-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaFine-Mapping and Initial Characterization of QT Interval Loci in African Americans
The QT interval (QT) is heritable and its prolongation is a risk factor for ventricular tachyarrhythmias and sudden death. Most genetic studies of QT have examined European ancestral populations; however, the increased genetic diversity in African Americans provides opportunities to narrow association signals and identify population-specific variants. We therefore evaluated 6,670 SNPs spanning eleven previously identified QT loci in 8,644 African American participants from two Population Architecture using Genomics and Epidemiology (PAGE) studies: the Atherosclerosis Risk in Communities study and Women's Health Initiative Clinical Trial. Of the fifteen known independent QT variants at the eleven previously identified loci, six were significantly associated with QT in African American populations (P≤1.20×10−4): ATP1B1, PLN1, KCNQ1, NDRG4, and two NOS1AP independent signals. We also identified three population-specific signals significantly associated with QT in African Americans (P≤1.37×10−5): one at NOS1AP and two at ATP1B1. Linkage disequilibrium (LD) patterns in African Americans assisted in narrowing the region likely to contain the functional variants for several loci. For example, African American LD patterns showed that 0 SNPs were in LD with NOS1AP signal rs12143842, compared with European LD patterns that indicated 87 SNPs, which spanned 114.2 Kb, were in LD with rs12143842. Finally, bioinformatic-based characterization of the nine African American signals pointed to functional candidates located exclusively within non-coding regions, including predicted binding sites for transcription factors such as TBX5, which has been implicated in cardiac structure and conductance. In this detailed evaluation of QT loci, we identified several African Americans SNPs that better define the association with QT and successfully narrowed intervals surrounding established loci. These results demonstrate that the same loci influence variation in QT across multiple populations, that novel signals exist in African Americans, and that the SNPs identified as strong candidates for functional evaluation implicate gene regulatory dysfunction in QT prolongation.
Published in the journal: . PLoS Genet 8(8): e32767. doi:10.1371/journal.pgen.1002870
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002870Summary
The QT interval (QT) is heritable and its prolongation is a risk factor for ventricular tachyarrhythmias and sudden death. Most genetic studies of QT have examined European ancestral populations; however, the increased genetic diversity in African Americans provides opportunities to narrow association signals and identify population-specific variants. We therefore evaluated 6,670 SNPs spanning eleven previously identified QT loci in 8,644 African American participants from two Population Architecture using Genomics and Epidemiology (PAGE) studies: the Atherosclerosis Risk in Communities study and Women's Health Initiative Clinical Trial. Of the fifteen known independent QT variants at the eleven previously identified loci, six were significantly associated with QT in African American populations (P≤1.20×10−4): ATP1B1, PLN1, KCNQ1, NDRG4, and two NOS1AP independent signals. We also identified three population-specific signals significantly associated with QT in African Americans (P≤1.37×10−5): one at NOS1AP and two at ATP1B1. Linkage disequilibrium (LD) patterns in African Americans assisted in narrowing the region likely to contain the functional variants for several loci. For example, African American LD patterns showed that 0 SNPs were in LD with NOS1AP signal rs12143842, compared with European LD patterns that indicated 87 SNPs, which spanned 114.2 Kb, were in LD with rs12143842. Finally, bioinformatic-based characterization of the nine African American signals pointed to functional candidates located exclusively within non-coding regions, including predicted binding sites for transcription factors such as TBX5, which has been implicated in cardiac structure and conductance. In this detailed evaluation of QT loci, we identified several African Americans SNPs that better define the association with QT and successfully narrowed intervals surrounding established loci. These results demonstrate that the same loci influence variation in QT across multiple populations, that novel signals exist in African Americans, and that the SNPs identified as strong candidates for functional evaluation implicate gene regulatory dysfunction in QT prolongation.
Introduction
The QT interval (QT), as measured by the resting 12-lead electrocardiogram (ECG), reflects the duration of ventricular depolarization and repolarization, providing a non-invasive assessment of an average ventricular action potential. QT prolongation is an established risk factor for ventricular tachyarrhythmias [1], coronary heart disease [2], and sudden cardiovascular as well as all-cause death [3]. Although numerous factors influencing QT have been identified, including heart rate [4], structural heart disease [5], [6], gender [7], [8], age [9], and medication use [10]–[12], a large portion of the variance in QT remains unexplained.
Several lines of evidence support a genetic contribution to QT. Initial evidence was provided by studies of inherited cardiac arrhythmias including long - and short-QT syndromes, which identified rare and highly penetrant mutations in ion channel and ion channel associated genes associated with QT [13]. Family studies have also reported that ventricular repolarization (as measured by QT) is heritable [14]–[17]. In addition, recent genome-wide association (GWA) studies performed in populations of predominantly European descent have identified common SNPs in twelve loci, including NOS1AP, KCNH2, and PLN [18]–[23], that influence the distribution of QT.
To date, the majority of published GWA studies examining QT have been performed in populations of European descent, although one study also examined an Indian Asian population [23]. It is therefore unclear whether previously identified QT loci are relevant in other racial groups such as African Americans or whether there are population-specific SNPs influencing QT. Furthermore, the increased genetic diversity in populations of African ancestry provides opportunities for the narrowing and fine-mapping of loci identified in European and Indian Asian populations [24]. Fine-mapping, which includes the dense genotyping of common and rare SNPs at already established loci, is a helpful next step in the identification of functional polymorphisms underlying the QT distribution. For example, dense genotyping can capture rarer SNPs that may be inadequately represented on frequently used genome-wide genotyping arrays, and which may have large effects, thereby potentially helping to explain a larger fraction of the QT heritability [25], which is estimated to range from 35% to 52% [14]–[17].
In this study, we evaluated eleven QT loci previously identified in populations of European and Indian Asian descent in 8,644 African American participants from the Atherosclerosis Risk in Communities study (ARIC) and Women's Health Initiative clinical trial (CT). In addition to testing the previously reported QT index SNPs at the known loci, we also searched for stronger markers of the index signal and investigated evidence for independent, novel SNPs influencing QT in African Americans. For loci associated with QT in African Americans, we also investigated whether patterns of linkage disequilibrium (LD) within African Americans could narrow the regions likely to harbor the biologically relevant variant. Finally, we queried bioinformatic databases and performed related in silico analyses to identify potential candidate polymorphisms for follow-up functional evaluation.
Results
Participants are drawn from two separate studies (Methods), which together comprise 9,702 individuals. After applying the exclusion criteria (Methods), phenotypic data from 8,644 participants are included in this analysis. The majority of participants are female (86%) and ages range from 45–79 years (Table S1). Estimates of mean QT and heart rate are consistent across the studies. Approximately 60% of participants are directly genotyped on the Metabochip; the remaining 40% have genotypes imputed from the Affymetrix 6.0 panel (Methods).
