-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
The ecdysone receptor is a heterodimer of two nuclear receptors, the Ecdysone receptor (EcR) and Ultraspiracle (USP). In Drosophila melanogaster, three EcR isoforms share common DNA and ligand-binding domains, but these proteins differ in their most N-terminal regions and, consequently, in the activation domains (AF1s) contained therein. The transcriptional coactivators for these domains, which impart unique transcriptional regulatory properties to the EcR isoforms, are unknown. Activating transcription factor 4 (ATF4) is a basic-leucine zipper transcription factor that plays a central role in the stress response of mammals. Here we show that Cryptocephal (CRC), the Drosophila homolog of ATF4, is an ecdysone receptor coactivator that is specific for isoform B2. CRC interacts with EcR-B2 to promote ecdysone-dependent expression of ecdysis-triggering hormone (ETH), an essential regulator of insect molting behavior. We propose that this interaction explains some of the differences in transcriptional properties that are displayed by the EcR isoforms, and similar interactions may underlie the differential activities of other nuclear receptors with distinct AF1-coactivators.
Published in the journal: . PLoS Genet 8(8): e32767. doi:10.1371/journal.pgen.1002883
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002883Summary
The ecdysone receptor is a heterodimer of two nuclear receptors, the Ecdysone receptor (EcR) and Ultraspiracle (USP). In Drosophila melanogaster, three EcR isoforms share common DNA and ligand-binding domains, but these proteins differ in their most N-terminal regions and, consequently, in the activation domains (AF1s) contained therein. The transcriptional coactivators for these domains, which impart unique transcriptional regulatory properties to the EcR isoforms, are unknown. Activating transcription factor 4 (ATF4) is a basic-leucine zipper transcription factor that plays a central role in the stress response of mammals. Here we show that Cryptocephal (CRC), the Drosophila homolog of ATF4, is an ecdysone receptor coactivator that is specific for isoform B2. CRC interacts with EcR-B2 to promote ecdysone-dependent expression of ecdysis-triggering hormone (ETH), an essential regulator of insect molting behavior. We propose that this interaction explains some of the differences in transcriptional properties that are displayed by the EcR isoforms, and similar interactions may underlie the differential activities of other nuclear receptors with distinct AF1-coactivators.
Introduction
Nuclear receptors are multifunctional transcription factors that mediate responses to steroids and other small hydrophobic signaling molecules. Most nuclear receptors have two transcriptional activation functions (AF1 and AF2). AF2 is formed by ligand-induced folding of the ligand-binding domain, and the structural basis of its interaction with coactivators is becoming known. AF1 designates a second, ligand-independent activation function often present in the N-terminal region of the receptor. AF1 sequences are not conserved, and the existence of an AF1 must be inferred from functional assays. Although the AF1s are of considerable interest, because they often differentiate receptor isoforms and because some have been shown to interact with general transcription factors, comparatively few AF1-coactivator interactions have been characterized. The relative contributions of AF1 and AF2 to transcriptional activation vary among receptors, and for any given receptor the relative contributions may depend upon the promoter context [1].
The three isoforms of EcR (FlyBase ID: FBgn0000546) have unrelated AF1 regions, each capable of mediating transcriptional activation in some contexts [2]–[6]. Although several coactivators and corepressors for the AF2 of EcR have been identified [7]–[13], the interacting factors for the unique AF1 domains remain unknown. The 17-residue AF1 region of isoform B2 is capable of strong transcriptional activation on a standard test promoter and is required for ecdysone-regulated differentiation in a few fly tissues [2], [4]. Here, we show that the bZIP transcription factor, CRC (FBgn0000370), binds the AF1 of isoform B2 to promote steroid-dependent expression of the peptide molting hormone, ETH (FBgn0028738; [14]).
Results
The AF1 of EcR-B2 Bound to the Leucine Zipper of CRC
We performed a yeast two-hybrid screen using the N-terminal region of EcR-B2 as bait and recovered a plasmid containing the complete coding sequence of the predominant CRC isoform, CRC-A [15]. Figure 1 illustrates the salient features of these assays. Interaction (as judged by reporter activation) required the presence of both CRC-A and the AF1 of EcR-B2 (Figure 1A). The EcR-B2 mutation E9K, which sharply reduces transcriptional activation in vivo [4], also abolished the two-hybrid interaction (Figure 1A). The interaction surface provided by CRC was contained within the C-terminal three-quarters of the protein, a region that includes its bZIP and PEST domains, and C-terminal truncation of the protein to remove just the leucine zipper domain abolished the interaction with EcR-B2 (Figure 1B).
Fig. 1. Interaction of the EcR-B2 N-terminus with CRC in yeast two-hybrid assay. 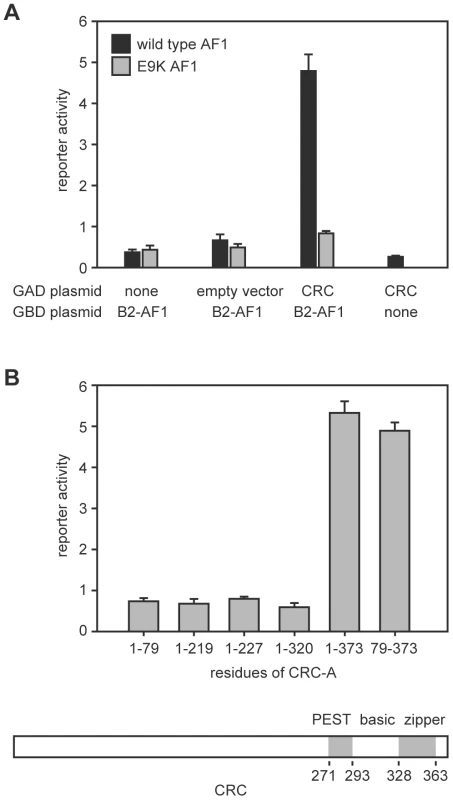
(A) In yeast two-hybrid assays, activation of a UAS-lacZ reporter (reporter activity) required the presence of both EcR-B2 and CRC-A. This interaction was abolished by the EcR-B2 mutation E9K. In these assays, a fusion of the GAL4 DNA-binding domain to either the 17-residue AF1 domain of EcR-B2 (black bars), or the same fragment containing the mutation E9K (gray bars) served as bait. Full length CRC-A was used as the prey. Error bars indicate standard deviations for 4 independent assays. (B) The indicated CRC residues were substituted for full-length CRC-A and tested for binding to the wild-type EcR-B2 fusion fragment in yeast two-hybrid assays. Coordinates of the conserved domains of CRC are indicated in the drawing below. Binding of CRC to the amino-terminal region of EcR-B2 was confirmed in vitro (Figure 2A). Radiolabeled CRC-A bound to the EcR-B2 amino-terminus and to full-length EcR-B2, but not to EcR-B1 or to EcR-B2 carrying the E9K mutation. The AF1 region of the EcR-B2 N-terminus contains sequences suggestive of a short amphipathic helix, and it is known that the acidic-to-basic substitution E9K (within the proposed helix) abolishes AF1 function in vivo [4]. We think it likely that this helix is unstructured in solution and infer that both ionic and hydrophobic interactions play roles in its dimerization, probably with the leucine zipper region of CRC. To test this idea further, we made several individual basic-to-acidic mutations within the CRC leucine zipper – at sites predicted to determine the dimerization specificity of the bZIP domain [16] – and tested the binding of the mutant CRC proteins to wild-type and E9K mutant EcR-B2 (Figure 2B). The binding properties of CRC-R347E and CRC-R353E were indistinguishable from those of wild-type CRC, but CRC-R361E bound EcR-B2-E9K. That an alteration in CRC reversed the effect of an EcR mutation strongly implies direct interaction.
