patterns follow those listed in Table 1. We also asked whether signatures of ERC between these new candidates and the rest of the large sets of seminal fluid or female proteins might identify additional network proteins (Figure S1C). To this end, we used RNAi to test two additional female genes that showed highly significant ERC levels with at least one new candidate protein (Table 2). One of these genes, epidermal stripes and patches (Esp), showed a highly significant effect on female remating receptivity. Taken together with the results above, these data suggest that ERC has strong sensitivity to detect new candidate members of the SP network.
Additional RNAi lines confirm the SP network phenotypes
To confirm that the receptivity and fertility effects we observed in the above RNAi experiments were not due to RNAi off-target effects and/or insertions of RNAi-triggering constructs into essential genes, we first used UP-TORR [66] to analyze each line's RNAi-triggering sequence against all current D. melanogaster gene annotations. No off-target transcripts were predicted for any RNAi construct used. We then performed receptivity and long-term fertility assays (see Methods and below) on additional RNAi lines, where available, that controlled for either the site of the UAS-RNAi construct insertion (for CG5630 and Esp) or both the insertion site and the hairpin sequence used to trigger RNAi (for CG30488 and CG3239). (No additional RNAi lines exist for CG14061 or CG12558). These tests (summarized in Table S3) confirmed the receptivity and fertility phenotypes seen with the first lines tested for CG30488 and Esp. Likewise, knockdown of CG5630 by a second hairpin showed a strong effect on fertility and a marginally significant effect on receptivity. Knockdown of CG3239 by a second hairpin also replicated a strong effect on fertility, but showed no significant effect on receptivity. However, RT-PCR revealed that with this hairpin, CG3239 transcript levels were only partially knocked down, which could explain the less severe phenotype. Because of the high degree of replication, results reported below come from experiments performed on the original lines (details of which are described in Table S1).
ERC-identified candidates show additional receptivity and fertility phenotypes consistent with SP network function
To evaluate whether each of these six genes was required only for extended female non-receptivity, we next tested each positive candidate for effects on remating receptivity at 1 day after an initial mating. As shown in Table 3, in no case did knockdown of a candidate gene cause an increase in short-term receptivity. Thus, rather than having general effects on female post-mating behavior, each candidate is required specifically for the long-term loss of female receptivity to remating. This phenotype is consistent with a malfunction in the SP network [20], [21]. In females mated to SP network knockdown males, SP transferred at mating but not bound to sperm is sufficient for full fertility and non-receptivity 1 day after mating. However, if SP cannot bind to sperm, it is no longer detected in the reproductive tract by 4 days after mating [19].
Tab. 3. Tests of female remating receptivity 1 day after an initial mating. 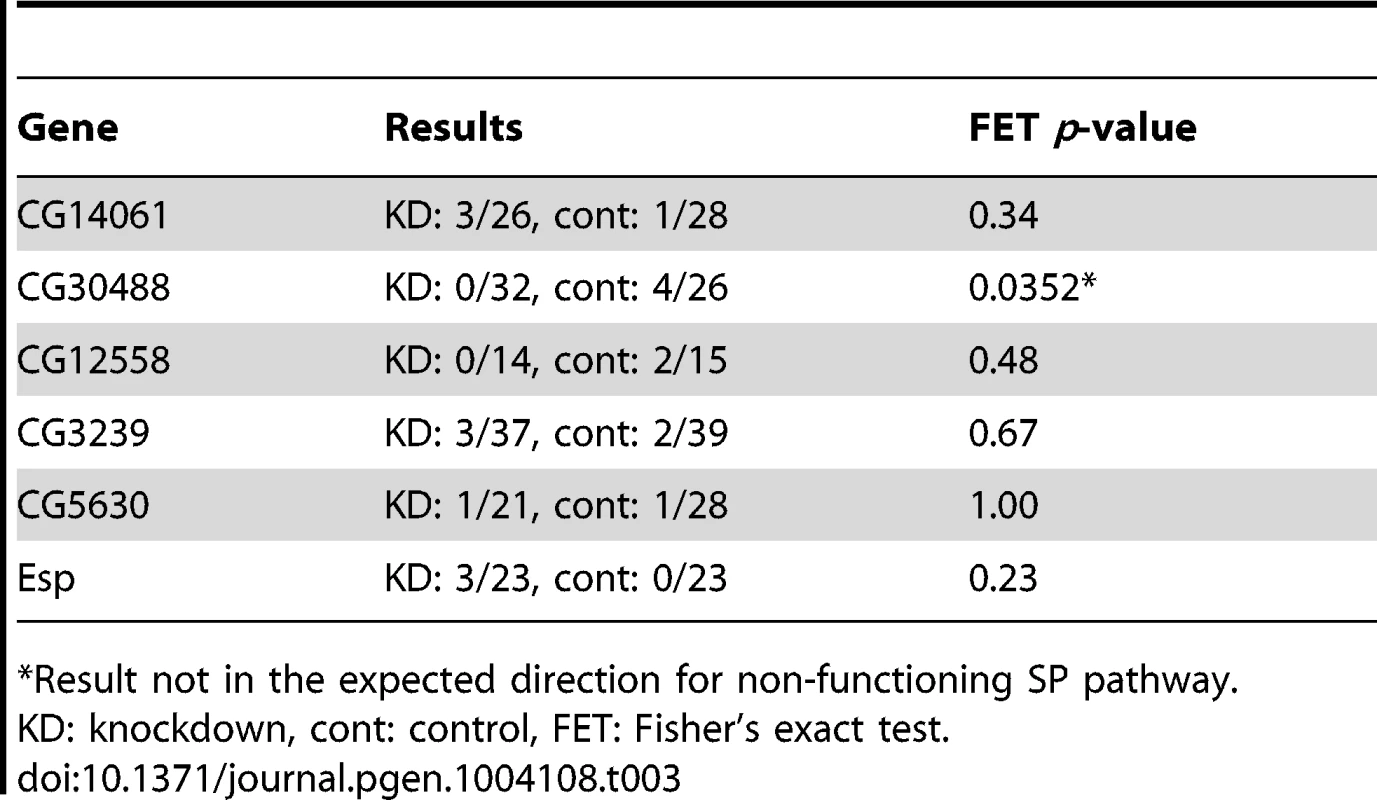
Result not in the expected direction for non-functioning SP pathway. We reasoned that if these six positive candidates affect the function of the SP network, they should also affect long-term fertility, which requires the long-term storage and utilization of SP [17], [18], [20], [26], [28]. Consistent with a role in the SP network, each new protein was required for full fertility over the course of a 10-day assay (Figure 2). Males knocked down for CG14061, CG30488 or CG12558 induced normal levels of egg-laying and progeny production in females for the first day after mating, but these measures declined relative to controls as early as the second day after mating. Females knocked down for CG5630 or Esp showed the same pattern of normal fertility on day 1 after mating, but reduced fertility in the following days. Females knocked down for CG3239 had significantly reduced egg-laying and progeny production even on the first day after mating, mimicking the effects of knocking down SPR (Figure 2, Figure S2). These knockdown females then continued to have lower egg and progeny production throughout the assay. We further observed that knockdown of any male gene or of the female gene Esp had no significant effect on egg-hatchability, while knockdown of the remaining female genes caused hatchability to be significantly lower (Figures S3, S4). This effect was most pronounced in CG3239 knockdown females, and much less severe in CG5630 and SPR knockdown females. Effects on hatchability were unlikely to be due primarily to reduced viability of offspring inheriting both the UAS-RNAi construct and the GAL4 driver (see Text S1).
