-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
Integrative and conjugative elements (ICEs) are agents of horizontal gene transfer and have major roles in evolution and acquisition of new traits, including antibiotic resistances. ICEs are found integrated in a host chromosome and can excise and transfer to recipient bacteria via conjugation. Conjugation involves nicking of the ICE origin of transfer (oriT) by the ICE–encoded relaxase and transfer of the nicked single strand of ICE DNA. For ICEBs1 of Bacillus subtilis, nicking of oriT by the ICEBs1 relaxase NicK also initiates rolling circle replication. This autonomous replication of ICEBs1 is critical for stability of the excised element in growing cells. We found a conserved and previously uncharacterized ICE gene that is required for conjugation and replication of ICEBs1. Our results indicate that this gene, helP (formerly ydcP), encodes a helicase processivity factor that enables the host-encoded helicase PcrA to unwind the double-stranded ICEBs1 DNA. HelP was required for both conjugation and replication of ICEBs1, and HelP and NicK were the only ICEBs1 proteins needed for replication from ICEBs1 oriT. Using chromatin immunoprecipitation, we measured association of HelP, NicK, PcrA, and the host-encoded single-strand DNA binding protein Ssb with ICEBs1. We found that NicK was required for association of HelP and PcrA with ICEBs1 DNA. HelP was required for association of PcrA and Ssb with ICEBs1 regions distal, but not proximal, to oriT, indicating that PcrA needs HelP to progress beyond nicked oriT and unwind ICEBs1. In vitro, HelP directly stimulated the helicase activity of the PcrA homologue UvrD. Our findings demonstrate that HelP is a helicase processivity factor needed for efficient unwinding of ICEBs1 for conjugation and replication. Homologues of HelP and PcrA-type helicases are encoded on many known and putative ICEs. We propose that these factors are essential for ICE conjugation, replication, and genetic stability.
Published in the journal: . PLoS Genet 9(1): e32767. doi:10.1371/journal.pgen.1003198
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003198Summary
Integrative and conjugative elements (ICEs) are agents of horizontal gene transfer and have major roles in evolution and acquisition of new traits, including antibiotic resistances. ICEs are found integrated in a host chromosome and can excise and transfer to recipient bacteria via conjugation. Conjugation involves nicking of the ICE origin of transfer (oriT) by the ICE–encoded relaxase and transfer of the nicked single strand of ICE DNA. For ICEBs1 of Bacillus subtilis, nicking of oriT by the ICEBs1 relaxase NicK also initiates rolling circle replication. This autonomous replication of ICEBs1 is critical for stability of the excised element in growing cells. We found a conserved and previously uncharacterized ICE gene that is required for conjugation and replication of ICEBs1. Our results indicate that this gene, helP (formerly ydcP), encodes a helicase processivity factor that enables the host-encoded helicase PcrA to unwind the double-stranded ICEBs1 DNA. HelP was required for both conjugation and replication of ICEBs1, and HelP and NicK were the only ICEBs1 proteins needed for replication from ICEBs1 oriT. Using chromatin immunoprecipitation, we measured association of HelP, NicK, PcrA, and the host-encoded single-strand DNA binding protein Ssb with ICEBs1. We found that NicK was required for association of HelP and PcrA with ICEBs1 DNA. HelP was required for association of PcrA and Ssb with ICEBs1 regions distal, but not proximal, to oriT, indicating that PcrA needs HelP to progress beyond nicked oriT and unwind ICEBs1. In vitro, HelP directly stimulated the helicase activity of the PcrA homologue UvrD. Our findings demonstrate that HelP is a helicase processivity factor needed for efficient unwinding of ICEBs1 for conjugation and replication. Homologues of HelP and PcrA-type helicases are encoded on many known and putative ICEs. We propose that these factors are essential for ICE conjugation, replication, and genetic stability.
Introduction
Integrative and conjugative elements (ICEs), also known as conjugative transposons, are mobile genetic elements that play a significant role in bacterial evolution and the acquisition of new traits [1]. They contribute significantly to the spread of antibiotic resistances in pathogenic bacteria. ICEs or putative ICEs are found in all major bacterial clades [2]. They reside integrated in a host genome and are propagated along with the host chromosome. Under certain conditions, an ICE can excise from the chromosome, form a double-stranded DNA (dsDNA) circle, and transfer to a recipient. Like conjugative plasmids, ICEs encode a multi-component mating pore complex that mediates their transfer from donors to recipients. Most ICEs are thought to transfer linear ssDNA. Transfer is through a type IV secretion system in Gram negative bacteria [3], [4], or its counterpart in Gram positive bacteria [5]. ICEs that transfer ssDNA were generally thought to lack the ability to undergo autonomous replication. However, recent work [6]–[8] and findings presented here indicate that autonomous replication is a property of many ICEs and that the mechanisms are conserved.
ICEBs1 is approximately 20 kb and normally found integrated in the tRNA gene trnS-leu2 of Bacillus subtilis [9], [10]. ICEBs1 gene expression and excision can be induced in >90% of cells in a population by overproduction of the activator and cell signaling regulator RapI [9]. Following induction, ICEBs1 undergoes autonomous plasmid-like rolling circle replication [6]. Replication of ICEBs1 is needed for stability of the element after excision [6].
ICEBs1 replication and conjugation both begin with nicking of the ICEBs1 origin of transfer, oriT, by the ICEBs1-encoded relaxase NicK. The nicked DNA is then unwound by the host-encoded helicase PcrA, rather than the replicative helicase (B. subtilis DnaC) [6]. During rolling-circle plasmid replication, the free 3′-OH of the nicked strand acts as a primer for replication by the host DNA polymerase, followed by recircularization of and complementary strand synthesis from the unwound single strand. By analogy to other conjugative systems, the single strand of ICEBs1 DNA covalently attached to the relaxase can also be targeted to the mating machinery by the putative coupling protein ConQ and transferred into recipient cells. Although unwinding of ICEBs1 DNA by the PcrA helicase is essential for both replication and conjugation of ICEBs1, replication of the element in donor cells is not required for its transfer to recipients [6].
PcrA-type helicases (PcrA from Gram positive bacteria and UvrD and Rep from E. coli) are required for rolling circle replication of many different plasmids and phages. The PcrA-type proteins are efficient and processive DNA translocases, but typically have poor helicase activity. For many of the characterized phages and plasmids, the element-encoded relaxase that is needed for initiation of replication interacts with the PcrA-type helicase to stimulate DNA unwinding {reviewed in [11], [12], [13]}.
Unlike these other relaxases, we found that the ICEBs1 relaxase NicK was not sufficient for ICEBs1 replication. In addition to nicK, a second ICEBs1 gene (helP, previously ydcP) was necessary for replication from ICEBs1 oriT. Expression of both nicK and helP in B. subtilis was sufficient to support replication from oriT. helP encodes a protein of previously unknown function and is conserved in many ICEs. We found that HelP is required for both mating and replication of ICEBs1, and that it stimulates the function of the helicase PcrA. We also found that the E. coli helicase UvrD (a homologue of PcrA) can substitute for PcrA in B. subtilis, to support both cell viability and ICEBs1 conjugation and replication. Based on in vivo and in vitro analyses, HelP is a helicase processivity factor that is needed for efficient unwinding of ICEBs1.
helP homologues are found in many ICEs, often in a module with genes encoding the relaxase and the putative coupling protein, indicating that these ICEs may also be capable of autonomous replication. PcrA homologues are also found on many extrachromosomal elements, either separately or as a helicase domain attached to the relaxase, indicating that these elements all share a need for DNA unwinding that is met in different ways.
Results
Two ICEBs1 genes, nicK and helP, are sufficient for autonomous replication from the ICEBs1 oriT in B. subtilis
Studies of several rolling-circle plasmids have shown that only one plasmid gene, encoding the plasmid relaxase, is required for replication from the cognate origin of replication {reviewed in [12]}. In contrast, we found that the ICEBs1 relaxase NicK was not sufficient for autonomous replication from the ICEBs1 origin of replication oriT. Instead, a second ICEBs1 gene, helP, was also needed.
To determine which ICEBs1 genes are needed for replication from oriT, we constructed a series of plasmids that carry the ICEBs1 oriT along with various candidate ICEBs1 genes. We then tested each plasmid for its ability to replicate in B. subtilis. The parent plasmid, pUS19 [14], carries a pUC-derived origin of replication that is not functional in B. subtilis, but is functional in E. coli, allowing purification of each test plasmid from E. coli. pUS19 also carries spcE, which allowed us to transform each plasmid into B. subtilis and select for spectinomycin-resistant transformants that stably acquired the plasmid. Transcription of the ICEBs1 genes was driven from the ICEBs1 promoter Pxis that was cloned onto the plasmid. Pxis is derepressed in cells lacking ICEBs1.
After analyzing various plasmids containing different combinations of candidate ICEBs1 genes (data not shown), we found that helP and nicK were sufficient to support replication from ICEBs1 oriT (Table 1). A plasmid containing oriT, nicK, and helP (pCAL1255) was capable of transforming a B. subtilis strain lacking ICEBs1 (Table 1, pCAL1255). The plasmid copy number was between 25–30 (Table 1) as indicated by the amount of spcE DNA (plasmid) relative to ydbT, a chromosomal gene adjacent to the ICEBs1 attachment site attB.
Tab. 1. NicK and HelP are the only ICE–encoded factors required for autonomous replication. 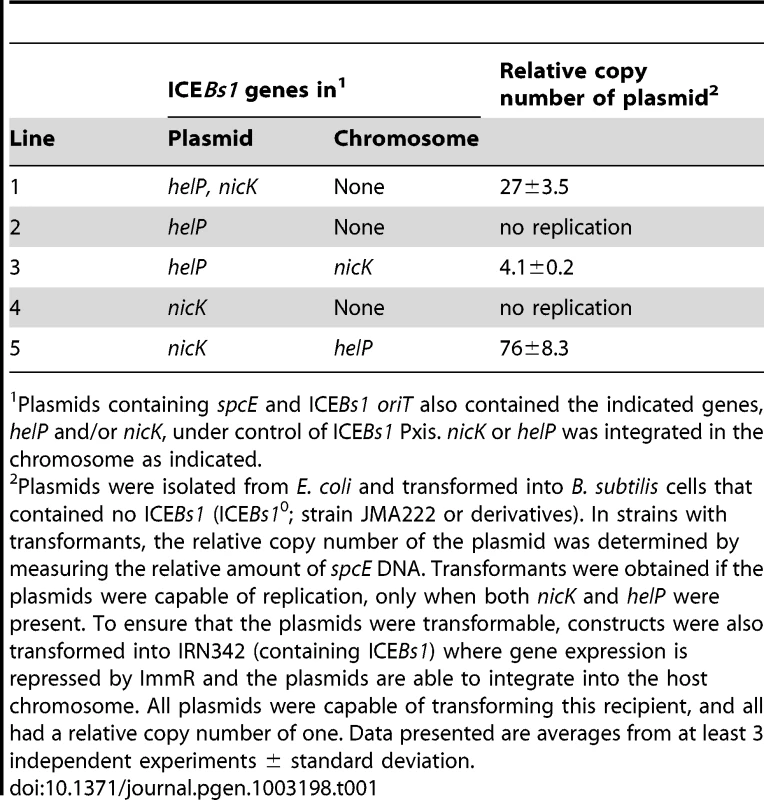
Plasmids containing spcE and ICEBs1 oriT also contained the indicated genes, helP and/or nicK, under control of ICEBs1 Pxis. nicK or helP was integrated in the chromosome as indicated. Replication of pCAL1255 (oriT, Pxis-helP-nicK) in B. subtilis was dependent on expression of helP and nicK from Pxis. We repressed expression from Pxis by transforming pCAL1255 into B. subtilis carrying an intact integrated ICEBs1, which expresses the ICEBs1 repressor ImmR. We were still able to obtain spectinomycin-resistant transformants of pCAL1255 in the ICEBs1-containing cells. However, in these transformants, the plasmid copy number was one, indicating that the plasmid was likely integrated into the chromosomal ICEBs1 by homologous recombination.
