-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaIs a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
Heterozygous mutations in the PRPF31 gene cause autosomal dominant retinitis pigmentosa (adRP), a hereditary disorder leading to progressive blindness. In some cases, such mutations display incomplete penetrance, implying that certain carriers develop retinal degeneration while others have no symptoms at all. Asymptomatic carriers are protected from the disease by a higher than average expression of the PRPF31 allele that is not mutated, mainly through the action of an unknown modifier gene mapping to chromosome 19q13.4. We investigated a large family with adRP segregating an 11-bp deletion in PRPF31. The analysis of cell lines derived from asymptomatic and affected individuals revealed that the expression of only one gene among a number of candidates within the 19q13.4 interval significantly correlated with that of PRPF31, both at the mRNA and protein levels, and according to an inverse relationship. This gene was CNOT3, encoding a subunit of the Ccr4-not transcription complex. In cultured cells, siRNA–mediated silencing of CNOT3 provoked an increase in PRPF31 expression, confirming a repressive nature of CNOT3 on PRPF31. Furthermore, chromatin immunoprecipitation revealed that CNOT3 directly binds to a specific PRPF31 promoter sequence, while next-generation sequencing of the CNOT3 genomic region indicated that its variable expression is associated with a common intronic SNP. In conclusion, we identify CNOT3 as the main modifier gene determining penetrance of PRPF31 mutations, via a mechanism of transcriptional repression. In asymptomatic carriers CNOT3 is expressed at low levels, allowing higher amounts of wild-type PRPF31 transcripts to be produced and preventing manifestation of retinal degeneration.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003040
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003040Summary
Heterozygous mutations in the PRPF31 gene cause autosomal dominant retinitis pigmentosa (adRP), a hereditary disorder leading to progressive blindness. In some cases, such mutations display incomplete penetrance, implying that certain carriers develop retinal degeneration while others have no symptoms at all. Asymptomatic carriers are protected from the disease by a higher than average expression of the PRPF31 allele that is not mutated, mainly through the action of an unknown modifier gene mapping to chromosome 19q13.4. We investigated a large family with adRP segregating an 11-bp deletion in PRPF31. The analysis of cell lines derived from asymptomatic and affected individuals revealed that the expression of only one gene among a number of candidates within the 19q13.4 interval significantly correlated with that of PRPF31, both at the mRNA and protein levels, and according to an inverse relationship. This gene was CNOT3, encoding a subunit of the Ccr4-not transcription complex. In cultured cells, siRNA–mediated silencing of CNOT3 provoked an increase in PRPF31 expression, confirming a repressive nature of CNOT3 on PRPF31. Furthermore, chromatin immunoprecipitation revealed that CNOT3 directly binds to a specific PRPF31 promoter sequence, while next-generation sequencing of the CNOT3 genomic region indicated that its variable expression is associated with a common intronic SNP. In conclusion, we identify CNOT3 as the main modifier gene determining penetrance of PRPF31 mutations, via a mechanism of transcriptional repression. In asymptomatic carriers CNOT3 is expressed at low levels, allowing higher amounts of wild-type PRPF31 transcripts to be produced and preventing manifestation of retinal degeneration.
Introduction
The penetrance of a disease-causing mutation corresponds to the proportion of individuals who carry such variant and develop clinical symptoms. In the majority of Mendelian disorders penetrance is 100%, but incomplete penetrance is far from being uncommon [1]. Although in medical genetics penetrance is still largely uncharacterized at the molecular level, it is usually determined by genetic or epigenetic factors, and sometimes even by environmental modifiers [2].
Retinitis pigmentosa (RP) is a group of inherited degenerative diseases of the retina that cause the progressive death of photoreceptors, the neurons of the eye that are sensitive to light. Typically, patients affected by RP first suffer from night blindness, most often during adolescence. Rod and cone photoreceptor cells start to degenerate from the mid periphery to the far periphery and the center of the retina, resulting in the so-called tunnel vision. Later in life, central vision is also lost, leading to legal or complete blindness [3]. Clinically, RP is a highly-heterogeneous disease, reflecting not only genetic heterogeneity (mutations in different genes), but also inter-individual diversity (penetrance and expressivity) [4].
The PRPF31 gene encodes in humans a pre-mRNA processing factor. In autosomal dominant RP (adRP) due to mutations in PRPF31 penetrance of the disease can be incomplete. Specifically, in families with PRPF31 mutations it is not uncommon to observe the presence of asymptomatic individuals who have affected parents, affected children, or both [5]–[8]. Although they carry the same PRPF31 mutation as their affected relatives, asymptomatic subjects show no visual impairment, even at older ages, and normal to slightly reduced electroretinographic recordings [7].
PRPF31 mutations causing adRP are largely null alleles, such as deletions, nonsenses, or DNA changes leading to premature termination codons and to mRNA degradation [9]–[14]. Patients are therefore hemizygotes for PRPF31, suggesting that the molecular pathophysiology of the disease is due to the functional loss of one allele and to haploinsufficiency [10], [12], [15]. The ubiquitous expression of PRPF31 has allowed a number of functional studies to be performed in immortalized lymphoblastoid cell lines (LCLs) from patients and asymptomatic carriers of mutations [16]–[18]. In particular, it has been shown that penetrance of mutations is due to the differential expression of the PRPF31 allele that is not inactivated by mutations, in both symptomatic and asymptomatic individuals. Unlike affected persons, asymptomatic carriers naturally express high amounts of functional PRPF31 mRNA, a phenomenon that compensates for the mutation-induced loss of one allele and prevents manifestation of symptoms [16]–[18].
