-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaBlockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
Piwi-interacting RNAs are a diverse class of small non-coding RNAs implicated in the silencing of transposable elements and the safeguarding of genome integrity. In mammals, male germ cells express two genetically and developmentally distinct populations of piRNAs at the pre-pachytene and pachytene stages of meiosis, respectively. Pre-pachytene piRNAs are mostly derived from retrotransposons and required for their silencing. In contrast, pachytene piRNAs originate from ∼3,000 genomic clusters, and their biogenesis and function remain enigmatic. Here, we report that conditional inactivation of the putative RNA helicase MOV10L1 in mouse spermatocytes produces a specific loss of pachytene piRNAs, significant accumulation of pachytene piRNA precursor transcripts, and unusual polar conglomeration of Piwi proteins with mitochondria. Pachytene piRNA–deficient spermatocytes progress through meiosis without derepression of LINE1 retrotransposons, but become arrested at the post-meiotic round spermatid stage with massive DNA damage. Our results demonstrate that MOV10L1 acts upstream of Piwi proteins in the primary processing of pachytene piRNAs and suggest that, distinct from pre-pachytene piRNAs, pachytene piRNAs fulfill a unique function in maintaining post-meiotic genome integrity.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003038
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003038Summary
Piwi-interacting RNAs are a diverse class of small non-coding RNAs implicated in the silencing of transposable elements and the safeguarding of genome integrity. In mammals, male germ cells express two genetically and developmentally distinct populations of piRNAs at the pre-pachytene and pachytene stages of meiosis, respectively. Pre-pachytene piRNAs are mostly derived from retrotransposons and required for their silencing. In contrast, pachytene piRNAs originate from ∼3,000 genomic clusters, and their biogenesis and function remain enigmatic. Here, we report that conditional inactivation of the putative RNA helicase MOV10L1 in mouse spermatocytes produces a specific loss of pachytene piRNAs, significant accumulation of pachytene piRNA precursor transcripts, and unusual polar conglomeration of Piwi proteins with mitochondria. Pachytene piRNA–deficient spermatocytes progress through meiosis without derepression of LINE1 retrotransposons, but become arrested at the post-meiotic round spermatid stage with massive DNA damage. Our results demonstrate that MOV10L1 acts upstream of Piwi proteins in the primary processing of pachytene piRNAs and suggest that, distinct from pre-pachytene piRNAs, pachytene piRNAs fulfill a unique function in maintaining post-meiotic genome integrity.
Introduction
Piwi-interacting RNAs (piRNAs) are a diverse class of gonad-specific small interfering RNAs that bind to members of the Piwi subfamily of Argonaute proteins. One common function of piRNAs in all species studied so far is the silencing of transposable elements, which is essential for the protection of genome integrity during germ cell development [1]–[3]. Distinct from miRNAs and siRNAs in origin, length, structure, and biogenesis, piRNAs are generated by dicer-independent processing of long precursor transcripts, however, the precise mechanisms of their biogenesis remain largely unclear [4], [5]. In mice, the Piwi family has three members: Miwi (Piwil1), Mili (Piwil2), and Miwi2 (Piwil4). These Piwi genes exhibit different developmental expression patterns in testis. While Miwi2 is expressed in fetal and perinatal germ cells [6], the expression of Miwi is restricted to pachytene spermatocytes and round spermatids in adult testes [7]. Mili is expressed from the fetal germ cell stage onwards through the round spermatid stage [8]. Two developmentally distinct populations of piRNAs are expressed in mouse male germ cells at the pre-pachytene and pachytene stages. Pre-pachytene piRNAs are mostly derived from transposable elements and are associated with MILI and MIWI2 in fetal and perinatal male germ cells [6], [9], [10]. Pachytene piRNAs originate from ∼3000 genomic clusters [11] and bind to both MILI and MIWI [12]–[17]. Interestingly, more than 90% of MILI - and MIWI-bound pachytene piRNAs shared identical 5′end sequences [18]. As a result, most MILI - and MIWI-bound pachytene piRNAs map to the same genomic clusters [18].
The biogenesis of piRNAs involves primary and secondary processing mechanisms [1], [2]. Pre-pachytene piRNAs derive from precursor transcripts that are cleaved into putative primary piRNA intermediate molecules by a yet unknown primary processing mechanism, followed by loading onto MILI for further processing. In embryonic germ cells, the endonuclease (slicer) activity of MILI is required for the secondary piRNA processing mechanism, which amplifies MILI-bound piRNAs through an intra-MILI ping-pong loop and generates all MIWI2-bound secondary piRNAs [19]. In this feed-forward ping-pong model, Piwi proteins with piRNAs complimentary to retroelement-derived transcripts drive transcript cleavage and piRNA amplification [6], [9], [10], [19]. In contrast, the biogenesis of pachytene piRNAs only engages the primary processing mechanism, i.e. the presumptive cleavage by an unknown nuclease and eventual processing of the precursor transcript into mature piRNAs [5], [17], [20], [21]. Therefore, pachytene piRNAs provide a simple and ideal system for dissecting the mysterious primary processing mechanism in mammals [11], [13]–[16].
We and others previously demonstrated that MOV10L1, a putative RNA helicase, interacts with all mouse Piwi proteins and is required for biogenesis of pre-pachytene piRNAs [22], [23]. MOV10L1 homologues are evolutionarily conserved among insects (Armi in Drosophila melanogaster), plants (SDE3 in Arabidopsis thaliana), and vertebrates (MOV10 and MOV10L1). Arabidopsis SDE3 is required for post-transcriptional gene silencing [24]. Drosophila Armi is essential for the maturation of RISC (RNA-induced silencing complex) and miRNA-mediated silencing [25], [26]. Armi is also relevant to the piRNA pathway, evident from the loss of specific piRNAs and the activation of retrotransposons in armi mutants [27], [28]. Specifically, Armi plays an essential role in the primary piRNA processing pathway [29]. In contrast to Drosophila and Arabidopsis with a single Mov10l1 homologue, the vertebrate genome encodes two genes (Mov10 and Mov10l1), which apparently arose by gene duplication. MOV10 is ubiquitously expressed and associates with Ago proteins, forming part of the purified human RISC [30], [31]. Depletion of MOV10 in cultured cells leads to reduced miRNA-mediated silencing [30]. We initially identified MOV10L1 as a putative RNA helicase that is specifically expressed in mouse germ cells [32], [33]. Disruption of Mov10l1 leads to meiotic arrest, de-repression of transposable elements, and depletion of both MILI - and MIWI2-associated perinatal piRNAs [22], [23]. Apparently, MOV10 and MOV10L1 function in the miRNA and the piRNA pathway, respectively, due to specialization after gene duplication during vertebrate evolution.