Fig. 2. Fertility assays for new candidate SP network proteins identified by ERC. 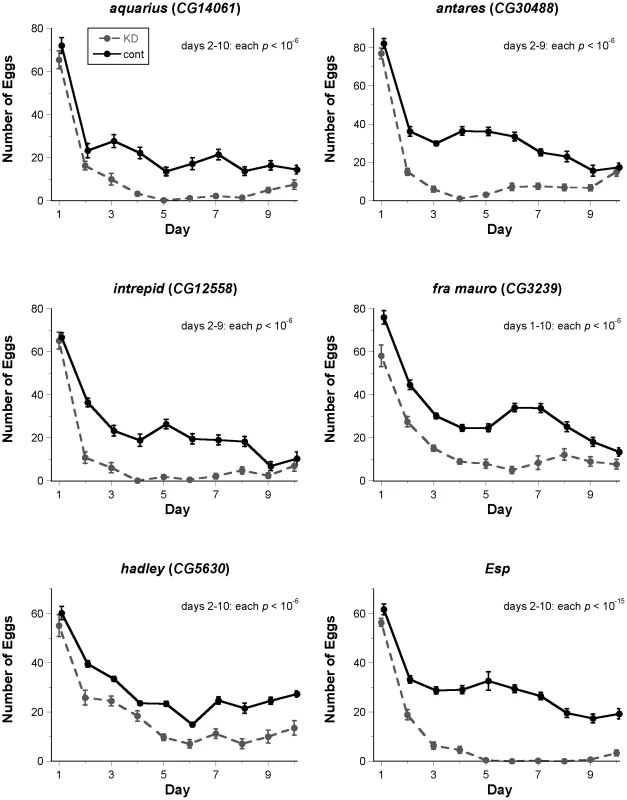
Each graph depicts the mean (± SE) number of eggs laid on each day of a 10-day fertility assay (knockdown: KD, dashed line; control: cont, solid line). For each male-expressed gene, knockdown or control males were mated to wild-type females. For each female-expressed gene, wild-type males were mated to knockdown or control females. Knockdown of each gene shown had a highly significant effect (corrected p<10−6 in all cases) on overall fertility; results of statistical testing for fertility on each day of the assay are shown on each graph. Control data points are offset horizontally from knockdown data points to facilitate comparison, but all flies in each experiment were transferred from one vial to the next at the same time each day. Samples sizes for each treatment range from 11 to 28. One representative biological replicate (out of 2–3 for each gene) is shown. Thus, each of these six candidates identified by ERC is required for both the long-term loss of remating receptivity and the long-term maintenance of fertility. In our subsequent results and discussion, we adopt new names for the previously unnamed genes: male-expressed genes are named after lunar modules used in the Apollo space program (CG14061: aquarius; CG30488: antares; CG12558: intrepid), and female-expressed genes are named after sites on the moon at which Apollo missions landed (CG3239: fra mauro; CG5630: hadley).
The new male genes encode proteins predicted to belong to functional classes often found in insect and mammalian seminal fluid [33],[34],[67]–[69] and already represented in the SP network. Like CG9997, aquarius and intrepid encode serine protease homologs [70]; like CG17575, antares encodes a cysteine-rich secretory protein. In females, fra mauro encodes a protein that contains a partial, predicted neprilysin protease domain. Neprilysins are a class of protease that preferentially cleave prohormones and neuropeptides and are important for male and female fertility in mammals [71]–[73] and Drosophila (J. Sitnik et al. submitted). Neither annotated isoform of fra mauro is predicted by SignalP [74] to be secreted or extracellular, raising the question of how this protein could interact with SP network proteins. Inspection of the 5′ untranslated region of fra mauro revealed the presence of a potential alternative initiation codon, which is followed by a region predicted by SignalP to encode a functional secretion signal sequence. RT-PCR analysis on female cDNA found that a product could be amplified when a forward primer is placed in this region (data not shown), raising the possibility that an alternative isoform of the protein may be secreted and thus more accessible to other network proteins. In addition, we found this alternative start codon and secretion signal to be conserved in at least 11 of 12 Drosophila species analyzed (the D. willistoni genome sequence contains a sequencing gap in this region), which provides strong evidence that this secreted protein isoform is functionally important (Figure S5). The hadley protein is predicted to be secreted, but its potential functional class remains unknown, as neither conserved domain searching [75] nor three-dimensional structural modeling [76] could identify a conserved protein domain. The Esp gene was initially identified as a target of homeotic genes [77] but is, otherwise, poorly characterized. While the Esp protein is not predicted to be secreted, it shows homology to transmembrane sulfate transporters. In adults, Esp is expressed predominantly in the spermathecae [64], with additional expression reported in the seminal receptacle [39].
Molecular characterization of new SP network proteins
We next sought to position these six new proteins in the SP network. To do so, we first used Western blotting to test whether SP was successfully stored over the long-term in mates of knockdown males or in knockdown females. In wild-type matings, SP is readily detectable from dissected female seminal receptacles (SRs) 4 days after a mating. However, knockdown of any of the known SP network proteins eliminates this retention [21], [28]. We observed that wild-type females mated to males knocked down for aquarius, antares or intrepid showed little or no SP at 4 days after mating (Figure 3). These reduced levels of SP were not due to less SP having been transferred at mating (see Figure 4). These results suggested that male proteins aquarius, antares and intrepid are each required for network function at a step upstream of SP binding sperm in the SR. By contrast, when wild-type males were mated to fra mauro, hadley or Esp knockdown females, normal levels of SP were observed at 4 days after mating (Figure 3). Thus, these female proteins may be necessary for the utilization of SP after it becomes stored in the SR or may be required for proper SP-SPR signaling.
Fig. 3. SP retention in mated females, 4 days after mating. 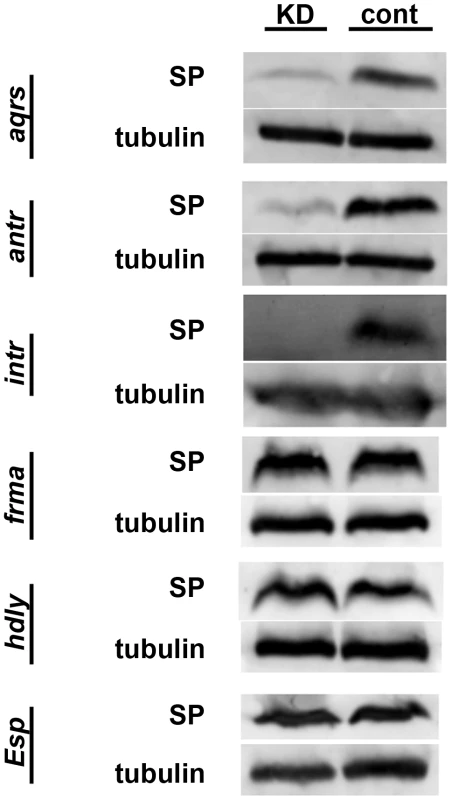
Western blots probed with antibodies to SP or alpha-tubulin (loading control). Proteins were isolated from lower female reproductive tracts 4 days after mating. Gene names to the left of each pair of blots indicate which gene was (KD) or was not (cont) knocked down in the mating pair. Across all experiments, the number of female reproductive tract (RT) equivalents used for each condition ranged from 13 to 20; however, for any given gene, the number of RT equivalents compared between KD and control was within 2 RTs. Fig. 4. Production, transfer and processing of SP network proteins in males knocked down for aquarius, antares or intrepid. 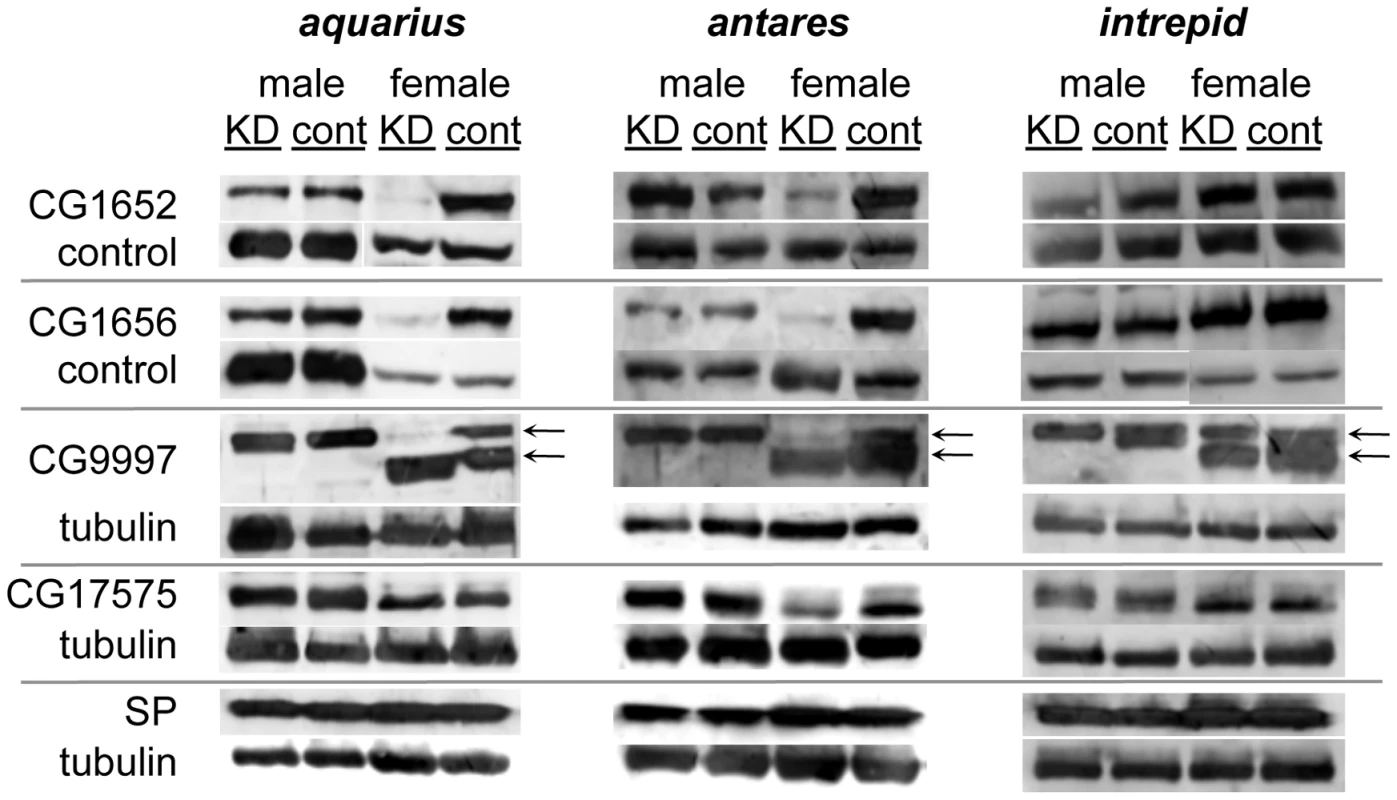
