-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSpatiotemporal Regulation of a T4SS Substrate by the Metaeffector SidJ
A key attribute of many pathogens is their ability to survive and replicate within eukaryotic host cells. One such pathogen, Legionella pneumophila, is able to grow within macrophages in the lungs, thereby causing a form of pneumonia called Legionnaires’ Disease. L. pneumophila causes disease by translocating several hundred proteins into the host cell. These proteins are typically referred to as ‘‘effectors’’, as they function as toxins to alter normal host cell function. However, since L. pneumophila remains within the host cells for approximately one day, continual poisoning of the eukaryotic cells by the bacterial effectors will result in the premature death of the host cell, thus restricting the growth of the pathogen. Previously the L. pneumophila secreted protein LubX was described as a “metaeffector”, which has been defined as an effector that acts directly on another effector to modulate its function inside the host cell. LubX accomplishes this task by directing the degradation of another effector, SidH. Here we report a second L. pneumophila metaeffector, SidJ, acts in a similar manner to neutralize SidE family effectors by removing them from the intracellular compartment that contains the bacterium. This further establishes the concept of metaeffectors, which are likely to be critical to how Legionella and many other pathogens cause disease.
Published in the journal: . PLoS Pathog 11(3): e32767. doi:10.1371/journal.ppat.1004695
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004695Summary
A key attribute of many pathogens is their ability to survive and replicate within eukaryotic host cells. One such pathogen, Legionella pneumophila, is able to grow within macrophages in the lungs, thereby causing a form of pneumonia called Legionnaires’ Disease. L. pneumophila causes disease by translocating several hundred proteins into the host cell. These proteins are typically referred to as ‘‘effectors’’, as they function as toxins to alter normal host cell function. However, since L. pneumophila remains within the host cells for approximately one day, continual poisoning of the eukaryotic cells by the bacterial effectors will result in the premature death of the host cell, thus restricting the growth of the pathogen. Previously the L. pneumophila secreted protein LubX was described as a “metaeffector”, which has been defined as an effector that acts directly on another effector to modulate its function inside the host cell. LubX accomplishes this task by directing the degradation of another effector, SidH. Here we report a second L. pneumophila metaeffector, SidJ, acts in a similar manner to neutralize SidE family effectors by removing them from the intracellular compartment that contains the bacterium. This further establishes the concept of metaeffectors, which are likely to be critical to how Legionella and many other pathogens cause disease.
Introduction
Legionella pneumophila, the causative agent of Legionnaires' disease, is a facultative intracellular bacterial pathogen that can replicate within fresh water amoeba and mammalian alveolar macrophages [1–3]. L. pneumophila survives and replicates within host cells by inhibiting the host endocytic pathway and creating a novel replicative compartment designated as the Legionella containing vacuole (LCV) [4–7]. Alteration of host function is mediated by the injection of a large number of proteins into the host cell by the L. pneumophila Dot/Icm type IV secretion system (T4SS) [8–12]. However, inactivation of individual (or even combinations of) Dot/Icm substrates in genetically engineered mutant strains rarely has a strong effect on the intracellular growth of L. pneumophila, consistent with extensive functional redundancy between effectors [13–15].
One notable exception to this generalization is the L. pneumophila SuperΔP170 mutant, which exhibited a substantial growth defect in the amoebae Acanthamoeba castellanii [16]. The SuperΔP170 was constructed while studying a locus that encodes multiple Dot/Icm substrates [16] and consists of two deletions: the first removes five adjacent genes (sdeC, lpg2154, sidJ, sdeB, and sdeA) and the second deletes the unlinked gene sidE. Four of the encoded proteins, SidE, SdeC, SdeB and SdeA, share extensive homology with each other and are all ∼170 kDa in size, thus they have been referred to as “P170s” [16]. In addition, they are called the “SidE family”, as SidE was the founding member of this related group of proteins [17]. The SidE proteins are Dot/Icm substrates that are translocated into the host cell and reside on the cytoplasmic face of the LCV (Legionella containing vacuole) [16], although their molecular function is not known. As the intracellular growth defect of the SuperΔP170 mutant could be complemented by expression of just one SidE family protein, SdeA, it was proposed that the SidE-like proteins were functionally redundant and the other two genes, lpg2154 and sidJ, must be dispensable for growth within host cells [16]. However, subsequently it was shown that inactivation of sidJ alone conferred an intracellular growth defect on L. pneumophila [18], suggesting the situation is more complicated than initially perceived.
Consistent with this observation is the increasingly appreciated paradigm in pathogenesis that secreted effectors are often subjected to spatiotemporal regulation and that there can be a complex interplay between the functions of different effectors. For example, the Salmonella T3SS substrates SopE and SptP, which possess opposing biochemical activities, act at different stage of infection to first induce bacterial uptake and then to down-modulate this effect in order to prevent host cell death [19]. Similarly, the Legionella pneumophila Dot/Icm T4SS effectors SidM/DrrA and LepB exhibit opposing functions. SidM/DrrA recruits and activates Rab1 to mediate fusion of ER microsomes with the LCV (Legionella containing vacuole). At later points, LepB inactivates Rab1 resulting in the removal of the GTPase from the LCV [20–22]. A third example is represented by the L. pneumophila effectors SidH and LubX. SidH is a homolog of the effector SdhA, which is required to maintain the integrity of the LCV [23,24]. LubX is a member of the U-box family of E3 ubiquitin ligases and functions as a metaeffector to inactivate SidH by promoting its proteolysis [25].
Due to their genetic proximity, the surprising phenotype of the ΔsidJ mutant, and the existing precedents of complex interplay between other L. pneumophila secreted substrates, we hypothesized that there may be a connection between the SidE proteins and SidJ. To test this hypothesis, we examined the phenotypes of a strain lacking just the four SidE proteins (ΔsdeC ΔsdeB ΔsdeA ΔsidE) and of an individual ΔsidJ mutant and discovered that overexpression of an individual SidE family protein in the absence of SidJ is toxic to host cells. This result, and the experiments that followed, have led to a model wherein SidJ functions as a metaeffector to regulate the activity of the SidE family of toxins.
Results
SidJ and the SidE family are required for optimal intracellular growth of L. pneumophila
The sdeC-sdeA locus encodes SdeC, Lpg2154, SidJ, SdeB, and SdeA (Fig. 1A). A related protein, SidE, is encoded at a separate site on the chromosome. SidE, SdeC, SdeB, and SdeA are each ∼170 kDa in size, share greater than 40% identity to each other, and are substrates of the L. pneumophila Dot/Icm T4SS (S1 Fig.) [16,17]. The sdeC-sdeA locus also encodes another Dot/Icm substrate, SidJ, which has no homology to the SidE family (S1 Fig.). Although the L. pneumophila strain Philadelphia I encodes a homolog to SidJ, SdjA (Lpg2508), it was previously shown to be dispensable for virulence and therefore was not characterized further [18].
Fig. 1. Genetic analysis of the SidE family and SidJ. 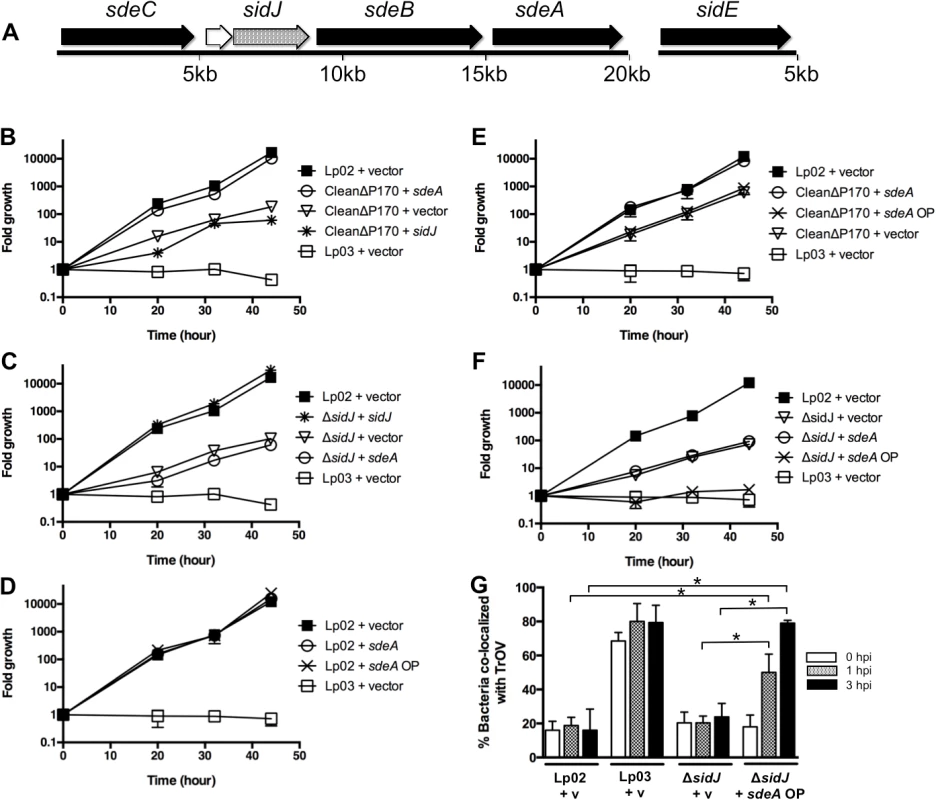
(A) sdeC-sdeA locus includes five genes (sdeC, lpg2154, sidJ, sdeB, and sdeA), whereas the sidE gene is located at a separate location. The SidE family consists of the related proteins SidE, SdeC, SdeB, and SdeA (shown with black arrows). (B-F) Replication of various L. pneumophila strains in A. castellanii was determined at the indicated time points post infection and expressed as fold growth. (B) Suppression of the growth defect of the CleanΔP170 mutant by expression of low amounts of SdeA. (C) Complementation of the ΔsidJ mutant by expression of SidJ. (D-F) Overexpression of SdeA does not inhibit the growth of the wild type strain Lp02 (D) or the CleanΔP170 mutant (E) but does inhibit the replication of the ΔsidJ mutant (F). (G) Overproduction of SdeA causes Legionella to traffic into the endocytic pathway of amoebae. A. castellanii pre-incubated with Texas Red Ovalbumin (TrOV), which labels their endocytic pathway and vacuole, were infected with Lp02, a dotA mutant (Lp03) and a ΔsidJ mutant containing vector (v) or a plasmid over-expressing SdeA (sdeA OP). The percent of bacteria that co-localized with TrOV was quantitated at 0, 1, and 3 hours post infection. Data are means ± SEM of three independent experiments. Approximately 100 bacteria were scored per condition and asterisks indicate statistical difference (P<0.05). To examine the connection between SidJ and the SidE family of proteins, we constructed two additional mutant strains. One mutant lacked only sidJ and the other mutant contained deletions in each of the four sidE-related genes, which we refer to as the “CleanΔP170” mutant. We then compared the intracellular replication properties of the ΔsidJ, the CleanΔP170, and the original SuperΔP170 mutant [16], which does not express SidJ or any member of the SidE family. Growth was assayed by infecting the amoebae A. castellanii for various amounts of time, lysing the cells and determining the fold bacterial growth based on the number of CFUs (colony forming units). In this assay, a wild-type strain of L. pneumophila was able to replicate greater than 1000-fold in forty-four hours whereas a T4SS-deficient strain, Lp03, was unable to grow (Fig. 1B). In contrast, the CleanΔP170 strain exhibited a 100-fold growth defect similar to that previously observed for the SuperΔP170 mutant (Figs. 1B and S2). Likewise, a strain lacking sidJ exhibited a similar intermediate intracellular replication deficiency (Fig. 1C) [18]. The CleanΔP170 growth defect could be complemented by expression of just one SidE family protein, SdeA, consistent with the redundancy previously observed between members of this family (Fig. 1B) [16]. As expected, the replication defect of the CleanΔP170 mutant could not be complemented by expression of SidJ from a plasmid as this strain already makes SidJ (Fig. 1B). Similarly, the strain lacking sidJ could be complemented by expression of sidJ, but not by sdeA (Fig. 1C).