Fig. 2. Binding of CRC to EcR-B2 in vitro. 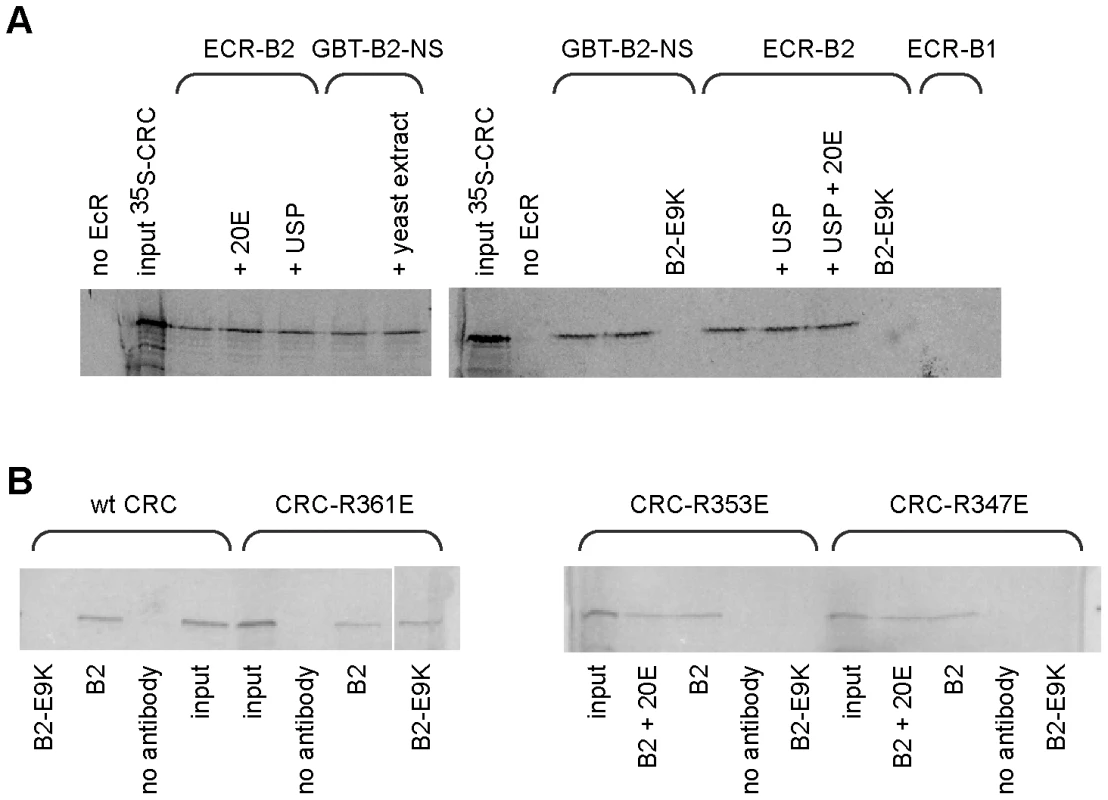
(A) CRC-A bound in vitro to the 17 amino acid amino-terminus of EcR-B2 and full length EcR-B2, but not to EcR-B1 or EcR-B2-E9K. This interaction was not dependent upon USP or ecdysone. Gels from two experiments are shown; the labeled protein was 35S-GAD-CRC-A in the left gel, 35S-CRC-A in the right gel. Lanes labeled “input” contained 100% of the input labeled protein. The remaining samples contained the EcR or EcR fragment shown above, and other components as indicated. GBT-B2-NS is a fusion of the DNA-binding domain of GAL4 with the amino-terminal 17 amino acids of EcR-B2. In lanes labeled B2-E9K, E9K mutant EcR-B2 or E9K mutant GBT-B2-NS was substituted 1∶1 for the corresponding wild-type protein. Full-length EcRs were precipitated with a mixture of two EcR-common region monoclonal antibodies, and GBT-B2-NS was precipitated with an antibody to the GAL4 DNA-binding domain. (B) A similar experiment, showing the binding of wild-type and mutant CRCs to full-length EcR-B2. Each incubation mixture contained the 35S-radiolabeled CRC protein listed above and the other components listed below, and precipitations were performed with the same mix of EcR common region antibodies as in (A). Wild-type CRC, CRC-R347E, and CRC-R353E bound to EcR-B2 but not EcR-B2-E9K. However, the basic-to-acidic mutation in CRC-R361E permitted binding to EcR-B2-E9K. Neither USP (FBgn0003964) nor the hormone ecdysone affected the CRC-EcR-B2 interaction as measured in our biochemical tests (Figure 2A). While AF1 activity is hormone-dependent in vivo, that is probably due to the effects of corepressors (e.g. SMRTER) that bind unliganded EcR/USP and suppress the activity of AF1 [4].
Loss of CRC Enhanced Phenotypes in Tissues Requiring EcR-B2
Both the crc mutant phenotype, which includes molting defects that result in supernumerary mouthparts in larvae and failure to evert the adult head at pupal ecdysis, and the pattern of crc expression suggest a role for CRC in the ecdysone response [15]. We used a genetic interaction test to determine whether CRC functions as a modulator of EcR-B2 function in flies. We examined the effects of a single copy of crc1 (a spontaneous mutation, Q171R; FBal0001818) [15], [17] on the phenotype produced by targeted expression of the dominant-negative mutant EcR-B1-F645A [2], [4]. EcR-B1-F645A is normal in transcriptional repression (the effect of unliganded receptor), but it fails to mediate transcriptional activation. Because the EcR isoforms do not display isoform specificity in DNA binding [5], the EcR-B1-F645A mutant is thought to competitively inhibit all three endogenous EcR isoforms [18]. Hence, a reduction in EcR-B2 coactivator titer should selectively enhance the effects caused by EcR-F645A expression only in tissues requiring the EcR-B2 isoform.
