-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSeizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
Epilepsy is a common disabling disorder characterized by seizures with complex genetic and environmental components. The absence of a definitive pathophysiology for epilepsy stymies the development of effective treatment strategies. In a small fraction of epilepsy cases however, single gene mutations may illuminate seizure-causing mechanisms, which may open the door to the discovery of broader, more effective therapeutic strategies. We have previously shown that disruption of Prickle genes in multiple species including humans, results in a predisposition to seizures. Those findings support Prickle in a seizure-preventing role and presents a possible anti-seizure therapeutic target. We identified the deubiquitinase Usp9x (ubiquitin-specific peptidase 9 X-linked) as a new Prickle binding partner which stabilized Prickle by preventing its degradation. In mice lacking the Usp9x protein in their forebrains, Prickle2 was barely detectable. In seizure-prone prickle mutant Drosophila, reducing fat facets (Drosophila usp9x) genetically or by a small-molecule usp9x inhibitor (Degrasyn/WP1130) suppressed the seizures. We also found 2 epilepsy patients harboring mutations in USP9X. Our findings demonstrate that inhibition of Usp9x can arrest prickle-mediated seizures, and variations in USP9X may predispose to seizures. From these studies, we have elucidated a seizure-inducing mechanism, identified a potential anti-seizure target, and a potential anti-seizure drug.
Published in the journal: . PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005022
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005022Summary
Epilepsy is a common disabling disorder characterized by seizures with complex genetic and environmental components. The absence of a definitive pathophysiology for epilepsy stymies the development of effective treatment strategies. In a small fraction of epilepsy cases however, single gene mutations may illuminate seizure-causing mechanisms, which may open the door to the discovery of broader, more effective therapeutic strategies. We have previously shown that disruption of Prickle genes in multiple species including humans, results in a predisposition to seizures. Those findings support Prickle in a seizure-preventing role and presents a possible anti-seizure therapeutic target. We identified the deubiquitinase Usp9x (ubiquitin-specific peptidase 9 X-linked) as a new Prickle binding partner which stabilized Prickle by preventing its degradation. In mice lacking the Usp9x protein in their forebrains, Prickle2 was barely detectable. In seizure-prone prickle mutant Drosophila, reducing fat facets (Drosophila usp9x) genetically or by a small-molecule usp9x inhibitor (Degrasyn/WP1130) suppressed the seizures. We also found 2 epilepsy patients harboring mutations in USP9X. Our findings demonstrate that inhibition of Usp9x can arrest prickle-mediated seizures, and variations in USP9X may predispose to seizures. From these studies, we have elucidated a seizure-inducing mechanism, identified a potential anti-seizure target, and a potential anti-seizure drug.
Introduction
Mutations in the PRICKLE genes can cause seizures in humans, zebrafish, mice, and flies, suggesting the seizure-suppression pathway is evolutionarily conserved.[1–5]. Prickle binding partners have been studied extensively only in either non-neuronal vertebrate cell lines or non-neuronal tissues in the fly. (In both cases Prickles were shown to bind other WNT/PCP proteins.[6, 7]) Such targeted approaches showed Prickles interact with REST, some kinases (including BCR), and post-synaptic density proteins, including TANC1 and TANC2.[4, 6] To identify neuronal proteins that bind Prickles, recent work by our group and others showed PRICKLE1 also binds to Smurf1 (a ubiquitin ligase),[8] and Synapsin1, (a gene implicated in both epilepsy and autism)[1]; and PRICKLE2 also binds PSD-95 and p150Glued.[9] To identify other PRICKLE binding partners in neuronal-like cells, we used mass spectroscopy: a more global, unbiased approach.
Results
The interaction was monitored in a subclone of rat pheochromocytoma PC12 cells which, when treated with Nerve Growth Factor (NGF), assume a sympathetic neuron-like phenotype.[10] Clonal PC12 lines that overexpressed doxycycline-inducible GFP, GFP-PRICKLE1 (GFP-PK1), or GFP-PRICKLE2 (GFP-PK2) were produced (S1 Fig). Protein complexes immunoprecipitated with anti-GFP beads from whole-cell lysates (S1C Fig) were analyzed by mass spectrometry (IP-MS, S2 Fig).[1] Prickle interactors were considered candidates only if they recovered >10 peptide matches with both GFP-PRICKLE1 and GFP-PRICKLE2, but not with GFP alone (S1 Table). This dataset recovered both known Prickle-interactors (e.g., Tanc2, and Bcr[6]) and novel Prickle interactors, including Usp9x,[11–15] a substrate-specific de-ubiquitinase. The putative Prickle-Usp9x interaction was of particular interest because Usp9x physically interacts with Smurf1 (to date, one of the few Prickle interactors identified in neural tissues[13] and both are implicated in neurite extension).[16] Moreover, since ubiquitination plays a role in cancer pathogenesis, a variety of reagents that modulate this system are already commercially available and in clinical trials.[17] The combination of previously identified Prickle-interacting partners with the present studies are depicted in the Prickle-interactome (Fig. 1) that we utilize to identify new seizure-modifying targets.
