-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEpigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
Meiosis is a specialized eukaryotic cell division that generates haploid gametes required for sexual reproduction. During meiosis, homologous chromosomes pair and undergo reciprocal genetic exchange, termed crossover (CO). Meiotic CO frequency varies along the physical length of chromosomes and is determined by hierarchical mechanisms, including epigenetic organization, for example methylation of the DNA and histones. Here we investigate the role of DNA methylation in determining patterns of CO frequency along Arabidopsis thaliana chromosomes. In A. thaliana the pericentromeric regions are repetitive, densely DNA methylated, and suppressed for both RNA polymerase-II transcription and CO frequency. DNA hypomethylated methyltransferase1 (met1) mutants show transcriptional reactivation of repetitive sequences in the pericentromeres, which we demonstrate is coupled to extensive remodeling of CO frequency. We observe elevated centromere-proximal COs in met1, coincident with pericentromeric decreases and distal increases. Importantly, total numbers of CO events are similar between wild type and met1, suggesting a role for interference and homeostasis in CO remodeling. To understand recombination distributions at a finer scale we generated CO frequency maps close to the telomere of chromosome 3 in wild type and demonstrate an elevated recombination topology in met1. Using a pollen-typing strategy we have identified an intergenic nucleosome-free CO hotspot 3a, and we demonstrate that it undergoes increased recombination activity in met1. We hypothesize that modulation of 3a activity is caused by CO remodeling driven by elevated centromeric COs. These data demonstrate how regional epigenetic organization can pattern recombination frequency along eukaryotic chromosomes.
Published in the journal: . PLoS Genet 8(8): e32767. doi:10.1371/journal.pgen.1002844
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002844Summary
Meiosis is a specialized eukaryotic cell division that generates haploid gametes required for sexual reproduction. During meiosis, homologous chromosomes pair and undergo reciprocal genetic exchange, termed crossover (CO). Meiotic CO frequency varies along the physical length of chromosomes and is determined by hierarchical mechanisms, including epigenetic organization, for example methylation of the DNA and histones. Here we investigate the role of DNA methylation in determining patterns of CO frequency along Arabidopsis thaliana chromosomes. In A. thaliana the pericentromeric regions are repetitive, densely DNA methylated, and suppressed for both RNA polymerase-II transcription and CO frequency. DNA hypomethylated methyltransferase1 (met1) mutants show transcriptional reactivation of repetitive sequences in the pericentromeres, which we demonstrate is coupled to extensive remodeling of CO frequency. We observe elevated centromere-proximal COs in met1, coincident with pericentromeric decreases and distal increases. Importantly, total numbers of CO events are similar between wild type and met1, suggesting a role for interference and homeostasis in CO remodeling. To understand recombination distributions at a finer scale we generated CO frequency maps close to the telomere of chromosome 3 in wild type and demonstrate an elevated recombination topology in met1. Using a pollen-typing strategy we have identified an intergenic nucleosome-free CO hotspot 3a, and we demonstrate that it undergoes increased recombination activity in met1. We hypothesize that modulation of 3a activity is caused by CO remodeling driven by elevated centromeric COs. These data demonstrate how regional epigenetic organization can pattern recombination frequency along eukaryotic chromosomes.
Introduction
During meiosis homologous chromosomes pair and undergo reciprocal exchange, to produce crossovers (COs). COs are initiated by SPO11-catalyzed DNA double strand breaks (DSBs), which are resected to generate single-stranded 3′ tails on either side of the break (ssDNA) [1]. The ssDNA can invade a non-sister chromatid to form an intermediate D-loop structure, which may proceed to form a double Holliday junction that can be resolved into a CO [1], [2], [3]. The D-loop can also participate in an alternative pathway to form non-crossovers (NCOs), which in Saccharomyces cerevisiae involves synthesis dependent strand annealing [1], [2], [3]. Concurrently with DSB generation a chromosome axis forms and physically connects the homologues with loops of chromatin projecting laterally [4], [5], [6]. DSBs arise on chromatin loops tethered to the axis, and changes to axis structure can dramatically alter recombination patterns [4], [6], [7]. A greater number of DSBs are generated than mature into COs, with the excess DSBs repaired as NCOs, some of which can be detected as gene conversions [8], [9]. COs occurring between homologous chromosomes can show distance-dependent interference causing them to be more widely spaced than expected by chance [9], [10]. For example, in A. thaliana 85–90% of COs form via the MSH4-dependent interfering pathway (type-I) and the remaining 10–15% form via the MUS81-dependent non-interfering pathway (type-II) [11], [12], [13], [14], [15]. Additional CO pathways must also exist in A. thaliana since residual COs or chiasmata have been observed in msh4 mus81 double mutants [11], [14]. A process related to interference, called homeostasis, maintains CO frequency when DSBs are reduced [16]. Interference and homeostasis cause CO number per chromosome to be distributed closer to a mean than expected from the Poisson distribution [13], [17]. Tight control of CO frequency is thought to be important because balanced homologue segregation at meiosis-I is dependent, in most organisms, on each pair of homologues having at least one CO [18].
CO frequency is variable along the length of A. thaliana chromosomes, for example the centromeres are CO suppressed, whereas gene-dense regions are active [19], [20], [21], [22]. A. thaliana chromosomes also display region-specific epigenetic modifications of DNA and histones that are associated with differential transcription [23], [24], [25], [26], [27], [28], [29], [30]. DNA cytosine methylation is an epigenetic modification that can be heritably maintained through DNA replication and in A. thaliana occurs in two major epigenomic contexts. First, the majority of DNA methylation overlaps with RNA polymerase II (Pol II) repressed repetitive sequences including transposons and also with histone H3K9me2, H3K27me1, H4K20me1 (me = methylation) [23], [24], [25], [26], [29], [30], [31], [32]. Repeats are DNA methylated in all sequence contexts (CG, CHG and CHH) and show a marked increase in density towards the centromeres [23], [24], [25], [29], [30] (Figure 1). In the second context, the open reading frames of Pol II transcribed genes contain CG methylation, coincident with overlapping peaks of histone H3K4me, me2, me3, H3K36me3, H3K56ac and H2Bub (ac = acetylation, ub = ubiquitination) [26], [27], [29], [30], [33], [34]. Epigenetic information is known to influence patterns of meiotic recombination. For example, in S.cerevisiae and mammals CO hotspots associate with ‘accessible’ chromatin modifications, including histone H3K4me3 [35], [36], [37], [38], [39], [40], [41], and DNA methylation can directly repress COs in Ascobolus immersus [42]. Here we investigate the role of DNA methylation in organizing patterns of meiotic recombination frequency in the A. thaliana genome.
Fig. 1. Epigenomic organisation and CO frequency in the A. thaliana genome. 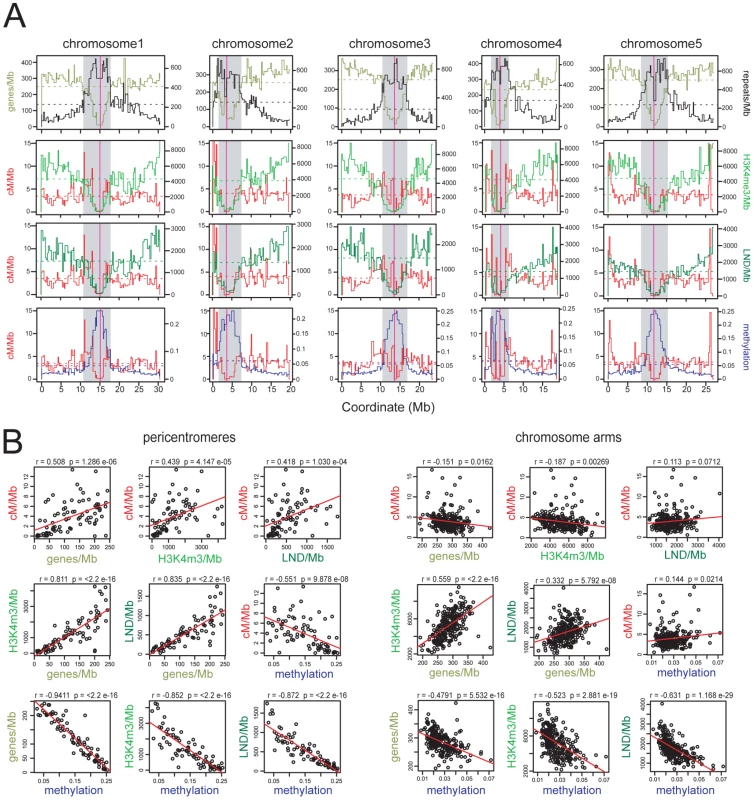
(A) Physical maps of A. thaliana chromosomes showing genes/Mb (olive green), repeats/Mb (black), cM/Mb (red), H3K3me3/Mb (light green), LND/Mb (dark green) and DNA methylation density (blue). Dotted horizontal lines indicate the means weighted by intermarker distance. Vertical magenta lines indicate the centromeres. Grey shaded areas indicate the pericentromeres. (B) Pairwise correlations between cM/Mb, genes/Mb, H3K4me3/Mb, LND/Mb and DNA methylation in either chromosome arms or pericentromeres. Pearson's correlation coefficients (r) and associated p-values (p) are shown and regression lines are plotted in red. See also Tables S1 and S2. Maintenance of CG DNA methylation in A. thaliana requires the cytosine methyltransferase METHYLTRANSFERASE1 (MET1) [43], [44], [45], [46]. A. thaliana met1 mutants show dramatic loss of DNA methylation and associated histone modifications, leading to increased Pol II transcription of repetitive sequences [25], [29], [30], [44], [45], [46], [47], [48]. Gene body DNA methylation is also lost in met1, though expression of these genes is maintained [25], [29], [30]. Self-fertilization and inbreeding of met1 mutants leads to stochastic generation of epialleles and transposon mobilization [44], [45], [46], [49], [50], [51], [52], [53], [54]. Epigenetic divergence is observed in within met1+/− segregating populations, even without met1 homozygosity, as plant haploid gametophytes undergo post-meiotic DNA replication and in met1 gametophytes this causes cytosine demethylation [44], [46], [52]. Here we demonstrate extensive remodeling of CO distributions in met1 mutants, with elevated centromere-proximal COs coupled to pericentromeric decreases and distal increases. Importantly total CO numbers are similar between wild type and met1, suggesting that interference and homeostasis may act to drive regional changes. We generate a fine-scale map of euchromatic recombination frequency close to the telomere of chromosome 3 and identify a novel, intergenic CO hotspot 3a. We observe an elevated recombination topology across this region in met1 and higher 3a CO frequency, consistent with remodeling modulating hotspot activity. Together this work reveals the importance of domains of epigenetic organization in determining chromosomal patterns of meiotic CO frequency.