We found that autonomous plasmid replication from oriT was dependent on both helP and nicK. We did not obtain any spectinomycin-resistant transformants in B. subtilis cells lacking ICEBs1 from plasmids containing nicK without helP (Table 1, pJT151) or helP without nicK (Table 1, pCAL1260). However, we were able to transform these plasmids into strains that expressed the missing gene from a chromosomal locus, showing that the replication defects could be complemented. pJT151 (oriT, Pxis-nicK) was able to replicate and had a copy number of 70–80 in a strain expressing helP from the chromosome. pCAL1260 (oriT, Pxis-helP) was able to replicate and had a copy number of approximately 4 in a strain expressing nicK from the chromosome (Table 1). The different copy numbers of pCAL1255 (oriT, Pxis-helP-nicK) and the plasmids missing helP or nicK but complemented from a chromosomal copy of the gene may be due to different expression levels of HelP and NicK from the plasmid versus the chromosome, or the effects of the different plasmid sequences on replication efficiency. The low copy number when nicK was expressed from the chromosome is also consistent with the notion that the relaxase functions preferentially in cis [15].
Based on these results, we conclude that, unlike many rolling circle plasmid replicons, the ICEBs1 relaxase NicK is not the only element-encoded protein needed for autonomous replication. ICEBs1-encoded NicK and HelP are both needed to support replication from oriT in the absence of other ICEBs1 products.
helP is required for autonomous replication of ICEBs1
We found that helP is required for replication of ICEBs1. We constructed an in-frame markerless deletion of helP (ΔhelP) that removed its entire coding sequence from ICEBs1 (Figure 1B). After inducing ICEBs1 gene expression, we measured ICEBs1 copy number by quantitative real time PCR (qPCR). The copy number of ICEBs1 relative to the chromosome is expressed as the relative amount of DNA from ICEBs1 oriT compared to ydbT, as described previously [6]. Consistent with previous findings, the relative copy number of ICEBs1 oriT was 3–4 per cell one hour after induction of ICEBs1 gene expression (Table 2, line 1). Under similar conditions, the copy number of the ICEBs1 ΔhelP mutant was approximately 0.5 (Table 2, line 2). A copy number less than one is consistent with previous findings that replication-defective ICEBs1 is progressively lost from a population of dividing cells [6].
Fig. 1. Map of ICEBs1 and derivatives. 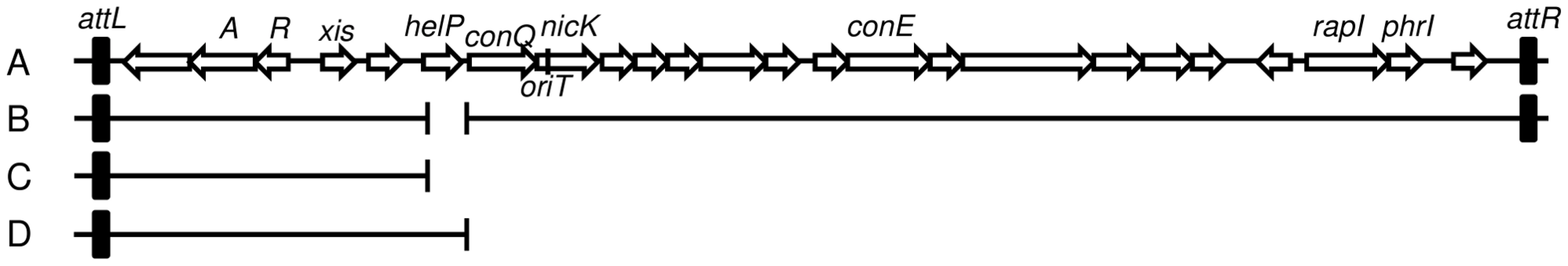
A–B. Schematic of ICEBs1 (A) and the helP mutant (B) integrated at the normal attachment site, attB. The antibiotic resistance marker (kan) that is inserted in rapI-phrI is not shown. A. Genetic map of ICEBs1. The linear integrated form of ICEBs1 is shown. Open arrows indicate open reading frames and the direction of transcription. The black rectangles at the ends of ICEBs1 represent the 60 bp direct repeats that contain the site-specific recombination sites in the left and right attachment sites, attL and attR. Some gene names are indicated. The genes encoding the ICEBs1 repressor immR and anti-repressor immA are indicated by R and A, respectively. The origin of transfer (oriT) is indicated by a black line in the 5′ end of nicK. oriT is needed for ICEBs1 transfer [15] and replication [6]. Primers used in ChIP-PCR experiments hybridize to nicK (oriT), and conE. B. Schematic of the ΔhelP155 mutation in ICEBs1. Thin horizontal lines represent the regions of ICEBs1 that are present in the mutant. The gap in the line represents the in-frame markerless deletion of helP. C–D. Diagram of the truncated ICEBs1 derivatives that were used to test complementation of ΔhelP donors. Both constructs are integrated at thrC and contain ICEBs1 genes represented by the horizontal lines, from attL to ydcO (C) or helP (D). Both derivatives contain cat (chloramphenicol resistance) in place of the part of ICEBs1 that is deleted, and neither can excise from the chromosome due to the absence of attR. Tab. 2. HelP is required for ICEBs1 transfer and replication. 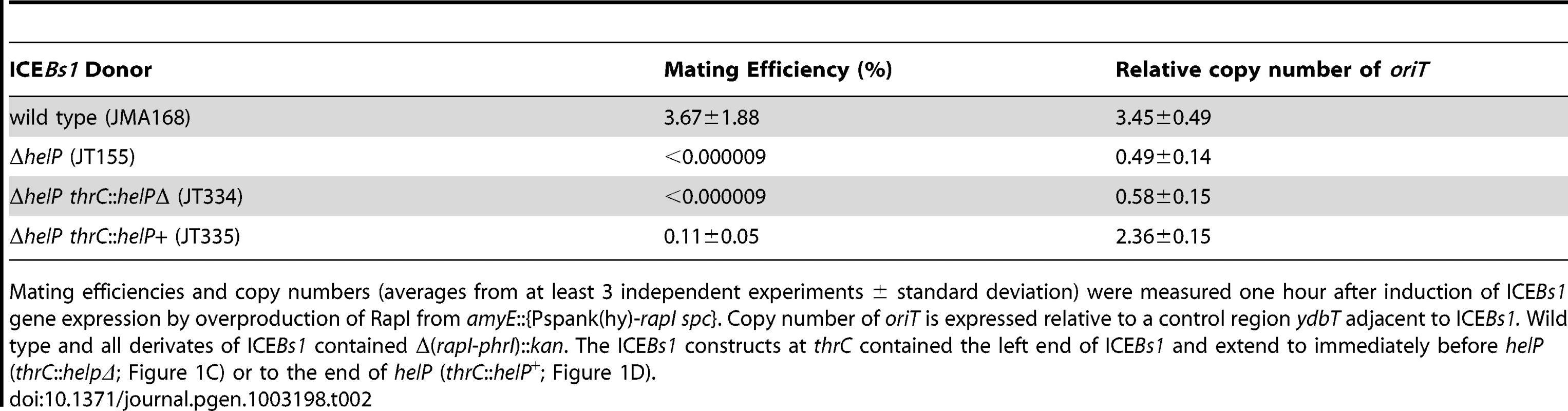
Mating efficiencies and copy numbers (averages from at least 3 independent experiments ± standard deviation) were measured one hour after induction of ICEBs1 gene expression by overproduction of RapI from amyE::{Pspank(hy)-rapI spc}. Copy number of oriT is expressed relative to a control region ydbT adjacent to ICEBs1. Wild type and all derivates of ICEBs1 contained Δ(rapI-phrI)::kan. The ICEBs1 constructs at thrC contained the left end of ICEBs1 and extend to immediately before helP (thrC::helpΔ; Figure 1C) or to the end of helP (thrC::helP+; Figure 1D). helP is required for ICEBs1 conjugation and functions primarily in donor cells
In order to determine the stage at which HelP functions in ICEBs1 replication, we tested whether helP is required for transfer of ICEBs1 into a recipient. Although ICEBs1 replication is not required for mating, the initial steps of nicking and unwinding of ICEBs1 DNA are common to both mating and replication [6], [15]. Wild type ICEBs1 had a mating efficiency of 3.7% (3.7 transconjugants per 100 donors) (Table 2, line 1), similar to previous results [9], [16]–[18]. In contrast, transfer of the ICEBs1 ΔhelP mutant was undetectable (Table 2, line 2). Expression of helP from a truncated ICEBs1 integrated into the chromosome at thrC (strain JT335; Figure 1) largely restored conjugation, indicating that the primary mating defect was due to loss of helP and not due to polarity on downstream genes (Table 2 line 3).
Although HelP primarily functions in the donor, it also appears to play a role in the recipient. We found that expression of helP in the recipient from the IPTG-inducible promoter Pspank(hy) increased the mating efficiency to 1.4%, from the 0.11% efficiency when helP was provided only in the donor. There was no detectable transfer of the ICEBs1 ΔhelP mutant unless helP was also expressed in the donor, and no increase in mating efficiency of wild type ICEBs1 when helP was also expressed in the recipient. We suspect that HelP is required for autonomous replication and increases the stability of ICEBs1 in recipient cells prior to its integration into the recipient chromosome.
Since HelP was required for both conjugation and replication of ICEBs1, we expected it to be involved in nicking or unwinding of ICEBs1 DNA. There was still nicking of the proper site in oriT in the absence of helP (data not shown), consistent with previous findings that NicK is the only ICEBs1-encoded protein needed for nicking of oriT [15]. Therefore we suspected that HelP was involved in the unwinding of ICEBs1 DNA by the host-encoded helicase PcrA, a DNA translocase that has very limited processivity as a helicase [19]. The solution structure of a HelP homologue, SAG0934 from Streptococcus agalactiae [20], which is identical to Orf22 of Tn916 from Enterococcus faecalis, indicates that HelP contains an oligonucleotide/oligosaccharide binding fold (OB-fold) that is present in many ssDNA binding proteins [21], consistent with a possible role in binding ICEBs1 ssDNA. Although HelP homologues and some Ssb proteins share the OB-fold, there appear to be no other significant sequence similarities between these proteins.
