-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaIs Non-Homologous End-Joining Really an Inherently Error-Prone Process?
DNA double-strand breaks (DSBs) are harmful lesions leading to genomic instability or diversity. Non-homologous end-joining (NHEJ) is a prominent DSB repair pathway, which has long been considered to be error-prone. However, recent data have pointed to the intrinsic precision of NHEJ. Three reasons can account for the apparent fallibility of NHEJ: 1) the existence of a highly error-prone alternative end-joining process; 2) the adaptability of canonical C-NHEJ (Ku - and Xrcc4/ligase IV–dependent) to imperfect complementary ends; and 3) the requirement to first process chemically incompatible DNA ends that cannot be ligated directly. Thus, C-NHEJ is conservative but adaptable, and the accuracy of the repair is dictated by the structure of the DNA ends rather than by the C-NHEJ machinery. We present data from different organisms that describe the conservative/versatile properties of C-NHEJ. The advantages of the adaptability/versatility of C-NHEJ are discussed for the development of the immune repertoire and the resistance to ionizing radiation, especially at low doses, and for targeted genome manipulation.
Published in the journal: . PLoS Genet 10(1): e32767. doi:10.1371/journal.pgen.1004086
Category: Review
doi: https://doi.org/10.1371/journal.pgen.1004086Summary
DNA double-strand breaks (DSBs) are harmful lesions leading to genomic instability or diversity. Non-homologous end-joining (NHEJ) is a prominent DSB repair pathway, which has long been considered to be error-prone. However, recent data have pointed to the intrinsic precision of NHEJ. Three reasons can account for the apparent fallibility of NHEJ: 1) the existence of a highly error-prone alternative end-joining process; 2) the adaptability of canonical C-NHEJ (Ku - and Xrcc4/ligase IV–dependent) to imperfect complementary ends; and 3) the requirement to first process chemically incompatible DNA ends that cannot be ligated directly. Thus, C-NHEJ is conservative but adaptable, and the accuracy of the repair is dictated by the structure of the DNA ends rather than by the C-NHEJ machinery. We present data from different organisms that describe the conservative/versatile properties of C-NHEJ. The advantages of the adaptability/versatility of C-NHEJ are discussed for the development of the immune repertoire and the resistance to ionizing radiation, especially at low doses, and for targeted genome manipulation.
DNA double-strand breaks (DSBs) are highly toxic lesions. However, in certain essential physiological processes, DSBs are used to promote genetic diversity. Programmed DSBs generated by cellular enzymes are repaired by the same mechanisms as those used for stress-induced DSBs. Thus, DSB repair stands at the crossroads between genetic variability and instability.
DSB repair uses two primary strategies: non-homologous end-joining (NHEJ), which is generally considered to be error-prone, and homologous recombination (HR), which is considered to be error-free. However, this view is too simplistic. Herein, we discuss several pieces of data that challenge the fallibility of NHEJ.
Canonical NHEJ versus Alternative End-Joining
The canonical C-NHEJ pathway joins double-strand DNA ends in a Ku - and Xrcc4/ligase IV–dependent manner. This pathway has been extensively described and is summarized in Figure 1A.
Fig. 1. End-joining models and competition between C-NHEJ and A-EJ for DSB repair. 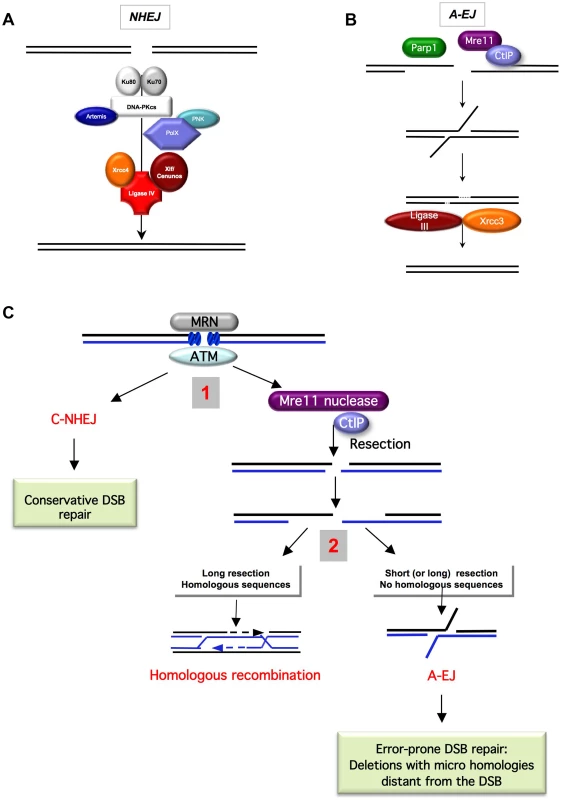
A) The canonical C-NHEJ. The heterodimer Ku80-Ku70 binds to the DNA ends, which then recruit DNA-PKcs. Note that DNA-PK is absent from yeast. Several proteins, including Artemis, the polynucleotide kinase (PNK), and members of the polymerase X family, process the DNA ends for subsequent steps [86]–[90]. In the last step, ligase IV, associated with its co-factors Xrcc4 and Cernunos/Xlf, joins the ends [91]–[93]. B) A-EJ. Parp1 plays a role in the initiation process [4], [17], [94], [95]. Without the protection by Ku70/Ku80, the DNA ends are resected in a reaction favored by the nuclease activity of Mre11 and CtIP [11], [13]. It has been proposed that a single-strand DNA resection reveals complementary microhomologies (two to four nt or more) that can anneal; gap filling completes the end-joining. Subsequently, Xrcc1 and ligase III (which can be substituted by ligase I) complete A-EJ [4], [9], [38]. A-EJ is always associated with deletions at the junctions and frequently (but not systematically) involves microhomologies that are distant from the DSB. The histone H1 has also been shown to act in A-EJ [96]. C) Two-step model for the choice of the DSB repair pathway [3], [11]. The MRN complex and ATM are involved in the early steps of DSB signaling and can activate both C-NHEJ and A-EJ. 1) Binding of Ku80/Ku70 protects from ssDNA resection, leading to a conservative DSB repair outcome through C-NHEJ. The nuclease activity of Mre11 and CtIP can initiate ssDNA resection. 2) A short ssDNA resection allows A-EJ but not homologous recombination. A long ssDNA resection allows A-EJ and HR, but HR requires the presence of homologous sequences. A-EJ results in error-prone repair associated with deletions at the repair junctions with frequent use of microhomologies distant from the DSB. The existence of alternative end-joining pathways has been recently reported (Figure 1B). This alternative end-joining process, which can be unmasked in the absence of functional C-NHEJ genes, is referred to as A-EJ or alt-NHEJ (alternative end-joining), B-NHEJ (backup NHEJ), and MMEJ (microhomology-mediated end-joining) [1]–[11]. Herein, to clearly distinguish it from C-NHEJ and because some repair events do not use microhomologies, it will be referred to as A-EJ. A-EJ is far from being fully characterized and might correspond to different molecular processes [12], but the common points are that it does not require extended sequence homologies, is independent of Ku80 or Xrcc4, and is associated with deletions at the repair junctions, frequently (but not systematically) using microhomologies distant from the DSB. This signature led to the model in Figure 1B, which proposes that A-EJ is initiated by a single-stranded (ssDNA) resection. Consistent with this view are the involvement of the nuclease activities of MRE11 and CtIP/Sae2 [11], [13], [14] and the fact that 53BP1, in association with RIF1 and BLM, protects against long deletions at the A-EJ repair junctions [15]. Consequently, A-EJ is highly mutagenic, typically generating deletions at the repair junction. Because HR is also initiated by a ssDNA resection, a two-step model has been proposed for the choice of the DSB repair pathway [3], [11]. The first alternative is the choice between C-NHEJ and the initiation of the resection; the second alternative is HR versus A-EJ (Figure 1C). Consistent with the first alternative, Ku represses both HR and A-EJ [1], [2], [7], [8], [16], [17]. A defect in Ku leads to extended DNA degradation at the DSBs and to increased deletion sizes at the A-EJ junctions [2], [6], [18]–[20]. Note that a defect in Ku does not significantly decrease, whereas the absence of Xrcc4 leads to a strong decrease in the total efficiency of end-joining [1], [21]. In both cases, the remaining events exhibit the signature of A-EJ at the repair junction (deletions). This shows that the absence of Ku is compensated by A-EJ. In the absence of Xrcc4, Ku is still present and able to repress A-EJ, thus independently of the late steps of C-NHEJ. These data support the concept that Ku protects against initiation of A-EJ. Because A-EJ is exclusively mutagenic, Ku favors the maintenance of genetic stability.