Results
Epigenetic organization and CO frequency in the A. thaliana genome
Because CO frequency is decreased close to the A. thaliana centromeres we investigated its relationship with DNA methylation in these regions [19], [20], [21], [22]. To obtain a genome-wide map of CO frequency we analyzed published genotype data for 17 F2 populations, providing a total dataset of 55,497 COs [22], [55]. Genetic maps for individual populations were created using R/qtl and merged using MergeMap, which yielded map lengths comparable to those previously published (Table S1) [21], [22], [56], [57], [58], [59]. We then calculated recombination frequency (cM/Mb), gene, H3K4me3, LND (low nucleosome density), repeat and DNA methylation densities within marker intervals of the merged map. Meiosis-specific epigenomic maps are not currently available in A. thaliana, so bisulfite sequencing data (DNA methylation) and ChIP-chip data (H3K4me3 and LND) generated from somatic tissues were used [23], [27], [28] (Table S2).
We defined pericentromeres as the intervals flanking the genetically defined centromeres that showed gene densities lower than the chromosome average, and defined the remaining regions as chromosome arms (Figure 1 and Table S2) [19]. The pericentromeres contain fewer genes, higher repetitive DNA content and denser DNA methylation compared to the chromosome arms (averages for chromosome arms vs pericentromeres are 286.9 vs 123.6 genes/Mb, 153.4 vs 556.4 repeats/Mb, 0.027 vs 0.147 for methylation). Gene density is positively correlated with H3K4me3 and LND density in all regions, consistent with the known function of these chromatin features in promoting gene expression (Figure 1B) [27], [28]. Gene, H3K4me3 and LND density are negatively correlated with DNA methylation, most strongly in the pericentromeres, consistent with dense DNA methylation associating with Pol II silenced repeats (Figure 1B) [23], [24], [25], [29], [30]. Mean CO frequencies within the chromosome arms (3.95 cM/Mb) and pericentromeres (3.83 cM/Mb) were similar, though within the pericentromeres CO frequency was strongly elevated towards the region boundaries (Figure 1A), and showed positive correlations with genes/Mb (r = 0.508, p = 1.29×10−06), H3K4me3/Mb (r = 0.439, p = 4.15×10−05), LND/Mb (r = 0.418, p = 1.03×10−04) and a negative correlation with DNA methylation (r = −0.551, p = 9.88×10−08) (Figure 1B). In contrast, cM/Mb in the chromosome arms was weakly correlated with genes/Mb, H3K4me3/Mb, LND/Mb and methylation (Figure 1B). This indicates that pericentromeres represent chromosomal domains with distinct patterns of epigenetic information and CO frequency control relative to the chromosome arms. Given the negative correlation between DNA methylation and CO frequency within the pericentromeres we decided to test CO patterns in hypomethylated met1–3 mutants [23], [25], [29], [30].
Elevated centromeric CO frequency in met1–3
To measure COs in proximity of the centromeres in met1 we analyzed the segregation of polymorphic markers (Figure 2A). We backcrossed the null met1–3 allele from the Columbia (Col) accession into Landsberg erecta (Ler) for 8 generations, maintaining met1–3 as a heterozygote to limit epigenetic divergence. met1–3+/− Ler and met1–3+/− Col heterozygotes were crossed to generate F1 individuals homozygous for met1–3 and heterozygous for Col/Ler polymorphisms. To generate recombinant populations these F1 individuals were backcrossed as males to Col, as were wild type Col/Ler heterozygotes (Figure 2A). We designed insertion-deletion Col/Ler PCR markers to centromere proximal positions that show CO suppression and dense DNA methylation (Figure 2B and 2C). We observed significantly elevated centromere-proximal CO frequency in the mutant met1–3−/− population relative to wild type (1.21 cM/Mb vs 0.38 cM/Mb, pmod = 2.0×10−4) (Figure 2C). As expected wild type recombination rates within these densely DNA methylated regions were lower than the chromosome averages (Figure 2C and Table S1). These data demonstrate elevated centromere-proximal COs in met1–3−/−, correlating with extensive DNA demethylation and increased Pol II transcription previously observed in these regions [23], [25], [29], [30].
Fig. 2. Elevated centromeric crossovers in met1–3. 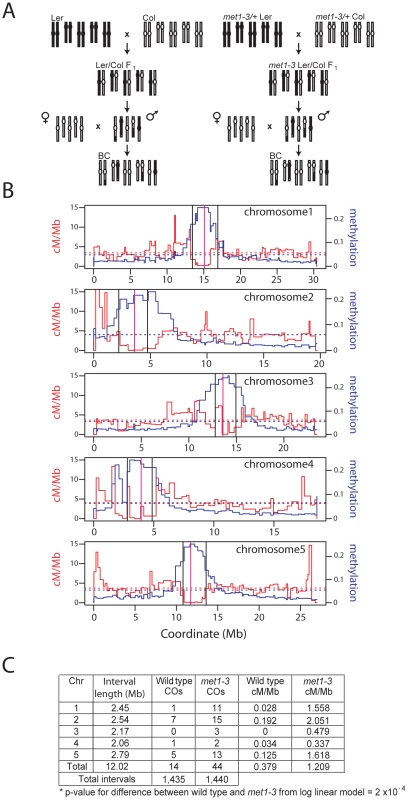
(A) Schematic diagram illustrating generation of wild type and met1–3−/− recombinant male backcross populations from Col and Ler homozygous parents. (B) Chromosome physical maps with overlaid cM/Mb (red) and DNA methylation (blue) plots; black vertical lines indicate the position of polymorphic Col/Ler markers tested for segregation frequency. Vertical magenta lines indicate centromeres. (C) Segregation data and centromeric CO measurements in wild type and met1–3−/− male backcross populations. Stochastic decrease of pericentromeric CO frequency in met1–3
We sought to test CO frequency in wild type and met1–3−/− across a wider pericentromeric interval. The FTL system uses segregation of heterozygous transgenes expressing distinct colors of fluorescent proteins in pollen to measure COs between insertion sites [60] (Figure 3). FTL segregation in the quartet1–2 (qrt1–2) mutant background, where sister pollen grains remain physically attached, allows tetrad analysis for male meioses [60] (Figure 3B and 3C). We used FTL lines located on chromosome 3 defining a 5.405 Mb interval that we call CEN3, which spans the centromere and includes the region previously measured in the backcross populations, in addition to flanking pericentromeric DNA (Figure 3A). CEN3 is repeat and methylation dense (650.8 repeats/Mb, 0.183 methylation) and gene-poor (75.1 genes/Mb) compared to the chromosome 3 averages (240.7 genes/Mb, 273.7 repeats/Mb, 0.056 methylation). In Col that has never been crossed to met1–3 (naïve wild type) CEN3 has a genetic distance of 11.04 cM, corresponding to 2.05 cM/Mb, compared to the 4.76 cM/Mb chromosome 3 male average (Figure 3D and Tables S1 and S3) [21]. Although CEN3 is relatively suppressed for COs, this interval shows increasing CO frequency towards its boundaries, correlating with higher gene densities and lower DNA methylation (Figure 3A).
