-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaHDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
Huntington’s disease (HD) is a neurodegenerative disorder in which the mutation results in an extra-long tract of glutamines that causes the huntingtin protein to aggregate. It is characterized by neurological symptoms and brain pathology, which is associated with nuclear and cytoplasmic protein aggregates and with transcriptional deregulation. Despite the fact that HD has been recognized principally as a neurological disease, there are multiple studies indicating that peripheral pathologies including cardiac dysfunction and skeletal muscle atrophy, contribute to the overall progression of HD. To unravel the cause of the skeletal muscle dysfunction, we applied a wide range of molecular and physiological methods to the analysis of two well established genetic mouse models of this disease. We found that symptomatic animals developed muscle dysfunction characterised by a change in the contractile characteristics of fast twitch muscles and a decrease in twitch and tetanic force of hindlimb muscles. In addition, there is a significant decrease in the number of motor units innervating the EDL muscle, and this motor unit loss progresses during the course of the disease. These changes were accompanied by the re-expression of contractile transcripts and markers of muscle denervation such as the HDAC4-Dach2-myogenin axis, as well as the apparent deterioration in energy metabolism and decreased oxidation. Therefore, we conclude, that the HD-related skeletal muscle atrophy is accompanied by progressive loss of functional motor units.
Published in the journal: . PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005021
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005021Summary
Huntington’s disease (HD) is a neurodegenerative disorder in which the mutation results in an extra-long tract of glutamines that causes the huntingtin protein to aggregate. It is characterized by neurological symptoms and brain pathology, which is associated with nuclear and cytoplasmic protein aggregates and with transcriptional deregulation. Despite the fact that HD has been recognized principally as a neurological disease, there are multiple studies indicating that peripheral pathologies including cardiac dysfunction and skeletal muscle atrophy, contribute to the overall progression of HD. To unravel the cause of the skeletal muscle dysfunction, we applied a wide range of molecular and physiological methods to the analysis of two well established genetic mouse models of this disease. We found that symptomatic animals developed muscle dysfunction characterised by a change in the contractile characteristics of fast twitch muscles and a decrease in twitch and tetanic force of hindlimb muscles. In addition, there is a significant decrease in the number of motor units innervating the EDL muscle, and this motor unit loss progresses during the course of the disease. These changes were accompanied by the re-expression of contractile transcripts and markers of muscle denervation such as the HDAC4-Dach2-myogenin axis, as well as the apparent deterioration in energy metabolism and decreased oxidation. Therefore, we conclude, that the HD-related skeletal muscle atrophy is accompanied by progressive loss of functional motor units.
Introduction
Huntington’s disease (HD) is neurodegenerative disorder in which the mutation results in the increased length of a tract of glutamines that causes the huntingtin protein (HTT) to aggregate. It is characterized by neurological symptoms and neurodegeneration that is prominent in the basal ganglia and cerebral cortex [1]. In mammals, HTT is expressed in many tissues and organs [2,3] and is involved in many critical cellular processes such as transcription, protein trafficking and vesicle transport [4]. HTT is predicted to form an elongated superhelical solenoid structure due to a large number of HEAT motifs, suggesting that it plays a scaffolding role for protein complex formation [5]. More than 200 HTT interacting partners have been identified which can be classified according to their function and include proteins that are involved in gene transcription, intracellular signalling, trafficking, endocytosis, and metabolism [6]. In mice, HTT deletion is embryonically lethal, leading to defects in all germ layers [7]. The process of mutant HTT self-aggregation is an early event in HD progression which may lead to the pathological features of HD. Insoluble polyQ aggregates are a hallmark of HD pathology and can be detected at the presymptomatic stage in HD post mortem brain [8] and can also be found in many non-central nervous system tissues in HD mouse models [9,10]. Recently, it has been shown that there are a number of factors to indicate that HD patients experience an HD-related heart pathology [11]. A recent study in the R6/2 and HdhQ150 knock-in mouse models showed that the HD-related cardiomyopathy is caused by altered central autonomic pathways and is not due to the accumulation of toxic HTT aggregates as had previously been proposed [12–14]. This was accompanied by the re-expression of foetal genes, apoptotic cardiomyocyte loss and a moderate degree of interstitial fibrosis [13]. There is also growing evidence to indicate that peripheral pathologies such as weight loss and skeletal muscle atrophy may not be a consequence of neurological dysfunction or neurodegeneration and might make a significant contribution to the disease presentation and progression [15]. Therefore it is important to identify the peripheral abnormalities that may contribute to disease progression as they may present targets for new treatment strategies.
To date, our knowledge of skeletal muscle pathology in HD is very limited as outlined in a recent review [16]. A case-study report showed that a semi-professional marathon runner (43 CAGs) developed signs of a slowly progressing myopathy with elevated creatine kinase levels many years before first signs of chorea were detected. A muscle biopsy revealed a mild myopathy with mitochondrial pathology including a complex IV deficiency [17]. Transcriptional deregulation is a typical feature of HD pathology in the brain [18] and a similar transcriptional profile in skeletal muscles (quadriceps) from R6/2 mice, HdhQ150 homozygous knock-in mice and HD patients has been identified that was consistent with a transition from fast-twitch to slow-twitch muscle fiber types [19]. Some of the molecular and physiological changes in HD muscles can even be detected in pre-symptomatic HD individuals [20–22]. At the molecular level, mitochondrial dysfunction, PPAR alpha signalling and HSF1 activation were identified as major players in skeletal muscle HD-related pathology [23,24]. Proof of concept studies have suggested that the progression of disease onset could be delayed and lifespan extended by improving muscle function in HD mouse models [25,26]. However, many aspects of HD neuromuscular transmission and muscle physiology remain unanswered and need to be studied more extensively. In this study, we have investigated the molecular and pathological features of the skeletal muscle dysfunction that develops with disease progression in mouse models of HD at the physiological level.
Results
To test the hypothesis that mutant HTT leads to the skeletal muscle atrophy through compensatory remodelling, including the HDAC4-myogenin axis, we used two well-established HD mouse models. R6/2 mice are transgenic for a mutated N-terminal exon 1 HTT fragment [27], while the HdhQ150 mice have an expanded CAG repeat knocked-in to the mouse huntingtin gene (Htt) [28,29], which is partially mis-spliced with the result that these mice express mutant versions of both an exon 1 HTT and a full length HTT protein [30]. For R6/2 mice, we studied skeletal muscle abnormalities at symptomatic (12 weeks) and end-stage (14 weeks) disease while HdhQ150 homozygotes were compared to wild type (WT) at 22 months (end-stage disease).
We began by quantifying the change in the weight of various skeletal muscles with disease progression (Fig. 1). There was a significant decrease in the muscle mass of all muscles examined types including quadriceps, gastrocnemius/plantaris complex (G/P), tibialis anterior (TA), extensor digitorum longus (EDL) and soleus at 12 weeks of age in R6/2 mice (Fig. 1A). The HdhQ150 knock-in model showed a strikingly similar muscle mass decrease at 22 months of age (Fig. 1B). We have previously shown that HTT inclusions can be detected throughout the periphery of the R6/2 and HdhQ150 mouse models by immunohistochemistry [10,31]. More recently, we have developed the seprion-ligand ELISA, a highly quantitative method with good statistical power that can be used to measure changes in aggregate load that occur in vivo in response to pharmacological or genetic manipulations [9]. Using this assay, we were able to detect mutant HTT aggregates in the different skeletal muscle types from either R6/2 at 12 and 14 weeks of age (Fig. 1C) or HdhQ150 (Fig. 1D) mice at 22 months. Surprisingly, we observed a higher accumulation of toxic aggregates in the TA muscles in comparison to G/P and quadriceps in both HD mouse models (Fig. 1C and D). Subsequently, we used Taqman qPCR to demonstrate that the expression of the exon-1 HTT mRNA was uniform in the different types of R6/2 skleletal muscles at 12 (S1A Fig.) and 14 weeks of age (S1B Fig.) or HdhQ150 muscle at 22 months (S1C Fig.) and was not therefore the reason for the increased level of aggregates in the TA muscles in both of these models.
