-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaFunction and Regulation of , a Gene Implicated in Autism and Human Evolution
Nucleotide changes in the AUTS2 locus, some of which affect only noncoding regions, are associated with autism and other neurological disorders, including attention deficit hyperactivity disorder, epilepsy, dyslexia, motor delay, language delay, visual impairment, microcephaly, and alcohol consumption. In addition, AUTS2 contains the most significantly accelerated genomic region differentiating humans from Neanderthals, which is primarily composed of noncoding variants. However, the function and regulation of this gene remain largely unknown. To characterize auts2 function, we knocked it down in zebrafish, leading to a smaller head size, neuronal reduction, and decreased mobility. To characterize AUTS2 regulatory elements, we tested sequences for enhancer activity in zebrafish and mice. We identified 23 functional zebrafish enhancers, 10 of which were active in the brain. Our mouse enhancer assays characterized three mouse brain enhancers that overlap an ASD–associated deletion and four mouse enhancers that reside in regions implicated in human evolution, two of which are active in the brain. Combined, our results show that AUTS2 is important for neurodevelopment and expose candidate enhancer sequences in which nucleotide variation could lead to neurological disease and human-specific traits.
Published in the journal: . PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003221
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003221Summary
Nucleotide changes in the AUTS2 locus, some of which affect only noncoding regions, are associated with autism and other neurological disorders, including attention deficit hyperactivity disorder, epilepsy, dyslexia, motor delay, language delay, visual impairment, microcephaly, and alcohol consumption. In addition, AUTS2 contains the most significantly accelerated genomic region differentiating humans from Neanderthals, which is primarily composed of noncoding variants. However, the function and regulation of this gene remain largely unknown. To characterize auts2 function, we knocked it down in zebrafish, leading to a smaller head size, neuronal reduction, and decreased mobility. To characterize AUTS2 regulatory elements, we tested sequences for enhancer activity in zebrafish and mice. We identified 23 functional zebrafish enhancers, 10 of which were active in the brain. Our mouse enhancer assays characterized three mouse brain enhancers that overlap an ASD–associated deletion and four mouse enhancers that reside in regions implicated in human evolution, two of which are active in the brain. Combined, our results show that AUTS2 is important for neurodevelopment and expose candidate enhancer sequences in which nucleotide variation could lead to neurological disease and human-specific traits.
Introduction
Autism spectrum disorders (ASDs) are common (1/88 in the United States) [1] childhood neurodevelopmental disorders known as pervasive developmental disorders (reviewed in [2]). ASDs are highly heritable, signifying a substantial genetic etiology [3]. A balanced translocation involving the autism susceptibility candidate 2 (AUTS2; GenBank NM_001127231.1) gene in a pair of monozygotic twins with ASD was the first to link this gene to autism [4] (Figure 1). Following this finding, thirty-six additional unrelated individuals with ASD, intellectual disability, or developmental delay were found to have distinct heterozygous structural variants disrupting the AUTS2 region [5]–[13], four exclusively in noncoding regions [5], [12] (Figure 1). Additional structural variants in AUTS2, some of which are only intronic, were also shown to be associated with attention deficit hyperactivity disorder (ADHD) [14], epilepsy [12], [15], dyslexia [11], motor delay, language delay, visual impairment, microcephaly and others [12]. In addition, a genome-wide association meta-analysis study identified SNP rs6943555 within the fourth intron of AUTS2 to be the most statistically significant SNP associated with alcohol consumption [16] (Figure 1). These various AUTS2-associated phenotypes suggest this gene has an important neurological function. It is worth noting though that some individuals with disrupted AUTS2 and mental retardation or autism have additional, potentially non-neuronal phenotypes, such as hypotonia, short stature, urogenital abnormalities, and skeletal abnormalities [4], [6].
Fig. 1. Schematic of the AUTS2 genomic region. 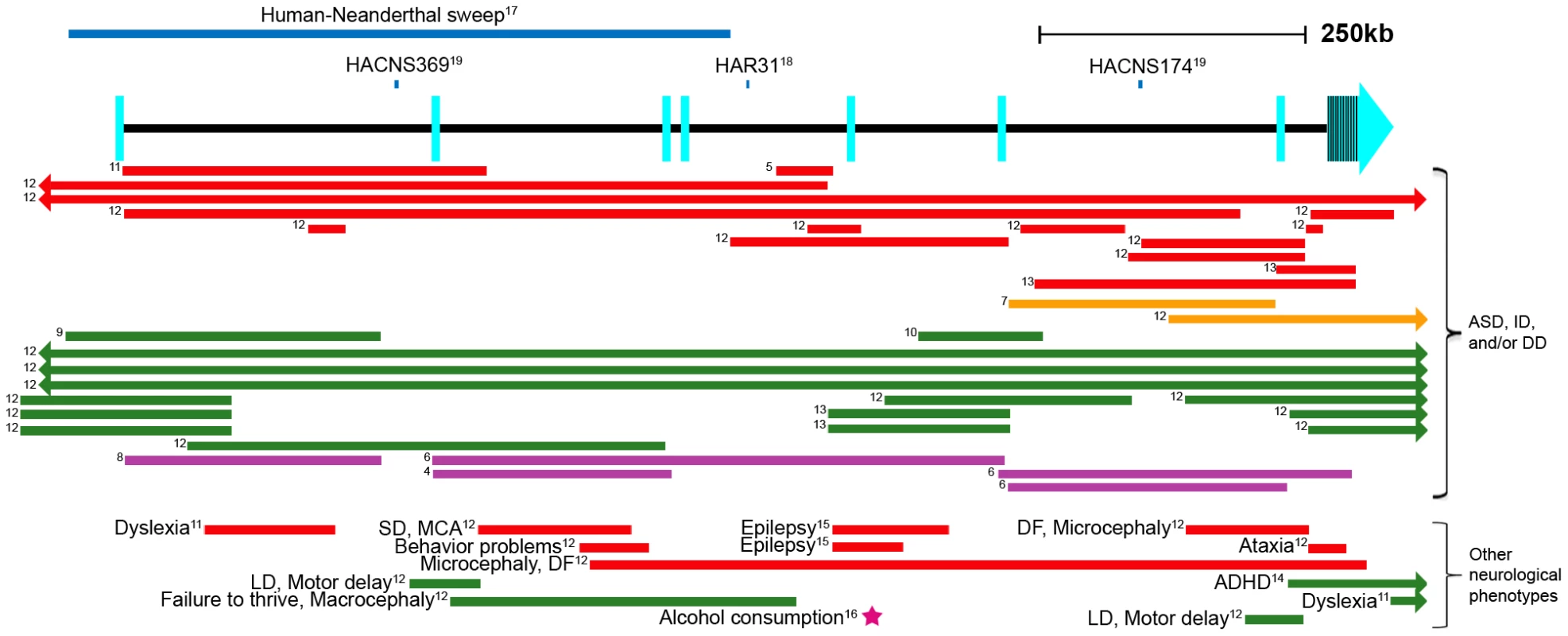