Fig. 3. Decreased pericentromeric crossovers in met1–3. 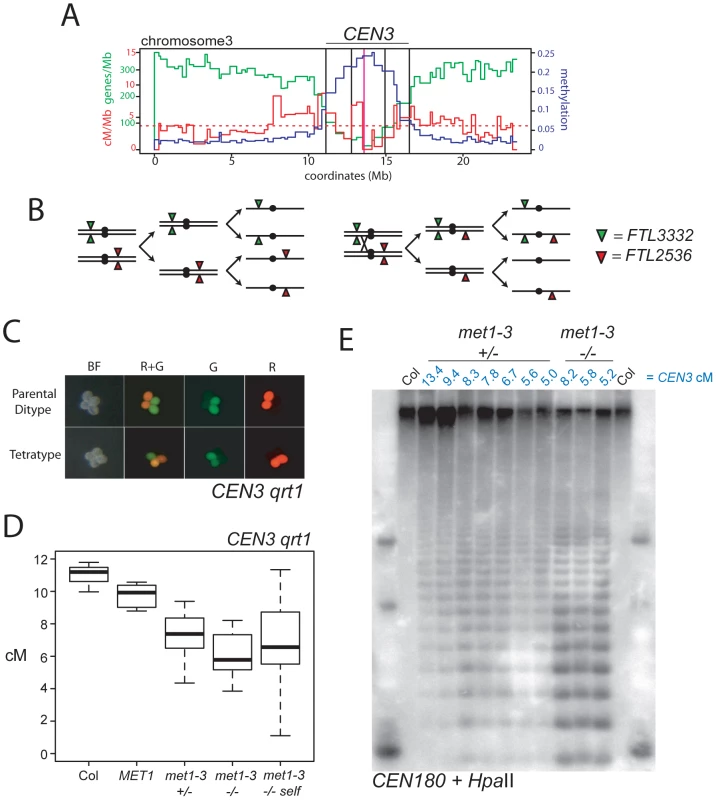
(A) Physical map of chromosome 3 with overlaid genes/Mb (green), cM/Mb (red) and DNA methylation (blue) plots. The dotted, horizontal red line indicates the cM/Mb weighted mean. Outer vertical black lines indicate the position of FTL transgene insertions that define CEN3. Inner vertical black lines indicate the position of centromeric markers analyzed in Figure 2. The vertical magenta line indicates the centromere. (B) Chromosomes heterozygous for trans-linked FTL332 (eYFP) and FTL2536 (DsRed) transgenes, which flank the centromere (black circle) segregating through meiosis-I and –II in the absence (left) or presence (right) of a CO within CEN3. (C) Fluorescence micrographs of qrt1–2 pollen showing patterns of inheritance associated with (tetratype) or without (parental ditype) a CO within CEN3. BF shows bight field illumination and R and G indicate red and green UV fluorescence. (D) CEN3 genetic map lengths for naïve wild type (Col), MET1, met1–3+/−, met1–3−/− segregants and self-fertilized met1–3−/− measured by qrt1–2−/− tetrad counting. (E) Southern blotting and hybridization analysis of CEN180 following digestion of genomic DNA using DNA methylation sensitive HpaII. DNA was prepared from CEN3 qrt1–2−/− individuals whose measured genetic distance in cM is indicated above the blot in blue in addition to their met1–3 genotype. See also Table S3. We self-fertilized CEN3/− − met1–3+/− qrt1–2−/− plants to generate populations segregating for met1–3 and measured CEN3 COs in MET1, met1–3+/− and met1–3−/− individuals. We observed significant decreases in CEN3 genetic distance in all groups relative to naïve wild type, with mean distances of MET1 9.76 cM (pt = 0.01), met1–3+/− 7.32 cM (pt = 4.31×10−5) and met1–3−/− 6.68 cM (pt = 0.002) (Figure 3D and Table S3). After self-fertilization met1–3−/− maintained a significantly decreased CEN3 mean genetic distance of 6.37 cM (pt = 0.001) (Figure 3D and Table S3). The met1–3+/− and met1–3−/− self-fertilized segregant groups also exhibited significantly greater variability in CO frequency compared to naïve wild type (F-test: met1–3+/− p = 0.0152 and met1–3−/− p = 4.32e-3) (Figure 3D and Table S3). Increased variance is consistent with stochastic epigenetic divergence observed in segregating met1 and ddm1 populations [44], [46], [50], [52], [61], [62], [63]. These data are consistent with increased centromere-proximal COs in met1–3−/− (met1–3−/− 1.21 cM/Mb vs wild type 0.38 cM/Mb) decreasing CO frequency in pericentromeric regions (met1–3−/− 1.24 cM/Mb vs wild type 2.05 cM/Mb), potentially via CO interference.
We investigated whether centromeric DNA methylation correlates with CEN3 genetic distance in this population. To analyze centromeric DNA methylation we used methyl-sensitive restriction digestion of genomic DNA with HpaII followed by Southern blotting and hybridization with the A. thaliana 180-bp satellite repeat CEN180 (Figure 3E) [48]. The 180-bp satellite repeats occur in tandem arrays of megabase length within centromeres and are densely DNA methylated in wild type [19], [48], [64]. In met1–3−/− mutants the satellite repeats lose methylation and are digested by HpaII, whereas wild type Col DNA is undigested (Figure 3E). We analyzed leaf DNA from met1–3+/− and met1–3−/− individuals for which we had measured CEN3 genetic distance. We observed that greater satellite demethylation was associated with decreased CEN3 recombination, though two met1–3+/− individuals (5.6 cM and 5.0 cM) deviated from this trend (Figure 3E). This may be explained by chromosome 3 being demethylated to a greater extent than other chromosomes in these lines. These data demonstrate decreased pericentromeric CO frequency in met1–3 mutants, coincident with DNA demethylation of the satellite repeats. This is consistent with CO interference from elevated centromere-proximal COs reducing events closer to the boundaries of CEN3.
Total genetic map length is similar between wild type and met1–3
Total CO numbers in A. thaliana do not follow the Poisson distribution, indicating homeostatic control [12], [13], [21], [57]. We therefore tested whether total genetic map length in met1–3−/− was different from wild type, given our observations that regional frequencies close to the centromeres were altered. To measure map length we genotyped 95 male backcross individuals, generated from wild type or met1–3−/− Col/Ler heterozygotes, for 35 Col/Ler SNPs spaced across the 5 chromosomes using KASPar technology (Figure 4A and Table S4) [65], [66]. Total CO numbers were not significantly different between wild type and met1–3−/− populations (pmod = 0.13) (Figure 4A and Table S4). Therefore, despite regional alterations in CO frequency, total genetic map length is similar between met1–3−/− and wild type.
Fig. 4. Total crossover numbers are similar between wild type and met1–3. 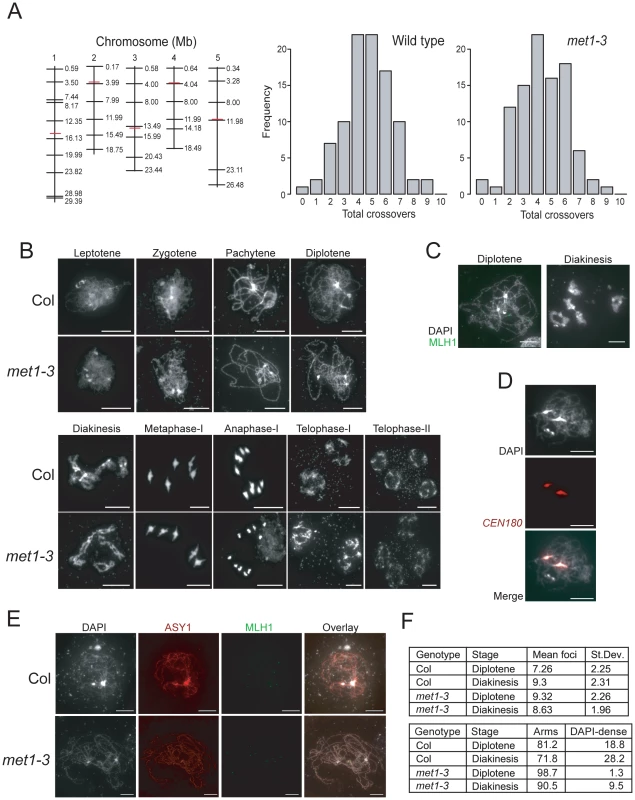
(A) Physical maps of chromosomes (vertical black lines) with KASPar marker (horizontal black lines) and centromere (horizontal red lines) positions indicated. Histograms showing the frequency of total CO numbers identified in male backcross individuals from either Col/Ler F1 (wild type) or met1–3−/− Col/Ler F1 (met1–3) parents. (B) Micrographs of DAPI-stained anther meiocytes showing the labeled stage of meiosis in Col and met1–3−/−. (C) Micrographs of diplotene and diakineses stage male meiocytes stained with DAPI (white) and immunostained for MLH1 (green). (D) Micrographs showing co-localisation of dense-DAPI staining and in situ hybridization with the CEN180 satellite repeat (red). (E) Micrographs of male meiocytes stained with DAPI (white) and immunostained with MLH1 (green) and the axis component ASY1 (red). (F) The upper table lists mean MLH1 foci numbers in wild type and met1–3−/− at diplotene or diakinesis with standard deviation (+/−). The lower table lists the relative proportions (%) of MLH1 foci localizing to chromosome arm regions (arms) vs densely-DAPI staining regions (DAPI-dense). All scale bars represent 10 µM. See also Table S4 and S5. To investigate meiotic progression in more detail we performed DAPI staining of anther meiocytes in wild type and met1–3−/−. The major cytological stages of meiosis in met1–3−/− lacked dramatic alterations to chromosome morphology or segregation (Figure 4B). At leptotene replicated chromosomes were present as thin threads, which condensed during zygotene, and became fully synapsed by pachytene (Figure 4B). At pachytene the centromeres, pericentromeres and nucleolar organizing regions (NORs) cluster into densely DAPI-staining regions, which remain evident in met1–3−/− [67] (Figure 4B). During diplotene desynapsis occurs and homologues begin to separate, which further condense during diakinesis, when chiasma connecting the homologues are evident (Figure 4B). At metaphase-I bivalents are maximally condensed with homologous centromeres segregating to opposite cell poles. Segregation forms cell dyads, each containing 5 homologues, which partially decondense at telophase-I (Figure 4B). The second meiotic division separates chromatids, which decondense to form haploid tetrads at telophase-II (Figure 4B). This analysis demonstrates that overall meiotic chromosome morphology and segregation are similar between wild type and met1–3−/−.
