-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCompartmentalized Replication of R5 T Cell-Tropic HIV-1 in the Central Nervous System Early in the Course of Infection
Early HIV-1 CNS replication likely provides a foundation for brain injury and a potentially important tissue reservoir. To explore the character and timing of emergence of early HIV-1 CNS replication, we examined paired cerebrospinal fluid (CSF) and blood samples from 72 ART-naïve adults, with one-half having longitudinal samples, during the first two years following HIV-1 subtype B infection. In a cross sectional analysis over the first two years of infection, 10–25% of subjects had evidence of either local viral replication in the CNS, defined by the presence of CSF compartmentalization, or a robust inflammatory response, and in approximately 16% of subjects this CNS involvement persisted over time. In some subjects, one of two transmitted viruses replicated predominantly within the CNS, providing insight into how HIV-1 can establish independently replicating populations early in different parts of the body. Based on their entry phenotype, all viruses were selected for replication in CD4+ T cells, although this phenotype was slightly altered in the compartmentalized virus. Overall, we suggest four states to model the nature of HIV-1 CNS infection, which imply distinct mechanisms of virus/host interaction within the CNS during early infection.
Published in the journal: . PLoS Pathog 11(3): e32767. doi:10.1371/journal.ppat.1004720
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004720Summary
Early HIV-1 CNS replication likely provides a foundation for brain injury and a potentially important tissue reservoir. To explore the character and timing of emergence of early HIV-1 CNS replication, we examined paired cerebrospinal fluid (CSF) and blood samples from 72 ART-naïve adults, with one-half having longitudinal samples, during the first two years following HIV-1 subtype B infection. In a cross sectional analysis over the first two years of infection, 10–25% of subjects had evidence of either local viral replication in the CNS, defined by the presence of CSF compartmentalization, or a robust inflammatory response, and in approximately 16% of subjects this CNS involvement persisted over time. In some subjects, one of two transmitted viruses replicated predominantly within the CNS, providing insight into how HIV-1 can establish independently replicating populations early in different parts of the body. Based on their entry phenotype, all viruses were selected for replication in CD4+ T cells, although this phenotype was slightly altered in the compartmentalized virus. Overall, we suggest four states to model the nature of HIV-1 CNS infection, which imply distinct mechanisms of virus/host interaction within the CNS during early infection.
Introduction
While HIV-1 can be detected in both the cerebrospinal fluid (CSF) and brain tissue during the weeks after initial exposure [1–7], it is unknown when the virus actually begins replicating independently in the central nervous system (CNS). Independent viral replication within the CNS has two important implications. First, HIV-1 replication can lead to CNS dysfunction and injury, and while combination antiretroviral therapy (cART) has markedly reduced the incidence of HIV-associated dementia (HAD), the prevalence of milder HIV-associated neurological disorders (HAND) has increased [8,9] in the cART era. Second, independent CNS replication may also provide a reservoir distinct from that found in CD4+ T cells in the blood and lymphoid tissue. We do not know the time course of the virologic events that lead to neurological dysfunction and the potential establishment of a CNS reservoir, or the extent to which these long-term outcomes are predicted by the initial aspects of virus-host interaction.
While extensive independent, or compartmentalized, CSF/CNS replication is associated with severe HIV-1 clinical CNS dysfunction [1,10–13], genetically distinct virus can be detected in the CNS throughout the course of infection [4,10]. Thus far, two types of compartmentalization have been defined: one in which a few variants are rapidly expanded giving a CSF viral population of low complexity (clonal amplification) consisting of variants that require high levels of CD4 for entry (R5 T cell-tropic). The second type is characterized by a complex CSF viral population consisting of variants that can enter cells expressing low levels of CD4 (macrophage-tropic), indicative of a more prolonged period of isolated replication and evolution of the entry phenotype. Additionally, we have recently shown that after vertical transmission to children, CNS compartmentalization can be established via two mechanisms: the early sequestration of one of multiple transmitted variants in the CNS, or the later establishment of compartmentalized CNS virus originating from the periphery [14]. In a previous study, we demonstrated CSF HIV-1 compartmentalization in human subjects during the first year of HIV-1 infection in adults [4]. However, that study only examined viral population sequences from CSF and plasma in a small number of subjects (eight), limiting the generalizability of our findings. Furthermore, the previous study had sparse assessment of longitudinal samples, and included no analysis of sources of compartmentalized HIV-1 within the CSF.
For the current study, we used single genome amplification (SGA) and phylogenetic analysis to assess viral populations in the CSF in the presence and absence of cellular inflammation (i.e. pleocytosis) in cross-sectional and often longitudinal paired blood plasma and CSF samples obtained during the first two years of HIV-infection in ART-naïve subjects. We also extended our analytical approach to include Bayesian Evolutionary Analysis by Sampling Trees (BEAST) to assess time to most recent common ancestor (TMRCA) of CSF and blood HIV-1 populations [15], providing new insights on the timing of establishment of early compartmentalized populations, and we assessed the entry phenotypes of selected clones, further confirming the nature of the transmitted virus as R5 T cell-tropic [16]. Based upon a complex interplay between HIV-1 RNA concentration, viral compartmentalization, and CSF white blood cell (WBC) count, we suggest at least four different patterns to characterize the relationship between virus in the blood and virus in the CSF/CNS during the early period after infection. The current study considerably enhances the generalizability of our results and provides an unprecedented view of the early interactions of HIV-1 in the CNS.
Results
Study population and analysis parameters
We analyzed the HIV-1 RNA concentrations in paired blood plasma and CSF samples collected from 72 adult subjects enrolled in an observational neurological study of primary HIV-1 infection, defined as within one year of initial infection. All subjects were infected with HIV-1 subtype B and were ART-naïve at all study intervals, except for one subject, 9018, who was treated with tenofovir, emtricitabine, and atazanavir between the first and second analyzed time points. Paired follow-up samples were assessed for 37 subjects with longitudinal samples available within the initial two years post infection (p.i.). In total, 144 paired samples were available for analysis. Baseline demographic and clinical characteristics at enrollment for the entire cohort (n = 72) and for the subset that had sufficient CSF viral RNA concentrations (defined as greater than 1,000 copies of viral RNA/ml) to allow adequate sampling of the viral population via SGA (n = 33) are shown in Table 1. For the 33 subjects whose samples were analyzed by SGA, a total of 55 blood plasma/CSF sample pairs were analyzed including the longitudinal samples (Table 2; three time points for subject 9018 and one time point for subject 9040 beyond 2 years p.i. were analyzed but were not included in any overall population analysis.)
Tab. 1. Background demographic and clinical characteristics of study participants at baseline. 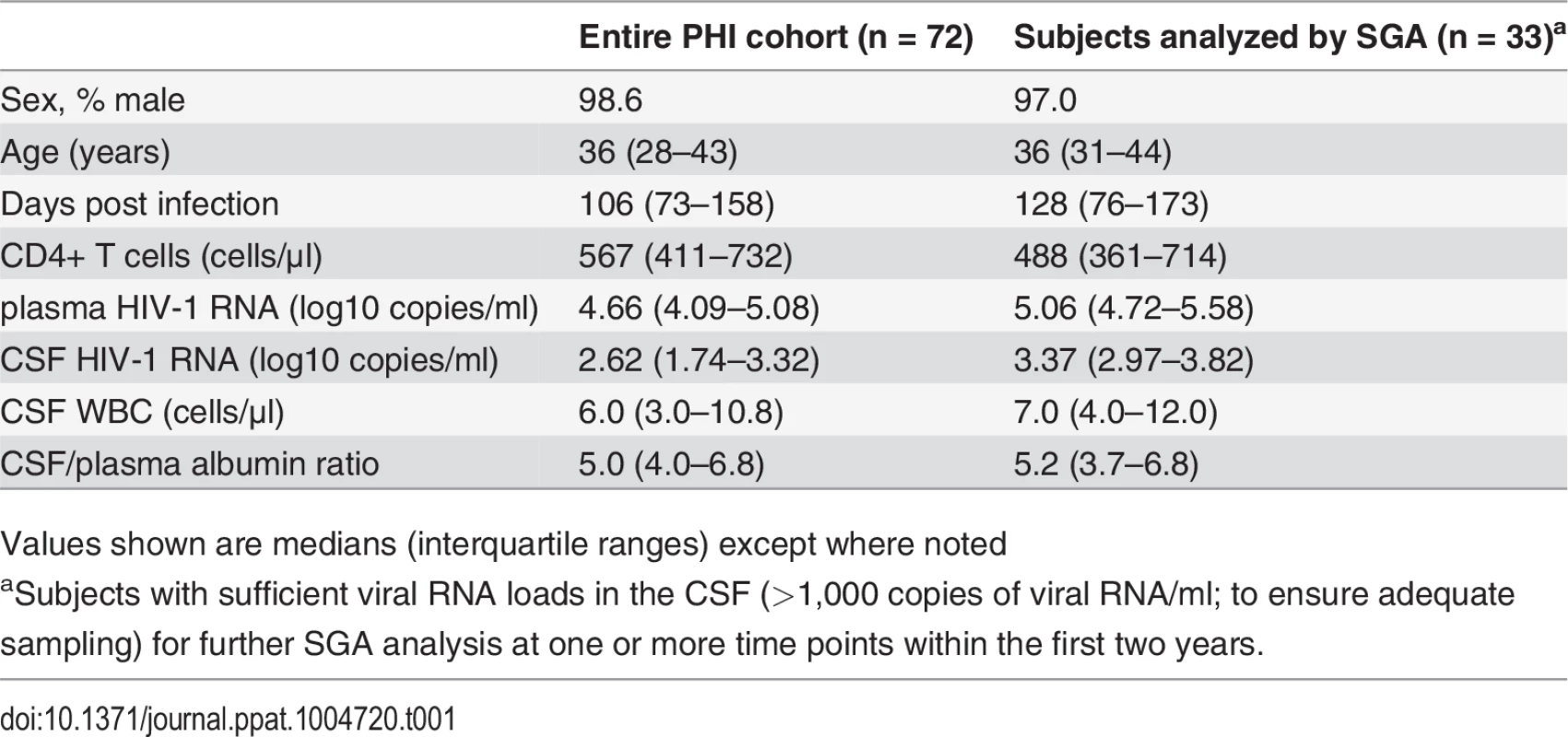
Values shown are medians (interquartile ranges) except where noted Tab. 2. Subject population virologic, clinical and phylogenetic characteristics. 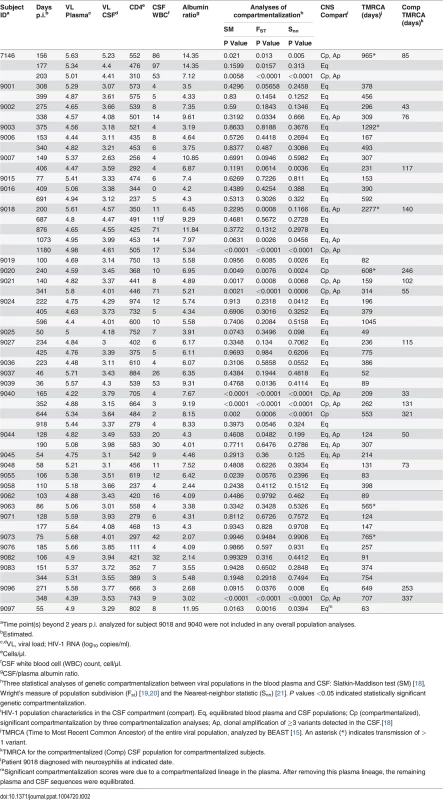
aTime point(s) beyond 2 years p.i. analyzed for subject 9018 and 9040 were not included in any overall population analyses. Following SGA and phylogenetic analysis, compartmentalization was assessed by three approaches. The choice to use multiple approaches was based on the recent findings of Zarate et al. [17] illustrating that different methods of assessing compartmentalization often yield divergent results. Thus, we assessed CNS compartmentalization using three methods—the tree-based Slatkin-Maddison test (SM) [18] and two distance-based methods, Wright’s measure of population subdivision (Fst) [19,20] and the Nearest-neighbor statistic (Snn) [21]. CNS lineages were interpreted as being significantly compartmentalized if all three tests were significant (P values < 0.05). We define CNS phylogenetic states as (Fig. 1): i) equilibrated, with similar populations between the blood and CSF, with no evidence of independent CNS replication (Fig. 1A); and ii) compartmentalized, with a genetically distinct CSF population as indicated by three statistically significant measures of compartmentalization (Fig. 1B). In our analyses, compartmentalized populations typically included clonally amplified variants often with more genetically diverse variants (Fig. 1B, Sub. 9040), and sometimes with the presence of recombinants between two clonally amplified variants (Fig. 1B, Sub. 9096). In order to determine how often our statistical assessment of CNS compartmentalization was due to the presence of clonally amplified viruses, we performed additional analyses in which we collapsed clonally amplified sequences into a single sequence and repeated the tests of compartmentalization. We determined that of the eight sample pairs that displayed both clonal amplification and CNS compartmentalization, three were significantly compartmentalized after collapsing the clonal sequences. This suggests that clonal amplification may drive the statistical assessment of compartmentalization or may be a symptom of ongoing CNS replication. Under this latter scenario, we propose that ongoing viral replication in the CNS may produce diverse, CNS-specific lineages and trigger an influx of T cells that amplify some of these lineages. Table 2 shows the clinical and virologic assessment for each subject who had at least one time point analyzed by SGA, and S1 Table shows this information for the 39 subjects who had no time points analyzed by SGA.
