-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaRECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
Recombinational DNA repair plays an important role in the maintenance of genomic stability by repairing DNA double-strand breaks and stalled replication forks. However, recombination between nonallelic similar sequences such as dispersed repeated sequences results in genomic instability. Plant plastid and mitochondrial genomes are compact (generally approximately 100–500 kb in size), but they contain essential genes. A substantial number of repeats are dispersed in these genomes, particularly in the mitochondrial genome. In this study, we showed that a knockout mutation of the newly identified plant-specific homolog of bacterial RecG DNA helicase RECG caused some defects in plastids and significant defects in the mitochondria. The organelle genomes in these mutants were destabilized by induced aberrant recombination between short (<100 bp) dispersed repeats. Recombination was induced at repeats as short as 8 bp. This suggests that RECG maintains plastid and mitochondrial genome stability by suppressing aberrant recombination between short dispersed repeats. Because such a phenomenon, to our knowledge, has not been observed in bacterial recG mutants, our results suggest an organelle-specific genome maintenance system distinct from that of bacteria.
Published in the journal: . PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005080
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005080Summary
Recombinational DNA repair plays an important role in the maintenance of genomic stability by repairing DNA double-strand breaks and stalled replication forks. However, recombination between nonallelic similar sequences such as dispersed repeated sequences results in genomic instability. Plant plastid and mitochondrial genomes are compact (generally approximately 100–500 kb in size), but they contain essential genes. A substantial number of repeats are dispersed in these genomes, particularly in the mitochondrial genome. In this study, we showed that a knockout mutation of the newly identified plant-specific homolog of bacterial RecG DNA helicase RECG caused some defects in plastids and significant defects in the mitochondria. The organelle genomes in these mutants were destabilized by induced aberrant recombination between short (<100 bp) dispersed repeats. Recombination was induced at repeats as short as 8 bp. This suggests that RECG maintains plastid and mitochondrial genome stability by suppressing aberrant recombination between short dispersed repeats. Because such a phenomenon, to our knowledge, has not been observed in bacterial recG mutants, our results suggest an organelle-specific genome maintenance system distinct from that of bacteria.
Introduction
Plants have two organelles, plastid and mitochondrion, that possess their own genomic DNA. The organelle genomes have become compact due to the endosymbiotic transfer of ancestral bacterial genes into the nucleus throughout evolution [1]. However, their genomes still encode components essential for photosynthesis, respiration and gene expression in organelles [2]. Since electron transport in photosynthesis and respiration produce reactive oxygen species (ROS), a harmful factor that damages DNA, plant organelle DNA is exposed to more severe conditions than nuclear DNA. Ultraviolet (UV) radiation from sunlight can also damage organelle DNA. However, the mechanism of how plant organelle DNA stability is maintained remains largely unknown.
Nuclear genes involved in mtDNA stability have been identified through the analyses of mutants displaying variegated leaves or by mutating genes that were predicted to be involved in organelle DNA metabolism [3]. The bryophyte P. patens has two functional bacterial-type RecA homologs, RECA1 and RECA2, which localize to mitochondria and plastids, respectively [4,5]. A RECA1 KO strain exhibits defects in growth and mitochondrial morphology, and results in lower rate of the recovery of damaged mtDNA [4,6]. Moreover, the RECA1 KO mutant displays gross rearrangements due to aberrant recombination between short repeats ranging from 62 to 84 bp scattered throughout mtDNA, which suggests that RECA1 maintains mtDNA stability by suppressing gross rearrangements [6].
In the angiosperm Arabidopsis thaliana, a mutation in the MutS homolog 1 (MSH1) causes mtDNA instability due to aberrant recombination between short dispersed repeats ranging in size from 108 to 556 bp [7–10]. Similarly, mutations in plant-specific single strand DNA-binding proteins, WHY2 from the whirly family of proteins [11] and organellar single-stranded DNA binding protein 1 (OSB1; [12]), lead to aberrant recombination between repeats. In the OSB1 mutant, repeats ranging in size from 249 to 556 bp are involved in the recombination [12], while in the WHY2 mutant, the recombination occurs between short repeats (<30 bp) and is gyrase inhibitor-dependent [11]. Mutations in RECA3, a RecA homolog, also cause mtDNA instability due to aberrant recombination between a few pairs of repeats (∼200 bp) [13].
A few genes are reported to be involved in the maintenance of plastid DNA (ptDNA) stability. Double mutations in WHY1 and WHY3, whirly family genes in A. thaliana, induce recombination between 10–18 bp ptDNA repeats. Thus, WHY1 and WHY3 protect ptDNA against illegitimate recombination [14]. A recent report showed that a mutation in A. thaliana MSH1 also induced rearrangements of plastid loci containing short repeats [15].
Bacterial RecG protein is a double-stranded DNA helicase that unwinds a variety of branched DNAs modeled after Holliday junctions and replication forks [16,17]. Analyses of a recG mutant suggest that RecG plays a role in homologous recombination and replication fork repair in vivo, similar to the proposed role of RecA [18]. In vitro studies also suggest the role of RecG in the repair of stalled replication forks [17,19,20] Recent reports suggest that RecG has an important function in the control of chromosomal replication and segregation in E. coli [21–23]. In this report, we analyzed a nuclear-encoded homolog of bacterial DNA helicase RecG, named RECG, which localized to both plastid and mitochondrial nucleoids in P. patens. We found that both organelle genomes of the RECG KO mutant were destabilized due to recombination between repeated sequences within a broad range in size (8–79 bp) and that the induced mtDNA recombination in RECG and RECA1 KO mutants partly differed. Here, we propose a vital role for RECG in the maintenance of plastid and mitochondrial genome stability.
Results
P. patens RECG protein localizes to both plastid and mitochondrial nucleoids
We identified a homolog of E. coli RecG in the P. patens nuclear genomic sequence [24] and named it RECG. Homologs of RecG are found in other plants, but not in fungi or animals, like bacterial-type RecA homologs [25]. Based on its cDNA sequence which we determined by rapid amplification of cDNA ends (RACE), the RECG protein is predicted to be 1152 amino acids in length and shares a high degree of sequence similarity with E. coli RecG, except for its extended N-terminal region (S1A Fig.). The extended N-terminal region, which is assumed to be a signal peptide that targets the protein to organelles, is potentially sufficient for localization to both plastid and mitochondrion (S1B Fig.) as judged by TargetP [26]. A similar N-terminal extension also exists in an annotated version of the RecG homolog in A. thaliana with a potential for localizing to both plastid and mitochondria (S1B Fig.). Fluorescent microscopy of protoplast cells expressing green fluorescent protein (GFP) gene fused to downstream of the 5’UTR and the N-terminus of RECG cDNA showed that the RECG-GFP localized to both plastids and mitochondria (Fig. 1A). Similar analysis with GFP gene fused to full-length RECG coding sequence demonstrated GFP fluorescence foci in both plastids and extra-plastid cytoplasmic space (Fig. 1B). 4′,6-diamidino-2-phenylindole (DAPI) staining of the cell showed that the GFP foci sometimes corresponded to some plastid and mitochondrial nucleoids (Fig. 1C and D), suggesting that RECG protein associates with these nucleoids.
Fig. 1. Subcellular localization of the RECG-GFP protein in P. patens protoplast cells. 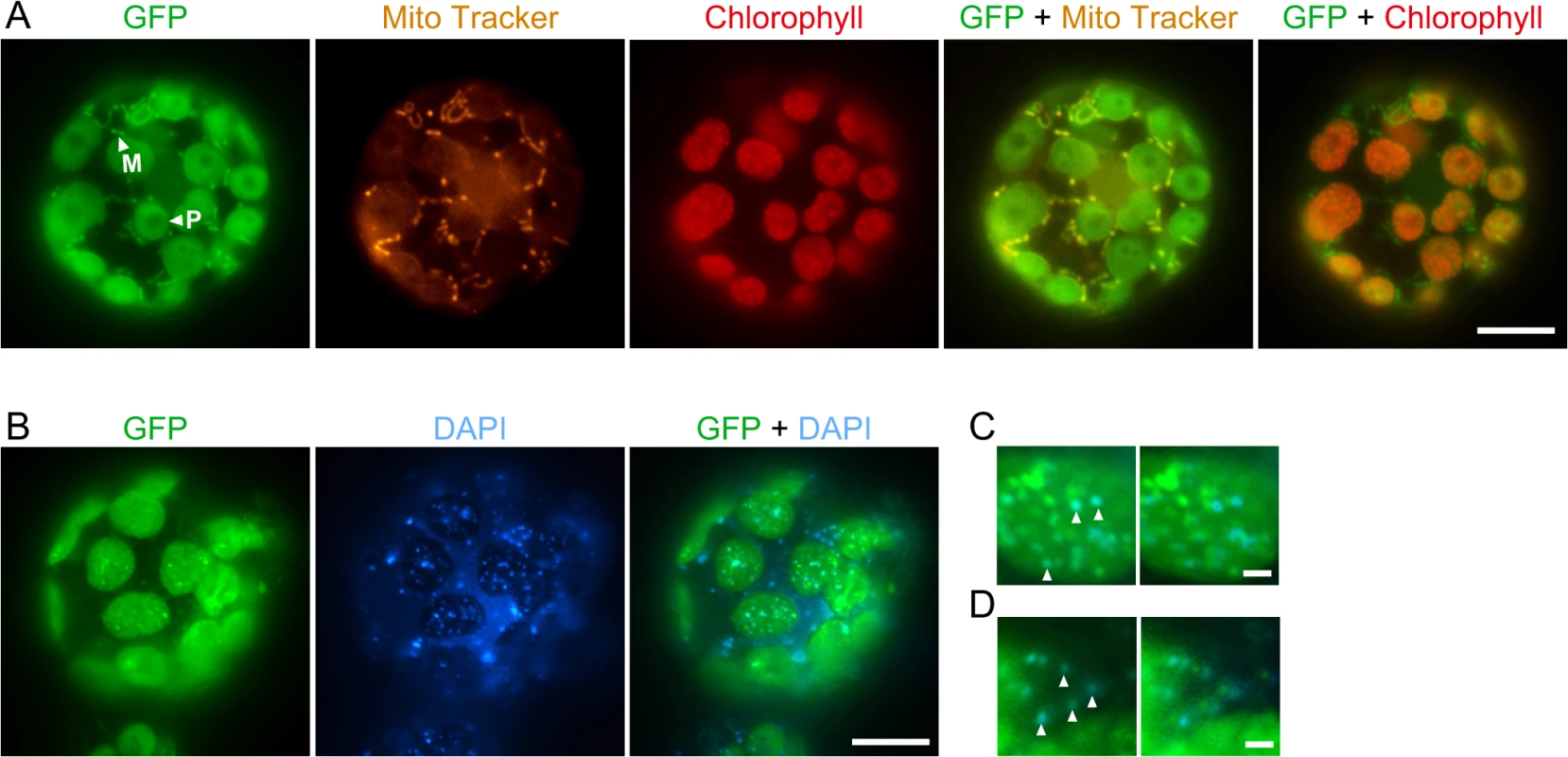
A. Subcellular localization of GFP fused to RECG N-terminal region. The fluorescence of GFP was merged with Mito Tracker or chlorophyll autofluorescence. Plastids were distinguished by the distribution of their chlorophyll autofluorescence, while mitochondria were detected by staining with Mito Tracker Orange. The arrowheads with P and M denote examples of RECG-GFP localized to plastid and mitochondrion, respectively. B. Subcellular localization of GFP fused to full-length RECG. The fluorescence of GFP was merged with DAPI fluorescence. C and D. Localization of full-length RECG-GFP to plastid (C) and mitochondrial (D) nucleoids. GFP fluorescence was merged with DAPI fluorescence (left panels) and then GFP fluorescence was shifted to left (right panels). The arrowheads denote examples of correspondence between GFP and DAPI signals. Bars = 10 μm in A and B, and 1 μm in C and D. RECG partially complements the defects of E. coli recG cells
To characterize the function of RECG, we examined whether RECG could complement the defects of an E. coli recG-deficient strain. E. coli recG-deficient strains harboring P. patens RECG lacking the signal peptide, intact E. coli recG, or no recG were subjected to UV irradiation after the induction of these genes. As reported by Ishioka et al. [27], the recG-deficient strain exhibited greater sensitivity to UV than the strain harboring the recG gene (Fig. 2), which implies that E. coli RecG participates in the recovery from UV damage. Expression of P. patens RECG conferred more than 10-fold greater resistance to UV in the recG-deficient cells, although not to the same degree as E. coli recG (Fig. 2). Therefore, RECG can partially complement the defects of E. coli recG-deficient cells.