Several parameters affect the second choice, such as the presence of a homologous sequence. Moreover, long resections are required for HR (hundreds of nucleotides), whereas short resections (a few tens of nucleotides) are sufficient for A-EJ, as estimated by the deletion sizes at the repair junctions. Nevertheless, long deletions can also lead to A-EJ. The cell cycle can also affect the DSB repair pathway choice; HR is only active in the S and G2 phases [22]–[26], whereas both C-NHEJ and A-EJ are active throughout the cell cycle [23], [24], but A-EJ is more active in the S phase [23].
C-NHEJ Is a Conservative but Versatile DSB Repair Process
Genetic instability can be evaluated at two levels: at the chromosome level or at the nucleotide level, at the DSB repair scar.
At the Chromosome Level
C-NHEJ can be involved in translocations and rearrangements [2], [27] and in programmed rearrangements (generating the immune repertoire). Whole genome sequencing of tumors has revealed complex inter - and intra-chromosomal rearrangements in a phenomenon named chromothripsis. Both C-NHEJ and A-EJ have been proposed to be involved in chromothripsis, but they cannot account for events involving sequence duplication (for review see [28], [29]).
Nevertheless, a defect in Ku or Xrcc4/lig IV leads to profound genome rearrangements, underlying the fact that NHEJ is essential for the maintenance of genomic stability [10], [30]–[33]. Additionally, NHEJ prevents trinucleotide repeat fragility and expansion [34]. Conversely, A-EJ is involved in chromosome translocation in mouse cells, Drosophila, and yeast cells [35]–[37]. Particularly, both CtIP and ligase III have been shown to be involved in translocations by A-EJ [38], [39].
The mobility of the DNA ends is a prerequisite to generate profound genome rearrangements. Remarkably, Ku80 protects broken DNA ends against mobility within the nucleus [40]. In addition, atomic force and electron microscopy studies have shown that Ku, in conjunction with DNA-PKcs, tethers DNA ends in vitro [41], [42], maintaining them in close proximity. Ku-mediated tethering could account for the protective role of Ku80 against the mobility of DNA ends and consequently against translocations. In addition, increased mobility of DSBs has been associated with DNA end resection in yeast, thus favoring the search for homology during HR [43], [44]. Because A-EJ is also initiated by DNA end resection, DSB mobility might also increase the risk of chromosome rearrangements promoted by A-EJ. Because Ku impairs both DSB mobility and DNA end resection, it likely plays a doubly protective role against chromosome rearrangements.
At the Nucleotide Level, at the Repair Scar
At the repair junctions, the apparent infidelity of end-joining should be reevaluated because, in many studies, A-EJ was not distinguishable from C-NHEJ. In addition, the repair of DSBs induced by ionizing radiation (IR) or V(D)J recombination requires processing of the DNA ends prior to ligation. Thus, it can be argued that mutagenesis is generated by DNA end processing rather than by the end-joining machinery per se. Thus using biological systems that do not require DNA end processing is necessary to address the question of the actual accuracy of C-NHEJ.
The end-joining accuracy of directly ligatable DNA ends
We will first discuss NHEJ in two biological models, Paramecium and mammalian cells, in which this pathway is of particular importance. Paramecium provides a physiological example of the efficient contribution of C-NHEJ to the precise repair of thousands of developmentally programmed DSBs [45]. In mammalian cells, C-NHEJ is a prominent DSB repair mechanism, and it is essential in fundamental processes establishing the immune repertoire. The accuracy of NHEJ will then be addressed in yeast, bacteria, and plants, and during cut-and-paste transposition.
Similar to other ciliates, Paramecium harbors two different nuclei in its cytoplasm. During vegetative growth, the diploid micronucleus (MIC) divides through mitosis but remains transcriptionally silent, whereas the highly polyploid macronucleus (MAC) ensures gene expression. During sexual processes, the MAC is fragmented and eventually lost. Subsequent divisions of the zygotic nucleus produce the new MICs and MACs of the next sexual generation (Figure 2A). During the MAC development, the germline genome is amplified to a final ploidy of ∼800 n. Concomitantly, massive genome rearrangements occur [46]: i) repeated sequences, including transposons or minisatellites, are eliminated in a heterogeneous manner and ii) at least 45,000 short, non-coding intervening sequences, the IESs (Internal Eliminated Sequences), are excised [47] (Figure 2B). IESs excision generates one chromosomal DSB every 1–2 kb within a defined time window [48]. Thus, because endoduplication occurs during rearrangements, an estimated 106 DSBs must be repaired in each developing MAC [49]. Despite this huge number, DSB repair preserves the linear organization of the MAC chromosomes. The highly precise repair of the IES excision sites occurs through the C-NHEJ pathway, as evidenced by the absolute requirement for ligase IV and Xrcc4 [50], but requires limited processing of DSBs (Figure 2C). Because 47% of the genes are interrupted by at least one IES in the MIC [47], the precision of end-joining is essential for the recovery of functional genes in the new MAC and, therefore, for cell survival.