Fig. 1. Prickle interactome. 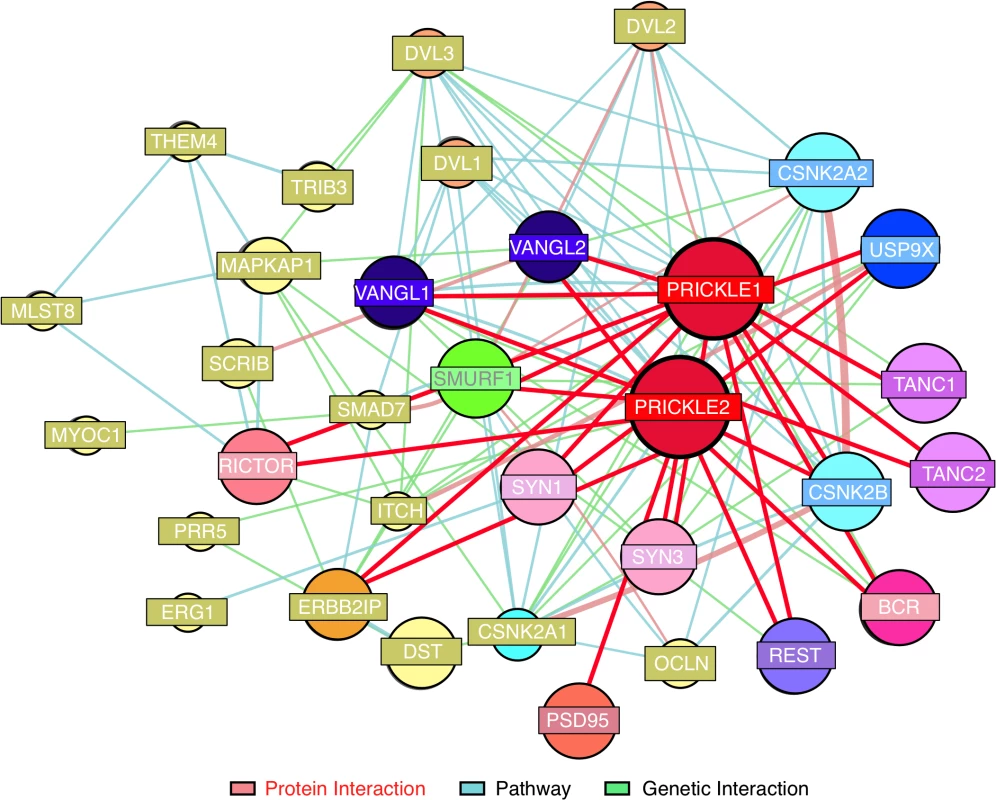
We combined findings from our proteomics interaction experiment and public databases to generate a prickle interactome. We used the MetaCore (MetaCore, GeneGO Inc., St. Joseph, MI, USA) networking function and String database 9.1 to curate interaction maps of the proteins identified. Information for identified interactions is obtained from several sources including but not limited to genomic context, database imports (PPI and pathway databases), high-throughput experiments, co-expression, and text mining. We uploaded our lists of proteins from LC-MS/MS into the software programs and exported the networks into Cytoscape 2.7.0 for manipulation of the network appearance. (Nodes, circles; Edges, lines). Red lines correspond to interactions observed by our labs using yeast-2-hybrid and IP-MS approaches. The extended interactome was generated as we have previously described. [1, 6–9, 18, 19] Prickle1 and Prickle2 interact with known synaptic proteins. The interaction with USP9X is novel. To evaluate further USP9X as a PRICKLE binding partner, coimmunoprecipitation (co-IP) assays were carried out in the original PC12 GFP, GFP-PRICKLE1, and GFP-PRICKLE2 cells lines, utilizing Tanc2 and Bcr as positive controls. To detect the respective endogenous proteins, GFP-immunoprecipitates from the differentiated cell lines were immunoblotted with anti-BCR, anti-TANC2, and anti-USP9X. Fig. 2A demonstrates interaction by all three proteins in GFP-PRICKLE1 and GFP-PRICKLE2 immunoprecipitates. To validate this interaction, co-IPs were carried out in a different cell line. Flag-tagged PRICKLE1 and PRICKLE2 were immunoprecipitated from human embryonic kidney (HEK293T) cells and subjected to anti-USP9X Western blot analysis; Fig. 2B shows both PRICKLE1 and PRICKLE2 interact with endogenous USP9X in HEK293T cells.
Fig. 2. PRICKLE interacts with USP9X via its carboxyl terminal. 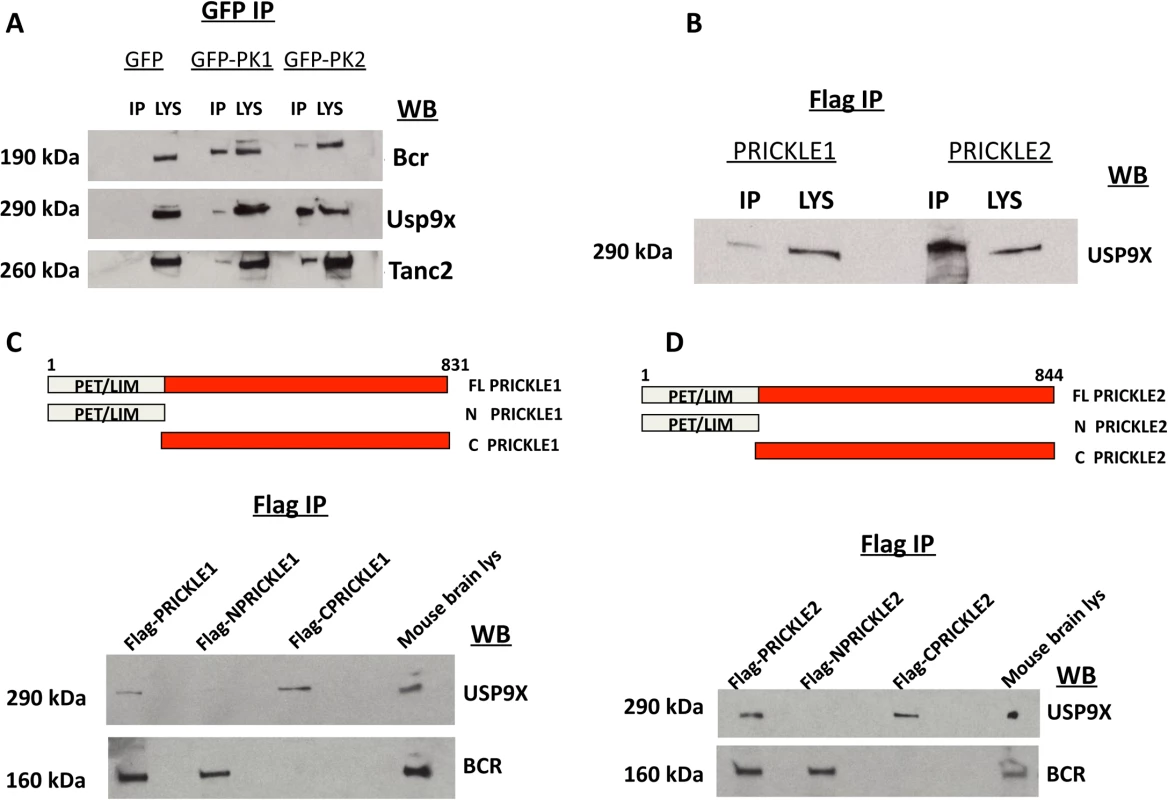
A. PRICKLE1 and PRICKLE2 interact with endogenous Bcr, Tanc2, and Usp9x in NGF-differentiated PC12 cells. GFP immunoprecipitates from stable lines expressing GFP, GFP-PRICKLE1 or GFP-PRICKLE2 confirm that PRICKLE interacts with Bcr, Usp9x and Tanc2. B. Flag immunoprecipitates, from HEK293T cells overexpressing flag-tagged PRICKLE1 or PRICKLE2, show endogenous USP9X physically interacts with PRICKLE. C, D. Schematic of PRICKLE1(C) and PRICKLE2(D) constructs. Flag-immunoprecipitates from HEK293T cells overexpressing the indicated constructs were analyzed by anti-USP9X Western blotting. Both PRICKLE 1 and 2 interact with USP9X via their C-termini while BCR binding mapped to their N-termini. To map the PRICKLE1-USP9X interacting domain, recombinant vectors expressing full-length, N-terminal or C-terminal portions of PRICKLE1 (schematic diagrams in Fig. 2C, D) were transfected into HEK293T cells and immunoprecipitated via the Flag tag. Fig. 2C showed that endogenous USP9X interacted specifically with the PRICKLE1 C-terminus. On the other hand, BCR interacted with the N-terminus, a region that includes the PET/LIM domains, previously known to mediate Prickle1 protein-protein interactions.[20] The same approach defined the PRICKLE2-USP9X interacting domain. As with PRICKLE1, while the interaction with BCR mapped to the N-terminus, the PRICKLE2–USP9X interaction mapped to the C-terminus of PRICKLE2 (Fig. 2D).