Taken together, these data suggest that expression of SidJ and at least one member of the SidE family is required for virulence of L. pneumophila within the environmental host A. castellanii. But this raised the question of how the original SuperΔP170 mutant strain, which is missing all four members of the SidE family and SidJ, could be fully complemented by expression of only SdeA (S2 Fig.) [16]. We hypothesized that this discordance might be related to expression levels from the sdeA expression plasmid. However, the sdeA complementing clone, pJB3556, did not synthesize aberrant amounts of SdeA and instead made a similar amount to what is normally expressed in Legionella (S3 Fig.).
Although pJB3556 expresses the appropriate level of SdeA in a cell, it is worth noting that this represents only a portion of the total amount of SidE family proteins expressed in a wild type cell. Therefore, we assayed the effect of expressing higher amounts of SdeA from a new complementing clone, SdeA OP (SdeA “over production”) (S3 Fig.). Expression of SdeA from this new construct had no effect on the growth of the wild type strain Lp02 (Fig. 1D) or the CleanΔP170 mutant (Fig. 1E). On the other hand, over-expression of SdeA remarkably completely inhibited the growth of the ΔsidJ mutant to levels similar to that of the T4SS-deficient dotA mutant (Fig. 1F). This extent of inhibition was shared with the SuperΔP170 mutant, which also does not express SidJ (S2 Fig.). Thus, inhibition of replication by SdeA over-production occurs only in strains lacking sidJ. Interestingly, this inhibitory effect appears to be specific to Legionella virulence, as over-expression of SdeA in the ΔsidJ mutant results in increased mis-targeting of Legionella to a late endocytic/lysosomal compartment (Fig. 1G). In summary, the absence of either the SidE family or SidJ results in diminished growth of L. pneumophila within amoebae. Furthermore, the partial growth defect of a ΔsidJ strain, but not of a wild type strain, can be exacerbated by the over-expression of SdeA, thus establishing a link between SidJ and the SidE family.
SidJ and SdeA do not affect each other’s secretion into host cells
One possible connection between SidJ and SdeA is that they might modulate each other’s secretion into host cells. To test this hypothesis, we measured the export of the reporter proteins CyaA-SidJ and CyaA-SdeA in different genetic backgrounds. Successful export of Bordetella pertussis CyaA fusions into the host cell cytoplasm is measured by increased cAMP production, since this version of CyaA is activated when it is bound by eukaryotic calmodulin [26,27]. Expression of either CyaA-SidJ or CyaA-SdeA in the wild-type strain Lp02 generated a large increase in host cell cAMP as compared to expression in T4SS-deficient Lp03 cells, mock infected cells, or Lp02 expressing only CyaA (S4 Fig.). As previously observed [16,28], SdeA secretion was strongly dependent on the type IV adaptor IcmS whereas SidJ secretion was only partially dependent. However, export of SdeA was not affected by the absence of sidJ nor was SidJ secretion diminished in a strain that did not express any of the SidE family (S4 Fig.). Therefore, SidJ does not appear to regulate the secretion of SidE family members and SidE proteins do not affect the export of SidJ, suggesting that the molecular connection between SidJ and SidE family members likely occurs within host cells.
SdeA toxicity in eukaryotic cells can be suppressed by SidJ
Since over-expression of SdeA is toxic to amoebae in the absence of SidJ, we chose to explore this phenomenon in more detail within the model eukaryote Saccharomyces cereviseae [29,30]. We began these yeast studies by expressing SdeA and SidJ under the control of the strong, regulated Pgal promoter. Yeast cells transformed with Pgal vectors carrying sdeA or sidJ were grown in the presence of glucose, diluted, and spotted onto selective media containing glucose (repressing conditions) or galactose (inducing conditions). Galactose-induced expression of either SdeA or SidJ was extremely toxic to yeast cells (Fig. 2A, rows 2–3) consistent with previous results [29,30]. To further evaluate this toxicity, we expressed each protein at lower levels using the constitutively expressed, weak promoter Pcyc. Expression of sidJ under the weaker promoter had only a subtle effect on yeast (Fig. 2A, row 5) whereas it was not possible to construct a yeast strain harboring Pcyc-sdeA, presumably due to SdeA-mediated toxicity. Strikingly, expression of low amounts of SidJ (Pcyc-sidJ) was able to partially suppress the toxicity caused by expression of high amounts of SdeA (Pgal-sdeA) (Fig. 2A, row 7).
Fig. 2. SidJ suppresses SdeA toxicity in yeast and mammalian cells. 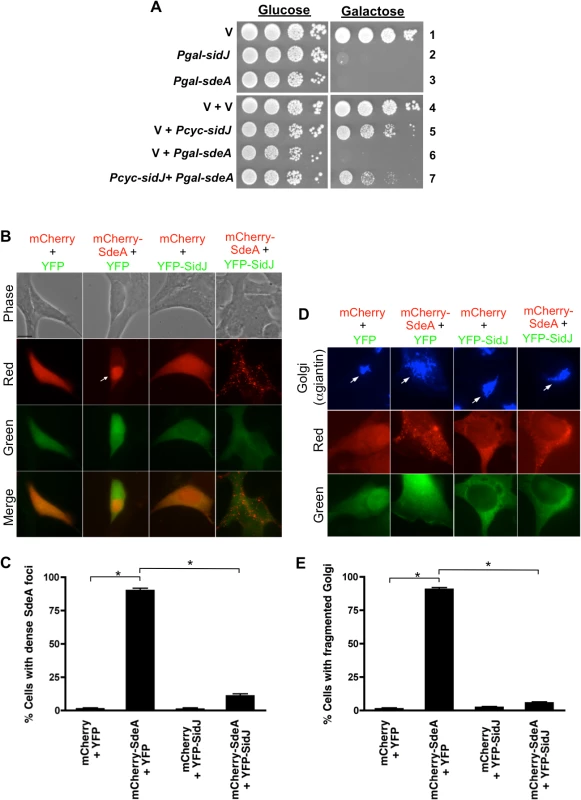
(A) Tenfold serial dilution of yeast strains expressing vector (v), Pgal-sidJ, Pgal-sdeA or Pcyc-sidJ were spotted and grown on selective plates containing glucose (repressing conditions) or galactose (inducing conditions). (B and C) Localization of SdeA and SidJ in HEK293 cells as observed by transfecting cells for 40-hours with mCherry, YFP, mCherry-SdeA and YFP-SidJ. Channels used to acquire images are indicated on the left of each row and merged images are shown in the bottom row. Arrow indicates a cell containing a dense mCherry-SdeA foci. Bar, 10 μm. (C) Scoring data of cells with dense SdeA loci observed in (B). (D) Shorter transfection time (20-hour) reveals mCherry-SdeA-induced Golgi fragmentation in HEK293 cells. Cells were fixed and stained with anti-giantin (Golgi marker). Bar, 10 μm. Arrows indicate Golgi. (E) Scoring data of Golgi fragmentation in (D). Data are means ± SEM of three independent experiments. Approximately 100 cells/condition were counted and asterisks indicate statistical difference (P<0.05). We then attempted to recapitulate this result in mammalian cells. Transient transfection of HEK293 cells for 40 hours with mCherry-SdeA resulted in the protein localizing in dense foci in ∼90% of the cells (Fig. 2B and 2C). This result was specific to the SdeA fusion as expression of only mCherry resulted in diffuse, cytoplasmic staining. The longer mCherry-SdeA was expressed in cells, the more toxic it became eventually causing the cells to round up (S5 Fig.). In contrast, expression of YFP-SidJ was not toxic and the protein localized diffusely in the cytoplasm similar to YFP alone (Fig. 2B and 2C). Remarkably, the accumulation and toxicity caused by mCherry-SdeA expression was suppressed by co-transfection with YFP-SidJ resulting in dispersal of mCherry-SdeA into smaller foci (Fig. 2B and 2C).