Fig. 2. End-joining accuracy of ligation-compatible ends. 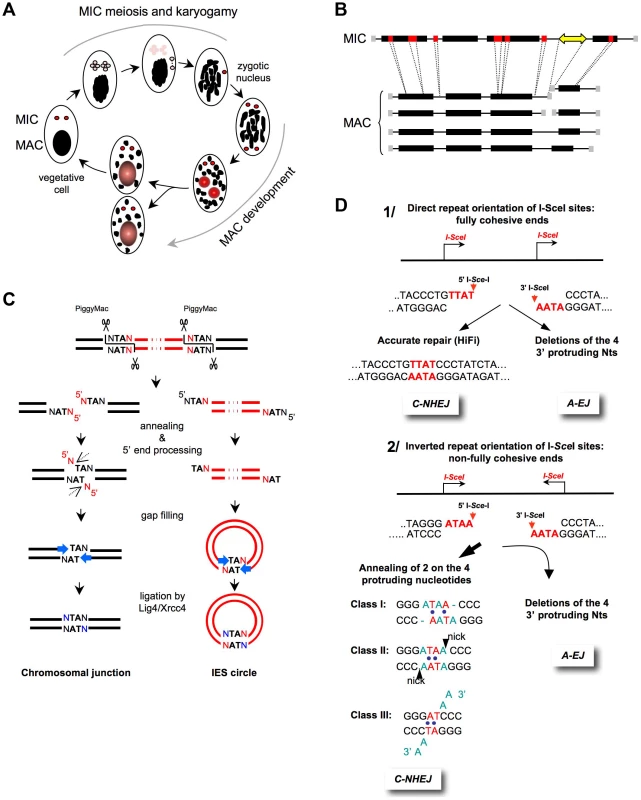
A) The Paramecium sexual cycle. Two types of sexual processes are induced through starvation in Paramecium: autogamy, a self-fertilization process (shown in the figure), and conjugation between compatible mating types (not shown). During autogamy, the two germline diploid MICs (red) undergo meiosis to generate eight haploid nuclei (pink), and a single nucleus migrates to a specialized cell compartment, dividing once to produce two identical gametic nuclei. The remaining seven meiotic products are degraded, and the old MAC (black) becomes fragmented. During karyogamy, the two gametic nuclei fuse to form a diploid zygotic nucleus. The zygotic nucleus subsequently undergoes two successive mitotic divisions; after the second division, the two nuclei become the new MICs of the sexual progeny (red), whereas the other two differentiate into new developing MACs (red and gray) and undergo programmed genomic rearrangements. At the first cell division, the new MICs divide mitotically, and each of the two developing new MACs segregates into a daughter cell where it continues to amplify the rearranged genome to a final ploidy of ∼800 n. During conjugation, MIC meiosis is triggered through the mating of two compatible sexual partners, which undergo a reciprocal exchange of their haploid gametic nuclei. Consequently, the zygotic nucleus in each partner is formed through the fusion of a resident and a migratory haploid nucleus. Exconjugants separate between the first and second divisions of the zygotic nucleus, and MAC development occurs as described for autogamous cells. B) General structure of MIC and MAC chromosomes. On the MIC chromosomes, genes (black boxes) and non-coding regions (thin lines) are interrupted by short internal eliminated sequences (IESs in red). Repeated germline sequences (e.g., transposons and minisatellites) are indicated with a yellow double-headed arrow. During MAC development, each MIC chromosome is amplified ∼400-fold to generate a population of heterogeneous MAC chromosomes. The imprecise elimination of repeated DNA is associated with the following alternative rearrangements: i) chromosome fragmentation and telomere addition to new MAC chromosome ends (gray squares) and ii) imprecise joining of the two chromosome arms that flank the eliminated germline region. C) Mechanism of IES excision. The successive DNA intermediates formed during IES excision are displayed, with IESs shown in red and flanking MAC-destined DNA shown in black. The first step of the reaction is the introduction of 4-base staggered double-strand breaks at each IES end, depending upon the PiggyMac domesticated transposase. The molecular steps that lead to the repair of the chromosomal junction are shown on the left, which might occur within a paired-end intermediate through the annealing of the central TAs within each 5′ overhang. The removal of the 5′-terminal nucleotide was demonstrated in vivo (dotted arrow), but the nuclease(s) involved has not been identified. For the 3′-processing step, ligase IV (Lig4) recruits or activates a gap-filling DNA polymerase, which adds one nucleotide to the recessive 3′-end prior to the final ligation. A similar mechanism has been proposed for the circularization of excised linear IES molecules (right), provided that these molecules are sufficiently long. IES circles do not replicate and are actively degraded. D) End-joining of fully versus non–fully complementary ends. 1) I-SceI sites in direct orientation (arrows). The cleavage generates 3′-overhangs (red nt), which are fully complementary. C-NHEJ promotes accurate ligation (left panel), and A-EJ deletes the four protruding nucleotides, leading to the deletion of at least 4 bp at the resealed junction (right panel) [1], [2], [11]. 2) I-SceI sites in an inverted orientation (arrows). The cleavage generates 3′overhangs (red nt), which are not fully complementary. Similarly, A-EJ deletes the 3′-protruding nt, resulting in the deletion of at least 4 bp at the resealed junction (left panel). C-NHEJ anneals two of the four protruding nt (red nt), according to three classes of events (right panel). This imperfect annealing generates gaps (in blue in class I), mismatches (in blue in classes I and II), or 3′-single-stranded tails (in blue in class III) [1], [2], [11]. In mammalian cells, different studies analyzing the end-joining of plasmids that are cleaved by restriction endonucleases in either acellular extracts or in transfected cells have all concluded that NHEJ is accurate [6], [51]–[53]. Defects in any C-NHEJ component resulted in error-prone end-joining [6], [52], [53], corroborating that mutagenic end-joining results, at least in part, from A-EJ. However, in these studies, the DSB repair was not monitored in a chromosomal context. Therefore, different systems, based on the use of intrachromosomal substrates containing cleavage sites for the meganuclease I-SceI, have been studied. Notably, these experiments facilitated the characterization of A-EJ at a precise molecular level in the context of chromosomes in living cells [1], [2], [11], [21], [23], [54], [55]. The conclusions drawn from these studies (see below) were confirmed in vivo in mice in the context of physiological processes, such as class switch or V(D)J recombination [5], [10].
Because the I-SceI cleavage site is not palindromic, the use of two cis sites (Figure 2D.1) has been informative [2]; with the sites in direct orientation, the I-SceI–mediated cleavage generates two fully complementary ends that can be readily ligated, whereas in the inverted orientation, only partially complementary ends are generated (Figure 2B). Note that in these cases, the DNA ends are not chemically modified and, thus, are competent for the ligation machinery; the differences arise from the annealing of the four protruding nucleotides, which are fully complementary or not. In the latter case, end-joining cannot restore the initial sequence, generating an apparently mutagenic event. Notably, the efficiency of the joining of imperfectly complementary ends is similar to that of fully complementary ends, underlying the adaptation capabilities of NHEJ [2].
With fully complementary ends (I-SceI sites in direct orientation), an error-free event restores one I-SceI cleavage site. Thus, with these substrates, the frequency of error-free end-joining is underestimated because residual I-SceI protein can re-cleave the repaired junction, increasing the possibility for error-prone repair and introducing a bias in favor of inaccurate repair. The frequency of error-free events in wild-type cells consistently varies from 35% to 75%, according to the level of I-SceI expression and the half-life of the expressed I-SceI protein [1], [2], [11]. Nevertheless, the high frequency of error-free events (up to 75%, which is likely underestimated) shows that C-NHEJ should not be primarily error-prone in mammalian cells. Deficiencies in Ku80 or Xrcc4 abolish error-free events, showing that accurate end-joining events result from C-NHEJ. The remaining end-joining events (i.e., A-EJ) correspond to deletions at the junctions, with the frequent use of microhomologies distant from the DSB site [1], [2], [11]. Mutagenic events exhibit a similar signature in wild-type cells, suggesting that they result from A-EJ and, thus, that C-NHEJ is not responsible for error-prone DSB repair.
