-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaChromosome Territories Meet a Condensin
article has not abstract
Published in the journal: . PLoS Genet 8(8): e32767. doi:10.1371/journal.pgen.1002939
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1002939Summary
article has not abstract
In human diploid cells, 23 pairs of chromosomes become recognizable as a discrete set of rod-shaped entities during the mitotic stage of the cell cycle. Upon exit from mitosis, they decondense and are packed together into the nucleus, in which individual chromosomes are no longer discernible. Nonetheless, evidence accumulated during past decades suggests that the decondensed chromosomes are not randomly located within the nucleus, but instead occupy distinct subnuclear domains known as chromosome territories (CTs) [1]. This raises a number of obvious questions. What protein factor(s) is(are) involved in the formation of CTs? Is there any mechanistic link between mitotic chromosome structures and interphase chromosome territories? In this issue of PLOS Genetics, Bauer et al. provide a remarkable answer to these questions by demonstrating that condensin II, one of the major chromosome condensation factors in mitosis [2], also makes an important contribution to the formation of chromosome territories during interphase [3].
To address the question of how interphase chromosomes might be organized, the authors focused on nurse cells in the ovary of the fruit fly Drosophila melanogaster, which display a highly characteristic mode of chromosome organization and dynamics. The chromosomes in these cells are amplified by repeated rounds of DNA replication without intervening mitoses (i.e., endoduplication), giving rise to polytene chromosomes in which many copies of homologous chromosomes and chromatids are paired along their entire lengths. After the fifth cycle of endoduplication, however, the pairing of homologous chromatids is disrupted, allowing each set of chromosome arms to occupy a discrete globular territory that is reminiscent of a CT observed in mammalian cells. A previous study from the same group demonstrated that condensin II function is required for the disassembly of polytene chromosomes occurring at this stage [4]. In the current study, Bauer et al. followed the fate of these chromosomes further and found that CTs are not properly formed in the condensin II mutants [3]. The authors also noticed that chromosomes in these mutants maintain their initial orientation (known as the Rabl configuration) in which the centromeric regions of all chromosomes cluster near the nuclear periphery and the telomeres cluster near the opposite pole of the nucleus (Figure 1A).
Fig. 1. A model for how condensin II might contribute to the formation of chromosome territories in diploid cells. 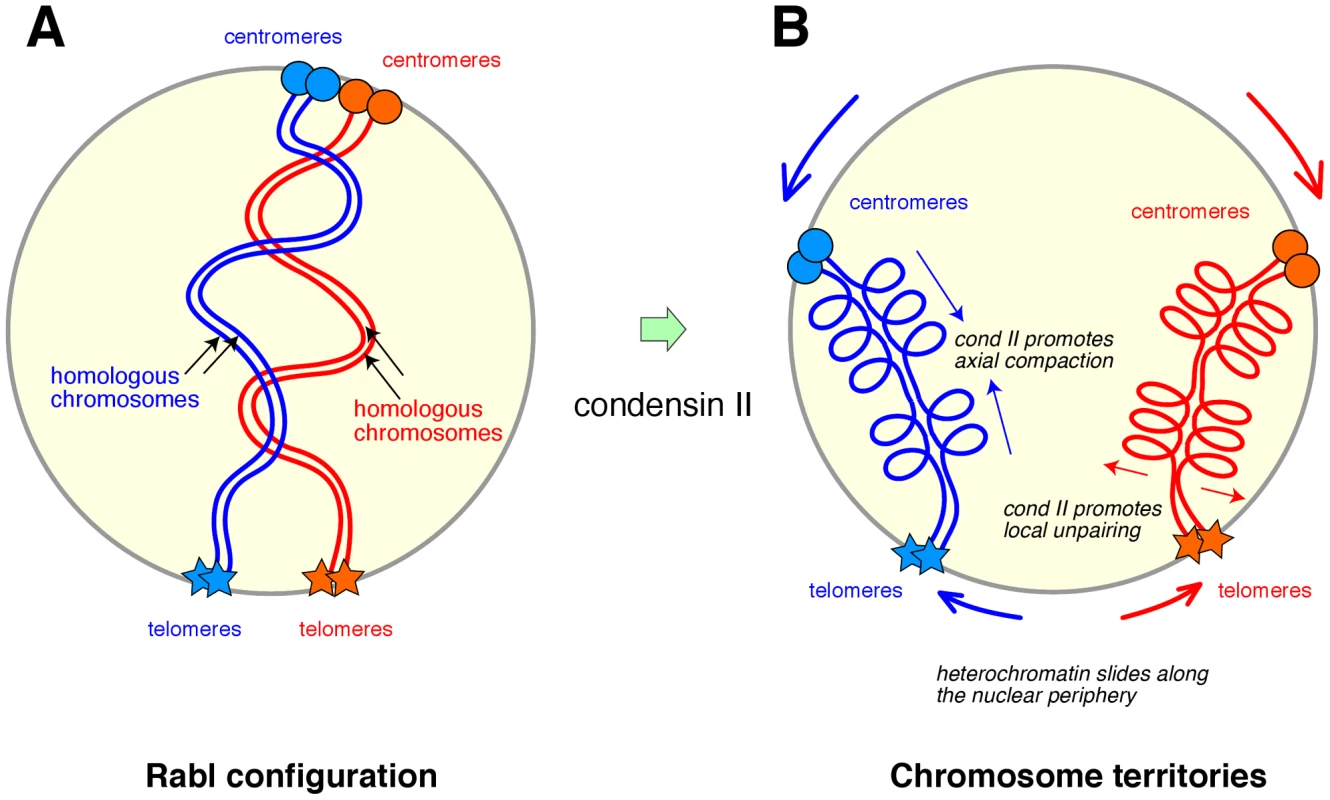