Fig. 1. Morphometric and mutant huntingtin aggregation analysis of HD mouse model skeletal muscle. 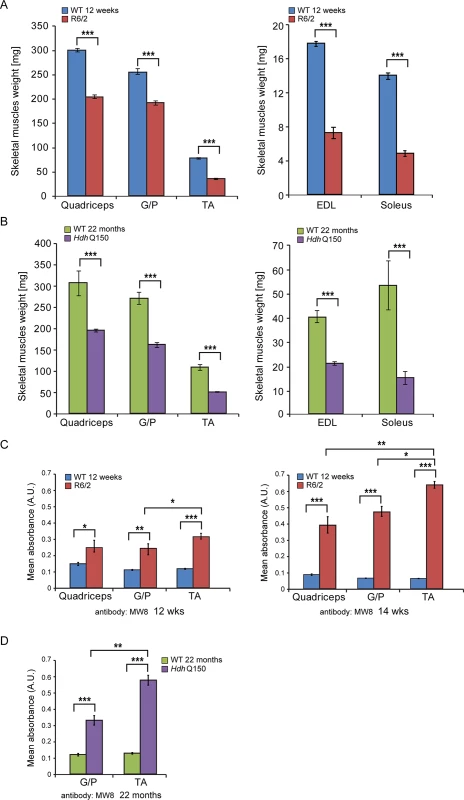
(A) wet skeletal muscle weight of R6/2 mice at 12 weeks of age and (B) HdhQ150 mice at 22 months of age. Quadriceps, gastrocnemius/plantaris complex (G/P), tibialis anterior (TA), extensor digitorum longus (EDL) and soleus. (C, D) The seprion-ligand ELISA was used to identify and quantify the aggregate load in skeletal muscle lysates from R6/2 and HdhQ150 mice. All values are mean ± SEM (n = 6/genotype/age). Student’s t test: *p < 0.05, **p < 0.01, ***p < 0.001. Next, to determine whether HD mice develop functional contractile abnormalities, we undertook isometric muscle tension experiments on TA and EDL muscles of the R6/2 mouse model and their respective WT littermates, as previously described [32], at 12 and 14 weeks of age. Both of these muscles exhibited a significant degree of atrophy as indicated above (Fig. 1) and significant alterations in their contractile function were revealed (Fig. 2A and B). The time taken for muscles to reach maximum force (time-to-peak, TTP) was significantly higher in the EDL at both time points in R6/2 mice, while TA showed a normal TTP in R6/2 mice in comparison to their WT littermates at both time points (Fig. 2A). The time taken for muscles to relax to a half of the maximum force (1/2 relaxation time) following a twitch-stimulus was significantly prolonged in both EDL and TA muscles in the R6/2 mice at 12 and 14 weeks of age in comparison to their WT littermates (Fig. 2B).
Fig. 2. Skeletal muscle contractile dysfunction in the HD mouse models. 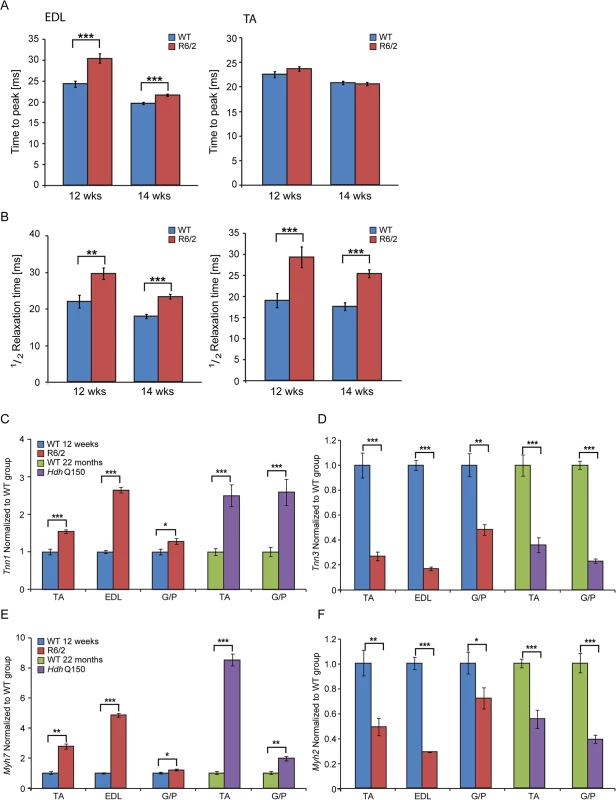
(A) Twitch stimuli were delivered to elicit TTP (time to peak) contraction of the hind limb EDL and TA skeletal muscles. A significant increase in TTP in the R6/2 skeletal muscles was observed for EDL but not TA at 12 and 14 weeks of age. (B) Twitch stimuli were delivered to elicit a half-relaxation time of the hind limb EDL and TA muscles. A significant increase in the half-relaxation time in R6/2 skeletal muscles was observed for both EDL and TA at 12 and 14 weeks of age. Error bars are SEM (n = 10). ONE-WAY ANOVA with Bonferroni post-hoc test: *p < 0.05, **p < 0.01; ***p < 0.001. Transcriptional deregulation of contractile transcripts: (C) Tnn1 (Troponin 1, slow), (D) Tnn3 (Troponin3, fast), (E) Myh7 (myosin heavy light chain 7), (F) Myh2 (myosin heavy light chain 2) mRNAs were significantly deregulated in the muscle of R6/2 and HdhQ150 mice. All Taqman qPCR values were normalized to the geometric mean of three housekeeping genes: Atp5b, Yhwaz and Rpl13a. Error bars are SEM (n = 6). Student’s t-test: *p < 0.05, **p < 0.01; ***p < 0.001. In order to corroborate the physiological findings described above, we used Taqman qPCR to quantify contractile transcipt levels that are representtive of the fast or slow type fibers. Given that global transcriptional dysregulation is a pathogenic characteristic of HD, we first performed a systematic study to identify suitable reference genes for use in the expression analysis of different skeletal muscles types from HD mouse models. We used the geNorm™ Housekeeping Gene Selection Mouse Kit and associated software to identify the three most stably expressed genes in specific muscles from R6/2 (S2 Fig.) and HdhQ150 (S3 Fig.) mice. Our relative quantification methods then used the geometric mean of these three selected reference genes for normalization, to accurately determine gene expression levels in WT, R6/2 and HdhQ150 skeletal muscle tissue. We found a significant up-regulation of slow-type contractile proteins such as Tnn1 (Troponin 1, slow) and Myh7 (myosin heavy light chain 7) in TA, EDL and G/P muscles from both HD mouse models (Fig. 2C and E). Consequently, a pronounced down-regulation of the fast-type contractile proteins like Tnn3 (Troponin3, fast) and Myh2 (myosin heavy light chain 2) was also observed in TA, EDL and G/P muscles from both HD mouse models (Fig. 2D and F). These findings indicate that there is a loss of fast-twitch muscle fibres in the EDL and TA of both models. Subsequently, we determined the expression levels of additional genes that are attributed to be altered in fast to slow twitch remodelling. TEA domain (TEAD) transcription factors and their co-activators serve important functional roles during embryonic development as well as in striated muscle gene expression and muscle regeneration [33–36]. It has been shown that striated muscle-restricted TEAD-1 expression induced a transition toward a slow muscle contractile protein phenotype, slower shortening velocity with longer contraction and relaxation times in the adult fast twitch EDL muscles [33]. We found that Tead-2 (TEA domain family member 2) (Fig. 3B) and Tead-4 (TEA domain family member 4) (Fig. 3D) were significantly up-regulated in the all diseased HD muscles in both mouse models, while Tead-1 (TEA domain family member 1) (Fig. 3A) and Tead-3 (TEA domain family member 3) (Fig. 3C) transcripts remained un-changed. The transcriptional activity of TEAD family members is highly dependent on the presence of their co-activators [37–39] and therefore, we used Taqman-qPCR to asses their transcriptional profile in the HD diseased muscles. We established that Vgll-2 (vestigial related factor 2) (Fig. 3E), Vgll-3 (vestigial related factor 3) (Fig. 3F), Vgll-4 (vestigial related factor 4) (Fig. 3G) and Yap-65 (Yes associated protein 65) (Fig. 3H) were significantly up-regulated in the TA, EDL and G/P muscles of R6/2 and HdhQ150 mice.
Fig. 3. Transcriptional deregulation of TEAD family members and their co-activators involved in the skeletal muscle atrophy. 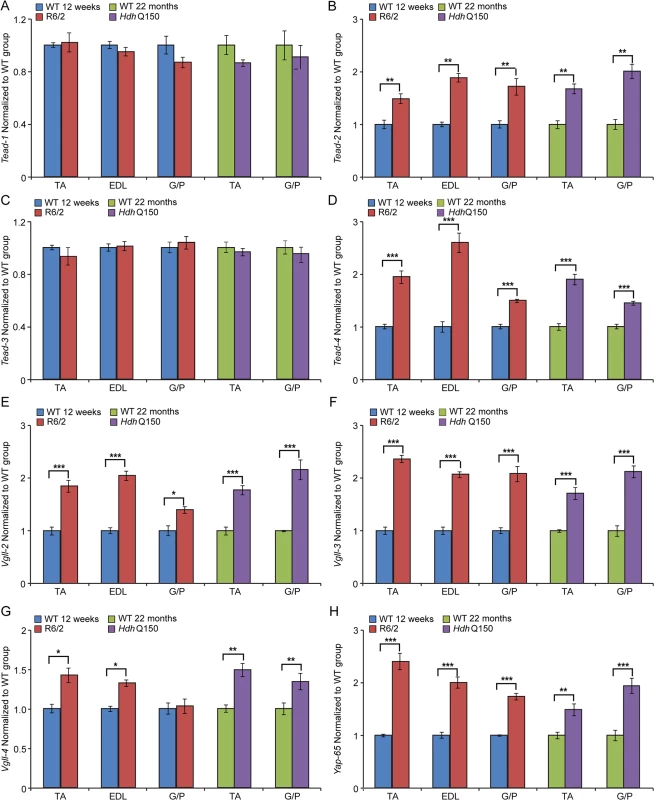
(A) Tead-1 (TEA domain family member 1), (B) Tead-2 (TEA domain family member 2), (C) Tead-3 (TEA domain family member 3), (D) Tead-4 (TEA domain family member 4). Tead-2 and Tead-4 transcripts were deregulated in various muscles of the HD models. (E) Vgll-2 (vestigial related factor 2), (F) Vgll-3 (vestigial related factor 3), (G) Vgll-4 (vestigial related factor 4) and (H) Yap-65 (Yes associated protein 65) were significantly up-regulated in skeletal muscles of the HD models. All Taqman qPCR values were normalized to the geometric mean of three housekeeping genes: Atp5b, Yhwaz and Rpl13a. Error bars are SEM (n = 6). Student’s t-test: *p < 0.05, **p < 0.01; ***p < 0.001. We also determined the maximum muscle force of TA and EDL muscles by physiological determination of single twitch (Fig. 4A) and tetanic force (Fig. 4B) in R6/2 mice. Twitch and tetanic force recordings showed that R6/2 TA muscles at 12 and 14 weeks of age were approximately 50% weaker than in their WT littermates. Moreover, physiological assessment of functional motor unit survival, revealed that there was a significant loss of motor units in R6/2 mice, which progressed from a 25% reduction at 12 weeks to over 60% loss at 14 weeks, compared to WT mice (Fig. 4C and D). These findings suggest that there is also likely to be a significant degeneration of spinal motor neurons during this period.