This variable expression of PRPF31 seems to be present within the general population [16] and therefore asymptomatic carriers of mutations would be individuals that by chance are “high expressors”. Furthermore, protection from PRPF31 mutations (and therefore variable PRPF31 expression) is itself an inheritable character [16], [19]. In an elegant meta-analytic study, McGee et al. [19] have shown that protective alleles, named isoalleles, are inherited by carriers of PRPF31 mutations from the parent who does not transmit the mutation (i.e. they are in trans with respect to the mutation). Furthermore, such isoalleles would be responsible for the majority of incomplete penetrance cases, and map to chromosome 19q13.4, in proximity to PRPF31 itself [19]. The same study also indicated that these isoalleles were not the only modulators of PRPF31 penetrance, since some individuals with discordant phenotypes carried an identical wild-type haplotype for the isoalleles on chromosome 19. Another genetic element potentially capable of influencing the penetrance of PRPF31 mutation was later mapped to chromosome 14q21–23 [16].
In this study, we search for and identify the major modifier gene responsible for penetrance of PRPF31 mutations, through the analysis of LCLs from a very large family with adRP due to a PRPF31 microdeletion [6], [20].
Results
CNOT3 expression is inversely proportional to that of PRPF31 in asymptomatic and affected carriers of mutations
The region on chromosome 19q13.4 harboring the main modifier gene for PRPF31 penetrance was determined by McGee et al. to lie between microsatellite markers D19S572 and D19S926 [19]. This interval contains 118 genes, including 50 protein-coding genes, 50 miRNAs and 18 pseudogenes.
Based on data from lymphoblast studies describing the nature and the possible mechanism of action of the penetrance modifier gene [16]–[18], we selected protein-coding genes that were consistently expressed in LCLs, as detected by q-PCR (18 genes). We also excluded some of the genes that in this region belong to the leukocyte receptor cluster (LRC) and are implicated exclusively in leukocyte functions. We were left with 10 sequences, namely: NDUFA3, TFPT, CNOT3, LENG1, MBOAT7, TSEN34, RPS9, LILRB3, ILT7, and NALP2. We then measured by q-PCR the mRNA expression levels of these genes in LCLs from 4 asymptomatic and 6 affected individuals from the RP856/AD5 family (Table S1 and Figure S1). All genes showed consistent expression across the family members. Of these, only CNOT3 showed a statistically significant difference in mRNA expression between the two groups of individuals (p<0.01) (Figure 1 and Figure S1). Unexpectedly, CNOT3 trend of expression was the opposite to that of PRPF31, as it showed lower expression in asymptomatic than in the affected carriers of PRPF31 mutations (Figure 1B). This phenomenon was particularly clear when expression of CNOT3 and PRPF31 were paired by cell lines and the relevant regression lines calculated (Figure 1C).
Fig. 1. CNOT3 shows an opposite trend of expression with respect to that of PRPF31 between the asymptomatic (AS) and affected (AF) individuals of the AD5 family. 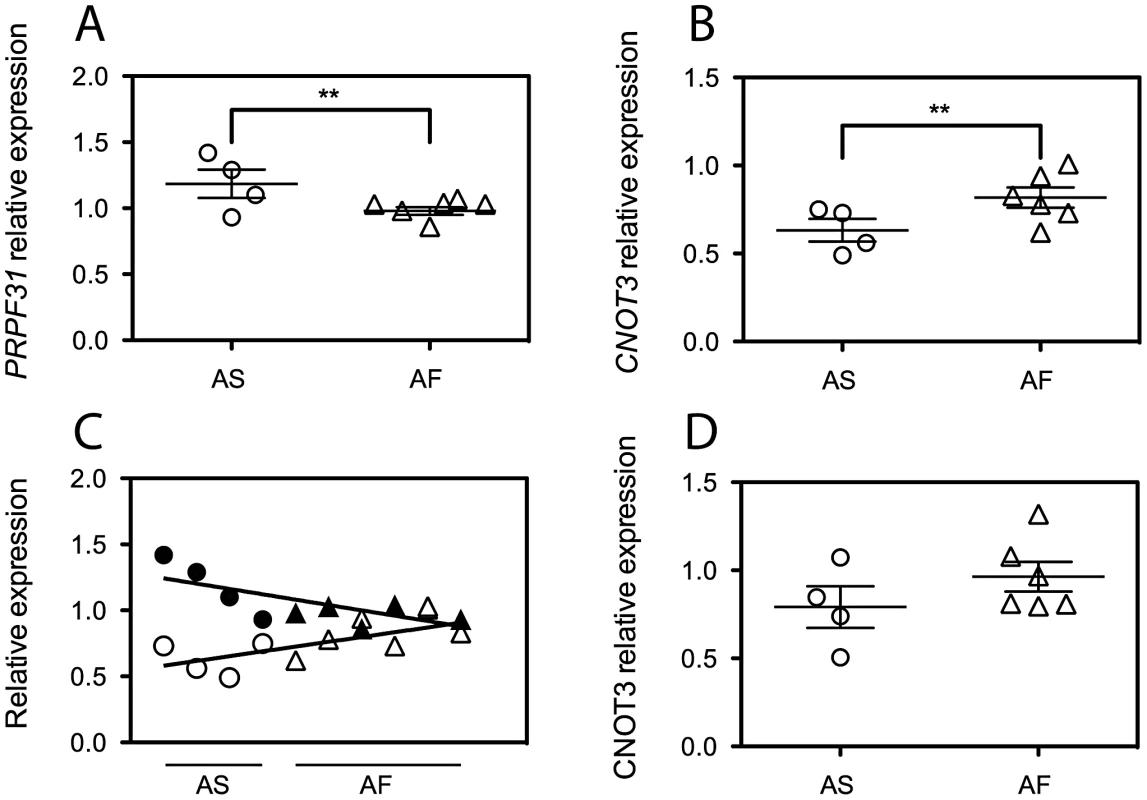
(A) PRPF31 mRNA expression normalized to the housekeeping gene GAPDH. Error bars refer to the standard deviation of the mean for 5 independent experiments for each group. (B) CNOT3 mRNA expression from the same 5 experiments used to generate PRPF31 data. **, p<0.01. (C) Linear regression analysis of PRPF31 and CNOT3 mRNA expression, which shows an inverse trend of the two genes in each cell line. Circles, asymptomatic subjects; triangles, affected individuals; open symbols, CNOT3 expression; Filled symbols, PRPF31 expression. Data having the same value for the x axis have been obtained from the same individual. (D) Quantification of CNOT3 protein abundance relative to β-actin from 3 independent SDS-PAGE gels, after simultaneous detection of the two proteins by quantitative LI-COR western blot. Assessment of CNOT3 protein by quantitative western blotting confirmed the differential expression detected by q-PCR (Figure 1D).
