-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamamalERA: An updated research agenda for basic science and enabling technologies in malaria elimination and eradication
Dyann F.
Wirth and colleagues propose a research agenda for basic science and enabling technologies in malaria elimination and eradication.
Published in the journal: . PLoS Med 14(11): e32767. doi:10.1371/journal.pmed.1002451
Category: Collection Review
doi: https://doi.org/10.1371/journal.pmed.1002451Summary
Dyann F.
Wirth and colleagues propose a research agenda for basic science and enabling technologies in malaria elimination and eradication.Summary points
The recent development of multiple in vitro systems for studying malaria biology has helped deepen our understanding of the disease. Nevertheless, research remains hampered by a lack of in vitro models that can probe key aspects of malaria (e.g., gametocyte development in Plasmodium vivax, fertilization, ookinete biology, parasite–midgut interactions, human hepatocyte infection) and generate biological materials (i.e., infectious sporozoites) for laboratory study. Developing the necessary cell lines and other in vitro culture tools to propel these studies represent important areas for future research.
With the emergence of widespread insecticide resistance in mosquito populations, there is a strong need to bring basic research in mosquito biology back into the malaria eradication agenda to strengthen current insecticide-based control campaigns and generate alternate vector control strategies.
Driven by the development and accessibility of large-scale research tools and technologies, the scientific community can systematically tackle key questions in malaria, such as the following. What are the genes that contribute to antimalarial drug resistance (thereby defining the full parasite “resistome”)? What are the functions of key Plasmodium genes (providing much-needed annotation of key Plasmodium genes)? What are the genes and gene mutations that drive resistance in mosquito populations?
Continued exploration of the potential of enabling technologies is needed. Important areas of future research include the use of gene-drive strategies and other gene-manipulation technologies; metabolomics-based approaches for biomarker discovery; structural vaccinology, novel technology platforms, and the use of novel adjuvants to improve vaccine design; and high-throughput approaches to facilitate drug discovery and screening.
Background
Since the first agenda for malaria eradication was published in 2011 [1], there have been many significant developments in basic science, including an enhanced understanding of parasite biology (both gametocyte and liver stages) as well as mosquito biology (Table 1). Some of these advances could not have been predicted 5 years ago, such as the use of mouse models engrafted with human liver to advance the biology of liver-stage parasites (including the quiescent P. vivax hypnozoite stage) and the development of powerful genome-editing capabilities based on clustered regularly interspaced short palindromic repeats/associated protein-9 nuclease (CRISPR/Cas9) technology. In contrast, little progress has been achieved in several key research areas that were previously prioritized and, as such, they remain important stumbling blocks on the road to eradication.
We focus here on these and other crucial areas—deficiencies in basic science research and the lack of enabling technologies—that currently limit our progress towards malaria elimination and eradication. Importantly, this analysis highlights specific aspects of the Plasmodium life cycle in both the human host and the Anopheles vector. Our integrated approach aims to combine research efforts and expertise across human immunology, parasitology, and entomology to introduce powerful new ideas and technologies from other fields, provide a multifaceted view of disease biology, and accelerate progress toward eradication.
Tab. 1. A listing of the important research areas highlighted in malERA 2011, the progress made since then, and the remaining areas that require additional research. 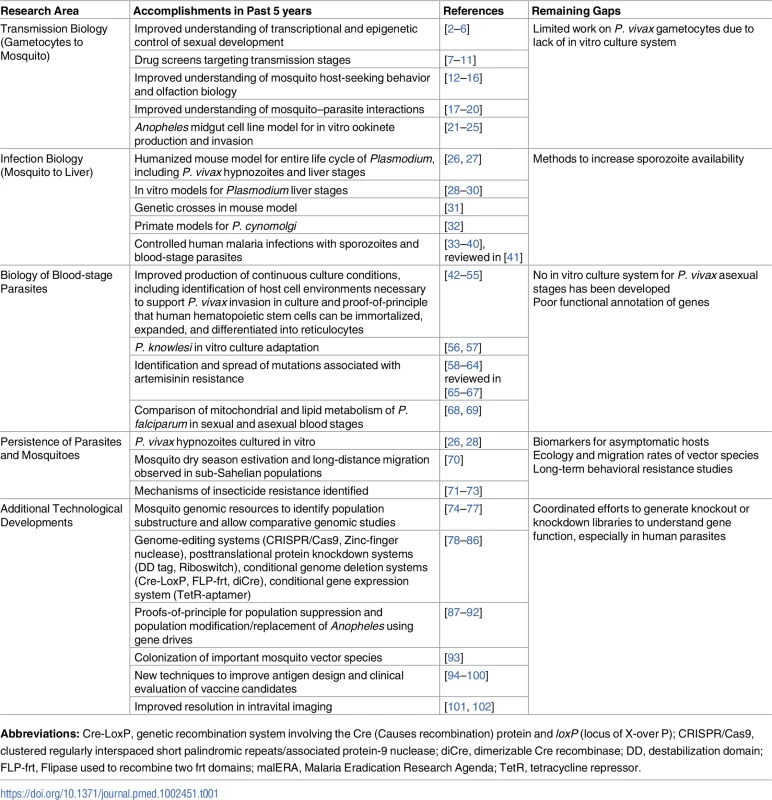
Abbreviations: Cre-LoxP, genetic recombination system involving the Cre (Causes recombination) protein and loxP (locus of X-over P); CRISPR/Cas9, clustered regularly interspaced short palindromic repeats/associated protein-9 nuclease; diCre, dimerizable Cre recombinase; DD, destabilization domain; FLP-frt, Flipase used to recombine two frt domains; malERA, Malaria Eradication Research Agenda; TetR, tetracycline repressor. Methods
The findings presented in this paper result from an extensive literature review of published and unpublished materials and the deliberations of the 2015 Malaria Eradication Research Agenda (malERA) Refresh Consultative Panel on Basic Science and Enabling Technologies. Electronic databases were systematically searched for published literature between January 1, 2010, and July 2, 2016, without language limitations. Panelists were invited to recommend additional literature and additional ongoing research projects. A 2-day workshop was held with the majority of the panel members, including field researchers, specialists from basic science, malaria genomics and epigenomics, regenerative medicine, and National Institutes of Health representatives. The panel broke into 6 breakout sessions to identify the problems that need to be solved in asexual blood stages, liver stage and mosquito, mosquito, P. vivax, population genetics and resistance, and transmission. The panel discussed what research is needed to address these problems and considered 6 crosscutting themes in CRISPR technologies, immunology and malaria vaccines, genomics tools for malaria, metabolism and malaria, structural biology, and diagnostics for malaria. Each group fed back to plenary session, where further robust discussions and input occurred. This helped refine the opportunities and gap areas in which research is needed. The final findings were arrived at with inputs from all panelists and several iterations of the manuscript.
Advances, challenges, and opportunities in transmission biology
Gametocytes
Plasmodium transmission begins with the development of sexual forms of the parasite (known as gametocytes) in an infected human host and their subsequent transfer to an anopheline mosquito following a blood meal (Fig 1). This stage represents a key bottleneck in the parasite life cycle and thus is an attractive opportunity for disrupting disease transmission. As shown in Box 1, in the past 5 years significant and exciting progress has been made in understanding gametocyte development, including insights into the transcriptional and epigenetic control of sexual differentiation and evidence for bone marrow sequestration [2–6, 103]. In the case of P. falciparum, newly available in vitro systems for gametocyte maturation have been used in small molecule screening, antibody reagent development, and transcriptional and metabolomics analyses [7–11].
Fig. 1. Schematic depicting the human and mosquito life cycles of <i>Plasmodium</i>, highlighting critical questions at specific points within the life cycle. 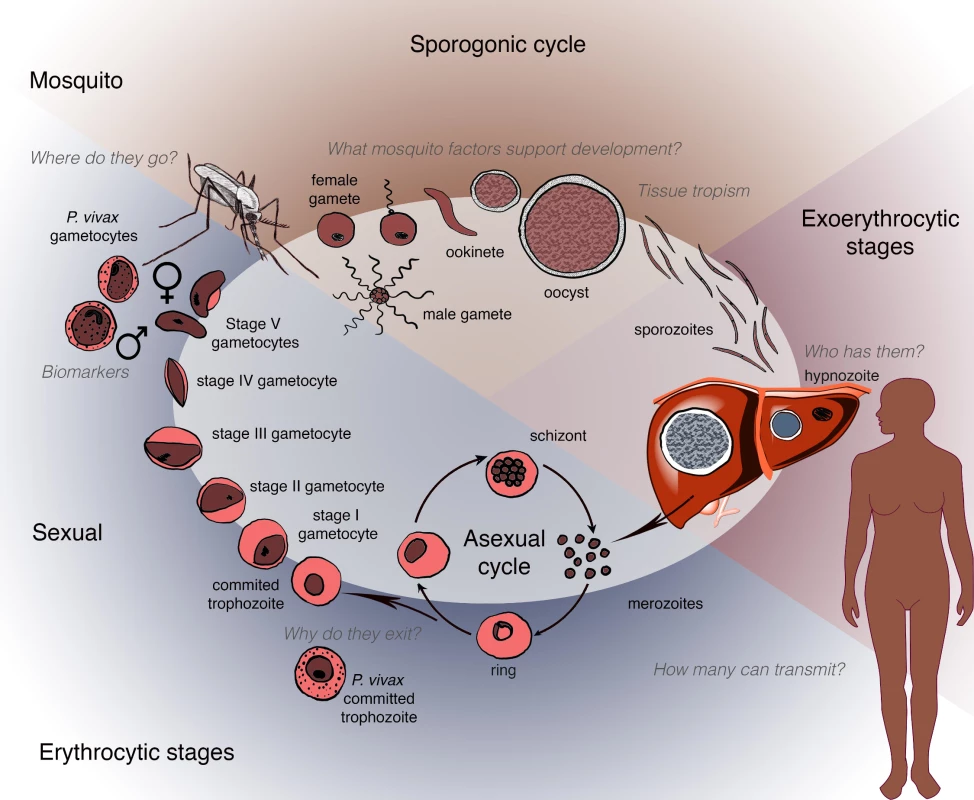
Box 1. Opportunities for the next 5 years
Functional genomics
Identification of regulatory sequences within the parasite genome, similar to the human Encyclopedia Of DNA Elements (ENCODE) project,
Genome wide annotation of gene function in human parasites to identify sets of genes involved in discrete cellular processes, including drug resistance,
Improved scalability of CRISPR/Cas9 technology in asexual parasites to allow for both pooled, genome-wide approaches (large scale) and single cell transformation (microscale),
Greater collaboration between researchers to avoid overlapping gene annotation efforts.
Advances in mosquito biology
Generation of a mosquito consortium to evaluate promising gene drive-based strategies for efficacy at scale and/or over time and share knockout and/or transgenic strains,
Greater understanding of mosquito behavior and ecology,
Colonization of important vector species,
Development of in vitro mosquito infection models.
New vaccine approaches
Improved adjuvants and identification of new targets, including better structures for existing (and new) targets to improve structural approaches,
Development of novel approaches with the potential to generate sterilizing immunity (i.e., cognate antigens),
Coordinated functional annotation of asexual-stage parasites to enable prioritization of functional vaccine antigens,
Greater access to samples and data from both human challenge studies and patient samples demonstrating natural immunity,
Application of gene-editing technologies to systematically understand the function of hypothetical genes.
Biomarkers and diagnostics
Indicators of transmissible gametocytes,
Markers of liver-stage infection, in particular, hypnozoites,
Markers/assays to identify asymptomatic carriers,
Identification of metabolic signatures of different stages of the life cycle.
Greater understanding of resistance to antimalarials and insecticides
Identification of genes and pathways (i.e., the “resistome”) involved in resistance,
Development of alternatives to insecticides,
Use evolutionary approaches to prevent resistance.
Greater accessibility to P. vivax gametocytes
Development of a P. vivax in vitro culture system (e.g., ookinetes to validate transmission-blocking vaccine targets),
Greater collaboration between groups to improve access to existing sporozoite sources. This would be coupled with advances in cryopreservation to improve access to sporozoites globally.
In contrast, the mechanisms of P. vivax gametocyte development remain largely unknown. Gametocyte biology within this species is quite distinct—development takes just 2 to 3 days and unfolds prior to any clinical symptom. P. vivax gametocytes appear susceptible to existing antimalarial drugs that are not effective against P. falciparum gametocyte stages [104–106]. Progress in this area has been hampered by the absence of a comparable in vitro culture system for asexual P. vivax parasites, which is an urgent priority, as it would enable the generation of gametocytes for laboratory study, mosquito infections, and sporozoite production.
Another major area for discovery is the elucidation of the biological determinants of gametocyte transmissibility, especially in areas of low endemicity. Does the success of transmission depend on gametocyte quantity and/or quality? Are there mosquito-specific factors that actively recruit gametocytes to the biting site or do gametocytes preferentially sequester near the skin? What factors and mechanisms enable male and female gametes to find one another in the mosquito midgut? Biomarkers for transmission competency could enable a broader understanding of the heterogeneity in natural infections.
Mosquito biology and host seeking
Transmission success also depends upon the interactions of the mosquito vector with both its human host and ingested parasites. Since 2011, there have been major advances in understanding the biology of olfaction and host-seeking behavior in mosquitoes via a combination of behavioral assays, electrophysiology, and functional genomic approaches [12–16]. High-throughput screens have identified new classes of attractants and repellents that are currently being tested in mosquito traps and spatial repellent trials ([107–110], also see MESA Track at http://www.malariaeradication.org/mesa-track). Moving forward, the identification of oviposition cues and the role of olfaction and taste in larval stages could facilitate the development of additional tools for vector control. Comparative genomic analysis of odorant receptor pathways that differ between anthropophilic and zoophilic species will help to elucidate the molecular basis of host-seeking behavior. Recent studies have shown that the composition of the human skin microbiota influences host attractiveness to mosquitoes [111] and identified volatile substances produced by parasites in human hosts thought to preferentially attract mosquitoes to infected individuals [112]. Nevertheless, gaps remain in our knowledge regarding the potential for gametocyte-seeking behavior by the mosquito and parasite-induced changes to the human host that may influence mosquito behavior to enhance biting and transmission.
