-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
Reklamaα-Hemolysin Counteracts the Anti-Virulence Innate Immune Response Triggered by the Rho GTPase Activating Toxin CNF1 during Bacteremia
The pathogenic potentials of most microbes depend on a repertoire of virulence factors. Despite major progress in the understanding of the molecular mechanisms underlying the activities of bacterial effectors, little is known about how they cooperate during infection to overcome host immune defenses and promote microbial persistence. Here, we investigated the roles of two uropathogenic Escherichia coli (UPEC) effectors that are co-ordinately expressed, α-hemolysin (HlyA) and cytotoxic necrotizing factor 1 (CNF1). We demonstrated that the HlyA toxin is critical for bacterial stability in the blood and showed that one important role of HlyA is to inhibit the CNF1-induced host response. Collectively, these findings reveal why the coordinated activities of HlyA and CNF1 are necessary for the full virulence of UPEC. Moreover, they unravel a HlyA-driven counter-defense mechanism used by bacteria to facilitate their survival.
Published in the journal: . PLoS Pathog 11(3): e32767. doi:10.1371/journal.ppat.1004732
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004732Summary
The pathogenic potentials of most microbes depend on a repertoire of virulence factors. Despite major progress in the understanding of the molecular mechanisms underlying the activities of bacterial effectors, little is known about how they cooperate during infection to overcome host immune defenses and promote microbial persistence. Here, we investigated the roles of two uropathogenic Escherichia coli (UPEC) effectors that are co-ordinately expressed, α-hemolysin (HlyA) and cytotoxic necrotizing factor 1 (CNF1). We demonstrated that the HlyA toxin is critical for bacterial stability in the blood and showed that one important role of HlyA is to inhibit the CNF1-induced host response. Collectively, these findings reveal why the coordinated activities of HlyA and CNF1 are necessary for the full virulence of UPEC. Moreover, they unravel a HlyA-driven counter-defense mechanism used by bacteria to facilitate their survival.
Introduction
Bacteremia caused by extraintestinal strains of pathogenic Escherichia coli is a leading cause of death worldwide [1,2]. Among these pathogens, uropathogenic E. coli (UPEC) is a major etiological agent of bacteremia [1,2]. Therefore, it is essential to define the mechanisms by which virulence factors of UPEC and innate immune signaling pathways control the bacterial burden in the blood.
The major virulence factors of UPEC have been characterized at the molecular level [3–6]. These factors include the presence of a specialized adhesive appendage and specific metabolic pathways as well as protein toxins; together, these features enable UPEC to efficiently colonize the urinary tract and translocate into the bloodstream of the host [7–9]. Two highly prevalent bacterial toxins, α-hemolysin (HlyA) and cytotoxic necrotizing factor-1 (CNF1), work together to damage and disrupt the cohesion of the uroepithelium, which additionally leads to the worsening of the host inflammatory reaction [10–12]. The high prevalences of hlyA and cnf1 in uroseptic strains of UPEC suggest the possible functions of both of these toxins during bacteremia [9,13,14]. In contrast, it has not been determined whether both toxins contribute to the pathogen burden during bacteremia. HlyA belongs to a group of pore-forming leukotoxins that contain RTX repeats [13–15]. Depending on its concentration and on the type of cell intoxicated, HlyA either displays cytolytic activity or hijacks innate immune signaling pathways [13,16–18]. However, its role during bacteremia remains to be determined. The gene encoding the CNF1 toxin is located downstream from the α-hemolysin operon and is co-expressed with HlyA [19,20]. All CNF1-positive uroseptic strains display a hemolytic phenotype [9]. The CNF1 toxin possesses an enzymatic activity that is responsible for the posttranslational deamidation of a specific glutamine residue in a subset of small Rho GTPases, namely, Rac, Cdc42 and RhoA [21]. This type of modification increases the flux of activated Rho proteins and augments signaling through their downstream signaling pathways [21].
The activation of small Rho GTPases by virulence factors is a common trait of various enteric and extraintestinal Gram-negative pathogens. This activation of Rho GTPases confers upon bacteria the property to invade epithelial cells and tissues as well as to hijack inflammatory cell responses [22–25]. Emerging studies have indicated that cells are capable of perceiving the abnormal activation of Rac/Cdc42 induced by virulence factors of pathogens and translating this information via NOD1 and RIP1/RIP2 kinase signaling pathways into danger signals [26,27]. This innate immune mechanism involving the sensing of pathogens is here referred to as anti-virulence immunity (AVI), and it shares similarities with effector-triggered immunity (ETI), the mechanism by which plants sense the activities of bacterial effectors [28,29]. It will be important to define whether and how AVI triggers pathogen destruction in collaboration with the recognition of conserved microbial-associated patterns by pattern recognition receptors (PRR).
Inflammatory caspases, such as caspase-1 and caspase-11, drive innate immune responses to a variety of bacterial stimuli, such as microbe-associated molecular patterns (MAMPs) [e.g., lipopolysaccharide (LPS) or muramyl dipeptide (MDP)], as well as toxin-driven membrane damage [30]. Pathogen perception by NOD-like receptors triggers the assembly of inflammatory caspases in an operational ASC-dependent inflammasome complex that carries out the processing and release of the pro-inflammatory cytokine IL-1β [31,32]. The activation of inflammatory caspases by various type III injected effectors of Salmonella, notably those activating the small GTPase Rac, largely accounts for the induction of inflammatory responses triggered by enteric epithelial cells [33–35]. The means by which pathogenic bacteria overcome inflammatory responses, notably those driven by caspases, and succeed in infecting their hosts largely remains to be elucidated.
Here, we investigate the manner by which virulence factors of UPEC and innate immune signaling pathways impact the outcome of bacteremia. To this end, we focused on the role of CNF1 and HlyA, two toxins produced by UPEC, on pathogen burden in the bloodstream and on animal survival.
Results
CNF1 activity decreases pathogen load and favors host survival during bacteremia
We first assessed the role of the CNF1 toxin in determining E. coli burden during the course of bacteremia in the absence of interference from the other toxin, HlyA. For this purpose, we generated both an hlyA- deletion mutant (referred to as E. coli CNF1+) and an hlyA-cnf1- double deletion mutant (referred to as E. coli CNF1-) from E. coli WT UTI89. By characterizing the strains at the genetic and functional levels, we determined that the two mutants and the wild-type strain had identical growth properties (S1 and S2 Figs). BALB/c mice were then infected intravenously with E. coli CNF1+ or E. coli CNF1- isogenic strains, and the pathogen load was monitored by the serial dilution of blood samples and the enumeration of CFUs (Fig. 1A). We found that the kinetics of clearance from the bloodstream of the E. coli CNF1+ strain was very different (Fig. 1A). The E. coli CNF1+ strain was rapidly cleared, with no bacteria detectable at 48 h p.i. compared with E. coli WT and E. coli CNF1-, which produced 104 and 103 CFU/mouse, respectively, at 48 h p.i. (Fig. 1A). We next assessed whether the rapid clearance of the E. coli CNF1+ strain was actually due to the enzymatic activity of CNF1. We tested this hypothesis by complementing the E. coli CNF1- strain with either an expression vector of wild-type CNF1 (E. coli CNF1 - pcnf1) or an expression vector of the catalytically inactive mutant CNF1 C866S (E. coli CNF1 - pcnf1 C866S) (Fig. 1B). We found that E. coli CNF1 - pcnf1 bacteria were cleared more rapidly from the blood than E. coli CNF1 - pcnf1 C866S (Fig. 1B). Together, these results indicate that CNF1 activity promotes the eradication of bacteria from the bloodstream.
