-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPlant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
The procambium and cambium are meristematic tissues from which vascular tissue is derived. Vascular initials differentiate into phloem towards the outside of the stem and xylem towards the inside. A small peptide derived from CLV-3/ESR1-LIKE 41 (CLE41) is thought to promote cell divisions in vascular meristems by signalling through the PHLOEM INTERCALLATED WITH XYLEM (PXY) receptor kinase. pxy mutants, however, display only small reductions in vascular cell number, suggesting a mechanism exists that allows plants to compensate for the absence of PXY. Consistent with this idea, we identify a large number of genes specifically upregulated in pxy mutants, including several AP2/ERF transcription factors. These transcription factors are required for normal cell division in the cambium and procambium. These same transcription factors are also upregulated by ethylene and in ethylene-overproducing eto1 mutants. eto1 mutants also exhibit an increase in vascular cell division that is dependent upon the function of at least 2 of these ERF genes. Furthermore, blocking ethylene signalling using a variety of ethylene insensitive mutants such as ein2 enhances the cell division defect of pxy. Our results suggest that these factors define a novel pathway that acts in parallel to PXY/CLE41 to regulate cell division in developing vascular tissue. We propose a model whereby vascular cell division is regulated both by PXY signalling and ethylene/ERF signalling. Under normal circumstances, however, PXY signalling acts to repress the ethylene/ERF pathway.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1002997
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002997Summary
The procambium and cambium are meristematic tissues from which vascular tissue is derived. Vascular initials differentiate into phloem towards the outside of the stem and xylem towards the inside. A small peptide derived from CLV-3/ESR1-LIKE 41 (CLE41) is thought to promote cell divisions in vascular meristems by signalling through the PHLOEM INTERCALLATED WITH XYLEM (PXY) receptor kinase. pxy mutants, however, display only small reductions in vascular cell number, suggesting a mechanism exists that allows plants to compensate for the absence of PXY. Consistent with this idea, we identify a large number of genes specifically upregulated in pxy mutants, including several AP2/ERF transcription factors. These transcription factors are required for normal cell division in the cambium and procambium. These same transcription factors are also upregulated by ethylene and in ethylene-overproducing eto1 mutants. eto1 mutants also exhibit an increase in vascular cell division that is dependent upon the function of at least 2 of these ERF genes. Furthermore, blocking ethylene signalling using a variety of ethylene insensitive mutants such as ein2 enhances the cell division defect of pxy. Our results suggest that these factors define a novel pathway that acts in parallel to PXY/CLE41 to regulate cell division in developing vascular tissue. We propose a model whereby vascular cell division is regulated both by PXY signalling and ethylene/ERF signalling. Under normal circumstances, however, PXY signalling acts to repress the ethylene/ERF pathway.
Introduction
Organised cell division and differentiation are required throughout nature for development of ordered body plans. The annual rings of trees which result from seasonal differences in radial growth are a widely recognisable example of the highly regulated nature of this process. Radial growth is achieved by generation of new vascular tissue that occurs via ordered cell divisions in the vascular meristem known as the cambium. Divisions in the cambium result in displacement of older cells to its periphery where they subsequently differentiate into xylem towards the inside of the stem or phloem towards the outside. Cambial cells divide in a highly ordered manner along their long axis giving rise to files of cells in a process that is most apparent in the growth rings of trees but also apparent in most higher plants such as Arabidopsis [1]. The ordered nature of this cell division is required for vascular tissue organisation and consequently is essential for both primary and secondary vascular development [2].
The receptor kinase PHLOEM INTERCALATED WITH XYLEM (PXY) was identified as being essential for ordered, coordinated cell divisions in the procambium [2] and has been shown to bind a peptide derived from CLV-3/ESR1-LIKE 41 (CLE41) and CLE44 [3], which was originally identified as TDIF, a peptide that represses tracheary element formation in transdifferentiation assays [4]. CLE41, and related CLE42 [5], [6] also function through the PXY receptor to provide positional information required for orientation of the cell division plane in the procambium [7]. CLE41 and CLE42 over-expression lines have more cells in vascular bundles than those of wild type counterparts [7] and an increased diameter of the hypocotyl vascular cylinder [3], [8]. These increases in vascular cell number and hypocotyl diameter are completely abolished in pxy 35S::CLE41 and pxy 35S::CLE42 lines [7]. Consequently, CLE41/42 induced vascular cell divisions occur in a PXY dependent manner demonstrating that PXY signalling, in addition to setting the division plane, also promotes the divisions themselves [3], [7]. A downstream target of PXY, the WUSCHEL-RELATED HOMEOBOX (WOX) gene, WOX4 is thought to be required for the promotion of these divisions [9] and wox4 mutants have been shown to have defects in vascular proliferation [10], [11].
Given that PXY signalling promotes vascular cell division, it might be expected that pxy mutants demonstrate a reduction in cell division, however in inflorescence stems of 5 week old plants no defects in the rate of cell division were reported [2]. Furthermore, pxy mutant hypocotyls exhibit only a small reduction in diameter at senescence suggesting only a small decrease in the total number of vascular cell divisions [7]. One explanation for this apparent contradiction is that a compensatory pathway exists that may be activated in the absence of pxy.
The gaseous hormone ethylene, has been shown to promote radial growth in several tree species [12], [13], [14], and more recently, radial growth and increased cambial cell division in tension wood of poplar was shown to be ethylene-induced [15]. Here we demonstrate that pxy and wox4 work together with several ETHYLENE RESONSE FACTOR (ERF) transcription factors and ethylene signalling to regulate cell divisions during Arabidopsis vascular development. We propose that in pxy mutants, cell numbers are maintained by the up-regulation of an ethylene pathway that increases expression of these ERFs. We present evidence for a model whereby vascular cell division is promoted by an interaction between PXY and ethylene signalling. Consequently, in addition to its role in mediating stress responses including the development of tension wood [15], our results suggest a more general role for ethylene in regulation of vascular cell division.
Results
Ethylene Response Factors are upregulated in pxy mutants
There are apparent contradictory observations with regard to the role of PXY/CLE41 in the regulation of the rate of vascular cell division. While CLE41 overexpression results in more cells [7], loss of PXY has little effect on vascular cell number [2]. One possible explanation is that an alternative pathway that also promotes vascular cell division is upregulated in pxy mutant plants. To test this hypothesis, we generated microarray expression data for the central part of pxy-3 mutant inflorescence stems and compared it to comparable data from wild type (Experiment E-MEXP-2420, http://www.ebi.ac.uk/arrayexpress). Intriguingly 12 members of the AP2/ERF family of transcription factors, predominantly from classes VIII-X [16] were found to be expressed at higher levels in pxy than wild type (Table S1). ERF109 (At4g34410; also known as RRTF [17]), ERF11 (At1g28370), ERF104 (At5g61600), and ERF018 (At1g74930; also known as ORA47 [18]) were increased 4.3, 3.0, 2.8 and 2.8, -fold, respectively (Table S1). Four further AP2/ERF family members AtERF1 (At4g17500), ERF2 (At5g47220), ERF5 (At5g47230), and ERF6 (At4g17490) demonstrated between 1.5 and 2-fold increases in expression. To confirm that the expression changes identified in array experiments were robust, we used qRT-PCR to retest expression levels of ERF018, ERF109 and AtERF1 in wild type and pxy-3 plants using RNA isolated from similar tissue to that used in microarrays. We observed similar fold changes in qRT-PCR to those previously identified in arrays when relative expression levels were normalised to that of ACT2 or 18s rRNA (Figure S1).
Arabidopsis inflorescence stems represent a developmental series as vascular tissue at the top of stems is newly initiated in contrast to more mature vasculature at the base of stems. To further investigate the expression pattern of genes differentially expressed in pxy, we assayed expression of four of the most upregulated ERF's (AtERF1, ERF11, ERF109 and ERF018) at both the top (2–4 cm below the shoot apex) and the base (1–3 cm above the rosette) of inflorescence stems from 5 week old plants using qRT-PCR (Figure 1A). The base of stems demonstrated larger fold changes in gene expression in pxy than was observed in the middle of stems (Table S1; Figure S1) as ERF109, ERF11, AtERF1 and ERF018 expression was increased 20, 7, 7 and 3-fold, respectively. In contrast, at the top of stems significant changes were only observed for AtERF1 and ERF11 suggesting that expression of these genes is upregulated in newly formed pxy mutant stems and this upregulation is progressively increased as vascular tissue matures (Figure 1A). Similar increases in ERF expression were also observed in pxy hypocotyls compared to wild type counterparts (Figure 1A).