Fig. 4. Neuromuscular function in the R6/2 mouse model of HD. 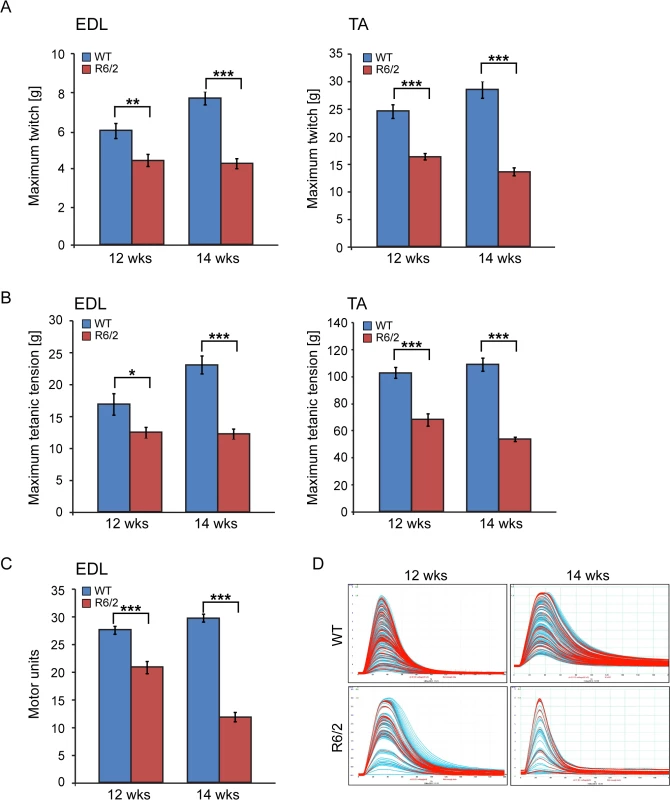
(A) A significant decrease in the maximum twitch tension was observed for EDL and TA at 12 and 14 weeks of age. (B) A significant decrease in the maximum tetanic tension was observed for EDL and TA at 12 and 14 weeks of age. (C) The motor unit output for each experimental group is summarised in the bar chart. A significant decrease in functional motor units was observed for EDL at 12 and 14 weeks of age. (D) Examples of the motor unit trace recordings of the WT and R6/2 hind limb EDL muscles at 12 and 14 weeks of age. Error bars are SEM (n = 10). ONE-WAY ANOVA with Bonferroni post-hoc test: *p < 0.05, **p < 0.01; ***p < 0.001). Physiological changes in skeletal muscle are often caused or associated with metabolic alterations. Therefore, we analysed two aspects of metabolism in the EDL and TA muscles. First we estimated the steady-state concentration of the major components of energy equilibrium that include creatine metabolites and adenine nucleotides. Analysis of ATP, phosphocreatine and related metabolites revealed a substantial depletion of the energy equilibrium in EDL and TA in both HD mouse models (Fig. 5A and B and Table 1). The phosphocreatine/creatine ratio as well as ADP and AMP levels were significantly decreased (Table 1) in both types of muscle in R6/2 and HdhQ150 mice. Besides the energy equilibrium, the total pools of the adenine nucleotides were also consistently depleted (Fig. 5 and Table 1) while changes in the redox status were less evident (Fig. 5B). A similar pattern of metabolic changes was found in the slow type soleus muscles of the HD mouse models (S1 Table). The second metabolic aspect concerned the evaluation of the substrate preference shift in these muscles. To address this, glycolysis was assessed by measuring the 13C alanine enrichment while the changes in Krebs cycle were estimated based on 13C glutamate levels after administration of 1–13C glucose. The analysis revealed that the EDL muscle showed a slower glycolytic flux from exogenous glucose as well less oxidation of glucose in both HD mouse models (Fig. 5C and D) while the TA the muscle remained unchanged (Fig. 5C and D).
Fig. 5. Deterioration of the energy metabolism and decrease in oxidation in the EDL and TA of HD mouse models. 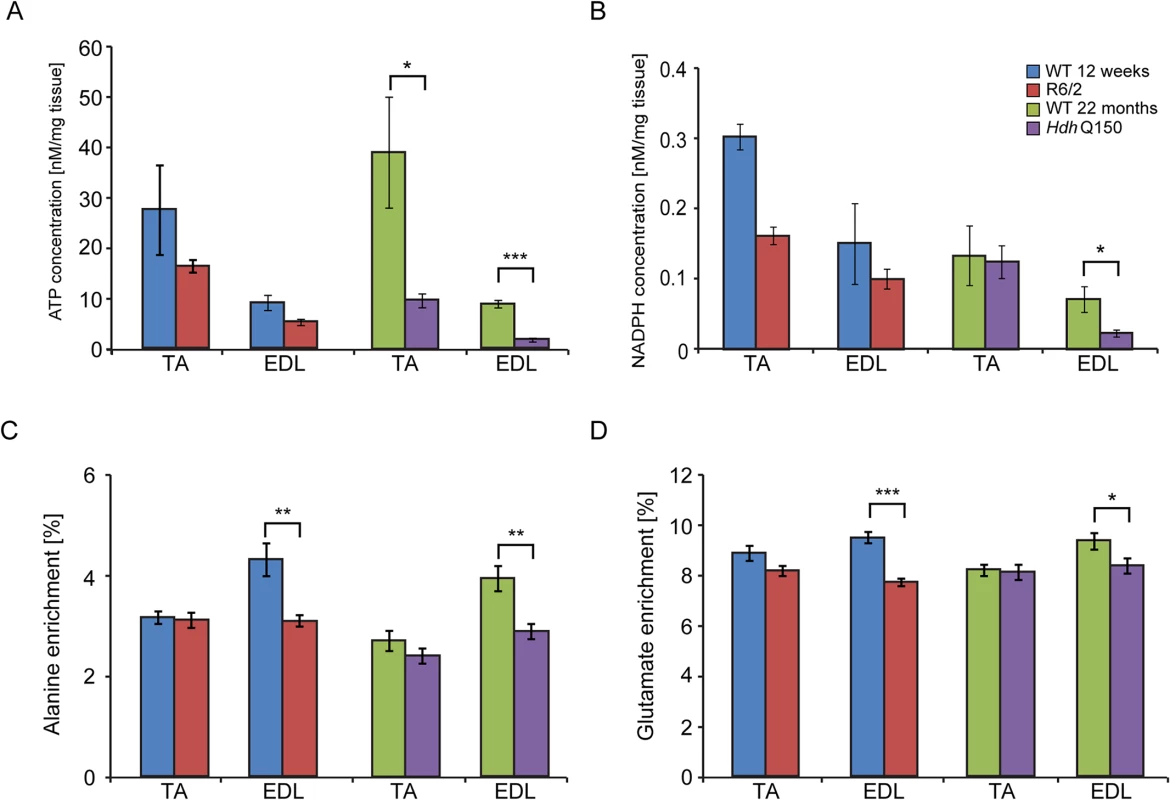
Concentrations of (A) ATP (B) NADH (C) alanine and (D) glutamate 13C enrichment in skeletal muscle extracts after administration of 1–13C glucose in WT and HD mice. Error bars are SEM (n = 6). Student’s t-test: *p < 0.05, **p < 0.01; ***p < 0.001. Tab. 1. Deterioration of the energy metabolism in the skeletal muscle (EDL and TA) of HD mouse models. 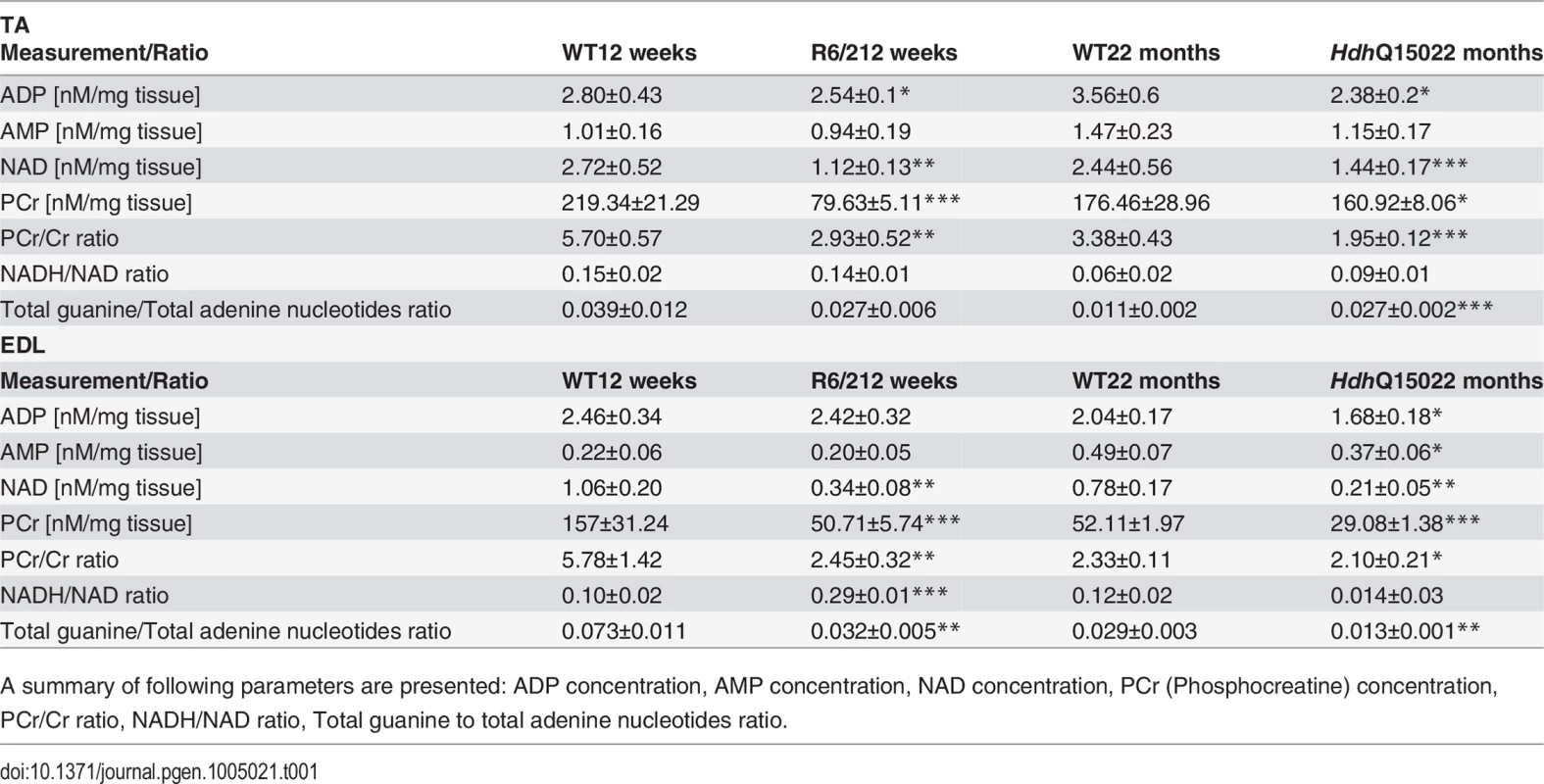
