-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaStarvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
High levels of antibiotic tolerance are a hallmark of bacterial biofilms. In contrast to well-characterized inherited antibiotic resistance, molecular mechanisms leading to reversible and transient antibiotic tolerance displayed by biofilm bacteria are still poorly understood. The physiological heterogeneity of biofilms influences the formation of transient specialized subpopulations that may be more tolerant to antibiotics. In this study, we used random transposon mutagenesis to identify biofilm-specific tolerant mutants normally exhibited by subpopulations located in specialized niches of heterogeneous biofilms. Using Escherichia coli as a model organism, we demonstrated, through identification of amino acid auxotroph mutants, that starved biofilms exhibited significantly greater tolerance towards fluoroquinolone ofloxacin than their planktonic counterparts. We demonstrated that the biofilm-associated tolerance to ofloxacin was fully dependent on a functional SOS response upon starvation to both amino acids and carbon source and partially dependent on the stringent response upon leucine starvation. However, the biofilm-specific ofloxacin increased tolerance did not involve any of the SOS-induced toxin–antitoxin systems previously associated with formation of highly tolerant persisters. We further demonstrated that ofloxacin tolerance was induced as a function of biofilm age, which was dependent on the SOS response. Our results therefore show that the SOS stress response induced in heterogeneous and nutrient-deprived biofilm microenvironments is a molecular mechanism leading to biofilm-specific high tolerance to the fluoroquinolone ofloxacin.
Published in the journal: . PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003144
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003144Summary
High levels of antibiotic tolerance are a hallmark of bacterial biofilms. In contrast to well-characterized inherited antibiotic resistance, molecular mechanisms leading to reversible and transient antibiotic tolerance displayed by biofilm bacteria are still poorly understood. The physiological heterogeneity of biofilms influences the formation of transient specialized subpopulations that may be more tolerant to antibiotics. In this study, we used random transposon mutagenesis to identify biofilm-specific tolerant mutants normally exhibited by subpopulations located in specialized niches of heterogeneous biofilms. Using Escherichia coli as a model organism, we demonstrated, through identification of amino acid auxotroph mutants, that starved biofilms exhibited significantly greater tolerance towards fluoroquinolone ofloxacin than their planktonic counterparts. We demonstrated that the biofilm-associated tolerance to ofloxacin was fully dependent on a functional SOS response upon starvation to both amino acids and carbon source and partially dependent on the stringent response upon leucine starvation. However, the biofilm-specific ofloxacin increased tolerance did not involve any of the SOS-induced toxin–antitoxin systems previously associated with formation of highly tolerant persisters. We further demonstrated that ofloxacin tolerance was induced as a function of biofilm age, which was dependent on the SOS response. Our results therefore show that the SOS stress response induced in heterogeneous and nutrient-deprived biofilm microenvironments is a molecular mechanism leading to biofilm-specific high tolerance to the fluoroquinolone ofloxacin.
Introduction
Formation of bacterial biofilms on medical implants is a major health threat due to their high levels of tolerance to multiple antibiotics [1]. Biofilm-associated antibiotic tolerance is mainly attributed to two distinct processes: persistence and drug indifference, which both characteristically disappear once multicellular conditions subside [2], [3]. Persistence occurs in subpopulations of slow or non-growing bacteria, whereas drug indifference is exhibited by the entire population [4], [5]. Although the molecular bases of persistence are under active investigation [5], [6], drug indifference is far less well understood and is hypothesized to be multifactorial and to result from reduced antibiotic diffusion to slow growth rate of many cells within biofilms [7]. Alternatively, local gradients of nutrients, oxygen, pH, signalling molecules and waste products, as well as genetic heterogeneity, which can arise through mutations, recombination, and stochastic gene expression, could lead to physiological adaptation and drug indifference in heterogeneous biofilms [7]–[12]. Biofilm heterogeneity creates specialized niches in which bacteria respond to local cues, leading to genetically and metabolically distinct subpopulations exhibiting high tolerance to extracellular stresses such as antibiotics.
Since physical isolation of biofilm subpopulations is technically challenging [13], [14], analyses of antibiotic tolerance have thus far relied mainly on isolation of mutants with decreased ability to form tolerant biofilms. However, biofilm heterogeneity and physiologically specialized subpopulations limit the power of these strategies. As an alternative approach to investigating the mechanisms of biofilm-associated tolerance to antibiotics, we mutagenized a biofilm-forming Escherichia coli strain to identify mutants forming biofilms with increased tolerance towards two bactericidal antibiotics, ticarcillin, a ß-lactam-targeting peptidoglycan and ofloxacin, a fluoroquinolone targeting DNA gyrase. We reasoned that mutants forming highly antibiotic tolerant biofilms could correspond to mutations causing all biofilm bacteria to adopt physiological states usually only transiently expressed by biofilm subpopulations.
In this study, we found that biofilms formed by amino acid auxotroph mutants display high antibiotic tolerance and we demonstrated that amino acid starvation strongly increases the antibiotic tolerance of biofilm bacteria. As expected, tolerance to the ß-lactam ticarcillin correlated with slow growth displayed by all tested amino acid auxotrophs. However, we demonstrated that starvation for most amino acids induced tolerance towards ofloxacin only under biofilm conditions. While this biofilm-specific tolerance was shown to depend partially on a functional stringent response in leucine-starved biofilms, we demonstrated that carbon source starvation also mediates ofloxacin tolerance in biofilm and that the SOS response appears to be necessary for the ofloxacin tolerance exhibited upon both amino acid and carbon source starvation. However, we showed that SOS-dependent biofilm-specific ofloxacin tolerance is independent of toxin-antitoxin systems induced by the SOS response and previously associated with bacterial persistence. Consistently, we observed that SOS-dependent ofloxacin tolerance increases with biofilm age. Since local nutrient deprivation is a characteristic of aging and heterogeneous biofilms, our study therefore reveals a general mechanism linking nutritional stress and reversible biofilm-specific tolerance to the fluoroquinolone ofloxacin.
Results
An in vitro model for studying antibiotic tolerance in E. coli biofilms
In order to study antibiotic tolerance in biofilms, we grew static biofilms in M63B1Gluc minimal medium in 96-well PVC microtiter plates using TG1, a previously described highly adherent E. coli K-12 strain expressing the F-conjugative pilus, involved in both conjugation and biofilm formation [15]. Twenty-four-hour E. coli TG1 biofilms (Figure S1) were treated with either ticarcillin, a β-lactam carboxypenicillin with bactericidal activity only against rapidly growing cells [16], or ofloxacin, a fluoroquinolone active against both growing and non-growing cells [17], [18]. We determined the viability of antibiotic-treated TG1 biofilm bacteria using both viable cells counts (CFUs) and bacterial metabolic activity (XTT-reduction assay) as survival read-outs. Although the antibiotic-tolerant population appeared greater when quantified using the XTT-reduction assay compared to the CFU count, both methods gave similar survival profiles in 24 h E. coli TG1 biofilms exposed to up to 80 - and 100-times the MIC values for ofloxacin and ticarcillin, respectively (Figure 1A–1D). These surviving bacteria, which tolerated either ticarcillin (100× MIC) or ofloxacin (80× MIC), did not correspond to antibiotic-resistant mutants, since they displayed wild-type resistance profiles once re-inoculated and grown under classical planktonic conditions (data not shown). Similar results were obtained using a different biofilm-forming E. coli K-12 strain constitutively expressing type 1 fimbriae (data not shown), therefore demonstrating that biofilm formation per se, rather than the nature of the surface adhesin promoting biofilm formation, is involved in antibiotic tolerance of E. coli K-12 biofilms in our model.
Fig. 1. Antibiotic-tolerant populations of E. coli K-12 TG1 biofilms. 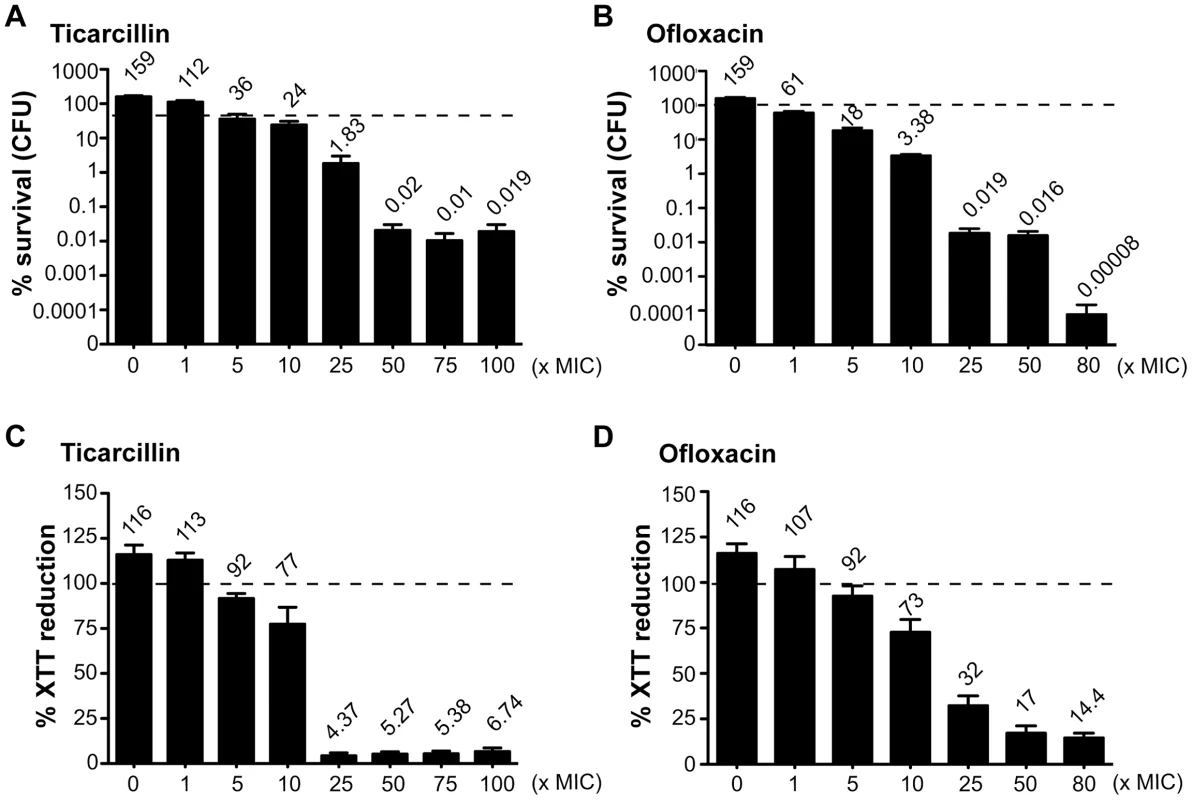
Twenty-four-hour biofilms of E. coli TG1 were preformed in M63B1Gluc and treated for a period of 24 h with concentrations up to 100- and 80-times the MIC values for ticarcillin and ofloxacin, respectively. Untreated controls represent 48 h biofilms in which fresh M63B1Gluc without antibiotic was added after 24 h, explaining their values above 100%, represented by a dotted line in each panel. Viable cells of the treated biofilm population were quantified by viable cell counts (A and B) and the XTT-reduction assay (C and D) and were compared to numbers obtained prior to antibiotic treatment. % survival (CFU) values are indicated on top of each bar and are means ± standard error of the means (SEM) of at least six replicates. Amino acid auxotroph mutants exhibit increased antibiotic tolerance in biofilms
To identify mutations leading to high antibiotic tolerance transiently displayed by E. coli biofilm bacteria, we performed random mariner transposon mutagenesis on E. coli TG1 derivative strain TG1gfp. In our in vitro biofilm model, we screened approximately 10,000 mutants for homogeneous increased tolerance to ticarcillin (100× MIC) or ofloxacin (50× MIC) using the XTT-reduction assay as a high throughput survival read-out. We identified a total of 18 transposon mutants with increased biofilm tolerance to these two antibiotics when compared to their parental strain, and 16 of these mutants displayed auxotrophy to various amino acids (Table 1). Transposon mapping performed on 10 of these auxotrophic mutants indeed showed mutations in genes involved in amino acid biosynthesis, including four insertions in leuC and two in proA, indicative of saturated mutagenesis (Table 1). These 10 mapped mutants displayed amino acid auxotrophies to leucine (leuB; leuC), proline (proA), arginine (argE), isoleucine/valine (ilvC) and aromatic amino acids (aroE) when grown on minimal medium containing glucose as sole carbon source (data not shown). In addition, we determined that six of the remaining unmapped mutants also displayed auxotrophies to five different amino acids (proline, threonine, histidine, cysteine, and tyrosine). Our screen also identified two prototrophic transposon mutants (rseC and pnp) displaying slight increased tolerance in biofilms, which were not further investigated in this study.