CNOT3 is a negative regulator of PRPF31 expression
CNOT3 belongs to the Ccr4-Not complex, a conserved multi-protein structure involved in the regulation of gene expression [21].
To investigate if CNOT3 could influence PRPF31 expression, we silenced its expression in ARPE-19 cell lines, by using two different siRNA sequences. Suppression of CNOT3 resulted in significant increase of PRPF31 mRNA and protein (p<0.001, Figure 2). This effect was very specific, as no influence was observed in negative controls and in TFPT expression, a neighboring gene sharing part of the promoter with PRPF31 (Figure S2).
Fig. 2. CNOT3 silencing stimulates PRPF31 expression in ARPE-19 cells. 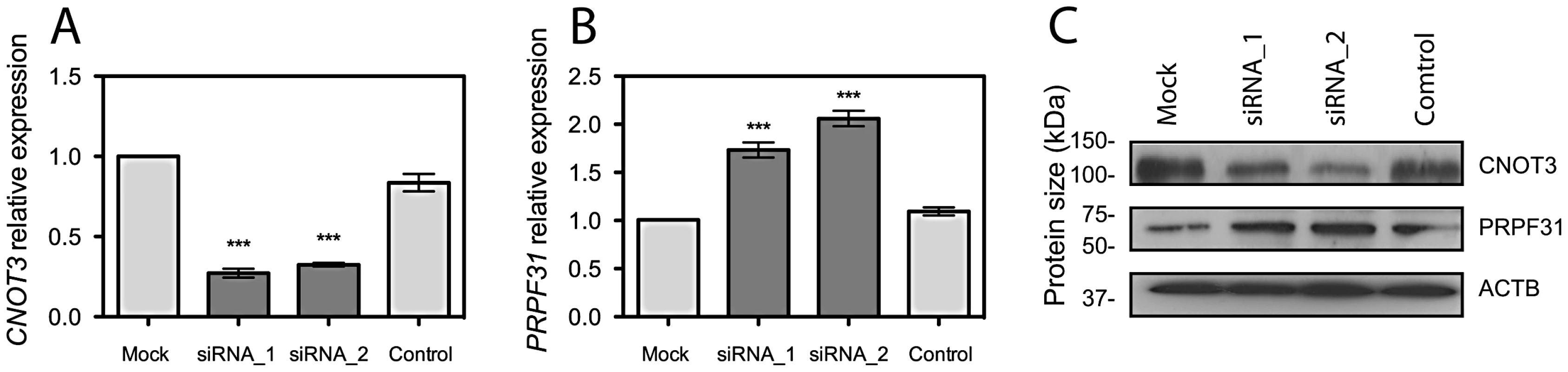
(A) CNOT3 mRNA depletion by 2 different siRNA sequences and its effect on PRPF31 mRNA expression (B). ***, p<0.001. (C) Representative western blot of CNOT3 silencing and effect on PRPF31 protein expression. siRNA_1 and siRNA_2, different CNOT3-specific siRNA sequences; Control, treatment with transfection reagent with no siRNA; Mock, treatment with transfection reagent and scrambled siRNA. CNOT3-dependent modulation of PRPF31 expression is achieved at the transcriptional level
CNOT3 can negatively regulate transcription by either directly binding to the promoter of target genes or by affecting their mRNA rate of degradation [22], [23].
To understand which could be the mechanism through which CNOT3 modulates PRPF31 expression, we incubated LCLs from two asymptomatic-affected pairs with Actinomycin D, a drug that inhibits de novo transcription, and then measured the rate of decay of PRPF31 mRNA. No statistically significant difference was observed between the asymptomatic and affected individuals (Figure S3), suggesting that the modulation of PRPF31 expression happens most probably at the transcriptional level.
CNOT3 binds directly to the PRPF31 promoter
To test this hypothesis, we performed a Chromatin ImmunoPrecipitation (ChIP) assay in LCLs from 3 healthy individuals, using an anti-CNOT3 antibody and serum IgG as a negative control. To confirm that CNOT3 enrichment of a target DNA region was due to a specific immunoprecipitation rather than to a random precipitation of DNA, we designed primers targeting genomic regions that were not supposed to be bound by CNOT3. Primers targeting CNOT3 promoter were used as a positive control, since it has been previously shown that CNOT3 self-regulates its expression by binding to its own promoter [23]. Both qualitative and quantitative PCR showed a statistically significant enrichment in PRPF31 promoter sequences in DNA that was immunoprecipitated by the CNOT3 antibody, compared to that exposed to serum IgG (Figure 3A, 3B).