With non–fully complementary ends, end-joining cannot restore a cleavable I-SceI site; therefore, this substrate monitors a single cleavage/joining event. Interestingly, 90–95% of the end-joining events involve 3′-protruding nucleotides that are generated by I-SceI cleavage (Figure 2D.2) [2]. Strikingly, although the four 3′-protruding nucleotides at each end are not complementary, the annealing of two out of the four 3′-protruding nucleotides is observed, corresponding to the maximum possible complementarities. Thus, C-NHEJ adapts to imperfectly complementary ends with minimal genetic modifications. A systematic in vitro analysis of most of the DNA end possibilities in human cell extracts consistently yielded similar results [6]. Importantly, in Ku80 - or Xrcc4-deficient mammalian cells, the repair events involve none of the 3′-protruding nucleotides, all the resulting products exhibiting deletions at the repair junctions with the frequent use of microhomologies that are distant from the DSB.
These data can be summarized as follows: regardless of the structure of the DNA ends (fully complementary or not), A-EJ removes at least all of the 3′-protruding nucleotides (and generally more), whereas C-NHEJ retains at least one of the 3′-protruding nucleotides, therefore accounting for 90–95% of the events using the 3′-protruding ends.
Importantly, these analyses have revealed that there are two different types of microhomologies (MHs): 1) MHs at the DSB itself that guide the annealing process of imperfectly complementary ends; end-joining is then processed by C-NHEJ in a conservative manner; and 2) MHs distant from the break that are involved in A-EJ, generating extended deletions at the repair junctions (Figure 1B).
These combined data show that C-NHEJ is not error-prone per se but is rather versatile and capable of adapting to non–fully complementary ends, maximizing the annealing process of potentially complementary nucleotides, which, in turn, limits genetic alterations. Thus, at the repair junction, C-NHEJ is conservative and the precision of end-joining is dictated by the structure of the DNA ends.
In Saccharomyces cerevisiae, sequence analysis of the end-joining events on transfected linearized plasmids revealed that NHEJ is very accurate. In contrast, extended deletions are recovered in yku70 mutant strains [19], [56], [57]. An alternative end-joining pathway (MMEJ), which increases upon Ku loss, has also been described in a chromosomal context [7]. In addition, NHEJ can generate reciprocal translocations, but in the absence of yKu80, the breakpoint junctions are associated with deletions [58]. An alternative end-joining pathway has also been identified in fission yeast [59].
The continuous expression of endonucleases, such as HO or I-SceI, consistently leads to multiple cycles of cleavage/repair in an essential chromosome, resulting in only 0.1% survival. This result suggests that NHEJ is at least 99.9% error-free because it restores a re-cleavable site [60]–[62]. NHEJ is also adaptable in S. cerevisiae. Indeed, the large majority of ends generated by HO are repaired by events involving the four 3′-protruding nucleotides, and the ligation of imperfect overhangs acts in a Ku-dependent manner [62], [63]. Finally, Tdp1, a yeast DNA 3′-phosphatase, has been proposed to increase the accuracy of the NHEJ machinery by preventing the modification of DNA ends [64].
C-NHEJ and A-EJ have also been described in bacteria [65]. In Mycobacterium smegmatis, the vast majority of Ku-independent junctions harbor microhomology-mediated deletions, indicating that A-EJ substituted for C-NHEJ during DSB repair [66]. Ku and ligase D are absent in the classical bacterial model Escherichia coli, and A-EJ is the most active end-joining pathway in this species [67]. Finally, evidence for conservative C-NHEJ and mutagenic A-EJ pathways has also been presented in plants [12], [68]–[70].
A large number of class II transposons transpose through a cut-and-paste mechanism in which the transposon is excised from its donor site and integrates into another locus where a target site duplication (TSD) is generated on both sides of the newly integrated element. Transposon excision leaves a DSB at the donor site with one copy of the initial TSD at each broken end; DSB repair through end-joining generally yields a characteristic footprint in which the two TSDs are separated by a few bp from the transposon (reviewed in [71]). The excision of cut-and-paste transposons, such as Sleeping Beauty [72], [73], Mos1 [74], or the P element [75], has been used in different hosts to induce DSBs at defined genomic loci. These studies have revealed that, in the absence of Ku, large deletions of the flanking sequences are recovered at transposon excision sites. This result confirms that Ku-dependent C-NHEJ is a conservative but versatile repair pathway in mammals, C. elegans, and, to a certain extent, Drosophila. In the latter, however, a chromosomal assay indicated that the most active end-joining pathway is independent of ligase IV [35].
End-joining requiring DNA end processing: The importance of being versatile
An efficient immune response absolutely requires genetic diversity at the immunoglobulin gene locus. The first level of diversity is generated through the rearrangement of the (V), (D), and (J) segments induced by the lymphoid-specific Rag1 and Rag2 proteins associated with the ubiquitous C-NHEJ machinery [76]–[79]. V(D)J recombination generates the coding and reciprocal signal joints (Figure 3A), and two steps increase the diversity at the coding joints. First, Rag1/Rag2-mediated cleavage produces hairpins at the broken coding ends (not on the signal ends), and hairpin resolution generates a combination of different sequences at the ends. Second, the addition of N (non-templated) nucleotides by the terminal deoxynucleotidyl transferase (TdT) adds junctional diversity to the coding joints [80]–[82]. Note that the diversity is not generated through C-NHEJ itself but rather through accessory mechanisms (i.e., via a hairpin resolution and TdT). The requirement for additional processes to generate diversity supports the notion that C-NHEJ is not, per se, sufficiently mutagenic at the coding joints. Moreover, the repair of signal joints, which results from the direct ligation of blunt ends, is largely error-free [76], [77]. This result shows that when the DNA ends are directly suitable for ligation, C-NHEJ is error-free.