The existing piRNA pathway mouse mutants either fail to deplete all pachytene piRNAs or exhibit meiotic arrest prior to the pachytene stage, leaving the biogenesis and role of pachytene piRNAs largely unexplored. Inactivation of either Mili or Miwi2 causes postnatal meiotic arrest at the leptotene/zygotene stage in the male germline [8], [34]. Similarly, other piRNA pathway mutants, such as Ddx4 (Vasa), Mael, Gasz, Tdrd9, Mov10l1, and Mitopld, also exhibit early meiotic arrest in males [22], [35]–[40]. Inactivation of Miwi leads to spermiogenic arrest at the round spermatid stage [7]. However, MILI-associated pachytene piRNAs are abundant in Miwi-deficient testes [17], [18]. Therefore, a mouse mutant containing pachytene spermatocytes, but lacking all pachytene piRNAs (both MILI - and MIWI-bound piRNAs) has not been available to specifically study the function of pachytene piRNAs. In this study, we have specifically and completely depleted the pachytene piRNA population in the male germline of Mov10l1 mutant mice, uncovering a novel function for pachytene piRNAs in maintaining post-meiotic genome integrity.
Results
MOV10L1 Is Essential for Spermiogenesis
MOV10L1, a putative RNA helicase, interacts with all three mouse Piwi proteins, and is an essential component of the piRNA pathway [22]. To explore the biogenesis and function of pachytene piRNAs, we disrupted MOV10L1 function specifically during and after male meiosis using Cre-mediated inactivation of a conditional Mov10l1 allele (Mov10l1fl) (Figure 1A and Figure S1) at the following stages: after postnatal day 7 (Neurog3-Cre) [41], at the pachytene stage (Hspa2-Cre) [42], and in post-meiotic spermatids (Prm-Cre) [43].
Fig. 1. Post-natal disruption of Mov10l1 leads to post-meiotic spermiogenic arrest. 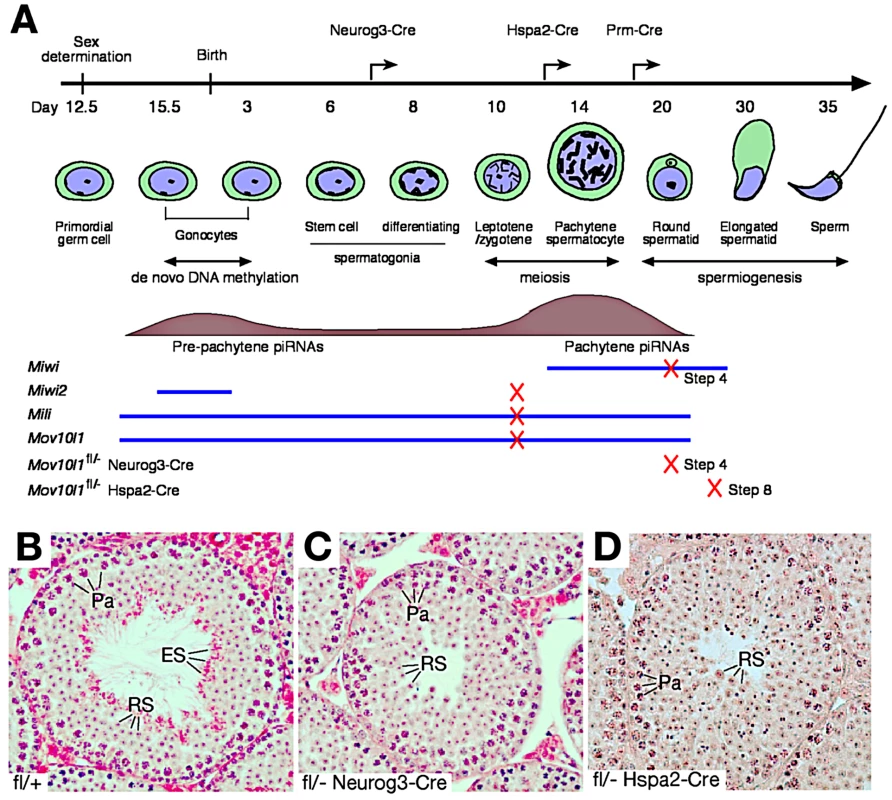
(A) Timeline of mouse spermatogenesis, with blue lines and tan histograms representing developmental expression patterns of three mouse Piwi proteins (MIWI, MIWI2, MILI) and MOV10L1, and pre-pachytene and pachytene piRNA populations, respectively. Crosses on lines mark the time point of spermatogenic arrest in the respective mouse mutant (4 ubiquitous null mutants and 2 Mov10l1 conditional mutants). Arrows on the timeline bar indicate the onset of Cre expression in the different Mov10l1 mutants generated. Disruption of Mov10l1 by Prm-Cre did not cause spermiogenic arrest (Table S1 and Figure S3). (B–D) Histology of testes from adult wild-type (B), Mov10l1fl/- Neurog3-Cre (C), and Mov10l1fl/- Hspa2-Cre (D) mice. H&E staining was performed on testis sections as described in the Materials and Methods. Abbreviations: Pa, pachytene spermatocytes; RS, round spermatids; ES, elongated spermatids. Cre-mediated recombination of the conditional Mov10l1 allele deletes the RNA helicase domain, producing a truncated protein termed MOV10L1Δ. In male Mov10l1fl/- Neurog3-Cre mice resulting from intercrosses of Mov10l1fl/fl mice with Neurog3-Cre mice [41], Cre-mediated disruption of Mov10l1 was first detected in testes at postnatal day 9 (leptotene/zygotene spermatocytes), with a decrease in the abundance of the full-length MOV10L1 protein in the mutant testes compared with the wild type (Figure S2A). Mov10l1fl/- Neurog3-Cre males were sterile, with substantially smaller testes (140±10.5 mg/pair at 2–4 months of age) compared to age-matched wild-type mice (189±18.4 mg/pair) (Student's t test, p<0.0008). In contrast to seminiferous tubules from wild-type mice (Figure 1B), tubules from Mov10l1fl/- Neurog3-Cre mutant mice lacked elongated spermatids, while earlier germ cell stages including pachytene spermatocytes and round spermatids were present (Figure 1C). Acrosome staining with the anti-ACRV1 antibody identified spermiogenic arrest at the step 4 spermatid stage. Therefore, very different to the meiotic arrest observed in male germ cells with ubiquitous deletion of Mov10l1 [22], [23], postnatal disruption of Mov10l1 mediated by Neurog3-Cre causes post-meiotic spermiogenic arrest (Figure 1C), revealing that MOV10L1 plays an essential role during the post-meiotic stages of spermatogenesis.
To distinguish consequences of inactivation of MOV10L1 during the pachytene stage from those resulting from disruption at earlier stages such as in differentiating spermatogonia, we generated Mov10l1fl/- Hspa2-Cre mice in which Cre is expressed specifically in spermatocytes, particularly pachytene cells (Figure 1A) [42]. Deletion of MOV10L1 in Mov10l1fl/- Hspa2-Cre mice occurred by postnatal day 14, apparent from a decrease in the abundance of the full-length MOV10L1 protein in the mutant testes (Figure S2B). Notably, Mov10l1fl/- Hspa2-Cre males were also sterile. Although testes (159±24 mg/pair) from 2–3 month old Mov10l1fl/- Hspa2-Cre mice were slightly smaller than those from Mov10l1+/− males (182±26 mg/pair) (Student's t test, p<0.2), histological analysis revealed spermiogenic arrest at the round spermatid stage (Figure 1D). The most advanced spermatids in Mov10l1fl/- Hspa2-Cre males were late round spermatids at step 8. The arrest of spermiogenesis at early and late round spermatid stages in Mov10l1fl/- Neurog3-Cre and Mov10l1fl/- Hspa2-Cre mutant mice, respectively, demonstrates that MOV10L1 is required for the differentiation of post-meiotic germ cells. The temporal delay in the spermiogenic arrest in Mov10l1fl/- Hspa2-Cre testes is likely due to the late onset of Hspa2-Cre expression, which may allow residual MOV10L1 to persist longer.