Fig. 3. CNOT3 binds to the PRPF31 promoter in cells. 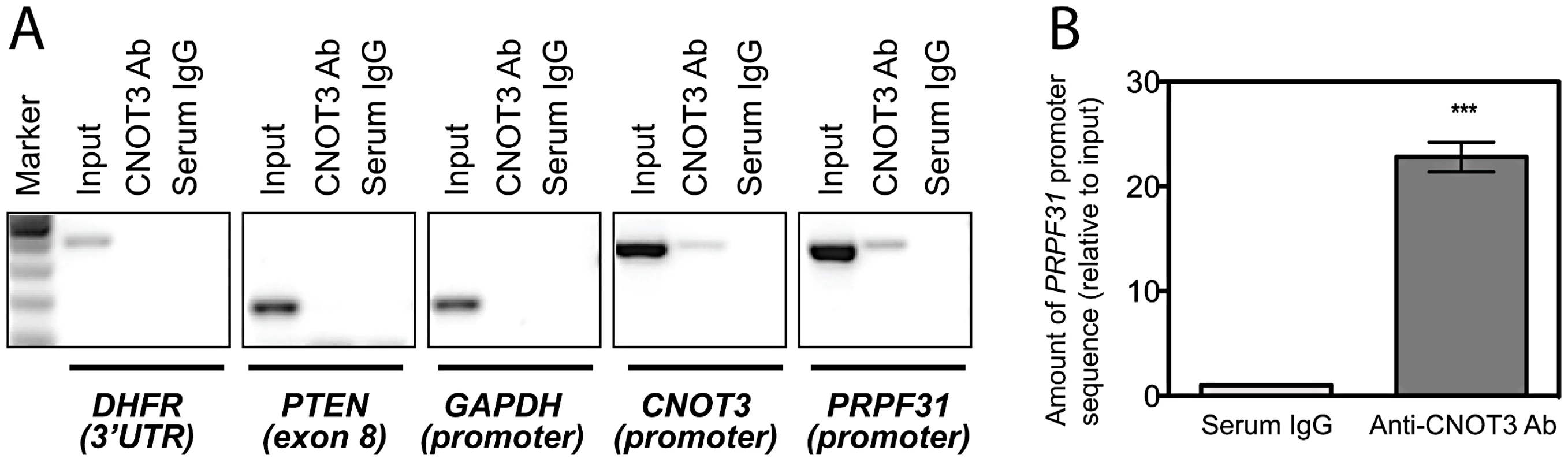
(A) CNOT3 ChIP-PCRs on different target sequences. Enrichment is visible only for PRPF31 promoter and CNOT3 promoter (positive control); DHFR 3′UTR, PTEN exon8, and GAPDH promoter sequences are all negative controls. (B) CNOT3 ChIP-q-PCR on PRPF31 promoter sequence. Error bars indicate the standard deviation of the mean for three independent ChIP-qPCR experiments. Serum IgG is used as IP negative control. ***, p<0.001. CNOT3 rs4806718 alleles are associated with the clinical manifestation of the disease
In order to identify genetic markers that could be associated with variable expression of CNOT3 and therefore with penetrance of PRPF31 mutations, we sequenced the entire CNOT3 genomic region by next-generation sequencing (NGS) in one asymptomatic-affected sibling pair. We identified five polymorphic variants (rs36643, rs56079424, rs36661, rs4806718, rs1055234) that differed between the two subjects. These five variants were subsequently analyzed in a second asymptomatic-affected sibling pair from the same pedigree, showing that only alleles of rs4806718, lying in intron 17 of CNOT3, segregated with the trait.
This SNP was then sequenced in a total of 38 asymptomatic and affected individuals from the RP856/AD5 family, as well as from an unrelated family for which the modifier gene for PRPF31 penetrance was also found to be linked to chromosome 19q13.4 [24] (Figure 4). Association between the C allele of rs4806718 with the affected status and the T allele with the asymptomatic status was moderately significant (p = 0.04, by Fisher exact test).
Fig. 4. Analysis of rs4806718 alleles in two unrelated pedigrees. 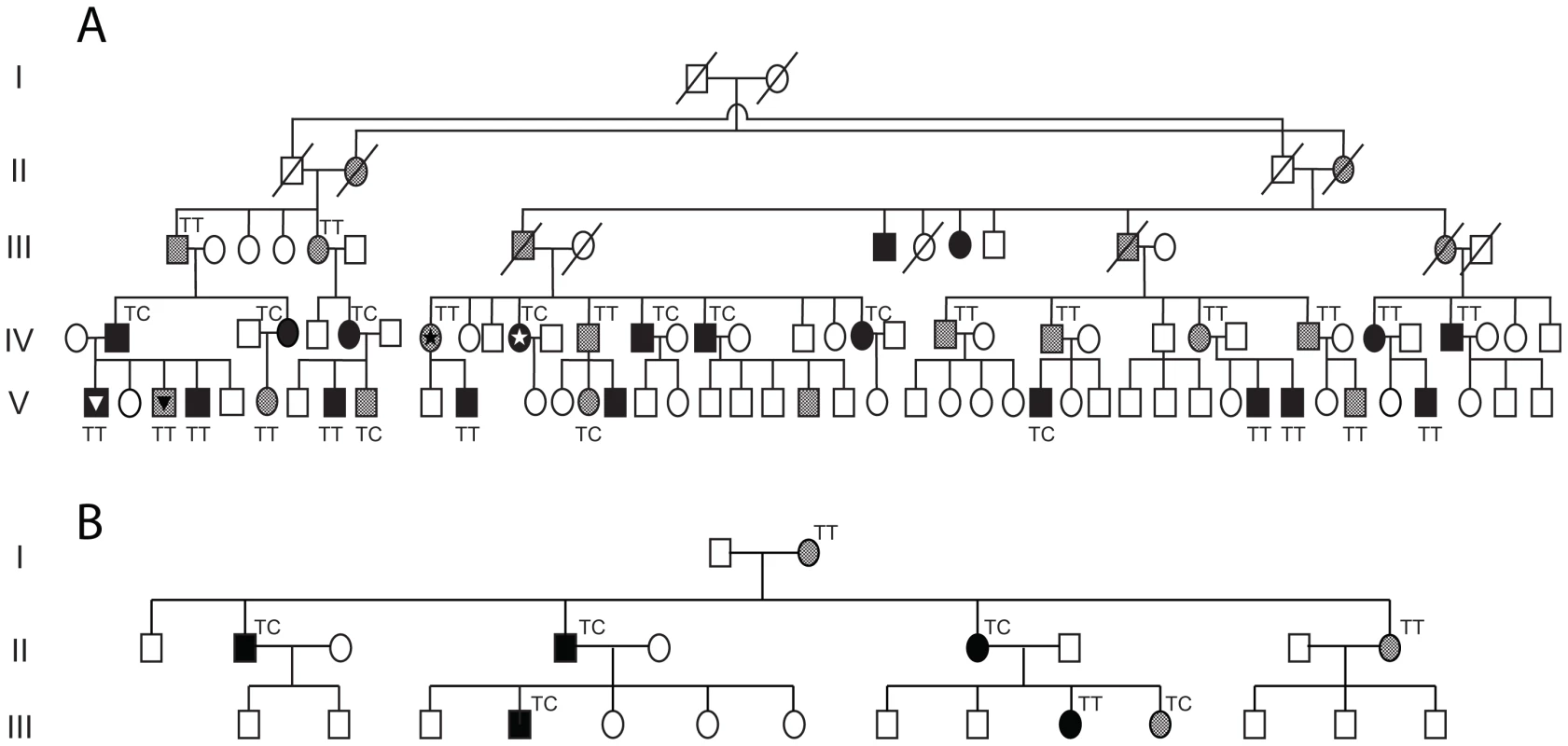
(A) Family RP856/AD5. The individuals initially tested with NGS are marked with a star. The individuals marked with a triangle belong to a sibship pair, which was previously shown by McGee et al. to have the same isoallele haplotype but different phenotypes. (B) Family ADB1, a Bulgarian gypsy family carrying a heterozygous splice site mutation in PRPF31 (NM_015629.3:c.527+1G>T, or IVS6+1G>T). In both pedigrees carriers of mutations are either in black (affected individuals) or in grey (asymptomatic individuals). Discussion
Despite penetrance being an old concept in genetics, little is known about its molecular causes, especially in inherited human diseases. Notable positive examples include dominant erythropoietic protoporphyria, caused by mutations in the FECH gene, and dominant elliptocytosis, due to mutations in SPTA1. In these disorders, an imbalance of expression between the wild-type and the mutated alleles causes the manifestation of the symptoms [25]–[27].