To further examine SdeA toxicity in mammalian cells, mCherry-SdeA was expressed for a shorter amount of time (20 hours) and the cells were analyzed by fluorescence microscopy using markers to detect the endoplasmic reticulum, mitochondria, Golgi, endosome/lysosome, actin, and tubulin. Shorter expression of mCherry-SdeA only affected the Golgi resulting in its fragmentation (Fig. 2D-2E). In contrast, expression of mCherry alone, YFP alone, or YFP-SidJ had no effect on the Golgi. Similar to the redistribution of SdeA, YFP-SidJ was able to suppress the Golgi fragmentation caused by mCherry-SdeA (Fig. 2D and 2E). In summary, SdeA expression was toxic to both yeast and mammalian cells and SdeA-toxicity could be suppressed by co-expression of SidJ, implying that SidJ may regulate SdeA function.
SidJ modulates SdeA localization on Legionella containing vacuoles
A number of Dot/Icm substrates localize to the outside of the Legionella containing vacuole (LCV) at various stages of infection, including members of the SidE family [16,17,22,31]. For example, the substrate SidM/DrrA associates with the LCV early during infection but cannot be detected at later time points [22]. In contrast, the substrate LepB has limited association with the LCV at early points but displays increased co-localization over time. Similar to SidM/DrrA, SidE family proteins can be detected in proximity to the LCV early on but then disappear at later points during infection [16].
Based on these results, we hypothesized that SidJ may modulate SidE family association with the LCV as a means of regulating their activity. We began by confirming the observation that SidE family proteins can be detected on the LCV only at the initial stages of infection [16]. Bone marrow macrophages (BMMs) were infected for increasing amounts of time with the wild-type L. pneumophila strain Lp02, the T4SS-deficient strain Lp03, and the ΔsidJ mutant. The cells were fixed and stained with antibodies specific for three Dot/Icm substrates including the SidE family [16], SidM/DrrA, and a protein known to remain on the LCV, LidA [31]. As shown in Fig. 3, SidE staining could be detected on wild-type LCVs adjacent to the bacterial poles at 1-hour post infection. The detected SidE signal at 1 hour represented secreted protein, as it was not detected in Lp03-infected macrophages. As previously observed [16], SidE staining decreased as the infection proceeded resulting in less than 10% co-localization with the LCV after 4 hours (Fig. 3B). SidM/DrrA exhibited a similar pattern of localization over time (Fig. 3C) [22]. In contrast to SidE and SidM/DrrA, the secreted effector LidA was retained on the LCV throughout the infection (Fig. 3D), thus demonstrating that the disappearance of SidE and SidM/DrrA is not a general phenomenon.
Fig. 3. SidJ mediated the disappearance of SidE proteins from the LCV at later time points of infection. 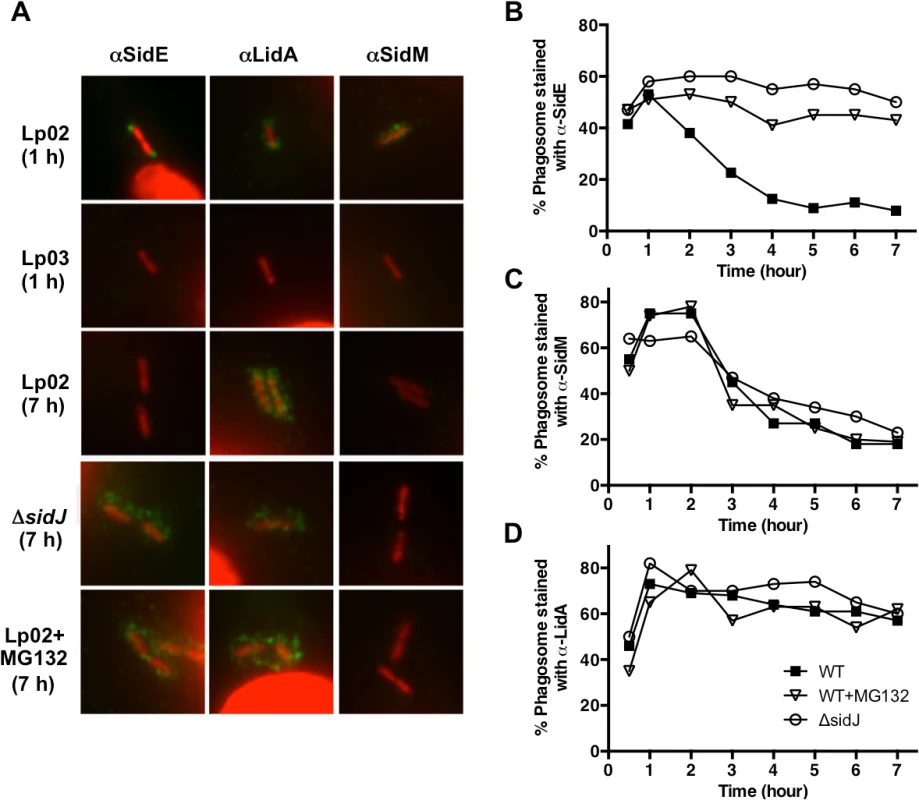
(A) BMMs were infected with the wild-type strain Lp02, the T4SS-deficient strain Lp03 or the ΔsidJ mutant, fixed, and stained with antibodies specific for the SidE family, LidA, or SidM (green). DNA (L. pneumophila and host nuclei) was stained with propidium iodide (red). Representative images are shown for 1 hour or 7-hour infections. Cells were treated with MG132 (10 μM) for 15 min prior to infection as needed. (B-D) Co-localization of SidE, SidM, and LidA with the LCV were scored and recorded as a percentage. Percentage of co-localization is plotted over time for SidE proteins (B), SidM (C), and LidA (D) for the wild-type strain Lp02 (filled squares), Lp02 + MG132 (open triangles), and the ΔsidJ mutant (open squares). Approximately 75 LCVs were counted for each Dot/Icm substrate at each point. Two independent experiments were conducted and the data is representative of both. To test our hypothesis that SidJ might mediate SidE family localization, we examined the intracellular position of the three Dot/Icm substrates when secreted from a ΔsidJ mutant. The absence of SidJ had no effect on the location of SidM/DrrA or LidA (Fig. 3). However, the localization of SidE was significantly altered in the ΔsidJ strain in a time-dependent manner resulting in retention of SidE on the LCV at later time points of infection (Fig. 3B). The disappearance of SidE proteins from the LCV in a wild-type infection could be due to their degradation and/or their dissociation from the L. pneumophila phagosome. To test these possibilities, we examined the presence of LCV-associated SidE proteins during an infection where the host cells were first treated with the proteasome inhibitor MG132. Interestingly, MG132 treatment resulted in a significant increase in the amount of retained SidE proteins at later time points (Fig. 3B), similar to that seen with the ΔsidJ mutant. In contrast, MG132 had no effect on SidM localization (Fig. 3C), thus demonstrating specificity. In summary, the disappearance of SidE proteins from the LCV at later points of infection is dependent on both SidJ and the proteasome.
SidE disappearance from the LCV is not due to general proteolysis of the protein
Based on the MG132 result, it is possible that SidJ removes SidE family proteins from the LCV via induced proteolysis, similar to the action of the metaeffector LubX on the substrate SidH [25]. To test this theory, we developed a method using the detergent digitonin to measure the total amount of secreted SidE family proteins. Digitonin extracts proteins from mammalian membranes without disrupting bacterial membranes due to the absence of cholesterol in the latter membranes. We then proceeded to infect the human monocytic cell line U937 with wild-type Legionella, gently lysed the cells using a dounce homogenizer and removed unbroken host cells via low speed centrifugation. This generated a post-nuclear supernatant (PNS) fraction that was then separated by a high-speed centrifugation step into a pellet fraction, containing host organelles and the LCV, and a soluble cytoplasmic fraction.
In the absence of digitonin, the majority of SidE-like proteins co-localized in the pellet fraction with DotF, an inner membrane protein of the Dot/Icm T4SS that was used as a marker for the LCV (Fig. 4A). In contrast, SidJ could be detected in the soluble fraction without digitonin, indicating that the majority of the protein was secreted into the host cytoplasm and not retained on the LCV (Fig. 4A). Inclusion of digitonin in the reaction resulted in the solubilization of a large percentage of the SidE proteins, indicating they were secreted into the host cell. DotF could not be extracted and therefore was retained within the digitonin-resistant, bacterial cell wall (Fig. 4A).