A summary of following parameters are presented: ADP concentration, AMP concentration, NAD concentration, PCr (Phosphocreatine) concentration, PCr/Cr ratio, NADH/NAD ratio, Total guanine to total adenine nucleotides ratio. To further examine the degree of skeletal muscle pathology, we determined the expression levels of additional genes that are typically altered in atrophied muscles. We found that AChR (nicotinic acetylcholine receptor) (Fig. 6A) was significantly up-regulated in all muscle types examined from mouse models. Usually, muscle atrophy is accompanied by a significant up-regulation of caspases [40]. Indeed, we found Caspase8 transcripts significantly up-regulated in the aged HdhQ150 muscles but not in those from the R6/2 mice (Fig. 6B). Similarly, we found Foxo-3 (Forkhead box O3) transcripts (Fig. 6D) to be markedly up-regulated, while Mck (muscle creatinine kinase) mRNA (Fig. 6C) was decreased in all of the muscle types examines from the R6/2 and HdhQ150 mice.
Fig. 6. Transcriptional deregulation of gene markers involved in skeletal muscle atrophy. 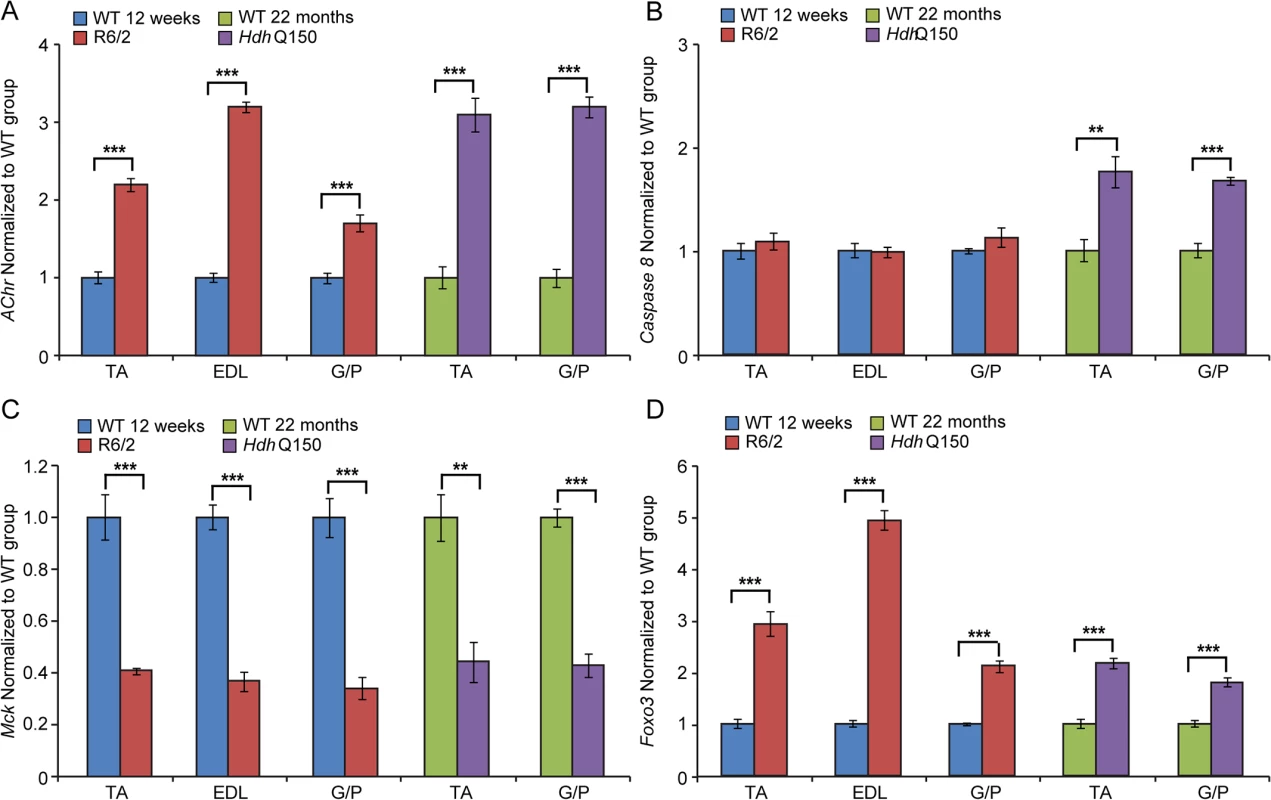
(A) AChR (nicotinic acetylcholine receptor), (B) Caspase8, (C) Mck (muscle creatine kinase) and (D) Foxo-3 (Forkhead box O3) transcripts were significantly deregulated in various muscles of the HD mouse models. All Taqman qPCR values were normalized to the geometric mean of three housekeeping genes: Atp5b, Yhwaz and Rpl13a. Error bars are SEM (n = 6). Student’s t-test: *p < 0.05, **p < 0.01; ***p < 0.001. Previous studies have established HDAC4 as a critical factor that connects neural activity to the muscle remodelling program [41,42] and inactivation of HDAC4 suppressed denervation-induced muscle atrophy while increasing re-innervation [43–45]. HDAC4 up-regulation was found to be significantly greater in patients with rapidly progressive ALS (amyotrophic lateral sclerosis) and was negatively correlated with the extent of muscle re-innervation and functional outcome [46]. Similarly, an increased level of HDAC4 has been found in SMA (spinal muscular atrophy) model mice and in SMA patient muscles [47]. Consistent with this, we found that Hdac4 transcripts were significantly up-regulated in the TA, EDL an G/P muscles in the HD mouse models as compared to WT littermates (Fig. 7A). Hdac4 up-regulation was accompanied by down-regulation its direct target Dach2 (Dachshund homolog 2) (Fig. 7B) that is a negative regulator of Myogenin. Consequently, we observed a very significant up-regulation of Myogenin (Fig. 7C) and its direct target Fbxo32 (F-box only protein 32) (Fig. 7D) in HD-related muscle atrophy. Thus, one might conclude that HD-related skeletal muscle atrophy displays the typical characteristics of a denervation like muscle phenotype.
