-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
article has not abstract
Published in the journal: . PLoS Genet 8(8): e32767. doi:10.1371/journal.pgen.1002860
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1002860Summary
article has not abstract
Transcriptional regulation in eukaryotes requires the transcriptional machinery to negotiate the complexities of DNA packaged into chromatin. Specific modifications of the core histone proteins serve to regulate transcription and ensure that genes are expressed at the right place and the right time [1]. Not surprisingly, the interaction between the transcriptional apparatus and chromatin is a tango in which the partners are in a close embrace. Cyclin-dependent kinase 9 (CDK9) and its orthologs control both transcription and transcription-coupled chromatin modifications in a variety of species [2], [3]. However, the mechanisms that intertwine CDK9 function with chromatin appear to be distinct in different organisms. A new manuscript in this issue of PLoS Genetics sheds light onto how Cdk9 function is interconnected with the monoubiquitination of histone H2B in the fission yeast Schizosaccharomyces pombe [4].
The Dance Partners: CDK9 and H2Bub1
CDK9 controls transcriptional elongation, transcription-coupled mRNA processing, and histone modification [2], [5]. It accomplishes this by phosphorylating a number of central transcriptional regulatory proteins, including RNA Polymerase II (RNAPII) (reviewed in [6]), Suppressor of Ty-5 (Spt5; or its metazoan ortholog SUPT5H) [7], [8], Rad6 (metazoan UBE2A) [9], [10], and Negative Elongation Factor-E (NELF-E) [11], [12]. Notably, phosphorylation of Ser2 within the repeated YSPTSPS heptapeptide motif of the RNAPII carboxy-terminal domain (CTD) and Thr1 within the Spt5 C-terminal repeat (CTR) are conserved across eukaryotes and responsible for various aspects of CDK9 function.
One post-translational histone modification emerging as a key player in numerous processes is histone H2B monoubiquitination (H2Bub1). In mammals this modification appears to serve as an important tumor suppressor [13]. H2Bub1 is associated with the transcribed region of active genes [14], [15] and promotes transcriptional elongation in vitro [16]. Interestingly, the trimethylation of histone H3 lysine 4 (H3K4me3) and lysine 79 (H3K79me3) also depend on H2Bub1 17–19, although its importance to mammalian H3K4me3 may be more limited [20], [21].
New Insights into the CDK9–H2Bub1 Tango
The new study by Sansó et al. has unraveled additional details about the complex dance between the fission yeast ortholog of CDK9 (spCdk9, also called Pch1) and H2Bub1 [4]. The authors utilized a genome-wide approach to characterize the effects of a loss of H2Bub1 on RNAPII occupancy and mRNA levels and observed a surprising disconnect: while RNAPII occupancy was significantly impacted at the majority of active genes, only a subset were affected at the mRNA level. In the absence of H2Bub1, RNAPII levels within the transcribed region decreased, and its typical accumulation at the 3′ end of yeast genes was even more pronounced. Consistent with a potential role in transcriptional elongation, Sansó et al. show that H2B monoubiquitination depends on spCdk9 activity, but not on the related RNAPII CTD kinase Lsk1. They further establish that, unlike in human cells [22], [23], this effect is independent of RNAPII CTD phosphorylation and instead requires spCdk9-mediated Spt5 phosphorylation. However, the relationship between CDK9 and H2Bub1 is not a linear pathway. In fact, blocking H2B monoubiquitination by replacing the ubiquitinated lysine in H2B with an arginine (K119R) leads to decreased recruitment of spCdk9 and reduced levels of Spt5 phosphorylation.
Does this mean that spCdk9 and H2Bub1 play opposing roles in fission yeast? Phenotypic studies suggest that this may be at least partially true. For example, while H2B K119R mutation or deletion of the S. pombe H2B ubiquitin ligase Brl2 both lead to septation defects, these can be rescued by blocking spCdk9. Furthermore, compound mutants of Spt5 and Set1 suggest that the phenotypic effects of H2Bub1 loss may be due to multifaceted downstream effects of H2Bub1 on both Spt5 (through spCdk9 recruitment) and H3K4me3 (through Set1). Further support for a homeostatic feedback mechanism is provided by studies of RNAPII occupancy in fission yeast with reduced spCdk9 activity. Consistent with opposing roles of H2Bub1 and spCdk9, the authors observed increased RNAPII occupancy in transcribed regions and decreased occupancy at gene 3′ ends. Importantly, the combined mutation of spCdk9 and H2Bub1 rescued the H2B K119R mutation and displayed RNAPII occupancy similar to mutant spCdk9 alone.