The round spermatid arrest in Mov10l1fl/- Neurog3-Cre and Mov10l1fl/- Hspa2-Cre testes could be due to disruption of MOV10L1 function during the pachytene stage of meiosis, or at early spermatid stages. To define the requirement for MOV10L1 more precisely, we disrupted Mov10l1 with Cre recombinase under the control of the protamine 1 (Prm) promoter, which is only expressed in post-meiotic spermatids [43]. Mov10l1fl/- Prm-Cre males exhibited normal fertility but a slight reduction in testis weight (Table S1). Histological analysis of testes from Mov10l1fl/- Prm-Cre males revealed normal spermiogenesis (Figure S3). These genetic studies demonstrate that disruption of MOV10L1 function at the pachytene stage causes spermiogenic arrest.
Blockade of Pachytene piRNA Biogenesis in the Mov10l1 Mutant
Isolation and radiolabeling of total testicular small RNAs from adult Mov10l1fl/- Neurog3-Cre testes showed that mutant testes were devoid of pachytene piRNAs (Figure 2A). Immunoprecipitation experiments further revealed that both MILI - and MIWI-associated pachytene piRNAs were absent in the mutant (Figure 2B, 2C). As Mov10l1 mutant testes contained less MIWI protein than wild-type testes, we performed serial dilutions of immunoprecipitated complexes to rule out the possibility that the observed loss of MIWI-bound piRNAs was due to the detection limit of the assay. However, MIWI-associated piRNAs were detectable in wild-type testes even when MIWI protein was not detectable (Figure 2C, lane 4), indicating a specific depletion of pachytene piRNAs in the testes from Mov10l1fl/- Neurog3-Cre mice. Moreover, the abundance of pachytene piRNAs was sharply reduced in Mov10l1fl/- Hspa2-Cre testes (Figure 2D). In addition, Northern blotting showed that individual pachytene piRNAs (piR1, piR2, and piR3) were absent in testes from Mov10l1fl/- Neurog3-Cre mice and dramatically reduced in abundance in testes from Mov10l1fl/- Hspa2-Cre mice (Figure 3). As expected, the abundance of individual pachytene piRNAs was not affected in the testes from Mov10l1fl/- Prm-Cre mice (Figure 3). Therefore, MOV10L1 function is essential for the biogenesis of all pachytene piRNAs.
Fig. 2. Mov10l1 is required for biogenesis of pachytene piRNAs. 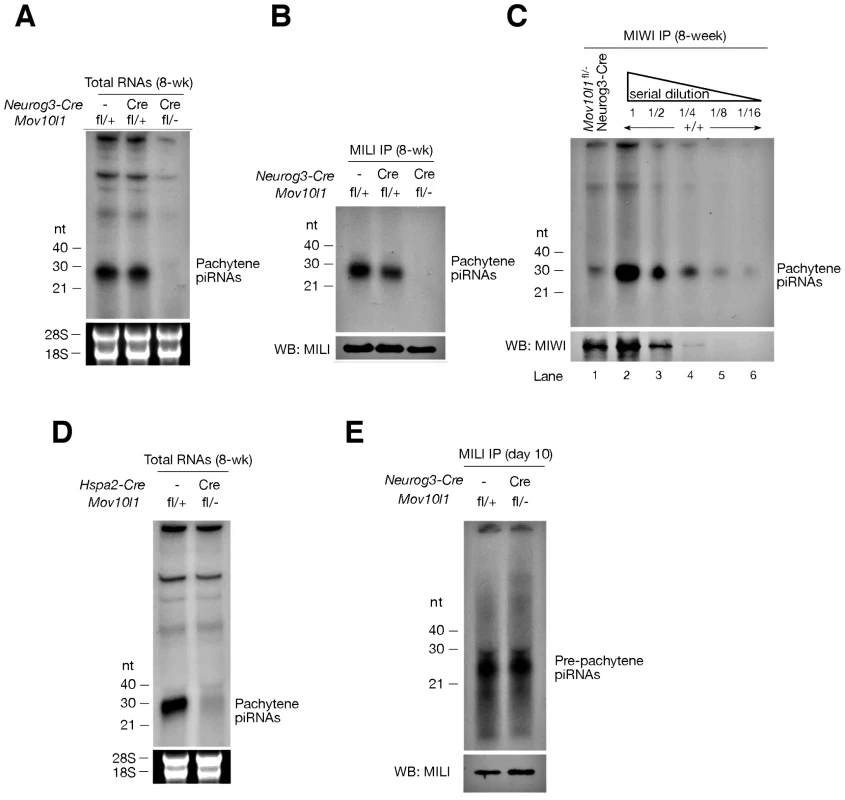
(A) Depletion of pachytene piRNAs in mutant testes from Mov10l1fl/- Neurog3-Cre adult mice. Total RNAs were 32P-end-labelled and separated by denaturing polyacrylamide gel electrophoresis. 18S and 28S ribosomal RNAs served as a loading control. (B) MILI is devoid of pachytene piRNAs in Mov10l1fl/- Neurog3-Cre testes. MILI was immunoprecipitated from testicular extracts. One-tenth of the immunoprecipitated material was used for detection of associated piRNAs, and the remainder was used for Western blotting (WB) analysis of MILI. (C) MIWI immunoprecipitation on Mov10l1fl/- Neurog3-Cre and wild-type (+/+) testes, and serial dilutions (1∶2) of MIWI IP complexes. (D) Depletion of pachytene piRNAs in mutant testes from Mov10l1fl/- Hspa2-Cre adult mice. (E) MILI is loaded with pre-pachytene piRNAs in postnatal day 10 Mov10l1fl/- Neurog3-Cre testes. Fig. 3. Northern blot analysis of individual pachytene piRNAs in conditional Mov10l1 mutant testes. 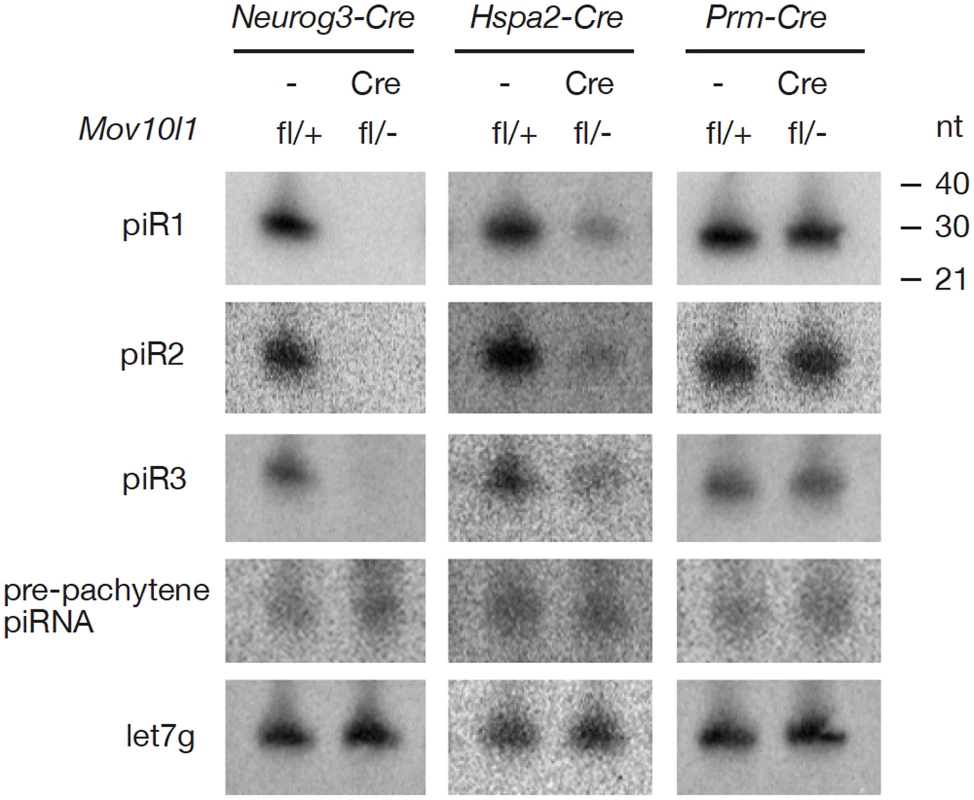
RNAs were prepared from adult testes and hybridized with radiolabeled probes specific for pachytene piR1, piR2, piR3, and a pre-pachytene piRNA. Hybridization with a let7g miRNA probe served as a loading control. The three individual pachytene piRNAs were absent in Mov10l1fl/- Neurog3-Cre testes, sharply reduced in abundance in Mov10l1fl/- Hspa2-Cre testes, and present in Mov10l1fl/- Prm-Cre testes, while the level of the pre-pachytene piRNA was not affected in either of these mutant testes. Production of Pre-Pachytene piRNAs Is Not Affected in Mov10l1fl/- Neurog3-Cre Testes