HelP associates with ICEBs1 oriT
Using crosslinking and immunoprecipitation (ChIP), we found that HelP was associated with ICEBs1 oriT in vivo (Figure 2). Following induction of ICEBs1 gene expression, protein and DNA were crosslinked with formaldehyde and HelP was immunoprecipitated with polyclonal anti-HelP antibodies. Preliminary experiments with DNA microarrays (ChIP-chip) indicated that HelP was strongly associated with the excised and replicating ICEBs1 DNA and that there was little or no specific association with the chromosome (data not shown). We then measured association of HelP with ICEBs1 DNA following ChIP using quantitative real time PCR (ChIP-PCR) with primers specific to ICEBs1 oriT and normalized to the amount of DNA from a chromosomal region (Materials and Methods). oriT DNA was enriched >500-fold in the anti-HelP immunoprecipitates (Figure 2A) indicating association of HelP with ICEBs1 oriT in vivo. The immunoprecipitation was specific to HelP as ICEBs1 oriT was not significantly enriched in a strain deleted for helP (2.4-fold relative association in ΔhelP mutant compared to >500-fold in wild type).
Fig. 2. HelP, PcrA, and Ssb-GFP association with ICEBs1 DNA. 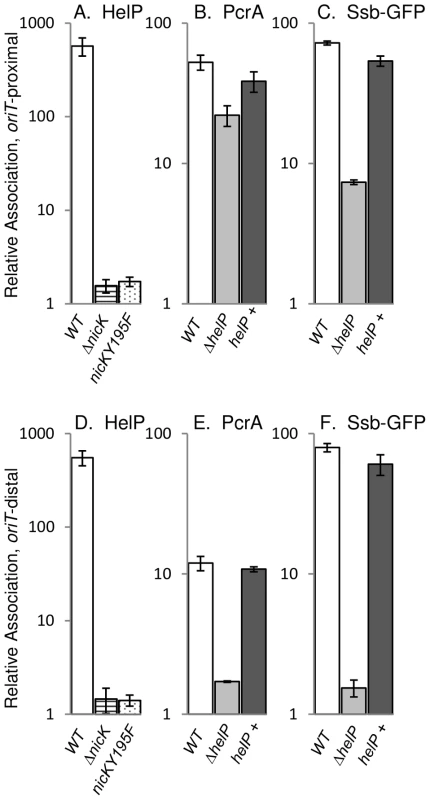
ICEBs1 gene expression was induced by overproduction of RapI from amyE::{Pspank(hy)-rapI spc} for one hour followed by DNA-protein crosslinking with formaldehyde. Association of HelP (A, D), PcrA (B, E), and Ssb-GFP (C, F) with DNA sequences near oriT (A–C) and conE (D–F) were measured by immunoprecipitation of lysates with polyclonal anti-HelP, anti-PcrA and anti-GFP antibodies respectively followed by qPCR (ChIP-PCR), relative to ydbT, adjacent to ICEBs1. When Ssb-GFP association was measured, strains contained the lacA::{PrpsF-ssb-GFPm tet} allele expressing GFP-tagged Ssb [6]. Strains included: wild type (JMA168 for HelP and PcrA; MMB834 for Ssb-GFP), white bars; ΔnicK (CAL306), horizontally striped bars; nicKY195F (JT340), dotted bars; ΔhelP (JT155 for HelP and PcrA; JT252 for Ssb-GFP), light grey bars; and ΔhelP, complemented with helP+ (JT335 for HelP and PcrA; JT398 for Ssb-GFP), dark grey bars. The c-Myc tag in the nicKY195F allele did not affect HelP association with conE which was unchanged in the NicK-Myc (JT308) control strain (421±96) compared to wild type (551±100) in three independent experiments. Error bars represent standard deviation. Association of HelP with ICEBs1 oriT depends on the ICEBs1 relaxase NicK
We found that in vivo, association of HelP with ICEBs1 oriT required the activity of the relaxase NicK. In a strain containing a nicK deletion, and a functional oriT [15], association of HelP with ICEBs1 was abolished (Figure 2A). To test if NicK nicking activity was required for association of HelP at oriT, we made a mutation in the catalytic site of NicK that changes the conserved tyrosine at position 195 to phenylalanine (nicKY195F). We also incorporated a C-terminal 3× c-Myc tag onto the wild type and Y195F mutant NicK proteins to use in ChIP experiments with monoclonal antibodies to c-Myc. Both NicKY195F-Myc NicK-Myc were associated with ICEBs1 oriT as determined by ChIP-PCR experiments (40±2-fold enrichment and 44±4-fold enrichment, respectively). As expected, there was no detectable nicking of oriT by NicKY195F-Myc (<0.05%±0.03% nicked oriT) whereas there was normal nicking of oriT by NicK-Myc (32%±3.7% nicked oriT), as determined by primer extension assays [15]. The NicK-Myc was also functional in conjugation (Materials and Methods).
We measured the association of HelP with ICEBs1 oriT in vivo in strains expressing either NicK-Myc or NicKY195F-Myc. As expected, HelP was associated with ICEBs1 oriT in the strain expressing functional NicK-Myc (data not shown). In contrast, HelP was not detectably associated with ICEBs1 oriT in the strain expressing NicKY195F-Myc (Figure 2A). These results indicate that the presence of NicK at oriT is not sufficient and that nicking of oriT is required for recruitment of HelP to oriT. We also found that HelP was associated with the rolling circle replicating plasmid pBS42 (data not shown), consistent with the notion that HelP does not require specific interaction with NicK.
HelP is not required for recruitment of the helicase PcrA to ICEBs1 oriT in vivo
The host-encoded helicase PcrA is required for both conjugation and replication of ICEBs1 [6]. Since HelP is also required for conjugation and replication of ICEBs1, we tested for effects of HelP on PcrA. We measured association of PcrA with ICEBs1 oriT by ChIP-PCR using polyclonal anti-PcrA antibodies and primers specific to oriT as described above. PcrA association with oriT was not significantly affected in a ΔhelP mutant, compared to wild type (Figure 2B). We conclude that association of PcrA with ICEBs1 oriT is not dependent on HelP.
In contrast, we found that association of PcrA with ICEBs1 oriT appeared to be largely, but not entirely, dependent on nicking of oriT by NicK. Enrichment of oriT in the PcrA immunoprecipitates was reduced but not abolished in the nicKY195F-myc mutant (approximately 6-fold enrichment in nicKY195F-myc vs 65–70-fold enrichment with nicK-myc). Association of PcrA with ICEBs1 DNA in the nicK catalytic site mutant indicates that there might be interactions between ICEBs1 NicK and the host PcrA, as there are with other replicative rolling circle relaxases and PcrA [22], [23]. The immunoprecipitation was specific for PcrA; in a pcrA recF double mutant [6], ICEBs1 sequences were not significantly enriched (approximately 1.1-fold relative enrichment of oriT compared to 12.5-fold enrichment in the recF parent). (Loss of recF suppresses the lethality caused by loss of pcrA [24], [25]).
HelP is required for progression of the helicase PcrA through ICEBs1 DNA in vivo
Unwinding and replication of ICEBs1 DNA by PcrA proceeds unidirectionally from the nicked oriT [6]. Replication of ICEBs1 is also accompanied by association of the host-encoded single stranded DNA binding protein (Ssb) to the ICEBs1 DNA [6]. To examine the location and role of HelP during unwinding and replication of ICEBs1 DNA from the nicked oriT, we compared association of HelP, PcrA and Ssb with an oriT-proximal (Figure 2A–2C) to an oriT-distal region (Figure 2D–2E).
We found that although HelP was not needed for the initial recruitment of PcrA to oriT (Figure 2B), it was required for the association of PcrA with oriT-distal regions (Figure 2E). Association of HelP and PcrA with the oriT-distal (conE) region of ICEBs1 DNA was readily detectable in wild type (helP+) cells (Figure 2D, 2E). In contrast, association of PcrA with ICEBs1 DNA in the ΔhelP mutant was greatly reduced in this region (Figure 2E). This reduction was due to loss of helP and not an unexpected secondary effect due to polarity or alterations in the DNA because association was restored when helP was expressed in trans from an ectopic locus (Figure 2E). Together, our results indicate that HelP is not needed for the initial association of PcrA with ICEBs1 oriT, but that HelP is needed for PcrA to become associated with distal regions, perhaps by affecting helicase processivity.
We monitored association of Ssb with ICEBs1 DNA (indicative of unwound ssDNA) using an Ssb-GFP fusion and ChIP-PCR with anti-GFP antibodies, essentially as described previously [6]. We found that Ssb-GFP was associated with both the oriT-proximal and oriT-distal (conE) region of ICEBs1 (Figure 2C, 2E). In contrast, there was little or no association of Ssb-GFP with the oriT-distal region in the helP mutant (Figure 2E), and association was reduced, but still appreciable, in the oriT region (Figure 2C). Association of Ssb with ICEBs1 DNA was restored by expression of helP from an ectopic locus (Figure 2C, 2E), indicating that the defect in the helP mutant was due to loss of helP and not some unexpected effect. Based on these results, we conclude that HelP is required for both PcrA and Ssb to associate with oriT-distal sequences in ICEBs1, and that HelP is likely needed for the processive unwinding of ICEBs1 DNA. This function would be sufficient to explain the requirement for helP in both conjugation and replication of ICEBs1.
HelP stimulates helicase activity of the PcrA homologue UvrD in vitro
We wished to test directly the ability of HelP to facilitate unwinding of duplex DNA by PcrA. Our many different preparations of B. subtilis PcrA were of low concentration and rapidly lost helicase activity. Since most structural and biochemical analyses of PcrA have been done with a homologue from another organism {reviewed in [26]}, we decided to test for effects of HelP on UvrD from E. coli. UvrD is a well-studied homologue of PcrA and has 41% identity with B. subtilis PcrA. Like PcrA, UvrD is required for replication of several rolling circle replicating plasmids [27]. We found that expression of E. coli uvrD from the IPTG-inducible promoter Pspank (Pspank-uvrD) in B. subtilis suppressed the lethality caused by loss of pcrA. This suppression occurred in both the absence and the presence of IPTG, indicating that, in the absence of induction, expression from Pspank was leaky and sufficient levels of UvrD were produced. In addition, UvrD was able to support replication and conjugation of ICEBs1 nearly as well as PcrA (Table 3). In cells missing pcrA but expressing uvrD, the mating efficiency was approximately 1% (transconjugants per donor) and ICEBs1 was capable of replication and had a copy number of 2–3 (Table 3 line 3). Based on these results, we conclude that E. coli uvrD can replace pcrA in B. subtilis and provides the functions of PcrA needed for cell viability and those needed for ICEBs1 conjugation and replication.