Fig. 4. SidE proteins are not degraded during infection. 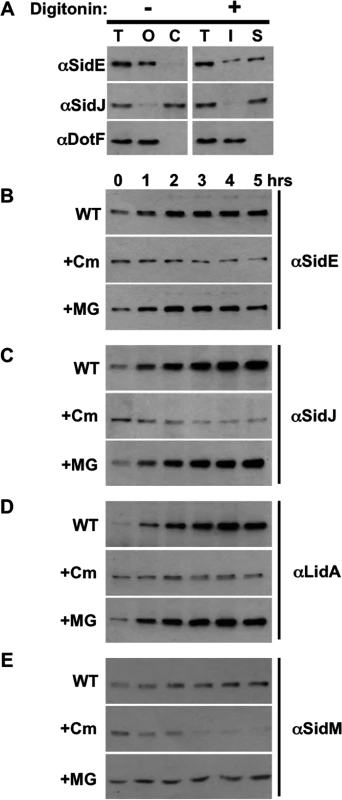
(A) Assay demonstrating secretion of Dot/Icm substrates SidE and SidJ. U937 cells were infected with wild-type Legionella, lysed by douncing, and post nuclear supernatant fractions (PNS) were prepared. Samples were processed in the absence (left column) or the presence of digitonin (right column). Fractions include total (T), organelles and LCV (O), cytoplasm (C), digitonin insoluble (I) and digitonin soluble (S), which contains secreted proteins. Fractions were analyzed by Western blot using antibodies specific for the indicated proteins. (B-E) U937 cells were infected with wild-type Legionella for 1 hour, washed, and the infection was allowed to proceed for the indicated times in the absence (WT) or presence of chloramphenicol (Cm) or MG132 (MG). The cells were processed as above including dounce lysis and digitonin treatment followed by centrifugation. The digitonin soluble data is shown for SidE (B), SidJ (C), LidA (D), and SidM (E) and is representative of three experiments. Using this PNS/digitonin assay, we measured the levels of secreted SidE and SidJ proteins by Western analysis. Rather than observing decreased quantity of digitonin-soluble SidE proteins consistent with proteolysis, their amounts actually increased over the first 5 hours of the infection (Fig. 4B, WT). The levels of secreted SidJ and LidA were elevated in a similar fashion, although the amount of SidM/DrrA did not significantly increase over time (Fig. 4B and 4C, WT). In order to more carefully examine if any of the initial, secreted protein was degraded, we examined the levels of the proteins after inhibition of protein synthesis using the antibiotic chloramphenicol. The initial wave of secreted SidE, SidJ, and LidA remained fairly stable in the presence of chloramphenicol (Fig. 4, +Cm). In contrast, chloramphenicol-treatment did affect the levels of SidM/DrrA as the infection proceeded, suggesting that continual secretion of SidM/DrrA protein was necessary to maintain the levels of the protein over time (Fig. 4E, +Cm).
Treatment with the proteasome inhibitor MG132 resulted in a slight stabilization of SidM/DrrA consistent with the decreased amounts of SidM in the presence of chloramphenicol being due to proteolysis by the proteasome (Fig. 4E, +MG). In contrast, MG132 surprisingly did not alter the levels of SidE proteins (Fig. 4B, +MG), indicating they were not subject to proteasomal degradation. This result was unexpected based on our previous data showing MG132 treatment prevented the disappearance of SidE from the LCV (Fig. 3). Therefore, the situation must be more complex than initially anticipated. For example, there could be two populations of SidE proteins within the cell, a small portion on the LCV and a distinct population located elsewhere, and SidJ mediates the proteasomal-degradation of only LCV-associated SidE proteins.
SidE disappears from the LCV and co-localizes with fractions containing host organelles at later time points of infection
To test this hypothesis, we separated the LCV from other cell organelles using discontinuous sucrose gradient analysis. Using this method, we were able to obtain fractions enriched for the cytoplasm (actin), endosomes/lysosomes (LAMP-1), mitochondria (PHB), Golgi (GM130), ER (calnexin), and the LCV (DotF) (Fig. 5A). We then examined the location of SidE proteins and SidJ at 1 hpi (hour post infection) and 3 hpi. Consistent with our immunofluoresence data in Fig. 3, we were able to detect SidE proteins that co-localized with the LCV at 1 hpi (Fig. 5B). However, we also observed a large amount of SidE proteins in fractions that contained host organelles, including endosomes/lysosomes, mitochondria and ER (Fig. 5). Strikingly, at 3-hours post infection there was a significant depletion of SidE proteins in the LCV fraction and instead there was an enrichment of these proteins in the organelle fractions distinct from the peak cytoplasmic fractions containing actin (Fig. 5A). In contrast to SidE proteins, SidJ was found in the cytoplasmic fractions at both 1 and 3-hours post infection (Fig. 5B). Taken together, our data suggests that the SidJ and proteasome-dependent removal of SidE proteins from the LCV is due to localized degradation of protein.
Fig. 5. SidE proteins removed from the LCV associate with host organelles. 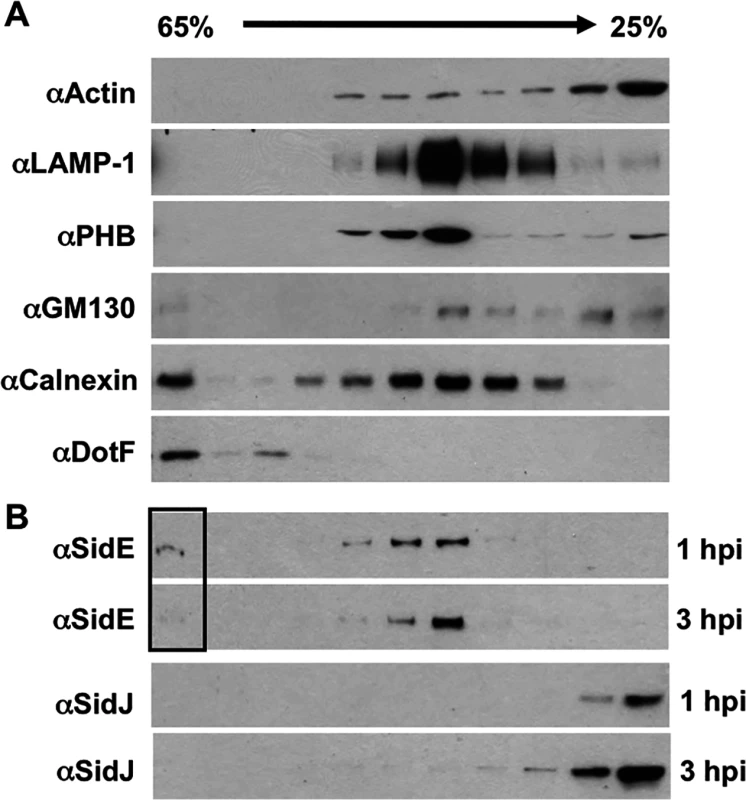
U937 cells were infected with wild-type Legionella for 1 hour or 3 hours, lysed by douncing, separated by sucrose gradient and analyzed by Western blot. (A) Western blots includes markers for various cell compartments including: actin (cytoplasm), LAMP-1 (endocytic compartments), PHB (mitochondria), GM130 (Golgi), calnexin (ER), and DotF (LCV). (B) Westerns for SidE proteins and SidJ at both 1 and 3 hour post infection (hpi). The box highlights the altered levels of SidE proteins in the LCV fraction as the infection progresses. SidJ directly mediates the disappearance of SidE proteins from the LCV
Although the removal of SidE proteins from the LCV requires SidJ, it is possible that this effect is indirect and perhaps due to maturation of the LCV over time. Therefore, we developed an assay to test if purified SidJ protein could remove SidE proteins from LCVs in vitro. This assay involved isolating post nuclear supernatants (PNSs) from bone marrow-derived macrophages (BMMs) infected for 1 hour or 4 hours with the L. pneumophila ΔsidJ mutant (Fig. 6). The PNSs were mock treated or incubated with purified wild-type SidJ, a SidJ mutant (SidJ DD), or BSA as a negative control. The SidJ DD mutant contains mutations in two conserved aspartate residues (D542A D545A) (S6B Fig.), was fortuitously identified based on a fallacious lead using a protein fold recognition server (Phyre), but resulted in a protein that nevertheless failed to complement the intracellular growth defect of a ΔsidJ mutant (S6C Fig.). As shown in Fig. 6A and 6B (mock), SidE proteins remained associated with the polar sites of the Legionella phagosome in the absence of SidJ at both 1-hour and 4-hour post infection. Addition of purified wild-type SidJ to the PNS displaced the SidE proteins from LCVs obtained from either the 1-hour or the 4-hour infection. In contrast, inclusion of an equivalent amount of the non-functional SidJ mutant, SidJ DD, or BSA had no effect on SidE co-localization with the LCV (Fig. 6A and 6B). The disappearance of SidE was specific as addition of SidJ did not cause removal of LidA from the LCV (Fig. 6C and 6D). SidJ-mediated removal of SidE was both concentration (Fig. 6E) and time-dependent (Fig. 6F) consistent with a catalytic mechanism.
Fig. 6. In vitro assay for SidJ removal of SidE proteins from the LCV. 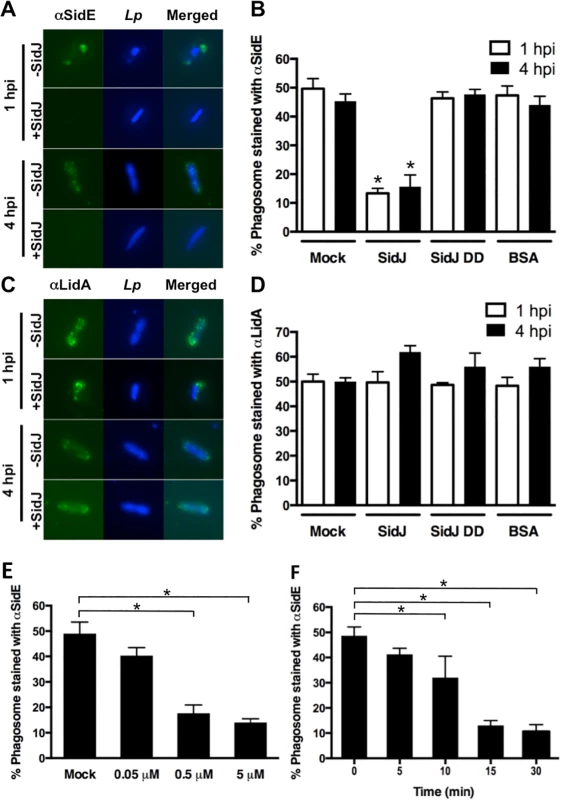
(A-D) BMMs were infected with a ΔsidJ mutant for the indicated time, lysed by gentle douncing, unbroken cells removed by centrifugation, and the PNS was incubated with 0.5 μM of purified SidJ, SidJ DD mutant or BSA. The reactions were stopped by addition of paraformaldehyde. The fixed cells were then stained with anti-SidE (A) or LidA (C) antibody. Representative images are shown with SidE and LidA in green and bacteria in blue. (B and D) Phagosomes were scored for co-localization with SidE proteins (B) or LidA (D). Removal of SidE proteins from the LCV is dependent on the concentration of SidJ (E) and the time of incubation (F). Data are means ± SEM of three independent experiments. Approximately 75 LCVs were counted for each reaction and asterisks indicate statistical difference among treatments in both B and D and in samples compared to the mock controls in both E and F, (P<0.05). The ability of SidJ to remove SidE proteins from the LCV in vitro using lysates prepared from either 1 hour or 4 hours infections indicates that their absence is not simply due to LCV maturation but rather is a direct consequence of SidJ action. In addition, concentration-dependence of the reaction is consistent with SidJ accumulation in the host cell cytoplasm at later time points of infection (Fig. 4). In summary, these data suggest that the SidE proteins function as toxins during early stages of infection and that SidJ inactivates them by mediating their active removal from the Legionella-containing vacuole.