Western blots were probed with either an antibody to an SP network protein or a loading control. Alpha-tubulin was used as the loading control for blots of CG9997, CG17575 and SP. Since CG1652 and CG1656 sometimes co-migrated with tubuiln, loading controls for these proteins were either a consistently observed cross-reactive band or tubulin. Proteins were isolated from male reproductive tracts (“male” columns) or lower female reproductive tracts dissected 1 hour after the start of mating (“female” columns). “KD” indicates males knocked down for aqrs, antr or intr or females mated to a knockdown male, while “cont” indicates control males or females mated to a control male. Arrows next to the blots for CG9997 indicate the ∼45 (top) and ∼36-kDa (bottom) forms of the protein [21]. Within each blot, the amount of RT equivalents loaded for each sex was equal. Across blots, male lanes contain 0.5–1 RT equivalents; female lanes contain 2–4 RT equivalents. To further determine where the new male proteins fit into the network, we examined the production of the known SP network proteins in males knocked down for aquarius, antares or intrepid (Figure 4). In all cases, we observed no difference in the production of SP, CG1652, CG1656, CG9997 and CG17575 between knockdown and control males (Figure 4; compare lanes for knockdown and control males). We then tested whether knockdown males could transfer these proteins to females and examined their processing in female reproductive tracts. Males knocked down for intrepid transferred all proteins at equivalent levels to controls, and females mated to these males showed normal CG9997 processing [21] in their reproductive tracts. Males knocked down for aquarius or antares transferred normal levels of SP, CG9997 and CG17575, but much lower levels of CG1652 and CG1656 (Figure 4; compare lanes for females mated to aquarius or antares knockdown or control males). Consistent with the absence of these proteins in females after mating [21], the post-mating processing of CG9997 was also disrupted, with mates of knockdown males showing an increased level of the 36-kDa form of CG9997 relative to the 45-kDa form of this protein. We also examined the production and transfer of seminase and observed no differences between knockdown and control flies for each gene (data not shown).
Because SP is required for the release of sperm from storage [27], we examined sperm storage and retention in the SRs of females mated to males knocked down for each of these genes (Figure 5). At 2 hours after mating, sperm from antares and intrepid males were present in the SR at equivalent levels to controls, while sperm from aquarius males were present at slightly lower levels. However, by 10 days after mating, mates of control males had largely depleted their stores of sperm in the SR, while mates of males knocked down for any of the three genes showed significantly higher numbers of sperm. Taken together with the lack of SP retention (see Figure 3), these data confirm that male proteins aquarius, antares and intrepid are each required for SP to become bound to sperm. Disruption of this binding, in turn, inhibits the ability of sperm to be released from the seminal receptacle. This inability to release sperm from storage likely contributes to the reduction in long-term fertility when each of these male genes is knocked down (Figure 2).
Fig. 5. Average number of sperm stored in the seminal receptacles (SR) of wild-type females mated to knockdown or control males for new SP network proteins. 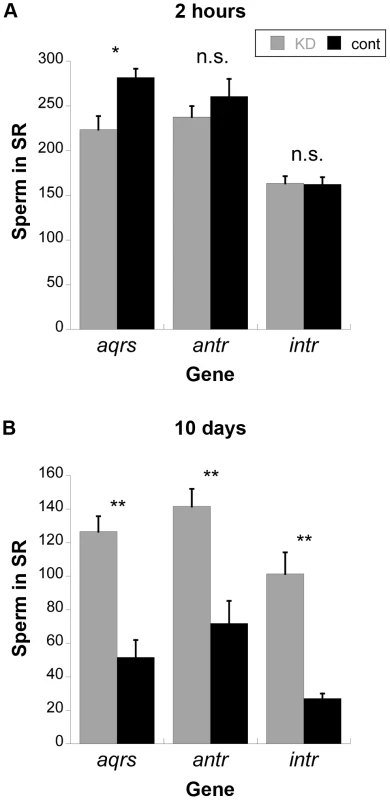
Average number of sperm in female SRs at 2(A) or 10 days (B) after mating to aqrs, antr or intr knockdown (KD, gray) or control (cont, black) males. Each bar indicates the mean; error bars indicate 1 standard error. *, p<0.01; **, p<0.002; n.s. = not significant. Samples sizes for each treatment range from 11 to 18. Taken together, our results allow us to place aquarius, antares, fra mauro, hadley and Esp into the SP network (Figure 6A). The male proteins aquarius and antares act at the same step of the network as CG9997, as each of these proteins is required for the transfer of CG1652 and CG1656. The female proteins fra mauro, hadley and Esp appear to act at the downstream end of the network, after SP has bound to sperm. At present, we are unable to position intrepid within the network, though its effect on SP retention (Figure 3) suggests that it acts upstream of SP-SPR signaling.
Fig. 6. An expanded network of proteins is required for SP to bind sperm and to be utilized in mated females. 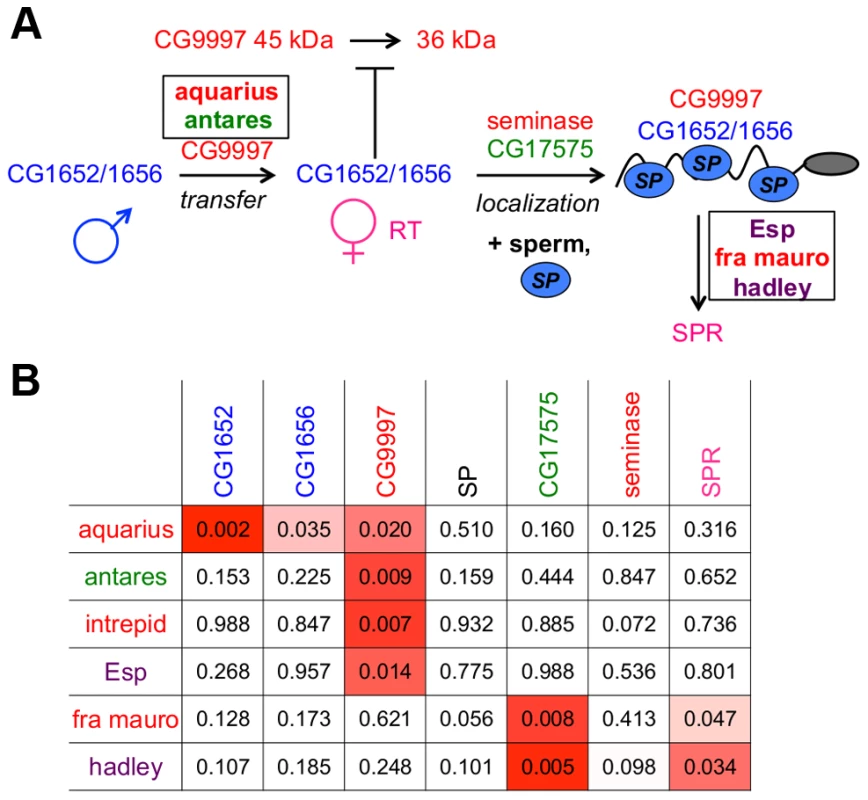
(A) The SP network. Colors of protein names indicate predicted protein functional classes: red = protease or protease homolog; green = cysteine-rich secretory protein (CRISP); dark blue = C-type lectin; light blue oval = SP; purple = unknown function. Boxes indicate proteins discovered by ERC; other proteins were described previously [21], [28]. Intrepid acts upstream of SP-SPR signaling, but at present we cannot position it further. (B) New members of the SP network function at steps consistent with their signals of ERC. New network proteins are shown in rows; known network proteins are shown in columns. Each cell indicates the empirical p-value associated with the protein's pair ERC value. P-values less than 0.05 are shaded in red; more intense shading indicates a stronger correlation. A protein's evolutionary correlations reflect its position in the SP network
When comparing the positioning of these six new proteins in the network to their patterns of ERC with the previous known seven network proteins (Figure 6B), we observed that the new male proteins showed their strongest correlations with the upstream players of the network. In particular, each new male protein showed a significant correlation with CG9997, which functions in the same step of the network (CG1652/CG1656 transfer) as aquarius and antares. At the downstream end of the pathway, two of the new female proteins showed their strongest correlations with downstream players in the network, including SPR, which is consistent with their potential functions. Thus, the patterns of ERC observed between new and established network proteins are consistent with the steps in the network in which these new proteins are found to act.