Fig. 2. Complementation of the E. coli recG defect by RECG. 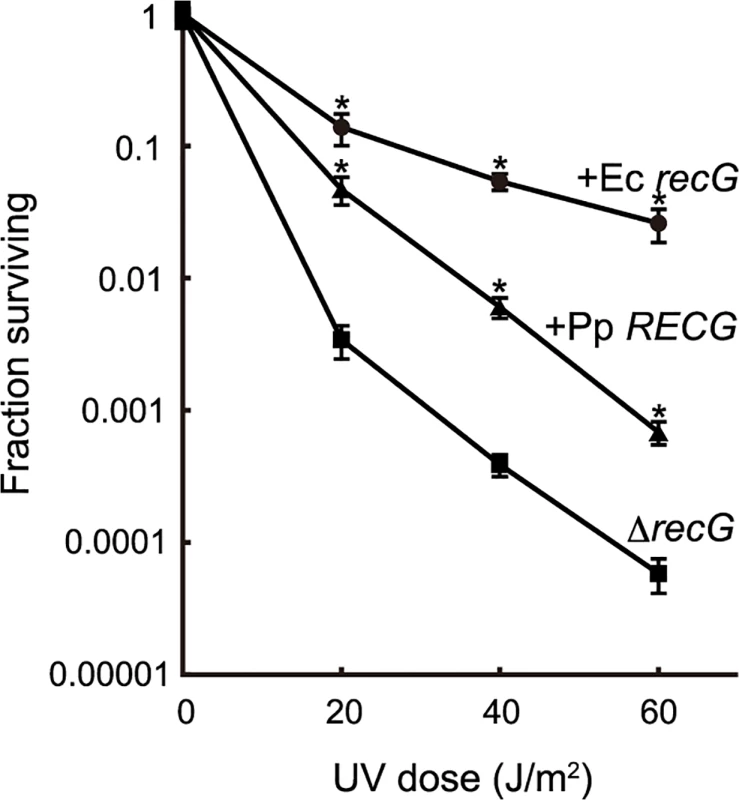
E. coli recG-deficient cells harboring a plasmid carrying the P. patens RECG (+Pp RECG, triangle), E. coli recG (+Ec recG, circle) or empty vector (ΔrecG, square) were subjected to UV irradiation, and the surviving fraction was calculated as described in Materials and Methods. Data from three independent experiments are expressed as mean ± SD. *p<0.01 (versus ΔrecG). Knock-out of the RECG gene causes growth and developmental defects
Efficient targeting of nuclear genes [28] and a sequenced nuclear genome [24] enable easy knock-out of nuclear genes in P. patens. Thus, we knocked out the RECG gene to analyze the in vivo role of RECG (S2 Fig.). To investigate the effect of RECG KO on the growth and development of P. patens, we compared the RECG KO lines (named recG-1 and recG-2) with wild type (WT). After inoculation on agar medium, we observed that P. patens initially formed colonies composed of filamentous protonemal cells, and gametophores subsequently developed in the colonies. The RECG KO colonies appeared small and had less developed gametophores, which indicates defects in growth and development, although the extent of the defects were milder than those of the RECA1 KO strain (Fig. 3A). The RECG KO colonies consisted of protonemal cells with heterogeneity in growth; relatively normal (recG-N) and atrophic (recG-A) protonemal cells. The recG-A protonemal cells were shorter and darker than the recG-N protonemal cells, while the recG-A cells are still shorter and darker than the WT (S3A Fig.). The atrophic protonemal cells of the RECG KO colonies were shorter than those of WT and were dense with plastids, likely due to a reduction in cell volume. Notably, the morphological abnormalities of the RECG KO and RECA1 KO colonies were similar (Fig. 3B). These morphological effects imply that RECG plays an important role in the growth and development of P. patens, and suggest that RECG and RECA1 share similar roles.
Fig. 3. Cell growth and morphology of RECG KO plants. 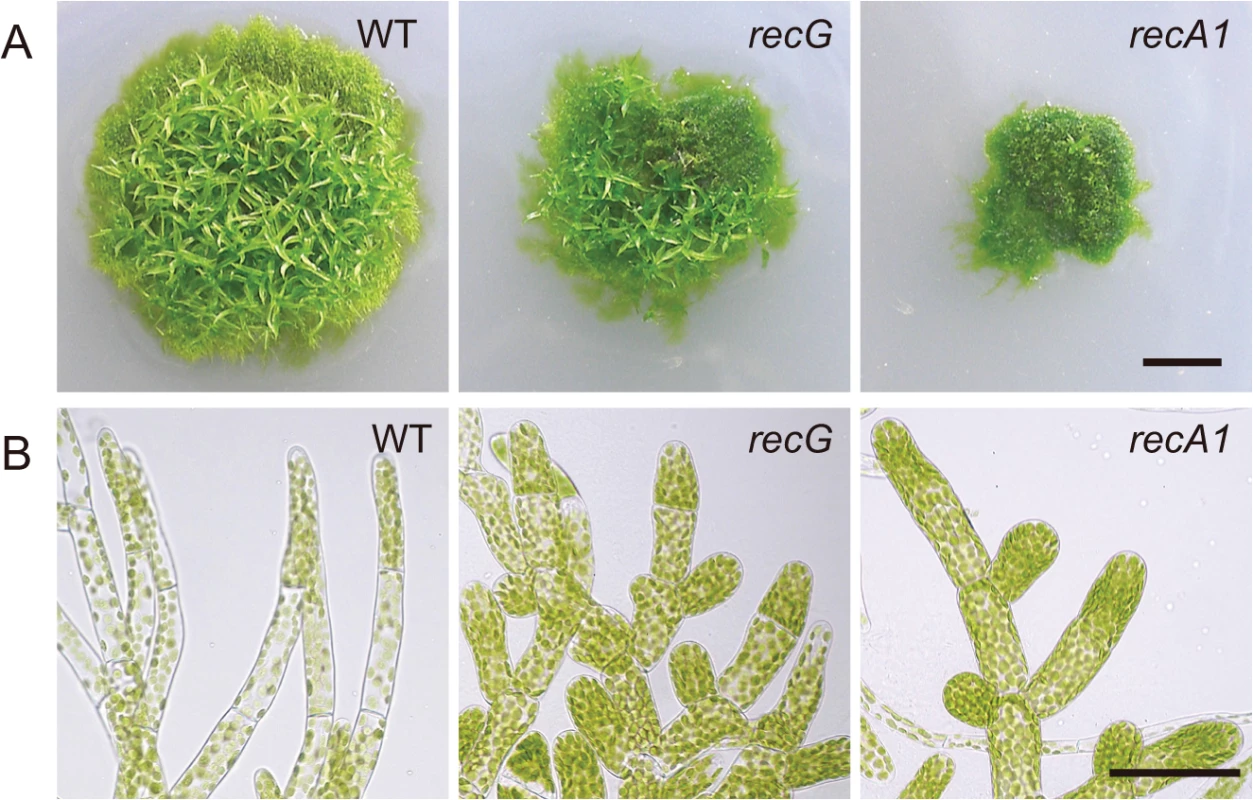
A. Colonies of wild type (WT), RECG KO (recG-2) or RECA1 KO plants cultivated on agar medium for four weeks. B. Protonemal cells. Bars = 5 mm in (A) and 50 μm in (B). Atrophic protonemal cells (S3 Fig.) are shown as RECG KO cells. Abnormal mitochondria and plastids in RECG KO cells
To analyze the effect of the RECG KO on the ultrastructure of subcellular components, especially on those of mitochondria and plastids, we observed RECG KO cells by transmission electron microscopy (TEM). Since the RECG KO plant appeared to be composed of recG-N and recG-A protonemal cells, these two cell types were analyzed separately. TEM analyses revealed that the RECG KO had various effects on the ultrastructure of mitochondria, plastids, and other cell components. Both recG-N and recG-A mitochondria had a lower number of cristae and cristae enlargement (Fig. 4D-F), and recG-A cell mitochondria showed weaker matrix staining, indicating a lower electron density of the mitochondrial matrix (Fig. 4F). Some RECG KO mitochondria were abnormally extended (Fig. 4B, C and G), and their sizes were sometimes comparable to those of plastids (S3B Fig.). The extended mitochondria were more frequently observed in recG-N cells than in recG-A cells. It is notable that these mitochondrial abnormalities, including a lower number of cristae, cristae disorganization, weaker matrix staining, and stretching, are also observed in RECA1 KO mitochondria [6]. We further analyzed the stretching by performing TEM on serial thin sections to elucidate the three-dimensional structure of the extended mitochondria, and found that one of these mitochondria penetrated 16 serial thin sections (S3C Fig.) and that the edge of each mitochondrion was swollen (S3D Fig.). This result suggested that the extended mitochondria were actually disc-shaped with thick edges.
