-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaStructural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
Histone lysine methylation is an important epigenetic modification in regulating chromatin structure and gene expression. Histone H3 lysine 4 methylation (H3K4me), which can be in a mono-, di-, or trimethylated state, has been shown to play an important role in gene expression involved in plant developmental control and stress adaptation. However, the resetting mechanism of this epigenetic modification is not yet fully understood. In this work, we identified a JmjC domain-containing protein, JMJ703, as a histone lysine demethylase that specifically reverses all three forms of H3K4me in rice. Loss-of-function mutation of the gene affected stem elongation and plant growth, which may be related to increased expression of cytokinin oxidase genes in the mutant. Analysis of crystal structure of the catalytic core domain (c-JMJ703) of the protein revealed a general structural similarity with mammalian and yeast JMJD2 proteins that are H3K9 and H3K36 demethylases. However, several specific features were observed in the structure of c-JMJ703. Key residues that interact with cofactors Fe(II) and N-oxalylglycine and the methylated H3K4 substrate peptide were identified and were shown to be essential for the demethylase activity in vivo. Several key residues are specifically conserved in known H3K4 demethylases, suggesting that they may be involved in the specificity for H3K4 demethylation.
Published in the journal: . PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003239
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003239Summary
Histone lysine methylation is an important epigenetic modification in regulating chromatin structure and gene expression. Histone H3 lysine 4 methylation (H3K4me), which can be in a mono-, di-, or trimethylated state, has been shown to play an important role in gene expression involved in plant developmental control and stress adaptation. However, the resetting mechanism of this epigenetic modification is not yet fully understood. In this work, we identified a JmjC domain-containing protein, JMJ703, as a histone lysine demethylase that specifically reverses all three forms of H3K4me in rice. Loss-of-function mutation of the gene affected stem elongation and plant growth, which may be related to increased expression of cytokinin oxidase genes in the mutant. Analysis of crystal structure of the catalytic core domain (c-JMJ703) of the protein revealed a general structural similarity with mammalian and yeast JMJD2 proteins that are H3K9 and H3K36 demethylases. However, several specific features were observed in the structure of c-JMJ703. Key residues that interact with cofactors Fe(II) and N-oxalylglycine and the methylated H3K4 substrate peptide were identified and were shown to be essential for the demethylase activity in vivo. Several key residues are specifically conserved in known H3K4 demethylases, suggesting that they may be involved in the specificity for H3K4 demethylation.
Introduction
Histone methylation is an important epigenetic modification for chromatin structure, genome function, and gene expression in eukaryotic cells [1]. Histone methylation can be reversed by histone demethylases [1]. Lysine Specific Demethylase 1 (LSD1) is the first identified histone demethylase characterized as a member of the flavin-dependent amine oxidase family [2]. The second class of histone demethylases featured with the jumonji C (JmjC) domain has been shown to catalyze histone lysine demethylation through ferrous ion (Fe(II)) and α-ketoglutaric acid (α-KG)-dependent oxidative reactions [3].
Structurally related JmjC domain-containing proteins are classified into 7 subgroups based on phylogenetic analysis of members from yeast and animal cells [4]. Among them, the JmjC domain-containing histone demethylase 1 (JHDM1) subgroup has been demonstrated to reverse mono - and dimethylated histone H3 lysine 36 (H3K36me1/me2) [3]. The JmjC domain-containing histone demethylase 2 (JHDM2) subgroup has been identified to reverse dimethylated histone H3 lysine 9 (H3K9me2) [5], whereas the JMJD2 (also called JHDM3) group specifically reverses methylated H3K9 and/or H3K36 [6]. UTX/UTY is involved in the reversal of histone H3 lysine 27 methylation (H3K27me) [7], whereas PHF8 which belongs to PHF2/PHF8 subfamily demethylates H3K9me1/2, H3K27me2 and monomethylated histone H4 lysine 20 (H4K20me1) [8]–[10]. Aside from these members, the JARID subgroup is responsible for H3K4 demethylation [11]. The JmjC domain-only subgroup has the smallest molecular architecture; members of which have been shown to catalyze divergent reactions, such as histone arginine demethylation [12] and asparagine protein hydroxylation [13]. More recently, JMJD6 is shown to catalyze protein lysyl-hydroxylation [14].
Plant JmjC proteins have been shown to play important roles in the regulation of epigenetic processes, growth and development [15]. Although conserved with yeast and animal homologues, plant JmjC proteins display several distinct features. For instance, the UTX subgroup proteins that exhibit H3K27 demethylase activity are not found in plants. Recent data show that RELATIVE OF EARLY FLOWERING 6 (REF6), a member of the JMJD2 subgroup, can demethylate H3K27 in Arabidopsis [16]. Conversely, there exists a subgroup of plant JmjC proteins that include additional protein modules that are missing from animal or yeast homologues [17]. The amino acid sequence of the JmjC domains of this subgroup is closely related to JARID, while the overall domain organization of the core protein is similar to that of the JMJD2 group [17], [18]. To study this plant specific group of JmjC proteins, we analyzed the developmental function, enzymatic activity, and crystal structure of JMJ703, a member of this subgroup in rice. Our results demonstrate that JMJ703 is essential for plant cell division and stem elongation and specifically demethylates mono-, di-, and trimethylated H3K4 in vivo and in vitro. The high resolution structure of the catalytic core of JMJ703 (c-JMJ703) in complex with cofactors and substrate peptide reveals that the overall folding of c-JMJ703 is similar to those defined in animal and yeast JMJD2 that are H3K9 and H3K36 demethylases. However, the crystal structure of c-JMJ703 displays a number of specific features in cofactor interaction and in substrate peptide binding. Substitution mutation analysis indicated that residues implicated in the specific structures are essential for the enzymatic activity, some of which are conserved within JARID proteins and plant H3K4 demethylases and may be involved in the specificity of this class of enzymes.
Results
JMJ703 is essential for rice stem elongation
To investigate the developmental function of JMJ703, we characterized a T-DNA insertion mutant and generated several RNAi lines of the gene (Figure 1A). Both knockout and knockdown plants were semi-dwarf and produced smaller seeds (Figure 1B, Figure S1). The phenotypes co-segregated with the T-DNA insertion or the transgene (Table S1). Histological study of stem epidermal tissues revealed no difference in cell length between wild type and the mutant (Figure 1C), suggesting that the shorter stem phenotype may be caused by a slower cell division rate in the jmj703 mutant plants. Quantitative RT-PCR analysis revealed that the expression of several cell cycle-related genes was unaffected by the mutation (not shown). However, analysis of the cytokinin oxidase (CKX) gene family that reduces active cytokinin levels revealed that several members were highly induced in the young stem of mutant plants (Figure 1D). Chromatin immunoprecipitation assays revealed that H3K4me3 was clearly increased over the promoter region of the CKX genes in the mutants (Figure 1D). These data suggested that JMJ703 might regulate H3K4me3 on CKX genes and that the mutant phenotype might be due to cytokinin deficiency caused by increased H3K4me3 and increased expression of CKX genes.