Human accelerated sequences are shown as blue lines above the gene [17]–[19]. Structural variants [4]–[12], [14], [15] are represented as colored lines (red: deletion, orange: inversion, green: duplication, purple: translocation). The rs6943555 SNP associated with alcohol consumption [16] is shown as a magenta star. Arrows in bars signify that the structural variant extends past the gene in that direction. Exons are depicted as light blue rectangles, as defined by the RefSeq genes track in the UCSC Genome Browser [52]. Numbers to the left of the lines correspond to a reference number. Human Accelerated Conserved Non-coding Sequence (HACNS), Human Accelerated Region (HAR), developmental delay (DD), intellectual disability (ID), dysmorphic features (DF), seizure disorder (SD), multiple congenital anomalies (MCA), language disability (LD). In addition to AUTS2's role in neurological disease, it was also shown to be important for human-specific evolution. The first half of AUTS2 displayed the strongest statistical signal in a genomic screen differentiating modern humans from Neanderthals [17]. This is attributed to a stretch of 293 consecutive SNPs, only two of which are coding variants: (a G to C nonsynonymous substitution at chr7 : 68,702,743 (hg18) only in the Han Chinese and a C to T synonymous change in chr7 : 68,702,866 (hg18) within the Yoruba and Melanesian populations). Other regions identified to have the most significant human-Neanderthal sweeps also include genes that are involved in cognition and social interaction, including DYRK1A, NRG3 and CADPS2 [17], reinforcing our interest in AUTS2's role in cognition and human-Neanderthal differences. In addition, three different evolutionary conserved noncoding intronic regions in AUTS2 (HAR31, HACNS174 and HACNS369) have been found to be significantly accelerated when compared to primates in two different studies [18], [19] (Figure 1). Combined, these data suggest that altered regulation of AUTS2 could be associated with human specific traits.
The functional role of AUTS2 is not well known, although some studies have identified a putative role in transcriptional regulation during neuronal development. The predicted AUTS2 protein contains a PY motif, a putative WW-domain-binding region [4] present in various transcription factors, implying that AUTS2 may be involved in transcriptional regulation [6]. In humans, AUTS2 is expressed in the brain, including the neocortex and prefrontal cortex [4], [20]. AUTS2 is also highly expressed in the skeletal muscle and the kidney, and in lower levels in the placenta, lung and leukocytes [4]. In the developing mouse, Auts2 is expressed in the forebrain, midbrain, hindbrain, olfactory bulb, olfactory epithelium, eye, neural tube and limb [21]. Among the regions that Auts2 was shown to be expressed in the brain are the neuronal nuclei in the developing cerebral cortex and cerebellum [22]. In the cortical preplate, Auts2 is activated by T-box brain 1 (Tbr1) [22], [23], a postmitotic projection neuron specific transcription factor that is critical for normal brain development. Tbr1 deficient mice display irregular laminar organization of cortical neurons [24]. Additionally, Cajal-Retzius cells in Tbr1 deficient mice have decreased levels of reelin (Reln) [23], a protein that is involved in neuronal migration in the developing brain and has been reported to be expressed at decreased levels in individuals with ASD [25].
In this study, we used zebrafish morpholinos to functionally characterize auts2. We show that knocking down this gene leads to an overall stunted developmental phenotype that includes a smaller head, body and reduced movement. Further characterization of morphant fish revealed a reduction in developing midbrain neurons and also in sensory and motor neurons. To characterize AUTS2 enhancers, we used both zebrafish and mouse transgenic enhancer assays. We identified three functional enhancers within an ASD-associated deletion and six brain enhancer in regions associated with human specific evolution. Combined, we found that AUTS2 is important for neuronal development and characterized several functional enhancers within this locus, where nucleotide changes could be associated with neurodevelopmental disease and human specific evolution.
Results
auts2 zebrafish expression
Zebrafish can be an effective tool to study ASD [26]. Using whole mount in situ hybridization, we determined that auts2 is expressed in zebrafish at 24 hours post fertilization (hpf) in the forebrain, midbrain and hindbrain (Figure S1A). Additionally, auts2 is expressed in the trunk (including the spinal cord), with stronger expression towards the caudal peduncle. At 48 hpf, auts2 is expressed in the brain and pectoral fin and from 72–120 hpf its expression is restricted primarily to the brain. auts2 is also weakly expressed in the eye from 24–120 hpf. Overall, we observed that the zebrafish expression largely correlates with the previously characterized mouse expression [21], [22].
