-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMultiple Roles and Interactions of and in Development of the Respiratory System
Normal development of the respiratory system is essential for survival and is regulated by multiple genes and signaling pathways. Both Tbx4 and Tbx5 are expressed throughout the mesenchyme of the developing lung and trachea; and, although multiple genes are known to be required in the epithelium, only Fgfs have been well studied in the mesenchyme. In this study, we investigated the roles of Tbx4 and Tbx5 in lung and trachea development using conditional mutant alleles and two different Cre recombinase transgenic lines. Loss of Tbx5 leads to a unilateral loss of lung bud specification and absence of tracheal specification in organ culture. Mutants deficient in Tbx4 and Tbx5 show severely reduced lung branching at mid-gestation. Concordant with this defect, the expression of mesenchymal markers Wnt2 and Fgf10, as well as Fgf10 target genes Bmp4 and Spry2, in the epithelium is downregulated. Lung branching undergoes arrest ex vivo when Tbx4 and Tbx5 are both completely lacking. Lung-specific Tbx4 heterozygous;Tbx5 conditional null mice die soon after birth due to respiratory distress. These pups have small lungs and show severe disruptions in tracheal/bronchial cartilage rings. Sox9, a master regulator of cartilage formation, is expressed in the trachea; but mesenchymal cells fail to condense and consequently do not develop cartilage normally at birth. Tbx4;Tbx5 double heterozygous mutants show decreased lung branching and fewer tracheal cartilage rings, suggesting a genetic interaction. Finally, we show that Tbx4 and Tbx5 interact with Fgf10 during the process of lung growth and branching but not during tracheal/bronchial cartilage development.
Published in the journal: . PLoS Genet 8(8): e32767. doi:10.1371/journal.pgen.1002866
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002866Summary
Normal development of the respiratory system is essential for survival and is regulated by multiple genes and signaling pathways. Both Tbx4 and Tbx5 are expressed throughout the mesenchyme of the developing lung and trachea; and, although multiple genes are known to be required in the epithelium, only Fgfs have been well studied in the mesenchyme. In this study, we investigated the roles of Tbx4 and Tbx5 in lung and trachea development using conditional mutant alleles and two different Cre recombinase transgenic lines. Loss of Tbx5 leads to a unilateral loss of lung bud specification and absence of tracheal specification in organ culture. Mutants deficient in Tbx4 and Tbx5 show severely reduced lung branching at mid-gestation. Concordant with this defect, the expression of mesenchymal markers Wnt2 and Fgf10, as well as Fgf10 target genes Bmp4 and Spry2, in the epithelium is downregulated. Lung branching undergoes arrest ex vivo when Tbx4 and Tbx5 are both completely lacking. Lung-specific Tbx4 heterozygous;Tbx5 conditional null mice die soon after birth due to respiratory distress. These pups have small lungs and show severe disruptions in tracheal/bronchial cartilage rings. Sox9, a master regulator of cartilage formation, is expressed in the trachea; but mesenchymal cells fail to condense and consequently do not develop cartilage normally at birth. Tbx4;Tbx5 double heterozygous mutants show decreased lung branching and fewer tracheal cartilage rings, suggesting a genetic interaction. Finally, we show that Tbx4 and Tbx5 interact with Fgf10 during the process of lung growth and branching but not during tracheal/bronchial cartilage development.
Introduction
The development of the respiratory system represents an evolutionary hallmark that allowed vertebrates to survive on land utilizing air as a source of oxygen. Because the respiratory system is dispensable for embryonic survival in mammals, defects in development of the respiratory system manifest at or after birth. Indeed, abnormal development of the respiratory system in humans is associated with multiple disorders such as tracheal/bronchial atresia, tracheoesophageal fistula, bronchogenic cysts, pulmonary/lobar atresia and pulmonary hypoplasia [1]. Thus, it is important to understand the genetic basis of development of the respiratory system.
In the mouse embryo, the endodermal foregut tube is patterned by signals from the lateral plate mesoderm leading to specification of the lung and trachea at embryonic day (E) 9.0 (19–24 somites). Nkx2.1 has been identified as the earliest marker of lung endoderm specification. At E9.25 (25–28 somites), the primary lung buds appear as ventro-lateral outpouchings of the foregut connected ventrally by the tracheal primordium. The lung buds grow in a ventro-posterior direction and continue to elongate until E11.5. The point of connection of the lung buds is thought to be the origin of the tracheal tube, which separates from the esophagus in a caudal to cranial direction by E11.5 [2], [3]. In the mouse, the left lung bud remains a single lobe and the right lung bud forms 4 lobes - cranial, medial, caudal and accessory [2]. The airways undergo a stereotypic pattern of branching beginning at E11.5 [4]; development and maturation of the alveoli occurs later.
Genes involved in different signaling pathways, including Wnt2, Fgf10, Bmp4, Shh and retinoic acid synthesis genes, have been shown to play important roles in lung specification and branch formation. Complete absence of both Wnt2 and Wnt2b in mesenchyme surrounding the anterior foregut or absence of β-catenin in the foregut epithelium leads to a loss of specification of lung primordia as seen by the absence of Nkx2.1 expression [5]–[7]. Embryos lacking Fgf10, which is normally expressed in mesenchyme surrounding the epithelial branching tips, form a short trachea but have no lungs [8], [9]. Inhibition of epithelial Bmp4 signaling by overexpression of Xnoggin leads to a decrease in lung size and irregularly shaped lung lobes [10]. Shh null mutant mice have only a rudimentary lung sac due to branching severe branching defect [11]. Additionally, conditional inactivation of Shh in lung epithelial cells leads to the formation of hypoplastic lungs with reduced branching of the peripheral tubules [12]. Retinoic acid receptor (RAR) α and RAR β2 double null mutants show left lung agenesis and a hypoplastic right lung at E18.5 [13]. Thus, genes expressed in both the mesenchyme and the epithelium are essential for correct lung bud specification and branching.
After E11.5, mesenchyme surrounding the dorsal aspect of the trachea differentiates into the trachealis smooth muscle. Mesenchyme surrounding the ventral aspect of the trachea and lateral aspect of the main stem bronchi segments and differentiates into C shaped rings composed of chondrocytes. Ventral tracheal cartilage is formed by migration of cells that undergo mesenchymal condensation [14]. Sox9 has been implicated as an important regulator of mesenchymal condensation and chondrocyte differentiation [15], [16]. In chondrocyte cultures it has been shown that in addition to Sox9, FGF2, Igf1, Tgfβ2 and Bmp2 enhance chondrocyte formation [14], [17]. Mutations in a number of genes including Shh, Sox2, retinoic acid synthesis genes and Fgf signaling pathway genes have been shown to affect cartilage ring formation [11]–[13], [18]–[21]. Fgf10 mutants form a partial tracheal tube in spite of the failure of lung formation [8], [9]. Recent evidence shows that loss of Fgf10 leads to defects in tracheal ring formation and that overexpression of Fgf10 between E11.5 and E13.5 disrupts tracheal rings by altering the periodic expression of Shh in the trachea [22].
The T-box transcription factor genes are important during embryonic development. All members of this gene family contain a conserved DNA-binding T-box domain, which binds to a conserved sequence, the T-box binding element, to activate or repress transcription of specific target genes [23]. All Tbx2 subfamily genes, Tbx2, Tbx3, Tbx4 and Tbx5 are expressed in the developing chick lung buds and trachea between stages 15–21 [24]. In the mouse, Tbx1 is expressed in lung epithelium at E12.5, Tbx2 and Tbx3 are expressed in lung mesenchyme at E11.5, and Tbx4 and Tbx5 are expressed in both lung and trachea mesenchyme at E12.5 and later [25]. Tbx1 homozygous null mutants die at birth due to severe heart defects; the lungs are never fully inflated [26] but lung development has not been further investigated. In Tbx4 homozygous mutants, lung buds form but the embryos die at E10.5 due to failure of allantois development and the subsequent lack of chorio-allantoic fusion leading to placental insufficiency [27]. Tbx5 mutants die around E10 due to defects in heart development [28]; lung development has not been previously investigated. Antisense oligonucleotide depletion of both Tbx4 and Tbx5, but not Tbx2 and Tbx3, in lung organ cultures results in inhibition of branching and loss of Fgf10 expression [29] suggesting a role for these factors in lung branching. In the chick embryo, interference with Tbx4 function leads to a reduction in Fgf10 expression in lung mesenchyme and inhibits lung bud formation. Ectopic expression of Tbx4 leads to ectopic expression of Fgf10 and Nkx2.1 and lung bud formation in the esophagus. Additionally, ectopic expression of Tbx4 at the boundary between the trachea and the esophagus can lead to lack of separation of these two structures, resulting in a tracheoesophageal fistula [30]. In humans, Tbx5 mutations cause Holt Oram syndrome characterized by heart and forelimb abnormalities. A single de-novo mutation in TBX5 has been linked to right lung agenesis [31].
To study the roles of Tbx4 and Tbx5 in lung and trachea development in the mouse, we made use of conditional alleles to bypass early embryonic lethality. We studied three distinct processes, namely 1) lung bud and trachea specification, 2) lung branching morphogenesis, and 3) tracheal/bronchial cartilage formation. We show that during early stages of development, Tbx5 is important for specification of the lung buds and the trachea. After specification, Tbx4 and Tbx5 interact during lung growth and branching and the regulation of branching is dependent on Fgf10 signaling. Additionally, Tbx4 and Tbx5 interact in the formation of mesenchymal condensations, which ultimately form the tracheal/bronchial cartilage rings independent of Fgf10 signaling.
Results
Expression of Tbx4 and Tbx5 in the developing lung and trachea
Tbx5 expression is first detected using in situ hybridization (ISH) at E9.0 (24 somites) in the mesenchyme of the lung and trachea primordia, concurrent with Nkx2.1 expression in the ventral foregut epithelium (Figure 1A–1D). The anterior extent of expression of both genes coincides with the posterior extent of the third pharyngeal pouch (red arrow in Figure 1A, 1B). Tbx4 expression is detected in the lung buds when they first appear a few hours later at E9.25 (28 somites) in a pattern similar to Tbx5 (Figure 1E, 1F). Tbx4 and Tbx5 are expressed at E11.5, E13.5 and E15.5 throughout the lung mesenchyme but not in the epithelium (Figure 1G–1L) [25].