Tab. 1. Auxotrophic transposon mutants with increased antibiotic tolerance in biofilms. 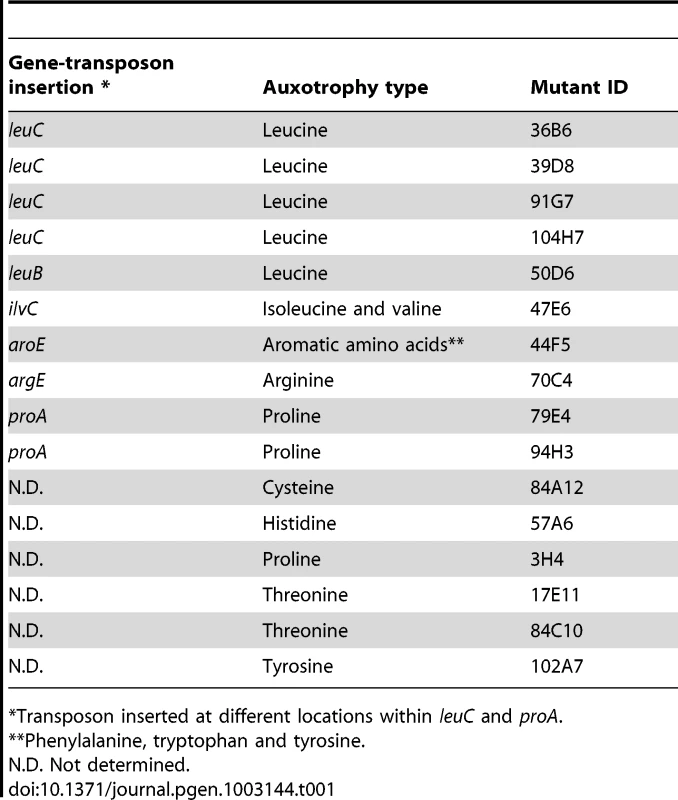
Transposon inserted at different locations within leuC and proA. Formation of antibiotic-tolerant biofilms in M63B1Gluc minimal medium by amino acid auxotrophs was intriguing, and suggested that use of overnight LB culture inoculum provided enough amino acids to support initial growth and biofilm development (data not shown). Indeed, we determined that supplementation of M63B1Gluc with as little as 25 µg ml−1 of the required amino acid was sufficient to promote growth and formation of antibiotic-tolerant biofilm by all tested E. coli TG1 auxotrophs when treated in amino-acid-free medium (M63B1Gluc) (see Materials and Methods and data not shown).
Starvation leads to increased antibiotic tolerance in biofilms
The identification, via our mutagenesis screen, of mutants auxotrophic for at least 12 amino acids suggested a link between biofilm antibiotic tolerance and amino acid starvation (Table 1). To confirm this relationship, we created E. coli TG1 amino acid auxotrophs that resulted from deletion of a single amino acid biosynthesis gene, including leucine, isoleucine, histidine, arginine, cysteine, methionine, lysine, proline, phenylalanine, tyrosine, tryptophan, glutamic acid, glycine, glutamine, serine and threonine (Table S1). However, auxotrophies for aspartic acid, asparagine, and alanine were not constructed due to the existence of multiple corresponding metabolic pathways. Similarly, we did not investigate auxotrophy for valine, since such mutants cannot be obtained without affecting isoleucine biosynthesis.
Among the 16 newly constructed amino acid auxotrophs, auxotrophy for glutamic acid, glycine, glutamine, serine and threonine showed reduced biofilm formation compared to the parent prototroph even when supplemented with 25 µg ml−1 of the required amino acid, and thus they could not be meaningfully tested for biofilm antibiotic tolerance (data not shown). Eleven of them, however, formed wild-type biofilms when supplemented with 25 µg ml−1 of the required amino acid (data not shown).
To evaluate the impact of starvation of a single amino acid upon the antibiotic tolerance of biofilm bacteria, we compared survival of starved auxotrophic biofilms formed by these 11 auxotrophs to that of the wild-type prototroph parental strain TG1. During the starvation process, bacterial growth was blocked due to the use of auxotrophic mutant strains in the absence of the required amino acid. In these experimental conditions, we observed that biofilm formed by histidine, methionine, phenylalanine, proline, tyrosine, isoleucine, arginine, cysteine, lysine and leucine auxotrophs survived significantly better than prototrophic TG1 biofilms exposed to either ticarcillin (100× MIC) or ofloxacin (80× MIC) (Figure 2A and B; P<0.05). In the case of the tryptophan auxotroph, increased survival was observed upon exposition to ticarcillin only (Figure 2A) indicating that growth arrest was not the only mechanism involved in ofloxacin tolerance upon starvation to the other amino acids (Figure 2B). The tolerance levels exhibited by starved biofilms (individual amino acid) were greater towards ticarcillin than ofloxacin confirming that fluoroquinolone antibiotics like ofloxacin kill bacterial cells independently of growth.
Fig. 2. Starvation leads to high antibiotic tolerance in biofilms of E. coli TG1. 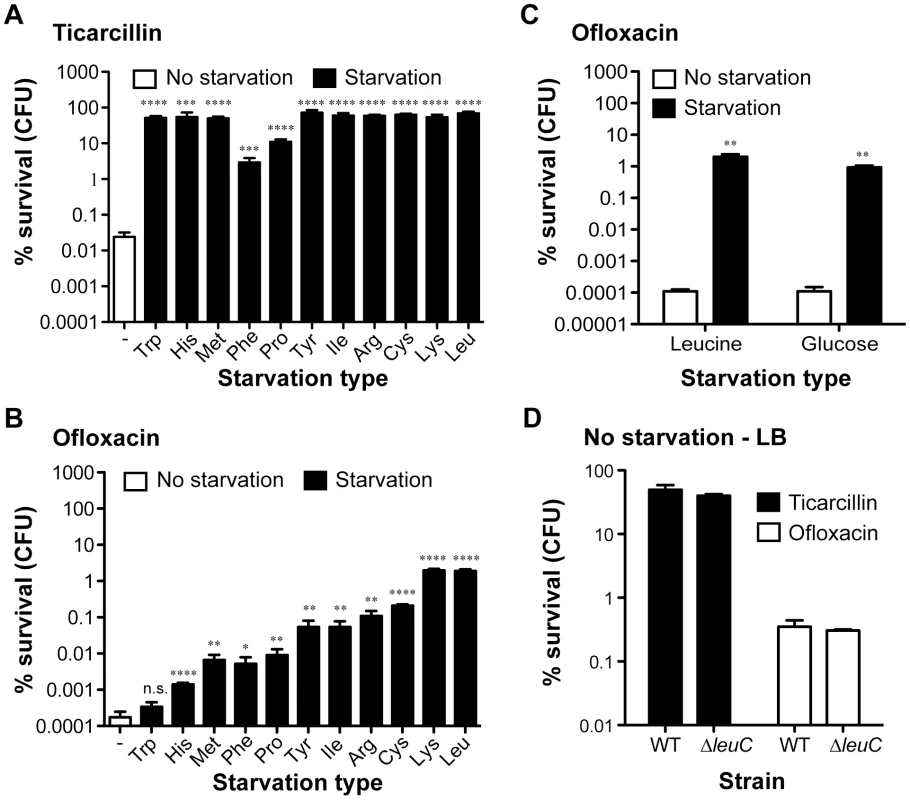
The tolerance of non-starved biofilms (white bars; wild-type prototroph TG1 (WT)) to (A) ticarcillin (100 µg ml−1, 100× MIC) or (B) ofloxacin (5 µg ml−1, 80× MIC) was compared to biofilms starved for individual amino acids (black bars; auxotrophic mutants). (C) Addition of exogenous leucine (10 µg ml−1) or glucose (0.4%) restored ofloxacin sensitivity to biofilms. (D) Antibiotic tolerance profiles of biofilms grown and treated in LB-rich medium were indistinguishable from a leucine auxotroph and its WT prototroph. For panels A to C, biofilms of all tested amino acid auxotrophs were grown in M63B1Gluc for 24 h with the addition of 25 µg ml−1 of the required amino acid, while WT was grown in M63B1Gluc only. Antibiotic treatments were performed for a period of 24 h on 24-h biofilms. Conditions of starvation during treatment were achieved by removing the required amino acids (M63B1Gluc) for the respective auxotrophic strains or glucose for the WT prototroph (M63B1). In panel D, biofilms were grown for 24 h and treated for 24 h in LB-rich medium. Surviving cells were quantified by viable cell counts. Percent survival represents viable cells after 24 h of treatment compared to untreated biofilm prior to addition of antibiotics. All compared biofilms had similar numbers of CFU prior to antibiotic treatment (data not shown). Data represented are means ± SEM of at least three replicates. Asterisks indicate values significantly different from conditions of no starvation by the two-tailed unpaired t test: * P≤0.05, ** P≤0.01, *** P≤0.001, **** P≤0.001 and n.s. (not significant). The genotypes of all amino acid auxotrophic mutants constructed in the WT prototroph TG1 genetic background are described in Table S1 or as follows: tryptophan (Trp; ΔtrpA::KmFRT), histidine (His; ΔhisG::KmFRT), methionine (Met; ΔmetA::KmFRT), phenylalanine (Phe: ΔpheA::KmFRT), proline (Pro: ΔproC::KmFRT), tyrosine (Tyr; ΔtyrA::KmFRT), isoleucine (Ile; ΔilvA::KmFRT), arginine (Arg; ΔargH::KmFRT), cysteine (Cys; ΔcysD::KmFRT), lysine (Lys; ΔlysA::KmFRT), and leucine (Leu: ΔleuC::GB). To further investigate the contribution of starvation to antibiotic tolerance in biofilms, we chose auxotrophy for leucine as a model of auxotrophy-induced tolerance. We observed that blocking starvation in leucine auxotroph biofilms by exogenous addition of leucine (5–10 µg ml−1) reverted both ofloxacin and ticarcillin tolerance to levels observed in non-starved wild-type TG1 biofilms (Figure 2C and Figure S2). To determine whether exposure to other nutritional stresses could lead to a similar antibiotic tolerance profile in starved prototroph biofilms, we exposed biofilms formed by wild-type TG1 to antibiotics in glucose-free medium (M63B1 - no carbon source). Under these conditions, we observed that deprivation of all carbon sources (glucose), and therefore absence of growth, increased the antibiotic tolerance of TG1 biofilms towards both ofloxacin and ticarcillin (Figure 2C and data not shown). Consistently, leucine auxotroph biofilms grown and treated in rich LB medium displayed wild-type sensitivity to both ticarcillin and ofloxacin, thereby demonstrating that amino acid shortage per se is involved in biofilm-associated antibiotic tolerance (Figure 2D). Biofilms grown in LB rich medium were more tolerant than those grown in minimal medium (M63B1Gluc) for both ofloxacin and ticarcillin (Figure 2A, 2C and 2D) suggesting the existence, in this rich medium, of other antibiotic tolerance mechanisms such as those mediated by indole production [19].