Fig. 7. The Hdac4-Myogenin axis displayed a typical denervation-like phenotype in the skeletal muscle of HD mouse models. 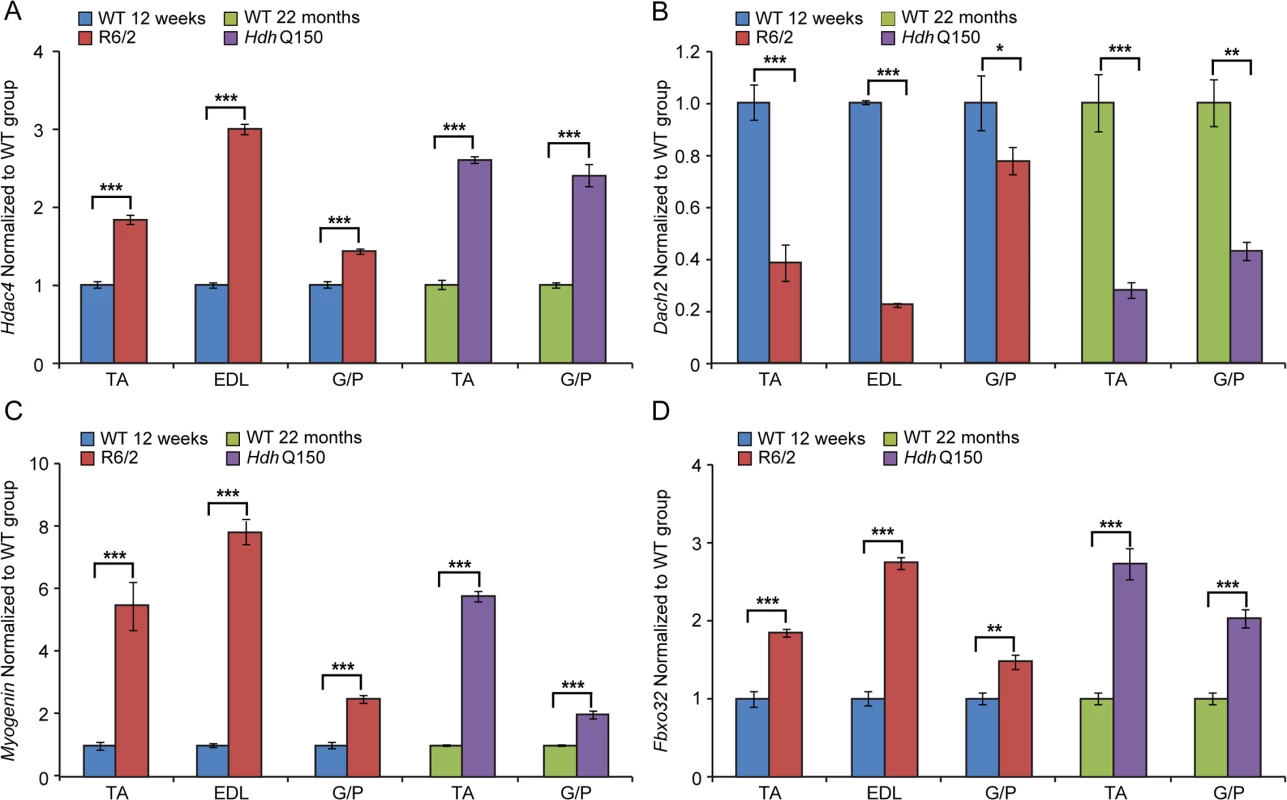
(A) Hdac4 (Histone deacetylase 4), (B) Dach2 (Dachshund homolog 2), (C) Myogenin and (D) Fbxo32 (F-box only protein 32) mRNAs were significantly deregulated in the skeletal muscles of HD mouse models. All Taqman qPCR values were normalized to the geometric mean of three housekeeping genes: Atp5b, Yhwaz and Rpl13a. Error bars are SEM (n = 6). Student’s t-test: *p < 0.05, **p < 0.01; ***p < 0.001. Discussion
Skeletal muscle is the most abundant tissue in the mammalian body accounting for approximately 40% of body weight, and is composed of multinucleated fibers that contract to generate force and movement. In addition, skeletal muscle possesses a remarkable ability to regenerate, and can go through rapid repair following severe damage caused by exercise, toxins or diseases. The atrophy caused by degeneration of myofibers and their replacement by fibrotic tissue is the major pathological feature in many genetic muscle disorders [48,49]. Skeletal muscle atrophy in HD is a comorbidity that is observed in catabolic disease and other conditions like cancer, congestive heart failure, sepsis, denervation and disuse [16,50]. Under normal physiological conditions muscle function is orchestrated by a network of intrinsic hypertrophic and atrophic signals linked to the functional properties of the motor units that are likely to be imbalanced in HD.
In this study we aimed to provide a broad spectrum of experimental insights into skeletal muscle-associated abnormalities that develop in the R6/2 transgenic and HdhQ150 knock-in HD mouse models, in which mutant Htt is expressed under the control of the Htt promoter. We found significant alterations at the physiological level in the contractile function of the EDL and TA R6/2 muscles at 12 and 14 weeks of age. The time taken for muscles to reach maximum force (time-to-peak, TTP) and time taken for muscles to relax to half the maximum force (1/2 relaxation time) were significantly changed in R6/2 mice, indicative of a loss of fast-twitch muscle fibres in the EDL and TA muscles. In addition, transcriptional deregulation is a typical feature of HD pathology in the brain [18] and a similar transcriptional profile in the skeletal muscles (quadriceps) from R6/2 mice, HdhQ150 homozygous knock-in mice and HD patients has been identified that was consistent with a transition from fast-twitch to slow-twitch muscle fiber types [19]. Although immunohistochemistry suggested that both type I (slow) and II (fast) muscles were atrophic [31], there were more type I fibers in the R6/2 skeletal muscles. Hence, a conversion of type II to type I fibers has occurred during the process of muscle atrophy [51], most likely as a result of the loss of motor units innervating type II fibres.
Indeed, our physiological findings were also supported by the quantification of contractile transcipt levels that are representative of fast or slow type fibers [52,53]. We found a significant up-regulation of the genes encoding slow-type contractile proteins like Tnn1 and Myh7 in the TA, EDL and G/P muscles of both HD mouse models. Consequently, the transcripts for fast-type contractile proteins like Tnn3 and Myh2 (myosin heavy light chain 2) were markedly down-regulated in these muscles. This was also accompanied by the up-regulation of members of the TEAD family and their co-activators. It is well established that MCAT elements are located in the promoter-enhancer regions of cardiac, smooth, and skeletal muscle-specific genes and play a key role in the regulation of these genes during muscle development and disease [33–36].
Following a significant decrease in the muscle mass of all of the muscle types that were examined in both HD mouse models, we found that the maximum twitch and tetanic force in TA and EDL hind limb muscles of R6/2 mice were significantly reduced at the symptomatic stage, indicative of motor neuron dysfunction in these mice. Moreover, the physiological assessment of functional motor units revealed that there was a progressive loss in the number of functional fewer motor units in the EDL muscle of R6/2 mice, from ∼25% loss at 12 weeks to more than 60% loss at 14 weeks of age, as compared to their WT littermates. This finding is supported by a previous study showing that skeletal muscles of R6/2 mice developed age-dependent denervation-like abnormalities, including reduced endplate area, supersensitivity to acetylcholine, decreased sensitivity to mu-conotoxin and anode-break action potentials [51]. Moreover, the miniature endplate potential (mEPP) amplitude was notably increased while mEPP frequency was significantly reduced in the R6/2 mice [51]. In contrast, the same study showed that severely affected R6/2 mice developed a progressive increase in the number of motor endplates that fail to respond to nerve stimulation but there was no constitutive sprouting of motor neurons, even in severely atrophic muscles [51]. In fact there was no age-dependent loss of regenerative capacity of motor neurons in R6/2 mice [51]. In line with our findings, a previous study showed that the action potentials in diseased muscles were more easily triggered and prolonged than in WT littermates. Furthermore, the expression of the muscle chloride channel (ClC-1) and Kcnj2 (Kir2.1 potassium channel) transcripts were significantly reduced and defects in mRNA processing were detected [54]. These dependent denervation-like abnormalities and the highly developed muscle atrophy could be partially explained by sciatic nerve degeneration [55]. A significant decrease in the axoplasm diameter of myelinated neurons and increased number of degenerating myelinated fibers were observed; although the myelin thickness and unmyelinated fiber diameter were not affected [55]. This might be also explained by the profound localisation of mutant HTT to the neuromuscular junctions as was previously published [56]. However, it is likely that the skeletal muscle denervation-like phenotype is linked to not only to spinal motor neuron loss, but also CNS dysfunction, as previously published pathological features in relevant brain regions in both mouse models might support this hypothesis [29]. In HD patient brains, a recent meta-analysis of morphometric MRI found degenerative changes in the amygdala and insular cortex, even in the prodromal form on the disease [57].