Discussion
The pathogen L. pneumophila serves as an excellent model system to study the interactions between secreted effector proteins, as it exports between 200–300 substrates via its Dot/Icm T4SS [32]. In this study, we examined the relationship between the L. pneumophila Dot/Icm substrates SidJ and the SidE family. Six lines of evidence support a functional connection between these proteins. First, SidJ and SdeC, SdeB, and SdeA are all expressed from the same locus (Fig. 1). Second, the SuperΔP170 mutant, the CleanΔP170 mutant, and the ΔsidJ mutant each have a similar growth defect within host cells (Figs. 1 and S2). Third, expression of low levels of SdeA was able to complement/suppress the intracellular growth defect of the SuperΔP170 mutant, which lacks both the sidE family genes and sidJ (Fig. 1). Fourth, overexpression of SdeA was detrimental for the growth of only strains lacking sidJ (Fig. 1). Fifth, SdeA toxicity to yeast and mammalian host cells could be suppressed by co-expression of SidJ (Fig. 2). Sixth, SidJ is able to promote the disappearance of SidE proteins from the LCV at later points of infection (Fig. 3) and this could be reproduced in an in vitro assay using purified SidJ (Fig. 6). Based on these results, we propose that the L. pneumophila SidJ protein functions as a metaeffector to regulate the activity of the SidE protein family.
The concept that intracellular pathogens must regulate the activity of their secreted effectors during an infection is not surprising, as unregulated toxin activity would lead to the premature demise of the host cell. One method of regulation might entail the spatiotemporal delivery and/or control of substrates with opposing activities. For example, Salmonella’s SopE and SptP toxins act antagonistically to activate and inactivate Rho-family GTPases CDC42 and Rac1 at different times of infection via a combination of differential activity and temporal stability [19]. Likewise L. pneumophila regulates the activity of Rab1 by using a GEF (SidM/DrrA) and a GAP (LepB) [20–22]. In addition to this general mode of GTPase regulation, L. pneumophila is able to stabilize Rab1 in an active form using ampylation by the effector SidM/DrrA and then reverse the effect via de-ampylation by SidD [33–36]. Legionella also employs two additional effectors with opposing activities, AnkX and Lem3/Lpg0696, to inactivate and then release a separate population of Rab1-GDP via cholination [33–36].
An even more elegant form of effector regulation was recently described, wherein the effector SidH was inactivated by LubX, a L. pneumophila secreted E3 ubiquitin ligase that marks SidH for proteasome-dependent proteolysis by polyubiquitination [25]. The key to effector regulation in this case was the differential translocation of LubX and SidH into host cells, with SidH being rapidly secreted followed by the slower intracellular accumulation of LubX. Based on these results, LubX was described as being a “metaeffector”, which was defined as an effector that regulates another effector protein [25]. Reminiscent of the differential regulation and secretion described for SidH and LubX, the expression of SidE proteins is induced in early stationary phase allowing export to occur immediately upon host cell infection [16]. In contrast, SidJ is expressed constitutively [18] and accumulates within the host cell at later time points of infection (Fig. 4). The gradual accumulation of intracellular SidJ during infection correlates with the decreased level of the SidE proteins on the LCV.
These observations prompted us to propose a model whereby SidJ functions as a metaeffector to modulate the activity of the SidE proteins (Fig. 7). In this model, SidE proteins are translocated into the host cells by the L. pneumophila Dot/Icm T4SS at early points of infection and localize on the cytoplasmic face of the immature LCV. Although the precise molecular function of the SidE proteins is not yet known, their early delivery into the host cell suggests they are involved in avoidance of the endocytic pathway and/or maturation of the LCV. As the infection proceeds, the SidJ protein begins to accumulate in the host cell, eventually reaching a critical threshold when it is competent to mediate the removal of the SidE proteins from the LCV (Fig. 7). Based on the inhibition by MG132, the simplest possibility is that SidJ directly targets SidE proteins for degradation by the proteasome (Fig. 7, top row). Alternatively, it is possible that SidJ mediates the degradation of another component that normally retains SidE proteins on the LCV surface. In the absence of this factor, the SidE proteins would no longer associate with the LCV and thus could redistribute and potentially associate with host organelles (Fig. 7, middle row).
Fig. 7. Model showing SidJ-mediated removal of SidE proteins from the Legionella containing vacuole (LCV). 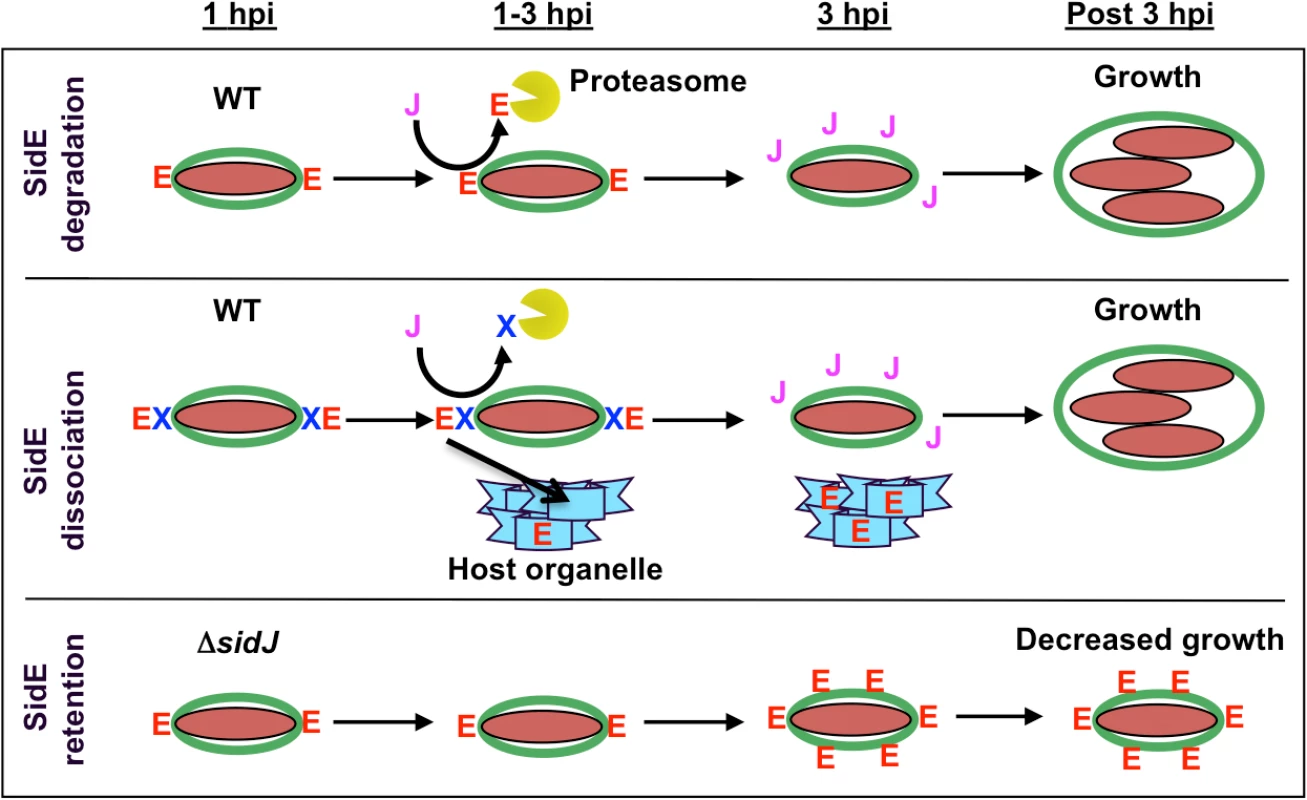
Shown is a time course for the initial hours of an infection by wild-type L. pneumophila and a ΔsidJ mutant. SidE proteins are indicated with a red letter E, SidJ protein with a purple letter J, a hypothetical protein necessary for retaining SidE proteins on the LCV with a blue letter X, the LCV membrane in green, L. pneumophila in maroon, the proteasome as a yellow Pac-man, and a host organelle in light blue. In a wild-type infection, SidE proteins localize to the LCV at early time points (1 hpi). At later time points, SidJ removes SidE proteins from the LCV either by localized degradation of SidE (top panel) and/or by degradation of a retention factor, thus leading to the relocalization of LCV-associated SidE proteins to a host organelle (middle panel). In a ΔsidJ infection, SidE proteins remain and accumulate on the LCV, which is detrimental for growth (lower panel). In the absence of SidJ, SidE proteins appear to localize normally to the LCV at early time points of infection (Fig. 7, bottom row). However, as the infection proceeds, the SidE proteins are no longer removed from the LCV, they accumulate to high levels, eventually inhibiting the growth of L. pneumophila. Overproduction of SdeA in the absence of SidJ was toxic to both yeast cells and HEK293 cells and inhibited the growth of L. pneumophila due to delivery of the LCV to the lysosome. The disruption of the Golgi in mCherry-SdeA transfected cells suggests that the target of SdeA is likely to be a component of the secretory pathway.