Fig. 3. Processing of DNA ends prior to ligation. 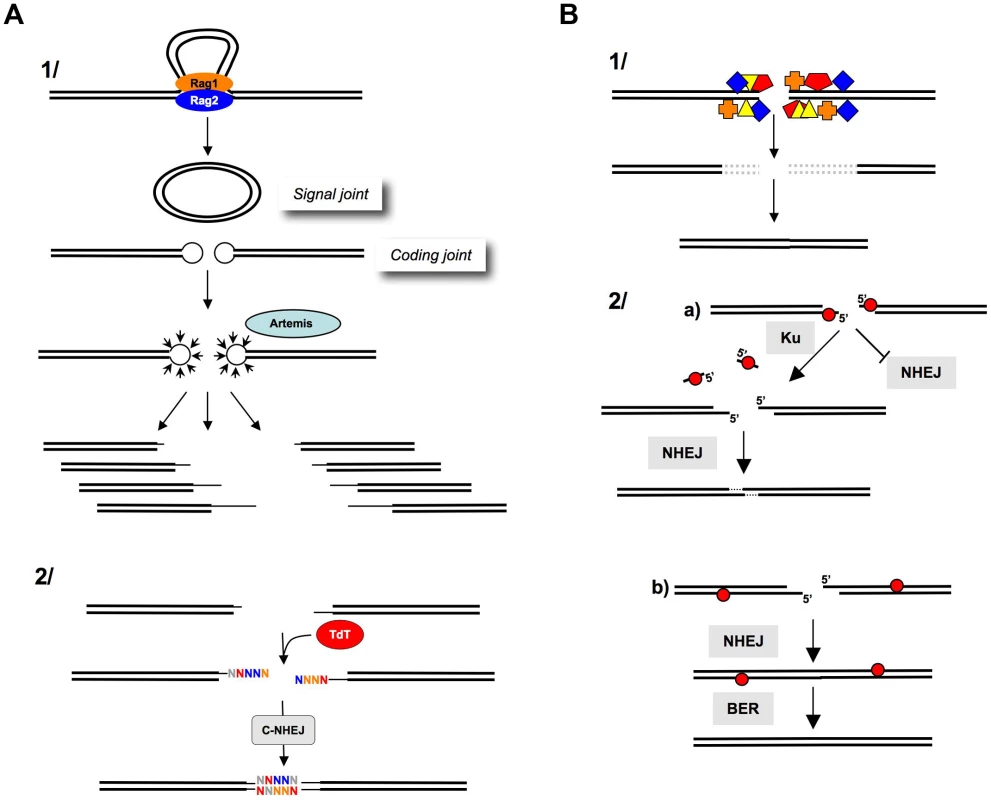
A) Junctional diversity through V(D)J recombination. 1) The Rag1-Rag2 proteins join the V(D)J recombination sites (synapsis step). The cleavage by Rag1-Rag2 generates a circular signal joint and a linear coding joint (containing the coding sequence); however, the cleavage generates hairpins at the extremities, which cannot be directly ligated. The opening of the hairpins (by Artemis) generates a combination of different DNA ends (thin lines), thereby, creating the first level of junctional diversity. 2) TdT subsequently adds N-nucleotides at the 3′ or blunt ends, creating a second level of junctional diversity. B) After IR. 1) IR generates multiple damages at DNA ends (colored boxes). These altered DNA ends are not compatible for enzymatic ligation by ligase IV. The excision of the damaged structures (dotted lines) results in nucleotide deletion after ligation. 2) Role of Ku at the DNA ends. a) Abasic sites (red circles) at the DSB inhibit NHEJ. Ku removes these sites, allowing the NHEJ of the processed DNA ends. This reaction results in a limited deletion (one to three nt at the resealed junction). b) Abasic sites (red circles) that are far from the DSB do not impair NHEJ. BER can then repair the abasic sites on the resealed molecule. Note that the reduced activity of Ku on these substrates prevents long deletions (from the abasic site to the end), which would result in large deletions at the resealed junction and would avoid the generation of new breaks in the resealed molecule (adapted from [97]). An end-joining process strictly restricted to fully complementary ends would be unable to ligate the coding joints. In contrast, a versatile but conservative process, such as C-NHEJ, is able to join these DNA ends and generate a highly diverse immune repertoire while protecting against side genomic instability. Notably, DNA ends are not complementary during class switch recombination, and the versatility of C-NHEJ is, therefore, essential to complete this process.
IR generates DSBs with chemically altered ends bearing complex lesions that are inept for enzymatic ligation. This situation is different from that of imperfectly complementary ends because the ligase is inactive on those types of chemically modified ends. Thus, IR-induced DSBs must be processed prior to ligation (Figure 3B). Consequently, mutagenesis at the resealed junctions of IR-induced DSBs results from this preliminary “cleaning” step rather than from C-NHEJ itself. Remarkably, Ku possesses a 5′-dRP/AP lyase activity, specialized for DSB, that restricts nucleotide loss at the ends (Figure 3B), therefore maintaining genomic stability [83].
The “cleaning” of IR-induced DSBs generates non-complementary ends. Thus, a non-versatile repair process would be unable to repair IR-induced DSBs, and the organism would be highly sensitive to IR, even at low doses. Therefore, the adaptability of C-NHEJ is essential for resistance to IR. This adaptability should have important consequences for the response to endogenous DSBs and to low exogenous doses, such as environmental or medical (radiological examination) exposures.
Genome Manipulation: Targeted Mutagenesis Induced by DSBs
The versatility of C-NHEJ has promising applications. Several strategies for targeting mutagenesis are based on mutagenic DSB end-joining. For example, targeted DSBs are generated through different types of nucleases, and unfaithful end-joining events are selected. One could argue that this strategy is primarily based on A-EJ–mediated events and that: i) A-EJ is accompanied by uncontrolled resection at the repaired junctions; ii) A-EJ favors translocations; and iii) in wild-type cells, A-EJ is less efficient than C-NHEJ [1], [2]. For these reasons, selecting strategies that act through C-NHEJ–dependent pathways should minimize the risks of side genomic instability, provided that controlled variability is introduced at the junction. Interestingly, ectopic expression of TdT efficiently adds a limited number of nucleotides at I-SceI-generated ends in a Ku - and ligase IV (C-NHEJ) –dependent manner, also in non-lymphoid cells [84]. One limitation is that TdT preferentially acts on 3′ overhangs or blunt ends. Consequently, TdT should be used in combination with nucleases that generate these types of ends. DNA end–modifying enzymes have also been shown to generate mutations at the resealed junctions of DNA ends generated by TAL endonucleases, but it is unknown whether they act through the C-NHEJ pathway [85].
Conclusion
C-NHEJ is a conservative end-joining process but permits controlled genetic variability required in essential physiological processes.
At the chromosome level, C-NHEJ protects against DSB movements and profound genome rearrangement. Note that C-NHEJ is involved in physiological processes leading to rearrangements, such as the development of the Paramecium macronucleus and V(D)J or class switch recombination. These processes are highly controlled, and the synapsis of the interacting DNA is frequently promoted by associated proteins but not the NHEJ machinery itself, as exemplified in the V(D)J recombination during which the Rag1-Rag2 proteins promote the synapsis of the distant interacting sequences before DNA cleavage. Thus, although C-NHEJ protects against chromosomal rearrangements, it should allow genetic diversity in highly controlled physiological processes.
At the junction sequence level, the previously proposed fallibility of the NHEJ pathway reflects a combination of factors: i) the involvement of the highly mutagenic A-EJ process, ii) the necessity of processing DNA ends prior to their joining, and iii) the versatility/adaptability of C-NHEJ. C-NHEJ is not intrinsically inaccurate, but is versatile and adaptable to imperfect ends, and the actual quality of the end-joining is dictated by the structure of the DNA ends rather than by the C-NHEJ machinery. Versatility/adaptability is paramount for certain essential processes and confers a key role for C-NHEJ in the balance between genetic stability and genetic diversity during the generation of the immune repertoire, molecular evolution, and when challenged with endogenous and environmental sources of DSBs.
Zdroje
1. Guirouilh-BarbatJ, RassE, PloI, BertrandP, LopezBS (2007) Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci U S A 104 : 20902–20907.
2. Guirouilh-BarbatJ, HuckS, BertrandP, PirzioL, DesmazeC, et al. (2004) Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell 14 : 611–623.