Parasite development in the mosquito
Fertilized zygotes develop into the motile ookinete, which in turn crosses the midgut wall. Major advances have been made in understanding midgut invasion and early mosquito anti-Plasmodium immune responses that target the ookinete stage. Several parasite genes that interact with the vector to enable its invasion of epithelial cells have been identified [17–19], and new insights have emerged regarding the role of epithelial responses to invasion and the corresponding epithelial interactions with the complement-like system to limit ookinete survival [113–117]. There is increasing evidence that the oocyst stage is also a target of innate immunity in the mosquito [118, 119]. Genome-wide association study (GWAS) mapping of Anopheles populations displaying different vector competence has identified mosquito genes that influence parasite development [120]. This list of potential targets to disrupt malaria transmission could be extended through functional screens using double-stranded ribonucleic acid (dsRNA)-mediated gene silencing in mosquitoes and synthetic approaches such as single-chain antibodies to block P. falciparum from infecting salivary glands.
A particular challenge for developing new interventions is the lack of culture systems to study fertilization, ookinete biology, and parasite–midgut interactions in human malaria parasites. Plasmodium species of rodents and birds have provided rapid proof-of-principle for new transmission-blocking strategies [121–123] and will likely continue to be critical for revealing the basic biology of sexual and mosquito stages. The development of mosquito midgut-derived cell lines (or organoids) supporting the in vitro culture of ookinetes and oocyst of human malaria parasites would enable high-throughput transcriptomic and metabolomic studies as well as high-resolution functional analysis of the parasite’s surface proteins and their interactions with mosquito cells. These assays could also be used to validate transmission-blocking drugs and vaccines.
Advances, challenges, and opportunities in infection biology
The past 5 years have seen rapid progress in understanding the biology of Plasmodium infection in the human liver. Increased availability of primary human hepatocytes has allowed the development of multiple in vitro platforms, all tailored toward the concept of a miniaturized experimental liver model [28, 29, 124]. Importantly, these innovations have allowed the liver stages of infection to be fully recapitulated outside the human host for the first time [26, 125]. They have also spurred the development of reagents to explore the biology of sporozoite infectivity and liver stage development and provided the first glimpse of the P. vivax hypnozoite [26, 28].
In parallel, the development of humanized mouse models of P. vivax and P. falciparum infection have opened up the potential for surrogate in vivo models of human liver infection [26] and allowed the first genetic crosses of parasites (P. falciparum) outside of a primate [31]. Studies in primates continue to play an important role; the P. cynomolgi monkey model of liver infection is the only in vivo relapse model of the P. vivax hypnozoite [30, 32, 126]. Combined with controlled human malaria infections [34, 35, 38, 127, 128] and in vitro models, these tools have highlighted key differences in the biology of different parasite species (specifically, P. vivax and P. falciparum) and paved the way for understanding the cellular biology of liver infection and the immune response and for performing high-throughput drug candidate screening.
To facilitate efforts aimed at eradication, we have identified a number of transformative actions in the field of infection biology. A transformative innovation would be the in vitro cultivation of large numbers of infectious P. falciparum and P. vivax sporozoites, bypassing the mosquito vector. This would not only facilitate basic research but also contribute to whole-parasite vaccine development. Alternatively, advances in the preservation of sporozoite viability and infectivity after mosquito dissection and/or the engineering of mosquitoes to produce sporozoites at high levels would increase the availability and distribution of infectious material for research purposes.
Improved liver-stage cell lines could also have a transformative effect on the pace of novel drug and vaccine development, especially for P. vivax [28–30]. Cell lines provide readily available, immortal, and genetically identical cells, allowing researchers to reliably obtain the same sensitivity measurements for each compound or antibody. This development could enable high-throughput drug screening for discovery of liver stage-specific compounds targeting either parasite functions [129] or human targets necessary for parasite development. Moreover, the availability of robust and inexpensive in vitro hepatocyte infection models for P. vivax and P. falciparum may allow the development of better in vitro assays for antibody-dependent inhibition of invasion (akin to virus neutralization assays) and cell-mediated killing of infected cells. This could allow the discovery of human monoclonal antibodies with broadly neutralizing activity, whose cognate antigens could then be used to create vaccines that give sterilizing immunity. Recent advances in proteomics and mass spectrometry may also support the identification of biomarkers for exoerythrocytic stages that are relevant in vivo.
Advances, challenges, and opportunities in asexual-stage biology
Defining the parasite “resistome”
Notable advances in asexual biology over the past 5 years include improvements in functional genomics, such as more robust RNA sequencing methods [130–132], a deeper understanding of transcription factors such as activator protein 2 (ap2) transcription factors [133] or alternative RNA splicing [134], and whole genome sequencing and genotyping of both field isolates and evolved cultures (see Table 1). Due to its rapidly decreasing cost and increasing accuracy, sequencing has accelerated our understanding of the mechanisms and modes of action of current and new antimalarials through drug-resistant parasite selection in vitro (reviewed in [135]) as well as population genetics of the parasite in vivo [62, 136]. Although numerous studies have described using in vitro evolution and whole genome analysis to both find targets of new antimalarial compounds and identify genes conferring resistance [62, 137, 138], in most cases, only a handful of genes were identified. Now that single cell sequencing is becoming a reality [139], we are in a position to identify every gene (and potentially allele) that contributes to drug resistance, thus defining the parasite “resistome.” The complete genetic basis of parasite drug resistance should provide better molecular markers of whether parasites have acquired resistance to drugs that may be used in elimination campaigns, informing drug or drug combination selections (See malERA Refresh paper on resistance [140]).
Systematic characterization of the asexual-stage parasite
The systematic knockout of genes in P. berghei has led to numerous advances in our understanding of fundamental asexual biology [141, 142], including the P. berghei identification of essential genes and pathways [143–146], greater understanding of merozoite invasion and egress [147–150], discovery of the parasite’s export machinery [145, 151, 152], and revealing how the red cell cytoplasm and membrane are remodelled [153, 154]. Such studies point to the critical nature of these processes and have opened the possibility of targeting them with drugs or vaccines.
Yet, major gaps remain in our knowledge of gene function in P. falciparum and, to an even greater extent, in other species (including P. vivax, P. ovale, and P. malariae) in which genetic diversity is also relatively uncharacterized. Although in many cases, genomic variants can be readily identified in sequencing data, poor annotations for predicted genes in the P. falciparum genome continue to slow progress. For example, we know little about the cellular function of the pfkelch13 gene, a major contributor to artemisinin resistance ([62, 155, 156], reviewed in [67]). Given that it is more efficient and inexpensive for the community to work together to functionally annotate the P. falciparum genome systematically rather than in a 1-researcher-1-gene fashion, coordinated large-scale projects with a focus on the easily accessible P. falciparum asexual blood stage should be considered. Such systematic data would also help in the interpretation of whole genome sequencing data from drug - or vaccine-resistant parasites. Desirable genomic annotations include the location of key transcription factor binding sites, transcriptional start and stops sites [157], epigenetic chromatin modifications, and the cellular localization of encoded proteins. These consortium-acquired data are critical to predict whether genetic variants discovered through genome sequencing of model organisms and humans are indeed functional and could also help prioritize antigens for vaccine development. In addition, if better in vitro culture systems can be developed for P. vivax (see “Advances, challenges, and opportunities in transmission biology”), these systematic approaches could be extended to this important species. A potential model for such a consortium-based effort is the human ENCODE project, which has identified functional elements in the human genome [158].
Using metabolomics to identify biomarkers and develop diagnostics
There have been major advances in the use of modern mass spectrometry-based methods for identifying and profiling metabolites from parasite-infected cells [159–161] as well as determining the mode of action of drugs through the metabolic perturbations of exposed parasites [162–165]. Two key areas in which metabolomics-based approaches have yet to make a significant impact are biomarkers and diagnostics. Given the difficulty and cost associated with identifying infected individuals (particularly those who are asymptomatic—see malERA Refresh paper on reservoir and transmission [166]), the development of effective metabolomic biomarkers with significant correlation to infection would represent a critical advance. Furthermore, to determine host markers of infection, field samples across a broad range of infectivities, including asymptomatic carriers, should be studied using metabolomic methods. Such analyses should also aim to span all Plasmodium parasite species as well, particularly P. vivax.
The question of persistence: Where do parasites—And mosquitoes—Hide?
In the drive towards elimination and eradication, a key question is how and where malaria infection persists in both humans and mosquitoes, both in individuals as well as populations. Recent genomic studies indicate that parasites may also persist in an additional zoonotic reservoir in nonhuman primates [167–169], although how this contributes to disease transmission in humans is currently unclear.
Persistence of malaria occurs in 2 modalities—asymptomatic carriers and latent liver stages. The asymptomatic carriers represent a significant threat to the reintroduction of malaria; thus, the identification of such carriers requires a heightened level of awareness and detection. The absence of symptoms in an individual may reflect the presence of disease-prevention host responses in the absence of sterilizing immunity, thereby allowing persistent parasitemia or the sequestration of parasites in sites (e.g., the liver or bone marrow) in which they are “hidden” from the immune system. Understanding the relative contributions of both human immune responses and parasite biology will be essential to maximize the efficacy of antimalarial interventions, particularly vaccines.
Parasite persistence in the liver is a major hurdle for elimination efforts, particularly for P. vivax, because of its rapid development of gametocytes in humans, enabling transmission before the onset of clinical symptoms. Insights have emerged from studies of nonhuman primate models and humanized mouse models [26] in which parasite forms resembling hypnozoites demonstrated some biologic activity. These findings imply that sensitive technologies, such as proteomics and metabolomics, may identify markers likely secreted at these stages. Such markers would require field validation but ultimately could be incorporated into point-of-care diagnostics, eliminating the need for primaquine or tafenoquine in mass drug administration campaigns and informing epidemiological studies of the load of hypnozoite infection in endemic regions.
The transmission of Plasmodium infections with low or submicroscopic levels of circulating gametocytes suggests the possibility of nonrandom sequestration of gametocytes at sites in peripheral skin that are accessible to mosquitoes. P. falciparum gametocytes have recently been found to have an extended maturation period in the bone marrow [103, 170]. A clear implication of this observation, however, is that gametocytes detected in the peripheral circulation may not accurately reflect overall or infectious gametocyte levels and that more sensitive assays are needed to identify potential sources of transmission.
Mosquito vector persistence
The aspects of vector biology that enable malaria persistence remain to be investigated and will be critical not only for informing and targeting current elimination and eradication strategies but also for the development and successful deployment of novel vector-based interventions. Recent data suggest that, in Africa, both mosquito estivation (dry season diapause) and long-distance migration contribute to the persistence of sub-Sahelian mosquito populations following a dry season, but in a species-specific manner [70]. New genomic resources have facilitated the understanding of fine-scale mosquito population structures [77, 171] suggesting large and stable populations [74–76]. The contribution of the observed genomic patterns to population persistence is unclear at this point, and a better understanding of the life history, ecology, and migration rates of vectors that result in the observed genomic patterns between populations is needed. Similar studies in non-African mosquito populations are needed.
Mosquitoes also persist through physiological resistance to insecticides (see malERA Refresh paper on resistance [140]), either through target site mutations, increased expression of detoxifying enzymes, or cuticular thickening. Genomic markers associated with resistance continue to be identified, yet together they do not adequately explain all the variation in insecticide resistance phenotypes observed in natural populations, and their relative functional impact in the field remains poorly understood.
Mosquito persistence may also occur due to heritable changes in behavior selected for by control interventions, so-called behavioral resistance. Recent work has captured mosquito interactions with bednets using mosquito-tracking cameras [172] and could be extended to other interventions (e.g., traps, sprays, repellents). Consistent longitudinal studies are also needed to track changes in mosquito biting behavior (e.g., outdoor versus indoor, evening versus night) after the use of interventions and to discriminate these changes from variation in species frequencies at specific sites. Subsequent genomic analyses could then reveal if there is a genetic component to these modified behaviors.
Technology and its application to malaria biology
Fundamental technologies: Genomics and transcriptomics
Whole genome sequencing has already had a major impact on multiple areas of parasite and vector research. It has transformed our understanding of parasite biology and drug resistance (see “Advances, challenges, and opportunities in asexual-stage biology”). In addition, it has been widely used to study the population genetics of mosquito species in the field [74–76, 173], and the genomes of 19 Anopheles species spanning 3 subgenera and including major and minor malaria vectors from diverse geographical locations have now been sequenced [77, 171]. These genomic resources have improved our understanding of the patterns of gene flow within and among mosquito populations. These “big data” resources available to the research community allow for powerful comparative functional and evolutionary analyses that will help elucidate the common basis of vector competence and identify effective vector control targets across multiple species. Recent work using these datasets has identified a reproductive trait with consequences for vectoral capacity that has evolved within the Anopheles genus and presents new potential targets to induce sterility in field populations [174–176]. Additional targets may be identified as our understanding of the biological coordination of simultaneous egg development and parasite transmission is improved. The declining cost of sequencing will make such studies more feasible in the future, such that a mosquito resistome—similar to the parasite resistome—may be compiled.
Further advances in genomic technology will enable a detailed analysis of natural populations of Plasmodium spp. at a worldwide scale. These include single cell technologies for genome sequencing and transcriptomic analyses, genotyping, and whole genome sequencing from dried blood spot samples. In addition, further comparative genomics [177] among all Plasmodium species infecting humans as well as those infecting nonhuman primates should identify key pathways in host switching. Genomic analysis of longitudinal samples will allow for the identification of population structure changes associated with changing epidemiology and emerging drug resistance. Coupled with gene-editing technologies, hypotheses generated by comparative genomics can be functionally tested.
Technical advances in RNA sequencing now make it feasible to interrogate the dynamic gene expression profiles of both the human host and the parasite during infection. This will provide new insights into the host response during infection and the potential adaptation of parasites during the infective process.
Gene-manipulation technologies: Genome editing and transgenics
Genome engineering tools, such as CRISPR/Cas9 systems (see glossary in the malERA Refresh Introductory paper [178]), have transformed the ability to manipulate the genomes of P. falciparum, P. berghei (reviewed in [179]), and Anopheles and understand gene function. CRISPR/Cas9-based genetic engineering of P. falciparum asexual blood stages has allowed for more complex genetic modifications within the parasite; for example, the tetracycline repressor protein (TetR) aptamer system to control gene expression [84] utilized CRISPR/Cas9 as an initial step to introduce the aptameric cassette. Beyond CRISPR/Cas9, however, there have been several other successful gene-editing technologies (see Table 1).