Fig. 1. Phenotype of jmj703 mutants. 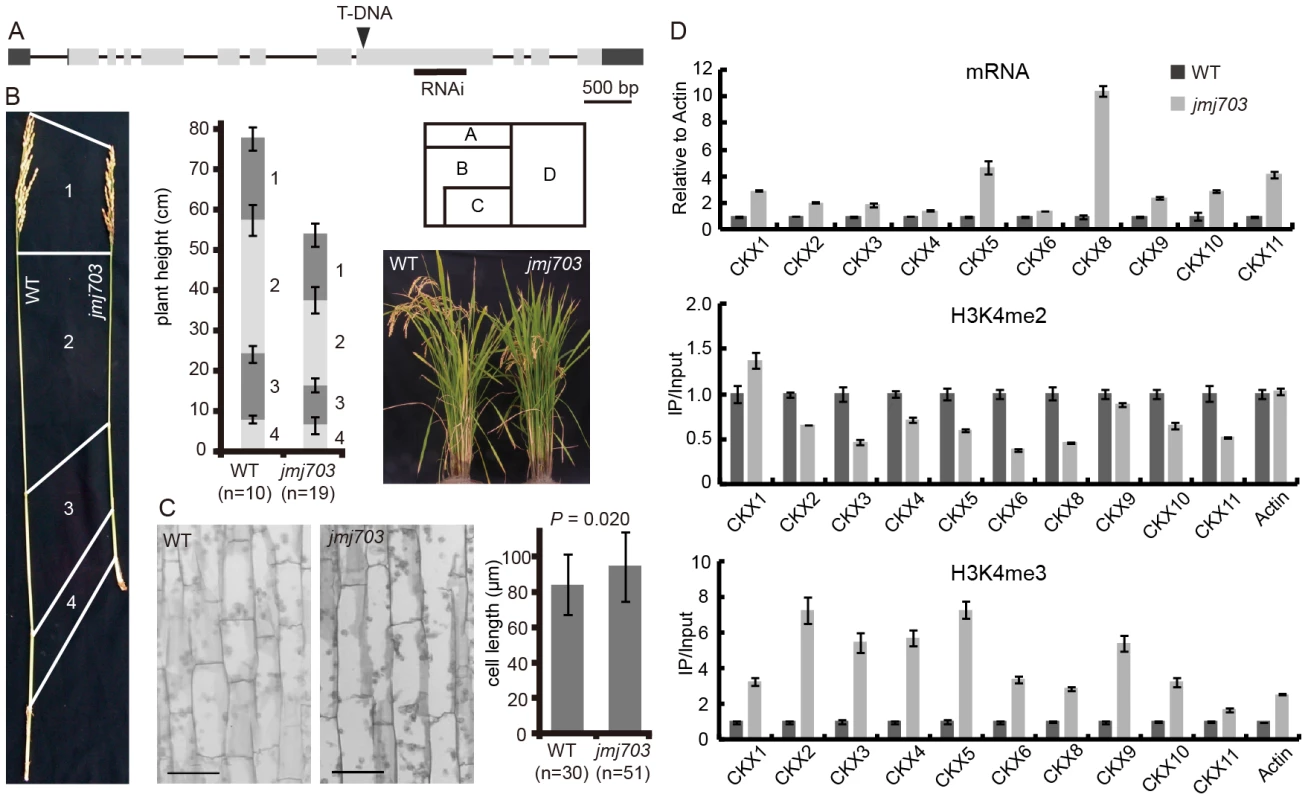
(A) T-DNA insertion and RNAi locations within the JMJ703 gene. (B) Comparison of the main panicle internodes between wild type and mutant plants. Lengths of the uppermost internodes were measured. (C) Comparison of stem epidermal cell lengths between wild type and mutant plants. No significant difference of cell lengths between wild type and mutant plants is detected by t-tests (p = 0.02). Bar = 50 µm. (D) Relative expression, H3K4me2 and H3K4me3 levels of cytokinin oxidase (CKX) genes in wild type and mutant plants. Expression levels in wild type are set as 1. JMJ703 is a histone H3K4 demethylase
JMJ703 protein contains several modules, including JmjN, JmjC, C5HC2 zinc finger, FYRN, and FYRC (Figure 2A). The JmjC domain is critical for histone demethylase activity [3]. To investigate the substrate specificity of JMJ703, the FLAG:HA-tagged JmjN-JmjC-zinc finger region (between amino acids 113 and 700, called FA-J3NCZ) of the protein was transiently over-expressed in tobacco leaves for in vivo histone demethylase assays. As shown in Figure 2B, nuclei expressing FA-J3NCZ showed a clear decrease of H3K4me1/2/3 compared to non-transfected nuclei, whereas no difference was observed for H3K27me3. FA-J3NCZ was then affinity-purified from transfected tobacco cells for in vitro histone demethylase assays. Consistent with the in vivo results, FA-J3NCZ could demethylate H3K4me1/2/3 in vitro (Figure 2C). By contrast, no activity of FA-J3NCZ to demethylate H3K9me1/2/3, or H3K36me1/2/3 was detected either in tobacco cells or in vitro (Figure S2A–S2B), indicating that JMJ703 is an H3K4-specific demethylase.
Fig. 2. JMJ703 is a histone H3K4 demethylase. 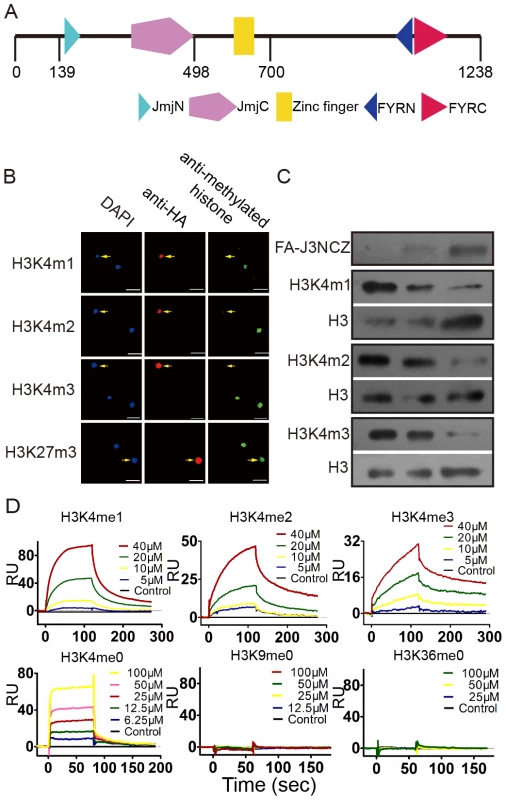
(A) Schematic presentation of JMJ703 protein structure. Fragment spanning amino acids 139–498 was obtained for crystal growth and structural determination. The region between amino acids 113–700 was used for enzymatic assays. (B) In vivo histone H3K4 demethylase activity of JMJ703. Constructs containing the JMJ703 fragment tagged by FLAG-HA (FA-J3NCZ) were transfected into tobacco leaf cells. Nuclei expressing FA-J3NCZ (stained with anti-HA) were examined for H3K4 methylation by anti-H3K4me1/2/3 and anti-H3K27me3 (indicated by arrows). At least 30 nuclei expressing JMJ703 per transfection were observed and imaged. Bar = 25 µm. (C) In vitro demethylase activity of JMJ703. Bulk histones were incubated with three quantities of FA-J3NCZ and analyzed by Western blots using antibodies of H3K4me1/2/3. The same blots were analyzed by anti-H3 to control loadings. FA-J3NCZ levels were revealed by anti-HA. (D) Characterization of c-JMJ703 binding to H3K4me0/1/2/3, H3K9me0 and H3K36me0 peptides using surface plasmon resonance. Curves for different concentration of peptide are differentially colored and labeled. Although the structures of a number of animal and yeast JmjC proteins have been defined [19]–[21], the structure of an H3K4 demethylase has not yet been reported. To study the structure of JMJ703, we made many attempts, including N - and C-terminal truncations and limited proteolysis, to obtain JMJ703 crystals. Finally, the fragment spanning amino acids 139 to 498 (termed c-JMJ703), covering the JmjN and JmjC domains, was deemed to be suitable for crystallographic investigation. We first performed surface plasmon resonance (SPR) experiments to measure the binding affinities of c-JMJ703 (J3NC) to H3K4 peptides with mono-, di-, or trimethylation. SPR results indicated that c-JMJ703 (J3NC) bound to H3K4me1, H3K4me2, and H3K4me3 peptides with dissociation constant Kd values of 28.9 µM, 19.3 µM, and 30.1 µM, respectively (Figure 2D). In addition, c-JMJ703 (J3NC) could also bind to H3K4me0 peptide, but with a much lower Kd value (76.6 µM), while no binding activity to H3K9me0 and H3K36me0 peptide was observed (Figure 2D). Meanwhile, the fragment corresponding to the JmjN-JmjC-zinc finger region (J3NCZ) showed a higher affinity to H3K4me1/2/3 peptides with a Kd value of 12.6 µM, 15.1 µM and 15.9 µM, respectively (Figure S2C). However, neither c-JMJ703 nor J3NCZ bound to H3K9me3 peptide, suggesting that the zinc finger enhances the substrate binding affinity of JMJ703 but is not essential for the binding specificity (Figure S2D).