SMURF1 and USP9X are known to physically interact via the second WW domain of SMURF1 and the USP9X carboxyl terminus (a fragment named C2)[13] so, hypothetically, the PRICKLE interaction should also map to the same region. To test this, a PRICKLE1 or PRICKLE2 c-terminal fragment was expressed with one of two flag-tagged Usp9x deletion fragments: C1Usp9x (amino acids 1216–2107) and C2Usp9x (the carboxy terminal amino acids 2102–2560; Fig. 3A). Complexes were immunoprecipitated via the Flag tag. Fig. 3A shows that the PRICKLE1 and PRICKLE2 interactions also map to the carboxyl terminus of Usp9x (C2). The C-terminal end of both PRICKLEs and USP9X are therefore crucial for the PRICKLE-USP9X interaction.
Fig. 3. USP9X stabilizes PRICKLE in HEK293T cells and the mouse brain. 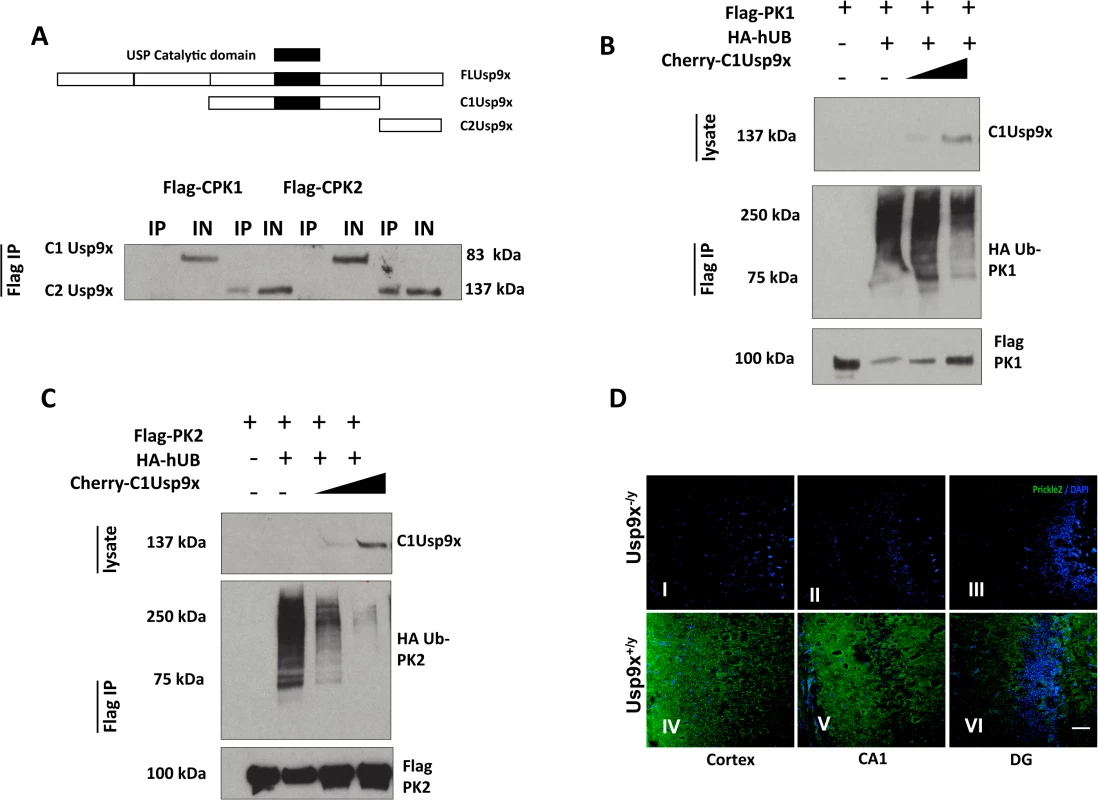
A. Flag-immunoprecipitates from HEK293T cells overexpressing the indicated constructs show PRICKLE and USP9X interact via their carboxyl termini. B, C. USP9X deubiquitinates PRICKLE. Immunoprecipitates from HEK293T cells overexpressing the indicated constructs show PRICKLE ubiquitination in the presence of HA-Ubiquitin and deubiquitination/stabilization by Usp9x. IP (immunoprecipitates), IN (input). D. Loss of Usp9X affects Prickle2 (green) expression. Deletion of Usp9x results in decreased Prickle2 expression in the cortex (I), CA1 (II) and dentate gyrus (III) of 4-week old mice when compared to controls (IV, V, VI). n = 2. Scale bar:20μM. Nuclear stain = DAPI. PRICKLE is ubiquitinated by the E3 ligase, SMURF2, which promotes PRICKLE degradation and turnover in HEK293T cells.[8] Ubiquitin tags from mono - and polyubiquitinated proteins can be removed by the C1-terminal catalytic motif of USP9X deubiquitinase.[12, 13, 21–23] We postulated the physical interaction between the PRICKLEs and USP9X might indicate that PRICKLE was a USP9X substrate, and that USP9X deubiquitinates and stabilizes both PRICKLE1 and PRICKLE2. To test this idea, PRICKLE substrates were monitored in USP9X deubiquitination assays.
In previous studies, overexpression of the C1Usp9x catalytic fragment alone was sufficient to deubiquitinate USP9X substrates.[23] Accordingly, the robust ubiquitination of Flag-PRICKLE1 (Fig. 3B, C) was antagonized by overexpression of C1Usp9x; and PRICKLE was stabilized (Fig. 3). Similar results were obtained with full-length USP9X (S3 Fig). In the presence of the proteasomal inhibitor LLNL (N-acetyl-L-leucyl-L-leucyl-L-norleucinal), polyubiquitinated PRICKLE1 or PRICKLE2 accumulated, indicating that degradation of ubiquitinated PRICKLE is mediated by the proteasome as opposed to the lysosome (S3 Fig). To determine if a Prickle-Usp9x interaction has a functional effect in mice, we generated mice with deletion of Usp9x in the forebrain (Emx1-Cre/Usp9x loxtemp)[12] and examined expression of Prickle2. In the absence of Usp9x in 4-week old mice, Prickle2 was not detected in the cortex, CA1 region of the hippocampus, and dentate gyrus (Fig. 3D). Taken together, these data suggest that Prickles are novel Usp9x substrates.
Genetic analysis was used to determine if Prickle and Usp9x interact genetically in fruit flies. We reduced the dosage of the USP9X orthologue (fat facets, or faf) in the context of a prickle mutation (pksple/+) which on its own promotes both behavioral and electrophysiologic seizure activity.[3, 5] Three separate loss-of-function faf alleles were used to create transheterozygotes, which were then assayed with the bang sensitivity behavioral assay to assess recovery time from seizure activity. For every faf allele used, reducing its level suppressed the seizures in the context of the pksple/+ flies (Fig. 4A-C). Adding the USP9X small-molecule inhibitor Degrasyn/WP1130[24] to the fly food inhibited seizure activity in pksple homozygous flies (Fig. 4D) using a “fly flip” assay (see Materials and Methods).