These results demonstrate the intricacy and complexity of the choreographed tango between CDK9 orthologs and chromatin, and illustrate significant differences between yeast and metazoans. This study shows that although H2Bub1 requires Cdk9 activity in fission yeast, Bur1 in budding yeast [24], and CDK9 in metazoans [5], [9], [20], [22], a different specific mechanism may operate in each species (Figure 1). In both yeasts, Spt5 appears to be the major Bur1/spCdk9 substrate responsible for controlling H2B monoubiquitination, and this occurs in a RNAPII CTD–independent manner [4], [8]. In human cells the situation appears more complex and involves the parallel effects of CDK9-dependent phosphorylation of the RNAPII CTD [20], [22], [23], as well as UBE2A [9] and probably also SUPT5H in some systems [5], [25]. The Sansó et al. study thus underscores the complexity of CDK9 function and the tight bilateral communication between chromatin and transcription. Further analyses will be necessary to address the various differences in CDK9 function in yeast and metazoans. For example, the existence of NELF-E and the prevalence of promoter proximal RNAPII pausing in metazoans likely represent key transcriptional elongation regulatory mechanisms that yeast lack. Furthermore, two additional closely related CDK9 orthologs, CDK12/CRKRS and CDK13/CDC2L5, have been identified and implicated in metazoan transcriptional elongation [26], [27]. Thus, determining the functions of the various CDK9 orthologs and identifying their substrates, and the interaction of these substrates with H2Bub1 and other chromatin modifications, represents an important challenge.
Fig. 1. Interconnections between CDK9 orthologs, their substrates, and downstream histone modifications in yeast and humans. 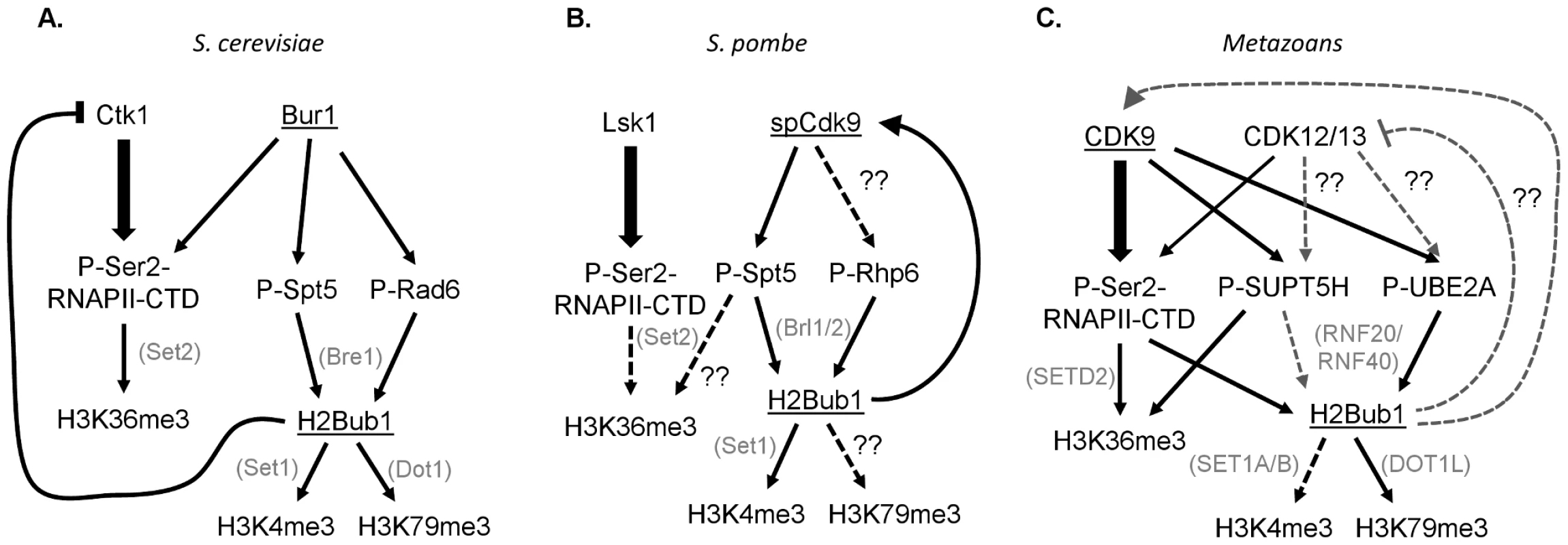
The CDK9 orthologs Bur1 in budding yeast (S. cerevisiae) and spCdk9 in fission yeast (S. pombe) are most closely related to human CDK9, while Ctk1 and Lsk1 are most homologous to CDK12 and 13. (A, B) In yeast the primary Ser2 RNAPII CTD kinases are Ctk1 and Lsk1. Furthermore, Spt5 phosphorylation, rather than Ser2 phosphorylation, is essential for H2Bub1 and its downstream histone modifications H3K4me3 and H3K79me. (C) In metazoans the separation of Ser2 and SUPT5H phosphorylation is less clear. Moreover, Ser2 phosphorylation is required for H2Bub1.