Fig. 1. Infection with E. coli encoding CNF1 triggers clearance of bacteria from blood and mouse survival. 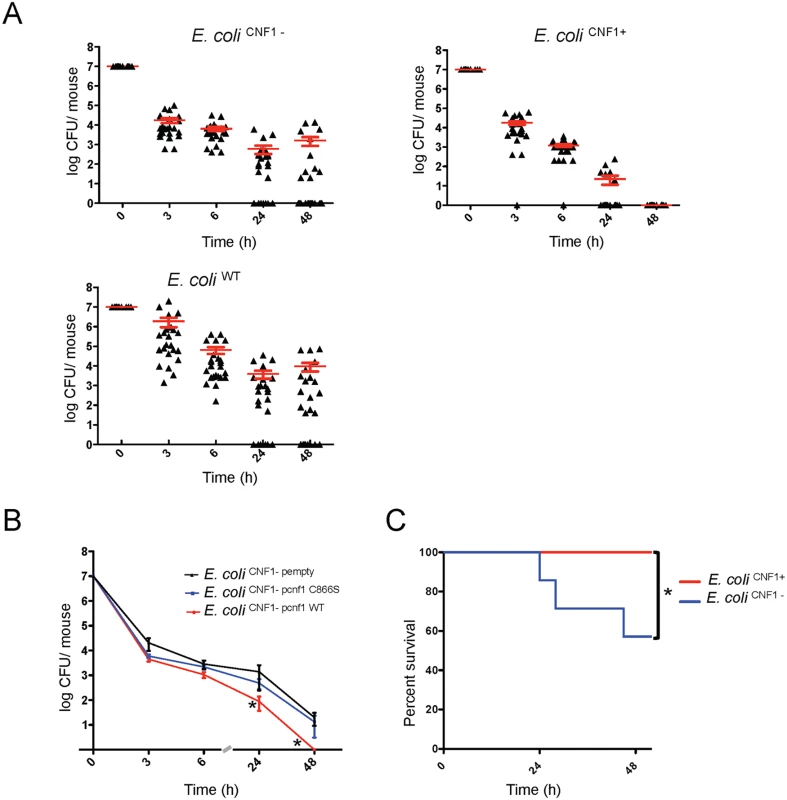
(A) Female BALB/c mice were intravenously infected with 107 CFUs of E. coli expressing CNF1, E. coli UTI89ΔhlyA (E. coli CNF1+), or with the isogenic mutant UTI89ΔhlyA/Δcnf1 (E. coli CNF1-) or the parental strain UTI89 (E. coli UTI89) prior to the collection of peripheral blood at 3, 6, 24 and 48 h for the measurement of bacteremia (n = 25–30). The red line indicates the mean values. (B) Female BALB/c mice were intravenously infected with 107 CFUs of the E. coli CNF1- strain transformed with a control empty plasmid (E. coli CNF1- pempty), with a plasmid encoding the CNF1-inactive mutant C866S (E. coli CNF1- pcnf1 C866S) or with a plasmid encoding E. coli CNF1 WT (E. coli CNF1-pcnf1 WT) prior to the collection of peripheral blood at 3, 6, 24 and 48 h for the measurement of bacteremia (n = 6–12). For both (A) and (B), the data are expressed as the mean ± SEM (n = 6–30) at a *p<0.05. (C) BALB/c mouse survival was monitored for 52 h after intravenous injection of 2.108 CFUs of E. coli CNF1+ or the isogenic mutant, E. coli CNF1- (n = 20). *p<0.05 using the Gehan-Breslow-Wilcoxon chi-squared test. To discern whether there is a link between the effects of CNF1 on pathogen burden and strain virulence, we monitored the deaths of infected animals. To this end, E. coli CNF1- bacteria were injected at a dose sufficient to kill half of the mice by 48 h p.i. Animal survival following injection with the different isogenic mutants was monitored (Fig. 1C). We found that all of the mice infected with E. coli CNF1+ survived, whereas the group of mice infected with E. coli CNF1- displayed only 57% survival (Fig. 1C).
Taken together, our data establish that CNF1 activity has a detrimental effect on the bacterial burden in the blood and that it protects against pathogen-induced animal death.
CNF1 potentiates the LPS-triggered secretion of IL-1β in an inflammatory caspase-dependent manner
We hypothesized that CNF1 activity has a negative impact on bacterial burden via the modulation of LPS-driven antimicrobial host responses. We assessed this conjecture by profiling the cytokines and chemokines secreted by primary monocytes isolated from the blood of mice after various experimental treatments. The monocytes were challenged with ultrapure LPS, with CNF1 alone, or with a combination of both factors. We used an unbiased approach that utilized an ELISArray semi-quantitative cytokine/chemokine screen to measure the levels of the following factors: IL-1β, TNFα, KC, IL-6, IL-1α, MIP1α, MIP1β, RANTES, MCP1, IL-12, MDC, MIG, IL17, IP10, TARC, EOTAXIN, IL-2, IL-4, IL-5, IL-10, IL-13, IL-23, INFγ, TNFβ1, GM-CSF, and G-CSF. Fig. 2A shows the panel of cytokines that were synergistically induced by LPS+CNF1 compared to ultrapure LPS or CNF1 alone. The results are presented as fold inductions compared to untreated monocytes for each treatment condition Figs. (2A and S3). The panel of cytokines produced by the monocytes treated with CNF1 alone is presented (S4A Fig). Other molecular mediators, such as the IL-4 and IL-10 anti-inflammatory cytokines, showed no significant induction in the monocytes treated with LPS+CNF1 (Fig. 2A). In support of our in vitro analysis results, we measured higher increases in these inflammatory mediators in the sera of mice infected with E. coli CNF1+compared with E. coli CNF1- (S4B Fig). Collectively, these results show that CNF1 activity potentiates the LPS-triggered secretion of the pro-inflammatory cytokines IL-1β, TNFα, and IL-6 primarily, as well as the secretion of the chemokines MCP1, MIP1α, MIP1β, and KC.
Fig. 2. CNF1 potentiates LPS-triggered immune responses. 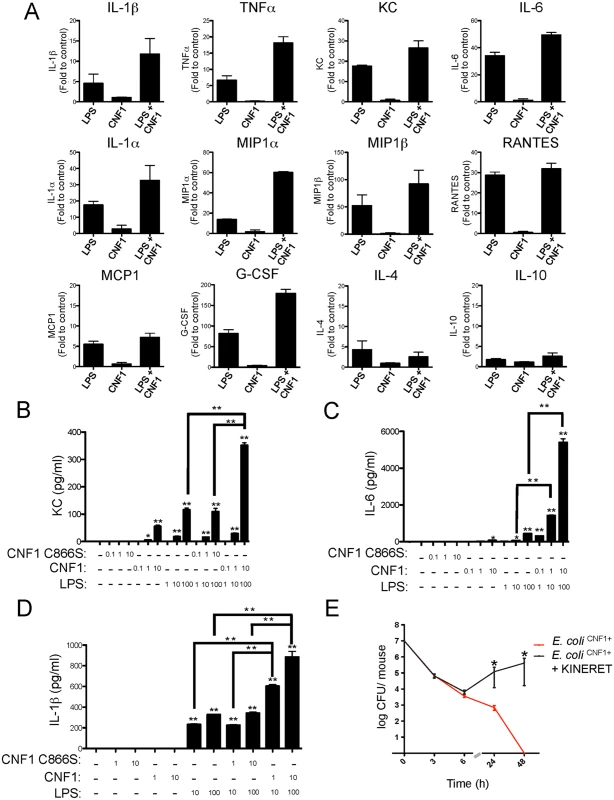
(A) Monocytes (5x105 cells per condition) isolated from mouse blood were treated with PBS (control) or with 1 μg/ml CNF1 toxin for 10 h with or without 100 ng/ml LPS. Cell culture supernatants were analyzed using mouse ELISArray kits (n = 3). The data are shown as fold inductions compared with the control condition. (B, C and D) Monocytes (5x105 cells per condition) purified from mouse blood were treated for 10 h. Cells were treated as indicated with 0.1, 1, or 10 μg/ml of CNF1 toxin or the CNF1 mutant C866S alone or in combination with ultrapure E. coli LPS at 1, 10 or 100 ng/ml (n = 3). (B) KC, (C) IL-6, and (D) IL-1β cytokine secretion was analyzed using ELISA (n = 4). (E) Female BALB/c mice were intravenously infected with 107 CFUs of E. coli expressing CNF1 (E. coli CNF1+) + PBS as a control or with an E. coli CNF1+ + IL-1β antagonist (Kineret; 1.5 mg/kg) prior to the collection of peripheral blood at 3, 6, 24 and 48 h for the measurement of bacteremia (n = 10). P-values <0.05 (*); and P-values <0.01 (**) were considered statistically significant. We next performed quantitative analysis of the impact of CNF1 activity on the monocyte responses to LPS. Primary monocytes isolated from the blood of naïve mice were treated with endotoxin-free CNF1 or with the catalytically inactive mutant CNF1 C866S. In the monocytes treated with recombinant purified CNF1, we observed the production of KC (75 +/ - 5 pg/ml), which was strictly dependent upon the activity of CNF1 (Fig. 2B). Control experiments with ultrapure LPS alone or in combination with the catalytically inactive mutant CNF1 C866S triggered the secretion of KC (120 +/-10 pg/ml). Strikingly, we observed a 3-fold increase in the production of KC (350 +/-10 pg/ml) in the cells treated with both LPS and CNF1 compared with those treated with ultrapure LPS alone (Fig. 2B). The co-stimulation of monocytes with CNF1 and LPS resulted in a 12-fold increase in IL-6 secretion and a 2-fold increase in IL-1β secretion compared with stimulation with LPS alone (Fig. 2C and D). This synergy was detected with doses of CNF1 as low as 10 ng/ml and was of the same magnitude as that observed with the ATP treatment (S4C Fig). We also observed that CNF1 acts synergistically with other Toll-like receptors (TLR) ligands, such as Pam3CSK4 (TLR1/TLR2 agonist) and FSL-1 (TLR2/TLR6 agonist) (S4D Fig). IL-1β is an important mediator of inflammatory responses and is notably important in enabling the host to mount an efficient antibacterial immune response [36,37]. As a first approach, to evaluate the role of IL-1β in the elimination of pathogens, we treated infected mice with an IL-1β antagonist (KINERET). We found that this treatment dramatically antagonized the clearance of E. coli CNF1+ (Fig. 2E).
Collectively, our data pointed for the importance of the synergic induction of IL-1β by LPS+CNF1 in promoting efficient bacterial clearance from the bloodstream.