Discussion
We have used signatures of covariation in protein evolutionary rates to investigate interactions between proteins that are required to maintain post-mating responses in Drosophila females. We first found that, as a group, proteins known to act in the SP network [20], [21], [26], [28] showed a significant signature of ERC. We then used ERC to screen 434 male Sfps and female reproductive tract proteins for those that correlated strongly with members of the SP network. RNAi functional testing of 16 top candidates identified five proteins that are each required for long-lasting SP responses in females, including reducing a female's willingness to remate and boosting female egg production. Additional tests of two candidates that showed high ERC with these new genes revealed a sixth network protein. The new male proteins, aquarius, antares and intrepid, act in the upstream part of the network: loss of any one of these proteins prevents SP from becoming bound to sperm, which in turn prevents sperm from being released from storage. Because SP binds to sperm in females knocked down for fra mauro, hadley or Esp, these proteins may affect the ability of SP to be used in females and/or may be required for normal SP-SPR signaling. Interestingly, the strongest evolutionary correlations between these new proteins and the known members of the network often occurred between pairs of proteins that appear to act in the same part of the pathway. These results verify the utility of ERC and suggest that this metric may be used prospectively to identify candidates acting in a particular part of a pathway.
ERC efficiently identifies new types of network proteins
Our results suggest that ERC successfully prioritized a large set of proteins for detailed functional testing; the observed success rate was six positive hits out of 18 candidates tested, and this rate could be higher if genetic redundancies or insufficient knockdown prevented positive results for some candidates. This rate likely represents a significant enrichment of network genes because if the same success rate were applied to the full list of 434 reproductive proteins, it would imply that there are 145 long-term mating response genes waiting to be discovered in that list alone. Although this is a formal possibility, this number seems high. Importantly, ERC allowed us to explore new functional classes of protein from the female reproductive tract. Previous studies [20], [28] chose male-expressed candidates based on molecular classes that were known to function in sperm storage and fertilization. In contrast, ERC directed us to proteins that unlikely would have been selected for screening, as fra mauro was not annotated to be extracellular and hadley had no predicted functional class. We can also prescribe a strategy to improve ERC analysis by retrospectively analyzing the positive candidates. Very strong correlations (p<0.01) tested positive more often, so future applications of this method could focus on single, strong correlations rather than those proteins that correlate more weakly (p<0.05) with multiple network members. Finally, we note that several reproductive proteins showed strong signals of ERC with the SP network but were not quickly testable because RNAi lines were not available. In cases like these, emerging technologies such as the CRISPR/Cas9 system that is now being optimized for Drosophila [78], [79] may in the future enable null mutants to be generated, which could potentially expand the SP network further.
Possible functions for new network proteins
By expanding the SP network to include new proteins from both sexes, our results provide a more complete picture of how SP controls female post-mating responses. Until now, SPR was the only known female regulator of SP action [26], but our results show that fra mauro, hadley and Esp are also necessary for sperm-bound SP to exert its long-term effects on females. In addition to their expression in the spermathecae, each of these female genes is expressed in regions other than the female reproductive tract [64]. SPR follows the same pattern: it is expressed in several reproductive regions [26], including the spermathecae, and elsewhere in the adult female. However, only six SPR-expressing neurons in the reproductive tract are required for the SP response [23]–[25]. It is also interesting to compare the fertility phenotypes for fra mauro, hadley, Esp and SPR knockdown females (Figure 2). Knockdown of fra mauro or SPR causes both a long-term fertility deficit and an immediate reduction in egg-laying in the first 24 hours after mating. In contrast, hadley or Esp knockdown females show normal fertility on day 1, but then have reduced fertility over the following days. Assuming that the extent of gene knockdown was sufficient to reveal null-like phenotypes, one possible model to explain these results could be that fra mauro is necessary to facilitate SP-SPR signaling, while hadley and Esp are necessary for the efficient release of SP from stored sperm. SP-SPR signaling is required for full fertility at all time points after mating (Figure S2 and [26]), but impaired release of SP from sperm affects fertility only after day 1 [19]. Another possibility is that fra mauro is required to coordinate temporally the release of sperm from storage when eggs are ovulated and ready to be fertilized. Furthermore, while knockdown of fra mauro, hadley and SPR each caused a reduction in egg hatchability, the magnitude of this effect was by far the greatest for fra mauro (Figures S3, S4). Thus, in addition to laying significantly fewer eggs than controls (Figure 2), fra mauro females also experience far lower egg-to-adult viability. Finally, it is interesting to observe that Esp is a predicted sulfate transporter. In mammalian systems, anion concentration in the female reproductive tract is critical for proper sperm function and fertility [80]. In Drosophila, it is possible that attenuation of extracellular levels of anions such as sulfate in the sperm storage organs affects Sfp-sperm binding, sperm storage, SP release, or another process required for SP network function.
Two observations suggest that interactions between SP network proteins may begin in the male. First, CG9997, aquarius and antares are each required for lectins CG1652 and CG1656 to be transferred efficiently to females [21] (Figure 4). It is possible that one or more of the former proteins may bind to either lectin protein as Sfps transit the male reproductive tract during mating. Such binding could protect the lectins from proteolysis or modification. For instance, CG9997 and aquarius both encode serine protease homologs that are predicted to have inactivating mutations in their active sites [70]. It has been speculated that such inactive proteases could act as competitive inhibitors of proteolytic processing by binding to processing targets, rendering them less accessible to the numerous active protease in the seminal fluid [81]. Second, it is presently unclear whether intrepid is transferred at mating, as previous proteomic experiments have not detected this protein in mated females [33]. While intrepid may be transferred but poorly detectable in mated females (e.g., due to low abundance or rapid degradation), it may, alternatively, act in males to modify or activate another network protein(s). Processing of Sfps within males is observed in other cases. For example, the Drosophila seminal metalloprotease CG11864 is processed in the male reproductive tract during transfer to females, and this processing is required for CG11864 to mediate the processing of additional Sfps in the female reproductive tract [28], [82] (B. LaFlamme, F. Avila et al., submitted). In nematodes, interactions between a protease, TRY-5, and a protease inhibitor, SWM-1, regulate the activation of sperm during transit through the male reproductive tract [83]–[85]. Thus, it will be interesting to determine whether any members of the SP network are the agents or targets of processing within the male reproductive tract. If network proteins are modified while still in the male, this process may be regulated spatially and temporally by the sequestration of interacting components into distinct compartments of the reproductive tract, including the ejaculatory bulb [86] and vesicles found in secondary cells of the accessory gland [87], [88]. Such compartmentalization could ensure that interacting proteins do not encounter each other until the appropriate time during or after mating.