Similar mechanisms determine penetrance of PRPF31 mutations, since asymptomatic carriers are individuals who display increased levels of wild-type mRNA alleles, which in turn compensate for the deficiency caused by the mutation [16]–[18]. However, unlike erythropoietic protoporphyria and elliptocytosis, in PRPF31-linked adRP the molecular causes of such beneficial hyper-expression have remained, up to now, unexplained. Previous mapping studies have shown that the penetrance and expression of PRPF31 is influenced by at least two loci: one, likely having a major effect, lies within the same chromosomal region as PRPF31 (proximal modifier), the other is on chromosome 14 (distant modifier) [16], [19]. Our previous work has also demonstrated that both modifiers would act through diffusible elements (e.g. transcription factors) since their effects on PRPF31 mRNA expression concerns equally both copies of the gene [16]. This observation probably explains the failure of previous attempts to identify the proximal modifier as a polymorphic variant of the PRPF31 sequence itself, according to the FECH or SPTA1 models.
Based on this previous knowledge, we reasoned that the expression of the proximal modifier of PRPF31 mutations should correlate with that of PRPF31. Therefore we started assessing mRNA levels of genes that reside within the mapped 19q13.4 interval, by using the same cellular model successfully used in previous studies of PRPF molecular genetics, and in particular of PRPF31 penetrance [10], [15]–[18], [28], [29]. Specifically, we studied cells derived from members of one of the largest pedigrees known to segregate a PRPF31 mutation, family RP856/AD5 [6], [20], for which incomplete penetrance could also be, at least in part, determined by the proximal modifier [19]. Following a filtering process based on both in silico analyses and on mRNA expression, we were left with only 10 candidates. Of these, only one, CNOT3, showed a pattern of expression that significantly correlated to that of PRPF31. Interestingly, its trend of expression was inverse to that of PRPF31, raising the possibility that CNOT3 may be a negative regulator of PRPF31 expression.
CNOT3 encodes a protein that is part of the Ccr4-Not multi-subunit complex, an evolutionary conserved multimeric structure involved in modulation of gene expression [21], [30]–[34]. Evidences that CNOT3 could be a negative regulator of transcription have been provided in yeast [31], and then confirmed in human cell lines, by the identification of a conserved motif at its C-terminus, called the Not-Box. This motif was originally identified in another subunit of the complex, CNOT2, where it was shown to repress reporter gene activity upon promoter targeting [35]. We confirmed the role of CNOT3 as a negative regulator of PRPF31 expression by siRNA-mediated silencing experiments in ARPE-19 cells. Specifically, we observed that 70% depletion of CNOT3 induced approximately a 2-fold increase in PRPF31 expression, but had no effects on TFPT, a gene that is contiguous to PRPF31 and shares with it part of the promoter [36].
CNOT3 can modulate transcription of its targets by the direct binding to their promoters [23] or by promoting the recruitment of deadenylases at the 3′ end of their transcripts [22]. Our data provide evidence showing that regulation of PRPF31 expression should be mainly at the transcriptional level. First, we observed that decay of PRPF31 mRNA was roughly the same in cells from individuals expressing different levels of CNOT3, disfavoring gene modulation through post-transcriptional mechanisms. Second, we showed by ChIP that CNOT3 could bind directly to the bona fide PRPF31 promoter.
In their work, McGee et al. identified the chromosomal interval containing the proximal modifier through linkage analysis, a technique that searches for relationships between phenotypes and physical elements on the DNA sequence [19]. This implies that variable expression of CNOT3 must be determined by a DNA variant that is present in this same region, possibly within CNOT3 itself. Given their supposedly high frequency within the general population, these isoalleles would very likely be polymorphic elements. Our search for CNOT3 DNA changes that would be present in asymptomatic but not in affected carriers of mutations (or vice versa) resulted in the identification of particular alleles of rs4806718.
Are these the isoalleles originally mapped by McGee et al.? Although statistically significant, the association between rs4806718's C allele and disease (and the T allele with an unaffected status) was not perfect. This phenomenon can be explained by the presence of additional factors capable of determining PRPF31 penetrance, such as the one mapped on chromosome 14 [16]. These modifiers could interfere with or even mask the effects of rs4806718 alleles, ultimately allowing the “wrong” rs4806718 variant to be associated with either phenotype. Such a hypothesis is in perfect agreement with the original data on PRPF31 isoalleles, as a few discordant phenotype-genotype associations concerning the mapped locus for the proximal modifier were also clearly recognized. Amongst other examples, 2 siblings from the last generation of RP856/AD5 had discordant phenotypes but concordant haplotypes [19], [37]. These same individuals, genotyped by us at the rs4806718 locus, were found indeed to share the same parental allele. Furthermore, if the modifier allele is truly inherited from the parent who does not transmit the mutation, then the chance that this does not forcibly correspond to an rs4806718 allele is relatively high in RP856/AD5, given the number of spouses external to the family who are present in this pedigree.