CNF1 anti-virulence immunity is mediated by caspase-1
Given the critical role of IL-1β signaling in the elimination of bacteria, we next aimed to precisely determine the components required for IL-1β maturation. IL-1β is expressed as a proform (proIL-1β) that is processed by caspases-1/11 to generate the mature p17-secreted active form [38,39]. We further assessed the effects of the interplay between LPS and CNF1 on IL-1β maturation. This interaction was evaluated by immunoblotting to determine the level of secretion of p17 IL-1β into the medium of monocytes upon co-stimulation with LPS+CNF1 (Fig. 3A and B). Our results confirmed that CNF1 acts at the level of IL-1β maturation/secretion rather than at the level of IL-1β expression (Fig. 3A and B). We observed the complete inhibition of the release of p17 IL-1β in the monocytes treated with the pan-caspase inhibitor QVD as well as in the monocytes isolated from caspase-1/11 (C1-C11)-impaired mice (Fig. 3A and B). Notably, these results indicate that CNF1 plays a critical role in promoting the caspase-1/11-dependent maturation/secretion of IL-1β by monocytes challenged with LPS. We next assessed the interplay between inflammatory caspases and CNF1 during UPEC-induced bacteremia. To this end, we measured bacterial loads in the blood of C1-C11-impaired mice infected with E. coli CNF1+ or E. coli CNF1-. We compared the kinetics of the bacterial burden in these animals with those of their wild-type congenic C57BL/6 littermates. In the wild-type animals, we measured a decrease in the bacterial load in the animals infected with E. coli CNF1+, with no bacteria detectable in the blood at 48 h, compared with the presence of 107 CFU/animal in the blood of the mice infected with E. coli CNF1- (Fig. 3C). In contrast with the wild-type mice, the E. coli CNF1+ burden in the C1-C11-impaired mice remained high, reaching 105 CFU/animal at 48 h p.i. (Fig. 3D). These results indicate that inflammatory caspases-1/11 play major roles in the clearance of bacteria from the bloodstream as triggered by CNF1.
Fig. 3. CNF1-triggered immunity requires inflammatory caspases. 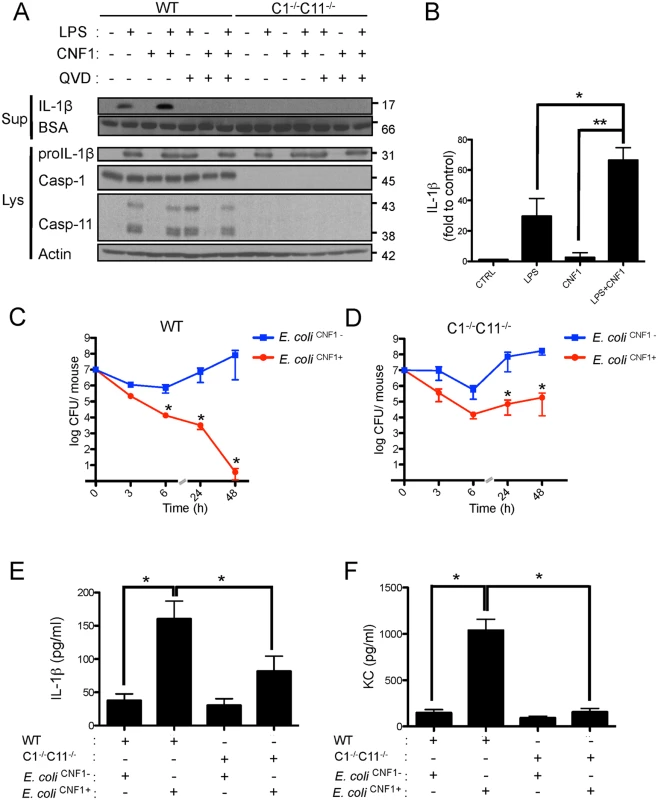
(A and B) IL-1β production and maturation/secretion after treatment with CNF1, LPS or CNF1 (1 μg/ml) + LPS (100 ng/ml) for 10 h. Actin and BSA were used as loading controls. (B) Graph showing the quantification of IL-1β secretion normalized to the control (n = 3). (C and D) Female C57BL/6 WT (C) or congenic C1-/-C11-/- mice (D) were intravenously infected with 107 CFUs of E. coli CNF1+ or with the isogenic CNF1-deleted mutant (E. coli CNF1-) prior to the collection of peripheral blood at 3, 6, 24 and 48 h for bacteremia measurements. The data are expressed as the mean ± SEM (n = 7–8). (E and F) Analysis of the circulating levels of IL-1β (E) and KC (F) in the mouse sera by ELISA. Serum samples from mice infected with 107 E. coli CNF1+ or with the isogenic mutant E. coli CNF1- were collected at 3 h after intravenous infection and analyzed by ELISA (n = 3). P-values<0.05 (*); and P-values<0.01 (**) were considered statistically significant. In an approach designed to be complementary to our functional approach, we analyzed the roles of inflammatory caspases in the initiation of the CNF1-dependent innate immune responses during bacteremia. To this end, we measured the levels of the IL-1β and KC cytokines in the sera of infected C1-C11-impaired mice and their congenic wild-type littermates. The wild-type infected with E. coli CNF1+ displayed higher levels of IL-1β and KC than the WT mice infected with E. coli CNF1- (Fig. 3E and F). Interestingly, in the C1–11-impaired mice infected with E. coli CNF1+, we measured dramatic decreases in the levels of KC and IL-1β in the sera compared with the wild-type mice (Fig. 3E and F). This finding is consistent with the fact that inflammatory caspases-1/11 are critical determinants in the CNF1-triggered cytokine response during bacteremia. Next, we aimed to determine whether the CNF1-triggered IL-1β secretion involved caspase-1 or caspase-11 using bone marrow-derived macrophages isolated from caspase-1 or caspase-11 single knock-out mice. Additionally, we analyzed the effect of CNF1 on macrophages of mice in which the inflammasome adaptor ASC was knocked out. We measured the inhibition of CNF1-triggered IL-1β secretion in macrophages isolated from caspase-1 or ASC but not caspase-11 knock-out mice (Fig. 4A). Thus, our analysis pinpointed the major roles of caspase-1 and ASC in the secretion of IL-1β triggered by CNF1+LPS. In addition, we measured that the activity of caspase-1 increased when monocytes were treated with LPS+CNF1 (Fig. 4B). We then investigated the role of the GTPase Rac in IL-1β secretion triggered by CNF1+LPS. To this aim, we expressed a mutant of Rac2 bearing the modification catalyzed by CNF1 (Rac2Q61E). We observed that the expression of Rac2Q61E in the macrophages challenged with LPS promoted the secretion of the p17 form of IL-1β (Fig. 4C). Finally, we were able to detect an association between Rac2 and caspase-1 upon CNF1+LPS treatment specifically by co-immunoprecipitation (Fig. 4D).
Fig. 4. CNF1-triggered IL-1β maturation requires activated Rac, ASC and caspase-1. 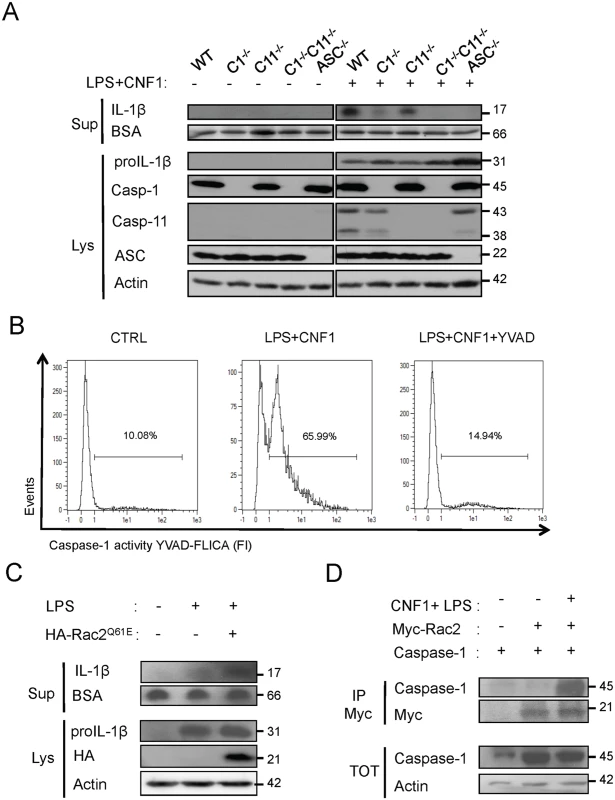
(A) Western blot analysis of the production and maturation/secretion of IL-1β by primary macrophages following treatment with CNF1, LPS or CNF1+LPS for 10 h. Actin and BSA were used as loading controls. (B) Quantification of caspase-1 activity in macrophages following treatment with CNF1+LPS for 6 h using YVAD-Fluorescent Labelled Inhibitor Caspase-1 Activity (FLICA). (C) Western blot analysis of macrophages IL-1β maturation/secretion upon transfection of HA-Rac2Q61E and LPS treatment. (D) Co-immunoprecipitation of Myc-Rac2 and caspase-1 using an anti-Myc antibody following the treatment of HEK 293T cells with CNF1+LPS for 6 h. Our results identify the Rac/caspase-1/IL-1β signaling axis as a major component of the CNF1-induced elimination of Escherichia coli from the bloodstream.