3. GrabarzA, BarascuA, Guirouilh-BarbatJ, LopezBS (2012) Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am J Cancer Res 2 : 249–268.
4. AudebertM, SallesB, CalsouP (2004) Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem 279 : 55117–55126.
5. CorneoB, WendlandRL, DerianoL, CuiX, KleinIA, et al. (2007) Rag mutations reveal robust alternative end joining. Nature 449 : 483–486.
6. FeldmannE, SchmiemannV, GoedeckeW, ReichenbergerS, PfeifferP (2000) DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res 28 : 2585–2596.
7. MaJL, KimEM, HaberJE, LeeSE (2003) Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol 23 : 8820–8828.
8. WangH, PerraultAR, TakedaY, QinW, IliakisG (2003) Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res 31 : 5377–5388.
9. WangH, RosidiB, PerraultR, WangM, ZhangL, et al. (2005) DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res 65 : 4020–4030.
10. YanCT, BoboilaC, SouzaEK, FrancoS, HickernellTR, et al. (2007) IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449 : 478–482.
11. RassE, GrabarzA, PloI, GautierJ, BertrandP, et al. (2009) Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol 16 : 819–824.
12. CharbonnelC, AllainE, GallegoME, WhiteCI (2011) Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair (Amst) 10 : 611–619.
13. BennardoN, ChengA, HuangN, StarkJM (2008) Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4: e1000110.
14. LeeK, LeeSE (2007) Saccharomyces cerevisiae Sae2 - and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics 176 : 2003–2014.
15. GrabarzA, Guirouilh-BarbatJ, BarascuA, PennarunG, GenetD, et al. (2013) A role for BLM in double-strand break repair pathway choice: prevention of CtIP/Mre11-mediated alternative nonhomologous end-joining. Cell Rep 5 : 21–28.
16. PierceAJ, HuP, HanM, EllisN, JasinM (2001) Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 15 : 3237–3242.
17. WangM, WuW, WuW, RosidiB, ZhangL, et al. (2006) PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 34 : 6170–6182.
18. BoultonSJ, JacksonSP (1996) Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res 24 : 4639–4648.
19. BoultonSJ, JacksonSP (1996) Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double - strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J 15 : 5093–5103.
20. LiangF, RomanienkoPJ, WeaverDT, JeggoPA, JasinM (1996) Chromosomal double-strand break repair in Ku80-deficient cells. Proc Natl Acad Sci U S A 93 : 8929–8933.
21. Schulte-UentropL, El-AwadyRA, SchlieckerL, WillersH, Dahm-DaphiJ (2008) Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease - and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res 36 : 2561–2569.
22. DelacoteF, LopezBS (2008) Importance of the cell cycle phase for the choice of the appropriate DSB repair pathway, for genome stability maintenance: the trans-S double-strand break repair model. Cell Cycle 7 : 33–38.
23. Guirouilh-BarbatJ, HuckS, LopezBS (2008) S-phase progression stimulates both the mutagenic KU-independent pathway and mutagenic processing of KU-dependent intermediates, for nonhomologous end joining. Oncogene 27 : 1726–1736.
24. RothkammK, KrugerI, ThompsonLH, LobrichM (2003) Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol 23 : 5706–5715.
25. SaintignyY, DelacoteF, BoucherD, AverbeckD, LopezBS (2007) XRCC4 in G1 suppresses homologous recombination in S/G2, in G1 checkpoint-defective cells. Oncogene 26 : 2769–2780.
26. Saleh-GohariN, HelledayT (2004) Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res 32 : 3683–3688.
27. PiganeauM, GhezraouiH, De CianA, GuittatL, TomishimaM, et al. (2013) Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res 23 : 1182–1193.
28. HollandAJ, ClevelandDW (2012) Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med 18 : 1630–1638.
29. JonesMJ, JallepalliPV (2012) Chromothripsis: chromosomes in crisis. Dev Cell 23 : 908–917.
30. BoboilaC, JankovicM, YanCT, WangJH, WesemannDR, et al. (2010) Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci U S A 107 : 3034–3039.
31. DifilippantonioMJ, ZhuJ, ChenHT, MeffreE, NussenzweigMC, et al. (2000) DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404 : 510–514.
32. DifilippantonioMJ, PetersenS, ChenHT, JohnsonR, JasinM, et al. (2002) Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J Exp Med 196 : 469–480.
33. GaoY, FergusonDO, XieW, ManisJP, SekiguchiJ, et al. (2000) Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404 : 897–900.
34. SundararajanR, GellonL, ZunderRM, FreudenreichCH (2010) Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics 184 : 65–77.
35. McVeyM, RadutD, SekelskyJJ (2004) End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics 168 : 2067–2076.
36. SimsekD, JasinM (2010) Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol 17 : 410–416.
37. WeinstockDM, BrunetE, JasinM (2007) Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol 9 : 978–981.
38. SimsekD, BrunetE, WongSY-W, KatyalS, GaoY, et al. (2011) DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet 7: e1002080.
39. ZhangY, JasinM (2011) An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol 18 : 80–84.
40. SoutoglouE, DornJF, SenguptaK, JasinM, NussenzweigA, et al. (2007) Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 9 : 675–682.
41. CaryRB, PetersonSR, WangJ, BearDG, BradburyEM, et al. (1997) DNA looping by Ku and the DNA-dependent protein kinase. Proc Natl Acad Sci U S A 94 : 4267–4272.
42. DeFazioLG, StanselRM, GriffithJD, ChuG (2002) Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J 21 : 3192–3200.
43. DionV, KalckV, HorigomeC, TowbinBD, GasserSM (2012) Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol 14 : 502–509.
44. Mine-HattabJ, RothsteinR (2012) Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol 14 : 510–517.
45. DuboisE, BischerourJ, MarmignonA, MathyN, RegnierV, et al. (2012) Transposon invasion of the Paramecium germline genome countered by a domesticated PiggyBac transposase and the NHEJ pathway. Int J Evol Biol 2012 : 436196.
46. BetermierM (2004) Large-scale genome remodelling by the developmentally programmed elimination of germ line sequences in the ciliate Paramecium. Res Microbiol 155 : 399–408.
47. ArnaizO, MathyN, BaudryC, MalinskyS, AuryJ-M, et al. (2012) The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genet 8: e1002984.
48. GratiasA, BétermierM (2003) Processing of double-strand breaks is involved in the precise excision of Paramecium IESs. Mol Cell Biol 23 : 7152–7162.
49. BetermierM, DuharcourtS, SeitzH, MeyerE (2000) Timing of developmentally programmed excision and circularization of Paramecium internal eliminated sequences. Mol Cell Biol 20 : 1553–1561.
50. KapustaA, MatsudaA, MarmignonA, KuM, SilveA, et al. (2011) Highly precise and developmentally programmed genome assembly in Paramecium requires Ligase IV-dependent end joining. PLoS Genet 7: e1002049.
51. KabotyanskiEB, GomelskyL, HanJO, StamatoTD, RothDB (1998) Double-strand break repair in Ku86 - and XRCC4-deficient cells. Nucleic Acids Res 26 : 5333–5342.