Zdroje
1. StrahlBD, AllisCD (2000) The language of covalent histone modifications. Nature 403 : 41–45.
2. PeterlinBM, PriceDH (2006) Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23 : 297–305 doi:10.1016/j.molcel.2006.06.014.
3. WoodA, ShilatifardA (2006) Bur1/Bur2 and the Ctk complex in yeast: the split personality of mammalian P-TEFb. Cell Cycle 5 : 1066–1068.
4. SansóM, LeeKM, ViladevallL, JacquesP-É, PagéV, et al. (2012) A positive feedback loop links opposing functions of P-TEFb/Cdk9 and histone H2B ubiquitylation to regulate transcript elongation in fission yeast. PLoS Genet 8: e1002822 doi:10.1371/journal.pgen.1002822.
5. PirngruberJ, ShchebetA, JohnsenSA (2009) Insights into the function of the human P-TEFb component CDK9 in the regulation of chromatin modifications and co-transcriptional mRNA processing. Cell Cycle 8 : 3636–3642.
6. EgloffS, MurphyS (2008) Cracking the RNA polymerase II CTD code. Trends Genet 24 : 280–288.
7. YamadaT, YamaguchiY, InukaiN, OkamotoS, MuraT, et al. (2006) P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 21 : 227–237.
8. ZhouK, KuoWH, FillinghamJ, GreenblattJF (2009) Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci U S A 106 : 6956–6961 doi:10.1073/pnas.0806302106.
9. ShchebetA, KarpiukO, KremmerE, EickD, JohnsenSA (2012) Phosphorylation by cyclin-dependent kinase-9 controls ubiquitin-conjugating enzyme-2A function. Cell Cycle 11 : 2122–2127.
10. WoodA, SchneiderJ, DoverJ, JohnstonM, ShilatifardA (2005) The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell 20 : 589–599.
11. FujinagaK, IrwinD, HuangY, TaubeR, KurosuT, et al. (2004) Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24 : 787–795.
12. YamaguchiY, InukaiN, NaritaT, WadaT, HandaH (2002) Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol 22 : 2918–2927.
13. JohnsenSA (2012) The enigmatic role of H2Bub1 in cancer. FEBS Lett 586 : 1592–1601 doi:10.1016/j.febslet.2012.04.002.
14. MinskyN, ShemaE, FieldY, SchusterM, SegalE, et al. (2008) Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol 10 : 483–488.
15. XiaoT, KaoCF, KroganNJ, SunZW, GreenblattJF, et al. (2005) Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol 25 : 637–651.
16. PavriR, ZhuB, LiG, TrojerP, MandalS, et al. (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125 : 703–717.
17. BriggsSD, XiaoT, SunZW, CaldwellJA, ShabanowitzJ, et al. (2002) Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418 : 498 doi:10.1038/nature00970.
18. NgHH, XuRM, ZhangY, StruhlK (2002) Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem 277 : 34655–34657 doi:10.1074/jbc.C200433200.
19. SunZW, AllisCD (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418 : 104–108.
20. KarpiukO, NajafovaZ, KramerF, HennionM, GalonskaC, et al. (2012) The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Mol Cell 46 : 705–713.
21. ShemaE, TiroshI, AylonY, HuangJ, YeC, et al. (2008) The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 22 : 2664–2676.
22. PirngruberJ, ShchebetA, SchreiberL, ShemaE, MinskyN, et al. (2009) CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′ end processing. EMBO Rep 10 : 894–900.
23. ZhangF, YuX (2011) WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell 41 : 384–397.
24. LaribeeRN, KroganNJ, XiaoT, ShibataY, HughesTR, et al. (2005) BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol 15 : 1487–1493.
25. ChenY, YamaguchiY, TsugenoY, YamamotoJ, YamadaT, et al. (2009) DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev 23 : 2765–2777 doi:10.1101/gad.1834709.
26. BartkowiakB, LiuP, PhatnaniHP, FudaNJ, CooperJJ, et al. (2010) CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24 : 2303–2316 doi:10.1101/gad.1968210.
27. BlazekD, KohoutekJ, BartholomeeusenK, JohansenE, HulinkovaP, et al. (2011) The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev 25 : 2158–2172 doi:10.1101/gad.16962311.