Fig. 1. HIV-1 blood and CSF populations early during infection can be equilibrated or compartmentalized. 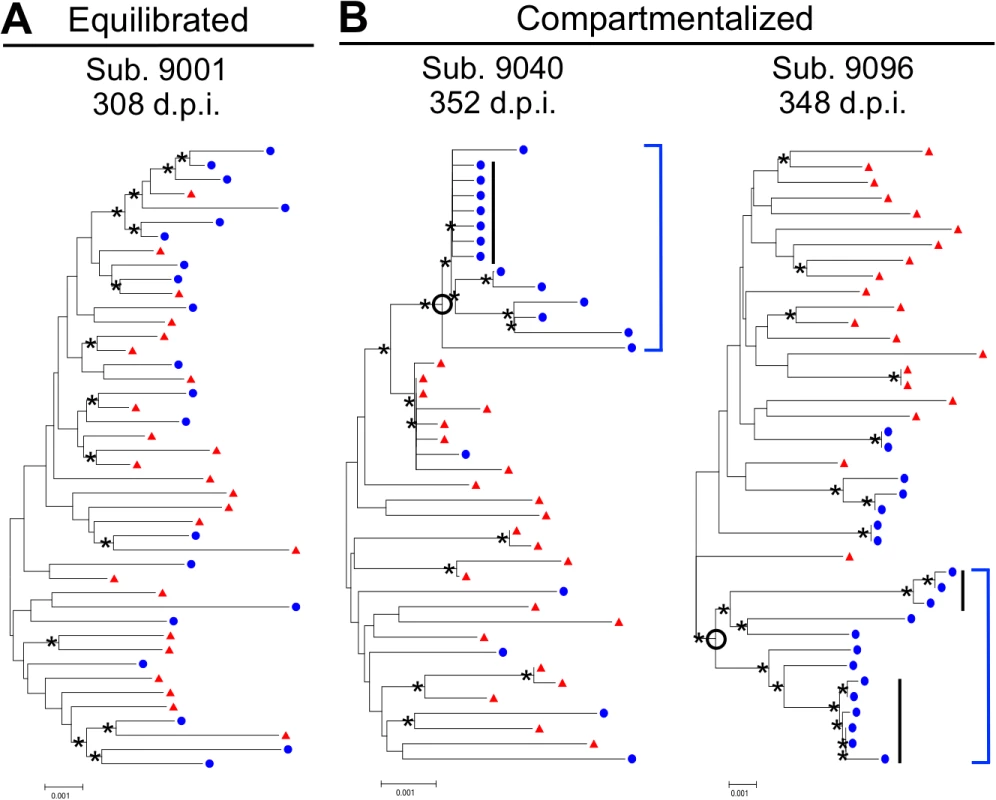
Neighbor-joining phylogenetic trees depicting sequence relationships from subjects with (A) equilibrated and (B) compartmentalized viral populations. env sequences from the CSF (blue circles) and blood plasma (red triangles) are shown. Bootstrap values ≥35 are included (*) at the appropriate nodes. Genetic distance is indicated at the bottom of each figure (0.001, number of nucleotide substitutions per site between env sequences). Compartmentalized CNS populations are designated by an open circle at the appropriate node and a solid blue bracket. Clonal amplification is indicated by a solid black bar. We also considered what role cellular inflammation (pleocytosis) might play in determining both the HIV-1 RNA concentrations and the nature of the viral population in the CSF, specifically whether an equilibrated population might exist with higher CSF HIV-1 RNA concentrations due to an influx of cells, including infected cells, during an inflammatory response. For our analyses, we chose a cut-off of 10 WBC/μl to define a state of CSF pleocytosis, as this is two-times the published upper limit of normal values of CSF WBC (5 cells/μl) [22] ensuring that the measured pleocytosis was a robust marker for an inflammatory response.
Approximately two-thirds of subjects have low CSF viral RNA concentrations during the first two years of infection
In order to assess temporal patterns of CNS viral replication and inflammation, we treated all 144 paired samples from the 72 subjects as independent observations (a limitation of the analysis but one that allowed us to categorize the samples by time post infection). We divided the samples into the following three windows: acute, 0–4 months p.i.; early, 5–12 months p.i.; or established, 13–24 months p.i.. The choice of these times also allowed us to bin the data into groups with reasonable sample sizes. Regardless of length of time since HIV-1 infection, the CSF viral RNA concentration was less than 1,000 copies/ml in approximately two-thirds of samples (Fig. 2A), suggestive of little local production of virus in the CNS, at least as assessed by viral load. Analysis of the viral populations in the remaining samples showed that in approximately 30% of the paired samples the sequence composition of the viral population in the CSF was similar to the viral population in the blood (i.e. equilibrated), and pleocytosis was detected in the CSF of one-quarter to one-half of the samples with these equilibrated populations (Fig. 2A). Compartmentalized viral populations were also detected but exclusively after the first 4 months, in 11% of samples in the early group (total n = 70) and 3% of the established group (total n = 30). However, those samples in which pleocytosis was detected were significantly less likely to have viral RNA levels in the CSF of less than 1,000 copies/ml compared to the total group of samples (see below). The percentages of samples in each phylogenetic group with pleocytosis (Fig. 2B) and without pleocytosis are noted (S1 Fig.).
Fig. 2. Correlates of CSF HIV-1 RNA levels, CNS inflammation, and CNS phylogenetic state. 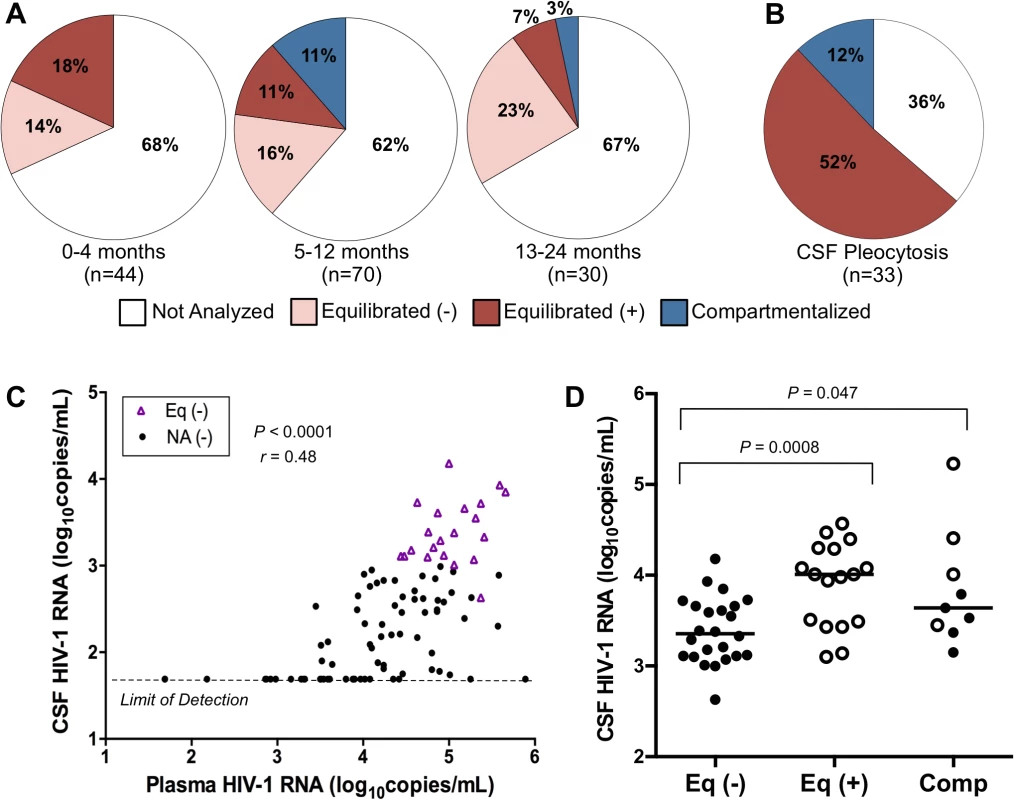
(A) Pie charts grouping the samples by days p.i. and showing the percentage of samples in each phylogenetic state as a function of time: 0–4 months (‘acute’), 5–12 months (‘early’), and 13–24 (‘established’). States represented include: Not Analyzed by SGA, due to CSF viral load <1,000 copies/ml; Equilibrated (−), CSF WBC <10 cells/μl; Equilibrated (+), CSF WBC ≥10 cells/μl; and Compartmentalized. We used a cut-off of 10 WBC/μl to define a state of substantial CSF pleocytosis because this is two-times the upper limit of normal value of CSF WBC (0 to 5 cells/μl) [22] ensuring that the measured pleocytosis was a robust marker for an inflammatory response. (B) Pie chart showing the percent of samples in each phylogenetic state exhibiting substantial CSF pleocytosis (CSF WBC ≥10 cells/μl). (C) Relationship between plasma and CSF HIV-1 RNA concentrations for not analyzed samples with minimal pleocytosis (NA (−); CSF WBC <10 cells/μl) and equilibrated samples with minimal pleocytosis (Eq (−), CSF WBC <10 cells/μl). Spearman’s rank correlation coefficient and P-value are indicated. Samples with undetectable CSF HIV-1 RNA (<50 copies/ml; limit of detection) were not included in the correlation analysis. (D) Relationship between CSF viral loads for equilibrated with minimal pleocytosis (Eq (−); CSF WBC <10 cells/μl); equilibrated with marked pleocytosis (Eq (+); CSF WBC ≥10 cells/μl); and compartmentalized (Comp) samples. Significant P values (Mann-Whitney Test) comparing relationships between groups are indicated. Samples with evidence of marked pleocytosis (CSF WBC ≥10 cells/μl) are indicated using open circles, while samples with minimal to no pleocytosis are indicated by solid circles. We next assessed factors associated with the CSF viral load. We first examined subjects without evidence of inflammation (i.e. no pleocytosis) and either low viral load in the CSF (less than 1,000 copies/ml) or an equilibrated population as evidence of no sustained local replication. In these subjects the HIV-1 RNA concentration in the CSF was proportionally 1–2% of the level in the blood (Spearman’s rank correlation coefficient = 0.48, P<0.0001) (Fig. 2C). We could not determine whether this proportional relationship was maintained for those samples in which the CSF viral load was undetectable (<50 copies/ml), although there was a trend for these samples to be from subjects with low plasma HIV-1 RNA concentrations (Fig. 2C). We hypothesize that this low level of virus in the CSF is due to the normal trafficking of T cells into the CNS, including infected CD4+ T cells that release the observed virus.