HlyA counteracts CNF1-triggered immunity
We sought to determine the manner by which pathogenic bacteria cope with anti-virulence immunity. Although HlyA has been shown to interfere with innate immune responses that occur during urinary tract infections, its role during bacteremia is still unknown. We experimentally addressed this question by analyzing the effect of HlyA on CNF1-triggered protection against bacteremia in mice. We observed that all E. coli strains expressing HlyA displayed increased stability in the blood compared with the other strains, independent of the presence or absence of CNF1 (Fig. 5A). We noticed a reduced bacterial burden in the E. coli CNF1- strain compared with E. coli HLY+ CNF1-. This reduction in the bacterial burden was the greatest in the E. coli CNF1+ strain. We next complemented the E. coli CNF1+ strain with a plasmid encoding HlyA (E. coli CNF1+ phlyA). We found that the complementation of E. coli CNF1+ with the HlyA expression plasmid stabilized the bacterial load in the blood (Fig. 5B). These results show that HlyA protects E. coli against host responses, particularly those triggered by CNF1, thereby promoting bacterial stability in the blood.
Fig. 5. HlyA counteracts the CNF1-induced host response. 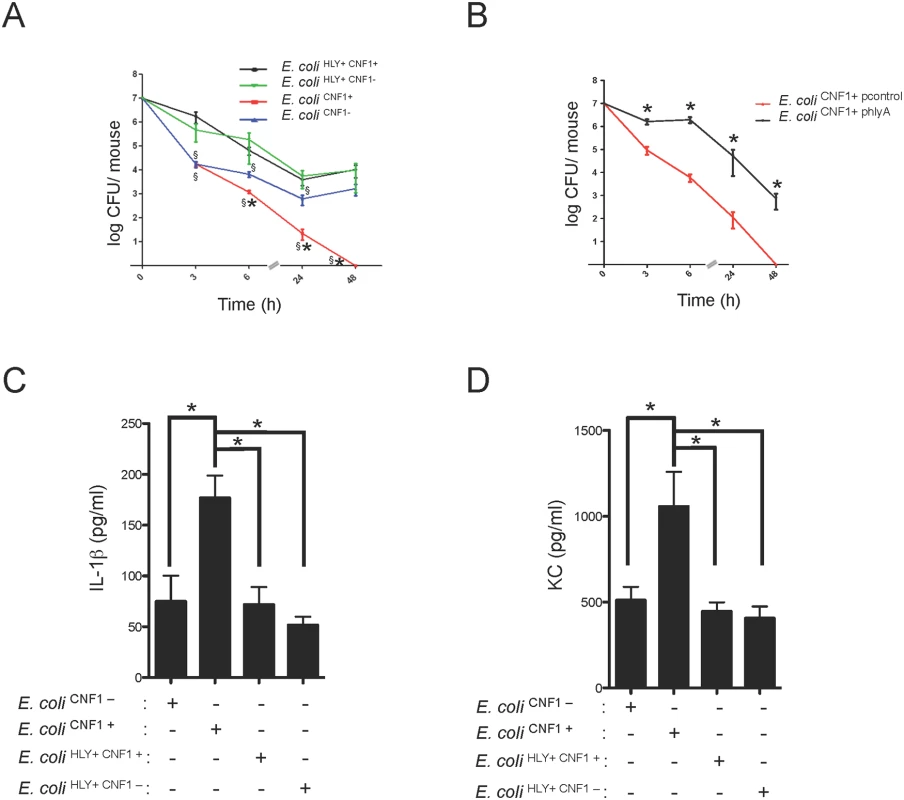
(A) Female BALB/c mice were intravenously infected with 107 CFUs of E. coli expressing CNF1 (E. coli CNF1+) or with 107 CFUs of one of the isogenic mutants, E. coli CNF1-, E. coli HLY+ CNF1+ or E. coli HLY+CNF1-, prior to the collection of peripheral blood at 3, 6, 24 and 48 h for bacteremia measurements. The data are presented as a line graph (n = 20–30; mean ± SEM). P-values<0.05 compared to E. coli CNF1- are indicated by (*); and P-values<0.05 compared to E. coli HLY+ CNF1+ are indicated by (§). (B) Female BALB/c mice were intravenously infected with 107 CFUs of the UTI89ΔhlyA strain transformed with a control empty plasmid (E. coli CNF1+ pcontrol) or with a plasmid encoding HlyA (E. coli CNF1+ phlyA) prior to the collection of peripheral blood at 3, 6, 24 and 48 h for bacteremia measurements (n = 6–12). (C and D) Analysis of the circulating levels of IL-1β and KC in the sera of C57BL/6 mice were measured by ELISA. Serum samples from mice infected with 107 CFUs of E. coli HLY+CNF1+ or with 107 CFUs of one of the isogenic mutants, E. coli CNF1+, E. coli HLY+CNF1-, or E. coli CNF1-, were collected at 3 h after intravenous infection and analyzed by ELISA (n = 6). P-values<0.05 (*); and P-values <0.01 (**) were considered statistically significant. We went on to determine at which level HlyA acts to prevent CNF1-triggered bacterial clearing. We first investigated whether HlyA has a blocking effect on the activation of the GTPase Rac by CNF1. We detected no modification in the CNF1-mediated activation of Rac when cells were co-incubated with HlyA (S5 Fig). Thus, we hypothesize that HlyA acts downstream of the activation of Rac at the level of cytokine production/secretion. Consistent with this hypothesis, we measured lower levels of IL-1β and KC in the bloodstream of the mice infected with HlyA-expressing E. coli strains (Fig. 5C and D).
Taken together, our data indicate that HlyA inhibits CNF1-induced pro-inflammatory cytokine responses.
Gr1+ cells are crucial effectors of anti-E. coli responses in the blood
Next, we aimed to further identify the key immune effector cells that control the rapid clearance of E. coli exacerbated by the Rho-activating toxin CNF1 during bacteremia. We monitored the levels of circulating innate immune cells in the blood at an early time period of infection with either the E. coli CNF1+ strain or the E. coli CNF1- strain. The data were analyzed as the percent of CD45-positive cells, a white blood cell marker, to exclude contamination by red blood cells. We first monitored circulating innate immune cells, including monocytes, neutrophils and granulocytes, using the CD11b marker. Interestingly, we measured higher percentages of CD11b+/CD45+ cells at both 3 h and 6 h p.i. in the blood of the mice infected with E. coli CNF1+ (3 h: 46% and 6 h: 64%) compared with E. coli CNF1- (3 h: 23% and 6 h: 43%). Control mice injected with PBS showed lower levels of CD11b+/CD45+ (3 h: 18% and 6 h: 22%) (Fig. 6A). We found that chemokines, such as MIP1α, MIP1β, MCP-1, and RANTES as well as KC were secreted by the monocytes treated with CNF1 in vitro and were increased in the sera of the mice at 3 h p.i. for the E. coli CNF1+ strain compared with E. coli CNF1- (S4B Fig). Because these chemokines are involved in chemotaxis as well as in the activation of neutrophils, we hypothesized that the clearance of the bacteria was due to cooperation between inflammatory monocytes and neutrophils. To test this hypothesis, we monitored the subpopulation of Gr1+ cells, including inflammatory monocytes and neutrophils, in the blood of infected mice. Mice infected with E. coli CNF1+ showed 37% and 58% Gr1+CD45+ cells at 3 h and 6 h, respectively, compared with only 21% and 40% when the mice were infected with E. coli CNF1- (Fig. 6B). These findings indicated that there was an increased recruitment of Gr1+ cells triggered by CNF1. The recruitment of Gr1+ cells was not reduced in the C1-C11 impaired mice (S6A Fig). Furthermore, the recruitment triggered by CNF1 was not impaired in HlyA-expressing E. coli (S6B Fig). To demonstrate the key role of Gr1+ cells in the clearance of E. coli expressing CNF1, we depleted this subpopulation, including inflammatory monocytes (Gr1+ F4/80+) and neutrophils (Gr1+ F4/80-), prior to infection. We measured an 80% reduction in the Gr1+ cell population following the injection of anti-Gr1+ (Ly-6G) monoclonal antibodies (RB6–8C5) (S7 Fig). We found that the depletion of Gr1+ cells was sufficient to block E. coli clearance during bacteremia and to prevent the rapid clearance of the E. coli CNF1+ strain (Fig. 6B).