Tab. 3. UvrD permits ICEBs1 replication in a pcrA-defective strain. 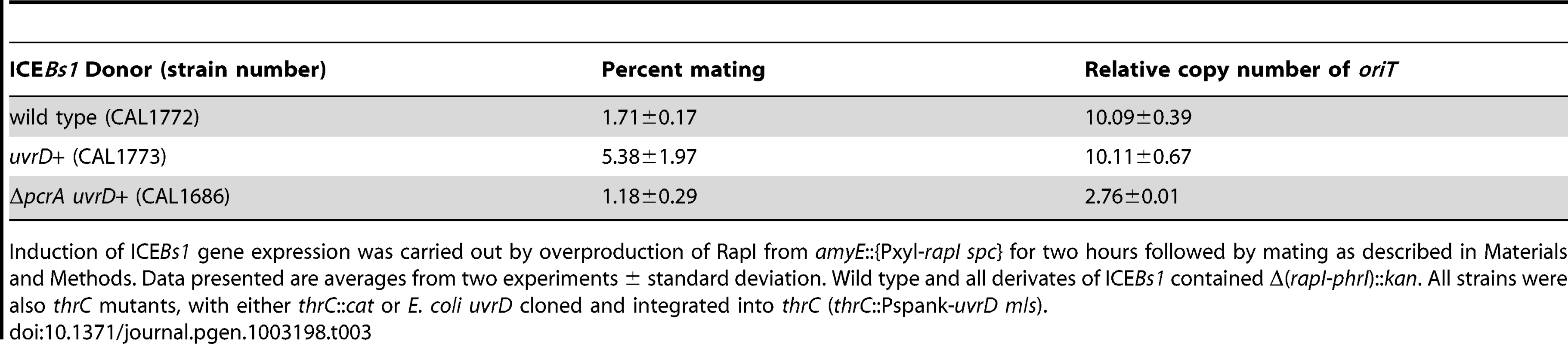
Induction of ICEBs1 gene expression was carried out by overproduction of RapI from amyE::{Pxyl-rapI spc} for two hours followed by mating as described in Materials and Methods. Data presented are averages from two experiments ± standard deviation. Wild type and all derivates of ICEBs1 contained Δ(rapI-phrI)::kan. All strains were also thrC mutants, with either thrC::cat or E. coli uvrD cloned and integrated into thrC (thrC::Pspank-uvrD mls). We found that HelP stimulated the ability of UvrD to unwind a partial duplex DNA substrate in vitro. We purified hexahistidine-tagged UvrD (his-UvrD) and HelP (his-HelP) and used two different partial duplex DNA substrates to monitor unwinding (Materials and Methods). The substrates had either a small (22 nucleotides) or large (81 nucleotides) fluorophore-labeled oligonucleotide hybridized to single-stranded M13mp8 DNA. Helicase activity was measured by release of the labeled oligonucleotide, detected by gel electrophoresis and fluorometry. HelP alone had no effect on the partial duplex substrate. In both cases, <0.5% of the substrate duplex was unwound after 45 minutes. UvrD alone was able to unwind the 22 bp duplex: approximately 20% of the substrate was unwound after 45 minutes (Figure 3A). The addition of HelP increased the amount of substrate that was unwound to approximately 40% (Figure 3A). In contrast, there was little or no release of the large (81-mer) oligonucleotide from the partial duplex by UvrD alone (<2% unwound after 45 minutes). In the presence of HelP and UvrD, >25% of the 81 bp duplex substrate was unwound by 45 minutes (Figure 3B). Based on the increase in unwinding and the difference in stimulation between the 22 bp and 81 bp duplex substrates, we conclude that HelP stimulates the ability of UvrD, and likely PcrA, to processively unwind duplex DNA. In addition, since there is no relaxase in this assay, relaxase, and specifically NicK, is not required, at least in vitro, for the function of HelP.
Fig. 3. DNA unwinding by UvrD is stimulated by HelP. 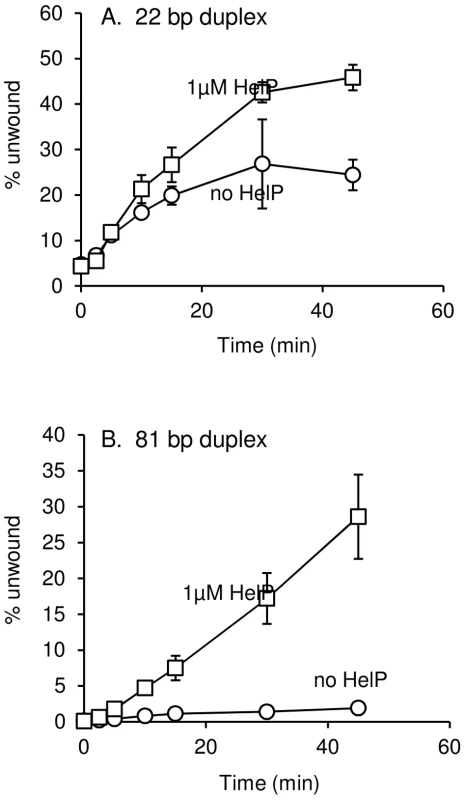
Substrates for unwinding were M13mp18 ssDNA annealed to either a 22 (A) or 81 (B) complementary fluorophore-labeled oligonucleotide. Data presented are the averages of three independent experiments ± standard deviation. Substrate unwinding was undetectable in the presence of HelP alone (<0.5% substrate unwound after 45 minutes) for both substrates. These controls were done with only the 45 minute time point. Conservation of helP in many ICEs
The well characterized ICE Tn916 encodes two HelP homologues, Orf22 and Orf23. Comparison of HelP to each of these indicates that HelP is more similar to each protein than they are to each other (Figure 4). In addition, Orf23 is about 20 amino acids shorter at the C-terminus than Orf22 and HelP.
Fig. 4. Comparison of HelP to Orf22 and Orf23 of Tn916. 
HelP (middle sequence), Orf22 of Tn916 (top sequence) and Orf23 of Tn916 (bottom sequence) were aligned using Clustal X. Markings above and below the HelP sequence indicate which amino acids are identical (asterisk), highly similar (colon) or weakly similar (period) to Tn916 Orf22 (marks above) and Tn916 Orf23 (marks below). Markings for the pairwise comparison of Orf22 to Orf23 are shown below the Tn916 Orf23 sequence. Needleman-Wunsch alignment scores indicate that HelP is more similar to Orf22 and Orf23 (N-W scores = 142 and 104, respectively) than Orf22 is to Orf23 (N-W score = 31). ICEBs1 HelP is 126 amino acids, Tn916 Orf22 is 128 amino acids, and Tn916 Orf23 is 104 amino acids. The gene organization in ICEBs1 near helP and in Tn916 near orf22 and orf23 is similar (Figure 5). helP and its homologues are usually grouped with two other genes: 1) a relaxase gene, nicK in ICEBs1 and orf20 in Tn916, and 2) a gene encoding the predicted coupling protein, conQ in ICEBs1 [18] and orf21 in Tn916 (Figure 5). The coupling protein targets the relaxosome complex that is linked to ssDNA to the mating pore [28]. This genetic arrangement reflects a functional relationship as the relaxase and likely HelP are part of the relaxosome that interacts with the cognate coupling protein. When present, the majority of helP homologues are found in pairs, although ICEBs1 has only one helP.
Fig. 5. Organization of genes for HelP, ConQ (coupling protein) and NicK homologues in ICEBs1 and Tn916. 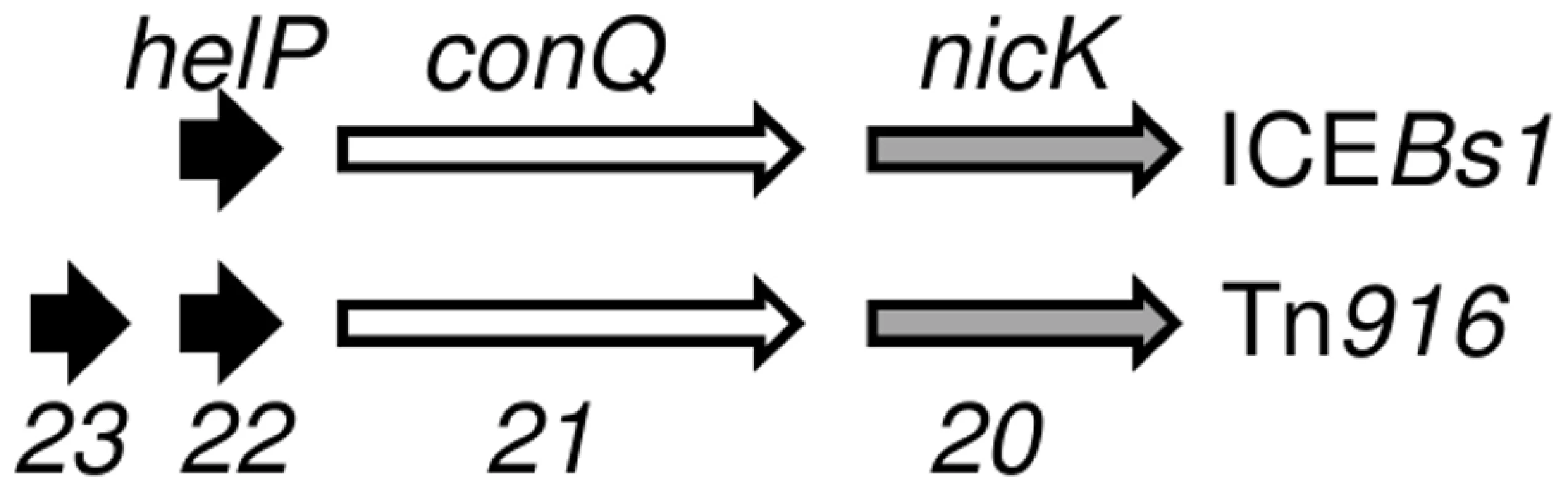
Schematic diagram showing the organization of genes encoding HelP, ConQ and NicK in ICEBs1 and their homologues Orf23, Orf22, Orf21, and Orf20 in Tn916. Most (>40) of the 72 ICEs in the ICEberg database that have helP homologues have this consecutive (helP)-helP-conQ-nicK configuration. Some ICEs have the same gene order but have an additional one or two genes located between the helP and conQ and/or between conQ and nicK. Not shown is a different gene organization found in ICEs related to ICE6013 in which the conQ is separate from helP and nicK. helP homologues are found in at least 72 ICEs or putative ICEs, primarily in firmicutes [29]. Phylogenic analysis revealed that these HelP homologues fall into seven clades (Figure 6). When an ICE encodes two HelP homologues, each one is in a separate clade: one clade contains the longer HelP homologue and other clade contains the shorter HelP homologue. For example the clade labeled “Tn916 Orf22” contains most of the longer HelP homologues and the clade labeled “Tn916 Orf23” contains most of the shorter HelP homologues. ICEBs1 HelP is similar in size to the longer Orf22, but appears to be almost equidistant from the Orf22 and Orf23 clades (Figure 6). Together, our results indicate that HelP proteins are encoded by many different ICEs from Firmicutes. If the function of these homologues is conserved, then HelP proteins act as helicase processivity factors for many ICEs and these ICEs likely undergo autonomous rolling circle replication.