Fig. 1. Expression of Tbx4 and Tbx5 in the developing lung and trachea. 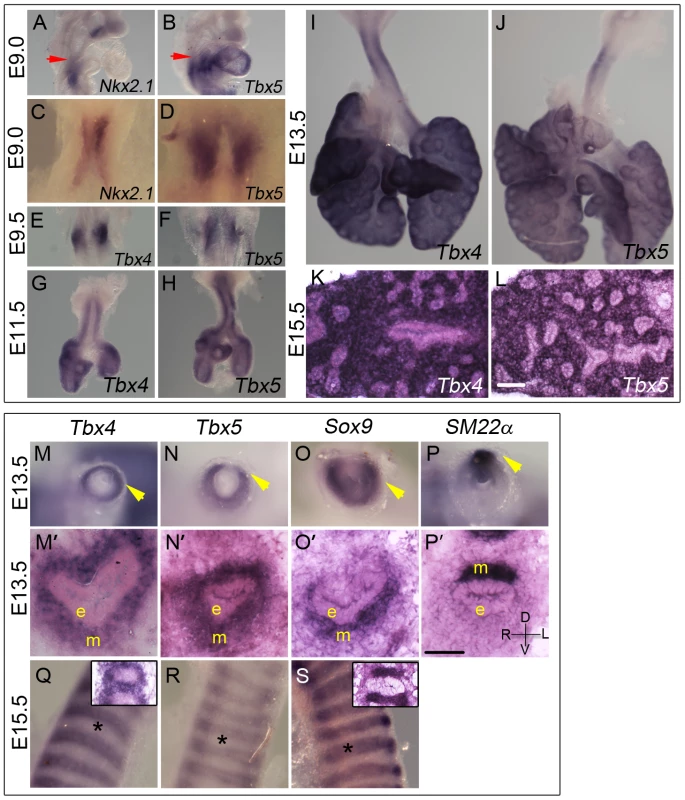
(A–L) Tbx4 and Tbx5 expression analyzed using ISH on lungs. Tbx5 is first expressed at E9.0 (B,D) when the specification of lung primordia occurs, as seen by Nkx2.1 expression (A,C). Red arrows point to the posterior extent of the third pharyngeal pouch which marks the anterior of the expression domain of both Nkx2.1 and Tbx5. Right views (A,B), ventral views (C,D). Tbx4 is first expressed at E9.5 along with Tbx5 in the newly formed lung buds (E,F). Expression is seen in lung whole mounts at E11.5 (G,H), E13.5 (I,J) and in lung mesenchyme in cryosections at E15.5 (K,L). (M–S) Tbx4 and Tbx5 expression analyzed using ISH on tracheas. Tbx4 and Tbx5 are expressed throughout tracheal mesenchyme (m) at E13.5 (M,M′,N,N′) but not in the epithelium (e) or the mesothelium (arrowheads). Caudal view of cut tracheas after whole mount ISH (M–P). ISH on cryosections (M′–P′). At E13.5, Sox9 is expressed in the mesenchyme on the ventral side (O,O′) and SM22α is expressed on the dorsal side (P,P′) of the trachea. D-dorsal; V-ventral; R-right; L-left. At E15.5, Tbx4 and Tbx5 are expressed around the condensing cartilage mesenchyme and in the intercartilage mesenchyme (Q,R). Sox9 is expressed in the condensing cartilage rings (S). Asterisks indicate areas of cartilage condensation. Insets in (Q) and (S) show ISH on E15.5 sagittal cryosections with Tbx4 and Sox9 probe, respectively. Scale bars represent 50 µm. Both Tbx4 and Tbx5 show a dynamic expression pattern in the developing trachea. At E11.5 and E13.5 both genes are expressed throughout the tracheal mesenchyme surrounding the epithelium but are excluded from the epithelium and the outermost layer, the mesothelium (Figure 1G, 1H, 1M, 1M′, 1N, 1N′). At these stages, Tbx4 and Tbx5 are expressed in ventral tracheal cells that also express Sox9 (Figure 1O, 1O′) [32] and in dorsal tracheal cells that also express SM22α (Figure 1P, 1P′) [33]. Within the ventral mesenchyme at E15.5, Tbx4 and Tbx5 expression is restricted to mesenchyme between and surrounding cartilage condensations (Figure 1Q, 1R) in a pattern complementary to that of Sox9, which is restricted to condensing mesenchyme of the cartilage rings at this stage (Figure 1S).
Efficiency of recombination of conditional alleles
The genotypes of embryos used in this study and the corresponding descriptive shorthand nomenclature are shown in Table 1. PCR genotyping was used to determine the efficiency of recombination of the conditional alleles. For embryos carrying the tamoxifen-inducible CreER transgene, a dose of 8 mg tamoxifen was injected into pregnant females at E9.0 and embryos were dissected at E12.5. Yolk sacs were analyzed as an estimate of recombination in the whole embryo. Both alleles of Tbx4fl/fl embryos were completely recombined at E12.5 to produce the mutant allele at this dose of tamoxifen (Figure S1A), but the single floxed allele of Tbx5fl/+ embryos was only partially recombined to the mutant form (Figure S1E). Doses of tamoxifen higher than 8 mg at E9.0 lead to a loss of pregnancy. When 7 mg tamoxifen was injected at E8.5, complete recombination of the Tbx4fl alleles (Figure S1B) and the single Tbx5fl allele (Figure S1F) was obtained at E13.5.
Tab. 1. Different allelic combinations and the descriptive nomenclature. 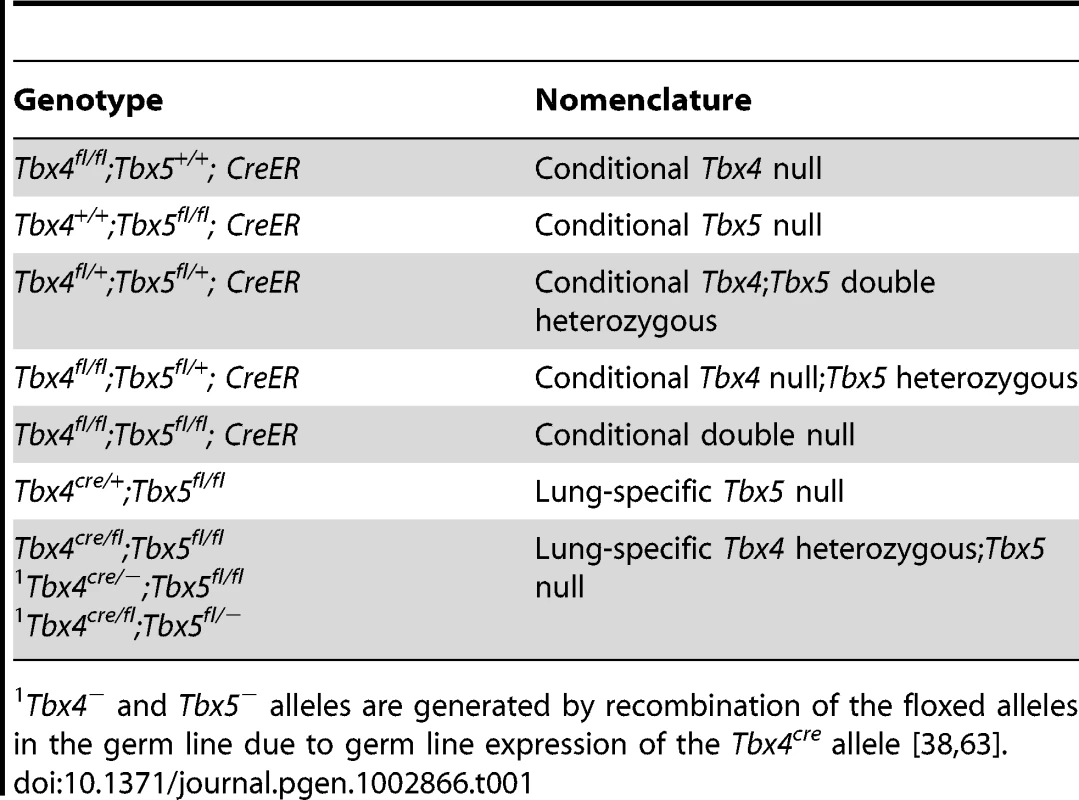
Tbx4− and Tbx5− alleles are generated by recombination of the floxed alleles in the germ line due to germ line expression of the Tbx4cre allele [38], [63]. In lung bud cultures a concentration of 1 µM 4-OH tamoxifen produced near-complete recombination of the Tbx4fl allele after 24 hours (Figure S1C) whereas the Tbx5fl allele was only partially recombined (Figure S1G). Virtually complete recombination of all floxed alleles was achieved after 4 days of culture (Figure S1D, S1H). These data suggest that the Tbx5fl allele has a lower efficiency of Cre-mediated recombination than the Tbx4fl allele. Thus we assume that there may be some residual Tbx5 activity from the Tbx5fl allele in the in vivo experiments, even in the presence of the CreER and Tbx4cre alleles.
Early loss of Tbx5, but not Tbx4, leads to a unilateral loss of lung bud specification and absence of tracheal specification
To explore the role of Tbx4 and Tbx5 in the earliest stages of lung and trachea specification, foregut culture [34] was used with Nkx2.1 as a marker of specification. This ex vivo technique allows for analysis of mutants in culture, circumventing early embryonic lethality of the Tbx4 homozygous mutants due to allantois defects and Tbx5 homozygous mutants due to heart defects. When foreguts and surrounding tissue are isolated at E8.75 (8–16 somites), the foregut tube is devoid of Nkx2.1 expression and the lung buds are not present [6]. At the end of 3 or 4 days of culture lung buds and trachea have formed as seen by Nkx2.1 expression (Figure 2A, 2D, 2G). Nkx2.1 is also expressed in the thyroid primordia at this stage (Figure 2D–2I and [35]). Expression of Tbx4 and Tbx5 was confirmed in control foreguts that were cultured for 4 days (Figure 2B, 2C). Foreguts from E8.5 embryos were cultured in the presence of 4-hydroxy (OH) tamoxifen and analyzed for Nkx2.1 expression. Reduction of Tbx5 alone lead to a lack of Nkx2.1 expression in one of the lung buds after 3 or 4 days of culture (Figure 2E and 2H, respectively) suggesting a unilateral loss of lung bud specification. Removal of Tbx4 alone did not affect Nkx2.1 expression after 3 days of culture (data not shown) and removal of Tbx4 in addition to Tbx5 did not exacerbate the Tbx5 phenotype (Figure 2F, 2I). Therefore, Tbx5 but not Tbx4 is important for the bilateral specification of lung buds ex vivo. Wnt2 and Wnt2b, genes essential for specification of respiratory primordia [5], were analyzed in the conditional Tbx5 null foreguts. Wnt2 expression was reduced (Figure 2J, 2K) and Wnt2b expression was absent (Figure 2L, 2M) suggesting that Tbx5 lies upstream of these genes in regulating the process of specification.