Pleocytosis can be associated with elevated viral load in the CSF and an influx of virus from the blood
Pleocytosis was detected in 36% of all subjects (Table 2 and S1). Consistent with our prior findings in HIV-infected subjects, lymphocytes were the predominant cell type in subjects with and without elevated levels of cellular inflammation [23]. When pleocytosis was detected, the HIV-1 RNA concentration was twice as likely (64%) to be above 1,000 copies/ml in the CSF compared to the overall population of which two-thirds were below 1,000 copies/ml. Additionally, when pleocytosis was present and the CSF viral load was high enough for SGA analysis, the viral populations were most often equilibrated (Fig. 2D, open circles), with a CSF viral load that was significantly higher than for equilibrated populations without pleocytosis (P = 0.0008, Mann-Whitney Test) (Fig. 2D). This suggests that the influx of infected CD4+ T cells associated with pleocytosis brings virus into the CSF/CNS from the blood. There was a trend toward increased CSF:blood albumin ratio (a marker for reduced blood-brain barrier integrity) in the presence of pleocytosis, which could contribute to an influx of virus (Table 2). However, there was a similar (and statistically significant) increase in CSF:blood albumin ratio in the compartmentalized subjects that did not account for the increase in CSF HIV-1 RNA concentration compared to the samples with equilibrated populations without pleocytosis (Fig. 2D; see below), which was instead due to local virus production. These data suggest that increased viral burden in the CNS can result from two factors: independent CNS replication, generating compartmentalized CSF populations, or an influx of infected cells as the result of the inflammatory response of pleocytosis, producing a viral population in the CSF that is similar to that in the blood. It is possible that much of the pleocytosis seen in these subjects is in response to HIV-1 replication in the CNS where the influx of infected cells from the blood produces elevated levels of virus that obscure the detection of a lower level of the locally produced and compartmentalized virus responsible for inducing the inflammation. Other agents that induce pleocytosis should have a similar effect on viral load in the CSF and this was seen in an incident of neurosyphilis in subject 9018 when sampled at day 687 p.i. (Table 2). However, we note that pleocytosis is not always associated with higher viral load (Table 2 and S1), indicating a more complex and perhaps dynamic relationship between pleocytosis and viral load.
Compartmentalized viral populations early are associated with higher CSF viral loads, and local replication and/or inflammation can persist or reoccur over time
Little evidence for local CNS replication was detected until after 4 months of infection (Fig. 2A), suggesting that detectable CNS-compartmentalized populations are present at greater frequency with a longer time since HIV-1 exposure. As noted above, CSF samples with compartmentalized populations had elevated viral RNA loads when compared to the samples with only equilibrated populations in the absence of pleocytosis (Fig. 2D), consistent with local production of virus with the detection of compartmentalization.
We used cross sectional analyses to examine when these markers of local replication and inflammation were observed. We estimated the percentage of samples with no evidence of viral involvement in the CNS from the number with a CSF viral RNA concentration less than 1,000 copies/ml (Fig. 2A; unanalyzed samples) and the number with equilibrated populations in their CSF and little to no pleocytosis (Fig. 2A; equilibrated (−)). In contrast, we estimated the percentage of samples with evidence of viral involvement in the CNS from the number with equilibrated populations in their CSF and pleocytosis (Fig. 2A; equilibrated (+)) and the number with compartmentalized populations in their CSF (Fig. 2A; compartmentalized). In these analyses we interpret equilibrated populations with pleocytosis/inflammation as an indirect marker of local viral replication, i.e. the inflammatory response to local viral replication. Based on these analyses we determined that the vast majority of samples collected during the three time periods (0–4, 5–12 and 12–24 months p.i.) had no evidence of CNS involvement (82, 78 and 90%, respectively), while the remaining samples (18, 22 and 10%, respectively) had evidence of CNS involvement. However, if viral replication and its associated markers (e.g. pleocytosis) are transient or produce small signals, these values will underestimate the actual proportion of subjects with viral replication in their CNS.
We used a longitudinal analysis to evaluate the persistence of viral replication in the CNS. Of the 37 subjects with longitudinal sampling, the majority (17 of 37) had low CSF viral loads at all sampling time points and were not analyzed by SGA (S1 Table). If we focus on the 20 subjects that had higher CSF viral loads (>1,000 copies of HIV-1 RNA/ml) and were analyzed by SGA, we observe that 9 (45%) had equilibrated populations with minimal to no pleocytosis at all time points analyzed (Fig. 3A). The remaining 11 subjects (55% or 30% of all longitudinally sampled subjects) all showed evidence of pleocytosis and/or compartmentalized viral replication at one or more time points within the initial two-year period (Fig. 3B) and 6 (30% or 16% of all longitudinally sampled subjects) had one of these states at two or more time points (Fig. 3B, subjects below dotted line). Thus, 55% of longitudinally sampled subjects analyzed by SGA showed evidence of viral replication and/or marked pleocytosis in the CNS at one or more time points, and 30% showed evidence of persistent viral replication in the CNS. Finally, if the initial sample showed evidence of compartmentalization or an equilibrated population with pleocytosis then subsequent samples were significantly more likely to also be compartmentalized or have pleocytosis than if the initial sample was not in one of these states (P = 0.001).
Fig. 3. An assessment of the viral populations in the two compartments longitudinally. 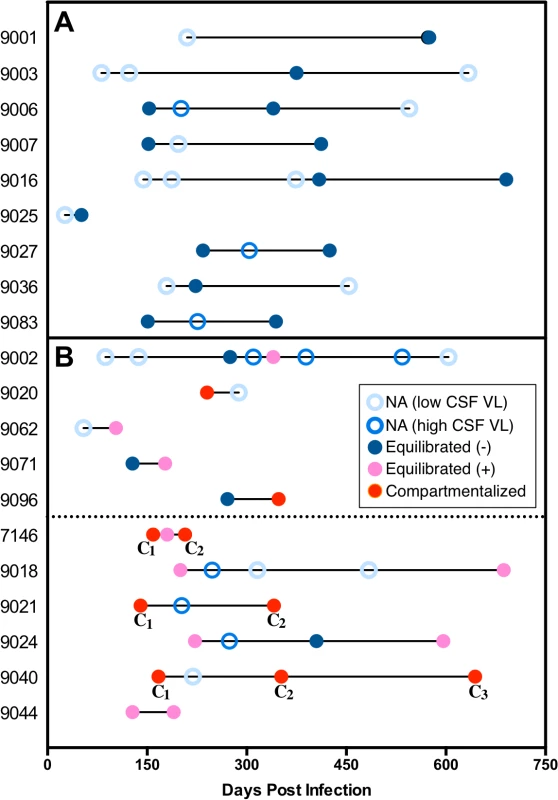
Examination of longitudinal relationships early during infection for 20 subjects with multiple time points where at least one time point was analyzed for viral populations. This group did not differ significantly from the 17 subjects with longitudinal samples available but not analyzed (due to low CSF viral load) in the average number of samples per person or the time span covered. Within individual subjects, the days post infection for each sample is indicated using circles, which represent the following: open light blue circle, sample not analyzed due to CSF viral load (VL) < 1,000 copies/ml (NA (low CSF VL)); open royal blue circle; sample not analyzed but CSF VL > 1,000 copies/ml (NA (high CSF VL)); closed dark blue circle, equilibrated with minimal CSF pleocytosis (CSF WBC <10 cells/μl) (Equilibrated (−)); closed pink circle, equilibrated with marked CSF pleocytosis (CSF WBC ≥10 cells/μl) (Equilibrated (+)); and closed red circle, compartmentalized. (A) Subjects with all analyzed time points equilibrated with minimal to no pleocytosis and remaining time points not analyzed. (B) Subjects with pleocytosis or compartmentalization in one time point (above dotted line) and subjects with pleocytosis, or compartmentalization in at least two time points (below dotted line). In every subject the compartmentalized (C) viral sequence population changed between sampling time points, indicated by different subscript numbers. Genetic evidence for the persistence of HIV-1 CNS replication
In two subjects, 9040 and 9021, a compartmentalized CNS population was observed at enrollment and for all analyzed longitudinal time points, spanning a period of 753 and 201 days p.i., respectively (one time point beyond 2 years p.i. was analyzed for subject 9040 but was not included in any overall population analysis). Further analysis of these subjects identified distinct trends of how HIV-1 becomes established in the CNS.
One pattern was seen in the viral evolution in subject 9040 (Fig. 4A). In this case a compartmentalized, clonally amplified population present at the initial time point (day 165) was replaced with a second compartmentalized, clonally amplified variant at day 352, but the sampling included a recombinant between the first and second clonally amplified variants. Several recombinants between these two early populations were maintained through day 918, however, the overall population was equilibrated at this last time point. Using BEAST analysis to estimate number of generations of the viral population, the time to most recent common ancestor (TMRCA) of the blood population at the initial time point was estimated to be 209 days, reasonably consistent with the reported date of transmission (165 days prior to sampling). We also estimated that the initial clonally amplified CSF population was established 33 days prior to sampling (approximately 170 days after transmission), followed by a subsequent clonal amplification event and recombination between the two lineages. These data showed the early establishment of a lineage within the CNS that persisted over a period of at least 2 years. To our knowledge, this was the first study to show the maintenance and evolution of a compartmentalized viral population within the CNS over a long duration of time starting during early infection.