With these powerful tools in place, we can now scale up the generation of conditional and/or complete knockout parasite libraries containing every single gene in the genome. Such an effort would greatly enhance our understanding of the biology of the parasite at all stages of development, as well as identify the functions of many hypothetical genes.
Gene-manipulation technologies: Gene drives
Mirroring the advances in gene-editing capabilities in the parasite, Anopheline spp. genomes can also now be engineered with unprecedented precision (see Table 1). Recent reports show that CRISPR/Cas9 gene-editing tools can be used for the generation of gene-drive systems [91, 92] that manipulate genetic inheritance in mosquitoes to spread anti-Plasmodium transgenes (population modification/replacement strategies) or lethality-inducing transgenes (population suppression strategies) through natural mosquito populations. Mendelian inheritance predicts 50% of offspring will inherit a transgene carried on one of a parent’s chromosomes. Genetic drive is the increased transmission of a genetic element to over 50% of offspring so that it increases in frequency in each generation. A gene drive typically refers to an artificial transgene that shows genetic drive by giving it the ability to trigger its own replication. A gene-drive transgene is copied from one chromosome to its homologous chromosome within germ line cells. With both chromosomes carrying a copy of the transgene (a homozygous germ line), all sperm or eggs derived from these cells will also carry the transgene, and if copying occurs in all germ cells, 100% of offspring will inherit the gene drive. This allows rapid spread of the gene drive (and its anti-Plasmodium cargo) into the mosquito population. A valuable debate on the safe use of gene drive systems has begun within the scientific community [180].
The feasibility of using gene drive strategies for mosquito control will need additional research efforts in 3 key areas. First, an understanding of mosquito mating biology and the determinants of male mating success and female mate choice will need to be developed. Colonization is likely to impact the mating ability of species that exhibit such a complicated mating behavior as swarming; mating competitiveness will be a key determinant of gene drive success. Second, effective, “evolution proof” gene-drive systems should be generated to preempt the selection of mosquitoes that are resistant to the drive mechanisms, which would otherwise reduce the efficiency of the drive. Gene drives will need to be optimized by testing different gene-drive architectures, especially if CRISPR/Cas9 mechanisms prove problematic. Third, effective antimalarial genes will need to be evaluated in a reliable and reproducible manner; many anti-Plasmodium factors have been identified and should be systematically tested in laboratory conditions for their ability to block parasite development within the mosquito host.
Consideration should be given to the formation of a consortium to evaluate and prioritize promising transgenic strategies and test these in multiple anopheline species and against a number of Plasmodium isolates. This represents an opportunity to avoid duplication of work; however, we would also argue for head-to-head comparison of transgenic strategies. Such a consortium could centralize resources, particularly in developing transgenic mosquitoes (e.g., injection service, mail-order mutants) and potentially a mutant library, but, currently, the space required for mosquito-line maintenance prevents this. As forward and reverse genetic screens become more realistic, we should develop methods to cryopreserve mosquito lines or, more realistically, store plasmids for injection to recreate lines as needed.
Cell - and tissue-based technologies
Since the discovery of malaria parasites by microscopy [181], imaging has played a central role in malaria research. However, recent advances in imaging techniques have allowed visualization of the parasite and its interactions with the mammalian host and insect vector at an unprecedented level of resolution [101] [182] [102]. We can expect that imaging will reveal other novel insights into the biology of human malaria parasites and play a major role in the science of malaria eradication.
New technologies to support tool development: Biomarkers and novel diagnostics
As our understanding of parasite biology advances—including insights into sequestration and dormancy—the potential to leverage emerging technologies to support the discovery of biomarkers of infection (see above) increases. Such insights into parasite biology are laying the foundation for novel diagnostic approaches based on more sensitive techniques to detect parasite byproducts (e.g., hemozoin) [183] or volatile substances [184]. When noninvasive, rapid, and inexpensive, these diagnostic approaches are likely to facilitate the identification of infected individuals who may be asymptomatic and/or functioning as reservoirs (see malERA Refresh papers on Tools [185] and the Reservoir and Transmission [166]).
Exosomes are key new players implicated in intercellular communication without direct cellular contact [186] and have a potential role as biomarkers [187]. The release of microparticles is augmented in human malaria [188, 189], and exosomes containing parasite proteins have been shown to be produced by infected cells [190] as well as by parasites [191, 192].
New technologies in vaccine development and leveraging existing human volunteer sample datasets
Protective immunity requires that human hosts recognize and respond appropriately to parasite-derived antigens and epitopes. Such immunity is complex, however, requiring both innate and acquired responses and biological regulation of such responses as well as ensuring the responses’ durability. Malaria parasites utilize a number of mechanisms to evade these immune responses, which infected hosts must then overcome. In this context, there is a fundamental gap in understanding the correlates of protective immunity in the human host that target exoerythrocytic-stage parasites in both P. falciparum and P. vivax. Multiple new technologies are now available to identify antigens and epitopes that are the targets of innate and acquired immune responses. Examples include high-throughput genomic sequencing, transcriptomics, and proteomics. Structural vaccinology [193–195] has proven immensely powerful in viral vaccine development through improved immunogen design and is now being applied to asexual blood stages [94–97]. Near-atomic resolution cryo-electron microscopy is now being used to inform antigen and drug target selection as well as the rational design of potent immunogens [196–198]. In addition, new technology platforms and novel adjuvants are being incorporated into vaccines to ensure appropriate immune responses are elicited. Approaches based on structural biology [98–100] and genomic sequencing [199] are now being introduced into the clinical evaluation of candidate malaria vaccines. These efforts provide an opportunity to further define the effective targets as well as the nature of protective immune responses.
An effective P. vivax vaccine strategy also needs to contend with the challenge of relapse infections. To prevent “relapse outbreaks,” antirelapse vaccines will need to be multistage and multivalent, including components to suppress blood-stage parasites emerging from the dormant liver stages as well as block transmission. There are relatively few P. vivax vaccine candidates progressing currently through the global pipeline [200].
Controlled human challenge studies are potentially transformational in enabling our understanding of the human immune response to malaria. Coupling controlled infections with technical advances for interrogating human immune cells in real time can give us new insights into both the temporal response and the contributions from innate and acquired immunity. Additionally, deeper interrogation of the immune profile of naturally acquired infections could also provide key insights. Providing access to them will require forethought in preparing future proposals, particularly with respect to human subject approvals, repository deposition, and community sharing. Harnessing available systems through existing networks as well as ongoing clinical trials could provide the necessary reagents and access to human samples.
Drug design and screening
The identification of potential targets through metabolomics and systems biology approaches coupled with advances in structural biology is now facilitating the design of compounds likely to interact with such targets. Moreover, high-throughput screening technologies are facilitating more rapid identification and prioritization of compounds for further investigation as potential leads, though corresponding techniques in high-throughput synthesis and characterization of small molecules require further development. In a reverse approach, high-throughput phenotypic screens are also enabling the selection of compounds whose structures can subsequently be used to inform the identification of potential molecular interactions and metabolic pathways for further analysis as targets for pharmacologic intervention (reviewed in [201]). It is important to note that because malaria primarily affects the developing world, the opportunity for profit is reduced. Malaria, with the assistance of the community and funders such as Medicines for Malaria Venture (MMV), has and will continue to function as a model for open source drug discovery [202–204].
Technologies targeting mosquito-based interventions: Paratransgenesis and genetically modified mosquitoes
Recent years have seen a focus toward the identification of microbial populations that can block parasite development in the mosquito vector [205–208]. Genetic modification of these bacterial populations (paratransgenesis) could be a key tool, particularly for the control of outdoor biting and resting mosquito populations that are not currently targeted by insecticide-based strategies. Advances in Wolbachia bacteria experiments in Anopheles mosquitoes are particularly promising. Wolbachia are intracellular endosymbiotic bacteria that, in some insects, spread through populations by maternal transmission and cytoplasmic incompatibility. These endosymbionts were shown to block malaria parasite development in artificial settings [209] and were negatively correlated with Plasmodium infections in natural A. coluzzii populations from Burkina Faso [210, 211]. Two key research priorities are the development of a method to transform Wolbachia to deliver effective antiplasmodial genes and understanding the role of natural Wolbachia infections in malaria transmission dynamics.
In light of widespread resistance to currently used insecticides, the identification of alternative, safe, active compounds that can extend the lifetime of long-lasting insecticide-treated nets (LLINs) and indoor residual spraying (IRS) is imperative. The study of key pathways in mosquito reproduction, susceptibility to infection, blood feeding behavior, and longevity that can be effectively targeted to reduce vectoral capacity is therefore a priority. For example, new sterilizing compounds that interfere with key hormonal reproductive pathways, such as those regulated by juvenile hormone and 20-hydroxyecdysone, could be incorporated into mosquito nets to reduce mosquito fertility, including insecticide-resistant mosquitoes that may survive exposure to the net.
A key issue in applying these novel strategies will be achieving effective colonization of anopheline species, as the lack of mosquito colonies is preventing studies on the biology of important malaria vectors. An important breakthrough has been the recent colonization of A. darlingi, the most important American vector [93]. On the road to eradication, a deeper understanding of the biology and behavior of these species will be essential.
Conclusions
As illustrated above, recent advances in basic science are providing deeper insights into the biology of the parasite, the mosquito vector, and the human host as well as their interactions at molecular, cellular, and organismic levels. Coupling these insights with recent technologies that help pinpoint potential methods to intervene or disrupt essential interactions can spur the use of novel tools to help eliminate and, ultimately, eradicate malaria.
Zdroje
1. The malERA Consultative Group on Basic Science and Enabling Technologies. A Research Agenda for Malaria Eradication: Basic Science and Enabling Technologies. PLOS Medicine. 2011;8(1):e1000399. doi: 10.1371/journal.pmed.1000399 21311584
2. Eksi S, Morahan BJ, Haile Y, Furuya T, Jiang H, Ali O, et al. Plasmodium falciparum gametocyte development 1 (Pfgdv1) and gametocytogenesis early gene identification and commitment to sexual development. PLoS pathogens. 2012;8(10):e1002964. Epub 2012/10/25. doi: 10.1371/journal.ppat.1002964 PPATHOGENS-D-12-01039 [pii]. 23093935; PubMed Central PMCID: PMC3475683.
3. Ikadai H, Shaw Saliba K, Kanzok SM, McLean KJ, Tanaka TQ, Cao J, et al. Transposon mutagenesis identifies genes essential for Plasmodium falciparum gametocytogenesis. Proc Natl Acad Sci U S A. 2013;110(18):E1676–84. Epub 2013/04/11. doi: 10.1073/pnas.1217712110 1217712110 [pii]. 23572579; PubMed Central PMCID: PMC3645567.
4. Brancucci NM, Bertschi NL, Zhu L, Niederwieser I, Chin WH, Wampfler R, et al. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell host & microbe. 2014;16(2):165–76. Epub 2014/08/15. doi: 10.1016/j.chom.2014.07.004 S1931-3128(14)00258-3 [pii]. 25121746.
5. Kafsack BF, Rovira-Graells N, Clark TG, Bancells C, Crowley VM, Campino SG, et al. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature. 2014;507(7491):248–52. Epub 2014/02/28. doi: 10.1038/nature12920 nature12920 [pii]. 24572369; PubMed Central PMCID: PMC4040541.
6. Sinha A, Hughes KR, Modrzynska KK, Otto TD, Pfander C, Dickens NJ, et al. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature. 2014;507(7491):253–7. Epub 2014/02/28. doi: 10.1038/nature12970 nature12970 [pii]. 24572359; PubMed Central PMCID: PMC4105895.
7. Duffy S, Avery VM. Identification of inhibitors of Plasmodium falciparum gametocyte development. Malaria journal. 2013;12 : 408. Epub 2013/11/12. doi: 10.1186/1475-2875-12-408 [pii]. 24206914; PubMed Central PMCID: PMC3842684.
8. Brancucci NM, Goldowitz I, Buchholz K, Werling K, Marti M. An assay to probe Plasmodium falciparum growth, transmission stage formation and early gametocyte development. Nature protocols. 2015;10(8):1131–42. doi: 10.1038/nprot.2015.072 26134953; PubMed Central PMCID: PMC4581880.
9. Duffy S, Loganathan S, Holleran JP, Avery VM. Large-scale production of Plasmodium falciparum gametocytes for malaria drug discovery. Nature protocols. 2016;11(5):976–92. doi: 10.1038/nprot.2016.056 27123949.
10. Lucantoni L, Fidock DA, Avery VM. Luciferase-Based, High-Throughput Assay for Screening and Profiling Transmission-Blocking Compounds against Plasmodium falciparum Gametocytes. Antimicrobial agents and chemotherapy. 2016;60(4):2097–107. doi: 10.1128/AAC.01949-15 26787698; PubMed Central PMCID: PMC4808229.
11. Plouffe DM, Wree M, Du AY, Meister S, Li F, Patra K, et al. High-Throughput Assay and Discovery of Small Molecules that Interrupt Malaria Transmission. Cell host & microbe. 2016;19(1):114–26. doi: 10.1016/j.chom.2015.12.001 26749441; PubMed Central PMCID: PMC4723716.
12. Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464(7285):66–71. Epub 2010/02/05. nature08834 [pii] doi: 10.1038/nature08834 20130575; PubMed Central PMCID: PMC2833235.
13. Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2010;107(9):4418–23. Epub 2010/02/18. doi: 10.1073/pnas.0913392107 0913392107 [pii]. 20160092; PubMed Central PMCID: PMC2840125.
14. Rinker DC, Pitts RJ, Zhou X, Suh E, Rokas A, Zwiebel LJ. Blood meal-induced changes to antennal transcriptome profiles reveal shifts in odor sensitivities in Anopheles gambiae. Proc Natl Acad Sci U S A. 2013;110(20):8260–5. doi: 10.1073/pnas.1302562110 23630291; PubMed Central PMCID: PMCPMC3657813.
15. Pellegrino M, Nakagawa T, Vosshall LB. Single sensillum recordings in the insects Drosophila melanogaster and Anopheles gambiae. J Vis Exp. 2010;(36):1–5. Epub 2010/02/19. doi: 10.3791/1725 1725 [pii]. 20164822; PubMed Central PMCID: PMC2830253.
16. Tauxe GM, MacWilliam D, Boyle SM, Guda T, Ray A. Targeting a dual detector of skin and CO2 to modify mosquito host seeking. Cell. 2013;155(6):1365–79. Epub 2013/12/10. doi: 10.1016/j.cell.2013.11.013 S0092-8674(13)01426-8 [pii]. 24315103; PubMed Central PMCID: PMC3899525.