Fig. 4. Ultrastructure of RECG KO protonemal cells. 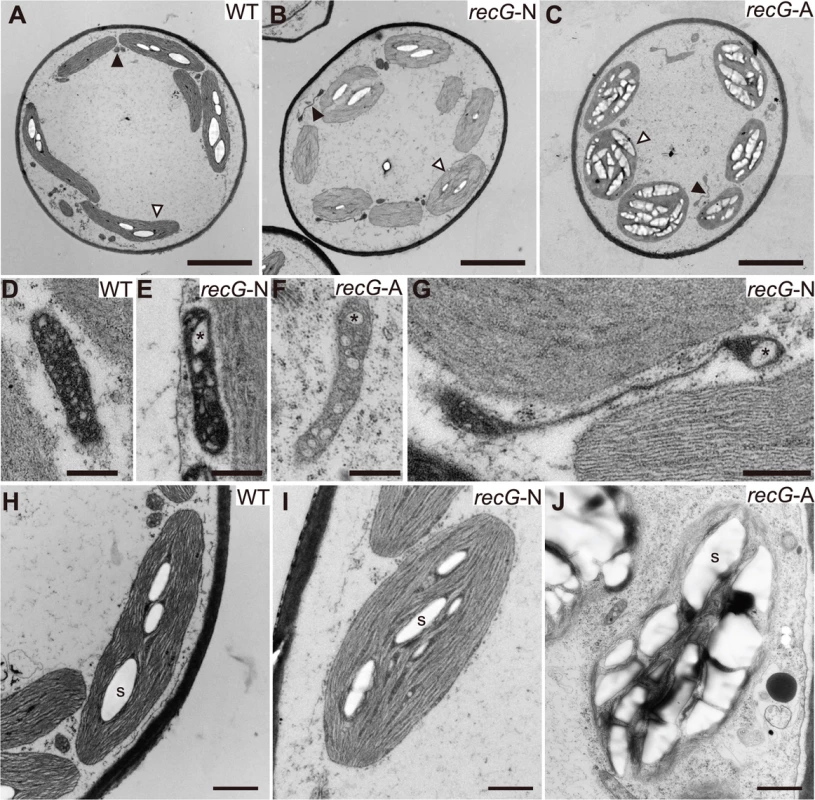
Protonemal cells of WT, RECG KO normal (recG-N), and RECG KO atrophic (recG-A) were analyzed by TEM. A to C. Images of transversion sections of WT (A), recG-6N (B), and recG-6A (C) cells. Filled and blank arrowheads denote examples of mitochondrion and plastid, respectively. D to G. Mitochondria of WT (D), recG-3N (E), recG-3A (F), and recG-6N (G) cells. Cristae were appeared as small regions with low electron density. Asterisks denote examples of enlarged cristae. H to J. Plastids of WT (H), recG-3N (I), and recG-3A (J) cells. Examples of starch grains are indicated by S. Bars = 5 μm in (A) to (C), 500 nm in (D) to (G), and 1 μm in (H) to (J). The RECG KO affected plastids. Although plastids in recG-N cells looked mostly normal (Fig. 4I), abnormally extended plastids were occasionally detected. Some of these plastids appeared to have a disturbance in cell division septum formation (S3E Fig.). Plastids in recG-A cell had abnormal shapes, and their membranes, including thylakoid, outer and inner membranes, appeared frail (Fig. 4J). Moreover, most plastids in recG-A cells abundantly accumulated starch and had an underdeveloped thylakoid membrane (Fig. 4C and J). The RECG KO also affected cell structure and cell components, especially in recG-A cells. Some recG-A protonemal cells accumulated oil (S3F Fig.), which is in contrast to wild type protonemal cells, which have few or no oil bodies [29]. These observations indicate that RECG KO had various effects on cell ultrastructure that were likely caused by functional defects in mitochondria and plastids.
Mitochondrial DNA rearrangements in RECG KO plants
In RECA1 KO mitochondria, aberrant recombination occurs frequently between repeats ranging in size from 62 to 84 bp and results in gross DNA rearrangements, which appear to be responsible for the phenotypic defects observed in RECA1 KO plants [6]. Therefore, the similar phenotype of RECG and RECA1 KO plants, as described above, predicts that RECG KO mtDNA also undergoes gross DNA rearrangements. To test this hypothesis, we carried out structural analyses of RECG KO mtDNA and compared the results with those of RECA1 KO mtDNA.
A product resulting from recombination between 69 bp direct repeats existing in nad2 and atp9 loci of mtDNA, which appears as a 1.8 kb EcoRI fragment on DNA gel blots [6], was confirmed in a blot hybridized with nad2 probe in RECA1 KO lines (Fig. 5A). The blot showed that the 1.8 kb EcoRI fragment of the nad2-atp9 recombination product was hardly detectable in the RECG KO lines, while a weak 1.9 kb band was detected in both RECG KO lines (Fig. 5A). Since the RECG KO-specific 1.9 kb DNA fragment strongly hybridized to both an atp9 probe, which does not hybridize to the nad2-atp9 recombination product, and a ccmF probe (Fig. 5A), this fragment is likely to be the result of recombination between the atp9 locus and ccmF locus. Forty-seven base pair repeats at both loci are suspected to be involved in recombination (Fig. 5B and S4D Fig.), and the size of the recombination product is consistent with the size of the observed 1.9 kb ccmF-atp9 product. Because the ccmF-atp9 recombination product includes the 69 bp nad2-atp9 repeats (Fig. 5B), it hybridized weakly to the nad2 probe (Fig. 5A). The ccmF-atp9 product also appeared in the RECG KO lines with the sizes of the DNA products resulting from recombination between the 47 bp repeats when the mtDNAs were digested with HindIII or NdeI (S4A–C Fig.). PCR amplification of the ccmF-atp9 product from RECG KO plants and direct sequencing analysis of the amplified fragments showed that almost all of the recombination junctions were within the 47 bp repeats, yet the sequence similarity extended to the region flanking the repeat (S4D Fig.). These results indicate that the ccmF-atp9 recombination product, but not the nad2-atp9 recombination product, accumulates in the RECG KO lines.
Fig. 5. mtDNA rearrangements in RECG KO lines. 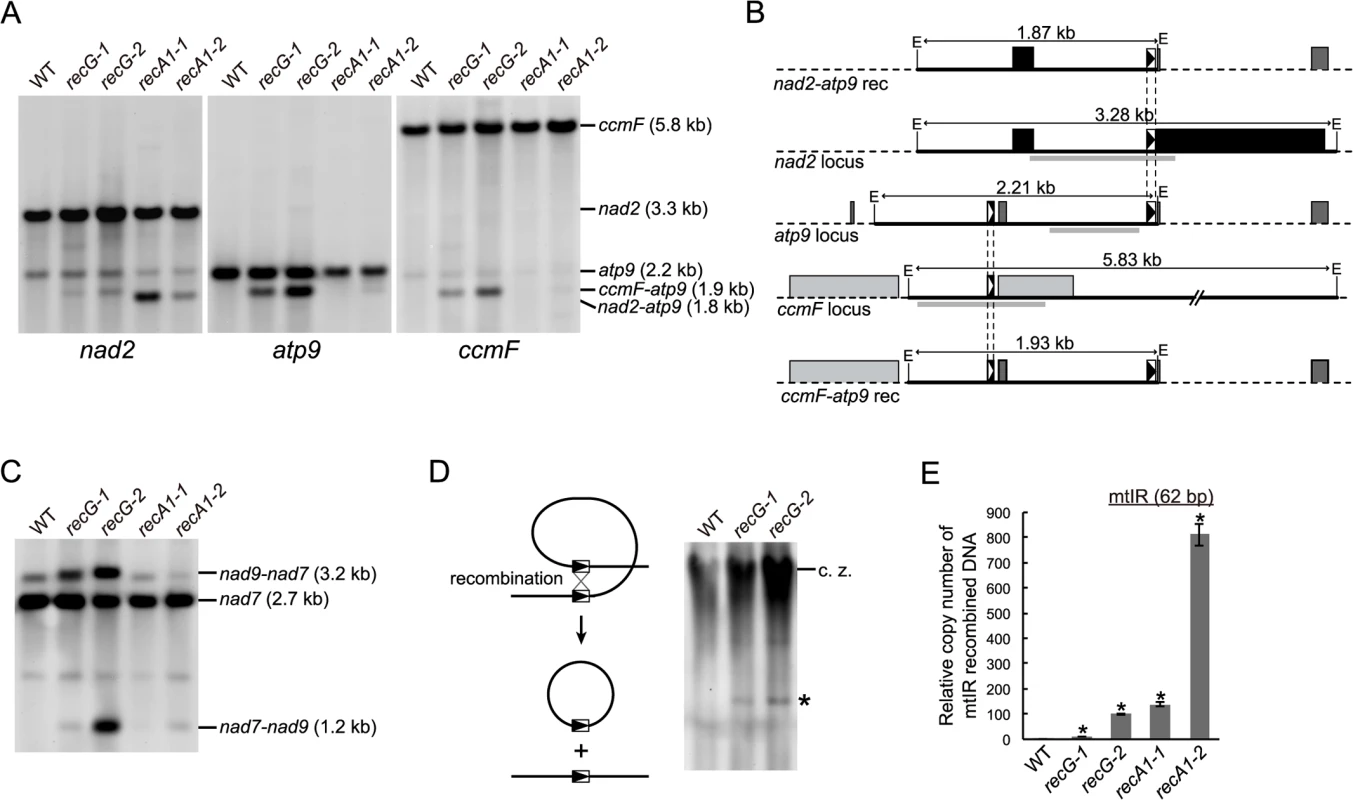
A. mtDNA configuration at nad2, atp9 and ccmF loci. DNA from WT, RECG KO and RECA1 KO strains digested with EcoRI was probed using nad2, atp9 and ccmF probes, as indicated below the blots. The predicted structure and length of the major bands are indicated on the right. B. Schematic representation of the DNA structures detected in (A). The EcoRI fragments corresponding to the bands on the blot in (A) and the flanking regions are represented by solid black lines and dashed black lines, respectively. The EcoRI recognition sites are indicated by E. The positions of the probes used in (A) are indicated by thick gray lines. The boxes represent exons, and the lines between boxes represent introns or noncoding flanking sequences. The 69 bp and 47 bp repeats are indicated by black triangles and white triangles in the boxes, respectively. C. mtDNA configuration at the nad7 locus containing 79 bp repeats. DNA from each of the indicated strains was digested with SacII and probed using nad7 probe. The structure of the fragments is detailed in Odahara et al. [6]. D. Production of deleted mitochondrial subgenome by recombination between repeats in nad7 and nad9. Left panel illustrates production of deleted subgenome by intramolecular recombination between direct repeats. Undigested DNA from WT and RECG KO strains was probed using nad7 probe. The asterisk denotes DNA corresponding to 11-kb subgenome. c.z., compression zone. E. The amount of DNA generated by recombination between mtIR (62 bp). Relative copy number of DNA resulting from recombination between mtIR per mitochondrial rpl2 DNA was measured by qPCR. WT was given a value of 1. The data represent mean of three replicates ± SD. *p<0.01 (versus WT). Next, we analyzed whether other hotspots identified in RECA1 KO mtDNA rearrangements [6] also induced recombination in the RECG KO plants. DNA gel blot analysis showed that recombination occurred between the 79 bp nad7-nad9 direct repeats in the RECG KO lines as well as the RECA1 KO lines (Fig. 5C). In the RECG KO lines, we also identified signals corresponding to 11 kb of deleted circular mtDNA (Fig. 5D), which is produced by recombination between the nad7-nad9 repeats, as reported in RECA1 KO plants [6]. In addition, we carried out quantitative PCR (qPCR) analyses to assess the copy number of DNA resulting from recombination between 62 bp inverted repeats (mtIR), another hot spot located in the intergenic region of mtDNA [6]. The results showed that the recombination at this locus was induced in the RECG KO lines and that the recombination level of one line was comparable to that of the RECA1 KO line (Fig. 5E). These results suggest that frequent mtDNA rearrangements, which associated with deletion in some cases, occur at multiple hot spots in RECG KO plants, and that some hot spots differ from those of the RECA1 KO plants.