Fig. 4. Compartmentalization can persist and evolve independently within the CSF over time. 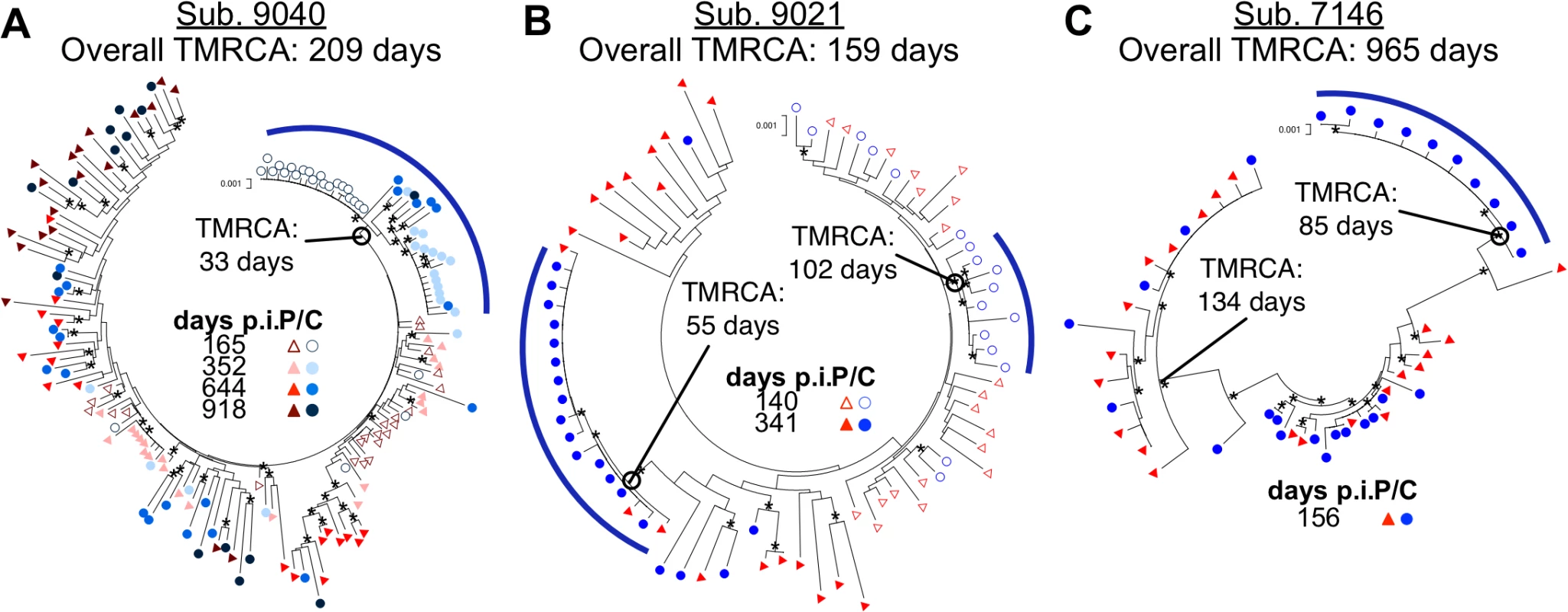
Neighbor-joining phylogenetic trees showing how compartmentalization can: (A) be persistent with multiple clonal expansions allowing recombination; (B) consist of sequential transient clonal expansions; and (C) be established with a transmitted variant. env sequences from the CSF are labeled with circles (C, colors designated in figure) and env sequences from the blood plasma are labeled with triangles (P, colors designated in figure). Days p.i. are noted. Bootstrap values ≥ 50 are indicated (*) at the appropriate nodes to highlight the more significant branch points. Genetic distance is indicated at the top of each phylogenetic tree (0.001, number of nucleotide substitutions per site between env sequences.) Compartmentalized populations are indicated at the appropriate node by an open circle and emphasized using a blue bar. BEAST-generated TMRCAs of the entire viral population are noted adjacent to the subject ID, and the TMRCAs of the different compartmentalized linages (subject 9040 and 9021) and transmitted parental lineages (subject 7146) are also noted. A second trend was seen for subject 9021 (Fig. 4B). Again, the reported date of transmission, 140 days prior to the first sampling date, was reasonably close to the transmission bottleneck estimated using BEAST at 159 days prior to sampling. In this subject one compartmentalized, clonally amplified lineage was detected in the CSF at the first sampling time point with an estimated age of 102 days, or starting 57 days post infection. This lineage was not present at the second sampling time point (at 341 days) but was replaced by a different compartmentalized, clonally amplified variant with an estimated age of 55 days. Thus for this subject we observed a permissive environment for viral replication in which variants were successively and independently amplified within the CNS.
Timing of introduction of HIV-1 into the CNS
We detected compartmentalization as early as 140 days p.i. (Table 2). Using BEAST to estimate the time to most recent common ancestor (TMRCA) we were able to show that often these CSF populations were established much earlier. Four subjects with longitudinal sampling (11 samples total) had a compartmentalized CSF population detected between 5–12 months post infection (Table 2); three of these subjects had an initial clonally amplified CSF population (7146, 9021 and 9040; Fig. 4). When we determined the estimated TMRCAs of these CSF populations, we estimate the initial clonally amplified populations were all established within approximately the first four months after infection and two were established within the first two months of infection (Table 2). These data show that the CNS compartment is permissive for HIV-1 replication in at least a subset of subjects from very early times after infection.
We have recently shown that after vertical transmission to children, CNS compartmentalization can be established early via the sequestration of one of multiple transmitted variants in the CNS [14]. When we reanalyzed data from a previously described subject (7146, Fig. 4C) [4] using BEAST, we showed that the phenomenon of transmission of two variants with one sequestered in the CSF/CNS shortly after transmission also occurs in adults. In this case the two transmitted variants diverged from each other in the donor (BEAST-estimated TMRCA of 965 days but with a reported transmission date of 156 days prior to sampling) while the transmitted variants diversified in the recipient. One lineage that was present in both the blood and CSF went through a bottleneck (presumably the transmission bottleneck) at 134 days prior to sampling, again consistent with the reported transmission date of 156 days prior to sampling. The other variant was sequestered in the CNS with an estimated bottleneck of 85 days, appearing early as a clonally amplified variant. In addition, a series of recombinants between these two lineages appeared in both the blood and CSF over time. This additional mechanism for establishing a compartmentalized viral population within the CNS early following transmission was also observed for subject 9018 (Table 2).
All transmitted variants are R5 T cell-tropic and are predominantly selected to use high levels of CD4 for entry
Macrophage-tropic HIV-1 variants can infect cells expressing low levels of CD4 while R5 T cell-tropic viruses are selected for replication in cells with high levels of CD4 for entry [24–31]. Macrophage-tropic HIV-1 is seen most reliably as a compartmentalized CSF/CNS population in a subset of individuals with HAD. To further our understanding of viral characteristics in the CNS early during infection, we analyzed the entry phenotype of viruses isolated from our adult primary infection cohort. Affinofile cells, on which CD4 and CCR5 surface expression can be differentially induced [32], are a more reproducible model for entry tropism analysis compared to primary cells [33]. We assessed entry phenotypes by measuring the ability of pseudotyped reporter viruses to enter Affinofile cells expressing either high or low levels of CD4.
Viruses pseudotyped with Env proteins derived from 24 subjects, representing all phylogenetic states and a wide range of days p.i., all required high levels of CCR5 and CD4 for efficient entry and were considered R5 T cell-tropic (Fig. 5A-B). However, Env proteins from CSF samples containing compartmentalized viral lineages (all collected more than 4 months post infection) were significantly better at entering cells expressing low CD4 than Env proteins derived from equilibrated CSF samples (ANOVA; P<<0.001). While this enhanced ability to enter low CD4 cells did not near the level of a macrophage-tropic Env protein (Fig. 5A-B, Ba-L), the detectably elevated levels suggest that compartmentalized lineages may be adapting to enter low CD4 cells in the CNS (discussed below). In contrast, viruses isolated from the CSF of adults within four months of infection are uniformly poor at infecting low CD4 cells, indicating that they have been selected for replication in T cells. This is consistent with multiple studies showing that macrophage-tropic viruses are not transmitted [16,27,28,34–39]. Together these results indicate that the CNS is initially exposed to R5 T cell-tropic variants and that viruses in the CNS remain R5 T cell-tropic for the first two years of infection, but the data provide suggestive evidence that the evolution of macrophage tropism may begin during this period.
Fig. 5. All viruses required high levels of CD4 for efficient entry, indicative of primarily being selected for replication in CD4+ T cells. 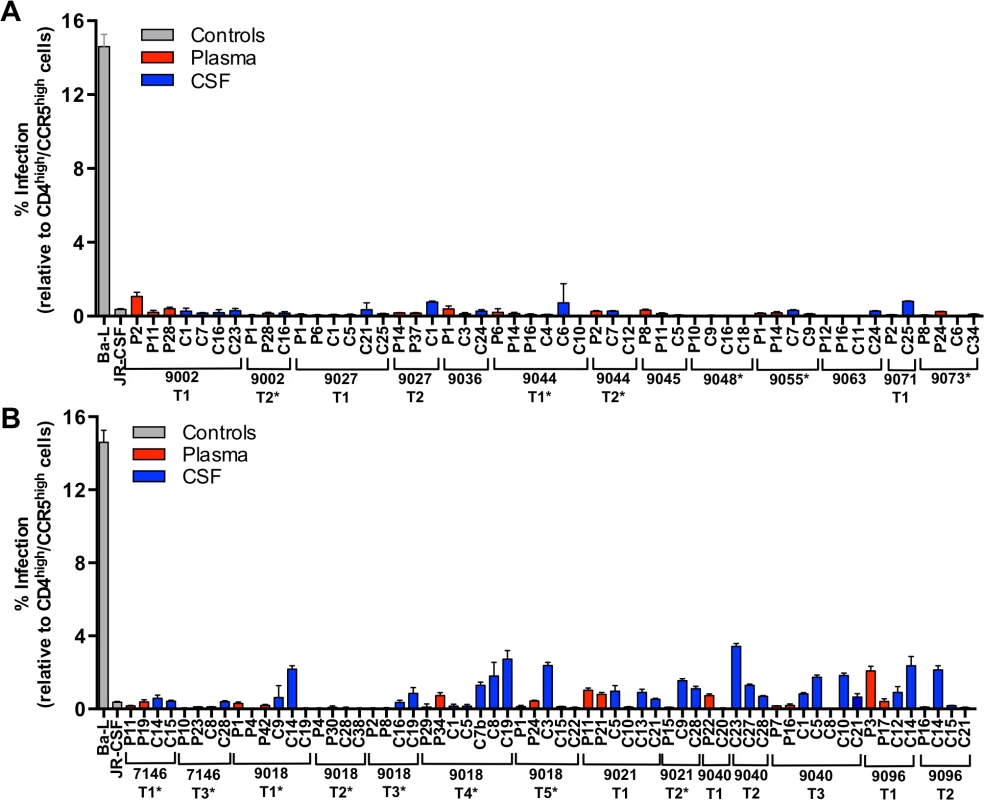
Single-cycle infection of HIV-1 Env-pseudotyped reporter viruses on CD4lowCCR5high 293-Affinofile cells [28]. Receptor expression was measured as follows: CD4low = 972 receptors/cell, CD4high = 72,041 receptors/cell, CCR5high = 15,636 receptors/cell. The data are averaged from triplicate wells for each of 2 to 3 env clones that were generated per indicated amplicon. Amplicons were selected for cloning to represent different portions of the phylogenetic tree. Subjects with no evidence of CNS compartmentalization (i.e. equilibrated) are shown in panel (A); and subjects with CNS compartmentalization at one or more time points are in panel (B). Longitudinal time points are indicated (T1, T2, T3, etc.) and samples with marked pleocytosis are noted (*). We also assessed the infectivity of a subset of the pseudotyped viruses on monocyte-derived macrophages (MDMs) generated from three separate donors. Infectivity was first assessed on Affinofile cells expressing high levels of CD4, with equal levels of infectious virus then added to each of the MDM preparations. There was general concordance between infectivity on Affinofile cells and infectivity on MDMs, although the level of infectivity differed significantly between the three donors (S2 Fig.). Viruses representing subjects with equilibrated populations and with low infectivity on Affinofile cells with low CD4 (Fig. 5A) had low infectivity on MDM (from subjects 9027, 9045, 9055, 9063, and 9073; S2 Fig.).