Crystal structure features of c-JMJ703
The crystal structure of c-JMJ703 alone or in complex with α-KG (termed as c-JMJ703-α-KG) or with NOG (N-oxalylglycine, a non-catalytic analog of α-KG) and H3K4me3 peptide (termed as c-JMJ703-NOG-H3K4me3) was determined via the molecular replacement method with the modified crystal structure of the core of human JMJD2A (PDB code 2OQ6) [22] as the initial searching model. The final model was best at a resolution of 2.35 Å with a final Rwork value of 19.3% (Rfree = 22.5%). The crystals belonged to the P63 space group with a slight change in unit cell parameters among the three crystals. One c-JMJ703 molecule was identified in the asymmetric unit with a Matthews coefficient of 2.3 Å3/Da (corresponding to 46% solvent content) [23]. There are five solvent-exposed regions in c-JMJ703 structure, including P195-K199, S224-R261, R288-S295, T329-Y349, and Q363-V377, which could not be built due to lack of interpretable electron density, suggesting their intrinsic structural flexibility. Moreover, although inter-molecular interactions were found in the c-JMJ703 crystal, gel filtration revealed that c-JMJ703 existed as a monomer in solution (Figure S3), suggesting that the monomer may be the biological unit.
The c-JMJ703 molecule presented a canonical overall folding of JMJD2 proteins and contained four of the five domains defined in the structure of c-JMJD2A: the JmjN domain (A139-K199), the long β-hairpin (D200-T271), the mixed domain (L272-V377) and the JmjC domain (L378-A498) (Figure 3A, Figure S4). The JmjC domain, sandwiched by the JmjN and the long β-hairpin with the mixed domain, adopted a jellyroll-like structure with two four-stranded β-sheets as a cupin fold (Figure 3A) [24].
Fig. 3. Overall structure of c-JMJ703. 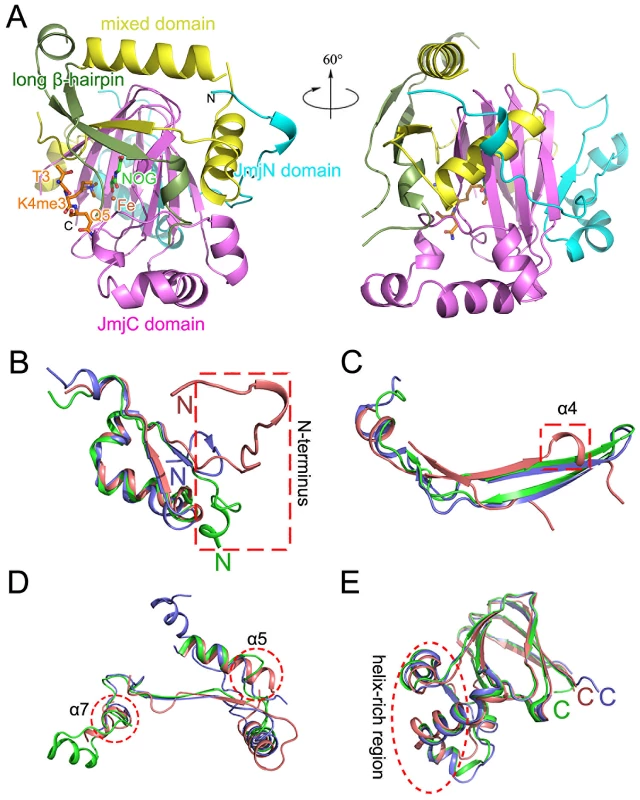
(A) Cartoon representation of the overall structure of c-JMJ703. Separate domains are colored differently and labeled. (B–E) Structural comparison of JmjN domain (B), long-beta sheet domain (C), mixed domain (D), and JmjC domain (E) among c-JMJ703 (deep salmon), c-JMJD2A (tv green), and c-Rph1 (slate). Although c-JMJ703 shared low primary sequence similarity (less than 25% of the sequence identities) with the reported structural homologues [20], [22], the core portion, especially the JmjC domain and the catalytic center of c-JMJ703, presented a topology similar to that of JMJD2 proteins with root mean square derivations of 1.75 Å and 1.81 Å relative to c-JMJD2A and c-Rph1, respectively (Figure 3B–3E). Nevertheless, c-JMJ703 displayed several significant structural differences. First, the JmjN domain presented a number of distinct features (Figure 3B). The orientation of the N-terminus of c-JMJ703 was opposite to that of c-JMJD2A and c-Rph1 (Figure 3B). Second, the long β-hairpin domain contained two β-strands (β2 and β3, aa 209–215 and aa 267–270) and a short α-helix (α4, aa 216–219). The two β-strands of c-JMJ703 were shorter than that of c-JMJD2A and c-Rph1, but had an extra long insertion between them. However, except for the short α-helix (α4), this insertion was mostly invisible in the structure (Figure 3C, Figure S4A). The short α-helix (α4) represented a sharp difference between c-JMJ703 and c-JMJD2A/c-Rph1. Moreover, the mixed domain of c-JMJ703, composed of several different structural elements, also showed clear structural differences. The first α-helix (α5, aa 272–286) of the mixed domain was two turns longer than that of c-JMJD2A and c-Rph1 (Figure 3D). The residues Q363 to V377 in the mixed domain of c-JMJ703 were structurally disordered and presented an uninterpretable electron density, whereas the corresponding region of c-JMJD2A is an α-helix. An additional short helix (α7, aa 356–361) was observed in c-JMJ703, whereas a loop was present in c-JMJD2A (Figure 3D). Several distinct differences could also be observed in the JmjC domain of JMJ703 compared to mammalian and yeast homologues, particularly in the helix-rich region (α8 to α11, aa 419–459) (Figure 3E, Figure S4A). Residues W381, C392, F437, Q440, L443, H445, L447 and V448 are conserved in and specific to JARID and plant H3K4 demethylases (Figure 4A). Most of these residues were located in the helix-rich region (α8 to α11) except W381 and C392 (Figure 4B). A potential extension path of H3K4me peptide toward its C-terminal was proposed based on the locations of these H3K4 demethylase-specific residues (Figure 4B). Substitution mutations of W381 and L447 abolished the H3K4 demethylase activities (Table 1, Figure S6), indicating that these residues are essential for the demethylation function of the protein.