Another important element to consider is whether rs4806718 alleles have a direct effect on CNOT3 expression, or whether the two factors are simply in linkage disequilibrium with other elements (e.g. transcription enhancers) lying somewhere else in the region. According to in silico prediction tools, the rs4806718 C variant, which has a frequency of 0.38 in the European population, could affect CNOT3 splicing by decreasing the binding energy for one acceptor splice site. Therefore, at least potentially, rs4806718 alleles could represent the true PRPF31 isoalleles.
Taken together, all our observations suggest that CNOT3 is the modifier gene on chromosome 19q13.4 that is responsible for penetrance of PRPF31 mutations. Through direct repression of PRPF31 transcription and in virtue of its own variable expression, CNOT3 would differentially reduce the amount of available PRPF31 mRNA, thus determining incomplete penetrance. Although further studies on the physiological role of CNOT3 in human cells and tissues are definitely needed, our data open the way for a possible treatment of PRPF31-linked RP through the inhibition of this transcriptional regulator.
Materials and Methods
Patients and cell lines
This study involved 10 individuals from the British family RP856/AD5, segregating an 11-bp deletion in exon 11 of PRPF31 (c.1115_1125del) [6], [20]. Our research has been conducted in accordance with the tenets of the Declaration of Helsinki and has been approved by the IRBs of our Institutions. Lymphoblastoid cell lines derived from peripheral blood leukocytes of each individual were either obtained from the Coriell Cell Repositories or through the immortalization of peripheral blood leukocytes. Cells were grown and maintained as previously described [18].
The human retinal pigment epithelial cell line ARPE-19 (kindly provided by Dr. Yvan Arsenijevic) was grown and maintained at 37°C with 5% CO2 in N1 medium (DMEM/F12 complemented with 2.5 mM L-glutamine, 56 mM NaHCO3, and 10% fetal bovine serum).
RNA extraction and cDNA synthesis
Lymphoblasts were harvested during their exponential growth phase (500,000–1,000,000 cells/ml) and RNA was isolated from 107 cells using the QIAGEN RNeasy Mini Kit, following the manufacturer's instructions. The only modification to the protocol concerned the DNase treatment, since we used double the amount of enzyme compared to the suggested quantity. RNA concentration was measured with the Dropsense 96 spectrophotometer (Trinean). cDNA synthesis was carried out as previously described [10].
q–PCR primer design and optimization
Most of the primer sequences used in this study were annotated in the qPrimerDepot database (http://primerdepot.nci.nih.gov/). These sequences are specifically designed to span exon-exon junctions, thus avoiding genomic DNA to be amplified during q-PCR. To design other primer sequences, which were not present in the qPrimerDepot database, we used the Primer Blast tool from NCBI (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). To validate each primer pair for q-PCR we first optimized the primer amounts (50–200 nM), and then loaded 10 µl of the q-PCR product obtained on a 1% agarose gel, in order to check the specificity of the amplification product. Finally, a standard curve using a control cDNA template was used to test each primer pair's efficiency. We considered as acceptable ranges of efficiency between 90 and 110%, corresponding to standard curve slopes between −3.6 and −3.1. All primer pairs used for this study are listed in Table S2. For GAPDH and PRPF31 amplification we used primers and probes previously described [16].
Real-time quantitative PCR
All genes but PRPF31 and GAPDH were amplified with the Sybr Green PCR Master Mix (Applied Biosystems). Q-PCR reactions were performed as published [16]. After having assessed that PCR efficiencies for all genes were comparable, mRNA expression of each of them was normalized with respect to GAPDH, using the ΔΔCt method.
Protein extraction
Total protein was extracted from lymphoblastoid cell lines in RIPA buffer as reported before [10]. ARPE-19 whole cell lysate was obtained by scraping the cells into 150 µl of lysis buffer (20 mM Tris HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 2 mM EDTA, 1% TritonX-100) complemented with protease and phosphatase inhibitors, and incubated on ice for 15 minutes followed by a centrifugation at 14.000 rpm for 30 minutes at 4°C. Proteins concentration was measured with the BCA protein assay kit (Pierce), using BSA to generate a standard curve.
Western blot
Anti-PRPF31 antibody was raised in rabbit as previously described [10]. Rabbit anti-CNOT3 antibody was purchased by Bethyl Laboratories. This targets residues 525 to 575 of the human CNOT3 protein (NP_055331.1), allowing detection of a 117-kDa protein. Mouse anti-β-actin antibody (Sigma) was used as a loading control.
Equal amounts of proteins were loaded and run on an 8% SDS-PAGE gel. Proteins were transferred to a nitrocellulose membrane and blocked in 5% milk overnight at 4°C or alternatively for 1 hour at room temperature. The incubation of all primary antibodies was performed for 1 hour at room temperature using the following dilutions: anti-PRPF31 (1∶500), anti-CNOT3 (1∶2,000), and anti-β-ACTIN (1∶2,500). The membrane was washed 3 times with 0.05% Tween-20 in TBS. Rabbit and mouse HRP-conjugated secondary antibodies were diluted 1∶1,000 in 2% milk and incubated for 1 hour at room temperature. Bands were detected using enhanced chemioluminescence (Pierce).
Signal detection via the Odyssey infrared imaging system (LI-COR) was performed by using fluorescently-labeled secondary antibodies provided by LI-COR, diluted 1∶5,000 in 0.5% milk and incubated in the dark, for 1 hour at room temperature. The membrane was then washed twice with 0.05% Tween-20 in TBS and once in PBS to remove residual Tween-20 prior to the laser scanning.