Since the recombination described above can cause deletion of mtDNA, it is possible that the copy number of RECG and RECA1 KO mtDNA loci altered. To test this, we measured the copy number of three mitochondrial loci, rps4, nad6 and rpl2, by qPCR. The results showed that the copy number of each mtDNA locus varied in the RECA1 KO lines whereas increased in the RECG KO lines (S4E Fig.).
Repeat-mediated genomic instability in RECG and RECA1 KO mitochondria
To analyze the effect of RECG or RECA1 KO on global structure of mtDNA, we performed a comprehensive analysis of DNA molecules resulting from recombination between repeats dispersed in the mtDNA. We first analyzed mtDNA repeat-mediated rearrangements in both KO mutants by DNA gel blot, and identified two DNA fragments that were most likely to be derived from recombination between nad4-nad1 direct repeats, named R4 (90 bp) or R6 (56 bp) [6], as judged by their sizes (Fig. 6A). Note that the DNA fragments were detected only in RECA1 KO lines, but not in RECG KO line as well as WT. We next carried out quantification of DNA resulting from recombination between other direct repeats (46–57 bp, R5, R11, R12, R13, R18, and R19) by qPCR. The results showed that recombined DNA from every tested repeats were apparently accumulated in both RECG and RECA1 KO lines; the level of accumulation was very high in the RECA1 KO lines regarding R5 and R13, and high in RECA1 KO lines regarding R11 and R18 (Fig. 6C). Collectively, these results suggest that the repeat-mediated recombination were induced in both RECG and RECA1 KO mitochondria at multiple loci, but the degree and the site of the recombination were somewhat different between them.
Fig. 6. Genomic instability in RECG and RECA1 KO mitochondria. 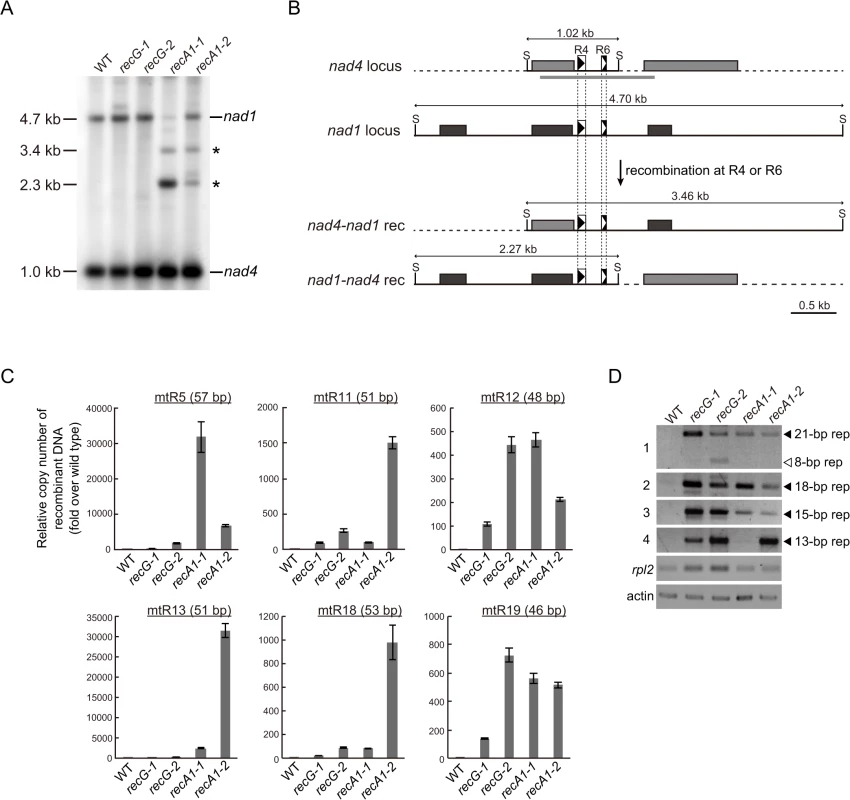
A. mtDNA configuration at the nad4 locus. DNA from WT, RECG KO, and RECA1 KO strains digested with SacII were probed with nad4 probe. The asterisks denote signals corresponding to DNA recombined between nad4-nad1 repeats. The length of the bands is indicated on the left. B. Schematic explanation of the DNA structures detected in (A). Reciprocal recombination at R4 (90 bp) or R6 (56 bp) at nad4 and nad1 loci produces both nad4-nad1 rec and nad1-nad4 rec products. The SacII recognition sites are indicated by S. The positions of the probes used in (A) are indicated by thick gray lines. For details of the scheme, see legend of Fig. 5B. C. The amount of DNA generated by recombination between several direct repeats 46 to 57 bp in length. Relative copy number of DNA resulting from recombination between direct repeats (R5, R11, R12, R13, R18, or R19) per mitochondrial rpl2 DNA was measured by qPCR. WT was given a value of 1. The data represent mean of three replicates ± SD. All the RECG KO and RECA1 KO values are significantly different from WT values (p<0.01). D. DNA generated by recombination between short (<35 bp) repeats. PCR reaction numbers indicated on the left correspond to those in S1 Table. Mitochondrial gene rpl2 and nuclear gene actin were amplified as a control. Filled and blank triangles indicate DNA with the expected and unexpected sizes, respectively. To understand further the effect of RECG and RECA1 KO on mtDNA stability, we examined whether shorter repeats were involved in the mtDNA instability. REPuter, a program that detects repeated sequences in DNA sequence [30], identified approximately 900 pairs of repeats (15–35 bp) in P. patens mtDNA [4]. Among the repeats, we analyzed selected repeats (S1 Table) by PCR with respect to the accumulation of recombination products, and the levels of DNA amplification at each repeat are shown in Fig. 6D. A greater level of amplification was observed in both KO lines than in the WT strain. Sequencing of the amplified DNA confirmed that they were the products of recombination between the 21, 18, 15 or 13 bp inverted or direct repeats (S1 Table and S8 Fig.). Moreover, the analysis further showed that an additional region of DNA, which was the product resulting from recombination between 8 bp repeats that are distantly positioned around the 21 bp repeats, was amplified in the RECG KO lines (Fig. 6D, panel 1). Therefore, DNA accumulates as a result of recombination between repeats ranging from 8 to 21 bp in the RECG KO, and from 13 to 21 bp in the RECA1 KO mitochondria.
Decrease in the levels of specific mitochondrial gene transcripts in RECG KO plants
The data presented here showed efficient rearrangements of RECG KO mtDNA at nad7, nad9, atp9, and ccmF loci caused by recombination between short repeats. Quantitative analysis using DNA gel blots revealed that the copy number of normal mtDNA bands (e.g., 1.5 kb of nad7 band and 1.7 kb nad9 band in S5A and B Fig.) in the RECG KO plants decreased to approximately 30%–45% and 35%–50% of WT at nad7 and nad9 loci, respectively. The copy number of normal mtDNA bands at atp9 and ccmF loci did not significantly change (S5C and D Fig.). To investigate the effect of the mtDNA rearrangements on mitochondrial transcripts, we analyzed the transcripts of these loci in the RECG KO plants. Quantitative RT-PCR (qRT-PCR) analysis demonstrated a significant reduction in the levels of transcripts from nad7 and nad9 for some of the primer pairs in the RECG KO mutants (Fig. 7A and B). We found a significant reduction (<10% of WT levels) in the levels of the transcript fragments when the primers were arranged to amplify a segment including a junction of exon 2 and 3 for nad7 or exon 1 and 2 for nad9 (Fig. 7A and B). Because the introns contain repeats involved in the mtDNA rearrangements (Fig. 7A and B), these results suggest that a substantial number of the nad7 and nad9 transcripts exist as chimeric transcripts of nad7 and nad9 and not as individual intact forms. RT-PCR analysis demonstrated the efficient amplification of nad7-nad9 chimeric transcripts from RECG KO mutants (Fig. 7C). We confirmed that the chimeric transcripts were precisely spliced between nad7 exon2 and nad9 exon2 (S5E Fig.). In contrast, similar qRT-PCR analysis demonstrated no significant differences in the levels of transcripts from ccmF and atp9 loci between the WT and RECG KO mutants (S5F Fig.). These results suggest that the efficient rearrangements of some mtDNA loci were associated with a decrease in the normal levels of mtDNA and a significant decrease in the number of the corresponding intact transcripts.