Representation of QT loci on the Metabochip and imputation
The Metabochip, a custom array containing approximately 195,000 SNPs, is designed to facilitate fine-mapping of loci associated with cardiovascular and metabolic traits, including QT, blood pressure, cholesterol, type 2 diabetes, and anthropometrics. To identify and fine-map signals associated with QT in African Americans, 6,670 SNPs from eleven previously identified QT loci represented on the Metabochip are examined (Table S2; Methods). The number of SNPs at each locus with minor allele frequency (MAF) estimates ≥1% ranges from 51 to 1,371, corresponding to regions spanning 67 Kb to 664 Kb in size (median size: 275 Kb).
On average, imputation quality for metabochip SNPs imputed in WHI SNP Health Association Resource (SHARe) participants (n = 3,531) is high across the 11 loci, with the exception of RNF207, KCNH2, and KCNQ1 (Table S2). As described in the Methods, SNPs with imputation quality scores <0.95 are discarded.
Association of QT loci in African Americans
The six published GWA studies of QT (five studies of European ancestral populations, one study of European and Indian Asian ancestral populations) reported 25 index SNPs (P≤5.0×10−8, Table 1; D' estimates provided in Table S3) across eleven loci, which together represent fifteen independent signals (at r2≥0.20 in European ancestry populations). The fifteen signals include three independent signals at NOS1AP and two independent signals at both PLN and KCNH2.
Tab. 1. Associations with common variants at fifteen previously reported QT loci across eleven chromosomes in n = 8,644 African American participants. 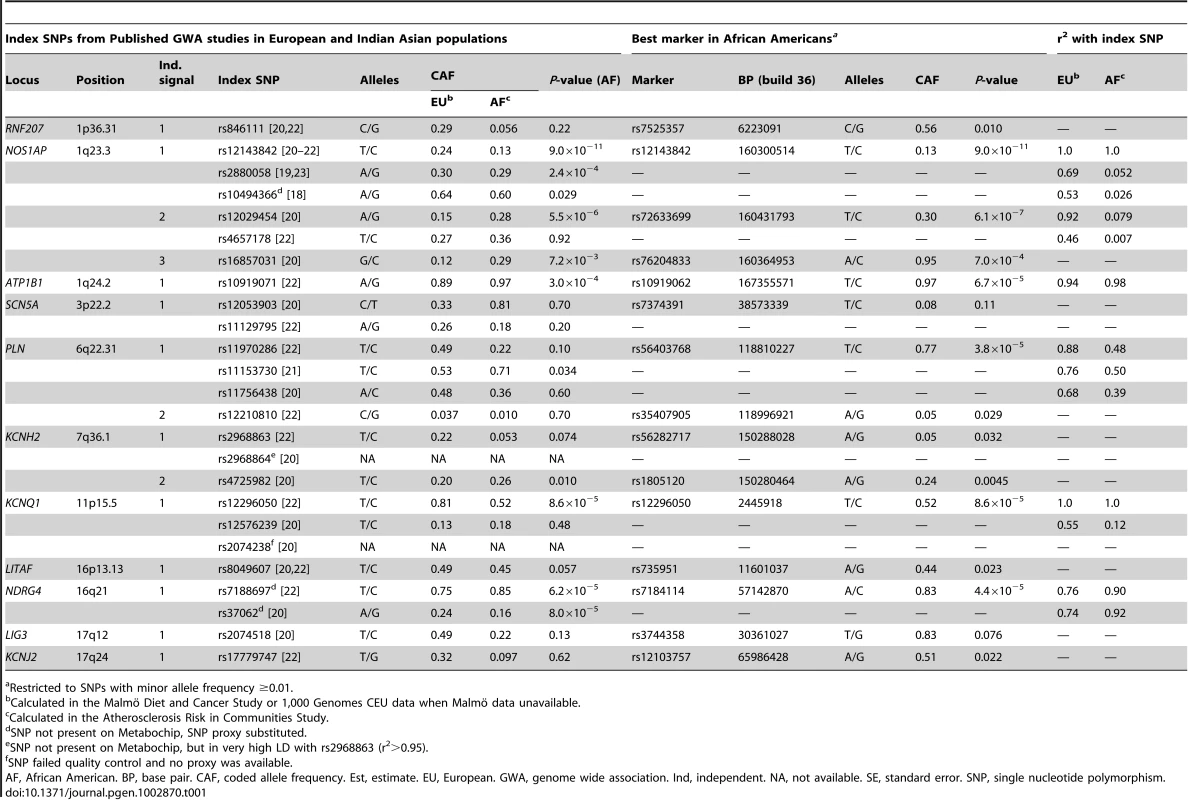
Restricted to SNPs with minor allele frequency ≥0.01. We first test the fifteen independent signals for association with QT in African Americans. Specifically, we evaluate the index SNPs for each of the fifteen independent signals as well as all SNPs in LD with the index SNPs (r2≥0.20 using European ancestral LD patterns) to determine whether any of the fifteen independent signals generalize to African Americans. The significance criterion, αa = 1.20×10−4, is based on the number of tag SNPs in African Americans that capture (r2≥0.80, using African American LD patterns) all SNPs that are correlated with the index SNPs (r2≥0.20; determined using European ancestral LD patterns; see Methods).
Six of the fifteen independent signals are significantly associated with QT in African Americans (Table 1): NOS1AP independent signals 1 and 2, ATP1B1, PLN independent signal 1, KCNQ1, and NDRG4. Of those that are not significantly associated with QT (P-value>1.20×10−4), estimates for index SNPs representing the two KCNH2 independent signals as well as the RNF207 index SNP show a consistent magnitude and direction of effect when compared to published estimates (Table S4).
The best marker in African Americans for the first NOS1AP independent signal (rs12143842) and KCNQ1 (rs12296050) is identical to the European index SNP. For the ATP1B1 and NDRG4 index SNPs as well as the second NOS1AP independent signal, the best marker in African Americans shows only a slightly more significant association than the index signal (<1 order of magnitude change in the P-value; Table 1). In contrast, at the first PLN independent signal, the three index SNPs are not significantly associated with QT in African Americans (P-value range: 0.034–0.60), although a substantially stronger marker of the index signal is detected (rs56403768; P-value = 3.8×10−5). In Europeans, rs56403768 is correlated with the three index SNPs (LD r2 range: 0.68–0.88). However, patterns of correlation are weaker in African Americans (LD r2 range: 0.39–0.50).