Pre-pachytene piRNAs are present in mitotic germ cells such as spermatogonia (Figure 1A). Because Neurog3-Cre initiated the disruption of Mov10l1 at post-natal day 9, we anticipated that the production of pre-pachytene piRNAs would not be affected in Mov10l1fl/- Neurog3-Cre testes. To test this hypothesis, we performed immunoprecipitation of postnatal day 10 testis lysates with anti-MILI antibodies. Postnatal day 10 testes do not contain pachytene spermatocytes, and express only MILI but not other Piwi proteins (Figure 1A). Consequently, all MILI-bound piRNAs in postnatal day 10 testes are pre-pachytene piRNAs [10]. We found that pre-pachytene piRNAs were present in postnatal day 10 Mov10l1fl/- Neurog3-Cre testes (Figure 2E). Furthermore, Northern blot analysis showed that abundance of a specific pre-pachytene piRNA was not reduced in adult testes from Mov10l1 mutant mice, regardless of whether deletion had been mediated by Neurog3-Cre, Hspa2-Cre, or Prm-Cre (Figure 3). These data demonstrate that pre-pachytene piRNA production is not affected in the Mov10l1 conditional mutant testes.
Polar Conglomeration of piRNA Pathway Components with Mitochondria in Pachytene piRNA-Deficient Spermatocytes
We next examined the consequences of the loss of pachytene piRNAs on the localization of piRNA pathway components such as MILI, MIWI, TDRD1, and GASZ. In wild-type pachytene spermatocytes, these proteins localize to cytoplasmic nuage granules (also called inter-mitochondrial cement) (Figure 4A, 4C, 4E, 4G) [7], [8], [37], [44]. Strikingly, in Mov10l1-deficient pachytene spermatocytes, these four proteins congregated to one extremely large novel perinuclear polar “granule” (Figure 4B, 4D, 4F, and 4H). Further analyses revealed immunoreactivity of the polar granule to a cocktail of antibodies against mitochondrial proteins (OXPHOS), demonstrating co-localization of mitochondria with MILI in polar granules of Mov10l1-deficient pachytene spermatocytes (Figure S4). Electron microscopy (EM) analysis confirmed that in Mov10l1-deficient pachytene spermatocytes, mitochondria form a single cluster (Figure 4J), in contrast to their random distribution in wild-type pachytene cells (Figure 4I). Consistent with a recently described role for MitoPLD, a mitochondrial surface protein, in the piRNA pathway [39], [40], these data strongly suggest a novel but yet unknown role for mitochondria in the biogenesis of pachytene piRNAs and/or a function for pachytene piRNAs in the cytoplasmic organization and distribution of mitochondria and piRNA pathway protein components.
Fig. 4. Polar conglomerate of piRNA pathway protein components and mitochondria in Mov10l1-deficient pachytene spermatocytes. 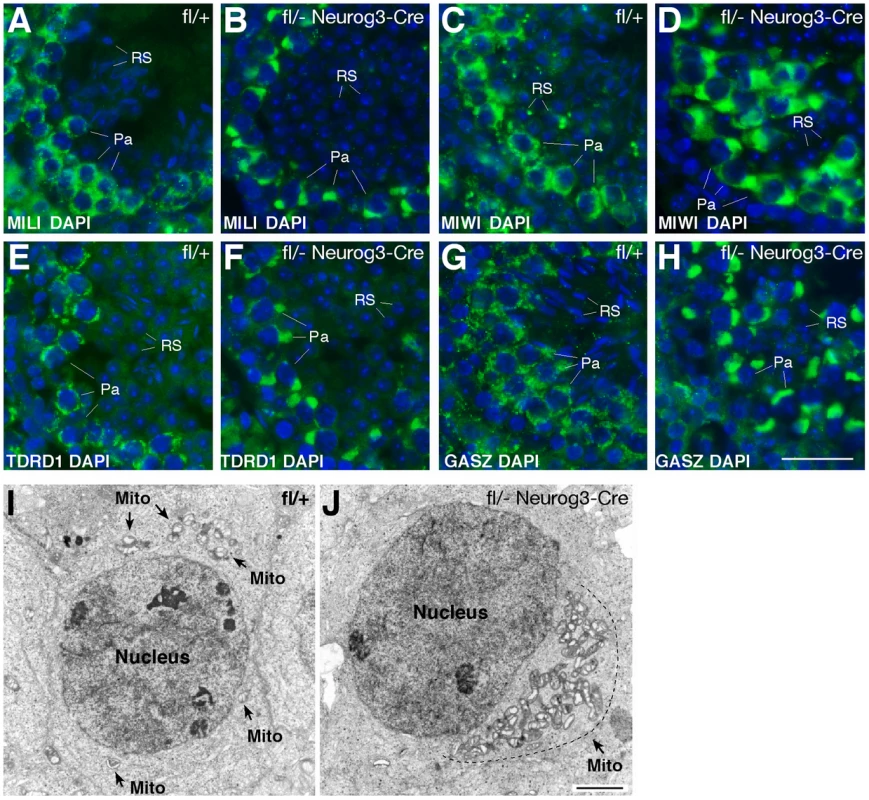
Adult testis sections were immunostained with antibodies (in green) against MILI (A, B), MIWI (C, D), TDRD1 (E, F), and GASZ (G, H). DAPI staining of DNA is shown in blue. (I) EM analysis of a wild-type pachytene spermatocyte. Note the random distribution of mitochondria (indicated by arrows). (J) EM image of a Mov10l1-deficient pachytene spermatocyte with a single mitochondrial cluster. Mito, mitochondria. Scale bars: A–H, 25 µm; I–J, 2 µm. We next examined the status of chromatoid bodies, which are large and dynamic ribonucleoprotein aggregates prominent in haploid spermatids. Chromatoid bodies contain various RNA regulatory proteins as well as piRNA pathway components, but their precise function remains unclear [45]. Wild-type round spermatids contained one prominent chromatoid body, visualized by EM as a multi-lobular electron-dense nuage (Figure S5). In Mov10l1-deficient spermatids, however, the chromatoid body was fragmented (Figure S5). A similar fragmentation of chromatoid bodies has been observed in other mouse mutants of RNA processing pathway proteins with male infertility phenotype, implying importance of their structural integrity (Miwi, Tdrd5, and Tdrd6) [7], [18], [46], [47].