As an independent measure for CO numbers we immunostained wild type and met1–3−/− meiocytes for MLH1, which is a homolog of bacterial MutL DNA repair proteins and localizes to foci corresponding to type-I (interference sensitive) COs (Figure 4C and Table S5) [17]. MLH1 foci are first detected at pachytene and increase to maximal numbers during diplotene and diakinesis (Figure 4C) [17]. MLH1 foci are closely associated with the chromosomes, visualized by either DAPI-staining or immunostaining for the axis component ASY1 (Figure 4C and 4E) [68]. We counted MLH1 foci from diplotene and diakinesis stage meiocytes in wild type and met1–3−/−. At diplotene there were significantly more MLH1 foci in met1–3−/− relative to wild type (wild type mean = 7.26, met1–3−/− mean = 9.32, pmod = 9.1 e-4) (Figure 4F and Table S5), though by diakinesis MLH1 numbers were not significantly different (wild type mean = 9.32, met1–3−/− mean = 8.63, pmod = 0.39) (Figure 4F and Table S5). These data are consistent with total MLH1 foci numbers being similar between met1–3−/− and wild type, though maximal numbers may be reached slightly earlier in met1–3−/−.
Previous work demonstrated that MLH1 foci show differential localization on chromosome arms (77%) versus DAPI-dense regions (23%) at diakinesis [17]. We confirmed that these DAPI-dense regions contain the centromeres using fluorescent in situ hybridization for the CEN180 satellite repeats (Figure 4D). We scored MLH1 foci distributions in wild type Col and observed similar results at diplotene (81.2% arms vs 18.8% DAPI-dense regions) and diakinesis (71.8% arms vs 28.2% DAPI-dense regions) (Figure 4E). In contrast, there were significantly fewer MLH1 foci in the DAPI-dense regions in met1–3−/− at both diplotene (98.7% arms vs. 1.3% DAPI-regions, chi-square p = 2.2 e-16) and diakinesis (90.5% arms vs. 9.5% DAPI-regions, chi-square p = 6.0 e-4) (Figure 4E). Together we interpret these data as indicating that although overall MLH1 foci numbers are similar between wild type and met1–3−/−, there are significantly fewer foci in the DAPI-dense regions in met1–3−/−. As DAPI-dense regions contain the pericentromeres, we interpret reduced MLH1 foci in these regions as reflecting the reduced pericentromeric genetic distance we observe over CEN3 (Figure 3).
As we propose that CO interference mediates CO frequency remodeling in met1–3−/− we investigating whether interference occurred to a similar degree between wild type and met1–3−/−. To compare CO interference strength we calculated the average distance between pairs of COs identified from marker segregation occurring on the same chromosome (Double COs, DCOs) in the male backcross population described above (Table S4). The inter-CO distances and therefore the strength of CO interference were not significantly different between wild type and met1–3−/− (pw = 0.67) (Table S4). As an additional measure of CO control we tested our MLH1 foci data for deviation from the Poisson distribution, which may indicate the action of CO interference [13], [69]. Using a goodness-of-fit test we observed significant deviations in all cases, with more MLH1 counts close to the mean than expected from the Poisson distribution (Table S5). This is consistent with interference acting in both wild type and met1–3−/−, supporting the idea that CO interference could contribute to the observed CO frequency remodeling in met1–3−/−. Together these data demonstrate that despite alteration of regional CO frequencies, total CO numbers and interference strength are similar between wild type and met1–3−/−. This is consistent with CO interference mediating inhibition of pericentromeric COs in met1–3−/−, due to elevated centromeric COs.
Elevated euchromatic CO frequency in met1–3
Given that we observed remodeling of centromere-associated CO frequencies in met1–3−/−, we next measured genetic distance in the euchromatic chromosome arms. The 1.85 Mb FTL I1b interval is relatively gene dense (310.8 genes/Mb) and repeat and methylation poor (84.3 repeats/Mb, 0.022 methylation) compared to the chromosome 1 averages (246.8 genes/Mb, 233.5 repeats/Mb, 0.048 methylation) (Figure 5A). I1b in naïve wild type measures 8.16 cM, and has a recombination rate in male meiosis of 4.41 cM/Mb, close to the chromosome 1 average (4.88 cM/Mb) (Figure 5D and Tables S1 and S6) [21]. In a population segregating for I1b and met1–3 we observed that met1–3−/− individuals showed significantly increased genetic distance of 11.00 cM (5.95 cM/Mb) compared to naïve wildtype, MET1 and met1–3+/− (pt = 0.001, 0.03 and 0.08 respectively) (Figure 5D and Table S6). Elevated CO frequencies were stable when met1–3−/− plants were self-fertilized and measured in the next generation (Figure 5D and Table S6). Mean I1b CO frequencies of met1–3+/− (9.07 cM) segregants were higher than naïve wild type, though not significantly (pt = 0.27). The met1–3+/−, met1–3−/− and met1–3−/− self-fertilized groups also had significantly higher variance relative to naïve wild type, consistent with epigenetic divergence (F-test: met1–3+/− p = 0.011, met1–3−/− p = 0.0447 and met1–3−/− self-fertilized p = 0.0445) (Figure 5D and Table S6).
Fig. 5. Elevated euchromatic crossovers in met1–3. 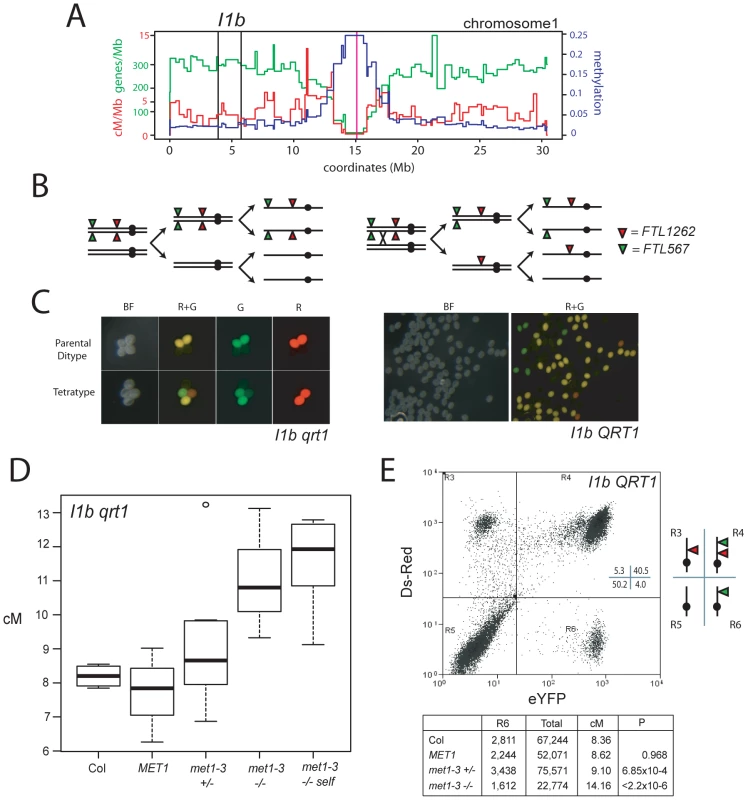
(A) Physical map of chromosome 1 with overlaid gene/Mb (green), DNA methylation (blue) and cM/Mb (red) plots. Black vertical lines indicate the I1b transgene insertions and the magenta vertical line indicates the centromere. (B) Schematic diagram showing homologous chromosomes (black lines) heterozygous for cis-linked FTL567 (eYFP) and FTL1262 (RFP) transgenes segregating through meiosis in the absence or presence of a CO. (C) Fluorescence micrographs showing qrt1–2−/− or QRT1 pollen from I1b cis-heterozygotes. (D) I1b genetic map length for naïve wild type (Col), MET1, met1–3+/− and met1–3−/− segregants and self-fertilized met1–3−/− (met1-self) measured by qrt1–2−/− tetrad counting. (E) I1b genetic map length for naïve wild type (Col) and MET1, met1–3+/− and met1–3−/− segregants measured by flow cytometry of individual pollen grains. A representative flow cytometry histogram from an I1b cis-heterozygote together with gate quadrant R6 counts, adjusted total pollen counts and cM. See also Table S6. We confirmed these observations after backcrossing I1bc qrt1–2−/− to either Col or met1–3−/− to complement with QRT1 and used flow cytometry to measure the fluorescence of individual pollen grains (Figure 5C, 5E and Figure S1). The I1b FTL transgenes are cis-linked, meaning pollen from I1b/−− heterozygotes expressing red-alone or yellow-alone represent single CO events (Figure 5E and Figure S1). The recombination rate is calculated by the ratio of yellow-alone pollen grains to an adjusted total (Text S1 and Figure S1). In naïve wild type this technique measured an I1b genetic distance (8.16 cM) close to that observed from qrt1–2−/− tetrad scoring (8.20 cM) (Figure 5D and 5E). Both met1–3+/− and met1–3−/− plants showed significantly increased genetic distances of 9.10 cM (pt = 6.85×10−4) and 14.16 cM (pt<2.20×10−16) respectively, whereas MET1 segregants were not significantly different from naïve wild type (Figure 5D and 5E). These results confirm that met1–3 CO frequency is elevated within I1b.