Altogether, these results demonstrated that starvation for essential growth nutrients such as amino acids and carbon sources increased the tolerance of E. coli K-12 biofilms to ticarcillin and ofloxacin. Further, we demonstrate that the use of auxotrophic mutant strains is an effective genetic strategy to adequately control and measure the impact of a specific starvation type such as a single amino acid.
Ofloxacin hypertolerance upon starvation is biofilm-specific
To determine whether increased antibiotic tolerance of starved amino acid auxotrophs was specific to the biofilm lifestyle, we compared the tolerance profiles of planktonic and biofilm bacteria upon starvation to glucose, leucine, lysine, and cysteine (Figure 3). To compare planktonic and biofilm lifestyles, planktonic cells were harvested from the same wells in which tolerant biofilms were formed, thereby growing both cell types under identical conditions. Interestingly, planktonic bacteria were more tolerant than biofilm bacteria to both ticarcillin and ofloxacin in non-starving conditions. In starvation conditions, with the exception of glucose, free-swimming planktonic bacteria harvested from the three chosen auxotrophs continued to display increased tolerance to ticarcillin like their biofilm counterparts (Figure 3A and 3B). In contrast with observations made under biofilm conditions (Figure 2B and Figure 3D), planktonic cells starving for glucose, leucine, lysine or cysteine did not exhibit increased tolerance to ofloxacin (Figure 3C). These results therefore indicated that, although ticarcillin tolerance displayed by starved bacteria in planktonic and biofilm conditions is likely a consequence of reduced growth or its absence, the increased tolerance to ofloxacin displayed by starved bacteria is biofilm-specific.
Fig. 3. Antibiotic tolerance profile of starved planktonic and biofilm bacteria. 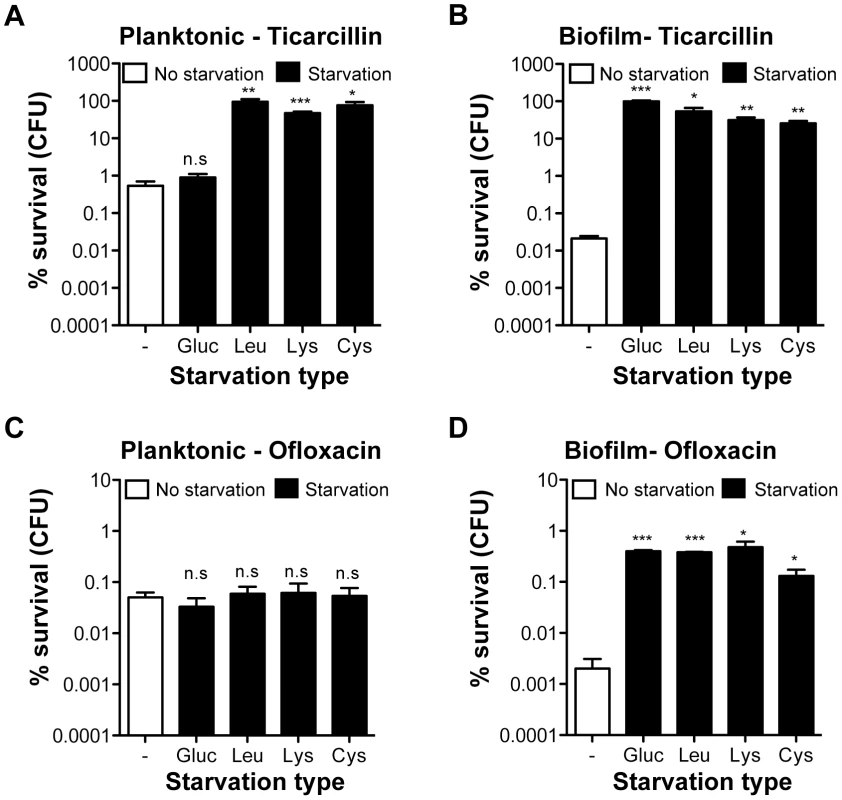
Static cultures were grown for 24 h in M63B1Gluc for wild-type prototroph TG1 (WT) and with the addition of 25 µg ml−1 of leucine, lysine, or cysteine for the corresponding auxotrophs. Planktonic bacteria from the 24-h static culture were removed from each well and therefore separated from the attached-biofilm cells. (A, C) Planktonic bacteria were then collected, spun, washed and treated in parallel with the biofilm bacteria (B, D) with ticarcillin (100 µg ml−1, 100× MIC) or ofloxacin (5 µg ml−1, 80× MIC) in the absence of glucose for WT prototroph TG1 (M63B1 medium) or amino acids for the different auxotrophic mutant strains (M63B1Gluc medium) for 24 h. Survivor cells were quantified by viable cell counts. Percent survival represents the tolerant population after 24 h of treatment compared to the total number of CFU prior to addition of antibiotics. Equivalent CFU were present in all compared planktonic populations before antibiotic treatment. Data represented are means ± SEM of at least three replicates. Asterisks indicate values significantly different from no starvation condition by the two-tailed unpaired t test: * P≤0.01, ** P≤0.001, *** P≤0.0001 and n.s. (not significant). The genotypes of the three amino acid auxotrophic mutants constructed in the WT prototroph TG1 genetic background are described in Table S1 or as follows: leucine (Leu: ΔleuC::GB), lysine (Lys; ΔlysA::KmFRT), and cysteine (Cys; ΔcysD::KmFRT). The biofilm-specific ofloxacin hypertolerance partially relies on the stringent response upon leucine starvation
The biofilm specificity of ofloxacin hypertolerance led us to speculate that a mechanism other than growth arrest could be involved in generating highly tolerant populations when starved of specific nutrients. Bacteria have evolved various general response mechanisms towards extracellular stresses with the potential of promoting antibiotic tolerance [20]. Starvation induces the so-called stringent response that is characterized by the synthesis of (p)ppGpp by both RelA and SpoT [21], which was recently linked to antibiotic tolerance in nutrient-limited biofilms [12]. The potential involvement of the stringent response in the biofilm-specific ofloxacin hypertolerance was first assayed by measuring the tolerance of ΔrelA in both wild-type TG1 and its derivative leucine auxotroph upon starvation for glucose (Figure S3A) or leucine (Figure S3B), respectively. The impairment of the stringent response by ΔrelA reduced the ofloxacin tolerance normally exhibited by biofilms starved for leucine without totally abolishing it (Figure S3B). As expected, since starvation to carbon sources like glucose is under the control of SpoT [22], the high ofloxacin tolerance exhibited upon glucose starvation was still displayed by ΔrelA biofilms supporting the absence of role for RelA upon glucose starvation (Figure S3A). Unfortunately, it was impossible to meaningfully determine the role of SpoT on the biofilms ofloxacin tolerance upon glucose starvation with our experimental conditions. Indeed, in E. coli K-12, a ΔspoT mutation is lethal in a wild-type background of E. coli K-12 and a (p)ppGpp0 strain (ΔrelAΔspoT) is itself auxotrophic to multiple amino acids [23].
These partial results, mainly due to genetic incompatibilities, at least suggest that stringent response through RelA partially contributes to ofloxacin hypertolerance exhibited by E. coli K-12 biofilms upon leucine starvation.
The SOS stress response is fully required for the starvation-induced biofilm ofloxacin tolerance
Although a ΔrelA mutation partially restored wild-type sensitivity to ofloxacin upon leucine starvation, their biofilms were still more tolerant than non-starved wild-type biofilms (Figure S3B). To determine whether a parallel mechanism was also involved in the biofilm-specific ofloxacin tolerance, we decided to focus our attention on the SOS response. The SOS response is known for its role in DNA repair and mutagenesis, however it was also shown to influence the formation of planktonic E. coli cells persistent to fluoroquinolone antibiotics [24]–[26]. Once induced, the SOS response is triggered upon RecA-dependent cleavage of LexA, the repressor of SOS response-regulated genes [27].