Fig. 4. H3K4 demethylase-specific residues within the JmjC domain. 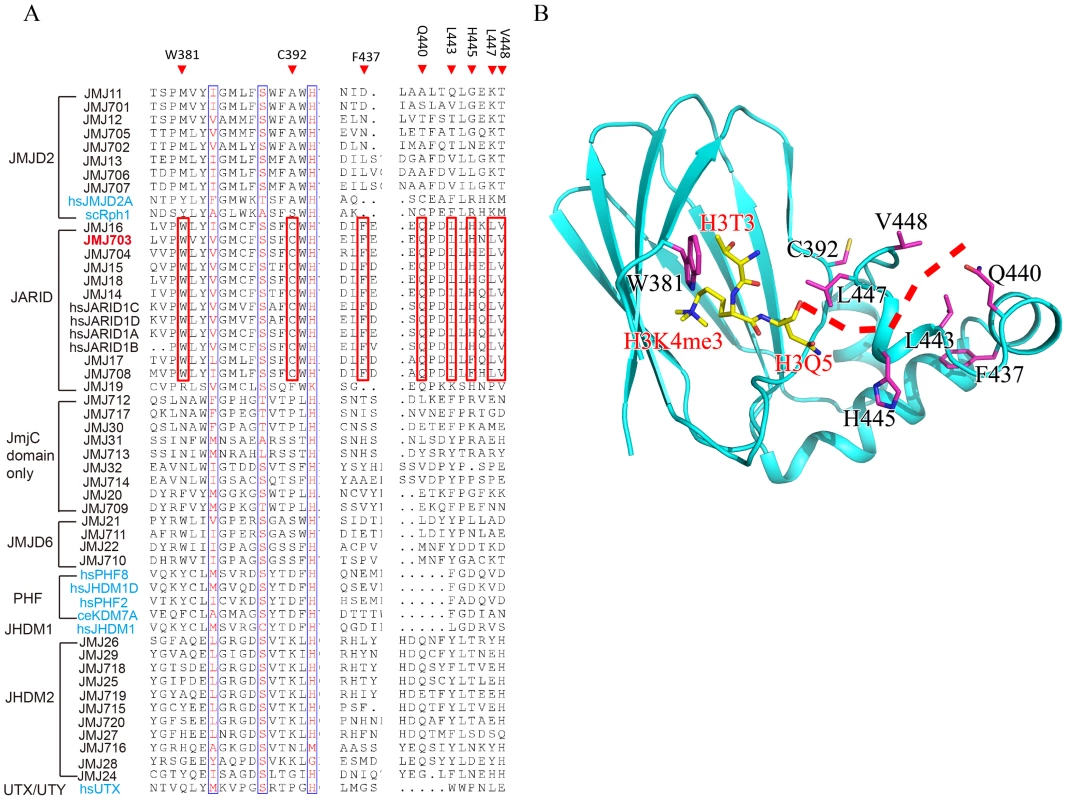
(A) Sequence alignment of the JmjC domain of JMJ703, plant JMJ proteins (in black) and representative human/yeast JMJ proteins (in blue). Residues conserved in and specific to H3K4 demethylases are boxed in red and labeled above the alignment. (B) Structure of JmjC domain of JMJ703 showing the locations of H3K4 demethylase-specific residues. H3K4me3 peptide and H3K4 demethylase-specific residues are shown in yellow sticks and purple sticks respectively. A proposed extension path of H3K4me3 peptide toward its C-terminal is shown in dashed red line. Tab. 1. Summary of H3K4 demethylation assays in tobacco cells of substitution mutants of key residues of JMJ703 catalytic domain. 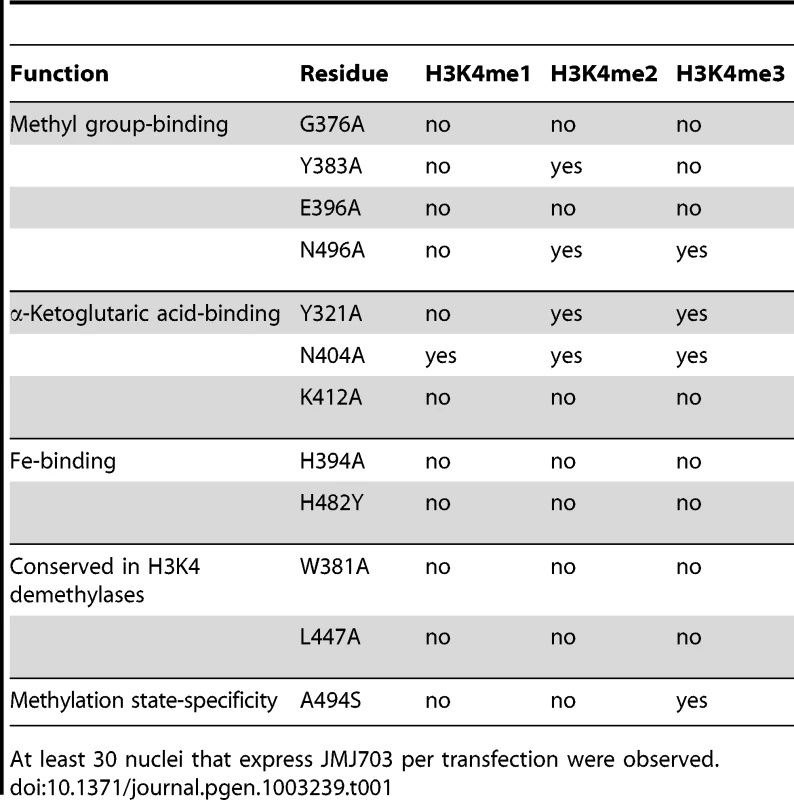
At least 30 nuclei that express JMJ703 per transfection were observed. Catalytic machinery of c-JMJ703
The unambiguous electron density denoted Fe(II) (determined by inductively coupled plasma mass spectrometry) bound to c-JMJ703 and identified the active site within the JmjC domain (Figure 5). Three key residues, H394, E396, and H482, are perfectly conserved in JMJD2 proteins. They chelated Fe(II) in the active site through their hydrophilic side chains. Fe(II) also interacted with the C-1 carboxyl and C-2 oxo groups of NOG (Figure 5). Substitution mutations H394A, E396A, and H482A abolished the activity of JMJ703 to demethylate H3K4me1/2/3 (Table 1, Figure S6), confirming their critical role in the enzymatic activity of the protein.
Fig. 5. The active site of c-JMJ703. 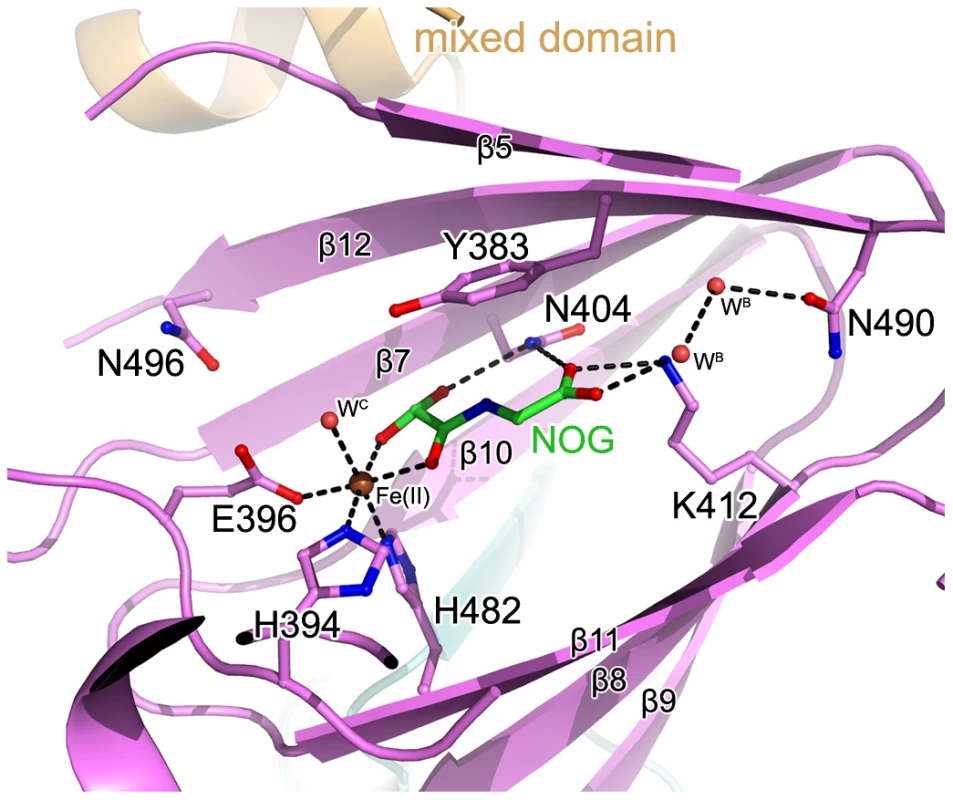
The interaction network of NOG (colored green), Fe(II) (colored brown), and c-JMJ703 residues (colored purple) involved in the interactions are shown. The H3K4me3 peptide is colored yellow. The secondary structure surrounding the active site is shown as a purple cartoon. The electron density indicated the presence of substrate peptide in the c-JMJ703-NOG-H3K4me3 complex (Figure 3, Figure 6). Three of the ten residues in the H3K4me3 peptide (ARTKme3QTARKS) were visible in the c-JMJ703-NOG-H3K4me3 complex structure (Figure 3, Figure 5, Figure 6). This complex structure provided a view of the substrate peptide-binding mode of an H3K4 demethylase. Compared with the structure of c-JMJD2A in complex with methylated H3K9/H3K36 peptide, the methylated H3K4 and its two flanking residues adopted a different binding conformation to c-JMJ703. H3K9me3/H3K36me3 and their two flanking residues stretched along the long axis of α-KG in c-JMJD2A-α-KG-H3K9me3/c-JMJD2A-NOG-H3K36me3 structure (PDB code 2Q8C/2Q8E, Figure 7B–7C), whereas H3K4me3 and its two flanking residues stretched along the short axis of NOG and was almost perpendicular to H3K9me3/H3K36me3 and their flanking residues (Figure 7A).