Severe DNA Damage in piRNA-Deficient Round Spermatids
The introduction of DNA double strand breaks (DSBs) into the germ cell genome takes place as part of the chromatin remodeling process occurring at the elongating spermatid stage (Figure 5A). This chromatin remodeling process is initiated by the replacement of canonical histones first with transition proteins and eventually by protamines. Concurrently, nucleosomal DNA supercoils must be resolved, presumably by topoisomerase IIB (TOP2B). TOP2B generates DNA double-strand breaks (DSBs), relaxes supercoils, and subsequently religates DNA ends [48]. DSBs trigger a DNA damage response, resulting in the phosphorylation of histone H2AX (γH2AX). In wild-type testis, histone H2AX phosphorylation is therefore detectable in several germ cell stages that undergo changes in their chromatin configuration, including elongating spermatids (Figure 5B), but it is absent from round spermatids. Intriguingly, round spermatids from Mov10l1fl/- Neurog3-Cre testes exhibit a high degree of DNA damage visualized by γH2AX (Figure 5C). This could be due to a developmental progression of piRNA-deficient round spermatids to the “elongating” spermatid stage without apparent morphological change. However, the absence of both TOP2B and PRM2 (protamine 2) in γH2AX-positive round spermatids from Mov10l1 mutant testes indicated that these round spermatids were not undergoing chromatin remodeling, excluding that γH2AX-positivity was due to TOP2B activity (Figure 5E, 5G). Secondly, DNA damage might be induced by de-repressed transposable elements active in piRNA-deficient round spermatids. Genetic studies have shown that the piRNA pathway is required for silencing of retrotransposons such as LINE1 and IAP in pre-pachytene germ cells [19]. However, quantitative RT-PCR analysis revealed no de-repression of LINE1 (Figure 6B, 6C) or IAP in Mov10l1fl/- Neurog3-Cre testes, confirmed by immunofluorescent analyses of testis sections with anti-LINE1 and anti-IAP antibodies (Figure S6). Therefore, pachytene piRNAs are not required for silencing of LINE1 and IAP retrotransposons, although we cannot rule out the possibility that other transposable elements might be de-repressed in Mov10l1-deficient round spermatids. Notably, we did not observe γH2AX foci in round spermatids from Rnf17-deficient mice, in which spermatogenesis is also arrested at the round spermatid stage [49] but piRNA biogenesis does not appear to be severely affected (data not shown). These results suggest that the DSBs observed in round spermatids from Mov10l1fl/- Neurog3-Cre testes and Mov10l1fl/- Hspa2-Cre testes are not a direct consequence of their developmental arrest. Rather, these observations suggest that the piRNA pathway, specifically MOV10L1 and pachytene piRNAs, play a yet undefined role in maintaining genome integrity in post-meiotic round spermatids.
Fig. 5. Massive DNA damage in piRNA-deficient round spermatids. 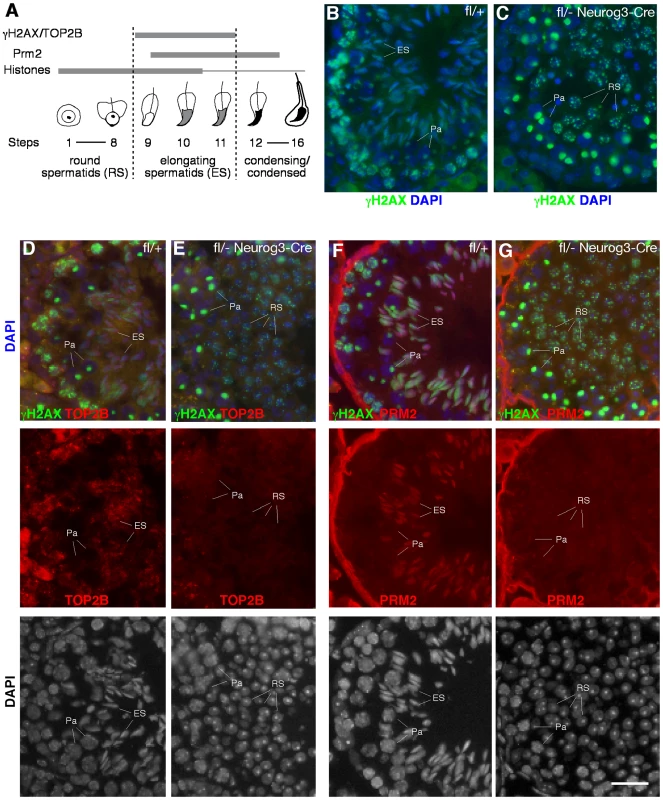
(A) Schematic representation of the expression of several proteins involved in chromatin remodeling during mouse spermiogenesis. (B) Phosphorylation of histone H2AX during normal spermatogenesis. Note the presence of γH2AX in three distinct types of germ cells: leptotene/zygotene spermatocytes (due to meiotic recombination), pachytene spermatocytes (XY body only, due to sex chromosome silencing), and elongating spermatids (due to chromatin remodeling). (C) Presence of γH2AX in round spermatids from Mov10l1fl/- Neurog3-Cre testes. (D, E) Double staining of γH2AX and TOP2B in seminiferous tubules from wild-type and Mov10l1fl/- Neurog3-Cre testes. (F, G) Double staining of γH2AX and PRM2 in seminiferous tubules from wild-type and Mov10l1fl/- Neurog3-Cre testes. Red channels and DAPI staining are also shown in separate panels (D–G). Pa, pachytene spermatocytes; RS, round spermatids; ES, elongating spermatids. Scale bar, 25 µm. Fig. 6. MOV10L1 is essential for the primary processing of pachytene piRNA precursors. 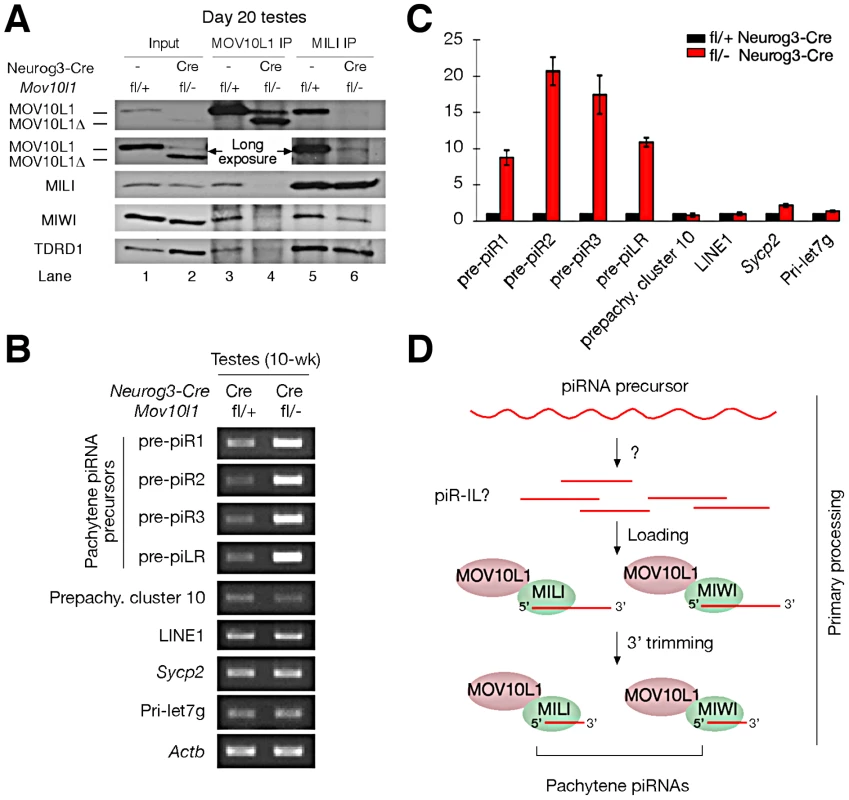
(A) The RNA helicase domain in MOV10L1 is required for interaction with MILI and MIWI. Input was 1/50th of extracts used for IP. TDRD1 is complexed with MILI and MOV10L1 and thus served as a positive control for immunoprecipitation. The same results were obtained from the repeat of IP experiments using independent samples. (B) Blockade of pachytene piRNA precursor processing in Mov10l1fl/- Neurog3-Cre testes. Total RNAs were pre-treated with DNase I. RT-PCR produced no products in controls without reverse transcriptase (data not shown). PCR primers and product sizes are listed in Table S2. (C) Quantitative RT-PCR analysis of piRNA precursor transcripts in adult testes. Numbers on the vertical axis represent fold increase of levels in mutant testis compared to levels in wild-type testes defined as 1. (D) Proposed model for the essential role of MOV10L1 in the primary processing of pachytene piRNAs. Single strand RNAs are transcribed from the piRNA clusters and are digested by an unknown nuclease into putative piRNA intermediate like molecules (piR ILs). Such piR ILs have been reported in Drosophila but not yet in mammals [20]. MOV10L1 and Piwi proteins select and bind to piR ILs for further processing into mature primary piRNAs through 3′ trimming [21]. The MOV10L1 RNA Helicase Domain Is Required for Its Interaction with Piwi Proteins