The failure to eliminate SidE proteins from the LCV in a ΔsidJ mutant does not appear to be due to an indirect effect of the LCV not maturing as we can induce removal of SidE proteins from 4-hour LCVs in vitro by the addition of wild type SidJ. Rather we prefer the idea that SidJ directly mediates removal of SidE proteins from the LCV, perhaps by some form of post-translational modification. Although we have been unable to reproducibly demonstrate a robust interaction between SidJ and SdeA, it is reasonable that the proteins interact based on the SidJ suppression of SdeA-mediated toxicity in yeast and HEK293 cells. It is also possible that SidJ, which is a large protein of ∼90 kDa, possesses multiple biochemical activities, particularly since a partial ER recruitment defect has been reported for a ΔsidJ mutant [18].
In summary, the Dot/Icm substrate SidJ functions as a metaeffector to regulate the activity of the SidE substrates. Similar to the metaeffector LubX, SidJ promotes the removal of Dot/Icm T4SS effectors from the LCV in a proteasome-dependent manner. The presence of dual effectors with opposing activities, and the existence of metaeffectors that modulate the activity of other effectors, may partially explain why L. pneumophila translocates such a vast repertoire of T4SS substrates into host cells. Moreover, the discovery of a second metaeffector in L. pneumophila suggests that the concept of metaeffectors is not unique to LubX and additional pathogens may use similar strategies to highjack host cells.
Materials and Methods
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Protocol 20120081 was approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine. All efforts were made to minimize suffering.
Bacterial strains, plasmids, media, and cell lines
Bacterial strains, plasmids, and primers are listed in S1 Table. Detailed plasmid construction is described in S2 Table. All L. pneumophila strains were cultured on ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered charcoal yeast extract agar (CYE) or in ACES-buffered yeast extract broth (AYE) [37]. Antibiotics and thymidine (100 μg/ml) were added as needed. Strain Lp02 (thyA hsdR rpsL) is a derivative of the clinical isolate L. pneumophila Philadelphia-1 [38]. E. coli strain XL1 Blue was grown in Luria-Bertani (LB) broth or on LB agar with antibiotics as needed. Yeast strains were grown in YPD medium or yeast minimal medium supplemented with amino acids as needed. A/J mice were obtained from Jackson Laboratories. Mouse bone marrow-derived macrophages (BMMs) were differentiated from stem cells isolated from the femurs of female A/J mice and cultured in RPMI-1640 containing 20% FBS, 1.6 mM glutamine, 30% L-cell culture medium, and penicillin (10,000 IU/ml)/streptomycin (10 mg/ml) for one week as previously described [6,39]. Acanthamoeba castellanii cultures were maintained in PYG broth as previously described [40]. HEK-293 cells (obtained from American Type Culture Collection, Manassas, VA) were maintained in DMEM supplemented with 10% heat-inactivated fetal calf serum (FBS) (HyClone, Logan, UT) in a humidified CO2 incubator at 5% CO2 concentration. Human monocytic cell line U937 [41] were cultured in RPMI-1640 supplemented with 10% FBS and 2 mM glutamine. To differentiate the cells, they were treated with phorbol 12-myristate 13-acetate (Sigma, St. Louis, MO) for 36 h before use.
Intracellular growth assay
Intracellular growth of L. pneumophila was assayed using A. castellanii as a host cell. A. castellanii was propagated using PYG medium. Cells were grown to near confluency, recovered, counted, and plated into 24-well culture dishes at a density of 6 x 105 per well. The following day, the cells were washed and equilibrated at 37°C for 1 hour in A. castellanii buffer [40,42–44]. The amoebae were infected with stationary phase L. pneumophila cells at a multiplicity of infection (MOI) of 0.2 for 1 hour, washed three times to remove extracellular bacteria, and incubated for two days. L. pneumophila growth was assayed at 0, 20, 32, and 44 hours. At each time point, infected amoebae were lysed with 0.05% saponin (Sigma, St. Louis, MO) in PBS, the lysate was serially diluted and plated on CYE plates to assess bacterial growth. All growth assays were performed in triplicate.
Expression of L. pneumophila proteins in yeast
Legionella sidJ and sdeA ORFs were cloned into the yeast expression vectors under control of the Pgal or Pcyc promoters (S1 and S2 Table). Plasmids were transformed into yeast cells (JY221, S1 Table) using the lithium acetate/PEG method [45]. Transformed cells were plated on yeast minimal media (US Biological, Massachusetts, MA), synthetic complete (SC) media lacking uracil (Ura) or leucine (Leu) in order to select for transformants. To determine the effect of SidJ or SdeA protein on yeast cell growth, strains were grown to saturation in SC minus Ura or SC minus Leu media containing 2% glucose. Cells were then adjusted to an A600 of 1.0, serially diluted 10-fold, and 5 μl of each dilution was spotted onto SC minus Ura or SC minus Leu containing either 2% glucose (non-induction) or 2% galactose (induction). Plates were incubated at 30° C for 48–72 hr and growth of recombinant strains was recorded.
Mammalian cell transfections and immunofluorescence
For transfection of YFP-SidJ and mCherry-SdeA, HEK293 cells were seeded onto glass coverslips in 24-well dishes, incubated for one day, then transfected with 0.2 μg plasmid DNA using FuGene6 (Invitrogen, Grand Island, NY) as described by the manufacturer. Transfections were allowed to proceed for 20–40 hr in DMEM supplemented with 10% FBS at 37°C with 5% CO2. Cells were then fixed with 4% paraformaldehyde for 20 min at room temperature, and fixed cells were analyzed by fluorescence microscopy. For the Golgi fragmentation assay, cells were permeabilized with 100% methanol for 10 sec, and then blocked for 10 min with 5% goat serum in PBS. Cells were then stained with anti-giantin antibody (1 : 400, Covance, Princeton, NJ), followed by Alexa blue-conjugated goat anti-rabbit IgG (Invitrogen, Grand Island, NY) as a secondary antibody. Coverslips were mounted using ProLong Gold antifade reagent (Invitrogen, Grand Island, NY) before examined by fluorescence microscopy.
Effector protein secretion assays using adenylate cyclase reporter
To quantitate effector protein secretion, we measured the adenylate cyclase activity of CyaA fusions (S1 Table). Differentiated U937 cells were plated into 24-well tissue culture plates at 2.5 x 106 per well. Legionella cultures, induced with IPTG at mid-log phase and grown two more hours to reach stationary phase, were harvested, washed, and diluted in RPMI-1640 supplemented with 10% FBS. 5 x 106 bacteria were added to each well for 1 hour, followed by washing three times with cold PBS to remove non-adherent cells, and lysis in 200 μl of lysis buffer (50 mM HCl and 0.1% Triton X-100) on ice. The lysates were transferred to 1.5 ml tubes, boiled for 5 min, and 12 μl of 0.5 M NaOH was added to neutralize the samples. cAMP was extracted using 2 volumes of 95% ethanol and collected after centrifugation at 12,000 g for 5 min to remove cell debris and then lyophilized. Total cAMP concentration was measured using an ELISA kit (GE Healthcare, Pittsburgh, PA).
Western blot analysis
Protein samples were collected and boiled for five minutes in Laemmli sample buffer and separated by SDS-PAGE gel electrophoresis, followed by transfer to PVDF membranes [46,47]. Membranes were blocked in BLOTTO (PBS containing 5% non-fat dry milk), washed with wash buffer (PBS containing 0.05% Tween 20) and incubated for 1 hour with antibody diluted in BLOTTO. Blots were washed with wash buffer followed by one hour incubation with secondary goat anti-rabbit antibody conjugated to horseradish peroxidase (Sigma, St. Louis, MO) diluted 1 : 10,000 in BLOTTO. Blots were subsequently washed with wash buffer prior to development using an ECL detection kit (GE Healthcare, Pittsburgh, PA) according to their protocol.
Immunofluoresence assay
Mouse BMM were seeded on glass coverslips at 1 x 105 cells in 24-well plates and incubated overnight. Legionella cells were grown to stationary phase in AYE, washed in sterile water, then adjusted to OD600 = 1.0. Cells were diluted in warmed RPMI-1640 and 5 x 105 bacteria were added to wells containing BMM attached to coverslip. BMMs were infected for 1 hour. After washing to remove uninfected bacteria, cells were fixed using Periodate-Lysine-Paraformaldehyde (PLP) [48] and then permeabilized with methanol for 10 seconds. For effector localization, cells were stained with the SidE family antibody that was raised against SdeC (1 : 1,000) [16], LidA (1 : 1,000) [31], or SidM (1 : 300) [49] antibodies followed by goat anti-rabbit secondary antibody conjugated to Oregon Green (1 : 1:1,000) (Molecular Probes, Eugene, OR). DNA (bacteria and host nuclei) was stained with propidium iodide (1 mg/ml, Invitrogen, Grand Island, NY). Coverslips were mounted using ProLong Gold antifade reagent (Invitrogen, Grand Island, NY) before being examined by fluorescence microscopy. Legionella containing vacuoles (LCVs) decorated with effectors were scored positive by the visual presence of foci of Oregon Green adjacent to bacterial-shaped propidium-iodide staining.
Fractionation of secreted SidJ and SidE family effectors
To detect the intracellular localization of SidJ and SdeA, differentiated U937 cells were plated at a density of 1 x 107 cells per well in a 6-well plate. The next day, U937 cells were infected for 1 hour with stationary phase cultures of L. pneumophila and washed three times with PBS to remove uninfected bacteria. Cells were harvested using a cell scraper, washed once with cold PBS and pelleted. To fractionate the lysates, harvested cells were dounced in cold PBS without digitonin. Unbroken cells were removed by centrifugation (3 min, 200 g) at 4°C. Pellets and supernatant were separated by centrifugation at 12,000 g for 10 min to collect the pellet and cytosolic fraction. To fractionate secreted effector proteins, cells were resuspended in lysis buffer (PBS containing 0.2% digitonin) and dounced. Unbroken cells were then removed by centrifugation (3 min, 200 g) at 4°C. The secreted effector proteins were collected from supernatant after removing cell pellets by centrifugation (10 min, 12,000 g). Samples were analyzed by Western blot with SidE, SidJ, DotF, LidA, and SidM specific antibodies as above.