Fig. 7. Mitochondrial transcripts in RECG KO plants. 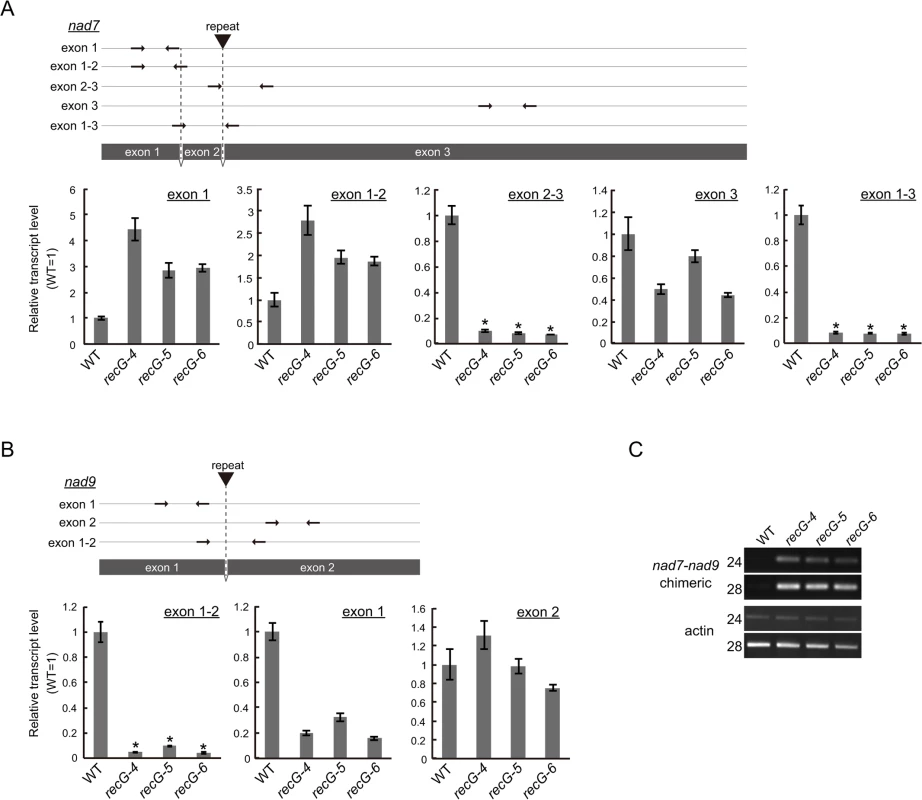
A and B. Detailed qRT-PCR analysis of nad7 (A) and nad9 (B) transcripts. Positions of the primers used in the qPCR are schematically represented in the upper parts of the panels. Coding regions are represented by grey boxes. Positions and directions of the primers are shown by arrows. Positions of the repeats involved in the rearrangements between nad7 and nad9 are indicated by triangles. Relative levels of segments of mitochondrial transcripts from nad7 and nad9 were normalized to reference of nuclear gene ST-P 2a transcript. WT was given a value of 1. Slight or no amplification was observed in no reverse-transcription controls (S5G Fig.). The data represent mean of three replicates ± SD. *p<0.01 (versus WT). C. RT-PCR analysis of nad7-nad9 chimeric transcripts. nad7-nad9 chimeric transcripts were amplified using cDNA from WT and RECG KO lines at cycles indicated on the left of the picture. Actin was amplified as an internal control. Repeat-mediated genomic instability in RECG KO plastids
The fact that RECG protein not only localizes to mitochondria, but also to plastids, raises the possibility that RECG plays a role in both plastids and mitochondria. We then analyzed the structure of RECG KO ptDNA, and focused on recombination between repeated sequences. We searched for repeats using REPuter and identified 16 pairs of repeats longer than 40 bp in the P. patens ptDNA sequence [31], most of which were located immediately downstream of genes as palindromic sequences (S2 Table), probably functioning in the stabilization of transcripts [32]. Among the repeats, we analyzed the level of recombination between inverted repeats-1 (ptIR-1, 63 bp long, shown as R6 in S2 Table), which are located in rpl16 and trnG, or direct repeats-1 (ptDR-1, 48 bp long, shown as R12 in S2 Table), which are located in psaA and psaB (S6A Fig.). DNA gel blot analysis using a plastid rpl16 probe showed accumulation of 4.3 kb DNA fragments in the RECG KO mutants (Fig. 8A). The size of these DNA fragments corresponds to that of a predicted product resulting from recombination between ptIR-1 (Fig. 8B). We next analyzed the IR-1 recombination product using qPCR (S6 Fig.). The analyses revealed that the product formed by recombination between ptIR-1 showed ∼160-fold increase in the RECG KO mutants (Fig. 8C). Similar qPCR analysis of a product formed by recombination between ptDR-1 showed a 6–16-fold increase in the RECG KO mutants compared with WT (Fig. 8C). These results showed increased accumulation of ptIR-1 and ptDR-1 recombination products in the RECG KO lines. To assess the effect of RECG KO on copy number of ptDNA, qPCR analysis of ptDNA loci was performed. The copy number of three plastidic loci rbcL, atpA and ndhH showed increases in the RECG KO lines compared with WT (S6B Fig.). As the RECG KO mutants showed increase of ptDNA in every tested locus, it is possible that plastid number was increased in the RECG KO mutants. However, no significant difference was observed in the number of plastids per cell between WT and RECG KO mutants (S6C Fig.).
Fig. 8. Genomic instability in RECG KO plastids. 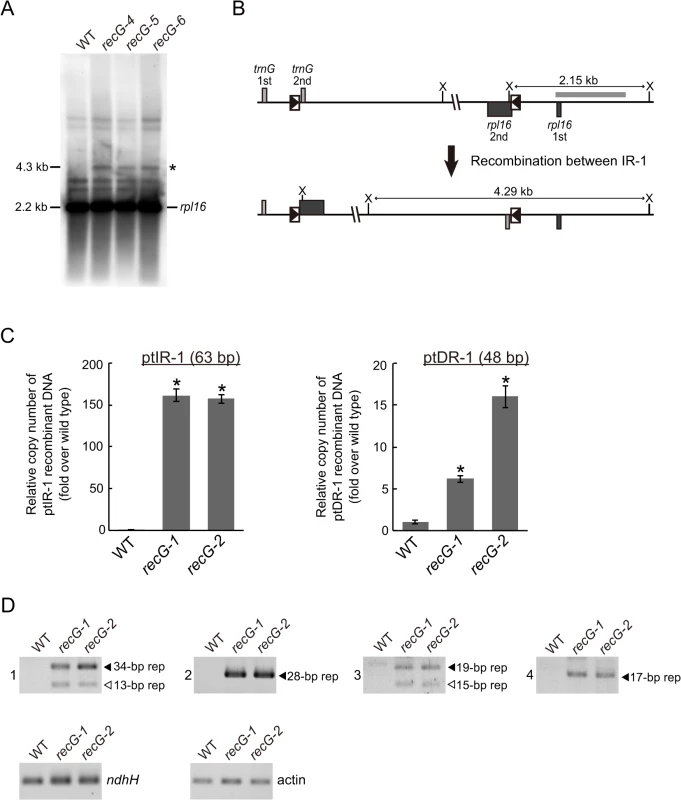
A. ptDNA configuration at the ptIR-1 (rpl16) locus. DNA from WT and RECG KO strains digested with XhoI were probed with rpl16 probe. The asterisk denotes signals corresponding to DNA recombined between ptIR-1 repeats. The length of the bands is indicated on the left. B. Schematic explanation of the DNA structures detected in (A). Intramolecular recombination between ptIR-1 causes inversion of a segment between ptIR-1. The XhoI recognition sites are indicated by X. The boxes represent exons, and the lines between boxes represent introns or noncoding flanking sequences. ptIR-1 are indicated by black triangles in the boxes. The positions of the probes used in (A) are indicated by thick gray line. C. The amount of DNA generated by recombination between ptIR-1 (63 bp) or ptDR-1 (48 bp). Relative copy number of DNA per plastid ndhH DNA was measured by qPCR. WT was given a value of 1. The data represent mean of three replicates ± SD. *p<0.01 (versus WT). D. DNA generated by recombination between short (<35 bp) repeats. PCR reaction numbers indicated on the left of the pictures correspond to those in S3 Table. Plastid gene ndhH and nuclear gene actin were amplified as a control. Filled and blank triangles indicate DNA with the expected and unexpected sizes, respectively. In the analyses described above, we applied PCR amplification for detection of DNA recombined between repeated sequences in the RECG or RECA1 KO lines. However, since such a recombined DNA can be created during PCR reaction, named PCR jumping, as reported by Alverson et al. [33], we quantified the amount of the artificially recombined DNA in our qPCR assay to evaluate the effect of PCR jumping. We first prepared two DNA fragments that contain copy1 or copy2 of ptIR-1, and then the two fragments were mixed so as to contain the amount of each IR-1 copy equivalent to that of the WT total genomic DNA (S6D Fig.). Next we quantified copy number of DNA recombined between the copy1 and copy2 of ptIR-1 using WT genomic DNA or the mixed DNA as templates. The amount of DNA recombined between IR-1 from the mixed DNA by PCR jumping was ∼1.5% of that from WT genomic DNA (S6D Fig.), indicating that recombination between IR-1 copies occurred during the qPCR reaction, but the efficiency was very low. Accordingly, these results suggest that the amplified recombined DNA that we observed was mostly derived from in vivo recombination of organelle DNA, and that the contribution of in vitro recombination, if it occurred, was very small.