17. Molina-Cruz A, DeJong RJ, Ortega C, Haile A, Abban E, Rodrigues J, et al. Some strains of Plasmodium falciparum, a human malaria parasite, evade the complement-like system of Anopheles gambiae mosquitoes. Proc Natl Acad Sci U S A. 2012;109(28):E1957–62. doi: 10.1073/pnas.1121183109 22623529; PubMed Central PMCID: PMC3396512.
18. Ghosh AK, Devenport M, Jethwaney D, Kalume DE, Pandey A, Anderson VE, et al. Malaria parasite invasion of the mosquito salivary gland requires interaction between the Plasmodium TRAP and the Anopheles saglin proteins. PLoS pathogens. 2009;5(1):e1000265. doi: 10.1371/journal.ppat.1000265 19148273; PubMed Central PMCID: PMC2613030.
19. Ghosh AK, Coppens I, Gardsvoll H, Ploug M, Jacobs-Lorena M. Plasmodium ookinetes coopt mammalian plasminogen to invade the mosquito midgut. Proc Natl Acad Sci U S A. 2011;108(41):17153–8. doi: 10.1073/pnas.1103657108 21949403; PubMed Central PMCID: PMC3193258.
20. Vega-Rodriguez J, Ghosh AK, Kanzok SM, Dinglasan RR, Wang S, Bongio NJ, et al. Multiple pathways for Plasmodium ookinete invasion of the mosquito midgut. Proc Natl Acad Sci U S A. 2014;111(4):E492–500. Epub 2014/01/30. doi: 10.1073/pnas.1315517111 1315517111 [pii]. 24474798; PubMed Central PMCID: PMC3910608.
21. Bounkeua V, Li F, Chuquiyauri R, Abeles SR, McClean CM, Neyra V, et al. Lack of molecular correlates of Plasmodium vivax ookinete development. Am J Trop Med Hyg. 2011;85(2):207–13. Epub 2011/08/05. doi: 10.4269/ajtmh.2011.10-0729 21813836; PubMed Central PMCID: PMC3144814.
22. Bounkeua V, Li F, Vinetz JM. In vitro generation of Plasmodium falciparum ookinetes. Am J Trop Med Hyg. 2010;83(6):1187–94. Epub 2010/12/02. doi: 10.4269/ajtmh.2010.10-0433 21118920; PubMed Central PMCID: PMC2990030.
23. Delves MJ, Sinden RE. A semi-automated method for counting fluorescent malaria oocysts increases the throughput of transmission blocking studies. Malaria journal. 2010;9 : 35. Epub 2010/02/02. doi: 10.1186/1475-2875-9-35 20113492; PubMed Central PMCID: PMC2824803.
24. Ghosh AK, Dinglasan RR, Ikadai H, Jacobs-Lorena M. An improved method for the in vitro differentiation of Plasmodium falciparum gametocytes into ookinetes. Malaria journal. 2010;9 (1)(194). http://dx.doi.org/10.1186/1475-2875-9-194. 2010408089.
25. Ghosh AK, Jacobs-Lorena M. In Vitro Differentiation of Plasmodium falciparum Gametocytes into Ookinetes. Methods in molecular biology. 2013;923 : 27–33. Epub 2012/09/20. doi: 10.1007/978-1-62703-026-7_3 22990769.
26. Mikolajczak SA, Vaughan AM, Kangwanrangsan N, Roobsoong W, Fishbaugher M, Yimamnuaychok N, et al. Plasmodium vivax liver stage development and hypnozoite persistence in human liver-chimeric mice. Cell host & microbe. 2015;17(4):526–35. doi: 10.1016/j.chom.2015.02.011 25800544.
27. Soulard V, Bosson-Vanga H, Lorthiois A, Roucher C, Franetich JF, Zanghi G, et al. Plasmodium falciparum full life cycle and Plasmodium ovale liver stages in humanized mice. Nat Commun. 2015;6 : 7690. Epub 2015/07/25. doi: 10.1038/ncomms8690 26205537; PubMed Central PMCID: PMCPMC4525212.
28. March S, Ng S, Velmurugan S, Galstian A, Shan J, Logan DJ, et al. A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax. Cell host & microbe. 2013;14(1):104–15. Epub 2013/07/23. doi: 10.1016/j.chom.2013.06.005 23870318; PubMed Central PMCID: PMC3780791.
29. Ng S, Schwartz RE, March S, Galstian A, Gural N, Shan J, et al. Human iPSC-derived hepatocyte-like cells support plasmodium liver-stage infection in vitro. Stem Cell Reports. 2015;4 (3):348–59. http://dx.doi.org/10.1016/j.stemcr.2015.01.002. doi: 10.1016/j.stemcr.2015.01.002 25660406.
30. Dembele L, Franetich JF, Lorthiois A, Gego A, Zeeman AM, Kocken CH, et al. Persistence and activation of malaria hypnozoites in long-term primary hepatocyte cultures. Nat Med. 2014;20(3):307–12. Epub 2014/02/11. doi: 10.1038/nm.3461 24509527.
31. Vaughan AM, Pinapati RS, Cheeseman IH, Camargo N, Fishbaugher M, Checkley LA, et al. Plasmodium falciparum genetic crosses in a humanized mouse model. Nature methods. 2015;12(7):631–3. Epub 2015/06/02. doi: 10.1038/nmeth.3432 26030447; PubMed Central PMCID: PMC4547688.
32. Voorberg-van der Wel A, Zeeman AM, van Amsterdam SM, van den Berg A, Klooster EJ, Iwanaga S, et al. Transgenic fluorescent Plasmodium cynomolgi liver stages enable live imaging and purification of Malaria hypnozoite-forms. PloS one. 2013;8(1):e54888. doi: 10.1371/journal.pone.0054888 23359816; PubMed Central PMCID: PMC3554669.
33. Herrera S, Solarte Y, Jordan-Villegas A, Echavarria JF, Rocha L, Palacios R, et al. Consistent safety and infectivity in sporozoite challenge model of Plasmodium vivax in malaria-naive human volunteers. Am J Trop Med Hyg. 2011;84(2 Suppl):4–11. Epub 2011/02/15. doi: 10.4269/ajtmh.2011.09-0498 84/2_Suppl/4 [pii]. 21292872; PubMed Central PMCID: PMC3032484.
34. Seder RA, Chang LJ, Enama ME, Zephir KL, Sarwar UN, Gordon IJ, et al. Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science. 2013;341(6152):1359–65. Epub 2013/08/10. doi: 10.1126/science.1241800 science.1241800 [pii]. 23929949.
35. Sheehy SH, Spencer AJ, Douglas AD, Sim BK, Longley RJ, Edwards NJ, et al. Optimising Controlled Human Malaria Infection Studies Using Cryopreserved P. falciparum Parasites Administered by Needle and Syringe. PloS one. 2013;8(6):e65960. Epub 2013/07/05. doi: 10.1371/journal.pone.0065960 PONE-D-12-35789 [pii]. 23823332; PubMed Central PMCID: PMC3688861.
36. Behet MC, Foquet L, van Gemert GJ, Bijker EM, Meuleman P, Leroux-Roels G, et al. Sporozoite immunization of human volunteers under chemoprophylaxis induces functional antibodies against pre-erythrocytic stages of Plasmodium falciparum. Malaria journal. 2014;13 : 136. Epub 2014/04/09. doi: 10.1186/1475-2875-13-136 1475-2875-13-136 [pii]. 24708526; PubMed Central PMCID: PMC4113136.
37. Talley AK, Healy SA, Finney OC, Murphy SC, Kublin J, Salas CJ, et al. Safety and comparability of controlled human Plasmodium falciparum infection by mosquito bite in malaria-naive subjects at a new facility for sporozoite challenge. PloS one. 2014;9(11):e109654. Epub 2014/11/19. doi: 10.1371/journal.pone.0109654 PONE-D-14-30972 [pii]. 25405724; PubMed Central PMCID: PMC4236046.
38. Gomez-Perez GP, Legarda A, Munoz J, Sim BK, Ballester MR, Dobano C, et al. Controlled human malaria infection by intramuscular and direct venous inoculation of cryopreserved Plasmodium falciparum sporozoites in malaria-naive volunteers: effect of injection volume and dose on infectivity rates. Malaria journal. 2015;14 : 306. Epub 2015/08/08. doi: 10.1186/s12936-015-0817-x 10.1186/s12936-015-0817-x [pii]. 26245196; PubMed Central PMCID: PMC4527105.
39. Ockenhouse CF, Regules J, Tosh D, Cowden J, Kathcart A, Cummings J, et al. Ad35.CS.01-RTS,S/AS01 Heterologous Prime Boost Vaccine Efficacy against Sporozoite Challenge in Healthy Malaria-Naive Adults. PloS one. 2015;10(7):e0131571. Epub 2015/07/07. doi: 10.1371/journal.pone.0131571 PONE-D-14-29774 [pii]. 26148007; PubMed Central PMCID: PMC4492580.
40. Schats R, Bijker EM, van Gemert GJ, Graumans W, van de Vegte-Bolmer M, van Lieshout L, et al. Heterologous Protection against Malaria after Immunization with Plasmodium falciparum Sporozoites. PloS one. 2015;10(5):e0124243. Epub 2015/05/02. doi: 10.1371/journal.pone.0124243 PONE-D-14-43731 [pii]. 25933168; PubMed Central PMCID: PMC4416703. 25933168
41. Spring M, Polhemus M, Ockenhouse C. Controlled human malaria infection. The Journal of infectious diseases. 2014;209 Suppl 2:S40–5. Epub 2014/05/30. doi: 10.1093/infdis/jiu063 jiu063 [pii]. 24872394.
42. Russell B, Suwanarusk R, Borlon C, Costa FT, Chu CS, Rijken MJ, et al. A reliable ex vivo invasion assay of human reticulocytes by Plasmodium vivax. Blood. 2011;118(13):e74–81. doi: 10.1182/blood-2011-04-348748 21768300; PubMed Central PMCID: PMC3438884.
43. Russell B, Suwanarusk R, Malleret B, Costa FT, Snounou G, Kevin Baird J, et al. Human ex vivo studies on asexual Plasmodium vivax: the best way forward. International journal for parasitology. 2012;42(12):1063–70. doi: 10.1016/j.ijpara.2012.08.010 23032102.
44. Borlon C, Russell B, Sriprawat K, Suwanarusk R, Erhart A, Renia L, et al. Cryopreserved Plasmodium vivax and cord blood reticulocytes can be used for invasion and short term culture. International journal for parasitology. 2012;42(2):155–60. doi: 10.1016/j.ijpara.2011.10.011 22240310; PubMed Central PMCID: PMC3438882.
45. Noulin F, Borlon C, van den Eede P, Boel L, Verfaillie CM, D'Alessandro U, et al. Cryopreserved reticulocytes derived from hematopoietic stem cells can be invaded by cryopreserved Plasmodium vivax isolates. PloS one. 2012;7(7):e40798. doi: 10.1371/journal.pone.0040798 22844411; PubMed Central PMCID: PMC3402485.
46. Tantular IS, Pusarawati S, Khin L, Kanbe T, Kimura M, Kido Y, et al. Preservation of wild isolates of human malaria parasites in wet ice and adaptation efficacy to in vitro culture. Tropical medicine and health. 2012;40(2):37–45. doi: 10.2149/tmh.2012-07o 23097618; PubMed Central PMCID: PMC3475313.
47. Noulin F, Borlon C, Van Den Abbeele J, D'Alessandro U, Erhart A. 1912–2012: a century of research on Plasmodium vivax in vitro culture. Trends in parasitology. 2013;29(6):286–94. doi: 10.1016/j.pt.2013.03.012 23623759.
48. Galinski MR, Meyer EV, Barnwell JW. Plasmodium vivax: modern strategies to study a persistent parasite's life cycle. Advances in parasitology. 2013;81 : 1–26. doi: 10.1016/B978-0-12-407826-0.00001-1 23384620.
49. Kurita R, Suda N, Sudo K, Miharada K, Hiroyama T, Miyoshi H, et al. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PloS one. 2013;8(3):e59890. doi: 10.1371/journal.pone.0059890 23533656; PubMed Central PMCID: PMC3606290.
50. Martin-Jaular L, Elizalde-Torrent A, Thomson-Luque R, Ferrer M, Segovia JC, Herreros-Aviles E, et al. Reticulocyte-prone malaria parasites predominantly invade CD71hi immature cells: implications for the development of an in vitro culture for Plasmodium vivax. Malaria journal. 2013;12 : 434. doi: 10.1186/1475-2875-12-434 24289105; PubMed Central PMCID: PMC4220676.
51. Zeeman AM, der Wel AV, Kocken CH. Ex vivo culture of Plasmodium vivax and Plasmodium cynomolgi and in vitro culture of Plasmodium knowlesi blood stages. Methods in molecular biology. 2013;923 : 35–49. doi: 10.1007/978-1-62703-026-7_4 22990770.
52. Furuya T, Sa JM, Chitnis CE, Wellems TE, Stedman TT. Reticulocytes from cryopreserved erythroblasts support Plasmodium vivax infection in vitro. Parasitology international. 2014;63(2):278–84. doi: 10.1016/j.parint.2013.11.011 24291603; PubMed Central PMCID: PMC3943572.
53. Noulin F, Manesia JK, Rosanas-Urgell A, Erhart A, Borlon C, Van Den Abbeele J, et al. Hematopoietic stem/progenitor cell sources to generate reticulocytes for Plasmodium vivax culture. PloS one. 2014;9(11):e112496. doi: 10.1371/journal.pone.0112496 25393299; PubMed Central PMCID: PMC4231068.
54. Roobsoong W, Tharinjaroen CS, Rachaphaew N, Chobson P, Schofield L, Cui L, et al. Improvement of culture conditions for long-term in vitro culture of Plasmodium vivax. Malaria journal. 2015;14 : 297. doi: 10.1186/s12936-015-0815-z 26243280; PubMed Central PMCID: PMC4524445.
55. Thomson-Luque R, Scopel KK. Immature reticulocytes as preferential host cells and the challenges for in vitro culture of Plasmodium vivax. Pathogens and global health. 2015;109(3):91–2. doi: 10.1179/2047772415Z.000000000264 25943155; PubMed Central PMCID: PMC4455358.