Fig. 2. Early loss of Tbx5 leads to aberrant lung bud and trachea specification. 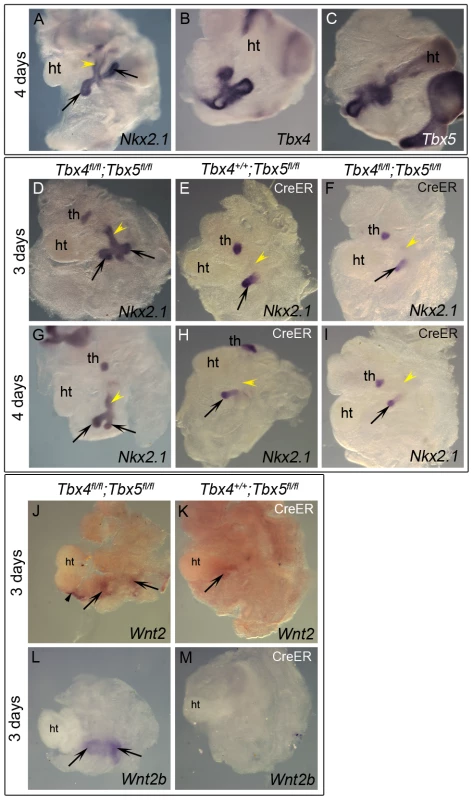
(A–I) Foreguts were isolated between 8–16 somite stages and analyzed by ISH following culture. Nkx2.1 (A), Tbx4 (B) and Tbx5 (C) expression in lung buds (arrows) and tracheal primordia (yellow arrowhead) was confirmed in control cultures. Excision of Tbx5 using 4-OH tamoxifen in conditional null foreguts with CreER leads to the loss of Nkx2.1 expression in one of the lung buds at 3 (E) or 4 days (H). Conditional double nulls (F,I) carrying CreER show a phenotype similar to the conditional Tbx5 nulls, suggesting additional removal of Tbx4 does not exacerbate the phenotype. Nkx2.1 expression was absent in the foregut tube of conditional Tbx5 null and conditional double null foreguts after 3 or 4 days of culture (yellow arrowheads in E,F,H,I) compared to controls (D,G). Conditional Tbx5 nulls show reduced Wnt2 expression (K) and absence of Wnt2b expression (M) in the developing lung buds as compared to controls (J,L). Black arrowheads (J) point to Wnt2 expression in the heart in the controls. ht, heart; th, thyroid primordia. With respect to tracheal specification, Nkx2.1 expression was not observed in the foregut tube after 3 or 4 days of culture in embryos lacking Tbx5 (arrowheads in Figure 2E, 2H) suggesting a lack of tracheal specification in the absence of Tbx5. Additional loss of Tbx4 alleles did not alter the phenotype (Figure 2F, 2I), supporting a role for Tbx5 in the specification of the trachea, independent of Tbx4.
Lung branching is severely affected by the loss of Tbx4 and Tbx5
To analyze the effect of loss of Tbx4 and Tbx5 on lung branching in vivo we made use of the tamoxifen-inducible CreER transgene. Tamoxifen was injected at E8.75 (8–16 somites), late enough to bypass lethality but well before lung branching begins. Embryos with CreER and different combinations of the Tbx4fl and Tbx5fl alleles were examined at E12.5 and E13.5. Conditional Tbx4 null lungs were similar to controls at E12.5 but had fewer branching tips at E13.5 (Figure 3A, 3B, 3E, 3F). Conditional Tbx4;Tbx5 double heterozygous lungs (Figure 3C) were smaller in size and had fewer branching tips than conditional Tbx4 null lungs at E13.5 (Figure 3B) but were more advanced developmentally (Figure 3E, 3F) than conditional Tbx4 null;Tbx5 heterozygous lungs (Figure 3D), which were severely retarded. Lobation in the right lung was disrupted in conditional Tbx4 null;Tbx5 heterozygous lungs: the accessory lobe was missing and only rudimentary cranial and medial lobes were present (Figure 3H). The lobes had a fused appearance (Figure 3H′) suggesting a failure of separation. Histologically, there were no obvious structural defects other than an overall reduction in size (Figure 3G, 3G′, 3H, 3H′).
Fig. 3. Loss of Tbx4 and Tbx5 causes reduced lung branching and lethality at birth. 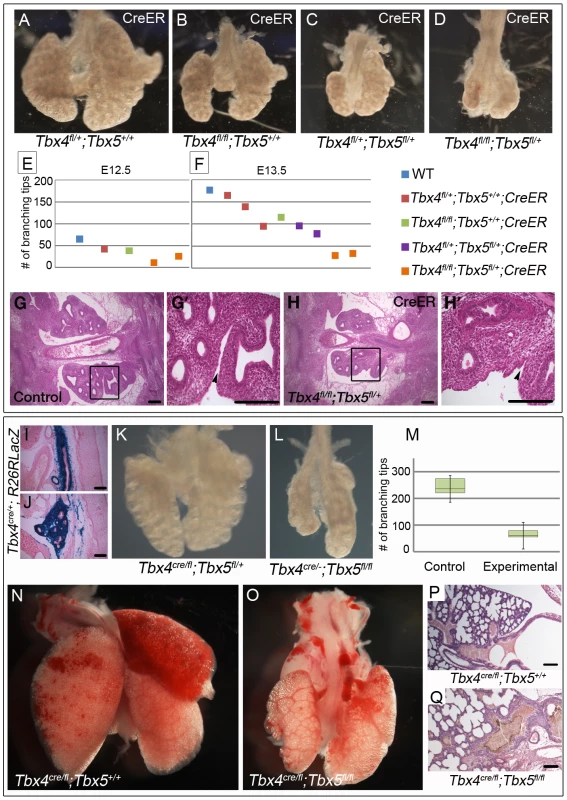
(A–H′) Tbx4fl and Tbx5fl alleles were excised using CreER by injecting tamoxifen at E8.75 and lungs were dissected at different time points. Loss of Tbx4 alone or both Tbx4 and Tbx5 leads to lung hypoplasia at E13.5 (A–D). The number of branching tips was quantitated following E-cadherin staining for different genotypes at E12.5 (E) and E13.5 (F). H & E on histological sections shows a decrease in lung size at E12.5 in the conditional Tbx4 null;Tbx5 heterozygous lungs (G,H,). (G′) and (H′) are magnified views of the boxed regions in (G) and (H), respectively. Black arrowheads indicate separation between the lobes of the right lung in the control (G′) and lack of separation in the mutants (H′). (I–Q) Tbx4cre was utilized for lung and trachea-specific excision of Tbx4fl and Tbx5fl alleles. Tbx4cre was expressed in most of the trachea mesenchyme (I) and lung mesenchymal cells (J) as seen by a R26RlacZ reporter expression at E13.5. Lung-specific Tbx4 heterozygous;Tbx5 null lungs show hypoplasia at E13.5 (K,L) and at birth (N,O). The number of branching tips in control lungs and the lung-specific Tbx4 heterozygous;Tbx5 null lungs, labeled as experimental in the box plot (M), are significantly different at E13.5. The green boxes contain 50% of the values; the median is indicated by a horizontal line in the box; bars represent the 5th and 95th percentiles. H & E staining on sections shows lung morphology for control (P) and mutants (Q) at birth. Scale bars represent 100 µm. Conditional Tbx4 null;Tbx5 heterozygous mutants could not be analyzed later than E13.5, due to hematopoietic defects caused by Cre-induced apoptosis [36] and it was not possible to analyze conditional Tbx5 null embryos using the inducible CreER in vivo due to lethal heart defects. To circumvent these limitations, we used the Tbx4cre allele, which is expressed in the lung and trachea but not in the heart [37]. From this allele, Cre is expressed in the majority of cells of the developing lung and trachea mesenchyme, as seen with lacZ reporter expression at E13.5 (Figure 3I, 3J) [38].
Lung-specific Tbx5 null mutants, carrying a single copy of Tbx4cre, showed a range of phenotypes from an apparently normal lung to a severe decrease in lung size (data not shown). We hypothesize that the variability in phenotype is due to variable extent of recombination of the Tbx5fl allele (Figure S1). All lung-specific Tbx4 heterozygous;Tbx5 null pups (n = 13/13 from 5 litters) became cyanotic at birth and died shortly thereafter due to respiratory distress. Unlike the variable lung size in the lung-specific Tbx5 null mutants, the lungs of these mutants were consistently smaller than controls at E13.5 (Figure 3K, 3L) and at birth (Figure 3N, 3O) and had significantly fewer branching tips at E13.5 (Figure 3M). At P0 histology of the lung-specific Tbx4 heterozygous;Tbx5 null lungs is comparable to controls although the mutant tracheas show accumulation of a mucus like substance (Figure 3P, 3Q). Epithelia of mutant lungs show expression of T1α and Pro-surfactant protein C (Pro-SPC) (Figure S2A–S2D), markers for alveolar cell differentiation [10], [19], suggesting that appropriate differentiation of the lung epithelium occurs in the lung-specific Tbx4 heterozygous;Tbx5 null lungs. These lungs show lobation defects very similar to the conditional Tbx4 null;Tbx5 heterozygous mutant lungs. The accessory lobe was absent in most embryos but when present showed less branching; the cranial and caudal lobes also showed decreased branching. The cranial, medial and caudal lobes were not separated (Figure S3A, S3B). In addition, although tertiary dorsal branches were present in these lungs, they were crowded together and the secondary lateral branches had outgrown a shorter distance compared to controls (Figure S3C, S3D)
Using Tbx4cre, it is not possible to study conditional double nulls as Tbx4 is also expressed from this allele. Thus, to further explore branching morphogenesis in conditions where both alleles of Tbx4 and Tbx5 could be deleted, a lung bud culture system was used in which lung buds from embryos with or without the CreER transgene were cultured in the presence of 4-OH tamoxifen. Conditional Tbx4 null;Tbx5 heterozygous, or conditional Tbx4 heterozygous;Tbx5 null lung buds showed reduced branching (Figure 4A, 4B, 4C) consistent with the reduced number of branching tips observed in vivo. The conditional double null lungs showed a complete branching arrest by 3 days of culture (Figure 4D, 4E, 4F). The existing branches continued to elongate as seen at 4 days of culture (arrow in Figure 4E). Therefore, Tbx4 and Tbx5 are essential for continuing branching morphogenesis ex vivo.