Targeted expression of the dominant-negative receptor EcR-B1-F645A permits an examination of the properties of EcR function in specific tissues in the context of an otherwise normal animal [2]. crc1 is a recessive mutation, and crc1/crc+ heterozygous cells are phenotypically normal. We generated sensitized tissues by targeting expression of EcR-B1-F645A to five different developmental domains, using specific GAL4 drivers [2]. As shown in Table 1, crc1 was a dominant enhancer of the EcR dominant negative phenotype in the Eip domain (largely larval epidermis) and in the slbo domain (specialized portions of the follicular epithelium of the egg chamber), but it had no significant effect in the GMR domain (primarily retinal epithelium), the dpp domain (primarily A/P disc boundaries), or the Lsp2 domain (fat body). We have previously described the EcR isoform requirements in each of these domains [2]. There was a remarkable correlation: Where EcR-B2 is required for development, wild-type crc function was also required, and where normal development does not require EcR-B2, reduction of the CRC titer had little or no effect.
Tab. 1. Dominant effects of crc1 on the phenotypes of EcR-B1-F645A expression. 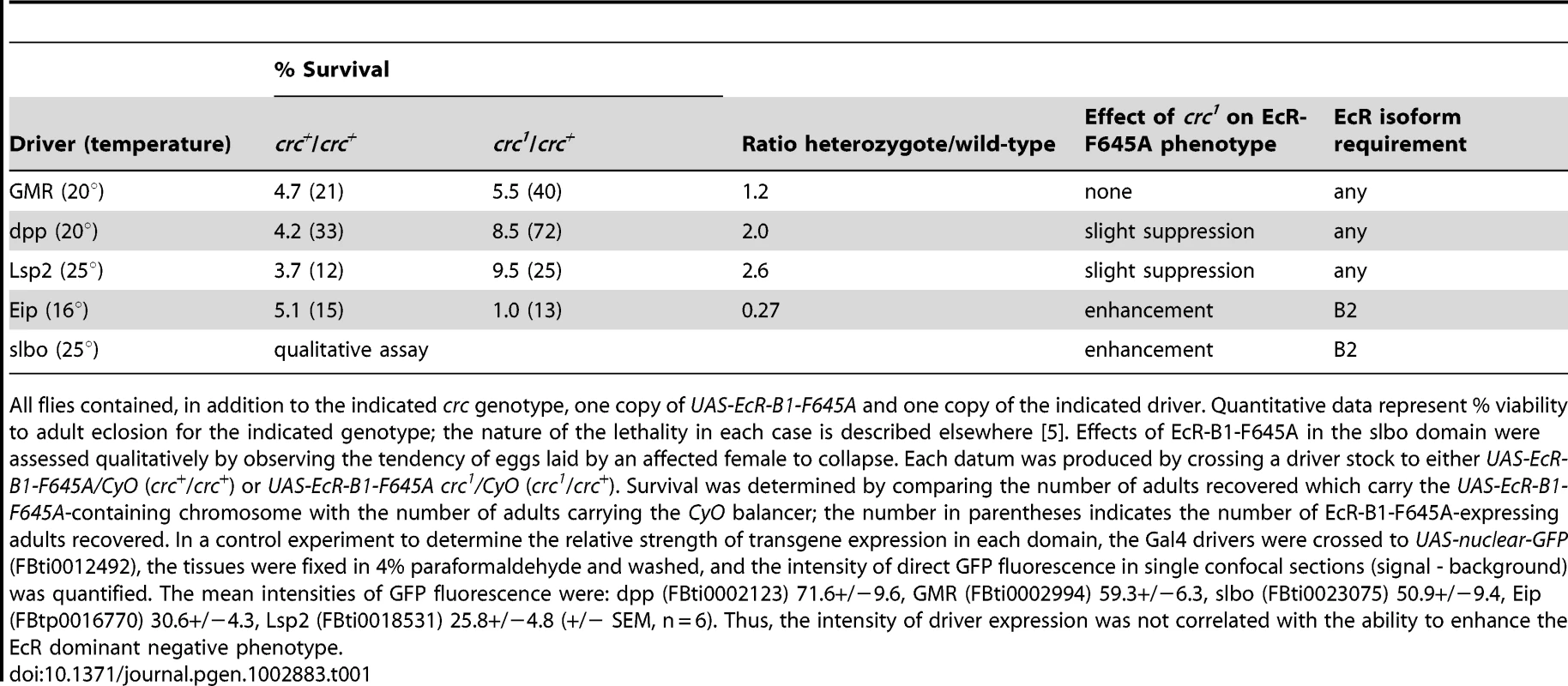
All flies contained, in addition to the indicated crc genotype, one copy of UAS-EcR-B1-F645A and one copy of the indicated driver. Quantitative data represent % viability to adult eclosion for the indicated genotype; the nature of the lethality in each case is described elsewhere [5]. Effects of EcR-B1-F645A in the slbo domain were assessed qualitatively by observing the tendency of eggs laid by an affected female to collapse. Each datum was produced by crossing a driver stock to either UAS-EcR-B1-F645A/CyO (crc+/crc+) or UAS-EcR-B1-F645A crc1/CyO (crc1/crc+). Survival was determined by comparing the number of adults recovered which carry the UAS-EcR-B1-F645A-containing chromosome with the number of adults carrying the CyO balancer; the number in parentheses indicates the number of EcR-B1-F645A-expressing adults recovered. In a control experiment to determine the relative strength of transgene expression in each domain, the Gal4 drivers were crossed to UAS-nuclear-GFP (FBti0012492), the tissues were fixed in 4% paraformaldehyde and washed, and the intensity of direct GFP fluorescence in single confocal sections (signal - background) was quantified. The mean intensities of GFP fluorescence were: dpp (FBti0002123) 71.6+/−9.6, GMR (FBti0002994) 59.3+/−6.3, slbo (FBti0023075) 50.9+/−9.4, Eip (FBtp0016770) 30.6+/−4.3, Lsp2 (FBti0018531) 25.8+/−4.8 (+/− SEM, n = 6). Thus, the intensity of driver expression was not correlated with the ability to enhance the EcR dominant negative phenotype. CRC Regulation of ETH Expression
In homozygous or hemizygous crc1 mutant larvae, expression of ETH is markedly reduced [19]. The loss of crc has similar effects on expression of an ETH-EGFP reporter gene, which contains 382 bp of the ETH promoter and precisely recapitulates the native pattern of ETH expression [14], [19]. Thus, CRC up-regulates ETH expression.
CRC is expressed in many larval tissues, including the endocrine source of ecdysone [15]. Therefore, to test for cell-autonomous regulation of ETH expression by CRC, we used an ETH-GeneSwitch driver to drive transgenic crc RNAi (UAS-crc-RNAi) specifically in the Inka cells (Figure S1), the site of ETH synthesis [14]. GeneSwitch is a conditional GAL4 protein that is activated by addition of the progesterone antagonist RU486 to the food [20]. Compared to the control larvae, larvae with crc RNAi showed a 15-fold or greater reduction in ETH transcript levels (Figure 3A). Thus, CRC was cell-autonomously required in the Inka cells for full ETH expression.