52. SmithJ, BaldeyronC, De OliveiraI, Sala-TrepatM, PapadopouloD (2001) The influence of DNA double-strand break structure on end-joining in human cells. Nucleic Acids Res 29 : 4783–4792.
53. SmithJ, RiballoE, KyselaB, BaldeyronC, ManolisK, et al. (2003) Impact of DNA ligase IV on the fidelity of end joining in human cells. Nucleic Acids Res 31 : 2157–2167.
54. WillersH, HussonJ, LeeLW, HubbeP, GazemeierF, et al. (2006) Distinct mechanisms of nonhomologous end joining in the repair of site-directed chromosomal breaks with noncomplementary and complementary ends. Radiat Res 166 : 567–574.
55. XieA, KwokA, ScullyR (2009) Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol 16 : 814–818.
56. HegdeV, KleinH (2000) Requirement for the SRS2 DNA helicase gene in non-homologous end joining in yeast. Nucleic Acids Res 28 : 2779–2783.
57. MoscarielloM, FlorioC, PulitzerJF (2010) Accurate repair of non-cohesive, double strand breaks in Saccharomyces cerevisiae: enhancement by homology-assisted end-joining. Yeast 27 : 837–848.
58. YuX, GabrielA (2003) Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics 163 : 843–856.
59. DecottigniesA (2007) Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics 176 : 1403–1415.
60. Frank-VaillantM, MarcandS (2002) Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell 10 : 1189–1199.
61. LeeSE, PaquesF, SylvanJ, HaberJE (1999) Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol 9 : 767–770.
62. MooreJK, HaberJE (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 16 : 2164–2173.
63. WilsonTE, LieberMR (1999) Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J Biol Chem 274 : 23599–23609.
64. BahmedK, NitissKC, NitissJL (2010) Yeast Tdp1 regulates the fidelity of nonhomologous end joining. Proc Natl Acad Sci U S A 107 : 4057–4062.
65. ShumanS, GlickmanMS (2007) Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol 5 : 852–861.
66. AniukwuJ, GlickmanMS, ShumanS (2008) The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev 22 : 512–527.
67. ChayotR, MontagneB, MazelD, RicchettiM (2010) An end-joining repair mechanism in Escherichia coli. Proc Natl Acad Sci U S A 107 : 2141–2146.
68. HuefnerND, MizunoY, WeilCF, KorfI, BrittAB (2011) Breadth by depth: expanding our understanding of the repair of transposon-induced DNA double strand breaks via deep-sequencing. DNA Repair (Amst) 10 : 1023–1033.
69. LloydAH, WangD, TimmisJN (2012) Single molecule PCR reveals similar patterns of non-homologous DSB repair in tobacco and Arabidopsis. PLoS ONE 7: e32255.
70. OsakabeK, OsakabeY, TokiS (2010) Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc Natl Acad Sci U S A 107 : 12034–12039.
71. FeschotteC, PrithamEJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41 : 331–368.
72. IzsvakZ, StuweEE, FiedlerD, KatzerA, JeggoPA, et al. (2004) Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol Cell 13 : 279–290.
73. YantSR, KayMA (2003) Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells. Mol Cell Biol 23 : 8505–8518.
74. RobertV, BessereauJL (2007) Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J 26 : 170–183.
75. BeallEL, RioDC (1996) Drosophila IRBP/Ku p70 corresponds to the mutagen-sensitive mus309 gene and is involved in P-element excision in vivo. Genes Dev 10 : 921–933.
76. DudleyDD, ChaudhuriJ, BassingCH, AltFW (2005) Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol 86 : 43–112.
77. JungD, AltFW (2004) Unraveling V(D)J recombination; insights into gene regulation. Cell 116 : 299–311.
78. LieberMR, MaY, PannickeU, SchwarzK (2004) The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 3 : 817–826.
79. RooneyS, ChaudhuriJ, AltFW (2004) The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev 200 : 115–131.
80. BenedictCL, GilfillanS, ThaiTH, KearneyJF (2000) Terminal deoxynucleotidyl transferase and repertoire development. Immunol Rev 175 : 150–157.
81. DesiderioSV, YancopoulosGD, PaskindM, ThomasE, BossMA, et al. (1984) Insertion of N regions into heavy-chain genes is correlated with expression of terminal deoxytransferase in B cells. Nature 311 : 752–755.
82. LewisSM (1994) P nucleotide insertions and the resolution of hairpin DNA structures in mammalian cells. Proc Natl Acad Sci U S A 91 : 1332–1336.
83. RobertsSA, StrandeN, BurkhalterMD, StromC, HavenerJM, et al. (2010) Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature 464 : 1214–1217.
84. Boubakour-AzzouzI, BertrandP, ClaesA, LopezBS, RougeonF (2012) Terminal deoxynucleotidyl transferase requires KU80 and XRCC4 to promote N-addition at non-V(D)J chromosomal breaks in non-lymphoid cells. Nucleic Acids Res 40 : 8381–8391.
85. CertoMT, GwiazdaKS, KuharR, SatherB, CuringaG, et al. (2012) Coupling endonucleases with DNA end-processing enzymes to drive gene disruption. Nat Methods 9 : 973–975.
86. CappJP, BoudsocqF, BertrandP, Laroche-ClaryA, PourquierP, et al. (2006) The DNA polymerase lambda is required for the repair of non-compatible DNA double strand breaks by NHEJ in mammalian cells. Nucleic Acids Res 34 : 2998–3007.
87. CappJP, BoudsocqF, BesnardAG, LopezBS, CazauxC, et al. (2007) Involvement of DNA polymerase mu in the repair of a specific subset of DNA double-strand breaks in mammalian cells. Nucleic Acids Res 35 : 3551–3560.
88. MahaneyBL, MeekK, Lees-MillerSP (2009) Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J 417 : 639–650.
89. Nick McElhinnySA, RamsdenDA (2004) Sibling rivalry: competition between Pol X family members in V(D)J recombination and general double strand break repair. Immunol Rev 200 : 156–164.
90. MoshousD, CallebautI, de ChassevalR, PoinsignonC, VilleyI, et al. (2003) The V(D)J recombination/DNA repair factor artemis belongs to the metallo-beta-lactamase family and constitutes a critical developmental checkpoint of the lymphoid system. Ann N Y Acad Sci 987 : 150–157.
91. AhnesorgP, SmithP, JacksonSP (2006) XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124 : 301–313.
92. BuckD, MalivertL, de ChassevalR, BarraudA, FondanecheMC, et al. (2006) Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124 : 287–299.
93. RevyP, MalivertL, de VillartayJP (2006) Cernunnos-XLF, a recently identified non-homologous end-joining factor required for the development of the immune system. Curr Opin Allergy Clin Immunol 6 : 416–420.
94. AudebertM, SallesB, WeinfeldM, CalsouP (2006) Involvement of polynucleotide kinase in a poly(ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J Mol Biol 356 : 257–265.
95. RobertI, DantzerF, Reina-San-MartinB (2009) Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J Exp Med 206 : 1047–1056.
96. RobbinsDJ, ColemanMS (1988) Initiator role of double stranded DNA in terminal transferase catalyzed polymerization reactions. Nucleic Acids Res 16 : 2943–2957.