We then evaluate evidence for additional independent signals at the eleven previously identified QT loci, focusing on SNPs that are uncorrelated with the index signals in European populations. Here, statistical significance is defined using an efficient Monte Carlo approach that accounts for LD between SNPs at the eleven previously identified QT loci (αb = 1.37×10−5; see Methods). Eleven SNPs at two loci that are uncorrelated with the index SNPs (r2≤0.20; evaluated using both African American and European ancestral LD patterns) exceed our significance threshold. Conditional analysis confirms that these eleven SNPs represent three novel associations, one flanking the NOS1AP locus and two residing within or nearby ATP1B1 (Table 2; Figures 1 and S1). Of note, the novel NOS1AP SNP is monomorphic in populations of European ancestry. Together, the six best markers in African Americans and the three novel SNPs explain 1.6% of the variance in heart rate-corrected QT.
Fig. 1. −Log P plot for common SNPs at the NOS1AP independent signal 1 and 2 loci. 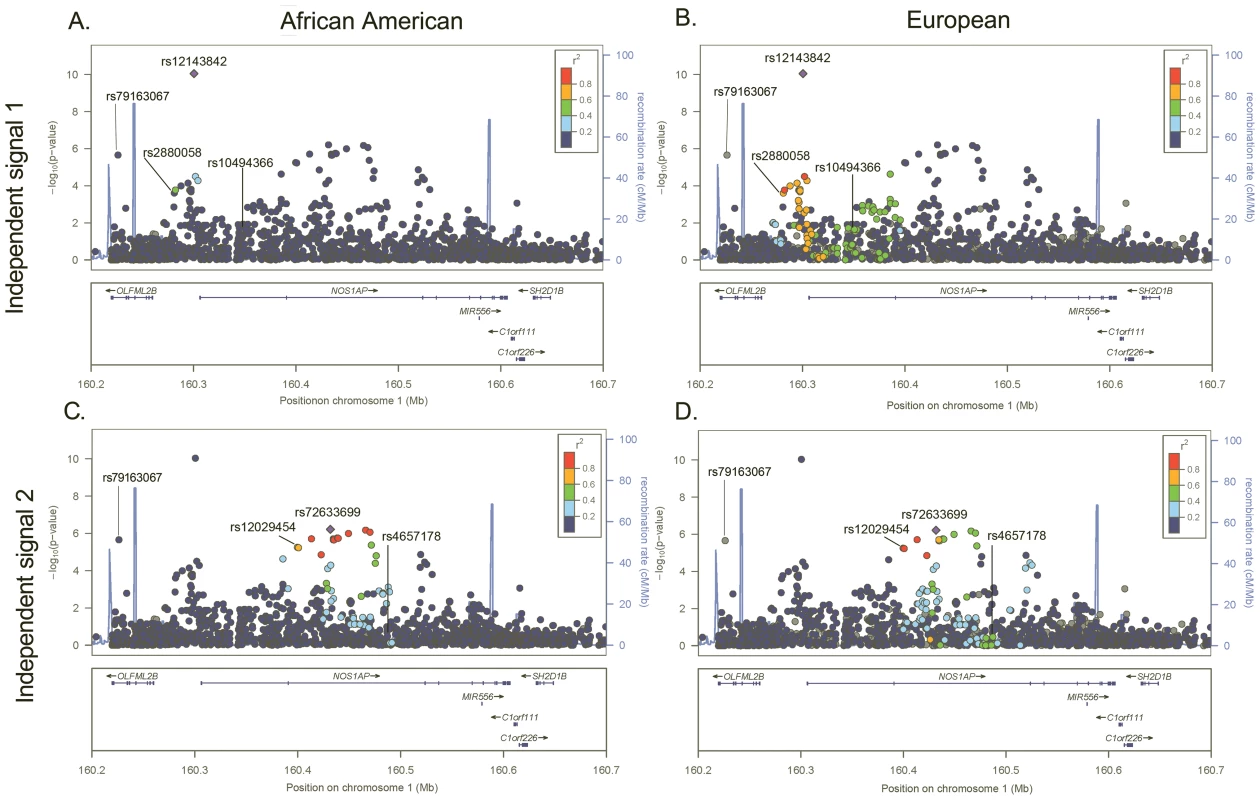
P-values are estimated in African Americans and are plotted using linkage disequilibrium estimates from African Americans (panels A and C) and Europeans (panels B and D). SNPs are represented by circles, lines indicate index SNPS previously identified in GWA studies of European and Indian Asian populations, and the large blue diamond is the best marker in African Americans. Circle color represents correlation with the best marker in African Americans: blue indicates weak correlation and red indicates strong correlation. Recombination rate is plotted in the background and annotated genes are shown at the bottom of the plot. Tab. 2. Novel and independent SNPs associated with QT at two previously identified QT loci in n = 8,644 African American participants. 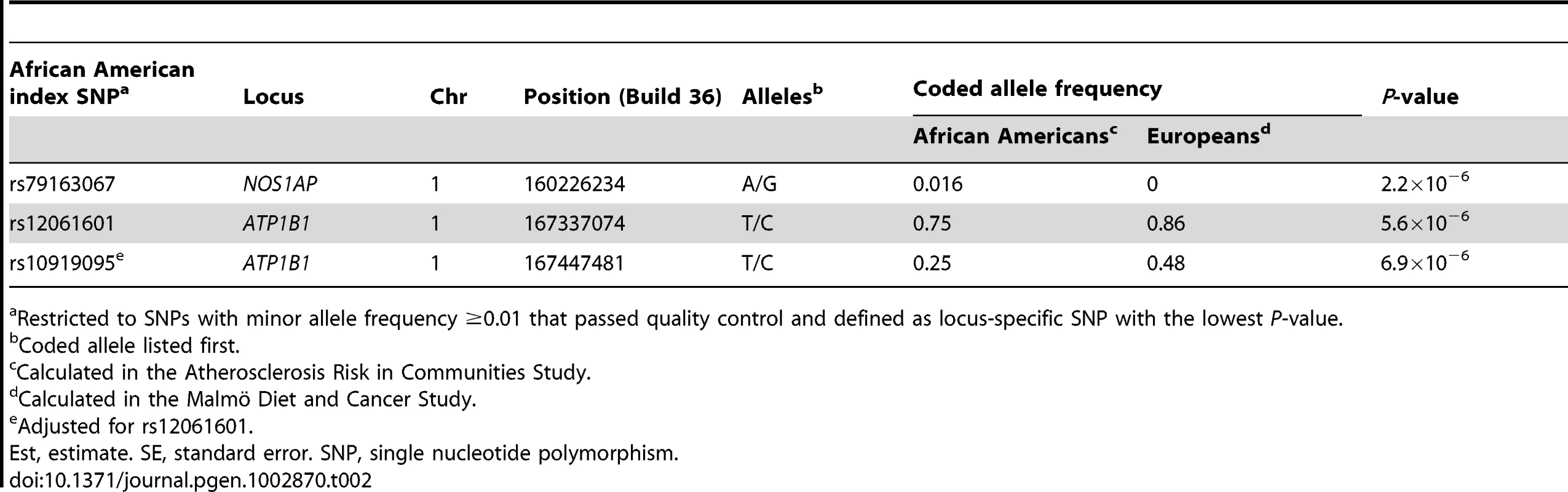
Restricted to SNPs with minor allele frequency ≥0.01 that passed quality control and defined as locus-specific SNP with the lowest P-value. Next, we examine whether African American LD patterns can assist with the narrowing of association signals at the six loci that generalized to African Americans. An example of variation in LD patterns by ancestral population is shown by the first NOS1AP independent signal. Among African Americans, 0 SNPs are correlated (r2≥0.50) with rs12143842, which is the best marker in African Americans and the index SNP reported by three prior QT GWA studies (Figure 1; Table 3) [20]–[22]. This is in contrast to LD patterns estimated in Europeans for NOS1AP independent signal 1, where 87 SNPs are correlated with the three index signals that characterize this independent signal (r2≥0.50), representing a region spanning 114.2 Kb. For the five remaining markers (Figures 1 and S1, S2, S3, S4), African American populations exhibit lower levels of LD as compared to European populations (mean narrowing = 48.2 Kb). Likewise, fine-mapping in African Americans helped to narrow the association signals for all six loci.