In vitro silencing experiments
We used two different siRNA sequences targeting CNOT3 (QIAGEN, FlexiTube siRNA, Hs_CNOT3_5 and Hs_CNOT3_8, 1 nmol) and a negative control siRNA for human genes (Santa Cruz Biotechnology). One day before transfection ARPE-19 cells were seeded at a concentration of 2×105 cells/well in a 6 well-plate, and transfection was achieved by using 5 µl Lipofectamine (Invitrogen) and 50 pmol siRNA. RNA was extracted 48 hrs after transfection.
Actinomycin D treatment of cells
Lymphoblasts grown at a concentration of ∼8 million cells in a T75 flask were treated with Actinomycin D (5 µg/ml in DMSO) (Sigma) by adding it directly to the medium. Cell pellets were collected at seven different time points (0–24 hrs) and total RNA was extracted and analyzed by q-PCR.
Chromatin immunoprecipitation (ChIP)
Three control lymphoblastoid cells from the Centre d'Etude du Polymorphisme Humain (CEPH) were grown to have 107 cells per ChIP experiment. DNA and proteins were cross-linked by adding 1% formaldehyde directly to the medium and by incubating the cells on a rotating hybridization oven at 37°C for 10 minutes. To quench cross-linking, we then added 125 mM glycine and incubated the cells at 37°C for 5 minutes. Cells were pelleted by centrifugation (800 g for 5 minutes at 4°C) and washed twice with cold PBS, supplemented with protease inhibitors. Optimization of the chromatin shearing was performed by using a Covaris sonicator, to obtain on average cross-linked DNA fragments of 150–400 bp. ChIP was performed using buffers provided with the Ep-iT Chromatin Immunoprecipitation kit (Bio-AAA). Immunoprecipitation was performed using three different antibodies: anti-CNOT3, anti-pol2 (Bio-AAA) as a positive control for IP, and serum IgG (Santa Cruz Biotechnology) as a negative control for IP. Antibody-protein-DNA complexes were collected on protein A agarose beads (2 hrs, 4°C), then washed with the low salt buffer, high salt buffer, LiCl buffer, and TE buffer (pH 8.0) provided in the kit to remove non-specific binding. Complexes were eluted from the beads by using the elution buffer (0.1 mM NaHCO3 and 1% SDS) in an orbital shaker. Cross-links were removed by an overnight incubation at 65°C. Ribonuclease and proteinase K digestion were added to remove specific contaminants, before the eluted DNA was extracted once in 25∶24∶1 phenol-chloroform-isoamyl alcohol and once in 24∶1 chloroform-isoamyl alcohol. DNA was ethanol precipitated, washed in 70% ethanol, and finally eluted in TE.
ChIP-PCR was performed using the GoTaq DNA Polymerase (Promega) and 0.5 µl of the ChIP DNA, by using standard cycling conditions and primers described in Table S3. GAPDH primer sequences are the ones provided by Millipore for the EZ-ChIP kit, while primers for DHFR have been previously described [38].
Two microliters of ChIP DNA were also amplified by q-PCR using Sybr Green PCR Master Mix (Applied Biosystems) and the PRPF31 promoter primer pair (Table S3).
Ultra-high-throughput sequencing
CNOT3 genomic region was amplified by 3 overlapping long-range PCRs (Table S4), for a total length of 34 Kb. PCR was performed in 20 µl using TaKaRa LA Taq and GC buffer I (Takara Bio Inc.). Final primers concentration was 1 µM, and 200 ng of genomic DNA were used as template. PCR amplification conditions were: an initial step at 94°C for 1 minute, 30 cycles of denaturation at 98°C for 5 seconds and annealing/extension at 68°C for 15 minutes, and a final extension step at 72°C for 10 minutes. Long-range PCR products were sequenced with an Illumina HiSeq 2000 machine, to obtain coverage values in the range of thousands of reads. Mapping of the reads and variant detection was performed by using the CLCbio Genomics Workbench software.
Statistical analysis
Differences of gene expression between asymptomatic and affected individuals were tested by t-test, and likelihood computed by 100 Monte Carlo label-swapping simulations per each gene.
One-way ANOVA followed by Bonferroni's multiple comparison tests was used to analyze the effect of CNOT3 silencing on the expression of the target genes. The enrichment of PRPF31 promoter sequence after CNOT3 immunoprecipitation compared to the serum IgG was evaluated by using the Mann Whitney non-parametric statistical hypothesis test.
In figures, p<0.05 is indicated by one star, p<0.01 by 2 stars, and p<0.001 by 3 stars.
Supporting Information
Zdroje
1. AhluwaliaJK, HariharanM, BargajeR, PillaiB, BrahmachariV (2009) Incomplete penetrance and variable expressivity: is there a microRNA connection? Bioessays 31 : 981–992.
2. ZlotogoraJ (2003) Penetrance and expressivity in the molecular age. Genet Med 5 : 347–352.
3. BersonEL (1993) Retinitis pigmentosa. The Friedenwald Lecture. Invest Ophthalmol Vis Sci 34 : 1659–1676.
4. HartongDT, BersonEL, DryjaTP (2006) Retinitis pigmentosa. Lancet 368 : 1795–1809.
5. EvansK, al-MaghthehM, FitzkeFW, MooreAT, JayM, et al. (1995) Bimodal expressivity in dominant retinitis pigmentosa genetically linked to chromosome 19q. Br J Ophthalmol 79 : 841–846.
6. MooreAT, FitzkeF, JayM, ArdenGB, InglehearnCF, et al. (1993) Autosomal dominant retinitis pigmentosa with apparent incomplete penetrance: a clinical, electrophysiological, psychophysical, and molecular genetic study. Br J Ophthalmol 77 : 473–479.
7. BersonEL, SimonoffEA (1979) Dominant retinitis pigmentosa with reduced penetrance. Further studies of the electroretinogram. Arch Ophthalmol 97 : 1286–1291.
8. BersonEL, GourasP, GunkelRD, MyrianthopoulosNC (1969) Dominant retinitis pigmentosa with reduced penetrance. Arch Ophthalmol 81 : 226–234.
9. RoseAM, MukhopadhyayR, WebsterAR, BhattacharyaSS, WaseemNH (2011) A 112 kb deletion in chromosome 19q13.42 leads to retinitis pigmentosa. Invest Ophthalmol Vis Sci 52 : 6597–6603.