Phenotypic characterization of auts2 morphants
We next used morpholinos (MOs) to knockdown auts2 in zebrafish during development. Fish injected with an auts2 translational blocking MO displayed a stunted developmental phenotype with smaller heads, eyes, body and pectoral fins (Figure 2A and Figure S1C). A second auts2 MO that disrupts the splice junction between intron two and exon three exhibited similar but less severe phenotypes (Figure S1D). These phenotypes appeared in 80–90% of injected fish and were rescued by co-injecting the full length human AUTS2 mRNA along with the translational blocking MO (68% of injected fish showed a partial to full rescue) (Figure S1E). Injection of a 5 base pair (bp) mismatch auts2 translational MO control did not show any phenotype (Figure 2A and Figure S1B), further validating the specificity of our MOs to effectively knockdown auts2 in zebrafish.
Fig. 2. auts2 48 hpf morphant phenotype. 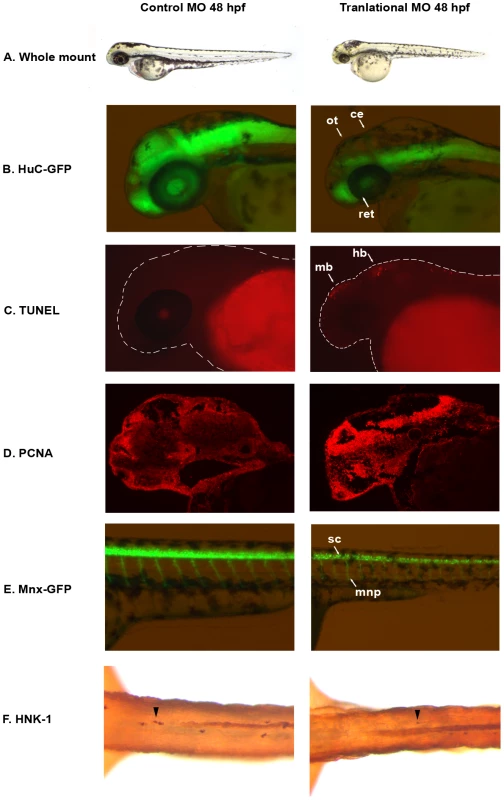
(A) Fish injected with the 5 base-pair translational MO mismatch control have similar morphology as wild type fish. Injection of the auts2 translational MO results in fish with a stunted development phenotype that includes a smaller head, eyes, body and fins. (B) HuC-GFP fish injected with the 5 bp control MO display normal levels of developing neurons in the brain. HuC-GFP translational MO injected fish display considerably less developing neurons in the optic tectum (ot), retina (ret), and cerebellum (ce). (C) 5 bp mismatch control injected fish have little to non-observable apoptosis in the brain as observed by TUNEL staining, while translational MO injected fish display high levels of apoptosis, primarily in the midbrain (mb) and hindbrain (hb). (D) PCNA cell proliferation assay in the 5 bp MO control injected fish shows lower levels of cell proliferation in the brain compared to the translational MO injected fish. (E) Tg(mnx1∶GFP) fish injected with the 5 bp MO control display normal levels of motor neurons versus the auts2 translational MO injected fish which have fewer motor neurons in the spinal cord (sc). In addition, motor neuron projections (mnp) are weaker and more perpendicular to the spinal cord. (F) Translational MO injected fish display fewer Rohon-Beard cells (arrowheads) in the spinal cord than morphants. All morphant fish are scaled to their 5 bp control counterparts. To further characterize the neurological function of auts2, we injected the translational MO into the HuC-GFP transgenic zebrafish line [27], where developing neurons express green fluorescent protein (GFP). Compared to the 5 bp mismatch control, translational MO injected fish showed a dramatic decrease in GFP at 48 and 72 hpf in the dorsal region of the midbrain, including the optic tectum, the midbrain-hindbrain boundary (which includes the cerebellum), the hindbrain and the retina (Figure 2B). This phenotype was also observed by staining neurons with Nissl at 48 hpf (Figure S2A). TUNEL staining of 48 hpf embryos revealed that morphant fish exhibit increased apoptosis in the midbrain in the same location where fewer neurons where observed (Figure 2C and Figure S2B). Anti-proliferating Cell Nuclear Antigen (PCNA) staining showed increased amounts of cell proliferation in morphant fish in the forebrain, midbrain and hindbrain (Figure 2D and Figure S2C–S2E). While seemingly contradictory, increased amounts of both TUNEL and PCNA positive cells has been previously shown, as cell death and proliferation could be coupled [28], [29]. It is conceivable that the increased PCNA positive cells are the result of morphant cells failing to differentiate into mature neurons, as seen in the HuC-GFP line. These results suggest that auts2 may be involved in the production and maintenance of neurons in the zebrafish brain.
Both the translational and splicing morphant fish also showed a decreased movement response when gently prodded with a pipette tip compared to controls that began at 48 hpf (Video S1 and Video S2). This phenotype was observed until 120 hours when the zebrafish were euthanized. In order to determine whether motor neuron defects could explain this phenotype, we injected the translational MOs into the Tg(mnx1∶GFP) zebrafish line, which expresses GFP in developing motor neurons [30]. At 48 hpf, morphant fish displayed fewer GFP labeled motor neuron cell bodies in the spinal cord. Additionally, motor neuron projections were weaker and perpendicular to the spinal cord, in contrast to the angled projections of the control injected fish (Figure 2E). This phenotype was also confirmed using the znp-1 antibody to mark motor neuron axons [31] in control and morphant fish. Morphant fish consistently showed more branching of axons compared to controls (Figure S3). To assess sensory neuron defects, Rohon-Beard neurons were stained with anti-HNK-1 in control and translational MO injected fish at 48 hpf. Morphant fish displayed on average 60% fewer sensory neurons in the spinal cord (Figure 2F). These results suggest that loss of auts2 in zebrafish could lead to motor and sensory neuron defects, which may play a role in their reduced movement and decreased response to touch.