Fig. 4. Genetic and pharmacological suppression of the seizure phenotype in prickle mutant flies. 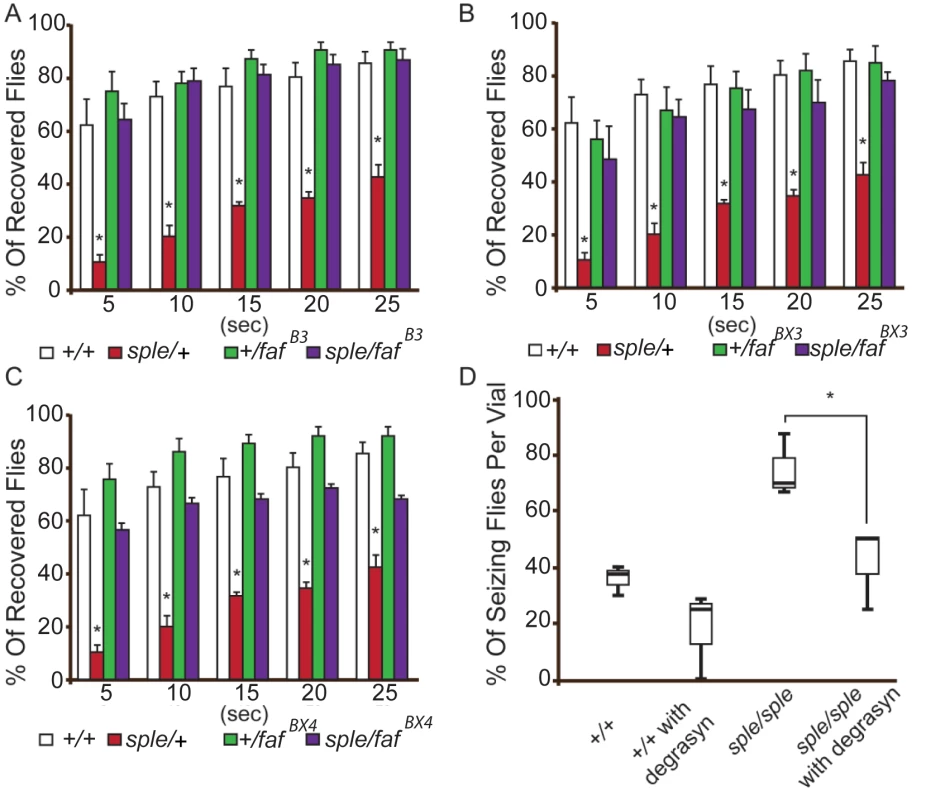
(A–C) Reducing faf dosage with three separate loss-of-function alleles ((fafB3, fafBX3, fafBX4; graphs A, B and C, respectively) suppresses the pksple-mediated seizure phenotype detected by the modified bang sensitivity assay. Six vials per genotype were assayed. D. Inhibition of faf activity with Degrasyn suppresses pksple-mediated seizure activity detected using the “fly flip” assay. Three vials per genotype were assayed. Note: +/+ = Oregon-R control flies; sple = pksple; sple/faf = pksple/+; +/faf. * = p < 0.05. Error bars = standard error. Each vial contains 5 males and 5 females. The demonstration that pharmacologic treatment with a USP9X inhibitor suppresses seizures in conjunction with genetic seizure suppression utilizing fly USP9X orthologue mutations suggests that USP9X mutations could protect humans from seizures. In contrast to this suggestion, in humans, USP9X has been postulated to be an epilepsy candidate gene. USP9X is X-linked; and in males has been associated with both autism spectrum disorder (ASD) and intellectual disability (which are frequently co-morbid with epilepsy). Moreover, mice with Usp9x-deficient neurons develop abnormal neuronal connectivity.[12, 14, 25, 26] A similar apparent discrepancy has been observed in the case of the SCN1A gene (which codes for the NAV1.1 channel) and its fly homologue para. Here, the great majority of 68 different mutations in para suppress seizures in the fly,[27–31] yet human SCN1A mutations are associated with severe epileptic encephalopathies, febrile seizure syndromes, and Dravet syndrome. Recent studies demonstrated that although loss-of-function para alleles suppress seizures, a few recently identified gain-of-function para alleles (e.g., parabss1) actually cause seizures in the fly.[27] Accordingly, many of the most severe human SCN1A mutations are missense or truncation, gain-of-function alleles (see web resources).[32]
To assess if USP9X coding variants might be associated with human seizures, the USP9X gene was resequenced in 284 male patients with epileptic encephalopathy. One male patient, T17133, presented with epileptic encephalopathy and was found to carry a de novo USP9X mutation (c.3034T>C, p.Ser1012Pro) at a highly conserved residue (GERP 5.62) that was predicted to be possibly damaging by PolyPhen (0.898). Another patient with infantile spasms (parental genotypes unavailable) harbored variant (T2587c.5669G>A, p.Gly1890Glu) a rare missense variant. Both mutations lie outside the identified Prickle-binding domain. The USP9X pSer1012 is very close to the Ubiquitin-like (UBL) domain; and the USP9X Gly1890 is in UCH (Ubiquitincarboxyl-terminal hydrolase) domain, just eleven amino acids from the proton acceptor in the active site, and in between the catalytic Cys and His motifs that form the catalytic domain, suggesting that both mutations likely alter normal USP9X function. Rare coding variants were sought in the cohort of 284 males with epilepsy and the male exomes in the NHLBI exome variant server EVS (see web resources) in a one-tailed Chi-squared analysis. For the combined African American and European American cohorts, we found a statistical association (p = 0.0350) between USP9X rare coding variants and epilepsy (S2 Table). This further supports the assertion that USP9X may be a novel epilepsy gene.