Fig. 6. Electron density of bound H3K4me3. 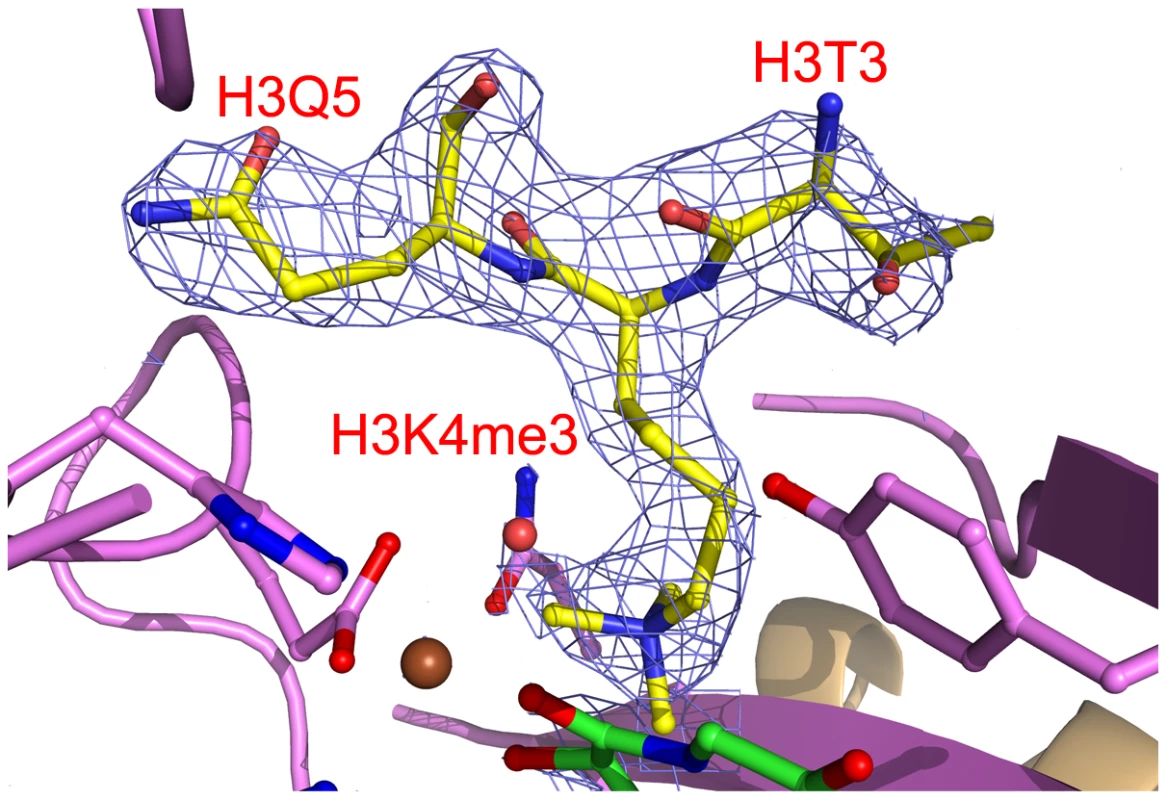
The H3K4me3 residues are shown in yellow sticks and labeled in red. Residues surrounding the peptide binding pocket are shown as cartoon and sticks and are colored as in Figure 3. The H3K4me3 peptide is covered by electron density (2Fo-Fc map at 1.0σ). Fig. 7. Comparison of the substrate peptide binding modes of c-JMJ703 and c-JMJD2A. 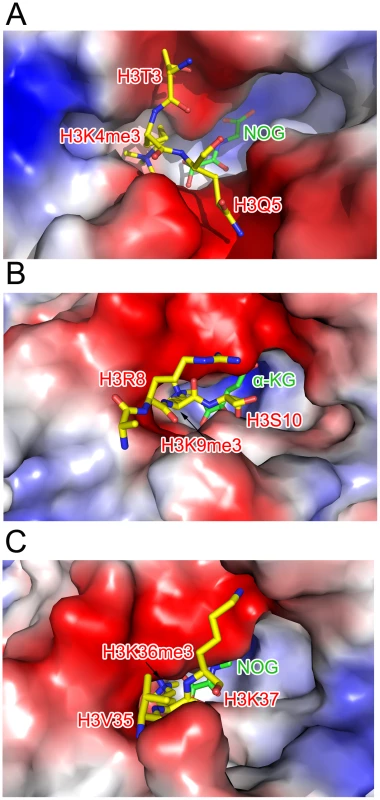
(A) Binding of H3K4me3 peptide by c-JMJ703. (B) Binding of H3K9me3 by c-JMJD2A. (C) Binding of H3K36me3 by c-JMJD2A. c-JMJ703 and c-JMJD2A are shown as electrostatic surface and the substrate peptides are shown as yellow sticks and the residues are labeled in red. NOG and α-KG are shown as green sticks and labeled. The methyl group binding pocket of JmjC domain is unique among methylated peptide - binding proteins due to the polar rather than hydrophobic environment [25]. The methyl groups could not be defined properly in the c-JMJ703-NOG-H3K4me3 complex structure because of low occupancy as indicated by obviously higher B-factor for atoms in H3K4me3 peptide. Based on the crystal structure of c-JMJD2A in complex with H3K9me3 (PDB code 2Q8C) [26], four of the five residues that have been shown to be important in methyl group binding in c-JMJD2A [25] were conserved in c-JMJ703, namely, G376, Y383, E396, and N496. S288 in c-JMJD2A (S335 in Rph1) is substituted by an alanine (A494) in c-JMJ703. G376 had no electron density and was invisible in structure, whereas Y383, E396, A494, and N496 were well-defined and adopted very similar conformations as in c-JMJD2A [26]. Substitution mutations of these potential methyl group-binding residues (G376A, Y383A, E396A, N496A) generally impaired the H3K4 demethylase activity of JMJ703 in tobacco cells, with the exceptions of Y383A, which retained a residual activity to demethylate H3K4me2, and N496A, which was still active to demethylate H3K4me2/3 (Table 1, Figure S6).
Distinct conformations of Y321 and N404 in stabilizing α-KG
In the crystal structure of c-JMJ703-NOG-H3K4me3, NOG formed three hydrogen bonds with the side chains of N404 and K412 (Figure 5) similar to the JMJD2 homologues. K412A mutation also abolished demethylation activity of H3K4 in all three methylation states (Table 1, Figure S6). Unexpectedly, the N404A mutation produced no clear effect on the demethylase activity of JMJ703, whereas the mutation of the counterpart in Rph1 abolished the H3K36me3 demethylase activity [20]. Moreover, NOG interacted indirectly with N490 through two water molecules (Figure 5), a finding that is not observed in mammalian and yeast homologues. Although most of the residues in α-KG/NOG stabilization showed high structural similarities with its homologues, several features, particularly those of Y321 and N404, distinguished JMJ703 from the JMJD2 members. Y321 and N404 showed significant conformational shifts compared with their counterparts in c-Rph1 and c-JMJD2A (Figure 8A–8C and Figure S5). Alternative conformation of the Tyr corresponding to Y321 in other JmjC proteins has been observed in JMJD2D (PDB code 3DXU). Interestingly, substitution of Y321 by alanine decreased H3K4me1 demethylase activity but did not affect that of H3K4me2 and H3K4me3 (Table 1, Figure S6), whereas substitution by alanine of the corresponding residue in Rph1 leads to loss of the H3K36me3 demethylase activity of the protein [20]. These data suggested a different NOG-binding mechanism between JMJ703 and Rph1.