To test the effect of met1–3 on a second euchromatic interval we used a seed reporter system (Col3–4/20, hereafter referred to as 420) [70] (Figure 6A, 6B and 6C). The 420 interval is defined by transgene insertions on chromosome 3 expressing GFP or RFP in seed from the NapA promoter [70] (Figure 6A). 420 spans 5.105 Mb and is relatively gene dense (311.9 genes/Mb) and repeat and methylation poor (71.5 transposons/Mb, 0.022 methylation) compared to the chromosome 3 averages (240.7 genes/Mb, 273.7 repeats/Mb, 0.056 methylation). In naïve, self-fertilised Col 420 has a mean genetic distance of 19.71 cM and recombination rate of 3.86 cM/Mb (chromosome 3 average 3.73 cM/Mb) (Figure 6D and Tables S1 and S7) [21]. We observed significant increases in mean 420 cM in met1–3+/− segregants to 23.32 cM (pt = 0.004), relative to naïve wild type (Figure 6D and Table S7). These data confirm that CO frequency is elevated in the distal chromosome arms in met1–3+/− populations.
Fig. 6. Elevated euchromatic recombination topology in wild type and met1–3. 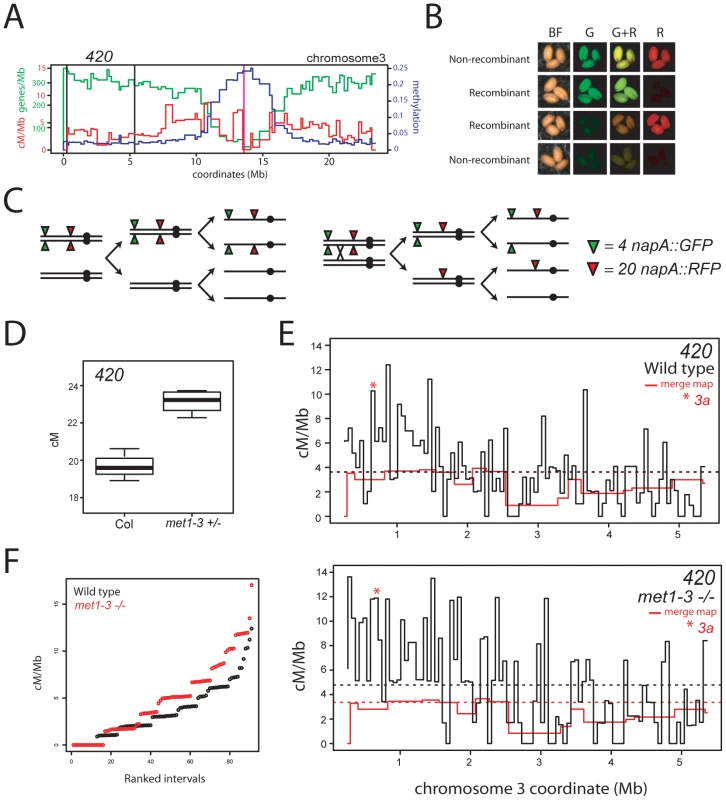
(A) Physical map of chromosome 3 with overlaid gene/Mb (green), DNA methylation (blue) and cM/Mb (red) plots. Black vertical lines indicate the positions of napA transgene insertions that define the 420 interval and the vertical magenta line indicates the centromere. (B) Fluorescence micrographs of seed expressing different combinations of napA transgenes. (C) Segregation diagram showing cis-heterozygous arrangement of 420 napA lines. (D) 420 genetic distance measured in naïve wild type (Col) and met1–3+/− segregants. (E) Black lines indicate recombination frequency (cM/Mb) maps of the 420 interval in wild type or met1–3−/− with horizontal dotted lines indicating weighted means. Red lines represent merged map recombination frequency data for the 420 interval. The red star indicates the interval containing the 3a CO hotspot. (F) Plots showing cumulative recombination value (cM/Mb) of ranked 420 mapping intervals in wild type (black) and met1–3−/− (red). See also Tables S7, S8 and S9. Elevated euchromatic recombination topology in met1–3
To compare wild type and met1–3 CO distributions at higher resolution we generated recombination frequency maps within the 420 interval. 420/−− Col/Ler F1 hybrids, that were wild type or met1–3−/−, were backcrossed to Col as males and seed expressing red or green fluorescence alone were selected to identify recombinants within the 420 interval (Figure 6B and 6C). The 420 interval is strongly heterochiasmic with significantly higher male CO frequency (4.82 cM/Mb) than female (2.57 cM/Mb) (pt = <2.20×10−8) (Table S8) [70], [71]. Male and female 420 genetic distances are reduced in Col/Ler heterozygotes compared to Col/Col homozygotes, potentially due to inhibition of recombination by polymorphisms (Table S8) [72]. CO frequency within 420 is significantly elevated by met1–3−/− in both Col/Col and Col/Ler backgrounds (Figure 6D, Tables S7 and S8), indicating that euchromatic remodeling is not dependent upon polymorphism levels.
We used an Illumina BeadArray to genotype 91 internal Col/Ler SNPs (average interval 56,067 bp) in 337 wild type and 268 met1–3−/− 420 recombinants (Table S9). Pronounced heterogeneity in cM/Mb was observed between intervals (range = 0–17.03 cM/Mb) with overall CO rate elevated in the met1–3−/− map relative to wild type (Figure 6E, 6F and Table S9). Recombination frequency topology was similar in both maps and showed significant correlation (r = 0.513, p = 1.95×10−7), and this correlation was stronger when comparisons were made over 255 Kb intervals (r = 0.789, p = 2.07×10−8). Elevated CO rates were observed towards the telomere in both populations (correlation between interval start coordinate and cM/Mb: wild type r = −0.496, p = 5.82×10−7; met1–3−/− r = −0.533, p = 5.21×10−7), consistent with higher telomeric CO rates observed in A. thaliana male meiosis relative to female (Figure 6A and Table S9) [20], [21], [57], [71], [73], [74], [75]. No significant correlations were detected between wild type cM/Mb and gene (r = −0.008, p = 0.94) or repeat (r = 0.005, p = 0.96) density at this scale. The similarity in overall recombination topology between wild type and met1–3−/− maps is consistent with remodeling acting to elevate existing CO patterns within 420.
Elevated CO hotspot 3a activity in met1–3
Mammalian and fungal meiotic recombination hotspots are typically ∼1–2 kb and display higher DSB and CO frequencies than surrounding regions [40], [41], [76], [77]. To identify CO hotspots within 420 we designed dCAPs PCR markers to define CO distributions at finer-scale within an active interval (interval 8, 10.37 cM/Mb) (Figure 7A) [78]. This defined a 6,708 bp sub-interval with a CO frequency of 76.15 cM/Mb (Figure 7A). To identify CO locations within this interval we used a ‘pollen-typing’ strategy, whereby nested allele-specific PCR primers are used to amplify CO molecules from Col/Ler F1 pollen genomic DNA (Text S1 and Figure S2) [79], [80]. We amplified and quantified parental versus CO molecules within a subinterval we call 3a (Figure 7B, 7C and Table S10). In naïve wild type 3a has a genetic distance of 0.164 cM (S.D. = 0.0171) and a CO rate of 28.24 cM/Mb (S.D. = 2.94) (Figure 7B, 7C and Table S10). We amplified single CO molecules and genotyped for internal Col/Ler polymorphisms to identify CO locations. Within the 3a amplicon we observe a complex distribution of CO frequency, with three distinct CO hotspots, each separated by at least one marker interval with 0 CO (hotspot #1 : 634109–636119 bp; hotspot #2 : 636199–638483 bp; hotspot #3 : 638687–639664 bp) (Figure 7B and Table S10). The hotspot peaks overlap with low nucleosome density regions located at the 5′ - and 3′-ends of a pair of convergently transcribed genes At3g02880 and At3g02885 (Figure 7B) [28]. The central hotspot has a width of 2,284 bp and a peak activity of 80.81 cM/Mb (Figure 7B, 7C and Table S10), which is 17 fold greater than the chromosome 3 male average (4.76 cM/Mb) (Table S1) [21]. We used epigenomic annotation of this region to investigate the presence of chromatin features associated with 3a (Figure 7B). The genes associated with 3a are Pol II transcribed and At3g02875, At3g02880 and At3g02890 posses H3K4me, me2 and me3 within their open reading frames (Figure 7B and 7D) [25], [27]. Low levels of DNA methylation are detected within 3a, though At3g02890 shows gene-body DNA methylation, consistent with active transcription (Figure 7B) [23].