Fig. 6. Phylogenic tree of HelP homologues. 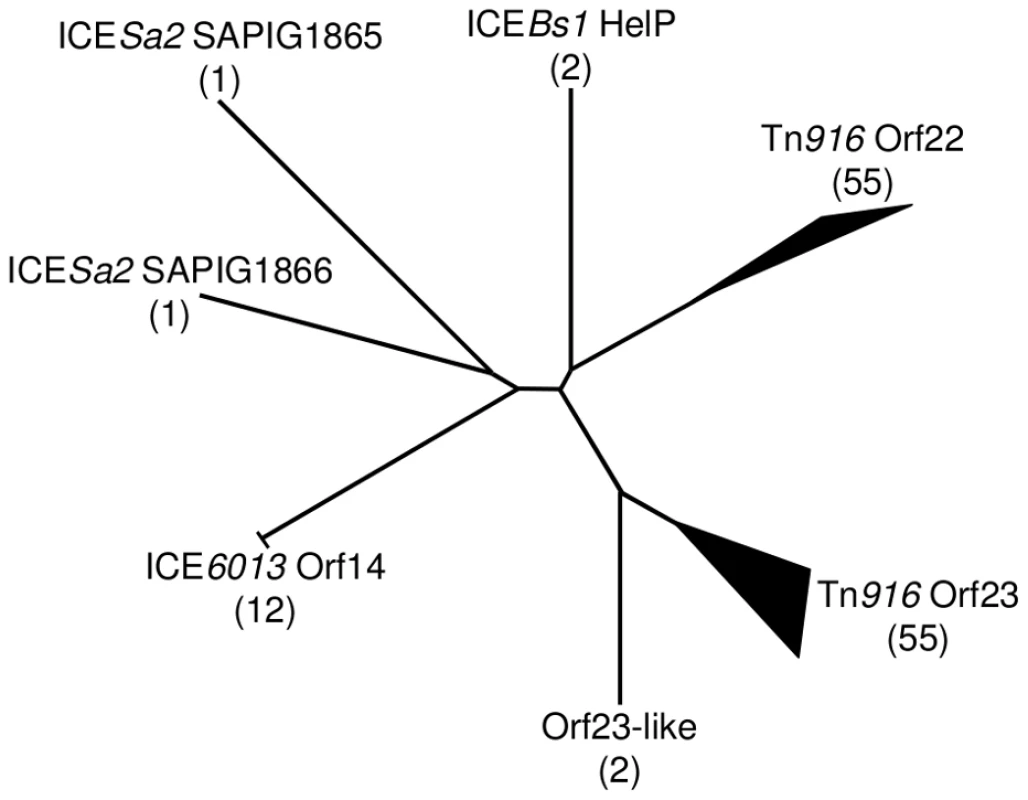
Using the ICEberg database and search tools (HMMER3 and WU-BLAST2) (http://db-mml.sjtu.edu.cn/ICEberg/), we identified 128 HelP homologues in 72 ICEs. The homologues were analyzed using CLUSTAL X and grouped into clades if they were within a distance of <0.22. The diagram is of an unrooted tree with the number of homologues in each clade shown in parentheses. Branch lengths indicate relative phylogenic distances. The width and length of branch tips indicate the number of homologues and the relative phylogenic distance between homologues in the clade, respectively. Of the seven clades, six are named for a representative ICE and its HelP homologue and one is named “Orf23-like” to reflect its close relationship to the Tn916 Orf23 clade. Twelve homologues in the ICE6013 Orf14 clade have the identical 106 amino acid sequence. The two homologues in the ICEBs1 HelP clade only differ by 5 of 126 amino acids. The two homologues in the Orf23-like clade only differ by 7 of 108 amino acids. Sixteen ICEs encode a single homologue of HelP. These sixteen homologues are from three clades - ICEBs1 HelP (2), ICE6013 Orf14 (12) and Tn916 Orf23 (2). The remaining 56 ICEs encode pairs of HelP homologues. The members of each pair of homologues fall into separate clades as exemplified by Tn916 Orf22 (128 aa) and Tn916 Orf23 (104 aa), and ICESa2 SAPIG1865 (141 aa) and ICESa2 SAPIG1866 (107 aa). Discussion
We found that the ICEBs1 helP gene product stimulates unwinding of ICEBs1 DNA by the helicase PcrA in vivo. In vitro, HelP stimulated processivity of the PcrA-like helicase UvrD from E. coli. Our findings indicate that HelP is a helicase processivity factor that is required for conjugation and replication of ICEBs1. HelP may represent a new family of helicase processivity factors as helP homologues are found in many other known and putative ICEs. Together, our results indicate that the order of events leading to ICEBs1 replication and conjugation include: 1) nicking by the ICEBs1-encoded relaxase NicK; 2) association of HelP and the host encoded helicase PcrA, independently of each other, with the nicked ICEBs1 DNA template; and 3) unwinding of ICEBs1 dsDNA by PcrA and association of the host Ssb with the ICEBs1 ssDNA (Figure 7).
Fig. 7. Model for association of the relaxase, helicase, HelP, and Ssb with ICEBs1 DNA. 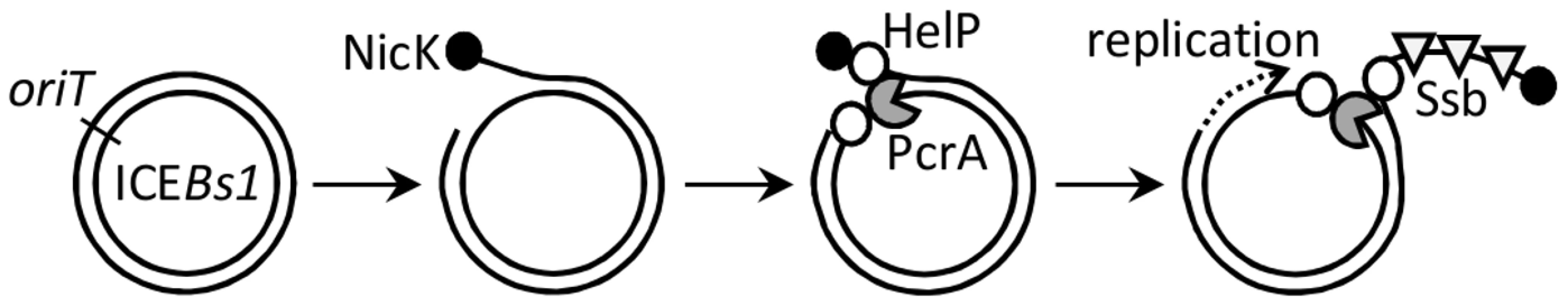
Following excision from the chromosome (not shown), the double-stranded circular ICEBs1 DNA is nicked at oriT by the relaxase NicK (filled circle). By analogy to related relaxases, NicK likely becomes covalently attached to the 5′ end of the nicked DNA on the strand that is transferred during conjugation. HelP (open circles) and the host-encoded helicase PcrA (gray packman) associate with the nicked ICEBs1 at oriT. HelP facilitates processive unwinding of ICEBs1 by PcrA and subsequent association of the host-encoded Ssb (open triangles). Unwinding of ICEBs1 by PcrA and HelP is required for replication and conjugation of ICEBs1. HelP is depicted as binding to both single strands of ICEBs1 DNA adjacent to PcrA, although it is possible that HelP associates only with one of the two single strands. DNA translocase and helicase activities of PcrA-family proteins
PcrA and its well-characterized homologues in E. coli, UvrD and Rep, are members of superfamily 1 (SF1) of non-hexameric helicases {reviewed in [13], [26]}. These proteins are highly processive 3′-5′ ssDNA translocases that bind to and move along strands of ssDNA. They are also weak 3′-5′ helicases that bind and destabilize dsDNA. They are involved in multiple cellular processes, including several types of DNA repair, replication restart, and clearing recombination proteins from DNA {reviewed in [26], [30]}. Strains lacking SF1 helicases are usually nonviable [24], [31], presumably due to loss of the DNA translocase activity of these proteins [31]–[33].
Although PcrA, Rep and UvrD are efficient and processive DNA translocases, they have very poor helicase activity on their own [19], [34], [35]. Helicase activity requires oligomerization [36]–[38], and these helicases interact with accessory factors that stimulate activity. For example, the DNA mismatch repair protein MutL facilitates loading and processivity of UvrD in E. coli [39] and the double-strand break repair protein Ku interacts with and stimulates processivity of UvrD1 in Mycobacterium smegmatis [40]. PcrA processivity is stimulated in vitro by YxaL from B. subtilis [41], although the role of YxaL in vivo is not known.
Plasmid - and phage-encoded helicase processivity factors
Although they are poorly processive, PcrA, UvrD, and Rep are used by many plasmids and phages that undergo rolling-circle replication. In E. coli, UvrD is needed for rolling circle replication of some plasmids [27], [42] and Rep is used for replication of several ssDNA phages {reviewed in [11]}. In some Gram positive bacteria, PcrA is used for replication of many different rolling circle plasmids [12], [31], [32]. In addition, PcrA is required for unwinding of ICEBs1 DNA for both conjugation and replication [6].
For rolling circle replicating elements that use a host-encoded SF1-type helicase, efficient unwinding of duplex DNA is often stimulated by interaction with the element-encoded replication relaxase. Relaxases introduce a nick into dsDNA and mark the site for recruitment of the helicase for unwinding and replication of the plasmid or phage DNA. Replicative relaxases of ssDNA phages from E. coli, including gpA (cisA) from øX174 and the product of gene II of the f1 family of phages, increase Rep helicase processivity {reviewed in [11]}. The replicative relaxases of rolling circle replicating plasmids pT181 and the related pC221, RepC and RepD, respectively, appear to interact with and stimulate PcrA [23], [43]–[47]. In the case of RepD, this stimulation is thought to occur by increasing the affinity of PcrA for its DNA substrate and potentially by decreasing its rate of dissociation, rather than by altering its kinetic properties [46], [48].