Fig. 1. Expression of ERF transcription factors in Col, pxy, and wox4 mutants. 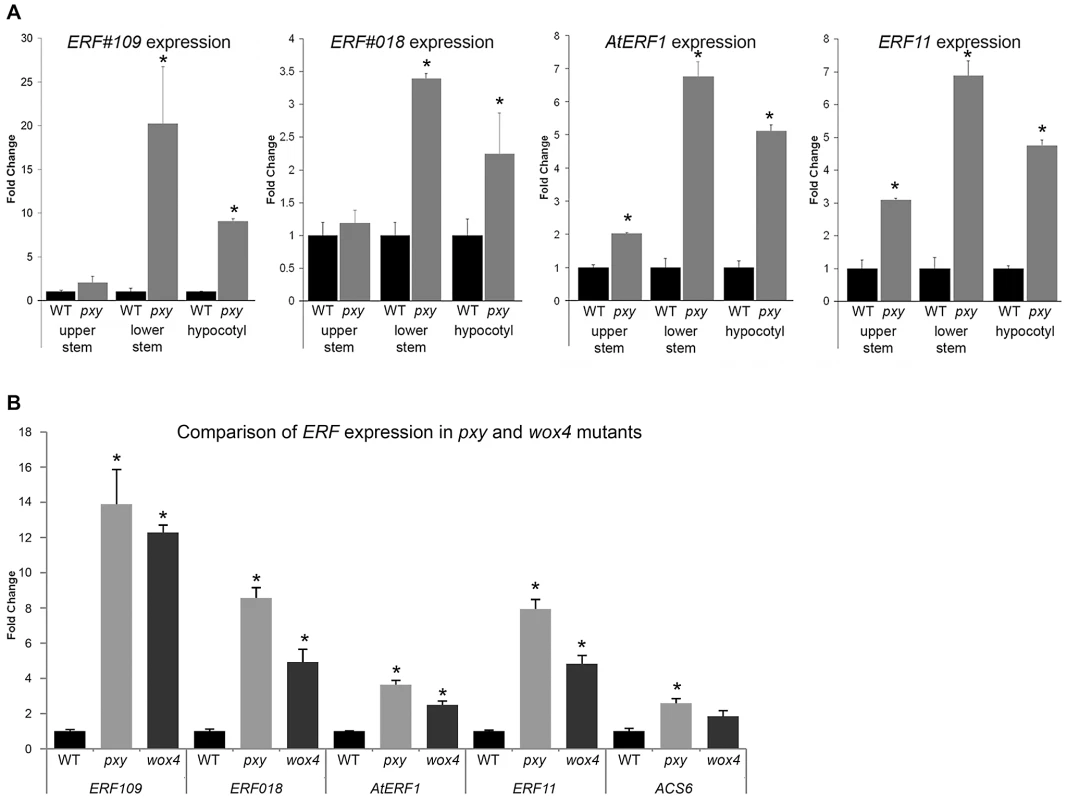
(A) qRT-PCR's showing ERF expression changes in upper inflorescence stem, lower inflorescence stem and hypocotyl of 5 week old plants, normalised to 18SrRNA. In all instances, higher ERF expression was observed in pxy mutants (grey) than wild type (black) for hypocotyls and stem bases. AtERF1 and ERF11 demonstrated expression increases in the upper inflorescence stem. (B) qRT-PCR's showing ERF expression changes in lower half of inflorescence stems of 5 week old plants, normalised to 18SrRNA. In all cases, expression was higher (p<0.0001) in pxy mutants (light grey bars) and wox4 (dark grey bars) than wild type (black). Error bars show standard error. *represents expression significantly different from wild type controls (p<0.0001). Samples were measured in technical triplicates on biological triplicates. WOX4 has been placed in a pathway downstream of the PXY receptor kinase [9] so we hypothesised that these genes up-regulated in pxy should also be up-regulated in wox4 mutants. qRT-PCR analysis of expression of the same ERF's upregulated in pxy mutants also demonstrated increases in expression in wox4 (Figure 1B). These observations suggest that ERF expression is suppressed by the pxy signalling pathway and that repression of ERF expression occurs downstream of WOX4.
We tested for vascular gene expression of two ERF transcription factors, ERF109 and ERF018, using in situ hybridization on sections of inflorescence stem from 5 week old plants 4 cm below the shoot apex (Figure 2) and found that Digoxigenin labelled antisense probes labelled many cell types. However, ERF109 and ERF018 expression was strongest in vascular bundles. Notably, in wild type, expression for both genes was most prominent in the procambium (arrows in Figure 2A, 2B) but absent from the phloem. In pxy mutant vascular tissue, ERF109 and ERF018 expression also appeared most prominently in the procambium and xylem (Figure 2). Sense negative controls for both genes did not label tissue above background levels but an antisense CLE41 positive control specifically labelled phloem tissue (Figure 2C) as previously reported [7]. Quantitative data from microarrays and qRT-PCR, combined with prominent vascular expression of these genes consequently suggests a role for ERF109 and ERF018 in vascular tissue.
Fig. 2. ERF109 and ERF018 expression determined by in situ hybridization in wild type, pxy, and eto1. 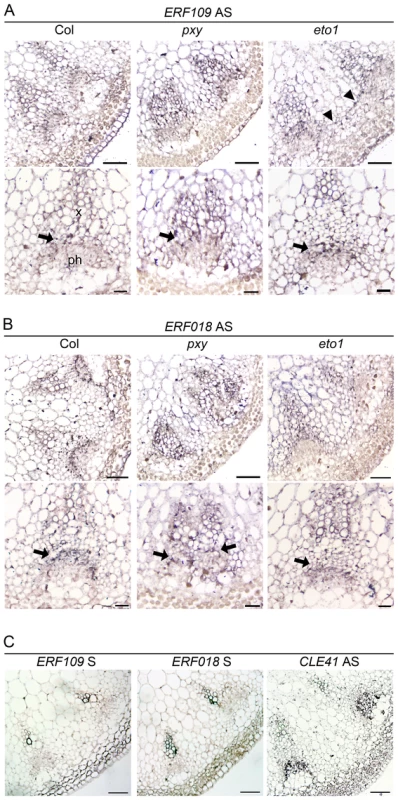
(A) Inflorescence stem ERF109 expression 4 cm below the shoot apex. Expression appears strongest in the vascular tissue division zone (arrows) in the three genotypes tested but absent from the phloem. Interfascicular ERF109 expression was apparent in eto1 mutants (arrowheads; right hand panel). Scales are 50 µm (upper panels) and 25 µm (lower panels). (B) ERF018 expression is similar to that observed for ERF109 (arrows show prominent expression in the procambium). Scales are 50 µm (upper panels) and 25 µm (lower panels). (C) Col wild type sections probed with ERF109 and ERF018 sense negative controls and CLE41 antisense positive control. Scales are 50 µm. erf mutants have fewer cells in vascular tissue
To determine the functional relevance of the gene expression changes observed in ERF's, we identified erf018 and erf109 loss-of-function mutants in publicly available T-DNA insertion libraries as these genes demonstrated relatively large increases in expression in pxy mutants. A confirmed T-DNA insertion within the coding sequence of ERF109 (Salk_150614) was renamed erf109-1, however, no insertion mutant was available that disrupted the coding sequence of ERF018. Salk_109440 line (erf018-1) was found to harbour a T-DNA insertion 142 base pairs upstream of the transcriptional start site and 249 base pairs upstream of the ATG. qPCR was used to analyse the expression of ERF018 in these lines and we found that expression was reduced to 60% of wild type levels (Figure S2) indicating that erf018-1 is a weak allele. Gross morphology of erf018, erf109 single and erf109 erf018 double mutants appeared identical to wild type counterparts (Figure 3) and the number of cells in erf018 and erf109 mutant vascular bundles was unchanged from wild type in 10 week old inflorescence stems (Figure 4A, Figure 5A). In contrast, erf109 erf018 double mutants demonstrated a small but significant reduction in the number of cells per vascular bundle (78% of wild type; Figure 4A, Figure 5A) suggesting that ERF109 and ERF018 act redundantly in promoting cell division in vascular bundles.