Fig. 6. Gr1+ cell subpopulations are necessary for the rapid clearance of the CNF1+ strain from the blood. 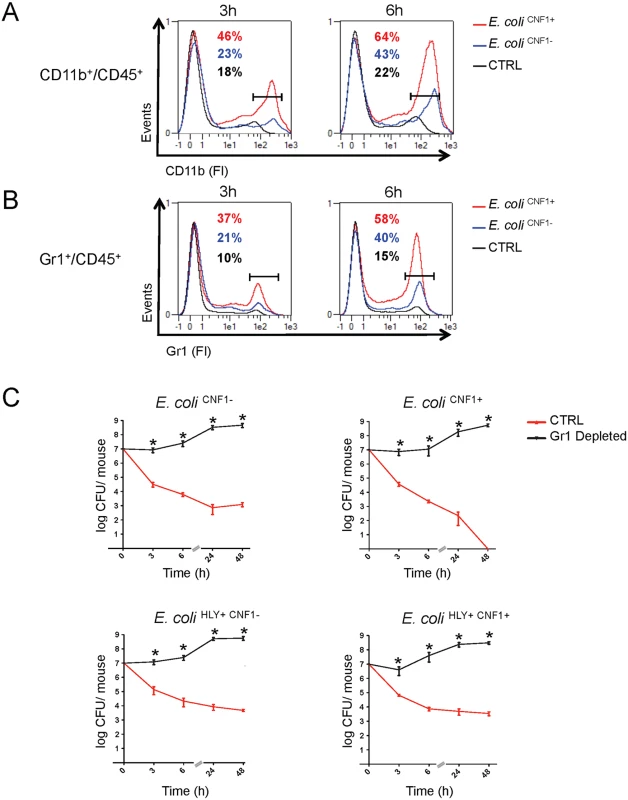
(A and B) Flow cytometry analysis of peripheral blood isolated from mice intravenously infected with 107 CFUs of E. coli CNF1+, E. coli CNF1- or PBS (control). Percentages of cells expressing (A) CD45 and CD11b or (B) CD45 and Gr1 are indicated (n = 5). (C) BALB/c mice were injected either with an anti-Gr1 antibody or vehicle, and after 48 h, they were inoculated intravenously with 107 CFUs of E. coli HLY+CNF1+ or with 107 CFUs of one of the isogenic mutants, E. coli CNF1+, E. coli HLY+CNF1-, or E. coli CNF1-, prior to the collection of peripheral blood at 3, 6, 24 and 48 h for the measurement of CFUs via the plating of serial dilutions. P-values<0.05 (*) were considered statistically significant. Altogether, we show that Gr1+ recruitment is required but is not sufficient for the anti-virulence immunity triggered by the Rho GTPase-activating toxin CNF1. In addition, we demonstrate that Gr1+ cells are critical for the clearance of E. coli from the bloodstream.
Discussion
The sensing of the activities of pathogen-encoded virulence factors is emerging as a paradigm of innate immune sensing. However, in vivo proof of the contribution of such sensing to mammalian immunity during infection is still scarce. Furthermore, the mechanisms by which pathogenic bacteria cope with the capacities of hosts to detect their virulence remain to be elucidated. As a major discovery, we demonstrated here the capacity of the host to control bacteremia through the exacerbation of LPS-driven IL-1β-mediated antimicrobial responses by CNF1 activity. This host feature relies on Rac/ASC/caspase-1 and the secretion of pro-inflammatory cytokines/chemokines, which in turn mobilize Gr1+ cells. Importantly, we described a yet unappreciated role of HlyA in impairing these innate immune responses downstream or in parallel with Gr1+ cell recruitment. We also established that pathogen burden and animal death were maximal for the HlyA-positive strains.
Inappropriate, excessive or absent innate immune responses have dramatic consequences on human health. Thus, it is critical to decipher the manner by which the host determines the pathogenic potential of a microbe and responds commensurately [40,41]. It is currently unclear how anti-virulence immunity (AVI) systems of detection work together with the recognition of MAMPs to control inflammation and bacterial virulence. Because CNF1 intoxicates cells without the involvement of additional bacterial factors, it can be used to address this critical question. In this work, we report that the detection of CNF1 activity amplifies the cellular LPS response to a large panel of pro-inflammatory cytokines, including IL-1β, by 2 - to 12-fold, thereby producing a more potent immune response. Analysis of the IL-1β level in the sera of mice infected with E. coli CNF1+ indicated that they exhibited a 3-fold higher response and better resistance to infection than those infected with isogenic E. coli CNF1-. Our study provides both in vitro and in vivo evidence that AVI works in concert with MAMP-triggered responses to amplify the innate immune response and ultimately improve host viability. We speculate that this cooperation between AVI and MAMP-triggered immunity is a means by which the host gauges the pathogenic potential of a microbe and tailors a response commensurate with the estimated threat level.
The sensing of the activities of bacterial virulence factors has recently emerged as a conserved means of detecting pathogens. Rho GTPases are targeted by various virulence factors encoded by pathogenic bacteria. These virulence factors either post-translationally modify Rho GTPases by deamidation, glucosylation, adenylylation, or ADP-ribosylation or mimic exchange factors or GTPase-activating proteins, thereby hijacking the GTP/GDP cycle and causing inappropriate activation or inactivation of the critical regulators of these cycles, which include Rho, Rac and Cdc42 GTPases [21]. Interestingly, recent studies have indicated that animal hosts have evolved dedicated strategies for detecting the activities of these virulence factors [26,27,42,43]. Indeed, based on our work focusing on CNF1 and studies of Salmonella typhimurium SopE/E2 virulence factors, we can speculate that the abnormal activation of Rac/Cdc42 triggers the assembly of an anti-virulence immune complex involving NOD1 and RIP kinases to promote NF-κB activation and in parallel to assemble a Rac/ASC/caspase-1 complex for the maturation of IL-1β during infections [27,35]. In addition, a recent study has implicated the NLR pyrin as a sensor of the inactivation of the Rho GTPase RhoA by virulence factors via a mechanism that leads to the activation of the pyrin inflammasome and inflammatory caspase-1 [42]. Taken together, these studies indicate that, in parallel with the PRR detection of MAMPS, the host monitors changes in the GTP/GDP cycles of Rho GTPases rather than monitoring each post-translational modification individually, a process that would require a large repertoire of receptors.
Our observation that CNF1 induces an immune response that is detrimental to bacteria raises the question of why CNF1 has been evolutionarily conserved in the UPEC genome. Several reports have established that the CNF1 toxin can trigger the disruption of epithelial cell junctions, promote cell migration and induce the internalization of bacteria into epithelial cells [25]. One hypothesis is that CNF1 has been evolutionarily conserved as an invasion factor to help bacteria cross epithelia during the early stages of infection. Our results led us to speculate that CNF1 has become genetically associated with HlyA to protect the bacteria from an otherwise detrimental CNF1-induced innate immune response. Indeed, the CNF1 and HlyA toxins are co-transcribed within a highly conserved PAI, and epidemiological studies have established that CNF1 is always expressed in association with HlyA [12,19,44,45]. Interestingly, the functional relationship between these toxins described here offers a new framework through which to understand the molecular basis of their tight genetic link and explains why E. coli that express both of these toxins are pathogenic to mammals. Consistent with this idea, our work sheds light on the manner by which pathogenic bacteria cope with AVI. We report a yet unappreciated role of HlyA in the impairment of innate immune responses. In this role, HlyA has major influences on bacterial burden and host viability. Our genetic analysis revealed that HlyA protects microbes from both CNF1-dependent and CNF1-independent detrimental effects. Mice infected with E. coli expressing CNF1, but not HlyA, showed an increase in the IL-1β and KC proinflammatory cytokine levels. Given that the bacterial load is minimal under these conditions, this finding cannot be ascribed to increased LPS exposure. It is possible that HlyA targets host immune cell signaling to prevent the production of inflammatory cytokines. This idea is supported by our findings that HlyA did not impair the CNF1-induced activation of Rac, although it blocked the secretion of IL-1β. Further, we showed that bacteria expressing HlyA, but not CNF1, showed an increase in persistence of one log unit in the blood compared with bacteria that were deficient in both toxins. Although HlyA acted primarily to counteract the host recognition of CNF1 activity in our model, it most likely has additional effects on other components of the innate immune response to E. coli, including phagocytosis or detection by the immune system of other bacterial components [18]. A common feature of pathogenic bacteria is the production of a wide range of pore-forming toxins of various sizes, which have specific ionic and molecular selectivities. It will be important to establish which types of pore-forming toxins are able to block innate immunity and to what extent HlyA blocks the recognition of other factors produced by E. coli.
Multicellular organisms have evolved sophisticated defense mechanisms to counter microbial attack. In turn, successful microbial pathogens have evolved strategies to overcome host defenses, leading to the occurrence of diseases or chronic infections [46]. In plants, a similar system of detection of the activities of virulence factors has been termed “effector-triggered immunity” [28]. Interestingly, in this model, the pathogen-evolved mechanism counteracting the innate immune defense response has been called a “counter-defense mechanism” [47]. In our model, HlyA counteracts the CNF1-induced host cytokine response. By analogy, if we consider that CNF1 is sensed by the innate immune defense system, HlyA must be considered as a counter-defense effector used by E. coli to counteract the CNF1-induced host response. The data presented in the present work support a model in which HlyA acts as a major virulence factor that protects microbes from both CNF1-dependent and CNF1-independent innate immune defenses during bacteremia. Based on this model, pore-forming toxins might represent viable drug targets for the treatment of UPEC bacteremia.
Materials and Methods
Ethics statement
This study was carried out in strict accordance with the guidelines of the Council of the European Union (Directive 86/609/EEC) regarding the protection of animals used for experimental and other scientific purposes. The protocol was approved by the Institutional Animal Care and Use Committee on the Ethics of Animal Experiments of Nice, France (reference: NCE/2012–64).