Fig. 8. Conformational shifts of α-KG/NOG interacting residues of c-JMJ703 and other members of JMJD2A. 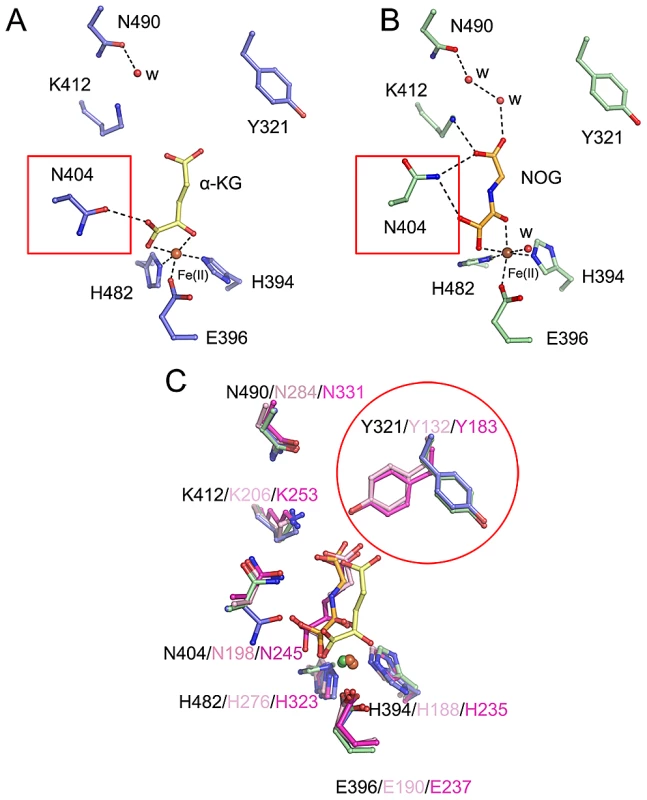
Residues interacting with (A) α-KG and (B) NOG in c-JMJ703 structures are represented as blue and green sticks, respectively. Dashed lines denote hydrogen bonds, including the solvent-mediated interactions formed by residues of c-JMJ703 and solvent molecules and bound α-KG/NOG. Fe(II) and solvent molecules are shown as deep red and red spheres, respectively. (C) Comparison of residues involved in binding to Fe(II) and α-KG/NOG in c-JMJ703, c-JMJD2A, and c-Rph1. Residues of c-JMJ703-α-KG, c-JMJ703-NOG-H3K4me3, c-JMJD2A, and c-Rph1 are colored in blue, green, light pink, and purple, respectively. Numbers for each residue in c-JMJ703, c-JMJD2A, and c-Rph1 are shown with black, light pink, and purple, respectively. Solvent molecules are not included in this panel. N404 and Y321, which adopt significant conformation shifts, are framed out by red squares and a circle. Discussion
In this work, we have provided evidence that JMJ703 is a histone demethylase specifically reversing the three forms of methylated H3K4. Arabidopsis JMJ14 that belongs to the same group as JMJ703 also has an H3K4 demethylase activity [27]. About two-thirds of plant genes contain at least one type of H3K4me, among which H3K4me3 is associated with gene activation in Arabidopsis and rice [28], [29]. Demethylation of the three forms of H3K4me implies that JMJ703 is important for rice gene expression. The phenotype caused by T-DNA insertion and RNAi and increased expression of CKX genes suggest that JMJ703 may regulate growth hormone metabolism in rice, which is different from JMJ14 that has functions in floral transition, DNA methylation, and RNA silencing in Arabidopsis [27], [30]–[33]. H3K4me3 on specific plant genes can be induced by external signals, such as light, drought, and submergence, among others [34]–[36]. H3K4me3 is suggested to be a mark of active genes and may play a role in plant adaptation to environmental cues. Whether JMJ703 is involved in resetting stress-induced gene expression requires further analysis.
Although JmjC domain of JMJ703 is closely related to that of the JARID proteins [17], the domain organization of the catalytic core of the enzyme (JmjN - long β-hairpin-mixed domain-JmjC) resembles that of JMJD2 members (c-JMJD2A and c-Rph1). However, our functional analysis has revealed that JMJ703 (i.e. FA-J3NCZ) was unable to demethylate H3K9me1/2/3 or H3K36me1/2/3 (Figure S2A–S2B). This suggests that the enzymatic specificity may be determined essentially in the JmjC domain. Our data indicate that the helix-rich region (α8–α11, aa 419–459) of the JmjC domain of JMJ703 is substantially divergent from that of JMJD2A and Rph1. For instance, K241 of c-JMJD2A, which is conserved in c-Rph1, is considered to be a key catalytic residue that recruits and positions O2 between Fe(II) and the methyl group to participate in the reaction [25]. Substitution mutation of K241L abolishes the activity of c-JMJD2A [25]. However, this position is replaced by a leucine in JMJ703 (L447) and in other plant homologues and JARID proteins (Figure 4). L447 was found to be essential for the H3K4 demethylase activity of JMJ703 (Table 1, Figure S6). This leucine together with the other residues that are specifically conserved in JARID/H3K4 demethylases and mostly found in the helix-rich region (α8–α11, aa 419–459) of c-JMJ703 (Table 1, Figure 4 and Figure S6), may be involved in the specificity of H3K4 demethylases.
Our data indicate that although most of the JMJ703 residues in α-KG/NOG stabilization showed a high structural similarity with JMJD2 proteins, the residues Y321 and N404 displayed specific features (Figure 8, Figure S5). The conformation of Y321 in c-JMJ703 indicates that the residue may not be in direct contact with α-KG or NOG. Substitution mutation of Y321 only affected H3K4me1 demethylase activity. In addition, N404A substitution did not clearly alter the H3K4 demethylase activity.
The methyl group binding pocket in c-JMJ703 differs also from that of c-JMJD2A and Rph1 in one key residue (A494 in JMJ703 compared to S288 in JMJD2A or S355 in Rph1). S288 of JMJD2A has been shown to be important to reverse di - and trimethylated H3K9 [25], [26], [37]. However, JMJD2D, which harbors an alanine in this position, is 7-fold and 60-fold more efficient than JMJD2A in the demethylation of H3K9me3 and H3K9me2, respectively. JMJD2D has residual activity to H3K9me1 that is absent from JMJD2A [26]. The presence of alanine at this position in JMJ703 (A494) supports the demethylation function of the protein of all three forms of methylated H3K4 (Figure 2). Substitution of this alanine by a serine abolished the demethylase activity of H3K4me1/2, but retained the activity toward H3K4me3 (Table 1, Figure S6). Interestingly, this alanine residue is conserved in most of the plant JMJD2 members (Figure S4), suggesting that these plant proteins may be able to demethylate the three methylated forms of relevant histone lysines.