Fig. 3. Morphology of plant lines used in this study. 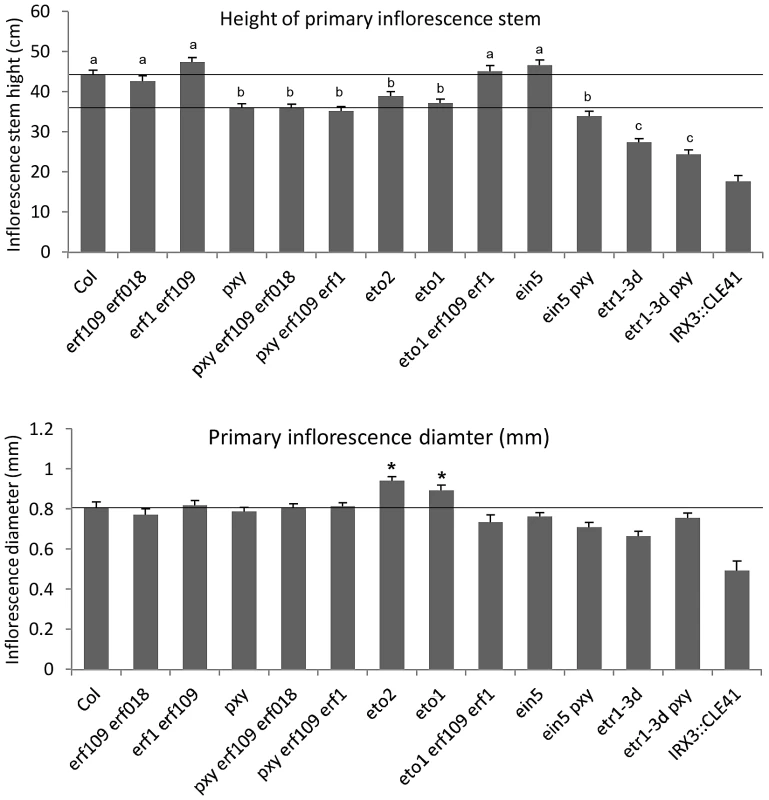
Upper graph shows height of plants at 10 weeks. Phenotype of pxy erf109 erf018, pxy erf109 erf1 and pxy ein5 vascular tissue is not due to a general growth defect as plants were indistinguishable from pxy single mutants. Similarly, etr1-3d and pxy etr1-3d lines were also indistinguishable. Bars marked (a) are similar to wild type; (b) is similar to pxy; (c) is similar to etr1-3d. Lower graph shows that inflorescence stem diameter is unchanged plant lines used in this study, except for eto lines which undergo secondary growth and are consequently larger than wild type (p<0.05). IRX3::CLE41 has more vascular cells than wild type but is of reduced height and stem diameter demonstrating that there is not a simple correlation between vascular cell division and overall plant morphology. Fig. 4. pxy interacts with erf transcription factors. 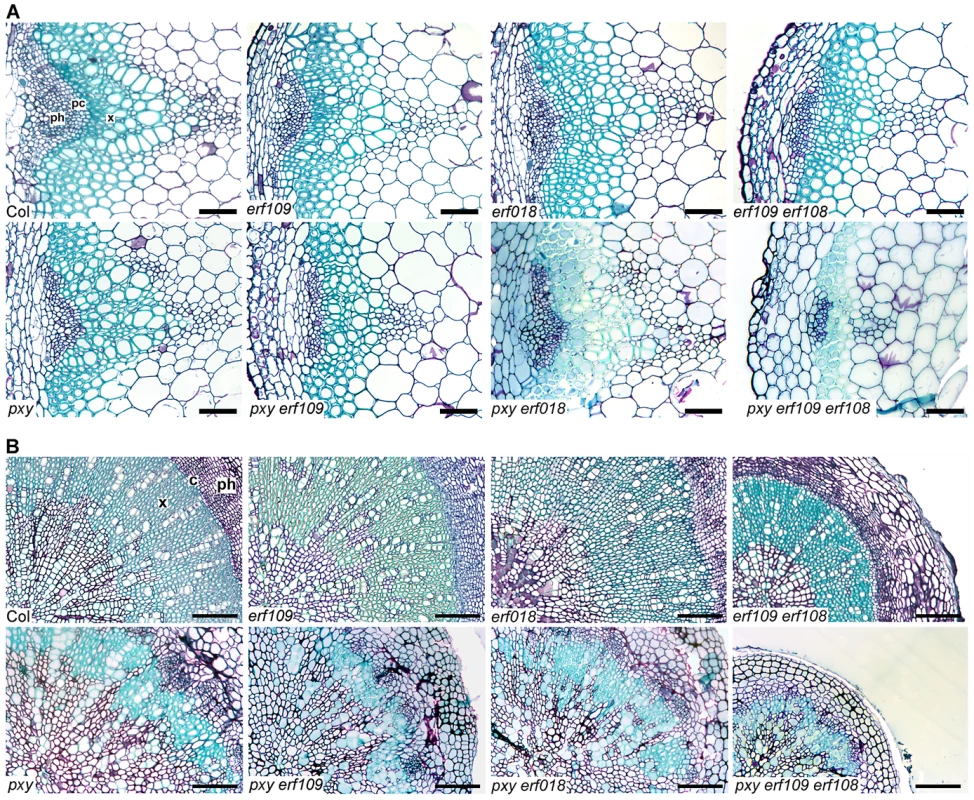
(A) Combinations of erf109, erf018 and pxy mutants. In comparison to wild type, erf109, erf018 and pxy vascular bundles have similar numbers of cells to wild type. erf109 erf018, pxy erf109 and pxy erf109 erf018 vascular tissue demonstrates a reduction in size compared to single mutants. Vascular bundles are from the base of 10 week inflorescence stems; scales are 50 µm. (B) erf109 erf018 hypocotyls are reduced in size compared to single mutant and wild type counterparts. pxy erf109 erf018 hypocotyls are smaller than those of parental lines. Images are from 10 week old hypocotyls; Scale bars are 100 µm. x is xylem, pc is procambium, c is cambium, ph is phloem. Fig. 5. Quantitative analysis of vascular cell division in erf109, erf018, erf1, and pxy mutant combinations. 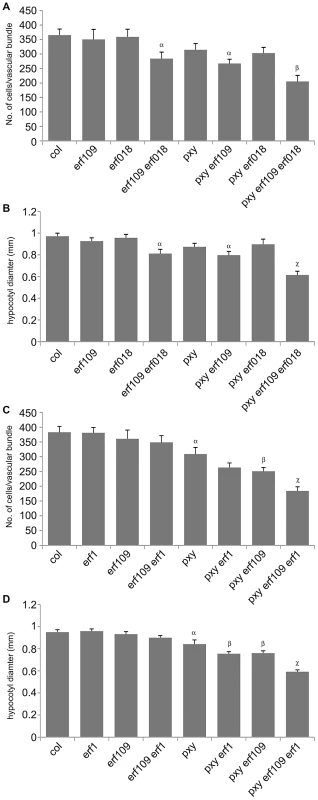
(A) and (B) erf109, erf018 and pxy mutant combinations. (A) Cells per vascular bundle at the base of the inflorescence stem of 10 week plants. (B) Hypocotyl diameter (mm) of plants at 10 weeks. (C) and (D) erf109, erf1 and pxy mutant combinations. (C) Cells per vascular bundle at the base of the inflorescence stem of 10 week plants. (D) Hypocotyl diameter (mm) of plants at 10 weeks. α is significantly smaller than Col; β is significantly smaller than pxy; χ is significantly smaller than respective double mutants. p<0.05. Error bars are standard error. The role of ERF109 and ERF018 in secondary growth was addressed in Arabidopsis hypocotyls. Several authors have used hypocotyl diameter as a measure of cell division during secondary growth [3], [7], [8], [19], [20], [21]. Consistent with our observation that erf109 erf018 lines had reductions in vascular cell division in stems, hypocotyl diameter was also reduced in erf109 erf018 double mutants to 83% of wild type diameter (Figure 4B, Figure 5B). Consequently, ERF109 and ERF018 are required for promotion of vascular cell division during both primary and secondary growth.
ERFs act in a parallel pathway to PXY signalling
It was clear from our analysis that ERF109 and ERF018 are required for promoting vascular cell divisions. Since these transcription factors are upregulated in pxy mutants it may be hypothesised that they represent a mechanism by which vascular cell division is maintained in the absence of PXY. pxy erf109 erf018 triple mutants were therefore generated with the expectation that if ERF transcription factors do compensate for loss of pxy then pxy erf109 erf018 lines would demonstrate a significant reduction in cell number when compared to pxy, erf109 erf018 or wild type. pxy mutant vascular bundles have been previously characterised with intercalated xylem and phloem, however, in inflorescence stems of 5 week old plants no differences in vascular cell number were observed [2]. We reasoned that differences in the number of cells in pxy vasculature may be observed in 10 week old tissue, particularly in hypocotyls which undergo continuous radial expansion, as subtle differences in the rate of cell division would have time to accumulate. All experiments on 10 week old tissue in this manuscript (see below) demonstrated a trend towards a reduction in cell number in pxy mutant vascular bundles compared to wild type (see below), however, in this instance, differences proved not to be statistically significant (Figure 4A, Figure 5A). Consistent with our hypothesis, vascular bundles of pxy erf109 mutants demonstrated a 27% reduction in cell number compared to wild type in contrast with pxy and erf109 which showed no significant difference (Figure 4A, Figure 5A). Consequently, clear defects in pxy mutant vascular cell number only became apparent when pxy was combined with an erf109 mutant. pxy erf018 and pxy erf018 erf109 were also generated to determine whether erf018 demonstrated a similar interaction with pxy. pxy erf018 double mutant inflorescence stem vascular tissue did not differ from parental lines (Figure 4A, Figure 5A), however, pxy erf018 erf109 lines demonstrated a 44% reduction in cells/vascular bundle demonstrating a significant enhancement of the pxy erf109 phenotype (Figure 4A, Figure 5A).