Fig. 3. EcR-B1, EcR-B2, and CRC regulated ETH expression. 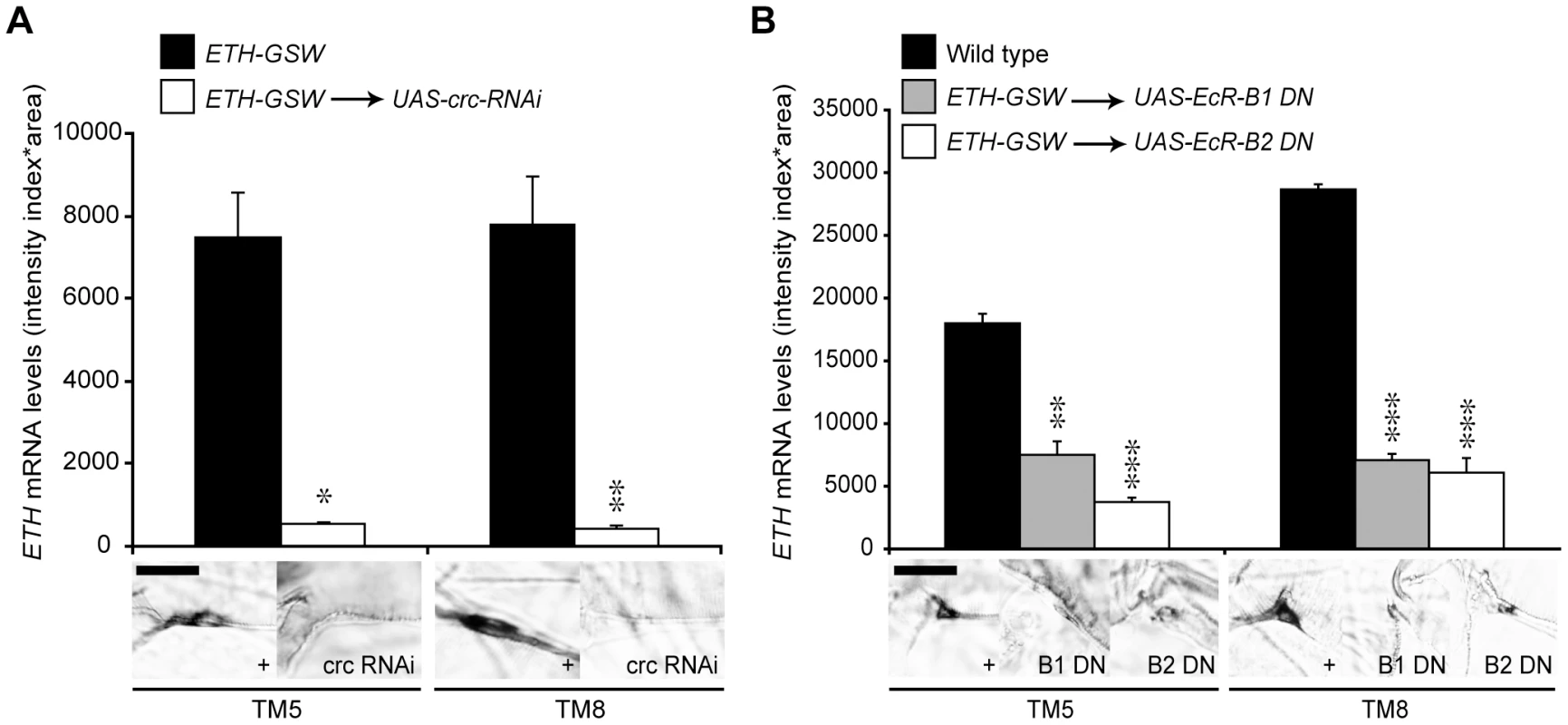
(A) Inka cell-specific crc RNAi lowered ETH transcript levels (n = 9, crc RNAi; n = 6, ETH-GSW>+controls). *, p<0.05; **, p<0.01 (One-way ANOVA: TM5, p = 0.01225; TM8, p = 0.001375). (B) Inka cell-specific expression of dominant negative EcR-B1-W650A or EcR-B2-W650A (EcR-B1 DN or EcR-B2 DN) lowered ETH transcript levels (n = 9, dominant negatives; n = 23, Oregon R controls). **, p<0.01; ***, p<0.001 [One-way ANOVA with Bonferroni (all-pairwise) multiple comparison test: TM5, p = 0.000265; TM8, p = 0.000044]. Bar = 20 µM. Regulation of ETH Expression by Ecdysone
The Drosophila melanogaster ETH promoter contains a putative ecdysone response element [19], [21], and ETH expression in the tobacco hawkmoth (Manduca sexta) fluctuates during the molts and is elevated in response to circulating ecdysteroids [22]. Therefore, we examined whether ETH expression is ecdysone-dependent. In larval and pupal Drosophila, expression of the ETH peptide hormone is restricted to 14 endocrine Inka cells located on the trachea [14]. We performed ETH in situ hybridization and found that ETH transcript levels increased gradually during the first few hours after metamorphosis was initiated (Figure 4A). The ETH transcript levels peaked 6–8 hr after the pulse of ecdysone that occurs at pupariation (Figure 4A), suggesting that ETH was transcribed in response to elevation of the circulating ecdysone titer.