Discussion
Our studies suggest USP9X-mediated de-ubiquitination stabilizes PRICKLE proteins in the nervous system. These results are consistent with results in other tissues, where faf RNAi reduced Prickle levels in Drosophila apicolateral junctions,[33] as well as studies showing USP9X stabilizes several proteins involved in various aspects of neuronal development and cancer.[12] Notably, USP9X was shown to stabilize oncoproteins (e.g., MCL-1 and p53), sparking interest in modulating USP9X in tumors, with promising results.[24]
Here, we showed that human USP9X mutations are associated with epilepsy. Our data corroborate other evidence showing USP9X mutations in males with co-morbid epilepsy conditions (e.g., intellectual disability and ASD). In addition to USP9X, several other members of our identified Prickle-interactome were already implicated in seizures, intellectual disability, and ASD. (For example, a TANC2 variant in a patient with intellectual disability and febrile seizures[34] and in a patient with ASD,[35] and a SMURF1 variant in a patient with epileptic encephalopathy).[36]
PRICKLE mutant humans, mice, zebrafish, and Drosophila all exhibit seizures.[3, 5] In flies, the prickle seizure phenotype can be genetically suppressed by reducing Faf (creating transheterozygotes) or pharmacologically rescued by treating pksple mutants with Degrasyn/WP1130. Although USP9X is genetically associated with seizures in both flies and humans, mutations are seizure-protective in the flies but seizure-inducing in humans. This opposing effect is likely because the identified human USP9X mutations are all protein-changing mutations that reside outside of the Prickle-interaction domain whereas the fly faf mutations that suppress seizures are all amorphic alleles. This suggests that, for USP9X, complete loss of function may have a different phenotype than protein coding alleles. Before our study, neither USP9X inhibition nor stimulation was identified as a potential anti-seizure pathway. Yet, given the evolutionarily conservation of the Prickle-pathway, our results suggest Degrasyn/WP1130 and other USP9X-modulating molecules should be pursued as novel therapeutic agents for patients suffering from seizures.
Materials and Methods
Antibodies and antibody-conjugated agarose beads
The antibodies employed were rabbit polyclonal anti-BCR (Santa Cruz), rabbit polyclonal anti-TANC2 (Bethyl), polyclonal rabbit-anti USP9X (Abcam), mouse monoclonal anti-β-actin (Sigma), mouse monoclonal anti-Flag (Sigma), rat monoclonal anti-HA-peroxidase (Roche), rabbit polyclonal anti-dsRed from Clontech (detects mCherry) mouse monoclonal anti-Myc (Santa Cruz), rabbit polyclonal anti-GFP (Santa Cruz). Anti-rabbit and anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibodies (Thermo Scientific). The agarose antibody-conjugated beads were protein A/G beads (Pierce), anti-Flag (Santa Cruz), and anti-GFP
Plasmids
Flag-PRICKLE1 plasmid was as previously described.[4] The N-terminus of PRICKLE1 (aa 1 to 313) and the C-terminus of PRICKLE1 (aa 314 to 831) were each Flag-tagged on the C-terminus and cloned into EcoRI (5’) and KpnI (3’) restriction sites of the pcDNA3.1 vector. The N-terminus (aa 1 to 317) and C-terminus of PRICKLE2 (aa 318 to 844) were also Flag-tagged on the C-termini and cloned into EcoRI (5’) and KpnI (3’) restriction sites of the same vector.
Using previous studies as a guide, we cloned deletion constructs of murine Usp9x.[23] C1Usp9x aa 1216–2107 (with the catalytic domain) and C2Usp9x aa 2102–2560 (the carboxyl terminus) were cloned into the BamH1 and BSpe1 restriction sites of the pmCherryC1 plasmid (Clontech). The previously described HA-UbiquitinC plasmids were kindly gifted by Pedro Gonzalez-Alegre (University of Iowa, Iowa City).
Tissue culture
HEK293T[37] cells were cultured in DMEM (Dulbecco’s modified eagle medium; Gibco) supplemented with 10% FBS (fetal bovine serum) and 1% penicillin-streptomycin (Gibco). PC12 cells were cultured in collagen-coated plates in RPMI (Roswell Park Memorial Institute medium) supplemented with 5% FBS, 10% HS (horse serum), 1% penicillin-streptomycin, hygromycin (100μg/ml), and blasticidin (5μg/ml). Cells were maintained in a humidified 37°C, 5% CO2 incubator. Transgene expression and differentiation were induced by 1.5μg/ml dox and 100 ng/ml NGF treatment in low serum RPMI medium (2% HS, 1% FBS) respectively.
Generation of PC12 inducible lines expressing GFP, GFP-PRICKLE1 or GFP-PRICKLE2
PC12 neuronal-like cell lines are used as a model system for investigating neuronal differentiation in culture.[10] As reported before, a PC6-3 (sub-clone of PC12 cells) clonal line, stably expressing the tet-repressor protein pcDNA6/TR (PC6-3/TR), was generously provided by Pedro Gonzalez-Alegre (University of Iowa, Iowa City).[1] Clonal cell lines inducibly expressing GFP, GFP-PRICKLE1 or GFP-PRICKLE2 were generated as previously described.[1, 38] GFP, GFP-PRICKLE1, and GFP-PRICKLE2 cDNAs were cloned into EcoRI and XhoI restriction sites of pcDNA5 TO (Invitrogen). Plasmids were transfected into the PC6-3/TR cells, with Lipofectamine 2000 (Qiagen), according to the manufacturer’s instructions. Clonal cells stably expressing the transgenes were selected (medium supplemented with hygromycin at 100μg/ml) and blasticidin (5μg/ml) and screened for doxycycline inducibility by fluorescence microscopy and anti-GFP Western blots.
Western blots
Dox-treated and untreated PC12 cells expressing GFP, GFP-PRICKLE1, or GFP-PRICKLE2 were lysed in ice-cold, NET-100 buffer (Tris 50mM, NaCl 100mM, EDTA 5mM supplemented with protease inhibitor (1X EDTA-free Complete Mini-tabs protease inhibitor cocktail from Roche). Equal amounts of proteins were resolved by sodium dodecyl sulfate-acrylamide gel electrophoresis (SDS-PAGE) in 4–20% acrylamide gels (pre-cast gels, Biorad) and transferred onto a Polyvinylidene fluoride (PVDF) membrane for 3 hours at 0.30 Amps. The membrane was blocked in 5% non-fat milk for 2 hours at room temperature followed by incubation in anti-GFP antibody at 1 : 1000 dilution overnight at 4°C. The membrane was then washed in TBST (Tris-buffered saline with Tween-20) at room temperature, followed by incubation in (horseradish) HRP-conjugated goat-anti-rabbit antibody (1 : 10 000) at room temperature for 2 hours. The blots were developed using ECL detection kit (Thermo Scientific) after washing, as per the manufacturer’s instructions. Signals were captured on x-ray films.
Immunoprecipitation
Immunoprecipitations were carried out as previously described.[1, 38] Differentiated, dox-treated GFP, GFP-PRICKLE1, and GFP-PRICKLE2 PC12 cell lines were lysed in ice-cold NET-100 buffer supplemented with a protease inhibitor. Lysates were immunoprecipitated overnight with GFP-conjugated agarose beads after 1 hour of pre-clearing in A/G agarose beads. After 5 X 5 minute washes in NET-100 buffer, immunoprecipitates were eluted in 2X Laemmli buffer at 100°C for 5 minutes. Equal volumes of immunoprecipitates were resolved by SDG-PAGE in 4–20% acrylamide gels and then silver-stained.