When analysing secondary growth in pxy erf109 double mutant hypocotyls, we found that the relationship between erf109 and pxy was similar to that observed in vascular bundles. pxy erf109 hypocotyls had the characteristic altered orientation of cell division associated with pxy mutants [7] but the hypocotyl diameters were narrower than either pxy or erf109 single mutants (Figure 4B). The decrease in hypocotyl diameter was most dramatic in pxy erf109 erf018 mutants where hypocotyl diameters were only 63% of that observed in wild type. These observations are consistent with fewer cell divisions having occurred in the triple mutant than the respective doubles, single mutants and wild type (Figure 4B, Figure 5B). As with our observation in vascular bundles, defects in vascular cell number are greatly enhanced when pxy mutants are combined with mutations in the ERF transcription factors erf018 and erf109.
We further examined ERF function in PXY signalling by analysing the function of AtERF1 (At1g17500; upregulated in both pxy and wox4 mutants; Figure 1B, Table S1). A T-DNA mutant (Salk_036267) was isolated and used to test whether AtERF1 acted similarly to ERF018 and ERF109. erf1 single and erf1 erf109 double mutants were indistinguishable from wild type, and although erf1 pxy double mutants were suggestive of a reduction in the size of vascular bundles compared to pxy single mutants, differences proved not significant (Figure 5C). In contrast, pxy erf1 erf109 triple mutants demonstrated a dramatic decrease in the number of cells/vascular bundle (48% of that observed in wild type), a significant reduction when compared to respective single and double mutants when assayed at the base of the inflorescence stems of 10 week old plants (Figure 5C). Similarly, pxy erf109 erf1 lines demonstrated reduced hypocotyl diameter (52% of wild type) when compared to control lines (≤80% of wild type; Figure 5D). erf1 therefore enhances vascular cell division defects of erf109 pxy mutants in both inflorescence and hypocotyl. These data are consistent with a role for AtERF1, ERF109 and ERF018 in promoting vascular cell division in the absence of PXY.
ERF expression in the context of ethylene signalling
Five of the ERF genes upregulated in pxy; AtERF1, ERF2, ERF5, ERF003/Atg525190 and ERF11 have previously been shown to be induced by ethylene [22], [23], [24], [25]. Furthermore, an enzyme responsible for catalysing the rate-limiting step of ethylene biosynthesis, ACS6 (At4g11280) was upregulated 2.5 fold in pxy mutants (Table S1; Figure 1B). Consequently, we hypothesised that the increase in expression of ERF transcription factors in pxy and wox4 mutants may be the result of an increase in ethylene signalling. To determine whether these genes also demonstrated elevated expression in stems of plants with higher levels of ethylene than wild type, their response to ethylene exposure was tested. We subjected five week old wild type Arabidopsis plants to ethylene stimuli of 3 hours, 16 hours and also made use of ethylene overproducer1 (eto1) mutants which produce more ethylene than wild type [26]. Expression levels of ERF's were compared in inflorescence stems using qRT-PCR (Figure 6). Expression was increased in response to exogenous ethylene treatment as in plants exposed to ethylene for 3 hours, AtERF1 and ERF11 underwent a 3-fold induction (Figure 6A) and following a 16 hour treatment 2 - and 5-fold inductions were observed (Figure 6B). Increased ERF109 and ERF018 expression of approximately 3-fold was observed in eto1. Consequently ERF109, ERF018, AtERF1 and ERF11 are ethylene responsive; however, the dynamics of induction varies in inflorescence stems. ERF1 and ERF11 demonstrated an early ethylene response and ERF018 and ERF109 expression was increased in response to a constitutive ethylene production (Figure 6).
Fig. 6. ERF expression responds to ethylene. 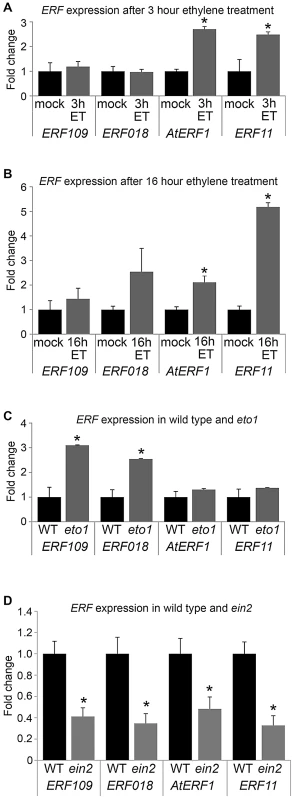
(A) qRT-PCR showing ERF expression in inflorescence stems in response to 3 hour ethylene exposure (grey bars) compared to mock treatment (black bars). (B) qRT-PCR showing ERF expression in inflorescence stems in response to 16 hour ethylene exposure (grey bars) compared to mock treatment (black bars). (C) Inflorescence stem ERF expression in plants that over-produce ethylene (eto1; grey bars) compared to wild type (black bars). (D) ERF expression in ein2 mutant inflorescence stems (grey bars) compared to wild type (black bars). *Expression significantly different from wild type controls (p<0.05). Samples were measured in technical triplicates on biological triplicates. To confirm the relationship between ERF expression and ethylene in stems, we carried out the converse experiment. ERF levels were determined in ethylene insensitive 2 (ein2) plants in which the ethylene signal transduction pathway is thought to be entirely abolished [27]. Consistent with ERF109, ERF018, AtERF1 and ERF11 acting downstream of the ethylene response in inflorescence stems, expression of the genes tested was reduced by half (Figure 6D). It is notable that ein2 mutants do not demonstrate reductions in vascular cell number (see below). Consequently, differences in ERF expression cannot be explained by phenotypic differences in vascular tissue and are likely the result of reduced ethylene signalling.
Ethylene promotes radial growth in Arabidopsis
Reports in poplar have demonstrated that ethylene promotes vascular cell division during secondary growth [15], so in order to determine whether ethylene, and therefore ERF's, function similarly in Arabidopsis we analysed the inflorescence stems of eto1 mutants at six weeks and found that they exhibited an increase in the number of procambial cells (Figure 7A–7B). eto1 mutants also demonstrated early onset of secondary growth as vascular cell divisions were observed in the interfascicular region prior to any divisions in wild type plants at an equivalent stage of development (Figure 7A–7B). This phenotype was particularly evident when eto1 mutant vascular sections were subjected to in situ hybridization with ERF109 antisense probes. In wild-type, labelling was absent from interfascicular tissue but present in eto1 (Figure 2). The phenotypic consequences of constitutive ethylene production were confirmed by analysis of eto2 mutants [28]. At ten weeks, as with eto1 mutants, eto2 plants had larger vascular bundles and cell divisions between vascular bundles indicative of secondary growth which were absent in wild type (Figure S3). To confirm that the observed differences in eto1 and eto2 were significant, the number of cells in vascular bundles of 10 week inflorescence stems and hypocotyl diameters were determined. Increases in vascular cell number of 21% for eto1 and 34% for eto2 in inflorescence stems and increases in hypocotyl diameter of 19% and 31%, respectively were apparent (Figure 7C–7D). Our data are therefore consistent with the idea that elevated levels of ethylene result in both an increase in vascular cell division and increased expression of ERF transcription factors which we have shown are required to promote vascular cell division in the absence of PXY.