Separation of Legionella containing vacuoles (LCV)
Separation of LCV by sucrose density gradient ultracentrifugation was performed as previously described [50]. Postnuclear supernatant (PNS) of infected cells was prepared as follows. Briefly, 1 x 107 differentiated U937 cells plated in 6-well plate were infected with stationary phase L. pneumophila at an MOI of 5. At the indicated time, the infected cells were suspended in 2 ml of homogenization buffer (20 mM HEPES pH 7.2, 250 mM sucrose, 0.5 mM EGTA) and were gently disrupted in a 7-ml dounce homogenizer. Unbroken cells and nuclei were pelleted by centrifugation at 4°C (3 min, 200 g). The PNS containing the L. pneumophila vacuoles were layered onto a 25–65% sucrose gradient and centrifuged at 100,000 g for 1 hour at 4°C. Fractions were collected from the bottom of the gradients and analyzed by SDS-PAGE followed by Western blotting. Separation of LCV from cell organelles was assessed by monitoring for the presence of DotF, a component of the Dot/Icm T4SS.
Protein purification and in vitro SidE family dissociation assay
The sidJ open reading frame was amplified and inserted into pQE-30 to express His-SidJ (S1 and S2 Table). E. coli strain XL1Blue, harboring the resulting plasmid, pJB5331, was used to purify His-tagged SidJ with Ni-NTA columns according to protocols suggested by the manufacturer (Qiagen, Valencia, CA). Dissociation of SidE proteins from PNS was performed as below. Briefly, 2 x 106 BMM cells plated in 6 well-plate were infected with stationary phase L. pneumophila at an MOI of 0.5, incubated for 1 hour, followed by washing with warm RPMI-1640 to remove uninfected bacteria. At the indicated time, the infected cells were suspended in two ml of homogenization buffer containing 250 mM sucrose in PBS and dounced with 3 strokes. PNS was collected by removing unbroken cells by centrifugation at 4°C (3 min, 200 g). Collected PNS were incubated with 0.5 μM of purified SidJ for 30 min at room temperature. The reaction was stopped by addition of equal volume of 4% paraformaldehyde in a stock PLP solution. The fixed cells were attached on lysine-coated glass slides and dissociation of SidE family from LCV was monitored by immunofluorescence detection using SidE antibody (1 : 1,000 dilution) or LidA (1 : 1,100 dilution), followed by goat anti-rabbit Oregon Green secondary antibody (Molecular Probes, Eugene, OR). DNA was stained with DAPI and coverslips were mounted using ProLong Gold antifade reagent (Invitrogen) before being examined by fluorescence microscopy. Legionella containing vacuoles (LCVs) decorated with effectors were scored positive by the visual presence of foci of Oregon Green adjacent to bacterial-shaped DAPI staining.
Statistical analysis
Statistical analysis was performed using the GLIMMIX procedure of SAS 9.4 (9.4 SAS Institute Inc.). Data are presented as means ± SEM from three independent experiments. Statistical significance was declared if P<0.05.
Supporting Information
Zdroje
1. Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, et al. (1977) Legionnaires' disease: description of an epidemic of pneumonia. N Engl J Med 297 : 1189–1197. 335244
2. McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, et al. (1977) Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med 297 : 1197–1203. 335245
3. Fields BS, Benson RF, Besser RE (2002) Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev 15 : 506–526. 12097254
4. Horwitz M (1983) Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med 158 : 1319–1331. 6619736
5. Kagan JC, Roy CR (2002) Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol 4 : 945–954. 12447391
6. Swanson MS, Isberg RR (1995) Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun 63 : 3609–3620. 7642298
7. Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR (2001) How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J Cell Sci 114 : 4637–4650. 11792828
8. Vogel JP, Andrews HL, Wong SK, Isberg RR (1998) Conjugative transfer by the virulence system of Legionella pneumophila. Science 279 : 873–876. 9452389
9. Segal G, Purcell M, Shuman HA (1998) Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Nat Acad Sci U S A 95 : 1669–1674. 9465074
10. Segal G, Feldman M, Zusman T (2005) The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol Rev 29 : 65–81. 15652976
11. Vincent CD, Friedman JR, Jeong KC, Buford EC, Miller JL, et al. (2006) Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol 62 : 1278–1291. 17040490
12. Vincent CD, Friedman JR, Jeong KC, Sutherland MC, Vogel JP (2012) Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol 85 : 378–391. doi: 10.1111/j.1365-2958.2012.08118.x 22694730
13. Dorer MS, Kirton D, Bader JS, Isberg RR (2006) RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog 2: e34. 16652170
14. O'Connor TJ, Adepoju Y, Boyd D, Isberg RR (2011) Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci U S A 108 : 14733–14740. doi: 10.1073/pnas.1111678108 21873199
15. O'Connor TJ, Boyd D, Dorer MS, Isberg RR (2012) Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science 338 : 1440–1444. doi: 10.1126/science.1229556 23239729
16. Bardill JP, Miller JL, Vogel JP (2005) IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol 56 : 90–103. 15773981
17. Luo ZQ, Isberg RR (2004) Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Nat Acad Sci U S A 101 : 841–846. 14715899
18. Liu Y, Luo ZQ (2007) The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun 75 : 592–603. 17101649
19. Kubori T, Galan JE (2003) Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. Cell 115 : 333–342. 14636560
20. Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, et al. (2006) The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol 8 : 971–977. 16906144
21. Machner MP, Isberg RR (2007) A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318 : 974–977. 17947549
22. Ingmundson A, Delprato A, Lambright DG, Roy CR (2007) Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450 : 365–369. 17952054
23. Creasey EA, Isberg RR (2012) The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc Natl Acad Sci U S A 109 : 3481–3486. doi: 10.1073/pnas.1121286109 22308473
24. Laguna RK, Creasey EA, Li Z, Valtz N, Isberg RR (2006) A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc Natl Acad Sci U S A 103 : 18745–18750. 17124169
25. Kubori T, Shinzawa N, Kanuka H, Nagai H (2010) Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog 6: e1001216. doi: 10.1371/journal.ppat.1001216 21151961
26. Glaser P, Ladant D, Sezer O, Pichot F, Ullmann A, et al. (1988) The calmodulin-sensitive adenylate cyclase of Bordetella pertussis: cloning and expression in Escherichia coli. Mol Microbiol 2 : 19–30. 2897067
27. Sory MP, Cornelis GR (1994) Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol 14 : 583–594. 7885236
28. Sutherland MC, Nguyen TL, Tseng V, Vogel JP (2012) The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Pathog 8: e1002910. doi: 10.1371/journal.ppat.1002910 23028312
29. Campodonico EM, Chesnel L, Roy CR (2005) A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol Microbiol 56 : 918–933. 15853880
30. Heidtman M, Chen EJ, Moy MY, Isberg RR (2009) Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol 11 : 230–248. doi: 10.1111/j.1462-5822.2008.01249.x 19016775
31. Conover GM, Derre I, Vogel JP, Isberg RR (2003) The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol 48 : 305–321. 12675793
32. Isaac DT, Isberg R (2014) Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future Microbiol 9 : 343–359. doi: 10.2217/fmb.13.162 24762308
33. Mukherjee S, Liu X, Arasaki K, McDonough J, Galan JE, et al. (2011) Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477 : 103–106. doi: 10.1038/nature10335 21822290
34. Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, et al. (2010) The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329 : 946–949. doi: 10.1126/science.1192276 20651120
35. Neunuebel MR, Chen Y, Gaspar AH, Backlund PS Jr., Yergey A, et al. (2011) De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333 : 453–456. doi: 10.1126/science.1207193 21680813
36. Tan Y, Luo ZQ (2011) Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475 : 506–509. doi: 10.1038/nature10307 21734656
37. Chatfield CH, Cianciotto NP (2013) Culturing, media, and handling of Legionella. Methods Mol Biol 954 : 151–162. doi: 10.1007/978-1-62703-161-5_7 23150393
38. Berger KH, Isberg RR (1993) Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7 : 7–19. 8382332
39. Sexton JA, Miller JL, Yoneda A, Kehl-Fie TE, Vogel JP (2004) Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect Immun 72 : 5983–5992. 15385502
40. Moffat JF, Tompkins LS (1992) A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect Immun 60 : 296–301. 1729191
41. Pearlman E, Jiwa AH, Engleberg NC, Eisenstein BI (1988) Growth of Legionella pneumophila in a human macrophage-like (U937) cell line. Microb Pathog 5 : 87–95. 3237054
42. Sexton JA, Pinkner JS, Roth R, Heuser JE, Hultgren SJ, et al. (2004) The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J Bacteriol 186 : 1658–1666. 14996796
43. Sexton JA, Vogel JP (2004) Regulation of hypercompetence in Legionella pneumophila. J Bacteriol 186 : 3814–3825. 15175295
44. Sexton JA, Yeo HJ, Vogel JP (2005) Genetic analysis of the Legionella pneumophila DotB ATPase reveals a role in type IV secretion system protein export. Mol Microbiol 57 : 70–84. 15948950
45. Sherman F (1991) Getting started with yeast. Methods Enzymol 194 : 3–21. 2005794
46. Vincent CD, Buscher BA, Friedman JR, Williams LA, Bardill P, et al. (2006) Identification of non-dot/icm suppressors of the Legionella pneumophila ΔdotL lethality phenotype. J Bacteriol 188 : 8231–8243. 16997951
47. Vincent CD, Vogel JP (2006) The Legionella pneumophila IcmS-LvgA protein complex is important for Dot/Icm-dependent intracellular growth. Mol Microbiol 61 : 596–613. 16803597