56. Moon RW, Hall J, Rangkuti F, Ho YS, Almond N, Mitchell GH, et al. Adaptation of the genetically tractable malaria pathogen Plasmodium knowlesi to continuous culture in human erythrocytes. Proc Natl Acad Sci U S A. 2013;110(2):531–6. Epub 2012/12/26. doi: 10.1073/pnas.1216457110 1216457110 [pii]. 23267069; PubMed Central PMCID: PMC3545754.
57. Gruring C, Moon RW, Lim C, Holder AA, Blackman MJ, Duraisingh MT. Human red blood cell-adapted Plasmodium knowlesi parasites: a new model system for malaria research. Cellular microbiology. 2014;16(5):612–20. doi: 10.1111/cmi.12275 24506567; PubMed Central PMCID: PMC4004062.
58. Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, et al. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336(6077):79–82. Epub 2012/04/12. doi: 10.1126/science.1215966 336/6077/79 [pii]. 22491853; PubMed Central PMCID: PMC3355473.
59. Takala-Harrison S, Clark TG, Jacob CG, Cummings MP, Miotto O, Dondorp AM, et al. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc Natl Acad Sci U S A. 2013;110(1):240–5. Epub 2012/12/19. doi: 10.1073/pnas.1211205110 1211205110 [pii]. 23248304; PubMed Central PMCID: PMC3538248.
60. Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis. 2013;13(12):1043–9. Epub 2013/09/17. doi: 10.1016/S1473-3099(13)70252-4 S1473-3099(13)70252-4 [pii]. 24035558.
61. Witkowski B, Khim N, Chim P, Kim S, Ke S, Kloeung N, et al. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrobial agents and chemotherapy. 2013;57(2):914–23. Epub 2012/12/05. doi: 10.1128/AAC.01868-12 AAC.01868-12 [pii]. 23208708; PubMed Central PMCID: PMC3553720.
62. Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505(7481):50–5. Epub 2013/12/20. doi: 10.1038/nature12876 24352242.
63. Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371(5):411–23. Epub 2014/07/31. doi: 10.1056/NEJMoa1314981 25075834; PubMed Central PMCID: PMC4143591.
64. Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, et al. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis. 2015;15(4):415–21. Epub 2015/02/24. doi: 10.1016/S1473-3099(15)70032-0 S1473-3099(15)70032-0 [pii]. 25704894; PubMed Central PMCID: PMC4374103.
65. Winzeler EA, Manary MJ. Drug resistance genomics of the antimalarial drug artemisinin. Genome Biol. 2014;15(11):544. Epub 2014/12/04. doi: 10.1186/s13059-014-0544-6 25470531; PubMed Central PMCID: PMCPMC4283579.
66. Fairhurst RM, Dondorp AM. Artemisinin-Resistant Plasmodium falciparum Malaria. Microbiology spectrum. 2016;4(3). Epub 2016/06/24. doi: 10.1128/microbiolspec.EI10-0013-2016 27337450; PubMed Central PMCID: PMCPMC4992992.
67. Tilley L, Straimer J, Gnadig NF, Ralph SA, Fidock DA. Artemisinin Action and Resistance in Plasmodium falciparum. Trends in parasitology. 2016;32(9):682–96. Epub 2016/06/13. doi: 10.1016/j.pt.2016.05.010 27289273; PubMed Central PMCID: PMCPMC5007624.
68. MacRae JI, Dixon MW, Dearnley MK, Chua HH, Chambers JM, Kenny S, et al. Mitochondrial metabolism of sexual and asexual blood stages of the malaria parasite Plasmodium falciparum. BMC Biol. 2013;11 : 67. Epub 2013/06/15. doi: 10.1186/1741-7007-11-67 23763941; PubMed Central PMCID: PMC3704724.
69. Gulati S, Ekland EH, Ruggles KV, Chan RB, Jayabalasingham B, Zhou B, et al. Profiling the Essential Nature of Lipid Metabolism in Asexual Blood and Gametocyte Stages of Plasmodium falciparum. Cell host & microbe. 2015;18(3):371–81. doi: 10.1016/j.chom.2015.08.003 26355219; PubMed Central PMCID: PMC4567697.
70. Dao A, Yaro AS, Diallo M, Timbine S, Huestis DL, Kassogue Y, et al. Signatures of aestivation and migration in Sahelian malaria mosquito populations. Nature. 2014;516(7531):387–90. doi: 10.1038/nature13987 25470038; PubMed Central PMCID: PMC4306333.
71. Mitchell SN, Stevenson BJ, Muller P, Wilding CS, Egyir-Yawson A, Field SG, et al. Identification and validation of a gene causing cross-resistance between insecticide classes in Anopheles gambiae from Ghana. Proc Natl Acad Sci U S A. 2012;109(16):6147–52. Epub 2012/03/31. doi: 10.1073/pnas.1203452109 1203452109 [pii]. 22460795; PubMed Central PMCID: PMC3341073.
72. Kwiatkowska RM, Platt N, Poupardin R, Irving H, Dabire RK, Mitchell S, et al. Dissecting the mechanisms responsible for the multiple insecticide resistance phenotype in Anopheles gambiae s.s., M form, from Vallee du Kou, Burkina Faso. Gene. 2013;519(1):98–106. Epub 2013/02/06. doi: 10.1016/j.gene.2013.01.036 S0378-1119(13)00077-2 [pii]. 23380570; PubMed Central PMCID: PMC3611593.
73. Riveron JM, Yunta C, Ibrahim SS, Djouaka R, Irving H, Menze BD, et al. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 2014;15(2):R27. Epub 2014/02/26. doi: 10.1186/gb-2014-15-2-r27 gb-2014-15-2-r27 [pii]. 24565444; PubMed Central PMCID: PMC4054843.
74. Reidenbach KR, Neafsey DE, Costantini C, Sagnon N, Simard F, Ragland GJ, et al. Patterns of genomic differentiation between ecologically differentiated M and S forms of Anopheles gambiae in West and Central Africa. Genome Biol Evol. 2012;4(12):1202–12. Epub 2012/11/08. doi: 10.1093/gbe/evs095 evs095 [pii]. 23132896; PubMed Central PMCID: PMC3542583.
75. Pinto J, Egyir-Yawson A, Vicente JI, Gomes B, Santolamazza F, Moreno M, et al. Geographic population structure of the African malaria vectorAnopheles gambiaesuggests a role for the forest-savannah biome transition as a barrier to gene flow. Evolutionary Applications. 2013;6(6):910–24. doi: 10.1111/eva.12075 24062800
76. O'Loughlin SM, Magesa S, Mbogo C, Mosha F, Midega J, Lomas S, et al. Genomic analyses of three malaria vectors reveals extensive shared polymorphism but contrasting population histories. Mol Biol Evol. 2014;31(4):889–902. Epub 2014/01/11. doi: 10.1093/molbev/msu040 msu040 [pii]. 24408911; PubMed Central PMCID: PMC3969563.
77. Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, et al. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science. 2015;347(6217):1258522. Epub 2015/01/03. doi: 10.1126/science.1258522 1258522 [pii] science.1258522 [pii]. 25554792; PubMed Central PMCID: PMC4380271.
78. van Schaijk BC, Vos MW, Janse CJ, Sauerwein RW, Khan SM. Removal of heterologous sequences from Plasmodium falciparum mutants using FLPe-recombinase. PloS one. 2010;5(11):e15121. doi: 10.1371/journal.pone.0015121 21152048; PubMed Central PMCID: PMC2994908.
79. Muralidharan V, Oksman A, Iwamoto M, Wandless TJ, Goldberg DE. Asparagine repeat function in a Plasmodium falciparum protein assessed via a regulatable fluorescent affinity tag. Proc Natl Acad Sci U S A. 2011;108(11):4411–6. Epub 2011/03/04. doi: 10.1073/pnas.1018449108 21368162; PubMed Central PMCID: PMC3060247.
80. O'Neill MT, Phuong T, Healer J, Richard D, Cowman AF. Gene deletion from Plasmodium falciparum using FLP and Cre recombinases: implications for applied site-specific recombination. International journal for parasitology. 2011;41(1):117–23. doi: 10.1016/j.ijpara.2010.08.001 20816845.
81. Straimer J, Lee MC, Lee AH, Zeitler B, Williams AE, Pearl JR, et al. Site-specific genome editing in Plasmodium falciparum using engineered zinc-finger nucleases. Nature methods. 2012;9(10):993–8. doi: 10.1038/nmeth.2143 22922501; PubMed Central PMCID: PMC3697006.
82. Collins CR, Das S, Wong EH, Andenmatten N, Stallmach R, Hackett F, et al. Robust inducible Cre recombinase activity in the human malaria parasite Plasmodium falciparum enables efficient gene deletion within a single asexual erythrocytic growth cycle. Molecular microbiology. 2013;88(4):687–701. doi: 10.1111/mmi.12206 23489321; PubMed Central PMCID: PMC3708112.
83. Prommana P, Uthaipibull C, Wongsombat C, Kamchonwongpaisan S, Yuthavong Y, Knuepfer E, et al. Inducible knockdown of Plasmodium gene expression using the glmS ribozyme. PloS one. 2013;8(8):e73783. doi: 10.1371/journal.pone.0073783 24023691; PubMed Central PMCID: PMC3758297.
84. Goldfless SJ, Wagner JC, Niles JC. Versatile control of Plasmodium falciparum gene expression with an inducible protein-RNA interaction. Nat Commun. 2014;5 : 5329. doi: 10.1038/ncomms6329 25370483; PubMed Central PMCID: PMC4223869.
85. Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nature biotechnology. 2014;32(8):819–21. doi: 10.1038/nbt.2925 24880488.
86. Wagner JC, Platt RJ, Goldfless SJ, Zhang F, Niles JC. Efficient CRISPR-Cas9-mediated genome editing in Plasmodium falciparum. Nature methods. 2014;11(9):915–8. doi: 10.1038/nmeth.3063 25108687; PubMed Central PMCID: PMC4199390.
87. Windbichler N, Menichelli M, Papathanos PA, Thyme SB, Li H, Ulge UY, et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature. 2011;473(7346):212–5. doi: 10.1038/nature09937 21508956; PubMed Central PMCID: PMC3093433.
88. Isaacs AT, Jasinskiene N, Tretiakov M, Thiery I, Zettor A, Bourgouin C, et al. Transgenic Anopheles stephensi coexpressing single-chain antibodies resist Plasmodium falciparum development. Proc Natl Acad Sci U S A. 2012;109(28):E1922–30. doi: 10.1073/pnas.1207738109 22689959; PubMed Central PMCID: PMC3396534.
89. Bernardini F, Galizi R, Menichelli M, Papathanos PA, Dritsou V, Marois E, et al. Site-specific genetic engineering of the Anopheles gambiae Y chromosome. Proc Natl Acad Sci U S A. 2014;111(21):7600–5. doi: 10.1073/pnas.1404996111 24821795; PubMed Central PMCID: PMC4040617.
90. Galizi R, Doyle LA, Menichelli M, Bernardini F, Deredec A, Burt A, et al. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat Commun. 2014;5 : 3977. doi: 10.1038/ncomms4977 24915045; PubMed Central PMCID: PMC4057611.
91. Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci U S A. 2015;112(49):E6736–43. doi: 10.1073/pnas.1521077112 26598698; PubMed Central PMCID: PMC4679060.
92. Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nature biotechnology. 2016;34(1):78–83. doi: 10.1038/nbt.3439 26641531.
93. Moreno M, Tong C, Guzman M, Chuquiyauri R, Llanos-Cuentas A, Rodriguez H, et al. Infection of laboratory-colonized Anopheles darlingi mosquitoes by Plasmodium vivax. Am J Trop Med Hyg. 2014;90(4):612–6. doi: 10.4269/ajtmh.13-0708 24534811; PubMed Central PMCID: PMC3973502.
94. Batchelor JD, Zahm JA, Tolia NH. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nature structural & molecular biology. 2011;18(8):908–14. doi: 10.1038/nsmb.2088 21743458; PubMed Central PMCID: PMC3150435.
95. Batchelor JD, Malpede BM, Omattage NS, DeKoster GT, Henzler-Wildman KA, Tolia NH. Red blood cell invasion by Plasmodium vivax: structural basis for DBP engagement of DARC. PLoS pathogens. 2014;10(1):e1003869. doi: 10.1371/journal.ppat.1003869 24415938; PubMed Central PMCID: PMC3887093.
96. Chen E, Paing MM, Salinas N, Sim BK, Tolia NH. Structural and functional basis for inhibition of erythrocyte invasion by antibodies that target Plasmodium falciparum EBA-175. PLoS pathogens. 2013;9(5):e1003390. doi: 10.1371/journal.ppat.1003390 23717209; PubMed Central PMCID: PMC3662668.
97. Chen E, Salinas ND, Ntumngia FB, Adams JH, Tolia NH. Structural Analysis of the Synthetic Duffy Binding Protein (DBP) Antigen DEKnull Relevant for Plasmodium vivax Malaria Vaccine Design. PLOS Neglected Tropical Diseases. 2015;9(3):e0003644. doi: 10.1371/journal.pntd.0003644 25793371
98. Thera MA, Doumbo OK, Coulibaly D, Laurens MB, Ouattara A, Kone AK, et al. A field trial to assess a blood-stage malaria vaccine. N Engl J Med. 2011;365(11):1004–13. Epub 2011/09/16. doi: 10.1056/NEJMoa1008115 21916638; PubMed Central PMCID: PMC3242358.
99. Ouattara A, Takala-Harrison S, Thera MA, Coulibaly D, Niangaly A, Saye R, et al. Molecular basis of allele-specific efficacy of a blood-stage malaria vaccine: vaccine development implications. The Journal of infectious diseases. 2013;207(3):511–9. Epub 2012/12/04. doi: 10.1093/infdis/jis709 jis709 [pii]. 23204168; PubMed Central PMCID: PMC3537449.
100. Graves SF, Kouriba B, Diarra I, Daou M, Niangaly A, Coulibaly D, et al. Strain-specific Plasmodium falciparum multifunctional CD4(+) T cell cytokine expression in Malian children immunized with the FMP2.1/AS02A vaccine candidate. Vaccine. 2016;34(23):2546–55. Epub 2016/04/19. doi: 10.1016/j.vaccine.2016.04.019 S0264-410X(16)30148-7 [pii]. 27087149.