(A) After mitosis, decondensed chromosomes adopt the Rabl configuration, in which the centromeric regions (circles) of all chromosomes cluster near the nuclear periphery and the telomeres (stars) cluster near the opposite side of the nucleus. This configuration reflects the orientation of chromosomes in previous anaphase. In Drosophila diploid cells, homologous chromosomes pair along their lengths. For simplicity, only one arm of each chromosome is drawn here. (B) According to the model proposed by Bauer at al. [3], condensin II promotes axial compaction of the chromosome arms. Coincidentally, centromere clustering is dissolved, allowing heterochromatic regions (represented by the centromeres and telomeres) to slide along the nuclear periphery. As a natural consequence of these two conditions, each pair of homologous chromosomes is sequestered into a discrete chromosome territory. Intrachromosomal folding mediated by condensin II helps disrupt interchromosomal interactions, thereby promoting unpairing of homologous chromosomes and antagonizing transvection. The basic idea presented here could potentially be applied to diploid cells in other organisms where the pairing of homologous chromosomes is not necessarily observed. Why, then, do chromosomes fail to form CTs when condensin II function is impaired? An extensive set of fluorescence in situ hybridization (FISH) data suggested that condensin II promotes not only axial compaction of chromosomes, but also dispersal of heterologous centromeres. Importantly, the dispersed pericentromeric heterochromatin nonetheless stayed near the nuclear periphery. Conversely, when one of the condensin II–specific subunits (Cap-H2) was overexpressed in salivary glands, their polytene chromosomes were disassembled, leading to axial shortening of chromosomes and CT formation. Based on these data, the authors propose a simple and appealing model in which the combination of two events, condensin II–mediated axial compaction and sustained interactions of heterochromatic regions with the nuclear periphery, drives each chromosome into a globular CT (Figure 1B). They also suggest that intrachromosomal folding promoted by condensin II disfavors interchromosomal interactions, thereby contributing to the disassembly of polytene chromosomes.
Although this is an interesting and plausible model, one could ask to what extent the insights obtained from the highly specialized polytene nuclei might be applicable to diploid nuclei. In Drosophila, unlike in other metazoans, homologous chromosomes are always paired in virtually all types of cells throughout development. It has long been known that the pairing of homologous chromosomes affects gene expression at a number of loci through a process known as transvection [5]. Notably, previous studies showed that condensin II antagonizes transvection in the imaginal wing disc [4], and also contributes to CT formation in diploid spermatocytes during meiosis [6]. Moreover, a more recent genome-wide screen identified condensin II subunits as factors that antagonize the pairing of homologous chromosomes in Drosophila tissue culture cells [7]. It will be of great importance to test whether condensin II–mediated axial shortening of chromosomes indeed underlies all these processes, although measurements of chromosome lengths are technically more challenging in diploid cells than in polyploid cells.