HelP, a conserved helicase processivity factor
As far as we are aware, all known examples of plasmid or phage-encoded helicase-stimulating proteins are relaxases. By analogy to the related plasmid relaxases, we expected that the only ICEBs1 product needed for ICEBs1 rolling circle replication would be its relaxase NicK. However, we found that the ICEBs1 gene product HelP is a helicase processivity factor that is required, in addition to NicK, for replication and conjugation of ICEBs1. We do not yet know how HelP stimulates helicase activity, but it appears to act at a step after association of PcrA with ICEBs1 oriT DNA. HelP could stimulate dimerization of PcrA since it is the dimer and not the monomer that has helicase activity [19]. HelP could also decrease the rate of dissociation of PcrA from DNA during unwinding, analogous to the activity demonstrated for RepD [46], [48]. HelP could accomplish one or a combination of these stimulatory functions by direct protein-protein contact with PcrA. Alternatively, HelP could remodel the DNA substrate during unwinding, promoting helicase activity.
helP homologues are found primarily in firmicutes. There appear to be >300 homologues in the non-redundant protein database, and at least 128 of these are found in known or putative ICEs that are included in the ICEberg database [29], and we suspect that most of the others are in unrecognized ICEs. Although ICEBs1 has only one helP, most, but not all, helP homologues are found in pairs, with one member of the pair approximately 20 amino acids longer than the other. It is possible that each member of a HelP pair stimulates processivity of a dissimilar range of helicases, thereby broadening the host range of the mobile element. If the function of HelP homologues is conserved, then the ICEs with helP homologues likely undergo autonomous replication and that HelP proteins are important for stability and transfer of many ICEs.
Other strategies for DNA unwinding of extrachromosomal elements
In contrast to the many ICEs and plasmids that utilize a host-encoded SF1-type helicase for replication and transfer and rely on an accessory factor to facilitate unwinding, some ICEs and plasmids encode their own SF1-type helicase. PcrA-like helicase domains are found attached to some plasmid relaxases. For example, TraI of the E. coli F plasmid and TrwC of R388, have a C-terminal PcrA-like helicase domain that is required for conjugation [49]–[51]. The helicase activity is highly processive, potentially because it is tethered to the relaxosome {reviewed in [52]}. In addition, many known and putative ICEs encode discrete PcrA-like helicases [29]. It is not known if they are processive or require accessory proteins, although some of those ICEs also contain a helP homologue. An element that encodes its own helicase might have a broader host range by avoiding reliance on a host-encoded helicase. That HelP homologues, relaxases that also function as processivity factors, and PcrA homologues are encoded on conjugative elements indicates that the need for DNA unwinding can be met in different ways.
Materials and Methods
Strains and alleles
All B. subtilis strains (Table 4) are derivatives of JH642 (trpC2 pheA1) and were constructed by natural transformation. Strains are either cured of ICEBs1 (designated ICEBs10) or contain ICEBs1 marked with the Δ(rapI-phrI)342::kan allele to monitor conjugative transfer [9]. ICEBs1 gene expression was induced by overproduction of rapI from amyE::{(Pspank(hy)-rapI) spc}, amyE::{(Pxyl-rapI) spc}, or lacA::{(Pxyl-rapI) tet}. Strains used as recipients in mating experiments contained the streptomycin resistance allele (str-84) [9]. Other mutations that were previously described include: the deletion-insertion ΔpcrA1021::cat [6]; thrC1165::cat [6], used as a control for threonine auxotrophy and chloramphenicol resistance; and thrC329::{(Pxis-nicK) mls} [15], used for ectopic expression of nicK.
helP
ΔhelP155 is an unmarked 413-bp deletion that removes the entire coding sequence and the 35 bp helP-ydcQ intergenic region (Figure 1), constructed using the same method as for the ΔnicK306 allele [15]. Complementation of ΔhelP155 was tested using a truncated ICEBs1 derivative integrated at thrC, thrC229::{(ICEBs1-1637 ΔconQ-attR::cat) mls} that is missing all of the ICEBs1 genes downstream from helP (Figure 1D), and contains helP and all the upstream ICEBs1 genes [18]. The control that does not complement ΔhelP was essentially the same ICEBs1 insertion at thrC, but with a deletion that removes helP (Figure 1C), thrC229::{(ICEBs1-1635 ΔhelP-attR::cat) mls}.
Expression of helP from LacI-repressible-IPTG-inducible promoter Pspank(hy) [53] in B. subtilis was from a fusion at thrC, thrC148::{(Pspank(hy)-helP) mls}. This was made by amplifying helP by PCR from B. subtilis genomic DNA, digesting with Nhe1 and ligating into Nhe1-cut pCAL215 [16] to give pCAL890. The Pspank promoter of pCAL890 was altered to Pspank(hy) by Quikchange mutagenesis to give the plasmid pJT146 which was then used to integrate Pspank(hy)-helP mls into the thrC locus by double-crossover recombination.
uvrD
E. coli uvrD was expressed in B. subtilis from Pspank inserted at thrC, thrC1642::{(Pspank-uvrD) mls}. uvrD was amplified from an E. coli MC1061-derived strain and then inserted between the HindIII and SphI sites of pCAL215 [16] using isothermal assembly [54].
pcrA is essential in B. subtilis [24], [31]. The presence of thrC1642::{(Pspank-uvrD) mls} suppressed the lethality caused by loss of pcrA. That is, ΔpcrA::cat could be transformed into competent cells containing the Pspank-uvrD, but could not be transformed into cells without uvrD. Growth of the Pspank-uvrD ΔpcrA mutant was independent of IPTG, indicating that the leaky (uninduced) expression from Pspank-uvrD was sufficient to compensate for the absence of pcrA.
nicK
The nicK-myc allele was constructed by allelic replacement using essentially the same method as for the ΔhelP155 allele. An approximately 2.1 kb DNA insert containing nicK, a C-terminal 3× c-myc tag and 897 bp of ICEBs1 sequence downstream of nicK were cloned into the plasmid pCAL1422 by isothermal assembly [54] to give the plasmid pJT296. pCAL1422 contains cat (chloramphenicol-resistance in B. subtilis) and E. coli lacZ under control of the constitutive promoter Ppen [55]. The tyrosine codon at position 195 of nicK-myc in pJT296 was changed to a phenylalanine codon, nicKY195F-myc, by quick-change mutagenesis (Stratagene), giving plasmid pJT318. pJT296 and pJT318 were used to replace nicK on ICEBs1 with nicK-myc and nicKY195F-myc, respectively, by first integrating by single crossover and then screening for loss of the plasmid by virtue of loss of lacZ, and then testing by PCR for introduction of the indicated allele, essentially as described [15].
Wild type NicK fused to the Myc-tag (NicK-Myc) was active as judged by normal conjugation efficiencies (approximately 1.2% transconjugants per donor). In contrast, the nicKY195F-myc mutant was defective in conjugation (<0.00008% transconjugants per donor).
Plasmids
Plasmids pCAL1255 (oriT, Pxis-helP-nicK), pCAL1260 (oriT, Pxis-helP), and pJT151 (oriT, Pxis-nicK) contain DNA segments from ICEBs1 inserted into the BamHI restriction site of pUS19, a spectinomycin-resistant derivative of pUC19 [14]. In addition to conferring resistance to spectinomycin in B. subtilis, each plasmid contains ICEBs1 oriT.
pCAL1255 (oriT, Pxis-helP-nicK) contains the xis promoter (Pxis) driving transcription of helP and nicK. The Pxis-helP-nicK insertion is comprised of three non-contiguous segments of ICEBs1: 1) a 527 bp segment containing 254 bp of the 5′-end of immR and the entire 273 bp intergenic region between immR and xis; 2) a 419 bp segment that contains helP and extends 35 bp downstream of the helP stop codon; and 3) a 1076 bp segment containing nicK (and oriT) and extending 17 bp downstream of the nicK stop codon. The junctions between the DNA segments were designed to allow translation initiation of helP from the xis ribosome binding site and translation initiation of nicK from the conQ ribosome binding site, as the start codons of helP and of nicK were placed the same distance downstream of the Pxis and the helP-conQ intergenic regions as the native start codons of xis and conQ, respectively. Transcription of the Pxis-helP-nicK insertion is in the same orientation as the spectinomycin - and ampicillin-resistance genes on the vector backbone.
pCAL1260 (oriT, Pxis-helP) contains the same Pxis-helP-nicK insertion present in pCAL1255, but with an additional T inserted between the 5th and 6th codon of nicK. The single base insertion in pCAL1260 leads to premature termination after the 10th codon of the nicK ORF. pJT151 is essentially pCAL1255 (oriT, Pxis-nicK) but with the entire helP coding sequence deleted.
N-terminal hexahistidine tagged HelP (his-HelP) and UvrD (his-UvrD) were overproduced in and purified from E. coli. helP was amplified by PCR from B. subtilis (strain JH642) chromosomal DNA, digested with BamHI and HindIII and ligated into the same sites of the T7-expression vector pET28b (Novagen) to give plasmid pCAL1297. uvrD was amplified from E. coli strain MC1061and cloned, by isothermal assembly, into the XbaI and BamHI sites of pET28b to give plasmid pJT370.
Conjugation experiments
For conjugation experiments, all donor strains contained ICEBs1 with the Δ(rapI-phrI)342::kan allele and ICEBs1 gene expression was induced by overproduction of the regulatory protein RapI from an ectopic locus [9]. When donors carried the Pspank(hy)-rapI allele, all strains were grown in rich medium and induction was for one hour with 1 mM IPTG. When the Pxyl-rapI was used, strains were grown in minimal S750 medium containing 1% arabinose and induction was for 2 hours after the addition of 1% xylose [56]. Recipients were streptomycin resistant (str-84). Donors and recipients were mixed in a 1∶1 ratio, filtered onto nitrocellulose membranes and incubated on agar containing minimal salts for 3 hours, essentially as described [9]. The mating efficiency was determined as the number of colony forming units (CFU) of streptomycin - and kanamycin - resistant transconjugants per CFU of the donor.
Chromatin immunoprecipitation, copy number, and qPCR
Association of HelP, PcrA, NicK-Myc and Ssb-GFP with ICEBs1 DNA was measured by chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) essentially as described [6], [57]. Polyclonal antibodies from rabbits (Covance) were used to precipitate HelP, PcrA and Ssb-GFP and monoclonal antibodies to c-Myc (Invitrogen) were used to precipitate NicK-Myc.
qPCR was used to measure the relative amount of DNA in immunoprecipitates and to measure relative plasmid copy number. Primers to the ICEBs1 nicK/oriT region, designated oriT, were CLO280, 5′-TGGCTACGTT GGCACGTATG-3′, and CLO281, 5′-AATTGACGGC AACCTTGACC-3′. Primers to ICEBs1 conE, approximately 6 kb downstream from oriT, were oMMB238, 5′-TGATGGTTCAAATCCTGCATTGTCAC-3′, and oMMB239, 5′-GAACTTACCT AGTGCAAACATGAC-3′.
Plasmid copy number was determined using primers oJT168, 5′-GTGGAATCAT CCTCCCAAAC-3′, and oJT169, 5′-AATGGCTCTT CTCACATCAG-3′, that are specific to spcE found on the plasmids and not in the chromosome.