Silver staining
Following electrophoresis, a gel with resolved GFP immunoprecipitates was incubated in fixing solution (50% methanol, 10% acetic acid) for 30 minutes at room temperature and washed overnight in water. The gel was then incubated in 100mL of sodium thiosulphate solution (0.33g sodium thiosulphate/ 1L water) for 120 seconds, followed by 3 X 30-second washes in water. This was followed by incubation in silver nitrate solution (0.2g silver nitrate/100mL water) for 30 minutes. The gel was then washed in distilled water for 3 X 60 seconds, and then incubated in developing solution (3g sodium carbonate, 50μl formaldehyde, 2mL sodium thiosulphate solution, 93mL water) until proteins bands became visible. The stop solution (7g EDTA/500ML water) was added to the gel and shaken for 10 minutes to stop further development.
Mass spectrometry LC-MS/MS
This was carried out as previously described.[1]
SDS-PAGE. 20 μL of clarified, soluble GFP, GFP-PRICKLE1, and GFP-PRICKLE2 immunoprecipitates were added to denaturing, SDS-PAGE, loading buffer (containing glycerin, beta-mercaptoethanol, and SDS in Tris buffer) and boiled for five minutes in preparation for electrophoresis. Bio-Rad precast 4–20% Tris-HCl gradient SDS-PAGE gels were run at 150 V for 45 minutes. Gels were then stained with Bio-Rad Flamingo fluorescent stain and imaged using a UVP PhotoDoc-It UV Imaging System (Upland, CA) followed by LC-MS/MS as described.
Data analysis
MS/MS data were analyzed and matched to rat protein sequences in the Swiss Prot and TrEMBL database, using the MASCOT 2.4 database search engine (Matrix Science, UK) with carbamidomethyl as a fixed modification and oxidation as a single variable modification. Mass window for the parent ions were set to 1.8 m/z and 0.4 m/z for MS/MS data. The same spectra were searched using similar restrictions with the SpectrumMill algorithm (Agilent) and alignments were merged and curated using Scaffold v3.6.4. A minimum peptide-ion score cut-off was set at 9 in SpectrumMill and the presence of at least six consecutive y - or b-ions was required. Alignments reported from Scaffold were restricted to a false discover rate of less than 1%, with peptide and protein confidence >90% and at least two unique peptides required.
Network analysis
We used the MetaCore (MetaCore, GeneGO Inc., St. Joseph, MI, USA) networking function and String database 9.1 to curate interaction maps of the proteins identified. Information for identified interactions is obtained from several sources including but not limited to genomic context, database imports (PPI and pathway databases), high-throughput experiments, co-expression, and text mining. We uploaded our lists of proteins from LC-MS/MS into the software programs and exported the networks into Cytoscape 2.7.0 for manipulation of the network appearance.
Co-immunoprecipitation
HEK293T[37] cells were transfected with Flag-PRICKLE1 or Flag-PRICKLE2 with Polyfect (Qiagen), according to the manufacturer’s protocol. Cells were lysed in ice-cold NET-100 buffer after 48 hrs of incubation and immunoprecipitated overnight with anti-Flag beads at 4°C. Beads were washed for 5 minutes x 5 times in ice-cold NET-100 buffer. Immunoprecipitates were resolved by SDS-PAGE on a 4–20% gel and then subjected to anti-USP9X (1 : 500) immunoblot analysis. HRP-conjugated anti-rabbit secondary antibody was used at a 1 : 2000 dilution. For mapping studies, HEK293T cells transfected with Flag-PRICKLE1, Flag-NPRICKLE1 or Flag-CPRICKLE1; Flag-PRICKLE2, Flag-NPRICKLE2 or Flag-CPRICKLE2 with Polyfect (Qiagen). Transfected cells were treated as described above. Lysates were also immunoprecipitated overnight with anti-Flag beads, eluates resolved by SDS-PAGE and then subjected to anti-USP9X Western blot analysis. Membranes were stripped and reprobed with anti-BCR antibodies (1 : 1000) overnight. HRP-conjugated anti-rabbit secondary antibody was used at a 1 : 10000 dilution. Blots were developed; and signals captured on X-ray films.
Ubiquitination/deubiquitination assays
Flag-PRICKLE1 only, Flag-PRICKLE1 + HA-UbiquitinC or Flag-PRICKLE1 + HA-UbiquitinC + mCherryC1Usp9x were transfected into HEK293T cells and incubated for 48 hours. Cells were lysed and PRICKLE1 immunoprecipitated overnight via the Flag tag and resolved by SDS-PAGE. Ubiquitinated and de-ubiquitinated PRICKLE1 was detected by anti-HA (1 : 1000) Western blot analysis. The membrane was incubated in anti-HA-peroxidase at room temperature for 2 hours and washed for 4 X 10 minutes in TBST. Blots were developed; and signals captured on X-ray films. The process was repeated for PRICKLE2 where Flag-PRICKLE2 only, Flag-PRICKLE2 + HA-UbiquitinC or Flag-PRICKLE2 + HA-UbiquitinC + mCherryC1Usp9x were transfected. The process was repeated with full-length USP9X with and without the proteasome inhibitor LLNL (N-acetyl-L-leucyl-L-leucyl-L-norleucinal) at a concentration of 50μm overnight.
Web resources
Ethics statement
Mice subjects
All mouse work was done under ethical clearance from the Griffith University, and The Women’s and Children’s Health Network, and the South Australian Pathology Department Animal Ethics Committees. All protocols used were in accordance with the policy and guidelines of the National Health and Medical Research Council of Australia. Personnel and investigators who handled the mice were properly trained and qualified. Animals were treated humanely, discomfort and distress were minimized during the experiments and animals were monitored for signs of pain and distress. All euthanasia was performed using cervical dislocation.
Human subjects
All human DNA samples were collected with proper informed consent and with approval from an institutional review board (IRB).
Generation of Emx1-Cre mouse
To generate mice in which Usp9X was conditionally deleted from the dorsal telencephalon, Usp9XloxP/loxP female mice were crossed with heterozygous Emx1-Cre males as described previously.8 Under this breeding regime, all male offspring receive a Usp9X loxP gene. Male offspring positive for Cre were used as Usp9X cKO and Cre negative male littermates were used as controls.[12]
Mouse genotyping
Mouse genotyping was performed on DNA extracted from tail tips and PCR was performed using RedExtract tissue PCR Kit (Sigma-Aldrich) as per the manufacturer’s instructions. Primers were designed to detect Cre-recombinase, F: CTGACCGTACACCAAAATTTGCCTG; R: GATAATCGCGAACATCTTCAGGTTC. Male embryos were identified using primers for the SRY region of the Y chromosome, F: GAGGCACAAGTTGGCCCAGCA; R: GGTTCCTGTCCCACTGCAGAAG.
Perfusion and Cryo-sectioning
Four-week old mice were anesthetized, perfused trans-cardially with 4% paraformaldehyde (PFA), then heads were drop-fixed in 4% PFA. Following fixation brains were cryo-protected in 15% sucrose at 4°C overnight, then in 30% sucrose at 4°C overnight, 1 : 1 ratio of 30% sucrose and O.C.T at 4°C overnight and frozen in Tissue-Tek O.C.T. compound (Sakura), then sectioned at 10 μm on a cryostat.