Evolution of the SP network
Our results, combined with previous work [20], [21], [26], [89], suggest that at least 13 proteins participate in the SP-mediated post-mating response in female Drosophila melanogaster. How did this complex network arise, and how have its members evolved? Orthologs of the sex peptide receptor (SPR) are found in diverse insect taxa, including mosquitoes, silkworms and moths, and these receptors are responsive to stimulation by D. melanogaster SP [26], [90]. However, SP has not been identified outside of Diptera; a putative SP ortholog was identified by bioinformatics in Anopheles [91], but the short length of SP makes it difficult to detect orthologs in other species, including some drosophilids. Furthermore, the female post-mating responses of insects with SPR orthologs often differ substantially from those of the melanogaster group of Drosophila. For example, D. mojavensis females re-mate more readily than D. melanogaster females [92], and while A. gambiae females become unreceptive to further courtship after a single mating, this behavioral change does not require the transfer of sperm [93].
Within the genus Drosophila, other members of the network show different levels of evolutionary conservation. We identified orthologs of CG1652, CG1656, CG9997 and CG17575 in 11 of 12 sequenced Drosophila species (all but the most distant species, D. grimshawi). Most of the new proteins we identified share this broad distribution throughout the genus. Hadley and fra mauro are found in all 12 species, but appear not to have orthologs in sequenced mosquito species (data not shown). Aquarius and antares show the same species distribution as CG1652, CG1656, CG9997 and CG17575. Esp orthologs are found in only ten species, but these include one member of the more distantly related Drosophila clade, D. mojavensis, suggesting an older origin for this protein. In contrast, intrepid and seminase appear to have evolved more recently, with orthologs detectable only in the Sophophora clade. Orthologs of intrepid were found in 9 of 12 species (all but D. virilis, mojavensis and grimshawi), while seminase orthologs were detected only in D. melanogaster-D. ananassae. Taken together, these varying degrees of evolutionary conservation suggest that the SP network, as it presently functions in D. melanogaster, may have evolved in pieces over time. Indeed, the emergence of the full SP network correlates with changes in remating rate. Frequent mating (daily or more than once per day) was inferred to be the ancestral condition for drosophilids, while less frequent mating is derived and appears in those species (D. melanogaster through D. pseudoobscura) that have all or nearly all of the SP pathway components [94].
Some reproductive proteins of many species have evolved under positive selection [95]–[97]. One proposed explanation for this pattern suggests that males and females may experience sexual conflict over some aspect of reproduction (e.g., the rate of female remating). Substantial evidence suggests that sexual conflict occurs in D. melanogaster [98]–[100] and is mediated by SP [101]. At the molecular level, the result of sexual conflict could be continual coevolution between male and female protein sequences. Population genetic studies have detected evidence of recent selective sweeps on SP [102] and CG9997 [103], but most other members of the network appear well conserved [33]. One possible explanation centers on the observation that SPR is sensitive to multiple ligands [26], [62], [63], which may constrain its ability to coevolve with SP and thus reduce the requirement for constant coevolution. It will also be instructive to examine the molecular evolution of all network members across the Drosophila phylogeny and to determine whether any have experienced bursts of positive selection on the same phylogenetic lineages, as might be predicted for proteins showing patterns of ERC [50].
Conclusions
We have shown that signatures of evolutionary rate covariation can be used prospectively to identify new members of a protein network. In the context of the Drosophila SP pathway, this genomic approach allowed us to efficiently screen hundreds of known reproductive proteins so as to prioritize candidates for functional analysis, thereby identifying new long-term mating response proteins from both males and females. Interestingly, male and female proteins appear to participate in distinct sections of the SP network, and this separation was reflected in their signatures of correlated evolution. We believe that the ERC approach will be broadly applicable to identifying new members of other protein networks in any taxa for which comparative genomic data are available.
Methods
Reproductive proteins data sets
We used a combination of published proteomic and transcriptomic data sets and genome-wide expression data to create three sets of reproductive genes used in the analysis: seminal fluid proteins (Sfps), female reproductive tract proteins, and sperm proteins. The first set consisted of 208 genes encoding Sfps that had been identified by mass spectrometry in the reproductive tracts of mated females [32], [33] or predicted secreted proteins from the male accessory gland [34]. The second set included 226 genes expressed in the female sperm storage organs. This set included the D. melanogaster orthologs of EST sequences identified from the spermathecae of D. simulans [36], [38] and EST sequences identified from the seminal receptacle of D. melanogaster [39]. We removed from these sets annotated housekeeping genes (e.g., ribosomal and mitochondrial proteins) since they were unlikely to interact with proteins in the SP network. Because EST sequencing may not sample all relevant genes, we then supplemented these genes with genes identified in FlyAtlas [64] to be predominantly expressed in the spermathecae (the only female sperm storage organ for which genome-wide expression data are available). The third set included 322 genes that encode proteins in the D. melanogaster sperm proteome [30], [31] and that were found in FlyAtlas to be predominantly expressed in the testis. This filtering was performed to enrich for proteins likely to function specifically in reproduction, since proteins involved in additional biological processes may interact with several partners and thus show dampened signals of ERC. While we used all three sets of genes (756 genes in total) for optimizing the ERC method (see below), we focused our further functional tests on ERC candidates identified from the seminal fluid and sperm storage organ gene sets (434 in total).
Alignment of orthologous protein coding sequences from 12 species
We identified orthologous genes from 12 Drosophila species using a combination of high-throughput and manual searching. Protein amino acid sequences were produced by the Drosophila 12 Genomes project and downloaded from FlyBase (http://flybase.org) [61]. The species were: Drosophila melanogaster, sechellia, simulans, yakuba, erecta, ananassae, pseudoobscura, persimilis, willistoni, grimshawi, virilis, and mojavensis. Orthologs were identified using InParanoid, and the resulting groups were aligned by MUSCLE [104], [105]. Many alignments were missing species either due to evolutionary loss or missed gene annotation. To increase the number of species and thereby improve our power, we manually searched for unannotated genes in the 11 non-melanogaster species using a combination of tBLASTn and BLAT. This effort added 81 previously unannotated sequences to a total of 31 alignments.
Genome-wide Evolutionary Rate Covariation (ERC) analysis across 12 Drosophila species
To perform ERC analysis, we first calculated the amount of amino acid divergence for each branch in the species tree for each of the 11,100 orthologous protein alignments produced above; this was done using ‘aaml’ of the PAML package [106]. Next, raw branch lengths were transformed into rates of evolution relative to the expected branch length. This projection operation, introduced by Sato et al. [58], removes the inherent correlation of all proteins due to the underlying species tree and improves the power of ERC to resolve functionally related protein pairs from unrelated pairs [55], [58]. Finally, we used these corrected branch-specific rates to calculate the correlations for all pairs of proteins, resulting in a proteome-by-proteome matrix of correlation coefficients, termed the ERC matrix. To limit the effect of outlier points, we limited all rates to 2 standard deviations from the mean.
In spite of our efforts (above) to improve species coverage, most alignments were missing at least one species. We set a minimum species threshold at 5, so species representation ranged from 5 to 12. This heterogeneity required us to create a flexible system to compare ERC results between different sets of species. A table of relative rates (projection operation, above) was produced for each unique set of species shared between protein pairs, resulting in 1,815 projections. Importantly, the distribution of ERC values varied depending on the particular set of species employed. For example, the variance of ERC values is consistently larger for smaller numbers of species (Figure S6). To correct for these effects we converted every observed ERC value in to an empirical p-value based on the observed distribution of ERC values for that particular set of species. The comparison of p-values allowed us to compare ERC results across all protein pairs. Hence, we report all ERC results as p-values ranging from 0 to 1, where a lower value indicates stronger evidence for rate correlation.
Significance testing for elevated ERC values in a set of proteins was performed using a proteome-wide permutation test (Figure S1A). The mean ERC value observed between all pairs in the tested set, such as the SP network, was compared to the mean ERC values of 10,000 sets of the same number of proteins randomly chosen from the entire proteome. A p-value for the tested set was computed as the proportion of random sets that had a mean ERC value equal to or greater than the tested set of proteins. Randomly chosen ERC values were taken from the same species-matched projections as in the observed set, which controlled for variation in ERC distributions due to different sets of species present in those genes.
The “reproductive protein only” analysis (Figure S1B–C) was performed as above, except that analysis was limited to the 756 Sfps, female proteins, and sperm proteins described above. We further limited this set to the 664 proteins that had detectable orthologs in at least 5 species. Significance testing for single pairs and for sets of proteins was performed as above, through empirical p-values. Calculations of pairwise correlations between pairs of known network proteins and between known network proteins and members of the sets of Sfps and female proteins were performed using this reproductive protein set.