Tab. 3. Comparison of linkage disequilibrium patterns between populations of African and European descent for six previously identified QT loci significantly associated with QT in n = 8,644 African American participants. 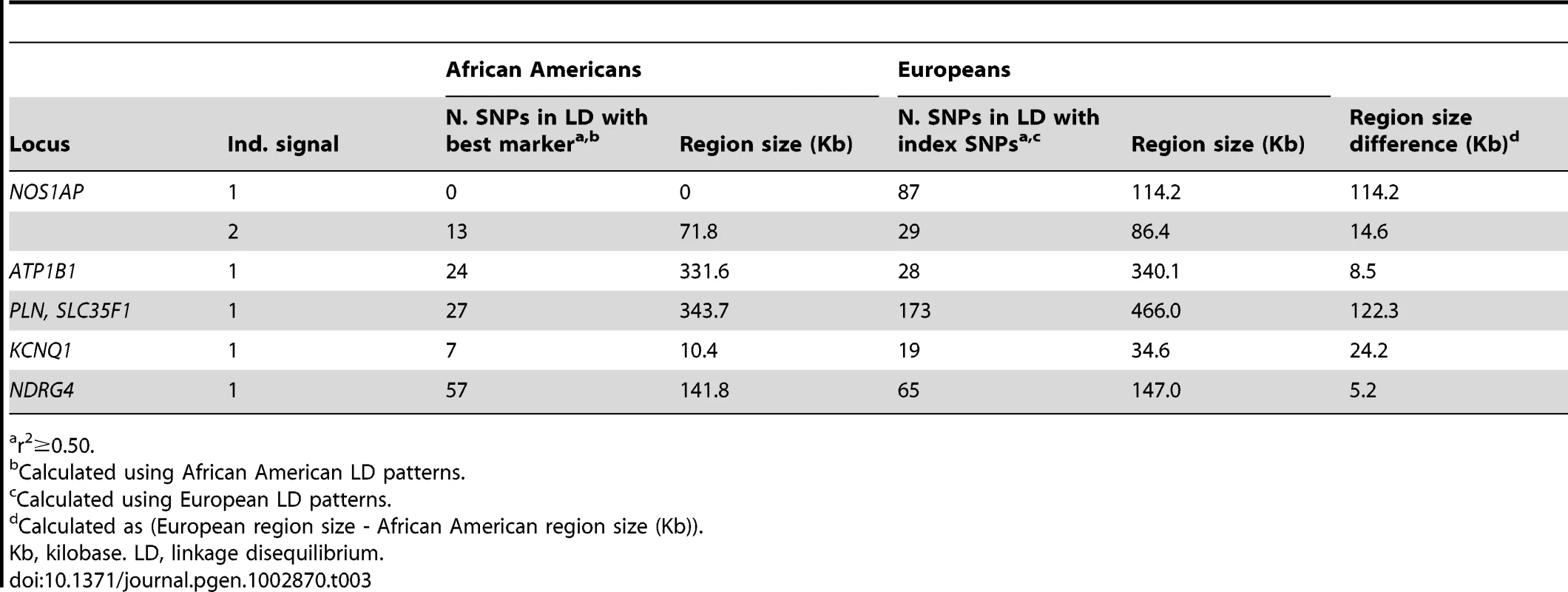
r2≥0.50. Bioinformatic-based functional characterization of QT loci
Bioinformatic analysis of the six best markers in African Americans and the three novel and independent SNPs (Table S5; Methods) does not identify any correlated non-synonymous coding variants; instead, all signals harbor variants that occur solely within non-coding regions with the potential for influencing cis-regulation (Table S6). Several variants occur within candidate regulatory elements (promoter regions and DNase hypersensitive sites in human cardiomyocytes), including three (rs3864884, rs1646010, and rs27097) that are predicted to have allele-specific binding affinities for various transcription factors of relevance to cardiac function (Table S7). Though further functional characterization is outside the scope of this study, rs3864884, rs1646010, rs27097, and rs37036 represent compelling candidates for follow-up evaluation.
Discussion
In this study composed of approximately 8,600 African American participants, we evaluated fifteen independent signals across eleven loci that were previously associated with QT in populations of European and Indian Asian ancestry at genome-wide significant levels. For five independent signals – the two NOS1AP independent signals, ATP1B1, NDRG4, and KCNQ1 – the best markers in African Americans were either identical to or only slightly more significant than the index signal. These five markers are therefore not considered better signals than the index SNP. However, for the first PLN locus, the three previously identified index SNPs were not significantly associated with QT in African Americans. This result suggests that rs56403768, the best marker in African Americans, is a better proxy of a biologically important PLN allele and may help improve localization of the true association.
In addition to generalizing six previously characterized QT loci, we also identified three novel and independent signals for NOS1AP and ATP1B1. Notably, rs79163067, the novel NOS1AP signal, was monomorphic in European populations. When these three novel and independent variants were combined with the fifteen independent QT loci previously identified in populations of European and Indian Asian ancestry, our results suggest that to-date at least 18 independent variants influence QT.