Values obtained in ChIP-PCR and plasmid copy number experiments were normalized to the chromosomal locus ydbT, approximately 15 kb upstream of the chromosomal attachment site for ICEBs1, with primers CLO284, 5′-CTTCCGCACA TGCTCCGAAC-3′ and CLO285, 5′-TCGGCAGCAG GATCACTGAC-3′.
Protein expression and purification
his-HelP and his-UvrD were purified using similar conditions. Expression strains were grown in 2 liters of 2×YT medium supplemented with 0.4% glycerol, 20 mM sodium phosphate buffer pH 7.0, and 10 mM MgSO4 at 30°C until they reached an OD600 of 0.8. Protein expression was induced by the addition of 0.2% arabinose and 1 mM IPTG followed by incubation for 3 hours. Cells were harvested by centrifugation, washed with 100 ml of phosphate-buffered saline, resuspended in 25 ml lysis buffer {50 mM Tris-HCl ph 8.0, 0.3 M NaCl, 10 mM imidazole, 1 mg/ml lysozyme, 5 µg/ml DNase I and 1× CellLytic Express (Sigma-Aldrich)} and lysed by incubation at room temperature with gentle inversions. The lysate was cleared by centrifugation at 15,000× g and the cleared lysate was applied to 1 ml Ni-NTA agarose resin, washed with Tris-NaCl (50 mM Tris-HCl pH 8.0, 0.3 M NaCl) containing 10 and 20 mM imidazole and eluted with the same buffer containing 250 mM imidazole. The eluted protein was dialyzed against 10 mM MOPS, pH 7.5, 200 mM NaCl and 1 mM tris(2-carboxyethyl)phosphine. his-UvrD was approximately 93% pure and his-HelP was approximately 97% pure.
Helicase assays
Helicase activity was measured using two different partial-duplex DNA substrates, designed by analogy to previously described templates [41]. The substrates were generated by annealing M13mp18 circular ssDNA (Affymetrix) with 1) a 22-bp oligonucleotide (oJT276, 5′-ACTCTAGAGGA TCCCCGGGTAC-3′), or 2) an 81-bp oligonucleotide (oJT278, 5′-GGCCAGTGCCA AGCTTGCATG CCTGCAGGTC GACTCTAGAG GATCCCCGGG TACCGAGCTC GAATTCGTAA TCATGGTCAT-3′). Each oligonucleotide was labeled on the 5′-end with an infrared fluorescent dye, IRDye800 for oJT276 and TYE705 for oJT278 (IDT).
Helicase assays were performed in 200 µl reactions at 37°C. Mixtures containing 0.5 nM DNA substrate and 0 or 1 µM his-HelP, in 20 mM MOPS, pH 7.5, 200 mM NaCl, 15 mM MgCl2, 2.5 mM ATP, 1 mM tris(2-carboxyethyl)phosphine were preincubated at 37°C for 10 min, and started by the addition of 25 nM his-UvrD. 20 µl aliquots were withdrawn at regular intervals and added to 5 µl stop buffer (5% Ficoll, 15% glycerol, 0.12% Orange G, 1% SDS and 50 mM sodium EDTA). Samples (10 µl) were analyzed by electrophoresis on a 15% TBE-polyacrylamide gel containing 2.5% glycerol followed by visualization and quantitation using the Odyssey Infrared imaging system (Li-Cor).
Identification and analysis of HelP homologues
We identified 128 homologues of ICEBs1 helP (including helP) in 72 bacterial ICEs using the HMMER3 and WU-BLAST2 search tools in ICEberg (http://db-mml.sjtu.edu.cn/ICEberg/), the web-based resource for bacterial ICEs . The ICEberg database (last updated on August 14, 2011) contains sequence information for 431 known and putative ICEs. All 130 homologues were aligned using ClustalX 2.1 [58] using multiple-alignment mode with default parameters. The neighbor-joining method was used to generate the phylogenic tree from the ClustalX PHYLIP output file at the Interactive Tree of Life (http://itol.embl.de/index.shtml) [59]. Seven clades were defined by grouping together homologues that had an average distance of <0.22.
An additional HMMER search [60] with default search settings (hmmer.janelia.org) identified >300 HelP homologues in the non-redundant protein database. All but five of these were in Firmicutes. The exceptions were one homologue in a Mycoplasma, Ureaplasma urealyticum, two in an uncultured bacterium MID12, and two homologues in Klebsiella pneumoniae which carries Tn916 (usually associated with Firmicutes) on the composite ICE Tn6009.
Global sequence similarities between HelP and the two HelP homologues from Tn916 were analyzed by the Needleman-Wunsch alignment method [61].
Zdroje
1. WozniakRA, WaldorMK (2010) Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8 : 552–563.
2. GuglielminiJ, QuintaisL, Garcillan-BarciaMP, de la CruzF, RochaEP (2011) The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7: e1002222 doi:10.1371/journal.pgen.1002222.
3. Alvarez-MartinezCE, ChristiePJ (2009) Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73 : 775–808.
4. ChristiePJ, VogelJP (2000) Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol 8 : 354–360.
5. AbajyMY, KopecJ, SchiwonK, BurzynskiM, DoringM, et al. (2007) A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in gram-positive bacteria. J Bacteriol 189 : 2487–2496.
6. LeeCA, BabicA, GrossmanAD (2010) Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol 75 : 268–279.
7. SitkiewiczI, GreenNM, GuoN, MereghettiL, MusserJM (2011) Lateral gene transfer of streptococcal ICE element RD2 (region of difference 2) encoding secreted proteins. BMC Microbiol 11 : 65.
8. CarraroN, LibanteV, MorelC, DecarisB, Charron-BourgoinF, et al. (2011) Differential regulation of two closely related integrative and conjugative elements from Streptococcus thermophilus. BMC Microbiol 11 : 238.
9. AuchtungJM, LeeCA, MonsonRE, LehmanAP, GrossmanAD (2005) Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A 102 : 12554–12559.
10. BurrusV, PavlovicG, DecarisB, GuedonG (2002) The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48 : 77–97.
11. BaasPD (1985) DNA replication of single-stranded Escherichia coli DNA phages. Biochim Biophys Acta 825 : 111–139.
12. KhanSA (2005) Plasmid rolling-circle replication: highlights of two decades of research. Plasmid 53 : 126–136.
13. LohmanTM, TomkoEJ, WuCG (2008) Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol 9 : 391–401.
14. BensonAK, HaldenwangWG (1993) Regulation of sigma-B levels and activity in Bacillus subtilis. J Bacteriol 175 : 2347–2356.
15. LeeCA, GrossmanAD (2007) Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol 189 : 7254–7261.
16. AuchtungJM, LeeCA, GarrisonKL, GrossmanAD (2007) Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol Microbiol 64 : 1515–1528.
17. BerkmenMB, LeeCA, LovedayEK, GrossmanAD (2010) Polar positioning of a conjugation protein from the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol 192 : 38–45.
18. LeeCA, ThomasJ, GrossmanAD (2012) The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J Bacteriol 194 : 3165–3172.
19. Niedziela-MajkaA, ChesnikMA, TomkoEJ, LohmanTM (2007) Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J Biol Chem 282 : 27076–27085.
20. Aramini J, Rossi P, Zhao L, Foote E, Jiang M, et al.. Solution NMR structure of sag0934 from Streptococcus agalactiae. Northeast structural genomics target sar32[1–108]. (PDB entry only; no PUBMED entry available) http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=66235.
21. FlynnRL, ZouL (2010) Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit Rev Biochem Mol Biol 45 : 266–275.
22. ZhangW, DillinghamMS, ThomasCD, AllenS, RobertsCJ, et al. (2007) Directional loading and stimulation of PcrA helicase by the replication initiator protein RepD. J Mol Biol 371 : 336–348.
23. AnandSP, MitraP, NaqviA, KhanSA (2004) Bacillus anthracis and Bacillus cereus PcrA helicases can support DNA unwinding and in vitro rolling-circle replication of plasmid pT181 of Staphylococcus aureus. J Bacteriol 186 : 2195–2199.
24. PetitMA, EhrlichD (2002) Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J 21 : 3137–3147.
25. AnandSP, ZhengH, BiancoPR, LeubaSH, KhanSA (2007) DNA helicase activity of PcrA is not required for the displacement of RecA protein from DNA or inhibition of RecA-mediated strand exchange. J Bacteriol 189 : 4502–4509.
26. DillinghamMS (2011) Superfamily I helicases as modular components of DNA-processing machines. Biochem Soc Trans 39 : 413–423.
27. BruandC, EhrlichSD (2000) UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol Microbiol 35 : 204–210.
28. LlosaM, Gomis-RuthFX, CollM, de la CruzF (2002) Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol 45 : 1–8.
29. BiD, XuZ, HarrisonEM, TaiC, WeiY, et al. (2012) ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res 40: D621–626.
30. MerrikhH, ZhangY, GrossmanAD, WangJD (2012) Replication-trancription conflicts in bacteria. Nat Rev Microbiol 10 : 449–458.
31. PetitMA, DervynE, RoseM, EntianKD, McGovernS, et al. (1998) PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol Microbiol 29 : 261–273.
32. IordanescuS (1993) Characterization of the Staphylococcus aureus chromosomal gene pcrA, identified by mutations affecting plasmid pT181 replication. Mol Gen Genet 241 : 185–192.
33. AnandSP, ChattopadhyayA, KhanSA (2005) The PcrA3 mutant binds DNA and interacts with the RepC initiator protein of plasmid pT181 but is defective in its DNA helicase and unwinding activities. Plasmid 54 : 104–113.
34. HaT, RasnikI, ChengW, BabcockHP, GaussGH, et al. (2002) Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature 419 : 638–641.
35. FischerCJ, MalufNK, LohmanTM (2004) Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J Mol Biol 344 : 1287–1309.
36. MalufNK, FischerCJ, LohmanTM (2003) A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J Mol Biol 325 : 913–935.
37. YangY, DouSX, RenH, WangPY, ZhangXD, et al. (2008) Evidence for a functional dimeric form of the PcrA helicase in DNA unwinding. Nucleic Acids Res 36 : 1976–1989.
38. ChengW, HsiehJ, BrendzaKM, LohmanTM (2001) E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J Mol Biol 310 : 327–350.
39. MatsonSW, RobertsonAB (2006) The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic Acids Res 34 : 4089–4097.
40. SinhaKM, StephanouNC, GaoF, GlickmanMS, ShumanS (2007) Mycobacterial UvrD1 is a Ku-dependent DNA helicase that plays a role in multiple DNA repair events, including double-strand break repair. J Biol Chem 282 : 15114–15125.
41. Noirot-GrosMF, SoultanasP, WigleyDB, EhrlichSD, NoirotP, et al. (2002) The beta-propeller protein YxaL increases the processivity of the PcrA helicase. Mol Genet Genomics 267 : 391–400.
42. VeauteX, DelmasS, SelvaM, JeussetJ, Le CamE, et al. (2005) UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J 24 : 180–189.