Viruses from four of five subjects with CNS compartmentalization were also assessed for their ability to enter Affinofile cells expressing low levels of CD4 and MDMs. Both blood - and CSF-derived viruses from one subject (subject 9096) were observed to have intermediate infectivity on low CD4 Affinofile cells (Fig. 5B) and MDMs (S2 Fig.). Similarly, two of the compartmentalized subjects (9018 and 9021) had CSF-derived viruses with elevated infectivity of low CD4 Affinofile cells (Fig. 5B) and MDMs (S2 Fig.). Finally, both blood - and CSF-derived viruses from one compartmentalized subject (subject 7146) were unable to efficiently enter both low CD4 expressing Affinofile cells and MDMs (S2 Fig.). Thus infectivity of MDMs is generally consistent with the conclusions derived from infection of Affinofile cells although the variability of infectivity of MDMs between donors precludes an accurate assessment of the range of CD4 entry phenotypes that can be observed using Affinofile cells.
Discussion
Independent HIV-1 replication in the CNS has been associated with neurological disorders [10,40,41] and may represent a distinct reservoir from that found in the blood and lymphoid tissue [42,43]. We examined the virologic characteristics associated with early CNS infection through analysis of paired cross-sectional and longitudinal blood plasma and CSF samples from a large cohort of 72 ART-naïve subjects infected with HIV-1 for less than two years. Our current study significantly builds upon a previous preliminary study by our group [4], enabling us to propose a model with four distinct states to describe the relationship between viral populations in the CSF/CNS and viral populations within the peripheral blood during early HIV-1 infection. These states are based upon details revealed by the current study on mechanisms of establishment of viral compartmentalization within the CNS, relationships between cellular inflammation, HIV-1 RNA levels and phylogenetic state, and insight into longitudinal maintenance and evolution of compartmentalization.
The first state (Fig. 6A) was observed in subjects with little evidence of CNS replication or pleocytosis, with CSF HIV-1 RNA concentrations proportionally 1–2% of the viral load in the periphery (Fig. 2B). In many of these subjects, the CSF HIV-1 RNA level was very low, below the limit of detection of standard assays. Minimal CSF viral burden has been observed in a prior report on a portion of this primary infection cohort [5]. With little or no pleocytosis, HIV-1 is likely entering the CSF/CNS at low levels via incomplete partitioning of virus at the blood-brain barrier, or low level trafficking of immune cells, including small numbers of infected CD4+ T cells. In this circumstance the viral population is very similar to the population in the blood. It is possible that some HIV-1 is replicating independently in the CNS at low levels in these subjects, but we were not able to detect these putative genetic variants above the low level background of virus recently imported from the periphery into the CSF/CNS. An argument in favor of even this low level viremia in the CSF being the result of T cell trafficking is the observation that in neuro-asymptomatic subjects with CD4+ T cells below 50 cells/ul in the blood the viral load in the CSF is on average lower than in subjects with higher CD4+ T cell counts [44,45].
Fig. 6. Four states can define the relationship between virus in the CSF/CNS and blood early during infection. 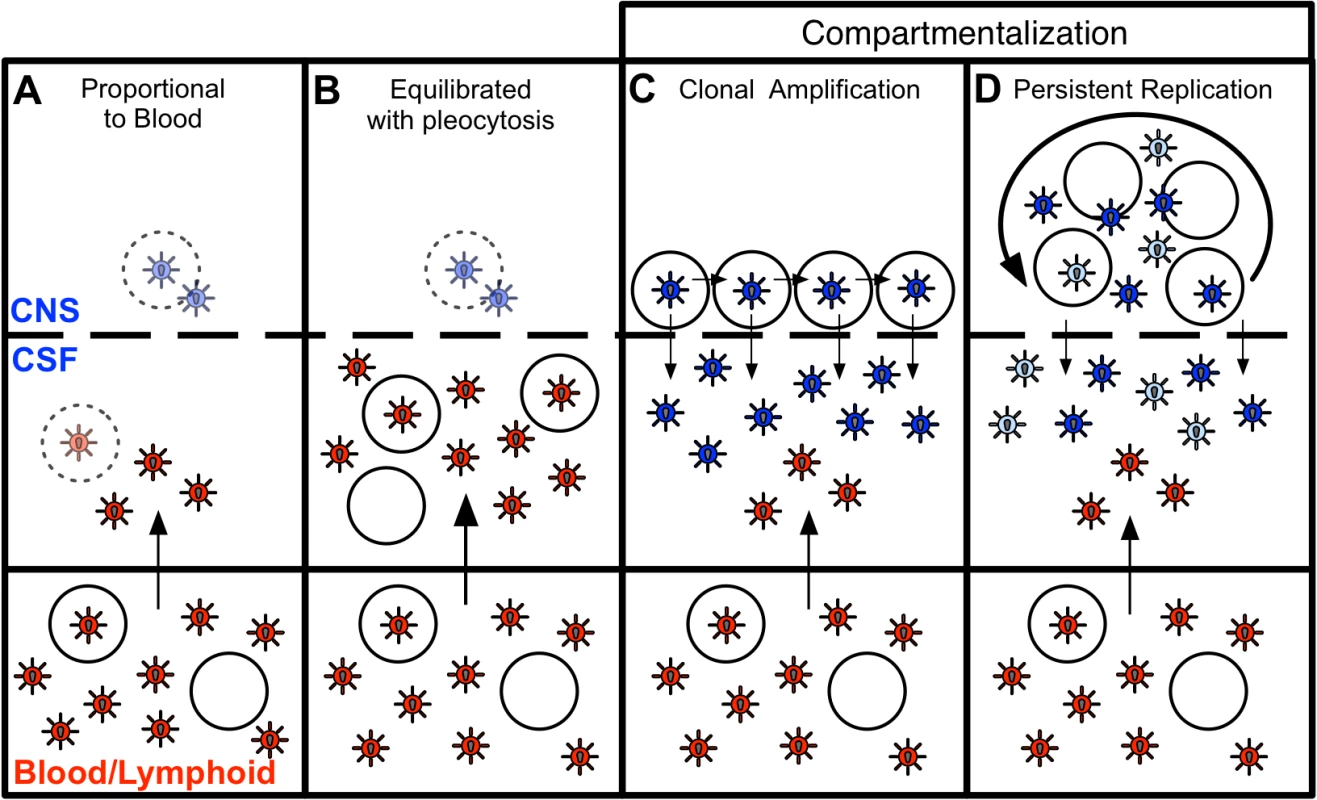
Blood and CSF/CNS compartments are indicated. Blood plasma viral variants are represented by the red virus particles; and CNS viral variants are represented by the blue virus particles. CD4+ T cells are represented by open circles. Arrows indicate direction of virus movement between compartments. (A) State with a CSF HIV-1 RNA level 1–2% of the viral load in the blood and defined by minimal to no local CNS replication or inflammation, resulting in an equilibrated state between the two compartments (when CSF viral load high enough for analysis). Transparent infected CD4+ T cell represents potential local CNS replication that may be obscured by the import of virus into the CNS from the periphery. (B) State of equilibration between CSF and blood accompanied by high levels of pleocytosis, potentially caused by local CNS replication. Transparent infected CD4+ T cell represents potential local CNS replication likely obscured by the high levels of virus secreted by the infiltrating CD4+ T cells. (C) State of CSF/CNS clonal amplification of identical or nearly identical variants within CD4+ T cells. These clonally amplified populations are characterized by low diversity, signified by all CSF viruses in a single shade of blue. (D) State of genetically complex, compartmentalized viral replication within CD4+ T cells in the CSF/CNS indicative of persistent replication beyond a single clonal amplification event; this complexity is signified by CNS viral variants in multiple shades of blue. In a second state (Fig. 6B), we observed a relationship between equilibrated viral populations with elevated viral load and high levels of pleocytosis (Fig. 2C). These equilibrated populations were most likely the result of the release of virus from increased numbers of infected CD4+ T cells trafficking from the periphery into the CNS. Though pleocytosis of > 10 cells/ul might have been due to a variety of inflammatory conditions (e.g. neurosyphilis as documented in one individual), it is most likely in response to HIV-1 replication in these PHI subjects. We screened for syphilis in this cohort, and detailed clinical and imaging assessment did not reveal other contributing causes of pleocytosis. Furthermore, other ‘background’ non-HIV causes of CSF WBC ≥ 10 cells/ul are unlikely in these subjects, as our parallel studies of 54 HIV-uninfected volunteers recruited from the similar local community demonstrated median CSF WBC counts of 1 cell/ul (IQR 0–2), and none of these 54 subjects had a CSF WBC as high as 10 [5]. In the setting of pleocytosis, while low levels of local CNS replication may have been occurring, the virus imported from the periphery by infected CD4+ T cells dominated the population as it raised the CSF HIV-1 RNA concentration by release of virus imported from the blood. If the inflammatory immune response was successful, pleocytosis might eventually result in low CSF viral loads, a condition observed in a small subset of subjects with pleocytosis but very low levels of virus in the CSF (S1 Table). Pleocytosis was also observed in several subjects with an intermediate viral population phenotype and half of the subjects with compartmentalized viral populations (Fig. 2D), suggesting pleocytosis may result in dynamic changes in the viral population in the CSF. An association between equilibrated compartments and high pleocytosis was also observed in a previous study analyzing four HIV-infected subjects during therapy interruption [46].
In a third state (Fig. 6C), we observed clonally amplified CSF populations of low complexity (Fig. 1 and 4) representing the recent expansion of identical or nearly identical variants that required high levels of CD4 for entry (R5 T cell-tropic; Fig. 5). High levels of pleocytosis were observed in approximately half of the subjects with clonally amplified CSF populations, making it possible that the influx of activated CD4+ T cells may also have provided cellular targets for further transient amplification of a CSF variant. We [47] (Dukhovlinova et al., in preparation) and others [48–50] have observed clonal amplification in the genital tract as well as in the CSF both early [4,14] and at later times in infection [41]. Clonal amplification appears to be a distinct type of virus-host interaction where infection of a population of CD4+ T cells in a compartment is a low probability event and when it occurs there is transient rapid expansion of the viral population. Due to the daily rapid turnover of the CSF viral RNA load, the elevated CSF viral RNA load that is often observed during clonal amplification, and the fact that these clonally amplified lineages generate their own diversity that can persist within the CNS, it is highly unlikely that clonally amplified virus represents virus produced from a single cell. The detection of clonally amplified populations in the CSF within the first year of infection has allowed us to estimate the establishment of these populations within the CNS to within the first 2–6 months (Table 2, Fig. 4), and such amplified populations were detected in 8% of subjects in this study within the first year.
In the final state, we observed more genetically complex compartmentalized viral replication within the CSF/CNS (Fig. 6D) indicative of persistent replication beyond a single clonal amplification event. In an effort to get a more complete view of the interaction of the virus within the CNS at these early times of infection we have interpreted the presence of persistent replication in the CNS based on four criteria: i) sequential clonal amplification events that indicated a permissive CNS environment for viral replication (Fig. 4B); ii) overlapping clonal amplification events that gave rise to compartmentalized recombinants showing continuous replication between the sampled time points (Fig. 4A); iii) intermittent compartmentalization and pleocytosis suggesting an inflammatory immune response to ongoing replication (Fig. 3B); and iv) sequestration of a transmitted variant within the CNS (Fig. 4C). Collectively these markers defined approximately 30% of subjects in the first two years as having evidence of viral replication in the CNS in at least one time point, and 16% having evidence of replication and/or inflammation at multiple time points within this period. This suggests that the CNS compartment is permissive for HIV-1 replication in at least a subset of subjects from a very early period after infection.