Štítky
Genetika Reprodukční medicína
Článek Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma GenomesČlánek Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail ExpansionČlánek Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid CellsČlánek Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug ActivationČlánek The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin DynamicsČlánek Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 8
-
Všechny články tohoto čísla
- Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma Genomes
- Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail Expansion
- Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid Cells
- Chromosome Territories Meet a Condensin
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans
- Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse ()
- A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins
- Advances in Quantitative Trait Analysis in Yeast
- Experimental Evolution of a Novel Sexually Antagonistic Allele
- Variation of Contributes to Dog Breed Skull Diversity
- , a Gene Involved in Axonal Pathfinding, Is Mutated in Patients with Kallmann Syndrome
- A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression
- Cryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
- Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug Activation
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
- A Sexual Ornament in Chickens Is Affected by Pleiotropic Alleles at and , Selected during Domestication
- Use of Allele-Specific FAIRE to Determine Functional Regulatory Polymorphism Using Large-Scale Genotyping Arrays
- Novel Loci for Metabolic Networks and Multi-Tissue Expression Studies Reveal Genes for Atherosclerosis
- The Genetic Basis of Pollinator Adaptation in a Sexually Deceptive Orchid
- Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
- Inheritance Beyond Plain Heritability: Variance-Controlling Genes in
- The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, and Anthropometric Traits
- Reprogramming to Pluripotency Can Conceal Somatic Cell Chromosomal Instability
- Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes
- PTEN Negatively Regulates MAPK Signaling during Vulval Development
- A Dynamic Response Regulator Protein Modulates G-Protein–Dependent Polarity in the Bacterium
- Population Genomics of the Facultatively Mutualistic Bacteria and
- Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
- Polysome Profiling in Liver Identifies Dynamic Regulation of Endoplasmic Reticulum Translatome by Obesity and Fasting
- Stromal Liver Kinase B1 [STK11] Signaling Loss Induces Oviductal Adenomas and Endometrial Cancer by Activating Mammalian Target of Rapamycin Complex 1
- Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
- Transgene Induced Co-Suppression during Vegetative Growth in
- Hox and Sex-Determination Genes Control Segment Elimination through EGFR and Activity
- A Quantitative Comparison of the Similarity between Genes and Geography in Worldwide Human Populations
- Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in and Mammals
- Comparative Analysis of Regulatory Elements between and by Genome-Wide Transcription Start Site Profiling
- Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome
- S Phase–Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
- Cell-Nonautonomous Signaling of FOXO/DAF-16 to the Stem Cells of
- The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin Dynamics
- Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus
- Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes
- Genetic Modulation of Lipid Profiles following Lifestyle Modification or Metformin Treatment: The Diabetes Prevention Program
- HAL-2 Promotes Homologous Pairing during Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors
- SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Germline
- Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
- Reduced Prostasin (CAP1/PRSS8) Activity Eliminates HAI-1 and HAI-2 Deficiency–Associated Developmental Defects by Preventing Matriptase Activation
- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
- and Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR
- CDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
- Rare Copy Number Variations in Adults with Tetralogy of Fallot Implicate Novel Risk Gene Pathways
- Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of
- NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing
- Human Developmental Enhancers Conserved between Deuterostomes and Protostomes
- A Luminal Glycoprotein Drives Dose-Dependent Diameter Expansion of the Hindgut Tube
- Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation
- Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration
- Chemical Genetics Reveals a Specific Requirement for Cdk2 Activity in the DNA Damage Response and Identifies Nbs1 as a Cdk2 Substrate in Human Cells
- Experimental Relocation of the Mitochondrial Gene to the Nucleus Reveals Forces Underlying Mitochondrial Genome Evolution
- Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome
- Mutations in a P-Type ATPase Gene Cause Axonal Degeneration
- A General G1/S-Phase Cell-Cycle Control Module in the Flowering Plant
- Multiple Roles and Interactions of and in Development of the Respiratory System
- UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex
- Epigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
- Modulating the Strength and Threshold of NOTCH Oncogenic Signals by
- Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer's Disease–Related Tau Phosphorylation Via PAR-1
- Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System
- ATXN2-CAG42 Sequesters PABPC1 into Insolubility and Induces FBXW8 in Cerebellum of Old Ataxic Knock-In Mice
- Cohesin Rings Devoid of Scc3 and Pds5 Maintain Their Stable Association with the DNA
- The MicroRNA Inhibits Calcium Signaling by Targeting the TIR-1/Sarm1 Adaptor Protein to Control Stochastic L/R Neuronal Asymmetry in
- Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength
- The Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Variation of Contributes to Dog Breed Skull Diversity
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