It is well established that pronounced skeletal muscles atrophy is accompanied by altered metabolism, reviewed in [58] and our demonstration that the energy equilibrium is depleted in the skeletal muscles of HD mouse models is an important and a novel finding. The decrease in the phosphocreatine/creatine ratio and ATP/ADP ratio directly translates into lower values for phosphorylation potential and the free energy of ATP hydrolysis that might decrease the efficiency of the muscle contraction [52]. Interestingly, our study is in line with previous observations in clinical settings, as muscle ATP/phosphocreatine and inorganic phosphate levels were significantly reduced in both symptomatic and presymptomatic HD subjects [20]. In addition, HD subjects displayed a deficit in mitochondrial oxidative metabolism that might support a role for mitochondrial dysfunction as a key factor involved in the HD-related muscle pathogenesis [21]. An important aspect of this study is the identification of the mechanism underlying the decreased energy equilibrium. One possible explanation is a lack of the trophic effect of nerve stimulation [51,55] that may down-regulate the expression of energy related proteins including factors responsible for mitochondrial biogenesis [58]. Consequently, this process might lead to a decreased oxidative and substrate phosphorylation efficiency translating into a shift of energy equilibrium. Alternatively, a direct local effect of genetic alterations in the skeletal muscle that are likely to be driven by mutant HTT directly [56,59] may deregulate energy metabolism. Interestingly, a similar metabolic profile has been found in mouse embryonic stem cell (mESC) lines: Htt(−/−), extended poly-Q (Htt-Q140/7) and wild-type mESCs (Htt-Q7/7) [60]. One might conclude that the HD-related skeletal muscle atrophy is caused by loss of function in HD mouse models.
At the pathological level, the HD-related skeletal muscle atrophy was accompanied by the deregulation of AChR, Foxo-3 and Mck, typical markers of muscle atrophy and denervation in both HD mouse models [48,61]. It has been also shown that inactivation of HDAC4 suppresses denervation-like induced muscle atrophy while increasing re-innervation [41,42,45]. These findings highlight a central regulatory role of HDAC4 in activity-dependent muscle remodelling. HDAC4 up-regulation was significantly greater in patients with rapidly progressive ALS (amyotrophic lateral sclerosis) and was negatively correlated with the extent of muscle re-innervation and functional outcome [46]. An increased level of HDAC4 has been found in SMA (spinal muscular atrophy) model mice and in SMA patient muscles [47]. We found an up-regulation of the HDAC4-Dach2-myogenin axis in both HD mouse models that might be indicative of a similar activity dependent muscle remodelling in HD to that observed in ALS or SMA.
In summary, mutant HTT results in the rapid development of pathological features that would be expected to lead to a skeletal muscle contractile dysfunction e.g. leading to fast to slow fibre twitch with aberrant deregulation of contractile protein transcripts and their up-stream transcriptional regulators. In addition, HD mouse models develop a notable decrease in the twitch and tetanic force of skeletal muscles and pronounced loss of motor units, which may contribute to deterioration of energy metabolism and decreased oxidation that is accompanied by the re-expression of HDAC4-Dach2-myogenin axis (Fig. 8). Importantly, our data connects gene alterations with physiological function in HD-related skeletal muscles atrophy and might have a therapeutic potential. Recently, two key signalling pathways, i.e. those driven by insulin like growth factor (IGF) and growth differentiation factor −8 (GDF-8), have emerged to be potent regulators of skeletal muscle size. In addition, our metabolomic profile of skeletal muscles in HD mouse models might be served as a biomarker platform for prospective pre - and clinical trials.
Fig. 8. A model depicting the mechanism by which mutant huntingtin causes skeletal muscle wasting in murine HD models. 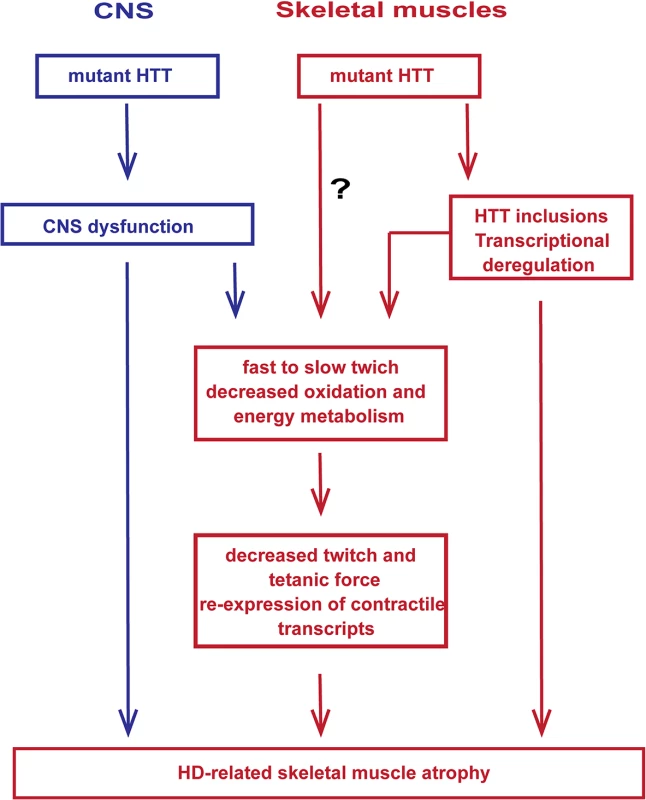
Muscle atrophy is likely driven by the aggregation of mutant HTT in muscle fibres and CNS dysfunction, the relative contribution of which has not been established. It is not possible to rule out that mHTT might exert additional direct detrimental effects within the muscle fibres. Materials and Methods
Ethics statement
All experimental procedures performed on mice were conducted under a project licence from the Home Office and approved by the King's College London Ethical Review Process Committee.
Mouse maintenance and breeding
Hemizygous R6/2 mice were bred by backcrossing R6/2 males to (CBA x C57BL/6) F1 females (B6CBAF1/OlaHsd, Harlan Olac, Bicester, UK). HdhQ150 homozygous mice on a (CBA x C57BL/6) F1 background were obtained by intercrossing HdhQ150 heterozygous CBA/Ca and C57BL/6J congenic lines as described previously [29]. All animals had unlimited access to water and breeding chow (Special Diet Services, Witham, UK), and housing conditions and environmental enrichment were as previously described [62]. Mice were subject to a 12-h light/dark cycle. All experimental procedures were performed according to Home Office regulations.
Genotyping
Genomic DNA was isolated from an ear-punch. R6/2 and HdhQ150 mice were genotyped by PCR and the CAG repeat length was measured as previously described [9] and listed in S2 Table. Dissected tissues were snap frozen in liquid nitrogen and stored at −80°C until further analysis.
RNA extraction and Taqman real-time PCR expression analysis
Total RNA from skeletal muscles was extracted with the mini-RNA kit according to the manufacturer instructions (Qiagen). The reverse transcription reaction (RT) was performed using MMLV superscript reverse transcriptase (Invitrogen) and random hexamers (Operon) as described elsewhere [63]. The final RT reaction was diluted 10-fold in nuclease free water (Sigma). All Taqman qPCR reactions were performed as described previously [64] using the Chromo4 Real-Time PCR Detector (BioRad). Stable housekeeping genes for qPCR profiling of various skeletal muscles for HD mouse models were determined using the Primer Design geNorm Housekeeping Gene Selection Mouse Kit with PerfectProbe software. Estimation of mRNA copy number was determined in triplicate for each RNA sample by comparison to the geometric mean of three endogenous housekeeping genes (Primer Design) as described [65]. Primer and probe sets for genes of interest were purchased from Primer Design or ABI.
Antibodies, Seprion ELISA
Aggregates were captured in Seprion ligand coated plates (Microsens) and detected using the MW8 mouse monoclonal antibody (1 : 4000) as described [9].
Nucleotides, alanine and glutamate concentrations in vivo
Mice were injected with glucose-13C subcutaneously as a 20% solution at a dose of 3ml/kg. Two hours after glucose administration, mice were deeply anesthetized with isoflurane and EDL and TA muscles were freeze-clamped in situ with aluminum clamps pre-cooled in liquid nitrogen. Freeze dried muscles were extracted with 0.4 M perchloric acid, extracts were neutralized with 2 M KOH as described previously [66] and analysed by liquid chromatography mass spectrometry [67] using TSQ Vantage triple quadrupole mass detector linked to Surveyor chromatography system. Mass detection was carried out in fragmentation mode (Tandem MS) and 13C isotopic enrichment of fragments containing C3 of alanine or C4 of glutamate were monitored.