RNA interference (RNAi)
To knock down expression of candidate genes, we used a variety of RNAi lines and drivers. Most lines were second-generation (KK) RNAi lines provided by the Vienna Drosophila RNAi Center (www.vdrc.at) [107]; several others were either provided by the Transgenic RNAi Project (TRiP; Harvard University) [108] or constructed in house using the pVALIUM20 vector [109], [110] provided by the TRiP. When possible, we used the tubulin-GAL4 driver to knockdown genes ubiquitously, but in some cases knockdown with this driver caused lethality. When ubiquitous knockdown of a male-expressed Sfp gene caused lethality, we first attempted to use the prd-GAL4 driver [111] to knockdown expression in the accessory glands. However, we observed phenotypes consistent with SP network malfunction when this driver was crossed to a control background strain that does not induce RNAi. Thus, we instead used the ovulin-GAL4 driver [17] to knock down male Sfp genes. To knockdown female genes expressed in the spermathecae, we used the Send1-GAL4 driver [112], sometimes in combination with a UAS-Dicer2 sequence to enhance RNA interference. The RNAi line numbers, specific crosses and genetic controls used are given in Tables S1 and S3. All flies were reared on a 12 hr/12 hr light-dark cycle. Most crosses were performed at room temperature (22°C±1°); some were instead performed at 25° to attempt to induce greater knockdown.
We determined the degree of knockdown by using RT-PCR [20], [28] to measure the expression level of each RNAi-targeted gene in knockdown flies and their respective controls, using amplification of the RpL32 transcript as a positive control (see Protocol S1 for further details). For tubulin-GAL4 knockdown, we analyzed RNA isolated from whole flies; for tissue-specific knockdown, we analyzed RNA isolated from dissected reproductive tracts. We qualitatively scored the degree of knockdown as “complete/near complete,” “partial,” or “no detectable knockdown”, and we chose for functional analyses only those genes (16 of 21 tested) that showed at least partial knockdown. Figure S7 shows knockdown levels for all positive candidates.
Screens for reproductive phenotypes
For several days after an initial mating, females are reluctant to remate in a one-hour, single-pair test, but only if the SP network is functioning properly [19], [20]. Thus, we initially screened each candidate gene for its effects on a female's willingness to remate within 1 hour, 4 days after an initial mating, using previously described methods [20]. Positive candidates were then evaluated by the same assay for remating receptivity at 1 day after mating, and for fertility, fecundity and egg hatchability over 10 days after an initial mating. These assays were performed according to previously described methods, with minor modifications. For more detail, see Protocol S1.
Confirmation of RNAi phenotypes
While all RNAi lines used above were designed to specifically minimize off-target effects [107], [108], we also confirmed that the phenotypes we observed were due specifically to the knockdown of the intended target. We first confirmed that all RNAi-triggering constructs had no predicted off-target effects against the most current D. melanogaster gene annotations [66]. We then tested an additional RNAi line for all genes for which such a line was available (antares, fra mauro, hadley and Esp). These tests controlled for either the insertion site of the RNAi-triggering construct or both the insertion site and the sequence of the RNAi-triggering construct, depending on which type of additional line was available. Details of these lines are given in Table S3. Finally, we note that our rate of positive hits in our screen (33 percent; 6 out of 18 ERC-identified candidates) is dramatically higher than previous estimates of RNAi effects on cell viability (maximum rate: 2.2 percent, including both true positive effects and potential off-targets) [113]. Thus, our results are unlikely to be due to off-target effects or general effects on cell viability.
Western blotting
To examine the production, transfer and processing of known SP network proteins in flies knocked down for a newly identified candidate, we performed Western blot experiments using available antibodies to SP, CG1652, CG1656, CG9997 and CG17575 as previously described [21]. For each positive candidate, we first tested whether SP was retained on sperm over the long term by dissecting 13–20 lower female reproductive tracts for each treatment at 4 days after the start of mating (ASM). While the number of female reproductive tracts per lane across experiments varied within this range, pairs of samples being compared never differed by more than 2 tracts. Extracted proteins were run on 15% acrylamide gels, transferred to membranes, and then probed for SP and alpha-tubulin (as a loading control) as previously described.
For candidates that caused a reduction of SP levels in females at 4 days ASM, we then evaluated the production, processing and transfer of the known network proteins by testing for their presence in male reproductive tracts and in mated females at 1 hr ASM. Proteins were separated on 10.6% acrylamide gels and then transferred and probed for as described previously. Approximately 0.5–1 male reproductive tract equivalents and 2–4 lower female reproductive tract equivalents were loaded in each lane. While the number of female reproductive tract equivalents per lane varied between blots for different SP network proteins, comparisons between knockdown and control flies for any given protein were performed with an equal number of reproductive tracts in each lane. As a loading control for each blot, we primarily used alpha-tubulin. In cases where CG1652 and CG1656 co-migrated with alpha-tubulin, we also examined a consistently observed cross-reactive band.
Supporting Information
Zdroje
1. PoianiA (2006) Complexity of seminal fluid: a review. Behavioral Ecology and Sociobiology 60 : 289–310.
2. Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF (2011) Insect seminal fluid proteins: Identification and function. In: Berenbaum MR, Carde RT, Robinson GE, editors. Annual Review of Entomology, Vol 56. pp. 21–40.
3. ChapmanT, DaviesSJ (2004) Functions and analysis of the seminal fluid proteins of male Drosophila melanogaster fruit flies. Peptides 25 : 1477–1490.
4. HerndonLA, WolfnerMF (1995) A Drosophila seminal fluid protein, Acp26Aa, stimulates egg-laying in females for 1 day after mating. Proceedings of the National Academy of Sciences of the United States of America 92 : 10114–10118.
5. SollerM, BownesM, KubliE (1997) Mating and sex peptide stimulate the accumulation of yolk in oocytes of Drosophila melanogaster. European Journal of Biochemistry 243 : 732–738.
6. SollerM, BownesM, KubliE (1999) Control of oocyte maturation in sexually mature Drosophila females. Developmental Biology 208 : 337–351.
7. AvilaFW, WolfnerMF (2009) Acp36DE is required for uterine conformational changes in mated Drosophila females. Proceedings of the National Academy of Sciences of the United States of America 106 : 15796–15800.
8. Bloch QaziMC, WolfnerMF (2003) An early role for the Drosophila melanogaster male seminal protein Acp36DE in female sperm storage. Journal of Experimental Biology 206 : 3521–3528.
9. NeubaumDM, WolfnerMF (1999) Mated Drosophila melanogaster females require a seminal fluid protein, Acp36DE, to store sperm efficiently. Genetics 153 : 845–857.
10. WongA, AlbrightSN, GiebelJD, Ravi RamK, JiS, et al. (2008) A role for Acp29AB, a predicted seminal fluid lectin, in female sperm storage in Drosophila melanogaster. Genetics 180 : 921–931.
11. PengJ, ZipperlenP, KubliE (2005) Drosophila sex peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Current Biology 15 : 1690–1694.
12. ShortSM, LazzaroBP (2010) Female and male genetic contributions to post-mating immune defence in female Drosophila melanogaster. Proceedings of the Royal Society B-Biological Sciences 277 : 3649–3657.
13. CarvalhoGB, KapahiP, AndersonDJ, BenzerS (2006) Allocrine modulation of feeding behavior by the sex peptide of Drosophila. Current Biology 16 : 692–696.
14. CognigniP, BaileyAP, Miguel-AliagaI (2011) Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell Metabolism 13 : 92–104.
15. IsaacRE, LiC, LeedaleAE, ShirrasAD (2010) Drosophila male sex peptide inhibits siesta sleep and promotes locomotor activity in the post-mated female. Proceedings of the Royal Society B-Biological Sciences 277 : 65–70.
16. Apger-McGlaughonJ, WolfnerMF (2013) Post-mating change in excretion by mated Drosophila melanogaster females is a long-term response that depends on sex peptide and sperm. Journal of Insect Physiology 59 : 1024–30.
17. ChapmanT, BanghamJ, VintiG, SeifriedB, LungO, et al. (2003) The sex peptide of Drosophila melanogaster: Female post-mating responses analyzed by using RNA interference. Proceedings of the National Academy of Sciences of the United States of America 100 : 9923–9928.
18. LiuHF, KubliE (2003) Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America 100 : 9929–9933.
19. PengJ, ChenS, BusserS, LiuHF, HoneggerT, et al. (2005) Gradual release of sperm bound sex-peptide controls female postmating behavior in Drosophila. Current Biology 15 : 207–213.
20. Ravi RamK, WolfnerMF (2007) Sustained post-mating response in D. melanogaster requires multiple seminal fluid proteins. Plos Genetics 3: e238.