Fig. 4. Loss of Tbx4 and Tbx5 leads to branching arrest ex vivo. 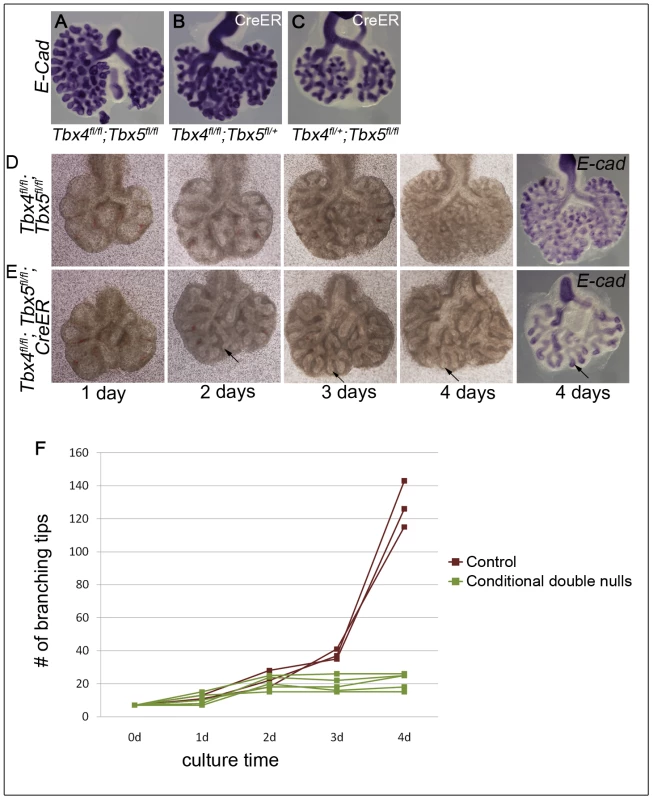
Lung buds were isolated at E11.5 and cultured in the presence of 4-OH tamoxifen. At the end of the culture airways were visualized using E-cadherin ISH. Conditional Tbx4 homozygous;Tbx5 heterozygous (B) and conditional Tbx4 heterozygous;Tbx5 null (C) lungs with CreER showed reduced branching after 4 days of culture compared to controls (A). Control, without CreER, (D) and conditional double null lung buds, with CreER, (E) were cultured for 4 days and photographed at 1 day, 2 days, 3 days and 4 days. Arrows in E show the progression of an elongating airway. A plot of the number of branching tips as a function of time for controls and the conditional double nulls (F), shows a branching arrest of the conditional double null lungs after 2 days of culture (Mann Whitney U test p = 0.036). Loss of Tbx4 and Tbx5 affects the Fgf10 signaling pathway and Wnt2 expression
Expression of the lung mesenchymal marker Fgf10 as well as epithelial targets of the Fgf10 signaling pathway, Bmp4, Spry2 and Etv5 [39], [40], [41], was analyzed in lungs with reduced Tbx4 and Tbx5 expression. Fgf10 is expressed in mesenchyme surrounding the distal epithelial tips that mark the site of future bud formation (Figure 5A) [42]. Consistent with the smaller overall lung size, there were fewer foci of Fgf10 expression in the conditional Tbx4 null;Tbx5 heterozygous lungs (Figure 5B) and in the lung-specific Tbx4 heterozygous;Tbx5 nulls (Figure 5C). The primary receptor for this pathway, Fgfr2 [43], is expressed normally in the epithelium of lung-specific Tbx4 heterozygous;Tbx5 null lungs (Figure 5D, 5E). Bmp4 (Figure 5F, 5G, 5H) and Spry2 (Figure 5I, 5J) were downregulated in Tbx4 and Tbx5-deficient lungs but Etv5 (Figure 5N, 5O) expression was not affected, although it was drastically reduced in conditional double nulls cultured ex vivo (see Figure 6J, 6K).
Fig. 5. Loss of Tbx4 and Tbx5 affects the Fgf10 signaling pathway and Wnt2 expression. 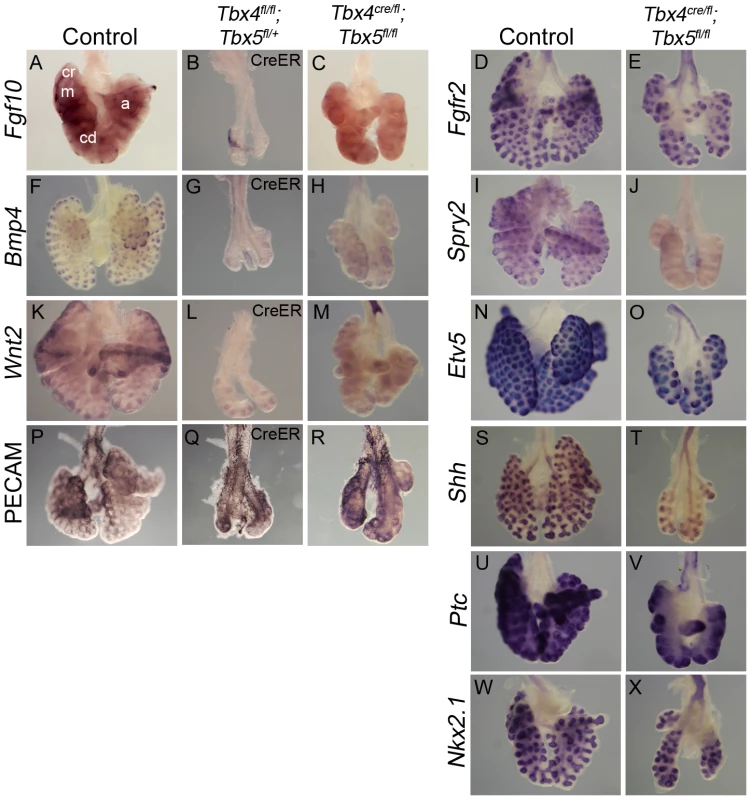
(A–X) Marker analysis of control and Tbx4- and Tbx5-deficient lungs using whole mount ISH (A–O, S–X) and IHC (P–R): Fewer foci of Fgf10 expression were seen in the Tbx4 and Tbx5-deficient lungs (B,C) compared to control (A). cr, cranial; m, medial; cd, caudal; a, accesory lobes. Fgf10 target genes Bmp4 (F,G,H) and Spry2 (I,J) and canonical Wnt2 (K,L,M) were downregulated in Tbx4 and Tbx5-deficient lungs compared to controls. Fgfr2 (D,E), Etv5 (N,O), PECAM (P,Q,R), Shh (S,T), Ptc (U,V) and Nkx2.1 (W,X) were expressed similarly in controls and Tbx4 and Tbx5-deficient lungs. Fig. 6. Tbx4 and Tbx5 interact with Fgf10, but FGF10 fails to rescue Tbx4- and Tbx5-deficient lungs. 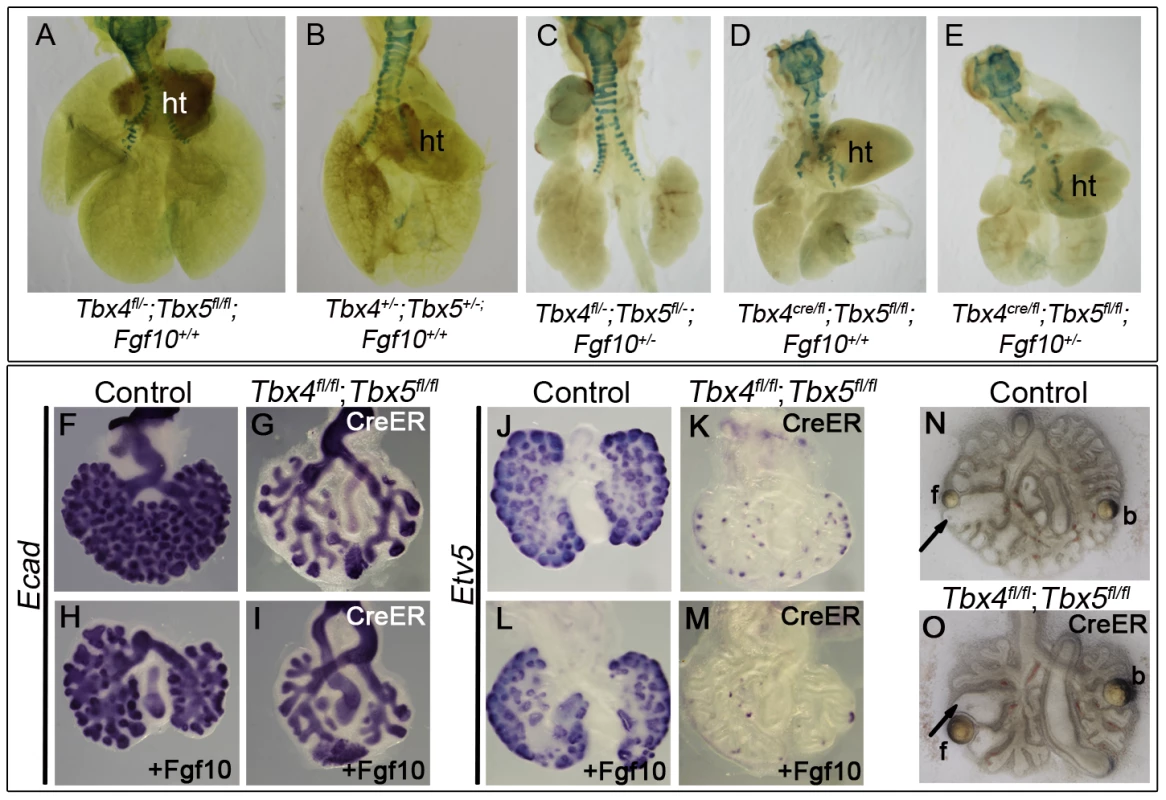
(A–E) Lungs of different genotypes at E18.5. Tbx4;Tbx5 double heterozygous lungs (B) are smaller than control lungs (A). Tbx4;Tbx5;Fgf10 triple heterozygous lungs (C) are smaller than the double heterozygous lungs (B) but comparable in size to the lung-specific Tbx4 heterozyous;Tbx5 null lungs (D). Removing an additional copy of Fgf10 from the lung-specific Tbx4 heterozyous;Tbx5 null lungs does not further affect lung size (E). ht, heart. (F–I) E-cad expression in epithelium of the control lungs and conditional double null lungs in the absence (F,G) and presence (H,I) of exogeneous Fgf10 showing a lack of rescue of branching morphogenesis. (J–M) Etv5 expression in control lungs and conditional double null lungs in the absence (J,K) and presence (L,M) of exogeneous Fgf10. Etv5 expression remains unchanged after addition of Fgf10. (N,O) Both control and conditional double null lungs respond to FGF10 coated beads (f) but not to BSA coated beads (b) as seen by swelling of tips (arrows) in proximity to the FGF10 bead. Wnt2, which is normally expressed in the developing lung mesenchyme (Figure 5K) [5], was greatly reduced at E13.5 in conditional Tbx4 null;Tbx5 heterozygous (Figure 5L) and lung-specific Tbx4 heterozygous;Tbx5 null lungs (Figure 5M). In addition to Fgf10 and Wnt2 signaling pathways, Shh signaling has also been implicated in branching morphogenesis in the lung [11]. Epithelial Shh (Figure 5S, 5T) and its mesenchymal receptor Ptc (Figure 5U, 5V) showed normal expression in lung-specific Tbx4 heterozygous;Tbx5 null lungs. The epithelial marker Nkx2.1, which is necessary for lung branching [44] and showed unilateral expression in the conditional Tbx5 null foreguts at the time of specification (Figure 2), showed expression similar to controls at E13.5 in the lung-specific Tbx4 heterozygous;Tbx5 null lungs (Figure 5W, 5X). At E13.5, expression of the vascular marker Pecam indicated normal development of vessels around individual bronchioles of Tbx4 and Tbx5-deficient mutants (Figure 5P, 5Q, 5R).
Tbx4 and Tbx5 interact with Fgf10 during lung development
Since mesenchymal Fgf10 expression and expression of epithelial targets was affected in mutants with reduced Tbx4 and Tbx5, we investigated the interactions between Tbx4, Tbx5 and Fgf10 during branching morhphogenesis using double and triple heterozygotus mutants. Tbx4;Tbx5 double heterozygous lungs were smaller than control lungs at E18.5 (Figure 6A, 6B). Tbx4;Tbx5;Fgf10 triple heterozygous lungs were smaller than Tbx4;Tbx5 double heterozygous lungs (Figure 6B, 6C), suggesting that Tbx4 and Tbx5 genetically interact with the Fgf10 signaling pathway during lung development. Removing one copy of Fgf10 from lung-specific Tbx4 heterozygous;Tbx5 nulls did not reduce lung size further (Figure 6D, 6E).