Fig. 7. Increased vascular cell divisions in eto1 and eto2 mutants. 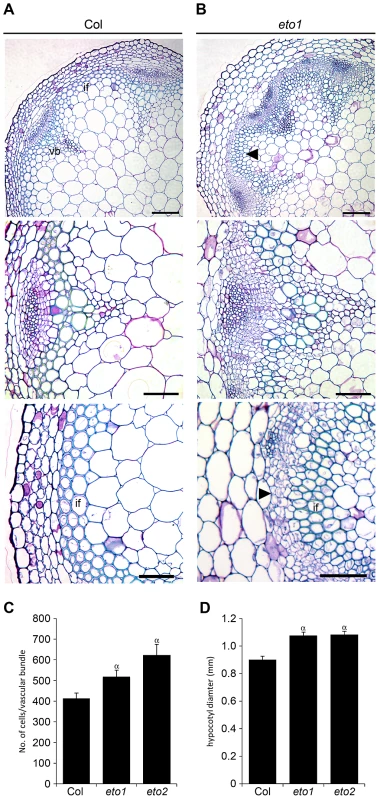
(A) Transverse sections of toluidine blue stained wild type 5 week inflorescence stems. (B) eto1 mutants inflorescence stems have more procambium than wild type (compare middle panels), and initiate secondary growth between vascular bundles at an early age (arrowheads in upper and lower panels). (A and B) Scales are 100 µm (upper), or 50 µm (middle and lower panels). vb is vascular bundle; if interfascicular region. (C) Graph showing cells per vascular bundle at the base of the inflorescence stem of 10 week Col, eto1 and eto2 plants. (D) Graph showing hypocotyl diameter (mm) of Col, eto1 and eto2 plants at 10 weeks. (C and D) α is significantly larger than Col (p<0.05); error bars are standard error. To confirm that ERF transcription factors upregulated in pxy mutants were required for vascular eto phenotypes and therefore ethylene mediated vascular expansion, we generated eto1 erf109 erf1 triple mutants. eto1 erf109 and eto1 erf1 double mutant lines were indistinguishable from eto1 single mutants, but in eto1 erf109 erf1 lines, vascular cell number was significantly smaller than that observed in eto1 (Figure 8). Furthermore interfascicular cell divisions that were sometimes present in eto1 lines were not observed in eto1 erf109 erf1 counterparts (Figure S4), and eto1 stems demonstrated an increase in diameter compared to those of eto1 erf109 erf1 (Figure 3), consistent with a requirement for ERF's in eto secondary growth phenotypes. Consequently, we have demonstrated that the ERF transcription factors that demonstrate increased expression in pxy mutants are upregulated in response to ethylene, their expression is reduced in ethylene signalling mutants and they are required for the phenotypic consequences of ethylene over-production in vascular tissue.
Fig. 8. ERF genes are required for the eto1 vascular phenotype. 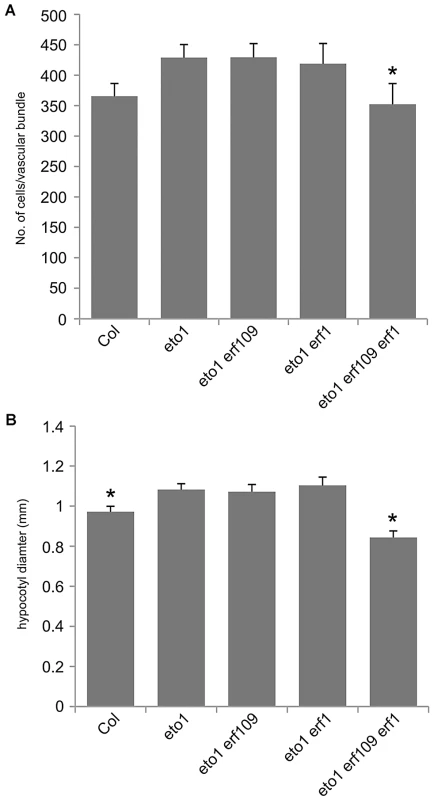
(A) Graph showing cells per vascular bundle at the base of the inflorescence stem of 10 week Col, eto1, eto1 erf109, eto1 erf1 and eto1 erf109 erf1 plants. eto1 erf109 erf1 plants are significantly smaller than eto1 single mutants. (B) Graph showing hypocotyl diameter (mm) of Col, eto1 erf109, eto1 erf1 and eto1 erf109 erf1 plants at 10 weeks. ERF109 and ERF1 are required for increases in hypocotyl diameter observed in eto1. (A and B) * significantly smaller than eto1 (p<0.05); error bars are standard error. Crosstalk between ethylene and pxy signalling
To directly address the relationship between PXY and ethylene signalling, we crossed mutants that are unable to respond to ethylene to pxy. ein2 encodes an integral membrane of unknown function that is essential for ethylene signal transduction [27] and is the only single mutant thought to entirely abolish ethylene signalling [27]. pxy ein2 double mutants developed normal rosettes and inflorescence stems were initiated normally, however, the plants senesced early so analysis of ten week plants, consistent with quantitative phenotypic analysis elsewhere in this manuscript was not possible. Analysis was carried out on six week old plants but at this developmental stage, wild type plants had similar numbers of cells in vascular bundles as present at ten weeks suggesting that vascular proliferation in the stem was complete (Figure 9A, 9C). Wild-type and ein2 vasculature in inflorescence stems were indistinguishable, with no significant difference in vascular cell number. pxy ein2 mutant vascular tissue demonstrated a dramatic reduction in vascular cell number (55% of wild type), having significantly fewer cells than pxy or ein2 single mutants (Figure 9A, 9C) and clearly demonstrating that ein2 is required for maintenance of vascular tissue in pxy mutants. Similar results were observed in the hypocotyl (Figure 9B, 9D) with pxy ein2 lines significantly smaller than ein2 or pxy single mutants.
Fig. 9. Analysis of vascular cell division in ein2 and pxy mutant combinations. 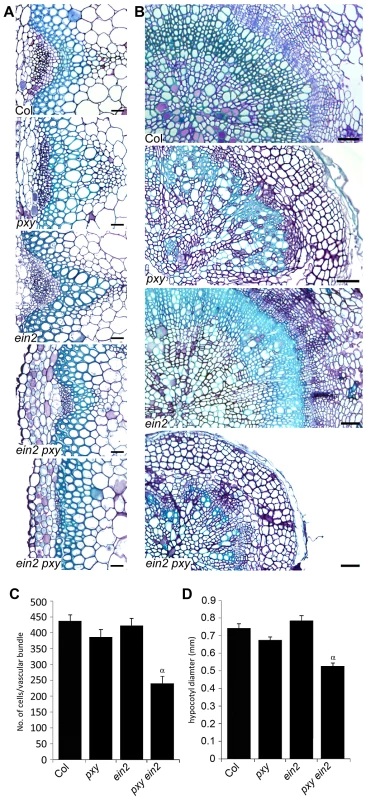
(A) Combinations of pxy and ein2 inflorescence stems. ein2 is similar to wild type. pxy ein2 is smaller than pxy and ein2 lines. In extreme cases (lower panel), pxy ein2 vascular bundles are extremely small. Scale bars are 25 µm. (B) Transverse sections through hypocotyls show that compared to wild type, pxy mutants have disrupted organisation due to loss of orientation of cell division. ein2 is indistinguishable from wild type. pxy ein2 hypocotyls are severely reduced in size and lack organisation like pxy mutants. Scale bars are 50 µm. (C) Cells per vascular bundle at the base of the inflorescence stem of 10 week plants. (D) Hypocotyl diameter (mm) of plants at 10 weeks. (C and D) α is significantly smaller than Col (p<0.0001); error bars are standard error. To confirm the results obtained with ein2, two further mutants in the ethylene signal transduction pathway were analysed. ethylene receptor1 (etr1) and ethylene insensitive5 (ein5) encode an ethylene receptor [29], and an exoribonuclease involved in ethylene signalling [30], [31], respectively. Neither etr1-3d nor ein5-1 exhibit the triple response and are partially ethylene insensitive [32]. In primary vascular tissue in inflorescence stems, in common with ein2 mutants, ein5 and etr1-3d were indistinguishable from wild type (Figures S5, S6) but in both cases a dramatic enhancement of the reduction in vascular cell number observed in pxy mutants (see above) was observed when pxy etr1-3d double mutants were analysed (Figures S5, S6).
Analysis of the role of etr1-3d and ein5 in hypocotyl secondary growth was also carried out. Hypocotyl diameters were measured at 10 weeks and ein5 was found not to differ from wild type, however etr1-3d demonstrated a small reduction (Figure S6). Although this differed from observations in ein2 and ein5, this is likely due to the age of plants tested with respect to ein2 and differences in the level of reduction of ethylene signalling with respect to ein5. In common with ein2, etr1-3d and ein5 both strengthened the pxy phenotype as double mutants were smaller than respective singles (Figures S5, S6).
If ERF109, ERF018 and ERF1 are targets of an ethylene-induced signalling mechanism that is upregulated in the absence of pxy, then pxy erf mutants should appear similar to those of pxy ein2, pxy etr1-3d and pxy ein5. As such, pxy erf109 erf018 and pxy erf109 erf1 vascular tissue was similar to that of pxy ein5, pxy etr1-3d and pxy ein2 as in all instances, the pxy cell division phenotype was enhanced. It is notable that ethylene signalling does not appear to greatly influence PXY signalling. Expression levels of CLE41, CLE42, PXY and WOX4 were unchanged in ein2 mutants or erf109 erf018 plants (Figure S7). CLE41, CLE42 and WOX4 expression was also unchanged in plants exposed to an ethylene stimulus, however, ethylene did promote PXY expression (Figure S7) suggesting that PXY is to some extent ethylene responsive.