Entry tropism analysis revealed that all compartmentalized variants required high levels of CD4 for entry. It is now widely described in the literature that macrophage-tropic variants utilize low levels of CD4 for entry [24–31], are not transmitted, and that the transmitted virus is R5 T cell-tropic [16,27,28,34–39], an understanding further supported by our phenotypic analysis (Fig. 5). Our finding that the viruses involved in this early persistent CNS replication were adapted to replication in CD4+ T cells is distinct from previous studies of individuals with HAD where genetically complex compartmentalized CSF populations that had been replicating as an isolated population had evolved to replicate in macrophages/microglia [41]. Thus, adaptation to use low levels of CD4 for entry, a hallmark of macrophage tropism, is not a feature of the transmitted virus and does not evolve during the early stages of CNS infection in adults, at least as reflected in the compartmentalized virus detected in the CSF. However, we do note that the compartmentalized viruses from the CSF show a small but statistically significant increase in the ability to enter cells with low levels of CD4 compared to CSF virus from equilibrated subjects (Fig. 5B). One explanation for this small difference is that the virus in the CNS is carrying out at least a portion of its replication in a cell with low levels of CD4 which allows for at least a low level of selection for a low CD4 entry phenotype. We found no consistent differences in glycosylation site count or positions, or consistent sequence changes in sites previously described as being associated with macrophage tropism in comparing the viral sequences from the plasma to the compartmentalized sequences in the CSF (S2 Table and S3).
The only CNS tissue that is readily sampled in volunteer human subjects is CSF, which, though not identical to brain, is produced within the brain in the choroid plexus, and reflects brain inflammation and infection in the context of CNS infections including HIV-1. Measures of immune activation, HIV-1 burden, and neural injury detected in CSF are markers of brain involvement in HIV-1 that correlate to clinical and pathologic disease [51]. While the cellular source of HIV-1 RNA detected in the CSF is not certain and may differ during different stages of infection [52], compartmentalization of HIV-1 detected in CSF associates with clinical dementia in humans [10] and immunopathology in the brain in rhesus macaques [53]. Despite limitations of generalizing CSF findings to those of the CNS more broadly, our studies have used the best methods available in living humans to assess HIV-1 populations derived from the CNS in a unique cohort of subjects enrolled during primary HIV-1 infection. Our results show that in cross-sectional analysis over the first two years of HIV-1 infection, 30% of subjects have evidence of either local viral replication in the CNS or a robust CNS inflammatory response, and that in approximately 16% of subjects this CNS involvement can persist over time. We have found that the viral population in the CSF is dynamic as the result of local replication and/or the influx of virus in infected CD4+ T cells as part of an inflammatory response. This early viral replication in a subset of subjects may represent an inability to protect the CNS from infection, potentially leading to HAND later in infection, and may also define a distinct reservoir of infected cells within the body. Longitudinal follow-up of these subjects to examine the long-term impact of the presence of early active HIV-1 replication in the CNS will help to define the significance of these findings for clinical neurologic disease outcomes and compartmentalized viral reservoirs in the setting of HIV-1.
Materials and Methods
Ethics statement
The study was approved by the Institutional Review Boards at UCSF, Yale University, and the University of North Carolina at Chapel Hill. All study participants were adults (age ≥ 18 years). Written informed consent was obtained from all participants.
Study design
We assessed samples obtained through an observational longitudinal neurological study of primary HIV-1 infection to determine viral characteristics associated with early HIV-1 CNS infection. Subjects were referred from the community and were eligible if they met prospectively determined criteria for laboratory confirmation of primary HIV-1 infection, as previously described [4]. Subjects were screened at study enrollment for systemic syphilis by blood RPR testing. Subsequent blood and CSF samples were tested for RPR and VDRL, respectively, if an outside test suggested syphilis exposure or CSF WBC was markedly elevated compared to that in an earlier longitudinal sample. No subjects had clinical evidence of other inflammatory neurologic disorders such as multiple sclerosis or CNS opportunistic infections based on interview and examination by an HIV-1 neurologist at each visit. A total of 57 of the 72 subjects volunteered to participate in magnetic resonance imaging of the brain for the overall study protocol; scans were reviewed by a neuroradiologist and none revealed evidence of encephalitis, tumor or opportunistic infection. Blood and CSF samples were collected at enrollment, six weeks, and every six months thereafter. This analysis included samples obtained up to two years post-infection from subjects enrolled prior to 4/1/2012. CSF and plasma HIV-1 RNA concentrations were determined as described [4]; paired samples were selected for further SGA analysis if CSF HIV-1 RNA was greater than1,000 copies/ml (to ensure adequate sampling). Samples with lower viral loads could be analyzed if larger volumes were committed to concentrate the virus, but we chose to use a cut-off of 1,000 viral RNA copies/ml for these studies. Primary study endpoints included SGA of the HIV-1 env gene for viral genetic compartmentalization and phenotypic analyses, CSF and blood HIV-1 RNA concentrations, measures of CSF cellular inflammatory response (white blood count, WBC) and blood brain barrier disruption (CSF:plasma albumin ratio).
Single genome amplification
When collected, CSF samples were initially placed on wet ice and delivered to the lab for processing within one hour. For virology analyses, CSF samples were centrifuged at 1200 x g for 10 min to remove contaminating cells or cellular debris, and the supernatant was subsequently aliquoted and stored at −70°C for later assay. Viral RNA was isolated as previously described from the CSF supernatant [4]. Briefly, RNA was isolated from samples (140 μl) with viral loads >10,000 copies/ml using the QIAmp Viral RNA Mini kit (Qiagen). To increase template number, samples with viral loads <10,000 copies/ml were first pelleted by ultracentrifugation. cDNA was generated using an oligo-d(T) primer. Single genome amplification/template endpoint dilution PCR [38] of the env gene through the 3’ LTR U3 end was conducted using the cDNA as template as previously described [4,14]. Sequences for full-length env were generated (samples analyzed previously were sequenced from the start of V1 through the ectodomain of gp41 [4]).
Phylogenetic analysis of env viral sequences
Phylogenetic analysis were conducted in a manner similar to our previous study [14]. In brief, DNA sequences were aligned (MUSCLE) [54–56] using EBI web tools [57], and phylogenetic trees were generated (neighbor-joining method, MEGA 4.0 [58]). Phylogenetic states were determined by statistical evaluation using the Slatkin-Maddison (SM) test [18] as previously described [14], Wright’s measure of population subdivision (Fst) [19,20] and the Nearest-neighbor statistic (Snn) [21]. CSF populations were defined as being compartmentalization if all three statistical tests (SM, Fst and Snn) yielded significant results (P values < 0.05) or equilibrated if one or more of tests yielded non-significant results. Low bootstrap values were used (≥35) because of the overall low diversity of the viral populations early after infection. Clonally amplified lineages (short branch lengths with bootstrap values ≥ 99 and a clade of ≥ 3 variants) were also identified. No contamination occurred between samples (S3 Fig.). Sequences for subjects 9002 (338 d.p.i.), 9007, 9018 (200 d.p.i.), 9025, 9037, 9039, 9040 (165 d.p.i.), and 7146 were generated in our previous study [4].
Bayesian analysis
A Bayesian Markov Chain Monte Carlo (MCMC) approach using BEAST v.1.6.1 [15] estimated the TMRCA for each viral population. A substitution rate of 1.5x10−5 substitutions/site/generation and standard deviation of 3.0x10−6 were fixed under a lognormal relaxed clock (uncorrelated) model. The rate was calculated via tip dating, using a consensus sequence (set as day 0), and the estimated days post infection. The HKY nucleotide substitution model had estimated base frequencies and a gamma-distributed rate heterogeneity (4 gamma categories). A coalescent Bayesian Skyline tree prior with a Piecewise-constant skyline model was used (10 groups). The MCMC algorithm was run for 30 million generations, logging every 1000 and with a 10% burn-in. The results from at least two independent runs were combined, and the effective sample size for all estimates was >200. A generation time of 1.0 day was used.
Construction of HIV-1 env clones
Full length HIV-1 env genes were re-amplified from the first-round SGA products as previously described [41]. The PCR product was cloned into the pcDNA3.1D/V5-His-TOPO expression vector (Invitrogen) using the pcDNA 3.1 directional TOPO expression kit (Invitrogen).
Cells
293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 mg/ml of penicillin and streptomycin. 293-Affinofile cells [32], generously provided by Dr. Ben-Hur Lee, were maintained in DMEM supplemented with 10% dialyzed FBS (12–14 kD dialyzed; Atlanta Biologicals) and 50 mg/ml blasticidin (D10F/B).
Env-pseudotyped viruses
Env-pseudotyped luciferase reporter viruses were generated as previously described [14]. Briefly, 293T cells were cotransfected with an env expression vector and the pNL4-3.LucR-E - HIV-1 backbone (obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) using the Fugene 6 transfection reagent and protocol (Roche). Transfection medium was replaced with fresh culture medium five hours post-transfection and the cells were incubated at 37°C for 48 hours, after which viral supernatants were filtered with 0.45 μM filters (Millipore) and stored at −80°C.
293-Affinofile cellular surface expression of CD4 and CCR5
293-Affinofile cell [32] CD4 and CCR5 receptor expression was induced with doxycycline (doxy; Invitrogen) and ponasterone A (ponA; Invitrogen), respectively, as previously described [14]. Briefly, cells were induced at two conditions: CD4high/CCR5high (6 ng/ml doxy and 5 μM ponA, respectively) and CD4low/CCR5high (0 ng/ml doxy and 5 μM ponA). CD4 and CCR5 receptor expression was measured using quantitative fluorescence-activated cytometry (qFACS) following staining with either phycoerythin (PE)-conjugated anti-human CD4 antibody (clone Q4120, BD Biosciences) or PE-conjugated mouse anti-human CCR5 antibody (clone 2D7, BD Biosciences), and surface levels were calculated using QuantiBRITE beads (BD Biosciences).
Single-cycle infection of 293-Affinofile cells
As described previously [14], Env-pseudotyped luciferase reporter viruses were first titered on 293-Affinofile cells [32] expressing CD4high/CCR5high. For viral infections, black tissue culture plates (96 wells) were coated with 10% poly-L-lysine and seeded with 293-Affinofile cells (1.85 x 104 cells/well). Eighteen to 24 hours later, expression of CD4 and CCR5 was induced at CD4high/CCR5high and CD4low/CCR5high. Eighteen to 24 hours later, the induction medium was removed and replaced with 100 μl of fresh, warmed culture medium containing Env-pseudotyped virus. The plates were spinoculated [59] at 2,000 rpm for 2 hours at 37°C, and incubated for 48 hours at 37°C. Infection medium was removed, cells were lysed, and luciferase activity was assayed using the luciferase assay system (Promega). Clone sequences were not compared to the original parental sequence prior to pseudotyping and Affinofile cell infection. Instead, entry tropism data for each parental amplicon included three replicates from 2–3 clones derived from the same parental amplicon. In this analysis we assume any PCR-introduced error would either not change the entry phenotype or would create a nonfunctional protein which would not be included in the subsequent analysis. The concordance of the entry phenotype of the replicate clones was taken to represent the phenotype of the amplicon sequence.
Statistical analysis
Compartmentalization was assessed statistically using the Slatkin-Maddison test for gene flow [18], Wright’s measure of population subdivision (Fst) [19,20] and the Nearest-neighbor statistic (Snn) [21]. Differences between groups were examined for statistical significance using the Mann-Whitney Test. The one exception was our analysis examining whether env genes derived from samples with and without CSF compartmentalized lineages differed in their ability to enter Affinofile cells expressing low levels of CD4 expression. We used a linear model to perform this analysis (performed in [R]) and used the stepAIC function to perform stepwise model selection. All correlations employed Spearman’s rank correlation coefficient. For all statistical tests, P values less than 0.05 were considered significant.