Despite the genetic interaction, the addition of exogenous Fgf10 to lung bud cultures of conditional double nulls did not rescue branching (Figure 6F–6I). The lack of change in Etv5 expression, an Fgf10 target, indicated that the Fgf10 signaling pathway was not activated in the presence of exogenous Fgf10 in the conditional double null lungs (Figure 6J–6M). To ensure that the Fgf10 used for the rescue experiments was active, Fgf10 coated heparin beads were placed near the branching tips of lung explants. The tips of both control and conditional double null lungs swelled up in response to Fgf10 beads but not BSA-coated heparin beads (Figure 6N, 6O) [45]. Since Fgfr2 is expressed normally in the Tbx4 and Tbx5-deficient lungs (Figure 5E) and the conditional double null lungs (data not shown), it is not surprising that the conditional double null lung tips can respond to Fgf10. However the conditional double null lungs fail to undergo branching in the presence of Fgf10. Thus, in addition to Fgf10 there must be other factors under the control of Tbx4 and Tbx5 important for activation of the Fgf10 signaling pathway leading to branching morphogenesis.
Loss of Tbx4 and Tbx5 causes tracheal/bronchial ring defects
To analyze the development of the tracheal/bronchial cartilage in embryos with reduced Tbx4 and Tbx5, cartilage rings were visualized at birth using alcian blue staining. Lung-specific Tbx5 nulls and lung-specific Tbx4 heterozygous;Tbx5 null embryos had defective cartilage ring development (Figure 7A–7C) with some normal rings (arrows in Figure 7B, 7C) and isolated foci of cartilage (black arrowheads in Figure 7B, 7C). The tracheal and bronchial lumen of newborn pups was expanded in the controls (Figure 7D, 7E) but collapsed in lung-specific Tbx4 heterozygous;Tbx5 nulls, and contained a mucus-like substance (Figure 7F, 7G). The tracheal epithelium of these mutants showed an increase in the number of mucus-producing cells, as seen by alcian blue staining (Figure 7D′, 7D″, 7F′, 7F″) [19]. To assess the development of cartilage rings at earlier stages, Sox9 expression was analyzed at E12.5 and E13.5 in lung-specific Tbx4 heterozygous;Tbx5 nulls. At E12.5, these mutant tracheas have Sox9 expression on the ventral aspect of the trachea similar to controls (Figure 7H, 7I) but fail to form mesenchymal condensations at E13.5 (Figure 7J, 7K). Expression of two genes genetically downstream of Sox9, Sox6 (Figure 7L, 7M) and Sox5 (Figure 7N, 7O), was downregulated at E13.5 whereas Col2α1, a Sox9 target, was expressed at apparently normal levels in those rings that were present (Figure 7P, 7Q). The smooth muscle marker SM22α was analyzed to assess the development of the dorsal trachealis muscle. SM22α was expressed in an expanded domain and there was a loss of the characteristic banding pattern in the mutant tracheas (Figure 7R, 7S) indicating a disruption in smooth muscle formation due to loss of Tbx4 and Tbx5.
Fig. 7. Loss of Tbx4 and Tbx5 disrupts tracheal/bronchial cartilage and smooth muscle formation. 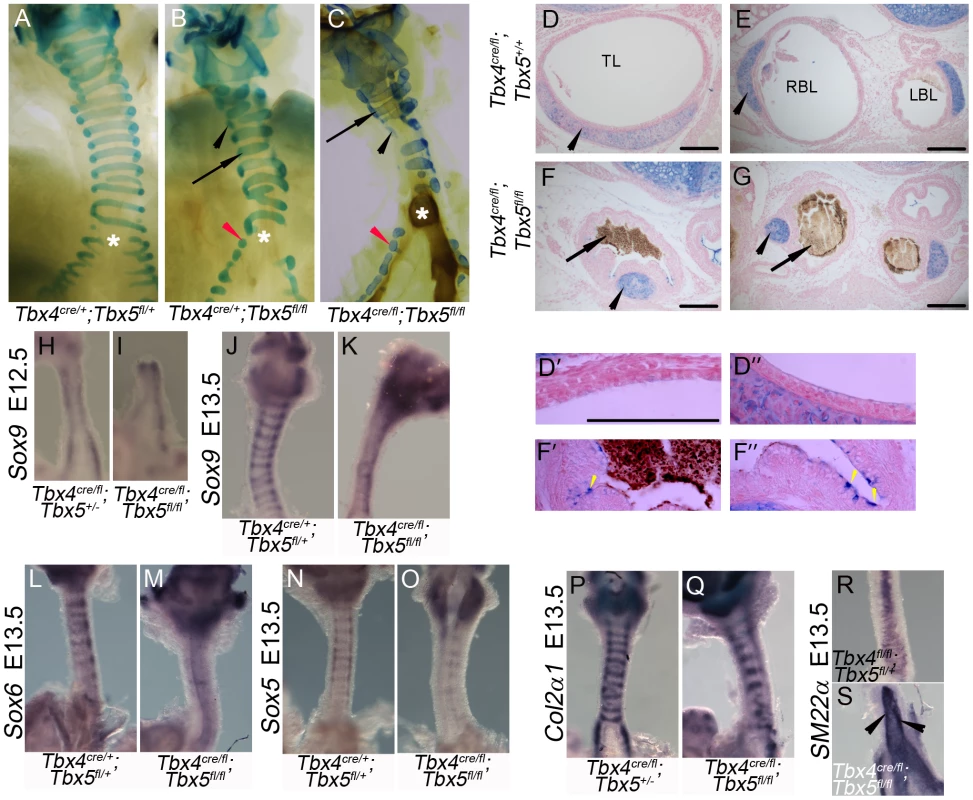
(A–G) Alcian blue staining was used to visualize tracheal/bronchial cartilage. Ten-eleven cartilage rings were seen in control tracheas (E18.5, A); normal (arrow) and incomplete rings were seen in tracheas and bronchi (black and red arrowheads respectively) of lung-specific Tbx5 null (E18.5, B) and lung-specific Tbx4 heterozygous;Tbx5 nulls (postnatal day (P) 0, C). Asterisks in (A–C) indicate the point of bronchial separation from the trachea. Alcian blue on P0 sections showed that the control trachea (D) and main stem bronchial lumens (E) were expanded and the chondrocytes were present as C shaped rings (arrowheads). The lung-specific Tbx4 heterozygous;Tbx5 null tracheal lumen (F) and main stem bronchial lumen (G) were collapsed and filled with a mucus like substance (arrows) and alcian blue positive foci were seen (black arrowheads). Higher magnification views of control (D′,D″) and lung-specific Tbx4 heterozygous;Tbx5 null (F′,F″) tracheas. Yellow arrowheads indicate alcian blue positive mucus-producing cells in the lung-specific Tbx4 heterozygous;Tbx5 null tracheas. TL, Tracheal lumen; RBL, Right bronchial lumen; LBL, Left bronchial lumen. (H–S) Genes important for chondrogenesis and smooth muscle development. Sox9 expression at E12.5 showed a comparable ventral expression pattern in control (H) and lung-specific Tbx4 heterozygous;Tbx5 null tracheas (I). At E13.5, Sox9 positive cells begin to condense and appear in a ring like pattern in controls (J) but not in the mutant tracheas (K). Sox6 (L,M) and Sox5 (N,O) expression was downregulated in the lung-specific Tbx4 heterozygous;Tbx5 null tracheas at E13.5. Col2α1 was expressed in its characteristic ring like pattern (P) similar to Sox9 in the control tracheas but there were fewer rings with apparently normal expression in the lung-specific Tbx4 heterozygous;Tbx5 null tracheas (Q). SM22α expression was analyzed on the dorsal trachea at E13.5 in the control (R) and in the lung-specific Tbx4 heterozygous;Tbx5 null tracheas (S). Arrowheads in (S) point to ectopic expression in an uncharacteristic pattern in the mutants. Scale bars represent 100 µm. Tbx4 and Tbx5 do not interact with Fgf10 during trachea development
Tracheas of controls (Figure 8A), Tbx4;Fgf10 double heterozygotes (Figure 8B) and Tbx5;Fgf10 double heterozygotes (Figure 8C) showed a normal pattern of 10–11 cartilage rings, suggesting a lack of genetic interactions between Tbx4 or Tbx5 and Fgf10 in trachea formation. Fgf10 null mutants do not form lungs but have a truncated trachea with 6–8 cartilage rings, some of which are aberrantly formed [22] (Figure 8D). Tbx4;Tbx5 double heterozygous mice also have tracheas with 6–8 cartilage rings but in addition have main stem bronchial cartilage rings (Figure 8E). Removing a copy of Fgf10 in these double heterozygotes did not alter the phenotype (Figure 8F) showing an Fgf10-independent role for Tbx4 and Tbx5 in the formation of tracheal/bronchial cartilage rings. Also, removing a copy of Fgf10 in the lung-specific Tbx4 heterozygous;Tbx5 nulls did not alter the phenotype of the tracheal/bronchial cartilage rings (Figure 8G, 8H). Thus, Tbx4 and Tbx5 affect lung development via control of Fgf10 expression but affect tracheal/bronchial cartilage development independently of the Fgf10 signaling pathway.