Bacterial strains and toxins
The E. coli UTI89 clinical isolate was originally obtained from a patient with cystitis [48] and was a kind gift from E. Oswald. The UTI89 streptomycin-resistant (SmR) evolved strain (WT) and isogenic mutants were grown in Luria-Bertani (LB) medium supplemented with streptomycin (200 μg/ml). The CNF1 strain was transformed with a pQE30 plasmid (Qiagen) (E. coli CNF1 - pempty), with pQE30-CNF1 (E. coli CNF1 - pcnf1 WT) or with pQE30-CNF1 C866S (E. coli CNF1 - pcnf1 C866S) and grown in LB supplemented with ampicillin (100 μg/ml) plus IPTG (200 μM) for the infection experiments. The E. coli CNF1+ strain was transformed with pBR322 (E. coli CNF1+ pcontrol) or with pEK50 (a plasmid bearing an operon encoding HlyA (hlyCABD)) (E. coli CNF1+ phlyA) and grown in LB supplemented with ampicillin (100 μg/ml). The DH10B K12 E. coli strain (Life technologies) was transformed with either pEK50 (K12-pHlyA) (a plasmid bearing the operon encoding HlyA (hlyCABD)) or the pCR2.1 (K12-pLacZ). The pEK50 plasmid and anti-HlyA antibody were a kind gift from V. Koronakis. The anti-TolC antibody was a kind gift from C. Wandersman. For the infections, a 1/50 dilution of an overnight culture of each strain was inoculated and grown to OD600 = 1.2. Bacteria were either washed in culture medium and diluted to obtain the corresponding MOI for the cell culture infection experiments or were harvested by centrifugation and washed twice in PBS before dilution in PBS to obtain the desired bacterial concentrations for the mouse infection experiments. Recombinant wild-type cytotoxic necrotizing factor-1 (CNF1) and its catalytically inactive form (CNF1-C866S; CNF1 CS) were produced and purified as previously reported [49]. The recombinant proteins were passed through a polymyxin B column (AffinityPak Detoxi-Gel, Pierce). The lack of endotoxin content was verified using a colorimetric LAL assay (LAL QCL-1000, Cambrex). Each stock of the CNF1 preparation (2 mg/ml) was shown to contain less than 0.5 endotoxin units/ml.
Generation of isogenic bacterial mutant strains
The multi-step procedure used to substitute the hlyA and cnf1 genes in the bacterial chromosome was performed as previously described [50]. Briefly, the pMLM135 plasmid (cat, rpsl+) was used to transform the UTI89 streptomycin-resistant (SmR) evolved strain. The integration of pMLM135 into the chromosome was selected by plating cells on chloramphenicol-containing medium at 42°C. Excision of the hlyA or cnf1 gene from the chromosome was selected by plating cells on medium containing streptomycin (200 μg/ml). The chromosomal deletions were verified by PCR and by the monitoring of the loss of HlyA and/or CNF1 activity in the deleted strains (S1 Fig). We verified that the isogenic mutant strains had growth properties that were identical to those of the UTI89 strain (S2 Fig). The sequences of the primers used in this study are available upon request.
Cell lines and primary monocytes
Murine monocytic cells were obtained from pooled blood from 5–10 mice. Monocytes were isolated using a Ficoll-Paque (GE Healthcare) gradient technique. Adherent cells were maintained in M medium [RPMI 1640 medium supplemented with 10% FCS (Lonza), 2 mmol/L L-glutamine, 1 mM pyruvate, 10 mM HEPES, penicillin (100 U/ml), and streptomycin (100 μg/ml)]. When indicated, M-CSF was added as previously described [51]. Cells were transfected using the Amaxa mouse macrophage nucleofector kit according to the manufacturer’s instructions with pCDNA3.1-HA-Rac2Q61E, pRK5-Myc-Rac2, pCAGGS-Caspase-1, pCDNA3.1 or pRK5. Monocyte isolation was confirmed by flow cytometry analysis using F4/80 and CD11b antibodies (Cedarlane). HEp-2 cells and HEK 293T cells were obtained from ATCC (CCL-23 and CRL-3216) and maintained according to ATCC instructions. HEK 293T cells were transfected using Lipofectamine 2000 (Life technologies) according to the manufacturer’s instructions.
Mouse model of infection
Female BALB/c and C57BL/6 mice (6–8 weeks old) were purchased from Janvier (Le Genest St Isle, France). Caspase-1/11-impaired (also designated as ICE KO) and congenic C57BL/6 mice have been previously described and were kindly provided by R. Flavell [52]. These mice are genetically identical to mice that are now also available from Jackson Laboratories (Stock #016621). Caspase-11 knock-out mice have been previously described by VM Dixit (Genentech). Caspase-1 knock-out mice were generated by D. Dieter and M. Lamkanfy. Their generation will be described elsewhere. Mice were injected i.v. with 107 CFUs of E. coli as previously described [8,53]. For the determination of bacteremia, blood was collected from the tail vein at the indicated times post-infection, serially diluted in sterile PBS and plated on LB plates containing streptomycin (200 μg/ml) or ampicillin (100 μg/ml) for the strains transformed with pQE30 - or the pBR322-derived plasmids, and the plates were incubated for 16 h at 37°C. Injection quality was controlled by plating blood samples obtained from the mice at 5 min after injection. Note that the kinetics for the experiments using the transformed strains were terminated after 24 h because we observed that without selective pressure, the plasmid was stable for up to 24 h. For cytokine analysis, plasma was collected (1200×g, 4°C, 5 min) and stored at −20°C.
In vivo Gr1+ cell depletion
Mice were injected intraperitoneally with a monoclonal anti-Gr1 antibody (RB6–8C5, 100 μg/20 g body weight). After 48 h, the depletion of Gr1+ cells was verified in four mice by analyzing F4/80 and/or Gr1-stained white blood cells by flow cytometry. The anti-Gr1-injected mice were then infected with either UTI89 or UTI89 isogenic mutants.
Cytokine assays, Caspase-1 assay and antibodies
ELISArrays were performed according to the manufacturer’s instructions (Qiagen, MEM-003A, MEM-004A, MEM-006A, MEM-008A, and MEM-009A). Cytokine concentrations were determined by ELISA and by IL-1β maturation and visualized by western blotting according to the manufacturer’s instructions (KC, TNFα and IL-6, R&D Systems, USA; IL-1β, Raybiotech, USA). Caspase-1 activity was measured using FAM-YVAD-FMK caspase-1 assay kit (ImmunoChemistry Technologies) according to the manufacturer’s instructions. Antibodies used in this study are: anti-IL-1β/IL-1F2 (R&D systems), anti-Caspase-1 (M-20, SantaCruz), anti-Caspase-11 (17D9, Novus), anti-ASC (AL177, Adipogen), anti-HA (16B12, Covance), anti-Myc (9E10, Roche), anti-Rac (102/Rac1, BD Biosciences), anti-GAPDH (FL335, SantaCruz), anti-β-actin (AC-74, Sigma).
Activated RacGTPase pull-down
Primary monocytes (5x106 cells per condition) were treated with 1 μg/ml of CNF1 toxin for 6 h with or without addition at identical MOI (MOI of 0.5) of live K12 E. coli expressing HlyA (K12-pHlyA) or LacZ (K12-pLacZ) as a control. Cells were lysed at 4°C using a lysis buffer (Tris 25mM pH 7.5, NaCl 150mM, MgCl2 5mM, EGTA 0.5mM, TritonX100 0.5%, glycerol 4%) and Pull-down assays were performed using 50 μg of GST-PAK70–106. Proteins were resolved on 12% SDS-PAGE followed by transfer on PVDF membranes. Equal amount of proteins engaged in the Pull-down assays was confirmed by immunoblotting anti-β-actin.
Statistical analyses
Statistical analyses were performed using Prism V5.0b software (GraphPad, La Jolla, CA). Unless stated otherwise, comparisons between two groups were performed using the Mann-Whitney nonparametric test, and comparisons among three or more groups were conducted with the Kruskal-Wallis test with Dunn's post-test. P-values<0.05 (*) and P-values<0.01 (**) were considered statistically significant.