Fig. 4. ETH expression was stimulated by ecdysone. 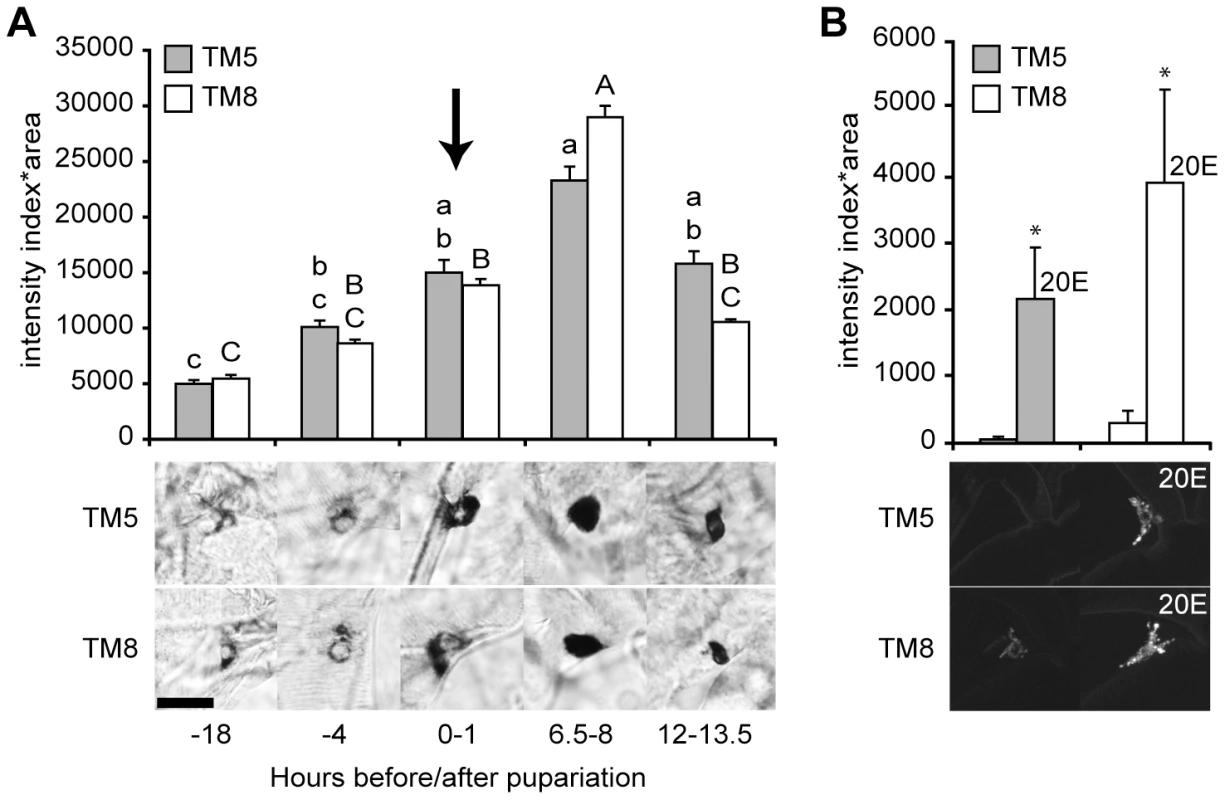
(A) ETH transcript levels mirrored the changes in the circulating ecdysone titer with a 6.5 to 8 hr delay. Transcript levels were measured semi-quantitatively in as the intensity of ETH in situ hybridization in Inka cells in tracheal metamere 5 (TM5) and TM8 at five times at the onset of metamorphosis (relative to pupariation). The arrow indicates the peak of the late-larval pulse of circulating 20E, which occurs at pupariation [23]. Means with the same lower case (TM5) or upper case (TM8) letters were not significantly different [p>0.05, n = 5–8; One-way ANOVA (TM5, p = 0.000776; TM8, p<0.000001) with Bonferroni (all-pairwise) multiple comparison test]. Insets below the histograms in (A) and (B) show representative images of cells at each time point. (B) The Inka cells of young third instar larvae fed with 20E for 12 hr showed higher expression levels for the fluorescent ETH reporter, ETH-EGFP (n = 4). *, p<0.05 (one-way ANOVA: TM5, p = 0.036; TM8, p = 0.044). Bar = 20 µM. We tested for direct ecdysone-dependence of ETH expression in young third instar stage larvae, when circulating ecdysone and ETH transcript and protein levels are low, by feeding them the major active form of ecdysone, 20-hydroxyecdysone (20E) [23]. These larvae carried the ETH-EGFP reporter gene. By 12 hr after the onset of the 20E treatment, the level of ETH-EGFP fluorescence was markedly elevated (Figure 4B). Thus, ETH expression was strongly up-regulated by circulating ecdysone.
To test for a direct, cell autonomous effect of ecdysone on ETH expression, we targeted EcR-F645A dominant negative proteins specifically to the Inka cells with the ETH-GeneSwitch driver. Following Inka cell expression of the EcR dominant negative proteins, ETH transcripts were still present but at levels that were 2–6 fold lower than in wild-type larvae (Figure 3B). Thus, ETH expression was strongly stimulated by ecdysone and required EcR expression in the Inka cells.
CRC and EcR-B2 Interacted to Boost ETH Expression In Vivo
The EcR-B1-F645A mutant is effective as a dominant negative when it is expressed in excess of the wild-type isoforms (Table 1) [2], [4]. However, the dominant negative EcR-B1-F645A protein competes poorly with wild-type EcR when both are expressed from identical promoters [2], [4]. Therefore, to determine which EcR isoforms support up-regulation of ETH expression in the Inka cells, we performed competition experiments in which EcR-B1-F645A and individual wild-type isoforms were coexpressed under the control of the ETH-GeneSwitch driver. The ability of a wild-type EcR isoform to mitigate the effects of the dominant negative is indirect evidence in support of transcriptional activation of the ETH promoter by that isoform.
Supernumerary mouthparts result when larvae fail to complete ecdysis to either the second or the third larval instar, and they are a characteristic feature of the ETH and crc mutant phenotypes [14], [15]. Over 95% of larvae with Inka cell-targeted EcR-B1-F645A expression had multiple mouthparts, and ∼70% of these animals died as larvae (Figure 5A). Simultaneous expression of wild-type EcR-B2 or EcR-B2-E9K with EcR-B1-F645A fully rescued lethality and ecdysis of the larval mouthparts, whereas EcR-B1 and EcR-A produced only partial rescue (Figure 5A). Within the Inka cells, ETH transcript levels were fully rescued by EcR-B2, but EcR-B1 and EcR-B2-E9K were ineffective at rescue (Figure 5B). Thus, of the three EcR isoforms, only wild-type EcR-B2 was capable of supporting full ETH expression and successful ecdysis. The E9K mutant of EcR-B2 failed to rescue ETH transcript levels, suggesting a model in which dimerization of EcR-B2 with CRC is required for ETH mRNA expression.