Discussion
Up regulation of the PXY signal transduction pathway by over expression of the CLE41 ligand results in massively increased vascular cell divisions, however, pxy mutants exhibit only limited reductions in cell division. We have identified a group of 12 ERF transcription factors that are upregulated in pxy mutants (Table S1; Figure 1). Loss of function analysis of three of these genes; ERF109, ERF018 and AtERF1 resulted in plants with inflorescence stems that were characterised by reduced numbers of vascular cells suggesting that these genes promote cell division in vascular meristems (Figure 4, Figure 5). This data suggests that these ERF transcription factors form part of a mechanism that is up-regulated in response to loss of pxy.
Previous authors have demonstrated that five of the genes identified have increased expression in response to ethylene in seedlings [22], [23], [24], [25]. We have demonstrated that several of the family members are upregulated in stems of ethylene overproducing eto1 mutants or in plants subjected to ethylene treatment (Figure 6). Furthermore these ERF's are required for the increased vascular tissue observed in eto1 plants (Figure 7, Figure 8). An involvement of ethylene in vascular cell division in pxy plants is supported by analysis of ein2, ein5 and etr1-3d mutants. EIN2, EIN5 and ETR1 are required for normal ethylene signal transduction [32] and pxy ein2, pxy ein5 and pxy etr1-3d plants had significant reductions in the number of vascular cells compared to single mutants or wild type (Figure 9; Figures S5, S6). Taken together, our results demonstrate that ERF transcription factors promote vascular cell division, that their expression is influenced by PXY-repression of ethylene signalling, and consequently, these signalling pathways interact to control the rate of cell division in plant vascular tissue (Figure 10).
Fig. 10. Model showing ethylene and PXY signalling act in parallel pathways in vascular development. 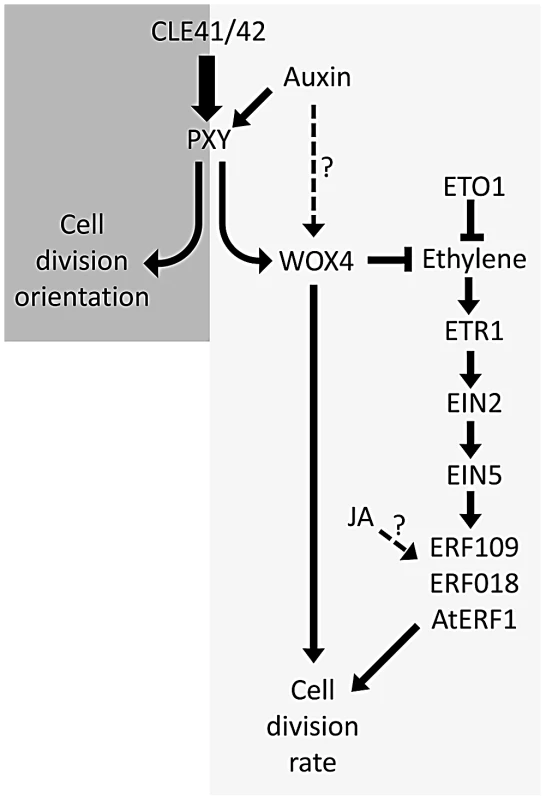
Crosstalk between these two signalling pathways contributes to levels of ERF transcription factors and control of the number of vascular cell divisions. Auxin and JA also influence this network. 1-aminocyclopropane-1-carboxylic acid synthase 6 (AtACS6) is also upregulated in pxy (Table S1; Figure 1B). ACS enzymes catalyse the rate-limiting step of ethylene biosynthesis [33], [34], i.e. conversion of S-adenosylmethionine to ACC [35]. Ethylene has previously been shown to promote cell division in the organising centre of Arabidopsis roots [36], and in the cambium of hybrid poplar [15]. In tree species, ethylene is produced in association with physical stress [13], and is known to have a role in promoting development of tension wood [15]. Our results suggest that it may have a more general role in regulating the rate of cell division in the cambium (Figure 10), particularly as etr1-3d mutants demonstrated significant reductions in radial growth compared to wild type in hypocotyls (Figure S6). These conclusions are also consistent with earlier studies that demonstrate that in trees ethylene levels are higher at the height of the growing season than towards the end [13].
It may be case that ERF109, ERF018, AtERF1 and other ERF's upregulated in pxy are essential components in the vascular developmental programme and their expression can be modulated by ethylene and other external factors. ERF transcription factors affect a variety of processes [17], , but strikingly, ERF018 and AtERF1 have also been described as being up-regulated by jasmonic acid [18], [25]. Jasmonic acid has recently emerged as a key modulator of cell division in the cambium [39] and consequently ERF018 and AtERF1 may be key integration points for vascular development. In support of this hypothesis (Figure 10), phenotypes were observed in erf018 mutants (in combination with erf109) despite the weak nature of the erf018 allele identified in this study. Our observations, and those of previous authors are consistent with an emerging picture that many of these transcription factors form part of a network [40] and how the plant responds to them is very much context dependent [41]. The ERF transcription factors analysed in this study have been suggested as having roles unrelated to vascular development [17], [18], [42], [43] and consequently, ERF109 and ERF018 have broad expression patterns (but are nevertheless expressed in vascular tissue), however, single mutants and neither erf109 erf1 nor erf109 erf018 double mutants demonstrated visible architectural defects, such as a change in height (Figure 3), suggesting that reductions in vascular cell number in these lines is not the consequence of a general disruption to plant growth.
The phytohormones brassinosteroid [44], cytokinin [45], [46], [47], strigolactone [48] and auxin [49], [50] have also been shown to have roles in Arabidopsis vascular development, and have been shown to be regulate each other's biosynthetic pathways [25]. Brassinosteroid upregulates genes required for ethylene biosynthesis and auxin up regulates genes involved in cytokinin biosynthesis. More complex interactions occur between ethylene and auxin, brassinosteroid and auxin as well as cytokinin and brassinosteroid, where phytohormone biosynthesis genes are both induced and repressed in response to respective phytohormone treatments [25]. A proper understanding of vascular cell division and differentiation will need to take into account interactions between these signalling pathways and their downstream targets.
One phytohormone in addition to ethylene that has been placed in a network with PXY signalling is auxin. WOX4 is positively regulated by auxin which has led to the suggestion that auxin lies upstream of PXY signalling in a hypothesis that is supported by the observation that part of the WOX4 response to auxin is PXY-dependent [10]. In support of this hypothesis, ERF genes upregulated in pxy mutants also demonstrated increased expression in wox4 lines (Figure 1B). However, WOX4 regulation may not depend entirely on PXY signalling and could also be regulated by an auxin-dependent, PXY-independent mechanism [51] because despite the observation that PXY is required for the WOX4 auxin response, WOX4 expression nevertheless increases in pxy mutants subjected to a 1 day auxin induction [10]. Further evidence for a WOX4, PXY-independent response comes from the observation that wox4 enhances pxy mutants [9]. If WOX4 expression was entirely controlled by PXY signalling wox4 pxy double mutants and respective single mutants would have identical phenotypes. Experiments presented here, and by previous authors therefore place PXY signalling in a network containing auxin, ethylene and JA signalling (Figure 10).
The procambium and cambium are meristematic tissues and as such, demonstrate similarities with the shoot apical meristem (SAM) and root apical meristem (RAM). All three structures require CLAVATA (CLV) -like, and phytohormone signalling mechanisms for their regulation. In the SAM, CLE41-related CLV3 is secreted from stem cells and binds to the PXY-related CLV1 receptor [52], . This precipitates a signal which culminates in negative regulation of WUSCHEL (WUS), a homeobox gene which promotes stem cell fate – and therefore cells that secrete CLV3. This feedback loop enables the plant to dynamically regulate the size of its apical stem cell population thus balancing organ generation with maintenance of its stem cells [56], [57]. WUS expression is also controlled by cytokinin signalling, which is thought to add robustness to the feedback mechanism [58]. It is tempting to speculate that the relationship between PXY and ethylene signalling acts similarly. In this case, interaction between the PXY and ethylene pathways culminates in appropriate regulation of downstream transcription factors required to regulate the rate of cell division and recruitment of daughter cells into xylem and phloem.