Supporting Information
Zdroje
1. Martin GS, Mannino DM, Eaton S, Moss M The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003; 348 : 1546–1554. 12700374
2. Russo TA, Johnson JR Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis. 2000; 181 : 1753–1754. 10823778
3. Wiles TJ, Kulesus RR, Mulvey MA Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol. 2008; 85 : 11–19. doi: 10.1016/j.yexmp.2008.03.007 18482721
4. Leimbach A, Hacker J, Dobrindt U E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol. 2013; 358 : 3–32. doi: 10.1007/82_2012_303 23340801
5. Hannan TJ, Totsika M, Mansfield KJ, Moore KH, Schembri MA et al. Host-pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic Escherichia coli bladder infection. FEMS Microbiol Rev. 2012; 36 : 616–648. doi: 10.1111/j.1574-6976.2012.00339.x 22404313
6. Welch RA, Burland V, Plunkett G, Redford P, Roesch P et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 2002; 99 : 17020–17024. 12471157
7. Nagy G, Altenhoefer A, Knapp O, Maier E, Dobrindt U et al. Both alpha-haemolysin determinants contribute to full virulence of uropathogenic Escherichia coli strain 536. Microbes Infect. 2006; 8 : 2006–2012. 16787757
8. Alteri CJ, Mobley HL Escherichia coli physiology and metabolism dictates adaptation to diverse host microenvironments. Curr Opin Microbiol. 2012; 15 : 3–9. doi: 10.1016/j.mib.2011.12.004 22204808
9. Dubois D, Delmas J, Cady A, Robin F, Sivignon A et al. Cyclomodulins in urosepsis strains of Escherichia coli. J Clin Microbiol. 2010; 48 : 2122–2129. doi: 10.1128/JCM.02365-09 20375237
10. Smith YC, Rasmussen SB, Grande KK, Conran RM, O'Brien AD Hemolysin of Uropathogenic Escherichia coli Evokes Extensive Shedding of the Uroepithelium and Hemorrhage in Bladder Tissue Within the First 24 Hours After Intraurethral Inoculation of Mice. Infect Immun. 2008;
11. Rippere-Lampe KE, Lang M, Ceri H, Olson M, Lockman HA et al. Cytotoxic necrotizing factor type 1-positive Escherichia coli causes increased inflammation and tissue damage to the prostate in a rat prostatitis model. Infect Immun. 2001; 69 : 6515–6519. 11553597
12. Real JM, Munro P, Buisson-Touati C, Lemichez E, Boquet P et al. Specificity of immunomodulator secretion in urinary samples in response to infection by alpha-hemolysin and CNF1 bearing uropathogenic Escherichia coli. Cytokine. 2007; 37 : 22–25. 17382555
13. Linhartova I, Bumba L, Masin J, Basler M, Osicka R et al. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev. 2010; 34 : 1076–1112. doi: 10.1111/j.1574-6976.2010.00231.x 20528947
14. Welch RA Pore-forming cytolysins of gram-negative bacteria. Mol Microbiol. 1991; 5 : 521–528. 2046545
15. Bhakdi S, Bayley H, Valeva A, Walev I, Walker B et al. Staphylococcal alpha-toxin, streptolysin-O, and Escherichia coli hemolysin: prototypes of pore-forming bacterial cytolysins. Arch Microbiol. 1996; 165 : 73–79. 8593102
16. Gadeberg OV, Orskov I, Rhodes JM Cytotoxic effect of an alpha-hemolytic Escherichia coli strain on human blood monocytes and granulocytes in vitro. Infect Immun. 1983; 41 : 358–364. 6345395
17. Gur C, Coppenhagen-Glazer S, Rosenberg S, Yamin R, Enk J et al. Natural Killer Cell-Mediated Host Defense against Uropathogenic E. coli Is Counteracted by Bacterial HemolysinA-Dependent Killing of NK Cells. Cell Host Microbe. 2013; 14 : 664–674. doi: 10.1016/j.chom.2013.11.004 24331464
18. Wiles TJ, Mulvey MA The RTX pore-forming toxin alpha-hemolysin of uropathogenic Escherichia coli: progress and perspectives. Future Microbiol. 2013; 8 : 73–84. doi: 10.2217/fmb.12.131 23252494
19. Landraud L, Gibert M, Popoff MR, Boquet P, Gauthier M Expression of cnf1 by Escherichia coli J96 involves a large upstream DNA region including the hlyCABD operon, and is regulated by the RfaH protein. Mol Microbiol. 2003; 47 : 1653–1667. 12622819
20. Leeds JA, Welch RA RfaH enhances elongation of Escherichia coli hlyCABD mRNA. J Bacteriol. 1996; 178 : 1850–1857. 8606157
21. Aktories K, Barbieri J Bacterial cytotoxins: targeting eukaryotic switches. Nat Rev Micro. 2005; 3 : 397–410. 15821726
22. Galan JE Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009; 5 : 571–579. doi: 10.1016/j.chom.2009.04.008 19527884
23. Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH et al. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 2009; 5: e1000538. doi: 10.1371/journal.ppat.1000538 19662166
24. Munro P, Flatau G, Doye A, Boyer L, Oregioni O et al. Activation and proteasomal degradation of rho GTPases by cytotoxic necrotizing factor-1 elicit a controlled inflammatory response. J Biol Chem. 2004; 279 : 35849–35857. 15152002
25. Lemonnier M, Landraud L, Lemichez E Rho GTPase-activating bacterial toxins: from bacterial virulence regulation to eukaryotic cell biology. FEMS Microbiol Rev. 2007; 31 : 515–534. 17680807
26. Boyer L, Magoc L, Dejardin S, Cappillino M, Paquette N et al. Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity. 2011; 35 : 536–549. doi: 10.1016/j.immuni.2011.08.015 22018470
27. Keestra AM, Winter MG, Auburger JJ, Frassle SP, Xavier MN et al. Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature. 2013; 496 : 233–237. doi: 10.1038/nature12025 23542589
28. Jones JD, Dangl JL The plant immune system. Nature. 2006; 444 : 323–329. 17108957
29. Stuart LM, Paquette N, Boyer L Effector-triggered versus pattern-triggered immunity: how animals sense pathogens. Nat Rev Immunol. 2013; 13 : 199–206. doi: 10.1038/nri3398 23411798
30. Broz P, Monack DM Molecular mechanisms of inflammasome activation during microbial infections. Immunol Rev. 2011; 243 : 174–190. doi: 10.1111/j.1600-065X.2011.01041.x 21884176
31. Keller M, Ruegg A, Werner S, Beer HD Active caspase-1 is a regulator of unconventional protein secretion. Cell. 2008; 132 : 818–831. doi: 10.1016/j.cell.2007.12.040 18329368
32. Kostura MJ, Tocci MJ, Limjuco G, Chin J, Cameron P et al. Identification of a monocyte specific pre-interleukin 1 beta convertase activity. Proc Natl Acad Sci U S A. 1989; 86 : 5227–5231. 2787508
33. Friebel A, Ilchmann H, Aepfelbacher M, Ehrbar K, Machleidt W et al. SopE and SopE2 from Salmonella typhimurium activate different sets of RhoGTPases of the host cell. J Biol Chem. 2001; 276 : 34035–34040. 11440999
34. Lara-Tejero M, Sutterwala FS, Ogura Y, Grant EP, Bertin J et al. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp Med. 2006; 203 : 1407–1412. 16717117
35. Muller AJ, Hoffmann C, Galle M, Van Den Broeke A, Heikenwalder M et al. The S. Typhimurium effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe. 2009; 6 : 125–136. doi: 10.1016/j.chom.2009.07.007 19683679
36. Dinarello CA Interleukin-1 beta, interleukin-18, and the interleukin-1 beta converting enzyme. Ann N Y Acad Sci. 1998; 856 : 1–11. 9917859
37. Garlanda C, Dinarello CA, Mantovani A The interleukin-1 family: back to the future. Immunity. 2013; 39 : 1003–1018. doi: 10.1016/j.immuni.2013.11.010 24332029
38. Franchi L, Munoz-Planillo R, Nunez G Sensing and reacting to microbes through the inflammasomes. Nat Immunol. 2012; 13 : 325–332. doi: 10.1038/ni.2231 22430785
39. Petrilli V, Dostert C, Muruve DA, Tschopp J The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol. 2007; 19 : 615–622. 17977705
40. Sansonetti PJ The innate signaling of dangers and the dangers of innate signaling. Nat Immunol. 2006; 7 : 1237–1242. 17110939
41. Vance RE, Isberg RR, Portnoy DA Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009; 6 : 10–21. doi: 10.1016/j.chom.2009.06.007 19616762
42. Xu H, Yang J, Gao W, Li L, Li P et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;
43. Wang X, Parashar K, Sitaram A, Bliska JB The GAP Activity of Type III Effector YopE Triggers Killing of Yersinia in Macrophages. PLoS Pathog. 2014; 10: e1004346. doi: 10.1371/journal.ppat.1004346 25165815
44. Caprioli A, Falbo V, Ruggeri FM, Minelli F, Orskov I et al. Relationship between cytotoxic necrotizing factor production and serotype in hemolytic Escherichia coli. J Clin Microbiol. 1989; 27 : 758–761. 2656748
45. Yamamoto S, Tsukamoto T, Terai A, Kurazono H, Takeda Y et al. Distribution of virulence factors in Escherichia coli isolated from urine of cystitis patients. Microbiol Immunol. 1995; 39 : 401–404. 8551971
46. Finlay BB, McFadden G Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006; 124 : 767–782. 16497587
47. Pumplin N, Voinnet O RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol. 2013; 11 : 745–760. doi: 10.1038/nrmicro3120 24129510
48. Mulvey MA, Schilling JD, Hultgren SJ Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun. 2001; 69 : 4572–4579. 11402001
49. Doye A, Boyer L, Mettouchi A, Lemichez E Ubiquitin-mediated proteasomal degradation of Rho proteins by the CNF1 toxin. Methods Enzymol. 2006; 406 : 447–456. 16472677
50. Lemonnier M, Bouet JY, Libante V, Lane D Disruption of the F plasmid partition complex in vivo by partition protein SopA. Mol Microbiol. 2000; 38 : 493–505. 11069673
51. Jacquel A, Obba S, Boyer L, Dufies M, Robert G et al. Autophagy is required for CSF-1-induced macrophagic differentiation and acquisition of phagocytic functions. Blood. 2012; 119 : 4527–4531. doi: 10.1182/blood-2011-11-392167 22452982
52. Kuida K, Lippke JA, Ku G, Harding MW, Livingston DJ et al. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995; 267 : 2000–2003. 7535475
53. Smith SN, Hagan EC, Lane MC, Mobley HL Dissemination and systemic colonization of uropathogenic Escherichia coli in a murine model of bacteremia. MBio. 2010; 1:
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole MembraneČlánek Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and FunctionČlánek Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 3- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- To Be or Not IIb: A Multi-Step Process for Epstein-Barr Virus Latency Establishment and Consequences for B Cell Tumorigenesis
- Is Antigenic Sin Always “Original?” Re-examining the Evidence Regarding Circulation of a Human H1 Influenza Virus Immediately Prior to the 1918 Spanish Flu
- The Great Escape: Pathogen Versus Host
- Coping with Stress and the Emergence of Multidrug Resistance in Fungi
- Catch Me If You Can: The Link between Autophagy and Viruses
- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- Evidence for Ubiquitin-Regulated Nuclear and Subnuclear Trafficking among Matrix Proteins
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Production of Anti-LPS IgM by B1a B Cells Depends on IL-1β and Is Protective against Lung Infection with LVS
- Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS
- α-Hemolysin Counteracts the Anti-Virulence Innate Immune Response Triggered by the Rho GTPase Activating Toxin CNF1 during Bacteremia
- Induction of Interferon-Stimulated Genes by IRF3 Promotes Replication of
- Intracellular Growth Is Dependent on Tyrosine Catabolism in the Dimorphic Fungal Pathogen
- HCV Induces the Expression of Rubicon and UVRAG to Temporally Regulate the Maturation of Autophagosomes and Viral Replication
- Spatiotemporal Analysis of Hepatitis C Virus Infection
- Subgingival Microbial Communities in Leukocyte Adhesion Deficiency and Their Relationship with Local Immunopathology
- Interaction between the Type III Effector VopO and GEF-H1 Activates the RhoA-ROCK Pathway
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
- Establishment of HSV1 Latency in Immunodeficient Mice Facilitates Efficient Reactivation
- XRN1 Stalling in the 5’ UTR of Hepatitis C Virus and Bovine Viral Diarrhea Virus Is Associated with Dysregulated Host mRNA Stability
- γδ T Cells Confer Protection against Murine Cytomegalovirus (MCMV)
- Rhadinovirus Host Entry by Co-operative Infection
- A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole Membrane
- Dermal Neutrophil, Macrophage and Dendritic Cell Responses to Transmitted by Fleas
- Elucidation of Sigma Factor-Associated Networks in Reveals a Modular Architecture with Limited and Function-Specific Crosstalk
- A Conserved NS3 Surface Patch Orchestrates NS2 Protease Stimulation, NS5A Hyperphosphorylation and HCV Genome Replication
- Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and Function
- Disruption of IL-21 Signaling Affects T Cell-B Cell Interactions and Abrogates Protective Humoral Immunity to Malaria
- Compartmentalized Replication of R5 T Cell-Tropic HIV-1 in the Central Nervous System Early in the Course of Infection
- Diminished Reovirus Capsid Stability Alters Disease Pathogenesis and Littermate Transmission
- Characterization of CD8 T Cell Differentiation following SIVΔnef Vaccination by Transcription Factor Expression Profiling
- Visualization of HIV-1 Interactions with Penile and Foreskin Epithelia: Clues for Female-to-Male HIV Transmission
- Sensing Cytosolic RpsL by Macrophages Induces Lysosomal Cell Death and Termination of Bacterial Infection
- PKCη/Rdx-driven Phosphorylation of PDK1: A Novel Mechanism Promoting Cancer Cell Survival and Permissiveness for Parvovirus-induced Lysis
- Metalloprotease NleC Suppresses Host NF-κB/Inflammatory Responses by Cleaving p65 and Interfering with the p65/RPS3 Interaction
- Immune Antibodies and Helminth Products Drive CXCR2-Dependent Macrophage-Myofibroblast Crosstalk to Promote Intestinal Repair
- Adenovirus Entry From the Apical Surface of Polarized Epithelia Is Facilitated by the Host Innate Immune Response
- The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family
- Neutrophils: Between Host Defence, Immune Modulation, and Tissue Injury
- CD169-Mediated Trafficking of HIV to Plasma Membrane Invaginations in Dendritic Cells Attenuates Efficacy of Anti-gp120 Broadly Neutralizing Antibodies
- Japanese Encephalitis Virus Nonstructural Protein NS5 Interacts with Mitochondrial Trifunctional Protein and Impairs Fatty Acid β-Oxidation
- Yip1A, a Novel Host Factor for the Activation of the IRE1 Pathway of the Unfolded Protein Response during Infection
- TRIM26 Negatively Regulates Interferon-β Production and Antiviral Response through Polyubiquitination and Degradation of Nuclear IRF3
- Parallel Epigenomic and Transcriptomic Responses to Viral Infection in Honey Bees ()
- A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication
- Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
- Exome and Transcriptome Sequencing of Identifies a Locus That Confers Resistance to and Alters the Immune Response
- The Role of Misshapen NCK-related kinase (MINK), a Novel Ste20 Family Kinase, in the IRES-Mediated Protein Translation of Human Enterovirus 71
- Chitin Recognition via Chitotriosidase Promotes Pathologic Type-2 Helper T Cell Responses to Cryptococcal Infection
- Activates Both IL-1β and IL-1 Receptor Antagonist to Modulate Lung Inflammation during Pneumonic Plague
- Persistence of Transmitted HIV-1 Drug Resistance Mutations Associated with Fitness Costs and Viral Genetic Backgrounds
- An 18 kDa Scaffold Protein Is Critical for Biofilm Formation
- Early Virological and Immunological Events in Asymptomatic Epstein-Barr Virus Infection in African Children
- Human CD8 T-cells Recognizing Peptides from () Presented by HLA-E Have an Unorthodox Th2-like, Multifunctional, Inhibitory Phenotype and Represent a Novel Human T-cell Subset
- Decreased HIV-Specific T-Regulatory Responses Are Associated with Effective DC-Vaccine Induced Immunity
- RSV Vaccine-Enhanced Disease Is Orchestrated by the Combined Actions of Distinct CD4 T Cell Subsets
- Concerted Activity of IgG1 Antibodies and IL-4/IL-25-Dependent Effector Cells Trap Helminth Larvae in the Tissues following Vaccination with Defined Secreted Antigens, Providing Sterile Immunity to Challenge Infection
- Structure of the Low pH Conformation of Chandipura Virus G Reveals Important Features in the Evolution of the Vesiculovirus Glycoprotein
- PPM1A Regulates Antiviral Signaling by Antagonizing TBK1-Mediated STING Phosphorylation and Aggregation
- Lipidomic Analysis Links Mycobactin Synthase K to Iron Uptake and Virulence in .
- Roles and Programming of Arabidopsis ARGONAUTE Proteins during Infection
- Impact of Infection on Host Macrophage Nuclear Physiology and Nucleopore Complex Integrity
- The Impact of Host Diet on Titer in
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Herpesviral G Protein-Coupled Receptors Activate NFAT to Induce Tumor Formation via Inhibiting the SERCA Calcium ATPase
- The Causes and Consequences of Changes in Virulence following Pathogen Host Shifts
- Small GTPase Rab21 Mediates Fibronectin Induced Actin Reorganization in : Implications in Pathogen Invasion
- Positive Role of Promyelocytic Leukemia Protein in Type I Interferon Response and Its Regulation by Human Cytomegalovirus
- NEDDylation Is Essential for Kaposi’s Sarcoma-Associated Herpesvirus Latency and Lytic Reactivation and Represents a Novel Anti-KSHV Target
- β-HPV 5 and 8 E6 Disrupt Homology Dependent Double Strand Break Repair by Attenuating BRCA1 and BRCA2 Expression and Foci Formation
- An O Antigen Capsule Modulates Bacterial Pathogenesis in
- Variable Processing and Cross-presentation of HIV by Dendritic Cells and Macrophages Shapes CTL Immunodominance and Immune Escape
- Probing the Metabolic Network in Bloodstream-Form Using Untargeted Metabolomics with Stable Isotope Labelled Glucose
- Adhesive Fiber Stratification in Uropathogenic Biofilms Unveils Oxygen-Mediated Control of Type 1 Pili
- Vaccinia Virus Protein Complex F12/E2 Interacts with Kinesin Light Chain Isoform 2 to Engage the Kinesin-1 Motor Complex
- Modulates Host Macrophage Mitochondrial Metabolism by Hijacking the SIRT1-AMPK Axis
- Human T-Cell Leukemia Virus Type 1 (HTLV-1) Tax Requires CADM1/TSLC1 for Inactivation of the NF-κB Inhibitor A20 and Constitutive NF-κB Signaling
- Suppression of RNAi by dsRNA-Degrading RNaseIII Enzymes of Viruses in Animals and Plants
- Spatiotemporal Regulation of a T4SS Substrate by the Metaeffector SidJ
- Antigenic Properties of the Human Immunodeficiency Virus Envelope Glycoprotein Gp120 on Virions Bound to Target Cells
- Dependence of Intracellular and Exosomal microRNAs on Viral Oncogene Expression in HPV-positive Tumor Cells
- Identification of a Peptide-Pheromone that Enhances Escape from Host Cell Vacuoles
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Dihydrobiopterin in Adult Falciparum Malaria: Association with Disease Severity, Impaired Microvascular Function and Increased Endothelial Activation
- Transgenic Expression of the Dicotyledonous Pattern Recognition Receptor EFR in Rice Leads to Ligand-Dependent Activation of Defense Responses
- Comprehensive Antigenic Map of a Cleaved Soluble HIV-1 Envelope Trimer
- Low Doses of Imatinib Induce Myelopoiesis and Enhance Host Anti-microbial Immunity
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Oxidized Biopterins in Pediatric Falciparum Malaria: Association with Disease Severity
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