48. McLean IW, Nakane PK (1974) Periodate-lysine-paraformaldehyde fixative. A new fixation for immunoelectron microscopy. J Histochem Cytochem 22 : 1077–1083. 4374474
49. Machner MP, Isberg RR (2006) Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell 11 : 47–56. 16824952
50. Howe D, Heinzen RA (2008) Fractionation of the Coxiella burnetii parasitophorous vacuole. Methods Mol Biol 445 : 389–406. doi: 10.1007/978-1-59745-157-4_25 18425464
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole MembraneČlánek Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and FunctionČlánek Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 3- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- To Be or Not IIb: A Multi-Step Process for Epstein-Barr Virus Latency Establishment and Consequences for B Cell Tumorigenesis
- Is Antigenic Sin Always “Original?” Re-examining the Evidence Regarding Circulation of a Human H1 Influenza Virus Immediately Prior to the 1918 Spanish Flu
- The Great Escape: Pathogen Versus Host
- Coping with Stress and the Emergence of Multidrug Resistance in Fungi
- Catch Me If You Can: The Link between Autophagy and Viruses
- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- Evidence for Ubiquitin-Regulated Nuclear and Subnuclear Trafficking among Matrix Proteins
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Production of Anti-LPS IgM by B1a B Cells Depends on IL-1β and Is Protective against Lung Infection with LVS
- Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS
- α-Hemolysin Counteracts the Anti-Virulence Innate Immune Response Triggered by the Rho GTPase Activating Toxin CNF1 during Bacteremia
- Induction of Interferon-Stimulated Genes by IRF3 Promotes Replication of
- Intracellular Growth Is Dependent on Tyrosine Catabolism in the Dimorphic Fungal Pathogen
- HCV Induces the Expression of Rubicon and UVRAG to Temporally Regulate the Maturation of Autophagosomes and Viral Replication
- Spatiotemporal Analysis of Hepatitis C Virus Infection
- Subgingival Microbial Communities in Leukocyte Adhesion Deficiency and Their Relationship with Local Immunopathology
- Interaction between the Type III Effector VopO and GEF-H1 Activates the RhoA-ROCK Pathway
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
- Establishment of HSV1 Latency in Immunodeficient Mice Facilitates Efficient Reactivation
- XRN1 Stalling in the 5’ UTR of Hepatitis C Virus and Bovine Viral Diarrhea Virus Is Associated with Dysregulated Host mRNA Stability
- γδ T Cells Confer Protection against Murine Cytomegalovirus (MCMV)
- Rhadinovirus Host Entry by Co-operative Infection
- A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole Membrane
- Dermal Neutrophil, Macrophage and Dendritic Cell Responses to Transmitted by Fleas
- Elucidation of Sigma Factor-Associated Networks in Reveals a Modular Architecture with Limited and Function-Specific Crosstalk
- A Conserved NS3 Surface Patch Orchestrates NS2 Protease Stimulation, NS5A Hyperphosphorylation and HCV Genome Replication
- Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and Function
- Disruption of IL-21 Signaling Affects T Cell-B Cell Interactions and Abrogates Protective Humoral Immunity to Malaria
- Compartmentalized Replication of R5 T Cell-Tropic HIV-1 in the Central Nervous System Early in the Course of Infection
- Diminished Reovirus Capsid Stability Alters Disease Pathogenesis and Littermate Transmission
- Characterization of CD8 T Cell Differentiation following SIVΔnef Vaccination by Transcription Factor Expression Profiling
- Visualization of HIV-1 Interactions with Penile and Foreskin Epithelia: Clues for Female-to-Male HIV Transmission
- Sensing Cytosolic RpsL by Macrophages Induces Lysosomal Cell Death and Termination of Bacterial Infection
- PKCη/Rdx-driven Phosphorylation of PDK1: A Novel Mechanism Promoting Cancer Cell Survival and Permissiveness for Parvovirus-induced Lysis
- Metalloprotease NleC Suppresses Host NF-κB/Inflammatory Responses by Cleaving p65 and Interfering with the p65/RPS3 Interaction
- Immune Antibodies and Helminth Products Drive CXCR2-Dependent Macrophage-Myofibroblast Crosstalk to Promote Intestinal Repair
- Adenovirus Entry From the Apical Surface of Polarized Epithelia Is Facilitated by the Host Innate Immune Response
- The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family
- Neutrophils: Between Host Defence, Immune Modulation, and Tissue Injury
- CD169-Mediated Trafficking of HIV to Plasma Membrane Invaginations in Dendritic Cells Attenuates Efficacy of Anti-gp120 Broadly Neutralizing Antibodies
- Japanese Encephalitis Virus Nonstructural Protein NS5 Interacts with Mitochondrial Trifunctional Protein and Impairs Fatty Acid β-Oxidation
- Yip1A, a Novel Host Factor for the Activation of the IRE1 Pathway of the Unfolded Protein Response during Infection
- TRIM26 Negatively Regulates Interferon-β Production and Antiviral Response through Polyubiquitination and Degradation of Nuclear IRF3
- Parallel Epigenomic and Transcriptomic Responses to Viral Infection in Honey Bees ()
- A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication
- Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
- Exome and Transcriptome Sequencing of Identifies a Locus That Confers Resistance to and Alters the Immune Response
- The Role of Misshapen NCK-related kinase (MINK), a Novel Ste20 Family Kinase, in the IRES-Mediated Protein Translation of Human Enterovirus 71
- Chitin Recognition via Chitotriosidase Promotes Pathologic Type-2 Helper T Cell Responses to Cryptococcal Infection
- Activates Both IL-1β and IL-1 Receptor Antagonist to Modulate Lung Inflammation during Pneumonic Plague
- Persistence of Transmitted HIV-1 Drug Resistance Mutations Associated with Fitness Costs and Viral Genetic Backgrounds
- An 18 kDa Scaffold Protein Is Critical for Biofilm Formation
- Early Virological and Immunological Events in Asymptomatic Epstein-Barr Virus Infection in African Children
- Human CD8 T-cells Recognizing Peptides from () Presented by HLA-E Have an Unorthodox Th2-like, Multifunctional, Inhibitory Phenotype and Represent a Novel Human T-cell Subset
- Decreased HIV-Specific T-Regulatory Responses Are Associated with Effective DC-Vaccine Induced Immunity
- RSV Vaccine-Enhanced Disease Is Orchestrated by the Combined Actions of Distinct CD4 T Cell Subsets
- Concerted Activity of IgG1 Antibodies and IL-4/IL-25-Dependent Effector Cells Trap Helminth Larvae in the Tissues following Vaccination with Defined Secreted Antigens, Providing Sterile Immunity to Challenge Infection
- Structure of the Low pH Conformation of Chandipura Virus G Reveals Important Features in the Evolution of the Vesiculovirus Glycoprotein
- PPM1A Regulates Antiviral Signaling by Antagonizing TBK1-Mediated STING Phosphorylation and Aggregation
- Lipidomic Analysis Links Mycobactin Synthase K to Iron Uptake and Virulence in .
- Roles and Programming of Arabidopsis ARGONAUTE Proteins during Infection
- Impact of Infection on Host Macrophage Nuclear Physiology and Nucleopore Complex Integrity
- The Impact of Host Diet on Titer in
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Herpesviral G Protein-Coupled Receptors Activate NFAT to Induce Tumor Formation via Inhibiting the SERCA Calcium ATPase
- The Causes and Consequences of Changes in Virulence following Pathogen Host Shifts
- Small GTPase Rab21 Mediates Fibronectin Induced Actin Reorganization in : Implications in Pathogen Invasion
- Positive Role of Promyelocytic Leukemia Protein in Type I Interferon Response and Its Regulation by Human Cytomegalovirus
- NEDDylation Is Essential for Kaposi’s Sarcoma-Associated Herpesvirus Latency and Lytic Reactivation and Represents a Novel Anti-KSHV Target
- β-HPV 5 and 8 E6 Disrupt Homology Dependent Double Strand Break Repair by Attenuating BRCA1 and BRCA2 Expression and Foci Formation
- An O Antigen Capsule Modulates Bacterial Pathogenesis in
- Variable Processing and Cross-presentation of HIV by Dendritic Cells and Macrophages Shapes CTL Immunodominance and Immune Escape
- Probing the Metabolic Network in Bloodstream-Form Using Untargeted Metabolomics with Stable Isotope Labelled Glucose
- Adhesive Fiber Stratification in Uropathogenic Biofilms Unveils Oxygen-Mediated Control of Type 1 Pili
- Vaccinia Virus Protein Complex F12/E2 Interacts with Kinesin Light Chain Isoform 2 to Engage the Kinesin-1 Motor Complex
- Modulates Host Macrophage Mitochondrial Metabolism by Hijacking the SIRT1-AMPK Axis
- Human T-Cell Leukemia Virus Type 1 (HTLV-1) Tax Requires CADM1/TSLC1 for Inactivation of the NF-κB Inhibitor A20 and Constitutive NF-κB Signaling
- Suppression of RNAi by dsRNA-Degrading RNaseIII Enzymes of Viruses in Animals and Plants
- Spatiotemporal Regulation of a T4SS Substrate by the Metaeffector SidJ
- Antigenic Properties of the Human Immunodeficiency Virus Envelope Glycoprotein Gp120 on Virions Bound to Target Cells
- Dependence of Intracellular and Exosomal microRNAs on Viral Oncogene Expression in HPV-positive Tumor Cells
- Identification of a Peptide-Pheromone that Enhances Escape from Host Cell Vacuoles
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Dihydrobiopterin in Adult Falciparum Malaria: Association with Disease Severity, Impaired Microvascular Function and Increased Endothelial Activation
- Transgenic Expression of the Dicotyledonous Pattern Recognition Receptor EFR in Rice Leads to Ligand-Dependent Activation of Defense Responses
- Comprehensive Antigenic Map of a Cleaved Soluble HIV-1 Envelope Trimer
- Low Doses of Imatinib Induce Myelopoiesis and Enhance Host Anti-microbial Immunity
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Oxidized Biopterins in Pediatric Falciparum Malaria: Association with Disease Severity
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