AUTS2 enhancer characterization
Due to the observations that noncoding regions in the AUTS2 locus are associated with neurological phenotypes and human-specific evolution, we set out to identify enhancers in this locus. To focus our search, we limited our candidates to be between the first exon and fifth intron, due to this region encompassing the human-Neanderthal sweep (exon 1–4; chr7 : 68,662,946-69,274,862 (hg18)) [17] and several noncoding nucleotide changes that have been associated with neurological phenotypes [5], [11], [12], [16]. AUTS2 enhancer candidate (AEC) sequences were selected based on evolutionary conservation, embryonic mouse forebrain and midbrain ChIP-seq datasets [32] and nucleotide variants that define the human-Neanderthal sweep [17] (see methods). We also tested the human accelerated region (HAR) in intron four, HAR31 [18], and the human accelerated conserved non-coding sequences (HACNS) in introns one and six, HACNS 369 and HACNS 174 respectively [19]. Using these criteria, 40 AECs were selected for zebrafish enhancer assays (Table S1). These human sequences were cloned into the E1b-GFP-Tol2 enhancer assay vector and injected into zebrafish [33]. Of the 40 candidates, 23 were found to be functional enhancers, 22 of which showed enhancer activity in locations that overlap auts2 expression in zebrafish and 10 that were active in the brain (Table S1 and Figure S4).
To further characterize the regulatory elements within a 33,519bp deletion associated with ASD in AUTS2 intron four [5], the three positive zebrafish enhancers in this region (AEC27, AEC29, AEC32) were analyzed in mice using a similar transgenic assay [34]. AEC27 showed enhancer expression in the somitic muscle in zebrafish, while examination of its enhancer activity at E11.5 (hs658;[34]) found it to be active in the midbrain and neural tube (Figure 3). At E12.5, AEC29 had enhancer activity in the olfactory epithelium similar to zebrafish and also displayed enhancer expression in the eye (Figure 3). AEC32 recapitulated the zebrafish enhancer expression in the midbrain and hindbrain with additional enhancer expression in the forebrain at E12.5. Histological sections of AEC32 showed enhancer activity in the mouse cerebellum (Figure 3), a region thought to play a role in ASD [2]. The removal of these three brain enhancers and potentially other functional sequences in this region could contribute to the neurological phenotypes in patients with deletions in this intron.
Fig. 3. Enhancers within an ASD–associated AUTS2 intronic deletion [5]. ![Enhancers within an ASD–associated <i>AUTS2</i> intronic deletion <em class="ref">[<b>5</b>]</em>.](https://www.prolekarniky.cz/media/cache/resolve/media_object_image_small/media/image/0c66efbd5348d885d667fa84e07aa607.png)
Three positive enhancers (AEC27, 29, 32) show positive enhancer activity in zebrafish (24 or 48 hpf) and in mice (E11.5 or 12.5). AEC27 shows enhancer expression in the somitic muscle in zebrafish, while in mouse at E11.5 (hs658; [34]) it is active in the midbrain, medulla, and neural tube at E11.5. The histological section below shows its enhancer activity in the pretectum and the pons. At E12.5, AEC29 shows enhancer activity in the olfactory epithelium (arrows in histological section) similar to zebrafish and in addition also displays enhancer expression in the eye. AEC32 recapitulates the zebrafish enhancer expression displaying strong enhancer activity in the midbrain (tectum) and hindbrain and in addition also displays enhancer expression in the forebrain at E12.5. Histological sections of AEC32 show enhancer activity in the mouse cerebellum (red arrowheads). We next set out to characterize enhancers in regions implicated in human-specific evolution. Four of the sixteen positive zebrafish enhancers identified in this region (Table S1 and Figure S4) were analyzed for enhancer activity in mice. These four sequences were positive mouse enhancers active in the brain, the otic vesicle, or eye (Figure 4 and Figure S5). Interestingly, two of these enhancers (AEC10 and 21) show enhancer expression in the developing tectum, a region in the brain that is thought to control auditory and visual responses.