In vivo muscle tension
Mice were deeply anesthetized with isoflurane and prepared for in vivo analysis of muscle function which was performed as previously described [68]. The distal tendons of the TA and EDL muscles in both hindlimbs were dissected free and attached by silk thread to isometric force transducers (Dynamometer UFI Devices, Welwyn Garden City, UK). The sciatic nerve was exposed and sectioned proximally. The length of the muscles was adjusted for maximum twitch tension. The muscles and nerve were kept moist with saline throughout the recordings and all experiments were carried out at room temperature. Isometric contractions were elicited by stimulating the nerve to TA and EDL using square-wave pulses of 0.02 ms duration at supra-maximal intensity, via silver wire electrodes. Contractions were elicited by trains of stimuli at frequencies of 40, 80 and 100 Hz. The maximum tetanic tension was measured using a computer and appropriate software Pico Technology,Cambridgeshire, UK. The number of motor units innervating the EDL muscles was also determined as previously described [69] by stimulating the motor nerve with stimuli of increasing intensity, resulting in stepwise increments in twitch tension due to successive recruitment of motor axons with increasing stimulus thresholds. The number of stepwise increments was counted to give an estimate of the number of functional motor units (MUNE) present in each muscle. Following recording of isometric tension, the contractile characteristics of EDL and TA muscles were determined. The time to peak (TTP) was calculated by measuring the time taken (ms) for the muscle to elicit peak twitch tension and the half relaxation time (the time taken for the muscle to reach half relaxation from peak contraction) was also calculated. The tetanic contractions were recorded on a Lectromed Multitrace 2 recorder (Lectromed Ltd, UK). All parameters were measured using a computer and Picoscope v5 and v6 software (Pico Technology,Cambridgeshire, UK).
Statistical analysis
All data were analysed with Microsoft Office Excel and Student's t-test (two tailed) or ONE-WAY ANOVA with Bonferroni post-hoc test.
Supporting Information
Zdroje
1. Bates GP, Tabrizi SJ, Jones AL (2014) Huntington's Disease. New York: Oxford University Press.
2. Strong TV, Tagle DA, Valdes JM, Elmer LW, Boehm K, et al. (1993) Widespread expression of the human and rat Huntington's disease gene in brain and nonneural tissues. Nat Genet 5 : 259–265. 8275091
3. Li SH, Schilling G, Young WS 3rd, Li XJ, Margolis RL, et al. (1993) Huntington's disease gene (IT15) is widely expressed in human and rat tissues. Neuron 11 : 985–993. 8240819
4. Li SH, Li XJ (2004) Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet 20 : 146–154. 15036808
5. Li W, Serpell LC, Carter WJ, Rubinsztein DC, Huntington JA (2006) Expression and characterization of full-length human huntingtin, an elongated HEAT repeat protein. J Biol Chem 281 : 15916–15922. 16595690
6. Harjes P, Wanker EE (2003) The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci 28 : 425–433. 12932731
7. Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A (1995) Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat Genet 11 : 155–163. 7550343
8. Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, et al. (1999) Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J Neurosci 19 : 2522–2534. 10087066
9. Sathasivam K, Lane A, Legleiter J, Warley A, Woodman B, et al. (2010) Identical oligomeric and fibrillar structures captured from the brains of R6/2 and knock-in mouse models of Huntington's disease. Hum Mol Genet 19 : 65–78. doi: 10.1093/hmg/ddp467 19825844
10. Moffitt H, McPhail GD, Woodman B, Hobbs C, Bates GP (2009) Formation of polyglutamine inclusions in a wide range of non-CNS tissues in the HdhQ150 knock-in mouse model of Huntington's disease. PLoS One 4: e8025. doi: 10.1371/journal.pone.0008025 19956633
11. Zielonka D, Piotrowska I, Mielcarek M (2014) Cardiac dysfunction in Huntington’s Disease. Exp Clin Cardiol 20 : 2547–2554.
12. Pattison JS, Sanbe A, Maloyan A, Osinska H, Klevitsky R, et al. (2008) Cardiomyocyte expression of a polyglutamine preamyloid oligomer causes heart failure. Circulation 117 : 2743–2751. doi: 10.1161/CIRCULATIONAHA.107.750232 18490523
13. Mielcarek M, Inuabasi L, Bondulich MK, Muller T, Osborne GF, et al. (2014) Dysfunction of the CNS-Heart Axis in Mouse Models of Huntington's Disease. PLoS Genet 10: e1004550. doi: 10.1371/journal.pgen.1004550 25101683
14. Mielcarek M, Bondulich MK, Inuabasi L, Franklin SA, Muller T, et al. (2014) The Huntington's Disease-Related Cardiomyopathy Prevents a Hypertrophic Response in the R6/2 Mouse Model. PLoS One 9: e108961. doi: 10.1371/journal.pone.0108961 25268775
15. van der Burg JM, Bjorkqvist M, Brundin P (2009) Beyond the brain: widespread pathology in Huntington's disease. Lancet Neurol 8 : 765–774. doi: 10.1016/S1474-4422(09)70178-4 19608102
16. Zielonka D, Piotrowska I, Marcinkowski JT, Mielcarek M (2014) Skeletal muscle pathology in Huntington's disease. Front Physiol 5 : 380 : 1–5.
17. Kosinski CM, Schlangen C, Gellerich FN, Gizatullina Z, Deschauer M, et al. (2007) Myopathy as a first symptom of Huntington's disease in a Marathon runner. Mov Disord 22 : 1637–1640. 17534945
18. Luthi-Carter R, Hanson SA, Strand AD, Bergstrom DA, Chun W, et al. (2002) Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum Mol Genet 11 : 1911–1926. 12165554
19. Strand AD, Aragaki AK, Shaw D, Bird T, Holton J, et al. (2005) Gene expression in Huntington's disease skeletal muscle: a potential biomarker. Hum Mol Genet 14 : 1863–1876. 15888475
20. Lodi R, Schapira AH, Manners D, Styles P, Wood NW, et al. (2000) Abnormal in vivo skeletal muscle energy metabolism in Huntington's disease and dentatorubropallidoluysian atrophy. Ann Neurol 48 : 72–76. 10894218
21. Saft C, Zange J, Andrich J, Muller K, Lindenberg K, et al. (2005) Mitochondrial impairment in patients and asymptomatic mutation carriers of Huntington's disease. Mov Disord 20 : 674–679. 15704211
22. Ciammola A, Sassone J, Sciacco M, Mencacci NE, Ripolone M, et al. (2011) Low anaerobic threshold and increased skeletal muscle lactate production in subjects with Huntington's disease. Mov Disord 26 : 130–137. doi: 10.1002/mds.23258 20931633
23. Quintanilla RA, Johnson GV (2009) Role of mitochondrial dysfunction in the pathogenesis of Huntington's disease. Brain Res Bull 80 : 242–247. doi: 10.1016/j.brainresbull.2009.07.010 19622387
24. Lin J, Handschin C, Spiegelman BM (2005) Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1 : 361–370. 16054085
25. Johri A, Calingasan NY, Hennessey TM, Sharma A, Yang L, et al. (2012) Pharmacologic activation of mitochondrial biogenesis exerts widespread beneficial effects in a transgenic mouse model of Huntington's disease. Hum Mol Genet 21 : 1124–1137. doi: 10.1093/hmg/ddr541 22095692
26. Fujimoto M, Takaki E, Hayashi T, Kitaura Y, Tanaka Y, et al. (2005) Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J Biol Chem 280 : 34908–34916. 16051598
27. Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, et al. (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87 : 493–506. 8898202
28. Lin CH, Tallaksen-Greene S, Chien WM, Cearley JA, Jackson WS, et al. (2001) Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum Mol Genet 10 : 137–144. 11152661
29. Woodman B, Butler R, Landles C, Lupton MK, Tse J, et al. (2007) The Hdh(Q150/Q150) knock-in mouse model of HD and the R6/2 exon 1 model develop comparable and widespread molecular phenotypes. Brain Res Bull 72 : 83–97. 17352931
30. Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, et al. (2013) Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci U S A 110 : 2366–2370. doi: 10.1073/pnas.1221891110 23341618
31. Sathasivam K, Hobbs C, Turmaine M, Mangiarini L, Mahal A, et al. (1999) Formation of polyglutamine inclusions in non-CNS tissue. Hum Mol Genet 8 : 813–822. 10196370
32. Mitchell JC, McGoldrick P, Vance C, Hortobagyi T, Sreedharan J, et al. (2013) Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age - and dose-dependent fashion. Acta Neuropathol 125 : 273–288. doi: 10.1007/s00401-012-1043-z 22961620