We extended the analyses to shorter repeats (15–35 bp), which are abundant (approximately 2000 pairs) in ptDNA. We carried out PCR analyses to estimate the amount of DNA that recombined between short repeats. Reliable DNA amplification occurred only in the RECG KO lines (Fig. 8D), and sequencing of the amplified DNAs demonstrated that they were the result of recombination between the 34, 28, 19 or 17 bp repeats (S3 Table and S8 Fig.). Moreover, some types of additional DNA, which were determined to be products of recombination between 13 bp or 15 bp repeats (S3 Table), were amplified only in the RECG KO lines (Fig. 8D). Collectively, these results suggest that genomic instability was induced in the RECG KO plastids by aberrant recombination among repeated sequences, ranging in size from 13 to 63 bp.
Increased accumulation of recombined mtDNA in recG-A cells
To investigate the relationship between the heterogeneity of atrophic phenotype of RECG KO plants appearing as recG-A and recG-N cells (S3A Fig.) and the stability of plastid and mitochondrial genomes, we compared the status of organelle DNA in recG-A and recG-N cells. We separately extracted total genomic DNA from protonemal cells mainly composed of recG-A cells or recG-N cells and measured the amount of mtDNA and ptDNA resulting from recombination between short repeats, using qPCR as described above (Fig. 6C and 8C). qPCR analyses of mtDNA showed that the number of the DNA molecules resulting from recombination between most of the tested repeats was higher in recG-A cells than that in recG-N cells. The levels of these recombination products in recG-N cells were still higher than those in WT cells (Fig. 9A). The levels of recombination product from ccmF-atp9 repeats (originally identified as repeats involved in the mtDNA instability, Fig. 5A) and R12 significantly increased in recG-A cells (Fig. 9A). However, the qPCR analysis of ptDNA showed no significant difference between the amounts of ptIR-1 or ptDR-1 recombination products in recG-A and recG-N cells (Fig. 9B).
Fig. 9. Status of organelle DNA in recG-N and recG-A cells. 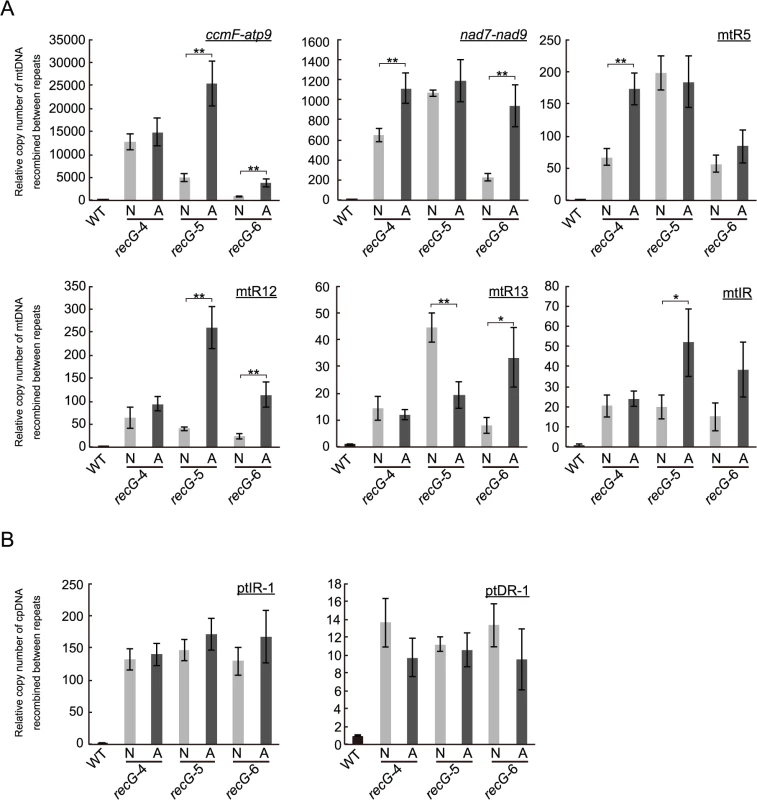
Relative copy number of DNA resulting from recombination between mitochondrial short repeats (ccmF-atp9, nad7-nad9, mtR5, mtR12, mtR13, and mtIR; A) per mitochondrial rpl2 and plastidic short repeats (ptIR-1 and ptDR-1; B) per plastid ndhH in cells mainly comoposed of recG-N or recG-A cells were measured by qPCR using three independent RECG KO lines. WT was given a value of 1. The data represent mean of three replicates ± SD. *p<0.05, **p<0.01. Discussion
In this report, we showed that knocking out a plant-specific RecG homolog RECG induced genomic instability due to repeat-mediated recombination in both mitochondrial genome and plastid genome of P. patens. The induction of organelle genome recombination by RECG KO implies that organelle genomes can potentially undergo repeat-mediated recombination under normal culture conditions. Indeed, plant mitochondrial genomes are occasionally rearranged by recombination between short (<1 kb, in most cases <200 bp) repeats [34], and double-strand breaks in the plastid genome are repaired efficiently by utilizing recombination between short (<100 bp) repeats [35,36], both of which could contribute to potential genome rearrangements in plant organelles. Repeat-mediated recombination similar to those described in this paper were not observed in the bacterial recG mutants as far as we know. We propose that an important role of RECG is to maintain organelle genome stability by suppressing recombination among dispersed repeats.
Analysis of RECG-GFP subcellular localization revealed that the product(s) from the RECG gene localized to both plastids and mitochondria (Fig. 1). Examples of product(s) from a single gene being targeted to both plastid and mitochondrion, named dual targeting, have been reported and classified into two types [37]: an ambiguous signal peptide that can be recognized by both plastid and mitochondrion, or multiple N-terminal signal peptides for different organelles that are produced by alternative translation initiation and/or alternative splicing. Either type may account for the dual targeting of RECG, since RECG has two in-frame AUG codons in its 5’part that could localize their products to both organelles (S1B Fig.). Since A. thaliana RecG homolog also has the potential to localize to both organelles, as determined by our in silico analysis, dual targeting of RecG homolog may be conserved in both P. patens and A. thaliana.
Full-length RECG-GFP formed foci in plastids and mitochondria, and the foci sometimes corresponded to organelle nucleoids. This suggests that RECG is not a constitutive nucleoid protein but associates with organelle nucleoids with a bias. The nucleoid-associating RECG-GFP may be the functional RECG interacting with organelle DNA to maintain genomic stability.
Intriguingly, the RECG KO plants exhibited similar defects in growth, cell morphology, and mitochondrial morphology/activity to those of RECA1 KO plants (Figs. 3 and 4). Mitochondrial defects including disorganized cristae, a lower electron density of the matrix, and an enlarged disc-shape, were also characteristic phenotypes of RECA1 KO plants. The lower electron density of the matrix suggests lower mitochondrial activity, and was frequently observed in recG-A cells, suggesting that mitochondrial dysfunction was the cause of the atrophic phenotype. On the other hand, disorganized cristae and the enlarged disc-shaped mitochondria were commonly observed in recG-A and recG-N cells. As some of the disc-shaped mitochondria exhibited a normal matrix electron density (Fig. 4 and S3 Fig.), mitochondrial dysfunction was probably not responsible for the disc-shape. Disc-shaped mitochondria have been reported to occur in tobacco cells in response to low oxygen pressure without loss of membrane potential [38]. Thus, the disc-shaped mitochondria we observed may reflect the dynamics of mitochondrial morphology rather than mitochondrial dysfunction. The mitochondria of RECG and RECA1 KO plants might change their shape to regulate their activity.
Our results revealed extensive mtDNA rearrangements due to recombination between short repeats in RECG KO plants (Figs. 5 and 6). Rearranged mtDNA produced by recombination between relatively long repeats (47–79 bp, most of which exist in introns as direct repeats) accumulated to a high level as detected by DNA gel blot. We found that in such mtDNA rearrangements, for some loci, there was a decrease in normal mtDNA and a significant decrease in intact transcript levels. The decrease in the production of intact transcripts (<10% of WT) was more extensive than the decrease in the levels of normal mtDNA (30%–50% of WT). On the other hand, a substantial amount of chimeric transcript derived from the rearranged mtDNA accumulated in the RECG KO mutants. Although the chimeric transcripts were properly spliced, the translation product from the chimeric transcripts had to be largely truncated due to the accidental appearance of stop codon; therefore, it should be defective or dominantly negative to the normal translation products. We also found that aberrant recombination between shorter repeats (<35 bp), which is abundantly scattered in the mitochondrial genome, was induced in both the RECG KO and RECA1 KO mtDNA. This type of recombination occurred at a low frequency, but could produce defective mtDNA with deletion of various loci. Collectively, the RECG KO mitochondria are thought to be in a pathological state that includes many kinds of defective mtDNA. These could cause mitochondrial defects via heteroplasmic effect if the normal mtDNA falls below a certain threshold level that is required for normal mitochondria function, as has been conceptually suggested in maize [39]. We found that some kinds of recombined defective mtDNA were highly accumulated in the recG-A cells than in recG-N cells (Fig. 9), proposing a relationship between the accumulation of the mutated mtDNA and atrophic phenotypes. The cytoplasmic segregation of heteroplasmic RECG KO cells with defective mtDNA may have resulted in the biased sorting of the defective mtDNA; when the population of the defective mtDNA exceeds the threshold for the normal mitochondrial function, the cell might exhibit atrophic phenotypes such as those displayed by recG-A cells.
A knock-out of the RECG gene caused genomic instability due to aberrant recombination among 13–63 bp dispersed repeats in the plastids (Fig. 8) as well as in the mitochondria. Since relatively long repeats ptIR-1 (63 bp) and ptDR-1 (48 bp) exist in the structural genes rpl16 and trnG, and psaA and psaB, respectively, recombination between the repeats causes truncation or chimerization in these genes (S6A Fig.). In addition, recombination between abundant dispersed short repeats (< 35 bp) causes deletions or inversions in various ptDNA loci. Thus, RECG KO plants heteroplasmically contain a substantial proportion of defective ptDNA. On the other hand, most of the plastidic longer repeats (> 40 bp), in which RECG KO is assumed to induce recombination more frequently than in shorter repeats, are characteristically located downstream of genes as palindromes (S2 Table), and thus recombinations between them do not disrupt genes directly nor produce defective ptDNA. This property of the plastidic long repeats might alleviate the effect of RECG KO on plastid function. The RECG KO cells, in particular recG-A cells, nevertheless exhibited defects in plastid structure (Fig. 4). Since the amount of the recombined ptDNA was not significantly different between the recG-A cells and recG-N cells, other ptDNA defect might account for the plastid defects as well as the atrophic phenotypes. Alternatively, the accumulation of starch and oil in the recG-A plastids might reflect reduced respiration activity due to mitochondrial dysfunction.