Fig. 4. Four positive zebrafish and mouse enhancers in regions implicated in human evolution. 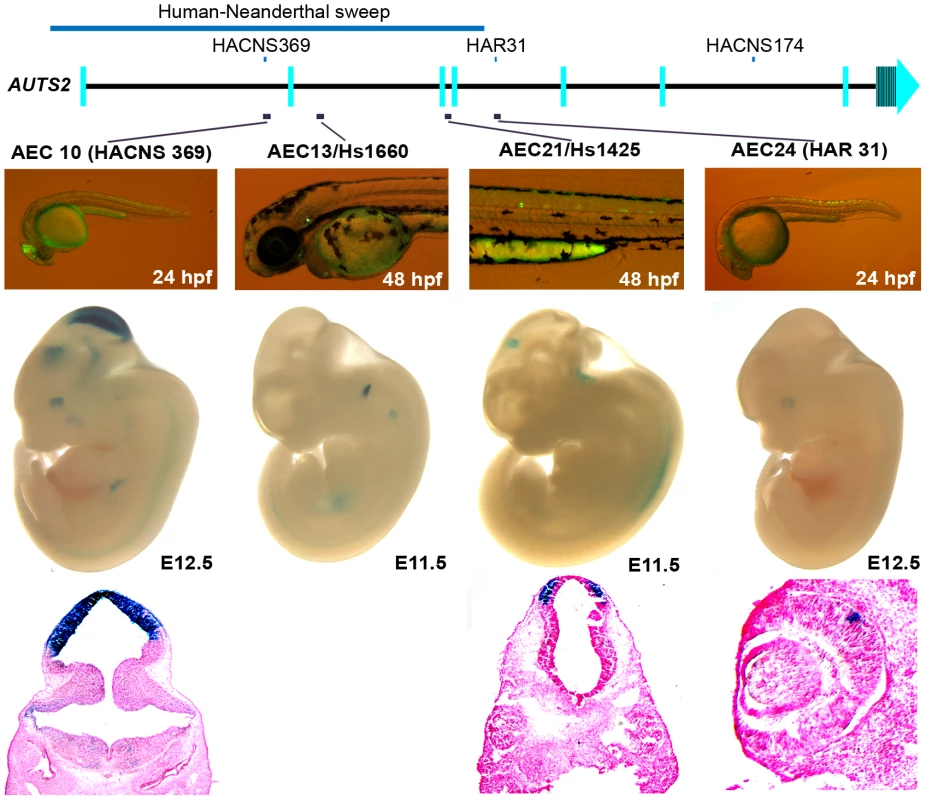
At E12.5, AEC10 shows zebrafish and mouse enhancer expression in the midbrain and eye. The histological section below highlights its expression in the tectum. AEC13, is expressed in the otic vesicle both in zebrafish and E11.5 mouse embryos (hs1660 ; [34]). AEC21 is expressed in the spinal cord in zebrafish, while in the mouse it showed midbrain expression at E11.5 (hs1425; [34]). Histological sections below show its expression in the pretectum of the midbrain. AEC24 was expressed in the spinal cord and hindbrain in zebrafish and in the eye in mouse at E12.5. Discussion
Using MOs to knockdown auts2, we observed an overall phenotype of stunted development, making it difficult to characterize discrete phenotypes. However, using neuronal-labeled zebrafish lines and immunohistochemistry, we showed a reduction in motor and sensory neurons in the spinal cord and developing neurons in regions that include the midbrain and cerebellum. The cerebellum is involved in cognitive and emotional function and has been repeatedly implicated in ASD [2]. In addition, the cerebellum plays a major role in motor control, and it is possible that the defects detected in cerebellar neurons could partially explain the reduced movement phenotype observed in morphant fish. It is worth noting that two individuals with AUTS2 structural variants had motor delay phenotypes (Figure 1) [12]. Given that the MO injected fish display additional phenotypes to the ones we focused on in this study, the effect of this gene on other tissues will need to be assessed in future experiments. Experiments such as mouse conditional knockouts should allow for a more complete understanding of AUTS2 function. Our auts2 MOs were designed to disrupt auts2 activity on chromosome 10 (build Zv9). It is worth noting, that there is also a putative, less characterized version of auts2 with an incomplete coding sequence located on zebrafish chromosome 15 (ENSDART00000012712). Knocking down this gene along with the auts2 gene that was assayed in our study may lead to more severe phenotypes.
Our enhancer search focused primarily on the first five introns due to the numerous reports of cognitive-related structural variations in that region [4]–[6], [8]–, along with the region's putative role in evolution. There could be numerous functional enhancers outside this region that we have not tested in this study. For example, there is an intragenic SNP (rs6961611) associated with processing speed [35] 1.6 mega bases downstream of AUTS2 which could be associated with a regulatory element for this gene. While the expression of our enhancers largely recapitulated Auts2 expression, it is possible that the enhancers we identified could regulate a neighboring gene. Future experiments such as chromatin interaction analyses [36], [37] could be able to distinguish what promoters our enhancers are interacting with.
Previous work has shown that human enhancer sequences can function as active enhancers in zebrafish, even without homologous sequences in zebrafish [38]–[40]. Our results confirm these findings for some of our enhancers. For example, AEC10, 13 and 29, which do not have homologous sequences in zebrafish, have similar enhancer expression patterns in zebrafish and mouse (Table S1). However, AEC21 and 27, which are conserved down to zebrafish, and AEC 24, which is conserved down to chicken, don't have matching expression patterns in zebrafish and mice.
We found three positive human enhancers in both zebrafish and mouse that reside within a 33,519 bp deletion detected in an individual with ASD, one of which, AEC32, is expressed in the cerebellum. This deletion was inherited from the individual's mother who was not diagnosed with ASD [5]. ASDs are likely caused by multiple genomic aberrations in combination with environmental factors. While it is possible that in this individual, this deletion leads to ASD due to the loss of these enhancers and potentially other functional sequences, it is also possible that the loss of these enhancers is one of multiple “hits” [41] or that the deletion is not causative. With the constantly growing number of individuals with ASDs or other neurological phenotypes that have AUTS2 mutations, some of which are purely noncoding, it is likely that improper regulation of this gene is involved in the progression of these disorders.
We also characterized enhancers in locations associated with other neurological phenotypes. In an 84 kb deletion in intron one of an individual with dyslexia, we identified four positive human enhancers in zebrafish (AEC3-6) (Table S1 and Figure S4), one of which is expressed in the midbrain. In addition, one of the candidates that was negative for zebrafish enhancer activity (AEC35) was a sequence that included the alcohol consumption associated SNP (rs6943555) [16]. It is possible that zebrafish is not a good model system for this region/phenotype or that the actual functional region/variant is further away from this tag SNP. By characterizing the regulatory landscape of this region we have obtained a better understanding of the functional units within this gene, which now pose as candidates for mutation analysis in individuals with various neurological phenotypes.