101. Gueirard P, Tavares J, Thiberge S, Bernex F, Ishino T, Milon G, et al. Development of the malaria parasite in the skin of the mammalian host. Proc Natl Acad Sci U S A. 2010;107(43):18640–5. doi: 10.1073/pnas.1009346107 20921402; PubMed Central PMCID: PMC2972976.
102. Gruring C, Heiber A, Kruse F, Ungefehr J, Gilberger TW, Spielmann T. Development and host cell modifications of Plasmodium falciparum blood stages in four dimensions. Nat Commun. 2011;2 : 165. doi: 10.1038/ncomms1169 21266965.
103. Joice R, Nilsson SK, Montgomery J, Dankwa S, Egan E, Morahan B, et al. Plasmodium falciparum transmission stages accumulate in the human bone marrow. Science translational medicine. 2014;6(244):244re5. doi: 10.1126/scitranslmed.3008882 25009232; PubMed Central PMCID: PMC4175394.
104. Pukrittayakamee S, Imwong M, Singhasivanon P, Stepniewska K, Day NJ, White NJ. Effects of different antimalarial drugs on gametocyte carriage in P. vivax malaria. Am J Trop Med Hyg. 2008;79(3):378–84. Epub 2008/09/12. 79/3/378 [pii]. 18784229.
105. Douglas NM, Simpson JA, Phyo AP, Siswantoro H, Hasugian AR, Kenangalem E, et al. Gametocyte dynamics and the role of drugs in reducing the transmission potential of Plasmodium vivax. The Journal of infectious diseases. 2013;208(5):801–12. Epub 2013/06/15. doi: 10.1093/infdis/jit261 23766527; PubMed Central PMCID: PMCPMC3733516.
106. Abdul-Ghani R, Basco LK, Beier JC, Mahdy MA. Inclusion of gametocyte parameters in anti-malarial drug efficacy studies: filling a neglected gap needed for malaria elimination. Malaria journal. 2015;14 : 413. Epub 2015/10/21. doi: 10.1186/s12936-015-0936-4 26481312; PubMed Central PMCID: PMCPMC4617745.
107. Okumu FO, Killeen GF, Ogoma S, Biswaro L, Smallegange RC, Mbeyela E, et al. Development and field evaluation of a synthetic mosquito lure that is more attractive than humans. PloS one. 2010;5(1):e8951. Epub 2010/02/04. doi: 10.1371/journal.pone.0008951 20126628; PubMed Central PMCID: PMC2812511.
108. Mukabana WR, Mweresa CK, Otieno B, Omusula P, Smallegange RC, van Loon JJ, et al. A novel synthetic odorant blend for trapping of malaria and other African mosquito species. J Chem Ecol. 2012;38(3):235–44. Epub 2012/03/20. doi: 10.1007/s10886-012-0088-8 22426893; PubMed Central PMCID: PMC3310138.
109. Menger DJ, Otieno B, de Rijk M, Mukabana WR, van Loon JJ, Takken W. A push-pull system to reduce house entry of malaria mosquitoes. Malaria journal. 2014;13 : 119. Epub 2014/03/29. 1doi: 10.1186/1475-2875-13-119 1475-2875-13-119 [pii]. 24674451; PubMed Central PMCID: PMC3986670.
110. Menger DJ, Van Loon JJ, Takken W. Assessing the efficacy of candidate mosquito repellents against the background of an attractive source that mimics a human host. Med Vet Entomol. 2014;28(4):407–13. Epub 2014/05/07. doi: 10.1111/mve.12061 24797537.
111. Verhulst NO, Qiu YT, Beijleveld H, Maliepaard C, Knights D, Schulz S, et al. Composition of Human Skin Microbiota Affects Attractiveness to Malaria Mosquitoes. PLoS One. 2011;6(12):e28991. doi: 10.1371/journal.pone.0028991 22216154
112. Kelly M, Su CY, Schaber C, Crowley JR, Hsu FF, Carlson JR, et al. Malaria parasites produce volatile mosquito attractants. MBio. 2015;6(2). Epub 2015/03/26. doi: 10.1128/mBio.00235-15 e00235-15 [pii] mBio.00235-15 [pii]. 25805727; PubMed Central PMCID: PMC4453533.
113. Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C. A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science. 2010;327(5973):1644–8. doi: 10.1126/science.1184008 20223948; PubMed Central PMCID: PMC3510679.
114. Molina-Cruz A, Garver LS, Alabaster A, Bangiolo L, Haile A, Winikor J, et al. The human malaria parasite Pfs47 gene mediates evasion of the mosquito immune system. Science. 2013;340(6135):984–7. Epub 2013/05/11. doi: 10.1126/science.1235264 science.1235264 [pii]. 23661646; PubMed Central PMCID: PMC3807741.
115. Oliveira Gde A, Lieberman J, Barillas-Mury C. Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science. 2012;335(6070):856–9. doi: 10.1126/science.1209678 22282475; PubMed Central PMCID: PMC3444286.
116. Ramphul UN, Garver LS, Molina-Cruz A, Canepa GE, Barillas-Mury C. Plasmodium falciparum evades mosquito immunity by disrupting JNK-mediated apoptosis of invaded midgut cells. Proc Natl Acad Sci U S A. 2015;112(5):1273–80. Epub 2015/01/02. doi: 10.1073/pnas.1423586112 1423586112 [pii]. 25552553; PubMed Central PMCID: PMC4321252.
117. Eldering M, Morlais I, van Gemert GJ, van de Vegte-Bolmer M, Graumans W, Siebelink-Stoter R, et al. Variation in susceptibility of African Plasmodium falciparum malaria parasites to TEP1 mediated killing in Anopheles gambiae mosquitoes. Sci Rep. 2016;6 : 20440. Epub 2016/02/11. doi: 10.1038/srep20440 srep20440 [pii]. 26861587; PubMed Central PMCID: PMC4748223.
118. Gupta L, Molina-Cruz A, Kumar S, Rodrigues J, Dixit R, Zamora RE, et al. The STAT pathway mediates late-phase immunity against Plasmodium in the mosquito Anopheles gambiae. Cell host & microbe. 2009;5(5):498–507. doi: 10.1016/j.chom.2009.04.003 19454353; PubMed Central PMCID: PMC2701194.
119. Smith RC, Eappen AG, Radtke AJ, Jacobs-Lorena M. Regulation of anti-Plasmodium immunity by a LITAF-like transcription factor in the malaria vector Anopheles gambiae. PLoS pathogens. 2012;8(10):e1002965. doi: 10.1371/journal.ppat.1002965 23093936; PubMed Central PMCID: PMC3475675.
120. Li J, Wang X, Zhang G, Githure JI, Yan G, James AA. Genome-block expression-assisted association studies discover malaria resistance genes in Anopheles gambiae. Proc Natl Acad Sci U S A. 2013;110(51):20675–80. doi: 10.1073/pnas.1321024110 24297936; PubMed Central PMCID: PMC3870758.
121. Ojo KK, Pfander C, Mueller NR, Burstroem C, Larson ET, Bryan CM, et al. Transmission of malaria to mosquitoes blocked by bumped kinase inhibitors. The Journal of clinical investigation. 2012;122(6):2301–5. doi: 10.1172/JCI61822 22565309; PubMed Central PMCID: PMC3366411.
122. Mathias DK, Pastrana-Mena R, Ranucci E, Tao D, Ferruti P, Ortega C, et al. A small molecule glycosaminoglycan mimetic blocks Plasmodium invasion of the mosquito midgut. PLoS pathogens. 2013;9(11):e1003757. doi: 10.1371/journal.ppat.1003757 24278017; PubMed Central PMCID: PMC3836724.
123. Sala KA, Nishiura H, Upton LM, Zakutansky SE, Delves MJ, Iyori M, et al. The Plasmodium berghei sexual stage antigen PSOP12 induces anti-malarial transmission blocking immunity both in vivo and in vitro. Vaccine. 2015;33(3):437–45. Epub 2014/12/03. doi: 10.1016/j.vaccine.2014.11.038 25454088.
124. Dembele L, Gego A, Zeeman AM, Franetich JF, Silvie O, Rametti A, et al. Towards an in vitro model of Plasmodium hypnozoites suitable for drug discovery. PloS one. 2011;6(3):e18162. doi: 10.1371/journal.pone.0018162 21483865; PubMed Central PMCID: PMC3069045.
125. Vaughan AM, Mikolajczak SA, Wilson EM, Grompe M, Kaushansky A, Camargo N, et al. Complete Plasmodium falciparum liver-stage development in liver-chimeric mice. The Journal of clinical investigation. 2012;122(10):3618–28. Epub 2012/09/22. doi: 10.1172/JCI62684 22996664; PubMed Central PMCID: PMC3461911.
126. Zeeman AM, van Amsterdam SM, McNamara CW, Voorberg-van der Wel A, Klooster EJ, van den Berg A, et al. KAI407, a potent non-8-aminoquinoline compound that kills Plasmodium cynomolgi early dormant liver stage parasites in vitro. Antimicrobial agents and chemotherapy. 2014;58(3):1586–95. doi: 10.1128/AAC.01927-13 24366744; PubMed Central PMCID: PMC3957848.
127. McCarthy JS, Griffin PM, Sekuloski S, Bright AT, Rockett R, Looke D, et al. Experimentally induced blood-stage Plasmodium vivax infection in healthy volunteers. The Journal of infectious diseases. 2013;208(10):1688–94. Epub 2013/08/03. doi: 10.1093/infdis/jit394 jit394 [pii]. 23908484; PubMed Central PMCID: PMC3888148.
128. McCarthy JS, Marquart L, Sekuloski S, Trenholme K, Elliott S, Griffin P, et al. Linking Murine and Human Plasmodium falciparum Challenge Models in a Translational Path for Antimalarial Drug Development. Antimicrobial agents and chemotherapy. 2016;60(6):3669–75. Epub 2016/04/06. doi: 10.1128/AAC.02883-15 AAC.02883-15 [pii]. 27044554; PubMed Central PMCID: PMC4879391.
129. Swann J, Corey V, Scherer CA, Kato N, Comer E, Maetani M, et al. High-Throughput Luciferase-Based Assay for the Discovery of Therapeutics That Prevent Malaria. ACS Infect Dis. 2016;2(4):281–93. Epub 2016/06/09. doi: 10.1021/acsinfecdis.5b00143 27275010; PubMed Central PMCID: PMC4890880.
130. Hoeijmakers WA, Bartfai R, Stunnenberg HG. Transcriptome analysis using RNA-Seq. Methods in molecular biology. 2013;923 : 221–39. Epub 2012/09/20. doi: 10.1007/978-1-62703-026-7_15 22990781.
131. Otto TD, Bohme U, Jackson AP, Hunt M, Franke-Fayard B, Hoeijmakers WA, et al. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 2014;12 : 86. Epub 2014/11/02. doi: 10.1186/s12915-014-0086-0 s12915-014-0086-0 [pii]. 25359557; PubMed Central PMCID: PMC4242472.
132. Yamagishi J, Natori A, Tolba ME, Mongan AE, Sugimoto C, Katayama T, et al. Interactive transcriptome analysis of malaria patients and infecting Plasmodium falciparum. Genome Res. 2014;24(9):1433–44. Epub 2014/08/06. doi: 10.1101/gr.158980.113 gr.158980.113 [pii]. 25091627; PubMed Central PMCID: PMC4158759.
133. Kaneko I, Iwanaga S, Kato T, Kobayashi I, Yuda M. Genome-Wide Identification of the Target Genes of AP2-O, a Plasmodium AP2-Family Transcription Factor. PLoS pathogens. 2015;11(5):e1004905. Epub 2015/05/29. doi: 10.1371/journal.ppat.1004905 PPATHOGENS-D-14-02008 [pii]. 26018192; PubMed Central PMCID: PMC4446032.
134. Eshar S, Altenhofen L, Rabner A, Ross P, Fastman Y, Mandel-Gutfreund Y, et al. PfSR1 controls alternative splicing and steady-state RNA levels in Plasmodium falciparum through preferential recognition of specific RNA motifs. Molecular microbiology. 2015;96(6):1283–97. Epub 2015/03/27. doi: 10.1111/mmi.13007 25807998.
135. Flannery EL, Fidock DA, Winzeler EA. Using genetic methods to define the targets of compounds with antimalarial activity. J Med Chem. 2013;56(20):7761–71. Epub 2013/08/10. doi: 10.1021/jm400325j 23927658; PubMed Central PMCID: PMC3880619.
136. Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47(3):226–34. Epub 2015/01/20. doi: 10.1038/ng.3189 25599401; PubMed Central PMCID: PMC4545236.
137. Baragana B, Hallyburton I, Lee MC, Norcross NR, Grimaldi R, Otto TD, et al. A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature. 2015;522(7556):315–20. Epub 2015/06/19. doi: 10.1038/nature14451 nature14451 [pii]. 26085270; PubMed Central PMCID: PMC4700930.
138. Bopp SE, Manary MJ, Bright AT, Johnston GL, Dharia NV, Luna FL, et al. Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet. 2013;9(2):e1003293. Epub 2013/02/15. doi: 10.1371/journal.pgen.1003293 PGENETICS-D-12-01478 [pii]. 23408914; PubMed Central PMCID: PMC3567157.
139. Nair S, Nkhoma SC, Serre D, Zimmerman PA, Gorena K, Daniel BJ, et al. Single-cell genomics for dissection of complex malaria infections. Genome Res. 2014;24(6):1028–38. Epub 2014/05/09. doi: 10.1101/gr.168286.113 gr.168286.113 [pii]. 24812326; PubMed Central PMCID: PMC4032849.
140. The malERA Refresh Consultative Panel on Insecticide and Drug Resistance. malERA: An updated research agenda for insecticide and drug resistance in malaria elimination and eradication. PLoS Med. 2017;14(11):e1002450 doi: 10.1371/journal.pmed.1002450
141. Gomes AR, Bushell E, Schwach F, Girling G, Anar B, Quail MA, et al. A genome-scale vector resource enables high-throughput reverse genetic screening in a malaria parasite. Cell host & microbe. 2015;17(3):404–13. Epub 2015/03/04. doi: 10.1016/j.chom.2015.01.014 S1931-3128(15)00034-7 [pii]. 25732065; PubMed Central PMCID: PMC4362957.