Immunohistochemistry
The coronal mouse brain sections were permeabilised in 1% sodium dodecyl sulphate (SDS) in 1X PBS for 4 minutes at room temperature. After washing 3 times with PBS, the samples were blocked in 2% Bovine serum albumin (BSA) in 1X PBS for 20 minutes, incubated with rabbit anti-Prickle2 antibody at 1/250 dilution in 2% BSA/PBS at 4°C overnight, then incubated with 1/200 dilution of goat anti-rabbit biotinylated antibody (Invitrogen) in 2% BSA/PBS for 1h and in 1/400 streptavidin-biotin-peroxidase complex (Invitrogen) in 2% BSA/PBS for 1h at room temperature, and mounted with Vectashield mounting medium with DAPI (Vector Laboratories). Images were obtained on Olympus FV1000 confocal microscope.
Fly lines
Oregon-R (OR) and outcrossed prickle-spiny-legs (pksple) lines are referred to in Ehaideb et al.[3] fat facets (faf) lines were obtained from the Bloomington Drosophila Stock Center at Indiana University (line numbers 25100 (w[*]; st1 fafB3/TM6B, Tb1), 25101 (w[*]; st1 fafBX3/TM6B, Tb1), 25107 (w[*]; st[1] faf[BX4]/TM6B, Tb[1])).
Modified bang-sensitivity behavioral assay
As previously described[3].
Drug treatment
Newly eclosed flies were collected and maintained on standard fly food for two days. On the third day, flies were starved for 6 hours by placing them in glass vials containing damp cotton. Three vials of each genotype were then transferred to vials containing 4 ml of standard fly food mixed with 40 μl of 100 mM Degrasyn (final concentration 1 mM; WP1130, Selleck Chemicals) and aged until the flies were 7 days old, before subjecting them to the fly-flip assay. Each vial contained ∼10 flies (5 males and 5 females) maintained at 25 degrees C.
“Fly flip” seizure assay
Seven-day old flies were transferred to an empty vial and the vial Flug (fly plug; Flystuff.com) was pushed downward until a gap of 2 cm (from the bottom of the vial to the Flug) was created, preventing flies from moving out of camera frame in order to keep track of individual flies. Six vials of flies of each genotype were then mechanically stimulated with a vortex mixer (Fisher Scientific Vortex Mixer; maximal setting of 10) for 5 seconds. After vortexing, flies were digitally recorded for 1 minute (from the start of the vortex); only flies that flipped on their back and remained at that position for at least one second were considered flies exhibiting muscle-jerk seizure activity. Tester was blinded to genotype for all tests. Flies were randomly sorted into placebo versus drug group.
Targeted resequencing of USP9X
We captured all USP9X exons and at least five base pairs of flanking sequence using molecular inversion probes, and performed next generation sequencing and data analysis as described previously.[39] Our resequencing cohort consisted of 284 males diagnosed with epileptic encephalopathy according to the International League Against Epilepsy (ILAE) classification criteria.[40] We considered only non-synonymous, frameshift or splice-site mutations that were rare (<2%) in the ESP6500 data set (see web resources) for further analysis. Segregation analysis using Sanger sequencing was performed where possible for variants that met these criteria, and paternity/maternity confirmed using the PowerPlex S5 system [Promega].
Characterization of USP9X variants in the exome variant server
USP9X variants were downloaded from the EVS website. Rare (<2%), hemizygous (male), non-synonymous variants considered damaging or possibly damaging by Polyphen were tabulated for the combined European-American and African-American populations and used for statistical analysis. In addition, one frameshift mutation not qualified by polyphen was included, for a total of four damaging protein-changing variants.
Protein accession numbers
Human-USP9X: Accession: NP_001034679.2 GI: 145309309, mouse-Usp9x; NP_033507.2 GI: 115511018; human-PRICKLE1 NP_001138354.1 GI: 23308518; human-PRICKLE2 NP_942559.1_GI: 38524620; Drosophila prickle NM_165508.2 GI: 442622668.
Statistics
To assess statistically enrichment for USP9X coding variants in the epilepsy cohort, a one-tailed chi-square test without Yates correction was performed using the number of mutant and wild-type alleles from male epilepsy patients and EVS male populations. For the EVS counts, the average number of individuals genotyped per variant in the database was used as the total genotype count in the chi-square test. Student’s t-tests were employed for the Drosophila ‘fly flip’ and modified bang sensitivity assays. Power to detect variants for both the human cohort and the fly assays was based on our previously published studies.[5]
Supporting Information
Zdroje
1. Paemka L, Mahajan VB, Skeie JM, Sowers LP, Ehaideb SN, Gonzalez-Alegre P, et al. PRICKLE1 interaction with SYNAPSIN I reveals a role in autism spectrum disorders. PLoS One. 2013; 8: e80737. doi: 10.1371/journal.pone.0080737 24312498
2. Sowers LP, Loo L, Wu Y, Campbell E, Ulrich JD, Wu S, et al. Disruption of the non-canonical Wnt gene PRICKLE2 leads to autism-like behaviors with evidence for hippocampal synaptic dysfunction. Mol Psychiatry. 2013; 18 : 1077–1089. doi: 10.1038/mp.2013.71 23711981
3. Ehaideb SN, Iyengar A, Ueda A, Iacobucci GJ, Cranston C, Bassuk AG, et al. prickle modulates microtubule polarity and axonal transport to ameliorate seizures in flies. Proc Natl Acad Sci U S A. 2014; 111 : 11187–11192. doi: 10.1073/pnas.1403357111 25024231
4. Bassuk AG, Wallace RH, Buhr A, Buller AR, Afawi Z, Shimojo M, et al. A homozygous mutation in human PRICKLE1 causes an autosomal-recessive progressive myoclonus epilepsy-ataxia syndrome. Am J Hum Genet. 2008; 83 : 572–581. doi: 10.1016/j.ajhg.2008.10.003 18976727
5. Tao H, Manak JR, Sowers L, Mei X, Kiyonari H, Abe T, et al. Mutations in prickle orthologs cause seizures in flies, mice, and humans. Am J Hum Genet. 2011; 88 : 138–149. doi: 10.1016/j.ajhg.2010.12.012 21276947
6. Daulat AM, Luu O, Sing A, Zhang L, Wrana JL, McNeill H, et al. Mink1 regulates beta-catenin-independent Wnt signaling via Prickle phosphorylation. Mol Cell Biol. 2012; 32 : 173–185. doi: 10.1128/MCB.06320-11 22037766
7. Shimojo M, Hersh LB. REST/NRSF-interacting LIM domain protein, a putative nuclear translocation receptor. Mol Cell Biol. 2003; 23 : 9025–9031. 14645515
8. Narimatsu M, Bose R, Pye M, Zhang L, Miller B, Ching P, et al. Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell. 2009; 137 : 295–307. doi: 10.1016/j.cell.2009.02.025 19379695