Finally, we showed that evaluating LD patterns in admixed populations such as African Americans assisted with the narrowing of intervals flanking the putative causal variants. This narrowing was particularly evident for the first NOS1AP locus that included index SNP rs12143842, the most frequently reported SNP in the QT GWA study literature to-date. Rs12143842 also was the SNP that explained the majority of variance in heart rate-corrected QT as well as the best marker in African Americans for the first NOS1AP independent signal. Rs12143842 has yet to be evaluated in any functional studies, and although our bioinformatics characterization did not identify any compelling functional candidates, the SNP resides less than five Kb from the annotated NOS1AP promoter. Future studies evaluating the functional relevance of rs12143842 are clearly indicated.
Of the nine QT SNPs we identified (i.e. the six best markers in African Americans and the three novel SNPs), all resided in or were in LD with SNPs residing in candidate long-range regulatory elements in human cardiomyocytes, annotated promoter regions, and highly conserved non-coding elements, and as such, strongly implicate gene regulatory dysfunction in QT prolongation. One of the more striking predictions was with rs3864884, where the major allele [C] is predicted to bind an entirely different set of transcription factors (TFs) than the minor allele [T]. Notably, only the major allele is predicted to bind Hairy-related TFs, which are involved in regulating cardiac morphogenesis [26]. The minor allele is predicted to bind TBX5 and AhR; the former has been linked to a number of cardiac phenotypes including Holt-Oram syndrome [27] and atrioventricular conduction [28] and the latter regulates cardiac size [29], a known risk factor for QT-prolongation and cardiac sudden death [30]. Consistent with this prediction, the minor allele is associated with an increased QT in this population.
We were unable to identify associations with RNF207, SCN5A, KCNH2, LITAF, LIG3, and KCNJ2 based on our statistical significance threshold. RNF207 was reported in two previous QT GWA studies. However, because of design priorities, only 51 SNPs with MAF≥1% were available for analysis. Future fine-mapping efforts in African Americans or other admixed populations that include denser genotyping of this locus may therefore be useful.
SCN5A, KCNH2, and LITAF were all reported by two prior GWA studies of QT. SCN5A also has also been implicated in GWA studies of PR in populations of European [31] and African ancestry [28] as well as QRS interval duration in populations of European descent [32]. Our inability to detect signals at these three loci may simply reflect inadequate power, especially for KCNH2, for which estimates of the index SNPs in African Americans were consistent with the published literature. KCNJ2 is a biologically plausible locus influencing QT, as it harbors mutations causing rare, familial forms of long QT syndrome [33]. Yet, the high P-values and the dense genotyping coverage of SNPs suggest that KCNJ2 does not influence QT in African American populations.
The genetic architecture of African Americans and other admixed populations is on average characterized by lower correlation between SNPs when compared to European populations. Such populations therefore are valuable for the fine-mapping of previously identified loci, as fewer SNPs are expected to be correlated with the underlying functional variant, which is expected to be the same in populations of different ethnicity. We therefore anticipate that future fine-mapping efforts that include populations with different ethnic backgrounds will be useful for the further refinement of loci influencing QT as well as the identification of population-specific variants, as demonstrated by the current report.
Several limitations of the present study warrant further consideration in order to inform future efforts for fine-mapping and functional characterization of QT loci. First, although the Metabochip includes dense genotyping of most QT loci, it is possible that the causative variants are not included on the Metabochip. Second, our functional characterization is based on in silico analyses and requires experimental validation. Third, the majority of study participants were female. It is unclear how a predominantly female population may have influenced the results presented herein, considering the well-known dependence of QT on gender [7], [8]. Finally, our results, which are consistent with prior studies [22], show that common SNPs only explain a very small fraction of the variance in QT, although heritability estimates suggest a substantial genetic component. These modest effect sizes corroborate the multifactorial etiology of QT and demonstrate that substantially greater efforts are required to explain the “missing heritability”. Future efforts with increased sample sizes that examine rare variants, gene-gene and gene-environment interactions, and structural variants poorly captured on existing arrays are clearly needed [34].
In conclusion, our findings provide compelling evidence that the same genes influence variation in QT across ancestral populations and that additional, independent signals exist in African Americans. Moreover, all SNPs identified as strong candidates for functional evaluation implicate gene regulatory dysfunction. Further characterization of these loci, including direct sequencing and large-scale genotyping in African Americans and other admixed populations, may provide more information on the genetic and molecular mechanisms underlying QT.
Materials and Methods
Ethics statement
The Institutional Review Board at all participating institutions approved the study protocol. This study was conducted according to the principles expressed in the Declaration of Helsinki.
Study populations
The Population Architecture using Genomics and Epidemiology (PAGE) study is a National Human Genome Research Institute funded effort examining the epidemiologic architecture of common genetic variants that have been reproducibly associated with human diseases and traits [35]. The PAGE study consists of a coordinating center and four consortia with access to large, diverse population-based studies including three National Health and Nutrition Examination Surveys, the Multiethnic Cohort, the WHI, the ARIC study, the Coronary Artery Risk Disease in Young Adults study, the Cardiovascular Health Study, the Hispanic Community Health Study/Study of Latinos, the Strong Heart Study, and the Strong Heart Family Study.
This PAGE Metabochip study included African American participants from the ARIC and WHI CT studies. Participants from the other PAGE studies were excluded from this effort due to the unavailability of ECG measures and/or genotype data. Genotypes of WHI CT participants were obtained in three phases: two sets of women were directly genotyped on the Metabochip platform by PAGE investigators during wave 1 (n = 797) and wave 2 (n = 1,128) and women (n = 3,531) with Metabochip variants imputed from previous genome-wide SNP data provided by the WHI SHARe [36]. Participants meeting the following criteria were excluded from the study: QT unavailable, atrial fibrillation/atrial flutter on ECG, left or right bundle branch block on ECG, QRS duration >120 milliseconds, intraventricular conduction delay on ECG, pacemaker implant antedating ECG, ancestry outlier, excessive heterozygosity, low call rate, or second member of first degree relative pair. Further details on the ARIC and WHI CT studies are provided in Text S1 (Participating Studies).
QT measurement
For each study, certified technicians digitally recorded resting, supine (or semi-recumbent), standard 12-lead ECGs at study baseline for each participant using Marquette MAC PC machines (GE Healthcare, Milwaukee, WI, USA). The ARIC and WHI CT studies used comparable procedures for preparing participants, placing electrodes, recording, transmitting, processing, and controlling quality of the ECGs. QT was measured electronically using the Marquette 12SL algorithm.