Nucleotide sequence accession numbers
The HIV-1 env nucleotide sequences determined in this study have been deposited in GenBank under accession numbers KM353586-KM355197
Supporting Information
Zdroje
1. Pilcher CD, Shugars DC, Fiscus SA, Miller WC, Menezes P, et al. (2001) HIV in body fluids during primary HIV infection: implications for pathogenesis, treatment and public health. AIDS 15 : 837–845. 11399956
2. Enting RH, Prins JM, Jurriaans S, Brinkman K, Portegies P, et al. (2001) Concentrations of human immunodeficiency virus type 1 (HIV-1) RNA in cerebrospinal fluid after antiretroviral treatment initiated during primary HIV-1 infection. Clin Infect Dis 32 : 1095–1099. 11264039
3. Tambussi G, Gori A, Capiluppi B, Balotta C, Papagno L, et al. (2000) Neurological symptoms during primary human immunodeficiency virus (HIV) infection correlate with high levels of HIV RNA in cerebrospinal fluid. Clin Infect Dis 30 : 962–965. 10880317
4. Schnell G, Price RW, Swanstrom R, Spudich S (2010) Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J Virol 84 : 2395–2407. doi: 10.1128/JVI.01863-09 20015984
5. Spudich S, Gisslen M, Hagberg L, Lee E, Liegler T, et al. (2011) Central nervous system immune activation characterizes primary human immunodeficiency virus 1 infection even in participants with minimal cerebrospinal fluid viral burden. J Infect Dis 204 : 753–760. doi: 10.1093/infdis/jir387 21844301
6. Davis LE, Hjelle BL, Miller VE, Palmer DL, Llewellyn AL, et al. (1992) Early viral brain invasion in iatrogenic human immunodeficiency virus infection. Neurology 42 : 1736–1739. 1513462
7. Valcour V, Chalermchai T, Sailasuta N, Marovich M, Lerdlum S, et al. (2012) Central nervous system viral invasion and inflammation during acute HIV infection. J Infect Dis 206 : 275–282. doi: 10.1093/infdis/jis326 22551810
8. Ances BM, Ellis RJ (2007) Dementia and neurocognitive disorders due to HIV-1 infection. Semin Neurol 27 : 86–92. 17226745
9. Fischer-Smith T, Rappaport J (2005) Evolving paradigms in the pathogenesis of HIV-1-associated dementia. Expert Rev Mol Med 7 : 1–26.
10. Harrington PR, Schnell G, Letendre SL, Ritola K, Robertson K, et al. (2009) Cross-sectional characterization of HIV-1 env compartmentalization in cerebrospinal fluid over the full disease course. Aids 23 : 907–915. doi: 10.1097/QAD.0b013e3283299129 19414991
11. Ohagen A, Devitt A, Kunstman KJ, Gorry PR, Rose PP, et al. (2003) Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J Virol 77 : 12336–12345. 14581570
12. Dunfee RL, Thomas ER, Gorry PR, Wang J, Taylor J, et al. (2006) The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc Natl Acad Sci U S A 103 : 15160–15165. 17015824
13. Schnell G, Spudich S, Harrington P, Price RW, Swanstrom R (2009) Compartmentalized human immunodeficiency virus type 1 originates from long-lived cells in some subjects with HIV-1-associated dementia. PLoS Pathog 5: e1000395. doi: 10.1371/journal.ppat.1000395 19390619
14. Sturdevant CB, Dow A, Jabara CB, Joseph SB, Schnell G, et al. (2012) Central nervous system compartmentalization of HIV-1 subtype C variants early and late in infection in young children. PLoS Pathog 8: e1003094. doi: 10.1371/journal.ppat.1003094 23300446
15. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7 : 214. 17996036
16. Ping LH, Joseph SB, Anderson JA, Abrahams MR, Salazar-Gonzalez JF, et al. (2013) Comparison of Viral Env Proteins From Acute and Chronic Infections of Subtype C Human Immunodeficiency Virus Type 1 Identifies Differences In Glycosylation and CCR5 Utilization and Suggests A New Strategy For Immunogen Design. J Virol.
17. Zarate S, Pond SLK, Shapshak P, Frost SDW (2007) Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. Journal of Virology 81 : 6643–6651. 17428864
18. Slatkin M, Maddison WP (1989) A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics 123 : 603–613. 2599370
19. Hudson RR, Boos DD, Kaplan NL (1992) A Statistical Test for Detecting Geographic Subdivision. Molecular Biology and Evolution 9 : 138–151. 1552836
20. Hudson RR, Slatkin M, Maddison WP (1992) Estimation of levels of gene flow from DNA sequence data. Genetics 132 : 583–589. 1427045
21. Hudson RR (2000) A new statistic for detecting genetic differentiation. Genetics 155 : 2011–2014. 10924493
22. Fishman RA (1992) Cerebrospinal fluid in diseases of the nervous system, 2nd ed. Philadelphia, PA: W. B. Saunders Company.
23. Ho EL, Ronquillo R, Altmeppen H, Spudich SS, Price RW, et al. (2013) Cellular Composition of Cerebrospinal Fluid in HIV-1 Infected and Uninfected Subjects. PLoS One 8: e66188. 23822975
24. Rossi F, Querido B, Nimmagadda M, Cocklin S, Navas-Martin S, et al. (2008) The V1-V3 region of a brain-derived HIV-1 envelope glycoprotein determines macrophage tropism, low CD4 dependence, increased fusogenicity and altered sensitivity to entry inhibitors. Retrovirology 5 : 89. doi: 10.1186/1742-4690-5-89 18837996
25. Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, et al. (2007) Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology 360 : 105–119. 17084877
26. Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, et al. (2002) Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol 76 : 6277–6292. 12021361
27. Alexander M, Lynch R, Mulenga J, Allen S, Derdeyn CA, et al. (2010) Donor and recipient envs from heterosexual human immunodeficiency virus subtype C transmission pairs require high receptor levels for entry. J Virol 84 : 4100–4104. doi: 10.1128/JVI.02068-09 20147398
28. Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, et al. (2012) Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. PLoS Pathog 8: e1002686. doi: 10.1371/journal.ppat.1002686 22693444
29. Dunfee R, Thomas ER, Gorry PR, Wang J, Ancuta P, et al. (2006) Mechanisms of HIV-1 neurotropism. Curr HIV Res 4 : 267–278. 16842080
30. Duenas-Decamp MJ, Peters PJ, Burton D, Clapham PR (2009) Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J Virol 83 : 2575–2583. doi: 10.1128/JVI.02133-08 19129457
31. Martin-Garcia J, Cao W, Varela-Rohena A, Plassmeyer ML, Gonzalez-Scarano F (2006) HIV-1 tropism for the central nervous system: Brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology 346 : 169–179. 16309726
32. Johnston SH, Lobritz MA, Nguyen S, Lassen K, Delair S, et al. (2009) A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J Virol 83 : 11016–11026. doi: 10.1128/JVI.01242-09 19692480
33. Joseph SB, Arrildt KT, Swanstrom AE, Schnell G, Lee B, et al. (2013) Quantification of Entry Phenotypes of Macrophage-Tropic HIV-1 Across a Wide Range of CD4 Densities. J Virol.
34. Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, et al. (2009) Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 206 : 1273–1289. doi: 10.1084/jem.20090378 19487424
35. Isaacman-Beck J, Hermann EA, Yi Y, Ratcliffe SJ, Mulenga J, et al. (2009) Heterosexual transmission of human immunodeficiency virus type 1 subtype C: Macrophage tropism, alternative coreceptor use, and the molecular anatomy of CCR5 utilization. J Virol 83 : 8208–8220. doi: 10.1128/JVI.00296-09 19515785
36. Li H, Bar KJ, Wang S, Decker JM, Chen Y, et al. (2010) High Multiplicity Infection by HIV-1 in Men Who Have Sex with Men. PLoS Pathog 6: e1000890. doi: 10.1371/journal.ppat.1000890 20485520
37. Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, et al. (2012) Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J Virol 86 : 2715–2728. doi: 10.1128/JVI.06157-11 22190722
38. Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, et al. (2008) Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105 : 7552–7557. doi: 10.1073/pnas.0802203105 18490657
39. Wilen CB, Parrish NF, Pfaff JM, Decker JM, Henning EA, et al. (2011) Phenotypic and immunologic comparison of clade B transmitted/founder and chronic HIV-1 envelope glycoproteins. J Virol 85 : 8514–8527. doi: 10.1128/JVI.00736-11 21715507
40. Ritola K, Robertson K, Fiscus SA, Hall C, Swanstrom R (2005) Increased human immunodeficiency virus type 1 (HIV-1) env compartmentalization in the presence of HIV-1-associated dementia. J Virol 79 : 10830–10834. 16051875
41. Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R (2011) HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog 7: e1002286. doi: 10.1371/journal.ppat.1002286 22007152
42. Alexaki A, Liu Y, Wigdahl B (2008) Cellular reservoirs of HIV-1 and their role in viral persistence. Curr HIV Res 6 : 388–400. 18855649
43. Pomerantz RJ (2003) Reservoirs, sanctuaries, and residual disease: the hiding spots of HIV-1. HIV Clin Trials 4 : 137–143. 12671782
44. Spudich SS, Nilsson AC, Lollo ND, Liegler TJ, Petropoulos CJ, et al. (2005) Cerebrospinal fluid HIV infection and pleocytosis: relation to systemic infection and antiretroviral treatment. BMC Infect Dis 5 : 98. 16266436
45. Price RW, Peterson J, Fuchs D, Angel TE, Zetterberg H, et al. (2013) Approach to Cerebrospinal Fluid (CSF) Biomarker Discovery and Evaluation in HIV Infection. J Neuroimmune Pharmacol 8 : 1147–1158. doi: 10.1007/s11481-013-9491-3 23943280
46. Smith DM, Zarate S, Shao H, Pillai SK, Letendre SL, et al. (2009) Pleocytosis is associated with disruption of HIV compartmentalization between blood and cerebral spinal fluid viral populations. Virology 385 : 204–208. doi: 10.1016/j.virol.2008.11.010 19100592
47. Anderson JA, Ping LH, Dibben O, Jabara CB, Arney L, et al. (2010) HIV-1 Populations in Semen Arise through Multiple Mechanisms. PLoS Pathog 6: e1001053. doi: 10.1371/journal.ppat.1001053 20808902
48. Shaheen F, Sison AV, McIntosh L, Mukhtar M, Pomerantz RJ (1999) Analysis of HIV-1 in the cervicovaginal secretions and blood of pregnant and nonpregnant women. J Hum Virol 2 : 154–166. 10413367
49. Kemal KS, Foley B, Burger H, Anastos K, Minkoff H, et al. (2003) HIV-1 in genital tract and plasma of women: compartmentalization of viral sequences, coreceptor usage, and glycosylation. Proc Natl Acad Sci U S A 100 : 12972–12977. 14557540
50. Bull ME, Heath LM, McKernan-Mullin JL, Kraft KM, Acevedo L, et al. (2013) Human immunodeficiency viruses appear compartmentalized to the female genital tract in cross-sectional analyses but genital lineages do not persist over time. J Infect Dis 207 : 1206–1215. doi: 10.1093/infdis/jit016 23315326
51. Gisslen M, Hagberg L, Rosengren L, Brew BJ, Cinque P, et al. (2007) Defining and evaluating HIV-related neurodegenerative disease and its treatment targets: a combinatorial approach to use of cerebrospinal fluid molecular biomarkers. J Neuroimmune Pharmacol 2 : 112–119. 18040834
52. Eden A, Price RW, Spudich S, Fuchs D, Hagberg L, et al. (2007) Immune activation of the central nervous system is still present after >4 years of effective highly active antiretroviral therapy. J Infect Dis 196 : 1779–1783. doi: 10.1086/523648 18190258