142. Schwach F, Bushell E, Gomes AR, Anar B, Girling G, Herd C, et al. PlasmoGEM, a database supporting a community resource for large-scale experimental genetics in malaria parasites. Nucleic Acids Research. 2015;43(Database issue):D1176–D82. doi: 10.1093/nar/gku1143. PMC4383951. 25593348
143. Guttery DS, Poulin B, Ramaprasad A, Wall RJ, Ferguson DJ, Brady D, et al. Genome-wide functional analysis of Plasmodium protein phosphatases reveals key regulators of parasite development and differentiation. Cell host & microbe. 2014;16(1):128–40. Epub 2014/07/11. doi: 10.1016/j.chom.2014.05.020 S1931-3128(14)00219-4 [pii]. 25011111; PubMed Central PMCID: PMC4094981.
144. Lin JW, Meireles P, Prudencio M, Engelmann S, Annoura T, Sajid M, et al. Loss-of-function analyses defines vital and redundant functions of the Plasmodium rhomboid protease family. Molecular microbiology. 2013;88(2):318–38. Epub 2013/03/16. doi: 10.1111/mmi.12187 23490234.
145. Matthews K, Kalanon M, Chisholm SA, Sturm A, Goodman CD, Dixon MW, et al. The Plasmodium translocon of exported proteins (PTEX) component thioredoxin-2 is important for maintaining normal blood-stage growth. Molecular microbiology. 2013;89(6):1167–86. Epub 2013/07/23. doi: 10.1111/mmi.12334 23869529.
146. Philip N, Waters AP. Conditional Degradation of Plasmodium Calcineurin Reveals Functions in Parasite Colonization of both Host and Vector. Cell host & microbe. 2015;18(1):122–31. Epub 2015/06/30. doi: 10.1016/j.chom.2015.05.018 S1931-3128(15)00250-4 [pii]. 26118994; PubMed Central PMCID: PMC4509507.
147. Burda PC, Roelli MA, Schaffner M, Khan SM, Janse CJ, Heussler VT. A Plasmodium phospholipase is involved in disruption of the liver stage parasitophorous vacuole membrane. PLoS pathogens. 2015;11(3):e1004760. Epub 2015/03/19. doi: 10.1371/journal.ppat.1004760 PPATHOGENS-D-14-02170 [pii]. 25786000; PubMed Central PMCID: PMC4364735.
148. Falae A, Combe A, Amaladoss A, Carvalho T, Menard R, Bhanot P. Role of Plasmodium berghei cGMP-dependent protein kinase in late liver stage development. The Journal of biological chemistry. 2010;285(5):3282–8. Epub 2009/11/27. doi: 10.1074/jbc.M109.070367 M109.070367 [pii]. 19940133; PubMed Central PMCID: PMC2823412.
149. Lehmann C, Heitmann A, Mishra S, Burda PC, Singer M, Prado M, et al. A cysteine protease inhibitor of plasmodium berghei is essential for exo-erythrocytic development. PLoS pathogens. 2014;10(8):e1004336. Epub 2014/08/29. doi: 10.1371/journal.ppat.1004336 25166051; PubMed Central PMCID: PMC4148452.
150. Rennenberg A, Lehmann C, Heitmann A, Witt T, Hansen G, Nagarajan K, et al. Exoerythrocytic Plasmodium parasites secrete a cysteine protease inhibitor involved in sporozoite invasion and capable of blocking cell death of host hepatocytes. PLoS pathogens. 2010;6(3):e1000825. Epub 2010/04/03. doi: 10.1371/journal.ppat.1000825 20361051; PubMed Central PMCID: PMC2845656.
151. Elsworth B, Matthews K, Nie CQ, Kalanon M, Charnaud SC, Sanders PR, et al. PTEX is an essential nexus for protein export in malaria parasites. Nature. 2014;511(7511):587–91. Epub 2014/07/22. doi: 10.1038/nature13555 nature13555 [pii]. 25043043.
152. Pasini EM, Braks JA, Fonager J, Klop O, Aime E, Spaccapelo R, et al. Proteomic and genetic analyses demonstrate that Plasmodium berghei blood stages export a large and diverse repertoire of proteins. Mol Cell Proteomics. 2013;12(2):426–48. Epub 2012/12/01. doi: 10.1074/mcp.M112.021238 M112.021238 [pii]. 23197789; PubMed Central PMCID: PMC3567864.
153. Ingmundson A, Nahar C, Brinkmann V, Lehmann MJ, Matuschewski K. The exported Plasmodium berghei protein IBIS1 delineates membranous structures in infected red blood cells. Molecular microbiology. 2012;83(6):1229–43. Epub 2012/02/15. doi: 10.1111/j.1365-2958.2012.08004.x 22329949; PubMed Central PMCID: PMC3502748.
154. Matz JM, Goosmann C, Brinkmann V, Grutzke J, Ingmundson A, Matuschewski K, et al. The Plasmodium berghei translocon of exported proteins reveals spatiotemporal dynamics of tubular extensions. Sci Rep. 2015;5 : 12532. Epub 2015/07/30. doi: 10.1038/srep12532 srep12532 [pii]. 26219962; PubMed Central PMCID: PMC4518229.
155. Mbengue A, Bhattacharjee S, Pandharkar T, Liu H, Estiu G, Stahelin RV, et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520(7549):683–7. Epub 2015/04/16. doi: 10.1038/nature14412 25874676; PubMed Central PMCID: PMCPMC4417027.
156. Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, et al. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347(6220):428–31. Epub 2014/12/17. doi: 10.1126/science.1260867 science.1260867 [pii]. 25502314; PubMed Central PMCID: PMC4349400.
157. Adjalley SH, Chabbert CD, Klaus B, Pelechano V, Steinmetz LM. Landscape and Dynamics of Transcription Initiation in the Malaria Parasite Plasmodium falciparum. Cell Rep. 2016;14(10):2463–75. Epub 2016/03/08. doi: 10.1016/j.celrep.2016.02.025 S2211-1247(16)30128-0 [pii]. 26947071; PubMed Central PMCID: PMC4806524.
158. ENCODE Project Consortium, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874 17571346; PubMed Central PMCID: PMC2212820.
159. Sana TR, Gordon DB, Fischer SM, Tichy SE, Kitagawa N, Lai C, et al. Global mass spectrometry based metabolomics profiling of erythrocytes infected with Plasmodium falciparum. PloS one. 2013;8(4):e60840. Epub 2013/04/18. doi: 10.1371/journal.pone.0060840 PONE-D-12-01745 [pii]. 23593322; PubMed Central PMCID: PMC3621881.
160. Sengupta A, Ghosh S, Basant A, Malusare S, Johri P, Pathak S, et al. Global host metabolic response to Plasmodium vivax infection: a 1H NMR based urinary metabonomic study. Malaria journal. 2011;10 : 384. Epub 2011/12/27. doi: 10.1186/1475-2875-10-384 1475-2875-10-384 [pii]. 22196439; PubMed Central PMCID: PMC3298531.
161. Teng R, Lehane AM, Winterberg M, Shafik SH, Summers RL, Martin RE, et al. 1H-NMR metabolite profiles of different strains of Plasmodium falciparum. Biosci Rep. 2014;34(6):e00150. Epub 2014/11/19. doi: 10.1042/BSR20140134 e00150 [pii] BSR20140134 [pii]. 25405893; PubMed Central PMCID: PMC4240024.
162. Allman EL, Painter HJ, Samra J, Carrasquilla M, Llinas M. Metabolomic Profiling of the Malaria Box Reveals Antimalarial Target Pathways. Antimicrobial agents and chemotherapy. 2016;60(11):6635–49. Epub 2016/08/31. doi: 10.1128/AAC.01224-16 27572391; PubMed Central PMCID: PMCPMC5075069.
163. Creek DJ, Chua HH, Cobbold SA, Nijagal B, MacRae JI, Dickerman BK, et al. Metabolomics-Based Screening of the Malaria Box Reveals both Novel and Established Mechanisms of Action. Antimicrobial agents and chemotherapy. 2016;60(11):6650–63. Epub 2016/08/31. doi: 10.1128/AAC.01226-16 27572396; PubMed Central PMCID: PMCPMC5075070.
164. Park YH, Shi YP, Liang B, Medriano CA, Jeon YH, Torres E, et al. High-resolution metabolomics to discover potential parasite-specific biomarkers in a Plasmodium falciparum erythrocytic stage culture system. Malaria journal. 2015;14 : 122. Epub 2015/04/19. doi: 10.1186/s12936-015-0651-1 10.1186/s12936-015-0651-1 [pii]. 25889340; PubMed Central PMCID: PMC4377044.
165. Tritten L, Keiser J, Godejohann M, Utzinger J, Vargas M, Beckonert O, et al. Metabolic profiling framework for discovery of candidate diagnostic markers of malaria. Sci Rep. 2013;3 : 2769. Epub 2013/09/27. doi: 10.1038/srep02769 srep02769 [pii]. 24067624.
166. The malERA Refresh Consultative Panel on Characterising the Reservoir and Measuring Transmission. malERA: An updated research agenda for characterizing the reservoir and measuring transmission in malaria elimination and eradication. PLoS Med. 2017;14(11):e1002452. doi: 10.1371/journal.pmed.1002452
167. Liu W, Sundararaman SA, Loy DE, Learn GH, Li Y, Plenderleith LJ, et al. Multigenomic Delineation of Plasmodium Species of the Laverania Subgenus Infecting Wild-Living Chimpanzees and Gorillas. Genome Biol Evol. 2016;8(6):1929–39. Epub 2016/06/12. doi: 10.1093/gbe/evw128 evw128 [pii]. 27289102; PubMed Central PMCID: PMC4943199.
168. Loy DE, Liu W, Li Y, Learn GH, Plenderleith LJ, Sundararaman SA, et al. Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. International journal for parasitology. 2017;47(2–3):87–97. Epub 2016/07/07. doi: 10.1016/j.ijpara.2016.05.008 27381764; PubMed Central PMCID: PMCPMC5205579.
169. Sundararaman SA, Plenderleith LJ, Liu W, Loy DE, Learn GH, Li Y, et al. Genomes of cryptic chimpanzee Plasmodium species reveal key evolutionary events leading to human malaria. Nat Commun. 2016;7 : 11078. Epub 2016/03/24. doi: 10.1038/ncomms11078 27002652; PubMed Central PMCID: PMCPMC4804174.
170. Aguilar R, Magallon-Tejada A, Achtman AH, Moraleda C, Joice R, Cistero P, et al. Molecular evidence for the localization of Plasmodium falciparum immature gametocytes in bone marrow. Blood. 2014;123(7):959–66. Epub 2013/12/18. doi: 10.1182/blood-2013-08-520767 24335496; PubMed Central PMCID: PMC4067503.
171. Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, et al. Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;347(6217):1258524. Epub 2014/11/29. doi: 10.1126/science.1258524 1258524 [pii] science.1258524 [pii]. 25431491; PubMed Central PMCID: PMC4380269.
172. Parker JE, Angarita-Jaimes N, Abe M, Towers CE, Towers D, McCall PJ. Infrared video tracking of Anopheles gambiae at insecticide-treated bed nets reveals rapid decisive impact after brief localised net contact. Sci Rep. 2015;5 : 13392. Epub 2015/09/02. doi: 10.1038/srep13392 srep13392 [pii]. 26323965; PubMed Central PMCID: PMC4642575.
173. The Ag1000G Consortium. Anopheles gambiae 1000 Genomes Project: Ag1000G 2014 [updated 2016, Accessed Nov 1st, 2017]. Available from: https://www.malariagen.net/projects/ag1000g.
174. Baldini F, Gabrieli P, South A, Valim C, Mancini F, Catteruccia F. The interaction between a sexually transferred steroid hormone and a female protein regulates oogenesis in the malaria mosquito Anopheles gambiae. PLoS Biol. 2013;11(10):e1001695. Epub 2013/11/10. doi: 10.1371/journal.pbio.1001695 24204210; PubMed Central PMCID: PMC3812110.
175. Gabrieli P, Kakani EG, Mitchell SN, Mameli E, Want EJ, Mariezcurrena Anton A, et al. Sexual transfer of the steroid hormone 20E induces the postmating switch in Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(46):16353–8. Epub 2014/11/05. 10.1073/pnas.1410488111. 25368171; PubMed Central PMCID: PMC4246312. doi: 10.1073/pnas.1410488111 25368171
176. Mitchell SN, Kakani EG, South A, Howell PI, Waterhouse RM, Catteruccia F. Mosquito biology. Evolution of sexual traits influencing vectorial capacity in anopheline mosquitoes. Science. 2015;347(6225):985–8. Epub 2015/02/28. 10.1126/science.1259435. 25722409; PubMed Central PMCID: PMC4373528. doi: 10.1126/science.1259435 25722409
177. Cai H, Zhou Z, Gu J, Wang Y. Comparative Genomics and Systems Biology of Malaria Parasites Plasmodium. Current bioinformatics. 2012;7(4). Epub 2013/12/04. doi: 10.2174/157489312803900965 24298232; PubMed Central PMCID: PMCPMC3844129.
178. Rabinovich RN, Drakeley C, Djimde AA, Hall BF, Hay SI, Hemingway J, et al. malERA: An updated research agenda for malaria elimination and eradication. PLoS Med. 2017;14(11):e1002456. doi: 10.1371/journal.pmed.1002456
179. de Koning-Ward TF, Gilson PR, Crabb BS. Advances in molecular genetic systems in malaria. Nature reviews Microbiology. 2015;13(6):373–87. doi: 10.1038/nrmicro3450 25978707.
180. Oye KA, Esvelt K, Appleton E, Catteruccia F, Church G, Kuiken T, et al. Biotechnology. Regulating gene drives. Science. 2014;345(6197):626–8. Epub 2014/07/19. doi: 10.1126/science.1254287 science.1254287 [pii]. 25035410.
181. Laveran A. Note sur un nouveau parasite trouvé dans le sang de plusieurs malades atteints de fièvre palustres. Bull Acad Med. 1880;9 : 1235–6.
182. Sturm A, Amino R, van de Sand C, Regen T, Retzlaff S, Rennenberg A, et al. Manipulation of host hepatocytes by the malaria parasite for delivery into liver sinusoids. Science. 2006;313(5791):1287–90. doi: 10.1126/science.1129720 16888102.
183. Zimmerman PA, Howes RE. Malaria diagnosis for malaria elimination. Curr Opin Infect Dis. 2015;28(5):446–54. Epub 2015/07/24. doi: 10.1097/QCO.0000000000000191 26203855.