The Metabochip
The Metabochip was a custom Illumina iSELECT array that contained approximately 195,000 SNPs and was designed to support large scale follow up of putative associations for cardiovascular and metabolic traits including QT, blood pressure, cholesterol, type 2 diabetes, and anthropometrics. Approximately 33% of the Metabochip SNPs were included as replication targets and 62% for fine-mapping. In total, 257 loci were selected for fine-mapping, with the surrounding regions totaling 45.5 Mb accounting for overlaps (14.2 Mb for the densest fine-mapping regions). Eleven QT loci identified in previous GWA studies in populations of European and Asian ancestry were represented on the Metabochip (Table 1). The only published QT locus that is not represented on the Metabochip is an intergenic region on 13q14 reported by Marroni et al [19], but not replicated by other published GWA studies of populations with similar ancestral backgrounds. SNPs reported in the literature but not genotyped on the Metabochip (NOS1AP, rs10494366; NDRG4, rs7188697, rs37062) were represented by proxies, defined as SNPs in high LD (r2≥0.90) with the index SNP using HapMap YRI data.
Genotyping and quality control assessment
Samples were genotyped at the Human Genetics Center of the University of Texas-Houston and the Translational Genomics Research Institute for ARIC and WHI, respectively, following each genotyping center's standard procedures. HapMap YRI (Yoruba in Ibadan, Nigeria) samples were also genotyped independently by each study to facilitate cross-study quality control. Genotypes were called separately for each study, albeit with a common protocol and common personnel, with GenomeStudio using the GenCall 2.0 algorithm. Because the Metabochip includes SNPs with much lower MAFs than are usually called with GenCall, SNPs were recalled using the GenoSNP genotype-calling algorithm [37]. SNPs with call rates <95%, Hardy-Weinberg equilibrium P<10−6, >1 Mendelian error (in 30 YRI trios), >2 replication errors, or >3.3% discordant calls in YRI across genotyping centers or against the HapMap database were considered quality control failures. Samples with call rates <0.95 or an inbreeding coefficient F>0.15 were excluded from further analysis [38].
Prior to analyses, related participants were identified using PLINK [39] by estimating identical-by-descent statistics for all pairs. When apparent first-degree relative pairs were identified, the member from each pair with the lower call rate was excluded from further analysis. Principal components of ancestry were determined using the Eigensoft software [40], [41] and apparent ancestral outliers were excluded from further analysis.
WHI SHARe imputation
Briefly, n = 1,962 WHI participants who were genotyped on both the Affymetrix 6.0 and Metabochip genotyping platforms were used to infer Metabochip genotypes to the n = 8,421 population of WHI participants genotyped on the Affymetrix 6.0 array [36]. Before phasing and imputation, Affymetrix 6.0 SNPs with genotype call rates <90%, Hardy-Weinberg P-values<10−6, or MAF<0.01 were removed. Participants with call rates <95%, those who demonstrated excess heterozygosity, were part of a first-degree relative pair, or who were identified as an ancestry outlier were excluded. This yielded a set of 987,749 SNPs for the 1,962 reference participants. Mean concordance rates for the 23,703 SNPs in common was 99.7%. Haplotypes were reconstructed using MaCH and were used as a reference to impute Metabochip data into the 6,459 WHI participants with only Affymetrix 6.0 data. Liu et al., (2012) demonstrated the ability to impute 99.9% (97.5%, 83.6%, 52.0%, 20.5%) of SNPs with MAF≥0.05 (0.03–0.05, 0.01–0.03, 0.005–0.01, and 0.001–0.005) with average dosage r2 = 94.7% (92.1%, 89.0%, 83.1%, and 79.7%), respectively. For this analysis, all imputed SNPs with r2<0.95 were excluded.
Statistical analysis
To interpret fine-mapping results, LD in our African American PAGE Metabochip sample was calculated in 500 Kb sliding windows using PLINK. In addition, Metabochip LD and frequency information (but not individual-level information) was provided by the Malmö Diet and Cancer Study on 2,143 control participants from a Swedish population [42] to facilitate LD and MAF comparisons to populations of European ancestry. HapMap CEU LD data were used for previously published GWA studies in European populations, as not all European index variants were represented on the Metabochip. Regional association plots use positions from NCBI build 36. Recombination rates were estimated from HapMap phase II data.
Linear regression models were used to study the association between QT and 6,670 SNPs from 11 regions fine-mapped for QT assuming an additive genetic model and including age, sex, study center, ancestry principal components, and heart rate as covariates. Study-specific association results were combined using an inverse variance meta-analysis approach as implemented in METAL [43].
For each QT locus, it is expected that SNPs associated with QT in African Americans will be correlated with the index SNP reported in Europeans. Therefore, we first identified and tested SNPs that are correlated (r2≥0.20) with the index signals in Europeans using LD statistics estimated in the Malmö Diet and Cancer Study. In order to determine the appropriate multiple testing threshold for declaration of whether the previously identified signals were significantly associated with QT in African Americans, i.e. generalizability, we then estimated the number of tag SNPs needed to capture all common alleles (r2≥0.80) using African American LD patterns. The multiple testing threshold for declaring generalization was αa = 0.05/415, where 415 = the total number of tags identified using African American LD patterns.
To identify significant population-specific SNPs influencing QT that were not correlated with the index signal in Europeans (i.e. r2<0.20, which was estimated in the Malmö Diet and Cancer Study), we used an efficient Monte Carlo approach that accounts for LD between SNPs at the previously identified QT loci (αb = 1.37×10−5) [44]. Conditional analyses were then performed to determine the number of independent signals the population-specific SNPs represent. Specifically, analyses were repeated for each locus including the SNP with the smallest P – value as a covariate. This approach was performed adjusting for successively less significant SNPs until no SNPs with P –values lower than αb = 1.37×10−5 were identified. To facilitate comparability with previous reports examining the proportion of variance in QT explained by common SNPs, heart rate-corrected QT [45] was regressed on the six best markers in African Americans and the three population-specific variants assuming an additive genetic model and including age, sex, study center, ancestry principal components as covariates.
Functional categorization of QT loci
For each of the nine QT SNPs (i.e. the six best markers in African Americans and the three novel SNPs), we identified all SNPs in LD (r2≥0.5) using the genotypes from the African American population described in this study. We refer to these SNP sets as Trait Associated SNP (TAS) blocks. We assigned each TAS to one or more of the functional annotation datasets listed in Table S5. These datasets are not mutually exclusive. For example, a TAS can reside in both a candidate regulatory element (dataset #7) and a CTCF binding site (dataset #10). For TASs that occur within predicted transcription factor binding sites (datasets #3 and #8), we calculated transcription factor binding affinity for each TAS allele using PWM-scan [46], as described previously [47]. For TASs that occur within 3′ untranslated regions, we used the TargetScanS algorithm to determine whether they disrupt likely microRNA target sites (dataset #5). To define candidate non-promoter regulatory elements of greatest relevance to QT (dataset #7), we restricted the analysis of DNase I hypersensitive sites (open chromatin loci) to only those present in human cardiomyocytes.