Fig. 7. Elevated crossover hotspot 3a1 activity in met1–3. 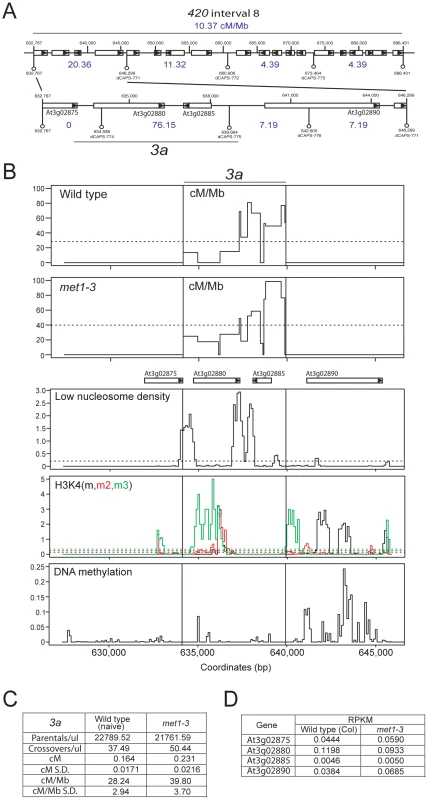
(A) CO frequency distributions (cM/Mb, blue) within 420 map interval 8 measured by dCAPs PCR marker segregation (white bars represent genes, with triangles indicating strand). (B) Plots of cM/Mb for the 3a CO hotspot shown for wild type and met1–3−/−. Vertical black lines indicate the position of the inner PCR primers used to amplify 3a. Epigenomic annotation of the 3a region with plots displaying low nucleosome density, histone H3K4m (black), H3K4m2 (red) H3K4m3 (green) and DNA methylation densities. (C) Table summarizing quantification of 3a parental and CO molecule amplifications from pollen genomic DNA and calculation of cM, cM/Mb and associated standard deviations (S.D.). (D) RNA-seq RPKM (total counts mapping to gene/length of gene×total mapped reads, multiplied by 106) for 3a associated genes in wild type (Col) and met1–3. See also Table S10. We next tested met1–3−/− Col/Ler F1 pollen genomic DNA and observed a significant increase in 3a CO frequency to 39.79 cM/Mb (S.D. = 3.70) compared to wild type 28.24 cM/Mb (S.D. = 2.94) (pt = 5.66×10−5) (Figure 7B, 7C and Table S10). Although hotspots locations are similar between wild type and met1–3−/− the relative proportions of COs observed between the three hotspots are significantly different (Figure 7B and Table S10). Hotspot #1 shows significantly more COs (chi-square p = 0.037), hotspot #2 showed significantly less COs (chi-square p = 0.011), whereas hotspot #3 showed no significant difference (chi-square p = 0.560). This demonstrates that although the 3a region has a significantly elevated overall CO frequency in met1–3−/−, the individual hotspots within this region respond differently. This may indicate compensatory interactions, related to observations in S.cerevisae where changes in local DSB frequency can alter DSB activity in adjacent regions [81], [82], [83], [84], [85], [86]. Importantly, the genes associated with 3a do not show significant expression changes in met1–3−/− relative to wild type in floral tissue, indicating that local Pol II accessibility is unlikely to be altered (Figure 7D) [25]. This is consistent with elevated 3a hotspot activity in met1–3−/− being mediated via remodeling driven by increased centromere-proximal COs.
Discussion
CO frequency is highly variable within the genomes of eukaryotes and local rates are determined by hierarchically interacting mechanisms. Here we demonstrate that domains of epigenetic information, specifically heterochromatic DNA methylation, are important for determining chromosomal patterns of CO frequency in A. thaliana. Wild type COs are less frequent in densely DNA methylated, transcriptionally silent regions close to the A. thaliana centromeres. These regions show dramatic elevations in Pol II transcription in met1–3−/− [23], [25], [29], [30]. We speculate that SPO11 accessibility similarly increases in met1–3−/−, leading to elevated DSBs and COs in the centromeric regions. Immunohistochemistry in A. thaliana indicates that SPO11 recruitment to the chromosome and the formation of DSBs, as indicated by γH2A.X foci, are temporally distinct [68]. This may reflect activation of the DSB machinery during axis maturation and tethering of chromatin loops [4], [6], [68]. Hence, it will be important to determine the dynamics of axis maturation to fully understand the changes in CO frequency observed in met1−/−. It is also possible that additional steps in the recombination pathway are sensitive to chromatin state. For example, if interhomolog strand invasion mediated by the recombinase DMC1 were inhibited by DNA methylation, this might lead to increased use of the homologous centromeric region as a repair template in met1−/−. Additionally, SPO11 is recruited to DNA following pre-meiotic S-phase and heterochromatin replicates later than euchromatin in A. thaliana mitotic cells [87], [88], [89]. Therefore, if heterochromatin also replicates earlier in met1–3−/− meiotic S-phase, SPO11 recruitment close to the centromeres may also advance, and thus altered temporal progression could contribute to CO remodeling. Hence, a complete understanding of the changes in CO frequency in met1–3−/− will require future study of many aspects of the meiotic recombination mechanism.
COs frequency and distribution are finely controlled. For example, the CO interference pathway inhibits the formation of adjacent CO events in a distance-dependent manner. In Caenorhabditis elegans strong interference leads to one CO per bivalent, independent of the physical length of the chromosome [90]. In A. thaliana the majority (85–90%) of COs (type-I) are derived from an interference-sensitive pathway, while the remaining events (type-II) are distributed randomly. In met1–3−/− we observe an increase in centromere-proximal COs, coupled to pericentromeric decreases and distal euchromatic increases, though total CO numbers are similar to wild type. As DNA methylation is most dramatically lost in the centromeric regions, we hypothesize that increases in recombination in these regions drive CO frequency remodeling. Specifically, increases in met1–3−/− centromeric COs would inhibit adjacent events in the pericentromeric regions via CO interference. In addition to interference, COs are known to be controlled by a homeostatic pathway. For example, reductions of DSB frequency in S.cerevisiae do not lead to proportional reductions in CO frequency, indicating compensatory mechanisms that maintain CO numbers close to a mean [16]. We hypothesize that increases in distal CO frequency in met1–3−/− arise as a consequence of related homeostatic mechanisms maintaining total CO numbers, at the expense of the pericentromeric regions. Extensive data in S.cerevisiae demonstrate that DSB frequency can also be influenced by changes in DSB activity in adjacent regions, over distances up to 60 kb [81], [82], [83], [84], [85], [86]. Similar effects could also contribute to the observed changes in met1–3−/− CO frequencies, driven by elevated DSB frequency in hypomethylated regions. Therefore, changes in met1–3−/− recombination frequency could be caused by both additional and redistributed DSBs. Although, DNA methylation, gene density and gene-associated chromatin strongly correlate with CO frequency in the pericentromeres, this is not the case in the chromosome arms. Other levels of meiotic chromosome organization may be dominant in the distal chromosome arms, for example the meiotic axis [4], [6], [7], [91]. However, it is also possible that loss of DNA methylation from gene bodies or local repeats contributes to changes in met1–3−/− CO frequency in the chromosome arms [23], [25], [29], [30].
Our 420 genetic maps provide evidence of pronounced heterogeneity of CO rate within A. thaliana gene-rich euchromatin. We identify a novel CO hotspot 3a within this region, which overlaps with intergenic regions of low nucleosome density. Although our hotspot comparisons are made with mitotic epigenomic datasets, in yeast and mammals the majority of low nucleosome density regions are similar between meiotic and mitotic cells [92], [93], [94]. The 3a hotspot shows elevated activity in met1–3−/−, though without local change in Pol II transcription. Elevated 3a activity is consistent with CO remodeling driven by increased centromere-proximal COs in met1–3−/−. The 3a hotspot shares many similarities with DSB hotspots defined in S.cerevisiae, which occur at LNDs with high SPO11 accessibility and active epigenetic modifications including H3K4me3 [37], [40], [41], [95]. However, low nucleosome density regions and H3K4 methylation are shared between 3a and many genes. Therefore, we predict that these features are necessary but not sufficient for hotspot activity. Specifically, regional factors such as axis structure or proximity to the telomere may predispose locally permissive chromatin to undergo CO. In humans and mice the PRDM9 zinc-finger H3K4 histone methyltransferase positions CO hotspots to specific cis-sequences [36], [96], [97], [98], [99], [100], [101]. As PRDM9 has yet to be identified outside of animals, CO hotspots in yeast and plants may represent a more ancestral pattern within eukaryotes [102]. Although the logic of epigenetic control is conserved throughout the eukaryotes, the distributions and uses of specific chromatin marks can vary. As meiosis originated early during eukaryotic evolution it will be interesting to determine similarities in hotspot specification and the relative contributions of epigenetic information to control of CO frequency within distinct lineages. Together our data demonstrates how epigenetic organization contributes to the hierarchy of CO control mechanisms in plant genomes.
Note added in proof: Decreased pericentromeric and elevated euchromatic CO frequencies have been observed in ddm1 and met1 mutant backgrounds, consistent with our observations [103], [104].
Materials and Methods
Statistical methods
The R Statistical Language was used for analysis and graphs [105]. Correlations were performed using Pearson's product moment correlation. Comparisons between groups were made using t-tests (pt) or, in the case of inter-CO distances, the Wilcoxon-rank sum test (pw). Comparisons between proportions were made using chi-square tests. Comparisons of variance between groups were made using F-tests. Using glm, a model was fitted to the counts in Figure 2C including the effects of genotype and chromosomes and with the number of plants and chromosome lengths as offsets. Backward elimination was used to arrive at a parsimonious model, which included the effect of genotype and chromosomes 3 and 4. The p-value for genotype from this final model is given in Figure 2C. The R function glm was used to fit a quasi-Poisson model to the data presented in Tables S4 and S5, using genotype as the predictor. The p-value (pmod) for genotype is presented in the tables. The fit of MLH1 count data to the Poisson distribution was performed using the R goodfit function within the vcd package.