10. Rio FrioT, WadeNM, RansijnA, BersonEL, BeckmannJS, et al. (2008) Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay. J Clin Invest 118 : 1519–1531.
11. WaseemNH, VaclavikV, WebsterA, JenkinsSA, BirdAC, et al. (2007) Mutations in the gene coding for the pre-mRNA splicing factor, PRPF31, in patients with autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci 48 : 1330–1334.
12. Abu-SafiehL, VithanaEN, MantelI, HolderGE, PelosiniL, et al. (2006) A large deletion in the adRP gene PRPF31: evidence that haploinsufficiency is the cause of disease. Mol Vis 12 : 384–388.
13. SullivanLS, BowneSJ, SeamanCR, BlantonSH, LewisRA, et al. (2006) Genomic rearrangements of the PRPF31 gene account for 2.5% of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci 47 : 4579–4588.
14. VithanaEN, Abu-SafiehL, AllenMJ, CareyA, PapaioannouM, et al. (2001) A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell 8 : 375–381.
15. TanackovicG, RansijnA, ThibaultP, Abou ElelaS, KlinckR, et al. (2011) PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum Mol Genet 20 : 2116–2130.
16. Rio FrioT, CivicN, RansijnA, BeckmannJS, RivoltaC (2008) Two trans-acting eQTLs modulate the penetrance of PRPF31 mutations. Hum Mol Genet 17 : 3154–3165.
17. RivoltaC, McGeeTL, Rio FrioT, JensenRV, BersonEL, et al. (2006) Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum Mutat 27 : 644–653.
18. VithanaEN, Abu-SafiehL, PelosiniL, WinchesterE, HornanD, et al. (2003) Expression of PRPF31 mRNA in patients with autosomal dominant retinitis pigmentosa: a molecular clue for incomplete penetrance? Invest Ophthalmol Vis Sci 44 : 4204–4209.
19. McGeeTL, DevotoM, OttJ, BersonEL, DryjaTP (1997) Evidence that the penetrance of mutations at the RP11 locus causing dominant retinitis pigmentosa is influenced by a gene linked to the homologous RP11 allele. Am J Hum Genet 61 : 1059–1066.
20. al-MaghthehM, InglehearnCF, KeenTJ, EvansK, MooreAT, et al. (1994) Identification of a sixth locus for autosomal dominant retinitis pigmentosa on chromosome 19. Hum Mol Genet 3 : 351–354.
21. CollartMA, PanasenkoOO (2012) The Ccr4–not complex. Gene 492 : 42–53.
22. MoritaM, OikeY, NagashimaT, KadomatsuT, TabataM, et al. (2011) Obesity resistance and increased hepatic expression of catabolism-related mRNAs in Cnot3+/ − mice. EMBO J 30 : 4678–4691.
23. HuG, KimJ, XuQ, LengY, OrkinSH, et al. (2009) A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 23 : 837–848.
24. ChakarovaCF, CherninkovaS, TournevI, WaseemN, KanevaR, et al. (2006) Molecular genetics of retinitis pigmentosa in two Romani (Gypsy) families. Mol Vis 12 : 909–914.
25. GratzerW (1994) Human genetics. Silence speaks in spectrin. Nature 372 : 620–621.
26. GouyaL, PuyH, RobreauAM, BourgeoisM, LamorilJ, et al. (2002) The penetrance of dominant erythropoietic protoporphyria is modulated by expression of wildtype FECH. Nat Genet 30 : 27–28.
27. GouyaL, PuyH, RobreauAM, LyoumiS, LamorilJ, et al. (2004) Modulation of penetrance by the wild-type allele in dominantly inherited erythropoietic protoporphyria and acute hepatic porphyrias. Hum Genet 114 : 256–262.
28. IvingsL, TownsKV, MatinMA, TaylorC, PonchelF, et al. (2008) Evaluation of splicing efficiency in lymphoblastoid cell lines from patients with splicing-factor retinitis pigmentosa. Mol Vis 14 : 2357–2366.
29. Rio FrioT, McGeeTL, WadeNM, IseliC, BeckmannJS, et al. (2009) A single-base substitution within an intronic repetitive element causes dominant retinitis pigmentosa with reduced penetrance. Hum Mutat 30 : 1340–1347.
30. WinklerGS, MulderKW, BardwellVJ, KalkhovenE, TimmersHT (2006) Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J 25 : 3089–3099.
31. CollartMA, StruhlK (1994) NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev 8 : 525–537.
32. LauNC, KolkmanA, van SchaikFM, MulderKW, PijnappelWW, et al. (2009) Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J 422 : 443–453.
33. AlbertTK, LemaireM, van BerkumNL, GentzR, CollartMA, et al. (2000) Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic Acids Res 28 : 809–817.
34. KerrSC, AzzouzN, FuchsSM, CollartMA, StrahlBD, et al. (2011) The Ccr4-Not complex interacts with the mRNA export machinery. PLoS ONE 6: e18302 doi:10.1371/journal.pone.0018302
35. ZwartjesCG, JayneS, van den BergDL, TimmersHT (2004) Repression of promoter activity by CNOT2, a subunit of the transcription regulatory Ccr4-not complex. J Biol Chem 279 : 10848–10854.
36. RoseAM, ShahAZ, WaseemNH, ChakarovaCF, AlfanoG, et al. (2012) Expression of PRPF31 and TFPT: regulation in health and retinal disease. Hum Mol Genet In press.
37. Al-MaghthehM, VithanaE, TarttelinE, JayM, EvansK, et al. (1996) Evidence for a major retinitis pigmentosa locus on 19q13.4 (RP11) and association with a unique bimodal expressivity phenotype. Am J Hum Genet 59 : 864–871.
38. OberleyMJ, InmanDR, FarnhamPJ (2003) E2F6 negatively regulates BRCA1 in human cancer cells without methylation of histone H3 on lysine 9. J Biol Chem 278 : 42466–42476.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