The outcome of these emerging studies strongly implies that chromosome–chromosome interactions in cis (i.e., intrachromosomal folding) and those in trans (interchromosomal pairing) compete with each other. Interestingly, a similar principle has already been put forward to delineate the dynamic interaction between sister chromatids within a duplicated chromosome. Upon entry into mitosis, bulk cohesin is lost from chromatid arms, and condensin II coincidentally initiates intrachromatid folding. As a consequence, sister–sister interactions are loosened, leading to the formation of metaphase chromosomes in which rod-shaped sister chromatids are juxtaposed along their lengths, a process known as sister chromatid resolution [8]. A recent study using Xenopus laevis egg extracts has demonstrated that this process is indeed controlled by an intricate balance between the cohesin-mediated cohesive forces and condensin II–mediated resolving forces [9]. Moreover, condensin II has been shown to contribute to axial compaction of chromosomes during mitosis in Xenopus egg extracts [9] and in chicken DT40 cells [10], implying that condensin II can perform essentially the same job during interphase and mitosis.
In summary, a unique and powerful experimental system, combined with a labor-intensive assay, enabled Bauer et al. to propose a model of how CTs might be formed in Drosophila interphase nuclei [3]. To what extent the current model might be applicable to organisms with more complex genomes remains to be determined. It is unlikely that condensin II is the sole factor contributing to CT formation. Searches for additional factors in different model systems would help advance our understanding of the organization of interphase chromosomes, and computational simulations could complement such efforts (e.g., [11]). The current work also reinforces the role of condensin II as one of the central organizers of chromosomes from interphase through mitosis. Future studies should be aimed at a mechanistic understanding of the fundamental principles of chromosome architecture and dynamics throughout the cell cycle as well as during the development of an organism.
Zdroje
1. CremerT, CremerM, DietzelS, MüllerS, SoloveiI, et al. (2006) Chromosome territories–a functional nuclear landscape. Curr Opin Cell Biol 18 : 307–316.
2. HiranoT (2012) Condensins, universal organizers of chromosomes with diverse functions. Genes Dev 26 : 1659–1678.
3. BauerCR, HartlTA, BoscoG (2012) Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes. PLoS Genet 8: e1002873 doi:10.1371/journal.pgen.1002873.
4. HartlTA, SmithHF, BoscoG (2008) Chromosome alignment and transvection are antagonized by condensin II. Science 322 : 1384–1387.
5. DuncanIW (2002) Transvection effects in Drosophila. Annu Rev Genet 36 : 521–556.
6. HartlTA, SweeneySJ, KneplerPJ, BoscoG (2008) Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet 4: e1000228 doi:10.1371/journal.pgen.1000228.
7. JoyceEF, WilliamsBR, XieT, WuC-T (2012) Identification of genes that promote or antagonize somatic homolog pairing ssing a high-throughput FISH-based screen. PLoS Genet 8: e1002667 doi:10.1371/journal.pgen.1002667.
8. ShintomiK, HiranoT (2010) Sister chromatid resolution: a cohesin releasing network and beyond. Chromosoma 119 : 459–467.
9. ShintomiK, HiranoT (2011) The relative ratio of condensin I to II determines chromosome shapes. Genes Dev 25 : 1464–1469.
10. GreenLC, KalitsisP, ChangTM, CipeticM, KimJH, et al. (2012) Contrasting roles of condensin I and II in mitotic chromosome formation. J Cell Sci 125 : 1591–1604.