Plant materials and growth conditions
All plants were cultivated on commercial soil and grown in controlled environment chambers at 20°C, 60% humidity with a long day photoperiod (16 hours light) with a light intensity of 150 µmols.
Pollen tetrad and seed fluorescent scoring
Pollen tetrad and seed fluorescence were assayed as described [60], [70]. For a detailed discussion of pollen flow cytometry see Text S1 and Figure S1.
PCR and bead array genotyping
Genomic DNA was extracted from leaves using the CTAB method and genotyped using either PCR, an Illumina Beadarray or KASPar technology. Pollen genomic DNA was extracted as described [79]. For a detailed discussion of pollen-typing experiments see Text S1, Figure S2 and Table S11.
Immunocytology
Meiotic cells were analyzed from staged anthers by immunostaining as described [17].
Supporting Information
Zdroje
1. KeeneySNealeMJ 2006 Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans 34 523 525
2. AllersTLichtenM 2001 Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106 47 57
3. SchwachaAKlecknerN 1994 Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76 51 63
4. KlecknerN 2006 Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115 175 194
5. PadmoreRCaoLKlecknerN 1991 Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66 1239 1256
6. PanizzaSMendozaMABerlingerMHuangLNicolasA 2011 Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146 372 383
7. MetsDGMeyerBJ 2009 Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139 73 86
8. BaudatFde MassyB 2007 Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res 15 565 577
9. YoudsJLBoultonSJ 2011 The choice in meiosis - defining the factors that influence crossover or non-crossover formation. J Cell Sci 124 501 513
10. BerchowitzLECopenhaverGP 2010 Genetic interference: don't stand so close to me. Curr Genomics 11 91 102
11. BerchowitzLEFrancisKEBeyALCopenhaverGP 2007 The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet 3 e132 doi:10.1371/journal.pgen.0030132
12. CopenhaverGPHousworthEAStahlFW 2002 Crossover interference in Arabidopsis. Genetics 160 1631 1639
13. HigginsJDArmstrongSJFranklinFCJonesGH 2004 The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev 18 2557 2570
14. HigginsJDBucklingEFFranklinFCJonesGH 2008 Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J 54 152 162
15. MercierRJolivetSVezonDHuppeEChelyshevaL 2005 Two meiotic crossover classes cohabit in Arabidopsis: one is dependent on MER3,whereas the other one is not. Curr Biol 15 692 701
16. MartiniEDiazRLHunterNKeeneyS 2006 Crossover homeostasis in yeast meiosis. Cell 126 285 295
17. ChelyshevaLGrandontLVrielynckNle GuinSMercierR 2010 An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet Genome Res 129 143 153
18. GrelonMVezonDGendrotGPelletierG 2001 AtSPO11-1 is necessary for efficient meiotic recombination in plants. Embo J 20 589 600
19. CopenhaverGPNickelKKuromoriTBenitoMIKaulS 1999 Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286 2468 2474
20. DrouaudJMercierRChelyshevaLBerardAFalqueM 2007 Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet 3 e106 doi:10.1371/journal.pgen.0030106
21. GirautLFalqueMDrouaudJPereiraLMartinOC 2011 Genome-Wide Crossover Distribution in Arabidopsis thaliana Meiosis Reveals Sex-Specific Patterns along Chromosomes. PLoS Genet 7 e1002354 doi:10.1371/journal.pgen.1002354
22. SalomePABombliesKFitzJLaitinenRAWarthmannN 2011 The recombination landscape in Arabidopsis thaliana F(2) populations. Heredity
23. CokusSJFengSZhangXChenZMerrimanB 2008 Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452 215 219
24. LippmanZGendrelAVBlackMVaughnMWDedhiaN 2004 Role of transposable elements in heterochromatin and epigenetic control. Nature 430 471 476
25. ListerRO'MalleyRCTonti-FilippiniJGregoryBDBerryCC 2008 Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133 523 536
26. RoudierFAhmedIBerardCSarazinAMary-HuardT 2011 Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 30 1928 1938
27. ZhangXBernatavichuteYVCokusSPellegriniMJacobsenSE 2009 Genome-wide analysis of mono-, di - and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10 R62
28. ZhangXClarenzOCokusSBernatavichuteYVPellegriniM 2007 Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5 e129 doi:10.1371/journal.pbio.0050129
29. ZhangXYazakiJSundaresanACokusSChanSW 2006 Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126 1189 1201
30. ZilbermanDGehringMTranRKBallingerTHenikoffS 2007 Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39 61 69
31. BernatavichuteYVZhangXCokusSPellegriniMJacobsenSE 2008 Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE 3 e3156 doi:10.1371/journal.pone.0003156
32. JacobYStroudHLeblancCFengSZhuoL 2010 Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 466 987 991
33. GuoLYuYLawJAZhangX 2010 SET DOMAIN GROUP2 is the major histone H3 lysine [corrected] 4 trimethyltransferase in Arabidopsis. Proc Natl Acad Sci U S A 107 18557 18562
34. TranRKHenikoffJGZilbermanDDittRFJacobsenSE 2005 DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr Biol 15 154 159
35. BerchowitzLEHanlonSELiebJDCopenhaverGP 2009 A positive but complex association between meiotic double-strand break hotspots and open chromatin in Saccharomyces cerevisiae. Genome Res 19 2245 2257
36. BergILNeumannRLamKWSarbajnaSOdenthal-HesseL 2010 PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet 42 859 863
37. BordeVRobineNLinWBonfilsSGeliV 2009 Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J 28 99 111
38. BuardJBarthesPGreyCde MassyB 2009 Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J 28 2616 2624
39. GreyCBarthesPChauveau-Le FriecGLangaFBaudatF 2011 Mouse PRDM9 DNA-Binding Specificity Determines Sites of Histone H3 Lysine 4 Trimethylation for Initiation of Meiotic Recombination. PLoS Biol 9 e1001176 doi:10.1371/journal.pbio.1001176
40. PanJSasakiMKniewelRMurakamiHBlitzblauHG 2011 A hierarchical combination of factors shapes the genome-wide topology of yeast meiotic recombination initiation. Cell 144 719 731
41. WuT-CLichtenM 1994 Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263 515 518
42. MaloiselLRossignolJL 1998 Suppression of crossing-over by DNA methylation in Ascobolus. Genes Dev 12 1381 1389
43. GollMGBestorTH 2005 Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74 481 514
44. KankelMWRamseyDEStokesTLFlowersSKHaagJR 2003 Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163 1109 1122
45. RonemusMJGalbiatiMTicknorCChenJDellaportaSL 1996 Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273 654 657
46. SazeHMittelsten ScheidOPaszkowskiJ 2003 Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34 65 69
47. TariqMSazeHProbstAVLichotaJHabuY 2003 Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci U S A 100 8823 8827
48. VongsAKakutaniTMartienssenRARichardsEJ 1993 Arabidopsis thaliana DNA methylation mutants. Science 260 1926 1928
49. JacobsenSEMeyerowitzEM 1997 Hypermethylated SUPERMAN epigenetic alleles in arabidopsis. Science 277 1100 1103
50. MathieuOReindersJCaikovskiMSmathajittCPaszkowskiJ 2007 Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130 851 862
51. MiuraAYonebayashiSWatanabeKToyamaTShimadaH 2001 Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411 212 214
52. ReindersJWulffBBMirouzeMMari-OrdonezADappM 2009 Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev 23 939 950
53. SazeHKakutaniT 2007 Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. Embo J 26 3641 3652
54. SoppeWJJacobsenSEAlonso-BlancoCJacksonJPKakutaniT 2000 The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6 791 802
55. SalomePABombliesKLaitinenRAYantLMottR 2011 Genetic Architecture of Flowering-Time Variation in Arabidopsis thaliana. Genetics 188 421 433
56. BromanKWWuHSenSChurchillGA 2003 R/qtl: QTL mapping in experimental crosses. Bioinformatics 19 889 890
57. CopenhaverGPBrowneWEPreussD 1998 Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc Natl Acad Sci U S A 95 247 252
58. ListerCDeanC 1993 Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant Journal 4 745 750
59. WuYCloseTJLonardiS 2008 On the accurate construction of consensus genetic maps. Comput Syst Bioinformatics Conf 7 285 296
60. FrancisKELamSYHarrisonBDBeyALBerchowitzLE 2007 Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Natl Acad Sci U S A 104 3913 3918
61. ChenMHaMLackeyEWangJChenZJ 2008 RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics 178 1845 1858
62. JohannesFPorcherETeixeiraFKSaliba-ColombaniVSimonM 2009 Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet 5 e1000530 doi:10.1371/journal.pgen.1000530
63. TeixeiraFKHerediaFSarazinARoudierFBoccaraM 2009 A role for RNAi in the selective correction of DNA methylation defects. Science 323 1600 1604
64. FranszPFArmstrongSde JongJHParnellLDvan DrunenC 2000 Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100 367 376
65. CuppenE 2007 Genotyping by allele-specific amplification (KASPar). CSH Protocols pdb.prot4841
66. WijnkerEvan DunKde SnooCBLeliveltCLKeurentjesJJ 2012 Reverse breeding in Arabidopsis thaliana generates homozygous parental lines from a heterozygous plant. Nat Genet 44 467 470
67. FranszPArmstrongSAlonso-BlancoCFischerTCTorres-RuizRA 1998 Cytogenetics for the model system Arabidopsis thaliana. Plant J 13 867 876
68. Sanchez-MoranESantosJLJonesGHFranklinFC 2007 ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev 21 2220 2233
69. LhuissierFGOffenbergHHWittichPEVischerNOHeytingC 2007 The mismatch repair protein MLH1 marks a subset of strongly interfering crossovers in tomato. Plant Cell 19 862 876
70. Melamed-BessudoCYehudaEStuitjeARLevyAA 2005 A new seed-based assay for meiotic recombination in Arabidopsis thaliana. Plant J 43 458 466
71. PecinkaAFangWRehmsmeierMLevyAAMittelsten ScheidO 2011 Polyploidization increases meiotic recombination frequency in Arabidopsis. BMC Biol 9 24
72. BortsRHHaberJE 1987 Meiotic recombination in yeast: alteration by multiple heterozygosities. Science 237 1459 1465
73. ArmstrongSJJonesGH 2001 Female meiosis in wild-type Arabidopsis thaliana and in two meiotic mutants. Sex Plant Reprod 13 177 183