Fig. 8. Lack of genetic interactions between Tbx4, Tbx5, and Fgf10 in trachea formation. 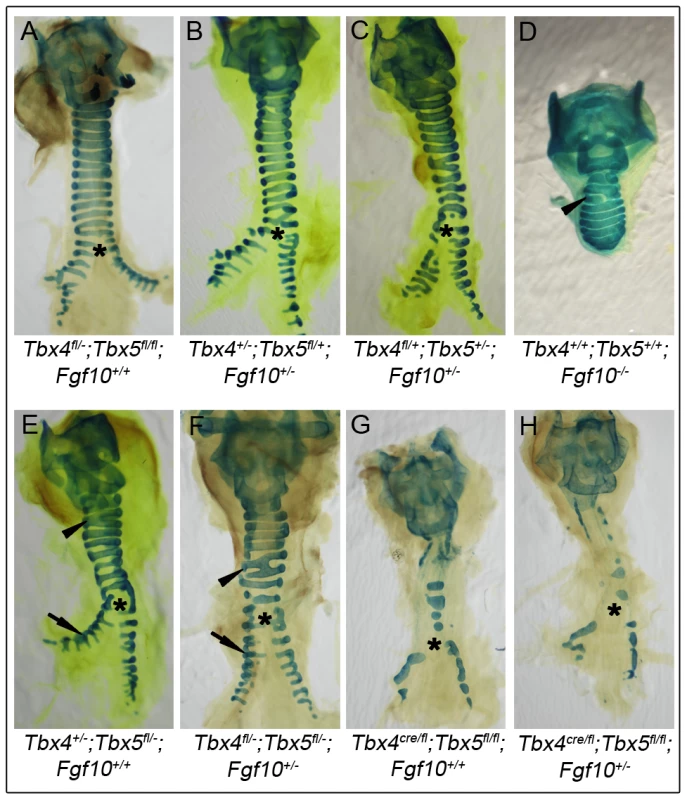
Alcian blue staining was used to analyze tracheal/bronchial cartilage development in tracheas from different genotypes at E18.5. Control tracheas (A), Tbx4;Fgf10 double heterozygous tracheas (B) and Tbx5;Fgf10 double heterozygous tracheas (C) have 10–11 C-shaped, ventral tracheal cartilage rings and the trachea forms the two main stem bronchi which have lateral C-shaped cartilage rings. Fgf10 homozygous mutants (D), Tbx4;Tbx5 double heterozygous mutants (E) and Tbx4;Tbx5;Fgf10 triple heterozygous mutants (F) each form fewer (6–8) tracheal cartilage rings, some irregularly shaped or incomplete (arrowheads). Additionally, Fgf10 mutants lack bronchi and hence any bronchial cartilage rings (D), but the double and triple heterozygotes form normal lateral bronchial cartilage rings (arrows in E and F). Lung-specific Tbx4 heterozygous;Tbx5 nulls (G) show severe disruptions in formation of tracheal/bronchial cartilage rings, a phenotype that is unchanged with the removal of an Fgf10 allele (H). Asterisk shows the point of bronchial separation from the trachea. Discussion
Loss of Tbx5 affects specification of the lung buds and trachea
Tbx5 is expressed around the lung/trachea primordia at the same time that Nkx2.1, a marker of specification, is first expressed in the primordia of the foregut endoderm. Tbx4 is expressed slightly later at the time of lung bud formation. In our study, loss of Tbx5, but not Tbx4, leads to unilateral loss of lung bud specification, indicating that Tbx5 has a distinct function in lung bud formation. In contrast, in the chick, although ectopic expression of Tbx4 in the esophagus can specify lung fate, expression of a dominant negative form of Tbx4 leads to a lack of primary budding in only a third of the mutants analyzed [30]. However, the dominant negative Tbx4 could also be affecting the expression of Tbx5 targets as the repressor construct utilized the complete T-box domain and Tbx4 and Tbx5 have 94% amino acid identity in their T-box domains [46]. This interpretation is compatible with our hypothesis that Tbx5 plays a distinct role in vertebrate lung primordia specification (Figure 9A). Further our results suggest that Tbx5 regulates specification by regulating the activity of Wnt2 and Wnt2b.
Fig. 9. Model for the role of Tbx4 and Tbx5 in lung and trachea development. 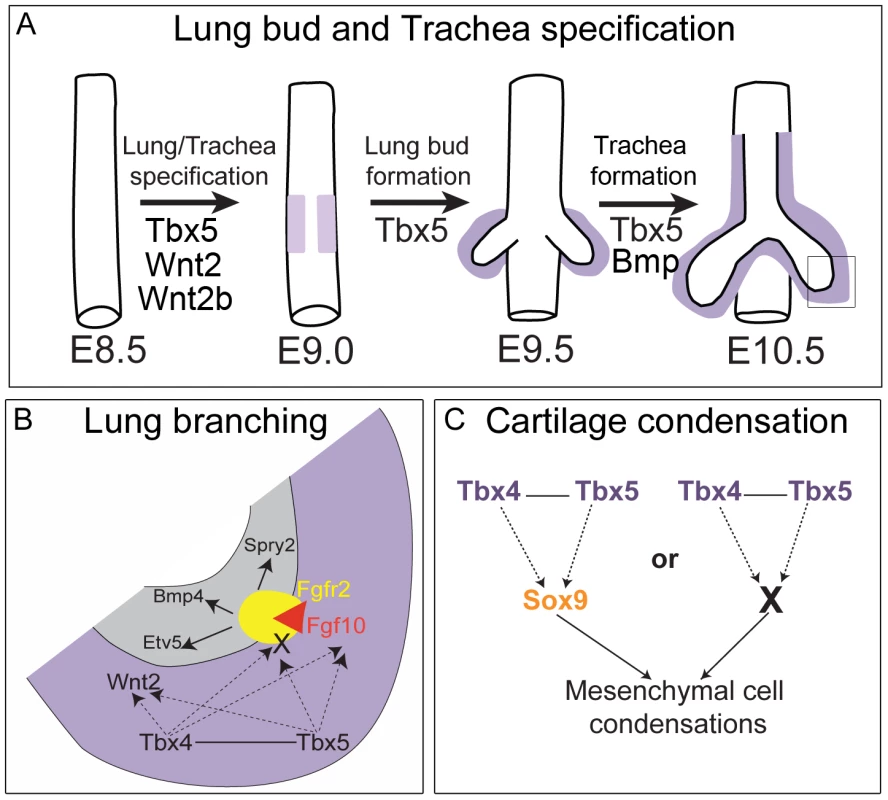
(A) Lung and trachea specification begins at E9.0 in the ventral foregut and at this time Tbx5 expression (light purple) is adjacent to the presumptive endoderm. Later, Tbx4 and Tbx5 expression (dark purple) is in mesenchyme associated with the lung and trachea. Tbx5 but not Tbx4 is important for specification of bilateral lung buds and the trachea. (B) Magnification of box shown in (A) representing the events in the growing tip during branching morphogenesis. Grey denotes epithelium and purple denotes mesenchyme. Tbx4 and Tbx5 interact with each other and act upstream of the Fgf10 signaling pathway. Decrease in Tbx4 and Tbx5 affects mesenchymal Fgf10 expression and expression of its targets in the epithelium – Bmp4, Spry2 and Etv5 – but not the expression of the epithelial Fgf10 receptor Fgfr2. In addition to Fgf10 expression in the mesenchyme, Tbx4 and Tbx5 also control the expression of an unknown factor(s) (X) that is essential for activation of the Fgf10 signaling pathway. Furthermore, Tbx4 and Tbx5 act upstream of Wnt2 in the mesenchyme. (C) In the trachea and the main stem bronchi Tbx4 and Tbx5 either control Sox9 expression, which in turn regulates cartilage condensation, or Tbx4 and Tbx5 regulate another factor (X) essential for chondrogenesis secondarily affecting Sox9 expression. Loss of Tbx5 alone leads to a lack of tracheal specification ex vivo, a phenotype similar to that of mice mutant for Bmpr1 and Bmpr2. In these mutants, although lung bud specification occurs, the ventral foregut fails to acquire tracheal identity [47]. Therefore, Tbx5 either acts in parallel with or in the BMP signaling pathway in specification of the foregut into a trachea (Figure 9A).
Tbx4 and Tbx5 interactions in lung development
In lung branching morphogenesis, Tbx4 and Tbx5 genetically interact with one another (Figure 9B). Although, neither Tbx5 (data not shown) nor Tbx4 single heterozygous lungs show a branching defect, double heterozygous lungs are smaller at E13.5 and E18.5 with a reduced number of branching tips at E13.5. Because these genes are closely related [46], they could potentially regulate the same target genes by binding to a similar T-box binding element, independently of one another. Alternatively, Tbx4 and Tbx5 could physically interact with each other as heterodimers to activate or repress transcription of downstream targets, as has been suggested for other T-box genes [48]. Even though Tbx4 and Tbx5 have a shared role in regulating lung branching, their relative contributions may not be equal, as Tbx4;Tbx5 double heterozygous lungs are smaller than conditional Tbx4 nulls. Unequal and distinct functions could be explained by domains outside of the T-box. For instance, the C terminal domain of both Tbx4 and Tbx5 has transcription activating capability which has been correlated to the shared limb outgrowth promoting activity of these genes [49] but Tbx4 also has a C terminal repressor domain which is proposed to be responsible for its distinct hind limb specific patterning activity [50]. In another example Tbx5 and Tbx4 both bind to a distinct LIM domain repeat of the LMP4 protein in the chick [51], illustrating distinct protein-protein interactions which could have functional implications.
In conditional Tbx4 nulls;Tbx5 heterozygous and lung-specific Tbx4 heterozygous;Tbx5 null lungs, reduction of Tbx4 or Tbx5 leads to a severe decrease in branching, a defect in formation of the lobes and failure of lobe separation, while absence of Tbx4 and Tbx5 leads to branching arrest. These results are in line with the published observations that antisense RNAs used against both Tbx4 and Tbx5 inhibit lung branching in culture and that this affect is explained by loss of Fgf10 expression [29]. Tbx5 binds and activates the Fgf10 promoter in vitro suggesting that Fgf10 is a direct downstream target [52]. The drastic effect on lung branching in our study is explained by loss of expression of the important regulatory genes Fgf10 and Wnt2 in the lung mesenchyme. The Fgf10 targets Bmp4 and Spry2 show very low levels of expression in the epithelium of Tbx4 and Tbx5-deficient mutants and Etv5, another Fgf10 target, shows greatly reduced expression in the conditional double null lungs, leading to the hypothesis that the Fgf10 signaling pathway is activated downstream of Tbx4 and Tbx5 in the developing lung and that Fgf10 genetically interacts with Tbx4 and Tbx5. Lungs that are triple heterozygous for Tbx4, Tbx5 and Fgf10 are reduced in size compared to Tbx4;Tbx5 double heterozygous lungs, which supports this hypothesis. Lung-specific Tbx4 heterozygote;Tbx5 null lungs are severely retarded but lung size is not affected by further loss of Fgf10, demonstrating epistasis and supporting the hypothesis that Fgf10 lies downstream of Tbx4 and Tbx5. Lack of rescue of branch formation and lack of activation of the Fgf10 signaling pathway by exogenously supplied Fgf10 in culture suggests that there are other factors downstream of Tbx4 and Tbx5 that affect Fgf10 signaling. Although, lungs deficient in Tbx4 and Tbx5 retain the ability to respond to Fgf10 coated beads (our study and [29]), they fail to activate additional factors necessary for branching morphogenesis (X in Figure 9B).
One possibility for such a factor is a mesenchymal signaling molecule that communicates with the epithelium and activates downstream target(s) required for the activation of the Fgf10 pathway. We examined expression of Ptc and Shh to determine whether the Shh pathway was involved, but Shh signaling is not affected in the lung-specific Tbx4 heterozygous;Tbx5 nulls or in the conditional double null cultures (data not shown). The extracellular matrix (ECM) molecules, heparan sulfate (HS) proteoglycans aid in Fgf10-Fgfr2 interactions during lung development [45] and hence are good candidates for factors missing in the Tbx4 and Tbx5-deficient mutants. Inhibition of heparanase decreases submandibular gland morphogenesis in culture due to deficiency of Fgf signaling [53]. Additionally, HS-deficient null mouse embryos fail to respond to Fgf signaling and the spatiotemporal expression of cell surface-tethered HS chains regulates the local reception of Fgf-signaling activity during embryonic development [54]. Tbx4 regulates ECM molecules in the developing allantois, specifically the chondroitin sulfate proteoglycan versican (R. Arora and V. E. Papaioannou unpublished observations), suggesting that ECM might be one of the targets for T-box genes in regulating development of other organs as well.