AUTS2 has been singled out as a gene that is rapidly evolving in humans in three different studies [17]–[19]. Using zebrafish enhancer assays, we identified sixteen different enhancers that lie within regions that were implicated in human evolution, six of which show expression in the brain. We tested four of the enhancers in mice and two of them had midbrain enhancer activity. Our enhancer results, combined with the observation that human-specific neurological disorders are associated with mutations in this gene, suggest that AUTS2 has an important role in the evolution of human cognitive traits.
Materials and Methods
Whole-mount in situ hybridization
Zebrafish embryos were collected from ABs or caspers [42] between 24 to 120 hpf and fixed in 4% paraformaldehyde buffered with 1× PBS (PFA). The zebrafish auts2 (Open Biosystems EDR1052-4681254) cDNA clone was used to generate digoxygenin labeled probes. Whole-mount in situ hybridizations were performed according to standard protocols [43].
Morpholino assays
Two morpholino (MO) antisense oligonucleotides targeting auts2 were designed by Gene-Tools. One MO was designed to target the translational start site of auts2 (GTGGAGAGTGTGTCAACACTAAAAT). The second was designed to target the splice junction between intron 2 and exon 3 of Ensembl Transcript ENSDART00000137928 (TCGACTACTGCTGTGAACAAAGAGA). A third 5 bp mismatch control for the translational MO (GTGGACACTGTGTGAAGACAAAAAT) was also designed. The MOs were diluted to 1 mM in deionized water and injected using standard techniques [44] into one cell-stage embryos. To rescue the morphant phenotypes, we transcribed full length human AUTS2 RNA (Open Biosystems MHS1010-9204165) using the T7 message machine (Ambion) and co-injected it along with the translational MO at a concentration of 168 ng/ul. The HuC line was generously donated by Dr. Su Guo (UCSF). The Tg(mnx1∶GFP) (AB) line (formerly known as hb9) was obtained from the Zebrafish International Resource Center (ZIRC; http://zebrafish.org/zirc/home/guide.php). Fish where injected with MOs as described above and annotated using the Leica M165 FC microscope. At least 50 translational MO injected fish and controls were compared in all zebrafish lines used.
Immunohistochemistry on zebrafish sections
AB zebrafish embryos injected with the auts2 translational MO or the 5 bp control were fixed at 48 hpf in 4% PFA overnight at 4°C, then washed for 15 minutes at room temperature in PBS. Zebrafish were frozen into blocks using Tissue-Tek O.C.T. (Sakura Finetek) then sectioned (10–20 microns) using a Leica CM1850 cryostat and stained with Nissl (FD NeuroTechnologies). Morphant and control sections represent comparable planes. Staining with PCNA (DAKO, Monoclonal Mouse PCNA clone PC10) was done according to the manufacturer's protocol. Cell nuclei were visualized using DAPI (Invitrogen). Staining sections with TUNEL (Roche, In Situ Cell Death Detection Kit, TMR red) was done according to the manufacturer's protocol. Zebrafish sections were analyzed using the Leica M165 FC or the Nikon Eclipse E800 microscope. At least 25 fish were analyzed in each condition. Control and morphant pictures were taken with identical exposures and are representative of each condition. For TUNEL staining on sections, criteria for amount of cell death was based on the number of individual TUNEL positive cells identified in the midbrain and eye, indicative of cell death in those regions. For PCNA staining (cell cycle marker) on sections, criteria for amount of proliferation in the forebrain, midbrain and hindbrain was qualitatively evaluated due to the larger number of PCNA positive cells in morphants compared to controls.
Zebrafish whole-mount immunohistochemistry
Casper zebrafish embryos injected with the auts2 translational MO or the 5 bp control were fixed at 48 hpf overnight at 4°C in 4% PFA. For TUNEL staining, embryos were transferred to methanol for 30 minutes followed by rehydration in methanol/PBST (PBS with 0.1% tween). They were then placed in Proteinase K (10 µg/ml) for 5 minutes and postfixed in 4% PFA for 20 minutes. Embryos were later placed in prechilled ethanol∶acetic acid (2∶1) at −20°C for 10 minutes and then washed in PBST for 20 minutes followed by TUNEL staining using the In Situ Cell Death Detection Kit, TMR red (Roche) according to the manufacturer's protocol. Sensory neurons were analyzed using anti-HNK-1 (Sigma) followed by the goat anti-mouse IgM HRP secondary antibody (abcam, ab5930) using previously described methods [45]. HNK-1 positive cells where manually counted in 6 different control and morphant fish. Fish were analyzed using the Leica M165 FC or the Nikon Eclipse E800 microscope. At least 25 fish were analyzed in each condition. Control and morphant pictures were taken with identical exposures and are representative of each condition. For TUNEL whole mount staining, criteria for amount of cell death was based on the number of viewable individual TUNEL positive cells in the forebrain, midbrain and hindbrain. For HNK-1 staining, criteria for amount of sensory neurons was based on the number of individual HNK-1 positive cells counted in equal lengths of the trunk. Motor neuron axons were analyzed using anti-znp-1 (Developmental Studies Hybridoma Bank) followed by anti-mouse IgG HRP (GE Healthcare) using previously described methods [46].