9. Shimojo M. Huntingtin regulates RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) nuclear trafficking indirectly through a complex with REST/NRSF-interacting LIM domain protein (RILP) and dynactin p150 Glued. J Biol Chem. 2008; 283 : 34880–34886. doi: 10.1074/jbc.M804183200 18922795
10. Pittman RN, Wang S, DiBenedetto AJ, Mills JC. A system for characterizing cellular and molecular events in programmed neuronal cell death. J Neurosci. 1993; 13 : 3669–3680. 8396168
11. Jolly LA, Taylor V, Wood SA. USP9X enhances the polarity and self-renewal of embryonic stem cell-derived neural progenitors. Mol Biol Cell. 2009; 20 : 2015–2029. doi: 10.1091/mbc.E08-06-0596 19176755
12. Stegeman S, Jolly LA, Premarathne S, Gecz J, Richards LJ, Mackay-Sim A, et al. Loss of Usp9x disrupts cortical architecture, hippocampal development and TGFbeta-mediated axonogenesis. PLoS One. 2013; 8: e68287. doi: 10.1371/journal.pone.0068287 23861879
13. Xie Y, Avello M, Schirle M, McWhinnie E, Feng Y, Bric-Furlong E, et al. Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J Biol Chem. 2013; 288 : 2976–2985. doi: 10.1074/jbc.M112.430066 23184937
14. Homan CC, Kumar R, Nguyen LS, Haan E, Raymond FL, Abidi F, et al. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am J Hum Genet. 2014; 94 : 470–478. doi: 10.1016/j.ajhg.2014.02.004 24607389
15. Xu J, Taya S, Kaibuchi K, Arnold AP. Spatially and temporally specific expression in mouse hippocampus of Usp9x, a ubiquitin-specific protease involved in synaptic development. J Neurosci Res. 2005; 80 : 47–55. 15723417
16. Chen H, Polo S, Di Fiore PP, De Camilli PV. Rapid Ca2+-dependent decrease of protein ubiquitination at synapses. Proc Natl Acad Sci U S A. 2003; 100 : 14908–14913. 14657369
17. Micel LN, Tentler JJ, Smith PG, Eckhardt GS. Role of ubiquitin ligases and the proteasome in oncogenesis: novel targets for anticancer therapies. J Clin Oncol. 2013; 31 : 1231–1238. doi: 10.1200/JCO.2012.44.0958 23358974
18. Fischer-Vize JA, Rubin GM, Lehmann R. The fat facets gene is required for Drosophila eye and embryo development. Development. 1992; 116 : 985–1000. 1295747
19. Darbro BW, Mahajan VB, Gakhar L, Skeie JM, Campbell E, Wu S, et al. Mutations in extracellular matrix genes NID1 and LAMC1 cause autosomal dominant Dandy-Walker malformation and occipital cephaloceles. Human mutation. 2013; 34 : 1075–1079. doi: 10.1002/humu.22351 23674478
20. Dawid IB, Breen JJ, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends in genetics: TIG. 1998; 14 : 156–162. 9594664
21. Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010; 463 : 103–107. doi: 10.1038/nature08646 20023629
22. Grou CP, Francisco T, Rodrigues TA, Freitas MO, Pinto MP, Carvalho AF, et al. Identification of ubiquitin-specific protease 9X (USP9X) as a deubiquitinase acting on ubiquitin-peroxin 5 (PEX5) thioester conjugate. J Biol Chem. 2012; 287 : 12815–12827. doi: 10.1074/jbc.M112.340158 22371489
23. Taya S, Yamamoto T, Kano K, Kawano Y, Iwamatsu A, Tsuchiya T, et al. The Ras target AF-6 is a substrate of the fam deubiquitinating enzyme. J Cell Biol. 1998; 142 : 1053–1062. 9722616
24. Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010; 70 : 9265–9276. doi: 10.1158/0008-5472.CAN-10-1530 21045142
25. Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009; 41 : 535–543. doi: 10.1038/ng.367 19377476
26. Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder: Supplementary Table 2. Nature. 2014.
27. Parker L, Padilla M, Du Y, Dong K, Tanouye MA. Drosophila as a model for epilepsy: bss is a gain-of-function mutation in the para sodium channel gene that leads to seizures. Genetics. 2011; 187 : 523–534. doi: 10.1534/genetics.110.123299 21115970
28. Suzuki DT, Grigliatti T, Williamson R. Temperature-sensitive mutations in Drosophila melanogaster. VII. A mutation (para-ts) causing reversible adult paralysis. Proc Natl Acad Sci U S A. 1971; 68 : 890–893. 5280526
29. Siddiqi O, Benzer S. Neurophysiological defects in temperature-sensitive paralytic mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1976; 73 : 3253–3257. 184469
30. Wu CF, Ganetzky B. Genetic alteration of nerve membrane excitability in temperature-sensitive paralytic mutants of Drosophila melanogaster. Nature. 1980; 286 : 814–816. 6250083
31. Kuebler D, Zhang H, Ren X, Tanouye MA. Genetic suppression of seizure susceptibility in Drosophila. J Neurophysiol. 2001; 86 : 1211–1225. 11535671
32. VIB Department of Molecular Genetics. SCN1A Variant Database Antwerp, Belgium: University of Antwerp; 2009 [updated 22 June 2011; cited 2014 13 Nov]. The Variation Database of SCN1A was made public in February 2009. The SCN2001A variation database aims at collecting all variations in the voltage-gated sodium channel Nav2001.2001. Variations are collected from the literature and by direct submission through this website.]. Available from: http://www.molgen.ua.ac.be/SCN1AMutations/Home/Default.cfm.
33. Strutt H, Searle E, Thomas-Macarthur V, Brookfield R, Strutt D. A Cul-3-BTB ubiquitylation pathway regulates junctional levels and asymmetry of core planar polarity proteins. Development. 2013; 140 : 1693–1702. doi: 10.1242/dev.089656 23487316
34. de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012; 367 : 1921–1929. doi: 10.1056/NEJMoa1206524 23033978
35. Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012; 74 : 285–299. doi: 10.1016/j.neuron.2012.04.009 22542183
36. De Novo Mutations in Synaptic Transmission Genes Including DNM1 Cause Epileptic Encephalopathies. American journal of human genetics. 2014; 95 : 360–370. doi: 10.1016/j.ajhg.2014.08.013 25262651
37. Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. The Journal of general virology. 1977; 36 : 59–74. 886304
38. Gonzalez-Alegre P, Paulson HL. Aberrant cellular behavior of mutant torsinA implicates nuclear envelope dysfunction in DYT1 dystonia. J Neurosci. 2004; 24 : 2593–2601. 15028751
39. Carvill GL, Heavin SB, Yendle SC, McMahon JM, O'Roak BJ, Cook J, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet. 2013; 45 : 825–830. doi: 10.1038/ng.2646 23708187
40. Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, van Emde Boas W, et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia. 2010; 51 : 676–685. doi: 10.1111/j.1528-1167.2010.02522.x 20196795
Štítky
Genetika Reprodukční medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 3
-
Všechny články tohoto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