21. Ravi RamK, WolfnerMF (2009) A network of interactions among seminal proteins underlies the long-term postmating response in Drosophila. Proceedings of the National Academy of Sciences of the United States of America 106 : 15384–15389.
22. Ravi RamK, JiS, WolfnerMF (2005) Fates and targets of male accessory gland proteins in mated female Drosophila melanogaster. Insect Biochemistry and Molecular Biology 35 : 1059–1071.
23. HasemeyerM, YapiciN, HeberleinU, DicksonBJ (2009) Sensory neurons in the Drosophila genital tract regulate female reproductive behavior. Neuron 61 : 511–518.
24. RezavalC, PavlouHJ, DornanAJ, ChanY-B, KravitzEA, et al. (2012) Neural circuitry underlying Drosophila female postmating behavioral responses. Current Biology 22 : 1155–1165.
25. YangCH, RumpfS, XiangY, GordonMD, SongW, et al. (2009) Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron 61 : 519–526.
26. YapiciN, KimYJ, RibeiroC, DicksonBJ (2008) A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature 451 : 33–U31.
27. AvilaFW, RamKR, QaziMCB, WolfnerMF (2010) Sex peptide is required for the efficient release of stored sperm in mated Drosophila females. Genetics 186 : 595–600.
28. LaFlammeBA, RamKR, WolfnerMF (2012) The Drosophila melanogaster seminal fluid protease “seminase” regulates proteolytic and post-mating reproductive processes. Plos Genetics 8: e1002435 doi:10.1371/journal.pgen.1002435
29. MuellerJL, LinklaterJR, Ravi RamK, ChapmanT, WolfnerMF (2008) Targeted gene deletion and phenotypic analysis of the Drosophila melanogaster seminal fluid protease inhibitor Acp62F. Genetics 178 : 1605–1614.
30. DorusS, BusbySA, GerikeU, ShabanowitzJ, HuntDF, et al. (2006) Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nature Genetics 38 : 1440–1445.
31. WasbroughER, DorusS, HesterS, Howard-MurkinJ, LilleyK, et al. (2010) The Drosophila melanogaster sperm proteome-II (DmSP-II). Journal of Proteomics 73 : 2171–2185.
32. FindlayGD, MacCossMJ, SwansonWJ (2009) Proteomic discovery of previously unannotated, rapidly evolving seminal fluid genes in Drosophila. Genome Research 19 : 886–896.
33. FindlayGD, YiX, MacCossMJ, SwansonWJ (2008) Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. Plos Biology 6 : 1417–1426.
34. Ravi RamK, WolfnerMF (2007) Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduuction. Integrative and Comparative Biology 47 : 427–445.
35. SwansonWJ, ClarkAG, Waldrip-DailHM, WolfnerMF, AquadroCF (2001) Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proceedings of the National Academy of Sciences of the United States of America 98 : 7375–7379.
36. AllenAK, SpradlingAC (2008) The Sf1-related nuclear hormone receptor Hr39 regulates Drosophila female reproductive tract development and function. Development 135 : 311–321.
37. ArbeitmanMN, FlemingAA, SiegalML, NullBH, BakerBS (2004) A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development 131 : 2007–2021.
38. ProkupekA, HoffmannF, EyunSI, MoriyamaE, ZhouM, et al. (2008) An evolutionary expressed sequence tag analysis of Drosophila spermatheca genes. Evolution 62 : 2936–2947.
39. ProkupekAM, EyunSI, KoL, MoriyamaEN, HarshmanLG (2010) Molecular evolutionary analysis of seminal receptacle sperm storage organ genes of Drosophila melanogaster. Journal of Evolutionary Biology 23 : 1386–1398.
40. ProkupekAM, KachmanSD, LadungaI, HarshmanLG (2009) Transcriptional profiling of the sperm storage organs of Drosophila melanogaster. Insect Molecular Biology 18 : 465–475.
41. ClarkNL, AlaniE, AquadroCF (2012) Evolutionary rate covariation reveals shared functionality and coexpression of genes. Genome Research 22 : 714–720.
42. DrummondDA, RavalA, WilkeCO (2006) A single determinant dominates the rate of yeast protein evolution. Molecular Biology and Evolution 23 : 327–337.
43. LarracuenteAM, SacktonTB, GreenbergAJ, WongA, SinghND, et al. (2008) Evolution of protein-coding genes in Drosophila. Trends in Genetics 24 : 114–123.
44. LiaoB-Y, ScottNM, ZhangJ (2006) Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Molecular Biology and Evolution 23 : 2072–2080.
45. McInerneyJO (2006) The causes of protein evolutionary rate variation. Trends in Ecology & Evolution 21 : 230–232.
46. MintserisJ, WengZP (2005) Structure, function, and evolution of transient and obligate protein-protein interactions. Proceedings of the National Academy of Sciences of the United States of America 102 : 10930–10935.
47. PalC, PappB, HurstLD (2001) Highly expressed genes in yeast evolve slowly. Genetics 158 : 927–931.
48. PalC, PappB, LercherMJ (2006) An integrated view of protein evolution. Nature Reviews Genetics 7 : 337–348.
49. RochaEPC, DanchinA (2004) An analysis of determinants of amino acids substitution rates in bacterial proteins. Molecular Biology and Evolution 21 : 108–116.
50. ClarkNL, GasperJ, SekinoM, SpringerSA, AquadroCF, et al. (2009) Coevolution of interacting fertilization proteins. Plos Genetics 5: e1000570.
51. HakesL, LovellSC, OliverSG, RobertsonDL (2007) Specificity in protein interactions and its relationship with sequence diversity and coevolution. Proceedings of the National Academy of Sciences of the United States of America 104 : 7999–8004.
52. KannMG, ShoemakerBA, PanchenkoAR, PrzytyckaTM (2009) Correlated evolution of interacting proteins: Looking behind the mirrortree. Journal of Molecular Biology 385 : 91–98.
53. LovellSC, RobertsonDL (2010) An integrated view of molecular coevolution in protein-protein interactions. Molecular Biology and Evolution 27 : 2567–2575.
54. ClarkNL, AlaniE, AquadroCF (2013) Evolutionary rate covariation in meiotic proteins results from fluctuating evolutionary pressure in yeasts and mammals. Genetics 193 : 529–538.
55. ClarkNL, AquadroCF (2010) A novel method to detect proteins evolving at correlated rates: identifying new functional relationships between coevolving proteins. Molecular Biology and Evolution 27 : 1152–1161.
56. GohCS, CohenFE (2002) Co-evolutionary analysis reveals insights into protein-protein interactions. Journal of Molecular Biology 324 : 177–192.
57. PazosF, ValenciaA (2001) Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Engineering 14 : 609–614.
58. SatoT, YamanishiY, KanehisaM, TohH (2005) The inference of protein-protein interactions by co-evolutionary analysis is improved by excluding the information about the phylogenetic relationships. Bioinformatics 21 : 3482–3489.
59. JuanD, PazosF, ValenciaA (2008) High-confidence prediction of global interactomes based on genome-wide coevolutionary networks. Proceedings of the National Academy of Sciences of the United States of America 105 : 934–939.
60. TabachY, BilliAC, HayesGD, NewmanMA, ZukO, et al. (2013) Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature 493 : 694–698.
61. ConsortiumDG (2007) Evolution of genes and genomes on the Drosophila phylogeny. Nature 450 : 203–218.
62. KimYJ, BartalskaK, AudsleyN, YamanakaN, YapiciN, et al. (2010) MIPs are ancestral ligands for the sex peptide receptor. Proceedings of the National Academy of Sciences of the United States of America 107 : 6520–6525.
63. PoelsJ, Van LoyT, VandersmissenHP, Van HielB, Van SoestS, et al. (2010) Myoinhibiting peptides are the ancestral ligands of the promiscuous Drosophila sex peptide receptor. Cellular and Molecular Life Sciences 67 : 3511–3522.
64. ChintapalliVR, WangJ, DowJAT (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nature Genetics 39 : 715–720.
65. ArnoneJT, Robbins-PiankaA, AraceJR, Kass-GergiS, McAlearMA (2012) The adjacent positioning of co-regulated gene pairs is widely conserved across eukaryotes. BMC Genomics 13 : 546 doi:10.1186/1471-2164-13-546
66. HuY, RoeselC, FlockhartI, PerkinsL, PerrimonN, et al. (2013) UP-TORR: Online tool for accurate and up-to-date annotation of RNAi reagents. Genetics 195 : 37–45.