Fig. 5. EcR-B2 rescued ecdysis and ETH expression following targeted expression of dominant negative EcR. 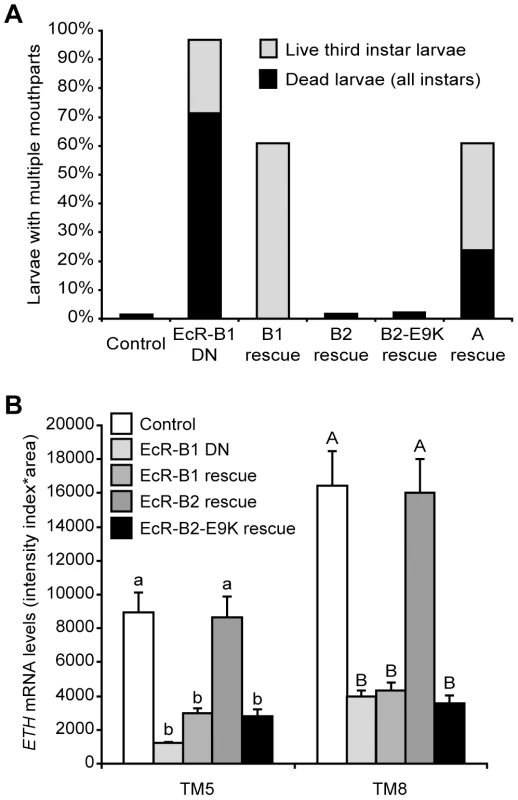
(A) Ecdysis in third instar larvae expressing dominant negative EcR-B1-F645A only in the Inka cells (n = 98; EcR-B1 DN) was rescued by expression of EcR-B2 (n = 165) or EcR-B2-E9K (n = 143), but not by expression of EcR-B1 (n = 91) or EcR-A (n = 59). The controls had ETH-GSW only (n = 148), and all larvae were fed RU486. The larval counts included all live third instar larvae (with or without multiple mouthparts), as well as all dead larvae (of all stages). All of the dead larvae found had multiple mouthparts. (B) ETH expression in Inka cells expressing dominant negative EcR-B1-F645A isoform (n = 11) was rescued by co-expression of wild-type EcR-B2 (n = 8) but not wild-type EcR-B1 (n = 9) or the E9K mutant of EcR-B2 (n = 8). The controls had ETH-GSW only (n = 8), and all larvae were fed RU486. All larvae were dissected at ∼12 hr after ecdysis to the third instar. Means with the same lower case (TM5) or upper case (TM8) letters were not significantly different [p>0.05, One-way ANOVA (TM5, p<0.000001; TM8, p = 0.000002) with Bonferroni (all-pairwise) multiple comparison test]. In M. sexta, 20E regulates ETH synthesis as well as the competency of the Inka cells to secrete ETH [24]. The ability of EcR-B2-E9K, and to a lesser extent EcR-B1 and EcR-A, to rescue ecdysis and lethality (Figure 5A) indicates that EcR likely regulates other Inka cell processes, such as ETH protein accumulation or secretory competence, that are necessary for signaling by ETH. We tested this hypothesis by performing ETH immunostaining in EcR-B2-E9K rescue animals both before and after secretion at pupal ecdysis. Although EcR-B2-E9K did not stimulate ETH transcription (Figure 5B), it drove ETH protein accumulation in the Inka cells (Figure S2A). Consistent with the predicted role of EcR in the development of secretory competence, we also observed a marked decrease in accumulated ETH at pupal ecdysis (Figure S2B). These results show that EcR—likely through different sets of EcR isoforms and transcriptional coactivators—regulates ETH protein accumulation independently of ETH mRNA expression.
Discussion
Our experiments suggest that the 17-residue B2-specific N-terminus binds to the bZIP region of CRC, that an ionic interaction between EcR-B2-E9 and CRC-R361 plays some role in the binding, and that the interaction of the two proteins plays a crucial role in those tissues where EcR-B2 is essential. These tissues include the endocrine Inka cells, which display ecdysone-dependent upregulation of ETH transcripts and which require EcR-B2 and CRC for full ETH expression. Taken together, these findings implicate CRC as an isoform-specific transcriptional activator for EcR-B2.
In diverse systems, bZIP proteins interact with dyadic or palindromic promoter sequences as homodimers or heterodimers with other bZIP partners [25]. Dimerization involves regularly spaced hydrophobic amino acids that form a coiled-coil between two leucine zipper domains [26]. Other bZIP transcription factors are known to interact with nuclear receptors, modulating the activities of either AF1 or AF2 [27]–[29], but in the cases reported previously, bZIP proteins bind either to the DNA-binding domain or to the hinge domain of the nuclear receptor. By contrast, CRC (through its bZIP domain) appears to bind directly to the EcR-B2 AF1 region, and its interaction is specific to one EcR isoform.
ATF4, the mammalian homolog of CRC, plays a central role in stress responses [30]. The role of CRC in ecdysone signaling suggests the possibility of interesting and unexpected connections between stress responses and the control of developmental timing and metamorphosis.
The ETH promoter contains sequences matching the consensus half-sites for binding of ATF4 and EcR to DNA. These half-sites are separated by 4 nucleotides, and they are located within a highly conserved sequence (comparing D. melanogaster to several other Drosophila species) that is 138–171 nucleotides upstream of the ETH transcriptional start site [19]. Since bZIP proteins may bind first sequentially as monomers and then dimerize while bound to DNA [26], [31], these observations suggest a model in which CRC participates in the stabilization of EcR-B2 binding to the ETH promoter. This interaction provides a basis for understanding some of the differences in transcriptional properties that are displayed by the EcR isoforms and perhaps other nuclear receptors with distinct AF1-coactivators.
Materials and Methods
Yeast Two-Hybrid Screening
Yeast two-hybrid assays were carried out using the Clontech Matchmaker yeast two-hybrid kit (Clontech, Mountain View, CA) and yeast strain Hf7C. The bait was a fusion of the GAL4 DNA-binding domain to either the 17-residue AF1 domain of EcR-B2, or the same fragment containing the mutation E9K. Binding was assayed as expression of β-galactosidase from a UAS-lacZ reporter.
In Vitro Protein Binding
Proteins were synthesized in vitro using the TNT reticulocyte lysate kit (Promega, Madison, WI); template plasmids were described previously or were generated by a similar procedure [4]. Binding reactions (50 µl) contained 3 µl of each indicated translation mix in buffer A (20 mM NaH2PO4, 150 mM NaCl, pH 8.0) were incubated at 4° for 30 min. Then, 25 µl of a 50% slurry of Sepharose-protein A (Sigma) loaded with the indicated antibody was added and the incubation continued for 30 min. Beads were washed 3 times in buffer A and then boiled in SDS-PAGE sample buffer. The eluted proteins were separated by SDS-PAGE and radiolabeled proteins detected by autoradiography. For precipitation of full length EcRs, a mixture of the EcR-common region monoclonal antibodies, AG 10.2 and DDA 2.7 [6], was used with each at a 1∶10,000 dilution of an ascites fluid. For precipitation of GBT-B2-NS, we used a commercial antibody to the GAL4 DNA-binding domain (1∶1000, Santa Cruz Biotechnology, Santa Cruz, CA).
Animals and Staging
Drosophila melanogaster were reared on standard cornmeal-yeast-agar media at 22–25° unless otherwise noted. Oregon-R was used as the wild-type strain. Larvae at the onset of metamorphosis were scored based on the blue color intensity observed in the gut of third instar larvae fed with cornmeal-yeast-agar food supplemented with 0.1% bromophenol blue. We collected blue gut larvae (18 hours before pupariation) and clear gut larvae (4 hours before pupariation) [32]. Prepupae and pupae were selected based on the criteria reported by Bainbridge and Bownes [33] at the following stages: white puparium (P1 stage; at puparium formation), buoyant prepupa (P4i stage; 6.5–8 hours after puparium formation), and moving bubble prepupa (P4ii stage; 12–13.5 hours after puparium formation).