To test the potential contribution of the SOS response to hypertolerance of biofilms starved for ofloxacin, we introduced a ΔrecA into selected auxotrophs (leucine, lysine, and tryptophan) and a lexAind3 mutation encoding an uncleavable LexA protein [28] into a leucine auxotroph. These mutations were also introduced into their prototrophic parental strain TG1 (Table S1). We first determined that these two loss-of-function mutations had no significant effect on ofloxacin tolerance displayed by non-starved wild-type E. coli TG1 biofilms (Figure 4A). We then observed that biofilm-associated ofloxacin tolerance of ΔrecA biofilm bacteria starved for glucose, leucine and lysine was reduced to levels comparable to those of non-starved biofilms (Figure 4A, 4B, and 4C). Moreover, ofloxacin hypertolerance was also reduced for lexAind3 biofilms starved for glucose or leucine (Figure 4A and 4B). In contrast, the ofloxacin tolerance of biofilms starved for tryptophan was not affected by a ΔrecA mutation, consistent with our previous observations showing that tryptophan starvation did not affect ofloxacin tolerance in biofilms (see Figure 2B). Consistently, biofilm-associated ofloxacin tolerance was fully restored to levels observed upon leucine starvation after introduction of a wild-type copy of recA in a leucine auxotroph background (Figure 4B). We also showed that overexpression of RecA in a ΔleuClexAind3 background failed to restore ofloxacin hypertolerance in biofilms formed by the leucine auxotroph, therefore demonstrating that this phenotype is SOS-response-dependent and not solely RecA-dependent (Figure 4B). Furthermore, a ΔrecA mutation had no effect on ticarcillin tolerance of biofilms starved for leucine, lysine or tryptophan (Figure 4D) suggesting that growth arrest is the likely mechanism involved in ticarcillin resistance upon starvation.
Fig. 4. The high ofloxacin tolerance exhibited by starved biofilms is SOS-response-dependent. 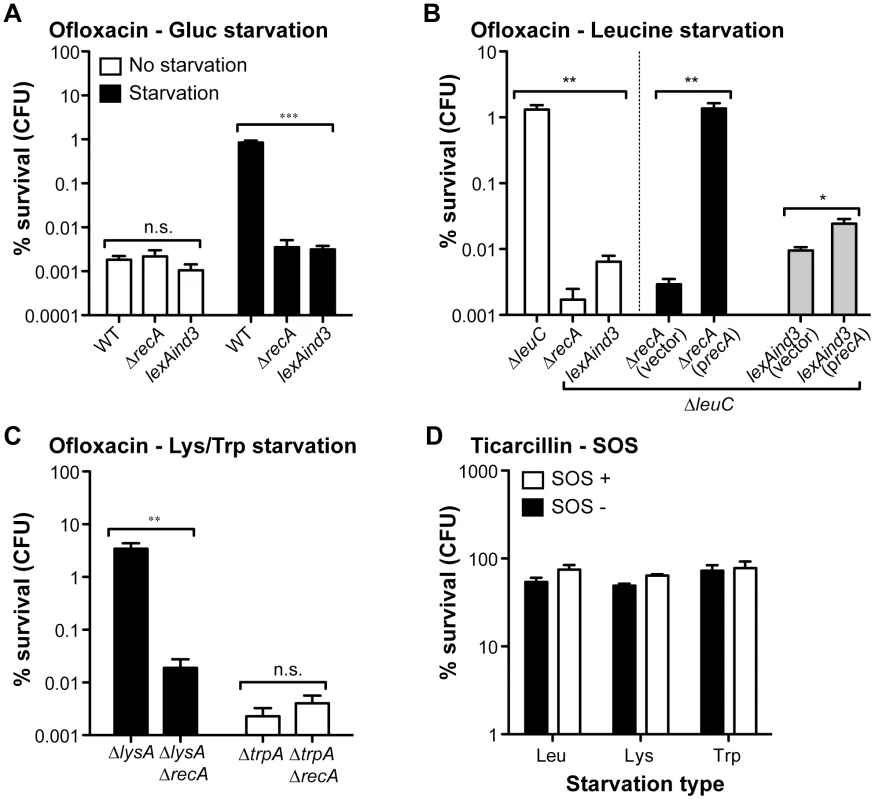
The impact of SOS response loss-of-function mutations ΔrecA and lexAind3 on the ofloxacin tolerance of starved biofilms for glucose (A), leucine (B), lysine and tryptophan (C). Briefly, all biofilms were grown for 24 h in M63B1Gluc for all prototrophic strains and with the addition of 25 µg ml−1 of leucine, lysine or tryptophan for corresponding auxotrophic strains. All biofilms were treated for 24 h in M63B1Gluc containing ofloxacin (5 µg ml−1). For the glucose starvation environment, M63B1 without glucose was used instead of M63B1Gluc (Panel A - black bars). Ofloxacin hypertolerance of ΔleuCΔrecA was not restored when adding the vector control plasmid pAM34 (vector), but complete restoration of high ofloxacin tolerance upon leucine starvation was observed by in trans complementation using pAM34recA (precA). Complementation of ΔleuClexAind3 with pAM34recA (precA) failed to fully restore ofloxacin hypertolerance back to leucine auxotroph levels. Both pAM34 and pAM34recA were maintained by the presence of 0.5 mM IPTG. (D) The SOS response mutation ΔrecA (SOS-) did not affect ticarcillin hypertolerance of starved biofilms for leucine (Leu - ΔleuCΔrecA), lysine (Lys – ΔlysAΔrecA) or tryptophan (Trp - ΔtrpAΔrecA). Viable cells of the treated biofilm population were quantified by viable cell counts. Percent survival represents the tolerant population after 24 h of treatment compared to untreated biofilm prior to addition of antibiotics. All compared biofilms had similar numbers of CFUs prior to antibiotic treatment (data not shown). Data represented are means ± SEM of at least three replicates. Asterisks indicate values significantly different by the two-tailed unpaired t test: * P≤0.05, ** P≤0.01, *** P≤0.001, and n.s. (not significant). All strains used here were originally constructed in the WT TG1 genetic background and are detailed in Table S1. The different SOS response mutant strains made in amino acid auxotrophic background are derivatives of ΔleuC::ΔFRT (Leu - leucine), ΔlysA::ΔFRT (Lys - lysine), and ΔtrpA::ΔFRT (Trp – tryptophan). We showed that the induced ofloxacin tolerance upon starvation was unique to the biofilm stage (Figure 3C and 3D). This was also confirmed by demonstrating that the SOS response, critical for biofilm bacteria (Figure 4), had no significant role in the ofloxacin tolerance of starved planktonic cells (Figure S4). Our genetic-based results confirmed that a functional SOS response system was fully required to tolerate high levels of ofloxacin for biofilm bacteria starving for glucose, leucine, or lysine.
The SOS-dependence of starved biofilms ofloxacin tolerance could potentially be due to an induction of the SOS response either during biofilm formation and/or by the ofloxacin treatment. In fact, fluoroquinolone antibiotics such as ofloxacin are known to induce the SOS response at subinhibitory and bactericidal concentrations, however, in conditions where bacteria exhibit growth [24], [29]. The ofloxacin treatment used in our experiments was performed in starvation condition, i.e. in absence of growth for the auxotrophic mutants or the prototrophic WT treated without glucose. Therefore, our current results would favour the idea that the SOS response cannot be induced during the ofloxacin treatment mainly because of a complete shutdown of both transcription and protein translation and thus pointing towards a role of the SOS response in ofloxacin tolerance during the process of biofilm formation itself. To provide evidence of this shut down of protein translation during starvation, here we used an E. coli strain containing a lacZ gene under the control of the PLtetO-1 promoter inducible by anhydrotetracycline (aTc). We showed that a 24-h biofilm of this strain subjected to glucose starvation was unable to translate lacZ in presence of aTc unlike unstarved 24-h biofilm (Figure S5).
Taken together, this strongly suggests that ofloxacin tolerance of biofilm is linked to the presence of a functional SOS response during biofilm formation and is only visible in condition where biofilms are subjected to a strong nutritional stress.
SOS-dependent toxin-antitoxin modules do not influence the ofloxacin hypertolerance of starved biofilms
The SOS system of E. coli involves the induction of at least 40 genes, for which the expression of specific toxin-antitoxin (TA) modules was previously associated with the SOS response, antibiotic tolerance and amino acid starvation [5], [25], [30]. To investigate whether TA modules that are part of the SOS regulon were directly responsible for the described biofilm-associated ofloxacin tolerance upon starvation, we first deleted the genes encoding the four currently known functional SOS-dependent TA modules TisAB, SymER, DinJ/YafQ and YafNO (the HokE toxin is inactive in E. coli K-12 [31]) in the biofilm-forming background of MG1655ΔleuC carrying the F episome (F'Tet; Table S1). Simultaneous deletion of these four SOS-dependent TA modules did not affect the ofloxacin tolerance profile of leucine-starved biofilms (Figure S6A). Although not regulated by the SOS response, we also evaluated the role of other well-characterized TA modules [32] some of which have been shown to be involved in antibiotic bacterial persistence as well as the Lon protease known to regulate TA protein stability [33]. However, individual deletion of the gene coding for the toxin of non-SOS TA modules (RelE, HipA, MazF, ChpB, YoeB, HicA as well as CcdB, located on the F plasmid) and for the Lon protease in a leucine auxotroph background of strain TG1 did not reduce ofloxacin tolerance of leucine-starved biofilms (Figure S6B).
These results showed that, although a functional SOS stress response is critical for the increased tolerance to ofloxacin displayed by starved biofilms, this tolerance is exerted independently of known SOS induced TA modules and is maintained in strains carrying single deletion of the best-characterized SOS-independent TA modules.