A. thaliana whirly mutant organelle DNA and A. thaliana MSH1 mutant ptDNA exhibit instability due to recombination between repeats shorter than ∼20 bp [11,14,15], and repeats with similar length are involved in the RECG KO organelle DNA instability. This suggests a possibility that RECG, whirly proteins and MSH1 suppress rearrangements in a similar way, however, whirly homologs do not seem to exist in the P. patens genome [24]. On the other hand, the length of the repeats (8–79 bp) involved in recombination of the mtDNA observed in RECG KO mutants differ from those of the A. thaliana MSH1, OSB1 and RECA3 mutants, in which repeats longer than 100 bp are involved [10,12,13]. As P. patens mtDNA has no repeats longer than 100 bp [6], it is impossible to analyze the effect of RECG KO mutants on recombination between repeats longer than 100 bp. Analysis of RECG and these genes in A. thaliana will be needed to elucidate the relationship between RECG and these genes.
Some repeated sequences were involved specifically in either RECG KO or RECA1 KO mutant mtDNA rearrangements, while others were involved in both (Fig. 5 and 6). This suggests that RECG and RECA1 suppress recombination in overlapping but also partially distinct ways in mitochondria. We previously proposed a model in which RECA1 prevents aberrant recombination during the process of repairing stalled or collapsed replication forks [6], based on the suggested role of E. coli RecA [40,41]. Similar to RecA, RecG is thought to function in the processing of stalled replication forks by reversing it in E. coli [17], which raises the possibility that P. patens RECG may also be involved in the repair of replication forks stalled by lesions on the template DNA. The lesions might be created by ROS or UV since they can cause replication fork stalling [42,43]. RECG may reverse stalled replication forks perhaps to prevent subsequent fork collapse and DNA double-strand breaks that can induce genomic instability such as aberrant recombination. Furthermore, RECG might also suppress aberrant recombination by stabilizing recombination intermediate between highly homologous sequences, as demonstrated in E. coli [44]. Differences between the mtDNA rearrangement caused by RECG KO and RECA1 KO might reflect differences in their roles in the repair of impaired replication forks, as studies in E. coli suggest that RecA and RecG function in replication fork repair in distinct situations [18,45]. On the other hand, similarities in organelle DNA defects of RECG, RECA1, MSH1, whirly, and OSB1 mutants might suggest that all of these genes are involved in the integrity of replication forks.
In conclusion, our results suggest that RECG suppresses the recombination between the scattered repeats in organelle genomes and that the pathway partly overlaps that of RECA1 in the mitochondria. A future genetic analysis of double KO mutants of these genes and comprehensive analysis of the mtDNA mutations induced in the single and double KO mutants may elucidate the exact relationship between RECA1 and RECG in the maintenance of mitochondrial genome stability. Our results for the RECG KO mutants indicate that the mechanisms of the suppression of aberrant recombination are probably common to plastids and mitochondria. Another RECA has been found in plastids of P. patens [5] and other plant species [25]. The investigation of the functional relationships between RECA and RECG may shed new light on the molecular mechanisms of plastid genome stability.
Materials and Methods
Plant materials, growth conditions, and preparation of nucleic acids
Physcomitrella patens Bruch & Schimp subsp. patens was used in this study. Protonemata of P. patens were cultured on BCDATG or BCDAT agar medium [46] at 25°C in white light. Growth rate comparisons were performed as described in Odahara et al. [6]. Genomic DNA was extracted by the CTAB method [47] from protonemata cultivated for four days after homogenization and inoculation on agar medium. Total RNA was extracted from protonemata using the RNeasy Plant Mini Kit (Qiagen).
Sequence determination of RECG cDNA
RACE was performed according to Hiwatashi et al. [48] using total RNA. RECG sequence data can be found in the GenBank/EMBL database under accession number AB646798.
Fluorescent microscopy
RECG cDNA corresponding to the full-length 5’UTR and the 284-amino acids of the N-terminal region was amplified by PCR using primers TCCGGATCCTTGCTACACCCTTCTTTCTGCTCCG and CGACCATGGTGCCATCCACAGTGGATTTGTACAGC and fused in frame to the GFP gene of p7133-sGFP [49], and the fused gene was expressed under control of E7133 promoter [50]. Similarly, full-length RECG coding sequence amplified by primers ACTACATCTAGAGGATCCCCGCGATGGCAATTAGAGGTTGTAG and AAGCTTTCCCATGGCCACCCAGTTTTGTTTATCTAGAGCCTCCAG was fused in frame to N-termini of GFP gene and expressed under control of the E7133 promoter. The resulting plasmid was introduced into P. patens protoplasts, and the protoplasts stained with 125 nM Mito Tracker Orange (Molecular Probes) or 1 μg/ml DAPI were observed with an epifluorescence microscope (AX80, Olympus; Axio Imager 2, Zeiss).
Complementation assay in E. coli cells
The RECG cDNA, except for a part corresponding to the N-terminal putative signal peptide (374 amino acids), was amplified by PCR using primers CCTCCATGGGAGCTGCTAACTACAAAGATTGTG and CCTCCCGGGCAACATTATCCTAAAGAACCCGGTG, and E. coli recG was amplified by primers CCCGAATTCACCATGAAAGGTCGGCTGTTAG and CCCCTGCAGTTACGCATTCGAGTAACGTTCCG. Both were placed under the inducible arabinose promoter of pBAD24 [51], and each of the resulting plasmids or pBAD24 was introduced into the E. coli recG deficient strain HRS2000 [27]. After cultivating the strains to an optical density at 600 nm of 0.5 in L broth medium [52] containing 0.5% arabinose, the cells were appropriately diluted and spread on L broth agar media. Then, they were subjected to UV (254 nm) irradiation and cultivated at 37°C. Colony-forming units (cfu) were counted and the surviving fraction was calculated by dividing the cfu in UV irradiated cells by that in nonirradiated cells.
Generating the RECG gene knockout
To prepare the RECG KO construct, fragments of the RECG gene were amplified using primers CAGAATGAGAGCTCTGAGTATGGTGTTC and GAGCCGCGGGAGAGGCTAAGCCTGAAACATTGGAG to obtain a 1.6-kb fragment from the 5’ region, and CAGATCGATGCGACAGTAACGCAAGAAGAAGCAC and GAAGGGCCCTGGGGATTAAGTCTTCTAGTTTGCCTG to obtain the 1.6-kb fragment from the 3’ region. Each of them was then introduced into either side of the nptII cassette of pTN3 [46]. The resulting plasmid, named pMSD202, was linearized before the transformation. Moss transformation was performed according to Nishiyama et al. [46]. The transformants were selected on medium containing G418. To confirm knockout of RECG gene, PCR was performed with primers TCCGGATCCTTGCTACACCCTTCTTTCTGCTCCG and GTTGCTCATCCACAACAGCC.
Transmission electron microscopy
Protonemata cultivated on BCDAT agar medium were fixed with 4% paraformaldehyde and 2% glutaraldehyde in sodium cacodylate buffer, pH 7.4, for overnight at 4°C. Then, cells were fixed with 1% osmium tetroxide in cacodylate buffer, pH 7.4, for 5 h at room temperature. After dehydration in a graded methanol series, the samples were embedded in Epon812 resin. Thin sections were stained with uranyl acetate and lead citrate and observed with JEM-1400 electron microscope (JEOL).
DNA gel blot analysis
Total genomic DNA was separated on a 0.7% agarose gel and blotted onto a nylon membrane. To detect nuclear-encoded and mitochondrial-encoded genes, 4 and 0.5 μg of total genomic DNA were used, respectively. Probes were prepared by PCR using the PCR DIG Probe Synthesis Kit (Roche) and the following primers: GCCTAGGAGGGCGCGTTTGGGAAGACG and CCCAGACACATAACTATAGTGCTAGCCG for nad2; AACACTTGGGTACATGCCAGCCA and GGACACACGGCTACTATGCGATT for atp9; GTGAATGAGTATAAGCTTCGCTGCTCAA and CTATGTATAGCCACTTTGGTAGTGCTTG for ccmF; CGGGTTAGGGGTACGACAGATAGCG and TAATACGACTCACTATAGGGCGAGTAGTTCTATCTATCTACCTCTCC for nad7; GCGCATGCACATTTCCAAGC and GTAGTTATGCTTCAGATGCTTTGC for nad7 in S5A Fig.; CTTGAGAAGCGCAACCTGTG and GCTGTGCCTTTGAAGCTTCG for nad9; ATCCCTGATCCCAGAATACGACTG and GGCTAAGAGCATGAAGACAGATCC for nad4; GTCTGCTGGCAGAAATTCATC and TTGCGCCTTGACCTGGATTC for rpl2; AACGAGTCGTACACTAAGC and ATTCGCGGTCGTTCGTATG for rpl16; and GGACGAATTTTCCATCTCCAAGG and GGAGGAGTTGCTGTAGATTTACC for ndhH. Hybridization of the probes was performed at 37°C, and the membranes were washed in 2x SSC with 0.1% SDS at 25°C and 0.5x SSC with 0.1% SDS at 65°C. Detection of the DIG-labeled probes was performed with Anti-DIG-Alkaline Phosphatase (Roche) and AttoPhos (Promega). All the DNA gel blots were re-hybridized to mitochondrial rpl2 or plastid ndhH probe to estimate organelle DNA copy number (S7 Fig.).
PCR analysis of organelle DNA
PCR analyses of DNA generated by recombination between repeated sequences were performed using total genomic DNA and the primers listed in S4 Table. Quantitative PCR was performed with the Applied Biosystems 7500 Fast Real-Time PCR System and POWER SYBR Green Master Mix (Applied Biosystems International). qPCR was performed with technical replicates of three independent reactions. Standard PCR analysis was performed in the exponential amplification phase, and the quantity of amplification products was compared using ethidium bromide staining.
RT-PCR analysis of mitochondrial transcripts
Total RNA extracted from protonemal cells was treated with TURBO DNase (Ambion) to remove residual genomic DNA and then reverse-transcribed using random hexamer. Quantitative RT-PCR was performed using primers listed in S5 Table with the Applied Biosystems 7500 Fast Real-Time PCR System and POWER SYBR Green Master Mix (Applied Biosystems International). Serine threonine protein phosphatase 2a, ST-P 2a, was used as a reference gene [53]. qRT-PCR was performed with technical replicates of three independent reactions. Standard RT-PCR analysis was performed in the exponential amplification phase.