Supporting Information
Zdroje
1. MossAJ (1999) The QT interval and torsade de pointes. Drug Saf 21 Suppl 1 : 5–10 discussion 81-17.
2. DekkerJM, CrowRS, HannanPJ, SchoutenEG, FolsomAR (2004) Heart rate-corrected QT interval prolongation predicts risk of coronary heart disease in black and white middle-aged men and women: the ARIC study. J Am Coll Cardiol 43 : 565–571.
3. ZhangY, PostWS, Blasco-ColmenaresE, DalalD, TomaselliGF, et al. (2011) Electrocardiographic QT Interval and Mortality: A Meta-analysis. Epidemiology 22 : 660–670.
4. BazettHC (1920) The time relations of the blood-pressure changes after excision of the adrenal glands, with some observations on blood volume changes. J Physiol 53 : 320–339.
5. KramerB, BrillM, BruhnA, KublerW (1986) Relationship between the degree of coronary artery disease and of left ventricular function and the duration of the QT-interval in ECG. Eur Heart J 7 : 14–24.
6. HartG (1994) Cellular electrophysiology in cardiac hypertrophy and failure. Cardiovasc Res 28 : 933–946.
7. BazettHC (1920) An analysis of the time-relations of electrocardiograms. Heart 7 : 353–357.
8. LepeschkinE, SurawiczB (1953) The duration of the Q-U interval and its components in electrograms of normal persons. Am Heart J 46.
9. MangoniAA, KinironsMT, SwiftCG, JacksonSH (2003) Impact of age on QT interval and QT dispersion in healthy subjects: a regression analysis. Age Ageing 32 : 326–331.
10. MeinertCL, KnatterudGL, ProutTE, KlimtCR (1970) A study of the effects of hypoglycemic agents on vascular complications in patients with adult-onset diabetes. II. Mortality results. Diabetes 19: Suppl: 789–830.
11. NajeedSA, KhanIA, MolnarJ, SombergJC (2002) Differential effect of glyburide (glibenclamide) and metformin on QT dispersion: a potential adenosine triphosphate sensitive K+ channel effect. Am J Cardiol 90 : 1103–1106.
12. RodenDM (2004) Drug-induced prolongation of the QT interval. N Engl J Med 350 : 1013–1022.
13. ShahM, AkarFG, TomaselliGF (2005) Molecular basis of arrhythmias. Circulation 112 : 2517–2529.
14. BusjahnA, KnoblauchH, FaulhaberHD, BoeckelT, RosenthalM, et al. (1999) QT interval is linked to 2 long-QT syndrome loci in normal subjects. Circulation 99 : 3161–3164.
15. HansonB, TunaN, BouchardT, HestonL, EckertE, et al. (1989) Genetic factors in the electrocardiogram and heart rate of twins reared apart and together. Am J Cardiol 63 : 606–609.
16. Newton-ChehC, LarsonMG, CoreyDC, BenjaminEJ, HerbertAG, et al. (2005) QT interval is a heritable quantitative trait with evidence of linkage to chromosome 3 in a genome-wide linkage analysis: The Framingham Heart Study. Heart Rhythm 2 : 277–284.
17. AkylbekovaEL, CrowRS, JohnsonWD, BuxbaumSG, NjemanzeS, et al. (2009) Clinical correlates and heritability of QT interval duration in blacks: the Jackson Heart Study. Circ Arrhythm Electrophysiol 2 : 427–432.
18. ArkingDE, PfeuferA, PostW, KaoWH, Newton-ChehC, et al. (2006) A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet 38 : 644–651.
19. MarroniF, PfeuferA, AulchenkoYS, FranklinCS, IsaacsA, et al. (2009) A genome-wide association scan of RR and QT interval duration in 3 European genetically isolated populations: the EUROSPAN project. Circ Cardiovasc Genet 2 : 322–328.
20. Newton-ChehC, EijgelsheimM, RiceKM, de BakkerPI, YinX, et al. (2009) Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet 41 : 399–406.
21. NolteIM, WallaceC, NewhouseSJ, WaggottD, FuJ, et al. (2009) Common genetic variation near the phospholamban gene is associated with cardiac repolarisation: meta-analysis of three genome-wide association studies. PLoS One 4: e6138.
22. PfeuferA, SannaS, ArkingDE, MullerM, GatevaV, et al. (2009) Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet 41 : 407–414.
23. ChambersJC, ZhaoJ, TerraccianoCM, BezzinaCR, ZhangW, et al. (2010) Genetic variation in SCN10A influences cardiac conduction. Nat Genet 42 : 149–152.
24. McCarthyMI, HirschhornJN (2008) Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet 17: R156–165.
25. SannaS, LiB, MulasA, SidoreC, KangHM, et al. (2011) Fine mapping of five Loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet 7: e1002198.
26. RutenbergJB, FischerA, JiaH, GesslerM, ZhongTP, et al. (2006) Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development 133 : 4381–4390.
27. LiQY, Newbury-EcobRA, TerrettJA, WilsonDI, CurtisAR, et al. (1997) Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet 15 : 21–29.
28. SmithJG, MagnaniJW, PalmerC, MengYA, SolimanEZ, et al. (2011) Genome-wide association studies of the PR interval in African Americans. PLoS Genet 7: e1001304.
29. ThackaberryEA, GabaldonDM, WalkerMK, SmithSM (2002) Aryl hydrocarbon receptor null mice develop cardiac hypertrophy and increased hypoxia-inducible factor-1alpha in the absence of cardiac hypoxia. Cardiovasc Toxicol 2 : 263–274.
30. KangYJ (2006) Cardiac hypertrophy: a risk factor for QT-prolongation and cardiac sudden death. Toxicol Pathol 34 : 58–66.
31. PfeuferA, van NoordC, MarcianteKD, ArkingDE, LarsonMG, et al. (2010) Genome-wide association study of PR interval. Nat Genet 42 : 153–159.
32. SotoodehniaN, IsaacsA, de BakkerPI, DorrM, Newton-ChehC, et al. (2010) Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet 42 : 1068–1076.
33. Tristani-FirouziM, JensenJL, DonaldsonMR, SansoneV, MeolaG, et al. (2002) Functional and clinical characterization of KCNJ2 mutations associated with LQT7 (Andersen syndrome). J Clin Invest 110 : 381–388.
34. ManolioTA, CollinsFS, CoxNJ, GoldsteinDB, HindorffLA, et al. (2009) Finding the missing heritability of complex diseases. Nature 461 : 747–753.
35. MatiseTC, AmbiteJL, BuyskeS, CarlsonCS, ColeSA, et al. (2011) The Next PAGE in understanding complex traits: design for the analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. Am J Epidemiol 174 : 849–859.