184. Berna AZ, McCarthy JS, Wang RX, Saliba KJ, Bravo FG, Cassells J, et al. Analysis of Breath Specimens for Biomarkers of Plasmodium falciparum Infection. The Journal of infectious diseases. 2015;212(7):1120–8. Epub 2015/03/27. doi: 10.1093/infdis/jiv176 jiv176 [pii]. 25810441; PubMed Central PMCID: PMC4559192.
185. The malERA Refresh Consultative Panel on Basic Science and Enabling Technologies. An updated research agenda for basic science and enabling technologies in malaria elimination and eradication. 2017. 10.1371/journal.pmed.1002451.
186. Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. Journal of extracellular vesicles. 2015;4 : 27066. doi: 10.3402/jev.v4.27066 25979354; PubMed Central PMCID: PMC4433489.
187. Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, et al. Exosomes: novel biomarkers for clinical diagnosis. TheScientificWorldJournal. 2015;2015 : 657086. doi: 10.1155/2015/657086 25695100; PubMed Central PMCID: PMC4322857.
188. Campos FM, Franklin BS, Teixeira-Carvalho A, Filho AL, de Paula SC, Fontes CJ, et al. Augmented plasma microparticles during acute Plasmodium vivax infection. Malaria journal. 2010;9 : 327. doi: 10.1186/1475-2875-9-327 21080932; PubMed Central PMCID: PMC2998527.
189. Nantakomol D, Dondorp AM, Krudsood S, Udomsangpetch R, Pattanapanyasat K, Combes V, et al. Circulating red cell-derived microparticles in human malaria. The Journal of infectious diseases. 2011;203(5):700–6. doi: 10.1093/infdis/jiq104 21282195; PubMed Central PMCID: PMC3072726.
190. Martin-Jaular L, Nakayasu ES, Ferrer M, Almeida IC, Del Portillo HA. Exosomes from Plasmodium yoelii-infected reticulocytes protect mice from lethal infections. PloS one. 2011;6(10):e26588. doi: 10.1371/journal.pone.0026588 22046311; PubMed Central PMCID: PMC3202549.
191. Mantel PY, Hoang AN, Goldowitz I, Potashnikova D, Hamza B, Vorobjev I, et al. Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell host & microbe. 2013;13(5):521–34. doi: 10.1016/j.chom.2013.04.009 23684304; PubMed Central PMCID: PMC3687518.
192. Regev-Rudzki N, Wilson DW, Carvalho TG, Sisquella X, Coleman BM, Rug M, et al. Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell. 2013;153(5):1120–33. doi: 10.1016/j.cell.2013.04.029 23683579.
193. Dormitzer PR, Grandi G, Rappuoli R. Structural vaccinology starts to deliver. Nature reviews Microbiology. 2012;10(12):807–13. doi: 10.1038/nrmicro2893 23154260.
194. Kulp DW, Schief WR. Advances in structure-based vaccine design. Current opinion in virology. 2013;3(3):322–31. doi: 10.1016/j.coviro.2013.05.010 23806515; PubMed Central PMCID: PMC4102719.
195. Welsh RM, Fujinami RS. Pathogenic epitopes, heterologous immunity and vaccine design. Nature reviews Microbiology. 2007;5(7):555–63. doi: 10.1038/nrmicro1709 17558423.
196. Li H, O'Donoghue AJ, van der Linden WA, Xie SC, Yoo E, Foe IT, et al. Structure - and function-based design of Plasmodium-selective proteasome inhibitors. Nature. 2016;530(7589):233–6. Epub 2016/02/13. doi: 10.1038/nature16936 nature16936 [pii]. 26863983; PubMed Central PMCID: PMC4755332.
197. Sun M, Li W, Blomqvist K, Das S, Hashem Y, Dvorin JD, et al. Dynamical features of the Plasmodium falciparum ribosome during translation. Nucleic Acids Res. 2015;43(21):10515–24. Epub 2015/10/04. doi: 10.1093/nar/gkv991 gkv991 [pii]. 26432834; PubMed Central PMCID: PMC4666399.
198. Wong W, Bai XC, Brown A, Fernandez IS, Hanssen E, Condron M, et al. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. Elife. 2014;3. Epub 2014/06/11. doi: 10.7554/eLife.03080 24913268; PubMed Central PMCID: PMC4086275.
199. Neafsey DE, Juraska M, Bedford T, Benkeser D, Valim C, Griggs A, et al. Genetic Diversity and Protective Efficacy of the RTS,S/AS01 Malaria Vaccine. N Engl J Med. 2015;373(21):2025–37. Epub 2015/10/22. doi: 10.1056/NEJMoa1505819 26488565; PubMed Central PMCID: PMCPMC4762279.
200. Mueller I, Shakri AR, Chitnis CE. Development of vaccines for Plasmodium vivax malaria. Vaccine. 2015;33(52):7489–95. Epub 2015/10/03. doi: 10.1016/j.vaccine.2015.09.060 S0264-410X(15)01336-5 [pii]. 26428453.
201. Hovlid ML, Winzeler EA. Phenotypic Screens in Antimalarial Drug Discovery. Trends in parasitology. 2016;32(9):697–707. Epub 2016/06/02. doi: 10.1016/j.pt.2016.04.014 27247245; PubMed Central PMCID: PMCPMC5007148.
202. Williamson AE, Ylioja PM, Robertson MN, Antonova-Koch Y, Avery V, Baell JB, et al. Open Source Drug Discovery: Highly Potent Antimalarial Compounds Derived from the Tres Cantos Arylpyrroles. ACS central science. 2016;2(10):687–701. Epub 2016/11/02. doi: 10.1021/acscentsci.6b00086 27800551; PubMed Central PMCID: PMCPMC5084075.
203. Van Voorhis WC, Adams JH, Adelfio R, Ahyong V, Akabas MH, Alano P, et al. Open Source Drug Discovery with the Malaria Box Compound Collection for Neglected Diseases and Beyond. PLoS pathogens. 2016;12(7):e1005763. Epub 2016/07/29. doi: 10.1371/journal.ppat.1005763 27467575; PubMed Central PMCID: PMCPMC4965013.
204. Spangenberg T, Burrows JN, Kowalczyk P, McDonald S, Wells TN, Willis P. The open access malaria box: a drug discovery catalyst for neglected diseases. PloS one. 2013;8(6):e62906. Epub 2013/06/27. doi: 10.1371/journal.pone.0062906 23798988; PubMed Central PMCID: PMCPMC3684613.
205. Bando H, Okado K, Guelbeogo WM, Badolo A, Aonuma H, Nelson B, et al. Intra-specific diversity of Serratia marcescens in Anopheles mosquito midgut defines Plasmodium transmission capacity. Sci Rep. 2013;3 : 1641. Epub 2013/04/11. doi: 10.1038/srep01641 23571408; PubMed Central PMCID: PMC3622076.
206. Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, Mulenga M, et al. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332(6031):855–8. doi: 10.1126/science.1201618 21566196; PubMed Central PMCID: PMC4154605.
207. Dennison NJ, BenMarzouk-Hidalgo OJ, Dimopoulos G. MicroRNA-regulation of Anopheles gambiae immunity to Plasmodium falciparum infection and midgut microbiota. Dev Comp Immunol. 2015;49(1):170–8. Epub 2014/12/03. doi: 10.1016/j.dci.2014.10.016 25445902; PubMed Central PMCID: PMC4447300.
208. Ramirez JL, Short SM, Bahia AC, Saraiva RG, Dong Y, Kang S, et al. Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PLoS pathogens. 2014;10(10):e1004398. doi: 10.1371/journal.ppat.1004398 25340821; PubMed Central PMCID: PMC4207801.
209. Bian G, Zhou G, Lu P, Xi Z. Replacing a native Wolbachia with a novel strain results in an increase in endosymbiont load and resistance to dengue virus in a mosquito vector. PLoS Negl Trop Dis. 2013;7(6):e2250. Epub 2013/06/12. doi: 10.1371/journal.pntd.0002250 23755311; PubMed Central PMCID: PMC3675004.
210. Shaw WR, Marcenac P, Childs LM, Buckee CO, Baldini F, Sawadogo SP, et al. Wolbachia infections in natural Anopheles populations affect egg laying and negatively correlate with Plasmodium development. Nat Commun. 2016;7 : 11772. doi: 10.1038/ncomms11772 27243367.
211. Baldini F, Segata N, Pompon J, Marcenac P, Shaw WR, Dabire RK, et al. Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nat Commun. 2014;5 : 3985. doi: 10.1038/ncomms4985 24905191; PubMed Central PMCID: PMC4059924.
Štítky
Interní lékařství
Článek Contemporary disengagement from antiretroviral therapy in Khayelitsha, South Africa: A cohort studyČlánek Bioequivalence of twice-daily oral tacrolimus in transplant recipients: More evidence for consensus?
Článek vyšel v časopisePLOS Medicine
Nejčtenější tento týden
2017 Číslo 11- Berberin: přírodní hypolipidemikum se slibnými výsledky
- Léčba bolesti u seniorů
- Příznivý vliv Armolipidu Plus na hladinu cholesterolu a zánětlivé parametry u pacientů s chronickým subklinickým zánětem
- Jak postupovat při výběru betablokátoru − doporučení z kardiologické praxe
- Červená fermentovaná rýže účinně snižuje hladinu LDL cholesterolu jako vhodná alternativa ke statinové terapii
-
Všechny články tohoto čísla
- Labour trafficking: Challenges and opportunities from an occupational health perspective
- The end of HIV: Still a very long way to go, but progress continues
- Contemporary disengagement from antiretroviral therapy in Khayelitsha, South Africa: A cohort study
- Bioequivalence of twice-daily oral tacrolimus in transplant recipients: More evidence for consensus?
- Treatment guidelines and early loss from care for people living with HIV in Cape Town, South Africa: A retrospective cohort study
- Perinatal mortality associated with induction of labour versus expectant management in nulliparous women aged 35 years or over: An English national cohort study
- Core Outcome Set-STAndards for Development: The COS-STAD recommendations
- Closing the gaps in the HIV care continuum
- Association between the 2012 Health and Social Care Act and specialist visits and hospitalisations in England: A controlled interrupted time series analysis
- HIV pre-exposure prophylaxis and early antiretroviral treatment among female sex workers in South Africa: Results from a prospective observational demonstration project
- Sexual exploitation of unaccompanied migrant and refugee boys in Greece: Approaches to prevention
- Child sex trafficking in the United States: Challenges for the healthcare provider
- The expanding epidemic of HIV-1 in the Russian Federation
- Cardiovascular disease (CVD) and chronic kidney disease (CKD) event rates in HIV-positive persons at high predicted CVD and CKD risk: A prospective analysis of the D:A:D observational study
- Validity of a minimally invasive autopsy for cause of death determination in maternal deaths in Mozambique: An observational study
- malERA: An updated research agenda for malaria elimination and eradication
- malERA: An updated research agenda for health systems and policy research in malaria elimination and eradication
- A combination intervention strategy to improve linkage to and retention in HIV care following diagnosis in Mozambique: A cluster-randomized study
- Bioequivalence between innovator and generic tacrolimus in liver and kidney transplant recipients: A randomized, crossover clinical trial
- malERA: An updated research agenda for basic science and enabling technologies in malaria elimination and eradication
- Human trafficking and exploitation: A global health concern
- Virological response and resistance among HIV-infected children receiving long-term antiretroviral therapy without virological monitoring in Uganda and Zimbabwe: Observational analyses within the randomised ARROW trial
- Postmenopausal hormone therapy and risk of stroke: A pooled analysis of data from population-based cohort studies
- Lansoprazole use and tuberculosis incidence in the United Kingdom Clinical Practice Research Datalink: A population based cohort
- malERA: An updated research agenda for insecticide and drug resistance in malaria elimination and eradication
- Safety, pharmacokinetics, and immunological activities of multiple intravenous or subcutaneous doses of an anti-HIV monoclonal antibody, VRC01, administered to HIV-uninfected adults: Results of a phase 1 randomized trial
- HIV prevalence and behavioral and psychosocial factors among transgender women and cisgender men who have sex with men in 8 African countries: A cross-sectional analysis
- Treatment eligibility and retention in clinical HIV care: A regression discontinuity study in South Africa
- malERA: An updated research agenda for characterising the reservoir and measuring transmission in malaria elimination and eradication
- Effectiveness of a combination strategy for linkage and retention in adult HIV care in Swaziland: The Link4Health cluster randomized trial
- The value of confirmatory testing in early infant HIV diagnosis programmes in South Africa: A cost-effectiveness analysis
- HIV self-testing among female sex workers in Zambia: A cluster randomized controlled trial
- The US President's Malaria Initiative, transmission and mortality: A modelling study
- Comparison of two cash transfer strategies to prevent catastrophic costs for poor tuberculosis-affected households in low- and middle-income countries: An economic modelling study
- Direct provision versus facility collection of HIV self-tests among female sex workers in Uganda: A cluster-randomized controlled health systems trial
- malERA: An updated research agenda for diagnostics, drugs, vaccines, and vector control in malaria elimination and eradication
- malERA: An updated research agenda for combination interventions and modelling in malaria elimination and eradication
- HIV-1 persistence following extremely early initiation of antiretroviral therapy (ART) during acute HIV-1 infection: An observational study
- Respondent-driven sampling for identification of HIV- and HCV-infected people who inject drugs and men who have sex with men in India: A cross-sectional, community-based analysis
- Extensive virologic and immunologic characterization in an HIV-infected individual following allogeneic stem cell transplant and analytic cessation of antiretroviral therapy: A case study
- Measuring success: The challenge of social protection in helping eliminate tuberculosis
- Prospects for passive immunity to prevent HIV infection
- Reaching global HIV/AIDS goals: What got us here, won't get us there
- Evidence-based restructuring of health and social care
- Extreme exploitation in Southeast Asia waters: Challenges in progressing towards universal health coverage for migrant workers
- PLOS Medicine
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Postmenopausal hormone therapy and risk of stroke: A pooled analysis of data from population-based cohort studies
- Bioequivalence between innovator and generic tacrolimus in liver and kidney transplant recipients: A randomized, crossover clinical trial
- HIV pre-exposure prophylaxis and early antiretroviral treatment among female sex workers in South Africa: Results from a prospective observational demonstration project
- Bioequivalence of twice-daily oral tacrolimus in transplant recipients: More evidence for consensus?
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