Assay for evaluation of PCR jumping in qPCR
DNA fragments harboring copy 1 or 2 of ptIR-1 was amplified by standard PCR using primers GGTCCATAAAGGAGCCGTATG and GGGTAGTGGGAATCGAACC for the copy 1 of ptIR-1; CATCCTTCCTCTATGTTGTTTACGA and TCACAAGAAGCCGGATGAAA for the copy 2 of ptIR-1. Relative copy number of the products was analyzed by qPCR using the same primer pairs, and the two fragments were mixed to contain the same copy number of copy 1 or 2 of ptIR-1 as the WT genomic DNA. Then copy number of the DNA recombined between copy 1 and 2 of ptIR-1 were analyzed by qPCR using primers GAATCGAACCCACATCATTAGCT and TATTCACAAGAAGCCGGATGAA and the WT genomic DNA or the mixed DNA fragments as templates.
Supporting Information
Zdroje
1. Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5 : 123–135. 14735123
2. Race HL, Herrmann RG, Martin W (1999) Why have organelles retained genomes? Trends Genet 15 : 364–370. 10461205
3. Marechal A, Brisson N (2010) Recombination and the maintenance of plant organelle genome stability. New Phytol 186 : 299–317. doi: 10.1111/j.1469-8137.2010.03195.x 20180912
4. Terasawa K, Odahara M, Kabeya Y, Kikugawa T, Sekine Y, et al. (2007) The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol Biol Evol 24 : 699–709. 17175527
5. Inouye T, Odahara M, Fujita T, Hasebe M, Sekine Y (2008) Expression and complementation analyses of a chloroplast-localized homolog of bacterial RecA in the moss Physcomitrella patens. Biosci Biotechnol Biochem 72 : 1340–1347. 18460812
6. Odahara M, Kuroiwa H, Kuroiwa T, Sekine Y (2009) Suppression of repeat-mediated gross mitochondrial genome rearrangements by RecA in the moss Physcomitrella patens. Plant Cell 21 : 1182–1194. doi: 10.1105/tpc.108.064709 19357088
7. Martinez-Zapater JM, Gil P, Capel J, Somerville CR (1992) Mutations at the Arabidopsis CHM locus promote rearrangements of the mitochondrial genome. Plant Cell 4 : 889–899. 1356535
8. Sakamoto W, Kondo H, Murata M, Motoyoshi F (1996) Altered mitochondrial gene expression in a maternal distorted leaf mutant of Arabidopsis induced by chloroplast mutator. Plant Cell 8 : 1377–1390. 8776901
9. Abdelnoor RV, Yule R, Elo A, Christensen AC, Meyer-Gauen G, et al. (2003) Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc Natl Acad Sci U S A 100 : 5968–5973. 12730382
10. Arrieta-Montiel MP, Shedge V, Davila J, Christensen AC, Mackenzie SA (2009) Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics 183 : 1261–1268. doi: 10.1534/genetics.109.108514 19822729
11. Cappadocia L, Marechal A, Parent JS, Lepage E, Sygusch J, et al. (2010) Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 22 : 1849–1867. doi: 10.1105/tpc.109.071399 20551348
12. Zaegel V, Guermann B, Le Ret M, Andres C, Meyer D, et al. (2006) The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell 18 : 3548–3563. 17189341
13. Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA (2007) Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 19 : 1251–1264. 17468263
14. Marechal A, Parent JS, Veronneau-Lafortune F, Joyeux A, Lang BF, et al. (2009) Whirly proteins maintain plastid genome stability in Arabidopsis. Proc Natl Acad Sci U S A 106 : 14693–14698. doi: 10.1073/pnas.0901710106 19666500
15. Xu YZ, Arrieta-Montiel MP, Virdi KS, de Paula WB, Widhalm JR, et al. (2011) MutS HOMOLOG1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell 23 : 3428–3441. doi: 10.1105/tpc.111.089136 21934144
16. Whitby MC, Ryder L, Lloyd RG (1993) Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell 75 : 341–350. 8402917
17. McGlynn P, Lloyd RG (2001) Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc Natl Acad Sci U S A 98 : 8227–8234. 11459957
18. Briggs GS, Mahdi AA, Weller GR, Wen Q, Lloyd RG (2004) Interplay between DNA replication, recombination and repair based on the structure of RecG helicase. Philos Trans R Soc Lond B Biol Sci 359 : 49–59. 15065656
19. McGlynn P, Lloyd RG, Marians KJ (2001) Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc Natl Acad Sci U S A 98 : 8235–8240. 11459958
20. Manosas M, Perumal SK, Bianco P, Ritort F, Benkovic SJ, et al. (2013) RecG and UvsW catalyse robust DNA rewinding critical for stalled DNA replication fork rescue. Nat Commun 4 : 2368. doi: 10.1038/ncomms3368 24013402
21. Rudolph CJ, Upton AL, Harris L, Lloyd RG (2009) Pathological replication in cells lacking RecG DNA translocase. Mol Microbiol 73 : 352–366. doi: 10.1111/j.1365-2958.2009.06773.x 19538444
22. Rudolph CJ, Upton AL, Lloyd RG (2009) Replication fork collisions cause pathological chromosomal amplification in cells lacking RecG DNA translocase. Mol Microbiol 74 : 940–955. doi: 10.1111/j.1365-2958.2009.06909.x 19818016
23. Rudolph CJ, Upton AL, Stockum A, Nieduszynski CA, Lloyd RG (2013) Avoiding chromosome pathology when replication forks collide. Nature 500 : 608–611. doi: 10.1038/nature12312 23892781
24. Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319 : 64–69. 18079367
25. Lin Z, Kong H, Nei M, Ma H (2006) Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci U S A 103 : 10328–10333. 16798872
26. Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300 : 1005–1016. 10891285
27. Ishioka K, Iwasaki H, Shinagawa H (1997) Roles of the recG gene product of Escherichia coli in recombination repair: effects of the delta recG mutation on cell division and chromosome partition. Genes Genet Syst 72 : 91–99. 9265736
28. Quatrano RS, McDaniel SF, Khandelwal A, Perroud PF, Cove DJ (2007) Physcomitrella patens: mosses enter the genomic age. Curr Opin Plant Biol 10 : 182–189. 17291824
29. Huang CY, Chung CI, Lin YC, Hsing YI, Huang AH (2009) Oil bodies and oleosins in Physcomitrella possess characteristics representative of early trends in evolution. Plant Physiol 150 : 1192–1203. doi: 10.1104/pp.109.138123 19420327
30. Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, et al. (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29 : 4633–4642. 11713313
31. Sugiura C, Kobayashi Y, Aoki S, Sugita C, Sugita M (2003) Complete chloroplast DNA sequence of the moss Physcomitrella patens: evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucleic Acids Res 31 : 5324–5331. 12954768
32. Stern DB, Gruissem W (1987) Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell 51 : 1145–1157. 3690662
33. Alverson AJ, Zhuo S, Rice DW, Sloan DB, Palmer JD (2011) The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS One 6: e16404. doi: 10.1371/journal.pone.0016404 21283772
34. Newton KJ, Gabay-Laughnan S, Paepe RD (2004) Mitochondrial mutation in plants.; Day DA, Millar AH, Whelan J, editors. Dordrecht, The Netherlands: Kluwer Academic Publishers. 121–141 p.
35. Odom OW, Baek KH, Dani RN, Herrin DL (2008) Chlamydomonas chloroplasts can use short dispersed repeats and multiple pathways to repair a double-strand break in the genome. Plant J 53 : 842–853. 18036204
36. Kwon T, Huq E, Herrin DL Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc Natl Acad Sci U S A 107 : 13954–13959. doi: 10.1073/pnas.1004326107 20643920
37. Peeters N, Small I (2001) Dual targeting to mitochondria and chloroplasts. Biochim Biophys Acta 1541 : 54–63. 11750662
38. Van Gestel K, Verbelen JP (2002) Giant mitochondria are a response to low oxygen pressure in cells of tobacco (Nicotiana tabacum L.). J Exp Bot 53 : 1215–1218. 11971932
39. Gu J, Miles D, Newton KJ (1993) Analysis of Leaf Sectors in the NCS6 Mitochondrial Mutant of Maize. Plant Cell 5 : 963–971. 12271093
40. Lusetti SL, Cox MM (2002) The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem 71 : 71–100. 12045091
41. Courcelle J, Hanawalt PC (2003) RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet 37 : 611–646. 14616075
42. Burhans WC, Weinberger M (2007) DNA replication stress, genome instability and aging. Nucleic Acids Res 35 : 7545–7556. 18055498
43. Courcelle J, Carswell-Crumpton C, Hanawalt PC (1997) recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci U S A 94 : 3714–3719. 9108043
44. Mawer JS, Leach DR (2014) Branch migration prevents DNA loss during double-strand break repair. PLoS Genet 10: e1004485. doi: 10.1371/journal.pgen.1004485 25102287
45. Robu ME, Inman RB, Cox MM (2004) Situational repair of replication forks: roles of RecG and RecA proteins. J Biol Chem 279 : 10973–10981. 14701860
46. Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res 7 : 9–17. 10718194
47. Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8 : 4321–4325. 7433111
48. Hiwatashi Y, Nishiyama T, Fujita T, Hasebe M (2001) Establishment of gene-trap and enhancer-trap systems in the moss Physcomitrella patens. Plant J 28 : 105–116. 11696191
49. Hara K, Sugita M, Aoki S (2001) Cloning and characterization of the cDNA for a plastid sigma factor from the moss Physcomitrella patens. Biochim Biophys Acta 1517 : 302–306. 11342113
50. Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, et al. (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37 : 49–59. 8720924
51. Guzman LM, Belin D, Carson MJ, Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177 : 4121–4130. 7608087
52. Yoshioka Y, Ohtsubo H, Ohtsubo E (1987) Repressor gene finO in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 into F finO. J Bacteriol 169 : 619–623. 3027040
53. Le Bail A, Scholz S, Kost B (2013) Evaluation of reference genes for RT qPCR analyses of structure-specific and hormone regulated gene expression in Physcomitrella patens gametophytes. PLoS One 8: e70998. doi: 10.1371/journal.pone.0070998 23951063
Štítky
Genetika Reprodukční medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 3
-
Všechny články tohoto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