SOS-dependent biofilm tolerance to ofloxacin increases with biofilm age
Biofilm specificity and SOS-dependence of this increased ofloxacin tolerance led us to speculate about the possible links between these two phenomena. Unlike planktonic bacteria, biofilms are heterogeneous environments in which bacteria can experience various stresses such as local transient nutritional deprivation, oxidative stress, local pH, and oxygen tension modification [7], [9], some of which may induce the SOS response [20], [34]–[37] and then influence biofilm antibiotic tolerance. Nutrient gradients or waste accumulation together with bacterial physiological heterogeneity are supposed to increase during biofilm maturation [7] and therefore potentially impact ofloxacin tolerance over time upon induction of the SOS response. To assess this hypothesis, we used aging biofilms to determine whether the SOS response was indeed induced over time. We monitored the expression of the SOS-regulated gene sulA using a lacZ transcriptional reporter fusion in an E. coli K-12 strain harbouring the biofilm-promoting factor F'tet. Transcription of sulA increased progressively over time in aging biofilms (Figure 5A) similarly to the increased ofloxacin tolerance exhibited in both the psulA::lacZ reporter strain and TG1 (Figure 5B). Although a little difference was observed in ofloxacin tolerance of 48-h biofilms between the two strains, the overall progressive tolerance was similar in both genetic backgrounds (Figure 5B). Therefore, the induction of the SOS response occurring in aging biofilms (Figure 5A) can confidently be extrapolated to strain TG1. Consistently, the overall increase ofloxacin tolerance demonstrated in aging biofilm of wild-type TG1 (Figure 5B) was abolished in both SOS response mutants ΔrecA and lexAind3 (Figure 6A). A significant increase of ofloxacin tolerance was, however, observed between 24 - and 48-h biofilms of strain lexAind3 but not in a recA mutant suggesting that unknown SOS-independent tolerance mechanisms could act during certain biofilm development steps.
Fig. 5. Induction of the SOS response and ofloxacin tolerance in aging biofilms. 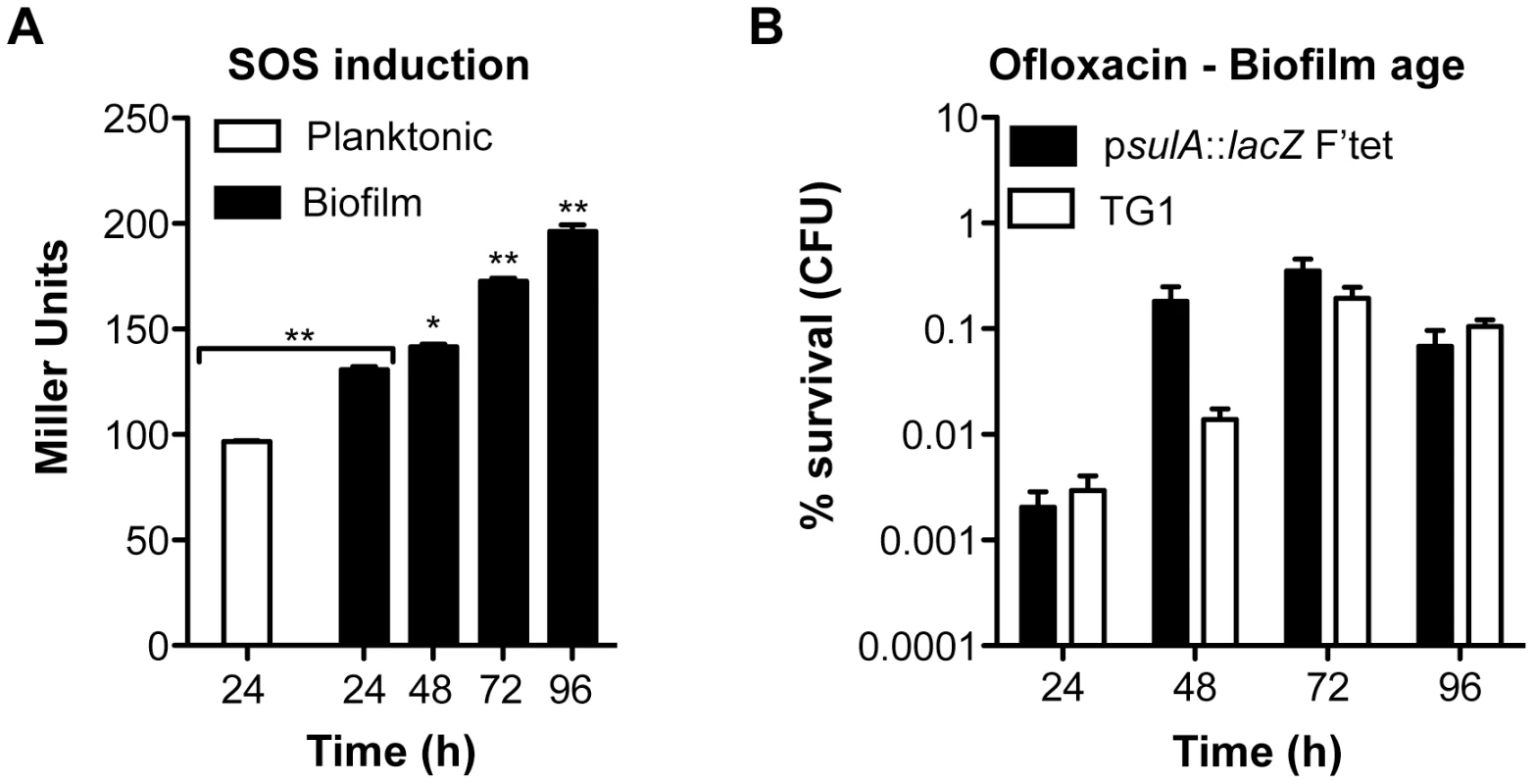
(A) Induction of the SOS response was monitored by β-galactosidase assay in aging biofilms and planktonic cells using an E. coli K-12 derivative carrying a psulA::lacZ transcriptional fusion and the biofilm promoting factor F'tet. (B) The influence of biofilm age on ofloxacin tolerance was evaluated in biofilms of the reporter strain psulA::lacZ F'tet (black bars) and wild-type TG1 (white bars). Briefly, all biofilms were grown for 24, 48, 72 and 96 h in M63B1Gluc with medium renewal every 24 h. In (B) biofilms were treated for 24 h with ofloxacin (5 µg ml−1, 80× MIC) in M63B1Gluc, therefore in absence of starvation. Survivors were quantified by viable cell counts. Percent survival represents the tolerant population after 24 h of treatment compared to untreated biofilm prior to addition of antibiotics. Data represented are mean ± SEM of at least three replicates. Asterisks indicate values significantly different than 24-h biofilms. Statistics were performed by the two-tailed unpaired t-test: * P≤0.001 and ** P≤0.0001. Fig. 6. Ofloxacin tolerance in aging biofilms is SOS-response-dependent. 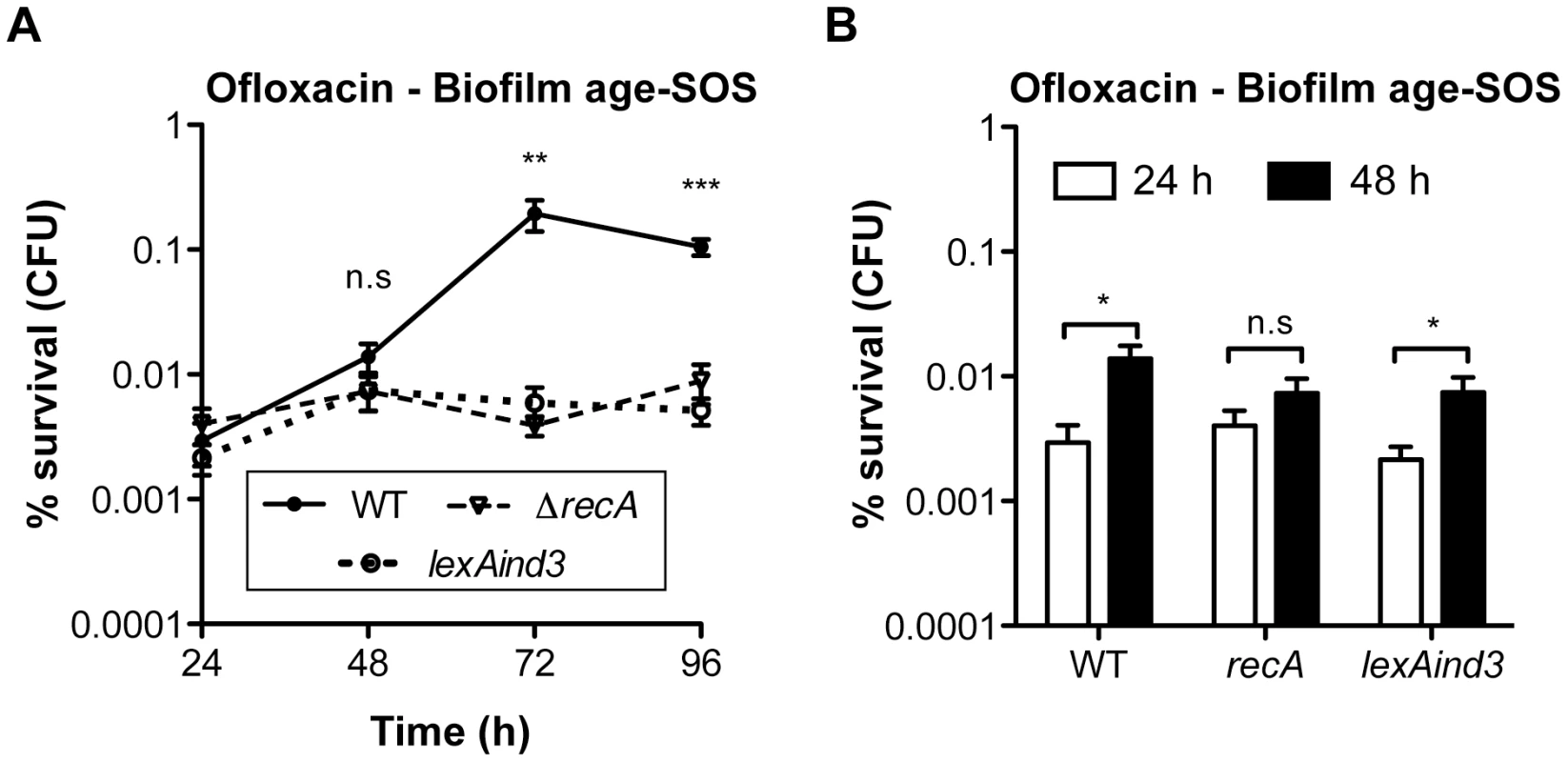
(A) The influence of the SOS response on the ofloxacin tolerance of aging biofilms was evaluated and compared between biofilms of wild-type TG1 (WT), ΔrecA, and lexAind3. (B) A close-up of the data obtained in panel A for the 24- and 48-h biofilms. Briefly, all biofilms were grown for 24, 48, 72 and 96 h in M63B1Gluc with medium renewal every 24 h and treated for 24 h with ofloxacin (5 µg ml−1, 80× MIC) in M63B1Gluc (no starvation). Survivors were quantified by viable cell counts. Percent survival represents the tolerant population after 24 h of treatment compared to untreated biofilm prior to addition of antibiotics. All compared biofilms had similar numbers of CFUs prior to antibiotic treatment (data not shown). Data represented are mean ± SEM of at least three replicates. Asterisks indicate values significantly different than WT (panel A). Statistics were performed by the two-tailed unpaired t-test: * P≤0.05, ** P≤0.01, *** P≤0.0001, and n.s. (not significant). The genotypes of the different strains used are detailed in Table S1 or as follows: WT (TG1), ΔrecA (TG1ΔrecA::KmFRT ), and lexAind3 (TG1lexAind3). Additionally, planktonic bacteria displayed a significant reduction of sulA expression compared to their 24-h biofilm counterparts (Figure 5A). This is in agreement with our previous observations demonstrating that, in planktonic bacteria, ofloxacin tolerance was not increased upon starvation (Figure 3C) and that the SOS response was not involved in the ofloxacin tolerance of planktonic bacteria independently of their starvation state (Figure S4), unlike biofilm bacteria (Figure 4). Taken together, these results demonstrated that the SOS response was induced in aging biofilms which increasing tolerance to fluoroquinolone ofloxacin is SOS-response-dependent.