The ETH-GeneSwitch (ETH-GSW) line was a kind gift from Michael Adams (University of California, Riverside) and Yoonseung Park (Kansas State University). It expresses a conditional, RU486-dependent GAL4 protein chimera [20] under the control of the 382 bp ETH promoter region [19], [21]. First instar larvae carrying ETH-GSW and selected UAS constructs were transferred after hatching to cornmeal-yeast-agar media supplemented with 500 mM RU486 [34]. In larvae, the expression of a reporter gene under ETH-GSW/RU486 control was restricted to just the Inka cells (Figure S1).
The CRC and EcR loss-of-function transgenes included UAS-Crc-RNAi (Vienna Drosophila RNAi Center (VDRC) line #2935, FBti0084038) [35], UAS-EcR-B1-W650A (FBti0026963), UAS-EcR-B2-W650A, UAS-EcR-B1-F645A (FBti0026961), and UAS-EcR-B2-E9K. The UAS-EcR-B1 (FBti0023086), UAS-EcR-B2 (FBti0023085), and UAS-EcR-A (FBti0023087) transgenes contain the three wild-type EcR isoforms [2].
20E Feeding
Freshly ecdysed ETH-EGFP (FBal0136020) third instar larvae were transferred on cornmeal-yeast-agar media supplemented with 0.08 mg/ml 20E [36] and collected 12 hours later for analysis of EGFP fluorescence in the Inka cells.
Tissue Preparation and Image Analysis
Digoxigenin-labeled DNA probe preparation, whole-mount larval in situ hybridization, ETH in situ hybridization, anti-PETH immunostaining, and ETH-EGFP imaging was performed as described [19]. In larvae, the Inka cells are identified by the tracheal metameres (TMs) on which they are located, and the TMs are numbered 1 to 10, starting with the anterior end of the animal. To quantify the intensity of EGFP, immunostaining, and in situ hybridization signals, we measured the Intensity Index*Area = S*[(I–B)/B] where (S) is the surface area covered by the signal, (I) is the mean pixel intensity of the signal within this area, and (B) is the background signal intensity [19], [37]. This method takes into consideration the density of the signal distributed over the cell area, and it therefore normalizes for the angle at which the Inka cell is photographed and for heterogeneity in the spatial distribution of the signal. The measurements were taken using Adobe Photoshop (San Jose, CA, USA).
Statistical Analysis
Statistical tests were performed using the NCSS 2001 software package (Kaysville, UT). Bonferroni corrections were performed to minimize type I errors in multiple pair-wise comparisons (Rice, 1989). We used parametric statistics because the data generally followed a normal distribution. All values are means ± s.e.m., except as indicated.
Supporting Information
Zdroje
1. WarnmarkA, TreuterE, WrightAP, GustafssonJA (2003) Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol 17 : 1901–1909.
2. CherbasL, HuX, ZhimulevI, BelyaevaE, CherbasP (2003) EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development 130 : 271–284.
3. Dela CruzFE, KirschDR, HeinrichJN (2000) Transcriptional activity of Drosophila melanogaster ecdysone receptor isoforms and ultraspiracle in Saccharomyces cerevisiae. J Mol Endocrinol 24 : 183–191.
4. HuX, CherbasL, CherbasP (2003) Transcription activation by the ecdysone receptor (EcR/USP): identification of activation functions. Mol Endocrinol 17 : 716–731.
5. MouilletJF, HenrichVC, LezziM, VogtliM (2001) Differential control of gene activity by isoforms A, B1 and B2 of the Drosophila ecdysone receptor. Eur J Biochem 268 : 1811–1819.
6. TalbotWS, SwyrydEA, HognessDS (1993) Drosophila tissues with different metamorphic responses to ecdysone express different ecdysone receptor isoforms. Cell 73 : 1323–1337.
7. GatesJ, LamG, OrtizJA, LossonR, ThummelCS (2004) rigor mortis encodes a novel nuclear receptor interacting protein required for ecdysone signaling during Drosophila larval development. Development 131 : 25–36.
8. BaiJ, UeharaY, MontellDJ (2000) Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell 103 : 1047–1058.
9. SedkovY, ChoE, PetrukS, CherbasL, SmithST, et al. (2003) Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature 426 : 78–83.
10. BadenhorstP, XiaoH, CherbasL, KwonSY, VoasM, et al. (2005) The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev 19 : 2540–2545.
11. ZhuJ, ChenL, SunG, RaikhelAS (2006) The competence factor betaFtz-F1 potentiates ecdysone receptor activity via recruiting a p160/SRC coactivator. Mol Cell Biol 26 : 9402–9412.
12. BecksteadR, OrtizJA, SanchezC, ProkopenkoSN, ChambonP, et al. (2001) Bonus, a Drosophila homolog of TIF1 proteins, interacts with nuclear receptors and can inhibit betaFTZ-F1-dependent transcription. Mol Cell 7 : 753–765.
13. TsaiCC, KaoHY, YaoTP, McKeownM, EvansRM (1999) SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol Cell 4 : 175–186.
14. ParkY, FilippovV, GillSS, AdamsME (2002) Deletion of the ecdysis-triggering hormone gene leads to lethal ecdysis deficiency. Development 129 : 493–503.
15. HewesRS, SchaeferAM, TaghertPH (2000) The cryptocephal gene (ATF4) encodes multiple basic-leucine zipper proteins controlling molting and metamorphosis in Drosophila. Genetics 155 : 1711–1723.
16. FasslerJ, LandsmanD, AcharyaA, MollJR, BonovichM, et al. (2002) B-ZIP proteins encoded by the Drosophila genome: evaluation of potential dimerization partners. Genome Res 12 : 1190–1200.
17. HadornE, GloorH (1943) Cryptocephal ein spat wirkender Letalfaktor bei Drosophila melanogaster. Revue suisse Zool 50 : 256–261.
18. BrownHL, CherbasL, CherbasP, TrumanJW (2006) Use of time-lapse imaging and dominant negative receptors to dissect the steroid receptor control of neuronal remodeling in Drosophila. Development 133 : 275–285.
19. GauthierSA, HewesRS (2006) Transcriptional regulation of neuropeptide and peptide hormone expression by the Drosophila dimmed and cryptocephal genes. J Exp Biol 209 : 1803–1815.
20. OsterwalderT, YoonKS, WhiteBH, KeshishianH (2001) A conditional tissue-specific transgene expression system using inducible GAL4. PNAS 98 : 12596–12601.
21. ParkY, ZitnanD, GillSS, AdamsME (1999) Molecular cloning and biological activity of ecdysis-triggering hormones in Drosophila melanogaster. FEBS Lett 463 : 133–138.
22. ZitnanD, RossLS, ZitnanovaI, HermesmanJL, GillSS, et al. (1999) Steroid induction of a peptide hormone gene leads to orchestration of a defined behavioral sequence. Neuron 23 : 523–535.
23. Riddiford LM (1993) Hormones and Drosophila development. In: The development of Drosophila melanogaster. Bate M, Martinez Arias A, editors. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. 899–939.