Materials and Methods
Generation of plant stocks
Plant growth conditions, and pxy alleles have been described previously [2]. T-DNA insertion lines in ERF018 (salk_109440), ERF109 (salk_150614), AtERF1 (salk_036267) and wox4-1 (GABI-Kat_462G01) were identified using the TAIR database [59] and confirmed using PCR. Insertion lines and eto1-1, ein5-1, and etr1-3d were obtained from NASC. erf109 erf018, pxy erf109, pxy erf018, pxy erf018 erf109, erf1 erf109, pxy erf1 and pxy erf1 erf109 lines were identified in segregating F2 populations. Primers for pxy genotyping have been described previously [7]. Oligos SALK_ERF109LB and SALK_ERF109RB (CGCGATGCTTTGTAGGAGTAG and TGTCAGGGTTTTTCCAGTGAC), SALK_ERF018LB and SALK_ERF018RB (TTCATGCTCATGATGATGAGC and ATCGACGGTGGATTATTAGGG) and salk-ERF1-F and salk-ERF1-R (CGTTCCTAACCAAACCCTAGC and TCCTACTCTTCTCCCTGCTCC) were used for the identification of erf109, erf018 and erf1 mutants. pxy ein2, pxy ein5 and pxy etr1-3d doubles were selected in the F3 generation from families that were ethylene insensitive as determined using a triple response screen [26].
Ethylene treatments of plants prior to measurement of ERF expression in inflorescence stems was carried out by placing Arabidopsis plants in a sealed container and generating ethylene gas to a maximal concentration of 500 µl l−1 of ethylene gas as described previously [60].
Gene expression analysis
For comparison of wild type and pxy transcriptomes, Col-0 and pxy-3 lines were used. For each replicate, plants were germinated on MS agar plates prior to transfer to soil (6 plants per 10 cm pot), where they were grown on for 5 weeks under long day (16/8 h light/dark) conditions at which point the inflorescence stem had 4–6 expanded siliques. Pots were randomised and rotated daily. For each replicate, the 6 primary inflorescence stems were taken from all the plants in a pot. Cauline leaves and side branches were removed. Stems were divided into 4 sections of equal size and RNA was isolated from the third section from the top using TRIzol Reagent (Invitrogen). RNA was sent to the University of Manchester Genomic Technologies Facility (http://www.ls.manchester.ac.uk/research/facilities/microarray/) where it was assessed for quality. ATH1 Affymetrix GeneChip oligonucleotide arrays were used to analyse the gene expression from each sample. Biotinylated cDNA samples from three biological replicates of pxy and wild type stems were synthesised and hybridized to Arabidopsis ATH1 genome oligonucleotide arrays. Technical quality control was performed with dChip (V2005; www.dchip.org), using the default settings [61]. Background correction, quantile normalization, and gene expression analysis were performed using RMA in Bioconductor [62]. Differential expression analysis was performed using Limma using the functions lmFit and eBayes [63]. Microarray data has been submitted in a MIAME compliant standard to the Array Express database (Experiment E-MEXP-2420, http://www.ebi.ac.uk/arrayexpress).
For RT-PCR, RNA was isolated using Trizol (Invitrogen). cDNA synthesis, following DNase treatment, was performed using Superscript III reverse transcriptase (Invitrogen). All samples were measured in technical triplicates on biological triplicates. The qRT-PCR reaction was performed using SYBR Green JumpStart Taq ReadyMix (Sigma) using an ABI Prism 7000 machine (Applied Biosystems) with the standard sybr green detection programme. A melting curve was produced at the end of every experiment to ensure that only single products were formed. Gene expression was determined using a version of the comparative threshold cycle (Ct) method. The average amplification efficiency of each target was determined using LinReg [64] and samples were normalised to 18S rRNA or ACT2. Primers for qRT-PCR are described in Table S2.
ERF109 and ERF018 probe templates for Digoxigenin-labelling of mRNA were generated by PCR amplification and subsequent cloning of products into pENTR-D-topo using primers (caccaacagagtcgcaaga and catgctttcttgttcttgttc for ERF109; caccaattcaaccaaaccgaat and ccagatttctccatgactcca for ERF018). The resulting plasmids were used with M13 forward and reverse primers to generate a template for antisense probes, and sense probe control templates were PCR amplified with a forward primer containing a T7 promoter site (taatacgactcactatagggatgcattatcctaac for ERF109; taatacgactcactatagggatggtgaagcaagcg for ERF018). Reverse primers were as above. CLE41 positive control and methods for probe labelling and in situ hybridization were as used in [7], and based on the method described in [65].
Analysis of vascular tissue
Analysis of vasculature tissue in thin sections, was carried out as described previously [66]. For hand cut sections, tissue was stained with either aqueous 0.02% Toluidine Blue or 0.05M Aniline blue in 100 mM Phosphate buffer (pH 7.2). Cell counts were carried out using thin sections of 10 week old stems on ≥10 biological replicates. Cells included were protoxylem (marking the inner part of the bundle), phloem cap cells (marking the outer part of the bundle) and all vascular cell types between. An area of secondary growth is reported to be present up to 2.4 mm above the upper rosette leaf [39]. Consequently, sections were taken 10 mm above the upper rosette leaf to avoid the secondary growth region. Statistical analysis (ANOVA) was carried using SPSS statistical analysis software using an LSD post-hoc test.
Accession numbers
AGI accession numbers for the genes used in this study are as follows: At3g24770 (CLE41), At5g61480 (PXY), At4g34410 (ERF109), At1g28370 (ERF11), At5g61600 (ERF104), and At1g74930 (ERF018), At4g17500 (AtERF1), At5g47220 (ERF2), At5g47230 (ERF5), At4g17490 (ERF6), At4g11280 (ACS6), At1g54490 (EIN5), At3g15770 (ETO1), At5g65800 (ETO2), At1g66340 (ETR1), At5g03280 (EIN2), At1g46480 (WOX4).
Supporting Information
Zdroje
1. ChaffeyN, CholewaE, ReganS, SundbergB (2002) Secondary xylem development in Arabidopsis: a model for wood formation. Physiologia Plantarum 114 : 594–600.
2. FisherK, TurnerS (2007) PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Current Biology 17 : 1061–1066.
3. HirakawaY, ShinoharaH, KondoY, InoueA, NakanomyoI, et al. (2008) Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proceedings of the National Academy of Sciences, USA 105 : 15208–15213.
4. ItoY, NakanomyoI, MotoseH, IwamotoK, SawaS, et al. (2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313 : 842–845.
5. OelkersK, GoffardN, WeillerG, GresshoffP, MathesiusU, et al. (2008) Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biology 8 : 1.
6. StrabalaTJ, O'DonnellPJ, SmitAM, Ampomah-DwamenaC, MartinEJ, et al. (2006) Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiology 140 : 1331–1344.
7. EtchellsJP, TurnerSR (2010) The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137 : 767–774.
8. WhitfordR, FernandezA, De GroodtR, OrtegaE, HilsonP (2008) Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proceedings of the National Academy of Sciences USA 105 : 18625–18630.
9. HirakawaY, KondoY, FukudaH (2010) TDIF Peptide Signaling Regulates Vascular Stem Cell Proliferation via the WOX4 Homeobox Gene in Arabidopsis. Plant Cell 22 : 2618–2629.
10. SuerS, AgustiJ, SanchezP, SchwarzM, GrebT (2011) WOX4 Imparts Auxin Responsiveness to Cambium Cells in Arabidopsis. The Plant Cell 23 : 3247–3259.
11. JiJ, StrableJ, ShimizuR, KoenigD, SinhaN, et al. (2010) WOX4 Promotes Procambial Development. Plant Physiology 152 : 1346–1356.
12. BrownKM, LeopoldAC (1973) Ethylene and the Regulation of Growth in Pine. Canadian Journal of Forest Research 3 : 142–145.
13. LeopoldAC, BrownKM, EmersonFH (1972) Ethylene in the Wood of Stressed Trees. Horticultural Science 7 : 175.
14. SavidgeRA (1988) Auxin and ethylene regulation of diameter growth in trees. Tree Physiology 4 : 401–414.
15. LoveJ, BjörklundS, VahalaJ, HertzbergM, KangasjärviJ, et al. (2009) Ethylene is an endogenous stimulator of cell division in the cambial meristem of Populus. Proceedings of the National Academy of Sciences, USA 106 : 5984–5989.
16. NakanoT, SuzukiK, FujimuraT, ShinshiH (2006) Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiology 140 : 411–432.
17. KhandelwalA, ElvitigalaT, GhoshB, QuatranoRS (2008) Arabidopsis Transcriptome Reveals Control Circuits Regulating Redox Homeostasis and the Role of an AP2 Transcription Factor. Plant Physiology 148 : 2050–2058.