Tbx4 and Tbx5 interactions in tracheal/bronchial cartilage development
Appropriate dorsal smooth muscle development and ventral tracheal/bronchial cartilage development is important for the normal functioning of the trachea. Smooth muscle provides tracheal flexibility and the cartilaginous rings prevent tracheal collapse. Defects in trachea formation may result in tracheomalacia or tracheal stenosis. Reduction of Tbx4 and Tbx5 causes defects in both formation of the tracheal cartilage rings and development of the trachealis smooth muscle indicating interactions between Tbx4 and Tbx5. Tbx4 and Tbx5 heterozygous tracheas have 10–11 cartilage rings but Tbx4;Tbx5 double heterozygous tracheas have 6–8 cartilage rings, some of which are incomplete. Additional reduction of Tbx4 and Tbx5 in the lung-specific Tbx4 heterozygous;Tbx5 null lungs leads to a complete disruption of cartilage ring formation supporting a genetic interaction between Tbx4 and Tbx5 in trachea formation.
Sox9, a master regulator of the process of chondrogenesis, is expressed normally at E12.5 throughout the ventral mesenchyme in lung-specific Tbx4 heterozygous;Tbx5 null tracheas but at E13.5 these tracheas show a lack of characteristic mesenchymal condensations and reduced Sox9 expression. Either Tbx4 and Tbx5 control of Sox9 expression becomes more sensitive to dosage at E13.5 or Tbx4 and Tbx5 control another factor, possibly an ECM molecule, which is important for formation of mesenchymal condensations and, in the absence of these condensations, there is a down regulation of Sox9 expression (Figure 9C). Expression of Sox5 and Sox6, genes genetically downstream of Sox9 and important for condensation and cartilage formation, is downregulated in lung-specific Tbx4 heterozygous;Tbx5 null tracheas, concordant with an aberration in the process of chondrogenesis.
While Tbx4 and Tbx5 regulate lung branching by controlling Fgf10 signaling, their control of tracheal/bronchial cartilage formation is independent of Fgf10 signaling. Fgf10 homozygous mutants have 6–8 tracheal cartilage rings, although the spacing between them is reduced and the rings do not always form the characteristic C shape [8], [9], [22]. Neither Tbx4;Fgf10 double heterozygous tracheas nor Tbx5;Fgf10 double heterozygous tracheas show cartilage condensation defects or a reduction in the number of cartilage rings. In contrast, Tbx4;Tbx5 double heterozygotes show a shorter trachea with 6–8 tracheal cartilage rings suggesting Tbx4 and Tbx5 interact with each other during trachea formation but do not interact with Fgf10. The triple heterozygous tracheas do not show an exacerbation of the Tbx4;Tbx5 double heterozygous trachea phenotype. Additionally, aberrant Fgf10 signaling has been shown to affect tracheal cartilage formation and loss of Fgf10 affects Shh expression but not Sox9 expression [22]. In contrast, in the Tbx4 and Tbx5-deficient mutants Shh expression appears to be unaffected whereas Sox9 expression is reduced at E13.5. Hence, Tbx4 and Tbx5 control tracheal/bronchial cartilage formation via Sox9, independent of the Fgf10 signaling pathway.
Materials and Methods
Mouse strains, crosses, and embryo collection
Mice carrying the following alleles were genotyped as previously described: a Tbx4 conditional ‘floxed’ allele, Tbx4tm1.2Pa [55], hereafter referred to as Tbx4fl; a Tbx5 conditional floxed allele, Tbx5tm1.2Jse [56], hereafter referred to as Tbx5fl; an Fgf10 null allele [8]; ROSA26CRE-ERT2, a ubiquitous tamoxifen-inducible cre transgene [57], hereafter referred to as CreER; Tbx4-cre, an insertion into the endogenous Tbx4 allele resulting in a bicistronic allele that expresses both cre and Tbx4 in all areas of Tbx4 expression including lung and trachea [37], [38], hereafter referred to as Tbx4cre; and a R26RlacZ reporter [58]. All lines of mice were kept on mixed genetic backgrounds. Embryos were dissected from timed matings and yolk sacs were removed for PCR genotyping. The dark period was 19.00 to 05.00 h and noon on the day of a mating plug was identified as E0.5. All mouse work was carried out under Columbia University Medical Center Institutional Animal Care and Use Committee guidelines.
Tamoxifen injections
Tamoxifen (Sigma) at a concentration of 20 mg/ml in sunflower oil (Sigma) was administered to pregnant females by intraperitoneal injection between 15.30 and 19.30 hours on E8.5 or between 23.00 and 24.00 hours on E9.0.
In situ hybridization, immunohistochemistry, and histology
Whole-mount ISH, immunohistochemistry (IHC), immunofluorescence (IF) and ISH on cryosections was performed as described previously [59], [60], [61]. Primary antibodies used were anti-PECAM (Pharmingen, catalog number 01951D), anti-E-cadherin (Takara clone ECCD-2), anti T1α (Developmental Studies Hybridoma Bank antibody 8.1.1) and anti Prosurfactant Protein C (Millipore catalog number AB3786). All secondary antibodies were either peroxidase-conjugated donkey IgG from Jackson Immunochemicals or Alexa Fluor 488 from Invitrogen.
For histology embryos were removed from the uterine horns, dissected out of the decidua and fixed in Bouin's fixative (Sigma). After dehydration in ethanol, embryos were embedded in paraffin wax, sectioned at 8 µm thickness and stained with hematoxylin and eosin (H & E).
Alcian blue staining
Alcian blue staining was performed according to standard protocols [62]. Lungs and trachea were dissected out at different stages, fixed in Bouin's fixative and washed with 70% ethanol. The tissue was then equilibrated in 5% acetic acid and stained with 0.05% alcian blue in 5% acetic acid for 2 hours. The tissue was washed in 5% acetic acid to remove excess stain and dehydrated in 100% methanol, cleared in BABB (benzyl alcohol, benzyl benzoate) and photographed. For alcian blue staining on sections, 8 µm paraffin sections were rehydrated, treated with 0.05% alcian blue in 5% acetic acid and then counterstained with nuclear fast red.
Foregut and lung bud culture
Foregut culture was carried out as described previously [34]. Foreguts were isolated from 8–16 somite stage embryos using tungsten needles and cultured at 37°C in the presence of 95% air and 5% CO2 on Transwell-Col filters (Fisher Scientific) containing 1.5 ml BGJb media (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 0.2 mg/ml vitamin C (Sigma) and 2 µM 4-OH tamoxifen (Sigma). For lung bud culture, lung buds were dissected at E11.5 in phosphate buffered saline (PBS) with 0.1% bovine serum albumin (Sigma) and cultured in media containing DMEM (Invitrogen) with 10% fetal bovine serum, 1% penicillin/streptomycin (Invitrogen) and 1 µM 4-OH tamoxifen on 3.0 µm filters (Millipore) or 0.4 µm Transwell filters (Fisher Scientific). Similar results were obtained using both types of filter; results reported are for experiments with Millipore filters. Where specified, Fgf10 (R&D) was added after 1 day of culture at a concentration of 500 ng/ml. In some experiments heparin beads coated with Fgf10 (100 µg/ml) or BSA (100 µg/ml) were placed near the branching tips of the explants after 1 day of culture. Transwell filters were used for the bead experiments.
Lung branching analysis
Lungs were stained with either E-cadherin antibody using IHC or E-cadherin RNA probe using ISH. In case of whole mount lungs, lobes were separated and photographed to count the number of branching tips. The cultured lungs were photographed and the branching tips were counted. For some experiments, after E-cadherin ISH, lungs were post fixed in 4% PFA, washed with PBT, dehydrated in 100% methanol, cleared in BABB and then photographed.
Supporting Information
Zdroje
1. WhitsettJAWertSETrapnellBC 2004 Genetic disorders influencing lung formation and function at birth. Hum Mol Genet 13 Spec No 2 R207 215
2. CardosoWVLuJ 2006 Regulation of early lung morphogenesis: questions, facts and controversies. Development 133 1611 1624
3. SpoonerBSWessellsNK 1970 Mammalian lung development: interactions in primordium formation and bronchial morphogenesis. J Exp Zool 175 445 454
4. MetzgerRJKleinODMartinGRKrasnowMA 2008 The branching programme of mouse lung development. Nature 453 745 750
5. GossAMTianYTsukiyamaTCohenEDZhouD 2009 Wnt2/2b and [beta]-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Developmental Cell 17 290 298
6. Harris-JohnsonKSDomyanETVezinaCMSunX 2009 beta-catenin promotes respiratory progenitor identity in mouse foregut. Proc Natl Acad Sci U S A 106 16287 16292
7. DomyanETSunX 2011 Patterning and plasticity in development of the respiratory lineage. Dev Dyn 240 477 485
8. MinHDanilenkoDMScullySABolonBRingBD 1998 Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev 12 3156 3161
9. SekineKOhuchiHFujiwaraMYamasakiMYoshizawaT 1999 Fgf10 is essential for limb and lung formation. Nat Genet 21 138 141
10. WeaverMYinglingJMDunnNRBellusciSHoganBL 1999 Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development 126 4005 4015
11. PepicelliCVLewisPMMcMahonAP 1998 Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr Biol 8 1083 1086
12. MillerLAWertSEClarkJCXuYPerlAK 2004 Role of Sonic hedgehog in patterning of tracheal-bronchial cartilage and the peripheral lung. Dev Dyn 231 57 71
13. MendelsohnCLohnesDDecimoDLufkinTLeMeurM 1994 Function of the retinoic acid receptors (RARs) during development (II) Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development 120 2749 2771
14. DeLiseAMFischerLTuanRS 2000 Cellular interactions and signaling in cartilage development. Osteoarthritis Cartilage 8 309 334
15. de CrombruggheBLefebvreVNakashimaK 2001 Regulatory mechanisms in the pathways of cartilage and bone formation. Curr Opin Cell Biol 13 721 727
16. AkiyamaHChaboissierMCMartinJFSchedlAde CrombruggheB 2002 The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 16 2813 2828
17. HardinghamTEOldershawRATewSR 2006 Cartilage, SOX9 and Notch signals in chondrogenesis. J Anat 209 469 480
18. ParkJZhangJJMoroAKushidaMWegnerM 2010 Regulation of Sox9 by Sonic Hedgehog (Shh) is essential for patterning and formation of tracheal cartilage. Dev Dyn 239 514 526
19. QueJLuoXSchwartzRJHoganBL 2009 Multiple roles for Sox2 in the developing and adult mouse trachea. Development 136 1899 1907
20. VermotJNiederreitherKGarnierJMChambonPDolleP 2003 Decreased embryonic retinoic acid synthesis results in a DiGeorge syndrome phenotype in newborn mice. Proc Natl Acad Sci U S A 100 1763 1768
21. TiozzoCDe LangheSCarraroGAlamDANagyA 2009 Fibroblast growth factor 10 plays a causative role in the tracheal cartilage defects in a mouse model of Apert syndrome. Pediatr Res 66 386 390
22. SalaFGDel MoralPMTiozzoCAlamDAWarburtonD 2011 FGF10 controls the patterning of the tracheal cartilage rings via Shh. Development 138 273 282
23. NaicheLAHarrelsonZKellyRGPapaioannouVE 2005 T-box genes in vertebrate development. Annu Rev Genet 39 219 239
24. Gibson-BrownJJAgulnikSISilverLMPapaioannouVE 1998 Expression of T-box genes Tbx2-Tbx5 during chick organogenesis. Mech Dev 74 165 169
25. ChapmanDLGarveyNHancockSAlexiouMAgulnikSI 1996 Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn 206 379 390
26. JeromeLAPapaioannouVE 2001 DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet 27 286 291
27. NaicheLAPapaioannouVE 2003 Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development 130 2681 2693
28. BruneauBGNemerGSchmittJPCharronFRobitailleL 2001 A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106 709 721