We previously found that MOV10L1 interacts with all three Piwi proteins (MILI, MIWI, and MIWI2) [22]. The low abundance of MOV10L1Δ in both Mov10l1−/− (ubiquitous null mutant) testes [22] and adult Mov10l1 conditional mutant testes (Figure S2A) precluded co-immunoprecipitation (IP) experiments to ascertain if deletion of the helicase domain affected interaction of the truncated protein with Piwi proteins in vivo. However, a peak expression of MOV10L1Δ protein in post-natal day 20 Mov10l1fl/- Neurog3-Cre testes, with a level highly exceeding that of the remaining wild-type MOV10L1 (Figure S2A), allowed us to perform co-immunoprecipitation of day 20 testicular extracts with anti-MILI and anti - MOV10L1 antibodies (Figure 6A). While wild-type MOV10L1 (due to the lack of Neurog3-Cre expression in spermatogonia) could be detected as a very faint band in the MILI immunoprecipitate as expected, the much more abundant MOV10L1Δ was absent (Figure 6A, Lane 6). Notably, MILI was not detectable in the MOV10L1/MOV10L1Δ immunoprecipitate, and the level of MIWI was extremely low (Figure 6A, Lane 4). These results suggest that, apart from its putative enzymatic activity, the RNA helicase domain of MOV10L1 is also essential for its association with MILI and MIWI, and that piRNA production could be affected by disruption of the MOV10L1-Piwi interactions.
Accumulation of Pachytene piRNA Precursors in Mov10l1 Mutant Testes
Pachytene piRNAs are derived from only one strand of genomic clusters [11], [13]–[16], prompting the hypothesis that a single long primary piRNA transcript is made from each cluster and is cleaved into intermediate RNAs by an unknown Dicer-independent mechanism [5], [20], [21]. Due to their large size and low abundance, detection of these precursor transcripts requires RT-PCR analysis, with the exception of a ∼10 kb piLR (piRNA like small RNA) transcript that can be visualized on Northern blots of testicular extracts [50]. As the depletion of pachytene piRNAs in Mov10l1fl/- Neurog3-Cre testes may be due to a blockade of pachytene piRNA precursor processing, we examined the abundance of precursors of four pachytene piRNAs (piR1, piR2, piR3, and piLR) by RT-PCR assays (Figure S7). All four precursors accumulated substantially in Mov10l1fl/- Neurog3-Cre testes, at 8 to 20 fold increased levels (Figure 6B, 6C). As expected, abundance of the pre-pachytene piRNA precursor (cluster 10) [10] and the miRNA precursor Pri-let7g remained constant (Figure 6B, 6C). These data suggest that MOV10L1 is required for the primary processing of precursor transcripts and thus plays an essential role in the early steps of the piRNA biogenesis pathway, i.e. primary processing and loading onto Piwi proteins (Figure 6D).
Discussion
We have identified MOV10L1 as the only factor known to date that is required for the production of all pachytene piRNAs in mouse. As the biogenesis of pachytene piRNAs only involves the primary processing pathway, our conditional Mov10l1 mutants provide a unique opportunity to delineate this enigmatic component of piRNA biogenesis in mammalian species. Presumably, long piRNA precursor transcripts are first cleaved into intermediate molecules, and then processed into mature piRNAs (Figure 6D). Observations that the Drosophila Armi-Piwi-Yb complex is associated with a population of 25–70 nt piRNA intermediate-like (piR-IL) molecules support this hypothesis [20]. Furthermore, recent biochemical studies using silkworm ovarian cell lysate have shown that intermediate piRNA molecules with 5′ U are specifically loaded onto Piwi proteins and then trimmed from the 3′end to generate mature piRNAs [21]. Here, we show that, in the mouse male germline, postnatal disruption of Mov10l1 does not affect the expression of Piwi proteins (MILI and MIWI) but causes a complete loss of pachytene piRNAs, demonstrating that MOV10L1 functions upstream of Piwi proteins in the piRNA biogenesis pathway. Consistent with its homology to Drosophila Armi [25], [26], MOV10L1 is therefore a master regulator of piRNA biogenesis in mouse. This notion is further supported by the dramatic accumulation of pachytene piRNA precursors in the Mov10l1 mutant testes. As MOV10L1 interacts with Piwi proteins, we postulate that MOV10L1 may facilitate the loading of intermediate piRNA molecules onto the Piwi proteins in mouse (Figure 6D).