11. CookPR, MarenduzzoD (2009) Entropic organization of interphase chromosomes. J Cell Biol 186 : 825–834.
Štítky
Genetika Reprodukční medicína
Článek Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma GenomesČlánek Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail ExpansionČlánek Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid CellsČlánek Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug ActivationČlánek The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin DynamicsČlánek Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 8
-
Všechny články tohoto čísla
- Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma Genomes
- Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail Expansion
- Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid Cells
- Chromosome Territories Meet a Condensin
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans
- Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse ()
- A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins
- Advances in Quantitative Trait Analysis in Yeast
- Experimental Evolution of a Novel Sexually Antagonistic Allele
- Variation of Contributes to Dog Breed Skull Diversity
- , a Gene Involved in Axonal Pathfinding, Is Mutated in Patients with Kallmann Syndrome
- A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression
- Cryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
- Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug Activation
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
- A Sexual Ornament in Chickens Is Affected by Pleiotropic Alleles at and , Selected during Domestication
- Use of Allele-Specific FAIRE to Determine Functional Regulatory Polymorphism Using Large-Scale Genotyping Arrays
- Novel Loci for Metabolic Networks and Multi-Tissue Expression Studies Reveal Genes for Atherosclerosis
- The Genetic Basis of Pollinator Adaptation in a Sexually Deceptive Orchid
- Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
- Inheritance Beyond Plain Heritability: Variance-Controlling Genes in
- The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, and Anthropometric Traits
- Reprogramming to Pluripotency Can Conceal Somatic Cell Chromosomal Instability
- Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes
- PTEN Negatively Regulates MAPK Signaling during Vulval Development
- A Dynamic Response Regulator Protein Modulates G-Protein–Dependent Polarity in the Bacterium
- Population Genomics of the Facultatively Mutualistic Bacteria and
- Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
- Polysome Profiling in Liver Identifies Dynamic Regulation of Endoplasmic Reticulum Translatome by Obesity and Fasting
- Stromal Liver Kinase B1 [STK11] Signaling Loss Induces Oviductal Adenomas and Endometrial Cancer by Activating Mammalian Target of Rapamycin Complex 1
- Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
- Transgene Induced Co-Suppression during Vegetative Growth in
- Hox and Sex-Determination Genes Control Segment Elimination through EGFR and Activity
- A Quantitative Comparison of the Similarity between Genes and Geography in Worldwide Human Populations
- Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in and Mammals
- Comparative Analysis of Regulatory Elements between and by Genome-Wide Transcription Start Site Profiling
- Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome
- S Phase–Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
- Cell-Nonautonomous Signaling of FOXO/DAF-16 to the Stem Cells of
- The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin Dynamics
- Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus
- Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes
- Genetic Modulation of Lipid Profiles following Lifestyle Modification or Metformin Treatment: The Diabetes Prevention Program
- HAL-2 Promotes Homologous Pairing during Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors
- SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Germline
- Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
- Reduced Prostasin (CAP1/PRSS8) Activity Eliminates HAI-1 and HAI-2 Deficiency–Associated Developmental Defects by Preventing Matriptase Activation
- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
- and Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR
- CDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
- Rare Copy Number Variations in Adults with Tetralogy of Fallot Implicate Novel Risk Gene Pathways
- Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of
- NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing
- Human Developmental Enhancers Conserved between Deuterostomes and Protostomes
- A Luminal Glycoprotein Drives Dose-Dependent Diameter Expansion of the Hindgut Tube
- Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation
- Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration
- Chemical Genetics Reveals a Specific Requirement for Cdk2 Activity in the DNA Damage Response and Identifies Nbs1 as a Cdk2 Substrate in Human Cells
- Experimental Relocation of the Mitochondrial Gene to the Nucleus Reveals Forces Underlying Mitochondrial Genome Evolution
- Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome
- Mutations in a P-Type ATPase Gene Cause Axonal Degeneration
- A General G1/S-Phase Cell-Cycle Control Module in the Flowering Plant
- Multiple Roles and Interactions of and in Development of the Respiratory System
- UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex
- Epigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
- Modulating the Strength and Threshold of NOTCH Oncogenic Signals by
- Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer's Disease–Related Tau Phosphorylation Via PAR-1
- Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System
- ATXN2-CAG42 Sequesters PABPC1 into Insolubility and Induces FBXW8 in Cerebellum of Old Ataxic Knock-In Mice
- Cohesin Rings Devoid of Scc3 and Pds5 Maintain Their Stable Association with the DNA
- The MicroRNA Inhibits Calcium Signaling by Targeting the TIR-1/Sarm1 Adaptor Protein to Control Stochastic L/R Neuronal Asymmetry in
- Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength
- The Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Variation of Contributes to Dog Breed Skull Diversity
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