74. BarthSMelchingerAEDevezi-SavulaBLubberstedtT 2001 Influence of genetic background and heterozygosity on meiotic recombination in Arabidopsis thaliana. Genome 44 971 978
75. VizirIYKorolAB 1990 Sex difference in recombination frequency in Arabidopsis. Heredity 65 379 383
76. JeffreysAJKauppiLNeumannR 2001 Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet 29 217 222
77. JeffreysAJNeumannR 2005 Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum Mol Genet 14 2277 2287
78. NeffMMNeffJDChoryJPepperAE 1998 dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J 14 387 392
79. DrouaudJMezardC 2011 Characterization of meiotic crossovers in pollen from Arabidopsis thaliana. Methods Mol Biol 745 223 249
80. KauppiLMayCAJeffreysAJ 2009 Analysis of meiotic recombination products from human sperm. Methods Mol Biol 557 323 355
81. FanQQXuFWhiteMAPetesTD 1997 Competition between adjacent meiotic recombination hotspots in the yeast Saccharomyces cerevisiae. Genetics 145 661 670
82. JessopLAllersTLichtenM 2005 Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics 169 1353 1367
83. RameshMAMalikSBLogsdonJMJr 2005 A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol 15 185 191
84. RobineNUematsuNAmiotFGidrolXBarillotE 2007 Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 27 1868 1880
85. WuT-CLichtenM 1995 Factors that affect the location and frequeny of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics 140 55 66
86. XuLKlecknerN 1995 Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J 14 5115 5128
87. CostasCde la Paz SanchezMStroudHYuYOliverosJC 2011 Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks. Nat Struct Mol Biol 18 395 400
88. LeeTJPascuzziPESettlageSBShultzRWTanurdzicM 2010 Arabidopsis thaliana chromosome 4 replicates in two phases that correlate with chromatin state. PLoS Genet 6 e1000982 doi:10.1371/journal.pgen.1000982
89. MurakamiHKeeneyS 2008 Regulating the formation of DNA double-strand breaks in meiosis. Genes Dev 22 286 292
90. HillersKJVilleneuveAM 2003 Chromosome-wide control of meiotic crossing over in C. elegans. Curr Biol 13 1641 1647
91. BlatYProtacioRUHunterNKlecknerN 2002 Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111 791 802
92. de CastroESorianoIMarinLSerranoRQuintalesL 2011 Nucleosomal organization of replication origins and meiotic recombination hotspots in fission yeast. EMBO J
93. GetunIVWuZKKhalilAMBoisPR 2010 Nucleosome occupancy landscape and dynamics at mouse recombination hotspots. EMBO Rep 11 555 560
94. ZhangLMaHPughBF 2011 Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res 21 875 884
95. NicolasATrecoDSchultesNPSzostakJW 1989 An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature 388 35 39
96. BaudatFBuardJGreyCFledel-AlonAOberC 2010 PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327 836 840
97. GreyCBaudatFde MassyB 2009 Genome-wide control of the distribution of meiotic recombination. PLoS Biol 7 e35 doi:10.1371/journal.pbio.1000035
98. HayashiKYoshidaKMatsuiY 2005 A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438 374 378
99. MyersSBowdenRTumianABontropREFreemanC 2010 Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327 876 879
100. ParvanovEDPetkovPMPaigenK 2010 Prdm9 controls activation of mammalian recombination hotspots. Science 327 835
101. SmagulovaFGregorettiIVBrickKKhilPCamerini-OteroRD 2011 Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472 375 378
102. PontingCP 2011 What are the genomic drivers of the rapid evolution of PRDM9? Trends Genet 27 165 171
103. Melamed-BessudoCLevyAA 2012 Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc Natl Acad Sci U S A 109 E981 988
104. MirouzeMLieberman-LazarovichMAversanoRBucherENicoletJ 2012 Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci U S A 109 5880 5885
105. R Development Core Team 2011 R: A language and environment for statistical computing. Vienna, Austria
Štítky
Genetika Reprodukční medicína
Článek Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma GenomesČlánek Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail ExpansionČlánek Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid CellsČlánek Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug ActivationČlánek The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin DynamicsČlánek Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 8
-
Všechny články tohoto čísla
- Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma Genomes
- Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail Expansion
- Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid Cells
- Chromosome Territories Meet a Condensin
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans
- Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse ()
- A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins
- Advances in Quantitative Trait Analysis in Yeast
- Experimental Evolution of a Novel Sexually Antagonistic Allele
- Variation of Contributes to Dog Breed Skull Diversity
- , a Gene Involved in Axonal Pathfinding, Is Mutated in Patients with Kallmann Syndrome
- A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression
- Cryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
- Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug Activation
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
- A Sexual Ornament in Chickens Is Affected by Pleiotropic Alleles at and , Selected during Domestication
- Use of Allele-Specific FAIRE to Determine Functional Regulatory Polymorphism Using Large-Scale Genotyping Arrays
- Novel Loci for Metabolic Networks and Multi-Tissue Expression Studies Reveal Genes for Atherosclerosis
- The Genetic Basis of Pollinator Adaptation in a Sexually Deceptive Orchid
- Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
- Inheritance Beyond Plain Heritability: Variance-Controlling Genes in
- The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, and Anthropometric Traits
- Reprogramming to Pluripotency Can Conceal Somatic Cell Chromosomal Instability
- Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes
- PTEN Negatively Regulates MAPK Signaling during Vulval Development
- A Dynamic Response Regulator Protein Modulates G-Protein–Dependent Polarity in the Bacterium
- Population Genomics of the Facultatively Mutualistic Bacteria and
- Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
- Polysome Profiling in Liver Identifies Dynamic Regulation of Endoplasmic Reticulum Translatome by Obesity and Fasting
- Stromal Liver Kinase B1 [STK11] Signaling Loss Induces Oviductal Adenomas and Endometrial Cancer by Activating Mammalian Target of Rapamycin Complex 1
- Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
- Transgene Induced Co-Suppression during Vegetative Growth in
- Hox and Sex-Determination Genes Control Segment Elimination through EGFR and Activity
- A Quantitative Comparison of the Similarity between Genes and Geography in Worldwide Human Populations
- Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in and Mammals
- Comparative Analysis of Regulatory Elements between and by Genome-Wide Transcription Start Site Profiling
- Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome
- S Phase–Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
- Cell-Nonautonomous Signaling of FOXO/DAF-16 to the Stem Cells of
- The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin Dynamics
- Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus
- Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes
- Genetic Modulation of Lipid Profiles following Lifestyle Modification or Metformin Treatment: The Diabetes Prevention Program
- HAL-2 Promotes Homologous Pairing during Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors
- SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Germline
- Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
- Reduced Prostasin (CAP1/PRSS8) Activity Eliminates HAI-1 and HAI-2 Deficiency–Associated Developmental Defects by Preventing Matriptase Activation
- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
- and Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR
- CDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
- Rare Copy Number Variations in Adults with Tetralogy of Fallot Implicate Novel Risk Gene Pathways
- Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of
- NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing
- Human Developmental Enhancers Conserved between Deuterostomes and Protostomes
- A Luminal Glycoprotein Drives Dose-Dependent Diameter Expansion of the Hindgut Tube
- Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation
- Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration
- Chemical Genetics Reveals a Specific Requirement for Cdk2 Activity in the DNA Damage Response and Identifies Nbs1 as a Cdk2 Substrate in Human Cells
- Experimental Relocation of the Mitochondrial Gene to the Nucleus Reveals Forces Underlying Mitochondrial Genome Evolution
- Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome
- Mutations in a P-Type ATPase Gene Cause Axonal Degeneration
- A General G1/S-Phase Cell-Cycle Control Module in the Flowering Plant
- Multiple Roles and Interactions of and in Development of the Respiratory System
- UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex
- Epigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
- Modulating the Strength and Threshold of NOTCH Oncogenic Signals by
- Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer's Disease–Related Tau Phosphorylation Via PAR-1
- Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System
- ATXN2-CAG42 Sequesters PABPC1 into Insolubility and Induces FBXW8 in Cerebellum of Old Ataxic Knock-In Mice
- Cohesin Rings Devoid of Scc3 and Pds5 Maintain Their Stable Association with the DNA
- The MicroRNA Inhibits Calcium Signaling by Targeting the TIR-1/Sarm1 Adaptor Protein to Control Stochastic L/R Neuronal Asymmetry in
- Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength
- The Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Variation of Contributes to Dog Breed Skull Diversity
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