In spermatocytes, proteins of the piRNA pathway such as MILI, MIWI, TDRD1, MAEL, and GASZ, localize to the nuage - inter-mitochondrial cement [7], [8], [36], [37], [44], [51]. However, the functional significance of the physical association of nuage with mitochondria in germ cells is poorly understood. MitoPLD, a mitochondrial signaling protein, is essential for nuage formation and piRNA production, suggesting an important role for mitochondria in these mechanisms [39], [40]. In this study, we find an unusual polar congregation of piRNA pathway proteins (such as MILI, MIWI, TDRD1, and GASZ). Similar to wild-type MOV10L1, truncated MOV10L1Δ is distributed diffusely through the cytoplasm of pachytene spermatocytes; therefore the polar coalescence of the other piRNA pathway components in MOV10L1-deficient pachytene cells is likely caused by the absence of pachytene piRNAs. However, as the association of Piwi-MOV10L1 is disrupted in the Mov10l1 mutant, it is also possible that the localization of Piwi proteins and their interacting partners has become perturbed as a consequence of this disruption. The unusual polar congregation of piRNA pathway proteins with mitochondria in Mov10l1 mutant spermatocytes suggests that MOV10L1 and/or pachytene piRNAs are essential for nuage formation and proper mitochondria distribution. Consistent with such a role, we find that the chromatoid body, a prominent nuage in spermatids, is fragmented in pachytene piRNA-deficient mutant cells. This previously unknown role in organelle distribution shows that pachytene piRNAs are intricately integrated in the inter-dependent relationships among piRNA production, nuage formation, and mitochondria organization that are essential for male germ cell maturation.
A recent study has shown that MIWI is an RNA-guided RNase with slicer activity that directly cleaves transcripts of the LINE1 retrotransposon [18]. Miwi-deficient and MiwiADH (slicer inactive) mutant testes, in which MIWI is either absent or lacks slicer activity, exhibit substantial accumulation of LINE1 transcripts and protein. In Mov10l1fl/- Neurog3-Cre testes, however, LINE1 RNA levels are not affected. One possible explanation for these differential effects on LINE1 abundance could be that, in Mov10l1fl/- Neurog3-Cre testes, MIWI is catalytically intact and may function as a slicer through pachytene piRNA-independent mechanisms. Moreover, MIWI directly binds to spermiogenic mRNAs, independent of piRNAs [17].
Although previous genetic studies of piRNA pathway mutants show that perturbation of pre-pachytene piRNAs causes meiotic arrest and de-repression of LINE1 and IAP retrotransposons, the functions of pachytene piRNAs have remained elusive. Our study on the role of Mov10l1 and the piRNA pathway during later stages of meiosis and spermiogenesis demonstrates that pachytene piRNAs fulfill distinct and essential functions during post-meiotic stages of male germ cell development. Most importantly, the massive DNA damage observed in piRNA-deficient round spermatids in the absence of de-repression of LINE1 and IAP transposable elements suggests that the integrity of the post-meiotic germ cell genome remains highly prone to damage, and that pachytene piRNAs fulfill a protective role at this stage by yet undefined mechanisms.
Materials and Methods
Ethics Statement
Mice were maintained and used for experimentation according to the guidelines of the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Mice and Antibodies
Neurog3-Cre, Hspa2-Cre, and Prm-Cre mice were purchased from the Jackson Laboratory (Stock numbers: Neurog3-Cre, 006333; Hspa2-Cre, 008870; Prm-Cre, 003328). Mov10l1fl/fl mice were generated previously [22]. Genotyping for Mov10l1 and Cre alleles was performed separately on genomic DNA isolated from tails. The anti-MOV10L1 antibody was generated previously [22]. Other antibodies used were MILI (Abcam), MIWI (Abcam, or gifts from R. Pillai), GASZ (M. M. Matzuk), LINE1 ORF1p (S. L. Martin), IAP (B. R. Cullen), TDRD1 (S. Chuma), TOP2B (Santa Cruz Biotechnology), PRM2 (SHAL), and ACTB (Sigma-Aldrich).
Immunoprecipitation and Detection of piRNAs
Mouse testicular extract preparation, immunoprecipitation, and 5′ end-labeling of piRNAs were performed as described previously [22]. Antibodies were described previously [22].
Northern Blot Analysis of piRNAs
Northern blot analyses were performed as previously described with modifications [14]. Total RNAs were isolated from mouse testes using Trizol reagent, separated by 15% denaturing polyacrylamide gel, and electro-blotted onto GeneScreen Plus hybridization membrane. Membranes were UV crosslinked and hybridized with 32P end-labeled oligonucleotide probes in Ultrahyb Oligo Buffer (Ambion Cat#8663) at 42°C. Probes for detecting pachytene piRNAs, a pre-pachytene piRNA, or microRNA were perfectly complementary to their sequences: probe-piR1: AAAGCTATCTGAGCACCTGTGTTCATGTCA; probe-piR2: ACCAGCAGACACCGTCGTATGCATCACACA; probe-piR3: ACCACTAAACATTTAGATGCCACTCTCA; probe-let7g: TACTGTACAAACTACTACCTCA; pre-pachytene piRNA probe (derived from sense SINE B1): 5′-TGGCTGTCCTGGAACTCACTYTGT [10]. After hybridization, membranes were washed three times at 42°C in 2×SSC buffer containing 0.5% SDS, or stripped by boiling in 0.1×SSC containing 0.1% SDS. Membranes were exposed to a phosphor imager screen for autoradiography.
Histological, Immunofluorescence, and Electron Microscopy (EM) Analyses
For histology, testes were fixed in Bouin's solution overnight, processed, sectioned, and stained with hematoxylin and eosin. Immunofluorescence was performed on frozen sections of testes fixed in 4% paraformaldehyde as previously described [52]. EM followed a standard protocol used at the Electron Microscopy Resource Laboratory of the University of Pennsylvania.
Expression Analysis of piRNA Precursor Transcripts
PCR primers for piRNA precursor transcripts were chosen from genomic clusters to which each piRNA was mapped [10], [14], [50]. PCR primers and PCR product sizes are listed in Table S2.
Supporting Information
Zdroje
1. SiomiMC, SatoK, PezicD, AravinAA (2011) PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol 12 : 246–258.
2. PillaiRS, ChumaS (2012) piRNAs and their involvement in male germline development in mice. Dev Growth Differ
3. AravinAA, NaumovaNM, TulinAV, VaginVV, RozovskyYM, et al. (2001) Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11 : 1017–1027.
4. BrenneckeJ, AravinAA, StarkA, DusM, KellisM, et al. (2007) Discrete small RNA-generating loci as master regulators of transposon activity in drosophila. Cell 128 : 1089–1103.
5. HaaseAD, FenoglioS, MuerdterF, GuzzardoPM, CzechB, et al. (2010) Probing the initiation and effector phases of the somatic piRNA pathway in drosophila. Genes Dev 24 : 2499–2504.
6. AravinAA, SachidanandamR, Bourc'hisD, SchaeferC, PezicD, et al. (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31 : 785–799.
7. DengW, LinH (2002) Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2 : 819–830.
8. Kuramochi-MiyagawaS, KimuraT, IjiriTW, IsobeT, AsadaN, et al. (2004) Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131 : 839–849.