53. Harrington PR, Connell MJ, Meeker RB, Johnson PR, Swanstrom R (2007) Dynamics of simian immunodeficiency virus populations in blood and cerebrospinal fluid over the full course of infection. J Infect Dis 196 : 1058–1067. 17763329
54. Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. Bmc Bioinformatics 5 : 1–19. 14706121
55. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32 : 1792–1797. 15034147
56. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 : 4673–4680. 7984417
57. McWilliam H, Li WZ, Uludag M, Squizzato S, Park YM, et al. (2013) Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Research 41: W597–W600. doi: 10.1093/nar/gkt376 23671338
58. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 : 1596–1599. 17488738
59. O'Doherty U, Swiggard WJ, Malim MH (2000) Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. Journal of Virology 74 : 10074–10080. 11024136
60. Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, et al. (1986) The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233 : 215–219. 3014648
61. Koyanagi Y, Miles S, Mitsuyasu RT, Merrill JE, Vinters HV, et al. (1987) Dual infection of the central-nervous-system by AIDS viruses with distinct cellular tropisms. Science 236 : 819–822. 3646751
62. Joseph SB, Arrildt KT, Swanstrom AE, Schnell G, Lee B, et al. (2014) Quantification of entry phenotypes of macrophage-tropic HIV-1 across a wide range of CD4 densities. Journal of Virology 88 : 1858–1869. doi: 10.1128/JVI.02477-13 24307580
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole MembraneČlánek Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and FunctionČlánek Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 3- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- To Be or Not IIb: A Multi-Step Process for Epstein-Barr Virus Latency Establishment and Consequences for B Cell Tumorigenesis
- Is Antigenic Sin Always “Original?” Re-examining the Evidence Regarding Circulation of a Human H1 Influenza Virus Immediately Prior to the 1918 Spanish Flu
- The Great Escape: Pathogen Versus Host
- Coping with Stress and the Emergence of Multidrug Resistance in Fungi
- Catch Me If You Can: The Link between Autophagy and Viruses
- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- Evidence for Ubiquitin-Regulated Nuclear and Subnuclear Trafficking among Matrix Proteins
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Production of Anti-LPS IgM by B1a B Cells Depends on IL-1β and Is Protective against Lung Infection with LVS
- Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS
- α-Hemolysin Counteracts the Anti-Virulence Innate Immune Response Triggered by the Rho GTPase Activating Toxin CNF1 during Bacteremia
- Induction of Interferon-Stimulated Genes by IRF3 Promotes Replication of
- Intracellular Growth Is Dependent on Tyrosine Catabolism in the Dimorphic Fungal Pathogen
- HCV Induces the Expression of Rubicon and UVRAG to Temporally Regulate the Maturation of Autophagosomes and Viral Replication
- Spatiotemporal Analysis of Hepatitis C Virus Infection
- Subgingival Microbial Communities in Leukocyte Adhesion Deficiency and Their Relationship with Local Immunopathology
- Interaction between the Type III Effector VopO and GEF-H1 Activates the RhoA-ROCK Pathway
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
- Establishment of HSV1 Latency in Immunodeficient Mice Facilitates Efficient Reactivation
- XRN1 Stalling in the 5’ UTR of Hepatitis C Virus and Bovine Viral Diarrhea Virus Is Associated with Dysregulated Host mRNA Stability
- γδ T Cells Confer Protection against Murine Cytomegalovirus (MCMV)
- Rhadinovirus Host Entry by Co-operative Infection
- A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole Membrane
- Dermal Neutrophil, Macrophage and Dendritic Cell Responses to Transmitted by Fleas
- Elucidation of Sigma Factor-Associated Networks in Reveals a Modular Architecture with Limited and Function-Specific Crosstalk
- A Conserved NS3 Surface Patch Orchestrates NS2 Protease Stimulation, NS5A Hyperphosphorylation and HCV Genome Replication
- Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and Function
- Disruption of IL-21 Signaling Affects T Cell-B Cell Interactions and Abrogates Protective Humoral Immunity to Malaria
- Compartmentalized Replication of R5 T Cell-Tropic HIV-1 in the Central Nervous System Early in the Course of Infection
- Diminished Reovirus Capsid Stability Alters Disease Pathogenesis and Littermate Transmission
- Characterization of CD8 T Cell Differentiation following SIVΔnef Vaccination by Transcription Factor Expression Profiling
- Visualization of HIV-1 Interactions with Penile and Foreskin Epithelia: Clues for Female-to-Male HIV Transmission
- Sensing Cytosolic RpsL by Macrophages Induces Lysosomal Cell Death and Termination of Bacterial Infection
- PKCη/Rdx-driven Phosphorylation of PDK1: A Novel Mechanism Promoting Cancer Cell Survival and Permissiveness for Parvovirus-induced Lysis
- Metalloprotease NleC Suppresses Host NF-κB/Inflammatory Responses by Cleaving p65 and Interfering with the p65/RPS3 Interaction
- Immune Antibodies and Helminth Products Drive CXCR2-Dependent Macrophage-Myofibroblast Crosstalk to Promote Intestinal Repair
- Adenovirus Entry From the Apical Surface of Polarized Epithelia Is Facilitated by the Host Innate Immune Response
- The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family
- Neutrophils: Between Host Defence, Immune Modulation, and Tissue Injury
- CD169-Mediated Trafficking of HIV to Plasma Membrane Invaginations in Dendritic Cells Attenuates Efficacy of Anti-gp120 Broadly Neutralizing Antibodies
- Japanese Encephalitis Virus Nonstructural Protein NS5 Interacts with Mitochondrial Trifunctional Protein and Impairs Fatty Acid β-Oxidation
- Yip1A, a Novel Host Factor for the Activation of the IRE1 Pathway of the Unfolded Protein Response during Infection
- TRIM26 Negatively Regulates Interferon-β Production and Antiviral Response through Polyubiquitination and Degradation of Nuclear IRF3
- Parallel Epigenomic and Transcriptomic Responses to Viral Infection in Honey Bees ()
- A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication
- Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
- Exome and Transcriptome Sequencing of Identifies a Locus That Confers Resistance to and Alters the Immune Response
- The Role of Misshapen NCK-related kinase (MINK), a Novel Ste20 Family Kinase, in the IRES-Mediated Protein Translation of Human Enterovirus 71
- Chitin Recognition via Chitotriosidase Promotes Pathologic Type-2 Helper T Cell Responses to Cryptococcal Infection
- Activates Both IL-1β and IL-1 Receptor Antagonist to Modulate Lung Inflammation during Pneumonic Plague
- Persistence of Transmitted HIV-1 Drug Resistance Mutations Associated with Fitness Costs and Viral Genetic Backgrounds
- An 18 kDa Scaffold Protein Is Critical for Biofilm Formation
- Early Virological and Immunological Events in Asymptomatic Epstein-Barr Virus Infection in African Children
- Human CD8 T-cells Recognizing Peptides from () Presented by HLA-E Have an Unorthodox Th2-like, Multifunctional, Inhibitory Phenotype and Represent a Novel Human T-cell Subset
- Decreased HIV-Specific T-Regulatory Responses Are Associated with Effective DC-Vaccine Induced Immunity
- RSV Vaccine-Enhanced Disease Is Orchestrated by the Combined Actions of Distinct CD4 T Cell Subsets
- Concerted Activity of IgG1 Antibodies and IL-4/IL-25-Dependent Effector Cells Trap Helminth Larvae in the Tissues following Vaccination with Defined Secreted Antigens, Providing Sterile Immunity to Challenge Infection
- Structure of the Low pH Conformation of Chandipura Virus G Reveals Important Features in the Evolution of the Vesiculovirus Glycoprotein
- PPM1A Regulates Antiviral Signaling by Antagonizing TBK1-Mediated STING Phosphorylation and Aggregation
- Lipidomic Analysis Links Mycobactin Synthase K to Iron Uptake and Virulence in .
- Roles and Programming of Arabidopsis ARGONAUTE Proteins during Infection
- Impact of Infection on Host Macrophage Nuclear Physiology and Nucleopore Complex Integrity
- The Impact of Host Diet on Titer in
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Herpesviral G Protein-Coupled Receptors Activate NFAT to Induce Tumor Formation via Inhibiting the SERCA Calcium ATPase
- The Causes and Consequences of Changes in Virulence following Pathogen Host Shifts
- Small GTPase Rab21 Mediates Fibronectin Induced Actin Reorganization in : Implications in Pathogen Invasion
- Positive Role of Promyelocytic Leukemia Protein in Type I Interferon Response and Its Regulation by Human Cytomegalovirus
- NEDDylation Is Essential for Kaposi’s Sarcoma-Associated Herpesvirus Latency and Lytic Reactivation and Represents a Novel Anti-KSHV Target
- β-HPV 5 and 8 E6 Disrupt Homology Dependent Double Strand Break Repair by Attenuating BRCA1 and BRCA2 Expression and Foci Formation
- An O Antigen Capsule Modulates Bacterial Pathogenesis in
- Variable Processing and Cross-presentation of HIV by Dendritic Cells and Macrophages Shapes CTL Immunodominance and Immune Escape
- Probing the Metabolic Network in Bloodstream-Form Using Untargeted Metabolomics with Stable Isotope Labelled Glucose
- Adhesive Fiber Stratification in Uropathogenic Biofilms Unveils Oxygen-Mediated Control of Type 1 Pili
- Vaccinia Virus Protein Complex F12/E2 Interacts with Kinesin Light Chain Isoform 2 to Engage the Kinesin-1 Motor Complex
- Modulates Host Macrophage Mitochondrial Metabolism by Hijacking the SIRT1-AMPK Axis
- Human T-Cell Leukemia Virus Type 1 (HTLV-1) Tax Requires CADM1/TSLC1 for Inactivation of the NF-κB Inhibitor A20 and Constitutive NF-κB Signaling
- Suppression of RNAi by dsRNA-Degrading RNaseIII Enzymes of Viruses in Animals and Plants
- Spatiotemporal Regulation of a T4SS Substrate by the Metaeffector SidJ
- Antigenic Properties of the Human Immunodeficiency Virus Envelope Glycoprotein Gp120 on Virions Bound to Target Cells
- Dependence of Intracellular and Exosomal microRNAs on Viral Oncogene Expression in HPV-positive Tumor Cells
- Identification of a Peptide-Pheromone that Enhances Escape from Host Cell Vacuoles
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Dihydrobiopterin in Adult Falciparum Malaria: Association with Disease Severity, Impaired Microvascular Function and Increased Endothelial Activation
- Transgenic Expression of the Dicotyledonous Pattern Recognition Receptor EFR in Rice Leads to Ligand-Dependent Activation of Defense Responses
- Comprehensive Antigenic Map of a Cleaved Soluble HIV-1 Envelope Trimer
- Low Doses of Imatinib Induce Myelopoiesis and Enhance Host Anti-microbial Immunity
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Oxidized Biopterins in Pediatric Falciparum Malaria: Association with Disease Severity
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