Discussion
Identification of antibiotic tolerance determinants that are unique to biofilms has proven to be an arduous task, due in part to the physiological heterogeneity of these bacterial communities [1], [5], [7]. Here we screened for E. coli mutants forming biofilms with increased antibiotic tolerance in order to reveal transient physiological states and genetic modifications as tolerance mechanisms potentially occurring within biofilm subpopulations.
This approach showed that most identified mutants were amino acid auxotrophs displaying strong tolerance to either ticarcillin or ofloxacin upon starvation. Biofilm heterogeneity through generation of amino acid auxotrophs had been previously observed. Pseudomonas aeruginosa amino acid auxotrophs are commonly isolated from sputa of cystic fibrosis patients and are generally more resistant to antibiotics than their prototrophic parental strains [9], [10], [38], [39]. In contrast to the planktonic cells and biofilm ticarcillin tolerance displayed by all tested starved amino acid auxotrophs, leucine, lysine, and cysteine auxotrophs exhibited increased tolerance to ofloxacin, which occurred only in starved biofilms. Since ofloxacin is active even against non-growing bacteria [17], [18], general growth arrest did not explain the observed increase in ofloxacin tolerance. Moreover, while bacteria starved for glucose, leucine, cysteine, and lysine were highly tolerant to ofloxacin in biofilms, tryptophan starvation had no significant effect and starvation for other amino acids led to intermediate levels of tolerance. While these results confirm that the observed tolerance was not due to growth arrest, but rather to an active mechanism, it also suggests that distinct starvation types may have different physiological consequences in biofilms, consistent with a recent study demonstrating that starvation for different amino acids resulted in variations in growth rates and ß-galactosidase activity [40].
We demonstrated that both a functional RecA and a cleavable LexA were essential for the ofloxacin tolerance phenotype exhibited by biofilms starved either for glucose, leucine or lysine using prototrophic and auxotrophic mutant strains. A RecA-mediated SOS response was not necessary for increased ticarcillin tolerance in biofilms suggesting growth arrest as tolerance determinant unlike ofloxacin. We previously showed that recA and other SOS response genes were significantly induced in mature biofilms compared to exponentially grown planktonic cells [41]. Consistently, our findings demonstrated that induction of the SOS response was significantly greater in biofilm bacteria than in their planktonic counterparts. This could account for the biofilm specificity of ofloxacin tolerance, in which RecA or SOS-regulated proteins reached a critical level that is not achieved in planktonic cells, which may explain the lack of ofloxacin tolerance induction in starved planktonic populations.
Interestingly, despite a lower level of SOS-response, non-starved planktonic bacteria were more tolerant than their biofilm counterparts to both ticarcillin and ofloxacin, which is in agreement with previous observations [3], [17]. The ticarcillin tolerance could be explained by the fact that planktonic stationary cells are under growth arrest and therefore highly tolerant to ticarcillin while biofilms are heterogeneous populations containing a mix of growing and non-growing bacterial cells. For ofloxacin, the hypothesis of slow or absence of growth cannot satisfactorily explain the higher planktonic cells tolerance since it kills independently of growth. Stationary phase cultures are known to generate more persister cells than exponentially growing cultures, even more than biofilm cells, therefore suggesting some level of planktonic and biofilm specificity involved in generation of persisters [17]. In absence of starvation, it is therefore possible to imagine that a higher level of persisters occurring in planktonic cells would explain their higher tolerance. Moreover, the ofloxacin tolerance of planktonic bacteria is likely due to a mechanism other than the SOS response since a ΔrecA did not significantly impair the overall tolerance of both non-starving and starving populations. The latter results strengthen the notion that the induction of ofloxacin tolerance in starving biofilms is likely to involve mechanisms different than those currently described in planktonic cells [5], [42], [43].
Although our study evaluates ofloxacin only, fluoroquinolones are known to induce the SOS response in E. coli [24], [29]. Here we showed that induction of the SOS response upon exposure to bactericidal concentrations of ofloxacin (80× MIC) was unlikely to occur in absence of carbon source, i.e. in full deprivation state, due to the absence of protein synthesis. Hence, both induction of the SOS response during biofilm formation and nutrient depletion (carbon source or amino acids) are necessary but not sufficient to lead to the observed biofilm-specific high tolerance to ofloxacin.
The SOS response is involved in bacterial adaptive responses and horizontal gene transfer potentially leading to the onset of antibiotic resistance in a broad range of bacterial species [44]–[49]. The SOS response was also previously shown to induce persistent cells in planktonic populations during treatment with the fluoroquinolone ciprofloxacin [24], [25]. “Persisters” are phenotypic variants that can revert to wild-type antibiotic sensitivity [4], [5]. They are believed to be the end result of stochastic endogenous stress leading to growth arrest, resulting in the shutdown of bactericidal antibiotic targets and therefore the creation of multidrug-tolerant cells [5], [6]. Among possible targets, the SOS response was shown to induce expression of the TA module TisAB [25]. However, TisAB-dependent ciprofloxacin persisters were only observed in exponentially growing bacteria [25], consistent with the fact that none of the TA modules tested in our study was required for the biofilm-associated increased tolerance to ofloxacin, including known SOS-induced TA modules TisAB, SymER, DinJ-YafQ and YafNO. Our results therefore indicate that starvation in biofilm bacteria induces a SOS-dependent ofloxacin tolerance, but independently of TA loci induced by the SOS response. As raised above, the ability in generating persisters differs greatly between planktonic and biofilms cells [17], therefore it can be speculated that ofloxacin-tolerant persisters in biofilms could be physiologically distinct from their planktonic persister counterparts. Future work should concentrate precisely on identifying which SOS-gene(s), among the over 40 known SOS-regulated genes [50], [51], is required to support this biofilm increased tolerance towards ofloxacin.
Since we showed that starvation, but not auxotrophy per se, promotes the high ofloxacin tolerance observed within biofilms, our results demonstrate a general link between nutrient limitation and formation of highly antibiotic-tolerant populations. Starvation to amino acids or carbon sources induces the stringent response through the synthesis of (p)ppGpp via both RelA and SpoT [21]. Indeed, while we could not assess the role of SpoT since a relA spoT double mutant, i.e. a ppGpp0 strain, is auxotroph to multiple amino acids [23], we demonstrated that relA plays a role in observed ofloxacin biofilm-specific hypertolerance. However, the role of this response seemed less important than that of the SOS response, since the stringent response was only partially involved following leucine starvation. Nguyen et al. recently demonstrated that antibiotic tolerance exhibited in nutrient-limited biofilms of P. aeruginosa depended on the stringent response [12]. It can therefore be imagined that multiple pathways might play a significant role in the biofilm-associated ofloxacin tolerance of starved biofilms, with their role depending on conditions prevailing within biofilms. The manner in which these two distinct responses become integrated at the molecular level to promote high biofilm-specific ofloxacin tolerance remains unclear at the moment.
Nutrient deprivation might occur in biofilm populations located in micro-niches of heterogeneous biofilms, leading to higher tolerance to antibiotics [7]. Alternatively, adverse physicochemical parameters such as oxygen rarefaction prevailing within deep biofilm layers could slow down bacterial metabolism, thereby causing bacteria to enter a quasi-auxotrophic state even in the presence of sufficient available carbon sources. In support of this hypothesis, we showed that ofloxacin tolerance increased with biofilm age, suggesting that nutrient-depleted pockets or starved layers of cells may increase in number or size with biofilm maturation, or else be accompanied by a reduction in nutrient flow. Interestingly, Allison et al. recently showed that reactivation of the metabolism of E. coli persister cells restored antibiotic sensitivity both in planktonic and biofilms cells [52]. In parallel with our study, the addition of exogenous leucine during antibiotic treatment of leucine auxotroph biofilms restored sensitivity to levels greater than those of the parent wild-type prototroph treated without leucine, suggesting that the addition of amino acids may also impact overall antibiotic tolerance. While the study by Allison et al. did not specifically involve ofloxacin, it clearly suggests that nutrient depletion might play a central role in the ability of biofilm-associated bacteria to tolerate otherwise lethal concentrations of antibiotics. Although lethal concentrations of ofloxacin induce the SOS response, we show here that increased tolerance of aging biofilms is SOS-dependent and is correlated with increased SOS induction in biofilm bacteria. While this result is consistent with SOS induction in aging biofilm-like E. coli colonies on agar plates [37], it also suggests that biofilm areas in which local nutrient depletion and/or various stresses (pH drop, reduction of oxygen, etc) occur may cause SOS induction and favour high antibiotic tolerance and thus persistence.
In conclusion, we show that starvation potentially occurring locally in heterogeneous and diffusion-limited biofilm microenvironments leads to biofilm-specific SOS-dependent tolerance to the fluoroquinolone ofloxacin. Identification of the SOS response as a key molecular determinant in antibiotic tolerance in nutrient-limited biofilms reveals a possible general mechanism leading to high, but transient tolerance to a medically relevant class of antibiotics. It underlines the importance of the SOS response in the different bacterial mechanisms counteracting the effects of antibiotics, and it raises the possibility of using the SOS response as a target for reducing the emergence of biofilm tolerance to antibiotics in clinical settings [53]–[55].