Transgenic enhancer assays
AUTS2 enhancer candidate (AEC) sequences were selected based on evolutionary conservation (sequences showing ≥70% identity for at least 100 bp between human and chicken), E1A binding protein p300 (EP300) forebrain or hindbrain ChIP-Seq datasets [32], and nucleotide variants that define the human-Neanderthal sweep [17] (Table S1). PCR was carried out on human genomic DNA (Qiagen) using primers designed to amplify the AEC sequences (Table S1). Primers were designed such that they will have additional flanking sequences to the conserved, ChIP-Seq or human-Neanderthal accelerated sequences based on previous experiments that have shown this to be a reliable method for obtaining positive enhancer activity [47]. PCR products were cloned into the E1b-GFP-Tol2 enhancer assay vector containing an E1b minimal promoter followed by GFP [33]. They were then injected following standard procedures [46], [48] into at least 100 embryos per construct along with Tol2 mRNA [49], to facilitate genomic integration. GFP expression was observed and annotated up to 48 hpf. An enhancer was considered positive if at least 15% of all fish surviving to 48 hpf showed a consistent expression pattern after subtracting out percentages of tissue expression in fish injected with the empty enhancer vector. Notably, the empty vector showed particularly high background for heart and somitic muscle and as described all enhancer results were obtained after deducting its expression pattern. Thus, in order to call positive somitic muscle enhancer activity, over 26% (24hpf) or 40% (48hpf) of alive fish needed to show positive enhancer activity. To call a positive heart enhancer, 32% (24hpf) or 50% (48hpf) of alive fish needed show positive heart activity. For each construct, at least 50 fish were analyzed for GFP expression at 48 hpf. For the mouse enhancer assays, the same human genomic fragment used in zebrafish was transferred into a vector containing the Hsp68 minimal promoter followed by a LacZ reporter gene [47], [50] and sequence verified to ensure the insert matched the human reference sequence. Sequences having rare variants were changed to the reference human genomic sequence by site-directed mutagenesis (Mutagenex or Quickchange II, Stratagene) and sequence verified for having the reference sequence. Transgenic mice were generated by Cyagen Biosciences using standard procedures [51]. Embryos were harvested at E12.5 and stained for LacZ expression using standard procedures [47]. Mouse embryos selected for sectioning were placed in an overnight cryoprotection stage using 30% sucrose in PBS. Mice were frozen into blocks using Tissue-Tek O.C.T. (Sakura Finetek) then sectioned (20 microns) using a Leica CM1850 cryostat and stained with Nuclear Fast Red Solution (Sigma-Aldrich) for one minute. There is no human subjects work involved in this article. All animal work was approved by the UCSF Institutional Animal Care and Use Committee (protocol number AN084690).
Supporting Information
Zdroje
1. BaioJ (2012) Prevalence of Autism Spectrum Disorders — Autism and Developmental Disabilities Monitoring Network , 14 Sites , United States , 2008. Centers for Disease Control and Prevention MMWR Surveillance Summaries 61.
2. PardoCa, EberhartCG (2007) The neurobiology of autism. Brain Pathol 17 : 434–447.
3. RischN, SpikerD, LotspeichL, NouriN, HindsD, et al. (1999) A Genomic Screen of Autism: Evidence for a Multilocus Etiology. Amer J Hum Genet 65 : 931.
4. SultanaR, YuC-E, YuJ, MunsonJ, ChenD, et al. (2002) Identification of a Novel Gene on Chromosome 7q11.2 Interrupted by a Translocation Breakpoint in a Pair of Autistic Twins. Genomics 80 : 129–134.
5. PintoD, PagnamentaAT, KleiL, AnneyR, MericoD, et al. (2010) Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466 : 368–372.
6. KalscheuerVM, FitzPatrickD, TommerupN, BuggeM, NiebuhrE, et al. (2007) Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation. Hum Genet 121 : 501–509.
7. BakkalogluB, RoakBJO, LouviA, GuptaAR, AbelsonJF, et al. (2008) Molecular Cytogenetic Analysis and Resequencing of Contactin Associated Protein-Like 2 in Autism Spectrum Disorders. Amer J Hum Genet 165–173 doi:10.1016/j.ajhg.2007.09.017.
8. HuangX-L, ZouYS, Maher Ta, NewtonS, MilunskyJM (2010) A de novo balanced translocation breakpoint truncating the autism susceptibility candidate 2 (AUTS2) gene in a patient with autism. Am J Med Genet A 152A: 2112–2114.
9. GlessnerJT, WangK, CaiG, KorvatskaO, KimCE, et al. (2009) Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459 : 569–573.
10. Ben-DavidE, Granot-HershkovitzE, Monderer-RothkoffG, LererE, LeviS, et al. (2011) Identification of a functional rare variant in autism using genome-wide screen for monoallelic expression. Hum Mol Genet 20 : 3632–3641.
11. GirirajanS, BrkanacZ, CoeBP, BakerC, VivesL, et al. (2011) Relative Burden of Large CNVs on a Range of Neurodevelopmental Phenotypes. PLoS Genet 7: e1002334 doi:10.1371/journal.pgen.1002334.
12. TalkowskiME, RosenfeldJA, BlumenthalI, PillalamarriV, ChiangC, et al. (2012) Sequencing Chromosomal Abnormalities Reveals Neurodevelopmental Loci that Confer Risk across Diagnostic Boundaries. Cell 149 : 525–537.
13. NagamaniSCS, ErezA, Ben-ZeevB, FrydmanM, WinterS, et al. (2012) Detection of copy-number variation in AUTS2 gene by targeted exonic array CGH in patients with developmental delay and autistic spectrum disorders. Eur J Hum Genet 1–4.
14. EliaJ, GaiX, XieHM, PerinJC, GeigerE, et al. (2010) Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry 15 : 637–646.
15. MeffordHC, MuhleH, OstertagP, Von SpiczakS, BuysseK, et al. (2010) Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet 6: e1000962 doi:10.1371/journal.pgen.1000962.
16. SchumannG, CoinLJ, LourdusamyA, CharoenP, BergerKH, et al. (2011) Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci U S A 108 : 7119–7124.
17. GreenRE, KrauseJ, BriggsAW, MaricicT, StenzelU, et al. (2010) A draft sequence of the Neandertal genome. Science 328 : 710–722.