18. PauwelsL, MorreelK, De WitteE, LammertynF, Van MontaguM, et al. (2008) Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proceedings of the National Academy of Sciences USA 105 : 1380–1385.
19. SiboutR, PlantegenetS, HardtkeCS (2008) Flowering as a Condition for Xylem Expansion in Arabidopsis Hypocotyl and Root. Current Biology 18 : 458–463.
20. RagniL, NieminenK, Pacheco-VillalobosD, SiboutR, SchwechheimerC, et al. (2011) Mobile Gibberellin Directly Stimulates Arabidopsis Hypocotyl Xylem Expansion. The Plant Cell Online 23 : 1322–1336.
21. OyamaT, ShimuraY, OkadaK (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes & Development 11 : 2983–2995.
22. FujimotoSY, OhtaM, UsuiA, ShinshiH, Ohme-TakagiM (2000) Arabidopsis Ethylene-Responsive Element Binding Factors Act as Transcriptional Activators or Repressors of GCC Box-Mediated Gene Expression. The Plant Cell 12 : 393–404.
23. SolanoR, StepanovaA, ChaoQ, EckerJR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes & Development 12 : 3703–3714.
24. AlonsoJM, StepanovaAN, LeisseTJ, KimCJ, ChenH, et al. (2003) Genome-Wide Insertional Mutagenesis of Arabidopsis thaliana. Science 301 : 653–657.
25. NemhauserJL, HongF, ChoryJ (2006) Different Plant Hormones Regulate Similar Processes through Largely Nonoverlapping Transcriptional Responses. Cell 126 : 467–475.
26. GuzmanP, EckerJR (1990) Exploiting the Triple Response of Arabidopsis To Identify Ethylene-Related Mutants. The Plant Cell 2 : 513–523.
27. AlonsoJM, HirayamaT, RomanG, NourizadehS, EckerJR (1999) EIN2, a Bifunctional Transducer of Ethylene and Stress Responses in Arabidopsis. Science 284 : 2148–2152.
28. KieberJJ, RothenbergM, RomanG, FeldmannKA, EckerJR (1993) CTR1, a negative regulator of the ethylene response pathway in arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72 : 427–441.
29. BleeckerAB, EstelleMA, SomervilleC, KendeH (1988) Insensitivity to Ethylene Conferred by a Dominant Mutation in Arabidopsis thaliana. Science 241 : 1086–1089.
30. PotuschakT, VansiriA, BinderBM, LechnerE, VierstraRD, et al. (2006) The Exoribonuclease XRN4 Is a Component of the Ethylene Response Pathway in Arabidopsis. Plant Cell 18 : 3047–3057.
31. OlmedoG, GuoH, GregoryBD, NourizadehSD, Aguilar-HenoninL, et al. (2006) ETHYLENE-INSENSITIVE5 encodes a 5′→3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proceedings of the National Academy of Sciences, USA 103 : 13286–13293.
32. EckerJR (1995) The ethylene signal transduction pathway in plants. Science 268 : 667–675.
33. VogelJP, WoesteKE, TheologisA, KieberJJ (1998) Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proceedings of the National Academy of Sciences USA 95 : 4766–4771.
34. ChaeHS, FaureF, KieberJJ (2003) The eto1, eto2, and eto3 Mutations and Cytokinin Treatment Increase Ethylene Biosynthesis in Arabidopsis by Increasing the Stability of ACS Protein. The Plant Cell 15 : 545–559.
35. YangSF, HoffmanNE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol Plant Mol Biol 35 : 155–189.
36. Ortega-MartínezO, PernasM, CarolRJ, DolanL (2007) Ethylene Modulates Stem Cell Division in the Arabidopsis thaliana Root. Science 317 : 507–510.
37. DietzK-J, VogelM, ViehhauserA (2010) AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245 : 3–14.
38. GuttersonN, ReuberTL (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Current Opinion in Plant Biology 7 : 465–471.
39. SehrEM, AgustiJ, LehnerR, FarmerEE, SchwarzM, et al. (2010) Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. The Plant Journal 63 : 811–822.
40. LorenzoO, PiquerasR, Sanchez-SerranoJJ, SolanoR (2003) ETHYLENE RESPONSE FACTOR1 Integrates Signals from Ethylene and Jasmonate Pathways in Plant Defense. The Plant Cell 15 : 165–178.
41. PauwelsL, InzéD, GoossensA (2009) Jasmonate-inducible gene: what does it mean? Trends in Plant Science 14 : 87–91.
42. PandeySP, RoccaroM, SchönM, LogemannE, SomssichIE (2010) Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. The Plant Journal 64 : 912–923.
43. KerchevPI, PellnyTK, VivancosPD, KiddleG, HeddenP, et al. (2011) The Transcription Factor ABI4 Is Required for the Ascorbic Acid–Dependent Regulation of Growth and Regulation of Jasmonate-Dependent Defense Signaling Pathways in Arabidopsis. The Plant Cell Online 23 : 3319–3334.
44. Cano-DelgadoA, YinYH, YuC, VafeadosD, Mora-GarciaS, et al. (2004) BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131 : 5341–5351.
45. PineauP, FreydierA, RanochaP, JauneauA, TurnerS, et al. (2005) hca: an Arabidopsis mutant exhibiting unusual cambial activity and altered vascular patterning. Plant Journal 44 : 271–289.
46. MahonenAP, BishoppA, HiguchiM, NieminenKM, KinoshitaK, et al. (2006) Cytokinin Signaling and Its Inhibitor AHP6 Regulate Cell Fate During Vascular Development. Science 311 : 94–98.
47. HejatkoJ, RyuH, KimG-T, DobesovaR, ChoiS, et al. (2009) The Histidine Kinases CYTOKININ-INDEPENDENT1 and ARABIDOPSIS HISTIDINE KINASE2 and 3 Regulate Vascular Tissue Development in Arabidopsis Shoots. The Plant Cell 21 : 2008–2021.
48. AgustiJ, HeroldS, SchwarzM, SanchezP, LjungK, et al. (2011) Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proceedings of the National Academy of Sciences
49. MattssonJ, SungZR, BerlethT (1999) Responses of plant vascular systems to auxin transport inhibition. Development 126 : 2979–2991.
50. GälweilerL, GuanC, MüllerA, WismanE, MendgenK, et al. (1998) Regulation of Polar Auxin Transport by AtPIN1 in Arabidopsis Vascular Tissue. Science 282 : 2226–2230.
51. SanchezP, NehlinL, GrebT (2012) From thin to thick: major transitions during stem development. Trends in plant science 17 : 113–121.
52. FletcherLC, BrandU, RunningMP, SimonR, MeyerowitzEM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 : 1911–1914.
53. ClarkSE, WilliamsRW, MeyerowitzEM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 : 575–585.
54. KondoT, SawaS, KinoshitaA, MizunoS, KakimotoT, et al. (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313 : 845–848.
55. OgawaM, ShinoharaH, SakagamiY, MatsubayashiY (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319 : 294–294.
56. BrandU, FletcherJC, HobeM, MeyerowitzEM, SimonR (2000) Dependence of stem cell fate in Arabidopsis an a feedback loop regulated by CLV3 activity. Science 289 : 617–619.
57. SchoofH, LenhardM, HaeckerA, MayerKFX, JurgensG, et al. (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 : 635–644.
58. GordonSP, ChickarmaneVS, OhnoC, MeyerowitzEM (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proceedings of the National Academy of Sciences, USA 106 : 16529–16534.
59. SwarbreckD, WilksC, LameschP, BerardiniTZ, Garcia-HernandezM, et al. (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucl Acids Res 36: D1009–1014.
60. TsangDL, EdmondC, HarringtonJL, NühseTS (2011) Cell Wall Integrity Controls Root Elongation via a General 1-Aminocyclopropane-1-Carboxylic Acid-Dependent, Ethylene-Independent Pathway. Plant Physiology 156 : 596–604.
61. LiC, WongWH (2001) Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proceedings of the National Academy of Sciences, USA 98 : 31–36.
62. BolstadBM, IrizarryRA, AstrandM, SpeedTP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 : 185–193.
63. SmythGK (2004) Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Statistical Applications in Genetics and Molecular Biology 3: Article 3.
64. HardstedtM, FinneganCP, KirchhofN, HylandKA, WijkstromM, et al. (2005) Post-transplant upregulation of chemokine messenger RNA in non-human primate recipients of intraportal pig islet xenografts. Xenotransplantation 12 : 293–302.
65. LongJA, MoanEI, MedfordJI, BartonMK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 : 66–69.
66. PinonV, EtchellsJP, RossignolP, CollierSA, ArroyoJM, et al. (2008) Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135 : 1315–1324.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