33. Tsika RW, Schramm C, Simmer G, Fitzsimons DP, Moss RL, et al. (2008) Overexpression of TEAD-1 in transgenic mouse striated muscles produces a slower skeletal muscle contractile phenotype. J Biol Chem 283 : 36154–36167. doi: 10.1074/jbc.M807461200 18978355
34. Yoshida T (2008) MCAT elements and the TEF-1 family of transcription factors in muscle development and disease. Arterioscler Thromb Vasc Biol 28 : 8–17. 17962623
35. Karasseva N, Tsika G, Ji J, Zhang A, Mao X, et al. (2003) Transcription enhancer factor 1 binds multiple muscle MEF2 and A/T-rich elements during fast-to-slow skeletal muscle fiber type transitions. Mol Cell Biol 23 : 5143–5164. 12861002
36. Zhao P, Caretti G, Mitchell S, McKeehan WL, Boskey AL, et al. (2006) Fgfr4 is required for effective muscle regeneration in vivo. Delineation of a MyoD-Tead2-Fgfr4 transcriptional pathway. J Biol Chem 281 : 429–438. 16267055
37. Mielcarek M, Gunther S, Kruger M, Braun T (2002) VITO-1, a novel vestigial related protein is predominantly expressed in the skeletal muscle lineage. Mech Dev 119 Suppl 1: S269–274. 14516696
38. Mielcarek M, Piotrowska I, Schneider A, Gunther S, Braun T (2009) VITO-2, a new SID domain protein, is expressed in the myogenic lineage during early mouse embryonic development. Gene Expr Patterns 9 : 129–137. doi: 10.1016/j.gep.2008.12.002 19118645
39. Gunther S, Mielcarek M, Kruger M, Braun T (2004) VITO-1 is an essential cofactor of TEF1-dependent muscle-specific gene regulation. Nucleic Acids Res 32 : 791–802. 14762206
40. Ehrnhoefer DE, Skotte NH, Ladha S, Nguyen YT, Qiu X, et al. (2014) p53 increases caspase-6 expression and activation in muscle tissue expressing mutant huntingtin. Hum Mol Genet 23 : 717–729. doi: 10.1093/hmg/ddt458 24070868
41. Cohen TJ, Barrientos T, Hartman ZC, Garvey SM, Cox GA, et al. (2009) The deacetylase HDAC4 controls myocyte enhancing factor-2-dependent structural gene expression in response to neural activity. FASEB J 23 : 99–106. doi: 10.1096/fj.08-115931 18780762
42. Cohen TJ, Waddell DS, Barrientos T, Lu Z, Feng G, et al. (2007) The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J Biol Chem 282 : 33752–33759. 17873280
43. Winbanks CE, Beyer C, Hagg A, Qian H, Sepulveda PV, et al. (2013) miR-206 represses hypertrophy of myogenic cells but not muscle fibers via inhibition of HDAC4. PLoS One 8: e73589. doi: 10.1371/journal.pone.0073589 24023888
44. Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, et al. (2012) microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest 122 : 2054–2065. doi: 10.1172/JCI62656 22546853
45. Williams AH, Valdez G, Moresi V, Qi X, McAnally J, et al. (2009) MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326 : 1549–1554. doi: 10.1126/science.1181046 20007902
46. Bruneteau G, Simonet T, Bauche S, Mandjee N, Malfatti E, et al. (2013) Muscle histone deacetylase 4 upregulation in amyotrophic lateral sclerosis: potential role in reinnervation ability and disease progression. Brain 136 : 2359–2368. doi: 10.1093/brain/awt164 23824486
47. Bricceno KV, Sampognaro PJ, Van Meerbeke JP, Sumner CJ, Fischbeck KH, et al. (2012) Histone deacetylase inhibition suppresses myogenin-dependent atrogene activation in spinal muscular atrophy mice. Hum Mol Genet 21 : 4448–4459. 22798624
48. Egerman MA, Glass DJ (2014) Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol 49 : 59–68. doi: 10.3109/10409238.2013.857291 24237131
49. Bassel-Duby R, Olson EN (2006) Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem 75 : 19–37. 16756483
50. Brooks NE, Myburgh KH (2014) Skeletal muscle wasting with disuse atrophy is multi-dimensional: the response and interaction of myonuclei, satellite cells and signaling pathways. Front Physiol 5 : 99. doi: 10.3389/fphys.2014.00099 24672488
51. Ribchester RR, Thomson D, Wood NI, Hinks T, Gillingwater TH, et al. (2004) Progressive abnormalities in skeletal muscle and neuromuscular junctions of transgenic mice expressing the Huntington's disease mutation. Eur J Neurosci 20 : 3092–3114. 15579164
52. Ciciliot S, Rossi AC, Dyar KA, Blaauw B, Schiaffino S (2013) Muscle type and fiber type specificity in muscle wasting. Int J Biochem Cell Biol 45 : 2191–2199. doi: 10.1016/j.biocel.2013.05.016 23702032
53. Schiaffino S, Reggiani C (2011) Fiber types in mammalian skeletal muscles. Physiol Rev 91 : 1447–1531. doi: 10.1152/physrev.00031.2010 22013216
54. Waters CW, Varuzhanyan G, Talmadge RJ, Voss AA (2013) Huntington disease skeletal muscle is hyperexcitable owing to chloride and potassium channel dysfunction. Proc Natl Acad Sci U S A 110 : 9160–9165. doi: 10.1073/pnas.1220068110 23671115
55. Wade A, Jacobs P, Morton AJ (2008) Atrophy and degeneration in sciatic nerve of presymptomatic mice carrying the Huntington's disease mutation. Brain Res 1188 : 61–68. 18062944
56. Rozas JL, Gomez-Sanchez L, Tomas-Zapico C, Lucas JJ, Fernandez-Chacon R (2011) Increased neurotransmitter release at the neuromuscular junction in a mouse model of polyglutamine disease. J Neurosci 31 : 1106–1113. doi: 10.1523/JNEUROSCI.2011-10.2011 21248135
57. Dogan I, Eickhoff SB, Schulz JB, Shah NJ, Laird AR, et al. (2013) Consistent neurodegeneration and its association with clinical progression in Huntington's disease: a coordinate-based meta-analysis. Neurodegener Dis 12 : 23–35. doi: 10.1159/000339528 22922585
58. Koopman R, Ly CH, Ryall JG (2014) A metabolic link to skeletal muscle wasting and regeneration. Front Physiol 5 : 32. doi: 10.3389/fphys.2014.00032 24567722
59. Ciammola A, Sassone J, Alberti L, Meola G, Mancinelli E, et al. (2006) Increased apoptosis, Huntingtin inclusions and altered differentiation in muscle cell cultures from Huntington's disease subjects. Cell Death Differ 13 : 2068–2078. 16729030
60. Ismailoglu I, Chen Q, Popowski M, Yang L, Gross SS, et al. (2014) Huntingtin protein is essential for mitochondrial metabolism, bioenergetics and structure in murine embryonic stem cells. Dev Biol 391 : 230–240. doi: 10.1016/j.ydbio.2014.04.005 24780625
61. Schips TG, Wietelmann A, Hohn K, Schimanski S, Walther P, et al. (2011) FoxO3 induces reversible cardiac atrophy and autophagy in a transgenic mouse model. Cardiovasc Res 91 : 587–597. doi: 10.1093/cvr/cvr144 21628326
62. Hockly E, Woodman B, Mahal A, Lewis CM, Bates G (2003) Standardization and statistical approaches to therapeutic trials in the R6/2 mouse. Brain Res Bull 61 : 469–479. 13679245
63. Mielcarek M, Benn CL, Franklin SA, Smith DL, Woodman B, et al. (2011) SAHA decreases HDAC 2 and 4 levels in vivo and improves molecular phenotypes in the R6/2 mouse model of Huntington's disease. PLoS One 6: e27746. doi: 10.1371/journal.pone.0027746 22140466
64. Mielcarek M, Landles C, Weiss A, Bradaia A, Seredenina T, et al. (2013) HDAC4 reduction: a novel therapeutic strategy to target cytoplasmic huntingtin and ameliorate neurodegeneration. PLoS Biol 11: e1001717. doi: 10.1371/journal.pbio.1001717 24302884
65. Mielcarek M, Seredenina T, Stokes MP, Osborne GF, Landles C, et al. (2013) HDAC4 does not act as a protein deacetylase in the postnatal murine brain in vivo. PLoS One 8: e80849. doi: 10.1371/journal.pone.0080849 24278330
66. Smolenski RT, Lachno DR, Ledingham SJ, Yacoub MH (1990) Determination of sixteen nucleotides, nucleosides and bases using high-performance liquid chromatography and its application to the study of purine metabolism in hearts for transplantation. J Chromatogr 527 : 414–420. 2387888
67. Soppa GK, Smolenski RT, Latif N, Yuen AH, Malik A, et al. (2005) Effects of chronic administration of clenbuterol on function and metabolism of adult rat cardiac muscle. Am J Physiol Heart Circ Physiol 288: H1468–H1476. 15528231
68. Acevedo-Arozena A, Kalmar B, Essa S, Ricketts T, Joyce P, et al. (2011) A comprehensive assessment of the SOD1G93A low-copy transgenic mouse, which models human amyotrophic lateral sclerosis. Dis Model Mech 4 : 686–700. doi: 10.1242/dmm.007237 21540242
69. Lu CH, Petzold A, Kalmar B, Dick J, Malaspina A, et al. (2012) Plasma neurofilament heavy chain levels correlate to markers of late stage disease progression and treatment response in SOD1(G93A) mice that model ALS. PLoS One 7: e40998. doi: 10.1371/journal.pone.0040998 22815892
Štítky
Genetika Reprodukční medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 3
-
Všechny články tohoto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