In addition, the zinc finger that overlaps partially with the JmjC domain of JMJD2A is not found in JMJ703. Although the residues involved in the zinc finger formation in JMJD2A are conserved in Rph1, no zinc ion is observed in its structure and this domain is disordered and invisible in the c-Rph1 structure [20]. By contrast, JMJ703 contains a distal zinc finger motif relative to the JmjC domain (about 100 amino acids downstream the JmjC domain), which is present in Rph1, but absent from JMJD2A. The distal zinc finger is found in a number of other JmjC proteins and is shown to be involved in nuclear localization of JMJ706 [17]. These observations together with our data showing that the inclusion of the motif increased the substrate peptide (H3K4me1/2/3) binding activity to two-folds but not alter the binding specificity, suggest that this zinc finger may not be a component of the catalytic core of JmjC proteins, but is important for their enzymatic activity. Our data also revealed a number of other clear structural differences in the plant JmjC protein. The N-terminal end of c-JMJ703 adopted a completely different conformation from that of c-JMJD2A and c-Rph1. The long β-hairpin domain and mixed domain of c-JMJ703 also showed significant divergence at the amino acid sequence level compared to c-JMJD2A and c-Rph1 (Figure S4). However, the divergent regions are mostly invisible in c-JMJ703 and c-Rph1, indicating intrinsic disorder.
In summary, we have shown that JMJ703 is an H3K4 demethylase that is important for rice plant development. Although JMJ703 displays an overall structure similar to that of JMJD2 H3K9 and H3K36 demethylases, it exhibits a number of important structural features which are conserved within JARID and plant H3K4 demethylases. In particular, our data suggest that the helix-rich region (α8–α11, aa 419–459) of the catalytic JmjC domain of JMJ703 may be involved in the determination of substrate specificity of the H3K4 demethylase.
Materials and Methods
T-DNA insertion identification of jmj703 mutant
A T-DNA insertion line of JMJ703 (3A-00550) was obtained from the Postech rice mutant database (http://www.postech.ac.kr/life/pfg/risd/). Insertion was confirmed by PCR using the primers muJ3-F and muJ3-R and a T-DNA-specific primer 2715L1 (Table S2).
RNAi vector construction and rice transformation
A cDNA fragment between nucleotides 2282 bp and 2834 bp relative to the translation start site was amplified using primers RiJ3-F and RiJ3-R and then cloned into T vector (Promega). The fragment was cloned into double strand RNAi vector pDS1301 [38]. Rice transformation with callus generated from the rice cultivar Zhonghua11 (ZH11) was performed as previously described [38].
Histone demethylation assay
The cDNA fragment of the J3NCZ region was amplified using primers FAJ3NCZ-F and FAJ3NCZ-R and then cloned into pFA121 vector modified from pBI121 (GenBank: AF485783.1), in which GUS was replaced by 2×FLAG∶2×HA tag. The pFA121-J3NCZ plasmid was transferred into Agrobacterium tumefaciens strain EHA105 cells for tobacco infection with the help of Agrobacterium cells harboring P19 plasmids [39]. For in vivo histone demethylation assay, tobacco infection and nuclei isolation were performed as previously described [27]. The immunostaining protocol was modified from http://sites.bio.indiana.edu/~pikaardlab. Briefly, the nucleus solution was placed on a poly-lysine coated slide, air dried, and then refixed with 4% formaldehyde in KPBS (1.28 M NaCl, 20 mM KCl, 80 mM Na2HPO4, 20 mM KH2PO4, pH7.2) containing 1% Triton X-100 for 20 min. After 3 washes with KPBS-1% Triton, slides were blocked with blocking solution (1% BSA in KPBS-1% Triton) and incubated at 37°C for 30 min. The blocking solution was washed off with KPBS-1% Triton and the slides were incubated with both anti-HA and histone méthylation specific antibodies diluted in blocking solution to 1∶300 at 4°C overnight. After washing with KPBS, slides were blocked and incubated with fluorescent-labeled antibodies at 37°C for 2 h. After washing with KPBS thrice, nuclei were stained with 5 µg/mL 4,6-diamidino-2-phenylindole (DAPI), coated with a drop of VECTASHIELD Mounting Medium (H-1000, Vector Laboratories), and then covered with a coverslip followed by confocal microscope (Leica) detection. In vitro histone demethylation assays were performed as previously described [6]. FA-J3NCZ was purified after overexpression with Anti-FLAG M2 magnetic beads according the manufacturer's instruction (Sigma).
Antibodies used in this study: anti-HA (M20003M, Ab-mart), anti-H3K4me1 (ab8895, Abcam), anti-H3K4me2 (04-790, Millipore), anti-H3K4me3 (07-473, Millipore), anti-H3K27me3 (ABE44, Millipore), anti-H3 (ab1791, Abcam), anti-H3K9me1 (ab9045, Abcam), anti-H3K9me2 (07-441, Millipore), anti-H3K9me3 (ab8898, Abcam), H3K36me1 (ab9048, Abcam), anti-H3K36me2 (ab9049, Abcam), anti-H3K36me3 (ab9050, Abcam), Goat Anti-Mouse secondary antibody (A-11029, Invitrogen), and Goat Anti-Rabbit secondary antibody (A-11036, Invitrogen).
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed as previously described [38]. Briefly, 2 g of 10 day-old rice seedlings were fixed with 1% formaldehyde, then chromatin was extracted and immunoprecipitated with anti-H3K4me3 (ab8580, Abcam) and anti-H3K4me2 (04-790, Millipore) antibodies. After reversing crosslink and protease K treatment, DNA was recovered for realtime PCR analysis with primers listed in Table S2.
Protein production for crystallization
DNA fragment encoding amino acids 139 to 498 of wild type JMJ703 was amplified by PCR, ligated into pET-28a expression vector using BamHI and XhoI and transformed into Escherichia coli Rosetta (DE3) cells. Transformed E. coli cells were cultured at 37°C in LB medium containing 50 mg/L kanamycin until the OD600 nm reached 0.8. The culture was cooled to 16°C and subsequently induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside. Cells were harvested after overnight induction by centrifugation at 5,000 g for 10 min at 4°C. The cell pellets were resuspended in lysis buffer containing 50 mM Tris (pH 8.0), 500 mM NaCl, and 10% (v/v) glycerol and then disrupted by an ultra-high pressure cell disrupter (JNBIO, Guangzhou, China) at low temperature. Cell debris was removed by centrifugation at 25,000 g for 30 min at 4°C. The 6×His tagged protein was purified by Ni-NTA affinity chromatography, cleaved with thrombin (Sigma) overnight at 4°C, and eluted with lysis buffer. The purified c-JMJ703 protein was concentrated and further purified by Hitrap Q (GE Healthcare) anion-exchange chromatography using a 0.05 M to 1 M NaCl gradient in 25 mM HEPES (pH 7.5). The target protein was confirmed to have purity over 95% by SDS-PAGE and concentrated to 10 mg/mL before crystal growth or storage.
Crystallization
Crystallization was performed at 18°C by the hanging-drop vapor-diffusion technique. Crystals were obtained by mixing 1 µL of the protein solution with an equal volume of a reservoir solution; the mixture drop was equilibrated against 500 µL of the reservoir solution. Crystals were obtained with a reservoir solution containing 0.05 M potassium phosphate monobasic, 25% (w/v) polyethylene glycol 8,000 and reached final dimensions of 50×50×100 µm3 within 2 days. Crystals of apo c-JMJ703 were initially obtained but showed poor diffraction at 3.0 Å resolution and poor reproducibility. Crystals of c-JMJ703 in complex with NOG displayed much better diffraction quality at 2.3 Å, indicating that NOG helps stabilize the conformation of the c-JMJ703 polypeptide. Crystals of the c-JMJ703 substrate complex were obtained by co-crystalizing c-JMJ703 with 10 mM peptide. Crystals were cryo-protected by soaking in a cryo-protectant consisting of the reservoir solution with additional 15% (v/v) glycol. Cryo-protected crystals were then flash-cooled in liquid nitrogen and transferred into a dry nitrogen stream at 100 K for X-ray data collection.