97. StrandeN, RobertsSA, OhS, HendricksonEA, RamsdenDA (2012) Specificity of the dRP/AP lyase of Ku promotes nonhomologous end joining (NHEJ) fidelity at damaged ends. J Biol Chem 287 : 13686–13693.
Štítky
Genetika Reprodukční medicína
Článek Unwrapping BacteriaČlánek A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome StabilityČlánek The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory OrgansČlánek The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking inČlánek Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma DevelopmentČlánek Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 MiceČlánek High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 1
-
Všechny články tohoto čísla
- How Much Is That in Dog Years? The Advent of Canine Population Genomics
- The Sense and Sensibility of Strand Exchange in Recombination Homeostasis
- Unwrapping Bacteria
- DNA Methylation Changes Separate Allergic Patients from Healthy Controls and May Reflect Altered CD4 T-Cell Population Structure
- Evidence for Mito-Nuclear and Sex-Linked Reproductive Barriers between the Hybrid Italian Sparrow and Its Parent Species
- Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA
- Relationship Estimation from Whole-Genome Sequence Data
- Genetic Models of Apoptosis-Induced Proliferation Decipher Activation of JNK and Identify a Requirement of EGFR Signaling for Tissue Regenerative Responses in
- ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Cells
- Loss and Recovery of Genetic Diversity in Adapting Populations of HIV
- Bioelectric Signaling Regulates Size in Zebrafish Fins
- Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions
- Loss of Histone H3 Methylation at Lysine 4 Triggers Apoptosis in
- Cell-Cycle Dependent Expression of a Translocation-Mediated Fusion Oncogene Mediates Checkpoint Adaptation in Rhabdomyosarcoma
- How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation
- A Nonsense Mutation in Encoding a Nondescript Transmembrane Protein Causes Idiopathic Male Subfertility in Cattle
- Deletion of a Conserved -Element in the Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription
- Developmental Link between Sex and Nutrition; Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles
- PP2A/B55 and Fcp1 Regulate Greatwall and Ensa Dephosphorylation during Mitotic Exit
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Comprehensive Functional Annotation of 77 Prostate Cancer Risk Loci
- Evolution of Chloroplast Transcript Processing in and Its Chromerid Algal Relatives
- A Chaperone-Assisted Degradation Pathway Targets Kinetochore Proteins to Ensure Genome Stability
- New MicroRNAs in —Birth, Death and Cycles of Adaptive Evolution
- A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism
- FGFR1-Frs2/3 Signalling Maintains Sensory Progenitors during Inner Ear Hair Cell Formation
- Regulation of Synaptic /Neuroligin Abundance by the /Nrf Stress Response Pathway Protects against Oxidative Stress
- Intrasubtype Reassortments Cause Adaptive Amino Acid Replacements in H3N2 Influenza Genes
- Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments
- WNT7B Promotes Bone Formation in part through mTORC1
- Natural Selection Reduced Diversity on Human Y Chromosomes
- In-Vivo Quantitative Proteomics Reveals a Key Contribution of Post-Transcriptional Mechanisms to the Circadian Regulation of Liver Metabolism
- The Candidate Splicing Factor Sfswap Regulates Growth and Patterning of Inner Ear Sensory Organs
- The Acid Phosphatase-Encoding Gene Contributes to Soybean Tolerance to Low-Phosphorus Stress
- p53 and TAp63 Promote Keratinocyte Proliferation and Differentiation in Breeding Tubercles of the Zebrafish
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
- The SET Domain Proteins SUVH2 and SUVH9 Are Required for Pol V Occupancy at RNA-Directed DNA Methylation Loci
- Down-Regulation of Rad51 Activity during Meiosis in Yeast Prevents Competition with Dmc1 for Repair of Double-Strand Breaks
- Multi-tissue Analysis of Co-expression Networks by Higher-Order Generalized Singular Value Decomposition Identifies Functionally Coherent Transcriptional Modules
- A Neurotoxic Glycerophosphocholine Impacts PtdIns-4, 5-Bisphosphate and TORC2 Signaling by Altering Ceramide Biosynthesis in Yeast
- Subtle Changes in Motif Positioning Cause Tissue-Specific Effects on Robustness of an Enhancer's Activity
- C/EBPα Is Required for Long-Term Self-Renewal and Lineage Priming of Hematopoietic Stem Cells and for the Maintenance of Epigenetic Configurations in Multipotent Progenitors
- The SPF27 Homologue Num1 Connects Splicing and Kinesin 1-Dependent Cytoplasmic Trafficking in
- Down-Regulation of eIF4GII by miR-520c-3p Represses Diffuse Large B Cell Lymphoma Development
- Genome Sequencing Highlights the Dynamic Early History of Dogs
- Re-sequencing Expands Our Understanding of the Phenotypic Impact of Variants at GWAS Loci
- Meta-Analysis Identifies Gene-by-Environment Interactions as Demonstrated in a Study of 4,965 Mice
- , a -Antisense Gene of , Encodes a Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas
- A Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration
- A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool
- A PNPase Dependent CRISPR System in
- Genomic Confirmation of Hybridisation and Recent Inbreeding in a Vector-Isolated Population
- Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution
- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development
- A Cell Cycle and Nutritional Checkpoint Controlling Bacterial Surface Adhesion
- High Risk Population Isolate Reveals Low Frequency Variants Predisposing to Intracranial Aneurysms
- E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses
- Evolutionary Rate Covariation Identifies New Members of a Protein Network Required for Female Post-Mating Responses
- 3′ Untranslated Regions Mediate Transcriptional Interference between Convergent Genes Both Locally and Ectopically in
- Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus
- Metabolic QTL Analysis Links Chloroquine Resistance in to Impaired Hemoglobin Catabolism
- Notch Controls Cell Adhesion in the Drosophila Eye
- AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes
- Genomes Reveal Evolution of Microalgal Oleaginous Traits
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Variation in Genome-Wide Levels of Meiotic Recombination Is Established at the Onset of Prophase in Mammalian Males
- Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan
- Lifespan Extension Conferred by Endoplasmic Reticulum Secretory Pathway Deficiency Requires Induction of the Unfolded Protein Response
- Is Non-Homologous End-Joining Really an Inherently Error-Prone Process?
- Vestigialization of an Allosteric Switch: Genetic and Structural Mechanisms for the Evolution of Constitutive Activity in a Steroid Hormone Receptor
- Functional Divergence and Evolutionary Turnover in Mammalian Phosphoproteomes
- A 660-Kb Deletion with Antagonistic Effects on Fertility and Milk Production Segregates at High Frequency in Nordic Red Cattle: Additional Evidence for the Common Occurrence of Balancing Selection in Livestock
- Comparative Evolutionary and Developmental Dynamics of the Cotton () Fiber Transcriptome
- The Transcription Factor BcLTF1 Regulates Virulence and Light Responses in the Necrotrophic Plant Pathogen
- Crossover Patterning by the Beam-Film Model: Analysis and Implications
- Single Cell Genomics: Advances and Future Perspectives
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- GATA6 Is a Crucial Regulator of Shh in the Limb Bud
- Large Inverted Duplications in the Human Genome Form via a Fold-Back Mechanism
- Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues
- Affects Plant Architecture by Regulating Local Auxin Biosynthesis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