67. DeanMD, ClarkNL, FindlayGD, KarnRC, YiXH, et al. (2009) Proteomics and comparative genomic investigations reveal heterogeneity in evolutionary rate of male reproductive proteins in mice (Mus domesticus). Molecular Biology and Evolution 26 : 1733–1743.
68. PilchB, MannM (2005) Large scale proteomic analysis of human seminal plasma. Molecular & Cellular Proteomics 4: S205–S205.
69. SirotLK, HardstoneMC, HelinskiMEH, RibeiroJMC, KimuraM, et al. (2011) Towards a semen proteome of the dengue vector mosquito: Protein identification and potential functions. Plos Neglected Tropical Diseases 5: e989 doi:10.1371/journal.pntd.0000989
70. RossJ, JiangH, KanostMR, WangY (2003) Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationships. Gene 304 : 117–131.
71. CarpentierM, GuillemetteC, BaileyJL, BoileauG, JeannotteL, et al. (2004) Reduced fertility in male mice deficient in the zinc metallopeptidase NL1. Molecular and Cellular Biology 24 : 4428–4437.
72. PintadoCO, PintoFM, PennefatherJN, HidalgoA, BaamondeA, et al. (2003) A role for tachykinins in female mouse and rat reproductive function. Biology of Reproduction 69 : 940–946.
73. PintoFM, ArmestoCP, MagranerJ, TrujilloM, MartinJD, et al. (1999) Tachykinin receptor and neutral endopeptidase gene expression in the rat uterus: Characterization and regulation in response to ovarian steroid treatment. Endocrinology 140 : 2526–2532.
74. PetersenTN, BrunakS, von HeijneG, NielsenH (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8 : 785–786.
75. Marchler-BauerA, LuS, AndersonJB, ChitsazF, DerbyshireMK, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Research 39: D225–D229.
76. KelleyLA, SternbergMJE (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols 4 : 363–371.
77. RocheN, KaufmanT (1996) Characterization of the homoeotic target gene Epidermal stripes and patches, a Drosophila homologue of the human diastrophic dysplasia gene. Molecular Biology of the Cell 7 : 699–699.
78. BassettAR, TibbitC, PontingCP, LiuJ-L (2013) Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Reports 4 : 220–228.
79. GratzSJ, CummingsAM, NguyenJN, HammDC, DonohueLK, et al. (2013) Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194 : 1029–1035.
80. ChanHC, RuanYC, HeQ, ChenMH, ChenH, et al. (2009) The cystic fibrosis transmembrane conductance regulator in reproductive health and disease. Journal of Physiology 587 : 2187–2195.
81. LaFlammeBA, WolfnerMF (2013) Identification and function of proteolysis regulators in seminal fluid. Molecular Reproduction and Development 80 : 80–101.
82. Ravi RamK, SirotLK, WolfnerMF (2006) Predicted seminal astacin-like protease is required for processing of reproductive proteins in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America 103 : 18674–18679.
83. SmithJR, StanfieldGM (2011) TRY-5 Is a sperm-activating protease in Caenorhabditis elegans aeminal fluid. Plos Genetics 7: e1002375 doi:10.1371/journal.pgen.1002375
84. StanfieldGM, VilleneuveAM (2006) Regulation of sperm activation by SWM-1 is required for reproductive success of C. elegans males. Current Biology 16 : 252–263.
85. ZhaoY, SunW, ZhangP, ChiH, ZhangM-J, et al. (2012) Nematode sperm maturation triggered by protease involves sperm-secreted serine protease inhibitor (Serpin). Proceedings of the National Academy of Sciences of the United States of America 109 : 1542–1547.
86. LungO, WolfnerMF (2001) Identification and characterization of the major Drosophila melanogaster mating plug protein. Insect Biochemistry and Molecular Biology 31 : 543–551.
87. GligorovD, SitnikJL, MaedaRK, WolfnerMF, KarchF (2013) A Novel Function for the Hox Gene Abd-B in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in Drosophila. Plos Genetics 9: e1003395 doi:10.1371/journal.pgen.1003395
88. LeiblichA, MarsdenL, GandyC, CorriganL, JenkinsR, et al. (2012) Bone morphogenetic protein - and mating-dependent secretory cell growth and migration in the Drosophila accessory gland. Proceedings of the National Academy of Sciences of the United States of America 109 : 19292–19297.
89. ChenPS, Stumm-ZollingerE, AigakiT, BalmerJ, BienzM, et al. (1988) A male accessory gland peptide that regulates reproductive behavior of female Drosophila melanogaster. Cell 54 : 291–298.
90. HaninO, AzrielliA, ZakinV, ApplebaumS, RafaeliA (2011) Identification and differential expression of a sex-peptide receptor in Helicoverpa armigera. Insect Biochemistry and Molecular Biology 41 : 537–544.
91. DottoriniT, NicolaidesL, RansonH, RogersDW, CrisantiA, et al. (2007) A genome-wide analysis in Anopheles gambiae mosquitoes reveals 46 male accessory gland genes, possible modulators of female behavior. Proceedings of the National Academy of Sciences of the United States of America 104 : 16215–16220.
92. MarkowTA (1996) Evolution of Drosophila mating systems. Evolutionary Biology, Vol 29 29 : 73–106.
93. ThailayilJ, MagnussonK, GodfrayHCJ, CrisantiA, CatterucciaF (2011) Spermless males elicit large-scale female responses to mating in the malaria mosquito Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America 108 : 13677–13681.
94. MarkowTA, O'GradyPM (2005) Evolutionary genetics of reproductive behavior in Drosophila: connecting the dots. Annual Review of Genetics 39 : 263–291.
95. ClarkNL, AagaardJE, SwansonWJ (2006) Evolution of reproductive proteins from animals and plants. Reproduction 131 : 11–22.
96. FindlayGD, SwansonWJ (2010) Proteomics enhances evolutionary and functional analysis of reproductive proteins. BioEssays 32 : 26–36.
97. TurnerLM, HoekstraHE (2008) Causes and consequences of the evolution of reproductive proteins. International Journal of Developmental Biology 52 : 769–780.
98. KuijperB, StewartAD, RiceWR (2006) The cost of mating rises nonlinearly with copulation frequency in a laboratory population of Drosophila melanogaster. Journal of Evolutionary Biology 19 : 1795–1802.
99. RiceWR (1996) Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature 381 : 232–234.
100. EdwardDA, FrickeC, GerrardDT, ChapmanT (2011) Quantifying the life-history response to increased male exposure in female Drosophila melanogaster. Evolution 65 : 564–573.
101. WigbyS, ChapmanT (2005) Sex peptide causes mating costs in female Drosophila melanogaster. Current Biology 15 : 316–321.
102. CireraS, AguadeM (1997) Evolutionary history of the sex peptide (Acp70A) gene region in Drosophila melanogaster. Genetics 147 : 189–197.
103. WongA, TurchinMC, WolfnerMF, AquadroCF (2008) Evidence for positive selection on Drosophila melanogaster seminal fluid protease homologs. Molecular Biology and Evolution 25 : 497–506.
104. OstlundG, SchmittT, ForslundK, KostlerT, MessinaDN, et al. (2010) InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Research 38: D196–D203.
105. EdgarRC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32 : 1792–1797.
106. YangZ (2007) PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24 : 1586–1591.
107. DietzlG, ChenD, SchnorrerF, SuKC, BarinovaY, et al. (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 : 151–U151.
108. NiJ-Q, ZhouR, CzechB, LiuL-P, HolderbaumL, et al. (2011) A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nature Methods 8 : 405–U446.
109. NiJ-Q, LiuL-P, BinariR, HardyR, ShimH-S, et al. (2009) A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182 : 1089–1100.
110. NiJ-Q, MarksteinM, BinariR, PfeifferB, LiuL-P, et al. (2008) Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nature Methods 5 : 49–51.
111. XueL, NollM (2002) Dual role of the Pax gene paired in accessory gland development of Drosophila. Development 129 : 339–346.
112. SchnakenbergSL, MatiasWR, SiegalML (2011) Sperm-storage defects and live birth in Drosophila females lacking spermathecal secretory cells. Plos Biology 9: e1001192 doi:10.1371/journal.pbio.1001192
113. BoutrosM, KigerAA, ArmknechtS, KerrK, HildM, et al. (2004) Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303 : 832–835.
Štítky
Genetika Reprodukční medicína
Článek Unwrapping BacteriaČlánek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 1
-
Všechny články tohoto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