Materials and Methods
Bacterial strains, plasmids, and growth conditions
Bacterial strains and plasmids used in this study are described in Table S1. All experiments were performed in 0.4% glucose M63B1 minimal medium (M63B1Gluc) or in lysogeny broth (LB) medium [56] at 37°C unless specified otherwise. Antibiotics were added when required at the following concentrations: kanamycin (Km), 50 µg ml−1; tetracycline (Tet), 15 µg ml−1; chloramphenicol (Cm), 25 µg ml−1; spectinomycin (Spec), 25 µg ml−1. Amino acids were added to M63B1Gluc when required to a final concentration varying from 1 to 100 µg ml−1. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Biofilm formation assay
A static biofilm formation assay was performed as previously described using 96-well polyvinyl chloride (PVC) microtiter plates (Falcon; Becton Dickinson Labware, Oxnard, CA) [57], [58]. Prototrophic bacteria: Overnight cultures grown in LB medium with required antibiotic(s) were diluted into M63B1Gluc to an OD600 of 0.05 and used as inoculum. Auxotrophic bacteria: One ml of an overnight culture grown in LB medium was washed twice in M63B1 (to remove excess amino acids), normalized to an OD600 of 0.05 in M63B1Gluc and supplemented with 25 µg ml−1 of the lacking amino acid corresponding to each evaluated auxotroph. Each microtiter well was then inoculated with 100 µl of the OD600 0.05 inoculum, with a minimum of three wells per bacterial strain for each assay, and incubated at 37°C for 24, 48, 72 or 96 hours with renewal of growth medium every 24 h.
Biofilm bacteria/biomass detection methods
Crystal violet staining
Biofilms attached to the sides of microtiter wells were thoroughly rinsed in water to remove unattached bacterial cells and stained with 125 µl of crystal violet (1% v/v) as previously described [57], [58]. Quantification of the stained biomass was achieved by adding 150 µl per well of a dissolving solution (ethanol (80%)/acetone (20%) (v/v)). Subsequently, dissolution of the bound crystal violet in each well was quantified by spectrometry at an absorbance of 595 nm.
Metabolic activity of biofilms in microtiter wells
Metabolic activity associated with biofilm cells was determined by the XTT (2,3-bis 2-methoxy-4-nitro-5-sulfo-phenyl-2H-tetra-zolium-5-carboxanilide) reduction assay as previously described [59]. Briefly, each well containing biofilms was washed once with 125 µl of phosphate-buffered saline (PBS: 10 mM sodium phosphate, pH 7.4, 0.9% NaCl) to remove unattached cells, and filled with 125 µl of a PBS solution containing 50 µg ml−1 XTT and 1 µM menadione. Plates were incubated at 37°C for 3.5 h in the dark, allowing metabolically active bacterial cells to reduce the XTT, which was then quantified colorimetrically by measuring absorbance at 492 nm.
Colony-forming units (CFU) of biofilms in microtiter wells
CFU of biofilms attached to the sides of microtiter wells were determined. Each well containing a biofilm was washed once with M63B1 to remove unattached cells and filled with 100 µl of the same medium enabling bacterial survival without growth. CFU determination was performed by serial dilutions on washed biofilms that were mechanically disrupted by pipetting.
Minimal inhibitory concentration (MIC) determination
MIC values of ticarcillin (Ticarpen; GlaxoSmithKline, Marly-le-Roi, France) and ofloxacin (Sigma-Aldrich) were determined by macrodilutions in M63B1Gluc as previously described [60]. MICs of ticarcillin and ofloxacin for TG1gfp were determined to be 1 and 0.0625 µg ml−1, respectively.
Antibiotic susceptibility assays
Biofilm bacteria
Biofilms were formed for 24 h as described above. Unattached and planktonic bacteria were first removed from 24 h pre-formed biofilms and wells were then filled with 100 µl of M63B1 containing a specific antibiotic concentration, unless noted otherwise. After 24 h of incubation at 37°C, the antibiotic susceptibility of the treated biofilm population versus a 24 h biofilm was determined by CFU counts and/or colorimetric differences using the XTT-reduction assay as mentioned above.
Planktonic bacteria
Bacterial cultures were grown statically using 96-well microtiter plates for 24 h at 37°C from a starting OD600 of 0.05 in 100 µl of M63B1Gluc containing 25 µg ml−1 of a specific amino acid if required. These bacterial cultures were moved by pipetting to new microtiter plates in order to have only the planktonic population and were then harvested from 12 wells in triplicate by centrifugation and resuspended in 100 µl of M63B1Gluc containing either ticarcillin (100 µg ml−1) or ofloxacin (5 µg ml−1). Antibiotic treatment was performed for 24 h at 37°C, following which the bacteria were centrifuged and washed once with M63B1 medium to remove any traces of antibiotics prior to performing dilutions to determine CFU.
Transposon mutagenesis
Mariner transposon mutagenesis was performed as previously described [61]. Briefly, plasmid pSC189 was conjugated from S17-1 λPir (pSC189) into recipient strain TG1gfp. The resulting transconjugants were selected on LB agar plates containing the required antibiotics (Km and Cm) and approximately 10,000 transposon mutants were transferred to 96-well microtiter plates.
Screening of the transposon library for antibiotic tolerance in biofilms
Our transposon library was used to identify mutants with increased biofilm tolerance to the antibiotics ticarcillin (100 µg ml−1 - 100 times the MIC) and ofloxacin (3.125 µg ml−1 - 50 times the MIC). Microtiter plates containing 100 µl/well of fresh LB with the required antibiotics (Km and Cm) were inoculated directly from each plate of the frozen library (in 15% glycerol) using a 96-pin replicator and incubated statically at 37°C overnight. Overnight cultures were then transferred with a 96-pin replicator to PVC microtiter plates containing 100 µl of M63B1Gluc to initiate growth of static biofilms for 24 h as described above. Biofilm inoculation via a 96-pin replicator reproduced biofilms and antibiotic susceptibility profiles similar to those normalized by optical density (data not shown). Each overnight plate was duplicated for biofilm assays in order to produce a set of plates for separately assessing the antibiotic susceptibilities to ticarcillin and ofloxacin. Following the 24-h biofilm formation period, each set of transposon mutant plates containing preformed biofilms was treated with either ticarcillin or ofloxacin in fresh M63B1Gluc medium for 24 h as mentioned earlier. Antibiotic susceptibility of each mutant towards the two antibiotics was determined by colorimetric differences from the wild-type parent using the XTT-reduction assay described below. Transposon mutants displaying a 2-fold increase in tolerance were first chosen for further characterization, but due to the relatively low number of such mutants, we subsequently decreased our selection criteria to 1.5 - and 1.25-fold tolerance increase to ticarcillin and ofloxacin, respectively. Using these arbitrary cut-off values likely led to the selection of false positive, however the antibiotic hypertolerance of all selected mutants based on these values were carefully validated by first transducing the mutations back into WT strain TG1 by P1vir prior to mapping their localization within the genome by arbitrary PCR.
Arbitrary PCR
Transposon insertion sites were determined as previously described [62]. Briefly, this method involves a first round of PCR using a primer specific for the right end of the transposon (IR2) and an arbitrary primer (ARB1 or ARB6). A second PCR was then performed on the product from the first PCR using a primer specific to the rightmost end of the transposon (IR2-60-5) and a primer identical to the 5′ end of the arbitrary primer (ARB2). Arbitrary PCR primer sequences are listed in Table S2.
Test of amino acid auxotrophy
One ml of an overnight LB culture of each putative auxotroph was washed twice in M63B1Gluc to remove all excess of amino acids and the resuspended pellets were used to inoculate either a minimal medium liquid culture (M63B1Gluc) or an agar plate of M63B1Gluc. Growth was monitored from an overnight incubation at 37°C. Absence of growth suggested amino acid auxotrophy, which was confirmed by the addition of the corresponding amino acid at 25 µg ml−1 for growth restoration in minimal medium (M63B1Gluc) using both liquid and agar plate cultures.
Construction of deletion mutant strains
The different E. coli mutant strains used in this study originated either from the Keio Collection [63], transferred to the appropriate genetic background by P1vir phage transduction, or were directly constructed using the λ-red linear DNA gene inactivation method [64], [65]. When required, kanamycin resistance markers flanked by two FRT sites were removed using Flp recombinase [66]. Primers used in this study are listed in Table S2. All mutations were confirmed by PCR and/or sequence analysis.
Transcriptional and translational analysis of the SOS response
Transcriptional analysis in biofilms
ß-galactosidase enzymatic activity was assessed to determine the induction of the SOS response under various conditions using the reporter strain psulA::lacZ F'tet. Biofilms were formed on the sides of microtiter wells as described above. Briefly, the reporter strain psulA::lacZ F'tet was initially grown in LB medium with required antibiotic(s), diluted into M63B1Gluc to an OD600 of 0.05 and used as inoculum. Each microtiter well was then inoculated with 100 µl of the OD600 0.05 inoculum, with a minimum of three wells and incubated at 37°C for 24, 48, 72 or 96 hours with renewal of growth medium every 24 h. To determine the expression of sulA in aging biofilms, planktonic bacteria were removed from each well and the ß-galactosidase activity was directly measured on the biofilm bacteria as previously described [67].
Transcriptional analysis of planktonic bacteria
As described above, planktonic bacteria sharing the same environment (microtiter well) as biofilm cells were used to determine the expression of the SOS response. Growth conditions were the same as those described above for biofilm growth, but instead of using the attached bacterial cells (biofilm), the free floating cells (planktonic – 24 h) from three independent pools of 12 wells were collected for the analysis. Bacteria from the planktonic population were then harvested by centrifugation, washed, and resuspended into 100 µl of M63B1 in which the ß-galactosidase activity was directly measured as previously described [67].
Translational analysis
Biofilms of a reporter strain MG1655KmRExTetlacZ_F'tet were grown M63B1Gluc for 24 h as described above. This reporter strain contains a lacZ gene under the control of an inducible promoter (PLtetO-1) by the addition of anhydrotetracycline (aTc) [68]. To determine whether lacZ was translated in conditions of starvation, 24-h biofilms were washed and exposed to 100 µl per well of M63B1Gluc (control) or M63B1 (glucose starvation) with increasing concentration of aTc (0, 5, 50 ng ml−1) during a period of 1 h after which biofilms were washed and resuspended in M63B1. Crude protein extracts were prepared and equivalent amounts of proteins were first loaded on a SDS-PAGE gel, proteins were transferred to a polyvinylidene difluoride membrane, and ß-galactosidase immunodetection was performed using a 1∶10,000 dilution of mouse antiserum raised against ß-galactosidase. A planktonic culture of the same reporter strain was used as a control to verify the efficiency of the induction system in non-starving conditions.