24. KinganTG, AdamsME (2000) Ecdysteroids regulate secretory competence in Inka cells. J Exp Biol 203 : 3011–3018.
25. MillerM (2009) The importance of being flexible: the case of basic region leucine zipper transcriptional regulators. Curr Protein Pept Sci 10 : 244–269.
26. VinsonC, AcharyaA, TaparowskyEJ (2006) Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta 1759 : 4–12.
27. BorukM, SavoryJG, HacheRJ (1998) AF-2-dependent potentiation of CCAAT enhancer binding protein beta-mediated transcriptional activation by glucocorticoid receptor. Mol Endocrinol 12 : 1749–1763.
28. TeyssierC, BelguiseK, GaltierF, ChalbosD (2001) Characterization of the physical interaction between estrogen receptor alpha and JUN proteins. J Biol Chem 276 : 36361–36369.
29. WardellSE, BoonyaratanakornkitV, AdelmanJS, AronheimA, EdwardsDP (2002) Jun dimerization protein 2 functions as a progesterone receptor N-terminal domain coactivator. Mol Cell Biol 22 : 5451–5466.
30. RutkowskiDT, KaufmanRJ (2003) All roads lead to ATF4. Dev Cell 4 : 442–444.
31. KohlerJJ, MetalloSJ, SchneiderTL, SchepartzA (1999) DNA specificity enhanced by sequential binding of protein monomers. Proc Natl Acad Sci U S A 96 : 11735–11739.
32. EmeryIF, BedianV, GuildGM (1994) Differential expression of Broad-Complex transcription factors may forecast tissue-specific developmental fates during Drosophila metamorphosis. Development 120 : 3275–3287.
33. BainbridgeSP, BownesM (1981) Staging the metamorphosis of Drosophila melanogaster. J Embryol Exp Morphol 66 : 57–80.
34. McGuireSE, MaoZ, DavisRL (2004) Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE 2004: pl6.
35. DietzlG, ChenD, SchnorrerF, SuKC, BarinovaY, et al. (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 : 151–156.
36. ColombaniJ, BianchiniL, LayalleS, PondevilleE, Dauphin-VillemantC, et al. (2005) Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310 : 667–670.
37. HewesRS, ParkD, GauthierSA, SchaeferAM, TaghertPH (2003) The bHLH protein Dimmed controls neuroendocrine cell differentiation in Drosophila. Development 130 : 1771–1781.
Štítky
Genetika Reprodukční medicína
Článek Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma GenomesČlánek Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail ExpansionČlánek Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid CellsČlánek Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug ActivationČlánek The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin DynamicsČlánek Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 8
-
Všechny články tohoto čísla
- Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma Genomes
- Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail Expansion
- Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid Cells
- Chromosome Territories Meet a Condensin
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans
- Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse ()
- A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins
- Advances in Quantitative Trait Analysis in Yeast
- Experimental Evolution of a Novel Sexually Antagonistic Allele
- Variation of Contributes to Dog Breed Skull Diversity
- , a Gene Involved in Axonal Pathfinding, Is Mutated in Patients with Kallmann Syndrome
- A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression
- Cryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
- Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug Activation
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
- A Sexual Ornament in Chickens Is Affected by Pleiotropic Alleles at and , Selected during Domestication
- Use of Allele-Specific FAIRE to Determine Functional Regulatory Polymorphism Using Large-Scale Genotyping Arrays
- Novel Loci for Metabolic Networks and Multi-Tissue Expression Studies Reveal Genes for Atherosclerosis
- The Genetic Basis of Pollinator Adaptation in a Sexually Deceptive Orchid
- Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
- Inheritance Beyond Plain Heritability: Variance-Controlling Genes in
- The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, and Anthropometric Traits
- Reprogramming to Pluripotency Can Conceal Somatic Cell Chromosomal Instability
- Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes
- PTEN Negatively Regulates MAPK Signaling during Vulval Development
- A Dynamic Response Regulator Protein Modulates G-Protein–Dependent Polarity in the Bacterium
- Population Genomics of the Facultatively Mutualistic Bacteria and
- Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
- Polysome Profiling in Liver Identifies Dynamic Regulation of Endoplasmic Reticulum Translatome by Obesity and Fasting
- Stromal Liver Kinase B1 [STK11] Signaling Loss Induces Oviductal Adenomas and Endometrial Cancer by Activating Mammalian Target of Rapamycin Complex 1
- Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
- Transgene Induced Co-Suppression during Vegetative Growth in
- Hox and Sex-Determination Genes Control Segment Elimination through EGFR and Activity
- A Quantitative Comparison of the Similarity between Genes and Geography in Worldwide Human Populations
- Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in and Mammals
- Comparative Analysis of Regulatory Elements between and by Genome-Wide Transcription Start Site Profiling
- Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome
- S Phase–Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
- Cell-Nonautonomous Signaling of FOXO/DAF-16 to the Stem Cells of
- The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin Dynamics
- Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus
- Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes
- Genetic Modulation of Lipid Profiles following Lifestyle Modification or Metformin Treatment: The Diabetes Prevention Program
- HAL-2 Promotes Homologous Pairing during Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors
- SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Germline
- Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
- Reduced Prostasin (CAP1/PRSS8) Activity Eliminates HAI-1 and HAI-2 Deficiency–Associated Developmental Defects by Preventing Matriptase Activation
- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
- and Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR
- CDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
- Rare Copy Number Variations in Adults with Tetralogy of Fallot Implicate Novel Risk Gene Pathways
- Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of
- NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing
- Human Developmental Enhancers Conserved between Deuterostomes and Protostomes
- A Luminal Glycoprotein Drives Dose-Dependent Diameter Expansion of the Hindgut Tube
- Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation
- Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration
- Chemical Genetics Reveals a Specific Requirement for Cdk2 Activity in the DNA Damage Response and Identifies Nbs1 as a Cdk2 Substrate in Human Cells
- Experimental Relocation of the Mitochondrial Gene to the Nucleus Reveals Forces Underlying Mitochondrial Genome Evolution
- Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome
- Mutations in a P-Type ATPase Gene Cause Axonal Degeneration
- A General G1/S-Phase Cell-Cycle Control Module in the Flowering Plant
- Multiple Roles and Interactions of and in Development of the Respiratory System
- UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex
- Epigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
- Modulating the Strength and Threshold of NOTCH Oncogenic Signals by
- Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer's Disease–Related Tau Phosphorylation Via PAR-1
- Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System
- ATXN2-CAG42 Sequesters PABPC1 into Insolubility and Induces FBXW8 in Cerebellum of Old Ataxic Knock-In Mice
- Cohesin Rings Devoid of Scc3 and Pds5 Maintain Their Stable Association with the DNA
- The MicroRNA Inhibits Calcium Signaling by Targeting the TIR-1/Sarm1 Adaptor Protein to Control Stochastic L/R Neuronal Asymmetry in
- Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength
- The Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Variation of Contributes to Dog Breed Skull Diversity
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