43. ChangTL, NaqviA, AnandSP, KramerMG, MunshiR, et al. (2002) Biochemical characterization of the Staphylococcus aureus PcrA helicase and its role in plasmid rolling circle replication. J Biol Chem 277 : 45880–45886.
44. IordanescuS (1993) Plasmid pT181-linked suppressors of the Staphylococcus aureus pcrA3 chromosomal mutation. J Bacteriol 175 : 3916–3917.
45. MachonC, LynchGP, ThomsonNH, ScottDJ, ThomasCD, et al. (2010) RepD-mediated recruitment of PcrA helicase at the Staphylococcus aureus pC221 plasmid replication origin, oriD. Nucleic Acids Res 38 : 1874–1888.
46. SlatterAF, ThomasCD, WebbMR (2009) PcrA helicase tightly couples ATP hydrolysis to unwinding double-stranded DNA, modulated by the initiator protein for plasmid replication, RepD. Biochemistry 48 : 6326–6334.
47. SoultanasP, DillinghamMS, PapadopoulosF, PhillipsSE, ThomasCD, et al. (1999) Plasmid replication initiator protein RepD increases the processivity of PcrA DNA helicase. Nucleic Acids Res 27 : 1421–1428.
48. ToselandCP, Martinez-SenacMM, SlatterAF, WebbMR (2009) The ATPase cycle of PcrA helicase and its coupling to translocation on DNA. J Mol Biol 392 : 1020–1032.
49. Abdel-MonemM, Taucher-ScholzG, KlinkertMQ (1983) Identification of Escherichia coli DNA helicase I as the traI gene product of the F sex factor. Proc Natl Acad Sci U S A 80 : 4659–4663.
50. LlosaM, GrandosoG, HernandoMA, de la CruzF (1996) Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J Mol Biol 264 : 56–67.
51. MatsonSW, SampsonJK, ByrdDR (2001) F plasmid conjugative DNA transfer: the TraI helicase activity is essential for DNA strand transfer. J Biol Chem 276 : 2372–2379.
52. de la CruzF, FrostLS, MeyerRJ, ZechnerEL (2010) Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev 34 : 18–40.
53. BrittonRA, EichenbergerP, Gonzalez-PastorJE, FawcettP, MonsonR, et al. (2002) Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol 184 : 4881–4890.
54. GibsonDG, YoungL, ChuangRY, VenterJC, HutchisonCA3rd, et al. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6 : 343–345.
55. YansuraDG, HennerDJ (1984) Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci U S A 81 : 439–443.
56. BoseB, AuchtungJM, LeeCA, GrossmanAD (2008) A conserved anti-repressor controls horizontal gene transfer by proteolysis. Mol Microbiol 70 : 570–582.
57. SmitsWK, GoranovAI, GrossmanAD (2010) Ordered association of helicase loader proteins with the Bacillus subtilis origin of replication in vivo. Mol Microbiol 75 : 452–461.
58. LarkinMA, BlackshieldsG, BrownNP, ChennaR, McGettiganPA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23 : 2947–2948.
59. LetunicI, BorkP (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–478.
60. FinnRD, ClementsJ, EddySR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39: W29–37.
61. NeedlemanSB, WunschCD (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48 : 443–453.
Štítky
Genetika Reprodukční medicína
Článek Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across PathogensČlánek TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association StudiesČlánek Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization inČlánek Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA ExpressionČlánek The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of GenesČlánek The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 1
-
Všechny články tohoto čísla
- A Model of High Sugar Diet-Induced Cardiomyopathy
- Comparative Genome Structure, Secondary Metabolite, and Effector Coding Capacity across Pathogens
- Emerging Function of Fat Mass and Obesity-Associated Protein (Fto)
- Positional Cloning Reveals Strain-Dependent Expression of to Alter Susceptibility to Bleomycin-Induced Pulmonary Fibrosis in Mice
- Genetics of Ribosomal Proteins: “Curiouser and Curiouser”
- Transposable Elements Re-Wire and Fine-Tune the Transcriptome
- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- MAML1 Enhances the Transcriptional Activity of Runx2 and Plays a Role in Bone Development
- Predicting Mendelian Disease-Causing Non-Synonymous Single Nucleotide Variants in Exome Sequencing Studies
- A Systematic Mapping Approach of 16q12.2/ and BMI in More Than 20,000 African Americans Narrows in on the Underlying Functional Variation: Results from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- Transcription of the Major microRNA–Like Small RNAs Relies on RNA Polymerase III
- Histone H3K56 Acetylation, Rad52, and Non-DNA Repair Factors Control Double-Strand Break Repair Choice with the Sister Chromatid
- Genome-Wide Association Study Identifies a Novel Susceptibility Locus at 12q23.1 for Lung Squamous Cell Carcinoma in Han Chinese
- Genetic Disruption of the Copulatory Plug in Mice Leads to Severely Reduced Fertility
- The [] Prion Exists as a Dynamic Cloud of Variants
- Adult Onset Global Loss of the Gene Alters Body Composition and Metabolism in the Mouse
- Fis Protein Insulates the Gene from Uncontrolled Transcription
- The Meiotic Nuclear Lamina Regulates Chromosome Dynamics and Promotes Efficient Homologous Recombination in the Mouse
- Genome-Wide Haplotype Analysis of Expression Quantitative Trait Loci in Monocytes
- TATES: Efficient Multivariate Genotype-Phenotype Analysis for Genome-Wide Association Studies
- Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in Rice
- The Ecm11-Gmc2 Complex Promotes Synaptonemal Complex Formation through Assembly of Transverse Filaments in Budding Yeast
- MCM8 Is Required for a Pathway of Meiotic Double-Strand Break Repair Independent of DMC1 in
- Comparative Genomic Analysis of the Endosymbionts of Herbivorous Insects Reveals Eco-Environmental Adaptations: Biotechnology Applications
- Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry
- Pharmacodynamics, Population Dynamics, and the Evolution of Persistence in
- A Hybrid Likelihood Model for Sequence-Based Disease Association Studies
- Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity
- Multiple Opposing Constraints Govern Chromosome Interactions during Meiosis
- Transcriptional Dynamics Elicited by a Short Pulse of Notch Activation Involves Feed-Forward Regulation by Genes
- Dynamic Large-Scale Chromosomal Rearrangements Fuel Rapid Adaptation in Yeast Populations
- Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability
- Gene Copy-Number Polymorphism Caused by Retrotransposition in Humans
- An Incompatibility between a Mitochondrial tRNA and Its Nuclear-Encoded tRNA Synthetase Compromises Development and Fitness in
- Secondary Metabolism and Development Is Mediated by LlmF Control of VeA Subcellular Localization in
- Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination
- Tbx2 Controls Lung Growth by Direct Repression of the Cell Cycle Inhibitor Genes and
- Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene
- A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element
- Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators
- Genome-Wide Analysis Reveals Selection for Important Traits in Domestic Horse Breeds
- Coordinated Degradation of Replisome Components Ensures Genome Stability upon Replication Stress in the Absence of the Replication Fork Protection Complex
- Nkx6.1 Controls a Gene Regulatory Network Required for Establishing and Maintaining Pancreatic Beta Cell Identity
- HIF- and Non-HIF-Regulated Hypoxic Responses Require the Estrogen-Related Receptor in
- Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages
- The Telomere Capping Complex CST Has an Unusual Stoichiometry, Makes Multipartite Interaction with G-Tails, and Unfolds Higher-Order G-Tail Structures
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Loci Associated with -Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers
- Switchgrass Genomic Diversity, Ploidy, and Evolution: Novel Insights from a Network-Based SNP Discovery Protocol
- Centromere-Like Regions in the Budding Yeast Genome
- Sequencing of Loci from the Elephant Shark Reveals a Family of Genes in Vertebrate Genomes, Forged by Ancient Duplications and Divergences
- Mendelian and Non-Mendelian Regulation of Gene Expression in Maize
- Mutational Spectrum Drives the Rise of Mutator Bacteria
- Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression
- The Roles of Whole-Genome and Small-Scale Duplications in the Functional Specialization of Genes
- Sex-Specific Signaling in the Blood–Brain Barrier Is Required for Male Courtship in
- A Newly Uncovered Group of Distantly Related Lysine Methyltransferases Preferentially Interact with Molecular Chaperones to Regulate Their Activity
- Is Required for Leptin-Mediated Depolarization of POMC Neurons in the Hypothalamic Arcuate Nucleus in Mice
- Unlocking the Bottleneck in Forward Genetics Using Whole-Genome Sequencing and Identity by Descent to Isolate Causative Mutations
- The Role of Autophagy in Genome Stability through Suppression of Abnormal Mitosis under Starvation
- MTERF3 Regulates Mitochondrial Ribosome Biogenesis in Invertebrates and Mammals
- Downregulation and Altered Splicing by in a Mouse Model of Facioscapulohumeral Muscular Dystrophy (FSHD)
- NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses
- Retroactive Maintains Cuticle Integrity by Promoting the Trafficking of Knickkopf into the Procuticle of
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network
- Genetic and Functional Modularity of Activities in the Specification of Limb-Innervating Motor Neurons
- A Population Genetic Model for the Maintenance of R2 Retrotransposons in rRNA Gene Loci
- A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in
- A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1)
- Mutation of the Diamond-Blackfan Anemia Gene in Mouse Results in Morphological and Neuroanatomical Phenotypes
- Life, the Universe, and Everything: An Interview with David Haussler
- Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome Oxidase Blockade and Limits Mitochondrial ROS Overproduction
- An Evolutionarily Conserved Synthetic Lethal Interaction Network Identifies FEN1 as a Broad-Spectrum Target for Anticancer Therapeutic Development
- The Flowering Repressor Underlies a Novel QTL Interacting with the Genetic Background
- Telomerase Is Required for Zebrafish Lifespan
- and Diversified Expression of the Gene Family Bolster the Floral Stem Cell Network
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development
- WNK1/HSN2 Mutation in Human Peripheral Neuropathy Deregulates Expression and Posterior Lateral Line Development in Zebrafish ()
- Synergistic Interaction of Rnf8 and p53 in the Protection against Genomic Instability and Tumorigenesis
- Dot1-Dependent Histone H3K79 Methylation Promotes Activation of the Mek1 Meiotic Checkpoint Effector Kinase by Regulating the Hop1 Adaptor
- A Heterogeneous Mixture of F-Series Prostaglandins Promotes Sperm Guidance in the Reproductive Tract
- Starvation, Together with the SOS Response, Mediates High Biofilm-Specific Tolerance to the Fluoroquinolone Ofloxacin
- Directed Evolution of a Model Primordial Enzyme Provides Insights into the Development of the Genetic Code
- Genome-Wide Screens for Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Function and Regulation of , a Gene Implicated in Autism and Human Evolution
- An Insertion in 5′ Flanking Region of Causes Blue Eggshell in the Chicken
- Comprehensive Methylome Characterization of and at Single-Base Resolution
- Susceptibility Loci Associated with Specific and Shared Subtypes of Lymphoid Malignancies
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání