-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaRapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
Xylose is an important component of lignocellulose hydrolysates used for the production of bioethanol, but the yeast Saccharomyces cerevisiae is unable to utilize xylose. Insertion of a bacterial xylose isomerase gene and improvement of growth on xylose by evolutionary adaptation resulted in amplification of this gene and efficient xylose fermentation capacity. Further analysis of the final and intermediate strains from the evolutionary adaptation process revealed interesting features about the mechanisms involved in gene amplification events, which have occurred frequently in natural evolution. We now show that a circular DNA element was spontaneously created by the yeast, encompassing the xylose isomerase gene and an ARS element, present by coincidence adjacent of the inserted xylose isomerase gene. ARS elements are the sites where DNA polymerase initiates duplication of DNA. Interestingly, this has revealed for the first time in yeast that circular DNA plasmids can be created from genomic DNA in the absence of flanking repetitive sequences.
Published in the journal: . PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005010
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005010Summary
Xylose is an important component of lignocellulose hydrolysates used for the production of bioethanol, but the yeast Saccharomyces cerevisiae is unable to utilize xylose. Insertion of a bacterial xylose isomerase gene and improvement of growth on xylose by evolutionary adaptation resulted in amplification of this gene and efficient xylose fermentation capacity. Further analysis of the final and intermediate strains from the evolutionary adaptation process revealed interesting features about the mechanisms involved in gene amplification events, which have occurred frequently in natural evolution. We now show that a circular DNA element was spontaneously created by the yeast, encompassing the xylose isomerase gene and an ARS element, present by coincidence adjacent of the inserted xylose isomerase gene. ARS elements are the sites where DNA polymerase initiates duplication of DNA. Interestingly, this has revealed for the first time in yeast that circular DNA plasmids can be created from genomic DNA in the absence of flanking repetitive sequences.
Introduction
Microbial evolutionary experiments have received considerable attention in recent years for various reasons. First they allow in depth understanding of the fundamental process of evolution in a rapid and rigorously controlled way [1]. Second, microbial evolution raises great interest in various fields such as in medicine and industrial applications [2–4]. Using nature’s evolutionary principle of variation and selection, microbial evolution has been used for development and optimization of several production host organisms in industrial applications. The speed of fitness gain in a new environment depends on the rate of genetic changes as well as their advantage [5]. Genetic changes that occur during evolution include point mutations, gene deletions or amplifications, and often gene rearrangements involving transposable elements, which in turn might generate deletions or amplifications.
In a broader context, gene duplications and amplifications have played a crucial role in the evolution and genetic diversity of species, in particular for adaptation to restrictive environmental conditions [6,7]. Segmental duplications and amplifications are common in eukaryotes. In the yeast Saccharomyces cerevisiae genome, about 1 out of 5 genes have been identified as duplicates [8]. Moreover, nearly 2% of the coding sequences in S. cerevisiae are tandem gene arrays [9]. Tandem repetitive DNA sequences that include ribosomal DNA (rDNA) and the telomeric loci are very prone to copy number alterations as a consequence of homologous recombination (HR). Such regions play a significant role in the plasticity of the genome. Other repetitive elements like Ty elements and solo Long Terminal Repeats (LTRs) that are widely dispersed in the yeast genome are potential substrates for HR between the short repeats flanking a DNA segment. In spite of the major contribution of repetitive DNA sequences in elevated rates of genome plasticity, segmental amplifications are not restricted to regions with repetitive sequences. However, the generation of tandem gene amplifications from originally single copy sequences is not well understood. The creation of extrachromosomal circular DNA (eccDNA) has been proposed as a possible mechanism for the origin and plasticity of tandem gene repeats [10]. The formation of eccDNA has been attributed to the circularization of a DNA segment from a chromosome during HR between preexisting closely located homologous sequences such as LTRs, resulting in the excision of the DNA segment [11]. There has only been little experimental evidence for the formation of eccDNA in the absence of repeat sequences [12].
The yeast S. cerevisiae has a very long proven record of industrial application, due to its efficient conversion of glucose into ethanol with high productivity, and its substantial tolerance to various inhibitory compounds, including ethanol [13,14]. However, it is unable to efficiently metabolize D-xylose into ethanol. Typically, D-xylose accounts for about one-third of the sugars in lignocellulosic biomass [15,16]. Due to the recent interest in biofuel production with biomass from waste streams and bioenergy crops, engineering S. cerevisiae for efficient D-xylose to ethanol conversion has become an important research focus [17]. Expression of the heterologous structural genes responsible for D-xylose to ethanol conversion in S. cerevisiae did not lead by itself to sufficient productivity for industrial scale application [18].
Recently, using a combination of metabolic and evolutionary engineering strategies, we have developed a robust industrial yeast strain that displayed the highest D-xylose to ethanol conversion rate and yield compared to any other recombinant yeast strain reported previously [19]. Here, we report the elucidation of one of the crucial genetic changes responsible for the rapid D-xylose utilization rate in this strain. Using whole genome sequence comparison of the evolved strain with that of the parent strain, we identified a major copy number variation in the heterologous gene XylA, encoding Clostridium phytofermentans XI (xylose isomerase)), that correlated with the high enzymatic activity measured in crude cell extracts. In addition, we investigated the evolutionary process that resulted in stable integration of this gene in tandem high copy number into the genome using several intermediate strains with varying xylose fermentation rate. We confirmed the formation of self-replicating eccDNA carrying the gene XylA during adaptive evolution in the absence of any homologous sequence flanking the repeat DNA segment. During later stages of the adaptive evolution, the generated eccDNA had integrated into the locus of origin in the genome, generating increasing numbers of tandem repeats first in one of the chromosomes and later in the second chromosome in the diploid strain. We propose that formation of eccDNA can occur in the absence of HR and can serve as a rapid means of adjustment to selection pressure during evolutionary adaptation, especially when higher gene dosage serves as a selective advantage for proliferation or survival.
Results
Whole genome sequence analysis
Recently, we reported the development of an industrial D-xylose utilizing strain of S. cerevisiae, GS1.11–26, using a combination of metabolic engineering, genome shuffling and evolutionary adaptation [19]. Briefly, we integrated the gene XylA coding for Xylose Isomerase from the bacterium Clostridium phytofermentans together with all the known genes important for D-xylose and L-arabinose metabolism into an industrial bioethanol production strain, ER, (Ethanol Red). However, the recombinant strain named HDY.GUF5 was unable to utilize any D-xylose. We then mutagenized this strain by EMS and selected a mutant isolate M315 that was able to grow slowly on D-xylose but had poor D-xylose fermentation capacity. Genome shuffling of this mutant with its parent HDY.GUF5, followed by selection for faster D-xylose growth resulted in little improvement. Subsequently, we performed evolutionary adaptation in serial batch cultures containing D-xylose as main carbon source. A striking observation during this adaptation process was a drastic gain in D-xylose fermentation capacity at the second serial batch culture. We proposed that a crucial genetic change had happened at that step, which resulted in a rapid gain in performance. To elucidate this change, we sequenced the genome of the original parent strain HDY.GUF5 and the final evolved strain GS1.11–26 and performed a global genome sequence comparison.
Detection and evaluation of copy-number variations (CNVs)
Since the coverage depth of the sequence reads reveals CNVs among genomes of different strains [20], the sequence coverage of all 16 nuclear chromosomes was analyzed at the nucleotide level with an average window of 500 bp. The log2 ratio of the read depth between the evolved and the parent strains was then calculated and plotted over the length of the chromosomes (Fig. 1). Accordingly, chromosome IX and chromosome XVI showed a 50% higher coverage in the evolved strain compared to the parent strain, indicating duplication of one of the two homologues for each chromosome in the evolved strain.
Fig. 1. Comparison of the genome sequence coverage between the parent strain HDY.GUF5 and evolved strain GS1.11–26. 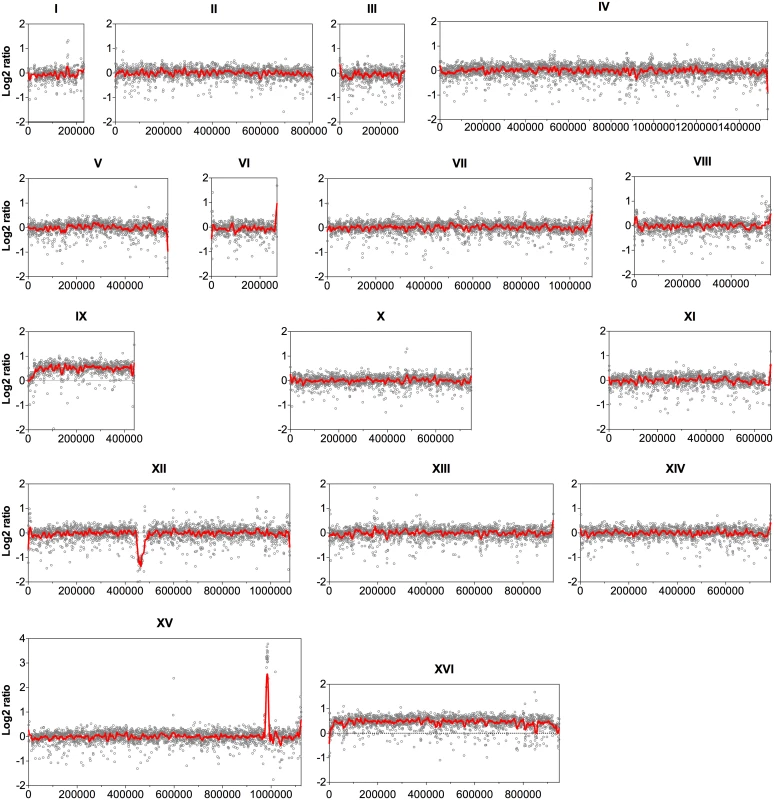
Log2 ratio depicted from whole genome sequence coverage between the parental and the evolved strain is presented for each of the 16 chromosomes. Each grey circle represents the value of the log2 ratio obtained from the sequence coverage calculated for averaged sliding windows of 500 nucleotide positions. The red line indicates the smoother trend calculated by moving average values of 10,000 bp. In chromosome XII, the coverage at the ribosomal DNA locus was reduced in the evolved strain by about 50% relative to the parent strain, indicating the loss of several copies of the ribosomal DNA (rDNA) genes. rDNA genes are present in about 150 to 200 tandem copies in the S. cerevisiae genome [21]. The possible effect of the reduction of the rDNA copies in our evolved strain was not investigated further. However, large deletions of multiple rDNA copies are common spontaneous phenomena in the yeast genome. Strains with up to 50% reduction in the number of rDNA copies compared to wild type laboratory strains did not show any noticeable defect in mitotic growth and meiotic reproduction [22].
The most prominent CNV occurred in a region at the right arm of chromosome XV. This was the region where the D-xylose and L-arabinose metabolism gene cassette had been integrated in the genome of the parent strain HDY.GUF5 [19]. Part of the integrated gene cassette, containing the XylA gene, and a sequence upstream of the integrated cassette, that includes the gene REV1, the tRNA gene tP(UGG)O3 and the autonomously replicating sequence ARS1529, were amplified about 9 times (estimated from the log2 ratio) in the evolved strain compared to the parent strain (Fig. 2). XylA encodes xylose isomerase that converts D-xylose to D-xylulose, the rate-limiting step in D-xylose fermentation. We previously showed that the evolved strain GS1.11–26 displayed significantly higher (about 17 times) XI activity than the parent strain, which displayed only moderate activity [19]. The high copy number of XylA in the evolved strain is therefore consistent with its high XI activity, though the fold increase in the XI activity was higher than that of the copy number.
Fig. 2. Comparison of the xylose cassette sequence coverage between the parent strain HDY.GUF5 and evolved strain GS1.11–26. 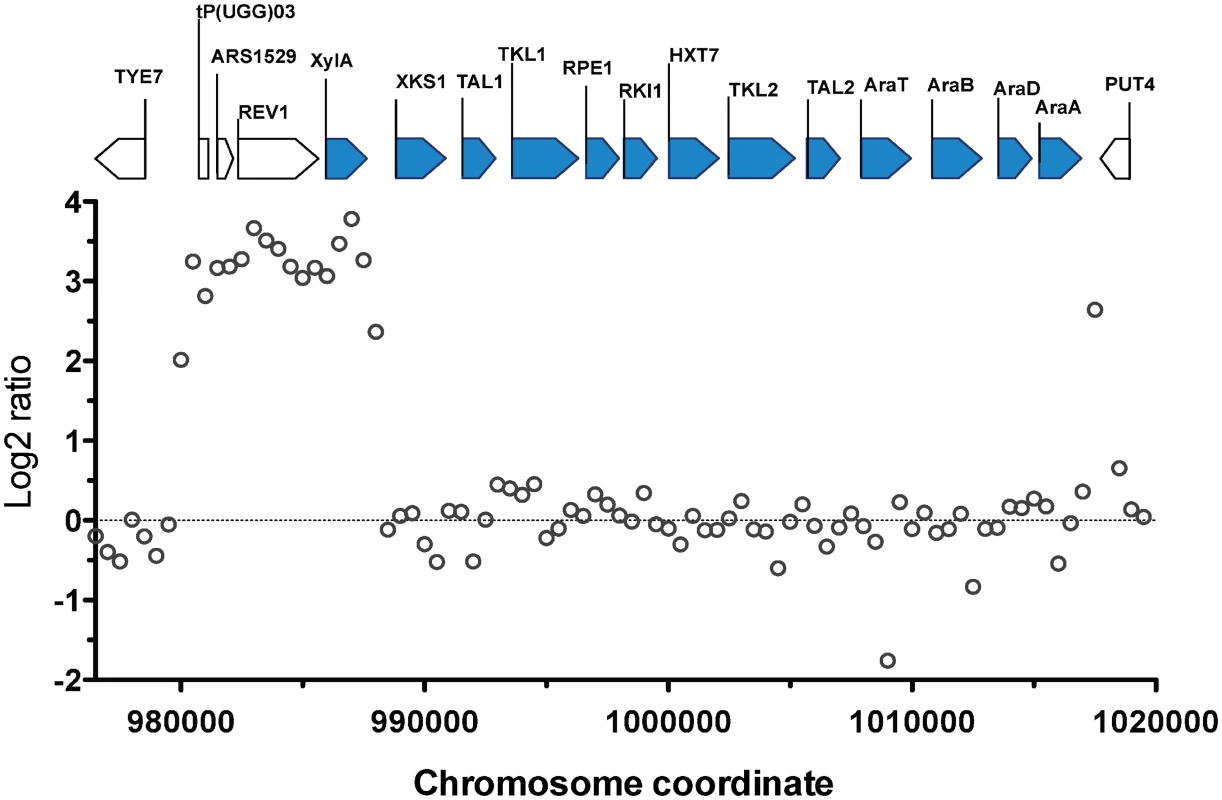
Log2 ratio of sequence coverage between the evolved and parent strain in the locus of chromosome XV, where the D-xylose and arabinose gene cassette has been integrated, is plotted over the length of the chromosomal fragment. Annotations present in the locus are indicated by bars at the top of the figure. Bars shaded in blue correspond to the heterologous genes that were inserted into the chromosome, while the unshaded bars represent genes present in part of the original yeast chromosome. The coverage was computed for averaged sliding windows of 500 nucleotide positions. Amplification of the XylA-locus as tandem repeat in GS1.11–26
We sought to understand the structural arrangement of the amplification of the XylA-locus. The presence of the autonomous replication sequence ARS1529 in the amplified XylA-locus made us consider the possibility that this region got amplified as a self-replicating eccDNA. This idea was supported by the observation of break points on either end of the amplified region when the Illumina sequence reads were mapped to the reference genome (Fig. 3A). The sequence reads at one end of the break point contained partially unmatched sequences that matched with the sequence of the opposite end. This condition implies either circular DNA or tandem repeat formation.
Fig. 3. Sequence analysis at the borders of the amplified XylA-locus, and verification of the presence of circular or tandem repeats. 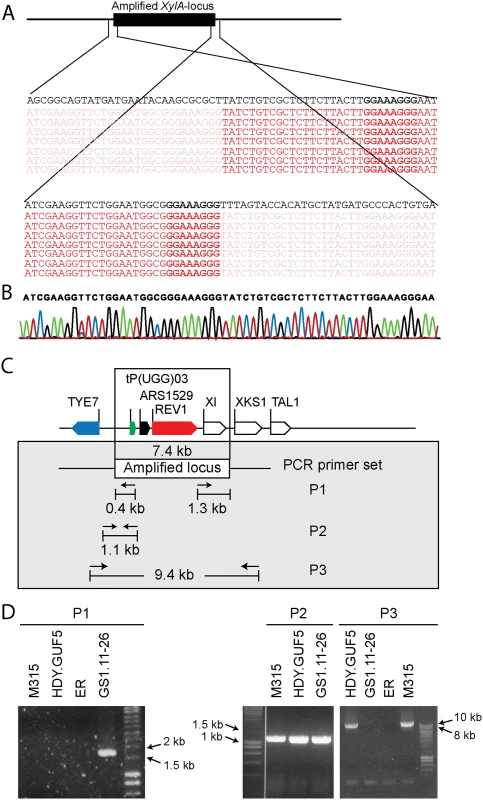
(A) Illumina sequence reads mapped to the reference sequence at both ends of the amplified XylA-locus. Part of the reads that match to the reference sequence are shown in dark red, while sub-reads that do not match to the reference sequence are indicated in faded red color. The reference sequence is shown in black at the top of the reads. Microhomologies located on either ends of the amplified locus are shown in black rectangular boxes. (B) Chromatogram from Sanger sequencing of the amplified locus, showing continuous reads through the break point of the Illumina sequence reads alignment. The Sanger sequencing was performed using a PCR product obtained by amplification with primers that anneal outwards from both ends of the locus. The same result was obtained when the plasmid isolated from strain GS1.2–6 was sequenced. (C) Schematic representation of the amplified XylA-locus and PCR primer sets used. Horizontal arrows stand for annealing sites and direction of the PCR primers used to verify circular or tandem repeat formation (primer set P1), the presence of the XylA locus at the right position (primer set P2) and the possibility of deletion or single copy of the XylA-locus (P3). The size of the expected PCR product is given in kb. (D) Agarose gel electrophoresis picture of the PCR products obtained using the three sets of primers (P1, P2 and P3) with DNA samples from different strains. For primer set P3, the result obtained with a long extension time (8 min) is shown. ER (Ethanol Red) is the original industrial strain in which the xylose/arabinose gene cassette has been inserted into the genome. To validate this assumption, we performed PCR, (polymerase chain reaction) using a primer set P1, which consisted of a pair of primers inside the amplified region that project outwards in opposite direction (Fig. 3C). A PCR product of 1.7kb was expected only if a circular or tandem repeat sequence had been generated. The evolved strain GS1.11–26 gave a PCR product of the expected size, while no PCR product was obtained from the parent strain HDY.GUF5, the mutant M315 and the original industrial strain Ethanol Red that does not have the cassette (Fig. 3D). The PCR product obtained was then sequenced using conventional Sanger sequencing. The resulting contigs were shown to read through the break point that was obtained from the alignment of Illumina sequence reads at either end of the amplified region (Fig. 3B), indicating a continuous DNA sequence, which in turn points to either a circular form or a tandem repeat.
To differentiate between circular DNA and tandem repeat, we first evaluated if the chromosomal copy of the amplified region had been deleted in the evolved strain. Deletion of the locus would contradict the possibility of tandem amplification of the locus. For this purpose, two sets of primers were used (Fig. 3C). The primer set P2 contained a forward primer annealing upstream of the amplified locus and a reverse primer annealing inside the amplified locus, and detects the presence of the XylA-locus at the right position in the chromosome. The primer set P3, contained a forward primer upstream of the amplified locus and a reverse primer downstream of the amplified locus, and was expected to give a PCR product only upon deletion of the XylA locus since the locus is too large to be amplified with the PCR conditions used (2 min extension time). A band of the expected 1.1 kb size with the PCR set P2 (Fig. 3D) and a negative result with the PCR set P3 with the 2 min extension time was obtained for both the parent HDY.GUF5, the M315 mutant and the evolved strain GS1.11–26. The positive band with PCR set P2 was expected since the whole genome sequencing data indicated that at least one of the alleles was present in the locus. However, the absence of a PCR product using primer set P3 indicated that neither of the two alleles of the chromosomal XylA-locus was deleted. Therefore, neither tandem amplification nor eccDNA could be excluded on the basis of this PCR analysis.
We then performed PCR using the primer set P3 under conditions that allow amplification of the whole amplified XylA-locus (long extension time). A single copy of the XylA-locus in the genome was expected to produce a 9.4 kb PCR product while chromosomal duplication or amplification of the locus should not produce any PCR product since it would be too large to be amplified. The parent HDY.GUF5 and the mutant M315 strains gave rise to a PCR product with the correct size of 9.4 kb but the evolved strain GS1.11–26 did not give rise to any band after several attempts (Fig. 3D). The HDY.GUF5 positive control gave rise to the expected band in all repetitions. This indicates that only one copy of the XylA locus is present in each of the two alleles in the parent strain and the M315 mutant, but that the evolved strain might have multiple copies in both alleles. To confirm this assumption, Southern blot analysis was performed with genomic DNA digested with two different restriction enzymes. First, the DNA was digested with HindIII that cuts only once inside the amplified XylA-locus. A unique probe that hybridizes in the XylA sequence was used to visualize the band. In the presence of only a single copy of the XylA-locus, a single band of 4.3 kb was expected while a circular or tandem repeat sequence should give two bands of 4.3 kb and 7.4 kb (Fig. 4A). Two bands of the expected size were detected for the evolved strain, GS1.11–26. The intensity of the 7.4 kb band was estimated to be about 8 fold higher than the intensity of the lower band, which closely correlated to the amplification level deduced from the whole genome sequence analysis. In the parent strain HDY.GUF5 and the mutant M315, only the 4.3 kb band was detected indicating a single copy of each allele (Fig. 4B). No band was detected in the control strain Ethanol Red, which does not contain the gene cassette in the genome.
Fig. 4. Evaluation of the amplified XylA-locus by Southern blot analysis. 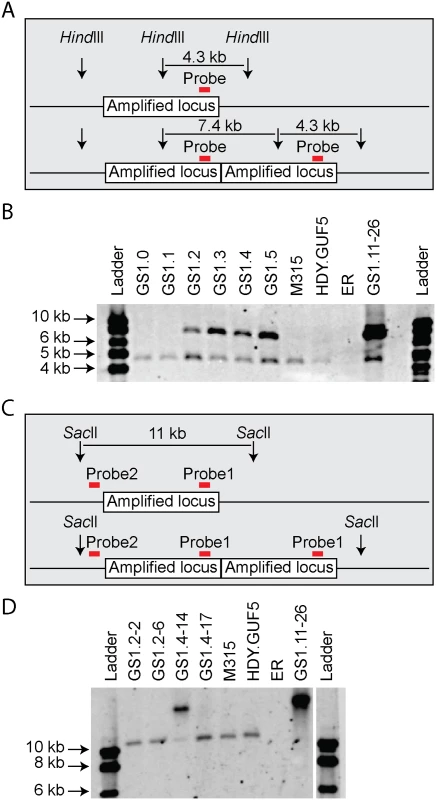
(A) and (C) show a schematic representation of the amplified XylA-locus in GS1.11–26. Vertical arrows represent cutting sites of the restriction enzymes; red horizontal bar indicates the locus of the unique site for probe hybridization. The amplified locus is shown once in the upper part and twice in the lower part. (B) Image of the Southern blot analysis after HindIII digestion. Two bands of expected size 4.3 kb and 7.4 kb were detected in GS1.11–26 and in cultures obtained from the second step of evolutionary adaptation onwards (GS1.2 to GS1.5). The parent strain HDY.GUF5, the mutant M315 and the culture from GS1.0 and GS1.1 showed only one band representing a single copy of the locus. The negative control ER, (Ethanol Red), which does not have the gene cassette, showed no band. (D) Southern blot image after digestion with SacII, that cuts only outside the amplified XylA-locus. Two single cell isolates from GS1.2 (GS1.2–2 and GS1.2–6) and two from GS1.4 (GS1.4–14 and GS1.4–17) were evaluated. The band of about 11kb represents a single copy of the XylA locus, and is present in all the tested strains except the final evolved strain GS1.11–26, which showed only a high molecular weight band. GS1.4–14 showed both the 11kb and a higher molecular weight band indicating the presence of multiple copies of the locus in one allele and a single copy in the other allele. GS1.2–6 apparently lost the circular DNA during growth in non-selective condition. We then digested the genomic DNA with SacII, which cuts only outside the amplified XylA-locus, and hybridized with two different probes annealing either inside (same as the previous probe, Fig. 4C), or outside the amplified locus (between the left SacII restriction site and the amplified locus). An 11 kb band was expected if a single copy of the XylA-locus was present in the chromosomal locus. Accordingly, the presence of the expected 11 kb fragment in the strains HDY.GUF5 and M315 using both probes hybridizing inside (Fig. 4D) or outside the amplified XylA-locus confirmed the existence of a single copy of the XylA-locus in both alleles. On the other hand, the evolved strain showed only a higher molecular weight band, both with the inside (Fig. 4D) and outside probes confirming the presence of multiple copies of the XylA-locus in both chromosomal alleles. This result together with the PCR amplification using PCR set P1 (primers directed outwards on either side of the amplified locus) clearly indicates that the amplification of the locus in GS1.11–26 had occurred in the form of a tandem repeat.
Amplification of the XylA-locus during evolutionary adaptation with eccDNA intermediate
As described in our previous report [19], the evolutionary adaptation step used to obtain the strain GS1.11–26 involved a series of 11 sequential batch cultures in D-xylose medium. To verify at which stage of the evolutionary adaptation process the amplification of the XylA-locus had occurred, a sample from the culture before the evolutionary adaption (GS1.0), and samples at the end of the first 5 serial transfers during the evolutionary adaptation (GS1.1, GS1.2, GS1.3, GS1.4 and GS1.5) were tested by PCR for the presence of the tandem amplification or circular DNA formation using PCR primer set P1. Interestingly, a positive PCR result was obtained in all the samples derived from the second culture (GS1.2) onwards, whereas isolates from GS1.0, GS1.1, as well as the original strains used for the genome shuffling step (HDY.GUF5 and M315) did not give rise to the PCR product (Fig. 5). Southern blot analysis of the same samples after HindIII digestion also confirmed the presence of either circular or multiple copies of the locus in the samples obtained from GS1.2 onwards (Fig. 4B). This strongly suggests that amplification of the XylA-locus had occurred at the second step of the evolutionary adaptation process (GS1.2). As we anticipated, the sharp rise in the rate of D-xylose fermentation in the second culture [19], correlated with amplification of the XylA-locus. Although XylA was expressed from a strong promoter in the parent strain HDY.GUF5, the level of expression was not high enough to confer strong D-xylose fermentation capacity. Amplification of the gene likely increased the expression of XI, which in turn alleviated the rate limiting bottleneck for fermentation of D-xylose.
Fig. 5. Evaluation of the XylA-locus in cultures and single cell clones obtained from the various stages of the evolutionary adaptation process. 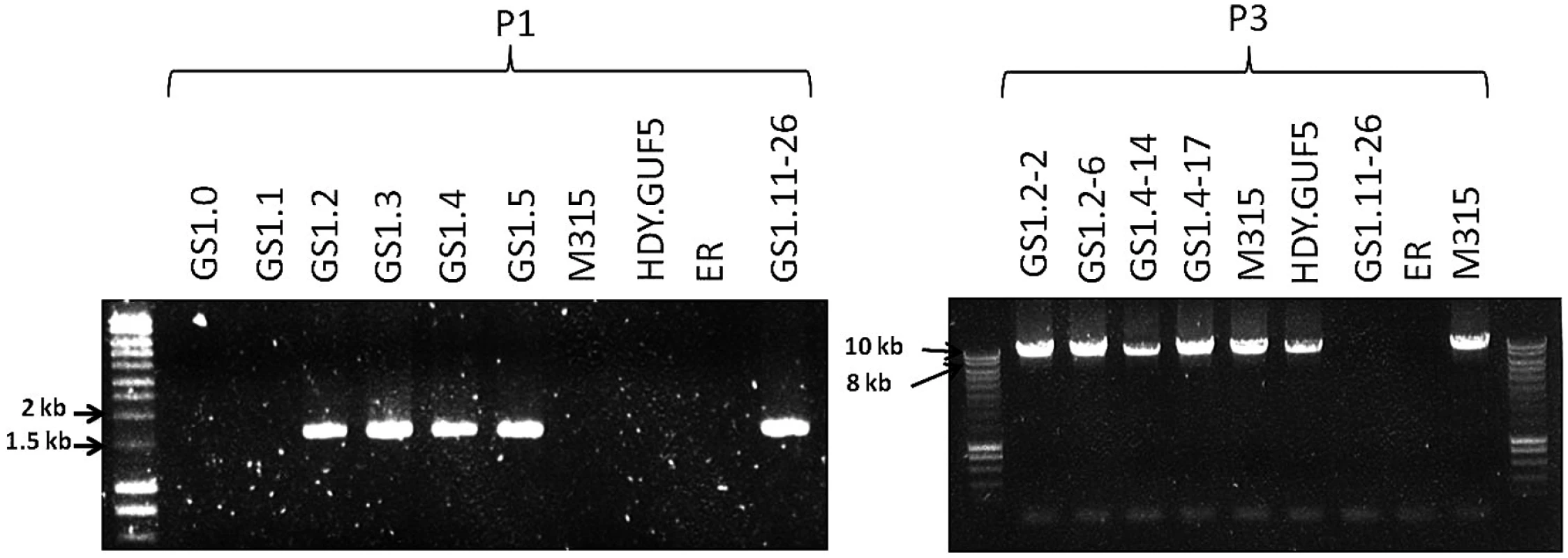
Agarose gel electrophoresis of PCR products obtained using primer sets P1 and P3 are shown (see Fig. 3C for the primer sets). DNA was isolated from a sample of the whole culture obtained just after the genome shuffling step (GS1.0) and after each batch of the first 5 cultures of the evolutionary adaptation process (GS1.1 to GS1.5). GS1.2–2 and GS1.2–6 are single cell isolates from the culture GS1.2, while GS1.4–14 and GS1.4–17 are single cell isolates from the culture GS1.4. The strains M315, HDY.GUF5, ER, (Ethanol Red) and GS1.11–26 have been included for comparison. Remarkably, the chromosomal tandem amplification of the XylA locus in GS1.11–26 was not detected in two D-xylose fermenting single cell clones obtained from GS1.2 (second culture) that showed a positive PCR using the primer set P1. When the Southern blot was performed after SacII digestion on these single cell isolates, called GS1.2–2 and GS1.2–6, only the 11kb band was obtained for both strains, which excludes the possibility of chromosomal amplification (Fig. 4D). This was supported by the result of the PCR amplification of the whole amplified XylA-locus using primer set P3, which gave only the expected 9.4 kb band (Fig. 5). Since no smaller PCR band was obtained when this primer set was used (indicating that the XylA-locus was not deleted), and only a single band was obtained with the Southern blot assay (Fig. 4D), both chromosomes should have a single copy of the XylA-locus in these two strains. Given the positive PCR result obtained using primer set P1 (Fig. 5) that indicates either circular or tandem copies of XylA in all the cultures obtained from GS1.2 onwards, and the absence of chromosomal tandem repeats in the culture of GS1.2 clearly indicate that a circular DNA was generated at the second stage (GS1.2) of the evolutionary adaptation process.
We also performed the Southern blot assay with SacII digested DNA using genomic DNA of two other single cell isolates from GS1.4 (4th culture) to test for the presence of the eccDNA. The first isolate GS1.4–14 had the highest D-xylose fermentation rate among all the isolates obtained from the culture GS1.4. Another isolate GS1.4–17 (with only moderate D-xylose fermentation capacity) was also used for comparison. Accordingly, GS1.4–14 showed both the 11kb and a higher molecular weight band (Fig. 4D). Together with the result of the PCR assay shown in Fig. 5, this result clearly indicates the presence of multiple copies of the locus in one of the alleles and a single copy in the other allele in strain GS1.4–14. The strain GS1.4–17 showed only the 11kb band in the Southern blot assay, which was also consistent with the PCR amplification of the whole XylA-locus using primer set P3 (Fig. 5), indicating the presence of a single copy of the XylA-locus in both alleles. Similar to GS1.2–6, strain GS1.4–17 gave a positive PCR result using primer set P1, indicating the presence of a circular DNA in this strain. This result also suggested a correlation between the multiple integration of the XylA-locus in the genome and the faster D-xylose fermentation.
Eventually, we had obtained clear indications that amplification of the XylA-locus had arisen through a circular intermediate in an early stage of the evolutionary adaptation process, and subsequently recombined in tandem array at the same locus in one of the chromosomes. Later, unequal crossover or other mechanisms might have led the tandem array to be copied into the second chromosome, since GS1.11–26 carried the amplified locus in both alleles.
Stability of eccDNA in intermediate strain GS1.2–6
Next, we evaluated the stability of the high xylose fermentation capacity phenotype in GS1.2–6. If the strain GS1.2–6 carried only the circular plasmid and not the genomic XylA amplification, the loss of the plasmid should cause loss of its high D-xylose growth capacity. To allow for loss of the plasmid, GS1.2–6 was grown in rich medium with glucose (YPD, Yeast extract peptone dextrose) for about 25 generations. The culture was spread for single colonies and 43 single cell isolates were tested for growth in liquid YPX (Yeast extract peptone D-xylose) medium. All isolates except one colony lost the ability to efficiently grow in D-xylose medium, consistent with loss of the XylA carrying circular DNA from GS1.2–6 (Fig. 6A). All the 43 colonies were further tested by PCR for the presence or absence of the eccDNA using primer set P1. Accordingly, the eccDNA could be detected in none of the colonies that lost the D-xylose growth capacity except in the one colony that kept the high growth efficiency in D-xylose (Fig. 6B). This indicates that the GS1.2–6 carried only the circular DNA and not the chromosomal amplification of XylA. The rapid D-xylose fermentation capacity by the final strain GS1.11–26 was previously shown to be stable for more than 50 generations [19]. Consequently, we concluded that the stability of the phenotype in GS1.11–26 is due to the integration of the circular DNA into the genome.
Fig. 6. Evaluation of the stability of the XylA-carrying circular DNA in GS1.2–6. 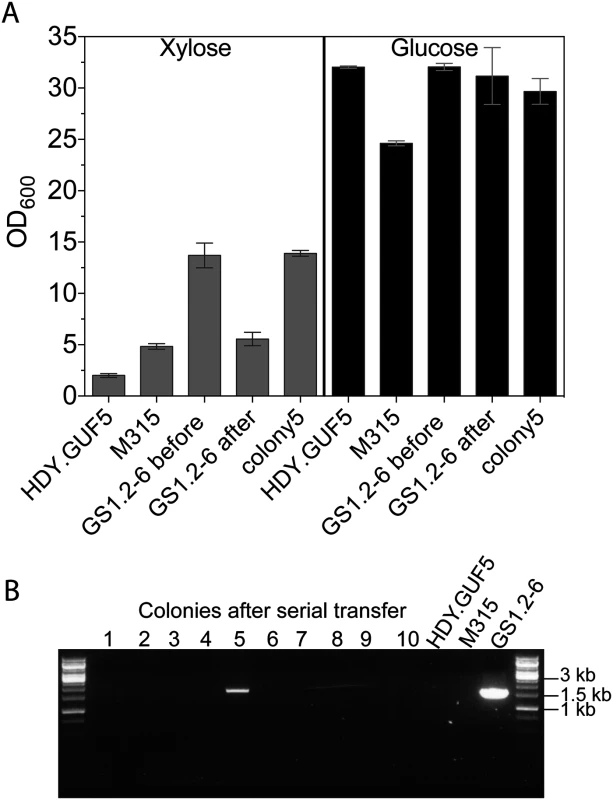
(A) A single cell isolate, GS1.2–6, from the GS1.2 culture, was grown in YPD medium for about 25 generations and spread for single colonies on YPD. Forty-three isolates from the serially transferred culture were tested for growth performance in YPX medium or YPD medium (as control). For comparison, the original GS1.2–6 before serial transfer in YPD, the parent HDY.GUF5 and the mutant M315 were included. The cells were inoculated into YPX medium at an initial OD600 of 0.5. Optical density was measured after 48 h at 30°C. From the 43 colonies tested, only colony 5 displayed efficient growth and is shown separately. The average OD value of the remaining 42 is shown as ‘GS1.2–6 after’. Error bars represent standard deviation from an average of 42 colonies for GS1.2–6 after, and at least triplicate values for the other strains. (B) Agarose gel electrophoresis picture of the PCR assay using primer set P1. The results for 10 colonies out of the 43, and for the control strains are shown. No band was detected for the remaining colonies. The bands in colony 5 and the control GS1.2–6 indicate the presence of the XylA carrying circular DNA. Isolation of eccDNA from intermediate strain GS1.2–6
To further confirm the presence of the eccDNA, plasmid DNA isolation was performed from the strain GS1.2–6, GS1.4–14 and GS1.11–26, using a protocol modified from Singh and Weil [23]. Cells were pre-grown in 100 ml YPX medium for 24 h to enrich the pXI2–6 plasmid content in the cells. The whole 100 ml culture was used for plasmid isolation (see material and methods). As a result, a substantial amount of pXI2–6 plasmid DNA (more than 1 μg) was obtained from GS1.2–6 (Fig. 7A). On the other hand, the amount of pXI2–6 plasmid DNA obtained from GS1.11–26 and GS1.4–14 was too low to be conclusive. This is probably due to the loss of the pXI2–6 plasmid in the later steps of the evolutionary adaptation process, since there was no longer a need for the strain to maintain the plasmid when enough copies of the essential gene XylA had been integrated in the genome sustaining rapid D-xylose utilization. When the pXI2–6 plasmid isolated from GS1.2–6 was sequenced, a 7483 bp circular sequence was obtained, matching the size of the amplified XylA-locus. The complete sequence of the pXI2–6 plasmid has been provided as supplementary information (S1 Dataset). Though there were several polymorphisms compared to the corresponding sequences in the reference S288c genome, the pXI2–6 plasmid sequence was identical to that of the original parent strain obtained by Illumina sequencing. Restriction analysis using two different enzymes also confirmed the correct size of the isolated pXI2–6 plasmid (Fig. 7A).
Fig. 7. Restriction analysis and xylose fermentation conferring capacity of plasmid pXI2–6. 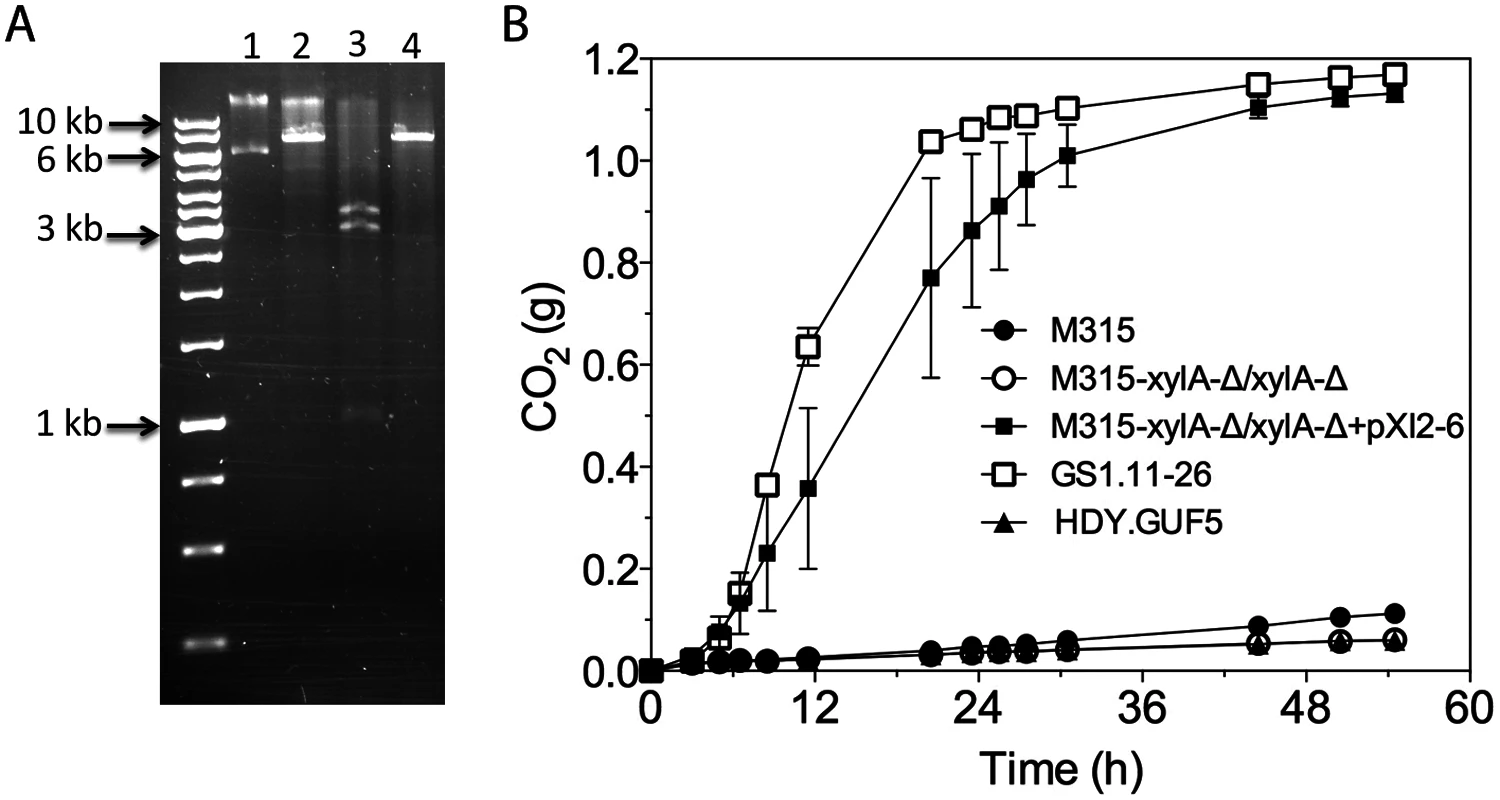
(A) Restriction analysis of pXI2–6. Lane 1, undigested plasmid showing two bands that stand for the supercoiled (lower band) and nicked (upper band) structure; lane 2, XhoI digestion which has single restriction site but shows incomplete digestion; lane 3, SphI and SacI double digestion showing the expected 1kb, 3kb and 3.4kb band; lane 4, HindIII which has a single restriction site and produced the expected 7.4 kb band. (B) D-xylose fermentation performance of the mutant strain M315 expressing the plasmid pXI2–6 after deletion of the chromosomal XylA copies. Batch fermentation was performed in YP medium containing 4% xylose using standing fermentation bottles. The CO2 production was estimated from the weight loss due to conversion of xylose to ethanol and CO2. Error bars represent standard deviation from the mean values of triplicate experiments. We further tested if the isolated pXI2–6 plasmid could be transferred to a new strain and confer the strain with the ability to ferment xylose. For that purpose, strain M315 was chosen since this strain was shown to be able to ferment xylose when XylA was over-expressed [19]. We first deleted both chromosomal copies of XylA from strain M315. Deletion of XylA in M315 completely abolished its growth ability on xylose. Subsequently the isolated pXI2–6 plasmid was transformed into the M315 deletion mutant. We were able to select transformants based on growth on plates containing xylose as a sole carbon source. No colonies were obtained with a control plasmid devoid of the XylA gene. We then evaluated three independent transformants for xylose fermentation capacity. Interestingly, all three transformants carrying the isolated pXI2–6 plasmid were able to efficiently ferment xylose (Fig. 7B) indicating that the isolated pXI2–6 plasmid can be transferred to a different strain and is sufficient to confer the ability to ferment xylose efficiently. To rule out the possibility of integration of the pXI2–6 plasmid in the genome, the three transformants were then grown in YPD medium for about 20 generations and 10 independent colonies from each strain were subsequently checked for growth on xylose as sole carbon source. All of the colonies lost the pXI2–6 plasmid and the ability to grow on xylose, indicating that the XylA gene had not been integrated into the genome and could easily be lost in the absence of selection pressure.
ARS1529 is a functional origin of replication in S. cerevisiae
ARS1529 was previously shown to be a functional replication site in yeast [24]. However, compared to the reference S288c genomic sequence, the ARS1529 sequence in the industrial parent as well as in the evolved strains contained a 5 bp deletion just 13 bp downstream of the ARS consensus sequence (ACS) and also 6 SNPs in an AT-rich region downstream of the ACS (Fig. 8). In order to validate the functionality of the modified ARS1529 version in the eccDNA intermediate, we assessed the ability of this sequence to confer self-replication. We first amplified the region containing ARS1529 together with the tRNA coding sequence and the XylA gene from the genomic DNA of the evolved strain GS1.11–26. The PCR product was then cloned into a yeast integrative vector containing kanMx as a selection marker. After transformation of this construct (pXI-ARS) into the mutant yeast strain M315 and selection for growth in the presence of geneticin, we obtained several transformants, with similar transformation efficiency as that of the 2μ-based plasmid, showing that the plasmid was able to propagate using ARS1529. To confirm that ARS1529 alone is sufficient to render replication capacity, we deleted all the sequences originating from the yeast genome, including the tRNA(UGG) and the XylA sequence, except the 236 bp ARS1529 sequence. Transformation of this plasmid into M315 and selection in the presence of geneticin, resulted in several transformants, which were confirmed by PCR for the presence of the plasmid. No transformants could be obtained when the same plasmid devoid of the ARS1529 sequence was transformed into M315. This resulted confirmed that the ARS1529 sequence is sufficient for plasmid replication in yeast.
Fig. 8. ARS1529 sequence comparison between the evolved strain GS1.11–26 and the sequence in the reference strain S288c. 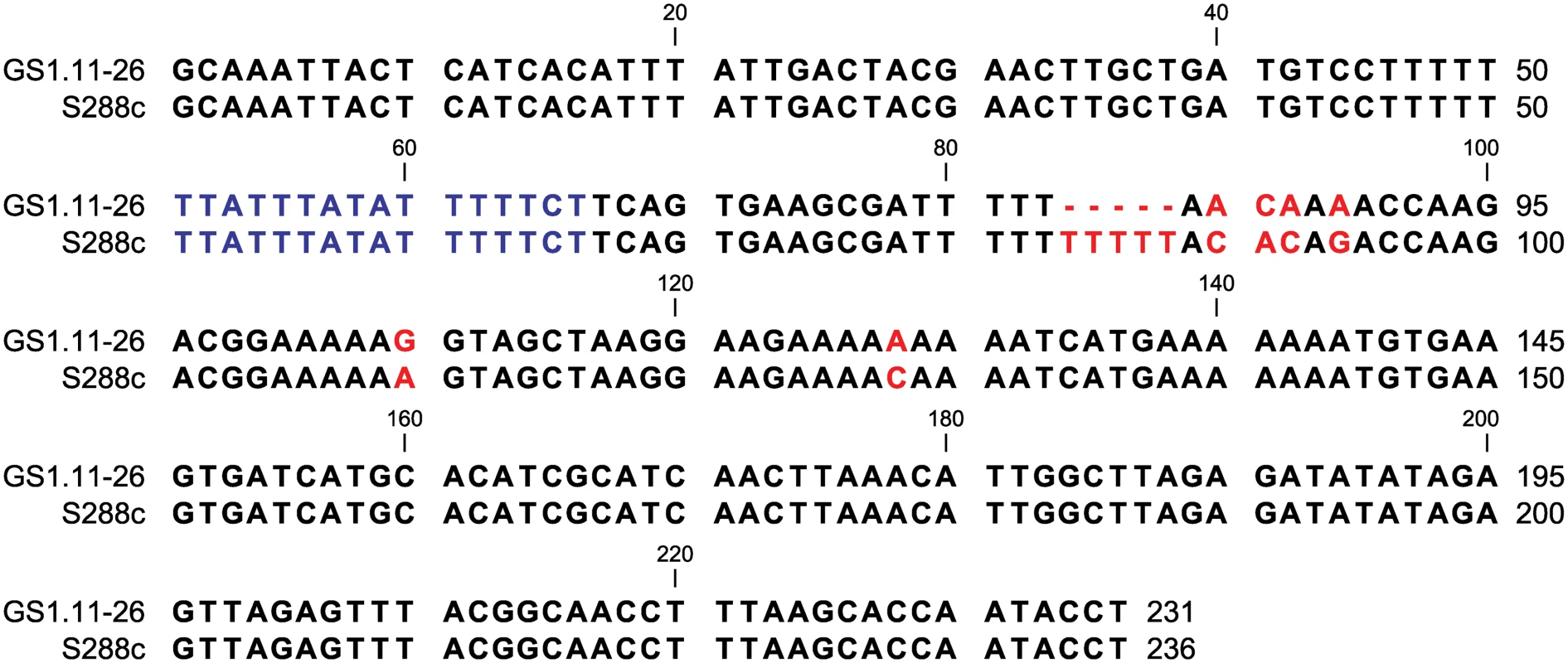
The nucleotide sequence in blue represents the ARS consensus sequence [24]. A gap of 5 base pairs in GS1.11–26 from bp 84 till 88 is indicated with red dashed lines. Other SNPs are also shown in red. The sequence from GS1.11–26 was confirmed by Sanger sequencing and was similar to the sequence obtained by Illumina sequencing. Discussion
Copy number variations are a major driving force for rapid genome evolution. The frequency of CNVs in eukaryotes is remarkably high. While many CNVs are detrimental or have no discernible effect, certain gene duplications or amplifications offer adaptive advantage under specific environmental conditions [6]. Typical examples include the copy number increase of the human salivary amylase gene AMY1, that is advantageous in a high starch diet [25] and duplication of genes coding for pepsin in Antarctic fish that allows the fish to efficiently digest at low temperature [26]. In the yeast S. cerevisiae, adaptation to new ecological niches has also been associated with several gene copy number variations and other chromosomal rearrangements, both in natural conditions [27,28] and in artificial evolution experiments [20,29].
In our previous report, we presented a strikingly rapid gain in D-xylose fermentation capacity observed during evolutionary adaptation of a recombinant industrial yeast strain for D-xylose utilization [19]. Here, we showed that amplification of a heterologous gene XylA in the form of an eccDNA, followed by reintegration in multiple tandem repeats in the genome has acted as a mechanism for rapid gain of function.
We compared the whole genome sequence of the original parent strain HDY.GUF5 that was unable to ferment D-xylose with its derivative strain GS1.11–26 that efficiently ferments D-xylose to ethanol. The strain GS1.11–26 was developed by mutagenesis, genome shuffling and evolutionary adaptation from the parent HDY.GUF5. Analysis of CNVs between the two strains revealed amplification of a chromosomal segment where the XylA gene had been integrated. The amplification of XylA correlated with elevated activity of XI. The inherently low activity of XI in the recombinant strains developed previously was a limiting factor for efficient D-xylose utilization [18]. Therefore, the high D-xylose utilization rate of the evolved strain GS1.11–26 is due, at least in part, to the high copy number of XylA, which resulted in high XI activity. A similar, elevated D-xylose utilization rate due to multi-copy integration of XylA, has been reported recently in a strain developed by expression of the Piromyces sp. XI and further evolutionary adaptation [30]. In that report, the original recombinant strain was constructed through expression of XI and xylulose kinase (XK) from a multi-copy plasmid. Since both genes coding for XI and XK present in the plasmid were under the same (but separate) promoter and terminator sequences (TDH3p and CYC1t), it was suggested that duplication and further copy number increase of the XylA gene might have been initiated though unequal crossover of homologous regions in the plasmid. However, the recombined plasmid carrying multiple copies of the XylA gene could not be isolated from the evolved strains due to integration of the plasmid into the genome. In our study, however, the original recombinant strain HDY.GUF5 had been constructed through direct integration of the gene cassette into the genome using an integrative vector. Moreover, we have confirmed the presence of a single copy of the XylA gene in both alleles of the target chromosomal locus in the strain HDY.GUF5 by whole genome sequencing and Southern blot assays. Therefore the amplification of the XylA locus did not start from the original plasmid that was used to construct the recombinant strain.
On the other hand, we have clearly demonstrated the occurrence of self-replicating eccDNA carrying the XylA gene in the course of the evolutionary adaptation process. The eccDNA carried not only the heterologous XylA but also a sequence upstream of the genomic locus in which it had been integrated. It included the gene REV1 that encodes for a Deoxycytidyl transferase, an autonomous replication sequence ARS1529, and a tRNA gene tP(UGG)O3. The presence of these genes in the eccDNA indicates that the eccDNA was generated from the chromosomal locus. During adaptation in a serial batch culture containing D-xylose as main carbon source, a dramatic increase occurred in the rate of D-xylose fermentation just after the second transfer. The eccDNA carrying XylA was detected at this step in which the rapid gain of function was observed, while it was not present in the cultures of the preceding steps, indicating that the eccDNA was apparently formed in that second culture. The dramatic increase in the rate of D-xylose fermentation can now be explained by the gain of this crucial genetic change, an increased copy number of the gene XylA sustaining higher XI activity. We believe that formation of self-replicating eccDNA intermediates that can be maintained in the cell for several generations allows enough time (and therefore, a higher chance) for recombination to happen into the genome in multiple copies. Our results provide convincing evidence for the role of self-replicating eccDNA in the generation of tandem repeat sequences originating from a previous single copy sequence.
Selection pressure that causes a high demand for the product of a specific gene apparently results in a high chance of amplification of the locus in which the gene is located. Examples of this phenomenon have been reported previously. They include amplification of the genes HXT6, SUL1 and GAP1 in response to glucose, sulfur and nitrogen limitation, respectively [11,31–34]. In those and other studies, the explanation for the mechanism of this gene amplification has commonly been attributed to recombination between repetitive sequences flanking the gene. However, such repetitive sequences were not always present, as in the case of SUL1 amplification under sulfur starvation [33,34]. Evidence for amplification of a DNA segment without involvement of repetitive flanking sequences was also reported in natural yeast populations and suggested to occur in the course of natural evolution [35]. The high frequency of such amplification events that do not involve repetitive sequences is not well understood.
A possible mechanism for formation of eccDNA is the involvement of transposons. Comparing the genome sequence of both the parent and evolved strains with the sequence of reference strain S288c, we noted that the reference sequence has a transposon sequence flanked by two LTRs just 3322 bp upstream of the amplified locus. However, this sequence was not present in our strain HDY.GUF5 nor in the evolved strain GS1.11–26. We also confirmed the absence of this sequence by PCR. A second solo LTR sequence inside the amplified XylA-locus is also present in the S288c genome but not in the strain background we used. Hence, we found no evidence for transposon mediated gene rearrangements.
Another possible mechanism involved is initial duplication by unequal sister chromatid exchange or microhomology mediated break induced repair mechanism (MMBIR). In yeast, microhomology mediated segmental duplications have been suggested to commonly occur by BIR mechanisms [36]. MMBIR occurs during DNA repair using single-stranded DNA template carrying a microhomology sequences as short as 5 to 25bp [37]. The single stranded DNA templates might be generated on several occasions: during replication, from stalled transcription complexes, or in promoter regions [36]. Analysis of the sequence of the amplified XylA-locus in our strain background revealed two potential sequences with microhomologies. The first 8 bp sequence homology (GGAAAGGG) is located just at the junction of the 3’ end of the amplified locus and 21 bp downstream of the 5’ end of the amplified locus (Fig. 3A). A second 7 bp homology (TATGATG) flanking the amplified locus is located 14 bp upstream and 15 bp downstream of the amplified locus. The former sequence homology is more likely to be the recombination point as it is located exactly at the break point where the DNA segment has been amplified. Since MMBIR mechanisms are known to induce duplications [36], this region might have first been duplicated and subsequently, one of the duplicated copies might be excised as a circular DNA element.
Though formation of eccDNA frequently involves terminal repeat sequences flanking a mobile DNA region [35], there was no evidence for such repeat sequences in the parent strain HDY.GUF5 nor in the evolved strain GS1.11–26 at the XylA-locus, that might have resulted in HR and excision. In addition, lack of evidence for deletion of the locus in any of the cultures or several single cell isolates indicated that the circular DNA was not formed by initial excision from the genome.
A common feature of previously identified gene amplifications is the presence of an ARS element in the amplified fragment. The genes HXT6, SUL1, GAP1 and CUP1, of which amplification under the appropriate selective conditions has been documented [31,33,34,38], all have an ARS element adjacent to the gene. For instance, in separate evolutionary engineering experiments under sulfur limitation [33,34] all clones characterized contained an amplification of SUL1 and none of SUL2 (which lacks an adjacent ARS element), while both genes encode a high-affinity sulfate permease [39]. This is generally explained by the ARS element being required for eccDNA maintenance. Our work here presented clear evidence for the formation and maintenance of ARS based circular DNA during adaptive evolution.
Chromosomal integration of the eccDNA in the form of a tandem repeat occurred in our work during further evolutionary adaptation. As a consequence, the high D-xylose fermentation phenotype was completely stable for several generations without selection pressure. Multiple integration of the XylA-locus as tandem array in the genome seemed to further improve the D-xylose fermentation rate compared to presence of the eccDNA form. This was evident from the fact that GS1.2–6 containing only the eccDNA still fermented much slower than GS1.4–14, which has the tandem amplification in one of the alleles of the chromosomal locus. In addition, the strain GS1.11–26 that contains the amplified XylA-locus in both alleles, showed still faster D-xylose fermentation than GS1.4–14. This suggests that higher copy numbers of XylA improve D-xylose fermentation capacity. On the other hand, the possibility that other genetic changes might have arisen during the subsequent evolutionary adaptation process that contribute to the higher D-xylose fermentation rate cannot be excluded.
Gene amplification has profound effects on rapid adaptation to new ecological niches. Several of the gene amplifications documented to date are associated with preexisting duplication that involves HR. Gene amplifications arising from single copy genes without flanking homologous sequences have been proposed to be generated by MMBIR mechanisms. Given the high frequency of gene amplification events arising from initially single copy sequences, the presence of an ARS element in many of the amplified sequences reported so far, and the possibility of recombination events with only little sequence homology, self-replicating eccDNA formation followed by tandem gene amplification probably serves as a general, rapid means of adaptation to novel environments that require high expression of a specific gene.
Materials and Methods
Strains and growth conditions
The S. cerevisiae strains used in this study are listed in Table 1. Yeast cells were propagated in yeast extract peptone (YP) medium (10 g/L yeast extract, 20 g/L bacteriological peptone) supplemented with either 20 g/L D-xylose (YPX) or 20 g/L D-glucose (YPD). For solid plates, 15 g/L Bacto agar was added. For batch fermentation, synthetic complete (SC) medium (1.7 g/L Difco yeast nitrogen base without amino acids and without ammonium sulfate, 5 g/L ammonium sulfate, 740 mg/L CSM-Trp and 100 mg/L L-tryptophan) supplemented with 40 g/L D-xylose was used. For selection of strains expressing the KanMX resistance marker, 200 mg/L geneticin was added to the medium. Yeast strains were maintained at -80°C in stock medium composed of YP and 26% glycerol.
Tab. 1. <i>S. cerevisiae</i> strains used in this study. 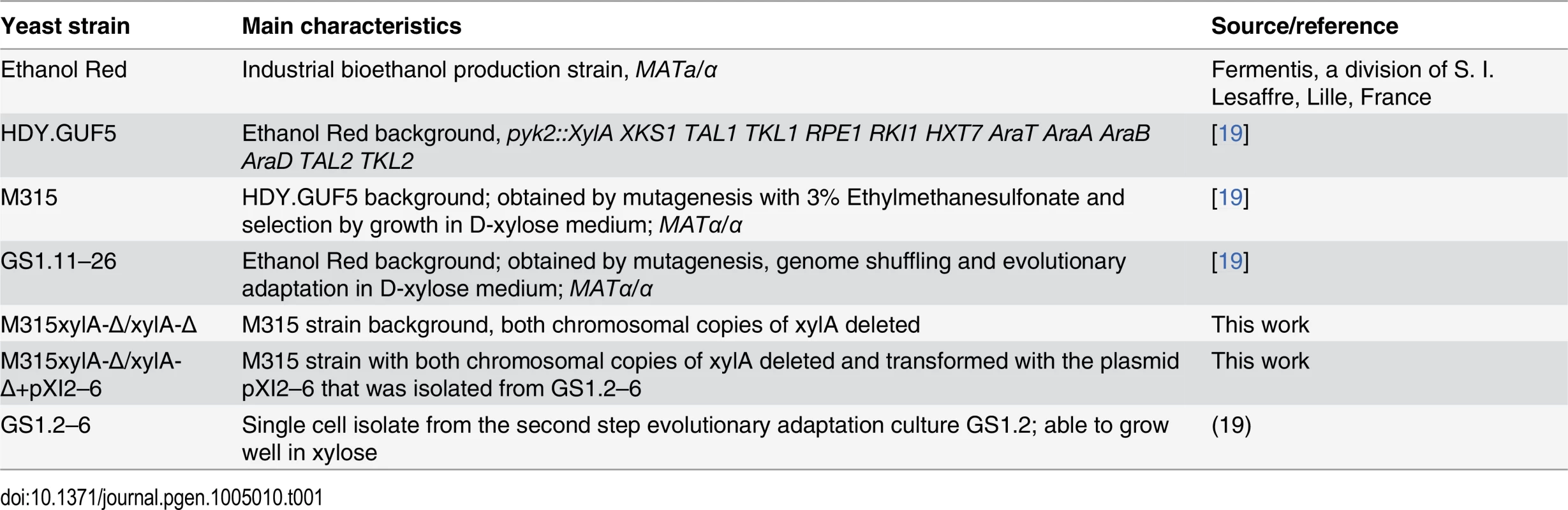
Small-scale fermentations
Semi-anaerobic batch fermentations were performed in 100 mL SC medium containing 40 g/L D-xylose as carbon source, in cylindrical tubes with cotton plugged rubber stopper. The strains were pre-grown for 24 hours in 5ml YPD medium. For strains carrying plasmid pXI-ARS, geneticin (200 mg/L) was added to the YPD to maintain the plasmid in the strains. The pre-culture was transferred to 50 ml YPD (+ geneticin) and grown to early stationary phase. Cells were harvested and fermentation was started by inoculating the pellet to an initial OD600 value of 5 into 100 ml SC + 4% xylose. The fermentation cultures were continuously stirred magnetically at 120 rpm and incubated at 35°C. The profile of the fermentation was followed based on the rate of CO2 production at different time intervals during the fermentation period. The CO2 production rate was estimated by measuring the weight loss of the fermentation tubes due to CO2 release.
Molecular biology methods
Yeast cells were transformed with the LiAc/SS-DNA/PEG method [40] or by electroporation [41]. Genomic DNA from yeast for PCR amplification was extracted using the PCI [phenol/chloroform/isoamyl-alcohol (25 : 24 : 1)] method [42]. PCR was performed with Phusion DNA polymerase (New England Biolabs) for construction of the vectors and sequencing purposes, and ExTaq polymerase (Takara) for diagnostic purposes. Sanger sequencing was performed by the Genetic Service Facility of the VIB, Belgium.
Genomic DNA isolation and whole genome sequencing
The genomic DNA was extracted using a standard protocol [43]. About 6 μg high quality DNA samples were sent for sequencing to BGI (Hong Kong). The sequencing was conducted by the facility using high-throughput Illumina sequencing technology. A paired end sequence library of 500 bp was constructed and sequence reads of 90 bp were generated. The sequencing reads provided from BGI were aligned to the reference S288c genome sequence using CLC Genomics Workbench5. Out of the 6 million reads with average length of 89.2 bp, 99% matched to the reference sequence when a 93% sequence similarity parameter was used. Additionally, 98% of the reference sequence has been covered with an average coverage depth of 44. The coverage depth per nucleotide position was extracted from the alignment and plotted using GraphPad prism software.
Southern blot analysis
Genomic DNA digested with the appropriate restriction enzyme was run on 0.8% agarose gel overnight at 50 V. A specific probe was prepared by PCR amplification from genomic DNA. The probe was labeled using Amersham Gene Images AlkPhos Direct labeling and detection system (GE Healthcare). The labeled probe was immediately used to hybridize the DNA that was blotted on a nylon membrane. Chemifluorescent signal was generated and detected using CDP-Star as a substrate in conjugation with LAS-4000 luminescent image analyzer.
Isolation of eccDNA from intermediate strain GS1.2–6
Plasmid DNA isolation was performed from the strain GS1.2–6, GS1.4–14 and GS1.11–26, using a modified protocol from [23]. Cells were pre-grown in 100 ml YPX medium for 24 h to enrich for the plasmid. The pellet from the 100 ml culture was divided into two and each pellet was resuspended in 5 ml buffer P1 from the QIAGEN plasmid purification kit. Freshly prepared lyticase solution (1.2 M sorbitol, 0.1 M Na3PO4 buffer pH 7.4 and 1 mg/ml lyticase) was added to the mixture and incubated for 45 min at 37°C. Once the cell lysate was obtained at this step, the protocol from the QIAGEN plasmid Maxi kit was followed.
Data access
All sequence data have been deposited in the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (NCBI) and can be accessed with references SRX651886 for Samplen56 (HDY-GUF5) and SRX647780 for Samplen57 (GS1.11–26).
Supporting Information
Zdroje
1. Barrick JE, Lenski RE (2009) Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb Symp Quant Biol 74 : 119–129. doi: 10.1101/sqb.2009.74.018 19776167
2. Frank SA (2013) Microbial evolution: regulatory design prevents cancer-like overgrowths. Curr Biol 23: R343–346. doi: 10.1016/j.cub.2013.03.046 23660352
3. Sauer U (2001) Evolutionary engineering of industrially important microbial phenotypes. Adv Biochem Eng Biotechnol 73 : 129–169. 11816810
4. Cakar ZP, Turanli-Yildiz B, Alkim C, Yilmaz U (2012) Evolutionary engineering of Saccharomyces cerevisiae for improved industrially important properties. FEMS Yeast Res 12 : 171–182. doi: 10.1111/j.1567-1364.2011.00775.x 22136139
5. Elena SF, Lenski RE (2003) Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet 4 : 457–469. 12776215
6. Katju V, Bergthorsson U (2013) Copy-number changes in evolution: rates, fitness effects and adaptive significance. Front Genet 4 : 273. doi: 10.3389/fgene.2013.00273 24368910
7. Kondrashov FA (2012) Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci 279 : 5048–5057. doi: 10.1098/rspb.2012.1108 22977152
8. Ames RM, Rash BM, Hentges KE, Robertson DL, Delneri D, et al. (2010) Gene duplication and environmental adaptation within yeast populations. Genome Biol Evol 2 : 591–601. doi: 10.1093/gbe/evq043 20660110
9. Despons L, Baret PV, Frangeul L, Louis VL, Durrens P, et al. (2010) Genome-wide computational prediction of tandem gene arrays: application in yeasts. BMC Genomics 11 : 56. doi: 10.1186/1471-2164-11-56 20092627
10. Cohen S, Segal D (2009) Extrachromosomal circular DNA in eukaryotes: possible involvement in the plasticity of tandem repeats. Cytogenet Genome Res 124 : 327–338. doi: 10.1159/000218136 19556784
11. Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, et al. (2010) Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. Proc Natl Acad Sci U S A 107 : 18551–18556. doi: 10.1073/pnas.1014023107 20937885
12. van Loon N, Miller D, Murnane JP (1994) Formation of extrachromosomal circular DNA in HeLa cells by nonhomologous recombination. Nucleic Acids Res 22 : 2447–2452. 8041604
13. Lau MW, Gunawan C, Balan V, Dale BE (2010) Comparing the fermentation performance of Escherichia coli KO11, Saccharomyces cerevisiae 424A(LNH-ST) and Zymomonas mobilis AX101 for cellulosic ethanol production. Biotechnol Biofuels 3 : 11. doi: 10.1186/1754-6834-3-11 20507563
14. Olsson L, Linden T, Hahn-Hägerdal B (1992) Performance of microorganisms in spent sulfite liquor and enzymatic hydrolysate of steam-pretreated salix. Appl Biochem Biotech 34–35 : 359–368.
15. de Souza AP, Leite DCC, Pattathil S, Hahn MG, Buckeridge MS (2013) Composition and structure of sugarcane cell wall polysaccharides: implications for second-generation bioethanol production. BioEnergy Res 6 : 564–579.
16. Limayem A, Ricke SC (2012) Lignocellulosic biomass for bioethanol production: current perspectives, potential issues and future prospects. Prog Energ Combust 38 : 449–467.
17. Alper H, Stephanopoulos G (2009) Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat Rev Microbiol 7 : 715–723. doi: 10.1038/nrmicro2186 19756010
18. Brat D, Boles E, Wiedemann B (2009) Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl Environ Microbiol 75 : 2304–2311. doi: 10.1128/AEM.02522-08 19218403
19. Demeke MM, Dietz H, Li Y, Foulquie-Moreno MR, Mutturi S, et al. (2013) Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels 6 : 89. doi: 10.1186/1754-6834-6-89 23800147
20. Araya CL, Payen C, Dunham MJ, Fields S (2010) Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomics 11 : 88. doi: 10.1186/1471-2164-11-88 20128923
21. Petes TD (1980) Molecular genetics of yeast. Annu Rev Biochem 49 : 845–876. 6996573
22. Michel AH, Kornmann B, Dubrana K, Shore D (2005) Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev 19 : 1199–1210. 15905408
23. Singh MV, Weil PA (2002) A method for plasmid purification directly from yeast. Anal Biochem 307 : 13–17. 12137773
24. Nieduszynski CA, Knox Y, Donaldson AD (2006) Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev 20 : 1874–1879. 16847347
25. Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, et al. (2007) Diet and the evolution of human amylase gene copy number variation. Nat Genet 39 : 1256–1260. 17828263
26. Carginale V, Trinchella F, Capasso C, Scudiero R, Parisi E (2004) Gene amplification and cold adaptation of pepsin in Antarctic fish. A possible strategy for food digestion at low temperature. Gene 336 : 195–205. 15246531
27. Carreto L, Eiriz MF, Gomes AC, Pereira PM, Schuller D, et al. (2008) Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics 9 : 524. doi: 10.1186/1471-2164-9-524 18983662
28. Liti G, Louis EJ (2005) Yeast evolution and comparative genomics. Annu Rev Microbiol 59 : 135–153. 15877535
29. Kao KC, Sherlock G (2008) Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet 40 : 1499–1504. doi: 10.1038/ng.280 19029899
30. Zhou H, Cheng JS, Wang BL, Fink GR, Stephanopoulos G (2012) Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng 14 : 611–622. doi: 10.1016/j.ymben.2012.07.011 22921355
31. Brown CJ, Todd KM, Rosenzweig RF (1998) Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol 15 : 931–942. 9718721
32. Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, et al. (2002) Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 99 : 16144–16149. 12446845
33. Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, et al. (2008) The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet 4: e1000303. doi: 10.1371/journal.pgen.1000303 19079573
34. Payen C, Di Rienzi SC, Ong GT, Pogachar JL, Sanchez JC, et al. (2014) The dynamics of diverse segmental amplifications in populations of Saccharomyces cerevisiae adapting to strong selection. G3 (Bethesda) 4 : 399–409. doi: 10.1534/g3.113.009365 24368781
35. Galeote V, Bigey F, Beyne E, Novo M, Legras JL, et al. (2011) Amplification of a Zygosaccharomyces bailii DNA segment in wine yeast genomes by extrachromosomal circular DNA formation. PLoS One 6: e17872. doi: 10.1371/journal.pone.0017872 21423766
36. Hastings PJ, Ira G, Lupski JR (2009) A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327. doi: 10.1371/journal.pgen.1000327 19180184
37. Hastings PJ, Lupski JR, Rosenberg SM, Ira G (2009) Mechanisms of change in gene copy number. Nat Rev Genet 10 : 551–564. doi: 10.1038/nrg2593 19597530
38. Fogel S, Welch JW (1982) Tandem gene amplification mediates copper resistance in yeast. Proc Natl Acad Sci U S A 79 : 5342–5346. 6291039
39. Cherest H, Davidian JC, Thomas D, Benes V, Ansorge W, et al. (1997) Molecular characterization of two high affinity sulfate transporters in Saccharomyces cerevisiae. Genetics 145 : 627–635. 9055073
40. Gietz RD, Schiestl RH, Willems AR, Woods RA (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11 : 355–360. 7785336
41. Thompson JR, Register E, Curotto J, Kurtz M, Kelly R (1998) An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast 14 : 565–571. 9605506
42. Hoffman CS, Winston F (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57 : 267–272. 3319781
43. Johnston JR (1994) Molecular genetics of yeast: A practical approach. New York: Oxford University Press.
Štítky
Genetika Reprodukční medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 3
-
Všechny články tohoto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