29. Cebra-ThomasJABromerJGardnerRLamGKSheipeH 2003 T-box gene products are required for mesenchymal induction of epithelial branching in the embryonic mouse lung. Dev Dyn 226 82 90
30. SakiyamaJYamagishiAKuroiwaA 2003 Tbx4-Fgf10 system controls lung bud formation during chicken embryonic development. Development 130 1225 1234
31. TsengYRSuYNLuFLJengSFHsiehWS 2007 Holt-Oram syndrome with right lung agenesis caused by a de novo mutation in the TBX5 gene. Am J Med Genet A 143A 1012 1014
32. ElluruRGWhitsettJA 2004 Potential role of Sox9 in patterning tracheal cartilage ring formation in an embryonic mouse model. Arch Otolaryngol Head Neck Surg 130 732 736
33. BadriKRZhouYSchugerL 2008 Embryological origin of airway smooth muscle. Proc Am Thorac Soc 5 4 10
34. ChenFCaoYQianJShaoFNiederreitherK 2010 A retinoic acid-dependent network in the foregut controls formation of the mouse lung primordium. J Clin Invest 120 2040 2048
35. DesaiTJMalpelSFlentkeGRSmithSMCardosoWV 2004 Retinoic acid selectively regulates Fgf10 expression and maintains cell identity in the prospective lung field of the developing foregut. Dev Biol 273 402 415
36. NaicheLAPapaioannouVE 2007 Cre activity causes widespread apoptosis and lethal anemia during embryonic development. Genesis 45 768 775
37. LuriaVKrawchukDJessellTMLauferEKaniaA 2008 Specification of motor axon trajectory by ephrin-B:EphB signaling: symmetrical control of axonal patterning in the developing limb. Neuron 60 1039 1053
38. NaicheLAAroraRKaniaALewandoskiMPapaioannouVE 2011 Identity and fate of Tbx4-expressing cells reveal developmental cell fate decisions in the allantois, limb, and external genitalia. Dev Dyn 240 2290 2300
39. WeaverMDunnNRHoganBL 2000 Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development 127 2695 2704
40. HorowitzASimonsM 2008 Branching morphogenesis. Circ Res 103 784 795
41. FirnbergNNeubuserA 2002 FGF signaling regulates expression of Tbx2, Erm, Pea3, and Pax3 in the early nasal region. Dev Biol 247 237 250
42. BellusciSGrindleyJEmotoHItohNHoganBL 1997 Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development 124 4867 4878
43. ArmanEHaffner-KrauszRGorivodskyMLonaiP 1999 Fgfr2 is required for limb outgrowth and lung-branching morphogenesis. Proc Natl Acad Sci U S A 96 11895 11899
44. KimuraSHaraYPineauTFernandez-SalgueroPFoxCH 1996 The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev 10 60 69
45. IzvolskyKIShoykhetDYangYYuQNugentMA 2003 Heparan sulfate-FGF10 interactions during lung morphogenesis. Dev Biol 258 185 200
46. PapaioannouVEGoldinSN 2008 Introduction to the T-box genes and their roles in develoopmental signaling pathways. EpsteinCJEricksonRPWynshaw-BorisA Inborn Errors of Development. The Molecular Basis of Clinical Disorders of Morphogenesis, 2nd Edition Oxford Oxford University Press 852 861
47. DomyanETFerrettiEThrockmortonKMishinaYNicolisSK 2011 Signaling through BMP receptors promotes respiratory identity in the foregut via repression of Sox2. Development 138 971 981
48. GoeringLMHoshijimaKHugBBisgroveBKispertA 2003 An interacting network of T-box genes directs gene expression and fate in the zebrafish mesoderm. Proc Natl Acad Sci U S A 100 9410 9415
49. DubocVLoganMP 2011 Regulation of limb bud initiation and limb-type morphology. Dev Dyn 240 1017 1027
50. OuimetteJFJolinMLL'HonoreAGifuniADrouinJ 2010 Divergent transcriptional activities determine limb identity. Nat Commun 1 35
51. KrauseAZachariasWCamarataTLinkhartBLawE 2004 Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev Biol 273 106 120
52. AgarwalPWylieJNGalceranJArkhitkoOLiC 2003 Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development 130 623 633
53. PatelVNKnoxSMLikarKMLathropCAHossainR 2007 Heparanase cleavage of perlecan heparan sulfate modulates FGF10 activity during ex vivo submandibular gland branching morphogenesis. Development 134 4177 4186
54. ShimokawaKKimura-YoshidaCNagaiNMukaiKMatsubaraK 2011 Cell surface heparan sulfate chains regulate local reception of FGF signaling in the mouse embryo. Dev Cell 21 257 272
55. NaicheLAPapaioannouVE 2007 Tbx4 is not required for hindlimb identity or post-bud hindlimb outgrowth. Development 134 93 103
56. MoriADZhuYVahoraINiemanBKoshiba-TakeuchiK 2006 Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev Biol 297 566 586
57. de LucaCKowalskiTJZhangYElmquistJKLeeC 2005 Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. J Clin Invest 115 3484 3493
58. SorianoP 1999 Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21 70 71
59. WilkinsonDGNietoMA 1993 Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol 225 361 373
60. DavisCA 1993 Whole-mount immunohistochemistry. Methods Enzymol 225 502 516
61. GrieshammerULeMPlumpASWangFTessier-LavigneM 2004 SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev Cell 6 709 717
62. NagyAGertsensteinMVinterstenKBehringerR 2009 Alcian blue staining of the mouse fetal cartilaginous skeleton. Cold Spring Harbor Protocols 2009 pdb.prot5169
63. DouglasNCHengKSauerMVPapaioannouVE 2011 Dynamic expression of Tbx2 subfamily genes in development of the mouse reproductive system. Dev Dyn 241 365 375
Štítky
Genetika Reprodukční medicína
Článek Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma GenomesČlánek Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail ExpansionČlánek Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid CellsČlánek Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug ActivationČlánek The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin DynamicsČlánek Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 8
-
Všechny články tohoto čísla
- Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma Genomes
- Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail Expansion
- Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid Cells
- Chromosome Territories Meet a Condensin
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans
- Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse ()
- A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins
- Advances in Quantitative Trait Analysis in Yeast
- Experimental Evolution of a Novel Sexually Antagonistic Allele
- Variation of Contributes to Dog Breed Skull Diversity
- , a Gene Involved in Axonal Pathfinding, Is Mutated in Patients with Kallmann Syndrome
- A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression
- Cryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
- Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug Activation
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
- A Sexual Ornament in Chickens Is Affected by Pleiotropic Alleles at and , Selected during Domestication
- Use of Allele-Specific FAIRE to Determine Functional Regulatory Polymorphism Using Large-Scale Genotyping Arrays
- Novel Loci for Metabolic Networks and Multi-Tissue Expression Studies Reveal Genes for Atherosclerosis
- The Genetic Basis of Pollinator Adaptation in a Sexually Deceptive Orchid
- Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
- Inheritance Beyond Plain Heritability: Variance-Controlling Genes in
- The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, and Anthropometric Traits
- Reprogramming to Pluripotency Can Conceal Somatic Cell Chromosomal Instability
- Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes
- PTEN Negatively Regulates MAPK Signaling during Vulval Development
- A Dynamic Response Regulator Protein Modulates G-Protein–Dependent Polarity in the Bacterium
- Population Genomics of the Facultatively Mutualistic Bacteria and
- Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
- Polysome Profiling in Liver Identifies Dynamic Regulation of Endoplasmic Reticulum Translatome by Obesity and Fasting
- Stromal Liver Kinase B1 [STK11] Signaling Loss Induces Oviductal Adenomas and Endometrial Cancer by Activating Mammalian Target of Rapamycin Complex 1
- Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
- Transgene Induced Co-Suppression during Vegetative Growth in
- Hox and Sex-Determination Genes Control Segment Elimination through EGFR and Activity
- A Quantitative Comparison of the Similarity between Genes and Geography in Worldwide Human Populations
- Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in and Mammals
- Comparative Analysis of Regulatory Elements between and by Genome-Wide Transcription Start Site Profiling
- Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome
- S Phase–Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
- Cell-Nonautonomous Signaling of FOXO/DAF-16 to the Stem Cells of
- The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin Dynamics
- Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus
- Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes
- Genetic Modulation of Lipid Profiles following Lifestyle Modification or Metformin Treatment: The Diabetes Prevention Program
- HAL-2 Promotes Homologous Pairing during Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors
- SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Germline
- Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
- Reduced Prostasin (CAP1/PRSS8) Activity Eliminates HAI-1 and HAI-2 Deficiency–Associated Developmental Defects by Preventing Matriptase Activation
- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
- and Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR
- CDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
- Rare Copy Number Variations in Adults with Tetralogy of Fallot Implicate Novel Risk Gene Pathways
- Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of
- NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing
- Human Developmental Enhancers Conserved between Deuterostomes and Protostomes
- A Luminal Glycoprotein Drives Dose-Dependent Diameter Expansion of the Hindgut Tube
- Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation
- Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration
- Chemical Genetics Reveals a Specific Requirement for Cdk2 Activity in the DNA Damage Response and Identifies Nbs1 as a Cdk2 Substrate in Human Cells
- Experimental Relocation of the Mitochondrial Gene to the Nucleus Reveals Forces Underlying Mitochondrial Genome Evolution
- Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome
- Mutations in a P-Type ATPase Gene Cause Axonal Degeneration
- A General G1/S-Phase Cell-Cycle Control Module in the Flowering Plant
- Multiple Roles and Interactions of and in Development of the Respiratory System
- UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex
- Epigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
- Modulating the Strength and Threshold of NOTCH Oncogenic Signals by
- Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer's Disease–Related Tau Phosphorylation Via PAR-1
- Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System
- ATXN2-CAG42 Sequesters PABPC1 into Insolubility and Induces FBXW8 in Cerebellum of Old Ataxic Knock-In Mice
- Cohesin Rings Devoid of Scc3 and Pds5 Maintain Their Stable Association with the DNA
- The MicroRNA Inhibits Calcium Signaling by Targeting the TIR-1/Sarm1 Adaptor Protein to Control Stochastic L/R Neuronal Asymmetry in
- Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength
- The Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Variation of Contributes to Dog Breed Skull Diversity
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