Statistical analysis
Analyses were performed using Prism 5.0 for Mac OS X (GraphPad Software, Inc.). Each experiment was performed at least three times.
Supporting Information
Zdroje
1. StewartP, William CostertonJ (2001) Antibiotic resistance of bacteria in biofilms. The Lancet 358 : 135–138.
2. AnwarH, van BiesenT, DasguptaM, LamK, CostertonJW (1989) Interaction of biofilm bacteria with antibiotics in a novel in vitro chemostat system. Antimicrob Agents Chemother 33 : 1824–1826.
3. EvansDJ, AllisonDG, BrownMR, GilbertP (1991) Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: effect of specific growth rate. J Antimicrob Chemother 27 : 177–184.
4. LevinBR, RozenDE (2006) Non-inherited antibiotic resistance. Nat Rev Micro 4 : 556–562.
5. LewisK (2010) Persister cells. Annu Rev Microbiol 64 : 357–372.
6. GefenO, BalabanNQ (2009) The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol Rev 33 : 704–717.
7. StewartPS, FranklinMJ (2008) Physiological heterogeneity in biofilms. Nat Rev Microbiol 6 : 199–210.
8. AnderlJN, ZahllerJ, RoeF, StewartPS (2003) Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother 47 : 1251–1256.
9. BolesBR, SinghPK (2008) Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci U S A 105 : 12503–12508.
10. BolesBR, ThoendelM, SinghPK (2004) Self-generated diversity produces “insurance effects” in biofilm communities. Proc Natl Acad Sci U S A 101 : 16630–16635.
11. LewisK (2007) Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5 : 48–56.
12. NguyenD, Joshi-DatarA, LepineF, BauerleE, OlakanmiO, et al. (2011) Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334 : 982–986.
13. LenzAP, WilliamsonKS, PittsB, StewartPS, FranklinMJ (2008) Localized gene expression in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 74 : 4463–4471.
14. Pérez-OsorioAC, WilliamsonKS, FranklinMJ (2010) Heterogeneous rpoS and rhlR mRNA levels and 16S rRNA/rDNA (rRNA gene) ratios within Pseudomonas aeruginosa biofilms, sampled by laser capture microdissection. J Bacteriol 192 : 2991–3000.
15. GhigoJM (2001) Natural conjugative plasmids induce bacterial biofilm development. Nature 412 : 442–445.
16. TuomanenE, CozensR, ToschW, ZakO, TomaszA (1986) The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol 132 : 1297–1304.
17. SpoeringAL, LewisK (2001) Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183 : 6746–6751.
18. ZhaoX, MalikM, ChanN, Drlica-WagnerA, WangJY, et al. (2006) Lethal action of quinolones against a temperature-sensitive dnaB replication mutant of Escherichia coli. Antimicrob Agents Chemother 50 : 362–364.
19. VegaNM, AllisonKR, KhalilAS, CollinsJJ (2012) Signaling-mediated bacterial persister formation. Nat Chem Biol 8 : 431–433.
20. PooleK (2012) Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria. Trends Microbiol 20 : 227–234.
21. PotrykusK, CashelM (2008) (p)ppGpp: still magical? Annu Rev Microbiol 62 : 35–51.
22. SrivatsanA, WangJD (2008) Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol 11 : 100–105.
23. XiaoH, KalmanM, IkeharaK, ZemelS, GlaserG, et al. (1991) Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266 : 5980–5990.
24. DorrT, LewisK, VulicM (2009) SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet 5: e1000760 doi:10.1371/journal.pgen.1000760.
25. DorrT, VulicM, LewisK (2010) Ciprofloxacin Causes Persister Formation by Inducing the TisB toxin in Escherichia coli. PLoS Biol 8: e1000317 doi:10.1371/journal.pbio.1000317.
26. FungDK, ChanEW, ChinML, ChanRC (2010) Delineation of a bacterial starvation stress response network which can mediate antibiotic tolerance development. Antimicrob Agents Chemother 54 : 1082–1093.
27. ButalaM, Zgur-BertokD, BusbySJ (2009) The bacterial LexA transcriptional repressor. Cell Mol Life Sci 66 : 82–93.
28. BierneH, SeigneurM, EhrlichSD, MichelB (1997) uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol Microbiol 26 : 557–567.
29. KaldaluN, MeiR, LewisK (2004) Killing by ampicillin and ofloxacin induces overlapping changes in Escherichia coli transcription profile. Antimicrob Agents Chemother 48 : 890–896.
30. Van MelderenL (2010) Toxin-antitoxin systems: why so many, what for? Curr Opin Microbiol 13 : 781–5.
31. PedersenK, GerdesK (1999) Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol 32 : 1090–1102.
32. YamaguchiY, ParkJ-H, InouyeM (2011) Toxin-Antitoxin Systems in Bacteria and Archaea. Ann Rev Genet 45 : 61–79.
33. GerdesK, MaisonneuveE (2012) Bacterial persistence and toxin-antitoxin Loci. Ann Rev Microbiol 66 : 103–123.
34. ErillI, CampoyS, BarbeJ (2007) Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev 31 : 637–656.
35. JanionC, SikoraA, NowosielskaA, GrzesiukE (2002) Induction of the SOS response in starved Escherichia coli. Environ Molecular Mutagenesis 40 : 129–133.
36. PetrosinoJF, GalhardoRS, MoralesLD, RosenbergSM (2009) Stress-induced beta-lactam antibiotic resistance mutation and sequences of stationary-phase mutations in the Escherichia coli chromosome. J Bacteriol 191 : 5881–5889.
37. TaddeiF, MaticI, RadmanM (1995) cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc Natl Acad Sci U S A 92 : 11736–11740.
38. AgarwalG, KapilA, KabraSK, DasBK, DwivediSN (2005) Characterization of Pseudomonas aeruginosa isolated from chronically infected children with cystic fibrosis in India. BMC Microbiol 5 : 43.
39. ThomasSR, RayA, HodsonME, PittTL (2000) Increased sputum amino acid concentrations and auxotrophy of Pseudomonas aeruginosa in severe cystic fibrosis lung disease. Thorax 55 : 795–797.
40. BodiniS, NunziangeliL, SantoriF (2007) Influence of amino acids on low-density Escherichia coli responses to nutrient downshifts. J Bacteriol 189 : 3099–3105.
41. BeloinC, ValleJ, Latour-LambertP, FaureP, KzreminskiM, et al. (2004) Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol 51 : 659–674.
42. GirgisHS, HarrisK, TavazoieS (2012) Large mutational target size for rapid emergence of bacterial persistence. Proc Natl Acad Sci U S A 109 : 12740–12745.
43. HansenS, LewisK, VulicM (2008) Role of Global Regulators and Nucleotide Metabolism in Antibiotic Tolerance in Escherichia coli. Antimicrob Agents Chemother 52 : 2718–2726.
44. BaharogluZ, MazelD (2011) Vibrio cholerae Triggers SOS and Mutagenesis in Response to a Wide Range of Antibiotics: a Route towards Multiresistance. Antimicrob Agents Chemother 55 : 2438–2441.
45. BlázquezJ, Gómez-GómezJ-M, OliverA, JuanC, KapurV, et al. (2006) PBP3 inhibition elicits adaptive responses in Pseudomonas aeruginosa. Mol Microbiol 62 : 84–99.
46. MaiquesE, UbedaC, CampoyS, SalvadorN, LasaI, et al. (2006) beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol 188 : 2726–2729.
47. BaharogluZ, BikardD, MazelD (2010) Conjugative DNA Transfer Induces the Bacterial SOS Response and Promotes Antibiotic Resistance Development through Integron Activation. PLoS Genet 6: e1001165 doi:10.1371/journal.pgen.1001165.
48. BeaberJW, HochhutB, WaldorMK (2004) SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427 : 72–74.
49. GuerinE, CambrayG, Sanchez-AlberolaN, CampoyS, ErillI, et al. (2009) The SOS response controls integron recombination. Science 324 : 1034.
50. CourcelleJ, KhodurskyA, PeterB, BrownPO, HanawaltPC (2001) Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158 : 41–64.
51. Fernandez De HenestrosaAR, OgiT, AoyagiS, ChafinD, HayesJJ, et al. (2000) Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol 35 : 1560–1572.
52. AllisonKR, BrynildsenMP, CollinsJJ (2011) Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473 : 216–220.
53. LuTK, CollinsJJ (2009) Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci U S A 106 : 4629–4634.
54. SextonJZ, WigleTJ, HeQ, HughesMA, SmithGR, et al. (2010) Novel Inhibitors of E. coli RecA ATPase Activity. Curr Chem Genomics 4 : 34–42.
55. WigleTJ, SextonJZ, GromovaAV, HadimaniMB, HughesMA, et al. (2009) Inhibitors of RecA Activity Discovered by High-Throughput Screening: Cell-Permeable Small Molecules Attenuate the SOS Response in Escherichia coli. J Biomol Screening 14 : 1092–1101.
56. BertaniG (2004) Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol 186 : 595–600.
57. O'TooleGA, KolterR (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28 : 449–461.
58. RouxA, BeloinC, GhigoJM (2005) Combined inactivation and expression strategy to study gene function under physiological conditions: application to identification of new Escherichia coli adhesins. J Bacteriol 187 : 1001–1013.
59. RamageG, Vande WalleK, WickesBL, Lopez-RibotJL (2001) Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother 45 : 2475–2479.
60. HacekDM, DresselDC, PetersonLR (1999) Highly reproducible bactericidal activity test results by using a modified National Committee for Clinical Laboratory Standards broth macrodilution technique. J Clin Microbiol 37 : 1881–1884.
61. ChiangSL, RubinEJ (2002) Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296 : 179–185.
62. Da ReS, GhigoJM (2006) A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J Bacteriol 188 : 3073–3087.
63. BabaT, AraT, HasegawaM, TakaiY, OkumuraY, et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2 : 2006 0008.
64. ChaverocheMK, GhigoJM, d'EnfertC (2000) A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res 28: E97.
65. DerbiseA, LesicB, DacheuxD, GhigoJM, CarnielE (2003) A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol Med Microbiol 38 : 113–116.
66. CherepanovPP, WackernagelW (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158 : 9–14.
67. Miller JH (1992) A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press,.
68. Da ReS, Le QuereB, GhigoJM, BeloinC (2007) Tight modulation of Escherichia coli bacterial biofilm formation through controlled expression of adhesion factors. Appl Environ Microbiol 73 : 3391–3403.
Štítky
Genetika Reprodukční medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 1
-
Všechny články tohoto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