9. Kuramochi-MiyagawaS, WatanabeT, GotohK, TotokiY, ToyodaA, et al. (2008) DNA methylation of retrotransposon genes is regulated by piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22 : 908–917.
10. AravinAA, SachidanandamR, GirardA, Fejes-TothK, HannonGJ (2007) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316 : 744–747.
11. GanH, LinX, ZhangZ, ZhangW, LiaoS, et al. (2011) piRNA profiling during specific stages of mouse spermatogenesis. RNA 17 : 1191–1203.
12. WatanabeT, TakedaA, TsukiyamaT, MiseK, OkunoT, et al. (2006) Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 20 : 1732–1743.
13. LauNC, SetoAG, KimJ, Kuramochi-MiyagawaS, NakanoT, et al. (2006) Characterization of the piRNA complex from rat testes. Science 313 : 363–367.
14. GirardA, SachidanandamR, HannonGJ, CarmellMA (2006) A germline-specific class of small RNAs binds mammalian piwi proteins. Nature 442 : 199–202.
15. GrivnaST, BeyretE, WangZ, LinH (2006) A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20 : 1709–1714.
16. AravinA, GaidatzisD, PfefferS, Lagos-QuintanaM, LandgrafP, et al. (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442 : 203–207.
17. VourekasA, ZhengQ, AlexiouP, MaragkakisM, KirinoY, et al. (2012) Mili and miwi target RNA repertoire reveals piRNA biogenesis and function of miwi in spermiogenesis. Nat Struct Mol Biol 19 : 773–781.
18. ReuterM, BerningerP, ChumaS, ShahH, HosokawaM, et al. (2011) Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480 : 264–267.
19. De FazioS, BartonicekN, Di GiacomoM, Abreu-GoodgerC, SankarA, et al. (2011) The endonuclease activity of mili fuels piRNA amplification that silences LINE1 elements. Nature 480 : 259–263.
20. SaitoK, IshizuH, KomaiM, KotaniH, KawamuraY, et al. (2010) Roles for the yb body components armitage and yb in primary piRNA biogenesis in drosophila. Genes Dev 24 : 2493–2498.
21. KawaokaS, IzumiN, KatsumaS, TomariY (2011) 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell 43 : 1015–1022.
22. ZhengK, XiolJ, ReuterM, EckardtS, LeuNA, et al. (2010) Mouse MOV10L1 associates with piwi proteins and is an essential component of the piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci U S A 107 : 11841–11846.
23. FrostRJ, HamraFK, RichardsonJA, QiX, Bassel-DubyR, et al. (2010) MOV10L1 is necessary for protection of spermatocytes against retrotransposons by piwi-interacting RNAs. Proc Natl Acad Sci U S A 107 : 11847–11852.
24. DalmayT, HorsefieldR, BraunsteinTH, BaulcombeDC (2001) SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in arabidopsis. EMBO J 20 : 2069–2078.
25. TomariY, DuT, HaleyB, SchwarzDS, BennettR, et al. (2004) RISC assembly defects in the drosophila RNAi mutant armitage. Cell 116 : 831–841.
26. CookHA, KoppetschBS, WuJ, TheurkaufWE (2004) The drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116 : 817–829.
27. VaginVV, SigovaA, LiC, SeitzH, GvozdevV, et al. (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313 : 320–324.
28. MaloneCD, BrenneckeJ, DusM, StarkA, McCombieWR, et al. (2009) Specialized piRNA pathways act in germline and somatic tissues of the drosophila ovary. Cell 137 : 522–535.
29. OlivieriD, SykoraMM, SachidanandamR, MechtlerK, BrenneckeJ (2010) An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in drosophila. EMBO J 29 : 3301–3317.
30. MeisterG, LandthalerM, PetersL, ChenPY, UrlaubH, et al. (2005) Identification of novel argonaute-associated proteins. Curr Biol 15 : 2149–2155.
31. ChendrimadaTP, FinnKJ, JiX, BaillatD, GregoryRI, et al. (2007) MicroRNA silencing through RISC recruitment of eIF6. Nature 447 : 823–828.
32. WangPJ, McCarreyJR, YangF, PageDC (2001) An abundance of X-linked genes expressed in spermatogonia. Nat Genet 27 : 422–426.
33. WangPJ, PageDC, McCarreyJR (2005) Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum Mol Genet 14 : 2911–2918.
34. CarmellMA, GirardA, van de KantHJ, Bourc'hisD, BestorTH, et al. (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12 : 503–514.
35. TanakaSS, ToyookaY, AkasuR, Katoh-FukuiY, NakaharaY, et al. (2000) The mouse homolog of drosophila vasa is required for the development of male germ cells. Genes Dev 14 : 841–853.
36. SoperSF, van der HeijdenGW, HardimanTC, GoodheartM, MartinSL, et al. (2008) Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell 15 : 285–297.
37. MaL, BucholdGM, GreenbaumMP, RoyA, BurnsKH, et al. (2009) GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet 5: e1000635 doi:10.1371/journal.pgen.1000635.
38. ShojiM, TanakaT, HosokawaM, ReuterM, StarkA, et al. (2009) The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 17 : 775–787.
39. WatanabeT, ChumaS, YamamotoY, Kuramochi-MiyagawaS, TotokiY, et al. (2011) MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell 20 : 364–375.
40. HuangH, GaoQ, PengX, ChoiSY, SarmaK, et al. (2011) piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 20 : 376–387.
41. SchonhoffSE, Giel-MoloneyM, LeiterAB (2004) Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol 270 : 443–454.
42. InselmanAL, NakamuraN, BrownPR, WillisWD, GouldingEH, et al. (2010) Heat shock protein 2 promoter drives cre expression in spermatocytes of transgenic mice. Genesis 48 : 114–120.
43. O'GormanS, DagenaisNA, QianM, MarchukY (1997) Protamine-cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci U S A 94 : 14602–14607.
44. ChumaS, HosokawaM, KitamuraK, KasaiS, FujiokaM, et al. (2006) Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci U S A 103 : 15894–15899.
45. MeikarO, Da RosM, KorhonenH, KotajaN (2011) Chromatoid body and small RNAs in male germ cells. Reproduction 142 : 195–209.
46. YabutaY, OhtaH, AbeT, KurimotoK, ChumaS, et al. (2011) TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J Cell Biol 192 : 781–795.
47. VasilevaA, TiedauD, FiroozniaA, Muller-ReichertT, JessbergerR (2009) Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol 19 : 630–639.
48. LeducF, MaquennehanV, NkomaGB, BoissonneaultG (2008) DNA damage response during chromatin remodeling in elongating spermatids of mice. Biol Reprod 78 : 324–332.
49. PanJ, GoodheartM, ChumaS, NakatsujiN, PageDC, et al. (2005) RNF17, a component of the mammalian germ cell nuage, is essential for spermiogenesis. Development 132 : 4029–4039.
50. RoS, ParkC, SongR, NguyenD, JinJ, et al. (2007) Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA 13 : 1693–1702.
51. ChumaS, HosokawaM, TanakaT, NakatsujiN (2009) Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: Germinal granules in mammals. Mol Cell Endocrinol 306 : 17–23.
52. ZhengK, WuX, KaestnerKH, WangPJ (2009) The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev Biol 9 : 38.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