36. LiuEY, BuyskeS, AragakiAK, PetersU, BoerwinkleE, et al. (2011) Genotype Imputation of Metabochip SNPs Using a Study Specific Reference Panel of ∼4,000 Haplotypes in African Americans from the Women's Health Initiative. Genetic Epidemiology 36 : 107–117.
37. GiannoulatouE, YauC, ColellaS, RagoussisJ, HolmesCC (2008) GenoSNP: a variational Bayes within-sample SNP genotyping algorithm that does not require a reference population. Bioinformatics 24 : 2209–2214.
38. BuyskeS, WuY, CartyCL, AssimesTL, DumitrescuL, et al. (2012) Evaluation of the Metabochip Genotyping Array in African Americans and Implications for Fine Mapping of GWAS-Identified Loci: The PAGE Study. PLoS One In press.
39. PurcellS, NealeB, Todd-BrownK, ThomasL, FerreiraMA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics 81 : 559–575.
40. PattersonN, PriceAL, ReichD (2006) Population structure and eigenanalysis. PLoS genetics 2: e190.
41. PriceAL, PattersonNJ, PlengeRM, WeinblattME, ShadickNA, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics 38 : 904–909.
42. BerglundG, ElmstahlS, JanzonL, LarssonSA (1993) The Malmo Diet and Cancer Study. Design and feasibility. Journal of internal medicine 233 : 45–51.
43. WillerCJ, LiY, AbecasisGR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26 : 2190–2191.
44. LinDY (2005) An efficient Monte Carlo approach to assessing statistical significance in genomic studies. Bioinformatics 21 : 781–787.
45. RautaharjuPM, ZhangZM (2002) Linearly scaled, rate-invariant normal limits for QT interval: eight decades of incorrect application of power functions. J Cardiovasc Electrophysiol 13 : 1211–1218.
46. LevyS, HannenhalliS (2002) Identification of transcription factor binding sites in the human genome sequence. Mamm Genome 13 : 510–514.
47. SethupathyP, GiangH, PlotkinJB, HannenhalliS (2008) Genome-wide analysis of natural selection on human cis-elements. PLoS One 3: e3137.
Štítky
Genetika Reprodukční medicína
Článek Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma GenomesČlánek Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail ExpansionČlánek Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid CellsČlánek Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug ActivationČlánek The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin DynamicsČlánek Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 8
-
Všechny články tohoto čísla
- Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma Genomes
- Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail Expansion
- Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid Cells
- Chromosome Territories Meet a Condensin
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans
- Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse ()
- A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins
- Advances in Quantitative Trait Analysis in Yeast
- Experimental Evolution of a Novel Sexually Antagonistic Allele
- Variation of Contributes to Dog Breed Skull Diversity
- , a Gene Involved in Axonal Pathfinding, Is Mutated in Patients with Kallmann Syndrome
- A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression
- Cryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
- Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug Activation
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
- A Sexual Ornament in Chickens Is Affected by Pleiotropic Alleles at and , Selected during Domestication
- Use of Allele-Specific FAIRE to Determine Functional Regulatory Polymorphism Using Large-Scale Genotyping Arrays
- Novel Loci for Metabolic Networks and Multi-Tissue Expression Studies Reveal Genes for Atherosclerosis
- The Genetic Basis of Pollinator Adaptation in a Sexually Deceptive Orchid
- Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
- Inheritance Beyond Plain Heritability: Variance-Controlling Genes in
- The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, and Anthropometric Traits
- Reprogramming to Pluripotency Can Conceal Somatic Cell Chromosomal Instability
- Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes
- PTEN Negatively Regulates MAPK Signaling during Vulval Development
- A Dynamic Response Regulator Protein Modulates G-Protein–Dependent Polarity in the Bacterium
- Population Genomics of the Facultatively Mutualistic Bacteria and
- Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
- Polysome Profiling in Liver Identifies Dynamic Regulation of Endoplasmic Reticulum Translatome by Obesity and Fasting
- Stromal Liver Kinase B1 [STK11] Signaling Loss Induces Oviductal Adenomas and Endometrial Cancer by Activating Mammalian Target of Rapamycin Complex 1
- Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
- Transgene Induced Co-Suppression during Vegetative Growth in
- Hox and Sex-Determination Genes Control Segment Elimination through EGFR and Activity
- A Quantitative Comparison of the Similarity between Genes and Geography in Worldwide Human Populations
- Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in and Mammals
- Comparative Analysis of Regulatory Elements between and by Genome-Wide Transcription Start Site Profiling
- Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome
- S Phase–Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
- Cell-Nonautonomous Signaling of FOXO/DAF-16 to the Stem Cells of
- The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin Dynamics
- Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus
- Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes
- Genetic Modulation of Lipid Profiles following Lifestyle Modification or Metformin Treatment: The Diabetes Prevention Program
- HAL-2 Promotes Homologous Pairing during Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors
- SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Germline
- Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
- Reduced Prostasin (CAP1/PRSS8) Activity Eliminates HAI-1 and HAI-2 Deficiency–Associated Developmental Defects by Preventing Matriptase Activation
- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
- and Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR
- CDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
- Rare Copy Number Variations in Adults with Tetralogy of Fallot Implicate Novel Risk Gene Pathways
- Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of
- NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing
- Human Developmental Enhancers Conserved between Deuterostomes and Protostomes
- A Luminal Glycoprotein Drives Dose-Dependent Diameter Expansion of the Hindgut Tube
- Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation
- Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration
- Chemical Genetics Reveals a Specific Requirement for Cdk2 Activity in the DNA Damage Response and Identifies Nbs1 as a Cdk2 Substrate in Human Cells
- Experimental Relocation of the Mitochondrial Gene to the Nucleus Reveals Forces Underlying Mitochondrial Genome Evolution
- Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome
- Mutations in a P-Type ATPase Gene Cause Axonal Degeneration
- A General G1/S-Phase Cell-Cycle Control Module in the Flowering Plant
- Multiple Roles and Interactions of and in Development of the Respiratory System
- UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex
- Epigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
- Modulating the Strength and Threshold of NOTCH Oncogenic Signals by
- Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer's Disease–Related Tau Phosphorylation Via PAR-1
- Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System
- ATXN2-CAG42 Sequesters PABPC1 into Insolubility and Induces FBXW8 in Cerebellum of Old Ataxic Knock-In Mice
- Cohesin Rings Devoid of Scc3 and Pds5 Maintain Their Stable Association with the DNA
- The MicroRNA Inhibits Calcium Signaling by Targeting the TIR-1/Sarm1 Adaptor Protein to Control Stochastic L/R Neuronal Asymmetry in
- Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength
- The Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Variation of Contributes to Dog Breed Skull Diversity
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