18. PollardKS, SalamaSR, KingB, KernAD, DreszerT, et al. (2006) Forces shaping the fastest evolving regions in the human genome. PLoS Genet 2: e168 doi:10.1371/journal.pgen.0020168.
19. PrabhakarS, NoonanJP, PääboS, RubinEM (2006) Accelerated evolution of conserved noncoding sequences in humans. Science 314 : 786.
20. ZhangYE, LandbackP, VibranovskiMD, LongM (2011) Accelerated Recruitment of New Brain Development Genes into the Human Genome. PLoS Biol 9: e1001179 doi:10.1371/journal.pbio.1001179.
21. ViselA, ThallerC, EicheleG (2004) GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res 32: D552–6.
22. BedogniF, HodgeRD, NelsonBR, Frederick Ea, ShibaN, et al. (2010) Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr Patterns 10 : 9–15.
23. BedogniF, HodgeRD, ElsenGE, NelsonBR, Daza R aM, et al. (2010) Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci U S A 107 : 13129–13134.
24. HevnerRF, ShiL, JusticeN, HsuehY, ShengM, et al. (2001) Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29 : 353–366.
25. FatemiSH, SnowAV, StaryJM, Araghi-NiknamM, ReutimanTJ, et al. (2005) Reelin signaling is impaired in autism. Biol Psychiatry 57 : 777–787.
26. TropepeV, SiveHL (2003) Can zebrafish be used as a model to study the neurodevelopmental causes of autism? Genes Brain Behav 268–281 doi:10.1046/j.1601-183X.2003.00038.x.
27. ParkHC, KimCH, BaeYK, YeoSY, KimSH, et al. (2000) Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol 227 : 279–293.
28. EvanG, LittlewoodT (1998) A Matter of Life and Cell Death. Science 281 : 1317–1322.
29. AlenziFQB (2004) Links between apoptosis, proliferation and the cell cycle. Br J Biomed Sci 61 : 99–102.
30. Flanagan-SteetH, Fox Ma, MeyerD, SanesJR (2005) Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development 132 : 4471–4481.
31. GordonLR, GribbleKD, SyrettCM, GranatoM (2012) Initiation of synapse formation by Wnt-induced MuSK endocytosis. Development 139 : 1023–1033.
32. ViselA, BlowMJ, LiZ, ZhangT, AkiyamaJa, et al. (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457 : 854–858.
33. LiQ, RitterD, YangN, DongZ, LiH, et al. (2010) A systematic approach to identify functional motifs within vertebrate developmental enhancers. Dev Biol 337 : 484–495.
34. ViselA, MinovitskyS, DubchakI, Pennacchio La (2007) VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–92.
35. LucianoM, HansellNK, LahtiJ, DaviesG, MedlandSE (2012) UKPMC Funders Group Whole genome association scan for genetic polymorphisms influencing information processing speed. Biol Psychol 86 : 193–202 doi:10.1016/j.biopsycho.2010.11.008.Whole.
36. FullwoodMJ, LiuMH, PanYF, LiuJ, HanX, et al. (2010) NIH Public Access. Nature 462 : 58–64 doi:10.1038/nature08497.An.
37. Lieberman-AidenE, Van BerkumNL, WilliamsL, ImakaevM, RagoczyT, et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326 : 289–293.
38. NavratilovaP, FredmanD, Hawkins Ta, TurnerK, LenhardB, et al. (2009) Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes. Dev Biol 327 : 526–540.
39. McgaugheyDM, VintonRM, HuynhJ, Al-saifA, BeerMA, et al. (2008) Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res 18 : 252–260 doi:10.1101/gr.6929408.1.
40. FisherS, GriceEa, VintonRM, BesslingSL, McCallionAS (2006) Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312 : 276–279.
41. Poot M, Smagt JJ Van Der, Brilstra EH, Bourgeron T (2011) Disentangling the Myriad Genomics of Complex Disorders , Specifically Focusing on Autism , Epilepsy, and Schizophrenia. Cytogenet Genome Res. doi:10.1159/000334064.
42. WhiteRM, SessaA, BurkeC, BowmanT, LeBlancJ, et al. (2008) Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell stem cell 2 : 183–189.
43. ThisseB, HeyerV, LuxA, AlunniV, DegraveA, et al. (2004) Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol 77 : 505–519.
44. Naseviciusa, EkkerSC (2000) Effective targeted gene “knockdown” in zebrafish. Nat Genet 26 : 216–220.
45. HolderN, HillJ (1991) Retinoic acid modifies development of the midbrain-hindbrain border and affects cranial ganglion formation in zebrafish embryos. Development (Cambridge, England) 113 : 1159–1170.
46. Westerfield M (2007) The Zebrafish Book. 5th ed. Eugene, Oregon: University of Oregon Press.
47. Pennacchio La, AhituvN, MosesAM, PrabhakarS, Nobrega Ma, et al. (2006) In vivo enhancer analysis of human conserved non-coding sequences. Nature 444 : 499–502.
48. Nusslein-Volhard C and RD(2002) Zebrafish. Oxford: Oxford University Press.
49. KawakamiK (2005) Transposon tools and methods in zebrafish. Dev Dyn 234 : 244–254 Av.
50. KotharyR, ClapoffS, BrownA, CampbellR (1988) PA& RJ (1988) A transgene containing lacZ inserted into the dystonia locus is expressed in neural tube. Nature 335 : 435–437.
51. Nagy A., Gertsenstein M., Vintersten K BR (2003) Manipulating the Mouse Embryo: A Laboratory Manual (Third Edition). Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
52. PruittKD, TatusovaT, MaglottDR (2005) NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 33: D501–4.
Štítky
Genetika Reprodukční medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 1
-
Všechny články tohoto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