X-Ray data collection, processing, and structure determination
Diffraction data for apo c-JMJ703 crystals were collected at a resolution of 3.0 Å at 100 K using a MAResearch M165 CCD detector in beamline 1W2A at the Beijing Synchrotron Radiation Facility. The data set for c-JMJ703-α-KG and c-JMJ703-NOG-H3K4me3 were collected at 2.3 Å and 2.4 Å in beamline BL17A (Photon Factory, Japan) and BL17U1 (SSRF, Shanghai, China), respectively, with an ADSC Q315 CCD detector at the wavelength of 1.0000 Å. All data sets were indexed, integrated, and scaled using the HKL2000 package [40]. The crystals belonged to space group P63 with cell parameters a = 53.0 Å, b = 86.1 Å, c = 87.3 Å, and β = 90.7°. All crystals showed very small shifts in their cell parameters.
The molecular replacement method was used to calculate the phases using the PHASER program [41] and the modified crystal structure of a catalytic core of the human c-JMJD2A (PDB code 2OQ6) as the initial searching model. Manual model building and refinement were performed with the programs COOT [42] and PHENIX [43]. Solvent molecules were located from stereochemically reasonable peaks in the σA-weighted 2Fo–Fc difference Fourier electron density map (1.2 σ). Model geometry was verified using the program PROCHECK (Table S3) [44]. Coordinates have been deposited in PDB with accession 4IGP, 4IGO and 4IGQ for apo c-JMJ703, c-JMJ703-α-KG and c-JMJ703-NOG-H3K4me3, respectively.
Surface plasmon resonance binding analysis
Analyses were carried out at 25°C with the BIAcore 3000 system. Ten µg/mL c-JMJ703 in 10 mM sodium acetate buffer (pH 5.0) was covalently coupled to a CM5 chip (Biacore) using an Amine Coupling Kit (Biacore) according to the manufacturer's instructions. Using HEPES buffer (10 mM HEPES pH 7.5, 150 mM NaCl, 0.005% Tween20), various concentrations of the H3K4me1/2/3 peptide were injected through a flow cell that was not activated and then through another flow cell containing c-JMJ703 at a rate of 10 µL/min for 2 min. The c-JMJ703 surface was regenerated between two injections by running 15 µL of 5 mM NaOH twice through the flow cell at 30 µL/min. The sensorgram obtained for the inactivated flow cell was subtracted to correct for nonspecific binding and the bulk signal from the peptide in solution. Data were analyzed using BIAevaluation 4.1 software.
Supporting Information
Zdroje
1. MosammaparastN, ShiY (2010) Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem 79 : 155–179.
2. ShiY, LanF, MatsonC, MulliganP, WhetstineJR, et al. (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119 : 941–953.
3. TsukadaY, FangJ, Erdjument-BromageH, WarrenME, BorchersCH, et al. (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439 : 811–816.
4. KloseRJ, KallinEM, ZhangY (2006) JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7 : 715–727.
5. YamaneK, ToumazouC, TsukadaY, Erdjument-BromageH, TempstP, et al. (2006) JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125 : 483–495.
6. WhetstineJR, NottkeA, LanF, HuarteM, SmolikovS, et al. (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125 : 467–481.
7. AggerK, CloosPA, ChristensenJ, PasiniD, RoseS, et al. (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449 : 731–734.
8. LiuW, TanasaB, TyurinaOV, ZhouTY, GassmannR, et al. (2010) PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466 : 508–512.
9. QiHH, SarkissianM, HuGQ, WangZ, BhattacharjeeA, et al. (2010) Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466 : 503–507.
10. YatimA, BenneC, SobhianB, Laurent-ChabalierS, DeasO, et al. (2012) NOTCH1 Nuclear Interactome Reveals Key Regulators of Its Transcriptional Activity and Oncogenic Function. Mol Cell 48 : 445–458.
11. SewardDJ, CubberleyG, KimS, SchonewaldM, ZhangL, et al. (2007) Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat Struct Mol Biol 14 : 240–242.
12. ChangB, ChenY, ZhaoY, BruickRK (2007) JMJD6 is a histone arginine demethylase. Science 318 : 444–447.
13. LandoD, PeetDJ, GormanJJ, WhelanDA, WhitelawML, et al. (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev 16 : 1466–1471.
14. WebbyCJ, WolfA, GromakN, DregerM, KramerH, et al. (2009) Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325 : 90–93.
15. ChenX, HuY, ZhouDX (2011) Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim Biophys Acta 1809 : 421–426.
16. LuF, CuiX, ZhangS, JenuweinT, CaoX (2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43 : 715–719.
17. SunQ, ZhouDX (2008) Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci U S A 105 : 13679–13684.
18. LuF, LiG, CuiX, LiuC, WangXJ, et al. (2008) Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 50 : 886–896.
19. HouH, YuH (2010) Structural insights into histone lysine demethylation. Curr Opin Struct Biol 20 : 739–748.
20. ChangY, WuJ, TongXJ, ZhouJQ, DingJ (2010) Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism. Biochem J 433 : 295–302.
21. SengokuT, YokoyamaS (2011) Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev 25 : 2266–2277.
22. ChenZ, ZangJ, WhetstineJ, HongX, DavrazouF, et al. (2006) Structural insights into histone demethylation by JMJD2 family members. Cell 125 : 691–702.
23. MatthewsBW (1968) Solvent content of protein crystals. J Mol Biol 33 : 491–497.
24. ClissoldPM, PontingCP (2001) JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem Sci 26 : 7–9.
25. ChenZ, ZangJ, KapplerJ, HongX, CrawfordF, et al. (2007) Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc Natl Acad Sci U S A 104 : 10818–10823.
26. CoutureJF, CollazoE, Ortiz-TelloPA, BrunzelleJS, TrievelRC (2007) Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol 14 : 689–695.
27. LuF, CuiX, ZhangS, LiuC, CaoX (2010) JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res 20 : 387–390.
28. ZhangX, BernatavichuteYV, CokusS, PellegriniM, JacobsenSE (2009) Genome-wide analysis of mono-, di - and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10: R62.
29. HeG, ZhuX, EllingAA, ChenL, WangX, et al. (2010) Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22 : 17–33.
30. SearleIR, PontesO, MelnykCW, SmithLM, BaulcombeDC (2010) JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev 24 : 986–991.
31. YangW, JiangD, JiangJ, HeY (2010) A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J 62 : 663–673.
32. JeongJH, SongHR, KoJH, JeongYM, KwonYE, et al. (2009) Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS ONE 4: e8033 doi:10.1371/journal.pone.0008033.
33. DelerisA, GreenbergMV, AusinI, LawRW, MoissiardG, et al. (2010) Involvement of a Jumonji-C domain-containing histone demethylase in DRM2-mediated maintenance of DNA methylation. EMBO Rep 11 : 950–955.
34. CharronJB, HeH, EllingAA, DengXW (2009) Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21 : 3732–3748.
35. TsujiH, SaikaH, TsutsumiN, HiraiA, NakazonoM (2006) Dynamic and reversible changes in histone H3-Lys4 methylation and H3 acetylation occurring at submergence-inducible genes in rice. Plant Cell Physiol 47 : 995–1003.
36. KimJM, ToTK, IshidaJ, MorosawaT, KawashimaM, et al. (2008) Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol 49 : 1580–1588.
37. NgSS, KavanaghKL, McDonoughMA, ButlerD, PilkaES, et al. (2007) Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 448 : 87–91.
38. HuangL, SunQ, QinF, LiC, ZhaoY, et al. (2007) Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol 144 : 1508–1519.
39. VoinnetO, RivasS, MestreP, BaulcombeD (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33 : 949–956.
40. Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In: Carter Jr CW, Sweet RM, editors. Macromolecular Crystallography, part A: Academic Press. pp. 307–326.
41. McCoyA, Grosse-KunstleveR, AdamsP, WinnM, StoroniL, et al. (2007) Phaser crystallographic software. J Appl Cryst 40 : 658–674.
42. EmsleyP, CowtanK (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 : 2126–2132.
43. AdamsPD, Grosse-KunstleveRW, HungLW, IoergerTR, McCoyAJ, et al. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58 : 1948–1954.
44. LaskowskiR, MacArthurM, MossD, ThorntonJ (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26 : 283–291.
Štítky
Genetika Reprodukční medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 1
-
Všechny články tohoto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



